- 1School of Agriculture, Meiji University, Kawasaki, Japan
- 2Forest Bio-Research Center, Forestry and Forest Products Research Institute, Hitachi, Japan
Pine wilt disease (PWD), which is caused by the pine wood nematode Bursaphelenchus xylophilus, is among the most serious tree diseases worldwide. PWD is thought to be initiated by sequential excessive hypersensitive responses to B. xylophilus. Previous studies have reported candidate pathogenic molecules inducing hypersensitive responses in pine trees susceptible to B. xylophilus. The functions of some of these molecules have been analyzed in model plants using transient overexpression; however, whether they can induce hypersensitive responses in natural host pines remains unclear due to the lack of a suitable functional analysis method. In this study, we established a novel functional analysis method for susceptible black pine (Pinus thunbergii) seed embryos using transient overexpression by the Apple latent spherical virus vector and investigated five secreted proteins of B. xylophilus causing cell death in tobacco to determine whether they induce hypersensitive responses in pine. We found that three of five molecules induced significantly higher expression in pathogenesis-related genes ( p < 0.05), indicating hypersensitive response in pine seed embryos compared with mock and green fluorescence protein controls. This result suggests that tobacco-based screening may detect false positives. This study is the first to analyze the function of pathogenic candidate molecules of B. xylophilus in natural host pines using exogenous gene expression, which is anticipated to be a powerful tool for investigating the PWD mechanism.
Introduction
Pine wilt disease (PWD), which is caused by the pine wood nematode Bursaphelenchus xylophilus, is a serious threat to pine forests worldwide (Kiyohara and Tokushige, 1971; Yi et al., 1989; Burgermeister et al., 1999; Abelleira et al., 2011). The mechanism by which B. xylophilus kills host pine trees has been extensively studied during the past 50 years. Based on physiological and histological observation, sequential excessive hypersensitive responses to the spread of B. xylophilus through the tree are thought to lead, eventually, to the death of susceptible pine trees (Myers, 1988; Futai, 2013). The expression levels of pathogenesis-related (PR) genes, that is the landmark of hypersensitive responses, have been found to be much higher in susceptible trees inoculated with nematodes than in resistant trees (Hirao et al., 2012). Furthermore, Yamaguchi et al. (2019) showed that all of pine seedling, that increased the number of B. xylophilus and expression levels of PR genes after nematodes inoculation, exhibited external symptoms. Accordingly, identifying B. xylophilus molecules that induce hypersensitive responses in host pine trees is important for predicting PWD occurrence, and the range of candidate molecules has been narrowed down using genomic, transcriptome, and proteome methods (Kikuchi et al., 2011; Shinya et al., 2013, 2021; Palomares-Rius et al., 2015; Cardoso et al., 2016, 2021; Ding et al., 2016; Espada et al., 2016, 2018; Filipiak et al., 2020; Silva et al., 2021). Recent functional analyses of these candidate molecules have mainly applied RNAi in B. xylophilus or examined the expression of its virulence factors in plants (Ma et al., 2011; Huang et al., 2019; Qiu et al., 2019). Because B. xylophilus RNAi effects are generally transient and weak, functional analysis using plants appears to be a more suitable approach for this research (Park et al., 2008).
The overexpression of foreign genes in plants is a major biotechnological tool in molecular functional analysis. For example, leaf-disk assays based on transient overexpression in the model plant Nicotiana benthamiana have been applied to investigate whether pathogenic candidate molecules of B. xylophilus induce cell death (Hu et al., 2019; Kirino et al., 2020; Shinya et al., 2021). Such studies have successfully narrowed the field of candidate molecules; however, whether they induce hypersensitive responses in natural pines remains unclear. Therefore, the next challenge is to develop a method for expressing B. xylophilus protein in pine trees and to perform functional analysis of the pathogenic candidate molecules. Because transgenic pine seedling production requires substantial time and effort, they are unsuitable for screening B. xylophilus pathogenic candidates. Therefore, in this study, we focused on pine seed embryos instead of pine seedlings. Exogenous proteins such as green fluorescent protein (GFP) can be transiently expressed in pine seed embryos using the apple latent spherical virus (ALSV) vector (Konagaya, 2017). ALSV is a small spherical virus (diameter, 25 nm) originally isolated from apple trees (Li et al., 2000, 2004) and has two ssRNA species (pEALSR1 and pEALSR2). ALSV-based vectors can be used to infect experimentally a broad range of plant species without causing symptoms, effectively inducing stable virus-induced expression of foreign genes in plants (Takahashi and Yoshikawa, 2008). Therefore, we hypothesized that an ALSV-based method is suitable for identifying pathogenic molecules in B. xylophilus.
To evaluate the effectiveness of this method using pine seed embryos with ALSV for the functional analysis of pathogenic molecules in B. xylophilus, we selected five proteins secreted by B. xylophilus causing cell death in tobacco (Kirino et al., 2020; Shinya et al., 2021) and expressed them in susceptible black pine (Pinus thunbergii) seed embryos through transient overexpression using the ALSV vector. Because B. xylophilus protein expression induced using the ALSV vector is transient and insufficient for inducing cell death, genetic markers are more suitable for evaluating host responses. Then, we examined whether hypersensitive response could be induced through the expression of each B. xylophilus protein based on expression of PR-1b (enigmatic biochemical function), PR-2 (beta-1,3-glucanase), PR-3 (classes I and IV chitinase), PR-4 (chitinase types I and II), PR-5 (thaumatin-like protein), and PR-6 (type II proteinase inhibitor family protein) in pine seed embryos because the expression of these genes was induced more quickly and to a higher level in susceptible than in resistant trees following infection by a virulent isolate of B. xylophilus, suggesting that the expression of these PR genes is good indicators of hypersensitive response that precede pine wilt (Hirao et al., 2012). To our knowledge, this is the first report of functional analysis using recombinant pathogenic candidate molecules of B. xylophilus in host pines. This method is anticipated to be a powerful tool for investigating the PWD mechanism.
Materials and Methods
Nematode Strains and Culturing
We used the virulent Ka4 isolate of B. xylophilus, which was propagated on the BC-3 strain of the fungus Botrytis cinerea, grown at 25°C on malt extract agar (MEA) plates (Difco Laboratories) containing 100 μg/ml chloramphenicol in 90-mm petri dishes.
Nematode Sterilization
Nematodes grown for 3–5 days in mixed culture were collected into 15-mL tubes containing sterile water. The tubes were centrifuged at 2,000 rpm for 2 min. The supernatant was discarded, with nematodes retained at the bottom of the tube. We added 5 ml of a solution consisting of 0.002% mercuric chloride, 0.0025% sodium azide, and 0.001% Triton X-100 in sterile water; the solution was allowed to stand for 10 min. The nematodes were washed with sterile water three times. Then, a nematode solution in 5 ml sterile water containing 5 μl of 1,000 ppm streptomycin was incubated at 12°C overnight. The nematodes were again washed with sterile water three times and then re-suspended in sterile water and used for inoculation into pine seed embryos.
Pine Seed Sterilization
We used a PWD-susceptible black pine Taga-1 isolate of P. thunbergii in this study. Pine seeds were immersed in 70% ethanol and shaken for 3 min. Floating seeds were discarded. The remaining seeds were immersed in 5% hydrogen peroxide solution in sterile water containing a drop of Tween 20 and shaken for 10 min. The seeds were washed with sterile water five times, immersed in sterile water at 4°C for 1 week, and then used for nematode or biolistic inoculation.
