
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 17 May 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.878001
This article is part of the Research TopicFunctional Genomics in Fruit Trees: from ‘Omics to Sustainable Biotechnologies, Volume IIView all 11 articles
 Molly Clemens1,2,3
Molly Clemens1,2,3 Michele Faralli1*†
Michele Faralli1*† Jorge Lagreze1
Jorge Lagreze1 Luana Bontempo1
Luana Bontempo1 Stefano Piazza1
Stefano Piazza1 Claudio Varotto1
Claudio Varotto1 Mickael Malnoy1
Mickael Malnoy1 Walter Oechel2,4
Walter Oechel2,4 Annapaola Rizzoli1
Annapaola Rizzoli1 Lorenza Dalla Costa1*
Lorenza Dalla Costa1*Epidermal Patterning Factor Like 9 (EPFL9), also known as STOMAGEN, is a cysteine-rich peptide that induces stomata formation in vascular plants, acting antagonistically to other epidermal patterning factors (EPF1, EPF2). In grapevine there are two EPFL9 genes, EPFL9-1 and EPFL9-2 sharing 82% identity at protein level in the mature functional C-terminal domain. In this study, CRISPR/Cas9 system was applied to functionally characterize VvEPFL9-1 in ‘Sugraone’, a highly transformable genotype. A set of plants, regenerated after gene transfer in embryogenic calli via Agrobacterium tumefaciens, were selected for evaluation. For many lines, the editing profile in the target site displayed a range of mutations mainly causing frameshift in the coding sequence or affecting the second cysteine residue. The analysis of stomata density revealed that in edited plants the number of stomata was significantly reduced compared to control, demonstrating for the first time the role of EPFL9 in a perennial fruit crop. Three edited lines were then assessed for growth, photosynthesis, stomatal conductance, and water use efficiency in experiments carried out at different environmental conditions. Intrinsic water-use efficiency was improved in edited lines compared to control, indicating possible advantages in reducing stomatal density under future environmental drier scenarios. Our results show the potential of manipulating stomatal density for optimizing grapevine adaptation under changing climate conditions.
Drought is a threat to the quality and yield of grapevine in the world’s important wine grape growing regions (Mosedale et al., 2016; Van Leeuwen and Destrac-Irvine, 2017; Van Leeuwen et al., 2019). These regions are expected to have decreased precipitation with associated risks of developing soil water deficit in coming years (IPCC, 2014; Sherwood and Fu, 2014; Scholasch and Rienth, 2019). One adaptation strategy seen in plants to tolerate water limitation involves stomatal regulation of water loss (Hunt et al., 2010; Hughes et al., 2017; Bertolino et al., 2019; Caine et al., 2019; Dayer et al., 2020; Gambetta et al., 2020). Stomata are pores mainly located in the leaf epidermis. The opening of these pores controls leaf gas exchange (CO2 uptake for photosynthesis and water loss via transpiration) and is regulated by changes in turgor pressure in the guard cells surrounding these pores. The two guard cells respond to a range of environmental signals, often in conflict with each other, and sometimes rapidly changing (e.g., humidity, CO2 concentration, light). In drought-stressed grapevine, stomatal closure is triggered by hydraulic signals and maintained by abscisic acid following re-watering (Lovisolo et al., 2010; Tombesi et al., 2015). Genotypic variation for stomatal sensitivity to reduced water availability has been shown to exist in grapevine (Schultz, 2003; Soar et al., 2006; Bota et al., 2016; Villalobos-González et al., 2019; Faralli et al., 2021).
Stomatal density and distribution in the epidermal tissue also plays a critical role in determining transpiration rate per unit of leaf area (Hunt et al., 2010). Previous work focusing on natural variation for stomatal anatomical features provided evidence of a close negative relationship between plant water-use efficiency and stomatal density (Bertolino et al., 2019; Faralli et al., 2019). According to extensive studies carried out in Arabidopsis (Doheny-Adams et al., 2012; Franks et al., 2015; Hepworth et al., 2015; Lee et al., 2015), stomatal density and distribution are under the control of small cysteine-rich peptides (CRP) called epidermal patterning factors (EPFs) highly conserved in a wide range of higher plants (Lu et al., 2019). Three members of this family play a key role in the formation of stomata: EPF1, EPF2 and EPFL9. EPF2 and EPF1 are expressed in the epidermis, in the earlier and later stages of leaf development, respectively. EPF2 inhibits the formation of cells considered the precursors of stomata guard cells, while EPF1 inhibits the subsequent differentiation of these same precursors and induces asymmetric cell division (Hara et al., 2009). Epidermal Patterning Factor Like 9 (EPFL9), also known as STOMAGEN, plays an antagonist role with respect to EPF1 and EPF2 as it induces stomata formation (Kondo et al., 2010). EPF-peptides interact with two transmembrane receptors of epidermal cells, ERECTA and Too Many Mouths (TMM). While EPF1 and EPF2 activate the receptor complex which in turn induces a MAPKs (Mitogen-Activated Protein Kinases) cascade (Morales-Navarro et al., 2018; Zoulias et al., 2018) leading to the destabilization of important transcription factors involved in the formation of stomata (SPEECHLESS, MUTE, FAMA; Pillitteri et al., 2007; Chen et al., 2020), STOMAGEN inactivates it. STOMAGEN is the only known positive regulator of stomata produced in mesophyll, and was confirmed to act independently of EPF1 and EPF2 (Hunt et al., 2010; Kondo et al., 2010; Sugano et al., 2010; Ohki et al., 2011). Its activity is antagonized by that of EPF2, however, it is not well understood if the antagonistic action is due to the sharing of an identical binding site in the common receptor or to other mechanisms (Ohki et al., 2011). An evolutionary model suggests that EPFL9 may derive from the duplication of EPF1/2 with a subsequent alteration in the function (Shimada et al., 2011). This is confirmed by the fact that EPF1/2 are more widespread in higher plants compared to EPFL9 (Lu et al., 2019). Despite the different amino acid composition among the CRP different sub-classes and across species, the members of CRPs have in common a small size, a conserved N-terminal region that include an apoplast secretion signal and a functional C-terminal domain containing cysteine residues (Marshall et al., 2011).
Several functional genomics studies, based on the ectopic expression or silencing of EPF1, EPF2, or EPFL9, have recently demonstrated a highly conserved functional paradigm in Arabidopsis and cereals. In barley, Hughes et al. (2017) proved that HvEPF1 overexpression limits stomatal development. In a hexaploid bread wheat, Dunn et al. (2019) decreased stomatal density (SD) via the overexpression of TaEPF1 and TaEPF2 orthologues and demonstrated improvements in water-use efficiency without affecting yield when SD reduction was moderate. Similarly, in rice Caine et al. (2019) and Mohammed et al. (2019) elucidated the function of OsEPF1 adopting an over-expression approach. Adding to the studies on rice, Lu et al. (2019) confirmed the role of OsEPF1, OsEPF2 and OsEPF9 by a dual strategy, both over-expression and down-regulation via RNA interference. Yin et al. (2017) were the first to apply the genome editing technology in rice to disrupt OsEPFL9.
Gene editing via the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) (Jinek et al., 2012) is to date the most powerful tool for functional genomics studies in plants (Liu et al., 2016). CRISPR/Cas9 system can efficiently produce nucleotide mutations into precise positions in the genome through the combined action of a specific guide RNA and the Cas9 nuclease which cleaves the DNA eliciting the non-homologous end-joining (NHEJ) pathway for DNA repair (Podevin et al., 2013). NHEJ may produce knock-out (KO) mutants with random insertion or deletion (indels) of variable lengths at the Cas9 cleavage site causing frameshift mutations or loss of amino acids in protein-coding sequences. These KO mutants are perfect systems to prove the function of a candidate gene (Jain, 2015). This technology is steadily boosting (Hess et al., 2017; Anzalone et al., 2019) and, coupled with the advancements of in-vitro culture practices, represents a knowledge-based strategy for the genetic improvements of cultivated plants, with relevant advantages compared to traditional breeding (Chen et al., 2019).
In grapevine, CRISPR/Cas9 technology has been successfully applied to evaluate the function of genes involved in susceptibility or tolerance to diseases, mainly caused by fungal pathogens (Malnoy et al., 2016; Giacomelli et al., 2019; Li et al., 2020; Wan et al., 2020; Chen et al., 2021; Scintilla et al., 2021), or to enhance tolerance to cold stress (Wang et al., 2021).
In this study, we inactivated VvEPFL9-1 in a grapevine table grape variety, ‘Sugraone’, adopting a genome editing approach based on CRISPR/Cas9 technology. Different edited lines with a significant reduction in stomatal density were produced and three of them were analyzed to investigate how reducing stomatal density affects grapevine physiological performance under different environmental conditions.
AtEPFL9 sequence (AT4G12970) was used as a query to interrogate the publicly available genomic databases of Vitis spp. (Supplementary Table 1). To experimentally confirm the presence of two VvEPFL9 paralogs in a set of grapevine genotypes, DNA was extracted from leaf tissue of ‘Chardonnay’, ‘Merlot’, ‘Syrah’, ‘Cabernet Sauvignon’, ‘Touriga National’, ‘Pinot Noir clone Entav 115’, ‘Pinot Noir PN40024’, ‘Sugraone’ and ‘Riparia Glorie de Montpellier’ using Nucleospin Plant II kit (Macherey–Nagel, Düren, Germany) following the manufacturer’s instruction. Genomic DNA was quantified using Nanodrop 8,800 (Thermo Fischer Scientific, Waltham, MA, United States) and diluted to a final concentration of 30 ng/μL. Two PCR reactions were performed in 25 μl final volume containing 1 × PCR BIO (Resnova, Rome, Italy), 30 ng of genomic DNA and 0.5 μM of primers in order to amplify VvEPFL9-1 (primer VvEPFL9-1_fw and VvEPFL9-1_rv, see Supplementary Table 2) and VvEPFL9-2 (primer VvEPFL9-2_fw and VvEPFL9-2_rv, see Supplementary Table 2). Amplification products were checked on agarose gel, purified using CleanNGS magnetic beads (CleanNA, Waddinxveen, Netherlands) and sequenced by Sanger sequencing (FEM Sequencing Platform Facility, San Michele all’Adige, Italy). Sequencing outputs were analyzed with Blast online tool1 and for the alignment of the sequences the software MEGAX (Kumar et al., 2018) was used.
The CRISPR/Cas9 binary vector with the customized sgRNA was purchased from DNA Cloning Service (Hamburg, Germany). The nucleotide sequence of SpCAS9 and of NPTII genes were codon optimized for the plant expression system and their sequences are available on the company website.2 The sequence of the guide RNA carried by the vector was designed with CRISPR-P 2.0 software3 and recognizes a region of 20 bp in the third exon of VvEPFL9-1 (GCACATACAATGAATGCAAA, on-score = 0.7058). Agrobacterium tumefaciens (A.t.)-mediated gene transfer was performed on embryogenic calli of ‘Sugraone’ according to Dalla Costa et al. (2022). NPTII was used as selectable marker to confer resistance to kanamycin. Regenerated plants were screened by PCR for the presence of SpCAS9 (to select plants which integrated T-DNA) in 20 μl final volume containing 1 × PCR BIO (Resnova, Rome, Italy), 0.5 μM of each primer (SpCAS9_Fw and SpCAS9_Rv, see Supplementary Table 2) and 30 ng of genomic DNA. DNA was extracted from freshly frozen leaf tissue (approximately 100 mg) using Nucleospin Plant II kit (Macherey–Nagel, Düren, Germany) following the manufacturer’s instruction, quantified using Nanodrop 8,800 (Termo Fischer Scientific, Waltham, MA, United States) and diluted to a final concentration of 30 ng/μL.
Edited lines and WT control were propagated in-vitro in sterilized jars containing WP medium (McCown and Lloyd, 1981) in a growth chamber at 100 photosynthetic photon flux density (PPFD) ± 20 (μmol m−2 s−1), 24°C and a 16/8 light/dark photoperiod. Four biological replicates of healthy developed edited lines and of the WT control were acclimatized in the greenhouse using 0.25 l plastic pots with three holes in the bottom to allow for water drainage, filled with a similar amount of growing substrate (Extra quality - Semina, TerComposti, Calvisano, Italy) and covered by parafilm on the top. Plants were kept in a growth chamber (PPFD 100 +/− 20 μmol m−2 s−1, 24°C, 16/8 light/dark photoperiod) and after 1 week, holes were gradually made in the top of the parafilm over the course of 2 weeks. After 17 days, plants were repotted into 0.75 l pots all containing growing substrate (Extra quality - Special Cactus, TerComposti, Calvisano, Italy). Pots were kept in the same growth chamber for a subsequent 10 days before moving to the greenhouse. In the greenhouse, plants were grown under natural light supplemented by high-pressure sodium lamps system (PPFD 200–250 μmol m−2 s−1) with a 16-h/8-h light–dark photoperiod. Environmental conditions including temperature and humidity during the growth chamber and greenhouse cultivation are shown in Supplementary Figure 1.
