
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 26 April 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.863789
This article is part of the Research Topic Reproductive Barriers and Gene Introgression in Rice Species, Volume II View all 14 articles
The narrow base of genetic diversity of modern rice varieties is mainly attributed to the overuse of the common backbone parents that leads to the lack of varied favorable alleles in the process of breeding new varieties. Introgression lines (ILs) developed by a backcross strategy combined with marker-assisted selection (MAS) are powerful prebreeding tools for broadening the genetic base of existing cultivars. They have high power for mapping quantitative trait loci (QTLs) either with major or minor effects, and are used for precisely evaluating the genetic effects of QTLs and detecting the gene-by-gene or gene-by-environment interactions due to their low genetic background noise. ILs developed from multiple donors in a fixed background can be used as an IL platform to identify the best alleles or allele combinations for breeding by design. In the present paper, we reviewed the recent achievements from ILs in rice functional genomics research and breeding, including the genetic dissection of complex traits, identification of elite alleles and background-independent and epistatic QTLs, analysis of genetic interaction, and genetic improvement of single and multiple target traits. We also discussed how to develop ILs for further identification of new elite alleles, and how to utilize IL platforms for rice genetic improvement.
Introgression lines (ILs) are a set of lines constructed by using a combination of continuous backcrossing and selfing to replace chromosome fragments of the recipient parent (RP) with chromosome fragments of the donor parent (DP), which can directly transfer the desired traits of interest from exotic varieties to adapted varieties. IL sets containing overlapping introgression segments covering the entire genome of DP in the RP background are referred to as chromosome segment substitution lines (CSSLs).
The number of backcrosses, number of backcrossed lines in each generation and number of selfings vary in different rice ILs construction programs, depending on the purpose of ILs construction, crossing methodology, genetic distance between parents, compatibility of genotypes, and number or size of target chromosome segment introgressions (Cavanagh et al., 2008; Balakrishnan et al., 2019). The backcrossed progenies can be selected either by phenotyping or by genotyping during the ILs construction, and then are continuously backcrossed with the RP to produce ILs. When screening progenies of backcrosses based on phenotype, if the trait of interest is controlled by a recessive gene, selfing is required before each backcross to identify plants with the recessive allele of interest (Vogel, 2009). With the development of rice functional genomics and molecular markers, marker-assisted selection (MAS) provides a more direct and effective way to obtain the desired rice ILs (Agarwal et al., 2008), and a series of IL/CSSL libraries have been constructed for QTL identification, gene cloning and variety improvement (Zhang et al., 2021c). The rapid development of high-throughput and high-density SNP genotyping technologies has facilitated the precise identification of introgression segments in ILs (Michael, 2014).
ILs are not only powerful prebreeding tools for broadening the genetic base of existing cultivars, but also valuable resources for quantitative trait loci (QTLs) mapping, gene effects evaluation, favorable alleles identification and genetic interactions analysis. The genetic background noise of ILs with a few chromosome-segment substitutions is low, so ILs with significant differences from the recurrent parents can be rapidly identified by evaluating the phenotype of target trait. Although the construction of ILs is time-consuming and labor-intensive, ILs have been widely reported in cereals, oil crops, vegetables, and commercial fiber crops due to their advantages (Balakrishnan et al., 2019). Harlan and Pope (1922) first used the backcrossing to transfer the smooth awn trait to an elite barley cultivar a century ago. Since then, backcross breeding has gradually developed into a widely used crop breeding method, which is often used to enhance crop resistance to disease and insects to ensure high and stable yield. Rice breeding has undergone two important revolutions including semidwarf breeding and heterosis utilization, both of which are based on gene introgression (Rao et al., 2014; Wu et al., 2018).
Cultivated rice, especially Asian rice, mainly consists of two subspecies, namely indica and japonica (Kovach et al., 2007; Wang et al., 2018). The two subspecies are further classified into 9 subpopulations with substantial genetic divergence and geographical distribution differences, including XI-1A, XI-1B, XI-2, XI-3, GJ-trp, GJ-sbtrp, GJ-tmp, cA, and cB (Wang et al., 2018). Inter- and intra-specific genetic diversity in mapping and breeding populations is fundamental. To date, a large number of rice ILs have been developed with distant hybridizations or intraspecific crosses (Balakrishnan et al., 2019; Zhang, 2021). The characteristics and construction processes of ILs/CSSLs were well described in previous reviews (Cavanagh et al., 2008; Ali et al., 2010; Balakrishnan et al., 2019). In this paper, we discussed the importance of parental selection during ILs construction, reviewed the recent achievements from rice ILs in functional genomics and molecular breeding, and proposed how to develop ILs for further exploring novel alleles and facilitating genetic improvement of rice.
The selection of parents is important for ILs construction because genetic differences between parents are the basis of breeding and functional genomics studies using ILs. A traditional backcross scheme for ILs construction with MAS is showed in Figure 1A. Rice varieties with excellent comprehensive traits but defects in one or few traits are often used as RPs for ILs construction with the goal of breeding improvement, and varieties that excel in these traits are selected as DPs. To date, the excellent indica rice varieties, including Huajingxian 74, Zhenshan 97B, 93-11, IR24, and IR64, and the elite japonica rice varieties, including Nipponbare, Koshihikari and Taichung 65, have been used as RPs (Table 1). Of course, these varieties are also used as donors in ILs construction. With the reference genomes of parents, ILs/CSSLs are ideal materials with which to comprehensively dissect the genetic basis of agronomic traits, and any genomic variations that cause phenotypic changes can be verified by these materials. Several sets of CSSLs have been constructed using parents with reference genomes (Table 1), such as CSSLs derived from the crosses between Zhenshan 97B and Nipponbare (Li et al., 2011), between Zhenshan 97B and Minghui 63 (Shen and Xing, 2014), and between 93 and 11 and Nipponbare (Zhu et al., 2009; Yuan et al., 2019).
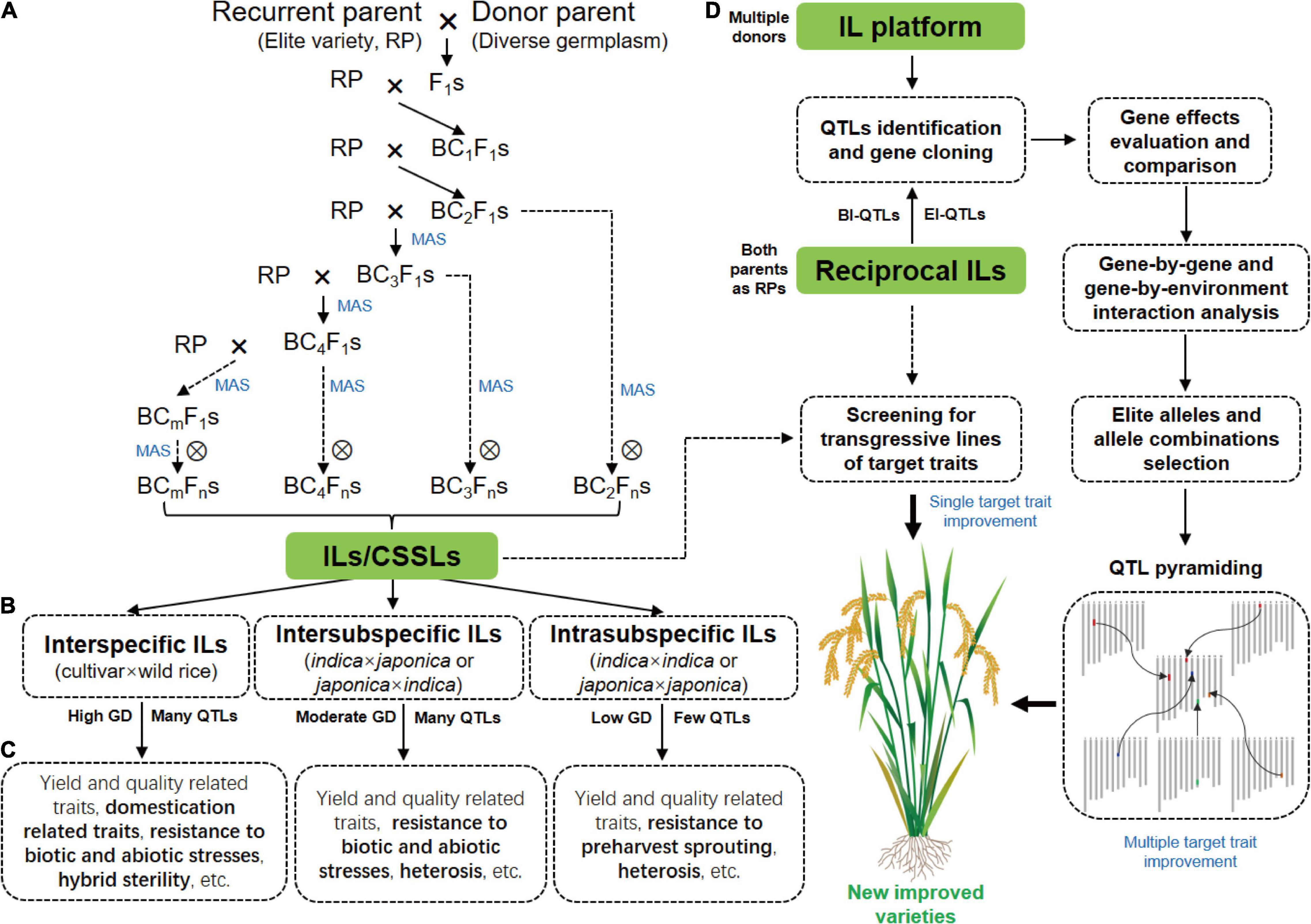
Figure 1. Construction of rice ILs and their application in functional genomics research and breeding. (A) A traditional backcross scheme for ILs/CSSLs development with MAS. (B) ILs with different crossing types. (C) The main traits for genetic dissection by ILs derived from different crosses. The specific traits for the genetic dissection by different ILs are highlighted in bold. (D) The strategy of rice breeding by using ILs or IL platform. GD, genetic diversity; BI-QTLs, background-independent QTLs; E-QTLs, epistatic QTLs.
Wild species of rice contain many useful allelic variations or haplotypes for improving cultivated rice yield and resistance to biotic and abiotic stresses (Kovach et al., 2007). Using wild rice as donors can significantly increase the genetic diversity of cultivars. Although a number of genes from wild species, especially biotic stress resistance genes, have been used in various breeding programs, most of the elite allelic variations in wild rice remains to be exploited (Ali et al., 2010; Li and Zheng, 2013). The genus Oryza is composed of 2 domesticated (O. sativa and O. glaberrima) and 22 wild species (Sanchez et al., 2013). All of six AA-genome wild species and 5 distantly related species with CC, BBCC, CCDD, EE, and FF genomes have been successfully used to construct ILs (Ali et al., 2010; Sanchez et al., 2013; Zhang et al., 2022). To date, more than twenty sets of ILs derived from crosses between wild relatives and cultivated rice have been developed (Ali et al., 2010; Balakrishnan et al., 2019), and some public resource platforms, such as CSSLs created with introgression from four wild rice into the japonica rice variety Taichung 65 (Yamagata et al., 2019) and ILs developed from crosses between two varieties (IR64 and Cybonnet) and three wild accessions (Singh et al., 2020), have been initially established.
Although hybrids between indica and japonica subspecies commonly exhibit varying degrees of hybrid sterility and hybrid breakdown, many researchers have focused on constructing ILs by using indica-japonica crosses to perform genetic dissection and improvement of important agronomic traits due to the large degree of genetic divergence between the two subspecies (Balakrishnan et al., 2019). Japonica accessions experience harsher high-altitude and/or high-latitude environments, compared with the less harsh but more diverse environments experienced by indica rice, resulting in a great difference in agronomic traits between the two subspecies (Wang et al., 2018). Some favorable alleles introgressed between subpopulations, such as the long-grain haplotype of GS3 (Fan et al., 2006), the wide-grain haplotype of qSW5/GW5 (Shomura et al., 2008; Weng et al., 2008), the low-amylose haplotype of Wx (Tian et al., 2009) and the haplotype of OsNRT1.1B with high nutrient use efficiency (Hu et al., 2015), are of great significance for the improvement of rice agronomic traits.
Rice subpopulations originating in specific regions, such as cA and cB, have a larger number of “private” alleles than other subpopulations (Wang et al., 2018). Subpopulation cA encompasses the Aus, Boro and Rayada ecotypes from Bangladesh and India and subpopulation cB comprises the famous Basmati and Sadri aromatic varieties (Wang et al., 2018). The two subpopulations show many elite characteristics, such as the early maturity and drought and heat resistance of Aus varieties and superb fragrance and good cooking quality of aromatic varieties (Civáò et al., 2015). Several Aus varieties, such as Nagina 22 (Lin et al., 2015) and Chuan 7 (Zhang et al., 2021a), have been used as donors to construct ILs, while the aromatic varieties preferred for consumption, such as Khao Dawk Mali 105 (Nounjan et al., 2016; Chutimanukul et al., 2018), Basmati (Grover et al., 2020; Yadav et al., 2020), and Mushk Budji (Khan et al., 2018), are usually used as RPs.
In addition, African cultivated varieties (Oryza glaberrima) with an independent origin of domestication are also a valuable exotic gene pool for Asian cultivated rice and have been used as DPs for IL development (Li et al., 2015). The interspecific upland New Rice for Africa (NERICA) varieties have been developed by introgression of chromosome segments from O. glaberrima (CG 14) into three japonica rice varieties. Blending traits from the African traditional and Asian varieties, NERICA varieties have superior agronomic characteristics such as better weed resistance, resilience against major African biotic and abiotic stresses, high fertilizer returns, and high yields (Jones et al., 1997; Kijima et al., 2006; Fukuta et al., 2012).
