
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 September 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.871331
This article is part of the Research Topic Mechanisms of Abiotic Stress Responses and Tolerance in Plants: Physiological, Biochemical and Molecular Interventions, volume II View all 37 articles
Rice (Oryza sativa) is a human staple food and serves as a model organism for genetic and molecular studies. Few studies have been conducted to determine the effects of ultraviolet-B (UV-B) stress on rice. UV-B stress triggers morphological and physiological changes in plants. However, the underlying mechanisms governing these integrated responses are unknown. In this study, we conducted a proteomic response of rice leaves to UV-B stress using two-dimensional gel electrophoresis and identified the selected proteins by mass spectrometry analysis. Four levels of daily biologically effective UV-B radiation intensities were imposed to determine changes in protein accumulation in response to UV-B stress: 0 (control), 5, 10, and 15 kJ m−2 d−1in two cultivars, i.e., IR6 and REX. To mimic the natural environment, we conducted this experiment in Sunlit Soil-Plant-Atmosphere-Research (SPAR) chambers. Among the identified proteins, 11% of differentially expressed proteins were found in both cultivars. In the Rex cultivar, only 45% of proteins are differentially expressed, while only 27.5% were expressed in IR6. The results indicate that REX is more affected by UV-B stress than IR6 cultivars. The identified protein TSJT1 (spot 16) in both cultivars plays a crucial role in plant growth and development during stress treatment. Additionally, we found that UV-B stress altered many antioxidant enzymes associated with redox homeostasis and cell defense response. Another enzyme, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH), has been identified as spot 15, which plays an essential role in glycolysis and cellular energy production. Another vital protein identified is glycosyl hydrolase (GH) as spot 9, which catalyzes the hydrolysis of glycosidic bonds in cell wall polymers and significantly affects cell wall architecture. Some identified proteins are related to photosynthesis, protein biosynthesis, signal transduction, and stress response. The findings of our study provide new insights into understanding how rice plants are tailored to UV-B stress via modulating the expression of UV-B responsive proteins, which will help develop superior rice breeds in the future to combat UV-B stress. Data are available via ProteomeXchange with identifier PXD032163.
Sunlight emits a broad spectrum of radiation: visible light, ultraviolet (UV), and infrared radiation, which reaches the Earth's surface (Kumar et al., 2016). Among these radiations, UV rays have direct and indirect effects at every level of the biological organization of plant molecules, affecting all ecosystems. The UV is divided into three different forms based on their wavelengths such as UV-A (320–400 nm), UV-B (280–320 nm), and UV-C (100–280 nm). Among them, UV-C is the most dangerous radiation, entirely absorbed by the ozone layer in the atmosphere. In contrast, UV-A radiation is unaffected, though such radiation does not harm plants. However, the UV-B radiation intensity is affected mainly by the thickness of the stratospheric ozone layer and is most harmful to plants (Sztatelman et al., 2015). The amount of UV-B radiation has increased due to stratospheric ozone (O3) depletion caused by anthropogenic chlorofluorocarbons. The increased doses of UV-B radiation cause severe biological effects on plants and other organisms. UV-B damages the cells at many levels, including altering DNA, proteins, and lipids (Brown and Jenkins, 2008). The UV-B also damages the photosynthetic pigments in plants because DNA and proteins can absorb the UV spectrum, declining the plants' photosynthetic efficiency (Wu et al., 2011). Besides these, a high level of UV-B enhances the production of reactive oxygen species (ROS), which activates stress signaling pathways (Brown and Jenkins, 2008). Mainly, UV-B radiation directly damages the DNA by forming dimers between adjacent pyrimidines in DNA strands, blocking DNA polymerase's progress and leading to cell death (Rastogi et al., 2010; Santos et al., 2012). Some genes are identified as UV-B dependent genes and are responsible for synthesizing UV screening compounds, DNA repairs, and activation of oxidative enzymes (Brosche and Strid, 2003). In addition, UV radiation causes downregulation of photosynthesis-related proteins, such as light-harvesting Chl a/b binding protein, whereas it upregulates protective proteins such as pathogen-related protein-1 (PR-1) at the mRNA level (Surplus et al., 1998). UV-B radiation also negatively affects leaf, hypocotyl, and seedling growth (Robson and Aphalo, 2012; Biever et al., 2014; Fina et al., 2017). The effects of UV-B have been studied in different crops like Glycine max (Liang et al., 2006; Peng and Zhou, 2009; Iii et al., 2013), Triticum aestivum (Agrawal and Rathore, 2007), Hordeum vulgare (Fedina et al., 2003), Pisum sativum (Qu et al., 2006), Zea mays (Casati et al., 2005; Reddy et al., 2013), and Arabidopsis thaliana (Sztatelman et al., 2015). Rice (Oryza sativa L.) is one of the most important staple food crops for human consumption and a model plant for genetic and molecular study. Some studies have been conducted on the physiological response of UV-B stress on rice (He et al., 1993; Dai et al., 1994; Roy and Roy, 2013); however, only a few studies (Du et al., 2011; Wu et al., 2011) have been conducted on the UV-B stress on protein levels. Therefore, the UV-B-induced proteomic study on rice will provide new insight into understanding the mechanisms underlying injurious and protective responses of crops during UV-B stress.
Nowadays, a proteomic approach to revealing the mechanisms of plants under stress has become the method of choice. Mass spectrometry is an essential tool in proteomic research (Li and Assmann, 2000). Innovation in mass spectrometry has significantly improved the study of protein structure and function. Especially when the genome sequence is unavailable, amino acid sequencing is required for protein identification. Amino acid sequence information can be determined by mass spectrometry, which allows homology searching and cloning or database identification of the corresponding gene (Shevchenko et al., 1997). In the past, Li and Assmann showed that the de novo peptide sequencing of a low-abundance broad bean (Vicia faba) protein isolated from two-dimensional gels illustrates the power of mass spectrometry for protein identification (Li and Assmann, 2000). In this study, the proteome analysis of the rice leaves was determined for two different varieties (Japonica REX and Indica IR6) irradiated by artificial UV-B radiation. IR6 and REX are more popular than other Indica and Japonica rice cultivars and are cultivated widely, especially in the United States. Both were considered tolerant stress to temperature and drought. Previously, we studied the effect of day/night temperatures on rubisco activity and electron transport in these two varieties (Singh et al., 2019). Moreover, our previous studies showed that IR6 performed very well under low and high temperatures (Reddy et al., 2021). Thus, we want to see how both these varieties perform under UV-B stress. Both varieties showed significant changes in the number and expression of various proteins using mass spectrometry analysis. Specific novel UV-B proteins were identified, and their role in UV-B stress was implicated. The findings of this study may provide new insights into understanding how rice plants adapt to UV-B stress by modulating the expression of UV-B responsive proteins and the relevant genes, which may provide information for cultivating superior rice breeds in the future.
This experiment used two varieties of rice: Japonica Rex and Indica IR6. The rice varieties were obtained from Dr. Ed Rodona, Rice Breeder, Delta Research and Extension Center, Stoneville, MS, USA. This study was conducted in sunlit Soil-Plant-Atmosphere-Research (SPAR) chambers located at the Rodney Foil Plant Research Center, Mississippi State University, Mississippi State (33°28′ N, 88°47′ W), USA. All the SPAR chambers are made up of a steel soil bin (1-m deep by 2 m long by 0.5 m wide) to accommodate the root system and a Plexiglas chamber (2.5 m tall by 2.5 m long by 1.5 m wide) to accommodate aerial plant parts, a heating and cooling system, and an environmental monitoring and control system. The Plexiglas allows 97% of the visible solar radiation to pass without spectral variability in absorption and is opaque to solar UV-B radiation (280–320 nm) but transmits 12% of UV-A radiation (wavelength 320–400 nm). The lamps were wrapped with pre-solarized 0.07 mm cellulose diacetate film (JCS Industries Inc., La Mirada, CA) to avoid the germicidal effects of UV-C radiation. During these experiments, the measured minimum, maximum, and mean of daily solar radiation were 7.96, 31.22, and 22.34 ± 0.63 MJ m−2 d−1, respectively. Temperatures in all SPAR units were maintained at 29/21°C (day/night) and CO2 410 μmol mol−1 during the experimental period. Air temperature in all the SPAR units was documented and adjusted every 10 seconds within set points of 30/22°C (day/night) ± 0.5°C. The daytime temperature was initiated at sunrise and returned to the nighttime temperature 1 h after sunset (Reddy et al., 2013).
Two varieties of rice seeds (Rex and IR6) were sown on May 10, 2017, in polyvinyl chloride pots (6” diameter by 12” high) filled with sand with a layer of gravel at the bottom. Each pot had a small hole at the bottom for excess water drainage. A total of 48 pots were used in the study. Six replicates per treatment for each variety were used. For each treatment, different SPAR chambers were used. Six pots per cultivar were arranged as a completely randomized design in six rows with two pots per row in each SPAR chamber (Supplementary Figure 1). Initially, four seeds were sown in each pot, and 11 days after emergence, the plants were thinned to one plant per pot. To ensure promising water and nutrients for growth, plants were irrigated three times a day with full-strength Hoagland's nutrient solution delivered at 08:00, 12:00, and 17:00 h through an automated and computer-controlled drip system.
