
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 25 April 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.868874
This article is part of the Research Topic Mechanisms of Abiotic Stress Responses and Tolerance in Plants: Physiological, Biochemical and Molecular Interventions, volume II View all 37 articles
Environmental stress is one of the major restrictions on plant development and foodstuff production. The adaptive response in plants largely occurs through an intricate signaling system, which is crucial for regulating the stress-responsive genes. Myelocytomatosis (MYC) transcription factors are the fundamental regulators of the jasmonate (JA) signaling branch that participates in plant development and multiple stresses. By binding to the cis-acting elements of a large number of stress-responsive genes, JA-responsive transcription factors activate the stress-resistant defense genes. The mechanism of stress responses concerns myriad regulatory processes at the physiological and molecular levels. Discovering stress-related regulatory factors is of great value in disclosing the response mechanisms of plants to biotic or abiotic stress, which could guide the genetic improvement of plant resistance. This review summarizes recent researches in various aspects of MYC2-mediated JA signaling and emphasizes MYC2 involvement in plant growth and stress response.
Phytohormones are a group of signaling molecules that are essential to perceive changes in the external environment, regulate their growth status, defend themselves against insects and maintain survival (Santner and Estelle, 2009; Dubois et al., 2018). JA is often used as a plant growth regulator as well as a signaling molecule for plant defense responses (Mao et al., 2017). Biotic and abiotic stresses have always been the vital factors in restricting the yield of food products. The adaptive responses are mainly achieved by simulating the transcription of a large number of downstream genes through complex signaling pathways (Sabagh et al., 2022). JA mediated plant development and a variety of biotic stresses caused by physical injury, pathogen attack, and insect feeding, as well as several abiotic challenges such as low temperature, drought, excessive salinity, and UV radiation (Verma et al., 2016; Yang et al., 2019). To protect against external invasion, plants perceive and transmit JA cascade signals through a variety of regulatory factors that are over-defensive and may hamper plant growth and development (Zander et al., 2020). MYC transcription factors could bind to the promoter regions of several targeted-genes, which affects their expression that help plants deal with stresses. In Arabidopsis, MYC2-mediated activation of JA signaling is precisely regulated by its repressor, the JASMONATE-ZIM DOMAIN (JAZ) protein, and a subunit of the Mediator complex (MED25) protein (Wu et al., 2020). Jasmonic acid-isoleucine conjugate (JA-Ile), as an active molecule, binds to the subunit of the Skp1/Cullin/F-box (SCFCOI1) ubiquitin ligase and transmits downstream signaling (Wasternack and Xie, 2010). In the absence of external stress, JAZ protein binds to MYC2 and other transcription factors, rendering them inactive and devoid of triggering the transcription of downstream genes. MYC2 is repressed by JAZ, resulting in the closure of the downstream regulatory switch. Once JA-Ile is present as an activating signal, CORONATINE-INSENSITIVE 1 (COI1), JA-Ile and JAZ proteins form a co-receptor complex. COI1 regulates ubiquitin-dependent JAZ degradation via the 26S proteasome, releasing MYC2 from repression (Thines et al., 2007). Some studies demonstrated that MYC2-mediated JA signaling had an influence on the transcription reprogramming of a variety of stress-responsive genes (Zander et al., 2020). Given the critical role of MYC transcription factors, this review highlights the regulatory functions of MYCs at multiple levels and dwells on the activation and reduction mechanisms of JA signaling.
MYC proteins are the core transcription factors in JA signaling pathway. As a branch of the basic helix-loop-helix (bHLH) superfamily, the MYC family is widely present in bryophytes and angiospermae (Peñuelas et al., 2019). The MYC-related TFs between Arabidopsis thaliana and the liverwort Marchantia polymorpha had an early-diverging lineage. The evolutionary relationship suggested MYC function originally occurred in charophycean algae and hence predates the evolutionary appearance of any other jasmonate pathway component. Plants have a large number of bHLH family members that mainly involved in plant growth, stress response, and secondary metabolic processes (Heim et al., 2003; Toledo-Ortiz et al., 2003). Their bHLH genes are more closely related to the group B in the animal members. AtbHLHs were categorized into 12 families and then further classified based on conserved domains, with 14 new members found (Toledo-Ortiz et al., 2003). Currently, 158 and 173 bHLH transcription factors have been identified in A. thaliana and O. sativa, respectively (Pires and Dolan, 2010).
MYC-like transcription factors have three subfamilies, namely IIId, IIIe, and IIIf bHLHs. MYC2 belongs to the IIIe bHLH clade. In A. thaliana, IIIe bHLH members consist of AtMYC2, AtMYC3, AtMYC4, and AtMYC5. AtMYC2 was discovered from the screening of jasmonate-insensitive (jin) mutants. Earlier study indicated that jin1 and jin4 mutant showed insensitivity to exogenous JA stimulation (Berger et al., 1996). AtMYC2 positively regulated both JA-mediated wounding response and negatively regulates JA-mediated resistance to saprophytic pathogens (Lorenzo et al., 2004). In addition to its high affinity for the canonical G-box, MYC2 binds to the G-box mutants 5′-CACATG-3′ and 5′-CACGTT-3′ with low affinity (Dombrecht et al., 2007). The base types at both ends of the G-box affect MYC2 binding, and the number of thymine residing at the C-terminus of the G-box could affect the binding ability of MYC2 (Figueroa and Browse, 2012).
MYC2 included a 60-amino acid conserved bHLH domain at the C-terminus, which was composed of a Basic and an HLH domain, along with a JID domain and a TAD domain at the N-terminus (Goossens et al., 2015). The Basic domain consists of 15–20 basic amino acids and is mainly responsible for binding the G-box. The HLH domain consists of a loop region connecting two α-helices composed of hydrophobic amino acids, which are responsible for forming homo- or heterodimers. The JID domain is responsible for interacting with JAZ proteins, while the TAD domain is responsible for transcriptional activation and interactions with MED25 (Figure 1). MYC2 functions by binding to DNA through its basic domain (Lian et al., 2017). The crystal structure of the bHLH domain of MYC2 protein combined with DNA shows that two homodimers form parallel shoulder-to-shoulder tetramers reversely arranged in the MYC2-DNA structure, while MYC3 can only form homodimers (Lian et al., 2017). In contrast to MYC3, MYC2 has a stronger activation capacity. This is because the tetramerized MYC2 can mediate the formation of DNA looping structures, which in turn increases the ability of MYC2 to bind DNA and activate gene expression (Kazan and Manners, 2013).
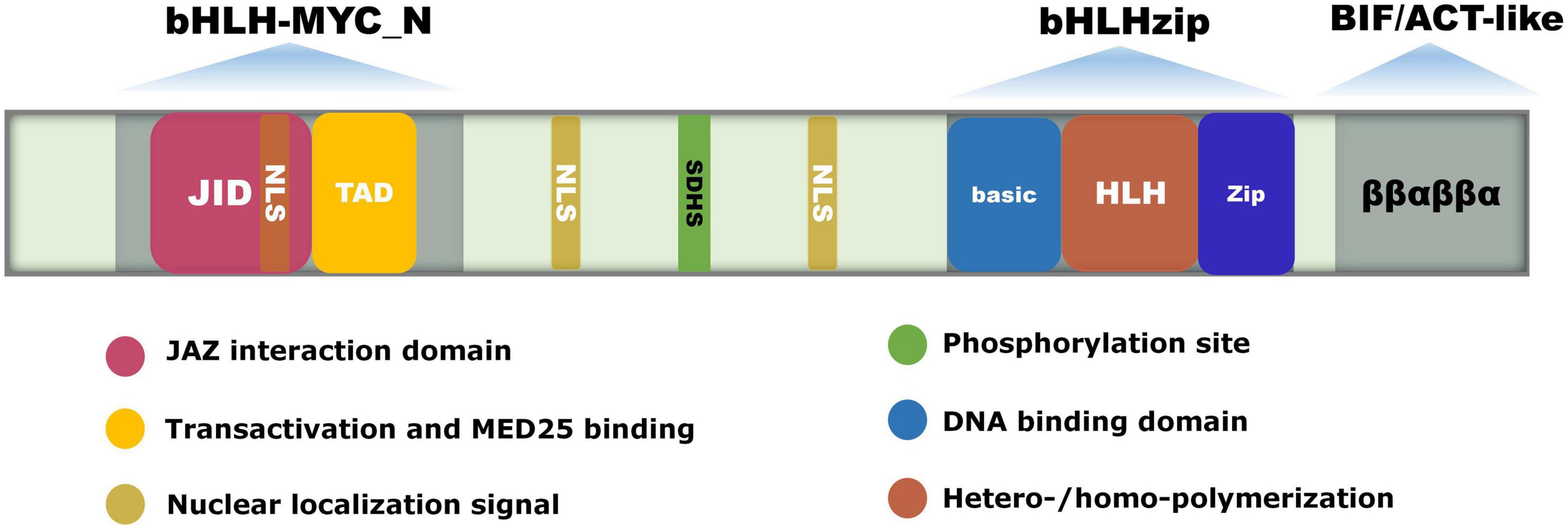
Figure 1. The gene structure and conserved domains of MYC2. bHLH-MYC_N, bHLH-zip, and BIF/ACT-like are the three conserved domains of MYC2. The bHLH domain at the C-terminus is composed of the Basic and HLH domains, and the N-terminus contains the JID and TAD domains. The Basic domain is responsible for binding to the G-box in the promoter of the targeted gene. The HLH domain is responsible for forming homo- or heterodimers. The JID domain binds to the Jas domain of the JAZ protein, and the TAD domain is responsible for transactivation and interactions with the MED25 subunit. The BIF structural domain contains a ββαβββα structure similar to the ACT domain, and is responsible for transactivation and homodimer formation. NSL is the nuclear localization signaling structure. The conserved amino acid SDHS is the phosphorylation site.
Although JA is the initiator that activates the JA signaling pathway, the real functional component is JA-Ile (Suza and Staswick, 2008). JA-Ile is synthesized by an ATP-dependent jasmonate-amino acid amidase encoded by JASMONATE RESISTANT 1 (JAR1) and whose product, (3R,7S)-jasmonoyl-L-isoleucine, is the most biologically active JA component. JAZ proteins, as a type of repressors, could interact with other transcription factors or co-repressors to block downstream signaling. The TIFY family included three subfamilies JAZ, Zinc-finger protein expressed in Inflorescence Meristem (ZIM) and PEAPOD (PPD), among which JAZ did not contain DNA binding domains and required reciprocal interactions to perform its functions (Bai et al., 2011). There were three conserved domains within JAZ, including NT, ZIM, and Jas (Zhang et al., 2017). The NT domain at the N-terminus interacted with the GIBBERELLIC ACID INSENSITIVE (DELLA) protein (Wager and Browse, 2012). The ZIM domain contained the TIFY conserved motif TIF[F/Y]XG, which was responsible for homo- or heterodimer formation of the JAZ protein and interacts with the co-repressor Novel Interactor of JAZ (NINJA) (Pauwels et al., 2010). The C-terminal Jas domain could directly interact with the JID domain of MYC2 or COI1, which was responsible for the degradation of JAZ. Some JAZ members also contained an EAR motif at the N-terminus, which interacted directly with its co-repressor TOPLESS (TPL). Those proteins that did not have an EAR motif could not directly interact with TPL and required the participation of the NINJA protein. Due to the EAR motif in NINJA, JAZ, and TPL formed a JAZ-NINJA-TPL complex to exert its transcriptional repression. TPL protein is a member of the Gro/Tup1 family. Sixteen Groucho/Thymidine uptake 1 (Gro/Tup1) members from Arabidopsis are divided into two major groups. As co-repressors of MYC-regulon, TOPLESS/TOPLESS-Related/WUSCHELINTERACTING PROTEIN (TPL/TPR/WSIP) and LEUNIG/LEUNIG_HOMOLOG (LUG/LUH) participated in organ development, stem cell maintenance, environmental physiology, and so on (Ma et al., 2017). Although JAZ probably suppressed MYC function by interacting with MYC, some studies demonstrated that JAZ–TF transcriptional modules could control discrete branches of the JA-responsive genes in an MYC-independent manner (Cheng et al., 2011; Major et al., 2017). JASMONATE-ASSOCIATED MYC2-LIKE (JAM) and MYC2-TARGETED BHLH (MTB) from IIId bHLHs inhibited JA response by binding competitively to the promoters of MYC2-targeted genes (Sasaki-Sekimoto et al., 2013; Song et al., 2013; Wasternack, 2019).
