- 1Department of Biotechnology, National Agri-Food Biotechnology Institute, Mohali, India
- 2CSIR-National Botanical Research Institute, Lucknow, India
- 3Institute of Botany, Heinrich Heine University Düsseldorf, Düsseldorf, Germany
- 4Cluster of Excellence on Plant Sciences, Heinrich Heine University Düsseldorf, Düsseldorf, Germany
Our knowledge of iron (Fe) uptake and mobilization in plants is mainly based on Arabidopsis and rice. Although multiple players of Fe homeostasis have been elucidated, there is a significant gap in our understanding of crop species, such as wheat. It is, therefore, imperative not only to understand the different hurdles for Fe enrichment in tissues but also to address specifically the knowns/unknowns involved in the plausible mechanism of Fe sensing, signaling, transport, and subsequent storage in plants. In the present review, a unique perspective has been described in light of recent knowledge generated in wheat, an economically important crop. The strategies to boost efficient Fe uptake, transcriptional regulation, and long-distance mobilization in grains have been discussed, emphasizing recent biotechnological routes to load Fe in grains. This article also highlights the new elements of physiological and molecular genetics that underpin the mechanistic insight for the identified Fe-related genes and discusses the bottlenecks in unloading the Fe in grains. The information presented here will provide much-needed resources and directions to overcome challenges and design efficient strategies to enhance the Fe density in wheat grains.
Introduction
Iron (Fe) is an essential micronutrient for all life forms that participate as catalytic cofactors in several vital processes; including phosphorylation, photosynthesis, and chlorophyll biosynthesis in plants (Yadavalli et al., 2012; Kroh and Pilon, 2020; Schmidt et al., 2020). The connection between Fe deficiency and its effect on human health is an important research subject nowadays. Improving the grain Fe content has been considered a unique way to tackle the hidden hunger and Fe deficiency ailments in humans (micronutrient biofortification). Wheat is an important source of energy nutrition in developing countries and has been a target crop for addressing multiple nutritional traits, including micronutrient enhancement, such as Fe and Zinc (Zn) (Borg et al., 2012; Sui et al., 2012; Chattha et al., 2017; Sazawal et al., 2018). The limited genetic variation for Fe content and its low bioavailability in improved adapted wheat varieties have posed challenges to the breeders (Velu et al., 2014). In addition to this, multiple bottlenecks have been identified for the uptake and mobilization of Fe in plants. After the uptake of Fe, it has to be distributed into plant organelles through various transporters, tissues via the cell-to-cell communication network and different organs through phloem and xylem mediated long-distance transport systems. Research on Fe homeostasis has been largely focused on the model plant Arabidopsis thaliana, which represents a non-graminaceous plant system. Earlier, the lack of wheat gene resources and information about detailed mechanisms involved in Fe homeostasis was a major obstacle for devising biofortification approaches. Therefore, it is imperative to discuss and explore new target areas that could be conserved or specific to wheat crops to deploy biotechnological routes to generate high Fe in grain.
Herein, we primarily provide a glimpse of the genetic components involved in the uptake and mobilization of Fe from roots to the grains and the potential gene targets. Although Fe loading in cereals’ grain is a multistep process, few reliable strategies for grain micronutrient enrichment can be devised by drawing parallels with other plants, including the model plant Arabidopsis and Oryza sativa. Some of these genetic components have been specifically identified and reported recently in wheat (Kaur et al., 2019; Wang M. et al., 2019). In the present review, some of the important candidate genes that could be used by biotechnological interventions are described and innovative measures have also been discussed. We have provided an insight into the comparative account of genes involved in Fe homeostasis from well-studied plants, such as Arabidopsis and O. sativa, and presented an overview of the gene regulatory network. We have also emphasized the anatomical bottlenecks and the key areas that need immediate attention to finally generate new genotypes with high Fe in grains for developing viable and sustainable approaches to overcome low Fe in wheat grain. In turn, the challenges of grain Fe enrichment and bottleneck through genetic engineering are discussed in this review with emphasis on possible solutions as well.
Conserved Molecular Components of Fe Uptake in Wheat
Fe is found to be the fourth most abundant metal in soil. However, the majority of Fe in the soil is present in the form that plants cannot assimilate. In aerobic soil, Fe concentration and bioavailability depends on inorganic forms, such as oxides, hydroxides, and phosphate. Trivalent form (Fe3+) is the predominant form of Fe present in soil and the alkaline pH of soil favors the formation of hydrous Fe3+ oxides (a solid and insoluble state of Fe), hence becomes inaccessible to the plants (Lindsay, 1991). To acquire this form, strategy I plants reduce Fe3+ to ferrous (Fe2+) and then facilitate its transport whereas strategy II plants export phytosiderophores (PS) that chelate Fe3+ and make this form bio-available to plants (Kobayashi and Nishizawa, 2012).
Wheat utilizes a chelation-based strategy (strategy-II) of Fe uptake. The Fe transport mechanism in wheat is coupled with the production of Fe chelators, such as 2-deoxymugineic acid (DMA) and mugineic acid (MA), and their subsequent secretion into the rhizosphere. Among these PS, DMA is the predominating Fe chelator secreted by wheat roots (Tolay et al., 2001) into the rhizosphere through the transporter of mugineic acid (TOM) and facilitates the formation of PS-Fe3+ complex (Nozoye et al., 2011). This bioavailable PS-Fe complex is further taken up in the plant root through yellow stripe-like transporters (YSL; Curie et al., 2001). The gene families that constitute the strategy-II pathway of Fe transport have been characterized in wheat. The wheat genome encodes for 21 NAS, 6 NAAT, 3 DMAS, 5 TOM, and 26 YSL genes (Bonneau et al., 2016; Beasley et al., 2017; Kumar et al., 2019; Sharma et al., 2019). Table 1 summarizes the number of genes reported in Arabidopsis, rice, and wheat. The expression analysis of these identified genes under Fe limiting conditions showed significant induction in wheat root and shoot (Kaur et al., 2019; Wang M. et al., 2019). Although the differential expression pattern of these genes indicates their potential involvement in Fe transport, their site of action and functional studies in wheat are yet to be explored. Interestingly, the number of genes for each family shows the highest number of genes in hexaploid wheat compared to rice and other crops.
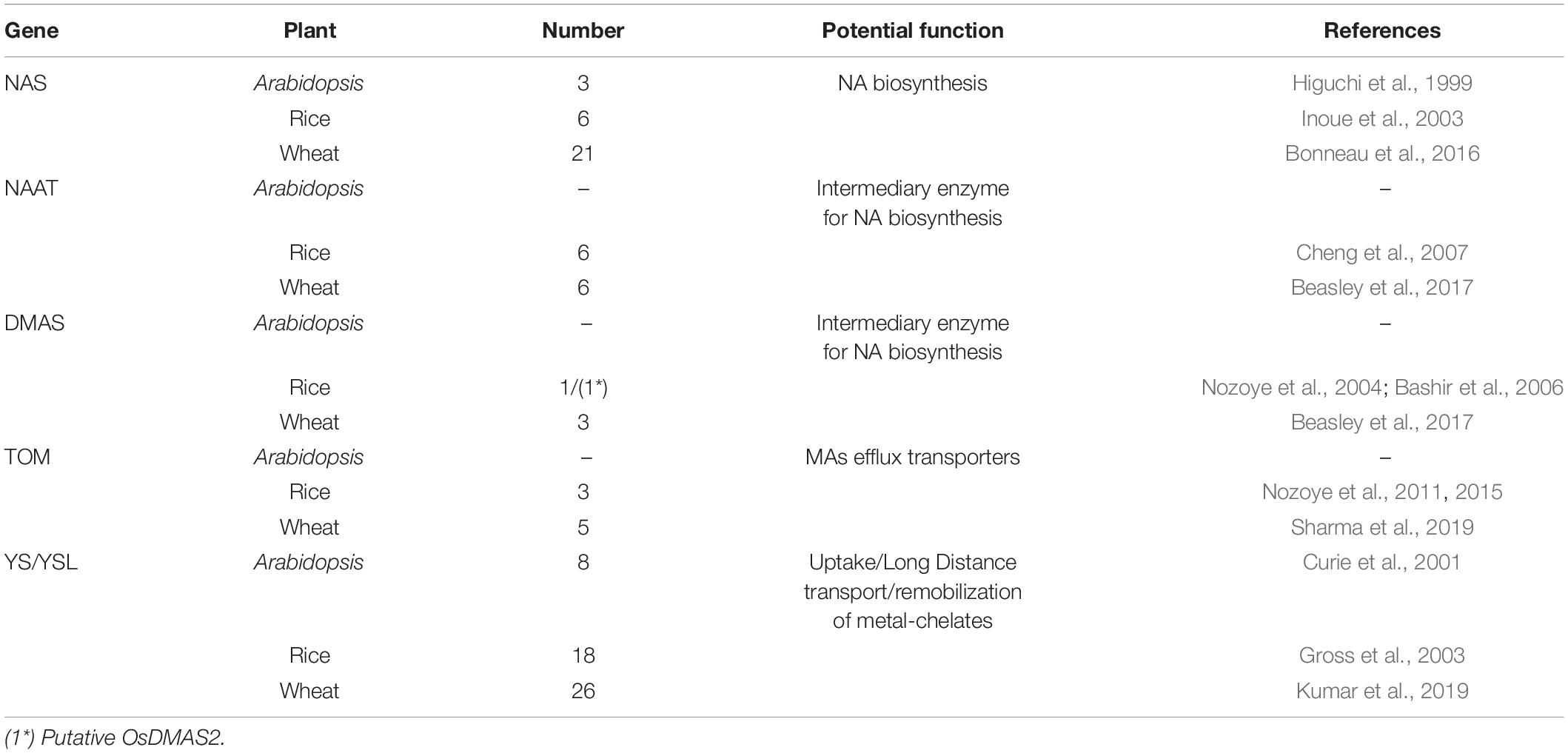
Table 1. Putative orthologs/homologs and their number of genes for the Fe uptake reported from Arabidopsis, rice, and wheat.
Interestingly, transcriptome data showed the presence of strategy I components, including Iron-Regulated Transporter 1 (IRT1) and Ferric Reduction Oxidase (FRO) genes, in wheat. The functional studies of IRT1 and FRO2 in Arabidopsis and other crops are already known for Fe transport via strategy I (Eide et al., 1996; Wu et al., 2005). However, rice, barley, and other prominent graminaceous cereals retain both strategies for Fe acquisition simultaneously (Ishimaru et al., 2006). The expression analysis showed significant induction of IRT1 orthologs in wheat under Fe limiting conditions, whereas strategy 1 genes were unaltered (Kaur et al., 2019). Our previous study also showed that as many as 51 genes encoding putative IRT/ZIP Fe and Zn transporters are present in wheat (Kaur et al., 2019). This may suggest that the IRT1 genes could function independently of strategy II component, which is the predominant route of Fe acquisition in wheat.
Moreover, the production and release of various metabolites are paired with changes in root phenotype under Fe deficiency in wheat. In the recent past, numerous reports have described that the exudation of phenylpropanoids and flavin-derived metabolites from roots are essential for efficient Fe uptake in calcareous soil and alkaline conditions (Rodríguez-Celma et al., 2013; Schmid et al., 2014; Sisó-Terraza et al., 2016). These derivatives may help in the solubilization of (Fe3+) Fe precipitates and subsequent reduction of Fe3+ to Fe2+ for direct import (Mladìnka et al., 2010). Metabolic analysis of wheat under Fe limiting conditions revealed the enhanced production of organic acids and polyhydroxy acids, such as fumarate, malonate, succinate, and xylofuranose, along with PS release in wheat (Garnica et al., 2018; Kaur et al., 2019).
