- 1Qinghai Provincial Key Laboratory of Crop Molecular Breeding, Laboratory for Research and Utilization of Qinghai Tibet Plateau Germplasm Resources, Northwest Institute of Plateau Biology, Innovation Academy for Seed Design Chinese Academy of Sciences (CAS), Xining, China
- 2Hongshan Laboratory, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 3International Maize and Wheat Improvement Center (CIMMYT), Texcoco, Mexico
- 4International Maize and Wheat Improvement Center (CIMMYT), Nairobi, Kenya
- 5Borlaug Institute for South Asia (BISA), New Delhi, India
- 6Campo Experimental Valle de México, Instituto Nacional de Investigacion Forestales Agricolas y Pecuarias (INIFAP), Texcoco, Mexico
- 7Commonwealth Scientific and Industrial Research Organization (CSIRO) Plant Industry, Canberra, ACT, Australia
Developing wheat varieties with durable resistance is a core objective of the International Maize and Wheat Improvement Center (CIMMYT) and many other breeding programs worldwide. The CIMMYT advanced wheat line “Mucuy” displayed high levels of resistance to stripe rust (YR) and leaf rust (LR) in field evaluations in Mexico and several other countries. To determine the genetic basis of YR and LR resistance, 138 F5 recombinant inbred lines (RILs) derived from the cross of Apav#1× Mucuy were phenotyped for YR responses from 2015 to 2020 at field sites in India, Kenya, and Mexico, and LR in Mexico. Seedling phenotyping for YR and LR responses was conducted in the greenhouse in Mexico using the same predominant races as in field trials. Using 12,681 polymorphic molecular markers from the DArT, SNP, and SSR genotyping platforms, we constructed genetic linkage maps and QTL analyses that detected seven YR and four LR resistance loci. Among these, a co-located YR/LR resistance loci was identified as Yr29/Lr46, and a seedling stripe rust resistance gene YrMu was mapped on the 2AS/2NS translocation. This fragment also conferred moderate adult plant resistance (APR) under all Mexican field environments and in one season in Kenya. Field trial phenotyping with Lr37-virulent Puccinia triticina races indicated the presence of an APR QTL accounting for 18.3–25.5% of the LR severity variation, in addition to a novel YR resistance QTL, QYr.cim-3DS, derived from Mucuy. We developed breeder-friendly KASP and indel molecular markers respectively for Yr29/Lr46 and YrMu. The current study validated the presence of known genes and identified new resistance loci, a QTL combination effect, and flanking markers to facilitate accelerated breeding for genetically complex, durable rust resistance.
Introduction
Wheat (Triticum aestivum L.) is grown on about 215 M ha globally and stands as an indispensable staple food for over 7.5 billion people and an important source of daily calories and protein (http://www.fao.org/home/en/). Biotic stresses, particularly diseases such as wheat rusts, significantly reduce crop yields and quality, particularly where varieties are susceptible and favorable conditions exist. Stripe rust (also known as yellow rust, YR) and leaf rust (LR), caused by Puccinia striiformis f. sp. tritici (Pst) and P. triticina (Pt), respectively, can cause total crop loss when an early infection strikes in susceptible varieties (Chen, 2005; Bolton et al., 2008). YR generally occurs in cool and moist environments, whereas LR is more adapted to warmer environments coupled with ideal moisture conditions (Zadoks, 1961), but migrating and evolving YR races have infected wheat crops in previously unaffected areas (Ali et al., 2014; Hovmøller et al., 2016). The rusts can be curtailed using fungicides and other measures, but the best control is to grow wheat varieties that carry genetically complex and thus durable disease resistance.
There are three common rust resistance mechanisms in wheat, depending on the host response, the crop growth stage at which the mechanism activates, and the type of resistance gene: (1) race-specific seedling/all-stage resistance, (2) race-specific adult plant resistance (APR), and (3) race non-specific APR (Lan et al., 2017a). The race-specific genes (R-genes) governing seedling/all-stage or adult plant resistance can provide relatively high resistance and are thus easier to select for in breeding. However, given the rapid evolution of the pathogen, R-genes tend to succumb quickly to new rust races, especially when deployed singly, resulting in “boom and bust” cycles of high productivity followed by widespread and potentially disastrous disease outbreaks. In contrast, race-nonspecific APR genes confer partial but broad-spectrum resistance against multiple rust races (Kolmer, 1996) and, when deployed in combinations in wheat varieties, can present a genetically complex resistance that pathogen mutations will not readily overcome. So far, 83 YR and 80 LR resistance genes have been cataloged in wheat (McIntosh et al., 2017; Kumar et al., 2021). Most of these are R-genes, against several of which virulence already exists in the pathogen population. But three pleiotropic multi-pathogen APR genes, Lr34/Yr18/Sr57/Pm38/Ltn1 (Singh et al., 2012), Lr67/Yr46/Sr55/Pm46/Ltn3 (Herrera-Foessel et al., 2014), and Lr46/Yr29/Sr58/Pm39/Ltn2 (Singh et al., 1998), confer partial resistance to LR, YR, stem rust (SR), and powdery mildew (PM) diseases. The first two genes have already been cloned and characterized (Krattinger et al., 2009; Moore et al., 2015), These pleiotropic genes condition partial levels of resistance and a combination of 4–5 APR genes can result in near-immune levels of resistance to rust diseases in CIMMYT wheat germplasm (Singh et al., 2000a).
High-throughput genotyping platforms provide dense coverage of markers, which have enabled the identification of molecular markers linked to resistance genes that are now widely used in wheat breeding programs (Chen, 2013; Rosewarne et al., 2013). Over the last two decades, more than 200 quantitative trait loci (QTL) for YR and LR resistance have been mapped on the 21 wheat chromosomes using diversity arrays technology (DArT), single sequence repeats (SSRs), and single-nucleotide polymorphisms (SNP) marker platforms (McIntosh et al., 2017). In addition, 11 potential co-located APR QTL conferring pleiotropic resistance to YR, LR, and PM on chromosomes 1BS, 1BL, 2AL, 2BS (two QTL), 2DL, 4DL, 5BL, 6AL,7BL, and 7DS were identified through comparative mapping (Li et al., 2014).
When distributed for international testing in 2013, the advanced CIMMYT breeding line “Mucuy” showed high levels of resistance to both YR and LR in several countries. Mucuy was released in 2017 as “Super 272” in the Northwestern Plain Zone of India, where YR is prevalent. The line also resisted YR races in Kenya and Ethiopia, where PstS2 and PstS11 are the predominant race groups, prompting further studies to understand the genetics of rust resistance in Mucuy. The current study sought to (1) investigate the genetic bases of YR and LR resistance using an F5 recombinant inbred line (RIL) population derived from the cross of “Apav#1” × “Mucuy;” (2) identify loci conferring resistance at both the seedling and adult plant stages using molecular markers; and (3) understand the QTL combinations effects on YR and LR, among identified resistance loci.
Materials and methods
Plant materials
We used 138 F4-derived F5 RILs from the cross of Apav#1 × Mucuy. The susceptible parent, Apav#1 (CIMMYT Germplasm ID: 1853706), derived from a cross of “Avocet-YrA” × “Pavon 76,” was susceptible to YR, LR, and stem rust (SR) at all growth stages, against predominant Pst and Pt races used in various trials in Mexico. In contrast, Mucuy (CIMMYT Germplasm ID: 5663955), derived from the cross “Mutus”*2 × “Akuri,” showed intermediate resistance during seedling evaluations in the greenhouse but high resistance to both YR and LR at the adult plant stage in field trials, in both cases against races Pst and Pt. The RILs were developed following a bulk advancement of the population until the F4 generation and then harvesting random plants individually to obtain F5 RILs (Basnet et al., 2014a). We multiplied the RIL seed and used it in all studies.
Seedling evaluations
Stripe rust
For YR seedling evaluations, the parents and F5 RILs were grown in a greenhouse, and seedlings inoculated at the two-leaf stage with Pst isolate Mex14.191 (Avirulence/virulence: Yr1, 4, 5a, 10, 15, 24, 26, 5b, Poll/Yr2, 3, 6, 7, 8, 9, 17, 27, 31, A; Randhawa et al., 2018). A set of 30 differential lines possessing the known YR resistance genes, mostly in the Avocet background, were also included. An atomizer was used to spray urediniospores suspended in light-weight mineral oil Soltrol 170 (Chempoint.com) at the two-leaf stage. Inoculated plants were moved to a dew chamber at 7°C for 24 h after mineral oil had evaporated from the leaf surface to facilitate spore germination and initiate infection, and then transferred back to greenhouse benches for disease development. The minimum, maximum, and average temperatures of the greenhouse were 9.4, 20.8, and 16.1°C. Infection type (IT) data was recorded 2 weeks post-inoculation using the modified 0-9 scale (McNeal et al., 1971), where 0 = no visible infection, 1 = necrotic/chlorotic flecks without sporulation, 2 = necrotic/chlorotic stripes without sporulation, 3 = necrotic/chlorotic stripes with trace sporulation, 4 = necrotic/chlorotic stripes with light sporulation, 5 = necrotic/chlorotic stripes with intermediate sporulation, 6 = chlorosis stripes with abundant sporulation, 7 = chlorotic stripes with abundant sporulation, 8 = stripe without chlorosis, moderate sporulation, 9 = stripes without chlorosis and abundant sporulation. Infection types “7,” “8,” and “9” were considered susceptible; all others were recorded as resistant.
Leaf rust
Parents and RILs were evaluated at seedling stage with Pt races MBJ/SP (isolate MEX96.560) [Avirulence/virulence: Lr2a, 2b, 2c, 3ka, 9, 16, 19, 21, 24, 25, 28, 29, 30, 32, 33, 36/1, 3, 3bg, 10, 11, 12, 13, 14a, 14b, 15, 17a, 18, 20, 23, (26), 27+31, 37; Herrera-Foessel et al., 2012; Huerta-Espino et al., 2020] and TBD/TM [isolate MEX91.28A; Avirulence/virulence: Lr3ka, 11, 16, (23), 24, 26, 37/1, 2a, 2b, 2c, 3, 3bg, 10, 13, 14a, 15, 17, 18, 27+31, 28; Singh, 1991]. The latter race was used for better expression of Lr16 resistance, postulated to be present in Mucuy and segregated in the RIL population. A set of 48 lines with known LR genes, mostly in the “Thatcher” background, were also included. Inoculation was performed as for YR but with overnight misting at room temperatures and the minimum, maximum, and average temperatures were maintained at 9.0, 23.0, and 18.1°C, for disease development with both Pt races. LR ITs were recorded 11 days post-inoculation using the 0–4 scale (Roelfs et al., 1992), where 0 = no visible symptoms,; = necrotic/chlorotic flecks, 1 = small uredinia surrounded by necrosis, 2 = small-to-medium uredinia surrounded by chlorosis or necrosis, X = random distribution of variable-sized uredinia, 3 = medium-sized uredinia without chlorosis, 4 = large uredinia without chlorosis, and + and – indicated a bit larger or smaller uredinia than normal for the infection type. Infection types “3” and “4” were considered susceptible while all other infection types were considered resistant.
