- 1College of Agronomy, Anhui Agricultural University, Key Laboratory of Wheat Biology and Genetic Improvement on Southern Yellow and Huai River Valley, Ministry of Agriculture and Rural Affairs, Hefei, China
- 2National Key Laboratory for Crop Genetics and Germplasm Enhancement, Jiangsu Plant Gene Engineering Research Center, Nanjing Agricultural University, Nanjing, China
Wheat is one of the most widely cultivated food crops worldwide, and the safe production of wheat is essential to ensure food security. Soil salinization and drought have severely affected the yield and quality of wheat. Valine-glutamine genes play important roles in abiotic stress response. This study assessed the effect of the gene TaVQ14 on drought and salt stresses resistance. Sequence analysis showed that TaVQ14 encoded a basic unstable hydrophobic protein with 262 amino acids. Subcellular localization showed that TaVQ14 was localized in the nucleus. TaVQ14 was upregulated in wheat seeds under drought and salt stress. Under NaCl and mannitol treatments, the percentage of seed germination was higher in Arabidopsis lines overexpressing TaVQ14 than in wild-type lines, whereas the germination rate was significantly lower in plants with a mutation in the atvq15 gene (a TaVQ14 homolog) than in WT controls, suggesting that TaVQ14 increases resistance to salt and drought stress in Arabidopsis seeds. Moreover, under salt and drought stress, Arabidopsis lines overexpressing TaVQ14 had higher catalase, superoxide dismutase, and proline levels and lower malondialdehyde concentrations than WT controls, suggesting that TaVQ14 improves salt and drought resistance in Arabidopsis by scavenging reactive oxygen species. Expression analysis showed that several genes responsive to salt and drought stress were upregulated in Arabidopsis plants overexpressing TaVQ14. Particularly, salt treatment increased the expression of AtCDPK2 in these plants. Moreover, salt treatment increased Ca2+ concentrations in plants overexpressing TaVQ14, suggesting that TaVQ14 enhances salt resistance in Arabidopsis seeds through calcium signaling. In summary, this study demonstrated that the heterologous expression of TaVQ14 increases the resistance of Arabidopsis seeds to salt and drought stress.
Introduction
Wheat (Triticum aestivum L.) is a widely cultivated grain crop and one of the three most important cereal grains worldwide (Brenchley et al., 2012). Wheat is a good energy source and provides essential nutrients such as protein, vitamins, and trace elements. Therefore, the safe production of wheat is crucial to ensure food security globally.
Droughts, soil salinization, and other environmental stresses are the main factors limiting the increase in wheat yield and quality. Resistance to drought and salinity is a complex biological trait involving multiple molecular, physiological, biochemical, and morphological changes (Zhou et al., 2018; Wang and Huang, 2019; Zelm et al., 2020). Therefore, identifying key genes and elucidating the mechanisms regulating crop resistance to drought and salinity stress are useful for increasing food production while allowing the sustainable management of ecological resources (Ingram and Bartels, 1996; Xiong et al., 2002; Jakab et al., 2005; Verslues et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006).
Valine-glutamine-motif proteins have attracted increasing attention because of their interaction with WRKY transcription factors (Lai et al., 2011). A total of 34, 40, and 61 VQ proteins have been identified in Arabidopsis, rice, and maize, respectively (Cheng et al., 2012; Li N. et al., 2014; Wang et al., 2014). These proteins regulate plant growth and development and the response to biotic and abiotic stresses (Andreasson et al., 2005; Petersen et al., 2010; Wang et al., 2010; Fill and Petersen, 2011; Hu P. et al., 2013; Hu Y. et al., 2013; Kim et al., 2013; Li Y. et al., 2014; Wang et al., 2014; Pascal et al., 2014; Wang H. et al., 2015; Wang M. et al., 2015; Song et al., 2016; Li et al., 2020). For instance, AtVQ14 (IKU1) is strongly expressed in the embryo and endosperm and controls endosperm development and seed size. Given that the nutrients required for seed germination are affected by seed size, thus it also affects resistance to adverse environments (Wang et al., 2010). AtVQ29 is involved in the photomorphogenesis of Arabidopsis seedlings. The hypocotyls of plants overexpressing AtVQ29 are longer than those of wild-type (WT) plants under far-red light or low light and regulate flowering time (Li Y. et al., 2014). AtVQ21 (MSK1) transgenic plants positively regulate the resistance of the pathogen Pseudomonas syringae and negatively mediate the resistance of the pathogen Botrytis cinerea (Petersen et al., 2010; Fill and Petersen, 2011). AtVQ22 improves JA-mediated disease resistance, mutant plants overexpressing this gene are more resistant to necrotizing pathogens, and transgenic lines were extremely sensitive to pathogen infection. In addition, the analysis of the rice transcriptome showed that VQ22 expression increased after infection with Magnaporthe grisea, indicating that this gene plays an important role in disease resistance (Hu Y. et al., 2013). AtCaMBP25 (AtVQ15) reduced osmotic stress during seed germination and growth in Arabidopsis thaliana. Under salt and osmotic stress, transgenic lines are highly sensitive to seed germination, growth, and development (Wang M. et al., 2015). Seed germination and seedling growth were inhibited in plants overexpressing AtVQ9 under salt stress (Hu Y. et al., 2013). ZmVQ54, ZmVQ19, OsVQ2, OsVQ16, and OsVQ20 were highly expressed under drought stress (Kim et al., 2013). GmVQ6 and GmVQ53 were highly expressed in roots and stems under low-nitrogen conditions. Arabidopsis lines overexpressing (Wang et al., 2014). PeVQ28 were salt tolerant (Cheng et al., 2020). These data demonstrate that VQ genes have multiple roles in regulating plant growth and development and resistance to biotic and abiotic stresses.
Little is known about the functions of VQ genes in wheat. Our previous study showed that TaVQ14 was related to salt stress response (Cheng et al., 2021). Therefore, TaVQ14 was selected as the target gene for further functional analysis. First, the subcellular location of TaVQ14 was determined. Second, Arabidopsis plants overexpressing TaVQ14 were obtained by genetic transformation, and molecular, physiological, and phenotypic analyses were carried out. This study elucidated the functions of VQ genes and provided useful information for genetically improving salt and drought resistance in wheat crops.
Materials and Methods
Experimental Materials and Stress Treatment
Wheat varieties Jing 411 (J411) and Hongmangchun 21 (HMC21) were provided by Shihe Xiao from the Chinese Academy of Agricultural Sciences, both of which were moderately salt and drought tolerant (Ren, 2012). Wheat seeds were treated with 300 mM NaCl or 300 mM mannitol. Samples were collected at 0, 4, 6, 10, 48, and 72 h, frozen in liquid nitrogen and stored immediately at −80°C.
Tobacco (Nicotiana tabacum) and Colombian ecotype Arabidopsis [Columbia-0 (Col-0), wild-type (WT)] seeds were provided by the State Key Laboratory of Crop Resistance of Anhui Agricultural University and were expanded and preserved in our laboratory. The seeds of WT and mutant Arabidopsis lines were cultivated in Murashige and Skoog (MS) medium at 24°C under a 16-h light/8-h dark cycle and transferred to square pots (diameter 6 cm) containing black soil and verstone (1:3, v/v) (Chen D. et al., 2017; Gao et al., 2017). Tobacco seeds were planted in square pots (6 cm in diameter) containing black soil and verstone (1:2, v/v).
Bioinformatics Analysis of TaVQ14
TaVQ14 sequences were obtained from the Ensembl Plants database. The isoelectric point (pI), molecular weight (MW), and other properties of TaVQ14 protein were analyzed using ExPASy (Cheng et al., 2018). The number of exons and introns of TaVQ14 was analyzed using Gene Structure Display Server1 (Zhu et al., 2018). The phylogenetic analysis of TaVQ14 was performed using MEGA 7.0 (Cheng et al., 2019a). The promoter region of TaVQ14 was analyzed using PlantCARE (Toufighi et al., 2005; Cheng et al., 2019b).
Total RNA Extraction and Real-Time PCR Analysis
Total RNA was extracted from wheat grains using RNzol Universal Total RNA Extraction Reagent (Tiangen, Beijing, China). Primers were designed using Primer Premier 6, and TaActin was used as a reference gene (Supplementary Table 1; Ji et al., 2011). RT-PCR analysis was performed using the TransStart Tip Green qPCR SuperMix kit (Transgen, Beijing, China). Each treatment included three biological replicates and three technical replicates. Data were transferred to Excel spreadsheets and analyzed using GraphPad Prism version 6.0 (Livak and Schmittgen, 2001; Zhao et al., 2012; Wang et al., 2017).
Cloning and Expression of TaVQ14
Specific primers were designed to clone the coding sequence of TaVQ14 (Supplementary Table 1).
The vector pCAMBIA1305 (p1305) containing a GPF reporter gene was used for subcellular localization. Primers containing XbaI and BamHI restriction sites were designed, and the stop codon was removed to construct the p1305-CaMV35S-TaVQ14-GFP expression vector, which served as a control (Dai et al., 2007).
The vector pCAMBIA1301a (p1301a) was used in overexpression experiments. Primers containing BamHI and XbaI restriction sites were designed, and the p1301a-TaVQ14 expression vector was constructed.
Subcellular Localization of TaVQ14
The subcellular localization of TaVQ14 was predicted using CELLO version 2.5 (Yu et al., 2004, 2006). The Agrobacterium suspension containing the p1301a vector was injected into tobacco leaves, and tissue sections were observed under a confocal microscope.
