
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 08 April 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.864986
This article is part of the Research TopicSignal Transduction of Plant Organ Senescence and Cell DeathView all 11 articles
Leaf senescence is a developmentally programmed cell death process that is influenced by a variety of endogenous signals and environmental factors. Here, we report that MPK3 and MPK6, two Arabidopsis mitogen-activated protein kinases (MAPKs or MPKs), and their two upstream MAPK kinases (MAPKKs or MKKs), MKK4 and MKK5, are key regulators of leaf senescence. Weak induction of constitutively active MAPKKs driven by steroid-inducible promoter, which activates endogenous MPK3 and MPK6, induces leaf senescence. This gain-of-function phenotype requires functional endogenous MPK3 and MPK6. Furthermore, loss of function of both MKK4 and MKK5 delays leaf senescence. Expression profiling leads to the identification of matrix metalloproteinases (MMPs), a family of zinc- and calcium-dependent endopeptidases, as the downstream target genes of MPK3/MPK6 cascade. MPK3/MPK6 activation-triggered leaf senescence is associated with rapid and strong induction of At3-MMP and At2-MMP. Expression of Arabidopsis MMP genes is strongly induced during leaf senescence, qualifying them as senescence-associated genes (SAGs). In addition, either constitutive or inducible overexpression of At3-MMP is sufficient to trigger leaf senescence. Based on these findings, we conclude that MPK3/MPK6 MAPK cascade and MMP target genes further downstream are involved in regulating leaf senescence in Arabidopsis.
Senescence at the final stage of plant growth season or life cycle is a programmed cell death process that is influenced by both endogenous developmental programs and environmental factors (Gan and Amasino, 1997; Lim et al., 2007; Woo et al., 2013). Leaf yellowing as a result of the disruption in photosynthesis and the loss of chlorophyll due to the breakdown of the structural integrity of chloroplasts is a typical phenotypic symptom of leaf senescence. This is accompanied by other dramatic changes in cellular metabolisms, such as degradation of membrane lipids, proteins, nucleic acids, and other macromolecules (Gan and Amasino, 1997; Lim et al., 2007). Nutrients released from the senescing leaves are recycled and used by younger growing leaves, developing seeds, or storage tissues to ensure an optimal condition for the next generation or growing season (Gan and Amasino, 1997; Guo and Gan, 2005; Lim et al., 2007; Woo et al., 2016). In addition to age, the onset and progression of leaf senescence are influenced by various internal signals including abscisic acid, salicylic acid, ethylene, and jasmonate, as well as environmental stresses such as darkness, extreme high or low temperature, nutrient deficiency, oxidative stress, and pathogen infections (Gan, 2003; Lim et al., 2007). Although many genes have been identified as senescence-associated genes (SAGs), their exact functions in the senescence remain largely unclear (Li et al., 2012, 2014).
Mitogen-activated protein kinase (MAPK or MPK) cascades are major pathways by which extracellular stimuli are transduced into cellular responses in eukaryotic cells (Ichimura et al., 2002; Pedley and Martin, 2005; Pitzschke et al., 2009; Andreasson and Ellis, 2010; Rodriguez et al., 2010; Tena et al., 2011; Xu and Zhang, 2015; Zhang et al., 2018). A basic MAPK cascade is composed of three interconnected kinases, a MAPK (MPK), which is activated by its upstream MAPK kinase (MAPKK, MKK, or MEK), via phosphorylation of the TXY activation motif. MAPKK activity is in turn regulated by the topmost member of the module, MAPKK kinase (MAPKKK, or MEKK), via phosphorylation. A MAPKKK receives signals from receptors/sensors either directly or indirectly. In Arabidopsis, there are 20 MAPKs, 10 MAPKKs, and approximately 60 putative MAPKKKs (Ichimura et al., 2002). MPK3 and MPK6, two Arabidopsis MAPKs with the highest homology, share two redundant upstream MAPK kinases, MKK4 and MKK5 (Wang et al., 2007; Su et al., 2017, 2018). In plant defense, MAPKKK3 and MAPKKK5 function redundantly upstream of MKK4/MKK5-MPK3/MPK6 in plant PAMP-triggered immunity (Bi et al., 2018; Sun et al., 2018). In plant development, YDA is upstream of the MKK4/MKK5-MPK3/MPK6 module in plant inflorescence architecture and stomatal development (Bergmann et al., 2004; Wang et al., 2007; Meng et al., 2012). Several reports have revealed the important roles of MAPK signaling cascade in regulating leaf senescence. MKK9-MPK6 cascade was shown to play a positive role in regulating leaf senescence in Arabidopsis (Zhou et al., 2009). MPK6-WRKY6-NPR1 and MPK6-EIN2-EIN3-ORE9 modules were implicated in SA- and MeJA-induced leaf senescence, respectively (Chai et al., 2014; Zhang et al., 2016). MAPKKK18-MKK3-MPK1/2/7, another MAPK cascade, involved in ABA-triggered leaf senescence (Matsuoka et al., 2015). Recently, MKK4/MKK5-MPK1/MPK2 cascade was reported to regulate SA-induce leaf senescence via phosphorylation of NPR1 (Zhang et al., 2020). These findings suggest a complex regulation network involved the MAPK signaling network during leaf senescence.
Matrix metalloproteinases (MMPs) are a family of zinc- and calcium-dependent endopeptidase (Rawlings et al., 2010). As a well-known proteolytic enzyme in animals, MMPs are involved in extracellular matrix remodeling, cell migration, cell proliferation, adhesion, and cellular signaling by limited proteolytic processing of their substrate proteins (Sternlicht and Werb, 2001; Overall, 2002; Parks et al., 2004; Page-McCaw et al., 2007). MMPs have also been identified in plants (Pak et al., 1997). Based on their expression patterns, plant MMPs have been implicated in plant growth and development (Delorme et al., 2000; Liu et al., 2001; Frick and Schaller, 2002; Ratnaparkhe et al., 2009; Schiermeyer et al., 2009; Zimmermann et al., 2016; Das et al., 2018). Soybean SEMP1/Gm1-MMP was shown to be expressed only in mature leaves, suggesting a role in tissue remodeling during leaf expansion (Pak et al., 1997). In cucumber, Cs1-MMP was found to be associated with senescence and cell death in cotyledon development (Delorme et al., 2000). A subtilisin-like proteinase p69b was identified as a substrate of tomato Sl-MMPs in cell death control (Zimmermann et al., 2016). More recently, Rice OsMMP1 was reported to play pleiotropic roles in plant development and symplastic-apoplastic transport by modulating cellulose and callose depositions (Das et al., 2018). In Arabidopsis, the MMP family consists of five members named At1-MMP to At5-MMP (Maidment et al., 1999). All five At-MMP genes display distinct tissue/organ development-specific expression patterns, suggesting differential physiological functions for each enzyme (Maidment et al., 1999; Flinn, 2008).
In this study, we found that a weak and long-lasting activation of MPK3/MPK6 after a low-level induction of the constitutively active MAPKKs upstream (MKK4, MKK5, and their tobacco ortholog NtMEK2) is sufficient to trigger leaf senescence, which is associated with At3-MMP and At2-MMP gene activation. In contrast, loss of function of both MKK4 and MKK5 delays leaf senescence. Arabidopsis MMP gene expression is strongly induced during leaf senescence, qualifying them as senescence-associated genes (SAGs). Over-expression of At3-MMP, either constitutively or under the control of an inducible promoter, is sufficient to trigger leaf senescence. Collectively, this study reveals a signaling pathway involving MPK3/MPK6 cascade and MMP target genes in leaf senescence in Arabidopsis thaliana.
After surface-sterilized and vernalization at 4°C for 3–5 days, seeds were sown in half-strength Murashige and Skoog (MS) medium with 0.45% Phytagar and grown in a growth chamber at 22°C with continuous light (80 μE/m–2s–1). Seven-day-old seedlings were transplanted to soil and grown at 22°C and a 14-h-light/10-h-dark cycle. Col-0 ecotype was used as the wild type. T-DNA insertion mutants including at1-mmp (SALK_205145C), at2-mmp (SM_3_5305), at3-mmp (SM_3_28404), at4-mmp (GABI_075C07), at5-mmp (SAIL_390_c06), mpk3-1(SALK_151594), mpk6-2 (SALK_073907), mkk7 (SM_3_21961), and mkk9 (SAIL_60_H06) were obtained from the Arabidopsis Biological Resource Center (ABRC). The mkk4 and mkk5 single tilling mutants (Zhao et al., 2014) were backcrossed with Col-0 to remove the er-105 mutant allele and then were crossed to generate a mkk4 mkk5 double mutant (Li et al., 2018). Transgenic plant GVG:NtMEK2DD (abbreviated as DD), in which dexamethasone (DEX)-inducible promoter-driven constitutively active NtMEK2DD transgene, was previously described (Yang et al., 2001; Ren et al., 2002). DD mpk3 (Wang et al., 2007) and DD mpk6 (Liu and Zhang, 2004) were generated by genetic cross between DD and mpk3-1, DD and mpk6-2, respectively. The mmp2 mmp3 double mutant, mmp2 mmp3 mmp5 triple mutant, at1;2;3;4;5-mmp pentuple mutant, and mkk7 mkk9 double mutant were generated by genetic cross and homozygous mutant plants were identified using T-DNA border primers and gene-specific primers (listed in Supplementary Table 1).
To generate DEX-inducible promoter-driven At-3MMP construct (GVG:At3-MMP-dHA) and constitutive overexpression 35S:At3-MMP-dHA construct, we amplified the full-length At-3MMP cDNA fragment using primers LP1/RP1 and LP2/RP2 and cloned the PCR fragment into a modified pBlueScript II vector with a double HA tag at the 3′-end. The 3MMP-dHA fragment was subsequently cloned into the XhoI/SpeI sites of the pTA7002 vector and a modified pBI121 vector with 35S double enhancer and XhoI/SpeI restriction sites (pBId vector), respectively. These two binary vectors were transformed into Agrobacterium tumefaciens strain GV3101. Arabidopsis transformation was performed by the floral dip procedure (Clough and Bent, 1998), and transformants were identified by screening for hygromycin (pTA7002 vector) or kanamycin (pBId vector) resistant T1 seedlings. Independent lines with At-3MMP transgene induction or expression were identified based on immunoblot analysis. From these transformations, two independent lines with a single copy of T-DNA insertion (based on the 3:1 segregation of antibiotic resistance in T2 progeny) were isolated, and homozygote transgenic plants were further identified in the progeny based on segregation of antibiotic resistance.
