- 1Key Laboratory of Biology of Crop Pathogens and Insects of Zhejiang Province, Institute of Biotechnology, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 2Centre of Analysis and Measurement, Zhejiang University, Hangzhou, China
- 3Hainan Institute, Zhejiang University, Sanya, China
Rapid alkalinization factors (RALFs) were recently reported to be important players in plant immunity. Nevertheless, the signaling underlying RALF-triggered immunity in crop species against necrotrophic pathogens remains largely unknown. In this study, RALF family in the important oil crop oilseed rape (Brassica napus) was identified and functions of BnRALF10 in immunity against the devastating necrotrophic pathogen Sclerotinia sclerotiorum as well as the signaling underlying this immunity were revealed. The oilseed rape genome carried 61 RALFs, half of them were atypical, containing a less conserved YISY motif and lacking a RRXL motif or a pair of cysteines. Family-wide gene expression analyses demonstrated that patterns of expression in response to S. sclerotiorum infection and DAMP and PAMP treatments were generally RALF- and stimulus-specific. Most significantly responsive BnRALF genes were expressionally up-regulated by S. sclerotiorum, while in contrast, more BnRALF genes were down-regulated by BnPep5 and SsNLP1. These results indicate that members of BnRALF family are likely differentially involved in plant immunity. Functional analyses revealed that BnRALF10 provoked diverse immune responses in oilseed rape and stimulated resistance to S. sclerotiorum. These data support BnRALF10 to function as a DAMP to play a positive role in plant immunity. BnRALF10 interacted with BnFER. Silencing of BnFER decreased BnRALF10-induced reactive oxygen species (ROS) production and compromised rape resistance to S. sclerotiorum. These results back BnFER to be a receptor of BnRALF10. Furthermore, quantitative proteomic analysis identified dozens of BnRALF10-elicited defense (RED) proteins, which respond to BnRALF10 in protein abundance and play a role in defense. Our results revealed that BnRALF10 modulated the abundance of RED proteins to fine tune plant immunity. Collectively, our results provided some insights into the functions of oilseed rape RALFs and the signaling underlying BnRALF-triggered immunity.
Introduction
As sessile organisms, plants have evolved sophisticated communication systems to deal with constantly changing environmental conditions. Under this scenario, small secreted peptides serve as signal molecules to orchestrate a plethora of plant processes such as development and immune responses (Olsson et al., 2019). Rapid alkalinization factors (RALFs) are small cysteine-rich secreted peptides that are involved in multiple physiological and developmental processes, ranging from cell elongation to modulation of immune responses, and thus are also referred to as plant peptide hormones (Blackburn et al., 2020). RALFs were first discovered over 20 years ago due to their ability to cause rapid alkalinization of the extracellular compartment of tobacco cells (Pearce et al., 2001). Since then, RALFs were found to be ubiquitous in plants and microbes. Genome-wide analyses have identified 37 RALFs in Arabidopsis (Abarca et al., 2021), 765 in 51 plant species (Campbell and Turner, 2017), 124 in 7 Rosaceae species (Zhang H. et al., 2020), 17 in Fusarium species and 18 in 6 nematode species (Thynne et al., 2017; Zhang X. et al., 2020). The wide distribution of RALFs highlighted their functional importance. RALFs are well known pivotal contributors to plant growth and development (Haruta et al., 2014; Du et al., 2016; Dressano et al., 2017; Abarca et al., 2021; Zhu et al., 2021). However, relatively, role and mechanisms of RALFs in plant immunity are much less studied. Recently, two studies in Arabidopsis revealed that RALFs affect immune responses such as reactive oxygen species (ROS) generation and pathogen associated molecular pattern (PAMP)-triggered immunity (PTI) (Stegmann et al., 2017; Abarca et al., 2021). RALFs are recognized by its receptor FERONIA (FER) (Haruta et al., 2014). They form a complex together with LLGs (Xiao et al., 2019), and negatively modulate flg22-triggered immunity to Pseudomonas syringae (Stegmann et al., 2017). In contrast to the recognition of RALFs, the signaling downstream the recognition is still largely unclear.
Typically, RALF proteins are small molecules with an average full length of 80–120 amino acids. They execute their functions in their mature form released via cleaving by proteases (Matos et al., 2008; Srivastava et al., 2009). Arabidopsis RALF22 and RALF23 are cleaved at a di-basic RRXL site by site-1 protease (AtS1P), a plant subtilisin-like serine protease. This processing contributes to the function of these two RALFs on salt tolerance and plant immunity regulation, respectively (Stegmann et al., 2017; Zhao et al., 2018). Fourteen out of 37 Arabidopsis RALFs display the predicted S1P cleavage site, suggesting the conservation of this processing mechanism in the RALF family (Abarca et al., 2021). In addition to the RRXL cleavage site, the mature RALFs also contain other conserved motifs, such as the YISY motifs near the N-terminus and the four cysteine residues at the C-terminus (Campbell and Turner, 2017). The YISY motif of RALFs is crucial for their biological functions and receptor binding. Replacing the isoleucine with alanine significantly compromised their ability of alkalization and the root growth inhibition (Pearce et al., 2010; Xiao et al., 2019). The four conserved cysteines form two intramolecular disulfide bonds, which play a key role in maintaining the three-dimensional conformation and biological activity of RALFs. Mutation of the first two cysteine residues of Arabidopsis RALF1 influenced its root growth and PTI inhibition activity (Zhang X. et al., 2020), while replacement of the four cysteines of RALF4 with alanine weakened its binding to LRR extensin proteins, thereby affects pollen tube germination (Moussu et al., 2020). Nevertheless, it is noticeable that atypical RALFs that do not carry canonical RRXL, YISY and/or four conserved cysteines exist widely in plants (Campbell and Turner, 2017). Functions and mechanisms of these atypical RALFs remain largely unknown.
Sclerotinia sclerotiorum (Lib.) de Bary is a devastating soil-borne necrotrophic fungal pathogen. It causes diseases on a broad range of hosts including economically important crops such as oilseed rape (Brassica napus), sunflower and soybean, resulting in substantial yield losses (Ding et al., 2021). Oilseed rape is one of the most important oil crops globally. White mold disease caused by S. sclerotiorum is the most severe disease in oilseed rape. Nevertheless, the mechanisms underlying immunity to S. sclerotiorum in oilseed rape remain largely unexplored. In this study, we identified the RALF gene family in oilseed rape, and defined the expression patterns of the whole BnRALF family in response to S. sclerotiorum infection and PAMP/DAMP treatments. Furthermore, we conducted a series of analyses to reveal the functions and mechanisms of BnRALF10, one of the BnRALFs which highly responded to S. sclerotiorum in oilseed rape. Our results demonstrated that BnRALF10 acted as a DAMP molecule to elicit multiple typical immune responses and triggered immunity against S. sclerotiorum. It modulated accumulation level of various defense regulators to stimulate immunity against S. sclerotiorum. This RALF exhibited potential for breeding for improved disease resistance against necrotrophic pathogens.
Materials and Methods
Identification of Putative Rapid Alkalinization Factor Proteins in Brassica napus
Protein sequences of Arabidopsis RALFs were downloaded from TAIR database1. Arabidopsis RALF protein sequences were used as queries to search their orthologs in B. napus genomes using BLASTp program in NCBI database2 with default settings. All retrieved protein sequences were examined for the presence of conserved motifs and redundant sequences were removed. The physico-chemical properties of BnRALF proteins were predicted using ExPASy Compute pI/Mw tool3 (Bjellqvist et al., 1993).
Construction of Rapid Alkalinization Factor Alignments and Phylogenetic Trees
Multiple sequence alignment of the full-length RALF protein sequences were created using the MUSCLE algorithm with the MEGA X software (Kumar et al., 2018). Phylogenetic tree was constructed with the MEGA X software by using the unweighted pair-group method with arithmetic means (UPGMA) algorithm following the Jones-Taylor-Thornton (JTT) model. Bootstrapping was performed 1,000 times. All other parameters were left as default. The inferred tree were visualized using iTOL4 (Letunic and Bork, 2019). The multiple mature peptides alignment was displayed using the GeneDoc2.7.0 and the results were manually edited5. WebLogo36 was used to provide a visual summary of conserved residues within the alignments.
Plant Materials and Pathogen Inoculation Analysis
Brassica napus plants were grown in growth cabinets at 23°C under a 14 h/10 h light/dark photoperiod. Fresh sclerotia of S. sclerotiorum strain UF1 (Xu T. T. et al., 2018) were cultured at 23°C on potato dextrose agar medium (PDA) to produce mycelia, which were transferred to new PDA plates and grown for 2 days. The PDA plugs containing young S. sclerotiorum mycelia were punched to inoculate the plant leaves. Area of disease lesions was measured using the ImageJ image analysis software7. For disease resistance evaluation, at least five plants for each genotype were examined and the experiments were conducted three times independently.
Peptides
BnRALF10 (AQKYVSYGAMRKNSVPCSRGASYNCQRGAQNP YRGCSTIRCR), BnPep5 (SLNVSSKLTRKLPVSSGKRGGVN) and S. sclerotiorum necrosis and ethylene-inducing peptide 1 (SsNep1)-like peptide 1 (SsNLP1) (GIMYAWYFPKDQPAAGN VVGGHRHDWE) were synthesized by Chinapeptides, Suzhou, China.
RNA Isolation and Gene Expression Analysis
For gene expression analysis, the oilseed rape leaves were sampled at 7 h post inoculation with S. sclerotiorum and 4 h post infiltration with 1 μM SsNLP1 and 200 nM BnPep5. Total RNA was extracted using Trizol reagent (Vazyme, Nanjing, China) following the manufacturer’s procedure. Quantitative real-time PCR (qRT-PCR) was performed using the StepOne Real-Time PCR system (Applied Biosystems, United States) with SYBR Green PCR Master Mix (TaKaRa, Dalian, China). The relative fold changes were calculated using the 2–ΔΔCt method as previously described (Cao et al., 2016), with two technical replicates for each of the three biological replicates. The housekeeping gene BnActin7 was used as an internal control. The heatmaps were created by TBtools (Chen et al., 2020) on the basis of the log2-fold-transformed data. Primers used for qRT-PCR are listed in Supplementary Table 1.
Turnip Yellow Mosaic Virus-Induced Gene Silencing Manipulation Procedure
Turnip yellow mosaic virus-based VIGS experiment was performed as previously described with some modifications (Pflieger et al., 2008). The TYMV-derived construct pTY-S (controlled by 35S promoter) contained the full length cDNA of TYMV, of which a unique SnaBI restriction site was inserted into the coat protein. Both sense and antisense versions of a specific 40 bp fragment from the coding region of BnRALF10 were designed to form self-hybridized palindromic oligonucleotide according to the reported method (Pflieger et al., 2008). The pTY-BnRALF10 constructs were obtained by inserting the self-hybridized palindromic oligonucleotide of BnRALF10 into the SnaBI site of pTY-S. The synthesis of the self-hybridized palindromic oligonucleotide and construction of pTY-BnRALF10 recombinant vector were completed by the company (Tsingke, Hangzhou, China). pTY-BnPDS (B. napus phytoene desaturase) vector was constructed to monitor the silencing efficiency. The sequence information of the oligonucleotides was listed in Supplementary Table 1. Two-week-old B. napus cultivar Zhongshuang 11 (ZS11) seedlings were used for VIGS experiments with pTY, pTY-BnPDS and pTY-BnRALF10 plasmid (6 μg). Plants were placed in the dark 24 h prior to inoculation, and were mechanically inoculated by rubbing the upper surface of three rosette leaves with 8 μL of inoculum, using a gloved finger and celite (Sigma-Aldrich, St. Louis, MO, United States). The inoculated B. napus seedlings were then transferred into the chamber for silencing. Plants were checked for BnRALF10 gene silencing by qRT-PCR, and meanwhile the same plants were used for the pathogen inoculation analysis.
Reactive Oxygen Species Assay
Leaf disks (3 mm in diameter) of B. napus were collected and dipped in 96-well plates containing sterile water in the dark overnight. The next day, the water was replaced by a solution containing 11 μM L-012 (FUJIFILM Wako Pure Chemical, Osaka, Japan), 20 μg/mL horseradish peroxidase (HRP, Sigma-Aldrich) and 1 μM BnRALF10 peptide. Luminescence was measured for the indicated time period using a Microplate Luminometer (Titertek Berthold, Bad Wildbad, Germany). ROS production is either displayed as the integration of total photon counts or as the progression of photon counts.
Measurement of Cytosolic Calcium
Arabidopsis thaliana plants expressing cytosolic apoaequorin used for [Ca2+]cyt measurements were kindly provided by Prof. Yan Liang (Zhejiang university, China). Seventy microliters of ddH2O containing 10 mM coelenterazine h (Sigma-Aldrich, St. Louis, MO, United States) was added per well in a 96-well plate. Vertically grown 5-day-old transgenic seedlings were individually transferred to each well and incubated overnight in the dark at room temperature. At the next day, 30 μL of ddH2O containing BnRALF10 peptide was added to each well, the final concentration of BnRALF10 was 1 mM, and the chemiluminescent signal was immediately recorded using a Microplate Luminometer (Titertek Berthold, Bad Wildbad, Germany). Photon counts were converted to calcium concentration.
Mitogen-Activated Protein Kinases Phosphorylation Assay
Seven-day-old Arabidopsis Col-0 seedlings were immersed in liquid 1/2MS medium overnight. BnRALF10 peptide was then added to a final concentration of 1 μM for 5–30 min induction. After induction, the seedlings were snap-frozen in liquid nitrogen and ground to a fine powder, from which total protein was extracted by suspension in the extraction buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, and 1 × protease inhibitor cocktail (HuaBio, Hangzhou, China). An anti-phospho p44/p42 mitogen-activated protein kinases (MAPK) antibody (Cell Signaling Technology, Danvers, MA, United States) was used to detect active MPK6 and MPK3 via immunoblotting.
Yeast Two-Hybrid Assay
The extracellular domain of BnFER (XP_013734398.1) and the full-length BnRALF10 were cloned into the bait vector pGBKT7 and the prey vector pGADT7, respectively. The resulting plasmids BD-BnFER and AD-BnRALF10 were co-transformed into the yeast strain Y2H Gold, while pGBKT7 and AD-BnRALF10 were co-transformed into the same strain to serve as a negative control. Presence of the transgenes was confirmed by growth on an SD-Leu-Trp agar plate. To assess protein interactions, the transformed yeast cells were suspended in liquid SD-Leu-Trp medium to an optical density at 600 nm of 1. Five microliters of suspended yeast cells was dropped onto an SD-Ade-His-Leu-Trp agar plate. The resulting agar plate was incubated at 30°C and observed for yeast growth. The primers used for the yeast two-hybrid (Y2H) assay are listed in Supplementary Table 1.
Bimolecular Fluorescence Complementation Assay
For the bimolecular fluorescence complementation (BiFC) assay, the coding sequences of BnRALF10 and BnFER were cloned into the vectors cYFP and nYFP, respectively. Agrobacterium suspensions carrying each plasmid were mixed at 1:1 ratio and were then infiltrated into Nicotiana benthamiana leaves. Confocal microscopy images were captured at 2 days after infiltration. The primers used for the BiFC are listed in Supplementary Table 1.
Luciferase Complementation Imaging Assay
A luciferase complementation imaging (LCI) assay was conducted. The full-length BnFER and BnRALF10 were cloned into the vectors NLuc and CLuc, respectively. Paired constructs of BnRALF10-CLuc and BnFER-NLuc were transiently co-expressed in the leaves of N. benthamiana through Agrobacterium-mediated co-infiltration. The primers used for this assay are listed in Supplementary Table 1.
