- Ecology and Evolution, Research School of Biology, Australian National University, Canberra, ACT, Australia
Sexually deceptive plants secure pollination by luring specific male insects as pollinators using a combination of olfactory, visual, and morphological mimicry. Flower color is a key component to this attraction, but its chemical and genetic basis remains poorly understood. Chiloglottis trapeziformis is a sexually deceptive orchid which has predominantly dull green-red flowers except for the central black callus projecting from the labellum lamina. The callus mimics the female of the pollinator and the stark color contrast between the black callus and dull green or red lamina is thought to enhance the visibility of the mimic. The goal of this study was to investigate the chemical composition and genetic regulation of temporal and spatial color patterns leading to visual mimicry, by integrating targeted metabolite profiling and transcriptomic analysis. Even at the very young bud stage, high levels of anthocyanins were detected in the dark callus, with peak accumulation by the mature bud stage. In contrast, anthocyanin levels in the lamina peaked as the buds opened and became reddish-green. Coordinated upregulation of multiple genes, including dihydroflavonol reductase and leucoanthocyanidin dioxygenase, and the downregulation of flavonol synthase genes (FLS) in the callus at the very young bud stage underpins the initial high anthocyanin levels. Conversely, within the lamina, upregulated FLS genes promote flavonol glycoside over anthocyanin production, with the downstream upregulation of flavonoid O-methyltransferase genes further contributing to the accumulation of methylated flavonol glycosides, whose levels peaked in the mature bud stage. Finally, the peak anthocyanin content of the reddish-green lamina of the open flower is underpinned by small increases in gene expression levels and/or differential upregulation in the lamina in select anthocyanin genes while FLS patterns showed little change. Differential expression of candidate genes involved in specific transport, vacuolar acidification, and photosynthetic pathways may also assist in maintaining the distinct callus and contrasting lamina color from the earliest bud stage through to the mature flower. Our findings highlight that flower color in this sexually deceptive orchid is achieved by complex tissue-specific coordinated regulation of genes and biochemical pathways across multiple developmental stages.
Introduction
The majority of angiosperms are pollinated by animals (Ollerton et al., 2011), with floral signaling key to this crucial interaction. Flowers not only contain the reproductive structures, but also convey long-range visual and olfactory signals for pollinator attraction and the simultaneous deterrence of unwanted visitors (Raguso, 2004; Borghi et al., 2017). Brightly colored flowers are the norm in diurnally pollinated plants, ensuring visibility to pollinators against the background, with flower colors spanning the UV and visible spectrum. In addition to bright colors, many flowers display contrasting color markings, spots, and lines, which serve as “nectar guides” for pollinators, with some markings only visible to pollinators that can perceive UV light (Davies et al., 2012; Glover et al., 2013; Nadot and Carrive, 2021).
A diverse range of pigment compounds are responsible for the various colors and markings of flowers. Anthocyanins typically confer the orange, red, violet, and blue colors to flowers, while chalcones, aurones, flavonols, and flavones are often associated with different degrees of pale yellow coloration. Chlorophylls and carotenoids can further extend the color palette to include green and yellow-to-red colors (Grotewold, 2006; Tanaka and Tanaka, 2006; Tanaka et al., 2008). Importantly, the color of flowers is determined not only by the specific pigment compounds present, but also by their interactions with cellular pH, metal ions, and other co-pigments (Tanaka et al., 2008).
The anthocyanin and flavonol glycoside pathway is highly conserved across the flowering plants (Grotewold, 2006; Tanaka et al., 2008; Davies et al., 2012), and thus provides a helpful framework for any new investigation of the molecular basis of flower color. Dramatic floral color changes can be mediated by simple genetic changes affecting the regulation, expression, or function of just a few genes, or via more complex coordinated changes over many genes within the anthocyanin and flavonol pathway (Wessinger and Rausher, 2012; Tanaka and Brugliera, 2013; Zhao and Tao, 2015). Floral color changes can also be due to changes in the downstream genes that modify anthocyanins and flavonol glycosides (Morita et al., 2015). Even further downstream, changes in transport genes (Cheng et al., 2015; Zhao et al., 2020; Lu et al., 2021), and genes associated with the pH of vacuole storage (Fukada-Tanaka et al., 2000; Faraco et al., 2014) can also impact flower color. A case for non-catalytic proteins that can influence anthocyanin production, and so alter flower colors, is also known (Morita et al., 2014).
It is also now well-established that a highly conserved protein complex consisting of MYB–basic-helix-loop-helix (bHLH)–WD40 repeat (WDR) proteins (MBW) are key regulatory determinants of floral pigmentation and patterning in numerous plants (Wong et al., 2016; Allan and Espley, 2018; LaFountain and Yuan, 2021; Rodrigues et al., 2021). However, the precise components of this complex and its transcriptional control are poorly understood in non-model plant species (Davies et al., 2012; Wessinger and Rausher, 2012). Recent work has nonetheless highlighted the complexity of the anthocyanin gene regulatory network, which integrates distinct hierarchical, feedback, and repressor activities, as well as intercellular mobility of some MBW components that execute various developmental stage, tissue-specific, and stress-responsive pigmentation programs (Albert et al., 2014). Due to these complexities, it is impossible to accurately predict the pigments present and the mechanisms involved in pigmentation of a flower based on its color alone. Therefore, chemical and molecular investigations are necessary, particularly for unusually colored flowers.
Uniformly black flowers are very rare in nature, although human interest has resulted in the development of some black-flowered ornamental cultivars of dahlias, lilies, pansies, and tulips (Davies, 2008). Due to the relative rarity of black flowers, the chemical and genetic basis of black coloration in flowers remains poorly understood (Grotewold, 2006; Davies, 2008; Tanaka et al., 2008; Davies et al., 2012; Iwashina, 2015). However, it is clear that an overaccumulation of anthocyanins is largely responsible for the black colors of flowers (Markham et al., 2004; Thomas et al., 2009; Thill et al., 2012; Kellenberger et al., 2019), leaves (Hatier and Gould, 2007; Zheng et al., 2019), grains (Abdel-Aal et al., 2006; Yang et al., 2019), and fruits (Boss et al., 1996; Fan-Chiang and Wrolstad, 2006; Liu et al., 2018).
Examples of naturally black flowers include Lisianthius nigrescens where the uniformly dark colored petals contain extraordinary high levels of delphinidin-based anthocyanins (Markham et al., 2004). In the alpine orchid Gymnadenia rhellicani, higher amounts of cyanidin-based anthocyanins were observed in the petals of the black compared to red or white floral morphs (Kellenberger et al., 2019). This high anthocyanin content is linked to differential expression of an anthocyanidin synthase gene, GrANS1 brought by loss-of-function mutation of its regulator, GrMYB1 (Kellenberger et al., 2019). Local tissue-specific upregulation is also involved in the formation of specific dark markings and spots that generate strong contrasting colors on the flower (Martins et al., 2013, 2017; Li et al., 2014; Zhang et al., 2015). For example, in several subspecies of Clarkia gracilis (e.g., the basal-spotted ssp. albicaulis and the central-spotted ssp. sonomensis), the appearance of reddish-purple petal spots containing cyanidin and peonidin pigments is largely the result of spatially restricted dihydroflavonol reductase (DFR) expression in the early stages of flower development (Martins et al., 2013, 2017).
Competition between flavonol glycoside and anthocyanin biosynthesis is predicted to be another common mechanism underpinning variation in the spatial pattern of coloration, although the details remain poorly understood (Sheehan et al., 2016; Yuan et al., 2016). Flavonol synthase (FLS) and DFR use the same dihydroflavanol substrates, but produce flavonol and anthocyanin biosynthetic products, respectively (Tanaka and Brugliera, 2013). Flavonols are uncolored or only faintly colored compared to the vibrant colors of anthocyanins, so tissue- or region-specific coloration can be mediated by the differential expression of these two genes. For example, the absence of anthocyanins in the white areas surrounding the otherwise pink corolla of bumblebee-pollinated Mimulus lewisii flowers is due to high FLS expression, while low FLS expression abolishes this spatial patterning in red hummingbird-pollinated Mimulus cardinalis flowers (Yuan et al., 2016). Another study showed that constitutive overexpression of three distinct FLS from various plant species (Rosa rugosa, Prunus persica, and Petunia hybrida) in tobacco resulted in white flowers compared to pale pink flowers in control plants (Luo et al., 2016). Conversely, constitutive overexpression of DFR genes in transgenic tobacco plants led to increased anthocyanin accumulation and a deep red flower phenotype.
Sexually deceptive plants secure pollination by luring specific male insects as pollinators using a combination of olfactory, visual, and morphological mimicry (Bohman et al., 2016). Within the orchids, pollination by sexual deception has evolved independently on four continents and is employed by hundreds of species spanning multiple lineages of the Orchidaceae (Peakall et al., 2020). The types of pollinators involved are also diverse, and include ants, bees and wasps (Hymenoptera), fungus gnats and other flies (Diptera), and beetles (Coleoptera) (Gaskett, 2011; Bohman et al., 2016; Peakall et al., 2020; Cohen et al., 2021; Hayashi et al., 2021). In these cases, pollination is achieved during attempted copulation with the labellum, an often highly modified petal of the orchids. Furthermore, in the multiple cases now chemically characterized, sex pheromone mimicry, often involving unusual compounds or unusual blends, is critical for the long range attraction of the pollinator (Bohman et al., 2016; Wong et al., 2017b; Peakall et al., 2020). Rather than being brightly colored, sexually deceptive orchid flowers are most often dull red and green in color, with the labellum often exhibiting starkly contrasting dark structures and markings which are thought to be visual cues that improve the insect mimicry (Figure 1, see also photos in Bohman et al., 2016; Johnson and Schiestl, 2016; Peakall et al., 2020). Outside of the Orchidaceae, there are two other known cases of pollination by sexual deception: Gorteria diffusa (Figure 1G) in the Asteraceae (Ellis and Johnson, 2010) and Iris paradoxa (Figure 1H) in the Iridaceae (Vereecken et al., 2012) both characterized by flowers that include some very dark pigmentation.

Figure 1. An illustration of the diversity of sexually deceptive orchid and non-orchid species. Orchids: (A) The floral structure of Chiloglottis trapeziformis with (B) its male pollinator Neozeleboria cryptoides. The arrows point to the very dark, three dimensional “callus” structure on the dull green and red labellum lamina of Chiloglottis trapeziformis, (C) Drakaea glyptodon, (D) Disa atricapilla, (E) Cryptostylis leptochila, and (F) Ophrys speculum. Non-orchids: (G) Gorteria diffusa, and (H) Iris paradoxa. All images have been reproduced with permission from the respective copyright holders. Please refer to the Section “Acknowledgments” for image credits.
Beyond the potential role of female visual mimicry, where a close match to the color of the female of the pollinator is expected (de Jager and Peakall, 2016), the tissue-specific pigmentation patterns of sexually deceptive flowers may also provide strong color contrasts that improve detectability. For example, Drakaea livida shares the same male thynnine wasp pollinator as the unrelated, morphologically distinct, and differently colored Caladenia decora, C. pectinata, and C. procera. Yet, across this set of species, tissue-specific pigmentation provides strong achromatic contrast against a chromatic background, thus, potentially enhancing the detectability by the pollinator (Gaskett et al., 2017).
The very dark, three dimensional “calli” on the dull green and red labellum lamina of sexually deceptive Chiloglottis orchids are crucial to their sexual mimicry (Figure 1). In the most well-studied case of C. trapeziformis, the central black callus structure is the source of the unique volatile compound chiloglottone 1, which is the sex pheromone of their specific thynnine wasp pollinator, Neozeleboria cryptoides (Schiestl et al., 2003; Franke et al., 2009). The structure also serves as a visual and tactile mimic of the female, providing a gripping point for male wasps as they attempt to copulate with the tip of the labellum (de Jager and Peakall, 2016). Furthermore, the distance from callus to labellum tip is a close match to the length of the female wasp, and experimentally shortened or elongated labella result in significantly reduced durations of attempted copulation, indicating the importance of morphological mimicry (de Jager and Peakall, 2016, 2019). Finally, spectral reflectance measurements revealed strong similarity between the perceived color of the black callus and the female wasp abdomen in hymenopteran visual models. Both female and orchid structures are predicted to be perceived as uncolored or achromatic to male wasps, consistent with the hypothesis that the dark callus structure acts as a visual mimic of the female (de Jager and Peakall, 2016). Conversely, the surrounding lamina of the labellum is chromatic, indicating morphological adaptations to produce within-flower chromatic/achromatic contrast (Figure 1).
As the first step toward understanding the chemical and molecular basis of floral color adaptations in Chiloglottis trapeziformis, we leveraged targeted metabolite and transcriptome analysis of a flower tissue and developmental series to ask the following questions: (1) What anthocyanins and flavonol glycoside co-pigments are present in the flower? (2) What are the spatiotemporal patterns of anthocyanin and flavonol glycoside levels across flower development and tissue types? (3) Is the unique spatiotemporal distribution of color in the flower explained by differential expression of a few anthocyanin and flavonol glycoside pathway-related genes at specific developmental stages, or coordinated regulation at many genes and multiple stages? We propose possible mechanisms for the tissue-specific pigmentation and its regulation and discuss the evolutionary implications of these findings more broadly for Chiloglottis and other sexually deceptive plants.
Materials and Methods
Sample Collection
This study builds on a larger effort to understand the biochemistry, biosynthesis and regulation of chiloglottones, the unique semiochemicals used by sexually deceptive Chiloglottis orchids (Falara et al., 2013; Amarasinghe et al., 2015; Wong et al., 2017a,2018, 2019). Here, we turn attention to the chemistry and biosynthesis of flower color in Chiloglottis trapeziformis. Flowers at five developmental stages (very young buds, vyb; young bud, yb; mature buds, mb; very mature bud, vmb; and mature flowers open to the sun, or sun flowers, sflw) were sampled from the Australian National Botanic Gardens (Canberra, ACT, Australia) in September 2014. All tissues were immediately snap-frozen in liquid nitrogen at the point of collection and stored at –80°C until further use. Dissection of floral tissues into callus and labellum remains were performed in liquid N2.
