- 1College of Science & Engineering, Flinders University, Bedford Park, SA, Australia
- 2College of Science, Australian National University, Canberra, ACT, Australia
Stress-responsive components of the mitochondrial alternative electron transport pathway have the capacity to improve tolerance of plants to abiotic stress, particularly the alternative oxidase AOX1A but also external NAD(P)H dehydrogenases such as NDB2, in Arabidopsis. NDB2 and AOX1A can cooperate to entirely circumvent the classical electron transport chain in Arabidopsis mitochondria. Overexpression of AOX1A or NDB2 alone can have slightly negative impacts on plant growth under optimal conditions, while simultaneous overexpression of NDB2 and AOX1A can reverse these phenotypic effects. We have taken a global transcriptomic approach to better understand the molecular shifts that occur due to overexpression of AOX1A alone and with concomitant overexpression of NDB2. Of the transcripts that were significantly up- or down- regulated in the AOX1A overexpression line compared to wild type (410 and 408, respectively), the majority (372 and 337, respectively) reverted to wild type levels in the dual overexpression line. Several mechanisms for the AOX1A overexpression phenotype are proposed based on the functional classification of these 709 genes, which can be used to guide future experiments. Only 28 genes were uniquely up- or down-regulated when NDB2 was overexpressed in the AOX1A overexpression line. On the other hand, many unique genes were deregulated in the NDB2 knockout line. Furthermore, several changes in transcript abundance seen in the NDB2 knockout line were consistent with changes in the AOX1A overexpression line. The results suggest that an imbalance in AOX1A:NDB2 protein levels caused by under- or over-expression of either component, triggers a common set of transcriptional responses that may be important in mitochondrial redox regulation. The most significant changes were transcripts associated with photosynthesis, secondary metabolism and oxidative stress responses.
1 Introduction
The importance of alternative oxidase (AOX) in mitigating oxidative stress is well documented and the mechanisms underlying this are popular research topics (Maxwell et al., 1999; Giraud et al., 2008; Watanabe et al., 2008; Florez-Sarasa et al., 2011; Panda et al., 2013; Vishwakarma et al., 2015; Del-Saz et al., 2016; Demircan et al., 2020; Jayawardhane et al., 2020). Transgenic Arabidopsis thaliana lines with modified expression of AOX1A, generated in the laboratory of Jim Siedow (Fiorani et al., 2005; Umbach et al., 2005), together with similar transgenic tobacco lines (Amirsadeghi et al., 2006) have been a cornerstone for understanding the roles of AOX (Smith et al., 2009; Skirycz et al., 2010; Wang et al., 2011; Cvetkovska and Vanlerberghe, 2012; Dahal et al., 2014; Liu et al., 2014; Dahal et al., 2015; Dahal and Vanlerberghe, 2017). These and other studies have confirmed the role of AOX in minimising the production of reactive oxygen species (ROS) in mitochondria by preventing over-reduction of the ubiquinone pool (for a review, see Vanlerberghe, 2013). Studies with AOX knockdown lines have noted an extra strain on photosynthesis and growth when these plants are exposed to stress (Giraud et al., 2008; Yoshida et al., 2011; Gandin et al., 2012; Dahal et al., 2014; Jiang et al., 2019), while AOX overexpression can protect photosynthetic machinery by utilising excess reducing equivalents and preventing ROS accumulation in the chloroplast (Dahal et al., 2014; Dahal et al., 2015; Dahal and Vanlerberghe, 2017). AOX isoforms have even been shown to functionally replace plastid localised PTOX in the immutans variegation mutant (Fu et al., 2012). Interrupting AOX expression affects the transcription of extramitochondrial proteins (Umbach et al., 2005; Clifton et al., 2006) and evidence for an intricate network of mitochondrial and chloroplast retrograde signaling factors is emerging (for a recent review, see Wang et al., 2020).
Relatively few studies have assessed the effects of modifying the expression of type II NAD(P)H dehydrogenases (NDs) on plant growth and stress tolerance, a task that is complicated by the dual targeting of NDAs, NDC, and NDB1 to other organelles in the cell as well as the mitochondrion (Carrie et al., 2008). Various Arabidopsis thaliana lines with knockdown or knockout of ND genes have now been reported for all except the non-expressed AtNDB3 (Smith et al., 2011; Wallström et al., 2014a; Wallström et al., 2014b; Fatihi et al., 2015; Sweetman et al., 2019). Simultaneous knockdown of both NDA proteins, or NDB1, by RNA interference resulted in delayed growth without affecting photosynthesis per se (Wallström et al., 2014a; Wallström et al., 2014b). Knockdown of AtNDB4 by RNA interference and TDNA insertion also resulted in growth delays during early development, however this disappeared during subsequent growth and the plants ultimately showed improved growth and resilience to salinity stress, attributed to the increase in endogenous NDB2 and AOX1a in these lines (Smith et al., 2011). Knockout of NDB2 by TDNA insertion had no obvious growth phenotype but caused increased sensitivity to a combined drought and high light stress (Sweetman et al., 2019). Knockout of NDC1 had no obvious effect on growth nor photosynthesis under optimal growth conditions (Piller et al., 2011), but growth and photosystem II efficiency were restricted upon exposure to high light (Fatihi et al., 2015). Interestingly, the latter studies also showed that NDC1 is a key enzyme for vitamin K1 biosynthesis.
The five AOX and seven ND isoforms of A. thaliana are expressed differently according to location, life cycle and external stimuli (Michalecka et al., 2003; Escobar et al., 2004; Clifton et al., 2006; Carrie et al., 2008). Each transcript has been identified as responsive to at least one stressor or treatment (Clifton et al., 2005; Elhafez et al., 2006; Feng et al., 2013) and some appear to be co-regulated in response to stress, particularly AOX1A, NDA2 and NDB2 (Clifton et al., 2005; Ho et al., 2008; Smith et al., 2009; Vijayraghavan and Soole, 2010). Physical association of some AOX and ND proteins has been observed in non-denaturing PAGE experiments (Senkler et al., 2017). Functional cooperation of AOX1A and NDB2 was also demonstrated recently in isolated mitochondria: over-expression of NDB2 alone resulted in high levels of largely inactive protein, which became active when co-expressed with AOX1A (Sweetman et al., 2019). Such cooperation between ND and AOX isoforms provides a complete bypass of the classical ETC. Measurements using these A. thaliana lines with modified AOX1A and NDB2 expression highlighted differences in their ability to grow under non-limiting conditions, and to survive and recover from a combined drought and increased light stress (Sweetman et al., 2019). Specifically, plants over-expressing AOX1A alone or AOX1A and NDB2 together, were more stress-resilient and capable of recovery than wild type plants, or plants overexpressing NDB2 alone, while plants lacking either AOX1A or NDB2 were severely impacted. However, the molecular mechanisms behind these phenotypes have yet to be determined. In addition, plants over-expressing AOX1A alone exhibited a small growth delay under standard growth conditions (not an isolated incident, see Skirycz et al., 2010) that was rectified by concomitant NDB2 overexpression.
In the present study, a global approach has been used to determine the effects of altered AOX1A and NDB2 proteins on growth-, metabolism- and stress-related gene transcripts in plants grown under standard conditions. The plant lines investigated included one of the original AOX1A-overexpression lines (AOX1A-OEX) generated in the Siedow laboratory (Umbach et al., 2005), an AOX1A and NDB2 dual overexpression line (dual-OEX) developed recently by overexpressing NDB2 in the AOX1A overexpression line (Sweetman et al., 2019), and an NDB2 knockout line (ndb2) from the same study. We address the following question: what mechanisms, driven by transcriptional changes, could cause the growth delay phenotype in plants over-expressing AOX1a alone, and how is this rectified by concomitant NDB2 overexpression? A second question arose during the analysis: namely, is there a common transcriptional response to alterations in the ratio of AOX1A:NDB2?
2 Methods
2.1 Plant materials and growth conditions
Four plant lines were used in this study; wild type (WT; A. thaliana Col0), an AOX1A overexpression line kindly provided by Jim Siedow’s laboratory many years ago (AOX1A-OEX; XX1; Umbach et al., 2005), an AOX1A and NDB2 dual overexpression line (dual-OEX; 5.2; Sweetman et al., 2019) and an NDB2 TDNA insertion line (ndb2; SALK_036330; Sweetman et al., 2019). The “single” NDB2 overexpression lines reported by Sweetman et al. (2019) were not analysed here, as enhanced NDB2 activity could not be detected in those lines (unless AOX1A was also overexpressed). A. thaliana plants were grown in a controlled temperature cabinet at 20°C with 16h day length and PAR of 80-120 μmols.m-2.sec-1 using custom-made panels of red and blue LED light modules (Phoenix Biosystems, Australia). A coco-peat soil mix (PIRSA-SARDI, Australia) was supplemented with slow-release fertiliser with trace elements (Osmocote, Australia) and used to fill ~6 cm diameter pots, each harbouring a single A. thaliana plant. Seedlings were germinated directly on soil, watered every 2-3 days and positions rotated regularly within the cabinet. Whole rosettes were harvested at 44 days after sowing, flash frozen in liquid nitrogen, pulverised using a mortar and pestle and stored at -80°C until use.
2.2 RNA extraction and sequencing
Total RNA was extracted from frozen tissue powder using the Isolate II RNA Plant kit (Bioline, NSW). RNA quality was assessed using the Perkin Elmer LabChip GX Touch 24 with the RNA quality score cut off set at >8. mRNA was purified and cDNA prepared using the Truseq Stranded mRNA HT Sample Prep Kit (Illumina).
Flinders Genomics Facility, Adelaide, Australia, provided RNA sequencing services. cDNA libraries were analysed with the Perkin Elmer LabChip GX Touch 24 using the High Sensitivity DNA kit producing an average fragment size of 268 bp. Libraries were sequenced using the Illumina NextSeq 500 to generate single-end reads of 75 bp. Raw sequence reads were then processed using the following pipeline: (1) remove reads with sequence quality scores less than Q30; (2) remove adapter/overrepresented sequence and cross species contamination; (3) remove reads with final lengths less than 30 bp. The quality of the cleaned reads was assessed using FastQC. The cleaned sequence reads were then aligned and mapped against the Arabidopsis genome (Ensembl, TAIR10) using hisat2 aligner (v2.2.1). Read counts were obtained using FeatureCounts (Liao et al., 2014) with default parameters except for –C (exclude chimeric fragments) option enabled. A summary of read data are included in Table S1.
DESeq2 was used for statistical analysis of differential expression. Pre-filtering of lowly-expressed genes was enabled prior to differential expression comparisons for each line relative to WT. Differentially Expressed Genes were defined as log2 fold changes greater than 1 or less than -1, with p-adjusted values <0.05. All associated expression (e.g. normalised counts, RPKM) and differential expression results tables (containing the baseMean, log2FoldChange and FDR) are included in Table S2.
2.3 Functional enrichment analysis
Assessment of enriched functional categories in up- and downregulated DEGs in various lines relative to the WT were performed using BiNGO, using the hypergeometric distribution-adjusted Benjamini & Hochberg false discovery rate (FDR) for multiple hypothesis correction (Maere et al., 2005). Plant Gene Ontology (GO) slim categories and in-depth biological process (BP) were evaluated and an FDR-adjusted p value < 0.05 indicates enriched GO functional categories for a given comparison. Enrichment of upstream TFs were identified using EAT-UpTF based off the relative enrichment of TF-target genes (Shim and Seo, 2020). A FDR-adjusted p value < 0.01 indicates significantly enriched TFs in up- and downregulated DEGs. All associated visualisation of enriched categories were performed with ggplot2 (https://ggplot2.tidyverse.org/). Commonalities and differences of up- and downregulated DEGs between various lines were identified using Jvenn (Bardou et al., 2014). Transcriptomic data (log2 FC, FDR-adjusted p-value <0.05) were visualized using MAPMAN 3.0.0 (Thimm et al., 2004).
2.4 Malondialdehyde measurements
Lipid peroxidation levels were estimated using the spectrophotometric TBARS method for measuring malondialdehyde (MDA) equivalents, as described by Hodges et al. (1999) and Singh et al. (2012). Approximately 50 mg of frozen leaf powder was extracted in 1 ml of 80% (v/v) ethanol and centrifuged at 16,000 g for 15 minutes. The supernatant was divided into two aliquots. Each aliquot was mixed with an equal volume of assay mix (20% (w/v) trichloroacetic acid and 0.01% (w/v) B-hydroxybutyric acid) either with or without 0.01% (w/v) thiobarbituric acid. Samples were heated to 96°C for 30 minutes and then quickly cooled on ice. After centrifugation at 9,500 g for 10 minutes, absorbance was measured at 440, 532 and 600 nm, and MDA equivalents calculated according to Hodges et al. (1999).
3 Results and discussion
3.1 Transcriptomic effects of AOX1A and NDB2 manipulation
Principal Component Analysis (PCA) (Figure 1) showed that the different lines were transcriptionally distinct but generally consistent between replicates, with PC1 and PC2 explaining 38 and 27% of total variation, respectively. Replicate 2 of ndb2 showed separation from other replicates, but remained distinct from the other lines, and clear differences in transcript levels of ndb2 compared to wild type were still observed. Clustering patterns suggested that (1) the transcriptome of AOX1A-OEX was very different from the transcriptome of WT plants; (2) concomitant AOX1A and NDB2 overexpression partially caused a reversion towards the WT transcriptome and (3) the ndb2 transcriptome was different from all other lines on the PC1 axis, but similar to that of the AOX1A-OEX line on the PC2 axis. To further explore these relationships, differentially expressed genes (DEGs) were determined for the AOX1A-OEX, dual-OEX and ndb2 lines relative to WT plants (|log2FC| > 1, FDR-adjusted p-value < 5%).
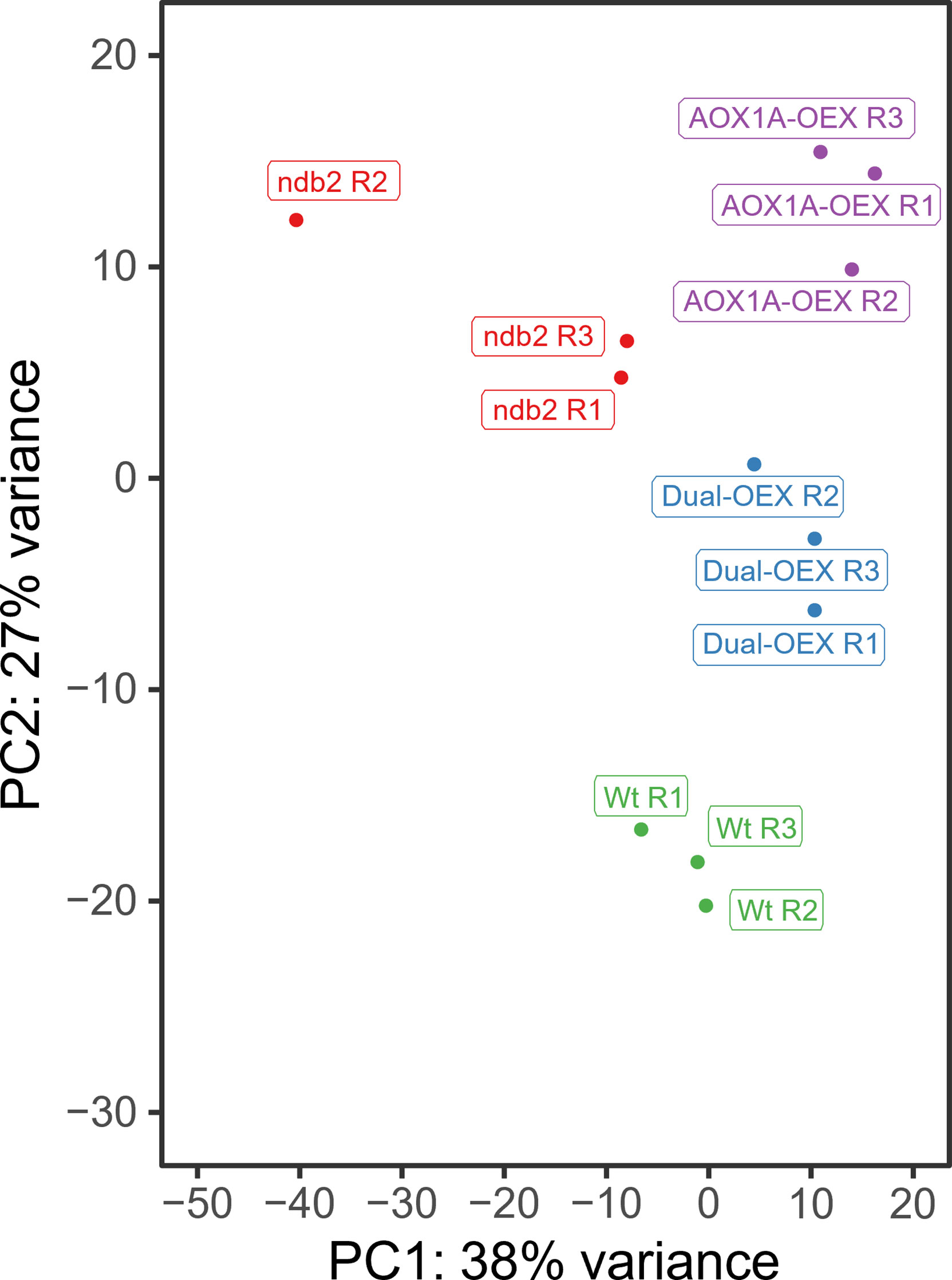
Figure 1 PCA plot of RNAseq data. Each point represents an individual replicate of wild type (Wt, green), AOX1A-OEX (purple), dual-OEX (blue) and ndb2 (red). The proportion of variance explained by PC1 and PC2 are indicated in the axis titles.
To reduce “wasteful” respiratory flux, overexpressed AOX1A protein might be maintained in an inert state when not required, via post-translational controls. Nonetheless, a total of 818 (410 up- and 408 down-regulated) DEGs were observed in the AOX1A-OEX line (Figure 2). Therefore, it is likely that the excess protein is at least partially active and alters respiratory properties of the tissue. Conversely, overexpression of both AOX1A and NDB2 in the dual-OEX line led to fewer DEGs, with only 48 and 89 transcripts up- and down-regulated, respectively. That is, almost 90% of up-regulated gene transcripts and almost 80% of down-regulated transcripts in the AOX1A-OEX line were no longer differentially expressed in the dual-OEX line. Only 10 up-regulated and 18 down-regulated DEGs in the dual-OEX line were unique changes: all others were also found in the single AOX1A-OEX line. (Figure 2; Table S3). This suggests that overexpression of NDB2 together with AOX1A caused a major reversion of the AOX1A-OEX transcriptome, in agreement with the PCA, and overexpression of NDB2 in the AOX1A background led to only a few novel transcriptional changes. Of the latter, a nicotianamine synthase gene and an ABC transporter involved in auxin efflux were up-regulated, while an UMAMI transporter and a QQS gene involved in regulating carbon/nitrogen balance were down-regulated.