Interaction of Fe With Phosphorus and Sulfur
Despite the varying concentrations of nutrients in soil, plants maintain a cellular nutrient balance for optimal growth as well as avoid toxic effects. This is achieved by the interwoven nutrient homeostasis and synergistic/antagonistic interactions among the nutrients. Any change in soil nutrients is efficiently sensed by the root, which initiates a transcriptional network that eventually helps the plant to deal with the deficiency/excess response. Depending on the charge, some nutrients interact with others in the rhizosphere and determine their bioavailabilities. For instance, phosphorus (P) and sulfur (S) display interconnected homeostasis with Fe in the rhizosphere (Hirsch et al., 2006; Ward et al., 2008). P exists in many chemical forms in soil, but plants acquire mainly inorganic phosphate (Pi) and organic phosphorus (Po) for their growth and development. The strong interaction between Fe and Pi in soil forms precipitates, affecting the availability of both nutrients to the plant. It has been observed that a high-affinity root Fe2+ uptake system which is usually activated under Fe-deficiency, is also induced under excess Pi environment (Ward et al., 2008). Similarly, under Pi-deficient conditions, Arabidopsis accumulated more Fe and heavy metals (Misson et al., 2005; Hirsch et al., 2006). Studies showed that increased accumulation of Fe in Pi-deficient medium causes Fe-toxicity leading to primary root inhibition (Ward et al., 2008; Ticconi et al., 2009). In Arabidopsis, the increase in Fe availability after Pi-deficiency was thought to be correlated at the molecular level by induction of AtFER1, which encodes ferritin (a Fe storage protein) (Hirsch et al., 2006). However, other ferritin genes, such as AtFER3 and AtFER4 lack PHR1-binding sites, in their promoters and are induced under high Fe but non-responsive to Pi-starvation (Petit et al., 2001; Bournier et al., 2013). In hexaploid wheat, the physiological effects were accounted for the molecular changes that involve the regulation of a subset of downstream signaling genes potentially cross-regulated by Fe and Pi (Kaur et al., 2021). The data set from this study reveals that PS release and metabolic pathway for phenyl-propanoid plays an important role for Fe mobilization in the roots under single and dual deficiency of these nutrients (Kaur et al., 2021).
By contrast, many studies have reported a synergistic interaction between Fe and S in the rhizosphere. The S and Fe are highly essential for the optimal functioning of chlorophyll biosynthesis, thereby controlling the plant’s photosynthetic ability through modulating the assembly of Fe-S cluster proteins (Couturier et al., 2013). The genes involved in S assimilation, namely TdSultr1.1, TdSultr1.3, TdAPR, and TdSiR showed similar overlapping expressions under Fe and S deficiency in the durum wheat (Ciaffi et al., 2013). Fe uptake and assimilation are hampered under the S depleted condition (Astolfi et al., 2003, 2018; Ciaffi et al., 2013). Further, when plants are subjected to an S limiting environment, a reduction of NA in the shoot is observed even under Fe sufficient conditions resulting in the low mobilization of micronutrients (Fe and Zn) toward the sink tissue. Thus, it is conclusive that elemental interactions in the rhizosphere also create multiple nutrient deficiencies for plants and dramatically affect plant growth, and could modulate the Fe availability in roots for its directional uptake.
Transcriptional Regulation of Fe Homeostasis in Wheat
Although few molecular components, including transporters, are largely known, the entire transcriptional regulation of genes involved in Fe deficiency response remains elusive mainly in wheat. Under Fe-deficiency, hexaploid wheat revealed a perturbation of the sub-set of transcription factors (TFs), including basic helix-loop-helix (bHLH), basic leucine zipper (bZIP), WRKY, C2H2, MYB, NAC, and homeobox-leucine zipper (Figure 1). Among these, bHLHs were found to be the most predominant TFs suggesting their conserved role in regulating Fe deficiency-related responses (Kaur et al., 2019). The role of bHLHs mediated transcriptional regulation has been extensively investigated in Arabidopsis and to some extent in rice. Given the large role of bHLH in Fe-homeostasis, we will be focusing on the homologs of the candidate bHLH genes in wheat. We have summarized the paralogs of bHLHs in wheat-related with Fe-homeostasis based on orthologous sequence similarity with Arabidopsis and rice (Table 2). The bHLH family in plants is clustered into 26 different groups out of which six sub-groups are reported to regulate Fe homeostasis or deficiency response in Arabidopsis (Pires and Dolan, 2010). The FIT (Fer-Like Fe Deficiency-Induced Transcription Factor), a homolog of the FER transcription factor, has been demonstrated to play a central role in adapting and controlling the amount of Fe uptake in Arabidopsis (Ling et al., 2002; Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005; Mai et al., 2015; Schwarz and Bauer, 2020). The FIT is shown to interact with members of the bHLH subgroup Ib (bHLH38, 39, 100, 101) leading to stabilization of FIT and activation of other subgroup Ib bHLHs (Naranjo-Arcos et al., 2017; Cui et al., 2018; Trofimov et al., 2019). These transcriptional networks, in turn, provide the adaptive and sustainable condition for regulating the expression of multiple downstream genes committed for Fe uptake and mobilization in plants (Mai et al., 2016; Schwarz and Bauer, 2020). Further, OsIRO2, a Fe related bHLH TF, is thought to be a master regulator of Fe deficiency response in rice and five orthologs of IRO2 are present in the hexaploid wheat genome (Table 2; Ogo et al., 2007). IRO2 and OsbHLH156 regulate the expression of a subset of root-specific Fe deficiency responsive genes, thus helping Fe mobilization. Feedback regulation of FIT can also be tightly controlled by other bHLH TFs as FIT interaction with IVa subgroup members (bHLH18, bHLH19, bHLH20, bHLH25) leads to proteasomal degradation (Cui et al., 2018). The MYC2, a bHLH of subgroup IIIe, regulates the expression of the other genes representing the IVa subgroup (Li et al., 2016; Cui et al., 2018). The bHLH121/URI (member of IVb subgroup) and bHLH034, bHLH104, bHLH105/ILR3, and bHLH115 (members of IVc subgroup) were shown to be involved in the regulation of Fe homeostasis in both rice and Arabidopsis (Li et al., 2016; Liang et al., 2017; Wang et al., 2017; Kim et al., 2019; Tissot et al., 2019; Gao et al., 2020). Similarly, OsbHLH057/PRI4, OsbHLH058/PRI2, OsbHLH059/PRI3, and OsbHLH060/PRI1 act upstream to bHLH genes namely OsIRO2, PYE, and IRO3 by binding to their promoters under Fe deficiency (Zhang et al., 2017, 2020; Kobayashi et al., 2019). It is important to mention that bHLHs do not always positively activate Fe homeostasis-related genes expression but AtPYE and OsIRO3 are characterized as negative regulators of certain Fe-deficiency-induced genes (Long et al., 2010; Zheng et al., 2010). OsIRO3 was found to repress OsIRO2 transcripts to regulate Fe uptake in rice (Long et al., 2010; Zheng et al., 2010).
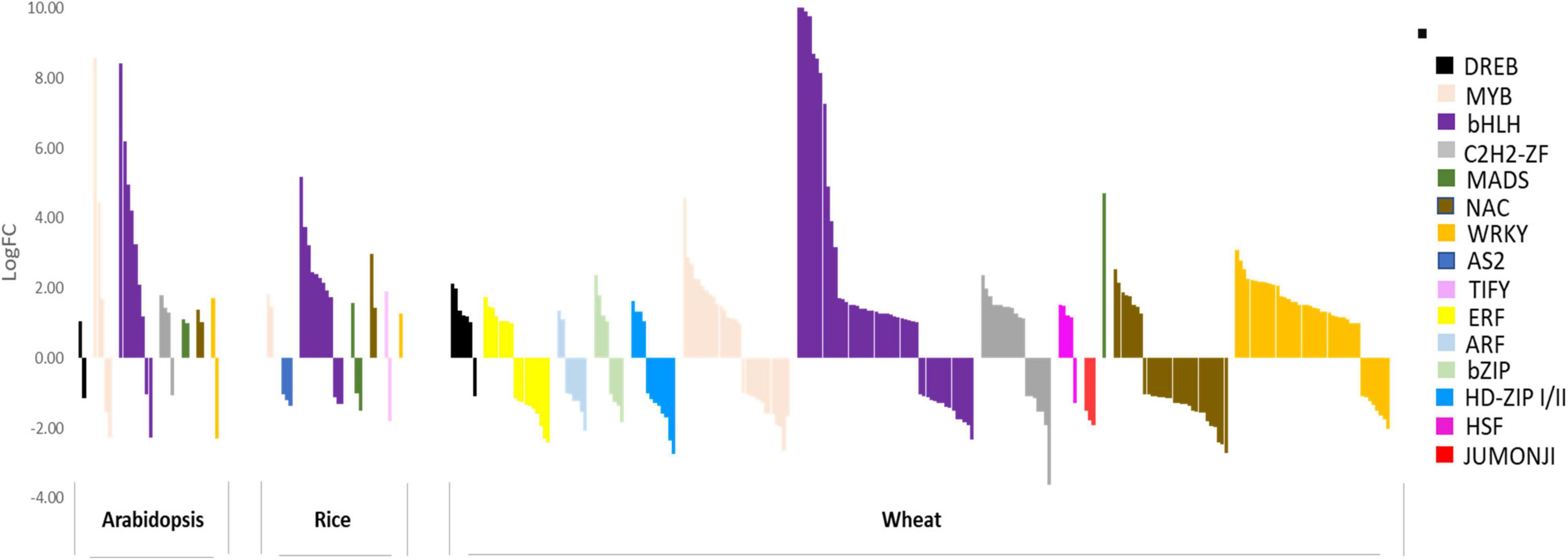
Figure 1. TF subfamilies differentially expressed (DE) in response to Fe starvation in Arabidopsis, rice, and wheat. Expression bar plot representing transcription factor subfamilies differentially expressed in response to Fe starvation in Arabidopsis, rice and wheat. X-axis depicts the TF genes in respective species and Y-axis shows the log fold change (logFC) for the respective genes under Fe starvation with reference to the respective control samples. TF subfamilies are color-coded according to the legend on the right. Respective datasets were downloaded and analyzed for DEGs using the Kallisto-DESeq2 pipeline after getting high-quality, adapter trimmed reads using Trimmomatic. The DEGs for respective species were annotated for TF subfamilies using Mercator4 v2.0 (Schwacke et al., 2019) (TF subfamilies with only 1 gene DE in At and rice; less than 5 genes DE for wheat are not depicted in this graph).
