
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 May 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.867503
This article is part of the Research Topic Molecular and Genetic Perspectives of Cold Tolerance in Plants View all 13 articles
The mitochondrial calcium uniporter (MCU), as an important component of the Ca2+ channel uniporter complex, plays a regulatory role in intracellular Ca2+ signal transduction. However, only a few studies to date have investigated plant MCU genes. In this study, we identified the MCU family genes in wild soybean and investigated their expression under cold and carbonate alkaline stresses. Eleven Glycine soja MCU genes (GsMCUs) were identified and clustered into two subgroups (subgroups I and II), and subgroup II could be further divided into two branches (MCU5 and MCU6). A total of 21 pairs of GsMCUs were characterized as duplicated genes, and displayed a similar exon-intron architecture. All GsMCU proteins contained one conserved MCU domain, within which two transmembrane domains were found. An analysis of the conserved motifs further supported that the GsMCUs showed high conservation in protein sequence and structure. Moreover, we found that all GsMCUs were expressed ubiquitously in different tissues and organs, and GsMCUs from the same subgroup displayed varied tissue expression profiles. In addition, based on RNA-seq and qRT-PCR assays, six and nine GsMCUs were differentially expressed under cold and carbonate alkaline stress, respectively. Promoter analysis also uncovered the existence of two canonical cold-related cis-acting elements, LTR and DRE/CRT, as well as stress-related phytohormone-responsive elements. Our results provide valuable information about the MCU family in soybean responses to cold and carbonate alkaline stress, which will be helpful in further characterizing their biological roles in response to abiotic stress.
In the 1970s, calcium ions were proposed to be important messenger ions in cells. Over the years, a large number of studies have demonstrated that many functions of cells are inextricably linked to Ca2+ (Clapham, 2007). Mitochondria absorb Ca2+ mainly through mitochondrial calcium uniporters (MCU) located on the inner membrane (Deluca and Engstrom, 1961; Vasington and Murphy, 1962). A typical MCU protein harbors two transmembrane domains within a conserved MCU domain (PF04678), with the N- and C-terminus facing the mitochondrial matrix. The N-terminal domain is essential for the Ca2+ uptake ability of MCU proteins, while the formation of a functional channel requires the pentamerization of transmembrane regions. By interacting with MICU1 (mitochondrial calcium uptake 1), MCUs can transport Ca2+ along electrochemical gradients (Perocchi et al., 2010), and play key roles in mitochondrial Ca2+ homeostasis, regulation and signaling (Baughman et al., 2011; De Stefani et al., 2011; Drago et al., 2011). In plants, MCUs are important components for sensing environmental stress because they modulate stromal Ca2+ in chloroplasts (Teardo et al., 2019). In recent decades, an increasing number of MCUs have been functionally characterized in mammals (Mallilankaraman et al., 2012; Bisbach et al., 2020; Liu et al., 2020), but little has been reported regarding plant MCUs.
Taking advantage of whole genome sequencing and public expression data, researchers could genome-widely identify MCU family genes, and systemically investigate their expression characteristics, which will provide hints to deeply study the function of MCUs in diverse physiological and biological processes. However, compared with other gene families, fewer studies have been reported on MCU family. To date, six, five, seven, and four MCUs have been identified in Arabidopsis thaliana (Stael et al., 2012), Oryza sativa (Gu, 2020), Pyrus seratina (Wang et al., 2018), and Sorghum bicolor (Gu, 2020), respectively. Among the six MCU family genes in Arabidopsis, AtMCU1 and AtMCU2 were found to regulate pollen tube germination and growth (Selles et al., 2018), while AtMCU6, a chloroplast-localized mitochondrial calcium uniporter, negatively modulated resistance to long-term water deficits (Teardo et al., 2019). In pears, MCUs have been suggested to play an important role in fruit ripening and senescence (Wang et al., 2018). However, there have been no reports on soybean MCU proteins.
Compared with cultivated soybean (Glycine max L.), wild soybean (Glycine soja) exhibits very high adaptability to harsh environments and extensive genetic diversity (Kofsky et al., 2018). It will be of great help to identify the soybean MCU genes, and screen the stress responsive MCUs in wild soybean. Therefore, in this study, we systematically characterized the MCU family genes in wild soybean and investigated their expression profiles in different tissues and under abiotic stress. Our findings suggest the potential regulatory roles of wild soybean MCU genes in response to cold and carbonate saline-alkaline stresses, which will facilitate the molecular breeding of soybean cultivars with higher tolerance to cold and carbonate saline-alkaline stresses.
To identify the candidate MCU genes in the wild soybean genome, we obtained the MCU proteins sequences which have been identified in Arabidopsis thaliana (Stael et al., 2012), Pyrus seratina (Wang et al., 2018), Oryza sativa and Sorghum bicolor (Gu, 2020) from Phytozome database.1 These protein sequences were used for generating a hidden Markov Model,2 which was then used to search against the wild soybean (Glycine soja v1.1) and cultivated soybean (Glycine max Wm82.a2.v1) proteomes. The amino acid sequences of the candidates were manually checked to determine whether they harbored the MCU domain (PF04678) using the NCBI Conserved Domain Database3 and Pfam4 program.
Detailed information on the GsMCUs, including their locus ID and genomic and protein sequences, was obtained from the Phytozome database. Isoelectric point values and molecular weights were estimated using ExPASy.5 The transmembrane (TM) region was predicted by using the TMHMM.6
The chromosomal locations of the GsMCUs were obtained from the Phytozome database. Duplicated segmental blocks of soybean genome were downloaded from the Plant Genome Duplication Database (PGDD). The criteria for identifying duplicated gene pairs were: (a) the length of the shorter aligned sequence covered > 70% of the longer sequence; and (b) the similarity of the two aligned sequences were > 70%. Orthologous soybean genes were downloaded from Ortho Venn2.7 The chromosomal locations and syntenic information were visualized using TBtools software. Ka (non-synonymous rate), Ks (synonymous rate), and Ka/Ks ratios were calculated using TBtools according to their coding sequences. The timing of the genome-wide duplication event was calculated using the formula: Time = Ks/2r (r = 6.98 × 10–9).
For phylogenetic analysis, multiple alignments were performed with full-length amino acid sequences of MCU proteins in rice, Arabidopsis, and soybean using ClustalW. A neighbor-joining phylogenetic tree was constructed using MEGA5.1 software with the following parameters: Poison correction, pairwise deletion, uniform rates, and 1,000 bootstrap replicates.
The exon/intron structure of the GsMCU genes was analyzed using the Gene Structure Display Server (GSDS).8 Functional domain information was derived from the NCBI Batch-CDD database.9 The conserved motifs were predicted using MEME10 with the following parameters: number of motifs = 15 and zero or one occurrence per sequence. The functional domains and conserved motifs of the GsMCUs were illustrated using TBtools.
The protein-interacting network of the GsMCUs was generated using the STRING database.11
The 2,000 bp promoter sequences upstream of the transcript start sites of the GsMCU genes were obtained from the Phytozome database (see text footnote 1). The cis-acting elements were analyzed using New PLACE.12
The accession numbers of the public expression data are as follows: NaHCO3 stress (GSE17883), and cold stress (GSE117686) (Yamasaki and Randall, 2016; Robison et al., 2019).
Glycine soja (G07256) seeds were treated with 98% sulfuric acid for 10–15 min, and washed five times with distilled water, and then kept in complete darkness with humidity for 1 day to promote germination. The seedlings were transferred and grown in 1/2 Hoagland’s nutrient solution at 26–28°C under 16-h light/8-h dark conditions.
To explore the gene expression patterns in different tissues, freshly germinated seeds, roots, stems, and leaves of 21-day-old seedlings and the flowers were harvested. For the cold stress, 2-week-old seedlings were exposed to 4°C. For the carbonate saline-alkaline treatment, 21-day-old seedlings were transferred to a Hoagland’s nutrient solution containing 50 mM NaHCO3 (pH 8.5). Equal amounts of seedling roots were harvested at the indicated time points, and the samples were snap-frozen in liquid nitrogen and stored at −80°C.
The total RNA was isolated using the TriPure Isolation Reagent (Roche, United States), and cDNAs were synthesized using HiScript ® III RT SuperMix for qPCR (Vazyme, Nanjing, China) according to the manufacturer’s instructions. Quantitative real-time PCR assays were performed using TransStart ® Top Green qPCR SuperMix according to the manufacturer’s protocol. The F_box gene in wild soybeans (GlysoPI483463.12G046600) was used as an internal control (Le et al., 2012; Bansal et al., 2015). All experiments were performed using three independent biological replicates for statistical analyses. The primers used are listed in Supplementary Table 3.