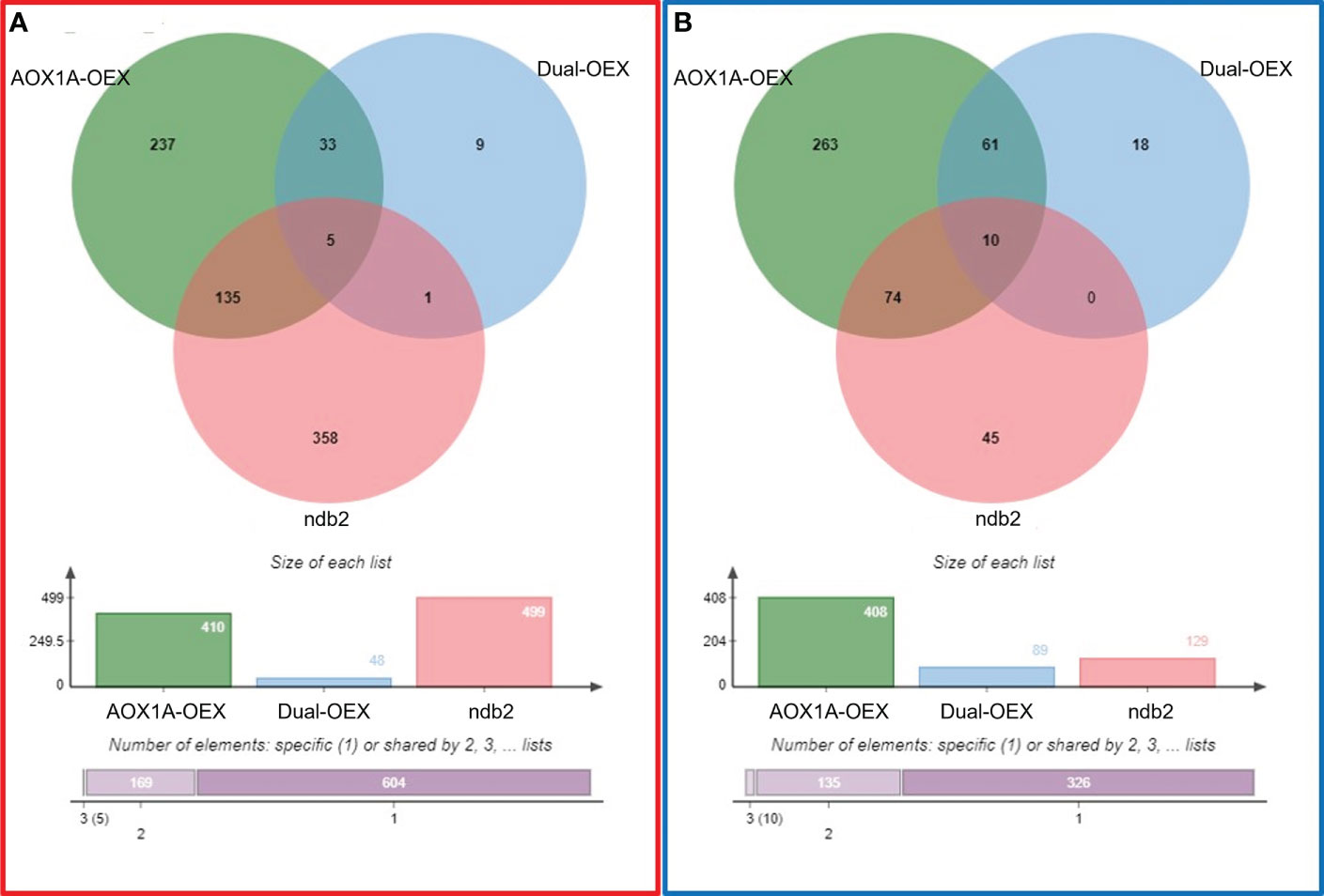
Figure 2 Commonalities and differences between (A) up-regulated and (B) down-regulated DEGs, relative to wild type. AOX1A-OEX (green), dual-OEX (blue) and ndb2 (red).
Of the 628 DEGs in the ndb2 line, 80% (499 transcripts) were up-regulated and only 20% (129 transcripts) down-regulated. A surprising and potentially important finding was the level of commonality between DEGs in lines with alternative pathway genes knocked out (ndb2) and overexpressed (AOX1A-OEX): approximately 28% and 65% of transcripts were commonly up- and down-regulated in these lines (224 transcripts in total). However, there was also a large set of unique responses in both lines (Figure 2). The set of commonly up- and down-regulated transcripts featured genes involved in development and growth (e.g. ARGOS-like genes), photosynthesis (e.g. photosystem II reaction centre proteins), respiration (e.g. a succinate dehydrogenase subunit), lipid metabolism (e.g. oleosin genes), secondary metabolism (e.g. dihydroflavonol reductase), and abiotic and biotic stress responses (e.g. defensin and DREB genes). These are described in more detail in later sections.
The relationships between AOX1A-OEX, dual-OEX and ndb2 were further explored through Gene Ontology profiling, MapMan visualisation and a survey of the most affected transcripts in each line.
3.2 Gene ontology enrichment profiles
Comparisons between transgenic lines enabled identification of ontologies that were significantly over-represented relative to WT. While there was a considerable level of overlap, particularly between AOX1A-OEX and ndb2 lines, a number of ontologies were also represented uniquely in individual lines (Figure 3). Enrichment of “signal transduction” and “respone to stress” terms were seen in up-regulated transcripts of the ndb2 line, relating to both biotic and abiotic stimuli. Cell death also featured in this line, as well as protein modification and metabolism and a significant enrichment of “metabolic process” and “cellular process”. On the other hand, the AOX1A-OEX line demonstrated a decrease in “cellular process”, while “response to stress” terms were enriched in both up- and down-regulated transcripts, particularly the latter, and could be mostly related to “response to abiotic stimulus”. The AOX1A-OEX line generally showed a stronger enrichment of ontologies among down-regulated transcripts compared to up-regulated transcripts, including terms related to development and stress. Enhanced AOX expression has been linked previously with resistance to several different abiotic stress conditions (Smith et al., 2009; Cvetkovska and Vanlerberghe, 2013; Dahal et al., 2015; Yang et al., 2019) and our results suggest that constitutive expression of AOX may prepare the plant for such stress responses. The down-regulation of development-related transcripts could relate to the delayed growth phenotype that was observed in this line but not in the other two lines (Sweetman et al., 2019).
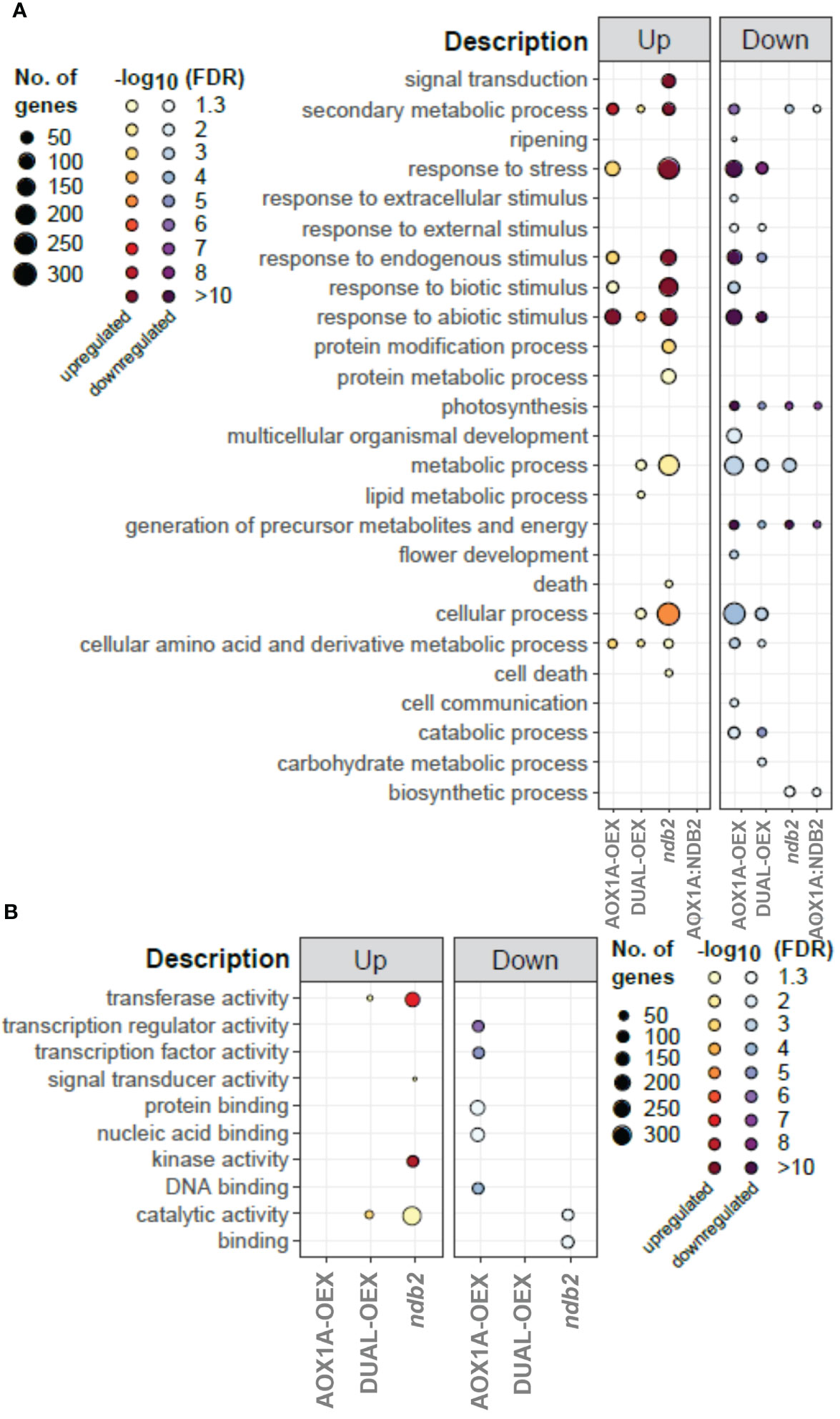
Figure 3 Enrichment of gene ontology terms between lines. BinGO was used to profile the enrichment of GO SLIM terms of individual lines, relative to wild type. The set of DEGs that are potentially regulated by AOX1A:NDB2 were also profiled separately, relative to wild type. Larger data points represent a larger number of genes within that category, while greater colour intensity represents greater significance (-log10FDR). (A) biological processes, (B) molecular functions.
The only unique enrichment terms seen for the dual-OEX line were an increase in lipid metabolic process and a decrease in carbohydrate metabolic process. Due to the lack of growth phenotype in this line (Sweetman et al., 2019), changes in transcript levels of the genes within these functional categories either do not affect protein activity, or if they do, the changes do not affect growth under non-limiting conditions. Knocking out NDB2, on the other hand, caused a large enrichment of genes involved in the response to stress and cell death, which, while not manifesting in growth changes under standard conditions, could have led to the drought and light stress sensitivity of this line (Sweetman et al., 2019).
3.3 The largest transcriptional responses to altered AOX1A and NDB2 expression
3.3.1 The top DEGs of the AOX1A-OEX line
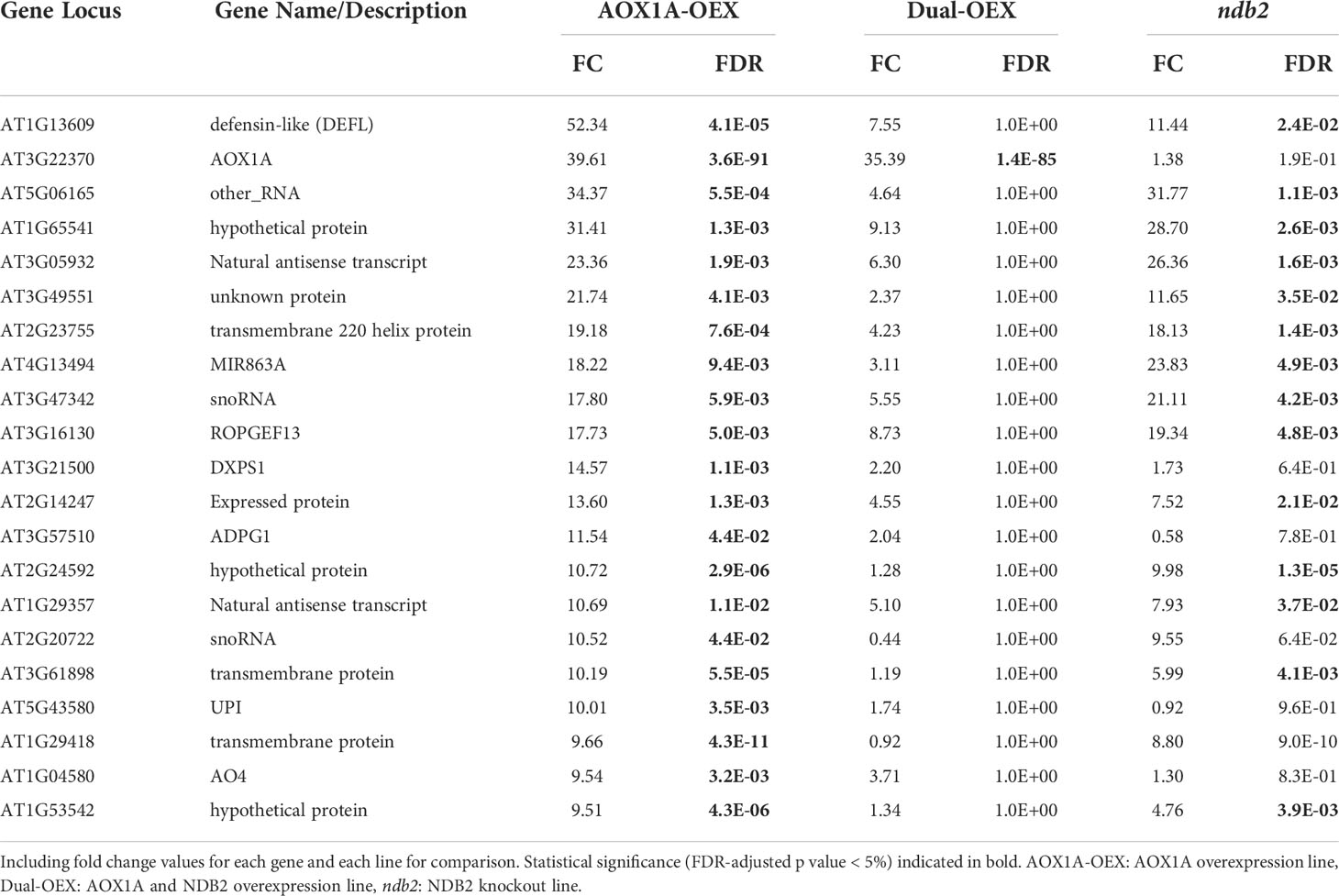
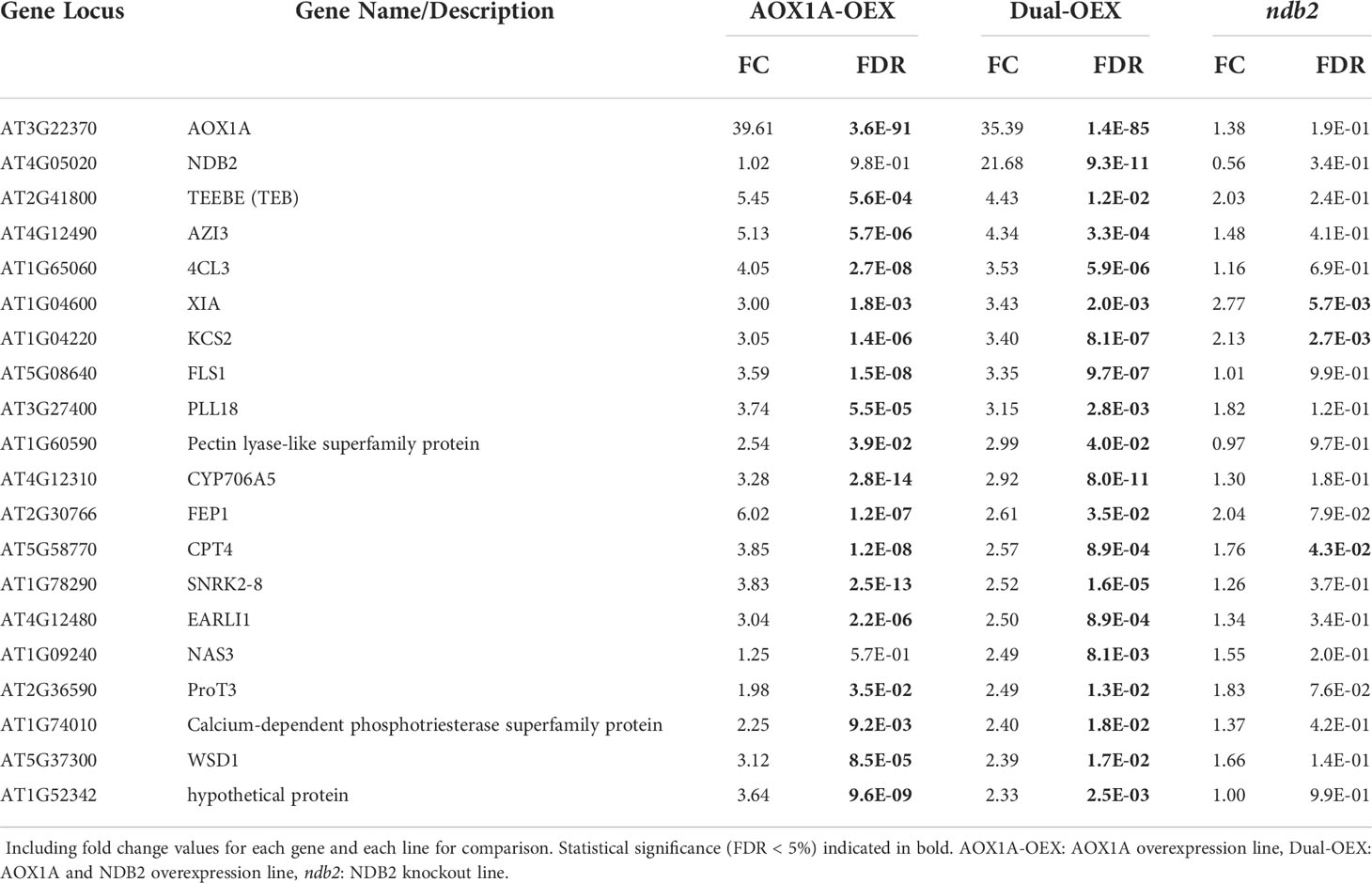
The top 20 up-regulated DEGs in the AOX1A-OEX line were expressed 9.5 to 50-fold higher than wild type transcript levels. None of these, except the AOX1A transgene, were up-regulated in the dual-OEX line. The largest transcript induction in the AOX1A-OEX line was for a defensin-like (DEFL) family protein gene (over 50-fold, Table 1), which suggested a strong defense response when AOX1A was over-expressed alone. Consistent with this, MIR863A, a microRNA thought to be involved in biotic defense (Niu et al., 2016), UNUSUAL SERINE PROTEASE INHIBITOR (UPI) and the POLYGALACTURONASE ABSCISSION ZONE A. THALIANA (PGAZAT) gene ADPG1, also featured within the top 20 most up-regulated transcripts. ADPG genes are thought to be involved in releasing pathogenesis-related signaling molecules from the plant cell wall (Ogawa et al., 2009; Gallego-Giraldo et al., 2020). AOX expression is induced also by biotic stress (Zhang et al., 2012) and constitutive over expression of AOX seems to prime the plant for increased stress resilience. Other top 20 up-regulated genes were: two small nucleolar RNAs (snoRNA); ROP (RHO OF PLANTS) GUANINE NUCLEOTIDE EXCHANGE FACTOR 13 (ROPGEF13), which may be a GDP/GTP exchange protein for a Rho of plants signal switch (Berken et al., 2005; Shin et al., 2010); 1-DEOXY-D-XYLULOSE 5-PHOSPHATE SYNTHASE 1 (DXPS1), which belongs to a gene family involved in the non-mevalonate pathway of isprenoid biosynthesis (Lange and Ghassemian, 2003), and AO4, an aldehyde oxidase.
