- 1Department of Genetics and Biotechnology, Graduate School of Biotechnology, College of Life Sciences, Kyung Hee University, Yongin, South Korea
- 2Division of Life Sciences, Bio-Resource and Environmental Center, Incheon National University, Incheon, South Korea
The methylerythritol 4-phosphate (MEP) pathway is responsible for providing common precursors for the biosynthesis of diverse plastidial terpenoids, including chlorophylls, carotenoids, and phytohormones, in plants. In rice (Oryza sativa), the last-step genes encoding 4-hydroxy-3-methylbut-2-enyl diphosphate reductase [HDR/isoprenoid synthesis H (IspH)] have been annotated in two genes (OsIspH1 and OsIspH2) in the rice genome. The spatial transcript levels indicated that OsIspH1 is highly expressed in all tissues at different developmental stages, whereas OsIspH2 is barely expressed due to an early stop in exon 1 caused by splicing error. OsIspH1 localized into plastids and osisph1, a T-DNA inserted knockout mutant, showed an albino phenotype, indicating that OsIspH1 is the only functional gene. To elucidate the role of OsIspH1 in the MEP pathway, we created two single (H145P and K407R) and double (H145P/K407R) mutations and performed complementation tests in two hdr mutants, including Escherichia coli DLYT1 strains and osisph1 rice plants. The results showed that every single mutation retained HDR function, but a double mutation lost it, proposing that the complementary relations of two residues might be important for enzyme activity but not each residue. When overexpressed in rice plants, the double-mutated gene, OsIspH1MUT, reduced chlorophyll and carotenoid biosynthesis in the leaves and seeds. It confirmed the crucial role of OsIspH1 in plastidic terpenoid biosynthesis, revealing organ-specific differential regulation of OsIspH1 in rice plants.
Introduction
Plants synthesize an enormous variety of terpenoids that serve as growth regulators (cytokines, gibberellins, abscisic acids, strigolactones, and brassinosteroids), photosynthetic and respiratory components (chlorophylls, carotenoids, prenylquinones, and ubiquinone), membranous sterols, and other secondary metabolites (Nagegowda and Gupta, 2020). All terpenoids are derived by the condensation of two universal precursors, namely, isopentenyl diphosphate (IPP, C5) and dimethylallyl diphosphate (DMAPP, C5). While most organisms retain one pathway for the biosynthesis of these isoprene building units, plants have two independent pathways, namely, the cytosolic mevalonate (MVA) and plastidic methylerythritol 4-phosphate (MEP) pathways (Hemmerlin et al., 2012). The latter has been evolutionally acquired due to endosymbiosis with prokaryotes and consists of seven enzymatic steps that start with the condensation of pyruvate (C3) and glyceraldehyde 3-phosphate (C3) derived from glycolysis into 1-deoxy-D-xylulose 5-phosphate (DXP, C5) by the action of DXP synthase (DXS) and end with the production of IPP and DMAPP from 4-hydroxy-3-methylbut-2-enyl diphosphate (HMBPP, C5) by the catalyst of HMBPP reductase (HDR), also known as isoprenoid synthesis H (IspH) or lysis-tolerant B (LytB; Vranova et al., 2013).
The MEP genes have been reported mainly in photosynthetic organisms, such as cyanobacteria, algae, and plants (Hemmerlin et al., 2012). However, Apicomplexa (malaria causative Plasmodium spp.), a non-photosynthetic protist, possesses MEP genes, although its vestigial chloroplasts and apicoplasts result from convergent reduction (Mathur et al., 2021). Bacterial MEP genes have also been discovered in Eubacteria, such as Escherichia coli and Streptomyces species, Chlamydia (obligate intracellular bacteria causing a sexually transmitted infection), and Mycobacterium tuberculosis (tuberculosis causative bacteria) (Rohmer et al., 1993; Grieshaber et al., 2004; Eoh et al., 2009). Therefore, MEP genes, including IspH, have been identified as promising targets for developing algicides, herbicides, anti-malaria drugs, and bactericides to combat infectious diseases, such as Chlamydia and tuberculosis (Singh et al., 2007; Masini and Hirsch, 2014; Wang and Dowd, 2018).
At present, several IspH genes have been characterized in diverse species, including cyanobacteria (Synechocystis strain PCC 6803, Cunningham et al., 2000), E. coli (Altincicek et al., 2002), Aquifex (Aquifex aeolicus, Wang et al., 2010), green microalgae (Botryococcus braunii, Uchida et al., 2018), ginkgo (Ginkgo biloba, Kim et al., 2008), red pine (Pinus densiflora, Kim et al., 2009), tobacco (Nicotiana benthamiana, Page et al., 2004), tomato (Lycopersicon esculentum, Botella-Pavia et al., 2004), Arabidopsis (Arabidopsis thaliana, Hsieh and Goodman, 2005), corn (Zea mays, Lu et al., 2012), danshen (Salvia miltiorrhiza, Hao et al., 2013), melon (Cucumis melo, Saladie et al., 2014), sweet wormwood (Artemisia annua, Ma et al., 2017), and rice (Oryza sativa, Liu et al., 2020). Among them, the protein structures were first examined in bacterial systems, such as A. aeolicus and E. coli, to discover IspH inhibitors that have potential for novel cancer immunotherapy because the substrate of IspH, HMBPP, has been known as a potent phosphoantigen that activates γδ T cells to kill tumor cells (Hintz et al., 2001; Grawert et al., 2004; Rao and Oldfield, 2016). IspH is identified as a 4Fe-4S cluster-containing protein with a trefoil-like structure, and its key residues are elucidated: the Fe-S cluster acts as an electron transfer cofactor in the active site, with direct involvement of the fourth Fe in substrate binding and catalysis, and it requires coordination with three Cys-residues for ligand binding (Wang et al., 2010; Hsieh et al., 2014; Rao and Oldfield, 2016).
In planta IspH functions have been reported for diverse terpenoid production in several plants. The overexpression of the L. esculentum HDR gene (LeHDR) increased carotenoids in tomato fruits during ripening and Arabidopsis seedlings during de-etiolation, and co-expression with taxadiene synthase gene increased taxadiene levels, indicating synergistic effects (Botella-Pavia et al., 2004). The overexpression of the S. miltiorrhiza HDR gene (SmHDR1) enhanced tanshinone production in cultured hairy roots of S. miltiorrhiza Bge. F. alba (Hao et al., 2013). The overexpression of the A. annua HDR gene (AaHDR1) increased the contents of artemisinin, arteannuin B, other sesquiterpenes, and monoterpenes, while its antisense-induced suppression exhibited opposite effects (Ma et al., 2017). Recently, the overexpression of the G. biloba HDR2 gene (GbHDR2/GbIspH2) in Nicotiana tabacum cv. Xanthi resulted in an increased photosynthetic rate through the upregulation of biosynthetic genes for photosynthetic pigments, including chlorophylls and carotenoids, as well as the elevated contents of duvatrienediol, the major diterpene of the N. tabacum leaf surface (Kim et al., 2021). However, the loss-of-function studies on IspH genes have been reported in several plants. Tobacco plants infected with the tobacco rattle virus posttranscriptionally silence the expression of several MEP genes, including IspG, IspH, and IPI, resulting in an albino phenotype (Page et al., 2004). T-DNA knockout mutant of Arabidopsis, atisph, exhibited an albino phenotype, whereas transgenic-induced gene silencing of AtIspH exhibited variegated albino phenotypes (Hsieh and Goodman, 2005). In the case of Z. may IspH (ZmIspH), an ethyl methanesulfonate (EMS)-induced recessive mutant and foxtail mosaic virus-induced gene silencing both showed albino phenotypes, suggesting that it is fatal to chlorophyll biosynthesis (Lu et al., 2012; Liu et al., 2016).
Interestingly, in the rice genome, the existence of two IspH alleles is predicted unlike in corn that has one IspH gene. Although a recent study using the CRISPR-Cas9 system presented an albino phenotype in seedlings caused by defective chloroplast biogenesis of a rice lethal albinic seedling 1 (las1) mutant, corresponding to the osisph1 (Liu et al., 2020), the rice IspH function has not been fully understood. In this study, we analyzed the molecular characteristics of OsIspH1 and OsIspH2, performed the complementation assays of OsIspH1 in two hdr mutant backgrounds, namely, E. coli DLYT1 strains and rice T-DNA insertional osisph1 mutant plants, identified key residues crucial to HDR activity, and investigated the IspH effects on the biosynthesis of terpenoids, including chlorophylls and carotenoids, in the source and sink organs of rice plants.
Materials and Methods
Plant Materials and Growth Conditions
Two commercial varieties of Korean rice, O. sativa L. cv. Dongjin and Ilmi, were used to analyze endogenous gene expression, amplify the genes, isolate protoplasts, and perform a stable transformation. The Crop Biotech Institute in Kyung Hee University (Yongin, South Korea) generously provided a T-DNA-insertional rice mutant. Mature seeds were dehulled, surface-sterilized by 70% ethanol for 2 min and 2% sodium hypochlorite for 40 min, washed five times with distilled water, germinated for a week on Murashige and Skoog agar medium (Duchefa, Haarlem, Netherlands) in a plant growth chamber, transplanted into soil, and cultivated in a greenhouse under the conditions of 14-h light/10-h dark cycle at 28°C or in the paddy field until maturity during the summer. Mature seeds were harvested 60 days after flowering, and their endosperm color was visually compared after dehusking (TR-200 Electromotion rice husker, Kett, Tokyo, Japan) and polishing (Pearlest Polisher, Kett).
RNA and DNA Analysis
Rice samples of diverse organs obtained at different developmental stages and 2-week-old seedlings treated with six phytohormones, including 100-μM abscisic acid (ABA), 100-μM gibberellic acid (GA3), 100-μM auxin (IAA), 100-μM kinetin (KT), 100-μM methyl jasmonic acid (MeJA), and 200-μM salicylic acid (SA), were used for total RNA extraction using mainly RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) and PureLink® Plant RNA Reagent (Invitrogen, Waltham, MA, United States) for seed case. The real-time quantitative reverse transcriptase PCR (qRT-PCR) was performed using iQ™ SYBR® Green Supermix (Bio-Rad) and CFX Connect™ Real-Time System (Bio-Rad), with PCR conditions and data calculated relative to the rice ubiquitin 5 gene (Os01g22490) to normalize RNA amounts, as previously described (You et al., 2020), and each qRT-PCR was performed in three technical repeats. Semiquantitative RT-PCR was performed under the following conditions: a cycle at 95°C for 3 min, 30 cycles at 95°C for 15 s, 55°C for 15 s, 72°C for 1 min/kb, followed by 3 min at 72°C with KOD FX DNA polymerase (Toyobo, Osaka, Japan).
