- 1Xinjiang Key Laboratory of Biological Resources and Genetic Engineering, College of Life Science and Technology, Xinjiang University, Urumqi, China
- 2State Key Laboratory of Desert and Oasis Ecology, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Urumqi, China
- 3University of Chinese Academy of Sciences, Beijing, China
Salinity is a major limiting factor in crop productivity. Dehydration-responsive element-binding protein (DREB) transcription factors have been widely identified in a variety of plants and play important roles in plant stress responses. Studies on DREBs have primarily focused on the A-1 and A-2 DREB groups, while few have focused on the A-5 group. In this study, we concentrated on ScDREB5, an A-5b type DREB gene from the desiccation-tolerant moss Syntrichia caninervis. ScDREB5 is a transcription factor localized to the nucleus that exhibits transactivation activity in yeast. Ectopic ScDREB5 expression in Arabidopsis thaliana increased seed germination and improved seedling tolerance under salt stress. ScDREB5-overexpression transgenic Arabidopsis lines showed lower methane dicarboxylic aldehyde (MDA) and hydrogen peroxide (H2O2) contents, but higher peroxidase (POD), superoxide dismutase (SOD), and catalase (CAT) activities compared to wild plants. Moreover, the transcriptional levels of stress marker genes, including RD29B, COR47, LEA6, LEA7, ERD1, P5CS1, and salt overly sensitive (SOS) genes (SOS1, SOS2, and SOS3), were upregulated in the transgenic lines when subjected to salt treatment. Transcriptome and real-time quantitative PCR (RT-qPCR) analyses indicated that transgenic lines were accompanied by an increased expression of jasmonic acid (JA) biosynthesis genes, as well as a higher JA content under salt stress. Our results suggest that ScDREB5 could improve salt tolerance by enhancing the scavenging abilities of reactive oxygen species (ROS), increasing JA content by upregulating JA synthesis gene expression, regulating ion homeostasis by up-regulating stress-related genes, osmotic adjustment, and protein protection, making ScDREB5 a promising candidate gene for crop salt stress breeding.
Introduction
In nature, plants are constantly challenged by adverse abiotic environmental conditions, such as drought, heat, cold, nutrient deficiencies, and excess salt or toxic metal levels in the soil. These abiotic stresses limit the global utilization of arable lands and negatively affect crop productivity (Zhang et al., 2021). Salinity is one of the most deteriorating of these stresses because it affects approximately 20% of irrigated agricultural land and one-third of the agricultural productivity globally (Shah et al., 2021). High salinity causes ionic, osmotic, and oxidative stresses to plants (Song et al., 2021). To protect against the detrimental effects of environmental stresses, plants have gradually evolved appropriate response mechanisms, in which gene expression regulation plays an important role (Gururani et al., 2015). Transcription factors are critical abiotic stress tolerance regulators (Sakuma et al., 2006a); for example, APETALA2/ethylene responsive factor (AP2/ERF), one of the largest plant transcription factor families containing a highly conserved AP2 domain, plays a key role in plant development and stress responses (Xu et al., 2011; Chandler, 2018). Based on the number of AP2 and B3 domains, the AP2/ERF family is comprehensively divided into the AP2, ERF, related to ABI3 and VP1 (RAV), dehydration-responsive element-binding protein (DREB), and Soloist subfamilies (Sakuma et al., 2002; Nakano et al., 2006; Giri et al., 2014). Among these, the DREB subfamily is mainly involved in drought, high salinity, cold, and heat stress responses (Sakuma et al., 2006b; Qin et al., 2007; Matsukura et al., 2010; Mizoi et al., 2013) and has been extensively studied in many plants (Mizoi et al., 2012). The DREB subfamily is further divided into six subgroups, A-1–A-6. The functions of A-1 and A-2 have been thoroughly studied (Agarwal et al., 2017), while the functions and mechanisms of A-5 type genes have received limited attention and remain unclear, although A-5 type DREB has great potential.
In recent years, most reports have shown that A-5 DREB transcription factors can improve stress resistance in plants. In potato plants, low temperature, drought, and abscisic acid (ABA) induced StDREB2, an overexpression of which could enhance the salt stress tolerance in transgenic Arabidopsis thaliana (Bouaziz et al., 2012). Subsequent studies have shown that an overexpression of StDREB2 enhanced drought stress tolerance in cotton (El-Esawi and Alayafi, 2019); in soybean plants, GmDREB2 was induced by drought, salt, low temperature, and ABA treatments, which showed drought and salt stress tolerances in transgenic Arabidopsis (Chen et al., 2007); overexpression of GmDREB3 enhanced the resistance of transgenic Arabidopsis to cold, drought, and salt stresses (Chen et al., 2009); overexpression of PpDBF1 has increased salt, drought, and cold stress tolerances in transgenic tobacco plants (Liu et al., 2007); and overexpression of HhDREB2 in Arabidopsis resulted in enhanced tolerance to salt and drought stresses (Ma et al., 2014). All these studies indicated that A-5 DREB genes are good resistance gene candidates for crop resistance breeding; thus, it is meaningful to study the biological functions of A-5 DREB genes from different plants.
Mosses are one of the oldest species in the transition from aquatic to terrestrial plants (Oliver et al., 2000), and because of their high oxygen content, intense atmospheric radiation, and drastic temperature changes on land, mosses have evolved special physiological and biochemical characteristics to tolerate multiple stresses (Wu et al., 2012; Pan et al., 2016). S. caninervis, a desiccation-tolerant moss, is a dominant species in biological soil crusts in the Gurbantunggut Desert of Northwestern China (Wang et al., 2020a), and was proven to be a category 1 (A) desiccation tolerance (DT; Wood, 2007). To adapt to harsh desert environments, S. caninervis has evolved multiple stress tolerances. Many stress-related genes have been identified from S. caninervis, making it a well-studied model for plant DT mechanism studies and stress-related gene identification (Yang et al., 2012, 2015; Li et al., 2015, 2016, 2017).
In a previous study, we found that A-5 DREB genes were largely expanded in S. caninervis, indicating their importance in stress responses (Li et al., 2017). Ten A-5 DREB genes (ScDREB1–ScDREB10) were screened and cloned based on the S. caninervis dehydration-rehydration transcriptome (Li et al., 2016). Among these, an overexpression of ScDREB8 could improve salt tolerance in Arabidopsis (Liang et al., 2017), while an overexpression of ScDREB10 significantly increased the drought and salt tolerance of transgenic Arabidopsis at both the germination and seedling stages (Li X. et al., 2019). In the present study, we further characterized the function of ScDREB5. Our results showed that ScDREB5 is located in the nucleus and possesses transactivation activity. We overexpressed ScDREB5 in transgenic Arabidopsis to evaluate its role in salt stress responses. Our findings suggest that ScDREB5 is a positive regulator of the salt-stress response that improves salt tolerance by enhancing reactive oxygen species (ROS) scavenging abilities, upregulating stress-responsive genes, and modulating jasmonic acid (JA) biosynthesis in plants under salt stress.
Materials and Methods
Plant Samples and Treatments
Ecotype colombia of Arabidopsis (Col-0) was used as the wild type (WT), and the genetic background for transgenic plants. Plants were grown in a growth chamber at 22°C using 16/8 h light/dark photocycles (approximately 100 μmol m–2s–1 constant light, 60% relative humidity).
Seven-day-old seedlings, grown under normal conditions, were carefully transferred to a Murashige and Skoog (MS) medium (control) or a MS medium containing 150 mM NaCl and incubated for 7 d. Two transgenic lines with three replicates were used for salt stress experiments, with at least 20 seedlings were used for each replicate. Whole plants were harvested for physiological index analysis, JA content measurement, real-time quantitative PCR (RT-qPCR), and RNA sequencing.
DNA/Protein Sequence and Phylogenetic Analyses
The protein sequences of classic DREBs from the model plants A. thaliana, Oryza sativa, Vaccinium vitis-idaea, Malus sieversii, Zea mays, Glycine max, and Gossypium hirsutum were retrieved from the National Center for Biotechnology Information (NCBI) Entrez database1. Multiple sequence alignments of amino acid sequences from different species were performed using ClustalW, and phylogenetic trees were constructed with the neighbor-joining (NJ) method and a bootstrap test with 1000 replicates using MEGA7 software for Poisson correction and pairwise deletion (Kumar et al., 2016). Motif prediction of ScDREB5 and other classic DREB proteins was performed using MEME (Bailey et al., 2015).
Subcellular Localization Analysis
The open reading frame (ORF) for ScDREB5 without a stop codon was amplified from pMD18-T-ScDREB5 positive plasmids using gene-specific primers containing an SmaI restriction site, and the PCR product was fused to the N-terminus of the green fluorescent protein (GFP) gene driven by the CaMV 35S promoter in the pBI121 vector using an in-fusion PCR cloning system (Clontech, Mountain View, CA, United States). The primers used in this experiment are listed in Supplementary Table 1. Both the transient expression of the 35S:ScDREB5-GFP fusion and 35S:GFP control plasmids were introduced into living onion epidermal cells via Agrobacterium-mediated infiltration for transient transformation (Sun et al., 2007). The transformed cells were cultured in 1/2 Murashige and Skoog (MS) medium at 22°C ± 2°C in the dark for 24 h and visualized using confocal laser scanning microscopy LSM800 (Zeiss, Jena, Germany).