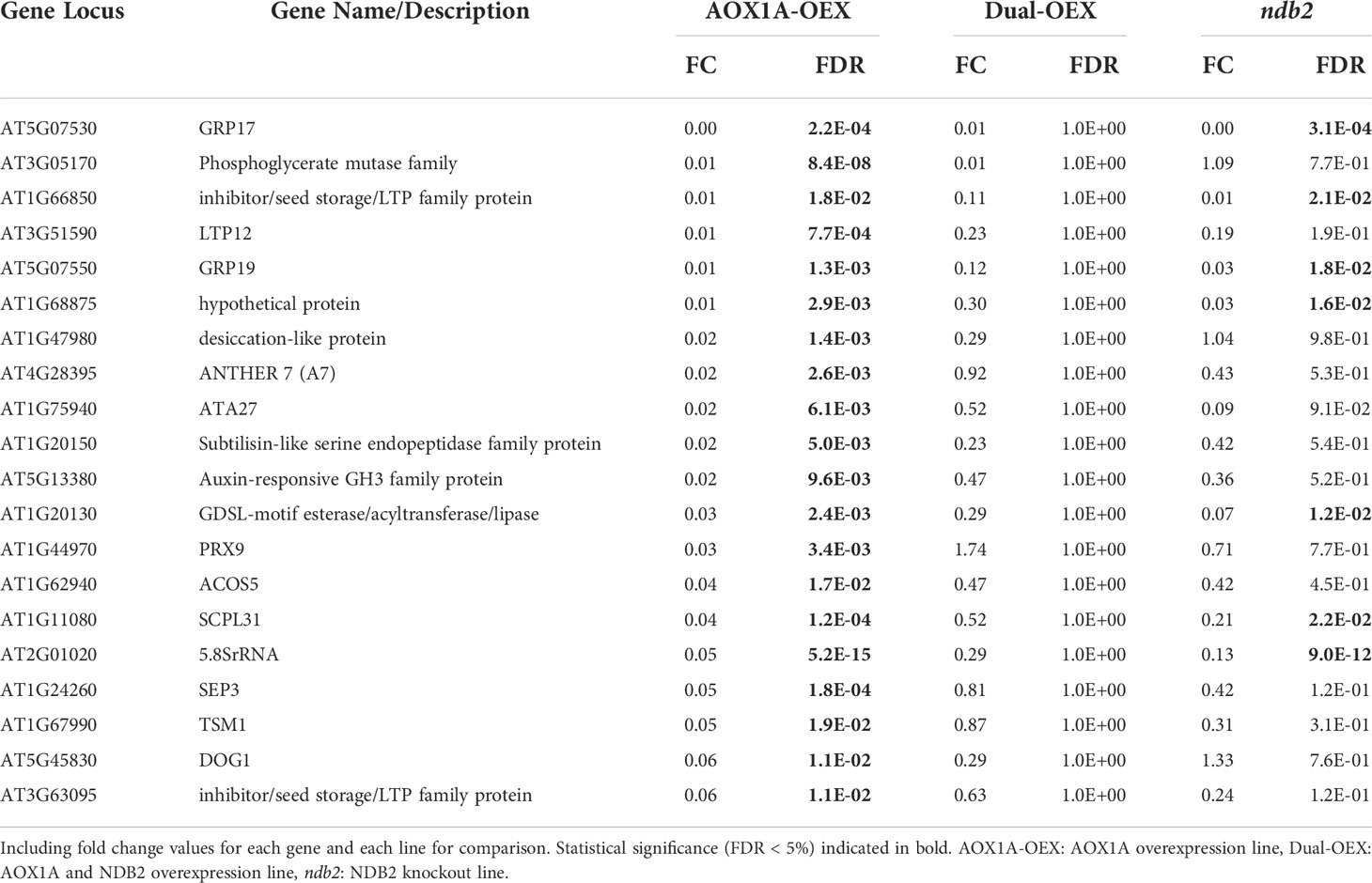
The most strongly down-regulated transcript in the AOX1A-OEX line was GLYCINE RICH PROTEIN 17 (GRP17), expressed at 0.5% of wild type levels (Table 2). This gene encodes an oleosin, important for maintaining the structure of oil bodies during seed dessication. In fact, six of the eight most strongly down-regulated genes (all transcribed to less than 2% of wild type levels) are involved in lipid transfer, seed storage, oil bodies and dessication, including another oleosin gene, GRP19. Both GRP17 and GRP19 are expressed during floral development and the corresponding proteins are found on mature pollen coats (Mayfield et al., 2001), although evidently the transcripts can also accumulate in the rosettes. Other genes that potentially relate to seeds (and all transcribed to less than 6% of wild type levels), included an esterase/acyltransferase/lipase that may be involved in reactions with lipids; an acyl-CoA synthetase that is active against medium- to long-chain fatty acids and involved in pollen formation; a tapetum-specific O-methyltransferase that may be involved with seed development and; DOG1, a QTL that is involved in controlling seed dormancy and is typically expressed in seed only. A class III peroxidase and SEPALLATA3, a MADS box transcription factor, both involved in floral development, were also strongly down-regulated to 3% and 5% of wild type levels, respectively. It is unclear why so many genes involved in flower and seed processes were expressed in the rosettes of Arabidopsis, and further, why they were down-regulated so strongly in the AOX1A-OEX line. Down-regulation of mitochondrial ROS might be involved, as ROS play integral roles in vegetative and reproductive development (see recent review by Huang et al., 2019). We have observed that seeds of the AOX-OEX lines tended to be smaller than those from WT plants (C. Sweetman unpublished results) and this may partially explain the growth delay seen in this line.
3.3.2 The top DEGs of the NDB2 knockout line
The ndb2 line showed a small and non-significant reduction in NDB2 transcript. This contradicts our previously described qPCR results (Sweetman et al., 2019) but can be explained by the RNAseq detection of a truncated mRNA. The first seven exons form a truncated transcript that cannot produce functional enzyme (Figure S1), while the qPCR assay targeted a region across the tenth and eleventh exons (Sweetman et al., 2019).
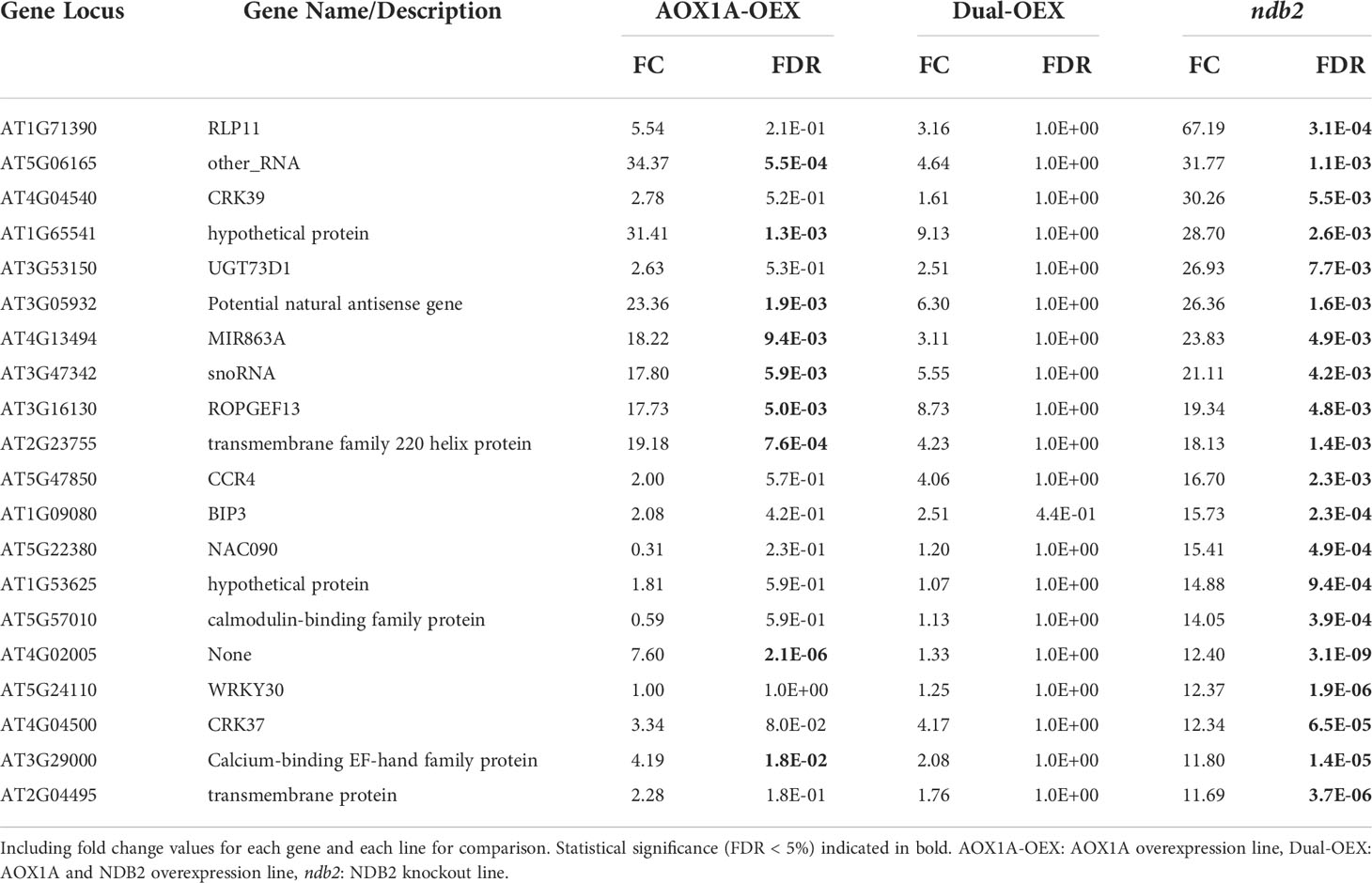
The ndb2 transcriptome showed similarities to the AOX1A-OEX transcriptome, with 9 of the top 20 up-regulated genes in the ndb2 line also up-regulated in the AOX1A-OEX line, along with 15 of the top 20 down-regulated genes (Table 3). This is consistent with the PCA clustering patterns and considerable overlap in Gene Ontology terms between lines. The most highly up-regulated genes unique to ndb2 were largely involved in signal reception, including RECEPTOR LIKE PROTEIN 11 (RLP11); CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASE 39 (CRK39) and 37 (CRK37); and CRINKLY4 RELATED 4 (CCR4), which were up-regulated between 16-60-fold. An unknown calmodulin-binding protein and transmembrane protein were also uniquely up-regulated in ndb2, along with two transcription factors, NAC DOMAIN CONTAINING PROTEIN 90 (NAC090) and WRKY DNA-BINDING PROTEIN 30 (WRKY30), plus UDP-GLUCOSYL TRANSFERASE 73D1 (UGT73D1) and a heat shock protein.
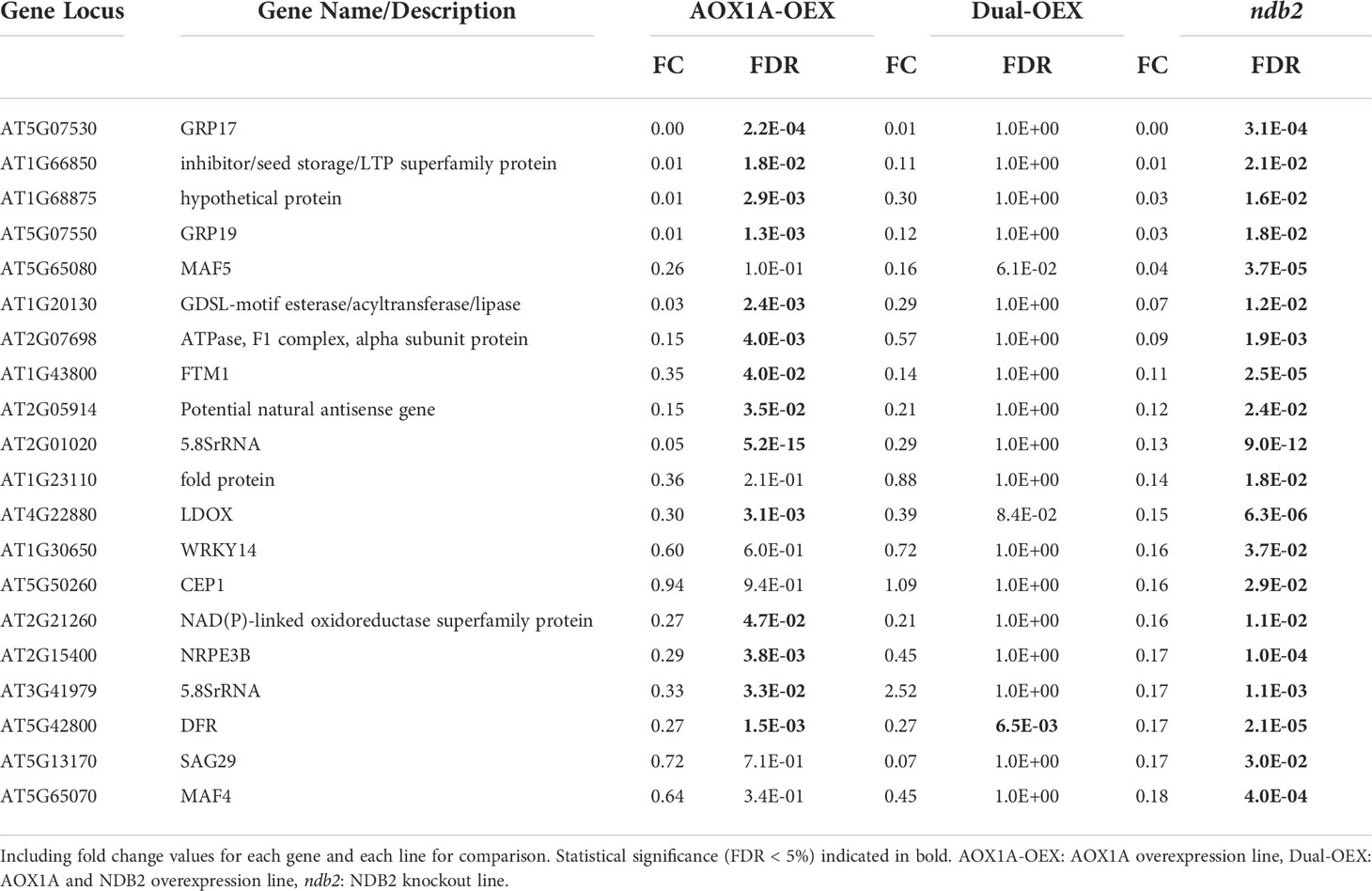
Transcripts that were uniquely down-regulated in the ndb2 line included: MADS AFFECTING FLOWERING 5 (MAF5) and 4 (MAF4), which regulate flowering time, WRKY DNA-BINDING PROTEIN 14 (WRKY14), CYSTEINE ENDOPEPTIDASE 1 (CEP1), which is likely involved in cell death, vacuolar rupture and/or cell wall thickening (Han et al., 2019), and SENESCENCE-ASSOCIATED GENE 29 (SAG29), which encodes a SWEET sucrose efflux transporter family protein (Table 4). All of these transcripts were down-regulated to less than 18% of wild type values and may be involved in regulating growth and development. Other down-regulated transcripts included an oil storage protein FLORAL TRANSITION AT THE MERISTEM1 (FTM1), two 5.8SrRNA-like genes and two genes involved in flavonoid biosynthesis, DIHYDROFLAVONOL 4-REDUCTASE (DFR) and LEUCOANTHOCYANIDIN DIOXYGENASE (LDOX).
3.3.3 The top DEGs of the dual-OEX line
The vast majority of top-20 DEGs in the dual-OEX line were also differentially expressed in the AOX1A-OEX line. Only three were uniquely up-regulated in the dual-OEX line: the NDB2 transgene, over 20-fold higher than wild type levels, ProT3, a proline transporter gene that was up-regulated 3.5-fold but was also very close to being significantly up-regulated in the AOX1A-OEX line, and NAS3, a nicotianamine synthase gene that was up-regulated 2.5-fold (Table 5). Interestingly, both NDB2 and NAS3 were up-regulated in response to UV-B treatments and this up-regulation was lost in cop1 and hy5 knockout plants, suggesting a COP1/HY5-mediated mechanism of transcription for NDB2 and NAS3 (Oravecz et al., 2006). COP1 is a positive regulator of transcriptional responses to low levels of UV-B and it acts upstream of HY5, which is involved in photomorphogenic development (Cluis et al., 2004). Neither COP1 nor HY5 were differentially expressed in the dual-OEX line, but a homolog of HY5, HYH, was up-regulated almost 4-fold. The possibility of COP1/HY5 acting as a regulator of NDB2 transcription in response to changes in light quantity and quality warrants further investigation.
All of the other top 20 up-regulated transcripts in the dual-OEX line increased by only 2.3- to 4.4-fold and many of these were up-regulated to a larger degree in the AOX1A-OEX line (Table 5). These included several genes related to cell walls, lipids and membranes (often related to defense), secondary metabolite pathway genes and a stress-related SUCROSE NON-FERMENTING 1-RELATED PROTEIN KINASE (SNRK2-8) that is activated by salt, osmotic and drought stresses but also biotic stresses (Kim et al., 2012; Lee et al., 2015; Lei et al., 2020). The FE-UPTAKE-INDUCING PEPTIDE1 (FEP1) was also highly up-regulated in both dual-AOX and AOX1A-OEX lines. This protein is responsible for activating iron deficiency response genes, although no cognate downstream transcripts were significantly up-regulated.
