- 1Rice Research Institute, Fujian High Quality Rice Research and Development Center, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 2College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Faculty of Agriculture, Department of Agronomy, Assiut University, Assiut, Egypt
Plant height is one of the most important agronomical traits in rice (Oryza sativa L.). Introducing the semidwarf rice in the 1960s significantly enhanced the rice yield potential in Asia. Implementing near-isogenic lines (NILs) is the most powerful tool for the identification and fine mapping of quantitative trait loci (QTLs). In this study, 176 NILs were produced from the crossing and back-crossing of two rice cultivars. Specifically, the indica rice cultivar Jiafuzhan served as a recipient, and the restorer japonica cultivar Hui1586 served as a donor. Using the 176 NILs, we identified a novel major QTL for reduced plant height in the NIL36 line. The qph12 QTL was mapped to a 31 kb genomic region between the indel markers Indel12-29 and Indel12-31. The rice genome annotation indicated the presence of three candidate genes in this genomic region. Through gene prediction and cDNA sequencing, we confirmed that LOC_Os12g40890 (qPH12) is the target gene in the NIL36 line. Further analysis showed that the qph12 QTL is caused by a 1 bp deletion in the first exon that resulted in premature termination of the qPH12. Knockout experiments showed that the qph12 QTL is responsible for the reduced plant height phenotype of the NIL36 line. Although the qph12 gene from the NIL36 line showed a shorter panicle length, fewer spikelets per panicle and a lower plant grain yield, the plant also exhibited a lower plant height. Taken together, our results revealed that the qph12 have good specific application prospects in future rice breeding.
Introduction
Plant height is an important factor that determines the architecture and grain yield of cereal plants (Wang et al., 2018; Liao et al., 2019). The semidwarf genes, which result in a shortened culm, improved lodging resistance and an increased harvest index, contributed to the “Green Revolution” in wheat and rice (Peng et al., 1999; Spielmeyer et al., 2002). However, the wide application of dwarf germplasm resources and their narrow genetic range coupled with the excessive use of pesticides and fertilizers have led to serious environmental problems (Pimentel, 1996; Deng et al., 2019), and these problems have encouraged the study of genetic and molecular mechanisms for establishing an “ideal” plant structure through regulating plant height.
Several QTLs associated with plant height have been identified in rice (Li et al., 2003; Zhang et al., 2006; Lee et al., 2014). Moreover, using RILs, Han et al. (2017) identified three QTLs designated qPh3.1, qPh1 and qPh7.1 for plant height. Furthermore, using the CSSL population, Shearman et al. (2019) identified plant height QTLs on chromosomes 1 and 4. Most of the identified genes related to plant height, such as semidwarf1 (sd1; Sasaki et al., 2002), GA-insensitive dwarf1 (gid1; Ueguchi-Tanaka et al., 2005), GA-insensitive dwarf2 (gid2; Hirano et al., 2010), BR-deficient dwarf1 (brd1; Mori et al., 2002), BR-insensitive mutant (d61; Hong et al., 2003), and BR-deficient mutant (osdwarf4-1; Sakamoto et al., 2006), are related to the metabolism or signaling of the phytohormones gibberellin (GA) and brassinosteroid (BR; Deng et al., 2019).
Although the GA- and BR-related genes associated with plant height have been extensively studied, an increasing number of novel plant height-related genes that rely on pathways other than the GA and BR pathways have been identified. For instance, the carotenoid-derived phytohormone strigolactone has become a focus of research on plant architecture patterning (Zhou et al., 2013). Recent studies have shown that OsCKX9, which encodes a cytokinin oxidase that catalyzes the degradation of cytokinin, functions as a primary strigolactone-responsive gene that regulates rice tillering, plant height, and panicle size, likely via the secondary response gene OsRR5, which encodes a cytokinin-inducible rice type-A response regulator. This pathway demonstrates that strigolactone regulates the rice shoots architecture by enhancing cytokinin catabolism through modulating the expression of OsCKX9 (Duan et al., 2019).
Auxin exerts pleiotropic effects on plant cell elongation, cell division and differentiation, root initiation, apical dominance, and tropic responses by regulating the expression of the early auxin-responsive auxin/indoleacetic acid (Aux/IAA) genes (Jain et al., 2006). By screening the publicly available databases, Jain et al. (2006) identified 31 Aux/IAA genes in rice and found that these genes had different functions. OsIAA1 and OsIAA3 play important roles in the crosstalk between the auxin and brassinosteroid signaling pathways and plant morphogenesis (Thakur et al., 2001; Nakamura et al., 2006). The gain-of-function mutation in OsIAA11 inhibits lateral root development in rice (Zhu et al., 2012). OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice (Kitomi et al., 2012). OsIAA6 is involved in drought tolerance and tiller outgrowth (Jung et al., 2015), and the OsIAA10 protein directly targets the rice dwarf virus P2 protein and enhances viral infection and disease development (Jin et al., 2016). Near-isogenic lines (NILs) carried one or more donor chromosome segments provide distinct advantages for QTL identification (Yang et al., 2016). Moreover, NILs can block background genetic noise, undoubtedly enhance our understanding of complex traits and promote plant genomic studies (Henry et al., 2015; Yang et al., 2016).
The current study was carried out to: (i) develop a rice NIL population through the crossing and back-crossing of two rice cultivars, i.e., the japonica cultivar Hui1586 that served as a donor, and the indica cultivar Jiafuzhan that served as a recipient, (ii) implementing 176 NILs for fine mapping of major QTLs and cloning of genes underlying plant height using, (iii) performing a functional analysis of the cloned genes using CRISPR/Cas9 genome editing.
Materials and Methods
Plant Materials
The indica rice cultivar Jiafuzhan and the japonica rice cultivar Hui1586 were preserved at the Rice Research Institute, Fujian Academy of Agricultural Sciences, China. Hui1586 was derived from a cross between Suxiu867 and Minghui86, and the detailed selection process was as follows: Suxiu867 (Food Crops Research Institute, Jiangsu Academy of Agricultural Sciences), a japonica cultivar, was used as a recipient, and Minghui86 (Rice Research Institute, Fujian Academy of Agricultural Sciences), a restorer indica cultivar, was used as a donor. The F1 plants were generated from Suxiu867 as the female parent and Minghui86 as the male parent. The F1 plants were backcrossed to the Suxiu867 parent to produce the BC1F1 generation. Six BC1F1 plants were backcrossed to the Suxiu867 parent to produce 6 BC2F1 plants, which were self-pollinated to produce 72 BC2F2 lines (12 individuals from each of the six BC2F1 plants was sown). The 72 individuals were self-pollinated for six generations. In this process, shorter plants, higher seed setting rate and better comprehensive agronomic traits were selected for to continue planting single plant and eliminate the remaining lines. Then a stable line designated Hui1586 was obtained.
NILs Development
For the development of the NILs, the indica cultivar Jiafuzhan was used as a recipient, and the restorer japonica cultivar Hui1586 was used as a donor. The F1 plants were generated from the Jiafuzhan as the female parent and Hui1586 as the male parent. The F1 plants were back-crossed to the Jiafuzhan parent to produce the BC1F1 generation. These BC1F1 plants were then backcrossed to the Jiafuzhan parent to produce BC2F1 plants. Using the same approach, 118 BC3F1 individuals were obtained, which were self-pollinated to produce the BC3F2 lines. Based on their characteristics, we selected one or two individual plants from each line. As a result, 176 NILs were obtained (Supplementary Figure 1).