ILs constructed by using interspecific and intersubspecific crosses can significantly expand the gene pools of RPs and the large genetic differences between parents also increase the available molecular markers for MAS, which is conducive to mining QTLs. However, the cross incompatibility and F1 hybrid sterility increase the difficulties of interspecific and intersubspecific ILs/CSSLs construction (Ali et al., 2010). In contrast, ILs from intrasubspecific crosses are easier to construct but have lower genetic diversity. A number of intrasubspecific ILs have been developed by using elite indica or japonica varieties as the RPs (Table 1).
ILs are powerful tools with which to dissect complex traits into a set of monogenic loci, and are appropriate for detecting QTLs with both large and small effects (Xi et al., 2006). Over the past three decades, a large number of IL sets derived-from different crosses have been constructed for mining QTLs/genes for agronomic traits (Figures 1B,C and Table 1).
Wild species have acquired many elite alleles for resistance to biotic and abiotic stresses during the long period of natural selection (Atwell et al., 2014; Mammadov et al., 2018). A large number of IL libraries constructed from wild and cultivated rice were used to detect QTLs for cold (Yuan R. et al., 2020), heat (Lei et al., 2013; Prasanth et al., 2016; Cao et al., 2020), drought (Zhang et al., 2006; Zhou et al., 2006), and salt (Tian et al., 2011; Yang et al., 2012) tolerance. For example, a set of 132 CSSLs were developed from a cultivated rice, 93–11, and the Guangxi common wild rice (GXCWR), and the cold-tolerance QTL qCT2.1 on chromosome 2 was identified using the secondary mapping population (Yuan R. et al., 2020). Using a set of 90 ILs derived from the cross between Teqing and Yuanjiang common wild rice (YJCWR), five QTLs related to heat response (qHST1-1, qHST1-2, qHST2, qHST3, and qHST8) in the seedling stage were detected, and O. rufipogon-derived alleles at one locus reduced sensitivity to heat (Lei et al., 2013). QTLs for resistance to biotic stress, such as that imposed by the green rice leafhopper (Thein et al., 2019), rice stripe necrosis virus (Gutiérrez et al., 2010) and rice blast fungus (Xu et al., 2015), were also mapped using ILs carrying wild rice donor fragments in cultivated rice backgrounds.
ILs developed from wild and cultivated rice are valuable materials for the genetic dissection of domestication-related traits such as an erect growth habit, inflorescence architecture, and awn length. Several domestication genes have been cloned using such IL-derived populations (Table 2). PROG1, a prostrate growth gene, was identified in a set of ILs developed from Teqing and YJCWR. Its non-functional allele determines the critical transition of rice from prostrate to erect growth (Tan et al., 2007, 2008). Using the same IL sets, a domestication gene, OsLG1, controlling inflorescence architecture was cloned (Zhu et al., 2013). Using a set of 354 ILs derived from the cross between 93 and 11 and YJCWR, an IL with long and barbed awns was identified. Further fine-mapping indicated that a frameshift deletion in LABA1 of cultivated rice reduces the cytokinin concentration in awn primordia, disrupting barb formation and awn elongation (Fu et al., 2010; Hua et al., 2015). Another domestication gene controlling plant architecture, TIG1, was also cloned in an IL population with the wild rice (W2014) segments in the 93–11 background (Zhang et al., 2019d).
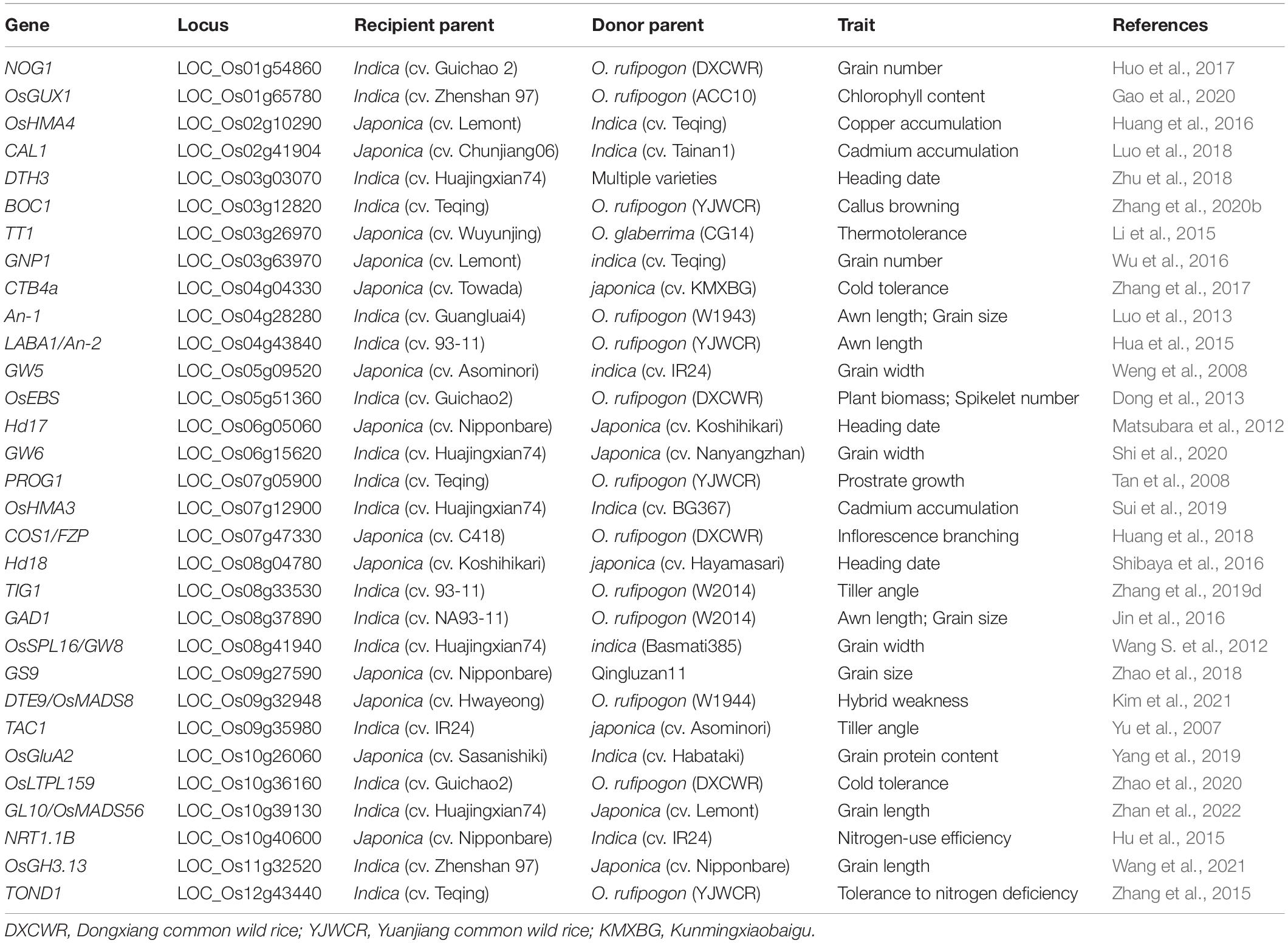
Table 2. List of genes with natural variations identified by ILs and cloned by IL-derived populations in rice.
There has also been a burst of studies on mapping novel genomic regions and QTLs for yield-related traits (Furuta et al., 2014, 2016; Ma et al., 2016; Qiao et al., 2016; Rao et al., 2018) and quality traits (Garcia-Oliveira et al., 2009; Fasahat et al., 2012; Yun et al., 2016; Qi et al., 2017) by using IL populations derived from the crosses between wild and cultivated rice, generally with the elite varieties as RPs. For example, using 33 CSSLs of O. rufipogon (W0106) in the background of the elite japonica rice variety Koshihikari, a total of 15 major QTLs for eight yield-related traits were detected, and a novel QTL controlling the number of grains per panicle were identified on chromosome 10 (Furuta et al., 2014). In the same background of Koshihikari, a set of 26 CSSLs with segments from O. nivara were constructed, and new QTLs associated with yield-related traits were identified (Furuta et al., 2016). A set of 198 CSSLs were developed by introgressing O. rufipogon segments into the background of the elite indica rice variety 93–11, and a new QTL associated with the heading date was detected in a 78-kb region on chromosome 10 (Qiao et al., 2016). Similarly, a set of 131 ILs were developed by introducing O. nivara segments into the 93–11 background, and 65 QTLs for 13 yield-related traits were detected by bin-map, with ∼36.9% of the alleles at the detected QTLs from O. nivara leading to improved yield-associated traits (Ma et al., 2016). QTLs associated with 12 grain quality traits were detected using 96 ILs developed from an interspecific cross between the Korean elite japonica rice variety Hwaseong and O. rufipogon (IRGC 105491). A total of 48 QTLs for these traits were identified, and most wild alleles of these detected QTLs had negative effects on the traits (Yun et al., 2016).
In addition to the traits mentioned above, IL sets developed from wild and cultivated rice have also been used for genetic dissection of other traits, such as interspecific hybrid sterility (Yang et al., 2016; Li et al., 2018a; Zhang et al., 2018), anther length (Ogami et al., 2019), the stigma exsertion rate (Tan et al., 2020), out-crossing rate (Prahalada et al., 2021), and photosynthetic efficiency (Rao et al., 2018).
Multiple IL sets derived from indica-japonica crosses have been developed to mine QTLs for rice yield-related traits (Ando et al., 2008; Sun et al., 2017; Mitsuya et al., 2019), quality-related traits (Hao et al., 2009; Sun et al., 2015; Wang et al., 2021), and resistance to biotic (Park et al., 2007) and abiotic (Zang et al., 2008; Zhao et al., 2016; Feng et al., 2018) stresses. Several QTLs for these traits have been detected and causal natural variations for the detected QTLs have been identified by using IL-derived F2 populations (Table 2). For example, two sets of reciprocal ILs derived from a japonica rice variety Lemont and an indica variety Teqing were constructed, and multiple QTLs for grain number per panicle (GNP) were identified form these two ILs. Among these detected QTLs, a stable QTL GNP1 was further finely mapped to a 33.7-kb region and a GA biosynthesis gene GA20-oxidase 1 was identified as the candidate gene for GNP1 (Wu et al., 2016). QTL analysis of seven panicle- and grain-related traits was performed in a set of genotype-defined CSSLs derived from a cross of two genome-sequenced varieties, Nipponbare and Zhenshan 97, and a total of 43 QTLs for these traits were identified. Furthermore, the novel locus qGL11 for grain length and thousand-grain weight was finely mapped to a 25-kb region, and the IAA-amido synthetase gene OsGH3.13 was verified as the causal gene for qGL11 (Wang et al., 2021).
Heterosis is one of the most important characteristics of F1 plants derived from indica-japonica crosses (Dan et al., 2014). Many ILs and IL-derived backcross and/or testcross populations have been used to dissect the genetic basis of heterosis (Xin et al., 2011; Wang Z. et al., 2012; Tao et al., 2016; Xiong et al., 2021; Yang et al., 2021c). For example, a set of 70 ILs and corresponding testcross F1 populations were developed from a cross between the indica rice variety IR24 (RP) and japonica rice variety Asominori to investigate heterotic loci (HLs) associated with six yield-related traits (Xin et al., 2011). A total of 41 HLs were detected on the basis of mid-parent heterosis values with single-point analysis, and most of QTLs were overdominant, suggesting that heterotic effects at the single-locus level are mainly overdominant in rice (Xin et al., 2011). Tao et al. (2016) constructed a set of 128 CSSLs derived from a cross between the indica rice variety 93–11 and japonica rice variety Nipponbare to investigate the genetic basis of heterosis. Using the testcross populations derived from CSSLs and three P-TGMS lines, a total of 97 HLs associated with yield components were detected. Genetic analysis revealed that the contribution of different genetic effects to heterosis might vary among traits (Tao et al., 2016; Yang et al., 2021c). In a recent study, Ghd8 was identified and verified as a major HL for yield components by using a set of intersubspecific CSSLs and two test populations (Xiong et al., 2021). This HL has also been identified in the two-line rice hybrid system (Li et al., 2016), indicating its important role in hybrid breeding.
QTLs have been mapped in ILs derived from indica-japonica crosses for other traits, including hybrid sterility (Zhao et al., 2011), nutrient utilization (Kabange et al., 2021), seed germination and dormancy (Li et al., 2011; Zhang et al., 2020a), mature seed culturability (Zhao et al., 2009), carbon isotope discrimination (Takai et al., 2009), lodging resistance (Mulsanti et al., 2018), and cell wall characteristics (Xu et al., 2017).
ILs developed from intrasubspecific crosses are favored by breeders because there are no incompatibility barriers in these crosses. Although the genetic diversity within rice subspecies is lower than that between subspecies, there are still large phenotypic differences among varieties from different regions with different genealogical relationships in the same subspecies. Several IL libraries from intrasubspecific crosses have been developed in rice to genetically dissect complex traits such as yield-related traits (Shen and Xing, 2014; Hori et al., 2021a; Kato and Hirayama, 2021), quality-related traits (Yang et al., 2021b), root traits (Anis et al., 2019), resistance to preharvest sprouting (Hori et al., 2010; Mizuno et al., 2018), and heterosis (Lin et al., 2020). For example, a set of 202 CSSLs of an elite hybrid, Shanyou 63, were developed in the Zhenshan 97 background, and QTLs for heading date and plant height were detected (Shen and Xing, 2014; Shen et al., 2014). A total of 15 partial dominance QTLs for plant height were identified in 15 CSSL-derived F2 populations, and these QTLs were further identified as HLs for plant height that acted dominantly and epistatically (Shen et al., 2014). Furthermore, multiple QTLs and HLs for yield and spikelets per panicle were identified in these CSSLs, and the hybrids pyramiding these detected HLs in the combined parental genome background showed yield performance similar to that of Shanyou 63, indicating that heterosis might be successfully achieved by manipulating several major dominant HLs (Shen et al., 2022).