Of the top-20 down-regulated transcripts in the dual-OEX line, only three were unique (Table 6). First, an unnamed gene typically expressed in the root cortex (Denyer et al., 2019). This gene is down-regulated in cold (Zhu et al., 2003) but up-regulated in cellulose synthase-deficient mutants as part of a defense response (Hernandez-Blanco et al., 2007). Second, a MtN21-like amino acid transporter family protein USUALLY MULTIPLE ACIDS MOVE IN AND OUT TRANSPORTERS 33 (UMAMIT33). Third, a QUA-QUINE STARCH (QQS) gene, important for carbon-nitrogen balance (Li et al., 2015) and involved in pest defense (Qi et al., 2019).
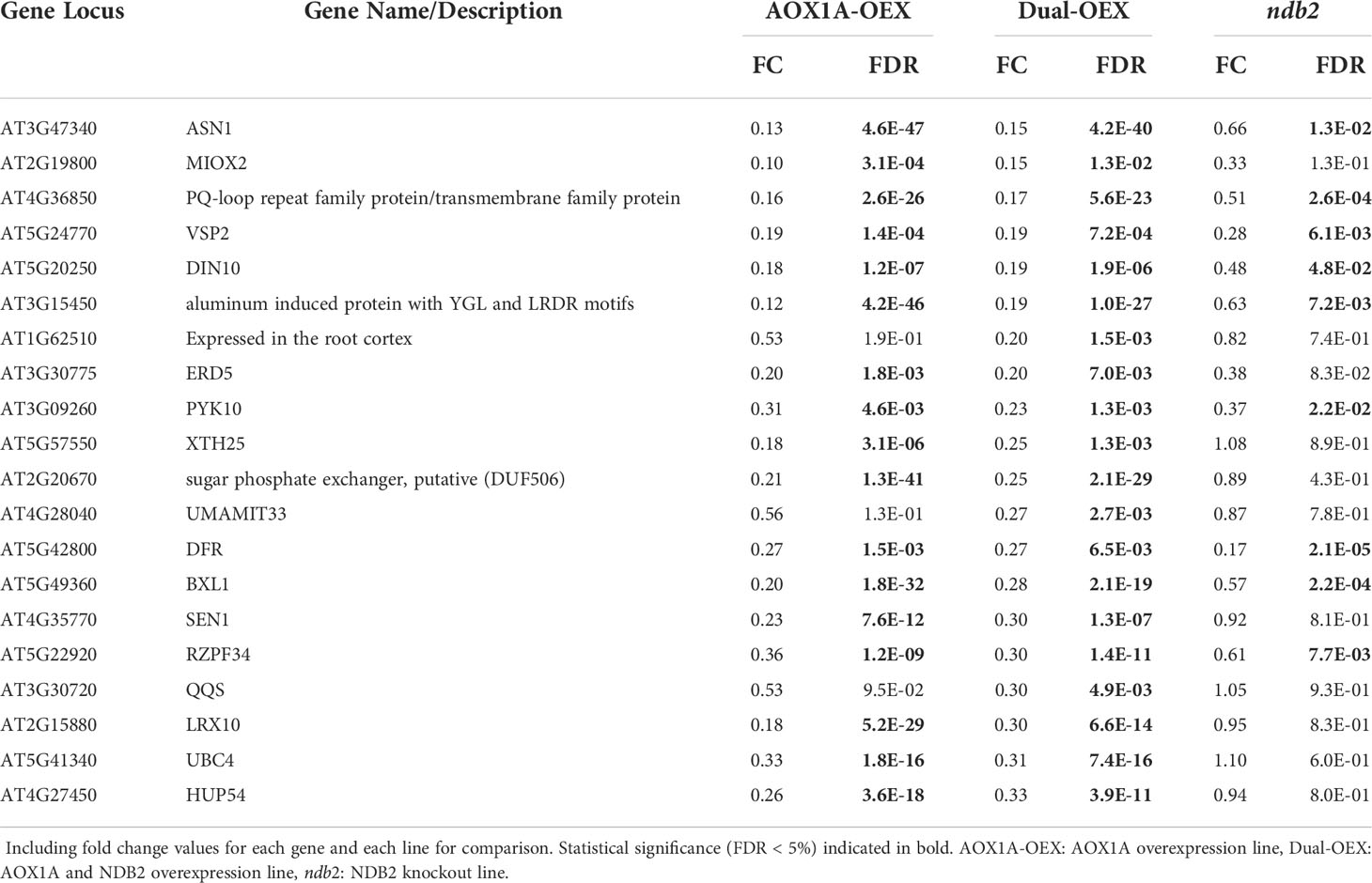
Table 6 Top 20 down-regulated DEGs for dual AOX1A and NDB2 overexpression line relative to wild type.
Several of the other top 20 down-regulated transcripts in the dual-OEX line were related to energy metabolism: GLUTAMATE-DEPENDENT ASPARAGINE SYNTHASE (ASN1) and a glycosyl hydrolase family member DARK INDUCIBLE 10 (DIN10), both of which may be suppressed by high sugar (Lam et al., 1994; Fujiki et al., 2001); a PUTATIVE SUGAR PHOSPHATE EXCHANGER (DUF506); a MYO-INOSITOL OXYGENASE (MIOX2); and EARLY RESPONSIVE TO DEHYDRATION 5 (ERD5), which is a proline oxidase that is thought to be localised to the inner mitochondrial membrane, induced by oxidative stress and high L-proline concentrations and important for sustaining growth during stress (Verbruggen et al., 1996; Sharma et al., 2011). Other down-regulated transcripts were potentially involved in abiotic stress responses, including VEGETATIVE STORAGE PROTEIN 2 (VSP2), SENESCENCE 1 (SEN1); RING ZINC-FINGER PROTEIN 34 (RZPF34) and HYPOXIA RESPONSE UNKNOWN PROTEIN 54 (HUP54). Also down-regulated in both AOX1A-OEX and dual-OEX lines were A beta-glucosidase, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 25 (XTH25) and a bifunctional (beta)-D-xylosidase/(alpha)-L-arabinofuranosidase, all involved in loosening cell walls and secondary cell wall thickening.
3.4 Transcript levels of a select set of genes may be regulated by the AOX1A:NDB2 ratio
Genes were classified into a group called “regulated by AOX1A:NDB2”, if they fulfilled two conditions: their transcript must be either up- or down-regulated in the AOX1A-OEX line but expressed at wild type levels in the dual-OEX line; and the transcript must be commonly up- or down-regulated in both the AOX1A-OEX and ndb2 lines. That is, DEGs that overlapped between the AOX1A-OEX and ndb2 lines but not the dual-OEX line. This included 135 up-regulated transcripts (Figure 2A) and 74 down-regulated transcripts (Figure 2B), as listed in Table S4. Up-regulated transcripts could not be ascribed to particular biological processes using GO SLIM analysis, likely because a large proportion of these genes were either unnamed/hypothetical proteins or RNA molecules. On the other hand, down-regulated transcripts were strongly enriched for “photosynthesis” and “secondary metabolic process”.
Of the 209 genes in this group, 18 were chloroplast-encoded. Without exception, all chloroplast-encoded transcripts that were found to be down-regulated in AOX1A-OEX were also down-regulated in the ndb2 line and unaffected in the dual-OEX line. This included several reaction center genes, ATP synthase subunits, a photosynthetic electron transfer transcript required for ATP synthesis, and the large subunit of RuBisCO (Table S4). This lends support to the idea that increasing AOX1A relative to NDB2 (either by over-expressing AOX1A or deleting expression of NDB2) drives a unique subset of transcriptional responses. In this case the down-regulated photosynthetic transcripts do not explain the growth phenotype of AOX1A-OEX, because the same changes were seen in ndb2 in the absence of any growth phenotype. Previous studies with AOX1A knockdown or knockout lines highlighted the extramitochondrial effect of AOX1A manipulation on the expression of nuclear genes whose proteins localize to other cellular compartments (Umbach et al., 2005; Clifton et al., 2006; Giraud et al., 2008). Others have found that simultaneously inhibiting electron transport through mitochondrial complex IV and AOX leads to decreased transcription in the chloroplast (Zubo et al., 2014; Adamowicz-Skrzypkowska et al., 2020). Our results extend these observations, whereby enhanced AOX1A expression or loss of NDB2 expression in the mitochondria led to modified transcription of genes directly within the chloroplast.
Only 6 genes in this group were mitochondrially-encoded, including two Complex I subunits that were down-regulated in both AOX1A-OEX and ndb2. Together with the down-regulation of a nuclear-encoded anchor subunit of succinate dehydrogenase (SDH3-2), an imbalance in AOX1A:NDB2 may affect classical mitochondrial electron transport chain components via internal and retrograde signaling mechanisms. Two up-regulated calmodulin proteins, CML47 and CML23, are also of interest because NDB2 activity is partially regulated by calcium and could be involved in calcium sensing or signaling (Geisler et al., 2007; Wagner et al., 2016). CML23 also regulates nitric oxide levels, which inhibits cytochrome c oxidase but not AOX (Millar and Day, 1996).
AOX1A and NDB2 are commonly co-expressed in response to stresses (Clifton et al., 2005), therefore it is interesting that RARE COLD-INDUCIBLE 2B (RCI2B) and another protein of this family, F-BOX STRESS INDUCED 3 (FBS3), as well as the cold-responsive DREB1A, are among the group of genes affected by the AOX1A:NDB2 ratio. Several transcription factors and hormone-responsive genes were also affected, many with roles in development and differentiation, including ANGUSTOFOLIA 3 (AN3), INFLORESCENCE DEFICIENT IN ABSCISSION-LIKE 2 (IDL2), LITTLE ZIPPER 1 (ZPR1), ARGOS and ARGOS-LIKE (ARL), and TEOSINTE BRANCHED 1, CYCLOIDEA AND PCF (TCP3). A NEET group protein that is important for plant development, senescence, reactive oxygen homeostasis and Fe metabolism is interesting in relation to the delayed growth phenotype observed in the AOX1A-OEX line (Sweetman et al., 2019). The presence of ABA4 in this set of genes was also relevant, as it is essential for neoxanthin biosynthesis and protection of photosystem II and there is building evidence that mitochondrial alternative respiration can protect photosynthetic activities during prolonged stress (Vanlerberghe et al., 2016). It should be noted also that expression of AOX1A in Arabidopsis is regulated by ABA response factors: in particular, AOX1A expression is repressed by the ABSCISIC ACID INSENSITIVE 4 (ABI4) transcription factor that also acts downstream of chloroplast retrograde signalling pathways (Giraud et al., 2009). The large number of photosynthesis-related genes belonging to this group suggests that altering the AOX1A:NDB2 ratio has a dramatic effect on the composition of the photosynthetic apparatus and presumably photosynthetis in the leaves.
3.5 Functional responses to AOX1A and NDB2 manipulation
Increased transcript levels of AOX1A and NDB2 confirmed that these genes were indeed overexpressed in the AOX1A-OEX and dual-OEX lines, by at least 35-fold and 20-fold, respectively (Table S5). However, there were no significant changes in other alternative pathway transcripts in any line, relative to wild type.
3.5.1 Overviews of metabolism, cellular responses and regulation
To gauge the overall effect of altered AOX1A and NDB2 expression on various processes and metabolic pathways, the data were used to populate “overview” MapMan maps (Thimm et al., 2004). As mentioned previously, many transcriptional changes were observed in the AOX1A-OEX line, and these either returned to normal in the dual-OEX line, or were retained (but often only partially), while the ndb2 line also shared many transcriptional effects with the AOX1A-OEX line. These responses are clearly demonstrated in “Metabolism Overview” maps (Figure S2), especially under the sub-sections of “Light Reactions”, “Tetrapyrrole”, “minor CHO” and “Cell wall”.
The “Regulation Overview” map (Figure S3) shows that many transcription factors were up- and down-regulated in the AOX1A-OEX line, but far fewer were altered in the dual-OEX line. Of the hormone pathways, the AOX1A-OEX line showed general up-regulation of genes related to SA and ABA, and general down-regulation of genes related to GA, cytokinins and JA. IAA and Ethylene-related genes were both up- and down-regulated in AOX1A-OEX. The down-regulated IAA-related genes were conserved in dual-OEX, suggesting that these genes may respond to increased AOX1A regardless of NDB2 expression. The story was quite different for the ndb2 line, which showed a clear up-regulation of genes related to protein degradation, receptor kinases and calcium regulation. This may suggest a greater sensitivity to external stimuli such as pathogens, consistent with the “Cellular Response Overview” map (Figure S4), which showed remarkable up-regulation of genes involved in biotic stress and heat stress responses. There were also strong up- and down-regulation of genes involved in biotic stress, plant development, heat and drought/salt stress in the AOX1A-OEX line (Figure S4). Similar profiles were seen in the dual-OEX line, but again, these were generally fewer and less pronounced. The ndb2 line again shared similarities with the AOX1A-OEX line, particularly for drought/salt stress and “Misc. abiotic stress”.
3.5.2 Mitochondrial electron transport chain components
A decrease in ATP synthesis might be expected in the AOX1A-OEX line, not only due to a smaller proton motive force across the inner mitochondrial membrane as a result of enhanced AOX1A (assuming it is active), but also via up-regulation of mitochondrial ATP SYNTHASE INHIBITOR FACTOR 1 (IF1) and via down-regulation of Complex I and II subunit transcripts (NAD9, NAD4, SDH3-2) (Table S6; Figure S5). The ndb2 line showed a decrease in the same Complex I and II transcripts, but no effect on ATP synthase nor its inhibitor was evident. Intriguingly, there was also a 2.6-fold induction of a putative mitochondrial phosphate transporter in the ndb2 line. Further studies with purified mitochondria from these lines together with oxygen discrimination and flux analyses are necessary to determine whether there are any changes to mitochondrial activities in vivo.
3.5.3 Primary respiratory pathways and carbon usage
Transcripts of sugar- suppressed genes ASN1, DIN10 and MYO-INOSITOL OXYGENASE 2 (MIOX2) were strongly down-regulated in both the AOX1A-OEX and dual-OEX lines and featured in the top 20 down-regulated DEGs of the dual-OEX line. Other transcripts related to sugar metabolism, transport and regulation (SWEET1, 2, 10 and AKINBETA1) were also affected although some of these occurred only in the AOX1A-OEX line. These changes may reflect an increase in mobilisation of stored carbohydrates, perhaps in response to decreased ATP synthesis in the mitochondria (see above). However, these genes cannot explain the different growth responses because most changes were observed in both the AOX1A-OEX and dual-OEX lines.
Transcripts of core glycolytic and TCA cycle enzymes were generally unaffected (Figures S6, S7, Table S7), consistent with previous studies with AOX1A antisense plants (Umbach et al., 2005; Clifton et al., 2006). This may indicate that glycolysis and the TCA cycle do not rate limit sugar metabolism under our growth conditions. However, strong and opposing regulation of phosphoglycerate mutase isoforms were seen, with one putative isoform down-regulated in the AOX1A-OEX line (by 99%) and another isoform up-regulated in the ndb2 line (7-fold). Phosphoglycerate mutase catalyses a reversible, non-rate-limiting step but transcript level changes of this magnitude could affect carbohydrate metabolism.
3.5.4 Photosynthesis
Several photosynthesis-related ontologies were significantly affected in the AOX1A-OEX line, including members of light harvesting complexes for photosystem I and II, chlorophyll-binding proteins, reaction centres, antennae systems, electron transfer mediators, complex assembly proteins and RuBisCO subunits (Table S8). The MapMan “Photosynthesis” map (Figure S8) also indicated strong down-regulation of eleven PSI and PSII light-harvesting complex genes in the AOX1A-OEX line as well as two PSI reaction center genes. Five PSII reaction center genes were down-regulated, although another four were up-regulated. The J subunit of PSI was also up-regulated, as well as a protein required for assembly of the thylakoid NAD(P)H complex, PQL3. At least one PSII light-harvesting gene and several PSI and PSII reaction centre genes were among the few transcripts that showed a conserved response in the AOX1A-OEX and dual-OEX lines but not in the ndb2 line. Therefore, while many photosynthesis-related genes may be responsive to changes in AOX1A:NDB2, the impact of overexpression of AOX1A alone on some photosynthetic genes was not reversed by rebalancing AOX1A and NDB2 expression (Figure S8). If these transcript changes cause changes in protein abundance, we might have expected plant growth to be more affected than previously observed in these lines (see Sweetman et al., 2019); perhaps photosynthesis was not limiting under our growth conditions. On the other hand, decreased expression of light-harvesting complexes is consistent with the ability of the AOX1A-OEX and the dual over expressing lines to better tolerate an increase in light intensity (Sweetman et al., 2019).
Knockout aox1a lines have impaired PSII function, especially under high light and drought conditions (Bartoli et al., 2006; Giraud et al., 2008; Yoshida et al., 2011). Watanabe et al. (2016) suggested that when the cytochrome pathway becomes limited or inhibited, excess reductants can flow through the AP, minimising excess ROS/RNS production. Zhang et al. (2011) suggested that AOX inhibition leads to build-up of excess reducing equivalents and PSI becomes over-reduced. Our experiments suggest that overexpression of AOX1A can alter signalling that affects the expression of both PSI and PSII in the nucleus and chloroplast, and that this effect can be largely (but not completely) reversed if NDB2 is overexpressed as well (Figure S8). Overexpression of both NDB2 and AOX1A together could allow more rapid oxidation of excess reducing power exported from the chloroplast, especially under photo-inhibitory conditions such as combined drought and high light stress.
3.5.5 Antioxidants and osmoprotectants
In a previous study, AOX-overexpressing lines of A. thaliana (S5 and S9) showed higher ascorbate content in leaf extracts, and enhanced production of ascorbate in detached leaves and isolated mitochondria (Bartoli et al., 2006). Galactonolactone dehydrogenase (GLDH) is the only mitochondrial enzyme of the ascorbate biosynthesis pathway, while GDP-L-galactose phosphorylase and the others are dual localized to the cytosol and nucleus (Fenech et al., 2021). Furthermore, GLDH associates with Complex I of the mETC, and donates electrons directly to cytochrome c during the catalysis of galactonolactone to ascorbate (Millar et al., 2003). While the transcript and activity of GLDH was unchanged in AOX1A-overexpressing lines (Table S9; Bartoli et al., 2006), an increase in AOX activity might increase the availability of oxidized cytochrome c. Interestingly, in the present study, the gene encoding GDP-L-galactose phosphorylase (VITAMIN C DEFECTIVE 2; VTC2), was strongly up-regulated in AOX1A-OEX. This is a potentially rate-limiting step (Dowdle et al., 2007; Fenech et al., 2021) and enhanced VTC2 transcript in AOX1A-OEX could lead to increased ascorbate biosynthesis. Given the increase in ascorbate levels and VTC2 transcript in separate AOX1A-OEX lines, it is possible that over-expression of AOX1A can drive increased ascorbate production. Meanwhile in the ndb2 line, an ascorbate oxidase gene involved in the oxidation of ascorbate to monodehydroascorbate was strongly up-regulated, suggesting an increase in oxidative stress signaling.