Field experiments
Stripe rust
We conducted YR field evaluations at the CIMMYT research station at Toluca, State of Mexico, Mexico, during the 2015, 2016, and 2017 seasons (hereafter referred to as YrMV15, YrMV16, and YrMV17), at Kenya Agriculture and Livestock Research Organization (KALRO) research station in Njoro, Kenya, during the main-season of 2016 and off-seasons of 2016, 2019, and 2020 (referred as YrKE16M, YrKE16O, YrKE19, and YrKE20), and at the research station of Borlaug Institute for South Asia (BISA) in Ludhiana, India, during the 2018–2019 and 2019–2020 seasons (referred as YrIN19 and YrIN20). Each genotype was sown in 0.7-m paired rows with a 0.3-m pathway between rows. Field trials were unreplicated, given rust resistance's highly heritability when phenotyping is conducted under managed epidemics at hot spot sites where disease pressure is maximum. At Toluca, the YR spreader rows consisted of a mixture of susceptible wheat lines (Yr27-carrying lines derived from the “Avocet” × “Attila” cross, “Morocco” and “Avocet+Yr31”). This spreader mixture was planted both as hill plots in the middle of the 0.3-m pathway and around the experimental nursery. The same Pst isolate (Mex14.191) used for seedling evaluations was sprayed onto YR spreaders about 4 weeks post-germination and this was repeated three times to initiate artificial epidemics. At the KALRO station, YR evaluations were carried out under natural epidemics and the causal race was identified as PstS11 [Avirulence/virulence: Yr1, 3, 5, 9, 10, 15, 24, 25, 26, Sp, Amb/2, (4), 6, 7, 8, 17, 27, 32, AvS] by the GRRC (Global Rust Reference Center), Denmark. Inoculation at Ludhiana was carried out using spreader rows of the variety PBW343, known to carry Yr27, inoculated with a mixture of races 110S84, 46S119, 110S119, and 238S119 (Supplementary Table 1) that are predominant in this region. Even though the inoculated races dominate in the screening nurseries, the presence of natural inoculum carrying other races at low frequencies cannot be ruled out. The key difference in Pst populations in Mexico vs. Kenya and India is the avirulence for resistance gene Yr4 in Mexico but virulence for it in the races of Kenya and India, based on the response of the “Avocet+Yr4” tester line.
Leaf rust
In the 2015–2016, 2016–2017, and 2017–2018 growing seasons, the parents and RILs were evaluated for APR to LR at Norman E. Borlaug Research Station (CENEB), Ciudad Obregón, State of Sonora, Mexico (hereafter referred to as LrY16, LrY17, and LrY18). The field experiment design was similar to that for YR. The susceptible LR spreader lines included Morocco and Avocet near-isolines carrying Yr24/26. The mixture of Pt races MCJ/SP [isolate MEX94.47; Avirulence/virulence: Lr2a, 2b, 2c, 3ka, 9, 16, 19, 21, 24, 25, 28, 29, 30, 32, 33, 36/1, (3), 3bg, 10, 11, 12, 13, 14a, 14b, 15, 17a, 18, 20, 23, 26, 27+31, 37; Herrera-Foessel et al., 2012] and MBJ/SP (isolate MEX96.560, same as MCJ/SP except virulent on Lr3 and avirulent on Lr26) was suspended in Soltrol 170 and sprayed on the spreaders to cause artificial epidemics.
Disease severity evaluation and statistical analyses
Disease severity (DS) of the parents and RILs were recorded on 3 occasions using the modified Cobb's Scale (Peterson et al., 1948). Initial data were recorded when the DS of Apav#1 was around 80% and repeated after 7 days. The last data set was recorded when it reached 100%. For multiple disease readings, the area under the disease progress curve (AUDPC) was calculated as per Bjarko and Line (1988). The correlation analysis of final disease severity (FDS) in each environment was conducted using SAS 9.4 software (SAS Institute, Cary, NC).
Genetic linkage map construction and QTL mapping
DNA was extracted using the CTAB method (Dreisigacker et al., 2012) from one-week-old seedlings of the parents and RILs grown in a greenhouse and genotyped with the DArT-GBS platform (Reference for SAGA). In total, we genotyped 40,519 GBS-in-Silico and 39,849 GBS-SNP, in combination with closely linked molecular markers for different rust genes (Xgwm210 for Lr16, cslv46G22 for Lr46/Yr29, two Yr17-linked markers WGGB156 and WGGB159 by Wang et al. (2018), and one Yr17-linked marker provided by Evans Lagudah), in the entire RIL population. Genetic linkage maps were constructed using Joinmap 4.1 (Van Ooijen, 2006) with 12,681 polymorphic markers, and 56 linkage groups were constructed. In addition, QTL analysis was performed by inclusive composite interval mapping (ICIM) using IciMapping 4.2 (Meng et al., 2015) with the DS of each tested environment and the mean of FDS (referred to as YrM for YR and LrM for LR). The logarithm of odds (LOD) score was determined based on the 1,000 permutation test and a significance level of α = 0.05. The percentages of phenotypic variance explained (PVE) were determined using stepwise regression at the LOD peaks (Somers et al., 2004; Francki et al., 2009; Huang et al., 2012; Wilkinson et al., 2012).
Results
Seedling responses
Stripe rust
Apav#1 and Mucuy showed seedling infection type (IT) responses of “8” and “1,” respectively, against Pst isolate Mex14.191. Seedling evaluation of the RILs identified 67 resistant lines (ITs ranging from 1 to 6) and 63 susceptible lines (ITs ranging from 67 to 9); segregating lines were excluded from the analysis (Figure 1A). Chi-squared analysis of goodness of fit suggested segregation of a single resistance gene in this population (χ2 = 0.069, P = 0.73). It was temporarily named as YrMu, mapped on wheat chromosome 2AS at 16.6–19.1 Mb (International Wheat Genome Sequencing Consortium, 2018), and co-segregated with 79 molecular markers including WGGB156, WGGB159, and InD_hzau_MuYLr-2AS (Supplementary Figure 1A). WGGB156 and WGGB159 had previously been confirmed as closely linked to Yr17 (Wang et al., 2018). In addition, we phenotyped the Avocet+Yr17 isoline against Mex14.191 and the IT response was “8.” Thus, we speculated that YrMu might be a new stripe rust resistance gene in Mucuy at the seedling stage against Mex14.191, although the possibility of an enhanced expression of Yr17 due to the genetic background cannot be ruled out, due to the continuous variation for resistance phenotypes included in the resistance category.
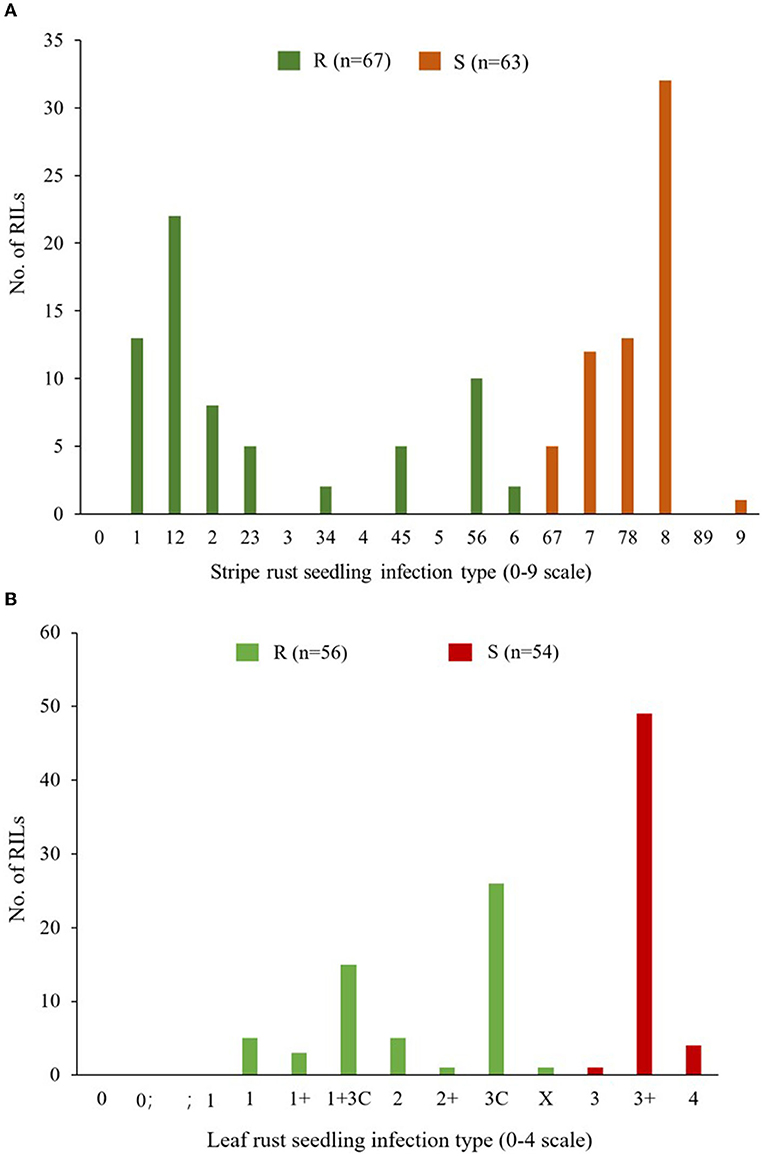
Figure 1. Frequency distributions of the Apav#1×Mucuy recombinant inbred lines (RILs) for stripe rust (YR) responses (A) and leaf rust (LR) responses (B) grouped as resistant (R) and susceptible (S) at the seedling stage. For stripe rust, IF ≤ 6 is in the R group, while the rest lines with IF ≥ 67 are in the S group. For leaf rust, IF ≤ X is the R group and IF ≥ 3 is the S group.
Leaf rust
Seedling evaluation of parents under LR showed IT responses “4” for Apav#1 and “3C” for Mucuy, against the Pt race MBJ/SP. The distribution of 56 resistant and 54 susceptible RILs conformed to the segregation of a single resistance gene (χ2 = 0.009, P = 0.85) that mapped on the short arm of chromosome 2B in the interval of molecular markers 2325486 and 4405950 (Figure 1B, Supplementary Figure 1B). The infection types of Mucuy and resistant RILs were similar to the tester line for gene Lr16, hence this gene might be Lr16.
Adult plant response
The FDS and host response to YR for Apav#1 ranged from 70 to 100 S and for Mucuy ranged from 0 to 5 MS at the adult plant stage over 9 environments. The continuous distribution of YR DS for RILs in each environment indicated the polygenic inheritance of APR (Figures 2A–C).
