- 1State Key Laboratory Breeding Base of Dao-di Herbs, National Resource Center for Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
- 2School of Traditional Chinese Medicine, Guangdong Pharmaceutical University, Guangzhou, China
- 3Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, China
Root size is a key trait in plant cultivation and can be influenced by the cultivation environment. However, physical evidence of root size change in a secular context is scarce due to the difficulty in preserving ancient root samples, and how they were modified during the domestication and cultivation stays unclear. About 100 ancient root samples of Panax notoginseng, preserved as tribute in the Palace Museum (A.D. 1636 to 1912, Qing dynasty), provided an opportunity to investigate the root size changes during the last 100 years of cultivation. The dry weight of ancient root samples (~120 tou samples, tou represents number of roots per 500 g dry weight) is 0.22-fold of the modern samples with the biggest size (20 tou samples). Transcriptome analysis revealed that PnGAP and PnEXPA4 were highly expressed in 20 tou samples, compared with the 120 tou samples, which might contribute to the thicker cell wall and a higher content of lignin, cellulose, and callose in 20 tou samples. A relatively lower content of dencichine and higher content of ginsenoside Rb1 in 20 tou samples are also consistent with higher expression of ginsenoside biosynthesis-related genes. PnPHL8 was filtrated through transcriptome analysis, which could specifically bind the promoters of PnGAP, PnCYP716A47, and PnGGPPS3, respectively. The results in this study represent the first physical evidence of root size changes in P. notoginseng in the last 100 years of cultivation and contribute to a comprehensive understanding of how the cultivation environment affected root size, chemical composition, and clinical application.
Introduction
Plant root size is a key trait for improving water and nitrogen uptake efficiency. In cultivation, temperature (Teskey and Hinckley, 2010; Chathurika et al., 2018), precipitation (Ba Rraclough et al., 2010; Ghaffari et al., 2021), light transmittance (Cheon et al., 2004; Kuang et al., 2015), fertilization (Ba Rraclough et al., 2010; Goodsman et al., 2010), and agrotype (Hiroyoshi et al., 2004; Chen, 2017) can lead to deformation of plant root phenotype. Except for the environmental factors, the genotype is the main factor determining the root size, and a number of quantitative trait loci (QTL) or genes associated with root size have been identified (Jeong et al., 2013; Tamirisa et al., 2014; Yao et al., 2014; Cheng et al., 2016; Ding et al., 2016). It was reported that glycosylphosphatidylinositol (GPI)-anchored protein (GAP) has diverse function on root architecture, by affecting cell wall architecture (Macmillan et al., 2010), cell elongation (Niu et al., 2018), cytoderm thickness, and content of lignin, cellulose, and callose (Liu, 2009; Bundy et al., 2016; Zhao et al., 2020). High temperature decreased the expression of OsGAP18, leading to a thinner cell wall (Zhao et al., 2020), while several GAPs were prominently upregulated during cold acclimation (Daisuke et al., 2016) and nitrogen supply (Engelsberger and Schulze, 2012). Expansin (EXP) also plays significant role in root architecture, and the expression of EXPAs could be induced under cold acclimation, water stress, and higher application of fertilizer (Bian, 2006; Kozbial et al., 2010; Li et al., 2013, 2019b; Sun, 2013; Han et al., 2014; Ren et al., 2019).
Panax notoginseng is a popular functional food and traditional medicine, whose benefits are considered to be represented by the root size and ginsenoside content. The main root of P. notoginseng cultivated at higher altitude is significantly larger (Zheng et al., 2014). Root diameter is directly proportional to light transmittance within limit (Kuang et al., 2014; Wang et al., 2018a), and the application of phosphorus and potassium can increase root weight and promote root thickening (Wang et al., 2008; Zhang et al., 2008). A suitable soil texture possesses great fertilizer preserving capability and abundant mineral elements, promoting the growth of root system (Cui et al., 2005; Li et al., 2016).
Cultivation also affects the content of ginsenosides and amino acids, as well as the transcriptional level of corresponding biosynthetic genes in P. notoginseng. High precipitation inhibits the accumulation of total ginsenosides, while low temperature induced upregulation of HMGR, SS, and SE and increased the ginsenoside content (Liu et al., 2016b; Ma et al., 2021). Increasing the concentration of potassium, nitrogen, magnesium, and calcium brings a remarkable boost to the ginsenoside content, in a certain range (Konsler et al., 1990; Yu et al., 2001). As a key enzyme in dencichine biosynthesis, activity of serine acetyltransferase (SAT) can be decreased by drought stress (Ahmad et al., 2016; Yang et al., 2021).
The cultivation of P. notoginseng can be tracked back to Jiaqing year in Qing dynasty, which is around A.D. 1800 (Wu, 1963). The Palace Museum (Beijing, China) has abundant and well-preserved P. notoginseng samples from Qing dynasty, and those ancient root samples were tributes from Guangxi or Yunnan provinces to the emperor, which represented the best quality of P. notoginseng at that time. The cultivation environments of P. notoginseng showed a great improvement toward planting temperature and humidity (Hua, 1967; He and Deng, 1981; China Association of Chiniese Medicine., 2019), light transmittance (Chen, 1958; Huang et al., 2007; China Association of Chiniese Medicine., 2019), soil type (He and Deng, 1981; China Association of Chiniese Medicine., 2019), and fertilizer application (Chen, 1958; He and Deng, 1981; China Association of Chiniese Medicine., 2019) during past 100 years, with root size also showing an enlargement since 1950s (Jin et al., 1996; Xu, 2016). Tributes from Qing dynasty ought to be an excellent material to investigate how long cultivation influenced root development.
In this study, we compared the root length, diameter, and weight of P. notoginseng from the Palace Museum (Beijing, China) and modern market and performed detailed RNA-seq, UPLC-QQQ-MS, and desorption electrospray ionization analyses of two types of root size (SRW samples, 120 tou with small root weight, representing substitutes for Qing dynasty tribute; and LRW samples, 20 tou with large root weight, representing for highest benefits in modern market). We used these datasets to explore how domestication and cultivation lead to deformation of P. notoginseng root size and to investigate the gene-level regulatory mechanisms that control a better-architected cell wall obtained in LRW samples, providing guidance on artificial cultivation of P. notoginseng.
Materials and Methods
Plant Materials
About 100 root samples of P. notoginseng in Qing dynasty were obtained from The Palace Museum, Beijing. Six types of dry root samples with different sizes (20 tou, 40 tou, 60 tou, 80 tou, 120 tou, and >120 tou, tou represents the number of roots per 500 g dry weight) were purchased from Anhui Tienho Herbal Source Company. Fresh root samples were collected in Wenshan, Yunnan Province, and stored at −80°C (Zheng et al., 2017). According to the drying rate (Xu, 2016), 21 fresh roots of P. notoginseng were divided into two groups: (1) Seven LRW samples that have fresh weight of 14.27 ± 2.54 g and are equivalent to 20 tou samples, and (2) 14 SRW samples that have fresh weight of 5.17 ± 0.62 g and are equivalent to 120 tou samples (Supplementary Table 1).
Analysis of RNA Sequencing Profiles
Raw data from transcript database of P. notoginseng (Zheng et al., 2017) were used for further analysis. Clean data were obtained by removing reads with low quality from raw data. Filtered reads were aligned to the P. notoginseng genome (Jiang et al., 2020) using STAR (Valencia, 2014) and counted using RSEM (Dewey and Li, 2011). Differentially expressed genes (DEGs) were obtained by comparing the gene expression in LRW and SRW P. notoginseng using DESeq2 (Love et al., 2014). Genes with padj below 0.05 and log2 (fold change) >1 were considered as DEGs.
Microscope Observation
Root material was cut into 3-mm tissue samples and dehydrated with a gradient of 70, 80, 95, and 100% of ethanol. The tissue was treated with xylene twice for cell permeabilization and then was soaked and embedded with paraffin. After that, the tissue was cut into 5-μm slice, which was heated and dewaxed in water. Microsection was observed using an Olympus BX51 microscope. The diameter of vessel and the thickness of cell wall were measured using DP2-BSW software. The diameters of vessels in one field of view were measured and averaged, and six fields of view of each five biological replicates were obtained for t-test. Measurement of cell wall thickness in each tissue was same as that of the vessel diameter, and the thickness of 10 cells in one field of view was measured and averaged.
Cell Wall Component Measurement
Sulfuric acid hydrolysis method was used to determine the lignin content (Xiong et al., 2005; Chen et al., 2010). LRW and SRW samples were freeze-dried and ground into powder. About 100 mg of powder was weighed and extracted with 1% acetic acid solution twice. The precipitate was soaked in a mixture of ethanol and diethyl ether (1:1) for three times and evaporated to residue. About 3 mL of 72% (w/v) sulfuric acid was mixed up with the precipitate and stood for 16 h at room temperature. Then, 10 mL of distilled water was added and placed in boiling water bath for 5 min. After cooling, 5 mL of distilled water and 0.5 mL of 10% (w/v) barium chloride solution were added, and the residue was washed with distilled water subsequently. About 10 mL of 10% (w/v) sulfuric acid solution and 10 mL of 0.1 mol·L−1 potassium dichromate solution were added to the residue and heated in boiling water for 15 min. The supernatant after cooling was transferred to flask and then mixed up with 5 mL of 20% (w/v) KI solution and 1 mL of 0.5% (w/v) starch solution for titration. The titrant was 0.2 mol·L−1 of sodium thiosulfate.
Determination of cellulose was performed using enzyme-linked immunosorbent assay (ELISA), according to the manufacturer's instructions (Jiangsu Jingmei Biological Technology Co. Ltd., China. Item number, JM-110113P1). Homogenized LRW and SRW samples were extracted in 900-μL PBS buffer (pH 7.4), and then, supernatant was obtained by centrifugation at 2,000 rpm for 20 min. Microtitration plates were coated with purified cellulose antibody. After 5 times dilution, the sample solution was added to coated micropore, which subsequently bound with HRP-labeled cellulose antibody at 37°C for 30 min. Tetramethylbenzidine (TMB) was added as substrate and then incubated in the dark at 37°C for 10 min. The reaction was terminated using 1 mol·L−1 of sulfuric acid, and the absorbance value was determined at 450 nm. Calibration curve was obtained using cellulose standard solution of 400, 200, 100, 50, and 25 ng·L−1.