Strong up-regulation of transcripts encoding two proline transporters (PROT2 and 3) and a membrane protein potentially involved in drought-induced proline accumulation (AFL1; (Kumar et al., 2015), might suggest accumulation of proline in the AOX1A-OEX and dual-OEX lines (Table S9). This is supported by decreased transcript levels of proline catabolism enzymes, the mitochondrial PROLINE DEHYDROGENASE 1 (PRODH1), also known as ERD5 (Kiyosue et al., 1996) and PRODH2, which is colocalized to the mitochondria and chloroplasts (Van Aken et al., 2009). However, the ERD5 promoter contains a proline-inducible element (Nakashima et al., 1998), therefore down-regulation of this transcript could also point to decreased availability of proline in the cell. Recently, it was revealed that AOX activity can facilitate the catabolism of stress-accumulated proline (Oh et al., 2021), and therefore a lower level of available proline might be expected in plants over-expressing AOX1A.
3.5.6 Secondary metabolism – flavonoids, pigments, phenylpropanoid pathway
Both the AOX1A-OEX and dual-OEX lines showed increased expression of genes related to chalcones and flavonols, with a down-regulation of anthocyanin metabolism genes (Figure S9). The ndb2 line showed fewer changes but strong up- and down-regulation of dihydroflavonol- and anthocyanin-related transcripts. At the gene ontology level, flavonoid biosynthetic process was over-represented in all three lines compared to wild type. Representative genes of this group included CHIL, FLS1, TT4, TT5, UGT84A2, FLA15 and PDE339. FLS1 (FLAVONE SYNTHASE 1) was one of the most significantly altered transcripts in the dual OEX line. Balance between TT4 (a chalcone synthase) and TT7 (a flavonoid 3-hydroxylase) can determine the relative abundance of quercitin and kaempferol in Arabidopsis (Lewis et al., 2011). This may be relevant considering the inhibitory effects of quercitin, but not kaempferol, on alternative respiration (Shimoji and Yamasaki, 2005). The increase in TT4 transcript in both AOX1A-OEX and dual-OEX lines (and in a separate AOX1A overexpression line; Fiorani et al., 2005) may increase kaempferol biosynthesis as a result of elevated AOX. Meanwhile, a decrease in TT7 transcript only in the AOX1A-OEX line, suggests that the inhibitory role of quercitin, if indeed it occurs in vivo, may be by-passed when AOX1A is overexpressed, but potentially regained when NDB2 and AOX1A levels are rebalanced (Figure 4). This raises the possibility that in vivo AOX1A activity may be greater in the AOX1A-OEX line relative to the dual overexpression line, and this in turn may directly affect energy state and growth in the AOX1A-OEX line. This requires further experimentation using oxygen discrimination to determine AOX activity. Transcripts of DIHYDROFLAVONOL 4-REDUCTASE (DFR) and LEUCOANTHOCYANIDIN DIOXYGENASE (LDOX) were down-regulated strongly in all three lines, suggesting decreased capacity for further metabolism of kaempferol (e.g. to anthocyanins). Based on these transcriptional changes, levels of kaempferol may be higher in both lines overexpressing AOX1A, which could ready the cell for rapid biosynthesis of quercitin and inhibition of the alternative respiration, as necessary.
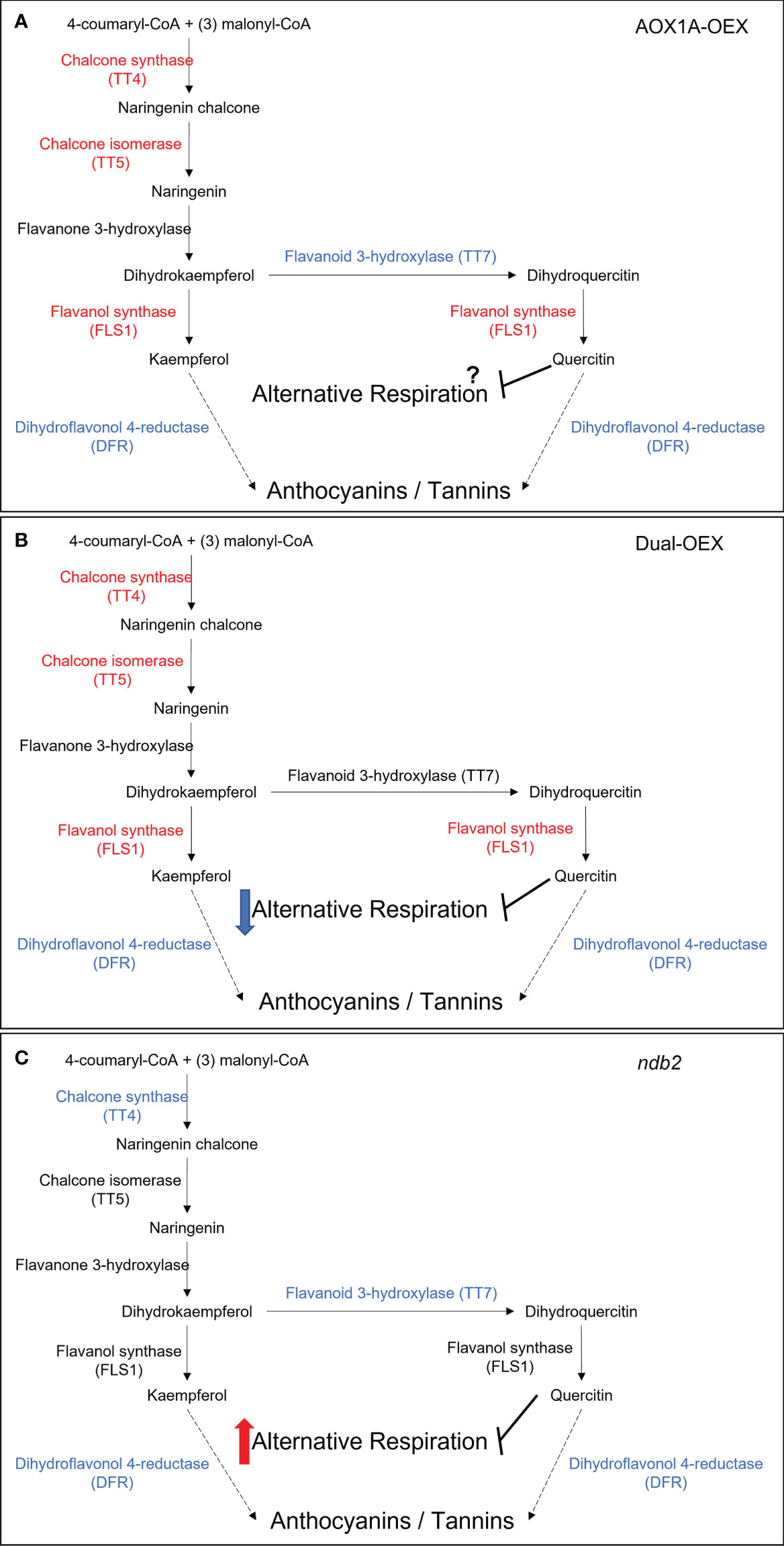
Figure 4 Flavonoid biosynthesis pathways and the potential effects on alternative pathway regulation in (A) AOX1A overexpression line, (B) dual overexpression line and (C) NDB2 knockout line. Major metabolite and enzyme steps in black. Red indicates an up-regulated transcript, blue indicates a down-regulated transcript. Red and Blue arrows represent potential down- and up-regulation of AOX activity. Dotted arrow indicates a multiple-step pathway with details omitted.
Flavonoids often accumulate in response to abiotic or biotic stresses, to assist the scavenging of ROS that escape from the chloroplast or mitochondria during oxidative stress, thereby improving tolerance to stress (Fini et al., 2011; Nakabayashi et al., 2014). This may explain the very strong upregulation of flavonoid and anthocyanin-related transcripts in aox1a lines exposed to a combined drought and light stress (Giraud et al., 2008), which coincided with increased anthocyanin content, but does not explain the accumulation of transcripts when AOX1A is overexpressed in the absence of a stress (Figure S9). It is feasible that the regulators of flavonoid genes, including MYB transcription factors, which are known to be affected by redox status (Dubos et al., 2010), including photosynthetic redox status (Akhtar et al., 2010), may also be responsive to changes in the expression of alternative respiration pathway components. Transcript levels of several MYB transcription factors, including MYB111, which is known to positively affect flavonol biosynthesis (Stracke et al., 2007) were significantly up-regulated in AOX1A-OEX and dual-OEX, giving credence to this hypothesis.
Phenylpropanoid biosynthesis genes were up- and down-regulated in the AOX1A-OEX line and also, but to a lesser extent, in the dual-OEX line (Figure S10). Strongly up-regulated transcripts included a feruloyl-CoA transferase (RWP1), two cinnamyl alcohol dehydrogenases (CAD4 and CAD9), a fatty acid reductase (FAR5), a 3-ketoacyl-CoA synthase (KCS2) and a 4-coumarate:CoA ligase (4CL3). The latter was also one of the most strongly up-regulated transcripts in the dual-OEX line. Sharma et al. (2019) suggested that the role of 4CL3 was to alter the activity and transcription of numerous genes in the phenylpropanoid pathway. Links between AOX and phenylpropanoid metabolism have been demonstrated previously (Macedo et al., 2012; Sircar et al., 2012). Many of the genes involved in the phenylpropanoid pathway are stress responsive and react to a broad range of stressors (Dixon and Paiva, 1995; Sharma et al., 2019). Alteration to the alternative pathway of respiration, with its broad ranging effects, may modify the signaling between mitochondria and chloroplasts, resulting in adjustments to downstream secondary metabolite pathways such as flavonoid and phenylpropanoid biosynthesis. This also may explain, in part, why these plants were able to recover when exposed to a combined moderate light and drought stress (Sweetman et al., 2019).
3.5.7 Hormone metabolism/regulation
Transcripts of genes involved in various hormone biosynthesis and regulation pathways were affected, particularly in the AOX1A-OEX and ndb2 lines (Figure S3). However, the transcriptional similarities typically observed between AOX1A-OEX and ndb2 in other pathways, were less conserved here. For example, the AOX1A-OEX line showed strong down-regulation of abscisic acid (ABA) biosynthesis genes and up-regulation of the ABA catabolism gene, CYP707A1 (Table S10), supporting the negative relationship between AOX1A and ABA, while ndb2 showed little regulation of the same transcripts. For the jasmonate biosynthesis pathway, AOX1A-OEX showed strong down-regulation of several transcripts, while ndb2 did not (Table S10, Figure S3). This suggests that the overexpression of AOX1A and knockdown of NDB2 may lead to different hormonal signals despite similar transcriptional responses in other pathways.
Transcripts of several genes involved in ethylene metabolism and signaling, salicylic acid metabolism and responses, auxin transport and responses, gibberellin biosynthesis and modifications, ABA biosynthesis and jasmonate biosynthesis, were variously up- and down-regulated (Figure S3; Table S10). Overall, increased AOX1A expression may (i) influence ethylene- and jasmonate-directed ROS responses, cell death and root morphology pathways (ii) induce the accumulation of SA via ROS signaling (Zhang et al., 2012), which may sacrifice growth to up-regulate stress priming mechanisms, (iii) support the antagonistic relationship previously observed between auxin signaling and mitochondrial signaling for AOX1A expression (Ivanova et al., 2014), and (iv) lead to a general increase in gibberellins but a proportional decrease in GA4, which is important for shoot elongation and flower initiation (Eriksson et al., 2006). Many of these factors could contribute to the delayed growth phenotype of AOX1A-OEX (Sweetman et al., 2019), but are yet to be tested experimentally.
3.5.8 Stress responses
A defensin-like protein showed the strongest up-regulation of any transcript in the AOX1A-OEX line, and numerous other transcripts related to abiotic and biotic stress featured in the top 20 up- and down-regulated transcript lists (Figure S11; Tables 1, 2). Some examples are given below, and in many cases these changes were absent, or at least dampened in the dual-OEX line. Most notable, though, was the response of the ndb2 line, with a striking up-regulation of transcripts relating to abiotic and biotic stress, R proteins, PR proteins, heat shock and signaling (Figures S4, S11) and receptor kinases (Figures S3, S12). These receptor kinases, particularly DUF26 members, have been implicated in biotic and abiotic stress responses and are regulated by growth effectors such as SA (Wrzaczek et al., 2010). Considering the vulnerability of ndb2 plants to a combined drought and moderate light treatment (Sweetman et al., 2019), this up-regulation of stress response pathways under standard growth conditions does not appear to improve tolerance to stress.
A major role proposed for AOX is in the prevention of ROS production during exposure to environmental stress (Maxwell et al., 1999; Vanlerberghe, 2013). Therefore, it was not surprising to see strong down-regulation of several peroxidase genes in the AOX1A-OEX line, including a 97% decrease in PEROXIDASE9 (PRX9) transcript. Transcription factors such as REDOX RESPONSIVE TRANSCRIPTION FACTOR 1 (RRTF1), ETHYLENE-RESPONSIVE ELEMENT BINDING PROTEIN (EBP) and ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 6 (ERF6) respond to ROS and mitigate oxidative stress by decreasing ROS levels, ultimately also protecting against cell death. Each of these transcripts was strongly down-regulated in the AOX1A-OEX line. That is, AOX1A overexpression led to altered ROS-signaling networks, likely via a decrease in ROS generation in the mitochondria. Indeed, there was a significant decrease in levels of malondialdehyde equivalents in the AOX1A-OEX line, suggesting a decrease in ROS-related lipid peroxidation (Figure 5). The dual-OEX and ndb2 lines also showed similar decreases but were not statistically significant (Figure 5).
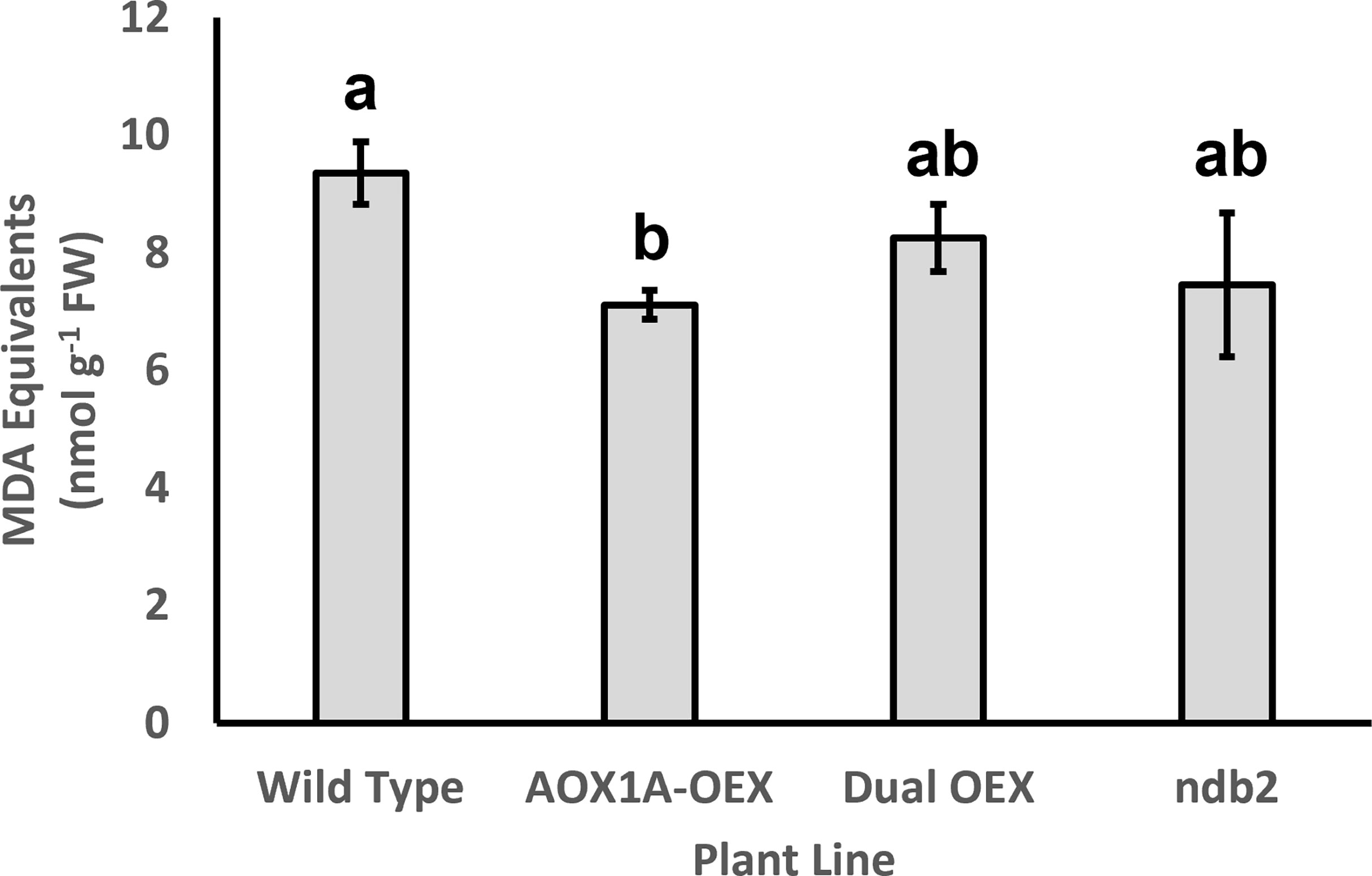
Figure 5 Effect of altered AOX1A and NDB2 expression on lipid peroxidation levels. Malondialdehyde equivalents were measured using a TBARS assay (Hodges et al., 1999), from aliquots of the powdered leaf samples used for RNA extractions. Columns represent means (n = 3 ± S.D.). Statistical significance indicated by different letters, based on one-way ANOVA with post-hoc Bonferonni-corrected pairwise comparison (corrected p < 0.05).
Several stress-related transcripts that were up-regulated in the AOX1A-OEX line also have roles in the regulation of growth and morphology. For example (1) a Cytochrome P450 involved directly in ABA catabolism and post-germination growth (CYP707A1; Okamoto et al., 2006), (2) a TF involved in integrating light and hormone signaling to regulate internode elongation (ATH1; Ejaz et al., 2021), (3) a coiled-coil protein involved in chloroplast movement in response to excess light (PMI2; Kodama et al., 2010), (4) a tandem zinc knuckle protein that negatively regulates hypocotyl growth in the mornings (TZP; (Loudet et al., 2008), and (5) all four “suppressor of phyA-105” protein family genes (SPA1-4), which suppress photomorphogenesis in seedlings and elongation in mature plants (Laubinger et al., 2004). These changes could contribute to both the stress tolerance feature of AOX1A-OEX lines and the delayed growth phenotype observed under conventional growth conditions (Sweetman et al., 2019).
3.5.9 Biotic stress
Glucosinolates in the Brassicales order are typically associated with biotic stress defence and innate immune response (Sonderby et al., 2010). Glucosinolate biosynthesis genes were significantly downregulated and “glucosinolate metabolic process” featured in GO profiling for down-regulated transcripts in the AOX1A-OEX and ndb2 lines (Table S12). An antagonistic relationship between AOX and glucosinolates has been hypothesised before, whereby repression of AOX by glucosinolates could either enable mitochondrial ROS accumulation, triggering a defense response via mitochondrial retrograde signaling pathways, or otherwise enable prioritisation of alternative stress responses (Zhang et al., 2017). Similarities between the transcriptome responses to mitochondrial perturbation and biotic stress have also been noted previously (Clifton et al., 2006; Schwarzlander et al., 2012; Umbach et al., 2012).