We extracted the rice genomic DNAs using a Genomic DNA Preparation Kit (QIAGEN). The genotyping PCRs were performed using an EmeraldAmp® PCR Master Mix (TaKaRa Bio, Shiga, Japan). To examine the transgene homozygosity, TaqMan PCR was performed to detect Bar as a transgene with an internal reference in the rice genome, i.e., the α-tubulin gene (Os11g14220), as previously described (Ha et al., 2019). Supplementary Table 10 contains all sequence information of primers.
Cloning and Subcellular Localization Using Rice Protoplasts
The coding region of OsIspH1 (Os03g0731900) was amplified using gene-specific primer pair from the total RNA of 10-day-old seedlings of rice (O. sativa L. cv. Ilmi) and cloned into the pDONR221 vector using the Gateway® BP® Clonase II Enzyme Mix (Invitrogen), yielding pDONR221-OsIspH1. To determine the subcellular localization, OsIspH1 was fused to the N-terminus of superfolder green fluorescent protein (sGFP) and cloned into the pB2GW7 binary vector using Gateway® LR Clonase® II Enzyme Mix (Invitrogen). Protoplast isolation, DNA transfection, and confocal microscopy analysis were conducted, as previously described (You et al., 2016). The fluorescence was detected and imaged using a confocal laser scanning microscope (Carl Zeiss LSM 880, Jena, Germany).
Site-Directed Mutagenesis and Complementation Assay in E. coli DLYT1 Strain
Using the pDONR221-OsIspH1 as a template, two non-synonymous substitution mutations (H145P and K407R) in OsIspH1 were created to have one or both of each through site-directed mutagenesis with sequence-substituted primers (Supplementary Table 10). Original and mutated genes were cloned into pMW118 (Nippon Gene, Tokyo, Japan) and transformed into the E. coli DLYT1 strain, which was disrupted in LytB/EcIspH encoding E. coli HDR gene (Kim et al., 2008). An E. coli DLTY1 strain transformed with pTTQ18-EcIspH/LytB was used as a positive control for complementation analysis. The complementation tests were performed on the Luria-Bertani (LB) medium containing 50 μg/ml kanamycin, 100 μg/ml spectinomycin, and 0.01% (w/v) (±)-mevalonolactone (MVA) (Sigma-Aldrich, St. Louis, MO, United States).
Transformation and Cross-Fertilization of Rice Plants
After amplification of a double mutated gene using the pMW118-OsIspH1H145P/K407R as a template, a constitutive expression cassette was prepared by consecutive cloning into the pDONR221 vector and the p600-PGD1 vector containing a rice phosphogluconate dehydrogenase 1 promoter using the Gateway procedures (Park et al., 2010), resulting in pIspH1MUT. It was further introduced into pGlb:stPAC vector, in which endosperm specifically produces β-carotene (Jeong et al., 2017), resulting in pIspH1MUT-stPAC. Two final binary vectors were transformed via triparental mating of Agrobacterium LBA4404 harboring pSB1 plasmids and cocultivated with embryogenic calli differentiated from matured rice seeds (O. sativa L. cv. Ilmi), as previously described (You et al., 2020). The putative transgenic plants were generated on a selection medium containing phosphinothricin (4 mg/L) and cefotaxime (500 mg/L) using shooting and rooting procedures, according to a previously described method (Ko et al., 2018).
In planta complementation assay was performed using conventional interbreeding during the flowering season at the T3 generation between heterozygous plants of osisph1 as the female parent and homozygous pIspH1MUT-overexpressed plants as the male parent. Filial (F) 1 progenies were self-pollinated in the paddy field for three more generations to obtain homozygous seeds for transgene traits of both parents.
Analyses of Chlorophyll and Carotenoid Metabolites
Rice leaves and dehusked seeds were ground to a fine powder using a pestle in liquid nitrogen. Total chlorophylls were extracted from rice leaf powders with 100% methanol at 70°C for 30 min using ThermoMixer Comfort (Eppendorf AG, Hamburg, Germany) at 500 rpm and centrifuged for 10 min at 4°C and 3,000 rpm. A spectrophotometer was used to measure the absorbance of the supernatant at 666 and 653 nm (Mecasys Co., Daejeon, South Korea). The chlorophyll content was calculated using the formula stated by Wellburn (1994).
Carotenoids were extracted from 100 mg powders of each rice leaf and seed, following the previous report with minor modifications (Kim et al., 2010). Notably, 0.1% ascorbic acid-containing ethanol was added to 100 mg powder to be incubated at 85°C for 5 min, after vortex-mixing for 20 s. After 10 min of saponification with potassium hydroxide (80%, w/v) in an 85°C water bath, samples were immediately placed on ice, and we added cold deionized water with β-apo-8′-carotenal (25 μg/ml) as an internal standard. Hexane-extracted carotenoid layers were separated two times by centrifugation at 1,200 × g. Aliquots were dried under a nitrogen stream and redissolved in 50:50 (v/v) dichloromethane/methanol solution. High-performance liquid chromatography (HPLC) was performed on a C30 YMC column (4.6 mm × 250 mm, 3 μm; Waters Corporation, Milford, MA, United States) using gradient elution at 1 ml/min with solvent A [methanol/water (92.8, v/v) with 10 mM ammonium acetate] and B (100% methyl tert-butyl ether) under the following conditions: 0 min, 83% A/17% B; 23 min, 70% A/30% B; 29 min, 59% A/41% B; 35 min, 30% A/70% B; 40 min, 30% A/70% B; 44 min, 83% A/17% B; 55 min, 83% A/17% B. Chromatograms were generated at 450 nm. The calibration curves were set using 5 concentrations of carotenoid standards, except for lycopene, from 0.3 to 5.0 μg/ml based on the peak area ratios obtained with the internal standard. Linearity was tested by least-squares regression analysis of the corrected peak area ratios against increasing weight ratios. The calibration curves of lutein, zeaxanthin, α-carotene, and β-carotene were linear (r2 = 0.9988–0.9998). Lycopene undergoes degradation via isomerization and oxidation. Thus, the quantitative calculation of lycopene was based on the peak area of 5.0 μg/ml of lycopene standard (Kim et al., 2017).
Results
Phylogenetic Tree Analysis of IspH Family
To investigate the evolutionary relationship of the rice IspH family, the predicted amino acid sequences of 79 IspHs were first collected from GreenPhylDB1, confirmed whether they contained a full-length open reading frame (ORF) in the National Center for Biotechnology Information Web BLAST2, and finally selected for the phylogenetic tree analysis using the maximum-likelihood method (Figure 1). As a result, the tree classified them into six clades, namely, bacteria, mosses, ferns, gymnosperms, monocotyledons, and dicotyledons, as they showed a sequential relationship based on general evolutionary processes. All bacteria have a single IspH gene, whereas plants possess one to three copies. Considering the closer relationship between cyanobacteria and plants, it was proposed that plants obtain IspH genes from cyanobacteria via endosymbiosis, which might have evolved independently with non-photosynthetic bacteria from the common prokaryotic ancestor, as previously reported (Hsieh and Hsieh, 2015). Particularly, family genes belonging to the same species were mostly grouped into the same clade or closely related, indicating that they might be multiplicated during evolutionary processes.
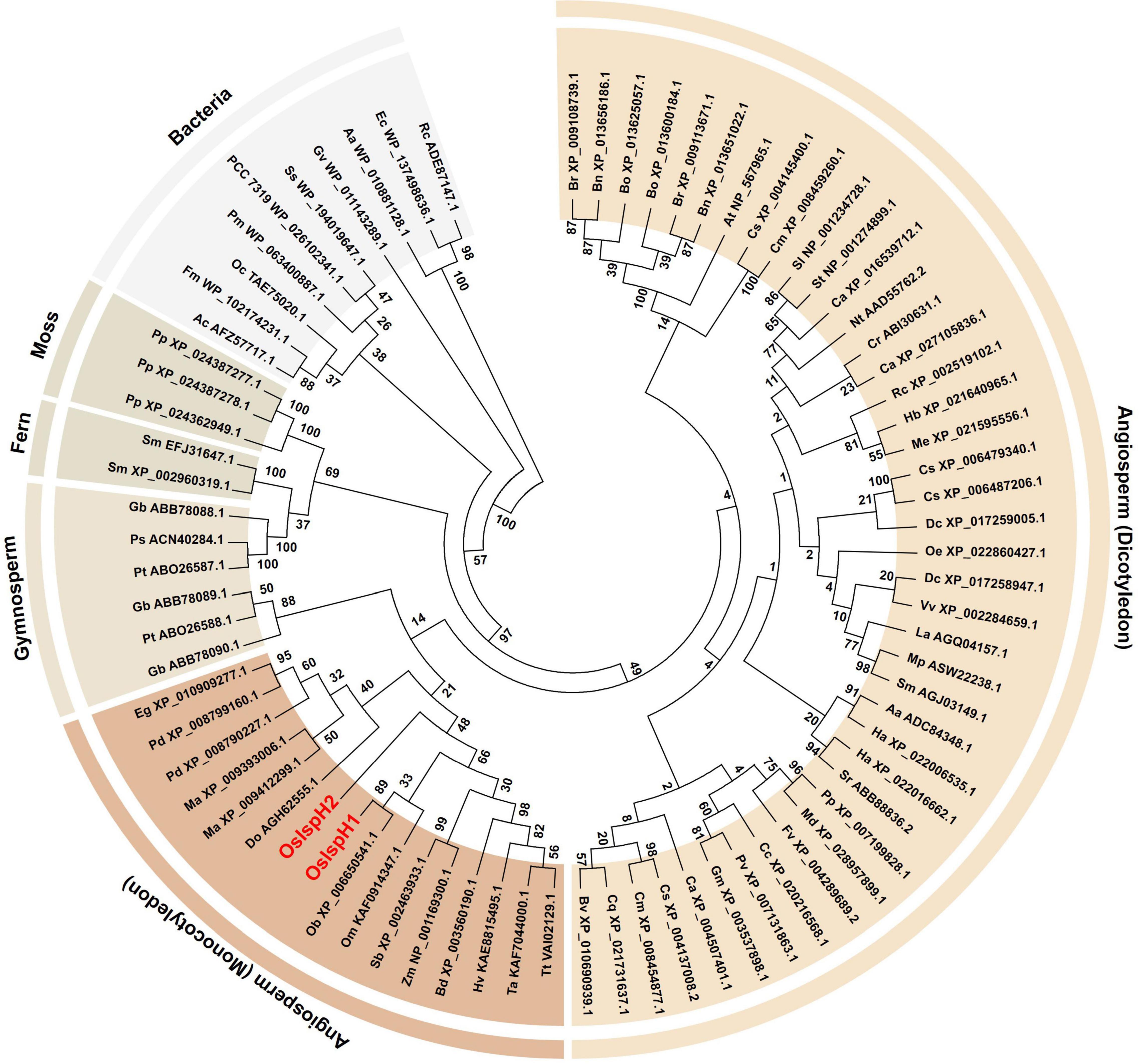
Figure 1. Evolutionary relationships among isoprenoid synthesis H (IspH) families. The phylogenetic tree was inferred using the maximum-likelihood method based on the JTT matrix-based model. The bootstrap consensus tree was inferred from 100 replicates. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The IspH family is named by a combination of a species abbreviation and an accession number, with rice IspHs marked in red letters. Evolutionary analyses were conducted on 79 amino acid sequences using MEGA7, and Supplementary Table 1 contains all information of the IspH family used in this study.
