- 1National Bureau of Plant Genetic Resources, Pusa Campus, New Delhi, India
- 2Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi, India
- 3National Bureau of Plant Genetic Resources, Thrissur, India
- 4Indian Agricultural Research Institute, Pusa Campus, New Delhi, India
- 5National Institute for Plant Biotechnology, Pusa Campus, New Delhi, India
- 6Department of Plant Breeding and Genetics, Punjab Agricultural University, Punjab, India
- 7International Crops Research Institute for the Semi-Arid Tropics, Hyderabad, India
- 8The UWA School of Agriculture and Environment, The UWA Institute of Agriculture, The University of Western Australia (UWA), Perth, WA, Australia
Climate change is shifting agricultural production, which could impact the economic and cultural contexts of the oilseed industry, including sesame. Environmental threats (biotic and abiotic stresses) affect sesame production and thus yield (especially oil content). However, few studies have investigated the genetic enhancement, quality improvement, or the underlying mechanisms of stress tolerance in sesame. This study reveals the challenges faced by farmers/researchers growing sesame crops and the potential genetic and genomic resources for addressing the threats, including: (1) developing sesame varieties that tolerate phyllody, root rot disease, and waterlogging; (2) investigating beneficial agro-morphological traits, such as determinate growth, prostrate habit, and delayed response to seed shattering; (3) using wild relatives of sesame for wide hybridization; and (4) advancing existing strategies to maintain sesame production under changing climatic conditions. Future research programs need to add technologies and develop the best research strategies for economic and sustainable development.
Introduction
Sesame (Sesamum indicum L., Pedaliaceae) is the oldest known oilseed crop, domesticated nearly 3,000 years ago, and the first known oil consumed by man. It is known as the “Queen of oilseeds,” and an orphan crop because it receives little research attention. However, the demand for sesame seeds has increased in the last two decades due to high oil quality, protein content, antioxidant content, and wide adaptability in extreme climatic and edaphic environments (Myint et al., 2020). The genus, Sesamum comprises 30 “accepted” species and 17 species with “unresolved” status (Kalwij, 2012). The basic chromosome numbers for the family, Pedaliaceae are x = 8 or 13, with some tetraploids and octaploids (Kobayashi, 1991). Among the Sesamum species, S. indicum (2n = 26) is the only widely cultivated species across tropical regions, particularly for its oil and protein. Among the wild species, Sesamum angustifolium and Sesamum radiatum are cultivated infrequently in the regions of Africa (Ashri, 1998). Occasionally, sesame behaves as a perennial species (Ashri, 2007). Seeds are an important source of oil (44–57%), proteins (18–25%), carbohydrates (13.5%), and ash (5%); they also have medicinal and nutritional value (Myint et al., 2020). Sesame consumption is increasing globally due to changes in consumer lifestyle and health awareness programs. Sesame seed consumption will reach approximately $7,245 million by the end of 2024 (∼$6,559 million in 2018).1 Globally, the total annual sesame consumption as food and oil is approximately 35 and 65%, respectively. India ranks fourth in the world in sesame seed consumption after Tanzania, China, and Myanmar (Myint et al., 2020). Tanzania remains as the largest producer of the sesame seed worldwide (14.6%), followed by Myanmar (12.78%) and India (12.4%).
Globally, sesame is cultivated on 11.7 million hectares (Mha), producing 6.02 million tons (Mt) with an average seed yield of 512 kg ha–1. Continent-wise, Asia and Africa are the largest producers of sesame seeds (97%), followed by North America and Europe. European countries demand high imports of sesame seeds from other countries (Islam et al., 2012). India is the largest producer of sesame seeds, followed by Myanmar and China. Myanmar contributes approximately 9.5% of sesame exports globally. Recent (Food and Agriculture Organization Statistical Databases [FAOSTAT], 2018) data showed that China (1,223 kg ha–1) has the highest sesame seed yields, followed by Nigeria (729 kg ha–1) and Tanzania (720 kg ha–1) (Myint et al., 2020).
Global oil production [214 Million Metric Tons (MMT)] from sesame (5.5 MMT) lags behind groundnut, soybean, rapeseed, sunflower, and linseed.2 By 2030, sesame oil consumption is estimated to be approximately 100 MMT globally (Troncoso-Ponce et al., 2011). However, the global cultivated area of sesame has recently declined, mainly due to shifts in cultivation to other cash crops and due to the exposure of sesame crops to many biotic and abiotic stresses. Sesame cultivation is currently limited by low yields due to lack of production strategies. Thus, there is an urgent need to investigate the potential of sesame as a genetic resource for improving sesame production. This study reviews the challenges for sesame production and reveals its potential uses and prospects.
Conservation of Sesame Germplasm Worldwide
Improving sesame yield and quality is possible by exploring and using its genetic diversity. Sesame germplasm resources offer a broad genetic foundation for breeding (Dossa et al., 2017a). However, studies on the genetic diversity of sesame germplasm are not available, with only a few studies available on developing phenotypically desirable variable mutants through induced mutagenesis (Benson et al., 2013). As the primary center of origin, Indian sesame cultivars have broad variability for different agro-morphological traits; nevertheless, there is limited information on the genetic diversity in Indian sesame. Developing a core set of sesame germplasm (generally 10% of an entire collection) is efficient for accessing available genetic diversity (Iqbal et al., 2018). However, a major limiting factor for productivity is the narrow genetic base with single genotypes subjected to mutation and selection to develop cultivars. An alternative might be to use existing variability conserved in gene banks rather than inducing variability in existing cultivars. Such variability in crop germplasm, including wild relatives, could be used in pre-breeding programs to broaden the genetic base of various desirable traits for introgression into existing cultivars.
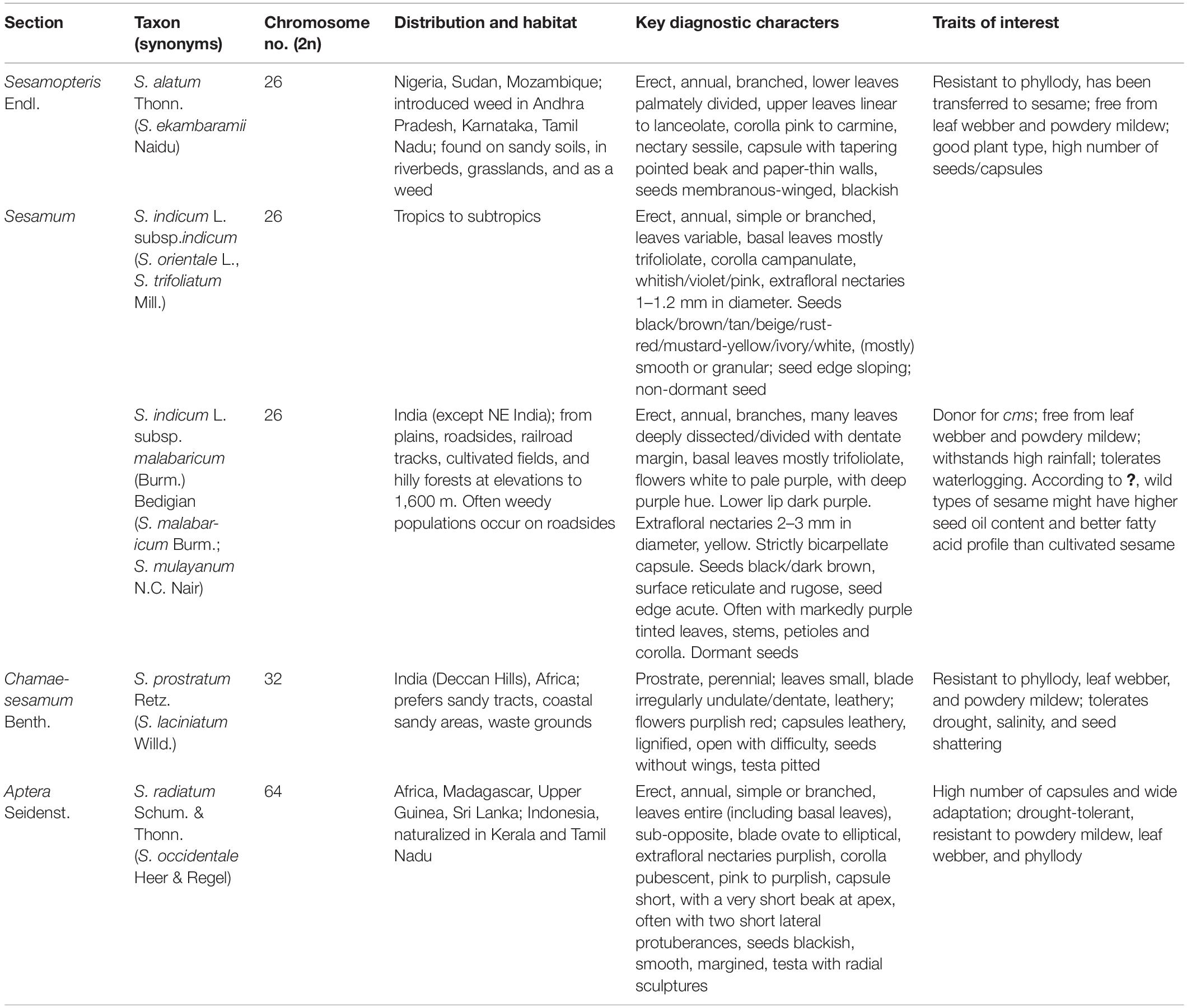
Earlier studies classified the genus, Sesamum based on morphological and microscopic observations. Sesamum orientale var. malabaricum and Sesamum mulayanum belong to the same taxon. A wild progenitor of sesame, Sesamum indicum subsp. malabaricum, is only found on the Indian subcontinent, evidenced by morphological, cytogenetical, and molecular closeness (Hiremath and Patil, 1999; Kawase, 2000; Nanthakumar et al., 2000). In 2004, the International Plant Genetic Resource Institute (IPGRI) and National Bureau of Plant Genetic Resources (NBPGR) recognized 17 species (23 taxa, including intraspecific) in the genus Sesamum, mostly African species (IPGRI and NBPGR, 2004). Later, Sesamum laciniatum Willd. was merged with Sesamum prostratum Retz., reducing the species rank of Sesamum malabaricum to subspecies under S. indicum (Bedigian, 2015).
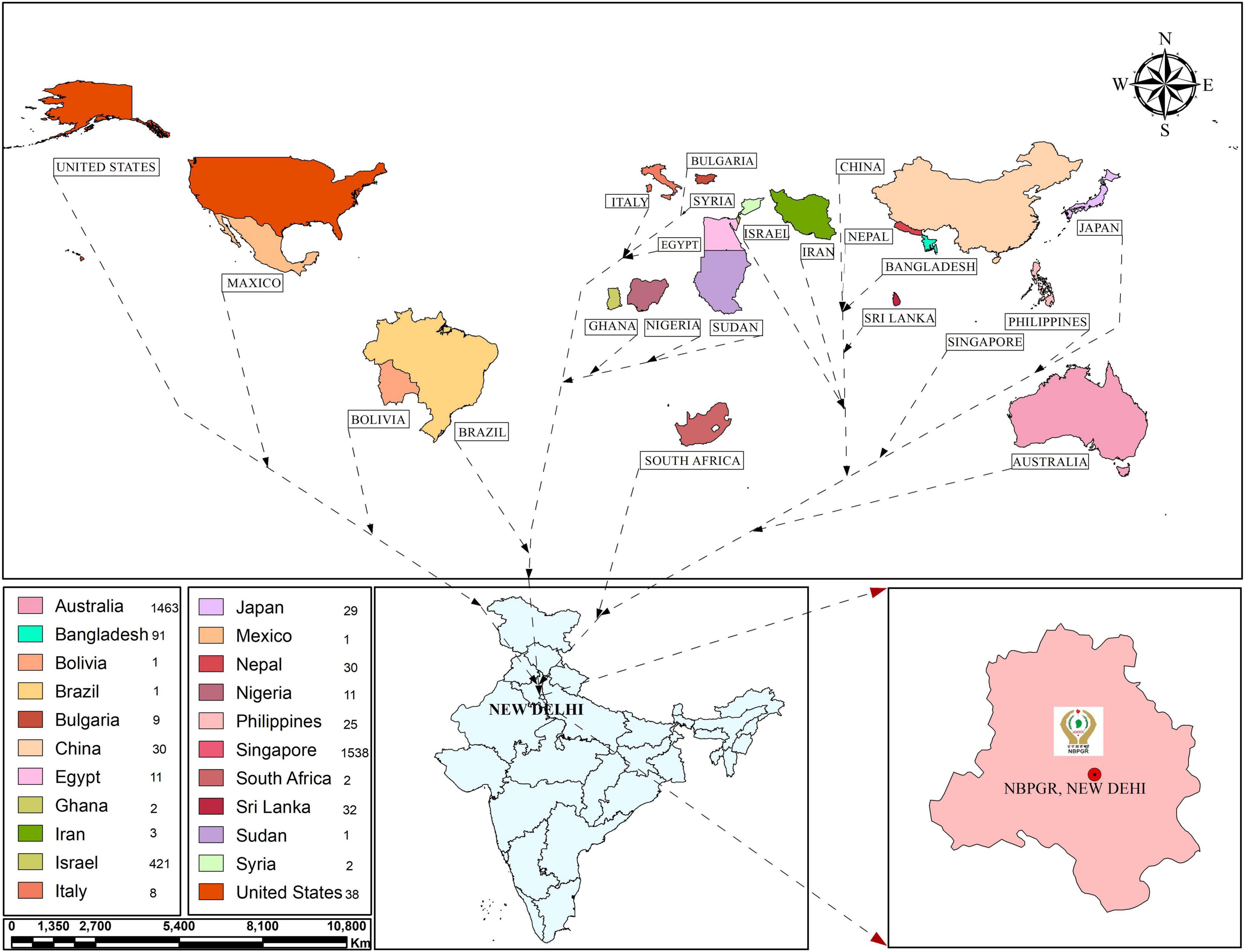
Assessing genetic diversity is paramount in crop improvement programs. Studies involving fewer than a few thousand cultivars/landraces/genotypes have been reported, especially for morphological traits. The earlier study investigated 12 agronomic traits in 2,246 sesame accessions from ten agroclimatic zones preserved in the Rural Development Administration (RDA) Genebank in Korea, establishing a core collection of 475 accessions (Kang et al., 2006). To date, 7,698 sesame accessions are stored at RDA (Park et al., 2015). Park et al. (2015) established a core collection of 278 sesame accessions at RDA Genebank from 2,751 accessions collected from 15 countries. The core set was prepared based on 10 quantitative and five qualitative traits. Similarly, Laurentin and Karlovsky (2006) carried out genetic diversity relationship studies in Venezuelan sesame germplasm (32 accessions collected from five countries). The ICAR-NBPGR, New Delhi, India, has 9,625 conserved accessions (as of December 2, 2021) of sesame germplasm in the National Genebank (NGB), including 7,217 indigenous collections (ICs) and 2,408 exotic collections (ECs) (Figure 1). These accessions were procured from various sources, mainly through the International Board of Plant Genetic Resources, Rome, Italy (now Bioversity International) supported program and world collection based in Israel. Bisht et al. (1998) reported diversity in an Indian sesame collection (3,129 accessions), stratifying the accessions into seven diversity groups, developing a core set of 172 accessions. Similarly, other workers on sesame collected 2,168 sesame germplasm from 14 countries and developed a core set of 343 accessions (Mahajan et al., 2007). Wide variation has been reported for quantitative variables, such as yield per plant, capsules per plant, seeds per capsule, height, locule number and arrangement, and the number of primary and secondary branches. Another study revealed substantial variability in S. indicum and S. mulayanum (now S. indicum subsp. malabaricum) germplasm (72 accessions) based on agro-morphological and molecular characters (Sarkar and Saha, 2013). The key traits associated with sesame and its closely related wild species are listed in Table 1. Pandey et al. (2015) reported morphological and genetic diversity in 60 sesame accessions collected from different parts of the world (India, Bangladesh, Bulgaria, and United States) based on molecular and phenotypic data. The global genebanks/seed banks procuring sesame accessions are provided in Figure 1, with India, China, and South Korea among the largest sesame germplasm holders. Further, exploration is needed to collect locally available sesame accessions for efficient genetic diversity conservation.
