- 1Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2Institute of Biotechnology Research, Beijing Key Laboratory of Agricultural Genetic Resources and Biotechnology, Beijing, China
- 3Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
- 4Institute of Vegetable Research, Beijing Key Laboratory of Vegetable Germplasm Improvement, National Engineering Research Center for Vegetables, Beijing, China
- 5Institute of Millet Crops, Hebei Academy of Agriculture and Forestry Sciences, Shijiazhuang, China
CCT transcription factors are involved in the regulation of photoperiod and abiotic stress in Arabidopsis and rice. It is not clear that how CCT gene family expand and regulate heading date in foxtail millet. In this study, we conducted a systematic analysis of the CCT gene family in foxtail millet. Thirty-nine CCT genes were identified and divided into four subfamilies based on functional motifs. Analysis showed that dispersed duplication played a predominant role in the expansion of CCT genes during evolution. Nucleotide diversity analysis suggested that genes in CONSTANS (COL)-like, CCT MOTIF FAMILY (CMF)-like, and pseudoresponse response regulator (PRR)-like subfamilies were subjected to selection. Fifteen CCT genes were colocalized with previous heading date quantitative trait loci (QTL) and genome-wide association analysis (GWAS) signals. Transgenic plants were then employed to confirm that overexpression of the CCT gene SiPRR37 delayed the heading date and increased plant height. Our study first investigated the characterization and expansion of the CCT family in foxtail millet and demonstrated the role of SiPRR37. These results lay a significant foundation for further research on the function of CCT genes and provide a cue for the regulation of heading date.
Introduction
Gene duplication is prevalent and supplies raw genetic material for evolution (Ohno, 1970; Zhang, 2003). There are multiple ways for genes to become duplicated, including unequal crossing over, retroposition, and chromosomal (or genome) duplication, which result in different consequences (Ramsey and Schemske, 1998; Zhang, 2003). As per the region’s size, duplications are classified as tandem, segmental duplication, retrotransposition, or whole-genome duplication (Vision et al., 2000; Simillion et al., 2002; Cannon et al., 2004). Polyploidy has been widely appreciated and acknowledged as a significant mechanism of adaptation and speciation, in part because polyploidy duplicates the members of the entire regulatory network (Ramsey and Schemske, 1998; Bowers et al., 2003; Shoemaker et al., 2006). In Arabidopsis\ thaliana, two or more rounds of polyploidy were identified by comparative genomic analysis (Ku et al., 2000). About 70 million years ago (mya), an ancient polyploidization event occurred before the divergence of the major cereals from one another, and a subsequent polyploid event happened over 11 mya in maize (Gaut et al., 2000; Paterson et al., 2004). The latest research indicates that the polyploidization in Poaceae occurred at ~96 mya, while the split of Andropogoneae (containing sorghum and maize) and Setaria occurred at ~51 mya (Wang et al., 2015). Compared to whole-genome duplication, segmental duplications are large, interspersed duplications that involve 1,000–200,000 nucleotides (Samonte and Eichler, 2002). Segmental duplication plays an essential role in promoting the generation of new resistance gene specificities in the plant (Cannon et al., 2004). In addition, segmentally duplicated genes may have divergent expression patterns; for example, two myb-homologous genes in maize, p1 and p2, had a differential expression, generating tissue specificities (Zhang and Peterson, 2000). Tandem duplications usually result from unequal crossing-over; therefore, tandemly duplicated genes are linked in a chromosome (Zhang, 2003). As recent technology advances, genome structural variations, particularly copy-number variations (CNVs), have become the focus of attention. CNVs could be considered processes underlying gene duplication and loss, which tend to influence specific functional genes, such as environmental response-related genes (Korbel et al., 2008).
Under the model contributed to Ohno (1970), after duplication, one copy retains the ancestral function. In contrast, the other copies will be pseudogenized due to adverse selection or the acquisition of new tasks via the accumulation of substitutions. The subfunctionalization model and the duplication–degeneration–complementation model presume that two copies experienced degenerate mutations during evolution (Hughes, 1994; Force et al., 1999). Subneofunctionalization postulates rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicated genes (He and Zhang, 2005). The above models have been demonstrated in specific organisms. Gene duplication contributes not only to genomic and organismal evolution but also to the evolution of a sophisticated gene network (Wagner, 1994; Zhang, 2003). Without gene duplication, genomic evolution would be limited. In addition, gene duplication has also been recognized as a mechanism of genome adaptation in the course of adaptation to stressful environments, such as the identification of CNVs in species adapting to novel conditions (Kondrashov, 2012).
The important switch in flowering plants is the transition from vegetative to reproductive development, which is regulated by multiple environmental and endogenous factors (Simpson and Dean, 2002). The CCT [CONSTANS (CO), CO-like, and TIMING OF CAB EXPRESSION1 (TOC1)] domain-containing protein family has been intensively studied in flowering plants, such as A. thaliana, rice, and maize, which mainly affect flowering time (Putterill et al., 1995; Strayer et al., 2000; Robson et al., 2001; Liu et al., 2020). In rice, a total of 41 CCT genes have been identified, 18 of which have been shown to regulate heading date (Zhang et al., 2015, 2021). Hd1, an orthologue gene of CO in rice, could promote the heading date under short-day conditions and function as an inhibitor under long-day conditions; Hd1 accumulation was mediated by a RING-figure ubiquitin ligase (Yano et al., 2000; Yang et al., 2015). TOC1 encodes a circadian clock-associated factor containing the pseudoresponse response regulator (PRR) domain apart from the CCT domain (Strayer et al., 2000). According to the functional domains, the CCT gene family in rice was divided into three subgroups: the CCT MOTIF FAMILY (CMF) subgroup, the CONSTANS-like (COL) subgroup, and the PRR subgroup, while another subgroup TIFY was also subdivided in maize (Cockram et al., 2012; Jin et al., 2018).
A thorough study of flowering time has been conducted in Arabidopsis and rice. The CCT genes play an important role in the molecular regulatory network of flowering time in plants. FLOWERING LOCUST (FT) is a molecular hub in flowering time pathways that encodes mobile florigen in leaves (Abe et al., 2005). The transcription of FT could be activated by CO, and the latter was activated by Gigantea (GI) in Arabidopsis under short-day conditions (Hayama et al., 2003; Hayama and Coupland, 2003). In addition, there was another signaling pathway with an early heading date 1 (Ehd1)-centered specific pathway under long-day conditions in monocots. OsMADS50 activates the expression of Ehd1, and in turn, Ehd1 activates RFT1 (the long-day specific florigen) expression (Tsuji et al., 2011). In this pathway, an important CCT-motif gene, Grains Height Date 7 (Ghd7), functions as a repressor of Ehd1 to delay flowering (Xue et al., 2009). Many CCT family genes have been identified by QTL mapping and comparative genomic analysis in Setaria, such as the homologous genes of OsPRR95 and OsPRR59 (Mauro-Herrera et al., 2013).
Recently, map-based cloning and genome-wide association analysis (GWAS) have been utilized to perform genetic analysis of important traits in foxtail millet. Based on 916 accessions of foxtail millet, 13 signals related to heading date and early flowering were identified, including the homologous genes of known photoperiodic pathway genes Heading Date 1 (Hd1) and OsPRR37 (Jia et al., 2013). The homolog gene of Hd1 in foxtail millet with a splicing variation from “GT” to “AT” was correlated with heading date, and Hd1 was also under parallel domestication in rice, sorghum, and foxtail millet (Liu et al., 2015). Quantitative trait loci (QTL) mapping of 182 recombinant inbred lines (derived from a cross between foxtail millet accession B100 and green foxtail accession A10) under short-day (8 or 12 h light) and long-day (16 h light) conditions showed that the 8 and 12 h photoperiods shared more QTL, while 16 h light conditions only shared one QTL, which indicated that Setaria possessed secondary long-day genetic regulation apart from the short-day pathway (Doust et al., 2017). Another recombinant inbred line from a cross between foxtail millet accessions “Zhanggu” and “A2” also uncovered different QTLs under field conditions and short- and long-day photoperiods (Mauro-Herrera et al., 2013; Ni et al., 2017; Zhang et al., 2017). Although a significant number of QTLs have been identified by forward genetics, QTL intervals are still large, and only a few genes for heading date have been isolated in foxtail millet (Mauro-Herrera et al., 2013; Yoshitsu et al., 2017).
The high synteny and conservation of flowering time-related genes in plants provide insights into the regulatory mechanism of heading date in foxtail millet. The CCT family has been comprehensively analyzed in multiple plants, such as rice (Zhang et al., 2015, 2021), maize (Jin et al., 2018), wheat (Zheng et al., 2017), and soybean (Mengarelli and Zanor, 2021). However, the underlying expansion model and molecular mechanisms of CCT domain proteins remain largely unexplored in foxtail millet. Foxtail millet (Setaria italica) is an ancient cereal, whose attributes of small diploid genomes, self-fertilization, and strong abiotic stress accentuated foxtail millet as an ideal model for functional genomic studies (Doust et al., 2009; Li and Brutnell, 2011; Muthamilarasan and Prasad, 2015). Now, under pandemic, the foxtail millet has the potential to become a new staple crop (Muthamilarasan and Prasad, 2021). In this study, we identified and analyzed the molecular evolution and expansion patterns of the CCT family from the foxtail millet. The results showed that 15 CCT genes localized with the identified heading date QTLs. Combining genome-wide association analysis signals with the heading date, we verified the function of SiPRR37 in the heading date regulation network. These results provide clues for governing the heading date and future genetic improvements.
Materials and Methods
Identification of CCT Family Genes in Foxtail Millet
The foxtail millet genome (version 2.2, Bennetzen et al., 2012) was downloaded from the plant genome database Phytozome.1 The Hidden Markov Model (HMM) program hmmsearch was applied to identify CCT proteins with the parameter “--cut_tc” to control model-specific thresholding. The HMMER profile of the CCT domain (PF06203) was downloaded from the PFAM protein family dataset (Finn et al., 2010).2 The basic properties of the CCT genes, namely, the length of amino acids, isoelectric point (PI), and molecular weights (MW), were estimated using the ExPASy program.3
Phylogenetic Analysis and Structure of CCT Proteins
To analyze the evolutionary relationship, the reference genome sequences of Setaria viridis (V2.1), Sorghum bicolor (V3.1.1), Oryza sativa (V7.0), and Zea mays (RefGen V4) were downloaded from Phytozome. Similarly, we identified the putative CCT genes in green foxtail, rice, maize, and sorghum by HMM profiles corresponding to the CCT domain. The primary protein of CCT family from these four species was aligned with clustalW and the phylogenetic tree was constructed by MEGA-X using the neighbor-joining (NJ) method with 1,000 bootstraps (Kumar et al., 2018). Finally, the evolutionary tree was visualized and illuminated by the software iTOL (Letunic and Bork, 2007).4 The conserved motifs of CCT proteins in foxtail millet were determined using the Pfam 33.1 database. Promoter sequences (2 kb upstream of the transcription start site) were extracted based on the genome sequence and annotation file, and promoter cis-acting elements were identified by Plantcare.5 The Gene Structure Display Service (GSDS 2.0; Hu et al., 2015) was used to draw the main protein domain and cis-regulatory elements distribution.