Nematode Inoculation to Pine Seed Embryos
For each sterilized pine seed, the seed coat and endosperm were removed and the embryo was extracted using tweezers and knife. These pine seed embryos were sown on a 60-mm mGD plate (Konagaya et al., 2020) containing 10 μg/ml benomyl and 100 μg/ml chloramphenicol. Twelve pine seed embryos were inoculated approximately 500 nematodes per plant under aseptic conditions, and six were not inoculated nematodes (mock control) with one replicate per experiment. The embryos were placed into a growth chamber (25°C, 24 h light).
Nematode Total RNA Extraction and cDNA Synthesis
Mixed-stage nematodes grown for 5 days on MEA plates were collected with sterile water, sedimented by centrifugation at 2,500 rpm for 2 min, and re-suspended in fresh sterile water. This procedure was repeated three times. After the final wash, the nematode pellet was homogenized using a mortar and pestle after freezing with liquid nitrogen. Total RNA was extracted from homogenized nematodes and purified using the RNeasy Mini Kit (Qiagen), according to the manufacturer’s protocol. Reverse transcription was performed using 1 μg of total RNA and the PrimeScript II first-strand cDNA Synthesis Kit (Takara Bio Inc.), according to the manufacturer’s protocol. The transcribed cDNA was adjusted to a concentration of 200 ng/μL and subjected to PCR amplification.
Construction of ALSV Vectors
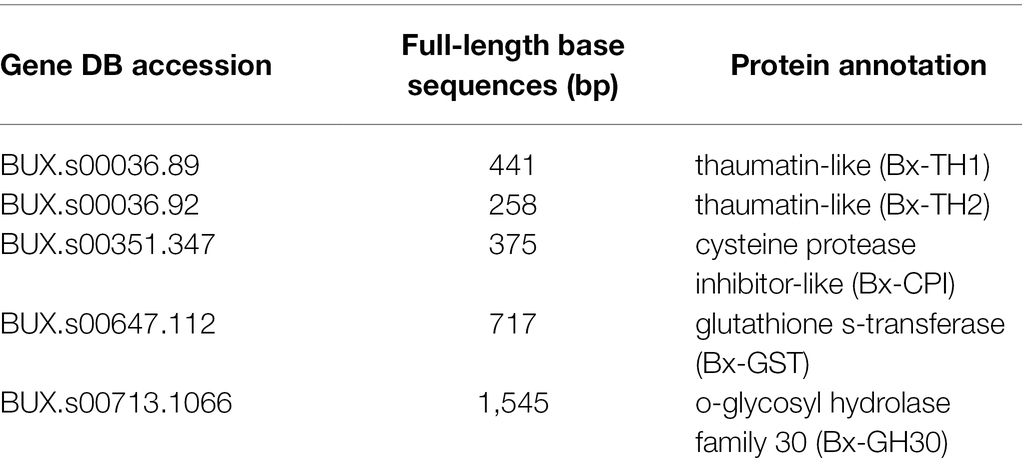
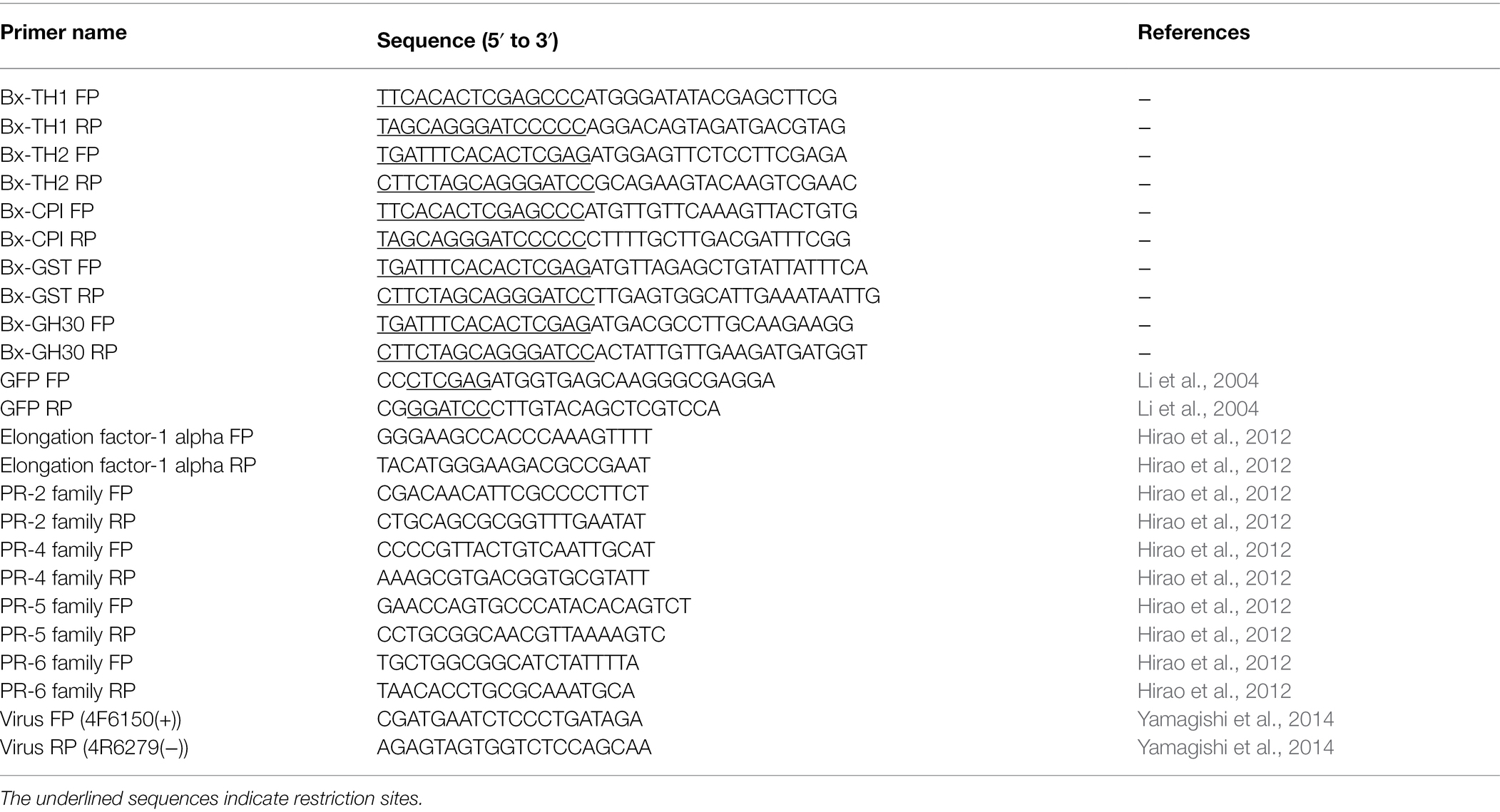
The candidate proteins (Table 1) were proteins secreted by B. xylophilus that were found to cause tobacco cell death in our previous studies (Kirino et al., 2020; Shinya et al., 2021). The sequence of Bx-CPI included signal peptide because it induced cell death in N. benthamiana only when signal peptide was present. Signal peptides of the other proteins were removed. All sequences including the GFP control were amplified by PCR from cDNA extracted from B. xylophilus or pRSET-EmGFP (Thermo Fisher Scientific). All PCR assays were performed using Takara PrimeSTAR GXL DNA polymerase, according to the manufacturer’s protocol (Takara Bio Inc.). The PCR components were as follows: 10 μl 5 × Prime STAR GXL buffer, 4 μl dNTP Mixture (2.5 mM each), 1 μl forward primer (10 μM), 1 μl reverse primer (10 μM), 1 μl Prime STAR GXL Polymerase, 1 μl cDNA template (200 ng/μL), and 32 μl H2O. The cycling conditions were as follows: 35 cycles at 98°C for 10 s, 55°C for 15 s, and 68°C for 60 s/kb. The primers were designed using the Primer 3 Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). The primers are listed in Table 2. The DNA products from B. xylophilus were inserted into the pEALSR2L5R5 vector (Yamagishi and Yoshikawa, 2013) using the In-Fusion HD Cloning Kit (Takara Bio Inc.) under the following conditions: 37°C for 15 min and 42°C for 15 min. The amplified GFP was digested with Xho I and Bam HI, and ligated to pEALSR2L5R5 vectors restricted with the same enzymes following previous research (Li et al., 2004). The resulting clones were designated pEALSR2-BxTH1, pEALSR2-BxTH2, pEALSR2-BxCPI, pEALSR2-BxGH30, and pEALSR2-BxGST. Gene sequences were confirmed by sanger sequence analysis using primer pair; Virus FP and Virus RP in Table 2 (Yamagishi et al., 2014). Both pEALSR1 and the constructed pEALSR2 that ALSV consists of were transformed in Escherichia coli JM109 and purified from large-scale cultures using the Plasmid Mini Kit (Qiagen); each was mixed at a concentration of 0.5 μg/μL and then mechanically inoculated into Chenopodium quinoa leaves at the eight true leaf stage for propagating ALSV vectors (25°C, 16 h:8 h light:dark). Three independent plants were inoculated per vectors. Leaves with chlorotic spots were homogenized by shaking twice at 1,500 rpm for 3 min with 15 iron balls (diameter, 5 mm) in 15-mL tubes, and RNA was extracted using the RNeasy Mini Kit.