JA-mediated MYC2 activation is fine-tuned by JAZ and the mediator complex MED25 (Figure 2). In addition to interacting with MYC2, MED25 also interacts with the receptor COI1 and a subset of JAZ repressors like NINJA and TPL (Zhai and Li, 2019; Zhai et al., 2020). In the repression phase, JAZ and its co-repressors compete with MED25 for the interaction with MYC2. In the activation phase, JA-Ile acts as molecular docking to facilitate the COI1–JAZ conjugation, which leads to proteasome-dependent degradation of JAZ repressors (Zhang F. et al., 2015). In the response phase, MED25 interacts with MYC2 and recruits HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY1 (HAC1) as well as RNA polymerase II (Pol II) to the promoters of MYC2-targeted genes, thereby activating their expression (An et al., 2017). HAC1 regulates the transcription of JA-responsive genes by epigenetically controlling the H3K9 acetylation of MYC2-target genes. The Gro/Tup1 family members LUG and LUH activate MYC2-mediated transcription of JAZ2 and LOX2 through the mediator complex and HAC1 (You et al., 2019). LUH promotes JA-dependent enhancement of protein interactions between MYC2 and its coactivators MED25 and HAC1. MED25 recruits LUH to the promoter of the MYC2-targeted gene. MED25 and MYC2 can activate the expression of MYC2-TARGETED BHLH (MTB), which reduces the expression of JA-responsive genes. MTBs negatively regulate JA-responsive transcriptional reprogramming by weakening the stability of MYC2-MED25 complex (Liu et al., 2019). Accompanied by the deactivation of the JA signal, the status returns to the repression phase. MYC2-mediated transcription of downstream targets is not only dependent on the activation of MED25, but also associated with enhancers that assist the transcription of promoters, such as JA enhancers (JAEs) (Wang H. et al., 2019). JA mediates MED25 to regulate the chromatin looping formation between JAE and its promoter, while MYC2 uses the JAE element to regulate its own expression. The JAE (ME2 element) in the MYC2 locus positively regulates MYC2 expression in the short-term JA response and negatively regulates MYC2 in the persistent JA response. Recent studies had emerged CUL3-based E3 ubiquitin ligases(CUL3BPM) roles in the ubiquitination of MYC proteins (Chico et al., 2020). JA stabilized BPM3 to construct a negative feedback regulatory loop to mediate the expression of MYC.
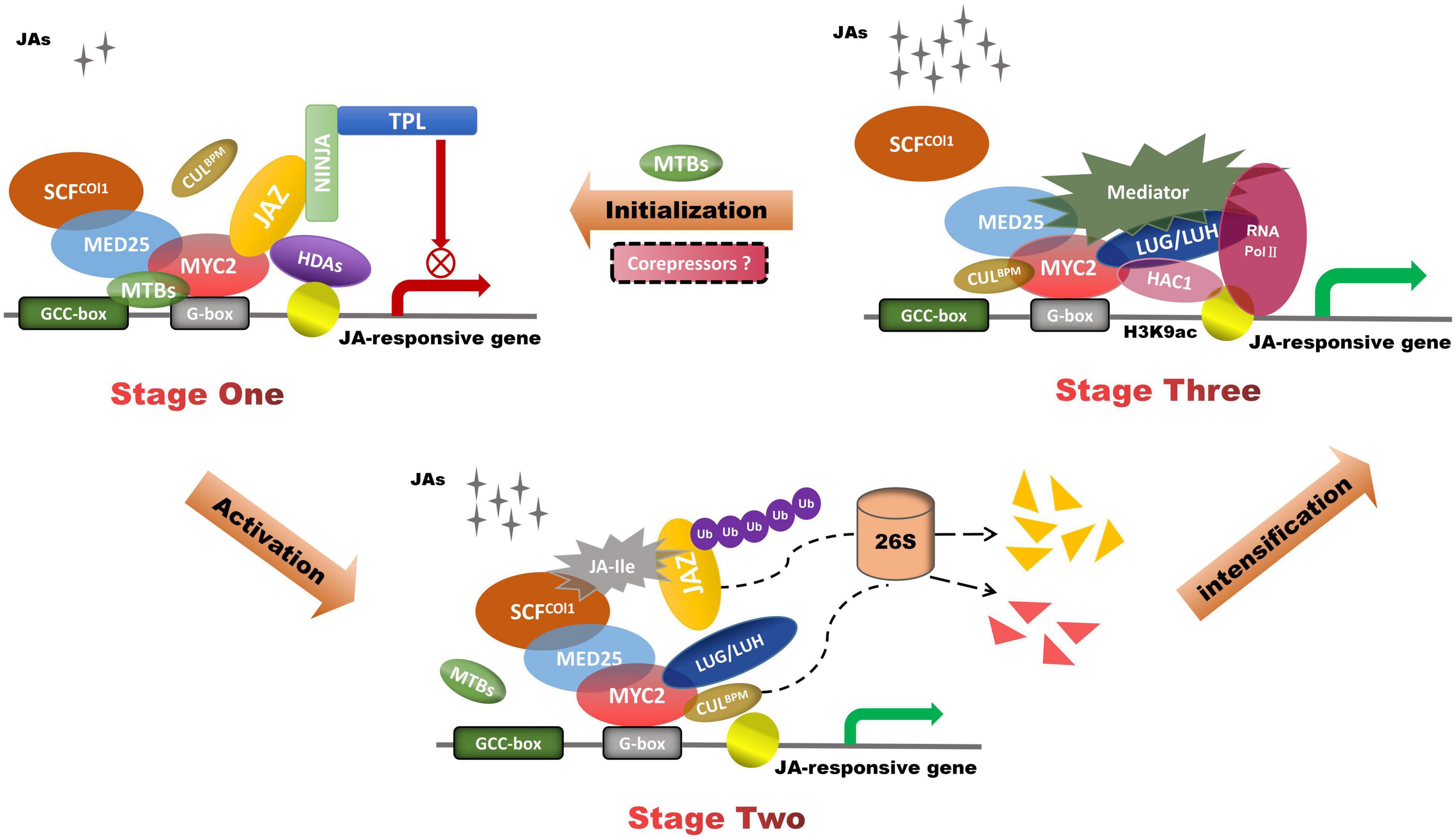
Figure 2. JAZ and MED25 mutually regulate JA-induced transactivation of MYC2-targeted genes. In Stage one, the MED25–MYC2 interaction is relatively weak because JAZ competes with MED25 for interaction with MYC transcription factors. In Stage two, COI1–JAZ co-receptor complex leads to proteasome-dependent degradation of JAZ repressors, which participate in the degradation of JAZ by 26S proteasome. In Stage three, MED25 recruits Pol II and LUH to the promoter of the MYC2-targeted gene, thereby regulating their expression. LUH enhanced the interactions of MYC2 coupled with its co-activators MED25 and HAC1. MTBs competitively interact with MYC2-targeted genes and impair the MED25–MYC2 complex. CUL3BPM reduce MYC levels by the ubiquitination of MYC TFs.
Depending on the JID domain, AtMYC2 can interact with most JAZs, and SlMYC2 can interact with all JAZs in tomato (Fernández-Calvo et al., 2011; Du et al., 2017). MYC3 interacted with JAZ9 not only in the JID domain but also in the TAD domain. This suggests that MYC2 utilizes the same and overlapping regions to interact with JAZ and MED25. The result that JAZ9 competitively interacted with MED25 and MYC2 explained why JAZ and MED25 had both antagonistic and synergistic effects on MYC2 transcription (Zhang F. et al., 2015). Changes in conserved amino acids would bring MYC2 and MYC3 out of repression and increased their transcriptional activities (Goossens et al., 2015). The MYC2D105N mutant was constructed by introducing aspartic acid at position 105 of the MYC2 amino acid, which merely interacted with JAZ1 and JAZ10 with significantly reduced binding capacity. Compared with MYC2, MYC3, and MYC4 have overlapping roles and their uniqueness in response to JA signaling and downstream regulation. MYC3 and MYC4 could form heterodimers and tetramers with MYC2, respectively. The functional deletion of MYC3 and MYC4 partially attenuated the capacities of JA response, while increased the insensitivity in myc2 mutants (Fernández-Calvo et al., 2011). In contrast to MYC3, MYC2 had a faster and higher levels of inducible response to JA (Lian et al., 2017). Taken together, MYC3 and MYC4 may play a part in the activation of JA signaling, which requires collaboration with MYC2 to regulate JA-mediated stress responses at different levels.
MYC2 not only regulates JA-mediated growth, development and resistance, but also participates in light signaling, circadian rhythm and cross-talk with other hormones (Figure 3). MYC2 restrained leaf vein development by reducing auxin biosynthesis (Dombrecht et al., 2007; Huang et al., 2017). MYC2 negatively regulates the development of leaf veins. JA also mediates MYC2 to inhibit the cell division of the primary root meristem (Chen et al., 2011). MYC2 binds to the G-box promoter of PLETHORA (PLT) in response to JA signaling, resulting in the reduction of MYC2-targeted genes. JA-mediated root growth is suppressed by MYC2. MYC2 is also involved in seed size, seed weight, and the formation of seed storage proteins. AtMYC2 has been shown to be positively correlated with seed storage rate at various stages (Gao et al., 2016). A recent study has demonstrated that MYC2 could inhibit adventitious root initiation by suppressing CYTOKININ OXIDASE/DEHYDROGENASE1 (CKX1) and enhancing the transcription of APETALA2.6 LIKE (RAP2.6L) (Dob et al., 2021). MYC2 and ETHYLENE INSENSITIVE3 (EIN3) synergistically controlled plant development and JA response (Zheng et al., 2017). MYC2 physically interacted with EIN3 to reduce HOOKLESS1 (HLS1) expression or binding to the promoter of EIN3 BINDING F-BOX PROTEIN1 (EBF1) to induce its expression (Zhang et al., 2014).
