- 1Department of Biotechnology and Bioinformatics, School of Life Sciences, University of Hyderabad, Hyderabad, India
- 2Department of Systems and Computational Biology, School of Life Sciences, University of Hyderabad, Hyderabad, India
- 3Department of Plant Sciences, School of Life Sciences, University of Hyderabad, Hyderabad, India
- 4Department of Botany, Deshbandhu College, University of Delhi, New Delhi, India
In higher plants, alternative oxidase (AOX) participates in a cyanide resistant and non-proton motive electron transport pathway of mitochondria, diverging from the ubiquinone pool. The physiological significance of AOX in biotic/abiotic stress tolerance is well-documented. However, its structural and biophysical properties are poorly understood as its crystal structure is not yet revealed in plants. Also, most of the AOX purification processes resulted in a low yield/inactive/unstable form of native AOX protein. The present study aims to characterize the purified rAtAOX1A protein and its interaction with inhibitors, such as salicylhydroxamic acid (SHAM) and n-propyl gallate (n-PG), as well as pyruvate (activator), using biophysical/in silico studies. The rAtAOX1A expressed in E. coli BL21(DE3) cells was functionally characterized by monitoring the respiratory and growth sensitivity of E. coli/pAtAOX1A and E. coli/pET28a to classical mitochondrial electron transport chain (mETC) inhibitors. The rAtAOX1A, which is purified through affinity chromatography and confirmed by western blotting and MALDI-TOF-TOF studies, showed an oxygen uptake activity of 3.86 μmol min−1 mg−1 protein, which is acceptable in non-thermogenic plants. Circular dichroism (CD) studies of purified rAtAOX1A revealed that >50% of the protein content was α-helical and retained its helical absorbance signal (ellipticity) at a wide range of temperature and pH conditions. Further, interaction with SHAM, n-PG, or pyruvate caused significant changes in its secondary structural elements while retaining its ellipticity. Surface plasmon resonance (SPR) studies revealed that both SHAM and n-PG bind reversibly to rAtAOX1A, while docking studies revealed that they bind to the same hydrophobic groove (Met191, Val192, Met195, Leu196, Phe251, and Phe255), to which Duroquinone (DQ) bind in the AtAOX1A. In contrast, pyruvate binds to a pocket consisting of Cys II (Arg174, Tyr175, Gly176, Cys177, Val232, Ala233, Asn294, and Leu313). Further, the mutational docking studies suggest that (i) the Met195 and Phe255 of AtAOX1A are the potential candidates to bind the inhibitor. Hence, this binding pocket could be a ‘potential gateway' for the oxidation-reduction process in AtAOX1A, and (ii) Arg174, Gly176, and Cys177 play an important role in binding to the organic acids like pyruvate.
Introduction
The plant mitochondrial electron transport chain (mETC) possesses two terminal oxidases for the reduction of molecular oxygen into water (Rich and Moore, 1976). The cytochrome oxidase (COX) pathway is found in all eukaryotes. Electron transport through the COX pathway generates a proton gradient, which drives the ATP synthase for ATP production. The alternative oxidase (AOX) pathway is present in all plants, fungi, some protists, and a few animal species (Vanlerberghe and McIntosh, 1997; McDonald et al., 2009). AOX pathway, which branches at the ubiquinol pool, was identified during the thermogenic respiration studies on arum lilies (Meeuse, 1975; Rich and Moore, 1976). It is involved in a non-phosphorylating electron transport mechanism and dissipates excess energy as heat (Wagner and Moore, 1997; Siedow and Umbach, 2000; Millar et al., 2011; Moore et al., 2013). In higher plants, salicylhydroxamic acid (SHAM) and n-propyl gallate (n-PG) are frequently used to inhibit the activity of AOX, and, thereby, AOX pathway under both in vitro and in vivo conditions to reveal its physiological function(s) during normal growth, as well as biotic/abiotic stress conditions (Diethelm et al., 1990; Padmasree and Raghavendra, 1999a,b; Yoshida et al., 2006; Giraud et al., 2008; Dinakar et al., 2010; Florez-Sarasa et al., 2011; Zhang et al., 2011).
Mitochondrial AOX is encoded by a nuclear gene, and in many plants, it is known to exist as a multi-gene family and is regulated by retrograde signaling mechanisms (Whelan et al., 1996; Saisho et al., 1997; Liu and Butow, 2006; Rhoads and Subbaiah, 2007; Giraud et al., 2009; Ng et al., 2013). For instance, Arabidopsis thaliana has five AOX genes: AOX1A-D and AOX2. However, the isoforms corresponding to these genes cannot compensate for each other's function even under stress conditions (Considine et al., 2002; Clifton et al., 2006; Costa et al., 2017). A remarkable increase in the expression of AOX1A in Arabidopsis thaliana at a wide range of stress conditions and during the impairment of respiratory metabolism indicates its primary role in stress response as compared to other AOX genes (Clifton et al., 2005, 2006; Ho et al., 2008; Vishwakarma et al., 2015). In non-thermogenic plants, the AOX pathway is known to play a vital role in regulating the cellular redox balance when the cytochrome pathway is over-reduced or chemically inhibited and during abiotic stresses, such as high light, drought, temperature, UV-B stress, and high levels of greenhouse gasses (Berthold et al., 2000; Moore et al., 2002, 2013; Giraud et al., 2008; Vanlerberghe, 2013; Vishwakarma et al., 2014, 2015; Dahal and Vanlerberghe, 2017; Florez-Sarasa et al., 2020; Garmash et al., 2020). The AOX pathway is known to benefit plants by lowering ROS production, either by maintaining electron transport chain components upstream of the ubiquinone pool in a more oxidized state or by preventing the over-reduction of the COX pathway during stress conditions (Maxwell et al., 1999; Vishwakarma et al., 2015). The plants deficient in AOX1A showed acute sensitivity in response to light, drought, and antimycin A treatment than wild-type plants (Giraud et al., 2008). In contrast, overexpression of AtAOX1A induced tolerance to salt (Smith et al., 2009) and hypoxia (Vishwakarma et al., 2018) in A. thaliana plants. Similarly, plants lacking AOX were found to be more vulnerable to bacterial pathogens, sucking insects, and chewing herbivores as compared to wild-type plants (Zhang et al., 2012). Furthermore, infection with pathogens caused an increase in AOX transcript, as well as protein levels, thereby, AOX respiration in plants, substantiating the importance of AOX in biotic stress tolerance (Liao et al., 2012, 2021; Zhu et al., 2015).
The AOX is located on the inner membrane of mitochondria facing toward the mitochondrial matrix. Post-translational modifications in AOX, such as the disulfide bond formation, dimerization through non-covalent linkage, and its interaction with α-keto acids, such as pyruvate, are known to regulate its activity. The reduced form of AOX is found to be more active than the oxidized form as it interacts with α-keto acids through a thiohemiacetal linkage (Millar et al., 1993; Umbach and Siedow, 1993; Rhoads et al., 1998; Vanlerberghe et al., 1998; Umbach et al., 2002, 2006; Moore et al., 2013; Selinski et al., 2018; Xu et al., 2021). Besides, plant AOX isoforms expressed in E. coli membrane are known to be activated differently by 2-oxo acids/TCA cycle intermediates such as pyruvate, glyoxylate, citrate, oxaloacetate, malate, succinate, and α-ketoglutarate (Crichton et al., 2005, 2010; Carré et al., 2011; Ito et al., 2011; Moore et al., 2013; Selinski et al., 2016, 2017; Xu et al., 2021). Also, the addition of pyruvate is found to be essential to maintain the AOX activity during the harvesting of cells and purification of recombinant AOX from the E. coli membrane (Kido et al., 2010; Elliott et al., 2014).
Structural studies on the plant and trypanosomal AOX revealed that it possessed a non-heme diiron carboxylate active site (Berthold et al., 2000, 2002; Affourtit et al., 2002; Moore and Albury, 2008; Moore et al., 2008, 2013; Albury et al., 2009; Maréchal et al., 2009). The studies on the crystal structure of trypanosomal AOX (TAO) revealed that it exists as a homodimer and each monomer consists of a four-helix bundle (α2, α3, α5, and α6) ligated to a diiron core by highly conserved four glutamate and two histidine residues. Overall, there are six long and four short α-helices in each monomer, along with two hydrophobic cavities in TAO. The α1 and α4 helices of TAO were embedded in a large hydrophobic region that might be involved in membrane-binding. Furthermore, the binding of ubiquinol to Tyr220 of TAO leads to the catalysis of O2 reduction (Moore et al., 2013; Shiba et al., 2013; May et al., 2017). Besides, the studies of Elliott et al. (2014) revealed the secondary structure of a thermogenic rSgAOX, while such secondary structure is not yet unveiled for AOX from non-thermogenic plants. Also, despite the identification of several inhibitors and activators for AOX, the information on how these molecules interact with it is not yet known (Xu et al., 2021).
Several attempts have been made by various scientists to purify AOX from mitochondria of different plant sources (Huq and Palmer, 1978; Rich, 1978; Kay and Palmer, 1985; Bonner et al., 1986; Elthon and McIntosh, 1986, 1987; Berthold and Siedow, 1993; Zhang et al., 1996; Affourtit and Moore, 2004). However, the low yield and thermal instability of AOX impeded the studies related to its structural and biophysical properties. In this context, the rDNA technology allowed the expression of plant AOX in E. coli. The AOX from A. thaliana and Trypanosoma were expressed in a ΔhemA E. coli strain, which is found to be deficient in the cytochrome pathway (Kumar and Söll, 1992; Nihei et al., 2003). Besides, the supplementation of iron in the form of Fe2+/Fe3+ was in ambiguity during heterologous expression of AOX to acquire it in pure and active form (Minagawa et al., 1990; Ajayi et al., 2002; Affourtit and Moore, 2004).
Thus, this study aims to acquire an adequate amount of AtAOX1A pure protein by expressing it in a routinely used laboratory strain [BL21(DE3)] of E. coli and understand its secondary structure and stability toward temperature and pH. Also, the present study intends to examine the interaction of AtAOX1A with well-known inhibitors (SHAM and n-PG) and activators (pyruvate) under in vitro conditions by employing CD spectroscopy, SPR, and molecular docking studies. Besides, respiratory and growth inhibitory studies were performed in E. coli transformed with AtAOX1A to ensure its functional expression.
Materials and Methods
Reagents and Chemicals
Sodium pyruvate, tris base, imidazole, duroquinone, diethyl ether, sodium dithionite, n-dodecyl β-D maltoside (DDM), octyl-β-D glucopyranoside (OG), isopropyl β-D-1-thiogalactopyranoside (IPTG), phenylmethylsulfonyl fluoride (PMSF), n-PG, SHAM, coomassie brilliant blue R-250, and TRIzol reagent were purchased from Sigma Aldrich, St Louis, Mo, USA. The protease inhibitor ‘cocktail' was purchased from Roche. CM5 sensor chips, PBS, surfactant P20, glycine-HCl pH 2.5, sodium acetate pH 5, and amine coupling kit were purchased from GE Healthcare Bio-Sciences Corp., USA. Verso cDNA synthesis kit, 50 bp DNA ladder, bicinchoninic acid (BCA) protein estimation kit, protein molecular mass standard, and 3 kDa cut-off snakeskin dialysis membrane were purchased from Thermo Fisher Scientific, USA. Talon cobalt metal affinity resin was purchased from Clontech, Takara Bio, Japan. His-Tag mouse monoclonal antibody (HRP conjugated) was purchased from Cell Signaling Technologies. ECL Western blotting reagents were purchased from GE Healthcare Bio-Sciences Corp., USA. Polymerase chain reaction (PCR) components and restriction endonucleases were purchased from New England Biolabs. Luria-Bertani medium, agar-agar, and kanamycin were purchased from Hi-Media, Mumbai, India. Ferrous sulfate and glycerol were purchased from SRL. All the compounds used were of the biochemical grade.
Strains and Plasmid Construction
The Escherichia coli (E. coli) strain DH5α was used to maintain the clone, while the strain BL21(DE3) (InvitrogenTM, Waltham, MA, USA) was used for the expression of recombinant protein AOX, which has a His6-tag on it. Further details on cloning of AtAOX1A and plasmid construction are described in Vishwakarma et al. (2016).
Method of Measuring Cell Growth
The BL21(DE3) cells transformed with pET28a (E. coli/pET28a) and recombinant pET28a with AtAOX1A (E. coli/pAtAOX1A) were inoculated into 3 ml of LB medium containing 50 μg/ml of kanamycin and incubated overnight at 37°C (pre-culture). The pre-culture was inoculated into 20 ml of LB medium with 50 μg/ml of kanamycin and allowed to grow. As the OD600 of the culture reached 0.15, IPTG (0.1 mM) was added into the culture medium to induce rAtAOX1A. To ascertain the functions of rAtAOX1A in E. coli, the cultures were treated with each of the following mitochondrial inhibitors at a wider range of concentrations: 0.05–1 mM KCN (inhibitor of cytochrome oxidase pathway), 0.025–0.5 mM n-PG and 0.5–2 mM SHAM (inhibitors of alternative oxidase pathway). Subsequently, both control (E. coli/pET28a) and transformed (E. coli/pAtAOX1A) cells were incubated for 5 h at 37°C, and OD600 was monitored at regular intervals of 30 min (Kumar and Söll, 1992; Berthold, 1998; Fukai et al., 1999).
Assay of Cell Respiration
After induction of rAtAOX1A with IPTG as described above, E. coli were extracted and suspended in a suitable volume of LB medium to achieve a cell density of 20 at OD600. The cellular respiration of both E. coli/pET28a and E. coli/pAtAOX1A were measured in terms of rates of oxygen consumption with or without the addition of KCN, n-PG, and SHAM. Respiration rates were measured with a total of 4.8 × 108 cells/ml for each reaction using the Clark-type oxygen electrode (Kirimura et al., 1999; Ajayi et al., 2002; Oxygraph plus, Hansatech instruments, UK).
Preparation of E. coli/pAtAOX1A Membrane Sample
The E. coli carrying recombinant AtAOX1A was pre-cultured overnight at 37°C in 10 ml of LB medium containing 50 μg/ml of kanamycin. About 5 ml of pre-culture was inoculated into 500 ml of LB medium containing 50 μg/ml kanamycin and 0.1 mM ferrous sulfate and grown at 37°C with 200 rpm. The culture was allowed to grow until OD600 reached 0.4 and IPTG (0.1 mM) was added to induce the expression of rAtAOX1A. Subsequently, the culture was allowed to grow at 28°C for 4 h, and cells were harvested by centrifugation at 5,000 rpm for 7 min at 4°C. The pellet obtained was resuspended in 50 mM Tris-HCl containing 10 mM pyruvate at pH 7.5.
E. coli cells were lysed by sonication with an amplitude of 35 for 20 min by switching on/off of the probe at regular intervals of 30 s each, in the presence of a protease inhibitor cocktail and PMSF (1 mM). After lysis, cell debris was removed in a single step by centrifugation at 12,000 g. The supernatant was centrifuged at 200,000 g for 1 h at 4°C to separate cytoplasm from membrane fraction. The membrane pellet was resuspended in a minimal volume of 50 mM Tris-HCl (pH 7.5) containing 10 mM of pyruvate.
Solubilization of rAtAOX1A From E. coli Membranes
The solubilization (10 ml) buffer, which contains 1% (w/v) DDM in 50 mM of Tris-HCl, along with 10 mM of pyruvate and 20% (v/v) glycerol at pH 7.5, was used to solubilize the membrane fraction obtained from E. coli/pAtAOX1A as described above. The buffer was added dropwise with gentle mixing and immediately centrifuged for 1 h at 200,000 g. All the steps were performed at 4°C. The supernatant enriched in rAtAOX1A is labeled as DDM extract.