Biolistic Inoculation of Pine Seed Embryos With ALSV
Biolistic inoculation of pine seed embryos with ALSV-RNAs was performed as described previously (Yamagishi et al., 2010), with modifications. We added 2 mg of gold particles (diameter, 0.6 μm) and 12.5 μl RNase-free H2O into a 2-mL microcentrifuge tube. The tube was then vortexed vigorously using a MicroMixer E-36 (Taitec) for 5 min and then sonicated for at least 5 min. During vortexing, 33 μl of 1.5 μg/μL total RNA solution extracted from ALSV-infected C. quinoa leaves, 11 μl of 10 M ammonium acetate, and 113 μl of 2-propanol were added to the tube sequentially. After continual vortexing for 7–10 min, gold particles coated with the RNAs were stored at −20°C for 1 h to overnight. The pellet of gold particles was washed gently three times with 1 ml of 100% ethanol and the pellet was re-suspended in 66 μl of 100% ethanol. Pine seed embryos extracted from sterilized seeds were placed on a 90-mm mGD plate containing 10 μg/ml benomyl and 0.4 M maltose and inoculated biolistically with ALSV on the following day. Fifteen pine seed embryos were bombarded with gold particles coated with total RNAs from ALSV-infected leaves per different ALSV vectors, and nine were bombarded with non-coated gold particles (mock control) at a pressure of approximately 1,100 psi using the PDS-1000/He Particle Delivery system (Bio-Rad) with one replicate per experiment. Pine seed embryos were bombarded in two shots, each containing 0.3 mg of particles. Next, the pine seed embryos were placed in a growth chamber (25°C, 24 h light). The next day, pine seed embryos were placed on a 90-mm mGD plate containing 10 μg/ml benomyl and returned to the growth chamber. GFP fluorescence was observed using a MS FLIII fluorescence stereomicroscope (Leica Microsystems) with a GFP2 Plus filter system (excitation filter, 480/40 nm; emission filter, 510 nm). The GFP signal was imaged using a DC300 F digital camera system (Leica Microsystems).
Quantitative Reverse-Transcription Polymerase Chain Reaction and Normalization
For quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis, samples were collected from pine seed embryos at 3 and 5 days post-B. xylophilus infection (dpi), or at 3, 5, and 7 dpi after particle bombardment with ALSV vectors. Total RNA was isolated from pine seed embryos using the RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s protocol. RT-PCR was performed using total pine seed embryo RNA. Total RNA was reverse-transcribed using the PrimeScript II first-strand cDNA synthesis kit (TaKaRa) according to the manufacturer’s protocol and qRT-PCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems) on the StepOnePlus Real-Time PCR System (Applied Biosystems). The PCR mixtures were prepared according to the manufacturer’s instructions and contained 300 nM each of the forward and reverse gene-specific primers and 4 μl of the 20-fold diluted reverse-transcription reaction (total, 5 ng) for a final volume of 20 μl. All reactions were heated to 95°C for 10 min; this denaturation step was followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The PCR products were subjected to melting curve analysis under the following conditions: 60–95°C at a temperature increment of 1.6°C/s. Elongation factor 1-alpha (FY842441.1) was used as the reference gene for normalizing the transcript profiles. Primer pairs were designed as described previously (Hirao et al., 2012; Table 2). Real-time PCR data were calibrated against the transcript levels of mock samples; the 2−ΔΔCt method was used to quantify relative transcript abundance. Pine seed embryos expressing GFP were used as a control sample to distinguish specific and non-specific induced expression changes. All PR gene expression levels in pine seed embryos expressing foreign genes were normalized by detecting the amount of viral RNA. Primer pairs for detecting the amount of viral RNA were designed as described previously (Yamagishi et al., 2014; Table 2). Figure 1 shows an outline of the experiment method.
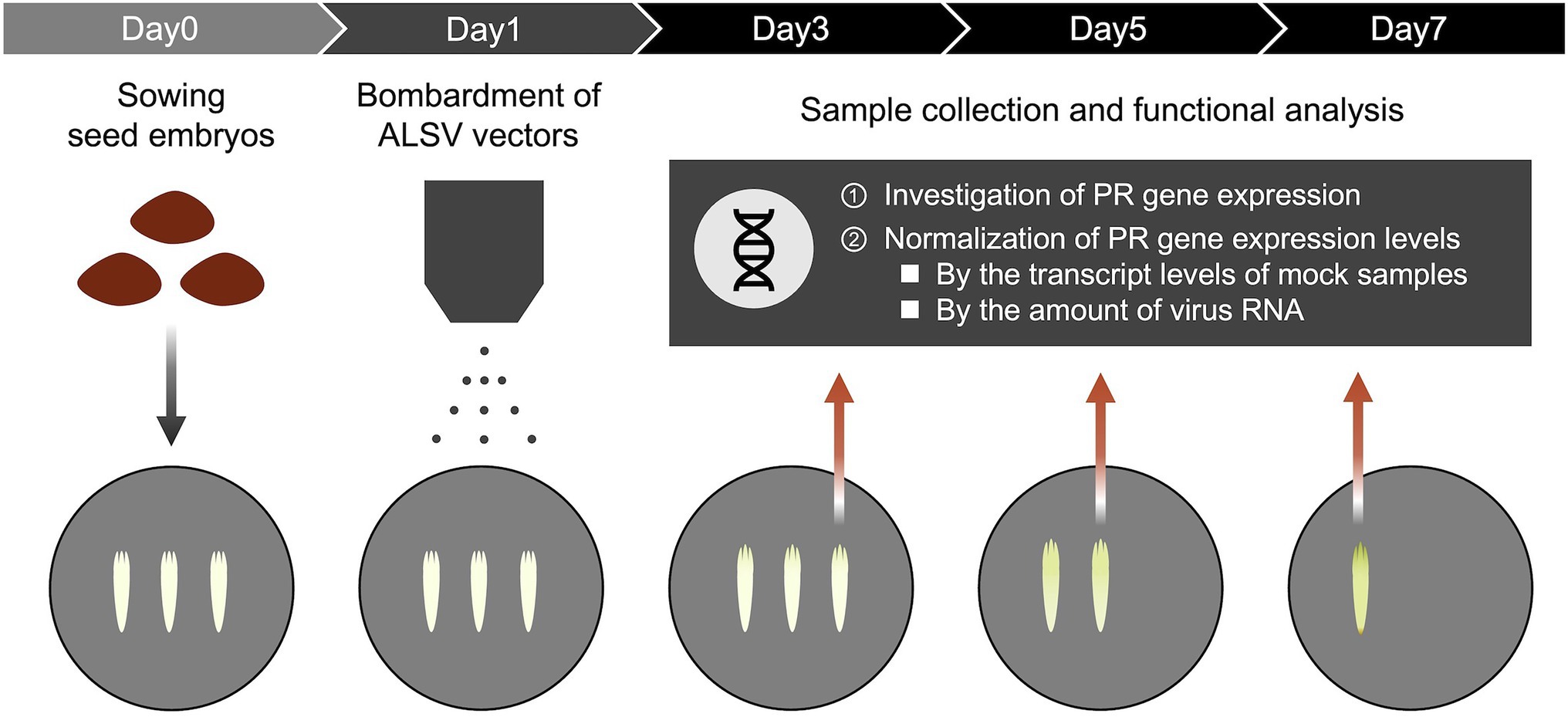
Figure 1. Functional analysis for pathogenic proteins of Bursaphelenchus xylophilus in pine seed embryos using a virus vector. Pine seed embryos extracted from sterilized seeds were placed on mGD plate and bombarded with gold particles coated with ALSV or non-coated gold particles (mock control) on the following day. The expression of PR genes was investigated at 3, 5, and 7 dpi after particle bombardment of ALSV vectors using quantitative real-time polymerase chain reaction (qRT-PCR). All PR gene expression levels in pine seed embryos expressing foreign genes were normalized by transcript levels of mock samples and detecting the amount of viral RNA.
