- Fujian Provincial Key Laboratory of Plant Functional Biology, College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
Alternative splicing (AS) is a common post-transcriptional regulatory process in eukaryotes. AS has an irreplaceable role during plant development and in response to environmental stress as it evokes differential expression of downstream genes or splicing factors (e.g., serine/arginine-rich proteins). Numerous studies have reported that loss of AS capacity leads to defects in plant growth and development, and induction of stress-sensitive phenotypes. A role for post-translational modification (PTM) of AS components has emerged in recent years. These modifications are capable of regulating the activity, stability, localization, interaction, and folding of spliceosomal proteins in human cells and yeast, indicating that PTMs represent another layer of AS regulation. In this review, we summarize the recent reports concerning ubiquitin and ubiquitin-like modification of spliceosome components and analyze the relationship between spliceosome and the ubiquitin/26S proteasome pathway in plants. Based on the totality of the evidence presented, we further speculate on the roles of protein ubiquitination mediated AS in plant development and environmental response.
Introduction
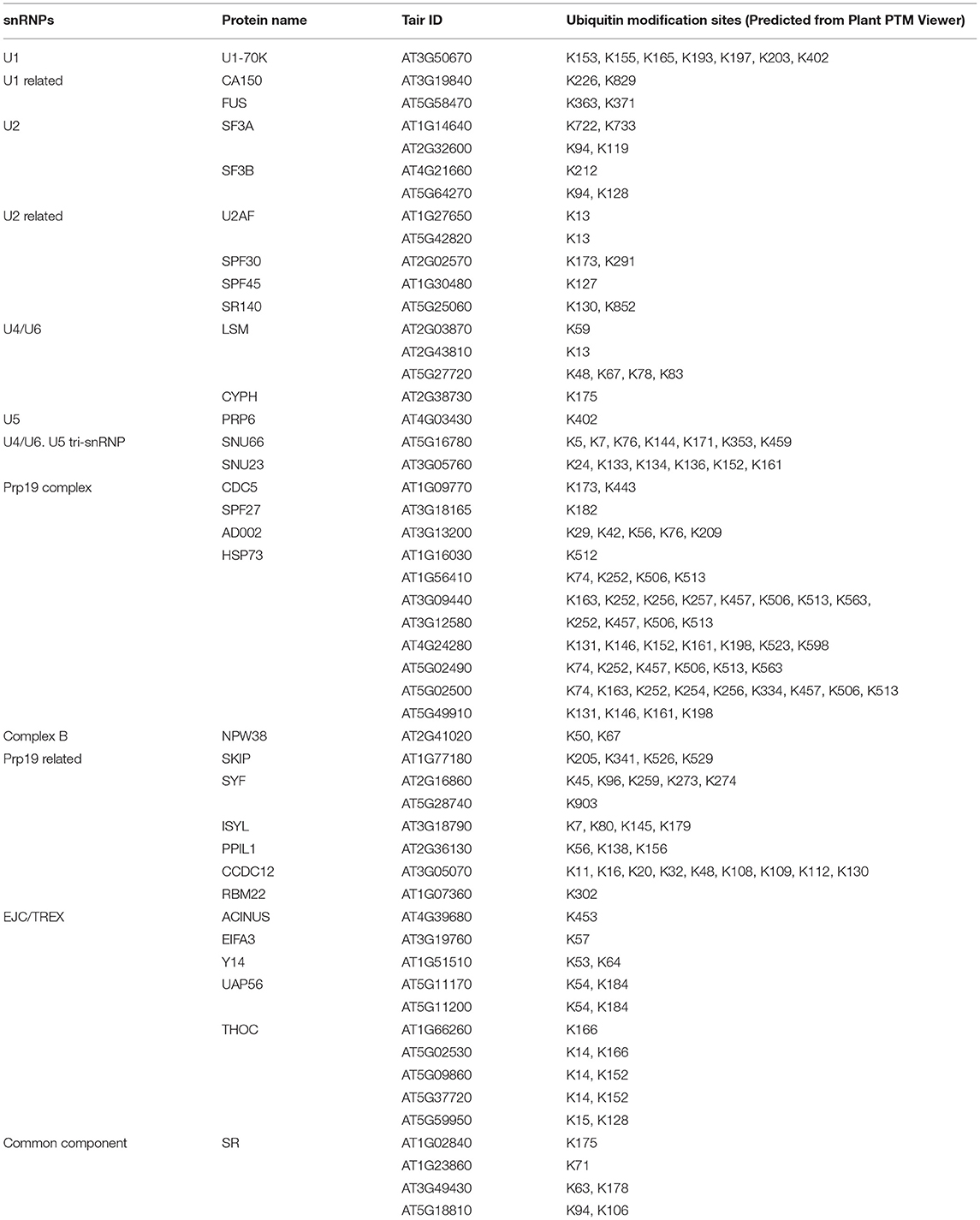
Removal of introns through pre-mRNA splicing is a key regulatory step in eukaryotic gene expression, a process which was initially characterized in viruses (Berget et al., 1977; Chow et al., 1977). During this process, introns are removed via the spliceosome, a high molecular weight complex that is assembled at each intron. The spliceosome is made up of five small nuclear ribonucleoprotein particles (snRNPs)—U1, U2, U4, U5, and U6 snRNPs—and over 200 additional proteins (Figure 1). The five snRNPs and their associated proteins coordinate splice-site recognition, spliceosome assembly, and catalysis of the two transesterification steps (Mckay and Johnson, 2010; Koncz et al., 2012; Reddy et al., 2013; Carvalho et al., 2016). Contained within these five snRNPs are uridine-rich small nuclear RNAs, and their core particles are formed by Sec1/Munc18 (SM) or like-SM (LSM) proteins. In addition to these snRNPs, other spliceosomal components include pre-mRNA processing factor 19 (PRP19) complex, exon junction complex (EJC)/transcription and export (TREX) complex, U1 related, U2 related, U4/U6, U5 tri-snRNP, and more (Figure 1B) (Tharun, 2009). These components are assembled on pre-mRNAs in a stepwise manner to form active spliceosomes with highly dynamic and tightly regulated conformation and composition (Figure 1A) (Tharun, 2009; Mckay and Johnson, 2010).
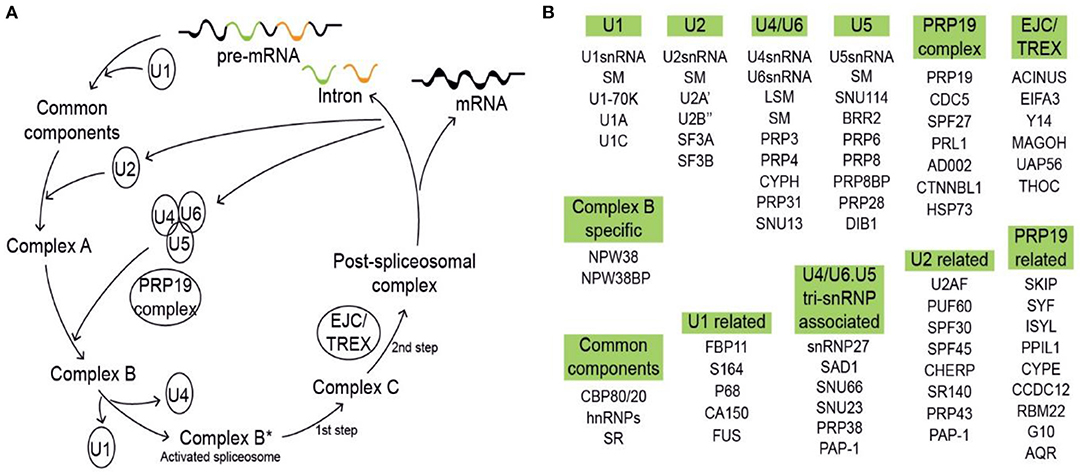
Figure 1. Model of pre-mRNA splicing process (drawn based on https://www.kegg.jp/pathway/map03040). (A) The process of pre-mRNA splicing. The initial step of splice-site recognition comprises U1 small nuclear ribonucleoprotein particle (snRNP) binding to the 5′ splice site and U2 auxiliary factor (U2AF) binding to the 3′ splice site. U2AF35, the small subunit of U2AF, binds to the intron/exon border, whereas the large subunit U2AF65 binds to a region rich in pyrimidines designated the polypyrimidine tract. Subsequently, U2 snRNP binds to the branch point, and a preformed complex of U4, U5, and U6 snRNPs is recruited to the intron. After major rearrangements and release of the U1 and U4 snRNPs, the splicing reaction takes place. (B) All spliceosomal components are grouped by category, including U1, U2, U4/U6, U5, PRP19 complex, EJC/TREX, U1 related, U2 related, PRP19 related, U4/U6, U5 tri-snRNP associated, and common components.