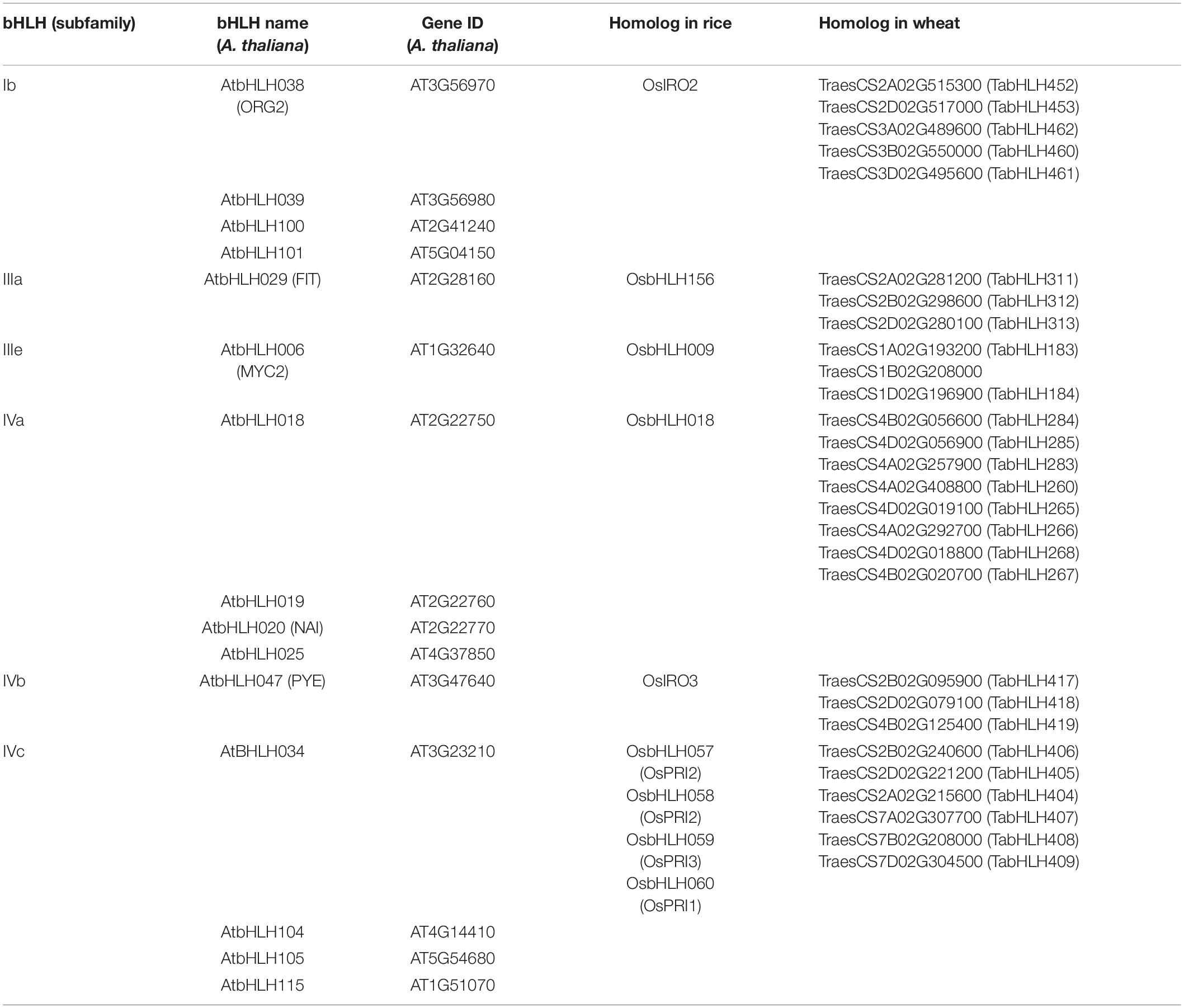
Table 2. Fe homeostasis-related regulatory bHLH protein from A. thaliana and their homologs from rice and wheat.
Although the role of bHLHs has been systematically characterized in Arabidopsis and rice, their distinct or conserved function in wheat is still largely unaddressed. Thus, we decided to generate a transcriptional network of bHLHs in wheat based on their annotation corresponding to homologs in Arabidopsis and rice (Figure 2). This network depicts the categorization of the identified bHLHs of wheat into five subgroups and explains how subgroups interact with each other to evoke the gene expression of Fe deficiency (Figure 2). The functional validation of candidate bHLH using genome editing tools or overexpressing wheat lines could be the appropriate way to investigate their role in Fe-homeostasis. It is worthy to mention here that cis-regulatory elements (CRE) are the binding sites for TFs regulating the expression of Fe homeostasis-related genes in plants. A machine-learning-based computational method identified novel CREs (∼100 putative CREs) involved in Fe deficiency regulation have been identified in rice and Arabidopsis (Schwarz et al., 2020). Similarly, several Fe homeostasis-related orthologs in wheat show the presence of multiple important CREs, including E-box/G-box, in the promoter regions suggesting the occurrence of bHLHs transcriptional hub in wheat (Beasley et al., 2017; Kumar et al., 2019; Sharma et al., 2019, 2020). Altogether, the network of TFs especially the bHLHs hub defines the integration of different regulatory nodes for controlling the downstream target genes involved in Fe uptake and mobilization. Upon functional characterization, these bHLH TFs could be utilized to enhance the mobilization of Fe to foliar parts mainly in grain.
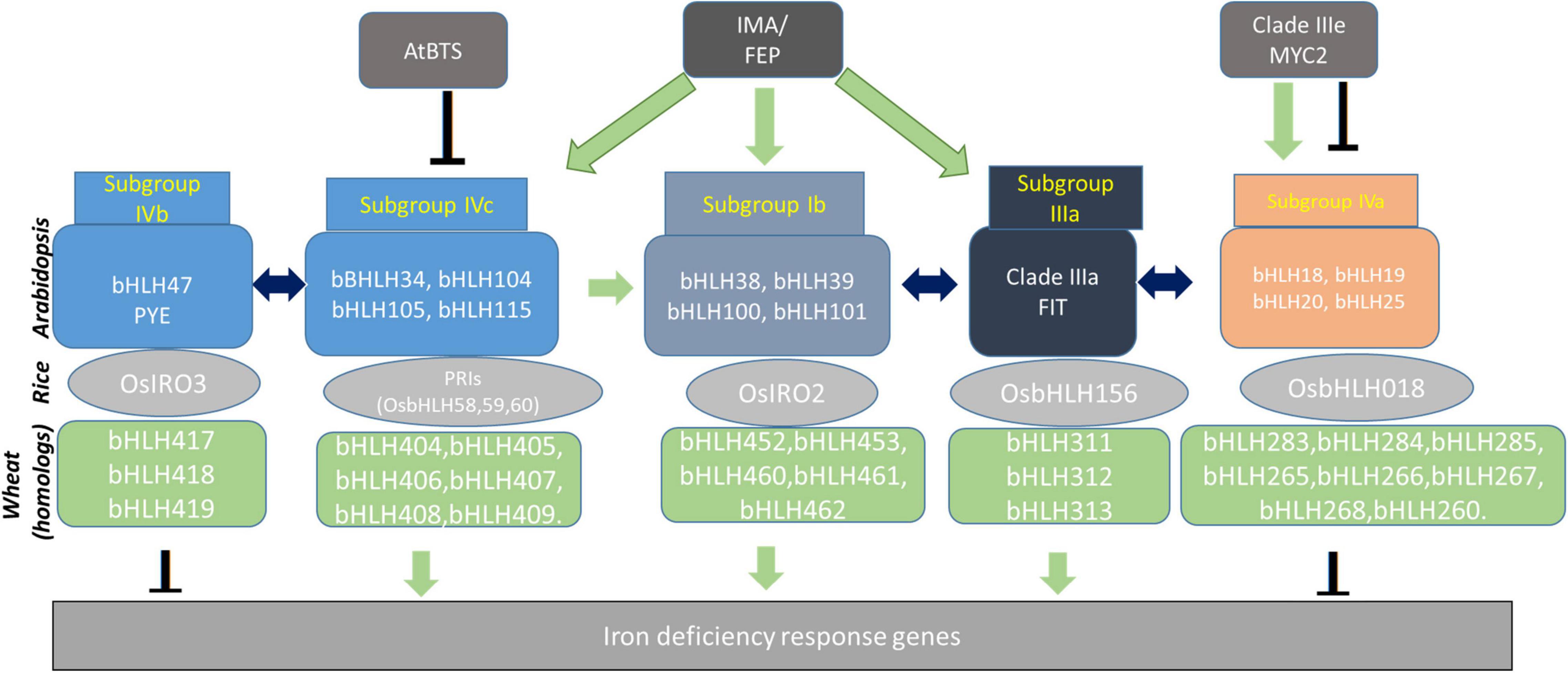
Figure 2. Transcriptional homologs of Arabidopsis and rice bHLH proteins reported for their functional activity for Fe homeostasis. The respective homologs from wheat were identified and placed in the network as a probable candidate for function. Their conserved role in Fe-related phenomenon remains to be tested and validated. Many of these bHLH transcription factors are in turn regulated by BRUTUS, IRON MAN: IMA, and bHLH-like MYC2. Schematic arrangement of bHLH dependent interaction and control at the transcription level is indicated by respective arrow signs: Green arrow: transcriptional activation; Inverted T: gene repression; Both side arrow: protein-protein interaction.
Biotechnological Routes for Fe Fortification and Accomplishments in Wheat
In the past few years, limited attempts have been made to increase Fe content or its bioavailability in wheat grain using a biotechnological route. The criteria for the selection of genes were mainly based on overlapping functions in rice, maize, barley, and Arabidopsis. At this point, it is important to consider the loading of Fe in the grains and enhance its bioavailability for better human nutrition. Therefore, biotechnological routes to enhance these two traits is challenging in crops. Therefore researchers have opted for different strategies to target these traits.
Targeting Genes for Uptake and Loading in Grains
The first successful report to achieve high Fe in cereals was demonstrated by transferring the soybean ferritin gene driven under rice glutelin-1 promoter resulted in threefold higher Fe content as compared to wildtype rice (cv. Kita-ake) (Goto et al., 1999). Subsequently, many research groups have attempted to improve Fe content in the rice grain using SoyFer1 in different rice cultivars under the control of different promoters (Drakakaki et al., 2000; Qu et al., 2005; Oliva et al., 2014). The genes involved in Fe uptake like OsIRT1, OsTOM1, OsYSLs (YSL15, YSL2, and YSL13), and long-distance Fe translocation namely OsNAS1, OsNAS2, OsNAS3 have been targeted to achieve a significant increase (up to 40 μg/g) of Fe levels in rice (Lee and An, 2009; Lee et al., 2009a,b, 2012; Ishimaru et al., 2010; Nozoye et al., 2011; Díaz-Benito et al., 2018; Zhang et al., 2018). The transfer of genes related to Fe uptake, such as NAS and YSL from Hordeum vulgare, has also been able to increase Fe content in rice varieties (Masuda et al., 2012; Boonyaves et al., 2016; Banakar et al., 2017). A study reported that OsIRO2, a gene responsible for Fe utilization and regulatory response, improves growth and yield in calcareous soil as well as accumulated Fe in rice grain (Ogo et al., 2011). Simultaneously, the gene silencing approach was also employed to enhance Fe levels in rice by targeting the intercellular/intracellular transporters comprising OsVIT1, OsVIT2, OsYSL9, and OsDMAS1 (Zhang et al., 2012; Bashir et al., 2017; Senoura et al., 2017).
