
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 May 2022
Sec. Functional Plant Ecology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.864085
This article is part of the Research Topic Patterns, Functions, and Processes of Alpine Grassland Ecosystems under Global Change. View all 76 articles
 Yu Li1
Yu Li1 Shikui Dong2,3*
Shikui Dong2,3* Qingzhu Gao4*
Qingzhu Gao4* Chun Fan1
Chun Fan1 Moses Fayiah5
Moses Fayiah5 Hasbagan Ganjurjav4
Hasbagan Ganjurjav4 Guozheng Hu4†
Guozheng Hu4† Xuexia Wang6†
Xuexia Wang6† Yulong Yan7
Yulong Yan7 Xiaoxia Gao8
Xiaoxia Gao8 Shuai Li8†
Shuai Li8†Grazing is a substantial threat to the sustainability of grassland ecosystems, while it is uncertain about the variety of plant and soil microbial community and the linkages between them limit the comprehensive understanding of grazing ecology. We conducted an experiment on the effects of the grazing regimes rotational grazing (RG), continuous grazing (CG), and grazing exclusion (GE) on an alpine meadow in Qinghai-Tibetan Plateau. The differences of plant community composition, soil microbial community assembly mechanism, and taxonomic and functional composition between grazing regimes were examined, and the relationship between plant species and the soil microbes was assessed by constructing a co-occurrence network. The results showed that the plant community composition varied with the grazing regimes, while the soil microbial community composition did not vary with the grazing regimes. The soil bacterial functional composition was similar under RG and CG, while the soil fungal functional composition was similar under GE and RG. The soil microbial community under all grazing regimes was assembled mainly according to stochastic rather than deterministic mechanisms, and RG and CG reduced the relative importance of the stochastic ratio. At the microbial phylum level, CG and GE increased the relative abundance of Acidobacteria and Armatimonadetes and CG and RG increased the relative abundance of Elusimicrobia. In the network of plant species and soil microbial classes, plants and bacteria themselves were mainly positively linked (symbiosis and promotion), while plants and soil microbes were mainly negatively linked (competition). There were five microbial generalists in the network, which connected with many microbes, and four showed no difference in their abundance among the grazing regimes. Overall, the stable key microbes in the network and the fact that many of the plants are unconnected with microbes weakened the impact of grazing-induced changes in the plant community on soil microbes, probably resulting in the stable soil microbial community composition. Moreover, there was still a dominant and tolerant plant species, Kobresia pygmaea, that connected the plant and microbial communities, implying that the dominant plant species not only played a crucial role in the plant community but also acted as a bridge between the plants and soil microbes; thus, its tolerance and dominance might stabilize the soil microbial community.
Grassland ecosystems, accounting for approximately 40% of the terrestrial surface in China, are facing remarkable sustainability challenges from livestock grazing (Zhou et al., 2017; Tang et al., 2019). Therefore, an in-depth understanding of the impact of grazing on the grassland ecosystem is urgently needed to deal with and recover the grazing-induced grassland degradation. Grazing has substantial influences on grassland ecosystems such as the deposition of herbivore urine and dung, general decreases in soil porosity through trampling, changes in litter quality, and stimulated root exudates via selective defoliation (Zhong et al., 2018; Tang et al., 2019); these factors affect the nutrient sources and living environment of soil microbes. Conversely, the soil microbial community mediates the energy and material fluxes of ecosystems and alters soil biogeochemical properties (Chapin III et al., 2000), thus affecting the soil nutrients needed by the plant community. These linkages form a feedback response between the plant and soil microbial communities to grazing. In addition, a given plant species may preferentially associate with distinct soil microbial taxa (Fierer, 2017). As a result, understanding the effects of grazing on grasslands should stress not only the changes of the plant community or soil microbial community separately but also the linkages between them.
At present, the respective effects of grazing on plant community and soil microbial community are well understood (Shen et al., 2013; Yang et al., 2013; Beck et al., 2015; Macdonald et al., 2015; Wang et al., 2016; Xun et al., 2018; Tang et al., 2019). However, the relationship between plant and soil microbial community in grazed grasslands remains unclear, hence it hinders the systematic understanding of the effect of grazing on grassland ecosystems. To fill this knowledge gap, we conducted a grazing experiment on the grassland ecosystem of the Qinghai-Tibetan Plateau (QTP), the highest plateau in the world with a total area of 2.5 × 106 km2 and an average altitude of over 4,000 m. As one of the key types of grassland on the QTP, the alpine meadow contributes considerably to global soil C and N pools (Li et al., 2014; Ding et al., 2016), and this grassland is also a very sensitive eco-region for climate change and anthropogenic disturbances (Tang et al., 2019) and is highly threatened by grazing (Li et al., 2020). The key objectives of the study were to explore (1) the effect of grazing on plant community composition; (2) the effect of grazing on soil microbial diversity, taxonomic and functional composition, and community assembly; (3) the relationship between plant community and soil microbial community in grazed grassland.
The grazing experiment was conducted in Nagqu city, Tibetan Autonomous Region of China, which is situated at the central QTP with latitude 31.441°N, longitude 92.017°E, and altitude 4,500 m a.s.l. The region has an arid and cold climate. The annual average temperature is 0.03°C and the annual average rainfall is 474.7 mm. In the growing season (May–September), the average rainfall is 414.7 mm and the average temperature is 7.8°C. The soil agro-type is alpine meadow soil with high proportions of clay. The main vegetation is an alpine meadow, with the plant community dominated by Kobresia pygmaea and accompanied by Kobresia humilis, Potentilla humilis, Potentilla saundersiana, Potentilla bifurca, Astragalus membranaceus, Leontopodium leontopodioides, Stipa capillata, and so on.
During the growing season of 2014–2017, the grazing experiment was carried out in a 30,000 m2 paddock of the alpine meadow, which was enclosed with wire mesh and divided into 12 50 × 50 m plots (Supplementary Figure S1). Nine plots were randomly placed on the paddock for rotational grazing (RG) and were further divided into groups A, B, and C, with each group having three replicates. The other three plots which were subjected to grazing before 2014 were used as three replicates of grazing exclusion (GE). At the beginning of July 2014, 2015, 2016, and 2017, two yaks with an average weight of 105±10 kg were grazed in each replicate plot of group A for 7 days (six yaks in three replicates). Then, these yaks grazed in groups B and C in turn in the same way. Three rotation cycles lasted 63 days each year except for 2015, in which only two rotation cycles lasted 42 days due to drought, which led to the earlier withering of plants. Three plots of the same size as the RG and GE plots were randomly selected as continuous grazing (CG) plots outside the fence in 2016 and 2017. The area outside the fence contained a 37.5-ha paddock of the alpine meadow that was permanently grazed by approximately 300 yaks. In the RG and CG plots, the grazing intensity was eight young yaks per hectare.