Chromosomal Localization and Duplication of CCT Family Genes
Chromosomal localization information for CCT genes in foxtail millet was identified from the reference genome database, and the genes were mapped to nine chromosomes using MapChart software according to the manual (Voorrips, 2002).6 To investigate the evolutional mechanism of the CCT family genes, five modes of duplication as whole-genome duplicates (WGD), tandem duplicates (TD), proximal duplicates (less than 10 gene distance on the same chromosome: PD), transposed duplicates (transposed gene duplications: TRD), or dispersed duplicates (DSD) occurred in CCT genes were obtained from PlantDGD database (Qiao et al., 2019).7 Gene duplication and synteny analysis were also performed using MCScanX software (Wang et al., 2012). The whole-genome duplication pattern was visualized using Circos software (Krzywinski et al., 2009).
Expression Patterns and Alternative Splicing Event Identification of CCT Family Genes
The expression profile of some CCT genes in foxtail millet cultivar “Jingu 21” was extracted from the multi-omics database for S. italica (MDSi; Yang et al., 2020),8 which contained 23 samples from several tissues (including seeds, leaf, panicle, stem, and root) at vegetative and reproductive stages. The expression data (fragments per kilobase of transcript per million fragments mapped, FPKM values) were normalized into log2 (1 + FPKM), and the heat map was constructed using the R package “pheatmap” (version 1.0.12). To identify alternative splicing (AS) event, the genomic annotation file of the foxtail millet was used to extract all transcripts of CCT family genes. Then, AS events identification was performed by using ASTALAVISTA (Foissac and Sammeth, 2007).
Variation Distribution of CCTs and Nucleotide Diversity
To characterize the natural variation of CCT genes in foxtail millet, single-nucleotide polymorphisms (SNPs) in the coding region of CCT genes were identified based on the haplotype variation map from whole-genome resequencing of 312 accessions (Li et al., 2021). In addition, we extracted the SNPs from the genomic sequence, which referred to the sequence from the transcription initiation site to the transcription stop site. SNPs from the promoter region (within 2 kb upstream of the transcription start site) were also extracted. The average number of nucleotide differences per site between the two random sequences (π), the Watterson estimator (θ), and Tajima’s D were calculated using DnaSP v5.0 according to the manual (Librado and Rozas, 2009). The π ratio of genetic diversity in landraces to that in improved cultivars of foxtail millet was calculated for selection signals related to screening improvement.
Functional Identification of SiPRR37 by Transformation
We previously found that SiPRR37 had different haplotypes with or without the Tc1-Mariner transposon (Li et al., 2021). To validate the function of the CCT gene SiPRR37, we cloned the functional SiPRR37 haplotype and performed overexpression assays to generate the OE-SiPRR37 transgenic lines. Total RNA was extracted from the leaf tissue of foxtail millet landrace “Daqingmiaoyuci” using an RNAprep Prue plant kit (Tiangen Biotech Co., Ltd.). After isolation, first-strand cDNA synthesis was performed using the Transcriptor First Strand cDNA synthesis kit (Takara Biotech Co., Ltd) according to the manufacturer’s protocol. The 2.2-kb coding sequence of SiPRR37 was amplified using KOD FX (TOYOBO Life Science) and cloned into the binary vector pCUbi1390 driven by the ubiquitin promoter. The specific primers were designed using Primer-BLAST (National Center for Biotechnology Information, Maryland, United States) and are listed in Supplementary Table S7. The resultant plasmid was transformed into Agrobacterium tumefaciens strain EHA105. Agrobacterium-mediated ProUbi::SiPRR37 transformation was performed according to a previous study with some modifications (Yang et al., 2020). First, the palea and lemma of Ci846 mature seeds were removed and surface sterilized. The rinsed seeds were cultured on a medium for callus induction at 28°C in the dark for 8–10 weeks. Strain EHA105 harboring the ProUbi::SiPRR37 vector was cultured overnight and resuspended to a density of 600 nm (OD600) = 0.5. High-quality calli were infected in resuspended cells for 5 min and transferred into a co-cultivation medium at 22°C in the dark. Then, calli were transferred to callus induction medium and selection medium with hygromycin B successively. Resistant calli were transferred to the shoot induction medium and cultured for root formation. Putative transgenic plants with healthy roots were moved to pots and detected by PCR. In addition, the expression level of positive transgenic plants was measured by qPCR.
The T2 transgenic lines with a higher expression level of SiPRR37 were chosen to observe the phenotype change. Wild-type Ci846 and overexpressing transgenic lines were grown under field conditions in Beijing (40°13′N, 116°13′E). Heading date was scored by means ± SDs (n = 20) in the field conditions.
Subcellular Localization and Transcriptional Activity Assay
To examine the subcellular localization, Agrobacterium GV3101 containing pCAMBIA1305-GFP ligated to two haplotypes of SiPRR37 was transformed into Nicotiana benthamiana leaves. After 2 days of incubation, the infected leaves were harvested and used to observe GFP fluorescence using a confocal laser scanning microscope Nikon A1R. For the transcriptional activity assay, the functional and mutational SiPRR37 protein was amplified and cloned into the GAL4BD vector. The plasmids containing GAL4BD-SiPRR37/GAL4BD-Siprr37, pRTL, and 35sLUC were transformed into Arabidopsis protoplast cells as previously described with modifications (Yoo et al., 2007). The positive and negative controls were GAL4BD-VP16 and GAL4BD, respectively. Due to the transcriptional inhibitory activity of SiPRR37, the CDS of SiPRR37 and Siprr37 was cloned and introduced into GAL4BD-VP16. After incubation in the dark overnight, the protoplast cells were harvested and used to measure luciferase activity according to the manufacturer’s instructions for the Dual-Luciferase Reporter (DLRTM) Assay System (Promega).
RNA Sequencing
RNA sequencing was performed on T2 OE-SiPRR37 plants under long-day conditions. The leaf tissues with three biological replicates were harvested at the vegetative stage for T2 plants. Six samples were prepared for 150 bp paired-end sequencing using an Illumina HiSeq X Ten platform. The raw data were trimmed and mapped to the foxtail millet reference genome using HISAT2 (hierarchical indexing for spliced alignment of transcripts; Kim et al., 2015). The FPKM of all genes was calculated using StringTie (v2.1.4, Pertea et al., 2015). Differentially expressed genes (DEGs) were detected using the R package edgeR (Robinson et al., 2010) following the criteria [|log2(fold-change)| > 1 and false discovery rate < 0.01]. The DEGs were validated through qRT-PCR. MapMan bins of the foxtail millet genome were assigned (Thimm et al., 2004).9 Open-source MapMan software was used to identify the categories of differentially expressed genes. The RNA-seq raw data were submitted to the National Center for Biotechnology Information (NCBI) Bioproject under accession number PRJNA797949.
Results
Identification and Phylogenetic Analysis of CCT Proteins in Foxtail Millet
A total of 39 putative CCT genes were identified in the S. italic reference genome (Supplementary Table S1). The amino acid number of these CCT proteins varied greatly, ranging from 122 (Seita.8G159000) to 760 (Seita.9G445200), and the molecular weight ranged from 13.82 to 82.67 kDa. CCT family genes were distributed on each chromosome (Supplementary Figure S1). There were nine CCT genes on chromosome 9; chromosomes 1 and 4 harbored seven, and five CCT genes, respectively; chromosomes 2 and 3 each contained four CCT genes; chromosomes 6, 7, and 8 each contained three CCT genes; and chromosome 5 carried only one CCT gene.
To investigate the evolutionary relationship of CCT proteins in S. viridis, S. italic, O. sativa, S. bicolor, and Z. mays, a phylogenetic tree was constructed. A total of 208 CCT genes were identified from five Poaceae plants, with 39 CCT genes in foxtail millet, 39 CCT genes in green foxtail, 40 CCT genes in rice, 35 CCT genes in sorghum, and 55 CCT genes in maize. The number of genes in rice and maize was similar to previous studies (Figure 1; Zhang et al., 2015; Jin et al., 2018). Based on the domains they contained, all CCT genes were also divided into four subfamilies, with the largest CMF-like subfamily containing 108 members. The COL-like subfamily was composed of 54 members, the PRR-like subfamily had 27 members, and the TIFY-like subfamily involved 19 members. In each subfamily, CCT genes from the five species were closely clustered, suggesting that CCT gene families may have evolved before species differentiation.