ILs derived from advanced backcrosses generally carry only one or a few chromosome fragments from the donor, which greatly minimizes the genetic background noise and facilitates the evaluations of the genetic effects of QTLs/genes on corresponding traits. The target introgressed segment of an IL is homozygous, conferring its genomic stability, so ILs can be repeatedly planted in multiple sites to evaluate the genetic effects of QTLs across different environments (Zhou et al., 2017). A set of ILs covering the entire DP genome can be applied to identify multiple QTLs associated with the trait of interest, and the genetic effects of these detected QTLs can be compared based on their additive effect in the same background, especially using single segment substitution lines (SSSLs) (Ye et al., 2010; Zhu et al., 2018). ILs are not only beneficial for comparing the genetic effects of different QTLs in the same background but also beneficial for comparing the effects of different alleles at one QTL in the fixed background, so as to identify the favorable alleles or allele combinations. For example, the heading date QTL qHD-3 was identified by using a library consisting of 1123 CSSLs from multiple donors in the same genetic background, Huajingxian 74 (HJX74). Compared with the HJX74 allele, the qHD-3 allele from donor variety Lemont delayed rice heading, while qHD-3 from another donor parent, IR64, promoted rice heading. Further sequencing analysis revealed that different variants of DTH3 were identified between Lemont and IR64 (Zhu et al., 2018). To investigate the interaction effect of Ghd8 on yield mid-parent heterosis (MPH), Xiong et al. (2021) constructed five near-isogenic lines (NILs) with each carrying an introgression segment covering Ghd8 from a particular donor in the same background of ZS97, and identified the combination (Ghd8ACC10/Ghd8MH63) with the highest MPH for both spikelet number and grain yield per plant using a half-diallel mating design.
Reciprocal ILs developed in both parental backgrounds have the advantage of enabling evaluation of differences in allelic effects of QTLs in both genetic backgrounds (Kubo et al., 2002). Reciprocal ILs are not only conducive to the detection and fine mapping of QTLs that have both large and small effects but also appropriate for evaluating gene activity as a single factor or in epistatic interactions (Takai et al., 2014). Reciprocal ILs have been used to identify background-independent QTLs (BI-QTLs) and epistatic QTLs (E-QTLs) for grain yield- and quality-related traits (Takai et al., 2014; Qiu et al., 2017; Hori et al., 2021b), salt tolerance (Cheng et al., 2012), lodging resistance (Ookawa et al., 2016), the leaf net photosynthetic rate (Adachi et al., 2019), and sink- and source-related traits (Wang et al., 2020). For example, using the reciprocal CSSLs derived from a cross between MH63 and 02428, a total of nine BI-QTLs for appearance quality were identified. Thirteen and 10 stably expressed QTLs (SE-QTLs) were also detected in the MH63 and 02428 backgrounds, respectively (Qiu et al., 2017). Compared with E-QTLs, BI-QTLs, and SE-QTLs are more easily applied for rice genetic improvement because their function does not depend on background or the environment.
Most agronomic traits of crops are complex traits controlled by multiple genes and affected by environmental factors. Understanding the genetic and environmental bases of QTL × QTL and gene-by-environment (G × E) interactions is of fundamental importance in plant breeding (Monteverde et al., 2019). As in recombinant inbred lines (RILs), genetic interactions between QTLs/genes, such as additive-by-dominance and dominance-by-dominance interactions, also cannot be analyzed using ILs because all sites in ILs are homozygous. However, the offspring-separated populations derived from the crosses between ILs or backcrosses between ILs and RPs are ideal populations for genetic interaction analysis due to their low genetic background noise. Genetic interaction analysis among four major rice heading date genes, Ghd7, Ghd8, OsPRR37, and Hd1, was performed in a 4-gene segregating population in the near-isogenic background under both natural long-day (NLD) and natural short-day (NSD) conditions (Zhang et al., 2019a). Tetragenic, trigenic and digenic interactions among these four genes for heading date were observed under both conditions but were more significant under NLD conditions. Further analysis showed that the differences in digenic interactions between Ghd7 and OsPRR37 under different conditions were essential to the alternative function of OsPRR37 (Zhang et al., 2019a). Genetic interaction analysis between QTLs for other agronomic traits, such as plant height (Shen et al., 2014), grain size (Xia et al., 2018), tillering (Zhou et al., 2020), cold tolerance (Liang et al., 2018), and drought tolerance (Yadav et al., 2019), was also performed in IL-derived populations.
Most the traits with low heritability are easily affected by the environment, and there is an obvious interaction between gene and the environment. Understanding G × E interactions can help breeders in deciding which QTL to use in their breeding programs while tailoring crop cultivars for specific or more diverse environments (Liu et al., 2008). G × E interactions were analyzed in ILs or IL-derived populations planted in a variety of environments. To dissect the genetic basis of the genetic main effect (G) and G × E interaction effect (GE) for panicle number (PN) in rice, a population consisting of 35 SSSLs derived from originating from crosses between the RP, HJX74, and 17 DPs was grown in six cropping season environments (Liu et al., 2008). The total genetic effect was partitioned into G and GE by using the mixed linear-model approach, and then QTL analyses of these effects were conducted separately. A single QTL effect was divided into two components: The additive effect (a) and additive × environment interaction effect (ae). A total of 18 QTLs for PN were identified, and three types of QTLs were suggested according to their effects expressed. Two QTLs expressed stably across environments due to the association with only a, nine QTLs with only ae were unstable, and the remaining seven QTLs were identified with both a and ae, which were also unstable across environments (Liu et al., 2008). Notably, when QTLs have large ae values in opposing directions in different environments, selection for an allele with favorable effects in certain environments could lead to undesired results in other environments.
Rice breeding is based on the use of valuable alleles and elite germplasm resources. Fortunately, rice has a large number of germplasms containing all kinds of excellent genes for high yield, good quality, resistance to biotic and abiotic stresses, and etc. (Tanksley and McCouch, 1997). Transferring these excellent genes into modern varieties absolutely could broaden the gene pool and greatly improve the productivity. However, many of these valuable alleles have linkage drags (Ali et al., 2010). IL populations have the advantage of breaking the linkage between valuable genes and linkage drags. To date, many traits in rice have been greatly improved by developing ILs in the backgrounds of modern varieties (Hu et al., 2016; Balakrishnan et al., 2019; Zhang, 2021; Zhang et al., 2021c).
Each line of ILs carries only one or a few donor chromosome segments, which usually affects only one or a few traits of the RP. Therefore, using ILs to improve single-target trait in rice is efficient. In the elite genetic backgrounds, IL with improved agronomic performance can be released as a new variety.
Increasing rice yield to meet the need for a rapidly growing global population is one of the most important goals of rice breeding. Many yield-related QTLs/genes have been introgressed into elite backgrounds to increase rice yield. For example, GN4-1, a QTL for grain number per panicle, was introgressed into ZH8006 from WYJ6, and grain yield increased by more than 10% (Zhou et al., 2018). To achieve the high yield potential in Kongyu 131, a minute chromosome fragment carrying the favorable Gn1a allele from the donor parent GKBR was introgressed into the genome of Kongyu 131, which resulted in larger panicles and a subsequent yield increase in the new Kongyu 131 (Feng et al., 2017). In addition, introgression of a functional epigenetic OsSPL14WFP allele into elite indica cultivars can greatly improve panicle traits and grain yield (Kim et al., 2018).
With the increasing development of people’s living standards, rice quality has received attention from both rice producers and consumers. There have been many successful cases of improving rice grain quality through ILs. For example, to improve the eating and cooking quality of Zhenshan 97 and its hybrid Shanyou 63, the Wx/Waxy gene region from Minghui 63, a restorer line with medium amylose content (AC), soft gel consistency (GC), and high gelatinization temperature (GT), was introgressed into Zhenshan 97B and then transferred into Zhenshan 97A (Zhou et al., 2003). In recent years, the discovery of different alleles of Wx and ALK has provided more choices for rice grain quality improvement via gene introgression (Chen et al., 2020; Huang et al., 2021; Zhang et al., 2021b; Zhou et al., 2021). GS9 is an important gene for both grain shape and chalkiness, and appearance quality could be largely improved by developing an IL for this gene (Zhao et al., 2018, 2021).
Biotic and abiotic stresses are large threats to rice production, leading to serious yield losses every year. ILs constructed by introgressing novel genes into modern varieties are an effective tool for solving this problem. For example, by combining a backcross breeding strategy combined with MAS, a total of 12 blast resistance genes were introgressed into one maintainer line of cytoplasmic male sterility (CMS) and three photo-thermo genetic male sterility (P-TGMS) lines. The blast resistance of these lines was significantly improved, while other traits were not changed (Jiang et al., 2012, 2015, 2019). Using the same strategy, brown planthopper resistance has been improved by introgressing Bph3, Bph14, Bph15, Bph18, Bph20, Bph21, and Bph33 into 93–11 and Jin23B (Hu et al., 2013, 2018; Jiang et al., 2018), and bacterial blight resistance has been improved by pyramiding Xa4, xa5, xa13, and Xa21 (Huang et al., 1997). In the two IL populations developed with C258 and ZGX1 as RPs and IR75861 as the DP, 12 and 8 lines had higher salt tolerance than the RPs, respectively, and they could be directly planted into environments characterized by salt stress (Qiu et al., 2015).
The excess application of fertilizers in rice production not only increases cost, but also causes severe environmental problems (Zhang et al., 2020c). Breeding rice varieties with high nitrogen-use efficiency (NUE) is of great significance to the practice of Green Super Rice (Yu et al., 2020). GRF4 promotes and integrates nitrogen assimilation, carbon fixation, and growth, whereas DELLA inhibits these processes. Introgression of GRF4 into the elite variety 93–11 carrying sd1 can significantly improve its NUE, thereby increasing yield (Li et al., 2018b). NAL1 and the gain-of-function mutation dep1-1 have been reported to play important roles in N-responsive tillering regulation, and pyramiding of the dep1-1 and NAL1NJ6 alleles can achieve sustainable improvements in NUE and grain yield in japonica rice breeding (Sun et al., 2014; Xu et al., 2019). The introgression of favorable alleles of other NUE-related genes such as OsTCP19 and OsSBM1 also showed great potential for genetic improvement of high NUE in rice (Liu et al., 2021; Xu et al., 2021).
To meet people’s diverse demands for rice, modern rice varieties should be improved mainly by multiple locus introgression to improve the overall features of rice in terms of grain yield and quality, resistance to biotic and abiotic stresses and nitrogen use efficiency, etc. For example, Luan et al. (2019) used four SSSLs with HJX74 as the RP to develop a new maintainer named H131B with a suitable heading date, good appearance and eating and cooking quality, and fragrant smell by pyramiding MADS50, gs3, fgr, Wxgl, and ALK. Dixit et al. (2020) developed seven ILs possessing a combination of seven to ten QTLs for resistance to different biotic and abiotic stresses by using the MAS breeding method in the background of Swarna. These ILs were superior to the respective RPs in terms of agronomic performance and possessed superior grain quality. Luo et al. (2016) successfully developed a new restorer line, WH6725, with disease resistance to rice blast and bacterial blight, tolerance to submergence and an aromatic fragrance by pyramiding Xa27, Pi9, Sub1A, and badh2.1. Through genotyping-based identification and backcrossing with MAS, Zeng et al. (2017) introgressed 21 favorable alleles from NIP and 9311 into Teqing and then developed three lines that exhibited higher yield potential and better grain quality than their parental varieties and the super-hybrid rice Liang-you-pei-jiu. Guangzhan 63-4S is an elite two-line thermos-sensitive genic male sterile line. However, it is highly susceptible to blast and bacterial blight. Two BC2 ILs were developed to introgress Xa7 for bacterial blight and Pi-2 for blast resistance. Then, the ILs were crossed and pyramided with these two genes. A new two-line thermosensitive genic male-sterile line named Hua1228S that was resistant against both rice blast and bacterial blight was developed (Mi et al., 2018).
IL platform developed by multiple donors in an elite genetic background facilitates the discovery of new QTLs and identification of multiple alleles of a gene. IL platform can be used to identify the optimal allele or allele combination by evaluating their genetic effects in the fixed background and then improve varieties by QTL pyramiding (Zhang, 2021; Figure 1D). In addition, new varieties can be directly developed by screening transgressive lines of target traits in the IL platform (Figure 1D). HJX74 is an elite variety from South China. A single segment IL platform including more than 2000 ILs was developed with HJX74 as the RP and more than 40 varieties as DPs (Luan et al., 2019; Zhang, 2021). A large number of important genes were identified using this platform (Wang S. et al., 2012; Wang et al., 2015; Yang et al., 2021a). In addition, a series of varieties have been bred by pyramiding different favorable genes in this platform, such as three-line maintainer lines (H121B, H131B, HZB, HBB, E5-HXB, and E5-HBB) (Luan et al., 2019), black rice (Huaxiaohei1 and Huaxiaohei 2), red rice (Huaxiaohong1 and Huaxiaohong 2), and high-quality rice (Huabiao1 and Huabiao 3) (Zhang, 2021). Huanghuazhan (HHZ) is an elite rice variety widely planted in the middle and downstream regions of the Yangtze River and in South China. An IL platform including 496 ILs were developed using HHZ as the RP and 8 varieties as DPs (Ali et al., 2017). Most ILs showed significantly higher yields than the parents under both abiotic stress conditions (salt and drought) and non-abiotic stress conditions (submergence). Ultimately, at least six ILs were directly released as new green super rice varieties in the Philippines and Pakistan and have been grown on more than 1 million ha of land in different ecosystems (Ali et al., 2017).