QTL Analysis
The WinQTLCart 2.5 software (Wang et al., 2012) implementing the composite interval mapping (CIM) was employed for QTL detection (Zeng, 1993). The confidence interval was defined as the 1-LOD reducing region around the locus of a peak LOD value of the identified QTL. LOD value of ≥ 2.5 was set as a threshold for QTL identification.
Identification of Major QTLs for Plant Height
In autumn 2019, Jiafuzhan, Hui1586 and 176 NILs were planted under natural conditions in the paddy fields of Sanya Experimental Station, Hainan Province, China (18°14′N, 109°31′E, 7.0 m asl). Forty-eight plants of each of the parents and the NILs were planted in six rows. Four plants from the center of each plot were picked to evaluate their plant height characteristics. Major QTLs associated with plant height were identified on the basis of significant differences in plant height between parents and each of the NILs, as determined by t-test. Moreover, panicle length, effective panicle number, spikelets per panicle, seed setting rate and 1, 000-grain weight were also estimated at maturity stage for the parents, NIL36 and knockout lines.
All plants were planted in accordance with standard commercial practices. Plants were grown in the field at 13.3 cm plant to plant distance and 26.40 cm row to row distance. Agronomical practices were performed according to the normal agricultural practices locally recommended for rice production.
Construction of a Mapping Population for Major QTLs Identification
The NIL36 line was crossed with the Jiafuzhan cultivar to develop a mapping population. The F2 population was constructed by self-crossing of the F1 hybrid. A primary linkage of the QTLs for plant height was obtained using 45 recessive plants from the F2 population. Furthermore, 1264 recessive plants from the F2 population were selected for fine mapping of major plant height QTLs.
PCR Amplification and Marker Detection Analysis
The CTAB method (Murray and Thompson, 1980) with minor modifications was used for the extraction of plant DNA from frozen leaves of the rice plants. For PCR amplification, each 20-μl reaction mixture contained 30 ng DNA, 0.4 μm primers and 2× Es Tag MasterMix (Dye). The amplification program includes the following procedures: 2 min at 94°C; 33 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C; and a final extension at 72°C for 2 min. The amplified PCR products underwent 3% agarose gel electrophoresis and were stained with ethidium bromide (Panaud et al., 1996).
Genetic Mapping of Plant Height QTLs
We used the obtained phenotypic data and SSR markers for the identification of QTLs. Genetic distance was estimated using MapDraw V2.1 (Liu and Meng, 2003). The genetic linkage map obtained in this study is basically consistent with that reported by Rahman et al. (2007).
Physical Mapping and Bioinformatics Analysis of the Major Plant Height QTL qph12
The physical map of QTLs for plant height was constructed through a bioinformatics analysis using the published sequences of BAC and P1-derived artificial chromosome (PAC) clones of cv. Nipponbare released by the International Rice Genome Sequencing Project.1 Target gene linkage markers were used to clone, and sequence alignment was performed using the matching Basic Local Alignment Search Tool. According to the existing sequence annotation database,2 candidate genes based on the existing sequence annotation database analysis were identified.
Targeted Knockout of Candidate Genes in Jiafuzhan Using the CRISPR/Cas9 Approach
The first exon of the qPH12 gene in the Jiafuzhan cultivar was targeted with one gRNA spacer. Highly specific gRNA spacer sequences were designed using CRISPR plant database and website (gRNA: ggctgacgaccgggagaagaagg; Xie et al., 2014). The primer sequence for vector construction were showed (Supplementary Table 1). Genome editing mutations of target genes in regenerated plants were analyzed. The deletion and insertion within targeted genes were detected by PCR. PCR products were selected from transgenic CRISPR-edited strains for sequencing to identify specific mutations. The degradation sequence decoding method was used to analyze the double peaks (Ma et al., 2015). The primers used in CRISPR/Cas9 experiments are shown in Supplementary Table 1.
Measuring the Levels of Phytohormones
Stems of Jiafuzhan (CK), NIL36, qPH12KO-line1, qPH12KO-line2 and qPH12KO-line3 were sampled during the rice jointing stage. The auxin (IAA) contents were measured using MetWare3 based on the AB Sciex QTRAP4500 LC–MS/MS platform. For the determination of auxin content, plants were sampled at the heading and jointing stage of rice with 3 replicates were sampled from each genotype. Samples were treated with methanol dissolved as a solvent and stored at −20°C. Samples were diluted into different gradient concentrations before mass spectrometry. The Ultra Performance Liquid Chromatography was used for separation. The Multiple Reaction Monitoring was implemented for the analysis.
Expression Analysis of the qPH12
Total RNA was extracted from rice leaves according to the instructions of the extraction kit (TRIzol, Invitrogen, United States), and DNase treated. An aliquot of about 1.5 μg of RNA was reverse transcribed using the first strand cDNA synthesis kit (Bao Bioengineering Co., LTD.), and the cDNA was 10-times diluted for RT-qPCR. Primers for RT-qPCR were designed and optimized to >95% amplification efficiency. Fluorescence of the Real-Time Fluorescence Quantitative kit (Bao Bioengineering Co., Ltd.) was measured in an CFX96 real-time PCR apparatus (Bio-Rad, Munich, Germany) according to the RT-qPCR procedure described by Bustin et al. (2009). Expression levels were measured using three independent biological replicates and three technical replicates and normalized against the reference gene UBIQUITIN. Primer sequences of related genes are shown in Supplementary Table 1.
Results
Identification and Analysis of Major QTLs for Plant Height in the NILs
To evaluate the potential advantages of the NILs for major QTLs detection, the phenotypic variations in plant height were observed in 176 NILs, with the NIL36 line exhibited a lower plant height compared to the Jiafuzhan cultivar, however, no significant differences were observed in plant height among the remaining NILs. QTL analysis using the WinQTLCart 2.5 software showed that the LOD score value of the plant height QTL for the NIL36 reached 7.84, with an explained phenotypic variance (R2) of 17.52%. Further investigations and analyses showed that the plant height of the Jiafuzhan plants was 116.22 cm, whereas that of the NIL36 line was 79.52 cm. The differences in plant height between these lines reached a highly significant level, as revealed by the t-test (Table 1; Figure 1).
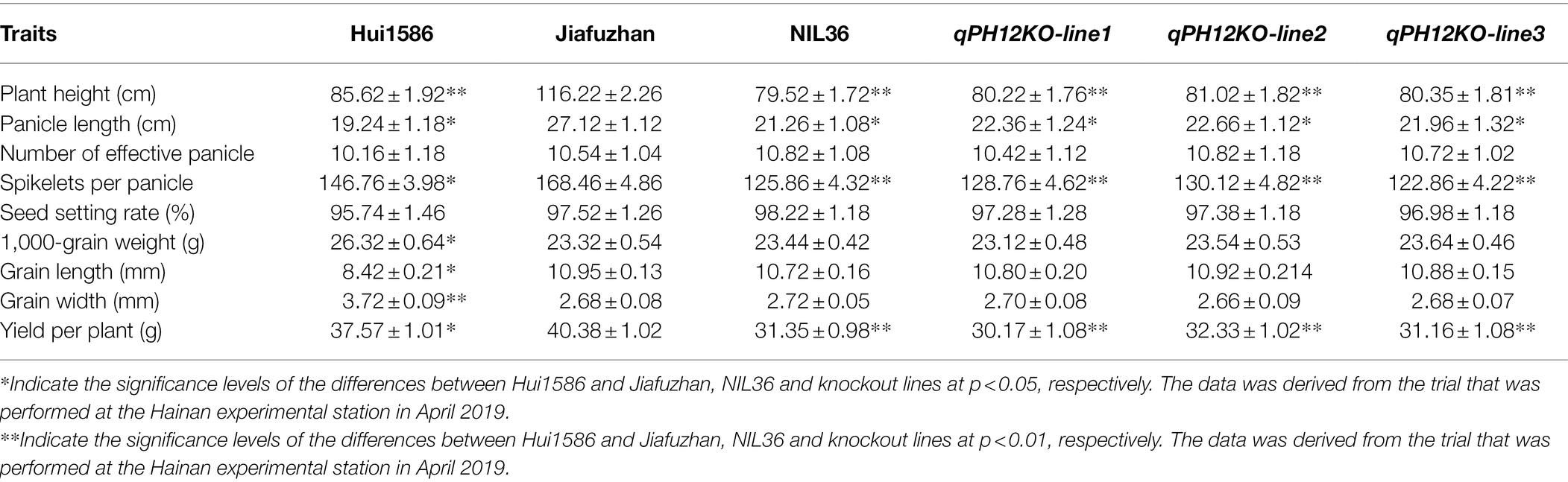
Table 1. Comparison of the main agronomical traits of Hui1586, Jiafuzhan, NIL36 and the qPH12KO knockout mutant lines.

