- 1Chengdu Academy of Agricultural and Forestry Sciences, Chengdu, China
- 2Chengdu Research Branch, National Rapeseed Genetic Improvement Center, Chengdu, China
- 3Agricultural College, Sichuan Agricultural University, Chengdu, China
- 4Maize Research Institute, Sichuan Agricultural University, Chengdu, China
- 5Key Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu, China
- 6Rice Research Institute, Sichuan Agricultural University, Chengdu, China
When homozygously fertile plants were induced using doubled haploid (DH) induction lines Y3380 and Y3560, the morphology of the induced F1 generation was basically consistent with the female parent, but the fertility was separated, showing characteristics similar to cytoplasmic male sterile (CMS) and maintainer lines. In this study, the morphology, fertility, ploidy, and cytoplasm genotype of the induced progeny were identified, and the results showed that the sterile progeny was polima cytoplasm sterile (pol CMS) and the fertile progeny was nap cytoplasm. The molecular marker and test-cross experimental results showed that the fertile progeny did not carry the restorer gene of pol CMS and the genetic distance between the female parent and the offspring was 0.002. This suggested that those inductions which produced sterile and fertile progeny were coordinated to CMS and maintainer lines. Through the co-linearity analysis of the mitochondrial DNA (mtDNA), it was found that the rearrangement of mtDNA by DH induction was the key factor that caused the transformation of fertility (nap) into sterility (pol). Also, when heterozygous females were induced with DH induction lines, the induction F2 generation also showed the segregation of fertile and sterile lines, and the genetic distance between sterile and fertile lines was approximately 0.075. Therefore, the induction line can induce different types of female parents, and the breeding of the sterile line and the maintainer line can be achieved through the rapid synchronization of sister crosses and self-crosses. The induction of DH inducer in B. napus can provide a new model for the innovation of germplasm resources and open up a new way for its application.
Introduction
Brassica napus belongs to the Cruciferae (Chalhoub et al., 2014) and evolved as an allo-tetraploid crop from Brassica rapa (AA, 2n = 20) and Brassica oleracea (CC, 2n = 18) through interspecific hybridization and natural doubling (NU, 1935; Palmer et al., 1983; Bancroft et al., 2011; Sun et al., 2017). Since B. napus has two sets of heterologous chromosome sets (A/C; Olsson, 1960; Leflon et al., 2006; Allender and King, 2010; Chalhoub et al., 2014; Hu et al., 2021), the cycle of selfing to obtain homozygous lines is long. Although anther or microspore culture techniques can obtain homozygous line plants (Keller and Armstrong, 1978; Lichter, 1982; Kott et al., 1998; Xu et al., 2007; Shariatpanahi and Ahmadi, 2016) and have the potential to develop improved varieties (Custers, 2003; Ferrie and Caswell, 2011), they are influenced by some conditions such as culture temperature (Custers et al., 1994), plant development stage (Takahata et al., 1991), and genotype of donor plants (Takahata et al., 1996; Barro and Martín, 1999; Li and Devaux, 2001). B. napus is an early crop to use CMS lines for heterosis utilization (Fu et al., 1990). In 1973, Prof. Fu Tingdong from Huazhong Agricultural University in China discovered polima cytoplasm male sterile line (pol CMS) (Fu, 1981), which opened the prelude of heterosis utilization of B. napus (Witt et al., 1991). Although the traditional cytoplasmic three lines technology (maintainer line, CMS line, restorer line) has been applied to rapeseed production on a large scale (Fu et al., 1989, 1990; Li et al., 1997), the breeding and improvement of CMS had a long and difficult cycle, requiring 6–12 generations. Generally, the new CMS lines are bred by backcrossing stable or largely stable maintainer lines in pairs for multiple generations with the original CMS lines. The application of biotechnology, such as gene editing, has enabled the rapid creation of male-sterile lines and maintainers, but the technology is difficult at the research stage because of the stability and transgenic safety issues (Du et al., 2020).
Since the discovery of haploids in Datura stramonium in the early 1920s (Blakeslee et al., 1922), the methods to obtain pure lines by haploid induction lines and distant hybridization have been widely used in maize (Pollacsek, 1992; Zhao et al., 2013), rice (Niizeki and Oono, 1968), wheat (Laurie and Bennett, 1986; Laurie, 1989), Arabidopsis (Ravi et al., 2014), and other plants (Burk et al., 1979; Forster et al., 2007; Chen et al., 2011b). The induction efficiency of haploid induction lines represented by maize “Stock 6” was about 2% (Coe, 1959). Now the efficiency of haploid induction has been increased to more than 10% by improvement (Eder and Chalyk, 2002; Geiger and Gordillo, 2009; Rotarenco et al., 2010). Therefore, in vivo haploid induction and artificial chromosome doubling can obtain homozygous lines more quickly and easily (Chase, 1952; Palmer et al., 1996; Jacquier et al., 2020). In recent years, Fu et al. (2018) artificially synthesized two allo-octoploid (AAAACCCC, 2n = 8X ≈ 76) rapes of doubled haploid induction lines, Y3560 and Y3380, to provide a new way for obtaining stable pure lines (maintainer line or restorer line) in rapeseed. Y3560, Y3380, and their parents P3-2 have changes in the methylation patterns of pollen (Yin et al., 2021) and have abnormal meiosis (Yin et al., 2020) to produce aneuploid gametes, which exhibited strong induction ability (Luo et al., 2021). When the DH induction lines were used as the male parent to pollinate different genotypes of B. napus, the induction effect was found to be influenced by maternal karyogene and cytoplasmic genotype (Zhang et al., 2021). Therefore, it suggests differences in the induction mechanism of DH induction lines in B. napus by haploid induction in maize and Arabidopsis. Meanwhile, DH inducer can be used to quickly obtain interspecific hybrid offspring and broaden genetic resources (Zhou et al., 2022), and it can realize multiple gene homoeologs editing of B. napus and B. oleracea using the DH induction line Y3380-mediated editing system, which brings hope to rapeseed and cruciferous vegetable breeding that are difficult to be gene-edited through genetic transformation (Li et al., 2021). In the process of induction, it was found that when the DH induction line Y3380 and Y3560 induced homozygously fertile parents, the morphology of the induced F1 generation plants was almost similar to that of the female parent, but sterile and fertile segregation occurred, showing similar characteristics of CMS and maintainer lines. During the induction process, it was found that the morphology of the F1 generation plants induced by the DH induction lines Y3380 and Y3560 when inducing homozygous fertile parents was basically similar to that of the female parents (Zhang et al., 2021). However, the F1 generation showed segregation of sterile and fertile plants in terms of fertility, while exhibiting similar characteristics of CMS and maintainer lines. Based on these phenomena, we performed morphological and cytological observations combined with molecular markers to identify the fertility and cytoplasm genotypes between parents and offspring before and after induction. We further performed mitochondrial DNA co-linearity analysis with genome resequencing and mitochondrial genome assembly. These data will provide a theoretical basis for rapid and simultaneous breeding of CMS lines and maintainer lines, and provide a new insight for accelerating and enriching the use of hybrid advantage in B. napus.
Materials and Methods
Plants Materials and Cultivation Conditions
Information on the main plant materials for this study is presented in Table 1, in which the control male parent 20–2,386 was a normal tetraploid pol restorer line (F22). The female parent 0933A was pol CMS (BC15). Z1732 was their hybrid F1. The rape DH induction lines Y3560 and Y3380 were bred by Fu Shaohong of Chengdu Academy of Agriculture and Forestry Sciences through artificial synthesis in 2011 and were identified as allo-octoploid rapes by cytology and flow cytometry (AAAACCCC, 2n = 8x≈ 76; Fu et al., 2018). The 0933B is the 0933A maintainer line and is a normal tetraploid pol maintainer line (F20). ZY21-1 and ZY21A-1 are the induced progeny with 0933B as the female parent and Y3560 as the male parent. The female parent L0933A is normal ogu CMS (BC10) and the maintainer line is 0933B, which is the same as pol 0933A maintainer line. ZY26A-1 is a hybrid hexaploid progeny with L0933A as the female parent and the induction line Y3380 as the male parent (Table 1). To avoid mixing during planting, all planting material were planted in pots at a young stage and each plant was marked. Plants were subsequently transplanted to the field after the determination of plant ploidy by flow cytometry. All plants were hand-pollinated at flowering and seeds were obtained by single threshing. The plant material in this experiment was grown under the same conditions in October 2019 in an experimental field in Wenjiang, Chengdu, China (E103.83, N30.70).
Field Identification and Sampling of Plant Morphology and Fertility
Based on the fertility characteristics, fertile or sterile was identified as a marker trait by manual observation in the field. The morphology of pol CMS plants was not significantly different from that of the normal fertile plants, but the anthers were shrunken and there was no pollen or only a small amount of pollen at low temperature, while ogu types of sterile lines did not produce pollen. According to the results of fertility identification, the individual plants with the same morphology as the female parent were sampled and stored at −80°C for future use.
DNA Extraction and PCR Amplification
All parents and offspring associated with this study were identified by molecular markers. The total DNA was extracted by referring to Doyle’s classical CTAB method. PCR amplification procedure: (1) restorer gene amplification procedure (Zhang et al., 2017): 94°C, 5 min, 94°C, 30 s, Tm, 30 s, 72°C, 45 s (Goto Step2, 35 cycles), 72°C, 8 min, 10°C for holding. (2) Cytoplasmic primer amplification program 94°C, 4 min, 94°C, 30 s, Tm, 30 s, 72°C, 1 min (Goto Step2, 24 cycles), 72°C, 8 min, 4°C keep warm. The setting of Tm was according to different primers (Heng et al., 2015). Among the cytoplasmic identification primers (Table 1), MSS4, MSS8, and MSS14 identified nap, pol, and ogu cytoplasm, respectively (Heng et al., 2015).
Chromosome Number Identification in Somatic Cells
Young flower buds of F1-generation (ZY21-1, ZY21A-1, and ZY26A-1) plants were treated with 8-hydroxyquinoline solution, placed in the dark for 3 h, and then fixed by Carnot fixative (ethanol: glacial acetic acid, 3:1) for 24 h and stored in 70% ethanol. Alcohol was removed with distilled before observation followed by immersion in 1 mol/L hydrochloric acid in a water bath at 60°C for 8–10 min. Carbol magenta staining was used to determine chromosome numbers under the microscope. Cytogenetic observations were made according to the process detailed in Fu et al. (2018).
