- 1College of Pharmacy, Ningxia Medical University, Yinchuan, China
- 2Shaanxi Academy of Traditional Chinese Medicine, Shaanxi Traditional Chinese Medicine Hospital, Xi’an, China
- 3Laboratory Animal Center, Ningxia Medical University, Yinchuan, China
- 4Ningxia Engineering and Technology Research Center of Regional Characterizistic Traditional Chinese Medicine, Ningxia Collaborative Innovation Center of Regional Characterizistic Traditional Chinese Medicine, Key Laboratory of Ningxia Minority Medicine Modernization, Ministry of Education, Yinchuan, China
The aim of this study was to evaluate the effect of Bacillus cereus (B. cereus) on the seedling growth and accumulation of medicinal ingredients of Glycyrrhiza uralensis Fisch. (G. uralensis) under control and salt stress conditions. Our results revealed the different effects of B. cereus on the seedling growth and accumulation of medicinal ingredients particularly in different conditions based on the transcriptome and polymerase chain reaction (PCR) analysis. Under the control condition, B. cereus significantly increased the expression level of the β-AS, SQS, CHS, LUS, UGAT, CYP72A154, and CYP88D6 genes and liquiritigenin content. Under salt stress, B. cereus significantly increased root length and lateral root number of G. uralensis seedlings, the expression level of HMGR, β-AS, CHS, LUS, UGAT, CYP72A154, CYP88D6, and SE genes, and the contents of glycyrrhizic acid and glycyrrhetinic acid. Notably, the effect of B. cereus on the seedling growth and the medicinal ingredient biosynthesis was different under control and salt stress conditions. Specifically, the effect of B. cereus on the seedling growth under salt stress was greater than that under the control condition. Moreover, B. cereus increased liquiritigenin content under the control condition, which is closely related to flavone and flavonol biosynthesis, while it increased the contents of glycyrrhizic acid and glycyrrhetinic acid under salt stress, which is closely related to phenylpropanoid biosynthesis, and the MVA pathway is also involved. All in all, endophytes B. cereus could be used as a sustainable tool to develop effective bioinoculants to enhance the contents of medicinal ingredients in G. uralensis.
Introduction
Glycyrrhiza uralensis Fisch. (G. uralensis), used as a common Chinese medicinal herb, contains abundant pharmacological active substances, which have attracted more attention (Hosseinzadeh and Nassiri-Asl, 2016). In the desert, although often suffering from drought and salt stresses, it is still an important sand fixation and windbreak plant (Zhou et al., 2020). In recent years, the wild resources are sharply declining and turning endangered, so the market demand has been markedly increasing for the cultivated G. uralensis (Chen et al., 2017). Therefore, improving the quality, yield, and stress tolerance of G. uralensis cultivated is the key task.
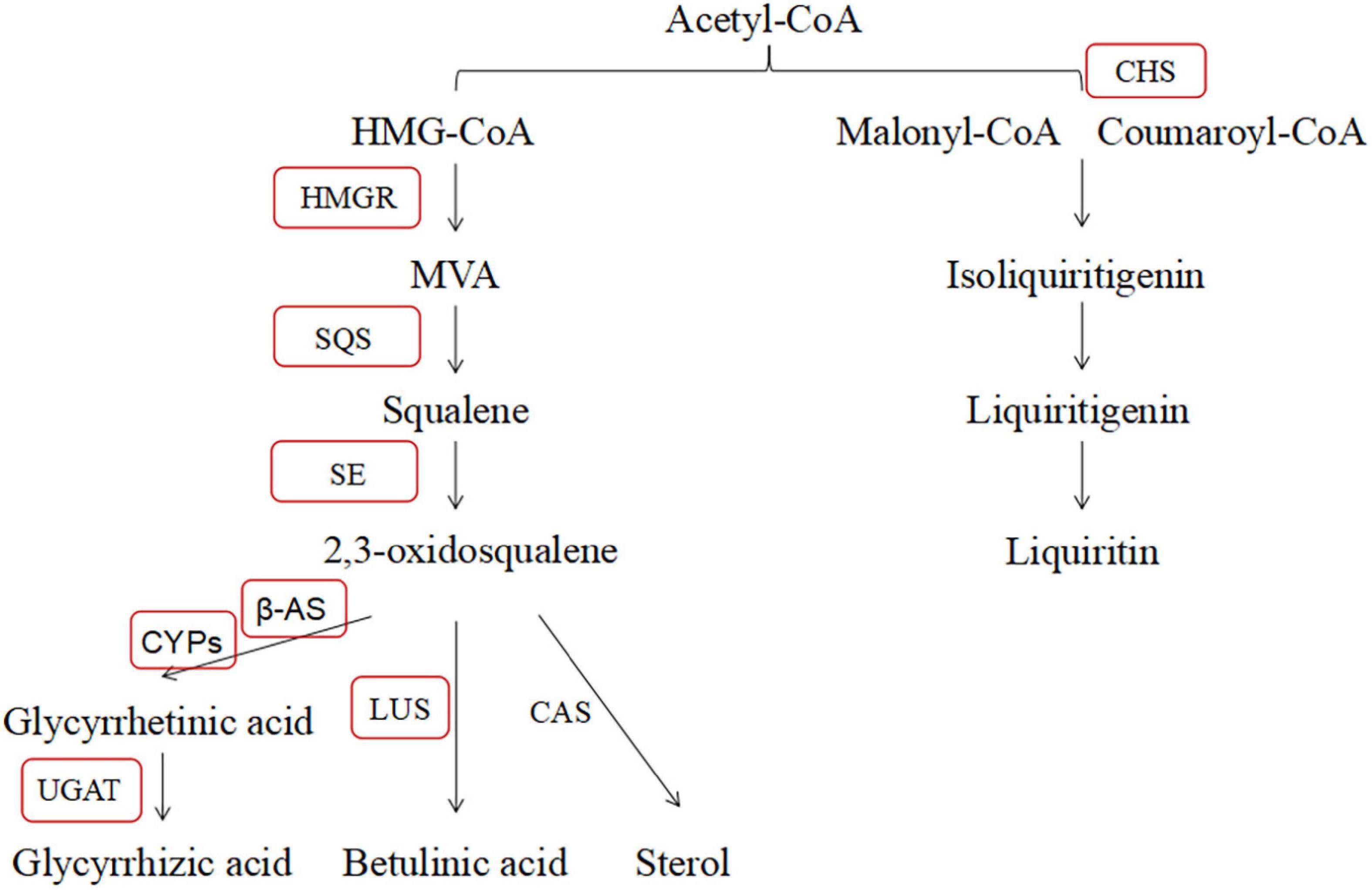
The glycyrrhizic acid and liquiritin biosynthesis pathway of G. uralensis is well known. In this pathway, HMGR catalyzes HMG-CoA and NADPH into MVA first, and farnesyl diphosphate (FDP), squalene synthase enzyme (SQS), and squalene epoxidase (SE) were involved in the subsequent reaction (Yan et al., 2014; Zheng et al., 2014). Then, β-amyrin synthase (β-AS), lupeol synthase (LUS), and cycloartenol synthase (CAS) are responsible for the synthesis of triterpene saponins, betulinic acid, and phytosterols, respectively (Mochida et al., 2017). Specifically, cytochromes P450s, UGAT, and CHS play crucial roles in the final step of major medicinal ingredient biosynthesis in G. uralensis (Xu et al., 2016; Chen et al., 2017). The content of compounds and the expression level of key genes have significantly changed in glycyrrhizic acid and liquiritin biosynthesis pathway during abiotic stress, salt stress specifically caught our attention (Wang et al., 2015, 2017; Zhou et al., 2017; Yeo et al., 2018; Kaur and Ganjewala, 2019). Shirazi et al. (2019) reported that salt stress significantly upregulated the expression level of SQS, β-AS, CAS, LUS, and some CYPs genes and increased the content of glycyrrhizin in Glycyrrhiza glabra, which illustrated that salt stress inhibited in the glycyrrhizin biosynthesis pathway. In conclusion, we provided a basis for further studies concerned with the relation between salt stress and glycyrrhizic acid pathway and liquiritin pathway in G. uralensis.
Endophytes colonized the internal tissues of host plants, promoted growth conditions, improved stress tolerance, and so on, so it is regarded as a low-cost and eco-friendly technology to enhance crop productivity under salt stress (Vaishnav et al., 2019). In promoting the growth of plant aspect, Mesorhizobium sp. and Pseudomonas extremorientalis endophytes could improve yield, nodule numbers, and nitrogen contents of shoot and root in salt-stressed G. uralensis (Egamberdieva et al., 2016). In promoting compound biosynthesis of plant aspect, endophyte Methylobacterium oryzae regulated ethylene metabolism in salt-stressed Oryza sativa by improving H+ ATPase activity and decreasing 1-Aminocylopropane-1-carboxylic acid (ACC) accumulation and ACC oxidase activity (Chatterjee et al., 2019). Also, endophytes increased withanolide A content in Withania somnifera, and significantly upregulated the expression level of genes in withanolide and sterol biosynthetic pathways including HMGR, DXR, FPPS, SQS, SQE, CAS, SMT1, STE1, and CYP710A1 (Kushwaha et al., 2019). In conclusion, endophytes can effectively mitigate high-salinity stress and improve the stress tolerance of plants, but the effect of Bacillus cereus on G. uralensis is still not fully reported, particularly in the glycyrrhizic acid and liquiritin biosynthesis pathway aspect.
Consequently, in this study, we hypothesized that B. cereus can promote the growth of seedlings and increase the contents of medicinal ingredients (liquiritin, liquiritigenin, isoliquiritigenin, glycyrrhizic acid, and glycyrrhetinic acid) of G. uralensis under control and salt stress conditions. Furthermore, we want to explain the effect that B. cereus increased the contents of medicinal ingredients under different conditions based on the molecular level. Finally, we expected to unravel the underlying effect for the interactions of B. cereus and salt stress, provide an efficient solution for promoting the quality of G. uralensis cultivation, and set a solid foundation for B. cereus promoting the biosynthesis of glycyrrhizic acid and liquiritin.