Ferritin is a Fe storage protein exclusively targeted to plastids and mitochondria in plants. The ferritin encoding genes are well conserved in the plant kingdom, and the genome of hexaploid wheat contains two ferritin genes. The two separate pioneering studies reported that expression of the ferritin gene in endosperm enhanced Fe up to 1.25–1.35 fold and 1.04–1.64 fold, respectively, in the wheat grain (Borg et al., 2012; Sui et al., 2012). The Vacuolar Iron Transporter (VIT), which transports Fe into vacuoles, is involved in Fe loading in the Arabidopsis seeds (Kim et al., 2006). The overexpression of TaVIT2 in endosperm caused efficient vacuolar Fe transport in the endosperm which ultimately resulted in a significant ∼2-fold increase in Fe content in wheat grain (Connorton et al., 2017). In an elegant study, Narayanan et al. (2019) combined three genes from the Arabidopsis (VIT, IRT1, and FER1) into one expression cassette and developed transgenic lines in cassava (Manihot esculenta Crantz), a staple crop in the West African human population. The field data of transgenic lines suggested an elevated Fe and Zn in cassava roots that may benefit the long-awaited goal of improved nutritional status of consumers (Narayanan et al., 2019). Therefore, multiplexing of important genes with different combinations might be a viable option to channelize more Fe in the wheat endosperm. Such a strategy has also been attempted in rice, for instance, overexpression of AtIRT1, PvFerritin, and AtNAS1 led to an increment in grain Fe content (2–10 μg/g) (Boonyaves et al., 2016), and a combination of AtNAS1, PvFer, and AtNRAMP3 resulted in the rise of Fe from 2 to 13 μg/g (Wu et al., 2019). Using OsNAS2, PvFERRITIN, and in a combination of both genes, Singh et al. (2017) developed wheat transgenic lines containing enough Fe in grain to meet the recommended dietary allowances for humans (Singh et al., 2017). Moreover, there are several genotypes available with higher Fe and Zn concentrations that could be important resources for breeding (Khokhar et al., 2020). Such wheat lines can also serve as excellent germplasm for the introgression of this trait in elite varieties via conventional breeding.
Enhancing Grain Fe Bioavailability
The cereals accumulate phytic acid (phytate) in the vacuoles which chelate Fe and its metal ions, thereby drastically reducing its bioavailability. The phytate is thus considered an anti-nutrient and low phytate (lpa) could be another promising strategy for improving Fe accessibility in cereals (García-Estepa et al., 1999; Bhati et al., 2016). The lpa can be attained by repressing or inducing mutation in the genes either responsible for PA biosynthesis or transport to seed. The previous study of our research group identified putative PA-related genes, such as TaIPK1, which catalyzes the last step of PA biosynthesis and TaABCC13 in wheat (Bhati et al., 2014). The silencing of TaIPK1 using RNAi significantly lowered PA production and led to an increased Fe and Zn content in wheat grain (Bhati et al., 2016; Aggarwal et al., 2018). However, pleiotropic developmental changes, including low crop yield, were observed in wheat RNAi lines. Moreover, Abid et al. (2017) have used phytase gene from Aspergillus japonicus for the degradation of stored PA in endosperm leading to enhanced bioavailability of both Fe and Zn in wheat (Abid et al., 2017). More recently, using the genome editing-based tool, TaIPK1 was chosen as a target (Ibrahim et al., 2021). The indel mutations increased the accumulation of both Fe and Zn contents in genome-edited Borlaug 2016 wheat variety wheat compared to the non-edited. Both these studies highlight the IPK1 gene target as a major factor contributing to the bioavailability of Fe in cereal crops.
Furthermore, the constitutive overexpression of the rice OsNAS2 gene resulted in an increased Fe and Zn content in wheat endosperm. Moreover, the same study also showed an enhanced bioavailability of both metals (Beasley et al., 2019). Recent studies suggest that Fe-bioavailability and its absorption are independent of the enhanced Fe-content in grains. The foremost reason for this is that even though Fe content in the whole seed is increased, it is stored in the aleurone layer which gets removed while milling, leaving only the endosperm for consumption. Therefore, both Fe content and bioavailability should be taken into account while addressing the biofortification of wheat (Eagling et al., 2014; Wright et al., 2021). To further address this, the wheat genome was studied for the identification of the QTLs that could be responsible for both high Fe-content and bioavailability (Wright et al., 2021). Very recently, a two-gene strategy to overexpress VIT-NAS in wheat endosperm showed high Fe concentration and also improves mineral bioaccessibility (Harrington et al., 2022). Such innovative approaches directed at content and its bioavailability is a feasible approach to generate nutrition-rich grains. Our insight about genes and rate-limiting steps of Fe homeostasis in wheat is increasing day by day. The recent characterization of TaYSL and TaZIFL/TaTOM genes has filled the knowledge gap of the Fe-transport mechanism in wheat (Kumar et al., 2019; Sharma et al., 2019). In addition, transcriptional activators, namely NAM-B1 and GPCs have been found to control Fe and Zn remobilization to sink organs during senescence and grain filling (Uauy et al., 2006; Pearce et al., 2014). We assume that these genes can be potential candidates for gene manipulation/editing efforts either independently or via multiplexing to meet the ultimate goal of Fe enriched wheat grain in the future.
Challenges of Fe Enrichment in Wheat Grain and Integrating Novel Approaches
Being the second most produced cereals in the world, wheat can be the most promising crop for Fe fortification to combat the hidden hunger caused by Fe deficiency in human beings. In the past few years, some efforts have been made in this direction; however, limited success is achieved using genetic engineering. It is noteworthy to mention here that the use of orthologous genes is not sufficient to mobilize Fe in wheat grain up to a certain level. In this section, we describe the main hurdles of increasing Fe content in wheat grain via transgenic technology and discuss the future strategies to address limitations as well.
Loading in the Grains: The Real Bottleneck of Fe Fortification
The main bottleneck of Fe fortification using the transgenic approach appears to be the lack of molecular details of its homeostasis and transcriptional regulators of Fe mobilization in wheat. Upon uptake, micronutrients enter into the root xylem and reach the spikelets where it gets discontinued at the base of the grain (Zee and O’Brien, 1970). At this site, phloem takes a central role to mobilize the macro-and micronutrients by providing passage for unloading into the grain. Studies exhibited that NA is required for phloem unloading and loss of function mutant of NAS led to sterility whereas RNAi lines of NAS resulted in low nutrients in Arabidopsis seeds (Klatte et al., 2009; Schuler et al., 2012).
There is an almost complete lack of knowledge about genes, transporters, and transcriptional activators responsible for the root to shoot distribution (xylem loading), phloem loading and unloading of Fe at the time of grain filling in wheat (Figure 3). It should be noted that transfer cells (TCs) play a central role in nutrient distribution by facilitating high rates of transport for apo-/symplastic solute exchange during nutrient loading in phloem and unloading in the endosperm. The TCs found near the endosperm, are specialized cells having unique wall architecture with abundant transporters to enhance membrane transport capacity (Offler et al., 2003). The mechanical removal of TCs reduced in vitro photo-assimilate transport rates suggesting that they are responsible for 70–80% of sucrose imported by the filial tissues in wheat (Wang et al., 1995). TCs are also present near fine veins in leaf and nodes where phloem loading takes place and at the interface of maternal and filial tissues which form symplastic solute transport routes in connection with vascular conduits. Even though all imported solutes, including Fe and Zn, have to be transported through TCs in the endosperm cavity, there is a complete lack of understanding about the mode of transport and genes in TCs.
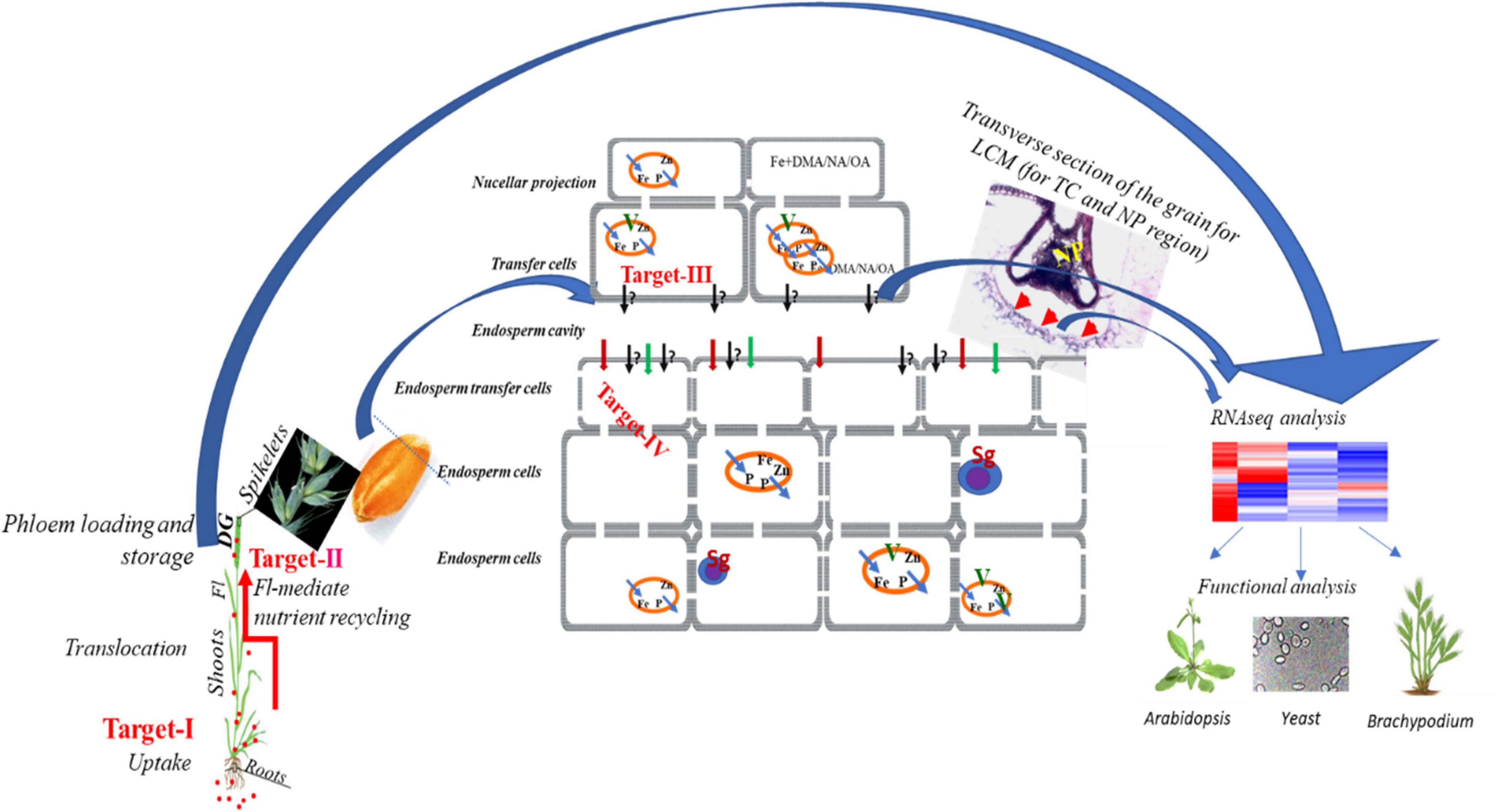
Figure 3. Schematic representation of the primary bottleneck for Fe acquisition, uptake and mobilization in wheat shoot and grain. Multiple target areas have been identified to enhance the mobilization of Fe from soil to roots, from roots to shoots, and subsequently their loading in the grains through specific conducting tissue. The first is referred as Target-I, and Target-II depict the uptake of Fe in roots and then to shoots and eventually loading to the edible grains. In wheat, primarily at this stage, the bottleneck includes the transfer and mobilization of Fe from discontinued xylem to the transfer cells and subsequently to the endodermal cells (Target-III and IV). The target areas Target-III and IV remained unexplored in hexaploid wheat. The area that needs attention is to perform grain or Tissue or single cell-specific-omics approach that will help in identifying key transporters and regulators. These identified key genes could be tested for their functional validation in yeast and subsequent proof-of-function in Arabidopsis, Brachypodium, or in wheat. The red arrow indicates the process in wheat, and the blue arrow indicates the means to further characterize wheat genes in different model species.