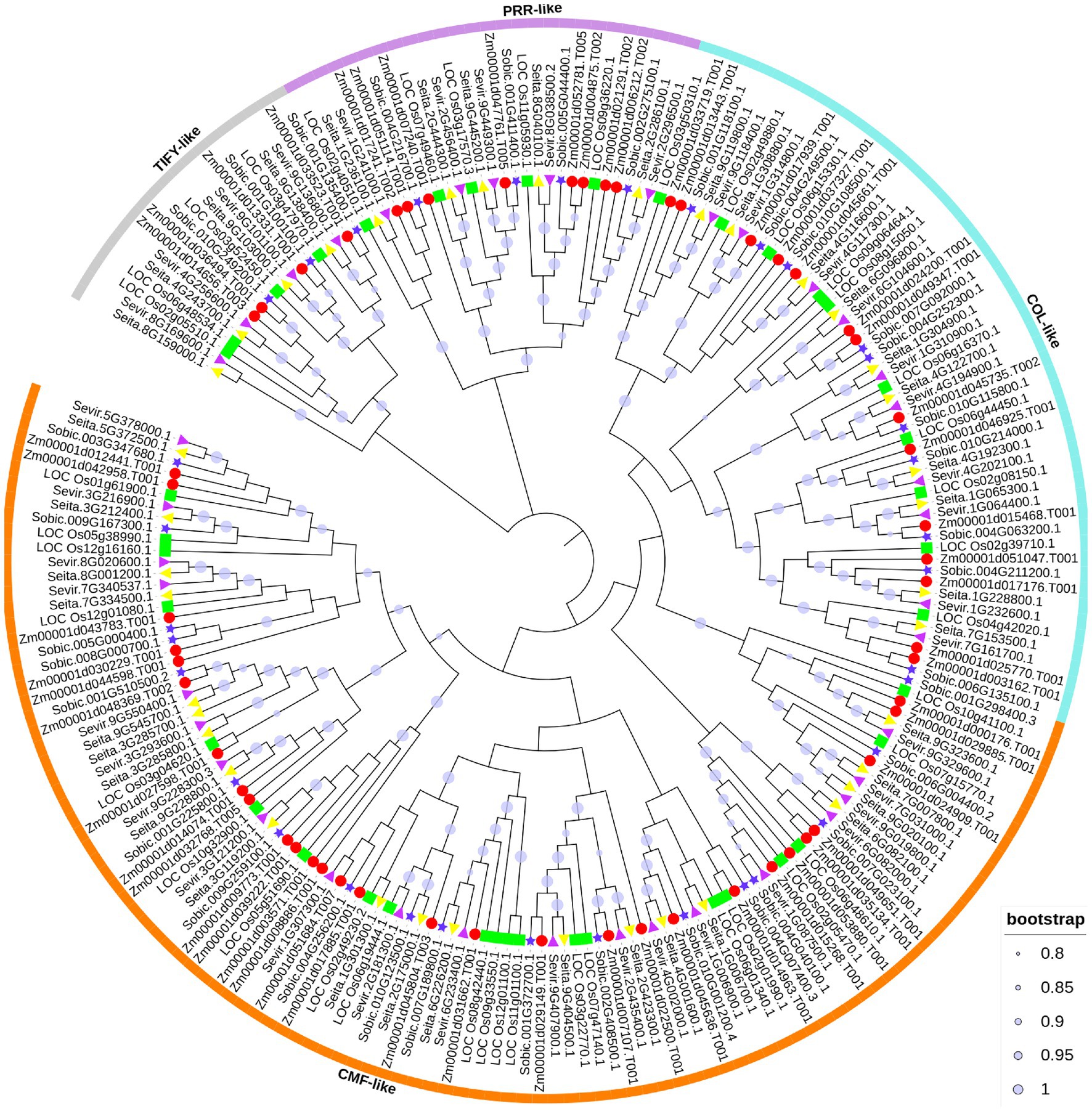
Figure 1. Phylogenetic relationship of CCT family genes. The putative CCT genes from rice, maize, sorghum, green foxtail, and foxtail millet were identified to construct the neighbor-joining phylogenetic tree. Solid lines with different colors enclose predominantly CONSTANS (COL)-like, CCT MOTIF FAMILY (CMF)-like, pseudoresponse response regulator (PRR)-like, and TIFY-like subfamilies. To distinguish the species, the red circle, blue pentagram, green square, purple triangle, and yellow triangle represent the CCT proteins from maize, sorghum, rice, green foxtail and foxtail millet, respectively.
Based on the topology of CCT genes in four Poaceae plants, there were 10 COL-like genes, 20 CMF-like genes, five PRR-like genes, and four TIFY-like genes in foxtail millet (Figure 2A). The analysis of these CCT protein structures showed a similar pattern with the CCT family divergence (Figure 2B). It also indicated that different subfamilies encoded proteins of various lengths and that the PRR subfamily was the longest. In addition, four genes only including the CCT domain were clustered in the COL subfamily or PRR subfamily, and one gene with zf-B box domain was clustered in the CMF-like subfamily. Partial CCT genes also exhibited a similar phenomenon in maize (Jin et al., 2018). It could be inferred that these genes lost the functional B-box type zinc finger domain during CCT gene family expansion.
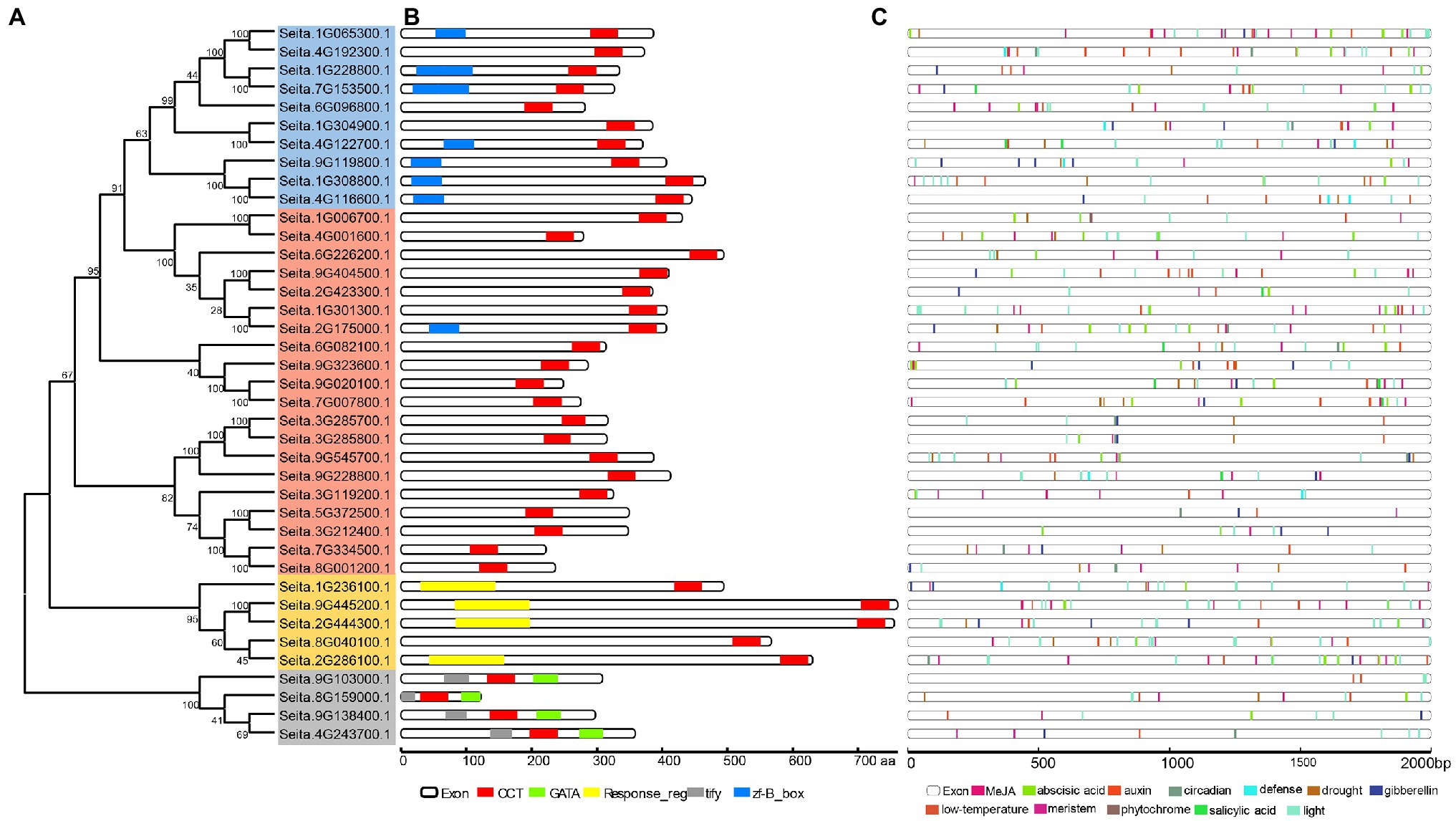
Figure 2. Phylogenetic relationship, conserved motifs, and promoter regulatory element analysis of foxtail millet CCT genes. (A) The full-length protein sequences of CCT genes were used to construct the neighbor-joining phylogenetic tree with 1,000 bootstrap replicates. The blue, orange, yellow, and gray shadow color indicated the COL-like, CMF-like, PRR-like, and TIFY-like subfamilies, respectively. (B) The main motif composition of CCT proteins is shown. Each motif is represented in a colored box. Protein length is scaled at the bottom. (C) The main response elements in the promoters are shown. Different regulatory elements are displayed in different colors.
The promoter sequences of CCT genes were analyzed in the PlantCARE database. The result showed that the promoters contained over 20 conserved elements, such as G-box, sp1, and TGA-element, with the highest number of G-box and ABRE elements (Figure 2C; Supplementary Table S2). These conserved elements were related to light responsiveness, plant hormones (abscisic acid, gibberellin, MeJA, and salicylic acid), and stress (low temperature and drought). Almost each CCT gene included a light responsiveness element, which inferred that the CCT domain genes were related to photoperiod.
Expression Profiles and AS Events of CCT Family Genes
To further study the functions of CCT genes, we detected the expression profile of partial CCT genes in different tissues and multiple growth stages. Overall, CCT genes in the same subfamily displayed similar expression patterns (Supplementary Figure S2). For example, most of the CMF subfamily genes were not expressed or had a lower expression level in all 23 tissues/stages, while the PRR subfamily genes showed a higher expression level. Moreover, we compared the expression patterns of segmentally duplicated genes. Some duplicated genes, including Seita.1G006700/Seita.4G001600, had similar expression patterns, while largely duplicated genes exhibited different patterns in most tissues. It was speculated that the newly duplicated genes played a powerful role in functional innovation.