Analysis of Transactivation Activity of ScDREB5
The coding sequence of ScDERB5 was fused in frame with the GAL4 DNA-binding domain (pGBKT7 vector) to produce the BD-ScDREB5 fusion construct using the in-fusion PCR cloning system (Clontech), and the full-length ORF of ScDREB5 was amplified from pMD18-T plasmids by PCR using gene-specific primers containing EcoRI and BamHI restriction sites, and the PCR product was inserted into the EcoRI/BamHI pGBKT7 vector. The construct was subsequently introduced into yeast Y2H gold cells (Clontech). The yeast positive transformants were adjusted to an OD600 of 2.0, and the yeast cells were then serially diluted 10-fold and dropped with 2 μL of synthetic dropout (SD) medium without tryptophan (SD/-Trp), without tryptophan and histidine (SD/-Trp/-His), and with SD/-Trp/-His plates containing x-α-gal (SD/-Trp/-His + x-α-gal). Yeast cells expressing the pGBKT7 empty vector or expressing GAL4 were used as negative and positive controls, respectively. The plates were incubated at 30°C for 2–4 d before photographing. Primer information is listed in Supplementary Table 1.
Generation of ScDREB5-Overexpressing Arabidopsis
The 35S:ScDREB5-GFP was introduced into WT Arabidopsis plants using the floral dip method (Clough and Bent, 1998). T1 to T3 generation seeds of transgenic plants were selected on MS medium containing 80 mg L–1 hygromycin. The hygromycin-resistant T1 seedlings were tested by PCR analysis and sequencing, and the T3 homozygous lines produced from the T1 plants expressing the ScDREB5 gene. We obtained 18 transgenic lines in total (the positive tests of all the T3 seedings lines by PCR analysis were provided and showed in Supplementary Figure 1A). The ScDREB5 had the higher expression in line 2 and line 3 in different T3 transgenic lines (Supplementary Figure 1B), which were selected for further functional analysis.
Evaluation of the Salt Stress Tolerance of Transgenic Arabidopsis at Germination Stage
To determine the sensitivity of the seed germination process to salt stress, Arabidopsis seeds of WT and transgenic plants were surface-sterilized and placed on MS agar plates (control) or MS agar plates supplied with 150 mM NaCl (salt stress). Fifty seeds per line with three replicates were used for the stress germination experiments. The seeds were incubated at 4°C for 2 d and then transferred to a growth chamber at 22 ± 2°C under a 16/8 h light/dark cycle. For germination rate statistics, the number of seeds that germinated were recorded every 24 h for 7 d. Seeds were considered to have germinated when the radicles were 1 mm long. Germination percentages were recorded as the number of seeds that germinated out of the total number of seeds tested.
Phenotype and Physiological Index Analysis of Transgenic Arabidopsis
After 7 d of salt stress treatment, the phenotype of plants was photographed under normal and stress conditions, and the root length, fresh weight, and number of lateral roots were measured. Then, these seedlings were collected and used for diaminobenzidine (DAB) and nitrotetrazolium blue chloride (NBT) staining, and seedlings were harvested by immediate flash freezing in liquid nitrogen to measure physiological indices. WT and transgenic plants (100 mg fresh weight) were ground with ice-cold 0.1 mol L–1 potassium phosphate buffer (pH 7.4; 1:9 w/v) and clarified by centrifugation at 8000 × g for 10 min at 4°C. The supernatants were harvested for further physiological analysis, and hydrogen peroxide (H2O2) levels, methane dicarboxylic aldehyde (MDA) content, superoxide dismutase (SOD, EC 1.15.1.1), catalase (CAT, EC 1.11.1.6), and peroxidase (POD, EC 1.11.1.7) activities were measured using assay kits (kit Numbers. A064-1, A003-3, A001-3, A084-3, A007-1, Nanjing Jian Cheng Bioengineering Institute, China), respectively.
Jasmonic acid was extracted and quantified by Suzhou Comin Biotechnology Co., Ltd., by high-performance liquid chromatography apparatus Rigol L3000 (Rigol, Beijing, China).
Transcriptomic Analysis
The L2 was chosen for the transcriptomic profiling analysis due to better stress tolerant phenotypes (great fresh weight, more number of lateral roots). L2 and WT plant samples were collected before and after salt stress with three biological replicates, total twelve samples were used for RNA isolation. RNA purity and concentration were then examined using a NanoPhotometer® spectrophotometer; RNA integrity and quantity were measured using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system. Total RNA was used as the input material for library preparation. RNA sequencing analysis was performed by Novogene Corporation (Beijing, China). After the unique molecular identifier (UMI) library preparation and pooling of different samples, the samples were subjected for Illumina sequencing. The pearson’s correlation between samples showed in Supplementary Figure 2. Then, the fragments per kilobase of exon model per million mapped reads (FPKM) of each gene was calculated based on the length of the gene and read counts mapped to this gene. Differential expression analysis was performed using DESeq R package (1.10.1). Genes with | log2(fold change) | > 1 and an adjusted P-value < 0.05, were considered differentially expressed.
RT-qPCR Assay
RNA was extracted using a Plant RNA Kit (OMEGA, Guangzhou, China). First-strand cDNA was synthesized using 1 μg of RNA with the PrimeScript RT reagent Kit (TaKaRa, Tokyo, Japan). RT-qPCR was performed on a CFX-96 Real-Time System (Bio-Rad, Hercules, CA, United States) with SYBR Premix Ex Taq II (TaKaRa). The AtTubulin (AT1G50010) gene of Arabidopsis was used as a reference for RT-qPCR normalization. The PCR conditions were as follows: initial denaturation step of 30 s at 95°C, 40 cycles of PCR (95°C for 5 s, 58–60°C for 30 s). Three biological and technological replicates were performed for each sample. The relative expression of the detected genes was calculated using the 2–ΔΔCt method. The expression levels of classic stress-responsive genes (AtRD29B, AtCOR47, AtLEA6, AtLEA7, AtERD1, and AtP5CS1), salt stress-related salt overly sensitive (SOS) genes (AtSOS1, AtSOS2, and AtSOS3), and JA biosynthesis genes (AtDAD1, AtDGL, AtDALL3, AtLOX3, AtLOX4, AtAOS, AtAOC3, AtOPR3, AtJMT, AtYCYP94C1, and AtCYP9483) were investigated using RT-qPCR. The primers used for the RT-qPCR analysis are listed in Supplementary Table 2.
Statistical Analysis
One-way analysis of variance (ANOVA) was used for statistical analyses with the SPSS software (version 19.0; IBM Corp., Armonk, NY, United States). All data in this study represent the mean values ± standard error (SE) of at least three replicates. We used the least significant difference (LSD) multiple comparison tests to identify the significance level, and *p < 0.05 was considered significant, while **p < 0.01 was considered highly significant. All figures were generated using GraphPad Prism 9 and Adobe Illustrator CS software. Heatmaps were generated using TBtools (Chen et al., 2020).
Results
Characterization and Phylogenetic Analysis of ScDREB5
The ScDREB5 gene has been cloned previously (Li et al., 2016), full length of ScDREB5 is 889 bp, containing an ORF of 612 bp and encoded a predicted polypeptide of 203 amino acids with a putative molecular mass of 22.29 kDa with an isoelectric point of 5.97. ScDREB5 shared the highest sequence similarity with AT1G77640 in Arabidopsis (55% identity) with full-length predicted protein. Phylogenetic analysis showed that ScDREB5 clustered with the A-5 DREB proteins (Figure 1A). Sequence alignment analysis showed that ScDREB5 shared a conserved AP2 domain with three β-sheets and an α-helix, while having little sequence similarity outside the AP2 domain compared to other DREBs (Figure 1B). Altogether, 18 out of the 59 amino acids in AP2 domains, including the residues G-4, R-6, R-8, W-13, V-14, E-16, R-18, P-20, R-25, W-27, L-28, G-29, A-37, A-38, A-40, D-42, A-44 and N-59, were completely conserved among the 21 representative A-1–A-6 type DREB proteins from various plants. The 12th amino acid is an S in ScDREB5 and AT1G77640 proteins; however, it is a K for other DREB proteins. Motif analysis results showed that, similar to other classic DREB A-5 proteins, ScDREB5 has motif 1 (three β-sheets) and motif 2 (α-helix), which is composed of the AP2 domain, motif 3 seems unique for A-6 group DREBs, motif 4 exists in A-2 and A-3 DREBs, and motif 5 is predominately located in A-5 type DREBs and in some of the A-1 and A-4 DREBs (Figure 1C).