Results
PR-2, PR-4, PR-5, and PR-6 Reacted to B. xylophilus in Pine Seed Embryos
To identify suitable marker genes to react to B. xylophilus invasion in pine seed embryos, we assayed the expression of six PR genes in pine seed embryos using qRT-PCR. The expression of these PR genes has been suggested to be a good indicator of hypersensitive response that precede pine wilt (Hirao et al., 2012). In this study, the expression levels of four genes (PR-2, PR-4, PR-5, and PR-6) were markedly upregulated following inoculation with B. xylophilus into pine seed embryos (Figure 2). The expression levels of PR-2 and PR-6 in pine seed embryos infected with B. xylophilus were > 10-fold higher than those of the mock control (non-inoculated) at 3 and 5 days post-B. xylophilus infection (dpi; p < 0.05). The expression levels of PR-4 and PR-5 were significantly higher than those of the mock control only at 5 dpi (p < 0.05). Expression data for PR-1b and PR-3 were omitted from Figure 2 due to their unstable expression in pine seed embryos, irrespective of the presence of B. xylophilus. Therefore, we used PR-2, PR-4, PR-5, and PR-6 as marker genes for subsequent molecular functional analyses.
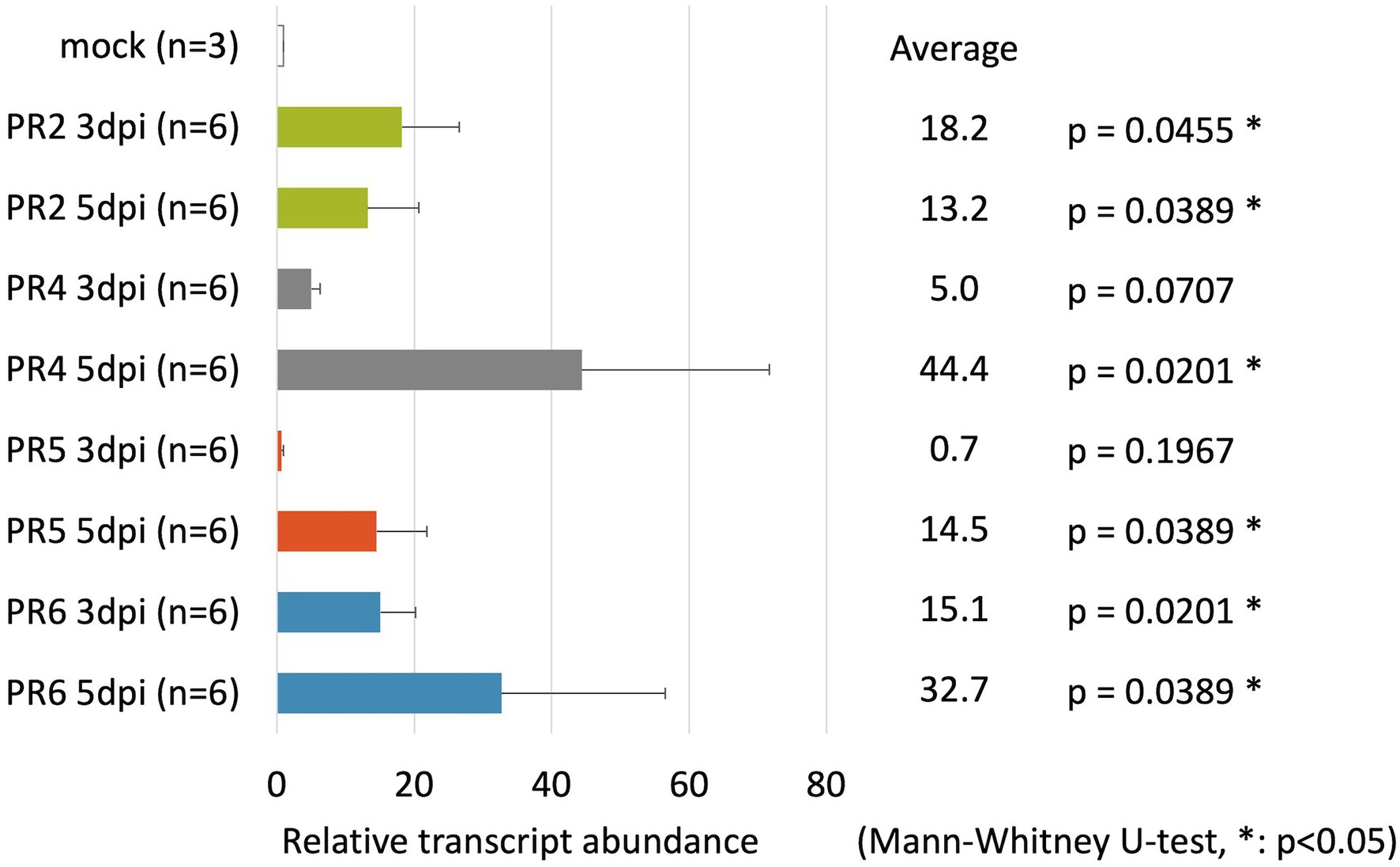
Figure 2. Relative transcript abundance of pathogenesis-related (PR) genes in pine seed embryos following pine wood nematode inoculation. Relative transcript abundance of PR-2, PR-4, PR-5, and PR-6 genes in pine seed embryos at 3 and 5 days post-infection with Bursaphelenchus xylophilus.