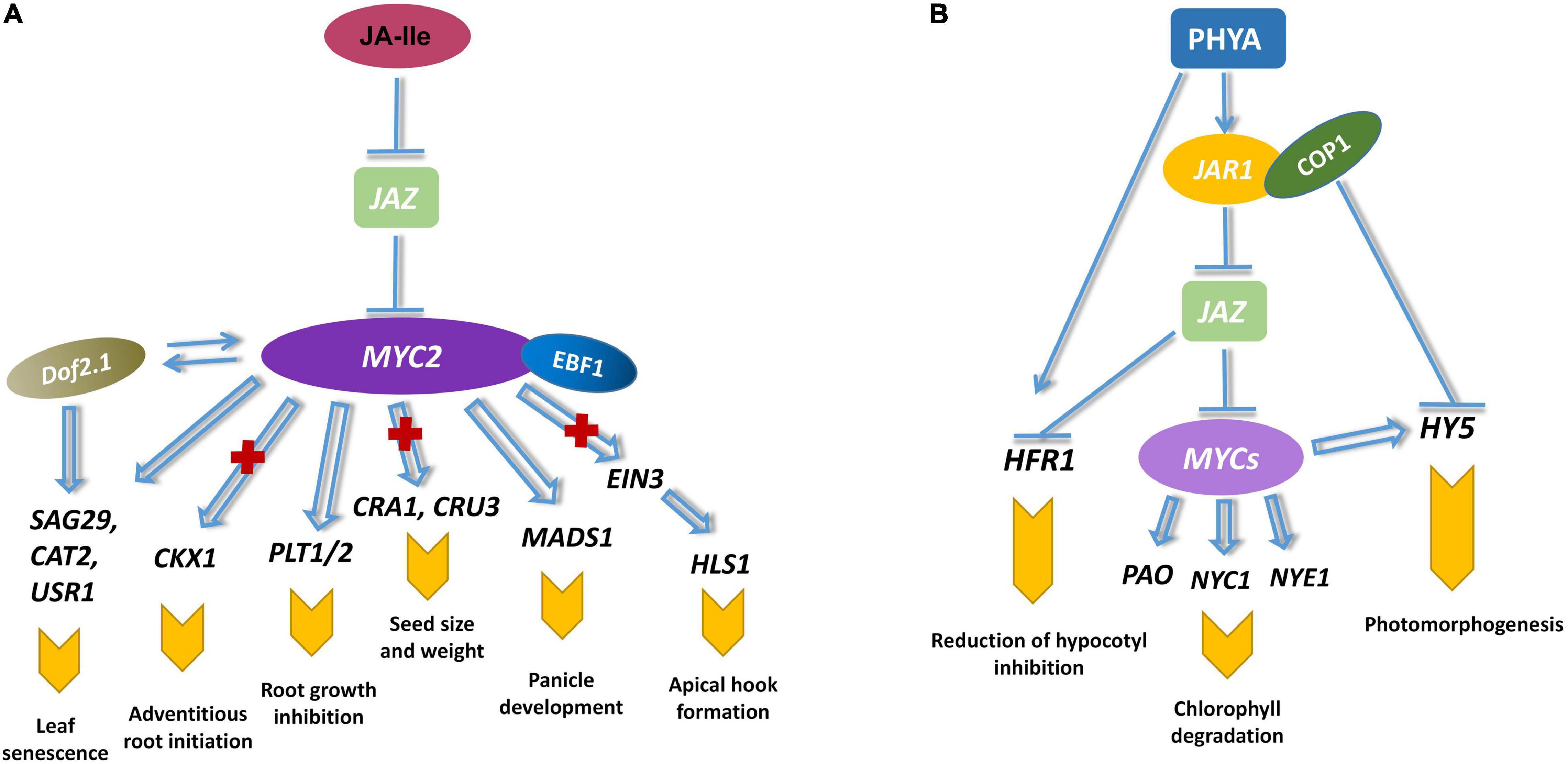
Figure 3. MYC2 regulates plant growth and photomorphogenesis. (A) MYC2 mediates JA-regulated root growth and meristem development. MYC2 inhibits adventitious root initiation by reducing the expression of CKX1 (Dob et al., 2021). JA-induced division and stem cell differentiation in meristematic tissue is mediated by MYC2, which interacts with PLT to suppress the expression of PLT-targeted genes (Chen et al., 2011). MYC2 restrains seed development by regulating the expression of seed storage protein CRA1 and CRU3 (Gao et al., 2016). MYC2 and EIN3 additionally controlled apical hook development by interacting with EBF1 and repressing the expression of HLS1 (Zhang et al., 2014). Senescence-associated genes are mutually enhanced by MYC2 and Dof proteins (Zhuo et al., 2020). MYC2 accelerates leaf senescence by raising the transcription of SAG29, CAT2, and USR1 (Qi et al., 2015b; Zhang Z. et al., 2020). (B) MYCs are involved in the photomorphogenesis of plants. PHYA regulates the downstream of JA signaling and ultimately affects the activation of MYCs (Chico et al., 2014). PHYA control the hypocotyl inhibition and chlorophyll reduction through JA-mediated HFR1 and HY5 (Chakraborty et al., 2019). MYCs regulate JA-induced Chl degradation by directly activating some Chl catabolic genes (PAO, NYC1, NYE1) (Zhu et al., 2015).
Light is indispensable for plant development and morphogenesis. MYC2 participated in photomorphogenesis by activating phytochrome A (phyA), which led to JA biosynthesis that recruited COI1 from the degradation of JAZ (Hsieh and Okamoto, 2014). Degradation of JAZ repressors results in the release of MYC transcription factors. At low R/FR ratios, phyA and myc2 mutants both showed decreased hypocotyl inhibition, which suggested MYC2 was responsible for far-red light response. Additionally, myc2 and phyA mutants exhibited a significant hypocotyl elongation of sheltering effect (Chico et al., 2014). HYPOCOTYL 5 (HY5) is a basic leucine-zipper (bZIP) transcription factor involved in photomorphogenesis at different wavelengths. hy5 mutants exhibit reduced hypocotyl elongation and chlorophyll accumulation. MYC2 and HY5 regulate seedling development under blue light irradiation (Chakraborty et al., 2019). MYC2 negatively regulates the expression of HY5 by binding to the promoter E-box. HY5 acts toward MYC2 in different patterns, implying that they coordinate seedling growth in a mutually antagonistic way. Chlorophyll (Chl) degradation is a characteristic event during green organ senescence or maturation. JA-induced Chl degradation could be mediated by directly activating some Chl catabolic genes, such as pheophorbide a oxygenase (PAO), NON-YELLOW COLORING 1 (NYC1), and NON-YELLOWING 1 (NYE1) (Zhu et al., 2015). The myc and phyB mutants exhibit similar phenotypes in delaying seed development and hypocotyl growth. Overexpression of MYC2 in phyB mutants partially reduces the prolonged hypocotyl phenotype (Ortigosa et al., 2020). By binding to the promoter of HY5, MYC2 and MYC3 activate the expression of the red light-responsive gene HY5. In addition to light signaling, the expression of MYC2 is regulated by circadian rhythm. MYC2 interacts with TIME FOR COFFEE (TIC) resulting in its degradation by the ubiquitination of 26S proteasome. The tic and myc2 mutants exhibited opposite JA-responsive phenotypes (Shin et al., 2012).
Floral initiation is a sophisticated and complex process, and the timing of flowering is synergistically regulated by a variety of factors such as temperature, light, and hormones. MYC2 is involved in JA-mediated inhibition of flowering (Wang et al., 2017). The myc2/3 and myc2/4 double mutants exhibited an earlier flowering phenotype than the wild type under long-day conditions, whereas the myc2, myc3, and myc4 single mutants and the myc3/4 double mutant had similar flowering times to the wild type. MYC2 is also involved in the developmental regulation of the calyx (Qi et al., 2015a). The myc2, myc3, myc4, and myc5 mutants displayed a failure to dehisce anther phenotype, delayed dehiscence, and low pollen grain survival, whereas the quadruple mutants had a delayed calyx development phenotype. Floral spike is an important component of the floral organ that affects reproductive growth and crop yield (Cai et al., 2014). EXTRA GLUME 1 and 2 (EG1 and EG2) were identified from spike deficient mutants of rice, where EG2 worked as a negative regulator to reduce MYC2-mediated floral morphogenesis. In the presence of JA signals, MYC2 promotes normal flowering spike by activating the flowering-related MADS1.
Ethylene is a regulatory molecule that promotes fruit ripening and organ senescence. In apples, JA induction and MYC2 can accelerate the expression of 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE (ACO) and 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE (ACS), on the other hand, ethylene production (Li et al., 2017). MYC2 not only bond to the promoters of ACS1 and ACO1 to increase their expression levels, but also simultaneously increased the activity of ERF3, which mediated the transcription regulation of ACS1 (Hu et al., 2022). MYC2 suppressed the repressive effect of ERF2 on the ACS1 promoter by interacting with ERF2, which prevented ERF2 from binding to ERF3 and produced large quantities of free ERF3 to activate the transcription of ACS1 (Kazan, 2015; Li et al., 2016). Interestingly, the IIIe bHLHs and the IIIb bHLH transcription factors bHLH3, bHLH13, bHLH14, and bHLH17 have antagonistic roles in regulating physiological processes such as chlorophyll accumulation, photochemical efficiency of PHOTOSYSTEM II (PSII), and membrane ion permeation (Qi et al., 2015b).
Leaf senescence is regulated by a large number of internal and environmental factors. MYC2, MYC3, and MYC4 are capable of activating JA-induced leaf senescence and regulating the senescence process by binding to the promoter of SENESCENCE-ASSOCIATED GENE29 (SAG29), whereas IIIb bHLHs bind to the SAG29 promoter to reduce its expression (Qi et al., 2015b). H2O2 scavenging significantly inhibits JA-induced leaf senescence and the expression of SAGs. Some studies have shown the myc2 mutant exhibits delayed leaf senescence with increased expression of CATALASE 2 (CAT2) and lower H2O2 accumulation (Zhang Y. et al., 2020). Arabidopsis DNA binding-with-one-finger (Dof) proteins are associated with leaf senescence. Dof2.1, a JA-inducible gene, accelerated leaf senescence by increasing the expression of MYC2. MYC2 was also responsible for the JA-inducible expression of Dof2.1 (Zhuo et al., 2020). U-box Senescence Related 1 (USR1) is involved in both age-dependent and dark-induced leaf senescence in Arabidopsis. The usr1 mutant exhibited a delay in the age-dependent and dark-induced leaf senescence. JA treatments promoted the accumulation of AtUSR1 by the activation function of MYC2 (Zhang Z. et al., 2020).
MYC2, MYC3, and MYC4 play analogous roles in regulating plant resistance to insects (Figure 4). The myc2 myc3 myc4 triple mutant is more susceptible to Spodoptera mauritia and Spodoptera exigua Hiibner than the wild plant (Fernández-Calvo et al., 2011; Schweizer et al., 2013; Song et al., 2014). MYC2 regulates the associated mechanical injury caused by herbivorous insects to positively regulate the expression of JASMONIC ACID2 (JA2) and JASMONIC ACID2-LIKE (JA2L), therefore controlling ABA biosynthesis and stomatal closure (Du et al., 2014). MYC2 promotes the transcription of the IAA biosynthesis gene YUCCA induced by the bite of the two-spotted spider mite Tetranychus urticae by binding to the tandem G-box in the promoter of YUC8 and YUC9 (Pérez-Alonso et al., 2021). MYC2, MYC3, and MYC4 promote the content of glucosinolates to enhance insect resistance. Glucosinolates are a class of thioglucosides containing nitrogen and sulfur, whose biosynthesis were governed by six R2R3-MYB transcription factors: MYB28, MYB29, MYB76, MYB34, MYB51, and MYB122. The interaction of MYCs and MYBs promotes the transcription of glucosinolate-related genes, resulting in increased GS levels (Schweizer et al., 2013). By forming a transcription factor complex with several MYB transcription factors, MYC2, MYC3, and MYC4 bond to the promoters of more than twenty glucosinolate biosynthesis genes, and directly regulated their expressions (Schweizer et al., 2013). In addition, MYC2 binds directly to the promoters of the terpene synthases TPS10, TPS11, and TPS21, and triggers their transcription. This could boost terpenoid production and provide independent resistance to insect attacks (Hong et al., 2012). Wounding-induced JA biosynthesis was relevant to the transcriptional reprogramming on the downstream of JA biosynthesis. JA catabolic genes, such as allene oxide synthase (AOS), allene oxide cyclase1 (AOC1), cis-(+)-12-oxo- phytodienoic acid reductase 3(OPR3), lipoxygenase 2 (LOX2), JASMONATE RESISTANT 1 (JAR1), jasmonic acid carboxyl methyltransferase (JMT), jasmonate-induced oxygenases 2 (JOX2), and cytochrome P450 94B1 (CYP94B1), etc. are upregulated by MYC2 at different levels (Zhang C. et al., 2020). ET-induced ERF15 and ERF16 also regulate JA biosynthesis in tomato. ET triggered the transcription of MYC2 and ERF16 by regulating JA biosynthesis genes (Hu et al., 2021).