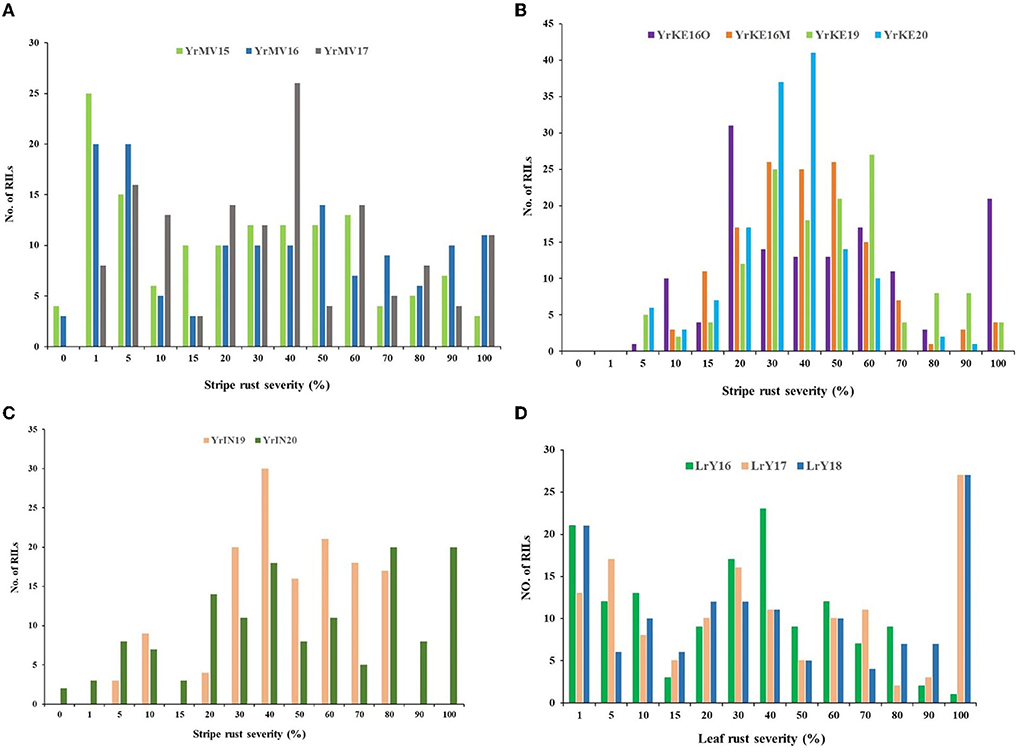
Figure 2. Frequency distributions of Apav#1×Mucuy recombinant inbred lines (RILs) for the final stripe rust severities in three environments in Mexico (A), four environments in Kenya (B), and two environments in India (C), along with final leaf rust severities in three environments in Mexico (D).
LR FDS and host responses were 100 S for Apav#1 and 0 for Mucuy over all 3 seasons. The LR DS frequency distributions were distorted and skewed toward the resistance (Figure 2D), indicating the segregation of at least one large-effect LR resistance locus in the population.
Correlation coefficients
The correlation coefficients of FDS for RILs varied from 0.23 to 0.93 in the nine YR environments (Table 1), while the phenotypic correlation coefficient was 0.90 to 0.93 across the three Mexican environments. Low phenotypic correlation coefficients among Mexico, Kenya, and India were attributed to the presence of different rust isolates in these locations. For LR FDS, the correlation coefficients among the three test environments in Mexico were high, ranging from 0.86 to 0.89 (Table 1). In addition, significant phenotypic correlations, ranging from 0.30 to 0.75, occurred in all environments between LR and YR (Table 1), indicating the presence of pleiotropic/co-located resistance loci in the population.
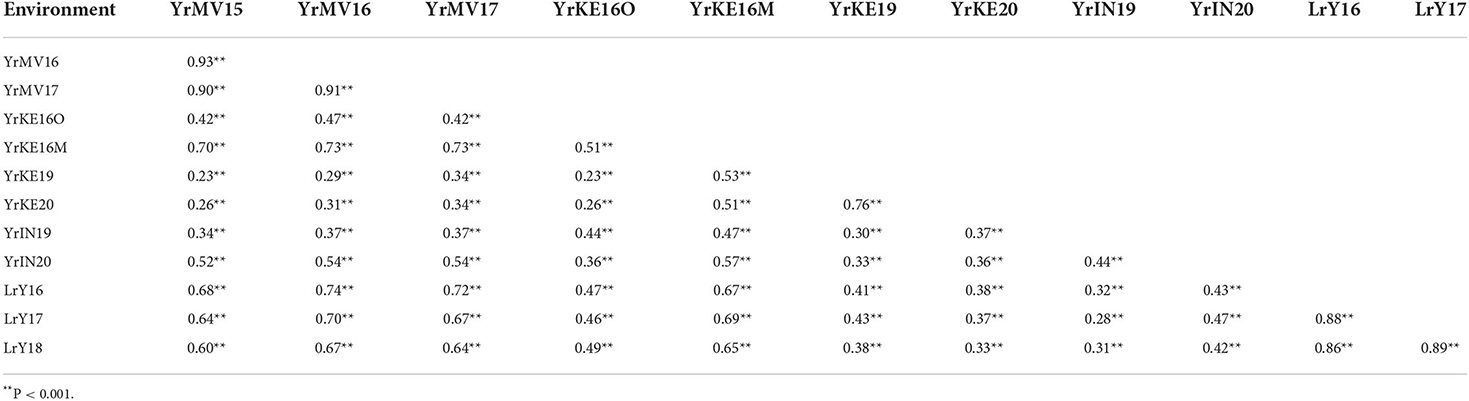
Table 1. Phenotypic correlations among final stripe rust severities in nine environments (Toluca YrMV15, YrMV16, and YrMV17; Kenya YrKE16O, YrKE16M, YrKE19, YrKE20; India YrIN19, YrIN20) and final leaf rust severities in three environments (Ciudad Obregón LrY16, LrY17, and LrY18), all in Mexico.
Co-located resistance loci
We identified two co-located resistance loci for YR and LR in the RIL population. The first locus was located on chromosome 1BL and designated QYr.cim-1BL/QLr.cim-1BL. This locus was detected in all tested YR and LR environments and accounted for 10.4–33.3% of YR phenotypic variation and 20.6–33.6% of LR phenotypic variation (Table 2, Figure 3A). Based on the closely linked molecular markers, we developed a KASP marker, such as Kasp_hzau_MuYLr-1BL, and genotyped the entire RIL population (Supplementary Figure 2, Supplementary Table 2). The single marker analysis showed highly significant mean differences of both YR and LR for RILs carrying the positive allele and those lacking it (Supplementary Table 3). Kasp_hzau_MuYLr-1BL was one of the flanking markers of QLr.cim-1BL (Table 2). Because the known pleiotropic multi-pathogen slow-rusting resistance gene Lr46/Yr29/Sr58/Pm39 is also located on 1BL, we genotyped the RIL population with the closely linked molecular marker cslv46G22. The result showed that cslv46G22 was one of the flanking markers of QYr.cim-1BL/QLr.cim-1BL as well.
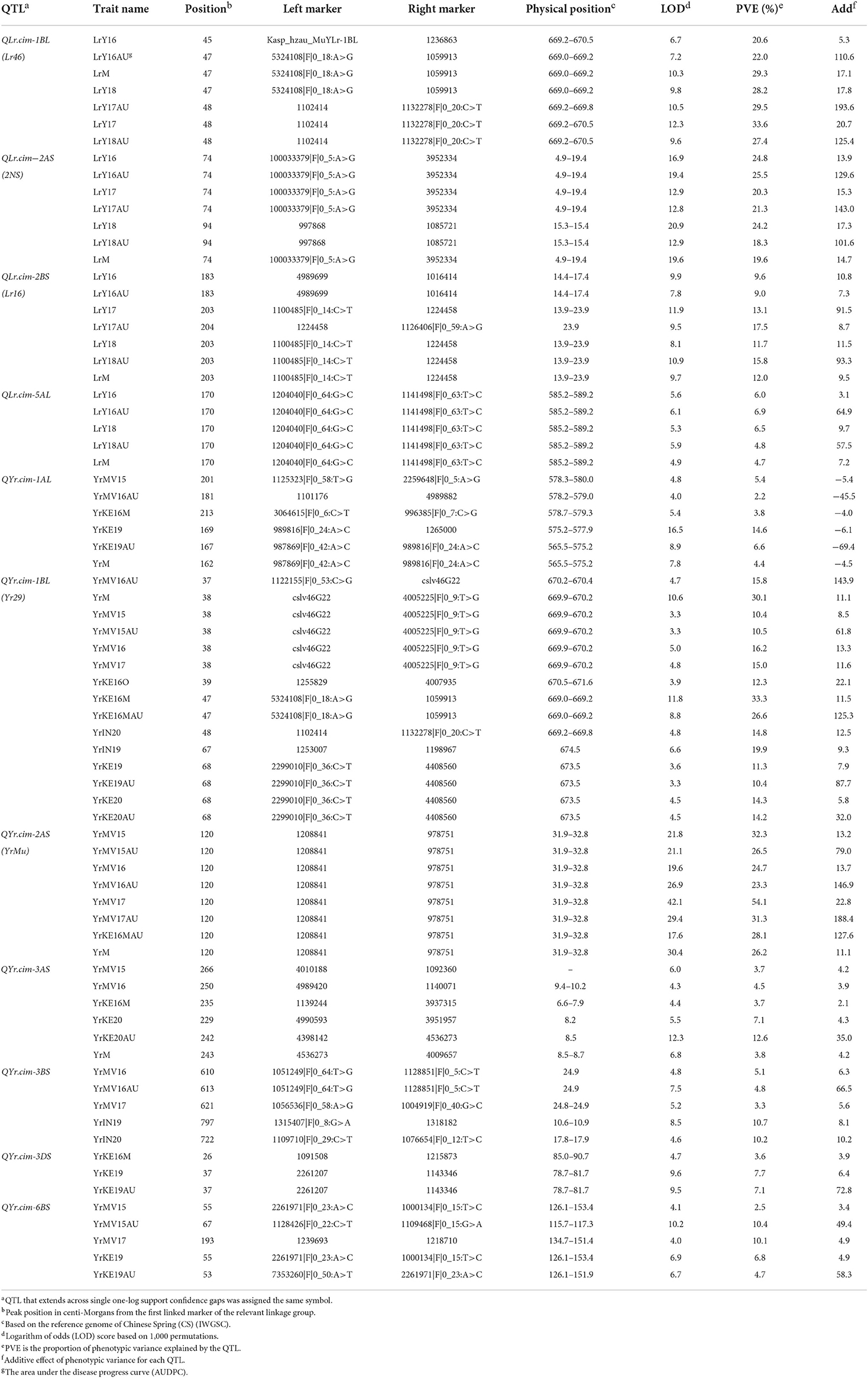
Table 2. Position and effects of quantitative trait loci (QTL) for adult plant resistance (APR) to stripe rust, leaf rust, and mean of final stripe and leaf rust severities over all tested environments (YrM and LrM), using inclusive composite interval mapping (ICIM) by IciMapping 4.2 in the 138 Apav#1 × Mucuy F5 RIL population.