Analysis of TaVQ14 Overexpression in Arabidopsis
Arabidopsis ecotype Col-0 (WT) was transformed with the p1301-TaVQ14 vector using the floral dip method to obtain mutant T0 seeds. Transformed seeds were screened on MS medium supplemented with hygromycin. SYBR Green I fluorescence quantitative PCR was used to detect the copy number of the exogenously introduced TaVQ14 gene relative to the dxr gene, the dxr gene encoding terpenoid synthase is a single copy in the Arabidopsis genome, setting the internal reference gene (Supplementary Table 1). Positive plants were obtained by cloning GUS gene fragments (Supplementary Table 1) and GUS staining and propagated to T3 overexpressed Arabidopsis plants. Seeds from WT and transgenic plants were grown on MS medium supplemented with mannitol (0, 150, or 300 mM) or NaCl (0, 100, or 150 mM) under the same experimental conditions, and the rates of germination were calculated. The calculation formula is: germination rate (GP) = number of normal germinated seeds at the end of germination/number of tested seeds × 100%. The effect of TaVQ14 overexpression on stress resistance was assessed.
Analysis of TaVQ14 Homologs
The atvq15 mutant (SALK_005722), with a mutation in position 17114641 of AtVQ15 (AT2G41010, chr2: 17113798-17115047), was obtained from the Arashare platform. Mutants were screened by three-primer PCR to obtain homozygous plants. Seeds from WT and transgenic plants were grown on MS medium supplemented with mannitol (0, 150, or 300 mM) or NaCl (0, 100, or 150 mM) under the same experimental conditions, and the rates of germination were calculated.
Measurement of Stress-Related Physiological Indexes and Gene Expression Analysis
Seedlings of WT and transgenic lines were grown in a greenhouse for 24 h in plates containing MS medium supplemented with 300 mM mannitol or 150 mM NaCl, and plates containing MS medium were used as controls. Relative water content (RWC) (Bates et al., 1973; Zhao et al., 2014), and the levels of catalase (CAT) (Rorth and Jensen, 1967; Goldstein, 1968; Del Rio et al., 1977), proline (PRO) (Bates et al., 1973), malondialdehyde (MDA) (Heath and Packer, 1968), superoxide dismutase (SOD) (Sequeira and Mineo, 1966; Kochba et al., 1977), Ascorbate peroxidase (APX) (Zhao et al., 2018), Ascorbic acid (AsA) (Ji et al., 2019), Glutathion reductases (GR) (Zhang et al., 2018), L-Glutathione (GSH) (Xin et al., 2019), and L-Glutathione oxidized (GSSG) (Zhang et al., 2019) were determined, and the expression levels of genes induced by drought stress (AtRD29A, AtRD29B, AtP5CS1, AtOST1, AtDi19-3, and AtWRKY46) and salt stress (AtSHM1, AtSOS2, AtCDPK2, AtDi19-3, and AtPP2C49) were quantified (Huang et al., 2012; Qin et al., 2014; Chen J. et al., 2017; Chu et al., 2021).
Seedlings of WT and transgenic lines were grown in square pots for 20 days and treated with 150 mM NaCl or 300 mM mannitol every 3 days. The concentrations of Na+, K+, and Ca2+ were measured on treatment day 12.
Statistical Analysis
Statistical analysis was performed using SPSS version 19.0 and Origin software.
Results
Bioinformatics Analysis of TaVQ14
TaVQ14 has 789 base pairs and encodes a 262-amino acid protein. The protein has a pI of 9.164, MW of 27481.07 Da, hydrophobicity index of 64.39, hydrophilicity index of -0.234, and instability index (II) of 74.16. The percentage of alanine, serine, and proline was 17.9, 14.5, and 11.8%, respectively. In addition, the most common cis-acting elements in the promoter region were associated with response to light (64.32%), abscisic acid (21.4%), and auxin (7.14%), and with zein metabolism regulation (7.14%) (Supplementary Table 2).
Fourteen homologous genes were obtained from the Ensembl Plants database, and a phylogenetic tree was constructed. The results showed that TaVQ14 was highly homologous with sequences from Aegilops tauschii, Triticum turgidum, Hordeum vulgare, and Brachypodium distachyon (Supplementary Table 3 and Figure 1).
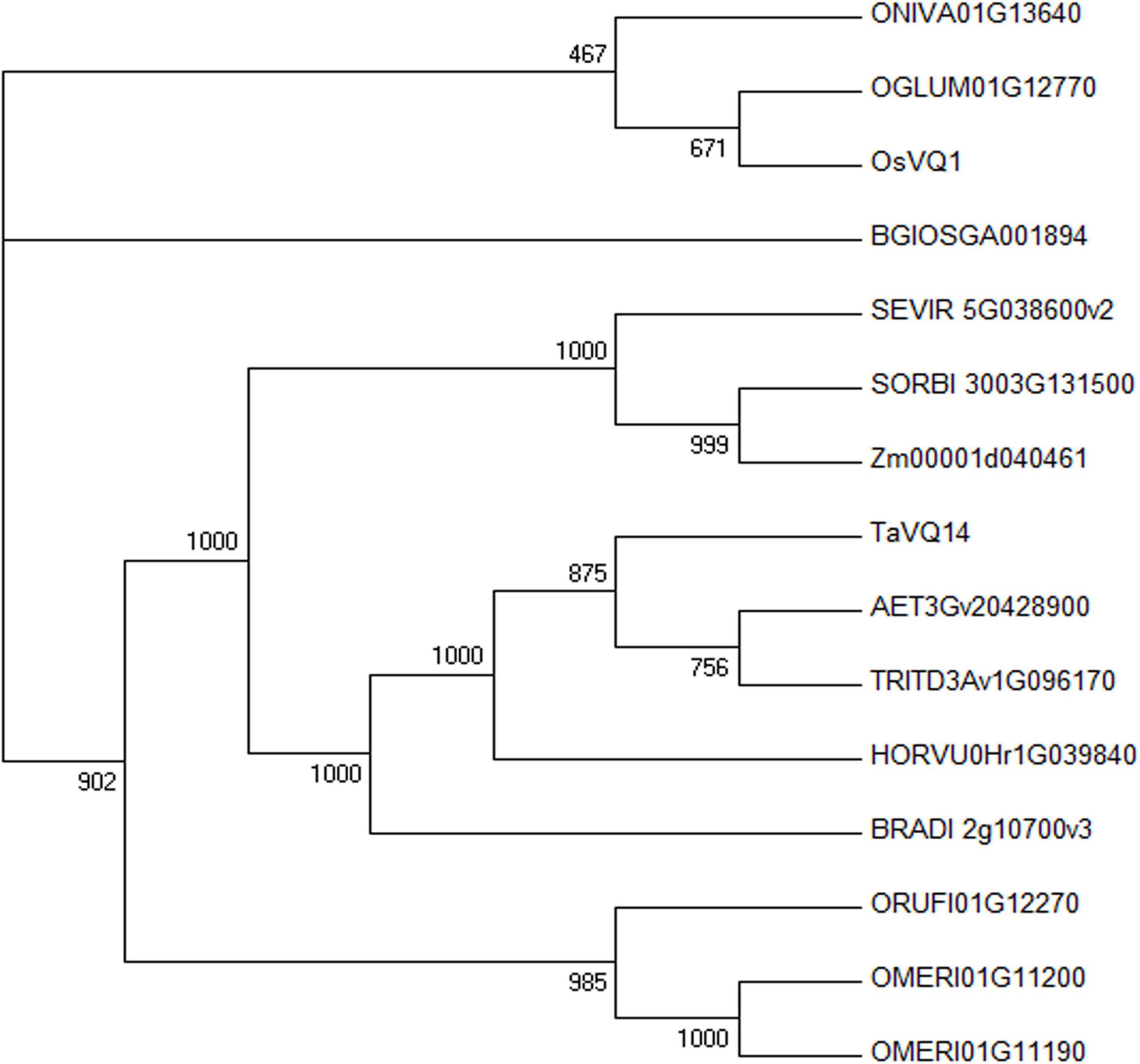
Figure 1. Phylogenetic analysis of TaVQ14 and its homologs. Homologs: Aegilops tauschii, AET3Gv20428900; Brachypodium distachyon, BRADI_2g10700v3; Hordeum vulgare, HORVU0Hr1G039840; Oryza glumipatula, OGLUM01G12770; Oryza sativa Indica Group, BGIOSGA001894; Oryza meridionalis, OMERI01G11190; Oryza meridionalis, OMERI01G11200; Oryza nivara, ONIVA01G13640; Oryza rufipogon, ORUFI01G12270; Oryza sativa Japonica Group, OsVQ1 (Os01g0278000); Setaria viridis, SEVIR_5G038600v2; Sorghum bicolor, SORBI_3003G131500; Triticum turgidum, TRITD3Av1G096170; Zea mays, Zm00001d040461.
Expression Pattern Analysis of TaVQ14
Valine-glutamine genes are involved in abiotic stress response in plants (Hu Y. et al., 2013; Kim et al., 2013; Wang M. et al., 2015; Cheng et al., 2020). Our previous study showed that TaVQ14 was associated with salt stress response (Cheng et al., 2021). Based on these findings, wheat seeds were treated with NaCl or mannitol to assess the role of TaVQ14 in salinity and drought stress. The results showed that salt and mannitol treatment increased the expression of TaVQ14 in a time-dependent manner (Figure 2). Furthermore, the expression levels of TaVQ14 after treatment were similar between the two wheat varieties. These results indicated that TaVQ14 expression was induced by NaCl and mannitol, suggesting that TaVQ14 might play essential roles in salinity and drought stress.
