- 1Plant Molecular Biology Laboratory, Department of Botany, Sri Krishnadevaraya University, Anantapur, India
- 2Department of Biotechnology, St. Josephs College (Autonomous), Bengaluru, India
Cluster bean (Cyamopsis tetragonoloba L.) is one of the multipurpose underexplored crops grown as green vegetable and for gum production in dryland areas. Cluster bean is known as relatively tolerant to drought and salinity stress. To elucidate the molecular mechanisms involved in the drought tolerance of cluster bean cultivar RGC-1025, RNA sequencing (RNA-seq) of the drought-stressed and control samples was performed. De novo assembly of the reads resulted in 66,838 transcripts involving 203 pathways. Among these transcripts, differentially expressed gene (DEG) analysis resulted in some of the drought-responsive genes expressing alpha dioxygenase 2, low temperature-induced 65 kDa protein (LDI65), putative vacuolar amino acid transporter, and late embryogenesis abundant protein (LEA 3). The analysis also reported drought-responsive transcription factors (TFs), such as NAC, WRKY, GRAS, and MYB families. The relative expression of genes by qRT-PCR revealed consistency with the DEG analysis. Key genes involved in the wax biosynthesis pathway were mapped using the DEG data analysis. These results were positively correlated with epicuticular wax content and the wax depositions on the leaf surfaces, as evidenced by scanning electron microscope (SEM) image analysis. Further, these findings support the fact that enhanced wax deposits on the leaf surface had played a crucial role in combating the drought stress in cluster beans under drought stress conditions. In addition, this study provided a set of unknown genes and TFs that could be a source of engineering tolerance against drought stress in cluster beans.
Introduction
Cyamopsis tetragonoloba (L.) Taub. (Cluster bean) is a drought-adapted annual legume crop with lower water requirements than many other dryland legume crops. Cluster beans can grow in marginal soils because of their high water use efficiency, deep tap rooting system, etc. In India, cluster bean is cultivated for its green vegetables, foraging cattle, green manure, and dry pods for guar gum production (Rao and Shahid, 2011; Global Agricultural Information Network [GAIN], 2014). Globally, India ranks first and produces about 80% of the world’s cluster beans, and Rajasthan is the top state, making 75% of the total production in India. Due to high prices and export demand for guar gum, the cultivation of cluster bean is gradually increased in India from the year 2010 onward, and the total area is about 5,345 ha with 615 kg per ha yield during the agricultural year 2018–2019 (DoA, Government of India, Annual Report). The guar gum produced from cluster beans is rich in galactomannan, which is 78–82% of the seed’s endosperm. Guar gum is an essential non-toxic agrochemical, mostly an export product, and a source of polysaccharide emulsifier used primarily in the food, cosmetic, and pharmaceutical sectors (Mudgil et al., 2014). In addition, it is also used in the oil and gas industry as a gelling agent and as an additive in the milling industry (Coveney et al., 2000). The co-products for guar are guar meal and guar bagasse used to produce biofuels and other value co-products (Gresta et al., 2017).
Although cluster bean is considered a highly valued crop, its productivity is lesser than other legume crops; consequently, a significant gap exists between demand and export of guar. Biotic and abiotic factors are major limiting factors for guar yield enhancement. Thus, there is a need to increase the productivity of cluster beans to meet the demand-supply gap of cluster beans and their derivatives through genetic enhancement (Kumar et al., 2020). To increase guar output, it is necessary to produce cluster bean cultivars with improved abiotic stress tolerance, particularly drought resistance, for growing in the semi-arid tropics. The genetic improvement of cluster beans for enhanced drought resistance is not achieved due to insufficient genomic resources and inadequate germplasm availability. C. tetragonoloba genome size has been estimated to be approximately 580 Mbp using flow cytometry (Tyagi et al., 2019). The present study adopted next-generation sequencing (NGS) technologies to understand the detailed information of the drought-stressed transcriptome of cluster beans. This technology enables the identification of differentially expressed genes (DEGs), the deciphering of metabolic pathways involved in drought resistance, gum biosynthesis, and the identification of DNA-based markers, all of which may open up new avenues for molecular breeding to improve cluster bean production, gum quality, and stress resistance.
For the past few decades, extensive research has been carried out on applying omics technologies to identify many candidate genes, proteins, and metabolic pathways of various crop species under different stress conditions (Panda et al., 2021; Raza et al., 2021). In recent years, transcriptome technology has become an essential tool for analyzing the molecular mechanisms of abiotic stresses in plants (Cai et al., 2019; Hasan et al., 2019). Global transcriptome profiling of drought-stressed grain legumes, such as chickpea (Garg et al., 2011; Hiremath et al., 2011; Kumar et al., 2019), groundnut (Brasileiro et al., 2015; Zhao et al., 2018), and lentil (Singh et al., 2017; Morgil et al., 2019), revealed a set of DEGs involved in various metabolic pathways under stress conditions. Following transcriptome analysis, Wu et al. (2016) have identified 22 NAC TFs from drought-tolerant and drought-sensitive genotypes of common bean. Transcriptome analysis of drought-tolerant and drought-sensitive genotypes of wheat showed significant induction or repression of genes involved in secondary metabolism, nucleic acid synthesis, protein synthesis, and transport in the tolerant genotype when compared with the sensitive genotype (Kumar et al., 2018). RNA sequencing (RNA-Seq) analysis has been employed to elucidate drought-tolerance molecular mechanisms in other crops, such as cotton (Hasan et al., 2019), buckwheat (Hou et al., 2019), and Proso millet (Zhang et al., 2019). To date, very few studies on the transcriptome analysis of C. tetragonoloba have been published. For instance, Rawal et al. (2017) reported an RNA-Seq-based transcriptome from the leaf, shoot, and flower tissues of Guar; Tanwar et al. (2017) detailed the transcriptome of leaf tissues from two leaf tissue guar varieties M-83 and RGC-1066. Al-Qurainy et al. (2019) published a transcriptome of guar, accession BWP 5595 under various treatments, such as drought, salinity, and heat stress. The present study was focused on targeted transcriptome deep sequencing of a drought-adapted cultivar RGC-1025 to characterize the genes responsible for the drought resistance. The gene information thus obtained would pave the way for using DEGs in developing strategies for drought resistance through various approaches. Moreover, transcriptome data sets could be valuable for novel gene discovery and the marker-assisted selective breeding of cluster bean species.
Materials and Methods
Screening Cluster Bean Cultivars for Drought Tolerance
Initially, four cluster bean cultivars, namely, RGC-1025, RGC-1038, RGC-1055, and RGC-1066, were screened for their drought tolerance based on various parameters, such as germination, seedling growth, biomass, relative water content (Barrs and Weatherley, 1962), cell membrane injury (Leopold et al., 1981), malondialdehyde (MDA) (Hodges et al., 1999), total chlorophylls (Arnon, 1949), and total proline content (Bates et al., 1973).
Plant Samples, Processing, and Sequencing
Seeds of cluster bean (C. tetragonoloba L.) cultivar RGC-1025 were sterilized in 0.5% (W/V) sodium hypochlorite solution for 5 min, then rinsed thoroughly, and soaked in distilled water for 30 min. Seeds were sown in earthen pots containing soil and farmyard manure in a 3:1 proportion maintained in the departmental botanical garden. After 20 days post-sowing, drought stress was induced by withholding water to one set of pots, and respective fully watered controls were maintained in another set of pots. Ten days after stress imposition, fresh leaf samples from five plants were collected, pooled, flash-frozen in liquid nitrogen, and transported immediately to the sequencing facility.
For total RNA-seq, total RNA was extracted using the Qiagen RNeasy Plant Mini Kit with DNAse treatment (Thermo Fisher Scientific, United States) as per the manufacturer’s instructions. The quality and quantity of the RNA were estimated using a NanoDrop Spectrophotometer (Thermo Fisher Scientific, United States) and Qubit Fluorometer (Thermo Fisher Scientific, United States). The integrity of the RNA samples was analyzed using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). RNA-seq libraries were prepared with Illumina-compatible NEB Next® Ultra™ II Directional RNA Library Prep Kit (New England BioLabs, MA, United States). In total, 500 ng of total RNA was taken for mRNA isolation, fragmentation, and priming. Fragmented and primed mRNA was subjected to first-strand synthesis followed by second-strand synthesis. The double-stranded cDNA was purified using JetSeq Clean Beads (Bioline Meridian Bioscience, Australia). Purified cDNA was end-repaired, adenylated, and Illumina multiplex barcode adapters were ligated as per NEBNext® Ultra™ II Directional RNA Library Prep protocol, followed by second-strand excision using USER enzyme at 37°C for 15 min. Adapter-ligated cDNA was purified using JetSeq Beads and was subjected to 10 cycles for indexing (98°C for 30 s, cycling (98°C for 10 s, 65°C for 75 s) and 65°C for 5 min) to enrich the adapter-ligated fragments. The final PCR product (sequencing library) was purified with JetSeq Beads, followed by a library-quality control check using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). A total of 7,629,816 short reads were obtained, 150 bp length paired-end (P.E.) reads and average fragment size of ∼400 bp were used, and three biological replicates were processed for sequencing analysis with Illumina HiSeq™ 4000, which was out-sourced at Genotypic Technologies, Bengaluru, India.
Read Quality Control, Adapter Removal, de novo Assembly, and Clustering
The reads were processed for quality assessment and low-quality filtering before the FastQC tool assembly. The reads were then processed by removing the adapter sequences and low-quality bases (<q30) using the Cutadapt tool. Processed reads were assembled using a graph-based approach by the rnaSPAdes program. The characteristic properties, such as N50 length, average length, maximum length, and a minimum length of the assembled contigs, were calculated. De novo transcriptome assembly of the processed reads from all the libraries was done using Bowtie2 with end-to-end parameters. In the second step of the assembly procedure, clustering of the assembled transcripts based on sequence similarity is performed using the Cluster Database at High Identity with Tolerance (CD-HIT)-EST program1 with 95% similarity between the sequences. This reduces the redundancy without excluding sequence diversity used for further transcript annotation and the DEG analysis.
Functional Annotation of Transcripts
All unigenes were annotated using the BLASTX search tool on Viridiplantae transcripts from the UniProt database containing 8,058,045 protein sequences and the NCBI non-redundant database (N.R.). The cutoff e-value was 10––5, and the minimum similarity was more significant than 40%. Gene ontology annotation was carried out using the Blast2go program and visualized using Web Gene Ontology Annotation Plot (WEGO).2
Differentially Expressed Gene Analysis and Pathway Analysis
DESeq, an R package, was used for differential expression analysis. Sequencing (variable library size/depth) bias among the samples was removed by library normalization using size factor calculation in DESeq. DESeq normalized expression values were used to calculate fold change for a given transcript. The regulation for each transcript was assigned based on log2-fold change. The transcripts that show a log2-fold change less than −1 are represented as downregulated. The values greater than one are upregulated and between −1 and 1 are termed neutrally regulated. Gene Ontology (GO) enrichment analysis and pathway analysis for DEG were done against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. KAAS server was used to analyze and characterize associated pathways. To obtain the highly significant differential expression genes, the criterion of the absolute value of reads per kilobase of transcript per million reads (RPKM) ratio > 1,000 was used.