Pollen Viability Identification
Fresh flower bud of different fertile progeny (ZY21-1, ZY21A-1) of the F1 generation at the blooming stage was collected separately. The anthers were placed on slides, crushed with forceps, and then 1–2 drops of magenta acetate solution were placed on coverslips, gently pressed, and observed under the microscope. When pollen grains are dark red, they are viable and light red indicates partial loss of viability; colorless, hollow, or deformed are dead and sterile pollen grains. Cytogenetic observations were made according to the process detailed in Yin et al. (2020).
Anther Freezing Section
Three flower buds of different lengths from different fertility plants (ZY21-1, ZY21A-1) in the induced F1 were selected separately. The anthers were separated from the flower buds and dehydrated. The dehydrated anthers were placed in an embedding box containing 1/3 of the embedding solution and frozen at −20°C. After complete solidification, 1/3 of the embedding solution was added and frozen at −20°C for complete solidification and then prepared for use. The slides were stained with eosin staining solution for about 15 s. The slides were washed with water to remove excess staining solution, dried, and placed under a microscope for observation and storage.
Flow Cytometry Detection
Fresh young leaves of the tested plants (all induced offspring) were quantitatively taken with a 5-mm diameter punch and placed in a pre-chilled culture dish. Then 0.5 ml of pre-chilled cell lysis buffer was added and the leaves were quickly chopped and filtered through a 300-mesh filter in a 2-ml EP tube. A total of 1.5 ml PI (propidium canonical) staining solution (50 μg/mL PI, 50 μg/mL RNase) was added and stored for 30 min in a place protected from light. The samples were analyzed using a flow cytometer (Accuri® C6 Plus, BD) and analyzed using the accompanying Accuri C6plus software. The specific operation procedure is described in Yin et al. (2020).
Whole-Genome Resequencing
Sampling was performed by manually identifying the fertility of the plants (a total of 32 materials are involved). DNA was extracted from the collected plant leaves, and the DNA samples of acceptable quality were sent to Biomarker Technologies Co., Ltd. for next-generation sequencing at a depth of 10X. Sequencing data were used to calculate Nei genetic distance for SNP data and to perform inter-sample cluster analysis.
Assembly and Annotation of the Mitochondrial Genome
The analysis of the mitochondrial genome mainly involved a total of 10 materials (Y3380, Y3560, 0933A, 0933B, L0933A, ZY21-1, ZY21A-1, ZY26A-1, 20-2386, and Z1732). Based on the raw data obtained from whole-genome resequencing, sequences were analyzed and extracted according to the mitochondrial genome characteristics (Albertsen et al., 2013; Zhou et al., 2013), and finally, the assembly and annotation (Supplementary Figure 3 and Figures 1A,B) of the mitochondrial genome was completed by Wuhan Yiersan Biotechnology Co., Ltd. The primers (Supplementary Table 1) were designed to amplify and sequence at both ends of the linked overlapping cluster contig to verify the splice region and obtain the complete mtDNA (Supplementary Figures 2B–D). The covariance between mitochondrial genomes was analyzed using NCBI Blast,1 co-linearity analysis software Mauve. DNAman was also used to detect the identity between mitochondrial genomes. Among them, the induced lines Y3560, Y3380, 20-2386, and Z1732 unified the cut positions to compare the sequence alignment among mitochondrial genomes more clearly (Figures 1C–F).
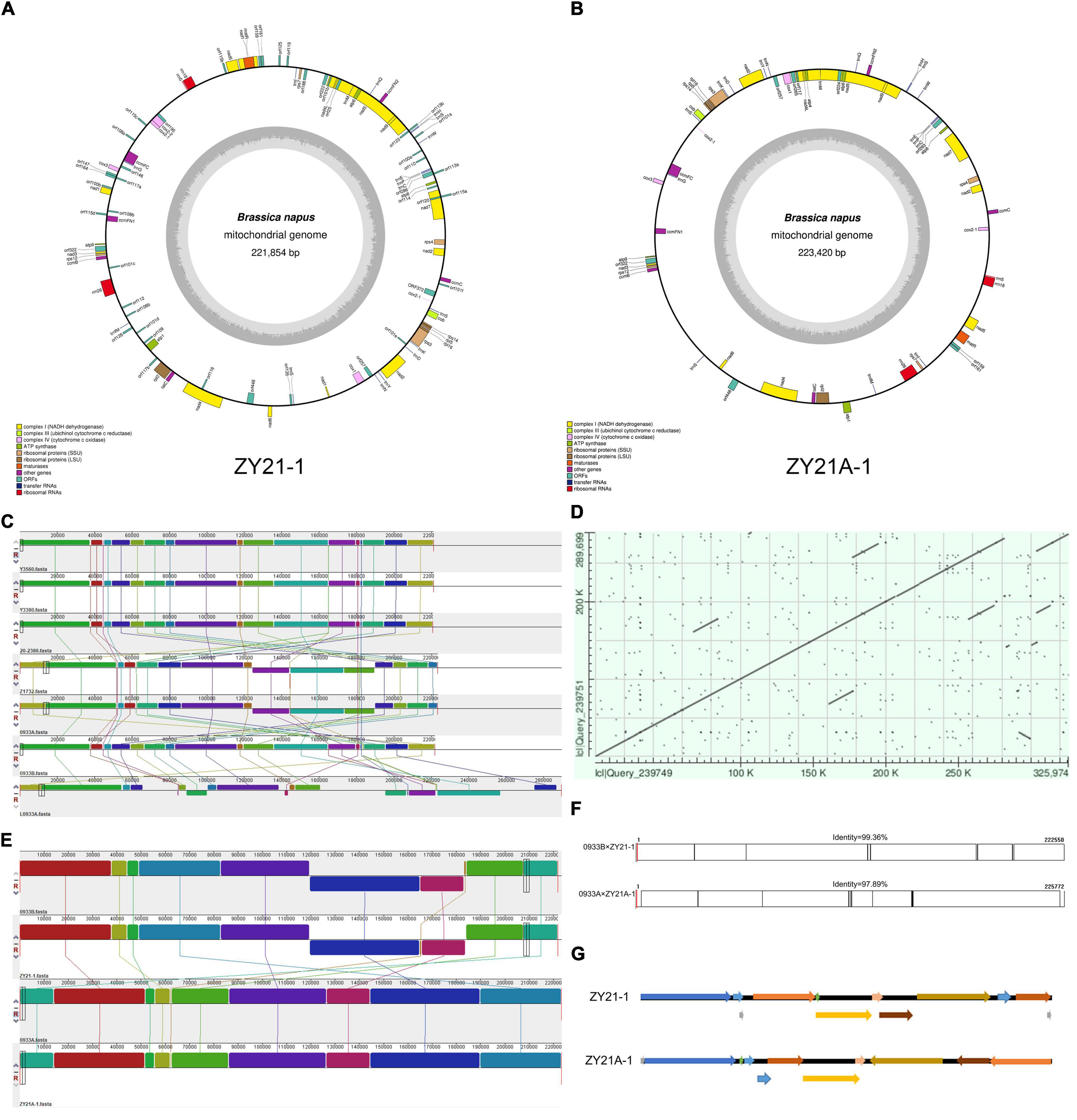
Figure 1. Results of mitochondrial genome co-linearity analysis for different samples. (A) Annotated map of the mitochondrial genome of induced F1 generation fertile progeny ZY21-1. (B) Annotated map of the mitochondrial genome of induced F1 generation sterile progeny ZY21A-1. (C) Schematic diagram of mitochondrial genome co-linearity between parental and hybrid F1 generation before and after induction, from top to bottom, Y3560, Y3380, 20-2386, Z1732, 0933A, 0933B, L0933A. (D) Comparison of the mitochondrial genome between hybrid hexaploid ZY26A-1 and its female parent L0933A. (E) Schematic diagram of mitochondrial genomic covariance between induced F1 generation and 0933A and 0933B, from top to bottom, 0933B, ZY21-1, 0933A, ZY21A-1. (F) Comparison of mitochondrial sequences between 0933B and ZY21-1; comparison of mitochondrial sequences between 0933A and ZY21-1. (G) Comparison of mitochondrial genome arrangement of different fertile single plants in the induced F1 generation ZY21-1and ZY21A-1.
Functional Annotation and Enrichment
The mutation loci were screened in whole-genome re-sequencing and ZS11 was used as a reference to annotate and enrich the genes where the mutation loci are located. Subsequently, GO and KEGG annotation information2 were integrated using Perl language. The KEGG annotation results were also enriched through the KEGG online website.3
Results
Flower Morphology and Cytoplasm Genotype Identification
It was found that the induced F1 generation plants produced when the induction lines induced the homozygous female parent were consistent with the female parent in morphology and ploidy, but the segregation for fertility had occurred (Zhang et al., 2021). Therefore, the induced F1 generation ZY21-1 (fertile) and ZY21A-1 (sterile) with different fertility were identified for the homozygous maintainer line (0933B, fertile) as the female parent and the induction line Y3560 (fertile) as the male parent. The floral organs of the fertile progeny ZY21-1, including sepals, petals, stamens, and nectaries were normally developed (Figures 2A–C). They had high pollen fertility (Figure 2D) and the tapetum was able to develop normally to produce spores (Figures 2I–K). In contrast, the sterile progeny ZY21A-1 had reduced floral organ size, slightly ruffled petals, reduced anther length, shrunken stigma (Figures 2E–G), and only a small amount of pollen (Figure 2H), which might be due to the degeneration of the tapetum resulting in abnormal microspore development (Figures 2L–N); consequently no pollen or only a small amount of pollen was produced (Figures 2F–H). Since the DH induction line Y3560 is an allo-octoploid B. napus, pollination to the tetraploid fertile plant 0933B could produce ploidy hybridization, resulting in chromosome increase or elimination, leading to the development of sterility. Further, the ploidy and chromosome observation of the sterile and fertile progeny after induction showed that the ploidy of the induced F1 generation fertile progeny ZY21-1 (Figure 2O) and sterile progeny ZY21A-1 (Figure 2Q) was consistent, and they were both tetraploid (400.0–500.0D thousand lines) with 38 chromosomes (Figures 2P,R). The results indicated that the appearance of sterile plants has not resulted from chromosome gain or loss after ploidy hybridization. To further clarify whether the cytoplasm genotypes of the induced F1 generation were changed, the cytoplasm genotypes of the parents and the offspring before and after induction were identified by cytoplasm identification primers (Heng et al., 2015). The results showed that the induction lines Y3560 and Y3380 were nap cytoplasm and the female parental 0933B was nap cytoplasm, 0933A was pol cytoplasm (0933B was the maintainer line of the high generation self-cross of 0933A), and L0933A was ogu cytoplasm (Figure 3A). Further, the hybrid F1 generation Z1732 (fertile) and ZY26A-1 (sterile) were identified (Figure 3A), and the results showed that the cytoplasm of the hybrid progeny (Z1732, ZY26A-1) was the same as the cytoplasm of the female parent (0933A, L0933A). The fertile progeny ZY21-1 of the induced F1 generation was nap cytoplasm, and the sterile progeny ZY21A-1 was pol CMS (Figure 3A), indicating that the CMS progeny ZY21A-1 that appeared in the induced F1 generation was consistent with the speculation of the flower morphology observation.