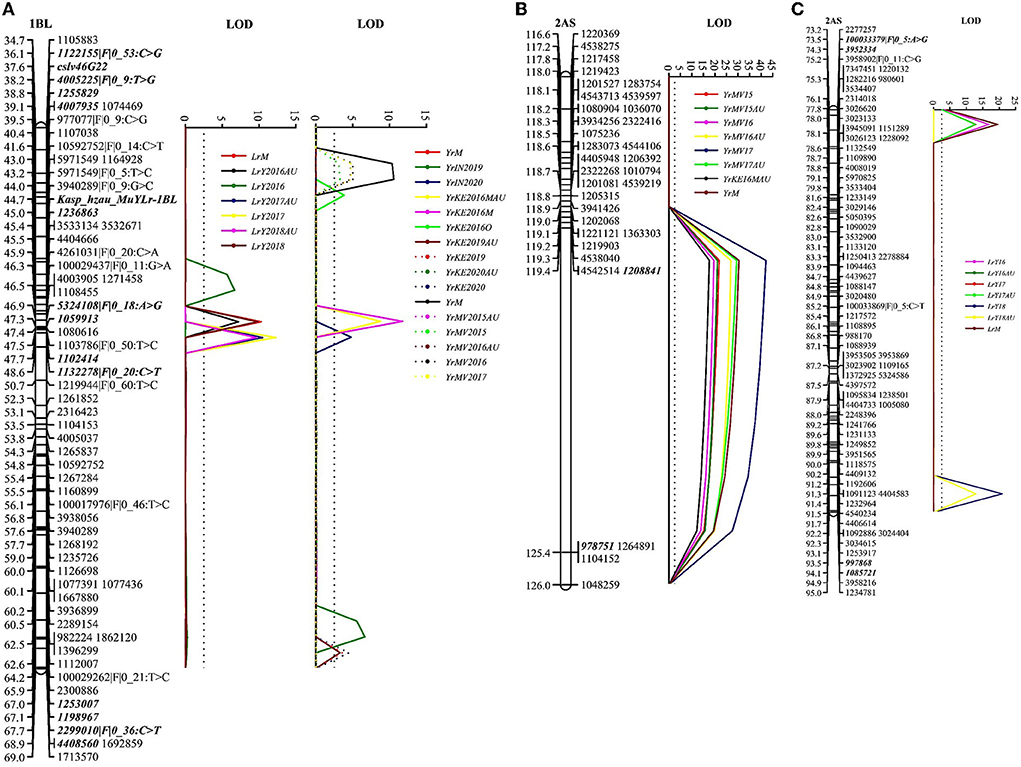
Figure 3. The logarithm of odds (LOD) plot of quantitative trait loci (QTL) for adult plant resistance to both stripe rust and leaf rust on chromosomes 1BL (A), 2AS for YR (B), and 2AS for LR (C) in the Apav#1×Mucuy RIL population. Positions (cM) of the molecular markers on chromosomes are shown on the vertical axes; cumulative genetic distances of linkage groups are also shown. QTL flanking markers are in bold.
The second QTL was QYr.cim-2AS/QLr.cim-2AS, located on the short arm of chromosome 2A and which explained 23.3–54.1 and 18.3–25.5% of YR and LR DS variations, respectively. QYr.cim-2AS was detected in all Mexican YR environments and 1 year of the Kenyan environment and was located in the interval of (DArT)-GBS markers 1208841 and 978751 (Table 2, Figure 3) within 2.1 cM from the seedling resistance gene YrMu. We also mapped a leaf rust resistance QTL, QLr.cim-2AS, on 2AS, which was flanked by molecular markers 100033379|F|0_5:A>G, 3952334, 997868 and 1085721 (Table 2, Figure 3C). We developed an InDel marker named InD_hzau_MuYLr-2AS that was genotyped on the F5 RIL population (Supplementary Table 2); the single marker analysis showed that it was significantly correlated with both YR and LR phenotypes and co-located with YrMu (Supplementary Table 4). Thus, the InDel marker InD_hzau_MuYLr-2AS can be used in wheat breeding to select for QTL QYr.cim-2AS/QLr.cim-2AS.
Other QTL conferring APR to YR or LR
In addition to the 2 co-located resistance loci mentioned above, we found 4 more resistance loci derived from Mucuy that confer APR to YR, named QYr.cim-3AS, QYr.cim-3BS, QYr.cim-3DS and QYr.cim-6BS, in combination with a locus, QYr.cim-1AL, contributed by Apav#1. Identified at both the Mexican and Kenyan testing locations in 2 years, QYr.cim-3AS was associated with molecular markers 4010188, 1092360, 4989420, 1140071, 1139244, 3937315, 4990593, 3951957, 4398142, 4536273, and 4009657 (Table 2). Its corresponding physical locations on the Chinese Spring (CS) reference genome (International Wheat Genome Sequencing Consortium, 2018) ranged from 6.6 to 10.2 Mb and it explained 3.7–12.6% of the YR phenotypic variation. QYr.cim-3BS was in vicinity of the molecular markers 1051249|F|0_64:T>G, 1128851|F|0_5:C>T, 1056536|F|0_58:A>G, 1004919|F|0_40:G>C, 1315407|F|0_8:G>A, 1318182, 1109710|F|0_29:C>T, and 1076654|F|0_12:T>C, and the corresponding physical locations on the CS reference genome (International Wheat Genome Sequencing Consortium, 2018) spanned from 10.6 to 24.9 Mb. It was detected in 2 years at each of the Mexican and Indian locations and accounted for 3.3–10.7% of YR phenotypic variation (Table 2). QYr.cim-3DS was detected in 2 years at the Kenyan location, explained 3.6–7.7% of YR phenotypic variation, and was linked to markers 1091508, 1215873, 2261207, and 1143346 in physical positions spanning from 78.7 Mb to 90.7 Mb (International Wheat Genome Sequencing Consortium, 2018; Table 2). QYr.cim-6BS was located near markers 2261971|F|0_23:A>C, 1000134|F|0_15:T>C, 1128426|F|022:C>T, 1109468|F|0_15:G>A, 1239693, 1218710, 7353260|F|0_50:A>T and 2261971|F|0_23:A>C, with physical positions ranging from 115.7 to 153.4 Mb (International Wheat Genome Sequencing Consortium, 2018). QYr.cim-6BS was detected for 2 years at the Mexican location and 1 year at the Kenyan location, and it explained 2.5–10.4% of the YR phenotypic variation. QYr.cim-1AL for YR resistance was the only locus identified that was derived from Apav#1. It explained 2.2–14.6% of the phenotypic variation, had a physical position ranging from 565.5 to 580.0 Mb (International Wheat Genome Sequencing Consortium, 2018), and was flanked by molecular markers 1125323|F|0_58:T>G, 22599648|F|0_5:A>G, 1101176, 4989882, 3064615|F|0_6:C>T, 996385|F|0_7:C>G, 989816|F|0_24:A>C, 126500, 987869|F|0_42:A>C and 989816|F|0_24:A>C (Table 2).
We found 2 more resistance loci derived from Mucuy that conferred APR to LR, named QLr.cim-2BS and QLr.cim-5AL. Both loci were consistently identified in all LR environments except for QLr.cim-5AL in LrY17. QLr.cim-2BS explained 9.0–17.5% of LR phenotypic variation and was located on the short arm of chromosome 2B. It was flanked by molecular markers 4989699, 1016414, 1100485|F|0_14:C>T, 1224458, and 1126406|F|0_59:A>G (Table 2). The seedling LR resistance gene Lr16 was also identified on 2BS. The physical location of Lr16 was 13.7–23.9 Mb, based on the CS reference genome (International Wheat Genome Sequencing Consortium, 2018), meaning it overlapped with QLr.cim-2BS at 13.9–23.9 Mb. This confirmed that QLr.cim-2BS and Lr16 should be the same gene that provided all-stage resistance to LR in the RIL population. QLr.cim-5AL was located in the interval of molecular markers 1204040|F|0_64:G>C and 1141498|F|0_63:T>C and explained 4.7–6.9% of LR phenotypic variation (Table 2).
Phenotypic effects of QTL combinations
Due to the differences among rust races in different countries, resistance effects provided by individual loci varied greatly at different locations. For example, QYr.cim-2AS provided significant resistance in Mexico, could not be detected in India and was identified in Kenya only in 2016. Therefore, we analyzed the phenotypic effects of QTL combinations among 4 stably detected QTLs according to the average DS in the three countries.
The F5 RILs were divided into 16 groups according to the genotypes of the 4 stable YR resistance QTL on chromosomes 1Bl, 2AS, 3AS, and 6BS derived from Mucuy. The presence of resistance alleles for the QTL in each RIL was inferred with the QTL combination based on the flanking molecular markers. In Mexico, the disease severity of lines with QYr.cim-2AS ranged from 4.5 to 21.3% (Figure 4A). In addition, QYr.cim-1BL played a great role in reducing YR severity: the mean DS of YR was 63.3% with QYr.cim-1BL present alone, whereas YR DS ranged from 4.5 to 52.7% when QYr.cim-1BL was present with other QTL (Figure 4A). Although the resistance effects of QYr.cim-3AS and QYr.cim-6BS were not significantly different from lines without any resistance QTL, they conferred a significantly lower YR DS when combined with QYr.cim-2AS or QYr.cim-1BL. In Kenya and India (Figure 4A), QYr.cim-1BL showed a higher effect than QYr.cim-2AS, when both were present alone but the presence of both loci reduced DS in the line, overall, with the highest average DS of 40.6%.
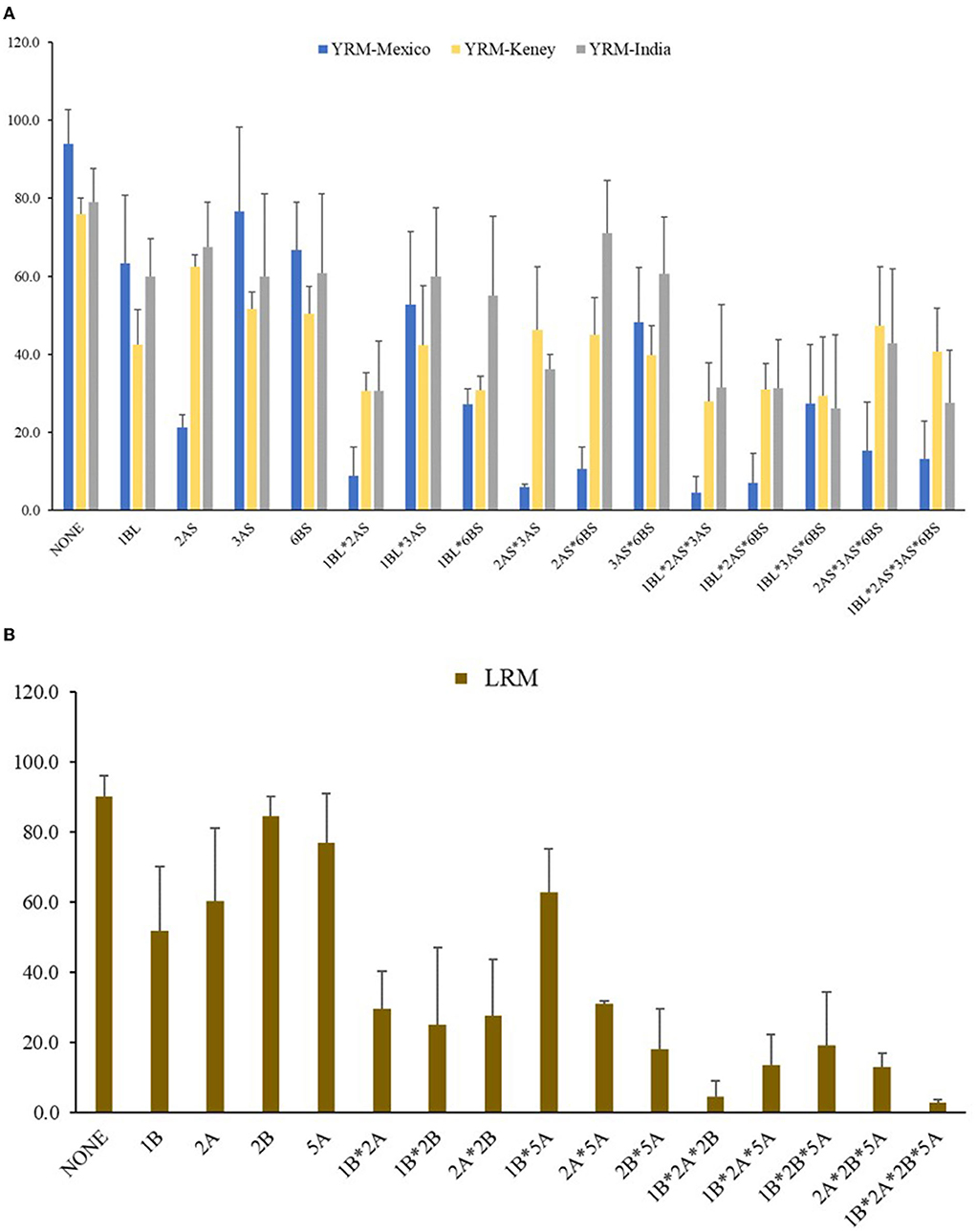
Figure 4. Mean stripe rust severity in three counties (A) and leaf rust severity in Mexico (B) for lines carrying different QTL combinations, based on the flanking molecular marker of each identified locus from the Apav#1× Mucuy F5 RIL population.