To investigate the precise information and potential function of Glycine soja MCU (GsMCU) genes, we carried out systematic genome-wide identification of putative MCU genes in the wild soybean genome. To do this, we generated a hidden Markov model based on protein sequences of the reported Arabidopsis (Stael et al., 2012), Pyrus seratina (Wang et al., 2018), and Sorghum bicolor (Gu, 2020) MCU proteins, respectively, and then searched for MCU candidates against the soybean proteome. After manually checking protein sequences for to ensure the existence of the MCU domain, a total of 11 genes were identified as GsMCU family members and named according to Arabidopsis orthologs.
Detailed information about wild soybean (Glycine soja v1.1) GsMCUs was obtained from the Phytozome database and is listed in Table 1. Notably, GsMCU5.2 had only 264 amino acids (aa), and did not possess the N-terminal extension, compared to other members. The other GsMCU members shared similar protein sequence lengths (from 317 to 357 aa). Correspondingly, the molecular weight of the GsMCU proteins (except GsMCU5.2) varied from 36.2 to 40.6 kDa, and the predicted isoelectric points ranged between 8.65 and 9.51. Furthermore, all GsMCU proteins had two predicted transmembrane regions, with both the N-and C-terminus facing the matrix. These results imply that the GsMCU family possesses a conserved protein structure.
To investigate the phylogenetic relationships among the GsMCUs, we generated a neighbor-joining tree using the full-length amino acid sequences of wild soybean, Arabidopsis (Stael et al., 2012) and rice (Gu, 2020) MCU family proteins (Figure 1). As shown in Figure 1, MCU proteins were clustered into two subgroups: subgroup I and subgroup II. Subgroup I contained nine members, including four GsMCUs, two AtMCUs, and three OsMCUs, whereas subgroup II included seven GsMCUs, four AtMCUs, and two OsMCUs (Supplementary Table 1). In each subgroup, the OsMCUs were first separated from the other members and clustered into one branch. This finding implies that the appearance of these two subgroups occurred before the division of monocots and dicots during evolution. Moreover, the soybean possessed more MCU genes than Arabidopsis and rice, and GsMCUs appeared in pairs in the phylogenetic tree, which might be a consequence of whole genome duplication of soybean during evolution. This result also indicates possible gene duplication within the GsMCU family, as well as the conservation of GsMCUs in gene architecture and protein structure.
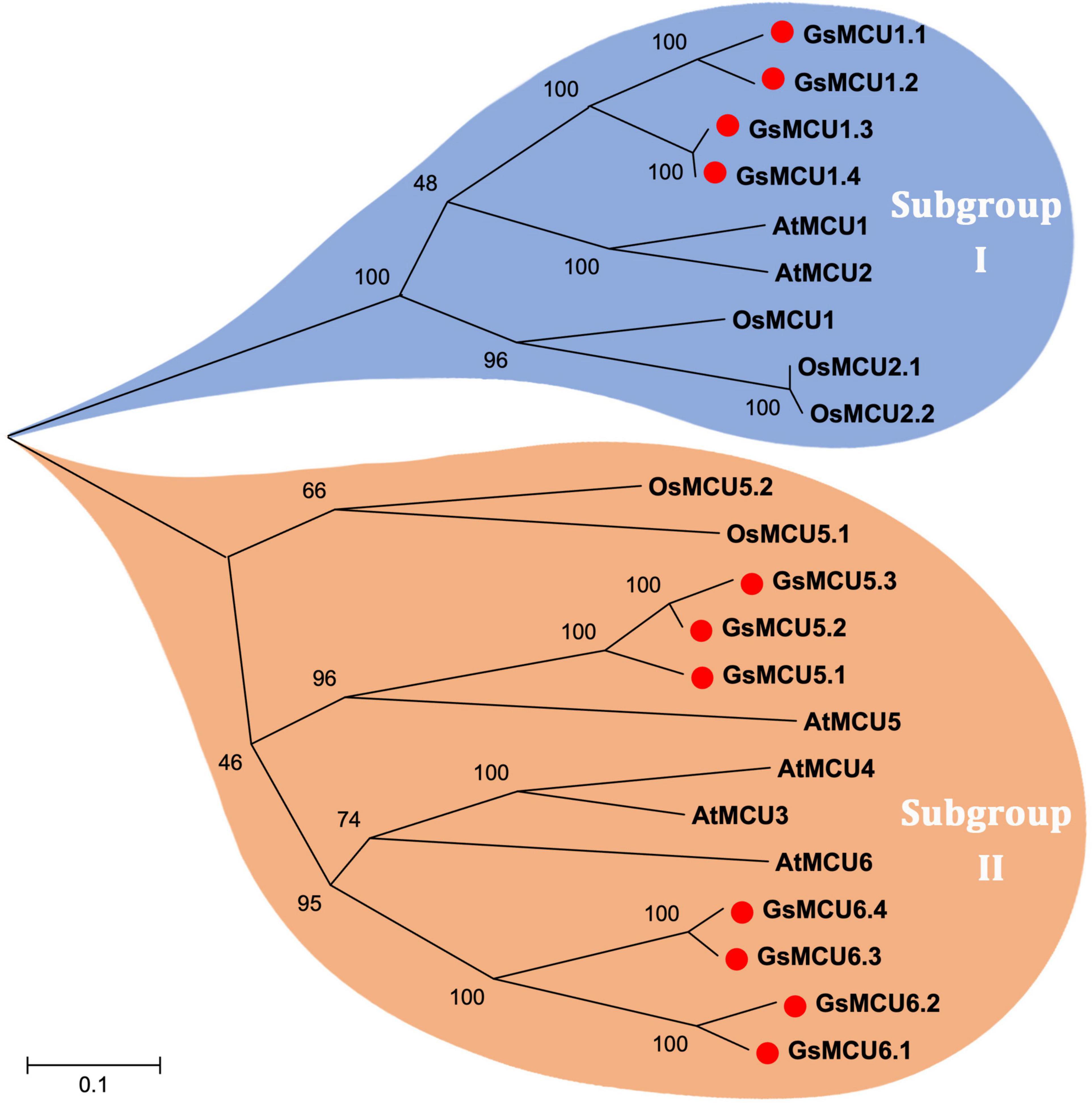
Figure 1. Phylogenetic relationship of MCU proteins from Arabidopsis, rice and soybean. The phylogenetic tree was constructed using MEGA 5.10 software using the neighbor-joining method, and the bootstrap values were set at 1,000.
To confirm the existence of gene duplication within the GsMCU family, we performed chromosomal location and collinearity analyses. As shown in Figure 2, 11 GsMCUs were randomly distributed on seven chromosomes. Chromosomes 2, 11, 14, and 17 each possessed two GsMCUs, whereas chromosomes 1, 16, and 18 each contained one. To obtain the collinearity of GsMCUs, we retrieved duplicated gene pairs of the MCU family from the Plant Genome Duplication Database and identified 21 pairs among 11 GsMCU genes (Figure 2). Furthermore, we found that the Ka/Ks values of the duplicated pairs were less than 1, indicating purification of GsMCU genes during evolution (Supplementary Table 2). In addition, the duplication of the GsMCUs occurred 10.5246–322.3279 Mya (million years ago).