Following the initial discovery and characterization of spliceosomal introns, subsequent work revealed their functional roles in gene expression. In particular, eukaryotic genes can generate multiple transcripts by selective removal of specific introns, a process called alternative splicing (AS) which was initially hypothesized by Gilbert (1978). AS does not occur in isolation, and it is performed in conjunction with other pre-mRNA editing events to ensure the diversity of gene transcripts in response to various cues. The selection of splice sites in response to various cues is determined by splicing factors (SFs), which guide spliceosomal components to proper assembly on their respective splice sites (Matlin et al., 2005; Nilsen and Graveley, 2010; Wachter et al., 2012). Major SF families include serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoprotein particle (hnRNP) proteins (Figure 1B). These proteins bind specific pre-mRNA sequences called intronic- or exonic-splicing enhancer or suppressor sequences (Witten and Ule, 2011). Clearly, changes in SF level or activity can lead to subtle or profound effects on the splicing outcomes of downstream target genes (Stamm et al., 2005; Kalyna et al., 2006; Stauffer et al., 2010; Saltzman et al., 2011; Thomas et al., 2012). Overall, pre-mRNA splicing is a highly dynamic and tightly regulated post-transcriptional process which occurs in a stepwise manner (Wahl et al., 2009; Mckay and Johnson, 2010; Will and Luhrmann, 2011). Significant strides have been made in describing the dynamics of spliceosome machinery in recent years, yet the mechanisms by which spliceosomal proteins mediate the ordered rearrangements within the spliceosome remain elusive.
Post-translational modifications (PTMs), including phosphorylation, acetylation, O-GlcNAcylation, methylation, ubiquitination, ubiquitin-like modification, and more, work in every cellular process to reversibly adjust and fine-tune protein activity and to direct subcellular localization. PTMs play important roles in the splicing cycle and in ensuring splicing fidelity by facilitating the rearrangements of the spliceosome, controlling the timing of spliceosome rearrangements, regulating splicing factor activities, or altering the mRNP composition assembled on a particular transcript (Mckay and Johnson, 2010). Among these PTMs, ubiquitination or ubiquitin-like modification functions as a key regulator of various processes in eukaryotic cells (Smalle and Vierstra, 2004). These two forms of PTMs modify target proteins via covalent or non-covalent conjugation with small molecules including ubiquitin, small ubiquitin-related modifier (SUMO), and homologous to ubiquitin 1 (HUB1). These modifications can cause the target proteins to either be sequentially translocated to the proteasome for degradation, or to acquire new interactors which can change subcellular localization, affinity, and function (Smalle and Vierstra, 2004). Many proteins undergo modification in this manner, including transcription factors, various enzymes, and functional proteins, and also some spliceosome-associated proteins (Mckay and Johnson, 2010; Hu and Sun, 2016; Chanarat and Ishra, 2018; Xu, 2020). Indeed, PTMs of spliceosome components have recently emerged as major regulatory factors in yeast and human cells (Mckay and Johnson, 2010; Hu and Sun, 2016; Chanarat and Ishra, 2018; Pozzi et al., 2018; Xu, 2020; Li et al., 2021; Mulvaney et al., 2021).
In this review, we summarize recent reports about ubiquitin and ubiquitin-like modification of AS components and analyze the relationship between spliceosome and the ubiquitin (Ub)/26S proteasome pathway in plants. We further provide speculation about the mechanism of these PTMs in controlling plant development and response to environmental cues.
The Regulation of as Through PTMs
As discussed above, PTMs are involved in the regulation of protein folding, activity, stability, localization, and interactions with other proteins/molecules through covalent or non-covalent modification of the protein by functional chemical groups, including phosphate, acetyl, methyl, carbohydrate, ubiquitin, and ubiquitin-like modifications (Deribe et al., 2010; Mckay and Johnson, 2010; Lin and Begley, 2011; Chiang and Ack, 2017). Recent evidence suggests that PTM of AS components may represent another layer of AS control. Specifically, recent reports have revealed that ubiquitin and ubiquitin-like modifications of spliceosome play a role in regulating eukaryotic pre-mRNA splicing in mammalian cells. These modifications have been shown to promote splicing by generating new surfaces for intermolecular interactions, thereby refining gene expression (Chanarat and Ishra, 2018).
Biochemical Aspects of Ubiquitin and Ubiquitin-Like Modification
Ubiquitin and ubiquitin-like modification is a dynamic and reversible process involving the covalent attachment of a conserved 76-amino acid peptide ubiquitin (ub) to the ε-amino groups of lysine residues in target proteins. Ubiquitin addition is catalyzed by an enzymatic cascade involving an E1 ub-activating enzyme, followed by the sequential activities of E2 ub-conjugating enzyme and E3 ub ligase (Smalle and Vierstra, 2004; Lan and Miao, 2019). This process is reversible as a result of deubiquitinating enzymes (DUBs). DUBs are responsible for the removal of ubiquitin from their target proteins, rescuing them from the degradative pathway (Reyes-Turcu et al., 2009). Mono-ubiquitination has emerged to have an important signaling role which is distinct from polyubiquitination. Specifically, mono-ubiquitination of both histones and histone-associated proteins has been shown to regulate transcription, DNA repair, signal transduction, and receptor internalization (reviewed in Welchman et al., 2005). Ubiquitin and ubiquitin-like modifications have also been shown to modulate protein–protein interactions, often serving as an initiating signal for subsequent downstream phosphorylation and methylation in splicing events (Li et al., 2021; Mulvaney et al., 2021).
Ubiquitin-Like Modification of Spliceosomal Components
Since the initial discovery of ubiquitin, a number of ubiquitin-like protein modifications (e.g., SUMO and HUB1) have been identified (Jentsch and Pyrowolakis, 2000). SUMO proteins are covalently attached to targets in a stepwise manner through a process called SUMOylation (small ubiquitin-related modifier modification). The consequences of SUMOylation are target-dependent and include altering protein activity, subcellular localization, and protection from ubiquitination (Melchior, 2000). The first study on the roles of SUMOylation in pre-mRNA splicing and mRNA metabolism is the discovery that SUMO E2-conjugating enzyme UBC9 is located in nuclear speckles. These are discrete subnuclear structures that are responsible for pre-mRNA splicing and mRNA export, often co-localizing with SC35 (Ihara et al., 2008; Spector and Lamond, 2011). In addition, SUMO proteomics of human cell lines and yeast have shown that the SR proteins, members of the hnRNP family, and many SFs are sumoylated upon exposure to stress (Golebiowski et al., 2009; Psakhye and Jentsch, 2012; Pozzi et al., 2017, 2018; Riboldi et al., 2021). Although there is limited evidence of spliceosomal SUMOylation in plants to this point, according to the public database Biological General Repository for Interaction Datasets (BioGRID) (https://thebiogrid.org/) (Altmann et al., 2020; Oughtred et al., 2021), the SUMO E2, AtSCE1 (AT3G57870), can interact with six spliceosome components, including AT1G09760/U2A' and AT1G20960/AtBRR2A (Figure 2B). In addition to AtSCE1, the two SUMOs AT4G26840/AtSUMO1 and AT5G55170/AtSUMO3 are capable of interacting with several splicing components (Figure 2B). Taken together, this in silico analysis suggests that SUMOylation is likely to regulate pre-mRNA splicing in plants.
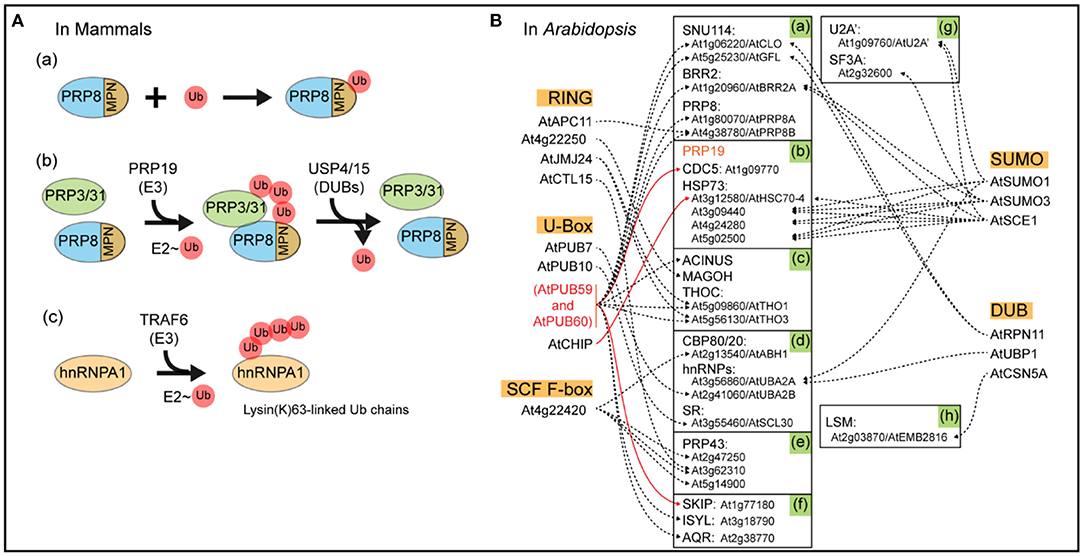
Figure 2. Schematic diagram of ubiquitination in mammals and Arabidopsis. (A) Ubiquitination and deubiquitination of mammalian spliceosomal components. MPN, the MPN domain, a variant of the Jab1/MPN domain found in a class of deubiquitinating enzymes, located near the C terminus of PRP8p; Ub, Ubiquitin. (B) The ubiquitin-associated interacting proteins. Shown in the orange box are RING E3s, U-Box E3s, SCF F-box E3s, SUMOylation components, and DUBs of Arabidopsis spliceosomal components (https://www.kegg.jp/pathway/ath03040). Shown in the green highlighted boxes are components of U5 (a), PRP19 complex (b), EJC/TREX (c), common components (d), U2 related (e), Prp19 related (f), U2 (g), and U4/U6 (h). The solid line denotes established interactions; the broken line denotes putative interaction. These interactome networks were downloaded from the BioGRID4.4 database (https://thebiogrid.org/).