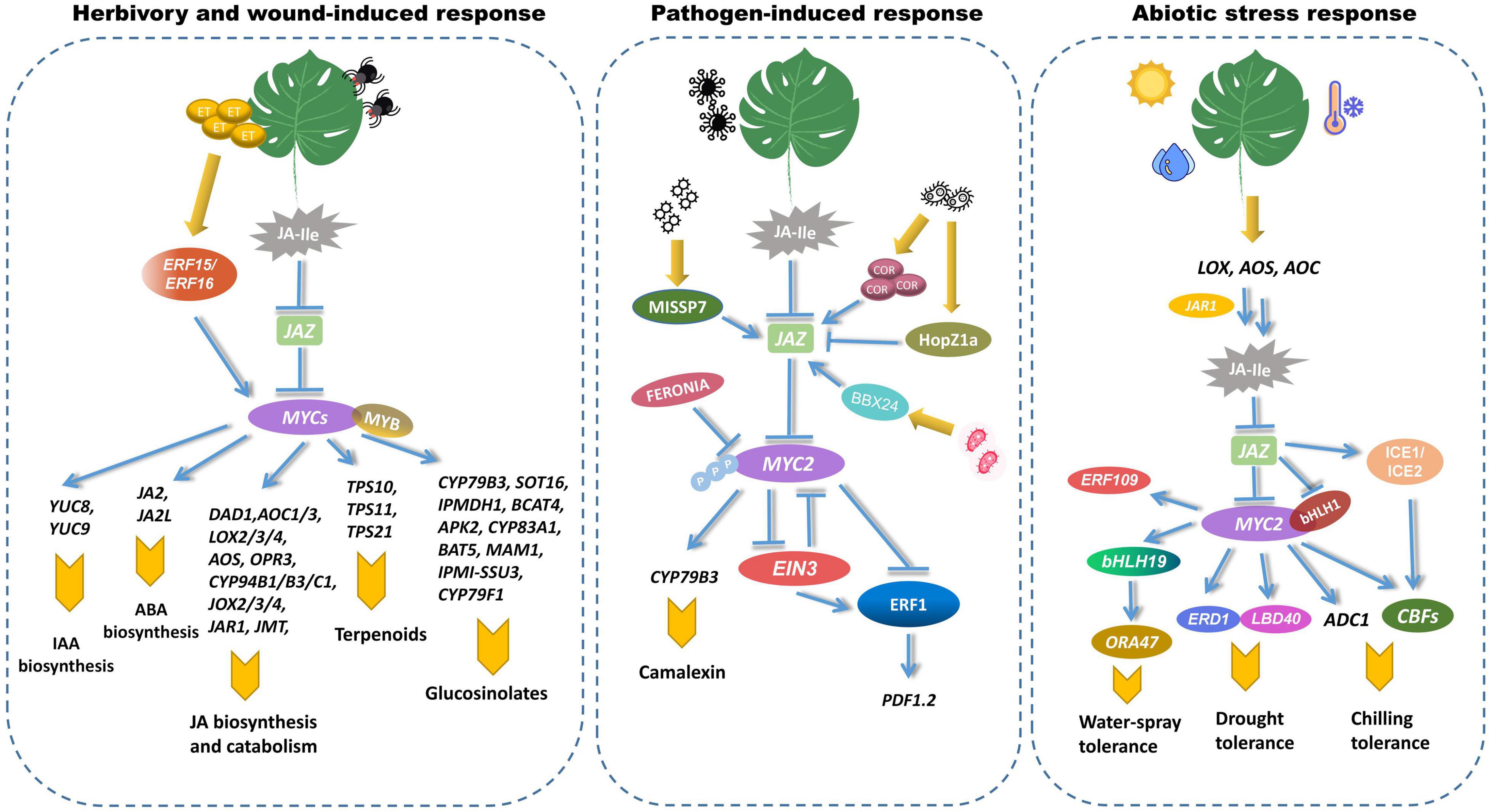
Figure 4. MYCs regulate biotic and abiotic stress responses in plants. MYCs participate in the process of pathogen infection and wound responses. MYCs interact with MYB to control the expression of several glucosinolate-biosynthetic genes, such as CYP79B3, SOT16, IPMDH1, and BCAT4, etc. (Mikkelsen et al., 2003; Piotrowski et al., 2004; Knill et al., 2008; He et al., 2011). MYC2 promotes the production of terpenoid phytolexin by upregulating the expression of TPS10, TPS11, and TPS21 (Hong et al., 2012). MYC2 regulates the associated mechanical injury of herbivorous insects by positively regulating the expression of NAC019 and JA2L, which increase the expression level of VSP1 and TD (Du et al., 2014, 2017). MYC2 enhanced the transcription of the IAA biosynthesis genes YUC8 and YUC9 (Pérez-Alonso et al., 2021). The transcription of multiple JA biosynthesis and catabolism genes are also mediated by MYC2 under wounding response. MYC2 interacts with EIN3 in ethylene signaling to antagonistically regulate the expression of ERF, which in turn regulates the expression of pathogen-related PDF1.2 (Song et al., 2014). MiSSP7 and BBX24 inhibit MYC2-targeted gene expression by increasing their interaction with JAZ (Daguerre et al., 2020; Zhang H. et al., 2020). HopZ1a triggered AtJAZ1 degradation and increased the expression of JA-responsive genes (Jiang et al., 2013). MYC2 function is inhibited by FER via its phosphorylation (Guo et al., 2018). MYCs are engaged in abiotic stresses such as low temperature, water-spray and drought. Environmental stress can increase the transcription of MYC2 and bHLH1, which in turn upregulate the expression of low-temperature stress-related CBFs and drought stress-related ERD1 (Li Y. et al., 2019; Wang Y. et al., 2019). ICE1 and ICE2 upregulate the expression of CBFs (Kashyap and Deswal, 2019). MYC2 increased ADC1 levels, which resulted in increased putrescine biosynthesis (Ding et al., 2021). Water spray-inducible MYC-regulon creates a hierarchical transcriptional network via interacting with bHLH19 and ERF109 (van Moerkercke et al., 2019). bHLH19 in turn directly activates the promoter of ORA47 (Chen et al., 2016).
In contrast to its regulatory role in insect resistance, MYC2 serves as a negative regulator of plant pathogens (Zheng et al., 2017). MYC2 blocked the function of EIN3 by decreasing ethylene-mediated pathogen response. MYC2 interacts with EIN3 to antagonistically regulate the expression of ERF, which enhanced the expression of pathogen-induced PDF1.2 (Song et al., 2014). The myc2 mutants are more resistant to pathogens like Botrytis cinerea than the wild type, probably attributed to MYC2 reciprocally inhibiting the function of EIN3. Cytochrome P450 enzymes are required for camalexin biosynthesis. CYP79B2 or CYP79B3 convert Trp to indole-3-acetaldoxime. Pathogen infection or abiotic stresses could induce the expression of CYP71A12 and CYP71A13, which dehydrate indole-3-acetaldoxime to produce indole-3-acetonitrile (IAN). IAN is subsequently activated, probably to indole cyanohydrin, and conjugated with glutathione to form GS-IAN. GS-IAN is degraded to Cys (IAN) via glutamyl peptidase 1 (GGP1), and Cys (IAN) is then converted to camalexin via CYP71B15 (Koprivova et al., 2019; Mucha et al., 2019). MiSSP7, a fungal symbiotic effector from Laccaria bicolor, blocked the dimerization of Populus JAZs, which facilitated the release of MYC2-targeted genes (Plett et al., 2014; Daguerre et al., 2020). The jin1 mutants are resistant to both P. syringae pv. tomato DC3000 and COR. The JIN gene is necessary for the inhibition of salicylic acid-dependent defenses in P. syringae infection. COR activates NAC transcription factors via MYC2, impairing the production of salicylic acid (SA) and weakening host immunity (Laurie-Berry et al., 2006). By phosphorylating and degrading MYC2, the receptor kinase FERONIA (FER) suppresses JA and COR signaling, hence positively regulating immunity (Guo et al., 2018). Transcriptional activation of the SA hormone defense sector acts as a critical barrier to biotrophic infections. In Arabidopsis effector-triggered immunity, the EDS1 complex in combination with PAD4 inhibits MYC2 to enhance the SA defense (Cui et al., 2018). P. syringae HopZ1a induced the degradation of AtJAZ1, and fostered JA-responsive gene expression during bacterial infection (Jiang et al., 2013). HopZ1a acetylated JAZs through a putative acetyltransferase catalysis. In recent studies, MYC2 has been implicated as a negative regulator of SA-mediated defense against bacterial infections. Overexpression of MYC2 confers hyper-resistance to a wide range of virulent and avirulent bacterial pathogens (Gautam et al., 2021). IbBBX24, a transcription factor from the B-box (BBX) family, modulated JA responses in sweet potato by antagonistically binding to IbJAZ10, which interacted with IbMYC2 (Zhang H. et al., 2020). Overexpression of IbBBX24 dramatically enhanced resistance to Fusarium wilt disease. Using the ChIP-seq approach, 665 MYC2-targeted binding genes in tomato were discovered (Du et al., 2017). These genes were found to be highly abundant under early JA stress, and one type of MYC2-targeted transcription factor (MTF) directly regulated the transcription of late defense genes.
Beyond modulating biotic stress responses, MYC2 is engaged in abiotic resistance, such as cold and drought stress (Figure 4). To avoid freezing injury or death, plants bolster the expression of C-repeat-binding factor (CBF), cold-regulated (COR), cold-induced (KIN), and low-temperature-induced (LTI). The INDUCER OF CBF EXPRESSION (ICE)-CBF transcriptional cascade is critical for the plant cold stress response (Kashyap and Deswal, 2019). ICE1 and ICE2, two bHLH TFs from A. thaliana, upregulate the expression of CBF by binding to CANNTG motif in the promoter. At the normal temperature, JAZ1/4 interacts with ICE1 and ICE2 to block the ICE-CBF pathway. However, Low-temperature conditions promote the expression of JA biosynthesis-related genes such as AOS1, DAD1, AOC, and LOX2, resulting in the production of JA-Ile and stimulation of COI1 to interact with JAZ1 (Zhu, 2016; Hu et al., 2017). Putrescine alleviated the oxidative damage caused by cold stress. Some research has demonstrated that inhibition of MYC2 decreased the expression of arginine decarboxylase 1 (ADC1) and lowered putrescine accumulation (Ding et al., 2021). JA regulated chilling response in apple (Wang Y. et al., 2019). Exogenous application of MeJA could increase the expression of bHLH1, CBF1, CBF2, and CBF3. bHLH1 regulated the chilling resistance by interacting with JAZ1/4 and MYC2. MYC2 sharply increased the expression level of CBFs, whereas JAZ1/4 decreased the facilitation of MYC2 on chilling resistance. Furthermore, MYC2 participated in JA-mediated drought tolerance (Li Y. et al., 2019). In A. thaliana, the coi1-2, JAZ1-Jas, and myc2-2 mutants exhibit drought sensitivity and regulate the stress caused by water deficit through the induction of EARLY RESPONSIVE TO DEHYDRATION 1 (ERD1). MYC2 modulates drought-responsive genes by binding to the promoter of ERD1, which in turn enhances drought tolerance. JA and 12-OPDA could increase stomatal closure. The elevation in OPDA levels coordinated with the decreased stomatal aperture and the increased drought tolerance (Savchenko et al., 2014). OsbHLH148 binds to OsJAZ1 to induce the expression of OsDREB1, thereby enhancing the drought resistance in rice (Sarwat and Tuteja, 2017). Recent studies have demonstrated that the Arabidopsis BBD proteins are essential in the ABA-mediated interplay of ABA and JA against drought stresses. Overexpression of AtBBD1 increased ABA sensitivity considerably and enhanced the expression of critical genes involved in the ABA-mediated drought cascade, particularly those related to ABA biosynthesis and downstream transcription factors (Mahmudul Huque et al., 2021). The LATERAL ORGAN BOUNDARIES DOMAIN (LBD)-containing protein isolated from tomato participates in JA-mediated drought tolerance (Liu et al., 2020). The knockout of SlLBD40 improved the water-holding ability of tomatoes. Water spray-inducible MYC-regulon creates a hierarchical transcriptional network via interacting with bHLH19 and ERF109, which demonstrated that spray-induced jasmonate accumulation was transcriptionally regulated by an MYC2-controlled positive-feedback loop (van Moerkercke et al., 2019).