Leaves from 4-week-old plants were used for the leaf senescence assay. For detached leaf senescence analysis, fully expanded leaves (from the fifth to eighth leaf position) were detached and their petioles were inserted into 0.6% agar plates with DEX or EtOH solvent with the adaxial side facing up. The plates were kept under light (60 μE/m–2s–1) at 22°C. For observation of leaf senescence in whole plants, 4-week-old soil-grown plants were sprayed with 30 μM DEX. Whole plants were photographed at indicated times after treatment.
Chlorophyll was extracted with 80% (v/v) acetone from detached leaves (Arnon, 1949). Chlorophyll contents were measured at 645 and 663 nm and chlorophyll concentrations were calculated as (20.2 × A645 + 8.02 × A663)/g fresh weight. Maximal quantum yield of PSII photochemistry (Fv/Fm) was determined using a PAM 2000 portable chlorophyll fluorimeter in the dark-adapted leaf samples.
Total RNA was extracted using Trizol reagent (Invitrogen). RNA concentration was measured using a NanoDrop (Model 2000C). After an additional ethanol precipitation and DNase treatment, 1 μg of total RNA was used for reverse transcription. Quantitative PCR was conducted using a real-time PCR machine (Eppendorf, Germany). EF1α was used for internal control. The relative gene expression was calculated using the double ΔCt method. The absolute copy numbers were calculated based on the standard curve for each MMP gene for better assessment of the potential contribution of each MMP gene. The primers pairs used for qPCR are listed in Supplementary Table 1.
Total protein was extracted from leaf tissues by grinding with 3x volume of 1xSDS sample buffer [60 mM Tris/HCl, pH 6.8, 10% (v/v) glycerol, 1% (w/v) SDS] followed by boiling for 15 min. Protein extracts for immunoblot detection of phosphorylated MAPKs were prepared by grinding with extraction buffer [100 mM HEPES, pH 7.5, 5 mM EDTA, 10 mM DTT, 10 mM Na3VO4, 10 mM NaF, 50 mM β-glycerophosphate, 1 mM phenylmethylsulfony fluoride (PMSF), 5 μg/mL antipain, 5 μg/mL aprotinin, 5 μg/mL leupeptin, and 10% (v/v) glycerol]. Protein concentration was determined by using the Bio-Rad protein assay kit with BSA as a standard. Total proteins (10 μg) were separated in 10% SDS-PAGE gels. At-3MMP and DD protein induction were detected by immunoblot analysis using anti-HA (Sigma, dilution 1:10,000) and anti-FLAG (Sigma, dilution 1:10,000), respectively. Activation of MPK3 and MPK6 was detected by using anti-pTEpY (Cell Signaling, dilution 1:3,500). The blots were incubated with horseradish peroxidase-conjugated goat anti-mouse (for anti-HA and anti-FLAG) or goat anti-rabbit (for anti-pTEpY) secondary antibodies. Coomassie brilliant blue staining of duplicated gels was used to confirm equal loading.
At least two independent repetitions were performed for experiment with multiple time points. For single time point experiments, at least three independent repetitions were done. Results from one of the independent repeats that gave similar results were shown. Two-way ANOVA assay was used to determine whether the difference between two groups of data at a specific time point is statistically significant (P < 0.05). Statistically different data groups are indicated by using asterisks placed above the columns in the graphs.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At1-MMP (At4G16640), At2-MMP (At1G70170), At3-MMP (At1G24140), At4-MMP (At2G45040), At5-MMP (At1G59970), MPK3 (At3G45640), MPK6 (At2G43790), MKK4 (At1G51660), MKK5 (At3G21220), EF1α (At5G60390, and SAG12 (At5G45890).
To understand the function of MPK3 and MPK6 in Arabidopsis and their orthologs in other plant species, we previously generated conditional gain-of-function systems by expressing the constitutively active MAPKK (Arabidopsis MKK4 and MKK5, and tobacco NtMEK2) variants under the control of a steroid-inducible promoter (Yang et al., 2001; Ren et al., 2002). In this study, DD plants refer specifically to GVG:NtMEK2DD transgenic Arabidopsis (abbreviated as DD for the substitution of two Ser/Thr residues in the activation loop of MAPKK with Asp to make it constitutively active). Full induction of transgene expression after dexamethasone (DEX) treatment induces high-level activation of endogenous MPK3/MPK6 in Arabidopsis, which triggers a hypersensitive response (HR)-like cell death (Figure 1; Yang et al., 2001; Ren et al., 2002). However, when these plants were treated with lower concentrations of DEX (equal or less than 100 nM), the plants showed a leaf-yellowing phenotype similar to senescence (Figure 1). Similarly, lower concentrations of DEX treatment of GVG:AtMKK4DD and GVG:AtMKK5DD Arabidopsis plants also lead to leaf-yellowing phenotype (Supplementary Figure 1). This phenotype is dependent on functional MPK3 and MPK6. As shown in Figure 2A, in either mpk3 or mpk6 single mutant background, the gain-of-function DD-induced leaf yellowing phenotype was attenuated.
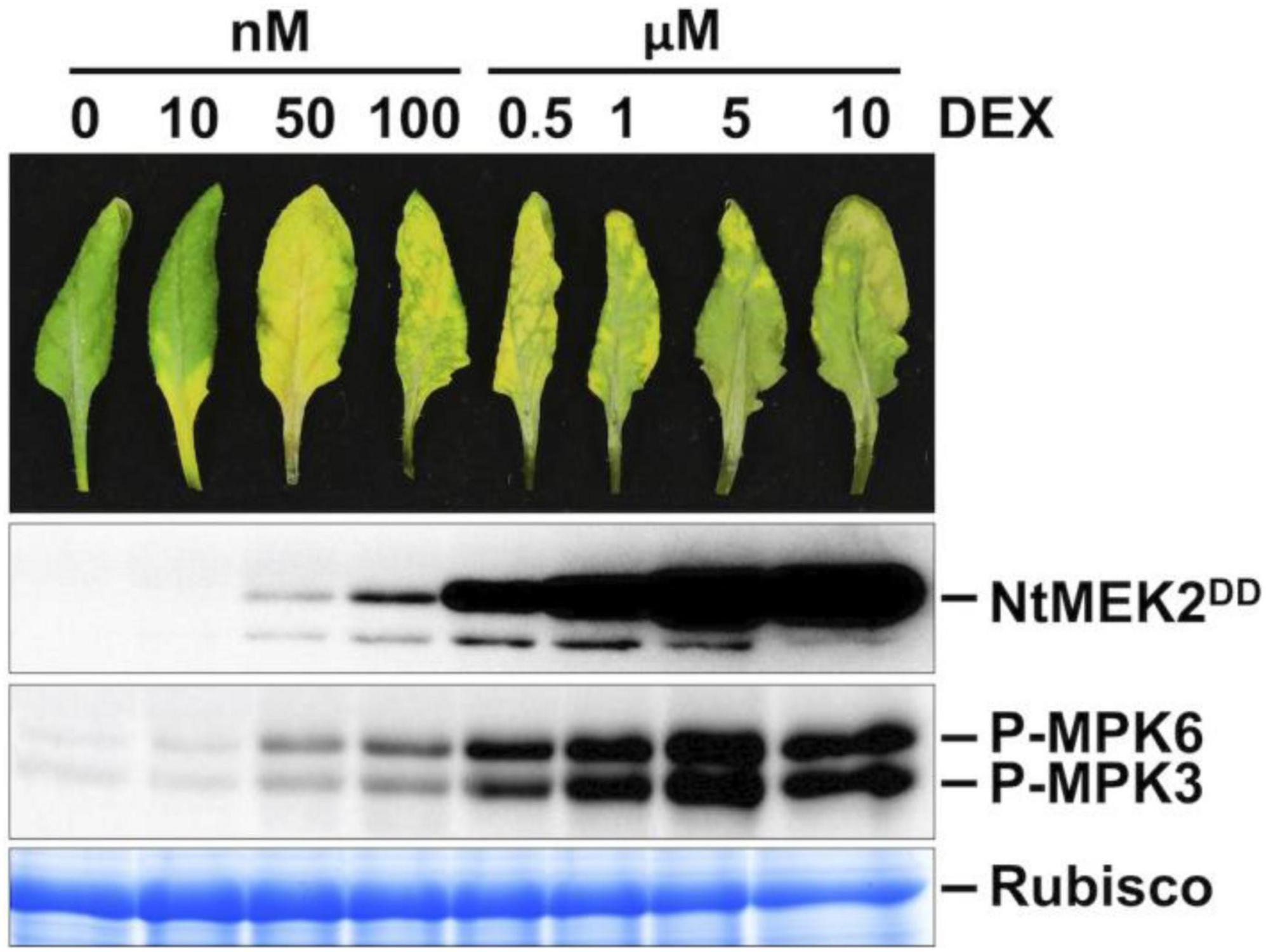
Figure 1. Weak induction of constitutively active MAPKK promotes leaf senescence. Treatment of DD plants with different concentrations of DEX can control the amplitude of MPK3/MPK6 phosphorylation/activation. Fully expanded leaves from 4-week-old soil-grown plants were detached and their petioles were inserted into 0.6% agar medium with different concentrations of DEX. Photos were taken at 3 days after DEX treatment (first panel). Leaves treated with DEX at 0.5 μM or above showed rapid cell death and stayed green. Leaf samples were collected at 1 day after DEX treatment. DD protein induction was detected by immunoblot assay using an anti-flag antibody (second panel). MAPK activation was detected by immunoblot assay using an anti-pTEpY antibody (third panel). Coomassie brilliant blue staining of a duplicated gel was used to confirm equal loading (fourth panel).