Figure 1. Phenotypic comparison of Jiafuzhan and NIL36. The phenotypes of Jiafuzhan and NIL36 at the mature period are shown.
Phenotypic comparisons between the NIL36 and the Jiafuzhan plants are presented in Table 1. The results showed some significant differences in major agronomical traits, including plant height, panicle length, spikelets per panicle and yield per plant, between the NIL36 line and the Jiafuzhan cultivar. However, no significant differences observed in the number of effective panicles, seed setting rate, 1,000-grain weight, grain length or grain width (Table 1).
Genetic Analysis of the Plant Height Phenotype in the NIL36
To determine whether plant height in the NIL36 line is controlled by a single gene or not, NIL36 was crossed to the Jiafuzhan cultivar. F1 plants showed the plant height phenotype of the Jiafuzhan cultivar, meanwhile the F2 population showed Mendelian segregation (Table 2). The segregation between the Jiafuzhan and NIL36 phenotypes fit the 3:1 segregation ratio in the two F2 populations (χ2 = 0.134 ~ 0.456, p > 0.5). The results showed that the plant height phenotype in the NIL36 is controlled by a single recessive gene.
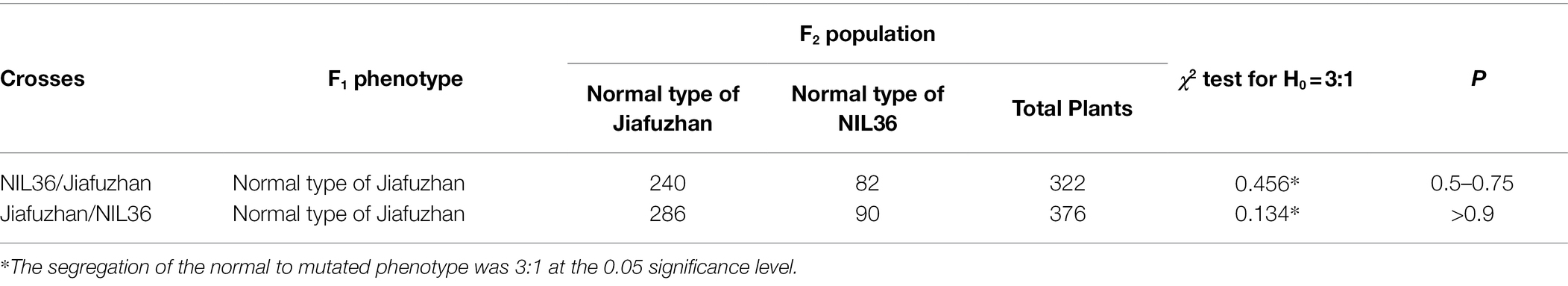
Table 2. Phenotypic segregation for plant height in the F2 populations derived from crosses between the Jiafuzhan cultivar and the NIL36 lines.
Co-segregation Analysis of Plant Height Phenotype and the Major QTL in the NIL36
To identify the gene responsible for the NIL36 plant height phenotype, we located the plant height QTL in the NIL36 and a total of 506 SSR markers from the rice molecular map were selected for polymorphism surveys between Hui1586 and Jiafuzhan (Mccouch et al., 2002). Out of those 506 SSR markers, 296 exhibited polymorphisms between Hui1586 and Jiafuzhan cultivars. Based on the genotypic data of these 296 SSR markers, 45 recessive plants from the F2 population (NIL36/Jiafuzhan) were used for co-segregation analysis between the SSR markers and the plant height phenotype. One of these SSR markers, RM3326, that located on chromosome 12, showed a complete co-segregation with the plant height phenotype in the selected 45 F2 recessive individuals. The major QTL was therefore designated qph12.
Based on 296 polymorphisms exhibited between Hui1586 and Jiafuzhan cultivars, the genetic background of NIL36 homozygous material was analyzed using 145 SSR primer pairs distributed evenly on the 12 chromosomes of rice. The results showed that four markers, i.e., RM6832 on chromosome 3, RM3498 on chromosome 6, RM3496 on chromosome 8, and RM2584 on chromosome 12, exhibited the homozygous alleles of the Hui1586 cultivar., whereas the remaining 292 markers showed the genetic background of the Jiafuzhan cultivar (Supplementary Figure 2). Therefore, the NIL36 basically restores the genetic background of the recipient parent.
Initial Localization of the qph12 for Plant Height
Publicly available molecular markers around RM3326 marker were used to initially locate the qph12 QTL. A genetic linkage analysis revealed that the qph12 QTL is located between the molecular markers RM2854 and RM235, which are located at a distance of 7.7 cM (Figure 2A). To delimit the genomic region of the qph12, 1264 recessive plants from the Jiafuzhan/NIL36 F2 population were genotyped using six polymorphic indel markers selected from 18 newly developed indel markers (Table 3). Indel markers from the open rice genome sequences were designed and tested to predict the likelihood of polymorphism between the NIL36 line and the Jiafuzhan cultivar by comparing sequences from Nipponbare4 and the indica cultivar 93–11.5 The genotyping of all recombinant genes was performed using six polymorphic markers. The results showed that the qph12 QTL was located within a 295 kb region between the molecular markers Indel12-7 and Indel12-9 on chromosome 12 (Figure 2B; Table 3).