Significant Advances in Genomics and Genetics
Genome Sequence Resources
Sesame is a member of the family, Pedaliaceae with 16 genera. The genus, Sesamum comprises nearly 37 species distributed across Africa, India, East Indies, and Australia. Cultivated sesame, S. indicum L. (syn. Sesamum orientale L.), is a diploid species (2n = 2x = 26) with an annual crop duration of 70–150 days. Four Sesamum species have been characterized as tetraploids (2n = 4x = 32) [Sesamum angolense, S. angustifolium, S. laciniatum, and Sesamum prostratum] and three as octaploid (2n = 8x = 64) [S. radiatum, Sesamum occidentale, and Sesamum schinzianum]. The octoploid species were studied cytologically for their genomic relatedness—S. radiatum and S. occidentale were similar and homologous, with 32 bivalents and fertile hybrids. Closely related species of Sesamum are Ceratotheca sesamoides and C. triloba (2n = 32) (Hiremath and Patil, 2002). The chromosome number of Sesamothamnus busseanus is unknown, while S. triphyllum and S. capense are 2n = 26. S. tavakarii is a new species from Maharashtra, India, with further investigations required in this context. For C-values estimated from the Kew Database, the genome size (1C value) of diploid species (2n = 26) ranged from 950 Mb (S. indicum) to 1,651 Mb (S. alatum), while tetraploids and octaploids (2n = 32 and 64) ranged from 933 to 1,154 Mb and 1,306 to 1,551 Mb, respectively. The release of sesame whole-genome sequence information triggered the functional analysis of candidate genes. De novo-based assembly of the sesame genome (29 sesame accessions from 12 countries) revealed 27,148 genes and high genetic diversity in genes related to oil biosynthesis, resulting in variation in sesame oil content (Wang S. et al., 2014).
However, whole-genome sequencing of black-seeded sesame (variety GT-10) using ABI SOLiD and ion torrent estimated the genome size at 375 Mb (Vaja and Golkiya, 2016). Whole-genome sequence information from a single genotype or cultivar will not indicate genetic diversity. Hence, a pangenome is required to understand the extent of existing genomic variation and associated phenotypic variability (Golicz et al., 2016). Recently, genome assemblies have become available for five sesame varieties, including two landraces (S. indicum cv. Baizhima and Mishuozhima) and three modern cultivars (S. indicum var. Zhongzhi13, Yuzhi11, and Swetha), providing a valuable resource for comparative genomics and gene discovery (Yu et al., 2019). Pangenome studies revealed nearly 81.52, 81.87, 85.95, and 91.14% of the genome sequence in sesame cultivars, Yuzhi11, Baizhima, Mishuozhima, and Swetha, respectively (Yu et al., 2019). Two draft genomes (both at scaffold level) for Indian sesame are available in the public database—one for cv. Swetha (ASM97556v1; PRJNA219369)3 and the other for cv. GT-10 (PRJNA 284826; SAMN 3733426) (Wang S. et al., 2014) (Figure 2). Wei et al. (2017) collected genetic information and combined it with comprehensive phenotypic information in a web-based database (SesameFG) for molecular breeding and genetic improvement in sesame. Other sesame-related databases are listed in Figure 2. Therefore, studies are needed to reveal the full sesame genome sequence; in-depth knowledge of the sesame genome will facilitate the genetic improvement of sesame.
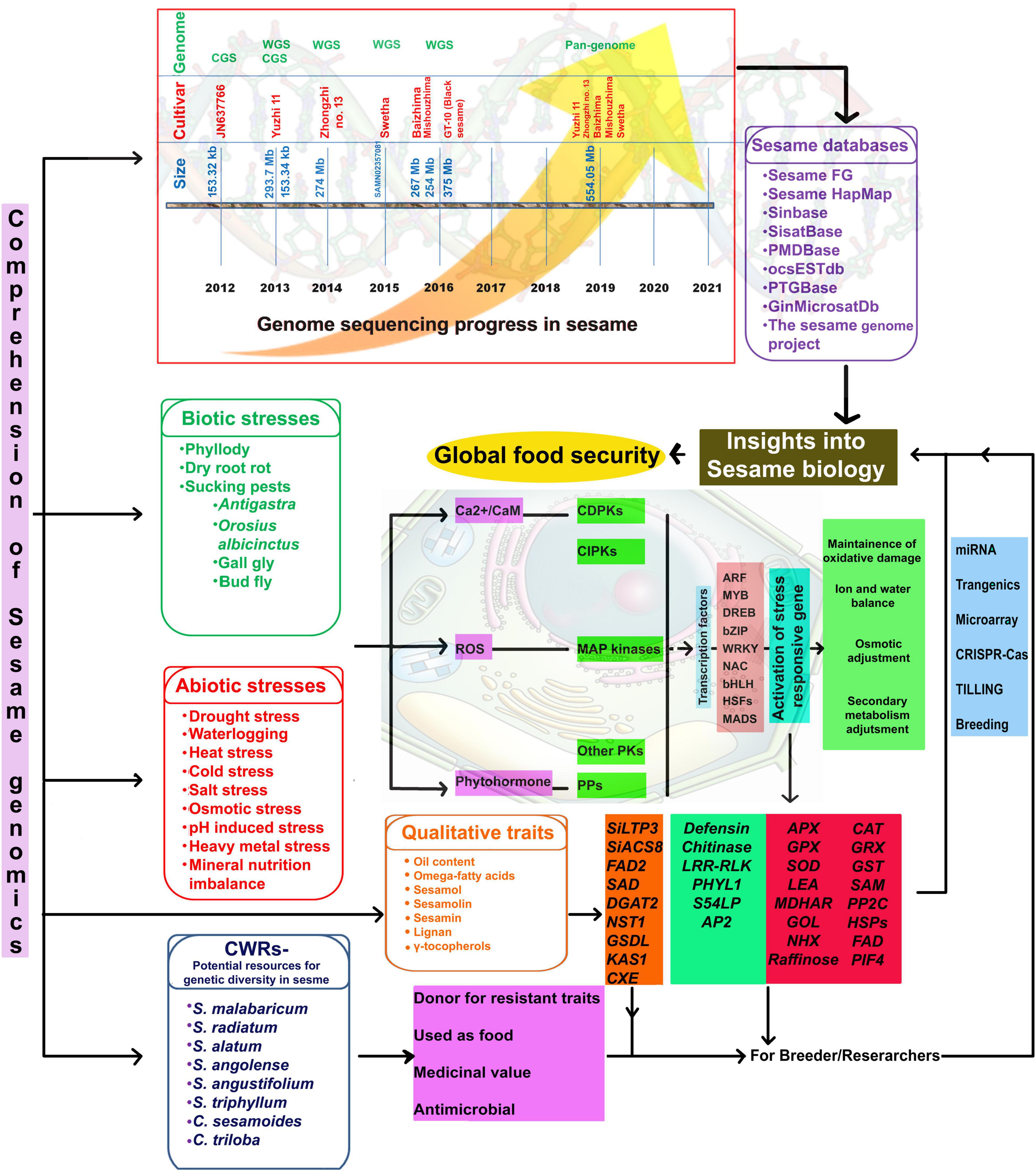
Figure 2. Comprehension of sesame genomics for global food security. CGS, Chloroplast genome sequencing; WGS, Whole-genome sequencing; CWRs, Crop wild relatives; Ca2+, Calcium ion; CaM, Calmoldulin; ROS, Reactive oxygen species; CDPKs, Calcium-dependent protein kinases; CIPKs, CBL-interacting protein kinases PPs; MAP kinases, Mitogen activated protein kinases; PKs, Protein kinases; PPs, Protein phosphatases; ARF, Auxin response factors; MYB, Myeloblastoma; DREB, Dehydration responsive elemental binding; bZIP-Basic leucine zipper domain; NAC, NAM [No apical meristem, ATAF, CUC (Cup-shaped cotyledon)]; bHLH, Basic helix loop helix; Hsfs, Heat stress specific transcription factors; MADS-M, minichromosome maintenance factor 1 from Saccharomyces cerevisiae; A for Agamous (AG) from Arabidopsis thaliana (from now on, Arabidopsis); D for deficient from Antirrhinum majus; and S for serum response factor (SRF); APX, Ascorbate peroxidase; GPX, Glutathione peroxidase; SOD, Super oxide dismutase; LEA, Late embryogenesis abundant protein; MDHAR, Monodehydro ascorbate reductase; GOL, Galactinol synthase; CAT, Catalase; GRX, Glutathione reductase; GST, Glutathione-S-transferase; SAMe, S’adenosyl-l-methionine; PP2C, Protein phosphatase 2C; HSPs, Heat shock proteins; FAD, Fatty acid desaturase; PIF4, Phytochrome-interacting transcription factor 4.
Molecular Markers in Sesame
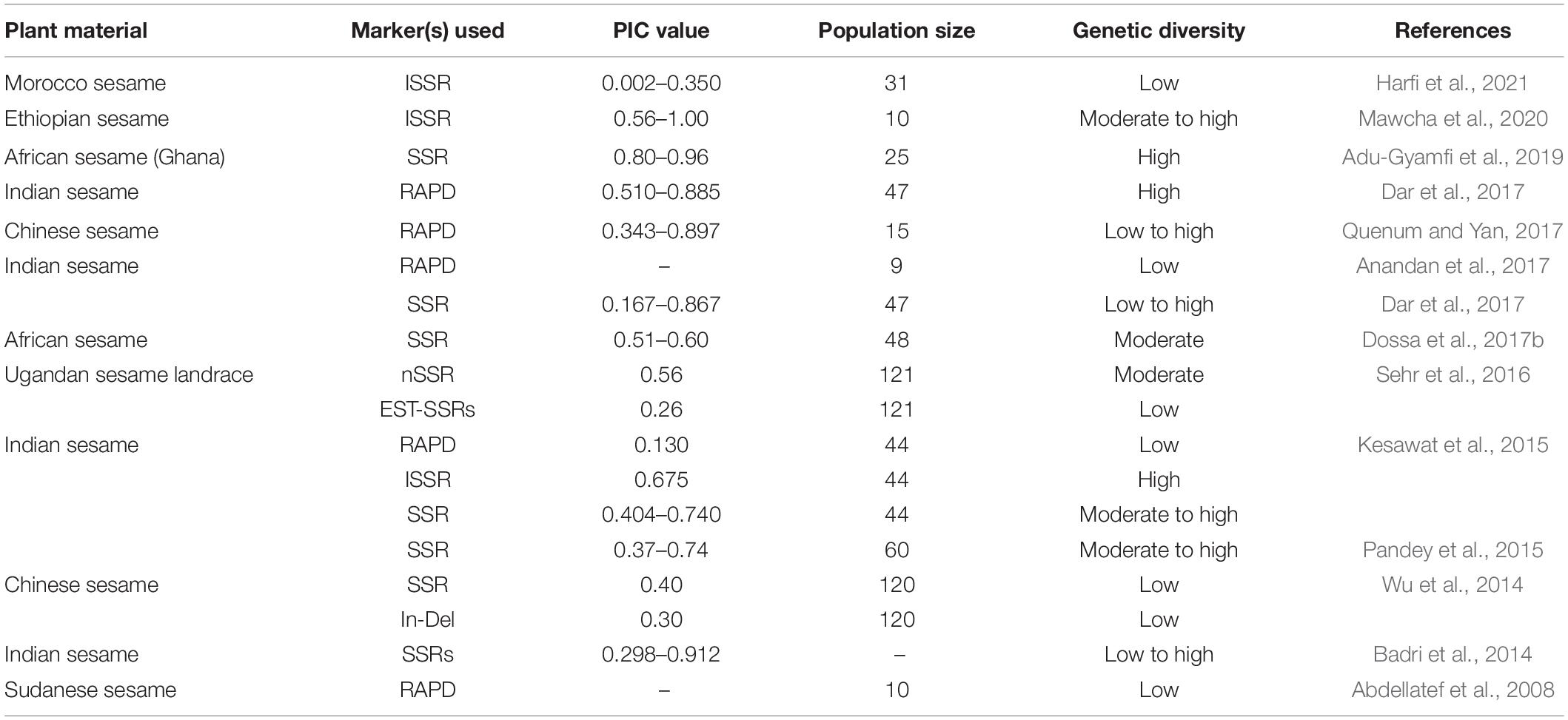
Genetic markers play an important role in identifying genetic diversity and important traits present in wild and cultivated species of sesame. This technology has geared plant breeding for genetic gain in many oilseed crop species. The randomly amplified polymorphic DNA (RAPD) technique pioneered in identifying sesame molecular markers (Bhat et al., 1999). RAPD and other markers (AFLP, SSR, and ISSR) had since then used genetic diversity analysis and molecular breeding for association mapping. Genetic diversity studies of sesame have been conducted using RAPD and inter-simple sequence repeats (ISSR) markers (Parsaeian et al., 2011). The amplified fragment length polymorphism (AFLP) technique identified molecular markers associated with the capsule indehiscence trait in a closed capsule mutant (indehiscent) derived from induced mutation (Uzun et al., 2003). High throughput techniques, such as restriction site-associated DNA sequencing (RAD-seq), specific length amplified fragment sequencing (SLAF-seq), RNA-seq, genotyping by sequencing (GBS), are being employed in sesame research for single nucleotidepolymorphisms (SNPs) discovery (Wu et al., 2014; Zhang et al., 2016). Genetic diversity assessments using molecular markers have been undertaken in various sesame-growing countries, including India, Uganda (Sehr et al., 2016), Turkey, China, Cambodia, and Vietnam (Myint et al., 2020). Table 2 lists the different molecular markers used for genetic diversity studies in sesame. Simple sequence repeats (SSRs) are robust molecular markers for genetic diversity analysis, including expressed sequenced tag-SSRs (EST-SSRs), chloroplast SSRs (cp-SSRs) (Sehr et al., 2016), genome sequence SSRs (gSSRs) (Wei et al., 2013; Dossa et al., 2016a), cDNA SSRs (Wang et al., 2012), and ISSRs (Kim et al., 2002). Only approximately 7% of the total available markers (∼100,000) have been validated, with a large proportion available for research. Studies related to SSRs in sesame are in progress. Recently, with the availability of whole-genome sequence (WGS) information, SNP markers have been used to survey sesame populations for marker-assisted breeding (Uncu et al., 2016). Next-generation sequencing data identified SNPs with bi-allelic, co-dominant, and inheritance characteristics. High-density arrays can rapidly genotype several common SNPs across samples for genetic analysis and breeding application, as seen in maize (Unterseer et al., 2014), palm oil (Kwong et al., 2016), pigeon pea (Singh et al., 2020), rice (Singh et al., 2015), soybean (Song et al., 2013), sunflower (Bachlava et al., 2012), and wheat (Wang L. et al., 2014).