In the O. sativa genome, two IspH genes with 10 introns, OsIspH1 (Os03t0731900-01) and OsIspH2 (Os03t0732000-00), designated in this study are annotated in Rice Annotation Project Database (RAP-DB). They have 68% amino acid sequence homology and are adjacent genes about 2.8 Mbp apart in the same chromosome 3. In monocotyledon plants, IspH genes are analyzed to exist as two copies in date palm (Phoenix dactylifera), banana (Musa acuminata), and a domesticated-rice (O. sativa; AA genotype) genomes and as just a single copy in other wild rice species (Oryza brachyantha, FF genotype; Oryza meyeriana, GG genotype), sorghum, corn, barley, wheat, and so on (Figure 1). As shown in the phylogenetic tree, IspH genes exist mostly as two or three copies in moss, fern, and gymnosperm genomes, one or two copies in monocotyledon genomes of angiosperm plants, and mostly a single copy in bacteria, dicotyledon genomes of angiosperm plants, indicating that the copy number of IspH genes has evolved to increase from a single copy in bacteria to two or three copies in gymnosperms but has evolved to decrease back to a single copy in angiosperms, during the genomic evolution. Of two rice IspH genes, an OsIspH1 was evolutionarily grouped with a single-copy IspH gene of other monocotyledon plants in a phylogenetic tree, but OsIspH2 was distant from them (Figure 1), supposing that OsIspH1 might have a conservative IspH function in angiosperms, rather than OsIspH2 having it.
Molecular Identification and Characterization of Rice IspH Genes
To define the structures of two rice IspH genes, we extracted genomic DNA sequence information from the RAP-DB3 and constructed their genome structures, which are equally composed of ten exons and nine introns (Figure 2A). The 1st and 3rd introns of OsIspH2 have unusually longer sequences, 1,061 bp and 13.5 kbp, respectively, compared with 652 and 409 bp of OsIspH1 when predicted only with the genomic DNA sequence because the cDNA of OsIspH2 has never been reported. It meant that the functionality of OsIspH2 was largely unknown.
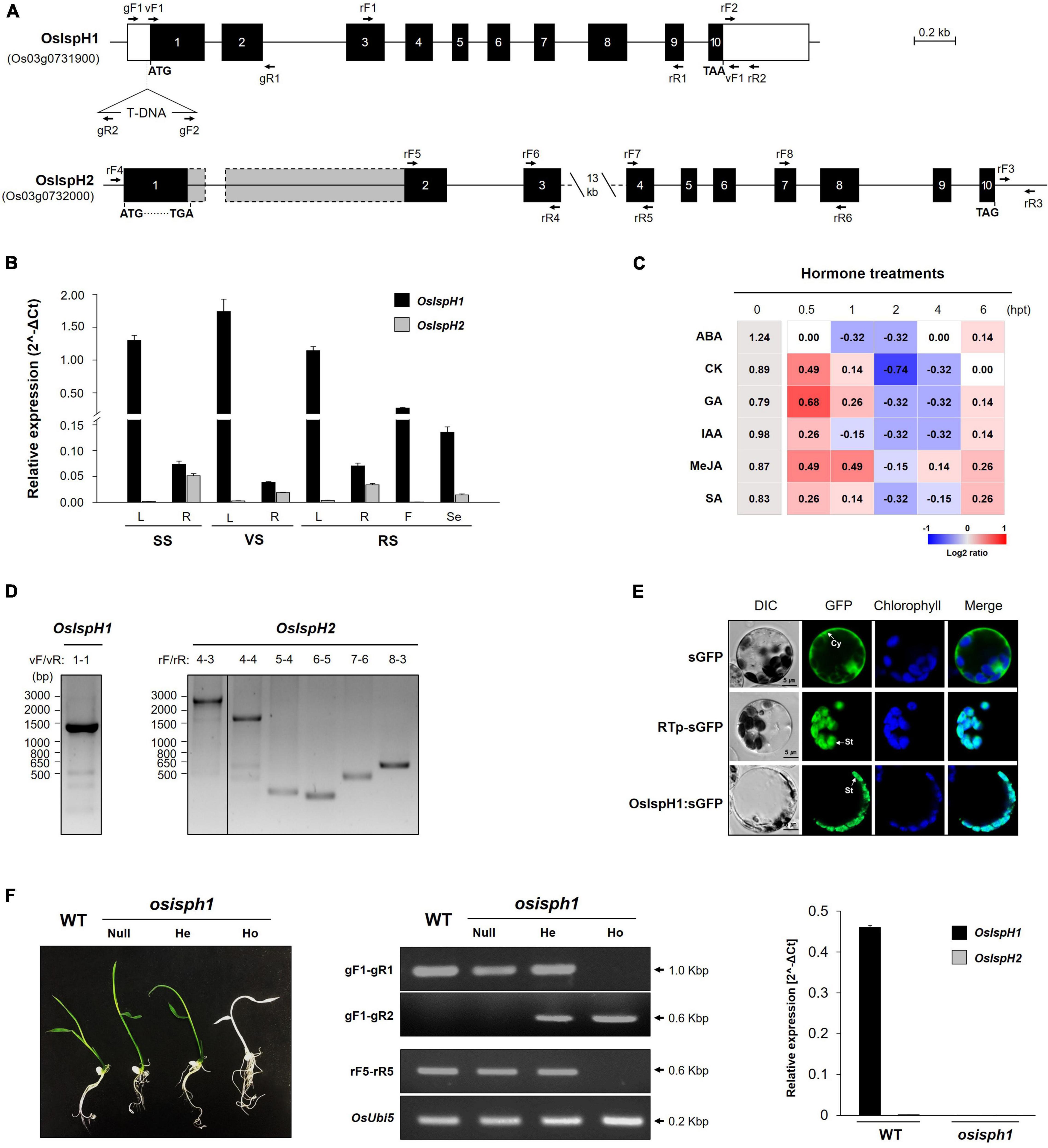
Figure 2. Molecular characterization of rice IspH genes. (A) Genomic structures of the OsIspH1 and OsIspH2 genes and verification of T-DNA position in knockout mutant osisph1. Solid boxes represent exon (black) and untranslated regions (white); the actual sizes of exons 1 and 2 of OsIspH2 are represented by gray boxes with dotted line, and solid lines indicate introns. Triangle indicates the position of T-DNA insertion. (B) Spatial and temporal expression of OsIspH1 and OsIspH2 in various tissues, including leaves (L), roots (R) of the seedling stage (SS), vegetative stage (VS), and reproductive stage (RS) tissues, and florets (F) of RS and seeds (Se) harvested at 60 days after flowering. (C) Expression profiles of endogenous OsIspH1 in response to different hormone treatments in 10-day-old leaves. The heatmap graph is presented by the relative log2 fold change values of each time point, which were obtained by normalizing the ΔCt values of each 0 h time point (gray-colored boxes) as individual control, and the ΔCt value of each 0 h time point is presented in column 0 hpt. (D) Reverse transcriptase PCRs (RT-PCRs) verify the predicted transcript sizes of OsIspH1 (left panel) and OsIspH2 (right panel) using rice young seedling roots. (E) Subcellular localization of the OsIspH1:superfolder green fluorescent protein (sGFP) fusion protein with sGFP and RTp-sGFP, which are used as cytosol (Cy)- and stroma (St)-localized markers, respectively. A confocal microscope was used to detect the sGFP fluorescence and chlorophyll autofluorescence. (F) Characterization of the loss-of-function mutant for OsIspH1 using 5-day-old seedling phenotypes (left panel), genotyping and semiquantitative reverse-transcriptase PCRs (middle panel), and real-time quantitative reverse transcriptase PCR (qRT-PCR, right panel). WT, Oryza sativa Japonica cv. Dongjin as a wild-type control; null, nullizygous plants; He, heterozygous plants; Ho, homozygous plants. The used primer locations are indicated with arrows in (A) and Supplementary Table 10 shows the sequence information. All qRT-PCRs were triplicated and calculated using the ΔCt equation against the OsUbi5 gene as an internal control. The results are expressed as the mean ± standard error (SE), and Supplementary Tables 3, 4 show the original values.
To compare the expression patterns of two rice IspH genes, we examined them in various tissues at different developmental stages with the gene-specific primers located in a 3′-untranslated region (Figure 2B and Supplementary Table 3). The spatial expression pattern revealed that OsIspH1 transcripts were 20–40 times higher in the leaves than roots at all developmental stages, indicating a preference for leaves. They were also expressed in florets and seeds. OsIspH2 was not expressed in any leaf tissue and florets but was found in low levels in roots and seeds, indicating that OsIspH1 might be mainly functional in all rice tissues. Furthermore, we examined the expression patterns of two rice IspH genes against the treatments of six phytohormones, namely, ABA, CK, GA, IAA, MeJA, and SA (Figure 2C and Supplementary Table 4). Transcript levels of OsIspH1 were slightly suppressed by ABA but were induced by other five hormones in early time posttreatment, proposing different responses. Those of OsIspH2 were barely detectable before and after treatment.
To simultaneously identify the cDNAs of two rice IspH genes, total RNAs were prepared from roots at the seedling stage using the spatial expression pattern in Figure 2B, and RT-PCRs were performed using diverse primer combinations (Figure 2D). OsIspH1 exhibited a single 1,410-bp-long band (including 1,380 bp encoding 459 amino acids) as predicted in amplification, including the full-length ORFs; however, OsIspH2 unexpectedly showed a longer single 2,705-bp band than the predictable 1,644 bp (including 1,449 bp encoding 482 amino acids). Through further RT-PCRs of OsIspH2 with different combinations of exon-specific primers and sequencing of corresponding amplicons, we confirmed the only splicing error in the 1st intron, generating the longer 1st exon, shorter 1st intron, and quite longer 2nd exon than expected size, as indicated by the dotted line in Figure 2A. Interestingly, the extended 1st exon region contained the early stop codon, indicating that OsIspH2 could be a non-functional gene by causing a disabled small ORF of 324 bp encoding 107 amino acids.