Materials and Methods
Preparation of Bacillus cereus
The obtained target strain was isolated and selected from the early stage G. uralensis and identified by partial sequencing of 16SrDNA as B. cereus by Bioengineering (Shanghai) Co., Ltd. This strain was deposited at the China General Microbiological Culture Collection Center (No. CGMCC No. 16671). The strain was stored in 25% glycerol solution at −70°C until use. The bacterial culture suspension was incubated at 28°C for 2 days in 180 rpm, on a shaking incubator until bacteria density reached 108 cfu ml–1.
Plant Growth Condition and Treatment
Glycyrrhiza uralensis seeds were obtained from Ningxia Academy of Agriculture and Forestry Sciences, China, in September 2018. The seeds were steeped with 85% H2SO4 for 2.5 h, then 0.1% H2O2 surface-sterilized for 15 min, distilled water rinsed three times, and distilled water imbibed for 8 h at room temperature. Then, 50 seeds were sown in a 500 ml flowerpot (contains 400 g of sand) not containing any nutrients and applied to different treatment fluids separately. Then, pre-irrigated with distilled water (300 ml) as the control group and distilled water containing 75 mM NaCl as the salt stress group. Germination experiments were carried out in an incubator maintained at 28°C/20°C (day/night) with a 12 h photoperiod at a light intensity of 37.5 μmol m–2 s–1 PAR. After 23 days, B. cereus treatment was initiated. The bacteria culture suspension with B. cereus was centrifuged for 15 min at 8,000 rpm and washed with sterile distilled water, and then the optical density of the B. cereus at 600 nm was adjusted to 1 (∼108 cfu ml–1). Then, both CK and S groups were divided into two subgroups and were watered with either 300 ml distilled water or 300 ml of distilled water containing B. cereus (108-cfu ml–1). Six replications of all treatments were used.
At 7 days after B. cereus treatment, all the seedlings were collected from the flowerpot. Then, growth indicators were recorded, some were oven-dried at 60°C for 48 h and weighed, and some samples were used for HPLC analysis. Some seedlings were taken out and soil particles adhering to the root surface were gently removed with distilled water, for monitoring the colonization of B. cereus. Additionally, some samples were immediately stored at −80°C for transcriptomic and real-time PCR.
Observation of Colonization on Bacillus cereus
Samples of root segments (1 cm) were fixed in a mixture of 3% (v/v) glutaraldehyde and 2% (w/v) paraformaldehyde in 0.1 M phosphate buffer (pH = 7), overnight at room temperature. Samples were subsequently postfixed with 1% (w/v) osmium tetroxide in the same buffer for 1 h at 4°C and dehydrated in a graded ethanol series before being embedded in SPURR resin. Ultrathin sections were contrasted with uranyl acetate and lead citrate and examined with a Hitachi H-7650 TEM at 80 kV (Vigani et al., 2010).
Measurement of Content on Major Active Compounds
The standard of all active compounds was purchased from Shanghai Jianglai Biological Technology Co., Ltd. Preceding HPLC analysis, extracts were dissolved in a small volume of 70% ethyl alcohol and filtered using a 0.22 μm microporous membrane. A 20 μl aliquot of each sample extract was analyzed via HPLC (Agilent 1260 Infinity II) at 25°C. The determination of liquiritin, liquiritigenin, isoliquiritigenin, glycyrrhizic acid, and glycyrrhetinic acid was performed according to the Zheng et al. (2013) method. Acetonitrile–0.1% aqueous solution of phosphoric acid was used as the mobile phase for gradient elution, the eluent was collected with the gradient-wavelength assay, and the flow rate was 0.6 ml min–1, followed by measurement of UV absorbance at 254 nm. Analysis of variance was performed on the data using SPSS software (version 18.0), and significant differences compared with the control values were determined using the Duncan’s multiple range test.
Transcriptome Sequencing
Of note, 1 μg total RNA was prepared for cDNA libraries using the protocol provided by Oxford Nanopore Technologies (ONT). In brief, the SuperScript IV First-Strand Synthesis System (Invitrogen) was used for full-length mRNA reverse transcription and following cDNA PCR for 14 circles with LongAmp Tag (NEB). The PCR products were then subjected to FFPE DNA repair and end-repair (NEB) steps and following adaptor ligation using T4 DNA ligase (NEB). Agencourt XP beads were used for DNA purification according to the ONT protocol. The final cDNA libraries were added to FLO-MIN109 flow cells and were run on the PromethION platform at Biomarker Technology Company (Beijing, China).
Transcriptome Assembly and Annotation
Raw reads of fastq format were first processed through in-house Perl scripts. In this step, clean reads were obtained by removing reads containing adapter, reads containing ploy-N, and low-quality reads from raw data. At the same time, Q30 and sequence duplication levels of the clean data were calculated. All the downstream analyses were based on clean data with high quality. The adaptor sequences and low-quality sequence reads were removed from the data sets. Raw sequences were transformed into clean reads after data processing. These clean reads were then mapped to the reference genome sequence. Only reads with a perfect match or one mismatch were further analyzed and annotated based on the reference genome. Hisat2 tools soft were used to map with the reference genome. Gene function was annotated based on the following databases: all the plant proteins in the NCBI NR database in performing the homology search, and for each sequence, we selected the closest match for analysis. We used KOBAS software to test the statistical enrichment of differential expression genes in KEGG pathways (Kanehisa et al., 2004).
Differential Expression Analysis
The threshold of p-value < 0.05 and log2 (fold change)| > 1.5 were set as the two criteria for significantly differential expression.
Real-Time PCR
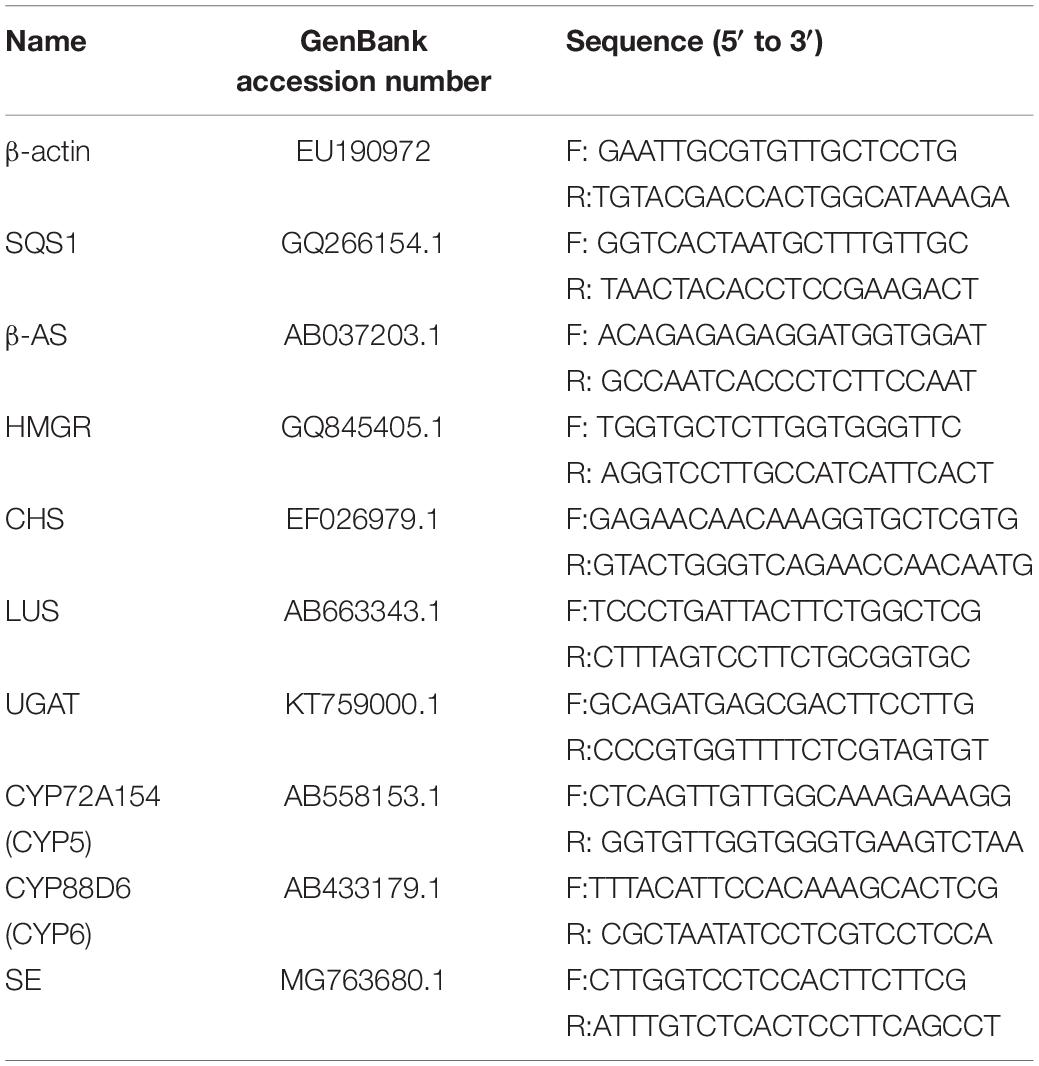
The total RNA was extracted using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China) from G. uralensis seedlings in each experimental group. Then, 200 μg of total RNA from each sample was reverse transcribed to single-stranded cDNA using the FastQuant RT Kit (TIANGEN) according to the manufacturer’s protocol. The primers were designed using the Primer Premier 5 software (premier biosoft, Palo Alto, CA, United States; Table 1). For RT-PCR analysis, the target genes were amplified using the PCR amplification system (ABI 2720, Carlsbad, CA, United States) under the following conditions: 95°C for 4 min followed by 30 cycles consisting of 95°C for 20 s, 58°C for 20 s, 72°C for 20 s, and, 72°C for 10 min. The PCR products were analyzed using the FR-980B gel imaging analysis system (FuRi Science & Technology Co., Ltd., Shanghai, China) after 1% agarose gel electrophoresis. β-Actin gene was used as the reference gene.
Statistical Analysis
All experimental data were analyzed using ANOVA, SPSS 17.0 software (SPSS Inc., Chicago, IL, United States). Significant differences were tested using the least significant difference (LSD) test at p < 0.05. Mean values and standard errors (SEs) were presented. Pearson’s correlation analysis was performed when the variables were approximately normally distributed and had no outliers.