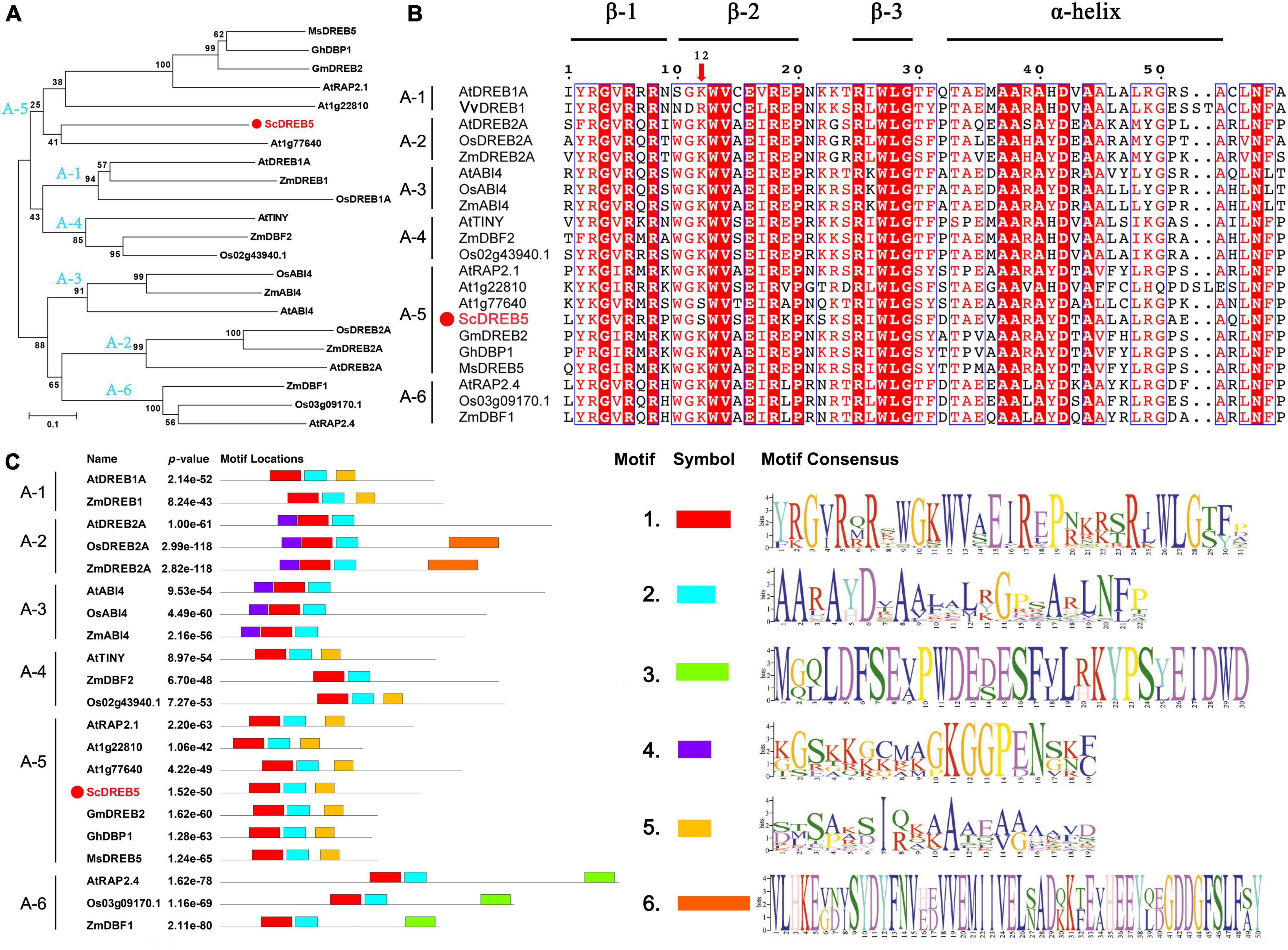
Figure 1. Phylogenetic analysis, sequence alignment and motif prediction of ScDREB5. (A) Phylogenetic analysis of ScDREB5 with DREBs from different plant species. A phylogenetic tree was constructed using the neighbor-joining method with Poisson correction (pairwise deletion), and bootstrap values from 1000 replicates were used to assess the robustness of the tree. (B) Multiple sequence alignment of ScDREB5 with other classic DREB proteins from different plants. (C) Motif prediction of ScDREB5 with other classic DREB proteins. The red color highlights the same amino acid sequence. All amino acid sequences were retrieved from GenBank: S. caninervis ScDREB5 (AMT92113.1); A. thaliana AtDREB1A (BAA33791), AtDREB2A (BAA33794), AtABI4 (ABE65896.1), AtTINY (NP_197953.1), AtRAP2.1 (OAP00017.1), AT1G22810 (ABD57508.1) AT1G77640 (AAT06448.1), AtRAP2.4 (NP_177931.1); O. sativa, OsDREB2A (AFB77198), OsABI4 (C7J2Z1.1), Os02g43940.1 (XP_015626317.1), Os03g09170.1 (XM_015773644.2); V. vitis-idaea VvDREB1 (AFC89543.1); Z. mays ZmDREB2A (PWZ08831.1), ZmABI4 (KJ727241.1), ZmDBF2 (AAM80485.1), ZmDBF1 (AAM80486.1); M. sieversii MsDREB5 (AFM84627.1), G. max GmDREB2 (ABB36645.1); G. hirsutum GhDBP1 (AAO43165.1).
Subcellular Localization and Transactivation Activity Analysis of the ScDREB5 Protein
To understand the characteristics of ScDREB5 as a transcription regulator, we determined its localization and transactivation activity. The subcellular location of the ScDREB5 protein was analyzed by transient expression of the GFP fusion protein introduced into onion epidermal cells. The results revealed that ScDREB5-GFP fusion was co-localized to the nucleus with DAPI stain. In contrast, the control GFP protein was distributed in the nucleus and cytoplasm, indicating that ScDREB5 is a nuclear-localized protein (Figure 2A).
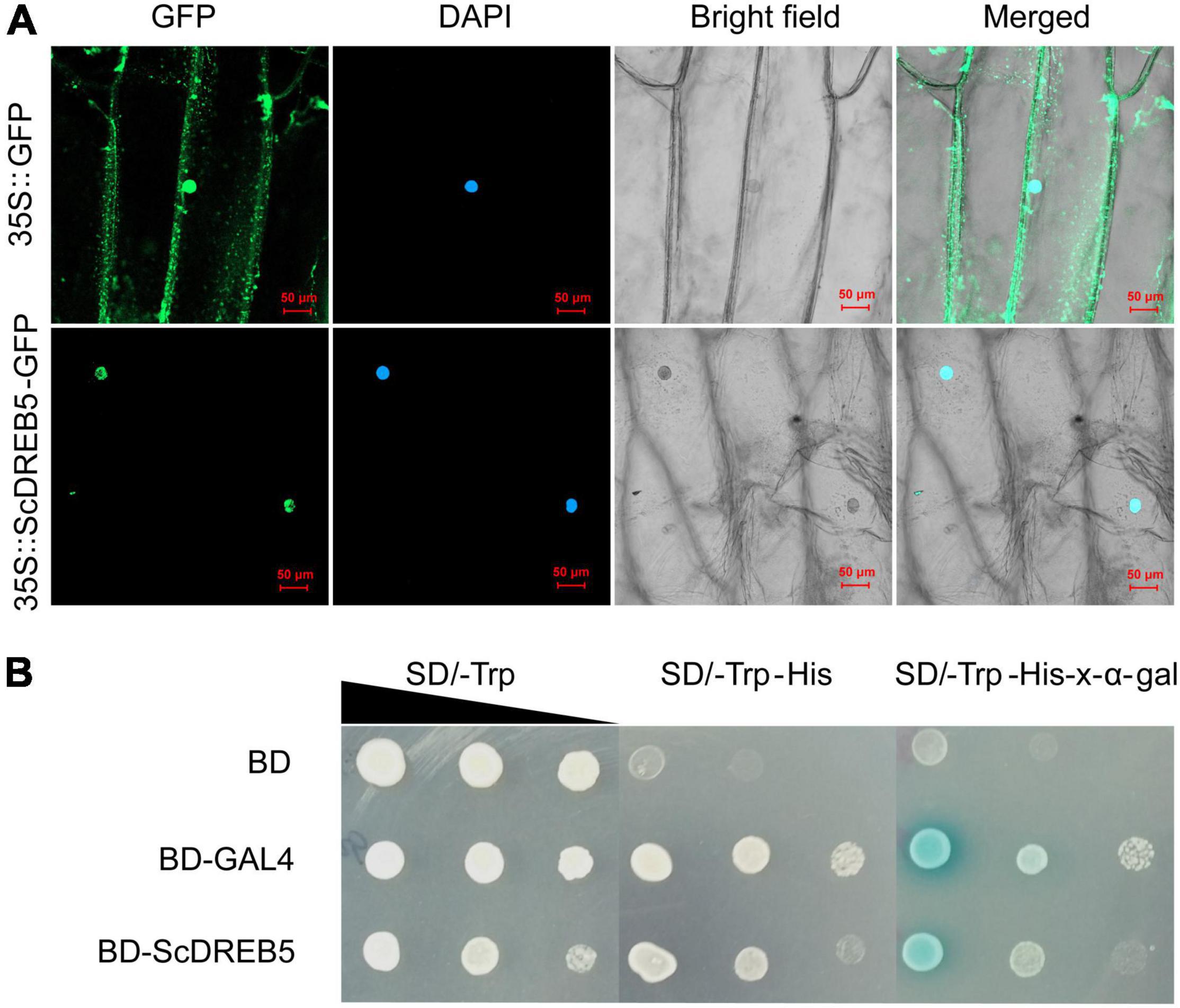
Figure 2. Subcellular localization and transactivation activity analyses of ScDREB5 protein. (A) 35S:ScDREB5-GFP and 35S:GFP fusion protein were transiently expressed in onion epidermal cells. After DAPI staining, the transformed cells were examined using confocal microscopy to simultaneously capture GFP (488 nm) and DAPI (405 nm) signals. Left to right: green, GFP fluorescence; blue, nucleus stained with DAPI; bright field; merged, combined fluorescence from GFP and DAPI (scale bar: 50 μm). (B) The transcriptional activation ability of ScDREB5 was examined using the yeast one-hybrid assay. Yeast cells Y2H expressing the fusion proteins were cultured and adjusted to an OD600 of 2.0, then series diluted and dropped with 2 μL on nutritional selective mediums SD/-Trp, SD/-Trp/-His and SD/-Trp/-His + x-α-gal. Yeast cells expressing pGBKT7-BD empty vector served as the negative control and those expressing pGBKT7-GAL4 served as the positive control. Photos were taken after incubating at 30°C for 2–4 d.
To examine the transactivation activity of ScDREB5, the Y2H system was transformed with the fusion construct pGBKT7-BD-ScDREB5, the negative control pGBKT7-BD, and the positive control pGBKT7-BD-GAL4. The yeast cells containing pGBKT7-BD-ScDREB5 or pGBKT7-BD-GAL4 grew well in the SD/-Trp/-His medium, whereas yeast cells containing the negative control pGBKT7-BD did not grow (Figure 2B). Furthermore, in the presence of x-α-gal, the yeast cells harboring pGBKT7-BD-ScDREB5 and pGBKT7-BD-GAL4 that grew well in the SD/-Trp/-His medium turned blue (Figure 2B). These results indicate that ScDREB5 has transactivation activity in yeast.