Targeting Key Agronomic and Quality Traits Using Functional Genomics
Sesame oil is a sesame seed product with higher unsaturated fatty acids and natural antioxidants, with medicinal, organoleptic, nutritional properties, and superior oxidation stability than other vegetable oils (Takahashi et al., 2016). The oil yield from sesame seeds generally ranges from 35 to 55%, with some cultivars up to 63.2% (Wang S. et al., 2014). Sesame seed oil is rich in omega-6 fatty acids but lacks omega-3 fatty acids, warranting the production of more omega-3 fatty acids, such as alpha-linolenic acids. A comparative study on the fatty acid composition of sesame seed oil from different geographical regions (Morocco, Sudan, Congo, Turkey, and Egypt) revealed variation in palmitic acid (8.5–12.9%), stearic acid (3.0–5.5%), oleic acid (38.9–47.5%), linoleic acid (36.4–46.2%), linolenic acid (0.2–0.4%), saturated fatty acids (14.0–16.3%), and unsaturated fatty acids (83.9–85.1%) (Gharby et al., 2017). Characterizing key genes involved in the metabolic pathway would be important to gain deep insights into sesame oil diversity and other traits. Wei et al. (2015) identified key genes associated with sesame oil composition, pigmentation, and cell wall lignification in 705 accessions. A genome-wide association study (GWAS) identified the novel genes, SiLTP3 and SiACS8 related to seed yield traits, such as capsule length and number, but such studies are limited (Wang S. et al., 2014; Zhou et al., 2018). Variation in oil content may arise from seed color (Wei et al., 2015). Similarly, a recent GWAS study on sesame (336 lines in 12 environments) identified 92 potential genes associated with sesame seed coat color (Cui et al., 2021); late-maturing and white-seeded sesame cultivars had higher oil contents than early maturing or black-seeded genotypes, indicating a possible link between oil content and seed coat color. Another study reported increased antioxidant activity in white seeds compared to black seeds (Vishwanath et al., 2012). Capsules at different positions on the same plant can have varying oil contents, with basal capsules containing more oil than apex or branch capsules (Mosidis and Yermanos, 1985). In general, capsules with more carpels would produce more seeds and yield. Germplasm accessions originating from Japan and Far East countries generally exhibit tetracarpellary capsules, while those from other Asian countries mostly have bicarpellary capsules (Ali et al., 2007). Variation in the agro-morphological traits of sesame germplasm is shown in Figure 3. Cultivars with a determinate growth type generally have higher stearic and oleic acid contents and lower linoleic acid than indeterminate types. Environmental factors, especially temperature, strongly influence the fatty acid composition. Determinate sesame genotypes generally have higher oleic acid content and lower linoleic acid content than indeterminate genotypes (Uzun et al., 2002). Thus, non-branching phenotypes might be a good choice for sustained oil content. A combined research effort involving germplasm from different countries could produce sesame genotypes with desired traits.

Figure 3. Variability in sesame germplasm for agro-morphological traits. (A) Internode stem length, (B) branching pattern, (C) leaf shape, (D) locules per sesame capsule, (E) capsule length, and (F) seed coat color.
There is limited information on the biochemical composition of cultivated sesame germplasm. Fifty-four indigenous elite varieties evaluated for oil and fatty acid content contained oleic acid (38–50%) and linoleic acid (18–43%) as major fatty acids but had very low α-linolenic acid (ALA, C18:3, ω-3) (Bhunia et al., 2016). Indian sesame varieties, such as TC-289, PB Til 2, and Var-9 had the highest amount of oleic (50%), linoleic (43%), and α-linolenic acids (1.3%), respectively. Interestingly, Krishna, T-13, and Var-9 contained approximately 1:1 proportions of oleic and linoleic acid. Another study reported a similar percentage range for fatty acid composition using Indian S. indicum, S. mulayanum, and S. radiatum species (Mondal et al., 2010). NCRIBEN-01M, NCRIBEN-O2M, and NCRIBEN-03L were released in Nigeria for high oil content and white seed color (Olowe and Adeoniregun, 2010). Important genes related to oil biosynthesis in sesame are listed in Table 3. These genes are likely to play a central role in sesame and other oilseed crops and could be used to study the oil biosynthesis pathway, offering new opportunities to investigate novel genes.
Previous reports identified genetic variation in sesame using agro-morphological traits, such as plant height, primary and secondary branch numbers per plant, internode length, stem height to first branch, day to flowering, capsule number per plant, seed number per capsule, and yield per plant. Several agro-morphologically important traits of sesame are listed in Table 4. Seed color, seed size, and seed shattering are traits associated with domestication syndrome (Bartel and Fink, 1995), defined as the loss of the seed dispersal mechanism in plants, where seeds remain attached to the parent plant for easy harvest. However, mature sesame capsules shatter seeds on their own—a genetically controlled mechanism—limiting crop production. The anatomical basis of capsule shattering has been reported for 32 sesame varieties (Hiremath and Patil, 2002); two important enzymes [cellulase and polygalacturonase (PG)] partly control dehiscence in sesame. Thus, selecting genotypes with low cellulase and PG activity has the potential for breeding genotypes with the non-shattering trait.
Sesame is considered a small factory of potent molecules. Sesame oil contains important bioactive metabolites, including tocopherols and lignans, that contribute to human health (Mancebo-Campos et al., 2014). Tocopherols prevent the oxidation of sesame oil. γ-tocopherol is a major compound in sesame oil (∼90.5%), while other tocopherols (α, β, δ) account for less than 5% of the total content (Li et al., 2011). Another important sesame metabolite is lignan, formed by the oxidative coupling of p-hydroxyphenylpropane; the major lignans in sesame seeds are sesamin, sesamolin, and sesamol (Shi et al., 2017) (Table 5). Sesamol protects cell membranes from oxidative damage (Chennuru and Saleem, 2013). γ-tocopherol and lignans have cholesterol-lowering, neuroprotective, anticarcinogenic properties, and coronary protective effects (Kim et al., 2016). γ-tocopherol and lignans prevent peroxidation of cell membranes, increasing the rate of fatty acid oxidation in peroxisomes. Sesamin reduces the neurotoxic effect of microglial activation (mediated through lipopolysaccharide), increases the vitality of neuronal cells, and helps mitigate brain injury by lowering the levels of the mediator compound responsible for inflammation (Udomruk et al., 2018). Thus, sesamin could be used to treat serious brain diseases, such as Parkinson’s and Alzheimer’s disease. Other studies have reported the role of sesamin in curing disorders, such as osteoarthritis, ischemic brain stroke, acute hepatic injury, hepatic failure, and diabetic retinopathy, using rats as the model organisms (Wu et al., 2019). By 2035, the estimated number of cancer cases will be approximately 24 million per year, with 14.6 million associated deaths (Stewart et al., 2016). Therefore, sesamin is a potential adjuvant therapeutic agent for developing tumors and could be used to treat/prevent various types of cancer (Majdalawieh et al., 2017). Limited progress has been made using desirable traits through plant breeding due to the lack of appropriate genomic tools. However, breakthroughs in “omics” may advance sesame breeding.
Strategies for Enhancing Stress Resistance in Sesame
Enhanced Abiotic Stress Resistance in Sesame
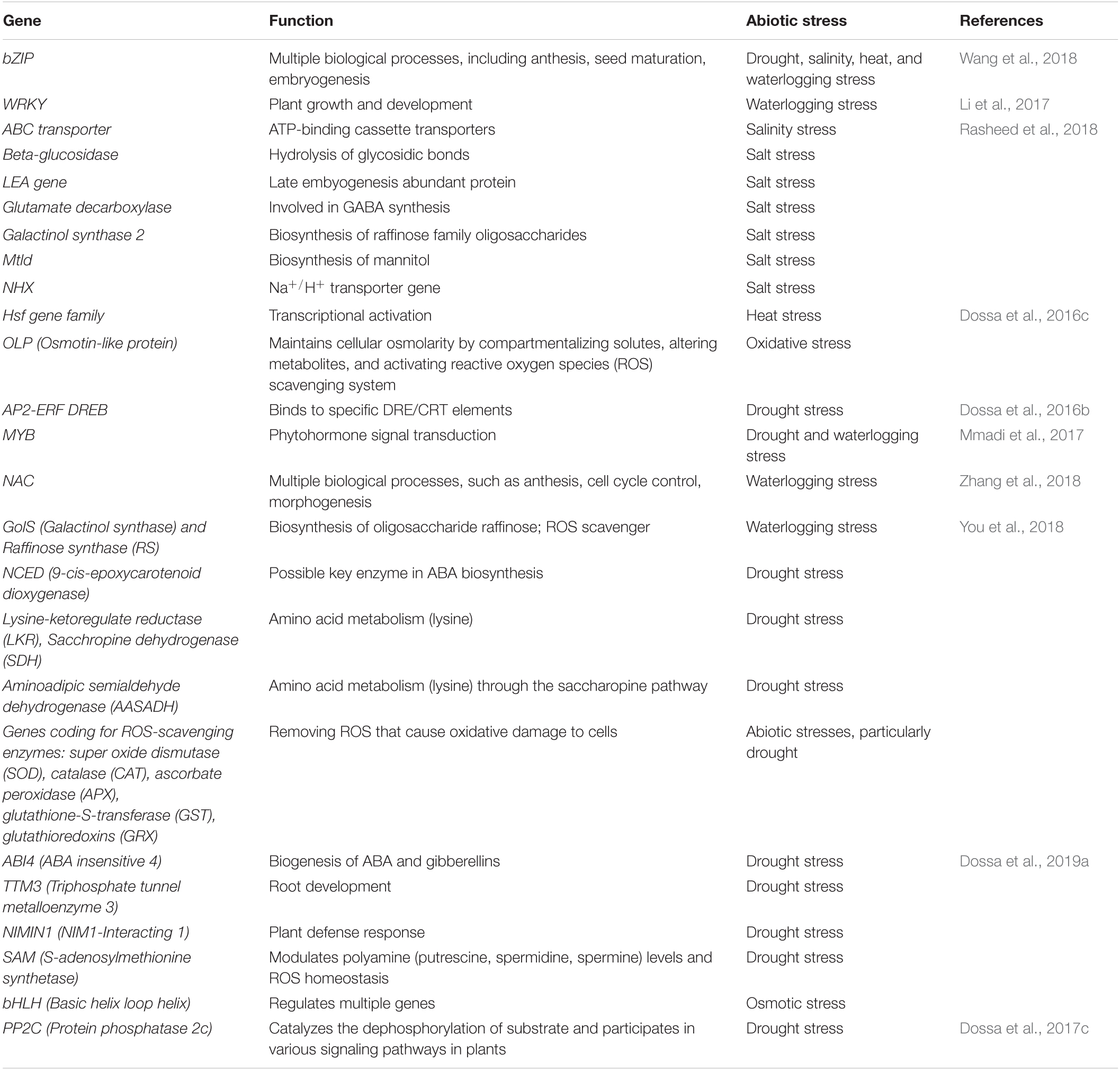
Various abiotic stresses threaten sesame production, including drought, waterlogging, salinity, and high temperature. Stress signals are received by specific receptors present on the plasma membrane, including ion channels, G-protein coupled receptors (GPCRs), and kinases, such as receptor-like kinases and histidine kinases. GPCRs are considered the most prominent signal, activating downstream signaling. Once the receptor is activated upon stress or signal induction, it regulates calcium ion concentration inside the cell and secondary messengers, such as abscisic acid (ABA), reactive oxygen species (ROS), and inositol phosphates. This ultimately activates various transcription factors (TFs), such as MYB/MYC, dehydration responsive element binding protein (DREB), basic leucine zipper domain (bZIP), WRKY by turning on protein kinases, phosphatases, and calcium-dependent protein kinases (CDPKs). The TFs activate myriad novel/unknown and known genes, such as ascorbate peroxidase (APX) (Padaria et al., 2014), heat shock proteins (HSPs) (Vishwakarma et al., 2018; Vishwakarma et al., 2019), and osmolytes (e.g., proline), to save the cell from desiccation. Table 6 describes various genes/TFs involved in the abiotic stress tolerance of sesame. Biochemical changes in stressed plants can be detected by measuring relative water content, total chlorophyll content, malondialdehyde (MDA) content, proline content, hydrogen peroxide (H2O2) levels, superoxide dismutase (SOD) activity, among others. A few studies have attempted to understand sesame tolerance to different abiotic stresses by systematically screening germplasm and the core collections in Korea (Park et al., 2015), India (Bisht et al., 1998), and China (Zhang et al., 2000). Various states in India have released 89 sesame varieties, with particular phenotypic traits and biotic and abiotic stress tolerance. Madhya Pradesh contributed 11 varieties, followed by Orissa (10), Tamil Nadu (9), Rajasthan, Gujarat, Maharashtra, and Andhra Pradesh (8 each), Kerala (7), Uttar Pradesh, Punjab, West Bengal, and Karnataka (4 each), Haryana (2), and Bihar and Himachal Pradesh (1 each) (Ranganatha et al., 2013). Dossa et al. (2016a) emphasized the need for more phenotypic and molecular analysis of African germplasm for stress-tolerant traits. Several recent studies have used transcriptomics to identify TFs, genes, and miRNAs associated with various abiotic stresses in sesame (Zhang et al., 2020). Major abiotic stresses affecting sesame are discussed below.
Drought Stress
Sesame is a rainfed crop cultivated primarily in arid and semi-arid areas and is thus prone to terminal and intermittent drought. Drought stress (DS) mainly occurs in the arid and semi-arid areas of Africa, America, and Asia. Sesame is typically considered a drought-tolerant crop compared to other oilseed crops (Langham, 2007). However, intense and prolonged drought stress during anthesis has deleterious effects on flowering, capsule and seed formation, seed yield, and ultimately oil quality (Dossa et al., 2017b). Prolonged DS at the seedling stage can increase plant mortality. For sesame, long drought stress at the vegetative stage is the most devastating factor, reducing the average yield. There are few systematic studies on tolerant and susceptible sesame genotypes. In a recent study, integrated transcriptomics and metabolomics were used to study the differential expression pattern of metabolites (e.g., proline, cGMP, adenine, guanine, benzoic acid, and putrescine) in drought-tolerant and drought-susceptible sesame genotypes (Dossa et al., 2017d; You et al., 2019). Several earlier studies revealed the role of micronutrients, such as silica, molybdenum, and boron, in providing drought stress tolerance to plants (Rana et al., 2020). A recent study showed that selenium applied to sesame leaves (50-day-old plants) improved DS tolerance by enhancing proline accumulation (Thuc et al., 2021a). In another report, exogenous application of phytohormones [salicylic acid (SA) and kinetin] on sesame plants induced DS resistance by increasing the endogenous phytohormone levels (Hussein et al., 2015). Therefore, studies related to metabolomics should find key differences between drought-sensitive and drought-tolerant genotypes to establish links between phenotypic (morphological level) and genotypic (DNA level) changes in plants under DS (Llanes et al., 2018). DS can be induced artificially using polyethylene glycol (PEG) solution to study morphological and physiological changes in sesame. Earlier studies revealed the role of PEG (in varying concentrations) in examining DS tolerance in sesame plants (Liang et al., 2021). However, only one report has evaluated sesame genotypes under DS (varying PEG concentration) using mutant plants at the seedling stage, establishing two sesame mutants “ML2-5” and “ML2-10” tolerant of severe DS (Kouighat et al., 2021).
Very few molecular studies have been conducted in sesame under DS. Stress-related TFs are essential for plant survival under DS. Evolutionary analysis of important TFs families, such as heat stress-specific transcription factors (Hsfs) and AP2/ERF, revealed that most of the genes in these families were retained during evolution. The conservation of these families is linked to high drought tolerance of sesame. The use of RNA-seq technologies led to the discovery of 61 candidate genes and 722 known genes involved in the drought response (Dossa et al., 2017c). More studies are being implemented to dissect the DS response pathways in sesame.
Waterlogging Stress
Sesame is highly susceptible to waterlogging, a major threat to sesame production in South and East Asia, particularly China and Korea (Wang et al., 2016). Even short periods of waterlogging reduce sesame growth and development, leaf area index, chlorophyll content, plant dry matter accumulation, and capsule number, and increase the number of aborted seeds, thus decreasing the yield (Sarkar et al., 2016). Saha et al. (2010) reported that sesame is more sensitive to waterlogging at the reproductive stage than the vegetative stage, reducing seed yield by approximately 44% (3.6–6.4 g plant–1) compared to control conditions (6.5–9.9 g plant–1).