The quantification of SpCAS9 copy number (CN) in grapevine lines was carried out according to real-time PCR method developed by Dalla Costa et al. (2009). Reactions were performed in a 96-well plate on a C1000 thermal cycler (Bio-Rad, Hercules, United States) equipped with CFX96 real-time PCR detection system (Bio-Rad, Hercules, United States). The real-time PCR singleplex reaction was carried out in a 10 μl final volume containing 1 × SsoAdvanced Universal Probes Supermix (Bio-Rad, Hercules, United States), 40 ng of genomic DNA, 0.3 μM primers (Sigma, Haver hill, UK) and a 0.2 μM specifc Taqman probe (Sigma, Haverhill, UK). The thermal protocol was as follows: polymerase activation for 3 min at 95°C followed by 40 cycles of denaturation of 10 s at 95°C, annealing of 5 s at 58°C and 5 s at 60°C and an elongation of 30 s at 72°C. Primers and Taqman probes used to amplify grapevine endogenous VvCHI (VvChiRT_fw; VvChiRT_rv; VvChiRT_Probe) and SpCAS9 (SpCas9RT_fw; SpCas9RT_rv; SpCas9RT_Probe) were reported in Supplementary Table 2. The standard curves (four points, starting from 106 plasmid molecules and adopting a serial dilution of 1:5) were built with a plasmid pGEM-T easy (Promega, Madison, Wisconsin, United States), in which we cloned a fragment of VvCHI and SpCAS9. For each sample, the SpCAS9 CN was calculated using the following formula: (transgene total copies / endogenous gene total copies) × 2. The total copies of transgene and endogenous gene were calculated on the basis of the mean values of the quantification cycles (Cq) of two technical replicates.
In the grapevine lines integrating T-DNA, a region of the gene VvEPFL9-1 containing the site targeted by the sgRNA/Cas9 complex, was amplified with primers VvEPFL9-1_fw and VvEPFL9-1_rv (see Supplementary Table 2) both elongated with overhang Illumina adapters. PCR was carried out in 20 μl final volume containing 1 × PCR BIO (Resnova, Rome, Italy), 0.4 μM of each primer and 30 ng of genomic DNA. The Illumina library was sequenced on an Illumina MiSeq (PE300) platform at the Sequencing Platform Facility of Fondazione Edmund Mach (San Michele all’Adige, Italy). CRISPResso2 pipeline4 (Clement et al., 2019) was used to process the raw paired end reads with default parameters and to visualize the mutations profiles in the target sequences. For the analysis of the off-target site in the gene VvEPFL9-2, a PCR was carried out in 25 μl final volume containing 1 × PCR BIO (Resnova, Rome, Italy), 0.5 μM of each primer (VvEPFL9-2_fw and VvEPFL9-2_rv, see Supplementary Table 2) and 30 ng of genomic DNA. Amplification products were checked on agarose gel, purified using CleanNGS magnetic beads (CleanNA, Waddinxveen, Netherlands) and sequenced by Sanger sequencing (FEM Sequencing Platform Facility). Sequencing outputs were analyzed with Blast online tool.5
T-DNA integration points (IP) were determined following the method described in Dalla Costa et al. (2020). The library was sequenced by Illumina MiSeq (PE300) platform at the Sequencing Platform Facility of Fondazione Edmund Mach (San Michele all’Adige, Italy). The putative genomic regions identified were validated by PCR amplification. PCR was performed in a 20 μl final volume containing 1 × PCR BIO (Resnova, Rome, Italy), 40 ng of genomic DNA and 0.5 μM of the primers reported in Supplementary Table 2. Amplification products were checked on agarose gel, purified using PureLink Quick Gel Extraction (Invitrogen, Carlsbad, CA, United States) and sequenced by Sanger sequencing (FEM Sequencing Platform Facility). Sequencing outputs were analyzed with the Blast sequence server (using the database PN40024.v4_REF_genome) available online at the European network INTEGRAPE website.6
Biological replicates of edited lines S-epfl9KO1 (n = 4) and S-epfl9KO2 (n = 4), and of ‘Sugraone’ WT (n = 4) kept in a greenhouse for 2 months were used. Pots were covered in aluminum foil and wrapped in plastic to limit soil evaporation (Supplementary Figure 2). All plants were measured daily for 14 days at the same time each morning for mass of water loss.
The same plants used in Experiment 1 were used in Experiment 2. Control pots (soil-filled pots without plants) were placed at the end of each row in randomized positions, weighed by balance and returned to the same positions every day to assess soil evaporation. Pots dried down naturally for a subsequent 15 days.
Biological replicates of the edited line S-epfl9KO6 (n = 6) and ‘Sugraone’ WT (n = 4), maintained in greenhouse for 12 months, with a height range of 60–70 cm and a weight brought to 3,000 g (in 5 l pots) were used. Plants were moved inside the phenotyping platform (WIWAM, Ghent, Belgium) at the Plant Phenotyping Facility of Fondazione Edmund Mach where temperature was set to 28/25°C, photoperiod to 16/8 h and average PPFD to 300 μmol m−2 s−1 at apical leaf level. Plants were automatically watered every day at 6:00 AM to target weight (3,000 g) and pot weight was evaluated before and after watering for 12 days.
In Experiment 1 and 2, total transpirable soil water (TTSW) was calculated as the difference between pot mass at day 1, fully watered (100% capacity), and the pot mass at the end of the natural dry down when transpiration reached a minimum. Fully watered plants (100% relative soil water content) were weighted after watering to capacity and allowing pots to drain for 2 h. The fraction of transpiration soil water (FTSW) was calculated as a daily ratio between the amount of soil water remaining in the pot left for transpiration and the TTSW using the equation: FTSW = (PMn – PMfinal)/TTSW, where PMn is the pot mass for each day, and PMfinal is the pot mass at the end of the day 11. FTSW data were reported in Supplementary Figure 3. At day 12 (i.e., after Experiment 1), plants were unwrapped from the aluminum and plastic coverings, re-watered to 100% of their initial weight using syringes and weighed as a starting mass for the stress application. In both Experiment 1 and 2, transpiration (g/cm2) was measured as the grams of water lost daily, normalized by the relative leaf area for each individual [T = (mass 0 - mass 1)/relative leaf area, where 0 and 1 represent the days in consecutive order]. Growth was measured as a relative leaf area every other day for a period of 28 days using RGB imaging. The software Easy Leaf Area (Easlon and Bloom, 2014) was used for analysis. Photos of the plants were taken at the same distance and tripod angle (45°) to provide uniform and consistent assessment of relative leaf area (example in Supplementary Figure 4A). A biomass-leaf area estimated curve was constructed using eight plants of varying sizes validating the non-destructive approach (Supplementary Figure 5). In Experiment 3, daily water-use was automatically calculated as daily pot weight loss (g). In addition, projected leaf area (pixels) was calculated at the beginning and at the end of the experiment (day 1 and day 12 respectively) as the average green pixels in four RGB images collected at different pot angles and analyzed with the WIWAM software (example in Supplementary Figure 4B).
Samples for stomatal characterization were taken under well-watered conditions as well as at the end of the drought treatment (i.e., Experiment 1 and 2). Leaves were chosen with the same size and position, typically leaf three, unless abnormal. Clear gel nail polish was applied to the abaxial and adaxial surfaces of the leaf to create an imprint of the leaf surface and allowed to dry. Clear tape was used to peel off the nail polish, and the tape was mounted on a microscope slide. Slides were imaged using a compound microscope (DM, Leica Microsystems, Wetzlar, Germany) at 40x and at five different technical positions of the same area (0.3 mm2) on the four biological replicates for a total of twenty measurements of stomata density per individual. Stomatal size (SS) was characterized from three technical replicates from three biological replicates for a total of 9 replicates per individual. These 9 replicates were averaged to create an average radius (r) for reach individual, and the stomatal size was subsequently calculated as ; stomatal size is equal to 0.5 multiplied by the average length of stomata squared multiplied by
For Experiment 1, 2 and 3, gas-exchange measurements were carried out using a portable infra-red gas analyzer and a 2 cm2 leaf cuvette with an integral blue–red LED light source (LiCOR 6,400-40XT, Lincoln, NE, United States). Inside the cuvette, flow rate was set at 400 μmol s− 1, leaf temperature at 24°C, PPFD to 1,500 μmol m−2 s−1 and Ca of 400 μmol mol−1. In Experiment 1, measurements of the response of photosynthesis (A) to sub-stomatal CO2 concentrations (Ci) curves (A/Ci) were performed between 9:00 and 12:00, on the most expanded leaf from each plant. For A/Ci, Ca was sequentially decreased to 300, 200, 150, 75 and 50 μmol mol−1 before returning to the initial concentration of 400 μmol mol−1. This was followed by a sequential increase to 500, 700, 900, 1,100, 1,300, and 1,500 μmol mol−1. Readings were recorded when A reached steady state. The maximum velocity of Rubisco for carboxylation (Vcmax) and the maximum rate of electron transport demand for Ribulose 1,5-bisphosphate (RuBP) regeneration (Jmax) were estimated as described by (Duursma, 2015; Easlon and Bloom, 2014). Asat represents CO2 assimilation rate at saturating PPFD while gs represents stomatal conductance at ambient CO2 (Ca). Intrinsic water-use efficiency ( iWUE) was calculated as = Asat / gs. During Experiment 2, measurements of A and gs were taken every day on fully expanded leaves for the first 3 days to record a baseline gas-exchange before water stress was applied. Subsequently gas-exchange data were recorded every 2 days in fully expanded leaves. In Experiment 3, gas-exchange parameters (A and gs), leaf temperature and leaf chlorophyll content were measured at day 5 on the same leaves, respectively with LiCOR 6,400-40XT (Lincoln, NE, United States), an infra-red thermometer (62 MAX+, FLUKE Corporation, Everett, Washington, United States) and a SPAD (Minolta SPAD 502).
Carbon isotope composition was estimated in leaves with the same leaf size and position, count as leaf three unless abnormal. Samples for stomatal characterization were taken first, and the remaining fresh leaf tissue was dried at 80°C for 2 days to be used for δ13C determination. δ13C was analyzed in 2 mg aliquots of leaf sample weighed in tin capsules. Samples were combusted in an elemental analyzer (Thermo Flash EA 1112 Series, Bremen, Germany), CO2 was separated by chromatography and directly injected into a continuous-flow isotope ratio mass spectrometer (Thermo Finnigan Delta V, Bremen, Germany) through the interface ConFlo IV dilutor device (Thermo Finningan, Bremen, Germany). Samples were measured in duplicate. The isotope ratios were expressed in δ‰ against Vienna-Pee Dee Belemnite for δ13C according to the following equation: ‰ = ( where RSA is the isotope ratio measured for the sample and RREF is the international standard isotope ratio. The isotopic values were calculated using a linear equation against working in-house standards, which were themselves calibrated against the international reference materials L-glutamic acid USGS 40 (US Geological Survey, Reston, VA, United States), fuel oil NBS-22 and IAEA-CH-6. The uncertainty of measurement (calculated as 2 standard deviations) was 0.1‰.
Statistical analyses were performed using R software (R Core Team, 2020). A one-way ANOVA was used to compare differences in cumulative transpiration, conductance, photosynthesis, and water use efficiency between edited and WT lines for each day of measurement. Post hoc comparisons using Fisher’s LSD test were carried out to assess group differences. p values lower than 0.05 were considered significant.
Two VvEPFL9 gene variants (hereinafter VvEPFL9-1 and VvEPFL9-2) were found in contigs of publicly available genomes of different Vitis vinifera varieties and of some other species within the same genus (Vitis sylvestris, Vitis arizonica, Vitis riparia; Supplementary Table 1). In the last annotation of the PN40024 grapevine reference genome (PN40024.v4.1,7 genome assembly version 12X.v4) VvEPFL9-1 (Vitvi05g01370) was localized on chromosome 5 (position 20,461,188–20,461,813) while VvEPFL9-2 (Vitvi07g04390) on chromosome 7 (position 17,537,397–17,536,742). Interestingly, before the new version of reference genome and related annotation was made publicly available (INTEGRAPE Workshop, 2021) in November 2021, only VvEPFL9-1 was localized on the genome while the position of VvEPFL9-2 was not assigned (VCost.v3 annotation). According to gene prediction, VvEPFL9-1/−2 coding sequence have a length of about 330/315 bp and are composed of three exons encoding for: an N-terminal region with a secretion signal for the apoplast [i.e., first 27 amino acid according to SignalP-5.0 software (Almagro Armenteros et al., 2019)8 a central region likely involved in the processing of the mature peptide and a C-terminal domain of 45 amino acids containing 6 conserved cysteines, that is the functional peptide. A check on genomic DNA extracted from a panel of genotypes (i.e., ‘Pinot Noir PN40024’, ‘Riparia Glorie de Montpellier’, ‘Pinot Noir clone Entav 115’, ‘Cabernet Sauvignon’, ‘Chardonnay’, ‘Merlot’, ‘Sugraone’, ‘Syrah’ and ‘Touriga National’), confirmed the presence of both gene variants in all the analysed samples with a very high conservation among genotypes (Supplementary Table 3). In all the genotypes no SNPs were detected between the two alleles of both isoforms in the region coding for the functional domain, except in Cabernet Sauvignon where an allelic polymorphism in position 25 was detected in VvEPFL9-1, which leads to two different amino acids after the first cysteine of the array (serine or threonine, both polar uncharged). Considering only the region encoding for the C-terminal domain (135 bp), the identity between the two variants was 74%, with a large part of polymorphism leading to synonymous codons (Figure 1A). At the protein level, the alignment of the C-terminal domains encoded by the two variants showed an identity of 82%, with 8 out of 45 different amino acids (Figure 1B). In five positions (14, 25, 28, 40, and 42) substitutions are conservative, i.e., the pair of amino acids belong to the same class, while in the remaining three positions (5, 18, and 34) the substitutions are non-conservative. A comparison with AtEPFL9 mature peptide revealed that the identity between VvEPF9-1 and AtEPFL9 is 82% while the identity between VvEPF9-2 and AtEPFL9 is 95% (Supplementary Figure 6). Moreover, the relationship of VvEPFL9-1/−2 with the orthologues of some di- and monocotyledonous plant species including some perennial fruit trees (retrieved from Ensembl Plants genomic database),9 is shown in Figure 1C.
