- 1Plant Genetics Laboratory, National Institute of Genetics, Mishima, Japan
- 2Plant Breeding Laboratory, Faculty of Agriculture, Kyushu University, Fukuoka, Japan
Gene duplication plays an important role in genetic diversification, adaptive evolution, and speciation. Understanding the mechanisms and effects of postzygotic isolation genes is important for further studies of speciation and crop breeding. The duplicate recessive genes hwe1 and hwe2 cause hybrid breakdown, characterized by poor vegetative growth and reproductive dysgenesis in intersubspecific crosses between Oryza sativa ssp. indica and japonica. Using a map-based cloning strategy, we found that HWE1 and HWE2 encode the Esa1-associated factor 6 (EAF6) protein, a component of histone acetyltransferase complexes. The indica hwe1 and japonica hwe2 alleles lacked functional EAF6, demonstrating that the double recessive homozygote causes hybrid breakdown. Morphological and physiological observations showed that weak plants with double recessive homozygotes had serious morphological defects with a wide range of effects on development and organs, leading to leaves with reduced chlorophyll content, flower and pistil malformation, and anomalies of gametogenesis. These findings suggest that EAF6 plays a pivotal role in the transcriptional regulation of essential genes during the vegetative and reproductive development of rice.
Introduction
In eukaryotic cells, histone acetylation regulates the chromatin structure, affecting gene transcription, DNA replication, and DNA damage repair. Nucleosome acetyltransferase of histone 4 (NuA4), a histone acetyltransferase (HAT) complex, is composed of multiple proteins and preferentially acetylates histones H4 and H2A on the nucleosome. The components of NuA4 are highly conserved in yeast and human (Doyon et al., 2004). Yeast NuA4 consists of 13 subunits, with two independent NuA4 sub-complexes, namely, piccolo-NuA4, composed of Esa1, Epl1, Yng2, and Eaf6, and the TINTIN triad of Eaf5/7/3 (Wang X. et al., 2018). Piccolo-NuA4, which is thought to also exist alone, contains the catalytic subunit protein essential Sas2-related acetyltransferase-1 (Esa1) (Ohba et al., 1999; Boudreault et al., 2003). Esa1 alone can acetylate free histones but cannot acetylate nucleosomal histones (Doyon et al., 2004). This protein also plays a crucial role in cell cycle progression and DNA double-strand break repair (Clarke et al., 1999; Bird et al., 2002) and is essential for yeast cell viability (Doyon et al., 2004). Another component, Esa1-associated factor 6 (EAF6, known as MEAF6 in mammals), interacts with Piccolo-NuA4 through Yng2 in yeast (Mitchell et al., 2008). Unlike the catalytic subunit Esa1, the yeast eaf6Δ mutant is viable without detectable changes, indicating that the NuA4 subunit is not essential for yeast cellular processes (Lafon et al., 2007). However, a study in human showed that a fusion protein of MEAF6 with PHD finger protein 1 generated by chromosomal translocation caused endometrial stromal tumors in human (Panagopoulos et al., 2012). These findings suggest that EAF6 (MEAF6) is important in cell proliferation. Yeast EAF6 is also a component of another HAT complex, nucleosome acetyltransferase of histone 3, which acetylates histone H3. This HAT complex was reported to be involved in transcriptional activation and cell cycle regulation (Lafon et al., 2007). Compared with the number of studies performed to characterize NuA4 components in yeast and mammals, studies in plants are limited. Two components of the NuA4 complex, namely, EAF1 and YAF9, were found to regulate flowering via histone H4 acetylation in Arabidopsis (Zacharaki et al., 2012; Bieluszewski et al., 2015). Double mutations in HAM1 and HAM2 (Arabidopsis ESA1 homologs) induced lethality in diploid plants and haploid gametophytes, suggesting a function in histone acetylation during mitotic cell division of gametogenesis in Arabidopsis (Latrasse et al., 2008). The plant EAF6 protein is an uncharacterized potential subunit of plant NuA4; less is known about its biological functions in plant development and growth.
Hybrid breakdown is defined as deleterious characteristics, such as sterility and non-viability, occurring only after F2 generations of crosses between distantly related species. Similar to other hybrid incompatibility mechanisms, hybrid breakdown contributes to speciation by restricting gene flow between diverging taxa. Although such phenomena are widely observed in numerous animal and plant species (Stebbins, 1958), few hybrid breakdown genes have been identified and characterized at the molecular level. Seminal studies in plants demonstrated that autoimmune responses involving nucleotide-binding site-leucine-rich repeat genes can cause hybrid weakness and breakdown (Bomblies et al., 2007; Alcazar et al., 2010). A further systematic study using a large diallel cross containing more than 6,400 cross combinations in Arabidopsis revealed that one hybrid necrosis gene, Dangerous Mix 2 (DM2), plays a central role in the epistatic network involving numerous independent loci related to hybrid necrosis (Chae et al., 2014). Such autoimmune systems cause hybrid incompatibility, including the hybrid breakdown in other plant species (Hannah et al., 2007; Jeuken et al., 2009; Yamamoto et al., 2010; Chen et al., 2014). Previous studies demonstrated that defense systems against biotic stress, including programmed cell death and nucleotide-binding site-leucine-rich repeats, are major and common causes of hybrid weakness and hybrid breakdown in plant species. Although progress has been made recently in identifying the genes involved in hybrid incompatibility, the molecular basis of hybrid breakdown other than the autoimmune response is poorly understood in plants. In many cases, the causal genes remain unknown. Whether other physiological mechanisms underlie hybrid weakness and how different genes contribute to genetic diversification and speciation are also unclear.
Our previous study demonstrated hybrid breakdown characterized by weak growth and complete sterility between Oryza sativa ssp. indica and japonica (2n = 24). Genetic analysis has revealed that this weakness is caused by double recessive genes, hwe1 and hwe2, which are localized on rice chromosomes 1 and 12, respectively (Kubo and Yoshimura, 2002). Although the phenomenon and basic genetics of this hybrid breakdown were characterized more than a decade ago, the molecular mechanism is not well-understood. The specific objectives of this study were to isolate hwe1 and hwe2, characterize the abnormal phenotype of the weak plant morphologically and physiologically, and identify the physiological function of the causal genes at the molecular level.
Materials and Methods
Plant Materials
The characterization of weak plants and high-resolution mapping was carried out using the backcross population derived from the population previously used for rough mapping of hwe1 and hwe2 (Kubo and Yoshimura, 2002). Additionally, we used two other indica/japonica populations, namely, BC2F2 derived from the Nipponbare/93-11 cross (Kubo et al., 2011) and a newly developed Nipponbare/IR8 F2 population.
Characterization of Morphological and Physiological Traits
Normal and weak segregants (BC3F6–7, n = 10) were evaluated to determine their seed fertility, column length, and number. The chlorophyll content was examined using the last fully opened leaf blades from each genotype (n = 5) at the tillering stage. Seed fertility was evaluated as previously described (Kubo et al., 2016). To evaluate leaf cell viability related to the autoimmune response, leaves from normal and weak plants during vegetative development were collected and stained with trypan blue. For staining, the detached leaves were completely submerged in lactic acid-phenol-trypan blue solution (0.5 mg/ml trypan blue, 25% phenol, 25% lactic acid, and 25% glycerol) and microwaved for 1.0 min in a domestic microwave oven. The tissue was destained by placing the samples in staining solution without trypan blue and overnight incubation. The tissue was transferred to 50% ethanol and observed under a stereomicroscope.