Callose determination was performed according to the published method (Khle et al., 1985) with some modification. Briefly, 100 mg of fresh plant materials was washed with ethanol for three times to eliminate autofluorescence, ground in liquid nitrogen, and extracted with 1 mL of 1 mol·L−1 NaOH at 80°C for 15 min. After centrifugation (10,000 × g, 15 min), the supernatant was mixed with 0.1% (w/v) aniline blue to produce a violet-red color. Then, 1 mol·L−1 of glycine/NaOH buffer (pH 9.5) was added and incubated at 50°C for 20 min, and then at room temperature for 20 min. Fluorescence was recorded using a HITACHI F-7000 spectrofluorometer (Tokyo, Japan) with the following parameters: excitation wavelength of 400 nm, emission wavelength of 510 nm, and slit width of 10 nm. Calibration curve was obtained using β-1,3-glucan in 1 mol·L−1 of NaOH.
qRT-PCR Analysis
Total RNA was extracted from root of P. notoginseng, using the Plant RNA Purification Reagent (Invitrogen, USA), according to the manufacturer's instructions. qRT-PCR was performed on a LightCycler 480 Real-Time PCR System (Roche Diagnostics, Basel, Switzerland), with primers listed in Supplementary Table 2.
Determination of Ginsenosides Using UPLC-QTRAP-MS/MS
The ginsenoside content was measured as described previously (Liu et al., 2020), with some modifications. P. notoginseng samples were first ground into powder, and 0.1 g powder was weighed accurately into 5-mL centrifuge tubes and extracted with 2 mL of 70% ethanol solution. The tubes were then sonicated for 30 min at room temperature. The supernatant was collected after being centrifuged for 10 min at 13,000 g. The test solution was obtained by filtrating the supernatant through 0.22-μm Millipore filter.
UPLC was performed on a Waters ACQUITY UPLC I-Class system. Chromatographic separations were performed on a ACQUITY BEH C18 column (2.1 × 100 mm, 1.7 μm) with a flow rate of 0.5 mL·min−1 at 40°C. The mobile phase was composed of 0.1% of formic acid–acetonitrile (A) and 0.05% of formic acid–water (B). Gradient elution program was as follows: 0–0.5 min, 20% A; 0.5–3 min, 20–80% A; 3–3.1 min, 80–98% A; 3.1–5 min, 98% A; 5–5.1 min, 98–20% A; 5.1–8 min 20% A. The injection volume was 1 μL for each sample.
Mass analysis was performed on a ABSCIEX 6500 QTRAP mass spectrometer. Mass spectrometer was performed in a positive ion mode using multiple reaction monitoring (MRM) mode. Optimized MS/MS parameters of saponins are shown in Supplementary Table 3. Ion spray voltage was set at 5500 eV, and turbo spray temperature was 550°C. Both gas 1 and gas 2 were set at 50 psi.
Quantitation of Dencichine Using UPLC-UV-MS
Determination of dencichine was performed as reported with some modification (Ju et al., 2015). About 0.1 g of root power was added with 5 mL of 70% methanol solution, and the mixture was sonicated for 2 h subsequently. The supernatant was obtained through centrifugation at 12,000 × g for 15 min and then diluted with 70% of methanol for 10 times before quantification. Dencichine was detected on a Waters ACQUITY UPLC I-Class system, equipped with a PDA detector under a UV wavelength of 213 nm. An ACQUITY BEH C18 column (2.1 × 100 mm, 1.7 μm) was used for separation, with a flow rate of 0.3 mL·min−1. Mobile phase A consisted of 0.05% phosphoric acid in water, while mobile phase B was methanol, and an isocratic elution of 53% A was used. Injection of samples was 1 μL.
Yeast One-Hybrid Assay
Y1H assay was performed as described previously (Zheng et al., 2021). The sequences of proPnEXPA4, proPnGGPPS3, proPnFPS, proPnCYP716A47, proPnCYP716A53v2, and proPnGAP were nested PCR-amplified according to genomic sequences (Jiang et al., 2020). The probable binding domains of promoters were inserted into pAbAi vector as baits and were integrated into Y1HGold, and then, the minimal inhibitory concentration of aureobasidin (AbA) was tested on SD/-Uracil (Ura) plates. A pGADT7-PnPHL8 recombinant plasmid was synthesized by RuiBiotech Co. Ltd., as prey. The pGADT7-PnPHL8 construct and a blank pGADT7 were introduced into bait reporter strains, while blank pGADT7 was used as control. Positive transformants were selected on SD/-Leucine (Leu)/-Ura plates with an appropriate concentration of AbA. Primers used for promotor nested PCR amplification and bait construction are listed in Supplementary Table 2.
Electrophoretic Mobility Shift Assay
EMSA was performed as described previously (Zheng et al., 2021). The full-length cDNA of PnPHL8 was first cloned into a pMAL-c2x vector and then transformed into a Rosetta (DE3) competent cell. Prokaryotic expression was performed at 20°C for 12 h, and then, recombinant protein was purified using Amylose Resin High Flow (NEB, Ipswich, MA, USA). EMSA was performed according to the manufacturer's instructions, using chemiluminescent EMSA kit (Beyotime, Item number, GS009). Primers and probes are listed in Supplementary Table 2.
Results
Transition of Root Size of P. notoginseng From Qing Dynasty to Modern Times
To investigate the morphological difference between Qing dynasty tributes and modern commodities, root length, diameter, and root weight of ancient samples (Figure 1A) from the Palace Museum and six types of root samples (20 tou, 40 tou, 60 tou, 80 tou, 120 tou, and >120 tou) bought from modern market (Figure 1B) were measured. The root of P. notoginseng in Qing dynasty had a similar diameter and dry weight with 120 tou samples, while the length of ancient samples was relatively shorter than the 120 tou samples (Figure 1C).
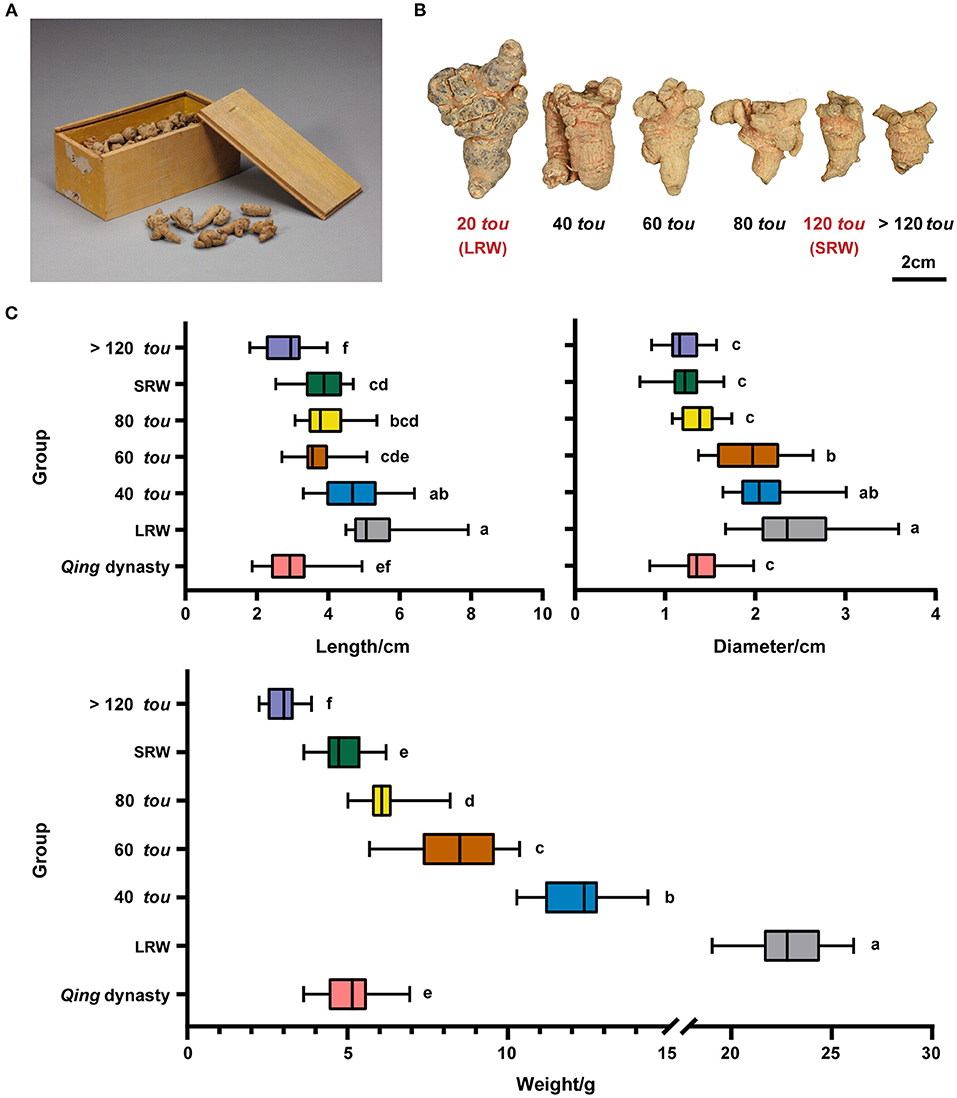
Figure 1. Morphological features of P. notoginseng in Qing dynasty and modern times. (A) P. notoginseng in Qing dynasty, relic number, Gu00172482-8/9. (B) P. notoginseng of different root sizes (20 tou, 40 tou, 60 tou, 80 tou, 120 tou, and >120 tou; tou is the number of roots per 500 g). (C) Length, diameter, and weight of P. notoginseng in Qing dynasty (n = 30) and in modern times (n = 20). Data are shown in box plot, and the different letters indicate values that vary significantly at P < 0.05 (one-way ANOVA).
After 1950s, artificial cultivation techniques, for example, the application of chemical fertilizers, reduced light transmittance and temperature has been applied to the cultivation of P. notoginseng (Supplementary Table 4), and the biggest root weight increased from 6.25 to 12.5 g in 1950s (Jin et al., 1996) to 25 g at present (Liu et al., 2016a; Xu, 2016). The dry weight of Qing root samples is around 5.17 g, which is 0.22-fold of the biggest of modern samples (20 tou samples), suggesting that a transition in root size of P. notoginseng has occurred following the change in cultivation practices and environment among Qing dynasty, 1950's, and modern time.