Altered cell wall biochemistry can be another indicator of altered biotic stress responses. Several transcripts relating to cell walls were up-regulated in the AOX1A-OEX line, and in many cases the ndb2 line, including a polygalacturonase protein required for release of cell-wall-derived PR elicitors (ADPG1; Gallego-Giraldo et al., 2020), a pectate lyase involved in the response to nematodes (PLL18; Wieczorek et al., 2014) and a DUF642 cell wall protein involved in cell replication and hypocotyl cell elongation (TEB; Salazar-Iribe et al., 2016). These responses, as well as the strong down-regulation of a gene involved in converting myo-inositol to D-glucuronic acid (MIOX2), suggests that there may be an increase in cell wall degradation and a decrease in the synthesis of new cell wall material in these lines.
3.5.10 AOX1A regulators and signaling pathways
WRKY transcription factors have been suggested as a nexus for mitochondrial and chloroplast stress responses (Van Aken and Whelan, 2012). WRKY40 in particular, has been recognized as a negative regulator of AtAOX1A and more generally as a modulator of stress-responsive genes that are encoded in the nucleus but whose proteins localize to the mitochondria or chloroplast (Van Aken et al., 2013). Transcript levels of WRKY40 and one of its targets, REDOX-REGULATED TRANSCRIPTION FACTOR (RRTF1), were both strongly down-regulated in the AOX1A-OEX line. This implies a decrease in stress-related ROS accumulation in the AOX1A-OEX line and a release of AOX1A repression. RRTF1 was one of five ethylene responsive factors that were strongly down-regulated in AOX1A-OEX (Table S10), which also included ERF4 and ERF11. Target genes of these transcription factors were enriched among down-regulated transcripts of the AOX1A-OEX line (Figure 6), therefore the decrease in WRKY40, RRTF1, ERF4 and ERF11 transcripts likely resulted in a decrease in their protein level and activity.
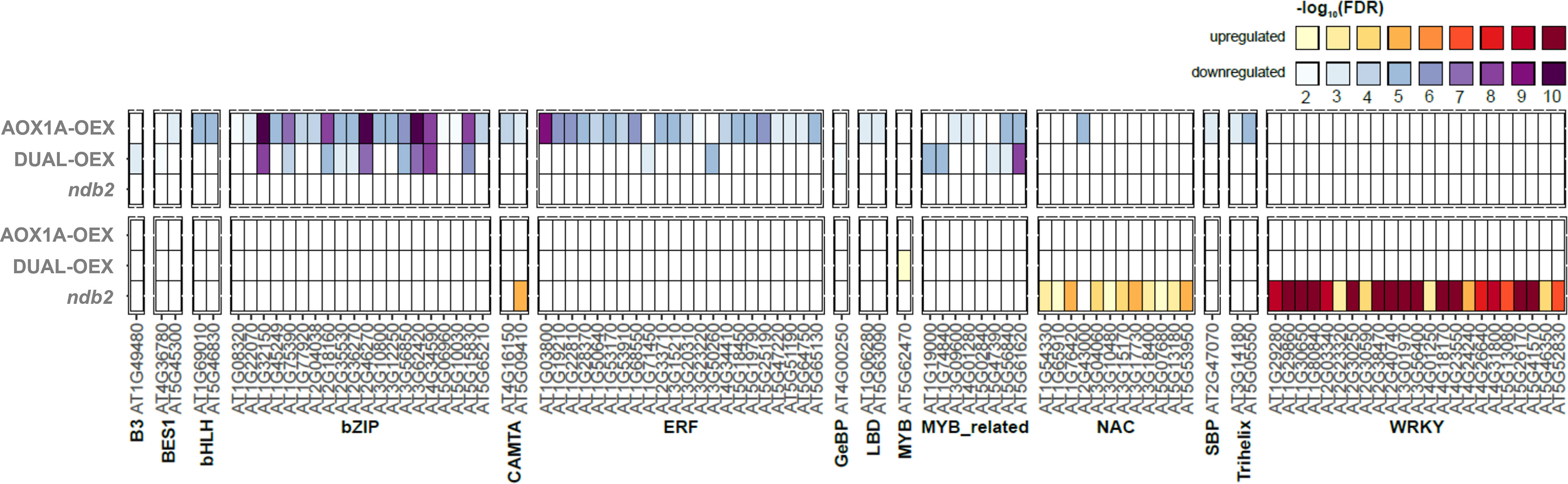
Figure 6 Enrichment of transcription factors based on differential expression of target genes. EAT-UpTF was used to profile the enrichment of transcription factor target genes among the DEGs of each line, relative to wild type. greater colour intensity represents greater significance (-log10FDR). Transcription factors were enriched in both down-regulated (top) and up-regulated (bottom) DEGs.
The ndb2 line also showed enhanced levels of transcripts relating to transcriptional control (Figure S13), particularly WRKY TFs. WRKY40, it’s binding partner WRKY18 and the target genes of these two (and other WRKY targets) were strongly up-regulated in the ndb2 line (Figure 6, Table S13), suggesting that NDB2 also can play a role in mitochondrial retrograde signaling. The WRKY40 gene is reportedly induced by pathogens including Pseudomonas syringae and Botrytis cinerea (Xu et al., 2006; Abeysinghe et al., 2019), and this transcript represents a rare example of opposing transcriptional responses between the AOX1A-OEX and ndb2 lines. While repression of AOX1A transcription by WRKY40 in the ndb2 line could be interpreted as an attempt to recreate balance between AOX1A and NDB2, there was no significant change in AOX1A transcript level in this line.
Many of the stress-responsive mitochondrial genes contain multiple WRKY binding sites within their promoters (Van Aken et al., 2013). In the ndb2 line, there was a 12-fold increase in WRKY30 (among the top 20 DEGs for the ndb2 line) and 2-3-fold increases in WRKY46, 47, 48 and 53. Target genes of many WRKY TFs were also strongly enriched in the up-regulated DEGs of the ndb2 line (Figure 6) indicating that the WRKY transcription factors were functional. Many WRKYs are responsive to oxidative stress, including WWRKY30, 46 and 48 (Scarpeci et al., 2008) and some can provide oxidative stress and salinity tolerance when overexpressed (Scarpeci et al., 2013). However, there was no evidence of oxidative stress in the ndb2 line under our standard growth conditions (Figure 5).
Transcript levels of AOX regulators NAC017 and NAC013 (De Clercq et al., 2013; Ng et al., 2013), their negative regulator RCD1 (Shapiguzov et al., 2019; Wang et al., 2020) and positive regulators CDKE;1 and KIN10 (Ng et al., 2013) remained unchanged in all lines. However, many other transcriptional regulators were affected in the AOX1A-OEX line, including down-regulation of EREBP, GRAS, HB, C2C2-CO-like, C3H and MADS transcription factors. CEJ1, which is an ethylene- and jasmonic acid-responsive DREB involved in defense and freezing stress responses, also showed down-regulation in transcript levels of both itself and its target genes. These ethylene response factors may be important for regulating the transcriptional responses to AOX1A overexpression. Any of these transcription factors and their target genes could also be involved in the delayed growth phenotype of the AOX1A-OEX line.
Several bZIP transcription factors were enriched in the down-regulated DEGs of the AOX1a-OEX line (Figure 6). This included almost the entire S1 subgroup of bZIPs (i.e. bZIP2, 11, 44 and 53), which can regulate key metabolic genes in response to cellular energy status, by forming heterodimers with group C bZIPs (for a recent review, see Droge-Laser and Weiste, 2018). Transcript levels of group C genes including BZIP63 were directly down-regulated, as well as several downstream targets of bZIP63 including e.g. DREB1A, ASN1, ERD5, DIN10 (Table S11; Baena-Gonzalez et al., 2007; Dietrich et al., 2011; Matiolli et al., 2011). bZIP63 is activated by phosphorylation through the activity of the SUCROSE NON-FERMENTING 1-RELATED KINASE 1 (SnRK1), whose regulatory subunit AKINbeta1 was also down-regulated in the AOX1A-OEX line. Overall, this suggests that the plant signaling network is responding to a high-energy status within the cell (i.e. high levels of free sugars) but this is unlikely to drive the delayed growth phenotype as discussed in a previous section. It is also possible that some of these genes respond to factors other than sugars or energy status, such as ROS-induced signals.
Overall, our results suggest that altered AOX1A and NDB2 expression causes changes in signaling pathways that extend far beyond the mitochondrion, resulting in many transcriptional changes. While some of these changes were shared between lines, many were also unique to each line, highlighting complexities in retrograde signaling that require further investigation.
3.5.11 Comparison of lines with differentially expressed AOX1A and NDB2
To date, we have compared transcriptome changes in lines overexpressing either AOX1A alone or with NDB2, as well as an ndb2 line, under standard growth conditions. The se lines behave quite differently when subjected to environmental stresses such as high light and drought (Sweetman et al., 2019): the AOX1A-OEX and dual-OEX lines show improved resilience, while the ndb2 line is more sensitive. Previous studies have focused on the transcriptomic responses of aox1a plants (e.g. Umbach et al., 2005; Giraud et al., 2008), rather than AOX1A-overexpressing plants. One manuscript has assessed the transcriptome profile of rapeseed seedlings over-expressing a typically seed-specific AOX1B gene, where it was found that transcript levels of stress-related genes were both up- and down- regulated, including genes involved in ABA, oxidative stress and osmotic stress responses, and the over-expressing plants were hypersensitive to ABA (Yang et al., 2019). Here in Arabidopsis, alterations in AOX1a and NDB2 expression also led to transcriptional changes of genes involved in stress responses and ABA. Over-expression of AOX1A led to downregulation of transcripts involved in ABA and JA biosynthesis, ABA-regulated bZIP transcription factors and ethylene response factors. Meanwhile, knockout of NDB2 led to up-regulation of WRKY and NAC signalling pathways that may involve an increase in abscisic acid and ethylene. Decreased functionality of ethylene response factors and bZIP TFs in the AOX1A-OEX line, and to a lesser degree in the dual-OEX line, suggests a general decrease in ethylene and abscisic acid signaling pathways in response to AOX1A overexpression that is somewhat reversed upon rebalancing AOX1A and NDB2 levels. Exposing plants of the AOX1A-OEX line to ethylene or ABA might therefore prevent some of the transcriptional changes, and potentially reverse the growth phenotype effect. Clearly, further growth analyses under various environmental conditions are needed.
WRKY40 was one of the few transcripts that showed opposing responses between AOX1A-OEX and ndb2 lines, and could be a significant point of difference, considering the importance of this transcription factor in stress signaling pathways (Van Aken et al., 2013). Knockout of NDB2 also led to a notable up-regulation of transcripts involved in biotic stress responses.
Our results suggest that changing the ratio of AOX expression to that of NDB2 has a particularly pronounced transcriptional response, and that the two enzyme activities need to be balanced to preserve wild-type gene expression patterns. The AOX1A-OEX line had over four times the number of significantly altered genes (relative to wild type), compared to the dual AOX1A and NDB2 overexpression line. The most likely explanation is that changes caused by overexpression of AOX1a alone were attenuated by co-overexpression of NDB2. This agrees with phenotyping data where growth delays seen in the single OEX were no longer present in the dual overexpression lines (Sweetman et al., 2019) and may also be related to altered ROS metabolism, based on MDA equivalents (Figure 5). Meanwhile, a subset of transcripts behaved similarly in both the AOX1A-OEX line and the ndb2 line, suggesting some common adjustment when AOX1A expression is high relative to NDB2.
Rebalancing the input (i.e. NDB2) and output (AOX1A) of the electron transport chain seems important for maintaining regular growth during non-limiting conditions while maintaining the potential stress benefit of an enhanced alternative respiration pathway. Altering this balance is likely to affect not only mitochondrial redox poise and ROS accumulation, but also the redox poise of other cell compartments. For example, decreasing the capacity to oxidise NADH generated by photorespiration of glycine, or excess reducing equivalents exported from the chloroplasts, could lead to over reduction of the chloroplast and accumulation of reactive oxygen species in the cell. This can certainly explain the sensitivity of the ndb2 and aox1a lines to photoinhibitory stress conditions (Giraud et al., 2008; Watanabe et al., 2016; Sweetman et al., 2019; Li et al., 2020). Redox regulation has been demonstrated for several transcription factors, affecting DNA binding and cellular distribution (Tron et al., 2002; Heine et al., 2004; Dietz, 2014). As an example, group G bZIPs such as bZIP16 contain a conserved cysteine residue which, when reduced, enables binding and transcriptional activation of the target DNA (Shaikhali et al., 2012). In AOX1A overexpressing plants, target genes from three such transcription factors, including bZIP16, were significantly down regulated despite little or no change in expression of the transcription factors themselves. As such, decreased transcript levels of the PSII light harvesting complex gene LHCB2.4 and other target genes in the AOX overexpression lines might be due to decreased binding efficiency of bZIP16 as a result of altered redox poise of the cell. Further comparisons between control and stressed plants may help to elucidate the role of bZIP16, WRKY40 and other signaling molecules in the altered stress tolerance of these lines.
It should be noted that some transcriptional changes observed between lines may be due to their proximity to transgene insertion sites, which are unknown for the AOX1A-OEX background and dual-OEX lines. However, at least three individual dual-OEX lines showed the same growth phenotype (Sweetman et al., 2019), therefore, if there is an insertional effect in dual-OEX, it is unlikely to affect growth. We can also assume that any transcriptional change in AOX1A-OEX that reverted towards WT expression in the dual-OEX line, is not due to an insertional effect in either line. Nevertheless, targeted experimental analyses are required to define the origin and downstream effects of the key transcriptional changes.
It should also be noted that a common problem in studies where plant growth is affected, is separating primary and secondary effects. That is, determining which, if any, of the transcriptional changes cause the growth phenotype and which are a consequence of the growth phenotype. Here we present all the significant transcript changes, to serve as a starting point for further research. Tracking transcriptional changes over time may help to distinguish between primary and secondary transcriptional effects, whereby transcripts affected very early in development may be causative but those that only appear later may be secondary effects.
4 Conclusions
In this report, we describe the detailed transcriptional profile of plants with genetically enhanced AOX1A, and of plant lines with modified NDB2 expression. A wide range of transcriptional changes were observed as a consequence of this genetic manipulation, even though plants were grown under non-limiting conditions. These include changes in the expression of:
1. Photosynthesis genes, with PSII reaction centre genes down-regulated in response to increased AOX1A:NDB2 and PSI and PSII light harvesting genes down-regulated in all lines with increased AOX1A transcript. Typically, a loss of AOX1 limits the efficiency of photosynthesis, but according to these transcriptional changes, increasing AOX1A expression by itself may also limit photosynthetic capacity.
2. Sugar signaling and transport genes e.g. DIN10, ASN3 and MIOX2 and SWEET1,2,1. These changes suggest increased mobilization of carbohydrate stores in lines overexpressing AOX1A.
3. Hormone-related genes including ethylene biosynthesis genes and response factors, small auxin upregulated RNAs, down-regulation of abscisic acid biosynthesis genes and up-regulation of abscisic acid catabolism genes, jasmonate biosynthesis genes, down-regulation of salicylic acid biosynthesis and responsive genes, and up-regulation of gibberellin biosynthesis genes but down-regulation of GA4 biosynthesis genes (A form of gibberellin). These implicate abscisic acid, ethylene, salicylic acid and gibberellins in AOX-regulated growth.
4. Secondary metabolism genes coordinating flavonoid, phenylpropanoid, lignin and glucosinolate metabolism
5. Abiotic and biotic stress-responsive genes, including defensins, heat shock protein genes, redox-responsive transcription factors, DREBs and cell wall related genes (potentially promoting cell wall degradation).
6. A suite of transcription factors, most notably WRKY40 and other WRKY TFs (e.g. WRKY8, 27 and 30).
Based on the above, the following analyses of these plants may be useful:
1. Gas exchange and chlorophyll fluorescence measurements under standard growth conditions, particularly looking at rates of photosynthesis and PSII efficiency.
2. Quantification of sugars and other metabolites.
3. Endogenous concentrations of hormones, and the effect of ethylene, abscisic acid, salicylic acid or gibberellin (especially GH4) treatments on the delayed growth phenotype.
4. Assessment of secondary metabolites using a metabolomics approach, or targeted towards phenylpropanoids and flavonoids.
5. Tolerance to additional stress treatments including starvation (i.e. extended darkness or low CO2) or pathogen attack.
6. Measurement of transcriptional effects during stress exposure.
Based on the above findings, it is surprising that growth was not more affected in the AOX1A-OEX and ndb2 lines under normal conditions (Sweetman et al., 2019). However, in this preliminary study we have focused on transcript levels, and these do not always correlate with protein abundance, nor enzyme activity, due to post-transcriptional and post-translational regulation. Further investigations of our various lines are required to determine how proteins and activities are affected by changes in AOX1A and NDB2 protein levels. Further studies with purified mitochondria from these lines together with oxygen discrimination and flux analyses are also necessary to determine whether there are any changes to mitochondrial activities in vivo.
AOX1A has been linked to retrograde signaling processes throughout plant cells (Van Aken et al., 2009). Our study confirms this, showing that expression of genes encoded not only in the mitochondria and nucleus, but the chloroplast as well, are changed when AOX expression is altered. For example, signaling processes that lead to down-regulation of chloroplast-encoded transcripts appear to be dependent on the relative abundance of AOX1A and NDB2. Over-expressing NDB2 together with AOX1A appears to reverse the general down-regulation of photosynthesis-related transcripts in response to increased AOX1A alone and this may account for the delayed growth phenotype seen in the latter line but not the former (Sweetman et al., 2019). It is well known that alternative respiration in the mitochondrion is fundamentally linked to photosynthetic performance, especially under stress conditions.
Overall, this report demonstrates that physiological effects of altered mitochondrial electron transport might not be due directly to the changes in activity of AOX1A and NDB2 per se. Rather, a change in AOX1A:NDB2 might initiate signalling processes that impact other compartments of the cell and consequential transcriptional changes might affect plant growth. While such signalling responses have been seen in other lines with decreased AOX1A expression (e.g. Giraud et al., 2008), this is the first study to look at effects of altering expression of both AOX1A and NDB2. Importantly, we have identified potential mechanisms for the observed physiological effects, which now need to be experimentally tested. Future work will elucidate the metabolic effect of disrupting the AOX1A:NDB2 balance and an investigation of the transcriptomic and metabolomic responses when these lines are exposed to a stress.
Dedication
This manuscript is dedicated to the memory of Jim Siedow. An inspirational mentor, colleague and friend. KLS.
Data availability statement
The data presented in the study are deposited in the Sequence Read Archive (NCBI) repository, accession number PRJNA896774.
Author contributions
KS and CJ conceived the project. CW carried out all experimental procedures. DW carried out all data processing. CS interpreted the data and drafted the manuscript with feedback from DD, KS and CJ. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by The Australian Research Council, Discovery Project DP140103090 and a Flinders University Internal Grant, Project 10683.