Map-Based Cloning
To perform high-resolution mapping of the hwe1 and hwe2 loci, seedlings of the segregating populations (approximately 2,387 BC3F5–6 plants for hwe2 and 383 BC3F5–6 individuals for hwe1) were genotyped using polymerase chain reaction (PCR)-based markers, and plants with recombination around the hwe1 and hwe2 loci were identified. PCR-based markers, insertion and deletion markers, and simple sequence repeat markers were identified using sequence polymorphism data for Nipponbare and 93-11 (MSU7.0)1. The primer sequences for the DNA markers are listed in Supplementary Table 1. For DNA marker genotyping, crude DNA extracts of seedling leaves were prepared using 0.25 M NaOH followed by neutralization with 0.1 M Tris–HCl. These DNA extracts were used in PCR with GoTaq polymerase (Promega, Madison, WI, United States) and the following cycling profile: 94°C for 2 min, followed by 30 cycles of 94°C for 20 s, 50–60°C for 20 s, and 72°C for 30 s.
Gene Cloning and Rice Transformation
Gene cloning and allelic diversity analyses were carried out using PCR analysis of purified DNA from rice varieties and wild accessions prepared using the CTAB method (Murray and Thompson, 1980), followed by sequencing analysis on an ABI Prism 3130xl genetic analyzer (Applied Biosystems, Foster City, CA, United States). For the complementation test, the Asominori genomic DNA fragment (7,023 bp) containing the LOC_Os12g20310 gene with flanking 5′ (2,872 bp) and 3′ (1,487 bp) regions was amplified with KOD-Plus-Neo DNA polymerase (Toyobo, Osaka, Japan) using the primer G20310 (Supplementary Table 1). For another candidate, LOC_Os12g20324, the Asominori genomic DNA fragment (7,689 bp) was amplified by PCR using the primer G20324. The amplified fragments were cloned into the pBluescript SK cloning vector and subcloned into the pPZP2H-lac binary vector (Fuse et al., 2001). The cloned genomic fragments and empty vector were transformed into HWE1 heterozygotes (Hwe1/hwe1hwe2/hwe2) via Agrobacterium tumefaciens-mediated transformation (Hiei et al., 1994; Nishimura et al., 2006). Complementation was examined based on the phenotype of the selfed progeny (T1 and T2) of the T0 transformant.
Reverse Transcription-PCR Analysis
Total RNA from Asominori plant tissues (i.e., leaf, stem, root, young, and flowering panicles) and NILs were prepared using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). cDNA was generated by reverse transcription of 2.0 μg of total RNA using the SuperScript III First-Strand Synthesis System (Life Technologies, Carlsbad, CA, United States). RT-PCR analysis was performed using two primer sets (i.e., eaf6sp and eaf6co) (Supplementary Table 1) to discriminate the products from the Nipponbare HWE1 and HWE2 loci. The cDNA of OsAct1 (rice Actin 1) was amplified using the Act1 primer and used as a standard control. RT-PCR was performed in a Biometra thermocycler with the following cycling profile: 94°C for 2 min, followed by 32 cycles at 94°C for 20 s, 58°C for 20 s, and 72°C for 30 s.
Histological Experiments
To observe the embryo sac, pre-flowering panicles were collected from normal and weak plants, fixed, and stored in FAA solution (45% ethanol, 5% formalin, and 5% acetic acid). After fixation, the samples were embedded in paraffin (Paraplast Plus; McCormick Scientific, St. Louis, MO, United States), sectioned, and stained with hematoxylin. To observe the morphology of the mature pollen grains, ethanol-fixed pollen grains were stained with 1.0% iodine-potassium iodide (I2-KI) and observed under a microscope. Male gametogenesis was analyzed using young panicles collected from normal and weak plants at different developmental stages. Panicles were fixed and stored in FAA solution. After fixation, the micropores were extracted from the anther using forceps, and released micropores were stained with hematoxylin solution as described by Kindiger and Beckett (1985).
Histochemical Analysis of Beta-Glucuronidase Expression
An Asominori genome fragment containing 2,872 bp of the upstream region of OsEAF6 (LOC_Os12g20310) was amplified using the primer set G20310pro (Supplementary Table 1) and cloned into the Kpn I-Spe I site of the binary vector pBGH2 (Ito and Kurata, 2008) to drive beta-glucuronidase (GUS) expression. The resulting construct, ProOsEAF6:GUS, was transformed into Asominori plants. For GUS staining, tissue samples (i.e., leaf, young spikelets, and stem) were vacuum-infiltrated with staining solution (50 mM Na3PO4, pH 7.0, 1.0 mg/ml 5-bromo-4-chloro-3-indolyl-β-glucuronide, and 0.5% Triton X-100) and incubated at 37°C for 16 h. The stained samples were fixed for 10 min in formaldehyde, acetic acid, and 22% ethanol (5:5:90, v/v) and then destained in 100% ethanol until the chlorophyll was removed.
Subcellular Localization Analysis
Transient expression assays using polyethylene glycol-mediated transformation were performed as previously described (Shen et al., 2014). The 35S:OsEAF6-mCherry construct was prepared by amplifying mCherry from the pmCherry-C1 vector (Takara) and inserting it into the plant binary vector pRI201-ON (Takara). The coding sequence (CDS) of OsEAF6 (LOC_Os12g20310) was cloned into the pCR-Blunt II TOPO vector (Life Technologies) and transferred into the mCherry-pRI201-ON vector. Rice Oc cells were provided by RIKEN BRC through the National Bio-Resource Project of MEXT, Japan. Protoplasts from Oc cells were adjusted to a concentration of 1.0–2.0 × 106 cells/ml; a 0.1-ml aliquot was transfected with 10–20 μg plasmids. After 16 h of incubation at 28°C, transformed cells were observed under an optical/fluorescence microscope (Biozero BZ-8000, Keyence, Osaka, Japan).
Genome Sequence and Haplotype Analysis
The genome sequences of cultivated and wild rice accessions were downloaded from the Gramene database2. Genome sequences of the japonica variety Nipponbare (IRGSP version 1.0) and indica varieties IR8 (IOMAP version 1) and 93-11 (ASM465v1) were used to compare chromosomal structural differences between indica and japonica around the HWE1 and HWE2 regions. The genomic sequences were identified using GenomeMatcher version 2.03 software (Ohtsubo et al., 2008). The genome sequences of the following species were also used: Oryza nivara, accession W0106 (version AWHD00000000); Oryza rufipogon, W1943 (PRJEB4137); Oryza barthii, IRGC105608 (ABRL00000000); Oryza glaberrima, IRGC96717 (AGI1.1); Oryza glumaepatula, GEN1233_2 (ALNU02000000); Oryza meridionalis, W2112 (Oryza_meridionalis_v1.3); Oryza punctata, IRGC105690 (AVCL01000000); Oryza brachyantha, IRGC101232 (AGAT00000000), and Leersia perrieri, IRGC105164 (version 1.4). Gene annotation of Leersia was based on the Gramene database, and the others were based on RiceGAAS (Sakata et al., 2002). Haplotype analysis of the HWE1 and HWE2 loci was performed based on published SNP data for cultivated and wild rice compared with the Nipponbare genome. SNP data for cultivars and wild rice were obtained from the International Rice Information System3 and OryzaGenome4, respectively.
Phylogenetic Analysis of EAF6 Protein Homologs
A search for EAF6 homologs in different organisms was performed using the BLASTP program (NCBI) website. Protein sequences of 25 plant species and 8 microbe and animal species were obtained from phytozome5 and the NCBI database (Supplementary Table 2). Protein sequences were aligned using ClustalW to construct a phylogenetic tree using the neighbor-joining method (Saitou and Nei, 1987) and MEGA6 software (Tamura et al., 2013). Bootstrap values were calculated with 1,000 replications.