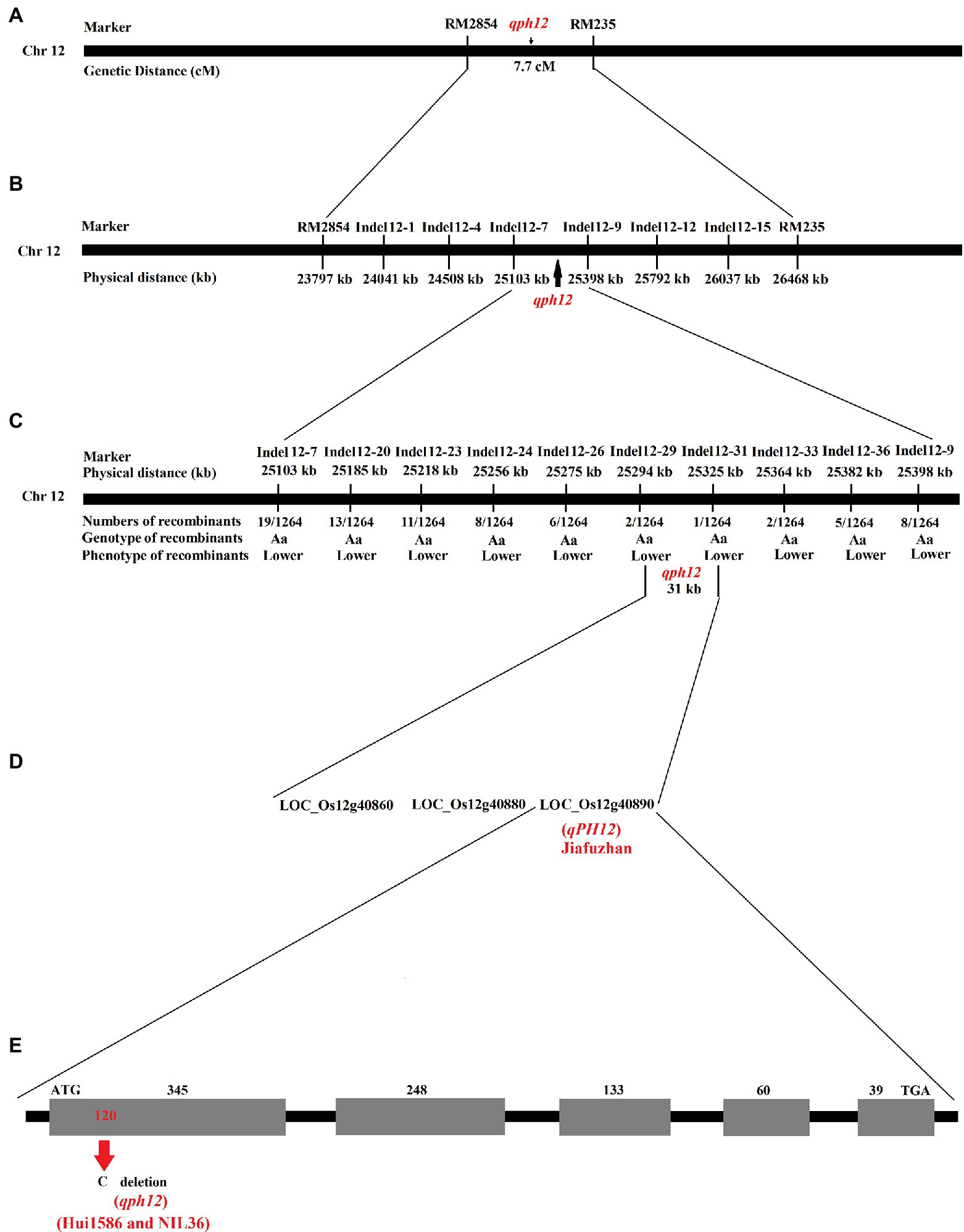
Figure 2. Physical maps and structural comparison of qPH12. (A) Primary mapping of qPH12. The gene was mapped to the region between the markers RM2854 and RM235. (B) Further mapping of qPH12. The gene was mapped to the region between markers Indel12-7 and Indel12-9. (C) Fine mapping of qPH12. qPH12 was localized to a 31 kb region between the markers Indel12-29 and Indel12-31, and the recombinant number between the markers and target genes is indicated under the linkage map. (D) Candidate genes in the 31 kb target region. (E) qPH12 has five exons, and qph12 exhibits a 1 bp deletion in the first exon.
Fine Mapping of the Plant Height QTL qph12
For fine mapping of the qph12 QTL, eight polymorphic indel markers were selected from 26 newly developed indel markers (Table 3). Recombinant screening with eight markers located in a more internal position within the target locus detected 13, 11, eight, six, two, one, two, and five recombinant plants, respectively (Figure 2C). Thus, the qph12 locus was precisely located within a 31 kb region between the molecular markers Indel12-29 and Indel12-31.
Candidate Genes Identification Within the 31 Kb Region of the qph12 Locus
According to the publicly available sequence annotation databases,6,7 three annotated genes with a corresponding full-length cDNA are located within the 31 kb region (Figure 2D). Among these genes, LOC_Os12g40860 encodes the leucine-rich repeat family protein, LOC_Os12g40880 encodes the uridine kinase family protein, and LOC_Os12g40890 is the auxin-responsive Aux/IAA gene family member OsIAA30.
Sequence Analyses of the Plant Height QTL qph12
To identify the gene responsible for the observed plant height phenotype, we sequenced the three candidate genes in the Jiafuzhan cultivar and the NIL36 line. A deletion of only 1 bp (120:C) was found in the LOC_Os12g40890 gene in the NIL36 (Figure 2E), and no further difference was observed between the sequences of the remaining two genes in the two genotypes. Thus, we hypothesized that the recessive gene corresponds to the major plant height QTL qph12 in the NIL36 line, and the dominant allele (LOC_Os12g40890) of the WT Jiafuzhan was designated qPH12. Since the fragment of qph12 was probably derived from the parent Hui1586, we sequenced the qph12 of Hui1586, and the results showed that the sequences of qph12 from both Hui1586 and NIL36 are identical (Figure 2E).
The analysis of the open reading frame (ORF) region showed that the qPH12 gene has five exons. qph12 exhibited a 1 bp deletion in the 120th bp of the first exon, which resulted in premature termination of the qPH12 (Figure 2E).
The qph12 Is Responsible for the Plant Height Phenotype in the NIL36
To confirm that qph12 confers the plant height phenotype, we examined whether the knockout of qPH12 in the Jiafuzhan cultivar would lead to the NIL36 phenotype. One sequence-specific guide RNA (sgRNA) was designed to knock out qPH12 using the CRISPR/Cas9 gene editing system. After resistance screening, 20 strains were randomly selected for sequencing. The results revealed four main types of mutations, i.e., insertion, deletion, complex variant and no mutation, from which insertion and deletion T1 homozygous genotypes were self-pollinated for two generations to produce the T3 generations, designated as qPH12KO-line1 and qPH12KO-line2, respectively. The complicated variant T1 replaced the heterozygous genotype, and the T2 generation was sequenced and screened, and self-pollinated for three generations to produce the T4 generation, qPH12KO-line3. A total of three plants from three independent events (qPH12KO-line1, qPH12KO-line2 and qPH12KO-line3) were obtained. Sequencing of the qPH12 gene in the three independent knockout lines confirmed that these plants carry mostly insertion or deletion in the targeted sites (Figure 3A). Evaluation of the plant height phenotype of these three homozygous lines at maturity and found that all three lines showed the NIL36 phenotype (Table 1; Figure 3B). Therefore, the targeted mutation of the qPH12 gene led to the NIL36 plant height phenotype, indicating that the loss of function of qPH12 was responsible for the reduced plant height phenotype. Notably, the three knockout lines qPH12KO-line1, qPH12KO-line2 and qPH12KO-line3 showed shorter panicle length, fewer number spikelets per panicle and lower plants yields compared to the Jiafuzhan cultivar (Table 1; Figures 3C–F). Therefore, we hypothesized that the qph12 QTL not only affects plant height but also underlies panicle length, spikelets per panicle and plant yield in rice. According to the standard commercial practices, qPH12KO-line1, qPH12KO-line2, qPH12KO-line3 and Jiafuzhan were grown in a paddy field under natural environmental conditions at transgenic experimental base in Fuzhou, China.