Protein Extraction
Total proteins from each sample were extracted by the modified method of trichloroacetic acid (TCA)-acetone precipitation (Cao et al., 2016). Approximately 1 g samples were pulverized using a mortar and pestle in liquid nitrogen, and the fine powder was suspended overnight at −20°C in 30 ml of ice-cold acetone containing 10% (w/v) TCA and 0.07% (w/v) DL-Dithiothreitol (DTT). The protein precipitate was pelleted by centrifugation at 35,000 × g for 1 h and resuspended in 30 ml of ice-cold acetone containing 0.07% (w/v) DTT for 1 h at −20°C. The protein precipitate was centrifuged again at 35,000 × g for 1 h (4°C) and washed three times with ice-cold acetone containing 0.07% (w/v) DTT. Protein was extracted by resuspending the final dry pellet in a dissolution buffer [8 M Urea, 50 mM triethyl ammonium bicarbonate (TEAB), pH 8]. The mixtures were heated in 30°C water bath for 1 h and subjected to ultrasonication at 50 W output with two bursts of 10 s each and the lysates were cooled on ice for 1 min between bursts. Following lysis, the protein extracts were clarified by centrifuging for 30 min at 25,000 × g (4°C). The supernatants were collected and quantified by Bradford assays, using BSA as a standard.
Protein Digestion and Sample Cleanup
Protein samples (100 μg) were solved in 8 M urea (pH 8.5) in a volume ratio of 1:4 and added the solution to a 10 K ultrafiltration tube, centrifuged at 12,000 × g at 4°C for 15 min. The 100 mM iodoacetamide (IAA) was then added to the protein supernatant, and the mixture was incubated for 30 min in the dark. The proteins were then diluted 3-folds using 50 mM NH4HCO3 and digested with trypsin (100:1) for 20 h at 37°C. Trypsin digestion was stopped by the addition of NH4HCO3. After centrifugating at 12,000 × g for 20 min, the supernatants were collected and were then desalted with C18 spin tips and freeze-dried. All prepared samples were stored at −80°C until LC–MS/MS analysis.
Liquid Chromatography–Mass Spectrometry Analyses
Liquid chromatography–mass spectrometry (LC–MS/MS) analysis was performed as previously described with some modifications (Cao et al., 2016). Each of the dried fractions was dissolved in 20 μL of 0.1% (v/v) formic acid and centrifuged at 20,000 × g for 10 min. The final concentration of the peptide solution was 0.4 μg/μL and the peptide (5 μL) was injected onto the trap column with a flow rate of 10 μL/min for 2 min using a Thermo Scientific Orbitrap Elite. The trap was equilibrated at a maximum pressure of 500 bar for 12 μL followed by column equilibration at a maximum of 500 bar for 3 μL before starting gradient elution of column. The peptide samples were subsequently eluted with a five-step linear gradient of A/B mixture (A: ddH2O with 0.1% formic acid, B: ACN with 0.1% formic acid): 0–10 min, 3–8% B; 10–120 min, 8–20% B; 120–137 min, 20–30% B; 137–143 min, 30–90% B; 143–150 min, 90% B. The column flow was maintained as 250 nL/min. The chromatographic system was composed of a trapping column (75 μm × 2 cm, nanoviper, C18, 3 μM, 100 A) and an analytical column (50 μm × 15 cm, nanoviper, C18, 2 μM, 100 A). Data collection was performed using Theromo Xcalibur Qual Browser and Proteome Discoverer 2.0 software.
Results
Identification and Phylogenetic Analysis of RALF Genes in Brassica napus Genome
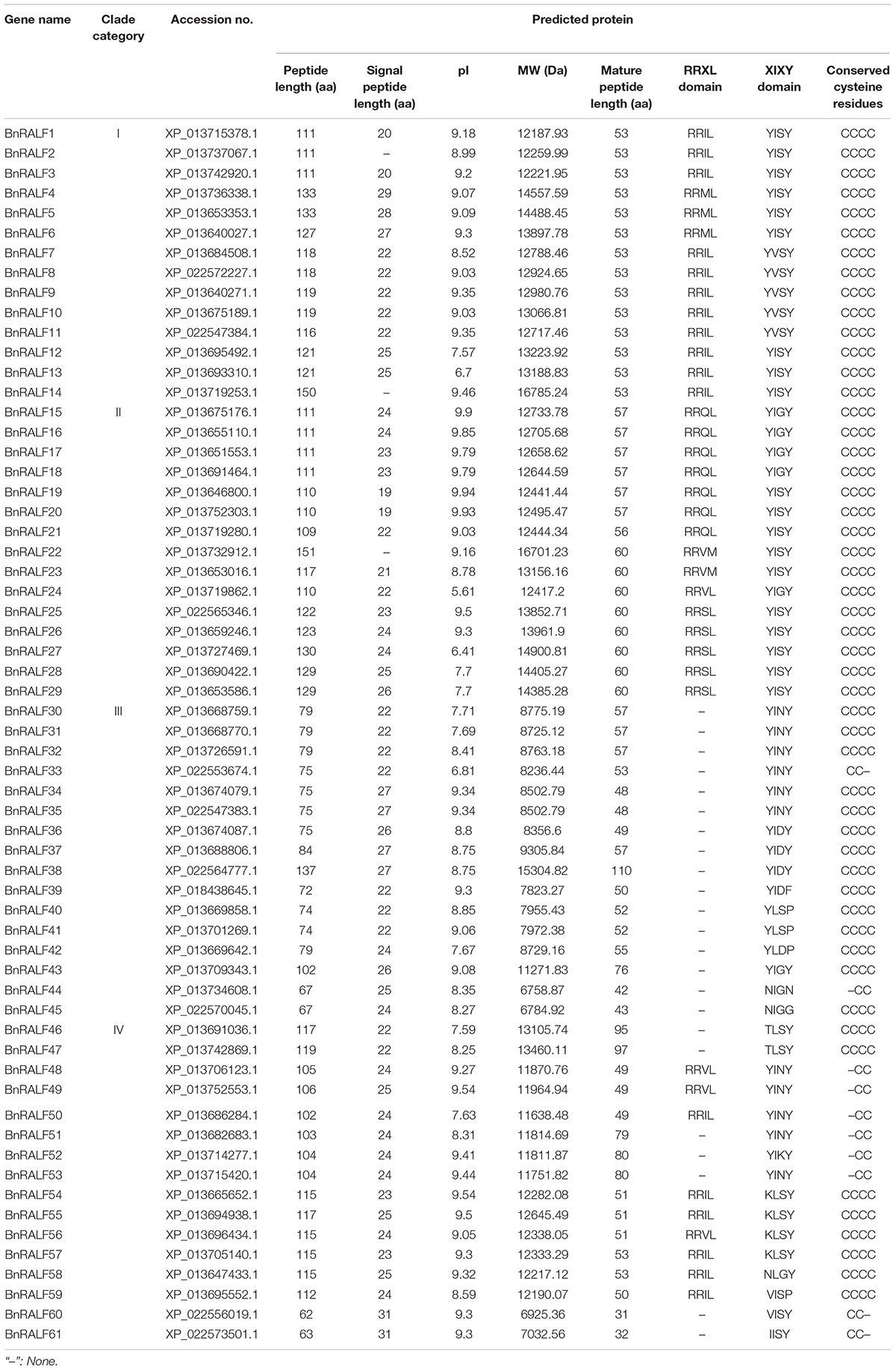
To investigate RALF genes in oilseed rape, a BLASTp search was performed against B. napus genome in NCBI database (see text footnote 2) using well-characterized Arabidopsis RALF protein sequences as query, which resulted in identification of 61 potential RALF sequences in the genome of B. napus (Table 1). To understand the evolutionary relationship of the RALFs, we constructed a phylogenetic tree based on the full-length BnRALF protein sequences employing the UPGMA method along with their 37 A. thaliana RALF homologs and two outgroup protein sequences (AT1G78000 and AT5G50920) from Arabidopsis (Figure 1). According to the phylogenetic analysis, these total 98 RALF protein sequences could be separated into four major clades (clades I–IV), which contained in turn 18, 21, 36, and 22 members. Each clade included at least one oilseed rape and one Arabidopsis RALFs, indicating that all clades evolved before the divergence of these two cruciferous lineages. However, uneven divergence existed between clades. BnRALFs accounted for over 70% in clades I [77% (14/18)], II [71% (15/21)], and IV [76% (16/22)], while only 44% (16/36) in clade III (Figure 1).
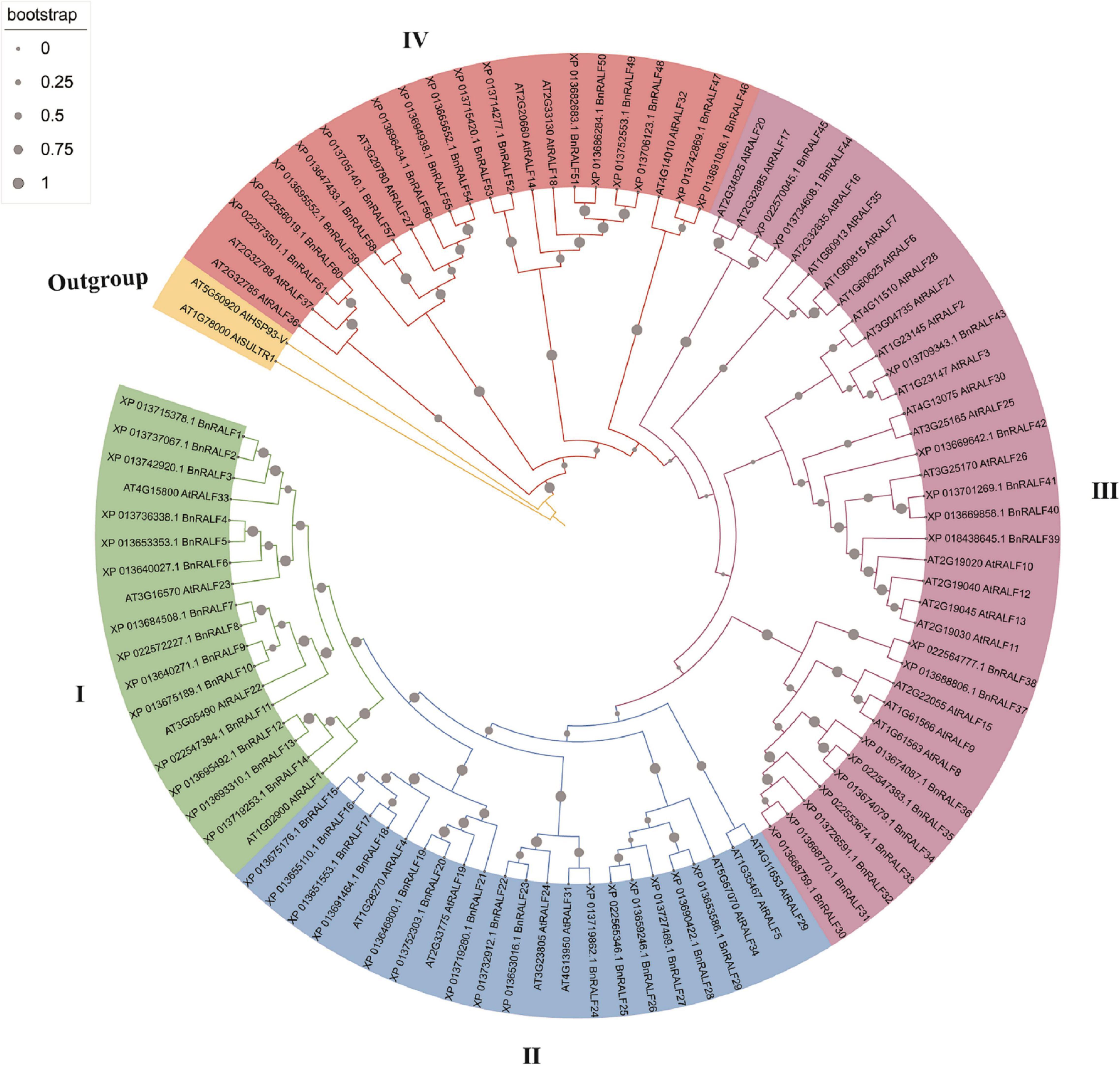
Figure 1. Phylogenetic tree of B. napus and A. thaliana RALF proteins. Full-length protein sequences were aligned using MUSCLE. The phylogenetic tree was constructed employing the UPGMA method with 1,000 bootstrap values in MEGA-X software, and was optimized with the iTOL online tool. The clades were marked with different colors. The gray filled circles in the branch indicate bootstrap value.
The retrieved BnRALF protein sequences were further analyzed for physico-chemical properties (Table 1). These 61 BnRALF proteins displayed diversities in full length, signal peptide length, theoretical isoelectric point (pI), molecular weight (MW) and putative mature peptide length. Specifically, the full length of the 61 BnRALF protein sequences varied from 62 to 151 amino acids (aa), among which the majority of members in clade III were short with a full-length of less than 100 aa. Additionally, 93% (57/61) RALF proteins exhibited a pI higher than 7 and 90% (55/61) contained a signal peptide, the hallmark of post-translationally modified secreted peptide precursors, indicating that these proteins mainly function outside the cell.
Conserved Motifs of BnRALF Proteins
To further characterize BnRALFs, we performed mature peptide sequence alignment using GeneDoc and motif prediction employing MEME tool (Bailey et al., 2009). The total sequence alignment and group-wide conservative sequences were shown in Figures 2A,B, respectively. The results showed that clades I, II, and IV all possessed the conserved RRXL (X represents any amino acid) motif, which is a protease cleavage site for the mature peptide process (Srivastava et al., 2009). Remarkably, this motif was entirely absent across the 16 clade III BnRALF proteins (Table 1 and Figure 2A). Given that the length of these proteins were already similar to that of mature proteins of other clades, these data implied that BnRALFs of the clade III likely required no further cleaving processing. This was likely also the case for two clade IV BnRALFs, BnRALF61 and BnRALF62. Uniquely, five BnRALFs of clade IV, BnRALF46, BnRALF47, BnRALF51, BnRALF52 and BnRALF53, did not harbor canonical RRXL motif either. They were comprised of 79–97 aa, which were much longer than the general size of mature BnRALF peptides (less than 60 aa). This indicated that these five BnRALFs required further maturation processing and were likely cleaved in a RRXL-independent mechanism.
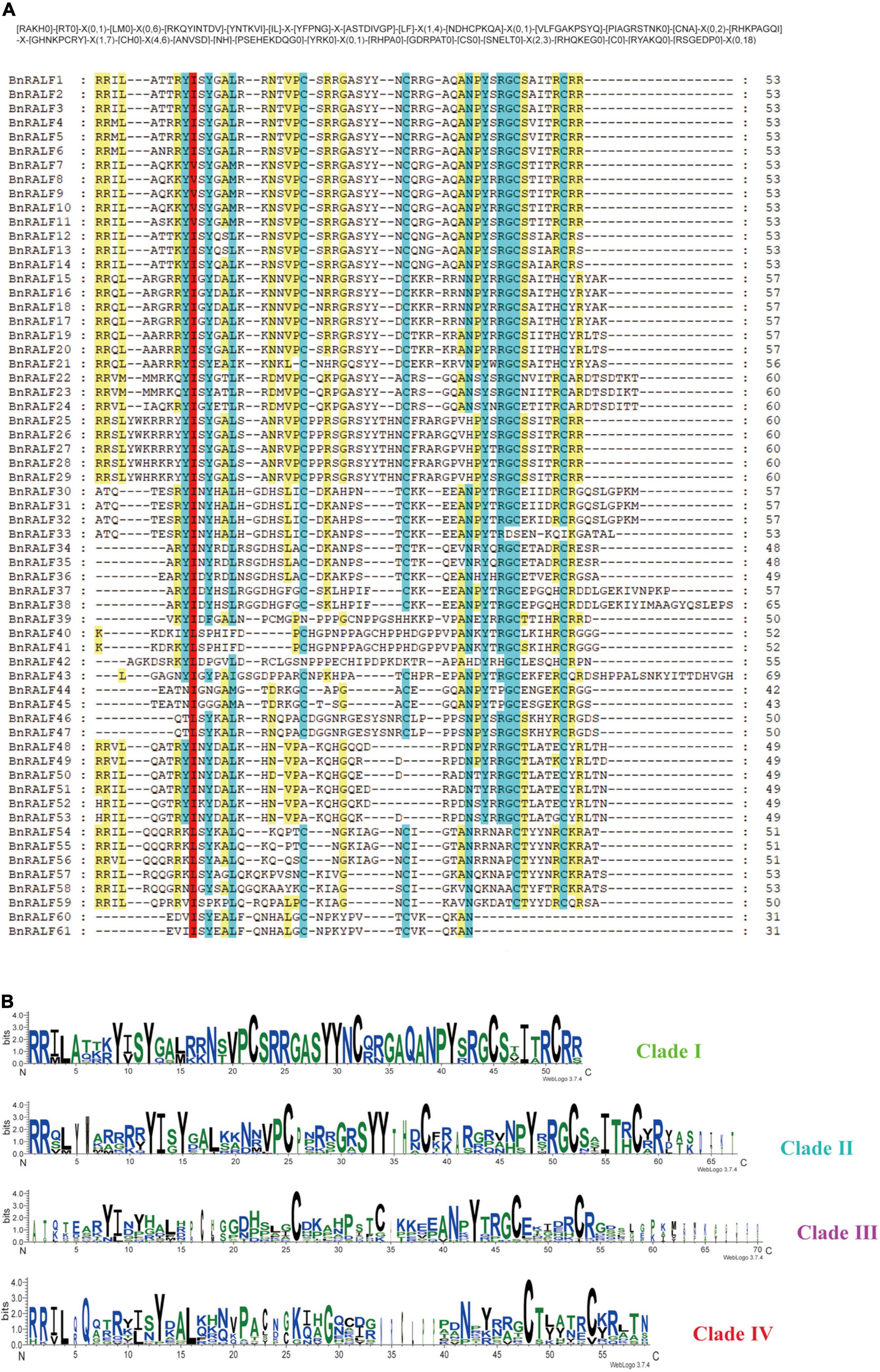
Figure 2. Conserved motifs of B. napus RALF proteins. (A) Multiple sequence alignment of BnRALF proteins for conserved motifs. Alignment was carried out using ClustalW and presented by GeneDoc. Residues with various conservation percentage were highlighted in different colors: 100% in red; 80∼100% in blue, 60∼80% in yellow. The square bracket ‘[ ]’ indicates the amino acids allowed in this position of motif; A ‘X’ represents any amino acid, while the round bracket ‘()’ denotes the number of amino acids. (B) Residue conservation of BnRALFs within the clades. The group-wise sequence logos showing the amino acid bias of the RALF mature peptides were generated from the WebLogo3 analysis. Y-axis expresses the bit score of each position in the sequence and the group names are indicated on the right.