Purification of rAtAOX1A
Purification of rAtAOX1A was done according to Kido et al. (2010) and Elliott et al. (2014), with minor modifications. The cobalt resin (selective for His-tag) was equilibrated with an equilibration buffer at a 1:3 ratio for 1 h. The equilibration buffer contained 0.5% (w/v) DDM and 0.5% (w/v) OG in 50 mM of Tris-HCl (pH 7.5), along with 10 mM of pyruvate, 20% (v/v) glycerol, and 100 mM of MgSO4. After equilibration, 10 ml of DDM extract was added to the resin and allowed to mix gently overnight at 4°C. Further, the resin which is bound with rAtAOX1A was washed with 10 ml of wash buffer, which contained 0.5% (w/v) DDM and 0.5% (w/v) OG in 50 mM Tris-HCl (pH 7.5), along with 10 mM of pyruvate, 20% (v/v) glycerol, 100 mM MgSO4, and 50 mM of imidazole. Subsequently, the resin was transferred onto a column and rAtAOX1A was eluted as 1-ml fractions using elution buffer, which contained 0.5% (w/v) DDM and 0.5% (w/v) OG, in 50 mM of Tris-HCl along with 10 mM of pyruvate, 250 mM of imidazole, 20% (v/v) glycerol, and 100 mM of MgSO4 at pH 7.5. Protein purification was visualized on 12.5% SDS-PAGE under reducing conditions, as described in Laemmli (1970). The gel was stained using Coomassie Brilliant Blue and the protein was estimated using the BCA kit.
Western Blot Analysis
The proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were electrophoretically transferred onto the polyvinylidene difluoride (PVDF) membrane using a Western blotting unit (Smart Scientific Instruments, Chennai), as described by Towbin et al. (1979). After transfer, the PVDF membrane was incubated at 4°C with a mouse monoclonal anti-His antibody (1:5,000 dilution) for 5–6 h, which is conjugated with horseradish peroxidase (Cell signaling technology). The blot was washed with tris buffered saline containing 0.2% of tween 20 (TBST) for 30–40 min. The blot was developed using Enhanced chemiluminescence (ECL) western blotting reagents and rAtAOX1A protein was detected with the help of the ChemiDoc imaging system (BioRad).
Oxygen Uptake Activity
The activity of rAtAOX1A was measured polarographically by monitoring the oxygen uptake rates using an S1 Clark-type oxygen electrode (Oxygraph plus, Hansatech Instruments, UK). The activity assay was performed by adding 0.7 μg of purified protein in 400 μl of air saturated reaction medium (50 mM of Tris-HCl, pH 7.5), and the reaction was started by the addition of 600 μM DQH2 as a substrate. The stocks of DQH2 were prepared on the day of use by suspending the solid DQH2 in acidified ethanol (Rich, 1978). The stock concentration of DQH2 was determined based on its molar extinction coefficient (2.15 mM−1 cm−1 at 283 nm in water) using a UV-Visible spectrophotometer (Shimadzu UV-1700). The acidified ethanol blank was subtracted from the assay while measuring the activity.
MALDI TOF/TOF Analysis
The purified recombinant protein band was excised with a sharp and sterile scalpel from the gel after performing SDS-PAGE and kept in a 1.5-ml tube. Later, the gel plug was reduced with DTT (10 mM) and alkylated with iodoacetamide (55 mM) before subjecting to digestion with trypsin (12.5 μg/μl). The tryptic digested protein sample was analyzed by using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). The method uses Autoflex III smart beam instrument (Bruker Daltonics, Bremen, Germany), equipped with a nitrogen laser (355 nm), and operated in a reflection mode for peptide mass and sequencing in the presence of α-cyano-4-hydroxycinnamic acid as a matrix, as described in Swathi et al. (2014). The spectra from MALDI-MS and MALDI-MS-MS ionization were searched with the MASCOT search engine. The biotools software (Bruker Daltonics, version 3.1) was used to analyze the lift spectra, while the Sequence similarity analysis was performed by the National Center for Biotechnology Information (NCBI) blast tool.
Circular Dichroism (CD) and Thermal Stability Studies
The purified rAtAOX1A present in the elution buffer described above was transferred into a 10-mM phosphate buffer (pH 7.5) through the process of step-down dialysis for performing CD. Three different dialysis buffers [Buffer 1: 20 mM Tris-HCl (pH 7.5) containing 10 mM of pyruvate, 0.03% of DDM, and 10% of Glycerol; Buffer 2: 10 mM of phosphate buffer (pH 7.5) containing 0.03% of DDM and 1% of glycerol and Buffer 3: 10 mM of phosphate buffer (pH 7.5)] were used at a ratio of 10:500 (sample volume to buffer volume). The sample was dialyzed in each buffer for a period of 4 h at 4°C under continuous stirring. Finally, the rAtAOX1A present in 10 mM of phosphate buffer (pH 7.5) was concentrated by lyophilization. The CD spectrophotometer (Jasco J-1500, Japan) coupled with a thermostat was used to analyze rAtAOX1A (0.8 mg/ml). The parameters used for the CD scan were as follows: wavelength, 190 to 260 nm; speed, 50 nm/min; step resolution, 1 nm; bandwidth, 1 nm; response, 3 s; temperature, 25°C, and sensitivity, 50 mdegrees. Similarly, the secondary structural stability of rAtAOX1A at different pH, from 2 to 12, was determined after incubating rAtAOX1A in different buffers [5 mM of Glycine-HCl (pH 2), 10 mM of sodium phosphate (pH 7.5)—control sample and 5 mM Glycine-NaOH (pH 10-12)] at 37°C for 1 h. Conversely, for the temperature melting studies, the parameters used were as follows: wavelength, 190 to 260 nm; response, 1 s; temperature, 4 to 90°C; speed, 50 nm/min; and sensitivity, 50 mdegrees (Kelly et al., 2005; Elliott et al., 2014). Furthermore, different concentrations (0.05, 0.1, and 0.5 mM) of SHAM and n-PG were added separately to 0.4 mg/ml of purified rAtAOX1A to analyze the secondary structure conformational changes induced in rAtAOX1A during its interaction with the inhibitors. Similarly, different concentrations (0.05, 0.1, 0.5, and 1 mM) of pyruvate was added to 0.4 mg/ml of purified rAtAOX1A to study the secondary structure conformational changes induced in rAtAOX1A during its interaction with the activator. The changes in secondary structural elements of rAtAOX1A were analyzed by submitting the raw CD data to the online server named Dichroweb (Whitmore and Wallace, 2008) using the CDSSTR algorithm with a reference set 4 (Sreerama and Woody, 2000).
Surface Plasmon Resonance (SPR) Studies
The purified rAtAOX1A protein sample was dialyzed into a 10-mM phosphate buffer (pH 7.5) by using above-mentioned step-down dialysis procedure and concentrated by lyophilization.
Immobilization of the rAtAOX1A on CM5 Sensor Chip
The following steps are performed to immobilize the rAtAOX1A on the CM5 sensor chip: (i) a mixture of 40 mM of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and 10 mM of N-hydroxysuccinimide (NHS) are passed through the reference (blank) flow cell (Fc-3), as well as the sample flow cell (Fc-4) of CM5 sensor chip at a flow rate of 30 μl/min for 300 s, to generate reactive succinimide esters; (ii) the rAtAOX1A (100 μg/ml) prepared in sodium acetate buffer (pH 5) was injected into the activated flow cell (Fc-4), where the NHS esters spontaneously react with uncharged amino groups of rAtAOX1A (ligand) and covalently link it to the dextran matrix; (iii) the residual active NHS esters in Fc-4 are blocked by passing 1 M of ethanolamine-HCl (pH 8.5) for 300 s, at a flow rate of 30 μl/min; (iv) all the active NHS esters of the reference (blank) flow cell (Fc-3), which is devoid of rAtAOX1A are blocked with ethanolamine-HCl (pH 8.5).
Sample Injection
The analytes SHAM and n-PG (1 to 5 mM) dissolved in double distilled water are injected independently into the flow cells (Fc-3 & Fc-4) in a running buffer composed of PBS (2 mM of KH2PO4, 10 mM of Na2HPO4, 2.7 mM of KCl, and 137 mM of NaCl at pH 7.4) containing 0.005% P20 surfactant, at a flow rate of 30 μl/min at 25°C. In the association phase (120 s), the analytes were allowed to bind to the rAtAOX1A, while in the dissociation phase (120 s), they are separated from each other. In the regeneration phase (30 s), the analytes are disposed of from the flow cells using 10 mM of Glycine-HCl (pH 2.5) and 0.5 M of NaCl, as described in the Biacore T-200 manual (GE Healthcare Life Sciences). The experiment was performed twice. In each experiment, three cycles of association, dissociation, and regeneration were carried out against each concentration of inhibitors (SHAM and n-PG) tested in the present study.
Kinetic Analysis
The acquired data were analyzed using BIA evaluation software (version 2.0, GE Healthcare Life Sciences) with the Langmuir fit model of 1:1 binding. To determine the kinetic constants (ka, kd, and KD), the sensorgrams were fitted to a 1:1 kinetic model using the pooled data analysis option. The ka represents the association constant, kd represents the dissociation constant, and KD represents the equilibrium dissociation constant. The affinity of inhibitor(s) with rAtAOX1A was derived by measuring the kinetic parameters at five different concentrations (1, 2, 3, 4, and 5 mM) of n-PG and SHAM.
Molecular Docking
The homology model of the AtAOX1A monomer (PMDB Accession number: PM0080189) was chosen as the target protein for the docking study (Pennisi et al., 2016). The 3-dimensional models of the ligands ubiquinol-1 (Q1H2), ubiquinone-1 (UQ1), duroquinol (DQH2), duroquinone (DQ), SHAM, n-PG, and pyruvate were generated using PubChem (Kim et al., 2021) and CHARMM-GUI (Jo et al., 2008) programs. The binding affinities of different ligands, such as Q1H2, UQ1, DQH2, DQ, SHAM, n-PG, and pyruvate with AtAOX1A are studied using the SwissDock program (Grosdidier et al., 2011a,b). To identify the potent residues that may have a direct impact on the binding of the ligands (DQ, SHAM, n-PG, and pyruvate), point mutations with alanine were performed.
Statistical Analysis
The data shown are the Mean ± SE of three replicates. The statistical differences were evaluated by one-way ANOVA with the Tukey test integrated into Sigma plot, version 12.0, Systat Software Inc., San Jose, CA, USA, at a significance level of P ≤ 0.05.
Results
Cloning and Expression of Arabidopsis thaliana AOX1A (AtAOX1A)
The complementary DNA (cDNA) of AtAOX1A coding for mature protein was amplified (Figure 1A) using AOX1A- specific primers (Vishwakarma et al., 2016) and cloned into a pET28a vector (Figure 1B). The pAtAOX1A construct was transformed into BL21(DE3) cells. Transformed positive colonies (E. coli/pAtAOX1A) were selected by colony PCR (Supplementary Figure 1A). Recombinant plasmids from these colonies were isolated and confirmed by restriction digestion with EcoRI and XhoI (Supplementary Figure 1B) and DNA sequencing using T7 primers (Supplementary Data S1). After confirming the AtAOX1A sequence, these colonies were used for recombinant protein synthesis. The expression of rAtAOX1A protein (~37 kDa) in E. coli/pAtAOX1A after induction with IPTG was visualized on 12.5% (w/v) SDS-PAGE under reducing conditions (Figure 1C).
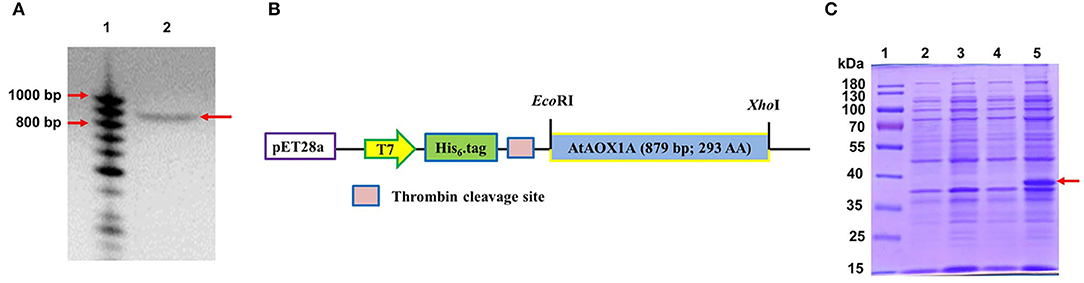
Figure 1. Cloning and expression of AtAOX1A in E. coli. (A) Polymerase chain reaction (PCR) amplification of 879 bp AtAOX1A using gene-specific primers. Lane 1, 50–1000 bp DNA ladder; lane 2, PCR amplified AtAOX1A (indicated with arrow). (B) Schematic diagram of the expression vector construction. The gene sequences were cloned into restriction endonuclease EcoRI and XhoI recognition sites of the pET28a vector to produce N-terminal His6-tag recombinant AtAOX1A protein. (C) Total protein of both E. coli/pET28a and E. coli/pAtAOX1A cells were separated on 12.5% (w/v) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for recombinant protein expression analysis. Each well was loaded with 40 μg of protein. Lane 1, 10–180 kDa protein marker; lane 2, E. coli/pET28a; lane 3, E. coli/pET28a induced with 0.1 mM IPTG; lane 4, E. coli/pAtAOX1A; lane 5, E. coli/pAtAOX1A induced with 0.1 mM IPTG. The induced rAtAOX1A is indicated with an arrow.
Functional Characterization of rAtAOX1A in E. coli
To identify that expressed rAtAOX1A is functionally active, both E. coli/pET28a and E. coli/pAtAOX1A were evaluated for their respiratory and growth rates in the presence of KCN, n-PG, and SHAM at a wide range of concentrations (Figure 2). In the absence of any inhibitor, both E. coli/pET28a (34.52 ± 0.5 μmoles O2 min−1) and E. coli/pAtAOX1A (32 ± 0.74 μmoles O2 min−1) showed comparable respiratory O2 uptake rates per 4.8 x 108 cells ml−1. However, supplementation of KCN at an increasing concentration from 0.05 to 1 mM caused a significant reduction in the O2 uptake (up to 90%) of E. coli/pET28a due to inhibition of COX-catalyzed respiration, whereas it only caused 37% reduction in respiratory rate of E. coli/pAtAOX1A, suggesting that remaining O2 uptake in transformed cells is contributed by AOX-catalyzed respiration (Figure 2A).