Plants have evolved over time to develop a variety of stress-responsive mechanisms, including the production of defense chemicals against external stimuli. When a plant is stressed or induced by hormones, numerous secondary metabolites arise and accumulate. JA regulates the expression of key enzymes involved in plant secondary metabolism by activating or suppressing the activity of relevant transcription factors, which influences the expression of key enzymes and, as a result, influences secondary metabolite accumulation (Chen et al., 2019). In general, different types of transcription factors specifically regulate the biosynthetic pathways of different secondary metabolites, and some transcription factors are usually capable of regulating multiple genes along the same route (Figure 5).
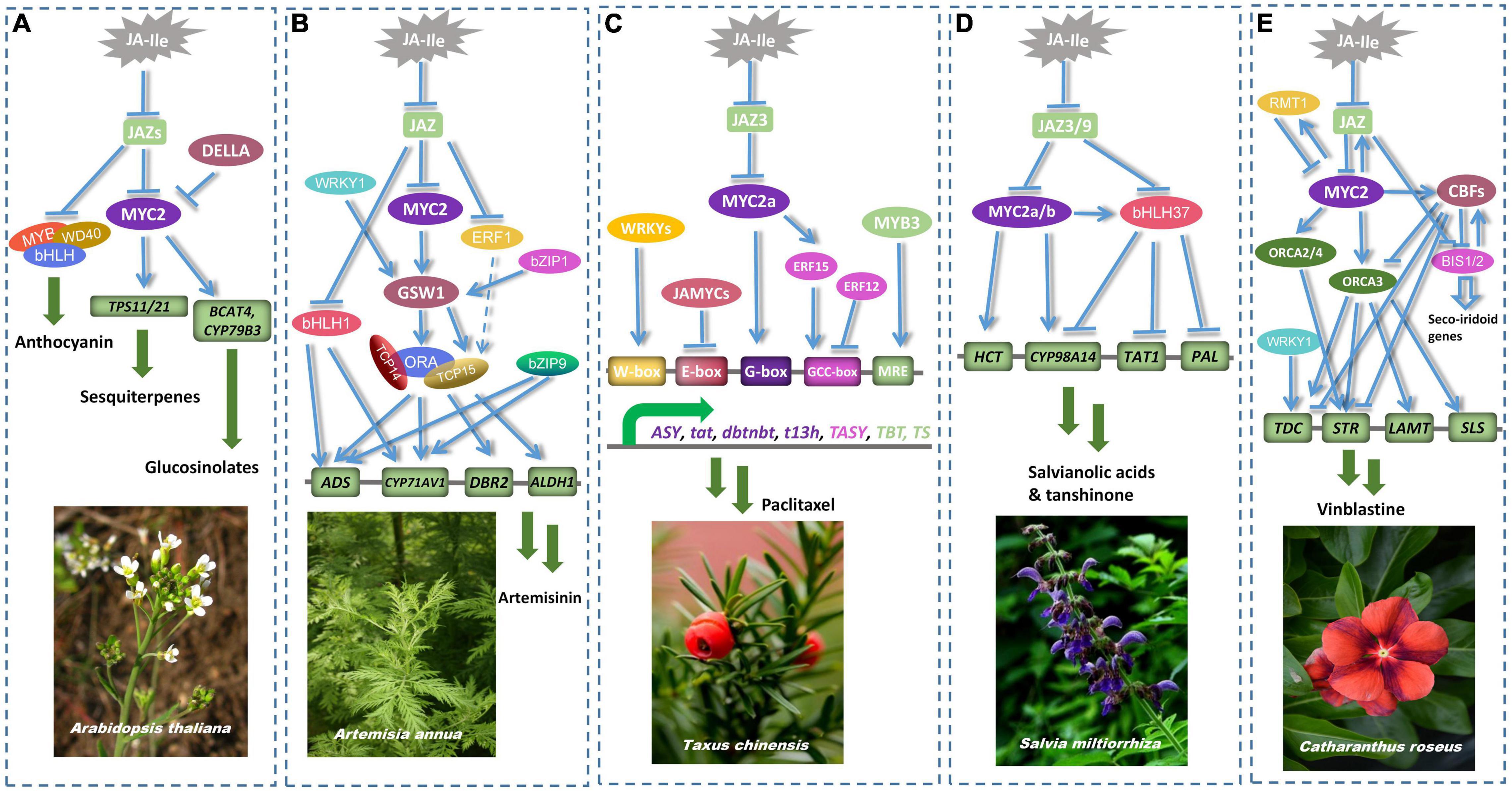
Figure 5. MYC2 plays an important regulatory role in the formation of secondary metabolites in model plants. (A) AtMYC2 alone or together with the MYB/bHLH/WD40 complex regulates the expression of CYP79B3, TPS11, TPS21 and anthocyanins accumulation (Xu et al., 2013; Wang L. et al., 2019). (B) AaMYC2 up-regulates the expression of CYP71AV1, by regulating the transcription of the glandular trichome-specific transcription factor AaGSW1 (Yu et al., 2012). AaMYC2 cooperates with ERFs, bHLHL1 and WRKY1 to jointly regulate the expression of ADS and DBR2 (Ma et al., 2018; Li L. et al., 2019). (C) TcMYC2a interacts with JAZ3 protein to regulate the expression of tat, dbtnbt, t13h, and coordinates with ERF12/15 to control the expression of TASY (Zhang M. et al., 2015; Zhang et al., 2018b). TcMYC2 and TcJAMYCs antagonistically regulate key genes of paclitaxel biosynthetic pathway (Lenka et al., 2015). (D) SmMYC2 interacts with SmJAZ3/9 to competitively control the expression of HCT6 and CYP98A14 (Zhou et al., 2016). bHLHL37 and MYC2 synergistically regulate the synthesis of salvianolic acid B. Unlike MYC2, bHLHL37 negatively regulates the expression of TAT1, PAL1, and CYP98A14 (Du et al., 2018). (E) CrMYC2 affects the expression of downstream genes STR and TDC by regulating the transcription of ORCA2 and ORCA3 (Van der Fits and Memelink, 2001; Zhang et al., 2011). In contrast, the negative regulators CBF1 and CBF2 competitively bind to MYC2-targeted genes thereby repressing the expression of TIA pathway genes (Sui et al., 2018).
MYC-like transcription factors are involved in the regulation of secondary metabolites such as terpenoids, alkaloids, flavonoids, and thioglycosides (Afrin et al., 2015). Paclitaxel, artemisinin, and tanshinone are terpenoids, which are classified into monoterpenes, sesquiterpenes, diterpenes, and triterpenes based on the number of isoprenoid units. AtTPS21 and AtTPS11 are responsible for volatile sesquiterpene biosynthesis. AtMYC2 directly binds to the promoters of TPS11 and TPS21 to regulate their expressions (Hong et al., 2012). DELLA can interact with MYC2 to repress the transactivation of TPS. Artemisinin is a sesquiterpene lactone whose biosynthesis is restricted to the specific glandular trichome cells (Olsson et al., 2009). Artemisinin biosynthesis genes are regulated by octadecanoid-derivative responsive AP2-domain protein (AaORA), which is regulated by the glandular trichome-specific transcription factor Artemisia annua GST-specific WRKY1 (AaGSW1). Overexpression of AaMYC2 resulted in large increases in cytochrome P450–dependent hydroxylase (CYP71AV1) and double bond reductase 2 (DBR2) levels, as well as an elevation in artemisinin content (Yu et al., 2012). MYC2 and bZIP1 synergistically regulate the expression of GSW1, while GSW1 binds to the promoter of TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL FACTOR15 (TCP15) to regulate its transcription. ORA interacts with TCP14/15 to enhance the expression and transactivation of DBR2 and ALDEHYDE DEHYDROGENASE 1 (ALDH1) via TCP14/15 (Ma et al., 2018, 2021). AabHLH1 was specifically expressed in glandular trichome, and overexpression of bHLH1 can increase the expression of AMORPHA-4, 11-DIENE SYNTHASE (ADS), and CYP71AV1 (Li L. et al., 2019). AabZIP9 binds to the ACGT element on the promoters of ADS and CYP71AV1 (Shen et al., 2019). Overexpression of AabZIP9 increased the expression of ADS, but had no significant effect on the expression of CYP71AV1, DBR2, and ALDH1.
Paclitaxel is a diterpene that is strongly triggered by JA. Extensive in vitro interaction studies revealed that MYCs were involved in the expression of several critical genes in the upstream of paclitaxel (Cui et al., 2019). TcMYC2a can interact with JAZ3 protein to mediate the expression of paclitaxel biosynthesis genes. By binding to T/G-box, G-box, and E-box, TcMYC2a affects the expression level of the GUS gene. Overexpression of MYC2a in the Taxus chinensis suspension cell line increases the expression of taxadiene synthase (TASY), taxadiene-5α-ol-acetyltransferase (tat), 30-N-dephenylated paclitaxel-N-benzoyltransferase (dbtnbt), and taxane 13-alpha-hydroxylase (t13h), whereas TcMYC2a inhibits the expression of TASY through its interaction with TcERF12/15 (Zhang M. et al., 2015; Zhang et al., 2018b). JA-inducible MYC transcription factors (TcJAMYC1, TcJAMYC2, and TcJAMYC4) are a group of negative regulators induced by JA whose domains are highly homologous to AtMYC2 (Lenka et al., 2015). JAMYCs regulate paclitaxel biosynthesis via binding to the E-box on the promoters of paclitaxel-biosynthetic genes TASY, taxadien-5α-ol by taxadien-5α-ol hydroxylase(T5aH), 10-deacetyl baccatinIII-10-O- acetyltransferase(DBAT), taxaneα-O-benzoyltransferase(DBBT), phenylalanine aminomutase (PAM), Baccatin III:3-amino, 3-phenylpropanoyl transferase(BAPT), 3-N-debenzoyl-2-deoxypaclitaxel-N-benzoyltransferase(DBTNBT). Overexpression of JAMYC negatively regulates the transcription of key genes related to paclitaxel biosynthesis. TcWRKYs regulate the expression of TcERF12/15 and paclitaxel-biosynthetic genes at different levels (Zhang et al., 2018a). Overexpression of WRKY8/20/26/47 positively regulates the expression of paclitaxel-biosynthetic genes. TmMYB3 increases the production of the paclitaxel precursor 10-deacetylbaccatin by binding to the MRE elements to upregulate the expression of TBT and TS (Yu et al., 2020).