The classifications of bud and flower stages follows Amarasinghe et al. (2015). Briefly, the vyb is characterized by a small and very tightly closed green bud. However, it is noteworthy that dissection of the buds reveals that the callus is already very dark in color at the vyb stage. The vmb stage is characterized by larger green buds that are about to open, with sepals and petals beginning to separate. By this stage the callus is black, and remains so for the duration of flowering. At the onset of flower opening, the lamina is green. However, within 2 to 3 days of exposure to sunlight, the lamina becomes reddish-green in color. It was at this stage that the sflw was sampled (Figure 2 inset).
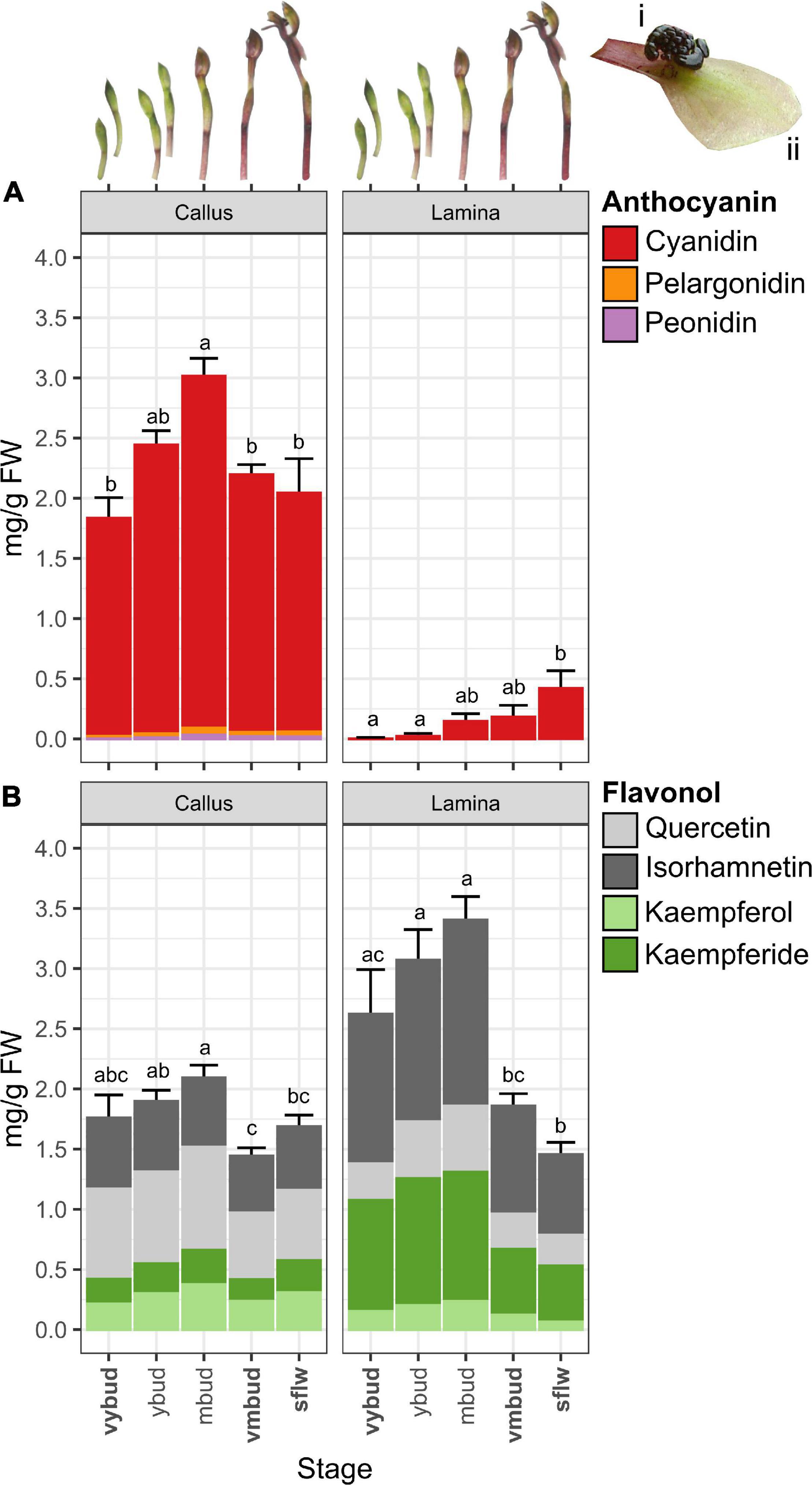
Figure 2. Changes in anthocyanins and flavonol glycoside co-pigments of Chiloglottis trapeziformis flowers. Bar graphs depict the total (A) anthocyanin and (B) flavonol glycoside content (average ± s.e.) in the callus and labellum lamina (see insets i and ii) of Chiloglottis trapeziformis flowers at different developmental stages (i.e., very young bud, vyb; young bud, yb; mature buds, mb; very mature bud, vmb; and naturally opened flowers in the field, sflw). Bold indicates developmental stages with corresponding transcriptome profiled. Labels not connected by the same letter are significantly different at P < 0.05 based on ANOVA and Tukey HSD test (see Supplementary Figure 7 for further information).
Ultra High-Performance Liquid Chromatography Mass Spectrophotometric Analysis
For the targeted metabolite analysis, a total of 54 separate labellum samples were used, encompassing replication for all of the five developmental stages (i.e., vyb, yb, mb, vmb, sflw). Each sample consisted of either the dissected callus or lamina (labellum remains after the stalked callus was removed) from several individual flowers. Pre-weighed samples were homogenized and extracted in 400 μL of solvent composed of 70:30:1 methanol:water:acetic acid with agitation at 4°C for 24 h. Samples were centrifuged at 13,000 rpm for 10 min and the supernatant was filtered with 0.2 μm PTFE SINGLE StEP filter vials (Thomson) and analyzed using a Thermo Q Extractive Plus UPLC-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, Waltham, MA, United States). Samples and standards (5 μL injection volumes) were separated chromatographically on an Agilent reversed-phase Zorbax Eclipse XDB-C18 column (2.1 × 50 mm, 1.8 μm particles) held at 40°C. The mobile phases used were water with 0.1% HPLC grade formic acid (solvent A), and methanol with 0.1% formic acid (solvent B). Samples and standards were eluted with a constant flow rate of 400 μL min–1, with a 25-min gradient program as follows: 0–1.5 min, 6% B; 1.5–2 min, 6–10% B; 2–14 min, 10–60% B; 14–15 min, 60–90% B; 15–19 min, 90% B; 19–20 min, 6% B; 20–25 min, 6% B. Eluted compounds were introduced to the MS via a HESI-II probe (Thermo Fisher Scientific, Waltham, MA, United States). Representative samples were also analyzed on a separate LC-MS instrument fitted with a diode array detector (DAD) operating at 520 nm and at 365 nm and using the same chromatographic parameters, to distinguish anthocyanins from flavonol glycosides.
For putative identification and relative quantification of anthocyanins and flavonols, the HESI was operated in the positive mode. Mass spectra were acquired using full MS and data-dependent MS/MS acquisition (DDA) modes at a scan range of 100 to 1500 m/z. Additionally, representative samples of both tissue types at various developmental stages were analyzed in negative ionization mode. In negative ionization mode, anthocyanins produce a distinctive [M-2H + H2O]− ion in addition to the [M-2H]− ion, while isomeric flavonol glycosides do not (Sun et al., 2012a). Thus, the presence of this ion is diagnostic of anthocyanins and can be used to distinguish them from isomeric flavonols with similar fragmentation patterns.
Data were acquired using Thermo Scientific XCALIBUR 4.0 and analyzed using Thermo Scientific FreeStyle software. Anthocyanins and flavonols were distinguished based on (i) accurate masses (4 decimal places) of molecular ions in full MS spectra in both ionization modes, (ii) comparisons of fragmentation patterns in the MS/MS spectra in both ionization modes with those available in online databases such as RIKEN tandem mass spectral database (Sawada et al., 2012) and MassBank of North America1, (iii) the presence or absence of the diagnostic [M-2H + H2O]− ion in the negative ionization analyses, and (iv) absorbance spectra at 520 and 365 nm. Putative anthocyanins and flavonols were quantified by integration of the molecular ion peak in full MS spectra and calculated from linear calibration curves of cyanidin 3-O-glucoside chloride (Sigma-Aldrich) and 7-hydroxycoumarin (Sigma-Aldrich), respectively.
An analysis of variance (ANOVA) and post hoc Tukey–Kramer Honestly Significant Difference test were performed to evaluate the differences in total anthocyanin and flavonol glycoside content in the callus and labellum during flower development, following the confirmation of normality by a Shapiro-Wilk test. Statistical analyses were conducted using R2.
De novo Transcriptome Assembly
RNA extraction, library construction, and RNA sequencing were performed as previously described (Wong et al., 2017a,2018). However, the assembly and downstream analysis of the sflw stage has not been previously reported. Therefore, paired-end reads from the callus and lamina tissues of vyb, vmb (Wong et al., 2017a,2018) and sflw (used for the first time in this study) stages were first pooled. Next, removal of the adaptor, sliding-window trimming, length (l = 40) and quality filtering, and base correction (-c) of the raw PE reads were performed with fastp v0.20.0 (Chen et al., 2018) using default settings unless otherwise specified. De novo transcriptome construction based on pooled tissue and developmental stages was performed using Trinity v2.11.0 (Haas et al., 2013) with default settings except the minimum contig length and k-mer size were set to 300 and 31, respectively. Protein-coding prediction was achieved using TransDecoder v5.5.03. To maximize the prediction sensitivity, homology searches against the UniProt Reference Clusters (UniRef90) and protein families (Pfam) databases were performed using DIAMOND (Buchfink et al., 2014) and HMMER (Mistry et al., 2013), and incorporated into the TransDecoder pipeline. Resolution of assembly redundancy, protein-coding region prediction, and identification of accurate gene sets was made using the EvidentialGene tr2aacds4 pipeline (Gilbert, 2013) using default settings.
Transcriptome Analysis of Anthocyanin and Flavonol Glycoside Pathway-Related Genes
Alignment of filtered PE reads toward the assembled transcriptome of C. trapeziformis were individually performed with bowtie2 (Langmead and Salzberg, 2012) using the local read alignment mode (–local). Read count matrices were obtained using FeatureCounts (Liao et al., 2014) with default parameters except for the -B (both ends must be aligned) and -C (exclude chimeric fragments) option enabled for each species. Transcripts having sufficiently large counts were retained for further downstream statistical analysis using edgeR with the filterByExpr option (Robinson et al., 2009). Differential expression (DE) analysis between groups/treatments of interests were performed using DESeq2 (Love et al., 2014). An absolute log2 fold change (| log2FC|) > 0.5 and a false discovery rate (FDR) threshold < 0.05 determines transcripts that are differentially expressed in each contrast. Transcript expression was expressed as Fragments Per Kilobase of transcript per Million mapped reads (FPKM). The assembled transcriptome was annotated with MapMan categories using Mercator (Lohse et al., 2014). Additional Pfam domains, putative homolog annotations, and transcripts sharing homology and functional domains to known and/or emerging flavonoid pathway components (Grotewold, 2006; Wessinger and Rausher, 2012; Tohge et al., 2017), were obtained via the TransDecoder’s prediction pipeline. Hierarchical clustering of expression and DE patterns of the callus and lamina across multiple developmental stages were performed using hclust and plotted using the ggplot2 package in R (see text footnote 2). A summary of developmental stage- and tissue-specific transcriptome analysis of Chiloglottis trapeziformis flowers is described in Supplementary Figure 1 and corresponding gene expression dataset and differential expression result tables are available in Supplementary Datas 1–3. New sequence reads obtained for the sflw stage has been added to the existing BioProject accession PRJNA390683 and SRA study accession SRP1093281.
Results
The Callus and Labellum Contain a Diverse Array of Anthocyanin and Flavonol Glycosides
A total of five anthocyanins and 14 flavonol glycosides were detected in Chiloglottis trapeziformis labella by UHPLC-MS/MS and LC-DAD-MS analysis (Table 1 and Supplementary Figure 2). Cyanidin-based anthocyanins were the dominant pigment, with peonidin- and pelargonidin-based anthocyanins also detected. Kaempferol (K), Quercetin (Q), and their methylated derivatives (Kaempferide, Kde; Isorhamnetin, IR) were the dominant flavonol glycosides. No delphinidin-based anthocyanins, myricetin flavonol glycosides nor common acyl modifications aside from malonyl groups (e.g., acetylation, coumarylation, caffeoylation) were detected (Table 1).