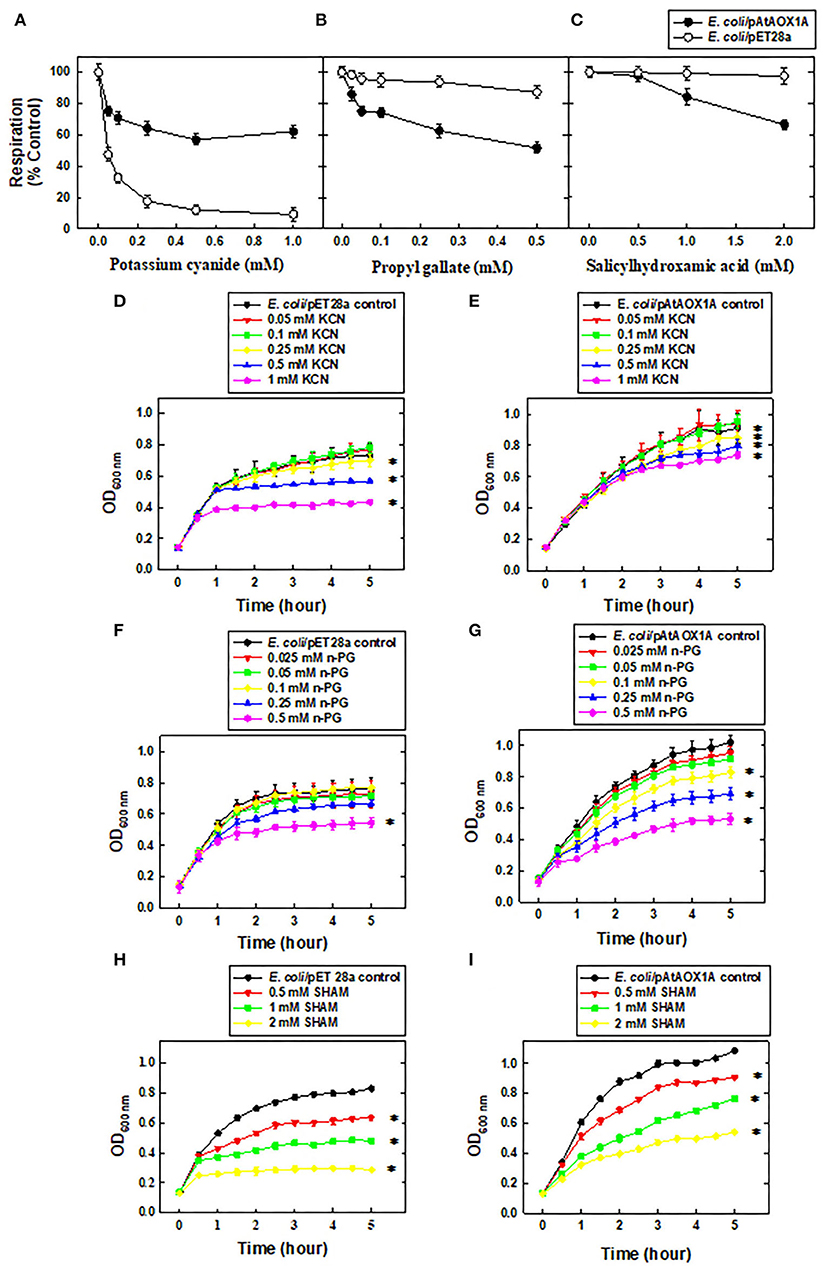
Figure 2. Functional characterization of rAtAOX1A in E. coli. Oxygen uptake rates (%) of E. coli/pET28a and E. coli/pAtAOX1A in the absence or presence of respiratory inhibitors: (A) KCN (0.05, 0.1, 0.25, 0.5, and 1 mM), (B) n-PG (0.025, 0.05, 0.1, 0.25, and 0.5 mM), and (C) SHAM (0.5, 1, and 2 mM). The growth pattern of E. coli cells was monitored for 5 h in the absence or presence of respiratory inhibitors: (D) Growth pattern of E. coli/pET28a and (E) E. coli/pAtAOX1A in the absence or presence of KCN; (F) Growth pattern of E. coli/pET28a and (G) E. coli/pAtAOX1A in the absence or presence of n-PG; (H) Growth pattern of E. coli/pET28a and (I) E. coli/pAtAOX1A in the absence or presence of SHAM. Each value represents the mean ± SD of three experiments. The statistical significance difference (P <0.05) was calculated for the endpoint of the growth curve and indicated with asterisks.
In contrast to KCN, supplementation of n-PG, a commonly used inhibitor of AOX, has shown only a marginal (<13%) effect on respiratory O2 uptake rates of E. coli/pET28a even at a concentration as high as 0.5 mM due to the lack of AOX-catalyzed respiration. Conversely, at this concentration, n-PG caused a 50% reduction in respiratory rates of E. coli/pAtAOX1A (Figure 2B). In the case of SHAM supplementation, another inhibitor of the AOX respiratory pathway decreased the respiratory rates of E. coli/pAtAOX1A remarkably up to 33%, as its concentration is increased from 0.5 to 2 mm. However, treatment of E. coli/pET28a with 2 mM SHAM caused only a 2.5% reduction in its respiratory rate (Figure 2C).
To examine the effect of respiratory inhibitors on the growth of the E. coli/pET28a and E. coli/pAtAOX1A, the bacterial growth was compared at the end of 5 h. Interestingly, E. coli/pET28a achieved a stationary phase after 4 h (Figures 2D,F,H), while E. coli/pAtAOX1A cells remained in a log phase even after 5 h of growth (Figures 2E,G,I), suggesting the role of AOX-catalyzed respiration in enhancing the metabolic activities of E. coli. The supplementation of KCN at an increasing concentration (0.05 to 1 mM) has suppressed the growth of E. coli/pET28a significantly by 51%. However, KCN has suppressed only 23% of growth in E. coli/pAtAOX1A (Figures 2D,E). When E. coli/pET28a was supplemented with n-PG, it caused only a trivial (14%) effect on its growth even at 0.25-mM concentration, whereas it suppressed the growth of E. coli/pAtAOX1A up to 38% at the same concentration. However, n-PG at higher concentration (0.5 mM) showed remarkable growth inhibitory effect on both E. coli/pET28a (34%) and E. coli/pAtAOX1A (54%), respectively (Figures 2F,G). On contrary, SHAM has suppressed the growth of E. coli/pET28a and E. coli/pAtAOX1A by 88 and 57%, respectively, as its concentration was raised from 0.5 to 2 mM (Figures 2H,I). However, the greater effect of SHAM on the growth of E. coli/pET28a, as compared to E. coli/pAtAOX1A, might be due to the non-specific inhibitory effect on other oxidases of E. coli, besides its specific effect on AOX. These results, taken together, suggest that rAtAOX1A expressed in E. coli/pAtAOX1A is functionally active.
Purification of rAtAOX1A and Its Analysis by MALDI TOF/TOF
As the expression cassette contains an N-terminal His6-tag, cobalt column affinity chromatography was used to purify rAtAOX1A from the soluble fraction of E. coli/pAtAOX1A membrane. To get active protein: (1) rAtAOX1A was induced by IPTG in the presence of FeSO4 (0.1 mM) and (2) after induction, the harvested cells were suspended in 50 mM Tris-HCl containing 10 mM of pyruvate at pH 7.5. The purified rAtAOX1A with a molecular mass of ~37 kDa was observed in reducing SDS-PAGE, which includes AtAOX1A mature protein (33.44 kDa) and pET28a (+) vector sequence (3.83 kDa) (Figure 3A). Furthermore, western blot analysis with anti-His antibody showed a single band correlating with the molecular mass observed in SDS-PAGE (Figure 3B). The rAtAOX1A has shown a specific activity of 3.86 μmol O2 min−1 mg−1 protein and a 15% recovery rate, where 600 μM of duroquinol was used as a substrate (Table 1).
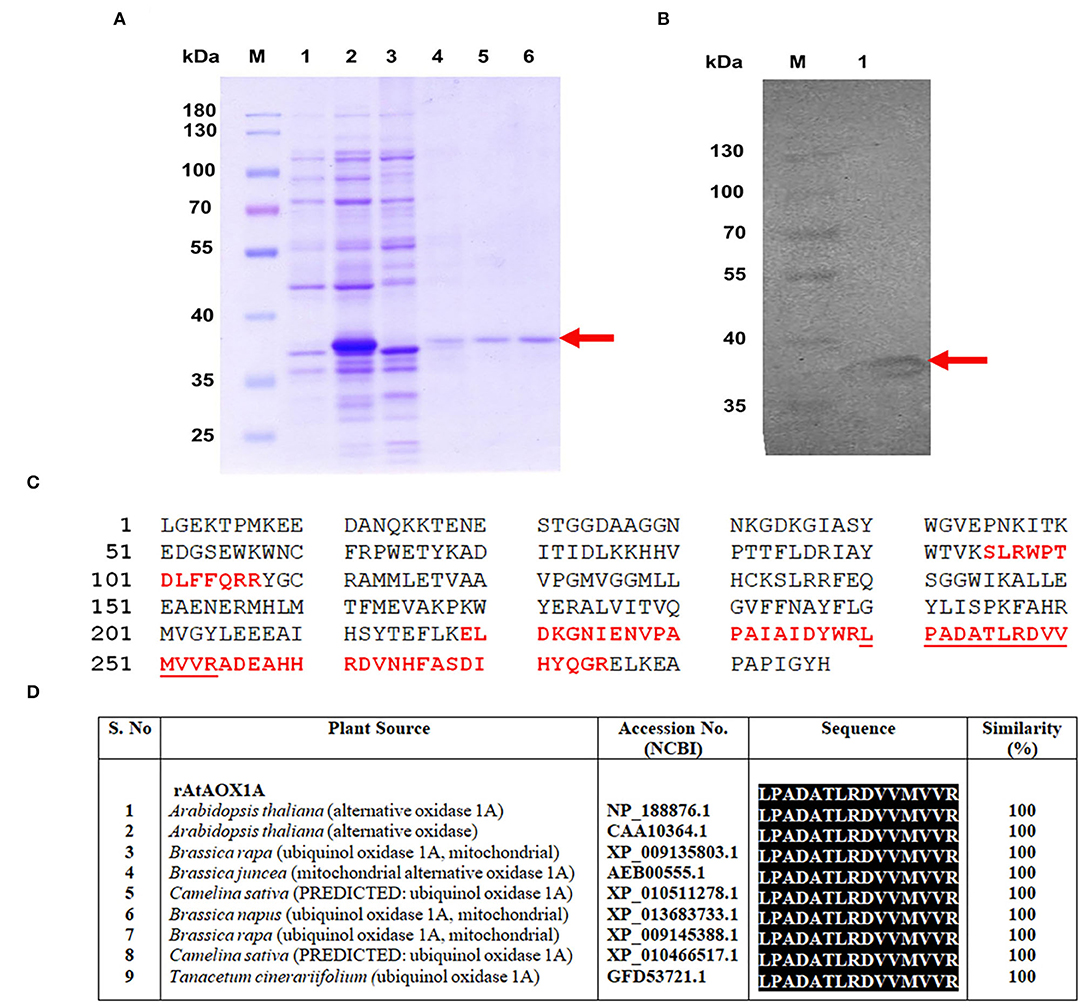
Figure 3. Purification and validation of rAtAOX1A protein using MALDI-MS-MS. (A) SDS-PAGE visualizing purification profile of rAtAOX1A from E. coli. Lane M, 10-180 kDa protein marker; lane 1, protein from E. coli/pAtAOX1A; lane 2, protein from E. coli/pAtAOX1A induced with IPTG; lane 3, flow-through; lane 4, washing fraction; lanes 5 and 6, elution fractions, arrow indicates purified rAtAOX1A with ~37 kDa molecular mass. (B) Western blot showing purified rAtAOX1A: lane M, 10-180 kDa protein marker; lane 1, 40 μg of purified rAtAOX1A. (C) A. thaliana partial AOX (gi/1872517) sequence from NCBI database. The sequence of MS peaks (1209.854, 1365.972, 1566.099, 1656.258, 2385.628, and 2476.697 Da) of rAtAOX1A are indicated in bold red font, and sequence coverage from these six major peaks is about 24%. The obtained sequence from peptide peak m/z 1656.258 (‘LPADATLRDVVMVVR') has shown 100% identity with AtAOX1A in BLAST search (D). The gel picture shown is the selective representation of three biological replicates.
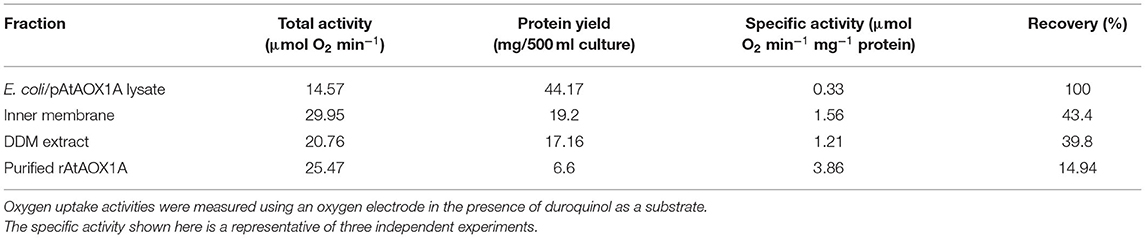
Table 1. The membrane fractions of E. coli were prepared from 500 ml of E. coli/pAtAOX1A culture and used for rAtAOX1A purification.
Moreover, the purified recombinant protein band (~37 kDa) from SDS-PAGE (Figure 3A) was digested with trypsin and subjected to MALDI-TOF/TOF analysis. The MS-MS ion search in the Mascot search database resulted in matching with partial A. thaliana AOX (ID: gi/1872517) with a significant score of 308 (Supplementary Data S2). The six major peptide peaks (m/z 1209.854, m/z 1365.972, m/z 1566.099, m/z 1656.258, m/z 2385.628, and m/z 2476.697) in peptide mass fingerprint (PMF) spectrum covered 24% of partial A. thaliana AOX (ID: gi/1872517) protein sequence from NCBI database (Figure 3C). The Biotools software revealed the following sequence ‘LPADATLRDVVMVVR' for lift spectra corresponding to the peak 1656.258 Da (Supplementary Figure 2). Blast search of the sequence (LPADATLRDVVMVVR) showed 100% identity with the A. thaliana AOX1A in the NCBI database (Figure 3D). These results confirm that the purified protein is rAtAOX1A.
CD Spectroscopic Analysis of rAtAOX1A and Its Interaction With SHAM, n-PG, and Pyruvate
The CD spectroscopy is frequently used to study the secondary structure and its conformational state in proteins. It detects the difference in the absorption of left and right-handed circularly polarized light in optically active molecules such as proteins, and differences in the absorbance are monitored in terms of ellipticity. In the CD spectroscopy graph, a combination of a particular wavelength of positive and negative peaks defines the secondary structure, such as: (1) A positive peak at 192 nm and negative peaks at 208 and 222 nm represent an α-helical structure; (2) A positive peak at 195 nm, and a negative peak at 216 nm indicates a β-sheet structure; (3) A positive peak at 205 nm represents a β-turn, and (4) A negative peak at 200 nm and a positive peak at 212 nm represent a random coil (Kelly and Price, 2000; Kelly et al., 2005).
The secondary structural analysis of rAtAOX1A in Far-UV (190–260 nm) CD spectra indicated that it possesses an α-helical structure (Figure 4A). The presence of two negative peaks at 208 nm and 222 nm is a characteristic feature of the typical α-helical structure. The high tension (HT) voltage trace was shown as an inset in Figure 4A. Further, any change in the pH (2–12) caused only a marginal variation in the ellipticity of rAtAOX1A, as compared to the ellipticity observed at optimal pH 7.5 (Figure 4B). Similarly, the thermal treatment of rAtAOX1A showed only a marginal decrease in its ellipticity with a gradual increase in the temperature from 4° to 90°C (Figure 4C). However, this effect was reversed by gradually cooling down the temperature from 90° to 4°C (Figure 4D), suggesting that rAtAOX1A is structurally stable to heat treatment. The axes for spectra in Figures 4B–D are enlarged between 205 to 230 nm to bring more clarity between different treatments (Supplementary Figures 3A–C). Also, the protein has retained its helical absorbance signal (222 nm) up to ~67% despite an increase in temperature from 25° to 90°C (Figure 4E). These results suggest that rAtAOX1A is highly stable at a wide range of temperature and pH conditions.