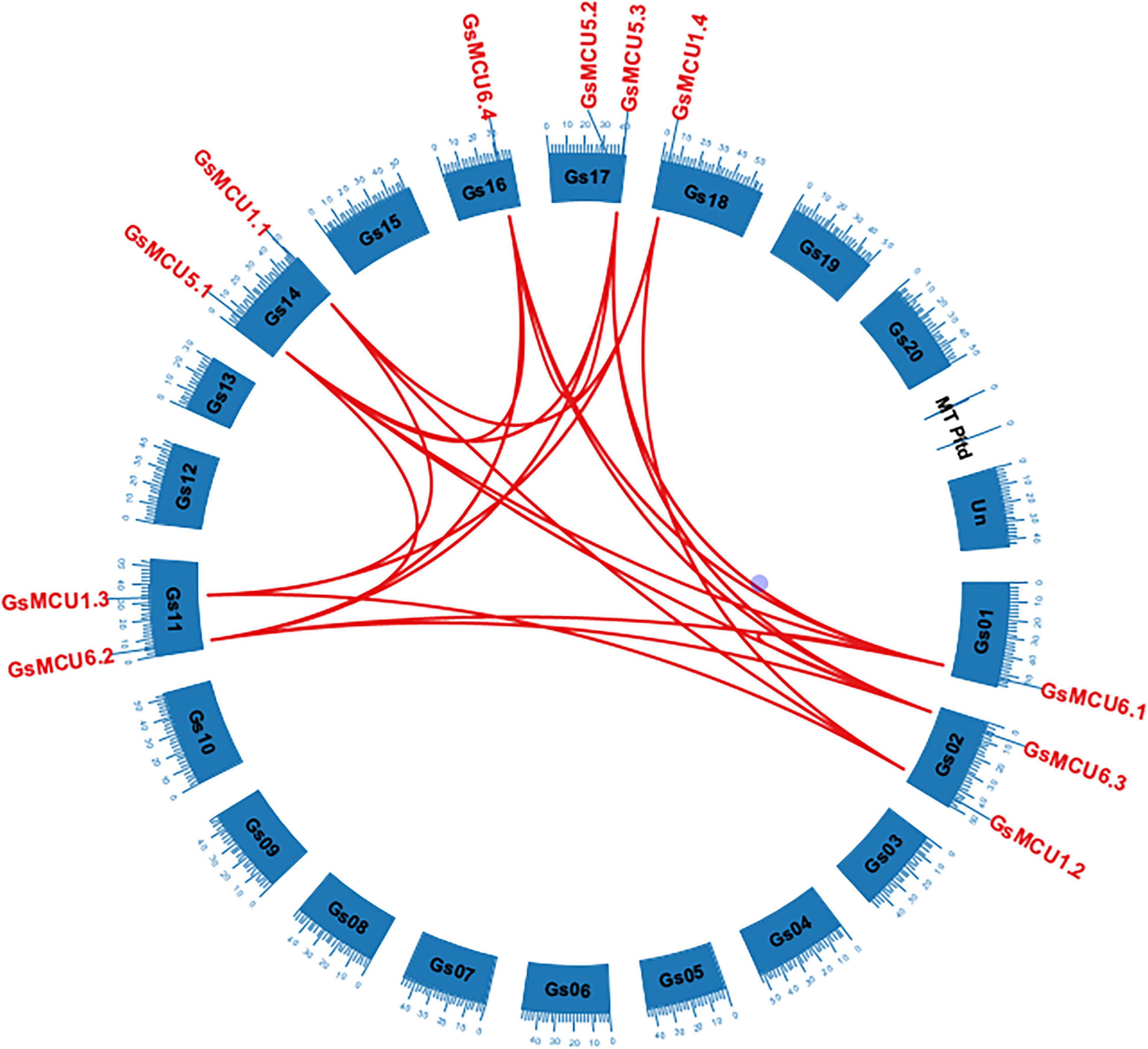
Figure 2. Distribution and duplication of the MCU genes in the wild soybean genome. Duplicated genes among the GsMCUs were downloaded from the PGDD database and are indicated by red lines.
In generally, duplicated gene pairs are highly conserved in terms of exon-intron organization. As shown in Figure 3, all GsMCUs contained only two exons of similar length, suggesting a highly conserved gene architecture. Moreover, synteny analysis of MCU genes from different dicot species identified a number of duplicated MCU gene pairs among soybean, alfalfa, and Arabidopsis (Supplementary Figure 1). These findings further support the idea that whole-genome duplication was the main contributor to expansion of the MCU family.
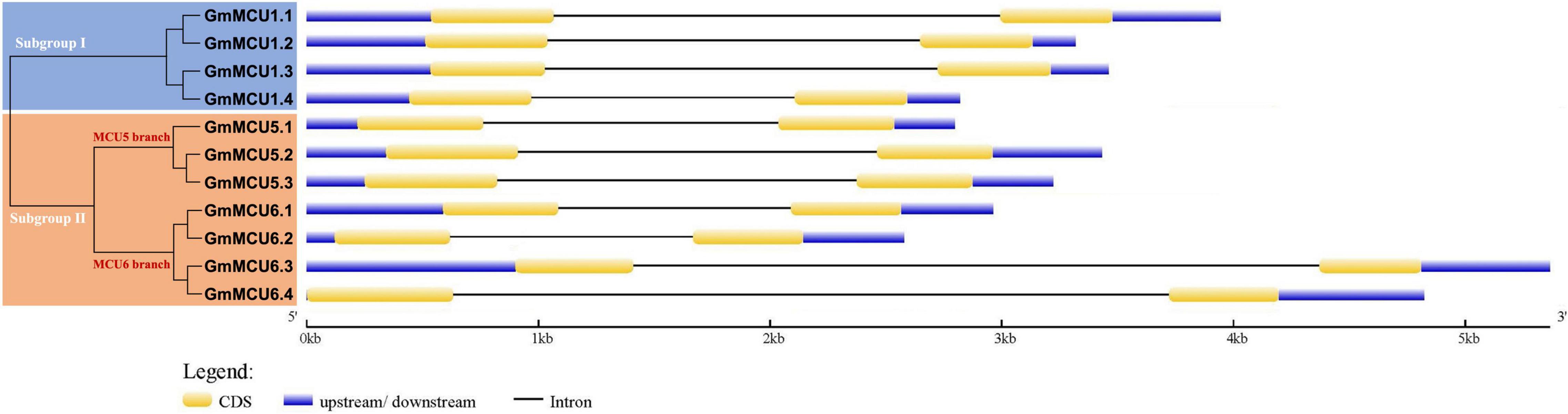
Figure 3. Gene structure analysis of GsMCU family. The position of exons, introns, and untranslated regions (UTRs) are indicated by yellow, black, and blue boxes, respectively. The scale bar at the bottom was used to estimate the sizes of the exons, introns, and untranslated regions.
To verify the conservation of the GsMCU family, we further analyzed the functional domains and conserved motifs. All GsMCUs harbored one conserved MCU domain (PF04678) (Figure 4A). MEME analysis of the conserved motifs also suggested that the GsMCU proteins possessed high similarity in motif organization (Figure 4B). Among the 15 motifs, motifs 1 to 4, representing the MCU domain, were present in all GsMCUs. Multiple alignments illustrated that the MCU domain contained two transmembrane regions (TMs) and displayed a high sequence similarity (Figure 5). These results further highlight the high conservation of GsMCUs in terms of protein structure.
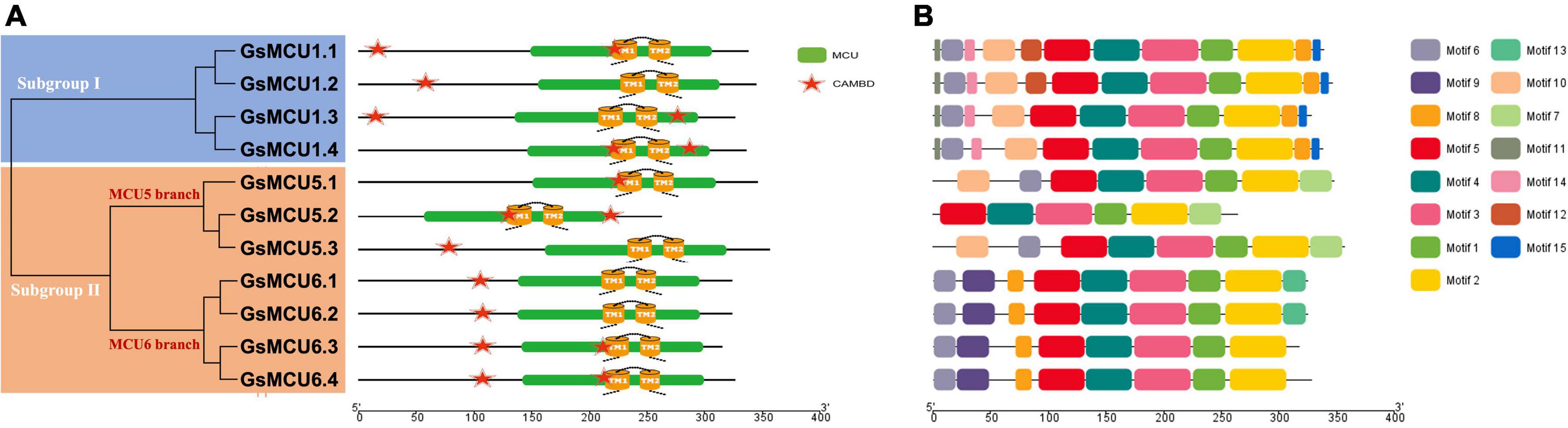
Figure 4. Functional domains and conserved motifs of GsMCU proteins. Functional domains (A) and conserved motifs (B) are indicated by green boxes and different colored boxes represent different motifs. The five-pointed stars represent the calmodulin-binding sites in the GsMCU proteins. Protein domains and conserved motifs were predicted using a CDD search and MEME.