Only a few studies have unraveled the genetic basis of the waterlogging stress response in sesame (Zhang et al., 2014). A comparative RNA-seq-based analysis between waterlogging-tolerant and susceptible sesame genotypes revealed 66 potential genes for improving sesame tolerance to waterlogging stress (Wang et al., 2016). Another study suggested different underlying molecular mechanisms due to the contrasting response of sesame genotypes under waterlogging stress, which were correlated further with DNA methylation patterns (Dossa et al., 2019b). Upregulation of genes in the bZIP family (e.g., SibZIP03, SibZIP04, SibZIP30, SibZIP44, and SibZIP62) might play an important role in metabolic reprogramming under waterlogging stress (Wang et al., 2018). Earlier studies have explored WRKY TFs to understand the structure and function of sesame WRKY genes, concluding that manipulating these genes could enhance waterlogging and DR (Li et al., 2017). Other genes related to waterlogging stress tolerance are listed in Table 6. Cultivar Zhongzhi No. 13 is a waterlogging-tolerant cultivar that could be used to genetically enhance sesame under waterlogging stress (Wang et al., 2016). A more comprehensive study identified 1,379 genes central to waterlogging stress tolerance, including 66 new candidate genes. A total of 13,307 genes were differentially regulated under waterlogging stress in sesame (Wang et al., 2012). SSR markers (ZM428) and QTL (qEZ10CHL07, qEZ10ZCL07, qWH10CHL09, qWH10ZCL09, qEZ09ZCL13, and qWH09CHL15) have been used in marker-assisted selection (MAS) for waterlogging stress tolerance (Wang et al., 2012). Combined transcriptomics and SSR studies will unveil sesame genomics associated with waterlogging stress tolerance.
High-Temperature Stress
High-temperature stress is a major constraint in sesame productivity, especially during anthesis and seed filling. Temperatures of 25 ± 2°C encourage rapid sesame seed germination, initial growth, and anthesis, while approximately 32°C is considered heat stress and greater than 40°C is considered severe heat stress for sesame, affecting fertilization and seed set, and ultimately reducing the sesame yield and quality. Late-sown sesame suffers yield losses of up to 40–50% (Rani and Kiranbabu, 2017). Therefore, thermotolerant sesame genotypes that can compensate for the yield penalty are urgently needed. Rani and Kiranbabu (2017) screened 442 sesame genotypes under high-temperature stress during anthesis, capsule formation, and seed set, identifying four genotypes (JCS 2846, JCS 2892, JCS 3102, and JCS 3258) with the potential to tolerate severe HS (>40°C). Such genotypes could be used in breeding programs to develop high-temperature stress-tolerant genotypes. Heat stress increases the length of the vegetative phase of sesame compared to plants grown under normal conditions (Firmansyah et al., 2012). Therefore, a systematic and comprehensive screening of sesame germplasm under suitable environmental conditions and hotspot locations for individual abiotic stress tolerance is needed.
Stress Perception and Signal Transduction Under Abiotic Stress
To cope with various biotic and abiotic stresses, plants have evolved mechanisms to bring changes at the physiological and biochemical levels that are translated into internal signals to activate stress-responsive genes through an interconnected network of signaling cascades. Thus, identifying genes and metabolites is necessary to understand abiotic stress response mechanisms in sesame plants. Abiotic and biotic stresses lead to oxidative bursts inside plant cells that accumulate ROS, such as hydroxyl radical (OH), hydroperoxyl radical (HO2), superoxide anion (O2–), and alkoxy radical (RO) (Gill and Tuteja, 2010). Plants deal with the increased ROS levels with different enzymes, such as ascorbate peroxidase (APX), glutathione reductase (GR), superoxide dismutase (SOD), catalase (CAT), monodehydroascorbate reductase (MDHAR), glutathione peroxidase (GPX), and glutathione S-transferase (GST) (Gill and Tuteja, 2010). The main sites of ROS production are chloroplasts in green plants and mitochondria in non-green parts (Singh et al., 2019) (Figure 2). Recent studies have compared two sesame genotypes (TS-5 and TH-6) under abiotic stress (salinity stress) by measuring oxidative markers, concluding that TS-5 tolerates abiotic stress better than TH-6. Increased membrane damage, as evidenced from MDA level (oxidative marker to measure stress), under salinity stress is also reported for other plant species, such as Medicago truncatula and Ailanthus altissima (Mhadhbi et al., 2013; Filippou et al., 2014). Anee et al. (2019) reported increased H2O2 and MDA levels in S. indicum cv. BARI Til-4 (waterlogging sensitive) with time (2, 4, 6, and 8 days) under waterlogging conditions. Similar results (enhanced oxidative markers) have been reported for species, such as the Antarctic plant (Park and Lee, 2019), Sorghum bicolor (Zhang et al., 2019), and Solanum lycopersicon (Rasheed et al., 2018), under waterlogging. In barley, the waterlogging-tolerant genotype (TF58) overexpressed SOD and GST compared to the sensitive genotype (TF57) under waterlogging stress (Luan et al., 2018). In sesame, increased SOD activity during waterlogging stress suggests that SOD acts as an ROS scavenger and thus plays an important role in maintaining cell membrane and protein stability. Using different biochemical markers, such as SOD, CAT, and GPX, studies have identified drought-tolerant (SI-1025, SI-205, SI2138-2, KM-13) and drought-sensitive (Prachi, SI-1926, ENT-78-301, Kanaka, IS-607-1-84) sesame genotypes (Hota et al., 2019). Traits related to yield enhancement under normal and stress conditions are listed in Table 7.
Involvement of Heat Shock Proteins and Heat Stress Specific Transcription Factors to Overcome Stress
Heat shock proteins (HSPs) or molecular chaperones are highly conserved proteins that protect plant cells under abiotic stress through protein folding, assembly, and translocation (Huttner and Strasser, 2012). There are five categories of HSPs based on molecular weight: HSP100, HSP90, HSP70, HSP60, and small HSPs. HSPs are major players enabling cell proteins to survive under stress conditions by stabilizing protein folding (Khan et al., 2019). In sesame under abiotic stress, tolerant genotypes have higher expression of HSPs than sensitive genotypes. Changes induced by plants under abiotic stress are controlled by gene expression, and TFs are important in regulating gene expression. Among TFs, heat stress specific transcription factors (Hsfs), MYB, WRKY, NAC, and bZIP are involved in complex and overlapping processes under abiotic stress (Gill and Tuteja, 2010). The Hsf gene family has been studied widely in many crop plants, with 30 Hsf genes reported in sesame (Dossa et al., 2016c), 25 in Arabidopsis, rice (Guo et al., 2008), maize (Lin et al., 2011), and Chinese pepper (Jin et al., 2014), 26 in tomato (Yang et al., 2016), 27 in potato (Tang et al., 2016), 29 in Chinese white pear (Qiao et al., 2015), and 35 in Chinese cabbage (Song et al., 2014). So, it can be concluded that the number of Hsf genes present is not linked to plant genome size. In a previous study, all 30 sesame Hsf genes were expressed in different parts of the sesame plant (the leaves, shoots, roots, and the flowers), suggesting a conserved functional role at various stages of development. Sesame Hsf genes were classified into three major groups (A, B, and C), with group B genes being most expressive under abiotic stress and thus could be used to improve the abiotic stress tolerance in sesame (Dossa et al., 2016c).
Enhanced Biotic Stress Resistance in Sesame
Biotic stress is induced by living organisms, such as fungi, viruses, bacteria, insects, and nematodes. Plants respond to biotic stresses with ROS-induced oxidative bursts, cell wall lignification, and chemical compound releases, such as benzothiadiazole (BTH) and β-aminobutyric acid (BABA). Various plant hormones, such as SA, jasmonic acid (JA), ethylene (ET), and ABA, play a central role in biotic stress signaling. Here, we mainly focus on two devastating diseases in sesame, i.e., phyllody and dry root rot caused by phytoplasma and fungi, respectively (Rao et al., 2015). Phyllody associated with phytoplasma is the major limiting factor affecting cultivation, described more than 100 years ago, first recorded in 1908 at Mirpur Khas, Pakistan (Vasudeva and Sahambi, 1955). Most cultivated sesame varieties are susceptible to phyllody disease, causing severe economic losses. Phyllody disease also occurs in related wild species, including S. alatıun, S. indicatum, S. occidentale, and S. radiatum, resulting in yield losses up to 34–100% (Ramanujam, 1944; Rao et al., 2015). Symptoms associated with phyllody disease in sesame include phyllody (floral parts replaced with leafy structures), flower virescence (abnormal development of green pigmentation), witches’ broom, shoot tip fasciation, vivipary (seeds or embryos begin to develop before being detached from the parent), and cracking of seed capsules. Phyllody disease has also been reported in the Asian and African countries, including Burkina Faso, Ethiopia, and India (Madhupriya et al., 2015; Rao et al., 2015), Iraq, Israel, Mexico, and Myanmar (Win et al., 2010), Nigeria, Oman, Pakistan, Sudan, and Taiwan (Tseng et al., 2014), Tanzania, Thailand, and Turkey (Catal et al., 2013), and Venezuela and Uganda (Singh et al., 2007).
Several diagnostic methods, including Dienes stains and TEM techniques, can detect phytoplasma bodies in phloem sieve tissues; however, PCR-based assays are more reliable for identifying phytoplasmas associated with the disease (Rao et al., 2015). Four phytoplasma ribosomal groups, 16SrI, 16SrII, 16SrVI, and 16SrIX, are associated with phyllody disease worldwide, of which 16SrI and 16SrII are the most widespread affecting sesame crops in India (Madhupriya et al., 2015). Complete phytoplasma genome sequencing has not been undertaken. The disease spreads via leafhopper species, with Orosius orientalis as the major vector. Recently a new vector, Hishimonus phycitis, was identified from India. Moreover, many weed species host sesame phyllody-associated phytoplasmas (Rao et al., 2015). The cure for disease phyllody is not known, so prevention is paramount. Developing cultivars with durable phyllody resistance would be the best management tool and should be an integral component of sesame breeding programs. Since most cultivated germplasm is susceptible to phyllody, exploitation of wild relatives as sources of resistance genes could be viable. Some studies have attempted to confer resistance to phyllody disease in sesame using intra- and interspecific crosses of sesame species, including S. mulayanum, S. indicum, and S. alatum (Singh et al., 2007).
Epidemiologic studies should also be undertaken to eradicate alternate plant hosts and control insect vectors, preventing the epidemic spread of phyllody in sesame. Identifying true phyllody resistance is important because symptomless infections are often undetectable, and disease-resistant genotypes selected in the field might respond differently under other environmental conditions (Atkinson and Urvin, 2012). Ustun et al. (2017) recently screened 542 sesame genotypes under field and greenhouse conditions to identify phyllody resistance; only two accessions, ACS38 and ACS102, were disease resistant. Phytoplasma infection (phyllody disease) in sesame has been identified and characterized by molecular analysis using the 16S rRNA region of phytoplasma (Junior et al., 2019). Phyllogen (a virulence factor) may induce phyllody in many plant species of angiosperms, gymnosperms, and ferns by inhibiting MTFs (MADS domain transcription factor), a key regulator of flower development (Kitazawa et al., 2017).
Another widespread and destructive disease of sesame is dry root rot, caused by Macrophomina phaseolina (Tassi) Goid, a soil-borne Deuteromycetes fungus (Islam et al., 2016). Although M. phaseolina is reported to be from North and South America, Asia, Africa, and Europe, this pathogen is more prevalent in subtropical and tropical countries having a semi-arid climate. Macrophomina exists in two alternative forms: saprophytic (Rhizoctonia bataticola) and pathogenic (M. phaseolina). The pathogen causes significant yield losses in sesame, ranging from 50 to 100% (Yan et al., 2021). A few studies have investigated dry root rot resistance or tolerance for commercial cultivar development in sesame (Cummings and Bergstrom, 2013). Bedway and Moharam (2019) screened 86 sesame genotypes and found highly significant variations between genotypes across two seasons for disease-infection percentage and seed yield. A recent transcriptomics comparison between resistant (Zhengzhi 13) and susceptible (Ji 9014) sesame genotypes revealed the disease-resistance mechanism against Macrophomina phaseolina, with 52 differentially expressed genes involved in different signaling pathways (Yan et al., 2021).
Another major limiting factor for sesame crop production is sucking pest insects. Larvae of Antigastra catalaunalis (sesame leaf rollers) feed on the topmost leaves of sesame, resulting in plant death. If infection occurs at later stages, the larvae penetrate the capsule and feed on the developing seeds. Orosius albicinctus (jassid) feeds on soft tissues, such as leaves, causing leaf curling; it is also the major vector for transmitting phyllody disease. Maggots, such as gall fly (Asphondylia sesami) and bud fly (Dasineura sesami) affect sesame floral buds by forming gall-like structures. Another pathogen affecting sesame growth is Phytopthora nicotianae (Kumari et al., 2019). Of the 186 sesame genotypes screened, 12 had high resistance to Phytophthora. Four P. nicotianae isolates (KACC48120, KACC48121, No2526, and No2040) were identified by the Korean National Agrobiodiversity (Oh et al., 2018), but there is little information on sesame resistance to Phytopthora blight. Identifying the molecular mechanisms involved in biotic stress tolerance in sesame would strengthen our understanding of the regulatory mechanisms against biotic stresses. In addition, identifying changes in gene expression would help predict sesame tolerance to biotic stresses. Genes related to biotic stress tolerance in sesame are listed in Table 8.
Insect and Pest Resistance in Sesame
Host plant resistance is a key component of integrated pest management as it is environmentally safe, sustainable, and easy to adopt. The attacking pests on sesame are classified into pathogens, predators, and parasitoids. Antigastra catalaunalis is a pest that causes leaf webbing in sesame (Gebregergis et al., 2016). Intercropping sesame with green gram, black gram, sorghum, pearl millet, cluster bean, and pigeon pea significantly reduced the damage caused by A. catalaunalis (Ahirwar et al., 2009). Common cultural practices assist pest management in sesame, including timely crop sowing, field sanitation, deep plowing during summer, destroying alternate host plants, sowing of guard/barrier crops, such as maize, jowar, and bajra. Studies have reported that releasing parasitoids (Trichoderma spp., Bracon brevicornis, Bracon heabator, and Campoplex spp.) and predators (Eocantheconia furcellata, Cicindella spp., red ant, spiders, and ladybird beetle) in sesame reduced the population growth of A. catalaunalis. Conserving existing natural parasitoids (Ichneumonidae and Braconidae) and predators (coccinellids, predatory stink bugs, mantids, and spiders) combined with appropriate microbial pesticides (e.g., Bacillus thuringiensis var. kurstaki) help manage leaf webber in sesame (Sasikumar and Kumar, 2014). Gall fly, Asphondylia sesami Feltis, is another significant crop pest, reducing sesame seed yield by up to 100% in susceptible genotypes (Mehalingam, 2012). The variety, N-32 released in India is tolerant to gall fly. Leafhopper, Orosius albicinctus, transmits phyllody disease caused by phytoplasma, infecting sesame crops under field conditions by 0–65.12% (Seemuller and Harries, 2009). The long-term solution for leafhopper control is the development of resistant genotypes.