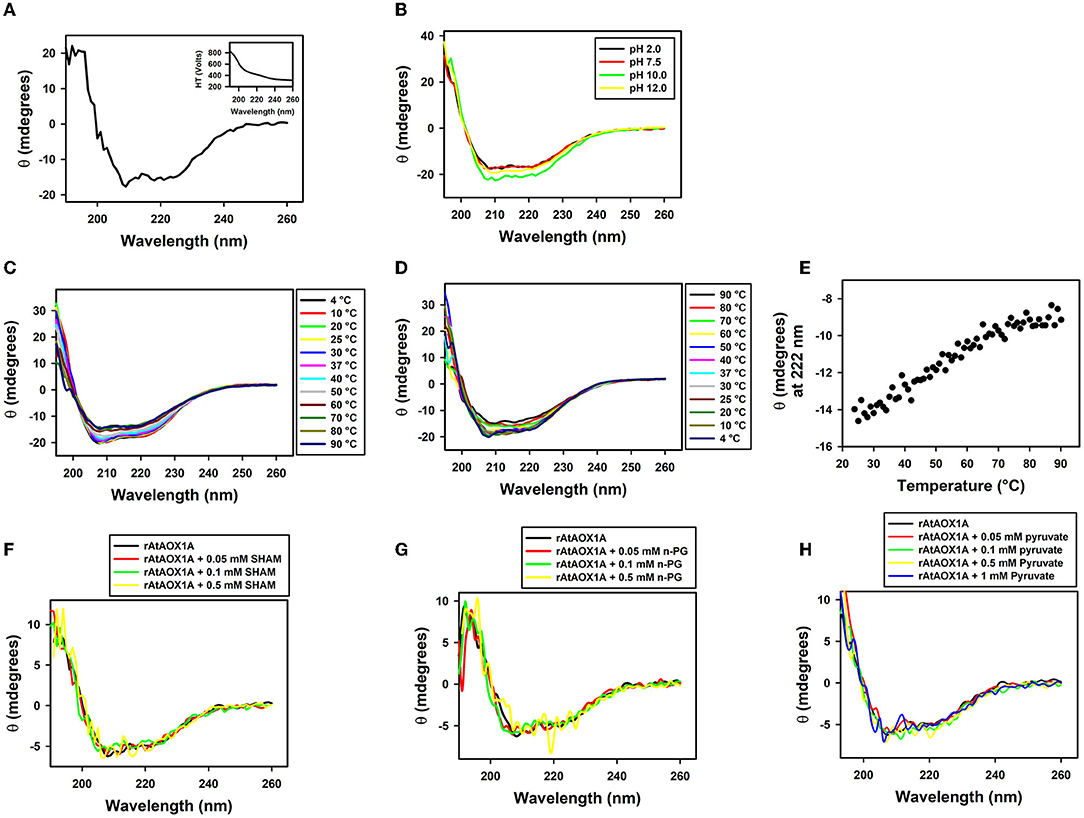
Figure 4. Secondary structural stability analysis of rAtAOX1A. (A) CD spectra of purified rAtAOX1A at far UV (190-260 nm) in 10-mM phosphate buffer. Ellipticity in θ (mdeg) of rAtAOX1A (0.8 mg/ml) was plotted against the wavelength. Inset: High Tension profile of voltage trace. The secondary structural stability was determined at different (B) pH from 2 to 12; (C) Temperature from 4 to 90°C; (D) Temperature from 90 to 4°C. (E) Ellipticity of rAtAOX1A at 222 nm between 25°C and 90°C. (F) CD spectra of rAtAOX1A at far UV (190–260 nm), with 0.05, 0.1, and 0.5 mM SHAM and, without inhibitor; (G) CD spectra of rAtAOX1A at far UV (190-260 nm), with 0.05, 0.1, and 0.5 mM n-PG, and without inhibitor; (H) CD spectra of rAtAOX1A at far UV (190-260 nm), with 0.05, 0.1, 0.5, and 1 mM of pyruvate, and without activator. Concentration of purified rAtAOX1A used to obtain the CD spectrum (SHAM, n-PG, and pyruvate) was 0.4 mg/ml. The final spectrum is an average of three scans as described in the Materials and Methods section.
Furthermore, the alterations in secondary structural elements of purified rAtAOX1A upon addition of SHAM, n-PG, and pyruvate were determined separately by CD spectroscopy. The addition of different concentrations of SHAM, n-PG, or pyruvate to rAtAOX1A has not shown any deviation in the helical absorbance signal in the CD spectrum observed between 190–260 nm. However, a marginal deviation in the negative peaks was observed (Figures 4F–H). The axes for spectra in Figures 4F–H are enlarged between 205 and 230 nm to bring more clarity between different treatments (Supplementary Figures 3D–F). Further, the changes in the secondary structure composition of the rAtAOX1A in the presence of SHAM, n-PG, and pyruvate were summarized in Table 2. The addition of both inhibitors (SHAM and n-PG) and activator (pyruvate) to the rAtAOX1A caused an increase in β-sheets (~7%) with a concomitant decrease in α-helical (~10%) content, while the changes in β-turns and random coils (together) are negligible (~3%).
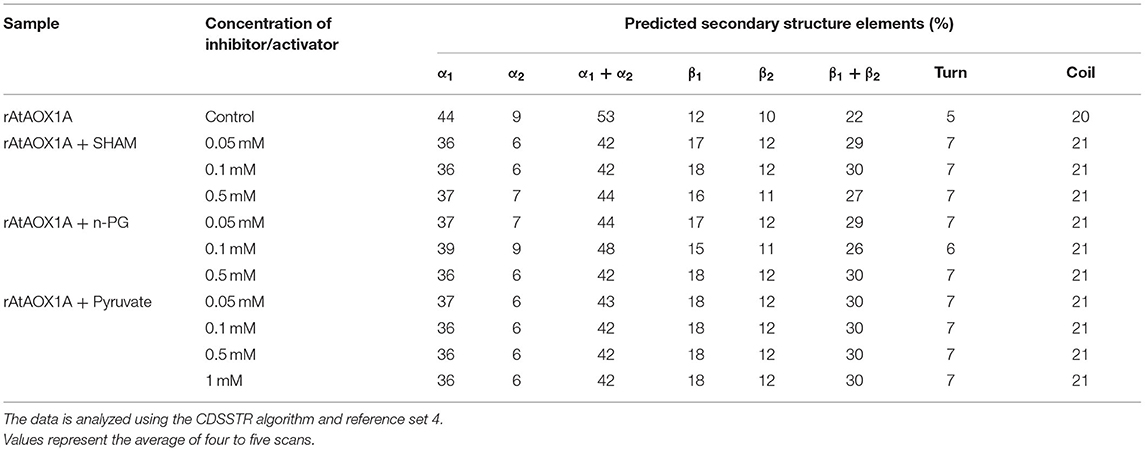
Table 2. Modulation in rAtAOX1A secondary structural elements during interaction with SHAM, n-PG, and pyruvate.
SPR Kinetic Analysis of rAtAOX1A With SHAM and n-PG
The SPR technique has been extensively used to study the molecular interaction of proteins with small-molecule(s) and in drug discovery due to its improved selectivity, stability, and sensitivity (Homola, 2003; Olaru et al., 2015). The binding events of the immobilized ligand and analytes are ascertained by monitoring the changes in the SPR signal (Response unit). As the analyte is passed on an immobilized ligand through a microfluidic channel, the interaction between ligand and analyte leads to the formation of a complex structure, which causes a change in the mass on the sensor chip surface and, thereby, a change in the SPR signal of the sensorgram. This change/difference in the signal is used to derive kinetic constants for both complex formation (association) and its dissociation in a particular molecular interaction between a ligand and an analyte (Nguyen et al., 2015).
The mechanism of rAtAOX1A interaction with SHAM and n-PG is characterized by immobilization of rAtAOX1A on a CM5 sensor chip and following its binding affinities (Figure 5). Initially, the immobilization of rAtAOX1A to the Fc-4 in the sensor chip is visualized by an increase in its RU by ~4,117 when compared with its reference (blank) cell (Fc-3) RU (Figures 5A,B). Later, the analytes (SHAM and n-PG) are allowed to pass independently through both Fc-3 (reference/blank flow cell) and Fc-4 (rAtAOX1A immobilized flow cell). The binding of SHAM and n-PG with immobilized rAtAOX1A at different concentrations (1 to 5 mM) was visualized in the sensorgrams as the rate of increase in RU of Fc-4 [note: RU of blank/reference flow cell (Fc-3) corresponding analytes is subtracted automatically] (Figures 5C,D). From these sensorgrams, association, dissociation, and steady-state interaction kinetics of SHAM and n-PG with rAtAOX1A were calculated. SHAM has shown an equilibrium dissociation constant KD = 3.08 x 10−9 M with the following association (ka = 1.11 x 105 M−1 s−1) and dissociation (kd = 3.40 x 10−4 s−1) rate constants. Similarly, the kinetic parameters measured for n-PG has shown equilibrium dissociation constant KD = 4.91 x 10−10 M with the following association (ka = 1.80 x 107 M−1 s−1) and dissociation (kd = 8.85 x 10−3 s−1) rate constants. These results suggest that the n-PG (~0.49 nM) has more affinity to rAtAOX1A than SHAM (~3 nM). However, both the inhibitors bind reversibly to the rAtAOX1A, which is evident by the changes in the RU observed during their association and dissociation in corresponding sensorgrams (Figures 5C,D).
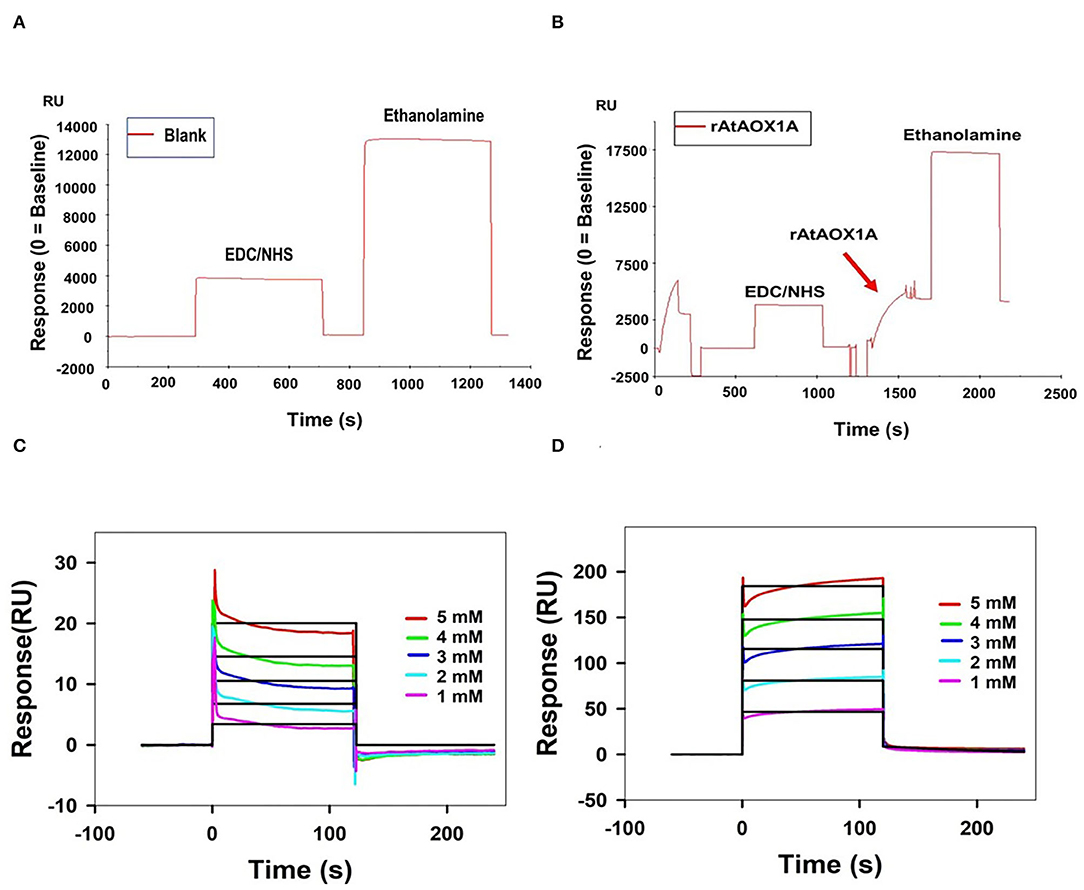
Figure 5. Purified rAtAOX1A is immobilized onto Series S Sensor Chip CM5. (A) Blank immobilization on flow cell Fc-3; (B) 100 μg/ml of purified rAtAOX1A (ligand) immobilization on flow cell Fc-4. Arrow indicates rAtAOX1A injection, which resulted in ~4,117 RU. SPR kinetics of SHAM and n-PG with rAtAOX1A: (C,D) Binding curves for SHAM and n-PG with rAtAOX1A at 25°C. The colored lines represent the concentrations of SHAM/n-PG (1, 2, 3, 4, and 5 mM), the black lines correspond to the fit lines to the data, and each fit line is a result of a global fit. The final sensorgram is representative of three cycles.
Molecular Docking of AtAOX1A With Its Ligands
The interaction of the following ligands with AtAOX1A was examined by in silico molecular docking and mutational studies: (i) Q1H2 (reduced form of ubiquinone), which plays a significant role in the transfer of electrons from complex I and complex II of mETC to AOX and its oxidized form (UQ1), which is released from the AOX after the donation of electrons by Q1H2 to its catalytic center under in vivo conditions; (ii) DQH2 (reduced form of duroquinone), an analog of ubiquinol and electron donor to AOX under in vitro conditions, and its oxidized form (DQ) that is released from AOX after electron donation by DQH2 to its catalytic center; (iii) n-PG and SHAM, which inhibit the transfer of electrons through the AOX pathway, and (iv) pyruvate, which activate the AOX and, thereby, electron transfer through the AOX pathway (Figures 6, 7).