Results
Characteristics of Hybrid Breakdown
A single substitution with indica (cv. IR24) chromosome 12 in a japonica (Asominori) genetic background causes hybrid breakdown characterized by poor growth and complete sterility (Kubo and Yoshimura, 2002). Simple epistasis with double recessive genes, named hwe1 and hwe2, was sufficient to explain this hybrid breakdown (Figures 1A–C). Similarly, genetic analysis showed that epistasis between hwe1 and hwe2 caused a hybrid breakdown in other cross combinations (Nipponbare/93-11 and Nipponbare/IR8) (Supplementary Figures 1, 2). The weak phenotype was characterized by shorter and smaller culm lengths, partially sheathed panicles, and both male and female sterility (Figures 1B–F and Supplementary Figures 3A,B). The weak segregant appeared as pale green compared with normal plants at the adult stage, likely due to the lower chlorophyll content in the leaves (approximately half of that in the normal segregant) (Figure 1F). Leaves from weak plants during the vegetative growth stage were not stained by trypan blue (Figures 1G,H), suggesting that this weak phenotype did not result from an autoimmune response by nucleotide-binding site-leucine-rich repeat or other related molecules, as previously reported (Bomblies et al., 2007; Alcazar et al., 2010). A much more severe phenotype was observed in the reproductive organs. Weak plants produced smaller panicles and yielded fewer spikelets compared with normal plants (Figures 1B,F). An abnormal phenotype was also found in the reproductive organs, such as depressed palea or palea-less flowers, degenerated anthers, and abnormal stigma formation (Supplementary Figures 4A–L). Microscopy revealed that pollen sterility was attributed to meiotic defects, including abnormal or incomplete cell division (Supplementary Figures 3C–M). Although the female gametes were also completely sterile, various ovules swollen by imbibition were observed without fertilization in weak plants (Supplementary Figure 4M).
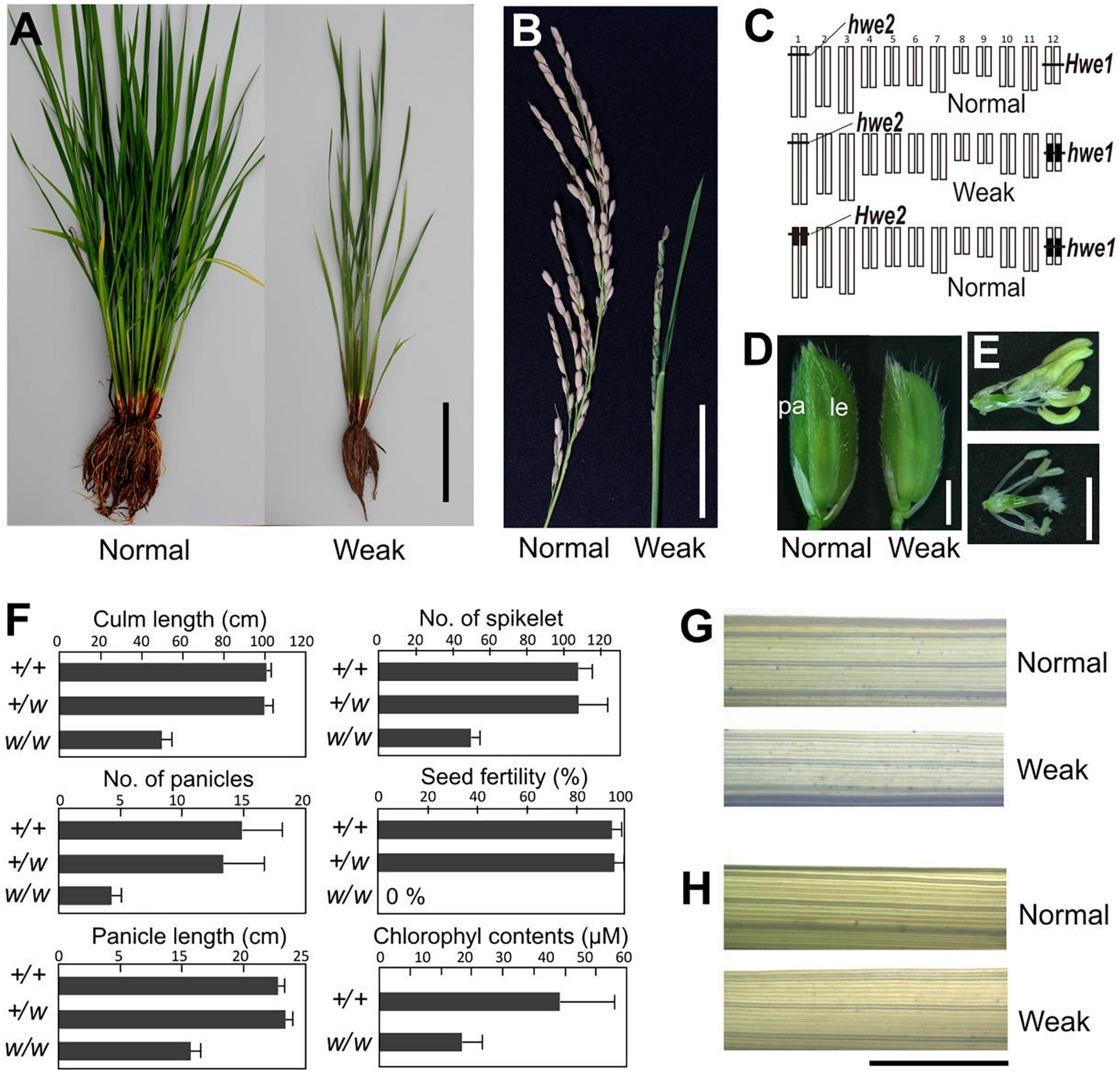
Figure 1. Morphology of weak plants. (A) Plant morphology at vegetative stage 3 weeks before heading. (B) Morphology of panicle at the mature seed stage. Weak plants exhibited a partially sheathed panicle. (C) Graphical genotype of weak plants. White and black bars represent Asominori and IR24 chromosomes, respectively. Only the double recessive homozygote showed the weakness phenotype. (D) Spikelets of normal (left) and weak plants (right). Le, lemma; pa, palea. (E) Flower organs of normal (upper) and weak plants (lower). (F) Characterization of weak plants. Column length, number of panicles, number of spikelets per panicle, seed fertility, and leaf chlorophyll contents at 3 weeks before flowering. +/+: Hwe1/Hwe1 hwe2/hwe2, +/w: Hwe1/hwe1, hwe2/hwe2, and w/w: hwe1/hwe1 hwe2/hwe2. N = 10 for each genotype excluding the chlorophyll contents (N = 5). (G,H) Trypan blue staining to assess viability leaf blade cell. Third youngest leaf blades from normal and weak plants at the developmental stage before heading [(G), 90 days after sowing] and after heading [(H), 120 days after sowing) were stained by trypan blue. Neither showed remarkable cell lethality. Scale bar = 10.0 cm in (A), 5.0 cm in (B), 2.0 mm in (D,E), and 5.0 mm in (G,H).
