- 1Laboratory of Plant Molecular Pathology and Functional Genomics, Department of Plant Biosciences, School of Applied Biosciences, College of Agriculture and Life Science, Kyungpook National University, Daegu, South Korea
- 2Laboratory of Cell Biology, Department of Entomology, Abdul Wali Khan University, Mardan, Pakistan
- 3Department of Horticultural Sciences, Kyungpook National University, Daegu, South Korea
The liaison between Nitric oxide (NO) and phytohormones regulates a myriad of physiological processes at the cellular level. The interaction between NO and phytohormones is mainly influenced by NO-mediated post-translational modifications (PTMs) under basal as well as induced conditions. Protein S-nitrosylation is the most prominent and widely studied PTM among others. It is the selective but reversible redox-based covalent addition of a NO moiety to the sulfhydryl group of cysteine (Cys) molecule(s) on a target protein to form S-nitrosothiols. This process may involve either direct S-nitrosylation or indirect S-nitrosylation followed by transfer of NO group from one thiol to another (transnitrosylation). During S-nitrosylation, NO can directly target Cys residue (s) of key genes involved in hormone signaling thereby regulating their function. The phytohormones regulated by NO in this manner includes abscisic acid, auxin, gibberellic acid, cytokinin, ethylene, salicylic acid, jasmonic acid, brassinosteroid, and strigolactone during various metabolic and physiological conditions and environmental stress responses. S-nitrosylation of key proteins involved in the phytohormonal network occurs during their synthesis, degradation, or signaling roles depending upon the response required to maintain cellular homeostasis. This review presents the interaction between NO and phytohormones and the role of the canonical NO-mediated post-translational modification particularly, S-nitrosylation of key proteins involved in the phytohormonal networks under biotic and abiotic stresses.
Introduction
Nitric oxide (NO) is a multi-tasked, gaseous signaling molecule. This is due to its smallest diatomic molecular stucture which makes it highly diffusible across biomembranes. It is a key signaling molecule involved in numerous biotic and abiotic stress responses in plants (Yu et al., 2012). In plants, it regulates a plethora of physiological processes ranging from seed germination to plant growth, reproduction (Kwon et al., 2012), and defense against biotic and abiotic stresses (Hussain et al., 2019; Khan et al., 2019; Nabi et al., 2019). The biological functions mediated by NO are mainly chemical reactions of different nitrogenous products known as reactive nitrogen species (RNS) which includes NO, nitroxyl (HNO/NO–), nitrosonium cation (NO+), peroxynitrite (ONOO–), S-nitrosothiols (RSNOs), and dinitrosyl iron complexes. The transformation of NO into other redox forms under physiological conditions, is due to the susceptibility of free NO radical to oxidation and reduction. One of the key mechanisms by which NO mediates various pathways is through post-translational modification of various proteins. Among these, protein S-nitrosylation is perhaps the most prominent. It is the selective but reversible redox-based covalent addition of a NO moiety to the sulfhydryl group of cysteine (Cys) molecule(s) on a target protein to form S-nitrosothiols. The role of NO in signal transduction was first established in the animal system wherein the smooth vascular muscle cells it was found to bind to the heme group of guanylate cyclase (Murad, 1986) and consequent protein kinase-mediated, cyclic guanosine monophosphate (cGMP-dependent activation of potassium channels) (Bolotina et al., 1994). In plants, S-nitrosylation regulates a myriad of pathophysiological and physiological processes as well as those involved in biotic and abiotic stresses. NO interacts with all the major phytohormones including abscisic acid, auxin, gibberellic acid, cytokinin, ethylene, salicylic acid, jasmonic acid, brassinosteroid, and strigolactone (Asgher et al., 2017) during various metabolic and physiological conditions and environmental stress responses. S-nitrosylation of key proteins involved in the phytohormonal network occurs during their synthesis, degradation, or signaling roles depending upon the response required to maintain cellular homeostasis. This review presents NO-mediated S-nitrosylation of key proteins involved in phytohormonal networks under biotic and abiotic stresses.
An Overview of Nitric Oxide-Mediated S-Nitrosylation, Transnitrosylation, and Denitrosylation
Post-translational modification by NO takes place by one of the three modification processes viz, S-nitrosylation (the formation of a nitrosothiol group on cysteine residues of target proteins), metal nitrosylation (interaction of NO with metalloproteins), and tyrosine nitration (covalent addition of NO to the tyrosine residues (as shown in Figure 1). Out of these three modifications, S-nitrosylation, also known as S-nitrosation plays crucial roles in various physiological and pathological processes by modulating protein activities. It is a highly conserved post-translational modification and is broadly studied and described in plants (Astier and Lindermayr, 2012). This dynamic and reversible process involves nitrosylation, transnitrosylation, and denitrosylation (Benhar et al., 2009; Astier and Lindermayr, 2012; Feng et al., 2019). S-nitrosylation or the more biochemistry-oriented term S-nitrosation is considered a non-enzymatic process, in which NO mediates the formation of S-nitrosothiols either directly or indirectly by higher nitrogen oxides (NOx), metal-NO intermediates, SNOs, or ONOO– (Wang et al., 2006). S-nitrosylation of cysteine residues in the tripeptide glutathione (GSH) leads to the formation of low molecular weight SNOs which in turn function as NO donors depending on their redox potential under physiological conditions (Hess et al., 2005). This transfer of NO group from one thiol to another has also been reported and termed transnitrosylation and is known to be an important enzymatic process (Wu et al., 2010; Qu et al., 2011; Choi, 2018; Feng et al., 2019). In this process, the donor protein that carries and transfers the NO moiety to its target is termed as a transnitrosylase (Stomberski et al., 2019; Chen et al., 2020). A major determinant for NO transfer is the difference between the redox potential of the cysteine residues of the interacting proteins (donor protein-SNO and target protein with a free thiol). The process of transnitrosylation involves overturning of the first SNO-mediated regulation of the donor protein and may also be termed as denitrosylation (Choi, 2018; Feng et al., 2019). Denitrosylation of target proteins is promoted enzymatically and non-enzymatically which tightly regulates cysteine modification (Benhar et al., 2008, 2009; Astier et al., 2011). Transnitrosylation is carried out by proteins, also known as transnitrosylases that carry and transfer the NO group to its target (Anand and Stamler, 2012; Stomberski et al., 2019). In bacterial and mammalian cells, several transnitrosylases like cytochrome c, cytoglobin, caspase 3, thioredoxin, and hemoglobins have been identified and functionally characterized (Anand and Stamler, 2012). However, in the case of plants, GSNO is considered to possess major transnitrosylase activity, modulating the total SNO content (Wang et al., 2006; Yu et al., 2012). Studies have reported that the transnitrosylation of a protein-SNO to another protein with free thiol plays role in the regulation of NO-mediated regulatory mechanisms of signaling pathways (Nakamura et al., 2010; Choi et al., 2014). For instance, selective denitrosylation of S-nitrosylated proteins by Trx-h3 and Trx-h5 were found to regulate plant immunity in Arabidopsis (Kneeshaw et al., 2014). In another study, S-nitrosylation/denitrosylation are reported to be influenced by auxins in Arabidopsis roots. Overall, NO-mediated S-nitrosylation is a crucial signaling mechanism. However, regulators of similar processes like transnitrosylation and denitrosylation can also modulate protein functionality in response to environmental stress in plants.