Overexpression of ScDREB5 Improved the Salt Stress Tolerance of Transgenic Arabidopsis
To further evaluate the function of ScDREB5 in plants, we introduced the ScDREB5 gene into Arabidopsis. We first analyzed the germination rate and phenotype of the transgenic lines (L2 and L3) and WT plants under drought and salt stress. Under drought stress, there was no significant difference in the germination rate and tolerance phenotype between transgenic and WT plants after drought stress (Supplementary Figure 3); therefore, we focused on the salt stress tolerance function in further investigations.
Under normal conditions, the seed germination rate and percentage of transgenic lines were faster and higher than those of WT plants during the first 2 days (Figure 3A). Under salt stress, the germination rate of both transgenic lines (L2 and L3) reached 100% on day 3; however, the germination rate of WT plants was only 87.74% until day 7 (Figure 3B). The results indicated that the overexpression of ScDREB5 increased salt tolerance in Arabidopsis at the germination stage. Additionally, we analyzed the growth of the transgenic lines and WT plants at the seedling stage. Under normal growth conditions, the WT plants and the two transgenic lines had similar phenotypes with comparable fresh weight, root length, and lateral root numbers (Figures 3D–F); however, under salt stress conditions, the transgenic plants had significant higher fresh weight, and the fresh weight of the WT plants was 3.63 mg, while for L2 and L3, they were 5.54 and 5.28 mg, respectively (**p < 0.01) (Figure 3D). Although the root length of L2 and L3 were not different from that of the WT plants under salt stress, the lateral root numbers of transgenic plants were significantly higher than that of WT plants (Figures 3E,F) (**p < 0.01). These results indicate that overexpression of ScDREB5 enhances tolerance to salt stress in transgenic plants.
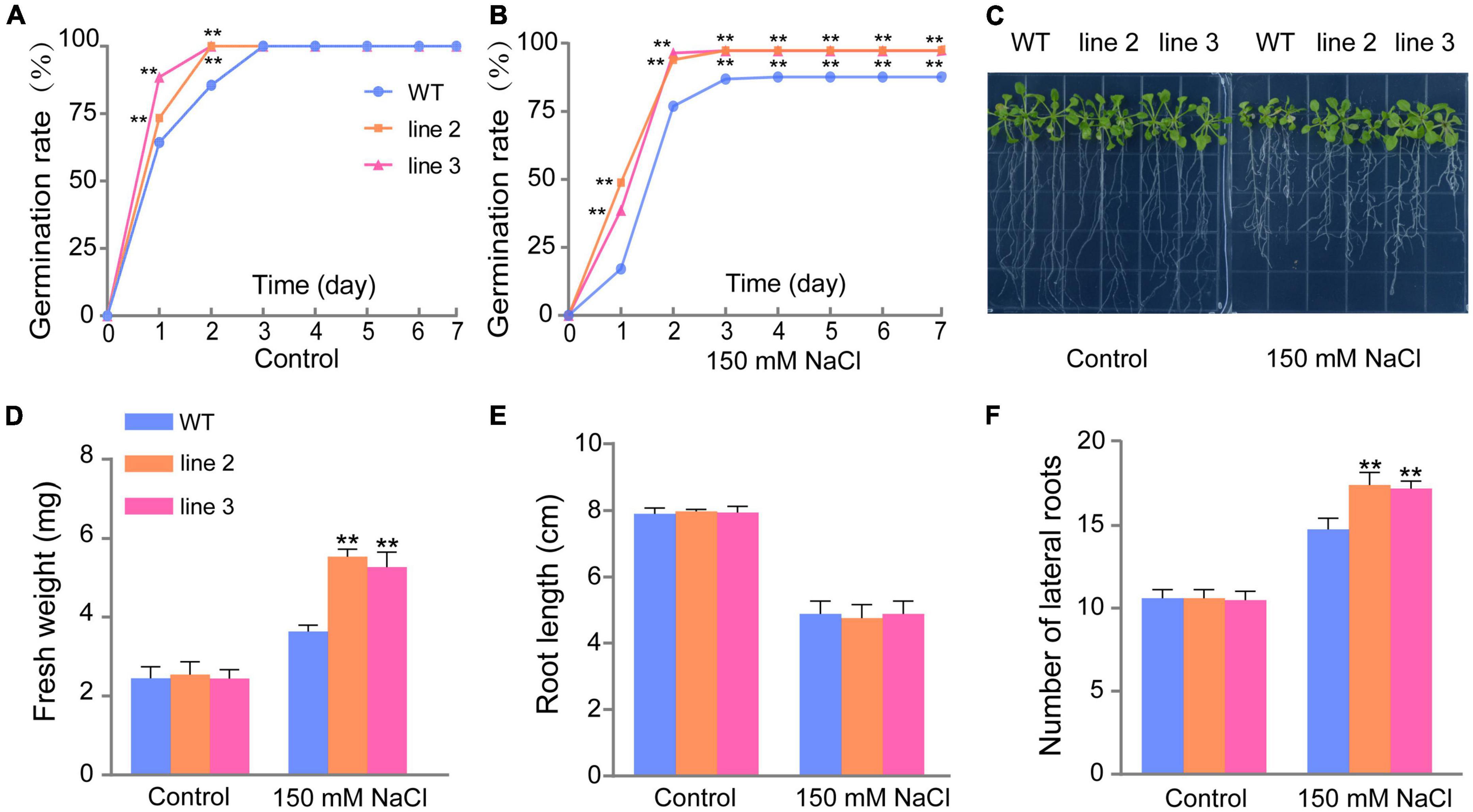
Figure 3. Germination rate, fresh weight, root length, and number of lateral roots comparisons of WT plants and two ScDREB5 transgenic Arabidopsis lines. (A,B) Germination percentages of WT and ScDREB5-transformed seeds under normal and salt stress conditions. The germination percentages were calculated as the number of germinated seeds divided by the total number of seeds. Values are means ± SE of three replicates (n = 40–60 seeds). Asterisks indicate statistically significant differences from WT (**P < 0.01). (C) Phenotype observation of WT and transgenic plants under normal and salt stress conditions. (D–F) Analysis of the fresh weight (mg), root length (cm), and number of lateral roots. Data are displayed with mean ± SE using at least 20 plants per replicates. Significant difference compared with WT plants with the same treatment. LSD multiple comparison test was used: *p < 0.05, **p < 0.01.
ScDREB5 Improved Reactive Oxygen Species Scavenging Capability of Transgenic Arabidopsis
The physiological changes in the transgenic lines and WT were analyzed after salt stress. Abiotic stress can lead to oxidative damage due to increased ROS production. Therefore, it is important for plants to activate antioxidative systems to cope with this oxidative damage. NBT and DAB staining were used to determine the two main ROS species (H2O2 and superoxide anion, O2–). As shown in Figure 4A, the staining results showed that there were lighter NBT and DAB staining in transgenic plants than in the WT plants under salt stress conditions; however, there were no significant differences between transgenic and WT plants under normal conditions. This means that transgenic plants have less H2O2 and O2– accumulation than WT plants under salt stress conditions. Accordingly, the H2O2 levels in ScDREB5 transgenic plants were significantly lower than those in the WT plants (Figure 4B). We compared the MDA contents between WT and transgenic plants and found that transgenic plants had significantly lower MDA contents after salt stress than WT plants (Figure 4C). Furthermore, transgenic and WT plants showed similar SOD, POD, and CAT activities under normal growth conditions; however, these activities were significantly higher in L2 and L3 than in WT plants under salt stress (Figures 4D–F). In conclusion, our results suggest that ScDREB5 may enhance the salt resistance of transgenic Arabidopsis by reducing the accumulation of ROS and inducing higher antioxidant activities.
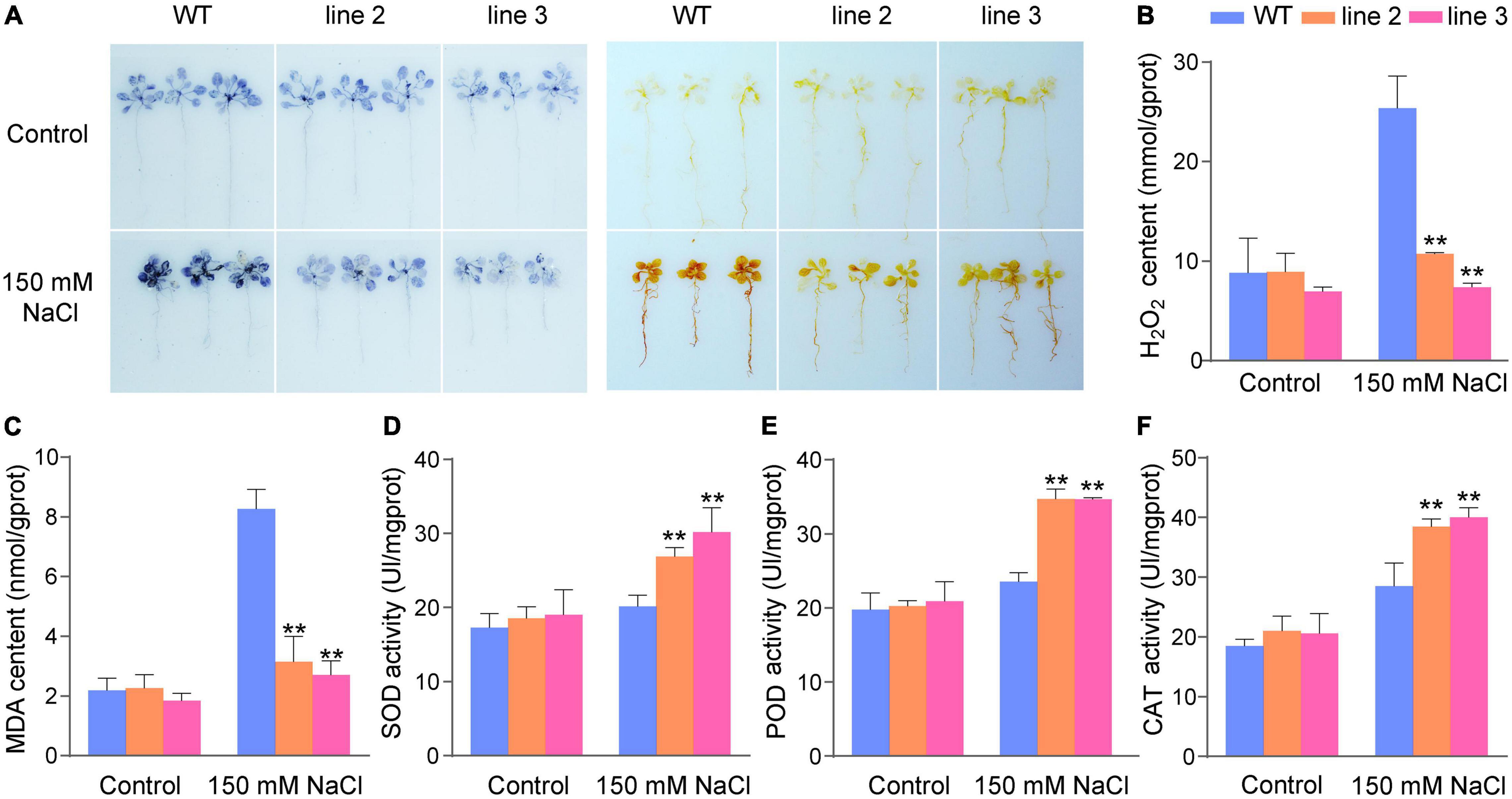
Figure 4. Analysis of ROS level and ROS scavenging ability of transgenic Arabidopsis under salt stress treatment. (A) Analysis of ROS production in intact plants by NBT and DAB staining. (B) Analysis of H2O2 level. (C–F) Analysis of MDA content, SOD, POD and CAT activities, respectively. All experiments were repeated three times. Data are means ± SE from three independent experiments. Significant difference compared with WT plants with the same treatment, LSD multiple comparison test was used: *p < 0.05, **p < 0.01.