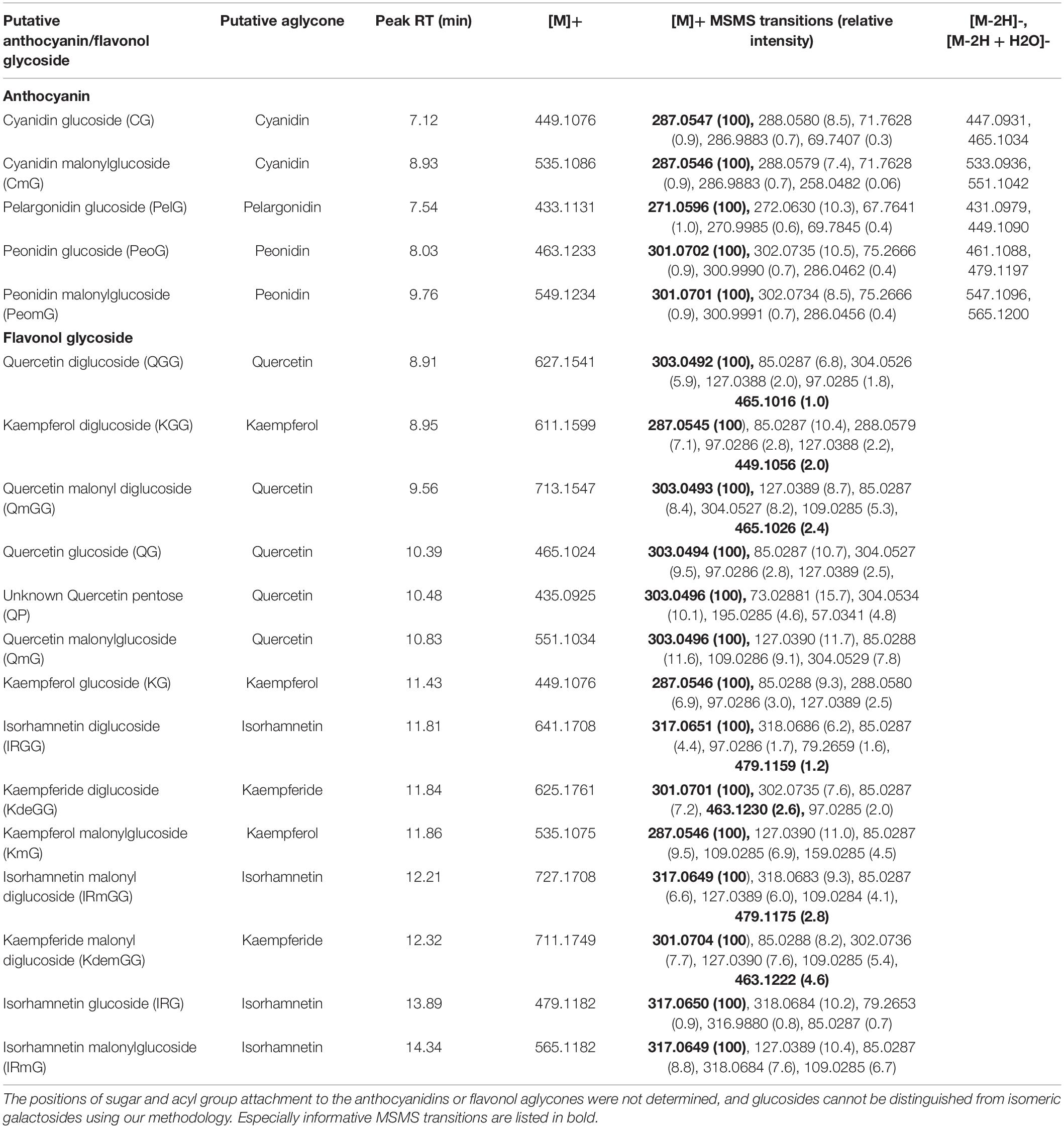
Table 1. Putative anthocyanins and flavonols in floral extracts of Chiloglottis trapeziformis calli and labellum lamina tissues.
Candidate Anthocyanin and Flavonol Glycoside Pathway-Related Genes
Our transcriptome analysis revealed a set of candidate gene homologous to well-known anthocyanin and flavonol glycoside pathway genes (Grotewold, 2006; Tanaka et al., 2008; Davies et al., 2012). This included three chalcone synthase (CtrCHS1–3), one chalcone isomerase (CtrCHI), two flavone 3-hydroxylase (CtrF3H1–2), and four flavonoid 3’-hydroxylase (CtrF3’H1–4) encoding enzymes involved in the early biosynthetic steps of general flavonoid biosynthesis. Notably, F3’H catalyzes hydroxylation at the 3’-position of naringenin and dihydrokaempferol to yield eriodictyol and dihydroquercetin, and also converts kaempferol to quercetin (Tohge et al., 2017). Three dedicated anthocyanidin biosynthesis genes (one CtrDFR1 and two leucoanthocyanidin dioxygenase syn. anthocyanidin synthase) (CtrLDOX1–2) and two flavonol glycoside biosynthesis genes (e.g., flavonol synthase, CtrFLS1–2) were also identified. The discovery of the candidate genes downstream of F3’H is in accord with the anthocyanins (two cyanidin and two peonidin anthocyanins) and flavonol glycosides (five Q and four IR flavonol glycosides) we detected. One pelargonidin anthocyanin, three K and three Kde flavonol glycosides which do not depend on F3’H activity were also found (Figure 2 and Table 1). Many of these genes also fall within the specific BIN9.2.2 category of secondary metabolism.phenolics.flavonoid biosynthesis (e.g., CtrCHS in BIN9.2.2.1, CtrCHI in BIN9.2.2.2, CtrF3H in BIN9.2.2.4, and CtrDFR and CtrLDOX in BIN9.2.2.9) or were reciprocal best BLAST hits of several plant CYP75B (i.e., CtrF3’H) enzymes (Supplementary Data 1).
Unlike the conserved early steps in the pathway, the downstream modifications of flavonoids with various glycosyl, methyl, and/or acyl groups are achieved by highly diverse and often family, genera-, and/or even species-specific sets of genes (Tanaka et al., 2008). Nonetheless, typically one or more flavonoid glycosyltransferases, methyltransferases, or acyltransferases catalyze these reactions. Three glycosyltransferase homologs potentially relevant to flavonoid metabolism were identified: two transcripts encoding Arabidopsis UDP-glucose:flavonoid 3-O-glucosyltransferase homolog (CtrUGT78D2a/b) potentially involved in the glycosylation of anthocyanidins and flavonols at the 3-position (Lim et al., 2004; Tohge et al., 2005), and one encoding an Arabidopsis UGT71B1 (Lim et al., 2004) homolog with specific flavonol 3-O-glucosyltransferase activity (CtrUGT71B1).
O-methyltransferases with both broad and narrow flavonoid substrate specificities have been identified in many plants. For example, the grape anthocyanin O-methyltransferase VviAOMT that catalyzes the methylation of anthocyanins is also active on flavonol glycosides compounds (Hugueney et al., 2009; Lücker et al., 2010; Provenzano et al., 2014). Here, four transcripts sharing homology toward VviAOMT (CtrCOMT1–4) were identified. Many plant flavonoid malonyltransferases characterized to date can catalyze the malonylation of anthocyanins but also flavonol glycosides (Bontpart et al., 2015) and one such enzyme is dahlia anthocyanin 3-glucoside malonyltransferase, Dv3MaT (Suzuki et al., 2002). Four C. trapeziformis homologs of Dv3MaT were identified (Ctr3MAT1–4).
Subcellular flavonoid transport homologs such as Arabidopsis glutathione-S-transferase, Transparent Testa 19 (TT19) involved in the transport of cyanidin anthocyanins from the cytosol to the tonoplast (Sun et al., 2012b), and various plant ABC and MATE transporters that also facilitate the vacuolar transport of anthocyanins and flavonoids such as grape ABCC1, A. thaliana ABCC2, and grape anthoMATEs (Gomez et al., 2009; Francisco et al., 2013; Behrens et al., 2019) were also identified (e.g., CtrTT19a/b, CtrABCC1, CtrAM1, and CtrABCC2a/b). These include transcripts encoding homologs of petunia H+ P-ATPase PH5 and Na+/H+ and K+/H+ antiporter NHX1 (Faraco et al., 2014) and Japanese morning glory NHX1 (Fukada-Tanaka et al., 2000) involved in the regulation of vacuolar pH were also identified (CtrPH5 and CtrNHX1). Finally, a homolog to the gene encoding CHIL that binds to CHS to serve as a rectifier, thus, facilitating the flux of phenylpropanoid pathway precursors to the flavonoid pathway (Waki et al., 2020), was also found (CtrCHIL).
Developmental- and Tissue-Specific Expression of Anthocyanin and Flavonol Glycoside Pathway-Related Genes
In this study, we considered four pairwise developmental stage comparisons between the vyb, vmb (devC1/L1) and sflw (devC2/L2) stages across the callus and lamina: Three tissue-specific comparisons between callus and lamina across the respective vyb (ts1), vmb (ts2), and sflw (ts3) developmental stages corresponding with the samples profiled for anthocyanin and flavonol glycosides. First, we clustered candidate anthocyanin and flavonol glycoside pathway-related genes based on their expression abundance (Supplementary Figure 3). Four clusters (cluster A–D) of shared expression intensities were identified. Most notably, CtrCHS1, CtrF3H1/2, and CtrLDOX1 were highly expressed (FPKM > 100) especially in the callus and/or lamina of vyb (Cluster D) while lowly expressed (FPKM < 1) transcripts such as CtrF3pH2/3, CtrMAT4, and CtrCHS2/3 were observed in Cluster C. Furthermore, hierarchical clustering based on the patterns of differential expression revealed clusters containing genes that were differentially expressed between callus and lamina in both specific and multiple developmental stages (Supplementary Figure 4). For example, several groups of genes were coordinately downregulated to varying degrees (i.e., cluster 3–5) during the transition from vyb to vmb and sflw stage regardless of tissue type. In cluster 5, there were many genes encoding enzymes involved in the formation of early anthocyanin and flavonol precursors such as chalcones, flavanones, and dihydroflavonols. These include two CHS (CtrCHS1/3), one CHI (CtrCHI1), two F3H (CtrF3H1/2), and two F3’H (CtrF3pH1/4) genes. Conversely, cluster 3 contained mostly downstream flavonoid modification genes and displayed stronger and more consistent developmental stage downregulation in both tissues compared to other clusters. Across several clusters in both tissue types over flower development, coordinated regulation was not limited to the upstream biosynthetic pathway but also included downstream modification and/or transport genes. For example, CtrAM1/2, CtrPH5, and two flavonoid-related glycosyltransferase homologs CtrUGT71B1 and CtrUGT78D2b were present in cluster 5, CtrTT19b in cluster 2, and CtrCOMT1/4 in cluster 4. However, cluster 1 and 3 mostly contained flavonoid modification or transport genes, while the majority of transcripts in cluster 1 encoded downstream transport genes and displayed developmental stage upregulation in both the callus and lamina.
The Chemical Basis and Patterns of Gene Expression in the Black Callus
The total anthocyanin content was substantially higher in callus tissue compared to the lamina across all developmental stages (Figure 2A). For example, at the vyb stage, total anthocyanin quantities were up to 1300 times higher in the callus than in the lamina. Even after the late peak of anthocyanin production in the sflw stage, the callus anthocyanin content remained 5 times greater than in the lamina. Across developmental stages, total anthocyanin content of the callus averaged 1.8 mg g–1 FW in vyb stage, before peaking in mb (3.0 mg g–1 FW) and then decreasing in vmb and sflw to comparable levels observed in vyb (2.1–2.2 mg g–1 FW). The majority of anthocyanins were cyanidin-based (96–97%) regardless of developmental stage, with relatively low amounts (0.02–0.06 mg g–1 FW) of peonidin- and pelargonidin-based anthocyanins (Supplementary Figure 5). Across the different developmental stages, the pattern of accumulation for individual anthocyanins were largely consistent with total anthocyanin content (Supplementary Figure 5).
Four main types of flavonol glycosides were found in the callus, with the proportions of quercetin (Q)-, isorhamnetin- (IR), kaempferol- (K), and kaempferide- (Kde) based flavonol glycosides ranging between 36–44%, 26–32%, 12–18%, and 12–16%, respectively (Supplementary Figure 6). Compared to the anthocyanins, less variation in total flavonol glycoside content was found across the different bud (1.4–2.1 mg g–1 FW) and flower (1.7 mg g–1 FW) stages (Figure 2B). Major flavonol glycosides such as QG and IRG were often most abundant in early developmental stages (i.e., pre mb before decreasing slightly in vmb and sflw) while others such as KG, K(mal)G, and KdeGG showed subtle increments from vyb to mb before decreasing in sflw (Supplementary Figure 6). However, the ratios of total anthocyanins to flavonol glycosides in the callus were broadly similar across developmental stages (approximately 1.5 to 1).
One key pattern of gene expression in the callus, especially at the vyb stage, was that genes related to anthocyanin and/or flavonol glycoside biosynthesis (e.g., CtrF3H1/2, CtrDFR1, CtrLDOX1/2), transport (e.g., CtrAM1/2, CtrTT19b), and vacuolar acidification (CtrPH5)-related pathways showed significantly higher gene expression when compared to the lamina. Most notably, CtrDFR1 and CtrLDOX1/2 transcripts peaked in the callus of vyb and were 10–35-fold higher compared to the lamina (Figure 3 and Supplementary Figure 4). Despite evidence for coordinated downregulation, and hence lower gene expression levels for many of these genes from vmb onward, some transcripts such as CtrCHS1, CtrFLS1/2, CtrMAT1, CtrCOMT2/3, CtrNHX1, CtrAM1/2, and CtrABCC2a/b continued to exhibit higher gene expression levels in the callus compared to the lamina in one or more stages post vyb.