The YISY motif, which is essential for activity and receptor binding of RALFs (Pearce et al., 2010; Xiao et al., 2019), existed in different forms in a clade-dependent manner, canonical YISY and its variant YVSY in clade I; canonical YISY and its variant YIGY in clade II; variants YIN/DY and YLS/DP dominated in clade III, while variants YINY and KLSY dominated in clade IV (Table 1 and Figure 2A). Whether the variants are as functional as the canonical YISY awaits further experimental verification.
Fifty-one out of 61 BnRALFs carried four conserved cysteines, which are supposed to be folded into intramolecular disulfide bridges for functions of RALFs (Pearce et al., 2001; Moussu et al., 2020; Zhang X. et al., 2020). The remaining 10 BnRALFs were distributed in the clades III and IV. They contained two cysteine residues at the first and second positions, or the third and fourth (Table 1 and Figure 2A), which may form one intramolecular disulfide bond.
Additionally, BnRALF38 and BnRALF43 of clade III contained a 53aa and 25aa extension at their C termini (Table 1 and Figure 2A). These extended sequences did not possess known motifs based on prediction analyses using SMART and Pfam tools. Whether these sequences play a role in the functions of these BnRALFs requires further study.
Collectively, BnRALF sequences are generally conserved within clades but diverse among clades, suggesting that they might be widely involved in a variety of biological processes.
Expression Patterns of BnRALF Genes in Response to Sclerotinia sclerotiorum Infection and BnPep5 and SsNLP1 Treatments
To obtain hints for functions of BnRALF genes in immunity, we systematically examined the expression patterns of all 61 BnRALF genes in response to pathogen (S. sclerotiorum) infection and DAMP (BnPep5) and PAMP (SsNLP1) treatments via quantitative real-time PCR (qRT-PCR) analysis. BnPep5, a B. napus plant elicitor peptide, was identified by our lab via BLASTp search using well-characterized Arabidopsis Peps as query sequences (unpublished data), and SsNLP1 was identified by our lab from S. sclerotiorum necrosis and ethylene-inducing peptide 1 (SsNep1) (Bashi et al., 2010, our unpublished data). Both BnPep5 and SsNLP1 could elicit immunity to S. sclerotiorum in B. napus (unpublished data). To compare the BnRALF expression patterns in response to different treatments, we converted the qRT-PCR gene expression values into a heatmap on the basis of the log2-fold change-derived data (Figure 3). Comparison of BnRALF expression profiles in response to S. sclerotiorum at 0 and 7 h post inoculation (hpi) showed that the expression of most BnRALFs were up-regulated by S. sclerotiorum infection. Among them, the expression level of BnRALF10, −15, −16, −19, and −51 was most up-regulated by over 10-fold. BnRALF6 was the only one whose expression was significantly down-regulated, by 6.6 folds (Figure 3). This result suggested that these BnRALF genes may be involved in the regulation of the interaction between B. napus and S. sclerotiorum.
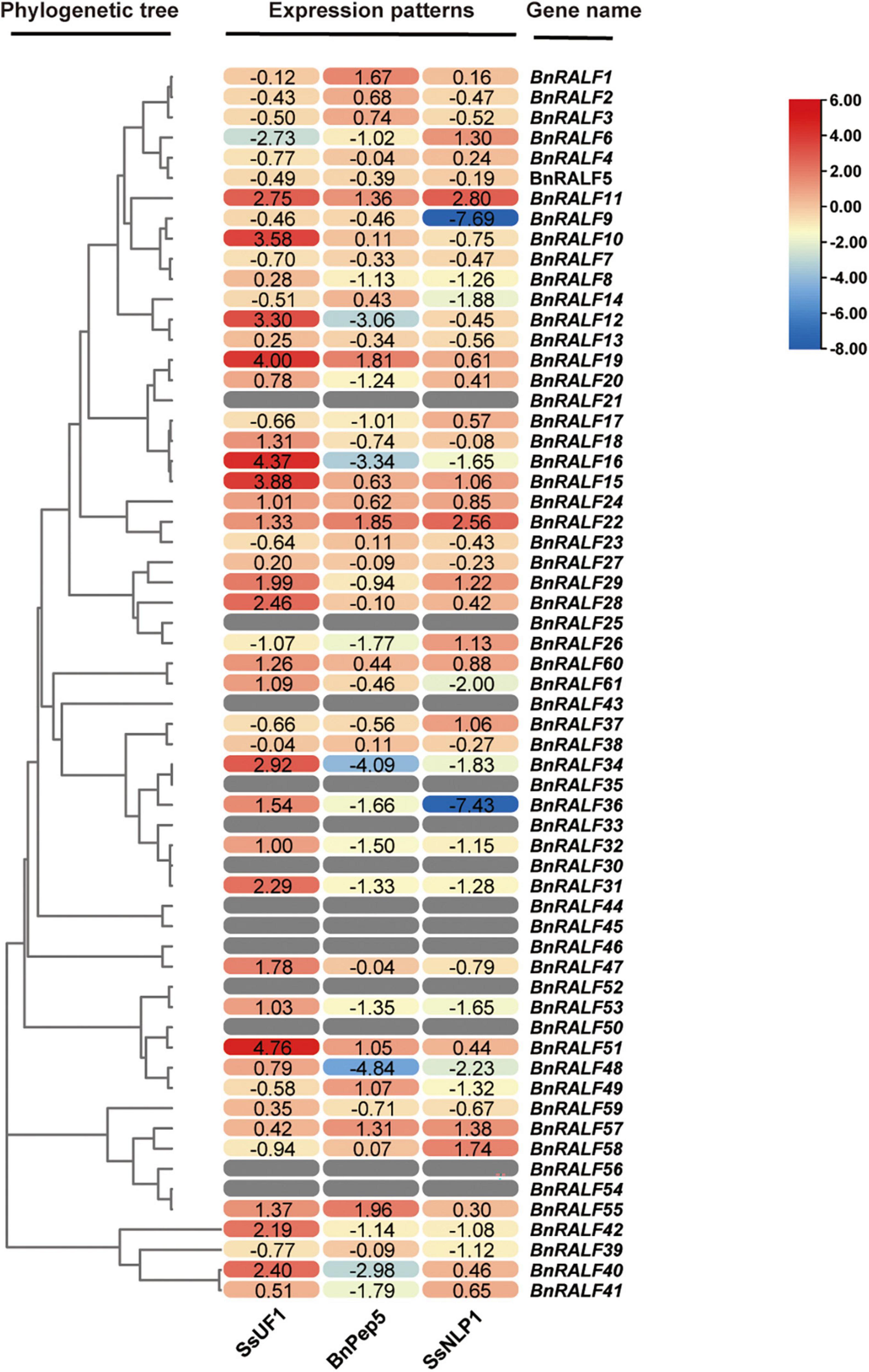
Figure 3. Expression patterns of the whole BnRALF gene family in response to different stimuli. The heatmap was constructed by TBtools to present expression patterns of the 61 BnRALF genes in oilseed rape leaves inoculated with S. sclerotiorum strain UF1 (SsUF1) or infiltrated with 0.2 μM BnPep5 (B. napus plant elicitor peptide 5) or 1 μM SsNLP1 [S. sclerotiorum ethylene-inducing peptide 1 (SsNep1)-like peptide 1] peptide solution (n = 3 for each group). PDA plug mock inoculation was served as the control for the SsUF1 inoculation, while ddH2O infiltration was conducted as the control for the BnPep5 and SsNLP1 infiltration treatments. The expression levels from low to high are indicated by a change in color from blue to red. The numbers in the figure are represented as log(2)-fold change of treated samples versus mock control. These experiments were performed three times, each yielding similar results.
In contrast, more BnRALF genes were expressionally down-regulated in response to DAMP and PAMP treatments. At 2 h post treatment with 200 nM BnPep5, only BnRALF1, −19, −22, and −55 were expressionally up-regulated by over threefold, while BnRALF12, −16, −34, −40, and −48 were expressionally decreased by 5∼28 folds. At 2 h post treatment with 1 μM SsNLP1, expression of BnRALF9 and BnRALF36 were extremely reduced by over 100-fold, while that of BnRALF11 and BnRALF22 were enhanced by fivefold and sevenfold, respectively (Figure 3). These genes were strongly responsive to DAMP/PAMP treatments and thus may play an important role in plant immunity to S. sclerotiorum.
The expression of 13 BnRALF genes, BnRALF21, −25, −30, −33, −35, −43, −44, −45, −46, −50, −52, −54, and −56 could not be detected no matter under normal growth conditions or after treated with three stimuli in our assays (Figure 3), implying their expression was extremely low, if any in leaves.
Taken together, the expression patterns of the whole BnRALF family indicate that BnRALF members might be differentially involved in immunity to S. sclerotiorum.
BnRALF10 Elicits Resistance to Sclerotinia sclerotiorum
Our observation that the expression of BnRALF10 was strongly up-regulated by 10-fold in response to S. sclerotiorum inoculation (Figure 3) and the reported fact that its Arabidopsis ortholog AtRALF22 stimulated immune responses such as ROS accumulation (Abarca et al., 2021) prompted us to perform functional analyses of BnRALF10 in immunity to S. sclerotiorum. The Turnip yellow mosaic virus (TYMV)-based virus induced gene silencing (VIGS) technology was employed to knock-down BnRALF10 in B. napus. A typical photobleaching symptom (caused by silence of the BnPDS gene) in B. napus plants bombarded with pTY-BnPDS was observed on the newly emerged leaves 14 days after treatment (Figure 4A), indicating that TYMV-mediated gene silencing was effective in B. napus. Meanwhile, the plants treated with pTY-S (empty vector) showed normal growth (Figure 4A), suggesting that the TYMV-based vector infection did not obviously affect plant vegetative growth. In plants bombarded with pTY-BnRALF10, the recombinant pTY vector containing both sense and antisense versions of a specific 40 bp fragment from the coding region of BnRALF10 so that to form its self-hybridized palindromic fragment, BnRALF10 expression was significantly reduced (Figure 4B). Additionally, expression level of the closest homologs of BnRALF10 (BnRALF8, BnRALF9, BnRALF11), which were clustered in the same clade with BnRALF10, but not less close homologs of BnRALF10 (BnRALF12 and BnRALF13), which were not clustered in the same clade with BnRALF10, was also decreased in the pTY-BnRALF10-silence treated plants (Supplementary Figure 1). Compared with the plants treated with pTY-S, the oilseed rape plants with decreased BnRALF10-clade gene expression exhibited more severe necrosis, larger disease areas and increased S. sclerotiorum biomass (Figure 4C). This indicated that silencing of BnRALF10 and its clade homologs increased susceptibility to S. sclerotiorum challenge.
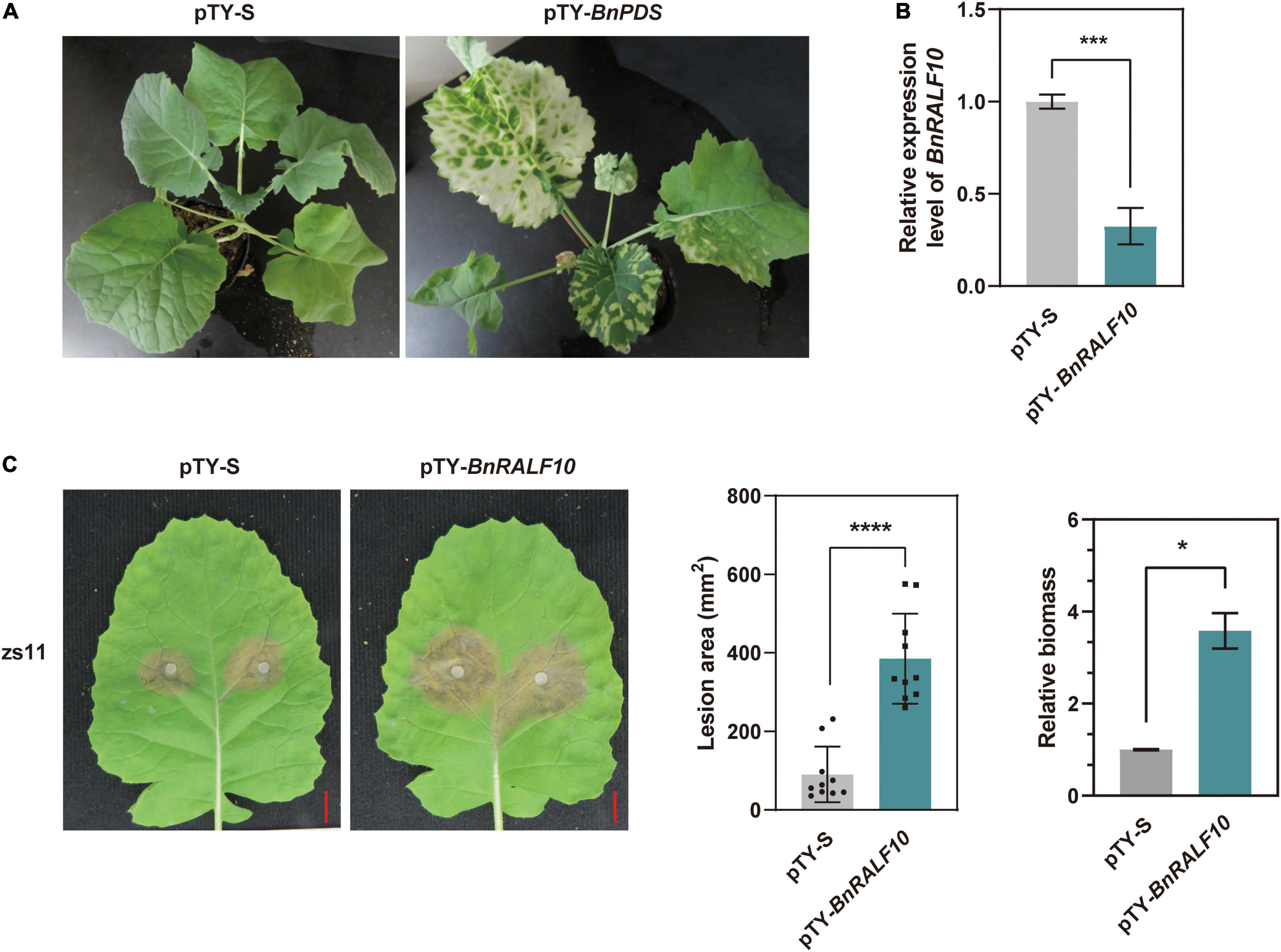
Figure 4. Silencing of BnRALF10 reduced resistance to S. sclerotiorum in oilseed rape. (A) Photobleaching phenotype of BnPDS silencing in oilseed rape employing pTY-based VIGS. Empty vector pTY-S was used as a control. (B) Efficient silencing of BnRALF10 in leaves as manifested by the dramatically reduced level of BnRALF10 transcript, which was detected by qRT-PCR analysis using B. napus actin (BnActin7) as reference gene. (C) Representative disease symptoms (left), lesion areas (middle) and fungal biomass (right) of BnRALF10-silenced leaves at 24 h post inoculation with S. sclerotiorum. Scale bar: 1 cm. Data are statistically analyzed by Student’s t-test (n = 10) and shown as the mean ± SE. The asterisks indicate significant differences (*P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001).