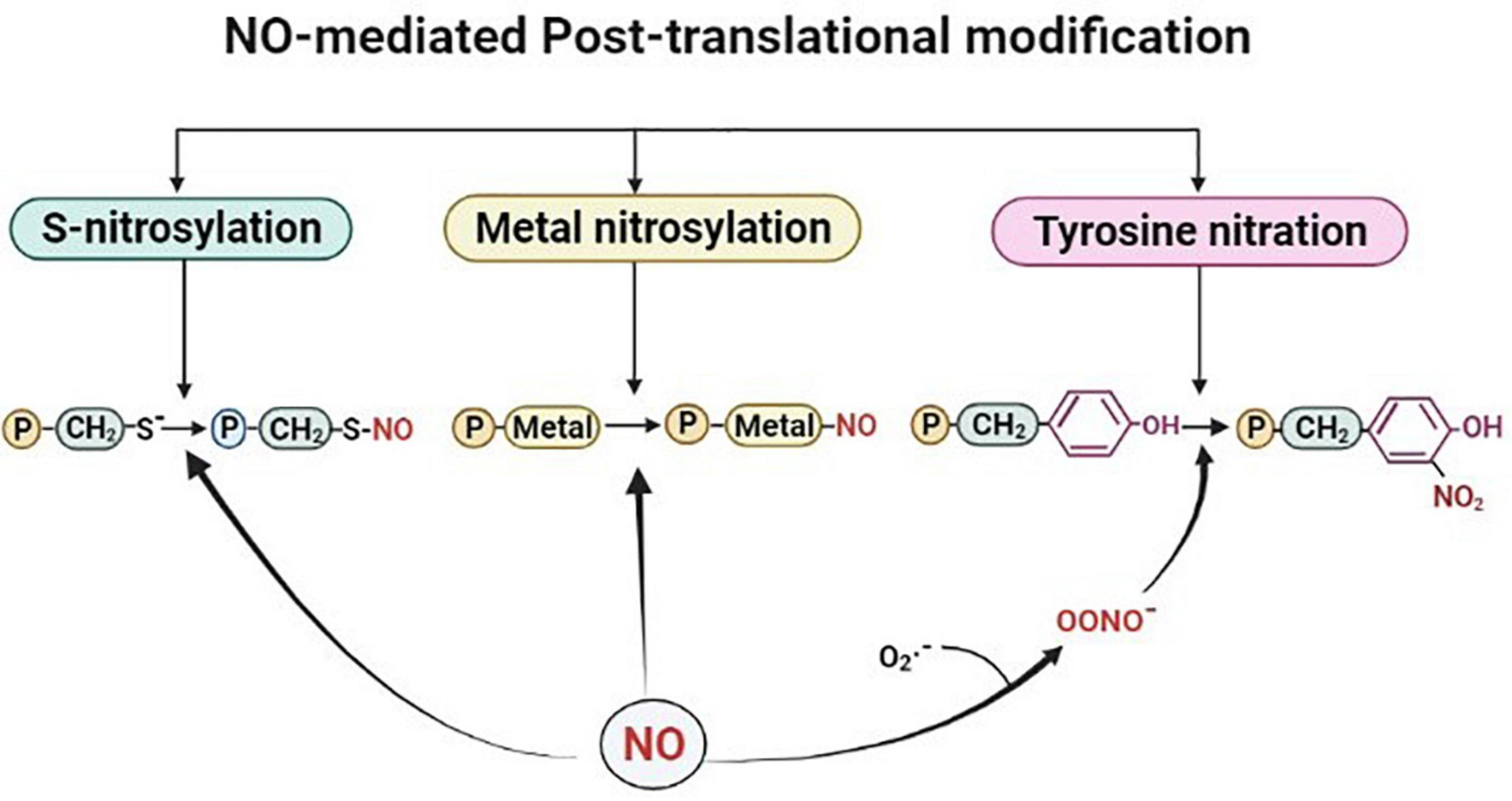
Figure 1. Outline of the nitric oxide (NO)-mediated post-translational modifications. Proteins are represented with letter “P.” Figure made in BioRender.com.
S-Nitrosylation of Target Proteins Involved in Phytohormonal Network
Plants synthesize and maintain delicate levels of phytohormones to promote normal growth in plants under optimal conditions. However, under changing environmental conditions or upon exposure to biotic/abiotic stress conditions, phytohormones also regulate plant adaptation and survival by controlling the production of various stress-responsive proteins, antioxidants, and ion transporters. The phytohormones that promote the growth and govern the plant survival under stressful conditions include auxins, gibberellins, jasmonic acid, salicylic acid, abscisic acid, cytokinins, ethylene, brassinosteroids, and strigolactones. These undergo coordinated interactions with various signaling molecules like nitric oxide and hydrogen peroxide to regulate their activity depending on the severity of the environmental stress. Largely, the liaison between NO and phytohormones regulates a myriad of physiological processes (as shown in Figure 2) at the cellular level. Under environmental stress conditions, NO and phytohormonal coordination regulates gene express and activities of antioxidative enzymes. The phytohormonal levels are regulated either at the biosynthesis, distribution, or signaling stage. NO can regulate the level of hormones at transcriptional (Bethke et al., 2007; Lozano-Juste and León, 2011) or post-translational levels (Lindermayr et al., 2005; Terrile et al., 2012; Feng et al., 2019). S-nitrosylation is a common signaling mechanism mediated by nitric oxide which regulates hormonal signaling at the post-translational level. Thus, S-nitrosylated proteins involved in phytohormonal activity are regulated at the pre-receptor level (as in the case of ethylene), receptor level (as in the case of auxin signaling where TIR1 binds auxin directly conferring an increased affinity for AUX/IAA proteins) and post-receptor levels (as in the case of cytokinin and abscisic acid signaling where hormonal binding to the receptor determines the response) of phytohormonal activity. Overall, we discuss below the interplay between NO and phytohormones and the S-nitrosylation of target proteins involved in the phytohormonal network at various levels.
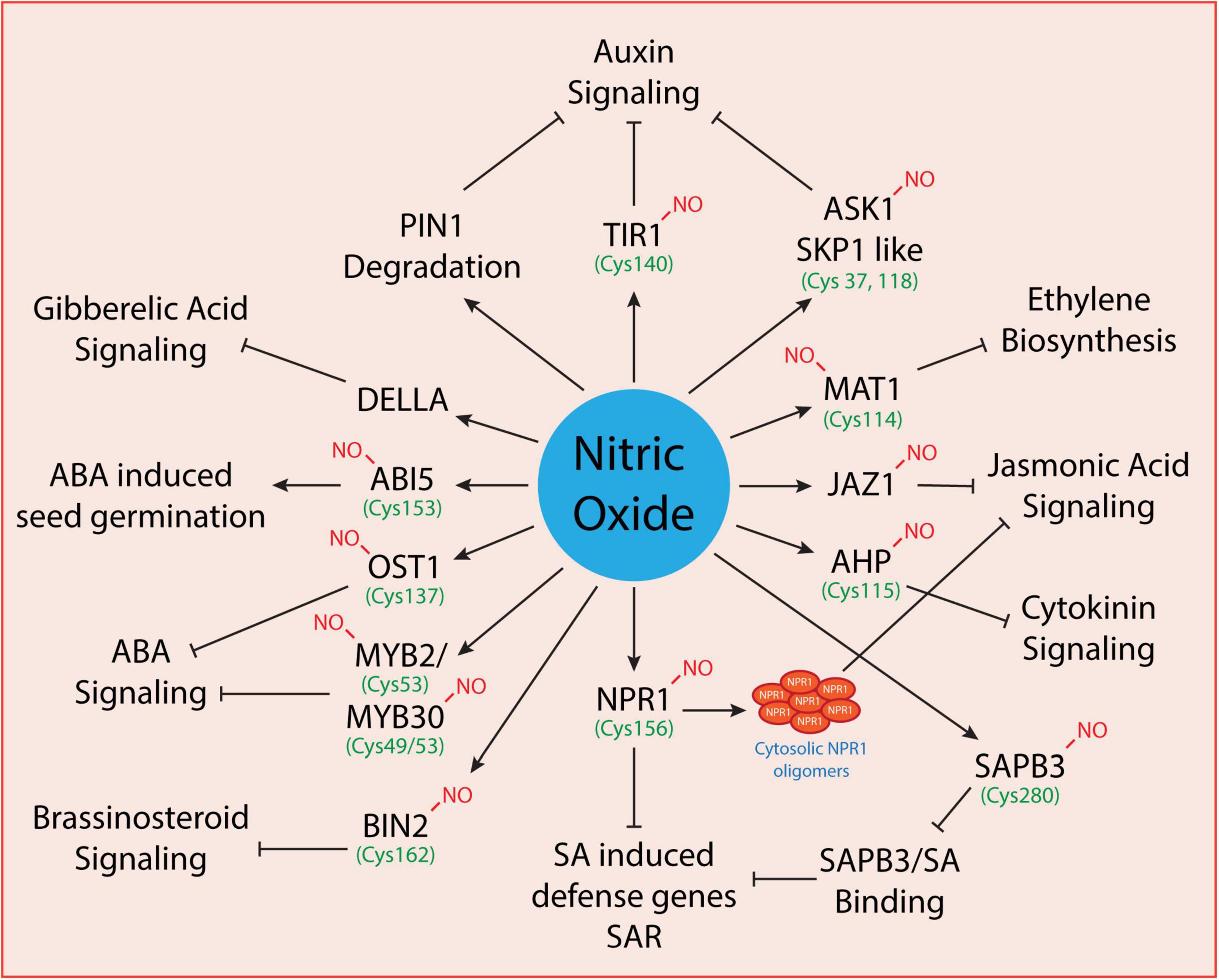
Figure 2. Regulation of phytohormonal network by nitric oxide. Some of the known proteins involved in phytohormonal signaling that are directly modulated by nitric oxide have been shown here. However, only a few are known to undergo S-nitrosylation (where the cysteine residues have been indicted with green color) while others need to be explored further.