On September 5, 2017, plant leaves of all species were collected following the method of Cornelissen et al. (2003) and Pérez-Harguindeguy et al. (2013). The sampling method has been specifically described previously (Li et al., 2019, 2020). In brief, at least 30 plants of each species were selected in each grazing regime; three to five fully expanded leaves of each individual were sampled for further leaf trait measurements. In the laboratory, the dry plant leaves of each species were ground by a ball mill (NM200; Retsch, Haan, Germany) for chemical analysis. Total carbon and nitrogen contents were measured by an elemental analyzer (Perkin-Elmer, Boston; MA, USA), and total phosphorus content was measured by an ICP-AES analyser (Thermo-Jarrell Ash Corp; MA, USA). In addition, six soil samples with a diameter of 5 cm were randomly collected at a depth of 0 to 15 cm in each plot, three samples were used to analyze the soil nutrient content and microbial community, and three samples were used to estimate the plant belowground biomass. The NH4-N and NO3-N of the soil samples were determined by a flow injection AutoAnalyser (AACE, Germany), and available phosphorus was determined by inductively coupled plasma spectrometry (SPECTRO ARCOS EOP, Germany). Soil total carbon content was measured using an elemental analyser (2400 II CHNS/O Elemental Analyser; Perkin-Elmer, Boston, MA, USA). Soil bulk density was determined with a cutting ring by sampling three cores in each plot at a depth of 0 to 15 cm. The detailed estimation of the aboveground and belowground biomass of the plant community can be found in our previous study (Li et al., 2020).
The soil microbial community was analyzed using high-throughput sequencing. Soil microbial DNA was extracted from each soil sample using a Fast DNA SPIN Kit for Soil (MP Biochemicals, Solon, OH, USA). The V4-V5 of bacterial 16S rRNA gene was amplified by polymerase chain reaction (95°C for 2 min, followed by 25 cycles at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s) using the universal primer 515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′). ITS rRNA of fungi was amplified by polymerase chain reaction (95°C for 2 min, followed by 32 cycles at 95°C for 30 s, annealing at 61°C for 30 s, and extension at 72°C for 45 s) with primer ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′). The PCR products were gel-purified with an AxyPre DNA Gel Extraction kit (Axygen Biosciences, Union City, CA, USA) and quantified by Qubit® 2.0 Fluorometer (Invitrogen Corp., Carlsbad, CA, USA), then a mixture of amplicons was used for sequencing on the Illumina MiSeq platform. The processes of controlling quality and trimming sequencing reads were performed following the method of Hong et al. (2015). Operational Taxonomic Units (OTUs) were built at 97% sequence similarity cutoff using UPARSE (Edgar et al., 2011). OTUs were assigned to taxonomic lineages using the Ribosomal Database Project (RDP) classifier within the Silva database (http://www.arb-silva.de). The total sequence number in each sample was rarefied to the minimum sequence number across all the samples. The rarefaction curves and the number of OTU reads of samples are shown in Supplementary Figure S6 and Table S3, respectively. The functional profiles of soil bacteria and fungi were assigned by FAPROTAX 1.2.3 (Louca et al., 2016) and FUNGuild (Nguyen et al., 2016), respectively.
Two-tailed paired t-tests and analysis of variance (ANOVA) were performed to test the grazing regime effect on plant species coverage and soil total carbon content. Permutational multivariate analysis of variance (PERMANOVA) based on Bray–Curtis dissimilarity indices were performed to examine the difference in plant community composition among grazing regimes. Kruskal–Wallis tests were performed to examine grazing regime effects on soil microbial diversity indices and microbial phyla relative abundance. Microbial community alpha diversity was represented by richness, Shannon, Simpson, PD whole tree, and Chao 1 indices. Richness was presented as the observed OTU number. Three nonparametric tests (multiple response permutation procedure, MRPP; PERMANOVA; and analysis of similarity, ANOSIM) based on Bray–Curtis dissimilarity indices were performed by R software to examine the difference in soil microbial taxonomic composition among grazing regimes. A phylogenetic tree was annotated and visualized in iTOL software (Letunic and Bork, 2019).
A null modeling-based approach by Ning et al. (2019) was performed to infer soil microbial community assembly mechanism, that is, calculating the normalized stochastic ratio for estimating the relative importance of stochasticity and determinism in shaping community structure. Specifically, the null model algorithm was used to qualify the significance of the observed difference in the microbial community from random expectation. If the observed microbial community is statistically different from the null expectation, the community is regarded as largely shaped by deterministic processes. Otherwise, it is considered to be dominated by stochastic processes. Also known as normalized stochasticity ratio with 50% as the boundary point between more deterministic (<50%) and more stochastic (>50%) assembly.
To explore the relationships among bacteria, fungi, archaea, and plant species, a co-occurrence network based on the relative abundance of microbial classes and plant species dominance data was constructed using the MENA pipeline (http://ieg2.ou.edu/MENA), which performs random matrix theory to identify thresholds for constructing a highly confident ecological network (Zhou et al., 2011; Deng et al., 2012), and the default settings of MENA were adopted.
The plant community composition under RG was significantly different compared to that under GE and CG (Supplementary Table S1). Specifically, Kobresia pygmaea was the dominant species among the 13 main species; it was a palatable species for yaks and its coverage under RG was higher than that under GE and CG (Figure 1). Another priming species was Stipa capillata, which was also a palatable species. However, its coverage under GE was higher than that under RG and CG.
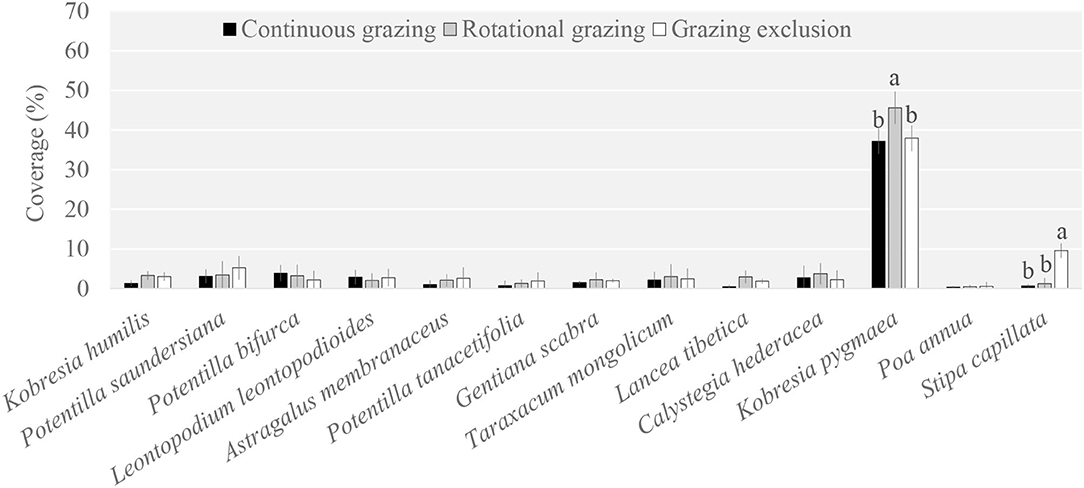
Figure 1. Plant species coverage among grazing regimes. Different lowercase letters indicate significant differences between grazing regimes (p < 0.05).