Identification of HWE1 and HWE2
HWE1 and HWE2 have been roughly mapped to rice chromosomes 12 and 1, respectively (Kubo and Yoshimura, 2002). Using the small-scale mapping population (N = 383), we identified the HWE1 locus within a 5.4-Mb region close to the centromere of chromosome 12 (Figure 2A). As meiotic recombination is repressed around the centromere and pericentromeric regions, we then focused on the partner gene HWE2. To isolate HWE2, we screened recombinant individuals from a large segregating population (N = 2,387). The result showed that the HWE2 locus was delimited within a 38.7-kb region between the PCR markers 1c215 and 1c219, which encoded four predicted genes (i.e., LOC_Os01g13210, LOC_Os01g13229, LOC_Os01g13250, and LOC_Os01g13260) (Figure 2A). Of these four genes, two genes (i.e., LOC_Os01g13250 and LOC_Os01g13260) shared homology with LOC_Os12g20310 and LOC_Os12g20324 in the HWE1 region, indicating a small segmental duplication between chromosomes 1 and 12 of the Nipponbare genome. In comparative sequence analysis, the DNA sequence of the indica Hwe2 allele showed much greater similarity to that of the japonica Hwe1 allele than to that of the japonica hwe2 allele (Supplementary Figure 5). However, there were no segmental blocks corresponding to LOC_Os12g20310 and LOC_Os12g20324 on indica chromosome 12 (93-11 and IR8 genomes). In the context of double recessive epistasis, these duplicated genes were considered good candidates for HWE1/2. The LOC_Os01g13250 and LOC_Os12g20310 loci encode EAF6, and the adjacent LOC_Os01g13260 and LOC_Os12g20324 encode the cyclin-A1 protein. We performed complementation analysis to determine whether one or both of these genes contribute to hybrid breakdown. Due to the complete sterility of weak segregants, we transformed Hwe1/hwe1 heterozygotes with Asominori genomic DNA containing LOC_Os12g20310 or LOC_Os12g20324 and then evaluated the complementation in their selfed progeny (T1 and T2 generations). The resultant transformants with LOC_Os12g20310 recovered the weak growth phenotype with complete sterility, whereas the other transformants with LOC_Os12g20324 did not (Supplementary Figure 6 and Supplementary Tables 3, 4). This result indicates that OsEAF6, encoded by HWE1/2, was responsible for the hybrid breakdown. A full-length cDNA of OsEAF6 has been previously cloned (GenBank, CT830710). The gene structure of the Nipponbare allele of LOC_Os01g13250 was predicted to encode a variant form of OsEAF6 with extra amino acids (14 extra amino acids) in the C-terminus (Figures 2B,C and Supplementary Figure 7). We predicted that this variant form (hwe2-j) was functionally defective.
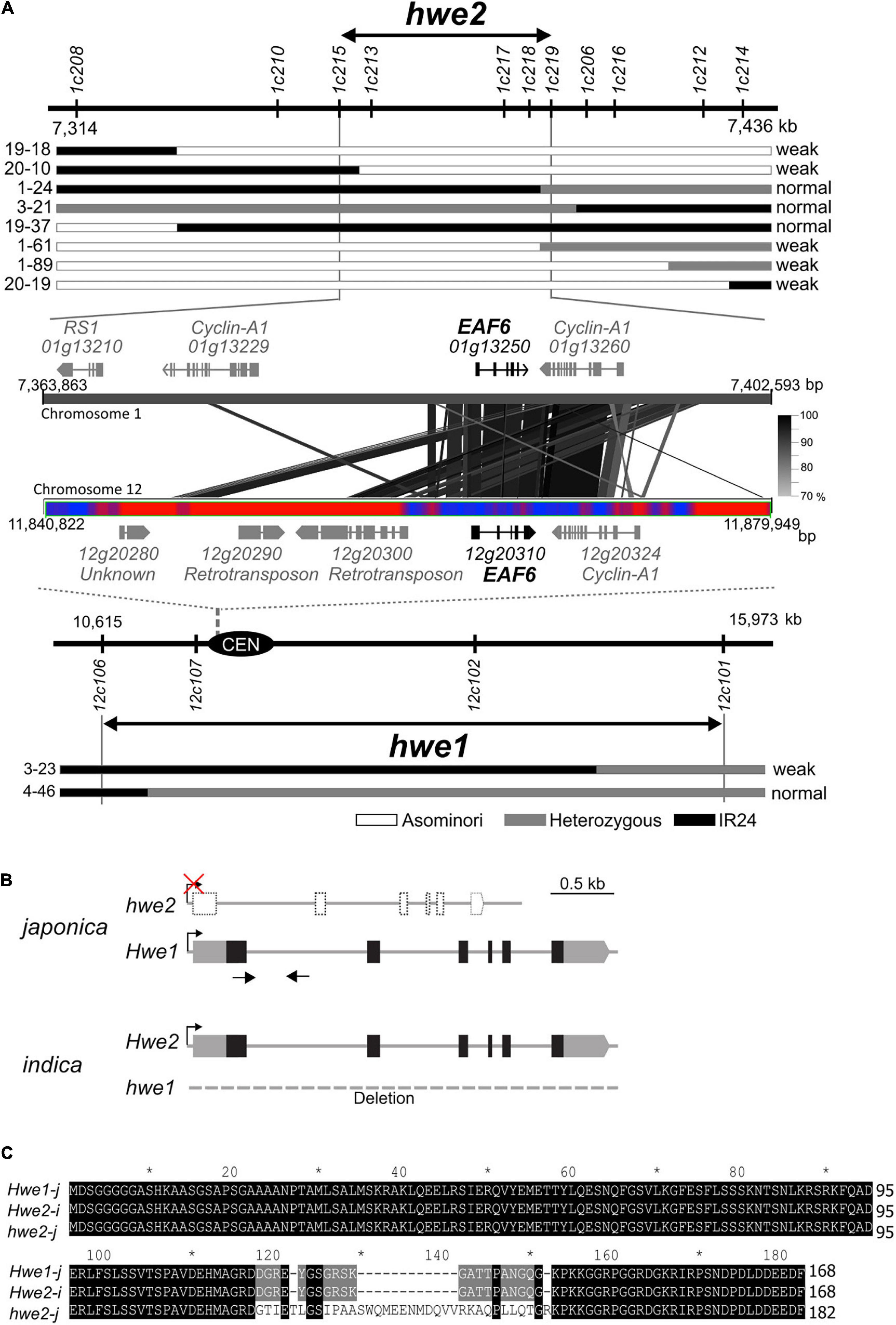
Figure 2. Identification of causal genes for hwe1 and hwe2. (A) Map locations of hwe2 (upper) and hwe1 (lower), and MSU7.0 gene annotation in candidate regions. The prefix “LOC_” in locus ID is omitted for convenience. The most informative recombinant genotypes are shown along with the maps. Similarity of DNA sequences between chromosomes 1 and 12 is shown as connected with black and gray lines. The bar with red and blue heat map represents the abundance of repetitive DNA sequences around the centromere (CEN) of chromosome 12. Red represents the repeat region. (B) Gene structure of HWE1 and HWE2 of japonica (Asominori) and indica (93-11). Gray and black boxes denote untranslated region (UTR) and coding sequence (CDS) of rice OsEAF6, respectively. The CDS sequences of OsEAF6 were identical between japonica and indica. (C) Multiple alignments of predicted OsEAF6 protein sequences of japonica (Hwe1-j and hwe2-j) and indica (Hwe2-i).
Hybrid Breakdown Attributed to Loss of OsEAF6 Expression
To verify this hypothesis, we examined the expression of LOC_Os12g20310 and LOC_Os01g13250 in Asominori and IR24 using RT-PCR with two primer sets, namely, eaf6sp and eaf6co. The first primer set, eaf6sp, was specific to Nipponbare LOC_Os12g20310 but not to LOC_Os01g13250, which encodes the valiant form. The second primer, eaf6co, matched the conserved identical sequence between LOC_Os12g20310 and LOC_Os01g13250 and was used to examine the presence of the variant mRNA from LOC_Os01g13250 (Figure 3A). The mRNA from LOC_Os12g20310 was ubiquitously expressed in all organs examined, except for the root, which showed faint expression (Figure 3B). OsEAF6 was not expressed in the weak plants, whereas co-introgression with the IR24 segment around the HWE1 and HWE2 loci recovered the expression (Figure 3C), indicating that active OsEAF6 mRNA was generated from the japonica Hwe1-j and indica Hwe2-i alleles. The absence of the mRNA signal from LOC_Os01g13250 with the eaf6co primer indicated that japonica hwe2-j is an OsEAF6 pseudogene (Figure 3C). Thus, these results support that defects were present in functional OsEAF6 on the recessive hwe2-j and hwe1-i alleles and indicate that the lack of OsEAF6 on the hwe1 and hwe2 alleles induces hybrid breakdown.