In the beginning, all the chambers were maintained at a 0 kJ m−2 d−1 UV-B level. Fifteen days after planting (15DAP), four levels of daily biologically effective UV-B radiation intensities, 0 (control), 5, 10, and 15 kJ m−2 d−1 were imposed and continued until harvest. The expected UV-B radiation dosage was provided by the square-wave UV-B supplementation system under near-ambient photosynthetically active radiation. The UV-B radiation was delivered daily from 08:00 to 16:00 h by eight fluorescent UV-313 lamps (Q-Panel Company, Cleveland, OH, USA) attached horizontally on a metal frame inside each chamber, powered by 40 W dimmable ballasts. However, the same setup was used in the control unit to simulate equal shading; however, the lamps were kept unilluminated. To filter out UV-C (<280 nm), we wrapped individual lamps with solarized 0.07 mm cellulose diacetate films (JCS Industries Inc., La Mirada, CA, USA), and these were replaced as needed to maintain the particular UV-B radiation treatment. The distance from the lamps to the plant canopy was maintained at 0.5 m and adjusted to match the plant height throughout the experiment. The anticipated UV-B level was supplied to the top of the plant canopy and was checked daily at 09:00 h with a UVX digital radiometer (UVP, Inc., San Gabriel, CA, USA) and standardized against an Optronic Laboratory Model 754 Spectroradiometer (Orlando, FL, USA) that was used initially to quantify lamp output. To ensure the plants received the exact UV-B dosage, we took the respective UV-B measurements at three locations, 50 cm apart, in each SPAR chamber. The measured average daily biologically effective UV-B radiation doses during the experimental period were 5.08 ± 0.13 for 5 KJ, 10.08 ± 0.14 for 10 kJ, and 15.9 ± 0.63 for 15 kJ of UV-B treatments.
The second uppermost leaf of the treated rice plants was collected on the 20th day after UV-B exposure, control treatment, and 35 days after transplanting (35DAP). The samples were immediately frozen in liquid nitrogen and stored at −80°C for further use. Samples were ground to a fine powder in liquid nitrogen, and total proteins were extracted from rice leaves using the method of Wang et al. (2006), with slight modification. First, the 0.3–0.4 g fine powder was transferred into a 2 ml Eppendorf tube and filled with chilled 10% trichloroacetic acid (TCA)/acetone (Sigma-Aldrich, ≥ 99.0%). The mixture was vortexed and then centrifugated at 4°C for 3 mins at 16,000 x g, and the supernatant was discarded without touching the pellet with a glass pipette. The pellet was washed by adding 80% methanol plus 0.1 M ammonium acetate (Sigma, ~98%) and further washed with 80% acetone at 4°C for 3 mins at 16,000 × g to remove residual TCA and contaminants. The pellet was air-dried in the hood for at least 10 mins to remove residual acetone. The pellet was suspended in a 1:1 ratio of phenol (pH 8.0) and sodium dodecyl sulfate (SDS) buffer [30% sucrose, 2% SDS (Bio-Rad, 1610302), 0.1 M Tris-HCl] supplemented with 5% 2-mercaptoethanol (Thermo-Fisher) and vortex for 30 s and then incubated at room temperature for 5–10 mins. The mixture was centrifuged at 16,000 × g for 3 mins at room temperature. The upper phenol phase (about 300 μl) was transferred into a new 2-ml Eppendorf tube, filled with five volumes of cold methanol plus 0.1 M ammonium acetate, and incubated at −20°C for about 2 h. After incubation, the mixture was centrifuged at 16,000 × g for 5 mins at 4°C, and the supernatant was discarded without disturbing the pellet. The tube containing the pellet was filled with methanol plus 0.1 M ammonium acetate and centrifuged at 16,000 × g for 5 mins at 4°C. After centrifugation, the supernatant was discarded without disturbing the pellet. Finally, the pellet was washed with 80% acetone by centrifugation at 16,000 × g for 5 mins. The supernatant was discarded and pellet were air-dried to remove any remaining acetone, and the dried pellet (proteins) was stored in the tube at −80°C for further use.
The protein pellet was resuspended in an appropriate volume (about 150 μl) of rehydration buffer [9% urea, 4% 3-[(3-cholamidopropyl)dimethylammonium]-1-propanesulfonate (CHAPS) (GoldBio, 75621033), 50 mM Dithiothreitol (DTT; GoldBio, 27565419) supplemented with BioLyte 3/10 Ampholyte (Bio-Rad, 1632094). After gentle mixing, the mixture was incubated for 1 h with agitation. The protein samples were centrifuged at 16,000 xg at room temperature for 3 mins. The protein content in the supernatant was quantified using the modified Bradford assay (Bradford, 1976).
The 2-DE was performed using the precast immobilized pH gradient (IPG) strips (Bio-Rad Laboratories) in the first dimension by using an isoelectric focusing (IEF) system (PROTEAN IEF Cell, Bio-Rad Laboratories) according to the manufacturer's instructions. Rice leaf proteins (400 μg) in rehydration buffer (9 M urea, 4% CHAPS, 50 mM dithiothreitol, 0.1% Bio-Lyte Ampholyte) were added to individual channels of an IPG focusing tray (Bio-Rad Laboratories). The Bio-Rad IPG strips (pH 4–7, 11 cm) were then placed over the protein solutions in the channels of the IPG focusing tray. The protein samples were rehydrated in the IPG focusing tray at 50 Volt for 1 h. After rehydration, the samples were run at 250 Volt for 15 mins, then ramped to 8,000 Volt over 2.5 h, and kept at 8,000 Volt for 13 to 15 h. After isoelectric focusing, the IPG strips were equilibrated in equilibration buffer I (6 M urea, 0.375 M Tris-HCl, pH 8.8, 2% SDS, 20% glycerol, and 100 mM dithiothreitol) for 10 mins at room temperature and then placed in equilibration buffer II (100 mg Iodoacetamide, 6 M urea, 0.375 M Tris-HCl, pH 8.8, 2%SDS, 20% glycerol, and 0.01% bromophenol blue) for 10 mins. After incubation, the strips were directly placed on 12% polyacrylamide-SDS slab gels for second-dimension separation. Second-dimension gels were run at 8 mA per gel using a vertical gel electrophoresis system (R. Shadel Inc.). The same gel was stained with colloidal Coomassie blue or silver nitrate, and then the sensitivity in detecting different proteins between colloidal Coomassie staining and silver staining was compared. The results also showed that silver staining is more sensitive than colloidal Coomassie staining and provides better resolution of the spots (Supplementary Figure 2). After that, all the gels were then stained with silver staining, as described by Shevchenko et al. (1996), with minor modifications.
After staining, the 2-DE images were obtained with a ChemiDoc Touch imaging system (Bio-Rad Laboratories), and protein spots were detected and analyzed with Image Lab 6.0 (Bio-Rad Laboratories). Silver-stained gels were stored in a 1% acetic acid solution at 4°C until further analysis.
Protein spots on stained 2-DE gels were cut into ~1 mm3 pieces. Gel pieces were then subjected to a modified in-gel trypsin digestion procedure (Shevchenko et al., 1996). The samples were reduced by using DTT (1 mM concentration in 50 mM ammonium bicarbonate) for 30 mins at 60°C. The samples were then cooled to room temperature, and iodoacetamide (5 mM) was added for 15 mins in the dark at room temperature. DTT (5 mM) was then added to quench the reaction. Then sequence grade trypsin (Promega, Madison, WI) 5 ng/μl was added. After 45 mins, the excess trypsin solution was removed and replaced with a 50 mM ammonium bicarbonate solution to cover the gel pieces. The digestion was performed overnight at 37°C. Peptides were later extracted by removing the ammonium bicarbonate solution, followed by one wash with a solution containing 50% acetonitrile and 1% formic acid. The extracts were then dried in a speed-vac (~1 h). The samples were then stored at 4°C until analysis. On the analysis day, the samples were reconstituted in 5–10 μl of HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). A nano-scale reverse-phase HPLC capillary column was created by packing 2.6 μm C18 spherical silica beads into a fused silica capillary (100 μm inner diameter × ~30 cm length) with a flame-drawn tip (Peng and Gygi, 2001). After equilibrating the column, each sample was loaded onto the column via a famous autosampler (LC Packings, San Francisco, CA). A gradient was formed, and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid). As peptides were eluted, they were subjected to electrospray ionization and then entered into an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein databases (http://www.uniprot.org/proteomes/?query=taxonomy:39947) with the acquired fragmentation pattern by the software program, Sequest (Thermo Fisher Scientific, Waltham, MA) (Eng et al., 1994). All databases include a reversed version of all the sequences, and the data were filtered to between a one and two percent peptide false discovery rate.