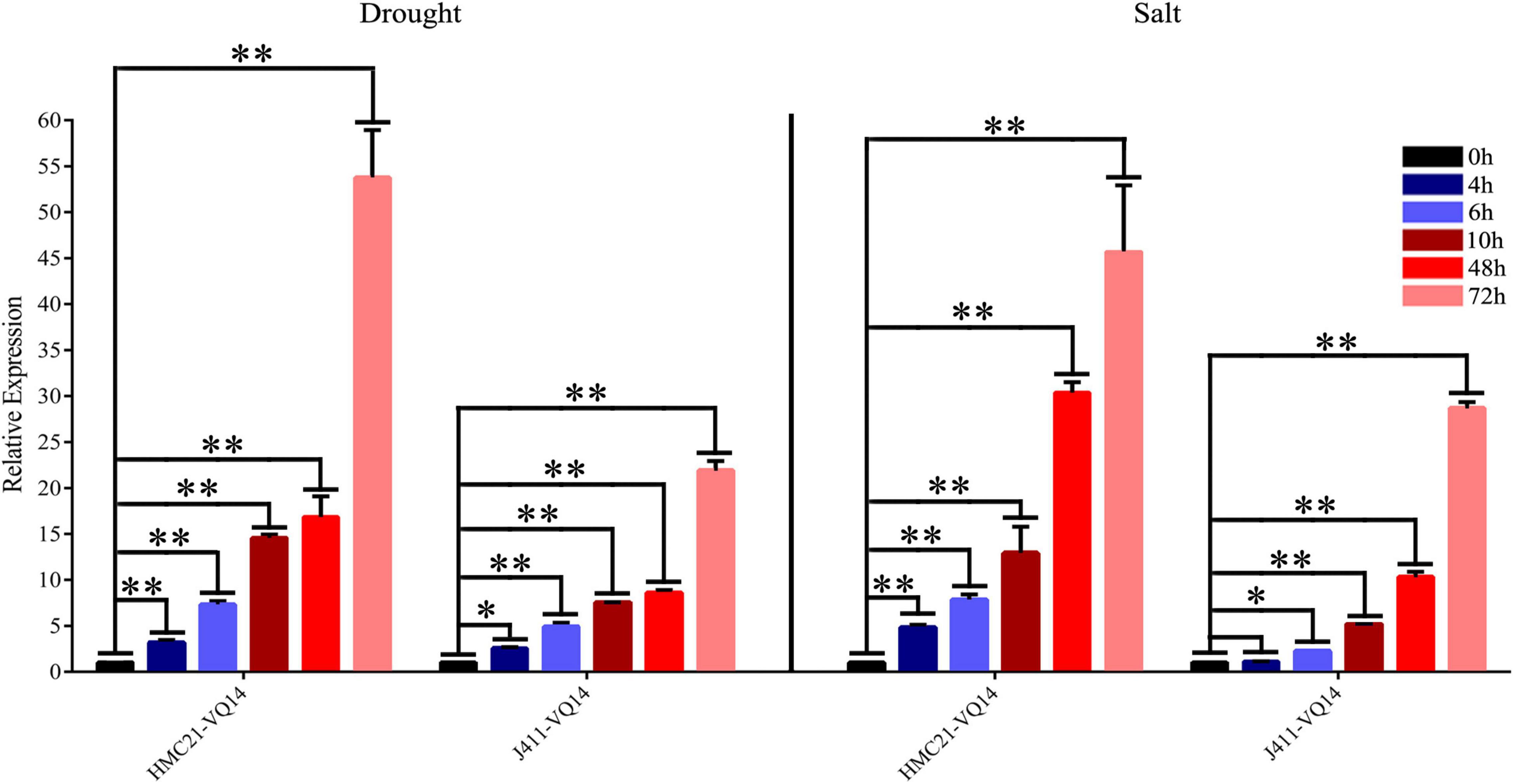
Figure 2. Analysis of TaVQ14 expression by reverse-transcription quantitative PCR in wheat seeds subjected to drought and NaCl stress. *P < 0.05, **P < 0.01.
Subcellular Localization Analysis of TaVQ14
TaVQ14 was predicted to be found in the nucleus. To confirm this prediction, tobacco leaves were transformed with Agrobacterium tumefaciens harboring the p1305-CaMV35S-TaVQ14-GFP fusion protein and observed on a confocal microscope. The results showed that the protein signal was detected in the cell nucleus, whereas the GFP control vector was distributed throughout the cell (Figure 3).
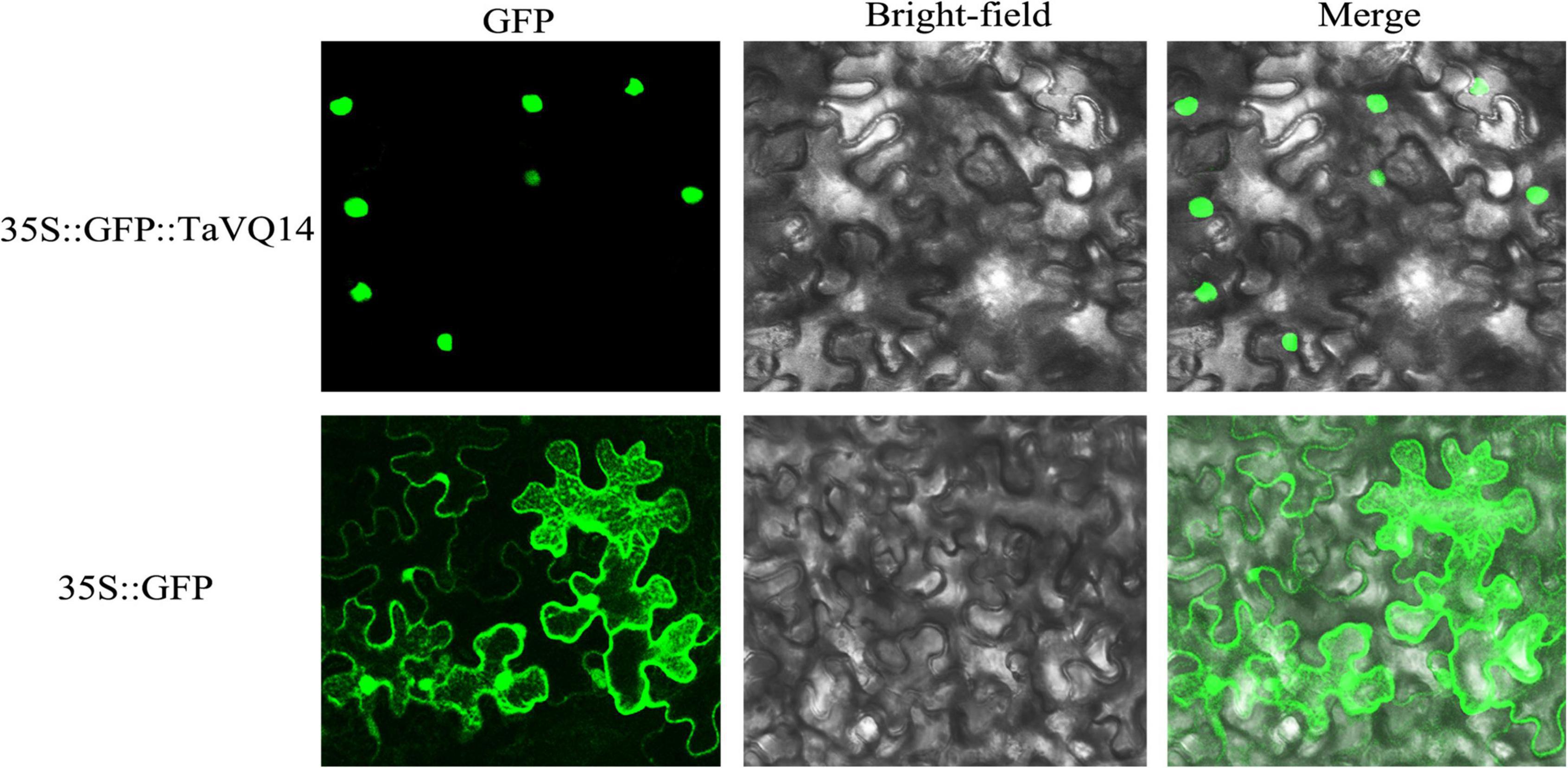
Figure 3. Nuclear localization of TaVQ14. The construct 35S:GFP:TaVQ14 and the control vector 1305 (35S:GFP) were transformed into Nicotiana tabacum leaves. The GFP signals in root cells were observed by confocal microscopy.
Obtaining of Arabidopsis Plants Overexpressing TaVQ14
Twenty T1 lines overexpressing TaVQ14 were obtained. GUS activity results showed that these lines were genetically transformed (stained blue), whereas WT lines (negative control) were not transformed and stained yellow (Supplementary Figure 1a). DNA was extracted from transgenic (T1) and WT plants. The results showed that the transgenic line and positive control, but not the WT line, presented a 650-bp band (Supplementary Figure 1b). We randomly selected five lines from the TaVQ14 transgenic Arabidopsis positive seedlings to detect the expression level of the target gene TaVQ14. The results showed that TaVQ14 was not expressed in WT, and highly expressed in four lines (Supplementary Figure 1c). In WT, and Lines 1/-2/-3, the copy number of TaVQ14 gene was detected. The results showed that the Ct value of TaVQ14 gene in WT was greater than 40, indicated that there was no TaVQ14 gene in this material. The TaVQ14 gene of Lines 1/-2/-3 were all single copy material (Supplementary Table 4). Transgenic and WT plants were self-crossed for three generations, and their seeds were harvested.
Resistance to Salt Stress
Seedlings were treated with NaCl (0, 100, or 150 mM), and the rates of germination were calculated. There was no significant difference in germination rate between transgenic and WT plants (100 vs. 99%) before treatment. Treatment with 100 and 150 mM NaCl decreased the rate of germination in WT and transgenic plants; nonetheless, germination was lower in the former (78 and 34% vs. 90 and 51%) (Figures 4A,B).