ILs simplify the genetic dissection of complex traits and accelerate whole-genome large-scale gene discovery in rice. To date, a large number of the natural variations associated with agronomic traits in rice have been identified from ILs and then cloned using IL-derived mapping populations (Li et al., 2018c; Zhang, 2021; Table 2). The primary mapping populations may be more cost-effective for large-effect QTLs mapping, but ILs have a greater advantage in detecting minor QTLs because of their low genetic background noise. The resolution of QTL mapping using ILs is low due to the large size of the introgression segments and the selective abandonment of some recombination events (lack of lines) during the construction of ILs (Cavanagh et al., 2008; Balakrishnan et al., 2019). Substitution mapping of QTLs using multiple ILs is an effective way to improve the mapping resolution (Zhang, 2021). In addition, any two ILs carrying different QTLs/genes controlling the same trait can be used for the epistatic interaction detection with their offspring-segregated population, which can provide useful information on how to select favorable alleles and allele combinations in rice molecular design breeding.
A large number of genetic dissections of traditional agronomic traits have been performed in rice (Li et al., 2018c; Wei et al., 2021), so it is difficult to further mine new QTLs/genes for these traits. While some rice subpopulations or distant relatives, such as cA, cB, and Africa cultivated rice, with narrow geographic or relatively independent origins, may carry many private (rare) genes and novel natural variations due to the differences in domestication and environmental pressures (Li et al., 2015; Bai et al., 2017; Wang et al., 2018). Accessions from these subpopulations were rarely used in the past, but are encouraged used as donors for constructing ILs in the future because more new QTLs might be detected using this kind of ILs. Many ILs have been constructed using AA-genome wild species as donors, but there are few successful cases of constructing ILs using other wild Oryza species as donors due to the strong incompatibility barriers (Sanchez et al., 2013). These wild species not only have strong biotic and abiotic stress resistance, but also have unique and excellent traits, such as the early morning flowering trait in CC-genome wild rice Oryza officinalis and the high biomass production in CCDD-genome wild species (Sanchez et al., 2013; Hirabayashi et al., 2015). Thus, using more different wild rice species as genetic resources to broaden the gene pool of cultivars by constructing ILs is of great significance, but it is also full of challenges. Moreover, multiple donors from different wild/cultivated rice species and subpopulations are also encouraged to develop IL platforms in one genetic background for new alleles detection. To date, the third-generation sequencing technology makes the cost of genome assembly dramatically decreased, a large number of rice varieties have been deeply sequenced, and reference genomes of dozens of varieties (including wild and cultivated rice) have been assembled (Wang et al., 2018; Wing et al., 2018; Qin et al., 2021). Thus, the accessions used for ILs development are suggested to generate the genome reference to facilitate the discovery of genetic variations between the recurrent and donor parents. Based on the difference of gene annotations between parents, the ILs contained the target genes can be used to precisely examine the potential target traits and rapidly verify the valuable natural variations. In addition, ILs combined with high-throughput phenotyping platform are encouraged to detect QTLs for traits measured dynamically and non-destructively, especially for the root associated traits and abiotic stress tolerance, which are hard to precisely measure by traditional ways (Yang et al., 2014).
As important prebreeding materials, ILs facilitate the genetic improvement of rice via gene transfer and pyramiding. However, QTL pyramiding for rice variety improvement requires multiple crosses and the avoidance of genetic drag, which usually requires several generations and large populations (Zhang et al., 2021c). Applying the IL platform in an elite background can accurately reveal multiple favorable alleles that can be used to improve the RP. This platform can also be used to evaluate the genetic effects of different alleles at the same locus and the effects of different allele combinations to identify the optimal allele or combination for target genetic improvement. In addition, the excellent allele combinations existing in the background will be kept in the process of QTL pyramiding, so target genetic improvement can be realized quickly. Moreover, the IL platform provides a chance to rapidly generate multi-line varieties by mixing the ILs with different biotic stress resistant genes, such as those for rice blast and brown planthopper resistance (Koizumi et al., 2004; Li et al., 2013). In some elite backgrounds, multiple ILs have been constructed by different institutions (Table 1), and global sharing of these resources would accelerate the construction of IL platforms and further facilitate the consortium of rice genetic improvement.
YX provided the idea for the review. BZ and XQ collected the data and wrote the manuscript. LM assisted in literature collection. YX, XQ, BZ, LM, and BW revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the National Natural Science Foundation of China (Grant Nos. 32171993 and 32101740) and the China Postdoctoral Science Foundation (Grant No. 2021M701360).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adachi, S., Yamamoto, T., Nakae, T., Yamashita, M., Uchida, M., Karimata, R., et al. (2019). Genetic architecture of leaf photosynthesis in rice revealed by different types of reciprocal mapping populations. J. Exp. Bot. 70, 5131–5144. doi: 10.1093/jxb/erz303
Agarwal, M., Shrivastava, N., and Padh, H. (2008). Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 27, 617–631. doi: 10.1007/s00299-008-0507-z
Ali, J., Xu, J. L., Gao, Y. M., Ma, X. F., Meng, L. J., Wang, Y., et al. (2017). Harnessing the hidden genetic diversity for improving multiple abiotic stress tolerance in rice (Oryza sativa L.). PLoS One 12:e0172515. doi: 10.1371/journal.pone.0172515
Ali, M. L., Sanchez, P. L., Yu, S.-B., Lorieux, M., and Eizenga, G. C. (2010). Chromosome segment substitution lines: a powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa). Rice 3, 218–234. doi: 10.1007/s12284-010-9058-3
Ando, T., Yamamoto, T., Shimizu, T., Ma, X. F., Shomura, A., Takeuchi, Y., et al. (2008). Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor. Appl. Genet. 116, 881–890. doi: 10.1007/s00122-008-0722-6
Anis, G. B., Zhang, Y., Islam, A., Zhang, Y., Cao, Y., Wu, W., et al. (2019). RDWN6XB, a major quantitative trait locus positively enhances root system architecture under nitrogen deficiency in rice. BMC Plant Biol. 19:12. doi: 10.1186/s12870-018-1620-y
Atwell, B. J., Wang, H., and Scafaro, A. P. (2014). Could abiotic stress tolerance in wild relatives of rice be used to improve Oryza sativa? Plant Sci. 21, 48–58. doi: 10.1016/j.plantsci.2013.10.007
Bai, X., Huang, Y., Hu, Y., Liu, H., Zhang, B., Smaczniak, C., et al. (2017). Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants 3, 885–893. doi: 10.1038/s41477-017-0042-4
Balakrishnan, D., Surapaneni, M., Mesapogu, S., and Neelamraju, S. (2019). Development and use of chromosome segment substitution lines as a genetic resource for crop improvement. Theor. Appl. Genet. 132, 1–25. doi: 10.1007/s00122-018-3219-y
Bessho-Uehara, K., Furuta, T., Masuda, K., Yamada, S., Angeles-Shim, R. B., Ashikari, M., et al. (2017). Construction of rice chromosome segment substitution lines harboring Oryza barthii genome and evaluation of yield-related traits. Breed. Sci. 67, 408–415. doi: 10.1270/jsbbs.17022
Bian, J. M., Jiang, L., Liu, L. L., Wei, X. J., Xiao, Y. H., Zhang, L. J., et al. (2010). Construction of a new set of rice chromosome segment substitution lines and identification of grain weight and related traits QTLs. Breed. Sci. 60, 305–313. doi: 10.1270/jsbbs.60.305
Cao, Z., Li, Y., Tang, H., Zeng, B., Tang, X., Long, Q., et al. (2020). Fine mapping of the qHTB1-1 QTL, which confers heat tolerance at the booting stage, using an Oryza rufipogon Griff. introgression line. Theor. Appl. Genet. 133, 1161–1175. doi: 10.1007/s00122-020-03539-7
Cavanagh, C., Morell, M., Mackay, I., and Powell, W. (2008). From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11, 215–221. doi: 10.1016/j.pbi.2008.01.002
Cheema, K. K., Bains, N. S., Mangat, G. S., Das, A., Vikal, Y., Brar, D. S., et al. (2008). Development of high yielding IR64 × Oryza rufipogon (Griff.) introgression lines and identification of introgressed alien chromosome segments using SSR markers. Euphytica 160, 401–409. doi: 10.1007/s10681-007-9560-x
Chen, Z., Lu, Y., Feng, L., Hao, W., Li, C., Yang, Y., et al. (2020). Genetic dissection and functional differentiation of ALKa and ALKb, two natural alleles of the ALK/SSIIa gene, responding to low gelatinization temperature in rice. Rice 13:39. doi: 10.1186/s12284-020-00393-5
Cheng, L., Wang, Y., Meng, L., Hu, X., Cui, Y., Sun, Y., et al. (2012). Identification of salt-tolerant QTLs with strong genetic background effect using two sets of reciprocal introgression lines in rice. Genome 55, 45–55. doi: 10.1139/g11-075
Chutimanukul, P., Kositsup, B., Plaimas, K., Buaboocha, T., Siangliw, M., Toojinda, T., et al. (2018). Data in support of photosynthetic responses in a chromosome segment substitution line of ‘Khao Dawk Mali 105’ rice at seedling stage. Data Brief 21, 307–312. doi: 10.1016/j.dib.2018.09.128
Civáò, P., Craig, H., Cox, C. J., and Brown, T. A. (2015). Three geographically separate domestications of Asian rice. Nat. Plants 1:15164. doi: 10.1038/nplants.2015.164
Dan, Z., Liu, P., Huang, W., Zhou, W., Yao, G., Hu, J., et al. (2014). Balance between a higher degree of heterosis and increased reproductive isolation: a strategic design for breeding inter-subspecific hybrid rice. PLoS One 9:e93122. doi: 10.1371/journal.pone.0093122
Dixit, S., Singh, U. M., Singh, A. K., Alam, S., Venkateshwarlu, C., Nachimuthu, V. V., et al. (2020). Marker assisted forward breeding to combine multiple biotic-abiotic stress resistance/tolerance in rice. Rice 13:29. doi: 10.1186/s12284-020-00391-7
Dong, X., Wang, X., Zhang, L., Yang, Z., Xin, X., Wu, S., et al. (2013). Identification and characterization of OsEBS, a gene involved in enhanced plant biomass and spikelet number in rice. Plant Biotechnol. J. 11, 1044–1057. doi: 10.1111/pbi.12097
Ebitani, T., Takeuchi, Y., Nonoue, Y., Yamamoto, T., Takeuchi, K., and Yano, M. (2005). Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed. Sci. 55, 65–73. doi: 10.1270/jsbbs.55.65
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Fasahat, P., Muhammad, K., Abdullah, A., and Wickneswari, R. (2012). Identification of introgressed alien chromosome segments associated with grain quality in Oryza rufipogon x MR219 advanced breeding lines using SSR markers. Genet. Mol. Res. 11, 3534–3546. doi: 10.4238/2012.September.26.10
Feng, B., Chen, K., Cui, Y., Wu, Z., Zheng, T., Zhu, Y., et al. (2018). Genetic dissection and simultaneous improvement of drought and low nitrogen tolerances by designed QTL pyramiding in rice. Front. Plant Sci. 9:306. doi: 10.3389/fpls.2018.00306
Feng, X., Wang, C., Nan, J., Zhang, X., Wang, R., Jiang, G., et al. (2017). Updating the elite rice variety Kongyu 131 by improving the Gn1a locus. Rice 10:35. doi: 10.1186/s12284-017-0174-1
Fu, Q., Zhang, P., Tan, L., Zhu, Z., Ma, D., Fu, Y., et al. (2010). Analysis of QTLs for yield-related traits in Yuanjiang common wild rice (Oryza rufipogon Griff.). J. Genet. Genomics 37, 147–157. doi: 10.1016/s1673-8527(09)60033-3
Fujita, D., Tagle, A. G., Ebron, L. A., Fukuta, Y., and Kobayashi, N. (2012). Characterization of near-isogenic lines carrying QTL for high spikelet number with the genetic background of an indica rice variety IR64 (Oryza sativa L.). Breed. Sci. 62, 18–26. doi: 10.1270/jsbbs.62.18
Fukuta, Y., Konisho, K., Senoo-Namai, S., Yanagihara, S., Tsunematsu, H., Fukuo, A., et al. (2012). Genetic characterization of rainfed upland New Rice for Africa (NERICA) varieties. Breed. Sci. 62, 27–37. doi: 10.1270/jsbbs.62.27
Furuta, T., Uehara, K., Angeles-Shim, R. B., Shim, J., Ashikari, M., and Takashi, T. (2014). Development and evaluation of chromosome segment substitution lines (CSSLs) carrying chromosome segments derived from Oryza rufipogon in the genetic background of Oryza sativa L. Breed. Sci. 63, 468–475. doi: 10.1270/jsbbs.63.468
Furuta, T., Uehara, K., Angeles-Shim, R. B., Shim, J., Nagai, K., Ashikari, M., et al. (2016). Development of chromosome segment substitution lines harboring Oryza nivara genomic segments in Koshihikari and evaluation of yield-related traits. Breed. Sci. 66, 845–850. doi: 10.1270/jsbbs.16131
Gao, D., Sun, W., Wang, D., Dong, H., Zhang, R., and Yu, S. (2020). A xylan glucuronosyltransferase gene exhibits pleiotropic effects on cellular composition and leaf development in rice. Sci. Rep. 10:3726. doi: 10.1038/s41598-020-60593-3
Garcia-Oliveira, A. L., Tan, L., Fu, Y., and Sun, C. (2009). Genetic identification of quantitative trait loci for contents of mineral nutrients in rice grain. J. Integr. Plant Biol. 51, 84–92. doi: 10.1111/j.1744-7909.2008.00730.x
Grover, N., Kumar, A., Yadav, A. K., Gopala Krishnan, S., Ellur, R. K., Bhowmick, P. K., et al. (2020). Marker assisted development and characterization of herbicide tolerant near isogenic lines of a mega Basmati rice variety, “Pusa Basmati 1121”. Rice 13:68. doi: 10.1186/s12284-020-00423-2
Guo, S.-B., Wei, Y., Li, X.-Q., Liu, K.-Q., Huang, F.-K., Chen, C.-H., et al. (2013). Development and identification of introgression lines from cross of Oryza sativa and Oryza minuta. Rice Sci. 20, 95–102. doi: 10.1016/S1672-6308(13)60111-0
Gutiérrez, A. G., Carabalí, S. J., Giraldo, O. X., Martínez, C. P., Correa, F., Prado, G., et al. (2010). Identification of a Rice stripe necrosis virus resistance locus and yield component QTLs using Oryza sativa x O. glaberrima introgression lines. BMC Plant Biol. 10:6. doi: 10.1186/1471-2229-10-6
Hao, W., Zhu, M. Z., Gao, J. P., Sun, S. Y., and Lin, H. X. (2009). Identification of quantitative trait loci for rice quality in a population of chromosome segment substitution lines. J. Integr. Plant Biol. 51, 500–512. doi: 10.1111/j.1744-7909.2009.00822.x
Harlan, H. V., and Pope, M. N. (1922). The use and value of back-crosses in small-grain breeding. J. Hered. 13, 319–322. doi: 10.1093/oxfordjournals.jhered.a102237
He, F., Xi, Z., Zeng, R., Akshay, T., and Zhang, G. (2005a). Mapping of heading date QTLs in rice (Oryza sativa) using single segment substitution lines. Zhongguo Nong Ye Ke Xue 38, 1505–1513.