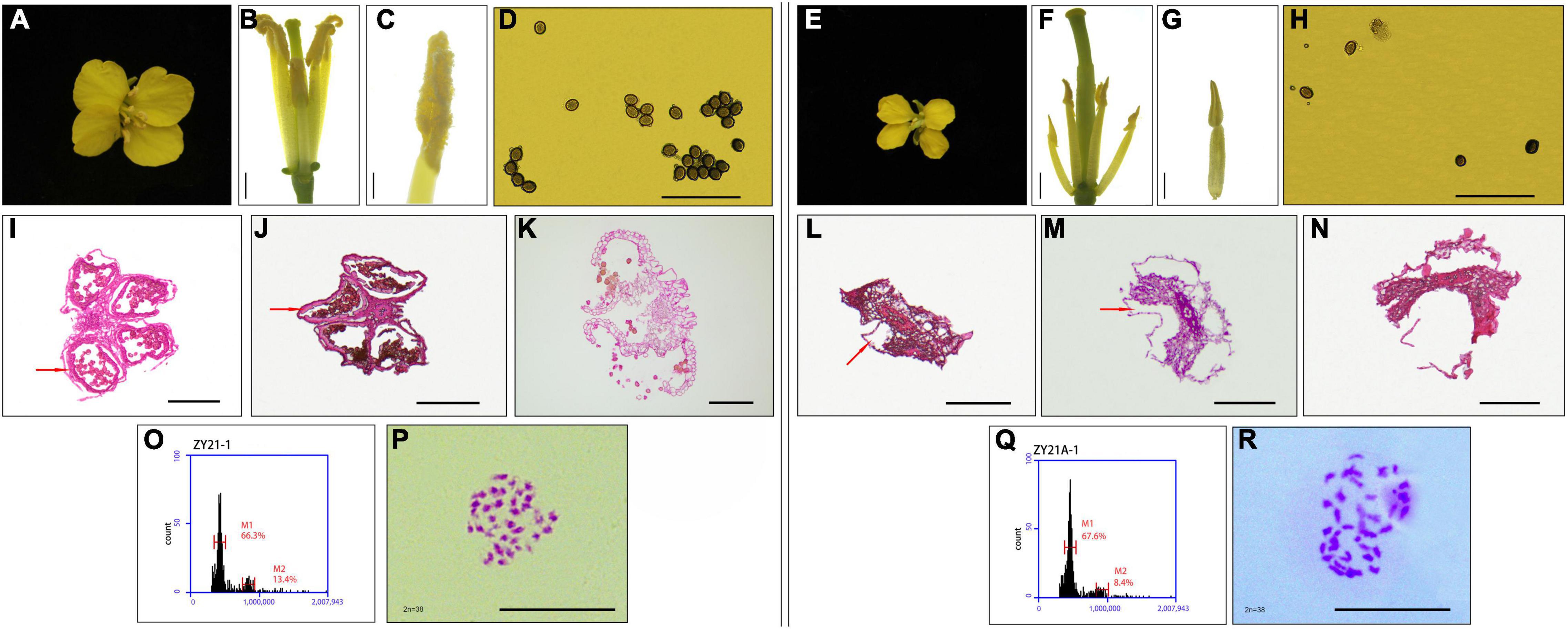
Figure 2. Fertility and ploidy identification of different fertility progeny (ZY21-1 and ZY21A-1) in the induced F1 generation. (A) Flower morphology of induced F1-generation fertile progeny ZY21-1. (B,C) Anther morphology of induced F1-generation fertile progeny (ZY21-1), (B) bar = 2mm, (C) bar = 1mm. (D) Pollen activity assay results of induced F1-generation fertile progeny (ZY21-1). (E) Flower morphology of induced F1 generation sterile progeny ZY21A-1. (F,G) Anther morphology of induced F1 generation sterile progeny (ZY21A-1), Bar = 2mm. (H) Pollen activity assay results of induced F1 generation sterile progeny (ZY21A-1). (I–K) Induced F1 fertile progeny (ZY21-1) with different flower bud lengths (0.2–0.25cm, 0.25–0.3cm, > 0.3cm) of anther slices, in which the red-brown arrow points to the tapetum layer. Bar = 100μm. (L–N) Induced F1 sterile progeny (ZY21A-1) with different flower bud lengths (0.25–0.3cm, 0.3–0.4cm, > 0.4cm) of anther slices. The position indicated by the red-brown arrow indicates that the tapetum layer has degenerated. Bar = 100μm. (O) Flow cytometry results of induced F1 generation fertile progeny ZY21-1. (P) Number of the chromosome of induced F1 generation fertile progeny ZY21-1, Bar = 10μm. (Q) Flow cytometry results of induced F1 generation sterile progeny ZY21A-1. (R) Number of the chromosome of induced F1 generation sterile progeny ZY21A-1, Bar = 10μm.

Figure 3. Identification results of cytoplasmic genotype and pol CMS recovery gene Rfp1. (A) Results of cytoplasmic identification with primers MSS-4 (nap), MSS-8 (pol) and MSS-14 (ogu). (B) Results of recovery gene identification, 1–9, respectively: 20-2386; 0933A control; Y3380-1, Y3380-2, Y3560-1, Y3560-2, 0933B, ZY21-1, ZY21A-1, and OPSNP7 are primers for the specific amplification of pol CMS restorer gene Rfp1.
In conclusion, the induction effect of the DH inducer was different from the crosses of parents with the same ploidy level. When the homozygous female parent 0933B with nap cytoplasm was induced, the flower morphology and cytoplasmic genotype of the sterile progeny were identified as pol CMS, that is, cytoplasmic changes occurred after induction.
Nei Genetic Distances Revealed a Relationship in the Nuclear Background of the Maternal Parent and Offspring Before and After Induction
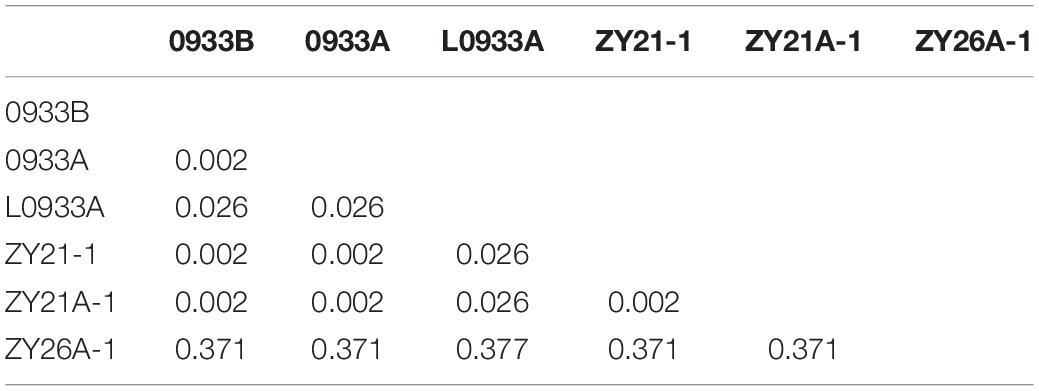
Due to the induction effect of the induction line, if 0933B converts nap cytoplasm into pol CMS, would the corresponding karyogene also change? Therefore, SNP loci (2296756 ∼ 3102779) of different samples from whole-genome resequencing data at 10x depth were examined and the Nei genetic distances (Table 2) were calculated by SNP loci data to assess the nuclear background between the maternal parent and offspring before and after induction. The results showed that the genetic distance between 0933B (maintainer lines) and 0933A (pol CMS) was only 0.002 (Table 2), while 0933B and 0933A were the maintainer (BC15) and CMS lines with the same nuclear background. However, 0933B and L0933A were genetically distant (0.026) (Table 2). The number of the back cross was less than that of 0933B and 0933A (BC10); therefore it was reliable to detect the genetic relationship between samples by Nei genetic distance. Subsequently, it was found that when 0933B was the female parent, the genetic distances between the induced F1 generation ZY21-1 and ZY21A-1 and 0933B were 0.002 (Table 2), indicating that the nuclear backgrounds between the induced F1 generation and the female parent were almost identical. However, the hybrid hexaploid progeny ZY26A-1 (Supplementary Figure 1) with L0933A as the female parent and the induction line Y3380 as the male parent had a genetic distance of 0.377 (Table 2) from the female parent.
The restorer genes corresponding to the pol CMS of the parents and offspring before and after induction were examined (Figure 3B and Supplementary Figure 2A), and it was found that the induction lines Y3560 and Y3380 carried the pol CMS restorer gene Rfp1 (Zhang et al., 2017), but there was segregation between different individual plants, probably due to genetic instability (Fu et al., 2018). The induced F1 generation fertile progeny ZY21-1 and the female parent 0933B did not carry the pol CMS restorer gene Rfp1. Meanwhile, the hybrid offspring Z1732, which was crossed with pol CMS restorer line 20-2386 and pol CMS 0933A, carried the restorer gene. These data indicate that the induction of the induction lines occurs and can cause changes in plant fertility and cytoplasm without affecting the nuclear background of the induced progeny, which will lay the foundation for the rapid and simultaneous acquisition of the maintainer lines and the corresponding sterile lines.