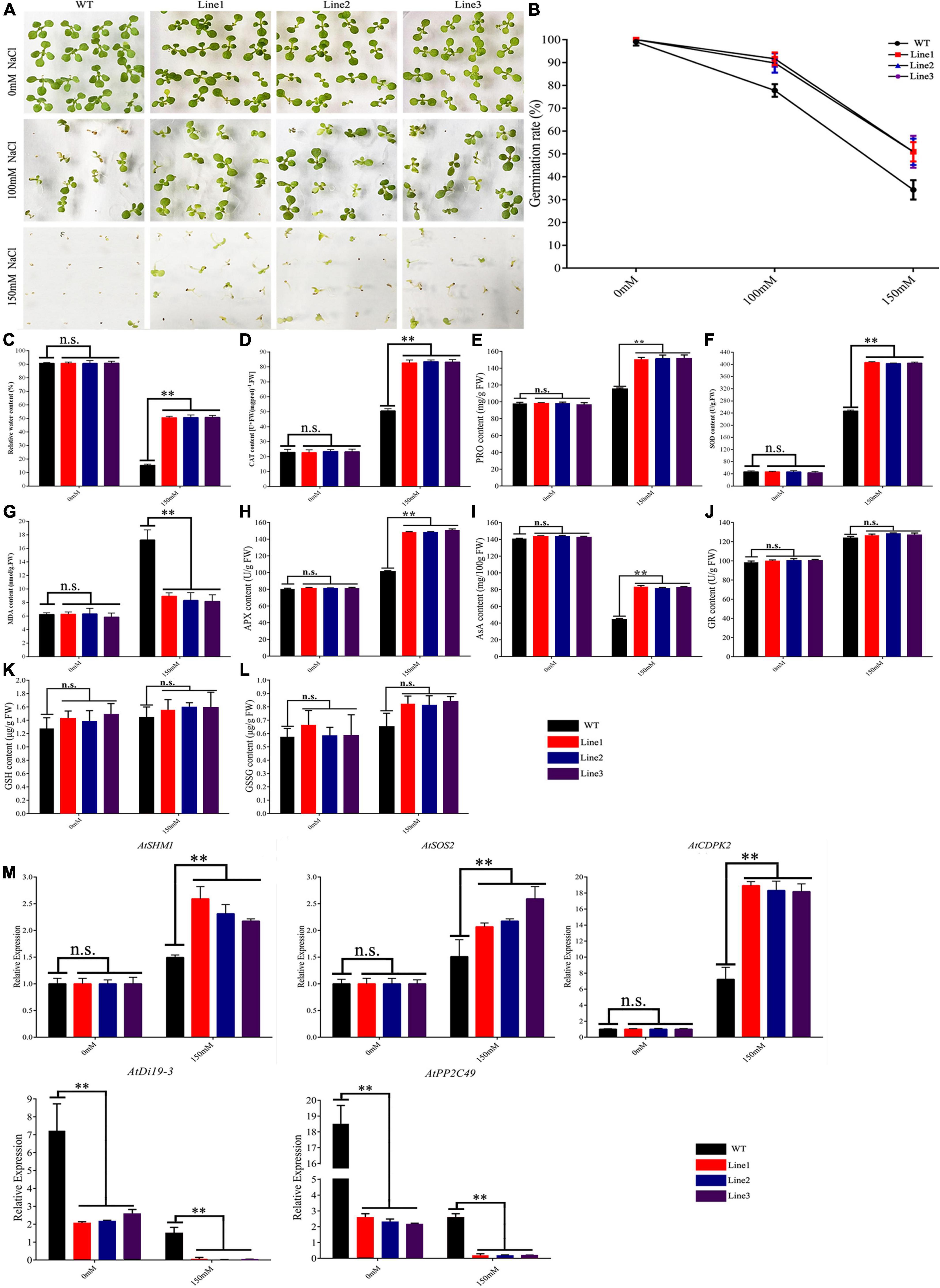
Figure 4. Germination phenotypes of TaVQ14 in transgenic Arabidopsis plants under salt stress. (A) Germination performance in TaVQ14-overexpressing and wild-type (WT) seeds grown on Murashige and Skoog medium containing 0, 100, or 150 mM of NaCl. (B) Rate of germination in transgenic and WT seeds. (C–L) Relative water content (C) and levels of catalase (D), proline (E), superoxide dismutase (F), malondialdehyde (G), Ascorbate Peroxidase (H), Ascorbic acid (I), Glutathion Reductases (J), L-Glutathione (K), and L-Glutathione oxidized (L) in transgenic and WT plants after salt treatment. Values are means ± SE (n = 3). *P < 0.05 **P < 0.01 (t-test). (M) Relative expression levels of salt-responsive genes in transgenic A. thaliana plants under normal salinity. Leaves of transgenic and WT plants were collected after salt stress. Y-axis: relative expression levels; X-axis: the time course of stress treatments; Error bars, 6 ± SE.
We measured several physiological indexes in seedlings treated with 150 mM NaCl. Treatment decreased the RWC of transgenic and WT plants; however, the rate of decline was significantly higher in WT plants. Treatment increased the concentrations of CAT, PRO, SOD, APX, GR, GSH, and GSSG in both lines. The increase was more significant in the transgenic line. The increase in MDA levels was less pronounced. The increase in GR, GSH, and GSSG was no obvious changed. Treatment reduced the concentrations of AsA in both lines, the reduce was more significant in WT (Figures 4C–L).
These results showed that transgenic plants had stronger salt tolerance than controls, indicating that the overexpression of TaVQ14 improved salt tolerance. To further assess the effect of TaVQ14 on salt tolerance in Arabidopsis, we analyzed the expression of salt stress genes (AtSHM1, AtSOS2, AtCDPK2, AtDi19-3, and AtPP2C49) (Huang et al., 2012; Qin et al., 2014; Chu et al., 2021). Salt treatment increased the expressions of AtSHM1, AtSOS2 and AtCDPK2, decreased the expression of AtDi19-3 and AtPP2C49 (Figure 4M). In addition, the relative expression of AtCDPK2 increased in both groups, suggesting that, as a crucial Ca2+ sensor, AtCDPK2 enhances salt tolerance in Arabidopsis seeds.
Resistance to Drought Stress
Seedlings were treated with mannitol (0, 150, or 300 mM), and the rates of germination were measured. There was no significant difference in the percentage of germination between transgenic and WT plants (100 vs. 99%). Treatment with 150 and 300 mM mannitol decreased the rate of germination in both groups; however, germination was less affected in transgenic plants (95 and 85% vs. 86 and 47%) (Figures 5A,B).
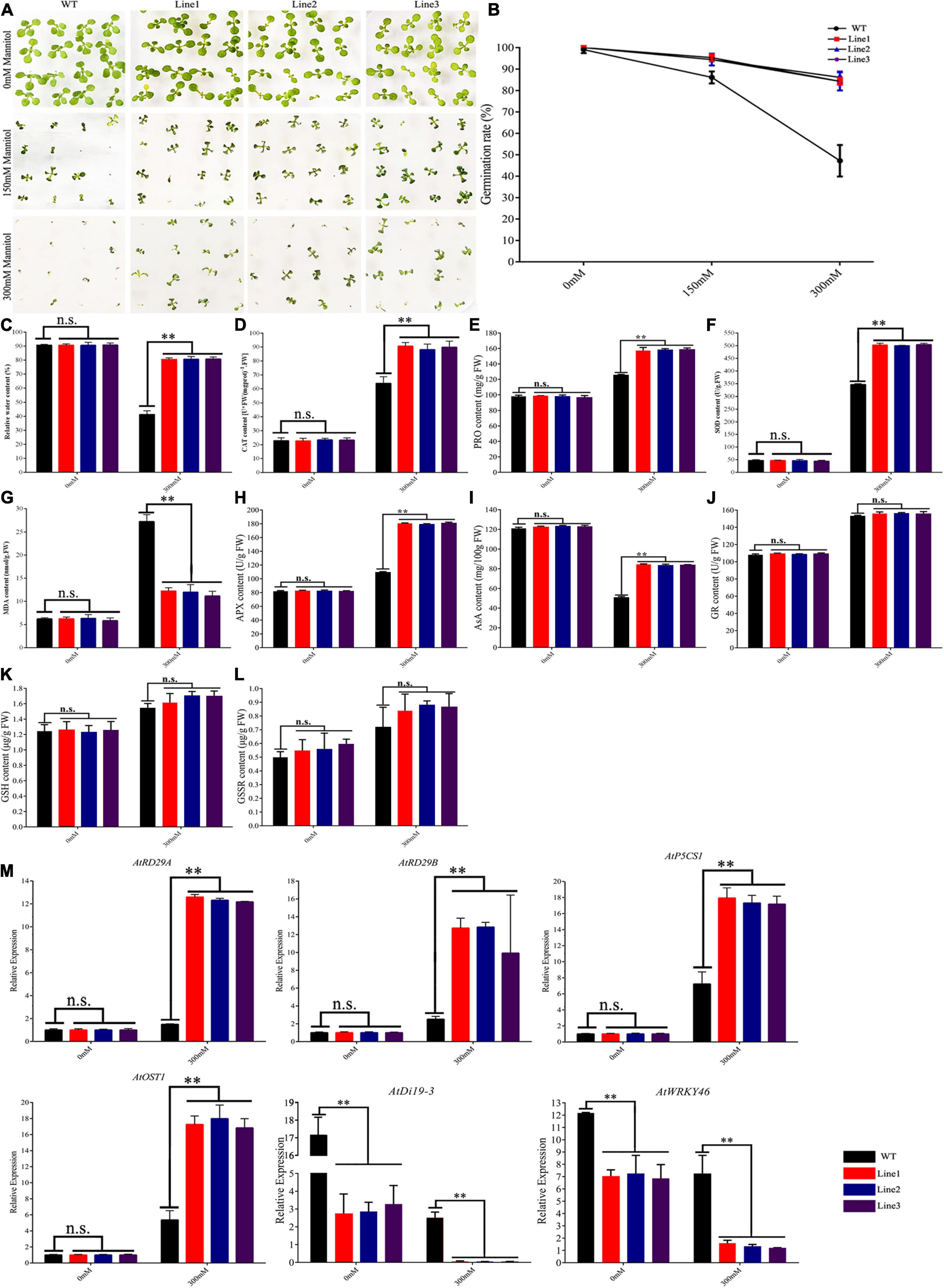
Figure 5. Germination phenotypes of TaVQ14 in transgenic Arabidopsis plants under drought stress. (A) Germination performance of TaVQ14-overexpressing and wild-type (WT) seeds on Murashige and Skoog medium containing 0, 150, or 300 mM of mannitol. (B) Rate of germination in transgenic and WT seeds. (C–L) Relative water content (C), and levels of catalase (D), proline (E), superoxide dismutase (F), malondialdehyde (G), Ascorbate Peroxidase (H), Ascorbic acid (I), Glutathion Reductases (J), L-Glutathione (K), and L-Glutathione oxidized (L) in transgenic and WT plants after drought treatment. Values are means ± SE (n = 3). *P < 0.05 **P < 0.01 (t-test). (M) Relative expression levels of drought-responsive genes in transgenic A. thaliana plants under normal water conditions. Leaves of transgenic and WT plants were collected after drought stress. Y-axis: relative expression levels; X-axis: the time course of stress treatments; Error bars, 6 ± SE.