Acknowledgments
RNA sequencing services were provided by Flinders Genomics Facility, Adelaide, Australia
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.876843/full#supplementary-material
References
Abeysinghe, J. K., Lam, K. M., Ng, D. W. K. (2019). Differential regulation and interaction of homoeologous WRKY18 and WRKY40 in arabidopsis allotetraploids and biotic stress responses. Plant J. 97 (2), 352–367. doi: 10.1111/tpj.14124
Adamowicz-Skrzypkowska, A., Kwasniak-Owczarek, M., Van Aken, O., Kazmierczak, U., Janska, H. (2020). Joint inhibition of mitochondrial complex IV and alternative oxidase by genetic or chemical means represses chloroplast transcription in Arabidopsis. Phil. Trans. R. Soc B 375, 20190409. doi: 10.1098/rstb.2019.0409
Akhtar, T. A., Lees, H. A., Lampi, M. A., Enstone, D., Brain, R. A., Greenberg, B. M. (2010). Photosynthetic redox imbalance influences flavonoid biosynthesis in lemna gibba. Plant Cell Environ. 33 (7), 1205–1219. doi: 10.1111/j.1365-3040.2010.02140.x
Amirsadeghi, S., Robson, C. A., McDonald, A. E., Vanlerberghe, G. C. (2006). Changes in plant mitochondrial electron transport alter cellular levels of reactive oxygen species and susceptibility to cell death signaling molecules. Plant Cell Physiol. 47 (11), 1509–1519. doi: 10.1093/pcp/pcl016
Baena-Gonzalez, E., Rolland, F., Thevelein, J. M., Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signaling. Nature 448 (7156), 938–U910. doi: 10.1038/nature06069
Bardou, P., Mariette, J., Escudie, F., Djemiel, C., Klopp, C. (2014). Jvenn: an interactive Venn diagram viewer. BMC Bioinf. 15, 293. doi: 10.1186/1471-2105-15-293
Bartoli, C. G., Yu, J. P., Gomez, F., Fernandez, L., McIntosh, L., Foyer, C. H. (2006). Inter-relationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J. Exp. Bot. 57 (8), 1621–1631. doi: 10.1093/jxb/erl005
Berken, A., Thomas, C., Wittinghofer, A. (2005). A new family of RhoGEFs activates the rop molecular switch in plants. Nature 436 (7054), 1176–1180. doi: 10.1038/nature03883
Carrie, C., Murcha, M. W., Kuehn, K., Duncan, O., Barthet, M., Smith, P. M., et al. (2008). Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 582 (20), 3073–3079. doi: 10.1016/j.febslet.2008.07.061
Clifton, R., Lister, R., Parker, K. L., Sappl, P. G., Elhafez, D., Millar, A. H., et al. (2005). Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58 (2), 193–212. doi: 10.1007/s11103-005-5514-7
Clifton, R., Millar, A. H., Whelan, J. (2006). Alternative oxidases in arabidopsis: A comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim. Et Biophys. Acta-Bioenerg. 1757 (7), 730–741. doi: 10.1016/j.bbabio.2006.03.009
Cluis, C. P., Mouchel, C. F., Hardtke, C. S. (2004). The arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 38 (2), 332–347. doi: 10.1111/j.1365-313X.2004.02052.x
Cvetkovska, M., Vanlerberghe, G. C. (2012). Alternative oxidase modulates leaf mitochondrial concentrations of superoxide and nitric oxide. New Phytol. 195 (1), 32–39. doi: 10.1111/j.1469-8137.2012.04166.x
Cvetkovska, M., Vanlerberghe, G. C. (2013). Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ. 36 (3), 721–732. doi: 10.1111/pce.12009
Dahal, K., Martyn, G. D., Vanlerberghe, G. C. (2015). Improved photosynthetic performance during severe drought in Nicotiana tabacum overexpressing a nonenergy conserving respiratory electron sink. New Phytol. 208 (2), 382–395. doi: 10.1111/nph.13479
Dahal, K., Vanlerberghe, G. C. (2017). Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol. 213 (2), 560–571. doi: 10.1111/nph.14169
Dahal, K., Wang, J., Martyn, G. D., Rahimy, F., Vanlerberghe, G. C. (2014). Mitochondrial alternative oxidase maintains respiration and preserves photosynthetic capacity during moderate drought in Nicotiana tabacum. Plant Physiol. 166 (3), 1560–1574. doi: 10.1104/pp.114.247866
De Clercq, I., Vermeirssen, V., Van Aken, O., Vandepoele, K., Murcha, M. W., Law, S. R., et al. (2013). The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in arabidopsis. Plant Cell 25 (9), 3472–3490. doi: 10.1105/tpc.113.117168
Del-Saz, N. F., Florez-Sarasa, I., Jose Clemente-Moreno, M., Mhadhbi, H., Flexas, J., Fernie, A. R., et al. (2016). Salinity tolerance is related to cyanide-resistant alternative respiration in Medicago truncatula under sudden severe stress. Plant Cell Environ. 39 (11), 2361–2369. doi: 10.1111/pce.12776
Demircan, N., Cucun, G., Uzilday, B. (2020). Mitochondrial alternative oxidase (AOX1a) is required for the mitigation of arsenic-induced oxidative stress in Arabidopsis thaliana. Plant Biotechnol. Rep. 14 (2), 235–245. doi: 10.1007/s11816-020-00595-9
Denyer, T., Ma, X., Klesen, S., Scacchi, E., Nieselt, K., Timmermans, M. C. P. (2019). Spatiotemporal developmental trajectories in the arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell 48, 840–852. doi: 10.1016/j.devcel.2019.02.022
Dietrich, K., Weltmeier, F., Ehlert, A., Weiste, C., Stahl, M., Harter, K., et al. (2011). Heterodimers of the arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23 (1), 381–395. doi: 10.1105/tpc.110.075390
Dietz, K. J. (2014). Redox regulation of transcription factors in plant stress acclimation and development. Antioxid. Redox Signaling 21 (9), 1356–1372. doi: 10.1089/ars.2013.5672
Dixon, R. A., Paiva, N. L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7 (7), 1085–1097. doi: 10.1105/tpc.7.7.1085
Dowdle, J., Ishikawa, T., Gatzek, S., Rolinski, S., Smirnoff, N. (2007). Two genes in arabidopsis thaliana encoding GDP-l-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 52 (4), 673–689. doi: 10.1111/j.1365-313X.2007.03266.x
Droge-Laser, W., Weiste, C. (2018). The C/S1 bZIP network: A regulatory hub orchestrating plant energy homeostasis. Trends in Plant Science 23 (5), 423–433. doi: 10.1016/j.tplants.2018.02.003
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in arabidopsis. Trends Plant Sci. 15 (10), 573–581. doi: 10.1016/j.tplants.2010.06.005
Ejaz, M., Bencivenga, S., Tavares, R., Bush, M., Sablowski, R. (2021). ARABIDOPSIS THALIANA HOMEOBOX GENE 1 controls plant architecture by locally restricting environmental responses. PNAS 118 (17), e2018615118. doi: 10.1073/pnas.2018615118
Elhafez, D., Murcha, M. W., Clifton, R., Soole, K. L., Day, D. A., Whelan, J. (2006). Characterization of mitochondrial alternative NAD(P)H dehydrogenases in arabidopsis: Intraorganelle location and expression. Plant Cell Physiol. 47 (1), 43–54. doi: 10.1093/pcp/pci221
Eriksson, S., Bohlenius, H., Moritz, T., Nilsson, O. (2006). GA(4) is the active gibberellin in the regulation of LEAFY transcription and arabidopsis floral initiation. Plant Cell 18 (9), 2172–2181. doi: 10.1105/tpc.106.042317
Escobar, M. A., Franklin, K. A., Svensson, A. S., Salter, M. G., Whitelam, G. C., Rasmusson, A. G. (2004). Light regulation of the arabidopsis respiratory chain. multiple discrete phatoreceptor responses contribute to induction of type IINAD(P)H dehydrogenase genes. Plant Physiol. 136 (1), 2710–2721. doi: 10.1104/pp.104.046698
Fatihi, A., Latimer, S., Schmollinger, S., Block, A., Dussault, P. H., Vermaas, W. F. J., et al. (2015). A dedicated type II NADPH dehydrogenase performs the penultimate step in the biosynthesis of vitamin K-1 in synechocystis and arabidopsis. Plant Cell 27 (6), 1730–1741. doi: 10.1105/tpc.15.00103
Fenech, M., Amorim-Silva, V., del Valle, A. E., Arnaud, D., Ruiz-Lopez, N., Castillo, A. G., et al. (2021). The role of GDP-l-galactose phosphorylase in the control of ascorbate biosynthesis. Plant Physiol. 185 (4), 1574–1594. doi: 10.1093/plphys/kiab010
Feng, H. Q., Guan, D. D., Sun, K., Wang, Y. F., Zhang, T. G., Wang, R. F. (2013). Expression and signal regulation of the alternative oxidase genes under abiotic stresses. Acta Biochim. Et Biophys. Sin. 45 (12), 985–994. doi: 10.1093/abbs/gmt094
Fini, A., Brunetti, C., Di Ferdinando, M., Ferrini, F., Tattini, M. (2011). Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signaling Behav. 6 (5), 709–711. doi: 10.4161/psb.6.5.15069
Fiorani, F., Umbach, A. L., Siedow, J. N. (2005). The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature. a study of arabidopsis AOX1a transgenic plants. Plant Physiol. 139 (4), 1795–1805. doi: 10.1104/pp.105.070789
Florez-Sarasa, I., Flexas, J., Rasmusson, A. G., Umbach, A. L., Siedow, J. N., Ribas-Carbo, M. (2011). In vivo cytochrome and alternative pathway respiration in leaves of Arabidopsis thaliana plants with altered alternative oxidase under different light conditions. Plant Cell Environ. 34 (8), 1373–1383. doi: 10.1111/j.1365-3040.2011.02337.x
Fujiki, Y., Yoshikawa, Y., Sato, T., Inada, N., Ito, M., Nishida, I., et al. (2001). Dark-inducible genes from arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant. 111 (3), 345–352. doi: 10.1034/j.1399-3054.2001.1110312.x
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in arabidopsis. Plant Cell Physiol. 50 (12), 2123–2132. doi: 10.1093/pcp/pcp147
Fu, A. G., Liu, H. Y., Yu, F., Kambakam, S., Luan, S., Rodermel, S. (2012). Alternative oxidases (AOX1A and AOX2) can functionally substitute for plastid terminal oxidase in arabidopsis chloroplasts. Plant Cell 24 (4), 1579–1595. doi: 10.1105/tpc.112.096701
Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. United States America 103 (6), 1988–1993. doi: 10.1073/pnas.0505667103
Gallego-Giraldo, L., Liu, C., Pose-Albacete, S., Pattathil, S., Peralta, A. G., Young, J., et al. (2020). ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1) releases latent defense signals in stems with reduced lignin content. PNAS 117, 6, 3281–3290. doi: 10.1073/pnas.1914422117
Gandin, A., Duffes, C., Day, D. A., Cousins, A. B. (2012). The absence of alternative oxidase Aox1a results in altered response of photosynthetic carbon assimilation to increasing CO2 in Arabidopsis thaliana. Plant Cell Physiol. 53 (9), 1627–1637. doi: 10.1093/pcp/pcs107
Geisler, D. A., Broselid, C., Hederstedt, L., Rasmusson, A. G. (2007). Ca2+-binding and Ca2+-independent respiratory NADH and NADPH dehydrogenases of Arabidopsis thaliana. J. Biol. Chem. 282 (39), 28455–28464. doi: 10.1074/jbc.M704674200
Giraud, E., Ho, L. H. M., Clifton, R., Carroll, A., Estavillo, G., Tan, Y. F., et al. (2008). The absence of alternative oxidase1a in arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 147 (2), 595–610. doi: 10.1104/pp.107.115121
Giraud, E., Van Aken, O., Ho, L. H. M., Whelan, J. (2009). The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150, 1286–1296. doi: 10.1104/pp.109.139782
Han, J. Y., Li, H., Yin, B., Zhang, Y. Z., Liu, Y. D., Cheng, Z. Y., et al. (2019). The papain-like cysteine protease CEP1 is involved in programmed cell death and secondary wall thickening during xylem development in arabidopsis. J. Exp. Bot. 70 (1), 205–215. doi: 10.1093/jxb/ery356
Heine, G. F., Hernandez, J. M., Grotewold, E. (2004). Two cysteines in plant R2R3 MYB domains participate in REDOX-dependent DNA binding. J. Biol. Chem. 279 (36), 37878–37885. doi: 10.1074/jbc.M405166200
Hernandez-Blanco, C., Feng, D. X., Hu, J., Sanchez-Vallet, A., Deslandes, L., Llorente, F., et al. (2007). Impairment of cellulose synthases required for arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19 (3), 890–903. doi: 10.1105/tpc.106.048058
Hodges, D. M., DeLong, J. M., Forney, C. F., Prange, R. K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207 (4), 604–611. doi: 10.1007/s004250050524
Ho, L. H. M., Giraud, E., Uggalla, V., Lister, R., Clifton, R., Glen, A., et al. (2008). Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in arabidopsis. Plant Physiol. 147 (4), 1858–1873. doi: 10.1104/pp.108.121384
Huang, H. L., Ullah, F., Zhou, D. X., Yi, M., Zhao, Y. (2019). Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00800
Ivanova, A., Law, S. R., Narsai, R., Duncan, O., Lee, J. H., Zhang, B. T., et al. (2014). A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates alternative Oxidase1a expression in arabidopsis. Plant Physiol. 165 (3), 1233–1254. doi: 10.1104/pp.114.237495
Jayawardhane, J., Cochrane, D. W., Vyas, P., Bykova, N. V., Vanlerberghe, G. C., Igamberdiev, A. U. (2020). Roles for plant mitochondrial alternative oxidase under normoxia, hypoxia, and reoxygenation conditions. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00566
Jiang, Z. X., Watanabe, C. K. A., Miyagi, A., Kawai-Yamada, M., Terashima, I., Noguchi, K. (2019). Mitochondrial AOX supports redox balance of photosynthetic electron transport, primary metabolite balance, and growth in Arabidopsis thaliana under high light. Int. J. Mol. Sci. 20 (12), 3067. doi: 10.3390/ijms20123067
Kim, M. J., Park, M. J., Seo, P. J., Song, J. S., Kim, H. J., Park, C. M. (2012). Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 448, 353–363. doi: 10.1042/bj20120244
Kiyosue, T., Yoshiba, Y., YamaguchiShinozaki, K., Shinozaki, K. (1996). A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in arabidopsis. Plant Cell 8 (8), 1323–1335. doi: 10.2307/3870304
Kodama, Y., Suetsugu, N., Kong, S. G., Wada, M. (2010). Two interacting coiled-coil proteins, WEB1 and PMI2, maintain the chloroplast photorelocation movement velocity in arabidopsis. Proc. Natl. Acad. Sci. United States America 107 (45), 19591–19596. doi: 10.1073/pnas.1007836107
Kumar, M. N., Hsieh, Y. F., Verslues, P. E. (2015). At14a-Like1 participates in membrane-associated mechanisms promoting growth during drought in arabidopsis thaliana. Proc. Natl. Acad. Sci. United States America 112 (33), 10545–10550. doi: 10.1073/pnas.1510140112
Lam, H. M., Peng, S. S. Y., Coruzzi, G. M. (1994). Metabolic-regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol. 106 (4), 1347–1357. doi: 10.1104/pp.106.4.1347
Lange, B. M., Ghassemian, M. (2003). Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol. Biol. 51 (6), 925–948. doi: 10.1023/a:1023005504702
Laubinger, S., Fittinghoff, K., Hoecker, U. (2004). The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell 16 (9), 2293–2306. doi: 10.1105/tpc.104.024216
Lee, H.-J., Park, Y.-J., Seo, P. J., Kim, J.-H., Sim, H.-J., Kim, S.-G., et al. (2015). Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in arabidopsis. Plant Cell 27 (12), 3425–3438. doi: 10.1105/tpc.15.00371
Lei, L., Stevens, D. M., Coaker, G. (2020). Phosphorylation of the pseudomonas effector AvrPtoB by arabidopsis SnRK2.8 is required for bacterial virulence. Mol. Plant 13 (10), 1513–1522. doi: 10.1016/j.molp.2020.08.018
Lewis, D. R., Ramirez, M. V., Miller, N. D., Vallabhaneni, P., Ray, W. K., Helm, R. F., et al. (2011). Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 156 (1), 144–164. doi: 10.1104/pp.111.172502
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 (7), 923–930. doi: 10.1093/bioinformatics/btt656
Li, Y. T., Liu, M. J., Li, Y., Liu, P., Zhao, S. J., Gao, H. Y., et al. (2020). Photoprotection by mitochondrial alternative pathway is enhanced at heat but disabled at chilling. Plant J. 104 (2), 403–415. doi: 10.1111/tpj.14931
Liu, J., Li, Z., Wang, Y. Q., Xing, D. (2014). Overexpression of ALTERNATIVE OXIDASE1a alleviates mitochondria-dependent programmed cell death induced by aluminium phytotoxicity in arabidopsis. J. Exp. Bot. 65 (15), 4465–4478. doi: 10.1093/jxb/eru222
Li, L., Zheng, W. G., Zhu, Y. B., Ye, H. X., Tang, B. Y., Arendsee, Z. W., et al. (2015). QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc. Natl. Acad. Sci. United States America 112 (47), 14734–14739. doi: 10.1073/pnas.1514670112
Loudet, O., Michael, T. P., Burger, B. T., Le Mette, C., Mockler, T. C., Weigel, D., et al. (2008). A zinc knuckle protein that negatively controls morning-specific growth in arabidopsis thaliana. Proc. Natl. Acad. Sci. United States America 105 (44), 17193–17198. doi: 10.1073/pnas.0807264105
Macedo, E. S., Sircar, D., Cardoso, H. G., Peixe, A., Arnholdt-Schmitt, B. (2012). Involvement of alternative oxidase (AOX) in adventitious rooting of olea europaea l. microshoots is linked to adaptive phenylpropanoid and lignin metabolism. Plant Cell Rep. 31 (9), 1581–1590. doi: 10.1007/s00299-012-1272-6
Maere, S., Heymans, K., Kuiper, M. (2005). BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21 (16), 3448–3449. doi: 10.1093/bioinformatics/bti551
Matiolli, C. C., Tomaz, J. P., Duarte, G. T., Prado, F. M., Del Bem, L. E. V., Silveira, A. B., et al. (2011). The arabidopsis bZIP gene AtbZIP63 is a sensitive integrator of transient abscisic acid and glucose signals. Plant Physiol. 157 (2), 692–705. doi: 10.1104/pp.111.181743
Maxwell, D. P., Wang, Y., McIntosh, L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. United States America 96 (14), 8271–8276. doi: 10.1073/pnas.96.14.8271