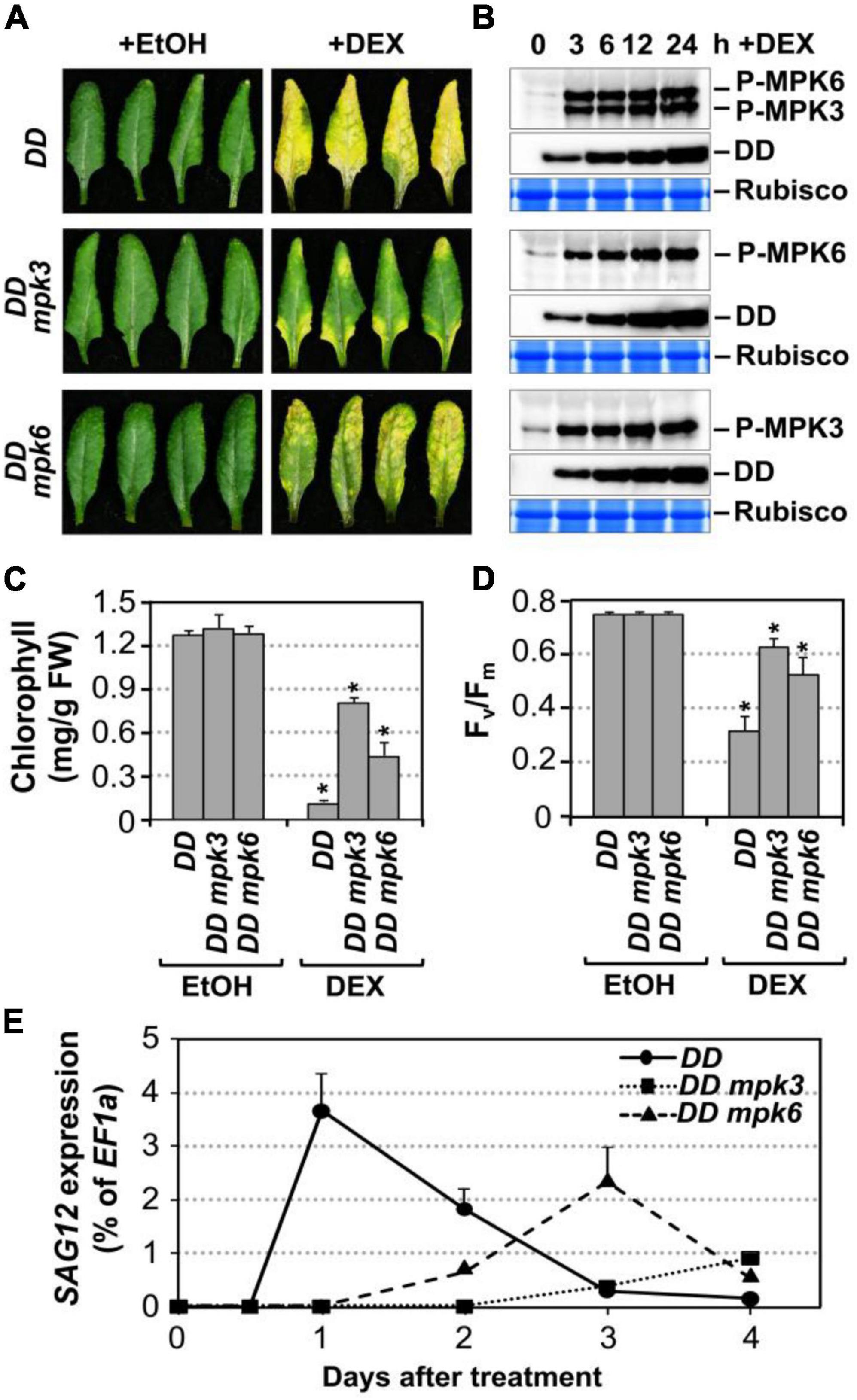
Figure 2. Leaf senescence in gain-of-function DD plant is dependent on endogenous MPK3 and MPK6. (A) Fully expanded leaves from 4-week-old DD, DD mpk3, DD mpk6 plants were detached and their petioles were inserted into 0.6% agar medium with DEX (100 nM) or EtOH (solvent control). Photos were taken 4 days after treatment. (B) MAPK activation (top) and DD protein induction (middle) in DD, DD mpk3, and DD mpk6 plants after DEX treatment were detected by immunoblot analysis using anti-pTEpY and anti-Flag antibody, respectively. Coomassie brilliant blue staining of duplicated gels were used to confirm equal loading (bottom). (C,D) Measurement of chlorophyll content (C) and Fv/Fm (D) in leaves shown in (A). Asterisks indicate a significant difference (P ≤ 0.001). Values are means ± SD, n = 10. (E) RT-qPCR analysis of SAG12 expression in leaves at indicated times after DEX treatment. Gene expression was calculated using the double ΔCt method and EF1α was used as a reference. Values were means ± SD, n = 3.
To confirm that the leaf yellowing phenotype in DD plants treated with low concentrations of DEX is indeed a senescence process, we measured typical senescence-associated physiological markers including chlorophyll content, photochemical efficiency of photosystem II (monitored as Fv/Fm), and the transcript levels of SAG12, a widely used molecular marker for leaf senescence (Noh and Amasino, 1999; Pontier et al., 1999). As shown in Figure 2C, the chlorophyll content decreased by approximately 92, 42, and 66% in the DD, DD mpk3, and DD mpk6 plants, respectively. Similarly, Fv/Fm value reduced by ∼56% in the DD plants at 4 days after DEX treatment. In contrast, in mpk3 or mpk6 mutant background, DD-induced Fv/Fm reduction was significantly attenuated (Figure 2D). Furthermore, SAG12 expression was strongly induced in DD plants, while the induction of SAG12 was partially compromised in DD mpk3 and DD mpk6 plants (Figure 2E). Taken together, these results suggested DD-induced leaf yellowing is a senescence process, which is dependent on functional downstream MPK3 and MPK6.
To further confirm that MPK3 and MPK6 were responsible for the induction of leaf senescence in DD plants, we measured the activation of MPK3/MPK6 using phospho-specific pTEpY antibody in DD, DD mpk3, and DD mpk6 plants. Immunoblot analysis using anti-flag antibody showed comparable DD protein induction in all three genotypes (Figure 2B). The delayed leaf senescence in DD mpk3 and DD mpk6 plants demonstrated that leaf senescence induction by DD required the activation of endogenous MPK3 and MPK6. The senescence phenotype in DD mpk3 leaves was much less severe than that in DD mpk6 leaves in comparison to the DD control, suggesting that MPK3 plays a more important role than MPK6 in this process.
Arabidopsis MKK4 and MKK5 function redundantly upstream of MPK3/MPK6 in several developmental processes (Xu and Zhang, 2015). In addition, expression of gain-of-function constitutively active DD form of these MAPKKs is sufficient to induce senescence. To gain loss-of-function evidence, we compared the leaf senescence in wild type, mkk4 single, mkk5 single, and mkk4 mkk5 double mutants in detached leaves under continuous light. Slight delay in leaf senescence was observed in mkk4 or mkk5 single mutant compared to wild type (Supplementary Figure 2A). In contrast, leaf senescence was significantly delayed in the mkk4 mkk5 double mutant (Figure 3A). Associated with the delayed phenotype, no induction of SAG12 gene expression was detected in mkk4 mkk5 double mutants 6 days after detachment. In wild type, the level of SAG12 transcript was detected in 4 days after detachment and increased greatly afterward (Figure 3B). Measurement of chlorophyll content and Fv/Fm showed that greater losses in chlorophyll content and Fv/Fm value in the wild type plants than those in the mkk4 mkk5 double mutant plants (Figures 3C,D). To further confirm whether MKK4 and MKK5 acted upstream of MPK3 and MPK6 during senescence, we evaluated the phosphorylation activation of MPK3 and MPK6 during different leaf senescence stage. As shown in Figure 3E, age-dependent leaf senescence-induced MPK3/MPK6 activation was impaired in the mkk4 mkk5 double mutant. These data suggested that MKK4 and MKK5 are involved in plant leaf senescence and play redundant function in activating MPK3/MPK6 during leaf senescence.