Accordingly, OsIspH1 turned out to be the only functional HDR gene in rice. Its localization was examined using a GFP-fused protein transformation in rice protoplasts prepared from green seedlings and confirmed in stroma inside chloroplasts, with a similar pattern to stroma marker, rice ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit transit peptide, but different from cytosol marker (Figure 2E).
To evaluate the role of OsIspH1 in rice plants, we obtained a rice mutant with a T-DNA insertion in the region of 5′-untranslated region of OsIspH1 (PFG_1B-11039)4, as depicted in Figure 2A. We confirmed that the osisph1 knockout mutant exhibits albinism in homozygous plants based on genotyping and qRT-PCR (Figure 2F), and the albino phenotypes were observed at a 3:1 segregation ratio to identify a single-copy insertion of T-DNA, supposing that the albino phenotype might be derived from the knockout of OsIspH1 gene functions. Similarly, another knockout mutant of an OsIspH1 gene showed also the lethal albinism and the defectiveness of chloroplast biogenesis, which was constructed by a CRISPR-Cas9 gene-editing system (Liu et al., 2020), supposing that the albinism phenotype of osisph1 might also be mediated by the defective chloroplast biogenesis. Taken together, OsIspH1 is suggested to be crucial in photosynthetic pigments, including each or both chlorophylls and carotenoids, and that OsIspH2 could not compensate for the loss of function of OsIspH1.
Elucidation of Key Residues of OsIspH1 by Functional Complementation in E. coli
To confirm the conservation in key residues of OsIspH1, we aligned amino acid sequences among 12 monocot plants, including rice (Figure 3). Previously known key residues, an N-terminal conserved domain (NCD), four Cys-residues consisting of Fe-S clusters at active sites, and eight substrate-binding sites are highly conserved among these IspHs, indicating that OsIspH1 could function as an HDR. Interestingly, a potential key residue, such as protein kinase C (PKC) phosphorylation site in the C-terminal region, was newly predicted to be conserved among IspHs in the corresponding position of OsIspH1, Ser-Tyr-Lys405––407, through a Motif Scan analysis5, and Lys407 was also predicted as one of the ubiquitination sites through computational prediction of ubiquitination site6.
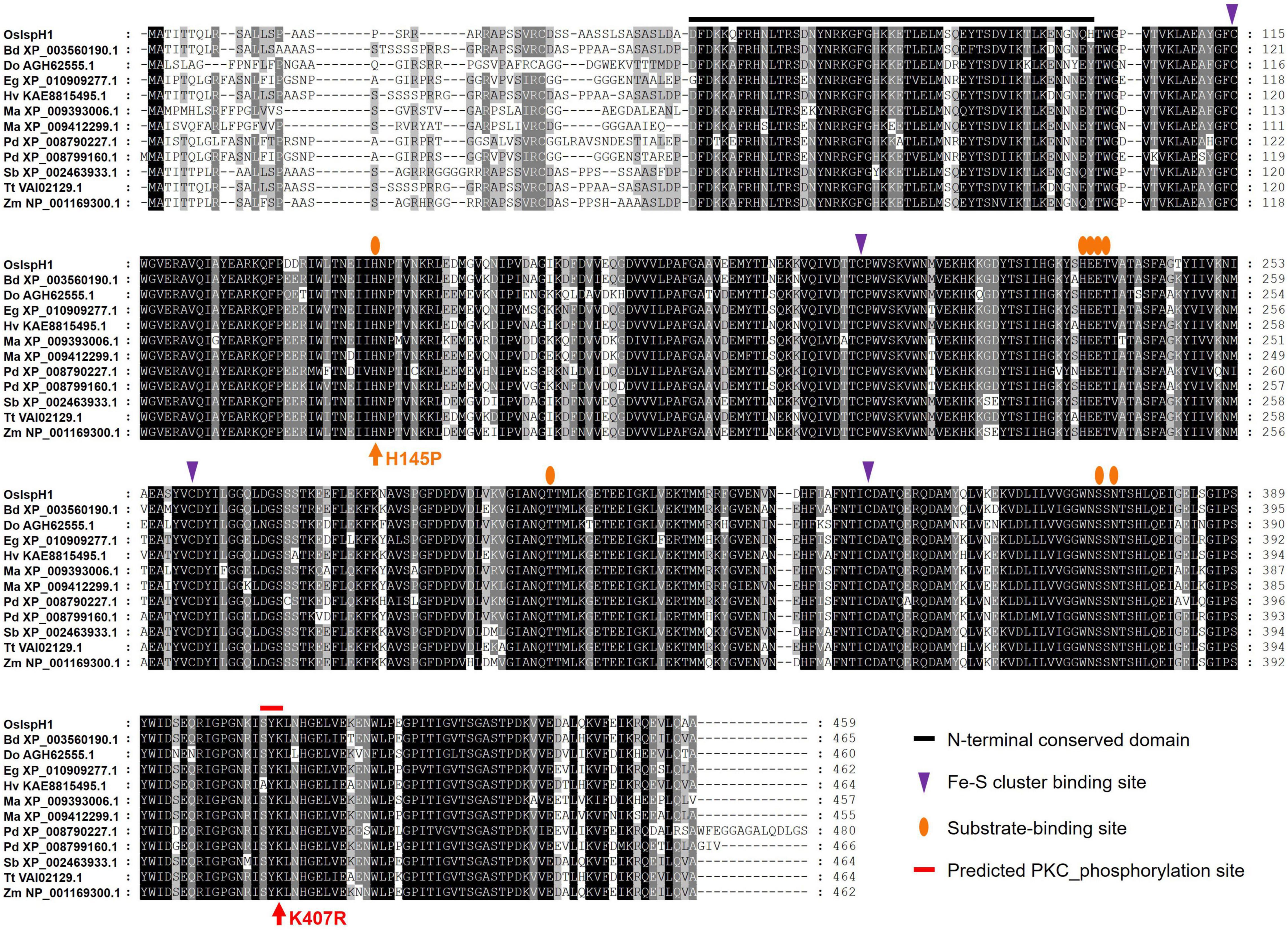
Figure 3. Amino acid sequence alignment among monocot plant IspHs. Supplementary Table 2 contains more information on IspH proteins.
Referring to the well-studied key residue of His152 in Arabidopsis (Hsieh et al., 2014), we mutated OsIspH1 to have non-synonymous substitutions in the corresponding His145 residue and newly predicted Lys407 residue into Pro145 and Arg407, respectively. Two single-mutated OsIspH1H145P and OsIspH1K407R genes and a double-mutated gene of OsIspH1H145P/K407R were transformed into an E. coli IspH1/LytB-lesion mutant strain, a DLYT1 (Kim et al., 2008). As a result, OsIspH1, OsIspH1H145P, and OsIspH1K407R successfully complemented the lethal phenotype of DLYT1 when EcIspH1/LytB was transformed as a positive control (Figure 4A). It confirmed that individual single mutation does not affect HDR activity. However, a double-substitution mutant, OsIspH1H145P/K407R (also called OsIspH1MUT hereafter), barely restored DLYT1 growth, indicating the complementary relationship of two residues for HDR enzyme activity. Additionally, to inquire whether substitution mutations of amino acids cause structural change in OsIspH1, we simulated the three-dimensional modeling based on PDB ID: 3DNF from A. aeolicus IspH using SWISS-MODEL7, as depicted in Figure 4B. According to a previous report, the first mutation from a positively charged His to a non-polar Pro is crucial for the “induced fit” effect to bind the substrate HMBPP (Hsieh et al., 2014). In this study, the predicted modeling showed the possibility that 1st key His145 residue might be faced to the Lys407 residue at nearly position (Figure 4B), supporting the possibility that the complementary relationship of two residues might be important for HDR enzyme activity in rice plants, although the mechanism is largely unknown.
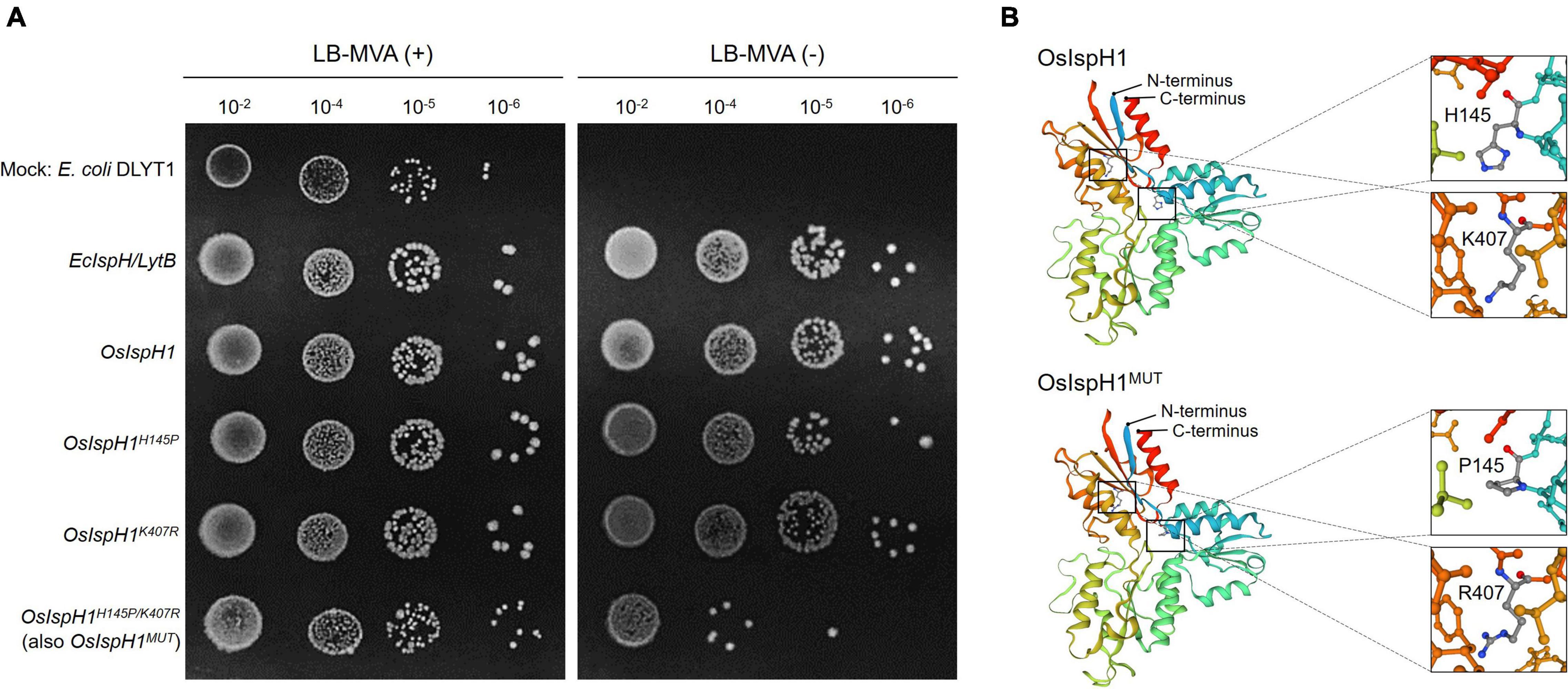
Figure 4. The 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR) enzyme assay in an Escherichia coli system and simulated protein structures of IspHs. (A) Functional complementation in E. coli DLYT1, an EcIspH lesion strain. An E. coli IspH gene, EcIspH/LytB, was used as a positive control. Mevalonate (MVA) (+) and (-) mean 0.01% and none of mevalonolactone, respectively. (B) Simulation of the 3D modeling to predict protein structure as rainbow ribbon drawing based on PDB ID: 3DNF from Aquifex aeolicus IspH built using SWISS-MODEL. Enlarged views showed the region with H145 and K407 residues as a ball and stick model with carbon (gray), nitrogen (blue), and oxygen (red) atoms.