Transcriptomic Profiling Analysis of Overexpression of ScDREB5 Plants After Salt Stress Treatment
To investigate the molecular regulation mechanism of salt tolerance in ScDREB5 transgenic plants, we performed RNA-seq of both WT and ScDREB5 transgenic plants under normal and salt stress conditions. For WT plants, 927 and 1071 differentially expressed genes (DEGs) were up- and downregulated, respectively, after salt stress treatment (WT-salt vs. WT-control); In ScDREB5 transgenic lines, 789 and 1811 DEGs were up- and downregulated, respectively (ScDREB5-salt vs. ScDREB5-control) (Figure 5A). Compared to WT plants, more DEGs (977 genes) were up-regulated and fewer DEGs (71 genes) were downregulated in ScDREB5 transgenic lines under normal conditions (ScDREB5-control vs. WT-control), and 333 and 301 DEGs were up- and downregulated in ScDREB5 transgenic lines under salt stress conditions (ScDREB5-salt vs. WT-salt) (Figure 5A). Overlapping analyses of the four comparisons are displayed in Figures 5B,C. Among them, 0 genes were downregulated together (Figure 5C), 23 genes were upregulated together (Figure 5B), and the results of Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that 23 genes were predominately enriched in “cyanoamino acid metabolism” and “plant hormone signal transduction”. To investigate the roles of ScDREB5 in salt stress tolerance, functional annotation of the upregulated DEGs under salt stress (ScDREB5-salt vs. WT-salt) was analyzed by gene ontology (GO) enrichment. For biological process GO terms, “regulation to salt stress”, “response to stimulus”, “response to stress”, “response to abiotic stimulus”, “response to oxygen-containing compound”, “response to JA”, and “response to hormone” were enriched (Figure 5D). Moreover, KEGG pathway analyses revealed that “plant hormone signal transduction”, “alpha-linolenic acid metabolism”, “linoleic acid metabolism”, “diterpenoid biosynthesis”, and “carotenoid biosynthesis” were significantly enriched (Figure 5E).
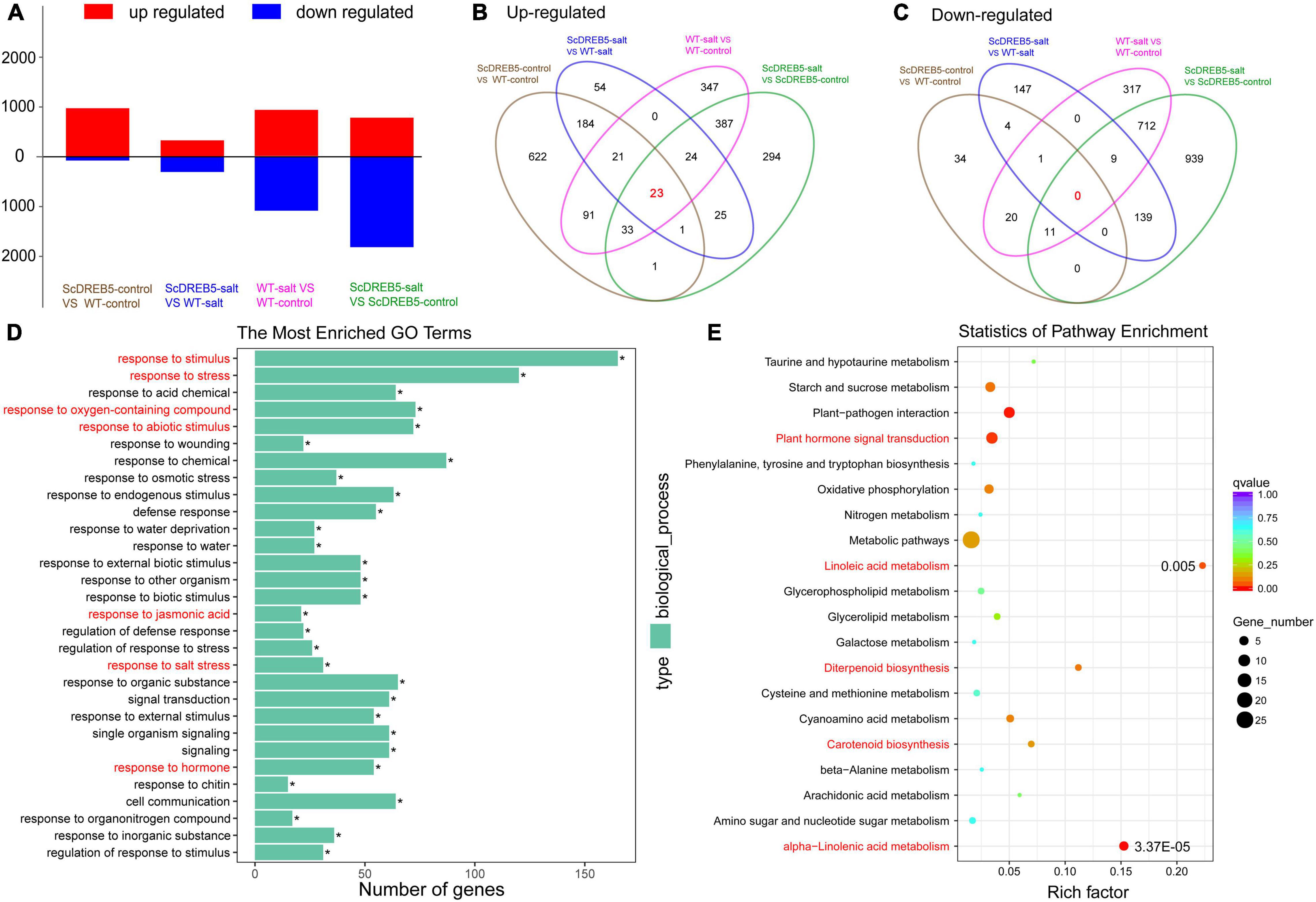
Figure 5. Differentially accumulated transcripts, GO enrichment, and KEGG pathway analysis between ScDREB5 transgenic and WT plants based on RNA-seq analysis. (A) Number of DEGs in WT and the transgenic ScDREB5 plants under normal and salt stress condition. (B,C) Overlapping analyses of up- and downregulated genes induced by ScDREB5 or salt stress. (D,E) GO term and KEGG analyses of the upregulated genes under salt stress (ScDREB5-salt vs. WT-salt).
Analysis of the Expression Level Changes of Classic Stress-Related Genes in ScDREB5 Transgenic Plants
The transcript abundance of classic stress-related genes, such as AtRD29B, AtLEA, and AtSOS, were significantly upregulated in ScDREB5 transgenic plants based on RNA-seq (Supplementary Figure 4). RT-qPCR analysis was used to confirm the expression levels of stress-related genes. Under normal conditions, all these genes showed no differences in the expression levels between the transgenic lines and WT plants (Figure 6A); however, after salt stress treatment, the expression of these genes increased significantly. In particular, the AtLEA7 gene was upregulated approximately 4.3 and 3.6 fold in L2 and L3 compared to the WT, respectively. The SOS pathway plays an essential role in conferring salt tolerance in Arabidopsis (Ji et al., 2013); therefore, we further investigated the expression levels of the SOS pathway genes (AtSOS1, AtSOS2, and AtSOS3). The results showed that the transcripts of AtSOS1, AtSOS2, and AtSOS3 were more abundant in response to salinity (both increased more than twofold) (Figure 6B). Overall, these results suggest that the upregulation of classic stress-related genes may partly explain the increased tolerance to salt stress in ScDREB5 transgenic Arabidopsis.