He, F., Xi, Z., Zeng, R., Talukdar, A., and Zhang, G. (2005b). Developing single segment substitution lines (SSSLs) in rice (Oryza sativa L.) using advanced backcrosses and MAS. Yi Chuan Xue Bao 32, 825–831.
He, F., Zhang, X., Zeng, R., Talukdar, A., and Zhang, G. (2005c). Identification of QTLs for plant height and its components by using single segment substitution lines in rice (Oryza sativa). Rice Sci. 12, 151–156.
He, N., Wu, R., Pan, X., Peng, L., Sun, K., Zou, T., et al. (2017). Development and trait evaluation of chromosome single-segment substitution lines of O. meridionalis in the background of O. sativa. Euphytica 213:281. doi: 10.1007/s10681-017-2072-4
Hirabayashi, H., Sasaki, K., Kambe, T., Gannaban, R. B., Miras, M. A., Mendioro, M. S., et al. (2015). qEMF3, a novel QTL for the early-morning flowering trait from wild rice, Oryza officinalis, to mitigate heat stress damage at flowering in rice, O. sativa. J. Exp. Bot. 66, 1227–1236. doi: 10.1093/jxb/eru474
Hori, K., Saisho, D., Nagata, K., Nonoue, Y., Uehara-Yamaguchi, Y., Kanatani, A., et al. (2021a). Genetic elucidation for response of flowering time to ambient temperatures in Asian rice cultivars. Int. J. Mol. Sci. 22:1024. doi: 10.3390/ijms22031024
Hori, K., Suzuki, K., Ishikawa, H., Nonoue, Y., Nagata, K., Fukuoka, S., et al. (2021b). Genomic regions involved in differences in eating and cooking quality other than Wx and Alk genes between indica and japonica rice cultivars. Rice 14:8. doi: 10.1186/s12284-020-00447-8
Hori, K., Sugimoto, K., Nonoue, Y., Ono, N., Matsubara, K., Yamanouchi, U., et al. (2010). Detection of quantitative trait loci controlling pre-harvest sprouting resistance by using backcrossed populations of japonica rice cultivars. Theor. Appl. Genet. 120, 1547–1557. doi: 10.1007/s00122-010-1275-z
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Hu, J., Chang, X., Zou, L., Tang, W., and Wu, W. (2018). Identification and fine mapping of Bph33, a new brown planthopper resistance gene in rice (Oryza sativa L.). Rice 11:55. doi: 10.1186/s12284-018-0249-7
Hu, J., Cheng, M., Gao, G., Zhang, Q., Xiao, J., and He, Y. (2013). Pyramiding and evaluation of three dominant brown planthopper resistance genes in the elite indica rice 9311 and its hybrids. Pest Manage. Sci. 69, 802–808. doi: 10.1002/ps.3437
Hu, J., Xiao, C., and He, Y. (2016). Recent progress on the genetics and molecular breeding of brown planthopper resistance in rice. Rice 9:30. doi: 10.1186/s12284-016-0099-0
Hua, L., Wang, D. R., Tan, L., Fu, Y., Liu, F., Xiao, L., et al. (2015). LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27, 1875–1888. doi: 10.1105/tpc.15.00260
Huang, L., Gu, Z., Chen, Z., Yu, J., Chu, R., Tan, H., et al. (2021). Improving rice eating and cooking quality by coordinated expression of the major starch synthesis-related genes, SSII and Wx, in endosperm. Plant Mol. Biol. 106, 419–432. doi: 10.1007/s11103-021-01162-8
Huang, N., Angeles, E. R., Domingo, J., Magpantay, G., Singh, S., Zhang, G., et al. (1997). Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor. Appl. Genet. 95, 313–320. doi: 10.1007/s001220050565
Huang, X. Y., Deng, F., Yamaji, N., Pinson, S. R., Fujii-Kashino, M., Danku, J., et al. (2016). A heavy metal P-type ATPase OsHMA4 prevents copper accumulation in rice grain. Nat. Commun. 7:12138. doi: 10.1038/ncomms12138
Huang, Y., Zhao, S., Fu, Y., Sun, H., Ma, X., Tan, L., et al. (2018). Variation in the regulatory region of FZP causes increases in secondary inflorescence branching and grain yield in rice domestication. Plant J. 96, 716–733. doi: 10.1111/tpj.14062
Huo, X., Wu, S., Zhu, Z., Liu, F., Fu, Y., Cai, H., et al. (2017). NOG1 increases grain production in rice. Nat. Commun. 8:1497. doi: 10.1038/s41467-017-01501-8
Jiang, H., Feng, Y., Bao, L., Li, X., Gao, G., Zhang, Q., et al. (2012). Improving blast resistance of Jin 23B and its hybrid rice by marker-assisted gene pyramiding. Mol. Breed. 30, 1679–1688. doi: 10.1007/s11032-012-9751-6
Jiang, H., Hu, J., Li, Z., Liu, J., Gao, G., Zhang, Q., et al. (2018). Evaluation and breeding application of six brown planthopper resistance genes in rice maintainer line Jin 23B. Rice 11:22. doi: 10.1186/s12284-018-0215-4
Jiang, H., Li, Z., Liu, J., Shen, Z., Gao, G., Zhang, Q., et al. (2019). Development and evaluation of improved lines with broad-spectrum resistance to rice blast using nine resistance genes. Rice 12:29. doi: 10.1186/s12284-019-0292-z
Jiang, J., Mou, T., Yu, H., and Zhou, F. (2015). Molecular breeding of thermo-sensitive genic male sterile (TGMS) lines of rice for blast resistance using Pi2 gene. Rice 8:11. doi: 10.1186/s12284-015-0048-3
Jin, J., Hua, L., Zhu, Z., Tan, L., Zhao, X., Zhang, W., et al. (2016). GAD1 encodes a secreted peptide that regulates grain number, grain length, and awn development in rice domestication. Plant Cell 28, 2453–2463. doi: 10.1105/tpc.16.00379
Jones, M. P., Dingkuhn, M., Aluko/snm, G. K., and Semon, M. (1997). Interspecific Oryza Sativa L. X O. Glaberrima Steud. progenies in upland rice improvement. Euphytica 94, 237–246. doi: 10.1023/A:1002969932224
Kabange, N. R., Park, S. Y., Lee, J. Y., Shin, D., Lee, S. M., Kwon, Y., et al. (2021). New insights into the transcriptional regulation of genes involved in the nitrogen use efficiency under potassium chlorate in rice (Oryza sativa L.). Int. J. Mol. Sci. 22:2192. doi: 10.3390/ijms22042192
Kato, K., and Hirayama, Y. (2021). Development and characterization of chromosome segment substitution lines derived from backcross between japonica donor rice cultivar Yukihikari and japonica recipient cultivar Kirara397. Breed. Sci. 71, 283–290. doi: 10.1270/jsbbs.20128
Khan, G. H., Shikari, A. B., Vaishnavi, R., Najeeb, S., Padder, B. A., Bhat, Z. A., et al. (2018). Marker-assisted introgression of three dominant blast resistance genes into an aromatic rice cultivar Mushk Budji. Sci. Rep. 8:4091. doi: 10.1038/s41598-018-22246-4
Kijima, Y., Sserunkuuma, D., and Otsuka, K. (2006). How revolutionary is the “Nerica Revolution”? Evidence from Uganda. Dev. Econ. 44, 252–267. doi: 10.1111/j.1746-1049.2006.00016.x
Kijoji, A. A., Nchimbi-Msolla, S., Kanyeka, Z. L., Serraj, R., and Henry, A. (2014). Linking root traits and grain yield for rainfed rice in sub-Saharan Africa: response of Oryza sativa×Oryza glaberrima introgression lines under drought. Field Crops Res. 165, 25–35. doi: 10.1016/j.fcr.2014.03.019
Kim, S. H., Ji, S. D., Lee, H. S., Jeon, Y. A., Shim, K. C., Adeva, C., et al. (2021). A novel embryo phenotype associated with interspecific hybrid weakness in rice is controlled by the MADS-domain transcription factor OsMADS8. Front. Plant Sci. 12:778008. doi: 10.3389/fpls.2021.778008
Kim, S. R., Ramos, J. M., Hizon, R. J. M., Ashikari, M., Virk, P. S., Torres, E. A., et al. (2018). Introgression of a functional epigenetic OsSPL14WFP allele into elite indica rice genomes greatly improved panicle traits and grain yield. Sci. Rep. 8:3833. doi: 10.1038/s41598-018-21355-4
Koizumi, S., Ashizawa, T., and Zenbayashi, K. S. (2004). “Durable control of rice blast disease with multilines,” in Rice Blast: Interaction with Rice and Control, ed. S. Kawasaki (Dordrecht: Springer), 191–199. doi: 10.1007/978-0-306-48582-4_23
Kovach, M. J., Sweeney, M. T., and McCouch, S. R. (2007). New insights into the history of rice domestication. Trends Genet. 23, 578–587. doi: 10.1016/j.tig.2007.08.012
Kovi, M. R., Zhang, Y., Yu, S., Yang, G., Yan, W., and Xing, Y. (2011). Candidacy of a chitin-inducible gibberellin-responsive gene for a major locus affecting plant height in rice that is closely linked to Green Revolution gene sd1. Theor. Appl. Genet. 123, 705–714. doi: 10.1007/s00122-011-1620-x
Kubo, T., Aida, Y., Nakamura, K., Tsunematsu, H., Doi, K., and Yoshimura, A. (2002). Reciprocal chromosome segment substitution series derived from japonica and indica cross of rice (Oryza sativa L.). Breed. Sci. 52, 319–325. doi: 10.1270/jsbbs.52.319
Lei, D., Tan, L., Liu, F., Chen, L., and Sun, C. (2013). Identification of heat-sensitive QTL derived from common wild rice (Oryza rufipogon Griff.). Plant Sci. 20, 121–127. doi: 10.1016/j.plantsci.2012.12.001
Li, D., Huang, Z., Song, S., Xin, Y., Mao, D., Lv, Q., et al. (2016). Integrated analysis of phenome, genome, and transcriptome of hybrid rice uncovered multiple heterosis-related loci for yield increase. Proc. Natl. Acad. Sci. U.S.A. 113, E6026–E6035. doi: 10.1073/pnas.1610115113
Li, M., Sun, P., Zhou, H., Chen, S., and Yu, S. (2011). Identification of quantitative trait loci associated with germination using chromosome segment substitution lines of rice (Oryza sativa L.). Theor. Appl. Genet. 123, 411–420. doi: 10.1007/s00122-011-1593-9
Li, X., Chao, D., Wu, Y., Huang, X., Chen, K., Cui, L., et al. (2015). Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 47, 827–833. doi: 10.1038/ng.3305
Li, X., Liu, Z., Lu, D., Liu, Y., Mao, X., Li, Z., et al. (2013). Development and evaluation of multi-genotype varieties of rice derived from MAGIC lines. Euphytica 192, 77–86. doi: 10.1007/s10681-013-0879-1
Li, J., Zhou, J., Xu, P., Deng, X., Deng, W., Zhang, Y., et al. (2018a). Mapping five novel interspecific hybrid sterility loci between Oryza sativa and Oryza meridionalis. Breed. Sci. 68, 516–523. doi: 10.1270/jsbbs.18001
Li, S., Tian, Y., Wu, K., Ye, Y., Yu, J., Zhang, J., et al. (2018b). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560, 595–600. doi: 10.1038/s41586-018-0415-5
Li, Y., Xiao, J., Chen, L., Huang, X., Cheng, Z., Han, B., et al. (2018c). Rice functional genomics research: past decade and future. Mol. Plant 11, 359–380. doi: 10.1016/j.molp.2018.01.007
Li, Z., and Zheng, T. (2013). “Utilization of exotic germplasm,” in Genetics and Genomics of Rice, eds Q. Zhang and R. A. Wing (New York, NY: Springer New York), 349–361. doi: 10.1007/978-1-4614-7903-1_23
Liang, Y., Meng, L., Lin, X., Cui, Y., Pang, Y., Xu, J., et al. (2018). QTL and QTL networks for cold tolerance at the reproductive stage detected using selective introgression in rice. PLoS One 13:e0200846. doi: 10.1371/journal.pone.0200846
Lin, Q., Wang, W., Ren, Y., Jiang, Y., Sun, A., Qian, Y., et al. (2015). Genetic dissection of seed storability using two different populations with a same parent rice cultivar N22. Breed. Sci. 65, 411–419. doi: 10.1270/jsbbs.65.411
Lin, T., Zhou, C., Chen, G., Yu, J., Wu, W., Ge, Y., et al. (2020). Heterosis-associated genes confer high yield in super hybrid rice. Theor. Appl. Genet. 133, 3287–3297. doi: 10.1007/s00122-020-03669-y
Liu, G., Zeng, R., Zhu, H., Zhang, Z., Ding, X., Zhao, F., et al. (2009). Dynamic expression of nine QTLs for tiller number detected with single segment substitution lines in rice. Theor. Appl. Genet. 118, 443–453. doi: 10.1007/s00122-008-0911-3
Liu, G., Zhang, Z., Zhu, H., Zhao, F., Ding, X., Zeng, R., et al. (2008). Detection of QTLs with additive effects and additive-by-environment interaction effects on panicle number in rice (Oryza sativa L.) with single-segment substitution lines. Theor. Appl. Genet. 116, 923–931. doi: 10.1007/s00122-008-0724-4
Liu, G. M., Li, W. T., Zeng, R. Z., Zhang, Z. M., and Zhang, G. Q. (2004). [Identification of QTLs on substituted segments in single segment substitution lines of rice]. Yi Chuan Xue Bao 31, 1395–1400.