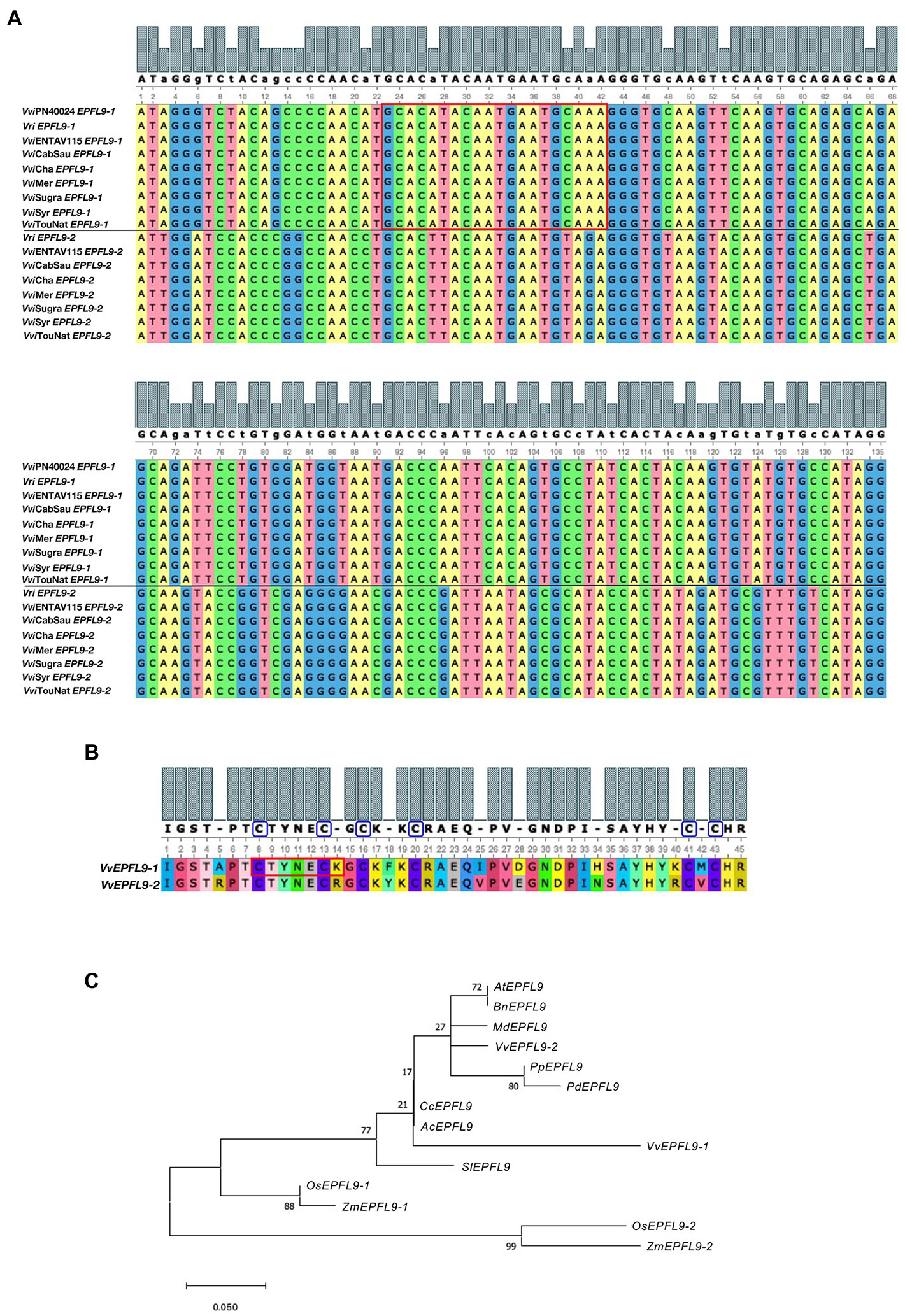
Figure 1. Analysis of VvEPFL9 paralogs. (A) Alignment of the nucleotide sequence encoding for the C-terminal domain (135 bp) obtained by Sanger sequencing of PCR fragments amplified on genomic DNA with primers VvEPFL9-1_fw; VvEPFL9-1_rv and VvEPFL9-2_fw; VvEPFL9-2_rv (see primer list in Supplementary Table 2). Genomic DNA was extracted from leaves of ‘Pinot Noir PN40024’, Vitis riparia ‘Riparia Glorie de Montpellier’, ‘Pinot Noir clone Entav 115’, ‘Cabernet Sauvignon’, ‘Chardonnay’, ‘Merlot’, ‘Sugraone’, ‘Syrah’, ‘Touriga National’. The red rectangle indicates the 20 bp-target site recognized by the sgRNA/Cas9 complex. (B) Alignment of the C-terminal protein domain of VvEPFL9-1 and VvEPFL9-2, translated from the 135 bp nucleotide sequences shown in (A). Cysteine residues are circled in blue. The red rectangle indicates the peptide region corresponding to the target site. (C) Phylogenetic tree of the Arabidopsis AtEPF9 mature peptide and its orthologs from some dicotyledonous (Brassica napus, Malus × domestica, Vitis vinifera, Prunus persica, Prunus domestica, Prunus dulcis, Citrus clementina, Actinidia chinensis, Solanum lycopersicum) and monocotyledonous (Orytia sativa, Zea mays) plant species. The alignments were generated with MUSCLE (MEGA X) and visualized with Unipro UGENE [http://ugene.net/faq.html (Okonechnikov et al., 2012)]. The phylogenetic tree was built with MEGA X using Maximum Likelihood (1,000 replicates bootstrap). Accession Numbers: VvEPFL9-1 (Vitis vinifera; Vitvi05g01370); VvEPFL9-2 (Vitis vinifera; contig VV78X057312.8. BioProject PRJEA18357); AtEPFL9 (Arabidopsis thaliana; AT4G12970); BnEPFL9 (Brassica napus; BnaA08g04900D-1); OsEPFL9-1 (Oryza sativa; BGIOSGA005039-TA); OsEPFL9-2 (Oryza sativa; BGIOSGA026626-TA); ZmEPFL9-2 (Zea mays; Zm00001d049795_T001); ZmEPFL9-1 (Zea mays; Zm00001d012079_T001); SlEPFL9 (Solanum lycopersicum; Solyc08g066610.3.1); MdEPFL9 (Malus domestica; mRNA:MD10G0128800); CcEPFL9 (Citrus clementina; ESR50459); PpEPFL9 (Prunus persica; ONH92727); PdEPFL9 (Prunus dulcis; VVA33635); AcEPFL9 (Actinidia chinensis; PSR86312).
A highly transformable genotype of Vitis vinifera, ‘Sugraone’ was used for gene transfer of the CRISPR/Cas9 machinery in order to obtain edited plants knocked-out for the VvEPF9-1 gene. The sgRNA was designed to target a region of 20 nucleotides in the third exon, spanning across “TGC” triplets coding for the first and the second cysteine of the functional C-terminal domain (Figures 1A,B; Supplementary Table 4). In particular, the cleavage operated by Cas9 was expected to affect the “TGC” triplet coding for the second cysteine, this being located 3 nucleotides upstream of the PAM site (i.e., GGG; Figure 1A). The corresponding region of VvEPF9-2 has 3 mismatches compared with the target site on VvEPF9-1, in positions 6, 18 and 20, the last two in the seed region close to the PAM site (Figure 1A). Several shoots were regenerated from somatic embryos after 7–10 months from Agrobacterium tumefaciens co-culture (Figure 2), and nine of them were selected for molecular characterization. The Cas9 integration copy number varied in the transgenic lines, ranging from 1 integration copy for line S-epfl9KO7 to 5 integration copies for line S-epfl9KO1, with the majority of lines showing values close to one or two copies (Figure 3A). A T-DNA integration site was identified for 5 lines: S-epfl9KO1 (chr18: position 2,096,753), S-epfl9KO2 (chr01: position 4,310,437), S-epfl9KO3 (Chr13: position 5,599,304), S-epfl9KO6 (chr04: position 6,948,780), S-epfl9KO7 (chr03: position 405,924). Concerning T-DNA rearrangements, all the lines showed a trimming of several bases at the LB border, ranging from 31 bp of S-epfl9KO7 to 110 bp of line S-epfl9KO3, and a T-DNA tandem repeat was detected in line S-epfl9KO1. The analysis of the genomic “on-target” site in VvEPF9-1 proved that all lines were edited, some completely while others showed a degree of wild-type target sequence, indicated as WT (Figure 3B; Supplementary Table 4). In general, the editing profile was highly heterogeneous, with a composite mutation profile for many lines (e.g., S-epfl9KO2, S-epfl9KO5, S-epfl9KO6, S-epfl9KO7, S-epfl9KO9), including deletions of increasing size (from 1 bp to more than 7 bp), insertions of 1 or 2 bp, and single base substitutions. The most frequent kind of mutations were deletions of 4 or 5 bp (Figure 3B). The resulting mutations in the protein sequence were frameshift mutations (FS) with or without the formation of premature stop codons (SC), or non-frameshift mutations with loss of the second cysteine due to deletion of 3 or 6 bp or to a single base substitution (Figure 3C). The analysis of stomatal density in leaves of greenhouse-cultivated plants (2 months old) showed a significant reduction in stomata number in transgenic lines compared to WT (Figure 3D). This reduction was significant even for the lines maintaining a remarkable rate of non-mutated VvEPF9-1 (i.e., S-epfl9KO1, S-epfl9KO5, S-epfl9KO7) and for lines that went through the loss of the second cysteine of the 6-Cys-array, highlighting the crucial role of such residue (i.e., S-epfl9KO5 and S-epfl9KO8). The editing in the potential “off-target” site in VvEPFL9-2 was assessed and no mutations were found in all the transgenic lines (Supplementary Figure 7). This proved that 3 mismatches with respect to the sgRNA, 2 of which close to the PAM site, were enough to avoid Cas9 unspecific cleavage at this site. In vitro and greenhouse edited plants did not show phenotypic defects due to pleiotropic effects (e.g., rate of growth, total leaf area, chlorophyll content) compared to the control plants (data not shown).
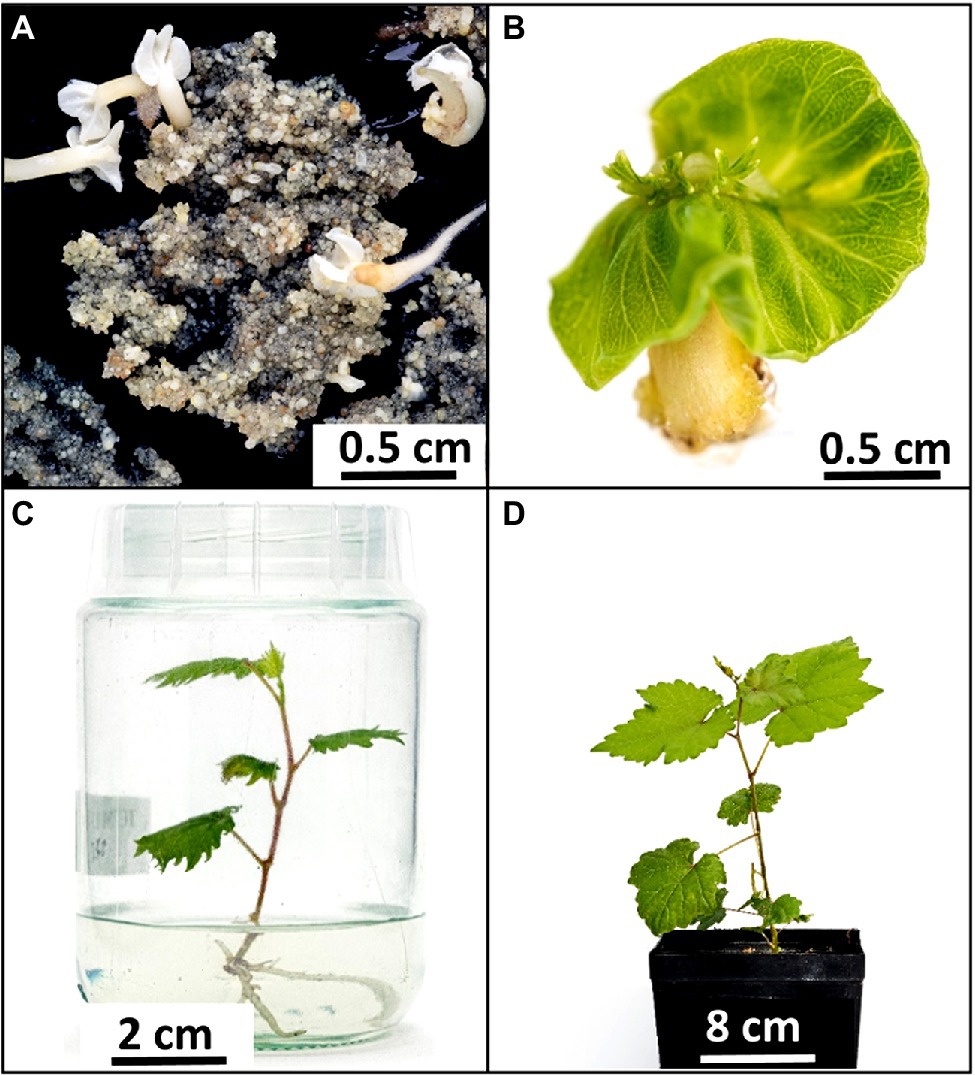
Figure 2. Pipeline to obtain epfl9-1 mutants for physiological characterization. (A) Embryogenic callus of ‘Sugraone’ 7 months after co-cultivation with Agrobacterium tumefaciens. Some embryos are developing on a homogeneous callus mainly formed by small globular embryos. (B) Embryo producing shoot. (C) In-vitro plantlet cultivated in baby jar. (D) Greenhouse plant after 2 months from acclimatization of an in-vitro plantlet.