Mannitol treatment decreased RWC in both groups, the rate of decline was higher in the WT group. Treatment increased the levels of CAT, SOD, PRO, MDA, APX, GR, GSH, and GSSG. The rate of increase in catalase and SOD was higher in transgenic plants, whereas the rate of increase in MDA was higher in WT plants. The increase in GR, GSH, and GSSG was no obvious changed. Treatment reduced the concentrations of AsA in both lines, the reduce was more significant in WT (Figures 5C–L).
These results showed that transgenic plants had stronger drought tolerance than WT plants, indicating that the overexpression of TaVQ14 increased drought resistance in Arabidopsis seeds. To further evaluate the effect of TaVQ14 on drought tolerance, the expression levels of drought-related genes (AtRD29A, AtRD29B, AtP5CS1, AtOST1, AtDi19-3, and AtWRKY46) were quantified (Huang et al., 2012; Qin et al., 2014; Chen J. et al., 2017). Drought treatment increased the expression of AtRD29A, AtRD29B, AtP5CS1, and AtOST1, decreased the expression of AtDi19-3 and AtWRKY46 (Figure 5M); suggesting that TaVQ14 improves drought tolerance by enhancing the expression of these genes.
Measurement of Ion Concentrations
The concentration of K+ and Ca2+ was significantly higher in TaVQ14-overexpressing lines, whereas Na+ content was similar between the groups (Figure 6A). Treatment with 150 mM NaCl increased Na+ contents in both groups; however, the increase was more pronounced in the WT group (Figure 6B). Treatment with 300 mM mannitol increased K+ and Ca2+ concentrations in both groups. However, the increase was higher in the transgenic group (Figure 6C).
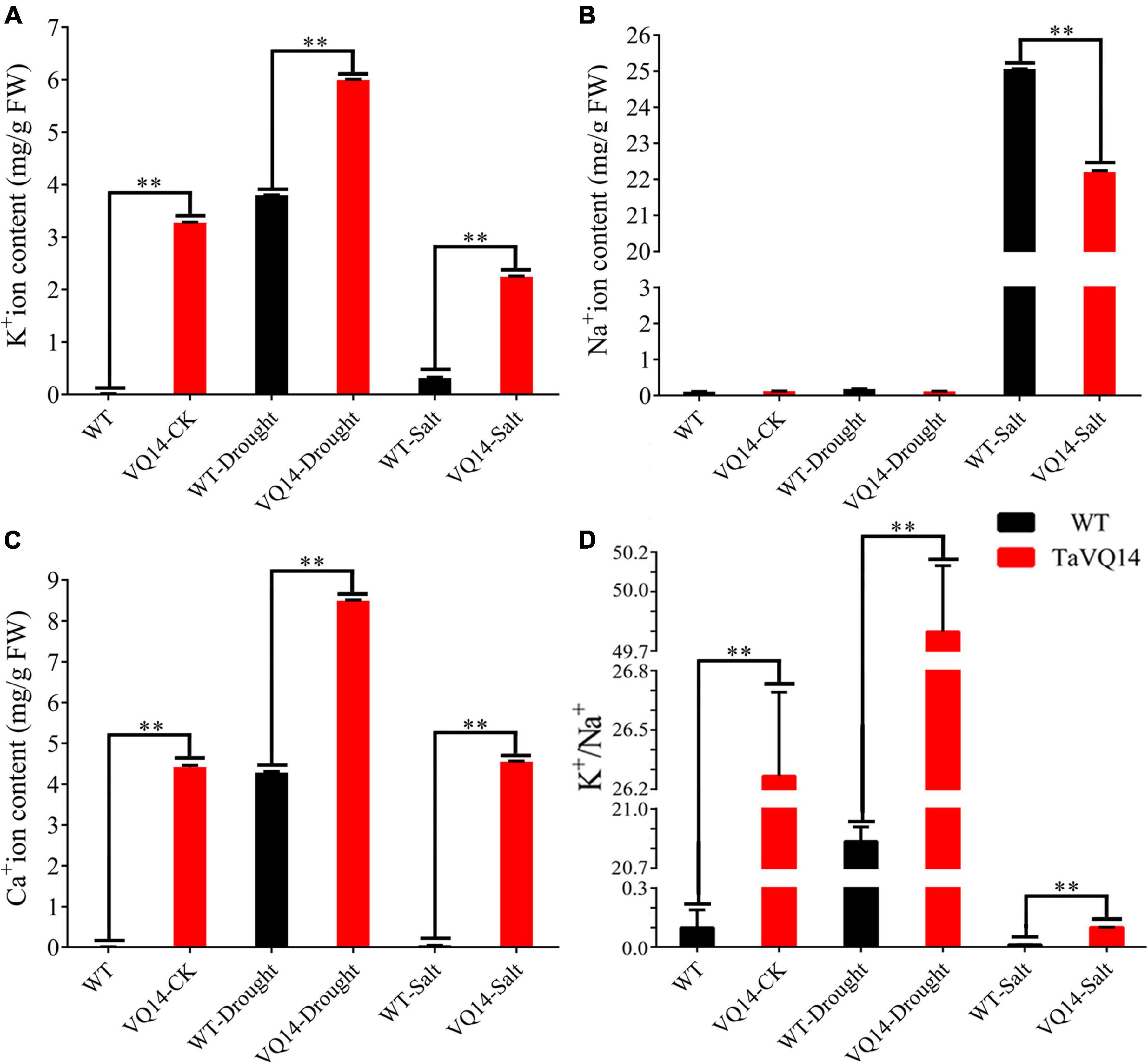
Figure 6. Concentration of Na+, K+, and Ca2+ in transgenic Arabidopsis thaliana plants under salt and drought stress. (A) K+ ion content in transgenic Arabidopsis thaliana plants under salt and drought stress. (B) Na+ ion content in transgenic Arabidopsis thaliana plants under salt and drought stress. (C) Ca2+ ion content in transgenic Arabidopsis thaliana plants under salt and drought stress. (D) K+/Na+ in transgenic Arabidopsis thaliana plants under salt and drought stress. *P < 0.05, **P < 0.01.
Response of atvq15 Mutants to Salt and Drought Stresses
Homologous genes have similar functions across species (Wang et al., 2017). To further investigate the role of TaVQ14 homologs in regulating salt and drought resistance, a phylogenetic analysis of TaVQ14 and VQ family members was performed (Supplementary Figure 2a). The atvq15 sequence was obtained from the Arashare platform, and detected homozygous plants of atvq15 mutant by screening leaf DNA (Supplementary Figure 2b). Seeds of atvq15 mutant and WT plants were treated with mannitol (0, 150, or 300 mM) or NaCl (0, 100, or 150 mM), and the rates of germination were calculated. There was no significant difference in the rate of germination rate between atvq15 mutant and WT plants (94% in both groups) before treatment (Figure 7). Treatment with 100 and 150 mM NaCl decreased the germination rate in both groups, but the rates were lower in atvq15 mutants (20 and 0% vs. 64 and 14%) (Figures 7A,B). Treatment with 150 and 300 mM Mannitol decreased the germination rate in both groups; nonetheless, the effect was stronger in atvq15 mutants (rates of 45 and 5% vs. 78 and 16%) (Figures 7C,D). These results support that AtVQ15 and TaVQ14 regulate salt and drought resistance in Arabidopsis seeds.