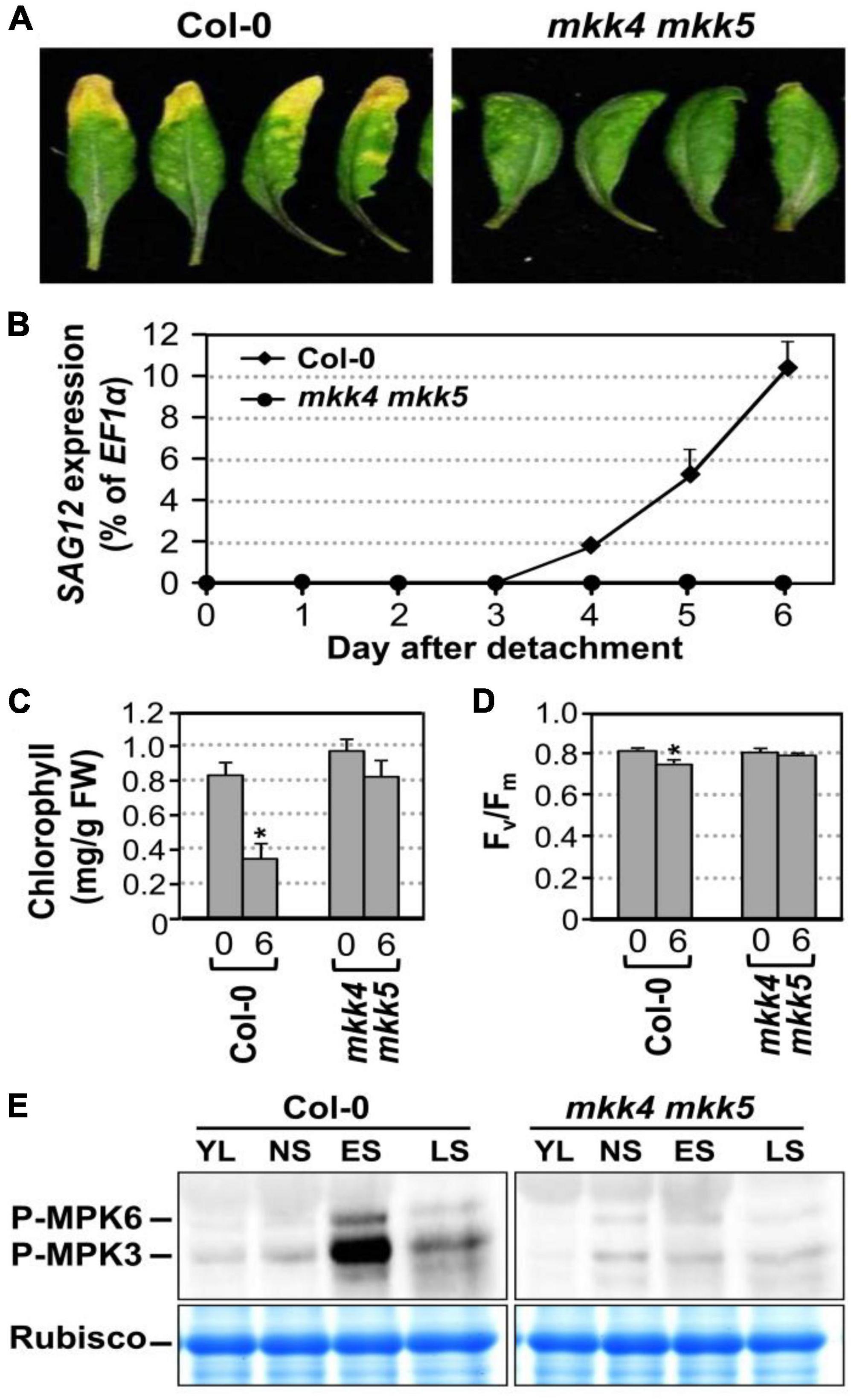
Figure 3. MKK4 and MKK5 play redundant function in leaf senescence. (A) Leaf senescence phenotype of detached leaves of wild type and mkk4 mkk5 double mutants. Fully expanded leaves from 4-week-old soil-grown plants were detached and their petioles were inserted to 0.6% agar medium under continuous light. Photos were taken 6 days after detachment. (B) RT-qPCR analysis of the transcript levels of SAG12 in wild type and mkk4 mkk5 double mutants at indicated times. The expression of EF1α was used as an internal reference. Values are means ± SD, n = 3. (C,D) Quantitative analysis of chlorophyll contents (C) and Fv/Fm (D) in leaves shown in (A). Values are means ± SD, n = 10. (E) Phosphorylation/activation of MPK3/MPK6 induced by age-dependent leaf senescence was detected by immunoblot analysis using anti-pTEpY antibody (upper). Equal loading of proteins was confirmed by Coomassie brilliant blue staining of Rubisco (lower). YL, young leaves; NS, fully expanded no senescence mature leaves; ES, early senescence leaves, with < 25% leaf yellowing; LS, lately senescence leaves, with > 50% leaf yellowing (please also see Figure 5A). Asterisks indicate a significant difference (P ≤ 0.01).
In addition to MKK4 and MKK5, MKK7 and MKK9 were also reported to be upstream of MPK3/MPK6 (Xu et al., 2008; Zhou et al., 2009; Jia et al., 2016). As a result, we also examined the leaf senescence in mkk7 single, mkk9 single, and mkk7 mkk9 double mutants under the same experimental condition. As shown in Supplementary Figure 2A, we did not see an obvious difference in leaf senescence in comparison to wild type. As a result, we conclude that MPK4/MPK5, but not MKK7/MKK9, are upstream of the MPK3/MPK6 in plant leaf senescence.
To identify unknown components downstream of MPK3/MPK6 cascade in leaf senescence, we mined the expression profiling data in DD transgenic plants after DEX treatment in our previous study (Su et al., 2018). At3-MMP is one of the highest induced genes. As a result, we quantified the expression of all five At-MMP genes in DD plants after DEX treatment and in leaves during senescence using RT-qPCR. As shown in Figure 4A, the expression of At2-MMP and At3-MMP was induced approximately 10 and 500-folds over its basal level, respectively. In contrast, At1-MMP, At4-MMP, and At5-MMP transcripts could be reliably detected, but were not induced after MPK3/MPK6 activation. To eliminate the influence of amplification efficiency during qPCR and better compare the expression levels of different MMP genes, we also generated the standard curves and calculated the absolute expression levels of each MMP after activation of DD. As shown in Figure 4B, we found that the expression levels of At2-MMP and At3-MMP were among the highest, while At1-MMP, At4-MMP, and At5-MMP were expressed at low levels. To confirm that MPK3 and MPK6 were responsible for the induction of At2-MMP and At3-MMP expression level in the DD plants, we also examined the expression of At2-MMP and At3-MMP in DD, DD mpk3, and DD mpk6 plants. As shown in Figures 4C,D, while At2-MMP and At3-MMP expression was highly induced in DD plants after DEX treatment, their expression in DD mpk3 and DD mpk6 plants was partially compromised, correlating with the delayed leaf senescence (Figure 2A). This finding demonstrated that the induction of At2-MMP and At3-MMP in DD seedlings after DEX treatment is a result of MPK3/MPK6 activation.
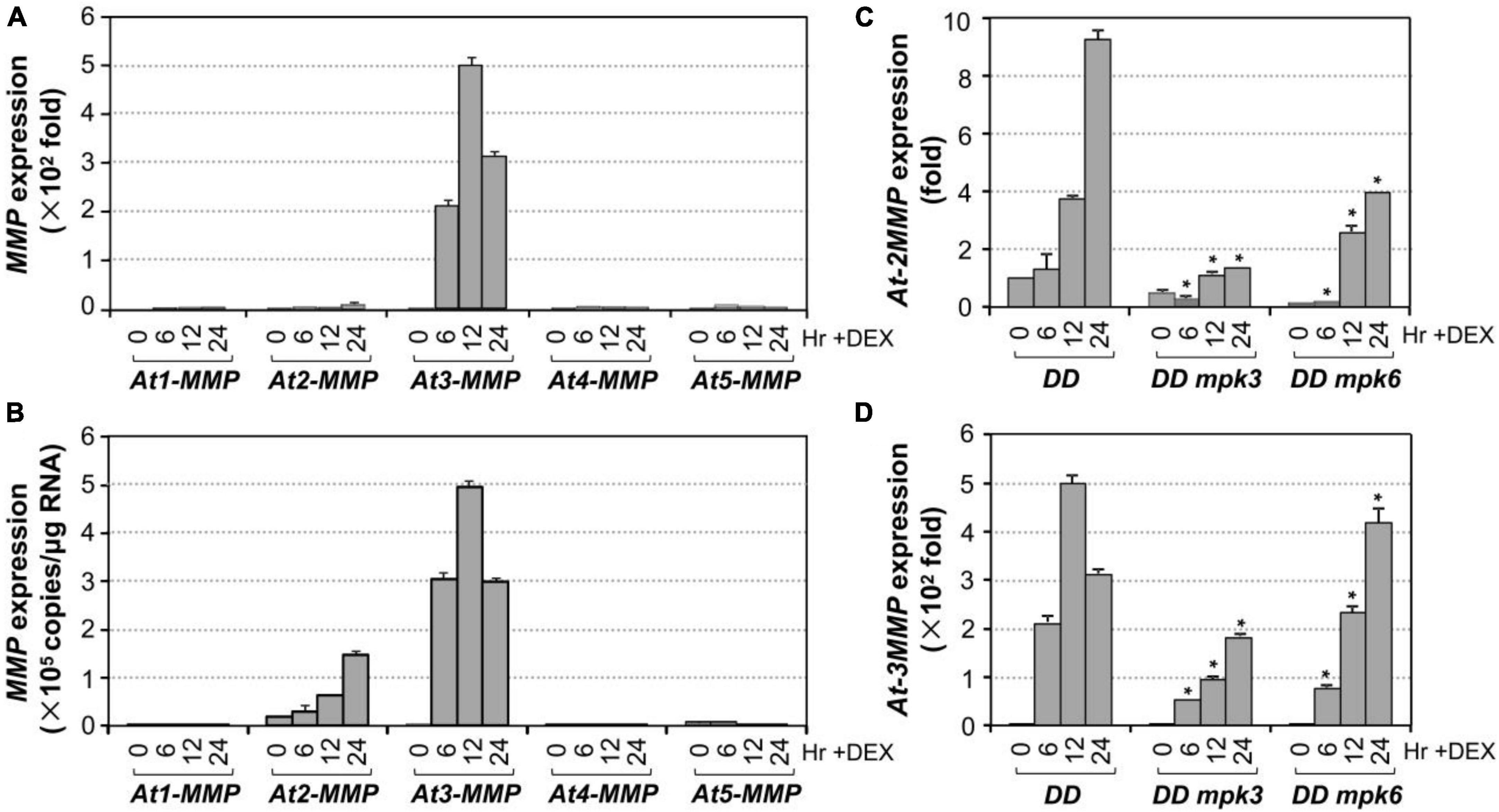
Figure 4. Induction of Arabidopsis MMP genes expression after MPK3/MPK6 activation in the gain-of-function DD seedlings. (A) The induction of MMP gene expression was quantified by RT-qPCR. MMP induction (fold of induction relative to the level before induction) was calculated using the double ΔCt method. The expression of EF1α was used to normalize the samples. (B) Absolute expression levels of MMP induction was calculated as copies numbers per μg of total RNA by the calibration curve established for each MMP gene, which allows comparison to expression levels between different MMP genes. Values were means ± SD, n = 3. (C,D) Activation of At2-MMP (C) and At3-MMP (D) expression in the DD seedlings were compromised in either mpk3 or mpk6 mutant background. Gene expression was quantified by RT-qPCR and calculated as in (A). Asterisks indicate a significant difference (P ≤ 0.001). Values were means ± SD, n = 3.
To assess whether MMP genes are involved in regulating leaf senescence, we first investigated their expression patterns during the senescence process (Figure 5A). RT-qPCR analysis revealed that the levels of all MMPs except At5-MMP transcripts increased during the progression of leaf development and senescence (Figures 5B–F). At3-MMP had the highest copy numbers, approximately 10 times more than the other four homologs during senescence (Figure 5D). Based on these results, we conclude that Arabidopsis MMPs are senescence-associated genes (SAGs) and might be involved in regulating leaf senescence downstream of MPK3/MPK6 cascade.