Gene ontology(GO) annotation was performed via the Gene Ontology Database at (http://systemsbiology.cau.edu.cn/agriGOv2/) (Tian et al., 2017). The GO function of the identified proteins was classified into three categories, i.e., biological process, molecular process, and cellular component. The annotated GO term was plotted via a web tool, WEGO 2.0 (https://wego.genomics.cn/) (Ye et al., 2018). The pathway analysis was performed using the database RiceNetDB (http://bis.zju.edu.cn/ricenetdb/). The annotation was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Liu et al., 2013). The gene functions were categorized based on databases, including the Rice Annotation Project Database (RAP-DB) build 5.0 (Sakai et al., 2013) by the International Rice Genome Sequencing Project (IRGSP), the MSU Rice Genome Annotation Database (Ouyang et al., 2007), and the Oryzabase Integrated Rice Science Database (https://shigen.nig.ac.jp/rice/oryzabase/).
In this study, the seedlings of two rice cultivars (REX, IR6) were treated with UV-B irradiation of 5, 10, and 15 kJ m−2 d−1 for 8 h daily. There is no morphological damage to plants, except for the fact that plants were shorter than the control (Figure 1). The plant height in both cultivars significantly differs between the treatments and cultivars. The UV-B effects on plant height in Rex cultivars are more severe than IR6 (Figure 1A). While increasing the UV-B dose, the plants become shorter in both varieties (Figure 1A). IR6 has more tillers than Rex, but there is no effect of UV-B stress on tiller number per plant in both cultivars compared to control (Figure 1B). The highly significant decreasing pattern in leaf numbers in both varieties is observed as UV-B stress dose increases compared to control (Figure 1C).
Proteomic analysis of rice leaves 2D-GE carried out the protein of two varieties modulated by UV-B stress. Each 2D-GE gel had ~1,000 protein spots covering a 4.0–7.0 pH range and a molecular weight of 4–120 KDa, based on their isoelectric points. Seventeen spots, labeled 1 to 17, showed significant changes in the IR6 varieties under three different levels of UV-B stress compared to their respective controls (Figure 2A). Similarly, 23 spots labeled 18 to 40 exhibited significant changes in the rice Rex varieties compared to control in UV-B stress (Figure 2B). Figure 2 shows the representative 2D-GE gel images of IR6 and Rex. By comparing the gels produced in the three different doses of UV-B treatment, the 18 spots in the IR6 variety displayed significant changes in expression. These protein spots exhibited altered expression profiles (1, 2, 4–11, 14–18, 21, 34, and 37; Figure 2) were successfully identified by mass spectrometry (Table 1). Similarly, there are 30 spots in Rex varieties in the different UV-B treatment, i.e., spot 1, 4–6, 14–16, 1–40 (Table 1). Among these spots, 11 spots were identified in both varieties. These spots were No. 1, 4–6, 14–16, 17, 21, 34, 37, and 40 (Table 1). Out of 11 spots, six spots (No. 4, 5, 14, 16, 21, and 37) showed a similar expression pattern in both the varieties in all the different levels of UV-B treatment (Table 2). Altogether, in both varieties, 40 spots were identified. The Venn diagram showed the differentially expressed proteins in both the cultivars (Figure 3A). A total of 45% of proteins were differentially expressed in the Rex cultivar, whereas only 27.5% were expressed in the IR6 cultivar. Moreover, 11% of differentially expressed proteins were common in both cultivars (Figure 3A). These results suggest that REX is more affected by UV-B stress than IR6 cultivars.
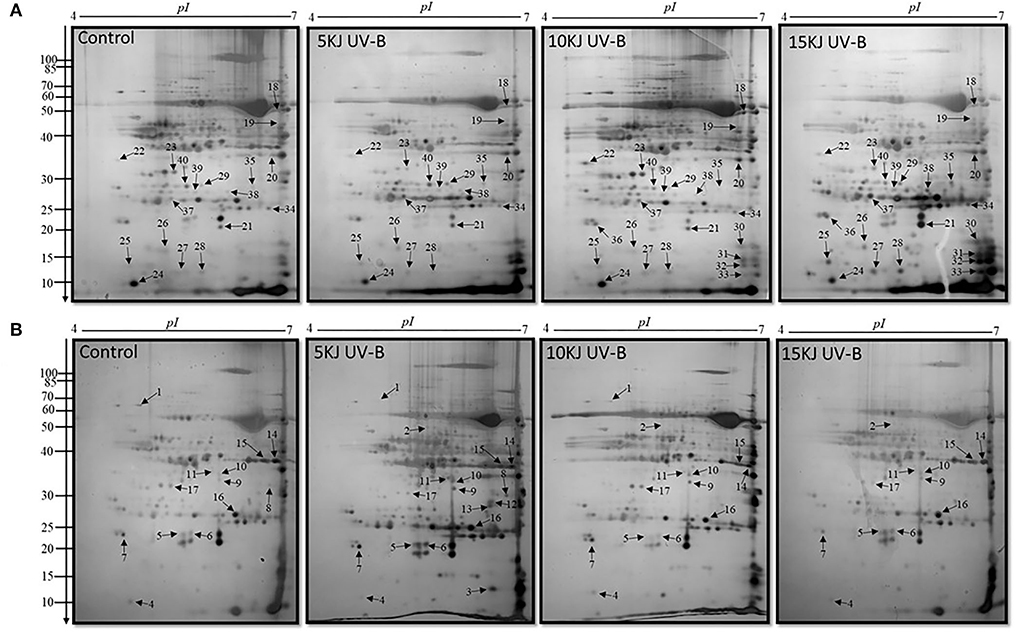
Figure 2. 2D-GE maps of rice leaves proteins during 5, 10, and 15 kJ m−2 d−1 UV-B exposure along with control (A) cultivar REX (B) cultivar IR6. Quantitative image analysis revealed 40 differential spots were up and downregulated with silver staining. The black arrowheads showed the number of proteins identified. The numbers at the left of each panel are molecular masses in kilodaltons.
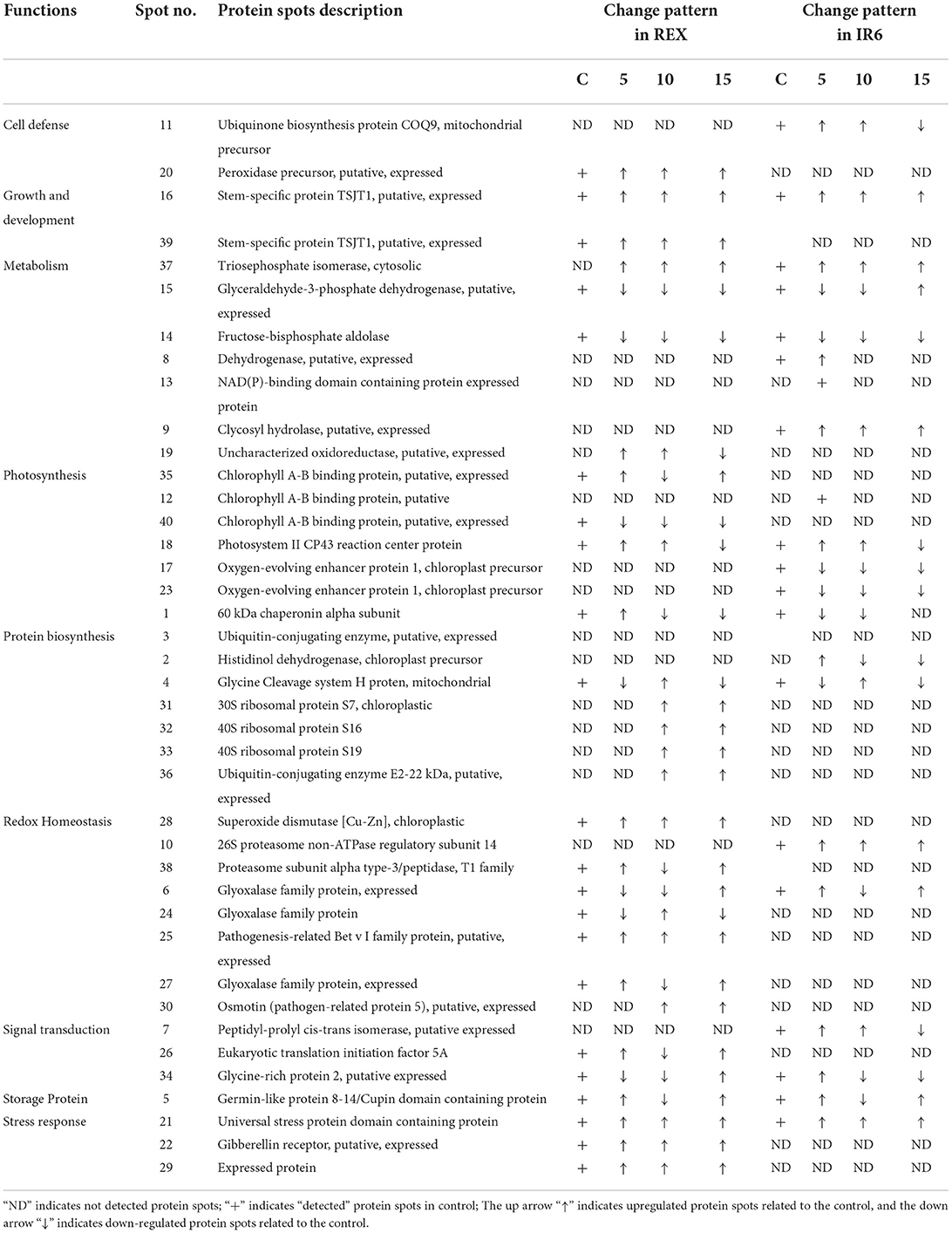
Table 2. Functional classification and expression pattern of proteins in rice leaves under different levels of UV-B response.