Tanshinone is a representative diterpenoid. Tanshinone and salvianolic acid are two important active components in Salvia miltiorrhiza, whose biosynthesis is induced by JA. SmMYC2a and SmMYC2b participate in JA-mediated biosynthesis of tanshinone and salvianolic acid (Zhou et al., 2016). Hydroxy-cinnamoyl transferase6 (SmHCT6) and cytochrome P450 monooxygenase (SmCYP98A14) are key enzymes of the salvianolic acid biosynthesis pathway, whose expressions are induced by JA. MYC2a activates the expression of HCT6 and CYP98A14 by attaching to the promoter E-box of HCT6 and CYP98A14. MYC2b regulates the expression of CYP98A14 by binding to the E-box of CYP98A14. On the contrary, overexpression of SmJAZ3 or SmJAZ9 in S. miltiorrhiza hairy roots resulted in a significant decrease in tanshinone content, which might be attributed to the interaction of JAZ proteins with MYC2 to inhibit their transactivation. Tyrosine aminotransferase1(TAT1), phenylalanine ammonialyase1(PAL1) and CYP98A14 are key genes for tanshinolate biosynthesis. SmbHLHL37 and SmMYC2 synergistically regulate the synthesis of salvianolic acid B (Du et al., 2018). The expression of bHLHL37 is regulated by MYC2. It was found that MYC2 regulates the transcriptional activity of salvianolic acid B biosynthesis genes by binding to their promoters. bHLHL37 can dimerize and interact with JAZ3/8. Overexpression of bHLHL37 decreases the synthesis of salvianolic acid B. bHLHL37 inhibits the production of salvianolic acid B by binding to the promoters of SmTAT1, SmPAL1, and SmCYP98A14 to suppress their transcriptional activities.
Terpene alkaloids, especially terpene indole alkaloids (TIAs), have received more attention in Catharanthus roseus. JA is primarily responsible for inducing the production of these alkaloids. CrMYC1 was altered by fungal elicitor and JA in C. roseus suspension cells, indicating that CrMYC1 may be involved in the regulation of genes involved in JA signaling (Chatel et al., 2003). CrMYC2 is an early JA-responsive factor and regulates the expression of a series of TIA biosynthetic genes, by modulating the transcription of octadecanoid-derivative responsive Catharanthus AP2-domain proteins (ORCA2 and ORCA3) (Zhang et al., 2011). Knockdown of CrMYC2 by RNA interference significantly reduced the expression of ORCA2 and ORCA3. MeJA induced the expression of ORCA2 and ORCA3 and was specifically bound to the JERE element in the strictosidine synthase (STR) promoter. This implies that CrMYC2 stimulates STR activity via binding to a G-box-like element on the promoter of ORCA2/3, triggering the production of ORCAs-targeted genes rather than directly to the STR promoter. CrMYC2 also controls the expression of TIA biosynthesis genes in collaboration with certain repressors. The G-box binding factors CrCBF1 and CrCBF2 act as negative regulators to repress the production of TIAs (Sui et al., 2018). Overexpression of CrMYC2 increased the expression of CrGBF and eventually led to a reduction in alkaloid accumulation. CrGBF1 and CrGBF2 can form homo- and heterodimers to repress the activation of TIA pathway genes. CrGBF dimerizes with CrMYC2 and suppresses the transactivation of alkaloid-bisynthetic genes by competitively binding to the T/G-box of CrMYC2 targeted genes. Like CrGBF, CrRMT1 acts as a repressor of CrMYC2 that negatively regulates the MYC2-mediated transactivation of TIA-related genes (Patra et al., 2018). Nicotine is another well-studied alkaloid, featuring NtCOI1, NtJAZ, and NtMYC2 involved in its production. Putrescine N-methyltransferase (NtPMT1a) is a key enzyme in nicotine production. NtMYC2 controls nicotine production via two regulatory mechanisms (Shoji and Hashimoto, 2011). The first directly binds to the G-box of the PMT1a promoter to regulate its transcriptional activation, and the second regulates the expression of PMT1a through interacting with ERF in the NIC2 region. NtMYC2a and NtMYC2b form a conjugated complex with NtJAZ1 that negatively regulates multiple steps of JA-induced nicotine biosynthesis (Zhang et al., 2012). Our research investigated the TIA-regulatory gene MYC2 in Dendrobium officinale (Zhu et al., 2017). DoMYC2 negatively regulates the upstream pathway of terpene alkaloids. The expression of DoMYC2 could be induced by MeJA and chitosan in a short period. Overexpression of DoMYC2 in Arabidopsis attenuated the expression levels of HMG-CoA reductase (AtHMGR2), farnesyl diphosphate synthase (AtFPS1/AtFPS2), and isopentenyl transferases (AtIPTs).
Since the discovery of JA receptor COl1, several regulatory factors in the JA signaling pathway have been uncovered one after another. JA plays an essential role in both maintaining normal development and protecting plants from environmental stresses. Plants have evolved a sophisticated negative feedback mechanisms to regulate the moderate onset of resistance responses during their continuous adaptation to the environment and various stresses. In this review, we encapsulate the diverse roles of MYC TFs and generalize a MYC-mediated JA signaling pathway. This gives our understanding of how JA works to maintain a balance between plant growth and adaptability to the external environment. The basic guidelines described here provide a framework for elucidating a cross-regulation between MYC2 and its co-regulons. As the core regulatory molecules of the JA signaling branch, MYC2 and its relatives directly target hundreds of JA-responsive genes, building a positive cascade of transcriptional reprogramming. The downstream TFs under MYC2 and its homologs directly targeted overlapping but separate cohorts of genes, indicating that they have different roles within the JA response. The intricacy within the MYC2-mediated JA signaling pathway in general is the presence of some feedforward and feedback loops that are simultaneously activated. However, relationships between these subnetworks through the dynamics and resilience of their regulatory effects on the broader network are not well understood. Uncoupling these subnetworks would be an effective way to assess how they socialize to drive a robust activation of JA signaling.
CS, YC, and HD discussed the writing plan. CS, YC, JD, and GL drafted the manuscript. CS, MM, and CC edited the manuscript. HD and CS acquired the funding. All authors have read, reviewed, and approved the submitted version.
This work was supported by the Natural Science Foundation of Anhui Province (1808085MH307), High-level Talents Research Initiation Funding Project of West Anhui University (WGKQ2022025), and Quality Engineering Project in Anhui Province (2020jyxm2137).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
An, C., Li, L., Zhai, Q., You, Y., Deng, L., Wu, F., et al. (2017). Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. U S A. 114, E8930–E8939. doi: 10.1073/pnas.1710885114
Afrin, S., Huang, J. J., and Luo, Z. Y. (2015). JA-mediated transcriptional regulation of secondary metabolism in medicinal plants. Sci. Bull. 60, 1062–1072. doi: 10.1007/s11434-015-0813-0
Bai, Y., Meng, Y., Huang, D., Qi, Y., and Chen, M. (2011). Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98, 128–136. doi: 10.1016/j.ygeno.2011.05.002
Berger, S., Bell, E., and Mullet, J. E. (1996). Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111, 525–531. doi: 10.1104/pp.111.2.525
Cai, Q., Yuan, Z., Chen, M., Yin, C., Luo, Z., Zhao, X., et al. (2014). Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5:3476. doi: 10.1038/ncomms4476
Chakraborty, M., Gangappa, S. N., Maurya, J. P., Sethi, V., Srivastava, A. K., Singh, A., et al. (2019). Functional interrelation of MYC2 and HY5 plays an important role in Arabidopsis seedling development. Plant J. 99, 1080–1097. doi: 10.1111/tpj.14381
Chatel, G., Montiel, G., Pré, M., Memelink, J., Thiersault, M., Saint-Pierre, B., et al. (2003). CrMYC1, a Catharanthus roseus elicitor- and jasmonate-responsive bHLH transcription factor that binds the G-box element of the strictosidine synthase gene promoter. J. Exp. Bot. 54, 2587–2588. doi: 10.1093/jxb/erg275
Chen, H. Y., Hsieh, E. J., Cheng, M. C., Chen, C. Y., Hwang, S. Y., and Lin, T. P. (2016). ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol. 211, 599–613. doi: 10.1111/nph.13914
Chen, Q., Sun, J., Zhai, Q., Zhou, W., Qi, L., Xu, L., et al. (2011). The basic helix-loop-helix transcription factor myc2 directly represses plethora expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23, 3335–3352. doi: 10.1105/tpc.111.089870
Chen, X., Wang, D. D., Fang, X., Chen, X. Y., and Mao, Y. B. (2019). Plant specialized metabolism regulated by jasmonate signaling. Plant Cell Physiol. 60, 2638–2647. doi: 10.1093/pcp/pcz161
Cheng, Z., Sun, L., Qi, T., Zhang, B., Peng, W., Liu, Y., et al. (2011). The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4, 279–288. doi: 10.1093/mp/ssq073
Chico, J. M., Fernández-Barbero, G., Chini, A., Fernández-Calvo, P., Díez-Díaz, M., and Solano, R. (2014). Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26, 1967–1980. doi: 10.1105/tpc.114.125047
Chico, J. M., Lechner, E., Fernandez-Barbero, G., Canibano, E., García-Casado, G., Franco-Zorrilla, J. M., et al. (2020). CUL3BPM E3 ubiquitin ligases regulate MYC2, MYC3, and MYC4 stability and JA responses. Proc. Natl. Acad. Sci. U. S. A. 117, 6205–6215. doi: 10.1073/pnas.1912199117
Cui, H., Qiu, J., Zhou, Y., Bhandari, D. D., Zhao, C., Bautor, J., et al. (2018). Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in Arabidopsis effector-triggered immunity. Mol. Plant 11, 1053–1066. doi: 10.1016/j.molp.2018.05.007
Cui, Y., Mao, R., Chen, J., and Guo, Z. (2019). Regulation mechanism of MYC family transcription factors in jasmonic acid signalling pathway on taxol biosynthesis. Int. J. Mol. Sci. 20:1843. doi: 10.3390/ijms20081843
Daguerre, Y., Basso, V., Hartmann-Wittulski, S., Schellenberger, R., Meyer, L., Bailly, J., et al. (2020). The mutualism effector MiSSP7 of Laccaria bicolor alters the interactions between the poplar JAZ6 protein and its associated proteins. Sci. Rep. 10:20362. doi: 10.1038/s41598-020-76832-76836
Ding, F., Wang, C., Xu, N., Wang, M., and Zhang, S. (2021). Jasmonic acid-regulated putrescine biosynthesis attenuates cold-induced oxidative stress in tomato plants. Sci. Hortic. (Amsterdam) 288:110373. doi: 10.1016/j.scienta.2021.110373
Dob, A., Lakehal, A., Novak, O., and Bellini, C. (2021). Jasmonate inhibits adventitious root initiation through repression of CKX1 and activation of RAP2.6L transcription factor in Arabidopsis. J. Exp. Bot. 72, 7107–7118. doi: 10.1093/jxb/erab358
Dombrecht, B., Gang, P. X., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245. doi: 10.1105/tpc.106.048017
Du, M., Zhai, Q., Deng, L., Li, S., Li, H., Yan, L., et al. (2014). Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 26, 3167–3184. doi: 10.1105/tpc.114.128272
Du, M., Zhao, J., Tzeng, D. T. W., Liu, Y., Deng, L., Yang, T., et al. (2017). MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 29, 1883–1906. doi: 10.1105/tpc.16.00953
Du, T., Niu, J., Su, J., Li, S., Guo, X., Li, L., et al. (2018). SmbHLH37 functions antagonistically with smMYC2 in regulating jasmonate-mediated biosynthesis of phenolic acids in salvia miltiorrhiza. Front. Plant Sci. 871:1720. doi: 10.3389/fpls.2018.01720
Dubois, M., Van den Broeck, L., and Inzé, D. (2018). The pivotal role of ethylene in plant growth. Trends Plant Sci. 23, 311–323. doi: 10.1016/j.tplants.2018.01.003
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Figueroa, P., and Browse, J. (2012). The Arabidopsis JAZ2 promoter contains a G-box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol. 53, 330–343. doi: 10.1093/pcp/pcr178
Gao, C., Qi, S., Liu, K., Li, D., Jin, C., Li, Z., et al. (2016). MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis. Plant Physiol. Biochem. 108, 63–70. doi: 10.1016/j.plaphy.2016.07.004
Gautam, J. K., Giri, M. K., Singh, D., Chattopadhyay, S., and Nandi, A. K. (2021). MYC2 influences salicylic acid biosynthesis and defense against bacterial pathogens in Arabidopsis thaliana. Physiol. Plantarum 173, 2248–2261. doi: 10.1111/ppl.13575
Goossens, J., Swinnen, G., Vanden Bossche, R., Pauwels, L., and Goossens, A. (2015). Change of a conserved amino acid in the MYC2 and MYC3 transcription factors leads to release of JAZ repression and increased activity. New Phytol. 206, 1229–1237. doi: 10.1111/nph.13398
Guo, H., Nolan, T. M., Song, G., Liu, S., Xie, Z., Chen, J., et al. (2018). FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr. Biol. 28, 3316–3324.e6. doi: 10.1016/j.cub.2018.07.078.