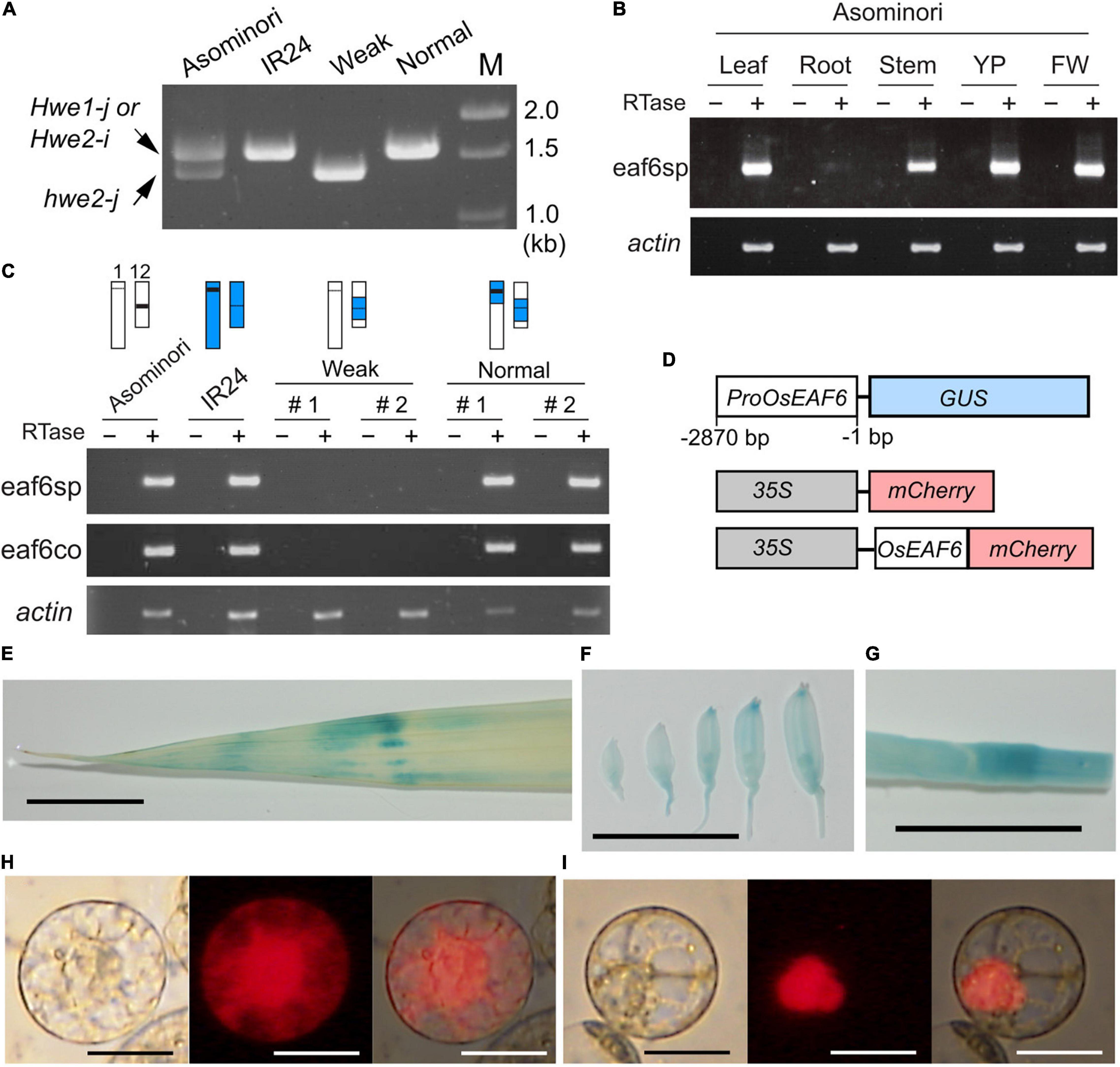
Figure 3. Expression pattern and cellular localization of the OsEAF6. (A) Amplified genomic DNA product obtained with the primer eaf6co. The expected sizes of DNA bands were 1,557 bp from Hwe1-j and Hwe2-i and 1,352 bp from hwe2-j. The primer eaf6co was designed for the identical sequence region between Hwe1-j (functional gene) and hwe2-j (pseudogene). It is noted that the eaf6co primer could amplify the product from genomic DNA but not from mRNA, as shown in (C). (B) mRNA expression of OsEAF6 in rice tissues detected using RT-PCR analysis. eaf6sp is a primer set specific to functional OsEAF6 [japonica Hwe1 (Hwe1-j) allele and indica Hwe2 (Hwe2-i) allele] but not the pseudogene on the hwe2-j allele. YP, young panicle; FW, flowering panicles (C) mRNA expression of OsEAF6 in the NILs and parents with the two different primer sets by RT-PCR analysis. Genotype of each sample for chromosomes 1 and 12 is shown on the upper part. White and blue bars represent Asominori and IR24 chromosomes, respectively. Two individuals for the weak and normal plants (#1 and 2) are shown. (D) Diagram of constructs designed for expression analysis of OsEAF6. (E–G) GUS expression analysis of OsEAF6 by using ProOsEAF6:GUS expresser in leaf (E), developing flowers (F), and stem (G) tissues. (H,I) Subcellular localization of OsEAF6 protein fused with mCherry (I) and mCherry alone (H) in rice protoplast cells. Transient expression of each fluorescent construct was observed. Left, DIC image; middle, mCherry channel image; right, merged image. Scale bar = 1.0 cm in (E–G), and 20 μm in (H–I).
Tissue and Subcellular Localization of OsEAF6
In higher plants, EAF6 proteins are 144–170 amino acids long and highly conserved throughout herbaceous and woody plants (Supplementary Figure 8 and Supplementary Table 2). Their sequences are partially conserved with those of yeast and animal species. Yeast (Saccharomyces cerevisiae) EAF6 is a small protein (113 amino acids) and subunit of the NuA4 HAT complex that is involved in transcriptional regulation through nuclear H4 acetylation (Mitchell et al., 2008). To gain insight into the function of OsEAF6, we examined the expression of OsEAF6 in transgenic rice plants with ProOsEAF6:GUS (Figure 3C). GUS expression was observed in vegetative organs, including the leaves and stems, and in developing spikelets (Figures 3D–G). This result is consistent with those of RT-PCR analysis. Subcellular localization analysis using rice Oc cells showed that OsEAF6 protein was present predominantly in the nucleus and, to a lesser extent, in the cytoplasm, whereas the control mCherry plasmid was detectable throughout the cell (Figures 3H,I). This result indicates that OsEAF6 functions in the nucleus.
Evolution of EAF6 in Oryza Species
In the rice genome sequencing project, 450 O. rufipogon accessions and 3,000 cultivars were sequenced using next-generation sequencing techniques (Huang et al., 2012; Wang W. et al., 2018). Based on published genome sequence data, we investigated the distribution of hwe1 and hwe2 alleles in cultivars and their wild relative O. rufipogon. The allelic diversity of duplicated hwe loci was examined based on eight SNPs at the 5′ and 3′ terminal regions of OsEAF6, which can discriminate between alleles in HWE1 and HWE2. The Nipponbare Hwe1 allele (called Nip-type) is GGAA-ATTT and is common among the O. sativa-O. rufipogon complex. The 93-11 hwe1 null allele (9311-type) appeared to be distributed in the indica ecotype (255 accessions) but was minor in O. rufipogon (Figures 4A,B and Supplementary Table 5). We characterized the Nipponbare hwe2 null allele as TCGC-ATTT (Nip-type) and 9311 Hwe2 allele as GGAA-GGCC. Most japonica subspecies varieties (99.6%, 250/251) and 30–44% of O. rufipogon Or-I and Or-III ecotypes carried the Nip-type hwe2 allele. Since Or-III has been reported as a progenitor of japonica, hwe2 of japonica rice may have originated from Or-III. Some O. rufipogon accessions and O. sativa ssp. indica “aus” ecotype had two copies of functional OsEAF6 on chromosomes 1 and 12 (Supplementary Figure 7).