The soil bacterial diversity indices showed no differences among the grazing regimes (Supplementary Figure S2). The Shannon, Chao 1, richness, and PD whole tree indices of the soil fungal community were all higher under CG than under RG, while the Simpson index showed no difference among the grazing regimes (Figure 2). The soil's whole microbial community composition did not show any difference among the grazing regimes (Table 1). Stochasticity dominated soil microbial community assembly, RG, and CG significantly decreased the relative importance of stochastic processes (Figure 3).
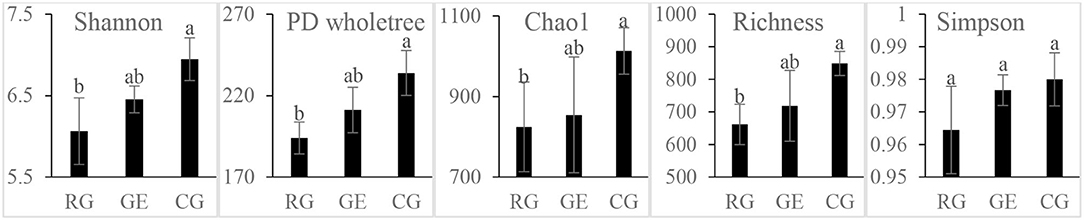
Figure 2. Soil fungal diversity among the grazing regimes. GE, grazing exclusion; RG, rotational grazing; and CG, continuous grazing. Different lowercase letters indicate significant differences between grazing regimes (p < 0.05).
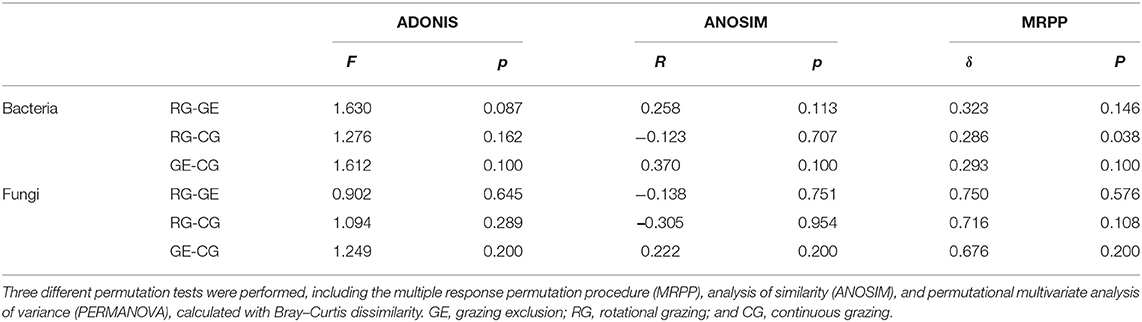
Table 1. Nonparametric analyses to test the dissimilarity of the soil microbial communities among the grazing regimes.
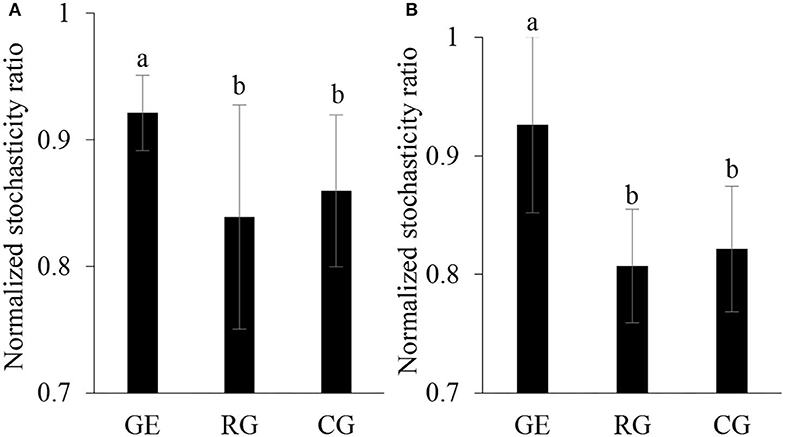
Figure 3. Normalized stochasticity ratio of community assembly of soil bacteria (A) and fungi (B). GE, grazing exclusion; RG, rotational grazing; and CG, continuous grazing. Different lowercase letters indicate significant differences between grazing regimes (p < 0.05).
The soil bacterial functional composition under RG and CG was similar (Figure 4A), while the soil fungal functional composition under GE and RG was similar (Figure 4B). Specifically, aerobic chemoheterotrophy and chemoheterotrophy were the main bacterial functional guilds, which include most OTU, and their relative abundances were higher under GE than those under RG and CG. Lichenized and Undefined Saprotroph were the main fungal functional guilds, while most OTU belonged to Arbuscular Mycorrhizal, with low relative abundance.