Bx-TH1, Bx-TH2, and Bx-GH30 Induced PR Gene Expression in Pine Seed Embryos
To determine whether B. xylophilus secreted proteins causing cell death in tobacco could induce the expression of PR genes in pine seed embryos, we constructed and transformed six ALSV vectors, coding sequences of Bx-TH1, Bx-TH2, Bx-CPI, Bx-GH30, Bx-GST, and GFP (control) in pine seed embryos. GFP fluorescence in pine seed embryos was observed from 1 dpi after particle bombardment with ALSV vectors (Figure 3), and almost no GFP signal was observed at 7 dpi. To analyze the function of pathogenic B. xylophilus candidate proteins, we investigated the expression of four PR genes (PR-2, PR-4, PR-5, and PR-6) at 3, 5, and 7 dpi after ALSV vector particle bombardment using qRT-PCR. Expression levels of the four PR genes in pine seed embryos expressing Bx-TH1, Bx-TH2, and Bx-GH30 were > 5-fold higher than those of mock following normalization by the reference gene elongation factor 1-alpha (Figure 4) and GFP controls following normalization of expression levels according to the amount of viral RNA detected (Figure 5). Though no discrepancies were found between the normalized results using the expression levels of the reference gene elongation factor 1-alpha (mock control) and the amount of viral RNA (GFP control), variation of relative transcript abundance among replicates was lower when normalized with GFP control. When we normalized the gene expression levels by the reference gene elongation factor 1-alpha, no statistically significant difference was observed in relative transcript abundance due to the large variations (Figure 4). When we normalized the gene expression levels by the amount of viral RNA, Bx-TH1 induced significantly higher expression levels of the four PR genes only at 7 dpi than GFP control (p < 0.05; Figure 5). Bx-TH2 induced high expression of PR-5 in pine seed embryos at all time points, and significantly higher expression of PR-2, PR-4, and PR-6 at 5 and 7 dpi than GFP control (p < 0.05; Figure 5). Bx-GH30 induced high expression of PR-4 at 5 and 7 dpi, and the other PR genes only at 5 dpi, although these differences were not significant than GFP control, except PR-6 expression at 7 dpi (p < 0.05; Figure 5). Bx-CPI and Bx-GST did not induce PR gene expression in pine seed embryos.
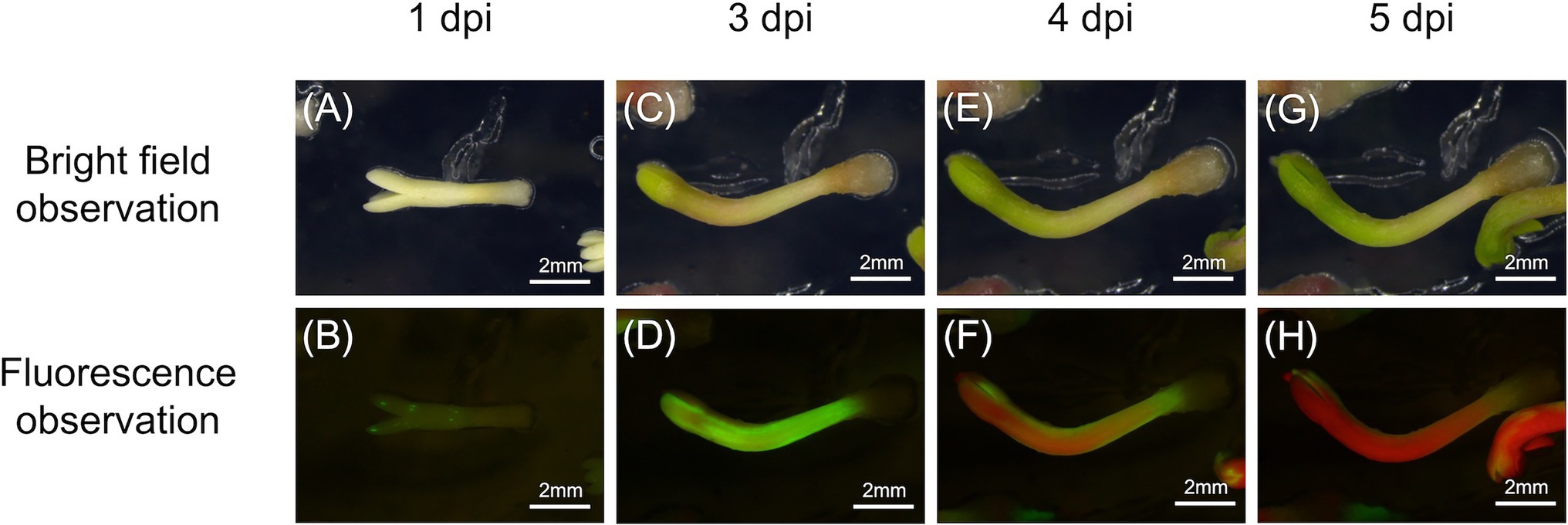
Figure 3. Green fluorescence protein (GFP) detection in pine seed embryos. GFP fluorescence in pine seed embryos at (A,B) 1, (C,D) 3, (E,F) 4, and (G,H) 5 days after particle bombardment with Apple latent spherical virus (ALSV) vectors. (A,C,E,G) Bright-field observations; (B,D,F,H) fluorescence observations.
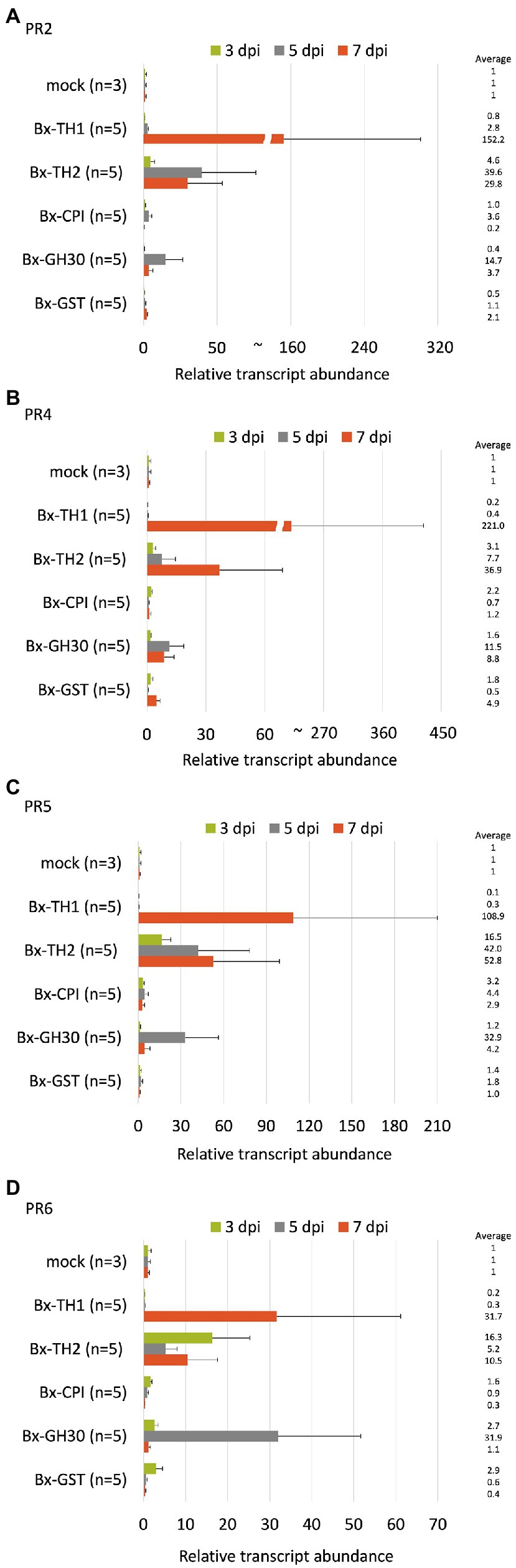
Figure 4. Relative transcript abundance of PR genes between pine seed embryos expressing pathogenic candidate proteins and mock control. Expression levels of (A) PR-2, (B) PR-4, (C) PR-5, and (D) PR-6 in pine seed embryos expressing pathogenic candidate proteins, compared with mock control, using the reference gene elongation factor 1-alpha to normalize results. No statistically significant difference was observed ( p > 0.05, Mann-Whitney U-test).