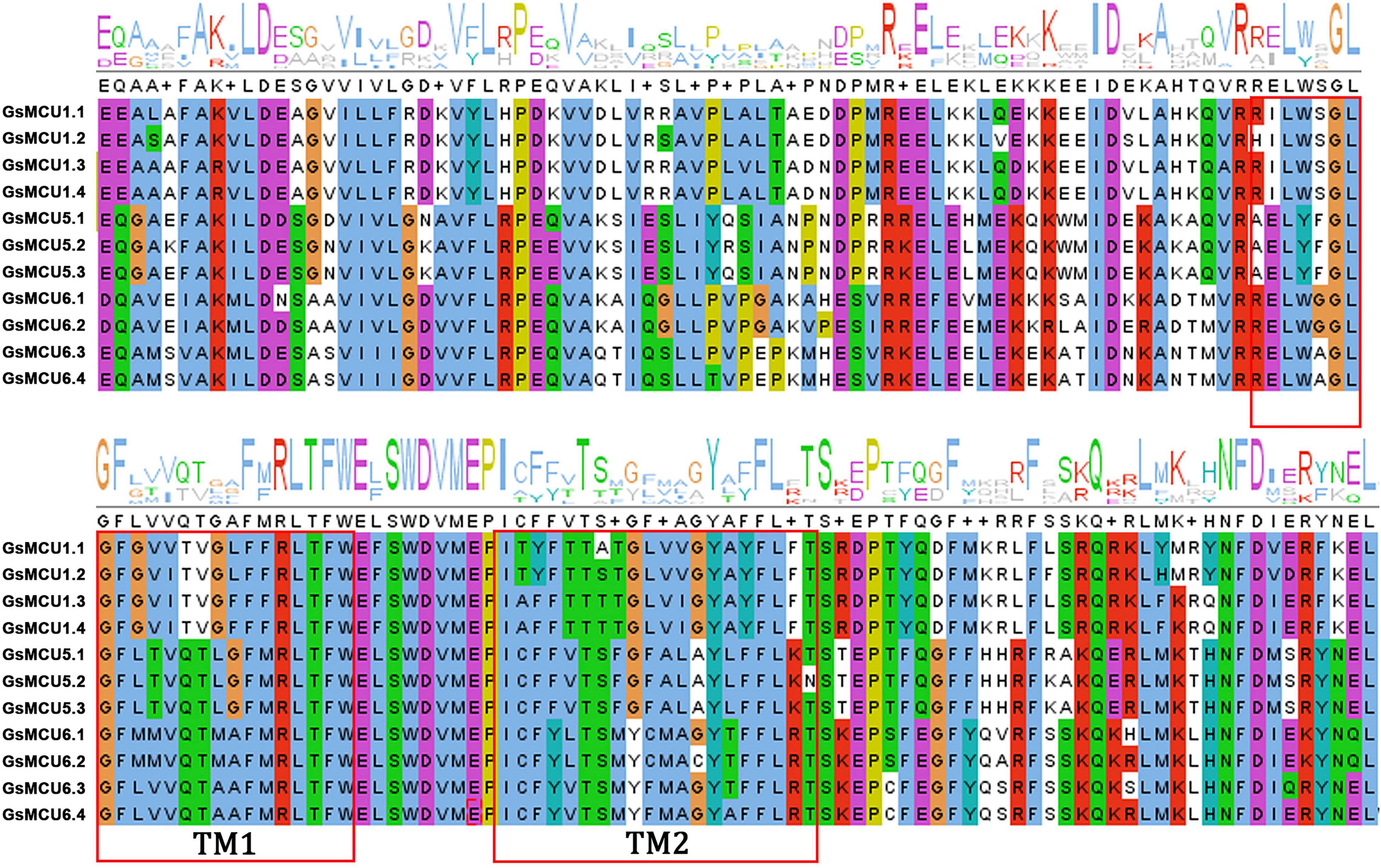
Figure 5. Multiple sequence alignment of the MCU domain within GsMCUs proteins. Two transmembrane regions (TM1 and TM2) are marked by red boxes.
Meanwhile, GsMCUs from different subgroups also showed sequence diversity at a certain degree (Figure 4B). For example, among the 15 motifs, motifs 11, 14, and 15 were specific to subgroup I GsMCUs. Motif 9 was only observed in the MCU6 branch of subgroup II, whereas motif 7 was observed in the MCU5 branch. Interestingly, motif 8 was located at the C-terminus of subgroup I, but at the N-terminus of the subgroup II MCU6 branch. In addition, one or two calmodulin-binding domains (CAMBD) were found in all GsMCU proteins. However, the location of the CAMBD varied among the different GsMCU proteins (Figure 4A). These differences imply possible different regulatory mechanisms of biochemical activity or functional specialization.
Several studies have reported the involvement of plant MCU genes in growth and development (Wang et al., 2018). To explore the potential function of MCU genes in soybean growth, we retrieved the expression profiles of GsMCUs in different tissues from Phytozome and verified these profiles via semiquantitative RT-PCR analysis (Figure 6). According to our RT-PCR results, the GsMCU genes were expressed in all detected tissues and organs (Figures 6B–D). For the four GsMCUs in subgroup I, two (GsMCU1.1 and GsMCU1.2) were expressed at a very low level, and the other two (GsMCU1.3 and GsMCU1.4) displayed a relatively high expression level, with the highest expression in leaves (Figure 6A). The RT-PCR results confirmed this pattern (Figure 6B). For members of the MCU5 branch in subgroup II, GsMCU5.1 was highly expressed in roots, flowers, and seeds, while GsMCU5.2 and GsMCU5.3 displayed high expression in seeds (Figure 6A). The RT-PCR analysis revealed a relatively high expression of the MCU5 branch genes in seeds and leaves (Figure 6C). This finding indicates a possible role for the MCU5 branch in seed development. In the MCU6 branch, GsMCU6.1 displayed the highest expression in roots, while the other three genes showed the highest expression levels in flowers (Figure 6A). As shown in Figure 6D, GsMCU6.1 showed a peak expression in leaves, GsMCU6.2 displayed a ubiquitous expression, with the highest level in the stem, while GsMCU6.3 and GsMCU6.4 had the highest transcript levels in flowers. In conclusion, these results suggest that GsMCUs from the same subgroup display variable tissue expression profiles, indicating their potentially divergent roles in soybean development.
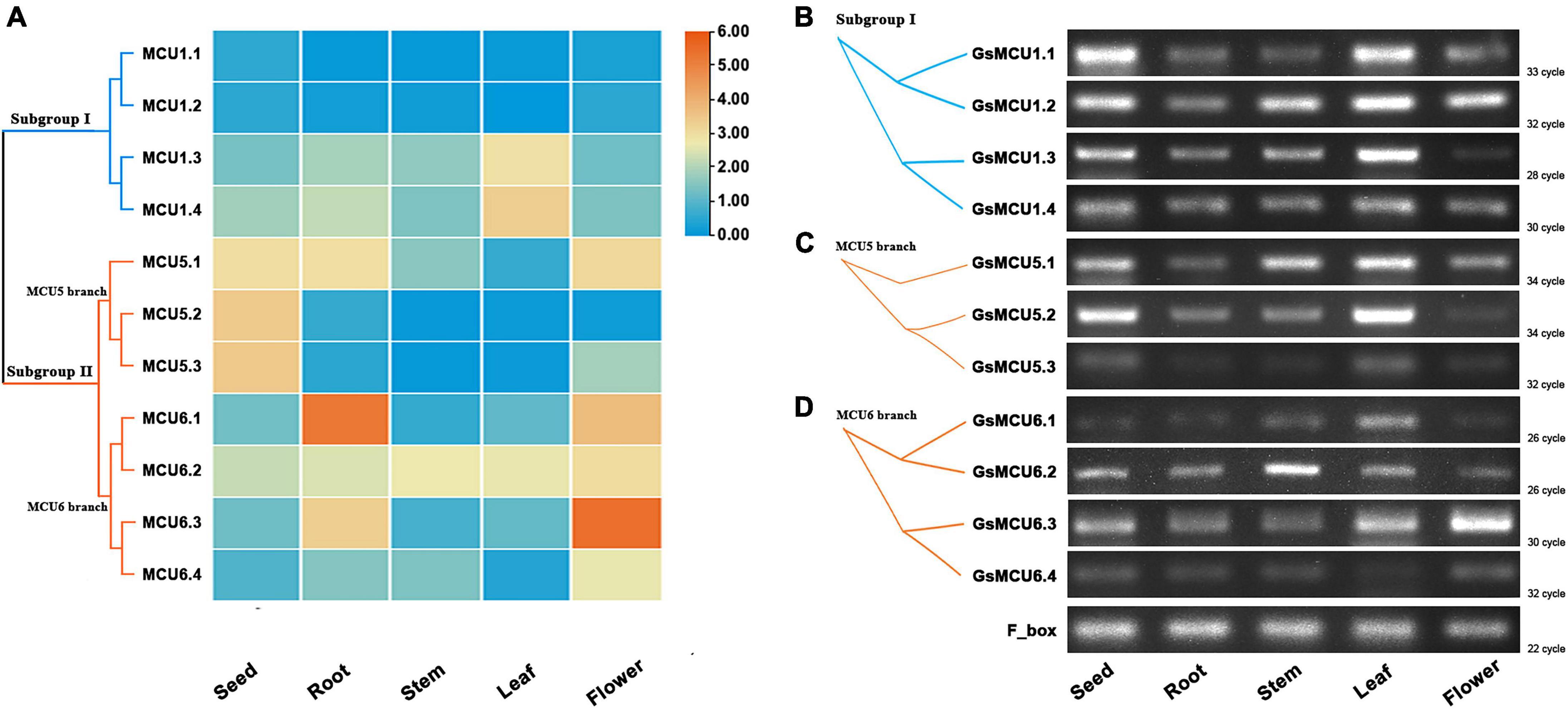
Figure 6. Expression of GsMCU genes in different tissues. (A) Heat map showing the expression levels of soybean MCUs in different tissues. The color scale represents the expression values: red indicates high levels and blue represents low levels. (B–D) RT-PCR assays for GsMCUs expression in different tissues. F_box (GlysoPI483463.12G046600) was used as an internal reference.