In contrast to SUMO, the ubiquitin-like protein HUB1 interacts with SNU66, a component of the U4/U6 complex. Furthermore, it has been reported that the U5 tri-snRNP complex is modified by HUB1 non-covalently binding to the N terminus of SNU66, thereby modulating pre-mRNA splicing in S. cerevisiae (Wilkinson et al., 2004; Mishra et al., 2011; Ammon et al., 2014). HUB1 promotes splicing of introns containing non-canonical 5′-splice sites and AS of the SRC1 gene (Mishra et al., 2011). Another report showed that AtUBL5/AtHUB1 is involved in root elongation, plant development, and auxin response through pre-mRNA splicing in Arabidopsis (Ammon et al., 2014; Sasaki et al., 2015; Haak et al., 2017; Park et al., 2021).
Ubiquitination and Deubiquitination of Spliceosomal Components in Mammalian Cells
Ubiquitination proceeds via a three-step conjugation cascades: ubiquitin becomes 'activated' when an E1 enzyme uses the energy supplied by ATP hydrolysis to generate a thioester bond between its active-site cysteine residue and the C-terminus of ubiquitin, then ubiquitin is transferred onto the active-site cysteine of an E2 enzyme. The charged E2 finally interacts with an E3 ligase that facilitates positioning and transfer of ubiquitin onto the substrate by one of several mechanisms dependent on the class of E3 enzyme (Smalle and Vierstra, 2004). These polyUb–protein conjugates are then either targeted to the 26S proteasome for degradation, or acquire new binding surfaces that alter the protein's subcellular localization, affinity, and function. Deubiquitination is a reverse reaction of ubiquitination, generating free ubiquitin from polyUb–protein conjugates via the action of deubiquitinating enzymes (DUBs) (Smalle and Vierstra, 2004). In Arabidopsis genome, there are >1,400 (or >5% of the proteome) ubiquitination and deubiquitination pathway components that can be connected to almost all biological processes (Vierstra, 2003). This includes 17 ubiquitin (Callis et al., 1990, 1995), 2 E1s, 45 E2s (Ramadan et al., 2015), >1,200 E3s (Gagne et al., 2002; Capron et al., 2003; Stone et al., 2005; Trujillo, 2018), 53 26S proteasome components (Fu et al., 1999; Kurepa and Smalle, 2008), and 45 DUBs (Yan et al., 2000; Isono and Nagel, 2014).
Initial reports on ubiquitin's function in pre-mRNA splicing are only ~10 years old, and subsequent work has revealed that ubiquitination controls a large number of diverse cellular processes. One study reported that a variant of the Jab1/MPN domain near the C-terminus of spliceosome component PRP8p directly binds to ubiquitin with an affinity comparable to other known ubiquitin-binding domains. This was one of the first reports suggesting that ubiquitin functionally regulates pre-mRNA splicing machinery (Figure 2A-a) (Bellare et al., 2006). In another study, the spliceosomal E3 ubiquitin ligase PRP19 recruits K63-linked ubiquitin chain to PRP3/PRP31 proteins, thereby stabilizing the association between PRP3/31 and the core spliceosomal protein PRP8 in the pre-catalytic mammalian spliceosome (Bellare et al., 2008; Song et al., 2010). PRP3 is a core SF of the U4/U6. Both the U5 tri-snRNP complex and PRP31 are components of the U4 snRNP. These associations need to be weakened in order to initiate catalytic activity of the spliceosome, which takes place through the removal of the ubiquitin chain from PRP3 by the deubiquitinating enzyme USP4 and from PRP31 by USP15 (Figure 2A-b) (Song et al., 2010; Park et al., 2016; Das et al., 2017). The TRAF6 E3 ligase regulates pre-mRNA splicing through ubiquitination of hnRNPA1, which leads to AS of ARHGAP1, an inhibitor of the GTP-binding Rho family protein CDC42. One report showed that this process led to activation of CDC42 and hematopoietic defects in TRAF6-expressing hematopoietic stem/progenitor cells (Figure 2A-c) (Fang et al., 2017).
Association Between Spliceosome and Ubiquitination Pathway During Arabidopsis Development
According to KEGG pathway analysis in Arabidopsis thaliana (https://www.kegg.jp/pathway/ath03040), there are more than 175 Arabidopsis spliceosomal protein components. Potential interacting proteins of the Arabidopsis spliceosomal components are then determined using BioGRID4.4 database analysis (Figure 2B) (Altmann et al., 2020; Oughtred et al., 2021). PRP19 is unique in that it is a component of the spliceosome and also one of the U-Box E3 ubiquitin ligases. In Arabidopsis, there are two homologies of PRP19—AtPUB59 (AT1G04510, also called MAC3A) and AtPUB60 (AT2G33340, also called MAC3B). According to BioGRID4.4, 11 spliceosome-associated proteins may interact with AtPUB59 and AtPUB60, including SUN114, BRR2, PRP8, CDC5, ACINUS, THOC, ISYL, and AQR (Figure 2B-a,f). At present, there is no evidence showing that AtPUB59 and AtPUB60 are involved in the ubiquitination of AtPRP3 and AtPRP31 in mammalian cells, but they are components of the multi-protein assembly MOS4-associated complex. This complex includes AtMOS4, AtCDC5, AtPRL1, AtPUB59, and AtPUB60 and is required for basal and R protein-mediated resistance in Arabidopsis (Monaghan et al., 2009). Arabidopsis MAC3A and MAC3B act redundantly in microRNA biogenesis, which targets many genes required for proper development (Li et al., 2018). Moreover, AtPUB59 and AtPUB60 physically and genetically interact with AtSKIP, mediating the AS of about 50% of expressed genes in Arabidopsis genome (Figure 2B-f) (Li et al., 2019). AtSKIP has previously been reported to be involved in 26S proteasome-mediated degradation (Li et al., 2016), affecting both the circadian clock and salt tolerance in plants (Wang et al., 2012; Feng et al., 2015).
With the exception of AtPUB59 and AtPUB60, there are 4 RING-type E3 ligases, 3 U-Box type E3 ligases, 1 SCF F-box E3 ligases, 3 DUBs, and 25 26S proteasome components which interact with spliceosomal components (Figure 2B), assuming that spliceosomal components may be ubiquitinated by E3 ligases and then degraded by 26S proteasome or deubiquitinated by DUBs. Although there is no direct biochemical evidence, in silico analysis of proteomic datasets indicates that the components of U5, ribosomal protein S5/Elongation factor G/III/V family proteins AtCLO and AtGFL, may interact with AtPUB59, AtPUB60 ligases, and multi-ubiquitin chain binding protein AtRPN11, a deubiquitinating enzyme. Collectively, this suggests that AtCLO and AtGFL may be ubiquitinated by AtPUB59 and AtPUB60 and deubiquitinated by AtRPN11 (Figure 2B-a).
AtBRR2 is a component of the spliceosome and highly conserved in eukaryotes. Arabidopsis BRR2a is ubiquitously expressed in all tissues and involved in the processing of gene transcripts which regulate flowering time, most notably FLC (Mahrez et al., 2016). It may be that FLC interacts with Ub-protein ligases AtPUB59 and AtPUB60, and SUMO ligase AtSCE1 (Figure 2B-a), which portends that AtBRR2A may be ubiquitinated and sumoylated. Furthermore, this implies that antagonism may exist between the effect of these two modifications on the targets. We have further observed that AtHSC70-4, a component of PRP19 complex, also interacts with Ub-protein ligase (AtCHIP) and SUMO ligase (AtSCE1) (Figure 2B-b).