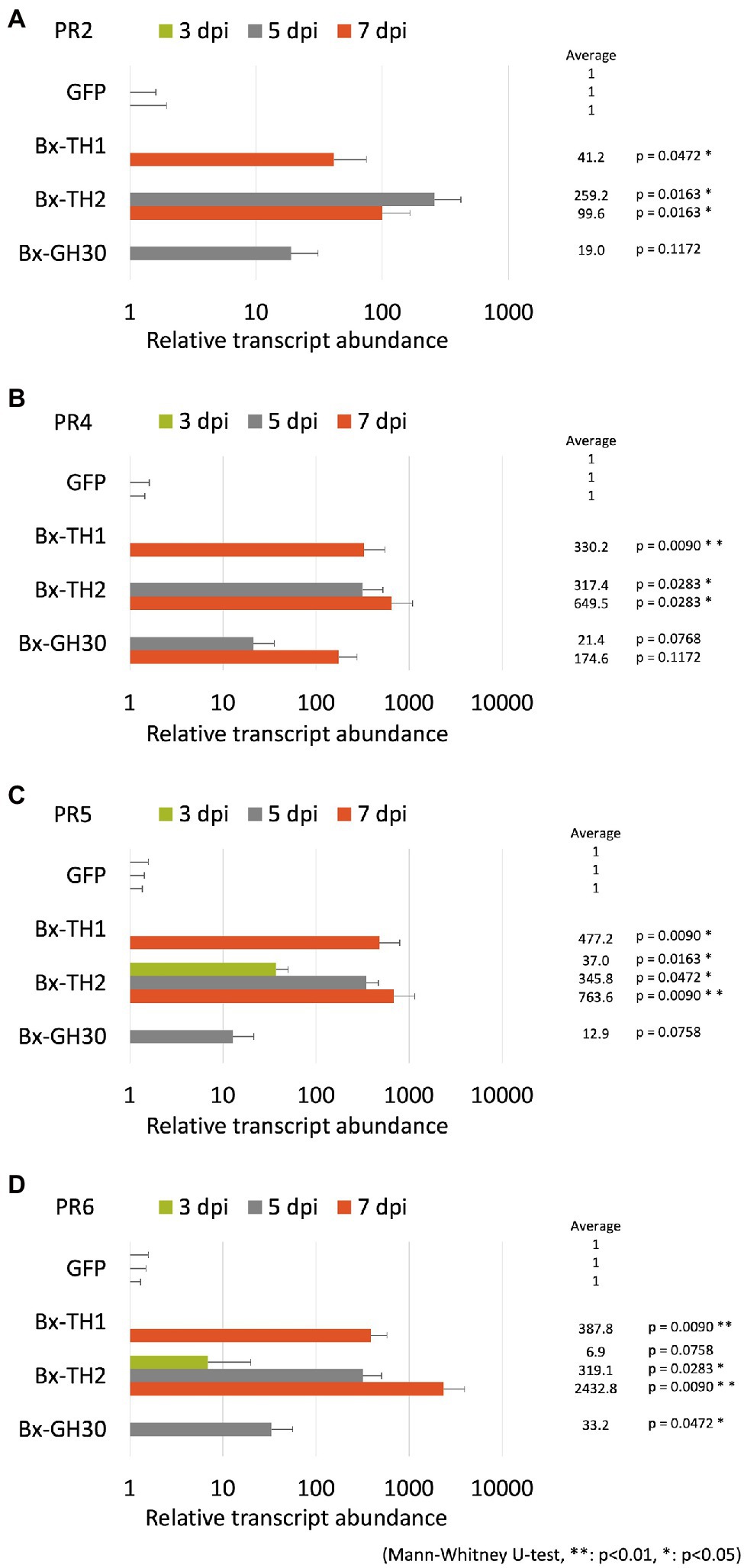
Figure 5. Relative transcript abundance of PR genes between pine seed embryos expressing pathogenic candidate proteins and GFP control. Expression levels of (A) PR-2, (B) PR-4, (C) PR-5, and (D) PR-6 in pine seed embryos expressing pathogenic candidate proteins, compared with GFP control, using the amount of viral RNA detected to normalize results. *p < 0.05 and **p < 0.01 (Mann-Whitney U-test).
Discussion
To develop a method for expressing B. xylophilus proteins in pine trees, which are its natural hosts, we conducted transient overexpression of five proteins secreted by B. xylophilus causing cell death in tobacco in susceptible black pine seed embryos using the ALSV vector and investigated whether a hypersensitive response could be induced by these proteins expression based on the transcript level of PR genes on pine seed embryos.
Previous studies using tobacco adopted cell death as an indicator for screening nematode pathogenic factors; however, exogenous gene expression was found to be transient and insufficient for inducing cell death during B. xylophilus protein expression in pine seed embryos using the ALSV vector. Instead, we constructed a GFP expression vector and bombarded ALSV particles into pine seed embryos and clearly observed the GFP signal at 1–7 dpi. This result suggests that pine embryos suppress the expression of foreign genes through post-transcription gene silencing (PTGS). PTGS is commonly observed when plants are transformed using foreign genes (Depicker and Van Montagu, 1997). For example, when ALSV with GFP or yellow fluorescent protein (YFP) was inoculated to tobacco leaves, both types of fluorescence gradually faded after 10 dpi (Takahashi et al., 2007; Yaegashi et al., 2007). Several pine species were transformed inefficiently and unstably using Agrobacterium, possibly due to gene silencing, including P. halepensis (Tzfira et al., 1996), P. taeda (Wenck et al., 1999), P. strobus (Levée et al., 1997), and P. pinaster (Trontin et al., 2002). Therefore, we focused on PR gene expression in host pine embryos as an indicator for screening nematode pathogenic factors. A previous study showed that the expression of antimicrobial peptides and putative PR genes (e.g., PR-1b, PR-2, PR-3, PR-4, PR-5, and PR-6) was higher in susceptible pine trees than in resistant trees after B. xylophilus infection (Hirao et al., 2012). Therefore, we used PR-1b, PR-2, PR-3, PR-4, PR-5, and PR-6 as candidate marker genes for B. xylophilus invasion into pine seed embryos. We found that the expression levels of PR-2, PR-4, PR-5, and PR-6 were significantly increased in susceptible pine seed embryos following B. xylophilus infection (p < 0.05). PR-1b and PR-3 expression levels were not stably detected, perhaps because they were too weak for detection by qRT-PCR. PR-2, PR-4, PR-5, and PR-6 showed similar responses to those in pine seedlings following B. xylophilus inoculation; therefore, we used these four genes as indicators of the pine hypersensitive response in subsequent analyses.
Next, we expressed candidate pathogenic B. xylophilus molecules in pine seed embryos and assessed their functions by detecting the expression patterns of PR-2, PR-4, PR-5, and PR-6. The expression levels of PR genes in pine seed embryos expressing Bx-TH1, Bx-TH2 and Bx-GH30 were > 5-fold higher than those of the mock and GFP controls. This result is consistent with our previous report that these three molecules induced cell death in N. benthamiana (Kirino et al., 2020). Further studies with many pathogenic molecules of B. xylophilus are necessary to make the consistency of the results obtained in tobacco and pine seed embryo more clear.
The two remaining pathogenic candidate proteins examined in this study (Bx-CPI and Bx-GST) did not increase PR gene expression in pine seed embryos relative to the mock and GFP controls. This result is inconsistent with the previous N. benthamiana functional screening result. Woody plants such as pines and herbaceous plants such as N. benthamiana differ in structure, lifespan, and defense systems. Generally, the defenses of slow-growing woody plants are typically quantitative and are affected by genetic variation, whereas those of fast-growing herbaceous plants are qualitative and influenced by environmental variation (Massad et al., 2011). This difference in defense systems between pine seed embryos and N. benthamiana may have influenced our functional screening results, suggesting that screening using N. benthamiana may result in false positives, although results can vary between tobacco and pine seed embryos depending on the type of gene expressed.