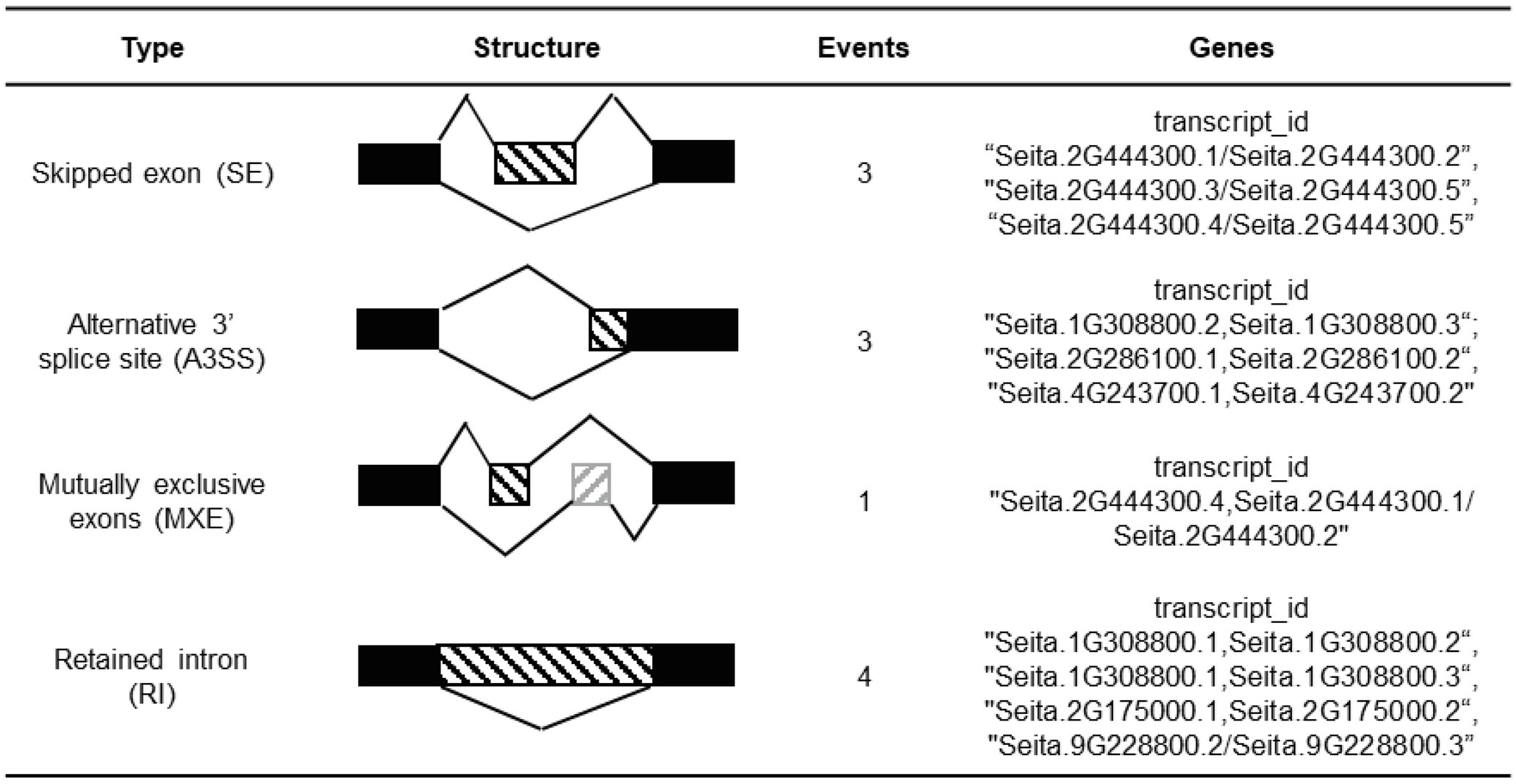
In total, 11 AS events were identified in the CCT family genes, which were mainly distributed in six genes containing two CMF-like subfamily genes, two PRR-like subfamily genes, one COL-like gene, and TIFY-like subfamily gene (Table 1). Among of six AS genes, Seita.2G444300 and Seita.1G308800 consisted of two types of AS events, while AS events did not occur in their highly homologous genes.
Gene Duplication of CCT Family Genes
Gene duplication is an important way for gene family expansion. To investigate the mode of duplication, we searched the PlantDGD database to extract all kinds of duplicates in foxtail millet CCT family genes. The results showed that a total of 48 duplication events occurred, including 11 WGD, 8 TRD, 28 DSD, and only one pair of tandem duplication genes, Seita.3G285700 and Seita.3G285800, closely clustering on chromosome 3 (Figure 3A; Supplementary Figure S1).
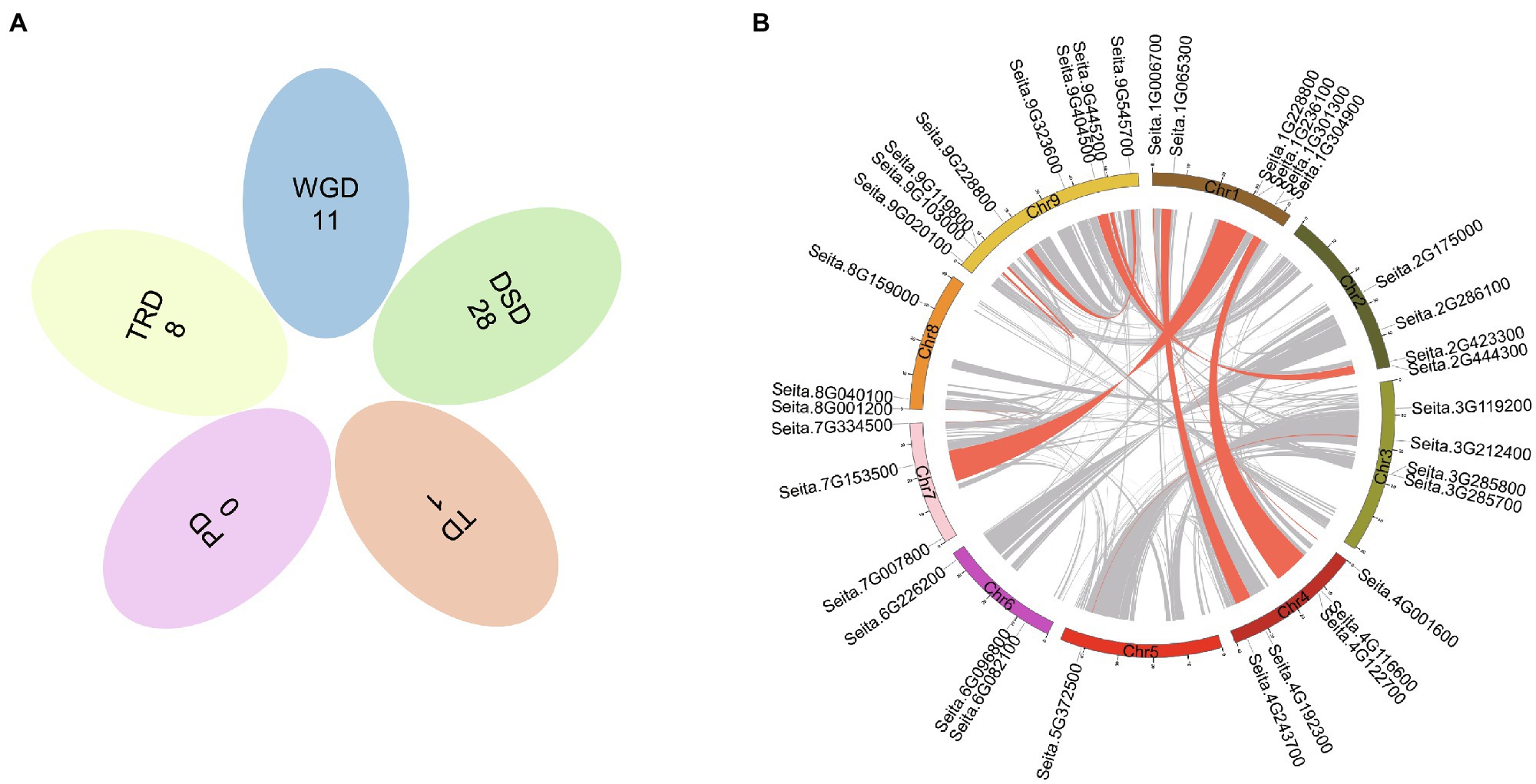
Figure 3. Possible modes of duplications in foxtail millet CCT genes. (A) Venn diagram shows the possible modes of duplication. WGD, whole-genome duplicates; DSD, dispersed duplicates; TRD, transposed duplicates; PD, proximal duplicates; and TD, tandem duplicates. (B) Circos plot showing the genomic distribution of CCT genes and the synteny blocks (gray lines) in foxtail millet. Red lines highlight the synteny blocks, including duplicated pairs of CCT genes.
Eleven WGD events were identified, including four duplication events in the COL-like subfamily, five events in the CMF-like subfamily, one event in the PRR subfamily, and one in the TIFY subfamily (Figure 3B). Interestingly, four WGD events (Seita.1G006700/ Seita.4G001600, Seita.1G065300/Seita.4G192300, Seita.1G304900/Seita.4G122700, and Seita.1G308800/Seita.4G116600) arose between chromosomes 1 and 4 (Figure 3B; Supplementary Table S3). Among eight TRD events, we found that the transposon jumping near Seita.2G444300 was highly active, which was associated with the other two genes (Seita.1G236100 and Seita.2G286100). The intricate network of DSD events played a predominant role in CCT gene family expansion (Supplementary Table S3). The result indicated that DSD events mainly happened in the same subfamily. And most genes participated in two or three rounds of dispersed duplication events. Extensive duplication facilitates the rapid evolution of the CCT gene family.
Nucleotide Diversity of CCT Family Genes in Foxtail Millet
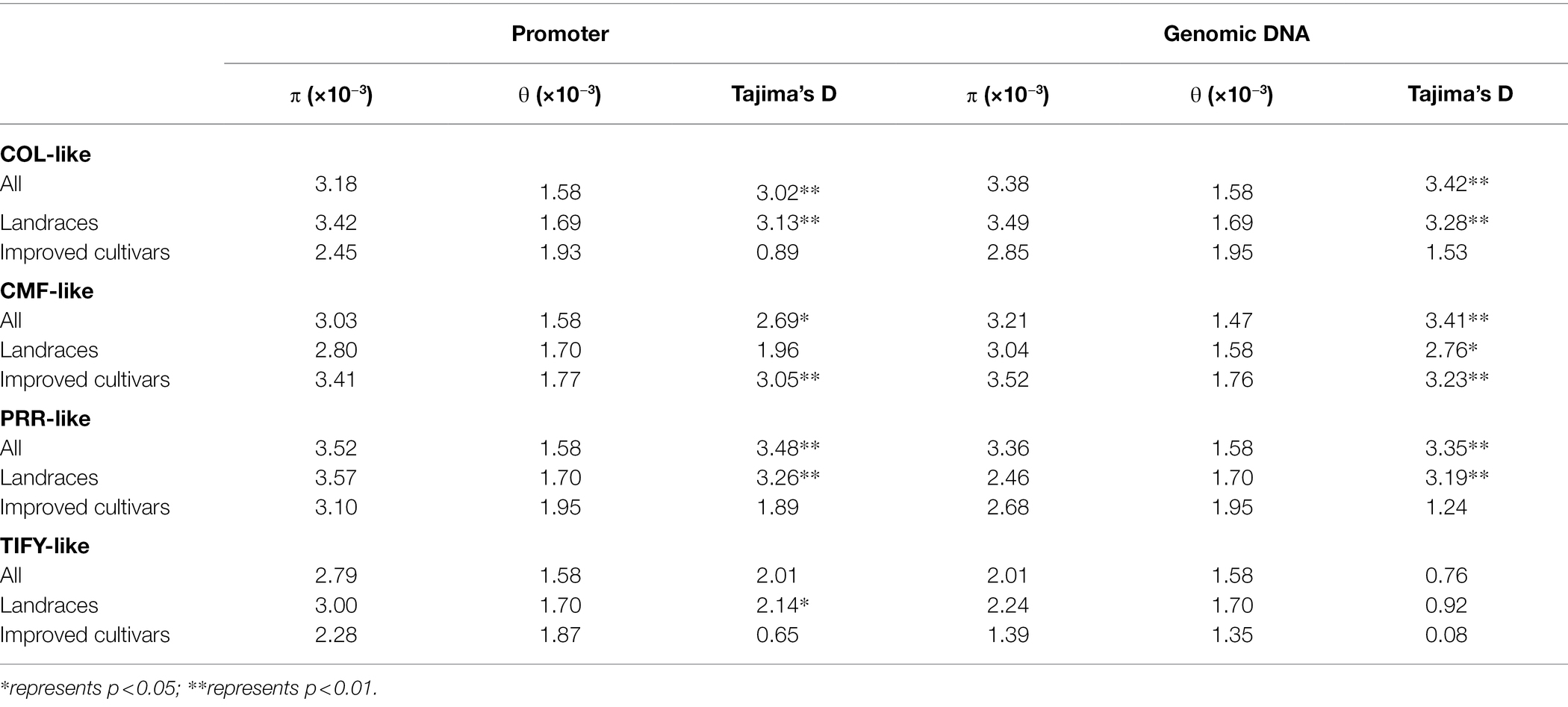
In an attempt to characterize the nucleotide diversity of CCT genes, we analyzed the genomic region and the promoter region of CCT family genes within 203 foxtail millet landraces and 96 improved cultivars. The results showed that the average nucleotide diversity of each CCT subfamily was higher than that of the whole genome in foxtail millet (π = 1.621 × 10−3 in traditional landraces and π = 1.568 × 10−3 in improved cultivars; Li et al., 2021; Table 2). In the genomic region, the TIFY subfamily showed lower nucleotide diversity (π = 2.01 × 10−3) than that of the other three subfamilies. Overall, the nucleotide diversity in the promoter regions exhibited a similar level to the genomic regions, except for the TIFY subfamily (Table 2). In addition, Tajima’s D of three subfamilies except for the TIFY subfamily reached a significant positive level. Interestingly, we also found that the diversity of the COL and TIFY subfamilies was lower in improved cultivars than in landraces, while the opposite pattern appeared in the CMF family. Therefore, four subfamilies underwent selection to different extents during improvement.