To further confirm the role of BnRALF10 in resistance to S. sclerotiorum. We synthesized the mature BnRALF10 peptide comprising the conserved YISY-motif and four conserved cysteines. The oilseed rape leaves were preinfiltrated with 1 μM BnRALF10 peptide solution, and were then challenged with S. sclerotiorum 12 h and 24 h later, respectively. BnRALF10 treatment for 12 h enhanced the resistance against S. sclerotiorum, as indicated by the weaker disease symptom and reduced lesion size in leaves treated with BnRALF10 compared to the control plant leaves (Figure 5). However, at 24 h post infiltration, the plants treated with BnRALF10 exhibited similar disease symptom and lesion size to the control plants (Figure 5). This result suggests that resistance elicited by BnRALF10 is not long-lasting.
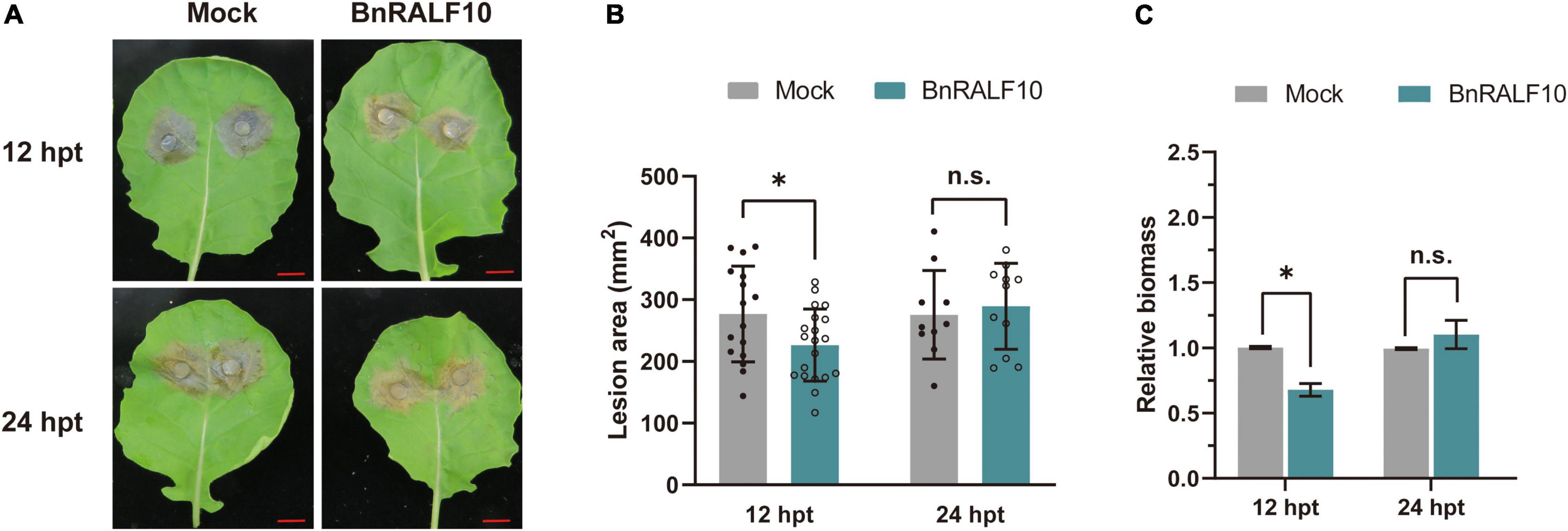
Figure 5. BnRALF10 peptide induced plant resistance to S. sclerotiorum. Representative disease symptoms (A), lesion area (B) and fungal biomass statistical analysis (C) of oilseed rape leaves pretreated with 1 μM BnRALF10 peptide solution or ddH2O (Mock) at 12 h and 24 h post inoculation with S. sclerotiorum. Scale: 1 cm. Data are shown as the mean ± SE. The asterisks indicate significant differences from the control, as determined by one-way ANOVA (n = 7 leaves, n.s., not significant, *P ≤ 0.05). The experiments were repeated three times with similar results.
Taken together, these results demonstrate that BnRALF10 stimulates resistance to S. sclerotiorum in B. napus.
BnRALF10 Activates Plant Immune Responses
Given that BnRALF10 expression was rapidly and strongly induced by S. sclerotiorum, and that BnRALF10 stimulated resistance against this pathogen, we inferred that BnPRORALF10 might encode a peptide that act as a DAMP. To confirm this, we analyzed the functions of BnRALF10 in activating early immune responses such as ROS production, [Ca2+]cyt promotion, MAPK activation and defense gene expression promotion. L-012-based leaf disk assay demonstrated that supply with 1 μM BnRALF10 peptide strongly induced extracellular ROS accumulation (Figure 6A). Further, we used the transgenic Arabidopsis seedlings carrying the calcium reporter aequorin to measure [Ca2+]cyt after treatment with 1 μM BnRALF10. A rapid [Ca2+]cyt elevation was observed after BnRALF10 treatment (Figure 6B). Moreover, MAPK assay in Arabidopsis seedlings revealed that MAPK3 and MAPK6 was promptly activated upon stimulation with BnRALF10 (Figure 6C). In addition, we examined transcriptional induction of genes closely linked to defense. Expression of BnWRKY33, BnACS6 and BnVSP1 was strongly induced by 12∼21-fold at 1 h post BnRALF10 treatment, while that of BnPDF1.2, BnWRKY70, BnPR1 and BnPAL was not obviously altered (Figure 6D).
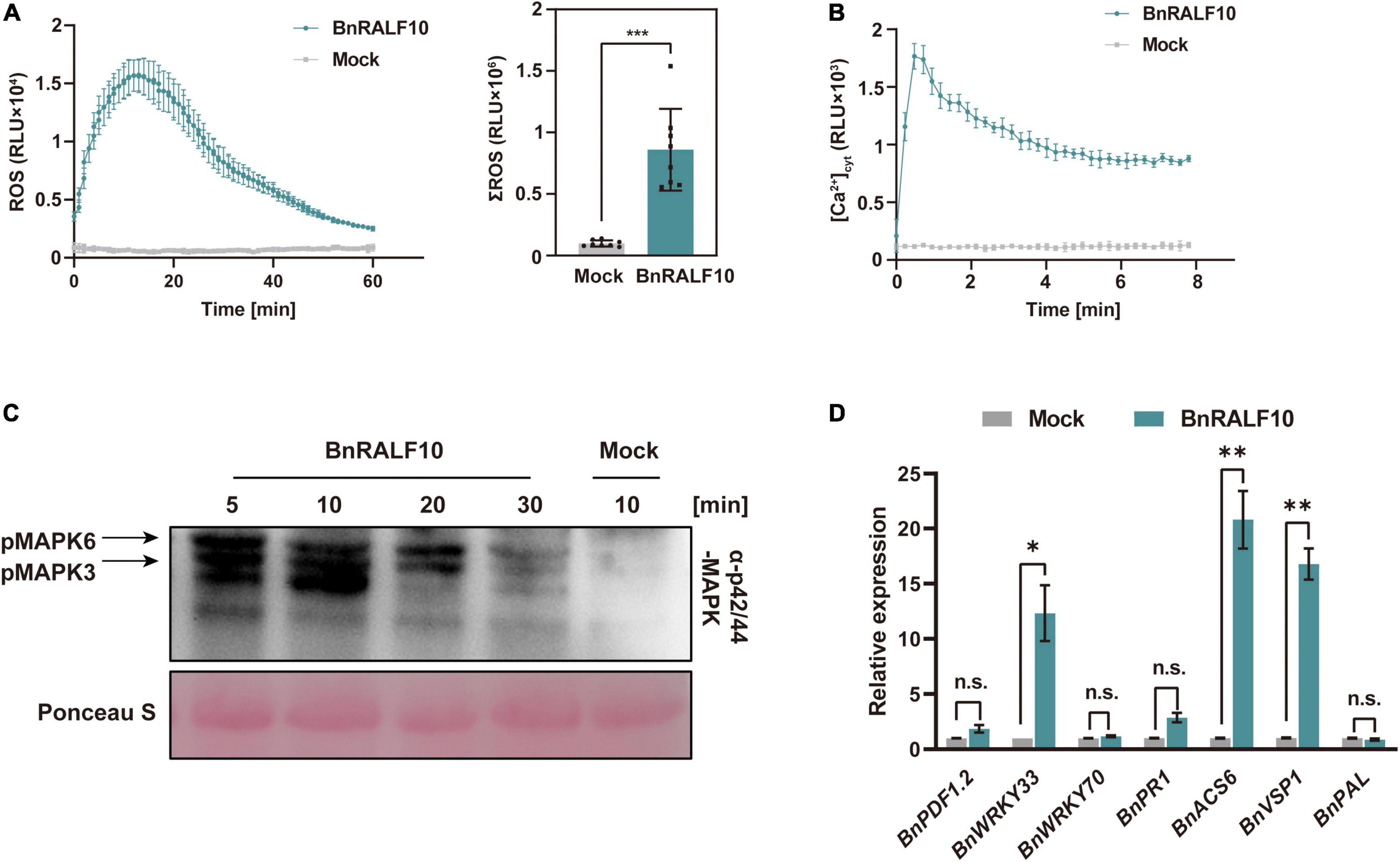
Figure 6. BnRALF10 elicited various immune responses. (A) Reactive oxygen species (ROS) measured in oilseed rape leaf disk assay after addition with 1 μM BnRALF10 peptide or water (Mock). Shown are the dynamics of ROS production (left) and their total amount (right) as mean values of total photon counts over 60 min. Data are shown as the mean ± SE (n = 6–8, ***P ≤ 0.001, Student’s t-test). RLU, relative light units. All experiments were repeated three times with similar results. (B) Dynamics in [Ca2+]cyt stimulated by 1 μM BnRALF10 peptide in Arabidopsis. The 5-day-old seedlings expressing Aequorin gene were treated with solutions containing 1 μM BnRALF10 peptide or mock. Signals were recorded for 8 min after treatment. Data are shown as the mean ± SE (n = 8). (C) MAPK activation induced by BnRALF10. Seven-day-old seedlings were exposed to 1 μM BnRALF10 peptide for 5, 10, 20 or 30 min. Western blot analysis was performed with the phospho-p44/42 MAPK antibody. Ponceau S was used as loading control. (D) Transcription of defense responsive genes in B. napus leaves treated with 1 μM BnRALF10 for 1 h. Data are shown as the mean ± SE (n = 3, *P ≤ 0.05, **P ≤ 0.01, Student’s t-test). n.s., not significant.
Taken together, these results indicate that BnRALF10 elicits diverse immune responses as a DAMP in B. napus, and triggers DTI (DAMP-triggered immunity) against the necrotrophic pathogen S. sclerotiorum.
BnFER Is a Receptor for BnRALF10
Holding ascertained that BnRALF10 modulates S. sclerotiorum resistance, we sought to investigate the molecular mechanism behind it. BnRALF10 is the close homolog of Arabidopsis RALF22 and RALF23, which are recognized by AtFER (Stegmann et al., 2017; Zhao et al., 2018). Therefore, we wondered whether the B. napus homolog of AtFER was the receptor of BnRALF10. To verify this, we identified BnFER and performed functional analyses for it. BLASTp search using AtFER protein sequence as query retrieved a B. napus homolog of AtFER (BnFER) in cultivar Zhongshuang 11 (ZS11) (Supplementary Figure 2). Domain composition analysis showed that like AtFER, BnFER consisted of two extracellular malectin domains, a transmembrane domain and an intracellular kinase domain (Supplementary Figure 2).
Next, we employed three methods to discern the physical interaction between BnRALF10 and BnFER. Y2H assay showed that BnRALF10 physically interacted with the extracellular domain of BnFER (ectoBnFER) in yeast (Figure 7A). We further conducted BiFC and LCI assay to validate the interaction between BnRALF10 and BnFER in planta. A strong Yellow Fluorescent Protein (YFP) signal was observed at the plasma membrane when BnFER-nYFP (BnFER fused with the N-terminal fragment of YFP) was co-expressed with BnRALF10-cYFP (BnRALF10 fused with the C-terminal fragment of YFP), whereas no YFP signal was detected in leaves expressing BnFER-nYFP and cYFP (Figure 7B). Moreover, co-expression of BnFER-NLuc (BnFER fused with the N-terminal fragment of Luciferase) with BnRALF10-CLuc (BnRALF10 fused with the C-terminal fragment of Luciferase), but not that of BnFER-NLuc and CLuc, NLuc and BnRALF10-CLuc, or NLuc and CLuc, resulted in strong LUC (Luciferase) activity (Figure 7C). These results verify that BnRALF10 indeed can interact with BnFER in the plasma membrane.