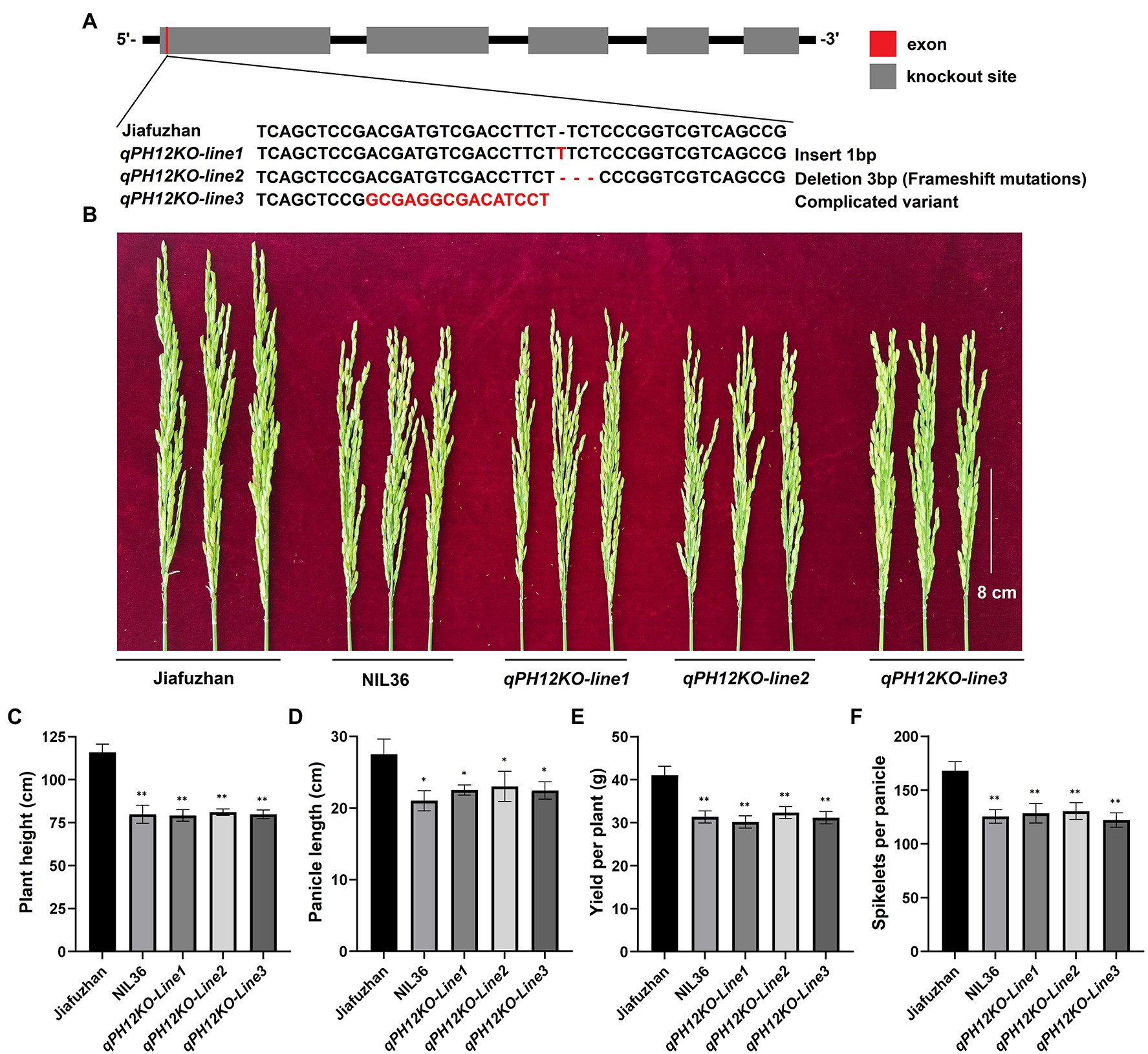
Figure 3. qPH12-knockout lines generated using CRISPR/Cas9 technology and showed the NIL36 phenotype. (A) Three independent events (designated qPH12KO-line1, qPH12KO-line2 and qPH12KO-line3) were generated using the CRISPR/Cas9 system and verified by sequencing. (B) Panicle differences of Jiafuzhan, NIL36 and knockout lines; (C–F) indicate the differences of plant height, panicle length, spikelets per panicle and yield per plant, respectively, across Jiafuzhan, NIL36 and knockout lines in Table 1. *Statistical significance (p < 0.05) determined using Student’s t-test. **Statistical significance (p < 0.01) determined using Student’s t-test.
Comparative Analysis of the Phytohormones Content Between the Jiafuzhan Cultivar and the Three qPH12KO Knockout Lines
To analyze whether the qph12 QTL affects the changes in the Aux/IAA levels, we measured the Aux/IAA content in the Jiafuzhan (CK) and the qPH12KO knockout lines. The results showed that the Aux/IAA content in qPH12KO-line1, qPH12KO-line2, qPH12KO-line3 and NIL36 line was significantly lower than that of the Jiafuzhan (CK; Figure 4).
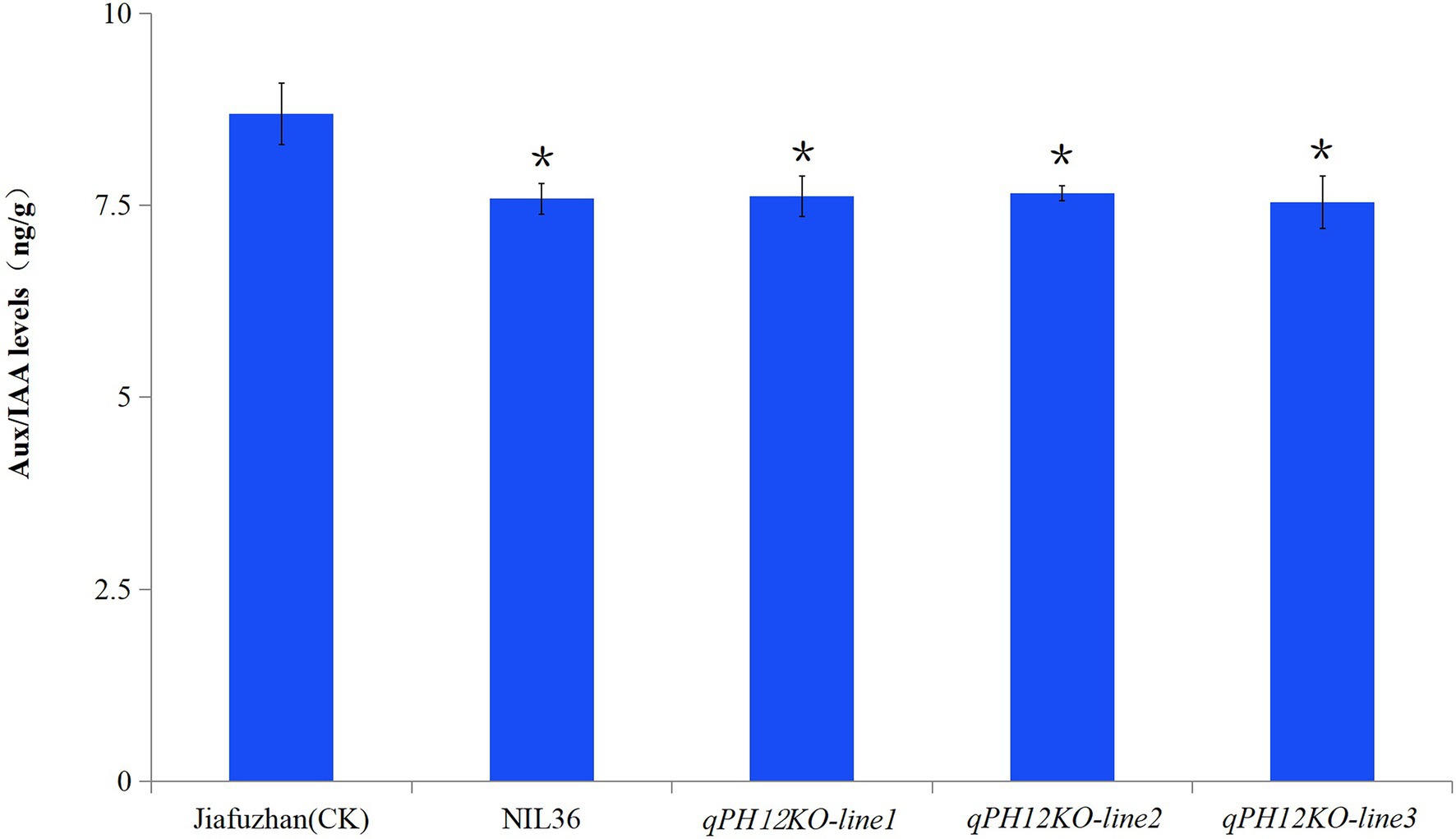
Figure 4. Comparison of Aux/IAA levels between Jiafuzhan and qPH12KO-lines. The Aux/IAA content of qPH12KO-line1, qPH12KO-line2, qPH12KO-line3 and NIL36 was significantly lower than that of the Jiafuzhan cultivar (CK). Three experimental replicates of each line were included. *Statistical significance (p < 0.05) determined using Student’s t-test.