In planta Effects of OsIspH1 on the Biosynthesis of Chlorophylls and Carotenoids
Based on the albino phenotype of the loss-of-function mutants, osisph1, as depicted in Figure 2F, we constitutively overexpressed an OsIspH1MUT to elucidate how an OsIspH1 affects chlorophyll and carotenoid biosynthesis in rice leaves and seeds. To learn more about its effects on carotenoid metabolism in seeds involved in the Golden Rice trait, the OsIspH1MUT cassette was further combined with a stPAC cassette, which produces β-carotene in rice endosperm (Jeong et al., 2017), as a single T-DNA vector (Figure 5A). Using three-independent homozygous T3 plants, which were selected by considering the lower copy number of one and two (Figure 5B), the expression patterns of a double-mutated transgene and an endogenous gene encoding rice HDR/IspH1 were examined in leaves (Figure 5C) and seeds (Figure 5D) of T4 rice plants. The overexpression of transgene, OsIspH1MUT, in both organs and seed-specific expression of stPAC was confirmed relative to non-transgenic (NT) rice plants. Interestingly, the ectopic expression of an OsIspH1MUT distinctly affected the expression patterns of endogenous OsIspH1 in both organs. In leaves of the IspH1MUT transgenic plants, the expression of endogenous OsIspH1 was significantly suppressed by an average of about 50% (P < 0.001 in IspH1MUT and P < 0.05 in IspH1MUT-stPAC), and the combined transcript levels of OsIspH1MUT and OsIspH1 were analyzed to be also significantly lower than the normal transcript level of endogenous OsIspH1 (P < 0.01 in IspH1MUT and P < 0.05 in IspH1MUT-stPAC), even under the ectopic expression of OsIspH1MUT transgene (Figure 5C and Supplementary Table 5). In contrast to the suppression patterns in leaves, the combined transcript levels of OsIspH1MUT and OsIspH1 were analyzed to be at least 4 times higher than the transcript level of endogenous OsIspH1 (P < 0.001 in IspH1MUT and P < 0.05 in IspH1MUT-stPAC), and the transcript levels of endogenous OsIspH1 were not suppressed in seeds of all transgenic plant (Figure 5D and Supplementary Table 6), indicating that OsIspH1MUT was expressed four times more than the intrinsic OsIspH1 gene. Finally, the distinct expression patterns between leaves and seeds were statistically significantly conservative in six independent transgenic plants of IspH1MUT and IspH1MUT-stPAC, and taken collectively, it was supposed that the expression of OsIspH1 might be differentially regulated between leaves and seeds.
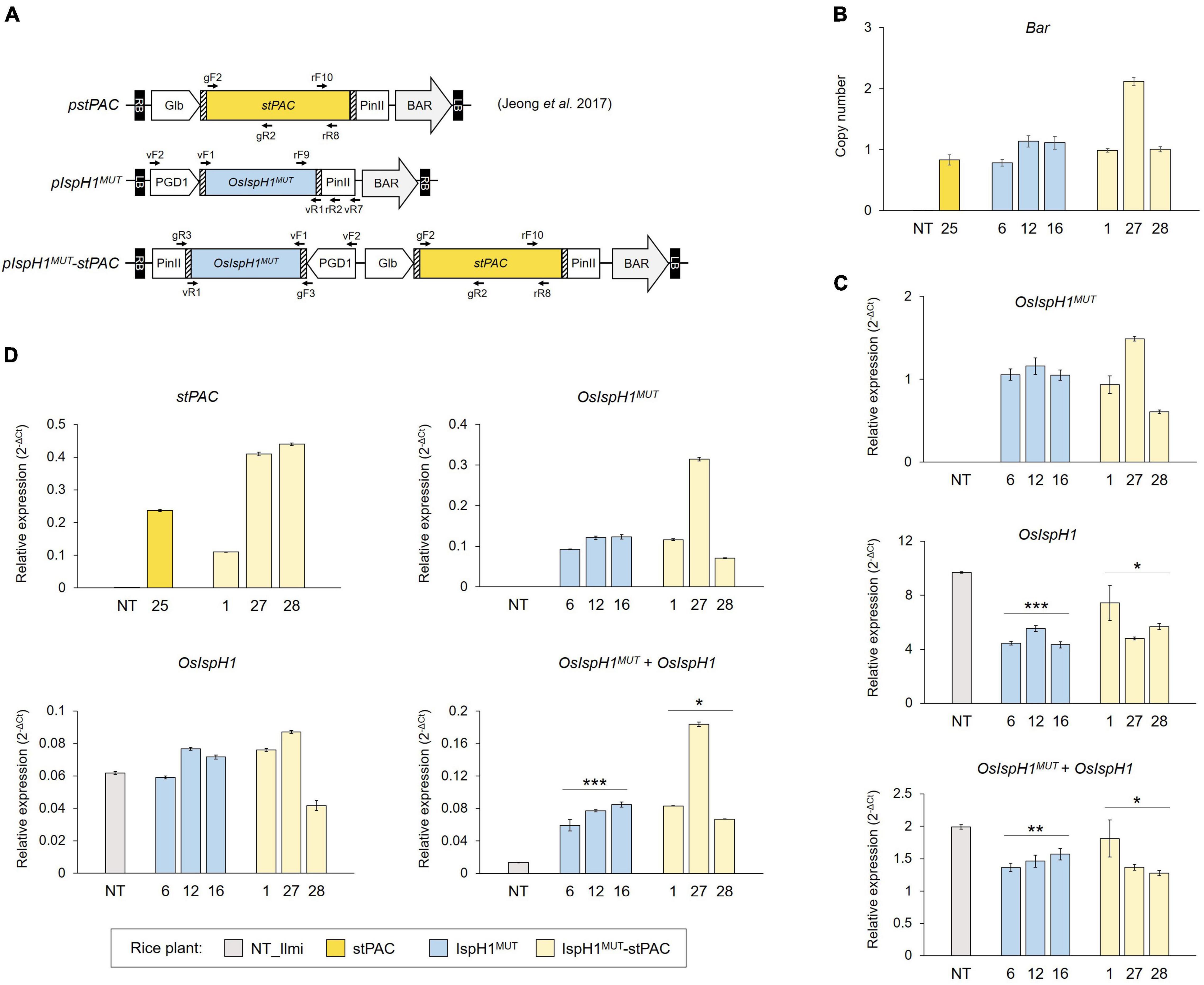
Figure 5. Generation of transgenic rice plants for in planta characterization. (A) Schematic representation of binary vectors for rice transformation with the indicated positions of primer sets listed in Supplementary Table 10. (B) TaqMan PCR to verify the transgene copy numbers using bialaphos-resistant (Bar) gene cassette. The previously reported homozygous stPAC 25 line was used as a reference for the single T-DNA insertion (Jeong et al., 2017). The qRT-PCR to examine transcript levels of transgenes, OsIspH1MUT or stPAC, endogenous OsIspH1, and combined genes, OsIspH1MUT and OsIspH1, in transgenic rice leaves (C) and seeds (D), respectively. The rice ubiquitin gene (Os01g22490) was used as an internal reference for quantitative normalization, and the values were represented as the mean ± SE of three independent measurements. Statistical analyses were performed using a one-tailed Student’s t-test, and significant differences were indicated by a p-value (*p < 0.05, **p < 0.01, and ***p < 0.001).
In the next step, we analyzed two terpene-derived photosynthetic pigments, including chlorophylls and carotenoids, using HPLC. In leaves, although there were differences between the independent transgenic plants, the total contents of chlorophylls (an average of 84.6% level, Supplementary Table 7) and carotenoids (an average of 84.3% level, Supplementary Table 8) were decreased in two transgenic plants with the significant differences compared to NT plants (Figure 6A). In addition to the functional complementation of OsIspH1MUT as HDR/IspH in E. coli (Figure 4A), in planta functional complementation was examined through cross-interbreeding between two transgenic rice plants, namely, osisph1 and IspH1MUT. In rice plants, the albinism of osisph1 was not restored by OsIspH1MUT overexpression at all, of which homozygosity was ascertained by genotyping (Figure 6B), indicating that OsIspH1MUT was a protein defective in HDR activity in rice plants. Collectively, it was supposed that the overexpression of OsIspH1MUT might cause the relative decrease in active HDR proteins through the competition of OsIspH1MUT with the endogenous OsIspHs and consequently decrease chlorophyll and carotenoid contents in the leaves. In the seeds, the carotenoid content of IspH1MUT (Figure 7A) and IspH1MUT-stPAC (Figure 7B) was statistically significantly reduced on the average of 61.4 and 54.2%, respectively, compared to NT, indicating a more severe pattern in seeds than leaves (Figure 7 and Supplementary Table 9). The results were considered to be closely related to the higher expression level of OsIspH1MUT than the endogenous OsIspH1 gene in the seeds.