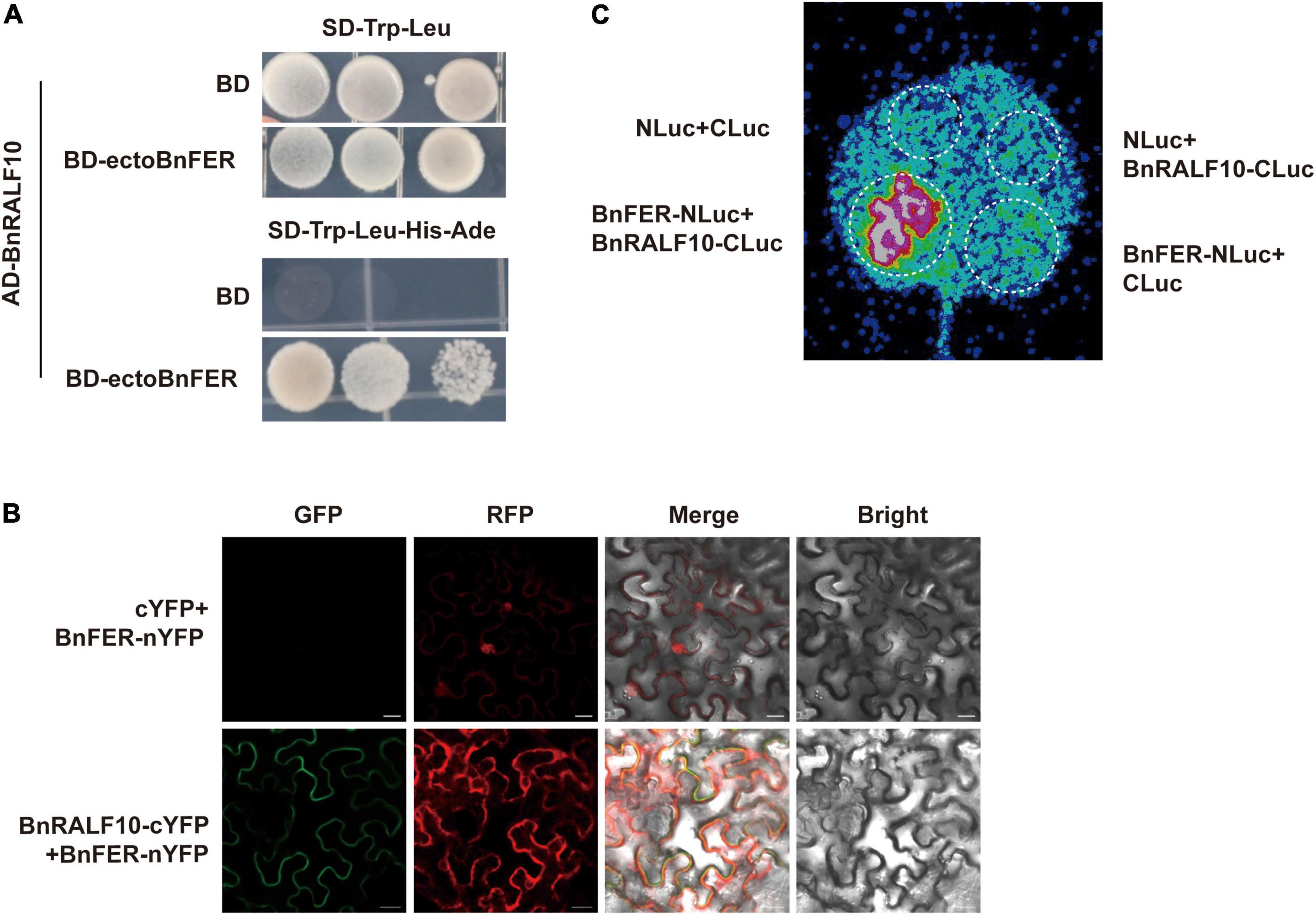
Figure 7. BnRALF10 physically interacted with BnFER. (A) BnRALF10 interacts with the extracellular domain of BnFER in yeast. Yeast (strain AH109) co-transformed with pGBKT7 (BD) or the BD-ectoBnFER bait vector and pGADT7 (AD)-BnRALF10 prey vector was grown on SD–Trp–Leu, SD–Trp–Leu–His–Ade plates for interaction confirmation. Only yeast co-transformed with BD-ectoBnFER and AD-BnRALF10 grew on the SD–Trp–Leu–His–Ade medium. BD + AD-BnRALF10 was used as negative control. (B) BiFC assay to detect BnRALF10–BnFER interaction in N. benthamiana. YFP fluorescence was detected in N. benthamiana leaves co-expressing BnRALF10-cYFP and BnFER-nYFP (Lower) but not in the control (cYFP + BnFER-nYFP; Upper). Scale bar: 20 μm. (C) LCI assay to monitor BnRALF10–BnFER interaction in N. benthamiana. LUC activity was detected in N. benthamiana leaves co-expressing BnRALF10-CLuc and BnFER-NLuc but not in the controls (BnRALF10-CLuc + NLuc, CLuc + BnFER-NLuc and CLuc + NLuc).
Further, we investigated whether BnFER is involved in BnRALF10 signaling and defense against S. sclerotiorum. pTY-S-based VIGS technology was used to silence BnFER in B. napus. In VIGS plants treated with pTY-BnFER, BnFER expression was significantly reduced (Figure 8A). These rape plants with decreased expression of BnFER accumulated only 45.5% of BnRALF10-elicited ROS compared with pTY-S-treated control plants (Figure 8B), suggesting that BnFER is required for BnRALF10 to stimulate immune responses including oxidative burst. When inoculated with S. sclerotiorum, these BnFER knock-down plants exhibited more severe symptoms with significantly larger lesions and two-fold increased pathogen biomass compared to the control plants (Figure 8C), indicating that BnFER is required for BnRALF10 to elicit resistance to S. sclerotiorum.
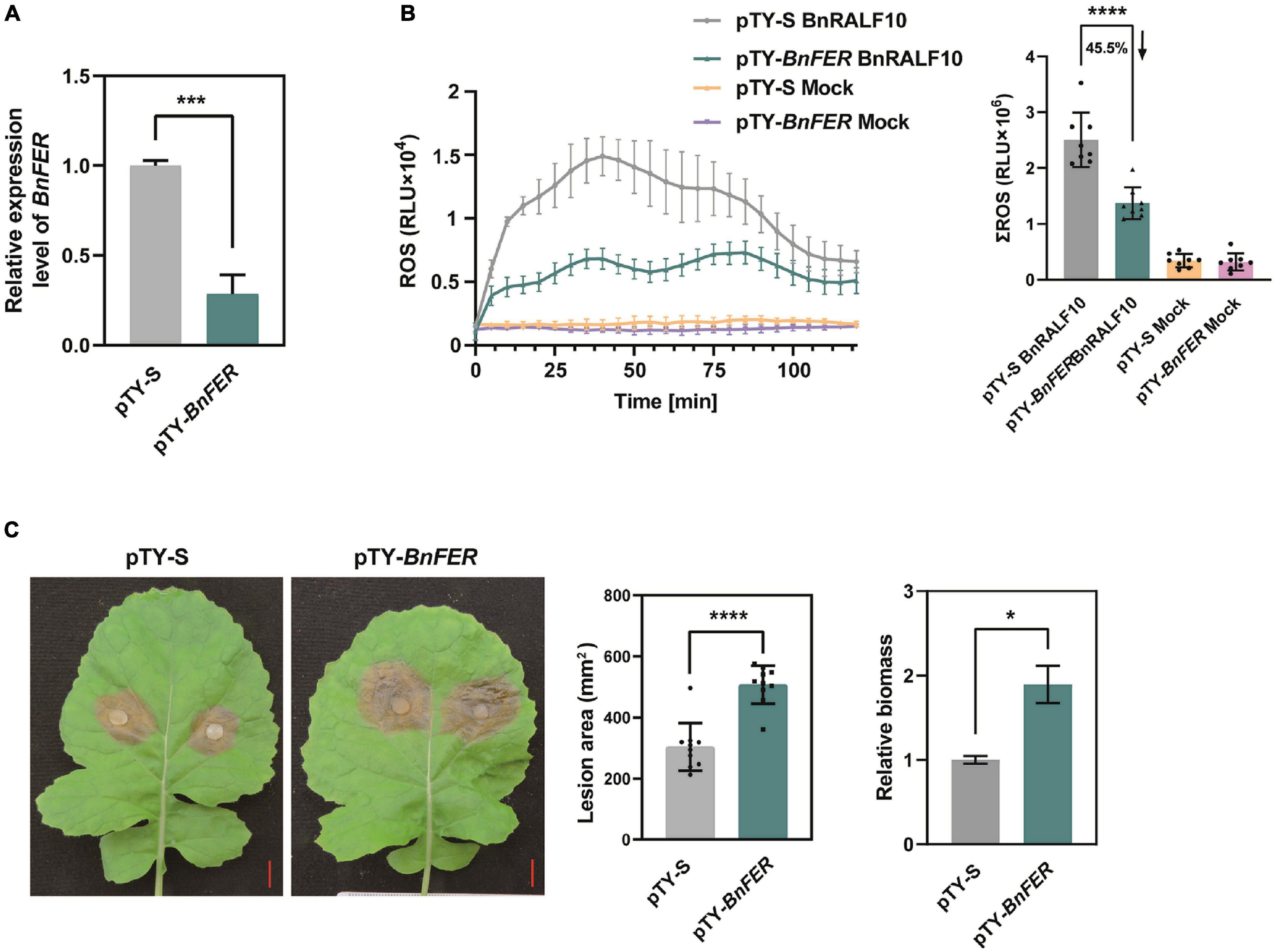
Figure 8. BnFER was required for BnRALF10 to function. (A) Efficient silencing of BnFER by pTY-BnFER treatment in rape leaves as manifested by the vastly reduced level of BnFER transcript, which was detected by qRT-PCR analysis using B. napus actin (BnActin7) as reference gene. Data are shown as the mean ± SE (n = 7, ***P ≤ 0.001, Student’s t-test). (B) Effect of BnFER-silencing on BnRALF10-induced ROS burst. ROS measured in rape leaf disks of BnFER-silenced and control plants after addition with 1 μM BnRALF10 peptide or water (mock). Shown are the dynamics of ROS production (left) and the integration (right) as mean values of total photon counts over 120 min. Data are shown as the mean ± SE (n = 8, ****P ≤ 0.0001, Student’s t-test). (C) Effect of BnFER-silencing on BnRALF10-induced resistance to S. sclerotiorum. Leaves of BnFER-silenced and control plants were inoculated with S. sclerotiorum. Disease symptoms (left), lesion area (middle) and fungal biomass in inoculated leaves (right) at 24 hpi were shown. Data are shown as the mean ± SE (n = 7, *P ≤ 0.05, ****P ≤ 0.0001, Student’s t-test).
Collectively, our results support BnFER to be a receptor for BnRALF10.
Proteomic Analysis Reveals the Molecular Basis of BnRALF10-Induced Plant Immunity
To reveal the possible mechanisms associated with the plant immunity stimulated by BnRALF10, we performed quantitative proteomic analysis to identify the proteins involved in this immunity. Total protein was extracted using the trichloroacetic acid (TCA)-acetone precipitation method for comparative proteomic analysis of the B. napus leaves at 4 h post infiltration with either 1 μM BnRALF10 or ddH2O as control. Principal components analysis (PCA) was performed to evaluate the test samples, and the result showed that the different groups of samples were well-distinguished, while three replicate samples of each group were clustered (Supplementary Figure 3), suggesting that the samples were highly qualified for proteomic analysis. We set the criteria for significantly differentially expressed proteins (DEPs) as that p < 0.05 and fold change >2 or <2 in three biological replicates. Based on these criteria, we identified 314 DEPs differentially expressed between leaves infiltrated with 1 μM BnRALF10 and ddH2O (Supplementary Table 2), including 163 up-regulated and 151 down-regulated proteins (Figure 9A).
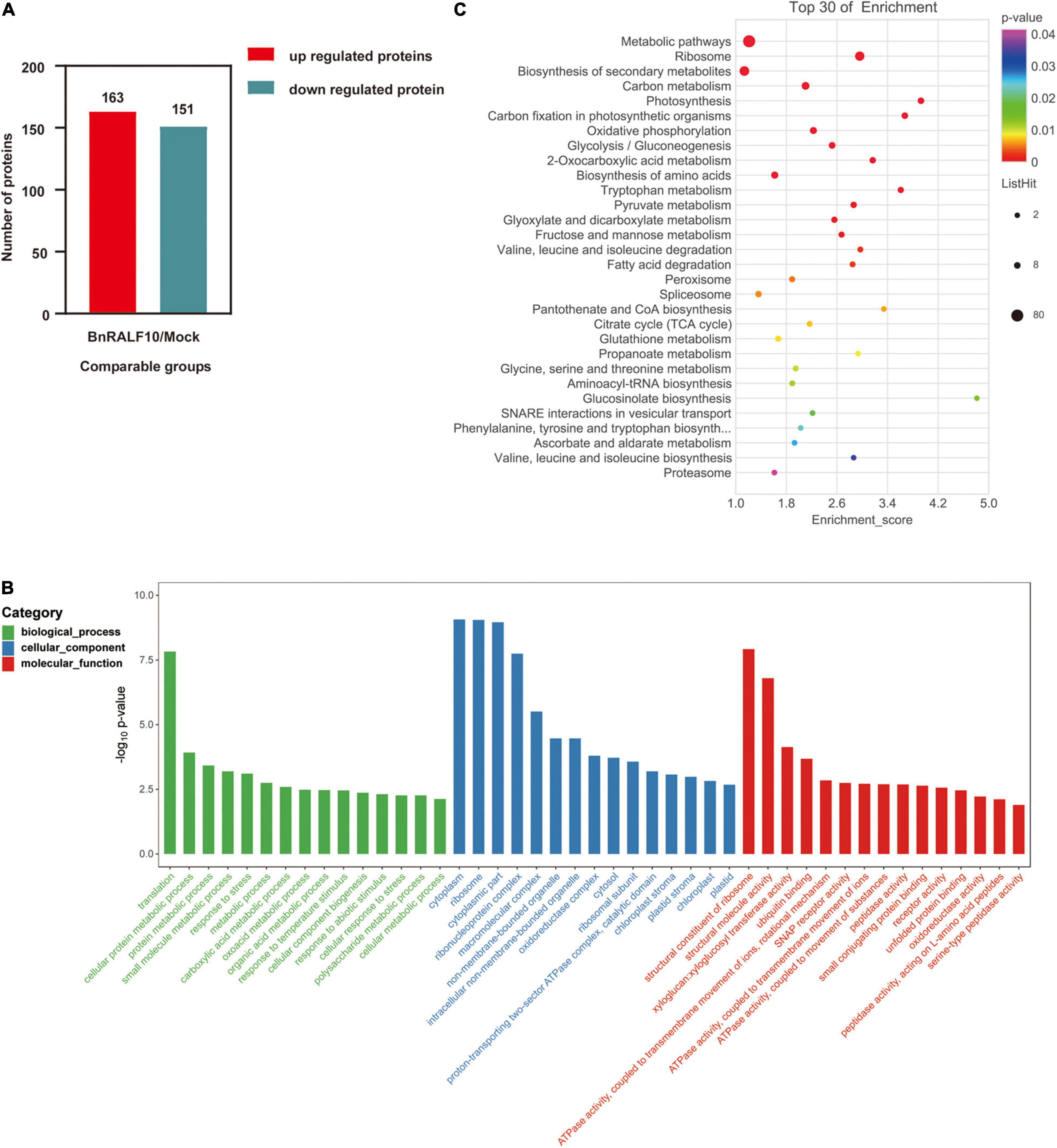
Figure 9. Functional categorization of proteins differentially expressed between oilseed rape leaves infiltrated with 1 μM BnRALF10 and ddH2O. The expression trend (A), GO and KEGG analyses of differentially expressed proteins (B,C) (p < 0.05 and fold change >2 or <2) were presented. Leaves of 4-week-old oilseed rape plants were sampled at 4 h after infiltration with ddH2O and 1 μM BnRALF10, respectively. The bubble size represents the number of proteins, and the bubble color means the p-value of the significance. The pathway enrichment statistical analysis was performed by Fisher’s exact test. Thirty pathways showing the lowest p-value were selected for this analysis.
GO annotation and KEGG pathways for DEPs were analyzed to understand the BnRALF10-stimulated response events and downstream biological processes. GO analysis showed that these DEPs belonged to 460 GO terms for 170 biological processes, 179 cellular components and 111 molecular functions (Supplementary Table 3). Among the main categories with the lowest p value in biological process (BP), the interesting terms included response to stress, response to abiotic stimulus and cellular response to stress, while the interesting molecular function (MF) comprised ubiquitin binding, ATPase activity coupled to transmembrane movement of ions and substances, SNAP receptor activity, receptor activity and peptidase activity (Figure 9B). KEGG pathway annotation analysis revealed that the DEPs were associated with 87 pathways (Supplementary Table 4). Among the 30 pathways with the lowest p value, the interesting pathways were the metabolic pathway, biosynthesis of secondary metabolites, peroxisome, glutathione metabolism, glucosinolate biosynthesis and proteasome (Figure 9C).