To date, few studies have reported the function of plant MCUs in abiotic stress response. Considering the severe effects of low temperature and saline-sodic stress on soybean yields, we further analyzed the expression patterns of GsMCUs under cold and carbonate alkaline treatments to explore their potential roles in these stress responses.
Previous RNA-seq data (Yamasaki and Randall, 2016; Robison et al., 2019) showed that six members of the soybean MCU family genes responded to cold treatment (Figure 7). To confirm the differential expression of the GsMCUs after cold treatment, we performed quantitative real-time PCR assays and found that the expression of three MCU6 branch genes (GsMCU6.1, −6.3, −6.4 in subgroup II) decreased at 1 h and then increased at 24 h after cold stress. However, the expression of GsMCU5.2 and GsMCU5.3 from the MCU5 branch in subgroup II was repressed by cold stress. Furthermore, in subgroup I, GsMCU1.2 expression was greatly increased at 24 h of cold treatment. These expression data imply that these GsMCUs may play different roles in cold stress response.
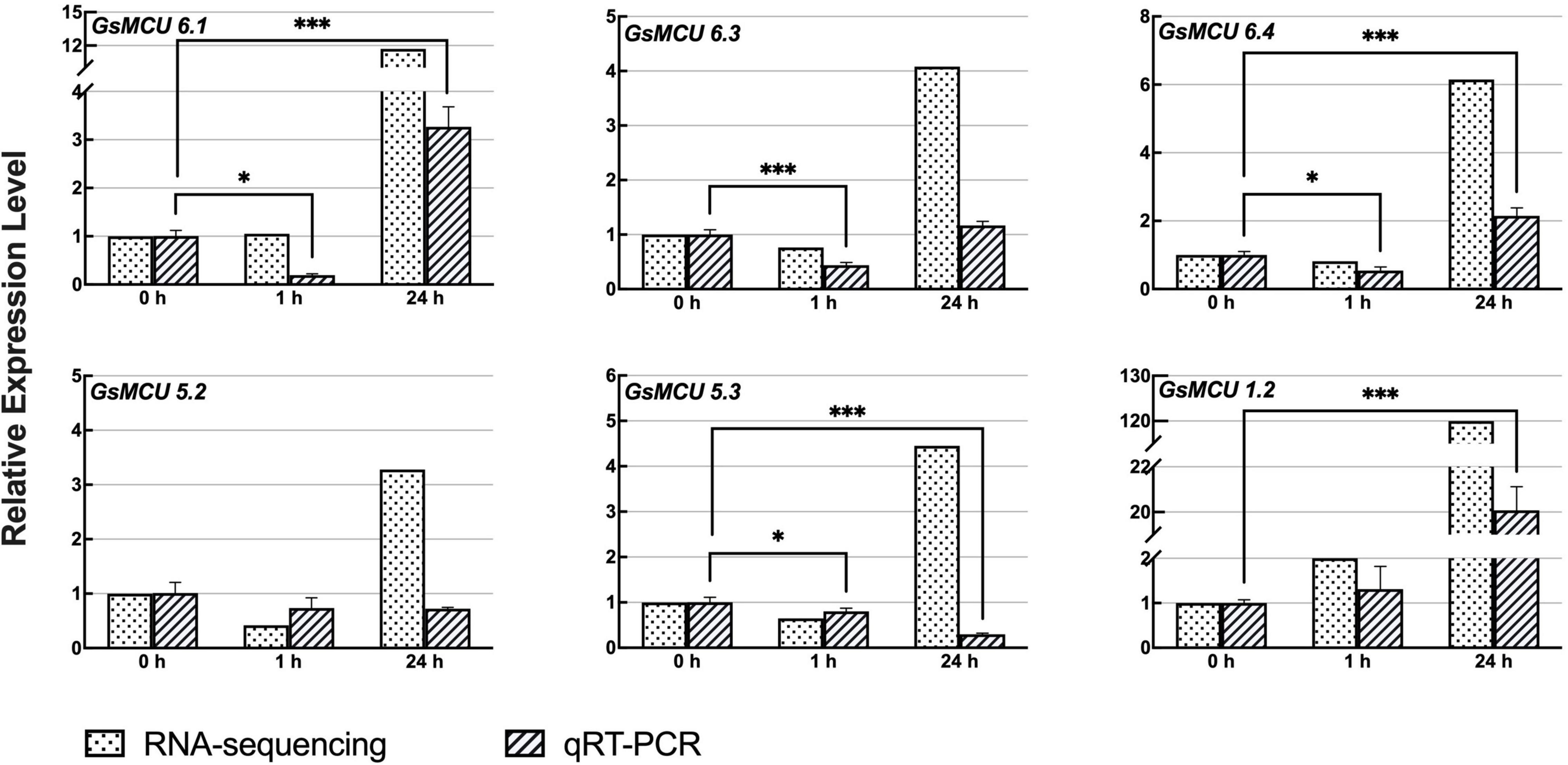
Figure 7. Expression analyses of wild soybean MCU family genes under cold stress. To induce cold stress, 2-week-old wild soybean seedlings were treated at 4°C for 0, 1, and 24 h. GlysoPI483463.12G046600 was used as an internal reference. Mean values (±SE) of the three fully independent biological repeats and three technical repeats are shown. Asterisks indicate significant differences (*p < 0.05, ***p < 0.001 using Student’s t-test).
According to our previous RNA-seq data, nine GsMCUs showed increased expression after carbonate alkaline treatment (50 mM NaHCO3, pH 8.5). As shown in Figure 8, qRT-PCR results showed that the expression of these nine GsMCUs was altered by carbonate alkaline treatment. In details, the expression of four genes (GsMCU6.1, −6.2, −1.1, and −1.3) was significantly up-regulated at all detected time points after NaHCO3 treatment. GsMCU5.2 and GsMCU5.3 displayed decreased expression at 1 h, followed by an increase. These results suggest that GsMCU genes may participate in soybean response to carbonate alkaline stress.