Co-linearity Analysis of Parental and Offspring Mitochondrial DNA
The Nei genetic distance indicated that the nuclear background between the female parent and offspring before and after induction was almost identical (Table 2), but the molecular marker identification results indicated that ZY21A-1 cytoplasm in the induced F1 generation was changed (Figure 3A). To further know whether the corresponding mtDNA controlling fertility is also changed and whether the production of sterile plants is caused by changes in the mitochondrial genome, we assembled and analyzed mtDNAs from a total of 10 samples before and after induction using whole-genome resequencing data (Figures 1A,B and Supplementary Figure 3). First, the mtDNAs of three female parents (0933B, 0933A, L0933A; Supplementary Figures 2B–D, 3A–C), the three male parents (DH induction lines Y3380, Y3560, pol restorer line 20-2386) (Supplementary Figures 3E–G), and one hybrid F1 generation (Z1732) (Supplementary Figure 3H) in this study were analyzed for co-linearity (Figure 1C), and it was found that the mtDNAs of the three male parents and the 0933B (nap) were almost identical, while the mtDNAs of the different female parents 0933B (nap), 0933A (pol,) and L0933A (ogu) differed significantly in the arrangement due to different cytoplasm genotypes (Figure 1A). However, the similarity in mitochondrial genome sequences between 0933A (pol) and 0933B (nap) was higher (Figure 1C), and the detection results were consistent with the molecular identification results and could be mutual verification.
Secondly, the hybrid F1 results showed that the mitochondrial genome of the F1 generation Z1732 (Supplementary Figure 3H) of the cross with 0933A (pol) (Supplementary Figure 3B) as the female parent and 20-2386 (nap) (Supplementary Figure 3G) as the male parent had the same mitochondrial genome sequence arrangement as that of the female parent 0933A (Supplementary Figure 3B). The results indicated that the inheritance of mtDNA in the normal crossing process followed maternal inheritance. In contrast, the mitochondrial genome of the hexaploid progeny ZY26A-1 (ogu) (Supplementary Figures 1, 3D), which was generated by crossing the induction line Y3380 (nap) (Supplementary Figure 3E) as the male and L0933A (ogu) (Supplementary Figure 3C) as the female parent, showed duplication of several fragments and partial sequence inversion compared with the female parent L0933A, which caused rearrangement of the ZY26A-1 mitochondrial genome and a significant increase in the genomic sequence length more than 36,275bp (Figure 1D and Supplementary Figures 3C,D). Subsequently, the mitochondrial genome of the induced F1 generation was compared with the same cytoplasmic genome, and it was found that the fertile progeny ZY21-1 had the same mitochondrial genome arrangement as the 0933B (nap) (Figure 1E), and the concordance was 99.36% (Figure 1F), while the mitochondrial genome arrangement of the sterile ZY21A-1 was the same as that of 0933A (pol) (Figure 1E) and the concordance was 97.89% (Figure 1F). These results showed that the genetic consequences of the crosses and the induction of plants with different ploidy levels and the hybridization of plants with the same ploidy level are different. Crossing and induction between different ploidy might cause changes in the mitochondrial genome, which might be affected by nucleo-cytoplasmic interactions. Together, the mitochondrial genome of the fertile F1 generation of ZY21-1 produced by induction of the inducer was the same as that of the female parent 0933B and did not carry the pol CMS restorer gene, while the CMS ZY21A-1 was the same as pol CMS (0933A). Therefore, since the nuclear background before and after induction is basically the same, it suggests that when the induced line is the male parent, the induced F1 generation is the relationship between the corresponding maintainer line (fertile F1 progeny) and the CMS line (sterile F1 progeny), and this process may be affected by the nuclear–cytoplasmic interaction.
Verification of Induction and Realization of Pol Cytoplasmic Male Sterile and Maintainer Line Synchronous Breeding
Through the application of various methods for the identification of induced F1 generation, it was finally shown that F1 generations produced by the inducer were correspondingly related to the CMS lines and the maintainer lines, and the reliability of the identification results by genetic correlation with plant morphology, ploidy, and molecular markers was confirmed. To further verify whether the differences generated by the DH induction lines could be inherited stably in the progeny, we studied self-pollinated progeny of fertile plants from induced F1 generation (ZY21-1, F2 generation) and the progeny of male–sterile plants obtained by sib-crossing with fertile plants (ZY21A-1 × ZY21A-1, BC1F1 generation). These progenies had similar growth phenotypes with different fertility (Figures 4A,B), and the ploidy was the same with the tetraploid (Figures 4C,D). The results showed that the cytoplasm genotypes of the induced F2 generation were the same as the induced F1 generation, and the cytoplasm of the fertile population was nap cytoplasm and the sterile population was pol cytoplasm (Figures 4E,F), and none of them contained the pol CMS restorer gene Rfp1. Therefore, the combination of morphology, ploidy, and molecular markers for the identification of the induced F2 and BC1F1 generations showed that the changes produced by the inducer could be inherited stably, and the breeding of the corresponding CMS lines could be achieved simultaneously and rapidly.

Figure 4. Identification results of different fertility populations (ZY21-1 F2 and ZY21A-1 × ZY21-1 (BC1F1) in the induced F2 generation. (A) Plant phenotypes of the induced F2 generation populations (ZY21-1 F2 and ZY21A-1 × ZY21-1 (BC1F1)) with different fertility populations. (B) Plant phenotypes of the different fertile progeny of the induced F2 generation population (ZY21-1 F2 and ZY21A-1 × ZY21-1 (BC1F1)), “F,” the fertile progeny ZY21-1 self-crossed F2 generation; “S,” the sterile progeny ZY21A-1 × ZY21-1 sister-crossed in the BC1F1 generation. (C) Flow cytometry results of induction of F2 generation plant of ZY21-1 self-crossed. (D) Flow cytometry results of induction of BC1F1 generation plant of ZY21A-1 × ZY21-1 sister-crossed. (E) Molecular markers result from induced F2 and BC1F1 generation with different fertility population cytoplasm genotypes. (F) Molecular markers result of induced F2 and BC1F1 generation pol restorer gene, and actin is internal reference gene.
Functional Annotation and Enrichment Analysis of the Genes in Which the Mutant Loci Are Located
To further explore the causes of cytoplasmic type changes, we screened SNP and Indel mutant loci between the parent and the offspring by re-sequencing and a total of 286,552 mutant loci were obtained. Among them, the fertile offspring ZY21-1 produced by induction had 1,064 mutant loci involving 211 genes, the sterile offspring ZY21A-1 had 1,100 mutant loci involving 258 genes, and the hybrid offspring ZY26A-1 had 11,842 mutant loci involving 1,319 genes. The genes where the mutant loci were located were also functionally annotated and enriched. The GO enrichment results showed that the differential loci were mainly enriched in protein binding (Supplementary Figures 4A–C) when the induced lines were used as male parents. However, compared to ZY21-1, ZY21A-1 and ZY26A-1 with significant changes in the mitochondrial genome had more genes related to protein kinase activity and other pathways, and more genes of the same pathway and a wider range of different pathways (Supplementary Figures 4A–C). Also, the KEGG Brite enrichment results showed that the genes corresponding to some mutant loci in the F1 generation sterile offspring ZY21A-1 and ZY26A-1 with significant changes in the mitochondrial genome were involved in the mitochondrial biogenesis, while the induced F1 generation fertile offspring ZY21-1 without significant changes in the mitochondrial genome was not found to have the same results. In conclusion, the functional annotation and enrichment of the genes at the mutant loci suggest that the induced lines as parents are different from conventional crosses in that they can cause changes in the plants to some extent through protein interactions and in the regulation of the metabolic pathways that eventually feed back to the plants.
Genetic Distance and Clustering Analysis Among the F2 Generation of the Induced Heterozygous Female Parent
The induction of homozygous female parents resulted in the rapid and simultaneous acquisition of both maintainer and sterile populations, but did this phenomenon apply only to the induction of homozygous female parents, can it also occur in heterozygous maternal parents of other cytoplasmic types? To verify whether the induction effect also existed in heterozygous female parent with different cytoplasm types (pol, ogu), we induced hybrid F1 of pol and ogu cytoplasm using Y3380 and Y3560 as the male parent. The results showed that the induced F1 generation was also segregated for fertility, but since the nuclear genes of the female parents were heterozygous, the F1 generation itself may also be heterozygous for nuclear genes. To exclude such differences, we self-crossed the induced F1 fertile plants from the heterozygous parents to obtain the induced F2 generation population. Through the fertility identification of the F2 generation population, the sterile plant was 15–34.21% (Supplementary Table 3). Based on these results, the fertile and sterile plants in the induced F2-generation population generated by inducing with pol and ogu cytoplasm types were randomly selected for whole-genome resequencing, and the number of SNP loci between 1857507 and 2273984 from resequencing were identified. The Nei genetic distance was calculated and clustered. The results showed that the genetic distance variation between the hybrid progeny Z1732 and other induced F2 generation ranged from 0.487 to 0.334 (Supplementary Table 2), with significant differences in the nuclear background. The genetic distance variation between paired fertile and sterile F2 individual plants from the same self-cross induced F1 plant ranged from 0.042 to 0.114, with an average of 0.075, and most of the genetic distances were 0.07–0.08 (Supplementary Table 2). The genetic distances between the induced progeny were closer than those between the hybrid progeny, and the nuclear backgrounds of these induced F2 generations are thought to be nearly identical between the individual plants of fertility and sterility (Supplementary Table 2). Also, the different fertility plants in the same induced F2 generation were clustered due to the same nuclear background, which was consistent with the actual lineage (Figure 5). These results indicated that the induction lines were capable of inducing fertility segregated from both homozygous and heterozygous maternal parents, and the segregated progeny could be rapidly stabilized by self and sister-crosses to eventually form the corresponding maintainer and CMS lines, thus the induction lines had a potential application in achieving simultaneous and rapid breeding of maintainers and CMS lines.