The C-terminus of heat shock protein (HSP) 70-interacting protein (CHIP), a chaperone-dependent and U-box-containing E3 ligase, is a key enzyme involved in protein quality control by recognizing misfolded proteins through its interacting chaperones and then directing their degradation. In plants, CHIP plays an important role in regulating responses to a broad spectrum of biotic and abiotic stresses. CHIP protects chloroplasts by coordinating protein quality control both outside and inside the photosynthetic organelles and also by modulating the activity of protein phosphatase 2A, a crucial component in plant signaling networks, including abscisic acid signaling (Zhang et al., 2021). Furthermore, the interaction between AtHSC70-4 and AtCHIP is not under the precondition that AtCHIP ubiquitinates AtHSC70-4, but is predicated on the basis that AtHSC70-4 and AtCHIP together mediate plastid precursor degradation (Lee et al., 2009). Therefore, the interaction between AtHSC70-4 and AtSCE1 mediates plastid precursor SUMOylation (Watson et al., 2021). There are multiple examples of Ub-protein ligases interacting with the same substrate, including anaphase-promoting complex AtAPC11, AtPUB59, and AtPUB60 interacting with AtPRP8B and AtJMJ24, AtPUB59, and AtPUB60 binding a nuclear matrix protein-like protein complex THOC (AtTHO1 and AtTHO3) and more. Indeed, as shown in Figure 2B-a,c, various Ub-protein ligases may support different kinds of ubiquitin chain modification for the same substrate.
In plants, the anaphase-promoting complex/cyclosome (APC/C) consists of at least 11 core subunits, each of which is encoded by a single gene, with exception of APC3, which is encoded by two genes, APC3a/CDC27a and APC3b/CDC27b/HOBBIT. The Arabidopsis APC/C activator cell division cycle 20.1(CDC20.1) is required for bivalent alignment and chromosome segregation during meiosis (Niu et al., 2015), suggesting that APC/C subunits may also have meiotic functions in plants. It was recently shown that Arabidopsis APC8 is necessary for male meiosis of plants (Liu et al., 2019). In addition to the APC complexes, THO/TREX complexes have been implicated in the transport of mRNA precursors in mammals. Mutants of THO3/TEX1, THO1, and THO6 in Arabidopsis accumulate reduced amounts of small interfering RNA, suggesting a role for the putative Arabidopsis THO/TREX in small interfering RNA biosynthesis and trafficking (Tao et al., 2016; Doll et al., 2018).
Taken together, the potential interaction between the spliceosome and ubiquitination or SUMOylation pathway in Arabidopsis indicates that these PTMs play an important role in controlling spliceosomal components (Figure 2B). As such, spliceosome PTMs are important for regulating RNA biogenesis, trafficking precursors of small RNAs (including microRNAs), and controlling protein quality, leading to proper plant development and adaptation to environmental stress.
Potential Ubiquitin Modification Sites in Arabidopsis Spliceosomal Components
The rise of modern proteomics has uncovered a vast array of ways in which proteins can be post-translationally modified. Here, we summarize the potential ubiquitination sites of Arabidopsis spliceosomal components based on the Plant PTM Viewer database (https://www.psb.ugent.be/webtools/ptm-viewer/index.php) (Table 1), which provides innovative tools to analyze the potential role of PTMs in regulating specific proteins or in a broader systems biology context (Wagner et al., 2011; Willems et al., 2019).
Although ubiquitination of plant spliceosomal components has not been well reported, a number of putative ubiquitination sites exist on lysine residue in these proteins, particularly the AtPRP19 complex and AtHSP73 proteins in Arabidopsis (Table 1) (Makarova et al., 2004; Grote et al., 2010; Slane et al., 2020). HSP70s are critical for maintaining cell viability in response to a large variety of cellular stresses, including high temperature, nutrient starvation, osmotic shock, oxidative stress, and DNA damage (Rosenzweig et al., 2019). In fact, recent work has uncovered a vast array of PTMs on HSP70 family proteins including phosphorylation, acetylation, ubiquitination, AMPylation, and ADP-ribosylation (Nitika et al., 2020). In addition, two interacting proteins of AtPUB59 and AtPUB60, AtCDC5 and AtSKIP, have potential ubiquitination sites, further suggesting that AtCDC5 and AtSKIP may be ubiquitinated by AtPUB59 and AtPUB60 or other Ub-protein ligases. These potential ubiquitination sites in spliceosomal components afford researchers with enormous opportunities to uncover novel post-translational regulatory mechanisms of spliceosome activity.
As ff Ub-Protein Ligase mRNAs
In addition to those spliceosome components discussed above, there is another aspect of pre-mRNA splicing which can potentially be regulated by ubiquitination modification. Recently, it was shown that salt-responsive alternatively spliced gene 1 (SRAS1), a RING-type E3s, undergoes salt-responsive AS generating two splicing variants—SRAS1.1 (the full-length SRAS1) and SRAS1.2 (a small-sized protein lacking 59 amino acids in the C-terminal RING finger domain). Interestingly, these variants exhibit reciprocal responses to salt stress—SRAS1.1 expression is enhanced, while SRAS1.2 is suppressed. Moreover, SRAS1.1 promotes degradation of the COP9 signalosome 5A, which plays an important role in plant development and stress responses via the 26S proteasome (Gusmaroli et al., 2004; Zhou et al., 2021). In contrast, SRAS1.2 preserves the COP9 signalosome 5A by competing with SRAS1.1 on the same binding site (Zhou et al., 2021). This finding underscores the complexity of the interaction between ubiquitin-related PTMs and AS and suggests mutual regulation between these processes.
We have recently observed that Arabidopsis HECT-type E3 ligases are co-expressed with spliceosomal components. To perform this analysis, we downloaded the top 200 co-expressed genes of spliceosomal components from ATTED-II (https://atted.jp/), including SF3A, SF3B, PRP6, PRP8, SNU114, BRR2, THOC, ACINUS, and SR (f) (Figures 3A–D). Interestingly, Furniss et al. found that the upl3 mutant shows impairment in transcriptome reprogramming following salicylic acid treatment and failed to establish immunity against a hemi-biotrophic pathogen (Downes et al., 2003; Furniss et al., 2018). Based on this finding, it is reasonable to suggest that this impaired reprogramming may be related to AS. In fact, our comparative proteomic analysis of the HECT E3 upl4/upl3 double mutant relative to wild type shows that most spliceosomal components are upregulated in upl4/upl3 double mutant with developmental defects and stress-sensitive phenotypes (Figure 3E), suggesting that there is a potential relationship between pre-mRNA splicing and HECT E3s.
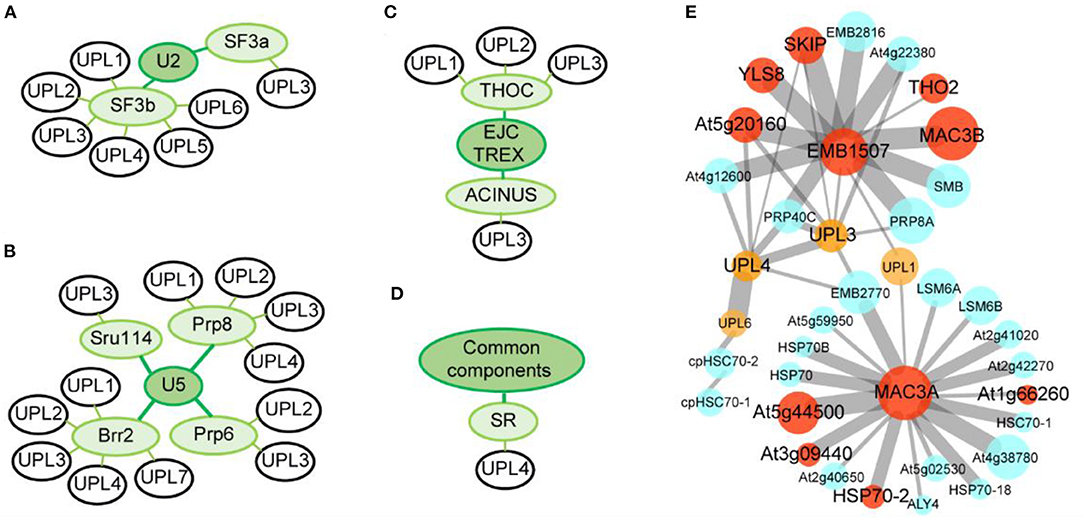
Figure 3. Relationship between spliceosomal components and HECT E3s. (A–D) Co-expression networks of HECT E3s family members with spliceosomal components. Green colored circles represent established spliceosomal components, and black-outlined circles represent HECT E3s. Proteomic analysis of spliceosomal components in the upl3 and the upl4/upl3 double mutants (Lan et al., 2022; unpublished data). (E) Red circles show upregulated genes, yellow circles show HECT E3s, while blue circles represent no change or undetected genes. Edge width indicates the strength of the relation between two proteins based on STRING database, (https://string-db.org/cgi/input?sessionId=HOnM9xTdLFXCandinput_page_active_form=multiple_identifiers), and editing through Cytoscape software.
The Important Roles of as for Plant Development and Stress Responses
It is now fully established that at least 95% of multi-exon genes in mammals undergo some form of AS (Pan et al., 2008; Wang et al., 2008), while only 40–70% of genes undergo AS in plants (Chamala et al., 2015; Zhu et al., 2017; Chaudhary et al., 2019). AS not only affects the abundance of mature mRNAs, but also intricately regulates transcription, translation, and downstream mRNA metabolic events including mRNA export and turnover in eukaryotes (Black, 2003; Moore and Proudfoot, 2009). In addition, splicing variants produce truncated proteins that may have different subcellular localization and/or functions (Samach et al., 2011; Shin et al., 2015; Wang et al., 2015; Ghelli et al., 2018). Thus, AS fulfills many vital biological functions in plants, including developmental regulation and adaptation to stress environments (Figure 4).