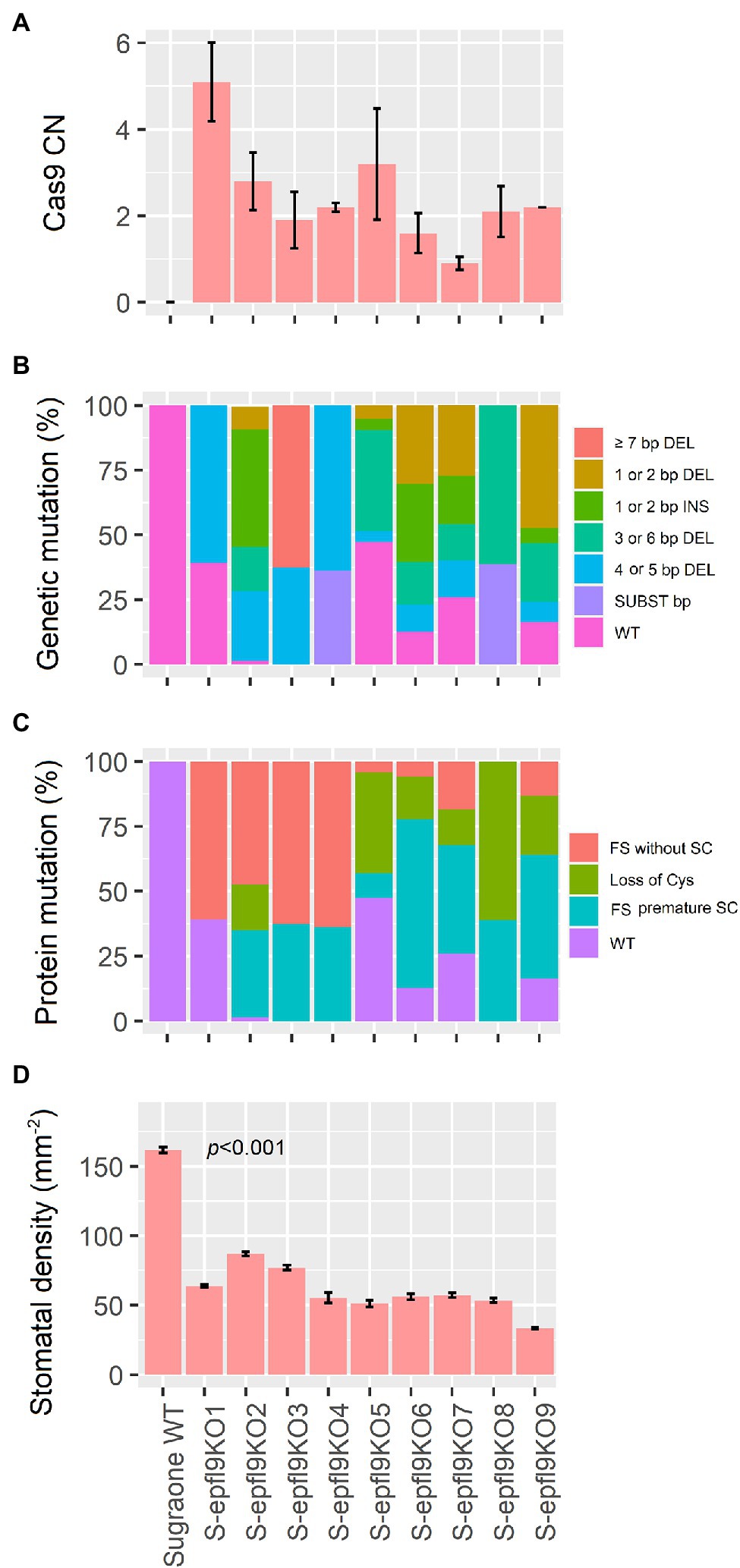
Figure 3. Characterization of 9 ‘Sugraone’ transgenic lines. (A) Quantification of SpCas9 copy numbers (CN) integrated in the plant genome. CN were calculated by Real-time PCR as the mean value of two measurements obtained for two in-vitro biological replicates (except for line S-epfl9KO9 for which only one value is available). (B) Bar plot indicating the mutation profile in the genomic target site on exon 3 of VvEPF9-1 after CRISPR/Cas9 editing. The mutation pattern and rate (%) of a specific mutation (IN/DEL, insertion/deletion and SUB, substitution) were determined by the number of reads calculated by Illumina sequencing (see Supplementary Table 4). Different kinds of mutations are indicated with a different color. WT = the wild-type sequence. (C) Bar plot indicating the resulting mutation profile in the functional mature VvEPF9-1 peptide, predicted according to the nucleotide mutations in B (see Supplementary Table 4). The different outcomes at protein level are indicated with a different FIGURE 3color. FS, frameshift mutations; SC, stop codons. (D) Measurements of stomatal density in the third leaf from the apex. For each plant, four leaves from four biological replicates were analyzed, each in five different technical positions of the same area for a total of twenty measurements.
Analysis of stomatal anatomical features confirmed the significant differences for stomatal density and pore length between the selected S-epfl9KO1 and S-epfl9KO2 knock-out mutants and WT (Figure 4). S-epfl9KO1 had an average SD of 65 stomata mm−2 while SD for S-epfl9KO2 was 95 stomata mm−2, both significantly lower values than that of ‘Sugraone’ WT (160 stomata mm−2) respectively by 60 and 40%. Conversely, pore length was significantly higher in S-epfl9KO1 and S-epfl9KO2 than ‘Sugraone’ WT, by up to 30%.
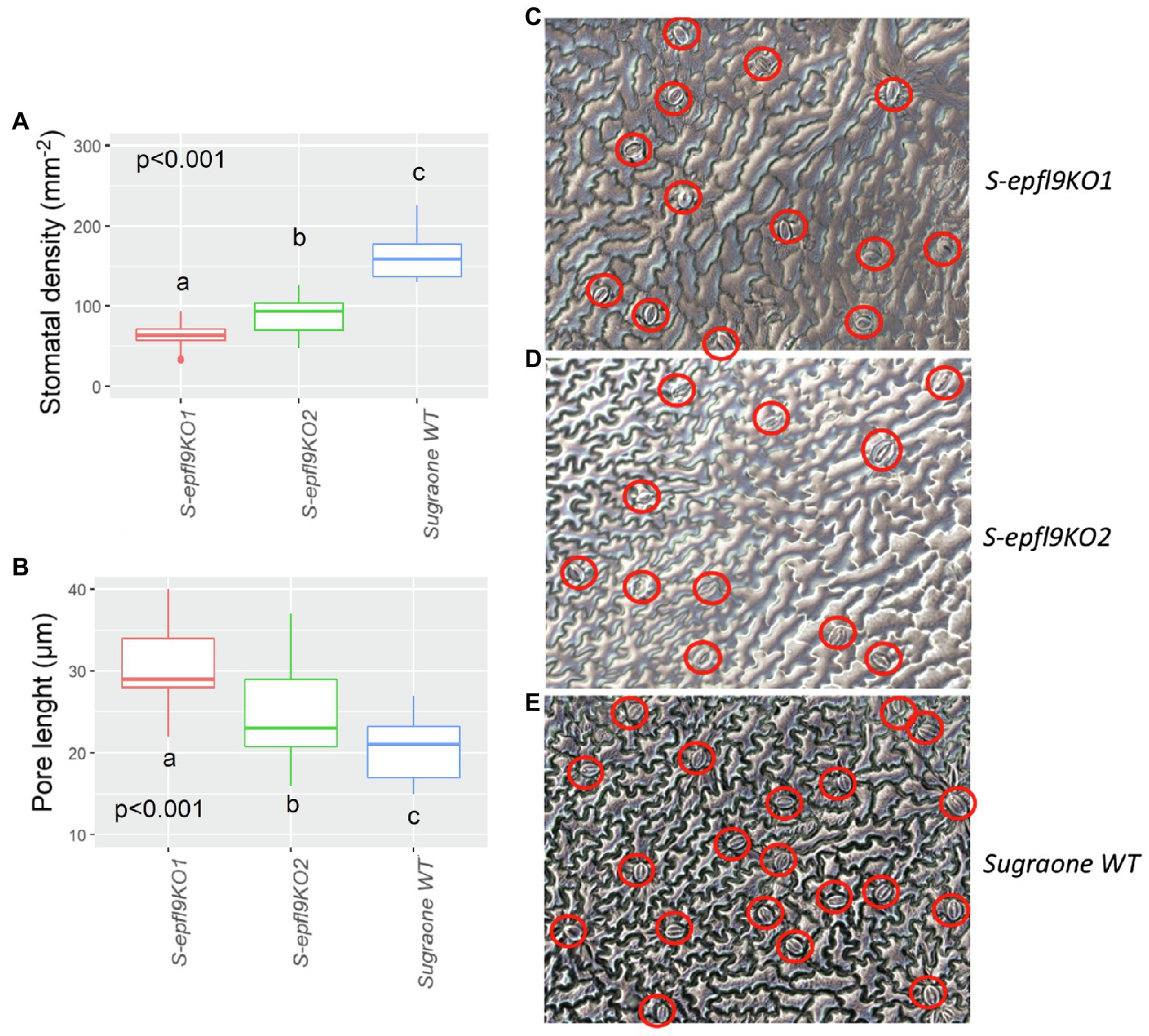
Figure 4. Characterization of stomata in selected epfl9-1 knock-out mutants. (A) Stomatal density for S-epfl9KO1, S-epfl9KO2 and Sugraone WT. (B) Pore length for S-epfl9KO1, S-epfl9KO2 and Sugraone WT. Whiskers indicate the ranges of the minimum and maximum values. Data were analysed with one-way ANOVA (n = 6–9). Different letters indicate significantly different values according to Fisher’s test. (C–E) Images of nail polish printing of leaf tissue, respectively, from S-epfl9KO1, S-epfl9KO2 and Sugraone WT.
A/Ci response curves (net CO2 assimilation rate, A, versus calculated substomatal CO2 concentration, Ci) were carried out under optimal environmental conditions and saturating light intensity assessed via light curves for selected edited lines S-epfl9KO1 and S-epfl9KO2 in Experiment 1 (Supplementary Figure 8). There were no significant differences for maximum rate of Rubisco-mediated carboxylation (Vcmax) between edited lines and WT control (p > 0.05, Figure 5A). Similarly, maximum electron transport rate for RuBP regeneration (Jmax) did not vary between edited lines and WT control (p > 0.05, Figure 5B). On the contrary, significant reductions in CO2 assimilation rate at saturating light (Asat) were detected for S-epfl9KO1 and, in particular, S-epfl9KO2 when compared to WT and up to 50% (p = 0.007, Figure 5C). S-epfl9KO1 and S-epfl9KO2 had significantly lower conductance (gs) than WT (p < 0.001) with S-epfl9KO2 showing the lowest values (0.030 mol m−2 s−1 on average, Figure 5D). This led to a significantly higher intrinsic water-use efficiency ( iWUE) for S-epfl9KO2 than ‘Sugraone’ WT (p = 0.024, Figure 5E). Accordingly, carbon isotope composition (δ13C) analysis detected for S-epfl9KO2 significant less negative δ13C values compared to ‘Sugraone’ WT (p = 0.046), indicating a higher iWUE (Figure 5F). Gravimetric assessments of transpired water normalized for leaf area highlighted significant differences in cumulative transpiration between edited and WT lines. In general, both S-epfl9KO1 and S-epfl9KO2 used less water throughout a 14 day experimental period, by up to 21%, compared to ‘Sugraone’ WT (Figure 5G). Moreover, to expand our data in well-watered conditions we evaluated the gas-exchange and transpiration performances of an additional line, S-epfl9KO6, maintained in greenhouse for 12 months (Experiment 3). S-epfl9KO6 showed similar SPAD values compared to ‘Sugraone’ WT (p = 0.607, Figure 6A) and trends were observed for leaf temperature with S-epfl9KO6 showing increased leaf temperature (p = 0.051, Figure 6B) compared to WT. This increase in leaf temperature was associated with a significant decrease in stomatal conductance (p = 0.042, Figure 6D) together with a non-significant difference for Asat (p = 0.125, Figure 6C). This led to a significant increase in iWUE for S-epfl9KO6 compared to control (p = 0.034, Figure 6E). No significant differences were observed for projected leaf area (PLA; Figures 6F,G) and water use (WU; Figure 6H) between S-epfl9KO6 and ‘Sugraone’ WT although a trend was present for WU (p = 0.088).