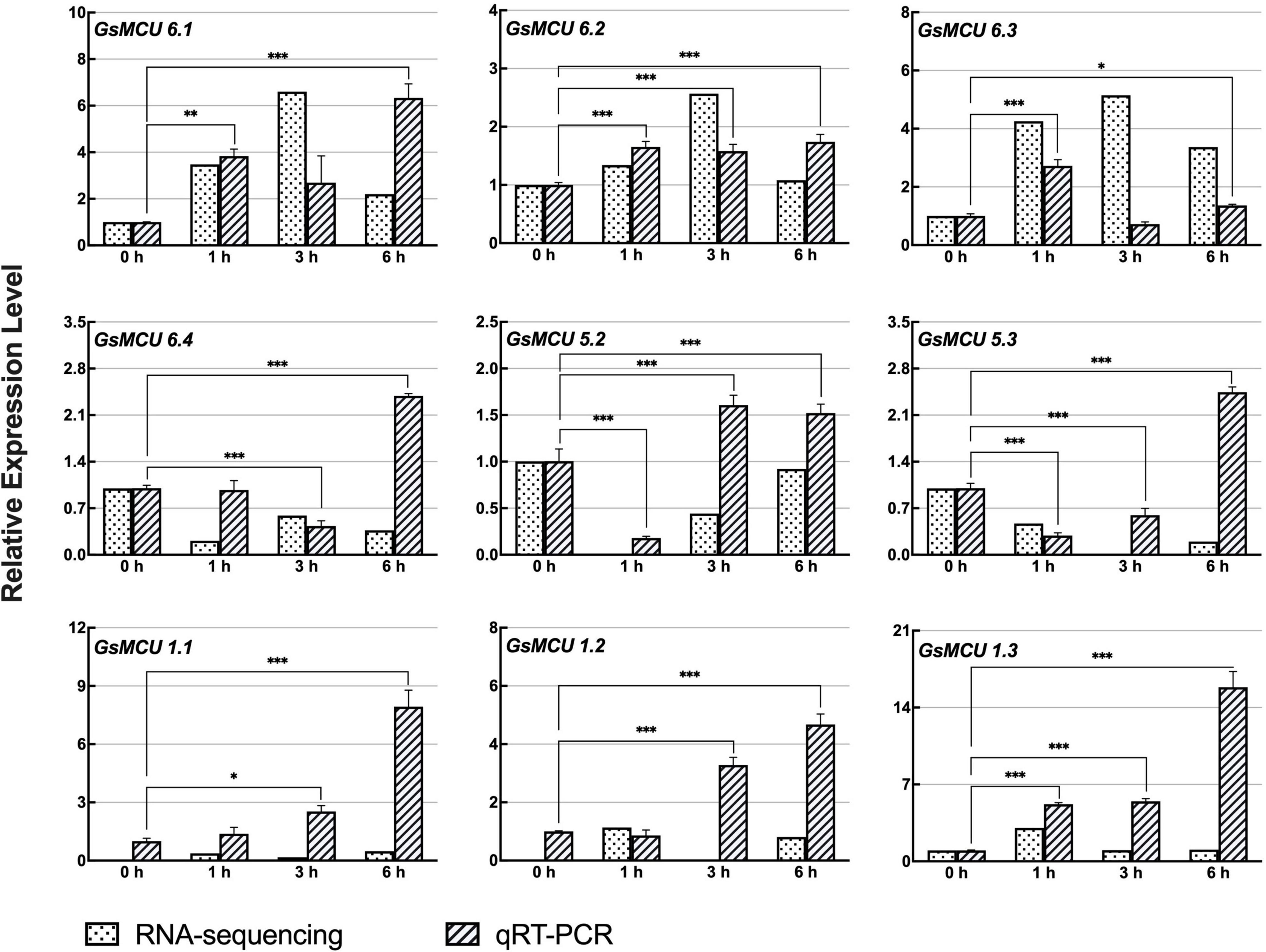
Figure 8. Expression analyses of wild soybean MCU family genes under NaHCO3 stress. For NaHCO3 stress, the 21-day-old wild soybean seedlings were treated with 50 mM NaHCO3 for 0, 1, 3, and 6 h. GlysoPI483463.12G046600 was used as an internal reference. The mean values (±SE) from three fully independent biological repeats and two technical repeats are shown. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t-test).
To further understand the changes in expression of GsMCUs under cold and carbonate alkaline stresses, we predicted stress-related cis-acting elements in their promoters (2 kb upstream of the transcription start site) based on the New PLACE database (Table 2). We found that two cold-related cis-acting elements, LTR (CCGAAA) and DRE/CRT (TACCGACAT), were observed in the promoters of GsMCU1.1, GsMCU5.1, GsMCU6.2, and GsMCU6.4. Moreover, phytohormones play well-documented and critical roles in stress responses (Fahad et al., 2015; Manasa et al., 2021). Several cis-elements related to phytohormones have also been found in GsMCU promoters, such as ABA (ABRE, core sequence ACGTG) (Gusta et al., 2005), JA (CGTCA, core sequence CGTCA) (Kosova et al., 2012), GA (GARE, core sequence TCTGTTG) (Manasa et al., 2021), and SA (TCA, core sequence CCATCTTTTT) (Miura and Ohta, 2010). Notably, cis-elements related to ABA and JA, which have been suggested to play key roles in cold stress, were enriched in the GsMCU promoters, indicating that GsMCU expression may respond to cold stress via ABA and JA signaling pathways.
To explore the potential function of GsMCUs, we used the STRING database to generate a protein interaction network for each GsMCU. Remarkably, the predicted interacting proteins of GsMCUs within each subgroup were identical. Therefore, we proposed two networks for the GsMCU family (Figure 9): network I (for subgroup I) and network II (for subgroup II). Each network contained 10 interacting partners, among which seven proteins were the same between networks I and II. The same interacting proteins between network I and network II further supported that the GsMCU family was a highly conserved gene family and possibly shared similar molecular and biological functions.
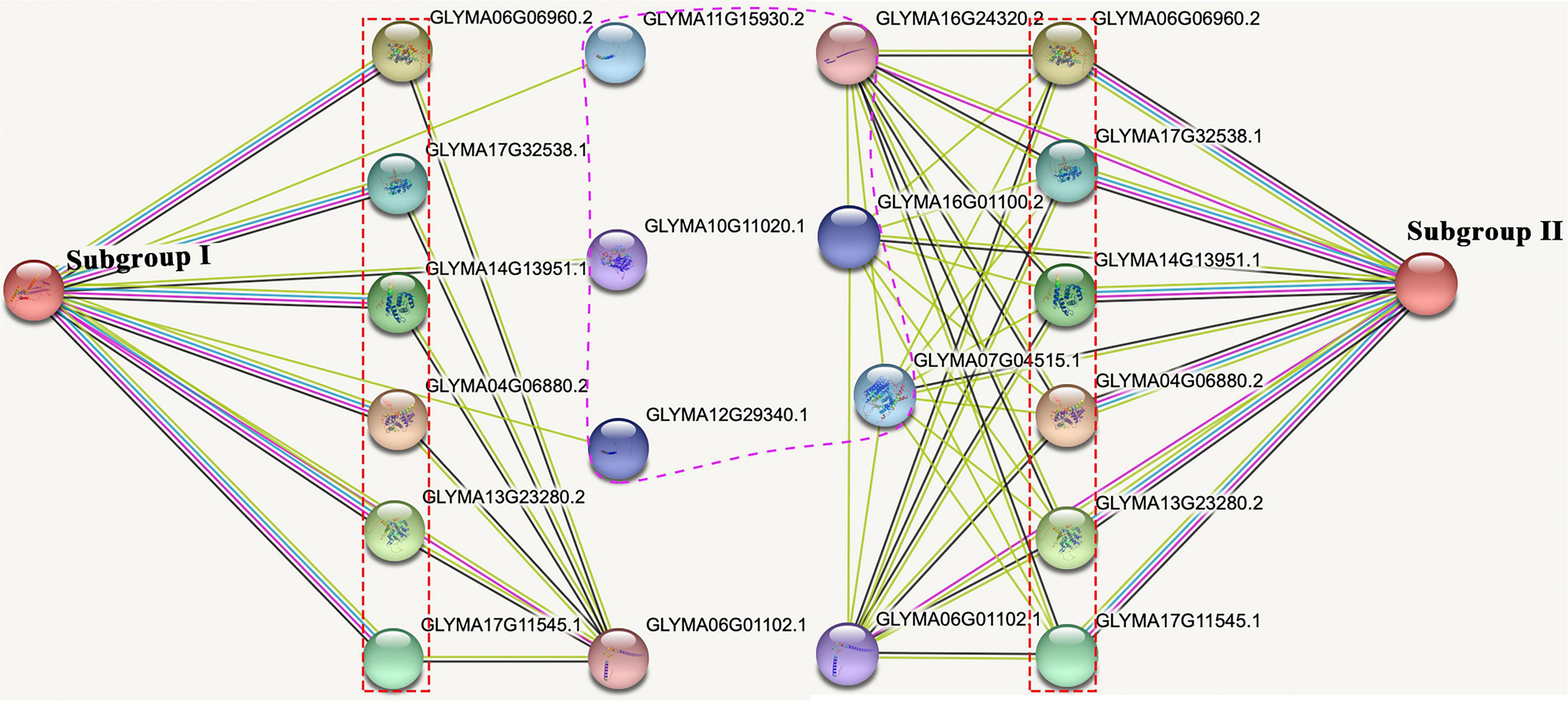
Figure 9. Protein-protein interaction network of soybean MCU proteins. The experimentally determined interacting proteins of the GsMCUs are marked with red boxes. Differences in interacting proteins between the two subgroups are marked by a purple dotted line.
Notably, among the seven proteins shared by networks I and II are six mitochondrial calcium uptake (MICU) proteins that help transport Ca2+ along electrochemical gradients by directly interacting with MCUs. Within network I, Glyma11g15930 and Glyma12g29340 encode transport protein Sec61 subunit beta, which forms the Sec61 complex for transporting newly synthesized polypeptides into the ER lumen and allows calcium leakage from the ER into the cytosol (Schmutz et al., 2010). Glyma10g11020 in network I encodes a calcium-dependent protein kinase that senses Ca2+ oscillations and mediates Ca2+ signal transduction. Within network II, Glyma16g01100 and Glyma07g04515 belong to the Ca2+ cation antiporter (CaCA) superfamily, which play a vital role in Ca2+ ion homeostasis (Table 3). In summary, the investigation of GsMCU-interacting proteins strongly suggests that GsMCUs play a regulatory role in intracellular Ca2+ signal transduction.