Mayfield, J. A., Fiebig, A., Johnstone, S. E., Preuss, D. (2001). Gene families from the arabidopsis thaliana pollen coat proteome. Science 292 (5526), 2482–2485. doi: 10.1126/science.1060972
Michalecka, A. M., Svensson, A. S., Johansson, F. I., Agius, S. C., Johanson, U., Brennicke, A., et al. (2003). Arabidopsis genes encoding mitochondrial type II NAD(P)H dehydrogenases have different evolutionary orgin and show distinct responses to light. Plant Physiol. 133 (2), 642–652. doi: 10.1104/pp.103.024208
Millar, A. H., Day, D. A. (1996). Nitric oxide inhibits the cytochrome oxidase but not the alternative oxidase of plant mitochondria. FEBS Lett. 398 (2-3), 155–158. doi: 10.1016/s0014-5793(96)01230-6
Millar, A. H., Mittova, V., Kiddle, G., Heazlewood, J. L., Bartoli, C. G., Theodoulou, F. L., et al. (2003). Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol. 133 (2), 443–447. doi: 10.1104/pp.103.028399
Nakabayashi, R., Yonekura-Sakakibara, K., Urano, K., Suzuki, M., Yamada, Y., Nishizawa, T., et al. (2014). Enhancement of oxidative and drought tolerance in arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77 (3), 367–379. doi: 10.1111/tpj.12388
Nakashima, K., Satoh, R., Kiyosue, T., Yamaguchi-Shinozaki, K., Shinozaki, K. (1998). A gene encoding proline dehydrogenase is nsot only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of arabidopsis. Plant Physiol. 118 (4), 1233–1241. doi: 10.1104/pp.118.4.1233
Nechushtai, R., Conlan, A. R., Harir, Y., Song, L. H., Yogev, O., Eisenberg-Domovich, Y., et al. (2012). Characterization of arabidopsis NEET reveals an ancient role for NEET proteins in iron metabolism. Plant Cell 24 (5), 2139–2154. doi: 10.1105/tpc.112.097634
Ng, S., Ivanova, A., Duncan, O., Law, S. R., Van Aken, O., De Clercq, I., et al. (2013). A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in arabidopsis. Plant Cell 25 (9), 3450–3471. doi: 10.1105/tpc.113.113985
Niu, D., Lii, Y. E., Chellappan, P., Lei, L., Peralta, K., Jiang, C., et al. (2016). miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun. 7 (1), 11324. doi: 10.1038/ncomms11324
Ogawa, M., Kay, P., Wilson, S., Swain, S. M. (2009). ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in arabidopsis. Plant Cell 21 (1), 216–233. doi: 10.1105/tpc.108.063768
Oh, G. G. K., O’Leary, B. M., Signorelli, S., Millar, A. H. (2021). Alternative oxidase (AOX) 1a and 1d limit proline-induced oxidative stress and aid salinity recovery in arabidopsis. Plant Physiol. 188 (3), 1521–1536. doi: 10.1093/plphys/kiab578
Okamoto, M., Kuwahara, A., Seo, M., Kushiro, T., Asami, T., Hirai, N., et al. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8 '-hydroxylases, are indispensable for proper control of seed dormancy and germination in arabidopsis. Plant Physiol. 141 (1), 97–107. doi: 10.1104/pp.106.079475
Oravecz, A., Baumann, A., Mate, Z., Brzezinska, A., Molinier, J., Oakeley, E. J., et al. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-b response in arabidopsis. Plant Cell 18 (8), 1975–1990. doi: 10.1105/tpc.105.040097
Panda, S. K., Sahoo, L., Katsuhara, M., Matsumoto, H. (2013). Overexpression of alternative oxidase gene confers aluminum tolerance by altering the respiratory capacity and the response to oxidative stress in tobacco cells. Mol. Biotechnol. 54 (2), 551–563. doi: 10.1007/s12033-012-9595-7
Piller, L. E., Besagni, C., Ksas, B., Rumeau, D., Brehelin, C., Glauser, G., et al. (2011). Chloroplast lipid droplet type II NAD(P)H quinone oxidoreductase is essential for prenylquinone metabolism and vitamin K-1 accumulation. Proc. Natl. Acad. Sci. United States America 108 (34), 14354–14359. doi: 10.1073/pnas.1104790108
Qi, M. S., Zheng, W. G., Zhao, X. F., Hohenstein, J. D., Kandel, Y., O'Conner, S., et al. (2019). QQS orphan gene and its interactor NF-YC4 reduce susceptibility to pathogens and pests. Plant Biotechnol. J. 17 (1), 252–263. doi: 10.1111/pbi.12961
Salazar-Iribe, A., Agredano-Moreno, L. T., Zuniga-Sanchez, E., Jimenez-Garcia, L. F., Gamboa-DeBuen, A. (2016). The cell wall DUF642 At2g41800 (TEB) protein is involved in hypocotyl cell elongation. Plant Sci. 253, 206–214. doi: 10.1016/j.plantsci.2016.10.007
Scarpeci, T. E., Zanor, M. I., Carrillo, N., Mueller-Roeber, B., Valle, E. M. (2008). Generation of superoxide anion in chloroplasts of arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol. Biol. 66 (4), 361–378. doi: 10.1007/s11103-007-9274-4
Scarpeci, T. E., Zanor, M. I., Mueller-Roeber, B., Valle, E. M. (2013). Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in arabidopsis thaliana. Plant Mol. Biol. 83 (3), 265–277. doi: 10.1007/s11103-013-0090-8
Schwarzlander, M., Konig, A. C., Sweetlove, L. J., Finkemeier, I. (2012). The impact of impaired mitochondrial function on retrograde signaling: a meta-analysis of transcriptomic responses. J. Exp. Bot. 63 (4), 1735–1750. doi: 10.1093/jxb/err374
Senkler, J., Senkler, M., Eubel, H., Hildebrandt, T., Lengwenus, C., Schertl, P., et al. (2017). The mitochondrial complexome of Arabidopsis thaliana. Plant J. 89 (6), 1079–1092. doi: 10.1111/tpj.13448
Shaikhali, J., Noren, L., Barajas-Lopez, J. D., Srivastava, V., Konig, J., Sauer, U. H., et al. (2012). Redox-mediated mechanisms regulate DNA binding activity of the G-group of basic region leucine zipper (bZIP) transcription factors in arabidopsis. J. Biol. Chem. 287 (33), 27510–27525. doi: 10.1074/jbc.M112.361394
Shapiguzov, A., Vainonen, J. P., Hunter, K., Tossavainen, H., Tiwari, A., Jarvi, S., et al. (2019). Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors. Elife 8, 35. doi: 10.7554/eLife.43284
Sharma, A., Shahzad, B., Rehman, A., Bhardwaj, R., Landi, M., Zheng, B. S. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24 (13), 2452. doi: 10.3390/molecules24132452
Sharma, S., Villamor, J. G., Verslues, P. E. (2011). Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiol. 157 (1), 292–304. doi: 10.1104/pp.111.183210
Shimoji, H., Yamasaki, H. (2005). Inhibitory effects of flavonoids on alternative respiration of plant mitochondria. Biol. Plant. 49 (1), 117–119. doi: 10.1007/s10535-005-7119-z
Shim, S., Seo, P. J. (2020). EAT-UpTF: Enrichment analysis tool for upstream transcription factors of a group of plant genes. Front. Genet. 11. doi: 10.3389/fgene.2020.566569
Shin, D. H., Cho, M. H., Kim, T. L., Yoo, J., Kim, J. I., Han, Y. J., et al. (2010). A small GTPase activator protein interacts with cytoplasmic phytochromes in regulating root development. J. Biol. Chem. 285 (42), 32151–32159. doi: 10.1074/jbc.M110.133710
Singh, S., Gupta, A., Kaur, N. (2012). Differential responses of antioxidative defence system to long-term field drought in wheat (Triticum aestivum l.) genotypes differing in drought tolerance. J. Agron. Crop Sci. 198 (3), 185–195. doi: 10.1111/j.1439-037X.2011.00497.x
Sircar, D., Cardoso, H. G., Mukherjee, C., Mitra, A., Arnholdt-Schmitt, B. (2012). Alternative oxidase (AOX) and phenolic metabolism in methyl jasmonate-treated hairy root cultures of daucus carota l. J. Plant Physiol. 169 (7), 657–663. doi: 10.1016/j.jplph.2011.11.019
Skirycz, A., De Bodt, S., Obata, T., De Clercq, I., Claeys, H., De Rycke, R., et al. (2010). Developmental stage specificity and the role of mitochondrial metabolism in the response of arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol. 152 (1), 226–244. doi: 10.1104/pp.109.148965
Smith, C., Barthet, M., Melino, V., Smith, P., Day, D., Soole, K. (2011). Alterations in the mitochondrial alternative NAD (P) h dehydrogenase NDB4 lead to changes in mitochondrial electron transport chain composition, plant growth and response to oxidative stress. Plant Cell Physiol. 52 (7), 1222–1237. doi: 10.1093/pcp/pcr073
Smith, C. A., Melino, V. J., Sweetman, C., Soole, K. L. (2009). Manipulation of alternative oxidase can influence salt tolerance in Arabidopsis thaliana. Physiol. Plant. 137 (4), 459–472. doi: 10.1111/j.1399-3054.2009.01305.x
Sonderby, I. E., Geu-Flores, F., Halkier, B. A. (2010). Biosynthesis of glucosinolates - gene discovery and beyond. Trends Plant Sci. 15 (5), 283–290. doi: 10.1016/j.tplants.2010.02.005
Stracke, R., Ishihara, H., Barsch, G. H. A., Mehrtens, F., Niehaus, K., Weisshaar, B. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the arabidopsis thaliana seedling. Plant J. 50 (4), 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Sweetman, C., Waterman, C. D., Rainbird, B. M., Smith, P. M. C., Jenkins, C. L. D., Day, D. A., et al. (2019). AtNDB2 is the main external NADH dehydrogenase in mitochondria and is important for tolerance to environmental stress. Plant Physiol. 181 (2), 774–788. doi: 10.1104/pp.19.00877
Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 (6), 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
Tron, A. E., Bertoncini, C. W., Chan, R. L., Gonzalez, D. H. (2002). Redox regulation of plant homeodomain transcription factors. J. Biol. Chem. 277 (38), 34800–34807. doi: 10.1074/jbc.M203297200
Umbach, A. L., Fiorani, F., Siedow, J. N. (2005). Characterization of transformed arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol. 139 (4), 1806–1820. doi: 10.1104/pp.105.070763
Umbach, A. L., Zarkovic, J., Yu, J. P., Ruckle, M. E., McIntosh, L., Hock, J. J., et al. (2012). Comparison of intact arabidopsis thaliana leaf transcript profiles during treatment with inhibitors of mitochondrial electron transport and TCA cycle. PloS One 7 (9), e44339. doi: 10.1371/journal.pone.0044339
Van Aken, O., Giraud, E., Clifton, R., Whelan, J. (2009). Alternative oxidase: a target and regulator of stress responses. Physiol. Plant. 137 (4), 354–361. doi: 10.1111/j.1399-3054.2009.01240.x
Van Aken, O., Whelan, J. (2012). Comparison of transcriptional changes to chloroplast and nnitochondrial perturbations reveals common and specific responses in arabidopsis. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00281
Van Aken, O., Zhang, B., Law, S., Narsai, R., Whelan, J. (2013). AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol. 162 (1), 254–271. doi: 10.1104/pp.113.215996
Vanlerberghe, G. C. (2013). Alternative oxidase: A mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 14 (4), 6805–6847. doi: 10.3390/ijms14046805
Vanlerberghe, G. C., Martyn, G. D., Dahal, K. (2016). Alternative oxidase: A respiratory electron transport chain pathway essential for maintaining photosynthetic performance during drought stress. Physiol. Plant. 157 (3), 322–337. doi: 10.1111/ppl.12451
Verbruggen, N., Hua, X. J., May, M., VanMontagu, M. (1996). Environmental and developmental signals modulate proline homeostasis: Evidence for a negative transcriptional regulator. Proc. Natl. Acad. Sci. United States America 93 (16), 8787–8791. doi: 10.1073/pnas.93.16.8787
Vijayraghavan, V., Soole, K. (2010). Effect of short- and long-term phosphate stress on the non-phosphorylating pathway of mitochondrial electron transport in Arabidopsis thaliana. Funct. Plant Biol. 37 (5), 455–466. doi: 10.1071/fp09206
Vishwakarma, A., Tetali, S. D., Selinski, J., Scheibe, R., Padmasree, K. (2015). Importance of the alternative oxidase (AOX) pathway in regulating cellular redox and ROS homeostasis to optimize photosynthesis during restriction of the cytochrome oxidase pathway in Arabidopsis thaliana. Ann. Bot. 116 (4), 555–569. doi: 10.1093/aob/mcv122
Wagner, S., De Bortoli, S., Schwarzlander, M., Szabo, I. (2016). Regulation of mitochondrial calcium in plants versus animals. J. Exp. Bot. 67 (13), 3809–3829. doi: 10.1093/jxb/erw100
Wallström, S. V., Florez-Sarasa, I., Araujo, W. L., Aidemark, M., Fernandez-Fernandez, M., Fernie, A. R., et al. (2014a). Suppression of the external mitochondrial NADPH dehydrogenase, NDB1, in Arabidopsis thaliana affects central metabolism and vegetative growth. Mol. Plant 7 (2), 356–368. doi: 10.1093/mp/sst115
Wallström, S. V., Florez-Sarasa, I., Araujo, W. L., Escobar, M. A., Geisler, D. A., Aidemark, M., et al. (2014b). Suppression of NDA-type alternative mitochondrial NAD(P)H dehydrogenases in Arabidopsis thaliana modifies growth and metabolism, but not high light stimulation of mitochondrial electron transport. Plant Cell Physiol. 55 (5), 881–896. doi: 10.1093/pcp/pcu021
Wang, J., Rajakulendran, N., Amirsadeghi, S., Vanlerberghe, G. C. (2011). Impact of mitochondrial alternative oxidase expression on the response of Nicotiana tabacum to cold temperature. Physiol. Plant. 142 (4), 339–351. doi: 10.1111/j.1399-3054.2011.01471.x
Wang, Y., Selinski, J., Mao, C. L., Zhu, Y. Q., Berkowitz, O., Whelan, J. (2020). Linking mitochondrial and chloroplast retrograde signaling in plants. Philos. Trans. R. Soc. B-Biol. Sci. 375 (1801), 20190410. doi: 10.1098/rstb.2019.0410
Watanabe, C. K., Hachiya, T., Terashima, I., Noguchi, K. (2008). The lack of alternative oxidase at low temperature leads to a disruption of the balance in carbon and nitrogen metabolism, and to an up-regulation of antioxidant defence systems in Arabidopsis thaliana leaves. Plant Cell Environ. 31 (8), 1190–1202. doi: 10.1111/j.1365-3040.2008.01834.x
Watanabe, C. K. A., Yamori, W., Takahashi, S., Terashima, I., Noguchi, K. (2016). Mitochondrial alternative pathway-associated photoprotection of photosystem II is related to the photorespiratory pathway. Plant Cell Physiol. 57 (7), 1426–1431. doi: 10.1093/pcp/pcw036
Wieczorek, K., Elashry, A., Quentin, M., Grundler, F. M. W., Favery, B., Seifert, G. J., et al. (2014). A distinct role of pectate lyases in the formation of feeding structures induced by cyst and root-knot nematodes. Mol. Plant-Microbe Interact. 27 (9), 901–912. doi: 10.1094/mpmi-01-14-0005-r
Wrzaczek, M., Brosche, M., Salojarvi, J., Kangasjarvi, S., Idanheimo, N., Mersmann, S., et al. (2010). Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in arabidopsis. BMC Plant Biol. 10, 95. doi: 10.1186/1471-2229-10-95
Xu, X. P., Chen, C. H., Fan, B. F., Chen, Z. X. (2006). Physical and functional interactions between pathogen-induced arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 (5), 1310–1326. doi: 10.1105/tpc.105.037523
Yang, H. L., Deng, L. B., Liu, H. F., Fan, S. H., Hua, W., Liu, J. (2019). Overexpression of BnaAox1b confers tolerance to osmotic and salt stress in rapeseed. G3-Genes Genomes Genet. 9 (10), 3501–3511. doi: 10.1534/g3.119.400366
Yoshida, K., Watanabe, C. K., Terashima, I., Noguchi, K. (2011). Physiological impact of mitochondrial alternative oxidase on photosynthesis and growth in Arabidopsis thaliana. Plant Cell Environ. 34 (11), 1890–1899. doi: 10.1111/j.1365-3040.2011.02384.x
Zhang, X. H., Ivanova, A., Vandepoele, K., Radomiljac, J., Van de Velde, J., Berkowitz, O., et al. (2017). The transcription factor MYB29 is a regulator of ALTERNATIVE OXIDASE1a(1). Plant Physiol. 173 (3), 1824–1843. doi: 10.1104/pp.16.01494
Zhang, L., Oh, Y., Li, H. Y., Baldwin, I. T., Galis, I. (2012). Alternative oxidase in resistance to biotic stresses: Nicotiana attenuata AOX contributes to resistance to a pathogen and a piercing-sucking insect but not Manduca sexta larvae. Plant Physiol. 160 (3), 1453–1467. doi: 10.1104/pp.112.200865
Zhang, L. T., Zhang, Z. S., Gao, H. Y., Xue, Z. C., Yang, C., Meng, X. L., et al. (2011). Mitochondrial alternative oxidase pathway protects plants against photoinhibition by alleviating inhibition of the repair of photodamaged PSII through preventing formation of reactive oxygen species in rumex K-1 leaves. Physiol. Plant. 143 (4), 396–407. doi: 10.1111/j.1399-3054.2011.01514.x
Zhu, M. Y. Y., Pan, J. W., Wang, L. L., Gu, Q., Huang, C. Y. (2003). Mutation induced enhancement of Al tolerance in barley cell lines. Plant Sci. 164 (1), 17–23. doi: 10.1016/s0168-9452(02)00317-5
Keywords: plant mitochondria, alternative oxidase, type II NAD(P)H dehydrogenase, overexpression, knockout, transcriptome, balancing act
Citation: Sweetman C, Waterman CD, Wong DCJ, Day DA, Jenkins CLD and Soole KL (2022) Altering the balance between AOX1A and NDB2 expression affects a common set of transcripts in Arabidopsis. Front. Plant Sci. 13:876843. doi: 10.3389/fpls.2022.876843
Received: 15 February 2022; Accepted: 24 October 2022;
Published: 15 November 2022.
Edited by:
Miquel A Gonzalez-Meler, University of Illinois at Chicago, United StatesReviewed by:
Marina Cvetkovska, University of Ottawa, CanadaIlya Velegzhaninov, Komi Scientific Center (RAS), Russia
Copyright © 2022 Sweetman, Waterman, Wong, Day, Jenkins and Soole. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kathleen L. Soole, S2F0aGxlZW4uc29vbGVAZmxpbmRlcnMuZWR1LmF1
 Crystal Sweetman
Crystal Sweetman Christopher D. Waterman1
Christopher D. Waterman1 Darren C.J. Wong
Darren C.J. Wong David A. Day
David A. Day Colin L.D. Jenkins
Colin L.D. Jenkins Kathleen L. Soole
Kathleen L. Soole