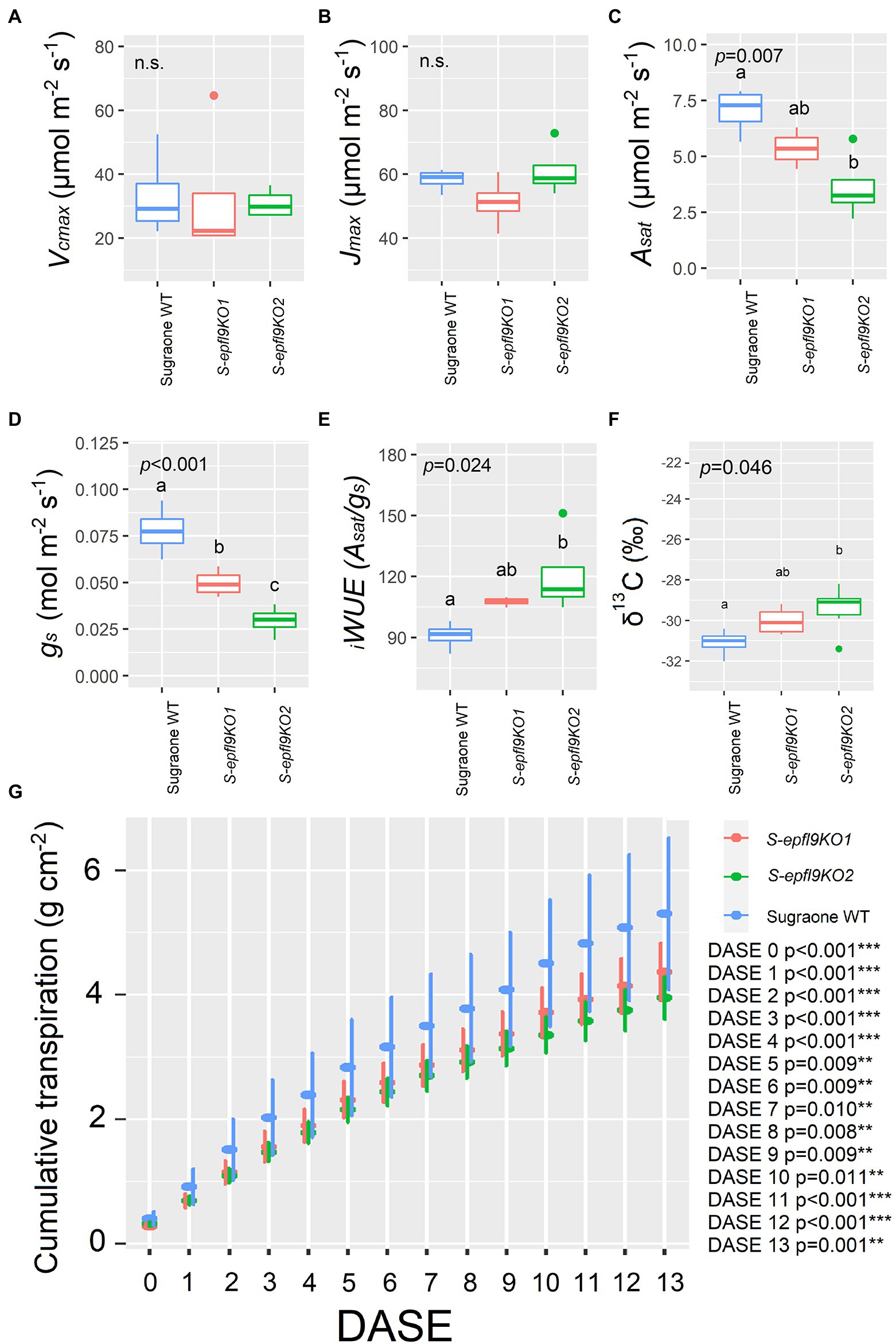
Figure 5. Trait assessment under well-watered (WW) conditions (Experiment 1). (A) Maximum velocity of Rubisco carboxylation (Vcmax). (B) Maximum electron transport rate for RuBP regeneration (Jmax) estimated with A/Ci curves and following curve fitting (Duursma, 2015; Easlon and Bloom, 2014). (C) CO2 assimilation rate at saturating light (Asat). (D) Stomatal conductance (gs) extrapolated from A/Ci curves at 400 ppm CO2 concentration and 1,500 μmol m−2 s−1. (E) Intrinsic water-use efficiency ( iWUE) calculated as iWUE = Asat/gs. (F) Carbon Isotope composition (δ13C) analysis. Data were collected on fully expanded leaves of 20 cm tall plants on the twelfth day from the start of the experiment and were elaborated with one-way ANOVA (n = 4 in A–E; n = 3–6 in F). Whiskers indicate the ranges of the minimum and maximum values and different letters indicate significantly different values according to Fisher’s test. (G) Cumulative water loss assessed gravimetrically and normalized for leaf area estimated via RGB imaging for a period of 14 days; DASE = Days After Start of the Experiment. Data were means ± standard error of the mean (n = 5–6). Data were elaborated with one-way ANOVA for each day (***p < 0.001, **p < 0.01, *p < 0.05). When present, different letters indicate significantly different values according to Fisher’s test.
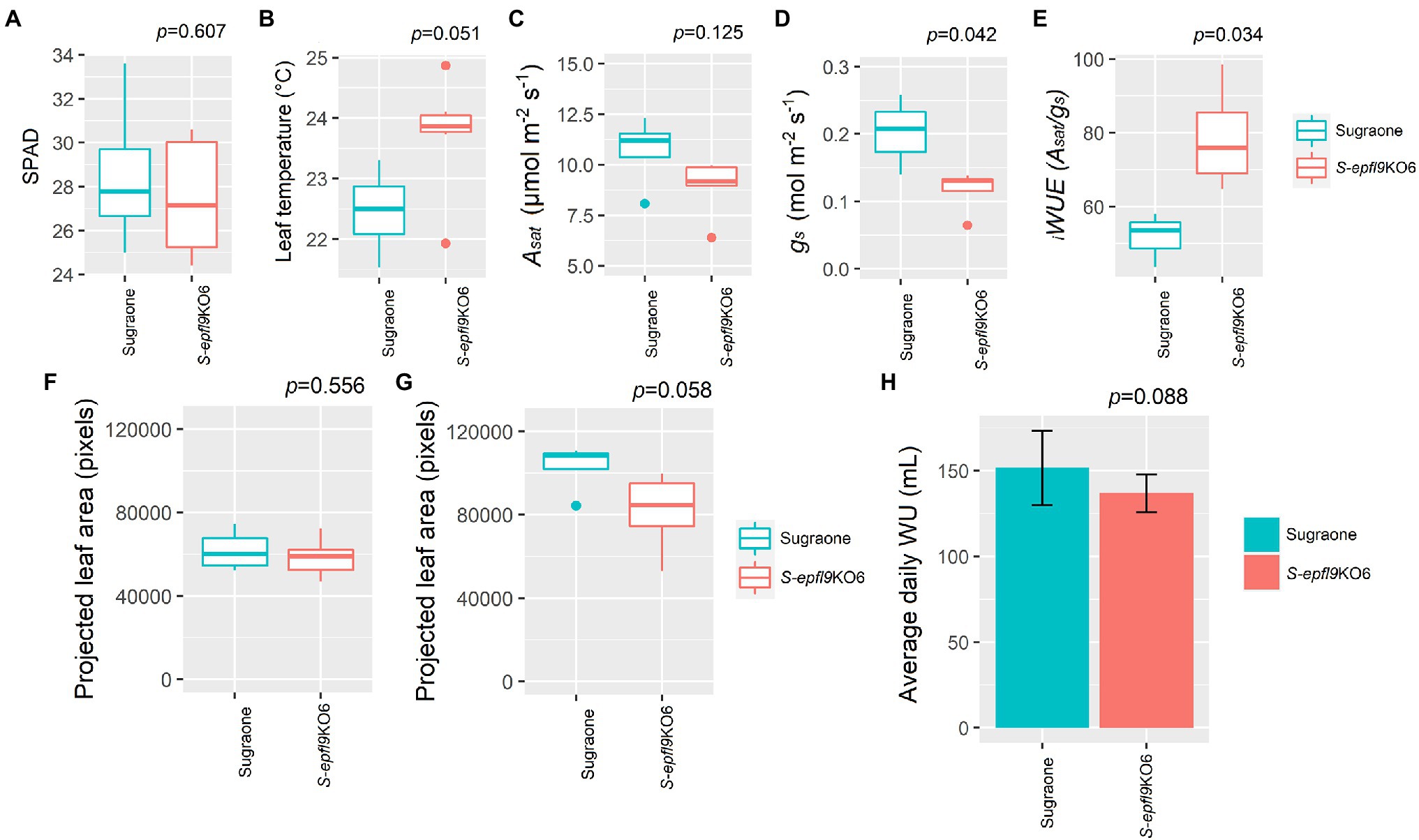
Figure 6. Dynamics of gas exchange, projected leaf area and water-use under well-watered (WW) conditions (Experiment 3) for S-epfl9KO6 (n = 6) and ‘Sugraone’ WT (n = 4). (A) SPAD values. (B) leaf temperature. (C) CO2 assimilation rate at saturating light (Asat). (D) stomatal conductance (gs). (E) Intrinsic water-use efficiency ( iWUE) calculated as iWUE = Asat/gs. (F,G) projected leaf area (PLA, pixels) collected at day 1 and at day 12, respectively, and (H) average daily water-use. For gas exchange measurements, data were collected on fully expanded leaves and were analysed with one-way ANOVA. Whiskers indicate the ranges of the minimum and maximum values.
In vivo gas-exchange measurements at saturating light were carried out throughout the dry down Experiment 2 (Figure 7). ANOVA output for each DASA (Day After Stress Application) is shown in Supplementary Table 5. In vivo CO2 assimilation rate (A) was significantly reduced by water stress (WS) in ‘Sugraone’ WT showing a steeper reduction than knock-out lines, although no significant differences were observed for each day and between lines (Figure 7A). S-epfl9KO1 and S-epfl9KO2 maintained a lower stomatal conductance (gs) than ‘Sugraone’ WT (p = 0.0276, DASA 5, Figure 7B) but intrinsic water-use efficiency iWUE resulted not significantly different between the analysed plants (Figure 7C). Transpiration normalized on leaf area was significantly reduced during the WS and for all the lines (Figure 7D). The average fraction of transpirable soil water (FTSW) during the dry down is shown in Supplementary Figure 3. There were significant differences (p < 0.05) between S-epfl9KO1 and ‘Sugraone’ WT, in particular in the first part of stress application (DASA 1 to 4). Trends (p < 0.1) were observed under severe WS (DASA 10 to 12) with S-epfl9KO2 having higher transpiration than ‘Sugraone’ WT. Carbon isotope composition (δ13C) analysis showed that water stress led to less negative values for all the lines (p < 0.001) although no significant differences were observed between edited lines and WT (p = 0.186; Supplementary Figure 9).