He, Y., Chen, L., Zhou, Y., Mawhinney, T. P., Chen, B., Kang, B. H., et al. (2011). Functional characterization of Arabidopsis thaliana isopropylmalate dehydrogenases reveals their important roles in gametophyte development. New Phytol. 189, 160–175. doi: 10.1111/j.1469-8137.2010.03460.x
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Hong, G. J., Xue, X. Y., Mao, Y. B., Wang, L. J., and Chen, X. Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24, 2635–2648. doi: 10.1105/tpc.112.098749
Hsieh, H. L., and Okamoto, H. (2014). Molecular interaction of jasmonate and phytochrome a signalling. J. Exp. Bot. 65, 2847–2857. doi: 10.1093/jxb/eru230
Hu, C., Wei, C., Ma, Q., Dong, H., Shi, K., Zhou, Y., et al. (2021). Ethylene response factors 15 and 16 trigger jasmonate biosynthesis in tomato during herbivore resistance. Plant Physiol. 185, 1182–1197. doi: 10.1093/PLPHYS/KIAA089
Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J., and Yu, D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. doi: 10.1093/jxb/erx004
Hu, Y., Sun, H., Han, Z., Wang, S., Wang, T., Li, Q., et al. (2022). ERF4 affects fruit ripening by acting as a JAZ interactor between ethylene and jasmonic acid hormone signaling pathways. Hortic. Plant J. doi: 10.1016/j.hpj.2022.01.002
Huang, C. F., Yu, C. P., Wu, Y. H., Lu, M. Y. J., Tu, S. L., Wu, S. H., et al. (2017). Elevated auxin biosynthesis and transport underlie high vein density in C4 leaves. Proc. Natl. Acad. Sci. U S A. 114, E6884–E6891. doi: 10.1073/pnas.1709171114
Jiang, S., Yao, J., Ma, K. W., Zhou, H., Song, J., He, S. Y., et al. (2013). Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog 9:e1003715. doi: 10.1371/journal.ppat.1003715
Kashyap, P., and Deswal, R. (2019). Two ICE isoforms showing differential transcriptional regulation by cold and hormones participate in Brassica juncea cold stress signaling. Gene 695, 32–41. doi: 10.1016/j.gene.2019.02.005
Kazan, K. (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20, 219–229. doi: 10.1016/j.tplants.2015.02.001
Kazan, K., and Manners, J. M. (2013). MYC2: the master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Knill, T., Schuster, J., Reichelt, M., Gershenzon, J., and Binder, S. (2008). Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. Plant Physiol. 146, 1028–1039. doi: 10.1104/pp.107.111609
Koprivova, A., Schuck, S., Jacoby, R. P., Klinkhammer, I., Welter, B., Leson, L., et al. (2019). Root-specific camalexin biosynthesis controls the plant growth-promoting effects of multiple bacterial strains. Proc. Natl. Acad. Sci. U S A. 116, 15735–15744. doi: 10.1073/pnas.1818604116
Laurie-Berry, N., Joardar, V., Street, I. H., and Kunkel, B. N. (2006). The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol. Plant-Microbe Interact. 19, 789–800. doi: 10.1094/MPMI-19-0789
Lenka, S. K., Ezekiel Nims, N., Vongpaseuth, K., Boshar, R. A., Roberts, S. C., and Walker, E. L. (2015). Jasmonate-responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1, TcJAMYC2, and TcJAMYC4. Front. Plant Sci. 6:115. doi: 10.3389/fpls.2015.00115
Li, L., Hao, X., Liu, H., Wang, W., Fu, X., Ma, Y., et al. (2019). Jasmonic acid-responsive AabHLH1 positively regulates artemisinin biosynthesis in Artemisia annua. Biotechnol. Appl. Biochem. 66, 369–375. doi: 10.1002/bab.1733
Li, T., Jiang, Z., Zhang, L., Tan, D., Wei, Y., Yuan, H., et al. (2016). Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 88, 735–748. doi: 10.1111/tpj.13289
Li, T., Xu, Y., Zhang, L., Ji, Y., Tan, D., Yuan, H., et al. (2017). The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29, 1316–1334. doi: 10.1105/tpc.17.00349
Li, Y., Yang, X., and Li, X. (2019). Role of jasmonate signaling pathway in resistance to dehydration stress in Arabidopsis. Acta Physiol. Plant. 41:100. doi: 10.1007/s11738-019-2897-2897
Lian, T. F., Xu, Y. P., Li, L. F., and Su, X. D. (2017). Crystal structure of tetrameric Arabidopsis MYC2 reveals the mechanism of enhanced interaction with DNA. Cell Rep. 19, 1334–1342. doi: 10.1016/j.celrep.2017.04.057
Liu, L., Zhang, J., Xu, J., Li, Y., Guo, L., Wang, Z., et al. (2020). CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 301:110683. doi: 10.1016/j.plantsci.2020.110683
Liu, Y., Du, M., Deng, L., Shen, J., Fang, M., Chen, Q., et al. (2019). Myc2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop[open]. Plant Cell 31, 106–127. doi: 10.1105/tpc.18.00405
Lorenzo, O., Chico, J. M., Sánchez-Serrano, J. J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. doi: 10.1105/tpc.022319
Ma, H., Duan, J., Ke, J., He, Y., Gu, X., Xu, T. H., et al. (2017). A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci. Adv. 3:e1601217. doi: 10.1126/sciadv.1601217
Ma, Y. N., Xu, D. B., Li, L., Zhang, F., Fu, X. Q., Shen, Q., et al. (2018). Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua. Sci. Adv. 4:eaas9357. doi: 10.1126/sciadv.aas9357
Ma, Y. N., Xu, D. B., Yan, X., Wu, Z. K., Kayani, S. I., Shen, Q., et al. (2021). Jasmonate- and abscisic acid-activated AaGSW1-AaTCP15/AaORA transcriptional cascade promotes artemisinin biosynthesis in Artemisia annua. Plant Biotechnol. J. 19, 1412–1428. doi: 10.1111/pbi.13561
Mahmudul Huque, A. K. M., So, W., Noh, M., You, M. K., and Shin, J. S. (2021). Overexpression of atbbd1, Arabidopsis bifunctional nuclease, confers drought tolerance by enhancing the expression of regulatory genes in aba-mediated drought stress signaling. Int. J. Mol. Sci. 22:2936. doi: 10.3390/ijms22062936
Major, I. T., Yoshida, Y., Campos, M. L., Kapali, G., Xin, X. F., Sugimoto, K., et al. (2017). Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 215, 1533–1547. doi: 10.1111/nph.14638
Mao, Y. B., Liu, Y. Q., Chen, D. Y., Chen, F. Y., Fang, X., Hong, G. J., et al. (2017). Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 8:13925. doi: 10.1038/ncomms13925
Mikkelsen, M. D., Petersen, B. L., Glawischnig, E., Jensen, A. B., Andreasson, E., and Halkier, B. A. (2003). Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 131, 298–308. doi: 10.1104/pp.011015
Mucha, S., Heinzlmeir, S., Kriechbaumer, V., Strickland, B., Kirchhelle, C., Choudhary, M., et al. (2019). The formation of a camalexin biosynthetic metabolon. Plant Cell 31, 2697–2710. doi: 10.1105/tpc.19.00403
Olsson, M. E., Olofsson, L. M., Lindahl, A. L., Lundgren, A., Brodelius, M., and Brodelius, P. E. (2009). Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 70, 1123–1128. doi: 10.1016/j.phytochem.2009.07.009
Ortigosa, A., Fonseca, S., Franco-Zorrilla, J. M., Fernández-Calvo, P., Zander, M., Lewsey, M. G., et al. (2020). The JA-pathway MYC transcription factors regulate photomorphogenic responses by targeting HY5 gene expression. Plant J. 102, 138–152. doi: 10.1111/tpj.14618
Patra, B., Pattanaik, S., Schluttenhofer, C., and Yuan, L. (2018). A network of jasmonate-responsive bHLH factors modulate monoterpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 217, 1566–1581. doi: 10.1111/nph.14910
Pauwels, L., Barbero, G. F., Geerinck, J., Tilleman, S., Grunewald, W., Pérez, A. C., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. doi: 10.1038/nature08854
Peñuelas, M., Monte, I., Schweizer, F., Vallat, A., Reymond, P., García-Casado, G., et al. (2019). Jasmonate-related MYC transcription factors are functionally conserved in marchantia polymorpha. Plant Cell 31, 2491–2509. doi: 10.1105/tpc.18.00974
Pérez-Alonso, M. M., Sánchez-Parra, B., Ortiz-García, P., Santamaría, M. E., Díaz, I., and Pollmann, S. (2021). Jasmonic acid-dependent myc transcription factors bind to a tandem g-box motif in the yucca8 and yucca9 promoters to regulate biotic stress responses. Int. J. Mol. Sci. 22:9768. doi: 10.3390/ijms22189768
Piotrowski, M., Schemenewitz, A., Lopukhina, A., Müller, A., Janowitz, T., Weiler, E. W., et al. (2004). Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279, 50717–50725. doi: 10.1074/jbc.M407681200
Pires, N., and Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. doi: 10.1093/molbev/msp288
Plett, J. M., Daguerre, Y., Wittulsky, S., Vayssierr̀es, A., Deveau, A., Melton, S. J., et al. (2014). Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. U S A. 111, 8299–8304. doi: 10.1073/pnas.1322671111
Qi, T., Huang, H., Song, S., and Xie, D. (2015a). Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 27, 1620–1633. doi: 10.1105/tpc.15.00116
Qi, T., Wang, J., Huang, H., Liu, B., Gao, H., Liu, Y., et al. (2015b). Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27, 1634–1649. doi: 10.1105/tpc.15.00110
Sabagh, A. E. L., Islam, M. S., Hossain, A., Battaglia, M., Ahmed, S., Rehman, A., et al. (2022). Phytohormones as growth regulators during abiotic stress tolerance in plants. Front. Plant Sci. 4:765068. doi: 10.3389/fagro.2022.765068
Santner, A., and Estelle, M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459, 1071–1078. doi: 10.1038/nature08122
Sarwat, M., and Tuteja, N. (2017). Hormonal signaling to control stomatal movement during drought stress. Plant Gene 11, 143–153. doi: 10.1016/j.plgene.2017.07.007
Sasaki-Sekimoto, Y., Jikumaru, Y., Obayashi, T., Saito, H., Masuda, S., Kamiya, Y., et al. (2013). Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol. 163, 291–304. doi: 10.1104/pp.113.220129
Savchenko, T., Kolla, V. A., Wang, C. Q., Nasafi, Z., Hicks, D. R., Phadungchob, B., et al. (2014). Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 164, 1151–1160. doi: 10.4161/psb.28304