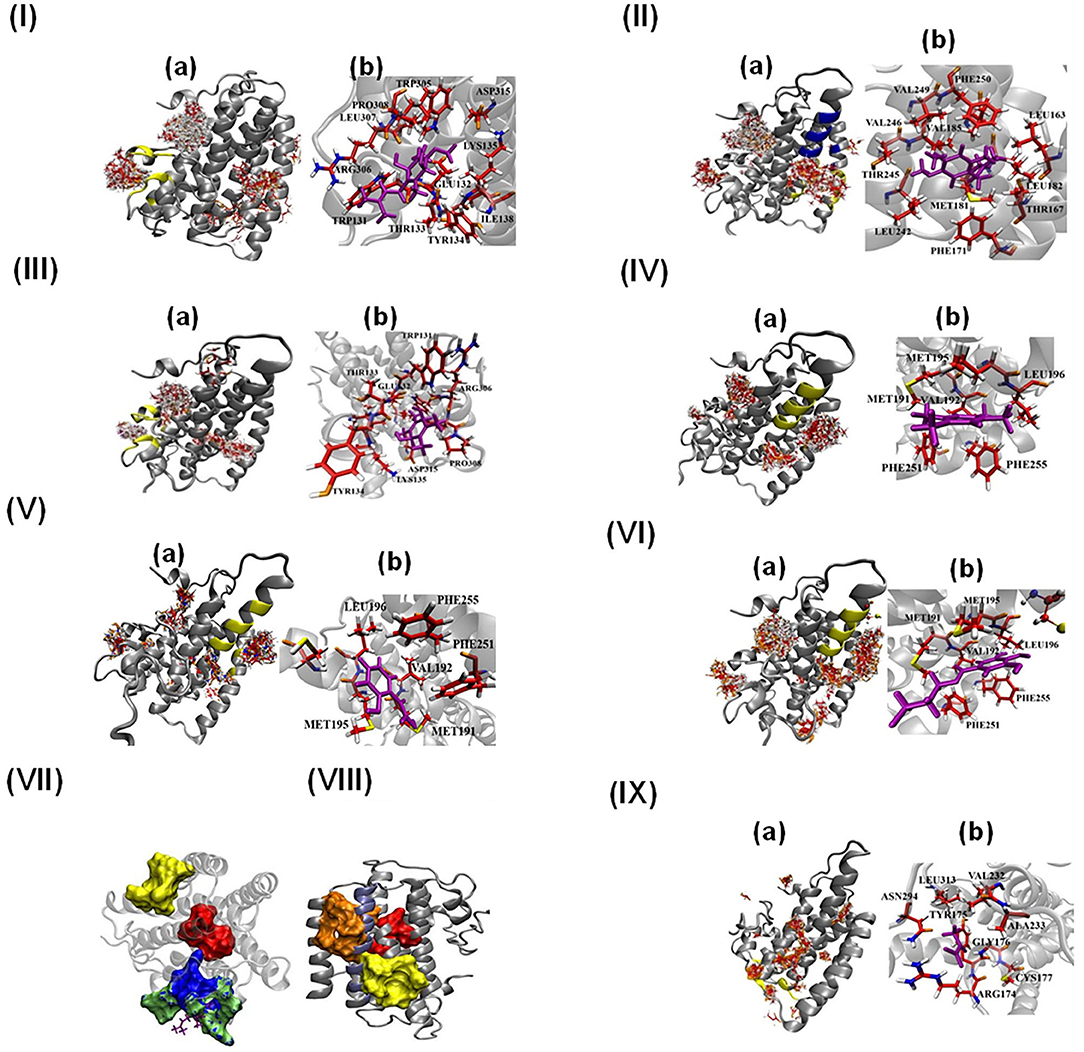
Figure 6. Molecular docking of AtAOX1A with different ligands (I-VI and IX). (a) All possible binding pockets of a specific ligand on AtAOX1A. The yellow region on AtAOX1A secondary structure indicates the best-fit binding pocket of a particular ligand. The Blue region in panel (IIa) represents the inhibitor binding pocket to compare with that of UQ, and (b) residues of AtAOX1A that are involved in forming the binding pocket of the best model (Cluster 0 Element 0) according to SwissDock results. The ligands are (I) Q1H2, (II) UQ1, (III) DQH2, (IV) DQ, (V) SHAM, (VI) n-PG, (VII) Binding pockets of inhibitor (yellow), DQH2 (green surface), Q1H2 (blue surface), and diiron binding cavity (red surface). DQH2 is shown with purple sticks. (VIII) Binding pockets of inhibitor (yellow surface), ubiquinone-1 (orange surface), and diiron cavity (red surface), (IX) Pyruvate. Q1H2, UQ1, DQH2, DQ, SHAM, n-PG, and pyruvate are shown in purple. Color scheme: carbon (red), oxygen (orange), sulfur (yellow), nitrogen (blue), and hydrogen (white).
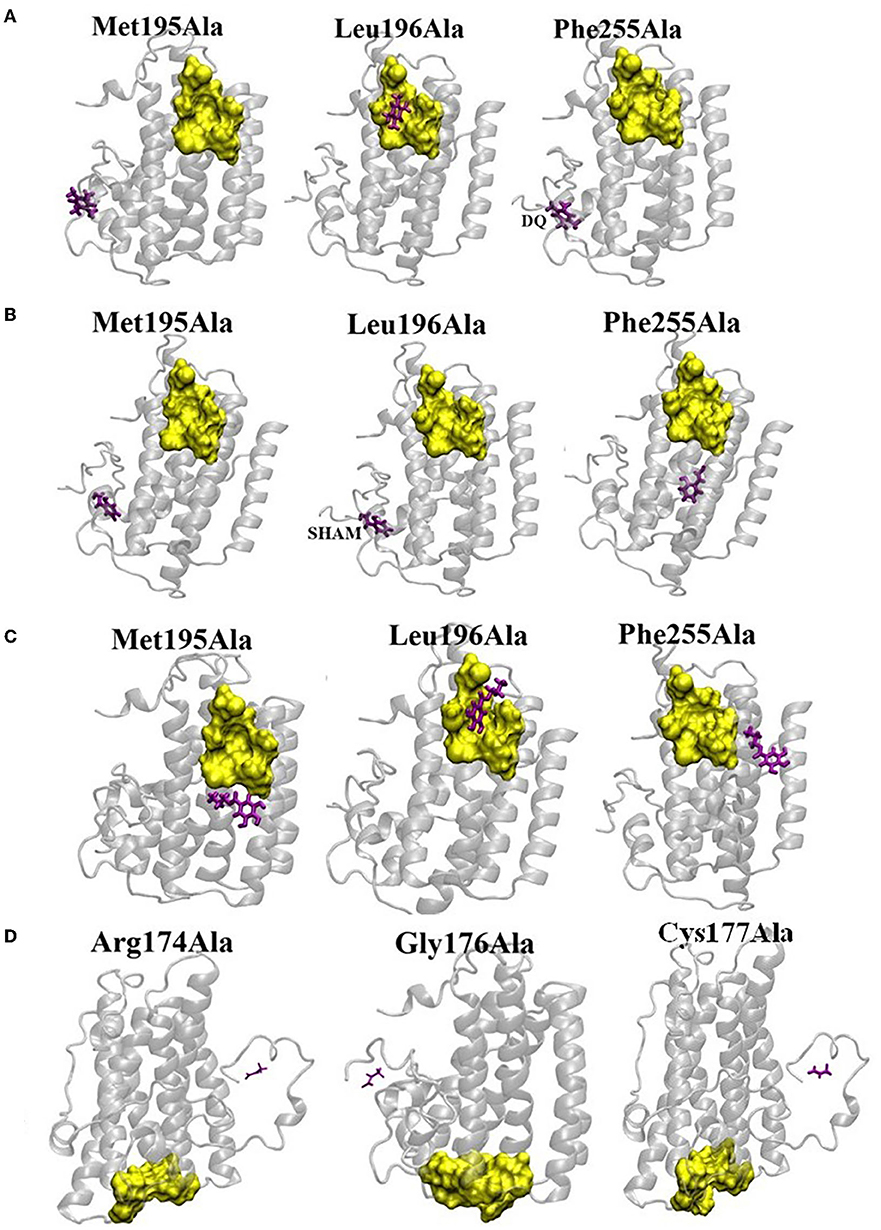
Figure 7. Effect of mutation on the binding affinities of DQ (A), SHAM (B), n-PG (C), and Pyruvate (D). The yellow surface indicates the location of the binding pocket in wild type (WT) AtAOX1A. The location of the ligand on the mutant AtAOX1A is shown in purple.
Panel (a) in Figures 6I–VI shows the most probable binding sites of the above-mentioned ligands on AtAOX1A. The Q1H2 binding site (Figure 6Ib) is composed of Trp131, Glu132, Thr133, Tyr134, Lys135, Ile138, Trp305, Arg306, Leu307, Pro308, and Asp315, while Q1H2 makes hydrogen bonds with Trp131 and Arg306, and forms a stable complex with ΔG = −7.05 kcal/mol. The ubiquinone or ubiquinone-1 (with one isoprenoid unit) binding pocket (Figure 6IIb) is made up of Leu163, Thr167, Phe171, Met181, Leu182, Val185, Leu242, Thr245, Val246, Val249, and Phe250, respectively, and the ligand is mostly hydrophobic due to the presence of five methyl groups. The binding energy of UQ1 in this pocket is ΔG = −6.74 kcal/mol. The binding pocket for DQH2 consists of Trp131, Glu132, Thr133, Tyr134, Lys135, Arg306, Pro308, and Asp315 (Figure 6IIIb). The binding energy of DQH2 to the AtAOX1A is ΔG = −6.63 kcal/mol. Interestingly, DQH2 and Q1H2 bind in the same location that is close to the diiron cavity (Figure 6VII). According to the SwissDock results, the binding pocket of DQ is made up of Met191, Val192, Met195, Leu196, Phe251, and Phe255 (Figure 6IVb). The binding energy of DQ to the AtAOX1A is ΔG = −6.11 kcal/mol. This surface exposed, hydrophobic region is located on the 2nd and 4th α-helices and is ~27Å away from the diiron center.
According to the SwissDock results, SHAM and n-PG bind to the same binding pocket (Figures 6Vb, VIb) as that of DQ. The binding energies of SHAM and n-PG are −6.01 and −6.13 kcal/mol, respectively. This observation confirms that the inhibitor binding pocket could be conserved in AtAOX1A. Figures 6VII,VIII compare the location of the diiron cavity with the inhibitor and Q1H2/DQH2 and UQ1 binding sites, respectively. It was observed that the UQ1 binding site bridges the diiron cavity and inhibitor binding site. Further, the pyruvate binding sites on the AtAOX1A are shown in Figure 6IXa. The most probable binding site for pyruvate (Figure 6IXb) is made of Arg174, Tyr175, Gly176, Cys177, Val232, Ala233, Asn294, and Leu313. The binding energy of pyruvate is ΔG = −6.87 kcal/mol.
Mutational Docking Studies of AtAOX1A With Different Ligands
To identify the potent residues on AtAOX1A that may have a direct impact on the binding of DQ, SHAM, and n-PG, point mutations were performed concerning three hydrophobic residues: (i) Met195 to Ala, (ii) Leu196 to Ala, and (iii) Phe255 to Ala in the DQ/inhibitor binding pocket (Figures 7A–C). The side chains of these residues lie within 3Å from the ligand as the hydrophobic interactions between the ligand and side chains hold the ligand in its binding pocket. Interestingly, upon mutations of Met195 and Phe255 to alanine, the DQ (Figure 7A), SHAM (Figure 7B), and n-PG (Figure 7C) binding locations are drastically altered in comparison to that of the wild type AtAOX1A. However, the mutation of Leu196 did not alter the binding pocket of DQ and n-PG. Similarly, in the case of pyruvate, three mutations were performed: Arg174 to Ala, Gly176 to Ala, and Cys177 to Ala (Figure 7D). All these mutations inhibited the activator from binding to the original binding site in the wild type AtAOX1A and, thus, may play primary roles in the binding of the activator.
Discussion
Differential Response of E. coli/pET28a and E. coli/pAtAOX1A to Mitochondrial Inhibitors
The electron transport chain components located in the inner membrane of E. coli possess two terminal oxidases, viz., ‘cytochrome oxidase bo' and ‘cytochrome oxidase bd,' to reduce molecular oxygen to water molecules and oxidize ubiquinol (Kita et al., 1984a; Anraku and Gennis, 1987; Mogi et al., 1994; Yap et al., 2010; Borisov et al., 2011). Cytochrome oxidase bo, which possesses heme-copper oxidase, shares functional similarities with mitochondrial cytochrome c oxidase of higher plants and is expressed during high oxygen concentrations. In contrast, cytochrome bd oxidase, which lacks both copper ion and the Fe–S cluster, is known to predominate in anoxic conditions and is not homologous to any other terminal oxidases, such as heme-copper oxidoreductases or AOX (Reid and Ingledew, 1979; Kita et al., 1984b; Poole and Cook, 2000; Borisov et al., 2011). The properties of both these oxidases (bo and bd) are quite distinct from the AOX, which contains a non-heme diiron carboxylate center (Berthold et al., 2002; Berthold and Stenmark, 2003; Moore et al., 2008, 2013). Therefore, in this study, BL21(DE3) E. coli strain was chosen to examine in vivo functional expression of pAtAOX1A and robust expression of rAtAOX1A protein for purification and in vitro characterization. AOX is known to be resistant to cyanide and sensitive to n-PG and SHAM, while E. coli oxidases are found to be sensitive to cyanide but not to AOX inhibitors (Bendall and Bonner, 1971; Siedow and Girvin, 1980; Fukai et al., 1999). Thus, consistent with the properties of different oxidases, the respiratory rates of E. coli/pAtAOX1A are resistant to KCN and sensitive to n-PG and SHAM when compared with E. coli/pET28a (Figures 2A–C).
In the absence of any inhibitor, after 5 h, E. coli/pAtAOX1A exhibited higher growth (OD600 = ~1) when compared with E. coli/pET28a (OD600 = ~0.8) due to the beneficial effects rendered by AtAOX1A expression in E. coli/pAtAOX1A. Consequently, the growth rates of E. coli/pAtAOX1A are found to be resistant to KCN and sensitive to n-PG and SHAM, which is not the case with the E. coli/pET28a (Figures 2D–I). Thus, the differential responses observed in respiration and growth rates of E. coli/pET28a and E. coli/pAtAOX1A, upon treatment with mitochondrial inhibitors, clearly demonstrate that the rAtAOX1A expressed in E. coli is functionally active (Berthold, 1998). The expression of AOX from A. thaliana in hemA E. coli strain (deficient in cytochrome-mediated aerobic respiration) allowed it to grow under aerobic conditions (Kumar and Söll, 1992). Also, the expression of TAO in E. coli and rAtAOX1A in yeast cells has shown cyanide-insensitive and ascofuranone/SHAM-sensitive growth (Fukai et al., 1999; Nihei et al., 2003; Vishwakarma et al., 2016). Nevertheless, the results from this study suggest the concentration of n-PG to be restricted to ≤0.25 mM, as treatment with higher concentrations, leads to non-specific inhibitory effects on E. coli oxidase(s) and/or cellular redox enzymes, as well as hydrolases (Boyd and Beveridge, 1979; Han and Park, 2009; Figure 2F). Similarly, a high concentration of SHAM (2 mM) showed an inhibitory effect on the growth of E. coli/pET28a (88%), possibly due to its non-specific effect on E. coli cytochrome oxidases (Figure 2H). However, the non-specific effects are exacerbated more with SHAM than n-PG. Further, the studies of Berthold (1998) reported similar results using cytochrome bd oxidase mutants where partial inhibition in the duroquinol oxidase activity of the E. coli membrane was observed with SHAM but not n-PG.
Purification of rAtAOX1A in Its Active Form
In the present study, the rAtAOX1A is overexpressed in E. coli BL21(DE3) and purified from E. coli membranes (Figure 3A). To obtain a pure, as well as active rAtAOX1A, it is essential to maintain the following conditions during its inductions and purification procedure.
Supplementation of Fe2+ Is Essential for the Expression of Active rAtAOX1A
The studies of Minagawa et al. (1990) and Ajayi et al. (2002) have reported that iron supplementation is essential during the heterologous expression of AOX. In this study, Fe2+ was added to the culture medium at the time of induction along with IPTG, which resulted in an active rAtAOX1A (Table 1). However, when rAtAOX1A was purified without Fe2+ supplementation, its activity was not detectable, despite the presence of pyruvate in all purification steps (data not shown). This could be due to a lack of formation of the hydroxo-bridged binuclear iron center, which is involved in the reduction of molecular oxygen to water, as evident by electron paramagnetic resonance studies (Berthold et al., 2002; Moore et al., 2008). Similarly, in this study, supplementation of Fe+3 ion to the culture medium could not retrieve AOX in pure form (data not shown).