Mining of Simple Sequence Repeats
Simple sequence repeats (SSRs) were identified using the MISA Perl script in each transcript (MIcroSAtellite identification tool).3 A simple repetition of motif length ranging from 1 to 6 bp was identified with recommended default parameters of MISA.
qRT-PCR Analysis of Gene Expression
To evaluate the gene expression pattern from the DEG analysis, the total RNA extracted, as mentioned earlier, from control and drought-stressed cluster bean variety RGC-1025 was used for the SYBR Green qRT-PCR assay. The qRT-PCR assay was performed for 16 different stress-responsive genes selected from DEG analysis. These genes include aldo-keto reductase 1 (AKR1), late embryogenesis Abundant14 (LEA14), non-specific lipid transfer protein, TFs MYB30, NAC4, scare crow-like protein1 (GRAS TF’s), BHLH, GATA, malate dehydrogenase (MDH), aquaporin, DNA helicase, nitrate reductase, proline dehydrogenase (PRODH), serine hydroxy methyltransferase (SHMT), and thaumatin, trehalose 6-phosphate phosphatase (TRE6PH) with actin and tubulin genes as an internal control. For cDNA synthesis, 1 μg of total RNA from control and drought-stressed cluster bean RGC-1025 samples was treated with a Turbo DNASE treatment kit (Thermo Fisher Scientific, United States) as per the manufacturer’s protocol to remove any DNA traces. cDNA was synthesized using Revert Aid M-MuLV Reverse Transcriptase (Thermo Fisher Scientific) as per the manufacturer’s instructions. qRT-PCR mix was comprised of 1× using Power SYBR Green Master Mix (Ambion, United States), 20 ng of cDNA, and 0.2 μM of forward and reverse primers. Supplementary Table 1 shows the primers used for the investigated genes. The RT-PCR analysis was done on Applied Biosystems Step One Real-Time PCR machine with standard cycling comprising 95°C for 30 s, 40 cycles of 95°C for 1 s, 60°C for 20 s, and a melt curve analysis. Relative quantification was studied using 2–Δ Δ CT method (Livak and Schmittgen, 2001). Each gene was analyzed in three biological samples, and three reaction replicates were performed for each biological sample.
Estimation of Epicuticular Wax and Scanning Electron Microscope Imaging of Leaf Surfaces for Wax Deposits
Leaf surface waxes exteriorly deposited were extracted and quantified by a colorimetric assay reported by Mamrutha et al. (2010). Carnauba wax was used as the standard for the wax quantification assay. The wax content is represented as μg/gm fresh weight.
An scanning electron microscope (SEM) examined the epicuticular wax crystals. The third and fourth leaves of drought-stressed and control plants were cut to 0.5 cm, mounted onto standard stubs, and coated with gold particles using a fully automated vacuum spotter smart coater (DII-29030SCTR, JOEL, United States). The surfaces of the coated samples were observed through an SEM (JOEL-JSM-IT500, Japan).
Results
Screening Cluster Bean Cultivars for Drought Tolerance
Cluster bean cultivars, namely, RGC-1025, RGC-1038, RGC-1055, and RGC-1066, were screened for drought tolerance and we found significant differences at the cultivar level in morphological and biochemical traits under stress treatments. Results revealed that among four cultivars evaluated; cultivar RGC-1025 showed a lesser decrease in seedling growth, better biomass, and relative water content; lesser extent of cell membrane injury, lesser MDA, total chlorophylls content, and significantly higher levels of osmoprotectant, proline when compared to other cultivars (Supplementary Tables), suggesting the relative tolerance of cultivar RGC-1025 over other cultivars to drought stress. Therefore, we further extended transcriptome studies to understand the molecular mechanisms conferring the drought tolerance of cultivar RGC-1025.
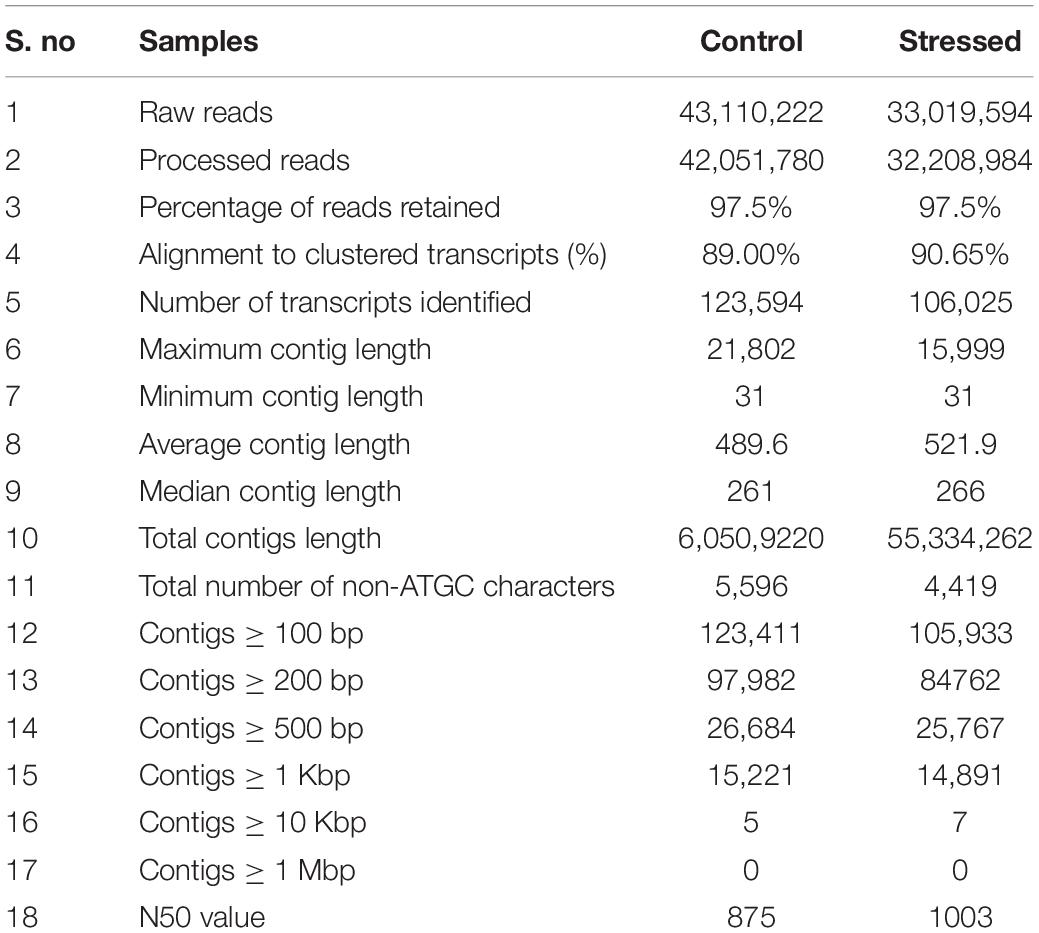
Reads and de novo Assembly
To construct the transcriptome of cluster bean cultivar RGC-1025, high-quality RNAs from three replicates of drought-stressed and unstressed conditions (control) were sequenced. An average of 18 million paired-end reads were used for the downstream analysis after pre-processing. In total, 76,129,816 short reads were obtained using Illumina HiSeq 4000 Technology (Table 1). Most of the reads had > 99.9% score. Around 97.5% of the reads were retained in both the samples post-filtering. The cleaned-up reads were assembled using the Bowtie2 tool. CD-HIT was used to cluster redundant and similar isoforms. Finally, 66,838 transcripts were clustered with an average length of 955 bp. The non-redundant transcripts were considered as unigenes and were further analyzed.
Characterization of Unigenes
All the obtained unigenes were annotated against the Uniprot Viridiplantae sequence database, NCBI non-redundant database, with a cutoff E-value of 10–5. Around 55.98% of the unigenes were found to have hits in the public databases. The transcripts with more than 30% identity were considered during the analysis. These unigenes were classified as 31 functional categories. The significant fall under the category of DNA templated transcriptional regulation (2.34%) among the biological processes, the category of integral component of the membrane (25.4%) under cellular components, and the category of adenosine 5′-triphosphate (ATP) binding (13.14%) under molecular function. The highest transcript matches during the functional annotation with members of the family Fabaceae, such as Glycine (10,131), Mucuna (5,826), Cajanus (4,533), Cicer (2,632), Vigna (2,183), Phaseolus (2,111), Arachis (1,806), Trifolium (1,597), and Medicago (1,524) (Figure 1).
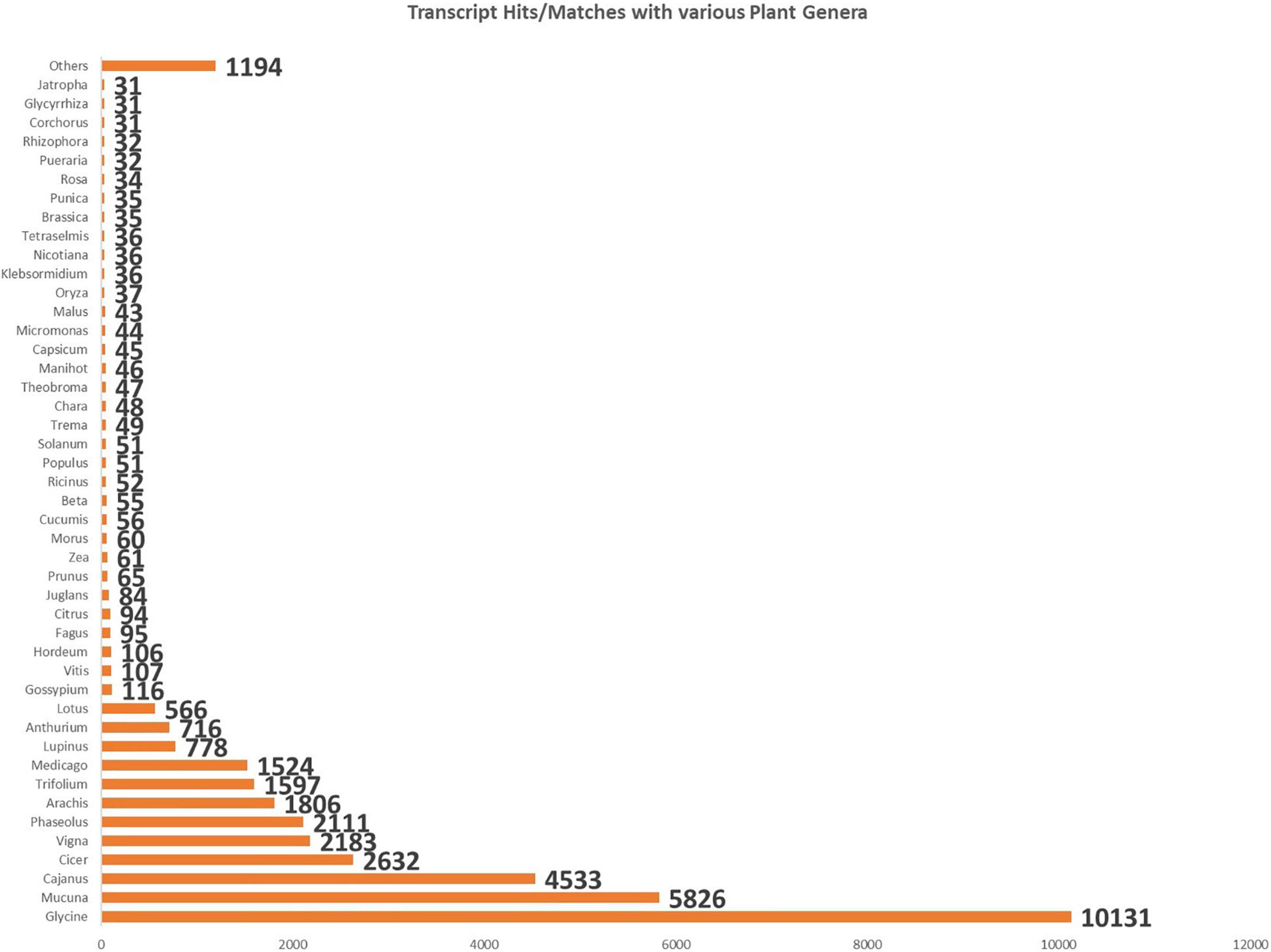
Figure 1. Bar chart shows the number of BlastX hits of cluster bean cultivar RGC-1025 with maximum hits from members of the family Fabaceae.