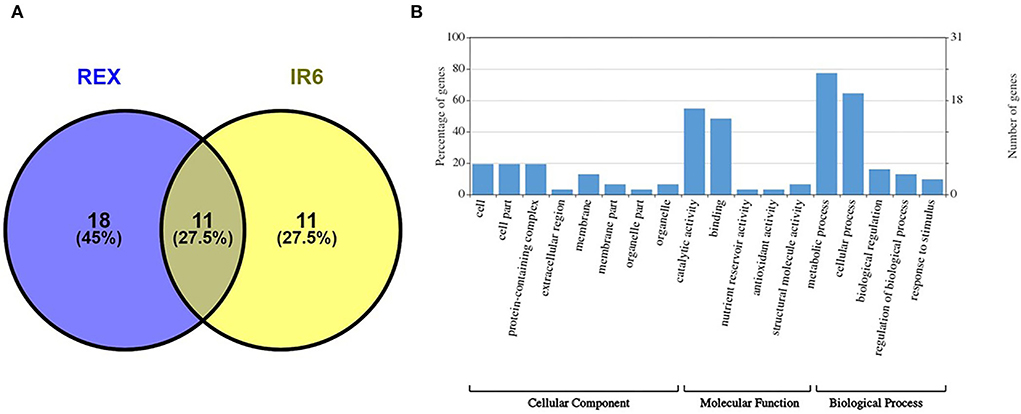
Figure 3. (A) Venn diagram showing the differentially expressed proteins in the cultivars REX and IR6 (B) GO enrichment analysis of identified proteins. The number of proteins enriched with each GO term in one of the three categories, including biological process, molecular function, and cellular component, is shown.
Forty protein spots exhibited altered expression profiles in both rice varieties (Figure 2). Among them, spot 12, 35, 16, and 39 were the same protein (Table 1). We next conducted a functional analysis of all identified proteins by mass spectrometry. Go enrichment analysis showed that proteins are involved in the metabolic and cellular processes in the subcategory of biological processes (Figure 3B). In addition, most of the proteins are also associated with molecular functions such as catalytic activity and binding (Figure 3B).
We did a pathway analysis further to understand the potential roles and significance of identified proteins. The KEGG analysis showed that most of the proteins are involved in metabolic pathways (Supplementary Table 2), which suggests that UV-B stress has a potential role in metabolic pathway regulation. Moreover, the cellular component analysis showed that the identified proteins mainly resided in the cell, cell part, or as a part of protein-containing complexes (Figure 3B).
Most identified proteins are involved in metabolism, photosynthesis, protein biosynthesis, and redox homeostasis (Table 2). Few proteins—such as peptidyl-prolyl cis-trans isomerase, eukaryotic translation initiation factor 5A, glycin-rich protein two—were involved in signal transduction. Three proteins are detected, i.e., universal stress protein domain-containing protein, gibberellin receptor, and expressed protein (Table 2) implicated in stress response. There are 7 spots identified in metabolism (i.e., spots 37, 15, 14, 8,13, 9, and 19). Spots 37 is identified as a triosephosphate isomerase (TPI), a glycolytic enzyme that catalyzes the reversible interconversion of dihydroxyacetone phosphate to glyceraldehyde-3-phosphate. In both the cultivars, TPI is upregulated at 5, 10, and 15 kJ m−2 d−1 UV-B levels compared to control (Table 2). Another important protein is identified as spot 15, which is glyceraldehyde-3-phosphate dehydrogenase (GAPDH). GAPDH expression is decreased in both the cultivars under 5 and 10 kJ m−2 d−1 UV-B levels compared to control, whereas the expression is high in IR6 under 15 kJ m−2 d−1 UV-B levels compared to control. Similarly, fructose-bisphosphate aldolase, putative, is identified as spot 14, which is downregulated in both the cultivars under all the UV-B dose levels compared to control. Dehydrogenase and NAD(P) binding domain-containing protein were only detected in IR6 at 5 kJ m−2 d−1 UV-B levels compared to control. Similarly, glycosyl hydrolases (i.e., spot 9) are upregulated only in IR6 under all the UV-B dose levels compared to control. Likewise, spot 19 is identified as an uncharacterized oxidoreductase, upregulated in IR6 under all the UV-B dose levels compared to control (Table 2).
Like in metabolism, seven proteins have been identified in the photosynthesis process. In this functional category, the same protein identified in two spots (i.e., spots 12 and 35) belongs to the family of light-harvesting complex chlorophyll a/b-binding proteins. Spots 35 were only detected in REX, not in the IR6 cultivar. Under 5 and 15 kJ m−2 d−1 UV-B level, chlorophyll a/b-binding protein is upregulated in REX cultivars, whereas the expression is decreased under 10 kJ m−2 d−1 UV-B level. However, in IR6, this protein is detected at the 5 kJ m-2 d-1 UV-B level, whose expression is also upregulated like in another cultivar (Table 2). Similarly, another protein in the same family is identified as spot 40, only detected in the REX cultivar. Photosystem II, CP43 reaction center protein, is recognized as spot 18. It is upregulated in both the cultivars at 5 and 10 kJ m−2 d−1 UV-B levels as compared to the control. However, this protein is downregulated in both the cultivars at the 15 kJ m−2 d−1 UV-B level as compared to the control (Table 2). Oxygen-evolving enhancer protein 1 is identified as two spots, i.e., 17 and 23, and expression is decreased under all levels of UV-B dose as compared to the control and only detected in the REX cultivar (Table 2). The 60 kDa chaperonin alpha sub-unit is identified as spot 1 and is noticed in both cultivars. At the 5 kJ m−2 d−1 UV-B level, expression of chaperonin protein is high in Rex but decreased in IR6; however, at the 10 kJ m−2 d−1 UV-B level, expression of this protein was reduced in both the cultivars (Table 2). Three proteins have been identified in the stress response category, i.e., spots 21, 22, and 29, and the expression level is high under all levels of UV-B dose in the REX cultivar, whereas only spot 21 has been detected in the IR6 cultivar, whose expression is high at the 5 and 10 kJ m−2 d−1 UV-B level but low at the 15 kJ m−2 d−1 UV-B levels as compared to control. UV-B also played a significant role in plant growth and development. Two spots (i.e., spots 16 and 39) have been identified in this category as the same protein, which is stem-specific protein TSJT1. TSJT1 expression levels are increased in both cultivars, which may explain why the plants look shorter after the UV-B treatment. Likewise, in stress response, there are three proteins identified in the signal transduction category as spots 7, 26, and 34 (Table 2). Another category is redox homeostasis, in which there are eight proteins identified, i.e., spots 6, 10, 24, 25, 27, 28, 30, and 38. Only one protein identified in the storage protein category, spot 5, is recognized as a germin-like protein. In addition, seven proteins have been identified in the protein biosynthesis category. In these categories, ribosomal proteins are the most predominant proteins (Table 2).
Unlike studies conducted in greenhouses, this research is carried out in (SPAR) chambers, which are closer to the natural environment. In this study, two varieties were used to compare the three levels of UV-B stress (5, 10, and 15 kJ m−2 d−1) to control. In this work, we discovered 38 distinct proteins that were differentially expressed in both cultivars at different UV-B exposure levels compared to controls. We found 38 proteins, including previously known proteins. We also identified some novel proteins. According to identified proteins and their expression levels, the REX cultivar is more sensitive to UV-B damage than IR6 (Figures 2, 3A).
Our studies identified the TSJT1 protein as spot 16 in both cultivars. After applying the UV-B stress, the expression of TSJT1 increases in both the cultivars compared to the control. Only a few studies have been conducted on the TSJT1 protein. In plants, internode elongation is correlated with cell elongation and cell division. Hu and coworkers reported that TSJT1 might act as a negative regulator in castor internode development (Hu et al., 2016). Another study demonstrated that the abundance of TSJT1 was decreased in the dwarf variety Zhebi 100 compared to Zhebi 26. Zhebi 26 suggests that TSJT1 plays a role in node development. Additionally, the expression of the TSJT1 protein demonstrated higher drought stress in roots and leaves in two cassava cultivars—SC124 and Arg7—which were identified in proteomic analysis (Zhao et al., 2015). Our study also observed that the plant height decreases after applying UV-B stress, and TSJT1 expression increases as UV-B stress is imposed. These results imply that TSJT1 inhibits plant growth and development when exposed to stress.