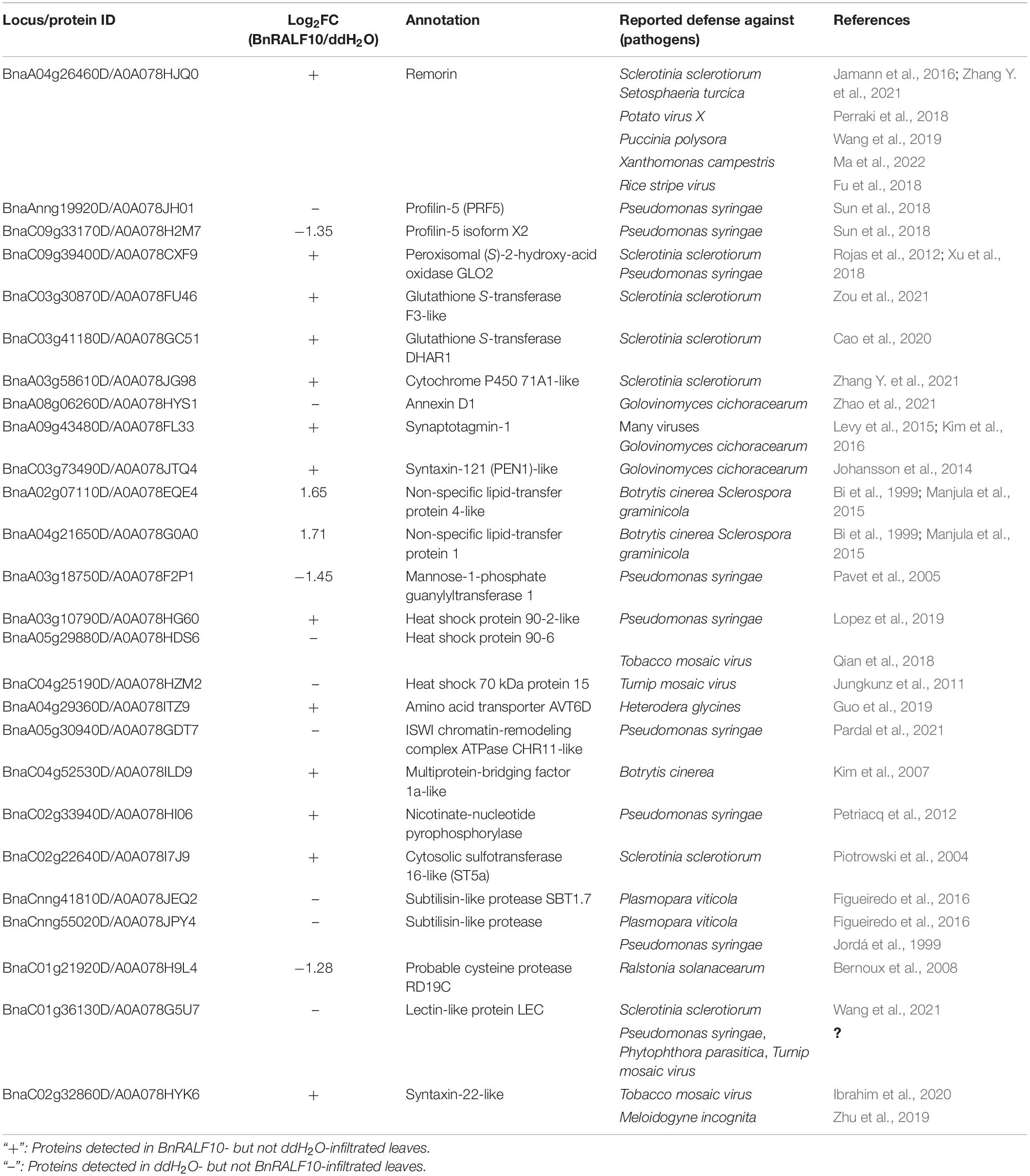
Intriguingly, dozens of DEPs have been reported to play a role in defense against various pathogens including S. sclerotiorum (Table 2). Here, we tentatively term the BnRALF10-elicited defense proteins as RED proteins. Among the REDs were a group of proteins involved in ROS generation and homeostasis, including ROS generator peroxisomal (S)-2-hydroxy-acid oxidase GLO2, and oxidoreduction regulators such as two glutathione S-transferase (GST) proteins and cytochrome P450 71A1. Remarkably, all these REDs were reported to function in plant defense against S. sclerotiorum (Table 2), demonstrating that BnRALF10 might tune GLOs- and GSTs-mediated ROS accumulation for immunity to S. sclerotiorum.
A remorin (REM) and two profilins (PRFs) are also among the RED proteins. REM positively while PRFs negatively regulate formin-mediated actin assembly to create membrane compartments for immune receptor complex formation and promote immune signal transduction thereby elicit plant immunity (Sun et al., 2018; Ma et al., 2022). Coincidentally, BnRALF10 increased REM accumulation while decreased PRFs accumulation (Table 2), and they are involved in plant immunity against diverse pathogens including S. sclerotiorum (Table 2), revealing that BnRALF10 likely fine-tunes REM- and PRFs-mediated actin cytoskeleton to stimulate plant immunity.
Some REDs exhibit Ca2+-dependent functions in plant defense. Among them were an annexin (ANN) and a synaptotagmin (SYT). AtSYT1 physically interacts with AtSYP121/PEN1, another RED, to modulate exocytosis for localization of immune proteins to plasma membrane (PM). AtANN8 and AtSYT1 regulate plant immunity to fungal and viral pathogens (Levy et al., 2015; Kim et al., 2016; Zhao et al., 2021). BnRALF10 strikingly changed the accumulation level of a variety of calcium sensors such as ANNs, SYTs, IQ-domain proteins (IQDs), CBL-interacting serine/threonine-protein kinase 4 (CIPK4), E3 ubiquitin-protein ligase HOS1-like protein, 14-3-3-like protein GF14 omega, serine/threonine-protein kinase STN7 and syntaxin 22-like protein (Supplementary Table 2). This result implies that BnRALF10 might rely heavily on calcium pathway to fine tune plant immunity.
Other RED proteins included two transcriptional regulators CHR11 (chromatin-remodeling protein 11) and MBF1a (multiprotein bridging factor 1a), two LTPs (lipid transfer proteins), three HSPs (heat shock proteins), two SBTs (subtilisin-like proteases), cysteine protease RD19C, sulfotransferase ST5a, mannose-1-phosphate guanylyltransferase 1, glucan endo-1,3-beta-glucosidase, syntaxin-22-like and lectin-like proteins (Table 2).
Collectively, the proteomic analysis results reveal that BnRALF10 likely modulates the abundance of RED proteins to fine-tune plant immunity against pathogens including S. sclerotiorum.
Discussion
Rapid Alkalinization Factor Family in Oilseed Rape
Genome-wide identification of RALFs in cruciferous species has been performed in Arabidopsis thaliana and Brassica rapa. Consequently, 37 and 32 RALF genes were identified in A. thaliana and B. rapa, respectively (Campbell and Turner, 2017; Abarca et al., 2021). In the present study, we identified 61 RALF genes in B. napus, another cruciferous species (Table 1). The RALF gene copy number in B. napus is much higher than that in A. thaliana and B. rapa although the three species belong to the same family. B. napus is an allotetraploid from crossing between B. oleracea and B. rapa, followed by chromosome doubling (Chalhoub et al., 2014). The copy number of RALF genes in B. napus almost doubles that in B. rapa, implying that the divergence of the RALF family may occurred as chromosome doubling. Brassica species underwent whole genome triplication (WGT) after the divergence of the Brassica ancestor and the genus Arabidopsis, which includes A. thaliana (Cheng et al., 2017), while the number of RALF genes in B. napus is not triple of that in A. thaliana. Considering that B. napus is tetraploid while A. thaliana is diploid, it is likely that the evolution of RALF genes was caused by segmental duplication.
Phylogenetic analysis for RALFs in B. napus and A. thaliana showed that these 98 RALFs diverged into four clades (Table 1 and Figure 1). Among them, clades I and II can be considered the canonical RALFs, since they contain the features previously depicted to be characteristic of the RALF family, including the N-terminal signal peptide cleavage site, S1P cleavage site RRXL, YISY motif and C-terminal four conserved cysteines (Campbell and Turner, 2017). BnRALF genes which strongly responded to S. sclerotiorum inoculation belonged to these two clades except BnRALF51 (Figure 3), indicating that the canonical RALF proteins may play a major role in the resistance to S. sclerotiorum. As matter of fact, BnRALF10, a member of clade I, indeed induced the resistance to S. sclerotiorum (Figures 4, 5), further supporting the important roles of canonical BnRALFs in immunity against the necrotrophic pathogen S. sclerotiorum.
RRXL dibasic site has been reported to be recognized by S1P and necessary for maturing of PRORALFs (Matos et al., 2008; Stegmann et al., 2017). Twenty-three BnRALFs lacked canonical RRXL site (Table 1 and Figure 2). They may execute either no further processing or one independent of S1P. All 16 clade III and two clade IV BnRALF proteins were significantly smaller than typical BnRALFs of clades I and II, rather similar to their mature peptides, suggesting that these 18 BnRALFs likely undergo no further cleaving processing. The remaining five, BnRALFs 46, 47, 51, 52 and 53, all belonging to clade IV, have a size similar to typical BnRALFs, indicating that they might perform further maturation processing and were likely cleaved in a RRXL-independent mechanism. Noticeably, they carry a single R in the corresponding RRXL position of typical BnRALFs, HRIL in BnRALF52 and BnRALF53, RKIL in BnRALF51, QRFT in BnRALF46 and QRLT in BnRALF47. Interestingly, monobasic cleavage sites have been found in proteins from vertebrates and insects (Veenstra, 2000), suggesting that these BnRALFs might be cleaved by proteases recognizing monobasic cleavage sites. Whether this is indeed the case as in vertebrates and insects awaits experimental verification. The S1P cleavage site seems to be important for the physiological function of RALFs. Arabidopsis RALF23 and its relative RALF33, with RRXL sites, are proteolytically cleaved by S1P and play negative roles in plant immunity, while non-S1P cleaved RALF peptide AtRALF17 is devoid of a propeptide region, positively regulates plant immunity (Stegmann et al., 2017). AtRALF22 regulates salt stress in a S1P-dependent way (Zhao et al., 2018). BnRALF10 contains the conserved RRXL site (Table 1). BLASTp search using AtS1P as query retrieved three B. napus orthologs of AtS1P (BnS1Ps) (Supplementary Figure 4A), which have the same domain composition with AtS1P, both containing the peptidase_S8 domain (Supplementary Figure 4B). The high similarity between AtS1P with the putative BnS1Ps implies that they may share the same functions. Together, it is highly possible that the precursor of BnRALF10 is processed in the RRXL site to release a C-terminal mature peptide by the putative BnS1Ps. Whether the maturation of BnRALF10 and its role in inducing resistance to S. sclerotiorum indeed depends on the S1P cleavage site and the putative BnS1Ps, is worth further study.
Rapid alkalinization factor was first associated with the plant defense response for its ability in MAPK activation (Pearce et al., 2001). Later, it was shown to be able to elicit ROS burst (Thynne et al., 2017), cytoplasmic Ca2+ spikes and apoplastic pH ascending in Arabidopsis root cells (Gjetting et al., 2020), which are hallmarks of defense activation by plants. Here we found that the BnRALF10 peptide, derived from the C-terminus of BnPRORALF10, could stimulate ROS burst, intracellular Ca2+ elevation, MAPK activation and defense-related gene expression induction (Figure 6). The transcriptional level of BnRALF10 was significantly enhanced by fungal infection (Figure 3), and pretreatment of BnRALF10 could induce resistance to S. sclerotiorum in oilseed rape leaves (Figure 5). These effects of BnRALF10 are comparable with AtPep1, the well-characterized DAMP (Bartels and Boller, 2015), indicating that BnRALF10 functions as a DAMP. Notably, the role of RALFs in plant defense reported to date is all negative. For example, AtRALF23 inhibits plant resistance to biotrophic pathogen Pseudomonas syringae through its receptor FER in a S1P-dependent manner (Stegmann et al., 2017); It has also been shown to stabilize the basic helix-loop-helix transcription factor MYC2 and promote JA signaling via FER, and play a negative role in plant immunity to P. syringae (Guo et al., 2018). Silencing of the Fragaria × ananassa RALF-33-like gene in red fruits of strawberry led to a delay in fruit colonization by fungal pathogen Colletotrichum acutatum (Merino et al., 2019). F-RALF from Fusarium oxysporum promotes fungal virulence and suppresses plant immune responses through the FER receptor kinase (Masachis et al., 2016); RALF-like peptides secreted by root-knot nematode (RKN) facilitate parasitism through the plant receptor FER (Zhang X. et al., 2020); RALF-like 1 from RKN uses soybean receptor kinase GmLMM1 as susceptible target to promote parasitism in soybean (Zhang X. et al., 2021). Intriguingly, here, we provide evidence that BnRALF10 interacts with BnFER and knock-down of BnFER renders BnRALF10 induce less ROS and lower resistance to S. sclerotiorum, revealing that BnRALF10, when recognized by BnFER, plays a positive role in resistance to the necrotrophic fungal pathogen S. sclerotiorum. Our results demonstrate that RALFs plays distinct roles in plant immunity.
Mechanisms Underlying BnRALF10-Stimulated Immunity
Although great insights have been unveiled for the maturation and recognition of RALFs (Haruta et al., 2014; Stegmann et al., 2017; Xiao et al., 2019), the signaling downstream the recognition remains largely unknown. To understand how BnRALF10 stimulates plant immunity to S. sclerotiorum, we performed quantitative proteomics analysis to detect the BnRALF10-elicited defense (RED) proteins, which significantly change in protein level between oilseed rape leaves pretreated with ddH2O and BnRALF10 peptide. Interestingly, among the 314 differentially expressed proteins (DEPs) (Supplementary Table 2), dozens of functionally proved RED proteins were identified and notably some novel potential defense mechanisms underlying BnRALF10-stimulated immunity against S. sclerotiorum were revealed (Table 2).
One of our interesting findings is that BnRALF10-stimulated immunity is associated with well positioning anchoring of immune proteins to plasma membrane (PM) via synergy of synaptotagmin 1 (SYT1) and syntaxin 121 (SYP121/PEN1). SYP121/PEN1 encodes a syntaxin localized at PM. As a member of the SNARE superfamily, it forms a complex with the PM-localized SNAP33, and the vesicle-residing VAMP721/722, which constitutes the exocytic secretion system to secrete immune molecules for immunity against a variety of pathogens (Johansson et al., 2014; Yun et al., 2016). SYT1 encodes a membrane trafficking proteins specifically localized to the ER-PM boundary. It regulates endocytosis endosome recycling at PM. The Arabidopsis SYT1 binds PEN1 and fine-tunes the PEN1-SNAP33-VAMP721/722 exocytic activity (Kim et al., 2016; Yun et al., 2016). In this context, it is interesting that BnRALF10 significantly altered the abundance of both SYT1 and SYP121 as well as other SYPs such as SYP21, 22, 61 and v-SNARE 13 (Table 2 and Supplementary Table 2). It is likely that BnRALF10 activates SYT1-SYP121-mediated secretion system to guarantee positioning anchoring and abundance of immune proteins at PM and probably other membranes thereby stimulate immune responses. To date, little is known about the immune molecules secreted by this system. In Arabidopsis, SNAREs SYP61 and SYP121 directly bind to and coordinate the trafficking of PM aquaporin PIP2;7, a water channel, to modulate the cell membrane water permeability (Hachez et al., 2014). Coincidently, BnRALF10 infiltration reduced abundance of the aquaporin PIP2;7 protein (Supplementary Table 2). This implies that BnRALF10 might regulate the water access to the infected pathogen to inhibit pathogen via SYP61-SYP121-mediated suppression of PIP2;7. Furthermore, the SYP121-SNAP33-VAMP721/722 exocytosis module directly interacts and controls PM localization and function of K+ channels (Grefen et al., 2015; Zhang et al., 2015). Whether calcium channels or transporters, like K+ channels, are regulated by SYT1 and SYP121-SNAP33-VAMP721/722 is a intriguing issue to be addressed, considering the pivotal role of calcium signaling pathways in BnRALF10-elicited immunity (refer to below for details, Table 2 and Supplementary Table 2). Additionally, homeostasis of some PM-localized immune receptors are regulated by PEN1 and other protein secretion systems. For instance, abundance of FLS2, the receptor of flg22, at PM is regulated by PEN1- and EXO70B1/EXO70B2-mediated exocyst pathways (Wang et al., 2020). Under this consideration, whether FER, the likely receptor of BnRALF10, is regulated by PEN1 and other exocyst pathways is worth further study. Moreover, it is also interesting to check whether other PM-localized RED proteins are modulated by the SYT1 and SYP121/PEN1-SNAP33-VAMP721/722 exocytosis systems.