It has been suggested that the MCU genes regulate Ca2+ signal transduction and participate in diverse physiological processes. Although mammalian MCU genes have been well characterized, the biological functions and regulatory roles of plant MCUs are unknown. Here, we systematically investigated the wild soybean MCU family genes and provided clues regarding their potential function in soybean development and abiotic stress responses.
In this study, we identified a total of 11 GsMCU genes and suggested that the GsMCU family was a highly conserved family. First, all GsMCUs contained one long intron at a similar position, indicating a conserved gene structure (Figure 3). Secondly, all GsMCU proteins harbored one conserved MCU domain, with two predicted transmembrane regions (Figure 4A). It has been suggested that the pentamerization of transmembrane regions is required for the formation of a functional channel (Oxenoid et al., 2016). MEME analysis further showed high similarity in conserved motif organization of the MCU domain, as well as the N- and C-terminal regions (Figure 4B), suggesting a conserved protein structure. Previous studies reported that the N-terminal domain was important for the Ca2+ uptake activity of the MCU proteins in human (Lee et al., 2015). Further studies are need to identify the function of conserved domains in Ca2+ transport activity. Third, based on the PGDD database, we identified a total of 21 duplicated pairs among 11 GsMCU genes, possibly due to whole-genome duplication of soybean during evolution (Figure 2). Synteny analysis also identified several duplicated MCU gene pairs in soybean, alfalfa, and Arabidopsis (Supplementary Figure 1). Similarly, whole-genome duplication is the main driving force for pear MCU family expansion (Wang et al., 2018). Moreover, MCUs from different plant species were clustered into two subgroups and showed similar phylogenetic relationships (Figure 1). In the phylogenetic tree, the GsMCUs appeared in pairs. Previous studies have also reported that MCUs from Arabidopsis and pear were clustered into two subgroups (Stael et al., 2012; Wang et al., 2018). Finally, GsMCUs within each subgroup shared the same interacting proteins in the STRING network prediction (Figure 9). In human, MICU1 is suggested to be associated with the mitochondrial inner membrane and is required for high-capacity mitochondrial Ca2+ uptake (Perocchi et al., 2010). In Arabidopsis, MICU was demonstrated to regulate mitochondrial Ca2+ dynamics in intact plants (Wagner et al., 2015). Therefore, it is important to validate the consensus protein interaction of GsMCU family members. Taken together, our results strongly demonstrate that the GsMCU family is highly evolutionarily conserved.
Although the MCU family is conserved, they display a certain degree of diversity in subcellular localization and tissue-specific expression characteristics. In Arabidopsis, AtMCU1–AtMCU5 localize in the mitochondria (Teardo et al., 2017; Nomura and Shiina, 2021) and AtMCU6 localizes in the chloroplast envelope (Carraretto et al., 2016; Teardo et al., 2019). Therefore, further studies are required to verify the mitochondria localization of GsMCU1s and GsMCU5s, as well as the chloroplast localization of GsMCU6s. Moreover, our studies also showed that GsMCU6.2 and GsMCU6.3 from the MCU6 branch were highly expressed in stems and flowers, respectively, while members from the MCU5 branch were highly expressed in seeds and leaves (Figure 6). Similarly, AtMCU genes also show specific expression characteristics in different tissues (Teardo et al., 2017, 2019). For example, AtMCU1 displayed the highest expression in roots and an intermediate expression in young leaves, fully expanded leaves, old leaves, and flowers, but no expression (below the detection limit) in seeds (Teardo et al., 2017). AtMCU4 also showed the highest expression in roots (Teardo et al., 2017). AtMCU6 is expressed in all tissues, with higher levels in the mature leaves (Teardo et al., 2019). Furthermore, PbrMCUs also exhibited different expression profiles in different parts of “Housui” pear fruit (Wang et al., 2018). The diversity of MCUs in terms of subcellular localization and tissue expression might be the main driving force for their potential function specialization in regulating plant growth and development.
Several studies have reported the role of MCU genes in plant development. Interestingly, both AtMCU1 knockout and overexpression caused shorter primary roots under restrictive growth conditions (Teardo et al., 2017). AtMCU2 knockout impairs Arabidopsis pollen tube germination and growth (Selles et al., 2018). Arabidopsis plants lacking AtMCU6 exhibit constitutive stomatal closure (Teardo et al., 2019). Furthermore, three MCU genes are involved in pear ripening (Wang et al., 2018). In future, genetic data regarding GsMCUs overexpression or knockout will be of great importance for understanding their biological roles during soybean growth and development.
Ca2+ signal transduction is of great importance in plant responses to various abiotic stresses including drought, salt and cold. In this study, we identified that six GsMCUs responded to cold stress (Figure 7), and nine GsMCUs showed increased expression under carbonate alkaline treatment (Figure 8). In particular, the MCU6 branch genes were upregulated under both cold and carbonate alkaline stresses, while GsMCU5s were downregulated. This difference indicates potential functional specialization of subgroup II MCUs. Members within the MCU6 and MCU5 branches, as well as three subgroup I members, showed roughly the same expression pattern under stress treatment, suggestive of possible functional redundancy. A recent study has reported that AtMCU6 regulates osmotic stress responses (Teardo et al., 2019). AtMCU6 knockout plants displayed increased resistance to long-term water deficits and improved recovery after re-watering. Therefore, functional validation of GsMCUs by using overexpressing or gene-editing soybean is particularly important for investigating their precise biological roles and molecular basis in regulating the cold and carbonate alkaline stress tolerance.
Furthermore, we observed several stress-related cis-acting elements in the promoters of GsMCUs, including two cold-related cis-acting elements, LTR (CCGAAA) and DRE/CRT (TACCGACAT), as well as ABA (ABRE, core sequence ACGTG) and JA (CGTCA, core sequence CGTCA) responsive cis-elements (Table 2). A previous study showed that PbrMCU expression is affected by another type of phytohormone, ethylene (Wang et al., 2018). Hence, more experiments are need to verify the hormone induced expression of GsMCUs, and the binding of key transcription factors, for example CBF/DREB1s, to GsMCUs promoters. Furthermore, the genetic evidence is also required in future to confirm whether GsMCUs participate in cold and carbonate alkaline stress via the ABA and JA signaling pathways.
In this study, we identified the MCU family genes in wild soybean and investigated their expression under cold and carbonate alkaline stresses. Eleven GsMCUs were identified and clustered into two subgroups. Gene duplication was detected in the GsMCU family, and the duplicated pairs displayed a similar exon-intron architecture. All GsMCU proteins contained one conserved MCU domain and two transmembrane helices, showing high conservation in protein sequence and structure. Moreover, all GsMCUs were expressed ubiquitously in different tissues and organs, and six and nine GsMCUs were differentially expressed under cold and carbonate alkaline stress, respectively. Our results provide valuable information about the soybean MCU family, which will facilitate functional analysis of GsMCUs in the future.
Publicly available datasets were analyzed in this study. This data can be found here: The datasets for the RNAseq experiments can be found on NCBI GEO (Accessions # GSE117686 and GSE17883) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117686 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17883).
XS and KY guided the design of the experiment and revised the manuscript. JL and MS conducted data analysis and manuscript writing. YL finished plant material handling. All authors reviewed and agreed to the published version of the manuscript.