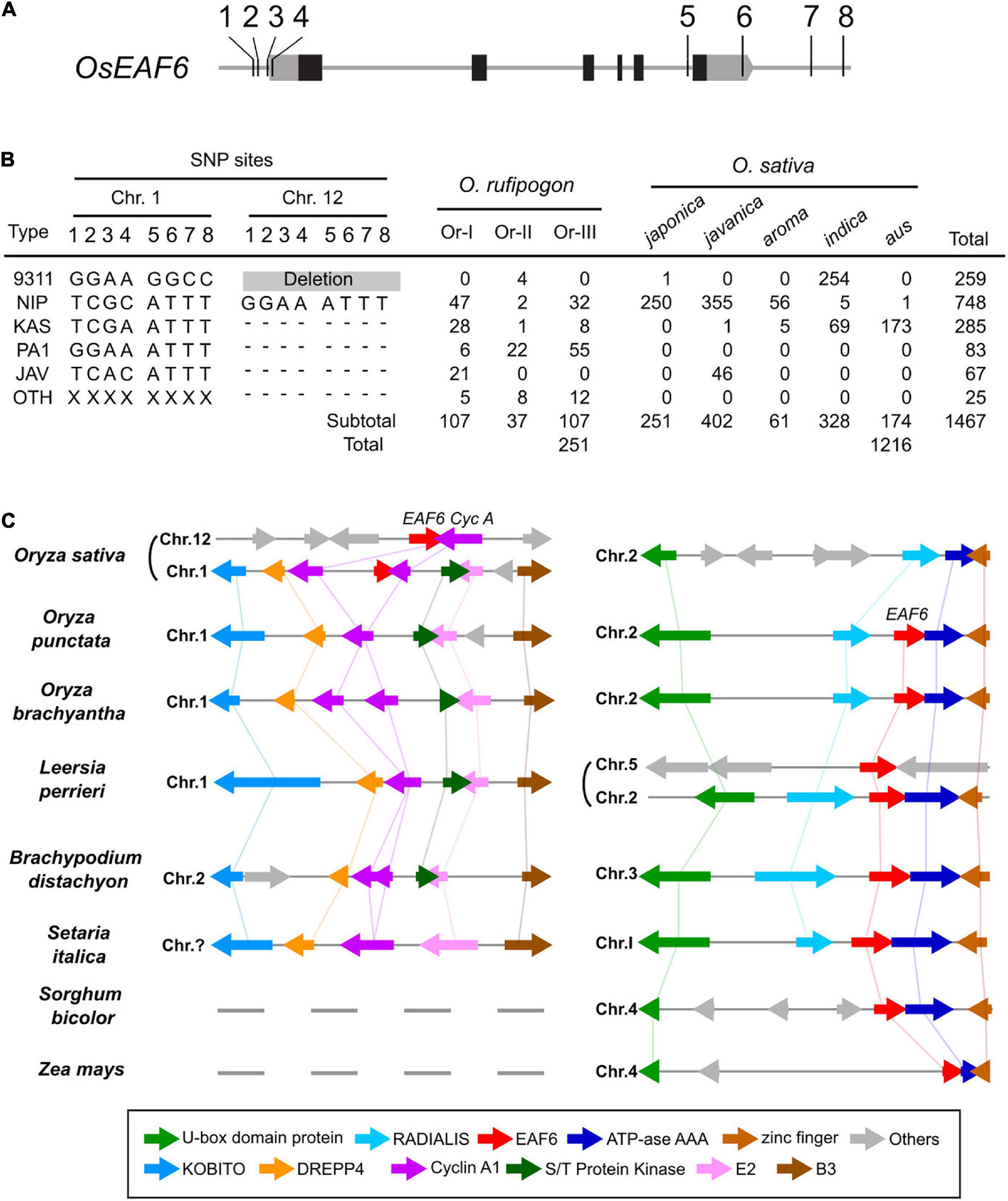
Figure 4. Allele distribution of OsEAF6 and syntenic gene analysis of OsEAF6 region. (A) Distribution of hwe1 and hwe2 alleles in Oryza sativa–Oryza rufipogon complex. Positions of eight SNPs (1–8) on the 5′ and 3′ regions of OsEAF6. Gray and black boxes denote UTR and CDS of rice OsEAF6, respectively. SNP site 1, -309 bp on chr.1 and -304 bp on chr.12; site 2, -299 bp on chr.1 and -294 bp on chr.12; site 3, -205 bp on chr.1 and -206 bp on chr.12; site 4, -157 bp on chr.1 and -158 bp on chr.12; site 5, +2,133 bp on chr.1 and +2,527 bp on chr.12; site 6, +2,494 bp on chr.1 and +2,890 bp on chr.12; site 7, +2,971 bp on chr.1, and site 8, +3,153 bp on chr.12 and +3,555 bp on chr.12. The positions of SNPs (bp) were based on the Nipponbare sequence [+1 refers to (A) in the ATG start codon]. (B) Distribution of six haplotypes of duplicated OsEAF6 in O. rufipogon and O. sativa. We evaluated 251 accessions for O. rufipogon and 1,216 accessions for O. sativa. The six haplotypes based on eight SNP sites consist of 93-11-type (9311), Nipponbare-type (NIP), Kasalath-type (KAS), PA1-type (PA1), javanica type (JAV), and other type (OTH). Hyphens represent identical SNPs with the Nipponbare SNP. (C) Conserved syntenic block harboring EAF6 and adjacent Cyclin-A1 loci. Gene annotation of the region surrounding the EAF6 (right) and Cyclin-A1 (left) loci in the cereal species. Homologous genes are shown in the same color connected by straight lines.
To determine the origin and timing of EAF6 duplication, EAF6 homologs were investigated in wild Oryza species. We first analyzed the AA genome species most closely related to O. sativa. A BLAST search of the LOC_Os12g20310 CDS showed hits on chromosome 1 of other AA genome species (Supplementary Figure 9). Then, we investigated a local synteny pattern around the EAF6 locus across distantly related Oryza species and five grass species. Oryza BB and FF genome species and other grass species (Leersia, Brachypodium, and Setaria) showed large blocks of homologous synteny around the HWE2 region of Nipponbare chromosome 1 but lacked the whole sequence of EAF6 on this syntenic block. Instead, these species contained a single copy of EAF6 on other syntenic chromosomes, which were syntenic to O. sativa chromosome 2 (Figure 4C). The closest genus Leersia, which carried two copies of EAF6, showed that the local gene order around the EAF6 locus on Leersia chromosome 5 did not differ among other species, indicating independent duplication events between rice and Leersia. The gene order and orientation around the Cyclin-A1 locus (LOC_Os01g13260) were conserved on chromosome 1 of O. punctata, O. brachyantha, and Leersia as a single copy segment. Based on the alignment of other AA genome species, this result suggests that EAF6 was initially transposed to chromosome 1 from chromosome 2 in the AA genome progenitor, followed by segmental duplication to chromosome 12 (Supplementary Figure 10).