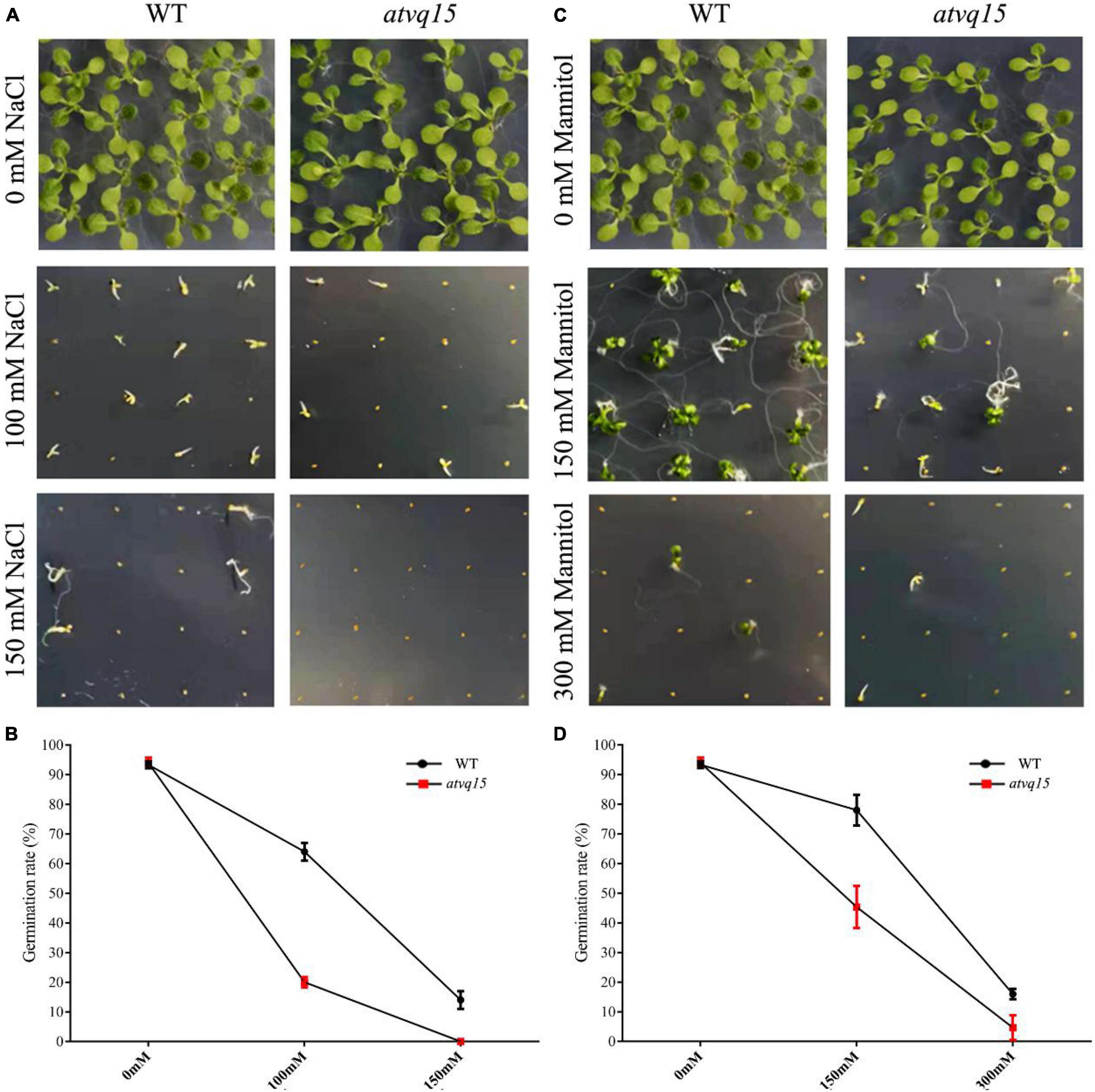
Figure 7. Germination phenotypes of Arabidopsis thaliana seeds with a mutation in the gene atvq15, a TaVQ14 homolog, under salt and drought stress. (A) Germination performance in atvq15 mutant and wild-type (WT) seeds grown on Murashige and Skoog (MS) medium containing 0, 100, or 150 mM of NaCl. (B) Rate of germination in atvq15 mutant and WT seeds. (C) Germination performance in atvq15 mutant and WT seeds grown on MS medium supplemented with 0, 150, or 300 mM of mannitol. (D) Rate of germination in atvq15 mutant and WT seeds. Relative water content, and levels of catalase, proline, superoxide dismutase, malondialdehyde, Ascorbate Peroxidase, Ascorbic acid, Glutathion Reductases, L-Glutathione, and L-Glutathione oxidized in atvq14 and WT plants after salt treatment. Relative water content, and levels of catalase, proline, superoxide dismutase, malondialdehyde, Ascorbate Peroxidase, Ascorbic acid, Glutathion Reductases, L-Glutathione, and L-Glutathione oxidized in atvq14 and WT plants after drought treatment.
Discussion
Valine-glutamine proteins are widely found in Arabidopsis, rice, maize, soybean, grapes, and other plant species (Cheng et al., 2012; Kim et al., 2013; Li N. et al., 2014; Wang et al., 2014; Wang M. et al., 2015). However, few studies have assessed the functions of these proteins. Wheat is one of the most widely cultivated crops; nonetheless, the functions of VQ genes in wheat are incompletely understood. Our previous study has shown that TaVQ14 encodes an unstable basic hydrophobic protein (Cheng et al., 2021). Therefore, this gene was selected for further functional analysis. Our results showed that TaVQ14 expression was significantly upregulated under high salinity and drought conditions, indicating that TaVQ14 was involved in salt and drought stress responses.
Arabidopsis thaliana is an excellent model for research in plant biology is an excellent model for research in plant biology, which can obtain transgenic plants in short time. Therefore, transgenic A. thaliana lines were used to assess the function of TaVQ14. Under drought and salt stress, the rate of germination in TaVQ14-overexpressing lines was significantly higher than that of WT plants, indicating that the tolerance of the former to drought and salinity stress was improved.
A plant mutant for AtVQ15, a TaVQ14 homolog, was produced. The results revealed that the percentage of seed germination was lower in these mutants than in WT controls under drought and salt stress. These findings indicate that the AtVQ15 mutation reduces stress tolerance and that TaVQ14 and its homolog AtVQ15 regulate tolerance to drought and salinity. Furthermore, gene expression analysis showed that several genes responsive to drought (AtRD29A, AtRD29B, AtP5CS1, AtOST1, AtWRKY46, and AtDi19-3) and salinity (AtSHM1, AtSOS2, AtCDPK2, AtPP2C29, and AtDi19-3) (Huang et al., 2012) were differentially expressed after treatment, suggesting that TaVQ14 enhances the resistance of Arabidopsis seeds to drought and salinity by regulating the expressions of these genes.
The concentrations of K+, Na+, and Ca2+ were measured in TaVQ14-overexpressing and WT lines. Ca2+ concentration and K+/Na+ ratio was significantly higher in the transgenic line before treatment, suggesting that TaVQ14 overexpression improves drought and salt resistance by increasing Ca2+ and K+ concentrations. Treatment with 300 mM NaCl increased Ca2+, K+, and K+/Na+ ratio in the transgenic line, suggesting that TaVQ14 increases resistance to salt stress by excreting Na+ and increasing the uptake of Ca2+ and K+ (Figures 6A–D). Treatment with 300 mM mannitol increased the uptake of K+ and Ca2+ in both groups; nonetheless, the increase was more pronounced in the transgenic line, suggesting that TaVQ14 overexpression improves drought resistance by increasing K+ and Ca2+ concentrations. AtCDPK2 was upregulated in the transgenic line under salt stress, suggesting that TaVQ14 improves salt tolerance by increasing AtCDPK2 expression through Ca2+ signaling. Salinity stress increases cytosolic Ca2+ levels. Calcium-dependent protein kinases (CPKs or CDPKs) are strongly implicated in Ca2+ signaling in plants and play an important role in salinity stress (Singh et al., 2017). In rice, OsCPK21 genes regulated the ABA-dependent salt stress signaling pathway (Asano et al., 2011). OsCPK12 conferred tolerance to salt stress through regulation of ROS homeostasis (Asano et al., 2012). In addition, salinity stress tolerance is stronger in plants overexpressing OsCPK4 (Campo et al., 2014). These findings reveal that Ca2+ signaling, together with ROS signaling and hormonal regulation, mediates the response to salinity stress. However, the mechanisms underlying the regulation of salt tolerance by TaVQ14 and CDPK need to be further investigated.
Stress resistance is improved by modulating gene expression and physiological and biochemical processes, including the accumulation of osmotic substances and the increase in active oxygen scavenging activity (Zhou et al., 2018; Zhang et al., 2021). Free proline levels in plants are low under normal conditions; nonetheless, under stress conditions, including drought, low temperature, high salinity, and high alkalinity, proline is stored in large quantities, and storage levels are positively correlated with stress resistance. Therefore, proline is used as a biochemical index of stress resistance in plants (Xiang et al., 2007; Hnilickova et al., 2021; Qian et al., 2021; Rajametov et al., 2021). Our results showed that proline level was significantly higher in transgenic plants than in WT controls. Consistent with our results, the large increase in proline concentration increases intracellular osmotic pressure and decreases water potential and water content (Song et al., 2011). MDA is the main product of membrane lipid peroxidation, leading to membrane damage and plant damage. Thus, MDA levels are positively correlated with the degree of membrane lipid peroxidation and can serve as an indicator of cellular reactive oxygen species stress (Xiong et al., 2002; Mittler et al., 2004). Our results showed that MDA concentration was significantly lower in transgenic plants than in WT controls, indicating that TaVQ14 overexpression increased resistance to oxidative stress. CAT, SOD, and APX are important protective enzymes and reduce oxidative stress by decreasing the production of active oxygen and hydrogen peroxide (May, 2008). In this study, CAT, SOD, and APX levels were significantly higher in the transgenic line than in WT plants, which may explain why the MDA concentration was lower in the former.