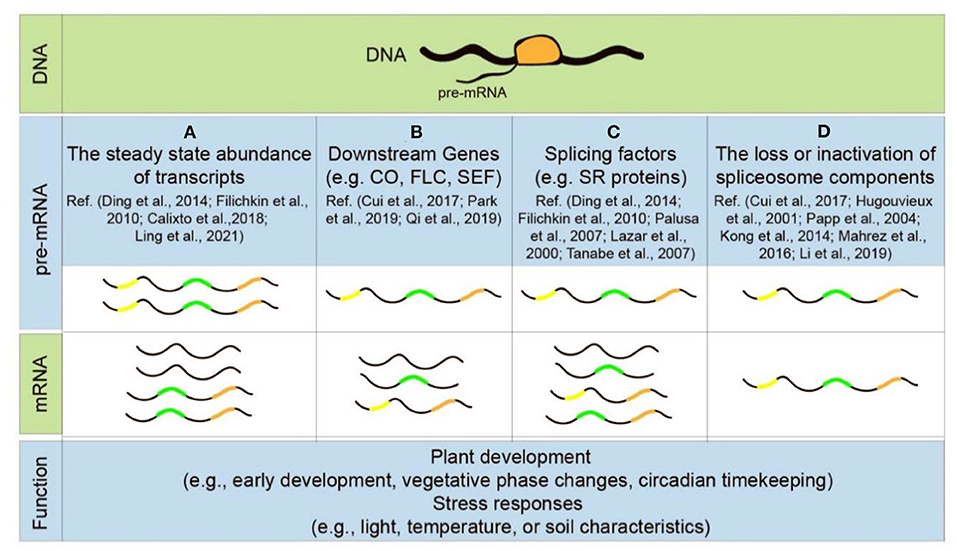
Figure 4. Patterns of alternative splicing during plant development and stress response. Each step of pre-mRNA splicing influences plant growth and adaptation to environments, such as the steady-state abundance of transcripts (A), the alternative splicing of downstream genes (B), splicing factors (C), and the loss or inactivation of spliceosomal components (D).
Specifically, AS has been implicated in the response to environmental cues in plants, including major deviations in ambient light, temperature, and soil characteristics (e.g., water/salt content) under normal or optimal conditions (Laloum et al., 2018; Punzo et al., 2020). It not only alters the steady-state abundance of many stress-induced gene transcripts but also evokes changes in AS patterns of these transcripts (Figures 4A,B) (Filichkin et al., 2010; Ding et al., 2014; Haak et al., 2017; Calixto et al., 2018; Ling et al., 2021). For example, about ~49% of all intron-containing genes are alternatively spliced under salt stress, 10% of which experience significant differential AS, a process significantly increased under salt stress conditions (Ding et al., 2014; Haak et al., 2017). It is also worth noting that at least 95 transcripts of SR proteins, produced from only 15 genes (Palusa et al., 2007), undergo AS themselves in response to stress (Figure 4C) (Lazar and Goodman, 2000; Palusa et al., 2007; Tanabe et al., 2007; Filichkin et al., 2010; Ding et al., 2014). Another classic example is AtSR45a protein, a homolog of metazoan TRA-2. There are six types of mRNA variants (AtSR45a-1a-e and AtSR45a-2) generated by alternative selection of transcriptional initiation sites and AS processing of introns in AtSR45a pre-mRNA. AtSR45a-1a and AtSR45a-2 expression is greatly increased by high-light irradiation (Tanabe et al., 2007; Carvalho et al., 2016). In addition, the loss or inactivation of spliceosomal components directly leads to plant growth defects and stress-sensitive phenotypes such as abh1, brr2a, and skip mutants (Figure 4D) (Hugouvieux et al., 2001; Papp et al., 2004; Kong et al., 2014; Mahrez et al., 2016; Cui et al., 2017; Li et al., 2019).
As an important factor in post-transcriptional gene regulation, AS is intimately involved in the regulation of developmental processes such as seedling development, juvenile vegetative phase, adult vegetative phase, senescence, flowering, and circadian timekeeping (Cui et al., 2017; Gil et al., 2017; Szakonyi and Duque, 2018; Bai et al., 2019; Park et al., 2019; Qi et al., 2019). For example, the floral activator CONSTANS (CO), which plays a crucial role in photoperiodic flowering (Gil et al., 2017), undergoes AS producing two isoforms—the full-size physiologically functional COα and the C-terminally truncated COβ (Qi et al., 2019). COβ acts as a competitive inhibitor of COα by forming non-functional heterodimers which have a significantly reduced DNA binding capability compared to the COα-COα homodimers (Qi et al., 2019). AS of polyadenylated ETHYLENE RESPONSE FACTOR4 (ERF4), a positive regulator of leaf senescence, results in two ERF4 isoforms: ERF4-R which contains the EAR-motif, and ERF4-A which lacks it (Lyons et al., 2013). ERF4-A acts as a transcriptional activator and ERF4-R as a repressor of their direct target gene CATALASE3, controlling the concentrations of reactive oxygen species (ROS) in cells and regulating leaf senescence (Riester et al., 2019). A minor spliceosome component U11-48 K, which is indispensable for correct splicing of U12 introns, is required for normal plant development (Gault et al., 2017; Bai et al., 2019).
Taken together, it is clear that pre-mRNA splicing plays a pivotal role in plant development and adaptation to stress environments. As such, the spliceosome activity in plants is indispensable during plant growth and stress responses.
Conclusions and Perspective
The AS fulfills vital biological functions, including control of plant development and adaptation to stress environments. AS processes not only affect the abundance and quality of mature mRNAs in eukaryotes, but are also intimately involved in transcription, translation, and downstream mRNA metabolic events including mRNA export and turnover. PTMs of spliceosomal components and SFs provide an effective platform for mechanistic fine-tuning of the pre-mRNA splicing process. Both the spliceosomal and ubiquitination systems contain a large number of components and are ubiquitous in eukaryotes, which indicate the possibility of co-evolutionary interaction between them.
Development of new and more sensitive mass spectroscopic techniques and artificial intelligence prediction, combined with recent structures of spliceosomal complexes, will allow researchers to address challenging questions regarding splicing regulation. Furthermore, the evolution of proteomics and systems biology presents an opportunity to address complex issues concerning splicing reactions and molecular machinery. Multi-omics analysis combined with classical genetic methods or CRISPR editing will provide even more genome-wide evidence to reveal the interaction between AS and PTMs, and their roles during plant development and stress response. Finally, structural biology, phase separation, and cryo-EM remain the most effective approaches to elucidate splicing machinery dynamics and timing of AS and their PTMs during plant development and in response to environmental cues.