Exploration of Hitherto Regulators of Fe Homeostasis in Wheat
Recent studies demonstrated that stem nodes in the graminaceous species are important transient sites for the mobilization of micronutrients, including Fe and Zn, to the above-ground organs (Yamaji and Ma, 2014). The genes encoding for plasma membrane-localized efflux transporter for citrate Ferric Reductase Defective like-1 (OsFRDL1) and a yellow stripe-like transporter (OsYSL) were specifically shown to be expressed in nodes, and demonstrated to be important for the distribution of Fe and Cu in rice grains (Zheng et al., 2012; Yokosho et al., 2016). It has been shown that Arabidopsis Ferric Reductase Defective-3 (AtFRD3) maintains citrate levels in the xylem of Arabidopsis, and mutation in this gene caused an accumulation of Fe in roots (Durrett et al., 2007). However, a distinct anatomical tissue arrangement has been reported in wheat, and multiple modes of efficient distribution of mineral elements have been speculated but the transporters and regulators involved at specific sites are largely unaddressed (Yamaji and Ma, 2014). Studies illustrated that the heterologous expression of Triticum polonicum genes namely TpSnRK2.10 and TpSnRK2.11 increased Fe accumulation in roots and influenced Fe distribution in Arabidopsis (Wang R. et al., 2019). Later studies reported the accumulation of IRT1 transcripts occurred in SnRK2.2/2.3 dependent manner in Arabidopsis (Fan et al., 2014). Furthermore, the plant hormone gibberellic acid (GA) is known to control the expression of IRT1 and FRO2 via DELLA protein in Arabidopsis (Matsuoka et al., 2014). The DELLA protein interacts with transcriptional activators, such as FIT, bHLH38, and bHLH39, and limits their transcriptional activities (Wild et al., 2016). The abundance of DELLA is regulated by Fe deficiency which ultimately favors Fe acquisition in roots. It should be worthy to mention here that semi-dwarf alleles of wheat were preferred during the green revolution and modern wheat varieties have been developed showing improved harvest index and resistance to water lodging. In general, modern wheat varieties are less sensitive to GA and contain less Fe in grain as well. Now, it is still unclear whether GA insensitivity and DELLA accumulation may be a reason for the low Fe in grains of commonly growing semi-dwarf modern wheat varieties. Furthermore, Fe interaction with other nutrients is a well-recognized phenomenon in soil for ensuring the bio-availability of Fe to plants which have been described in detail in previous sections of this review. The presence of excess/limited nutrients in the rhizosphere challenges the plant to integrate nutrient signals in the form of genetic shift which finally changes root architecture. The studies have shown that gaseous hormone ethylene is extremely required for the maintenance of primary and lateral root growth as well as Fe homeostasis in Arabidopsis (García et al., 2015; Li et al., 2015). Hence, focusing on unconventional hitherto regulators of Fe, such as SnRK, DELLA, components of ethylene signaling and others can boost our endeavor for not only Fe enrichment perspective but also enhanced crop productivity in the Fe deficient soil.
In this line, we describe four research directions that could be emphasized viz. Fe uptake, long-distance transport, remobilization, and unloading of Fe in developing wheat grains for dealing with Fe fortification. The schematic representation of these four main target areas has been illustrated in Figure 3. Targets I and II have been proposed to utilize a strategy wherein overexpression of genes encoding for NAS, NAAT, DMAS, TOM, and YSL could lead to an increase in Fe uptake by enhancing the PS production and release in soil. Next, the collective candidates of Target III and IV cover Fe transport from flag leaf to developing grains. The identification and characterization of transporters and TFs especially comprising Targets III and IV would provide molecular assets to be useful for Fe accumulation through genetic engineering or molecular breeding. Therefore, a concerted effort will be required to identify TCs/tissue-specific genes to cross the bottleneck, and which will serve as potential targets for developing wheat varieties having more Fe in grain.
Single-Cell RNA Sequencing
The evolution of high-throughput sequencing techniques is going very fast in recent years. The establishment of single-cell RNA sequencing (scRNA-seq) tools revolutionized the molecular studies of developmental processes in any organism. The single-cell transcriptome analysis is very helpful for the spatio-temporal characterization of genes and gene regulatory networks recruited in specific developmental trajectories. Nevertheless, the isolation of single-cell from plant tissues is the most crucial step and is a major constraint for the wide application of scRNA-seq analysis in crops. The technical advancements in this area, such as Drop-seq and InDrop-seq, will gear the implementation of scRNA-seq analysis in plant research. In recent years, scRNA-seq analysis has been successfully implemented to decipher cell type-specific information, for instance, developmental programs leading to meiosis in maize (Nelms and Walbot, 2019) and dynamics of gene expression in the root cells of Arabidopsis (Jean-Baptiste et al., 2019). We believe that scRNA-seq analysis of cell files involved in vascular loading and unloading of Fe in sink organs at the time of grain filling stage in wheat can be an ideal way to investigate the gene regulatory network of Fe mobilization. Furthermore, nucellar projection and TCs are the two main cells through which Fe has to pass before loading into the endosperm. The genes that facilitate Fe loading from these cells are still unknown not only in wheat but in model plants as well. Thus, sc RNA-Seq study either under varying Fe conditions or in contrasting Fe accumulating wheat varieties can potentially reveal these missing molecular links that will help in exploring new candidate genes to enhance Fe loading into seed.
Genome Expression Bias and Fe Homeostasis in Wheat
The modern wheat contains hexaploid genome (2n = 6x = 42) with much complex genome organization. Hence, the investigation of mechanisms, transcriptional regulators, and genes involved in Fe uptake and homeostasis in wheat seems to be a tough task. The wheat genome is derived from three related ancestral genomes from the genera Aegilops and Triticum. It is of high interest to understand how individual genomes interact with each other for determining the expression of highly related genes in wheat. The phenomenon of genome biasness has been reported earlier in polyploid plants, such as cotton and soybean (Roulin et al., 2013; Yoo et al., 2013). A homeolog-specific differential regulation of genes controlling growth and vigor was also observed in wheat (Akhunova et al., 2010). Subsequent studies pointed the homeolog induction bias under Fusarium infection in wheat (Powell et al., 2017). A large expression landscape study was recently conducted suggesting the existence of a tissue-specific co-expression network as well as coordination of homeologs during different developmental stages (Ramírez-González et al., 2018). Such induction bias is not only restricted to growth or biotic stresses but rather plays a significant role during Fe deficiency response in wheat (Kaur et al., 2019). Homeolog induction bias was observed under Fe deficiency for A and B genomes in wheat. Genes contributing toward induction bias comprises metal transporters, Zn transporters, and MYB transcription factors. Furthermore, it has been shown that Fe deficiency response, such as PS release, was more in tetraploid wheat with AABB sub-genome compared to DD genome (Ma et al., 1999). This explains why hexaploid wheat releases high PS simply due to retaining AABB genome biasness. The factors influencing such homeolog induction bias and genome bias remain largely unaddressed. A recent study demonstrates that varying chromatin accessibility of homeologs might be the reason for such genome bias in wheat (Jordan et al., 2020). Therefore, it will be interesting to pinpoint spatio-temporal genome biasness invoked due to Fe deficiency in wheat.
Conclusion and Future Perspectives
Improving the nutritional quality of cereals is of prime importance and yet challenging. The Fe biofortification in wheat has been an arduous task for many decades with limited success. Therefore, the means to design better strategies for improving Fe uptake by the roots, genetic screening of germplasm showing tolerance under limiting Fe conditions, and combining approaches of classical physiology with the omics data-driven system biological studies will certainly help in undermining the limiting steps involved in Fe remobilization into wheat grain. Also, homeolog specific expression analysis of Fe-regulated genes has revealed the expression biasness for A, B, and D subgenomes (Kaur et al., 2019). These studies have streamlined the selection of candidate gene/s and/or homeologs and also made it much more efficient and rational. The genetic engineering tools, genomic sequence information, gene mining, and functional genomics would help to carry forward the research for developing novel strategies of micronutrient enhancement in wheat. Moreover, controlling the transcriptional regulation of genes through gene editing could be an alternate strategy for trait improvement. Advancements in CRISPR/Cas9 technology would help to know gene expression regulation in a better way and provide an additional tool. Keeping in mind that excessive Fe concentration causes oxidative stress in plants, future attempts for the modulation of Fe homeostasis may consider both negative and positive regulators, including new players, such as IRO3, HRZ, and IMA, in wheat (Kobayashi et al., 2013; Grillet et al., 2018). Moreover, to control excessive Fe loading into the plant, the gene expressions can be fine-tuned with the use of CRISPRi and/or CRISPRa. For this, several vectors and toolkits with various monocot-specific strong promoters and gene regulatory elements (activators/repressors) are available in the Addgene repository (Kamens, 2015). The expansion of wheat genomic resources (Borrill et al., 2016; Appels et al., 2018; Ramírez-González et al., 2018) along with the modern gene-editing tools via CRISPR/Cas9 could offer a reliable potential to achieve Fe-rich wheat grains.
Author Contributions
AP and PB conceived and designed the review article. AK, PS, and MT contributed and collated information for the review article. GK, PS, SS, AP, and VM designed the figure and wrote the initial draft. GK, MT, and PB contributed for Tables. AP, MT, and PB edited the manuscript. All authors agreed with the final version.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC-2048/1 – project ID 390686111, Department of Biotechnology (Government of India) for extramural funding and NABI-CORE grants to AP and funding assistance in the form of Ramalingaswami Fellowship (Grant No. BT/HRD/35/02/2006) to MT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Executive Director, NABI, for facilities and support. We would also like to thank all the previous lab members for their contribution. DBT-eLibrary Consortium (DeLCON) is acknowledged for providing support and access to e-resources for this work.