Higher Expression of GPI-Anchored Protein and Thicker Cell Wall in Root With Larger Weight
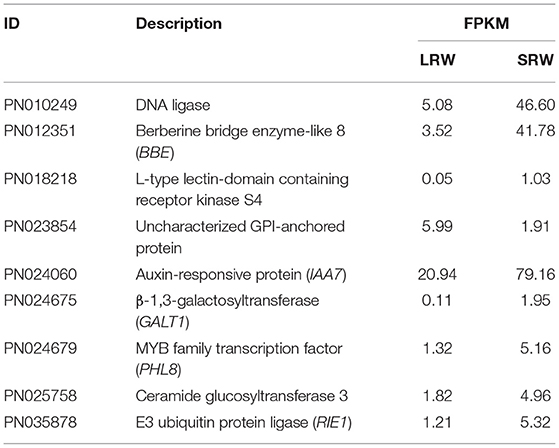
To further investigate the probable molecular mechanism behind the transition of root size, transcriptome sequencing was performed on 21 P. notoginseng root samples with two types of root size (LRW and SRW). A total of nine DEGs were identified between LRW and SRW samples, and the transcriptional levels of eight genes, namely a MYB-CC transcriptional factor, AUX/IAA, DNA ligase, ceramide glucosyltransferase, L-type lectin-domain containing receptor kinase, berberine bridge enzyme-like (BBE), β-1,3-galactosyltransferase (GALT), and E3 ubiquitin-protein ligase (RIE), were decreased in LRW samples (Table 1). Among the nine DEGs, only BBE was enriched into phenylpropanoid biosynthesis pathway in KEGG analysis, while no DEGs were enriched in GO analysis.
The expression of an uncharacterized GPI-anchored protein (GAP) was upregulated in LRW, with a 3.14-fold increase, compared with that in SRW. GAPs were reported to be associated with cell wall architecture (Macmillan et al., 2010; Niu et al., 2018), so we speculated that higher expression of PnGAP in LRW samples may lead to the thicker cell wall. We then measured the structure and component of cell wall in P. notoginseng root. Compared with SRW samples, cell wall in phloem and xylem of LRW samples was significantly thicker. An extremely significantly thicker cytoderm of vessel was detected in LRW, which was 1.41-fold of that in SRW. There was no remarkable difference of the cell wall thickness in cork and cortex between LRW and SRW samples (Figure 2A). In addition, LRW samples possessed a prominently larger vessel width than SRW samples (Figure 2B). As major components of cell walls, the content of lignin, cellulose, and callose was significantly higher in LRW samples (Figure 2C).
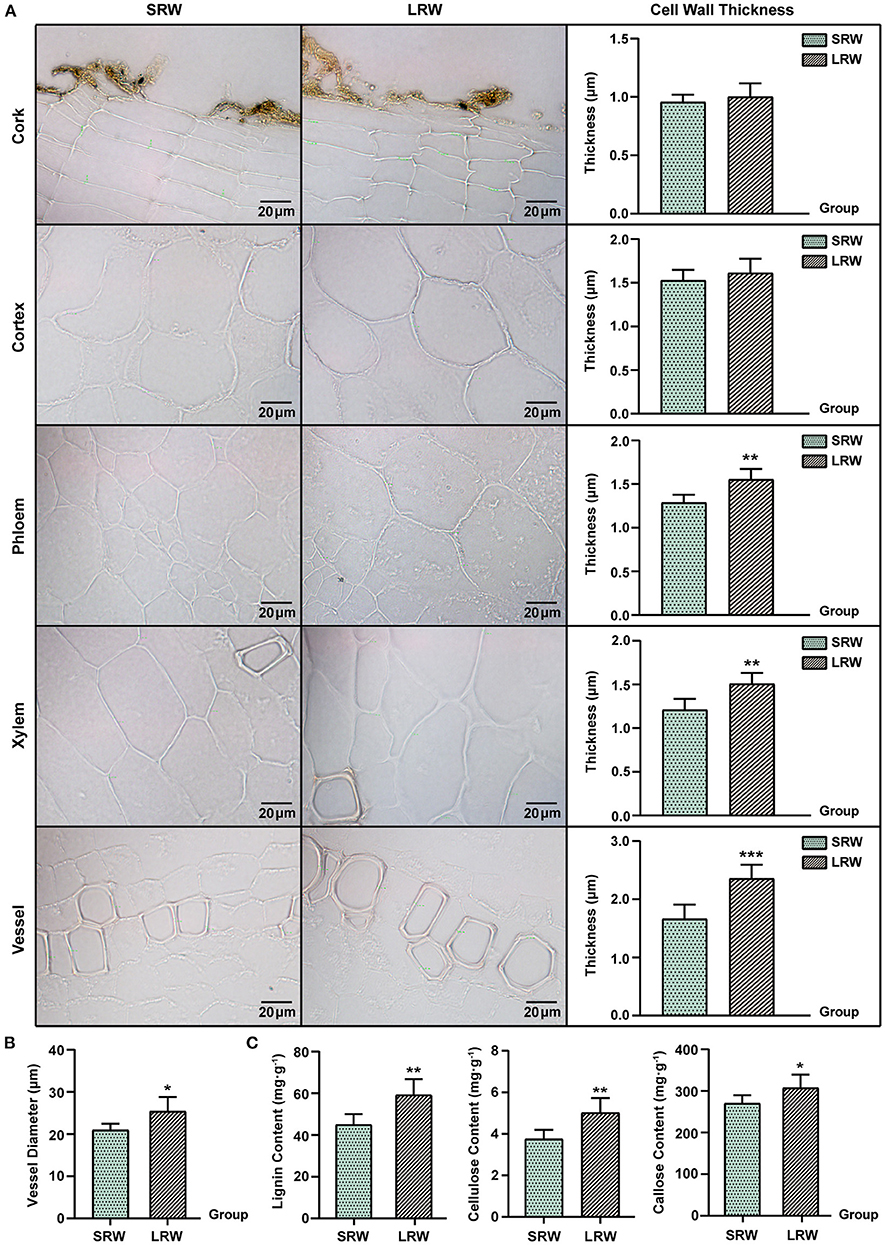
Figure 2. Different tissues and cell morphology in P. notoginseng. SRW was an abbreviation of small root weight, while LRW represented large root weight. (A) Microscope morphology of different tissues including cork, cortex, phloem, xylem, and vessel, and comparison of the cell wall thickness. (B) Comparison of the vessel diameter of P. notoginseng with different root weights. (C) Determination of lignin and callose in P. notoginseng with different root weights. The thickness of 10 cell walls in one field of view were measured and averaged, and then in accordance with this, a total of six fields of view from five biological replicates were obtained for t-test. Measurement of the vessel diameters was same as that of the cell wall thickness, while the diameter of seven vessels was measured and averaged. Asterisks denote Student's t-test significance: *P < 0.05 and **P < 0.01.
Expansin (EXP) and extension (EXT), which possess a membrane-binding mode of GPI-anchored, could be associated with thickening of P. notoginseng root through cell wall expansion pathway (Li et al., 2019b; Zhou, 2019). PnEXPA5 (PN022438) showed a prominently higher transcriptional level in SRW samples, while PnEXPA4 (PN017088) expressed significantly higher transcriptional level in LRW samples, which was 1.44-fold higher than that in SRW samples (Figure 3). Phylogenetic tree using 39 AtEXPs and multiple sequence alignments indicated that PnEXPA4 (PN017088) shows most similarity with AtEXPA4, and PnEXPA5 (PN022438) is homologous with AtEXPA5, both possessing a DPBB domain and a pollen allergen domain. In addition, a WCNP domain in front of a HFD motif was found in PN017088, which is particularly owned in alpha-expansin (Supplementary Figures 1A,B). Since AtEXPA4 had positively correlated with the cell wall thickness and root size (Ren et al., 2019), we speculated that PnEXPA4 (PN017088) is associated with root enlargement of P. notoginseng as an essential factor.
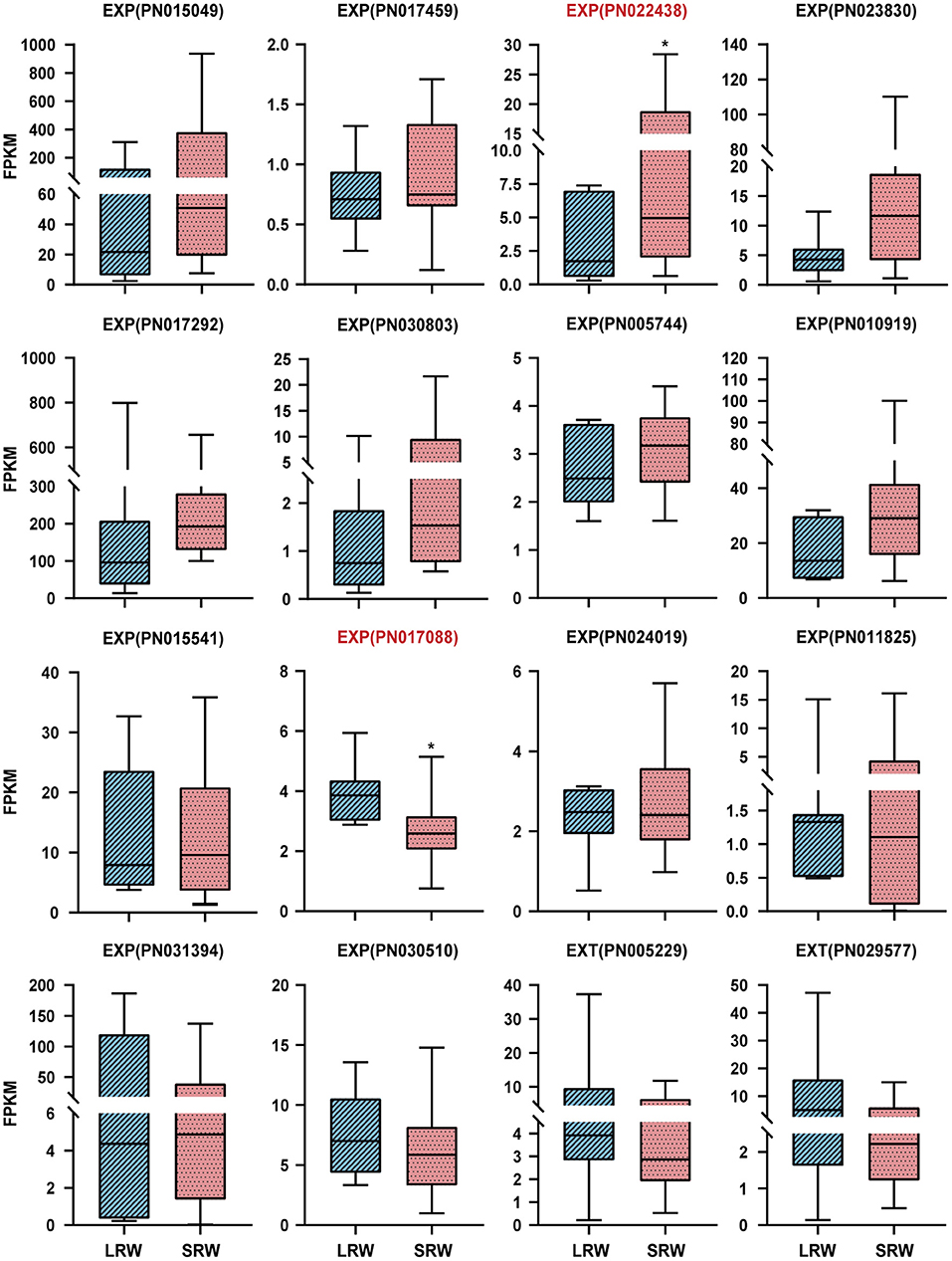
Figure 3. Expression pattern of EXPs and EXTs in LRW and SRW samples. EXPA, alpha-expansin; EXPB, beta-expansin; EXT, extension. Asterisks denote student's t-test significance: *P < 0.05.