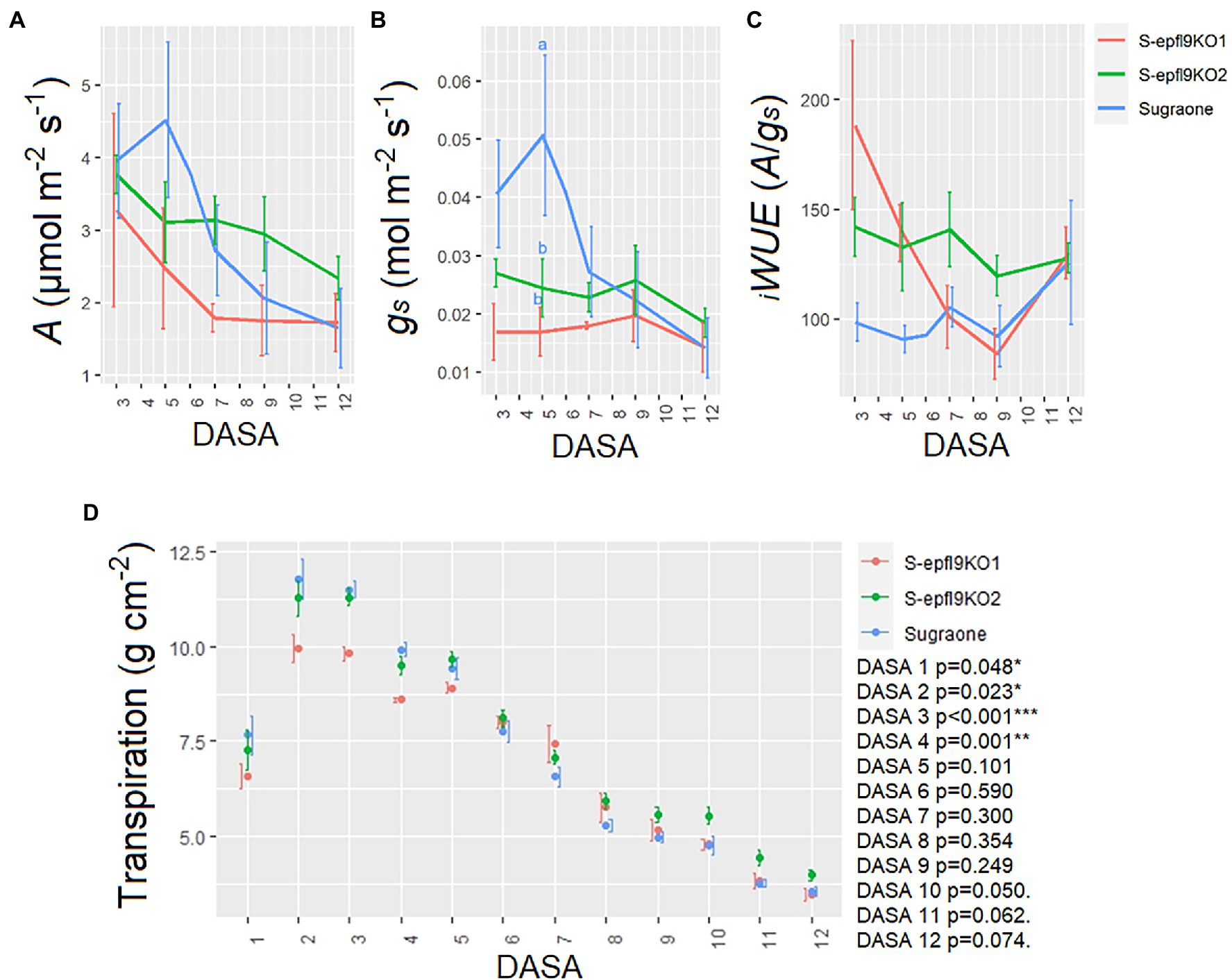
Figure 7. Trait assessment under water stress (WS) conditions. (A) In vivo CO2 assimilation rate at saturating light (Asat). (B) Stomatal conductance (gs). (C) Intrinsic water-use efficiency iWUE calculated as iWUE = Asat/gs. Data are the means ± standard error of the mean (n = 4–6). Data were analysed with one-way ANOVA (value of p in the Supplementary Table 5) while different letters indicate significant differences between lines according to Fisher’s test. (D) Transpiration assessed gravimetrically and normalized for leaf area estimated via RGB imaging. Data are means ± standard error of the mean (n = 5–6). Data were analysed with one-way ANOVA (***p < 0.001, **p < 0.01, *p < 0.05, p < 0.1) for each day. DASA, Days After Stress Application.
Crops worldwide will experience warmer conditions in the next decades, followed by limited water availability and increasing atmospheric CO2 concentration (McGranahan and Poling, 2018). Alteration of stomatal density and stomatal size through the genetic manipulation of epidermal patterning factors has been shown to be an effective approach to increase drought tolerance and reduce water loss in several species (Bertolino et al., 2019; Buckley et al., 2020). There is a lot of knowledge about EPF gene family in Arabidopsis and in domesticated grasses but in perennial crops, which present genetic and physiological differences compared to annual species due to ecological and agronomic peculiar features (Lundgren and Marais, 2020), no evidence has been collected on their role. The aim of our study was to shed light for the first time on the genetic basis of stomatal density traits in grapevine, a perennial woody fruit plant with a longer lifespan than previously studied crops (i.e., longer than 30 years).
Water conservation, higher iWUE and enhanced tolerance to multiple stresses (e.g., drought stress combined with heat stress) were achieved in Arabidopsis and grasses overexpressing EPF1/EPF2 or down-regulating EPFL9, due to a reduction in stomatal density (Franks et al., 2015; Hughes et al., 2017; Caine et al., 2019; Dunn et al., 2019; Lu et al., 2019). Between these two reverse genetics approaches, we have chosen the second, relying on the knock-out of VvEPFL9 by the powerful CRISPR/Cas9 gene editing technology.
In the grapevine genus we found two AtEPFL9 orthologs, we named VvEPFL9-1 and VvEPFL9-2, identical at 82% in the protein region corresponding to the functional peptide and, respectively, sharing 82 and 95% identity with the same region of AtEPFL9 peptide. So far, two EPFL9 paralogs have been found in maize and rice (Yin et al., 2017; Hepworth et al., 2018; Lu et al., 2019), showing, respectively, 84 and 73% (ZmEPFL9-1 and ZmEPFL9-2) and 82 and 73% (OsEPFL9-1 and OsEPFL9-2) identities to AtEPFL9 functional peptide. It has been suggested that EPFL9 paralogs in cereals might be functionally divergent (Lu et al., 2019) but definitive evidence indicating a different function has never been produced. In the study of Lu et al. (2019), the approach used to silence OsEPF9-1 was RNA interference with a 450 bp-long hairpin RNA, which hardly discriminated between the two variants. In our study, we decided to focus on VvEPFL9-1, since at the time the experiment was designed, VvEPFL9-2 was not anchored to any chromosome in the grapevine reference genome and this uncertainty oriented our choice on VvEPFL9-1. According to our data, the knock-out of VvEPFL9-1 can reduce stomatal density by up to 60%, leading to the hypothesis that VvEPFL9-1 and VvEPFL9-2 could be both involved in stomatal induction with a redundant function. A similar approach based on CRISPR/Cas9 technology to knock-out EPFL9 in rice achieved nearly 90% of stomatal density reduction compared to control by targeting a site on the first exon encoding for the signal peptide and thus not discriminating between OsEPFL9 paralogs (Yin et al., 2017).
Our study also confirms the crucial role of cysteine residues in the C-terminal functional peptide. This is demonstrated by the lines S-epfl9KO5 and S-epfl9KO8 in which the loss of the second cysteine (due to a 3 bp-deletion or single base substitution) resulted in a stomatal density reduction similar to the one gained by a full frameshift of the coding sequence. This is consistent with the finding of Ohki et al. (2011) who observed that impairing the formation of a disulphide bond prevented the correct protein folding and function. The design of a sgRNA that directed Cas9 cleavage next to the nucleotide triplet coding for the second cysteine proved to be a good choice for effective 3- and 6- bp deletions. Moreover, our data showed that the retention of almost 50% functional VvEPFL9-1 in some transgenic lines (S-epfl9KO1 and S-epfl9KO5) due to a partial editing of the target site, with a substantial maintenance of a WT peptide, still resulted in a significant decrease of SD, suggesting that a threshold amount of peptide may be required for EPFL9-1 to be functionally effective.
Reduction in stomatal density following VvEPFL9-1 knock-out was significant, although partially compensated by an increase in stomatal size (SS, inferred by pore length measurements). The negative yet non-linear association between SD and SS has been frequently reported in many species (Franks and Beerling, 2009) and often linked to an improved economy of epidermal space allocation with the combination of low SD and high SS as a preferable strategy when low stomatal conductance is required (Doheny-Adams et al., 2012; Lawson and McElwain, 2016). In our work, however, the reduction in SD was accompanied by only a partial compensation for SS.
Stomata are the main drivers of transpiration but at the same time are pivotal for CO2 uptake for mesophyll photosynthesis (Lawson and Blatt, 2014). For instance, in barley and wheat, a reduction in SD by 50% compared to WT led to a significant reduction in carbon assimilation (Asat) and conductance (gs) and to an enhanced water use efficiency ( iWUE) under optimal growth conditions (Hughes et al., 2017; Dunn et al., 2019). Similarly, in two-months-old ‘Sugraone’ at well-watered conditions (Experiment 1), we found that a 60% reduction in SD led to a reduced Asat for the edited lines compared to the WT. Additionally, the reduction in gs was even greater, leading to a higher value of iWUE (i.e., Asat/ gs) in edited versus WT lines. Moreover, the reduction in Asat was not concomitant to reductions in Rubisco velocity (Vcmax) or to impairment in electron transport chain (Jmax) suggesting that the knock-out of VvEPFL9-1 did not affect the photosynthetic machinery, at least at the conditions applied in this work. In an additional experiment (Experiment 3) carried out under well-watered conditions in a phenotyping platform on older plants than those used in experiment 1, iWUE confirmed to be significantly improved in the edited line (S-epfl9KO6) compared to ‘Sugraone’ WT while transpiration (WU) performances were not significantly different. The main sources of variation between the two experiments were plant age and environmental conditions. In Experiment 3 plants were older than in Experiment 1 (12- vs. 2-Months-old) and regarding light conditions, Experiment 1 was carried out under the natural fluctuating light of a greenhouse, while in Experiment 3 plants were subject to a steady-state light pattern. Our results suggest that canopy structure (over-saturation of apical leaves and basal leaves under the sub-saturating light intensities of the greenhouse) may play a role in defining the effectiveness of a reduced stomatal density phenotype. Furthermore, the conditions of dynamic light intensity such as those present in the greenhouse, may have contributed to accentuate the water saving behavior of the lines with lower stomatal density. Indeed, reducing stomatal density can limit stomatal clustering (Harrison et al., 2020) and therefore increase stomatal responsiveness to environmental cues (Faralli et al., 2021). Important differences for gs were also observed between Experiments 1 and 3, suggesting that plant age and pot-effect significantly influences operating gs, although the gs values are inside the ranges shown by Lavoie-Lamoureux et al. (2017) for pot-grown grapevine. Vitis vinifera genotypes with reduced SD and, in turn, limited Asat and greater iWUE, may be desirable to improve plant water conservation and to delay sugar accumulation under current and future climatic scenarios (Kuhn et al., 2014; Arrizabalaga-Arriazu et al., 2021). Sugars and organic acids along with various secondary metabolites (e.g., tannins, flavonols, anthocyanins, aroma compounds) are determinants of grape berry quality and their accumulation during berry ripening is the result of the interaction between genotype and environment, a relationship made vulnerable by climate change (Bobeica et al., 2015; Rienth et al., 2021). It is known that grapevine physiology will be impacted by elevated carbon dioxide, increasing temperatures, and extreme heat events during the growing season (De Cortázar-Atauri et al., 2017; Delrot et al., 2020). In particular, high temperature and increasing CO2 levels are already affecting viticulture (Cook and Wolkovich, 2016; Mosedale et al., 2016; Edwards et al., 2017; Droulia and Charalampopoulos, 2021) with an evident shift towards an earlier onset of phenological stages (Edwards et al., 2017; Alikadic et al., 2019) and accelerated berry ripening (Jones et al., 2005; Parker et al., 2020; Rienth et al., 2021). High temperatures and water stress slow down vine metabolism resulting in a lower accumulation of polyphenols and aromatic compounds in the berries (Tomasi et al., 2011; Jones, 2013; Pons et al., 2017; Venios et al., 2020). Thus, one of the consequences of a compressed phenology may be an earlier sugar accumulation in the berries that leads to anticipated harvest dates when the secondary metabolites content is sub-optimal (Palliotti et al., 2014; Edwards et al., 2017). Although currently several agronomic approaches of source-limitation (i.e., pre-flowering leaf removal, shading nets, anti-transpirant application, etc.) have been set up to delay sugar accumulation in ripening grapes in the field (Palliotti et al., 2014; Prats-Llinàs et al., 2020), stomatal manipulation may be a favorable genetic strategy for the future, that deserves to be further explored also under combined environmental stress and in field trials. In our study, we further applied a water stress experiment to test if and how a reduced stomatal density can affect plant behavior in drought conditions. During a progressive reduction in soil water availability, significant differences in transpiration rate were observed in edited lines compared to WT only under moderate water stress (i.e., DASA 3 and 4). Yet, under severe water stress (e.g., DASA 10–12), some trends (p < 0.1) were observed in edited lines showing higher transpiration rate followed by Asat and gs maintenance. Notably, the reduction in gs and Asat during the dry-down was evident for WT plants (p < 0.001) while this was not significant for edited lines. This conservative behavior induced by reduced SD has been previously associated with a longer period of transpiration maintenance during drought, leading to a prolonged carbon assimilation respect to WT (Caine et al., 2019). In rice, lines overexpressing the OsEPF1 gene had higher yield than WT when water-stressed at flowering stage (Caine et al., 2019) confirming that water conservation during key-stages of yield formation may be desirable for yield maintenance (Faralli et al., 2019). In addition, limiting plant transpiration could be an advantage for irrigated vineyards in terms of a reduction in water input demand (Keller et al., 2016). In view of an increase in the number of grapevine growing regions where water resources will become limited (Schultz, 2000; Santillán et al., 2020), genotypes with reduced stomatal density will require less units of irrigation water for cultivation area, thus increasing crop water productivity for farmers (Scholasch and Rienth, 2019).