Pyruvate Supplementation Is Essential During All the Steps of Purification
Pyruvate stabilizes the active enzyme conformation, although the specific mechanism of AOX activation by pyruvate is unclear (Carré et al., 2011; Elliott et al., 2014; Xu et al., 2021). In this study, the addition of pyruvate (10 mM) in all purification steps resulted in the retrieval of active rAtAOX1A from E. coli membranes (Table 1). However, the AOX activity was found to be insensitive to pyruvate when examined in mitochondria isolated from thermogenic plants, such as Arum italicum (Hoefnagel et al., 1997), S. guttatum (Crichton et al., 2005), and A. maculatum (Ito et al., 2011).
Detergent Choice to Solubilize the rAtAOX1A From E. coli Membranes
The choice of detergent usage also plays a major role in retaining AOX activity during the process of purification. For solubilizing the recombinant trypanosomal alternative oxidase (rTAO), OG was proven to be efficient (Kido et al., 2010). However, in the case of recombinant Sauromatum guttatum alternative oxidase (rSgAOX), OG caused a decrease in the enzyme activity, while DDM improved its activity (Elliott et al., 2014). In this study, DDM was used to solubilize rAtAOX1A from E. coli/pAtAOX1A membranes. The protocol used in this study resulted in a 15% recovery of rAtAOX1A. Further, the oxygen uptake activity of purified rAtAOX1A in the presence of duroquinol was found to be 3.8 μmol O2 min−1 mg−1 protein at pH 7.5 in Tris-HCl (Table 1). Also, the activity obtained in the present study was found to be comparable (4 μmol Q1 min−1.mg−1 protein) with that of rAtAOX1A, which is purified from the ΔhemA-deficient E. coli strain (FN102), which lacks the quinol oxidase activity for cytochrome bo and bd complexes (Xu et al., 2021). In contrast, the activity of rTAO measured as quinol oxidizing activity was found to be 207 μmol min−1 mg−1 protein (Kido et al., 2010), while rSgAOX measured as oxygen uptake activity was found to be 20 μmol min−1 mg−1 protein (Elliott et al., 2014), respectively. The lower activity levels of rAtAOX1A in the present study could be due to its origin in the non-thermogenic plant (Moore et al., 2013). The docking studies of May et al. (2017) and Xu et al. (2021) indicated that the differences in the polar residues surrounding the hydrophobic cavity of the quinol binding site might change the size of the cavity, which, in turn, may lead to the differences in the strength of attraction of quinol into the active site, and, thereby, affect the activity of AOX enzyme(s).
Structural Characterization of Purified rAtAOX1A and Its Interaction With Inhibitors and Activator
In this study, the structure of rAtAOX1A and the changes in the corresponding structure during interaction with its inhibitory (SHAM and n-PG) and activator (pyruvate) molecules are revealed by CD spectroscopy. Besides, the docking and SPR studies identified the specific binding pockets for these molecules on AtAOX1A, and revealed their binding energies/affinities during interaction with rAtAOX1A.
Analysis of far-UV (190–260 nm) CD spectra reveals the secondary structural elements such as α-helices and β-sheets present in a protein. The CD results from the present study have revealed that the purified rAtAOX1A possessed α-helices predominantly over β-sheets in its secondary structure (Figure 4A, Table 2), and this result is consistent with the secondary structural elements of rTAO and rSgAOX (Elliott et al., 2014). Besides, the following observations in this study, such as: (i) stability in the ellipticity of rAtAOX1A to a wide range of temperatures (Figures 4C,D) and (ii) retention of its helical absorbance signal up to 67% even at temperatures as high as 90°C (Figure 4E), indicated that the thermal stability of rAOX1A isoform from a non-thermogenic plant A. thaliana is comparable to that of rAOX from thermogenic plant S. guttatum and rTAO from a parasitic protozoan (Elliott et al., 2014). Further, the retention of the helical signal even at a wide range of pH conditions indicated the stability of the rAtAOX1A to changes in pH (Figure 4B). Besides, a decrease in the α-helical content and a rise in the β-sheets in rAtAOX1A upon interaction with SHAM, n-PG, and pyruvate demonstrated the conformational changes occurring in the protein, possibly due to a rearrangement in the hydrogen bond network of the secondary structural elements (Hebia et al., 2014; Yu et al., 2021; Table 2).
Furthermore, the SPR technique, which is known to provide real-time label-free binding kinetics, is used to analyze the interaction of rAtAOX1A with its inhibitors. The kinetic data obtained for n-PG and SHAM during their interaction with rAtAOX1A fitted well with the 1:1 Langmuir model. Also, n-PG (KD = 0.49 nM) showed higher affinity with rAtAOX1A than SHAM (KD = 3 nM) as the KD is inversely proportional to the binding affinity (Figures 5C,D). Also, a positive correlation was observed between the KD values (an indicator of binding affinity) obtained in this study with the IC50 values (an indicator of the inhibition potency) obtained for n-PG and SHAM against rAOX from different sources (Elliott et al., 2014; May et al., 2017; Xu et al., 2021). For example, the results from the studies of Xu et al. (2021) indicated that the n-PG inhibits rAtAOX1A at a concentration of ~1.58 μM (IC50), while SHAM inhibits at a concentration of ~33 μM (IC50).
In the case of plants, the crystal structure of AOX is not yet available, so far. Therefore, in the docking studies, we used the homology model generated for AtAOX1A (PMDB Accession number: PM0080189), using the crystal structure of TAO (PDB ID: 3VV9) as a template (Pennisi et al., 2016). The studies of Pennisi et al. (2016) also predicted the protein structure for N-terminal 31 residues (residues 63–93) of AtAOX1A by ab initio/threading program. Thus, irrespective of the origin, AOX possessed a common structural trend of forming a four-α-helix bundle with a diiron catalytic center.
The AOX1A from A. thaliana contained three domains: (i) a mitochondrial recognition signal peptide domain between residues 1 and 62, which detaches from AOX after mitochondrial recognition; (ii) a predicted region in the N-terminus between residues 63 and 93; and (iii) a catalytic domain between residues 94 and 354 that is responsible for the oxidation of ubiquinol, binding of activators and inhibitors, and reduction of an oxygen molecule to water. However, the model generated by Pennisi et al. (2016) includes only the catalytic domain of AtAOX1A, and the tunnel formed by four α-helices (α2, α3, α5, and α6) in this model has two prominent hydrophobic regions: (i) a catalytic cavity consisting of conserved glutamate and histidine residues (Glu183, Glu222, Glu273, Glu324, His225, and His327) hosting the diiron center and (ii) a second region with conserved residues Arg164, Asp168, Arg178, Leu182, Ala186, Leu272, Glu275, and Ala276 connects the first hydrophobic region with the lipid bilayer facing the mitochondrial matrix. Further, the monomer-monomer binding region lies between residues 94 and 127 of each monomer. However, this choice can have the least effect on the binding affinities of SHAM and n-PG as the inhibitor binding sites are far away from the monomer-monomer interacting region. Although the Cys127 residue of each monomer is involved in disulfide bond formation during the dimeric assembly of AtAOX1A, its interaction with pyruvate is not found to be important to keeping the AOX in its activation state, which is evident by the mutational studies performed by Selinski et al. (2017).
The docking results from the present study revealed that the pyruvate binding pocket contained Cys177, along with Arg174, Tyr175, Gly176, Val232, Ala233, Asn294, and Leu313 (Figure 6IX), substantiating the involvement of second cysteine (Cys177) residue in regulating the post-translational activation of AOX during its interaction with pyruvate (Polidoros et al., 2009; Moore et al., 2013). The studies of Crichton et al. (2005) identified four potential regions in AtAOX1A that played an important role in the interaction of organic acids. The Arg174, Gly176 and Cys177 present in the pyruvate binding pocket of the present study corroborated well with the residues identified in region 2, while Asn294 matched with the “N” in the ENV motif of region 3. Further, mutational docking studies performed with Arg174Ala, Gly176Ala, and Cys177Ala in AtAOX1A identified them as potential candidates for binding to pyruvate (Figure 7D). Further, the changes observed in the secondary structural elements of AtAOX1A upon interaction with pyruvate may not allow the protein to form an inactive dimeric form and might give more access to the substrate to bind, which, in turn, could lead to the full enzyme activity (Xu et al., 2021) (Figures 4H, 6IX, 7D, Table 2).
Furthermore, the analysis of binding sites for the molecules like Q1H2, UQ1, DQH2, DQ, SHAM, and n-PG on AtAOX1A using the molecular docking method indicated that because of the structural and functional similarities between Q1H2 and DQH2, the binding cavities of these two ligands are found to be identical to some extent (Figures 6I,III). As observed in TAO (Shiba et al., 2013), this binding cavity is near the diiron cavity formed by four glutamate residues (Glu183, Glu222, Glu273, and Glu324), but the inhibitor binding site is far from this Q1H2 and DQH2 binding cavity (Figure 6VII). Further, the UQ1 binding site (Figure 6II) is somewhat similar to that of the inhibitor (SHAM and n-PG) binding site as both are: (i) composed of hydrophobic residues and (ii) share α-2 and α-4 helices. Our results also show that the UQ1 binding site is slightly away from that of the inhibitor. The inhibitor and UQ1 binding pockets and diiron cavity have been shown in the surface model (Figure 6VIII). Besides, DQ binding site (Figure 6IV) is similar to that of the inhibitor (SHAM and n-PG) binding site. Thus, the binding of the inhibitors to a hydrophobic groove formed by the residues Met191, Val192, Met195, Leu196, Phe251, and Phe255 in AtAOX1A might block the electron transport through the AOX pathway (Figures 6V,VI). The mutational docking studies suggest that Met195 and Phe255 of AtAOX1A are the potential candidates to bind the inhibitors (Figures 7B,C). Hence, this binding pocket could be a potential “gateway” for the oxidation-reduction process in AtAOX1A.
According to the docking studies of Shiba et al. (2013), the inhibitor AF2279OH binds to the Q1H2 binding site on TAO, while the results from the present docking studies indicated that the Q1H2/DQH2 binding sites are different from that of inhibitors (SHAM and n-PG) binding pocket on AtAOX1A (Figure 6VII). Nevertheless, the UQ1 (oxidized form of Q1H2) binding site is slightly away from that of the inhibitor (Figure 6VIII). However, the inhibitors (SHAM and n-PG) bind at the DQ (oxidized form of DQH2) binding site. This difference in the inhibitor binding site in TAO and AtAOX1A could be due to the difference in the amino acid sequences, where the sequence similarity between them (TAO and AtAOX1A) is only 31.04% (Xu et al., 2021).
Taken together, the results obtained from CD, SPR, and docking studies suggest that binding of SHAM or n-PG to a specific hydrophobic groove associated with α2 and α4 helices on AtAOX1A might alter its α-helical conformation, which in turn may lead to the inhibition of AOX pathway.
Conclusion
In this study, the pAtAOX1A-transformed E. coli showed cyanide-resistant, as well as n-PG and SHAM-sensitive respiration, and growth characteristics, confirming the functional expression of rAtAOX1A in E. coli BL21(DE3) cells. CD spectroscopic analysis has revealed that the purified rAtAOX1A majorly possessed an α-helical structure that is stable against a wide range of pH (2 to 12) and temperature (4° to 90°C). These results indicate that the AOX protein remains stable in the non-thermogenic plant, even under extreme temperature(s). Further, the interaction of SHAM, n-PG, or pyruvate with the rAtAOX1A caused a significant reduction in its α-helical content while retaining its ellipticity. Besides, the in silico docking studies, together with mutational docking studies, revealed the following: (i) the inhibitors (SHAM and n-PG) and substrate (DQ) bind in the same hydrophobic binding site due to prominent interactions with Met195 and Phe255, (ii) Q1H2 and DQH2 binding sites are identical and close to the diiron center, and (iii) the activator (pyruvate) binding pocket contains Cys177, that regulates the post-translational activities in AtAOX1A. Arg174 and Gly176 also play an important role in the pyruvate interaction with AtAOX1A, (iv) UQ1 binding pocket connects the inhibitor binding site and diiron cavity.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
KP conceived the project. TVS, DS, and AV performed the wet lab experiments and interpreted the data and wrote the manuscript. MS performed docking studies. KP and MS edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
TVS gratefully acknowledges DBT for JRF/SRF and contingency grant [Ref. No.13/AL/241/2483 dt. 25/07/2013]. DS thanks NFST for JRF and contingency grant (202021-NFST-TEL-01185), and AV acknowledges CSIR for SRF and contingency grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We profusely thank Prof. Renate Scheibe, University of Osnabruck, Germany, for generously providing Arabidopsis thaliana (Columbia) seeds to our laboratory. We also thank Prof. A.S. Raghavendra and Prof. Saradadevi Tetali [Department of Plant Sciences, University of Hyderabad (UoH)] for extending critical suggestions on experimental protocols/results and their lab facilities during the execution of the project. We also thank Mrs. Monika Kannan (Proteomics Facility, School of Life Sciences, UoH) for helping in MALDI-TOF studies and Dr. Praveen (GE Healthcare Life Sciences) for helping in the analysis of SPR data. The Department(s)/School facilities of UoH were supported by grants received from DST-FIST-I, UGC-SAP-DRS, UPE-Phase I/II, UGC-SAP-CAS, DBT-CREBB, and DBT-BUILDER, all from New Delhi, India.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.871208/full#supplementary-material
Supplementary Figure 1. Confirmation of the clone (pAtAOX1A). (A) colony PCR: A single colony of E. coli cells was used as a DNA source to amplify using AtAOX1A primers. Lane 1, 50-1000 bp DNA ladder; lane 2, colony 1; lane 3, colony 2; lane 4, colony 3 and lane 5, non-template control. Amplified AtAOX1A is indicated with an arrow. (B) Restriction Digestion: Plasmid from pET28a and pAtAOX1A were digested with EcoRI and XhoI restriction endonucleases. Lane 1, 0.5-10 kb DNA ladder; lane 2, pET28a vector; lane 3, pAtAOX1A vector; lane 4, restriction digested pAtAOX1A; lane 5, 50–1000 bp DNA ladder. The inserted gene (AtAOX1A) released is indicated with an arrow. 20 μl of the sample was loaded in each well.
Supplementary Figure 2. The Biotools display of lift spectra corresponding to the peak 1656.258 Da.
Supplementary Figure 3. Secondary structural stability analysis of rAtAOX1A. The secondary structural stability was determined between 205 to 230 nm under different conditions: (A) pH from 2 to 12; (B) Temperatures from 4 to 90°C; (C) Temperatures from 90 to 4°C; (D) CD spectra of rAtAOX1A with 0.05, 0.1, and 0.5 mM of SHAM and without inhibitor; (E) CD spectra of rAtAOX1A with 0.05, 0.1, and 0.5 mM of n-PG, and without inhibitor; (F) CD spectra of rAtAOX1A with 0.05, 0.1, 0.5, and 1 mM of pyruvate, and without activator. Concentration of purified rAtAOX1A used to obtain the CD spectrum (SHAM, n-PG, and pyruvate) was 0.4 mg/ml. The final spectrum is an average of three scans as described in the Materials and Methods section.
Supplementary Data S1. The colony PCR positive clones were sequenced (Eurofins Pvt. Ltd.) with T7 forward primer (TAATACGACTCACTATAGGG). DNA sequence obtained from clone (pAtAOX1A) was aligned with original AtAOX1A gene using ClustalW Multiple alignment programs. DNA alignment results showed 100% similarity. The highlighted region represents His-tag sequences.
Supplementary Data S2. Matrix-assisted laser desorption- time of flight (MALDI-TOF) mascot search, National Center for Biotechology Information (NCBI) database protein view, Peptide mass fingerprinting (PMF) spectra and lift spectra indicated that the recombinant protein is Arabidopsis thaliana AOX1A.