Author Contributions
WL, YX, and YL are involved in writing—original draft preparation. WL, YQ, and YM are involved in writing—reviewing and editing. YM supervised the study and involved in project administration. YM and WL took a leading role in funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Scientific Research Foundation of Graduate School of Fujian Agriculture and Forestry University to WL and the National Natural Science Foundation of China (No. 324-1122yb049).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Altmann, M., Altmann, S., Rodriguez, P. A., Weller, B., Elorduy Vergara, L., Palme, J., et al. (2020). Extensive signal integration by the phytohormone protein network. Nature 583, 271–276. doi: 10.1038/s41586-020-2460-0
Ammon, T., Mishra, S. K., Kowalska, K., Popowicz, G. M., Holak, T. A., and Jentsch, S. (2014). The conserved ubiquitin-like protein Hub1 plays a critical role in splicing in human cells. J. Mol. Cell Biol. 6, 312–323. doi: 10.1093/jmcb/mju026
Bai, F., Corll, J., Shodja, D. N., Davenport, R., Feng, G., and Mudunkothge, J. (2019). RNA binding motif protein 48 is required for U12 splicing and maize endosperm differentiation. Plant Cell. 31, 715–733. doi: 10.1105/tpc.18.00754
Bellare, P., Small, E. C., Huang, X., Wohlschlegel, J. A., Staley, J. P., and Sontheimer, E. J. (2008). A role for ubiquitin in the spliceosome assembly pathway. Nat. Struct. Mol. Biol. 15, 444–451. doi: 10.1038/nsmb.1401
Bellare, P., Utach, A. K., Ines, A. K., Uthrie, C., and Ntheimer, E. J. (2006). Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA. 12, 292–302. doi: 10.1261/rna.2152306
Berget, S. M., Moore, C., and Sharp, P. A. (1977). Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA. 74, 3171–3175. doi: 10.1073/pnas.74.8.3171
Black, D. L. M. (2003). Mechanisms of alternative pre-messenger RNA splicing. Ann. Rev. Biochem. 72, 291–336. doi: 10.1146/annurev.biochem.72.121801.161720
Calixto, C. P. G., Guo, W., James, A. B., Tzioutziou, N. A., Entizne, J. C., Panter, P. E., et al. (2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell. 30, 1424–1444. doi: 10.1105/tpc.18.00177
Callis, J., Carpenter, T., Sun, C. W., and Vierstra, R. D. (1995). Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics. 139, 921–939. doi: 10.1093/genetics/139.2.921
Callis, J., Raasch, J. A., and Vierstra, R. D. (1990). Ubiquitin extension proteins of Arabidopsis thaliana. Structure, localization, and expression of their promoters in transgenic tobacco. J. Biol. Chem. 265, 12486–12493. doi: 10.1016/S0021-9258(19)38372-3
Capron, A., Okresz, L., and Genschik, P. (2003). First glance at the plant APC/C, a highly conserved ubiquitin-protein ligase. Trends Plant Sci. 8, 83–89. doi: 10.1016/S1360-1385(02)00028-6
Carvalho, R. F., Szakonyi, D., Simpson, C. G., Barbosa, I., Brown, J., Baena-González, E., et al. (2016). The Arabidopsis SR45 splicing factor, a negative regulator of sugar signaling, modulates SNF1-related protein kinase 1 stability. Plant Cell. 28, 1910–1925. doi: 10.1105/tpc.16.00301
Chamala, S., Feng, G., Chavarro, C., and Barbazuk, W. B. (2015). Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front. Bioeng. Biotechnol. 3, 33. doi: 10.3389/fbioe.2015.00033
Chanarat, S., and Ishra, S. K. (2018). Emerging roles of ubiquitin-like proteins in pre-mRNA splicing. Trends Biochem. Sci. 43, 896–907. doi: 10.1016/j.tibs.2018.09.001
Chaudhary, S., Abre, I., Reddy, A. S. N., Staiger, D., and Syed, N. H. (2019). Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci. 24, 496–506. doi: 10.1016/j.tplants.2019.02.006
Chiang, C., and Ack, M. U. (2017). Post-translational control of intracellular pathogen sensing pathways. Trends Immunol. 38, 39–52. doi: 10.1016/j.it.2016.10.008
Chow, L. T., Gelinas, R. E., Broker, T. R., and Roberts, R. J. (1977). An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 12, 1–8. doi: 10.1016/0092-8674(77)90180-5
Cui, Z., Tong, A., Huo, Y., Yan, Z., Yang, W., Yang, X., et al. (2017). SKIP controls flowering time via the alternative splicing of SEF pre-mRNA in Arabidopsis. BMC Biol. 15, 80. doi: 10.1186/s12915-017-0422-2
Das, T., Park, J. K., Park, J., Kim, E., Rape, M., Kim, E. E., et al. (2017). USP15 regulates dynamic protein-protein interactions of the spliceosome through deubiquitination of PRP31. Nucleic Acids Res. 45, 5010–5011. doi: 10.1093/nar/gkw1365
Deribe, Y. L., Pawson, T., and Dikic, I. (2010). Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672. doi: 10.1038/nsmb.1842
Ding, F., Cui, P., Wang, Z., Zhang, S., Ali, S., Xiong, L., et al. (2014). Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics. 15, 431. doi: 10.1186/1471-2164-15-431
Doll, S., Kuhlmann, M., Rutten, T., Mette, M. F., Scharfenberg, S., Petridis, A., et al. (2018). Accumulation of the coumarin scopolin under abiotic stress conditions is mediated by the Arabidopsis thaliana THO/TREX complex. Plant J. 93, 431–444. doi: 10.1111/tpj.13797
Downes, B. P., Stupar, R. M., Gingerich, D. J., and Vierstra, R. D. (2003). The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 35, 729–742. doi: 10.1046/j.1365-313X.2003.01844.x
Fang, J., Bolanos, L. C., Choi, K., Liu, X., Christie, S., Akunuru, S., et al. (2017). Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia. Nat. Immunol. 18, 236–245. doi: 10.1038/ni.3654
Feng, J., Li, J., Gao, Z., Lu, Y., Yu, J., Zheng, Q., et al. (2015). SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant. 8, 1038–1052. doi: 10.1016/j.molp.2015.01.011
Filichkin, S. A., Priest, H. D., Givan, S. A., Shen, R., Bryant, D. W., Fox, S. E., et al. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58. doi: 10.1101/gr.093302.109
Fu, H., Girod, P. A., Doelling, J. H., van Nocker, S., Hochstrasser, M., Finley, D., et al. (1999). Structure and functional analysis of the 26S proteasome subunits from plants. Mol. Biol. Rep. 26, 137–146. doi: 10.1023/A:1006926322501
Furniss, J. J., Grey, H., Wang, Z., Nomoto, M., Jackson, L., Tada, Y., et al. (2018). Proteasome-associated HECT-type ubiquitin ligase activity is required for plant immunity. PLoS Pathog. 14, e1007447. doi: 10.1371/journal.ppat.1007447
Gagne, J. M., Downes, B. P., Shiu, S. H., Durski, A. M., and Vierstra, R. D. T. (2002). F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA. 99, 11519–11524. doi: 10.1073/pnas.162339999
Gault, C. M., Martin, F., Mei, W., Bai, F., Black, J. B., and Barbazuk, W. B. (2017). Aberrant splicing in maize rough endosperm3 reveals a conserved role for U12 splicing in eukaryotic multicellular development. Proc. Natl. Acad. Sci. USA. 114, 2195–2204. doi: 10.1073/pnas.1616173114
Ghelli, R., Brunetti, P., Napoli, N., De Paolis Cecchetti, V., Tsuge, T., et al. (2018). A newly identified flower-specific splice variant of auxin response factor8 regulates stamen elongation and endothecium lignification in Arabidopsis. Plant Cell. 30, 620–637. doi: 10.1105/tpc.17.00840
Gil, K. E., Park, M. J., Lee, H. J., Park, Y. J., Han, S. H., Kwon, Y. J., et al. (2017). Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in Arabidopsis photoperiodic flowering. Plant J. 89, 128–140. doi: 10.1111/tpj.13351
Golebiowski, F., Matic, I., Tatham, M. H., Cole, C., Yin, Y., Nakamura, A., et al. (2009). System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2, ra24. doi: 10.1126/scisignal.2000282
Grote, M., Wolf, E., Will, C. L., Lemm, I., Agafonov, D. E., Schomburg, A., et al. (2010). Molecular architecture of the human Prp19/CDC5L complex. Mol. Cell Biol. 30, 2105–2119. doi: 10.1128/MCB.01505-09
Gusmaroli, G., Feng, S., and Deng, X. W. (2004). The CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell. 16, 2984–3001. doi: 10.1105/tpc.104.025999
Haak, D, Fukao, T, Grene, R., Hua, Z, Ivanov, R, Perrella, G, et al. (2017). Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 8, 1564. doi: 10.3389/fpls.2017.01564
Hu, H., and Sun, S. C. (2016). Ubiquitin signaling in immune responses. Cell Res. 26, 457–483. doi: 10.1038/cr.2016.40
Hugouvieux, V., Kwak, J. M., and Schroeder, J. I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 106, 477–487. doi: 10.1016/S0092-8674(01)00460-3
Ihara, M., Stein, P., and Schultz, R. M. (2008). UBE2I (UBC9), a SUMO-conjugating enzyme, localizes to nuclear speckles and stimulates transcription in mouse oocytes. Biol. Reprod. 79, 906–913. doi: 10.1095/biolreprod.108.070474