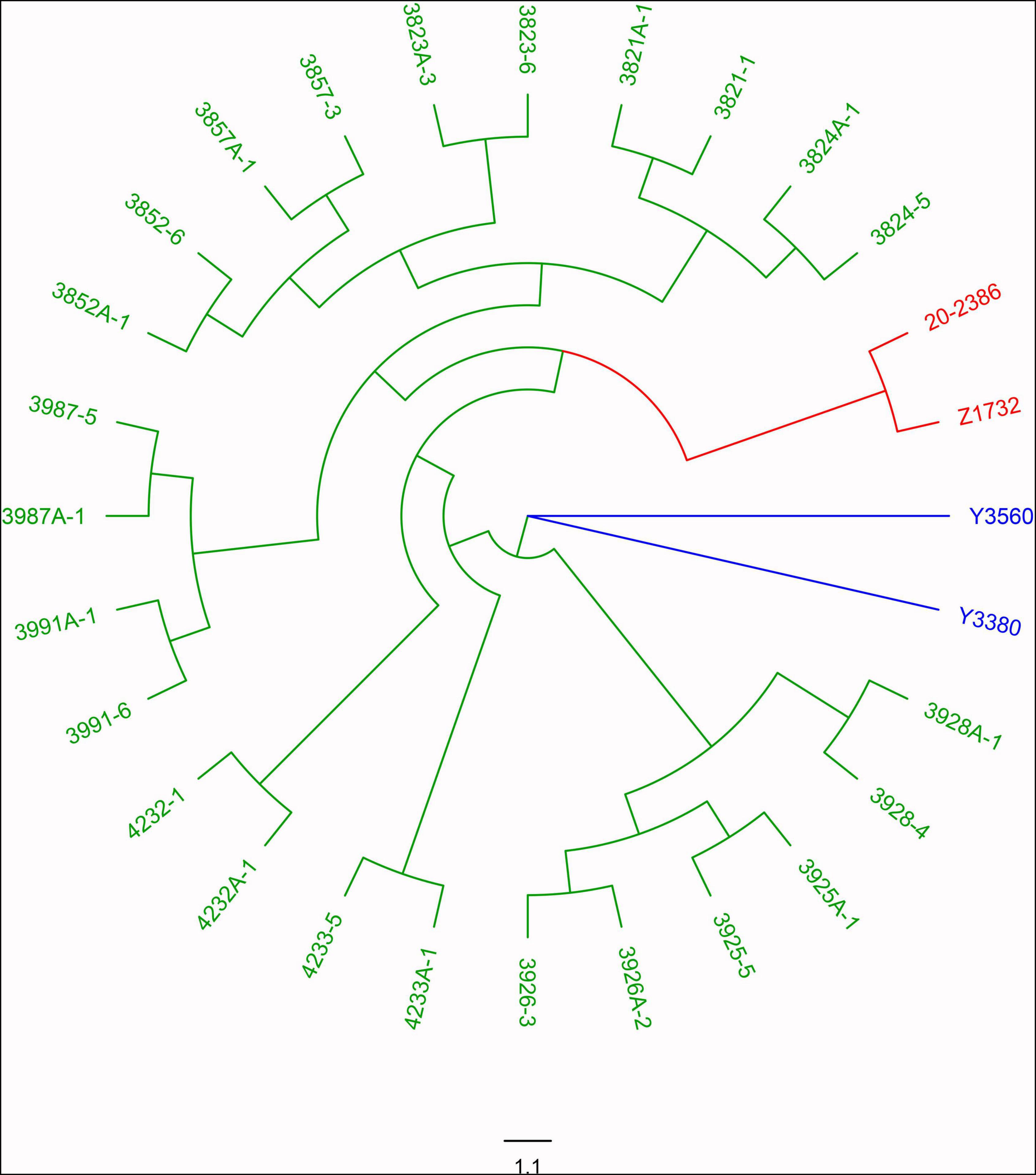
Figure 5. Genetic clustering analysis of F2 generations produced by induced heterozygous females. The induction lines Y3560 and Y3380 are in blue, the hybrid control is in red, and the induced F2 generation is in green.
Discussion
Rapid Acquisition of Cytoplasmic Male Sterile and Maintainer Lines by Rape Doubled Haploid Inducer
In this study, the parents and offspring were identified before and after induction, and the floral morphology of the sterile progeny ZY21A-1 in the F1 generation was highly similar to that of pol CMS (Figure 2). Further, molecular marker identification showed that the fertile progeny ZY21-1 in the induced F1 generation was nap cytoplasm (Figure 3A) and the sterile progeny ZY21A-1 was pol cytoplasm (Figure 3A). Also, both of them did not carry the pol CMS restorer gene Rfp1 (Figure 3B). The results of Nei genetic distance showed that the female parent 0933B and the induced F1 generation ZY21-1 and ZY21A-1 were all 0.002 (Table 2), and the nuclear background was almost identical. The mitochondrial genomes of the offspring before and after induction were compared with those of the parents. It was found that the mtDNA of the fertile progeny ZY21-1 was 99.36%, which is consistent with ZY21-1 (nap). The mtDNA of the sterile progeny ZY21A-1 was 97.89%, consistent with 0933A (pol) in the F1 generation (Figure 1F). These results are consistent with the identification results with molecular markers.
As a control, the mtDNA of the F1 generation of the normal hybrid Z1732 (pol) was identical to that of the female parent 0933A (Figure 1C). The genetic distance between the hybrid hexaploid ZY26A-1 and the female parent L0933A was 0.377, with the length of the mitochondrial genome increased by 36,275 bp and with multiple repetitive sequences appearing (Figure 1D and Supplementary Figure 2D). Y3380 and Y3560 were allo-octoploids synthesized by artificial chromosome doubling (Fu et al., 2018), and the size and genotype of the mitochondrial genome are the same as those of tetraploid (Figure 1C and Supplementary Figures 2E,F). This shows that hybridization between different ploidy is different from the same ploidy normal hybridization. The hybridization of plants with the same ploidy is the main hybridization type between karyogenes, and the cytoplasm genes are inherited from the female parent. In addition to karyogenes, cytoplasm genes may change in different ploidy hybridization, and there is an interaction between karyogene and cytoplasm (Zhang et al., 2021). Subsequently, by examining the F2 generation population formed from the induced homozygous female parent, it was found that the morphology, ploidy, and cytoplasm genotype in the induced F2 generation of different fertility populations were the same as those in the induced F1 generation (Figures 4A–F). Simultaneously, the induced F2 generation of different fertility plants produced from the induced heterozygous parent also tended to be stable. In summary, the different genotypes of female parents by the induction lines can produce induced progeny with only fertility difference, and the stable populations of maintainers and CMS lines with the same genetic background can be obtained rapidly through self and sister crosses (Figure 5 and Table 2).
Speculation and Analysis of Induced Cytoplasmic Changes
Numerous studies have shown that allopolyploidization in plants results in rapid genomic changes (Song et al., 1995; Ozkan et al., 2001; Comai et al., 2003; Pires et al., 2004; Soltis et al., 2004; Comai, 2005; Tate, 2006; Gaeta et al., 2007) and leads to chromosomal rearrangements (Pires et al., 2004; Pontes et al., 2004; Chen and Ni, 2006; Leitch and Leitch, 2008), altered DNA methylation (Lewis et al., 2006; Xu et al., 2009; Niederhuth et al., 2016), chromatin remodeling (Leitch and Leitch, 2008), altered gene expression (Chen and Pikaard, 1997; Osborn et al., 2003; Adams and Wendel, 2005; Gaeta et al., 2007), and activation of transposable elements (Kashkush et al., 2002; Ha et al., 2009). The rape DH induction lines by artificial synthesis have similar characteristics (Song et al., 1995; Lewis et al., 2006; Gaeta et al., 2007; Xiong et al., 2011). Also, are the CMS lines produced by the induction lines similar in mechanism to pol CMS in Brassica napus (Fu, 1981) and wild abortive lines in rice (Yuan, 1986)? In this study, the DH induction lines were used as the male parent and the allo-tetraploid B. napus plants were used as the female parent, while causing different genomes to be combined (Szadkowski et al., 2011; Fu et al., 2016). Since male parents have a much higher number of chromosomes than female parents, chromosomes from the male parents are gradually lost during embryonic development (Houben et al., 2011), and there is subsequent doubling of the chromosomes to reorganize the genome dose (Comai et al., 2003). As a result, the endosperm and embryo abortion rates are higher compared with normal crosses, resulting in the formation of a large number of defective seeds (Prigge et al., 2012; Xu et al., 2013; Qiu et al., 2014; Yang et al., 2019), which ultimately retain about 10% of the normal seeds in our experiment (data not shown) (Xu et al., 2013; Melchinger et al., 2017; Luo et al., 2021). The majority of these seeds completely lose the paternal chromosome during development (Houben et al., 2011; Tayeng et al., 2012; Chaudhary et al., 2013), leaving only the maternal chromosome before completing doubling, resulting in homozygous doubled haploid seeds. A small number of these seeds do not completely lose the paternal chromosome (Li et al., 2009; Newaskar et al., 2013; Zhao et al., 2013), resulting in hybrid offspring such as ZY26A-1. However, since the nuclear background between the offspring and the maternal parent is almost identical before and after induction, there is no infiltration or hybridization of large segments of the male parent, but the induced offspring will show partial functional changes and will involve multiple pathways through recombination or interactions. It was shown that transposons are involved in genome recombination in polyploids (Albertin et al., 2006; Ha et al., 2009; Chen, 2010; Zou et al., 2011; Shen et al., 2014), and so it is more reasonable to explain the gain (Gao et al., 2016) or loss of function of the induced progeny by jumping of polyploidy-induced transposons (Fu et al., 2016; Vicient and Casacuberta, 2017). It is further speculated that during induction, recombination genomes induce activation or jumping of nuclear genomic transposons while activating related genes in the maternal cytoplasm genome (Shen et al., 2015; Fu et al., 2016). Also compared to ogu cytoplasm, nap and pol cytoplasm undergo less rearrangement and recombination in the sequence and arrangement between mtDNAs (Figures 1A,B,G), and the similarity is also higher (Chen et al., 2011a). Therefore, a small change in mitochondrial genome can make nap cytoplasm change into pol sterile cytoplasm, resulting in the formation of CMS-inducing gene orf224 in the sterile plant (ZY21A-1, orf224 is the key regulatory gene of pol CMS). There was no orf224 in mitochondria before induction. During the induction, some fragments of mitochondria rearranged, resulting in the emergence of orf224, changing 0933B from fertile to sterile (ZY21A-1). This may explain why pol CMS in Brassica napus was discovered by Prof. Fu Tingdong in 1973 (Fu, 1981). Moreover, the same results occurred in multiple replicate experiments and were also stably inherited in the F2 generation after induction, while indicating that these transposon jumps were targeted and persistent. Although some speculations on the induction mechanism of DH induction lines were made according to current results, the induction mechanism of DH inducer is complex, and whether the real induction mechanism is the same as what we speculated will be explored in more depth.