References
Abid, N., Khatoon, A., Maqbool, A., Irfan, M., Bashir, A., Asif, I., et al. (2017). Transgenic expression of phytase in wheat endosperm increases bioavailability of iron and zinc in grains. Transgenic Res. 26, 109–122. doi: 10.1007/s11248-016-9983-z
Aggarwal, S., Kumar, A., Bhati, K. K., Kaur, G., Shukla, V., Tiwari, S., et al. (2018). RNAi-mediated downregulation of inositol pentakisphosphate kinase (IPK1) in wheat grains decreases phytic acid levels and increases Fe and Zn accumulation. Front. Plant Sci. 9:259. doi: 10.3389/fpls.2018.00259
Akhunova, A. R., Matniyazov, R. T., Liang, H., and Akhunov, E. D. (2010). Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genomics 11:505. doi: 10.1186/1471-2164-11-505
Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., Stein, N., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science (80) 361:eaar7191. doi: 10.1126/science.aar7191
Astolfi, S., Pii, Y., Terzano, R., Mimmo, T., Celletti, S., Allegretta, I., et al. (2018). Does Fe accumulation in durum wheat seeds benefit from improved whole-plant sulfur nutrition? J. Cereal Sci. 83, 74–82. doi: 10.1016/j.jcs.2018.07.010
Astolfi, S., Zuchi, S., Passera, C., and Cesco, S. (2003). Does the sulfur assimilation pathway play a role in the response to Fe deficiency in maize (Zea mays L.) plants. J. Plant Nutr. 26, 2111–2121. doi: 10.1081/PLN-120024268
Banakar, R., Alvarez Fernández, A., Abadía, J., Capell, T., and Christou, P. (2017). The expression of heterologous Fe (III) phytosiderophore transporter HvYS1 in rice increases Fe uptake, translocation and seed loading and excludes heavy metals by selective Fe transport. Plant Biotechnol. J. 15, 423–432. doi: 10.1111/pbi.12637
Bashir, K., Inoue, H., Nagasaka, S., Takahashi, M., Nakanishi, H., Mori, S., et al. (2006). Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J. Biol. Chem. 281, 32395–32402. doi: 10.1074/jbc.M604133200
Bashir, K., Nozoye, T., Nagasaka, S., Rasheed, S., Miyauchi, N., Seki, M., et al. (2017). Paralogs and mutants show that one DMA synthase functions in iron homeostasis in rice. J. Exp. Bot. 68, 1785–1795. doi: 10.1093/jxb/erx065
Beasley, J. T., Bonneau, J. P., and Johnson, A. A. T. (2017). Characterisation of the nicotianamine aminotransferase and deoxymugineic acid synthase genes essential to Strategy II iron uptake in bread wheat (Triticum aestivum L.). PLoS One 12:e0177061. doi: 10.1371/journal.pone.0177061
Beasley, J. T., Bonneau, J. P., Sánchez-Palacios, J. T., Moreno-Moyano, L. T., Callahan, D. L., Tako, E., et al. (2019). Metabolic engineering of bread wheat improves grain iron concentration and bioavailability. Plant Biotechnol. J. 17, 1514–1526. doi: 10.1111/pbi.13074
Bhati, K. K., Aggarwal, S., Sharma, S., Mantri, S., Singh, S. P., Bhalla, S., et al. (2014). Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticum aestivum L.). Plant Sci. 224, 74–85. doi: 10.1016/j.plantsci.2014.04.009
Bhati, K. K., Alok, A., Kumar, A., Kaur, J., Tiwari, S., and Pandey, A. K. (2016). Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 67, 4379–4389. doi: 10.1093/jxb/erw224
Bonneau, J., Baumann, U., Beasley, J., Li, Y., and Johnson, A. A. T. (2016). Identification and molecular characterization of the nicotianamine synthase gene family in bread wheat. Plant Biotechnol. J. 14, 2228–2239. doi: 10.1111/pbi.12577
Boonyaves, K., Gruissem, W., and Bhullar, N. K. (2016). NOD promoter-controlled AtIRT1 expression functions synergistically with NAS and FERRITIN genes to increase iron in rice grains. Plant Mol. Biol. 90, 207–215. doi: 10.1007/s11103-015-0404-0
Borg, S., Brinch-Pedersen, H., Tauris, B., Madsen, L. H., Darbani, B., Noeparvar, S., et al. (2012). Wheat ferritins: improving the iron content of the wheat grain. J. Cereal Sci. 56, 204–213. doi: 10.1016/j.jcs.2012.03.005
Borrill, P., Ramirez-Gonzalez, R., and Uauy, C. (2016). expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol. 170, 2172–2186. doi: 10.1104/pp.15.01667
Bournier, M., Tissot, N., Mari, S., Boucherez, J., Lacombe, E., Briat, J. F., et al. (2013). Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J. Biol. Chem. 288, 22670–22680. doi: 10.1074/jbc.M113.482281
Chattha, M. U., Hassan, M. U., Khan, I., Chattha, M. B., Mahmood, A., Nawaz, M., et al. (2017). Biofortification of wheat cultivars to combat zinc deficiency. Front. Plant Sci. 8:281. doi: 10.3389/fpls.2017.00281
Cheng, L., Wang, F., Shou, H., Huang, F., Zheng, L., He, F., et al. (2007). Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol. 145, 1647–1657. doi: 10.1104/pp.107.107912
Ciaffi, M., Paolacci, A. R., Celletti, S., Catarcione, G., Kopriva, S., and Astolfi, S. (2013). Transcriptional and physiological changes in the S assimilation pathway due to single or combined S and Fe deprivation in durum wheat (Triticum durum L.) seedlings. J. Exp. Bot. 64, 1663–1675. doi: 10.1093/jxb/ert027
Colangelo, E. P., and Guerinot, M. L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16, 3400–3412. doi: 10.1105/tpc.104.024315
Connorton, J. M., Jones, E. R., Rodríguez-Ramiro, I., Fairweather-Tait, S., Uauy, C., and Balk, J. (2017). Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol. 174, 2434–2444. doi: 10.1104/pp.17.00672
Couturier, J., Touraine, B., Briat, J. F., Gaymard, F., and Rouhier, N. (2013). The iron-sulfur cluster assembly machineries in plants: current knowledge and open questions. Front. Plant Sci. 4:259. doi: 10.3389/fpls.2013.00259
Cui, Y., Chen, C. L., Cui, M., Zhou, W. J., Wu, H. L., and Ling, H. Q. (2018). Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in arabidopsis. Mol. Plant 11, 1166–1183. doi: 10.1016/j.molp.2018.06.005
Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S. L., Briat, J. F., and Walker, E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349. doi: 10.1038/35053080
Díaz-Benito, P., Banakar, R., Rodríguez-Menéndez, S., Capell, T., Pereiro, R., Christou, P., et al. (2018). Iron and zinc in the embryo and endosperm of rice (oryza sativa l.) seeds in contrasting 2’-deoxymugineic acid/nicotianamine scenarios. Front. Plant Sci. 9:1190. doi: 10.3389/fpls.2018.01190
Drakakaki, G., Christou, P., and Stöger, E. (2000). Constitutive expression of soybean ferritin cDNA in transgenic wheat and rice results in increased iron levels in vegetative tissues but not in seeds. Transgenic Res. 9, 445–452. doi: 10.1023/A:1026534009483
Durrett, T. P., Gassmann, W., and Rogers, E. E. (2007). The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 144, 197–205. doi: 10.1104/pp.107.097162
Eagling, T., Neal, A. L., McGrath, S. P., Fairweather-Tait, S., Shewry, P. R., and Zhao, F. J. (2014). Distribution and speciation of iron and zinc in grain of two wheat genotypes. J. Agric. Food Chem. 62, 708–716. doi: 10.1021/JF403331P/SUPPL_FILE/JF403331P_SI_001.PDF
Eide, D., Broderius, M., Fett, J., and Guerinot, M. L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 5624–5628. doi: 10.1073/pnas.93.11.5624
Fan, S. K., Fang, X. Z., Guan, M. Y., Ye, Y. Q., Lin, X. Y., Du, S. T., et al. (2014). Exogenous abscisic acid application decreases cadmium accumulation in arabidopsis plants, which is associated with the inhibition of IRT1-mediated cadmium uptake. Front. Plant Sci. 5:721. doi: 10.3389/fpls.2014.00721
Gao, F., Robe, K., Bettembourg, M., Navarro, N., Rofidal, V., Santoni, V., et al. (2020). The transcription factor bHLH121 interacts with bHLH105 (ILR3) and its closest homologs to regulate iron homeostasis in Arabidopsis. Plant Cell 32, 508–524. doi: 10.1105/tpc.19.00541
García, M. J., Romera, F. J., Lucena, C., Alcántara, E., and Pérez-Vicente, R. (2015). Ethylene and the regulation of physiological and morphological responses to nutrient deficiencies. Plant Physiol. 169, 51–60. doi: 10.1104/pp.15.00708
García-Estepa, R. M., Guerra-Hernández, E., and García-Villanova, B. (1999). Phytic acid content in milled cereal products and breads. Food Res. Int. 32, 217–221. doi: 10.1016/S0963-9969(99)00092-7
Garnica, M., Bacaicoa, E., Mora, V., San Francisco, S., Baigorri, R., Zamarreño, A. M., et al. (2018). Shoot iron status and auxin are involved in iron deficiency-induced phytosiderophores release in wheat. BMC Plant Biol. 18:105. doi: 10.1186/s12870-018-1324-3
Goto, F., Yoshihara, T., Shigemoto, N., Toki, S., and Takaiwa, F. (1999). Iron fortification of rice seed by the soybean ferritin gene. Nat. Biotechnol. 17, 282–286. doi: 10.1038/7029
Grillet, L., Lan, P., Li, W., Mokkapati, G., and Schmidt, W. (2018). IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat. Plants 4, 953–963. doi: 10.1038/s41477-018-0266-y
Gross, J., Stein, R. J. Fett-Neto, A. G., and Fett, J. P. (2003). Iron homeostasis related genes in rice. Genet. Mol. Biol. 26, 477–497. doi: 10.1590/S1415-47572003000400012
Harrington, S. A., Connorton, J. M., Nyangoma, N. I. M., McNelly, R., Morgan, Y. M. L., Aslam, M. F., et al. (2022). A two-gene strategy increases the iron and zinc concentration of wheat flour and improves mineral bioaccesibility for human nutrition. bioRxiv [Preprint] 2022.02.15.480518. doi: 10.1101/2022.02.15.480518
Higuchi, K., Suzuki, K., Nakanishi, H., Yamaguchi, H., Nishizawa, N. K., and Mori, S. (1999). Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 119, 471–479. doi: 10.1104/pp.119.2.471
Hirsch, J., Marin, E., Floriani, M., Chiarenza, S., Richaud, P., Nussaume, L., et al. (2006). Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88, 1767–1771. doi: 10.1016/j.biochi.2006.05.007
Ibrahim, S., Saleem, B., Rehman, N., Zafar, S. A., Naeem, M. K., and Khan, M. R. (2021). CRISPR/Cas9 mediated disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J. Adv. Res. 37, 33–41. doi: 10.1016/j.jare.2021.07.006
Inoue, H., Higuchi, K., Takahashi, M., Nakanishi, H., Mori, S., and Nishizawa, N. K. (2003). Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J. 36, 366–381. doi: 10.1046/j.1365-313X.2003.01878.x
Ishimaru, Y., Masuda, H., Bashir, K., Inoue, H., Tsukamoto, T., Takahashi, M., et al. (2010). Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 62, 379–390. doi: 10.1111/j.1365-313X.2010.04158.x
Ishimaru, Y., Suzuki, M., Tsukamoto, T., Suzuki, K., Nakazono, M., Kobayashi, T., et al. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45, 335–346. doi: 10.1111/j.1365-313X.2005.02624.x
Jakoby, M., Wang, H. Y., Reidt, W., Weisshaar, B., and Bauer, P. (2004). FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 577, 528–534. doi: 10.1016/j.febslet.2004.10.062
Jean-Baptiste, K., McFaline-Figueroa, J. L., Alexandre, C. M., Dorrity, M. W., Saunders, L., Bubb, K. L., et al. (2019). Dynamics of gene expression in single root cells of arabidopsis thaliana. Plant Cell 31, 993–1011. doi: 10.1105/tpc.18.00785
Jordan, K. W., He, F., De Soto, M. F., Akhunova, A., and Akhunov, E. (2020). Differential chromatin accessibility landscape reveals structural and functional features of the allopolyploid wheat chromosomes. Genome Biol. 21:176. doi: 10.1186/s13059-020-02093-1
Kamens, J. (2015). The Addgene repository: an international nonprofit plasmid and data resource. Nucleic Acids Res. 43, D1152–D1157. doi: 10.1093/nar/gku893
Kaur, G., Shukla, V., Kumar, A., Kaur, M., Goel, P., Singh, P., et al. (2019). Integrative analysis of hexaploid wheat roots identifies signature components during iron starvation. J. Exp. Bot. 70, 6141–6161. doi: 10.1093/jxb/erz358
Kaur, G., Shukla, V., Meena, V., Kumar, A., Tyagi, D., Singh, J., et al. (2021). Physiological and molecular responses to combinatorial iron and phosphate deficiencies in hexaploid wheat seedlings. Genomics 113, 3935–3950. doi: 10.1016/j.ygeno.2021.09.019
Khokhar, J. S., King, J., King, I. P., Young, S. D., Foulkes, M. J., De Silva, J., et al. (2020). Novel sources of variation in grain Zinc (Zn) concentration in bread wheat germplasm derived from Watkins landraces. PLoS One 15:e0229107. doi: 10.1371/JOURNAL.PONE.0229107