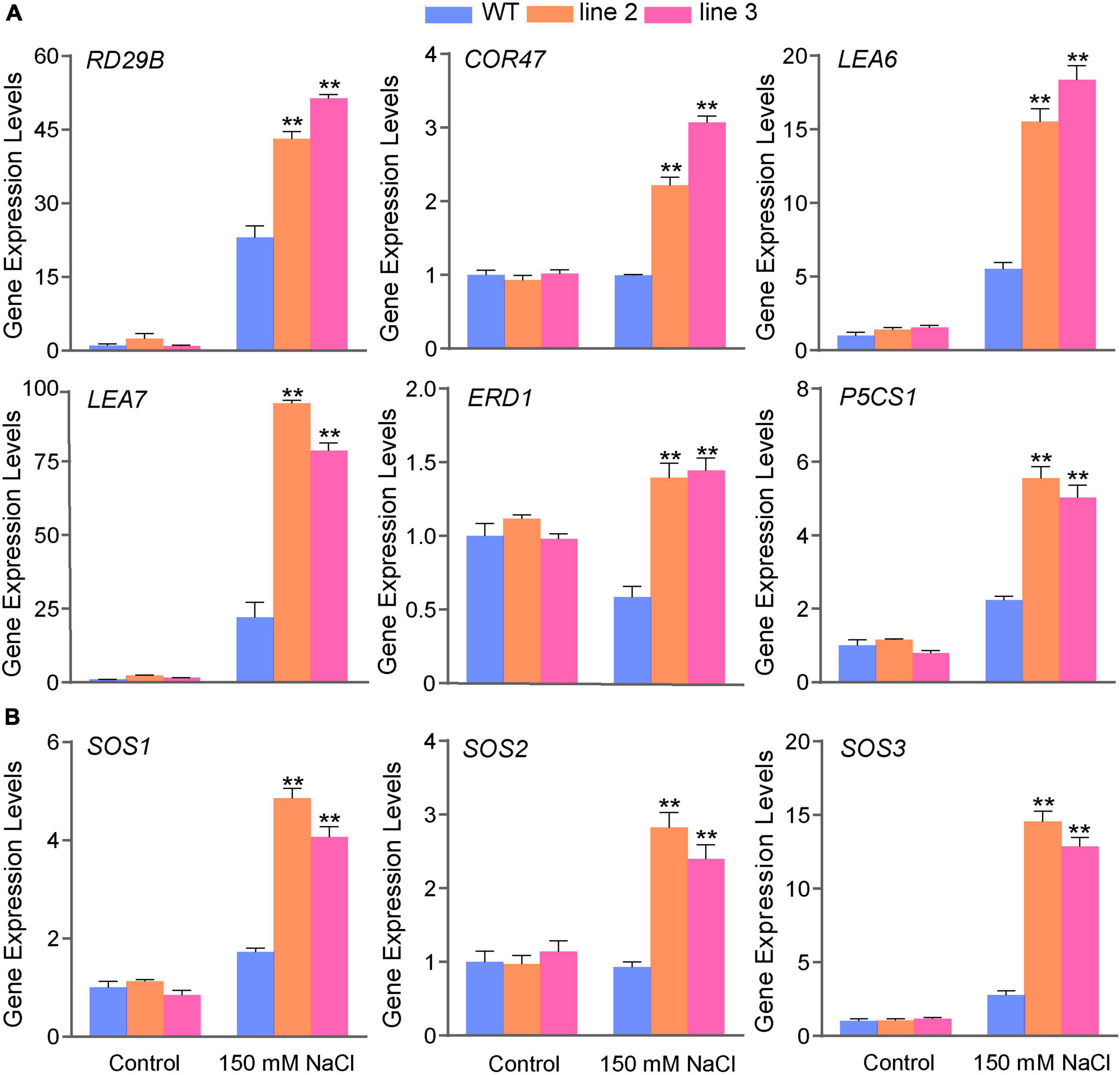
Figure 6. Gene expression level analysis of classic stress-related genes and SOS genes in ScDREB5 transgenic Arabidopsis response to salt stresses. (A) Abiotic stress-responsive genes were analyzed by RT-qPCR in WT and ScDREB5 transgenic plants under normal and salt stress conditions. (B) SOS genes (SOS1, SOS2, and SOS3) were analyzed by RT-qPCR in WT and ScDREB5 transgenic plants under normal and salt stress conditions. Total RNA was extracted from whole plants. The 2–ΔΔCt method was used for RT-qPCR normalization. The α-TUBULIN (AT1G50010) gene of Arabidopsis were used as the reference gene. Values are means ± SE of three replicates. Significant difference compared with WT plants with the same treatment. LSD multiple comparison test was used: *p < 0.05, **p < 0.01.
Overexpression of ScDREB5 Upregulated Jasmonic Acid Biosynthesis Related Genes Expression and Jasmonic Acid Content Under Salt Stress
From the KEGG pathway enrichment analysis, “alpha-linolenic acid metabolism” and “linoleic acid metabolism” had the highest enrichment categories: alpha-linolenic acid and linoleic acid are important precursors of the JA synthesis pathway. To further determine whether ScDREB5 improves the salt tolerance of transgenic plants by regulating JA, we examined the expression patterns of genes involved in JA biosynthesis based on RNA-Seq data. In total, 19 out of the 24 genes were differentially expressed in transgenic plants (Supplementary Figure 5), and their transcript abundance was higher than that of WT plants under salt stress. The transcript abundance of PLA1 (AtDAD1, AtDGL, and AtDALL3), LOX (AtLOX3 and AtLOX4), AOC (AtAOC3), OPR (AtOPR1 and AtOPR3), and CYP94 (AtCYP94C1 and AtCYP94B3) were significantly increased in ScDREB5 transgenic plants (Figure 7A), which were evaluated with RT-qPCR analysis to confirm the abundance change. Under salt stress conditions, all 11 genes had elevated transcript abundances in the transgenic plants compared to the WT plants (Figure 7B); among them, AtDAD1, a key gene for JA synthesis, were upregulated eightfold.
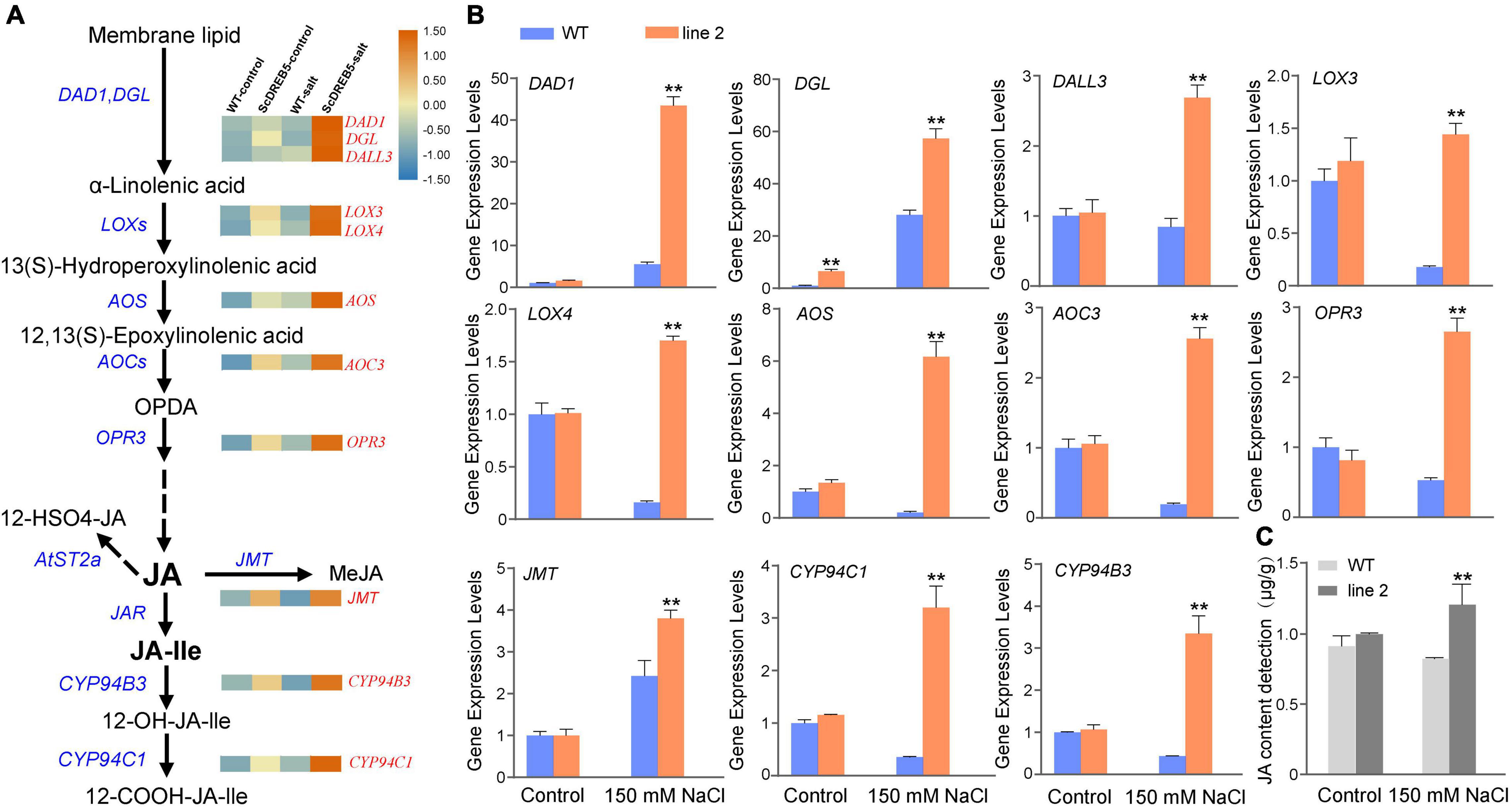
Figure 7. Transcriptome data were used to create heatmaps of the relative expression levels of JA biosynthesis, expression analysis of JA biosynthesis related genes, JA content detection. (A) Expression profiling of 11 JA biosynthesis genes which is significantly increased in transgenic plants. These heatmaps were created using TBTools. The FPKM values of the four samples are normalized by a row scale function, the expression levels from low (blue) to high (orange) indicate the minimum and maximum values for the same row. (B) Eleven JA biosynthesis related genes were selected for RT-qPCR analysis in WT and ScDREB5 transgenic plants under normal and salt stress conditions. The α-TUBULIN (AT1G50010) gene of Arabidopsis was used as the reference gene. (C) JA content detection. Values are means ± SE of three replicates. Significant difference compared with WT plants with the same treatment. LSD multiple comparison test was used: *p < 0.05, **p < 0.01.