S-Nitrosylated Protein in Auxin and Gibberellin Signaling
The phytohormone, auxin plays a crucial role in various developmental processes both under normal and stress conditions. The major function of auxin is the formation, development, and maintenance of roots (Overvoorde et al., 2010). However, for the efficient regulation of plant development under changing environmental conditions auxins interact with other phytohormones and signaling molecules like NO (Freschi, 2013). The NO-auxin interplay suggests an increase in NO after auxin is applied to the roots or in auxin overexpressing mutant lines which lead to the speculations that NO may act downstream of auxins (Correa-Aragunde et al., 2016). The interaction between NO and auxin has been studied under heavy metal stress conditions with the application of NO-donors (mostly sodium nitroprusside). However, the results have been contradictory suggesting that NO accumulation is responsible for an increase in auxin-dependent root elongation to overcome mercury-induced toxicity in rice (Chen et al., 2015) whereas it inhibits root meristem growth by repressing auxin signaling under cadmium stress in Arabidopsis (Yuan and Huang, 2016).
Auxin signaling response is suppressed due to the dimerization of AUXIN/INDOLEACETIC ACID (AUX/IAA) transcription factor with AUXIN RESPONSE FACTOR (ARF) transcriptional activators that are present on the auxin-responsive promoter elements (ARE). However, in the presence of auxin, this transcription factor is subjected to ubiquitin (Ub)-mediated proteasomal degradation with the help of SCF-E3 ligase complex (steps 1–3), and auxin response initiates (Ramos Báez and Nemhauser, 2021). The SCF-E3 complex comprises of four subunits: CULLIN1 (CUL1), S-PHASE KINASE-ASSOCIATED PROTEIN 1-LIKE1 (ASK1), RING BOX 1 (RBX1), and TRANSPORT INHIBITOR RESISTANT 1 (TIR1)/AUXIN SIGNALING F-BOX(AFB). S-nitrosylation of TIR1 and ASK1 in this pathway enhances the protein-protein interaction modulating the SCFTIR1/AFBs complex assembly (Iglesias et al., 2018), and hence auxin signaling response in plants, as shown in Figure 3A.
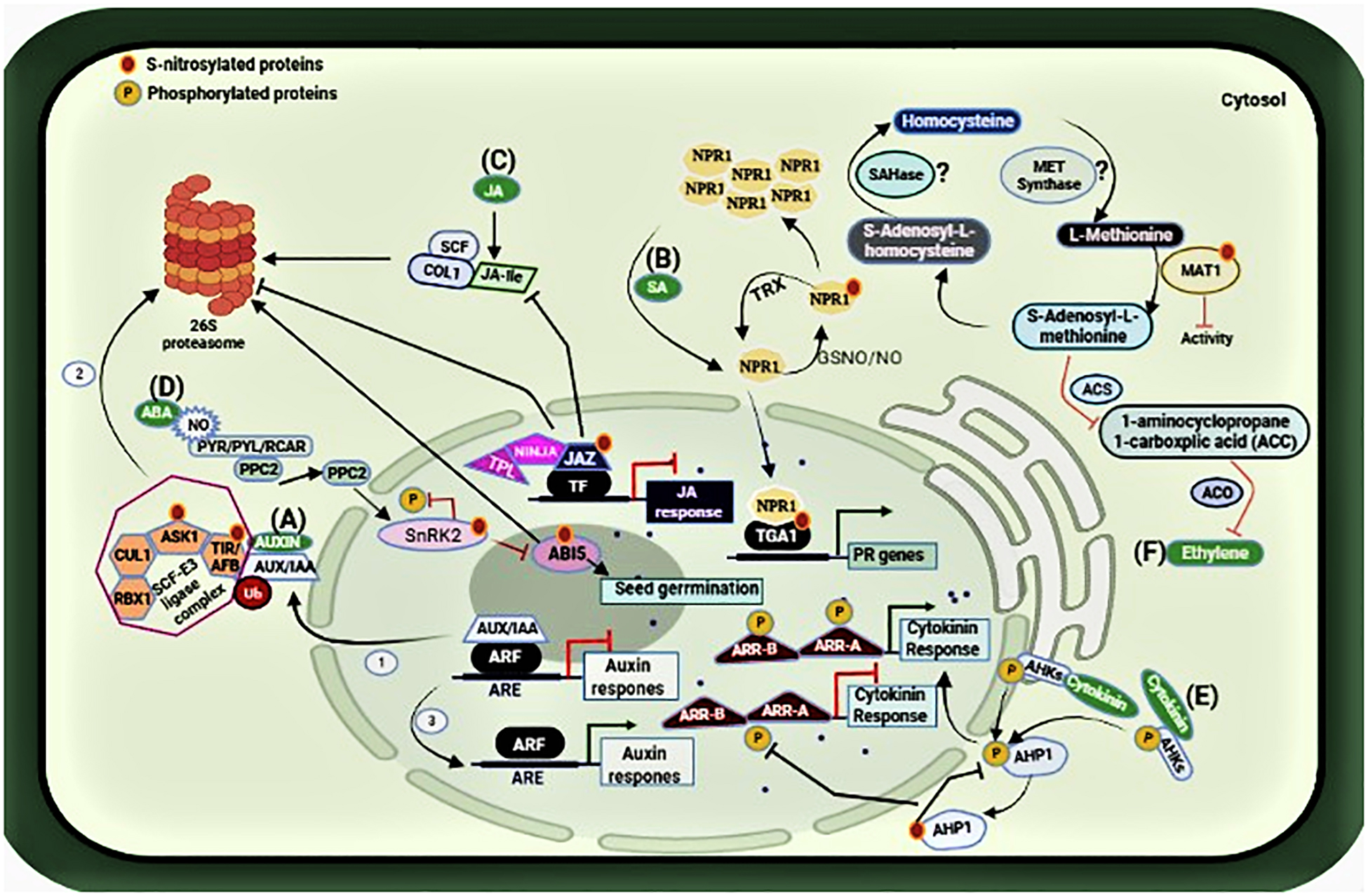
Figure 3. Representation of the S-nitrosylated proteins involved in the hormonal network. (A) S-nitrosylation of ASK1 and TIR1 lead to the proteolytic degradation of AUX/IAA, thus initiating auxin response (steps 1–3). (B) S-nitrosylation/glutathionylation of NPR1leads to its oligomerization while S-nitrosylation of TGA1 promotes its interaction with NPR1 enabling the expression of PR genes. (C) S-nitrosylation prevents the interaction between JAZ1 repressor proteins and COI1, which is a subunit of the SCF ubiquitin E3 ligase complex for its proteasomal degradation. Thus, allowing JAZ1 to recruit its co-repressors NINJA and TPL to repress JA signaling. (D) S-nitrosylation of ABI5 also leads to its proteasomal degradation, thus promoting seed germination in the presence of NO. (E) S-nitrosylation of AHP1 inhibits its phosphorylation, compromising cytokinin response. (F) S-nitrosylation of MAT1 suppresses the activity of 1-aminocyclopropane1-carboxplic acid (ACC) synthesis and ACC oxidase, thereby affecting the synthesis of ethylene Moreover, S-nitrosylation of SAHase and MET synthase are still under investigation (shown with question marks). Figure made in BioRender.com.