Results
Effect of Bacillus cereus on the Growth Condition of Glycyrrhiza uralensis Seedlings
The growth condition of G. uralensis seedlings and colonization of B. cereus are shown in Figures 1A,B. Salt stress significantly inhibited the root length of G. uralensis seedlings. The effect of B. cereus on seedling growth under salt stress was greater than that under the control condition. Under salt stress, B. cereus significantly increased the root length and the number of lateral roots (Figure 1A). In conclusion, B. cereus significantly improved the growth condition of G. uralensis seedlings under salt stress.
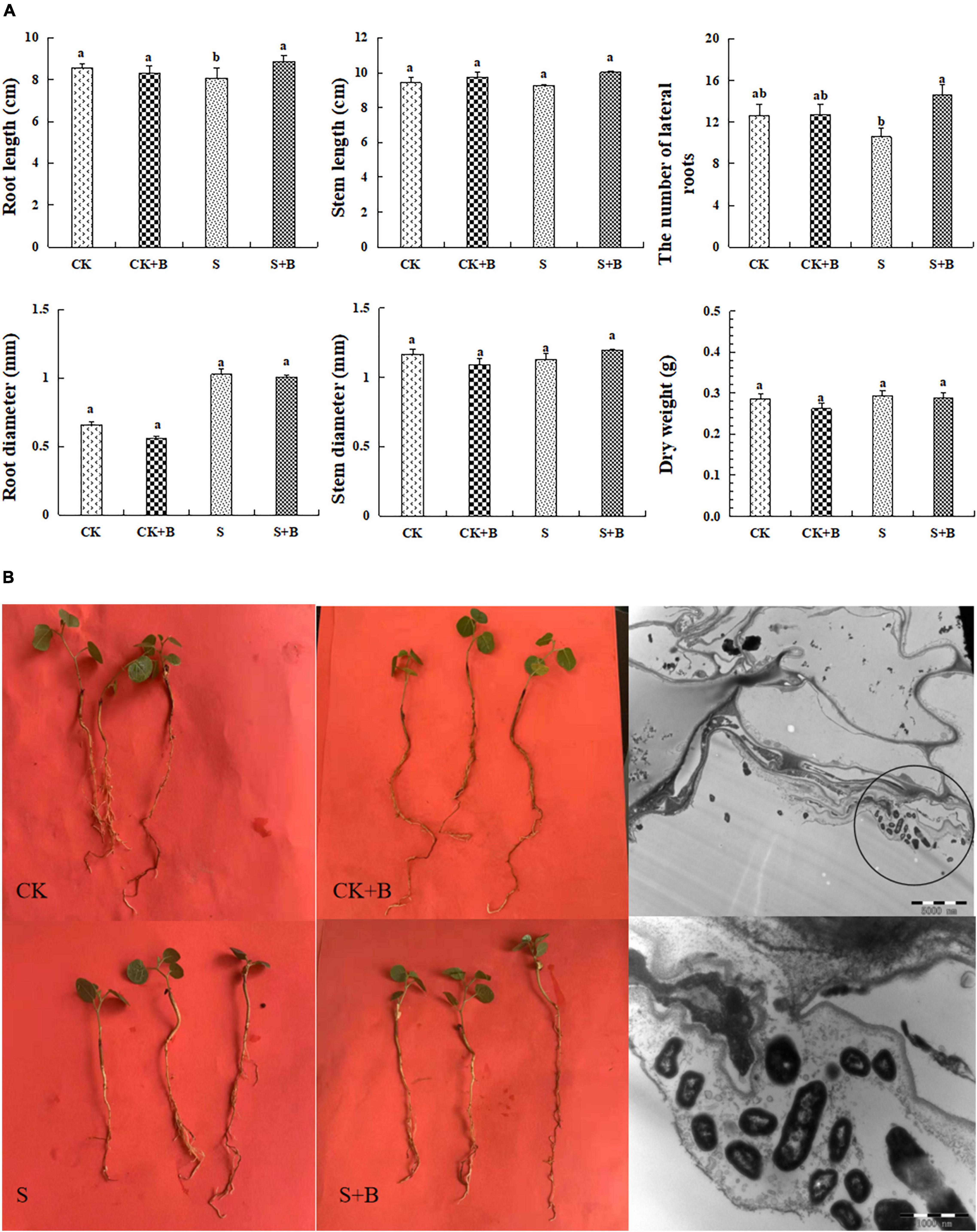
Figure 1. (A) Effect of B. cereus on seedlings growth of G. uralensis under control (CK) and salt stress (S) conditions. Data are expressed as the mean ± SE (n = 6). The different letters within the different treatments indicate the significant difference at P < 0.05. (B) The growth condition of seedlings under different treatments and colonization of B. cereus in the roots in G. uralensis.
Effect of Bacillus cereus on the Content of Major Active Compounds
As shown in Figure 2, compared with the control treatment, salt stress significantly increased the content of liquiritin and liquiritigenin. Compared with the salt stress, the content of glycyrrhetinic acid showed a higher level under the control treatment. The effect of B. cereus on the content of major active compounds under salt stress was greater than those under control conditions. Under the control condition, B. cereus significantly increased liquiritigenin content. Under salt stress, B. cereus significantly increased the contents of glycyrrhizic acid and glycyrrhetinic acid.
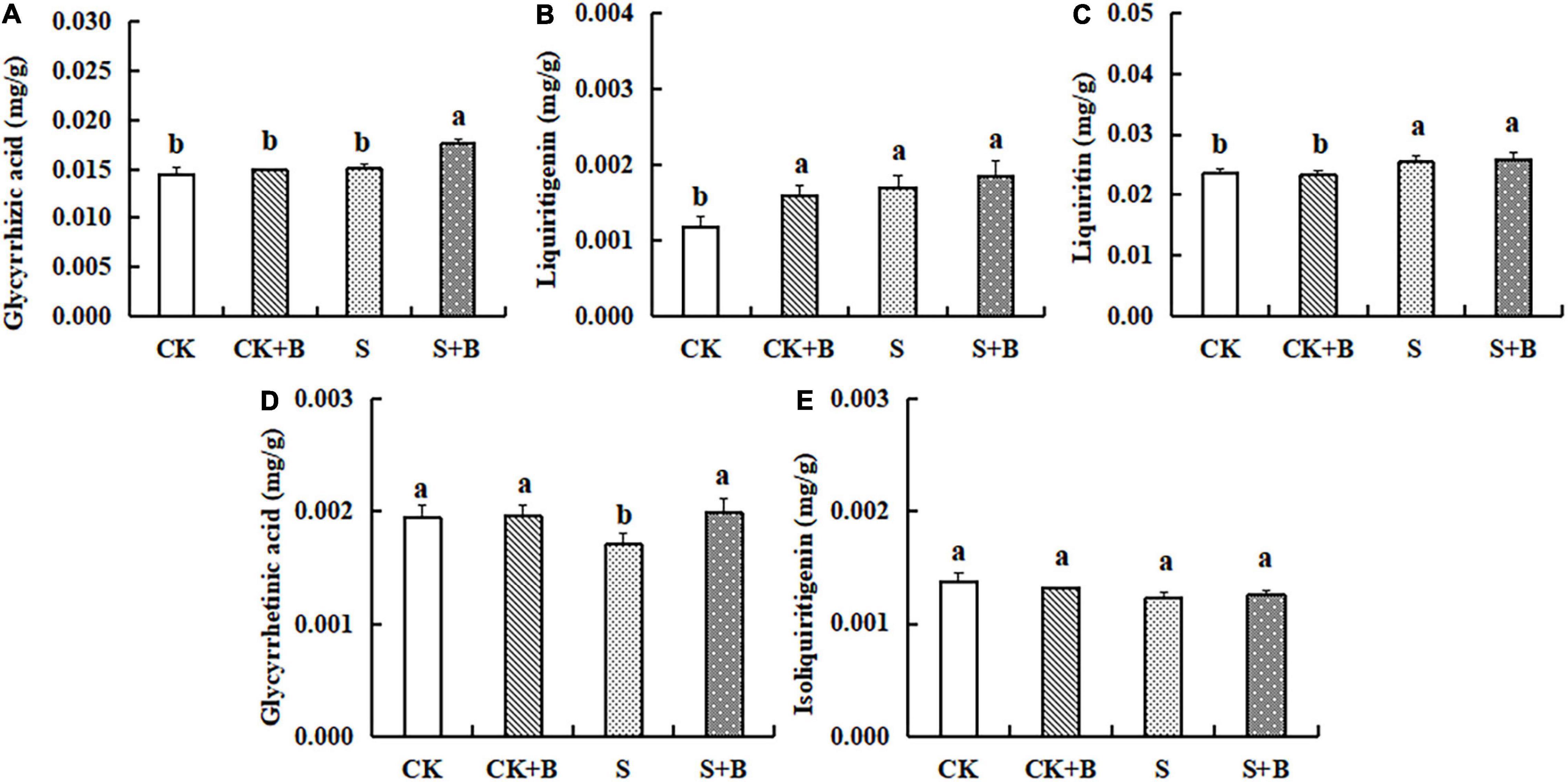
Figure 2. Effect of B. cereus on the content of (A) glycyrrhizic acid, (B) liquiritigenin, (C) liquiritin, (D) glycyrrhetinic acid, and (E) isoliquiritigenin of G. uralensis under control (CK) and salt stress (S) conditions. Data are expressed as the mean ± SE (n = 6). The different letters within the different treatments indicate the significant difference at P < 0.05.
Transcriptomic Data Analysis for the Effect of Bacillus cereus on Glycyrrhiza uralensis
As shown in Figure 3, the Venn diagram of the expression level of upregulated gene comparisons was found by caused B. cereus under control and salt stress conditions. B. cereus upregulated the expression level of 215 genes under the control condition and upregulated the expression level of 257 genes under salt stress. In the KEGG database, the upregulated DEGs by B. cereus under the control condition were mainly enriched in carbon metabolism (13.79%), glycolysis/gluconeogenesis (10.34%), flavone and flavonol biosynthesis (6.9%), flavonoid biosynthesis (6.9%), and diterpenoid biosynthesis (6.9%; Figure 4A). Additionally, the upregulated DEGs by B. cereus under salt stress were mainly enriched in phenylpropanoid biosynthesis (8.0%), plant hormone signal transduction (8.0%), starch and sucrose metabolism (8.0%), plant–pathogen interaction (6.0%), and glycerophospholipid metabolism (6.0%; Figure 4B).