Functional Classification
Differentially expressed genes were subjected to GO analysis to achieve functional classification. As a result, 37,418 DEGs fall into (i) molecular function, (ii) biological process, and iii) cellular components. In total, 30,900 (50.2%) DEGs were associated with molecular function terms, such as ATP binding encoding transcripts, followed by metal ion binding transcripts, DNA binding transcripts, zinc ion binding transcripts, nucleic acid binding transcripts, protein kinase activity transcripts, etc., and 16,410 (26.6%) DEGs were annotated with cellular component terms, represented by the integral component of the membrane encoding transcripts, followed by nucleus components transcripts, cytoplasm components transcripts, ribosome transcripts, plasma membrane transcripts, retrotransposon nucleocapsid transcripts, etc., and 14,191 (23%) DEGs were associated with biological process terms. This study also observed that identical DEG sequences could exist in more than one category. The most represented “biological process” subcategories identified were DNA-templated transcriptional regulation encoding transcripts followed by translation components, carbohydrate metabolic process transcripts, DNA integration transcripts, signal transduction transcripts, intracellular protein transcripts, etc. (Figure 2 and Supplementary Tables 2–4). In general, 12,866 genes were found to be upregulated, 16,177 genes were downregulated, and 27,782 genes had shown no change in their expression levels in cluster bean cultivar RGC-1025 due to drought stress. More interestingly, 3,745 transcripts were expressed only in the stressed sample.
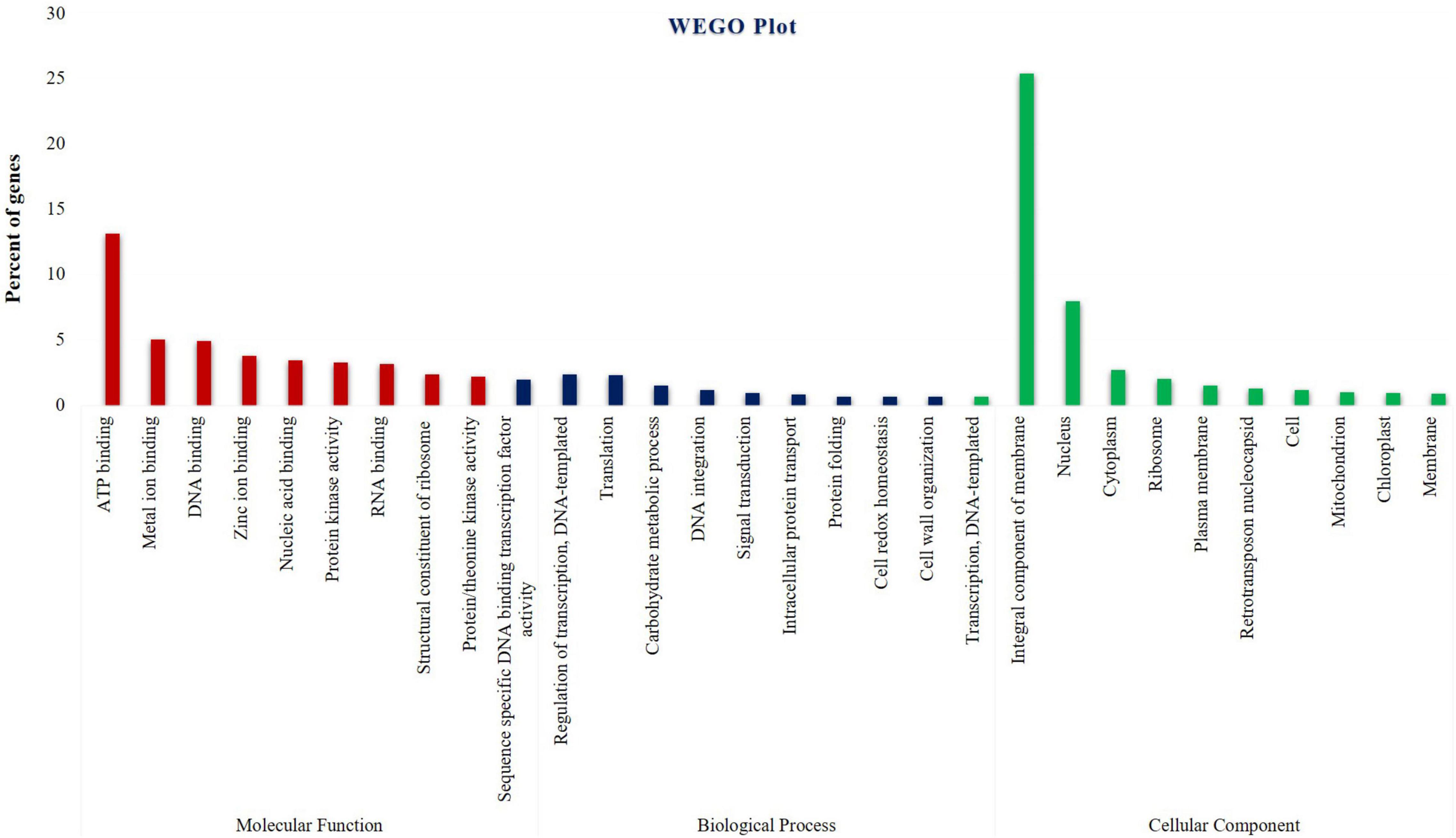
Figure 2. Pie chart shows drought-stressed cluster bean cultivar RGC-1025 Gene Ontology classification of transcripts into biological process, molecular functions, and cellular components.
Differentially Expressed Genes in Kyoto Encyclopedia of Genes and Genomes Pathways
Kyoto Encyclopedia of Genes and Genomes is an online database that deals with genomes and enzymatic pathways, and its identifiers were looked to predict biochemical pathways related to DEGs. Among 66,838 transcripts in unigene pathways, 17,211 transcripts against the KEGG pathways were identified. Of the 203 pathways identified, the top forty pathways are shown in Supplementary Table 5. DESeq analysis of the transcripts revealed that the enzymes with the most frequency of expression in the category of upregulated genes were alpha dioxygenase 2 (9.4-fold), low temperature-induced 65 kDa protein (LTI65; 9.2-fold), putative vacuolar amino acid transporter (9.05-fold), hexosyl transferase (EC 2.4.1.-; 7.95 fold), late embryogenesis abundant protein3 (LEA 3; 7.79-fold), Putative anthocyanidin 3-O-glucoside 2″-O-glucosyltransferase (EC 2.4.1.297; 7.44-fold), Glucosyltransferases, Rab-like GTPase Activators and Myotubularins (GRAM) domain protein/abscisic acid (ABA)-responsive-like protein (putative GRAM domain, P.H. domain-containing protein) (7.30-fold), and cytochrome P450 monooxygenase (EC:1.14.14.80; 7.14-fold). In the category of downregulated genes, the genes encoding the following proteins were found to be downregulated, such as putative CDP-alcohol phosphatidyl transferase class-I family protein 3 (EC 2.7.8.1; 0.5-fold), NEDD4-binding protein 2 (0.49-fold), dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex (EC 2.3.1.-; 0.49-fold), putative cyclic nucleotide-gated ion channel 14 (0.498), ATP binding cassette (ABC) transporter G family member 28 (0.49-fold), N-(5-phosphoribosyl) anthranilate isomerase (putative phosphoribosyl anthranilate isomerase) (EC 5.3.1.24; 0.49-fold), F-box/FBD/LRR-repeat protein (0.49-fold), and adenylate kinase (EC:2.7.4.3; 0.49-fold). The heat map of DEGs showed the genes expressed (Figure 3).
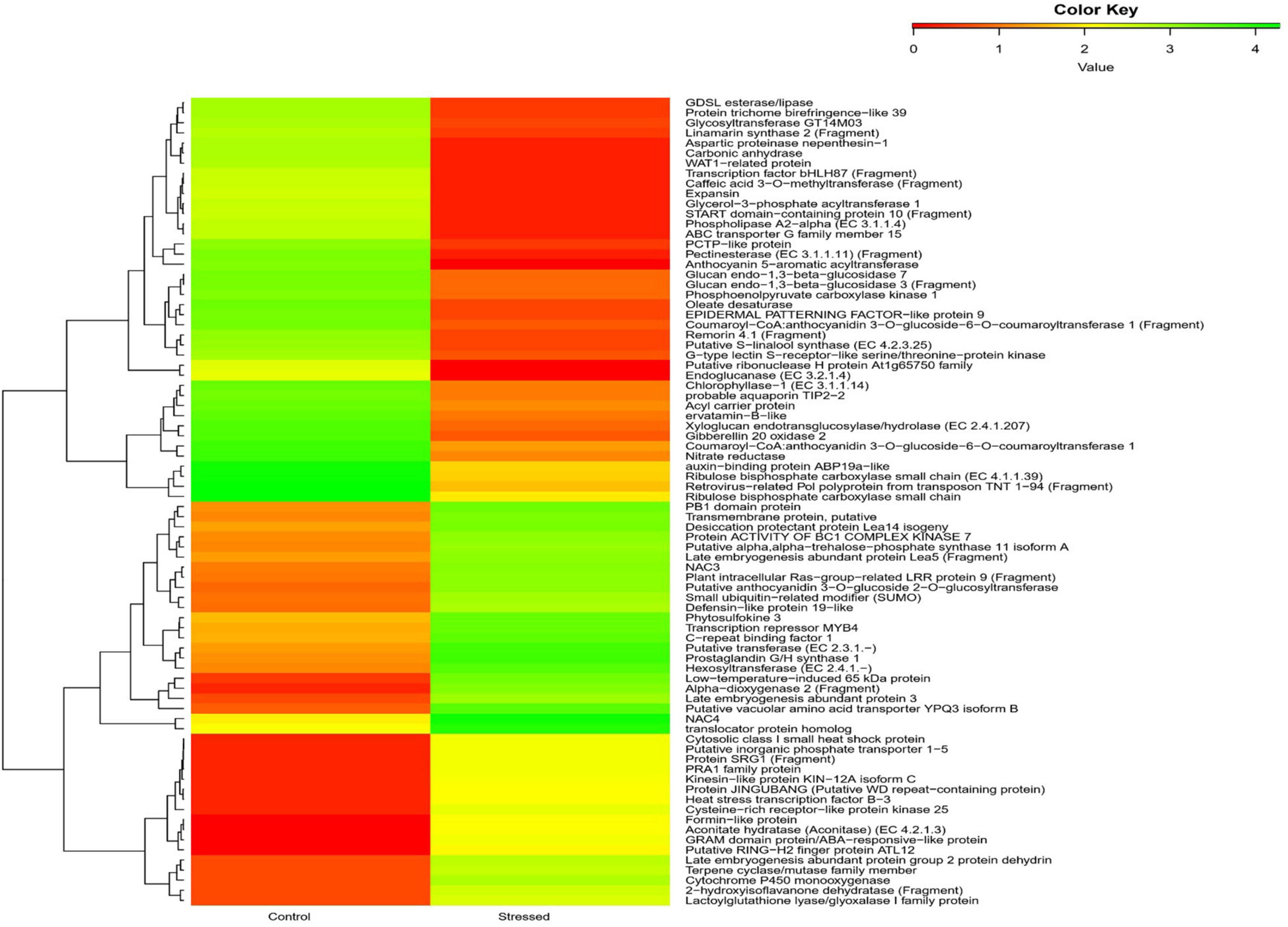
Figure 3. Heatmap shows the top 40 upregulated and downregulated genes of cluster bean cultivar RGC-1025 under control and drought stress.