Independent studies on the cross talk between NO and auxin in controlling plant development (probably through S-nitrosylation) has been reported by Fernández-Marcos et al. (2011) and Terrile et al. (2012). In Fernández-Marcos et al. (2011) reported that NO causes root apical meristem defects and growth inhibition in Arabidopsis by enhancing the degradation of the auxin efflux transporter PIN-FORMED 1 (PIN1), thereby reducing PIN1-dependent acropetal auxin transport. Later, Terrile et al. (2012) showed that NO also enhances the interaction between the auxin receptor TIR1 and Aux/IAA suppressor. NO directly targets the auxin receptor TIR1 for S-nitrosylation of its Cys140 residue which is critical for TIR1 function and its interaction with AUX/IAA repressor.
Nitric oxide also impacts auxin signaling pathway through SCFTIR1/AFB E3 ubiquitin ligase complex assembly via S-nitrosylation of the ARABIDOPSIS SKP1-LIKE1 (ASK1) at Cys37 and Cys118. S-nitrosylation of ASK1 influences its binding with TIR1/AFB2 and Cullin1 (CUL1) which in turn promotes SCFTIR1/ and SCFAFB2 assembly, resulting in an impaired auxin signaling activation (Iglesias et al., 2018).
Furthermore, an interesting study by Yang et al. (2015) reported that S-Nitrosylation of the NO scavenger ascorbate peroxidase (APX1) at Cys32 positively regulates its activity during stress. However, auxin induces the de-nitrosylation and partial inhibition of APX1 (Correa-Aragunde et al., 2013). This indicates the presence of an auxin-mediated APX1 S-nitrosylation/de-nitrosylation equilibrium at the cellular level that contributes to a fine-tuned control of reactive oxygen species (Fares et al., 2011).
Similarly, gibberellin constitutes a large family of tetracyclic diterpenoid phytohormones regulating plant growth and development. Nitric oxide regulates DELLA content and PIF expression to promote photomorphogenesis in Arabidopsis (Lozano-Juste and León, 2011). NO negatively regulates gibberellin signaling by promoting the accumulation of the gibberellin signaling repressor DELLA and decreasing the expression of phytochrome-interacting factors (PIFs) (Richter et al., 2010). This is further supported by studies involving the Arabidopsis loss of function mutant lines nox1 and gsnor1-3 that accumulate more NO/SNOs, and have abnormal auxin and gibberellin responses (Fernández-Marcos et al., 2011; Terrile et al., 2012). However, no S-nitrosylated target protein have been reported so far in the case of gibberellic acid.
S-Nitrosylated Proteins in Salicylic Acid and Jasmonic Acid Signaling
Salicylic Acid (SA), and Jasmonic Acid (JA) are key phytohormones that regulate plant responses to infection by a variety of pathogens including fungi, bacteria, viruses, and others (Fujita et al., 2006; Loake and Grant, 2007). NO exerts its role on these pathways to regulate plant defense during infection. Salicylic acid regulates plant responses to infection by pathogens and is essential for the establishment of resistance mechanisms such as host cell death and systemic acquired resistance (SAR). The S-nitrosoglutathione reductase (GSNOR) is a key enzyme regulating cellular S-nitrosothiol levels via denitrosylation (Malik et al., 2011). The Arabidopsis atgsnor1-3 line has significantly higher levels of cellular SNOs and perturbed SA biosynthesis and signaling. Tada et al. (2008) showed that plant immunity requires conformational changes of NPR1 via S-nitrosylation. NPR1, a master regulator of SA-mediated plant defense is sequestered in the cytoplasm as an oligomer. Upon infection, it monomerizes and is translocated to the nucleus to activate a battery of pathogenesis-related (PR) genes (Kinkema et al., 2000). However, in unchallenged plants, the oligomer to monomer switch is regulated by S-nitrosylation at Cys156 inhibiting its monomerization (Tada et al., 2008). Furthermore, NO accretion during the nitrosative burst after infection promotes S-nitrosylation of the Arabidopsis thaliana salicylic acid-binding protein 3 (ATSABP3) at Cys280 which not only suppresses its binding to SA but also inhibits its carbonic anhydrase activity (Wang et al., 2009). On the other hand, S-nitrosylation also regulates SAR by targeting the NPR1/TGA1 system. As described above, SA induces thioredoxin (TRX) which facilitates denitrosylation of NPR1 for its monomerization during plant immune response (Kneeshaw et al., 2014) and thus facilitating its translocation to the nucleus where they interact with the basic leucine zipper transcription factor TGA to promote TGA attachment to the promoters of PR genes (Tada et al., 2008). Notably, it has been demonstrated that S-nitrosylation (and S-glutathionylation) improves TGA1 binding to the PR1 promoter (Lindermayr et al., 2010) as shown in Figure 3B. Interestingly, the cytosolic NPR1 also contributes to the suppression of the JA pathway. Hence, it can be concluded that distinct redox signals work to maintain a cellular redox balance to regulate plant immunity and that S-nitrosylation is a key mediator of integrated phytohormonal networks concerning plant immunity.
Jasmonic acid (JA) is an important phytohormone involved in plant development and response to injury, attack by insects, and necrotrophic pathogens. Jasmonic acid (JA) signaling depends on the interaction between JASMONATE ZIM DOMAIN (JAZ1) repressor proteins and CORONATINE-INSENSITIVE 1 (COI1), which is a subunit of the SCF ubiquitin E3 ligase complex for its proteasomal degradation (Ghorbel et al., 2021). Ayyar (2016) investigated the relationship between NO and JA signaling during the plant immune response. Increasing NO content appeared to have a negative effect on JA signaling as the NO over-accumulating atgsnor1-3 line compromised JA signaling. Her results from the biotin switch assay indicated that JAZ1 is S-nitrosylated at CyS229 in vitro. Further investigations involving Flag-tagged JAZ1 over-expression in the atgsnor1-3 line also indicated the S-nitrosylation of JAZ1 in vivo. This implies that JA signaling during insect attack or infection by necrotrophs such as Botrytis cinerea is under redox control. She proposed that JAZ1 S-nitrosylation may block its interaction with the JA receptor component COI1, while enabling JAZ1 to recruit its co-repressors NINJA (Novel INteractor of JAZ) and TPL (TOPLESS) to inhibit turnover by the proteasome, consequently enabling prolonged JAZ1-mediated suppression of JA signaling, as shown in Figure 3C.