Based on the haplotype map of 312 accessions in foxtail millet, we cataloged the SNP variation profile of 39 CCT genes. The results showed that a total of 19 CCT genes had a missense mutation, including four CMF-like genes, nine COL-like genes, three PRR-like genes, and three TIFY-like genes (Supplementary Table S4). Population genetic variation provides the rich potential to facilitate adaptation to novel environments. To determine whether the CCT gene family could be associated with improvement selection, we utilized π to screen the selective signals. We found only one TIFY-like gene (Seita.9G103000) located in the improvement-related selective interval (Supplementary Figure S3).
Comparison of CCT Gene Localization and Heading Date QTLs
The protein family containing the CCT domain is mainly related to the flowering time pathway. To investigate the association between the CCT genes and heading date in foxtail millet, we collected heading date QTLs and GWAS signals according to previous studies and compared CCT gene localization with known heading date QTLs (Jia et al., 2013; Mauro-Herrera et al., 2013; Doust et al., 2017; Zhang et al., 2017; Li et al., 2021). Twenty-seven heading date-related QTLs and 40 GWAS signals were identified (Supplementary Tables S5, S6). Among 39 CCT family genes, 15 genes were located in the QTL regions or adjacent to the GWAS signals, including five COL-like genes, seven CMF-like genes, and three PRR-like genes (Supplementary Tables S5, S6). These genes may play an important role in regulating heading date.
SiPRR37 Is Related to Foxtail Millet Heading Date
In a previous study, we identified a significant association interval including SiPRR37 by GWAS (Li et al., 2021). SiPRR37 belongs to a member of the CCT transcription factor family. To validate the function of SiPRR37, we introduced the functional haplotype into the robust transformation receptor Ci846. The presence of the overexpression cassette in the transgenic lines was confirmed by PCR analysis and sequencing of the PCR fragment. Significantly enhanced expression of SiPRR37 in transgenic lines was also confirmed by real-time RT-PCR (Figure 4C). The positive transgenic lines showed an obvious difference in heading date compared with wild-type plants in the T1 generation under field conditions (Figure 4A). The SiPRR37-OE lines headed about 10 days later than the wild-type plant, indicating that SiPRR37 negatively regulated the heading date in foxtail millet (Figure 4B).
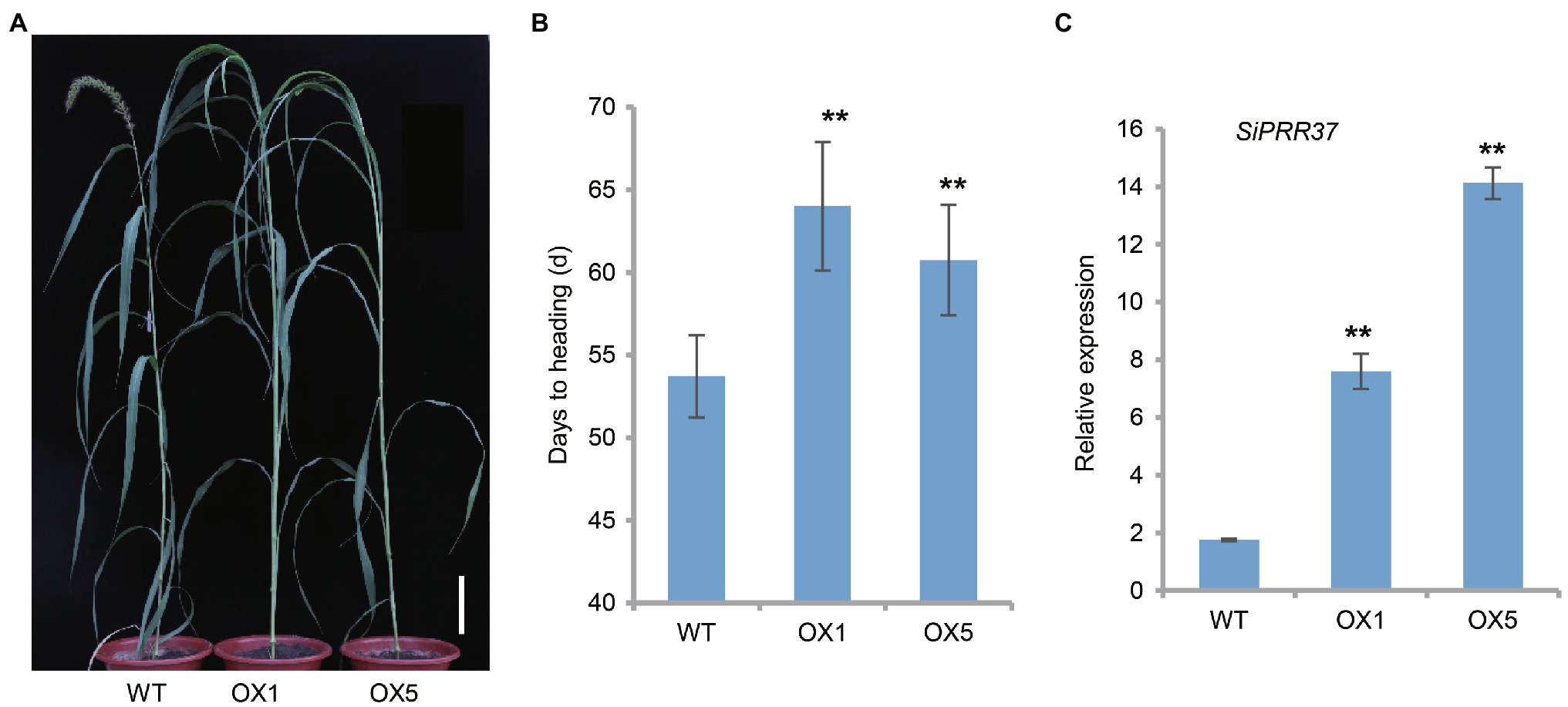
Figure 4. Phenotypical characterization of transgenic plants overexpressing the SiPRR37 gene in foxtail millet. (A) Representative graphic of the overexpressing transgenic plants at the heading date stage grown in the Beijing field in 2020. The scale bar indicates 10 cm. (B) Statistical analysis of days to heading of transgenic lines and wild type by Student’s t-test. Error bars represent ± SD (n = 10). ** indicates p < 0.01. (C) The transcriptional levels of SiPRR37 in the overexpressing plants and wild type. Data were shown as means ± SD (n = 3).
It was still unclear whether the SiPRR37-OE lines delayed the heading date under different conditions. To address this question, we planted the transgenic lines on June 22 and observed the phenotype in the greenhouse with a normal long-day photoperiod. We found that the transgenic line OX5 suppressed heading compared to the wild type (Supplementary Figure S4B). In addition, the SiPRR37-OE line OX5 influenced plant height simultaneously (Supplementary Figures S4A,C). Taken together, these results suggest that SiPRR37 is an important gene underlying heading date and plant architecture in foxtail millet.
SiPRR37 had two main haplotypes in foxtail millet of 312 accessions. In the mutant, transcription was truncated, and the CCT domain was lost due to Tc1-Marina transposon insertion (Li et al., 2021). Thus, we observed the sublocalization of two haplotypes. The two haplotypes (SiPRR37 and Siprr37) with GFP labels driven by the CaMV35S promoter were introduced into tobacco leaves. The SiPRR37-GFP signal appeared in the nucleus, while the Siprr37-GFP signal was detected in the nucleus and cytoplasm, similar to the localization of the blank control (Supplementary Figure S5A). These results validate that the CCT domain plays an important role in the nuclear localization of proteins (Robson et al., 2001). Thus, we evaluated the transcriptional activity of two haplotypes in Arabidopsis protoplasts. The results showed that SiPRR37 exhibited transcriptional inhibitory activity, which indicated that SiPRR37 functioned as a repressor in regulating heading date (Supplementary Figure S5B). In contrast, Siprr37 lost its transcriptional activity despite its nuclear localization.
To study the effect of SiPRR37 on the downstream genes, RNA-seq was performed on the leaves of the wild type and SiPRR37-OE transgenic line OX5 at the vegetative stage. Compared with the wild-type line, the SiPRR37-OE transgenic line has 1,295 DEGs, including 695 downregulated genes and 600 upregulated genes. DEGs were significantly enriched in MapMan categories, including signal, stress, protein, secondary metabolism, nucleotide metabolism, and cofactor metabolism, and the downregulated and upregulated gene compositions were different (Figure 5A). Among 695 downregulated genes, the most prominent subcategory was transcriptional regulation, which contained 36 transcription factors referring to the diverse aspects of development. In addition, an abundance of DEGs involved stress, which was also the target gene of PRR7 in Arabidopsis (Liu et al., 2013). This result indicated that SiPRR37 may participate in multiple regulation pathways besides heading date.