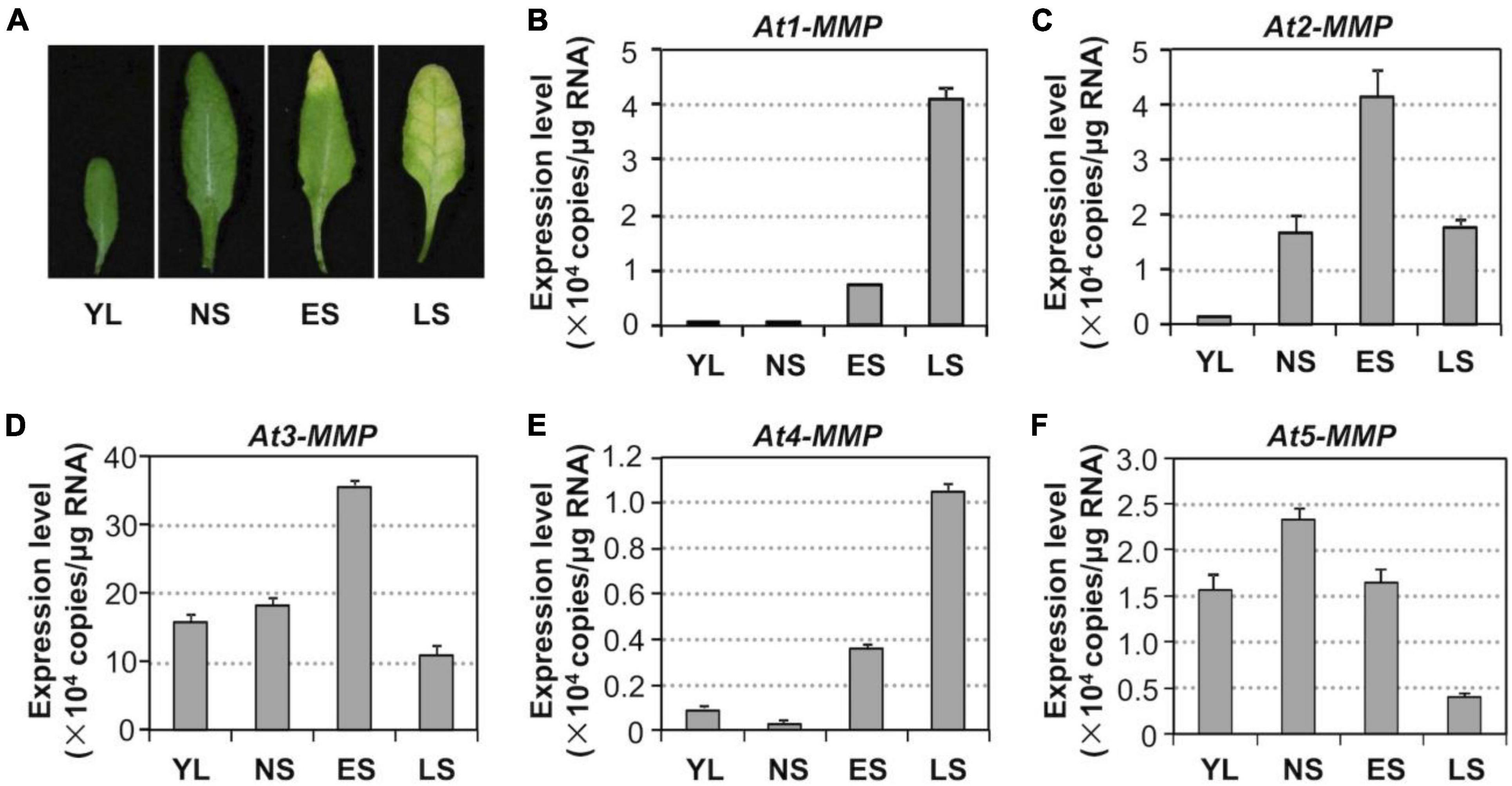
Figure 5. Activation of MMP expression during leaf senescence in Arabidopsis. (A) Leaf senescence in wild type plants at four different developmental stages. YL, young leaves; NS, fully expanded no senescence mature leaves; ES, early senescence leaves, with < 25% leaf yellowing; LS, lately senescence leaves, with > 50% leaf yellowing. (B–F) Transcript levels of MMPs increase in an age-dependent manner. Samples were collected from leaves shown in (A) at the indicated developmental stages. Gene expression was quantified by RT-qPCR and calculated as copy numbers per μg of total RNA. Values are means ± SD, n = 3.
Next, we tested whether an elevated expression of At3-MMP is sufficient to accelerate leaf senescence. Transgenic plants overexpressing a full-length At3-MMP gene with a C-terminal double HA tag driven by the constitutive 35S promoter (35S:At3-MMP-dHA) were generated. Two independent transgenic lines (#15 and #131) with different expression levels based on immunoblot analysis were selected for further analyses. We found that 35S:At3-MMP-dHA plants exhibited an early senescence phenotype compared with Col-0 (Supplementary Figure 3A). Next, we characterized the senescence of single leaves at different ages. As shown in Supplementary Figure 3B, the oldest seven leaves (numbered from the bottom; the first leaf is the oldest and the 12th leaf is the youngest) of the 35S:At3-MMP-dHA plants showed senescence, while only three leaves of the wild-type plants senesced. In addition, the cauline leaves of 35S:At3-MMP-dHA plants turned yellow earlier than the wild-type counterparts (Supplementary Figure 3C). Furthermore, there was a good correlation between the expression levels of At-3MMP protein and the severity of leaf senescence in different transgene lines (Supplementary Figures 4A,B). Clear acceleration of leaf senescence was also observed when the fully expanded leaves were excised from 35S:At3-MMP-dHA plants (Figure 6A). We also monitored the expression of SAG12, chlorophyll content, and Fv/Fm. As shown in Figure 6B, quicker/earlier SAG12 induction was associated with the early senescence symptoms in 35S:At3-MMP-dHA plants. The SAG12 transcript levels were induced significantly from 2 days in the 35S:At3-MMP-dHA plants. In contrast, the induction of SAG12 expression was undetectable in the first 4 days in wild type and only increased slightly afterward. Similarly, more severe reduction in chlorophyll content and Fv/Fm value happened in 35S:At3-MMP-dHA plants (Figures 6C,D). Taken together, these results suggested that constitutive overexpression of At-3MMP is sufficient to lead to early leaf senescence.
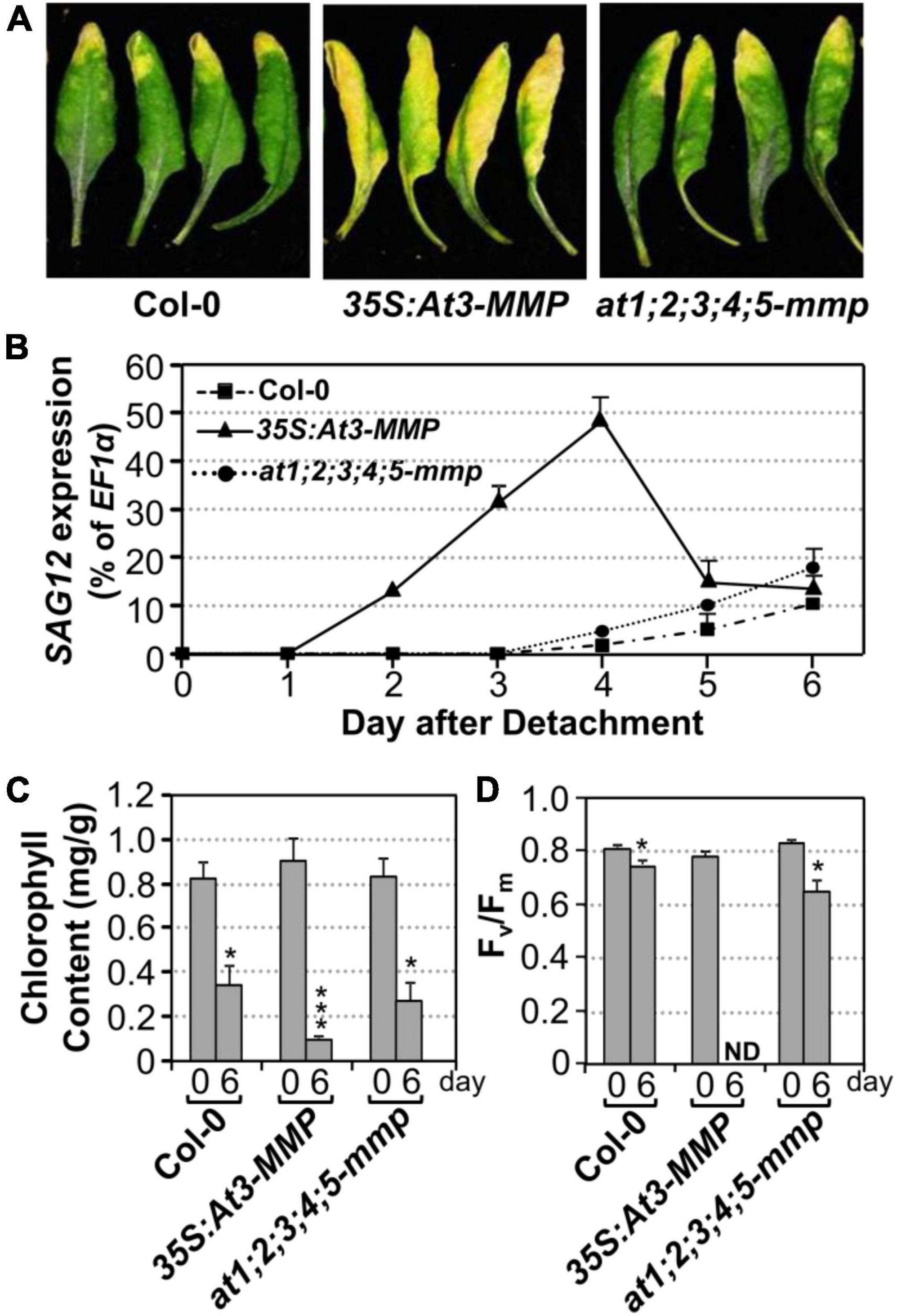
Figure 6. Constitutive overexpression of At3-MMP leads to leaf senescence under continuous light. (A) Fully expanded leaves from 4-week-old soil-grown plants were detached and their petioles were inserted into 0.6% agar medium. Photos were taken after detachment 6 days. (B) RT-qPCR analysis of the transcript levels of SAG12 in the leaves of plants shown in (A) at indicated times. The expression of EF1α was used as an internal reference. Values are means ± SD, n = 3. (C,D) Chlorophyll content (C) and Fv/Fm (D) in leaves shown in (A). Values are means ± SD, n = 10. ND, not detectable. Asterisks indicate a significant difference (* P < 0.05, *** P ≤ 0.001).