The F5 RILs were classified into 16 groups according to the genotype of 4 LR resistance QTL (Figure 4B). When present alone, QLr.cim-1BL, QLr.cim-2AS, QLr.cim-2BS and QLr.cim-5AL reduced the LR DS from 90.0% to 51.9, 60.3, 84.4, and 76.9%, respectively. Although the resistance effect provided by QLr.cim-2BS alone was small, it had a significant additive effect when combined with other loci. The DS ranged from 2.8 to 27.5% when QLr.cim-2BS was present with other QTL. In general, the number of QTL was negatively correlated with DS for RILs.
Discussion
Mucuy was highly resistant to YR in multi-year field trials in Mexico, Kenya, and India. Resistance in seedlings to Mexican Pst isolates and Pt races was based on YrMu and Lr16. In total, our molecular mapping identified 4 LR and 7 YR resistance loci. These loci together explained 74.4% of LR phenotypic variation at the adult plant stage, and 84.2, 43.2, and 58.9% of YR phenotypic variation for Mexican, Kenyan, and Indian Pst populations and field environments, respectively. All resistance loci/genes were derived from Mucuy, apart from QYr.cim-1AL. The newly developed molecular markers for the 2 co-located resistance loci will help wheat breeders to develop new varieties with more durable resistance to rusts.
Resistance loci on group 1 chromosomes
So far, chromosome 1AL lacks a formally designated Yr gene. In this study, the only resistance locus derived from susceptible parent Apav#1, QYr.cim-1AL, explained 2.2–5.4% of the YR phenotypic variation in Mexican environments and 3.8–14.6% of the YR phenotypic variation in Kenyan environments; however, no YR resistance conferred by this locus was detected in Indian environments, suggesting that QYr.cim-1AL provides either small race-specific resistance or is environmentally unstable. The physical location range of QYr.cim-1AL on the CS reference genome was 565.5–580.0 Mb (International Wheat Genome Sequencing Consortium, 2018). Several QTL have been reported near this interval (Ren et al., 2012a; Rosewarne et al., 2012; Basnet et al., 2014b). Thus, QYr.cim-1AL might be the same YR resistance locus from Pastor, Naxos, and TAM112, based on their physical positions of the CS reference genome (International Wheat Genome Sequencing Consortium, 2018). This needs to be further confirmed through gene cloning.
A co-located resistance locus QYr.cim-1BL/QLr.cim-1BL was identified on the long arm of wheat chromosome 1B. QYr.cim-1BL/QLr.cim-1BL showed stable and significant resistance effects in all tested environments for both YR and LR. We genotyped the RIL population with cslv46G22, a closely linked molecular marker for Lr46/Yr29/Sr58/Pm39 located on 1BL, which showed that cslv46G22 was loosely linked with QYr.cim-1BL/QLr.cim-1BL as one of the flanking markers. To further verify the relationship among QYr.cim-1BL/QLr.cim-1BL and Lr46/Yr29/Sr58/Pm39, we removed the effect of cslv46G22 and re-did the QTL analysis but were unable to detect any other resistance locus on 1BL. Therefore, we conclude that QYr.cim-1BL/QLr.cim-1BL should be the known APR gene Lr46/Yr29/Sr58/Pm39. Over the last two decades, many CIMMYT derived bread and durum wheat have been reported to possess Lr46/Yr29/Sr58/Pm39: for example, “Pavon 76” (William et al., 2006), “Saar” (Lillemo et al., 2008), “Pastor” (Rosewarne et al., 2012), “Quaiu 3” (Basnet et al., 2013), “Francolin#1” (Lan et al., 2014), “Sujata” (Lan et al., 2015), “Kundan” (Ren et al., 2017), “Bairds” (Lan et al., 2017b), “Chilero” (Ponce-Molina et al., 2018), and Arableu#1 (Yuan et al., 2020). Based on all reported studies, Lr46/Yr29/Sr58/Pm39 should be placed from 662.1 to 684.8 Mb, according to the CS reference genome (International Wheat Genome Sequencing Consortium, 2018). The relatively diffused localization of Lr46/Yr29/Sr58/Pm39 might be due either to the genetic background effect, phenotyping errors, different genotyping platforms, or population size. In addition, recent studies have reported that more than one pleiotropic APR locus could be present in the 1BL region (Yuan et al., 2020; Zhou et al., 2021), indicating that the 1BL region might carry a gene cluster composed of multiple APR genes, which will be confirmed by future gene cloning.
Resistance loci on group 2 chromosomes
Another co-located resistance locus, QYr.cim-2AS/QLr.cim-2AS, and the seedling YR resistance gene YrMu were located on 2AS (Supplementary Tables 5, 6). Yr17 is located on a translocation on wheat chromosome 2AS derived from the Aegilops ventricosa 2NvS segment, which is known to confer resistance against multiple wheat diseases but also plays role in increasing wheat yields (Gao et al., 2021) and has been used in CIMMYT wheat breeding. This translocation was initially reported to provide significant resistance to all three rusts due to the loci of Yr17, Lr37, and Sr38 (Bariana and McIntosh, 1993; Chen, 2005). However, races virulent to Yr17 rapidly evolved, once this gene was deployed in Europe (Bayles et al., 2000), Mexico (Randhawa et al., 2018), and in the Indian Pst population, due to the widespread cultivation of the Yr17-carrying variety HD2967 during the mid-2010's (unpublished results). However, we detected significant YR resistance on 2AS that was stable over 3 years in Mexico against the Pst isolates used in phenotyping, and this resistance was also identified in Kenya in 2016. PstS1 was an invasive strain that originated in East Africa in the early 1980's; PstS2 evolved from PstS1 and the two strains have become the dominant races in East Africa (Walter et al., 2016). As of 2019, the new genetic group PstS11 was reported as dominant in East Africa by the Global Rust Reference Center and, unlike PstS1 or PstS2, PstS11 is virulent to Yr17. Several reports have indicated that Yr17 might be considered a race-specific APR gene due to the difficulty and inconsistent seedling phenotypes, especially with the aggressive Pst1 lineage (Fang et al., 2011; Milus et al., 2015). In the present study, the seedling reaction of Mucuy was very low, while the single gene line of Yr17 (Avocet+Yr17) was susceptible to the Mexican Pst isolate Mex.14.191. However, YrMu was mapped on wheat chromosome 2AS at 16.6–19.1 Mb (International Wheat Genome Sequencing Consortium, 2018) and it co-segregated with 79 molecular markers including InD_hzau_MuYLr-2AS, WGGB156, and WGGB159, the latter of which were closely linked to Yr17 (Wang et al., 2018). Thus, YrMu should be the known YR resistance gene Yr17 and the enhanced expression of Yr17 in Mucuy seedlings might be due to the background effect of other APR loci, or due to the residual effect of the ineffective Yr17 on APR. Similarly, an LR APR QTL was also located on the translocation, in addition to Lr37, because Pt races MCJ/SP and MBJ/SP used in field trials are known to be virulent to Lr37 (Huerta-Espino et al., 2011).
Several previous studies have shown that chromosome 2BS possesses various race-specific and quantitative resistance loci to LR (Messmer et al., 2000; Xu et al., 2005). So far, six formally named LR resistance genes have been identified on chromosome 2BS, including Lr13 (Seyfarth et al., 1999), Lr16 (McCartney et al., 2005), Lr23 (Datta et al., 2008), Lr35 (Seyfarth et al., 1999), Lr48 (Bansal et al., 2008), and Lr73 (Park et al., 2014). In this study, polymorphism was found in both parents and RILs for Xgwm210, the closely linked molecular markers of Lr16 and the seedling reaction for Mucuy were similar to that of Thatcher+Lr16 against Pt races MBJ/SP. So, by comparing the physical locations, QLr.cim-2BS and Lr16 should be the same gene that conferred LR resistance in Mucuy. Lr16 was a widely deployed LR resistance gene and conferred moderate resistance at both seedling and adult plant stages, and also showed high additive effects with other resistance genes in the field (Lan et al., 2014), which can be considered useful for breeding.
Resistance loci on group 3 chromosomes
QYr.cim-3AS was detected in both Mexico and Kenya for multiple years, but not in India. Therefore, it was also considered an unstable or race-specific APR gene. So far, only one formally named gene, Yr76 (Xiang et al., 2016), is mapped on 3AS, but QYr.cim-3AS should be distinct from Yr76, given that the latter was a seedling resistance gene. For similar reasons, YrEDWL (Liu et al., 2017) and several YR resistance-linked SNPs (Jighly et al., 2015) were identified on 3AS but are unlikely to be QYr.cim-3AS. In addition, two APR QTL have been mapped on 3AS in the CIMMYT wheat line “Saar” (Lillemo et al., 2008) and the Swiss winter wheat cultivar Arina (Buerstmayr et al., 2014). QYr.cim-3AS might be the same as the two QTLs, according to its physical position based on the CS reference genome (International Wheat Genome Sequencing Consortium, 2018).
There are 4 officially named genes on 3BS: Yr4 (Bansal et al., 2010), Yr30 (Singh et al., 2000b), Yr57 (Randhawa et al., 2015), and Yr58 (Chhetri et al., 2016). QYr.cim-3BS should be different from Yr4 and Yr57 because the latter two provide resistance at the seedling stage and their physical positions are around 3.3 Mb (Xbarc75), which is at least 7.3 Mb away from QYr.cim-3BS. Yr58 starts to express and confer resistance at the four-leaf stage. Although Yr30 is a common APR gene in CIMMYT materials, the approximate physical interval of this gene is in the telomeric region distal to 10 Mb (William et al., 2006; Rosewarne et al., 2012; Basnet et al., 2014a; Lan et al., 2014; Wu et al., 2017; Jia et al., 2020), but QYr.cim-3BS is positioned at 10.6–24.9 Mb. Other YR resistance QTL located on 3BS, considered distinct from Yr30, include QYrhm.nwafu-3BS (Yuan et al., 2018), QYrsk.wgp-3BS (Liu et al., 2019), and Qyrto.swust-3BS (Zhou et al., 2019). The physical locations of QYrhm.nwafu-3BS and QYrsk.wgp-3BS overlapped with QYr.cim-3BS; it is possible that they are the same gene/allele, but this needs further verification. It seems that there is more than one APR gene with significant resistance to YR in the 3BS region, other than just Yr30, but we cannot rule out the possibility of the presence of Yr30 in Mucuy, since Yr30 is not cloned yet.