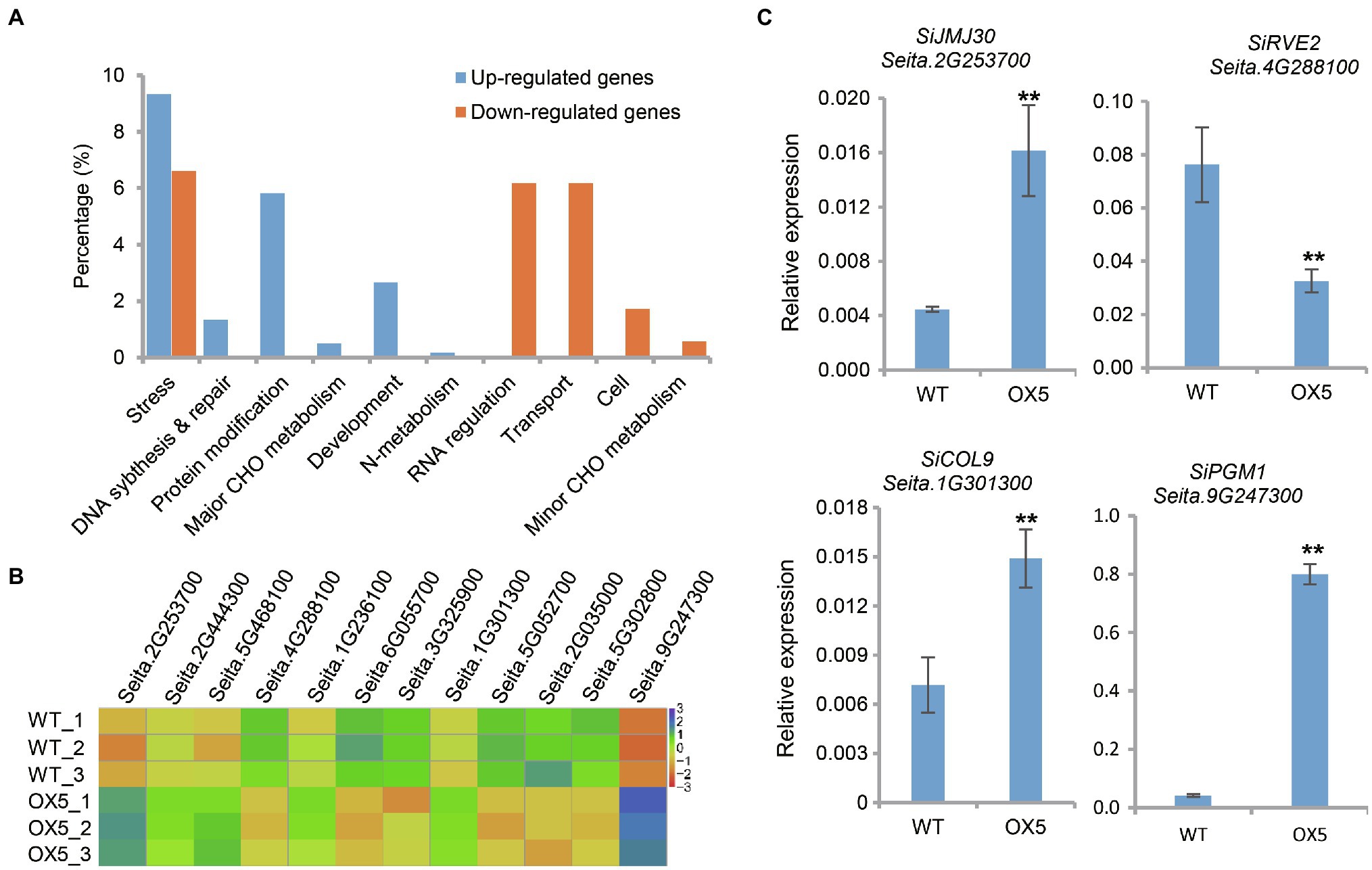
Figure 5. Differentially expressed genes (DEGs) analysis between overexpression transgenic plants OX5 and wild type. (A) Functional classification of DEGs according to Mapman bin categories. Upregulated and downregulated DEGs were displayed in blue and orange, respectively. (B) The heat map of 12 DEGs between wild type and OX5. These DEGs were homologous genes of known flowering-related genes in Arabidopsis. (C) The relative expression of partial DEGs was confirmed using real-time qPCR. The asterisks indicate a statistically significant difference (**p < 0.01, Student test).
Combined with the known flowering time genes from Arabidopsis, 12 DEGs were identified by comparative analysis, which were involved in the circadian clock, photoperiodism, sugar, and general pathways (Figure 5B). SiPRR37 (Seita.2G444300) is a circadian clock-related gene whose overexpression influenced the expression of other circadian clock-related genes, such as Seita.6G055700 (SiLHY, the homolog of LATE ELONGATED HYPOCOTYL), Seita.1G236100 (SiTOC1, the homolog of TIMING OF CAB EXPRESSION 1), Seita.4G288100 (SiRVE2, the homolog of REVEILLE 2), and Seita.5G468100 (SiPCL1, the homolog of PHYTOCLOCK 1). The expression profile of the clock genes demonstrated that Arabidopsis PRR7 can inhibit the expression of LHY, while LHY can bind to and activate the expression of PRR7 and PRR9 (Salomé et al., 2010). Notably, multiple photoperiodic genes were downregulated, including Seita.3G325900 (the homolog of AS1, ASYMMETRIC LEAVES 1), Seita.5G052700 (the homolog of CDF2, CYCLING DOF FACTOR 2), Seita.2G035000 (LATE, LATE FLOWERING), and Seita.5G302800 (the homolog of SPA3, SPA1-RELATED 3). Seita.1G301300 (the homolog of COL9, CONSTANS-LIKE 9) was upregulated. One glycometabolism-related gene, Seita.9G247300 (the homolog of PGM1, PHOSPHOGLUCOMUTASE), was also significantly upregulated. We also found two other DEGs belonging to the CCT family, Seita.2G175000 (the homolog of OsCCT22; Zhang et al., 2021) and Seita.9G228800 (a member of the CMF subfamily), were also upregulated. The qRT-PCR results showed that the expression patterns of most genes were consistent with the RNA-seq results (Figure 5C).
Discussion
Conservation and Divergence of CCT Family Genes
The important feature of the CCT family is the 43–45 conserved amino acids in the carboxy terminus, which could possess the function of a nuclear localization signal (Strayer et al., 2000; Robson et al., 2001). The CCT domain proteins participate in the signal cascade of environmental factors, such as light, gravity, and temperature, which may be involved in the regulation of the photoperiod pathway, circadian clock, and abiotic responses (Putterill et al., 1995; Kaczorowski and Quail, 2003; Salomé and McClung, 2005). In maize, the CCT genes can be classified into four subfamilies corresponding to the encoded domains, namely, the COL subfamily, the CMF subfamily, the PRR subfamily, and the TIFY subfamily (Jin et al., 2018). The COL-like genes possessed CCT and B-box zinc finger domains. The CMF genes only had a CCT domain. The PRR genes had CCT and response regulator receiver domains, while the TIFY genes held CCT, TIFY, and GATA zinc finger domains.
To date, at least 18 CCT domain-containing genes in rice have been isolated that participate in regulating heading date (Zhang et al., 2021). Hd1 was the first CCT family gene to be identified, and its expression reached a peak during the night under long- and short-day conditions (Yano et al., 2000; Hayama et al., 2003). Apart from Hd1, the other three genes (Ghd7, Ghd7.1/OsPRR37, and DTH2) were also obtained by a map-based cloning strategy. DTH2 (days to heading on chromosome 2) encodes a COL-like protein, which enhances adaptation for the northward expansion of rice cultivation (Wu et al., 2013). Ghd7 belongs to the CMF-like subfamily, while Ghd7.1/OsPRR37 belongs to the PRR-like subfamily, and both genes could cause pleiotropic effects, including delaying heading date, increasing plant height, and grain number per panicle (Xue et al., 2009; Yan et al., 2013; Gao et al., 2014). Other CCT genes, for example, OsCCT01, OsCCT03, and OsCCT41 were obtained through reverse genetics and functioned as repressors or activators in the flowering regulation network (Zhang et al., 2015, 2021). In addition, multiple genes are involved in abiotic stress responses. For example, overexpression of Ghd7 increased drought sensitivity, and Ghd7 could strongly respond to abscisic acid, jasmonic acid, and high-temperature stress (Weng et al., 2014). In Arabidopsis, AtCOL4 also participates in stress to salt through the ABA-dependent signaling pathway (Min et al., 2015).
In this study, a total of 39 genes of the CCT family, including CMF-like, COL-like, PRR-like, and TIFY-like subfamilies, were identified in foxtail millet. Among the different species, genes of the corresponding subfamily were clustered. This further proved that the CCT domain genes evolved before species divergence (Cockram et al., 2012). For example, genes OsCOL4, OsCOL10, and OsCOL9 belong to the COL subfamily, and the overexpression of OsCOL4 or OsCOL10 in rice delays heading date regardless of day length, while the overexpression of OsCOL9 also enhances rice blast resistance by interacting with the receptor for activated C-kinase 1 (Lee et al., 2010; Liu et al., 2016; Tan et al., 2016). In addition, the expression pattern of partially homologous genes in different tissues also exhibited vast diversity (Supplementary Figure S2).
Comparative genome analysis showed that the genes among foxtail millet, sorghum, and maize were highly conserved and had a higher identity in the panicoids (Bennetzen et al., 2012; Zhang et al., 2012). Considering that the CCT family genes evolved before the divergence between the monocots and dicots (Cockram et al., 2012), we speculated that the CCT genes in cereal crops possessed a conserved function. Thus, it will provide a reference for functional genome research of CCT genes in foxtail millet.
Expansion of the CCT Gene Family in Foxtail Millet
In this study, we comprehensively analyzed the expansion mode of the CCT gene family in foxtail millet, which has not been performed in maize (Jin et al., 2018), rice (Zhang et al., 2015, 2021), wheat (Zheng et al., 2017), and soybean (Mengarelli and Zanor, 2021). A large number of duplicated genes occurred after the pan-grass polyploidization ~96 mya (Wang et al., 2015). Until now, 11 whole-genome duplicated CCT gene pairs are still identified in the foxtail millet genome (Figure 3A), including genes from CMF-like, COL-like, PRR-like, and TIFY-like subfamilies, which further inferred CCT gene family evolved before Poaceae species differentiation. Among these WGD events, four duplication events took place between Chr1 and Chr 4, two happened between Chr2 and Chr 9, one occurred between Chr1 and Chr 7, and one appeared between Chr3 and Chr 5, which was consistent with long synteny blocks in foxtail millet genome (Zhang et al., 2012).