Higher Root Weight Was Accompanied by Higher Content of Ginsenoside Rb1 and Lower Content of Dencichine
To investigate whether the chemical composition is related to root size of P. notoginseng, we analyzed the expression of the genes related to ginsenosides biosynthesis, and the accumulation of ginsenosides and dencichine in LRW and SRW samples. In LRW, the expression level of GGPPS1 (PN000021), GGPPS3 (PN029682), GGPPS4 (PN016696), FPS (PN009896), CYP716A47 (PN011429), and CYP716A53v2 (PN006374) was 3.25-, 4.54-, 3.42-, 1.84-, 1.46-, and 1.68-fold higher than that in SRW, respectively (Figure 4A). The content of ginsenoside Rb1 was significantly lower in SRW samples than that in LRW samples, while the content of dencichine in SRW samples was 1.28-fold higher than that in LRW samples (Figure 4B). It was reported that the content of protopanaxadiol and ginsenoside Rb1 was considerably increased under overexpression of FPS and CYP716A47, respectively (Han et al., 2012; Yang et al., 2017; Li et al., 2019a), indicating that higher expression level of FPS and CYP716A47 led to remarkably higher content of ginsenoside Rb1 in LRW. In addition, WGCNA showed a significantly positive correlation between transcriptional level of PnEXPA4 and PnGGPPS3, which were positively associated with root weight (Supplementary Figures 2A,B), suggesting that LRW samples with the thicker cell wall may be related to a higher content of ginsenosides.
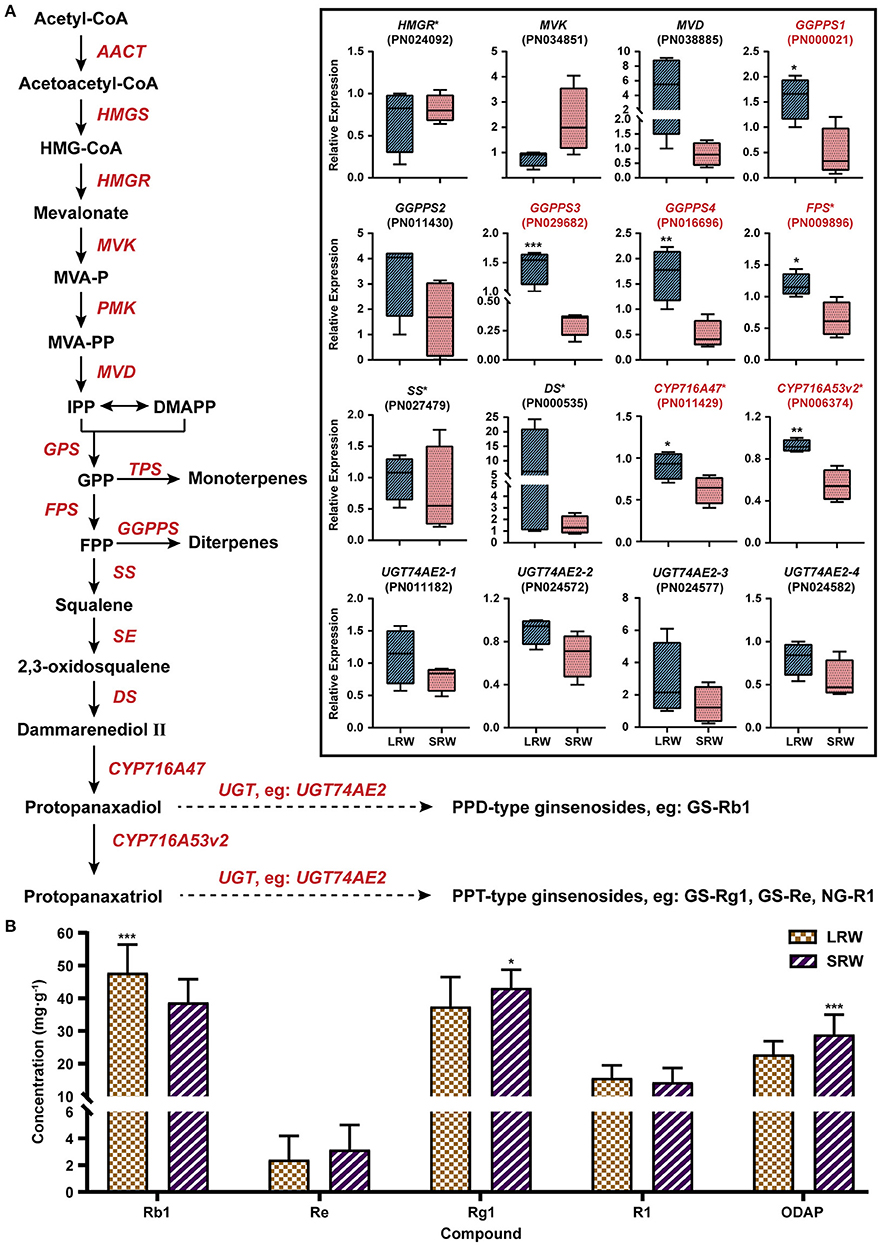
Figure 4. Expression pattern of genes related to biosynthesis of ginsenosides and content of active components. (A) Expression pattern of genes related to biosynthesis of ginsenosides in LRW and SRW groups. (B) Content of active components in LRW and SRW groups. AACT, acetyl-CoA C-acetyltransferase; HMGS, hydroxymethylglutaryl-CoA synthase; HMG-CoA, 3-hydroxy-3-methylglutaryl CoA; HMGR, hydroxymethylglutaryl-CoA reductase; MVK, mevalonate kinase; MVAP, mevalonate-5-phosphate; PMK, phosphomevalonate kinase; MVAPP, mevalonate-5-pyrophosphate; MVD, diphosphomevalonate decarboxylase; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPS, geranyl pyrophosphate synthase; GPP, geranyl pyrophosphate; FPS, farnesyl diphosphate synthase; FPP, farnesyl diphosphate; GGPPS, geranylgeranyl pyrophosphate synthase; SS, squalene synthase; SE, squalene epoxidase; DS, dammarenediol-II synthase; CYP, cytochrome P450 proteins; UGT, UDP-glycosyltransferase; GS, ginsenoside; NG, notoginsenoside. Genes obtained * on upper right were reported to be functional. Genes in red tag had statistical significance. Asterisks denote Student's t-test significance: *P < 0.05, **P < 0.01, and ***P < 0.001.
PnPHL8 Had Potential to Bind With PnGAP, PnCYP716A47, and PnGGPPS3 in Vitro
PHR transcription factor, belonging to MYB-CC family, participates in plant transcriptional responses to phosphate starvation (Wang et al., 2018b; Sega and Pacak, 2019). A PHR-like transcriptional factor PN024679 was filtrated through transcriptome analysis, which had a higher transcriptional level in SRW samples. PN024679 contains a 1074bp open reading frame (ORF) encoding 357 amino acids. A constructed phylogenetic tree using 14 AtPHLs indicated that PN024679 (named as PnPHL8) is homologous with AtPHL8 (Supplementary Figure 3A). Multiple sequence alignment showed PN024679 possessing a MYB DNA-binding domain and coiled-coil domain (Supplementary Figure 3B).
We investigated whether PnPHL8 regulates genes related to root size and ginsenosides biosynthesis in vitro by Y1H method. Promoter sequences of PnGAP, PnEXPA4, PnGGPPS3, PnFPS, PnCYP716A47, and PnCYP716A53v2 were PCR-amplified, all of which except proPnFPS and proPnCYP716A53v2 contained either a P1BS binding site (GNATATNC) or a P1BS-like element (Sun, 2015). MBS domain was also existed in proPnGAP, proPnFPS, proPnCYP716A53v2, and proPnGGPPS3 (Ding et al., 2017; Mabuchi et al., 2018). Specific P1BS domain and MBS domain of proPnGAP, proPnEXPA4, proPnGGPPS3, PnFPS, PnCYP716A47, and PnCYP716A53v2 were integrated into yeast, individually. After cotransformation with pGADT7-PnPHL8, we found that yeast strains carrying proPnGAP-MBS, proPnCYP716A47-P1BS, and proPnGGPPS3-P1BS could grow on SD minus leucine and uracil with aureobasidin (Figures 5A,B).