Reactive oxygen species (ROS) play a dual role as critical regulators of growth, development, and defense pathways and toxic byproducts of aerobic metabolism (Mittler et al., 2004). The production of ROS is associated with the loosening of the plant cell wall during cell elongation. Rapid plant growth is characterized by an increase in antioxidation capacity since excessive ROS generation might damage cells (Cui et al., 2012; Hu et al., 2016). An alternative method, such as ROS scavenging by superoxide dismutase, can control ROS levels in plants (Goossens et al., 2003). In our study, UV-B stress caused changes in a number of antioxidation enzymes related to redox homeostasis and cell defense response proteins. In this study, UV-B stress treatment in cultivar REX increased the abundance of superoxide dismutase (spot 28), pathogenesis-related Bet V I family protein (spot 25), osmotin (pathogen-related protein 5) (spot 30), and peroxidase precursor (spot 20) as compared to control. Hu et al. (2016) also reported that—in the castor bin (high stalk variety Zhebi 100)—peroxidase precursor, and superoxide dismutase abundance were increased. The results presented above, including previously published data, suggest that the enhanced abundance of these proteins may also imply that the anti-oxidative defense system and resistance to UV-B stress are increased in cultivar REX as compared to control. These proteins were not detected in IR6.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is an important enzyme involved in glycolysis and cellular energy production associated with plant development (Giegé et al., 2003). The primary function of GAPDH is to catalyze the conversion of glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate within the glycolytic pathways; therefore, it plays a crucial role in energy metabolism (Voss et al., 2007). Voss et al. (2007) used GAPDH as a model protein to study oxidative damage caused by irradiation such as UV-A, UV-B, and infrared (IR). They showed that only UV-B irradiation was more aggressive and that UV-B acted on specific amino acids, such as arginine, proline, and tyrosine (Voss et al., 2007). Our study identified that GAPDH (spot 15) and fructose-bisphosphate aldolase (spot 14) were decreased under UV-B stress. This agrees with an earlier report on cyanobacteria under UV-B stress using two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) (Babele et al., 2015). The enzyme fructose-bisphosphate aldolase occupies a central position in glycolysis and gluconeogenesis (Ziveri et al., 2017). Fructose-1,6 bisphosphate aldolase is an essential enzyme that converts fructose-1, 6-bis phosphate to glyceraldehyde-3-phosphate and dihydroxyacetone 3-phosphate. Under the stress conditions, glyceraldehyde-3-phosphate and fructose-1, 6 bis phosphate may be converted to glucose-6-phosphate to carry out the pentose phosphate pathway for NADPH synthesis (Nakahara et al., 2003). This may explain why both enzymes were downregulated in our study during UV-B stress. Fructose-bisphosphate aldolase is essential for bacterial multiplication and participates in the control of host redox homeostasis and the inflammatory immune response (Ziveri et al., 2017). This study also identified triosephosphate isomerase (OsTPI1.1) as spot 37. Interestingly, we found that OsTPI1.1 is upregulated in both cultivars under UV-B stress. The triosephosphate isomerase enzyme catalyzes the interconversion of dihydroxyacetone phosphate (DHAP) and GAPDH during glycolysis and gluconeogenesis. OsTPI1.1 also modulates ROS production as a resistance mechanism against Xanthomonas oryzae pv oryzae (Xoo) (Liu et al., 2018). Xoo causes severe damage to rice production worldwide (Liu et al., 2018). The above results, including our finding, suggest that UV-B plays a role in metabolic pathways, cell defense, and transcriptional regulation.
Moreover, in this study, we identified glycosyl hydrolase (GH) as spot 9, whose expression has increased in IR6. However, GH was not detected in the REX cultivar. The primary function of GH is to catalyze the hydrolysis of glycosidic bonds in cell wall polymers and can have significant effects on cell wall architecture (Sharma et al., 2013). In rice, 437 GH genes are classified into 34 families. They identified 138 GH genes that are highly diverged between monocots and dicots, 57 of which have diverged further in rice as compared with four monocot genomes. They found two-thirds of GH genes in rice are upregulated in response to biotic and abiotic stress treatments (Sharma et al., 2013). In our study, GH is also upregulated, suggesting that UV-B stress treatment plays a role in abiotic and biotic stress adaptation and cell wall architecture. So far, GH has not been identified in the UV-B stress study. GH is the new protein we identified in UV-B stress treatment, suggesting that it may have a new function in cell wall architecture under UV-B stress.
Photosynthesis and protein biosynthesis have a significant function in plant growth and development. UV-B responses on plant species and even genotypes within species are different (Yuan et al., 2000). Under UV-B stress, Oryza sativa cultivar Lemont, produced more water-soluble proteins without any harmful effects on rice leaf photosynthetic properties, which suggests that it might be related to its UV-B tolerance. However, in the same dose, it caused damage to pea plants (He et al., 1993). The net photosynthesis rate increased under moderate UV-B irradiation in Pseudotsuga menziesii (Bassman et al., 2002). The photosynthesis rate of Acorus calamus L. (sweet flag) plants was increased under low UV-B treatment, which led to increased biomass (Kumari et al., 2009). Similarly, all proteins associated with photosynthetic processes—such as oxygen evolution complex protein 1, RuBisCO, and carbonic anhydrase—were altered in Oryza sativa (Wu et al., 2011). In our study also, photosynthesis-related proteins such as chlorophyll A-B binding protein (12,35,40), photosystem II CP43 reaction center protein (spot 18), and oxygen-evolving enhancer protein (spot 17 and 23) were altered. Chlorophyll A-B binding protein is only detected in the cultivar REX. Spot 35 increases under lower and high doses but decreases under moderate doses. However, spot 40 proteins decreased under all levels of UV-B stress. Some proteins are represented by more than one spot. These spots may represent different products of two very closely related genes or post-translational modifications of a single protein. We also find that the expression level is also different, suggesting that there are probably different isoforms regulated in opposite ways or that one isoform is post-translationally regulated (Casati et al., 2011). Chlorophyll A-B binding proteins are part of the light-harvesting complex (LHC). The LHC functions as a light receptor that captures and delivers excitation energy to photosystems I and II. Chlorophyll A-B binding proteins are the most abundant membrane proteins in nature (Xu et al., 2011). The overexpression of chlorophyll A-B binding proteins enhances the stomatal sensitivity to ABA, whereas downregulation of chlorophyll A-B reduces the responsiveness of stomatal movement to ABA, which results in a decrease in plant tolerance to drought stress in Arabidopsis (Xu et al., 2011).
Similarly, we also identified photosystem II C43 reaction center protein (spot 18). CP43 is one of the components of the core complex of photosystem II (PSII). PSII is a large pigment-protein complex embedded in the thylakoid membrane of all oxygenic photosynthetic organisms. This pigment-protein complex contains the PSII core antenna proteins CP47 and CP43. These two proteins are the most dominant and long-lived proteins compared to other core RC proteins—D1 and D2 (Weisz et al., 2019). CP43 is increased in low and middle doses of UV-B but decrease in high dose of UV-B in both cultivars. We also identified oxygen-evolving enhancer protein 1 (OEE1) (spots 17 and 23) in both cultivars. The OEE1 is decreased in the cultivar IR6 but not detected in the REX cultivar. This protein's primary function is to stabilize the manganese cluster, the primary water-splitting site. Under drought and heat stress, OEE1 interacts with a dnaK family protein (HSP70, 4332413) in maize (Hu et al., 2015). The expression of OEE1 is also altered upon cytokine treatment (Talla et al., 2016). Recently, Zhao et al. (2021) reported that photoinactivation of the OEE regulates the photosynthetic strategy of the seagrass Zostera marina (Zhao et al., 2021). These data showed that UV-B plays a role in photosynthesis and ABA signaling partly by modulating ROS homeostasis.
In protein biosynthesis, the ubiquitin-proteasome pathway is essential for regulating cell physiology and a cellular signaling system because it removes most abnormal and short-lived peptides and proteins. Ubiquitin-conjugating enzymes (E2) play a significant role in transporting ubiquitin from the ubiquitin-activating enzyme (E1) to the ubiquitin-ligase enzyme (E3) and the substrate (Liu et al., 2020). This study identified ubiquitin-conjugating enzymes (E2), which are upregulated under UV-B stress. However, ubiquitin-conjugating enzyme (UBC) gene family expression was reduced in rice (E et al., 2015). Jeon et al. (2012) isolated a novel E2 ubiquitin-conjugating enzyme from wild rice plants and found it highly expressed in leaves treated with salicylic acid and UV-B. And they also showed that UBC1 confers disease resistance and UV-B tolerance in transgenic Arabidopsis plants (Jeon et al., 2012). In our study, E2 enzymes were only identified in REX cultivars—not in IR6. The expression of E2 is higher than wild type under moderate and high doses of UV-B. These results suggest that the REX cultivar has more UV-B tolerance than IR6.