Liu, X., Zhao, Z., Liu, L., Xiao, Y., Tian, Y., Liu, S.-J., et al. (2016). Construction of chromosomal segment substitution lines and genetic dissection of introgressed segments associated with yield determination in the parents of a super-hybrid rice. Plant Breed. 135, 63–72. doi: 10.1111/pbr.12329
Liu, Y., Wang, H., Jiang, Z., Wang, W., Xu, R., Wang, Q., et al. (2021). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590, 600–605. doi: 10.1038/s41586-020-03091-w
Luan, X., Dai, Z., Yang, W., Tan, Q., Lu, Q., Guo, J., et al. (2019). Breeding by design of CMS lines on the platform of SSSL library in rice. Mol. Breed. 39:126. doi: 10.1007/s11032-019-1028-x
Luo, J., Liu, H., Zhou, T., Gu, B., Huang, X., Shangguan, Y., et al. (2013). An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25, 3360–3376. doi: 10.1105/tpc.113.113589
Luo, J. S., Huang, J., Zeng, D. L., Peng, J. S., Zhang, G. B., Ma, H. L., et al. (2018). A defensin-like protein drives cadmium efflux and allocation in rice. Nat. Commun. 9:645. doi: 10.1038/s41467-018-03088-0
Luo, Y., Ma, T., Zhang, A., Ong, K. H., Li, Z., Yang, J., et al. (2016). Marker-assisted breeding of the rice restorer line Wanhui 6725 for disease resistance, submergence tolerance and aromatic fragrance. Rice 9:66. doi: 10.1186/s12284-016-0139-9
Ma, X., Fu, Y., Zhao, X., Jiang, L., Zhu, Z., Gu, P., et al. (2016). Genomic structure analysis of a set of Oryza nivara introgression lines and identification of yield-associated QTLs using whole-genome resequencing. Sci. Rep. 6:27425. doi: 10.1038/srep27425
Ma, X., Han, B., Tang, J., Zhang, J., Cui, D., Geng, L., et al. (2019). Construction of chromosome segment substitution lines of Dongxiang common wild rice (Oryza rufipogon Griff.) in the background of the japonica rice cultivar Nipponbare (Oryza sativa L.). Plant Physiol. Biochem. 144, 274–282. doi: 10.1016/j.plaphy.2019.09.041
Mammadov, J., Buyyarapu, R., Guttikonda, S. K., Parliament, K., Abdurakhmonov, I. Y., and Kumpatla, S. P. (2018). Wild relatives of maize, rice, cotton, and soybean: treasure troves for tolerance to biotic and abiotic stresses. Front. Plant Sci. 9:886. doi: 10.3389/fpls.2018.00886
Matsubara, K., Ogiso-Tanaka, E., Hori, K., Ebana, K., Ando, T., and Yano, M. (2012). Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 53, 709–716. doi: 10.1093/pcp/pcs028
Mi, J., Yang, D., Chen, Y., Jiang, J., Mou, H., Huang, J., et al. (2018). Accelerated molecular breeding of a novel P/TGMS line with broad-spectrum resistance to rice blast and bacterial blight in two-line hybrid rice. Rice 11:11. doi: 10.1186/s12284-018-0203-8
Michael, J. T. (2014). High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotechnol. 2, 195–212. doi: 10.9787/PBB.2014.2.3.195
Mitsuya, S., Murakami, N., Sato, T., Kazama, T., Toriyama, K., Skoulding, N. S., et al. (2019). Evaluation of rice grain yield and yield components of Nona Bokra chromosome segment substitution lines with the genetic background of Koshihikari, in a saline paddy field. AoB Plants 11:plz040. doi: 10.1093/aobpla/plz040
Mizobuchi, R., Sato, H., Fukuoka, S., Tsushima, S., Imbe, T., and Yano, M. (2013). Identification of qRBS1, a QTL involved in resistance to bacterial seedling rot in rice. Theor. Appl. Genet. 126, 2417–2425. doi: 10.1007/s00122-013-2145-2
Mizuno, Y., Yamanouchi, U., Hoshino, T., Nonoue, Y., Nagata, K., Fukuoka, S., et al. (2018). Genetic dissection of pre-harvest sprouting resistance in an upland rice cultivar. Breed. Sci. 68, 200–209. doi: 10.1270/jsbbs.17062
Monteverde, E., Gutierrez, L., Blanco, P., Pérez de Vida, F., Rosas, J. E., Bonnecarrère, V., et al. (2019). Integrating molecular markers and environmental covariates to interpret genotype by environment interaction in rice (Oryza sativa L.) grown in subtropical areas. G3 9, 1519–1531. doi: 10.1534/g3.119.400064
Mulsanti, I. W., Yamamoto, T., Ueda, T., Samadi, A. F., Kamahora, E., Rumanti, I. A., et al. (2018). Finding the superior allele of japonica-type for increasing stem lodging resistance in indica rice varieties using chromosome segment substitution lines. Rice 11:25. doi: 10.1186/s12284-018-0216-3
Munguambe, N. E., Inoue, S., Demeter, Z., Yamagata, Y., Yasui, H., Zheng, S. H., et al. (2021). Substitution mapping of a locus responsible for hybrid breakdown in populations derived from interspecific introgression line. Front. Plant Sci. 12:633247. doi: 10.3389/fpls.2021.633247
Murata, K., Iyama, Y., Yamaguchi, T., Ozaki, H., Kidani, Y., and Ebitani, T. (2014). Identification of a novel gene (Apq1) from the indica rice cultivar ‘Habataki’ that improves the quality of grains produced under high temperature stress. Breed. Sci. 64, 273–281. doi: 10.1270/jsbbs.64.273
Nounjan, N., Siangliw, J. L., Toojinda, T., Chadchawan, S., and Theerakulpisut, P. (2016). Salt-responsive mechanisms in chromosome segment substitution lines of rice (Oryza sativa L. cv. KDML105). Plant Physiol. Biochem. 103, 96–105. doi: 10.1016/j.plaphy.2016.02.038
Ogami, T., Yasui, H., Yoshimura, A., and Yamagata, Y. (2019). Identification of anther length QTL and construction of chromosome segment substitution lines of Oryza longistaminata. Plants 8:388. doi: 10.3390/plants8100388
Ookawa, T., Aoba, R., Yamamoto, T., Ueda, T., Takai, T., Fukuoka, S., et al. (2016). Precise estimation of genomic regions controlling lodging resistance using a set of reciprocal chromosome segment substitution lines in rice. Sci. Rep. 6:30572. doi: 10.1038/srep30572
Park, D. S., Lee, S. K., Lee, J. H., Song, M. Y., Song, S. Y., Kwak, D. Y., et al. (2007). The identification of candidate rice genes that confer resistance to the brown planthopper (Nilaparvata lugens) through representational difference analysis. Theor. Appl. Genet. 115, 537–547. doi: 10.1007/s00122-007-0587-0
Prahalada, G., Marathi, B., Vinarao, R., Kim, S., Diocton, R., Ramos, J., et al. (2021). QTL mapping of a novel genomic region associated with high out-crossing rate derived from Oryza longistaminata and development of new CMS lines in rice, O. sativa L. Rice 14:80. doi: 10.1186/s12284-021-00521-9
Prasanth, V. V., Basava, K. R., Babu, M. S., Vt, G. N. V., Devi, S. J., Mangrauthia, S. K., et al. (2016). Field level evaluation of rice introgression lines for heat tolerance and validation of markers linked to spikelet fertility. Physiol. Mol. Biol. Plants 22, 179–192. doi: 10.1007/s12298-016-0350-6
Qi, L., Sun, Y., Li, J., Su, L., Zheng, X., Wang, X., et al. (2017). Identify QTLs for grain size and weight in common wild rice using chromosome segment substitution lines across six environments. Breed. Sci. 67, 472–482. doi: 10.1270/jsbbs.16082
Qiao, W., Qi, L., Cheng, Z., Su, L., Li, J., Sun, Y., et al. (2016). Development and characterization of chromosome segment substitution lines derived from Oryza rufipogon in the genetic background of O. sativa spp. indica cultivar 9311. BMC Genomics 17:580. doi: 10.1186/s12864-016-2987-5
Qin, P., Lu, H., Du, H., Wang, H., Chen, W., Chen, Z., et al. (2021). Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184, 3542–3558.e16. doi: 10.1016/j.cell.2021.04.046
Qiu, X., Chen, K., Lv, W., Ou, X., Zhu, Y., Xing, D., et al. (2017). Examining two sets of introgression lines reveals background-independent and stably expressed QTL that improve grain appearance quality in rice (Oryza sativa L.). Theor. Appl. Genet. 130, 951–967. doi: 10.1007/s00122-017-2862-z
Qiu, X., Yuan, Z., Liu, H., Xiang, X., Yang, L., He, W., et al. (2015). Identification of salt tolerance-improving quantitative trait loci alleles from a salt-susceptible rice breeding line by introgression breeding. Plant Breed. 134, 653–660. doi: 10.1111/pbr.12321
Ramos, J., Furuta, T., Uehara, K., Chihiro, N., Angeles-Shim, R., Shim, J., et al. (2016). Development of chromosome segment substitution lines (CSSLs) of Oryza longistaminata A. Chev. & Röhr in the background of the elite japonica rice cultivar, Taichung 65 and their evaluation for yield traits. Euphytica 210, 151–163. doi: 10.1007/s10681-016-1685-3
Rao, Y., Li, Y., and Qian, Q. (2014). Recent progress on molecular breeding of rice in China. Plant Cell Rep. 33, 551–564. doi: 10.1007/s00299-013-1551-x
Rao, Y. V., Balakrishnan, D., Addanki, K. R., Mesapogu, S., Kiran, T. V., Subrahmanyam, D., et al. (2018). Characterization of backcross introgression lines derived from Oryza nivara accessions for photosynthesis and yield. Physiol. Mol. Biol. Plants 24, 1147–1164. doi: 10.1007/s12298-018-0575-7
Sanchez, P. L., Wing, R. A., and Brar, D. S. (2013). “The wild relative of rice: Genomes and genomics,” in Genetics and Genomics of Rice, eds Q. Zhang and R. A. Wing (New York, NY: Springer New York), 9–25. doi: 10.1007/978-1-4614-7903-1_2
Shen, G., Hu, W., Wang, X., Zhou, X., Han, Z., Sherif, A., et al. (2022). Assembly of yield heterosis of an elite rice hybrid is promising by manipulating dominant QTLs. J. Integr. Plant Biol. 64, 688–701. doi: 10.1111/jipb.13220
Shen, G., and Xing, Y. (2014). Two novel QTLs for heading date are identified using a set of chromosome segment substitution lines in rice (Oryza sativa L.). J. Genet. Genomics 41, 659–662. doi: 10.1016/j.jgg.2014.08.008
Shen, G., Zhan, W., Chen, H., and Xing, Y. (2014). Dominance and epistasis are the main contributors to heterosis for plant height in rice. Plant Sci. 215–216, 11–18. doi: 10.1016/j.plantsci.2013.10.004
Shi, C. L., Dong, N. Q., Guo, T., Ye, W. W., Shan, J. X., and Lin, H. X. (2020). A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 103, 1174–1188. doi: 10.1111/tpj.14793
Shibaya, T., Hori, K., Ogiso-Tanaka, E., Yamanouchi, U., Shu, K., Kitazawa, N., et al. (2016). Hd18, encoding histone acetylase related to Arabidopsis FLOWERING LOCUS D, is involved in the control of flowering time in rice. Plant Cell Physiol. 57, 1828–1838. doi: 10.1093/pcp/pcw105
Shim, R. A., Angeles, E. R., Ashikari, M., and Takashi, T. (2010). Development and evaluation of Oryza glaberrima Steud. chromosome segment substitution lines (CSSLs) in the background of O. sativa L. cv. Koshihikari. Breed. Sci. 60, 613–619. doi: 10.1270/jsbbs.60.613
Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S., et al. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028. doi: 10.1038/ng.169
Singh, N., Wang, D. R., Ali, L., Kim, H., Akther, K. M., Harrington, S. E., et al. (2020). A coordinated suite of wild-introgression lines in indica and japonica elite backgrounds. Front. Plant Sci. 11:564824. doi: 10.3389/fpls.2020.564824
Su, S., Hong, J., Chen, X., Zhang, C., Chen, M., Luo, Z., et al. (2021). Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice. Plant Biotechnol. J. 19, 2304–2318. doi: 10.1111/pbi.13661
Sui, F., Zhao, D., Zhu, H., Gong, Y., Tang, Z., Huang, X. Y., et al. (2019). Map-based cloning of a new total loss-of-function allele of OsHMA3 causes high cadmium accumulation in rice grain. J. Exp. Bot. 70, 2857–2871. doi: 10.1093/jxb/erz093