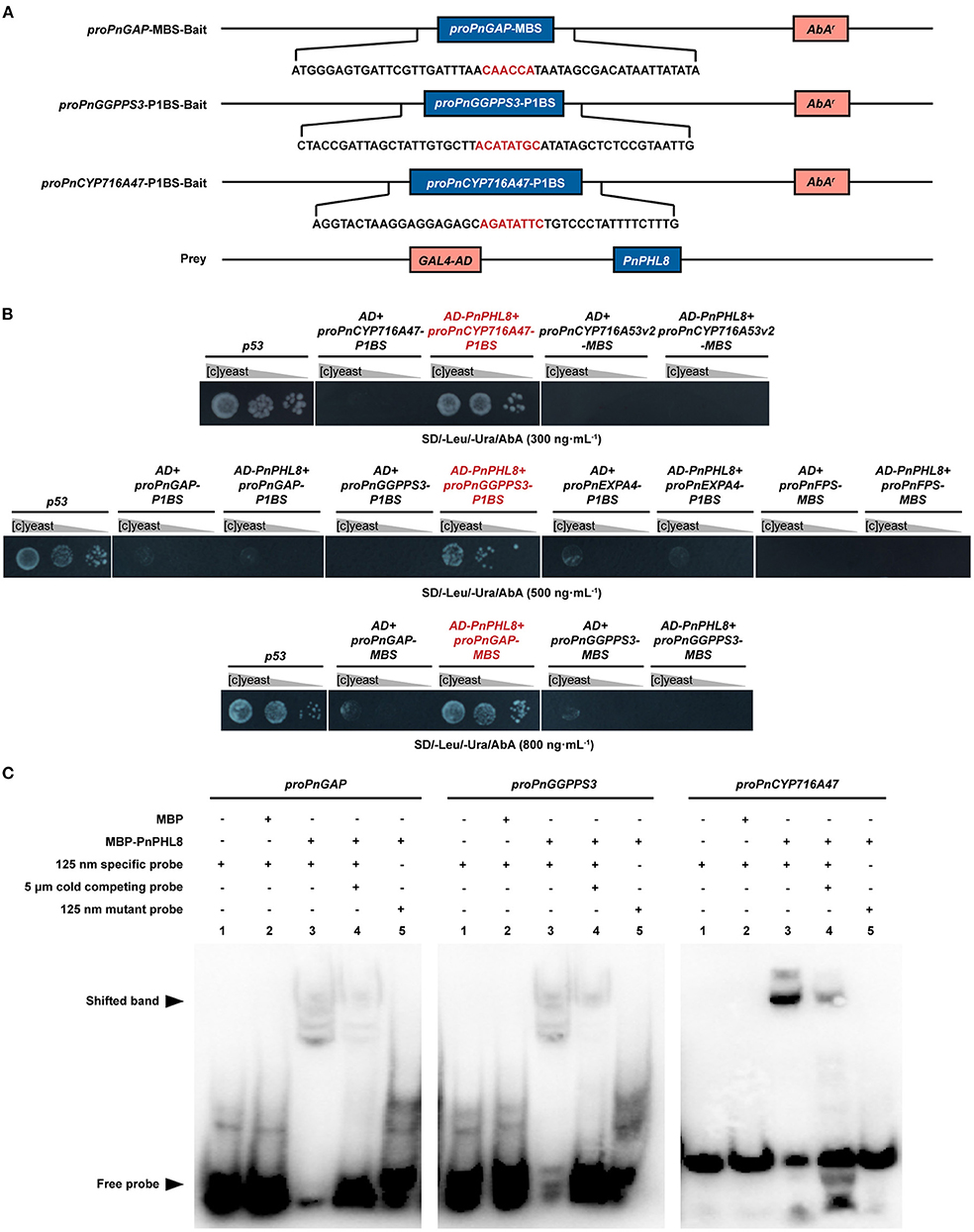
Figure 5. PnPHL8 binds with the promoter sequence of PnGAP, PnCYP716A47, and PnGGPPS3. (A) Structural schematics of baits and prey in yeast one-hybrid (Y1H) assay. (B) Y1H assay between PnPHL8 and PIBS domains as well as MBS domains of proPnGAP (promoter of PnGAP), proPnEXPA4 (promoter of PnEXPA4), proPnGGPPS3 (promoter of PnGGPPS3), proPnCYP716A47 (promoter of PnCYP716A47), proPnCYP716A53v2 (promotor of PnCYP716A53v2), and proPnFPS (promotor of PnFPS). Gray triangles represent dilution factor of the yeast concentration, while p53 served as a positive control. (C) EMSA among PnPHL8, proPnGAP, proPnCYP716A47, and proPnGGPPS3. GAP, glycosylphosphatidylinositol-anchored protein; PHL, phosphate starvation response transcription factor like; AbA, aureobasidin.
To avoid false-positive results caused by Y1H assay, we further performed EMSA to verify the interaction of PnPHL8 with the promoters of PnGAP, PnCYP716A47, and PnGGPPS3 in vitro. Fragments of proPnGAP, PnCYP716A47, and proPnGGPPS3, containing MBS or P1BS motif, were synthesized as specific probes for EMSA, while mutated probes were synthesized by replacing MBS/P1BS motif with poly-A/T. We found that MBP-PnPHL8 was able to bind to the promoter fragments of PnGAP, PnCYP716A47, and PnGGPPS3, but failed to bind to the mutated probes. Moreover, the cold competing probes, which contain high concentration of unlabeled promotor fragments, impaired the interaction between PnPHL8 and specific probes (Figure 5C). These results indicate that PnPHL8 specifically binds the promoters of PnGAP, PnCYP716A47, and PnGGPPS3 in vitro, suggesting that PnPHL8 may synergistically regulate biosynthesis of diterpenoid and triterpenoid, as well as cell wall architecture.
Discussion
Cell Wall Architecture Involved in Transition of Root Size of P. notoginseng
In this study, we revealed that the root of P. notoginseng exhibited a faster growth rate and better-developed root system during the past 100 years. Plant possessing larger diameter of vessels usually has a faster growth rate and water carrying capacity of vessel (Ian et al., 2008). Lignin also serves as a major component of vessel, which agreed with the significantly larger vessels and a higher content of lignin in LRW samples. In LRW samples, width of root, thickness of vessels and xylem cell wall, and the content of cellulose and callose were significantly higher than that in SRW samples, which might be due to the higher expression of PnGAP and PnEXPA4 in LRW. It was reported that GAPs related to cell wall architecture were mainly expressed in xylem vessels and adjacent parenchyma cells and then functioned in secondary wall deposition (Loopstra and No, 2000; Dahiya et al., 2006; Dai, 2015). In addition, the expression of EXPA4 and GAPs was positively related to the cell wall thickness, with GAPs also showing a positive correlation with the content of cellulose (Sun, 2013; Ben-Tov et al., 2015; Mcnair, 2015; Ren et al., 2019).
P. notoginseng is now usually cultivated at higher altitude, lower light transmittance, lower temperature, and lower precipitation condition, compared with earlier times. Lower temperature increased the gene expression level of PnGAP (Daisuke et al., 2016; Zhao et al., 2020), while cold acclimation, water stress, and phosphorus application led to higher expression of EXPA and EXT (Bian, 2006; Kozbial et al., 2010; Li et al., 2013; Han et al., 2014). The expression of EXPAs was upregulated under green shade within limits, but was strongly inhibited under dark treatment (Ping et al., 1994; Sasidharan et al., 2009; Liu et al., 2011). The vessel diameter and vessel wall thickness showed a significant increase under drought (Xu and Chen, 2012; Aref et al., 2013). Following the transition of cultivation measures, lower temperature and drought could increase the vessel diameter and vessel wall thickness, and a faster growth rate and then a bigger root size of P. notoginseng were formed. Higher expression of PnGAP in vessels and xylem parenchyma contributed to a higher content of cellulose and callose, while higher expression of PnEXPA4 led to better-architected cell wall. In addition, cell wall-mediated resistance is an important part of plant immune response system, to which the cell wall thickness was positively related (Aquije et al., 2010; Rachid et al., 2016), and a large root size was reportedly associated with resistance (Chloupek, 2010). The PHR transcription factor family participates in plant transcriptional responses to phosphate starvation (Wang et al., 2018b; Sega and Pacak, 2019). Lower application of phosphate in Qing dynasty might result in higher transcriptional level of PnPHL8, which subsequently influenced the expression of PnGAP and affected cell wall architecture and expansion of P. notoginseng. Based on this, we speculate that cell wall architecture played an important role in transition of root size from Qing dynasty to the present.
The Variation of Chemical Components Content and Root Size of P. notoginseng Might Lead to Transition in Clinical Usage
Ginsenoside Rb1 and dencichine, the major compounds in root of P. notoginseng, show diverse pharmacological activities. Ginsenoside Rb1 has great effects on vascular endothelial function improvement (Ohashi et al., 2006), cerebral ischemia protection (Yuan et al., 2007), myocardial preservation (Zhao et al., 2010), and neuroprotection (Jin et al., 2005; Liang et al., 2010), while dencichine was used for the treatment of injury induced trauma, and its hemostatic function was proven by clinical practice (Zhang and Yu, 2010). Modern cultivation condition, for example, lower temperature and drought, is beneficial to the accumulation of saponins in P. notoginseng (Konsler et al., 1990; Yu et al., 2001; Liu et al., 2016b; Ma et al., 2021). Here, we also found high content of ginsenosides Rb1 in LRW samples. In addition, the PHL transcription factor negatively regulates secondary metabolism such as carotenoid (Lu et al., 2021), indicating that the higher expression of PnPHL8 in SRW samples may lead to a lower content of secondary metabolism such as ginsenoside Rb1. In contrast, the content of dencichine was lower in LRW samples, which may be resulted from the decreased activity of SAT induced by drought (Ahmad et al., 2016; Yang et al., 2021). As recorded in herbal records, P. notoginseng in Qing dynasty was mostly applied externally or prepared into powder for hemostasis (Chen, 2009). However, prescription containing P. notoginseng with a higher content of ginsenoside Rb1 in recent years was mainly used to treat heart diseases and injuries (Chen et al., 2017). The change in root size, ginsenoside Rb1 and dencichine content of P. notoginseng from Qing dynasty to modern cultivation era, may also influence the clinical usage.
Triterpenoid is reported as a regulator of cell wall biosynthesis (Jozwiak et al., 2020), and ginsenoside Rb1 was localized to degrading primary cell wall of xylem in root of Panax ginseng (Yokota et al., 2011). In addition, a significantly positive correlation between the expression of PnEXPA4 and PnGGPPS3 was also observed by WGCNA, suggesting that cell wall architecture pathway and ginsenosides biosynthesis pathway may jointly participate in root enlargement of P. notoginseng during past 100 years.