The analysis of gene expression, physiological, biochemical, and phenotypic data demonstrated the role of TaVQ14 overexpression in Arabidopsis improving its salt tolerance and drought tolerance, indicating that TaVQ14 plays important roles in improving salt tolerance and drought resistance in wheat, and these data provide a basis for the functional analysis of TaVQ14 in wheat.
China’s Bohai Rim region has more than 40 million mu of medium and low yield farmland and more than 10 million mu of saline alkali wasteland, which has been suffering from drought, waterlogging and alkali disasters for a long time. This experiment proved that TaVQ14 had the functions of salt tolerance and drought tolerance, and was an excellent salt tolerance and drought tolerance gene. Overexpression and knockout of TaVQ14 gene in wheat is the direction of our subsequent experimental work, and we hope to widely apply this gene to the cultivation of new wheat varieties with salt tolerance and drought tolerance for the increase of agricultural production and income.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
XC and HY projected the study, put into effect the main bioinformatics analysis, and drew up the manuscript. ZC, CG, and WG carried out the software and helped to handle figures and tables. BT, JJC, and XP participated in the experimental test. SY processed experimental data and joined to amend the manuscript. JJC and XP took part in the software and draw up the manuscript. JL and CXM had a hand in the project of the study and helped to revamp the manuscript. CC and HPZ conceived and guided the experiment, were involved in its project and coordination, and helped to draw up the manuscript. All authors read and accepted the final manuscript.
Funding
This work was supported by grants from the China’s Agricultural Research System (CARS-03), Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP), and The Agriculture Research System of Anhui province (AHCYTX-02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.870586/full#supplementary-material
Abbreviations
VQ, valine-glutamine; ABA, abscisic acid; qRT-PCR, quantitative real-time PCR; RWC, plant water content; MDA, malondialdehyde; CAT, catalase; SOD, superoxide dismutase; PRO, proline; GUS, β -glucuronidase activity; APX, ascorbate peroxidase; AsA, ascorbic acid; GR, glutathion reductases; GSH, l-glutathione; GSSG, L-glutathione oxidized.
Footnotes
References
Andreasson, E., Jenkins, T., Brodersen, P., Thorgrimsen, S., Petersen, N. H. T., Zhu, S., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589. doi: 10.1038/sj.emboj.7600737
Asano, T., Hakata, M., Nakamura, H., Aoki, N., Komatsu, S., Ichikawa, H., et al. (2011). Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 75, 179–191. doi: 10.1007/s11103-010-9717-1
Asano, T., Hayashi, N., Kikuchi, S., and Ohsugi, R. (2012). CDPK-mediated abiotic stress signaling. Plant Signal. Behav. 7, 817–821. doi: 10.4161/psb.20351
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plan Soil 39, 205–207. doi: 10.1007/BF00018060
Brenchley, R., Spannagl, M., Pfeifer, M., Barker, G. L. A., Damore, R., Allen, A. M., et al. (2012). Analysis of the bread wheat genome using whole genome shotgun sequencing. Nature 491, 705–710. doi: 10.1038/nature11650
Campo, S., Baldrich, P., Messeguer, J., Lalanne, E., Coca, M., and San Segundo, B. (2014). Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704. doi: 10.1104/pp.113.230268
Chen, D., Chen, Z., Wu, M., Wang, Y., Wang, Y., Yan, H., et al. (2017). Genome-Wide Identification and Expression Analysis of the HD-Zip Gene Family in Moso Bamboo (Phyllostachys edulis). J. Plant Growth Regul. 36, 323–337. doi: 10.1007/s00344-016-9642-x
Chen, J., Nolan, T., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell 29, 1425–1439. doi: 10.1105/tpc.17.00364
Cheng, X. R., Gao, C., Liu, X., Xu, D. M., Pan, X., Gao, W., et al. (2021). Identification of the Wheat VQ protein family and expression analysis of candidate genes associated with seed dormancy and germination. BMC Plant Biol. 22:119. doi: 10.1186/s12870-022-03430-1
Cheng, X., Wang, S., Xu, D. M., Liu, X., Li, X., Xiao, W., et al. (2019a). Identification and analysis of the GASR gene family in Common Wheat (Triticum aestivum L.) and characterization of TaGASR34, a gene associated with seed dormancy and germination. Front. Genet. 10:980. doi: 10.3389/fgene.2019.00980
Cheng, X., Xiong, R., Yan, H., Gao, Y., Liu, H., Wu, M., et al. (2019b). The trihelix family of transcription factors: functional and evolutionary analysis in Moso bamboo (Phyllostachys edulis). BMC Plant Biol. 19:154. doi: 10.1186/s12870-019-1744-8
Cheng, X. R., Wang, Y., Xiong, R., Gao, Y., Yan, H., and Xiang, Y. (2020). A Moso bamboo gene VQ28 confers salt tolerance to transgenic Arabidopsis plants. Planta 251:99. doi: 10.1007/s00425-020-03391-5
Cheng, X. R., Xiong, R., Liu, H., Wu, M., Chen, F., Yan, H., et al. (2018). Basic helix-loop-helix gene family: genome wide identification, phylogeny, and expression in Moso bamboo. Plant Physiol. Biochem. 132, 104–119. doi: 10.1016/j.plaphy.2018.08.036
Cheng, Y., Zhou, Y., Yang, Y., Chi, Y. J., Zhou, J., Chen, J. Y., et al. (2012). Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159, 810–825. doi: 10.1104/pp.112.196816
Chu, M., Chen, P., Meng, S., Xu, P., and Lan, W. (2021). The Arabidopsis phosphatase PP2C49 negatively regulates salt tolerance through inhibition of AtHKT1;1. J. Integr. Plant Biol. 63, 528–542. doi: 10.1111/jipb.13008
Dai, X., Xu, Y., Ma, Q., Xu, W., Wang, T., Xue, Y., et al. (2007). Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 143, 1739–1751. doi: 10.1104/PP.106.094532
Del Rio, L. A., Ortega, M. G., Lopez, A. L., and Gorgo, J. L. (1977). A more sensitive modification of the catalase assay with the Clark oxygen electrode: Application to the kinetic study of the pea leaf enzyme. Anal. Biochem. 80, 409–415. doi: 10.1016/0003-2697(77)90662-5
Fill, B. K., and Petersen, M. (2011). Constitutive expression of MKS1confers susceptibility to Botrytis cinerea infection independent of PAD3 expression. Plant Signal. Behav. 6, 1425–1427. doi: 10.4161/psb.6.10.16759
Gao, Y., Liu, H., Wang, Y., Li, F., and Xiang, Y. (2017). Genome-wide identification of PHD-finger genes and expression pattern analysis under various treatments in moso bamboo (Phyllostachys edulis). Plant Physiol. Biochem. 123, 378–391. doi: 10.1016/j.plaphy.2017.12.034
Goldstein, D. B. (1968). A method for assay of catalase with the oxygen cathode. Anal. Biochem. 24, 431–437. doi: 10.1016/0003-2697(68)90148-6
Heath, R. L., and Packer, L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Hnilickova, H., Kraus, K., Vachova, P., and Hnilicka, F. (2021). Salinity stress affects photosynthesis, malondialdehyde formation, and proline content in Portulaca oleracea L. Plants 10:845. doi: 10.3390/plants10050845
Hu, P., Zhou, W., Cheng, Z., Fan, M., Wang, L., and Xie, D. (2013). JAV1 controls jasmonate-regulated plant defense. Mol. Cell 50, 504–515. doi: 10.1016/j.molcel.2013.04.027
Hu, Y., Chen, L., Wang, H., Zhang, L., Wang, F., and Yu, D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. doi: 10.1111/tpj.12159
Huang, X., Zhang, Y., Jiao, B., Chen, G., Huang, S., Guo, F., et al. (2012). Overexpression of the wheat salt tolerance-related gene TaSC enhances salt tolerance in Arabidopsis. J. Exp. Bot. 63, 5463–5473. doi: 10.1093/jxb/ers198
Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403. doi: 10.1146/annurev.arplant.47.1.377
Jakab, G., Zimmerli, L., Métraux, J. P., and Mauch-Mani, B. (2005). Enhancing Arabidopsis salt and drought Stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 139, 267–274. doi: 10.1104/pp.105.065698
Ji, X., Dong, B., Shiran, B., Shiran, B., Talbot, M. J., Edlington, J. E., et al. (2011). Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol. 156, 647–662. doi: 10.1104/pp.111.176164
Ji, Y., Zhang, P., Xing, X., Jia, L., Zhang, Y., Jia, T., et al. (2019). Effect of 1α, 25-dihydroxyvitamin D3 on the osteogenic differentiation of human periodontal ligament stem cells and the underlying regulatory mechanism. Int. J. Mol. Med. 43, 167–176. doi: 10.3892/ijmm.2018.3947
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev. Plant Biol. 57, 781–803. doi: 10.1146/ANNUREV.ARPLANT.57.032905.105444
Kim, D. Y., Kwon, S. I., Choi, C., Lee, H., Ahn, I., Park, S. R., et al. (2013). Expression analysis of rice VQ genes inresponse to biotic and abiotic stresses. Gene 529, 208–214. doi: 10.1016/j.gene.2013.08.023
Kochba, J., Lavee, S., and Spiegelroy, P. (1977). Differences in peroxidase activity and isoenzymes in embryogenic and non-embryogenic‘Shamouti’orange ovular callus lines. Plant Cell Physiol. 18, 463–467. doi: 10.1093/oxfordjournals.pcp.a075455