Role of Plant Growth Regulators in Combating Stresses
Phytohormones, including SA, JA, ET, and gibberellic acid (GA3), play a crucial role in regulating the immune responses of plants (Shah et al., 2021). Studies have shown that SA has a prominent role in limiting oxidative damage caused by ROS and stomatal conductance in sunflower and barley (Habibi, 2012). Treatment with phytohormones or PGRs improves plant growth and productivity. Exogenous application of PGRs, such as SA and kinetin, to sesame improved plant growth and other physiological activities (Llanes et al., 2018). GA3 improves the plant growth and yield in many crop plants, including sesame (Vekaria et al., 2017). Foliar application of GA3 at 30 ± 5 days after sowing increased seed number per capsule, plant yield, and percentage of oil content. Similarly, spraying GA3 on sesame plants increased the branch number and plant height (Thuc et al., 2021b). Similar effects of GA3 were observed in Vicia faba L. and other leafy vegetables (Ibrahim et al., 2007; Miceli et al., 2019). The growth regulator, Cycocel improved sesame growth better than Paclobutrazol. Paclobutrazol interferes with GA3 synthesis, with high concentrations retarding sesame growth (Sarkar et al., 2006), as reported for tomato (Pal et al., 2016). A high concentration of Paclobutrazol (300 mg L–1) decreased seed numbers in maize and canola (Kamran et al., 2018), but the same concentration applied to sesame improved dry matter accumulation and seed production and reduced seed shattering (Mehmood et al., 2021). Therefore, studies are needed to optimize the concentration of PGRs for increased sesame seed production under stress.
Role of Micro Rna in Sesame Yield and Oil Biosynthesis
Micro RNAs (miRNAs) are small (20–35 nucleotides long) non-coding RNAs that play a key role in various biological processes (anthesis, nutrient metabolism, light fluctuation, and biotic and abiotic stresses) by degrading targeted mRNAs (cleavage or translational repression) (Table 9). Usually, miRNAs are considered core components of the complex network-regulating gene expression. Studies have identified miRNAs in many plant species, such as Arabidopsis, wheat, cotton, eggplant, maize, radish, rice, sugarcane, and sesame. Before 2018, there were no reports of miRNAs in sesame (Joshi and Mandavia, 2018). Earlier studies identified a few novel miRNAs in sesame, such as mir447 and mir8140 (Marakali, 2018). However, deep sequencing has opened the door to identifying large numbers of miRNAs. Recent studies identified 302 miRNAs (including 19 novel and 283 known miRNAs) in sesame (Zhang et al., 2020). The miRNA studies combined with degradome and transcriptomics are unique, modern molecular platform for in-depth studies, which have been recently used to identify 65 novel and 220 previously known miRNAs that regulate oil biosynthesis during sesame germination (Zhang et al., 2021). Identifying novel miRNAs would offer insight into the biological functioning of miRNAs and their evolutionary relationship. Sesame miRNAs require further attention with relation to targeted genes involved in signaling and developmental pathways, especially leaf development, transmembrane transport, and biotic and abiotic stress responses.
Transgenic Sesame: A Brighter and Sustainable Future of the Crop
Production of transgenic sesame by integrating foreign genes was first reported by Taskin et al. (1999). Still, limited studies have been undertaken on developing the transgenic sesame. The genetic transformation (cotyledon explants) method used the intron interrupted Gus gene, with nptII as a selectable marker, with a transformation frequency of 1.01% (Yadav et al., 2010). In another study, sesame (6-day-old cotyledons) was transformed using the sonication-assisted Agrobacterium tumefaciens-mediated method (Debnath et al., 2014). Several authors have produced transgenic sesame with useful traits, such as increased biosynthesis of phenylpropane, improved abiotic stress tolerance, and increased oil content (Chowdhury et al., 2017; Bhunia et al., 2016) (Table 10). In most transformation protocols, Agrobacterium tumefaciens has been used to insert foreign DNA in sesame, with a few studies using Agrobacterium rhizogenes for hairy roots transformation (Jin et al., 2005). Increasing the expression level of key genes involved in oil biosynthesis is needed to increase sesame oil content. For example, manipulating genes, such as oleate Δ12-desaturase gene, FAD1, FAD2, DGAT1, and PDAT1 would enhance oil content in transgenic sesame. The genetic transformation of sesame would open new avenues for sesame crop improvement, especially increased oil content.
Sesamum Close Wild Relatives: Value Addition to Food and Medicine
Closely related genera of Sesamum include Ceratotheca sesamoides (2n = 32) or false sesame, indigenous to Africa. Leaves and flowers are consumed as vegetables, sauces, and soup preparation (Morakinyo et al., 2020). The slimy secretion from leaves (due to glandular trichomes) is used to treat eye conjunctivitis (Bedigian, 2015). Leaves of Ceratotheca triloba (Bernh.) Hook. are often used as a vegetable; the plant has medicinal value in Zimbabwe and South Africa. Young shoots and leaves of S. alatum are consumed as cooked vegetable in Africa and Sudan.4 Roots and leaves are used to treat insect (scorpion, wasps) bites. Root bark is used to treat stomach disorders in the northwestern Burkina Faso. There are no reports of S. angolense leaf consumption, but root extracts are used to treat coughs in Tanzania, and the leaves are used to treat smallpox, scabies, and vitiligo. All plant parts (leaves, stems, and flowers) of S. angustifolium are consumed in parts of Africa and Kenya. Leaves of S. radiatum are used to prepare thick soup, which is good for children’s teeth and bones, in the Ivory Coast, Benin, and Nigeria (Auwalu and Babatunde, 2007). Leaf extract is used to treat swellings, boils, insect bites, skin infections, and leprosy and facilitate childbirth in Nigeria and Ghana.5 People living in Darfur (western Sudan region) use the dried leaves of C. sesamoides, S. angustifolium, and S. radiatum in the preparation of meat and fish. Many mucilaginous polysaccharides from Pedaliaceae leaves are used as food and medicine in many countries (Bedigian, 2010), especially in Africa. Sesamothamnus busseanus leaves are used as a vegetable in Tanzania. S. triphyllum is used to treat epilepsy in South Africa. Leaves of Pedaliaceae members, especially C. sesamoides and Sesamum spp. are a rich source of carbohydrates and minerals (Ruffo et al., 2002). In India, people generally use sesame oil to help with anxiety and sleep disorders and as an antimicrobial mouthwash (Anilakumar et al., 2014). Roasted sesame seeds are consumed directly. Comprehensive studies are needed to save these beneficial species.
Proposed Strategies and Vision for Enhancing Sesame Production
In recent years, “omics” tools have revealed large numbers of functional genes associated with key agronomic traits. However, functional validation of these genes in model plant systems using genetic engineering approaches is lacking. Transgenic approaches offer a quick, easy, and cost-effective transformation of genes in sesame. Chowdhury et al. (2017) were the first to genetically transform sesame. Using a transgenic approach, potential genes related to yield enhancement, oil biosynthesis, and stress (biotic and abiotic) tolerance could be used to transform elite sesame cultivars for a deeper understanding of the molecular mechanisms involved. In general, transgenic sesame is not well accepted by the public. Biotechnology methods, such as genome editing/CRISPR-Cas and target-induced local lesions in genomes (TILLING) are needed to validate the available genes related to key agronomic traits. Wild relatives of sesame are a reservoir of candidate genes; therefore, there is an urgent need to characterize wild sesame germplasm, especially African species. Further, identifying genes related to the indehiscence trait and indeterminate growth habit—essential traits affecting the mechanization of sesame harvesting—should be prioritized. No studies have investigated the genes involved in the root system architecture (RSA) of sesame; there is an urgent need to dissect the RSA of sesame as it plays a vital role in adapting sesame to different climatic conditions. Various biotic and abiotic stresses and socio-economic and technological factors increase the gap between farmer yields and actual potential yields. Awareness programs are needed to teach farmers agricultural practices, such as proper sowing, plant spacings, intercropping practices, and reducing pests and diseases. Developing sesame varieties with traits, such as indehiscent capsules (non-shattering), photo-insensitivity, determinate growth habit, and monoculm stems (stems with no branching) for easy harvest would help enhance sesame productivity. Bulgaria has developed four sesame varieties (Victoria, Aida, Valya, and Nevena) for mechanized harvest, the United States has released several non-shattering varieties (Paloma, SW 16, SW17, Baco, and UCR3), and Myanmar has released early maturing (60–80 days) varieties (Ye-Kyaw, Pat-Le-War-Hmyaung, Man-Shwe-Wa, She-Ka-lay, and Mai-Thi-hla) (Myint et al., 2020). The useful bioactive compounds (sesamin, sesamol, and tocopherols) in sesame seeds add value to this crop. Therefore, a deep understanding of the genetic basis of such traits should be a priority for developing nutritionally superior sesame genotypes. Sesame is known to confer stress tolerance, but little information is available on this crop despite being a food security source for humans. Most studies related to biotic/abiotic stress tolerance in sesame have been undertaken in controlled conditions. Field studies would offer a more comprehensive understanding of stress responses, elaborating on the inter-connecting mechanisms involved in stress tolerance. Therefore, sesame crop mutants need to be generated using transgenic or breeding approaches to understand stress physiology in sesame. Limited micro RNA-related studies have targeted specific genes in sesame. Biodiesel is being produced from oilseed crops (such as castor, mustard, soybean); perhaps sesame should be investigated in this regard. Genetic transformation, gene stacking, and genome editing techniques could be used to reveal desirable sesame traits. In addition, hormonal crosstalk should focus on understanding the signaling behavior of SA and ET under waterlogging stress. Moreover, studies related to ribosomal proteins, such as the 30S subunit, should be undertaken to investigate translation-dependent stress-related protein synthesis in sesame, which may further improve sesame seed quality and the sustainability of sesame crops under harsh environmental conditions. Therefore, evaluating the available sesame germplasm for the aforementioned traits is a must for sustainable development. Wild relatives of sesame (S. alatum, S. radiatum, and Ceratotheca sp.) are resistant to various biotic and abiotic stresses; therefore, the utilization and conservation of such germplasm are urgently required.
Box 1. Future directions for enhanced yield and quality in sesame.
1. Genes related to indehiscence trait, monoculm stem, and determinate growth should be identified to improve the mechanization of sesame harvesting.
2. Root system architecture (RSA) of sesame should be investigated for adapting sesame to different abiotic stresses.
3. Farmers should be educated on sesame pests and diseases through awareness programs.
4. More sesame mutants are needed to gain deeper insights on sesame plant physiology.
5. Genetic transformation (transgenic sesame), gene stacking, and genome editing techniques should be used to reveal desirable sesame traits.
6. Exploitation of wild relatives of sesame to introgress resistant genes in cultivated species to produce climate-resilient sesame.
Conclusion and Future Prospects
Genes related to sesame resistance present a new exemplar in explicating gene interactions with diseases. Wild relatives of sesame are much more disease-resistant than sesame cultivars. Phyllody is a devastating disease in sesame, but the large genome size and polyploidy level in wild relatives make studies on sesame phyllody challenging. In contrast, the small diploid genome, higher oil content, and fewer linked genes make sesame (S. indicum) a potential model plant for studying disease resistance and oil biosynthesis. Developing sesame varieties with waterlogging tolerance and high water-use efficiency will benefit sesame production. The development of sesame hybrids carrying potential traits would reveal greater genetic variation. Therefore, hybrid development programs should be intensified to maximize the sesame yield. There have been few transgenic studies in sesame, with no standardized transformation protocol. However, the use of transgenic technology is limited in sesame; there is an urgent need to establish more precise protocols for the efficient transformation of sesame. Current approaches to raise transgenic sesame are not well accepted by the public. The large sesame germplasm available in different gene/seed banks needs to be explored for traits/genes which would improve seed quality and increase sustainability under harsh environmental conditions. Research on sesame is needed to improve yields, plant architecture, disease and pest resistance, and indehiscent capsules (non-shattering trait). Many wild sesame species are endowed with specific traits, such as resistance to biotic/abiotic stresses and other important agronomic traits, which breeders should use to develop sesame cultivars. A well-defined program should be discussed at the national and international levels for distributing sesame seeds developed from such highly concerted efforts.
Author Contributions
SK and KSin conceptualized the study. RY, PR, KP, GR, VK, RP, CV, SL, SS, BT, VRan, HV, ASh, and ASa wrote the original draft. AK and KSid contributed to writing, reviewing, and editing the manuscript. KSin and KSid contributed to supervision. RY, AK, and KSin contributed to project administration. RY, SK, and KSin contributed to funding acquisition. All authors contributed equally in their respective expertise and agreed to the published version of the manuscript.
Funding
This work was supported by the Department of Biotechnology (DBT), Government of India (Grant No: 801).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ https://www.mordorintelligence.com/industry-reports/sesame-seeds-market
- ^ https://www.statista.com/statistics/263978/global-vegetable-oil-production-since-2000-2001/
- ^ https://www.ncbi.nlm.nih.gov/biosample/SAMN02357081
- ^ https://www.cabdirect.org/cabdirect/abstract/20177200497 (accessed on December 1, 2020).
- ^ https://www.cabdirect.org/cabdirect/
References
Abdellatef, E., Sirelkhatem, R., Ahmed, M. M. M., Radwan, K. H., and Khalafalla, M. M. (2008). Study of genetic diversity in Sudanese sesame (Sesamum indicum L.) germplasm using random amplified polymorphic DNA (RAPD) markers. Afr. J. Biotechnol. 7, 4423–4427.
Adu-Gyamfi, R., Prempeh, R., and Zakaria, I. (2019). Diversity assessment of some sesame (Sesamum indicum L.) genotypes cultivated in northern Ghana using morphological and simple sequence repeat (SSR) markers. Adv. Agric. 3, 1–10. doi: 10.1155/2019/6067891
Ahirwar, R. M., Banerjee, S., and Gupta, M. P. (2009). Insect pest management in sesame crop by intercropping system. Ann. Plant Prot. Sci. 17, 225–226.
Ali, G. M., Yasumoto, S., and Seki-Katsuka, M. (2007). Assessment of genetic diversity in sesame (Sesamum indicum L.) detected by Amplified Fragment Length Polymorphism markers. Electron. J. Biotechnol. 10, 12–23.
Al-Shafeay, A. F., Ibrahim, A. S., Nesiem, M. R., and Tawfik, M. S. (2011). Establishment of regeneration and transformation system in Egyptian sesame (Sesamum indicum L.) cv Sohag1. GM Crops 2, 182–192. doi: 10.4161/gmcr.2.3.18378
Anandan, R., Deenathayalan, T., Prakash, M., Sunilkumar, B., and Narayanan, G. S. (2017). Assessment of genetic diversity among sesame (Sesamum indicum L.) germplasm as revealed by RAPD and SSR markers. Indian J. Biochem. Biophy. 55, 143–150.
Anee, T. I., Nahar, K., Rahman, A., Mahmud, J. A., Bhuiyan, T. F., Alam, M. U., et al. (2019). Oxidative damage and antioxidant defense in Sesamum indicum after different waterlogging durations. Plants 8:196. doi: 10.3390/plants8070196
Anilakumar, K. R., Pal, A., Khanum, F., and Bawa, A. S. (2014). Nutritional, medicinal and industrial uses in sesame (Sesamum indicum L.) seeds-An overview. Agric. Conspec. Sci. 75, 159–168.
Ashri, A. (2007). “Sesame (Sesamum indicum L.),” in Genetics Resources, Chromosome Engineering and Crop Improvement, ed. R. J. Singh (Boca Raton, FL: CRC Press), 231–289.