Although the assay system established in this study is useful, it has two limitations. First, this method tends to produce large variation in gene expression levels. Expression levels of the four selected PR genes differed among individuals and transgenes at each time point. When qRT-PCR is performed, individual variation in endogenous PR gene expression levels should be considered. Because ALSV vectors were introduced into pine seed embryos biolistically, the amount of vector introduced into each embryo is variable, leading to variation in B. xylophilus protein expression. The expression efficiency of each gene introduced into a pine seed embryo may also differ among genes. To reduce this variability in gene expression, we normalized PR gene expression based on the amount of viral RNA in each pine seed embryo (Figure 5), which produced more stable gene expression patterns. The second limitation consists of the differential characteristics between seed embryos and seedlings. Our previous studies showed that changes in the subcellular localization of candidate proteins can alter their ability to induce cell death in tobacco (Kirino et al., 2020; Shinya et al., 2021). As seed embryos are composed of undifferentiated tissue, the localization of B. xylophilus protein expressed in seed embryos may differ from that in seedlings. Therefore, to clarify whether candidate molecules that induced hypersensitive responses in seed embryos exhibit the same function in pine seedlings, it is necessary to compare their localization. Furthermore, though many reports have suggested the relationship between the hypersensitive response and pine wilt, there is still no direct evidence to prove it due to technical limitations. The method we have developed using ALSV vectors in this study may contribute to proving this in the future because ALSV vectors have been shown to effectively induce stable virus-induced gene silencing in various plants (e.g., Sasaki et al., 2011).
In summary, we report a novel functional analysis that uses recombinant pathogenic candidate molecules of B. xylophilus in host pines. The results of our investigation of tobacco and pine seed embryos suggest that tobacco-based screening for pathogenic B. xylophilus molecules may result in false positives. Although the proposed method has limitations that must be resolved in future research, it has potential as a powerful tool for screening pathogenic molecules of B. xylophilus in their native host.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material; further inquiries can be directed to the corresponding author.
Author Contributions
HK, KK, and RS designed the study and wrote the manuscript. HK and KK performed the research and analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by grants from JSPS Grant-in-Aid for Early-Career Scientists JP19K15853 (to RS), JSPS Grant-in-Aid for JSPS Research Fellow JP21J20926 (to HK), and JST PRESTO grant no. JPMJPR17Q5 (to RS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Tomonori Hirao (FFPRI) and Nobuyuki Yoshikawa (Iwate University) for generously providing me with pine seeds and ALSV vectors (pEALSR1 and pEALSR2L5R5), respectively. We thank the Forest Tree Gene Bank Program, Forest Tree Breeding Center, and Forest Products Research Institute in Japan for donating the black pine seeds.
References
Abelleira, A., Picoaga, A., Mansilla, J. P., and Aguin, O. (2011). Detection of Bursaphelenchus xylophilus, causal agent of pine wilt disease on Pinus pinaster in northwestern Spain. Plant Dis. 95:776. doi: 10.1094/PDIS-12-10-0902
Burgermeister, W., Braasch, H., Sousa, E., Penas, A. C., Mota, M., Metge, K., et al. (1999). First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology 1, 727–734. doi: 10.1163/156854199508757
Cardoso, J., Anjo, S., Fonseca, L., Egas, C., Manadas, B., and Abrantes, I. (2016). Bursaphelenchus xylophilus and B. mucronatus secretomes: a comparative proteomic analysis. Sci. Rep. 6:39007. doi: 10.1038/srep39007
Cardoso, J., Anjo, S., Manadas, B., Silva, H., Abrantes, I., Nakamura, K., et al. (2021). Virulence biomarkers of Bursaphelenchus xylophilus: a proteomic approach. Front. Plant Sci. 12:822289. doi: 10.3389/fpls.2021.822289
Depicker, A., and Van Montagu, M. (1997). Post-transcriptional gene silencing in plants. Curr. Opin. Cell Biol. 9, 373–382. doi: 10.1016/S0955-0674(97)80010-5
Ding, X., Ye, J., Lin, S., Wu, X., Li, D., and Nian, B. (2016). Deciphering the molecular variations of pine wood nematode Bursaphelenchus xylophilus with different virulence. PLoS One 11:e0156040. doi: 10.1371/journal.pone.0156040
Espada, M., Eves-van den Akker, S., Maier, T., Vijayapalani, P., Baum, T., Mota, M., et al. (2018). STATAWAARS: a promoter motif associated with spatial expression in the major effector-producing tissues of the plant-parasitic nematode Bursaphelenchus xylophilus. BMC Genom. 19:553. doi: 10.1186/s12864-018-4908-2
Espada, M., Silva, A. C., van den Akker, S. E., Cock, P. J. A., Mota, M., Jones, J. T., et al. (2016). Mol. Plant Pathol. 17, 286–295. doi: 10.1111/mpp.12280
Filipiak, A., Malewski, T., Matczyńska, E., and Tomalak, M. (2020). Molecular variation among virulent and avirulent strains of the quarantine nematode Bursaphelenchus xylophilus. Mol. Gen. Genomics. 296, 259–269. doi: 10.1007/s00438-020-01739-w
Futai, K. (2013). Pine wood nematode, Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 51, 61–83. doi: 10.1146/annurev-phyto-081211-172910
Hirao, T., Fukatsu, E., and Watanabe, A. (2012). Characterization of resistance to pine wood nematode infection in Pinus thunbergii using suppression subtractive hybridization. BMC Plant Bio. 12:13. doi: 10.1186/1471-2229-12-13
Hu, L. J., Wu, X. Q., Li, H. Y., Zhao, Q., Wang, Y. C., and Ye, J. R. (2019). An effector, BxSapB1, induces cell death and contributes to virulence in the pine wood nematode Bursaphelenchus xylophilus. Mol. Plant Microbe. Interact. 32, 452–463. doi: 10.1094/MPMI-10-18-0275-R
Huang, X., Hu, L., and Wu, X. (2019). Identification of a novel effector BxSapB3 that enhances the virulence of pine wood nematode Bursaphelenchus xylophilus. Acta. Bioch. Bioph. Sin. 51, 1071–1078. doi: 10.1093/abbs/gmz100
Kikuchi, T., Cotton, J. A., Dalzell, J. J., Hasegawa, K., Kanzaki, N., McVeigh, P., et al. (2011). Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 7:e1002219. doi: 10.1371/journal.ppat.1002219
Kirino, H., Yoshimoto, K., and Shinya, R. (2020). Thaumatin-like proteins and a cysteine protease inhibitor secreted by the pine wood nematode Bursaphelenchus xylophilus induce cell death in Nicotiana benthamiana. PLoS One 15:e0241613. doi: 10.1371/journal.pone.0241613
Kiyohara, T., and Tokushige, Y. (1971). Inoculation experiments of a nematode, Bursaphelenchus sp., onto pine trees. J. Jpn. For. Soc. 53, 210–218. doi: 10.11519/jjfs1953.53.7_210
Konagaya, K. (2017). A trial of forest tree breeding research by applying virus. Rinbokuikushu-joho (in Japanese) 23:5.