Differentially Expressed Transcription Factors
Among the drought stress-responsive upregulated TF families, NAC family TF was the most abundant (26%), followed by MYB TFs (12%) and WRKY TFs (9%) (Figure 4A). Figure 4B depicts the top 40 upregulated TFs under drought treatments in cluster bean cultivar RGC-1025. Most renowned drought stress-responsive TFs upregulated include NAC4, NAC3, NAC29, NAC 104, and NAC18 from the NAC family, followed by WRKY12, WRKY50, WRKY6, WRKY33, WRKY30, WRKY24, WRKY42, WRKY53, WRKY70, and WRKY7 from WRKY family, further followed by other TFs, such as homeobox domain TFs, scarecrow/GRAS TFs, and ethylene-responsive TFs. Among the downregulated TF families, the hemophagocytic lymphohistiocytosis (HLH) TF family was the most abundant (20%), followed by the cellular TF family (10%) and homeobox domains (∼8%) (Figures 4C,D).
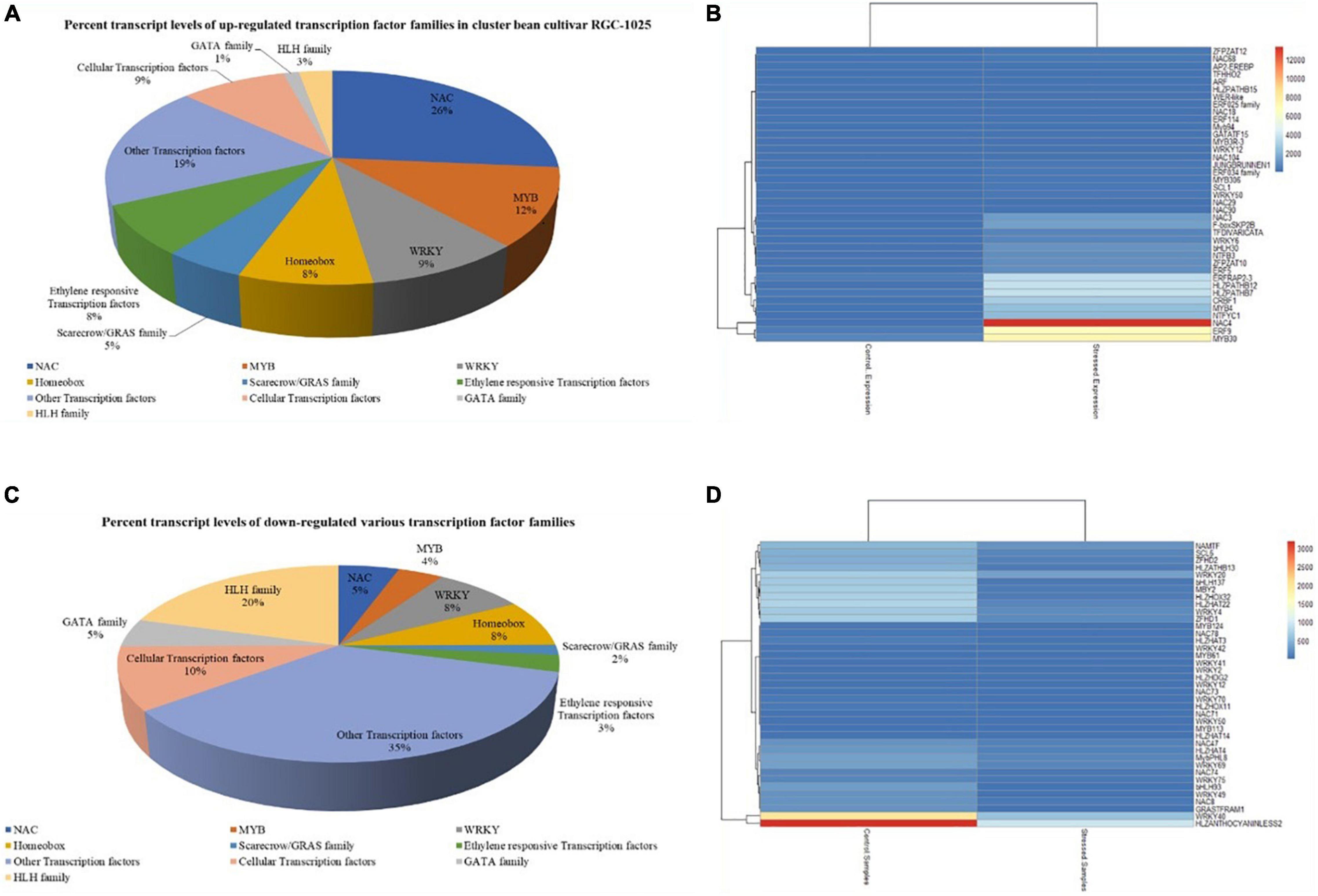
Figure 4. (A) Upregulated transcription factor families of drought-stressed cluster bean cultivar RGC-1025. (B) Heatmap of the top 40 upregulated transcription factors of cluster bean cultivar RGC-1025 in drought stress. (C) Downregulated transcription factor families of drought-stressed cluster bean cultivar RGC-1025. (D) Heatmap of the top 40 downregulated transcription factors of cluster bean RGC-1025 in drought stress.
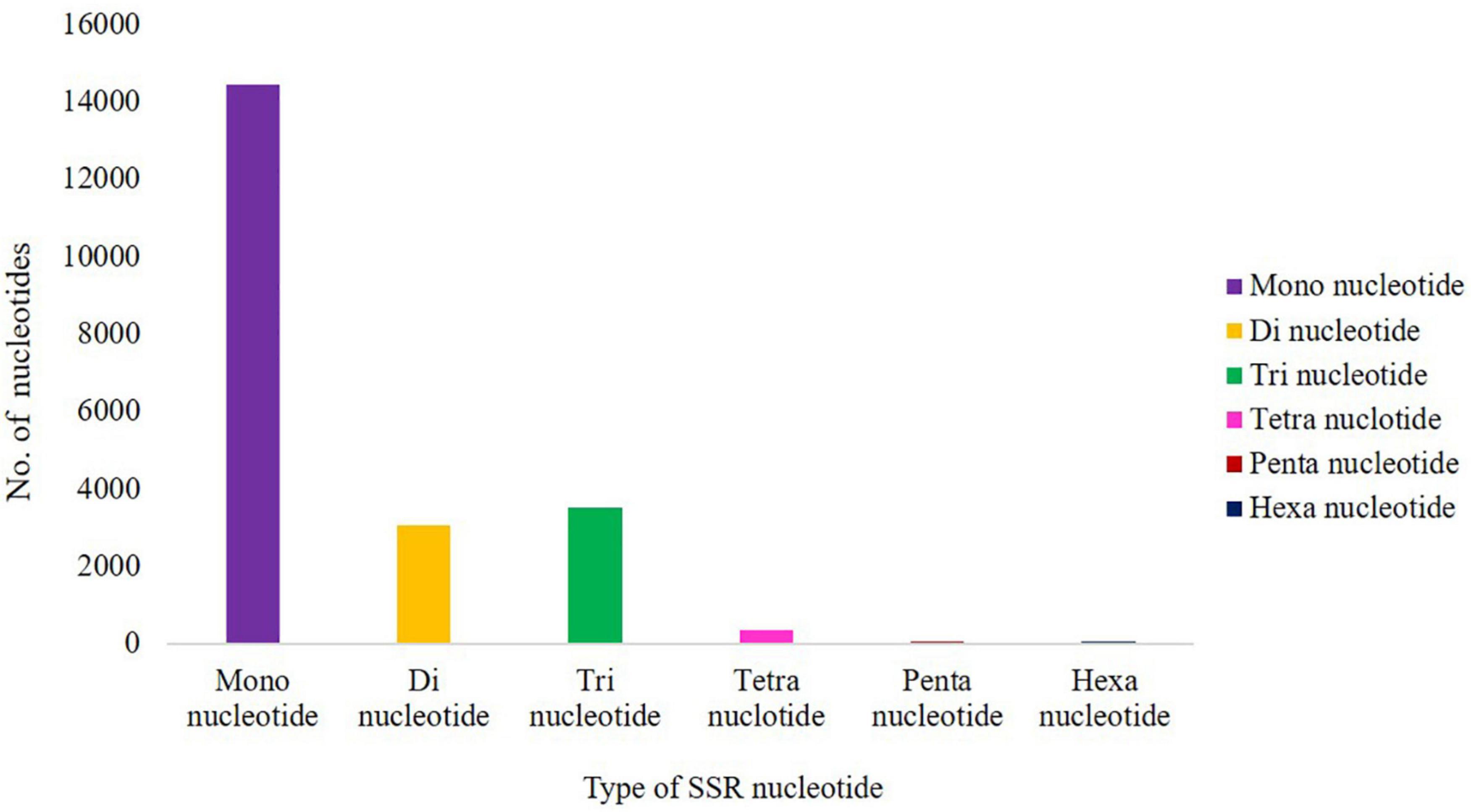
Simple Sequence Repeat Mining
A total of 21,494 SSRs were identified in the cluster bean data, 14,434 (67.15%) are mono-nucleotide repeats, 3,072 (14.29%) are di-nucleotide repeats, 3,516 (16.35%) are tri-nucleotide repeats, 341 (1.58%) tetra-nucleotide repeats, 69 (0.32%) pentanucleotide repeats, and 63 (0.25%) hexanucleotide repeats (Figure 5). The 21,494 potential SSRs identified from de novo transcriptome sequencing data represent a significant addition to the limited set of genic-SSR markers available in cluster bean cultivar RGC-1025.