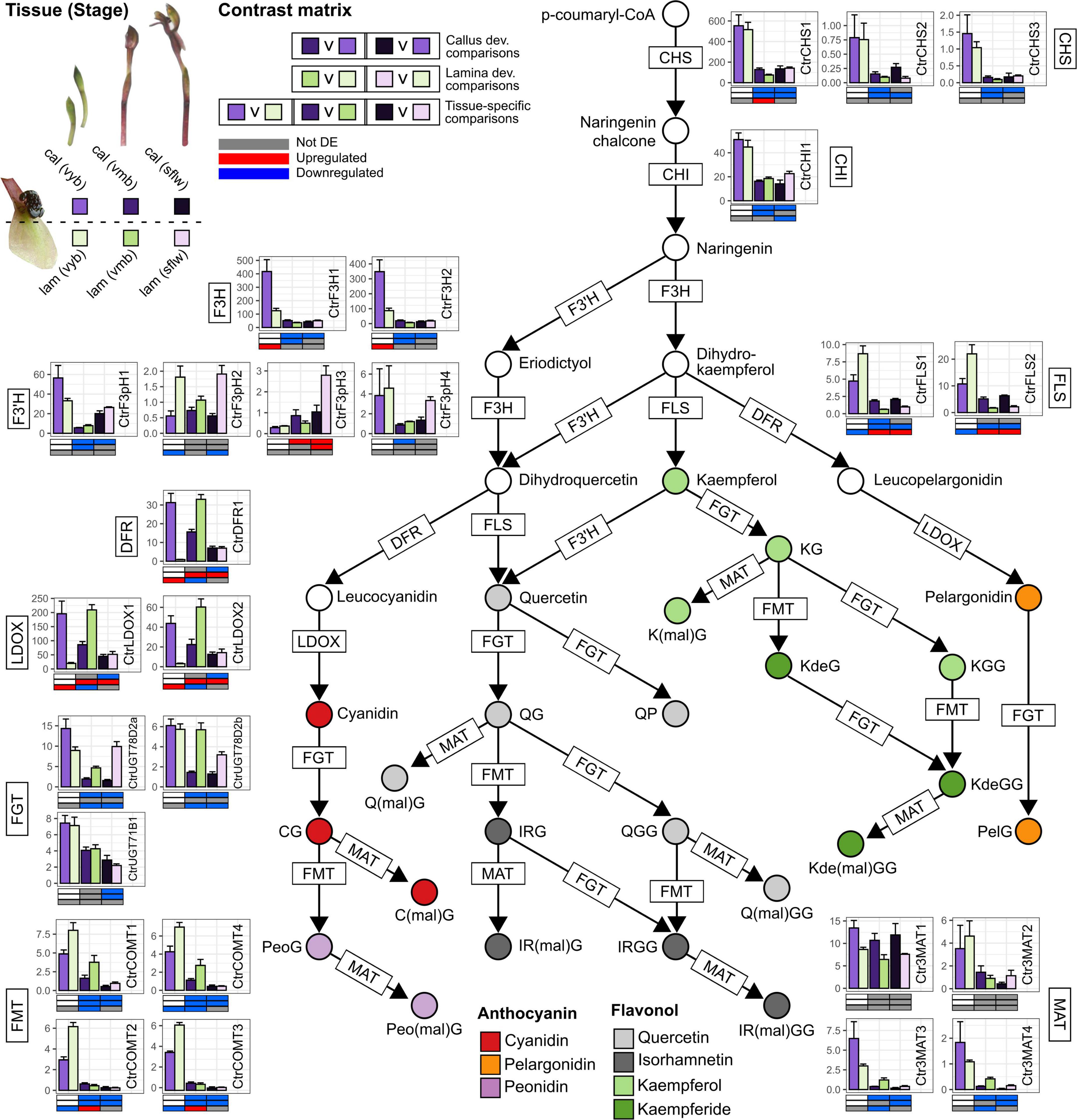
Figure 3. Anthocyanin and flavonol metabolism in the callus and labellum tissues of Chiloglottis trapeziformis during flower development. Bars depict the average (±s.e.) normalized expression values (FPKM) of anthocyanin and flavonol biosynthesis and modification pathway genes in the calli and labellum lamina tissues of very young buds (vyb), very mature buds (vmb), and mature sunflowers (sflw). Colored circles indicate putative anthocyanins and flavonols identified in Chiloglottis trapeziformis callus (cal) and labellum lamina (lam) tissues (Table 1). Various shades of purple and green depict the relevant color callus and labellum lamina tissues at various flower developmental stages. The contrast matrix indicates the differential expression outcomes based on comparisons in Supplementary Figure 1. Red, blue, and gray indicate significant upregulation (FDR < 0.05, log2FC > 0.5), downregulation (FDR < 0.05, log2FC < –0.5), and no significant differential expression in selected comparisons, respectively. CHS, Chalcone synthase; CHI, Chalcone isomerase; F3H, Flavanone 3-hydroxylase; F3′H, Flavonoid 3′-hydroxylase; F3′5′H, Flavonoid 3′5′-hydroxylase; DFR, Dihydroflavonol 4-reductase; LDOX, Leucoanthocyanidin dioxygenase; FLS, Flavonol synthase; FGT, Flavonoid/anthocyanin glucosyltransferase; FMT, Flavonoid/anthocyanin O-methyltransferase; MAT, Flavonoid/anthocyanin malonyltransferase.
The Chemical Basis and Patterns of Gene Expression in the Green Lamina
While the total anthocyanin content was much lower in the lamina than the callus, trace levels were nonetheless detected from the vyb stage (0.014 mg g–1 FW) with a steady increase to appreciable amounts at the sflw (0.42 mg g–1 FW) stage. These patterns match the observed transition from green buds to the reddish green mature flower. Like the callus, cyanidin-based anthocyanins were the major anthocyanin constituent of the lamina regardless of developmental stages (99–100%), with very low amounts of peonidin- and pelargonidin-based anthocyanins. Accordingly, CG and C(mal)G content increased steadily from vyb to sflw in the labellum. Interestingly, total flavonol glycoside content in the lamina closely mirrored the profile of total anthocyanins in the callus (Figure 1B). The average total flavonol glycoside content increased from 2.6 to 3.4 mg g–1 FW in the vyb and mb, before decreasing sharply (1.5 mg g–1 FW) in sflw. This decreasing pattern is concomitant with the sharp rise in total anthocyanins from vyb to sflw.
When compared with the callus, there was substantial variation in the ratio of total flavonol glycosides to anthocyanins across developmental stages of the lamina with the top two-most extremes observed in vyb (1800:1) and sflw (3.5:1). Notably, the total (and most individual) flavonol glycoside content in the lamina of sflw was the lowest of all developmental stages, directly contrasting with the total anthocyanin content which was greatest in this stage (Figure 2). Regardless of developmental stage, isorhamnetin (43–46%) and kaempferide (29–35%) flavonol glycosides were more prevalent in the lamina, while quercetin- (12–19%) and kaempferol-based flavonol glycosides (5–7%) were less prevalent relative to the callus (Supplementary Figure 6). Unlike the callus, in the lamina most major flavonol glycosides either displayed a marked increase in content (e.g., QG, IRGG) or remained constant [e.g., IRG, KdeGG, Kde(mal)GG] during the transition from vyb to mb, followed by a large decrease in the mature stages (Supplementary Figure 6).
Coordinated regulation of CtrFLS1/2 and CtrCOMT2/3 transcripts (cluster 4) was a striking feature of the lamina. Four transcripts were significantly upregulated in the lamina compared to callus of vyb, however, at later stages of vmb and sflw, higher callus expression was commonly observed. Coordinated downregulation during the transition from vyb to vmb and sflw stages was also common for many anthocyanin and flavonol glycoside pathway genes in the lamina (i.e., cluster 3–5). Another striking feature of the lamina was a strong (but lagging the callus) developmental upregulation of genes encoding CtrDFR1, CtrLDOX1/2, CtrAM1/2, and CtrTT19b (cluster 2). For example, CtrDFR1 was consistently upregulated by 12- to 55-fold in vmb and sflw compared to vyb. Similarly, CtrLDOX1/2 were upregulated by four- to 30-fold in vmb and flowers compared to vyb. Interestingly, these transcripts were only upregulated in the lamina compared to the callus at the vmb stage. Other notable transcripts include CtrUGT78D2a/b, CtrMAT3/4, CtrCOMT2/3, and CtrPH5, which were more highly expressed in the labellum than the callus in one or more developmental stages post vyb (Figure 3 and Supplementary Figure 4).
Discussion
Overview
The goal of this study was to combine targeted metabolite profiling and transcriptomic analysis to investigate the chemical composition and patterns of tissue- and developmental-specific color pathway gene expression in the labellum of Chiloglottis trapeziformis. In this sexually deceptive orchid (as in most others), the labellum is the center of the orchid-pollinator interaction, functioning as the olfactory, visual, and tactile mimic of the female of the pollinating species (de Jager and Peakall, 2016, 2019). Furthermore, the color contrast between the dark black 3D callus structure mimicking the female (Figure 1A) and the remainder of the reddish green labellum lamina is predicted to aid detectability to the male pollinator. In the discussion that follows, we explore in more detail hypotheses for the molecular basis of the contrasting labellum colors, and consider evolutionary implications and future research directions.
The Diversity of Anthocyanin and Flavonol Co-pigments in Chiloglottis trapeziformis
Cyanidin-based anthocyanins are widespread floral pigments across the flowering plants, and are also known from many orchid species (Iwashina, 2015). However, as one of the largest plant families, it is not surprising that anthocyanins have only been investigated in a fraction of the known genera and species of orchids. Pigment chemistry and biosynthesis has been most well studied in orchid genera of horticultural interest, such Phalaenopsis (Hsu et al., 2015; Liang et al., 2020), Vanda (Khunmuang et al., 2019), Cattleya (Li et al., 2020), and Cymbidium (Wang et al., 2014) but there have also been several studies of wild species which span a diversity of orchids with both rewarding and deceptive pollination strategies (George et al., 1973; Strack et al., 1989; Fossen and Øvstedal, 2003; Vignolini et al., 2012; Kellenberger et al., 2019; Zhang et al., 2020). Notably, cyanidin-based anthocyanins are major anthocyanin pigments of flowers in the European sexually deceptive genus Ophrys (Strack et al., 1989; Vignolini et al., 2012). Unlike anthocyanins, the constituent flavonol glycosides (or any other flavonoid classes) remain poorly understood across the Orchidaceae.
Despite the exceptional diversity (>1850 species) of the Australian orchid flora (Jones, 2021; Peakall et al., 2021), to the best of our knowledge only one previous study of floral anthocyanins has been published. This study, dating back to the 1970s, reported a qualitative survey of floral anthocyanins across some brightly colored and non-sexually deceptive species. The finding indicated the presence of cyanidin-, delphinidin-, petunidin- and malvidin-based anthocyanins (George et al., 1973). The present study is thus the first to investigate in detail the chemical and molecular basis of flower color in an Australian orchid. We have confirmed that cyanidin-based anthocyanins are the major pigment constituents of the labellum of Chiloglottis trapeziformis flowers. The labellum lamina also accumulates other mono- and di-hydroxylated anthocyanins and flavonol glycosides, but no trihydroxylated forms were detected. Additionally, among the many possible acyl modifications of the anthocyanin and flavonol glycosides, only the malonylated form was detected (Table 1). The composition of flavonol glycosides was more complex than the anthocyanin profile in both tissue types. In the callus, isorhamnetin- and quercetin-based compounds were equally dominant while the lamina accumulated more isorhamnetin- and kaempferide-based flavonol glycosides (Figure 2B and Supplementary Figures 5, 6).
Patterns of Anthocyanin and Flavonol Glycoside Levels in Chiloglottis trapeziformis
Dissection of the tightly closed green bud at the vyb stage shows that the callus is already darkly colored. By the mb stage the callus appears “black” in color, and it remains so for the lifetime of the flower (Amarasinghe et al., 2015). On the other hand, the color of the lamina changes from uniform green at vyb to greenish red at vmb, but only reaches its maximum reddish green color in flowers that have been open in sunlight for several days (Amarasinghe et al., 2015). Consistent with these observations, peak anthocyanin accumulation was observed in the vmb stage of the callus, compared with the final sflw stage of the lamina (Figure 2A). Furthermore, the lower levels of lamina anthocyanins are spread thinly across the larger surface area, compared with the smaller callus (Figure 1A). Reflectance measurements, projected into hymenopteran vision space, indicate that the dark achromatic callus should be readily distinguished by the pollinator from the contrasting lamina, even at full flower maturity (de Jager and Peakall, 2016).
By contrast with the anthocyanins, total flavonol glycoside levels were broadly similar across callus and lamina tissues and seemingly inversely correlated with the total anthocyanin levels. Nonetheless, some different patterns of composition and changes with development stage were evident (Figure 2B and Supplementary Figure 6). Critically, when compared to the lamina, lower levels of the methylated forms of Quercetin (Isorhamnetin) and Kaempferol (Kaempferide) were found in the callus (Figure 2B).
Patterns of Gene Expression in the Flower Color Pathway Genes of Chiloglottis trapeziformis
Figure 3 shows an overlay of the patterns of gene expression in the C. trapeziformis labellum partitioned by tissue type and development stage onto the general biosynthetic pathway for anthocyanin and flavonoid production in plants. The gene expression patterns in combination with the outcomes of the hierarchical clustering analysis (Supplementary Figures 3, 4) and knowledge of the anthocyanin and flavonol glycoside levels (Figure 2), provide insights into the probable genetic basis of the contrasting labellum colors of C. trapeziformis.
The two early pathway genes, CtrCHS1-3 and CtrF3H1-2, were equally highly expressed in both callus and lamina tissues of the vyb stage, indicating an efficient supply of precursors is available for both anthocyanin and flavonol glycosides biosynthesis in either tissue. Whereas, the dedicated anthocyanin pathway genes associated with the production of anthocyanins (cyanidin, peonidin, and pelargonidin), CtrDFR1, CtrLDOX1-2 were highly expressed in the vyb stage of the callus, but not the lamina. Conversely, the CtrFLS1-2 associated with the production of the flavanol glycosides, were significantly downregulated in the callus tissue. The methyltransferase, FMT (CtrCOMT1-4) genes mirrored the FLS gene expression patterns, consistent with the observed increasing accumulation of the methylated flavonol glycosides in the lamina from vyb to mb stages (Figure 3 and Supplementary Figure 6).
By the vmb stage, expression levels of the precursor supply genes, CHS and CHI, were significantly downregulated relative to the vyb stage in both tissue types, and remained low across the remaining developmental stages. In the callus, the intermediate anthocyanin pathway genes (F3H, F’3H, DFR and LDOX), generally showed a similar pattern. However, across the remaining developmental stages, there was a strong lag in the peak expression of CtrDFR1 and CtrLDOX1-2 between the lamina and callus. This suggests that the combination of upregulation of the anthocyanin genes in the lamina, coupled with a diminished precursor supply is likely linked to the increasing but much lower levels of anthocyanin production in the lamina with development stage.