Linked to the above point, another interesting finding is that BnRALF10-stimulated immunity is tightly associated with optimal membrane compartments creating for immune receptor complex formation and immune signal transduction via REMs and PRFs. REMs modulate PM nanodomain organization to promote formin condensation for actin remodeling in innate immune responses (Ma et al., 2022) and to enable the pathogen-derived outer membrane vesicles (OMVs) insert into the host PM, thereby alters host membrane properties to potentiate plant immune responses (Tran et al., 2022). In contrast, PRFs negatively regulate formin-mediated actin assembly (Sun et al., 2018). Intriguingly, BnRALF10 enhanced REMs abundance while reduced PRFs accumulation (Table 2). Together, these results reveal that BnRALF10 fine tunes REM- and PRFs-dependent actin cytoskeleton to promote plant immunity. In addition, REMs were reported to trigger immunity by polymerizing and physically interacting with receptor-like kinases (RLK) (Yu, 2020). Their phospho-status defines their PM nanodomain organization and activities in restricting viral cell-to-cell movement (Perraki et al., 2018) as well as in enhancing RBOHB-dependent ROS production (Cai et al., 2020). In this scenario, it is possible that BnRALF10 recognition somehow causes phosphorylation of REMs, which promotes their functions including creating optimal membrane compartments for immune complex formation and immune signal transduction, thereby induces immunity. Some RALFs modulate flg22-triggered immunity (Stegmann et al., 2017). It is likely that RALF recognition complex RALF-FER-LLGs localizes together with flg22 recognition complex flg22-FLS2-BAK1 in the same nanodomain on PM, which is separated from the nanodomain containing BRI1 complex for growth regulation and even other immunity receptor complexes (Bücherl et al., 2017).
Twelve proteins involved in redox homeostasis are enriched in the collection of proteins differentially expressed in between ddH2O and BnRALF10 peptide-treated leaves. These proteins include ROS generator peroxisomal (S)-2-hydroxy-acid oxidase GLO2 and a variety of antioxidants such as peroxidases, superoxide dismutase [Mn] 1, GSTs, thioredoxins and glutaredoxin (Table 2 and Supplementary Table 2). ROS is pivotal in plant-S. sclerotiorum interactions (Williams et al., 2011; Wang Z. et al., 2019; Ding et al., 2020). In this regard, it is noticeable that Arabidopsis GLO2 acts as a ROS producer in non-host resistance (Rojas et al., 2012). Intriguingly, its orthologs in Nicotiana benthamiana, NbGOX4, plays positive role in resistance to S. sclerotiorum (Xu Y. P. et al., 2018). These results suggest that GLO2 likely functions in BnRALF10-elicited ROS and plant immunity to S. sclerotiorum. This is an interesting reinforce on understanding the generation mechanisms of pattern-triggered ROS. Additionally, it has been reported that the activities of peroxidase and superoxide dismutase were higher in S. sclerotiorum-resistant B. napus genotypes compared with S. sclerotiorum-susceptible B. napus genotypes (Garg et al., 2013). Our previous proteomic analysis for oilseed rape inoculated with S. sclerotiorum and its avirulent strain EP-1PB showed that copper-zinc superoxide dismutase, thioredoxin 1 and glutaredoxin 3 were up-regulated in the latter, indicating the important role of antioxidant capacity of B. napus in defense against S. sclerotiorum (Cao et al., 2016). BnRALF10 elicits ROS burst in oilseed rape leaves (Figure 6). The fact that these ROS scavenging related enzymes are induced by BnRALF10 treatment may indicate their participation in the ROS homeostasis regulation during BnRALF10-stimulated immunity to S. sclerotiorum.
Rapid alkalinization factor-triggered immunity has been connected closely with Ca2+ signaling pathway. Previous reports have established that RALF peptides induce Ca2+ influx across the PM and the release of Ca2+ from intracellular reserves (Haruta et al., 2008; Shih et al., 2014). Here our results also verified this conclusion (Figure 6). Gjetting et al. (2020) showed that the rapid burst of intracellular Ca2+ preceded apoplastic alkalinization in roots triggered by RALFs, and the inhibition of H+-ATPase activity mediated by RALF involves an obligatory Ca2+ signal. Additionally, our previous studies reveal that Ca2+ signaling is essential to plant defense against S. sclerotiorum. Ca2+ signaling genes encoding calmodulins (CaMs), Ca2+-dependent protein kinase (CDPK)-related kinases (CRKs), calcium and calmodulin-dependent protein kinases (CCaMKs), cyclic nucleotide-gated channels (CNGCs), and calmodulin-binding transcription activator 3 (CAMTA3) significantly affect plant resistance to S. sclerotiorum (Zhao et al., 2013; Saand et al., 2015a,b; Wang et al., 2015, 2016; Rahman et al., 2016). Our current proteomics analysis demonstrates that annexin D1 (Ca2+-dependent phospholipid-binding protein) and synaptotagmin-1 (Ca2+ sensors), two important components of Ca2+ signaling pathway, were induced in B. napus infiltrated with BnRALF10 (Supplementary Table 4). Synaptotagmin-1 and annexins have been found to play a negative role in plant immunity to biotrophic pathogens (Levy et al., 2015; Kim et al., 2016; Zhao et al., 2021), while its role in plant defense against necrotrophic pathogens remains unknown. Therefore, it will be intriguing to further confirm the role of synaptotagmin-1 and annexin D1 in B. napus defense against S. sclerotiorum and dissect their functional mechanisms, especially in BnRALF10 evoked Ca2+ signaling.
Based on our results, it is proposed that BnRALF10 alters abundance of RED proteins, which ensures anchoring and abundance of immune proteins at PM via synergy of SYT1 and SYP121/PEN1, optimizes membrane compartments for immune receptor complex formation and immune signal transduction via REMs and PRFs, elicits various immune responses including ROS burst and cytosolic Ca2+ promotion, thereby stimulate plant immunity to pathogens including S. sclerotiorum.
Conclusion
The oilseed rape genome harbored 61 RALFs. Half of them (belonging to clades III and IV) were atypical, containing a less conserved YISY motif and lacking a RRXL motif or a pair of cysteines. Expression profiles of RALF family in response to pathogen and molecular patterns were RALF- and stimulus-dependent. In general, BnRALF genes were expressionally up-regulated by S. sclerotiorum, while down-regulated by BnPep5 and SsNLP1. Thus, BnRALF members are likely differentially involved in plant immunity. The canonical RALF, BnRALF10, induced diverse immune responses such as ROS accumulation, cytosolic Ca2+ promotion, MAPK activation, defense-related gene expression induction and stimulated resistance to S. sclerotiorum, and thus likely functioned as a DAMP to play a positive role in plant immunity. Moreover, BnFER was likely a receptor of BnRALF10. Quantitative proteomic analysis identified dozens of BnRALF10-elicited defense (RED) proteins. BnRALF10 modulated RED protein abundance to fine-tune plant immunity.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
X-ZC coordinated the project. Y-HH conducted the bioinformatics, gene expression, and immune response analyses. Y-HH and Z-RZ performed disease resistance evaluation. Y-HH, S-YC, and Y-PX conducted the proteomic analysis. X-ZC conceived of the study and participated in its design and coordination. X-ZC and Y-HH prepared the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by grants from the National Natural Science Foundation of China (Grant No. 31871947) and the Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding (Grant No. 2021C02064).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Prof. Yang Liang, College of Agriculture and Biotechnology, Zhejiang University, China, for providing seeds of A. thaliana plants expressing cytosolic apoaequorin.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.877404/full#supplementary-material
Footnotes
- ^ http://www.arabidopsis.org/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://au.expasy.org/tools/pi_tool.html
- ^ https://itol.embl.de/
- ^ http://nrbsc.org/gfx/genedoc
- ^ http://weblogo.threeplusone.com/
- ^ https://imagej.net
References
Abarca, A., Franck, C. M., and Zipfel, C. (2021). Family-wide evaluation of rapid alkalinization factor peptides. Plant Physiol. 187, 996–1010. doi: 10.1093/plphys/kiab308
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). Meme suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bartels, S., and Boller, T. (2015). Quo vadis, Pep? Plant elicitor peptides at the crossroads of immunity, stress, and development. J. Exp. Bot. 66, 5183–5193. doi: 10.1093/jxb/erv180
Bashi, D. Z., Hegedus, D. D., Buchwaldt, L., Rimmer, S. R., and Borhan, M. H. (2010). Expression and regulation of Sclerotinia sclerotiorum necrosis and ethylene-inducing peptides (NEPs). Mol. Plant Pathol. 11, 43–53. doi: 10.1111/j.1364-3703.2009.00571
Bernoux, M., Timmers, T., Jauneau, A., Brière, C., de Wit, P. J., Marco, Y., et al. (2008). RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 20, 2252–2264. doi: 10.1105/tpc.108.058685
Bi, Y. M., Cammue, B. P. A., Goodwin, P. H., KrishnaRaj, S., and Saxena, P. K. (1999). Resistance to Botrytis cinerea in scented geranium transformed with a gene encoding the antimicrobial protein Ace-AMP1. Plant Cell Rep. 18, 835–840. doi: 10.1007/s002990050670
Bjellqvist, B., Hughes, G. J., Pasquali, C., Paquet, N., Ravier, F., Sanchez, J. C., et al. (1993). The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14, 1023–1031. doi: 10.1002/elps.11501401163
Blackburn, M. R., Haruta, M., and Moura, D. S. (2020). Twenty years of progress in physiological and biochemical investigation of RALF peptides. Plant Physiol. 182, 1657–1666. doi: 10.1104/pp.19.01310
Bücherl, C. A., Jarsch, I. K., Schudoma, C., Segonzac, C., Mbengue, M., Robatzek, S., et al. (2017). Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 6:e25114. doi: 10.7554/eLife.25114
Cai, J., Chen, T., Wang, Y., Qin, G., and Tian, S. (2020). SLREM1 triggers cell death by activating an oxidative burst and other regulators. Plant Physiol. 183, 717–732. doi: 10.1104/pp.20.00120
Campbell, L., and Turner, S. R. (2017). A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front. Plant Sci. 8:37. doi: 10.3389/fpls.2017.00037
Cao, J. Y., Xu, Y. P., and Cai, X. Z. (2016). TMT-based quantitative proteomics analyses reveal novel defense mechanisms of Brassica napus against the devastating necrotrophic pathogen Sclerotinia sclerotiorum. J. Proteomics 143, 265–277. doi: 10.1016/j.jprot.2016.03.006
Cao, J. Y., Xu, Y. P., and Cai, X. Z. (2020). Integrated miRNAome and transcriptome analysis reveals argonaute 2-mediated defense responses against the devastating phytopathogen Sclerotinia sclerotiorum. Front. Plant Sci. 11:500. doi: 10.3389/fpls.2020.00500
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, F., Liang, J., Cai, C., Cai, X., Wu, J., and Wang, X. (2017). Genome sequencing supports a multi-vertex model for Brassiceae species. Curr. Opin. Plant Biol. 36, 79–87. doi: 10.1016/j.pbi.2017.01.006
Ding, L., Li, T., Guo, X., Li, M., Liu, X., Cao, J., et al. (2021). Sclerotinia stem rot resistance in rapeseed: recent progress and future prospects. J. Agric. Food Chem. 69, 2965–2978. doi: 10.1021/acs.jafc.0c07351
Ding, Y., Mei, J., Chai, Y., Yang, W., Mao, Y., Yan, B., et al. (2020). Sclerotinia sclerotiorum utilizes host-derived copper for ROS detoxification and infection. PLoS Pathog. 16:e1008919. doi: 10.1371/journal.ppat.1008919
Dressano, K., Ceciliato, P., Silva, A. L., Guerrero-Abad, J. C., Bergonci, T., Ortiz-Morea, F. A., et al. (2017). BAK1 is involved in AtRALF1-induced inhibition of root cell expansion. PLoS Genet. 13:e1007053. doi: 10.1371/journal.pgen.1007053
Du, C., Li, X., Chen, J., Chen, W., Li, B., Li, C., et al. (2016). Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 113, E8326–E8334. doi: 10.1073/pnas.1609626113
Figueiredo, J., Costa, G. J., Maia, M., Paulo, O. S., Malhó, R., Sousa, S. M., et al. (2016). Revisiting Vitis vinifera subtilase gene family: a possible role in grapevine resistance against Plasmopara viticola. Front. Plant Sci. 7:1783. doi: 10.3389/fpls.2016.01783
Fu, S., Xu, Y., Li, C., Li, Y., Wu, J., and Zhou, X. (2018). Rice stripe virus interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol. Plant 11, 269–287. doi: 10.1016/j.molp.2017.11.011
Garg, H., Li, H., Sivasithamparam, K., and Barbetti, M. J. (2013). Differentially expressed proteins and associated histological and disease progression changes in cotyledon tissue of a resistant and susceptible genotype of Brassica napus infected with Sclerotinia sclerotiorum. PLoS One 8:e65205. doi: 10.1371/journal.pone.0065205
Gjetting, S. K., Mahmood, K., Shabala, L., Kristensen, A., Shabala, S., Palmgren, M., et al. (2020). Evidence for multiple receptors mediating RALF-triggered Ca(2+) signaling and proton pump inhibition. Plant J. 104, 433–446. doi: 10.1111/tpj.14935
Grefen, C., Karnik, R., Larson, E., Lefoulon, C., Wang, Y., Waghmare, S., et al. (2015). A vesicle-trafficking protein commandeers Kv channel voltage sensors for voltage-dependent secretion. Nat. Plants 1:15108. doi: 10.1038/nplants.2015.108
Guo, H., Nolan, T. M., Song, G., Liu, S., Xie, Z., Chen, J., et al. (2018). FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr. Biol. 28, 3316–3324. doi: 10.1016/j.cub.2018.07.078
Guo, W., Zhang, F., Bao, A., You, Q., Li, Z., Chen, J., et al. (2019). The soybean RHG1 amino acid transporter gene alters glutamate homeostasis and jasmonic acid-induced resistance to soybean cyst nematode. Mol. Plant Pathol. 20, 270–286. doi: 10.1111/mpp.12753
Hachez, C., Laloux, T., Reinhardt, H., Cavez, D., Degand, H., Grefen, C., et al. (2014). Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26, 3132–3147. doi: 10.1105/tpc.114.127159
Haruta, M., Monshausen, G., Gilroy, S., and Sussman, M. R. (2008). A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in arabidopsis seedlings: identification of AtRALF1 peptide. Biochemistry 47, 6311–6321. doi: 10.1021/bi8001488
Haruta, M., Sabat, G., Stecker, K., Minkoff, B. B., and Sussman, M. R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. doi: 10.1126/science.1244454
Ibrahim, A., Yang, X., Liu, C., Cooper, K. D., Bishop, B. A., Zhu, M., et al. (2020). Plant SNAREs SYP22 and SYP23 interact with tobacco mosaic virus 126 kDa protein and SYP2s are required for normal local virus accumulation and spread. Virology 547, 57–71. doi: 10.1016/j.virol.2020.04.002