Validation of Differentially Expressed Genes by Quantitative Real-Time RT-PCR
To verify the reliability of the expression profiles from RNAseq data, qRT-PCR analysis was performed for DEG (Figure 6). Results of the qRT-PCR assay for 16 stress-responsive genes revealed consistency in gene expression patterns as compared to that of the DEG analysis of the transcriptome of RGC-1025. The relative fold change of both the qRT-PCR and NGS-DEG is represented (Figure 6). Upregulation of stress-responsive TFs, such as NAC4, MYB30, scarecrow-like protein (SCL-1), primary helix-loop-helix TF (SlbHLH22), and TF bHLH 22, and the overexpression of candidate stress-responsive functional genes, such as DNA helicase, MDH, AKR1, LEA14, PDH, and SHMT, during drought stress by improving the ROS scavenging system, increasing osmotic potential, stomatal regulation, pH stability, respiration, and β-oxidation of fatty acids could support further the drought tolerance of cluster bean cultivar RGC-1025 (Ahmad et al., 2017; Waseem et al., 2019). The correlation among the expression patterns of the genes in NGS and qRT-PCR represents the consistency of the data in the current study.
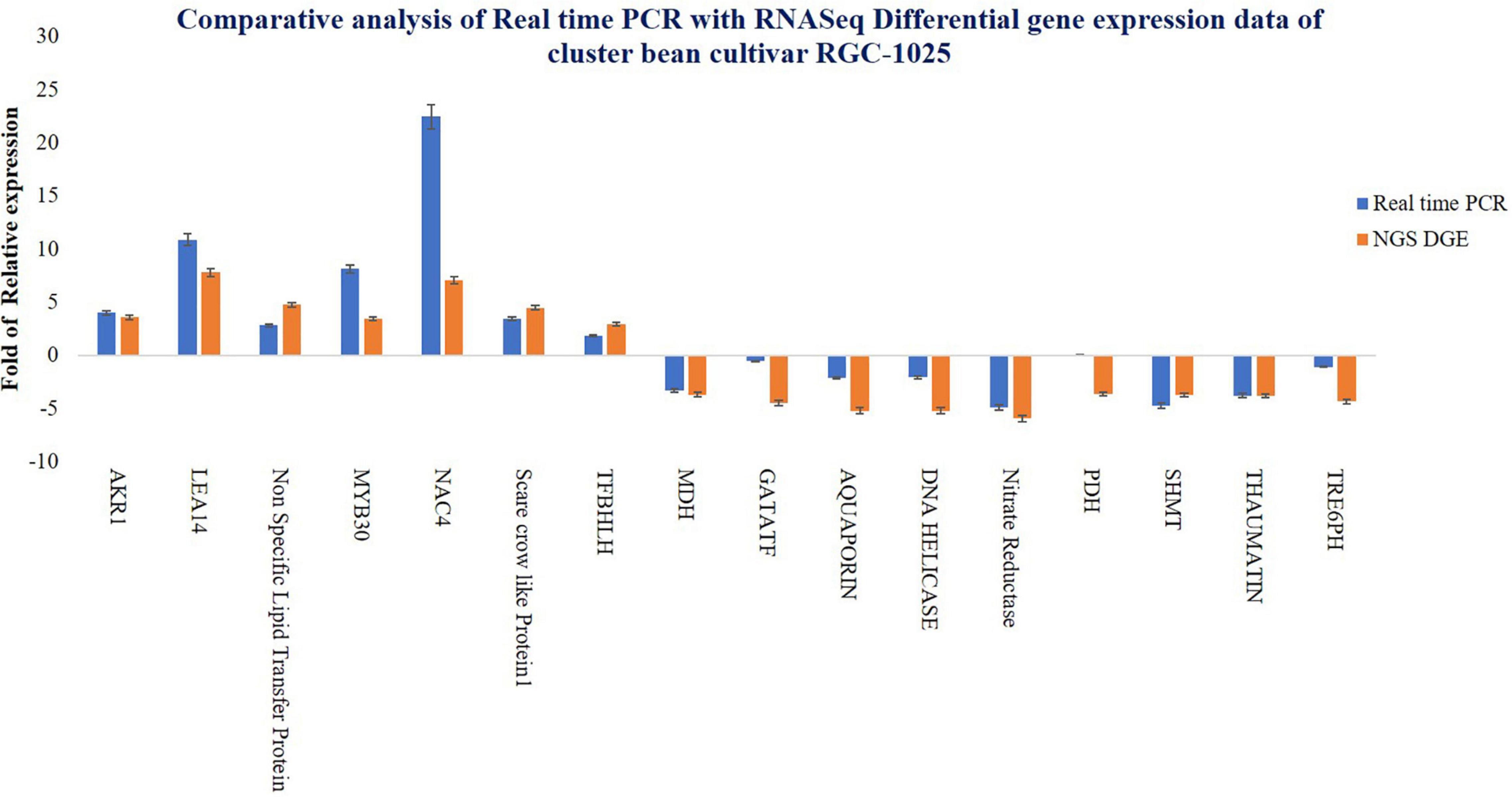
Figure 6. Comparison of real-time PCR data and RNA-Seq differential gene expression data of cluster bean cultivar RGC-1025.
Epicuticular Wax Content and Scanning Electron Microscope Imaging of Leaf Surfaces
The epicuticular wax content of the cluster bean cultivar RGC-1025 during the drought stress was 610.36 ± 0.53 μg/dm2 as compared to the control 432.41 ± 0.4 μg/dm2, which is 41.15% higher as compared to its control. These epicuticular wax data were supported by the SEM imaging of the leaf surfaces of control and drought-stressed plants. The SEM image of the leaf surface of RGC-1025 under drought stress showed enhanced wax crystal deposits (Figure 7).
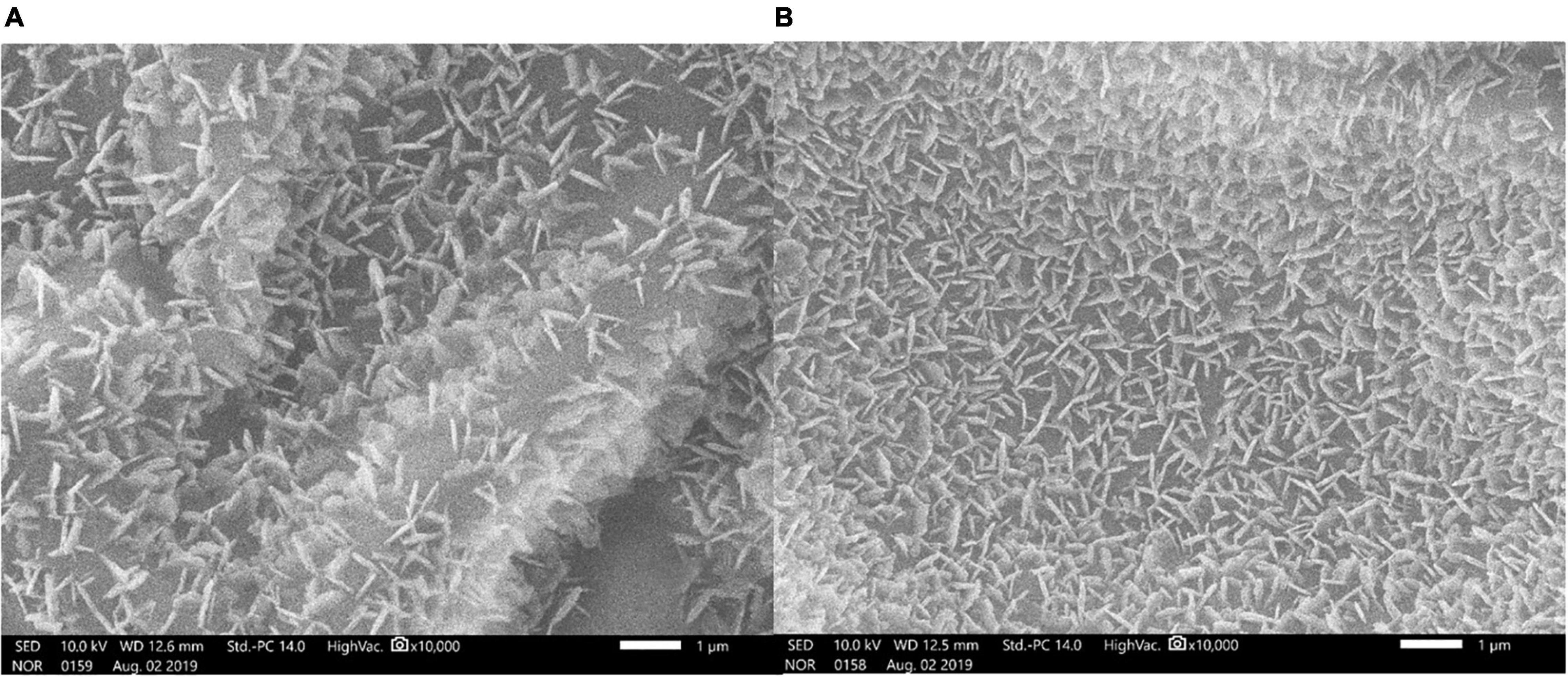
Figure 7. Scanning electron microscopy (SEM) analysis (2,000 × with 20 μm bar scale) of epicuticular wax depositions on the leaf surfaces of cluster bean cultivar RGC-1025 (A) control leaf and (B) drought-stressed leaf.