Abbreviations
AOX, Alternative oxidase; CD, Circular dichroism; DDM, n-dodecyl-β-D-maltopyranoside; DQH2, Duroquinol or Reduced duroquinone; DQ, Duroquinone; IPTG, Isopropyl, β-D-1-thiogalactoside; KD, Equilibrium dissociation constant; MALDI-TOF, Matrix-assisted laser desorption ionization time-of-flight; mETC, Mitochondrial electron transport chain; n-PG, n-propyl gallate; OD, Optical density; OG, n-octyl-β-D-glucopyranoside; rAtAOX1A, Recombinant Arabidopsis thaliana alternative oxidase; rSgAOX, Recombinant Sauromatum guttatum alternative oxidase; rTAO, Recombinant trypanosome alternative oxidase; RU, Resonance unit; SHAM, Salycylhydroxamic acid; SPR, Surface plasmon resonance; Q1H2, Ubiquinol-1 or Reduced ubiquinone; UQ1, Ubiquinone-1.
References
Affourtit, C., Albury, M. S., Crichton, P. G., and Moore, A. L. (2002). Exploring the molecular nature of alternative oxidase regulation and catalysis. FEBS Lett. 510, 121–126. doi: 10.1016/S0014-5793(01)03261-6
Affourtit, C., and Moore, A. L. (2004). Purification of the plant alternative oxidase from Arum maculatum: measurement, stability and metal requirement. Biochim. Biophys. Acta - Bioenerg. 1608, 181–189. doi: 10.1016/j.bbabio.2003.12.002
Ajayi, W. U., Chaudhuri, M., and Hill, G. C. (2002). Site-directed mutagenesis reveals the essentiality of the conserved residues in the putative diiron active site of the trypanosome alternative oxidase. J. Biol. Chem. 277, 8187–8193. doi: 10.1074/jbc.M111477200
Albury, M. S., Elliott, C., and Moore, A. L. (2009). Towards a structural elucidation of the alternative oxidase in plants. Physiol. Plant. 137, 316–327. doi: 10.1111/j.1399-3054.2009.01270.x
Anraku, Y., and Gennis, R. B. (1987). The aerobic respiratory chain of Escherichia coli. Trends Biochem. Sci. 12, 262–266. doi: 10.1016/0968-0004(87)90131-9
Bendall, D. S., and Bonner, W. D. (1971). Cyanide-insensitive respiration in plant mitochondria. Plant Physiol. 47, 236–245. doi: 10.1104/pp.47.2.236
Berthold, D. A.. (1998). Isolation of mutants of the Arabidopsis thaliana alternative oxidase (ubiquinol:oxygen oxidoreductase) resistant to salicylhydroxamic acid. Biochim. Biophys. Acta - Bioenerg. 1364, 73–83. doi: 10.1016/S0005-2728(98)00015-2
Berthold, D. A., Andersson, M. E., and Nordlund, P. (2000). New insight into the structure and function of the alternative oxidase. Biochim. Biophys. Acta - Bioenerg. 1460, 241–254. doi: 10.1016/S0005-2728(00)00149-3
Berthold, D. A., and Siedow, J. N. (1993). Partial purification of the cyanide-resistant alternative oxidase of skunk cabbage (Symplocarpus foetidus) mitochondria. Plant Physiol. 101, 113–119. doi: 10.1104/pp.101.1.113
Berthold, D. A., and Stenmark, P. (2003). Membrane -bound diiron carboxylate proteins. Annu. Rev. Plant Biol. 54, 497–517. doi: 10.1146/annurev.arplant.54.031902.134915
Berthold, D. A., Voevodskaya, N., Stenmark, P., Gräslund, A., and Nordlund, P. (2002). EPR studies of the mitochondrial alternative oxidase: evidence for a diiron carboxylate center. J. Biol. Chem. 277, 43608–43614. doi: 10.1074/jbc.M206724200
Bonner, W. D., Clarke, S. D., and Rich, P. R. (1986). Partial purification and characterization of the quinol oxidase activity of Arum maculatum mitochondria. Plant Physiol. 80, 838–842. doi: 10.1104/pp.80.4.838
Borisov, V. B., Gennis, R. B., Hemp, J., and Verkhovsky, M. I. (2011). The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta - Bioenerg. 1807, 1398–1413. doi: 10.1016/j.bbabio.2011.06.016
Boyd, I., and Beveridge, E. G. (1979). Relationship between the antibacterial activity towards Escherichia coli NCTC 5933 and the physico-chemical properties of some esters of 3,4,5-trihydroxybenzoic acid (Gallic acid). Microbios 24, 173–184. Available online at: https://europepmc.org/article/med/43458
Carré, J. E., Affourtit, C., and Moore, A. L. (2011). Interaction of purified alternative oxidase from thermogenic Arum maculatum with pyruvate. FEBS Lett. 585, 397–401. doi: 10.1016/j.febslet.2010.12.026
Clifton, R., Lister, R., Parker, K. L., Sappl, P. G., Elhafez, D., Millar, A. H., et al. (2005). Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58, 193–212. doi: 10.1007/sl
Clifton, R., Millar, A. H., and Whelan, J. (2006). Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim. Biophys. Acta - Bioenerg. 1757, 730–741. doi: 10.1016/J.BBABIO.2006.03.009
Considine, M. J., Holtzapffel, R. C., Day, D. A., Whelan, J., and Millar, A. H. (2002). Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol. 129, 949–953. doi: 10.1104/pp.004150
Costa, J. H., Santos, C. P., dos de Sousa e Lima, B., Moreira Netto, A. N., Saraiva, K. D., da, C., et al. (2017). In silico identification of alternative oxidase 2 (AOX2) in monocots: a new evolutionary scenario. J. Plant Physiol. 210, 58–63. doi: 10.1016/j.jplph.2016.12.009
Crichton, P. G., Affourtit, C., Albury, M. S., Carr,é, J. E., and Moore, A. L. (2005). Constitutive activity of Sauromatum guttatum alternative oxidase in Schizosaccharomyces pombe implicates residues in addition to conserved cysteines in α-keto acid activation. FEBS Lett. 579, 331–336. doi: 10.1016/j.febslet.2004.10.107
Crichton, P. G., Albury, M. S., Affourtit, C., and Moore, A. L. (2010). Mutagenesis of the Sauromatum guttatum alternative oxidase reveals features important for oxygen binding and catalysis. Biochim. Biophys. Acta - Bioenerg. 1797, 732–737. doi: 10.1016/j.bbabio.2009.12.010
Dahal, K., and Vanlerberghe, G. C. (2017). Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol. 213, 560–571. doi: 10.1111/nph.14169
Diethelm, R., Miller, M. G., Shibles, R., and Stewart, C. R. (1990). Effect of salicylhydroxamic acid on respiration, photosynthesis, and peroxidase activity in various plant tissues. Plant Cell Physiol. 31, 179–185. doi: 10.1093/oxfordjournals.pcp.a077890
Dinakar, C., Abhaypratap, V., Yearla, S. R., Raghavendra, A. S., and Padmasree, K. (2010). Importance of ROS and antioxidant system during the beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Planta. 231, 461–474. doi: 10.1007/s00425-009-1067-3
Elliott, C., Young, L., May, B., Shearman, J., Albury, M. S., Kido, Y., et al. (2014). Purification and characterisation of recombinant DNA encoding the alternative oxidase from Sauromatum guttatum. Mitochondrion. 19, 261–268. doi: 10.1016/j.mito.2014.03.002
Elthon, T. E., and McIntosh, L. (1986). Characterization and solubilization of the alternative oxidase of Sauromatum guttatum mitochondria. Plant Physiol. 82, 1–6. doi: 10.1104/pp.82.1.1
Elthon, T. E., and McIntosh, L. (1987). Identification of the alternative terminal oxidase of higher plant mitochondria. Proc. Natl. Acad. Sci. 84, 8399–8403. doi: 10.1073/pnas.84.23.8399
Florez-Sarasa, I., Fernie, A. R., and Gupta, K. J. (2020). Does the alternative respiratory pathway offer protection against the adverse effects resulting from climate change? J. Exp. Bot. 71, 465–469. doi: 10.1093/jxb/erz428
Florez-Sarasa, I., Flexas, J, Rasmusson, A. G., Umbach, A. L., Siedow, J. N., and Ribas-Carbo, M. (2011). In vivo cytochrome and alternative pathway respiration in leaves of Arabidopsis thaliana plants with altered alternative oxidase under different light conditions. Plant Cell Environ. 34, 1373–1383. doi: 10.1111/j.1365-3040.2011.02337.x
Fukai, Y., Amino, H., Hirawake, H., Yabu, Y., Ohta, N., Minagawa, N., et al. (1999). Functional expression of the ascofuranone-sensitive Trypanosoma brucei brucei alternative oxidase in the cytoplasmic membrane of Escherichia coli. Comp. Biochem. Physiol. - C Pharmacol. Toxicol. Endocrinol. 124, 141–148. doi: 10.1016/S0742-8413(99)00040-7
Garmash, E. V., Velegzhaninov, I. O., Ermolina, K. V., Rybak, A. V., and Malyshev, R. V. (2020). Altered levels of AOX1a expression result in changes in metabolic pathways in Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation. Plant Sci. 291, 110332. doi: 10.1016/j.plantsci.2019.110332
Giraud, E., Ho, L. H. M. M., Clifton, R., Carroll, A., Estavillo, G., Tan, Y. F., et al. (2008). The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 147, 595–610. doi: 10.1104/pp.107.115121
Giraud, E., van Aken, O., Ho, L. H. M., and Whelan, J. (2009). The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of alternative oxidase1a. Plant Physiol. 150, 1286–1296. doi: 10.1104/pp.109.139782
Grosdidier, A., Zoete, V., and Michielin, O. (2011a). Fast docking using the CHARMM force field with EADock DSS. J.Comput. Chem. 32, 2149–2159. doi: 10.1002/jcc.21797
Grosdidier, A., Zoete, V., and Michielin, O. (2011b). SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, 270–277. doi: 10.1093/nar/gkr366
Han, Y. H., and Park, W. H. (2009). Propyl gallate inhibits the growth of HeLa cells via regulating intracellular GSH level. Food Chem. Toxicol. 47, 2531–2538. doi: 10.1016/J.FCT.2009.07.013
Hebia, C., Bekale, L., Chanphai, P., Agbebavi, J., and Tajmir-Riahi, H. A. (2014). Trypsin inhibitor complexes with human and bovine serum albumins: TEM and spectroscopic analysis. J. Photochem. Photobiol. B Biol. 130, 254–259. doi: 10.1016/J.JPHOTOBIOL.2013.11.025
Ho, L. H. M., Giraud, E., Uggalla, V., Lister, R., Clifton, R., Glen, A., et al. (2008). Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in arabidopsis. Plant Physiol. 147, 1858–1873. doi: 10.1104/pp.108.121384
Hoefnagel, M. H. N., Rich, P. R., Zhang, Q., and Wiskich, J. T. (1997). Substrate kinetics of the plant mitochondrial alternative oxidase and the effects of pyruvate. Plant Physiol. 115, 1145–1153. doi: 10.1104/pp.115.3.1145
Homola, J.. (2003). Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 377, 528–539. doi: 10.1007/s00216-003-2101-0
Huq, S., and Palmer, J. M. (1978). Isolation of a cyanide-resistant duroquinol oxidase from Arum maculatum mitochondria. FEBS Lett. 95, 217–220. doi: 10.1016/0014-5793(78)80997-1
Ito, K., Ogata, T., Kakizaki, Y., Elliott, C., Albury, M. S., and Moore, A. L. (2011). Identification of a gene for pyruvate-insensitive mitochondrial alternative oxidase expressed in the thermogenic appendices in Arum maculatum. Plant Physiol. 157, 1721–1732. doi: 10.1104/pp.111.186932
Jo, S., Kim, T., Iyer, V. G., and Im, W. (2008). CHARMM-GUI a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865. doi: 10.1002/jcc.20945
Kay, C. J., and Palmer, J. M. (1985). Solubilization of the alternative oxidase of cuckoo-pint (Arum maculatum) mitochondria. Stimulation by high concentrations of ions and effects of specific inhibitors. Biochem. J. 228, 309–318. doi: 10.1042/bj2280309
Kelly, S., and Price, N. (2000). The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 1, 349–384. doi: 10.2174/1389203003381315
Kelly, S. M., Jess, T. J., and Price, N. C. (2005). How to study proteins by circular dichroism. Biochim. Biophys. Acta - Proteins Proteomics 1751, 119–139. doi: 10.1016/j.bbapap.2005.06.005
Kido, Y., Sakamoto, K., Nakamura, K., Harada, M., Suzuki, T., Yabu, Y., et al. (2010). Purification and kinetic characterization of recombinant alternative oxidase from Trypanosoma brucei brucei. Biochim. Biophys. Acta - Bioenerg. 1797, 443–450. doi: 10.1016/j.bbabio.2009.12.021
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., et al. (2021). PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 49, D1388–D1395. doi: 10.1093/nar/gkaa971
Kirimura, K., Yoda, M., and Usami, S. (1999). Cloning and expression of the cDNA encoding an alternative oxidase gene from Aspergillus niger WU-2223L. Curr. Genet. 34, 472–477. doi: 10.1007/s002940050422
Kita, K., Konishi, K., and Anraku, Y. (1984a). Terminal oxidases of Escherichia coli aerobic respiratory chain. I. Purification and properties of cytochrome b562-o complex from cells in the early exponential phase of aerobic growth. J. Biol. Chem. 259, 3368–3374. doi: 10.1016/s0021-9258(17)43304-7
Kita, K., Konishi, K., and Anraku, Y. (1984b). Terminal Oxidases of Escherichia coli Aerobic Respiratory Chain. II. Purification and properties of cytochrome b558-d complex from cells grown with limited oxygen and evidence of branched electron-carrying systems. J. Biol. Chem. 259, 3375-3381. doi: 10.1016/s0021-9258(17)43305-9
Kumar, A. M., and Söll, D. (1992). Arabidopsis alternative oxidase sustains Escherichia coli respiration. Proc. Natl. Acad. Sci. U. S. A. 89, 10842–10846. doi: 10.1073/pnas.89.22.10842
Laemmli, U. K.. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227, 680–685. doi: 10.1038/227680a0
Liao, Y., Cui, R., Xu, X., Cheng, Q., and Li, X. (2021). Jasmonic acid- and ethylene-induced mitochondrial alternative oxidase stimulates Marssonina brunnea defense in poplar. Plant Cell Physiol. 61, 2031–2042. doi: 10.1093/pcp/pcaa117
Liao, Y. W. K., Shi, K., Fu, L. J., Zhang, S., Li, X., Dong, D. K., et al. (2012). The reduction of reactive oxygen species formation by mitochondrial alternative respiration in tomato basal defense against TMV infection. Planta. 235, 225–238. doi: 10.1007/s00425-011-1483-z
Liu, Z., and Butow, R. A. (2006). Mitochondrial retrograde signaling. Annu. Rev. Genet. 40, 159–185. doi: 10.1146/annurev.genet.40.110405.090613
Maréchal, A., Kido, Y., Kita, K., Moore, A. L., and Rich, P. R. (2009). Three redox states of Trypanosoma brucei alternative oxidase identified by infrared spectroscopy and electrochemistry. J. Biol. Chem. 284, 31827–31833. doi: 10.1074/jbc.M109.059980
Maxwell, D. P., Wang, Y., and Mcintosh, L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. 96, 8271–8276. doi: 10.1073/pnas.96.14.8271