Konagaya, K., Nanasato, Y., and Taniguchi, T. (2020). A protocol for Agrobacterium-mediated transformation of Japanese cedar, Sugi (Cryptomeria japonica D. Don) using embryogenic tissue explants. Plant Biotechnol. 37, 147–156. doi: 10.5511/plantbiotechnology.20.0131a
Levée, V., Lelu, M. A., Jouanin, L., Cornu, D., and Pilate, G. (1997). Agrobacterium tumefaciens-mediated transformation of hybrid larch (Larix kaempferi × L. decidua) and transgenic plant regenerationn. Plant Cell Rep. 16, 680–685. doi: 10.1007/s002990050301
Li, C., Sasaki, N., Isogai, M., and Yoshikawa, N. (2004). Stable expression of foreign proteins in herbaceous and apple plants using apple latent spherical virus RNA2 vectors. Arch. Virol. 149, 1541–1558. doi: 10.1007/s00705-004-0310-2
Li, C., Yoshikawa, N., Takahashi, T., Ito, T., Yoshida, K., and Koganezawa, H. (2000). Nucleotide sequence and genome organization of apple latent spherical virus: a new virus classified into the family Comoviridae. J. Gen. Virol. 81, 541–547. doi: 10.1099/0022-1317-81-2-541
Ma, H. B., Lu, Q., Liang, J., and Zhang, X. Y. (2011). Functional analysis of the cellulose gene of the pine wood nematode, Bursaphelenchus xylophilus, using RNA interference. Genet. Mol. Res. 10, 1931–1941. doi: 10.4238/vol10-3gmr1367
Massad, T. J., Fincher, R. M., Smilanich, A. M., and Dyer, L. (2011). A quantitative evaluation of major plant defense hypotheses, nature versus nurture, and chemistry versus ants. Arthropod-Plant Interact. 5, 125–139. doi: 10.1007/s11829-011-9121-z
Myers, R. F. (1988). Pathogenesis in pine wilt caused by pinewood nematode, Bursaphelenchus xylophilus. J. Nematol. 20, 236–244.
Palomares-Rius, J. E., Tsai, I. J., Karim, N., Akiba, M., Kato, T., Maruyama, H., et al. (2015). Genome-wide variation in the pinewood nematode Bursaphelenchus xylophilus and its relationship with pathogenic traits. BMC Genom. 16:845. doi: 10.1186/s12864-015-2085-0
Park, J. E., Lee, K. Y., Lee, S. J., Oh, W. S., Jeong, P. Y., Woo, T., et al. (2008). The efficiency of RNA interference in Bursaphelenchus xylophilus. Mol. Cells 26, 81–86.
Qiu, X., Yang, L., Ye, J., Wang, W., Zhao, T., Hu, H., et al. (2019). Silencing of cyp-33C9 Gene affects the reproduction and pathogenicity of the pine wood nematode, Bursaphelenchus xylophilus. Int. J. Mol. Sci. 20:4520. doi: 10.3390/ijms20184520
Sasaki, S., Yamagishi, N., and Yoshikawa, N. (2011). Efficient virus-induced gene silencing in apple, pear and Japanese pear using apple latent spherical virus vectors. Plant Meth. 7:15. doi: 10.1186/1746-4811-7-15
Shinya, R., Kirino, H., Morisaka, H., Takeuchi-Kaneko, Y., Futai, K., and Ueda, M. (2021). Comparative secretome and functional analyses reveal glycoside hydrolase family 30 and cysteine peptidase as virulence determinants in the pinewood nematode Bursaphelenchus xylophilus. Front. Plant Sci. 12:640459. doi: 10.3389/fpls.2021.640459
Shinya, R., Morisaka, H., Kikuchi, T., Takeuchi, Y., Ueda, M., and Futai, K. (2013). Secretome analysis of the pine wood nematode Bursaphelenchus xylophilus reveals the tangled roots of parasitism and its potential for molecular mimicry. PLoS One 8:e67377. doi: 10.1371/journal.pone.0067377
Silva, H., Anjo, S. I., Manadas, B., Abrantes, I., Fonseca, L., and Cardoso, J. (2021). Comparative analysis of Bursaphelenchus xylophilus secretome under Pinus pinaster and P. pinea stimuli. Front. Plant Sci. 12:668064. doi: 10.3389/fpls.2021.668064
Takahashi, T., Sugawara, T., Yamatsuta, T., Isogai, M., Natsuaki, T., and Yoshikawa, N. (2007). Analysis of the spatial distribution of identical and two distinct virus populations differently labeled with cyan and yellow fluorescent proteins in coinfected plants. Phytopathology 97, 1200–1206. doi: 10.1094/PHYTO-97-10-1200
Takahashi, T., and Yoshikawa, N. (2008). “Analysis of cell-to-cell and long-distance movement of apple latent spherical virus in infected plants using green, cyan, and yellow fluorescent proteins,” in Plant Virology Protocols: From Viral Sequence to Protein Function. eds. G. D. Foster, I. E. Johansen, Y. Hong, and P. D. Nagy (Totowa, NJ: Humana Press), 545–554.
Trontin, J. F., Harvengt, L., Garin, E., Lopez-Vernaza, M., Arancio, L., Hoebeke, J., et al. (2002). Towards genetic engineering of maritime pine (Pinus pinaster Ait.). Ann. For. Sci. 59, 687–697. doi: 10.1051/forest:2002057
Tzfira, T., Yarnitzky, O., Vainstein, A., and Altman, A. (1996). Agrobacterium rhizogenes-mediated DNA transfer in Pinus halepensis mill. Plant Cell Rep. 16, 26–31. doi: 10.1007/BF01275443
Wenck, R. A., Quinn, M., Whetten, R. W., Pullman, G., and Sederoff, R. (1999). High-efficiency Agrobacterium-mediated transformation of Norway spruce (Picea abies) and loblolly pine (Pinus taeda). Plant Mol. Biol. 39, 407–416. doi: 10.1023/A:1006126609534
Yaegashi, H., Yamatsuta, T., Takahashi, T., Li, C., Isogai, M., Kobori, T., et al. (2007). Characterization of virus-induced gene silencing in tobacco plants infected with apple latent spherical virus. Arch. Virol. 152, 1839–1849. doi: 10.1007/s00705-007-1011-4
Yamagishi, N., Kishigami, R., and Yoshikawa, N. (2014). Reduced generation time of apple seedlings to within a year by means of a plant virus vector: a new plant-breeding technique with no transmission of genetic modification to the next generation. Plant Biotechnol. J. 12, 60–68. doi: 10.1111/pbi.12116
Yamagishi, N., Sasaki, S., and Yoshikawa, N. (2010). Highly efficient inoculation method of apple viruses to apple seedlings. J. K. Arch. 427, 226–229.
Yamagishi, N., and Yoshikawa, N. (2013). “Highly efficient virus-induced gene silencing in apple and soybean by apple latent spherical virus vector and biolistic inoculation,” in Virus-Induced Gene Silencing: Methods and Protocols. ed. A. Becker (Totowa, NJ: Humana Press), 167–181.
Yamaguchi, R., Matsunaga, K., Hirao, T., Tamura, M., and Watanabe, A. (2019). Spatiotemporal analysis of pine wilt disease: relationship between pinewood nematode distribution and defence response in Pinus thunbergii seedlings. Forest Pathol. 49:e12518. doi: 10.1111/efp.12518
Keywords: pine wilt disease, pine wood nematode, exogenous gene expression, hypersensitive responses, pathogenesis-related genes, apple latent spherical virus
Citation: Kirino H, Konagaya K and Shinya R (2022) Novel Functional Analysis for Pathogenic Proteins of Bursaphelenchus xylophilus in Pine Seed Embryos Using a Virus Vector. Front. Plant Sci. 13:872076. doi: 10.3389/fpls.2022.872076
Edited by:
Shahid Siddique, University of California, Davis, United StatesReviewed by:
Lee Robertson, National Institute of Agricultural and Food Research and Technology, SpainJoana M. S. Cardoso, University of Coimbra, Portugal
Copyright © 2022 Kirino, Konagaya and Shinya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryoji Shinya, c2hpbnlhQG1laWppLmFjLmpw
 Haru Kirino
Haru Kirino Ken-ichi Konagaya
Ken-ichi Konagaya Ryoji Shinya
Ryoji Shinya