Jamann, T. M., Luo, X., Morales, L., Kolkman, J. M., Chung, C. L., and Nelson, R. J. (2016). A remorin gene is implicated in quantitative disease resistance in maize. Theor. Appl. Genet. 129, 591–602. doi: 10.1007/s00122-015-2650-6
Johansson, O. N., Fantozzi, E., Fahlberg, P., Nilsson, A. K., Buhot, N., Tör, M., et al. (2014). Role of the penetration-resistance genes PEN1, PEN2 and PEN3 in the hypersensitive response and race-specific resistance in Arabidopsis thaliana. Plant J. 79, 466–476. doi: 10.1111/tpj.12571
Jordá, L., Coego, A., Conejero, V., and Vera, P. (1999). A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J. Biol. Chem. 274, 2360–2365. doi: 10.1074/jbc.274.4.2360
Jungkunz, I., Link, K., Vogel, F., Voll, L. M., Sonnewald, S., and Sonnewald, U. (2011). AtHsp70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J. 66, 983–995. doi: 10.1111/j.1365-313X.2011.04558.x
Kim, H., Kwon, H., Kim, S., Kim, M. K., Botella, M. A., Yun, H. S., et al. (2016). Synaptotagmin 1 negatively controls the two distinct immune secretory pathways to powdery mildew fungi in Arabidopsis. Plant Cell Physiol. 57, 1133–1141. doi: 10.1093/pcp/pcw061
Kim, M. J., Lim, G. H., Kim, E. S., Ko, C. B., Yang, K. Y., Jeong, J. A., et al. (2007). Abiotic and biotic stress tolerance in Arabidopsis overexpressing the multiprotein bridging factor 1a (MBF1a) transcriptional coactivator gene. Biochem. Biophys. Res. Commun. 354, 440–446. doi: 10.1016/j.bbrc.2006.12.212
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids. Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Levy, A., Zheng, J. Y., and Lazarowitz, S. G. (2015). Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr. Biol. 25, 2018–2025. doi: 10.1016/j.cub.2015.06.015
Lopez, V. A., Park, B. C., Nowak, D., Sreelatha, A., Zembek, P., Fernandez, J., et al. (2019). A bacterial effector mimics a host Hsp90 client to undermine immunity. Cell 179, 205–218. doi: 10.1016/j.cell.2019.08.020
Ma, Q. H., Tian, B., and Li, Y. L. (2010). Overexpression of a wheat jasmonate-regulated lectin increases pathogen resistance. Biochimie 92, 187–193. doi: 10.1016/j.biochi.2009.11.008
Ma, Z., Sun, Y., Zhu, X., Yang, L., Chen, X., and Miao, Y. (2022). Membrane nanodomains modulate formin condensation for actin remodeling in Arabidopsis innate immune responses. Plant Cell 34, 374–394. doi: 10.1093/plcell/koab261
Manjula, S., Murali, M., Shivamurthy, G. R., and Amruthesh, K. N. (2015). Non-specific lipid transfer proteins (ns-LTPs) from maize induce resistance in pearl millet against downy mildew disease. Phytoparasitica 43, 437–447. doi: 10.1007/s12600-014-0446-x
Masachis, S., Segorbe, D., Turra, D., Leon-Ruiz, M., Furst, U., El, G. M., et al. (2016). A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 1:16043. doi: 10.1038/nmicrobiol.2016.43
Matos, J. L., Fiori, C. S., Silva-Filho, M. C., and Moura, D. S. (2008). A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Lett. 582, 3343–3347. doi: 10.1016/j.febslet.2008.08.025
Merino, M. C., Guidarelli, M., Negrini, F., De Biase, D., Pession, A., and Baraldi, E. (2019). Induced expression of the Fragaria x ananassa rapid alkalinization factor-33-like gene decreases anthracnose ontogenic resistance of unripe strawberry fruit stages. Mol. Plant Pathol. 20, 1252–1263. doi: 10.1111/mpp.12837
Moussu, S., Broyart, C., Santos-Fernandez, G., Augustin, S., Wehrle, S., Grossniklaus, U., et al. (2020). Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. Proc. Natl. Acad. Sci. U. S. A. 117, 7494–7503. doi: 10.1073/pnas.2000100117/-/DCSupplemental
Olsson, V., Joos, L., Zhu, S., Gevaert, K., Butenko, M. A., and De Smet, I. (2019). Look closely, the beautiful may be small: precursor-derived peptides in plants. Annu. Rev. Plant Biol. 70, 153–186. doi: 10.1146/annurev-arplant-042817-040413
Pardal, A. J., Piquerez, S., Dominguez-Ferreras, A., Frungillo, L., Mastorakis, E., Reilly, E., et al. (2021). Immunity onset alters plant chromatin and utilizes EDA16 to regulate oxidative homeostasis. PLoS Pathog. 17:e1009572. doi: 10.1371/journal.ppat.1009572
Pavet, V., Olmos, E., Kiddle, G., Mowla, S., Kumar, S., Antoniw, J., et al. (2005). Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol. 139, 1291–1303. doi: 10.1104/pp.105.067686
Pearce, G., Moura, D. S., Stratmann, J., and Ryan, C. A. (2001). RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. U. S. A. 98, 12843–12847. doi: 10.1073/pnas.201416998
Pearce, G., Yamaguchi, Y., Munske, G., and Ryan, C. A. (2010). Structure–activity studies of RALF, rapid alkalinization factor, reveal an essential – YISY – motif. Peptides 31, 1973–1977. doi: 10.1016/j.peptides.2010.08.012
Perraki, A., Gronnier, J., Gouguet, P., Boudsocq, M., Deroubaix, A. F., Simon, V., et al. (2018). REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog. 14:e1007378. doi: 10.1371/journal.ppat.1007378
Petriacq, P., de Bont, L., Hager, J., Didierlaurent, L., Mauve, C., Guerard, F., et al. (2012). Inducible nad overproduction in arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst-avrRPM1. Plant J. 70, 650–665. doi: 10.1111/j.1365-313X.2012.04920.x
Pflieger, S., Blanchet, S., Camborde, L., Drugeon, G., Rousseau, A., Noizet, M., et al. (2008). Efficient virus-induced gene silencing in arabidopsis using a ‘one-step’. TYMV-derived vector. Plant J. 56, 678–690. doi: 10.1111/j.1365-313X.2008.03620.x
Piotrowski, M., Schemenewitz, A., Lopukhina, A., Müller, A., Janowitz, T., Weiler, E. W., et al. (2004). Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279, 50717–50725. doi: 10.1074/jbc.M407681200
Qian, L., Zhao, J., Du, Y., Zhao, X., Han, M., and Liu, Y. (2018). Hsp90 interacts with TM-2(2) and is essential for TM-2(2)-mediated resistance to tobacco mosaic virus. Front. Plant Sci. 9:411. doi: 10.3389/fpls.2018.00411
Rahman, H., Xu, Y. P., Zhang, X. R., and Cai, X. Z. (2016). Brassica napus genome possesses extraordinary high number of CAMTA genes and CAMTA3 contributes to PAMP triggered immunity and resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 7:581. doi: 10.3389/fpls.2016.00581
Rojas, C. M., Senthil-Kumar, M., Wang, K., Ryu, C. M., Kaundal, A., and Mysore, K. S. (2012). Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24, 336–352. doi: 10.1105/tpc.111.093245
Saand, M. A., Xu, Y. P., Li, W., Wang, J. P., and Cai, X. Z. (2015a). Cyclic nucleotide gated channel gene family in tomato: genome-wide identification and functional analyses in disease resistance. Front. Plant Sci. 6:303. doi: 10.3389/fpls.2015.00303
Saand, M. A., Xu, Y. P., Munyampundu, J. P., Li, W., Zhang, X. R., and Cai, X. Z. (2015b). Phylogeny and evolution of plant cyclic nucleotide-gated ion channel (CNGC) gene family and functional analyses of tomato CNGCs. DNA Res. 22, 471–483. doi: 10.1093/dnares/dsv029
Shih, H. W., Miller, N. D., Dai, C., Spalding, E. P., and Monshausen, G. B. (2014). The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr. Biol. 24, 1887–1892. doi: 10.1016/j.cub.2014.06.064
Srivastava, R., Liu, J. X., Guo, H., Yin, Y., and Howell, S. H. (2009). Regulation and processing of a plant peptide hormone. AtRALF23, in Arabidopsis. Plant J. 59, 930–939. doi: 10.1111/j.1365-313X.2009.03926.x
Stegmann, M., Monaghan, J., Smakowska-Luzan, E., Rovenich, H., Lehner, A., Holton, N., et al. (2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289. doi: 10.1126/science.aal2541
Sun, H., Qiao, Z., Chua, K. P., Tursic, A., Liu, X., Gao, Y. G., et al. (2018). Profilin negatively regulates formin-mediated actin assembly to modulate PAMP-triggered plant immunity. Curr. Biol. 28, 1882–1895. doi: 10.1016/j.cub.2018.04.045
Thynne, E., Saur, I. M. L., Simbaqueba, J., Ogilvie, H. A., Gonzalez-Cendales, Y., Mead, O., et al. (2017). Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Mol. Plant Pathol. 18, 811–824. doi: 10.1111/mpp.12444
Tran, T. M., Chng, C. P., Pu, X., Ma, Z., Han, X., Liu, X., et al. (2022). Potentiation of plant defense by bacterial outer membrane vesicles is mediated by membrane nanodomains. Plant Cell 34, 395–417. doi: 10.1093/plcell/koab276
Veenstra, J. A. (2000). Mono- and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch. Insect Biochem. Physiol. 43, 49–63. doi: 10.1002/(SICI)1520-6327(200002)43:2<49::AID-ARCH1<3.0.CO;2-M
Wang, J. P., Munyampundu, J. P., Xu, Y. P., and Cai, X. Z. (2015). Phylogeny of plant calcium and calmodulin-dependent protein kinases (CCaMKs) and functional analyses of tomato CCaMK in disease resistance. Front. Plant Sci. 6:1075. doi: 10.3389/fpls.2015.01075
Wang, J. P., Xu, Y. P., Munyampundu, J. P., Liu, T. Y., and Cai, X. Z. (2016). Calcium-dependent protein kinase (CDPK) and cdpk-related kinase (CRK) gene families in tomato: genome-wide identification and functional analyses in disease resistance. Mol. Genet. Genomics 291, 661–676. doi: 10.1007/s00438-015-1137-0
Wang, S., Chen, Z., Tian, L., Ding, Y., Zhang, J., Zhou, J., et al. (2019). Comparative proteomics combined with analyses of transgenic plants reveal ZmREM1.3 mediates maize resistance to southern corn rust. Plant Biotechnol. J. 17, 2153–2168. doi: 10.1111/pbi.13129
Wang, W., Liu, N., Gao, C., Cai, H., Romeis, T., and Tang, D. (2020). The Arabidopsis exocyst subunits EXO70B1 and EXO70B2 regulate FLS2 homeostasis at the plasma membrane. New Phytol. 227, 529–544. doi: 10.1111/nph.16515
Wang, Z., Ma, L. Y., Cao, J., Li, Y. L., Ding, L. N., Zhu, K. M., et al. (2019). Recent advances in mechanisms of plant defense to Sclerotinia sclerotiorum. Front. Plant Sci. 10:1314. doi: 10.3389/fpls.2019.01314
Wang, Z., Wan, L., Zhang, X., Xin, Q., Song, Y., Hong, D., et al. (2021). Interaction between Brassica napus polygalacturonase inhibition proteins and Sclerotinia sclerotiorum polygalacturonase: implications for rapeseed resistance to fungal infection. Planta 253:34. doi: 10.1007/s00425-020-03556-2
Williams, B., Kabbage, M., Kim, H. J., Britt, R., and Dickman, M. B. (2011). Tipping the balance: sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7:e1002107. doi: 10.1371/journal.ppat.1002107
Xiao, Y., Stegmann, M., Han, Z., DeFalco, T. A., Parys, K., Xu, L., et al. (2019). Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572, 270–274. doi: 10.1038/s41586-019-1409-7
Xu, T. T., Li, J. T., Yu, B. D., Liu, L., Zhang, X. H., Liu, J. L., et al. (2018). Transcription factor SsSte12 was involved in mycelium growth and development in Sclerotinia sclerotiorum. Front. Microbiol. 9:2476. doi: 10.3389/fmicb.2018.02476
Xu, Y. P., Yang, J., and Cai, X. Z. (2018). Glycolate oxidase gene family in Ncotiana benthamiana: genome-wide identification and functional analyses in disease resistance. Sci. Rep. 8:8615. doi: 10.1038/s41598-018-27000-4
Yu, Y. (2020). Remorins: essential regulators in plant-microbe interaction and cell death induction. Plant Physiol. 183, 435–436. doi: 10.1104/pp.20.00490
Yun, H. S., Kang, B. G., and Kwon, C. (2016). Arabidopsis immune secretory pathways to powdery mildew fungi. Plant Signal. Behav. 11:e1226456. doi: 10.1080/15592324.2016.1226456
Zhang, B., Karnik, R., Wang, Y., Wallmeroth, N., Blatt, M. R., and Grefen, C. (2015). The Arabidopsis R-SNARE VAMP721 interacts with KAT1 and KC1 K+ channels to moderate K+ current at the plasma membrane. Plant Cell 27, 1697–1717. doi: 10.1105/tpc.15.00305
Zhang, H., Jing, X., Chen, Y., Liu, Z., Xin, Y., and Qiao, Y. (2020). The genome-wide analysis of RALF-like genes in strawberry (wild and cultivated) and five other plant species (Rosaceae). Genes 11:174. doi: 10.3390/genes11020174
Zhang, X., Peng, H., Zhu, S., Xing, J., Li, X., Zhu, Z., et al. (2020). Nematode-encoded RALF peptide mimics facilitate parasitism of plants through the FERONIA receptor kinase. Mol. Plant 13, 1434–1454. doi: 10.1016/j.molp.2020.08.014
Zhang, X., Wang, D., Chen, J., Wu, D., Feng, X., and Yu, F. (2021). Nematode RALF-like 1 targets soybean malectin-like receptor kinase to facilitate parasitism. Front. Plant Sci. 12:775508. doi: 10.3389/fpls.2021.775508
Zhang, Y., Wang, Y., Zhou, W., Zheng, S., and Ye, R. (2021). Detection of candidate gene networks involved in resistance to Sclerotinia sclerotiorum in soybean. J. Appl. Genet. 63, 1–14. doi: 10.1007/s13353-021-00654-z
Zhao, C., Zayed, O., Yu, Z., Jiang, W., Zhu, P., Hsu, C. C., et al. (2018). Leucine-rich repeat extensin proteins regulate plant salt tolerance in arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 115, 13123–13128. doi: 10.1073/pnas.1816991115
Zhao, Y., Liu, W., Xu, Y. P., Cao, J. Y., Braam, J., and Cai, X. Z. (2013). Genome-wide identification and functional analyses of calmodulin genes in Solanaceous species. BMC Plant Biol. 13:70. doi: 10.1186/1471-2229-13-70
Zhao, Z. X., Xu, Y. J., Lei, Y., Li, Q., Zhao, J. Q., Li, Y., et al. (2021). ANNEXIN 8 negatively regulates RPW8.1-mediated cell death and disease resistance in Arabidopsis. J. Integr. Plant Biol. 63, 378–392. doi: 10.1111/jipb.13025
Zhu, S., Fu, Q., Xu, F., Zheng, H., and Yu, F. (2021). New paradigms in cell adaptation: decades of discoveries on the CrRLK1L receptor kinase signalling network. New Phytol. 232, 1168–1183. doi: 10.1111/nph.17683
Zhu, X. F., Liu, Y., Gai, X. T., Zhou, Y., Xia, Z. Y., Chen, L. J., et al. (2019). SNARE proteins SYP22 and VAMP727 negatively regulate plant defense. Plant Signal. Behav. 14:1610300. doi: 10.1080/15592324.2019.1610300
Zou, J., Li, W., Zhang, Y., Song, W., Jiang, H., Zhao, J., et al. (2021). Identification of glutathione transferase gene associated with partial resistance to Sclerotinia stem rot of soybean using genome-wide association and linkage mapping. Theor. Appl. Genet. 134, 2699–2709. doi: 10.1007/s00122-021-03855-6
Keywords: Brassica napus, FER, plant immunity, proteome, RALF, Sclerotinia sclerotiorum, signaling
Citation: He Y-H, Zhang Z-R, Xu Y-P, Chen S-Y and Cai X-Z (2022) Genome-Wide Identification of Rapid Alkalinization Factor Family in Brassica napus and Functional Analysis of BnRALF10 in Immunity to Sclerotinia sclerotiorum. Front. Plant Sci. 13:877404. doi: 10.3389/fpls.2022.877404
Received: 16 February 2022; Accepted: 11 April 2022;
Published: 03 May 2022.
Edited by:
Yuelin Zhang, University of British Columbia, CanadaReviewed by:
Jiasen Cheng, Huazhong Agricultural University, ChinaXiufang Xin, Institute of Plant Physiology and Ecology (CAS), China
Copyright © 2022 He, Zhang, Xu, Chen and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-Zhong Cai, eHpoY2FpQHpqdS5lZHUuY24=
 Yu-Han He1
Yu-Han He1 Zhuo-Ran Zhang
Zhuo-Ran Zhang You-Ping Xu
You-Ping Xu Song-Yu Chen
Song-Yu Chen Xin-Zhong Cai
Xin-Zhong Cai