S-Nitrosylated Proteins in Abscisic Acid Signaling
The isoprenoid phytohormone Abscisic acid (ABA) regulates a variety of physiological processes in plants including stomatal movement, protein storage, and adaptation to abiotic stresses such as cold, drought, and salt stresses. At the base of these events are complex signaling networks involving multiple components such as K+, Ca2+, MAP kinases (MAPK), H2O2, and others (Fan et al., 2004). NO is also known to be involved in various stress responses by regulating these components (Xu et al., 2015; Montilla-Bascón et al., 2017; Nabi et al., 2019; Lau et al., 2021). Ca2+ regulates stomatal movement during drought stress (Zou et al., 2010). On the other hand, NO exerts its effect on Ca2+ via S-nitrosylation of Ca2+ channels and transporters. Foresi et al. (2015) showed that NO triggers stomatal closure via regulation of the Ca2+- sensitive Cl– and K+ channels at the plasma membrane of the guard cells. However, some studies have also demonstrated a synergistic relationship between NO and Ca2+ in response to drought stress (Niu et al., 2017; Silveira et al., 2020). S-nitrosylation of transcription factors like the basic leucine zipper transcription factor ABI5, MYB2, and MYB30 is involved in abscisic acid (ABA) mediated regulation under drought stress.
The basic leucine zipper transcription factor ABI5 (ABA-INDUCED 5) regulates ABA-mediated seed germination and early seedling growth and is considered as a NO sensor, as Albertos et al. (2015) showed that NO regulates ABI5 at both transcription and translational levels. ABA signaling in the presence of NO involves tyrosine nitration of the PYRABACTIN RESISTANCE-LIKE REGULATORY COMPONENTS (PYR/PYL/RCAR) which inhibits its interaction with ABA thereby enabling the activity of type 2C protein phosphatases (PPC2), which inactivates SUCROSE NON-FERMENTING1 (SNF1)-RELATED PROTEIN KINASE2.6 (SnRK2.6) by dephosphorylation, which lead to the inhibition of ABI5. Moreover, S-nitrosylation of ABI5 at Cys153 facilitates ABI5 degradation by enhancing its interaction with the E3 ligase complex promoting seed germination and seedling growth (Albertos et al., 2015), as shown in Figure 3D. However, another study in the same year by Wang et al. (2015) reported that NO negatively regulates ABA signaling through S-nitrosylation of the open stomata 1 (OST1) at Cys137 in the guard cells. OST1 is a sucrose non-fermenting 1 (SNF1)-related protein kinase 2.6 (SnRK2.6). Loss of GSNOR1 function in the Arabidopsis atgsnor1-3 plants results in NO overaccumulation in the guard cells, leading to S-nitrosylation of SnRK2.6 abolishing ABA-dependent stomatal closure.
Transcription factors play an important role in regulating the expression of key genes involved in important physiological processes. The expression of the MYB2 transcription factor increases in response to ABA and water stress as Arabidopsis plants over-expressing MYB2 are ABA-hypersensitive indicating that MYB2 may be involved in ABA singling. On the other hand, MYB30 is involved in the hypersensitive response (HR) and is characterized by a vast production of ROS and NO. Both MYB2 and MYB30 have been shown to undergo S-nitrosylation at Cys53 and Cys49/53, respectively, which abolishes their DNA binding ability (Serpa et al., 2007; Tavares et al., 2014). Taken together, it can be concluded that both ABA and NO are key signaling molecules that regulate responses to drought stress.
S-Nitrosylated Proteins in Cytokinin and Ethylene Signaling
In plants, cytokinin is an essential phytohormone regulating plant growth and development. As Feng et al. (2013) describe, investigations indicate cytokinin promotes phosphorelay activity through the membrane-bound hybrid HISTIDINE PROTEIN KINASES (AHKs) to HISTIDINE PHOSPHOTRANSFER PROTEIN 1 (AHP1) and then to primary RESPONSE REGULATOR TYPE B (ARR-B) and primary RESPONSE REGULATOR TYPE A (ARR-A) to promote cytokinin signaling response. They showed that NO negatively regulates cytokinin signaling by inhibiting the phosphorelay system via S-nitrosylation of AHP1 at Cys115 rendering it unable to phosphorylate, as shown in Figure 3E. They also showed that a non-nitrosylatable mutant protein AHP1 partially relieves the negative effects of NO on cytokinin signaling. Their findings illustrate that cytokinin signaling and redox signaling via S-nitrosylation coordinated plant growth and development.
Similarly, ethylene is a versatile phytohormone that regulates both growth/development and senescence. Ethylene biosynthesis is regulated by NO through S-nitrosylation of enzymes involved in the Yang cycle (or the methyl-methionine cycle) (Pattyn et al., 2021). The activity of 1-aminocyclopropane1-carboxplic acid (ACC) synthesis, as well as ACC oxidase, is suppressed due to S-nitrosylation of METHIONINE ADENOSYLTRANSFERASE (MAT1) thereby affecting the synthesis of ethylene through ACC synthase (ACS) and (ACC) oxidase (ACO) (Lindermayr et al., 2006). Moreover, S-nitrosylation of S-adenosylhomocysteinase (SAHase) and cobalamin-independent methionine synthase (MET synthase) are still under investigation. Lindermayr et al. (2006) reported the inactivation of METHIONINE ADENOSYLTRANSFERASE (MAT1) due to its S-nitrosylation at Cys114. In addition, enzymes such as S-adenosylhomocysteinase and cobalamin-independent methionine synthase are also a part of the ethylene pathway and have been found to be the targets of S-nitrosylation by NO (Lindermayr et al., 2005; Abat et al., 2008). This indicates a multi-step control of ET biosynthesis and signaling in plants via S-nitrosylation as shown in Figure 3F. Table 1 below indicates the target proteins involved in phytohormonal network.
S-Nitrosylated Proteins Predicted in Brassinosteroid and Strigolactone Signaling
Brassinosteroids are a class of phytohormones that are polyhydroxylated and steroidal similar to the steroid hormones in animals. These steroidal hormones regulate a wide variety of physiological plant processes including growth, development, immunity, and stress responses (Ahammed et al., 2020; Hussain et al., 2020; Nolan et al., 2020; Ortiz-Morea et al., 2020). Brassinosteroid signaling involves a constitutively active kinase, BIN2 (Brassinosteroid-Insensitive2) which is suggested to be s-nitrosylated at the conserved Cys162 site as indicted by an in vitro assay (Mao and Li, 2020). It has been further suggested in this study that this modification could interfere with its structural assembly or its interaction with the two key transcription factors, BES1 (bri1-EMS suppressor1) and BZR1 (Brassinazole-Resistant1). However, to confirm these observations further in vivo investigations are required.
Strigolactones belong to a more recently studied group of phytohormones involved in growth and development processes mainly in symbiotic mycorrhizal plant-fungi association (Faizan et al., 2020). These are also involved in the regulation of stress responses in plants. In silico analysis of NO-mediated post-translational modification of proteins involved in strigolactones biosynthesis and signaling in A. thaliana and Oryza sativa predicted S-nitrosylation of MAX4/CCD8 (carotenoid cleavage dioxygenases) in Arabidopsis involved in biosynthesis while MAX2 (MORE AXILLARY GROWTH2 in Arabidopsis), and D53 (Dwarf 53) and D5 (Dwarf 5) in O. sativa involved in strigolactone signaling pathway (Kolbert, 2019). However, detailed studies are required to confirm such predictions in vitro and in vivo.