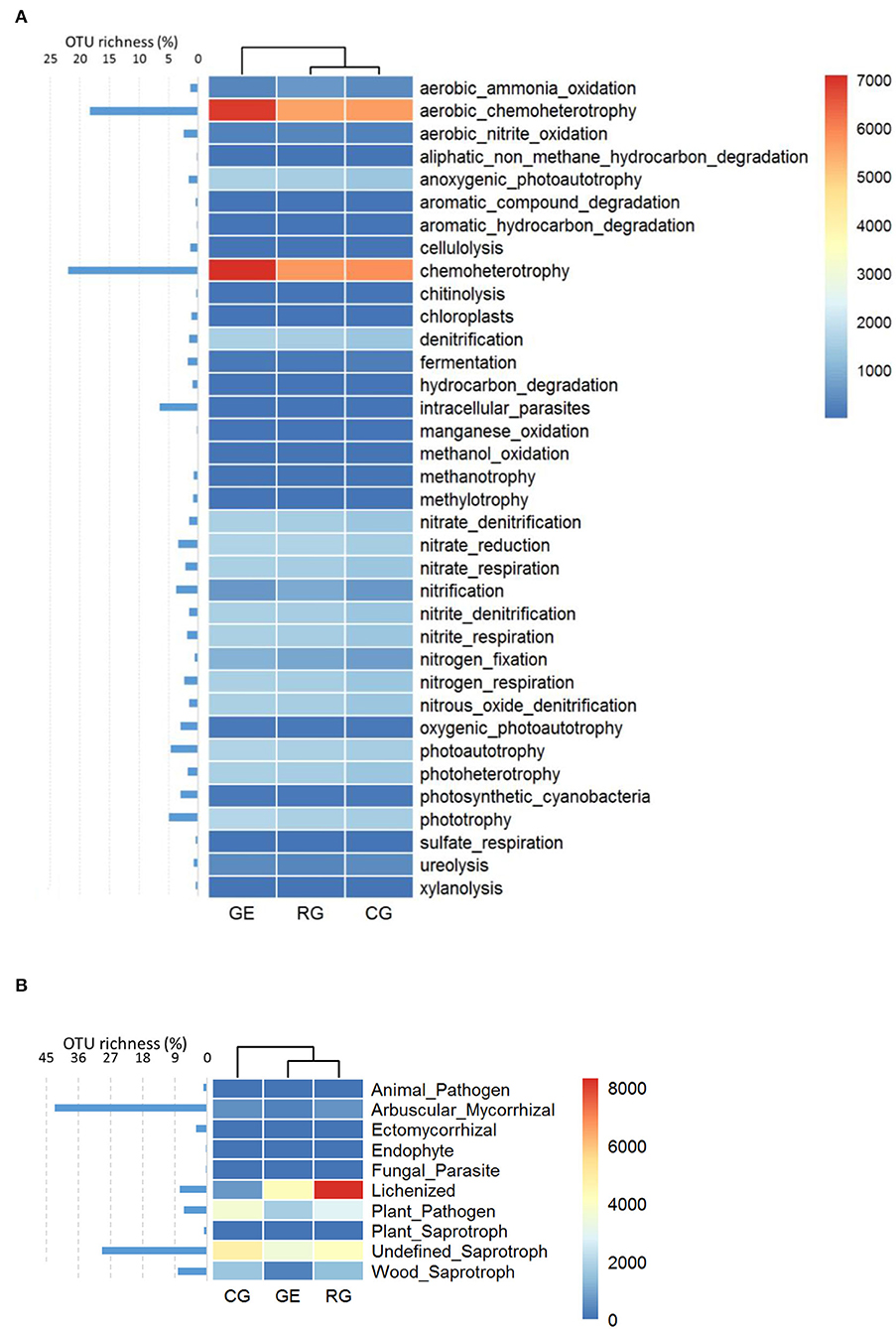
Figure 4. The relative abundance and OTU richness of bacterial (A) and fungal functional guilds (B). GE, grazing exclusion; RG, rotational grazing; and CG, continuous grazing.
To better characterize the grazing regime effect, we identified the dominant taxa (OTUs with a relative abundance ≥ 0.5%). Although 4684 bacterial OTUs and 5241 fungal OTUs were retrieved from all the grazed plots, only 42 (0.8%) and 84 (1.6%) OTUs were identified as the dominant taxa, respectively. For all the grazed plots, these dominant bacterial OTUs were mainly affiliated with Thermoleophilia (28.6%), Actinobacteria (23.8%), and Acidobacteria (26.2%) (Figure 5A); the dominant fungal OTUs were mainly affiliated with Sordariomycetes (26.2%) and Leotiomycetes (26.2%) (Figure 5B). The majority of the dominant taxa did not change with the grazing regime. Specifically, a greater relative abundance of nine dominant fungal taxa was observed under the CG than that under the RG, including OTU 52, OTU 5178, OTU 6, OTU 3067, OTU 5287, OTU 4, OTU 82, OTU 64, and OTU 27, which all belong to Ascomycota. However, a greater relative abundance of six dominant bacterial taxa was observed under the CG than that under the GE, including OTU 46, OTU 4, OTU 19, OTU 2050, OTU 7, and OTU 23.
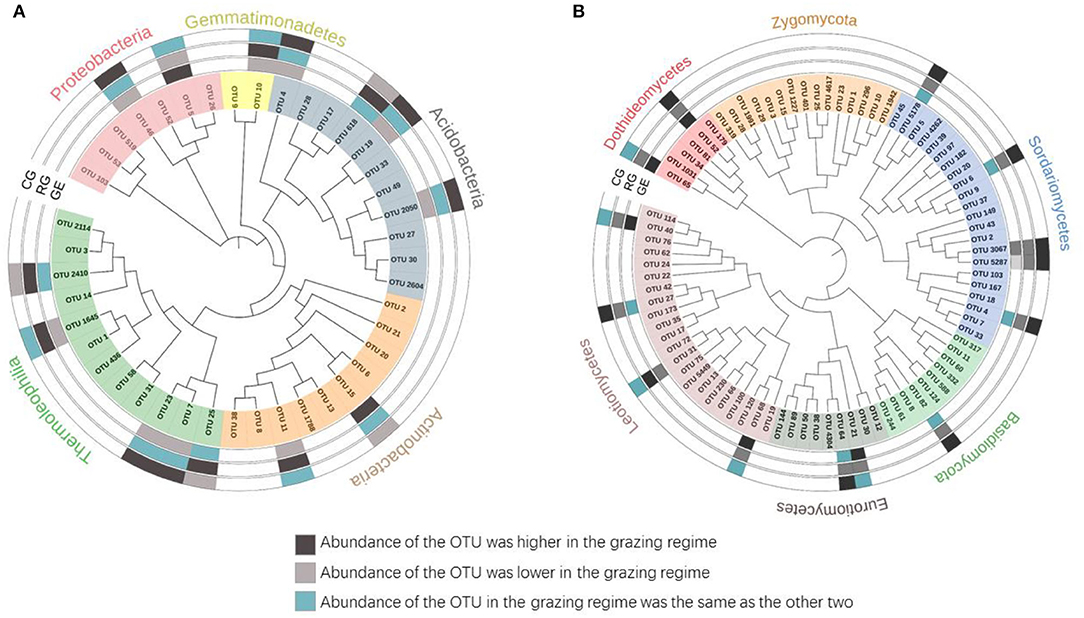
Figure 5. Phylogenetic tree of dominant bacterial (A) and fungal (B) taxa. GE, grazing exclusion; RG, rotational grazing; and CG, continuous grazing.
At the phylum level, soil bacteria were overwhelmingly dominated by Actinobacteria, Proteobacteria, and Acidobacteria; soil fungi were overwhelmingly dominated by Ascomycota, Zygomycota, and Basidiomycota (Supplementary Figure S3). The abundance of Acidobacteria and Armatimonadetes was higher under CG than that under GE, and the abundance of Elusimicrobia was higher under RG and CG than that under GE (Supplementary Figure S4).
The co-occurrence network showed 104 nodes, including 76 bacteria, 16 fungi, one archaea, and 11 plant species (Figure 6A). Node with a maximum degree and eigenvector centrality was Deltaproteobacteria, and node with maximum betweenness and stress centrality was Cytophagia. There were 288 links in the network, with 25 negative and 10 positive links between plants and bacteria; four negative links occurred between plants and fungi; one negative link occurred between plants (S. capillata) and archaea; 22 positive and 18 negative links occurred between bacteria and fungi; five positive links and one negative link occurred between bacteria and archaea; and one positive link occurred between fungi and archaea. In plant species, K. pygmaea is rarely linked with microbes, but it linked four plant specialists that were not directly linked to microbes. S. capillata had most links with microbes, 11 with bacteria, one with fungus, and one with archaea, while only two links of them were positive.