Schweizer, F., Fernández-Calvo, P., Zander, M., Diez-Diaz, M., Fonseca, S., Glauser, G., et al. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2,MYC3, andMYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25, 3117–3132. doi: 10.1105/tpc.113.115139
Shen, Q., Huang, H., Zhao, Y., Xie, L., He, Q., Zhong, Y., et al. (2019). The transcription factor Aabzip9 positively regulates the biosynthesis of artemisinin in Artemisia annua. Front. Plant Sci. 10:1294. doi: 10.3389/fpls.2019.01294
Shin, J., Heidrich, K., Sanchez-Villarreal, A., Parker, J. E., and Davis, S. J. (2012). TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24, 2470–2482. doi: 10.1105/tpc.111.095430
Shoji, T., and Hashimoto, T. (2011). Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-Locus ERF genes. Plant Cell Physiol. 52, 1117–1130. doi: 10.1093/pcp/pcr063
Song, S., Huang, H., Gao, H., Wang, J., Wu, D., Liu, X., et al. (2014). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26, 263–279. doi: 10.1105/tpc.113.120394
Song, S., Qi, T., Fan, M., Zhang, X., Gao, H., Huang, H., et al. (2013). The bHLH Subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 9:e1003653. doi: 10.1371/journal.pgen.1003653
Sui, X., Singh, S. K., Patra, B., Schluttenhofer, C., Guo, W., Pattanaik, S., et al. (2018). Cross-family transcription factor interaction between MYC2 and GBFs modulates terpenoid indole alkaloid biosynthesis. J. Exp. Bot. 69, 4267–4281. doi: 10.1093/jxb/ery229
Suza, W. P., and Staswick, P. E. (2008). The role of JAR1 in Jasmonoyl-l-isoleucine production during Arabidopsis wound response. Planta 227, 1221–1232. doi: 10.1007/s00425-008-0694-694
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Toledo-Ortiz, G., Huq, E., and Quail, P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
Van der Fits, L., and Memelink, J. (2001). The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 25, 43–53. doi: 10.1046/j.1365-313X.2001.00932.x
van Moerkercke, A., Duncan, O., Zander, M., Simura, J., Broda, M., Bossche, R., et al. (2019). A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. U S A. 116, 23345–23356. doi: 10.1073/pnas.1911758116
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wager, A., and Browse, J. (2012). Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front. Plant Sci. 3:41. doi: 10.3389/fpls.2012.00041
Wang, H., Li, Y., Pan, J., Lou, D., Hu, Y., and Yu, D. (2017). The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in Arabidopsis. Mol. Plant 10, 1461–1464. doi: 10.1016/j.molp.2017.08.007
Wang, H., Li, S., Li, Y., Xu, Y., Wang, Y., Zhang, R., et al. (2019). MED25 connects enhancer-promoter looping and MYC2-dependent activation of jasmonate signalling. Nat. Plants 5, 616–625. doi: 10.1038/s41477-019-0441-449
Wang, L., Tang, W., Hu, Y., Zhang, Y., Sun, J., Guo, X., et al. (2019). A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 99, 359–378. doi: 10.1111/tpj.14330
Wang, Y., Xu, H., Liu, W., Wang, N., Qu, C., Jiang, S., et al. (2019). Methyl jasmonate enhances apple’ cold tolerance through the JAZ-MYC2 pathway. Plant Cell. Tissue Organ Cult. 136, 75–84. doi: 10.1007/s11240-018-1493-1497
Wasternack, C. (2019). Termination in jasmonate signaling by MYC2 and MTBs. Trends Plant Sci. 24, 667–669. doi: 10.1016/j.tplants.2019.06.001
Wasternack, C., and Xie, D. (2010). The genuine ligand of a jasmonic acid receptor: improved analysis of jasmonates is now required. Plant Signal. Behav. 5, 337–340. doi: 10.4161/psb.5.4.11574
Wu, F., Deng, L., Zhai, Q., Zhao, J., Chen, Q., and Li, C. (2020). Mediator subunit MED25 couples alternative splicing of JAZ genes with fine-tuning of jasmonate signaling. Plant Cell 32, 429–448. doi: 10.1105/tpc.19.00583
Xu, W., Grain, D., Le Gourrierec, J., Harscoët, E., Berger, A., Jauvion, V., et al. (2013). Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 198, 59–70. doi: 10.1111/nph.12142
Yang, J., Duan, G., Li, C., Liu, L., Han, G., Zhang, Y., et al. (2019). The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 10:1349. doi: 10.3389/fpls.2019.01349
You, Y., Zhai, Q., An, C., and Li, C. (2019). Leunig_homolog mediates MYC2-dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25. Plant Cell 31, 2187–2205. doi: 10.1105/TPC.19.00115
Yu, C., Luo, X., Zhang, C., Xu, X., Huang, J., Chen, Y., et al. (2020). Tissue-specific study across the stem of Taxus media identifies a phloem-specific TmMYB3 involved in the transcriptional regulation of paclitaxel biosynthesis. Plant J. 103, 95–110. doi: 10.1111/tpj.14710
Yu, Z. X., Li, J. X., Yang, C. Q., Hu, W. L., Wang, L. J., and Chen, X. Y. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5, 353–365. doi: 10.1093/mp/ssr087
Zander, M., Lewsey, M. G., Clark, N. M., Yin, L., Bartlett, A., Saldierna Guzmán, J. P., et al. (2020). Integrated multi-omics framework of the plant response to jasmonic acid. Nat. Plants 6, 290–302. doi: 10.1038/s41477-020-0605-607
Zhai, Q., and Li, C. (2019). The plant Mediator complex and its role in jasmonate signaling. J. Exp. Bot. 70, 3415–3424. doi: 10.1093/jxb/erz233
Zhai, Q., Deng, L., and Li, C. (2020). Mediator subunit MED25: at the nexus of jasmonate signaling. Curr. Opin. Plant Biol. 57, 78–86. doi: 10.1016/j.pbi.2020.06.006
Zhang, C., Lei, Y., Lu, C., Wang, L., and Wu, J. (2020). MYC2, MYC3, and MYC4 function additively in wounding-induced jasmonic acid biosynthesis and catabolism. J. Integr. Plant Biol. 62, 1159–1175. doi: 10.1111/jipb.12902
Zhang, F., Ke, J., Zhang, L., Chen, R., Sugimoto, K., Howe, G. A., et al. (2017). Structural insights into alternative splicing-mediated desensitization of jasmonate signaling. Proc. Natl. Acad. Sci. U S A. 114, 1720–1725. doi: 10.1073/pnas.1616938114
Zhang, F., Yao, J., Ke, J., Zhang, L., Lam, V. Q., Xin, X. F., et al. (2015). Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273. doi: 10.1038/nature14661
Zhang, H. B., Bokowiec, M. T., Rushton, P. J., Han, S. C., and Timko, M. P. (2012). Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol. Plant 5, 73–84. doi: 10.1093/mp/ssr056
Zhang, H., Hedhili, S., Montiel, G., Zhang, Y., Chatel, G., Pré, M., et al. (2011). The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 67, 61–71. doi: 10.1111/j.1365-313X.2011.04575.x
Zhang, H., Zhang, Q., Zhai, H., Gao, S., Yang, L., Wang, Z., et al. (2020). IbBBX24 promotes the jasmonic acid pathway and enhances fusarium wilt resistance in Sweet Potato. Plant Cell 32, 1102–1123. doi: 10.1105/TPC.19.00641
Zhang, M., Jin, X., Chen, Y., Wei, M., Liao, W., Zhao, S., et al. (2018b). TcMYC2a, a basic helix-loop-helix transcription factor, transduces JA-signals and regulates taxol biosynthesis in taxus chinensis. Front. Plant Sci. 9:863. doi: 10.3389/fpls.2018.00863
Zhang, M., Chen, Y., Nie, L., Jin, X., Liao, W., Zhao, S., et al. (2018a). Transcriptome-wide identification and screening of WRKY factors involved in the regulation of taxol biosynthesis in Taxus chinensis. Sci. Rep. 8:5197. doi: 10.1038/s41598-018-23558-23551
Zhang, M., Li, S., Nie, L., Chen, Q., Xu, X., Yu, L., et al. (2015). Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis. Plant Mol. Biol. 89, 463–473. doi: 10.1007/s11103-015-0382-382
Zhang, X., Zhu, Z., An, F., Hao, D., Li, P., Song, J., et al. (2014). Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 26, 1105–1117. doi: 10.1105/tpc.113.122002
Zhang, Y., Ji, T. T., Li, T. T., Tian, Y. Y., Wang, L. F., and Liu, W. C. (2020). Jasmonic acid promotes leaf senescence through MYC2-mediated repression of CATALASE2 expression in Arabidopsis. Plant Sci. 299:110604. doi: 10.1016/j.plantsci.2020.110604
Zhang, Z., Xu, M., and Guo, Y. (2020). Ring/U-Box protein AtUSR1 functions in promoting leaf senescence through JA signaling pathway in Arabidopsis. Front. Plant Sci. 11:608589. doi: 10.3389/fpls.2020.608589
Zheng, Y., Lan, Y., Shi, T., and Zhu, Z. (2017). Diverse contributions of MYC2 and EIN3 in the regulation of Arabidopsis jasmonate-responsive gene expression. Plant Direct 1:e00015. doi: 10.1002/pld3.15
Zhou, Y., Sun, W., Chen, J., Tan, H., Xiao, Y., Li, Q., et al. (2016). SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 6:22852. doi: 10.1038/srep22852
Zhu, J.-K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Zhu, X., Chen, J., Xie, Z., Gao, J., Ren, G., Gao, S., et al. (2015). Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 84, 597–610. doi: 10.1111/tpj.13030
Zhu, Y., Meng, C., Zhu, L., Li, D., Jin, Q., Song, C., et al. (2017). Cloning and characterization of DoMYC2 from Dendrobium officinale. Plant Cell. Tissue Organ Cult. 129, 533–541. doi: 10.1007/s11240-017-1198-1193
Keywords: JA signaling, stress tolerance, transcriptional regulation, crosstalk, secondary metabolism
Citation: Song C, Cao Y, Dai J, Li G, Manzoor MA, Chen C and Deng H (2022) The Multifaceted Roles of MYC2 in Plants: Toward Transcriptional Reprogramming and Stress Tolerance by Jasmonate Signaling. Front. Plant Sci. 13:868874. doi: 10.3389/fpls.2022.868874
Received: 03 February 2022; Accepted: 17 March 2022;
Published: 25 April 2022.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Ali Raza, Fujian Agriculture and Forestry University, ChinaCopyright © 2022 Song, Cao, Dai, Li, Manzoor, Chen and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Deng, ZGh1cEBxcS5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.