Kim, S. A., LaCroix, I. S., Gerber, S. A., and Guerinot, M. L. (2019). The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc. Natl. Acad. Sci. U.S.A. 116, 24933–24942. doi: 10.1073/pnas.1916892116
Kim, S. A., Punshon, T., Lanzirotti, A., Li, A., Alonso, J. M., Ecker, J. R., et al. (2006). Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314, 1295–1298. doi: 10.1126/science.1132563
Klatte, M., Schuler, M., Wirtz, M., Fink-Straube, C., Hell, R., and Bauer, P. (2009). The analysis of arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol. 150, 257–271. doi: 10.1104/pp.109.136374
Kobayashi, T., Nagasaka, S., Senoura, T., Itai, R. N., Nakanishi, H., and Nishizawa, N. K. (2013). Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 4:2792. doi: 10.1038/ncomms3792
Kobayashi, T., and Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. doi: 10.1146/annurev-arplant-042811-105522
Kobayashi, T., Ozu, A., Kobayashi, S., An, G., Jeon, J. S., and Nishizawa, N. K. (2019). OsbHLH058 and OsbHLH059 transcription factors positively regulate iron deficiency responses in rice. Plant Mol. Biol. 101, 471–486. doi: 10.1007/s11103-019-00917-8
Kroh, G. E., and Pilon, M. (2020). Regulation of iron homeostasis and use in chloroplasts. Int. J. Mol. Sci. 21:3395. doi: 10.3390/ijms21093395
Kumar, A., Kaur, G., Goel, P., Bhati, K. K., Kaur, M., Shukla, V., et al. (2019). Genome-wide analysis of oligopeptide transporters and detailed characterization of yellow stripe transporter genes in hexaploid wheat. Funct. Integr. Genomics 19, 75–90. doi: 10.1007/s10142-018-0629-5
Lee, S., and An, G. (2009). Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 32, 408–416. doi: 10.1111/j.1365-3040.2009.01935.x
Lee, S., Chiecko, J. C., Kim, S. A., Walker, E. L., Lee, Y., Guerinot, M. L., et al. (2009a). Disruption of OsYSL15 leads to iron inefficiency in rice plants 1[C][W][OA]. Plant Physiol. 150, 786–800. doi: 10.1104/pp.109.135418
Lee, S., Jeon, U. S., Lee, S. J., Kim, Y. K., Persson, D. P., Husted, S., et al. (2009b). Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc. Natl. Acad. Sci. U.S.A. 106, 22014–22019. doi: 10.1073/pnas.0910950106
Lee, S., Kim, Y. S., Jeon, U. S., Kim, Y. K., Schjoerring, J. K., and An, G. (2012). Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol. Cells 33, 269–275. doi: 10.1007/s10059-012-2231-3
Li, G., Xu, W., Kronzucker, H. J., and Shi, W. (2015). Ethylene is critical to the maintenance of primary root growth and Fe homeostasis under Fe stress in Arabidopsis. J. Exp. Bot. 66, 2041–2054. doi: 10.1093/jxb/erv005
Li, X., Zhang, H., Ai, Q., Liang, G., and Yu, D. (2016). Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in arabidopsis Thaliana. Plant Physiol. 170, 2478–2493. doi: 10.1104/pp.15.01827
Liang, G., Zhang, H., Li, X., Ai, Q., and Yu, D. (2017). BHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 68, 1743–1755. doi: 10.1093/jxb/erx043
Lindsay, W. L. (1991). Iron oxide solubilization by organic matter and its effect on iron availability. Iron Nutr. Interact. Plants 130, 27–34. doi: 10.1007/978-94-011-3294-7_2
Ling, H. Q., Bauer, P., Bereczky, Z., Keller, B., and Ganal, M. (2002). The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc. Natl. Acad. Sci. U.S.A. 99, 13938–13943. doi: 10.1073/pnas.212448699
Long, T. A., Tsukagoshi, H., Busch, W., Lahner, B., Salt, D. E., and Benfey, P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in arabidopsis roots. Plant Cell 22, 2219–2236. doi: 10.1105/tpc.110.074096
Ma, J. F., Taketa, S., Chang, Y. C., Takeda, K., and Matsumoto, H. (1999). Biosynthesis of phytosiderophores in several Triticeae species with different genomes. J. Exp. Bot. 50, 723–726. doi: 10.1093/jxb/50.334.723
Mai, H. J., Lindermayr, C., von Toerne, C., Fink-Straube, C., Durner, J., and Bauer, P. (2015). Iron and FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR-dependent regulation of proteins and genes in Arabidopsis thaliana roots. Proteomics 15, 3030–3047. doi: 10.1002/pmic.201400351
Mai, H. J., Pateyron, S., and Bauer, P. (2016). Iron homeostasis in Arabidopsis thaliana: transcriptomic analyses reveal novel FIT-regulated genes, iron deficiency marker genes and functional gene networks. BMC Plant Biol. 16:211. doi: 10.1186/s12870-016-0899-9
Masuda, H., Ishimaru, Y., Aung, M. S., Kobayashi, T., Kakei, Y., Takahashi, M., et al. (2012). Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition. Sci. Rep. 2:543. doi: 10.1038/srep00543
Matsuoka, K., Furukawa, J., Bidadi, H., Asahina, M., Yamaguchi, S., and Satoh, S. (2014). Gibberellin-induced expression of fe uptake-related genes in arabidopsis. Plant Cell Physiol. 55, 87–98. doi: 10.1093/pcp/pct160
Misson, J., Raghothama, K. G., Jain, A., Jouhet, J., Block, M. A., Bligny, R., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. U.S.A. 102, 11934–11939. doi: 10.1073/pnas.0505266102
Mladìnka, P., Macáková, K., Zatloukalová, L., Øeháková, Z., Singh, B. K., Prasad, A. K., et al. (2010). In vitro interactions of coumarins with iron. Biochimie 92, 1108–1114. doi: 10.1016/j.biochi.2010.03.025
Naranjo-Arcos, M. A., Maurer, F., Meiser, J., Pateyron, S., Fink-Straube, C., and Bauer, P. (2017). Dissection of iron signaling and iron accumulation by overexpression of subgroup Ib bHLH039 protein. Sci. Rep. 7:10911. doi: 10.1038/s41598-017-11171-7
Narayanan, N., Beyene, G., Chauhan, R. D. Gaitán-Solís, E., Gehan, J., Butts, P., et al. (2019). Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin. Nat. Biotechnol. 37, 144–151. doi: 10.1038/s41587-018-0002-1
Nelms, B., and Walbot, V. (2019). Defining the developmental program leading to meiosis in maize. Science (80) 364, 52–56. doi: 10.1126/science.aav6428
Nozoye, T., Itai, R. N., Nagasaka, S., Takahashi, M., Nakanishi, H., Mori, S., et al. (2004). Diurnal changes in the expression of genes that participate im phytosiderophore synthesis in rice. Soil Sci. Plant Nutr. 50, 1125–1131. doi: 10.1080/00380768.2004.10408585
Nozoye, T., Nagasaka, S., Kobayashi, T., Sato, Y., Uozumi, N., Nakanishi, H., et al. (2015). The phytosiderophore efflux transporter TOM2 is involved in metal transport in Rice. J. Biol. Chem. 290, 27688–27699. doi: 10.1074/jbc.M114.635193
Nozoye, T., Nagasaka, S., Kobayashi, T., Takahashi, M., Sato, Y., Sato, Y., et al. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286, 5446–5454. doi: 10.1074/jbc.M110.180026
Offler, C. E., McCurdy, D. W., Patrick, J. W., and Talbot, M. J. (2003). Transfer cells: cells specialized for a special purpose. Annu. Rev. Plant Biol. 54, 431–454. doi: 10.1146/annurev.arplant.54.031902.134812
Ogo, Y., Itai, R. N., Kobayashi, T., Aung, M. S., Nakanishi, H., and Nishizawa, N. K. (2011). OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 75, 593–605. doi: 10.1007/s11103-011-9752-6
Ogo, Y., Nakanishi Itai, R., Nakanishi, H., Kobayashi, T., Takahashi, M., Mori, S., et al. (2007). The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 51, 366–377. doi: 10.1111/j.1365-313X.2007.03149.x
Oliva, N., Chadha-Mohanty, P., Poletti, S., Abrigo, E., Atienza, G., Torrizo, L., et al. (2014). Large-scale production and evaluation of marker-free indica rice IR64 expressing phytoferritin genes. Mol. Breed. 33, 23–37. doi: 10.1007/s11032-013-9931-z
Pearce, S., Tabbita, F., Cantu, D., Buffalo, V., Avni, R., Vazquez-Gross, H., et al. (2014). Regulation of Zn and Fe transporters by the GPC1 gene during early wheat monocarpic senescence. BMC Plant Biol. 14:368. doi: 10.1186/s12870-014-0368-2
Petit, J. M., Van Wuytswinkel, O., Briat, J. F., and Lobréaux, S. (2001). Characterization of an iron-dependent regulatory sequence involved in the transcriptional control of AtFer1 and ZmFer1 plant ferritin genes by iron. J. Biol. Chem. 276, 5584–5590. doi: 10.1074/jbc.M005903200
Pires, N., and Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. doi: 10.1093/molbev/msp288
Powell, J. J., Fitzgerald, T. L., Stiller, J., Berkman, P. J., Gardiner, D. M., Manners, J. M., et al. (2017). The defence-associated transcriptome of hexaploid wheat displays homoeolog expression and induction bias. Plant Biotechnol. J. 15, 533–543. doi: 10.1111/pbi.12651
Qu, L. Q., Yoshihara, T., Ooyama, A., Goto, F., and Takaiwa, F. (2005). Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 222, 225–233. doi: 10.1007/s00425-005-1530-8
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science (80) 361:eaar6089. doi: 10.1126/science.aar6089
Rodríguez-Celma, J., Lin, W. D., Fu, G. M., Abadía, J., López-Millán, A. F., and Schmidt, W. (2013). Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by arabidopsis and Medicago truncatula. Plant Physiol. 162, 1473–1485. doi: 10.1104/pp.113.220426
Roulin, A., Auer, P. L., Libault, M., Schlueter, J., Farmer, A., May, G., et al. (2013). The fate of duplicated genes in a polyploid plant genome. Plant J. 73, 143–153. doi: 10.1111/tpj.12026
Sazawal, S., Dhingra, U., Dhingra, P., Dutta, A., Deb, S., Kumar, J., et al. (2018). Efficacy of high zinc biofortified wheat in improvement of micronutrient status, and prevention of morbidity among preschool children and women - A double masked, randomized, controlled trial 11 Medical and Health Sciences 1117 Public Health and Health Se. Nutr. J. 17, 1–10. doi: 10.1186/s12937-018-0391-5
Schmid, N. B., Giehl, R. F. H., Döll, S., Mock, H. P., Strehmel, N., Scheel, D., et al. (2014). Feruloyl-CoA 6’-Hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol. 164, 160–172. doi: 10.1104/pp.113.228544
Schmidt, W., Thomine, S., and Buckhout, T. J. (2020). Editorial: iron nutrition and interactions in plants. Front. Plant Sci. 10:1670. doi: 10.3389/fpls.2019.01670
Schuler, M., Rellán-Álvarez, R., Fink-Straube, C., Abadía, J., and Bauera, P. (2012). Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell 24, 2380–2400. doi: 10.1105/tpc.112.099077
Schwarz, B., Azodi, C. B., Shiu, S. H., and Bauer, P. (2020). Putative cis-regulatory elements predict iron deficiency responses in Arabidopsis roots. Plant Physiol. 182, 1420–1439. doi: 10.1104/PP.19.00760
Schwacke, R. Ponce-Soto, G. Y., Krause, K., Bolger, A. M., Arsova, B., Hallab, A., et al. (2019). MapMan4: a refined protein classification and annotation framework applicable to multi-omics data analysis. Mol. Plant 12, 879–892. doi: 10.1016/j.molp.2019.01.003
Schwarz, B., and Bauer, P. (2020). FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and -independent gene signatures. J. Exp. Bot. 71, 1694–1705. doi: 10.1093/jxb/eraa012
Senoura, T., Sakashita, E., Kobayashi, T., Takahashi, M., Aung, M. S., Masuda, H., et al. (2017). The iron-chelate transporter OsYSL9 plays a role in iron distribution in developing rice grains. Plant Mol. Biol. 95, 375–387. doi: 10.1007/s11103-017-0656-y
Sharma, S., Kaur, G., Kumar, A., Meena, V., Kaur, J., and Pandey, A. K. (2019). Overlapping transcriptional expression response of wheat zinc-induced facilitator-like transporters emphasize important role during Fe and Zn stress. BMC Mol. Biol. 20:22. doi: 10.1186/s12867-019-0139-6
Sharma, S., Kaur, G., Kumar, A., Meena, V., Ram, H., Kaur, J., et al. (2020). Gene expression pattern of vacuolar-iron transporter-like (VTL) genes in hexaploid wheat during metal stress. Plants 9:229. doi: 10.3390/plants9020229
Singh, S. P., Keller, B., Gruissem, W., and Bhullar, N. K. (2017). Rice NICOTIANAMINE SYNTHASE 2 expression improves dietary iron and zinc levels in wheat. Theor. Appl. Genet. 130, 283–292. doi: 10.1007/s00122-016-2808-x
Sisó-Terraza, P., Rios, J. J., Abadía, J., Abadía, A., and Álvarez-Fernández, A. (2016). Flavins secreted by roots of iron-deficient Beta vulgaris enable mining of ferric oxide via reductive mechanisms. New Phytol. 209, 733–745. doi: 10.1111/nph.13633
Sui, X., Zhao, Y., Wang, S., Duan, X., Xu, L., and Liang, R. (2012). Improvement Fe content of wheat(Triticum aestivum) grain by soybean ferritin expression cassette without vector backbone sequence. J. Agric. Biotechnol. 20, 766–773.