Similarly, we identified histidinol dehydrogenase (spot 2). Histidinol dehydrogenase is the first enzyme characterized by plants. Histidinol dehydrogenase enzyme catalyzes the NAD-linked four-electron dehydrogenase reaction leading from histidinol to His (Nagai and Scheidegger, 1991). Histidinol dehydrogenase was upregulated in transplastomic Arabidopsis lines, suggesting that histidine should be accumulated in the transplastomic lines (Stavridou et al., 2019). Glutamate can also be metabolized to produce histidine via reactions catalyzed by histidinol phosphate phosphatase and histidinol dehydrogenase (Stepansky and Leustek, 2006). These results suggest that histidinol dehydrogenase plays a role in protein biosynthesis, which is also affected by UV-B stress. In this study, we also identified 3 different ribosomal proteins. Ribosomal proteins are well-known for their role in mediating protein synthesis and maintaining the stability of the ribosomal complex. All three identified proteins, 30S ribosomal protein S7 (spot 31), 40S ribosomal protein S16 (spot 32), and S19 (spot 33), are upregulated under UV-B stress in cultivar REX. These three proteins are novel and have not been identified under UV-B stress. Some other ribosomal proteins in maize and Arabidopsis, such as RPL10A and RPL10C, were significantly upregulated under UV-B stress (Casati and Walbot, 2003; Ferreyra et al., 2010). These results suggest that UV-B plays a role in protein biosynthesis. Additionally, this study provides a resource for subsequent exploitation of RPL genes to improve abiotic and biotic stress conditions in rice and some other crops in the future.
This study identified peptidyl-proyl cis-trans isomerase (PPIases) (spot 7). PPIases accelerate the folding of proteins. PPIases of rice OsIAA11 catalyzed by LATERAL ROOTLESS2 (LRT2), which is a cyclophilin-type peptidyl-prolyl cis/trans isomerase, directly regulates the stability of OsIAA11 (Jing et al., 2015). Knockdown of OsIAA11 expression partially rescues the LRT2 mutant phenotype in lateral root development. These results showed that cyclophilin-catalyzed peptidyl-prolyl isomerization promotes Aux/IAA degradation to regulate auxin signaling in rice (Jing et al., 2015; Acevedo et al., 2019). Plant cyclophilins have essential cellular functions in stress survival and the initiation of lateral root development (Kang et al., 2013). PPIase overexpressed under a low and moderate level of UV-B stress, whereas it was downregulated under a high level of UV-B doses in cultivar IR6 compared to control. So far, PPIase has not been identified in UV-B stress. Similarly, we also identified eukaryotic translation initiation factor 5A (eIF5A), which is upregulated in low and high doses of UV-B, whereas it was downregulated in the middle dose of UV-B. TaeIF5A1 from Tamarix androssowii, TaWRKY, and RAV protein share very similar expression patterns, and all stress-responsive genes are involved in the ABA signaling pathway (Wang et al., 2012). eIF5A-2 from Arabidopsis played a crucial role in plant growth and development by regulating cell division, cell growth, and cell death (Feng et al., 2007). This eIF5A has not been studied under UV-B stress. This study also identified glycine-rich protein 2, also called cold shock domain protein. In Arabidopsis, cold shock domain protein 2 negatively regulates seed germination by controlling ABA and GA levels (Sasaki et al., 2015). These results suggest that UV-B plays a role in signal transduction as well as the stress response.
In conclusion, we found that protein expression under different levels of UV-B stress seems to differ in both cultivars. The REX cultivar is more affected by UV-B stress than the IR6 cultivar. The differentially expressed proteins are related to plant growth and development, cell defense and redox homeostasis, metabolism, cell wall architecture, photosynthesis, signal transduction, stress response, and ABA signaling (Figure 4). Morphological and physiological studies are further required to understand the function of this protein in detail. This study helped develop the molecular marker in rice's molecular breeding to enhance the production during excessive UV-B exposure.
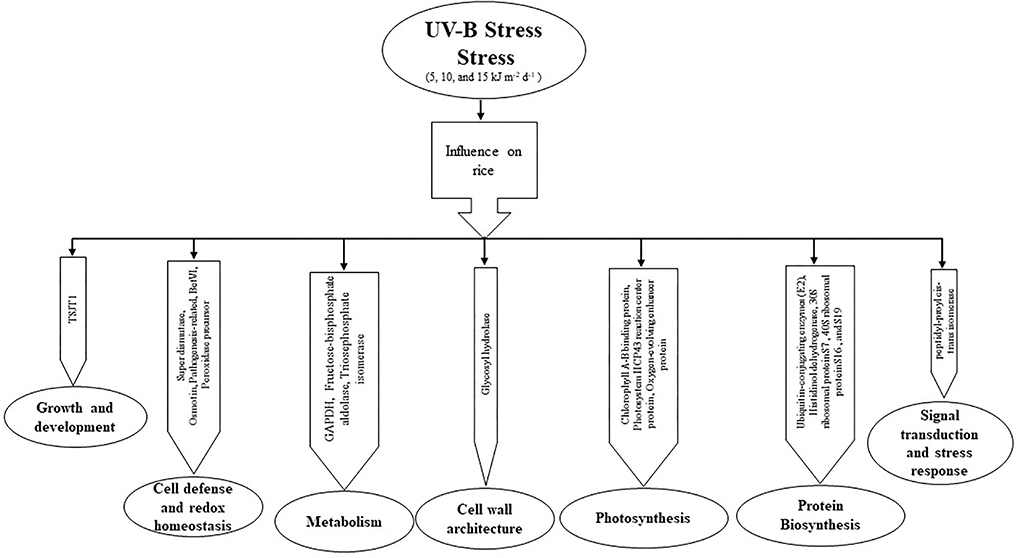
Figure 4. The pathway of UV-B induced protein expression in the proteomic response of rice leaves to cultivars IR6 and REX.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD032163.
SS: conceptualization, data curation, formal analysis, methodology, visualization, and writing–original draft and editing. SJ: data collection and review. JL: supervision, funding, review, and editing. KR: conceptualization, methodology, supervision, funding, review, and editing. All authors have read and approved the manuscript.
This research is partially funded by the National Institute of Food and Agriculture, NIFA 2019-34263-30552 and MIS 043050, and the MAFES SRI.
We thank David Brand for technical assistance and Dr. Ed Rodona, Rice Breeder, Delta Research, and Extension Center, Stoneville, MS, the USA, for providing the seed materials. SS was supported by a MAFES/FWRC Directors' Doctoral Fellowship awarded to JL and KR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.871331/full#supplementary-material
Supplementary Figure 1. Schematic illustration of the layout of experiments.
Supplementary Figure 2. Comparison of staining method in 2 D Gel analysis of control rice leaves samples (A) Colloidal coomassie staining and (B) Silver staining. The arrow indicates the protein spots detected only in silver staining, not coomassie staining.
Supplementary Table 1. GO enrichment analysis of identified proteins. The number of proteins enriched with each GO term in one of the three categories, including biological process, molecular function, and cellular component, is shown. The number in BG/RF refers to the annotated number in the background/reference. MSU7.0 gene ID (TIGR) was used as a reference.
Supplementary Table 2. Pathway analysis of identified proteins through mass spectrometry under different levels of UV-B stress.