Sun, H., Qian, Q., Wu, K., Luo, J., Wang, S., Zhang, C., et al. (2014). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46, 652–656. doi: 10.1038/ng.2958
Sun, W., Zhou, Q., Yao, Y., Qiu, X., Xie, K., and Yu, S. (2015). Identification of genomic regions and the isoamylase gene for reduced grain chalkiness in rice. PLoS One 10:e0122013. doi: 10.1371/journal.pone.0122013
Sun, Z., Yin, X., Ding, J., Yu, D., Hu, M., Sun, X., et al. (2017). QTL analysis and dissection of panicle components in rice using advanced backcross populations derived from Oryza sativa cultivars HR1128 and ‘Nipponbare’. PLoS One 12:e0175692. doi: 10.1371/journal.pone.0175692
Suralta, R. R., Inukai, Y., and Yamauchi, A. (2008). Utilizing chromosome segment substitution lines (CSSLs) for evaluation of root responses to transient moisture stresses in rice. Plant Prod. Sci. 11, 457–465. doi: 10.1626/pps.11.457
Takai, T., Ikka, T., Kondo, K., Nonoue, Y., Ono, N., Arai-Sanoh, Y., et al. (2014). Genetic mechanisms underlying yield potential in the rice high-yielding cultivar Takanari, based on reciprocal chromosome segment substitution lines. BMC Plant Biol. 14:295. doi: 10.1186/s12870-014-0295-2
Takai, T., Nonoue, Y., Yamamoto, S.-I., Yamanouchi, U., Matsubara, K., Liang, Z.-W., et al. (2007). Development of chromosome segment substitution lines derived from backcross between indica donor rice cultivar ‘Nona Bokra’ and japonica recipient cultivar ‘Koshihikari’. Breed. Sci. 57, 257–261. doi: 10.1270/jsbbs.57.257
Takai, T., Ohsumi, A., San-oh, Y., Laza, M. R., Kondo, M., Yamamoto, T., et al. (2009). Detection of a quantitative trait locus controlling carbon isotope discrimination and its contribution to stomatal conductance in japonica rice. Theor. Appl. Genet. 118, 1401–1410. doi: 10.1007/s00122-009-0990-9
Tan, L., Li, X., Liu, F., Sun, X., Li, C., Zhu, Z., et al. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364. doi: 10.1038/ng.197
Tan, L., Liu, F., Xue, W., Wang, G., Ye, S., Zhu, Z., et al. (2007). Development of Oryza rufipogon and O. sativa introgression lines and assessment for yield-related quantitative trait loci. J. Integr. Plant Biol. 49, 871–884. doi: 10.1111/j.1744-7909.2007.00497.x
Tan, Q., Zou, T., Zheng, M., Ni, Y., Luan, X., Li, X., et al. (2020). Substitution mapping of the major quantitative trait loci controlling stigma exsertion rate from Oryza glumaepatula. Rice 13:37. doi: 10.1186/s12284-020-00397-1
Tanksley, S. D., and McCouch, S. R. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066. doi: 10.1126/science.277.5329.1063
Tao, Y., Zhu, J., Xu, J., Wang, L., Gu, H., Zhou, R., et al. (2016). Exploitation of heterosis loci for yield and yield components in rice using chromosome segment substitution lines. Sci. Rep. 6:36802. doi: 10.1038/srep36802
Thein, H. W., Yamagata, Y., Van Mai, T., and Yasui, H. (2019). Four resistance alleles derived from Oryza longistaminata (A. Chev. & Roehrich) against green rice leafhopper, Nephotettix cincticeps (Uhler) identified using novel introgression lines. Breed. Sci. 69, 573–584. doi: 10.1270/jsbbs.19060
Tian, L., Tan, L., Liu, F., Cai, H., and Sun, C. (2011). Identification of quantitative trait loci associated with salt tolerance at seedling stage from Oryza rufipogon. J. Genet. Genomics 38, 593–601. doi: 10.1016/j.jgg.2011.11.005
Tian, Z., Qian, Q., Liu, Q., Yan, M., Liu, X., Yan, C., et al. (2009). Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. U.S.A. 106, 21760. doi: 10.1073/pnas.0912396106
Uga, Y., Kitomi, Y., Yamamoto, E., Kanno, N., Kawai, S., Mizubayashi, T., et al. (2015). A QTL for root growth angle on rice chromosome 7 is involved in the genetic pathway of DEEPER ROOTING 1. Rice 8:8. doi: 10.1186/s12284-015-0044-7
Ujiie, K., Yamamoto, T., Yano, M., and Ishimaru, K. (2016). Genetic factors determining varietal differences in characters affecting yield between two rice (Oryza sativa L.) varieties, Koshihikari and IR64. Genet. Resour. Crop Evol. 63, 97–123. doi: 10.1007/s10722-015-0237-3
Vogel, K. E. (2009). Backcross breeding. Methods Mol. Biol. 526, 161–169. doi: 10.1007/978-1-59745-494-0_14
Wan, J. L., Zhai, H. Q., Wan, J. M., Yasui, H., and Yoshimura, A. (2003). Mapping QTL for traits associated with resistance to ferrous iron toxicity in rice (Oryza sativa L.), using japonica chromosome segment substitution lines. Yi Chuan Xue Bao 30, 893–898.
Wang, D., Sun, W., Yuan, Z., Sun, Q., Fan, K., Zhang, C., et al. (2021). Identification of a novel QTL and candidate gene associated with grain size using chromosome segment substitution lines in rice. Sci. Rep. 11:189. doi: 10.1038/s41598-020-80667-6
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Wang, Y., Wang, J., Zhai, L., Liang, C., Chen, K., and Xu, J. (2020). Identify QTLs and candidate genes underlying source-, sink-, and grain yield-related traits in rice by integrated analysis of bi-parental and natural populations. PLoS One 15:e0237774. doi: 10.1371/journal.pone.0237774
Wang, Z., Yu, C., Liu, X., Liu, S., Yin, C., Liu, L., et al. (2012). Identification of indica rice chromosome segments for the improvement of japonica inbreds and hybrids. Theor. Appl. Genet. 124, 1351–1364. doi: 10.1007/s00122-012-1792-z
Wei, X., Qiu, J., Yong, K., Fan, J., Zhang, Q., Hua, H., et al. (2021). A quantitative genomics map of rice provides genetic insights and guides breeding. Nat. Genet. 53, 243–253. doi: 10.1038/s41588-020-00769-9
Weng, J., Gu, S., Wan, X., Gao, H., Guo, T., Su, N., et al. (2008). Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18, 1199–1209. doi: 10.1038/cr.2008.307
Wing, R. A., Purugganan, M. D., and Zhang, Q. (2018). The rice genome revolution: from an ancient grain to Green Super Rice. Nat. Rev. Genet. 19, 505–517. doi: 10.1038/s41576-018-0024-z
Wu, B., Hu, W., and Xing, Y. Z. (2018). The history and prospect of rice genetic breeding in China. Yi Chuan 40, 841–857. doi: 10.16288/j.yczz.18-213
Wu, Y., Wang, Y., Mi, X. F., Shan, J. X., Li, X. M., Xu, J. L., et al. (2016). The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 12:e1006386. doi: 10.1371/journal.pgen.1006386
Xi, Z. Y., He, F. H., Zeng, R. Z., Zhang, Z. M., Ding, X. H., Li, W. T., et al. (2006). Development of a wide population of chromosome single-segment substitution lines in the genetic background of an elite cultivar of rice (Oryza sativa L.). Genome 49, 476–484. doi: 10.1139/g06-005
Xia, D., Zhou, H., Liu, R., Dan, W., Li, P., Wu, B., et al. (2018). GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically Interacts with GS3 to produce extra-long grains in rice. Mol. Plant 11, 754–756. doi: 10.1016/j.molp.2018.03.006
Xin, X. Y., Wang, W. X., Yang, J. S., and Luo, X. J. (2011). Genetic analysis of heterotic loci detected in a cross between indica and japonica rice (Oryza sativa L.). Breed. Sci. 61, 380–388. doi: 10.1270/jsbbs.61.380
Xiong, Y., Zhang, C., Zhou, H., Sun, W., Wang, P., Wang, D., et al. (2021). Identification of heterotic loci with desirable allelic interaction to increase yield in rice. Rice 14:97. doi: 10.1186/s12284-021-00539-z
Xu, J., Shang, L., Wang, J., Chen, M., Fu, X., He, H., et al. (2021). The SEEDLING BIOMASS 1 allele from indica rice enhances yield performance under low-nitrogen environments. Plant Biotechnol. J. 19, 1681–1683. doi: 10.1111/pbi.13642
Xu, J., Zhao, Q., Du, P., Xu, C., Wang, B., Feng, Q., et al. (2010). Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.). BMC Genomics 11:656. doi: 10.1186/1471-2164-11-656
Xu, P., Dong, L., Zhou, J., Li, J., Zhang, Y., Hu, F., et al. (2015). Identification and mapping of a novel blast resistance gene Pi57(t) in Oryza longistaminata. Euphytica 205, 95–102. doi: 10.1007/s10681-015-1402-7
Xu, X., Wu, K., Xu, R., Yu, J., Wang, J., Zhao, Y., et al. (2019). Pyramiding of the dep1-1 and NAL1NJ6 alleles achieves sustainable improvements in nitrogen-use efficiency and grain yield in japonica rice breeding. J. Genet. Genomics 46, 325–328. doi: 10.1016/j.jgg.2019.02.009
Xu, Z., Li, S., Zhang, C., Zhang, B., Zhu, K., Zhou, Y., et al. (2017). Genetic connection between cell-wall composition and grain yield via parallel QTL analysis in indica and japonica subspecies. Sci. Rep. 7:12561. doi: 10.1038/s41598-017-12903-5
Yadav, A. K., Kumar, A., Grover, N., Ellur, R. K., Krishnan, S. G., Bollinedi, H., et al. (2020). Marker aided introgression of ‘Saltol’, a major QTL for seedling stage salinity tolerance into an elite Basmati rice variety ‘Pusa Basmati 1509’. Sci. Rep. 10:13877. doi: 10.1038/s41598-020-70664-0
Yadav, S., Sandhu, N., Majumder, R. R., Dixit, S., Kumar, S., Singh, S. P., et al. (2019). Epistatic interactions of major effect drought QTLs with genetic background loci determine grain yield of rice under drought stress. Sci. Rep. 9:2616. doi: 10.1038/s41598-019-39084-7
Yamagata, Y., Win, K. T., Miyazaki, Y., Ogata, C., Yasui, H., and Yoshimura, A. (2019). Development of introgression lines of AA genome Oryza species, O. glaberrima, O. rufipogon, and O. nivara, in the genetic background of O. sativa L. cv. Taichung 65. Breed. Sci. 69, 359–363. doi: 10.1270/jsbbs.19002
Yang, C., Zhang, T., Wang, H., Zhao, N., and Liu, B. (2012). Heritable alteration in salt-tolerance in rice induced by introgression from wild rice (Zizania latifolia). Rice 5:36. doi: 10.1186/1939-8433-5-36
Yang, W., Guo, Z., Huang, C., Duan, L., Chen, G., Jiang, N., et al. (2014). Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 5:5087. doi: 10.1038/ncomms6087
Yang, W., Zhang, F., Zafar, S., Wang, J., Lu, H., Naveed, S., et al. (2021c). Genetic dissection of heterosis of indica-japonica by introgression line, recombinant inbred line and their testcross populations. Sci. Rep. 11:10265. doi: 10.1038/s41598-021-89691-6
Yang, W., Xiong, L., Liang, J., Hao, Q., Luan, X., Tan, Q., et al. (2021b). Substitution mapping of two closely linked QTLs on chromosome 8 controlling grain chalkiness in rice. Rice 14:85. doi: 10.1186/s12284-021-00526-4
Yang, W., Liang, J., Hao, Q., Luan, X., Tan, Q., Lin, S., et al. (2021a). Fine mapping of two grain chalkiness QTLs sensitive to high temperature in rice. Rice 14:33. doi: 10.1186/s12284-021-00476-x
Yang, Y., Guo, M., Sun, S., Zou, Y., Yin, S., Liu, Y., et al. (2019). Natural variation of OsGluA2 is involved in grain protein content regulation in rice. Nat. Commun. 10:1949. doi: 10.1038/s41467-019-09919-y
Yang, Y., Zhou, J., Li, J., Xu, P., Zhang, Y., and Tao, D. (2016). Mapping QTLs for hybrid sterility in three AA genome wild species of Oryza. Breed. Sci. 66, 367–371. doi: 10.1270/jsbbs.15048
Yasui, H., Yamagata, Y., and Yoshimura, A. (2010). Development of chromosome segment substitution lines derived from indica rice donor cultivars DV85 and ARC10313 in the genetic background of japonica cultivar Taichung 65. Breed. Sci. 60, 620–628. doi: 10.1270/jsbbs.60.620