Conclusion
From Qing dynasty to modern times, cultivation increased the root size and changed the content of ginsenoside and dencichine of P. notoginseng. In this study, we revealed that large root size of modern P. notoginseng should be due to the high expression of PnGAP and PnEXPA4, by promoting better-architected cell walls and larger vessels. GGPPS, FPS, CYP716A47, and CYP716A53v2 involved in ginsenosides biosynthesis pathway are also induced to contribute to a relatively higher content of ginsenosides, while depressed expression of SAT in LRW sample affected dencichine biosynthesis, leading to transition toward clinical efficacy from Qing dynasty to cultivation era. PnPHL8 participates in transcriptional regulation of PnGAP, PnCYP716A47, and PnGGPPS3, modulating cell wall architecture and ginsenosides biosynthesis pathway (Figure 6). Our results toward P. notoginseng of 2 eras separated by 100 years provided enlightenment on how long cultivation affected root size, chemical composition, and clinical usage.
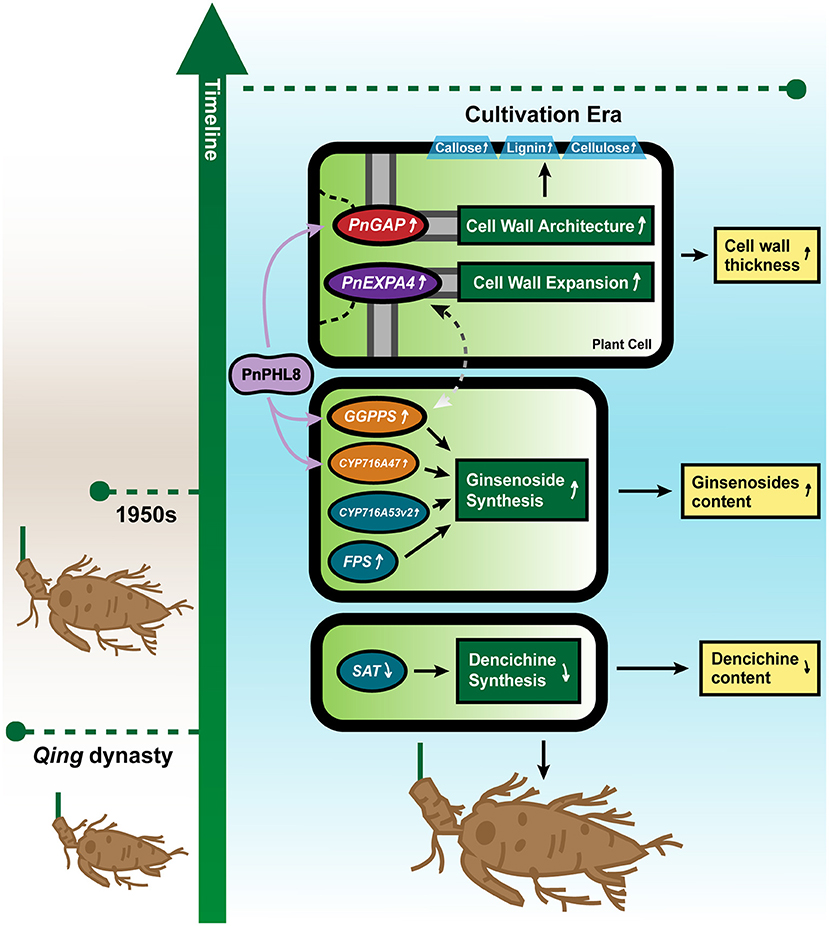
Figure 6. Probable molecular mechanism model for cultivation influencing root architecture and chemical component of P. notoginseng. Changes in root size and active component accumulation of P. notoginseng are influenced by related genes and affected by changes in the cultivation environment. Increased expression of PnGAP and PnEXPA4 results in the thicker cell wall and a higher content of lignin, cellulose, and callose in P. notoginseng of cultivation era. Higher expression of GGPPS, FPS, CYP716A47, and CYP716A53v2 contributes to a higher content of ginsenosides, while lower expression of SAT might result in a lower content of dencichine. A PnPHL8 transcriptional factor participates in transcriptional regulation of PnGAP, PnCYP716A47, and PnGGPPS3, while dotted line between PnEXPA4 and PnGGPPS3 indicates that a probable positive correlation may exist.
Data Availability Statement
The 10 P. notoginseng RNA-seq profiles (r9, r14, r20, r28, r29, r35, r36, r45, r48 and r55) are available in National Genomics Data Center under the GSA accession number CRA006118. The remaining 11 P. notoginseng RNA-seq profiles (r1, r13, r15, r18, r19, r23, r26, r32, r33, r53 and r59) have been deposited at DDBJ/EMBL/GenBank under the accession GFRX00000000 (Zheng et al., 2017).
Author Contributions
X-MC, L-QH, and YY designed the study. M-YY, Z-YH, P-RL, HZ, and YJ performed the experiments. M-YY, Z-YH, and H-SP analyzed data. M-YY, Z-YH, and YY wrote the manuscript. All authors discussed the results and commented on the manuscript.
Funding
This research was financially supported by the National Natural Science Foundation of China (NSFC) (81891013/81891010), the Scientific and Technological Innovation project of China Academy of Chinese Medical Science (C12021A041), and the Key project at the central government level for the ability to the establishment of sustainable use for valuable Chinese Medicine Resources (2060302).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.878796/full#supplementary-material
References
Ahmad, N., Malagoli, M., Wirtz, M., and Hell, R. (2016). Drought stress in maize causes differential acclimation responses of glutathione and sulfur metabolism in leaves and roots. BMC Plant Biol. 16, 247–261. doi: 10.1186/s12870-016-0940-z
Aquije, G., Zorzal, P. B., Buss, D. S., Ventura, J. A., Fernandes, P., and Fernandes, A. (2010). Cell wall alterations in the leaves of fusariosis-resistant and susceptible pineapple cultivars. Plant Cell Rep. 29, 1109–1117. doi: 10.1007/s00299-010-0894-9
Aref, I. M., Ahmed, A. I., Khan, P. R., and El-Atta, H. (2013). Drought-induced adaptive changes in the seedling anatomy of Acacia ehrenbergiana and Acacia tortilis subsp. raddiana. Trees 27, 959–971. doi: 10.1007/s00468-013-0848-2
Ba Rraclough, P. B., Kuhlmann, H., and Weir, A. H. (2010). The effects of prolonged drought and nitrogen fertilizer on root and shoot growth and water uptake by winter wheat. J. Agronomy Crop Sci. 163, 352–360. doi: 10.1111/j.1439-037X.1989.tb00778.x
Ben-Tov, D., Abraham, Y., Stav, S., Thompson, K., Loraine, A., Elbaum, R., et al. (2015). COBRA-LIKE2, a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE family, plays a role in cellulose deposition in arabidopsis seed coat mucilage secretory cells. Plant Physiol. 167, 711–724. doi: 10.1104/pp.114.240671
Bian, H. Y. (2006). Effect of the Key Enzymes and Exogenous Materials on Fiber Strength Formation in Low Temperature Condition. Nanjing: Nanjing Agricultural University.
Bundy, M. G., Kosentka, P. Z., Willet, A. H., Zhang, L., Miller, E., and Shpak, E. D. (2016). A mutation in the catalytic subunit of the glycosylphosphatidylinositol transamidase disrupts growth, fertility, and stomata formation. Plant Physiol. 171, 974–985. doi: 10.1104/pp.16.00339
Chathurika, W., Raja, R. K., Shankle, M. W., Stephen, M., and Gao, W. (2018). Low and high-temperature effects on sweetpotato storage root initiation and early transplant establishment. entia Horticulturae 240, 38–48. doi: 10.1016/j.scienta.2018.05.052
Chen, G., Liu, C., He, H. Q., Gao, J. L., Li, J., Xing, Y. W., et al. (2017). Prescription rules of Chinese patent medicines containing Notoginseng Radix et Rhizoma. Chin. J. Experi. Traditional Med. Formulae 23, 191−197. doi: 10.13422/j.cnki.syfjx.2017070191
Chen, M. Y. (2017). Effect of Different Irrigation Methods on Yield and Quality of Rice Under Different Soil Types. Yangzhou University.
Chen, N. L., Hu, M., Qiao, C. P., Nai, X. Y., and Wang, R. (2010). Effects of BTH, SA and SiO2 treatment on disease resistance and leaf HRGP and lignin contents of melon seedlings. Sci. Agri. Sinica 43, 535–541. doi: 10.4028/www.scientific.net/AMM.37-38.1549
Cheng, H., Chen, X., Zhu, J., and Huang, H. (2016). Overexpression of a hevea brasiliensis ErbB-3 binding protein 1 gene increases drought tolerance and organ size in Arabidopsis. Front. Plant Sci. 7, 1703–1713. doi: 10.3389/fpls.2016.01703
Cheon, S. K., Lee, T. S., Yoon, J. H., Lee, S. S., and Mok, S. K. (2004). Effect of light transmittance control on the root yield and quality during the growing season of panax ginseng. J. Ginseng Res. 4, 196–200. doi: 10.5142/JGR.2004.28.4.196
China Association of Chiniese Medicine. (2019). “Daodi herbs (in Chinese)”, in: Part 118: Sanqi. Beijing: China Standard Press.
Chloupek, O. (2010). The size of the root system of lucerne varieties grown for forage and seed, and its relation to yield. Plant Breeding 101, 169–172. doi: 10.1111/j.1439-0523.1988.tb00284.x
Cui, X. M., Xu, L. S., Wang, Q., and Chen, Z. J. (2005). Analysis on the geologic background and physicochemical properties of soil for the cultivation of Panax notoginseng in Yunnan province. China J. Chin. Materia Med. 30:332. doi: 10.3321/j.issn:1001-5302.2005.05.003
Dahiya, P., Findlay, K., Roberts, K., and Mccann, M. C. (2006). A fasciclin-domain containing gene, ZeFLA11, is expressed exclusively in xylem elements that have reticulate wall thickenings in the stem vascular system of Zinnia elegans cv Envy. Planta 223, 1281–1291. doi: 10.1007/s00425-005-0177-9
Dai, Y. J. (2015). Effects of Elevated Temperature During Flowering and Boll Formation Stage on Physiological Mechanisms of Cotton Leaves and Fiber. Nanjing Agricultural University.