Jiafuzhan Accumulates More qPH12 Transcript
To further investigate the expression abundance of qPH12 gene in Jiafuzhan, NIL36 and the three qPH12KO knockout mutant lines, we analyzed the expression profile of qPH12 in these materials using RT-qPCR. The qPH12 expression was significantly higher in the Jiafuzhan that showed higher plant height and grain yield, longer panicles and more spikelets per panicle compared to the NIL36 line and the three knockout mutant lines qPH12KO-line1, qPH12KO-line2 and qPH12KO-line3 that showed a reduced plant height phenotype (Figure 5).
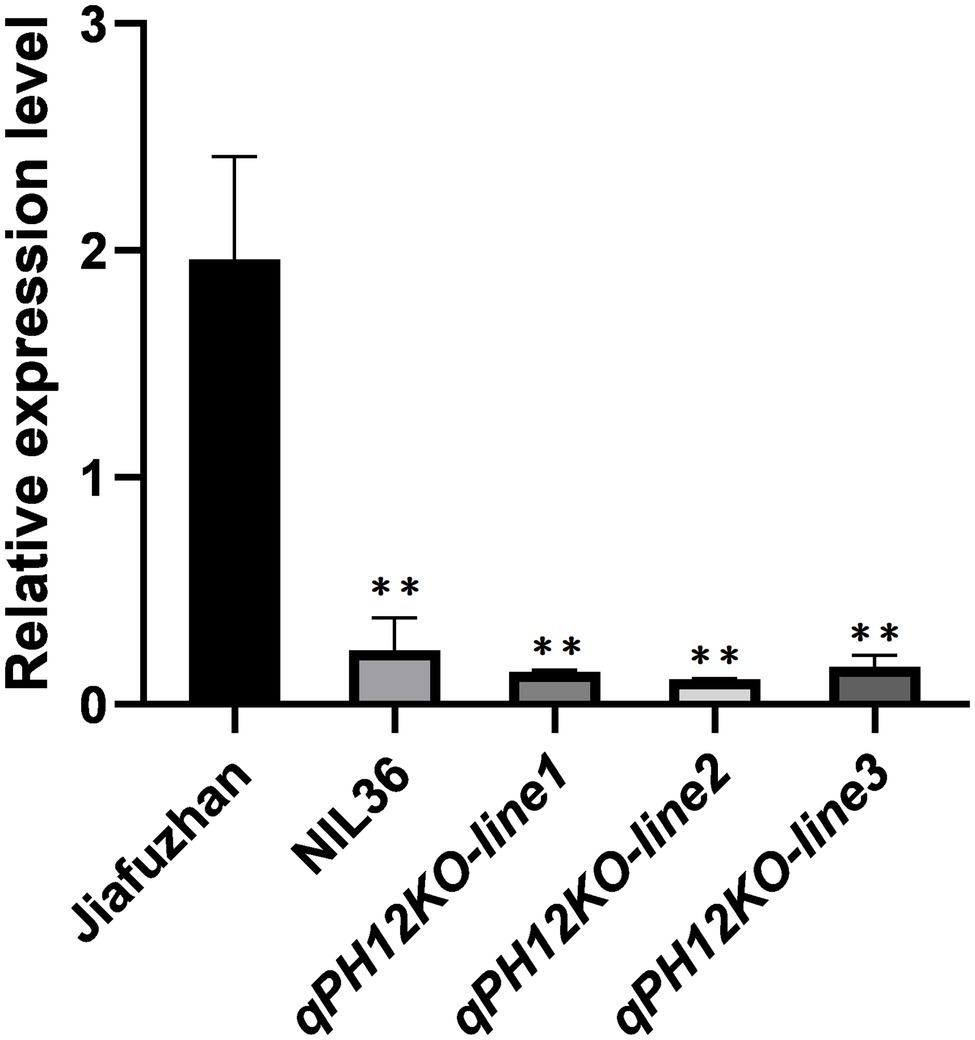
Figure 5. RT-qPCR expression analysis of LOC_Os12g40890 in Jiafuzhan, NIL36 and the three qPH12KO knockout mutant lines. The y-axis represents the relative expression value (log2 – transformed, mean ± SD, n = 3 biological replicates). **significance differences (p < 0.01) determined by the Student’s t-test; ns, non-significant difference.
Discussion
NIL populations have been developed and used for genetic studies and fine mapping of QTLs for genome-wide target traits (Yang et al., 2016). Each NIL carries one or more donor chromosome segments, which provide distinct advantages for QTLs identification, and a QTL can be visualized as a single Mendelian factor by blocking background genetic noise. Different types of QTLs have been identified and/or cloned using NIL populations, such as the drought resistance, qDTY 2.2 (Henry et al., 2015); the thermotolerance, TT1 (Li et al., 2015); the cold tolerance, QTLs (Zhou et al., 2012), the grain type, GS3 (Fan et al., 2006) and GW2 (Song et al., 2007); the heading stage, Ghd7 (Xue et al., 2008) and qHD19 (Yang et al., 2020); the spike type, DEP1 (Huang et al., 2009); the grain production, Cn1a (Ashikari et al., 2005) and 99 QTLs for different agronomic traits (Furuta et al., 2014).