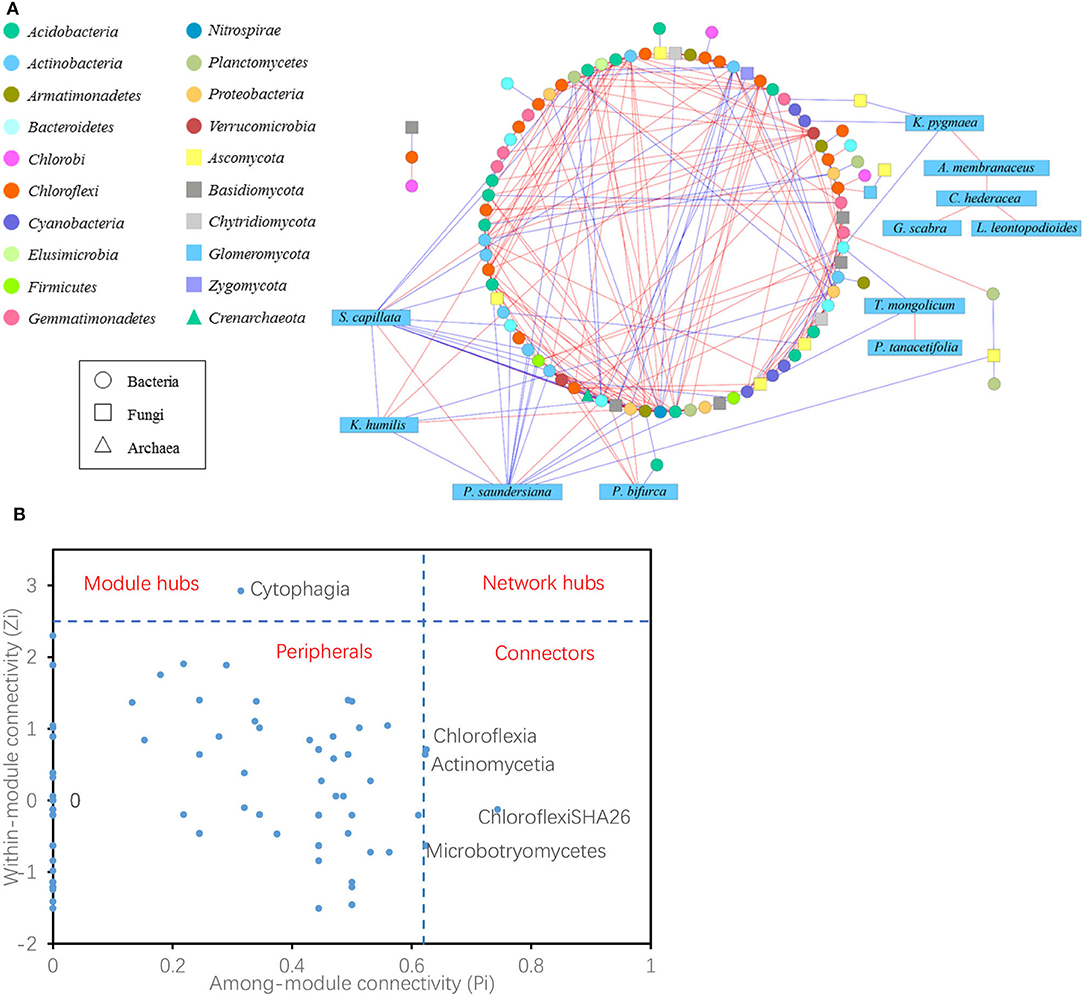
Figure 6. Network interactions of plant species and archaeal, bacterial, and fungal classes (A). A blue edge indicates a negative link between two individual nodes, while a red edge indicates a positive link. Z-P plot showing the distribution of the nodes based on their topological roles. The topological role of each node was determined according to the scatter plot of within-module connectivity (Zi) and among-module connectivity (Pi). The module hubs and connectors were labeled with microbial class names (B).
Within each group, six positive and two negative links were detected among 11 plant species, whereas 139 positive and 49 negative links were detected among 76 bacterial classes and two positive and two negative links were observed among 16 fungal classes. Overall, for bacteria, Chloroflexi had the most links of 79 among them 62 were positive; for fungi, Ascomycota had the most links of 23 among them 11 were positive.
The topological roles of the nodes identified in the network were shown as a Z-P plot in Figure 6B. The majority (95.2%) of the nodes were peripherals with most of their links inside their modules. Among these peripherals, 46.5% had no links outside their modules (i.e., Pi = 0). Approximately 4.8% of the nodes were generalists, including one module hub and four connectors. However, no network hubs (supergeneralists) were observed in the network. The module hub was Cytophagia, and the connectors were ChloroflexiSHA26, Actinomycetia, Chloroflexia, and Microbotryomycetes. These four of the five microbial generalists showed no difference in their abundances among the grazing regimes (Supplementary Figure S5).
In addition, the Mantel test showed that node connectivity was positively related to the correlation coefficient between plant aboveground biomass, belowground biomass, community coverage, and soil NO3-N, while other factors were irrelevant (Supplementary Table S2). Thus, nodes with high connectivity tended to be more sensitive to these factors.
Our results found that the aboveground plant community and belowground soil microbial community in the grassland ecosystem showed different responses to grazing disturbance, and the fungi and bacteria in the soil microbial community also behave differently. The grazing regime significantly affected plant community composition and is mainly reflected in two species, Kobresia pygmaea and Stipa capillata, which are both palatable species for yaks. The former is the dominant species, both GE and CG decreased their coverage compared to RG. However, RG and CG significantly decreased the coverage of Stipa capillata compared to GE; Stipa capillata therefore can be regarded as a species sensitive to grazing. In contrast, the grazing regime did not alter the soil microbial taxonomic composition, implying a stable soil microbial community in the grazed grassland. For soil microbial diversity, bacterial diversity indices were not affected by grazing regime, while only the Simpson index of the fungal diversity indices had no difference among grazing regimes; these results indicated that the diversity of the bacterial community was more tolerant, while the diversity of fungi was more sensitive to grazing. Previous studies obtained similar results showing that fungal community composition was more sensitive than bacterial community composition to disturbance or management regimes (Lauber et al., 2008) and vegetation change (Dassen et al., 2017) and that there is accumulating evidence that fungi are the first consumers of belowground inputs of plant-derived C (De Deyn et al., 2011; Ballhausen and de Boer, 2016).
The microbial community assembly process is important in coupling community composition with the ecosystem function, which includes two processes: determinism and stochasticity (Zhang et al., 2019). Deterministic processes are based on the theory of ecological niche, stressing the role of environmental filtering imposed by environmental factors in the assembly of the microbial community; stochastic process occurs through ecological drift, including random birth and death and dispersal events (Langenheder and Szekely, 2011). Both deterministic and stochastic processes occur simultaneously during the community assembly and the relative importance changes along with environmental conditions (Tang et al., 2021). We found stochastic mechanisms strongly structured soil microbial community assembly under all grazing regimes. Compared to grazing exclusion, rotational grazing and continuous grazing reduced the relative importance of stochastic ratio, indicating that grazing acted as an environmental filter that increased the deterministic soil microbial community assembly.