Lai, Z., Li, Y., Wang, F., Cheng, Y., Fan, B., Yu, J. Q., et al. (2011). Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23, 3824–3841. doi: 10.1105/tpc.111.090571
Li, N., Li, X., Xiao, J., and Wang, S. (2014). Comprehensive analysis of VQ motif-containing gene expression in rice defense responses to three pathogens. Plant Cell Rep. 33, 1493–1505. doi: 10.1007/s00299-014-1633-4
Li, Y., Jing, Y., Li, J., Xu, G., and Lina, R. (2014). Arabidopsis VQ-motif-containing protein 29 represses seedling de-etiolation by interacting with PIF1. Plant Physiol. 164, 2068–2080. doi: 10.1104/pp.113.234492
Li, X., Qin, R., Du, Q., Cai, L., Hu, D., Du, H., et al. (2020). Knockdown of GmVQ58 encoding a VQ motif-containing protein enhances soybean resistance to the common cutworm (Spodoptera litura Fabricius). J. Exp. Bot. 71, 3198–3210. doi: 10.1093/jxb/eraa095
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001
May, G. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. Cell Mol. Biol. 53, 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Mittler, R., Vanderauwera, S., Gollery, M., and Van, B. F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1007/978-90-481-3112-9_5
Pascal, P., Lennart, E. L., Siska, H., Katja, K., Kai, N., Gerit, B., et al. (2014). The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of VQ-motif-containing proteins to regulate immune responses. New Phytol. 203, 592–606. doi: 10.1111/nph.12817
Petersen, K., Qiu, J., Lutje, J., Katrine, F. B., Sidsel, H., John, M., et al. (2010). Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminaldomain for proper function. PLoS One 5:e14364. doi: 10.1371/journal.pone.0014364
Qian, M., Wang, L., Zhang, S., Sun, L., Luo, W., Posny, D., et al. (2021). Investigation of proline in superficial scald development during low temperature storage of ‘Dangshansuli’ pear fruit. Postharvest Biol. Technol. 181:111643. doi: 10.1016/j.postharvbio.2021.111643
Qin, L. X., Li, Y., Li, D. D., Xu, W. L., Zheng, Y., and Li, X. B. (2014). Arabidopsis drought-induced protein Di19-3 participates in plant response to drought and high salinity stresses. Plant Mol. Biol. 86, 609–625. doi: 10.1007/s11103-014-0251-4
Rajametov, S., Yang, E. Y., Cho, M. C., Chae, S. Y., and Chae, W. B. (2021). Heat-tolerant hot pepper exhibits constant photosynthesis via increased 4 transpiration rate, high proline content and fast recovery in heat stress 5 condition. Sci. Rep. 11:14328. doi: 10.20944/preprints202107.0348.v1
Ren, Y. (2012). Effects of salt stress on different wheat varieties at seedling stage. Hubei Agric. Sci. 51, 3702–3705. doi: 10.14088/j.cnki.issn0439-8114.2012.17.054
Rorth, M., and Jensen, P. K. (1967). Determination of catalase activity by means of the Clark oxygen electrode. Biochim. Biophys. Acta 139, 171–173. doi: 10.1016/0005-2744(67)90124-6
Sequeira, L., and Mineo, L. (1966). Partial purification and kinetics of indoleacetic acid oxidase from tobacco roots. Plant Physiol. 41, 1200–1208. doi: 10.1104/pp.41.7.1200
Singh, A., Sagar, S., and Biswas, D. K. (2017). Calcium dependent protein kinase, a versatile player in plant stress management and development. Crit. Rev. Plant Sci. 36, 336–352. doi: 10.1080/07352689.2018.1428438
Song, S., Chen, Y., Zhao, M., and Zhang, W. H. (2011). A novel Medicago truncatula HD-Zip gene, MtHB2, is involved in abiotic stress responses. Environ. Exp. Bot. 69, 1–9. doi: 10.1016/j.envexpbot.2012.02.001
Song, W., Zhao, H., Zhang, X., Lei, L., and Lai, J. (2016). Genome-wideidentification of VQ motif-containing proteins and their expressionprofiles under abiotic stresses in maize. Front. Plant Sci. 6:1177. doi: 10.3389/fpls.2015.01177
Toufighi, K., Brady, S. M., Austin, R. S., Ly, E., and Provart, N. J. (2005). The botany array resource: e-northerns, expression angling, and promoter analyses. Plant J. 43, 153–163. doi: 10.1111/j.1365-313X.2005.02437
Verslues, P. E., Agarwal, M., Surekha, K. A., Zhu, J. H., and Zhu, J. K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45, 523–539. doi: 10.1111/j.1365-313X.2005.02593.x
Wang, A., Garcia, D., Zhang, H., Feng, K., Chaudhury, A., Berger, F., et al. (2010). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63, 670–679. doi: 10.1111/j.1365-313X.2010.04271.x
Wang, H., Hu, Y., Pan, J., and Yu, D. (2015). Arabidopsis VQ motif-containing proteins VQ12 and VQ29 negatively modulatebasal defense against Botrytis cinerea. Sci. Rep. 5:14185. doi: 10.1038/srep14185
Wang, M., Vannozzi, A., Wang, G., Zhong, Y., Corso, M., Cavallini, E., et al. (2015). A comprehensive survey of the grapevine VQ gene family and its transcriptional correlation with WRKY proteins. Front. Plant Sci. 6:417. doi: 10.3389/fpls.2015.00417
Wang, J., and Huang, R. (2019). Modulation of ethylene and ascorbic acid on reactive oxygen species scavenging in plant salt response. Front. Plant Sci. 10:319. doi: 10.3389/fpls.2019.00319
Wang, X., Zhang, H., Sun, G., Jin, Y., and Qiu, L. (2014). Identification of active VQ motif-containing genes and the expression patterns under low nitrogen treatment in soybean. Gene 543, 237–243. doi: 10.1016/j.gene.2014.04.012
Wang, Y., Liu, H., Zhu, D., Gao, Y., Yan, H., and Yan, X. (2017). Genome-wide analysis of VQ motif-containing proteins in Moso bamboo (Phyllostachys edulis). Planta 246, 165–181. doi: 10.1007/s00425-017-2693-9
Xiang, Y., Huang, Y., and Xiong, L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144, 1416–1428. doi: 10.1104/pp.107.101295
Xin, G. S., Zhao, Y. Q., Jiang, B., Xin, Z. W., and Liu, X. Y. (2019). Inhibition of MUC1-C regulates metabolism by AKT pathway in esophageal squamous cell carcinoma. J. Cell. Physiol. 234, 12019–12028. doi: 10.1002/jcp.27863
Xiong, L., Schumaker, K. S., and Zhu, J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14, (Suppl), S165–S183. doi: 10.1105/TPC.000596
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Yu, C. S., Lin, C. J., and Huang, J. K. (2004). Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. doi: 10.1110/ps.03479604
Zelm, E. V., Zhang, Y., and Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 1–24. doi: 10.1146/annurev-arplant-050718-100005
Zhang, H., Zhu, J., Gong, Z., and Zhu, J. K. (2021). Abiotic stress responses in plants. Nat. Rev. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhang, J., Ahmad, S., Wang, L. Y., Han, Q., Zhang, J. C., and Luo, Y. P. (2019). Cell death induced by α-terthienyl via reactive oxygen species-mediated mitochondrial dysfunction and oxidative stress in the midgut of Aedes aegypti larvae. Free Radic. Biol. Med. 137, 87–98. doi: 10.1016/j.freeradbiomed.2019.04.021
Zhang, Z., Liu, H., Sun, C., Ma, Q., Bu, H., Chong, K., et al. (2018). A C2H2 zinc-finger protein OsZFP213 interacts with OsMAPK3 to enhance salt tolerance in rice. J. Plant Physiol. 43, 167–176. doi: 10.1016/j.jplph.2018.07.003
Zhao, H., Ma, H., Yu, L., Wang, X., and Zhao, J. (2012). Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS One 7:e49210. doi: 10.1371/journal.pone.0049210
Zhao, Y., Ma, Q., Jin, X., Peng, X., Liu, J., Deng, L., et al. (2014). A novel maize homeodomain-leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol. 55, 1142–1156. doi: 10.1093/pcp/pcu054
Zhao, Y., Yu, W., Hu, X., Shi, Y., Liu, Y., Zhong, Y., et al. (2018). Physiological and transcriptomic analysis revealed the involvement of crucial factors in heat stress response of Rhododendron hainanense. Gene 660, 109–119. doi: 10.1016/j.gene.2018.03.082
Zhou, Y. B., Liu, C., Tang, D. Y., Yan, L., Wang, D., Yang, Y. Z., et al. (2018). The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 30, 1100–1118. doi: 10.1105/tpc.17.01000
Keywords: wheat, salt, drought, subcellular localization, expression analysis, TaVQ14
Citation: Cheng X, Yao H, Cheng Z, Tian B, Gao C, Gao W, Yan S, Cao J, Pan X, Lu J, Ma C, Chang C and Zhang H (2022) The Wheat Gene TaVQ14 Confers Salt and Drought Tolerance in Transgenic Arabidopsis thaliana Plants. Front. Plant Sci. 13:870586. doi: 10.3389/fpls.2022.870586
Received: 07 February 2022; Accepted: 04 April 2022;
Published: 10 May 2022.
Edited by:
Rajeev K. Varshney, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaReviewed by:
Deepak Kumar, Banaras Hindu University, IndiaÁgnes Szepesi, University of Szeged, Hungary
Copyright © 2022 Cheng, Yao, Cheng, Tian, Gao, Gao, Yan, Cao, Pan, Lu, Ma, Chang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng Chang, aGFuZ3Rnd0AxNjMuY29t; Haiping Zhang, emhhbmdoYWlwaW5nQGFoYXUuZWR1LmNu
†These authors have contributed equally to this work
 Xinran Cheng
Xinran Cheng Hui Yao1†
Hui Yao1† Jiajia Cao
Jiajia Cao