Daisuke, T., Yukio, K., and Matsuo, U. (2016). Cold acclimation is accompanied by complex responses of glycosylphosphatidylinositol (GPI)-anchored proteins in Arabidopsis. J. Experi. Botany 17, 5203–5215. doi: 10.1093/jxb/erw279
Dewey, C. N., and Li, B. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323–323. doi: 10.1186/1471-2105-12-323
Ding, K., Pei, T., Bai, Z., Jia, Y., Ma, P., and Liang, Z. (2017). SmMYB36, a novel R2R3-MYB transcription factor, enhances tanshinone accumulation and decreases phenolic acid content in salvia miltiorrhiza hairy roots. Sci. Rep. 7:5104. doi: 10.1038/s41598-017-04909-w
Ding, W., Wang, Y., Fang, W., Gao, S., Li, X., and Xiao, K. (2016). TaZAT8, a C2H2-ZFP type transcription factor gene in wheat, plays critical roles in mediating tolerance to Pi deprivation through regulating P acquisition, ROS homeostasis and root system establishment. Physiol. Plant. 158, 297–311. doi: 10.1111/ppl.12467
Engelsberger, W. R., and Schulze, W. X. (2012). Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J. 69, 978–995. doi: 10.1111/j.1365-313X.2011.04848.x
Ghaffari, H., Tadayon, M. R., Bahador, M., and Razmjoo, J. (2021). Investigation of the proline role in controlling traits related to sugar and root yield of sugar beet under water deficit conditions. Agri. Water Manage. 243:e106448. doi: 10.1016/j.agwat.2020.106448
Goodsman, D. W., Lieffers, V. J., Landh?Usser, S. M., and Erbilgin, N. (2010). Fertilization of lodgepole pine trees increased diameter growth but reduced root carbohydrate concentrations. For. Ecol. Manage. 260, 1914–1920. doi: 10.1016/j.foreco.2010.08.041
Han, J. Y., Hwang, H. S., Choi, S. W., Kim, H. J., and Choi, Y. E. (2012). Cytochrome P450 CYP716A53v2 Catalyzes the Formation of Protopanaxatriol from Protopanaxadiol During Ginsenoside Biosynthesis in Panax Ginseng. Plant Cell Physiol. 53, 1535–1545. doi: 10.1093/pcp/pcs106
Han, Y. Y., Zhou, S., Chen, Y. H., Kong, X. Z., Xu, Y., and Wang, W. (2014). The involvement of expansins in responses to phosphorus availability in wheat, and its potentials in improving phosphorus efficiency of plants. Plant Physiol. Biochem. 78, 53–62. doi: 10.1016/j.plaphy.2014.02.016
He, Z. X., and Deng, X. Q. (1981). Investigation on cultivation of Panax notoginseng in Guangxi and Yunnan (in Chinese). Chin Tradit Herbal Drugs 12, 28–31.
Hiroyoshi, I., Satoshi, N., Seiji, M., Yasushi, T., and Yasuo, U. (2004). Interaction between genetic effects and soil type in diallel analysis of root shape and size of japanese radish (Raphanus sativus L.). Breed. Sci. 54, 313–318. doi: 10.1270/jsbbs.54.313
Huang, R. S., Yang, H. J., He, Z. J., Chen, C. J., Li, Z. W., and Zhang, C. L. (2007). Textual research on the origin in areas of Panax notoginseng. Lishizhen Med. Materia Med. Res. 18, 1610–1611. doi: 10.3969/j.issn.1008-0805.2007.07.035
Ian, C., Dodd, B. J., Ferguson, C., and Beveridge, A. (2008). Apical wilting and petiole xylem vessel diameter of the rms2 branching mutant of pea are shoot controlled and independent of a long-distance signal regulating branching. Plant Cell Physiol. 49, 791–800. doi: 10.1093/pcp/pcn052
Jeong, J. S., Kim, Y. S., Redillas, M., Jang, G., Jung, H., Bang, S. W., et al. (2013). OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114. doi: 10.1111/pbi.12011
Jiang, Z. Q., Tu, L. C., Yang, W. F., Zhang, Y. F., Hu, T. Y., Ma, B. W., et al. (2020). The chromosome-level reference genome assembly for Panax notoginseng and insights into ginsenoside biosynthesis. Plant Commun. 2:100113. doi: 10.1016/j.xplc.2020.100113
Jin, K. P., Namgung, U., Chang, J. L., Park, J. O., Jin, S. H., Kwon, O. B., et al. (2005). Calcium-independent CaMKII activity is involved in ginsenoside Rb1-mediated neuronal recovery after hypoxic damage. Life Sci. 76, 1013–1025. doi: 10.1016/j.lfs.2004.10.011
Jin, X. D., Qiao, R. Q., and Li, S. D. (1996). County Annal of Guangxi. Nanning: Guangxi People's Publishing House.
Jozwiak, A., Sonawane, P. D., Panda, S., Garagounis, C., and Aharoni, A. (2020). Plant terpenoid metabolism co-opts a component of the cell wall biosynthesis machinery. Nat. Chem. Biol. 740–748. doi: 10.1038/s41589-020-0541-x
Ju, Z. C., He, C. Y., Liu, Q., Yang, L., and Wang, Z. T. (2015). Determination of dencichine in Sanqi tablet by HILIC. China J. Chin. Materia Madica 40:4. doi: 10.4268/cjcmm20151320
Khle, H., Jeblick, W., Poten, F., Blaschek, W., and Kauss, H. (1985). Chitosan-elicited callose synthesis in soybean cells as a Ca-dependent process. Plant Physiol. 77, 544–551. doi: 10.1104/pp.77.3.544
Konsler, T. R., Zito, S. W., Shelton, J. E., and Staba, E. J. (1990). Lime and phosphorus effects on American ginseng: II. Root and leaf ginsenoside content and their relationship. J. Am. Soc. Horticultural Sci. 115, 575–580. doi: 10.21273/JASHS.115.4.575
Kozbial, P. Z., Jerzmanowski, A., Shirsat, A. H., and Kacperska, A. (2010). Transient freezing regulates expression of extensin-type genes in winter oilseed rape. Physiol. Plant. 103, 264–270. doi: 10.1034/j.1399-3054.1998.1030214.x
Kuang, S., Xu, X., Meng, Z., Zhang, G., and Chen, J. (2015). Effects of light transmittance on plant growth and root ginsenoside content of Panax notoginseng. Chin. J. Appl. Environ. Biol. 21, 279–286. doi: 10.3724/SP.J.1145.2014.08002
Kuang, S. B., Zhang, G. H.J C. Z, Wei, F. G., Yang, S. C., et al. (2014). Changes in morphological and growth indexes of Panax notoginseng seedling under different light conditions. J. Plant Resources Environ. 2014, 54–59. doi: 10.3969/j.issn.1674-7895.2014.02.08
Li, F., Han, Y. Y., Feng, Y. N., Xing, S. C., Zhao, M. R., Chen, Y. H., et al. (2013). Expression of wheat expansin driven by the RD29 promoter in tobacco confers water-stress tolerance without impacting growth and development. J. Biotechnol. 163, 281–291. doi: 10.1016/j.jbiotec.2012.11.008
Li, J., Ma, L., Zhang, S. T., Zuo, C. L., Song, N., Zhu, S. S., et al. (2019a). Transcriptome analysis of 1- and 3-year-old Panax notoginseng roots and functional characterization of saponin biosynthetic genes DS and CYP716A47-like. Planta 249, 1229–1237. doi: 10.1007/s00425-018-03083-1
Li, X. J., Yang, J. L., Hao, B., Lu, Y. C., and Yang, S. C. (2019b). Comparative transcriptome and metabolome analyses provide new insights into the molecular mechanisms underlying taproot thickening in Panax notoginseng. BMC Plant Biol. 19, 451–468. doi: 10.1186/s12870-019-2067-5
Li, Y. X., Chen, X. F., Li, Y., Liang, Q., Shi, F., Dou, M. M., et al. (2016). Correlation of agronomic character of Panax notoginseng in Guangxi with soil physical and chemical properties. J. Chin. Med. Mater. 39, 1982–1988. doi: 10.13863/j.issn1001-4454.2016.09.013
Liang, W., Ge, S., Yang, L., Yang, M., Ye, Z., Yan, M., et al. (2010). Ginsenosides Rb1 and Rg1 promote proliferation and expression of neurotrophic factors in primary Schwann cell cultures. Brain Res. 1357, 19–25. doi: 10.1016/j.brainres.2010.07.091
Liu, B. (2009). Biological Function Research of a GPI Anchored Protein Gene, OsGAP1 in Oryza sativa. Sun Yat-sen University.
Liu, D. H., Xu, N., Guo, L. P., Jin, Y., Cui, X. M., Yang, Y., et al. (2016a). Qualitative characteristics and classification study on commodity specification and grade standard of Panax notoginseng. China J. Chin. Mat. Med. 41, 776–785. doi: 10.4268/cjcmm20160504
Liu, J., Chen, T., Zhang, J., Li, C., Xu, Y., Zheng, H., et al. (2020). Ginsenosides regulate adventitious root formation in Panax ginseng via a PgCLE45-PgWOX11 regulatory module. J. Exp. Bot. 71, 6396–6407. doi: 10.1093/jxb/eraa375
Liu, J., Quan, X. L., Jiang, M. L., Li, X. G., Quan, L. H., and Wu, S. Q. (2016b). Effect of cold stress on expression characteristic of gene families of ginsenoside biosynthesis pathway. Chin. Traditional Herbal Drugs 47, 1956–1961. doi: 10.7501/j.issn.0253-2670.2016.11.024
Liu, W. G., Tao, J., Zhou, X. R., and Yang, W. Y. (2011). Characteristics of expansins in soybean (Glycine max) internodes and responses to shade stress. Asian J. Crop Sci. 3:34. doi: 10.3923/ajcs.2011.26.34
Loopstra, C. A., and No, P. (2000). Purification and cloning of an arabinogalactan-protein from xylem of loblolly pine. Planta 210, 686–689. doi: 10.1007/s004250050061
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, S., Ye, J., Zhu, K., Zhang, Y., and Deng, X. (2021). A citrus phosphate starvation response factor CsPHL3 negatively regulates carotenoid metabolism. Plant Cell Physiol. 62, 482–493. doi: 10.1093/pcp/pcab007
Ma, W.Q., Wang, H.Y., Zhang, W.J., Wang, S., and Guo, L.P. (2021). Effects of ecological factors on shape and ginsenoside of Panax ginseng. China J. Chin. Materia Med. 46, 1920–1926. doi: 10.19540/j.cnki.cjcmm.20210123.102
Mabuchi, K., Maki, H., Itaya, T., Suzuki, T., Nomoto, M., Sakaoka, S., et al. (2018). MYB30 links ROS signaling, root cell elongation, and plant immune responses. Proc. Natl. Acad. Sci. U.S.A. 115:E4710. doi: 10.1073/pnas.1804233115
Macmillan, C. P., Mansfield, S. D., Stachurski, Z. H., Evans, R., and Southerton, S. G. (2010). Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J. 62, 689–703. doi: 10.1111/j.1365-313X.2010.04181.x
Mcnair, G. (2015). COBRA-like4: a GPI-Anchored Protein Functioning as a Mediator of Cellulose Ultrastructure in Herbaceous and Woody Plants.