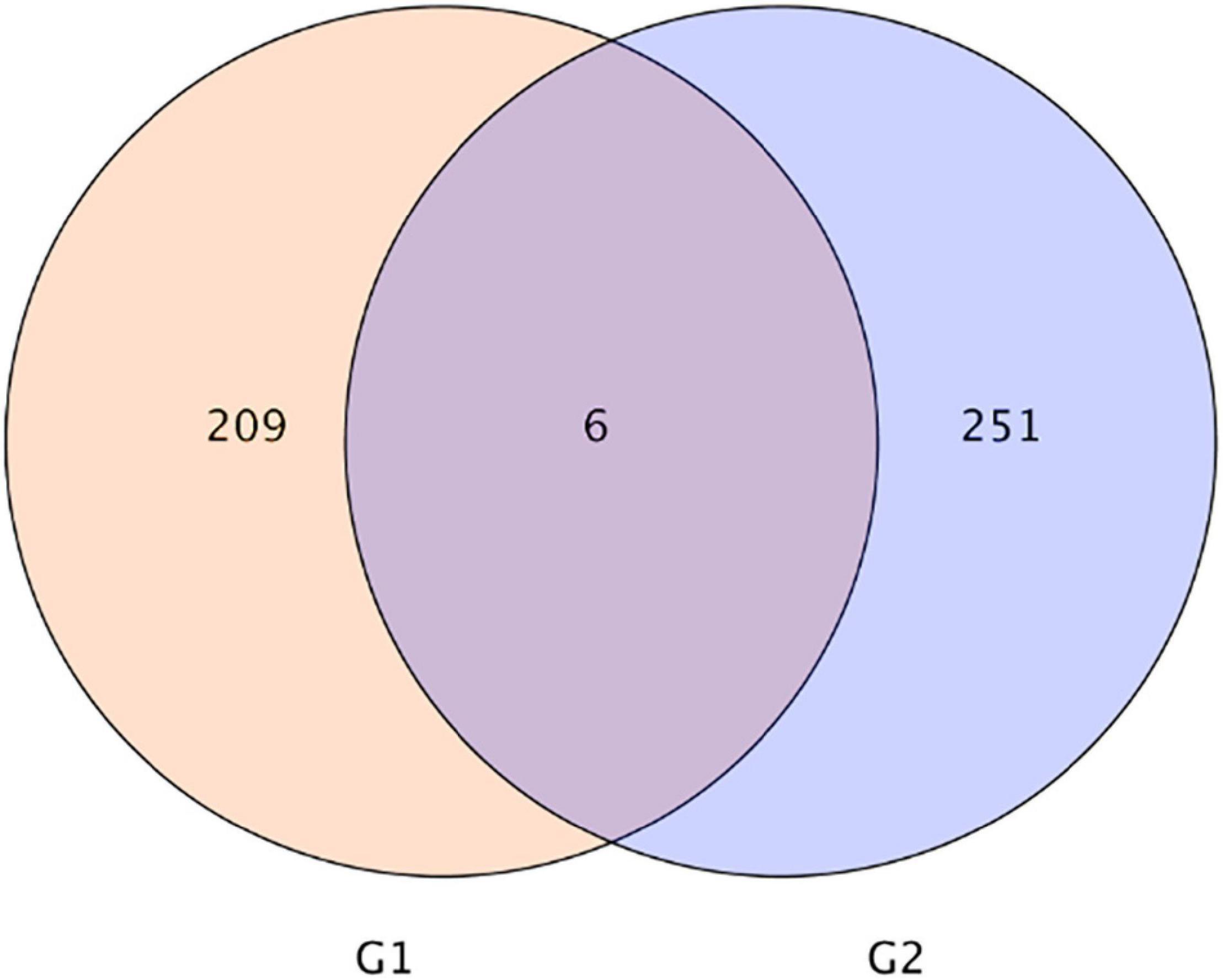
Figure 3. |Venn diagram of DEGs of control condition combined with B. cereus (CK+B) vs control (CK) and salt stress combined with B. cereus (S+B) vs salt stress (S) comparison in G. uralensis. G1, DEGs of CK+B vs CK; G2, DEGs of S+B vs S.
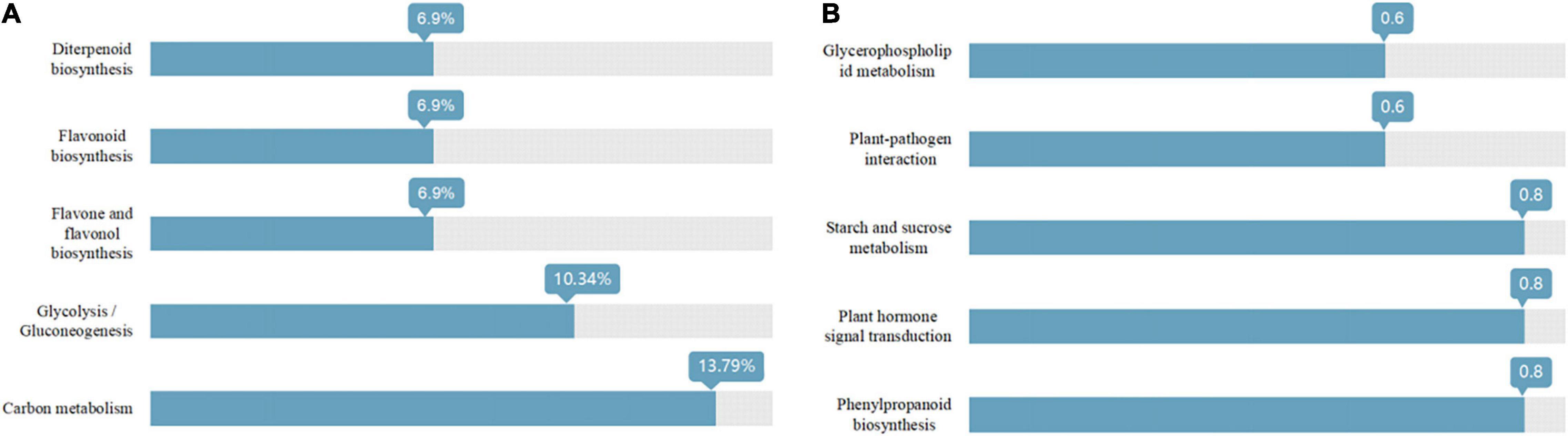
Figure 4. KEGG annotation of the DEGs (only up-regulated) in control condition combined with B. cereus (CK+B) vs control (CK) (A) and salt stress combined with B. cereus (S+B) vs salt stress (S) (B) comparisons in G. uralensis (Selected the top five items).
Effect of Bacillus cereus on Gene Expression of Glycyrrhizic Acid and Liquiritin Biosynthesis Pathway
As shown in Figure 5, compared with the control condition, the expression level of HMGR, β-AS, SQS, and SE genes was lower level, but CHS and CYP5 were higher under salt stress. B. cereus enhanced the gene expression level of glycyrrhizic acid and liquiritin biosynthesis pathway, but the effect of B. cereus was different under control and salt stress conditions. The expression level of SQS, CHS, and CYP5 genes was higher caused by B. cereus under the control condition compared with the control, and the expression level of HMGR, β-AS, and CYP5 genes was higher under salt stress compared with the salt stress.
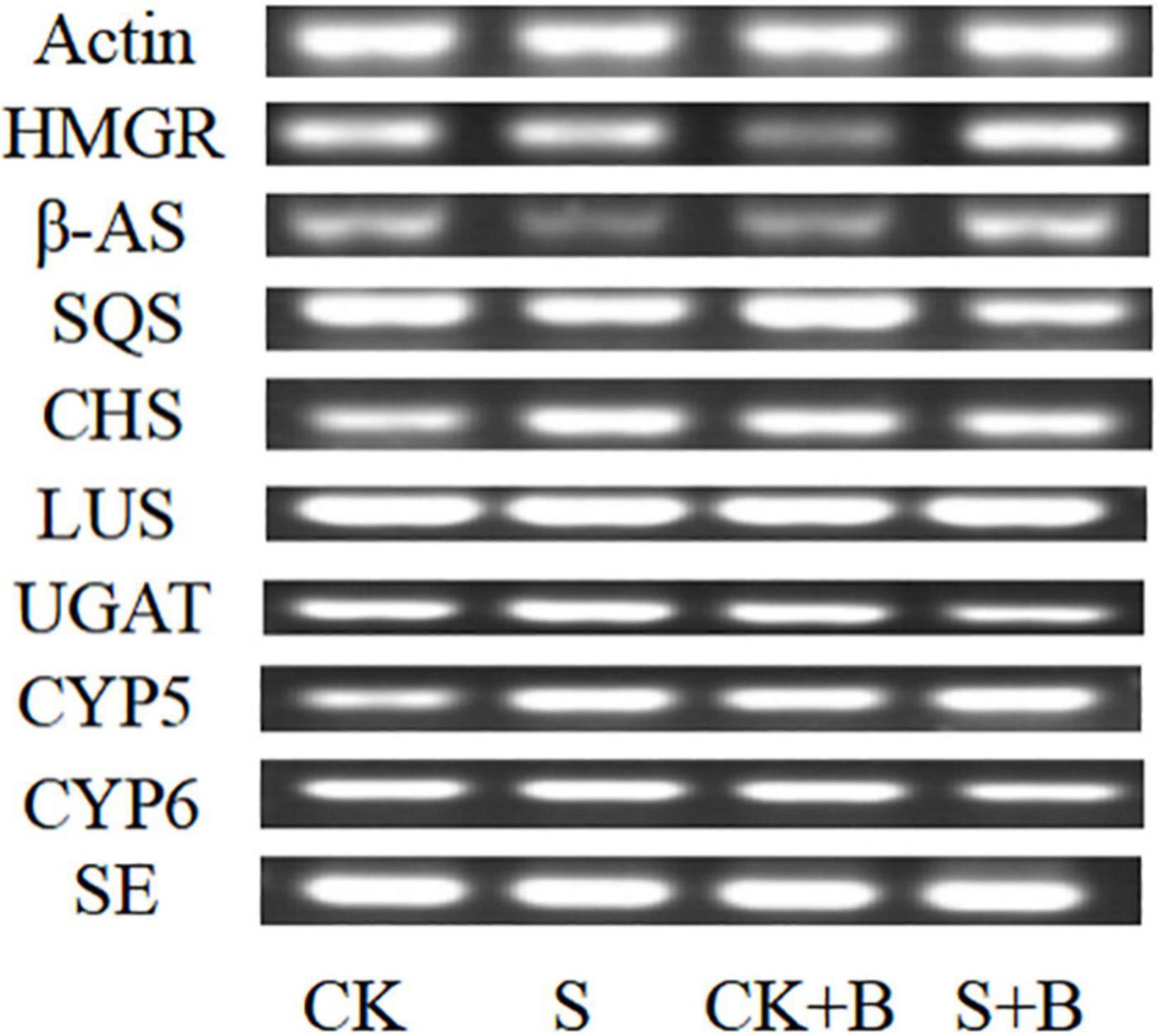
Figure 5. Expression level of glycyrrhizic acid and liquiritin biosynthesis pathway-related genes in G. uralensis seedlings under control (CK), salt stress (S), control condition combined with B. cereus (CK+B), and salt stress combined with B. cereus (S+B).