Doubled Haploid Induction Lines Provide a New Model for Innovative Germplasm in Brassica napus
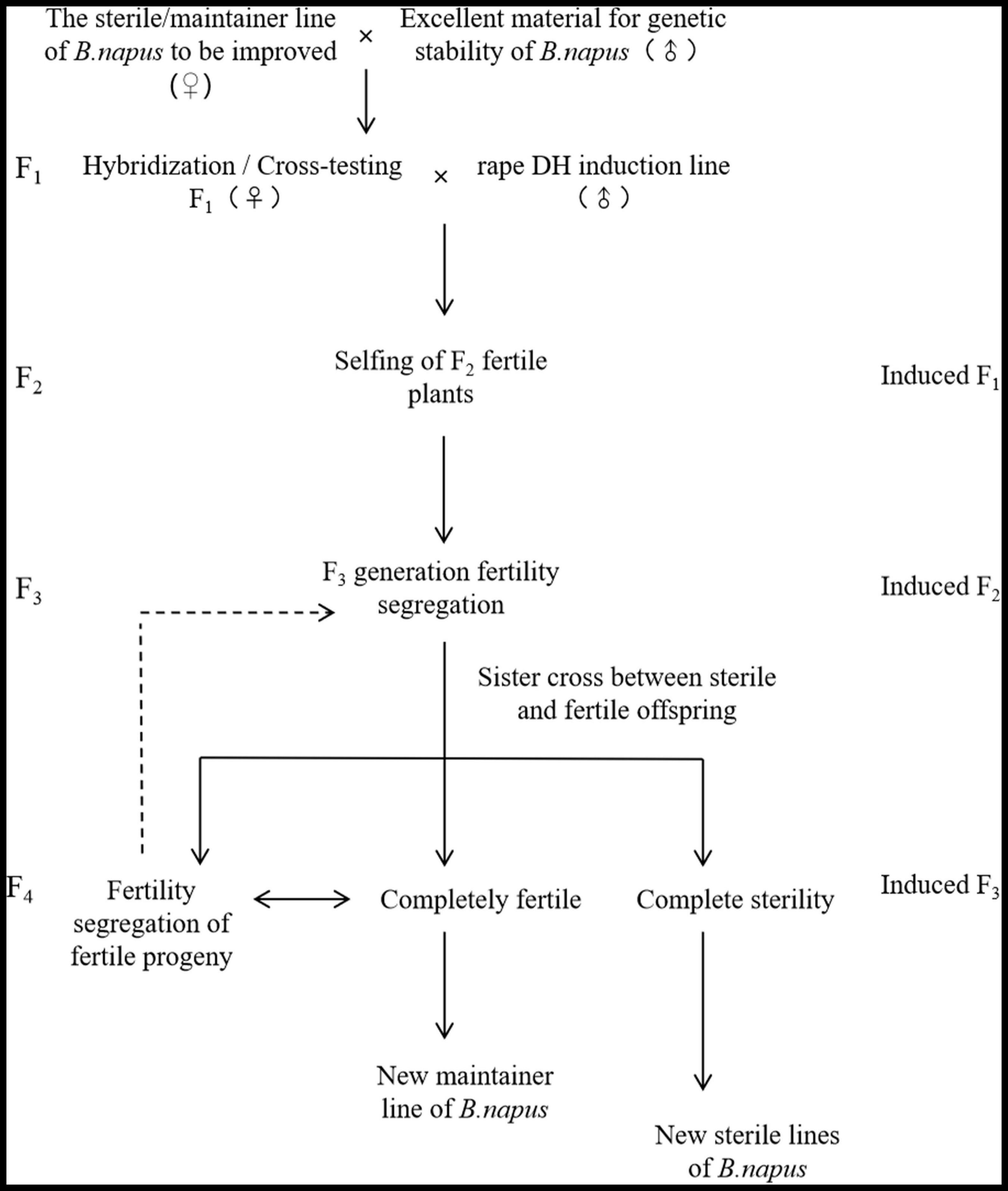
To ensure the reliability of the experiment, the planting process was strictly carried out with sorted harvesting and sorted seedling transplanting and planting. It excluded the possibility that the CMS plants in the induced F1 generation were produced by the mixed planting. Meanwhile by repeating the experiment, the CMS plants were also produced during maintainer induction of other genotypes. These results indicate that the phenomenon can be repeatedly realized, ensuring the realization path of the innovative CMS line selection method for DH induction lines. Meanwhile, because of the rapid and simultaneous breeding of maintainers and CMS lines when different genotypes of the female parents are induced by DH induction lines in B. napus, a new model for the innovation of germplasm resources is it provided. On the one hand, because the induction effect of the induction lines is influenced by the karyogene and cytoplasm genotype of the female parent, favorable traits are generated and rapid stabilization of traits is carried out by inducing female parents to gain or lose corresponding functions. On the other hand, the induction lines can enhance the SNP purity rate of the plants, and the induction progeny has the characteristics of fertility segregation for rapid and simultaneous breeding for the maintenance and CMS lines. First, the cytoplasmic maintainer or the CMS line of the known B. napus to be improved as the female parent and other excellent traits B. napus as the male parent to cross or test cross (Figure 6) was used and then DH induction lines were used to pollinate the generated F1 generation and induce it to produce karyogene homozygous F2 generation (induced F1). Subsequently, the excellent F2 generation (induced F1) to self-crosses were selected and then in the F3 generation (induced F2), fertile plants without restorer genes and CMS plants with a genetic similarity coefficient greater than 0.90 (genetic distance less than 0.1) to self-crossed and sister-crossed were screened out. Also, the sterility of fully fertile self-crossed plants and sister-crossed sterile lines, were observed when the sister-crossed sterile offspring are completely sterile and the corresponding fully fertile offspring are not separated from the sterile plants, the corresponding maintainer and CMS lines with the consistent nuclear background are formed. Meanwhile, when the offspring of a fully fertile individual plant is still a separated sterile and fertile offspring, the paired sister crosses of multiple individual plants are repeated until no longer separated (Figure 6). This not only enables rapid breeding of new inbred lines but also can obtain homozygous corresponding maintainer and CMS lines in about 2–4 generations at the earliest (Figure 6). This saves a lot of time and manpower compared to the traditional breeding method by saving the process of allelic purification of maintainer lines and backcrossing of maintainer lines with CMS lines for multiple generations and karyogene replacement. Therefore, the application of rape DH induction lines can accelerate and change the breeding pattern of rapeseed and create new ideas for the development of germplasm resources, which have great potential for application and practical value. There are also differences between rape DH induction lines and maize and Arabidopsis haploid induced lines in terms of induction function.
Conclusion
In this study, by examined the parents and offspring before and after induction, we found that when the inducer lines were pollinated as male parents to homozygous females, the nuclear background of the induced F1 generation was almost the same as that of the females, and the fertile progeny did not contain fertility restorer genes, but the cytoplasmic type of the sterile progeny changed and was rapidly stabilized in the F2 generation. Simultaneous induction of different types of heterozygous females resulted in similar results while indicating that the inducer lines have the ability to obtain both maintainers and sterile lines simultaneously and rapidly. Also, the induction mechanism of inducers may be related to chromosome elimination and nuclear-cytoplasmic interactions, and these findings provide new insights for the innovation of oilseed rape germplasm resources.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SF, WZ, and ZL conceived and designed the experiments. WZ, YZ, and XYL performed the experiments. WZ, HS, and XL carried out data analysis. YL, JY, PX, JW, WG, QZ, LT, ZK, and RT carried out field cultivation. WZ wrote the manuscript. SF and CX edited the manuscript. All authors read and approved the content.
Funding
This work was supported by the National Key Research and Development Program (2016YFD0101305), the Fundamental Research Funds for the Central Universities (SCU2021D006), the Rapeseed Breeding Program in Sichuan Province during the 14th Five-Year Plan Period (2021YFYZ0018), the Sichuan Rapeseed Innovation Team Program, the Major Science and Technology Program in Sichuan Province (2018NZDZX0003 and 2021YFN0014), and the Chengdu Applied Technology Research and Development Program (2021YF0500021SN).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.871006/full#supplementary-material
Footnotes
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://cbi.hzau.edu.cn/rape/download_ext/zs11.geneinformation.txt
- ^ https://www.genome.jp/kegg/ko.html
References
Adams, K. L., and Wendel, J. F. (2005). Novel patterns of gene expression in polyploid plants. Trends Genet. 21, 539–543. doi: 10.1016/j.tig.2005.07.009
Albertin, W., Balliau, T., Brabant, P., Chèvre, A. M., Eber, F., Malosse, C., et al. (2006). Numerous and rapid nonstochastic modifications of gene products in newly synthesized Brassica napus allotetraploids. Genetics 173, 1101–1113. doi: 10.1534/genetics.106.057554
Albertsen, M., Hugenholtz, P., Skarshewski, A., Nielsen, K.L., Tyson, G.W., and Nielsen, P.H. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538. doi: 10.1038/nbt.2579
Allender, C. J., and King, G. J. (2010). Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10:54. doi: 10.1186/1471-2229-10-54
Bancroft, I., Morgan, C., Fraser, F., Higgins, J., Wells, R., Clissold, L., et al. (2011). Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat. Biotechnol. 29, 762–766. doi: 10.1038/nbt.1926
Barro, F., and Martín, A. (1999). Response of different genotypes of Brassica carinata to microspore culture. Plant Breed. 118, 79–81. doi: 10.1046/j.1439-0523.1999.118001079.x
Blakeslee, A. F., Belling, J., Farnham, M. E., and Bergner, A. D. (1922). A haploid mutant in the jimson weed,” datura stramonium”. Science 55, 646–647. doi: 10.1126/science.55.1433.646
Burk, L. G., Gerstel, D. U., and Wernsman, E. A. (1979). Maternal haploids of Nicotiana tabacum L. from seed. Science 206:585. doi: 10.1126/science.206.4418.585
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chase, S. S. (1952). Production of homozygous diploids of maize from monoploids. Agron. J. 44, 263–267. doi: 10.2134/agronj1952.00021962004400050010x
Chaudhary, H. K., Tayeng, T., Kaila, V., and Rather, S. A. (2013). Use of asynchrony in flowering for easy and economical polyhaploid induction in wheat following Imperata cylindrica-mediated chromosome elimination approach. Plant Breed. 132, 155–158. doi: 10.1111/pbr.12036
Chen, J., Guan, R., Chang, S., Du, T., Zhang, H., and Xing, H. (2011a). Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L. PLoS One 6:e17662. doi: 10.1371/journal.pone.0017662
Chen, J. F., Cui, L., Malik, A. A., and Mbira, K. G. (2011b). Invitro haploid and dihaploid production via unfertilized ovule culture. Plant Cell Tissue Organ Cult. 104, 311–319. doi: 10.1007/s11240-010-9874-6
Chen, Z. J. (2010). Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 15, 57–71. doi: 10.1016/j.tplants.2009.12.003
Chen, Z. J., and Ni, Z. (2006). Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays 28, 240–252. doi: 10.1002/bies.20374
Chen, Z. J., and Pikaard, C. S. (1997). Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc. Natl. Acad. Sci. U.S.A. 94, 3442–3447. doi: 10.1073/pnas.94.7.3442
Coe, E. H. (1959). A line of maize with high haploid frequency. Am. Nat. 93, 381–382. doi: 10.1086/282098
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. doi: 10.1038/nrg1711
Comai, L., Madlung, A., Josefsson, C., and Tyagi, A. (2003). Do the different parental ‘heteromes’ cause genomic shock in newly formed allopolyploids? Philos. Trans. R Soc. Lond. B Biol. Sci. 358, 1149–1155. doi: 10.1098/rstb.2003.1305
Custers, J. B. M. (2003). “Microspore culture in rapeseed (Brassica napus L.),” in Doubled Haploid Production in Crop Plants: A Manual, eds M. Maluszynski, K. J. Kasha, B. P. Forster, and I. Szarejko (Dordrecht: Springer Netherlands), 185–193. doi: 10.1007/978-94-017-1293-4_29
Custers, J. B. M., Cordewener, J. H., Nöllen, Y., Dons, H. J., and Van Lockeren Campagne, M. M. (1994). Temperature controls both gametophytic and sporophytic development in microspore cultures of Brassica napus. Plant Cell Rep. 13, 267–271. doi: 10.1007/BF00233317
Du, M., Zhou, K., Liu, Y., Deng, L., Zhang, X., Lin, L., et al. (2020). A biotechnology-based male-sterility system for hybrid seed production in tomato. Plant J. 102, 1090–1100. doi: 10.1111/tpj.14678
Eder, J., and Chalyk, S. (2002). In vivo haploid induction in maize. Theor. Appl. Genet. 104, 703–708. doi: 10.1007/s00122-001-0773-4
Ferrie, A., and Caswell, K. L. (2011). Isolated microspore culture techniques and recent progress for haploid and doubled haploid plant production. Plant Cell Tissue Organ Cul. 104, 301–309. doi: 10.1007/s11240-010-9800-y
Forster, B. P., Heberle-Bors, E., Kasha, K. J., and Touraev, A. (2007). The resurgence of haploids in higher plants. Trends Plant Sci. 12, 368–375. doi: 10.1016/j.tplants.2007.06.007
Fu, S., Yin, L., Xu, M., Li, Y., Wang, M., Yang, J., et al. (2018). Maternal doubled haploid production in interploidy hybridization between Brassica napus and Brassica allooctaploids. Planta 247, 113–125. doi: 10.1007/s00425-017-2772-y
Fu, T. (1981). Production and research of rapeseed in the people’s republic of China. Eucarpia Cruciferae Newsl 6, 6–7.