May, B., Young, L., and Moore, A. L. (2017). Structural insights into the alternative oxidases: Are all oxidases made equal? Biochem. Soc. Trans. 45, 731–740. doi: 10.1042/BST20160178
McDonald, A. E., Vanlerberghe, G. C., and Staples, J. F. (2009). Alternative oxidase in animals: Unique characteristics and taxonomic distribution. J. Exp. Biol. 212, 2627–2634. doi: 10.1242/jeb.032151
Meeuse, B. J. D.. (1975). Thermogenic respiration in aroids. Annu. Rev. Plant Physiol. 26, 117–126. doi: 10.1146/annurev.pp.26.060175.001001
Millar, A. H., Whelan, J., Soole, K. L., and Day, D. A. (2011). Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 62, 79–104. doi: 10.1146/annurev-arplant-042110-103857
Millar, A. H., Wiskich, J. T., Whelan, J., and Day, D. A. (1993). Organic acid activation of the alternative oxidase of plant mitochondria. FEBS Lett. 329, 259–262. doi: 10.1016/0014-5793(93)80233-K
Minagawa, N., Sakajo, S., Komiyama, T., and Yoshimoto, A. (1990). Essential role of ferrous iron in cyanide-resistant respiration in Hansenula anomala. FEBS Lett. 267, 114–116. doi: 10.1016/0014-5793(90)80302-Y
Mogi, T., Nakamura, H., and Anraku, Y. (1994). Molecular structure of a heme-copper redox center of the Escherichia coli ubiquinol oxidase: evidence and model. J. Biochem. 116, 471–477. doi: 10.1093/oxfordjournals.jbchem.a124548
Moore, A. L., Albury, M. S., Crichton, P. G., and Affourtit, C. (2002). Function of the alternative oxidase: Is it still a scavenger? Trends Plant Sci. 7, 478–481. doi: 10.1016/S1360-1385(02)02366-X
Moore, A. L., Carré, J. E., Affourtit, C., Albury, M. S., Crichton, P. G., Kita, K., et al. (2008). Compelling EPR evidence that the alternative oxidase is a diiron carboxylate protein. Biochim. Biophys. Acta - Bioenerg. 1777, 327–330. doi: 10.1016/j.bbabio.2008.01.004
Moore, A. L., Shiba, T., Young, L., Harada, S., Kita, K., and Ito, K. (2013). Unraveling the heater: new insights into the structure of the alternative oxidase. Annu. Rev. Plant Biol. 64, 637–663. doi: 10.1146/annurev-arplant-042811-105432
Moore, A. L., and Albury, M. S. (2008). Further insights into the structure of the alternative oxidase: from plants to parasites. Biochem. Soc. Trans. 36, 1022–1026. doi: 10.1042/BST0361022
Ng, S., Ivanova, A., Duncan, O., Law, S. R., Van Aken, O., De Clercq, I., et al. (2013). A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25, 3450–3471. doi: 10.1105/tpc.113.113985
Nguyen, H. H., Park, J., Kang, S., and Kim, M. (2015). Surface plasmon resonance: a versatile technique for biosensor applications. Sensors (Switzerland). 15, 10481–10510. doi: 10.3390/s150510481
Nihei, C., Fukai, Y., Kawai, K., Osanai, A., Yabu, Y., Suzuki, T., et al. (2003). Purification of active recombinant trypanosome alternative oxidase. FEBS Lett. 538, 35–40. doi: 10.1016/S0014-5793(03)00120-0
Olaru, A., Bala, C., Jaffrezic-Renault, N., and Aboul-Enein, H. Y. (2015). Surface plasmon resonance (SPR) biosensors in pharmaceutical analysis. Crit. Rev. Anal. Chem. 45, 97–105. doi: 10.1080/10408347.2014.881250
Padmasree, K., and Raghavendra, A. S. (1999a). Importance of oxidative electron transport over oxidative phosphorylation in optimizing photosynthesis in mesophyll protoplasts of pea (Pisum sativum L.). Physiol. Plant. 105, 546–553. doi: 10.1034/j.1399-3054.1999.105321.x
Padmasree, K., and Raghavendra, A. S. (1999b). Response of photosynthetic carbon assimilation in mesophyll protoplasts to restriction on mitochondrial oxidative metabolism: Metabolites related to the redox status and sucrose biosynthesis. Photosynth. Res. 62, 231–239. doi: 10.1023/a:1006382518725
Pennisi, R., Salvi, D., Brandi, V., Angelini, R., Ascenzi, P., and Polticelli, F. (2016). Molecular evolution of alternative oxidase proteins: a phylogenetic and structure modeling approach. J. Mol. Evol. 82, 207–218. doi: 10.1007/s00239-016-9738-8
Polidoros, A. N., Mylona, P. V., and Arnholdt-Schmitt, B. (2009). Aox gene structure, transcript variation and expression in plants. Physiol. Plant. 137, 342–353. doi: 10.1111/j.1399-3054.2009.01284.x
Poole, R. K., and Cook, G. M. (2000). Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv. Microb. Physiol. 43, 165–224. doi: 10.1016/s0065-2911(00)43005-5
Reid, G. A., and Ingledew, W. J. (1979). Characterization and phenotypic control of the cytochrome content of Escherichia coli. Biochem. J. 182, 465–472. doi: 10.1042/bj1820465
Rhoads, D. M., and Subbaiah, C. C. (2007). Mitochondrial retrograde regulation in plants. Mitochondrion 7, 177–194. doi: 10.1016/j.mito.2007.01.002
Rhoads, D. M., Umbach, A. L., Sweet, C. R., Lennon, A. M., Rauch, G. S., and Siedow, J. N. (1998). Regulation of the cyanide-resistant alternative oxidase of plant mitochondria: Identification of the cysteine residue involved in α-keto acid stimulation and intersubunit disulfide bond formation. J. Biol. Chem. 273, 30750–30756. doi: 10.1074/jbc.273.46.30750
Rich, P. R.. (1978). Quinol oxidation in Arum maculatum mitochondria and its application to the assay, solubilisation and partial purification of the alternative oxidase. FEBS Lett. 96, 252–256. doi: 10.1016/0014-5793(78)80412-8
Rich, P. R., and Moore, A. L. (1976). The involvement of the protonmotive ubiquinone cycle in the respiratory chain of higher plants and its relation to the branchpoint of the alternate pathway. FEBS Lett. 65, 339–344. doi: 10.1016/0014-5793(76)80142-1
Saisho, D., Nambara, E., Naito, S., Tsutsumi, N., Hirai, A., and Nakazono, M. (1997). Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Mol. Biol. 35, 585–596. doi: 10.1023/A:1005818507743
Selinski, J., Hartmann, A., Höfler, S., Deckers-Hebestreit, G., and Scheibe, R. (2016). Refined method to study the posttranslational regulation of alternative oxidases from Arabidopsis thaliana in vitro. Physiol. Plant. 157, 264–279. doi: 10.1111/ppl.12418
Selinski, J., Hartmann, A., Kordes, A., Deckers-Hebestreit, G., Whelan, J., and Scheibe, R. (2017). Analysis of posttranslational activation of alternative oxidase isoforms. Plant Physiol. 174, 2113–2127. doi: 10.1104/pp.17.00681
Selinski, J., Scheibe, R., Day, D. A., and Whelan, J. (2018). Alternative oxidase is positive for plant performance. Trends Plant Sci. 23, 588–597. doi: 10.1016/j.tplants.2018.03.012
Shiba, T., Kido, Y., Sakamoto, K., Inaoka, D. K., Tsuge, C., Tatsumi, R., et al. (2013). Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc. Natl. Acad. Sci. U. S. A. 110, 4580–4585. doi: 10.1073/pnas.1218386110
Siedow, J. N., and Girvin, M. E. (1980). Alternative respiratory pathway. Plant Physiol. 65, 669–674. doi: 10.1104/pp.65.4.669
Siedow, J. N., and Umbach, A. L. (2000). The mitochondrial cyanide-resistant oxidase: Structural conservation amid regulatory diversity. Biochim. Biophys. Acta - Bioenerg. 1459, 432–439. doi: 10.1016/S0005-2728(00)00181-X
Smith, C. A., Melino, V. J., Sweetman, C., and Soole, K. L. (2009). Manipulation of alternative oxidase can influence salt tolerance in Arabidopsis thaliana. Physiol. Plant. 137, 459–472. doi: 10.1111/j.1399-3054.2009.01305.x
Sreerama, N., and Woody, R. W. (2000). Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260. doi: 10.1006/abio.2000.4880
Swathi, M., Lokya, V., Swaroop, V., Mallikarjuna, N., Kannan, M., Dutta-Gupta, A., et al. (2014). Structural and functional characterization of proteinase inhibitors from seeds of Cajanus cajan (cv. ICP 7118). Plant Physiol. Biochem. 83, 77–87. doi: 10.1016/j.plaphy.2014.07.009
Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Biotechnology. 24, 145–149. doi: 10.3389/fpls.2020.00566
Umbach, A. L., Gonzàlez-Meler, M. A., Sweet, C. R., and Siedow, J. N. (2002). Activation of the plant mitochondrial alternative oxidase: insights from site-directed mutagenesis. Biochim. Biophys. Acta - Bioenerg. 1554, 118–128. doi: 10.1016/S0005-2728(02)00219-0
Umbach, A. L., Ng, V. S., and Siedow, J. N. (2006). Regulation of plant alternative oxidase activity: A tale of two cysteines. Biochim. Biophys. Acta - Bioenerg. 1757, 135–142. doi: 10.1016/j.bbabio.2005.12.005
Umbach, A. L., and Siedow, J. N. (1993). Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol. 103, 845–854. doi: 10.1104/pp.103.3.845
Vanlerberghe, G. C.. (2013). Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 14, 6805–6847. doi: 10.3390/ijms14046805
Vanlerberghe, G. C., and McIntosh, L. (1997). Alternative oxidase: from gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 703–734. doi: 10.1146/annurev.arplant.48.1.703
Vanlerberghe, G. C., McIntosh, L., and Yip, J. Y. H. (1998). Molecular localization of a redox-modulated process regulating plant mitochondrial electron transport. Plant Cell 10, 1551–1560. doi: 10.1105/tpc.10.9.1551
Vishwakarma, A., Bashyam, L., Senthilkumaran, B., Scheibe, R., and Padmasree, K. (2014). Physiological role of AOX1a in photosynthesis and maintenance of cellular redox homeostasis under high light in Arabidopsis thaliana. Plant Physiol. Biochem. 81, 44–53. doi: 10.1016/j.plaphy.2014.01.019
Vishwakarma, A., Dalal, A., Tetali, S. D., Kirti, P. B., and Padmasree, K. (2016). Genetic engineering of AtAOX1a in Saccharomyces cerevisiae prevents oxidative damage and maintains redox homeostasis. FEBS Open Bio 6, 135–146. doi: 10.1002/2211-5463.12028
Vishwakarma, A., Kumari, A., Mur, L. A. J., and Gupta, K. J. (2018). A discrete role for alternative oxidase under hypoxia to increase nitric oxide and drive energy production. Free Radic. Biol. Med. 122, 40–51. doi: 10.1016/j.freeradbiomed.2018.03.045
Vishwakarma, A., Tetali, S. D., Selinski, J., Scheibe, R., and Padmasree, K. (2015). Importance of the alternative oxidase (AOX) pathway in regulating cellular redox and ROS homeostasis to optimize photosynthesis during restriction of the cytochrome oxidase pathway in Arabidopsis thaliana. Ann. Bot. 116, 555–569. doi: 10.1093/aob/mcv122
Wagner, A. M., and Moore, A. L. (1997). Structure and function of the plant alternative oxidase: Its putative role in the oxygen defence mechanism. Biosci. Rep. 17, 319–333. doi: 10.1023/A:1027388729586
Whelan, J., Millar, A. H., and Day, D. A. (1996). The alternative oxidase is encoded in a multigene family in soybean. Planta 198, 197–201. doi: 10.1007/BF00206244
Whitmore, L., and Wallace, B. A. (2008). Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400. doi: 10.1002/bip.20853
Xu, F., Copsey, A. C., Young, L., Barsottini, M. R. O., Albury, M. S., and Moore, A. L. (2021). Comparison of the kinetic parameters of alternative oxidases from Trypanosoma brucei and Arabidopsis thaliana—A Tale of Two Cavities. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.744218
Yap, L. L., Lin, M. T., Ouyang, H., Samoilova, R. I., Dikanov, S. A., and Gennis, R. B. (2010). The quinone-binding sites of the cytochrome bo3 ubiquinol oxidase from Escherichia coli. Biochim. Biophys. Acta - Bioenerg. 1797, 1924–1932. doi: 10.1016/j.bbabio.2010.04.011
Yoshida, K., Terashima, I., and Noguchi, K. (2006). Distinct roles of the cytochrome pathway and alternative oxidase in leaf photosynthesis. Plant Cell Physiol. 47, 22–31. doi: 10.1093/pcp/pci219
Yu, Y., Guan, Y., Liu, J., Hedi, W., Yu, Y., and Zhang, T. (2021). Molecular structural modification of egg white protein by pH-shifting for improving emulsifying capacity and stability. Food Hydrocoll. 121, 107071. doi: 10.1016/j.foodhyd.2021.107071
Zhang, L., Oh, Y., Li, H., Baldwin, I. T., and Galis, I. (2012). Alternative oxidase in resistance to biotic stresses: Nicotiana attenuata AOX contributes to resistance to a pathogen and a piercing-sucking insect but not Manduca sexta Larvae. Plant Physiol. 160, 1453–1467. doi: 10.1104/pp.112.200865
Zhang, L. T., Zhang, Z. S., Gao, H. Y., Xue, Z. C., Yang, C., Meng, X. L., et al. (2011). Mitochondrial alternative oxidase pathway protects plants against photoinhibition by alleviating inhibition of the repair of photodamaged PSII through preventing formation of reactive oxygen species in Rumex K-1 leaves. Physiol. Plant. 143, 396–407. doi: 10.1111/j.1399-3054.2011.01514.x
Zhang, Q., Hoefnagel, M. H. N., and Wiskich, J. T. (1996). Alternative oxidase from Arum and soybean: Its stabilization during purification. Physiol. Plant. 96, 551–558. doi: 10.1111/j.1399-3054.1996.tb00226.x
Keywords: Arabidopsis thaliana, alternative oxidase, respiratory inhibitors, mutational docking, AOX structure, recombinant protein, circular dichroism, surface plasmon resonance
Citation: Sankar TV, Saharay M, Santhosh D, Vishwakarma A and Padmasree K (2022) Structural and Biophysical Characterization of Purified Recombinant Arabidopsis thaliana's Alternative Oxidase 1A (rAtAOX1A): Interaction With Inhibitor(s) and Activator. Front. Plant Sci. 13:871208. doi: 10.3389/fpls.2022.871208
Received: 07 February 2022; Accepted: 27 April 2022;
Published: 16 June 2022.
Edited by:
Anthony L. Moore, University of Sussex, United KingdomReviewed by:
Fei Xu, Wuhan Institute of Bioengineering, ChinaAllison McDonald, Wilfrid Laurier University, Canada
Copyright © 2022 Sankar, Saharay, Santhosh, Vishwakarma and Padmasree. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kollipara Padmasree, a3Bzc2xAdW9oeWQuYWMuaW4=
 Tadiboina Veera Sankar
Tadiboina Veera Sankar Moumita Saharay
Moumita Saharay Dharawath Santhosh
Dharawath Santhosh Abhaypratap Vishwakarma
Abhaypratap Vishwakarma Kollipara Padmasree
Kollipara Padmasree