Isono, E., and Nagel, M. K. (2014). Deubiquitylating enzymes and their emerging role in plant biology. Front. Plant Sci. 5, 56. doi: 10.3389/fpls.2014.00056
Jentsch, S., and Pyrowolakis, G. (2000). Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 10, 335–342. doi: 10.1016/S0962-8924(00)01785-2
Kalyna, M., Lopato, S., Voronin, V., and Barta, A. (2006). Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 34, 4395–4405. doi: 10.1093/nar/gkl570
Koncz, C., Dejong, F., Villacorta, N., Szakonyi, D., and Koncz, Z. (2012). The spliceosome-activating complex: molecular mechanisms underlying the function of a pleiotropic regulator. Front. Plant Sci. 3, 9. doi: 10.3389/fpls.2012.00009
Kong, X., Ma, L., Yang, L., Chen, Q., Xiang, N., and Yang, Y. (2014). Quantitative proteomics analysis reveals that the nuclear cap-binding complex proteins Arabidopsis CBP20 and CBP80 modulate the salt stress response. J. Proteome Res. 13, 2495–2510. doi: 10.1021/pr4012624
Kurepa, J., and Smalle, J. A. (2008). Structure, function and regulation of plant proteasomes. Biochimie. 90, 324–335. doi: 10.1016/j.biochi.2007.07.019
Laloum, T., Martin, G., and Duque, P. (2018). Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23, 140–150. doi: 10.1016/j.tplants.2017.09.019
Lan, W., and Miao, Y. (2019). New aspects of HECT-E3 ligases in cell senescence and cell death of plants. Plants. 8, 483. doi: 10.3390/plants8110483
Lan, W., Ma, W., Zheng, S., Qiu, Y., Lu, H., Huang, C., and Miao, Y. (2022). Multi-omics analysis reveals a non-canonical regulatory pattern of UPL3 on carbon metabolism related cell senescence. Res. Square. doi: 10.21203/rs.3.rs-1264931/v1
Lazar, G., and Goodman, H. M. (2000). The splicing factor SR1 is regulated by alternative splicing. Plant Mol. Biol. 42, 571–581. doi: 10.1023/A:1006394207479
Lee, S., Lee, D. W., Lee, Y., Mayer, U., Stierhof, Y. D., Jurgens, G., et al. (2009). Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell. 21, 3984–4001. doi: 10.1105/tpc.109.071548
Li, S., Liu, K., Zhou, B., Li, M., Zhang, S., Zeng, L., et al. (2018). MAC3A and MAC3B, two core subunits of the MOS4-associated complex, positively influence miRNA biogenesis. Plant Cell. 30, 481–494. doi: 10.1105/tpc.17.00953
Li, W. J., He, Y. H., and Yang, J. J. (2021). Profiling PRMT methylome reveals roles of hnRNPA1 arginine methylation in RNA splicing and cell growth. Nat. Commun. 12, 1946. doi: 10.1038/s41467-021-21963-1
Li, Y., Xia, C., Feng, J., Yang, D., Wu, F., Cao, Y., et al. (2016). The SNW domain of SKIP is required for its integration into the spliceosome and its interaction with the Paf1 complex in Arabidopsis. Mol. Plant. 9, 1040–50. doi: 10.1016/j.molp.2016.04.011
Li, Y., Yang, J., Shang, X., Lv, W., Xia, C., Wang, C., et al. (2019). SKIP regulates environmental fitness and floral transition by forming two distinct complexes in Arabidopsis. New Phytol. 224, 321–335. doi: 10.1111/nph.15990
Lin, H., and Begley, T. (2011). Protein posttranslational modifications: chemistry, biology, and applications. Mol. Biosyst. 7, 14–15. doi: 10.1039/C0MB90037K
Ling, Y., Mahfouz, M. M., and Zhou, S. (2021). Pre-mRNA alternative splicing as a modulator for heat stress response in plants. Trends Plant Sci. 26, 1153–1170. doi: 10.1016/j.tplants.2021.07.008
Liu, Z., Chen, G., Gao, F., Xu, R., Li, N., Zhang, Y., et al. (2019). Transcriptional repression of the APC/C activator genes CCS52A1/A2 by the mediator complex subunit MED16 controls endoreduplication and cell growth in Arabidopsis. Plant Cell. 31, 1899–1912. doi: 10.1105/tpc.18.00811
Lyons, R., Iwase, A., Gänsewig, T., Sherstnev, A., Duc, C., and Barton, G. J. (2013). The RNA binding protein FPA regulates flg22-triggered defense responses and transcription factor activity by alternative polyadenylation. Sci Rep. 3, 2866. doi: 10.1038/srep02866
Mahrez, W., Shin, J., Munoz-Viana, R., Figueiredo, D. D., Trejo-Arellano, M. S., Exner, V., et al. (2016). BRR2a affects flowering time via FLC splicing. PLoS Genet. 12, e1005924. doi: 10.1371/journal.pgen.1005924
Makarova, O. V., Makarov, E. M., Urlaub, H., Will, C. L., Gentzel, M., Wilm, M., et al. (2004). A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23, 2381–2391. doi: 10.1038/sj.emboj.7600241
Matlin, A. J., Clark, F., and Smith, C. W. (2005). Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6, 386–398. doi: 10.1038/nrm1645
Mckay, S. L., and Johnson, T. L. (2010). A bird's-eye view of post-translational modifications in the spliceosome and their roles in spliceosome dynamics. Mol. Biosyst. 6, 2093–2102. doi: 10.1039/c002828b
Melchior, F. (2000). SUMO–nonclassical ubiquitin. Ann. Rev. Cell Dev. Biol. 16, 591–626. doi: 10.1146/annurev.cellbio.16.1.591
Mishra, S. K., Ammon, T., Popowicz, G. M., Krajewski, M., Nagel, R. J., and Ares, M. Jr. (2011). Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing. Nature. 474, 173–178. doi: 10.1038/nature10143
Monaghan, J., Xu, F., Gao, M., Zhao, Q., Palma, K., and Long, C. (2009). Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5, e1000526. doi: 10.1371/journal.ppat.1000526
Moore, M. J., and Proudfoot, N. J. (2009). Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 136, 688–700. doi: 10.1016/j.cell.2009.02.001
Mulvaney, K. M., Blomquist, C., Acharya, N., Li, R., Ranaghan, M. R., O'Keefe, M., et al. (2021). Molecular basis for substrate recruitment to the PRMT5 methylosome. Mol Cell. 81, 3481–3495. doi: 10.1016/j.molcel.2021.07.019
Nilsen, T. W., and Graveley, B. R. (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature. 463, 457–463. doi: 10.1038/nature08909
Nitika Porter, C. M., Truman, A. W., and Truttmann, M. C. (2020). Post-translational modifications of Hsp70 family proteins: expanding the chaperone code. J. Biol. Chem. 295, 10689–10708. doi: 10.1074/jbc.REV120.011666
Niu, B., Wang, L., Zhang, L., Ren, D., Ren, R., Copenhaver, G. P., et al. (2015). Arabidopsis cell division cycle 20.1 is required for normal meiotic spindle assembly and chromosome segregation. Plant Cell. 27, 3367–3382. doi: 10.1105/tpc.15.00834
Oughtred, R., Rust, J., Chang, C., Breitkreutz, B. J., Stark, C., Willems, A., et al. (2021). The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 30, 187–200. doi: 10.1002/pro.3978
Palusa, S. G., Ali, G. S., and Reddy, A. S. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49, 1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x
Pan, Q., Shai, O., Lee, L. J., Frey, B. J., and Blencowe, B. J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415. doi: 10.1038/ng.259
Papp, I., Mur, L. A., Dalmadi, A., Dulai, S., and Koncz, C. (2004). A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol. Biol. 55, 679–686. doi: 10.1007/s11103-004-1680-2
Park, H., Jung, H., Lee, A., Jo, S., Lee, H., Kim, H., et al. (2021). SUMO modification of OsFKBP20-1b is integral to proper Pre-mRNA splicing upon heat stress in rice. Int. J. Mol. Sci. 22, 9049. doi: 10.3390/ijms22169049
Park, J. K., Das, T., Song, E. J., and Kim, E. E. (2016). Structural basis for recruiting and shuttling of the spliceosomal deubiquitinase USP4 by SART3. Nucleic Acids Res. 44, 5424–5437. doi: 10.1093/nar/gkw218
Park, Y. J., Lee, J. H., Kim, J. Y., and Park, C. M. A. (2019). Splicing Expands the Developmental Plasticity of Flowering Transition. Front Plant Sci. 10, 606. doi: 10.3389/fpls.2019.00606
Pozzi, B., Bragado, L., Will, C., Mammi, P., Risso, G., Urlaub, H., et al. (2017). SUMO conjugation to spliceosomal proteins is required for efficient pre-mRNA splicing. Nucleic Acids Res. 45, 6729–6745. doi: 10.1093/nar/gkx213
Pozzi, B., Mammi, P., Bragado, L., Giono, L. E., and Srebrow, A. (2018). When met splicing. RNA Biol. 15, 689–695. doi: 10.1080/15476286.2018.1457936
Psakhye, I., and Jentsch, S. (2012). Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 151, 807–820. doi: 10.1016/j.cell.2012.10.021
Punzo, P., Grillo, S., and Batelli, G. (2020). Alternative splicing in plant abiotic stress responses. Biochem. Soc. Trans. 48, 2117–2126. doi: 10.1042/BST20200281
Qi, H. D., Lin, Y., Ren, Q. P., Wang, Y., Xiong, F., and Wang, X. L. (2019). RNA splicing of FLC modulates the transition to flowering. Front. Plant Sci. 10, 1625. doi: 10.3389/fpls.2019.01625
Ramadan, A., Nemoto, K., Seki, M., Shinozaki, K., Takeda, H., and Takahashi, H. (2015). Wheat germ-based protein libraries for the functional characterisation of the Arabidopsis E2 ubiquitin conjugating enzymes and the RING-type E3 ubiquitin ligase enzymes. BMC Plant Biol. 15, 275. doi: 10.1186/s12870-015-0660-9