Fu, T., Yang, G., and Yang, X. (1990). Studies on “three line” polima cytoplasmic male sterility developed in Brassica napus L. Plant Breeding 104, 115–120. doi: 10.1111/j.1439-0523.1990.tb00412.x
Fu, T., Yang, X., and Yang, G. (1989). Development and studies on polima cytoplasmic male sterile “three lines” in Brassica napus L. J. Huazhong Agric. 8, 201–207.
Fu, Y., Xiao, M., Yu, H., Mason, A. S., Yin, J., Li, J., et al. (2016). Small RNA changes in synthetic Brassica napus. Planta 244, 607–622. doi: 10.1007/s00425-016-2529-z
Gaeta, R. T., Pires, J. C., Iniguez-Luy, F., Leon, E., and Osborn, T. C. (2007). Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19, 3403–3417. doi: 10.1105/tpc.107.054346
Gao, C., Zhou, G., Ma, C., Zhai, W., Zhang, T., Liu, Z., et al. (2016). Helitron-like transposons contributed to the mating system transition from out-crossing to self-fertilizing in polyploid Brassica napus L. Sci. Rep. 6:33785. doi: 10.1038/srep33785
Geiger, H. H., and Gordillo, G. A. (2009). Doubled haploids in hybrid maize breeding. Maydica 54, 485–499.
Ha, M., Lu, J., Tian, L., Ramachandran, V., Kasschau, K. D., Chapman, E. J., et al. (2009). Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Nat. Acad. Sci. U.S.A. 106, 17835–17840. doi: 10.1073/pnas.0907003106
Heng, S., Shi, D., Hu, Z., Huang, T., Li, J., Liu, L., et al. (2015). Characterization and classification of one new cytoplasmic male sterility (CMS) line based on morphological, cytological and molecular markers in non-heading Chinese cabbage (Brassica rapa L.). Plant Cell Rep. 34, 1529–1537. doi: 10.1007/s00299-015-1804-y
Houben, A., Sanei, M., and Pickering, R. (2011). Barley doubled-haploid production by uniparental chromosome elimination. Plant Cell Tissue Organ Cul. 104, 321–327. doi: 10.1007/s11240-010-9856-8
Hu, D., Jing, J., Snowdon, R. J., Mason, A. S., Shen, J., Meng, J., et al. (2021). Exploring the gene pool of Brassica napus by genomics-based approaches. Plant Biotechnol. J. 19, 1693–1712. doi: 10.1111/pbi.13636
Jacquier, N. M. A., Gilles, L. M., Pyott, D. E., Martinant, J. P., Rogowsky, P. M., and Widiez, T. (2020). Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nat. Plants 6, 610–619. doi: 10.1038/s41477-020-0664-9
Kashkush, K., Feldman, M., and Levy, A. A. (2002). Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33, 102–106. doi: 10.1038/ng1063
Keller, W. A., and Armstrong, K. C. (1978). High frequency production of microspore-derived plants from Brassica napus anther cultures. Z Pflanzenzuchtg 80, 100–108.
Kott, L., Fletcher, R., and Coventry, J. (1998). Manual for Microspore Culture Technique for Brassica napus. Technical Bulletin-OAC Publication. Ontario: University of Guelph.
Laurie, D. A. (1989). The frequency of fertilization in wheat × pearl millet crosses. Genome 32, 1063–1067. doi: 10.1139/g89-554
Laurie, D. A., and Bennett, M. D. (1986). Wheat × maize hybridization. Can. J. Genet. Cytol. 28, 313–316. doi: 10.1139/g86-046
Leflon, M., Eber, F., Letanneur, J. C., Chelysheva, L., Coriton, O., Huteau, V., et al. (2006). Pairing and recombination at meiosis of Brassica rapa (AA) x Brassica napus (AACC) hybrids. Theor. Appl. Genet. 113, 1467–1480. doi: 10.1007/s00122-006-0393-0
Leitch, A. R., and Leitch, I. J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320, 481–483. doi: 10.1126/science.1153585
Lewis, L. N., Pires, J. C., Leon, E., Vogelzang, R., Oslach, L., and Osborn, T. (2006). Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140, 336–348. doi: 10.1104/pp.105.066308
Li, C., Sang, S., Sun, M., Yang, J., Shi, Y., Hu, X., et al. (2021). Direct modification of multiple gene homoeologs in Brassica oleracea and Brassica napus using doubled haploid inducer-mediated genome editing system. Plant Biotechnol. J. 19, 1889–1891. doi: 10.1111/pbi.13632
Li, D., Diao, P., Yong, R., and Huang, X. (1997). Development and studies on cytoplasmic male sterile “three lines” in Brassica napus L. of low ervcic acid and low giucosinolates. Seed 1, 40–42.
Li, H., and Devaux, P. (2001). Enhancement of microspore culture efficiency of recalcitrant barley genotypes. Plant Cell Rep. 20, 475–481. doi: 10.1007/s002990100368
Li, L., Xu, X., Jin, W., and Chen, S. (2009). Morphological and molecular evidences for DNA introgression in haploid induction via a high oil inducer CAUHOI in maize. Planta 230, 367–376. doi: 10.1007/s00425-009-0943-1
Lichter, R. (1982). Induction of haploid plants from isolated pollen of Brassica napus. Z. Pflanzenphysiol. 105, 427–434. doi: 10.1016/S0044-328X(82)80040-8
Luo, X., Yang, J., Zhu, Z., Huang, L., Ali, A., Javed, H. H., et al. (2021). Genetic characteristics and ploidy trigger the high inducibility of double haploid (DH) inducer in Brassica napus. BMC Plant Biol. 21:538. doi: 10.1186/s12870-021-03311-z
Melchinger, A. E., Munder, S., Mauch, F. J., Mirdita, V., Böhm, J., and Müller, J. (2017). High-throughput platform for automated sorting and selection of single seeds based on time-domain nuclear magnetic resonance (TD-NMR) measurement of oil content. Biosyst. Eng. 164, 213–220. doi: 10.1016/j.biosystemseng.2017.10.011
Newaskar, G. S., Chimote, V. P., Mehetre, S. S., Jadhav, A. S., and Jenkins, J. (2013). Interspecific hybridization in Gossypium L: characterization of progenies with different ploidy-confirmed multigenomic backgrounds. Plant Breed. 132, 211–216. doi: 10.1111/pbr.12031
Niederhuth, C. E., Bewick, A. J., Ji, L., Alabady, M. S., Kim, K. D., Li, Q., et al. (2016). Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17:194. doi: 10.1186/s13059-016-1059-0
Niizeki, H., and Oono, K. (1968). Induction of haploid rice plant from anther culture. Proc. Jpn. Acad. 44, 554–557. doi: 10.2183/pjab1945.44.554
Olsson, G. (1960). Species crosses within the genus Brassica napus L. II. Artificial Brassica napus L. Hereditas 46, 351–386. doi: 10.1111/j.1601-5223.1960.tb03092.x
Osborn, T. C., Pires, J. C., Birchler, J. A., Auger, D. L., Chen, Z. J., Lee, H. S., et al. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19, 141–147. doi: 10.1016/S0168-9525(03)00015-5
Ozkan, H., Levy, A. A., and Feldman, M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–Triticum) group. Plant Cell 13, 1735–1747. doi: 10.1105/tpc.010082
Palmer, C. E., Keller, W. A., and Arnison, P. G. (1996). “Utilization of Brassica haploids,” in In Vitro Haploid Production in Higher Plants: Volume 3 —Important Selected Plants, eds S. M. Jain, S. K. Sopory, and R. E. Veilleux (Dordrecht: Springer Netherlands), 173–192. doi: 10.1007/978-94-017-1858-5_10
Palmer, J. D., Shields, C. R., Cohen, D. B., and Orton, T. J. (1983). Chloroplast DNA evolution and the origin of amphidiploid Brassica species. TAG Theor. Appl. Genet. 65, 181–189. doi: 10.1007/BF00308062
Pires, J. C., Zhao, J., Schranz, M. E., Leon, E. J., Quijada, P. A., Lukens, L. N., et al. (2004). Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. 82, 675–688. doi: 10.1111/j.1095-8312.2004.00350.x
Pollacsek, M. (1992). Management of the ig gene for haploid induction in maize. Agronomie 12, 247–251. doi: 10.1051/agro:19920304
Pontes, O., Neves, N., Silva, M., Lewis, M. S., Madlung, A., Comai, L., et al. (2004). Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. U.S.A. 101, 18240–18245. doi: 10.1073/pnas.0407258102
Prigge, V., Xu, X., Li, L., Babu, R., Chen, S., Atlin, G. N., et al. (2012). New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics 190:781. doi: 10.1534/genetics.111.133066
Qiu, F., Liang, Y., Li, Y., Liu, Y., Wang, L., and Zheng, Y. (2014). Morphological, cellular and molecular evidences of chromosome random elimination in vivo upon haploid induction in maize. Curr. Plant Biol. 1, 83–90. doi: 10.1016/j.cpb.2014.04.001
Ravi, M., Marimuthu, M. P. A., Tan, E. H., Maheshwari, S., Henry, I. M., Marin-Rodriguez, B., et al. (2014). A haploid genetics toolbox for Arabidopsis thaliana. Nat. Commun. 5:5334. doi: 10.1038/ncomms6334
Rotarenco, V., Dicu, G., State, D., and Fuia, S. (2010). New inducers of maternal haploid maize. Maize Genet. Coop. Newsl Lett. 84, 1–7.