QYr.cim-3DS was identified only in Kenya for 2 consecutive years and accounted for 3.6–7.7% of the phenotypic variation. Singh et al. (2000b) and Boukhatem et al. (2002) both mapped a QTL on 3DS in the CIMMYT wheat “Opata 85.” The tightly linked marker, Xbcd532, is physically located at least 37.1 Mb away from QYr.cim-3DS. There are several other YR resistance genes identified on chromosome 3DS, such as Yr49 (McIntosh et al., 2017), Yr66 (McIntosh et al., 2013), YrY206 (Zhang et al., 2008), QYr.inra-3DS (Dedryver et al., 2009), YrS1 (Sun et al., 2019), and QYrsn.nwafu-3DS (Huang et al., 2020). Among them, QYrsn.nwafu-3DS provides resistance only in seedlings and YrY206 originated from Aegilops tauschii (Coss.) Schmal. According to the physical location of the flanking markers (International Wheat Genome Sequencing Consortium, 2018), Yr49 (Xgwm161) and YrS1 (Xcfd79) were mapped 71.7 and 65.6 Mb distal to QYr.cim-3DS, respectively. Yr66 (Xgwm341) and QYr.inra-3DS (Xgwm456) are at least 18.8 and 122.9 Mb away from QYr.cim-3DS, respectively. Based on the source of resistance, resistance characteristics, and physical location comparisons, QYr.cim-3DS might be a new YR APR QTL, in Kenyan environments.
Resistance loci on group 5 chromosomes
QLr.cim-5AL was located on 5AL, where no other LR resistance gene has been officially designated. Rosewarne et al. (2012) detected a QTL on 5AL derived from Avocet that provided APR to LR. The LOD peak of this locus was near wPt-0837, which corresponds to a physical location of 621.6 Mb, based on the CS reference genome (International Wheat Genome Sequencing Consortium, 2018). QLr.cim-5AL detected in the present study was located on 585.2–589.2 Mb and was derived from Mucuy, so the two QTL should be different, given the ~35 Mb gap between them. Recently, Zhang et al. (2019) identified QLr.hebau-5AL flanked by AX-110679506 and AX-110996595, which corresponded to 589.3–591.4 Mb of the CS reference genome (International Wheat Genome Sequencing Consortium, 2018). This locus explained 6.6–7.1% of the variation in the LR resistance response and was derived from a resistant cultivar SW 8588, whose pedigree includes the CIMMYT variety Milan. Therefore, this locus is likely to be QLr.cim-5AL. However, QLr.hebau-5AL also had an effect on YR across four environments in China, whereas no effect of QLr.cim-5AL on YR was identified in the present study. We speculate that this could be due to different Pst isolates present in China and Mexico or a genetic background effect.
Resistance loci on group 6 chromosomes
QYr.cim-6BS was identified on 6BS. There are three named YR resistance genes on this chromosome: Yr35 (Marais et al., 2003), Yr36 (Uauy et al., 2005), and Yr78 (Dong et al., 2017). Yr35 provides all-stage resistance and was transferred to wheat from T. turgidum ssp. dicoccoides. Yr36 also originated from T. dicoccoides and encodes a kinase-START protein that confers temperature-dependent broad-spectrum resistance (Fu et al., 2009). QYr.cim-6BS is unlikely to be Yr35 or Yr36, based on the source and its resistance characteristics. Yr78 provides APR only in the field (Dong et al., 2017). Comparing the genetic distance and physical position between the corresponding closely linked molecular markers (Somers et al., 2004), several QTL providing APR to YR that map on 6BS could likely be Yr78. These include QYrst.wgp-6BS.1 (Santra et al., 2008), QYr.inra-6B (Dedryver et al., 2009), QYr.caas-6BS (Lan et al., 2010), QYr.caas-6BS.3 (Ren et al., 2012b), and QYrMA.wgp-6BS (Liu et al., 2018). The physical locations of several markers closely linked to Yr78 are included in the range of QYr.cim-6BS detected in this study, such as Xwmc104 (149.1 Mb) and Xbarc136 (151.3 Mb). Therefore, it is likely that QYr.cim-6BS is the same as Yr78, but further verification of this is needed.
The evolution of new virulence and pathogen migration and adaptation to unconventional environments has been observed in the last decade. In addition to the rapid mutation from avirulence to virulence in rust fungi, global climate change and the limited use of resistance genes in complex combinations are important contributors. The average effective life of a race-specific resistance gene is 2–4 years, with the evolution of new races in Mexico. Breeding new varieties with durable resistance is the most effective way to control wheat diseases. Mucuy was distributed for international testing in 2013 and showed high resistance to both YR and LR in Mexico, India, Kenya, and China, suggesting that is a good choice as the donor for introducing resistance into other elite breeding materials. The new resistance loci identified in our study can be further studied to characterize their effects and interactions in other genetic backgrounds and thereby derive the best combinations of effective resistance genes to enhance durability.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
RS, CY, JH-E, MR, SB, EL, and CL conceived of the project. DL, CY, and CL wrote the article, performed QTL mapping, and SSR and KASP analyses. RS and CL performed the developmental analyses. JH-E, MR, SB, and EL evaluated the phenotyping of the RILs. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by International Cooperation and Exchange of the National Natural Science Foundation of China (Grant Nos. 31861143010 and 32101712), Hubei Hongshan Laboratory (Grant Nos. 2022hspy001 and 2021hskf008), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA24030102), Construction Project for Innovation Platform of Qinghai Province (Grant No. 2022-ZJ-Y04), Construction Project for Innovation Platform of Qinghai Province (Grant No. 2022-ZJ-Y01), Key R & D and Transformation Program of Qinghai Province (Grant No. 2022-NK-106), the Australian Grains Research and Development Corporation (GRDC) with funding to the Australian Cereal Rust Control Program (ACRCP), CGIAR Research Program WHEAT (CRPWHEAT), and the Foundation of Application of Basic Research Project of Qinghai Province (Grant No. 2022-ZJ-737).
Acknowledgments
We appreciate copy and content editing by CIMMYT communications consultant Mike Listman.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.880138/full#supplementary-material
Supplementary Figure 1. Genetic linkage maps of stripe rust (YR) seedling resistance gene YrMu on the chromosome 2NS/2AS (A) and of leaf rust (LR) seedling resistance gene Lr16 on wheat chromosome 2BS (B), after removing redundant markers. Locus names and corresponding locations on the genetic map are indicated on the right side. Map distances in cM are shown on the left side.
Supplementary Figure 2. Scatter plots for KASP marker Kasp_hzau_MuYLr-1BL genotyping in the “Apav#1 × Mucuy” F5 RIL population.
Supplementary Table 1. Avirulence/virulence information for rust races used at Ludhiana, India. Yrso, Suwon92Omar; Yrsp, Spalding Prolific; Yrsk, Selkirk; Yrsd, Strubes Dickkopf; Yrks, Kalyansona.
Supplementary Table 2. Sequences of newly developed primers used to detect the co-located resistance loci QYr.cim-1BL/QLr.cim-1BL and QYr.cim-2AS/QLr.cim-2AS.
Supplementary Table 3. Single marker analysis of KASP marker Kasp_hzau_MuYLr-1BL.
Supplementary Table 4. Single marker analysis of InDel marker InD_hzau_MuYLr-2AS.
Supplementary Table 5. The physical position of QYr.cim-2AS/YrMU/QLr.cim-2AS's flanking marker based on reference genomes of Chinese Spring and Jagger.
Supplementary Table 6. The physical location of QYr.cim-2AS's flanking marker showed that the actual genome sequence of Mucuy on Chromosome 2AS was different from that of Chinese Spring.
References
Ali, S., Gladieux, P., Rahman, H., Saqib, M. S., Fiaz, M., Ahmed, H., et al. (2014). Inferring the contribution of sexual reproduction, migration and off-season survival to the temporal maintenance of microbial populations: a case study on the wheat fungal pathogen Puccinia striiformis f.sp. Tritici. Mol. Ecol. 23, 603–617. doi: 10.1111/mec.12629
Bansal, U. K., Hayden, M. J., Gill, M. B., and Bariana, H. S. (2010). Chromosomal location of an uncharacterized stripe rust resistance gene in wheat. Euphytica 171, 121–127. doi: 10.1007/s10681-009-0007-4
Bansal, U. K., Hayden, M. J., Venkata, B. P., Khanna, R., Saini, R. G., and Bariana, H. S. (2008). Genetic mapping of adult plant leaf rust resistance genes Lr48 and Lr49 in common wheat. Theor. Appl. Genet. 117, 307–312. doi: 10.1007/s00122-008-0775-6
Bariana, H. S., and McIntosh, R. A. (1993). Cytogenetic studies in wheat. XV. Location of rust resistance genes in VPM1 and their genetic linkage with other disease resistance genes in chromosome 2A. Genome 36, 476–482. doi: 10.1139/g93-065
Basnet, B. R., Ibrahim, A. M. H., Chen, X. M., Singh, R. P., Mason, E. R., Bowden, R. L., et al. (2014b). Molecular mapping of stripe rust resistance in hard red winter wheat TAM 111 adapted to the U.S. High Plains. Crop Sci. 54, 1361–1373, doi: 10.2135/cropsci2013.09.0625
Basnet, B. R., Singh, R. P., Herrera-Foessel, S. A., Ibrahim, A. M. H., Huerta-Espino, J., Calvo-Salazar, V., et al. (2013). Genetic analysis of adult plant resistance to yellow rust and leaf rust in common spring wheat Quaiu 3. Plant Dis. 97, 728–736. doi: 10.1094/PDIS-02-12-0141-RE
Basnet, B. R., Singh, R. P., Ibrahim, A. M. H., Herrera-Foessel, S. A., Huerta-Espino, J., Lan, C., et al. (2014a). Characterization of Yr54 and other genes associated with adult plant resistance to yellow rust and leaf rust in common wheat Quaiu 3. Mol. Breed. 33, 385–399. doi: 10.1007/s11032-013-9957-2
Bayles, R. A., Flath, K., Hovmøller, M. S., and Vallavieille-Pope, C. de. (2000). Breakdown of the Yr17 resistance to yellow rust of wheat in Northern Europe. Agronomie 20, 805–811. doi: 10.1051/agro:2000176
Bjarko, M. E., and Line, R. F. (1988). Heritability and number of genes control-ling leaf rust resistance in four cultivars of wheat. Phytopathology 78, 457–461 doi: 10.1094/Phyto-78-457
Bolton, M. D., Kolmer, J. A., and Garvin, D. F. (2008). Wheat leaf rust caused by Puccinia triticina. Mol. Plant Pathol. 9, 563–575. doi: 10.1111/j.1364-3703.2008.00487.x
Boukhatem, N., Baret, P. V., Mingeot, D., and Jacquemin, J. M. (2002). Quantitative trait loci for resistance against yellow rust in two wheat-derived recombinant inbred line populations. Theor. Appl. Genet. 104, 111–118. doi: 10.1007/s001220200013
Buerstmayr, M., Matiasch, L., Mascher, F., Vida, G., Ittu, M., Robert, O., et al. (2014). Mapping of quantitative adult plant field resistance to leaf rust and stripe rust in two European winter wheat populations reveals co-location of three QTL conferring resistance to both rust pathogens. Theor. Appl. Genet. 127, 2011–2028. doi: 10.1007/s00122-014-2357-0
Chen, X. (2005). Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can. J. Plant Pathol. 27, 314–337. doi: 10.1080/07060660509507230
Chen, X. (2013). High-temperature adult-plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 4, 8–27. doi: 10.4236/ajps.2013.43080
Chhetri, M., Bariana, H., Kandiah, P., and Bansal, U. (2016). Yr58: a new stripe rust resistance gene and its interaction with Yr46 for enhanced resistance. Phytopathol. 106, 1530–1534. doi: 10.1094/PHYTO-04-16-0182-R
Datta, D., Nayar, S. K., Bhardwaj, S. C., Prashar, M., and Kumar, S. (2008). Detection and inheritance of leaf rust resistance in common wheat lines Agra Local and IWP94. Euphytica 159, 343–351. doi: 10.1007/s10681-007-9522-3
Dedryver, F., Paillard, S., Mallard, S., Robert, O., Trottet, M., Nègre, S., et al. (2009). Characterization of genetic components involved in durable resistance to stripe rust in the bread wheat ‘Renan'. Phytopathology 99, 968–973. doi: 10.1094/PHYTO-99-8-0968
Dong, Z. Z., Hegarty, J. M., Zhang, J. L., Zhang, W. J., Chao, S. M., Chen, X. M., et al. (2017). Validation and characterization of a QTL for adult plant resistance to stripe rust on wheat chromosome arm 6BS (Yr78). Theor. Appl. Genet. 130, 2127–2137. doi: 10.1007/s00122-017-2946-9
Dreisigacker, S., Sehgal, D., Luna, B., Reyes, A. E., and Mollins, J. (2012). CIMMYT Wheat Molecular Genetics Laboratory Protocols and Applications to Wheat Breeding. México-Veracruz: CIMMYT.