By the sflw stage, the formerly green color of the vmb stage, and the freshly opened flower was reddish-green in color. Consistent with this observation, anthocyanin levels in the lamina, while much lower than in the callus, peaked at the sflw stage (Figure 2A). Subtle gene expression shifts appear to be consistent with these flower color changes. For example, expression of CHS (CtrCHS1) and CHI (CtrCHI1) increased in both callus and lamina relative to the vmb stage. More importantly, callus expression was significantly upregulated relative to the lamina. Thus, it seems likely that precursor supply, while remaining diminished relative to the vyb stage, should increase somewhat in this final developmental stage (Figure 3).
Patterns of Gene Expression in Genes Potentially Modifying the Flower Color of Chiloglottis trapeziformis
Beyond the direct anthocyanin and flavonol metabolism genes (Figure 3), other genes such as those involved in modification, transport, and sequestration, as well as genes associated with chlorophyll production may influence the flower color of C. trapeziformis. Notably, some genes involved in the modification of anthocyanins and flavonol glycosides showed uniform levels of gene expression across tissue types and developmental stages. This included the flavonoid glucosyltransferases, FGT (CtrUGT78D2a/b and CtrUGT71B1) and malonyltransferases, MAT (Ctr3MAT1-4) (Figure 3). Thus, it is likely that these downstream modification steps are not rate-limiting during the active accumulation of peak anthocyanins in the callus and the flavonols in the lamina (vyb-mb stages), nor in the final stage of the lamina becoming reddish green (sflw). We further predict that one or more of the MAT transcripts (e.g., Ctr3MAT1-2) are involved in the production of the malonylated anthocyanins (such as cyanidin malonylglucoside). Such derivatives were abundant (ca. 31–33% of total anthocyanins) in both the callus and labellum (Figure 2A and Supplementary Figure 5), and their accumulation profiles closely mirrored their non-malonylated anthocyanin precursors. The high abundance of malonylated anthocyanin derivatives in the callus, may be relevant to the preservation of its distinct dark color across developmental stages. Processes downstream of anthocyanin biosynthesis, such as the acylation of anthocyanins (e.g., malonylation) can have important consequences for floral color. For example, acylation has many functional roles including the protection of glycosides from enzymatic degradation, anthocyanin structure stabilization, and assistance in vacuolar uptake and sequestration (Bontpart et al., 2015). Importantly, these various functions may all serve to aid the stabilization of pigments in flowers (Suzuki et al., 2002; Luo et al., 2007).
Among the predicted transporter gene homologs, TT19 (CtrTT19b) showed a pattern of gene expression which mirrored DFR and LDOX genes (cluster 2, in Supplementary Figure 4). For example, at the vyb stage there was high and differential expression in the callus relative to the lamina, with this pattern reversing at the vmb stage. Expression levels in the callus plateaued at the vmb stage, while they peaked a second time in the lamina at the sflw stage. Thus, high expression levels at this gene appear to correspond with the respective starting points of anthocyanin accumulation and color development which started first in the callus with a peak by mb, but lagged in the lamina peaking at sflw (Figure 2A). In peach, expression of a glutathione S-transferase gene, PpGST1 strongly correlates with anthocyanin accumulation in fruit tissues. Accordingly, transient overexpression of PpGST1 increased the anthocyanin content in the fruit of yellow-fleshed nectarine while silencing of PpGST1 in blood-fleshed (anthocyanin-rich) peach decreased anthocyanin accumulation resulting in white flesh (Zhao et al., 2020). Interestingly, PpGST1 is also a locus containing functional and non-functional alleles that co-segregated with white and non-white (i.e., pink and red) flowers (Lu et al., 2021). In the purple-flowered cyclamen, expression of an anthocyanin-related GST, CkmGST3 and upstream core biosynthetic genes such as CkmF3’5’H and CkmDFR2 were tightly co-regulated during flower development. These transcripts were strongly expressed in paler pigmented petals and correlated with the sharp rise in anthocyanin accumulation compared to fully pigmented purple petal (Kitamura et al., 2012).
The expression patterns of a homolog to a gene implicated in the regulation of vacuolar pH, PH5 (CtrPH5), also showed some similarity to those of DFR, LDOX and TT19 genes (CtrDFR1, CtrLDOX1/2, and CtrTT19b) (Supplementary Figures 3, 4). For example, peak expression occurred in the callus at the vyb stage with significant downregulation as the flower developed. By contrast, in the lamina expression peaked in the vmb stage. Interestingly, at a homolog of NHX1 (CtrNHX1), gene expression levels were high in both tissue types at vyb, and tended to remain high in the callus across developmental stages. However, in the lamina, gene expression levels were differentially downregulated relative to the callus. These findings may indicate that vacuolar pH is lowest in the vyb stage of the callus, and in the vmb stage of the lamina. Furthermore, we predict that vacuolar acidification in the callus during active anthocyanin accumulation could play a synergistic role in conferring the strong “black” color of the callus. Different levels of vacuolar acidification are known to cause color shifts in flowers. For example, a less acidic vacuolar pH is often associated with blue colored flowers (e.g., Petunia, Ipomea, Hydrangea, and Phalaenopsis spp.) that otherwise share similar anthocyanin composition to other non-blue colored flowers (Fukada-Tanaka et al., 2000; Yoshida et al., 2003; Verweij et al., 2008; Liang et al., 2020). Presently, genes implicated in the regulation of vacuolar pH with relevance to flower color have only been established for a few species (i.e., Petunia and Japanese morning glory). Notably, the loss-of-function mutation of petunia PH5 or the downregulation of PH5 using RNA interference reduces vacuolar acidification in petals, resulting in “blue-violet” flower color in mutants and transformants compared to “red-violet” wild-type flowers (Faraco et al., 2014). Conversely, a mutation of morning glory NHX1 impairs vacuolar alkalization, resulting in purple petals in fully bloomed flowers instead of bright blue colors in the wild-type (Fukada-Tanaka et al., 2000).
Chlorophyll is often present in petals during early developmental stages but usually decreases to trace levels in the typical brightly colored flowers of most plants. This loss of green coloration as the flower develops is considered crucial for making flowers more visible to animal pollinators against the green leafy background. However, examples of green colored flowers are nonetheless widespread across the flowering plants. Dull green and red flowers are particularly frequent among sexually deceptive plant species. These include hundreds of examples within the Australian genera Caladenia, Chiloglottis, Cryptostylis, Drakaea and Pterostylis, among others; tens of European Ophrys species, and several sexually deceptive species from South America (Bohman et al., 2016; Peakall et al., 2020).
Despite the widespread occurrence of green colored flowers and a thorough understanding of photosynthesis- and chlorophyll-related pathways (Hörtensteiner, 2006; Tanaka and Tanaka, 2006), little is known about how green colors occur in mature flowers. To date, the best clues are offered in carnations. Insights from carnation (Dianthus caryophyllus L.) floral transcriptomes reveal that expression of photosynthesis and chlorophyll biosynthesis genes are tightly associated with chlorophyll content, and thus coloration of green- and white-flowered cultivars (Ohmiya et al., 2014, 2017). Although the types and quantities of chlorophyll were not measured in this study, we predict that retention of chlorophyll pigments is pivotal for the green coloration of Chiloglottis trapeziformis flowers. When compared to the callus, the lamina was characterized by lower levels of anthocyanins and higher levels of colorless flavonol glycosides. Furthermore, in the lamina, consistently higher levels of gene expression were detected at photosynthesis- and chlorophyll-related genes across all developmental stages, indicative of enhanced chlorophyll biosynthesis in the lamina (Supplementary Figure 1 and Supplementary Data 1). Thus, the combination of low anthocyanins, high flavonol glycosides and sustained chlorophyll production are likely to be the basis of the predominant green coloration of the lamina from bud to flower opening.
Transcriptional Control of Color Pathways in the Flower
A detailed investigation of the transcriptional control of developmental and tissue-specific of pigmentation is beyond the scope of this study. Nonetheless, we highlight nine R2R3-MYB TF, two bHLH, and one WDR gene(s) potentially implicated in anthocyanin pigmentation in the flower of Chiloglottis trapeziformis (Figure 4 and Supplementary Data 4). The striking similarity in differential expression profile between CtrMYB2a and CtrF3H1/2, CtrDFR1 and CtrLDOX1/2 indicate that some shared and dedicated anthocyanin pathway genes are regulatory targets of CtrMYB2a. CtrMYB2a/b belongs to subgroup 5 of R2R3-MYB TFs that contains several known orchid tissue-specific floral pigmentation regulators (Chiou and Yeh, 2008; Hsu et al., 2015; Li et al., 2020). For example, the CtrMYB2a homolog in Phalaenopsis, PeMYB2 is correlated with anthocyanin abundance in the petals and sepals of several cultivars and is highly expressed in full-red pigmented flowers. Transient overexpression of PeMYB2 in white-flowered P. aphrodite ssp. formosana induced the expression of endogenous flavonoid and anthocyanin biosynthesis genes (e.g., F3H, F3’H, DFR, or ANS genes) and the development of reddish-pink pigmentation (Hsu et al., 2015).
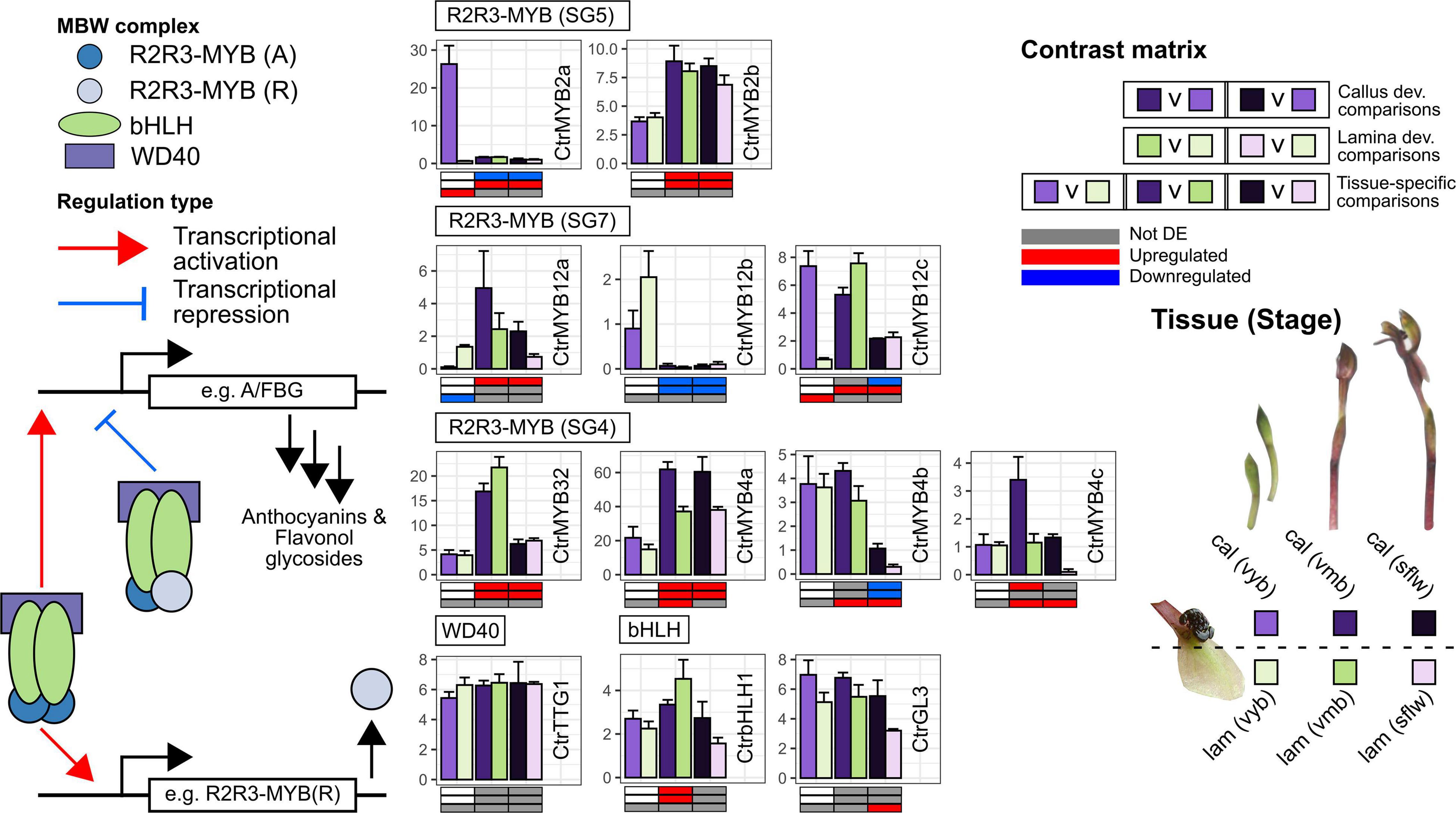
Figure 4. Transcriptional regulation of anthocyanin and flavonol glycoside pathway genes in the callus and labellum tissues of Chiloglottis trapeziformis during flower development. Bars depict the average (±s.e.) normalized expression values (FPKM) of putative transcriptional regulatory genes encoding relevant R2R3-MYB, basic-helix-loop-helix (bHLH), and WD40 repeat (WDR) proteins in the calli and labellum lamina tissues of very young buds (vyb), very mature buds (vmb), and mature sunflowers (sflw). A/FBG indicate shared flavonoid as well as anthocyanin/flavonol glycoside biosynthetic pathway genes. Various shades of purple and green depict the relevant color callus and labellum lamina tissues at various flower developmental stages. The contrast matrix indicates the differential expression outcomes based on comparisons in Supplementary Figure 1. Red, blue, and gray indicate significant upregulation (FDR < 0.05, log2FC > 0.5), downregulation (FDR < 0.05, log2FC < –0.5), and no significant differential expression in selected comparisons, respectively.