Acevedo, L. A., Kwon, J., and Nicholson, L. K. (2019). Quantification of reaction cycle parameters for an essential molecular switch in an auxin-responsive transcription circuit in rice. Proc. Natl. Acad. Sci. 116, 2589 LP–94. doi: 10.1073/pnas.1817038116
Agrawal, S., and Rathore, D. (2007). Changes in oxidative stress defense system in wheat (Triticum aestivum L.) and mung bean (Vigna radiata L.) cultivars grown with and without mineral nutrients and irradiated by supplemental ultraviolet-B. Environ. Exp. Bot. 59, 21–33. doi: 10.1016/j.envexpbot.2005.09.009
Babele, P. K., Singh, G., Kumar, A., and Tyagi, M. B. (2015). Induction and differential expression of certain novel proteins in Anabaena L31 under UV-B radiation stress. Front. Microbiol. 6, 1–14. doi: 10.3389/fmicb.2015.00133
Bassman, J. H., Edwards, G. E., and Robberecht, R. (2002). Long-term exposure to enhanced UV-B radiation is not detrimental to growth and photosynthesis in douglas-fir. New Phytol. 154, 107–120. doi: 10.1046/j.1469-8137.2002.00354.x
Biever, J. J., Brinkman, D., and Gardner, G. (2014). UV-B inhibition of hypocotyl growth in etiolated Arabidopsis thaliana seedlings is a consequence of cell cycle arrest initiated by photodimer accumulation. J. Exp. Bot. 65, 2949–2961. doi: 10.1093/jxb/eru035
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brosche, M., and Strid, A. (2003). Molecular events following perception of ultraviolet-B radiation by plants. Physiol. Plant. 117, 1–10. doi: 10.1034/j.1399-3054.2003.1170101.x
Brown, B. A., and Jenkins, G. I. (2008). UV-B Signaling pathways with different fluence-rate response profiles are distinguished in mature arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH 1. Plant Physiol. 146, 576–588. doi: 10.1104/pp.107.108456
Casati, P., Campi, M., Morrow, D. J., Fernandes, J. F., and Walbot, V. (2011). Transcriptomic, proteomic and metabolomic analysis of UV-B signaling in maize. BMC Genomics 12, 321. doi: 10.1186/1471-2164-12-321
Casati, P., and Walbot, V. (2003). Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol. 132, 1739–1754. doi: 10.1104/pp.103.022871
Casati, P., Zhang, X., Burlingame, A. L., and Walbot, V. (2005). Analysis of leaf proteome after UV-B irradiation in maize lines differing in sensitivity. Mol. Cell. Proteomics 4, 1673–1685. doi: 10.1074/mcp.M500173-MCP200
Cui, K., He, C., Zhang, J., Duan, A., and Zeng, Y. (2012). Temporal and spatial profiling of internode elongation-associated protein expression in rapidly growing culms of bamboo. J. Proteome Res. 11, 2492–2507. doi: 10.1021/pr2011878
Dai, Q., Shaobing, P., Chavez, A. Q., and Vergara, B. S. (1994). Intraspecific responses of 188 rice cultivars to enhanced UVB radiation. Environ. Exp. Bot. 34, 433–442. doi: 10.1016/0098-8472(94)90026-4
Du, H., Liang, Y., Pei, K., and Ma, K. (2011). UV radiation-responsive proteins in rice leaves: a proteomic analysis. Plant Cell Physiol. 52, 306–316. doi: 10.1093/pcp/pcq186
E, Z., Zhang, Y., Li, T., Wang, L., and Zhao, H. (2015). Characterization of the ubiquitin-conjugating enzyme gene family in rice and evaluation of expression profiles under abiotic stresses and hormone treatments. PLoS ONE 10, e0122621. doi: 10.1371/journal.pone.0122621
Eng, J. K., McCormack, A. L., and Yates, J. R. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989. doi: 10.1016/1044-0305(94)80016-2
Fedina, I. S., Grigorova, I. D., and Georgieva, K. M. (2003). Response of barley seedlings to UV-B radiation as affected by NaCl. J. Plant Physiol. 160, 205–208. doi: 10.1078/0176-1617-00760
Feng, H., Chen, Q., Feng, J., Zhang, J., Yang, X., and Zuo, J. (2007). Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A-2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death. Plant Physiol. 144, 1531–1545. doi: 10.1104/pp.107.098079
Ferreyra, M. L. F., Pezza, A., Biarc, J., Burlingame, A. L., and Casati, P. (2010). Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 153, 1878–1894. doi: 10.1104/pp.110.157057
Fina, J., Casadevall, R., AbdElgawad, H., Prinsen, E., Markakis, M. N., Beemster, G. T., et al. (2017). UV-B inhibits leaf growth through changes in growth regulating factors and gibberellin levels. Plant physiol. 174, 1110–1126. doi: 10.1104/pp.17.00365
Giegé, P., Heazlewood, J. L., Roessner-Tunali, U., Millar, A. H., Fernie, A. R., Leaver, C. J., et al. (2003). Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15, 2140–2151. doi: 10.1105/tpc.012500
Goossens, A., Häkkinen, S. T., Laakso, I., Seppänen-Laakso, T., Biondi, S., De Sutter, V., et al. (2003). A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. 100, 8595–8600. doi: 10.1073/pnas.1032967100
He, J., Huang, L., Chow, W., Whitecross, M., and Anderson, J. (1993). Effects of supplementary ultraviolet-B radiation on rice and pea plants. Aust. J. Plant Physiol. 20, 129. doi: 10.1071/PP9930129
Hu, W., Chen, L., Qiu, X., Lu, H., Wei, J., Bai, Y., et al. (2016). Morphological, physiological and proteomic analyses provide insights into the improvement of castor bean productivity of a dwarf variety in comparing with a high-stalk variety. Front. Plant Sci. 7, 1473. doi: 10.3389/fpls.2016.01473
Hu, X., Wu, L., Zhao, F., Zhang, D., Li, N., Zhu, G., et al. (2015). Phosphoproteomic analysis of the response of maize leaves to drought, heat and their combination stress. Front. Plant Sci. 6, 298. doi: 10.3389/fpls.2015.00298
Iii, D. C. G., Britz, S. J., and Sullivan, J. H. (2013). Effect of ambient UV-B on stomatal density, conductance and Isotope discrimination in four field grown soybean [Glycine max (L.) Merr.] isolines. Am. J. Plant Sci. 4, 100–108. doi: 10.4236/ajps.2013.412A3012
Jeon, E. H., Pak, J. H., Kim, M. J., Kim, H. J., Shin, S. H., Lee, J. H., et al. (2012). Ectopic expression of ubiquitin-conjugating enzyme gene from wild rice, OgUBC1, confers resistance against UV-B radiation and Botrytis infection in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 427, 309–314. doi: 10.1016/j.bbrc.2012.09.048
Jing, H., Yang, X., Zhang, J., Liu, X., Zheng, H., Dong, G., et al. (2015). Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat. Commun. 6, 7395. doi: 10.1038/ncomms8395
Kang, B., Zhang, Z., Wang, L., Zheng, L., Mao, W., Li, M., et al. (2013). OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. Plant J. 74, 86–97. doi: 10.1111/tpj.12106
Kumar, J., Babele, P. K., Singh, D., and Kumar, A. (2016). UV-B radiation stress causes alterations in whole cell protein profile and expression of certain genes in the rice phyllospheric bacterium enterobacter cloacae. Front. Microbiol. 7, 1440. doi: 10.3389/fmicb.2016.01440
Kumari, R., Singh, S., and Agrawal, S. B. (2009). Effects of supplemental ultraviolet-b radiation on growth and physiology of Acorus calamus L. (sweet flag). Acta Biol. Cracoviensia Ser. Bot. 51, 19–27.
Li, J., and Assmann, S. M. (2000). Mass spectrometry. an essential tool in proteome analysis. Plant Physiol. 123, 807–809. doi: 10.1104/pp.123.3.807
Liang, C. J., Huang, X. H., and Qingll, Z. (2006). Effect of cerium on photosynthetic characteristics of soybean seedling exposed to supplementary ultraviolet-B radiation. J. Environ. Sci. 18, 1147–1141. doi: 10.1016/S1001-0742(06)60053-3
Liu, L., Mei, Q., Yu, Z., Sun, T., Zhang, Z., and Chen, M. (2013). An integrative bioinformatics framework for genome-scale multiple level network reconstruction of rice. J. Integr. Bioinform. 10, 223. doi: 10.1515/jib-2013-223
Liu, W., Tang, X., Qi, X., Fu, X., Ghimire, S., Ma, R., et al. (2020). The ubiquitin conjugating enzyme: an important ubiquitin transfer platform in ubiquitin-proteasome system. Int. J. Mol. Sci. 21, 2894. doi: 10.3390/ijms21082894
Liu, Y., Cao, Y., Zhang, Q., Li, X., and Wang, S. (2018). A cytosolic triosephosphate isomerase is a key component in Xa3/XA26-medIated Resistance. Plant Physiol. 178, 923–935. doi: 10.1104/pp.18.00348
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Nagai, A., and Scheidegger, A. (1991). Purification and characterization of histidinol dehydrogenase from cabbage. Arch. Biochem. Biophys. 284, 127–132. doi: 10.1016/0003-9861(91)90274-M
Nakahara, K., Yamamoto, H., Miyake, C., and Yokota, A. (2003). Purification and characterization of class-I and class-II fructose-1,6-bisphosphate aldolases from the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 44, 326–333. doi: 10.1093/pcp/pcg044
Ouyang, S., Zhu, W., Hamilton, J., Lin, H., Campbell, M., Childs, K., et al. (2007). The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 35, D883–D887. doi: 10.1093/nar/gkl976
Peng, J., and Gygi, S. P. (2001). Proteomics: the move to mixtures. J. Mass Spectrom,. 36, 1083–1091. doi: 10.1002/jms.229
Peng, Q., and Zhou, Q. (2009). Influence of lanthanum on chloroplast ultrastructure of soybean leaves under ultraviolet-B stress. J. Rare Earths 27, 304–307. doi: 10.1016/S1002-0721(08)60238-0
Perez-Riverol, Y., Bai, J., Bandla, C., Hewapathirana, S., García-Seisdedos, D., Kamatchinathan, S., et al. (2022). The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. doi: 10.1093/nar/gkab1038
Qu, Y., Feng, H., Wang, Y., Zhang, M., Cheng, J., Wang, X., et al. (2006). Nitric oxide functions as a signal in ultraviolet-B induced inhibition of pea stems elongation. Plant Sci. 170, 994–1000. doi: 10.1016/j.plantsci.2006.01.003
Rastogi, R. P., Richa, K., A., Tyagi, M. B., and Sinha, R.P. (2010). Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J. Nucleic Acids 2010, 1–32. doi: 10.4061/2010/592980
Reddy, K. R., Seghal, A., Jumaa, S., Bheemanahalli, R., Kakar, N., Redoña, E. D., et al. (2021). Morpho-physiological characterization of diverse rice genotypes for seedling stage high-and low-temperature tolerance. Agronomy, 11, 112. doi: 10.3390/agronomy11010112
Reddy, K. R., Singh, S. K., Koti, S., Kakani, V. G., Zhao, D., Gao, W., et al. (2013). Quantifying the effects of corn growth and physiological responses to ultraviolet-b radiation for modeling. Publ. Agron. J. 105, 1367–1377. doi: 10.2134/agronj2013.0113
Robson, T. M., and Aphalo, P. J. (2012). Species-specific effect of UV-B radiation on the temporal pattern of leaf growth. Physiol. Plant 144, 146–160. doi: 10.1111/j.1399-3054.2011.01546.x
Roy, S. C., and Roy, T. (2013). Peptide mass fingerprinting of rice (Oryza sativa L.) leaves during UV-B induced stress at seedling stage: a proteom analysis. Indian J. Biotechnol. 12, 504–508.