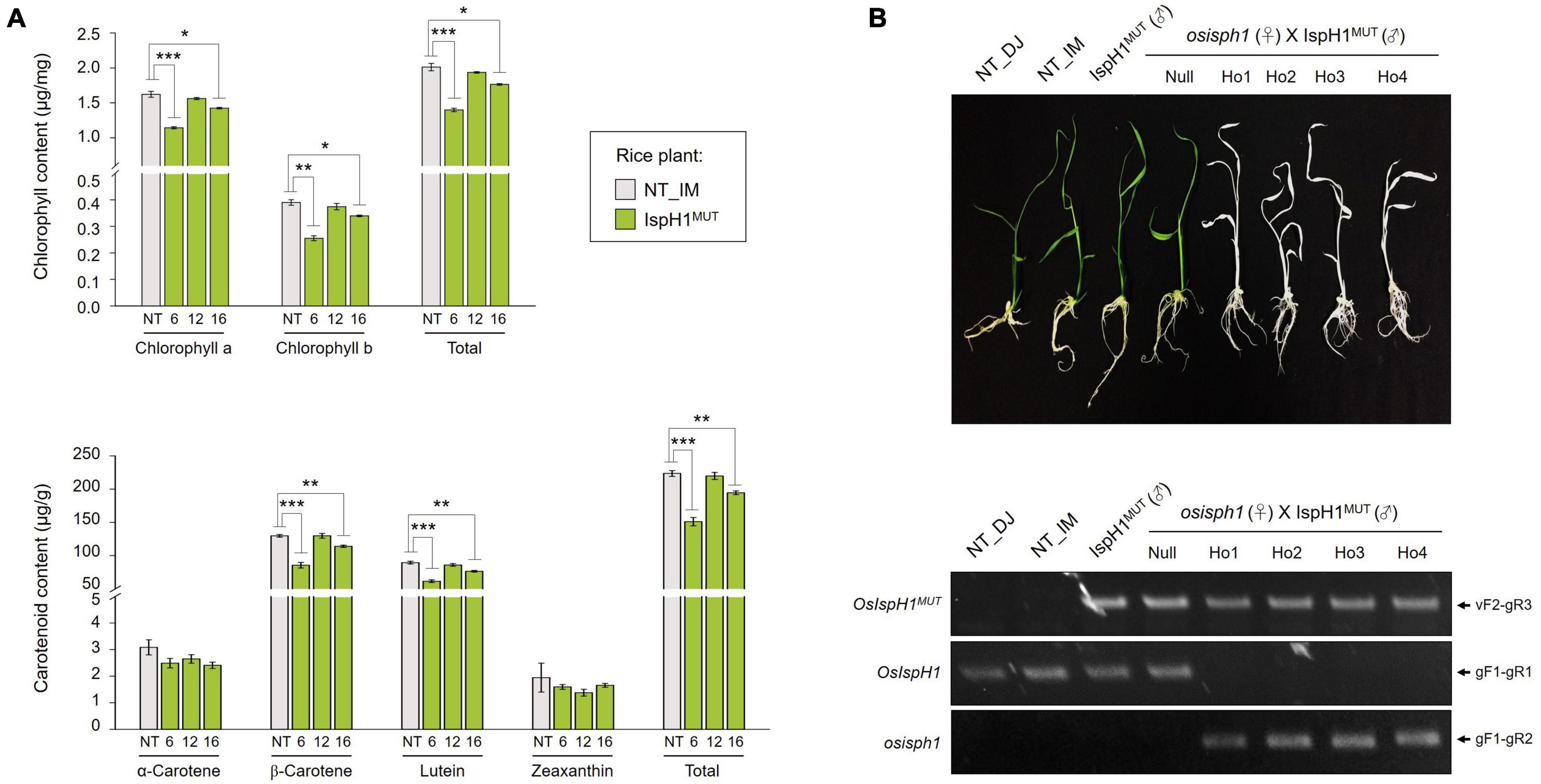
Figure 6. Effects of the OsIspH1MUT on rice leaves. (A) Contents and composition of chlorophylls and carotenoids in the leaves at T4 plants. Non-transgenic (NT) rice plants (O. sativa L. cv. Ilmi) and three independent IspH1MUT transgenic plants were used for metabolite analysis. All data are represented as the mean ± SE of three independent measurements. Statistical analyses were performed using a one-tailed Student’s t-test, and significant differences were indicated by a p-value (*p < 0.05, **p < 0.01, and ***p < 0.001). (B) Functional complementation in osisph1 rice plants. Phenotypes of 10-day-old seedlings by interbreeding between heterozygous osisph1 and IspH1MUT transgenic plants at F4 generation and genomic PCR to confirm genotypes using primer sets listed in Supplementary Table 10.
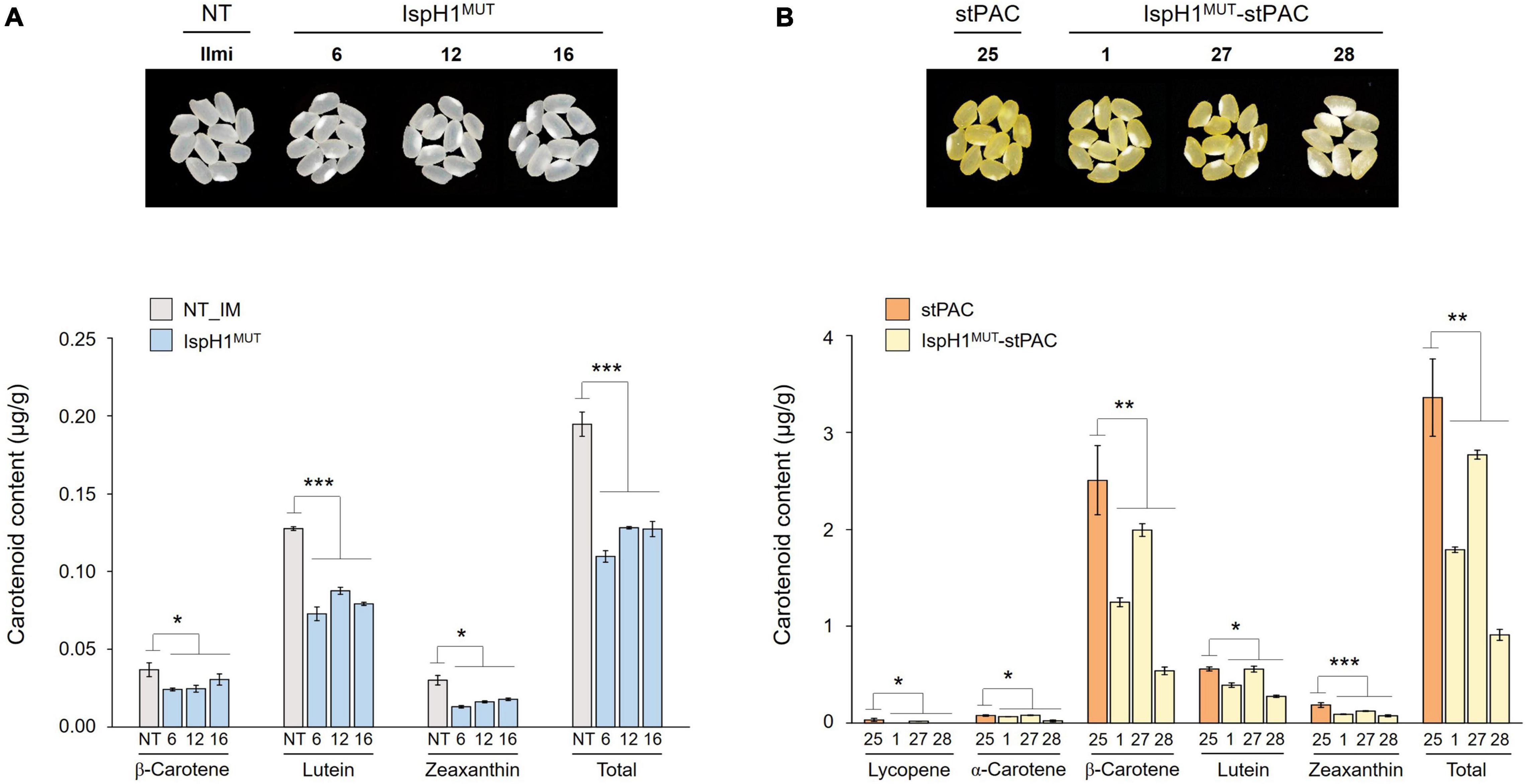
Figure 7. Color phenotypes and carotenoid levels in the rice seeds of IspH1MUT (A) and IspH1MUT-stPAC (B) transgenic lines. Colors of mature seed harvested 60 days after flowering were compared, dried for 2 weeks at room temperature, threshed, dehusked, and polished in the homozygous T4 generation. Carotenoid contents were analyzed using high-performance liquid chromatography (HPLC), and all data are represented as the mean ± SE of three independent measurements. Statistical analyses were performed using a one-tailed, and significant differences were indicated by a p-value (*p < 0.05, **p < 0.01, and ***p < 0.001).
Taken together, OsIspH1 is supposed to have crucial roles such as the rate-limiting regulation on carotenoid biosynthesis in seeds and photosynthetic pigments in leaves.
Discussion
OsIspH1 Is the Unique Functional HDR in Rice Plants
The MEP pathway consists of seven enzymatic steps that start with the catalysis of DXS and end with that of IspH. While the five intermediate enzymes are encoded by a single gene, DXS and IspH have been reported as one or multiple genes in diverse plants, proposing the potent roles as flux-controlling steps for terpenoid production. Plant DXSs have been classified into three clades, with different influences on the downstream pathways: DXS1s for chlorophyll biosynthesis with a housekeeping role, DXS2s for the production of specialized terpene metabolites, including artemisinins, ginkgolides, resins, and carotenoids, in different plants, and DXS3 for considerably understudied but still tentative roles in a few plants, as recently summarized (Cordoba et al., 2011; Zhang et al., 2018; de Luna-Valdez et al., 2021; Tian et al., 2021). Meanwhile, plant IspH has been reported as a single-gene family in most plant species but as a multiple-gene family of 2–3 copies in some species. In G. biloba and Pinus taeda of gymnosperm plants, GbIspH1 and PtIspH1 are for housekeeping roles, and GbIspH2 and PtIspH2 are for individually specialized ginkgolide and resin roles (Kim et al., 2008, 2009). In angiosperm plants, two C. melo genes showed different spatial expression: CmIspH1 for the most abundant in all organs and CmIspH2 in specialized contexts, such as de-etiolating seedlings and orange-fleshed fruit during ripening (Saladie et al., 2014). Thereby, it could be proposed that IspH1s play housekeeping roles, but IspH2s can contribute to plant species-specific terpene production. In the case of O. sativa, two IspH genes are predicted in the genome and have been previously presumed the different function: OsIspH1 as a predominantly light-induced gene but OsIspH2 as an upregulated gene in the dxr mutants without light response, proposing a compensating route awaiting validation (Jung et al., 2008). However, Liu et al. (2020) presented that the genome-edited OsIspH1 defective mutant showed the albino lethal phenotype, and similarly in this study, the T-DNA mutant of OsIspH1, osisph1, showed the albino lethal phenotype (Figure 6), supposing that OsIspH2 might not have any compensate function for it. Interestingly in this study, the sequence analysis of OsIspH2 mRNA presented that the extended 1st exon region had an early stop codon to consequentially generate the truncated-short polypeptide fragments, which was spliced at the quite different region from the predicted-splicing position provided by the RAP-DB (Figures 2A,D). Collectively, an OsIspH2 gene is supposed to be non-functional, indicating that an OsIspH1 is a unique functional HDR gene in rice.