Although a number of plant height related genes/QTLs have been identified, and these genes/QTLs tend to have a function in hormone biosynthesis pathways including gibberellic acid, brassinosteroid, and strigolactone (Shearman et al., 2019), there are also hundreds of QTLs where the underlying cause remains unknown (Lei et al., 2018). In the present study, we developed 176 NILs with a genetic background from the indica rice cultivar Jiafuzhan. Using these lines, we mapped a major plant height QTL designated as qph12, which encoded the auxin-responsive family protein Aux/IAA. Further analysis revealed that qph12 is a novel plant height QTL and its molecular function has not yet been identified. Map-based cloning and knockout experiments confirmed that the reduced plant height phenotype of the NIL36 line is caused by the loss of function of qPH12 that residing the major plant height QTL qph12. Further analyses revealed that the reduced plant height phenotype resulting from the qph12 QTL is caused by a functional deletion in the qPH12 gene. The sequencing results showed that the main cause of this mutation could be due to the mutation in the Hui1586 parent. Similarly, Shearman et al. (2019) reported that the reduced plant height phenotype resulted from the introgression segments from IR62266 into KDML 105 is due to that the IR62266 has a deletion in the Gibberellin 20-oxidase 2 gene that corresponds to the semi-dwarf 1 locus, which led to the identification of a plant height QTL.
Previous studies have showed that Aux/IAA genes play important roles in plant growth and development by regulating the expression of early auxin-responsive genes (Jain et al., 2006). For example, OsIAA1 and OsIAA3 affect plant morphogenesis (Thakur et al., 2001; Nakamura et al., 2006), OsIAA11 and OsIAA13 affect root development in rice (Kitomi et al., 2012; Zhu et al., 2012), and OsIAA6 is involved in tiller outgrowth (Jung et al., 2015). The present study showed that qph12 exhibits a 1 bp deletion in the first exon of qPH12 which is the auxin-responsive Aux/IAA gene family member OsIAA30. Further research showed that the qph12 QTL significantly reduces plant height in rice. Besides, the knockout experiments confirmed that qph12 QTL is responsible for the plant height phenotype. Three homozygous knockout lines also exhibited the phenotype of the NIL36 line, which includes a reduced plant height, a shorter panicle length, fewer spikelets per panicle and a lower yield per plant compared to the Jiafuzhan cultivar (Table 1). Analysis of the phytohormone content showed that the Aux/IAA contents in the NIL36 line, qPH12KO-line1, qPH12KO-line2, qPH12KO-line3 were significantly lower than that of the Jiafuzhan (CK) cultivar (Figure 4). These findings suggest that qph12 QTL simultaneously regulates plant height, panicle length, spikelets per panicle and yield per plant by manipulating the auxin levels in the plants.
In addition, the expression level of qPH12 was higher than that of NIL36 and the three knockout mutant lines (Figure 5). We speculated that this might be a feedback regulation. Under favorable conditions, the downstream signal would promote the expression of qPH12 in response to the activation of qPH12 signaling pathway. Meanwhile, the qPH12 mutant (qph12) would not activate the downstream signaling pathway, so the downstream components could not perceive this biological signal and the expression of qPH12 would no longer needed, and therefore the qPH12 expression level is reduced.
Although the qph12 and the qPH12 affects certain traits, such as plant height, panicle length, spikelets per panicle and yield per plant, the question should be answered, whether qph12 and qPH12 alleles have specific application prospects in the improvement of rice breeding? To address this question, we performed SNP (Single nucleotide polymorphisms) calling and haplotype analysis of the 3,000 sequenced rice genomes available in the CNCGB and CAAS databases (Li et al., 2014) and found 65 haplotypes for the qPH12 gene, including 6 haplotypes among more than 15 rice resource materials (Supplementary Table 2). Further analysis revealed that the Hap1 of the qPH12 contained 2453 rice resource materials, which showed that the qPH12 was predominantly existed in rice population. However, fewer haplotypes were found for the qph12 in the 3,000 sequenced rice genomes. Therefore, to breed a new hybrid rice variety with an ideal plant height, breeders can transfer qph12 into both restorer and sterile lines through molecular marker-assisted selection.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
DY planned and performed the experiments and data collection and wrote the manuscript with input from all authors. NH, GZ, and FH were involved in conducting the experiment, data collection, and analyses. SA-E revised the manuscript. All authors discussed the results and contributed to the final manuscript.
Funding
The work was supported by the Special Fund for Agro-scientific Research in the Public Interest of Fujian Province (no. 2020R1023003), the Fujian Provincial Natural Science Foundation of China (no. 2021J01471), Major Science and Technology Projects of Fujian Province (no. 2020NZ08016), Science and Technology Innovation Team (no. CXTD2021001), 5511 Collaborative Engineering Project (no. KXXYJBG0021), and the 100 Talent Plans of Fujian Province.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.878558/full#supplementary-material
Footnotes
1. ^http://rgp.dna.affrc.go.jp/IRGSP/index.html
2. ^http://rice.plantbiology.msu.edu/; http://www.tigr.org/
4. ^http://rgp.dna.affrc.go.jp/
5. ^http://rice.genomics.org.cn/
References
Ashikari, M., Sakakibara, H., Lin, S. Y., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Deng, W. J., Li, R. Q., Xu, Y. W., Mao, R. Y., Chen, S. F., Chen, L. B., et al. (2019). A lipid transfer protein variant with a mutant eight-cysteine motif causes photoperiod- and thermo-sensitive dwarfism in rice. J. Exp. Bot. 71, 1294–1305. doi: 10.1093/jxb/erz500
Duan, J. B., Yu, H., Yuan, K., Liao, Z. G., Meng, X. B., Jing, Y. H., et al. (2019). Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININOXIDASE/DEHYDROGENASE 9 in rice. Proc. Natl. Acad. Sci. U. S. A. 116, 14319–14324. doi: 10.1073/pnas.1810980116
Fan, C. C., Xing, Y. Z., Mao, H. L., Lu, T. T., Han, B., Xu, C. G., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Furuta, T., Uehara, K., Rosalyn, B., Angeles-Shim, R. B., Shim, J., Ashikari, M., et al. (2014). Development and evaluation of chromosome segment substitution lines (CSSLs) carrying chromosome segments derived from Oryza rufipogon in the genetic background of Oryza sativa L. Breed Sci. 63, 468–475. doi: 10.1270/jsbbs.63.468
Han, Z., Hu, W., Tan, C., and Xing, Y. (2017). QTLs for heading date and plant height under multiple environments in rice. Genetica 145, 67–77. doi: 10.1007/s10709-016-9946-6
Henry, A., Swamy, B. P. M., Dixit, S., Torres, R. D., Batoto, T. C., Manalili, M., et al. (2015). Physiological mechanisms contributing to the QTL-combination effects on improved performance of IR64 rice NILs under drought. J. Exp. Bot. 66, 1787–1799. doi: 10.1093/jxb/eru506
Hirano, K., Asano, K., Tsuji, H., Kawamura, M., Mori, H., Kitano, H., et al. (2010). Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22, 2680–2696. doi: 10.1105/tpc.110.075549
Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15, 2900–2910. doi: 10.1105/tpc.014712
Huang, X. Z., Qian, Q., Liu, Z. B., Sun, H. Y., He, S. Y., Luo, D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352
Jain, M., Kaur, N., Garg, R., Thakur, J. K., Tyagi, A. K., and Khurana, J. P. (2006). Structure and expression analysis of early auxin-responsive aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 6, 47–59. doi: 10.1007/s10142-005-0005-0
Jin, L., Qin, Q. Q., Wang, Y., Pu, Y. Y., Liu, L. F., Wen, X., et al. (2016). Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog. 12:e1005847. doi: 10.1371/journal.ppat.1005847
Jung, H., Lee, D. K., Choi, D. Y., and Kim, J. K. (2015). OsIAA6, a member of the rice aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 236, 304–312. doi: 10.1016/j.plantsci.2015.04.018
Kitomi, Y., Inahashi, H., Takehisa, H., Sato, Y., and Inukai, Y. (2012). OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Sci. 190, 116–122. doi: 10.1016/j.plantsci.2012.04.005
Lee, S., Jia, M. H., Jia, Y., and Liu, G. (2014). Tagging quantitative trait loci for heading date and plant height in important breeding parents of rice (Oryza sativa). Euphytica 197, 191–200. doi: 10.1007/s10681-013-1051-7
Lei, L., Zheng, H. L., Wang, J. G., Liu, H. L., Sun, J., Zhao, H. W., et al. (2018). Genetic dissection of rice (Oryza sativa L.) tiller, plant height, and grain yield based on QTL mapping and metaanalysis. Euphytica 214:109. doi: 10.1007/s10681-018-2187-2
Li, X. M., Chao, D. Y., Wu, Y., Huang, X. H., Chen, K., Cui, L. G., et al. (2015). Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 47, 827–833. doi: 10.1038/ng.3305
Li, Z. K., Fu, B. Y., Gao, Y. M., Wang, W. S., Xu, J. L., Zhang, F., et al. (2014). The 3,000 rice genomes project. Gigascience 3:7. doi: 10.1186/2047-217X-3-7
Li, Z. K., Yu, S. B., Lafitte, H. R., Huang, N., Courtois, B., Hittalmani, S., et al. (2003). QTL×environment interactions in rice. I. Heading date and plant height. Theor. Appl. Genet. 108, 141–153. doi: 10.1007/s00122-003-1401-2
Liao, Z. G., Yu, H., Duan, J. B., Yuan, K., Yu, C. J., Meng, X. B., et al. (2019). SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 10:2738. doi: 10.1038/s41467-019-10667-2
Liu, R. H., and Meng, J. L. (2003). MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25, 317–321.