As shown in Figures 6, 7, compared with the control condition, the highest fold-decrement in the β-AS was observed under salt stress (0.55-fold decrease), followed by HMGR (0.34-fold decrease) and UGAT (0.14-fold decrease). While the highest fold-increment in the LUS was observed (2.92-fold increase) under control condition inoculation with B. cereus, followed by β-AS (1.56-fold increase), SQS (1.01-fold increase), UGAT (0.57-fold increase), CYP72A154 (0.55-fold increase), CHS (0.53-fold increase), and CYP88D6 (0.12-fold increase). In contrast, the highest fold increment in the LUS was observed (0.67-fold increase) under salt stress inoculation with B. cereus, followed by CHS (0.50-fold increase), HMGR (0.37-fold increase), UGAT (0.29-fold increase), β-AS (0.19-fold increase), CYP88D6 (0.17-fold increase), SE (0.17-fold increase), and CYP72A154 (0.16-fold increase).
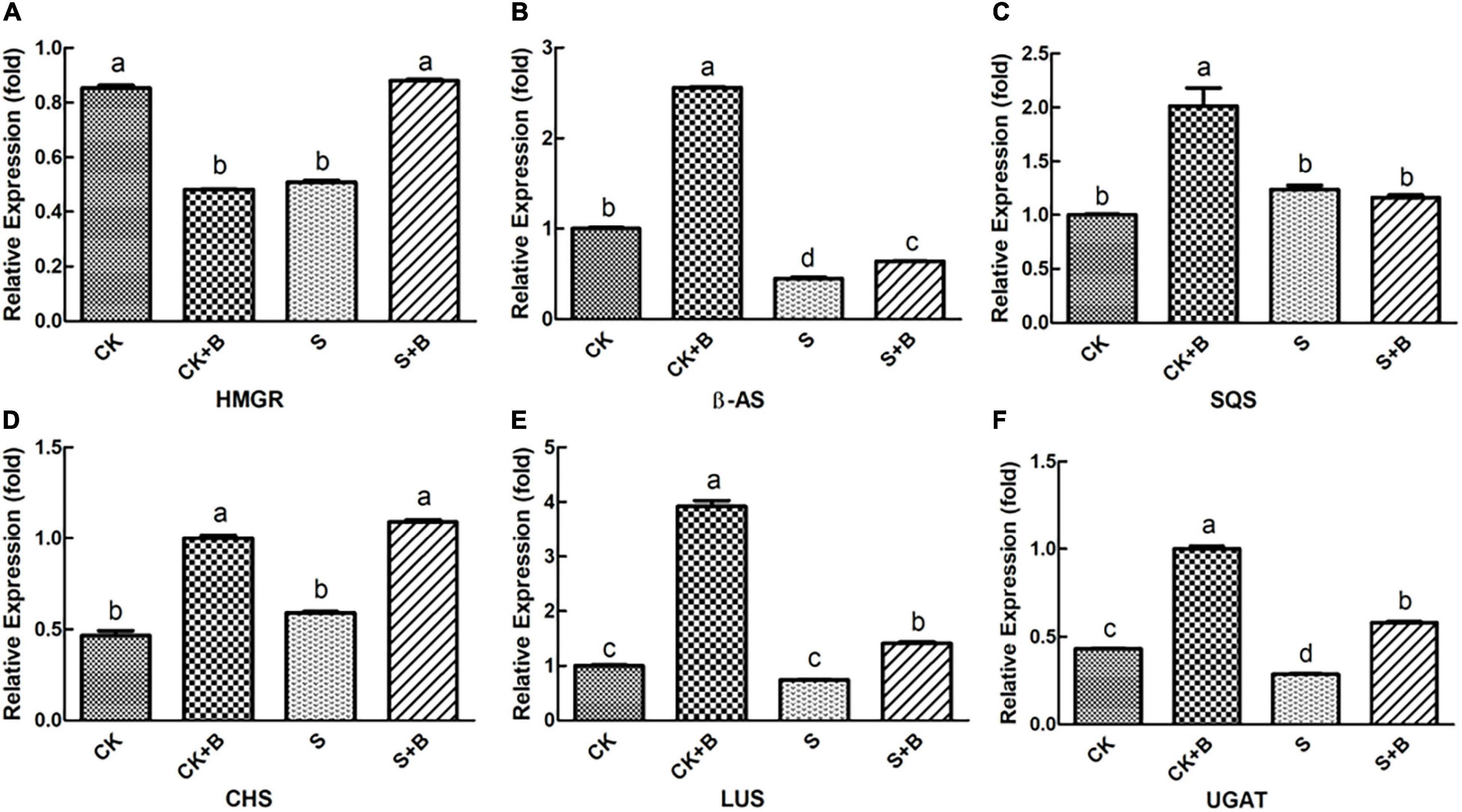
Figure 6. Effect of B. cereus on expression level of (A) HMGR, (B) β-AS, (C) SQS, (D) CHS, (E) LUS, and (F) UGAT genes of G. uralensis under control (CK) and salt stress (S) conditions. Data are expressed as the mean ± SE (n = 6). The different letters within the different treatments indicate the significant difference at P < 0.05.
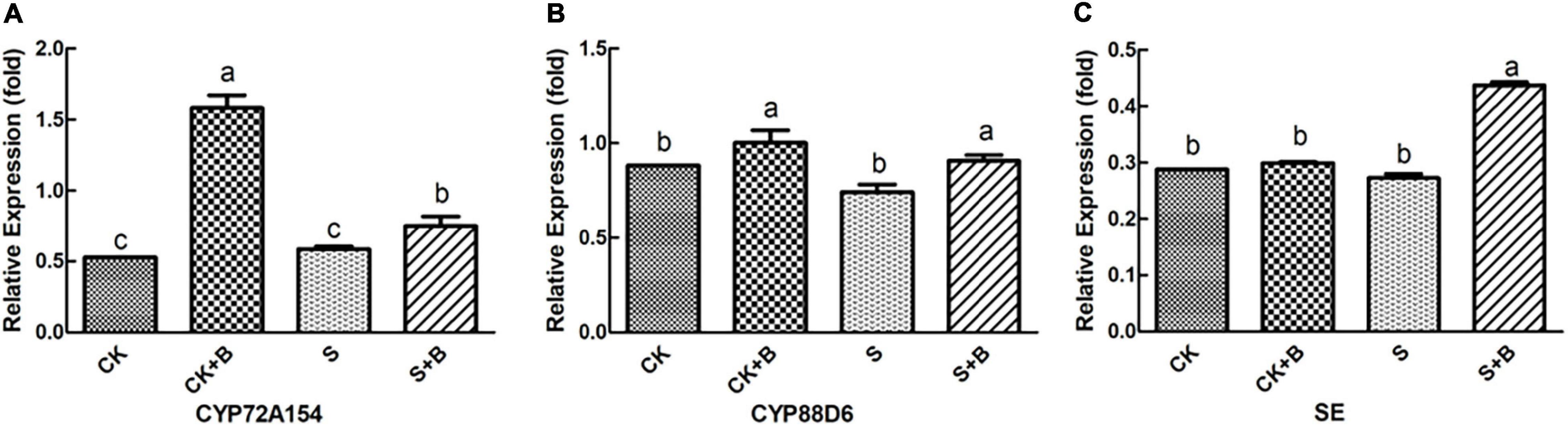
Figure 7. Effect of B. cereus on expression level of (A) CYP72A154, (B) CYP88D6, and (C) SE genes of G. uralensis under control (CK) and salt stress (S) conditions. Data are expressed as the mean ± SE (n = 6). The different letters within the different treatments indicate the significant difference at P < 0.05.