The soil bacterial functional composition under RG and CG was similar, while the soil fungal functional composition under GE and RG was similar. From GE to RG, then to CG, the interference increased gradually, implying that the type of interference determines the functional composition of bacteria, while the intensity of interference determines the functional composition of fungi. Aerobic chemoheterotrophy and chemoheterotrophy were the main bacterial functional guilds with rich OTUs, and their relative abundance was higher under GE than that under RG and CG, probably resulting from more organic matter under GE without grazing yak's intakes.
Only a few soil microbial phyla showed differences in abundance among the grazing regimes. The abundances of Acidobacteria and Armatimonadetes were higher under CG than that under GE, implying a more acidic environment under CG (Bardhan et al., 2012). Acidobacteria is mainly considered oligotrophs and are known to utilize a variety of carbon resources, even recalcitrant carbon substrates, and contribute to litter decomposition and carbon resource conversion, benefiting plant growth (Macdonald et al., 2015; Lladó et al., 2017). Armatimonadetes is related to carbohydrate transportation and metabolism and can also utilize a wide range of carbohydrates (Lee et al., 2014). Continuous removal of plant tissues by yaks under CG reduced plant carbon entering the soil (Supplementary Figure S7), probably stimulating soil microbial carbohydrate transportation and metabolism processes, and leading to the greater abundance of Acidobacteria and Armatimonadetes under CG. Different from our results, Yang et al. (2019) observed an equal relative abundance of Acidobacteria between grazing exclusion, winter grazing, and annual grazing on the Qinghai-Tibet Plateau. Macdonald et al. (2015) found rabbit grazing had a negative impact on the Acidobacteria and thought the result related to rabbit grazing increased C-allocation belowground through root exudation. Two other studies also observed a reduced relative abundance of Acidobacteria under grazing (Xun et al., 2018; Zhang and Fu, 2021). Different results may be related to climate, herbivore type, grazing method, and intensity, thus it is difficult to draw a generalized conclusion about the grazing effects on Acidobacteria. Armatimonadetes received little attention in the studies of grassland grazing, probably because it is not a dominant taxon. At the OTU level, nine dominant fungal OTUs were higher under CG than that under RG, as CG likely benefited slow-growing plant species that produce low-quality litter (high C/N), thus favoring soil fungi; six dominant bacterial OTUs were higher under CG than that under GE, as livestock fed on aboveground plants and returned nearly half of it as feces, thus favoring bacterial growth (Xun et al., 2018). In addition, the grazing increased Acidobacteria and Armatimonadetes that are related to C metabolism, while grazing did not change other microbial phyla.
For bacteria, Chloroflexi is diverse in terms of morphology, nutrition, and metabolic pathways, mediates the biogeochemical cycle of elements such as C, N, and S (Zarzycki et al., 2009; Sorokin et al., 2012, 2014), and had the most links in the network of plant species and soil microbial classes, including 62 positive and 17 negative links. For fungi, Ascomycota, which has the highest species diversity and can break down plant residues, had the most links in the network of plant species and soil microbial classes, including 11 positive and 12 negative links. The module hub of the network was Cytophagia (phylum Bacteroidia), which is an obligate anaerobic gut microbe in animals (Eckburg et al., 2005; Winter and Bäumler, 2014), and it was the hub likely due to the accumulation of dung and urine by grazing (Barik and Murugan, 2015); the connectors were ChloroflexiSHA26, Actinomycetia, Chloroflexia, and Microbotryomycetes. The abundance of four of the five microbial generalists was not affected by the grazing regime, and the little changes may have contributed to the stability of soil microbial composition.
Across plants, bacteria, fungi, and archaea, most of the links between plants and microbes were negative, and they may compete for nutrients, such as available nitrogen, in the infertile environment (Inselsbacher et al., 2010). Plant species were more linked to bacteria than fungi. There were 22 positive and 18 negative links between bacteria and fungi, five positive and one negative links occurred between bacteria and archaea, and only one positive link was detected between fungi and archaea, indicating that bacteria, fungi, and archaea exhibited cooperative interspecies relationships more than competitive interactions. Within each network component, there were six positive and two negative links in the plant community; 73.9% of the bacterial links were positive. Two positive and two negative links were found in 16 fungi, indicating that fungi were more linked with other components than themselves and that intra-group members were more independent. Overall, soil microbes exhibited cooperative behaviors such as cross-feeding, syntrophic interactions, mutualistic interactions, and competition with plant species, similar to intraspecific cooperation and interspecific competition. Microbial communities often act in consortia to synergistically degrade complex plant-derived compounds, with some microbes utilizing metabolites or taking advantage of broken-down products of extracellular enzymes produced by other taxa (Lynd et al., 2002; Alessi et al., 2017).
In the links between plants and microbes, two remarkable plant species were K. pygmaea and S. capillata. There were few links between K. pygmaea and microbes in the network, but it served as a bridge between microbes and four plant specialists that were not directly related to microbes. S. capillata was linked with 11 bacteria, one fungus, and one archaea, and most of the links were negative. Coincidentally, K. pygmaea and S. capillata were also the two special species that had different coverage among grazing regimes. K. pygmaea was a grazing-tolerant species and the dominant species in the grazed grassland, while S. capillata was a sensitive species that was rarely distributed in the grazed grassland but sharply increased after grazing was prohibited. K. pygmaea and S. capillata were the two extremes in the plant community, the dominant species K. pygmaea acted as a bridge between the plant community and microbes, and the sensitive species S. capillata was more independent. It was found that different species play distinct roles in the community and were generally divided into three types. One type was only widely linked to microbes; the other type was plant specialists, which were not directly linked with microbes; and the third type was the bridge between microbes and plant specialists.
Among the environmental factors, plant community aboveground biomass, belowground biomass, coverage, and soil NO3-N were closely related to the node connectivity of the network. Nodes with more links tended to be more sensitive to these factors but not to the other factors, such as plant community diversity, soil NH4-N, available P, and bulk density. This result implied a close relationship between the soil microbial community and plant community production, in line with previous findings (Van Der Heijden et al., 2008; Yang et al., 2017; Xun et al., 2018; Chen et al., 2020). NO3-N is much more mobile than NH4-N in soil and can be easily lost by leaching into groundwater and surface runoff (Scherer-Lorenzen et al., 2003); thus, NO3-N tends to be the object of competition between plants and microbes and maybe the major cause why many of the links between plants and microbes were negative.