We then measured endogenous JA content in WT and transgenic plants under normal and salt stress conditions. Consistent with the gene expression level changes in JA biosynthesis genes, under normal conditions, the JA content of ScDREB5 transgenic plants was comparable to that of WT plants; however, after salt stress, the JA contents of ScDREB5 transgenic plants were significantly higher than WT (Figure 7C). These findings, combined with the transcript profiling data, indicated that overexpression of ScDREB5 enhanced salt stress tolerance in Arabidopsis by regulating JA biosynthesis genes and increasing JA content.
Discussion
Increasing evidence has shown that A-5 type DREB genes in plants play an important role in plant responses to various stresses (Chen et al., 2007, 2009; Liu et al., 2007; Bouaziz et al., 2012; El-Esawi and Alayafi, 2019). In particular, in the desiccation-tolerant moss S. caninervis, A-5 DREBs were largely expanded and most of the ScDREBs showed tolerance to at least one kind of stress in transgenic plants (Li et al., 2016). ScDREB5, a novel A-5b type of DREB gene from S. caninervis, has the highest sequence identity with the AT1G77640 gene (55%), and the identities of ScDREB8 and ScDREB10 were 46 and 54%, respectively. Among these, the function of AT1G77640 has not yet been reported. ScDREB8, belonging to the A-5a type DREBs, improved salt stress tolerance in Arabidopsis (Liang et al., 2017). ScDREB10, an A-5c DREB, enhanced drought and salt tolerances in transgenic Arabidopsis (Li X. et al., 2019). Likewise, an A-5 DREB, PpDBF1, from desert moss Physcomitrella patens increased salt, drought, and cold tolerances in transgenic tobacco (Liu et al., 2007). These findings indicated that A-5 type DREB proteins in moss species may be the dominant stress response regulators, while A-2 type proteins may be such for Arabidopsis (Li et al., 2017). In this study, ScDREB5 improved salt resistance in transgenic Arabidopsis (Figure 3). It is well known that salt stress can lead to increased ROS production (Fichman and Mittler, 2020), particularly H2O2 and O2–, which can cause oxidative damage with MDA accumulation (Sofo et al., 2015; Yu et al., 2020). It is important for plants to activate antioxidative systems to cope with this oxidative damage (You and Chan, 2015), such as CAT, POD, and SOD, to scavenge ROS. The DAB staining, NBT staining, and MDA and H2O2 content analysis results showed that ScDREB5 transgenic lines had lower ROS damage than WT under salt stress (Figures 4A–C), which was consistent with the improved SOD, POD, and CAT activities (Figures 4D–F). The DEG enrichment in the responses to oxygen-containing compounds also indicated that the reactive oxygen system of ScDREB5 transgenic plants was more active than in that of WT plants under salt stress (Figure 5D). These results suggested that ScDREB5 alleviated oxidative damage by increasing antioxidant enzyme activities (Figure 8), then decreased H2O2 levels and MDA content under salt stress. ScDREB8 and ScDREB10 also showed enhanced ROS scavenging ability in transgenic Arabidopsis (Liang et al., 2017; Li X. et al., 2019), indicating that it is a common stress tolerance strategy for A-5 type DREBs.
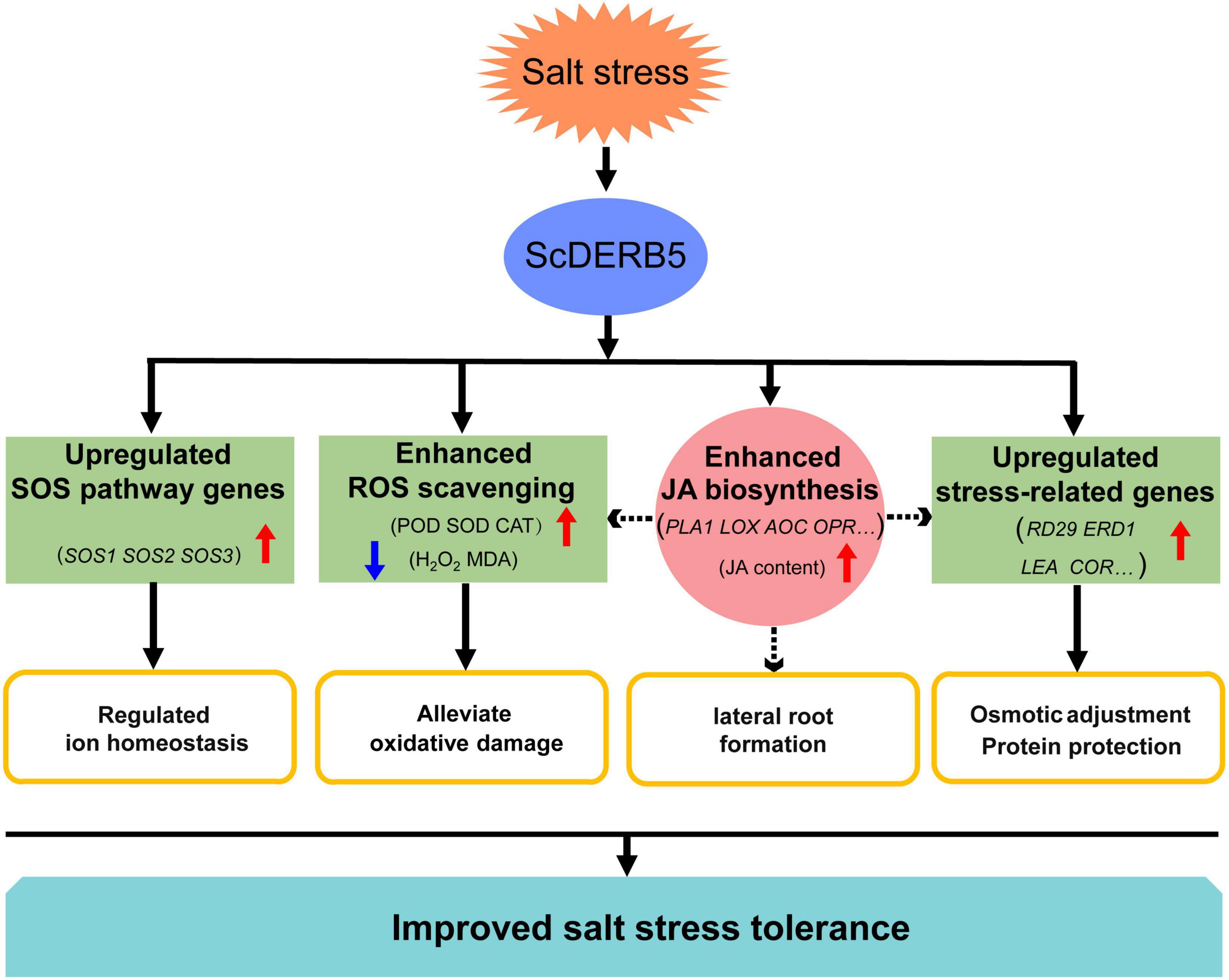
Figure 8. The model of the ScDREB5 regulatory network involved in salt stress responses. Enhanced activities of antioxidant enzymes in the ScDREB5-overexpression line resulting in decreased ROS accumulation, improving the expression of SOS pathway and stress-related genes, thus improving salt stress tolerances. ScDREB5 can activate the expression of JA biosynthesis related genes, modulating the JA content, thus improve salt stress tolerance in Arabidopsis. ↑, ↓ indicate upregulation, downregulation, respectively.
Dehydration-responsive element-binding protein transcription factors can induce the expression of a series of downstream genes, such as RD29, LEAs, CORs, ERDs, and P5CS, which alleviate osmotic damage and protect proline synthesis to enhance abiotic stress tolerances (Agarwal and Jha, 2010; Lata and Prasad, 2011). Similar to other DREBs, ScDREB5 has a highly conserved amino acid composition in the AP2 domain, with 18 out of the 59 identical amino acids in the AP2 domain (Figure 1B). In DREBs, amino acids at positions 14 and 19 were primarily valine (V) and glutamic acid (E), and V14 was almost completely conserved in all DREBs and played an important role in DNA binding (Sakuma et al., 2002; Li et al., 2017). In this study, transcripts of AtRD29B, AtERD1, AtLEA6, AtLEA7, AtCOR47/AtRD17, and AtP5CS1 genes were more upregulated in ScDREB5 overexpression lines than in WT plants under salt stress conditions (Figure 6A), and an increased expression of AtRD29, AtCOR47/AtRD17, and AtLEA genes were also found in transgenic ScDREB8 and ScDERB10 plants (Liang et al., 2017; Li X. et al., 2019). These results indicated that the genes regulated by an A-5 type DREB under stress had relatively high similarity. The SOS signal transduction pathway is very important for ionic homeostasis under salt stress (Yang and Guo, 2018). Wang et al. (2020b) recently reported that the A-2 type SlDREB2 can directly bind to SlSOS1 promoters, thus improving the expression level of SlSOS1 and promoting tomato tolerance to salt stress. ScDREB8 showed enhanced salt stress tolerance in transgenic Arabidopsis, with no significant difference in the expression levels of the SOS1, SOS2, and SOS3 genes (Liang et al., 2017), while their transcript abundances were significantly higher in ScDREB5 transgenic lines than in WT plants under salt stress (Figure 6B), suggesting that the ScDREB5 gene may enhance plant salt stress tolerance by regulating the SOS pathway (Figure 8). E19 was conserved in the A-1–A-4 DREB groups, while it was variable for the A-5 DREBs; ScDREB5 has a K, whereas AT1G77640, ScDREB8, and ScDREB10 have A, P, and I in this position, respectively (Figure 1B). The change in this amino acid residue within the AP2 domain suggested a different DNA binding preference in A-5 DREBs, leading to the expression of different target genes. Our study provides a case where A-5 type DREBs may also enhance the expression of SOS pathway genes, thus improving plant salt tolerance.