Ye, G., Liang, S., and Wan, J. (2010). QTL mapping of protein content in rice using single chromosome segment substitution lines. Theor. Appl. Genet. 121, 741–750. doi: 10.1007/s00122-010-1345-2
Yu, B., Lin, Z., Li, H., Li, X., Li, J., Wang, Y., et al. (2007). TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 52, 891–898. doi: 10.1111/j.1365-313X.2007.03284.x
Yu, S., Ali, J., Zhang, C., Li, Z., and Zhang, Q. (2020). Genomic breeding of Green Super Rice varieties and their deployment in Asia and Africa. Theor. Appl. Genet. 133, 1427–1442. doi: 10.1007/s00122-019-03516-9
Yuan, R., Zhao, N., Usman, B., Luo, L., Liao, S., Qin, Y., et al. (2020). Development of chromosome segment substitution lines (CSSLs) derived from Guangxi wild rice (Oryza rufipogon Griff.) under rice (Oryza sativa L.) background and the identification of QTLs for plant architecture, agronomic traits and cold tolerance. Genes 11:980. doi: 10.3390/genes11090980
Yuan, S., Wang, Y., Zhang, C., He, H., and Yu, S. (2020). Genetic dissection of seed dormancy using chromosome segment substitution lines in rice (Oryza sativa L.). Int. J. Mol. Sci. 21:1344. doi: 10.3390/ijms21041344
Yuan, Z., Fan, K., Xia, L., Ding, X., Tian, L., Sun, W., et al. (2019). Genetic dissection of seed storability and validation of candidate gene associated with antioxidant capability in rice (Oryza sativa L.). Int. J. Mol. Sci. 20:4442. doi: 10.3390/ijms20184442
Yun, Y. T., Chung, C. T., Lee, Y. J., Na, H. J., Lee, J. C., Lee, S. G., et al. (2016). QTL mapping of grain quality traits using introgression lines carrying Oryza rufipogon chromosome segments in japonica rice. Rice 9:62. doi: 10.1186/s12284-016-0135-0
Zang, J., Sun, Y., Wang, Y., Yang, J., Li, F., Zhou, Y., et al. (2008). Dissection of genetic overlap of salt tolerance QTLs at the seedling and tillering stages using backcross introgression lines in rice. Sci. China C Life Sci. 51, 583–591. doi: 10.1007/s11427-008-0081-1
Zeng, D., Tian, Z., Rao, Y., Dong, G., Yang, Y., Huang, L., et al. (2017). Rational design of high-yield and superior-quality rice. Nat. Plants 3:17031. doi: 10.1038/nplants.2017.31
Zhan, P., Ma, S., Xiao, Z., Li, F., Wei, X., Lin, S., et al. (2022). Natural variations in grain length 10 (GL10) regulate rice grain size. J. Genet. Genomics. [Epub ahead of print]. doi: 10.1016/j.jgg.2022.01.008
Zhang, F., Shi, Y., Ali, J., Xu, J., and Li, Z. (2021c). Breeding by selective introgression: theory, practices, and lessons learned from rice. Crop J. 9, 646–657. doi: 10.1016/j.cj.2021.03.006
Zhang, B., Qi, F., Hu, G., Yang, Y., Zhang, L., Meng, J., et al. (2021a). BSA-seq-based identification of a major additive plant height QTL with an effect equivalent to that of Semi-dwarf 1 in a large rice F2 population. Crop J. 9, 1428–1437. doi: 10.1016/j.cj.2020.11.011
Zhang, C., Yang, Y., Chen, S., Liu, X., Zhu, J., Zhou, L., et al. (2021b). A rare Waxy allele coordinately improves rice eating and cooking quality and grain transparency. J. Integr. Plant. Biol. 63, 889–901. doi: 10.1111/jipb.13010
Zhang, C., Yuan, Z., Wang, Y., Sun, W., Tang, X., Sun, Y., et al. (2020a). Genetic dissection of seed dormancy in rice (Oryza sativa L.) by using two mapping populations derived from common parents. Rice 13:52. doi: 10.1186/s12284-020-00413-4
Zhang, Z., Gao, S., and Chu, C. (2020c). Improvement of nutrient use efficiency in rice: current toolbox and future perspectives. Theor. Appl. Genet. 133, 1365–1384. doi: 10.1007/s00122-019-03527-6
Zhang, K., Su, J., Xu, M., Zhou, Z., Zhu, X., Ma, X., et al. (2020b). A common wild rice-derived BOC1 allele reduces callus browning in indica rice transformation. Nat. Commun. 11:443. doi: 10.1038/s41467-019-14265-0
Zhang, G. (2021). Target chromosome-segment substitution: a way to breeding by design in rice. Crop J. 9, 658–668. doi: 10.1016/j.cj.2021.03.001
Zhang, H., Zhao, Q., Sun, Z. Z., Zhang, C. Q., Feng, Q., Tang, S. Z., et al. (2011). Development and high-throughput genotyping of substitution lines carring the chromosome segments of indica 9311 in the background of japonica Nipponbare. J. Genet. Genomics 38, 603–611. doi: 10.1016/j.jgg.2011.11.004
Zhang, W., Tan, L., Sun, H., Zhao, X., Liu, F., Cai, H., et al. (2019d). Natural variations at TIG1 encoding a TCP transcription factor contribute to plant architecture domestication in rice. Mol. Plant 12, 1075–1089. doi: 10.1016/j.molp.2019.04.005
Zhang, B., Liu, H., Qi, F., Zhang, Z., Li, Q., Han, Z., et al. (2019a). Genetic interactions among Ghd7, Ghd8, OsPRR37 and Hd1 contribute to large variation in heading date in rice. Rice 12:48. doi: 10.1186/s12284-019-0314-x
Zhang, B., Shang, L., Ruan, B., Zhang, A., Yang, S., Jiang, H., et al. (2019b). Development of three sets of high-throughput genotyped rice chromosome segment substitution lines and QTL mapping for eleven traits. Rice 12:33. doi: 10.1186/s12284-019-0293-y
Zhang, H., Zhou, L., Xu, H., Wang, L., Liu, H., Zhang, C., et al. (2019c). The qSAC3 locus from indica rice effectively increases amylose content under a variety of conditions. BMC Plant Biol. 19:275. doi: 10.1186/s12870-019-1860-5
Zhang, X., Zhou, S., Fu, Y., Su, Z., Wang, X., and Sun, C. (2006). Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.). Plant Mol. Biol. 62, 247–259. doi: 10.1007/s11103-006-9018-x
Zhang, Y., Tan, L., Zhu, Z., Yuan, L., Xie, D., and Sun, C. (2015). TOND1 confers tolerance to nitrogen deficiency in rice. Plant J. 81, 367–376. doi: 10.1111/tpj.12736
Zhang, Y., Zhou, J., Li, J., Yang, Y., Xu, P., and Tao, D. (2018). Mapping of S56(t) responsible for interspecific hybrid sterility between Oryza sativa and Oryza glumaepatula. Breed. Sci. 68, 242–247. doi: 10.1270/jsbbs.17116
Zhang, Y., Zhou, J., Xu, P., Li, J., Deng, X., Deng, W., et al. (2022). A genetic resource for rice improvement: introgression library of agronomic traits for all AA genome Oryza species. Front. Plant Sci. 13:856514. doi: 10.3389/fpls.2022.856514
Zhang, Z., Li, J., Pan, Y., Li, J., Zhou, L., Shi, H., et al. (2017). Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 8:14788. doi: 10.1038/ncomms14788
Zhao, C., Zhou, L., Yu, X., Zhao, Q., and Wang, C. (2012). Identification of tiller angle quantitative trait loci based on chromosome segment substituted lines and high-density physical map in rice. Chin. Bull. Bot. 47, 594–601. doi: 10.3724/sp.j.1259.2012.00594
Zhao, D. S., Li, Q. F., Zhang, C. Q., Zhang, C., Yang, Q. Q., Pan, L. X., et al. (2018). GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9:1240. doi: 10.1038/s41467-018-03616-y
Zhao, D.-S., Liu, J.-Y., Ding, A.-Q., Zhang, T., Ren, X.-Y., Zhang, L., et al. (2021). Improving grain appearance of erect-panicle japonica rice cultivars by introgression of the null gs9 allele. J. Integr. Agric. 20, 2032–2042. doi: 10.1016/S2095-3119(21)63659-6
Zhao, J., Wang, S., Qin, J., Sun, C., and Liu, F. (2020). The lipid transfer protein OsLTPL159 is involved in cold tolerance at the early seedling stage in rice. Plant Biotechnol. J. 18, 756–769. doi: 10.1111/pbi.13243
Zhao, L., Lei, J., Huang, Y., Zhu, S., Chen, H., Huang, R., et al. (2016). Mapping quantitative trait loci for heat tolerance at anthesis in rice using chromosomal segment substitution lines. Breed. Sci. 66, 358–366. doi: 10.1270/jsbbs.15084
Zhao, L., Zhou, H., Lu, L., Liu, L., Li, X., Lin, Y., et al. (2009). Identification of quantitative trait loci controlling rice mature seed culturability using chromosomal segment substitution lines. Plant Cell Rep. 28, 247–256. doi: 10.1007/s00299-008-0641-7
Zhao, Z. G., Zhu, S. S., Zhang, Y. H., Bian, X. F., Wang, Y., Jiang, L., et al. (2011). Molecular analysis of an additional case of hybrid sterility in rice (Oryza sativa L.). Planta 233, 485–494. doi: 10.1007/s00425-010-1313-8
Zhou, H., Xia, D., Zhao, D., Li, Y., Li, P., Wu, B., et al. (2021). The origin of Wxla provides new insights into the improvement of grain quality in rice. J. Integr. Plant Biol. 63, 878–888. doi: 10.1111/jipb.13011
Zhou, H., Yang, W., Ma, S., Luan, X., Zhu, H., Wang, A., et al. (2020). Unconditional and conditional analysis of epistasis between tillering QTLs based on single segment substitution lines in rice. Sci. Rep. 10:15912. doi: 10.1038/s41598-020-73047-7
Zhou, P. H., Tan, Y. F., He, Y. Q., Xu, C. G., and Zhang, Q. (2003). Simultaneous improvement for four quality traits of Zhenshan 97, an elite parent of hybrid rice, by molecular marker-assisted selection. Theor. Appl. Genet. 106, 326–331. doi: 10.1007/s00122-002-1023-0
Zhou, S. X., Tian, F., Zhu, Z. F., Fu, Y. C., Wang, X. K., and Sun, C. Q. (2006). Identification of quantitative trait loci controlling drought tolerance at seedling stage in Chinese Dongxiang common wild rice (Oryza rufipogon Griff.). Yi Chuan Xue Bao 33, 551–558. doi: 10.1016/s0379-4172(06)60084-x
Zhou, Y., Dong, G., Tao, Y., Chen, C., Yang, B., Wu, Y., et al. (2016). Mapping quantitative trait loci associated with toot traits using sequencing-based genotyping chromosome segment substitution lines derived from 9311 and Nipponbare in rice (Oryza sativa L.). PLoS One 11:e0151796. doi: 10.1371/journal.pone.0151796
Zhou, Y., Tao, Y., Yuan, Y., Zhang, Y., Miao, J., Zhang, R., et al. (2018). Characterisation of a novel quantitative trait locus, GN4-1, for grain number and yield in rice (Oryza sativa L.). Theor. Appl. Genet. 131, 637–648. doi: 10.1007/s00122-017-3025-y
Zhou, Y., Xie, Y., Cai, J., Liu, C., Zhu, H., Jiang, R., et al. (2017). Substitution mapping of QTLs controlling seed dormancy using single segment substitution lines derived from multiple cultivated rice donors in seven cropping seasons. Theor. Appl. Genet. 130, 1191–1205. doi: 10.1007/s00122-017-2881-9
Zhu, H., Li, Y., Liang, J., Luan, X., Xu, P., Wang, S., et al. (2018). Analysis of QTLs on heading date based on single segment substitution lines in rice (Oryza Sativa L.). Sci. Rep. 8:13232. doi: 10.1038/s41598-018-31377-7
Zhu, W., Lin, J., Yang, D., Zhao, L., Zhang, Y., Zhu, Z., et al. (2009). Development of chromosome segment substitution lines derived from backcross between two sequenced rice cultivars, indica recipient 93-11 and japonica donor Nipponbare. Plant Mol. Biol. Rep. 27, 126–131. doi: 10.1007/s11105-008-0054-3
Keywords: introgression line, QTL detection, genetic interaction, elite allele identification, breeding by design
Citation: Zhang B, Ma L, Wu B, Xing Y and Qiu X (2022) Introgression Lines: Valuable Resources for Functional Genomics Research and Breeding in Rice (Oryza sativa L.). Front. Plant Sci. 13:863789. doi: 10.3389/fpls.2022.863789
Received: 27 January 2022; Accepted: 01 April 2022;
Published: 26 April 2022.
Edited by:
Dayun Tao, Yunnan Academy of Agricultural Sciences, ChinaReviewed by:
Zhang Yu, Yunnan Academy of Agricultural Sciences, ChinaCopyright © 2022 Zhang, Ma, Wu, Xing and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianjin Qiu, eGpxaXUyMTZAeWFuZ3R6ZXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.