We identified 48 duplication events that were classified into four categories: 11 WGD (22.9%), 28 DSD (58.3%), 8 TRD (2.1%), 1 TD (16.7%; Figure 3A). Our result suggested that DSD significantly contributed to CCT gene family expansion in foxtail millet. The duplication modes in different gene families or organisms may be discrepant. For example, the tandem duplication was the primary driving force for the expansion of the BpUGT gene family in Broussonetia papyrifera (Wang et al., 2021). While the dispersed duplication could directly affect the functional diversification by asymmetric regulatory divergence (Owens et al., 2013). Besides, alternative splicing is also an important regulatory mechanism for modulating gene expression and functional diversity (Lareau et al., 2004). However, AS events were only identified in six CCT genes (Table 1). One of the reasons may be that WGD-derived duplicated genes decrease the AS frequency (Shen et al., 2014). Therefore, the gene family could expand through duplication, while the duplicated genes maybe diverge over time to perform different functions.
SiPRR37 May Confer Diverse Functions
SiPRR37, which is the homologous gene of PRR7 in Arabidopsis, encodes a circadian rhythm protein. Two allelic mutants of prr7 exhibited reduced sensitivity to both continuous red and far-red light, and the stability of the PRR7 protein was regulated by light and the circadian clock (Kaczorowski and Quail, 2003; Farré and Kay, 2007). There were five members in the PRR family in Arabidopsis, namely, TOC1 (PRR1), PRR3, PRR5, PRR7, and PRR9, and there was a close relationship between them. TOC1 (PRR1) was the first PRR-like gene to be cloned and was the central regulator (Strayer et al., 2000). PRR3 could modulate the stability of TOC1, yet both genes exhibited partially overlapped expression patterns (Para et al., 2007). PRR5 could interact with TOC1 in vitro and in vivo through the N-terminus, and TOC1–PRR5 oligomerization enhanced the accumulation of TOC1 in the nucleus (Wang et al., 2010). Overexpression of PRR5 could inhibit the expression of PRR7 and PRR9 while enhancing the expression of PRR3 and TOC1 (Sato et al., 2002). The gene PRR5/PRR7/PRR9 played an overlapping and distinctive role in the central oscillator and could repress the expression of CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY; Nakamichi et al., 2005, 2010). In rice and Arabidopsis, five PRR members showed a similar expression order, and five members started accumulating sequentially after dawn with 2–3 h intervals in the order of PRR9, PRR7, PRR5, PRR3, and PRR1 (Matsushika et al., 2000; Murakami et al., 2003).
After the identification of PRR7 in Arabidopsis, homolog genes were gradually uncovered in many crops. Ppd-H1 was the major determinant of barley photoperiod response, which enhanced the adaptation of spring-sown varieties (Turner et al., 2005). OsPRR37 and SbPRR37 were involved in regulating heading date and yield, and the loss-of-function genotype contributed to cultivation at a wide latitude area (Murphy et al., 2011; Koo et al., 2013). Studies have shown that genes SbPRR37, OsPRR37, Ppd-H1, and PRR7 are all orthologous, possessing conserved functions in the photoperiodic pathway (Murphy et al., 2011; Gao et al., 2014). Here, we confirmed that overexpression of SiPRR37 delayed the heading date in natural long-day conditions, which suggests that SiPRR37 has a conservative function. The insertion of the transposon Tc1-Marina may be the result of adaptive selection.
The current research indicated that the PRR proteins also participate in abiotic stress regulation. Among the five PRR members in rice, OsPRR73 could inhibit OsHKT2;1 expression by binding the promoter and regulating the response to salt stress (Wei et al., 2021). In addition, PRR5 interacts with ABI5 and modulates expression to enhance ABA signal transduction during seed germination (Yang et al., 2021). In this study, we found that SiPRR37 may be involved in the stress-related pathway through transcriptome analysis. Therefore, we speculated that SiPRR37 also plays a role in the stress signal pathway, which requires further study.
Data Availability Statement
The datasets presented in this study can be found in the NCBI Sequence Read Archive under accession number PRJNA797949. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
LY, JW, and HuW designed the research. CL, JM, and GeW co-wrote the manuscript. HL, HaW, GuW, YJ, and YL performed the experiments and data analyses. GuiL, GuoL, and RC edited the article and were involved in many discussions. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Beijing Municipal Natural Science Foundation (6202011), the National Key R&D Program of China (2018YFD1000706/2018YFD1000700), the Hebei Modern Seed Industry Innovation Team of Hybrid and Superior Quality Foxtail Millet (21326302D), the HAAFS Agriculture Science and Technology Innovation Project (2022KJCXZX-GZS-1), and the HAAFS Institute of Millet Crops Open Project of Grain Research (ZLSYS2020KF01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.863298/full#supplementary-material
Footnotes
1. ^https://phytozome-next.jgi.doe.gov
3. ^http://us.expasy.org/tools/protparam.html
5. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
6. ^https://www.wur.nl/en/show/MapChart-2.32.htm
7. ^http://pdgd.njau.edu.cn:8080
8. ^http://foxtail-millet.biocloud.net/page/tools/expressionVisualization
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555–561. doi: 10.1038/nbt.2196
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Cockram, J., Thiel, T., Steuernagel, B., Stein, N., Taudien, S., Bailey, P. C., et al. (2012). Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS One 7:e45307. doi: 10.1371/journal.pone.0045307
Doust, A. N., Kellogg, E. A., Devos, K. M., and Bennetzen, J. L. (2009). Foxtail millet: a sequence-driven grass model system. Plant Physiol. 149, 137–141. doi: 10.1104/pp.108.129627
Doust, A. N., Mauro-Herrera, M., Hodge, J. G., and Stromski, J. (2017). The C4 model grass Setaria is a short day plant with secondary long day genetic regulation. Front. Plant Sci. 8:1062. doi: 10.3389/fpls.2017.01062
Farré, E. M., and Kay, S. A. (2007). PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. Plant J. 52, 548–560. doi: 10.1111/j.1365-313X.2007.03258.x
Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J., Hotz, H. R., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222. doi: 10.1093/nar/gkp985
Foissac, S., and Sammeth, M. (2007). ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 35, W297–W299. doi: 10.1093/nar/gkm311
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. doi: 10.1093/genetics/151.4.1531
Gao, H., Jin, M., Zheng, X. M., Chen, J., Yuan, D., Xin, Y., et al. (2014). Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 16337–16342. doi: 10.1073/pnas.1418204111
Gaut, B. S., Thierry d'Ennequin, M. L., Peek, A. S., and Sawkins, M. C. (2000). Maize as a model for the evolution of plant nuclear genomes. Proc. Natl. Acad. Sci. U.S.A. 97, 7008–7015. doi: 10.1073/pnas.97.13.7008
Hayama, R., and Coupland, G. (2003). Shedding light on the circadian clock and the photoperiodic control of flowering. Curr. Opin. Plant Biol. 6, 13–19. doi: 10.1016/S1369-5266(02)00011-0
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
He, X., and Zhang, J. (2005). Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169, 1157–1164. doi: 10.1534/genetics.104.037051
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hughes, A. L. (1994). The evolution of functionally novel proteins after gene duplication. Proc. Roy. Soc. B. Biol. Sci. 256, 119–124. doi: 10.1098/rspb.1994.0058
Jia, G. Q., Huang, X. H., Zhi, H., Zhao, Y., Zhao, Q., Li, W., et al. (2013). A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 45, 957–961. doi: 10.1038/ng.2673
Jin, M. L., Liu, X. G., Jia, W., Liu, H., Li, W., Peng, Y., et al. (2018). ZmCOL3, a CCT gene represses flowering in maize by interfering with the circadian clock and activating expression of ZmCCT. J. Integr. Plant Biol. 60, 465–480. doi: 10.1111/jipb.12632
Kaczorowski, K. A., and Quail, P. H. (2003). Arabidopsis PSEUDO-RESPONSE REGULATOR7 is a signaling intermediate in phytochrome-regulated seedling deetiolation and phasing of the circadian clock. Plant Cell 15, 2654–2665. doi: 10.1105/tpc.015065
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kondrashov, F. A. (2012). Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Roy. Soc. B. Biol. Sci. 279, 5048–5057. doi: 10.1098/rspb.2012.1108
Koo, B. H., Yoo, S. C., Park, J. W., Kwon, C. T., Lee, B. D., An, G., et al. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6, 1877–1888. doi: 10.1093/mp/sst088
Korbel, J. O., Kim, P. M., Chen, X., Urban, A. E., Weissman, S., Snyder, M., et al. (2008). The current excitement about copy-number variation: how it relates to gene duplications and protein families. Curr. Opin. Plant Biol. 18, 366–374. doi: 10.1016/j.sbi.2008.02.005
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Ku, H. M., Vision, T., Liu, J. P., and Tanksley, S. D. (2000). Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. U.S.A. 97, 9121–9126. doi: 10.1073/pnas.160271297
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lareau, L. F., Green, R. E., Bhatnagar, R. S., and Brenner, S. E. (2004). The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 14, 273–282. doi: 10.1016/j.sbi.2004.05.002
Lee, Y. S., Jeong, D. H., Lee, D. Y., Yi, J., Ryu, C. H., Kim, S. L., et al. (2010). OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 63, 18–30. doi: 10.1111/j.1365-313X.2010.04226.x
Letunic, I., and Bork, P. (2007). Interactive tree Of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Li, P., and Brutnell, T. P. (2011). Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses. J. Exp. Bot. 62, 3031–3037. doi: 10.1093/jxb/err096
Li, C. C., Wang, G. P., Li, H. Q., Wang, G., Ma, J., Zhao, X., et al. (2021). High-depth resequencing of 312 accessions reveals the local adaptation of foxtail millet. Theor. Appl. Genet. 134, 1303–1317. doi: 10.1007/s00122-020-03760-4
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, T., Carlsson, J., Takeuchi, T., Newton, L., and Farré, E. M. (2013). Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 76, 101–114. doi: 10.1111/tpj.12276
Liu, H., Dong, S. Y., Sun, D. Y., Liu, W., Gu, F. W., Liu, Y. Z., et al. (2016). CONSTANS-like 9 (OsCOL9) interacts with receptor for activated c-kinase 1(OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS One 11:e0166249. doi: 10.1371/journal.pone.0166249
Liu, H. H., Liu, H. Q., Zhou, L. N., Zhang, Z. H., Zhang, X., Wang, M. L., et al. (2015). Parallel domestication of the heading date 1 gene in cereals. Mol. Biol. Evol. 32, 2726–2737. doi: 10.1093/molbev/msv148