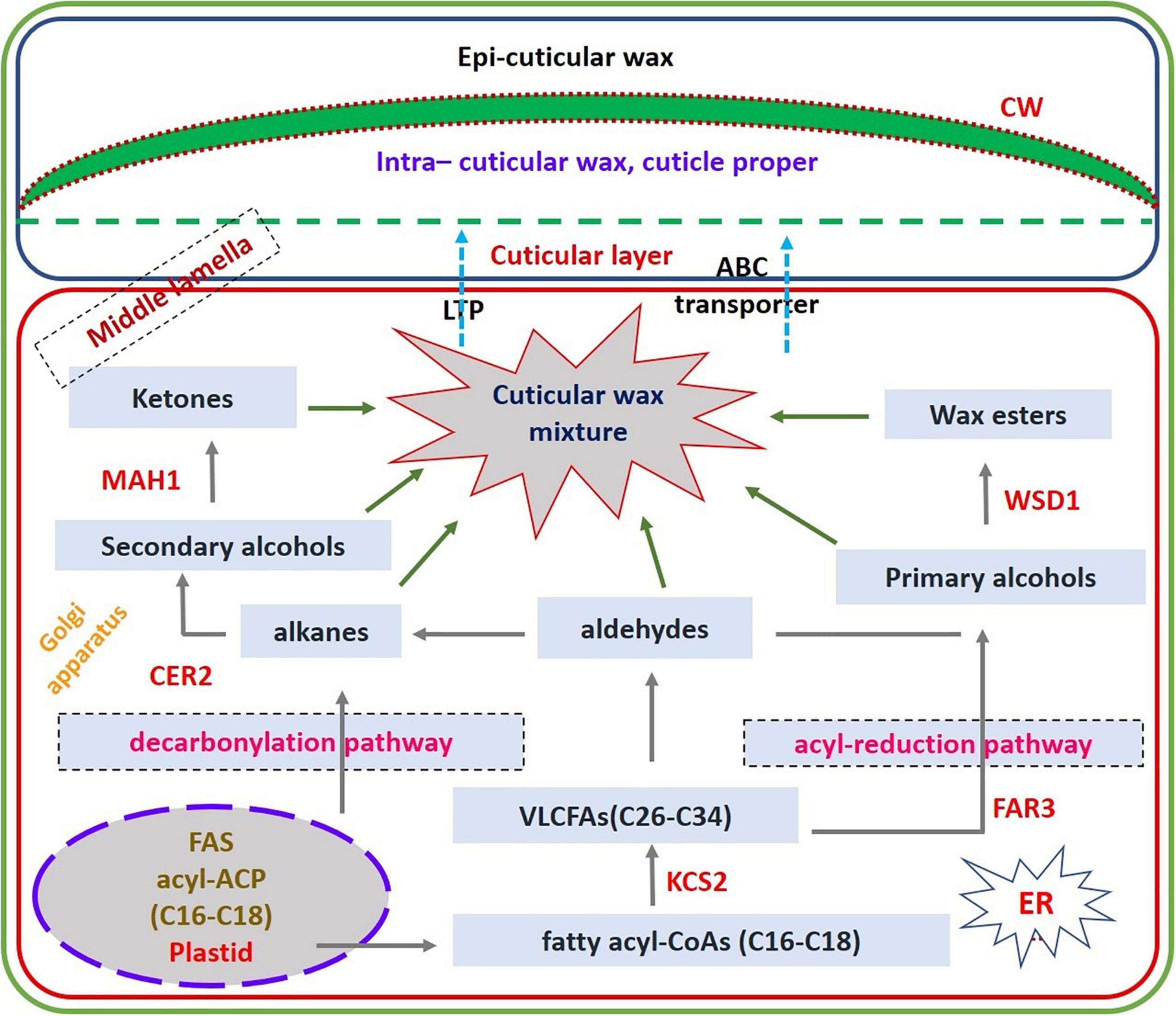
Mapping of the Wax Biosynthesis Pathway in Cluster Bean Using Differentially Expressed Gene Data
Among the genes differentially expressed in wax biosynthesis pathway, there were upregulated genes that include KCS1 that encodes β-ketoacyl-CoA synthase 1 that is involved in elongation of 24C fatty acids, WSD1 that encodes wax ester synthase/diacylglycerol acyl transferase, which involves in wax ester biosynthesis, KCR1 that encodes β-Ketoacyl-CoA reductase, which is involved in very long-chain fatty acid elongation (VLFCA elongation), FATB that encodes acyl-acyl carrier protein thioesterase, which is engaged in supply of saturated fatty acids for wax biosynthesis, CER4/FAR3 that encodes alcohol forming fatty acyl CoA reductase, which is involved in formation of C24:0 and C26:0 primary alcohols, protein WAX2 encoding gene, CER17 also called Eceriferum1, which encodes for acyl-CoA desaturase-like 4 protein that is involved in n-6 desaturation of very long-chain acyl-CoAs, ABC transporter G family member 11, which encodes ABC transporter proteins that is involved in secretion of surface waxes in interaction with CER5, which is an another ABC transporter protein, and lipid transfer protein gene that encodes a lipid transport protein, which has role in cuticular wax export or accumulation. Finally, the upregulation of these wax genes in the present study through transcriptome DEG data reveals that these gene products are responsible for accumulating or producing epicuticular wax in cluster bean cultivar RGC-1025 (Figure 8). qRT-PCR analysis of selective wax genes showed significant changes in the expression patterns and an increase in the expression of the KCS1 gene was 1.85-fold, WSD1 gene was 1.81-fold, KCR1gene was 3.73-fold, FATB gene was 1.8-fold, CER4/FAR3 gene was 0.4-fold, protein WAX2 gene was 2.36-fold, CER17 gene was 2.89-fold, ABC transporter G family member11 gene was 1.95-fold, and lipid transfer protein gene was 1.49-fold.
Discussion
Cluster bean (C. tetragonoloba L.) is an annual legume crop grown in arid and semiarid regions. Due to the lack of genomic resources, presently, conventional breeding is the only means of cluster bean improvement. In this regard, the availability of genomic resources can serve as a good platform for cluster bean improvement (Naoumkina et al., 2007; Tanwar et al., 2017). Cluster bean is known as relatively tolerant to abiotic stresses. Genotypic variation in stress tolerance exists in cluster bean cultivars (Alshameri et al., 2017), implying that it is a valuable repository for genes that are resistant to these abiotic stresses, and to use this genetic tank, the present study implemented a de novo transcriptome analysis of a drought-tolerant cluster bean cultivar RGC-1025.
The RNA-Seq (NGS) method offers a holistic view of the transcriptome, revealing many novel transcribed regions, splice isoforms, genic microsatellites, and the precise location of transcription boundaries (Cloonan et al., 2008; Wang et al., 2009; Li et al., 2010; Wilhelm et al., 2010). These technologies have been widely exploited in numerous plant species to produce molecular markers using transcriptome analysis (Dutta et al., 2011; Wang et al., 2014). In the present study, Illumina HiSeq 4000 Technology generated 76,129,816 short reads from the control and drought-stressed samples of cluster bean cultivar RGC-1025.
A cluster bean is a non-model plant without prior genome knowledge; BLASTX was used to search for sequence similarity and compare the assembled unigenes of the cluster bean transcriptome against multiple databases. Around 55.98% of the unigenes were obtained and annotated against the Uniprot Viridiplantae sequence database and NCBI non-redundant database, with a cutoff E-value of 10–5. According to species distribution analyses, many plant species have sequences that are homologous to cluster bean sequences. The highest transcript matches during the functional annotation with members of the family Fabaceae, such as Glycine (10131), Mucuna (5826), and Cajanus (4533).
Gene Ontology analysis provides a set of dynamically controlled and structured vocabularies for describing the roles of genes in any organism (Ashburner et al., 2000). Based on the sequence homology, 37,418 DEGs were assigned GO terms and classified into three categories, namely, molecular function, biological process, and cellular components. The results of this study agree with those of other plant leaf transcriptome investigations (Wu et al., 2015; Bose Mazumdar and Chattopadhyay, 2016). Alpha-dioxygenase (-DOX) is engaged in the catalysis of fatty acid oxygenation, resulting in the production of a recently found category of oxylipins, which plays a crucial role in shielding tissues from oxidative damage and cell death under drought stress (Tirajoh et al., 2005). Shi et al. (2015) reported that LTI30 protein positively regulates drought stress resistance in Arabidopsis through the modulation of ABA sensitivity, hydrogen peroxide levels, and proline accumulation. Yang et al. (2015) reported from their study that putative cationic amino acid transporter 9 (CAT9) mutation resulted in chlorotic leaves and overexpression resulted in the formation of stems and inflorescence transgenic Arabidopsis plants. Magwanga et al. (2018) also established the role of LEA proteins in cotton drought stress tolerance. Zheng et al. (2020) reported that the overexpressing phenotype of Oryza sativa ABA responsive protein 1 (OsABAR1), a GRAM protein-containing protein, showed resistance to drought and salinity. Identifying many DEGs in this study could help to gain in-depth knowledge of the diverse metabolic activities involved in the stress-resistant mechanisms of cluster beans. According to the gene function analysis, the KEGG database revealed that among 66,838 transcripts, 17,211 transcripts were allocated to 203 unigene pathways. A similar pattern was discovered in the transcriptome of Phyllanthus amarus leaves (Bose Mazumdar and Chattopadhyay, 2016).
Transcription factors are regulatory proteins involved in various regulatory processes, such as biotic and abiotic stress adaptation (Nakashima et al., 2014; Joshi et al., 2016). TF genes, such as NAC, WRKY, MYB, and bZIP, have been linked to drought stress responses (Gahlaut et al., 2016). NAC genes are TFs specific to plants and are involved in growth, development, and stress responses. Shi et al. (2018) reported that GmWRKY12 confers drought and salt tolerance in soybean. Auxins usually induce scarecrow-like genes and interact with histone deacetylase, resulting in chromatin modeling in drought stress (Gao et al., 2004; Sánchez et al., 2007). Similarly, Scarecrow-like protein 1, one of the GRAS proteins, was upregulated in this study during drought stress. Zhu et al. (2014) studied the role of the SlNAC4 TF in combating drought and salinity stress through RNAi-silenced transgenic tomato plants. Yu et al. (2016) also proved that Cicer arietinum NAC4 (CarNAC4) TF overexpression in Arabidopsis conferred resistance to drought and salinity stresses. Liu et al. (2013) reported enhanced dehydration and drought tolerance through overexpression of AhNAC3 in tobacco through enhanced superoxide scavenging. Tang et al. (2017) reported that the overexpression of the peanut NAC4 gene conferred drought tolerance in tobacco. These differentially expressed TFs propose their significant role in combating drought stress in cluster bean cultivar RGC-1025. SSRs are the most useful molecular markers for genetics and plant breeding applications (Hiremath et al., 2012). In the present study, 21,494 SSRs were identified in the cluster bean data, and the frequency distribution of SSR markers agrees with previous reports in guar (Kuravadi et al., 2014; Kumar et al., 2016).
Cuticular wax prevents non-stomatal water loss, allowing plants to adapt to water-limited conditions (Kerstiens, 1996; Buda et al., 2013). Cuticular waxes deposited on the plant’s organs play a critical role in sustaining harsh environmental conditions, such as drought (Jenks and Ashworth, 1999; Goodwin and Jenks, 2005). Drought stress enhances the increased deposition of waxes in many plants (Bondada et al., 1996; Samdur et al., 2003; Cameron et al., 2006). Lee and Suh (2013) and Mamrutha et al. (2017) reported that wax biosynthesis and its pathway genes are regulated at transcriptional, post-transcriptional, and translational levels. Guo et al. (2016) showed that drought-induced accumulation of wax biosynthesis positively correlated with drought-tolerant crops, such as wheat. In the present study, the ECERIFERUM1 was upregulated by 7.82-fold during the drought stress, revealing the upregulation of the wax biosynthesis pathway. Bourdenx et al. (2011) reported that overexpression of ECERIFERUM1 promotes wax’s very long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Xu et al. (2003) showed that an ABC transporter family gene, AtTGD1, is involved in the inter-organelle lipid transfer in Arabidopsis. Mizuno et al. (2013) reported that an ABC transporter gene, Sb06g023280, is responsible for epi-cuticular wax biosynthesis in Sorghum. Elango et al. (2020) assessed the epicuticular wax variability in the extensive genetic pool of Sorghum, and a genome-wide association mapping study showed genic regions associated with epicuticular wax production. Hence, the enhanced epicuticular wax content and the deposition of wax crystals on the leaf surfaces are essential components of plants for enhanced drought tolerance to overcome non-stomatal water loss.