Atkinson, N. J., and Urvin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Auwalu, B. M., and Babatunde, F. E. (2007). Analyses of growth, yield and fertilization of vegetable sesame (Sesamum radiatum Schum). J. Plant Sci. 2, 108–110. doi: 10.3923/jps.2007.108.112
Bachlava, E., Taylor, C. A., Tang, S., Bowers, J. E., Mandel, J. R., Burke, J. M., et al. (2012). SNP discovery and development of a high-density genotyping array for sunflower. PLoS One 7:e29814. doi: 10.1371/journal.pone.0029814.g002
Badri, J., Yepuri, V., Ghanta, A., Siva, S., and Siddiq, E. A. (2014). Development of microsatellite markers in sesame (Sesamum indicum L.). Turk. J. Agric. For. 38, 603–614. doi: 10.3906/tar-1312-104
Bartel, B., and Fink, G. R. (1995). ILR1. an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268, 1745–1748. doi: 10.1126/science.7792599
Bedigian, D. (2015). Systematics and evolution in Sesamum L. (Pedaliaceae), part 1: evidence regarding the origin of sesame and its closest relatives. J. Plant Taxon. Geogr. 70, 1–42. doi: 10.1080/00837792.2014.968457
Bedway, I., and Moharam, M. (2019). Reaction and performance of some sesame genotypes for resistance to Macrophomina phaseolina, the incitant of charcoal rot disease. Alex. Sci. Exch. J. 40, 12–18. doi: 10.21608/asejaiqjsae.2019.26294
Benson, O. N., Beatrice, A. W., Samuel, G., Otto, G. D., and Augustino, O. O. (2013). Genetic Diversity in cultivated Sesame and related wild species in East Africa. J. Crop Sci. Biotechnol. 16, 9–15. doi: 10.1007/s12892-012-0114-y
Bhat, K. V., Babrekar, P. P., and Lakhanpaul, S. (1999). Study of genetic diversity in Indian and exotic sesame (Sesamum indicum L.) germplasm using random amplified polymorphic DNA (RAPD) markers. Euphytica 110, 21–33.
Bhattacharyya, J., Chakraborty, A., Mitra, J., Chakraborty, S., Pradhan, S., Manna, A., et al. (2015). Genetic transformation of cultivated sesame (Sesamum indicum L.cv Rama) through particle bombardment using 5-day-old apical, meristematic tissues of germinating seedlings. Plant Cell Org. Tiss. Cult. 123, 455–466. doi: 10.1007/s11240-015-0848-6
Bhunia, R. K., Kaur, R., and Maiti, M. K. (2016). Metabolic engineering of fatty acid biosynthetic pathway in sesame (Sesamum indicum L.): assembling tools to develop nutritionally desirable sesame seed oil. Phytochem. Rev. 15, 799–811. doi: 10.1007/s11101-015-9424-2
Bisht, I. S., Mahajan, R. K., Loknath, T. R., and Agarwal, R. C. (1998). Diversity in Indian sesame collection and stratification of germplasm accessions in different diversity groups. Genet. Resour. Crop Evol. 45, 325–335. doi: 10.1023/A:1008652420477
Catal, M., Ikten, C., Yol, E., Ustun, R., and Uzun, B. (2013). First report of a 16SrIX group (pigeon pea witches’-broom) phytoplasma associated with sesame phyllody in Turkey. Plant Dis. 97:835. doi: 10.1094/PDIS-11-12-1100-PDN
Chandra, K., Sinha, A., and Arumugam, N. (2019). Gene isolation, heterologous expression, purification and functional confirmation of sesamin synthase from Sesamum indicum L. Biotechnol. Rep. 22:e00336. doi: 10.1016/j.btre.2019.e00336
Chennuru, A., and Saleem, M. T. (2013). Antioxidant, lipid lowering, and membrane stabilization effect of sesamol against doxorubicin-induced cardiomyopathy in experimental rats. BioMed. Res. Int. 2013:934239. doi: 10.1155/2013/934239
Chowdhury, S., Basu, A., and Kundu, S. (2017). Overexpression of a new osmotin like protein gene (SindOLP) confers tolerance against biotic and abiotic stresses in sesame. Front. Plant Sci. 8:410. doi: 10.3389/fpls.2017.00410
Cui, C., Liu, Y., Liu, Y., Cui, X., Sun, Z., Du, Z., et al. (2021). Genome-wide association study of seed coat color in sesame (Sesamum indicum L.). PLoS One. 16:e0251526. doi: 10.1371/journal.pone.0251526
Cummings, J. A., and Bergstrom, G. C. (2013). First report of charcoal rot caused by Macrophomina phaseolina in soybean in New York. Plant Dis. 97:1506. doi: 10.1094/PDIS-03-13-0318-PDN
Dar, A. A., Mudigunda, S., Mittal, P. K., and Arumugam, N. (2017). Comparative assessment of genetic diversity in Sesamum indicum L. using RAPD and SSR markers. 3 Biotech 7:10. doi: 10.1007/s13205-016-0578-4
Debnath, A. J., Basu, D., and Sikdar, R. S. (2014). An approach to standardize transient expression of GUS in sesame (Sesamum indicum L.) using the sonication-assisted Agrobacterium tumefaciens mediated transformation (SAAT) method. Israel J. Plant Sci. 61, 37–45. doi: 10.1080/07929978.2014.94827
Dossa, K., Diouf, D., Wang, L., Wei, X., Zhang, Y., Niang, M., et al. (2017a). The emerging oilseed crop Sesamum indicum enters the “Omics” era. Front. Plant Sci. 8:1154. doi: 10.3389/fpls.2017.01154
Dossa, K., Yehouessi, W. L., Likeng, B. C., Diouf, D., Liao, B., Zhang, X., et al. (2017b). Comprehensive screening of West and Central African sesame accessions for drought tolerance probing by agro-morphological, physiological, biochemical and nutritional traits. Agron 7:83. doi: 10.3390/agronomy7040083
Dossa, K., Li, D., Wang, L., Zheng, X., Yu, J., Wei, X., et al. (2017c). Dynamic transcriptome landscape of sesame (Sesamum indicum L.) under progressive drought and after rewatering. Genom. Data 11, 122–124. doi: 10.1016/j.gdata.2017.01.003
Dossa, K., Li, D., Wang, L., Zheng, X., Liu, A., Yu, J., et al. (2017d). Transcriptomic, biochemical and physio-anatomical investigations shed more light on responses to drought stress in two contrasting sesame genotypes. Sci. Rep. 7:8755. doi: 10.1038/s41598-017-09397-6
Dossa, K., Li, D., Zhou, R., Yu, J., Wang, L., Zhang, Y., et al. (2019a). The genetic basis of drought tolerance in the high oil crop Sesamum indicum. Plant Biotech. J. 17, 1788–1803. doi: 10.1111/pbi.13100
Dossa, K., Mmadi, M. A., Zhou, R., Zhang, T., Su, R., Zhang, Y., et al. (2019b). Depicting the core transcriptome modulating multiple abiotic stresses responses in Sesame (Sesamum indicum L.). Int. J. Mol. Sci. 20:3930. doi: 10.3390/ijms20163930
Dossa, K., Wei, X., Zhang, Y., Fonceka, D., Yang, W., Diouf, D., et al. (2016a). Analysis of genetic diversity and population structure of sesame accessions from Africa and Asia as major centers of its cultivation. Genes 7:14. doi: 10.3390/genes7040014
Dossa, K., Diouf, D., and Cissé, N. (2016c). Genome-wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Front. Plant Sci. 7:1522. doi: 10.3389/fpls.2016.01522
Dossa, K., Wei, X., Li, D., Zhang, Y., Wang, L., Fonceka, D., et al. (2016b). Insight into the AP2/ERF transcription factor superfamily in sesame (Sesamum indicum) and expression profiling of the DREB subfamily under drought stress. BMC Plant Biol. 16:171. doi: 10.1186/s12870-016-0859-4
Filippou, P., Bouchagier, P., Skotti, E., and Fotopoulos, V. (2014). Proline and reactive oxygen/nitrogen species metabolism is involved in the tolerant response of the invasive plant species Ailanthus altissima to drought and salinity. Environ. Exp. Bot. 97, 1–10. doi: 10.1016/j.envexpbot.2013.09.010
Firmansyah, F., Taryono, T., and Yudono, P. (2012). The dynamics of sesame (Sesamum indicum L.) growth type. Ilmu Pertanian (Agricul. Sci.) 15, 30–46. doi: 10.22146/ipas.2514
Food and Agriculture Organization Statistical Databases [FAOSTAT] (2018). Available online at: http://www.fao.org/faostat/en (accessed on February 5, 2020).
Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., et al. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. doi: 10.1093/emboj/18.17.4679
Gebregergis, Z., Dereje, A., and Fitwy, I. (2016). Assessment of incidence of sesame webworm Antigastra catalaunalis (Duponchel) in Western Tigray, North Ethiopia. J. Agric. Ecol. Res. Int. 9, 1–9. doi: 10.9734/JAERI/2016/28483
Gharby, S., Harhar, H., Bouzoubaa, Z., Asdadi, A., El Yadini, A., and Charrouf, Z. (2017). Chemical characterization and oxidative stability of seeds of oil sesame grown in Morocco. J. Saud. Soc. Agric. Sci. 16, 105–111. doi: 10.1016/j.jssas.2015.03.004
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Golicz, A. A., Batley, J., and Edwards, D. (2016). Towards plant pangenomics. Plant Biotechnol. J. 14, 1099–1105. doi: 10.1111/pbi.12499
Guo, J., Wu, J., Ji, Q., Wang, C., Luo, L., Yuan, Y., et al. (2008). Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genomics 35, 105–118. doi: 10.1016/S1673-8527(08)60016-8
Habibi, G. (2012). Exogenous salicylic acid alleviates oxidative damage of barley plants under drought stress. Acta Biol. Szeged. 56, 57–63. doi: 10.1016/j.chemosphere.2021.131827
Harfi, M. E., Charafi, J., Houmanat, K., Hanine, H., and Nabloussi, A. (2021). Assessment of genetic diversity in Moroccan sesame (Sesamum indicum) using ISSR molecular markers. OCL 28:8. doi: 10.1051/ocl/2020072
Hiremath, S. C., and Patil, C. G. (2002). Genome relations among octaploid species of Sesamum L. (Pedaliaceae). Cytologia 67, 403–409. doi: 10.1508/cytologia.67.403
Hiremath, S. C., and Patil, C. G. (1999). Genome homology and the putative progenitor of sesame. J. Cyt. Genet. 34, 69–74.
Hota, T., Pradhan, C., and Rout, G. R. (2019). Identification of drought tolerant Sesamum genotypes using biochemical markers. Indian J. Exp. Biol. 57, 690–699.
Hussein, Y., Amin, G., Azab, A., and Gahin, H. (2015). Induction of drought stress resistance in sesame (Sesamum indicum L.) plant by salicyclic acid and kinetin. J. Plant Sci. 10, 128–141. doi: 10.3923/jps.2015.128.141
Huttner, S., and Strasser, R. (2012). Endoplasmic reticulum-associated degradation of glycoproteins in plants. Front Plant Sci. 3:67. doi: 10.3389/fpls.2012.00067
Ibrahim, M. E., Bekheta, M. A., El-Moursi, A. D., and Gaafar, N. A. (2007). Improvement of growth and seed yield quality of (Vicia faba L.) plants as affected by application of some bioregulators. Aust. J. Basic Appl. Sci. 1, 657–666.
IPGRI, and NBPGR (2004). International Crop Network Series 5. Rome: International Plant Genetic Resources Institute, and Report of An International Workshop on Okra Genetic Resources. New Delhi: National Bureau of Plant Genetic Resources, 61.
Iqbal, A., Bhattacharyya, U., Akhtar, R., and Dasgupta, T. (2018). Genetic diversity computation in sesame genotypes using morphological traits and genic SSR markers. Indian J. Genet. Plant Breed. 78, 348–356. doi: 10.31742/IJGPB.78.3.12
Islam, F., Gill, R. A., Ali, B., Farrroq, M. A., Xu, L., Najeeb, U., et al. (2016). “Sesame,” in Sesame, Breeding Oilseed Crops for Sustainable Production: Opportunities and Constraints, ed. S. K. Gupta (Cambridge, MA: Academic Press), 135–147. doi: 10.1016/B978-0-12-801309-0.00006-9
Islam, S., Samiul, H., Islam, M. M., Emdad, E. M., Halim, A., Hossen, Q. M., et al. (2012). Tools to kill: genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genomics 13:493. doi: 10.1186/1471-2164-13-493
Jin, J., Zhang, H., Kong, L., Gao, G., and Luo, J. (2014). PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42, 1182–1187. doi: 10.1093/nar/gkt1016
Jin, U. H., Chun, J. A., Han, M. O., Li, J. W., Yi, Y. B. et al. (2005). Sesame hairy root cultures for extra-cellular production of a recombinant fungal phytase. Process Biochem. 40, 3754–3762
Joshi, H., and Mandavia, M. K. (2018). In silico identification and target prediction of microRNAs in sesame (Sesamum indicum l.) expressed sequence tags. Int. J. Curr. Microbiol. Appl. Sci. 7, 1275–1284. doi: 10.20546/ijcmas.2018.705.154
Junior, E. J. S., Segnana, L. G., Kitajima, E. W., and Bedendo, I. P. (2019). Sesame phyllody associated with a 16SrI-B phytoplasma, a‘Candidatus Phytoplasma asteris’-related strain, in Paraguay. Sci. Agric. 76, 47–50. doi: 10.1590/1678-992x-2017-0140
Kalwij, J. M. (2012). Review of ‘The Plant List, a working list of all plant species’. J. Veg. Sci. 23, 998–1002. doi: 10.1111/j.1654-1103.2012.01407.x
Kamran, M., Wennan, S., Ahmad, I., Xiangping, M., Wenwen, C., Xudong, Z., et al. (2018). Application of paclobutrazol affect maize grain yield by regulating root morphological and physiological characteristics under a semi-arid region. Sci. Rep. 8:4818. doi: 10.1038/s41598-018-23166-z
Kang, C. W., Kim, S. Y., Lee, S. W., Mathur, P. N., Hodgkin, T., Zhou, M. D., et al. (2006). Selection of a core collection of Korean sesame germplasm by a stepwise clustering method. Breed. Sci. 56, 85–91. doi: 10.1270/jsbbs.56.85
Kawase, M. (2000). Genetic relationships of the ruderal weed type and the associated weed type of Sesamum mulayanum Nair distributed in the Indian subcontinent to cultivated sesame, Sesamum indicum L. Jpn. J. Trop. Agri. 44, 115–122.
Kesawat, M. S., Das, B. K., Dey, S. K., Krishi, K. G., and Vidyalaya, V. (2015). Assessment of genetic diversity among Indian sesame (Sesamum indicum L.) accessions using RAPD, ISSR and SSR markers. Res. J. Biotechnol. 10, 35–47.
Khan, A., Ali, M., Khattak, A. M., Gai, W. X., Zhang, H. X., Wei, A. M., et al. (2019). Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 20:5321. doi: 10.3390/ijms20215321
Kim, D., Zur, G., Danin-Poleg, Y., Lee, S., Shim, K., Kang, C., et al. (2002). Genetic relationships of sesame germplasm collection as revealed by inter-simple sequence repeats. Plant Breed. 121, 259–262.