This work was financially supported by the Natural Science Foundation of Heilongjiang Province (JQ2021C002), the National Natural Science Foundation of China (32101672 and 31771692), Special Funds from the Central Finance to Support the Development of Local Universities (to XS), Fund Program for the Scientific Activities of Selected Returned Overseas Professionals in Heilongjiang Province (to XS), Funding for the Opening Project of Key Laboratory of Soybean Biology of Chinese Education Ministry (SBKF08), Fund Program for PHD student in Heilongjiang Bayi Agricultural University (YJSCX2018-Z11). The funders have no role in the study design, data analysis, and manuscript writing, but just provide the financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.867503/full#supplementary-material
Bansal, R., Mittapelly, P., Cassone, B. J., Mamidala, P., Redinbaugh, M. G., and Michel, A. (2015). Recommended reference genes for quantitative PCR analysis in soybean have variable stabilities during diverse biotic stresses. PLoS One 10:e0134890. doi: 10.1371/journal.pone.0134890
Baughman, J. M., Perocchi, F., Girgis, H. S., Plovanich, M., Belcher-Timme, C. A., Sancak, Y., et al. (2011). Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345. doi: 10.1038/nature10234
Bisbach, C. M., Hutto, R. A., Poria, D., Cleghorn, W. M., Abbas, F., Vinberg, F., et al. (2020). Mitochondrial Calcium Uniporter (MCU) deficiency reveals an alternate path for Ca2+ uptake in photoreceptor mitochondria. Sci Rep 10:16041. doi: 10.1038/s41598-020-72708-x
Carraretto, L., Teardo, E., Checchetto, V., Finazzi, G., Uozumi, N., and Szabo, I. (2016). Ion channels in plant bioenergetic organelles, chloroplasts and mitochondria: from molecular identification to function. Mol. Plant 9, 371–395. doi: 10.1016/j.molp.2015.12.004
Cheng, C. Y., Krishnakumar, V., Chan, A. P., Thibaud-Nissen, F., Schobel, S., and Town, C. D. (2017). Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89, 789–804. doi: 10.1111/tpj.13415
Consortium, A. I. M., Dreze, M., Carvunis, A.-R., Charloteaux, B., Galli, M., Pevzner, S. J., et al. (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607. doi: 10.1126/science.1203877
De Stefani, D., Raffaello, A., Teardo, E., Szabò, I., and Rizzuto, R. (2011). A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340. doi: 10.1038/nature10230
Deluca, H. F., and Engstrom, G. W. (1961). Calcium uptake by rat kidney mitochondria. Proc. Natl. Acad. Sci. U.S.A. 47, 1744–1750. doi: 10.1073/pnas.47.11.1744
Drago, I., Pizzo, P., and Pozzan, T. (2011). After half a century mitochondrial calcium in-and efflux machineries reveal themselves. EMBO J. 30, 4119–4125. doi: 10.1038/emboj.2011.337
Fahad, S., Hussain, S., Matloob, A., Khan, F. A., Khaliq, A., Saud, S., et al. (2015). Phytohormones and plant responses to salinity stress: a review. Plant Growth Regul. 75, 391–404. doi: 10.1007/s10725-014-0013-y
Fujikura, U., Ezaki, K., Horiguchi, G., Seo, M., Kanno, Y., Kamiya, Y., et al. (2020). Suppression of class I compensated cell enlargement by xs2 mutation is mediated by salicylic acid signaling. PLoS Genet. 16:e1008873. doi: 10.1371/journal.pgen.1008873
Gu, L. (2020). Identification and expression analysis of MCU protein family genes in Sorghum bicolor. Bot. Res. 9, 169–179. doi: 10.12677/br.2020.93021
Gusta, L., Trischuk, R., and Weiser, C. (2005). Plant cold acclimation: the role of abscisic acid. J. Plant Growth Regul. 24, 308–318. doi: 10.1007/s00344-005-0079-x
Kofsky, J., Zhang, H., and Song, B. H. (2018). The untapped genetic reservoir: the past, current, and future applications of the wild soybean (Glycine soja). Front. Plant Sci. 9:949. doi: 10.3389/fpls.2018.00949
Kosova, K., Prasil, I. T., Vitamvas, P., Dobrev, P., Motyka, V., Flokova, K., et al. (2012). Complex phytohormone responses during the cold acclimation of two wheat cultivars differing in cold tolerance, winter Samanta and spring Sandra. J. Plant Physiol. 169, 567–576. doi: 10.1016/j.jplph.2011.12.013
Le, D. T., Aldrich, D. L., Valliyodan, B., Watanabe, Y., Ha, C. V., Nishiyama, R., et al. (2012). Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS One 7:e46487. doi: 10.1371/journal.pone.0046487
Lee, Y., Min, C. K., Kim, T. G., Song, H. K., Lim, Y., Kim, D., et al. (2015). Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter. EMBO Rep. 16, 1318–1333.
Liu, Y., Jin, M., Wang, Y., Zhu, J., Tan, R., Zhao, J., et al. (2020). MCU-induced mitochondrial calcium uptake promotes mitochondrial biogenesis and colorectal cancer growth. Signal. Transduct. Target. Ther. 5:59. doi: 10.1038/s41392-020-0155-5
Mallilankaraman, K., Doonan, P., Cárdenas, C., Chandramoorthy, H. C., Müller, M., Miller, R., et al. (2012). MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca2+ uptake that regulates cell survival. Cell 151, 630–644. doi: 10.1016/j.cell.2012.10.011
Manasa, S. L., Panigrahy, M., Panigrahi, K. C. S., and Rout, G. R. (2021). Overview of cold stress regulation in plants. Bot. Rev. doi: 10.1007/s12229-021-09267-x
Miura, K., and Ohta, M. (2010). SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J. Plant Physiol. 167, 555–560. doi: 10.1007/10.1016/j.jplph.2009.11.003
Nomura, H., and Shiina, T. (2021). “Calcium transport systems in chloroplasts and mitochondria of plant cells,” in Calcium Transport Elements in Plants ed. S. K. Upadhyay (Cambridge, MA: Academic Press), 337–371. doi: 10.1016/b978-0-12-821792-4.00023-0
Oxenoid, K., Dong, Y., Cao, C., Cui, T., Sancak, Y., Markhard, A. L., et al. (2016). Architecture of the mitochondrial calcium uniporter. Nature 533, 269–273. doi: 10.1038/nature17656
Perocchi, F., Gohil, V. M., Girgis, H. S., Bao, X. R., McCombs, J. E., Palmer, A. E., et al. (2010). MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature 467, 291–296. doi: 10.1038/nature09358
Robison, J. D., Yamasaki, Y., and Randall, S. K. (2019). The ethylene signaling pathway negatively impacts CBF/DREB-regulated cold response in soybean (Glycine max). Front. Plant Sci. 10:121. doi: 10.3389/fpls.2019.00121
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Selles, B., Michaud, C., Xiong, T. C., Leblanc, O., and Ingouff, M. (2018). Arabidopsis pollen tube germination and growth depend on the mitochondrial calcium uniporter complex. New Phytol. 219, 58–65. doi: 10.1111/nph.15189
Shi, S., Li, S., Asim, M., Mao, J., Xu, D., Ullah, Z., et al. (2018). The Arabidopsis calcium-dependent protein kinases (CDPKs) and their roles in plant growth regulation and abiotic stress responses. Int. J. Mol. Sci. 19:1900. doi: 10.3390/ijms19071900
Stael, S., Wurzinger, B., Mair, A., Mehlmer, N., Vothknecht, U. C., and Teige, M. (2012). Plant organellar calcium signalling: an emerging field. J. Exp. Bot. 63, 1525–1542. doi: 10.1093/jxb/err394
Teardo, E., Carraretto, L., Moscatiello, R., Cendron, L., Cortese, E., Vicario, M., et al. (2019). A chloroplast-localized mitochondrial calcium uniporter transduces osmotic stress in Arabidopsis. Nat. Plants 5, 581–588. doi: 10.1038/s41477-019-0434-8
Teardo, E., Carraretto, L., Wagner, S., Formentin, E., Behera, S., De Bortoli, S., et al. (2017). Physiological characterization of a plant mitochondrial calcium uniporter in vitro and in vivo. Plant Physiol. 173, 1355–1370. doi: 10.1104/pp.16.01359
Vasington, F. D., and Murphy, J. V. (1962). Ca++ uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J. Biol. Chem. 237, 2670–2677. doi: 10.1016/s0021-9258(19)73805-8
Wagner, S., Behera, S., De Bortoli, S., Logan, D. C., Fuchs, P., Carraretto, L., et al. (2015). The EF-hand Ca2+ binding protein MICU choreographs mitochondrial Ca2+ dynamics in Arabidopsis. Plant Cell 27, 3190–3212. doi: 10.1105/tpc.15.00509
Wang, L., Wang, L., Chen, Q., Chen, G., Zhang, S., Wu, J., et al. (2018). Identification and comparative analysis of the MCU gene family in pear and its functions during fruit ripening. J. Plant Physiol. 229, 53–62. doi: 10.1016/j.jplph.2018.07.001
Wang, M., and Teng, Y. (2018). Genome-wide identification and analysis of MICU genes in land plants and their potential role in calcium stress. Gene 670, 174–181. doi: 10.1016/j.gene.2018.05.102
Keywords: wild soybean, mitochondrial calcium uniporter, cold stress, carbonate alkali stress, expression analysis
Citation: Li J, Sun M, Liu Y, Sun X and Yin K (2022) Genome-Wide Identification of Wild Soybean Mitochondrial Calcium Uniporter Family Genes and Their Responses to Cold and Carbonate Alkaline Stresses. Front. Plant Sci. 13:867503. doi: 10.3389/fpls.2022.867503
Received: 01 February 2022; Accepted: 13 April 2022;
Published: 03 May 2022.
Edited by:
Yingfang Zhu, Henan University, ChinaReviewed by:
Yuanhu Xuan, Shenyang Agricultural University, ChinaCopyright © 2022 Li, Sun, Liu, Sun and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Sun, c3VueGlhb2xpMjAxNkBieWF1LmVkdS5jbg==; Kuide Yin, eWlua3VpZGVAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.