Conclusion
In summary, the Cluster bean cultivar RGC-1025 is proved to have enhanced drought tolerance that was evident from DEGs and analyzed from the transcriptome sequencing. The transcriptome sequencing and analysis revealed that the differential expression of the different stress responsible and constitutive cellular TFs, the enhanced traits, such as enhanced wax biosynthesis, and the upregulation of various genes involved in wax biosynthesis played a key role in RGC-1025 to combat drought stress efficiently.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, #PRJNA669348.
Author Contributions
CS conceptualized and supervised this study and wrote the manuscript. BR performed the experiments. BR, AA, and MP analyzed the transcriptome data. NJ, BV, and NJ performed RT PCR analysis. All authors equally contributed to manuscript revision, read, and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We greatly acknowledge the Regional Agricultural Research Station, Rekulakunta, for providing cluster bean seed material and Yogi Vemana University, Kadapa for extending the SEM facility. CS acknowledges the UGC, GoI, New Delhi for the BSR (F.No. 26-13/2020-BSR) Faculty fellowship.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.868142/full#supplementary-material
Footnotes
- ^ http:www.bioinformatics.org/cd-hit/
- ^ http://wego.genomics.org.cn/cgi-bin/wego/index.pl
- ^ http://pgrx.ipk-gatersleben.de/misa/
References
Ahmad, J., Bashir, H., Bagheri, R., Baig, A., Al-Huqail, A., Ibrahim, M. M., et al. (2017). Drought and salinity induced changes in ecophysiology and proteomic profile of Parthenium hysterophorus. PLoS One 12:e0185118. doi: 10.1371/journal.pone.0185118
Al-Qurainy, F., Alshameri, A., Gaafar, A. R., Khan, S., Nadeem, M., Alameri, A. A., et al. (2019). Comprehensive stress-based de novo transcriptome assembly and annotation of guar (Cyamopsis tetragonoloba (L.) Taub.): an important industrial and forage crop. Int. J. Genomics 2019, 1–14. doi: 10.1155/2019/7295859
Alshameri, A., Al-Qurainy, F., Khan, S., Nadeem, M., Gaafar, A. R., and Tarroum, M. (2017). Appraisal of guar [Cyamopsis tetragonoloba (l.) Taub.] accessions for forage purpose under the typical Saudi Arabian environmental conditions encompassing high temperature, salinity and drought. Pakistan J. Bot. 49, 1405–1413.
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology. 24:1. doi: 10.1104/pp.24.1.1
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Gene. 25, 25–29. doi: 10.1038/75556
Barrs, H. D., and Weatherley, P. E. (1962). A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aus. J. Biol. Sci. 15, 413–428. doi: 10.1071/BI9620413
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant soil 39, 205–207. doi: 10.1007/BF00018060
Bondada, B. R., Oosterhuis, D. M., Murphy, J. B., and Kim, K. S. (1996). Effect of water stress on the epicuticular wax composition and ultrastructure of cotton (Gossypium hirsutum L.) leaf, bract, and boll. Environ. Experim. Bot. 36, 61–69. doi: 10.1016/0098-8472(96)00128-1
Bose Mazumdar, A., and Chattopadhyay, S. (2016). Sequencing, de novo assembly, functional annotation and analysis of Phyllanthus amarus leaf transcriptome using the Illumina platform. Front. Plant Sci. 6:1199. doi: 10.3389/fpls.2015.01199
Bourdenx, B., Bernard, A., Domergue, F., Pascal, S., Léger, A., Roby, D., et al. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156, 29–45. doi: 10.1104/pp.111.172320
Brasileiro, A., Morgante, C. V., Araujo, A. C., Leal-Bertioli, S., Silva, A. K., Martins, A. C., et al. (2015). Transcriptome profiling of wild Arachis from water-limited environments uncovers drought tolerance candidate genes. Plant Mol. Biol. Rep. 33, 1876–1892. doi: 10.1007/s11105-015-0882-x
Buda, G. J., Barnes, W. J., Fich, E. A., Park, S., Yeats, T. H., Zhao, L., et al. (2013). An ATP binding cassette transporter is required for cuticular wax deposition and desiccation tolerance in the moss Physcomitrella patens. Plant Cell 25, 4000–4013. doi: 10.1105/tpc.113.117648
Cai, X., Magwanga, R.O., Xu, Y., Zhou, Z., Wang, X., Hou, Y., et al. (2019). Comparative transcriptome, physiological and biochemical analyses reveal response mechanism mediated by CBF4 and ICE2 in enhancing cold stress tolerance in Gossypium thurberi. AoB Plants 11:lz045. doi: 10.1093/aobpla/plz045
Cameron, K. D., Teece, M. A., and Smart, L. B. (2006). Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 140, 176–183. doi: 10.1104/pp.105.069724
Cloonan, N., Forrest, A. R., Kolle, G., Gardiner, B., Faulkner, G. J., Brown, M. K., et al. (2008). Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods 5, 613–619. doi: 10.1038/nmeth.1223
Coveney, P. V., Silva, H. D., Gomtsyan, A., Whiting, A., and Boek, E. S. (2000). Novel approaches to cross-linking high molecular weight polysaccharides: application to guar-based hydraulic fracturing fluids. Mol. Simul. 25, 265–299. doi: 10.1080/08927020008024503
Dutta, S., Kumawat, G., Singh, B. P., Gupta, D. K., Singh, S., Dogra, V., et al. (2011). Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol. 11:17. doi: 10.1186/1471-2229-11-17
Elango, D., Xue, W., and Chopra, S. (2020). Genome wide association mapping of epi-cuticular wax genes in Sorghum bicolor. Physiol. Mol. Biol. Plants 26, 1727–1737. doi: 10.1007/s12298-020-00848-5
Gahlaut, V., Jaiswal, V., Kumar, A., and Gupta, P. K. (2016). Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor. Appl. Gen. 129, 2019–2042. doi: 10.1007/s00122-016-2794-z
Gao, M. J., Parkin, I., Lydiate, D., and Hannoufa, A. (2004). An auxin-responsive SCARECROW-like transcriptional activator interacts with histone deacetylase. Plant Mol. Biol. 55, 417–431. doi: 10.1007/s11103-004-0892-9
Garg, R., Patel, R. K., Tyagi, A. K., and Jain, M. (2011). De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 18, 53–63. doi: 10.1093/dnares/dsq028
Global Agricultural Information Network [GAIN] (2014). In: An Analysis of Guar Crop in India. Report Number IN4035. Geneva :GAIN
Goodwin, S. M., and Jenks, M. A. (2005). “Plant cuticlef unction as a barrier to water loss,” in Plant Abiotic Stress, eds M. A. Jenks and P. M. Hasegawa (Oxford: Blackwell Scientific Publishers), 14–36.
Gresta, F., Ceravolo, G., Presti, V. L., D’Agata, A., Rao, R., and Chiofalo, B. (2017). Seed yield, galactomannan content and quality traits of different guar (Cyamopsis tetragonoloba L.) genotypes. Indust. Crops Prod. 107, 122–129. doi: 10.1016/j.indcrop.2017.05.037
Guo, J., Xu, W., Yu, X., Shen, H., Li, H., Cheng, D., et al. (2016). Cuticular wax accumulation is associated with drought tolerance in wheat near-isogenic lines. Front. Plant Sci. 7:1809. doi: 10.3389/fpls.2016.01809
Hasan, M. M. U., Ma, F., Islam, F., Sajid, M., Prodhan, Z. H., Li, F., et al. (2019). Comparative transcriptomic analysis of biological process and key pathway in three cotton (Gossypium spp.) species under drought stress. Int. J. Mol. Sci. 20:2076. doi: 10.3390/ijms20092076
Hiremath, P. J., Farmer, A., Cannon, S. B., Woodward, J., Kudapa, H., Tuteja, R., et al. (2011). Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnol. J. 9, 922–931. doi: 10.1111/j.1467-7652.2011.00625.x
Hiremath, P. J., Kumar, A., Penmetsa, R. V., Farmer, A., Schlueter, J. A., Chamarthi, S. K., et al. (2012). Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 10, 716–732. doi: 10.1111/j.1467-7652.2012.00710.x
Hodges, D. M., Delong, J. M., Forney, C. F., and Prange, R. K. (1999). Improving the thiobarbitric acid reactive substance assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. doi: 10.1007/s004250050524
Hou, Z., Yin, J., Lu, Y., Song, J., Wang, S., Wei, S., et al. (2019). Transcriptomic analysis reveals the temporal and spatial changes in physiological process and gene expression in common buckwheat (Fagopyrum esculentum Moench) grown under drought stress. Agronomy 9:569. doi: 10.3390/agronomy9100569
Jenks, M. A., and Ashworth, E. N. (1999). Plant epicuticular waxes: function, production, and genetics. Horticult. Rev. 23, 1–68.