Liu, H. J., Zhou, X. C., Li, Q. P., Wang, L., and Xing, Y. Z. (2020). CCT domain-containing genes in cereal crops: flowering time and beyond. Theor. Appl. Genet. 133, 1385–1396. doi: 10.1007/s00122-020-03554-8
Matsushika, A., Makino, S., Kojima, M., and Mizuno, T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 41, 1002–1012. doi: 10.1093/pcp/pcd043
Mauro-Herrera, M., Wang, X. W., Barbier, H., Brutnell, T. P., Devos, K. M., and Doust, A. N. (2013). Genetic control and comparative genomic analysis of flowering time in Setaria (Poaceae). G3 3, 283–295. doi: 10.1534/g3.112.005207
Mengarelli, D. A., and Zanor, M. I. (2021). Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 253, 1–17. doi: 10.1007/s00425-020-03537-5
Min, J. H., Chung, J. S., Lee, K. H., and Kim, C. S. (2015). The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 57, 313–324. doi: 10.1111/jipb.12246
Murakami, M., Ashikari, M., Miura, K., Yamashino, T., and Mizuno, T. (2003). The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 44, 1229–1236. doi: 10.1093/pcp/pcg135
Murphy, R. L., Klein, R. R., Morishige, D. T., Brady, J. A., Rooney, W. L., Miller, F. R., et al. (2011). Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. U.S.A. 108, 16469–16474. doi: 10.1073/pnas.1106212108
Muthamilarasan, M., and Prasad, M. (2015). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128, 1–14. doi: 10.1007/s00122-014-2399-3
Muthamilarasan, M., and Prasad, M. (2021). Small millets for enduring food security amidst pandemics. Trends Plant Sci. 26, 33–40. doi: 10.1016/j.tplants.2020.08.008
Nakamichi, N., Kiba, T., Henriques, R., Mizuno, T., Chua, N. H., and Sakakibara, H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605. doi: 10.1105/tpc.109.072892
Nakamichi, N., Kita, M., Ito, S., Yamashino, T., and Mizuno, T. (2005). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46, 686–698. doi: 10.1093/pcp/pci086
Ni, X. M., Xia, Q. J., Zhang, H. B., Cheng, S., Li, H., Fan, G., et al. (2017). Updated foxtail millet genome assembly and gene mapping of nine key agronomic traits by resequencing a RIL population. GigaScience 6, 1–8. doi: 10.1093/gigascience/giw005
Owens, S. M., Harberson, N. A., and More, R. C. (2013). Asymmetric functional divergence of young, dispersed gene duplicates in Arabidopsis thaliana. J. Mol. Evol. 76, 13–27. doi: 10.1007/s00239-012-9530-3
Para, A., Farre, E. M., Imaizumi, T., Pruneda-Paz, J. L., Harmon, F. G., and Kay, S. A. (2007). PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19, 3462–3473. doi: 10.1105/tpc.107.054775
Paterson, A. H., Bowers, J. E., and Chapman, B. A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. U.S.A. 101, 9903–9908. doi: 10.1073/pnas.0307901101
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Qiao, X., Li, Q. H., Yin, H., Qi, K. J., Li, L. T., Wang, R. Z., et al. (2019). Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 20, 1–23. doi: 10.1186/s13059-019-1650-2
Ramsey, J., and Schemske, D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Evol. Syst. 29, 467–501. doi: 10.1146/annurev.ecolsys.29.1.467
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Robson, F., Costa, M. M. R., Hepworth, S. R., Vizir, I., Piñeiro, M., Reeves, P. H., et al. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28, 619–631. doi: 10.1046/j.1365-313x.2001.01163.x
Salomé, P. A., and McClung, C. R. (2005). Pseudo-response regulator 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17, 791–803. doi: 10.1105/tpc.104.029504
Salomé, P. A., Weigel, D., and McClung, C. R. (2010). The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22, 3650–3661. doi: 10.1105/tpc.110.079087
Samonte, R. V., and Eichler, E. E. (2002). Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3, 65–72. doi: 10.1038/nrg705
Sato, E., Nakamichi, N., Yamashino, T., and Mizuno, T. (2002). Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 43, 1374–1385. doi: 10.1093/pcp/pcf166
Shen, Y. T., Zhou, Z. K., Wang, Z., Li, W. Y., Fang, C., Wu, M., et al. (2014). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26, 996–1008. doi: 10.1105/tpc.114.122739
Shoemaker, R. C., Schlueter, J., and Doyle, J. J. (2006). Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 9, 104–109. doi: 10.1016/j.pbi.2006.01.007
Simillion, C., Vandepoele, K., Montagu, M. C. E. V., Zabeau, M., and Peer, Y. V. D. (2002). The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 13627–13632. doi: 10.1073/pnas.212522399
Simpson, G. G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time. Science 296, 285–289. doi: 10.1126/science.296.5566.285
Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Más, P., et al. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. doi: 10.1126/science.289.5480.768
Tan, J. J., Jin, M. N., Wang, J. C., Wu, F., Sheng, P., Cheng, Z., et al. (2016). OsCOL10, a CONSTANS-like gene, functions as a flowering time repressor downstream of Ghd7 in Rice. Plant Cell Physiol. 57, 798–812. doi: 10.1093/pcp/pcw025
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
Tsuji, H., Taoka, K. I., and Shimamoto, K. (2011). Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14, 45–52. doi: 10.1016/j.pbi.2010.08.016
Turner, A., Beales, J., Faure, S., Dunford, R., and Laurie, D. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. doi: 10.1126/science.1117619
Vision, T. J., Brown, D. G., and Tanksley, S. D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. doi: 10.1126/science.290.5499.2114
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wagner, A. (1994). Evolution of gene networks by gene duplications: a mathematical model and its implications on genome organization. Proc. Natl. Acad. Sci. U.S.A. 91, 4387–4391. doi: 10.1073/pnas.91.10.4387
Wang, L., Fujiwara, S., and Somers, D. E. (2010). PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 29, 1903–1915. doi: 10.1038/emboj.2010.76
Wang, F. F., Su, Y. L., Chen, N. Z., and Shen, S. H. (2021). Genome-wide analysis of the UGT gene family and identification of flavonoids in Broussonetia papyrifera. Molecules 26:3449. doi: 10.3390/molecules26113449
Wang, Y. P., Tang, H. B., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, X. Y., Wang, J. P., Jin, D. C., Guo, H., Lee, T.-H., Liu, T., et al. (2015). Genome alignment spanning major poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Mol. Plant 8, 885–898. doi: 10.1016/j.molp.2015.04.004
Wei, H., Wang, X. L., He, Y. Q., Xu, H., and Wang, L. (2021). Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2;1-mediated sodium homeostasis. EMBO J. 40:e105086. doi: 10.15252/embj.2020105086
Weng, X. Y., Wang, L., Wang, J., Hu, Y., Du, H., Xu, C. G., et al. (2014). Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 164, 735–747. doi: 10.1104/pp.113.231308
Wu, W. X., Zheng, X. M., Lu, G. W., Zhong, Z., Gao, H., Chen, L., et al. (2013). Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. U.S.A. 110, 2775–2780. doi: 10.1073/pnas.1213962110
Xue, W. Y., Xing, Y. Z., Weng, X. Y., Zhao, Y., Tang, W. J., Wang, L., et al. (2009). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yan, W. H., Liu, H. Y., Zhou, X. C., Li, Q., Zhang, J., Lu, L., et al. (2013). Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 23, 969–971. doi: 10.1038/cr.2013.43
Yang, Y., Fu, D. B., Zhu, C. M., He, Y. Z., Zhang, H. J., Liu, T., et al. (2015). The RING-finger ubiquitin ligase HAF1 mediates heading date 1 degradation during photoperiodic flowering in rice. Plant Cell 27, 2455–2468. doi: 10.1105/tpc.15.00320
Yang, M. L., Han, X., Yang, J. J., Jiang, Y. J., and Hu, Y. R. (2021). The Arabidopsis circadian clock protein PRR5 interacts with and stimulates ABI5 to modulate abscisic acid signaling during seed germination. Plant Cell 33, 3022–3041. doi: 10.1093/plcell/koab168
Yang, Z. R., Zhang, H. S., Li, X. K., Shen, H., Gao, J., Hou, S., et al. (2020). A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat Plants 6, 1167–1178. doi: 10.1038/s41477-020-0747-7
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2483. doi: 10.1105/tpc.12.12.2473
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsi mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yoshitsu, Y., Takakusagi, M., Abe, A., Takagi, H., Uemura, A., Yaegashi, H., et al. (2017). QTL-seq analysis identifies two genomic regions determining the heading date of foxtail millet, Setaria italica (L.) P.Beauv. Breed. Sci. 67, 518–527. doi: 10.1270/jsbbs.17061
Zhang, J. Z. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. doi: 10.1016/S0169-5347(03)00033-8
Zhang, J., Fan, X. W., Hu, Y., Zhou, X. C., He, Q., Liang, L. W., et al. (2021). Global analysis of CCT family knockout mutants identifies four genes involved in regulating heading date in rice. J. Integr. Plant Biol. 63, 913–923. doi: 10.1111/jipb.13013
Zhang, K., Fan, G. Y., Zhang, X. X., Zhao, F., Wei, W., du, G., et al. (2017). Identification of QTLs for 14 agronomically important traits in Setaria italica based on SNPs generated from high-throughput sequencing. G3 7, 1587–1594. doi: 10.1534/g3.117.041517
Zhang, L., Li, Q. P., Dong, H. J., He, Q., Liang, L., Tan, C., et al. (2015). Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci. Rep. 5:7663. doi: 10.1038/srep07663
Zhang, G. Y., Liu, X., Quan, Z. W., Cheng, S., Xu, X., Pan, S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30, 549–554. doi: 10.1038/nbt.2195
Zhang, P., and Peterson, C. T. (2000). A segmental gene duplication generated differentially expressed myb-homologous genes in maize. Plant Cell 12, 2311–2322. doi: 10.1105/tpc.12.12.2311
Keywords: gene duplication, transcriptional factor, CCT gene family, heading date, SiPRR37, foxtail millet
Citation: Li C, Ma J, Wang G, Li H, Wang H, Wang G, Jiang Y, Liu Y, Liu G, Liu G, Cheng R, Wang H, Wei J and Yao L (2022) Exploring the SiCCT Gene Family and Its Role in Heading Date in Foxtail Millet. Front. Plant Sci. 13:863298. doi: 10.3389/fpls.2022.863298
Edited by:
Xiaoming Song, North China University of Science and Technology, ChinaReviewed by:
Mehanathan Muthamilarasan, University of Hyderabad, IndiaJinpeng Wang, Institute of Botany (CAS), China
Copyright © 2022 Li, Ma, Wang, Li, Wang, Wang, Jiang, Liu, Liu, Liu, Cheng, Wang, Wei and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Yao, eWFvbGVpQGJhYWZzLm5ldC5jbg==; Jianhua Wei, d2Vpamlhbmh1YUBiYWFmcy5uZXQuY24=; Huan Wang, d2FuZ2h1YW5AY2Fhcy5jbg==
†These authors have contributed equally to this work and share first authorship
 Congcong Li
Congcong Li Jian Ma
Jian Ma Genping Wang
Genping Wang Haiquan Li5
Haiquan Li5 Hailong Wang
Hailong Wang Guoliang Wang
Guoliang Wang Guoqing Liu
Guoqing Liu Huan Wang
Huan Wang Jianhua Wei
Jianhua Wei