Shariatpanahi, M. E., and Ahmadi, B. (2016). “Isolated microspore culture and its applications in plant breeding and genetics,” in Plant Tissue Culture: Propagation, Conservation and Crop Improvement, eds M. Anis and N. Ahmad (Singapore: Springer Singapore), 487–507. doi: 10.1007/978-981-10-1917-3_21
Shen, E., Zou, J., Falk, H. B., Chen, L., Ye, C., Dai, S., et al. (2015). Identification, evolution, and expression partitioning of miRNAs in allopolyploid Brassica napus. J. Exp. Bot. 66, 7241–7253. doi: 10.1093/jxb/erv420
Shen, Y., Zhao, Q., Zou, J., Wang, W., Gao, Y., Meng, J., et al. (2014). Characterization and expression patterns of small RNAs in synthesized Brassica hexaploids. Plant Mol. Biol. 85, 287–299. doi: 10.1007/s11103-014-0185-x
Soltis, D. E., Soltis, P. S., Pires, J. C., Kovarik, A., Tate, J. A., and Mavrodiev, E. (2004). Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol. J. Linn. Soc. 82, 485–501. doi: 10.1111/j.1095-8312.2004.00335.x
Song, K., Lu, P., Tang, K., and Osborn, T. C. (1995). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Nat. Acad. Sci. U.S.A. 92, 7719–7723. doi: 10.1073/pnas.92.17.7719
Sun, F. M., Fan, G. Y., Hu, Q., Zhou, Y. M., Guan, M., Tong, C. B., et al. (2017). The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 92, 452–468. doi: 10.1111/tpj.13669
Szadkowski, E., Eber, F., Huteau, V., Lode, M., Coriton, O., Jenczewski, E., et al. (2011). Polyploid formation pathways have an impact on genetic rearrangements in resynthesized Brassica napus. New Phytol. 191, 884–894. doi: 10.1111/j.1469-8137.2011.03729.x
Takahata, Y., Brown, D. C. W., and Keller, W. A. (1991). Effect of donor plant age and inflorescence age on microspore culture of Brassica napus L. Euphytica 58, 51–55. doi: 10.1007/BF00035339
Takahata, Y., Komatsu, H., and Kaizuma, N. (1996). Microspore culture of radish (Raphanus sativus L.): influence of genotype and culture conditions on embryogenesis. Plant Cell Rep. 16, 163–166. doi: 10.1007/BF01890859
Tate, J. A. (2006). Evolution and expression of homeologous loci in tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 173, 1599–1611. doi: 10.1534/genetics.106.057646
Tayeng, T., Chaudhary, H. K., and Kishore, N. (2012). Enhancing doubled haploid production efficiency in wheat (Triticum aestivum L. em. Thell) by in vivo colchicine manipulations in Imperata cylindrica-mediated chromosome elimination approach. Plant Breed. 131, 574–578. doi: 10.1111/j.1439-0523.2012.01986.x
NU. (1935). Genome analysis in Brassica carinata with special reference to the experimental formation of Brassica napus, a peculiar mode of fertilization. Jpn. J. Bot. 7, 389–452. doi: 10.1007/s00122-009-1056-8
Vicient, C. M., and Casacuberta, J. M. (2017). Impact of transposable elements on polyploid plant genomes. Ann. Bot. 120, 195–207. doi: 10.1093/aob/mcx078
Witt, U., Hansen, S., Albaum, M., and Abel, W. O. (1991). Molecular analyses of the CMS-inducing ‘Polima’ cytoplasm in Brassica napus L. Curr. Genet. 19, 323–327. doi: 10.1007/BF00355063
Xiong, Z., Gaeta, R. T., and Pires, J. C. (2011). Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Nat. Acad. Sci. U.S.A. 108, 7908–7913. doi: 10.1073/pnas.1014138108
Xu, L., Najeeb, U., Tang, G. X., Gu, H. H., and Zhou, W. J. (2007). Haploid and doubled haploid technology. Adv. Bot. Res. 45, 181–216. doi: 10.1016/S0065-2296(07)45007-8
Xu, X., Liang, L., Dong, X., Jin, W., Melchinger, A. E., and Chen, S. (2013). Gametophytic and zygotic selection leads to segregation distortion through in vivo induction of a maternal haploid in maize. J. Exp. Bot. 64, 1083–1096. doi: 10.1093/jxb/ers393
Xu, Y., Zhong, L., Wu, X., Fang, X., and Wang, J. (2009). Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 229, 471–483. doi: 10.1007/s00425-008-0844-8
Yang, J., Li, H., Qu, Y., Chen, Q., Tang, J., lübberstedt, T., et al. (2019). Genetic dissection of haploid male fertility in maize (Zea mays L.). bioRxiv 138, 259–265. [Preprint]. doi: 10.1111/pbr.12688
Yin, L., Zhu, Z., Huang, L., Luo, X., Li, Y., Xiao, C., et al. (2021). DNA repair- and nucleotide metabolism-related genes exhibit differential CHG methylation patterns in natural and synthetic polyploids (Brassica napus L.). Hortic Res 8:142. doi: 10.1038/s41438-021-00576-1
Yin, L., Zhu, Z., Luo, X., Huang, L., Li, Y., Annaliese, S., et al. (2020). Genome-wide duplication of allotetraploid Brassica napus produces novel characteristics and extensive ploidy variation in self-pollinated progeny. G3 (Bethesda) 10, 3687–3699. doi: 10.1534/g3.120.401493
Zhang, H., Wu, J., Dai, Z., Qin, M., Hao, L., Ren, Y., et al. (2017). Allelism analysis of BrRfp locus in different restorer lines and map-based cloning of a fertility restorer gene, BrRfp1, for pol CMS in Chinese cabbage (Brassica rapa L.). Theor. Appl. Genet. 130, 539–547. doi: 10.1007/s00122-016-2833-9
Zhang, W., Ma, Y., Zhu, Z., Huang, L., Ali, A., Luo, X., et al. (2021). Maternal karyogene and cytoplasmic genotype affect the induction efficiency of doubled haploid inducer in Brassica napus. BMC Plant Biol. 21:207. doi: 10.1186/s12870-021-02981-z
Zhao, X., Xu, X., Xie, H., Chen, S., and Jin, W. (2013). Fertilization and uniparental chromosome elimination during crosses with maize haploid inducers. Plant Physiol. 163, 721–731. doi: 10.1104/pp.113.223982
Zhou, W., Hu, Y., Sui, Z., Fu, F., Wang, J., Chang, L., et al. (2013). Genome survey sequencing and genetic background characterization of Gracilariopsis lemaneiformis (Rhodophyta) based on next-generation sequencing. Plos One 8:e69909. doi: 10.1371/journal.pone.0069909
Zhou, Y., Yang, M., Zhao, S., Shi, H., Li, Y., Gong, W., et al. (2022). Rapid creation of interspecific hybrid progeny to broaden genetic distance through double haploid (DH) inducer in Brassica napus. Plants 11:695. doi: 10.3390/plants11050695
Zou, J., Fu, D., Gong, H., Qian, W., Xia, W., Pires, J. C., et al. (2011). De novo genetic variation associated with retrotransposon activation, genomic rearrangements and trait variation in a recombinant inbred line population of Brassica napus derived from interspecific hybridization with Brassica rapa. Plant J. 68, 212–224. doi: 10.1111/j.1365-313X.2011.04679.x
Keywords: double haploid inducer, rapid breeding, cytoplasmic male sterile lines (CMS), maintainer lines, mitochondrial DNA
Citation: Zhang W, Shi H, Zhou Y, Liang X, Luo X, Xiao C, Li Y, Xu P, Wang J, Gong W, Zou Q, Tao L, Kang Z, Tang R, Li Z, Yang J and Fu S (2022) Rapid and Synchronous Breeding of Cytoplasmic Male Sterile and Maintainer Line Through Mitochondrial DNA Rearrangement Using Doubled Haploid Inducer in Brassica napus. Front. Plant Sci. 13:871006. doi: 10.3389/fpls.2022.871006
Received: 07 February 2022; Accepted: 28 March 2022;
Published: 26 April 2022.
Edited by:
Sergio J. Ochatt, INRA UMR 1347 Agroécologie, FranceReviewed by:
Lev Aleksandrovich Elkonin, Agricultural Research Institute for South-East Region (ARISER), RussiaMohammad Reza Abdollahi, Bu-Ali Sina University, Iran
Copyright © 2022 Zhang, Shi, Zhou, Liang, Luo, Xiao, Li, Xu, Wang, Gong, Zou, Tao, Kang, Tang, Li, Yang and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuang Li, bGl6aHVhbmcyMDEyQHNpY2F1LmVkdS5jbg==; Jin Yang, eWpqaW5nQDE2My5jb20=; Shaohong Fu, aG9uem9uMjAxMEAxNjMuY29t
†These authors have contributed equally to this work
 Wei Zhang1,2,3†
Wei Zhang1,2,3† Chaowen Xiao
Chaowen Xiao Peizhou Xu
Peizhou Xu Wanzhuo Gong
Wanzhuo Gong Zhuang Li
Zhuang Li Shaohong Fu
Shaohong Fu