Fang, T., Campbell, K. G., Liu, Z., Chen, X., Wan, A., Li, S., et al. (2011). Stripe rust resistance in the wheat cultivar Jagger is due to Yr17 and a novel resistance gene. Crop Sci. 51, 2455–2465. doi: 10.2135/cropsci2011.03.0161
Francki, M. G., Walker, E., Crawford, A. C., Broughton, S., Ohm, H. W., Barclay, I., et al. (2009). Comparison of genetic and cytogenetic maps of hexaploid wheat (Triticum aestivum L.) using SSR and DArT markers. Mol. Genet. Genom. 281, 181–191. doi: 10.1007/s00438-008-0403-9
Fu, D., Uauy, C., Distelfeld, A., Blechl, A., Epstein, L., Chen, X. M., et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. doi: 10.1126/science.1166289
Gao, L., Koo, D.-H., Juliana, P., Rife, T., Singh, D., Silva, C. L., et al. (2021). The Aegilops ventricosa 2NvS segment in bread wheat: cytology, genomics and breeding. Theor. Appl. Genet. 134, 529–542. doi: 10.1007/s00122-020-03712-y
Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., Rosewarne, G. M., Periyannan, S. K., Viccars, L., et al. (2012). Lr68: a new gene conferring slow rusting resistance to leaf rust in wheat. Theor. Appl. Genet. 124, 1475–1486, doi: 10.1007/s00122-012-1802-1
Herrera-Foessel, S. A., Singh, R. P., Lillemo, M., Huerta-Espino, J., Bhavani, S., Singh, S., et al. (2014). Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 127, 781–789, doi: 10.1007/s00122-013-2256-9
Hovmøller, M. S., Walter, S., Bayles, R. A., Hubbard, A., Flath, K., Sommerfeldt, N., et al. (2016). Replacement of the European wheat yellow rust population by new races from the centre of diversity in the near-Himalayan region. Plant Pathol. 65, 402–411. doi: 10.1111/ppa.12433
Huang, B. E., George, A. W., Forrest, K. L., Kilian, A., Hayden, M. J., Morell, M. K., et al. (2012). A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Bio. J. 10, 826–839. doi: 10.1111/j.1467-7652.2012.00702.x
Huang, S., Liu, S. J., Zhang, Y. B., Xie, Y. Z., Wang, X. T., Jiao, H. X., et al. (2020). Genome-wide wheat 55K SNP-based mapping of stripe rust resistance loci in wheat cultivar Shaannong 33 and their alleles frequencies in current Chinese wheat cultivars and breeding lines. Plant Dis. 105, 1048–1056. doi: 10.1094/PDIS-07-20-1516-RE
Huerta-Espino, J., Singh, R. P., Crespo-Herrera, L. A., Villaseñor-Mir, H. E., Rodriguez-Garcia, M. F., and Dreisigacker, S. (2020). Adult plant slow rusting genes confer high levels of resistance to rusts in bread wheat cultivars from Mexico. Front. Plant Sci. 11, 824. doi: 10.3389/fpls.2020.00824
Huerta-Espino, J., Singh, R. P., Germán, S., McCallum, B. D., Park, R. F., and Chen, W. Q. (2011). Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 179, 143–160. doi: 10.1007/s10681-011-0361-x
International Wheat Genome Sequencing Consortium (2018). Shifting the limits in wheat research, and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Jia, M. J., Yang, L. J., Zhang, W., Rosewarne, G., Li, J. H., Yang, E., et al. (2020). Genome-wide association analysis of stripe rust resistance in modern Chinese wheat. BMC Plant Biol. 20, 491. doi: 10.1186/s12870-020-02693-w
Jighly, A., Oyiga, B. C., Makdis, F., Nazari, K., Youssef, O., Tadesse, W., et al. (2015). Genome-wide DArT and SNP scan for QTL associated with resistance to stripe rust (Puccinia striiformis f. sp. tritici) in elite ICARDA wheat (Triticum aestivum L.) germplasm. Theor. Appl. Genet. 128, 1277–1295. doi: 10.1007/s00122-015-2504-2
Kolmer, J. A. (1996). Genetics of resistance to wheat leaf rust. Ann Rev. Phytopathol. 34, 435–455. doi: 10.1146/annurev.phyto.34.1.435
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1362. doi: 10.1126/science.1166453
Kumar, S., Bhardwaj, S. C., Gangwar, O. P., Sharma, A., Qureshi, N., and Kumaran, V. V. (2021). Lr80: a new and widely effective source of leaf rust resistance of wheat for enhancing diversity of resistance among modern cultivars. Theor. Appl. Genet. 134, 849–858. doi: 10.1007/s00122-020-03735-5
Lan, C., Basnet, B. R., Singh, R. P., Huerta-Espino, J., Herrera-Foessel, S. A., and Ren, Y. (2017b). Genetic analysis and mapping of adult plant resistance loci to leaf rust in durum wheat cultivar Bairds. Theor. Appl. Genet. 130, 609–619. doi: 10.1007/s00122-016-2839-3
Lan, C., Hale, I. L., Herrera-Foessel, S. A., Basnet, B. R., Randhawa, M. S., Huerta-Espino, J., et al. (2017a). Characterization and mapping of leaf rust and stripe rust resistance loci in hexaploid wheat lines UC1110 and PI610750 under Mexican environments. Front. Plant Sci. 8, 1450. doi: 10.3389/fpls.2017.01450
Lan, C., Rosewarne, G. M., Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Basnet, B. R., et al. (2014). QTL characterization of resistance to leaf rust and stripe rust in the spring wheat line Francolin#1. Mol. Breed. 34, 789–803. doi: 10.1007/s11032-014-0075-6
Lan, C., Zhang, Y., Herrer-Foessel, S. A., Bhoja, R., Basnet, B. R., Huerta-Espino, J., et al. (2015). Identification and characterization of pleiotropic and co-located resistance loci to leaf rust and stripe rust in bread wheat cultivar Sujata. Theor. Appl. Genet. 128, 549–561. doi: 10.1007/s00122-015-2454-8
Lan, C. X., Liang, S. S., Zhou, X. C., Zhou, G., Lu, Q. L., Xia, X. C., et al. (2010). Identification of genomic regions controlling adult-plant stripe rust resistance in chinese landrace pingyuan 50 through bulked segregant analysis. Phytopathology 100, 313–318. doi: 10.1094/PHYTO-100-4-0313
Li, Z., Lan, C., He, Z., Singh, R. P., Rosewarne, G. M., Chen, X., et al. (2014). Overview and application of QTL for adult plant resistance to leaf rust and powdery mildew in wheat. Crop Sci. 54, 1907–1925. doi: 10.2135/cropsci2014.02.0162
Lillemo, M., Asalf, B., Singh, R. P., Huerta-Espino, J., Chen, X., He, Z., et al. (2008). The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theor. Appl. Genet. 116, 1155–1166. doi: 10.1007/s00122-008-0743-1
Liu, L., Wang, M. N., Feng, J. Y., See, D. R., Chao, S. M., and Chen, X. M. (2018). Combination of all-stage and high-temperature adult-plant resistance QTL confers high-level, durable resistance to stripe rust in winter wheat cultivar Madsen. Theor. Appl. Genet. 131, 1835–1849. doi: 10.1007/s00122-018-3116-4
Liu, L., Yuan, C. Y., Wang, M. N., See, D. R., Zemetra, R. S., and Chen, X. M. (2019). QTL analysis of durable stripe rust resistance in the North American winter wheat cultivar Skiles. Theor. Appl. Genet. 132, 1677–1691. doi: 10.1007/s00122-019-03307-2
Liu, W., Maccaferri, M., Rynearson, S., Letta, T., Zegeye, H., Tuberosa, R., et al. (2017). Novel sources of stripe rust resistance identified by genome-wide association mapping in Ethiopian Durum Wheat (Triticum turgidum ssp. durum). Front. Plant Sci. 8, 774. doi: 10.3389/fpls.2017.00774
Marais, G. F., Pretorius, Z. A., Marais, A. S., and Wellings, C. R. (2003). Transfer of rust resistance genes from Triticum species to common wheat. S. Afr. J. Plant Soil 20, 193–198. doi: 10.1080/02571862.2003.10634934
McCartney, C. A., Somers, D. J., McCallum, B. D., Thomas, J., Humphreys, D. G., Menzies, J. G., et al. (2005). Microsatellite tagging of the leaf rust resistance gene Lr16 on wheat chromosome 2BSc. Mol. Breed. 15, 329–337. doi: 10.1007/s11032-004-5948-7
McIntosh, R., Dubcovsky, J., and Rogers, W. (2013). Catalogue of Gene Symbols for Wheat: 2013−2014 Supplement [ER/OL]. Available online at: https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2013.pdf
McIntosh, R. A., Dubcovsky, J., Rogers, J., Morris, C., Appels, R., and Xia, X. (2017). Catalogue of Gene Symbols for Wheat: 2017 Supplement. Available online at: https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp
McNeal, F. H., Konzak, C. F., Smith, E. P., Tate, W. S., and Russell, T. S. (1971). A Uniform System for Recording and Processing Cereal Research Data. Washington, DC: USDA-ARS Bull, 34–121.