Discussion
We demonstrated that hybrid breakdown is caused by HWE1/2 encoding a rice homolog of the NuA4 HAT complex subunit protein EAF6. The NuA4 HAT complex is an essential transcriptional coactivator involved in gene regulation, cellular processes, and DNA double-strand break repair in eukaryotes. Yeast EAF6 interacts with another catalytic subunit protein, Esa1, via Yng2. The functional role of EAF6 protein in plants remains unclear. We found that the loss of the OsEAF6 protein exerted deleterious pleiotropic effects on both vegetative growth and reproductive development in rice. Particularly, it has a broad impact on reproductive development, ranging from inflorescence development to gametogenesis. During the preparation of this study, Zhou et al. (2022) found that the Arabidopsis eaf6 mutant shows growth inhibition and leaf yellowing and that the NuA4 complex is involved in transcriptional activation, specifically in light-responsive genes. Another research group reported that the loss of Arabidopsis Esa1-associated factor 1 (EAF1) inhibited growth and chloroplast development (Bieluszewski et al., 2022). These findings are consistent with our phenotypic observations, such as growth inhibition with reduced chlorophyll content during the vegetative phase in the double homozygote hwe1/2. It is suggested that such deleterious pleiotropic phenotypes occurred due to disorders of the universal chromatin state and transcriptional regulation caused by the loss of OsEAF6 protein.
Hypothetical Evolutionary History
Extensive genome sequencing and comparative studies revealed conserved microsynteny (gene order patterns) across different cereal species (Jaiswal et al., 2006). Similar to previously reported microsynteny, a conserved gene order around EAF6 was observed among the monocot crop species, including the wild rice relatives Leersia and sorghum (Figure 4C). However, the chromosome position of this synteny block containing EAF6 was not chromosome 12 or chromosome 1, but rather chromosome 2 in other wild rice species (O. punctata and O. brachyantha). We hypothesized the evolutionary history of EAF6 in Oryza genomes as follows: (1) EAF6 has resided on chromosome 2 of primitive Oryza species; (2) EAF6 was transposed to chromosome 1 in an early AA genome progenitor; (3) a segmental duplication occurred and was positioned on chromosome 12 in a subpopulation of O. rufipogon; and (4) one copy of the gene was lost in a progenitor population of O. sativa ssp. japonica (Supplementary Figure 10). This hypothetical scenario was based on the chromosome synteny and distribution of SNPs discriminating the two OsEAF6 copies in the cultivars and their close relatives (Figures 4A,C). Transposition to chromosome 1 was considered for the following reasons. First, Cyclin-A1 is not found on the corresponding region of chromosome 12 in primitive Oryza species. Second, the EAF6 CDS was localized on chromosome 1 according to recent next-generation sequencing analyses of other AA genome species (Supplementary Figure 7). Thus, in evolutionary history, OsEAF6/HWE1 was a copy of OsEAF6/HWE2 following the transposition from chromosome 2. Other grass species retain microsynteny around the EAF6 positions. The mechanisms of transposition and duplication of OsEAF6 remain unclear. Despite the positional differences in EAF6 in grass species, the protein sequence of EAF6 protein is largely conserved among plant species (Supplementary Figure 8), suggesting that it has an essential function in plant development. Therefore, duplicated EAF6 in other plant genomes may function as a reproductive isolation system.
Functional Role as the Reproductive Isolation System
In some animal studies, DNA-binding proteins, such as OdsH, PRDM9, and Zhr, were identified as causal molecules for hybrid sterility (Maheshwari and Barbash, 2011). These factors are likely associated with the dysfunction of chromatin remodeling in heterozygous hybrid progenies. Thus, abnormal chromatin formation during meiotic cell division in hybrids is a common factor responsible for reproductive isolation. From the perspective of the reproductive isolation mechanism, hybrid breakdown by hwe1/2 occurred due to the loss of gene function and differed from the disharmonious interactions in the animal cases mentioned above, although the mechanism of action targeting nucleosomes is similar. Since no remarkable changes were observed in the heterozygous state, we did not characterize the detailed phenotype of heterozygous plants. However, heterozygous plants for each single locus of HWE1 and HWE2 (i.e., Hwe1/hwe1 hwe2/hwe2 and hwe1/hwe1 Hwe2/hwe2) induced reduced transmission of the recessive alleles (hwe1 and hwe2) in the selfed progeny (Kubo and Yoshimura, 2002). Thus, hwe1/2 strongly impacts the elimination of the specific genotype around these genes in the hybrid population. Additionally, OsEAF6 may be involved in the haplotype gamete phase in rice. According to previous microarray data and laser capture microdissection of male and female gametes (Hobo et al., 2008; Kubo et al., 2013), OsEAF6 was substantially and constantly expressed in haploid organs, such as microspores and megaspores (Supplementary Figure 11). Furthermore, the involvement of NuA4 in gametogenesis has been previously reported in Arabidopsis (Latrasse et al., 2008). Therefore, we believe that OsEAF6 may regulate histone acetylation and transcription levels throughout the rice life-cycle including the diploid and haploid phases. Further studies are required to determine the functions of OsEAF6 as a subunit of the HAT complex in various developmental stages and tissues. This study demonstrated the involvement of EAF6 in plant development and reproductive isolation. These findings will provide a helpful clue to transcriptional regulation by histone acetylation in plant development and also aid to develop an efficient breeding program to overcome reproductive isolation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
TK, AY, and NK conceived and designed the experiments. TK performed the experiments, analyzed the data, and wrote the study, with input from AY and NK. All authors read and approved the final manuscript.
Funding
This study was partly supported by the JSPS KAKENHI Grant Numbers 18075009 (to TK and NK) and 20K05985 (to TK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank T. Makino, Y. Gonohe, and H. Kondo (National Institute of Genetics, Mishima, Japan) for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.866404/full#supplementary-material
Footnotes
- ^ http://rice.plantbiology.msu.edu/
- ^ http://ensembl.gramene.org/Oryza_sativa/Info/Index
- ^ http://oryzasnp.org/iric-portal/index.zul
- ^ http://viewer.shigen.info/oryzagenome/mapview/Top.do
- ^ https://phytozome-next.jgi.doe.gov
References
Alcazar, R., Garcia, A. V., Kronholm, I., de Meaux, J., Koornneef, M., Parker, J. E., et al. (2010). Natural variation at Strubbelig receptor kinase 3 drives immune-triggered incompatibilities between Arabidopsis thaliana accessions. Nat. Genet. 42, 1135–1139. doi: 10.1038/ng.704
Bieluszewski, T., Galganski, L., Sura, W., Bieluszewska, A., Abram, M., Ludwikow, A., et al. (2015). AtEAF1 is a potential platform protein for Arabidopsis NuA4 acetyltransferase complex. BMC Plant Biol. 15:75. doi: 10.1186/s12870-015-0461-1
Bieluszewski, T., Sura, W., Dziegielewski, W., Bieluszewska, A., Lachance, C., Kabza, M., et al. (2022). NuA4 and H2A.Z control environmental responses and autotrophic growth in Arabidopsis. Nat. Commun. 13:277. doi: 10.1038/s41467-021-27882-5
Bird, A. W., Yu, D. Y., Pray-Grant, M. G., Qiu, Q., Harmon, K. E., Megee, P. C., et al. (2002). Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419, 411–415. doi: 10.1038/nature01035
Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J. L., et al. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5:e236. doi: 10.1371/journal.pbio.0050236
Boudreault, A. A., Cronier, D., Selleck, W., Lacoste, N., Utley, R. T., Allard, S., et al. (2003). Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17, 1415–1428. doi: 10.1101/GAD.1056603
Chae, E., Bomblies, K., Kim, S.-T., Karelina, D., Zaidem, M., Ossowski, S., et al. (2014). Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159, 1341–1351. doi: 10.1016/j.cell.2014.10.049
Chen, C., Chen, H., Lin, Y. S., Shen, J. B., Shan, J. X., Qi, P., et al. (2014). A two-locus interaction causes interspecific hybrid weakness in rice. Nat. Commun. 5:3357. doi: 10.1038/ncomms4357
Clarke, A. S., Lowell, J. E., Jacobson, S. J., and Pillus, L. (1999). Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19, 2515–2526. doi: 10.1128/MCB.19.4.2515
Doyon, Y., Selleck, W., Lane, W. S., Tan, S., and Côté, J. (2004). Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24, 1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004
Fuse, A., Sasaki, T., and Yano, M. (2001). Ti-plasmid vector useful for functional analysis of rice genes. Plant Biotehcnol. 18, 219–222.