The Complementary Relationship of His145 and Lys407 Residues Is Crucial for the HDR Activity of OsIspH1 in Both Bacteria and Rice Plants
The X-ray crystal structure of IspH protein has been first reported in the hyperthermophilic eubacterium A. aeolicus (Rekittke et al., 2008), its critical residues have been elucidated mainly in bacterial systems, such as A. aeolicus, E. coli, and cyanobacteria (Rekittke et al., 2008; Grawert et al., 2009; Hsieh and Hsieh, 2015). Meanwhile, the plant-type IspH structure has also been reported in Arabidopsis and the same key residues to bacterial ones by complementation assay in E. coli isph mutant (Hsieh et al., 2014). By Hsieh et al. (2014), two bacterial critical residues showed the difference in the corresponding ones of Arabidopsis, His152 and His241, not individually but cooperatively critical for HDR activity with more significance of His152 than His241. In this study, we found another conserved motif, referred to as a predicted PKC phosphorylation site in Figure 3, and to determine whether it could be a novel crucial residue of OsIspH1, we mutated each or together at both positions of His145 and Lys407 to generate OsIspH1H145P and OsIspH1K407R, and OsIspH1H145P/K407R, and the former is the corresponding site to His152 of AtIspH, and the latter is a newly predicted site for PKC phosphorylation or ubiquitination in this study. The complementary experiments in the isph mutants of bacteria and rice plants showed that the simultaneous mutation of His145 and Lys407, an OsIspH1H145P/K407R, largely reduced the HDR activity of OsIspH1 not only in the bacteria expression system (Figure 4A) but also in the rice plant system (Figure 6B), but every single mutation did not have any effects on the HDR activity of it in bacteria system. These results supposed that the complementary relationship of two residues, namely, His145 and Lys407, might be crucial to the HDR activity of OsIspH1.
OsIspH1 Plays an Important Role in the Biosynthesis of Chlorophylls and Carotenoids, Through the Differential Expression Patterns Between Leaves and Seeds
The regulation of plant genes involved in MEP and MVA pathways occurs at multiple levels of transcription, posttranscription, translation, and posttranslation (Cordoba et al., 2009; Hemmerlin, 2013; Vranova et al., 2013). Particularly, plant IspHs contribute to the MEP pathway primarily through transcriptional regulation with tissue specificity and different responses against environmental conditions, such as light, circadian, mechanical wounding, and fungal elicitors in diverse plants (Kim et al., 2008, 2009; Cordoba et al., 2009; Ma et al., 2017). To scrutinize the intrinsic rice IspH regulation, we constitutively overexpressed the malfunctional OsIspH1MUT (Figure 5) in rice plants, which was identified in two hdr mutants, namely, E. coli DLYT1 strains (Figure 4) and rice osisph1 plants (Figure 6B). Interestingly, the effects of OsIspH1MUT-ectopic expression were observed to be quite differential between rice leaves and seeds. In leaves, not only the transcript level of an endogenous OsIspH1 was downregulated, but also the combined transcript level of endogenous OsIspH1 and exogenous OsIspH1MUT remained lower than that of NT (Figure 5C). In contrast, the expression of an endogenous OsIspH1 was not downregulated in the seeds and remained higher than that of NT (Figure 5D), suggesting that the expression of an intrinsic OsIspH1 gene was differentially regulated in leaves and seeds. Similarly, organ-specific differential roles of OsDXS2 and OsDXR on carotenoid accumulation between leaves and seeds have also been reported (You et al., 2020). Collectively, these results suggest that the DXS, DXR, and IspH steps belonging to the MEP pathway might be regulated to differentially affected carotenoid accumulation between leaves and seeds.
Despite such differential regulation, the ectopic expression of OsIspH1MUT simultaneously caused the decrease of chlorophyll and carotenoid content in leaves and seeds (Figures 6, 7), and the reduction patterns were proportional to the overexpression patterns of OsIspH1MUT, indicating the relative decrease in active HDR enzyme function by competition between endogenous OsIspH1 and OsIspH1MUT. In other words, the intrinsic OsIspH1 is supposed to play an important role in the biosynthesis of chlorophyll and carotenoid levels in rice leaves and seeds and to be a promising target to biofortify the functional terpene metabolites in rice plants.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
S-HH coordinated the project, supervised the manuscript, and was responsible for all contacts and correspondence. MY contributed to the interpretation of data for the work, and revised it critically for important intellectual content. YL performed all the experiments in planta and wrote the draft of the manuscript. JK and S-AB analyzed the contents of chlorophylls and carotenoids. J-SY contributed to the experiments in E. coli. All authors have read and approved the final manuscript.
Funding
This study was supported by two grants from the BioGreen21 Agri-Tech Innovation Program (PJ01567101 to S-HH), funded by the Rural Development Administration in South Korea, and National Research Foundation of Korea (NRF), funded by the Korean government (MSIT) (2021R1A2C2012227 to S-HH).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.861036/full#supplementary-material
Footnotes
- ^ https://www.greenphyl.org/cgi-bin/index.cgi
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://rapdb.dna.affrc.go.jp/
- ^ http://signal.salk.edu/cgi-bin/RiceGE
- ^ http://myhits.isb-sib.ch/cgi-bin/motif_scan
- ^ http://bdmpub.biocuckoo.org/prediction.php
- ^ https://swissmodel.expasy.org/
References
Altincicek, B., Duin, E. C., Reichenberg, A., Hedderich, R., Kollas, A. K., Hintz, M., et al. (2002). LytB protein catalyzes the terminal step of the 2-C-methyl-D-erythritol-4-phosphate pathway of isoprenoid biosynthesis. FEBS Lett. 532, 437–440. doi: 10.1016/s0014-5793(02)03726-2
Botella-Pavia, P., Besumbes, O., Phillips, M. A., Carretero-Paulet, L., Boronat, A., and Rodriguez-Concepcion, M. (2004). Regulation of carotenoid biosynthesis in plants: evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J. 40, 188–199. doi: 10.1111/j.1365-313X.2004.02198.x
Cordoba, E., Porta, H., Arroyo, A., Roman, C. S., Medina, L., Rodriguez-Concepcion, M., et al. (2011). Functional characterization of the three genes encoding 1-deoxy-D-xylulose 5-phosphate synthase in maize. J. Exp. Bot. 62, 2023–2038. doi: 10.1093/jxb/erq393
Cordoba, E., Salmi, M., and Leon, P. (2009). Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 60, 2933–2943. doi: 10.1093/jxb/erp190
Cunningham, F. X. Jr., Lafond, T. P., and Gantt, E. (2000). Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J. Bacteriol. 182, 5841–5848. doi: 10.1128/JB.182.20.5841-5848.2000
de Luna-Valdez, L., Chenge-Espinosa, M., Hernandez-Munoz, A., Cordoba, E., Lopez-Leal, G., Castillo-Ramirez, S., et al. (2021). Reassessing the evolution of the 1-deoxy-D-xylulose 5-phosphate synthase family suggests a possible novel function for the DXS class 3 proteins. Plant Sci. 310:110960. doi: 10.1016/j.plantsci.2021.110960
Eoh, H., Brennan, P. J., and Crick, D. C. (2009). The Mycobacterium tuberculosis MEP (2C-methyl-d-erythritol 4-phosphate) pathway as a new drug target. Tuberculosis (Edinb) 89, 1–11. doi: 10.1016/j.tube.2008.07.004
Grawert, T., Kaiser, J., Zepeck, F., Laupitz, R., Hecht, S., Amslinger, S., et al. (2004). IspH protein of Escherichia coli: studies on iron-sulfur cluster implementation and catalysis. J. Am. Chem. Soc. 126, 12847–12855. doi: 10.1021/ja0471727
Grawert, T., Rohdich, F., Span, I., Bacher, A., Eisenreich, W., Eppinger, J., et al. (2009). Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction. Angew. Chem. Int. Ed Engl. 48, 5756–5759. doi: 10.1002/anie.200900548
Grieshaber, N. A., Fischer, E. R., Mead, D. J., Dooley, C. A., and Hackstadt, T. (2004). Chlamydial histone-DNA interactions are disrupted by a metabolite in the methylerythritol phosphate pathway of isoprenoid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 101, 7451–7456. doi: 10.1073/pnas.0400754101
Ha, S. H., Kim, J. K., Jeong, Y. S., You, M. K., Lim, S. H., and Kim, J. K. (2019). Stepwise pathway engineering to the biosynthesis of zeaxanthin, astaxanthin and capsanthin in rice endosperm. Metab. Eng. 52, 178–189. doi: 10.1016/j.ymben.2018.11.012
Hao, G., Shi, R., Tao, R., Fang, Q., Jiang, X., Ji, H., et al. (2013). Cloning, molecular characterization and functional analysis of 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase (HDR) gene for diterpenoid tanshinone biosynthesis in Salvia miltiorrhiza Bge. f. alba. Plant Physiol. Biochem. 70, 21–32. doi: 10.1016/j.plaphy.2013.05.010
Hemmerlin, A. (2013). Post-translational events and modifications regulating plant enzymes involved in isoprenoid precursor biosynthesis. Plant Sci. 203-204, 41–54. doi: 10.1016/j.plantsci.2012.12.008
Hemmerlin, A., Harwood, J. L., and Bach, T. J. (2012). A raison d’etre for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog. Lipid Res. 51, 95–148. doi: 10.1016/j.plipres.2011.12.001