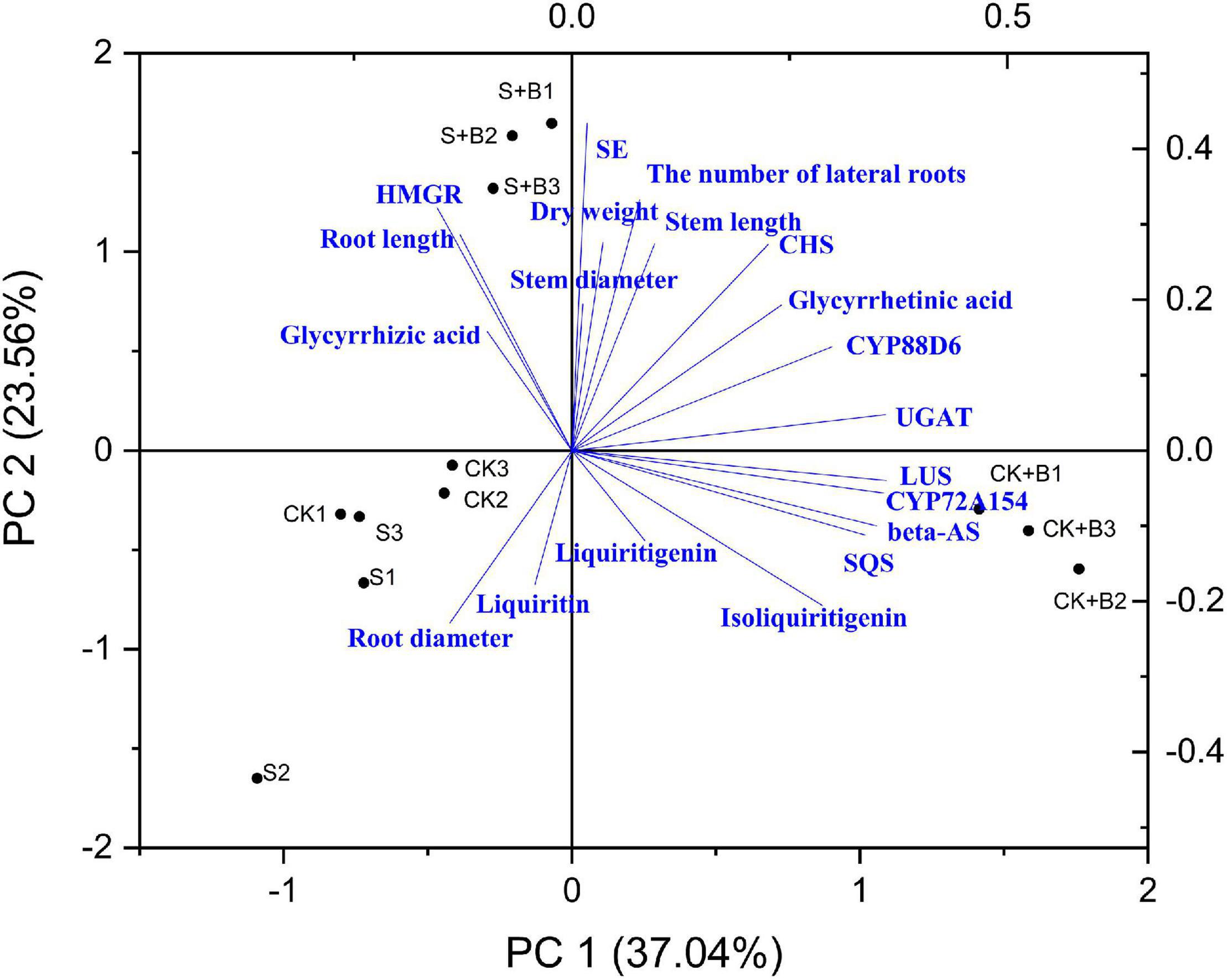
Correlation Analysis and Principal Component Analysis
As shown in Figures 7, 8, data of groups of very significant positive relations (p < 0.01) were observed. Additionally, data of 3 groups of significant positive relations (p < 0.05) were observed. In Figure 9, the first and second principal components (PCs), respectively, reflect 37.04 and 23.56% variation of the experimental data and altogether explain 60.60% variation. The biosynthesis pathway of glycyrrhizic acid majorly contributes to PC2 and PC1 formation. The sample distribution within the PC1/PC2 plane is heterogeneous, and PC2 clearly separated B. cereus-treated plants under the salt stress condition from control and salt-treated plants due to regulation of the biosynthesis pathway of glycyrrhizic acid under salt stress. PC1 significantly separated B. cereus-treated plants under the control condition from control and salt-treated according to the similar effect.
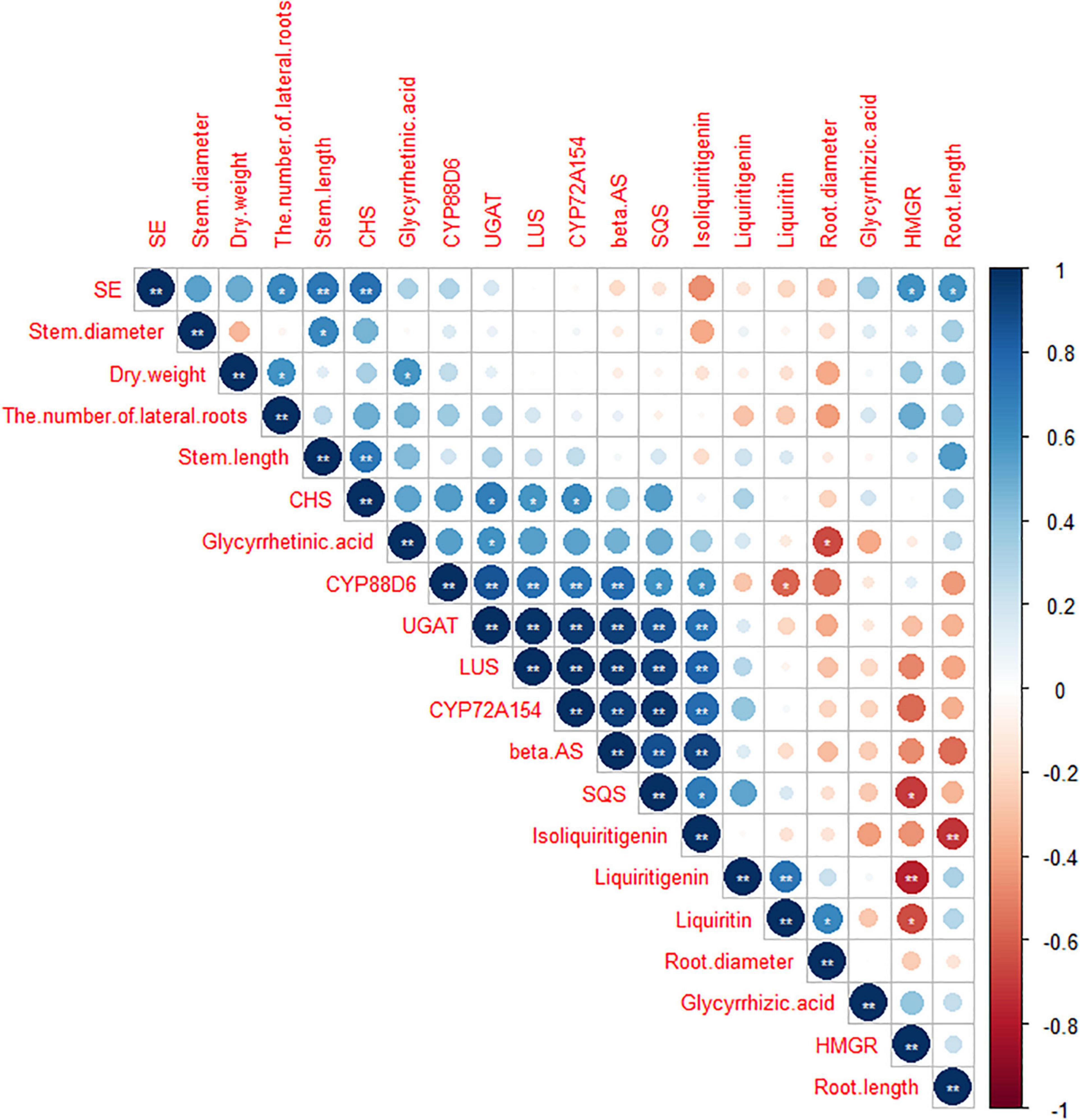
Figure 8. Correlation analysis among seedlings growth, medicinal ingredients, and secondary metabolites pathway in G. uralensis under four treatments. **Correlation is significant at the 0.01 level (two-tailed). *Correlation is significant at the 0.05 level (two-tailed).
Discussion
In general, salt is a necessary nutrient element for the normal growth of plants and also plays an important role in the physiological and biochemical functions of plants (such as osmotic adjustment, antioxidant system, and carbon and nitrogen metabolism), but excessive concentration will cause plant death (Jose et al., 2017; Kanagaraj and Sathish, 2017). Salt stress inhibited the root growth of plants, reduced the activity of root tip cells mitotic, and increased abnormalities such as chromosome breaks and bridges and formation of micronuclei, which belongs to the cytotoxic effect (Kiełkowska, 2017). In this experiment, salt stress significantly decreased the root length of seedlings. Salt stress would affect the absorption of water and nutrients by damaging the roots of plants and further affect the supply of water and nutrients above the ground (Guo et al., 2015). Surprisingly, one biological method is the application of plant growth-promoting bacteria to decrease the deleterious effect of salinity (Trdan et al., 2019). Under salt stress, B. cereus significantly increased the root length and the number of lateral roots, but have no significant effect on dry weight, which could attribute to need more time for accumulate the effect of inoculation B. cereus on dry weigh. Plant growth-promoting bacteria that can alleviate salt stress have been reported in pepper (Amor and Cuadra-Crespo, 2012), barley (Suarez et al., 2015), okra (Habib et al., 2016), and Medicago sativa L. (Ansari et al., 2019). In addition, we also found that the effect of B. cereus on regulating the growth condition was greater under salt stress than these under the control condition, probably because the microorganisms can secrete ACC deaminase, degrade ethylene synthesis precursors, and regulate ethylene concentration under salt stress, which is an important mechanism for microorganisms to enhance plant stress resistance (Pande et al., 2016). In summary, B. cereus can promote the growth of G. uralensis seedlings, but the effect was greater under salt stress than that under the control condition.
In the past, Ponce et al. (2004) observed inoculation of Glomus intraradices increased flavonoid content from roots and shoots of Trifolium repens through increasing the root colonization, which may refer to the plant metabolism to secrete different flavone derivatives. At present, it is difficult to explain meticulously the molecular mechanism of endophytes on active compound biosynthesis because it is difficult to evaluate the potential functional relationships in traditional Chinese medicine, so we will try to analyze the molecular effect of B. cereus on improving the content of active compounds in G. uralensis based on transcriptome and PCR results. First, we observed that B. cereus increased glycyrrhizin content under the control condition and the contents of glycyrrhizic acid and glycyrrhetinic acid under salt stress (Figure 2). Second, our KEGG annotation results revealed that B. cereus increased liquiritigenin content is closely related to regulate flavone and flavonol biosynthesis under the control condition (Figure 4A; Berim and Gang, 2016), and B. cereus increased contents of glycyrrhizic acid and glycyrrhetinic acid are closely related to the regulation of phenylpropanoid biosynthesis under salt stress (Figure 4B; Li et al., 2016).