Unlike orchid subgroup 5 R2R3-MYBs, the role of other subgroups (e.g., 6, 7, 20, and 4) in the regulation of flavonoid metabolism and pigmentation is unknown in the Orchidaceae. CtrMYB12a/b/c belongs to subgroup 7 R2R3-MYB TFs and is related to Arabidopsis MYB12 which commonly regulate flavonol glycoside accumulation via the activation of FLS and shared flavonoid biosynthetic pathway genes (Stracke et al., 2007). In particular, CtrMYB12a shares differential expression patterns with CtrFLS1/2 and CtrCOMT1-4. We predict that CtrMYB12a regulates flavonol glycoside accumulation by targeting CtrFLS1/2 and CtrCOMT1-4. Conversely, CtrMYB12c does not share expression similarities with CtrFLS1/2. Instead, CtrMYB12c expression coincides with CtrMYB2a, suggesting that CtrMYB12c is more relevant to the regulation of anthocyanin than flavonol glycoside biosynthesis. In plants, the transcriptional activation strength of FLS are known to differ between orthologs of AtMYB12 (Mehrtens et al., 2005) but also associated paralogs of certain species (Shan et al., 2020). In Freesia, all four AtMYB12 orthologs FhMYBF1–4 positively regulated several shared flavonoid pathway genes and FhFLS1 at different magnitudes when overexpressed, however FhMYBF4 lack the capacity to highly induce FhFLS1 expression (Shan et al., 2020). Some studies have even shown that AtMYB12 orthologs potentially regulate the anthocyanin biosynthetic pathway genes (Czemmel et al., 2009; Zheng et al., 2019; Zhong et al., 2020).
CtrMYB32 and CtrMYB4a/b/c belong to subgroup 4 R2R3-MYB TFs which are active repressors that fine tune flavonoid levels by balancing the inductive effects of the MBW complex (Chen et al., 2019; LaFountain and Yuan, 2021). In particular, CtrMYB32 and CtrMYB4a were consistently upregulated in vmb and sflw while the opposite was true for CtrMYB4b compared to vyb regardless of tissue type. Furthermore, CtrMYB4a/b/c showed significantly higher callus gene expression compared to the lamina in one or more stages post vyb. Since further anthocyanin pathway activation by the MBW complex does not benefit the establishment of color or contrast, in the callus, we predict that CtrMYB4a/b/c co-operatively repress the inductive effects of the MBW activity in vmb and sflw once the distinct black color has been established in vyb. Conversely, the weaker pathway repression in the labellum compared to the callus post vyb may signal the accumulation of anthocyanins.
Implications and Future Directions
In the last decade, considerable progress has been made toward understanding the diversity and distribution (Peakall et al., 2010) and the regulation of the chiloglottones which underpin the sex pheromone mimicry of Chiloglottis orchids (Falara et al., 2013; Amarasinghe et al., 2015). However, the chemical and molecular basis of the floral pigmentation in Chiloglottis, and indeed of any Australian sexually deceptive orchids, has remained unknown. Here, we demonstrate that the black callus of Chiloglottis trapeziformis flowers is characterized by a high abundance of cyanidin-based anthocyanins and diverse flavonol glycoside co-pigments concentrated in a small, highly localized area. Conversely, a high abundance of flavonol glycoside co-pigments and low anthocyanin content is characteristic of the lamina. Differential expression of specific genes controlling the branchpoints into the respective anthocyanin (e.g., DFR and LDOX) and flavonol glycoside biosynthesis (e.g., FLS) and its subsequent modification toward methylated derivatives (e.g., FMT) is likely pivotal for generating the contrasting dark color of the callus against the green to reddish green lamina (Figure 3).
Beyond Chiloglottis, inconspicuous dull colored flowers with tissue-specific dark maroon to black pigmentation is common in sexually deceptive species. In Australia, several orchid genera have some sexually deceptive species which exhibit this tissue-specific contrast (e.g., within Caladenia, Calochilus), and in some genera, this is exclusively the case (e.g., Drakaea, Cryptostylis, Paracaleana, Leporella, Caleana) (Gaskett, 2011). Other examples of tissue-specific dark colors in sexually deceptive species outside Australia are the dark labella of many Ophrys species (Bradshaw et al., 2010) such as Ophrys speculum (Figure 1F), the maroon-black callus structure on the pink labellum of Serapias lingua (Vereecken et al., 2012; Pellegrino et al., 2017), the large elongated dark maroon labellum of Asian beetle-pollinated Luisia teres (Arakaki et al., 2016), the triangular-shaped dark maroon pigmentation on the labellum of bee-pollinated Mormolyca ringens (Singer et al., 2004), the central dark maroon labellum and column on an otherwise bright yellow corolla of Telipogon peruvianus (Martel et al., 2016, 2019), a combination of dark maroon-colored floral structures including antennae-like petals, anther, and labellum of south African beetle-pollinated Disa forficaria orchids (Cohen et al., 2021), and the raised black spots on the yellow to bright orange ray florets of some fly pollinated Gorteria diffusa (Ellis and Johnson, 2010) morphotypes (Figure 1G). In these cases, the dark pigmentation likely serves several key purposes important for pollination, including mimicking the color of female insects, providing strong chromatic and achromatic contrast, and aiding the exploitation of pollinator sensory biases.
All of these examples showcase a strong correlation between sexual deception and dark colored flowers across many different evolutionary lineages. As such, understanding the chemical and genetic mechanisms of how such dark coloration is attained is pivotal for understanding how sexual deception evolves. We predict that similar chemical constituents and/or spatiotemporal gene expression of underlying pathway genes (e.g., DFR, LDOX, and FLS) will be pivotal for dark, potentially contrasting tissue-specific pigmentation in other sexually deceptive species. Currently, Ophrys is the best-characterized genus to support this prediction. In some confirmed sexually deceptive Ophrys orchids such as O. insectifera, O. sphegodes, and O. speculum (Schiestl et al., 1999; Vignolini et al., 2012), cyanidin-based anthocyanins are the major floral anthocyanin pigment present in whole flowers or the labellum (Strack et al., 1989). Furthermore, the dark labellum color of Ophrys speculum (Figure 1F) was demonstrated to contain a blend of anthocyanin and quercetin flavonol glycoside co-pigments (Vignolini et al., 2012). Transcriptomic surveys of O. exaltata, O. sphegodes, O. aymoninii, and O. garganica (Sedeek et al., 2013; Piñeiro Fernández et al., 2019) reveal that anthocyanin and flavonol glycoside biosynthesis pathway genes are active in the dark labella of these species. In Gorteria diffusa that possess dark tissue-specific pigmentation flanked by brightly colored orange ray florets, it will be of interest to determine the types of floral pigments, cellular chemistry, and genetic mechanisms involved. In this latter case, the relevance of carotenoid (e.g., xanthophylls) or flavonoid (e.g., chalcones and aurones) pigment pathways are predicted to be the driver of the contrast.
Future studies will also be required to determine the key transcription factors involved and how they regulate the expression of key flavonoid biosynthetic, modification, and transport pathway enzymes in the flowers of Chiloglottis and other sexually deceptive plants. Members of the R2R3-MYB, bHLH, and WD-Repeat proteins, including those that we have briefly highlighted in this study, are prime candidates given that they are key determinants of anthocyanin pigmentation intensity and also patterning in many flowers (Davies et al., 2012; Wessinger and Rausher, 2012). However, MADS-box transcription factors should also be investigated as some are involved in establishing proper floral organ identity programs, photosynthesis, chlorophyll metabolism, and pigmentation in orchids (Pan et al., 2014; Hsu et al., 2021). The integration of various omics approaches (e.g., large-scale transcriptomic, metabolomic, ionomics) is also anticipated to provide a holistic view of how structural genes, regulatory factors, and cellular constituents (e.g., pigments, metal ions, pH) interact and mediate the evolutionary color transitions between sexually deceptive systems.
Our findings in Chiloglottis trapeziformis indicate that its floral color adaptations for sexual deception require complex tissue-specific coordinated regulation at multiple developmental stages, involving many genes and diverse pigment metabolic and cellular constituent regulating pathways. It is further of interest that the genus Chiloglottis belongs to a subtribe (Drakaeinae) in which all genera are exclusively sexually deceptive (with the exception of some rare self-pollinating cases) and all fall within a well-resolved clade on a long branch of the diverse predominantly Australasian tribe the Diurideae (Peakall et al., 2021). Thus, it is likely that the complex chemical and genetic basis of floral color mimicry may have been finely tuned by a long evolutionary process in these sexually deceptive orchids (Weston et al., 2014; Peakall et al., 2021). This is in contrast with some of the well-known flower color polymorphisms found within natural populations (Streisfeld and Rausher, 2011; Kellenberger et al., 2019), and even the flower color changes associated with key pollinator shifts that are often mediated by relatively simple genetic changes restricted to a few genes within the anthocyanin and flavonol glycoside pathways (Wessinger and Rausher, 2012). It will be of interest in future research to determine whether sexually deceptive lineages of more recent evolutionary origin than Chiloglottis (e.g., Caladenia, Caladeniinae; Diurideae) (Peakall et al., 2021) exhibit floral color adaptions based on simple genetic changes, as one might expect at the early stages in the evolution of sexual deception.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: New sequence reads obtained for the sflw stage has been added to the existing BioProject accession PRJNA390683 and SRA study accession SRP1093281.
Author Contributions
DW and JP performed the experiments. RP and DW secured the funding, designed the study, and coordinated the experiments and data analysis. DW wrote the article with assistance from RP and JP. All authors have read and approved the manuscript.
Funding
This work was supported by the Australian Research Council projects DE190100249 to DW and DP150102762 to RP, and Australian Government Research Training Program to JP.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Thy Truong, Adam Carroll, and Joseph Boileau at the Joint Mass Spectrometry Facility for assistance with LC-MS operation, the Australian National Botanic Gardens, Canberra for permission and permits to take plant material from the gardens, and New South Wales (NSW) National Parks. Photographs were taken by Steven Johnson (Figures 1D,G), Jorun Tharaldsen (Figures 1F,H), and RP (Figures 1A–C,E).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.860997/full#supplementary-material
Footnotes
References
Abdel-Aal, E. S. M., Young, J. C., and Rabalski, I. (2006). Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J. Agric. Food Chem. 54, 4696–4704. doi: 10.1021/jf0606609
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, M., Brendolise, C., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
Allan, A. C., and Espley, R. V. (2018). MYBs drive novel consumer traits in fruits and vegetables. Trends Plant Sci. 23, 693–705. doi: 10.1016/j.tplants.2018.06.001
Amarasinghe, R., Poldy, J., Matsuba, Y., Barrow, R. A., Hemmi, J. M., Pichersky, E., et al. (2015). UV-B light contributes directly to the synthesis of chiloglottone floral volatiles. Ann. Bot. 115, 693–703. doi: 10.1093/aob/mcu262
Arakaki, N., Yasuda, K., Kanayama, S., Jitsuno, S., Oike, M., and Wakamura, S. (2016). Attraction of males of the cupreous polished chafer Protaetia pryeri pryeri (Coleoptera: Scarabaeidae) for pollination by an epiphytic orchid Luisia teres (Asparagales: Orchidaceae). Appl. Entomol. Zool. 51, 241–246. doi: 10.1007/s13355-016-0396-5
Behrens, C. E., Smith, K. E., Iancu, C. V., Choe, J. Y., and Dean, J. V. (2019). Transport of anthocyanins and other flavonoids by the arabidopsis ATP-Binding cassette transporter AtABCC2. Sci. Rep. 9, 1–15. doi: 10.1038/s41598-018-37504-8
Bohman, B., Flematti, G. R., Barrow, R. A., Pichersky, E., and Peakall, R. (2016). Pollination by sexual deception - it takes chemistry to work. Curr. Opin. Plant Biol. 32, 37–46. doi: 10.1016/j.pbi.2016.06.004
Bontpart, T., Cheynier, V., Ageorges, A., and Terrier, N. (2015). BAHD or SCPL acyltransferase? what a dilemma for acylation in the world of plant phenolic compounds. New Phytol. 208, 695–707. doi: 10.1111/nph.13498
Borghi, M., Fernie, A. R., Schiestl, F. P., and Bouwmeester, H. J. (2017). The sexual advantage of looking, smelling, and tasting good: the metabolic network that produces signals for pollinators. Trends Plant Sci. 22, 338–350. doi: 10.1016/j.tplants.2016.12.009
Boss, P. K., Davies, C., and Robinson, S. P. (1996). Anthocyanin composition and anthocyanin pathway gene expression in grapevine sports differing in berry skin colour. Aust. J. Grape Wine Res. 2, 163–170. doi: 10.1111/j.1755-0238.1996.tb00104.x
Bradshaw, E., Rudall, P. J., Devey, D. S., Thomas, M. M., Glover, B. J., and Bateman, R. M. (2010). Comparative labellum micromorphology of the sexually deceptive temperate orchid genus Ophrys: diverse epidermal cell types and multiple origins of structural colour. Bot. J. Linn. Soc. 162, 504–540. doi: 10.1111/j.1095-8339.2010.01033.x
Buchfink, B., Xie, C., and Huson, D. H. (2014). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Chen, L., Hu, B., Qin, Y., Hu, G., and Zhao, J. (2019). Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 136, 178–187. doi: 10.1016/j.plaphy.2019.01.024
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cheng, J., Liao, L., Zhou, H., Gu, C., Wang, L., and Han, Y. (2015). A small indel mutation in an anthocyanin transporter causes variegated colouration of peach flowers. J. Exp. Bot. 66, 7227–7239. doi: 10.1093/jxb/erv419
Chiou, C. Y., and Yeh, K. W. (2008). Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium gower ramsey. Plant Mol. Biol. 66, 379–388. doi: 10.1007/s11103-007-9275-3
Cohen, C., Liltved, W. R., Colville, J. F., Shuttleworth, A., Weissflog, J., Svatoš, A., et al. (2021). Sexual deception of a beetle pollinator through floral mimicry. Curr. Biol. 31, 1962–1969. doi: 10.1016/j.cub.2021.03.037
Czemmel, S., Stracke, R., Weisshaar, B., Cordon, N., Harris, N. N., Walker, A. R., et al. (2009). The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530. doi: 10.1104/pp.109.142059
Davies, K. M. (2008). “Modifying anthocyanin production in flowers,” in Anthocyanins, eds C. Winefield, K. Davies, and K. Gould (New York, NY: Springer), 49–80. doi: 10.1007/978-0-387-77335-3_3
Davies, K. M., Albert, N. W., and Schwinn, K. E. (2012). From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39, 619–638. doi: 10.1071/FP12195
de Jager, M. L., and Peakall, R. (2016). Does morphology matter? An explicit assessment of floral morphology in sexual deception. Funct. Ecol. 30, 537–546. doi: 10.1111/1365-2435.12517
de Jager, M. L., and Peakall, R. (2019). Experimental examination of pollinator-mediated selection in a sexually deceptive orchid. Ann. Bot. 123, 347–354. doi: 10.1093/aob/mcy083
Gilbert, D. (2013). EvidentialGene: Tr2aacds, mRNA Transcript Assembly Software. Available online at: http://arthropods.eugenes.org/EvidentialGene/trassembly.html (accessed May 1, 2021).