Sakai, H., Lee, S. S., Tanaka, T., Numa, H., Kim, J., Kawahara, Y., et al. (2013). Rice annotation project database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 54, e6. doi: 10.1093/pcp/pcs183
Santos, A. L., Gomes, N. C. M., Henriques, I., Almeida, A., Correia, A., and Cunha, Â. (2012). Contribution of reactive oxygen species to UV-B-induced damage in bacteria. J. Photochem. Photobiol. B Biol. 117, 40–46. doi: 10.1016/j.jphotobiol.2012.08.016
Sasaki, K., Kim, M. H., Kanno, Y., Seo, M., Kamiya, Y., and Imai, R. (2015). Arabidopsis cold shock domain protein 2 influences ABA accumulation in seed and negatively regulates germination. Biochem. Biophys. Res. Commun. 456, 380–384. doi: 10.1016/j.bbrc.2014.11.092
Sharma, R., Cao, P., Jung, K., Sharma, M. K., and Ronald, P. C. (2013). Construction of a rice glycoside hydrolase phylogenomic database and identification of targets for biofuel research. Front. Plant Sci. 4, 1–15. doi: 10.3389/fpls.2013.00330
Shevchenko, A., Wilm, M., and Mann, M. (1997). Peptide sequencing by mass spectrometry for homology searches and cloning of genes. J. Protein Chem. 16, 481–490. doi: 10.1023/A:1026361427575
Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. doi: 10.1021/ac950914h
Singh, B., Chastain, D. R., Jumaa, S., Wijewardana, C., Redoña, E. D., Gao, W., et al. (2019). Projected day/night temperatures specifically limits rubisco activity and electron transport in diverse rice cultivars. Environ. Exp. Bot. 159, 191–199. doi: 10.1016/j.envexpbot.2018.12.018
Stavridou, E., Michailidis, M., Gedeon, S., Ioakeim, A., Kostas, S., Chronopoulou, E., et al. (2019). Tolerance of transplastomic tobacco plants overexpressing a theta class glutathione transferase to abiotic and oxidative stresses. Front. Plant Sci. 9, 1–17. doi: 10.3389/fpls.2018.01861
Stepansky, A., and Leustek, T. (2006). Histidine biosynthesis in plants. Amino Acids 30, 127–142. doi: 10.1007/s00726-005-0247-0
Surplus, S. L., Jordan, B. R., Murphy, A. M., Carr, J. P., and Thomas, B. (1998). Ultraviolet-B-induced responses in Arabidopsis thaliana : role of salicylic acid and reactive oxygen species in the regulation of transcripts encoding photosynthetic and acidic pathogenesis-related proteins. Plant Cell Environ. 21, 685–694. doi: 10.1046/j.1365-3040.1998.00325.x
Sztatelman, O., Grzyb, J., Gabryś, H., and Banaś, A. K. (2015). The effect of UV-B on Arabidopsis leaves depends on light conditions after treatment. BMC Plant Biol. 15, 281. doi: 10.1186/s12870-015-0667-2
Talla, S. K., Panigrahy, M., Kappara, S., Nirosha, P., Neelamraju, S., and Ramanan, R. (2016). Cytokinin delays dark-induced senescence in rice by maintaining the chlorophyll cycle and photosynthetic complexes. J. Exp. Bot. 67, 1839–1851. doi: 10.1093/jxb/erv575
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). AgriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Voss, P., Hajimiragha, H., Engels, M., Ruhwiedel, C., Calles, C., Schroeder, P., et al. (2007). Irradiation of GAPDH: a model for environmentally induced protein damage. Biol. Chem. 388, 583–592. doi: 10.1515/BC.2007.068
Wang, L., Xu, C., Wang, C., and Wang, Y. (2012). Characterization of a eukaryotic translation initiation factor 5A homolog from Tamarix androssowii involved in plant abiotic stress tolerance. BMC Plant Biol. 12, 118. doi: 10.1186/1471-2229-12-118
Wang, W., Vignani, R., Scali, M., and Cresti, M. (2006). A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27, 2782–2786. doi: 10.1002/elps.200500722
Weisz, D. A., Johnson, V. M., Niedzwiedzki, D. M., Shinn, M. K., Liu, H., Klitzke, C. F., et al. (2019). A novel chlorophyll protein complex in the repair cycle of photosystem II. Proc. Natl. Acad. Sci. 116, 21907 LP−21913. doi: 10.1073/pnas.1909644116
Wu, X. C., Fang, C. X., Chen, J. Y., Wang, Q. S., Chen, T., Lin, W. X., et al. (2011). A Proteomic analysis of leaf responses to enhanced ultraviolet-b radiation in two rice (Oryza sativa L.) cultivars differing in UV sensitivity. J. Plant Biol. 54, 251–261. doi: 10.1007/s12374-011-9162-y
Xu, Y. H., Liu, R., Yan, L., Liu, Z. Q., Jiang, S. C., Shen, Y. Y., et al. (2011). Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 63, 1095–1106. doi: 10.1093/jxb/err315
Ye, J., Zhang, Y., Cui, H., Liu, J., Wu, Y., Cheng, Y., et al. (2018). WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 46, 71–75. doi: 10.1093/nar/gky400
Yuan, L., Yan-qun, Z., Haiyan, C., Jianjun, C., Jilong, Y., and Zhide, H. (2000). Intraspecific responses in crop growth and yield of 20 wheat cultivars to enhanced ultraviolet-B radiation under field conditions. Field Crop Res. 67, 25–33. doi: 10.1016/S0378-4290(00)00080-0
Zhao, P., Liu, P., Shao, J., Li, C., Wang, B., Guo, X., et al. (2015). Analysis of different strategies adapted by two cassava cultivars in response to drought stress: ensuring survival or continuing growth. J. Exp. Bot. 66, 1477–1488. doi: 10.1093/jxb/eru507
Zhao, W., Yang, X. Q., Zhang, Q. S., Tan, Y., Liu, Z., Ma, M. Y., et al. (2021). Photoinactivation of the oxygen-evolving complex regulates the photosynthetic strategy of the seagrass Zostera marina. J. Photochem. Photobiol. B Biol. 222, 112259. doi: 10.1016/j.jphotobiol.2021.112259
Keywords: Oryza sativa, ultraviolet-B stress, leaf-proteome, two-dimensional gel electrophoresis, mass spectrometry
Citation: Sah SK, Jumaa S, Li J and Reddy KR (2022) Proteomic analysis response of rice (Oryza sativa) leaves to ultraviolet-B radiation stress. Front. Plant Sci. 13:871331. doi: 10.3389/fpls.2022.871331
Received: 08 February 2022; Accepted: 05 August 2022;
Published: 15 September 2022.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Catherine Rayon, University of Picardie Jules Verne, FranceCopyright © 2022 Sah, Jumaa, Li and Reddy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saroj Kumar Sah, c2Fyb2oxMDIxQGdtYWlsLmNvbQ==; K. Raja Reddy, a3JyZWRkeUBwc3MubXNzdGF0ZS5lZHU=
†Present address: Saroj Kumar Sah, Biology Department, Brookhaven National Laboratory, Upton, NY, United States
Salah Jumaa, Department of Field Crop Science, Tikrit University, Tikrit, Iraq
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.