Hannah, M. A., Krämer, K. M., Geffroy, V., Kopka, J., Blair, M. W., Erban, A., et al. (2007). Hybrid weakness controlled by the dosage-dependent lethal (DL) gene system in common bean (Phaseolus vulgaris) is caused by a shoot-derived inhibitory signal leading to salicylic acid-associated root death. New Phytol. 176, 537–549. doi: 10.1111/j.1469-8137.2007.02215.x
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313x.1994.6020271.x
Hobo, T., Suwabe, K., Aya, K., Suzuki, G., Yano, K., Ishimizu, T., et al. (2008). Various spatiotemporal expression profiles of anther-expressed genes in rice. Plant Cell Physiol. 49, 1417–1428. doi: 10.1093/PCP/PCN128
Huang, X., Kurata, N., Wei, X., Wang, Z. X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Ito, Y., and Kurata, N. (2008). Disruption of KNOX gene suppression in leaf by introducing its cDNA in rice. Plant Sci. 174, 357–365. doi: 10.1016/j.plantsci.2007.12.007
Jaiswal, P., Ni, J., Yap, I., Ware, D., Spooner, W., Youens-Clark, K., et al. (2006). Gramene: a bird’s eye view of cereal genomes. Nucleic Acids Res. 34, D717–D723. doi: 10.1093/NAR/GKJ154
Jeuken, M. J. W., Zhang, N. W., McHale, L. K., Pelgrom, K., Den Boer, E., Lindhout, P., et al. (2009). Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21, 3368–3378. doi: 10.1105/TPC.109.070334
Kindiger, B., and Beckett, J. B. (1985). A hematoxylin staining procedure for maize pollen grain chromosomes. Biotech. Histochem. 60, 265–269. doi: 10.3109/10520298509113923
Kubo, T., Fujita, M., Takahashi, H., Nakazono, M., Tsutsumi, N., and Kurata, N. (2013). Transcriptome analysis of developing ovules in rice isolated by laser microdissection. Plant Cell Physiol. 54, 750–765. doi: 10.1093/PCP/PCT029
Kubo, T., Takashi, T., Ashikari, M., Yoshimura, A., and Kurata, N. (2016). Two tightly linked genes at the hsa1 locus cause both F1 and F2 hybrid sterility in rice. Mol. Plant 9, 221–232. doi: 10.1016/j.molp.2015.09.014
Kubo, T., and Yoshimura, A. (2002). Genetic basis of hybrid breakdown in a Japonica/Indica cross of rice, Oryza sativa L. Theor. Appl. Genet. 105, 906–911. doi: 10.1007/S00122-002-1059-1
Kubo, T., Yoshimura, A., and Kurata, N. (2011). Hybrid male sterility in rice is due to epistatic interactions with a pollen killer locus. Genetics 189, 1083–1092. doi: 10.1534/genetics.111.132035
Lafon, A., Chang, C. S., Scott, E. M., Jacobson, S. J., and Pillus, L. (2007). MYST opportunities for growth control: yeast genes illuminate human cancer gene functions. Oncogene 26, 5373–5384. doi: 10.1038/sj.onc.1210606
Latrasse, D., Benhamed, M., Henry, Y., Domenichini, S., Kim, W., Zhou, D. X., et al. (2008). The MYST histone acetyltransferases are essential for gametophyte development in Arabidopsis. BMC Plant Biol. 8:121. doi: 10.1186/1471-2229-8-121
Maheshwari, S., and Barbash, D. A. (2011). The genetics of hybrid incompatibilities. Annu. Rev. Genet. 45, 331–355. doi: 10.1146/annurev-genet-110410-132514
Mitchell, L., Lambert, J.-P., Gerdes, M., Al-Madhoun, A. S., Skerjanc, I. S., Figeys, D., et al. (2008). Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 Is essential for complex integrity. Mol. Cell. Biol. 28, 2244–2256. doi: 10.1128/mcb.01653-07
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Nishimura, A., Aichi, I., and Matsuoka, M. (2006). A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 1, 2796–2802. doi: 10.1038/nprot.2006.469
Ohba, R., Steger, D. J., Brownell, J. E., Mizzen, C. A., Cook, R. G., Côté, J., et al. (1999). A novel H2A/H4 nucleosomal histone acetyltransferase in Tetrahymena thermophila. Mol. Cell. Biol. 19, 2061–2068. doi: 10.1128/MCB.19.3.2061
Ohtsubo, Y., Ikeda-Ohtsubo, W., Nagata, Y., and Tsuda, M. (2008). GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9:376. doi: 10.1186/1471-2105-9-376
Panagopoulos, I., Micci, F., Thorsen, J., Gorunova, L., Eibak, A. M., Bjerkehagen, B., et al. (2012). Novel fusion of MYST/Esa1-associated factor 6 and PHF1 in endometrial stromal sarcoma. PLoS One 7:e39354. doi: 10.1371/journal.pone.0039354
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/OXFORDJOURNALS.MOLBEV.A040454
Sakata, K., Nagamura, Y., Numa, H., Antonio, B. A., Nagasaki, H., Idonuma, A., et al. (2002). RiceGAAS: an automated annotation system and database for rice genome sequence. Nucleic Acids Res. 30, 98–102. doi: 10.1093/nar/30.1.98
Shen, J., Fu, J., Ma, J., Wang, X., Gao, C., Zhuang, C. X., et al. (2014). Isolation, culture, and transient transformation of plant protoplasts. Curr. Protoc. Cell Biol. 63, 2.8.1–2.8.17. doi: 10.1002/0471143030.CB0208S63
Stebbins, G. L. (1958). The inviability, weakness, and sterility of interspecific hybrids. Adv. Genet. 9, 147–215. doi: 10.1016/S0065-2660(08)60162-5
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30:2725. doi: 10.1093/MOLBEV/MST197
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Wang, X., Ahmad, S., Zhang, Z., Côté, J., and Cai, G. (2018). Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex. Nat. Commun. 9:1147. doi: 10.1038/s41467-018-03504-5
Yamamoto, E., Takashi, T., Morinaka, Y., Lin, S., Wu, J., Matsumoto, T., et al. (2010). Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Mol. Genet. Genomics 283, 305–315. doi: 10.1007/s00438-010-0514-y
Zacharaki, V., Benhamed, M., Poulios, S., Latrasse, D., Papoutsoglou, P., Delarue, M., et al. (2012). The Arabidopsis ortholog of the YEATS domain containing protein YAF9a regulates flowering by controlling H4 acetylation levels at the FLC locus. Plant Sci. 196, 44–52. doi: 10.1016/j.plantsci.2012.07.010
Keywords: hybrid breakdown, histone acetyltransferase, rice, duplicate recessive gene, speciation
Citation: Kubo T, Yoshimura A and Kurata N (2022) Loss of OsEAF6, a Subunit of the Histone Acetyltransferase Complex, Causes Hybrid Breakdown in Intersubspecific Rice Crosses. Front. Plant Sci. 13:866404. doi: 10.3389/fpls.2022.866404
Received: 31 January 2022; Accepted: 08 February 2022;
Published: 08 March 2022.
Edited by:
Dayun Tao, Yunnan Academy of Agricultural Sciences, ChinaReviewed by:
Zhang Yu, Yunnan Academy of Agricultural Sciences, ChinaHee-Jong Koh, Seoul National University, South Korea
Copyright © 2022 Kubo, Yoshimura and Kurata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takahiko Kubo, dGFrdWJvQGFnci5reXVzaHUtdS5hYy5qcA==
†Present address: Takahiko Kubo, Plant Genetics Laboratory, Faculty of Agriculture, Kyushu University, Fukuoka, Japan
 Takahiko Kubo
Takahiko Kubo Atsushi Yoshimura2
Atsushi Yoshimura2