Conclusion and Future Prospects
Plants being sessile, are vulnerable to various environmental changes, challenging them to adjust and grow under such adversities. Plants can survive and grow under these challenging situations due to the coordinated signaling mechanisms involving phytohormones and other stress-responsive molecules like NO and H2O2. NO is an essential signaling molecule in signaling cascade interaction with almost all phytohormones. During the past few years, there is an extensive quest for search to explore the multiple and widely diverse mechanisms regarding plant hormones and NO interaction. Undoubtedly. NO has a dual role in the up and downregulation of plant hormones. The NO-phytohormonal interaction is mostly through NO-mediated post-translational modifications regulating the synthesis, distribution, degradation, and conjugation of the elements involved in the plant hormonal transport and signaling. However, there are still studies lagging in explaining how NO interacts with hormones and hormones-related proteins at translational and posttranslational levels. Moreover, the interplay among NO and phytohormones needs to be explored in future work. Overall, it requires extensive research and investigation to explore the coordination between NO and various phytohormonal signaling cascades operating in plants under the pressure of multiple stresses.
Author Contributions
AP contributed to the writing—original draft preparation and images. BM contributed to the review, editing and gathering resources. WR contributed to the preparation of the table and its contents. MK and TA contributed to the section on S-nitrosylation of target proteins involved in phytohormonal network. DL and GL contributed to the illustration and visualization of the images. AH revised and improved the manuscript and images. CK critically discussed and revised the manuscript. BY revised the final draft and supervised the entire manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Grant No. 2020R1I1A3073247), South Korea, and a project to train professional personnel in biological materials by the Ministry of Environment, South Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The figures were created using BioRender.com
References
Abat, J. K., Mattoo, A. K., and Deswal, R. (2008). S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata- ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 275, 2862–2872. doi: 10.1111/j.1742-4658.2008.06425.x
Ahammed, G. J., Li, X., Liu, A., and Chen, S. (2020). Brassinosteroids in plant tolerance to abiotic stress. J. Plant Growth Regul. 39, 1451–1464. doi: 10.1007/s00344-020-10098-0
Albertos, P., Romero-Puertas, M. C., Tatematsu, K., Mateos, I., Sánchez-Vicente, I., Nambara, E., et al. (2015). S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 6:8669. doi: 10.1038/ncomms9669
Anand, P., and Stamler, J. S. (2012). Enzymatic mechanisms regulating protein S-nitrosylation: implications in health and disease. J. Mol. Med. 90, 233–244. doi: 10.1007/s00109-012-0878-z
Asgher, M., Per, T. S., Masood, A., Fatma, M., Freschi, L., Corpas, F. J., et al. (2017). Nitric oxide signaling and its crosstalk with other plant growth regulators in plant responses to abiotic stress. Environ. Sci. Pollut. Res. 24, 2273–2285. doi: 10.1007/s11356-016-7947-8
Astier, J., and Lindermayr, C. (2012). Nitric oxide-dependent posttranslational modification in plants: an update. Int. J. Mol. Sci. 13, 15193–15208. doi: 10.3390/ijms131115193
Astier, J., Rasul, S., Koen, E., Manzoor, H., Besson-Bard, A., Lamotte, O., et al. (2011). S-nitrosylation: an emerging post-translational protein modification in plants. Plant Sci. 181, 527–533. doi: 10.1016/j.plantsci.2011.02.011
Ayyar, P. V. (2016). Uncovering the Role of S-Nitrosylation in Jasmonic Acid Signalling During the Plant Immune Response. Ph.D. thesis. Edinburgh: IMPS, The University of Edinburgh.
Backer, R., Naidoo, S., and Van den Berg, N. (2019). The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) and related family: mechanistic insights in plant disease resistance. Front. Plant Sci. 10:102. doi: 10.3389/fpls.2019.00102
Baek, D., Park, H. C., Kim, M. C., and Yun, D.-J. (2013b). The role of Arabidopsis MYB2 in miR399f-mediated phosphate-starvation response. Plant Signal. Behav. 8, 362–373. doi: 10.4161/psb.23488
Baek, D., Kim, M. C., Chun, H. J., Kang, S., Park, H. C., Shin, G., et al. (2013a). Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol. 161, 362–373. doi: 10.1104/pp.112.205922
Benhar, M., Forrester, M. T., and Stamler, J. S. (2009). Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 10, 721–732. doi: 10.1038/nrm2764
Benhar, M., Forrester, M. T., Hess, D. T., and Stamler, J. S. (2008). Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320, 1050–1054. doi: 10.1126/science.1158265
Bethke, P. C., Libourel, I. G., Aoyama, N., Chung, Y.-Y., Still, D. W., and Jones, R. L. (2007). The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 143, 1173–1188. doi: 10.1104/pp.106.093435
Bolotina, V. M., Najibi, S., Palacino, J. J., Pagano, P. J., and Cohen, R. A. (1994). Nitric oxide directly activates calcium-dependent potassium channels in vascular smooth muscle. Nature 368, 850–853. doi: 10.1038/368850a0
Chen, L., Wu, R., Feng, J., Feng, T., Wang, C., Hu, J., et al. (2020). Transnitrosylation mediated by the non-canonical catalase ROG1 regulates nitric oxide signaling in plants. Dev. Cell 53, 444–457.e445. doi: 10.1016/j.devcel.2020.03.020
Chen, Z., Zhang, L., and Zhu, C. (2015). Exogenous nitric oxide mediates alleviation of mercury toxicity by promoting auxin transport in roots or preventing oxidative stress in leaves of rice seedlings. Acta Physiol. Plant. 37, 1–9.
Choi, M. S. (2018). Pathophysiological role of s-nitrosylation and transnitrosylation depending on s-nitrosoglutathione levels regulated by s-nitrosoglutathione reductase. Biomol. Ther. 26:533. doi: 10.4062/biomolther.2018.179
Choi, M. S., Nakamura, T., Cho, S.-J., Han, X., Holland, E. A., Qu, J., et al. (2014). Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in Parkinson’s disease models. J. Neurosci. 34, 15123–15131. doi: 10.1523/JNEUROSCI.4751-13.2014
Correa-Aragunde, N., Foresi, N., and Lamattina, L. (2016). Auxin and nitric oxide: a counterbalanced partnership ensures the redox cue control required for determining root growth pattern. Adv. Bot. Res. 77, 41–54.