The grazing regime altered the composition of the plant community but did not change the soil microbial community composition. The network between plant and soil microbial community showed that many plant species were not directly connected with microbes but indirectly connected through dominant species K. pygmaea, thus weakening the effect of grazing-induced plant community composition variation on the soil microbial community. Four microbial generalists were not affected by the grazing, hence maintaining the stability of microbial community composition. The stable soil microbial composition is probably also due to the stochastic process dominated by its community assembly. Although soil microbial taxonomic composition was not affected by gazing, its functional composition was significantly changed and GE increased aerobic chemoheterotrophy and chemoheterotrophy in bacteria.
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA811340.
SD, QG, and CF planned and designed the research. YL, HG, YY, and XG performed experiments, conducted fieldwork, and analyzed data etc. YL and MF wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by grants from the National Natural Science Foundation of China (32101315), the China National Key R&D Program (2016YFC0501906 and 2016YFC0502003), the Qinghai Provincial Key R&D Program (2019-SF-145 & 2018-NK-A2), the CTBU high-level talent research start-up project (950319097), and the project of Scientific and Technological Research Program of Chongqing Municipal Education Commission (KJQN20200837).
YY was employed by China New Era Group Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to express gratitude to the reviewers and editors for their time and effort.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.864085/full#supplementary-material
Alessi, A. M., Bird, S. M., Bennett, J. P., Oates, N. C., Li, Y., Dowle, A. A., et al. (2017). Revealing the insoluble metasecretome of lignocellulose-degrading microbial communities. Sci. Rep. 7, 1–10. doi: 10.1038/s41598-017-02506-5
Ballhausen, M. B., and de Boer, W. (2016). The sapro-rhizosphere: carbon flow from saprotrophic fungi into fungus-feeding bacteria. Soil Biol. Biochem. 102, 14–17. doi: 10.1016/j.soilbio.2016.06.014
Bardhan, S., Jose, S., Jenkins, M. A., Webster, C. R., Udawatta, R. P., and Stehn, S. E. (2012). Microbial community diversity and composition across a gradient of soil acidity in spruce—fir forests of the southern Appalachian Mountains. Appl. Soil Ecol. 61, 60–68. doi: 10.1016/j.apsoil.2012.04.010
Barik, D., and Murugan, S. (2015). An artificial neural network and genetic algorithm optimized model for biogas production from co-digestion of seed cake of karanja and cattle dung. Waste Biomass valori. 6, 1015–1027. doi: 10.1007/s12649-015-9392-1
Beck, J. J., Hernández, D. L., Pasari, J. R., and Zavaleta, E. S. (2015). Grazing maintains native plant diversity and promotes community stability in an annual grassland. Ecol. Appl. 25, 1259–1270. doi: 10.1890/14-1093.1
Chapin, F. S. III, Zavaleta, E. S., Eviner, V. T., Naylor, R. L., Vitousek, P. M., et al. (2000). Consequences of changing biodiversity. Nature 405, 234–242. doi: 10.1038/35012241
Chen, Q. L., Ding, J., Zhu, Y. G., He, J. Z., and Hu, H. W. (2020). Soil bacterial taxonomic diversity is critical to maintaining the plant productivity. Environ. Int. 140, 105766. doi: 10.1016/j.envint.2020.105766
Cornelissen, J., Lavorel, S., Garnier, E., Díaz, S., Buchmann, N., Gurvich, D., et al. (2003). A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust. J. Bot. 51, 335–380. doi: 10.1071/BT02124
Dassen, S., Cortois, R., Martens, H., de Hollander, M., Kowalchuk, G. A., van der Putten, W. H., et al. (2017). Differential responses of soil bacteria, fungi, archaea and protists to plant species richness and plant functional group identity. Mol. Ecol. 26, 4085–4098. doi: 10.1111/mec.14175
De Deyn, G., Quirk, H., Oakley, S., Ostle, N., and Bardgett, R. (2011). Rapid transfer of photosynthetic carbon through the plant-soil system in differently managed species-rich grasslands. Biogeosciences 8, 1131–1139. doi: 10.5194/bg-8-1131-2011
Deng, Y., Jiang, Y. H., Yang, Y. F., He, Z., Luo, F., and Zhou, J. Z. (2012). Molecular ecological network analyses. BMC Bioinform. 13, 113. doi: 10.1186/1471-2105-13-113
Ding, J., Li, F., Yang, G., Chen, L., Zhang, B., Liu, L., et al. (2016). The permafrost carbon inventory on the Tibetan Plateau: a new evaluation using deep sediment cores. Glob. Change Biol. 22, 2688–2701. doi: 10.1111/gcb.13257
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fierer, N (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Hong, C., Si, Y., Xing, Y., and Li, Y. (2015). Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ. Sci. Pollut. Res. 22, 10788–10799. doi: 10.1007/s11356-015-4186-3
Inselsbacher, E., Umana, N. H. N., Stange, F. C., Gorfer, M., Schüller, E., Ripka, K., et al. (2010). Short-term competition between crop plants and soil microbes for inorganic N fertilizer. Soil Biol. Biochem. 42, 360–372. doi: 10.1016/j.soilbio.2009.11.019
Langenheder, S., and Szekely, A. J. (2011). Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J. 5, 1086–1094. doi: 10.1038/ismej.2010.207
Lauber, C. L., Strickland, M. S., Bradford, M. A., and Fierer, N. (2008). The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 40, 2407–2415. doi: 10.1016/j.soilbio.2008.05.021
Lee, K. C., Morgan, X. C., Dunfield, P. F., Tamas, I., Mcdonald, I. R., and Stott, M. B. (2014). Genomic analysis of Chthonomonas calidirosea, the first sequenced isolate of the phylum Armatimonadetes. ISME J. 8, 1522–1533. doi: 10.1038/ismej.2013.251
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucl. Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, Y., Dong, S., Gao, Q., Zhang, Y., Liu, S., Ganjurjav, H., et al. (2020). Rotational grazing promotes grassland aboveground plant biomass and its temporal stability under changing weather conditions on the Qinghai-Tibetan Plateau. Land Degrad. Dev. 31, 2662–2671. doi: 10.1002/ldr.3596
Li, Y., Dong, S., Gao, Q., Zhang, Y., Liu, S., Swift, D., et al. (2019). Grazing promotes plant functional diversity in alpine meadows on the Qinghai-Tibetan Plateau. Rangel. J. 41, 73–81. doi: 10.1071/RJ18091
Li, Y. Y., Dong, S. K., Wen, L., Wang, X. X., and Wu, Y. (2014). Soil carbon and nitrogen pools and their relationship to plant and soil dynamics of degraded and artificially restored grasslands of the Qinghai–Tibetan Plateau. Geoderma 213, 178–184. doi: 10.1016/j.geoderma.2013.08.022