Niu, E., Fang, S., Shang, X., and Guo, W. (2018). Ectopic expression of GhCOBL9A, a cotton glycosyl-phosphatidyl inositol-anchored protein encoding gene, promotes cell elongation, thickening and increased plant biomass in transgenic Arabidopsis. Mol. Genet. Genomics 293, 1191–1204. doi: 10.1007/s00438-018-1452-3
Ohashi, R., Yan, S., Hong, M., Hong, C., Yao, Q., Lin, P. H., et al. (2006). Effects of homocysteine and ginsenoside Rb1 on endothelial proliferation and superoxide anion production. J. Surg. Res. 133, 89–94. doi: 10.1016/j.jss.2005.09.016
Ping, W., Flores, H. E., and Humphrey, A. E. (1994). Production of expansin from light/dark growing Trichosanthes kirilowii var. Japonicum root cultures. Biotechnol. Lett. 16, 955–958. doi: 10.1007/BF00128632
Rachid, L., Saroj, K., Wang, L., Li, F., Nicole, S., Malgorzata, K., et al. (2016). Cell wall biomolecular composition plays a potential role in the host type II resistance to fusarium head blight in wheat. Front. Microbiol. 7, 910–921. doi: 10.3389/fmicb.2016.00910
Ren, H., Wen, L. Z., Guo, Y. H., Yu, Y. Y., Sun, C. H., Fan, H. M., et al. (2019). Expressional and functional verification of the involvement of CmEXPA4 in chrysanthemum root development. J. Plant Growth Regul. 38, 1375–1386. doi: 10.1007/s00344-019-09940-x
Sasidharan, R., Chinnappa, C. C., Voesenek, L., and Pierik, R. (2009). A molecular basis for the physiological variation in shade avoidance responses: A tale of two ecotypes. Plant Signal. Behav. 4, 528–529. doi: 10.4161/psb.4.6.8586
Sega, P., and Pacak, A. (2019). Plant PHR transcription factors: put on a map. Genes. 10, 1–14. doi: 10.3390/genes10121018
Sun, L. (2013). The Soybean Expansion GmEXPA4 Gene Transformed Arabidopsis Thaliana and Functional Verification.
Sun, L. (2015). Arabidopsis Transcription Factors PHL2 and PHR1 Regulate the Transcriptional Responses to Phosphate Starvation. Tsinghua University.
Tamirisa, S., Reddy, V. D., and Rao, K. V. (2014). Ectopic expression of pigeonpea cold and drought regulatory protein (CcCDR) in yeast and tobacco affords multiple abiotic stress tolerance. Plant Cell Tissue Organ Culture 119, 489–499. doi: 10.1007/s11240-014-0549-6
Teskey, R. O., and Hinckley, T. M. (2010). Influence of temperature and water potential on root growth of white oak. Physiol. Plant. 52, 363–369. doi: 10.1111/j.1399-3054.1981.tb06055.x
Wang, J., Kuang, S. B., Zhou, P., Fan, W., Long, G. Q., Zhang, G. H., et al. (2018a). Agronomic and quality traits of two-year old Panax notoginseng response to environmental light intensity. J. Trop Subtropical Botany. 26, 57–64.
Wang, Z., Zheng, Z., Song, L., and Liu, D. (2018b). Functional characterization of arabidopsis PHL4 in plant response to phosphate starvation. Other 9:e01432. doi: 10.3389/fpls.2018.01432
Wang, Z. L., Wei, M. L., Sun, Y. Q., Hunag, T. W., Wang, B. Y., and Chen, Z. J. (2008). Study on the effect of applying phosphate fertilizer to Panax notoginseng. Ginseng Res. 2, 29–30. doi: 10.3969/j.issn.1671-1521.2008.02.007
Xiong, S. M., Zuo, X. F., and Zhu, Y. Y. (2005). Determination of cellulose, hemi-cellulose and lignin in rice hull. Cereal Feed Industry 8:2. doi: 10.3969/j.issn.1003-6202.2005.08.018
Xu, N. (2016). Research of the Factors Influencing Commodity Quality of Notoginseng Radix et Rhizoma. Kunming University of Science and Technology.
Xu, Q., and Chen, Y. N. (2012). Response of anatomy hydraulic characteristics of xylem stem of Populus euphratica Oliv. to drought stress. Chin. J. Eco-Agricult. 8, 1059–1065. doi: 10.3724/SP.J.1011.2012.01059
Yang, Y., Ge, F., Sun, Y., Liu, D. Q., and Chen, C. Y. (2017). Strengthening triterpene saponins biosynthesis by over-expression of farnesyl pyrophosphate synthase gene and RNA interference of cycloartenol synthase gene in Panax notoginseng cells. Molecules 22, 581–592. doi: 10.3390/molecules22040581
Yang, Z. Y., Liu, G. Z., Zhang, G. H., Yan, J., Dong, Y., Lu, Y. C., et al. (2021). The chromosome: cale high quality genome assembly of Panax notoginseng provides insight into dencichine biosynthesis. Plant Biotechnol. J. 19, 869–871. doi: 10.1111/pbi.13558
Yao, Z. F., Liang, C. Y., Zhang, Q., Chen, Z. J., Xiao, B. X., Tian, J., et al. (2014). SPX1 is an important component in the phosphorus signalling network of common bean regulating root growth and phosphorus homeostasis. J. Experi. Botany 12, 3299–3310. doi: 10.1093/jxb/eru183
Yokota, S., Onohara, Y., Uto, T., Tanaka, H., and Shoyama, Y. (2011). Localization of ginsenoside-Rb1 in Panax ginseng revealed by immunofluorescence and immunoelectron microscopic techniques. J. Med. Plant Res. 5, 3176–3187. doi: 10.1002/cbic.201100002
Yu, K. W., Gao, W. Y., Hahn, E. J., and Paek, K. Y. (2001). Effects of macro elements and nitrogen source on adventitious root growth and ginsenoside production in ginseng (Panax ginseng C. A. Meyer). J. Plant Biol. 44, 179–184. doi: 10.1007/BF03030349
Yuan, Q. L., Yang, C. X., Xu, P., Gao, X. Q., Deng, L., Chen, P., et al. (2007). Neuroprotective effects of ginsenoside Rb1 on transient cerebral ischemia in rats. Brain Res. 1167, 1–12. doi: 10.1016/j.brainres.2007.06.024
Zhang, L. B., Sun, Y. Q., Wei, M. L., Wang, Z. L., and Chen, Z. J. (2008). Effect of potassium suppliment level on the growth and yield of Panax notoginseng. Special Wild Econ. Animal Plant Res. 4, 46–48. doi: 10.3969/j.issn.1001-4721.2008.04.014
Zhang, Y.P., and Yu, Q. (2010). Experimental study on hemostatic activity and neurotoxic effect of Panax notoginseng. Shandong J. Tradit. Chin. Med. 43–45.
Zhao, B., Tang, Y., Zhang, B., Wu, P., Li, M., Xu, X., et al. (2020). The temperature-dependent retention of introns in GPI8 transcripts contributes to a drooping and fragile shoot phenotype in rice. Int. J. Mol. Sci. 21, 299–315. doi: 10.3390/ijms21010299
Zhao, H., Lv, D., Zhang, W., Dong, W., Feng, J., Xiang, Z., et al. (2010). Ginsenoside-Rb1 attenuates dilated cardiomyopathy in cTnTR141W transgenic mouse. J. Pharmacol. Sci. 112, 214–222. doi: 10.1254/jphs.09314FP
Zheng, D. M., Wang, L., Ou, X. H., Guo, L. P., Hao, Q. X., Liu, D. H., et al. (2014). Comparison of agronomic traits of Panax notoginseng between traditional cultivated fields and new cultivated fields. China J. Chin. Materia Med. 39, 558–565. doi: 10.4268/cjcmm20140402
Zheng, H., Jing, L., Jiang, X., Pu, C., Zhao, S., Yang, J., et al. (2021). The ERF-VII transcription factor SmERF73 coordinately regulates tanshinone biosynthesis in response to stress elicitors in Salvia miltiorrhiza. New Phytol. 231, 1940–1955. doi: 10.1111/nph.17463
Zheng, Y., Chen, K., Xu, Z., Liao, P., Zhang, X., Liu, L., et al. (2017). Small RNA profiles from Panax notoginseng roots differing in sizes reveal correlation between miR156 abundances and root biomass levels. Rep 7:9418. doi: 10.1038/s41598-017-09670-8
Keywords: root size, cultivation, GPI-anchored, expansin, Panax notoginseng, cell wall
Citation: Yu M-Y, Hua Z-Y, Liao P-R, Zheng H, Jin Y, Peng H-S, Cui X-M, Huang L-Q and Yuan Y (2022) Increasing Expression of PnGAP and PnEXPA4 Provides Insights Into the Enlargement of Panax notoginseng Root Size From Qing Dynasty to Cultivation Era. Front. Plant Sci. 13:878796. doi: 10.3389/fpls.2022.878796
Received: 18 February 2022; Accepted: 07 April 2022;
Published: 20 May 2022.
Edited by:
Lei Zhang, Second Military Medical University, ChinaReviewed by:
Fangyuan Zhang, Southwest University, ChinaQingsong Shao, Zhejiang Agriculture and Forestry University, China
Copyright © 2022 Yu, Hua, Liao, Zheng, Jin, Peng, Cui, Huang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan Yuan, eV95dWFuMDczMkAxNjMuY29t; Lu-Qi Huang, aHVhbmdsdXFpMDFAMTI2LmNvbQ==; Xiu-Ming Cui, c2FucWkzN0B2aXAuc2luYS5jb20=
†These authors have contributed equally to this work
 Mu-Yao Yu
Mu-Yao Yu Zhong-Yi Hua
Zhong-Yi Hua Pei-Ran Liao
Pei-Ran Liao Han Zheng
Han Zheng Yan Jin1
Yan Jin1 Hua-Sheng Peng
Hua-Sheng Peng Lu-Qi Huang
Lu-Qi Huang Yuan Yuan
Yuan Yuan