Ellis, A. G., and Johnson, S. D. (2010). Floral mimicry enhances pollen export: the evolution of pollination by sexual deceit outside of the orchidaceae. Am. Nat. 176, E143–E151. doi: 10.1086/656487
Falara, V., Amarasinghe, R., Poldy, J., Pichersky, E., Barrow, R. A., and Peakall, R. (2013). The production of a key floral volatile is dependent on UV light in a sexually deceptive orchid. Ann. Bot. 111, 21–30. doi: 10.1093/aob/mcs228
Fan-Chiang, H.-J., and Wrolstad, R. E. (2006). Anthocyanin pigment composition of blackberries. J. Food Sci. 70, C198–C202. doi: 10.1111/j.1365-2621.2005.tb07125.x
Faraco, M., Spelt, C., Bliek, M., Verweij, W., Hoshino, A., Espen, L., et al. (2014). Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color. Cell Rep. 6, 32–43. doi: 10.1016/j.celrep.2013.12.009
Fossen, T., and Øvstedal, D. O. (2003). Anthocyanins from flowers of the orchids Dracula chimaera and D. cordobae. Phytochemistry 63, 783–787. doi: 10.1016/S0031-9422(03)00339-X
Francisco, R. M., Regalado, A., Ageorges, A., Burla, B. J., Bassin, B., Eisenach, C., et al. (2013). ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 25, 1840–1854. doi: 10.1105/tpc.112.102152
Franke, S., Ibarra, F., Schulz, C. M., Twele, R., Poldy, J., Barrow, R. A., et al. (2009). The discovery of 2,5-dialkylcyclohexan-1,3-diones as a new class of natural products. Proc. Natl. Acad. Sci. U. S. A. 106, 8877–8882. doi: 10.1073/pnas.0900646106
Fukada-Tanaka, S., Inagaki, Y., Yamaguchi, T., Saito, N., and Iida, S. (2000). Colour-enhancing protein in blue petals. Nature 407:581. doi: 10.1038/35036683
Gaskett, A. C. (2011). Orchid pollination by sexual deception: pollinator perspectives. Biol. Rev. 86, 33–75. doi: 10.1111/j.1469-185X.2010.00134.x
Gaskett, A. C., Endler, J. A., and Phillips, R. D. (2017). Convergent evolution of sexual deception via chromatic and achromatic contrast rather than colour mimicry. Evol. Ecol. 31, 205–227. doi: 10.1007/s10682-016-9863-2
George, A., Gonzales, C., Strauss, M. S., and Arditti, J. (1973). Chemotaxonomic and ecological implications of anthocyanins in Elythranthera. Biochem. Syst. Ecol. 1, 45–49. doi: 10.1016/0305-1978(73)90034-3
Glover, B. J., Walker, R. H., Moyroud, E., and Brockington, S. F. (2013). How to spot a flower. New Phytol. 197, 687–689. doi: 10.1111/nph.12112
Gomez, C., Terrier, N., Torregrosa, L., Vialet, S., Fournier-Level, A., Verries, C., et al. (2009). Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 150, 402–415. doi: 10.1104/pp.109.135624
Grotewold, E. (2006). The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 57, 761–780. doi: 10.1146/annurev.arplant.57.032905.105248
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hatier, J.-H. B., and Gould, K. S. (2007). Black coloration in leaves of Ophiopogon planiscapus “Nigrescens”. Leaf optics, chromaticity, and internal light gradients. Funct. Plant Biol. 34:130. doi: 10.1071/FP06220
Hayashi, T., Bohman, B., Scaffidi, A., Peakall, R., and Flematti, G. R. (2021). An unusual tricosatriene is crucial for male fungus gnat attraction and exploitation by sexually deceptive Pterostylis orchids. Curr. Biol. 31, 1954.e–1961.e. doi: 10.1016/j.cub.2021.01.095
Hörtensteiner, S. (2006). Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 57, 55–77. doi: 10.1146/annurev.arplant.57.032905.105212
Hsu, C.-C., Chen, Y.-Y., Tsai, W.-C., Chen, W.-H., and Chen, H.-H. (2015). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 168, 175–191. doi: 10.1104/pp.114.254599
Hsu, H. F., Chen, W. H., Shen, Y. H., Hsu, W. H., Mao, W. T., and Yang, C. H. (2021). Multifunctional evolution of B and AGL6 MADS box genes in orchids. Nat. Commun. 12, 1–12. doi: 10.1038/s41467-021-21229-w
Hugueney, P., Provenzano, S., Verries, C., Ferrandino, A., Meudec, E., Batelli, G., et al. (2009). A novel cation-dependent O-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol. 150, 2057–2070. doi: 10.1104/pp.109.140376
Iwashina, T. (2015). Contribution to flower colors of flavonoids including anthocyanins: a review. Nat. Prod. Commun. 10, 529–544. doi: 10.1177/1934578x1501000335
Johnson, S. D., and Schiestl, F. P. (2016). Floral Mimicry, 1st Edn. New York, NY: Oxford University Press. doi: 10.1093/acprof:oso/9780198732693.001.0001
Jones, D. L. (2021). A Complete Guide to Native Orchids of Australia, 3rd Edn. Wahroonga, NSW: New Holland Publishers.
Kellenberger, R. T., Byers, K. J. R. P., De Brito Francisco, R. M., Staedler, Y. M., LaFountain, A. M., Schönenberger, J., et al. (2019). Emergence of a floral colour polymorphism by pollinator-mediated overdominance. Nat. Commun. 10, 1–11. doi: 10.1038/s41467-018-07936-x
Khunmuang, S., Kanlayanarat, S., Wongs-Aree, C., Meir, S., Philosoph-Hadas, S., Oren-Shamir, M., et al. (2019). Ethylene induces a rapid degradation of petal anthocyanins in cut Vanda ‘Sansai Blue’ orchid flowers. Front. Plant Sci. 10:1004. doi: 10.3389/fpls.2019.01004
Kitamura, S., Akita, Y., Ishizaka, H., Narumi, I., and Tanaka, A. (2012). Molecular characterization of an anthocyanin-related glutathione S-transferase gene in cyclamen. J. Plant Physiol. 169, 636–642. doi: 10.1016/j.jplph.2011.12.011
LaFountain, A. M., and Yuan, Y. W. (2021). Repressors of anthocyanin biosynthesis. New Phytol. 231, 933–949. doi: 10.1111/nph.17397
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, B. J., Zheng, B. Q., Wang, J. Y., Tsai, W. C., Lu, H. C., Zou, L. H., et al. (2020). New insight into the molecular mechanism of colour differentiation among floral segments in orchids. Commun. Biol. 3, 1–13. doi: 10.1038/s42003-020-0821-8
Li, Q., Wang, J., Sun, H. Y., and Shang, X. (2014). Flower color patterning in pansy (Viola×wittrockiana Gams.) is caused by the differential expression of three genes from the anthocyanin pathway in acyanic and cyanic flower areas. Plant Physiol. Biochem. 84, 134–141. doi: 10.1016/j.plaphy.2014.09.012
Liang, C. Y., Rengasamy, K. P., Huang, L. M., Hsu, C. C., Jeng, M. F., Chen, W. H., et al. (2020). Assessment of violet-blue color formation in Phalaenopsis orchids. BMC Plant Biol. 20:212. doi: 10.1186/s12870-020-02402-7
Liao, Y., Smyth, G. K., and Shi, W. (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lim, E. K., Ashford, D. A., Hou, B., Jackson, R. G., and Bowles, D. J. (2004). Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol. Bioeng. 87, 623–631. doi: 10.1002/bit.20154
Liu, Y., Tikunov, Y., Schouten, R. E., Marcelis, L. F. M., Visser, R. G. F., and Bovy, A. (2018). Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: a review. Front. Chem. 6:52. doi: 10.3389/fchem.2018.00052
Lohse, M., Nagel, A., Herter, T., May, P., Schroda, M., Zrenner, R., et al. (2014). Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 37, 1250–1258. doi: 10.1111/pce.12231
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, Z., Cao, H., Pan, L., Niu, L., Wei, B., Cui, G. C., et al. (2021). Two loss-of-function alleles of the glutathione S-transferase (GST) gene cause anthocyanin deficiency in flower and fruit skin of peach (Prunus persica). Plant J. 107, 1320–1331. doi: 10.1111/tpj.15312
Lücker, J., Martens, S., and Lund, S. T. (2010). Characterization of a Vitis vinifera cv. Cabernet Sauvignon 3’,5’-O-methyltransferase showing strong preference for anthocyanins and glycosylated flavonols. Phytochemistry 71, 1474–1484. doi: 10.1016/j.phytochem.2010.05.027
Luo, J., Nishiyama, Y., Fuell, C., Taguchi, G., Elliott, K., Hill, L., et al. (2007). Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from arabidopsis thaliana. Plant J. 50, 678–695. doi: 10.1111/j.1365-313X.2007.03079.x
Luo, P., Ning, G., Wang, Z., Shen, Y., Jin, H., Li, P., et al. (2016). Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front. Plant Sci. 6, 1–12. doi: 10.3389/fpls.2015.01257
Markham, K. R., Bloor, S. J., Nicholson, R., Rivera, R., Shemluck, M., Kevan, P. G., et al. (2004). Black flower coloration in wild Lisianthius nigrescens: its chemistry and ecological consequences. zeitschrift fur naturforsch. - Sect. C J. Biosci. 59, 625–630. doi: 10.1515/znc-2004-9-1003
Martel, C., Cairampoma, L., Stauffer, F. W., and Ayasse, M. (2016). Telipogon peruvianus (Orchidaceae) flowers elicit pre-mating behaviour in Eudejeania (Tachinidae) males for pollination. PLoS One 11:e0165896. doi: 10.1371/journal.pone.0165896
Martel, C., Francke, W., and Ayasse, M. (2019). The chemical and visual bases of the pollination of the Neotropical sexually deceptive orchid Telipogon peruvianus. New Phytol. 223, 1989–2001. doi: 10.1111/nph.15902
Martins, T. R., Berg, J. J., Blinka, S., Rausher, M. D., and Baum, D. A. (2013). Precise spatio-temporal regulation of the anthocyanin biosynthetic pathway leads to petal spot formation in Clarkia gracilis (Onagraceae). New Phytol. 197, 958–969. doi: 10.1111/nph.12062
Martins, T. R., Jiang, P., and Rausher, M. D. (2017). How petals change their spots: cis-regulatory re-wiring in Clarkia (Onagraceae). New Phytol. 216, 510–518. doi: 10.1111/nph.14163
Mehrtens, F., Kranz, H., Bednarek, P., and Weisshaar, B. (2005). The arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096. doi: 10.1104/pp.104.058032
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., and Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41:e121. doi: 10.1093/nar/gkt263
Morita, Y., Ishiguro, K., Tanaka, Y., Iida, S., and Hoshino, A. (2015). Spontaneous mutations of the UDP-glucose:flavonoid 3-O-glucosyltransferase gene confers pale- and dull-colored flowers in the Japanese and common morning glories. Planta 242, 575–587. doi: 10.1007/s00425-015-2321-5
Morita, Y., Takagi, K., Fukuchi-Mizutani, M., Ishiguro, K., Tanaka, Y., Nitasaka, E., et al. (2014). A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. Plant J. 78, 294–304. doi: 10.1111/tpj.12469
Nadot, S., and Carrive, L. (2021). The colourful life of flowers. Bot. Lett. 168, 120–130. doi: 10.1080/23818107.2020.1839789
Ohmiya, A., Hirashima, M., Yagi, M., Tanase, K., and Yamamizo, C. (2014). Identification of genes associated with chlorophyll accumulation in flower petals. PLoS One 9:e113738. doi: 10.1371/journal.pone.0113738
Ohmiya, A., Sasaki, K., Nashima, K., Oda-Yamamizo, C., Hirashima, M., and Sumitomo, K. (2017). Transcriptome analysis in petals and leaves of chrysanthemums with different chlorophyll levels. BMC Plant Biol. 17:202. doi: 10.1186/s12870-017-1156-6
Ollerton, J., Winfree, R., and Tarrant, S. (2011). How many flowering plants are pollinated by animals? Oikos 120, 321–326. doi: 10.1111/j.1600-0706.2010.18644.x
Pan, Z. J., Chen, Y. Y., Du, J. S., Chen, Y. Y., Chung, M. C., Tsai, W. C., et al. (2014). Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 202, 1024–1042. doi: 10.1111/nph.12723
Peakall, R., Ebert, D., Poldy, J., Barrow, R. A., Francke, W., Bower, C. C., et al. (2010). Pollinator specificity, floral odour chemistry and the phylogeny of Australian sexually deceptive Chiloglottis orchids: implications for pollinator-driven speciation. New Phytol. 188, 437–450. doi: 10.1111/j.1469-8137.2010.03308.x
Peakall, R., Wong, D. C. J., Bohman, B., Flematti, G. R., and Pichersky, E. (2020). “Floral volatiles for pollinator attraction and speciation in sexually deceptive orchids,” in Biology of Plant, eds E. Pichersky and N. Dudareva (Boca Raton, FL: CRC Press), 271–295. doi: 10.1201/9780429455612-18