Hintz, M., Reichenberg, A., Altincicek, B., Bahr, U., Gschwind, R. M., Kollas, A. K., et al. (2001). Identification of (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate as a major activator for human gammadelta T cells in Escherichia coli. FEBS Lett. 509, 317–322. doi: 10.1016/s0014-5793(01)03191-x
Hsieh, M. H., and Goodman, H. M. (2005). The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol. 138, 641–653. doi: 10.1104/pp.104.058735
Hsieh, W. Y., and Hsieh, M. H. (2015). The amino-terminal conserved domain of 4-hydroxy-3-methylbut-2-enyl diphosphate reductase is critical for its function in oxygen-evolving photosynthetic organisms. Plant Signal. Behav. 10:e988072. doi: 10.4161/15592324.2014.988072
Hsieh, W. Y., Sung, T. Y., Wang, H. T., and Hsieh, M. H. (2014). Functional evidence for the critical amino-terminal conserved domain and key amino acids of Arabidopsis 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE. Plant Physiol. 166, 57–69. doi: 10.1104/pp.114.243642
Jeong, Y. S., Ku, H.-K., Kim, J. K., You, M. K., Lim, S.-H., Kim, J.-K., et al. (2017). Effect of codon optimization on the enhancement of the β-carotene contents in rice endosperm. Plant Biotechnol. Rep. 11, 171–179. doi: 10.1007/s11816-017-0440-0
Jung, K. H., Lee, J., Dardick, C., Seo, Y. S., Cao, P., Canlas, P., et al. (2008). Identification and functional analysis of light-responsive unique genes and gene family members in rice. PLoS Genet. 4:e1000164. doi: 10.1371/journal.pgen.1000164
Kim, J. K., Lee, S. Y., Chu, S. M., Lim, S. H., Suh, S. C., Lee, Y. T., et al. (2010). Variation and correlation analysis of flavonoids and carotenoids in Korean pigmented rice (Oryza sativa L.) cultivars. J. Agric. Food Chem. 58, 12804–12809. doi: 10.1021/jf103277g
Kim, S. M., Kuzuyama, T., Kobayashi, A., Sando, T., Chang, Y. J., and Kim, S. U. (2008). 1-Hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase (IDS) is encoded by multicopy genes in gymnosperms Ginkgo biloba and Pinus taeda. Planta 227, 287–298. doi: 10.1007/s00425-007-0616-x
Kim, T. J., Choi, J., Kim, K. W., Ahn, S. K., Ha, S. H., Choi, Y., et al. (2017). Metabolite profiling of peppers of various colors reveals relationships between tocopherol, carotenoid, and phytosterol content. J. Food Sci. 82, 2885–2893. doi: 10.1111/1750-3841.13968
Kim, Y. B., Kim, S. M., Kang, M. K., Kuzuyama, T., Lee, J. K., Park, S. C., et al. (2009). Regulation of resin acid synthesis in Pinus densiflora by differential transcription of genes encoding multiple 1-deoxy-D-xylulose 5-phosphate synthase and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase genes. Tree Physiol. 29, 737–749. doi: 10.1093/treephys/tpp002
Kim, Y. B., Kim, S. M., Sathasivam, R., Kim, Y. K., Park, S. U., and Kim, S. U. (2021). Overexpression of Ginkgo biloba Hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase 2 gene (GbHDR2) in Nicotiana tabacum cv. Xanthi. 3 Biotech 11:337. doi: 10.1007/s13205-021-02887-5
Ko, M. R., Song, M. H., Kim, J. K., Baek, S. A., You, M. K., Lim, S. H., et al. (2018). RNAi-mediated suppression of three carotenoid-cleavage dioxygenase genes, OsCCD1, 4a, and 4b, increases carotenoid content in rice. J. Exp. Bot. 69, 5105–5116. doi: 10.1093/jxb/ery300
Liu, N., Xie, K., Jia, Q., Zhao, J., Chen, T., Li, H., et al. (2016). Foxtail mosaic virus-induced gene silencing in monocot plants. Plant Physiol. 171, 1801–1807. doi: 10.1104/pp.16.00010
Liu, X., Cao, P. H., Huang, Q. Q., Yang, Y. R., and Tao, D. D. (2020). Disruption of a rice chloroplast-targeted gene OsHMBPP causes a seedling-lethal albino phenotype. Rice (N. Y.) 13:51. doi: 10.1186/s12284-020-00408-1
Lu, X. M., Hu, X. J., Zhao, Y. Z., Song, W. B., Zhang, M., Chen, Z. L., et al. (2012). Map-based cloning of zb7 encoding an IPP and DMAPP synthase in the MEP pathway of maize. Mol. Plant 5, 1100–1112. doi: 10.1093/mp/sss038
Ma, D., Li, G., Zhu, Y., and Xie, D. Y. (2017). Overexpression and suppression of Artemisia annua 4-hydroxy-3-methylbut-2-enyl diphosphate reductase 1 gene (AaHDR1) differentially regulate Artemisinin and terpenoid biosynthesis. Front. Plant Sci. 8:77. doi: 10.3389/fpls.2017.00077
Masini, T., and Hirsch, A. K. (2014). Development of inhibitors of the 2C-methyl-D-erythritol 4-phosphate (MEP) pathway enzymes as potential anti-infective agents. J. Med. Chem. 57, 9740–9763. doi: 10.1021/jm5010978
Mathur, V., Kwong, W. K., Husnik, F., Irwin, N. A. T., Kristmundsson, A., Gestal, C., et al. (2021). Phylogenomics identifies a new major subgroup of apicomplexans, Marosporida class nov., with extreme apicoplast genome reduction. Genome Biol. Evol. 13:evaa244. doi: 10.1093/gbe/evaa244
Nagegowda, D. A., and Gupta, P. (2020). Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 294:110457. doi: 10.1016/j.plantsci.2020.110457
Page, J. E., Hause, G., Raschke, M., Gao, W., Schmidt, J., Zenk, M. H., et al. (2004). Functional analysis of the final steps of the 1-deoxy-D-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virus-induced gene silencing. Plant Physiol. 134, 1401–1413. doi: 10.1104/pp.103.038133
Park, S. H., Yi, N., Kim, Y. S., Jeong, M. H., Bang, S. W., Choi, Y. D., et al. (2010). Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J. Exp. Bot. 61, 2459–2467. doi: 10.1093/jxb/erq076
Rao, G., and Oldfield, E. (2016). Structure and function of four classes of the 4Fe-4S protein, IspH. Biochemistry 55, 4119–4129. doi: 10.1021/acs.biochem.6b00474
Rekittke, I., Wiesner, J., Rohrich, R., Demmer, U., Warkentin, E., Xu, W., et al. (2008). Structure of (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase, the terminal enzyme of the non-mevalonate pathway. J. Am. Chem. Soc. 130, 17206–17207. doi: 10.1021/ja806668q
Rohmer, M., Knani, M., Simonin, P., Sutter, B., and Sahm, H. (1993). Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 295(Pt. 2), 517–524. doi: 10.1042/bj2950517
Saladie, M., Wright, L. P., Garcia-Mas, J., Rodriguez-Concepcion, M., and Phillips, M. A. (2014). The 2-C-methylerythritol 4-phosphate pathway in melon is regulated by specialized isoforms for the first and last steps. J. Exp. Bot. 65, 5077–5092. doi: 10.1093/jxb/eru275
Singh, N., Cheve, G., Avery, M. A., and McCurdy, C. R. (2007). Targeting the methyl erythritol phosphate (MEP) pathway for novel antimalarial, antibacterial and herbicidal drug discovery: inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) enzyme. Curr. Pharm. Des. 13, 1161–1177. doi: 10.2174/138161207780618939
Tian, S., Wang, D., Yang, L., Zhang, Z., and Liu, Y. (2021). A systematic review of 1-Deoxy-D-xylulose-5-phosphate synthase in terpenoid biosynthesis in plants. Plant Growth Regul. 96, 221–235. doi: 10.1007/s10725-021-00784-8
Uchida, H., Sumimoto, K., Oki, T., Nishii, I., Mizohata, E., Matsunaga, S., et al. (2018). Isolation and characterization of 4-hydroxy-3-methylbut-2-enyl diphosphate reductase gene from Botryococcus braunii, race B. J. Plant Res. 131, 839–848. doi: 10.1007/s10265-018-1039-4
Vranova, E., Coman, D., and Gruissem, W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700. doi: 10.1146/annurev-arplant-050312-120116
Wang, W., Wang, K., Liu, Y. L., No, J. H., Li, J., Nilges, M. J., et al. (2010). Bioorganometallic mechanism of action, and inhibition, of IspH. Proc. Natl. Acad. Sci. U.S.A. 107, 4522–4527. doi: 10.1073/pnas.0911087107
Wang, X., and Dowd, C. S. (2018). The methylerythritol phosphate pathway: promising drug targets in the fight against Tuberculosis. ACS Infect. Dis. 4, 278–290. doi: 10.1021/acsinfecdis.7b00176
Wellburn, A. R. (1994). The spectral determination of chlorophylls a and b, as well as total carotenoids, using various sovents with spectrophotometers of different resolution. J. Plant Physiol. 144, 307–313. doi: 10.1016/s0176-1617(11)81192-2
You, M. K., Kim, J. H., Lee, Y. J., Jeong, Y. S., and Ha, S. H. (2016). Plastoglobule-targeting competence of a putative transit peptide sequence from rice phytoene synthase 2 in plastids. Int. J. Mol. Sci. 18:18. doi: 10.3390/ijms18010018
You, M. K., Lee, Y. J., Kim, J. K., Baek, S. A., Jeon, Y. A., Lim, S. H., et al. (2020). The organ-specific differential roles of rice DXS and DXR, the first two enzymes of the MEP pathway, in carotenoid metabolism in Oryza sativa leaves and seeds. BMC Plant Biol. 20:167. doi: 10.1186/s12870-020-02357-9
Keywords: carotenoids and chlorophylls, duplicated pseudogene, functional complementation, methylerythritol 4-phosphate pathway, OsIspH1, OsIspH2, rice
Citation: Lee YJ, Kim JK, Baek S-A, Yu J-S, You MK and Ha S-H (2022) Differential Regulation of an OsIspH1, the Functional 4-Hydroxy-3-Methylbut-2-Enyl Diphosphate Reductase, for Photosynthetic Pigment Biosynthesis in Rice Leaves and Seeds. Front. Plant Sci. 13:861036. doi: 10.3389/fpls.2022.861036
Received: 24 January 2022; Accepted: 10 March 2022;
Published: 13 April 2022.
Edited by:
Guodong Wang, Institute of Genetics and Developmental Biology (CAS), ChinaReviewed by:
De-Yu Xie, North Carolina State University, United StatesRaimund Nagel, Leipzig University, Germany
Copyright © 2022 Lee, Kim, Baek, Yu, You and Ha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Kyoung You, bWlua3lvdUBraHUuYWMua3I=; Sun-Hwa Ha, c3VuaHdhQGtodS5hYy5rcg==
 Yeo Jin Lee
Yeo Jin Lee Jae Kwang Kim2
Jae Kwang Kim2 Min Kyoung You
Min Kyoung You Sun-Hwa Ha
Sun-Hwa Ha