Kim, M., Lee, Y. J., Jee, S. C., Choi, I., and Sung, J. S. (2016). Anti-adipogenic effects of sesamol on human mesenchymal stem cells. Biochem. Biophys. Res. Commun. 469, 49–54. doi: 10.1016/j.bbrc.2015.11.070
Kitazawa, Y., Iwabuchi, N., Himeno, M., Sasano, M., Koinuma, H., Nijo, T., et al. (2017). Phytoplasma-conserved phyllogen proteins induce phyllody across the Plantae by degrading floral MADS domain proteins. J. Exp. Bot. 68, 2799–2811. doi: 10.1093/jxb/erx158
Kobayashi, T. (1991). “Cytogenetics of Sesame (Sesamum indicum),” in Chromosome Engineering in Plants: Genetics, Breeding, Evolution, eds T. Suchiya and P. K. Gupta (Amsterdam: Elsevier), 581–592. doi: 10.1016/b978-0-444-88260-8.50036-7
Kouighat, M., Hanine, H., El Fechtali, M., and Nabloussi, A. (2021). First report of sesame mutants tolerant to severe drought stress during germination and early seedling growth stages. Plants 10:1166. doi: 10.3390/plants10061166
Kumari, V., Singh, A., Chaudhary, H. K., Kumar, A., Prasad, R., Jambhulkar, S., et al. (2019). Identification of Phytopthora blight resistant mutants through induced mutagenesis in sesame (Sesamum indicum L.). Indian Phytopathol. 72, 71–77. doi: 10.1007/s42360-018-0089-9
Kwong, Q. B., Teh, C. K., Ong, A. L., Heng, H. Y., Lee, H. L., Mohamed, M., et al. (2016). Development and validation of a high density SNP genotyping array for African oil palm. Mol. Plant 9, 1132–1141. doi: 10.1016/j.molp.2016.04.010
Langham, R. (2007). “Phenology of sesame,” in Issues in New Crop and New Uses, eds J. Janick and A. Whipley (Alexandria, VA: ASHS Press), 144–182. doi: 10.3389/fpls.2021.767150
Laurentin, H. E., and Karlovsky, P. (2006). Genetic relationship and diversity in a sesame (Sesamum indicum L.) germplasm collection using amplified fragment length polymorphism (AFLP). BMC Genet. 7:10. doi: 10.1186/1471-2156-7-10
Li, C., Yao, Y., Zhao, G., Cheng, W., Liu, H., Liu, C., et al. (2011). Comparison and analysis of fatty acids, sterols, and tocopherols in eight vegetable oils. J. Agric. Food. Chem. 59, 12493–12498. doi: 10.1021/jf203760k
Li, D., Liu, P., Yu, J., Wang, L., Dossa, K., Zhang, Y., et al. (2017). Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol. 17:152. doi: 10.1186/s12870-017-1099-y
Liang, J., Sun, J., Ye, Y., Yan, X., Yan, T., Rao, Y., et al. (2021). QTL mapping of PEG-induced drought tolerance at the early seedling stage in sesame using whole genome re-sequencing. PLoS One 16:e0247681. doi: 10.1371/journal.pone.0247681
Lin, Y. X., Jiang, H. Y., Chu, Z. X., Tang, X. L., Zhu, S. W., and Cheng, B. J. (2011). Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12:76. doi: 10.1186/1471-2164-12-76
Llanes, A., Andrade, A., Alemano, S., and Luna, V. (2018). “Metabolomic approach to understand plant adaptations to water and salt stress,” in Plant Metabolites and Regulation Under Environmental Stress (Cambridge, MA: Academic Press), 133–144. doi: 10.1016/B978-0-12-812689-9.00006-6
Luan, H., Shen, H., Pan, Y., Guo, B., Lv, C., and Xu, R. (2018). Elucidating the hypoxic stress response in barley (Hordeum vulgare L.) during waterlogging: a proteomics approach. Sci. Rep. 8:9655. doi: 10.1038/s41598-018-27726-1
Madhupriya, Rao, G. P., Kumar, A., and Baranwal, V. K. (2015). Classification of sesame phytoplasma strain in India at 16Sr subgroup level. J. Plant Pathol. 3, 523–528. doi: 10.4454/JPP.V97I3.032
Mahajan, R. K., Bisht, I., and Dhillon, B. S. (2007). Establishment of a core collection of world sesame (Sesamum indicum L.) germplasm accessions. SABRAO J. Breed. Genet. 39, 53–64.
Majdalawieh, A. F., Massri, M., and Nasrallah, G. K. (2017). A comprehensive review on the anti-cancer properties and mechanisms of action of sesamin, a lignan in sesame seeds (Sesamum indicum). Eur J. Pharmacol. 15, 512–521. doi: 10.1016/j.ejphar.2017.10.020
Mancebo-Campos, V., Salvador, M. D., and Fregapane, G. (2014). Antioxidant capacity of individual and combined virgin olive oil minor compounds evaluated at mild temperature (25 and 400C) as compared to accelerated and antiradical assays. Food Chem. 150, 374–381. doi: 10.1016/j.foodchem.2013.10.162
Marakali, S. (2018). Identification and functional analyses of new sesame miRNAs (Sesamum indicum L.) and their targets. Mol. Biol. Rep. 45, 2145–2155. doi: 10.1007/s11033-018-4373-7
Mawcha, K. T., Gebru, M. M., Assefa, M. K., Mesfin, M., and Gebre, G. G. (2020). Morphological characterization and genetic diversity of sesame (Sesamum indicum L.) varieties cultivated in Ethiopia. Open Agric. J. 14, 117–129. doi: 10.2174/1874331502014010117
Mehalingam, P. (2012). Morphological and anatomical studies of the ovary galls of Sesamum indicum L. induced by the gall midge, (Asphondylia sesami Felt.). J. Ornam. Hortic. Plants 2, 191–200.
Mehmood, M. Z., Qadir, G., Afzal, O., Din, A. M. U., Raza, M. A., Khan, I., et al. (2021). Paclobutrazol improves sesame yield by increasing dry matter accumulation and reducing seed shattering under rainfed conditions. Int. J. Plant Prod. 15, 337–349. doi: 10.1007/s42106-021-00132-w
Mhadhbi, H., Fotopoulos, V., Mylona, P. V., Jebara, M., Aouani, M. E., and Polidoros, A. N. (2013). Alternative oxidase 1 (AOX1) gene expression in roots of Medicago truncatula is a genotype-specific component of salt stress tolerance. J. Plant Physiol. 170, 111–114. doi: 10.1016/j.jplph.2012.08.017
Miceli, A., Moncada, A., Sabatino, L., and Vetrano, F. (2019). Effect of gibberellic acid on growth, yield, and quality of leaf lettuce and rocket grown in a floating system. Agron 9:382. doi: 10.3390/agronomy9070382
Mitsuma, S., Ishigaki, E., Sugiyama, R., Asamizu, T., Yamada, K., and Kurosaki, F. (2004). Activation of phenylpropanoid metabolism in sesame by over-expression of carrot Calmodulin gene. Biol. Pharm. Bull. 10, 1621–1625. doi: 10.1248/bpb.27.1621
Mmadi, M. A., Dossa, K., Wang, L., Zhou, R., Wang, Y., Cisse, N., et al. (2017). Functional characterization of the versatile MYB gene family uncovered their important roles in plant development and responses to drought and waterlogging in sesame. Gene 8:362. doi: 10.3390/genes8120362
Mondal, N., Bhat, V. K., and Srivastava, S. P. (2010). Variation in fatty acid composition in Indian germplasm of sesame. J. Am. Oil Chem. Soc. 87, 1263–1269. doi: 10.2478/v10045-008-0049-y
Morakinyo, T. A., Akanbi, C. T., and Adetoye, O. I. (2020). Drying kinetics and chemical composition of Ceratotheca sesamoides Endl leaves. Int. J. Eng. Appl. Sci. Technol. 5, 456–465. doi: 10.33564/ijeast.2020.v05i04.074
Mosidis, J. A., and Yermanos, D. M. (1985). Plant position effect on seed weight, oil content and oil composition in sesame. Euphytica. 34, 193–199. doi: 10.1007/bf00022879
Murata, J., Ono, E., Yoroizuka, S., Toyonaga, H., Shiraishi, A., Mori, S., et al. (2017). Oxidative rearrangement of (+)-sesamin by CYP92B14 co-generates twin dietary lignans in sesame. Nat. Commun. 8:2155. doi: 10.1038/s41467-017-02053-7
Muthulakshmi, C., Sivaranjani, R., and Selvi, S. (2021). Modification of sesame (Sesamum indicum L.) for triacylglycerol accumulation in plant biomass for biofuel application. Biotechnol. Rep. 32:e00668. doi: 10.1016/j.btre.2021.e00668
Myint, D., Gilani, S. A., Kawase, M., and Watanabe, K. N. (2020). Sustainable sesame (Sesamum indicum L.) production through improved technology: an overview of production, challenges and opportunities in Myanmar. Sustainability 12:3515. doi: 10.3390/su12093515
Nanthakumar, G. K., Singh, N., and Vaidyanathan, P. (2000). Relationship between cultivated sesame (Sesamum sp.) and the wild relatives based on morphological characters, isozymes and RAPD markers. J. Genet. Breed. 54, 5–12.
Oh, E., Kim, S., Asekova, S., Yoon, N. Y., and Lee, J. D. (2018). An improved screening method for evaluation of resistance to Phytophthora blight in sesame cultivars and accessions. Plant Breed. Biotechol. 6, 305–308. doi: 10.9787/pbb.2018.6.3.305
Olowe, V. I., and Adeoniregun, O. A. (2010). Seed yield, seed attributes and oil content of newly released sesame (Sesamum indicum L.) varieties. Arch. Agron. Soil Sci. 56, 201–210. doi: 10.1080/03650340903006176
Padaria, J. C., Vishwakarma, H., Biswas, K., Jasrotia, R. S., and Singh, G. P. (2014). Molecular cloning and in-silico characterization of high temperature stress responsive TapAPX gene isolated from heat tolerant Indian wheat cv. Raj 3765. BMC Res. Notes 7:713. doi: 10.1186/1756-0500-7-713
Pal, S., Zhao, J., Khan, A., Yadav, N. S., Batushansky, A., Barak, S., et al. (2016). Paclobutrazol induces tolerance in tomato to deficit irrigation through diversified effects on plant morphology, physiology and metabolism. Sci. Rep. 6:39321. doi: 10.1038/srep39321
Pandey, S. K., Das, A., Rai, P., and Dasgupta, T. (2015). Morphological and genetic assessment of sesame (Sesamum indicum L.) accessions differing in origin. Physiol. Mol. Biol. Plants 21, 519–529. doi: 10.1007/s12298-015-0322-2
Park, J., Sundan, S., Sebastin, R., Baek, H., Kim, C., Lee, S., et al. (2015). Development and evaluation of core collection using qualitative and quantitative trait descriptor in sesame (Sesamum indicum L.) germplasm. Korean J. Crop Sci. 60, 1–10. doi: 10.7740/kjcs.2014.60.1.075
Park, J. S., and Lee, E. J. (2019). Waterlogging induced oxidative stress and the mortality of the Antarctic plant, Deschampsia antarctica. J. Ecol. Environ. 43, 1–8. doi: 10.1186/s41610-019-0127-2
Parsaeian, M., Mirlohi, A., and Saeidi, G. (2011). Study of genetic variation in sesame (Sesamum indicum L.) using agro-morphological traits and ISSR markers. Genetica 47, 359–367.
Pathak, N., Rai, K. A., Kumari, R., Thapa, A., and Bhat, K. V. (2014). Sesame crop: an underexploited oilseed holds tremendous potential for enhanced food value. Agric. Sci. 5, 519–529. doi: 10.4236/as.2014.56054
Qiao, X., Li, M., Li, L., Yin, H., Wu, J., and Zhang, S. (2015). Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol. 15:12. doi: 10.1186/s12870-014-0401-5
Quenum, F. J., and Yan, Q. (2017). Assessing genetic variation and relationships among a mini core germplasm of sesame (Sesamum indicum L.) using biochemical and RAPD markers. Am. J. Plant Sci. 8, 311–327. doi: 10.4236/ajps.2017.83022
Ramanujam, S. (1944). The cytogenetics of an amphidiploid Sesamum orientale S. prostratum. Curr. Sci. 13, 40–41.
Rana, M. S., Bhantana, P., Sun, X., Imran, M., Shaaban, M., Moussa, M., et al. (2020). Molybdenum as an essential element for crops: an overview. Biomed J. Sci. Tech. Res. 24, 18535–18547. doi: 10.26717/bjstr.2020.24.004104
Ranganatha, A. R. G., Jyotishi, A., Deshmukh, M. R., Bisen, R., Panday, A. K., Gupta, K. N., et al. (2013). Improved Technology for Maximizing Production of Sesame. New Delhi: ICAR.
Rani, S. T., and Kiranbabu, T. (2017). Screening sesame (Sesamum indicum L.) germplasm for thermotolerance. Adv. Crop. Sci. Tech. 5:300. doi: 10.4172/2329-8863.1000300
Rao, G. P., Un, N. S., and MadhuPriya. (2015). Overview on a century progress in research on sesame phyllody disease. Phytopathogenic Mollicutes 5, 74–83. doi: 10.5958/2249-4677.2015.00064.X
Rasheed, R., Iqbal, M., Ashraf, M. A., Hussain, I., Shafiq, F., Yousaf, A., et al. (2018). Glycine betaine counteracts the inhibitory effects of waterlogging on growth, photosynthetic pigments, oxidative defence system, nutrient composition, and fruit quality in tomato. J. Hortic. Sci. Biotechnol. 93, 385–391. doi: 10.1080/14620316.2017.1373037
Ruffo, C. K., Birnie, A., and Tengnas, B. (2002). Edible Wild plants of Tanzania. RELMA Technical Handbook Series 27. Nairobi: Regional Land Management Unit (RELMA), Swedish International Development Cooperation Agency (SIDA), 766.
Saha, R. R., Ahmed, B., Aziz, M. A., and Hossain, M. A. (2010). Screening of sesame genotypes for water logging tolerance. Bangladesh Agron. J. 13:8393.
Sarkar, P. K., Khatun, A., and Singha, A. (2016). Effect of duration of waterlogging on crop stand and yield of sesame. Int. J. Innov. Appl. Stud. 14, 1–6.
Sarkar, S., Datta, J., and Bandhu, S. (2006). Influence of growth regulators on performance of sesame under varying irrigation levels. J. Crop Weed 2, 1–4. doi: 10.9734/ajrcs/2018/40304
Sarkar, S., and Saha, P. K. (2013). Assessment of phenotypic and genotypic diversity in Indian sesame. Nucleus 56, 107–115. doi: 10.1007/s13237-013-0089-1
Sasikumar, K., and Kumar, K. (2014). Laboratory evaluation of botanical, biopesticide and insecticides against the shoot and leaf webber, Antigastra catalaunalis Duponchel. J. Biopestic. 7, 67–69.
Sehr, E. M., Okello-Anyanga, W., Hasel-Hohl, K., Burg, A., Gaubitzer, S., Rubaihayo, P. R., et al. (2016). Assessment of genetic diversity amongst Ugandan sesame (Sesamum indicum L.) landraces based on agromorphological traits and genetic markers. J. Crop Sci. Biotechnol. 19, 117–129. doi: 10.1007/s12892-015-0105-x
Shah, A., Tyagi, S., Sartale, G. D., Guzik, H. A., and Mulla, S. I. (2021). A comprehensive review on influence of light on signaling cross-talk and molecular communication against phyto-microbiome interactions. Crit. Rev. Biotechnol. 41, 370–393. doi: 10.1080/07388551.2020.1844618
Shi, L. K., Liu, R. J., Jin, Q. Z., and Wang, X. G. (2017). The contents of lignans in sesame seeds and commercial sesame oils of China. J. Am. Oil. Chem. Soc. 94, 1035–1044. doi: 10.1007/s11746-017-3018-7
Singh, A., Kumar, A., Yadav, S., and Singh, I. K. (2019). Reactive oxygen species-mediated signaling during abiotic stress. Plant Gene 18:100173. doi: 10.1016/j.plgene.2019.100173
Singh, A., and Lakhanpaul, S. (2020). Detection, characterization and evolutionary aspects of S54LP of SP (SAP54 Like Protein of Sesame Phyllody): a phytoplasma effector molecule associated with phyllody development in sesame (Sesamum indicum L.). Physiol. Mol. Biol. Plants. 26, 445–458. doi: 10.1007/s12298-020-00764-8
Singh, N., Jayaswal, P. K., Panda, K., Mandal, P., Kumar, V., Singh, B., et al. (2015). Single-copy gene based 50K SNP chip for genetic studies and molecular breeding in rice. Sci. Rep. 5:11600. doi: 10.1038/srep11600
Singh, P. K., Akram, M., Vajpeyi, M., Srivastava, R. L., Kumar, K., and Naresh, R. (2007). Screening and development of resistant sesame varieties against phytoplasma. Bull. Insectol. 60, 303–304.