To further confirm the role of Arabidopsis MMPs in leaf senescence and avoid possible secondary complications associated with constitutive overexpression, we also performed a gain-of-function study using the glucocorticoid-inducible system to direct the expression of 3MMP-dHA transgene (GVG:At3-MMP-dHA). Two independent transgenic lines (#2 and #27) that accumulated At-3MMP protein at different levels were identified by immunoblot analysis and the T3 homozygous plants were selected for further experiments. We induced At-3MMP expression by treating 4-week-old plants with 30 μM DEX. As shown in Figure 7A, obvious leaf yellowing and leaf death were observed in the GVG: At3-MMP-dHA transgenic line #2 and #27 at 5 and 10 days, respectively. In contrast, no leaf senescence was observed in the wild type plants. No leaf senescence was detected in EtOH-treated control plants either. Associated with the leaf yellowing, chlorophyll and Fv/Fm loss and SAG12 gene expression were readily detectable in DEX-treated GVG: At3-MMP-dHA plants, but not solvent control-treated plants or wild type plants (Figures 7B–D). Immunoblot analysis showed that At3-MMP protein level was higher in line #2 compared to line #27 (Figure 7E). Correlating with the induction level, line #2 leaves became fully senescent or died, while line #27 leaves were still partially green (Figure 7A). Taken together, overexpression of At-3MMP, either constitutively under the control of 35S promoter or conditionally under the control of a steroid-inducible promoter, is sufficient to promote leaf senescence.
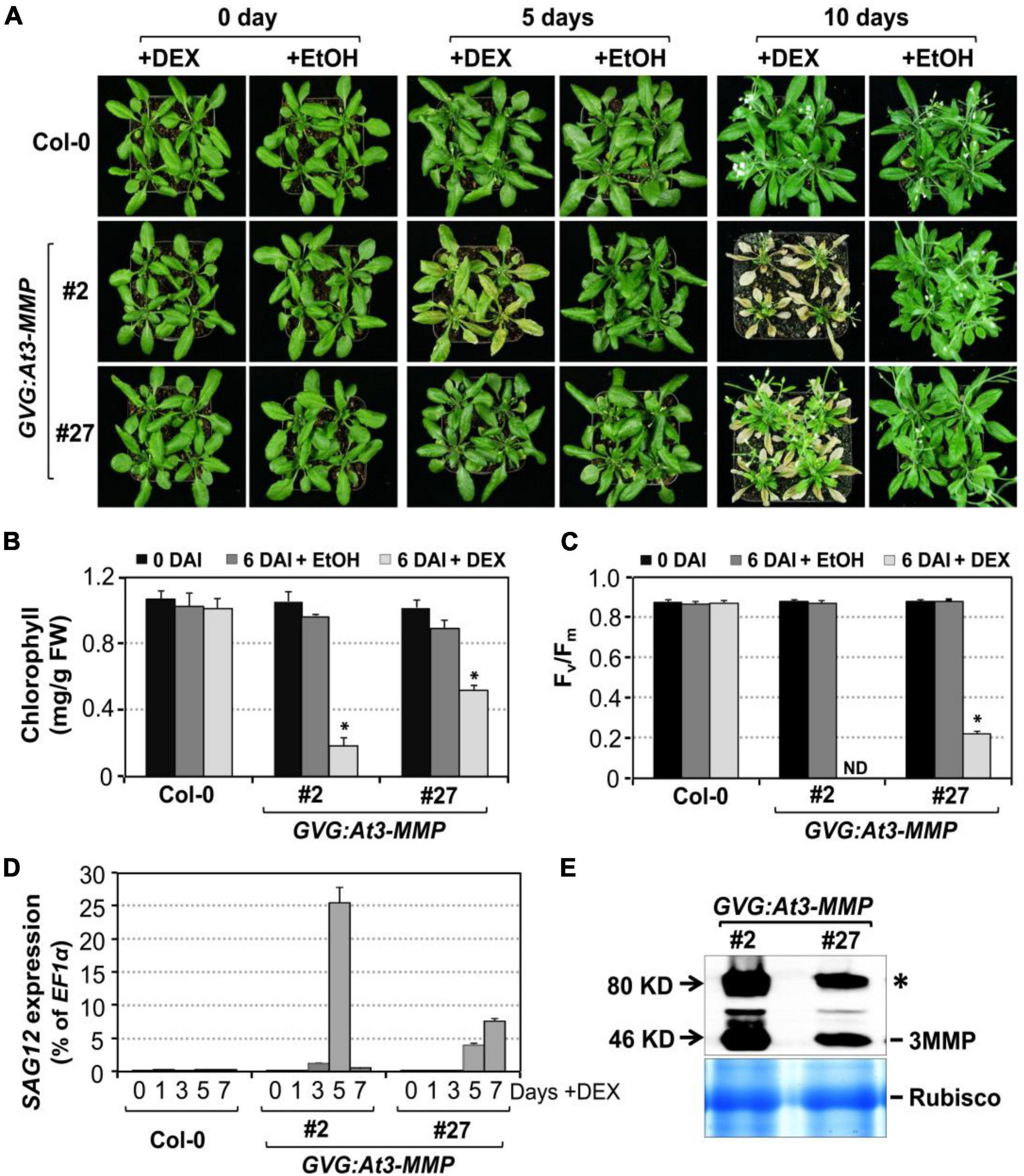
Figure 7. Conditional expression of At-3MMP is sufficient to induce leaf senescence. (A) The senescence phenotype of inducible At3-MMP transgenic lines (#2 and #27) and wild type plants after treatment with 30 μM DEX or EtOH (solvent control) for 0, 5, and 10 days. 4-week-old soil-grown plants were sprayed with DEX or EtOH. Photos were taken at the indicated time points. (B,C) Chlorophyll content (B) and Fv/Fm (C) of fifth to seventh leaves in wild-type and transgenic plants before and after DEX or EtOH treatment. Values are means ± SD, n = 10. ND: no detected. (D) RT-qPCR analysis of SAG12 transcript levels at indicated times after DEX treatment. Values are means ± SD, n = 3. (E) Induction of At3-MMP protein at 24 h after DEX (5 μM) treatment. HA-tagged At3-MMP protein was detected by immunoblot analysis using anti-HA antibody (upper). Coomassie brilliant blue staining of a duplicated gel was used to confirm equal loading (bottom). Asterisks indicate a significant difference (P ≤ 0.001).
To provide loss-of-functional evidence to support the role of At-MMPs in leaf senescence, we obtained loss-of-function T-DNA insertion mutants of all five Arabidopsis MMP genes from ABRC. Semi-quantitative RT-PCR analyses showed that At-MMPs transcripts were not detectable in the senescing leaves of the mutants, suggesting that these lines were all null knockout mutants (Supplementary Figure 5). No obvious growth/developmental or senescence phenotype was observed in at1-mmp to at5-mmp single mutant, mmp2 mmp3, mmp3 mmp5 double mutants, and mmp2 mmp3 mmp5 triple mutants (Supplementary Figure 6). Phylogenetic analysis revealed that all five Arabidopsis MMP genes shared high homology (Supplementary Figure 7). As a result, we generated pentuple at1;2;3;4;5-mmp mutant. Under our growth conditions, the at1;2;3;4;5-mmp high-order mutant was still wild-type like and no delayed senescence was observed (Supplementary Figure 6). We also examined the senescence of detached leaves of wild type and at1;2;3;4;5-mmp higher-order mutant under continuous light and found that there was no obvious difference between them in leaf yellowing, SAG12 gene activation, chlorophyll content, and Fv/Fm (Figure 6). Taken together, we conclude that although expression of At3-MMP is sufficient to induce leaf senescence, loss of MMPs is not sufficient to block leaf senescence, suggesting the presence of other redundant pathway(s) that are sufficient to signal/execute leaf senescence in the absence of MMPs.
In this study, we demonstrate that MPK3/MPK6, and their upstream MAPKKs, MKK4, and MKK5, are involved in regulating leaf senescence in Arabidopsis (Figures 1–3). Four of the five MMP family members are highly induced during leaf senescence (Figure 5), qualifying them as senescence-associated genes (SAGs). Among them, At3-MMP is induced to the highest level. Interestingly, At3-MMP is one of the top differentially expressed genes in DD plants (Su et al., 2018). More detailed qPCR analyses demonstrated that the expression of At-3MMP and, to a lesser extent, At-2MMP is rapidly induced by MPK3/MPK6 activation (Figure 4). Either constitutive or inducible overexpression of At3-MMP is sufficient to trigger leaf senescence (Figures 6, 7 and Supplementary Figure 3), suggesting a linear pathway from MPK3/MPK6 MAPK cascade to MMP target genes in leaf senescence.