Suzuki, K., Higuchi, K., Nakanishi, H., Nishizawa, N. K., and Mori, S. (2012). Cloning of nicotianamine synthase genes from Arabidopsis thaliana. Soil Sci. Plant Nutr. 45, 993–1002. doi: 10.1080/00380768.1999.10414350
Ticconi, C. A., Lucero, R. D., Sakhonwasee, S., Adamson, A. W., Creff, A., Nussaume, L., et al. (2009). ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc. Natl. Acad. Sci. U.S.A. 106, 14174–14179. doi: 10.1073/pnas.0901778106
Tissot, N., Robe, K., Gao, F., Grant-Grant, S., Boucherez, J., Bellegarde, F., et al. (2019). Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytol. 223, 1433–1446. doi: 10.1111/nph.15753
Tolay, I., Erenoglu, B., Römheld, V., Braun, H. J., and Cakmak, I. (2001). Phytosiderophore release in Aegilops tauschii and Triticum species under zinc and iron deficiencies. J. Exp. Bot. 52, 1093–1099. doi: 10.1093/jexbot/52.358.1093
Trofimov, K., Ivanov, R., Eutebach, M., Acaroglu, B., Mohr, I., Bauer, P., et al. (2019). Mobility and localization of the iron deficiency-induced transcription factor bHLH039 change in the presence of FIT. Plant Direct 3:e00190. doi: 10.1002/pld3.190
Uauy, C., Distelfeld, A., Fahima, T., Blechl, A., and Dubcovsky, J. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science (80) 314, 1298–1301. doi: 10.1126/science.1133649
Velu, G., Ortiz-Monasterio, I., Cakmak, I., Hao, Y., and Singh, R. P. (2014). Biofortification strategies to increase grain zinc and iron concentrations in wheat. J. Cereal Sci. 59, 365–372. doi: 10.1016/J.JCS.2013.09.001
Wang, C., Yao, X., Yu, D., and Liang, G. (2017). Fe-deficiency-induced expression of bHLH104 enhances Fe-deficiency tolerance of Arabidopsis thaliana. Planta 246, 421–431. doi: 10.1007/s00425-017-2703-y
Wang, H. L., Offler, C. E., and Patrick, J. W. (1995). The cellular pathway of photosynthate transfer in the developing wheat grain. II. A structural analysis and histochemical studies of the pathway from the crease phloem to the endosperm cavity. Plant. Cell Environ. 18, 373–388. doi: 10.1111/j.1365-3040.1995.tb00373.x
Wang, M., Kawakami, Y., and Bhullar, N. K. (2019). Molecular analysis of iron deficiency response in hexaploid wheat. Front. Sustain. Food Syst. 3:67. doi: 10.3389/fsufs.2019.00067
Wang, R., Wang, C., Yao, Q., Xiao, X., Fan, X., Sha, L., et al. (2019). The polish wheat (Triticum polonicum L.) TpSnRK2.10 and TpSnRK2.11 meditate the accumulation and the distribution of cd and Fe in transgenic Arabidopsis plants. BMC Genomics 20:210. doi: 10.1186/s12864-019-5589-1
Ward, J. T., Lahner, B., Yakubova, E., Salt, D. E., and Raghothama, K. G. (2008). The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 147, 1181–1191. doi: 10.1104/pp.108.118562
Wild, M., Davière, J. M., Regnault, T., Sakvarelidze-Achard, L., Carrera, E., Lopez Diaz, I., et al. (2016). Tissue-specific regulation of gibberellin signaling fine-tunes arabidopsis iron-deficiency responses. Dev. Cell 37, 190–200. doi: 10.1016/j.devcel.2016.03.022
Wright, T. I. C., Gardner, K. A., Glahn, R. P., and Milner, M. J. (2021). Genetic control of iron bioavailability is independent from iron concentration in a diverse winter wheat mapping population. BMC Plant Biol. 21:212. doi: 10.1186/S12870-021-02996-6/TABLES/3
Wu, H., Li, L., Du, J., Yuan, Y., Cheng, X., and Ling, H. Q. (2005). Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol. 46, 1505–1514. doi: 10.1093/pcp/pci163
Wu, T. Y., Gruissem, W., and Bhullar, N. K. (2019). Targeting intracellular transport combined with efficient uptake and storage significantly increases grain iron and zinc levels in rice. Plant Biotechnol. J. 17, 9–20. doi: 10.1111/pbi.12943
Yadavalli, V., Neelam, S., Rao, A. S. V. C., Reddy, A. R., and Subramanyam, R. (2012). Differential degradation of photosystem I subunits under iron deficiency in rice. J. Plant Physiol. 169, 753–759. doi: 10.1016/j.jplph.2012.02.008
Yamaji, N., and Ma, J. F. (2014). The node, a hub for mineral nutrient distribution in graminaceous plants. Trends Plant Sci. 19, 556–563. doi: 10.1016/j.tplants.2014.05.007
Yokosho, K., Yamaji, N., and Ma, J. F. (2016). OsFRDL1 expressed in nodes is required for distribution of iron to grains in rice. J. Exp. Bot. 67, 5485–5494. doi: 10.1093/jxb/erw314
Yoo, M. J., Szadkowski, E., and Wendel, J. F. (2013). Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity (Edinb) 110, 171–180. doi: 10.1038/hdy.2012.94
Yuan, Y. X., Zhang, J., Wang, D. W., and Ling, H. Q. (2005). AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 15, 613–621. doi: 10.1038/sj.cr.7290331
Zee, S. Y., and O’Brien, T. P. (1970). A special type of tracheary element associated with “xylem discontinuity” in the floral axis of wheat. Aust. J. Biol. Sci. 23, 783–792. doi: 10.1071/BI9700783
Zhang, C., Shinwari, K. I., Luo, L., and Zheng, L. (2018). OsYSL13 is involved in iron distribution in rice. Int. J. Mol. Sci. 19:3537. doi: 10.3390/ijms19113537
Zhang, H., Li, Y., Pu, M., Xu, P., Liang, G., and Yu, D. (2020). Oryza sativa POSITIVE REGULATOR OF IRON DEFICIENCY RESPONSE 2 (OsPRI2) and OsPRI3 are involved in the maintenance of Fe homeostasis. Plant Cell Environ. 43, 261–274. doi: 10.1111/pce.13655
Zhang, H., Li, Y., Yao, X., Liang, G., and Yu, D. (2017). POSITIVE REGULATOR OF IRON HOMEOSTASIS1, OsPRI1, facilitates Iron homeostasis. Plant Physiol. 175, 543–554. doi: 10.1104/pp.17.00794
Zhang, Y., Xu, Y. H., Yi, H. Y., and Gong, J. M. (2012). Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 72, 400–410. doi: 10.1111/j.1365-313X.2012.05088.x
Zheng, L., Yamaji, N., Yokosho, K., and Ma, J. F. (2012). YSL16 is a phloem-localized transporter of the copper-nicotianamine complex that is responsible for copper distribution in rice. Plant Cell 24, 3767–3782. doi: 10.1105/tpc.112.103820
Keywords: iron, wheat, transporter, grains, phloem, basic helix loop helix
Citation: Kumar A, Kaur G, Singh P, Meena V, Sharma S, Tiwari M, Bauer P and Pandey AK (2022) Strategies and Bottlenecks in Hexaploid Wheat to Mobilize Soil Iron to Grains. Front. Plant Sci. 13:863849. doi: 10.3389/fpls.2022.863849
Received: 27 January 2022; Accepted: 22 March 2022;
Published: 29 April 2022.
Edited by:
Felipe Klein Ricachenevsky, Federal University of Rio Grande do Sul, BrazilReviewed by:
Matthew John Milner, National Institute of Agricultural Botany (NIAB), United KingdomKaushal Kumar Bhati, Catholic University of Louvain, Belgium
Copyright © 2022 Kumar, Kaur, Singh, Meena, Sharma, Tiwari, Bauer and Pandey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra Bauer, UGV0cmEuQmF1ZXJAaGh1LmRl; Ajay Kumar Pandey, cGFuZGV5YWtAbmFiaS5yZXMuaW4=, cGFuZGV5YWsxOTc0QGdtYWlsLmNvbQ==, orcid.org/0000-0003-1064-139X
 Anil Kumar
Anil Kumar Gazaldeep Kaur
Gazaldeep Kaur Palvinder Singh1
Palvinder Singh1 Manish Tiwari
Manish Tiwari Petra Bauer
Petra Bauer Ajay Kumar Pandey
Ajay Kumar Pandey