Correa-Aragunde, N., Foresi, N., Delledonne, M., and Lamattina, L. (2013). Auxin induces redox regulation of ascorbate peroxidase 1 activity by S-nitrosylation/denitrosylation balance resulting in changes of root growth pattern in Arabidopsis. J. Exp. Bot. 64, 3339–3349. doi: 10.1093/jxb/ert172
Cortleven, A., Leuendorf, J. E., Frank, M., Pezzetta, D., Bolt, S., and Schmülling, T. (2019). Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 42, 998–1018. doi: 10.1111/pce.13494
Faizan, M., Faraz, A., Sami, F., Siddiqui, H., Yusuf, M., Gruszka, D., et al. (2020). Role of strigolactones: signalling and crosstalk with other phytohormones. Open Life Sci. 15, 217–228. doi: 10.1515/biol-2020-0022
Fan, L. M., Zhao, Z., and Assmann, S. M. (2004). Guard cells: a dynamic signaling model. Curr. Opin. Plant Biol. 7, 537–546. doi: 10.1016/j.pbi.2004.07.009
Fares, A., Rossignol, M., and Peltier, J.-B. (2011). Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem. Biophys. Res. Commun. 416, 331–336. doi: 10.1016/j.bbrc.2011.11.036
Feng, J., Chen, L., and Zuo, J. (2019). Protein S-nitrosylation in plants: current progresses and challenges. J. Integr. Plant Biol. 61, 1206–1223. doi: 10.1111/jipb.12780
Feng, J., Wang, C., Chen, Q., Chen, H., Ren, B., Li, X., et al. (2013). S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat. Commun. 4:1529. doi: 10.1038/ncomms2541
Fernández-Marcos, M., Sanz, L., Lewis, D. R., Muday, G. K., and Lorenzo, O. (2011). Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. U.S.A. 108, 18506–18511. doi: 10.1073/pnas.1108644108
Foresi, N., Mayta, M. L., Lodeyro, A. F., Scuffi, D., Correa-Aragunde, N., García-Mata, C., et al. (2015). Expression of the tetrahydrofolate-dependent nitric oxide synthase from the green alga Ostreococcus tauri increases tolerance to abiotic stresses and influences stomatal development in Arabidopsis. Plant J. 82, 806–821. doi: 10.1111/tpj.12852
Fousia, S., Tsafouros, A., Roussos, P., and Tjamos, S. (2018). Increased resistance to Verticillium dahliae in Arabidopsis plants defective in auxin signalling. Plant Pathol. 67, 1749–1757. doi: 10.1111/ppa.12881
Freschi, L. (2013). Nitric oxide and phytohormone interactions: current status and perspectives. Front. Plant Sci. 4:398. doi: 10.3389/fpls.2013.00398
Fujii, H., and Zhu, J.-K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385. doi: 10.1073/pnas.0903144106
Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014
Ghorbel, M., Brini, F., Sharma, A., and Landi, M. (2021). Role of jasmonic acid in plants: the molecular point of view. Plant Cell Rep. 40, 1471–1494. doi: 10.1007/s00299-021-02687-4
Gong, Q., Li, S., Zheng, Y., Duan, H., Xiao, F., Zhuang, Y., et al. (2020). SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis. Plant J. 102, 1157–1171. doi: 10.1111/tpj.14689
Hess, D. T., Matsumoto, A., Kim, S.-O., Marshall, H. E., and Stamler, J. S. (2005). Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166. doi: 10.1038/nrm1569
Hussain, A., Yun, B.-W., Kim, J. H., Gupta, K. J., Hyung, N.-I., and Loake, G. J. (2019). Novel and conserved functions of S-nitrosoglutathione reductase in tomato. J. Exp. Bot. 70, 4877–4886. doi: 10.1093/jxb/erz234
Hussain, M. A., Fahad, S., Sharif, R., Jan, M. F., Mujtaba, M., Ali, Q., et al. (2020). Multifunctional role of brassinosteroid and its analogues in plants. Plant Growth Regul. 92, 141–156. doi: 10.1007/s10725-020-00647-8
Iglesias, M. J., Terrile, M. C., Correa-Aragunde, N., Colman, S. L., Izquierdo-Álvarez, A., Fiol, D. F., et al. (2018). Regulation of SCF(TIR1/AFBs) E3 ligase assembly by S-nitrosylation of Arabidopsis SKP1-like1 impacts on auxin signaling. Redox Biol. 18, 200–210. doi: 10.1016/j.redox.2018.07.003
Jia, T., Zhang, K., Li, F., Huang, Y., Fan, M., and Huang, T. (2020). The AtMYB2 inhibits the formation of axillary meristem in Arabidopsis by repressing RAX1 gene under environmental stresses. Plant Cell Rep. 39, 1755–1765. doi: 10.1007/s00299-020-02602-3
Khan, M., Imran, Q. M., Shahid, M., Mun, B. G., Lee, S. U., Khan, M. A., et al. (2019). Nitric oxide- induced AtAO3 differentially regulates plant defense and drought tolerance in Arabidopsis thaliana. BMC Plant Biol. 19:602. doi: 10.1186/s12870-019-2210-3
Khan, N. A., Khan, M. I. R., Ferrante, A., and Poor, P. (2017). Ethylene: a key regulatory molecule in plants. Front. Plant Sci. 8:1782. doi: 10.3389/fpls.2017.01782
Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. doi: 10.1105/tpc.12.12.2339
Kneeshaw, S., Gelineau, S., Tada, Y., Loake, G. J., and Spoel, S. H. (2014). Selective protein denitrosylation activity of thioredoxin-h5 modulates plant immunity. Mol. Cell 56, 153–162. doi: 10.1016/j.molcel.2014.08.003
Kolbert, Z. (2019). Strigolactone-nitric oxide interplay in plants: the story has just begun. Physiol. Plant. 165, 487–497. doi: 10.1111/ppl.12712
Kwon, E., Feechan, A., Yun, B. W., Hwang, B. H., Pallas, J. A., Kang, J. G., et al. (2012). AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta 236, 887–900. doi: 10.1007/s00425-012-1697-8
Lau, S.-E., Hamdan, M. F., Pua, T.-L., Saidi, N. B., and Tan, B. C. (2021). Plant nitric oxide signaling under drought stress. Plants (Basel) 10:360. doi: 10.3390/plants10020360
Liao, C., Zheng, Y., and Guo, Y. (2017). MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 216, 163–177. doi: 10.1111/nph.14679
Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-Nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. doi: 10.1104/pp.104.058719
Lindermayr, C., Saalbach, G., Bahnweg, G., and Durner, J. (2006). Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J. Biol. Chem. 281, 4285–4291. doi: 10.1074/jbc.M511635200
Lindermayr, C., Sell, S., Müller, B., Leister, D., and Durner, J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907. doi: 10.1105/tpc.109.066464
Loake, G., and Grant, M. (2007). Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. doi: 10.1016/j.pbi.2007.08.008
Lozano-Juste, J., and León, J. (2011). Nitric oxide regulates DELLA content and PIF expression to promote photomorphogenesis in Arabidopsis. Plant Physiol. 156, 1410–1423. doi: 10.1104/pp.111.177741
Mabuchi, K., Maki, H., Itaya, T., Suzuki, T., Nomoto, M., Sakaoka, S., et al. (2018). MYB30 links ROS signaling, root cell elongation, and plant immune responses. Proc. Natl. Acad. Sci. U.S.A. 115, E4710–E4719. doi: 10.1073/pnas.1804233115
Malik, S. I., Hussain, A., Yun, B. W., Spoel, S. H., and Loake, G. J. (2011). GSNOR-mediated de-nitrosylation in the plant defence response. Plant Sci. 181, 540–544. doi: 10.1016/j.plantsci.2011.04.004
Mao, J., and Li, J. (2020). Regulation of three key kinases of brassinosteroid signaling pathway. Int. J. Mol. Sci. 21, 4340. doi: 10.3390/ijms21124340
Molla, K. A., Karmakar, S., Chanda, P. K., Sarkar, S. N., Datta, S. K., and Datta, K. (2016). Tissue-specific expression of Arabidopsis NPR1 gene in rice for sheath blight resistance without compromising phenotypic cost. Plant Sci. 250, 105–114. doi: 10.1016/j.plantsci.2016.06.005
Montilla-Bascón, G., Rubiales, D., Hebelstrup, K. H., Mandon, J., Harren, F. J. M., Cristescu, S. M., et al. (2017). Reduced nitric oxide levels during drought stress promote drought tolerance in barley and is associated with elevated polyamine biosynthesis. Sci. Rep. 7:13311. doi: 10.1038/s41598-017-13458-1
Murad, F. (1986). Cyclic guanosine monophosphate as a mediator of vasodilation. J. Clin. Invest. 78, 1–5. doi: 10.1172/JCI112536
Nabi, R. B. S., Tayade, R., Hussain, A., Kulkarni, K. P., Imran, Q. M., Mun, B.-G., et al. (2019). Nitric oxide regulates plant responses to drought, salinity, and heavy metal stress. Environ. Exp. Bot. 161, 120–133. doi: 10.1016/j.envexpbot.2019.02.003
Nakamura, T., Wang, L., Wong, C. C., Scott, F. L., Eckelman, B. P., Han, X., et al. (2010). Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol. Cell 39, 184–195. doi: 10.1016/j.molcel.2010.07.002
Nakashima, K., Fujita, Y., Kanamori, N., Katagiri, T., Umezawa, T., Kidokoro, S., et al. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2. 2, SRK2E/SnRK2. 6/OST1 and SRK2I/SnRK2. 3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363. doi: 10.1093/pcp/pcp083
Niu, L., Yu, J., Liao, W., Yu, J., Zhang, M., and Dawuda, M. M. (2017). Calcium and calmodulin are involved in nitric oxide-induced adventitious rooting of cucumber under simulated osmotic stress. Front. Plant Sci. 8:1684. doi: 10.3389/fpls.2017.01684
Nolan, T. M., Vukašinović, N., Liu, D., Russinova, E., and Yin, Y. (2020). Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32, 295–318. doi: 10.1105/tpc.19.00335
Ortiz-Morea, F. A., He, P., Shan, L., and Russinova, E. (2020). It takes two to tango–molecular links between plant immunity and brassinosteroid signalling. J. Cell Sci. 133:jcs246728. doi: 10.1242/jcs.246728
Overvoorde, P., Fukaki, H., and Beeckman, T. (2010). Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2:a001537.