Singh, S., Mahato, A. K., Jayaswal, P. K., Singh, N., Dheer, M., Goel, P., et al. (2020). A 62K genic-SNP chip array for genetic studies and breeding applications in pigeon pea (Cajanus cajan L. Millsp.). Sci. Rep. 10:4960. doi: 10.1038/s41598-020-61889-0
Song, Q., Hyten, D. L., Jia, G., Quigley, C. V., Fickus, E. W., Nelson, R. L., et al. (2013). Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One 8:e54985. doi: 10.1371/journal.pone.0054985
Song, X., Liu, G., Duan, W., Liu, T., Huang, Z., Ren, J., et al. (2014). Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Chinese cabbage. Mol. Genet. Genomics 289, 541–551. doi: 10.1007/s00438-014-0833-5
Stewart, B. W., Bray, F., Forman, D., Ohgaki, H., Straif, K., Ullrich, A., et al. (2016). Cancer prevention as part of precision medicine: plenty to be done. Carcinogenesis 37, 2–9. doi: 10.1093/carcin/bgv166
Takahashi, M., Nishizaki, Y., Sugimoto, N., Takeuchi, H., Nakagawa, K., Akiyama, H., et al. (2016). Determination and purification of sesamin and sesamolin in sesame seed oil unsaponified matter using reversed-phase liquid chromatography coupled with photodiode array and tandem mass spectrometry and high-speed countercurrent chromatography. J. Sep. Sci. 39, 3898–3905. doi: 10.1002/jssc.201600723
Tang, R., Zhu, W., Song, X., Lin, X., Cai, J., Wang, M., et al. (2016). Genome-wide identification and function analyses of heat shock transcription factors in potato. Front. Plant Sci. 7:490. doi: 10.3389/fpls.2016.00490
Taskin, K. M., Ercan, A. G., and Turgut, K. (1999). Agrobacterium tumefaciens mediated transformation of sesame (Sesamum indicum L.). Truk. J. Bot. 23, 291–295.
Thuc, L. V., Sakagami, J., Hung, L. T., Huu, T. N., Khoung, N. Q., and Vi, L. L. V. (2021a). Foliar selenium application for improving drought stress tolerance of sesame (Sesamum indicum L.). Open Agric. 6, 93–101. doi: 10.1515/opag-2021-0222
Thuc, L. V., Sakagami, J., Khuong, N. Q., Orgill, S., Huu, T. N., Lang, N. T. T., et al. (2021b). Effects of spraying gibberellic acid doses on growth, yield and oil content in black sesame (Sesamum indicum L.). Asian J. Crop Sci. 13, 1–8. doi: 10.3923/ajcs.2021.1.8
Troncoso-Ponce, M. A., Kilaru, A., Cao, X., Durrett, T. P., Fan, J., Jensen, J. K., et al. (2011). Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 68, 1014–1027. doi: 10.1111/j.1365-313X.2011.04751.x
Tseng, Y. W., Deng, W. L., Chang, C. J., Huang, J. W., and Jan, F. J. (2014). First Report on the Association of a 16SrII-A Phytoplasma with sesame (Sesamum indicum) exhibiting abnormal stem curling and phyllody in Taiwan. Plant Dis. 98:990. doi: 10.1094/PDIS-12-13-1212-PDN
Udomruk, S., Kaewmool, C., Pothacharoen, P., Phitak, T., and Kongtawelert, P. (2018). Sesamin suppresses LPS-induced microglial activation via regulation of TLR4 expression. J. Funct. Food. 49, 32–43. doi: 10.1016/j.jff.2018.08.020
Uncu, A. O., Frary, A., Karlovsky, P., and Doganlar, S. (2016). High-throughput single nucleotide polymorphism (SNP) identification and mapping in the sesame (Sesamum indicum L.) genome with genotyping by sequencing (GBS) analysis. Mol. Breed. 36:173. doi: 10.1007/s11032-016-0604-6
Unterseer, S., Bauer, E., Haberer, G., Seidel, M., Knaak, C., Ouzunova, M., et al. (2014). A powerful tool for genome analysis in maize: development and evaluation of the high density 600 k SNP genotyping array. BMC Genomics 15:823. doi: 10.1186/1471-2164-15-823
Ustun, R., Yol, E., Ikten, C., Catal, M., and Uzun, B. (2017). Screening, selection and real-time qPCR Validation for phytoplasma resistance in sesame (Sesamum indicum L.). Euphytica 213:159. doi: 10.1007/s10681-017-1936-y
Uzun, B., Lee, D., Donini, P., and Cagirgan, M. I. (2003). Identification of a molecular marker linked to the closed capsule mutant trait in sesame using AFLP. Plant Breed. 122, 95–97. doi: 10.1046/j.1439-0523.2003.00787.x
Uzun, B., Ulger, S., and Cagirgan, M. I. (2002). Comparison of determinate and indeterminate types of sesame for oil content and fatty acid compositions. Turk. J. Agric. For. 26, 269–274.
Vaja, M. B., and Golkiya, B. A. (2016). A draft genome sequencing using next generation sequencing technology in black sesame (Sesamum indicum L.). Res. J. Biotech. 11, 135–143.
Vasudeva, R. S., and Sahambi, H. S. (1955). Phyllody in Sesamum (Sesamum orientale L.). Indian Phytopathol. 8, 124–129.
Vekaria, G. B., Rakholiya, K. D., Vora, V. D., Patel, J. T., Sutaria, G. S., and Vekariya, P. D. (2017). Response of sesame (Sesamum indicum L.) to growth regulator under dry farming condition. Int. J. Curr. Microbiol. Appl. Sci. 6, 1113–1120. doi: 10.20546/ijcmas.2017.603.129
Vishwakarma, H., Junaid, A., Manjhi, J., Singh, G. P., Gaikwad, K., and Padaria, J. C. (2018). Heat stress transcripts, differential expression and profiling of heat stress tolerant gene TaHsp90 in Indian wheat (Triticum aestivum L.) cv C306. PLoS One 13:e0198293. doi: 10.1371/journal.pone.0198293
Vishwakarma, H., Sharma, J., Solanke, A., Singh, G. P., and Padaria, J. C. (2019). Isolation and characterization of stress inducible protein (TaSti/Hop) from heat tolerant wheat cultivar C306. Res. J. Biotech. 14, 111–121.
Vishwanath, H. S., Anilakumar, K. R., Harsha, S. N., Khanum, F., and Bawa, A. S. (2012). In vitro antioxidant activity of Sesamum indicum seeds. Asian J. Pharm. Clin. Res. 5, 56–60.
Wang, L., Li, D., Zhang, Y., Gao, Y., Yu, J., Wei, X., et al. (2016). Tolerant and susceptible sesame genotypes reveal water logging stress response patterns. PLoS One 11:e0149912. doi: 10.1371/journal.pone.0149912
Wang, L., Yu, S., Tong, C., Zhao, Y., Liu, Y., Song, C., et al. (2014). Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 15:R39. doi: 10.1186/gb-2014-15-2-r39
Wang, L., Zhang, Y., Qi, X., Li, D., Wei, W., and Zhang, X. (2012). Global gene expression responses to waterlogging in roots of sesame (Sesamum indicum L.). Acta Physiol. Plant. 34, 2241–2249.
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wang, Y., Zhang, Y., Zhou, R., Dossa, K., Yu, J., Li, D., et al. (2018). Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. PLoS One 13:e0200850. doi: 10.1371/journal.pone.0200850
Wei, W. L., Li, D. H., Wang, L. H., Ding, X., Zhang, Y. X., Gao, Y., et al. (2013). Morpho-anatomical and physiological responses to waterlogging of sesame (Sesamum indicum L.). Plant Sci. 208, 102–111. doi: 10.1016/j.plantsci.2013.03.014
Wei, X., Liu, K., Zhang, Y., Feng, Q., Wang, L., Zhao, Y., et al. (2015). Genetic discovery for oil production and quality in sesame. Nat. Commun. 6:8609. doi: 10.1038/ncomms9609
Wei, X., Gong, H., Yu, J., Liu, P., Wang, L., Zhang, Y., et al. (2017). Sesame FG: an integrated database for the functional genomics of sesame. Sci. Rep. 7:2342. doi: 10.1038/s41598-017-02586-3
Seemuller, E., and Harries, H. (2009). “Plant resistance,” in Phytoplasmas: Genomes, Plant Hosts and Vectors, eds P. G. Weintraub and P. Jones (Dossenheim: CABI), 147. doi: 10.1079/9781845935306.0147
Win, N. K. K., Back, C., and Jung, H. Y. (2010). Phyllody phytoplasma infecting sesame in Myanmar belongs to group 16SrI and subgroup 16SrI-B. Trop. Plant Pathol. 35, 310–313. doi: 10.1590/S1982-56762010000500006
Wu, K., Yang, M., Liu, H., Tao, Y., Mei, J., and Zhao, Y. (2014). Genetic analysis and molecular characterization of Chinese sesame (Sesamum indicum L.) cultivars using Insertion-Deletion (In Del) and Simple Sequence Repeat (SSR) markers. BMC Genetics 15:35. doi: 10.1186/1471-2156-15-35
Wu, M., Aquino, L. B. B., Barbaza, M. Y. U., Hsieh, C., Castro-Cruz, K. A., Yang, L., et al. (2019). Anti-inflammatory and anticancer properties of bioactive compounds from Sesamum indicum L.—A Review. Molecules 24:4426. doi: 10.3390/molecules24244426
Yadav, M., Chaudhary, D., Sainger, M., and Jaiwal, P. K. (2010). Agrobacterium tumefaciens-mediated genetic transformation of sesame (Sesamum indicum L.). Plant Cell Tiss. Org. Cult. 103, 377–386. doi: 10.1007/s11240-010-9791-8
Yan, W., Ni, Y., Liu, X., Zhao, H., Chen, Y., Jia, M., et al. (2021). The mechanism of sesame resistance against Macrophomina phaseolina was revealed via a comparison of transcriptomes of resistant and susceptible sesame genotypes. BMC Plant Biol. 21:159. doi: 10.1186/s12870-021-02927-5
Yang, X., Zhu, W., Zhang, H., Liu, N., and Tian, S. (2016). Heat shock factors in tomatoes: genome-wide identification, phylogenetic analysis and expression profiling under development and heat stress. Peer J. 4:e1961. doi: 10.7717/peerj.1961
You, J., Wang, Y., Zhang, Y., Dossa, K., Li, D., Zhou, R., et al. (2018). Genome-wide identification and expression analyses of genes involved in raffinose accumulation in sesame. Sci. Rep. 8:4331. doi: 10.1038/s41598-018-22585-2
You, J., Zhang, Y., Liu, A., Li, D., Wang, X., Dossa, K., et al. (2019). Transcriptomic and metabolomic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biol. 19:267. doi: 10.1186/s12870-019-1880-1
Yu, J., Golicz, A. A., Lu, K., Dossa, K., Zhang, Y., Chen, J., et al. (2019). Insight into the evolution and functional characteristics of the pan-genome assembly from sesame landraces and modern cultivars. Plant Biotechnol. J. 17, 881–892. doi: 10.1111/pbi.13022
Zhang, Y., Zhang, Y., Thakur, K., Zhang, F., Hu, F., Zhang, J., et al. (2021). Integration of miRNAs, degradome and transcriptome omics uncovers a complex regulatory network and provides insights into lipid and fatty acid synthesis during sesame seed development. Front. Plant Sci. 12:709197. doi: 10.3389/fpls.2021.709197
Zhang, R., Yu, Y., Hu, S., Zhang, J., Yang, H., Han, B., et al. (2016). Sesamin ameliorates hepatic steatosis and inflammation in rats on a high-fat diet via LXRα and PPARα. Nutr. Res. 36, 1022–1030.
Zhang, R., Zhou, Y., Yue, Z., Chen, X., Cao, X., Xu, X., et al. (2019). Changes in photosynthesis, chloroplast ultrastructure, and antioxidant metabolism in leaves of sorghum under waterlogging stress. Photosynthetica. 57, 1076–1083. doi: 10.32615/ps.2019.124
Zhang, X. (2018). The contrasting response to drought and waterlogging is underpinned by divergent DNA methylation programs associated with gene expression in sesame. Plant Sci. 277, 207–217. doi: 10.1111/pbi.13100
Zhang, X., Zhao, Y., Cheng, Y., Feng, X., Guo, Q., Zhou, M., et al. (2000). Establishment of sesame germplasm core collection in China. Genet. Resour. Crop Evol. 47, 273–279. doi: 10.1023/A:1008767307675
Zhang, Y., Gong, H., Li, D., Zhou, R., Zhao, F., Zhang, X., et al. (2020). Integrated small RNA and degradome sequencing provide insights into salt tolerance in sesame (Sesamum indicum L.). BMC Genomics 21:494. doi: 10.1186/s12864-020-06913-3
Zhang, Y., Li, D., Wang, Y., Zhou, R., Wang, L., Zhang, Y., et al. (2018). Genome-wide identification and comprehensive analysis of the NAC transcription factor family in Sesamum indicum. PLoS One. 13:e0136993. doi: 10.1371/journal.pone.0199262
Zhang, Y., Wang, L., Li, D., Gao, Y., Lu, H., and Zhang, X. (2014). Mapping of sesame waterlogging tolerance QTL and identification of excellent waterlogging tolerant germplasm. Sci. Agric. Sin. 47, 422–430. doi: 10.3864/j.issn.0578-1752.2014.03.002
Keywords: abiotic stresses, biotic stresses, core collection, genome assembly, germplasm, interspecific hybrids, phyllody, Sesamum indicum
Citation: Yadav R, Kalia S, Rangan P, Pradheep K, Rao GP, Kaur V, Pandey R, Rai V, Vasimalla CC, Langyan S, Sharma S, Thangavel B, Rana VS, Vishwakarma H, Shah A, Saxena A, Kumar A, Singh K and Siddique KHM (2022) Current Research Trends and Prospects for Yield and Quality Improvement in Sesame, an Important Oilseed Crop. Front. Plant Sci. 13:863521. doi: 10.3389/fpls.2022.863521
Received: 27 January 2022; Accepted: 16 March 2022;
Published: 06 May 2022.
Edited by:
Sezai Ercisli, Atatürk University, TurkeyCopyright © 2022 Yadav, Kalia, Rangan, Pradheep, Rao, Kaur, Pandey, Rai, Vasimalla, Langyan, Sharma, Thangavel, Rana, Vishwakarma, Shah, Saxena, Kumar, Singh and Siddique. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rashmi Yadav, cmFzaG1pLnlhZGF2MUBpY2FyLmdvdi5pbg==; Sanjay Kalia, c2FuamF5LmthbGlhQG5pYy5pbg==
 Rashmi Yadav
Rashmi Yadav Sanjay Kalia2*
Sanjay Kalia2* Parimalan Rangan
Parimalan Rangan Govind Pratap Rao
Govind Pratap Rao Vikender Kaur
Vikender Kaur Renu Pandey
Renu Pandey Vandna Rai
Vandna Rai Sapna Langyan
Sapna Langyan Virendra Singh Rana
Virendra Singh Rana Harinder Vishwakarma
Harinder Vishwakarma Anshuman Shah
Anshuman Shah Abhishek Saxena
Abhishek Saxena Ashok Kumar
Ashok Kumar Kuldeep Singh
Kuldeep Singh Kadambot H. M. Siddique
Kadambot H. M. Siddique