To our knowledge, this is the first study describing the function of VvEPFL9-1 in a perennial fruit crop as well as the physiological advantages of epfl9-1 knocked-out phenotype under different availability of soil water. In grapevine, reducing stomatal density via VvEPFL9-1 loss of function can induce water conservation and increase iWUE, although an impact of photosynthetic CO2 absorbance (Asat) was observed in some edited lines. While in several crops, reduced photosynthetic CO2 uptake can decrease yield and biomass, we speculate that reduced Asat and increased iWUE may be a favorable combination of physiological attributes in grapevine, especially under future climate change scenario. However, at this stage further trials in the field under standard management conditions are required as well as additional evaluations regarding the potential effects of reduced stomatal density under natural environmental fluctuations. To conclude, this work reinforces the concept that stomatal anatomical features constitute a promising target for designing climate change-resilient crops (Franks et al., 2015; Hughes et al., 2017; Bertolino et al., 2019; Caine et al., 2019; Dunn et al., 2019; Lu et al., 2019; Buckley et al., 2020) and provides evidence of this in grapevine, the most economically important fruit crop globally.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. The original contributions presented in the study are publicly available. This data can be found here: NCBI Sequence Read Archive, BioProject accession number: PRJNA820619.
MC performed plant transformation experiments, plant molecular analysis, phenotyping, statistical analysis, and wrote the paper. MF contributed to the dry-down experiment design, carried out Experiment 3, supervised physiological analysis and statistical elaboration of the data, and wrote the paper. JL carried out alignments and phylogenetic tree and revised the manuscript. LB performed carbon isotope composition analysis and revised the manuscript. SP performed the analysis for the T-DNA integration point determination. CV, MM, WO, and AR supervised and revised the manuscript. LDC conceived the project, designed vectors for gene editing, performed paralogs analysis, transformation experiments, plant molecular characterization, took care of the plants in greenhouse, and wrote the paper. All authors contributed to the article and approved the submitted version.
This research was funded by the Autonomous Province of Trento (Italy) in the framework of the Fondazione Edmund Mach (FEM) International PhD initiative and by the ERDF 2014–2020 Program of the Autonomous Province of Trento with EU co-financing (Fruitomics, CUP number: C49H18000000001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Valentino Poletti for his technical help with culture media preparation and Lisa Giacomelli for sharing with us embryogenic calli of ‘Sugraone’. We would like to thank Damiano Gianelle for lending the Li-Cor 6400. A special thanks to Claudio Moser for useful suggestions and discussion and to Nicola Busatto for taking photos for Figure 2.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.878001/full#supplementary-material
2. ^https://www.dna-cloning.com/
3. ^http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR
4. ^https://crispresso.pinellolab.partners.org/submission
6. ^https://integrape.eu/resources/genes-genomes/genome-accessions/
7. ^https://integrape.eu/resources/genes-genomes/genome-accessions/
Alikadic, A., Pertot, I., Eccel, E., Dolci, C., Zarbo, C., Caffarra, A., et al. (2019). The impact of climate change on grapevine phenology and the influence of altitude: A regional study. Agric. For. Meteorol. 271, 73–82. doi: 10.1016/j.agrformet.2019.02.030
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi: 10.1038/s41586-019-1711-4
Arrizabalaga-Arriazu, M., Morales, F., Irigoyen, J. J., Hilbert, G., and Pascual, I. (2021). Growth and physiology of four Vitis vinifera L. cv. Tempranillo clones under future warming and water deficit regimes. Aust. J. Grape Wine Res. 27, 295–307. doi: 10.1111/ajgw.12494
Bertolino, L. T., Caine, R. S., and Gray, J. E. (2019). Impact of stomatal density and morphology on water-use efficiency in a changing world. Front. Plant Sci. 10:225. doi: 10.3389/fpls.2019.00225
Bobeica, N., Poni, S., Hilbert, G., Renaud, C., Gomès, E., Delrot, S., et al. (2015). Differential responses of sugar, organic acids and anthocyanins to source-sink modulation in Cabernet Sauvignon and Sangiovese grapevines. Front. Plant Sci. 6:382. doi: 10.3389/fpls.2015.00382
Bota, J., Tomás, M., Flexas, J., Medrano, H., and Escalona, J. M. (2016). Differences among grapevine cultivars in their stomatal behavior and water use efficiency under progressive water stress. Agric. Water Manag. 164, 91–99. doi: 10.1016/j.agwat.2015.07.016
Buckley, C. R., Caine, R. S., and Gray, J. E. (2020). Pores for thought: can genetic manipulation of stomatal density protect future rice yields? Front. Plant Sci. 10:1783. doi: 10.3389/fpls.2019.01783
Caine, R. S., Yin, X., Sloan, J., Harrison, E. L., Mohammed, U., Fulton, T., et al. (2019). Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 221, 371–384. doi: 10.1111/nph.15344
Chen, T., Peng, J., Yin, X., Li, M., Xiang, G., Wang, Y., et al. (2021). Importin-αs are required for the nuclear localization and function of the Plasmopara viticola effector PvAVH53. Hortic. Res. 8, 46–12. doi: 10.1038/s41438-021-00482-6
Chen, K., Wang, Y., Zhang, R., Zhang, H., and Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Chen, L., Wu, Z., and Hou, S. (2020). SPEECHLESS speaks loudly in stomatal development. Front. Plant Sci. 11:114. doi: 10.3389/fpls.2020.00114
Clement, K., Rees, H., Canver, M. C., Gehrke, J. M., Farouni, R., Hsu, J. Y., et al. (2019). CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226. doi: 10.1038/s41587-019-0032-3
Cook, B. I., and Wolkovich, E. M. (2016). Climate change decouples drought from early wine grape harvests in France. Nat. Clim. Chang. 6, 715–719. doi: 10.1038/nclimate2960
Dalla Costa, L., Piazza, S., Pompili, V., Salvagnin, U., Cestaro, A., Moffa, L., et al. (2020). Strategies to produce T-DNA free CRISPRed fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci. Rep. 10:20155. doi: 10.1038/s41598-020-77110-1
Dalla Costa, L., Vaccari, I., Mandolini, M., and Martinelli, L. (2009). Elaboration of a reliable strategy based on real-time PCR to characterize genetically modified plantlets and to evaluate the efficiency of a marker gene removal in grape (Vitis spp.). J. Agric. Food Chem. 57, 2668–2677. doi: 10.1021/jf802740m
Dalla Costa, L., Vinciguerra, D., Giacomelli, L., Salvagnin, U., Piazza, S., Spinella, K., et al. (2022). Integrated approach for the molecular characterization of edited plants obtained via Agrobacterium tumefaciens-mediated gene transfer. Eur. Food Res. Technol. 248, 289–299. doi: 10.1007/s00217-021-03881-0
Dayer, S., Herrera, J. C., Dai, Z., Burlett, R., Lamarque, L. J., Delzon, S., et al. (2020). The sequence and thresholds of leaf hydraulic traits underlying grapevine varietal differences in drought tolerance. J. Exp. Bot. 71, 4333–4344. doi: 10.1093/jxb/eraa186
De Cortázar-Atauri, I. G., Duchêne, É., Destrac-Irvine, A., Barbeau, G., De Rességuier, L., Lacombe, T., et al. (2017). Grapevine phenology in France: from past observations to future evolutions in the context of climate change. Oeno One 51, 115–126. doi: 10.20870/oeno-one.2016.0.0.1622
Delrot, S., Grimplet, J., Carbonell-bejerano, P., Schwandner, A., Bert, P., Bavaresco, L., et al. (2020). “Genomic Designing of Climate-Smart Fruit Crops” in Genomic Designing of Climate-Smart Fruit Crops. ed. C. Kole (Switzerland: Springer International Publishing), 157–270.
Doheny-Adams, T., Hunt, L., Franks, P. J., Beerling, D. J., and Gray, J. E. (2012). Genetic manipulation of stomatal density influences stomatal size, plant growth and tolerance to restricted water supply across a growth carbon dioxide gradient. Philos. Trans. R. Soc. B Biol. Sci. 367, 547–555. doi: 10.1098/rstb.2011.0272
Droulia, F., and Charalampopoulos, I. (2021). Future climate change impacts on european viticulture: a review on recent scientific advances. Atmosphere (Basel). 12:495. doi: 10.3390/atmos12040495
Dunn, J., Hunt, L., Afsharinafar, M., Meselmani, M. Al, Mitchell, A., Howells, R., et al. (2019). Reduced stomatal density in bread wheat leads to increased water-use efficiency. J. Exp. Bot. 70, 4737–4748. doi: 10.1093/jxb/erz248
Duursma, R. A. (2015). Plantecophys - An R package for analysing and modelling leaf gas exchange data. PLoS One 10, 1–13. doi: 10.1371/journal.pone.0143346
Easlon, H. M., and Bloom, A. J. (2014). Easy leaf area: Automated digital image analysis for rapid and accurate measurement of leaf area. Appl. Plant Sci. 2:1400033. doi: 10.3732/apps.1400033
Edwards, E. J., Unwin, D., Kilmister, R., Treeby, M., and Ollat, N. (2017). Multi-seasonal effects of warming and elevated CO2 on the physiology, growth and production of mature, field grown, shiraz grapevines. J. Int. des Sci. Vigne Vin 51, 127–132. doi: 10.20870/oeno-one.2016.0.0.1586
Faralli, M., Bontempo, L., Bianchedi, P. L., Moser, C., Bertamini, M., Lawson, T., et al. (2021). Natural variation in stomatal dynamics drives divergence in heat stress tolerance and contributes to seasonal intrinsic water-use efficiency in Vitis vinifera (subsp. sativa and sylvestris). J. Exp. Bot. doi: 10.1093/jxb/erab552 [Epub Ahead of Print]
Faralli, M., Matthews, J., and Lawson, T. (2019). Exploiting natural variation and genetic manipulation of stomatal conductance for crop improvement. Curr. Opin. Plant Biol. 49, 1–7. doi: 10.1016/j.pbi.2019.01.003
Franks, P. J., and Beerling, D. J. (2009). Maximum leaf conductance driven by CO2 effects on stomatal size and density over geologic time. Proc. Natl. Acad. Sci. U. S. A. 106, 10343–10347. doi: 10.1073/pnas.0904209106
Franks, P. J., Doheny-Adams, W., Britton-Harper, Z. J., and Gray, J. E. (2015). Increasing water-use efficiency directly through genetic manipulation of stomatal density. New Phytol. 207, 188–195. doi: 10.1111/nph.13347
Gambetta, G. A., Herrera, J. C., Dayer, S., Feng, Q., Hochberg, U., and Castellarin, S. D. (2020). The physiology of drought stress in grapevine: towards an integrative definition of drought tolerance. J. Exp. Bot. 71, 4658–4676. doi: 10.1093/jxb/eraa245
Giacomelli, L., Zeilmaker, T., Malnoy, M., van der Rouppe Voort, J., and Moser, C. (2019). Generation of mildew-resistant grapevine clones via genome editing. Acta Hortic. 1248, 195–200. doi: 10.17660/ActaHortic.2019.1248.28
Hara, K., Yokoo, T., Kajita, R., Onishi, T., Yahata, S., Peterson, K. M., et al. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50, 1019–1031. doi: 10.1093/pcp/pcp068
Harrison, E. L., Arce Cubas, L., Gray, J. E., and Hepworth, C. (2020). The influence of stomatal morphology and distribution on photosynthetic gas exchange. Plant J. 101, 768–779. doi: 10.1111/tpj.14560
Hepworth, C., Caine, R. S., Harrison, E. L., Sloan, J., and Gray, J. E. (2018). Stomatal development: focusing on the grasses. Curr. Opin. Plant Biol. 41, 1–7. doi: 10.1016/j.pbi.2017.07.009
Hepworth, C., Doheny-Adams, T., Hunt, L., Cameron, D. D., and Gray, J. E. (2015). Manipulating stomatal density enhances drought tolerance without deleterious effect on nutrient uptake. New Phytol. 208, 336–341. doi: 10.1111/nph.13598
Hess, G. T., Tycko, J., Yao, D., and Bassik, M. C. (2017). Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol. Cell 68, 26–43. doi: 10.1016/j.molcel.2017.09.029
Hughes, J., Hepworth, C., Dutton, C., Dunn, J. A., Hunt, L., Stephens, J., et al. (2017). Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 174, 776–787. doi: 10.1104/pp.16.01844
Hunt, L., Bailey, K. J., and Gray, J. E. (2010). The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 186, 609–614. doi: 10.1111/j.1469-8137.2010.03200.x
INTEGRAPE Workshop (2021). In XIth International Symposium on Grapevine Physiology and Biotechnology. Stellenbosch, South Africa.
IPCC (2014). Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Switzerland: IPCC.