Reddy, A. S., Marquez, Y., Kalyna, M., and Barta, A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell. 25, 3657–3683. doi: 10.1105/tpc.113.117523
Reyes-Turcu, F. E., Ventii, K. H., and Wilkinson, K. D. (2009). Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397. doi: 10.1146/annurev.biochem.78.082307.091526
Riboldi, G., Faravelli, I., Kuwajima, T., Delestrée, N., Dermentzaki, G., Planell-Saguer, M., et al. (2021). Sumoylation regulates the assembly and activity of the SMN complex. Nat. Commun. 12, 5040. doi: 10.1038/s41467-021-25272-5
Riester, L., Koester-Hofmann, S., Doll, J., Berendzen, K. W., and Zentgraf, U. (2019). Impact of alternatively Polyadenylated isoforms of ETHYLENE RESPONSE FACTOR4 with activator and repressor function on senescence in Arabidopsis thaliana L. Genes. 10, 91. doi: 10.3390/genes10020091
Rosenzweig, R., Nillegoda, N. B., Mayer, M. P., and Bukau, B. (2019). The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 20, 665–680. doi: 10.1038/s41580-019-0133-3
Saltzman, A. L., Pan, Q., and Blencowe, B. J. (2011). Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 25, 373–384. doi: 10.1101/gad.2004811
Samach, A., Melamed-Bessudo, C., Avivi-Ragolski, N., Pietrokovski, S., and Levy, A. A. (2011). Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell. 23, 4266–4279. doi: 10.1105/tpc.111.091744
Sasaki, T., Kanno, T., Liang, S., Chen, P., Liao, W., Lin, W., et al. (2015). An Rtf2 domain-containing protein influences pre-mRNA splicing and is essential for embryonic development in Arabidopsis thaliana. Genetics. 200, 523–535. doi: 10.1534/genetics.115.176438
Shin, K. H., Yang, S. H., Lee, J. Y., Lim, C. W., Lee, S. C., Brown, J. W., et al. (2015). Alternative splicing of mini-exons in the Arabidopsis leaf rust receptor-like kinase LRK10 genes affects subcellular localisation. Plant Cell Rep. 34, 495–505. doi: 10.1007/s00299-014-1729-x
Slane, D., Lee, C. H., Kolb, M., Dent, C., Miao, Y., Franz-Wachtel, M., et al. (2020). The integral spliceosomal component CWC15 is required for development in Arabidopsis. Sci Rep. 10, 13336. doi: 10.1038/s41598-020-70324-3
Smalle, J., and Vierstra, R. D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Ann. Rev. Plant Biol. 55, 555–590. doi: 10.1146/annurev.arplant.55.031903.141801
Song, E. J., Werner, S. L., Neubauer, J., Stegmeier, F., Aspden, J., Rio, D., et al. (2010). The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 24, 1434–1447. doi: 10.1101/gad.1925010
Spector, D. L., and Lamond, A. I. (2011). Nuclear speckles. Cold Spring Harb Perspect Biol. 3, 646. doi: 10.1101/cshperspect.a000646
Stamm, S., Ben-Ari, S., Rafalska, I., Tang, Y., Zhang, Z., Toiber, D., et al. (2005). Function of alternative splicing. Gene. 344, 1–20. doi: 10.1016/j.gene.2004.10.022
Stauffer, E., Westermann, A., Wagner, G., and Wachter, A. (2010). Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J. 64, 243–255. doi: 10.1111/j.1365-313X.2010.04321.x
Stone, S. L., Hauksdottir, H., Troy, A., Herschleb, J., Kraft, E., and Callis, J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137, 13–30. doi: 10.1104/pp.104.052423
Szakonyi, D., and Duque, P. (2018). Alternative splicing as a regulator of early plant development. Front Plant Sci. 9, 1174. doi: 10.3389/fpls.2018.01174
Tanabe, N., Yoshimura, K., Kimura, A., Yabuta, Y., and Shigeoka, S. (2007). Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol. 48, 1036–1049. doi: 10.1093/pcp/pcm069
Tao, S., Zhang, Y., Wang, X., Xu, L., Fang, X., Lu, Z. J., et al. (2016). The THO/TREX complex active in miRNA biogenesis negatively regulates root-associated acid phosphatase activity induced by phosphate starvation. Plant Physiol. 171, 2841–2853. doi: 10.1104/pp.16.00680
Tharun, S. (2009). Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int. Rev. Cell Mol. Biol. 272, 149–189. doi: 10.1016/S1937-6448(08)01604-3
Thomas, J., Palusa, S. G., Prasad, K. V., Ali, G. S., Surabhi, G. K., Ben-Hur, A., et al. (2012). Identification of an intronic splicing regulatory element involved in auto-regulation of alternative splicing of SCL33 pre-mRNA. Plant J. 72, 935–946. doi: 10.1111/tpj.12004
Trujillo, M. (2018). News from the PUB: plant U-box type E3 ubiquitin ligases. J. Exp. Bot. 69, 371–384. doi: 10.1093/jxb/erx411
Vierstra, R. D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8, 135–142. doi: 10.1016/S1360-1385(03)00014-1
Wachter, A., Ruhl, C., and Stauffer, E. (2012). The role of polypyrimidine tract-binding proteins and other hnRNP proteins in plant splicing regulation. Front. Plant Sci. 3, 81. doi: 10.3389/fpls.2012.00081
Wagner, S. A., Beli, P., and Weinert, B. T. (2011). A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteomics 10, M111.013284. doi: 10.1074/mcp.M111.013284
Wahl, M. C., Will, C. L., and Luhrmann, R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell. 136, 701–718. doi: 10.1016/j.cell.2009.02.009
Wang, E. T., Sandberg, R., Luo, S., Khrebtukova, I., Zhang, L., Mayr, C., et al. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature. 456, 470–476. doi: 10.1038/nature07509
Wang, X., Wu, F., Xie, Q., Wang, H., Wang, Y., and Yue, Y. (2012). SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell. 24, 3278–3295. doi: 10.1105/tpc.112.100081
Wang, Z., Ji, H., Yuan, B., Wang, S., Su, C., Yao, B., et al. (2015). ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun. 6, 8138. doi: 10.1038/ncomms9138
Watson, S. J., Li, N., Ye, Y., Wu, F., Ling, Q., Jarvis, R. P., et al. (2021). Crosstalk between the chloroplast protein import and SUMO systems revealed through genetic and molecular investigation in Arabidopsis. Elife. 10, 960. doi: 10.7554/eLife.60960
Welchman, R. L., Gordon, C., and Mayer, R. J. (2005). Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6, 599–609. doi: 10.1038/nrm1700
Wilkinson, C. R., Dittmar, G. A., Ohi, M. D., Uetz, P., Jones, N., and Finley, D. (2004). Ubiquitin-like protein Hub1 is required for pre-mRNA splicing and localization of an essential splicing factor in fission yeast. Curr. Biol. 14, 2283–2288. doi: 10.1016/j.cub.2004.11.058
Will, C. L., and Luhrmann, R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect Biol. 3, 3707. doi: 10.1101/cshperspect.a003707
Willems, P., Horne, A., Van Parys, T., Goormachtig, S., De Smet, I, Botzki, A., et al. (2019). The Plant PTM Viewer, a central resource for exploring plant protein modifications. Plant J. 99, 752–762. doi: 10.1111/tpj.14345
Witten, J. T., and Ule, J. (2011). Understanding splicing regulation through RNA splicing maps. Trends Genet. 27, 89–97. doi: 10.1016/j.tig.2010.12.001
Xu, D. (2020). COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol. 228, 1748–1753. doi: 10.1111/nph.16296
Yan, N., Doelling, J. H., Falbel, T. G., Durski, A. M., and Vierstra, R. D. (2000). The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 124, 1828–43. doi: 10.1104/pp.124.4.1828
Zhang, Y., Xia, G., and Zhu, Q. (2021). Conserved and unique roles of chaperone-dependent E3 ubiquitin ligase CHIP in plants. Front. Plant Sci. 12, 699756. doi: 10.3389/fpls.2021.699756
Zhou, Y., Li, X., Guo, Q. H., Liu, P., Li, Y., Wu, C. A., et al. (2021). Salt responsive alternative splicing of a RING finger E3 ligase modulates the salt stress tolerance by fine-tuning the balance of COP9 signalosome subunit 5A. PLoS Genet. 17, e1009898. doi: 10.1371/journal.pgen.1009898
Keywords: ubiquitin and ubiquitin-like modification, pre-mRNA splicing, spliceosome components, plant development, stresses response
Citation: Lan W, Qiu Y, Xu Y, Liu Y and Miao Y (2022) Ubiquitination and Ubiquitin-Like Modifications as Mediators of Alternative Pre-mRNA Splicing in Arabidopsis thaliana. Front. Plant Sci. 13:869870. doi: 10.3389/fpls.2022.869870
Received: 05 February 2022; Accepted: 07 April 2022;
Published: 12 May 2022.
Edited by:
Konstantin V. Kiselev, Far Eastern Branch of the Russian Academy of Sciences, RussiaReviewed by:
Qingshun Quinn Li, Western University of Health Sciences, United StatesBorja Belda-Palazon, Gulbenkian Institute of Science (IGC), Portugal
Copyright © 2022 Lan, Qiu, Xu, Liu and Miao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Miao, eW1pYW9AZmFmdS5lZHUuY24=
 Wei Lan
Wei Lan Yuhao Qiu
Yuhao Qiu Ying Miao
Ying Miao