Meng, L., Li, H., Zhang, L., and Wang, J. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in bi-parental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Messmer, M. M., Seyfarth, R., Keller, M., Schachermayr, G., Winzeler, M., Zanetti, S., et al. (2000). Genetic analysis of durable leaf rust resistance in winter wheat. Theor. Appl. Genet. 100, 419–431. doi: 10.1007/s001220050055
Milus, E. A., Lee, K. D., and Brown-Guedira, G. (2015). Characterization of stripe rust resistance in wheat lines with resistance gene Yr17 and implications for evaluating resistance and virulence. Phytopathology 105, 1123–1130. doi: 10.1094/PHYTO-11-14-0304-R
Moore, J. W., Herrera-Foessel, S. A., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Park, R. F., Mohler, V., Nazari, K., and Singh, D. (2014). Characterization and mapping of gene Lr73 conferring seedling resistance to Puccinia triticina in common wheat. Theor. Appl. Genet. 127, 2041–2049. doi: 10.1007/s00122-014-2359-y
Peterson, R. F., Campbell, A. B., and Hannah, A. E. (1948). A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 26, 496–500. doi: 10.1139/cjr48c-033
Ponce-Molina, L. J., Huerta-Espino, J., Singh, R. P., Basnet, B. R., Alvarado, G., Randhawa, M. S., et al. (2018). Characterization of leaf rust and stripe rust resistance in spring wheat “Chilero.” Plant Dis. 102, 421–427. doi: 10.1094/PDIS-11-16-1545-RE
Randhawa, M. S., Bariana, H. S., Mago, R., and Bansal, U. K. (2015). Mapping of a new stripe rust resistance locus Yr57 on chromosome 3BS of wheat. Mol. Breed. 35, 65. doi: 10.1007/s11032-015-0270-0
Randhawa, M. S., Lan, C., Basnet, B. R., Bhavani, S., Huerta-Espino, J., Forrest, K. L., et al. (2018). Interactions among genes Sr2/Yr30, Lr34/Yr18/Sr57 and Lr68 confer enhanced adult plant resistance to rust diseases in common wheat (Triticum aestivum L.) line Arula. AJCS 12, 1023–1033. doi: 10.21475/ajcs.18.12.06.PNE1305
Ren, Y., He, Z. H., Li, J., Lillemo, M., Wu, L., Bai, B., et al. (2012a). QTL mapping of adult-plant resistance to stripe rust in a population derived from common wheat cultivars Naxos and Shanghai 3/Catbird. Theor. Appl. Genet. 125, 1211–1221. doi: 10.1007/s00122-012-1907-6
Ren, Y., Li, Z. F., He, Z. H., Wu, L., Bai, B., Lan, C. X., et al. (2012b). QTL mapping of adult-plant resistances to stripe rust and leaf rust in Chinese wheat cultivar Bainong 64. Theor. Appl. Genet. 125, 1253–1262. doi: 10.1007/s00122-012-1910-y
Ren, Y., Singh, R. P., Basnet, B. R., Lan, C., Huerta-Espino, J., Lagudah, E. S., et al. (2017). Identification and mapping of adult plant resistance loci to leaf rust and stripe rust in common wheat cultivar Kundan. Plant Dis. 101, 456–463. doi: 10.1094/PDIS-06-16-0890-RE
Roelfs, A. P., Singh, R. P., and Saari, E. E. (1992). Rust Diseases of Wheat: Concepts and Methods of Disease Management. Mexico, DF: CIMMYT.
Rosewarne, G. M., Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., Lan, C., and He, Z. (2013). Quantitative trait loci of stripe rust resistance in wheat. Theor. Appl. Genet. 126, 2427–2449. doi: 10.1007/s00122-013-2159-9
Rosewarne, G. M., Singh, R. P., Huerta-Espino, J., Herrera-Foessel, S. A., Forrest, K. L., Hayden, M. J., et al. (2012). Analysis of leaf and stripe rust severities reveals pathotype changes and multiple minor QTL associated with resistance in an Avocet × Pastor wheat population. Theor. Appl. Genet. 124, 1283–194. doi: 10.1007/s00122-012-1786-x
Santra, D. K., Chen, X. M., Santra, M., Campbell, K. G., and Kidwell, K. K. (2008). Identification and mapping QTL for high-temperature adult-plant resistance to stripe rust in winter wheat (Triticum aestivum L.) cultivar Stephens. Theor. Appl. Genet. 117, 793–802. doi: 10.1007/s00122-008-0820-5
Seyfarth, R., Feuillet, C., Schachermayr, G., Winzeler, M., and Keller, B. (1999). Development of a molecular marker for the adult plant leaf rust resistance gene Lr35 in wheat. Theor. Appl. Genet. 99, 554–560. doi: 10.1007/s001220051268
Singh, R. P. (1991). Pathogenicity variations of Puccinia recondita f. sp. tritici and P. graminis f. sp. tritici in wheat-growing areas of Mexico during 1988 and 1989. Plant Dis. 75, 790–794. doi: 10.1094/PD-75-0790
Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Bariana, H., Bansal, U., McCallum, B. C., et al. (2012). “Lr34/Yr18/Sr57/Pm38/Bdv1/Ltn1” confers slow rusting, adult plant resistance to stem rust,” in Proceedings of the 13th International Cereal Rusts and Powdery Mildews Conference (Beijing), 173.
Singh, R. P., Huerta-Espino, J., and Rajaram, S. (2000a). Achieving near immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta. Phytopathol. Entomol. Hung. 35, 133–139.
Singh, R. P., Mujeeb-Kazi, A., and Huerta-Espino, J. (1998). Lr46: a gene conferring slow rusting resistance to leaf rust in wheat. Phytopathol. 88, 890–894. doi: 10.1094/PHYTO.1998.88.9.890
Singh, R. P., Nelson, J. C., and Sorrells, M. E. (2000b). Mapping Yr28 and other Genes for resistance to stripe rust in wheat. Crop Sci. 40, 1148–1155. doi: 10.2135/cropsci2000.4041148x
Somers, D. J., Isaac, P., and Edwards, K. (2004). A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109, 1105–1114. doi: 10.1007/s00122-004-1740-7
Sun, C., Zhang, P., Fang, Z. W., Zhang, X., Yin, J. L., Ma, D. F., et al. (2019). Genetic analysis and molecular mapping of stripe rust resistance in an excellent wheat line Sanshumai1. J. Plant Pathol. 101, 235–241. doi: 10.1007/s42161-018-0166-z
Uauy, C., Brevis, J. C., Chen, X. M., Khan, I., Jackson, L., Chicaiza, O., et al. (2005). High-temperature adult-plant (HTAP) stripe rust resistance gene Yr36 from Triticum turgidum ssp. dicoccoides is closely linked to the grain protein content locus Gpc-B1. Theor. Appl. Genet. 112, 97–105. doi: 10.1007/s00122-005-0109-x
Van Ooijen, J. W. (2006). Join Map 4, Software for the Calculation of Genetic Linkage Maps in Experimental Population. Wageningen: Plant Research International.
Walter, S., Ali, S., Kemen, E., Nazari, K., Bahri, B. A., Enjalbert, J., et al. (2016). Molecular markers for tracking the origin and worldwide distribution of invasive strains of Puccinia striiformis. Ecol. Evol. 6, 2790–2804. doi: 10.1002/ece3.2069
Wang, Y., Zhang, H. Z., Xie, J. Z., Guo, B. M., Chen, Y. X., Zhang, H. Y., et al. (2018). Mapping stripe rust resistance genes by BSR-Seq: YrMM58 and YrHY1 on chromosome 2AS in Chinese wheat lines Mengmai 58 and Huaiyang 1 are Yr17. Crop J. 6, 91–98. doi: 10.1016/j.cj.2017.03.002
Wilkinson, P. A., Winfield, M. O., Barker, G. L. A., Allen, A. M., Burridge, A., Coghill, J. A., et al. (2012). CerealsDB 2.0: an integrated resource for plant breeders and scientists. BMC Bio. 13, 219. doi: 10.1186/1471-2105-13-219
William, H. M., Singh, R. P., Huerta-Espino, J., Palacios, G., and Suenaga, K. (2006). Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 49, 977–990. doi: 10.1139/g06-052
Wu, J. H., Wang, Q. L., Kang, Z. S., Liu, S. J., Li, H. Y., Mu, J. M., et al. (2017). Development and validation of KASP-SNP markers for QTL underlying resistance to stripe rust in common wheat cultivar P10057. Plant Dis. 101, 2079–2087. doi: 10.1094/PDIS-04-17-0468-RE
Xiang, C., Feng, J., Wang, M., Chen, X., See, D. R., Wan, A., et al. (2016). Molecular mapping of stripe rust resistance gene Yr76 in winter club wheat cultivar Tyee. Phytopathology 106, 1186–1193. doi: 10.1094/PHYTO-01-16-0045-FI
Xu, X., Bai, G., Carver, B., Shaner, G. E., and Hunger, R. M. (2005). Molecular characterization of slow leaf-rusting resistance in wheat. Crop Sci. 45, 758–765. doi: 10.2135/cropsci2005.0758
Yuan, C., Singh, R. P., Liu, D., Randhawa, M. S., Huerta-Espino, J., and Lan, C. (2020). Genome-wide mapping of adult plant resistance to leaf rust and stripe rust in CIMMYT wheat line “Arableu#1.” Plant Dis. 104, 1455–1463. doi: 10.1094/PDIS-10-19-2198-RE
Yuan, F. P., Zeng, Q. D., Wu, J. H., Wang, Q. L., Yang, Z. J., Liang, B. P., et al. (2018). QTL mapping and validation of adult plant resistance to stripe rust in Chinese wheat landrace Humai 15. Front. Plant Sci. 9, 968. doi: 10.3389/fpls.2018.00968
Zadoks, J. C. (1961). Yellow rust on wheat studies in epidemiology and physiologic specialization. Tijdschrift over Plantenziekten. 67, 69–256. doi: 10.1007/BF01984044
Zhang, H. Q., Lang, J., Ma, S. Q., and Zhang, B. S. (2008). Genetic analysis and SSR mapping on a new stem stripe rust resistance gene YrY206 in Aegilops tauschii. Chin. J. Biotech. 24, 1475–1479.
Zhang, P., Li, X., Gebrewahid, T. W., Liu, H., Xia, X., He, Z., et al. (2019). QTL mapping of adult-plant resistance to leaf and stripe rust in wheat cross SW 8588/Thatcher using the wheat 55K SNP array. Plant Dis. 103, 3041–3049. doi: 10.1094/PDIS-02-19-0380-RE
Zhou, X., Zhong, X., Roter, J., Li, X., Yao, Q., Yan, J., et al. (2021). Genome-wide mapping of loci for adult-plant resistance to stripe rust in durum wheat Svevo using the 90K SNP array. Plant Dis. 105, 879–888. doi: 10.1094/PDIS-09-20-1933-RE
Keywords: co-located resistance loci, Puccinia striiformis f. sp. tritici, P. triticina, QTL, adult plant resistance
Citation: Liu D, Yuan C, Singh RP, Randhawa MS, Bhavani S, Kumar U, Huerta-Espino J, Lagudah E and Lan C (2022) Stripe rust and leaf rust resistance in CIMMYT wheat line “Mucuy” is conferred by combinations of race-specific and adult-plant resistance loci. Front. Plant Sci. 13:880138. doi: 10.3389/fpls.2022.880138
Received: 21 February 2022; Accepted: 28 July 2022;
Published: 19 August 2022.
Edited by:
Christina Cowger, Plant Science Research Unit, Agricultural Research Service (USDA), United StatesReviewed by:
Volker Mohler, Bayerische Landesanstalt für Landwirtschaft (LfL), GermanyMatthew Rouse, Cereal Disease Laboratory, Agricultural Research Service (USDA), United States
Copyright © 2022 Liu, Yuan, Singh, Randhawa, Bhavani, Kumar, Huerta-Espino, Lagudah and Lan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ravi P. Singh, ci5zaW5naEBjZ2lhci5vcmc=; Caixia Lan, Y3hsYW5AbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Demei Liu
Demei Liu Chan Yuan
Chan Yuan Ravi P. Singh
Ravi P. Singh Mandeep S. Randhawa
Mandeep S. Randhawa Sridhar Bhavani
Sridhar Bhavani Uttam Kumar
Uttam Kumar Julio Huerta-Espino
Julio Huerta-Espino Evans Lagudah
Evans Lagudah Caixia Lan
Caixia Lan