Pattyn, J., Vaughan-Hirsch, J., and Van de Poel, B. (2021). The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 229, 770–782. doi: 10.1111/nph.16873
Qu, J., Nakamura, T., Cao, G., Holland, E. A., McKercher, S. R., and Lipton, S. A. (2011). S-Nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by β-amyloid peptide. Proc. Natl. Acad. Sci. U.S.A. 108, 14330–14335. doi: 10.1073/pnas.1105172108
Ramos Báez, R., and Nemhauser, J. L. (2021). Expansion and innovation in auxin signaling: where do we grow from here? Development 148, dev187120. doi: 10.1242/dev.187120
Richter, R., Behringer, C., Müller, I. K., and Schwechheimer, C. (2010). The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 24, 2093–2104. doi: 10.1101/gad.594910
Serpa, V., Vernal, J., Lamattina, L., Grotewold, E., Cassia, R., and Terenzi, H. (2007). Inhibition of AtMYB2 DNA-binding by nitric oxide involves cysteine S-nitrosylation. Biochem. Biophys. Res. Commun. 361, 1048–1053. doi: 10.1016/j.bbrc.2007.07.133
Shearer, H. L., Cheng, Y. T., Wang, L., Liu, J., Boyle, P., Després, C., et al. (2012). Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion. Mol. Plant Microbe Interact. 25, 1459–1468. doi: 10.1094/MPMI-09-11-0256
Silveira, N. M., Ribeiro, R. V., Prataviera, P. J. C., Pissolato, M. D., Pieretti, J. C., Seabra, A. B., et al. (2020). Germination and initial growth of common bean plants under water deficit as affected by seed treatment with S-nitrosoglutathione and calcium chloride. Theor. Exp. Plant Physiol. 32, 49–62. doi: 10.1007/s40626-020-00166-x
Skubacz, A., Daszkowska-Golec, A., and Szarejko, I. (2016). The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7:1884. doi: 10.3389/fpls.2016.01884
Stomberski, C. T., Hess, D. T., and Stamler, J. S. (2019). Protein S-nitrosylation: determinants of specificity and enzymatic regulation of S-nitrosothiol-based signaling. Antioxid. Redox Signal. 30, 1331–1351. doi: 10.1089/ars.2017.7403
Su, P., Zhao, L., Li, W., Zhao, J., Yan, J., Ma, X., et al. (2021). Integrated metabolo-transcriptomics and functional characterization reveals that the wheat auxin receptor TIR1 negatively regulates defense against Fusarium graminearum. J. Integr. Plant Biol. 63, 340–352. doi: 10.1111/jipb.12992
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z. L., Song, J. Q., Wang, C., et al. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
Tavares, C. P., Vernal, J., Delena, R. A., Lamattina, L., Cassia, R., and Terenzi, H. (2014). S-nitrosylation influences the structure and DNA binding activity of AtMYB30 transcription factor from Arabidopsis thaliana. Biochim. Biophys. Acta 1844, 810–817. doi: 10.1016/j.bbapap.2014.02.015
Terrile, M. C., París, R., Calderón-Villalobos, L. I., Iglesias, M. J., Lamattina, L., Estelle, M., et al. (2012). Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 70, 492–500. doi: 10.1111/j.1365-313X.2011.04885.x
Wang, P., Du, Y., Hou, Y. J., Zhao, Y., Hsu, C. C., Yuan, F., et al. (2015). Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. U.S.A. 112, 613–618. doi: 10.1073/pnas.1423481112
Wang, Y. Q., Feechan, A., Yun, B. W., Shafiei, R., Hofmann, A., Taylor, P., et al. (2009). S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 284, 2131–2137. doi: 10.1074/jbc.M806782200
Wang, Y., Yun, B.-W., Kwon, E., Hong, J. K., Yoon, J., and Loake, G. J. (2006). S-nitrosylation: an emerging redox-based post-translational modification in plants. J. Exp. Bot. 57, 1777–1784. doi: 10.1093/jxb/erj211
Wu, C., Liu, T., Chen, W., Oka, S.-i., Fu, C., Jain, M. R., et al. (2010). Redox regulatory mechanism of transnitrosylation by thioredoxin. Mol. Cell. Proteomics 9, 2262–2275. doi: 10.1074/mcp.M110.000034
Xu, W., Zhang, S. S., Wang, D. L., and Liu, J. Z. (2015). Drought tolerance of nitric oxide associated 1 mutant of Arabidopsis is mostly due to its reduced transpiration as a result of smaller stature. Acta Physiol. Plant. 37:134.
Yang, H., Mu, J., Chen, L., Feng, J., Hu, J., Li, L., et al. (2015). S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 167, 1604–1615. doi: 10.1104/pp.114.255216
Yoshida, R., Hobo, T., Ichimura, K., Mizoguchi, T., Takahashi, F., Aronso, J., et al. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43, 1473–1483. doi: 10.1093/pcp/pcf188
Yu, M., Yun, B.-W., Spoel, S. H., and Loake, G. J. (2012). A sleigh ride through the SNO: regulation of plant immune function by protein S-nitrosylation. Curr. Opin. Plant Biol. 15, 424–430. doi: 10.1016/j.pbi.2012.03.005
Yuan, H. M., and Huang, X. (2016). Inhibition of root meristem growth by cadmium involves nitric oxide-mediated repression of auxin accumulation and signalling in Arabidopsis. Plant Cell Environ. 39, 120–135. doi: 10.1111/pce.12597
Zhou, Q., Meng, Q., Tan, X., Ding, W., Ma, K., Xu, Z., et al. (2021). Protein phosphorylation changes during systemic acquired resistance in Arabidopsis thaliana. Front. Plant Sci. 12:748287. doi: 10.3389/fpls.2021.748287
Keywords: nitric oxide, phytohormones, S-nitrosylation, plant stress, proteins
Citation: Pande A, Mun BG, Rahim W, Khan M, Lee DS, Lee GM, Al Azzawi TNI, Hussain A, Kim CK and Yun BW (2022) Phytohormonal Regulation Through Protein S-Nitrosylation Under Stress. Front. Plant Sci. 13:865542. doi: 10.3389/fpls.2022.865542
Received: 30 January 2022; Accepted: 18 February 2022;
Published: 24 March 2022.
Edited by:
Anna Wawrzyńska, Institute of Biochemistry and Biophysics (PAN), PolandReviewed by:
Manda Yu, University of Wisconsin–Milwaukee, United StatesMaciej Ostrowski, Nicolaus Copernicus University in Toruń, Poland
Copyright © 2022 Pande, Mun, Rahim, Khan, Lee, Lee, Al Azzawi, Hussain, Kim and Yun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang Kil Kim, Y2traW1Aa251LmFjLmty; Byung Wook Yun, Ynd5dW5Aa251LmFjLmty
 Anjali Pande
Anjali Pande Bong Gyu Mun
Bong Gyu Mun Waqas Rahim
Waqas Rahim Murtaza Khan
Murtaza Khan Da Sol Lee1
Da Sol Lee1 Adil Hussain
Adil Hussain Chang Kil Kim
Chang Kil Kim Byung Wook Yun
Byung Wook Yun