Jain, M. (2015). Function genomics of abiotic stress tolerance in plants: a CRISPR approach. Front. Plant Sci. 6:375. doi: 10.3389/fpls.2015.00375
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jones, G. (2013). “Winegrape phenology,” in Phenology: An Integrative Environmental Science. ed. M. D. Schwartz (Dordrecht: Springer), 563–584.
Jones, G. V., White, M. A., Cooper, O. R., and Storchmann, K. (2005). Climate change and global wine quality. Clim. Chang. 73, 319–343. doi: 10.1007/s10584-005-4704-2
Keller, M., Romero, P., Gohil, H., Smithyman, R. P., Riley, W. R., Casassa, L. F., et al. (2016). Deficit irrigation alters grapevine growth, physiology, and fruit microclimate. Am. J. Enol. Vitic. 67, 426–435. doi: 10.5344/ajev.2016.16032
Kondo, T., Kajita, R., Miyazaki, A., Hokoyama, M., Nakamura-Miura, T., Mizuno, S., et al. (2010). Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 51, 1–8. doi: 10.1093/pcp/pcp180
Kuhn, N., Guan, L., Dai, Z. W., Wu, B. H., Lauvergeat, V., Gomès, E., et al. (2014). Berry ripening: recently heard through the grapevine. J. Exp. Bot. 65, 4543–4559. doi: 10.1093/jxb/ert395
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lavoie-Lamoureux, A., Sacco, D., Risse, P. A., and Lovisolo, C. (2017). Factors influencing stomatal conductance in response to water availability in grapevine: a meta-analysis. Physiol. Plant. 159, 468–482. doi: 10.1111/ppl.12530
Lawson, T., and Blatt, M. R. (2014). Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 164, 1556–1570. doi: 10.1104/pp.114.237107
Lawson, T., and McElwain, J. C. (2016). Evolutionary trade-offs in stomatal spacing. New Phytol. 210, 1149–1151. doi: 10.1111/nph.13972
Lee, J. S., Hnilova, M., Maes, M., Lin, Y. C. L., Putarjunan, A., Han, S. K., et al. (2015). Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522, 439–443. doi: 10.1038/nature14561
Li, M. Y., Jiao, Y. T., Wang, Y. T., Zhang, N., Wang, B. B., Liu, R. Q., et al. (2020). CRISPR/Cas9-mediated VvPR4b editing decreases downy mildew resistance in grapevine (Vitis vinifera L.). Hortic. Res. 7:149. doi: 10.1038/s41438-020-00371-4
Liu, D., Hu, R., Palla, K. J., Tuskan, G. A., and Yang, X. (2016). Advances and perspectives on the use of CRISPR/Cas9 systems in plant genomics research. Curr. Opin. Plant Biol. 30, 70–77. doi: 10.1016/j.pbi.2016.01.007
Lovisolo, C., Perrone, I., Carra, A., Ferrandino, A., Flexas, J., Medrano, H., et al. (2010). Drought-induced changes in development and function of grapevine (Vitis spp.) organs and in their hydraulic and non-hydraulic interactions at the whole-plant level: A physiological and molecular update. Funct. Plant Biol. 37, 98–116. doi: 10.1071/FP09191
Lu, J., He, J., Zhou, X., Zhong, J., Li, J., and Liang, Y. K. (2019). Homologous genes of epidermal patterning factor regulate stomatal development in rice. J. Plant Physiol. 235, 18–27. doi: 10.1016/j.jplph.2019.01.010
Lundgren, M. R., and Marais, D. L. (2020). Life history variation as a model for understanding trade-offs in plant – environment interactions. Curr. Biol. 30, R180–R189. doi: 10.1016/j.cub.2020.01.003
Malnoy, M., Viola, R., Jung, M.-H., Koo, O.-J., Kim, S., Kim, J.-S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7:1904. doi: 10.3389/fpls.2016.01904
Marshall, E., Costa, L. M., and Gutierrez-Marcos, J. (2011). Cysteine-rich peptides (CRPs) mediate diverse aspects of cell-cell communication in plant reproduction and development. J. Exp. Bot. 62, 1677–1686. doi: 10.1093/jxb/err002
McCown, B. H., and Lloyd, G. (1981). Woody plant medium (WPM) - a mineral nutrient formulation for microculture of woody plant-species. Hortic. Sci. 16:453
McGranahan, D. A., and Poling, B. N. (2018). Trait-based responses of seven annual crops to elevated CO2 and water limitation. Renew. Agric. Food Syst. 33, 259–266. doi: 10.1017/S1742170517000692
Mohammed, U., Caine, R. S., Atkinson, J. A., Harrison, E. L., Wells, D., Chater, C. C., et al. (2019). Rice plants overexpressing OsEPF1 show reduced stomatal density and increased root cortical aerenchyma formation. Sci. Rep. 9, 5584–5513. doi: 10.1038/s41598-019-41922-7
Morales-Navarro, S., Pérez-Díaz, R., Ortega, A., de Marcos, A., Mena, M., Fenoll, C., et al. (2018). Overexpression of a SDD1-like gene from wild tomato decreases stomatal density and enhances dehydration avoidance in arabidopsis and cultivated tomato. Front. Plant Sci. 9:940. doi: 10.3389/fpls.2018.00940
Mosedale, J. R., Abernethy, K. E., Smart, R. E., Wilson, R. J., and Maclean, I. M. D. (2016). Climate change impacts and adaptive strategies: lessons from the grapevine. Glob. Chang. Biol. 22, 3814–3828. doi: 10.1111/gcb.13406
Ohki, S., Takeuchi, M., and Mori, M. (2011). The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat. Commun. 2:512. doi: 10.1038/ncomms1520
Okonechnikov, K., Golosova, O., Fursov, M., Varlamov, A., Vaskin, Y., Efremov, I., et al. (2012). Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167. doi: 10.1093/bioinformatics/bts091
Palliotti, A., Tombesi, S., Silvestroni, O., Lanari, V., Gatti, M., and Poni, S. (2014). Changes in vineyard establishment and canopy management urged by earlier climate-related grape ripening: A review. Sci. Hortic. 178, 43–54. doi: 10.1016/j.scienta.2014.07.039
Parker, A. K., de Cortázar-Atauri, I. G., Trought, M. C. T., Destrac, A., Agnew, R., Sturman, A., et al. (2020). Adaptation to climate change by determining grapevine cultivar differences using temperature-based phenology models. Oeno One 54, 955–974. doi: 10.20870/OENO-ONE.2020.54.4.3861
Pillitteri, L. J., Sloan, D. B., Bogenschutz, N. L., and Torii, K. U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505. doi: 10.1038/nature05467
Podevin, N., Davies, H. V., Hartung, F., Nogué, F., and Casacuberta, J. M. (2013). Site-directed nucleases: A paradigm shift in predictable, knowledge-based plant breeding. Trends Biotechnol. 31, 375–383. doi: 10.1016/j.tibtech.2013.03.004
Pons, A., Allamy, L., Schüttler, A., Rauhut, D., Thibon, C., and Darriet, P. (2017). What is the expected impact of climate change on wine aroma compounds and their precursors in grape? Oeno One 51, 141–146. doi: 10.20870/oeno-one.2016.0.0.1868
Prats-Llinàs, M. T., Nieto, H., DeJong, T. M., Girona, J., and Marsal, J. (2020). Using forced regrowth to manipulate chardonnay grapevine (Vitis vinifera L.) development to evaluate phenological stage responses to temperature. Sci. Hortic. 262:109065. doi: 10.1016/j.scienta.2019.109065
Rienth, M., Vigneron, N., Darriet, P., Sweetman, C., Burbidge, C., Bonghi, C., et al. (2021). Grape berry secondary metabolites and their modulation by abiotic factors in a climate change scenario–a review. Front. Plant Sci. 12:643258. doi: 10.3389/fpls.2021.643258
Santillán, D., Garrote, L., Iglesias, A., and Sotes, V. (2020). Climate change risks and adaptation: new indicators for Mediterranean viticulture. Mitig. Adapt. Strateg. Glob. Chang. 25, 881–899. doi: 10.1007/s11027-019-09899-w
Scholasch, T., and Rienth, M. (2019). Review of water deficit mediated changes in vine and berry physiology; consequences for the optimization of irrigation strategies. Oeno One 53, 423–444. doi: 10.20870/oeno-one.2019.53.3.2329
Schultz, H. R. (2000). Climate change and viticulture: A European perspective on climatology, carbon dioxide and UV-B effects. Aust. J. Grape Wine Res. 6, 2–12. doi: 10.1111/j.1755-0238.2000.tb00156.x
Schultz, H. R. (2003). Differences in hydraulic architecture account for near-isohydric and anisohydric behaviour of two field-grown Vitis vinifera L. cultivars during drought. Plant Cell Environ. 26, 1393–1405. doi: 10.1046/j.1365-3040.2003.01064.x
Scintilla, S., Salvagnin, U., Giacomelli, L., Zeilmaker, T., Malnoy, M. A., van der Voort, J. R., et al. (2021). Regeneration of plants from DNA-free edited grapevine protoplasts. bioRxiv. doi: 10.1101/2021.07.16.452503
Shimada, T., Sugano, S. S., and Hara-Nishimura, I. (2011). Positive and negative peptide signals control stomatal density. Cell. Mol. Life Sci. 68, 2081–2088. doi: 10.1007/s00018-011-0685-7
Soar, C. J., Dry, P. R., and Loveys, B. R. (2006). Scion photosynthesis and leaf gas exchange in Vitis vinifera L. cv. Shiraz: mediation of rootstock effects via xylem sap ABA. Aust. J. Grape Wine Res. 12, 82–96. doi: 10.1111/j.1755-0238.2006.tb00047.x
Sugano, S. S., Shimada, T., Imai, Y., Okawa, K., Tamai, A., Mori, M., et al. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463, 241–244. doi: 10.1038/nature08682
Tomasi, D., Jones, G. V., Giust, M., Lovat, L., and Gaiotti, F. (2011). Grapevine phenology and climate change: relationships and trends in the Veneto region of Italy for 1964–2009. Am. J. Enol. Vitic. 62, 329–339. doi: 10.5344/ajev.2011.10108
Tombesi, S., Nardini, A., Frioni, T., Soccolini, M., Zadra, C., Farinelli, D., et al. (2015). Stomatal closure is induced by hydraulic signals and maintained by ABA in drought-stressed grapevine. Sci. Rep. 5, 1–12. doi: 10.1038/srep12449
Van Leeuwen, C., and Destrac-Irvine, A. (2017). Modified grape composition under climate change conditions requires adaptations in the vineyard. Oeno One 51, 147–154. doi: 10.20870/oeno-one.2016.0.0.1647
Van Leeuwen, C., Destrac-Irvine, A., Dubernet, M., Duchêne, E., Gowdy, M., Marguerit, E., et al. (2019). An update on the impact of climate change in viticulture and potential adaptations. Agronomy 9, 1–20. doi: 10.3390/agronomy9090514
Venios, X., Korkas, E., Nisiotou, A., and Banilas, G. (2020). Grapevine responses to heat stress and global warming. Plan. Theory 9, 1–15. doi: 10.3390/plants9121754
Villalobos-González, L., Muñoz-Araya, M., Franck, N., and Pastenes, C. (2019). Controversies in midday water potential regulation and stomatal behavior might result from the environment, genotype, and/or rootstock: evidence from Carménère and Syrah grapevine varieties. Front. Plant Sci. 10:1522. doi: 10.3389/fpls.2019.01522
Wan, D. Y., Guo, Y., Cheng, Y., Hu, Y., Xiao, S., Wang, Y., et al. (2020). CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic. Res. 7:116. doi: 10.1038/s41438-020-0339-8
Wang, Z., Wong, D. C. J., Wang, Y., Xu, G., Ren, C., Liu, Y., et al. (2021). GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol. 186, 1660–1678. doi: 10.1093/PLPHYS/KIAB142
Yin, X., Biswal, A. K., Dionora, J., Perdigon, K. M., Balahadia, C. P., Mazumdar, S., et al. (2017). CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 36, 745–757. doi: 10.1007/s00299-017-2118-z
Keywords: Vitis vinifera, stomata, genome editing, climate change, water-use efficiency
Citation: Clemens M, Faralli M, Lagreze J, Bontempo L, Piazza S, Varotto C, Malnoy M, Oechel W, Rizzoli A and Dalla Costa L (2022) VvEPFL9-1 Knock-Out via CRISPR/Cas9 Reduces Stomatal Density in Grapevine. Front. Plant Sci. 13:878001. doi: 10.3389/fpls.2022.878001
Received: 17 February 2022; Accepted: 11 April 2022;
Published: 17 May 2022.
Edited by:
Giorgio Gambino, Institute for Sustainable Plant Protection (CNR), ItalyReviewed by:
Claudio Lovisolo, University of Turin, ItalyCopyright © 2022 Clemens, Faralli, Lagreze, Bontempo, Piazza, Varotto, Malnoy, Oechel, Rizzoli and Dalla Costa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michele Faralli, bWljaGVsZS5mYXJhbGxpQHVuaXRuLml0; Lorenza Dalla Costa, bG9yZW56YS5kYWxsYWNvc3RhQGZtYWNoLml0
†Present address: Michele Faralli, Center Agriculture Food Environment (C3A), University of Trento, San Michele all’Adige, Italy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.