Liquiritigenin is one of the major active metabolites in G. uralensis, which belongs to flavonoid compounds and plays important physiological roles in protecting plants from biotic and abiotic stresses (Hosseinzadeh and Nassiri-Asl, 2016). Since liquiritigenin belongs to flavonoid compounds, our results explained that B. cereus increased the liquiritigenin content by upregulating the expression level of genes on the flavone and flavonol biosynthesis pathway under the control condition. Under the salt stress condition, B. cereus increased the contents of glycyrrhizic acid and glycyrrhetinic acid, this is a complicated process. These results are also consistent with our correlation analysis and PC analysis results. According to previous studies, glycyrrhizic acid and glycyrrhetinic acid are synthesized mainly by the MVA pathway, and this pathway is closely related to the cytochrome P450s (Pandian et al., 2020). Cytochrome P450s may account for the biosynthesis of triterpene saponins, phenylpropanoid, and fatty acids in plant-specific organs and further modified oxidation and glycosylation (Yendo et al., 2010; Liao et al., 2020). Phenylpropanoid compounds play a pivotal role particularly in signaling molecules both within the plant and in communications with other microbes, response to environmental conditions, and improving crop yield and quality (Ali and McNear, 2014). Also, we found that more than sixteen cytochrome P450s have been implicated in the phenylpropanoid metabolism pathway because most are derived from phenylalanine by the phenylpropanoid pathway leading to 4-coumaroyl-CoA (Pandian et al., 2020). In our study, B. cereus increased the contents of glycyrrhizic acid and glycyrrhetinic acid, upregulated the expression level of CYP88D6 and CYP72A154 genes, and upregulated the level of DEGs in phenylpropanoid biosynthesis under salt stress, which showed that glycyrrhizic acid and glycyrrhetinic acid, CYPs, and phenylpropanoid biosynthesis are closely related. Moreover, the complex relationships between B. cereus and glycyrrhizic acid and liquiritin biosynthesis pathway remain to be further studied.
In contrast, we interpreted the effect of B. cereus improving the contents of active compounds of G. uralensis based on PCR results. According to a large number of reports, we summarized the major active compounds and the enzymes of the secondary metabolic biosynthesis pathway in G. uralensis (Figure 10). In past studies, the effect of high salt concentration on the content of effective ingredients depends on different plant types, growth stages, and action sites (Wang et al., 2016). Behdad et al. (2020) found that 100 mM NaCl significantly upregulated the expression level of key genes (β-AS, CYP88D6, and CYP72A154) in Glycyrrhiza glabra L. involved in triterpenoid saponin biosynthesis and directly increased the content of glycyrrhizic acid. Shirazi et al. (2019) also found that salt stress upregulated the expression level of key genes (including SQS, β-AS, LUS, CAS, CYP88D6, and CYP93E6) in Iranian licorice. Moreover, Wang et al. (2020) reported that 200 mM NaCl upregulated the expression level of key enzymes (β-AS, CYP88D6, and CYP72A154) involved in the glycyrrhizic acid biosynthesis pathway in G. uralensis. Our results are different from theirs, and in this study, salt stress significantly downregulated the expression level of HMGR, β-AS, and UGAT genes, upregulated the expression level of CYP5 and CHS genes, significantly decreased glycyrrhetinic acid content, and significantly increased the contents of liquiritin and liquiritigenin, which is consistent with the biosynthesis pathway of active compounds in G. uralensis. Salt stress downregulated the expression level of HMGR, β-AS, and UGAT genes, reducing the content of compounds in glycyrrhizic acid synthesis pathway; at the same time, salt stress upregulated the expression level of CYP5 and CHS genes, increasing the content of compounds involved in the liquiritin synthesis pathway.
A large number of studies have found that endophytes can improve the accumulation of active compounds in plants. Native endophytes of Coleus forskohlii significantly promoted the plant growth (shoot weight, oil content, and oil yield) and increased the contents of davanone and ethyl cinnamate in Andrographis paniculata, Pelargonium graveolens, and Artemisia pallens (Mastan et al., 2019). Endophytes Phoma sp., Alternaria sp., Bipolaris sp., and Cladosporium sp. significantly increased seed germination, promoted plant growth, and increased the contents of flavonoid and phenolic in rice (Khan et al., 2017). Particularly in the salt stress, it has been reported that endophytes have many beneficial effects on salt-stressed plants including the production of scavengers, ACC deaminase enzyme, nitrogen fixation, production of compatible solutes, antibiotics, and phytohormones (Fouda et al., 2018). Amanifar et al. (2019) found that arbuscular mycorrhizal increased the content of glycyrrhizic acid and upregulated the expression level of β-AS and P450 genes in Glycyrrhiza glabra under salt stress. This is similar to our research results that B. cereus upregulated the expression level of key genes and the content of active compounds involved in the glycyrrhizic acid and liquiritin biosynthesis pathway, which indicated that B. cereus induced the accumulation of major active compounds in G. uralensis through the upregulation expression level of MVA and downstream pathway genes such as HMGR, β-AS, SQS, CHS, LUS, UGAT, CYP72A154, CYP88D6, and SE. Moreover, the regulatory effect of B. cereus was different under control and salt stress conditions. HMGR acts as the catalyst for the first committed reaction of the mevalonate pathway, which is critical for plant adaptations against harsh environmental conditions (Zheng et al., 2021), so the expression level of HMGR gene was higher by B. cereus under salt stress. Moreover, B. cereus upregulated the expression level of many key genes of G. uralensis in both control and salt stress conditions, but only increased the content of liquiritigenin under the control condition and increased the contents of glycyrrhizic acid and glycyrrhetinic acid under the salt stress condition. Interestingly, this result is consistent with the results of transcriptome analysis, which explained that the effect of B. cereus on the glycyrrhizic acid and liquiritin biosynthesis pathway of G. uralensis under control and salt stress conditions was different.
Conclusion
In this study, B. cereus improved the growth condition and the contents of major medicinal components of G. uralensis seedlings. Interestingly, the effect of B. cereus on the seedling growth and the glycyrrhizic acid and liquiritin biosynthesis pathway was different under control and salt stress conditions. In promoting the seedling growth aspect, the effect of B. cereus under the salt stress condition was greater than that under the control condition. Also, the effect of B. cereus on the accumulation of active compounds was different under control and salt stress conditions based on transcriptome and PCR results. B. cereus increased liquiritigenin content under the control condition, which is closely related to regulate flavone and flavonol biosynthesis, and increased the contents of glycyrrhizic acid and glycyrrhetinic acid involved in the phenylpropanoid biosynthesis pathway under the salt stress condition. Moreover, our study provided some theoretical basis for improving the contents of major active compounds and enhancing salt tolerance of G. uralensis and the development and utilization of B. cereus and molecular breeding of G. uralensis, which will help the further study in G. uralensis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE187003.
Author Contributions
YZ and XZ contributed to the experiment design. WZ and YZ performed the material preparation and data collection. DL and YZ analyzed the data. YZ wrote the first draft of the manuscript. All authors commented on previous versions of the manuscript, read and approved the final manuscript.
Funding
The authors are grateful for the financially supported by the project of the National Natural Science Foundation of China (82160724 and 31860343), the Ningxia Science and Technology Innovation Leader Program (2020GKLRLX12), and the Ningxia Natural Science Foundation (2020AAC03124).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ali, M. B., and McNear, D. H. (2014). Induced transcriptional profiling of phenylpropanoid pathway genes increased flavonoid and lignin content in Arabidopsis leaves in response to microbial products. BMC Plant Biol. 14:84. doi: 10.1186/1471-2229-14-84
Amanifar, S., Khodabandeloo, M., and Fard, E. M. (2019). Alleviation of salt stress and changes in glycyrrhizin accumulation by arbuscular mycorrhiza in liquorice (Glycyrrhiza glabra) grown under salinity stress. Environ. Exp. Bot. 160, 25–34. doi: 10.1016/j.envexpbot.2019.01.001
Amor, F. D., and Cuadra-Crespo, P. (2012). Plant growth-promoting bacteria as a tool to improve salinity tolerance in sweet pepper. Funct. Plant Biol. 39, 82–90. doi: 10.1071/FP11173
Ansari, M., Shekari, F., and Mohammadi, M. H. (2019). Salt-tolerant plant growth-promoting bacteria enhanced salinity tolerance of salt-tolerant alfalfa (Medicago sativa L.) cultivars at high salinity. Acta Physiol. Plant 41:195. doi: 10.1007/s11738-019-2988-5
Behdad, A., Mohsenzadeh, S., and Azizi, M. (2020). Salinity effects on physiological and phytochemical characteristics and gene expression of two Glycyrrhiza glabra L. populations. Phytochemistry 171:112236. doi: 10.1016/j.phytochem.2019.112236
Berim, A., and Gang, D. R. (2016). Methoxylated flavones: occurrence, importance, biosynthesis. Phytochem. Rev. 15, 363–390. doi: 10.1007/s11101-015-9426-0