Peakall, R., Wong, D. C. J., Phillips, R. D., Ruibal, M., Eyles, R., Rodriguez-Delgado, C., et al. (2021). A multitiered sequence capture strategy spanning broad evolutionary scales: application for phylogenetic and phylogeographic studies of orchids. Mol. Ecol. Resour. 21, 1118–1140. doi: 10.1111/1755-0998.13327
Pellegrino, G., Bellusci, F., and Palermo, A. M. (2017). Functional differentiation in pollination processes among floral traits in Serapias species (Orchidaceae). Ecol. Evol. 7, 7171–7177. doi: 10.1002/ece3.3264
Piñeiro Fernández, L., Byers, K. J. R. P., Cai, J., Sedeek, K. E. M., Kellenberger, R. T., Russo, A., et al. (2019). A phylogenomic analysis of the floral transcriptomes of sexually deceptive and rewarding European orchids. Ophrys and Gymnadenia. Front. Plant Sci. 10, 1–13. doi: 10.3389/fpls.2019.01553
Provenzano, S., Spelt, C., Hosokawa, S., Nakamura, N., Brugliera, F., Demelis, L., et al. (2014). Genetic control and evolution of anthocyanin methylation. Plant Physiol. 165, 962–977. doi: 10.1104/pp.113.234526
Raguso, R. A. (2004). Flowers as sensory billboards: progress towards an integrated understanding of floral advertisement. Curr. Opin. Plant Biol. 7, 434–440. doi: 10.1016/j.pbi.2004.05.010
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2009). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodrigues, J. A., Espley, R. V., and Allan, A. C. (2021). Genomic analysis uncovers functional variation in the C-terminus of anthocyanin-activating MYB transcription factors. Hortic. Res. 8, 1–14. doi: 10.1038/s41438-021-00514-1
Sawada, Y., Nakabayashi, R., Yamada, Y., Suzuki, M., Sato, M., Sakata, A., et al. (2012). RIKEN tandem mass spectral database (ReSpect) for phytochemicals: a plant-specific MS/MS-based data resource and database. Phytochemistry 82, 38–45. doi: 10.1016/j.phytochem.2012.07.007
Schiestl, F. P., Ayasse, M., Paulus, H. F., Löfstedt, C., Hansson, B. S., Ibarra, F., et al. (1999). Orchid pollination by sexual swindle. Nature 399, 421–421. doi: 10.1038/20829
Schiestl, F. P., Peakall, R., Mant, J. G., Ibarra, F., Schulz, C., Franke, S., et al. (2003). The chemistry of sexual deception in an orchid-wasp pollination system. Science 302, 437–438. doi: 10.1126/science.1087835
Sedeek, K. E. M., Qi, W., Schauer, M. A., Gupta, A. K., Poveda, L., Xu, S., et al. (2013). Transcriptome and proteome data reveal candidate genes for pollinator attraction in sexually deceptive orchids. PLoS One 8:e64621. doi: 10.1371/journal.pone.0064621
Shan, X., Li, Y., Yang, S., Yang, Z., Qiu, M., Gao, R., et al. (2020). The spatio-temporal biosynthesis of floral flavonols is controlled by differential phylogenetic MYB regulators in Freesia hybrida. New Phytol. 228, 1864–1879. doi: 10.1111/nph.16818
Sheehan, H., Moser, M., Klahre, U., Esfeld, K., Dell’olivo, A., Mandel, T., et al. (2016). MYB-FL controls gain and loss of floral UV absorbance, a key trait affecting pollinator preference and reproductive isolation. Nat. Genet. 48, 159–166. doi: 10.1038/ng.3462
Singer, R. B., Flach, A., Koehler, S., Marsaioli, A. J., and Amaral, M. D. C. E. (2004). Sexual mimicry in Mormolyca ringens (Lindl.) Schltr. (Orchidaceae: Maxillariinae). Ann. Bot. 93, 755–762. doi: 10.1093/aob/mch091
Strack, D., Busch, E., and Klein, E. (1989). Anthocyanin patterns in european orchids and their taxonomic and phylogenetic relevance. Phytochemistry 28, 2127–2139. doi: 10.1016/S0031-9422(00)97931-7
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Streisfeld, M. A., and Rausher, M. D. (2011). Population genetics, pleiotropy, and the preferential fixation of mutations during adaptive evolution. Evolution 65, 629–642. doi: 10.1111/j.1558-5646.2010.01165.x
Sun, J., Lin, L. Z., and Chen, P. (2012a). Study of the mass spectrometric behaviors of anthocyanins in negative ionization mode and its applications for characterization of anthocyanins and non-anthocyanin polyphenols. Rapid Commun. Mass Spectrom. 26, 1123–1133. doi: 10.1002/rcm.6209
Sun, Y., Li, H., and Huang, J. R. (2012b). Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 5, 387–400. doi: 10.1093/mp/ssr110
Suzuki, H., Nakayama, T., Yonekura-Sakakibara, K., Fukui, Y., Nakamura, N., Yamaguchi, M. A., et al. (2002). cDNA cloning, heterologous expressions, and functional characterization of malonyl-coenzyme a:anthocyanidin 3-O-glucoside-6′-O-malonyltransferase from dahlia flowers. Plant Physiol. 130, 2142–2151. doi: 10.1104/pp.010447
Tanaka, A., and Tanaka, R. (2006). Chlorophyll metabolism. Curr. Opin. Plant Biol. 9, 248–255. doi: 10.1016/j.pbi.2006.03.011
Tanaka, Y., and Brugliera, F. (2013). Flower colour and cytochromes P450. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 368:20120432. doi: 10.1098/rstb.2012.0432
Tanaka, Y., Sasaki, N., and Ohmiya, A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749. doi: 10.1111/j.1365-313X.2008.03447.x
Thill, J., Miosic, S., Ahmed, R., Schlangen, K., Muster, G., Stich, K., et al. (2012). Le rouge et le noir”: a decline in flavone formation correlates with the rare color of black dahlia (Dahlia variabilis hort.) flowers. BMC Plant Biol. 12:225. doi: 10.1186/1471-2229-12-225
Thomas, M. M., Rudall, P. J., Ellis, A. G., Savolainen, V., and Glover, B. J. (2009). Development of a complex floral trait: the pollinator-attracting petal spots of the beetle daisy. Gorteria diffusa (asteraceae). Am. J. Bot. 96, 2184–2196. doi: 10.3732/ajb.0900079
Tohge, T., De Souza, L. P., and Fernie, A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68, 4013–4028. doi: 10.1093/jxb/erx177
Tohge, T., Nishiyama, Y., Hirai, M. Y., Yano, M., Nakajima, J. I., Awazuhara, M., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42, 218–235. doi: 10.1111/j.1365-313X.2005.02371.x
Vereecken, N. J., Wilson, C. A., Hotling, S., Schulz, S., Banketov, S. A., and Mardulyn, P. (2012). Pre-adaptations and the evolution of pollination by sexual deception: cope’s rule of specialization revisited. Proc. R. Soc. B Biol. Sci 279, 4786–4794. doi: 10.1098/rspb.2012.1804
Verweij, W., Spelt, C., Di Sansebastiano, G. P., Pietro, Vermeer, J., Reale, L., et al. (2008). An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat. Cell Biol. 10, 1456–1462. doi: 10.1038/ncb1805
Vignolini, S., Davey, M. P., Bateman, R. M., Rudall, P. J., Moyroud, E., Tratt, J., et al. (2012). The mirror crack’d: both pigment and structure contribute to the glossy blue appearance of the mirror orchid. Ophrys speculum. New Phytol. 196, 1038–1047. doi: 10.1111/j.1469-8137.2012.04356.x
Waki, T., Mameda, R., Nakano, T., Yamada, S., Terashita, M., Ito, K., et al. (2020). A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat. Commun. 11, 1–14. doi: 10.1038/s41467-020-14558-9
Wang, L., Albert, N. W., Zhang, H., Arathoon, S., Boase, M. R., Ngo, H., et al. (2014). Temporal and spatial regulation of anthocyanin biosynthesis provide diverse flower colour intensities and patterning in Cymbidium orchid. Planta 240, 983–1002. doi: 10.1007/s00425-014-2152-9
Wessinger, C. A., and Rausher, M. D. (2012). Lessons from flower colour evolution on targets of selection. J. Exp. Bot. 63, 5741–5749. doi: 10.1093/jxb/ers267
Weston, P. H., Perkins, A. J., Indsto, J. O., and Clements, M. A. (2014). “Phylogeny of Orchidaceae tribe Diurideae and its implications for the evolution of pollination systems,” in Darwin’s Orchids: Then and Now, eds P. Bernhardt and R. Edens-Meier (Chicago, IL: University of Chicago Press), 91–154.
Wong, D. C. J., Amarasinghe, R., Falara, V., Pichersky, E., and Peakall, R. (2019). Duplication and selection in β-ketoacyl-ACP synthase gene lineages in the sexually deceptive Chiloglottis (Orchidaceace). Ann. Bot. 123, 1053–1066. doi: 10.1093/aob/mcz013
Wong, D. C. J., Amarasinghe, R., Pichersky, E., and Peakall, R. (2018). Evidence for the involvement of fatty acid biosynthesis and degradation in the formation of insect sex pheromone-mimicking chiloglottones in sexually deceptive Chiloglottis Orchids. Front. Plant Sci. 9:389. doi: 10.3389/fpls.2018.00839
Wong, D. C. J., Amarasinghe, R., Rodriguez-Delgado, C., Eyles, R., Pichersky, E., and Peakall, R. (2017a). Tissue-specific floral transcriptome analysis of the sexually deceptive orchid Chiloglottis trapeziformis provides insights into the biosynthesis and regulation of its unique UV-B dependent floral volatile, chiloglottone 1. Front. Plant Sci. 8:1260. doi: 10.3389/fpls.2017.01260
Wong, D. C. J., Pichersky, E., and Peakall, R. (2017b). The biosynthesis of unusual floral volatiles and blends involved in orchid pollination by deception: current progress and future prospects. Front. Plant Sci. 8:1955. doi: 10.3389/fpls.2017.01955
Wong, D. C. J., Schlechter, R., Vannozzi, A., Holl, J., Hmmam, I., Bogs, J., et al. (2016). A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res. 23, 451–466. doi: 10.1093/dnares/dsw028
Yang, X., Xia, X., Zhang, Z., Nong, B., Zeng, Y., Wu, Y., et al. (2019). Identification of anthocyanin biosynthesis genes in rice pericarp using PCAMP. Plant Biotechnol. J. 17, 1700–1702. doi: 10.1111/pbi.13133
Yoshida, K., Toyama-Kato, Y., Kameda, K., and Kondo, T. (2003). Sepal color variation of Hydrangea macrophylla and vacuolar pH measured with a proton-selective microelectrode. Plant Cell Physiol. 44, 262–268. doi: 10.1093/pcp/pcg033
Yuan, Y. W., Rebocho, A. B., Sagawa, J. M., Stanley, L. E., and Bradshaw, H. D. (2016). Competition between anthocyanin and flavonol biosynthesis produces spatial pattern variation of floral pigments between Mimulus species. Proc. Natl. Acad. Sci. U. S. A. 113, 2448–2453. doi: 10.1073/pnas.1515294113
Zhang, Y., Cheng, Y., Ya, H., Xu, S., and Han, J. (2015). Transcriptome sequencing of purple petal spot region in tree peony reveals differentially expressed anthocyanin structural genes. Front. Plant Sci. 6:964. doi: 10.3389/fpls.2015.00964
Zhang, Y., Zhou, T., Dai, Z., Dai, X., Li, W., Cao, M., et al. (2020). Comparative transcriptomics provides insight into floral color polymorphism in a Pleione limprichtii orchid population. Int. J. Mol. Sci 21, 247. doi: 10.3390/ijms21010247
Zhao, D., and Tao, J. (2015). Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 6:261. doi: 10.3389/fpls.2015.00261
Zhao, Y., Dong, W., Zhu, Y., Allan, A. C., Lin-Wang, K., and Xu, C. (2020). PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 18, 1284–1295. doi: 10.1111/pbi.13291
Zheng, J., Wu, H., Zhu, H., Huang, C., Liu, C., Chang, Y., et al. (2019). Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 223, 705–721. doi: 10.1111/nph.15807
Keywords: Chiloglottis, anthocyanin, flavonol glycoside, flower, orchids, transcriptome, sexual deception, mimicry
Citation: Wong DCJ, Perkins J and Peakall R (2022) Anthocyanin and Flavonol Glycoside Metabolic Pathways Underpin Floral Color Mimicry and Contrast in a Sexually Deceptive Orchid. Front. Plant Sci. 13:860997. doi: 10.3389/fpls.2022.860997
Received: 24 January 2022; Accepted: 17 February 2022;
Published: 23 March 2022.
Edited by:
Jen-Tsung Chen, National University of Kaohsiung, TaiwanReviewed by:
Carlos Martel, Royal Botanic Gardens, Kew, United KingdomSurendra Sarsaiya, Zunyi Medical University, China
Roman Tobias Kellenberger, University of Cambridge, United Kingdom
Copyright © 2022 Wong, Perkins and Peakall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Darren C. J. Wong, ZGFycmVuLndvbmdAYW51LmVkdS5hdQ==, d29uZ2RjakBnbWFpbC5jb20=
 Darren C. J. Wong
Darren C. J. Wong James Perkins
James Perkins