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7:1029. doi: 10.3389/fpls.2016.01029
Kerstiens, G. (1996). Cuticular water permeability and its physiological significance. J. Experim. Bot. 47, 1813–1832. doi: 10.1093/jxb/47.12.1813
Kumar, J., Gunapati, S., Kianian, S. F., and Singh, S. P. (2018). Comparative analysis of transcriptome in two wheat genotypes with contrasting levels of drought tolerance. Protoplasma 255, 1487–1504. doi: 10.1007/s00709-018-1237-x
Kumar, M., Chauhan, A. S., Yusuf, M. A., Sanyal, I., and Chauhan, P. S. (2019). Transcriptome sequencing of chickpea (Cicer arietinum L.) genotypes for identification of drought-responsive genes under drought stress condition. Plant Mol. Biol. Rep. 37, 186–203. doi: 10.1007/s11105-019-01147-4
Kumar, S., Palve, A. S., Patel, S. K., Selvanayagam, S., Sharma, R., and Rathore, A. (2020). Development of genomic microsatellite markers in cluster bean using next-generation DNA sequencing and their utility in diversity analysis. Curr. Plant Biol. 21:100134. doi: 10.1016/j.cpb.2019.100134
Kumar, S., Parekh, M. J., Patel, C. B., Zala, H. N., Sharma, R., Kulkarni, K. S., et al. (2016). Development and validation of EST-derived SSR markers and diversity analysis in cluster bean (Cyamopsis tetragonoloba). J. Plant Biochem. Biotechnol. 25, 263–269. doi: 10.1007/s13562-015-0337-3
Kuravadi, N. A., Tiwari, P. B., Tanwar, U. K., Tripathi, S. K., Dhugga, K. S., Gill, K. S., et al. (2014). Identification and Characterization of EST-SSR Markers in Cluster Bean (Cyamopsis spp.). Crop Sci. 54, 1097–1102. doi: 10.2135/cropsci2013.08.0522
Lee, S. B., and Suh, M. C. (2013). Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol. Plant 6, 246–249. doi: 10.1093/mp/sss159
Leopold, A. C., Musgrave, M. E., and Williams, K. M. (1981). Solute leakage resulting from leaf desiccation. Plant Physiol. 68, 1222–1225. doi: 10.1104/pp.68.6.1222
Li, B., Ruotti, V., Stewart, R. M., Thomson, J. A., and Dewey, C. N. (2010). RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26, 493–500. doi: 10.1093/bioinformatics/btp692
Liu, X., Liu, S., Wu, J., Zhang, B., Li, X., Yan, Y., et al. (2013). Overexpression of Arachis hypogaea NAC3 in tobacco enhances dehydration and drought tolerance by increasing superoxide scavenging. Plant Physiol. Biochem. 70, 354–359. doi: 10.1016/j.plaphy.2013.05.018
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Magwanga, R. O., Lu, P., Kirungu, J. N., Lu, H., Wang, X., Cai, X., et al. (2018). Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Gen. 19:6. doi: 10.1186/s12863-017-0596-1
Mamrutha, H. M., Mogili, T., Lakshmi, K. J., Rama, N., Kosma, D., Kumar, M. U., et al. (2010). Leaf cuticular wax amount and crystal morphology regulate post-harvest water loss in mulberry (Morus species). Plant Physiol. Biochem. 48, 690–696. doi: 10.1016/j.plaphy.2010.04.007
Mamrutha, H. M., Nataraja, K. N., Rama, N., Kosma, D. K., Mogili, T., Lakshmi, K. J., et al. (2017). Leaf surface wax composition of genetically diverse mulberry (Morus sp.) genotypes and its close association with expression of genes involved in wax metabolism. Curr. Sci. 112, 759–766. www.jstor.org/stable/24912577
Mizuno, H., Kawahigashi, H., Ogata, J., Minami, H., Kanamori, H., Nakagawa, H., et al. (2013). Genomic inversion caused by gamma irradiation contributes to downregulation of a WBC11 homolog in bloomless Sorghum. Theor. Appl. Gen. 126, 1513–1520. doi: 10.1007/s00122-013-2069-x
Morgil, H., Tardu, M., Cevahir, G., and Kavakli, I. H. (2019). Comparative RNA-seq analysis of the drought-sensitive lentil (Lens culinaris) root and leaf under short-and long-term water deficits. Funct. Integr. Genom. 19, 715–727. doi: 10.1007/s10142-019-00675-2
Mudgil, D., Barak, S., and Khatkar, B. S. (2014). Guar gum: processing, properties and food applications—a review. J. Food Sci. Technol. 51, 409–418. doi: 10.1007/s13197-011-0522-x
Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170. doi: 10.3389/fpls.2014.00170
Naoumkina, M., Torres-Jerez, I., Allen, S., He, J., Zhao, P. X., Dixon, R. A., et al. (2007). Analysis of cDNA libraries from developing seeds of guar (Cyamopsis tetragonoloba (L.) Taub). BMC Plant Biol. 7:62. doi: 10.1186/1471-2229-7-62
Panda, A., Rangani, J., and Parida, A. K. (2021). Unraveling salt responsive metabolites and metabolic pathways using non-targeted metabolomics approach and elucidation of salt tolerance mechanisms in the xero-halophyte Haloxylon salicornicum. Plant Physiol. Biochem. 158, 284–296. doi: 10.1016/j.plaphy.2020.11.012
Rao, N. K., and Shahid, M. (2011). Potential of cowpea [Vigna unguiculata (L.) Walp.] And guar [cyamopsis tetragonoloba (L.) Taub.] As alternative forage legumes for. Emir. J. Food Agric. 23, 147–156.
Rawal, H. C., Kumar, S., Mithra, S. V. A., Solanke, A. U., Nigam, D., Saxena, S., et al. (2017). High quality unigenes and microsatellite markers from tissue specific transcriptome and development of a database in clusterbean (Cyamopsis tetragonoloba, L. Taub). Genes 8, 313. doi: 10.3390/genes8110313
Raza, A., Razzaq, A., Mehmood, S. S., Hussain, M. A., Wei, S., He, H., et al. (2021). Omics: The way forward to enhance abiotic stress tolerance in Brassica napus L. G.M. Crops Food 12, 251–281. doi: 10.1080/21645698.2020.1859898
Samdur, M. Y., Manivel, P., Jain, V. K., Chikani, B. M., Gor, H. K., Desai, S., et al. (2003). Genotypic differences and water-deficit induced enhancement in epicuticular wax load in peanut. Crop Sci. 43, 1294–1299. doi: 10.2135/cropsci2003.1294
Sánchez, C., Vielba, J. M., Ferro, E., Covelo, G., Solé, A., Abarca, D., et al. (2007). Two SCARECROW-LIKE genes are induced in response to exogenous auxin in rooting-competent cuttings of distantly related forest species. Tree Physiol. 27, 1459–1470. doi: 10.1093/treephys/27.10.1459
Shi, H., Chen, Y., Qian, Y., and Chan, Z. (2015). Low temperature-induced 30 (LTI30) positively regulates drought stress resistance in Arabidopsis: effect on abscisic acid sensitivity and hydrogen peroxide accumulation. Front. Plant Sci. 6:893. doi: 10.3389/fpls.2015.00893
Shi, W. Y., Du, Y. T., Ma, J., Min, D. H., Jin, L. G., Chen, J., et al. (2018). The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. Int. J. Mol. Sci. 19, 4087. doi: 10.3390/ijms19124087
Singh, D., Singh, C. K., Taunk, J., Tomar, R. S. S., Chaturvedi, A. K., Gaikwad, K., et al. (2017). Transcriptome analysis of lentil (Lens culinaris Medikus) in response to seedling drought stress. BMC Genomics 18:1–20. doi: 10.1186/s12864-017-3596-7
Tang, G. Y., Shao, F. X., Xu, P. L., Shan, L., and Liu, Z. J. (2017). Overexpression of a peanut NAC gene, AhNAC4, confers enhanced drought tolerance in tobacco. Russian J. Plant Physiol. 64, 525–535. doi: 10.1134/S1021443717040161
Tanwar, U. K., Pruthi, V., and Randhawa, G. S. (2017). RNA-Seq of guar (Cyamopsis tetragonoloba, L. Taub.) leaves: de novo transcriptome assembly, functional annotation and development of genomic resources. Front. Plant Sci. 8:91. doi: 10.3389/fpls.2017.00091
Tirajoh, A., Aung, T. S., McKay, A. B., and Plant, A. L. (2005). Stress-responsive α-dioxygenase expression in tomato roots. J. Exper. Bot. 56, 713–723. doi: 10.1093/jxb/eri038
Tyagi, A., Sharma, P., Saxena, S., Sharma, R., Mithra, S. A., Solanke, A. U., et al. (2019). The genome size of clusterbean (Cyamopsis tetragonoloba) is significantly smaller compared to its wild relatives as estimated by flow cytometry. Gene 707, 205–211. doi: 10.1016/j.gene.2019.02.090
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Gen. 10, 57–63. doi: 10.1038/nrg2484
Wang, Z., Yu, G., Shi, B., Wang, X., Qiang, H., and Gao, H. (2014). Development and characterization of simple sequence repeat (SSR) markers based on RNA-sequencing of Medicago sativa and in silico mapping onto the M. truncatula genome. PLoS One 9:e92029. doi: 10.1371/journal.pone.0092029
Waseem, M., Rong, X., and Li, Z. (2019). Dissecting the role of a basic helix-loop-helix transcription factor, SlbHLH22, under salt and drought stresses in transgenic Solanum lycopersicum L. Front. Plant Sci. 10:734. doi: 10.3389/fpls.2019.00734
Wilhelm, B. T., Marguerat, S., Goodhead, I., and Bähler, J. (2010). Defining transcribed regions using RNA-seq. Nat. Prot. 5, 255–266. doi: 10.1038/nprot.2009.229
Wu, J., Wang, L., and Wang, S. (2016). Comprehensive analysis and discovery of drought-related NAC transcription factors in common bean. BMC Plant Biol. 16:193. doi: 10.1186/s12870-016-0882-5
Wu, Z. J., Li, X. H., Liu, Z. W., Li, H., Wang, Y. X., and Zhuang, J. (2015). Transcriptome-based discovery of AP2/ERF transcription factors related to temperature stress in tea plant (Camellia sinensis). Funct. Integr. Genom. 15, 741–752. doi: 10.1007/s10142-015-0457-9
Xu, C., Fan, J., Riekhof, W., Froehlich, J. E., and Benning, C. (2003). A permease-like protein involved in E.R. to thylakoid lipid transfer in Arabidopsis. EMBO J. 22, 2370–2379. doi: 10.1093/emboj/cdg234
Yang, H., Stierhof, Y. D., and Ludewig, U. (2015). The putative Cationic Amino Acid Transporter 9 is targeted to vesicles and may be involved in plant amino acid homeostasis. Front. Plant Sci. 6:212. doi: 10.3389/fpls.2015.00212
Yu, X., Liu, Y., Wang, S., Tao, Y., Wang, Z., Shu, Y., et al. (2016). CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant cell Rep. 35, 613–627.
Zhang, Y., Gao, X., Li, J., Gong, X., Yang, P., Gao, J., et al. (2019). Comparative analysis of proso millet (Panicum miliaceum L.) leaf transcriptomes for insight into drought tolerance mechanisms. BMC Plant Biol. 19:397. doi: 10.1186/s12870-019-2001-x
Zhao, X., Li, C., Wan, S., Zhang, T., Yan, C., and Shan, S. (2018). Transcriptomic analysis and discovery of genes in the response of Arachis hypogaea to drought stress. Mol. Biol. Rep. 45, 119–131. doi: 10.1007/s11033-018-4145-4
Zheng, C., Zhou, J., Zhang, F., Yin, J., Zhou, G., Li, Y., et al. (2020). OsABAR1, a novel GRAM domain-containing protein, confers drought and salt tolerance via an ABA-dependent pathway in rice. Plant Physiol. Biochem. 152, 138–146. doi: 10.1016/j.plaphy.2020.04.032
Keywords: drought stress, transcriptome, wax genes, cluster bean (Cyamopsis tetragonoloba L.), differentailly expressed genes
Citation: Reddy BM, Anthony Johnson AM, Jagadeesh Kumar N, Venkatesh B, Jayamma N, Pandurangaiah M and Sudhakar C (2022) De novo Transcriptome Analysis of Drought-Adapted Cluster Bean (Cultivar RGC-1025) Reveals the Wax Regulatory Genes Involved in Drought Resistance. Front. Plant Sci. 13:868142. doi: 10.3389/fpls.2022.868142
Received: 02 February 2022; Accepted: 02 June 2022;
Published: 28 June 2022.
Edited by:
Mostafa Abdelwahed Abdelrahman, Aswan University, EgyptReviewed by:
Yordan Muhovski, Walloon Agricultural Research Centre, BelgiumSai Shiva Krishna Prasad Vurukonda, University of Modena and Reggio Emilia, Italy
Copyright © 2022 Reddy, Anthony Johnson, Jagadeesh Kumar, Venkatesh, Jayamma, Pandurangaiah and Sudhakar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chinta Sudhakar, Y2hpbnRhc3VkaGFrYXJAeWFob28uY29t
 B. Manohara Reddy
B. Manohara Reddy A. M. Anthony Johnson
A. M. Anthony Johnson N. Jagadeesh Kumar
N. Jagadeesh Kumar Boya Venkatesh
Boya Venkatesh N. Jayamma
N. Jayamma Merum Pandurangaiah
Merum Pandurangaiah Chinta Sudhakar
Chinta Sudhakar