Lladó, S., López-Mondéjar, R., and Baldrian, P. (2017). Forest soil bacteria: diversity, involvement in ecosystem processes, and response to global change. Microbiol. Mol. Biol. Rev. 81, e00063. doi: 10.1128/MMBR.00063-16
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272. doi: 10.1126/science.aaf4507
Lynd, L. R., Weimer, P. J., Van Zyl, W. H., and Pretorius, I. S. (2002). Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577. doi: 10.1128/MMBR.66.3.506-577.2002
Macdonald, C. A., Crawley, M. J., Wright, D. J., Kuczynski, J., Robinson, L., Knight, R., et al. (2015). Identifying qualitative effects of different grazing types on below-ground communities and function in a long-term field experiment. Environ. Microbiol. 17, 841–854. doi: 10.1111/1462-2920.12539
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Ning, D., Deng, Y., Tiedje, J. M., and Zhou, J. (2019). A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. 116, 16892–16898. doi: 10.1073/pnas.1904623116
Pérez-Harguindeguy, N., Diaz, S., Gamier, E., Lavorel, S., Poorter, H., Jaureguiberry, P., et al. (2013). New handbook for standardised measurement of plant functional traits worldwide. Aust. J. Bot. 61:167–234. doi: 10.1071/BT12225
Scherer-Lorenzen, M., Palmborg, C., Prinz, A., and Schulze, E.-D. (2003). The role of plant diversity and composition for nitrate leaching in grasslands. Ecology 84, 1539–1552. doi: 10.1890/0012-9658(2003)084[1539:TROPDA]2.0.CO;2
Shen, H., Wang, S., and Tang, Y. (2013). Grazing alters warming effects on leaf photosynthesis and respiration in Gentiana straminea, an alpine forb species. J. Plant Ecol. 6, 418–427. doi: 10.1093/jpe/rtt010
Sorokin, D. Y., Lücker, S., Vejmelkova, D., Kostrikina, N. A., Kleerebezem, R., Rijpstra, W. I. C., et al. (2012). Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 6, 2245–2256. doi: 10.1038/ismej.2012.70
Sorokin, D. Y., Vejmelkova, D., Lücker, S., Streshinskaya, G. M., Rijpstra, W. I. C., Damste, J. S. S., et al. (2014). Nitrolancea hollandica gen. nov., sp. nov., a chemolithoautotrophic nitrite-oxidizing bacterium isolated from a bioreactor belonging to the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 64, 1859–1865. doi: 10.1099/ijs.0.062232-0
Tang, H., Zhang, N., Ni, H., Xu, X., and Liang, Y. (2021). Increasing environmental filtering of diazotrophic communities with a decade of latitudinal soil transplantation. Soil Biol. Biochem. 154, 108119. doi: 10.1016/j.soilbio.2020.108119
Tang, L., Zhong, L., Xue, K., Wang, S., Xu, Z., Lin, Q., et al. (2019). Warming counteracts grazing effects on the functional structure of the soil microbial community in a Tibetan grassland. Soil Biol. Biochem. 134, 113–121. doi: 10.1016/j.soilbio.2019.02.018
Van Der Heijden, M. G., Bardgett, R. D., and Van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Wang, M., Wang, S., Wu, L., Xu, D., Lin, Q., Hu, Y., et al. (2016). Evaluating the lingering effect of livestock grazing on functional potentials of microbial communities in Tibetan grassland soils. Plant Soil 407, 385–399. doi: 10.1007/s11104-016-2897-y
Winter, S. E., and Bäumler, A. J. (2014). Dysbiosis in the inflamed intestine: chance favors the prepared microbe. Gut Microbes 5, 71–73. doi: 10.4161/gmic.27129
Xun, W., Yan, R., Ren, Y., Jin, D., Xiong, W., Zhang, G., et al. (2018). Grazing-induced microbiome alterations drive soil organic carbon turnover and productivity in meadow steppe. Microbiome 6, 170. doi: 10.1186/s40168-018-0544-y
Yang, F., Niu, K., Collins, C. G., Yan, X., Ji, Y., Ling, N., et al. (2019). Grazing practices affect the soil microbial community composition in a Tibetan alpine meadow. Land Degrad. Dev. 30, 49–59. doi: 10.1002/ldr.3189
Yang, T., Adams, J. M., Shi, Y., He, J. S., Jing, X., Chen, L., et al. (2017). Soil fungal diversity in natural grasslands of the Tibetan Plateau: associations with plant diversity and productivity. New Phytol. 215, 756–765. doi: 10.1111/nph.14606
Yang, Y., Wu, L., Lin, Q., Yuan, M., Xu, D., Yu, H., et al. (2013). Responses of the functional structure of soil microbial community to livestock grazing in the Tibetan alpine grassland. Glob. Change Biol. 19, 637–648. doi: 10.1111/gcb.12065
Zarzycki, J., Brecht, V., Müller, M., and Fuchs, G. (2009). Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proc. Natl. Acad. Sci. 106, 21317–21322. doi: 10.1073/pnas.0908356106
Zhang, H., and Fu, G. (2021). Responses of plant, soil bacterial and fungal communities to grazing vary with pasture seasons and grassland types, northern Tibet. Land Degrad. Dev. 32, 1821–1832. doi: 10.1002/ldr.3835
Zhang, Z., Deng, Y., Feng, K., Cai, W., Li, S., Yin, H., et al. (2019). Deterministic assembly and diversity gradient altered the biofilm community performances of bioreactors. Environ. Sci. Technol. 53, 1315–1324. doi: 10.1021/acs.est.8b06044
Zhong, L., Li, F. Y., Wang, Y., Zhou, X., Zhou, S., Gong, X., et al. (2018). Mowing and topography effects on microorganisms and nitrogen transformation processes responsible for nitrous oxide emissions in semi-arid grassland of Inner Mongolia. J. Soils Sediment. 18, 929–935. doi: 10.1007/s11368-017-1819-9
Zhou, G., Zhou, X., He, Y., Shao, J., Hu, Z., Liu, R., et al. (2017). Grazing intensity significantly affects belowground carbon and nitrogen cycling in grassland ecosystems: a meta-analysis. Glob. Change Biol. 23, 1167–1179. doi: 10.1111/gcb.13431
Keywords: grazing regime, dominant species, functional composition, co-occurrence network, community assembly
Citation: Li Y, Dong S, Gao Q, Fan C, Fayiah M, Ganjurjav H, Hu G, Wang X, Yan Y, Gao X and Li S (2022) Grazing Changed Plant Community Composition and Reduced Stochasticity of Soil Microbial Community Assembly of Alpine Grasslands on the Qinghai-Tibetan Plateau. Front. Plant Sci. 13:864085. doi: 10.3389/fpls.2022.864085
Received: 28 January 2022; Accepted: 26 April 2022;
Published: 23 May 2022.
Edited by:
Junran Jimmy Li, University of Tulsa, United StatesReviewed by:
Roberta Pastorelli, Council for Agricultural and Economics Research (CREA), ItalyCopyright © 2022 Li, Dong, Gao, Fan, Fayiah, Ganjurjav, Hu, Wang, Yan, Gao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shikui Dong, ZG9uZ3NoaWt1aUBzaW5hLmNvbQ==; Qingzhu Gao, Z2FvcXpoQGFtaS5hYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.