Arabidopsis MPK6, along with MKK9, a MAPKK, has been implicated in leaf senescence in Arabidopsis (Zhou et al., 2009). It was reported that senescence is delayed in detached leaves of both mpk6 and mkk9 single mutant plants, while overexpression of MKK9 accelerates premature senescence in leaves. In this study, we tested both mpk3 and mpk6 single mutants. As shown in Supplementary Figure 2A, neither showed a clear senescence phenotype. We failed to observe any senescence phenotype in the chemical genetically rescued MPK3SR and MPK6SR double mutant either (Supplementary Figure 2B). The genotypes of MPK3SR and MPK6SR double mutant plants are mpk3 mpk6 proMPK3:MPK3TG and mpk3 mpk6 proMPK6:MPK6YG, respectively. In the presence of 4-amino-1-tert-butyl-3-(1′-naphthyl) pyrazolo [3,4-d] pyrimidine (NA-PP1), the activity of chemical-sensitized MPK3TG or MPK6YG is inhibited, making the MPK3SR and MPK6SR activity null mpk3 mpk6 double mutants (Xu et al., 2014; Su et al., 2017). The failure in observation of any senescence phenotype of MPK3SR and MPK6SR double mutant plants is likely because NA-PP1 can be metabolized in plant cells rather quickly, which makes it difficult to maintain a MPK3/MPK6 activity null condition for an extended period of time (over several days) to inhibit senescence. In contrast, we consistently observed a delayed senescence in mkk4 mkk5 double mutant leaves (Figure 3). In this double mutant, the mkk4 tilling allele has a substitution of the conserved Pro residue at the 240 position of the catalytic domain by Ser residue (CCT to TCT) and the mutated kinase carries ∼10% of the residual kinase activity. The mkk5 tilling allele is a null mutant with a premature stop codon (Arg72 to opal; CGA to TGA). As a result, the mkk4 mkk5 double mutant has about 5% of the residual activity if MKK4 and MKK5 contribute equally to activate downstream MPK3/MPK6 (Su et al., 2017). In addition to Arabidopsis MKK4 and MKK5 (two plant Group C MAPKKs), Arabidopsis MKK7 and MKK9, two members sharing closest homolog in the Group D MAPKKs (Ichimura et al., 2002), were also reported to be upstream of MPK3 and/or MPK6 (Xu et al., 2008; Jia et al., 2016). However, we did not observe any obvious senescence phenotype in mkk7 single, mkk9 single, and mkk7 mkk9 double mutants (Supplementary Figure 2A). As a result, we conclude that MKK4/MKK5, but not MKK7/MKK9, are upstream of the MPK3/MPK6 in plant leaf senescence.
In the gain-of-function DD transgenic system, our previous study showed that DD transgene expression is detectable within 2 h and HR-like cell death appears ∼24 h after the application of DEX. The dead leaves stay green and became brittle, similar to HR cell death triggered by pathogens (Ren et al., 2002). In this study, we confirm that strong and prolonged MPK3/MPK6 activation causes HR-like cell death in DD leaves after treatment with higher concentrations of DEX (>100 nM). In contrast, when DD leaves are treated with lower concentrations of DEX (<100 nM), leaf senescence is induced, which is associated with lower levels of MPK3/MPK6 phosphorylation and activation (Figure 1 and Supplementary Figure 1). These results provide another piece of evidence to support the hypothesis that MAPK signaling strength/duration could be important to the biological outcomes. Based on genetic evidence, we concluded that different developmental processes may have differential signal thresholds (strength and/or duration), which could specify the MAPK function in a quantitative way (Wang et al., 2007, 2008; Xu and Zhang, 2015).
Unlike mammalian MMP genes, which have been studied in great detail, the functions of MMPs in plants are mostly unknown. Based on the gene expression pattern, SMEP1, a plant matrix metalloproteinase gene isolated from soybean, was found to be expressed in mature leaves but not in young leaves and tissues (Pak et al., 1997). In cucumber, the Cs1-MMP was expressed in leaves during senescence and may be involved in programmed cell death (Delorme et al., 2000). In this study, we demonstrate that At-MMPs are SAGs, and play an important role in leaf senescence. Four of the five MMP transcripts are induced during the progression of leaf senescence, especially At3-MMP, which shows the greatest induction (Figure 5). Furthermore, constitutive or inducible overexpression of At3-MMP is sufficient to induce leaf senescence, as indicated by a loss/reduction in chlorophyll and photochemical efficiency, and the induction of SAG12 gene expression (Figures 6, 7 and Supplementary Figure 3). Collectively, these data suggest that plant MMPs are involved in regulating leaf senescence.
We obtained T-DNA insertion mutants of all five MMP genes from ARBC. Semi-quantitative PCR analysis confirms that all of them are complete knock-out mutants (Supplementary Figure 5). However, we failed to observe a difference in leaf senescence between wild type and the single, double, triple, and pentuple mmp mutants (Figure 6 and Supplementary Figure 6), suggesting parallel pathway(s) that is sufficient to execute leaf senescence. Transcriptome analyses of leaf senescence identified thousands of SAGs. However, most SAG mutants do not have altered leaf senescence, probably due to functional redundancy or the lack of a pronounced effect on leaf senescence (He et al., 2001). A large number of plant signaling molecules including plant hormones, kinases including receptor-like protein kinases, and transcription factors contribute to the regulation and execution of senescence at the cellular and subcellular levels, suggesting functionally redundant pathways (He et al., 2001; Guo et al., 2004; Lim et al., 2007; Schippers et al., 2007; Kim H. J. et al., 2016; Kim J. et al., 2016). Recently, MAPKKK18 was identified as a positive regulator of leaf senescence. It controls the timing of leaf senescence via its kinase activity in an ABA-dependent manner (Matsuoka et al., 2015). Phylogenetic analysis revealed that MAPKKK18 belongs to a different subgroup of MAPKKKs as YDA, MAPKKK3, and MAPKKK5, three MAPKKKs that have been functionally placed upstream of the MKK4/MKK5-MPK3/MPK6 module in various biological processes (Bergmann et al., 2004; Wang et al., 2007; Meng et al., 2012; Bi et al., 2018; Sun et al., 2018; Supplementary Figure 8). MEKK1, a MAPKKK that belongs to yet another subgroup of MAPKKKs, was reported to regulate leaf senescence by directly phosphorylating the WRKY53, a senescence-promoting transcription factor (Miao et al., 2007). At this stage, whether MAPKKK18 and/or MEKK1 are in the same MAPK cascade as MKK4/MKK5 and MPK3/MPK6 in leaf senescence is unknown.
In 35S:At3-MMP-HA and GVG:At3-MMP-HA transgenic plants, we detected two HA-tagged protein bands with molecular masses of 46 and 80 kDa by immunoblot analysis (Figure 7 and Supplementary Figure 4). The theoretical mass of At3-MMP based on its amino acid sequence is ∼43 kDa. A similar finding was reported in a previous study, in which the larger protein band was speculated to be glycosylated or dimer form of MMP protein (Schiermeyer et al., 2009). Substrate identification will provide further insights into how MMPs carry out their function(s) in plant leaf senescence, an important part of plant growth/development that is influenced by a wide variety of internal and environmental factors. Leaf senescence normally occurs at the final stage of the plant growth season or life cycle, and is essential for nutrient recycling and crop yield (Gan, 2003; Lim et al., 2007). In contrast, premature leaf senescence, as an exit strategy when plants are confronted with biotic/abiotic stresses, may reduce yield in crop by limiting the growth stage and causing post-harvest spoilage. As a result, understanding the regulation of leaf senescence will not only reveal insights into this fundamental developmental process but also shed light on ways of manipulating senescence for agriculture application.
Publicly available datasets were analyzed in this study. This data can be found here: NCBI Sequence Read Archive (SRP111959).
HW performed most of the experiments and analyzed the data. QS, JL, and LY provided technical assistance. HW, SZ, and JX conceived the project, designed the experiments, and wrote the manuscript. JX served as the author responsible for contact and ensure communication. All authors contributed to the article and approved the submitted version.