Chatterjee, P., Kanagendran, A., and Samaddar, S. (2019). Methylobacterium oryzae CBMB20 influences photosynthetic traits, volatile emission and ethylene metabolism in Oryza sativa genotypes grown in salt stress conditions. Planta 249, 1903–1919. doi: 10.1007/s00425-019-03139-w
Chen, M. L., Yang, G., Sheng, Y., Li, P. Y., Qiu, H. Y., Zhou, X. T., et al. (2017). Glomus mosseae inoculation improves the root system architecture, photosynthetic efficiency and flavonoids accumulation of liquorice under nutrient stress. Front. Plant Sci. 8:931. doi: 10.3389/fpls.2017.00931
Egamberdieva, D., Li, L., and Lindström, K. (2016). A synergistic interaction between salt-tolerant Pseudomonas and Mesorhizobium strains improves growth and symbiotic performance of liquorice (Glycyrrhiza uralensis Fish.) under salt stress. Appl. Microbiol. Biotechnol. 100, 2829–2841. doi: 10.1007/s00253-015-7147-3
Fouda, A., Hassan, S. E., Eid, A. M., and El-Din Ewais, E. (2018). “The interaction between plants and bacterial endophytes under salinity stress,” in Endophytes and Secondary Metabolites, ed. J. Jha (Cham: Springer), 1–17. doi: 10.1007/978-3-319-76900-4_15-1
Guo, R., Yang, Z., and Li, F. (2015). Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol. 15:170. doi: 10.1186/s12870-015-0546-x
Habib, S. H., Kausar, H., and Saud, H. M. (2016). Plant growth-promoting rhizobacteria enhance salinity stress tolerance in okra through ROS-scavenging enzymes. Biomed. Res. Int. 2016:6284547. doi: 10.1155/2016/6284547
Hosseinzadeh, H., and Nassiri-Asl, M. (2016). Pharmacological effects of Glycyrrhiza spp. and its bioactive constituents: update and review. Phytother. Res. 29, 1868–1886. doi: 10.1002/ptr.5487
Jose, A. M., Ortuño, M. F., Bernal-Vicente, A., Diaz-Vivancos, P., Sanchez-Blanco, M. J., and Hernandez, J. A. (2017). Plant responses to salt stress: adaptive mechanisms. Agronomy 7:18. doi: 10.3390/agronomy7010018
Kanagaraj, G., and Sathish, C. (2017). Salt stress induced changes in growth, pigments and protein contents in two horse gram [Macrotyloma uniflorum (Lam.) Verdc] varieties. J. Sci. Agri. 1, 08–18. doi: 10.25081/jsa.2017.v1i0.23
Kanehisa, M., Goto, S., Kawashima, S., and Okuno, Y. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, 277–280. doi: 10.1093/nar/gkh063
Kaur, G., and Ganjewala, D. (2019). “Stress protectant secondary metabolites and their metabolic engineering to enhance abiotic stress tolerance in plants,” in In Vitro Plant Breeding Towards Novel Agronomic Traits, eds M. Kumar, A. Muthusamy, V. Kumar, and N. Bhalla-Sarin (Singapore: Springer), 197–216. doi: 10.1007/978-981-32-9824-8_11
Khan, A. L., Gilani, S. A., and Waqas, M. (2017). Endophytes from medicinal plants and their potential for producing indole acetic acid, improving seed germination and mitigating oxidative stress. J. Zhejiang Univ. Sci. B 18, 125–137. doi: 10.1631/jzus.B1500271
Kiełkowska, A. (2017). Cytogenetic effect of prolonged in vitro exposure of Allium cepa L. root meristem cells to salt stress. Cytol. Genet. 51, 478–484. doi: 10.3103/S0095452717060068
Kushwaha, R. K., Singh, S., and Pandey, S. S. (2019). Fungal endophytes attune withanolide biosynthesis in Withania somnifera, prime to enhanced withanolide A content in leaves and roots. World J. Microbiol. Biotechnol. 35:20. doi: 10.1007/s11274-019-2593-1
Li, J., Wang, J., and Li, J. (2016). Aspergillus niger enhance bioactive compounds biosynthesis as well as expression of functional genes in adventitious roots of Glycyrrhiza uralensis fisch. Appl. Biochem. Biotechnol. 178, 576–593. doi: 10.1007/s12010-015-1895-5
Liao, W., Mei, Z., and Miao, L. (2020). Comparative transcriptome analysis of root, stem, and leaf tissues of Entada phaseoloides reveals potential genes involved in triterpenoid saponin biosynthesis. BMC Genomics 21:639. doi: 10.21203/rs.2.20018/v1
Mastan, A., Vivek Babu, C. S., and Hiremath, C. (2019). Treatments with native Coleus forskohlii endophytes improve fitness and secondary metabolite production of some medicinal and aromatic plants. Int. Microbiol. 23, 345–354. doi: 10.1007/s10123-019-00108-x
Mochida, K., Sakurai, T., Seki, H., Yoshida, T., Takahagi, K., Sawai, S., et al. (2017). Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J. 89, 181–194. doi: 10.1111/tpj.13385
Pande, A. M., Kulkarni, N. S., and Bodhankar, M. G. (2016). Effect of PGPR with ACC-deaminase activity on growth performance of wheat cultivated under stress conditions. Int. J. Appl. Res. 2, 723–726.
Pandian, B. A., Sathishraj, R., Djanaguiraman, M., Prasad, P., and Jugulam, M. (2020). Role of cytochrome p450 enzymes in plant stress response. Antioxidants 9:454. doi: 10.3390/antiox9050454
Ponce, M. A., Scervino, J. M., Erra, B. R., Ocampo, J. A., and Godeas, A. (2004). Flavonoids from shoots and roots of Trifolium repens (white clover) grown in presence or absence of the arbuscular mycorrhizal fungus glomus intraradices. Phytochemistry 65, 1925–1930. doi: 10.1016/j.phytochem.2004.06.005
Shirazi, Z., Aalami, A., and Tohidfar, M. (2019). Triterpenoid gene expression and phytochemical content in Iranian licorice under salinity stress. Protoplasma 256, 827–837. doi: 10.1007/s00709-018-01340-4
Suarez, C., Cardinale, M., Ratering, S., Steffens, D., Jung, S., Montoya, A. M. Z., et al. (2015). Plant growth-promoting effects of Hartmannibacter diazotrophicus on summer barley (Hordeum vulgare L.) under salt stress. Appl. Soil. Ecol. 95, 23–30. doi: 10.1016/j.apsoil.2015.04.017
Trdan, S., Vučajnk, F., Bohinc, T., and Vidrih, M. (2019). The effect of a mixture of two plant growth-promoting bacteria from Argentina on the yield of potato, and occurrence of primary potato diseases and pest-short communication. Acta. Agric. Scand. Sect. B. Soil Plant Sci. 69, 89–94. doi: 10.1080/09064710.2018.1492628
Vaishnav, A., Shukla, A. K., and Sharma, A. (2019). Endophytic bacteria in plant salt stress tolerance: current and future prospects. J. Plant Growth Regul. 38, 650–668. doi: 10.1007/s00344-018-9880-1
Vigani, G., Maffi, D., and Zocchi, G. (2010). Iron availability affects the function of mitochondria in cucumber roots. New Phytologist. 182, 127–136. doi: 10.1111/j.1469-8137.2008.02747.x
Wang, C., Chen, L., Cai, Z., Chen, C., and Mei, Y. (2020). Comparative proteomic analysis reveals the molecular mechanisms underlying the accumulation difference of bioactive constituents in Glycyrrhiza uralensis fisch under salt stress. J. Agr. Food Chem. 68, 1480–1493. doi: 10.1021/acs.jafc.9b04887
Wang, J. W., Tian, H., and Yu, X. (2017). Glucose-6-phosphate dehydrogenase plays critical role in artemisinin production of Artemisia annua under salt stress. Biol. Plant. 61, 529–539. doi: 10.1007/s10535-016-0674-7
Wang, L., Li, W., Ma, L., Chen, J., Lü, H., and Jian, T. (2016). Salt stress changes chemical composition in Limonium bicolor (Bag.) Kuntze, a medicinal halophytic plant. Ind. Crop. Prod. 84, 248–253. doi: 10.1016/j.indcrop.2016.01.050
Wang, Q., Eneji, A. E., Kong, X., Wang, K., and Dong, H. (2015). Salt stress effects on secondary metabolites of cotton in relation to gene expression responsible for aphid development. PLoS One 10:e0129541. doi: 10.1371/journal.pone.0129541
Xu, G., Cai, W., Gao, W., and Liu, C. (2016). A novel glucuronosyltransferase has an unprecedented ability to catalyse continuous two-step glucuronosylation of glycyrrhetinic acid to yield glycyrrhizin. New Phytol. 212, 123–135. doi: 10.1111/nph.14039
Yan, X., Fan, Y., Wei, W., Wang, P., Liu, Q., and Wei, Y. (2014). Production of bioactive ginsenoside compound k in metabolically engineered yeast. Cell Res. 24, 770–773. doi: 10.1038/cr.2014.28
Yendo, A. C., de Costa, F., Gosmann, G., and Fett-Neto, A. G. (2010). Production of plant bioactive triterpenoid saponins: elicitation strategies and target genes to improve yields. Mol. Biotechnol. 46, 94–104. doi: 10.1007/s12033-010-9257-6
Yeo, B. P. H., Bhave, M., and Hwang, S. S. (2018). Effects of acute salt stress on modulation of gene expression in a Malaysian salt-tolerant indigenous rice variety, Bajong. J. Plant Res. 131, 191–202. doi: 10.1007/s10265-017-0977-6
Zheng, T., Guan, L., and Yu, K. (2021). Expressional diversity of grapevine 3-Hydroxy-3-methylglutaryl-CoA reductase (HMGR) in different grapes genotypes. BMC Plant Biol. 21:279. doi: 10.1186/s12870-021-03073-8
Zheng, X., Xu, H., Ma, X., Zhan, R., and Chen, W. (2014). Triterpenoid saponin biosynthetic pathway profiling and candidate gene mining of the Ilex asprella root using RNA-Seq. Int. J. Mol. Sci. 15, 5970–5987. doi: 10.3390/ijms15045970
Zheng, Y. F., Wei, J. H., Qi, L. W., Cheng, J. M., and Peng, G. P. (2013). A green and efficient protocol for large-scale production of glycyrrhizic acid from licorice roots by combination of polyamide and macroporous resin adsorbent chromatography. J. Sep. Sci. 36, 809–816. doi: 10.1002/jssc.201200793
Zhou, S., Ma, Y. S., Hu, T., Zhang, X. D., Yin, Y. C., Gao, Z. Q., et al. (2017). Molecular cloning and sequence analysis of chalcone synthase cDNA in Glycyrrhiza uralensis. Lett. Biotech. 28, 812–817.
Keywords: Bacillus cereus, Glycyrrhiza uralensis, secondary metabolite pathways, transcriptome, PCR
Citation: Zhang Y, Lang D, Zhang W and Zhang X (2022) Bacillus cereus Enhanced Medicinal Ingredient Biosynthesis in Glycyrrhiza uralensis Fisch. Under Different Conditions Based on the Transcriptome and Polymerase Chain Reaction Analysis. Front. Plant Sci. 13:858000. doi: 10.3389/fpls.2022.858000
Received: 19 January 2022; Accepted: 21 April 2022;
Published: 02 June 2022.
Edited by:
Ying Ma, University of Coimbra, PortugalReviewed by:
Padmanabh Dwivedi, Banaras Hindu University, IndiaRupam Kapoor, University of Delhi, India
Copyright © 2022 Zhang, Lang, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinhui Zhang, zhang2013512@163.com
 Yu Zhang1,2
Yu Zhang1,2 Xinhui Zhang
Xinhui Zhang