Ma, X. L., Chen, L. T., Zhu, Q. L., Chen, Y. L., and Liu, Y. G. (2015). Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol. Plant 8, 1285–1287. doi: 10.1016/j.molp.2015.02.012
Mccouch, S. R., Teytelma, L., Xu, Y. B., Lobos, K. B., Clare, K., Walton, M., et al. (2002). Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9, 199–207. doi: 10.1093/dnares/9.6.199
Mori, M., Nomura, T., Ooka, H., Ishizaka, M., Yokota, T., Sugimoto, K., et al. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 130, 1152–1161. doi: 10.1104/pp.007179
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nakamura, A., Umemura, I., Gomi, K. J., Hasegawa, Y., Kitano, H., Sazuka, T., et al. (2006). Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J. 46, 297–306. doi: 10.1111/j.1365-313X.2006.02693.x
Panaud, O., Chen, X., and Mccouch, S. R. (1996). Development of microsatellite and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Gen. Genet. 252, 597–607. doi: 10.1007/BF02172406
Peng, J. F., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Pimentel, D. (1996). Green revolution agriculture and chemical hazards. Sci. Total Environ. 188, S86–S98. doi: 10.1016/0048-9697(96)05280-1
Rahman, M. L., Chu, S. H., Choi, M. S., Qiao, Y. L., Jiang, W. Z., Piao, R. H., et al. (2007). Identification of QTLs for some agronomic traits in rice using an introgression line from Oryaza minuta. Mol. Cell 24, 16–26.
Sakamoto, T., Morinaka, Y., Ohnishi, T., Sunohara, H., Fujioka, S., Ueguchi-Tanaka, M., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24, 105–109. doi: 10.1038/nbt1173
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). A mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. doi: 10.1038/416701a
Shearman, J. R., Vejchasarn, P., Naktang, C., Phansenee, Y., Jomchai, N., Lanceras-Siangliw, J., et al. (2019). Rice height QTLs in KDML105 chromosome segment substitution lines. Genomics 114, 482–487. doi: 10.1016/j.ygeno.2019.09.003
Song, X. J., Huang, W., Shi, M., Zhu, M. Z., and Lin, H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Spielmeyer, W., Ellis, M. H., and Chandler, P. M. (2002). Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. U. S. A. 99, 9043–9048. doi: 10.1073/pnas.132266399
Thakur, J. K., Tyagi, A. K., and Khurana, J. P. (2001). OsIAA1, an aux/IAA cDNA from rice, and changes in its expression as influenced by auxin and light. DNA Res. 8, 193–203. doi: 10.1093/dnares/8.5.193
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
Wang, S., Basten, C. J., Gaffney, P., and Zeng, Z. B. (2012). Windows QTL Cartographer Version 2.5. North Carolina: North Carolina State University Bioinformatics Research Center.
Wang, B., Smith, S. M., and Li, J. Y. (2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468. doi: 10.1146/annurev-arplant-042817-040422
Xie, K. B., Zhang, J. W., and Yang, Y. N. (2014). Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol. Plant 7, 923–926. doi: 10.1093/mp/ssu009
Xue, W. Y., Xing, Y. Z., Weng, X. Y., Zhao, Y., Tang, W. J., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yang, D. W., Cheng, C. P., Zheng, X. H., Ye, X. F., Ye, N., and Huang, F. H. (2020). Identification and fine mapping of a major QTL, qHD19, that plays pleiotropic roles in regulating the heading date in rice. Mol. Breed. 40:234. doi: 10.1007/s11032-020-1109-x
Yang, D. W., Ye, X. F., Zheng, X. H., Cheng, C. P., Ye, N., and Huang, F. H. (2016). Development and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of the whole wild rice genome. Front. Plant Sci. 7:1737. doi: 10.3389/fpls.2016.01737
Zeng, Z. B. (1993). Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. U. S. A. 90, 10972–10976. doi: 10.1073/pnas.90.23.10972
Zhang, Y. S., Luo, L. J., Xu, C. G., Zhang, Q. F., and Xing, Y. Z. (2006). Quantitative trait loci for panicle size, heading date and plant height co-segregating in trait-performance derived near-isogenic lines of rice (Oryza sativa). Theor. Appl. Genet. 113, 361–368. doi: 10.1007/s00122-006-0305-3
Zhou, F., Lin, Q. B., and Zhu, L. H. (2013). D14-SCF (D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. doi: 10.1038/nature12878
Zhou, L., Zeng, Y. W., Hu, G. L., Pan, Y. H., Yang, S. M., You, A. Q., et al. (2012). Characterization and identification of cold tolerant near-isogenic lines in rice. Breed Sci. 62, 196–201. doi: 10.1270/jsbbs.62.196
Keywords: rice (Oryza sativa L. subsp. indica), gene mapping, gene cloning, auxin, near-isogenic lines
Citation: He NQ, Zhan GP, Huang FH, Abou-Elwafa SF and Yang DW (2022) Fine Mapping and Cloning of a Major QTL qph12, Which Simultaneously Affects the Plant Height, Panicle Length, Spikelet Number and Yield in Rice (Oryza sativa L.). Front. Plant Sci. 13:878558. doi: 10.3389/fpls.2022.878558
Edited by:
Kejian Wang, China National Rice Research Institute (CAAS), ChinaReviewed by:
Joong Hyoun Chin, Sejong University, South KoreaZhiguo Zhang, Biotechnology Research Institute (CAAS), China
Copyright © 2022 He, Zhan, Huang, Abou-Elwafa and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dewei Yang, ZGV3ZWkteUAxNjMuY29t
†These authors have contributed equally to this work
 Niqing He
Niqing He Guanping Zhan
Guanping Zhan Fenghuang Huang
Fenghuang Huang Salah Fatouh Abou-Elwafa
Salah Fatouh Abou-Elwafa Dewei Yang
Dewei Yang