This research was supported by the grants from National Natural Science Foundation of China (31922005), the Fundamental Research Funds for the Central Universities (2021FZZX001-28), and 111 Project (B14027) to JX.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Arabidopsis Biological Resource Center for mutant seeds. In addition, HW especially would like to thank Lay Zhang, whose dance have given her powerful spiritual support during the Ph.D. period.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.864986/full#supplementary-material
Andreasson, E., and Ellis, B. (2010). Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 15, 106–113. doi: 10.1016/j.tplants.2009.12.001
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Bergmann, D. C., Lukowitz, W., and Somerville, C. R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497. doi: 10.1126/science.1096014
Bi, G., Zhou, Z., Wang, W., Li, L., Rao, S., Wu, Y., et al. (2018). Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 30, 1543–1561. doi: 10.1105/tpc.17.00981
Chai, J., Liu, J., Zhou, J., and Xing, D. (2014). Mitogen-activated protein kinase 6 regulates NPR1 gene expression and activation during leaf senescence induced by salicylic acid. J. Exp. Bot. 65, 6513–6528. doi: 10.1093/jxb/eru369
Clough, S. J., and Bent, A. F. (1998). Floral dip a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Das, P. K., Biswas, R., Anjum, N., Das, A. K., and Maiti, M. K. (2018). Rice matrix metalloproteinase OsMMP1 plays pleiotropic roles in plant development and symplastic-apoplastic transport by modulating cellulose and callose depositions. Sci. Rep. 8:2783. doi: 10.1038/s41598-018-20070-4
Delorme, V. G. R., McCabe, P. F., Kim, D. J., and Leaver, C. J. (2000). A matrix metalloproteinase gene is expressed at the boundary of senescence and programmed cell death in cucumber. Plant Physiol. 123, 917–927. doi: 10.1104/pp.123.3.917
Flinn, B. S. (2008). Plant extracellular matrix metalloproteinases. Funct. Plant Biol. 35, 1183–1193. doi: 10.1071/FP08182
Frick, U. B., and Schaller, A. (2002). cDNA microarray analysis of fusicoccin-induced changes in gene expression in tomato plants. Planta 216, 83–94. doi: 10.1007/s00425-002-0887-1
Gan, S. (2003). Mitotic and postmitotic senescence in plants. Sci. Aging Knowl. Environ. SAGE KE 38:RE7. doi: 10.1126/sageke.2003.38.re7
Gan, S., and Amasino, R. M. (1997). Making sense of senescence – Molecular genetic regulation and manipulation of leaf senescence. Plant Physiol. 113, 313–319. doi: 10.1104/pp.113.2.313
Guo, Y., and Gan, S. (2005). Leaf senescence: signals, execution, and regulation. Curr. Top. Dev. Biol. 71, 83–112. doi: 10.1016/S0070-2153(05)71003-6
Guo, Y., Cai, Z., and Gan, S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27, 521–549. doi: 10.1111/j.1365-3040.2003.01158.x
He, Y. H., Tang, W. N., Swain, J. D., Green, A. L., Jack, T. P., and Gan, S. S. (2001). Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol. 126, 707–716. doi: 10.1104/pp.126.2.707
Ichimura, K., Shinozaki, K., Tena, G., Sheen, J., Henry, Y., Champion, A., et al. (2002). Mitogen-activated protein kinase cascades in plants a new nomenclature. Trends Plant Sci. 7, 301–308. doi: 10.1016/s1360-1385(02)02302-6
Jia, W., Li, B., Li, S., Liang, Y., Wu, X., Ma, M., et al. (2016). Mitogen-activated protein kinase cascade MKK7-MPK6 plays important roles in plant development and regulates shoot branching by phosphorylating PIN1 in Arabidopsis. PLoS Biol. 14:e1002550. doi: 10.1371/journal.pbio.1002550
Kim, H. J., Nam, H. G., and Lim, P. O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56. doi: 10.1016/j.pbi.2016.06.002
Kim, J., Woo, H. R., and Nam, H. G. (2016). Toward systems understanding of leaf senescence: an integrated multi-omics perspective on leaf senescence research. Mol. Plant 9, 813–825. doi: 10.1016/j.molp.2016.04.017
Li, S., Han, X., Yang, L., Deng, X., Wu, H., Zhang, M., et al. (2018). Mitogen-activated protein kinases and calcium-dependent protein kinases are involved in wounding-induced ethylene biosynthesis in Arabidopsis thaliana. Plant Cell Environ. 41, 134–147. doi: 10.1111/pce.12984
Li, Z., Peng, J., Wen, X., and Guo, H. (2012). Gene network analysis and functional studies of senescence-associated genes reveal novel regulators of Arabidopsis leaf senescence. J. Integr. Plant Biol. 54, 526–539. doi: 10.1111/j.1744-7909.2012.01136.x
Li, Z., Zhao, Y., Liu, X., Peng, J., Guo, H., and Luo, J. (2014). LSD 2.0: an update of the leaf senescence database. Nucleic Acids Res. 42, D1200–D1205. doi: 10.1093/nar/gkt1061
Liu, Y., and Zhang, S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399. doi: 10.1105/tpc.104.026609
Liu, Y., Dammann, C., and Bhattacharyya, M. K. (2001). The matrix metalloproteinase gene GmMMP2 is activated in response to pathogenic infections in soybean. Plant Physiol. 127, 1788–1797. doi: 10.1104/pp.010593
Maidment, J. M., Moore, D., Murphy, G. P., Murphy, G., and Clark, I. M. (1999). Matrix metalloproteinase homologues from Arabidopsis thaliana – expression and activity. J. Biol. Chem. 274, 34706–34710. doi: 10.1074/jbc.274.49.34706
Matsuoka, D., Yasufuku, T., Furuya, T., and Nanmori, T. (2015). An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol. Biol. 87, 565–575. doi: 10.1007/s11103-015-0295-0
Meng, X., Wang, H., He, Y., Liu, Y., Walker, J. C., Torii, K. U., et al. (2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24, 4948–4960. doi: 10.1105/tpc.112.104695
Miao, Y., Laun, T. M., Smykowski, A., and Zentgraf, U. (2007). Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol. Biol. 65, 63–76. doi: 10.1007/s11103-007-9198-z
Noh, Y. S., and Amasino, R. M. (1999). Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 41, 181–194. doi: 10.1023/a:1006342412688
Overall, C. M. (2002). Molecular determinants of metalloproteinase substrate specificity. Mol. Biotechnol. 22, 51–86. doi: 10.1385/MB:22:1:051
Page-McCaw, A., Ewald, A. J., and Werb, Z. (2007). Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8, 221–233. doi: 10.1038/nrm2125
Pak, J. H., Liu, C. Y., Huangpu, J., and Graham, J. S. (1997). Construction and characterization of the soybean leaf metalloproteinase cDNA. FEBS Lett. 404, 283–288. doi: 10.1016/s0014-5793(97)00141-5
Parks, W. C., Wilson, C. L., and Lopez-Boado, Y. S. (2004). Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 4, 617–629. doi: 10.1038/nri1418
Pedley, K. F., and Martin, G. B. (2005). Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8, 541–547. doi: 10.1016/j.pbi.2005.07.006
Pitzschke, A., Schikora, A., and Hirt, H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12, 421–426. doi: 10.1016/j.pbi.2009.06.008
Pontier, D., Gan, S. S., Amasino, R. M., Roby, D., and Lam, E. (1999). Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol. Biol. 39, 1243–1255. doi: 10.1023/a:1006133311402
Ratnaparkhe, S. M., Egertsdotter, E. M., and Flinn, B. S. (2009). Identification and characterization of a matrix metalloproteinase (Pta1-MMP) expressed during Loblolly pine (Pinus taeda) seed development, germination completion, and early seedling establishment. Planta 230, 339–354. doi: 10.1007/s00425-009-0949-8
Rawlings, N. D., Barrett, A. J., and Bateman, A. (2010). MEROPS the peptidase database. Nucleic Acids Res. 28, 323–325. doi: 10.1093/nar/28.1.323
Ren, D., Yang, H., and Zhang, S. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277, 559–565. doi: 10.1074/jbc.M109495200
Rodriguez, M. C., Petersen, M., and Mundy, J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649.
Schiermeyer, A., Hartenstein, H., Mandal, M. K., Otte, B., Wahner, V., and Schillberg, S. (2009). A membrane-bound matrix-metalloproteinase from Nicotiana tabacum cv. BY-2 is induced by bacterial pathogens. BMC Plant Biol. 9:83. doi: 10.1186/1471-2229-9-83
Schippers, J. H. M., Jing, H. C., Hille, J., and Dijkwel, P. P. (2007). Developmental and hormonal control of leaf senescence. Senescence Process. Plants 26, 145–170. doi: 10.1002/9780470988855.ch7
Sternlicht, M. D., and Werb, Z. (2001). How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 17, 463–516. doi: 10.1146/annurev.cellbio.17.1.463
Su, J., Yang, L., Zhu, Q., Wu, H., He, Y., Liu, Y., et al. (2018). Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PLoS Biol. 16:e2004122. doi: 10.1371/journal.pbio.2004122
Su, J., Zhang, M., Zhang, L., Sun, T., Liu, Y., Lukowitz, W., et al. (2017). Regulation of Stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell 29, 526–542. doi: 10.1105/tpc.16.00577
Sun, T., Nitta, Y., Zhang, Q., Wu, D., Tian, H., Lee, J. S., et al. (2018). Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 19:e45324. doi: 10.15252/embr.201745324
Tena, G., Boudsocq, M., and Sheen, J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14, 519–529. doi: 10.1016/j.pbi.2011.05.006
Wang, H., Liu, Y., Bruffett, K., Lee, J., Hause, G., Walker, J. C., et al. (2008). Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20, 602–613. doi: 10.1105/tpc.108.058032
Wang, H., Ngwenyama, N., Liu, Y., Walker, J. C., and Zhang, S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73. doi: 10.1105/tpc.106.048298
Woo, H. R., Kim, H. J., Nam, H. G., and Lim, P. O. (2013). Plant leaf senescence and death – regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 126, 4823–4833. doi: 10.1242/jcs.109116
Woo, H. R., Koo, H. J., Kim, J., Jeong, H., Yang, J. O., Lee, I. H., et al. (2016). Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol. 171, 452–467. doi: 10.1104/pp.15.01929
Xu, J., and Zhang, S. (2015). Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 20, 56–64. doi: 10.1016/j.tplants.2014.10.001
Xu, J., Li, Y., Wang, Y., Liu, H., Lei, L., Yang, H., et al. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283, 26996–27006. doi: 10.1074/jbc.M801392200
Xu, J., Xie, J., Yan, C., Zou, X., Ren, D., and Zhang, S. (2014). A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidasemediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77, 222–234. doi: 10.1111/tpj.12382
Yang, K. Y., Liu, Y., and Zhang, S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. U.S.A. 98, 741–746. doi: 10.1073/pnas.98.2.741
Zhang, J., Gao, J., Zhu, Z., Song, Y., Wang, X., Wang, X., et al. (2020). MKK4/MKK5-MPK1/MPK2 cascade mediates SA-activated leaf senescence via phosphorylation of NPR1 in Arabidopsis. Plant Mol. Biol. 102, 463–475. doi: 10.1007/s11103-019-00958-z
Zhang, M., Su, J., Zhang, Y., Xu, J., and Zhang, S. (2018). Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 45, 1–10. doi: 10.1016/j.pbi.2018.04.012
Zhang, Y., Liu, J., Chai, J., and Xing, D. (2016). Mitogen-activated protein kinase 6 mediates nuclear translocation of ORE3 to promote ORE9 gene expression in methyl jasmonate-induced leaf senescence. J. Exp. Bot. 67, 83–94. doi: 10.1093/jxb/erv438
Zhao, C., Nie, H., Shen, Q., Zhang, S., Lukowitz, W., and Tang, D. (2014). EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet. 10:e1004389. doi: 10.1371/journal.pgen.1004389
Zhou, C., Cai, Z., Guo, Y., and Gan, S. (2009). An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 150, 167–177. doi: 10.1104/pp.108.133439
Keywords: MAPK cascade, matrix metalloproteinase, leaf senescence, Arabidopsis, transcription regulation
Citation: Wu H, Si Q, Liu J, Yang L, Zhang S and Xu J (2022) Regulation of Arabidopsis Matrix Metalloproteinases by Mitogen-Activated Protein Kinases and Their Function in Leaf Senescence. Front. Plant Sci. 13:864986. doi: 10.3389/fpls.2022.864986
Received: 29 January 2022; Accepted: 08 March 2022;
Published: 08 April 2022.
Edited by:
Ying Miao, Fujian Agriculture and Forestry University, ChinaReviewed by:
Yongfeng Guo, Tobacco Research Institute (CAAS), ChinaCopyright © 2022 Wu, Si, Liu, Yang, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Xu, eHVqdWFuQHpqdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.