Dehydration-responsive element-binding protein transcription factors are key regulators of various stress responses, in which they also respond to various plant hormones and improve plant survival under stress conditions (Nolan et al., 2017). For example, ABA and ethylene (ET) are involved in the regulation of abiotic stress in DREB-mediated plants (Xie et al., 2019). JA has been reported to play an important role in plant biotic stress responses, mainly in pathogens and insects (Chini et al., 2016; Bhavanam and Stout, 2021; Bian et al., 2021). Additionally, as cellular hubs for integrating informational cues from the environment, JA is also involved in plant responses to several abiotic stresses, such as salt, drought, heavy metals, and freezing stresses (Wang et al., 2020c). In this study, KEGG pathway analyses of DEG upregulation showed that DEGs were enriched in “alpha-linolenic acid metabolism” and “linoleic acid metabolism” (Figure 5E). The alpha-linolenic acid metabolism pathway can initiate JA biosynthesis (Wasternack and Song, 2017), and linoleic acid can be converted to alpha-linolenic acid through metabolic pathways (Wang et al., 2021), indicating that JA biosynthesis may partially contribute to salt stress tolerance. This hypothesis was supported by the significantly upregulated transcript abundance of JA biosynthesis-related genes in the transgenic lines compared with WT plants under salt stress (Figure 7B), which was also true for the increase in JA content (Figure 7C). JA has been shown to promote the formation of lateral roots (LR; Cai et al., 2014), which are important for water and nutrient acquisition (Yu et al., 2019) and their development plasticity plays an important role in improving plant survival in response to the external environment (Xu et al., 2020). We found that the number of LRs in the transgenic lines with increased JA content was significantly higher than that of WT plants under salt stress (Figure 3F). It may be that ScDREB5 can activate JA biosynthesis genes, thereby promoting LR formation in transgenic plants exposed to salt stress. Additionally, many transcription factors regulate JA biosynthesis in overexpressed Arabidopsis to improve plant stress tolerance (Verma et al., 2016; Raza et al., 2021). For instance, VvNAC26 from Vitis amurensis has been shown to regulate endogenous JA synthesis, thereby playing a positive role in drought tolerance in transgenic Arabidopsis (Fang et al., 2016), and IbMYB116 has been shown to enhance drought tolerance by regulating endogenous JA synthesis in transgenic Arabidopsis (Zhou et al., 2019); however, the relationship between DREBs and JA has not been extensively studied (Xie et al., 2019). A recent study reported that ORA47, an A-5 type DREB, plays an important role in JA biosynthesis and signaling (Chen et al., 2016); hence, JA might be involved in the ScDREB5-mediated plant salt stress resistance (Figure 8). JA can activate the ROS-scavenging system in plants to improve plant stress tolerance (Lim et al., 2015; Abouelsaad and Renault, 2018; Block et al., 2018). Some reports indicated that the JA contents of transgenic Arabidopsis containing TaAOC1, VaNAC26, and IBMYB116 genes increased, accompanied by improved ROS scavenging abilities under stress conditions, but the mechanism is still unclear. ScDREB5 transgenic lines also showed strong ROS scavenging abilities (Figures 4A–C). After salt stress, H2O2 levels in ScDREB5 transgenic Arabidopsis lines (L2 and L3) increased by approximately 1.2 fold and 1.04 fold, respectively, but increased by approximately 2.9 fold in the WT. Therefore, we speculate that the increase in JA content might enhance the ROS scavenging ability of ScDREB5 transgenic Arabidopsis under salt stress; however, the detailed relationship between ROS scavenging and JA accumulation requires further study. Interestingly, transcript levels of representative salinity-induced genes were induced less with a decrease in endogenous JA content (Mohamed et al., 2015). Li Y. et al. (2019) found that the mediation of the dehydration process by JA occurs via the induction of AtERD1 expression, which is an early response gene in dehydration stress (Nakashima et al., 2010). In our study, the stress marker genes (AtRD29, AtCOR47/AtRD17, AtLEA, AtERD1, and AtP5CS1) were significantly upregulated in ScDERB5 transgenic Arabidopsis under salt stress conditions. These results suggest that JA may participate in the ScDERB5-mediated process of enhancing salt tolerance by ROS scavenging and modulating stress response genes. In conclusion, JA may act as a crosstalk conjunction for the regulation mechanism in ScDREB5-mediated salt stress tolerance.
In conclusion, we identified a novel A-5 type DREB gene, ScDREB5, from S. caninervis, that could improve the salt tolerance of transgenic Arabidopsis by regulating the following processes (Figure 8): (1) Enhanced ROS scavenging ability: Overexpression of ScDREB5 in transgenic plants showed significantly lower ROS damage with significantly lower MDA and H2O2 contents, and significantly higher SOD, POD, and CAT activities compared to WT under salt stress. (2) Improved osmotic adjustment and protein protection abilities: ScDREB5-overexpression led to the upregulation of classic stress-related genes (RD29, LEA, COR47, ERD1, and P5CS1) under salt stress, which can alleviate the osmotic stress caused by salt stress and protein protection. (3) Regulated ion homeostasis: Overexpression of ScDREB5 increased the expression of SOS pathway genes (SOS1, SOS2, and SOS3) in transgenic plants under salt stress, thus regulating ion homeostasis and reducing ion detoxification in plants. (4) JA is a crosstalk conjunction that may be the core regulation mechanism for ScDREB5-mediated salt stress tolerance: ScDREB5 increased endogenous JA content in transgenic Arabidopsis by upregulating JA biosynthesis genes, improving lateral root formation, enhancing ROS scavenging ability, and upregulating stress-related genes under salt stress. However, the detailed biological function of increased JA content and the regulated relationship between ScDREB5 and SOS pathway genes in ScDREB5-mediated salt stress tolerance in transgenic Arabidopsis remains to be elucidated. Future studies should focus on ScDREB5 mechanisms under salt stress. In summary, we suggest that ScDREB5 is an excellent candidate gene for improving salt tolerance in plants.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repositories and accession number can be found below: http://www.ncbi.nlm.nih.gov/bioproject/798571, PRJNA798571.
Author Contributions
XL and YW conceived the ideas and designed the experiments. JL, RY, and YL performed the experiments and collected the data. JL reformed all the analyses and wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (Grant Nos. 31870318 and 31860061), Key Research Program of Frontier Sciences, Chinese Academy of Sciences (Grant No. ZDBS-LY-SM009), Youth Innovation Promotion Association of Chinese Academy of Sciences (Grant No. 2018478), and the Science Foundation for Excellent Young Scholars of Xinjiang Autonomous Region (Grant No. 2019Q013).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.857396/full#supplementary-material
Footnotes
References
Abouelsaad, I., and Renault, S. (2018). Enhanced oxidative stress in the jasmonic acid-deficient tomato mutant def-1 exposed to NaCl stress. J. Plant Physiol. 226, 136–144. doi: 10.1016/j.jplph.2018.04.009
Agarwal, P. K., Gupta, K., Lopato, S., and Agarwal, P. (2017). Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 68, 2135–2148. doi: 10.1093/jxb/erx118
Agarwal, P. K., and Jha, B. (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plant. 54, 201–212. doi: 10.1007/s10535-010-0038-7
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bhavanam, S., and Stout, M. (2021). Seed treatment with jasmonic acid and methyl jasmonate induces resistance to insects but reduces plant growth and yield in rice, Oryza sativa. Front. Plant Sci. 12:691768. doi: 10.3389/fpls.2021.691768
Bian, Z., Gao, H., and Wang, C. (2021). NAC transcription factors as positive or negative regulators during ongoing battle between pathogens and our food crops. Int. J. Mol. Sci. 22:81. doi: 10.3390/ijms22010081
Block, A., Christensen, S. A., Hunter, C. T., and Alborn, H. T. (2018). Herbivore-derived fatty-acid amides elicit reactive oxygen species burst in plants. J. Exp. Bot. 69, 1235–1245. doi: 10.1093/jxb/erx449
Bouaziz, D., Pirrello, J., Ben Amor, H., Hammami, A., Charfeddine, M., Dhieb, A., et al. (2012). Ectopic expression of dehydration responsive element binding proteins (StDREB2) confers higher tolerance to salt stress in potato. Plant Physiol. Biochem. 60, 98–108. doi: 10.1016/j.plaphy.2012.07.029
Cai, X., Xu, P., Zhao, P., Liu, R., Yu, L., and Xiang, C. (2014). Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 5:5833. doi: 10.1038/ncomms6833
Chandler, J. W. (2018). Class VIIIb APETALA2 ethylene response factors in plant development. Trends Plant Sci. 23, 151–162. doi: 10.1016/j.tplants.2017.09.016
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Hsieh, E. J., Cheng, M., Chen, C., Hwang, S. Y., and Lin, T. (2016). ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol. 211, 599–613. doi: 10.1111/nph.13914
Chen, M., Wang, Q., Cheng, X., Xu, Z., Li, L., Ye, X., et al. (2007). GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 353, 299–305. doi: 10.1016/j.bbrc.2006.12.027
Chen, M., Xu, Z., Xia, L., Li, L., Cheng, X., Dong, J., et al. (2009). Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). J. Exp. Bot. 60, 121–135. doi: 10.1093/jxb/ern269
Chini, A., Gimenez-Ibanez, S., Goossens, A., and Solano, R. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33, 147–156. doi: 10.1016/j.pbi.2016.07.005
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for a grobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
El-Esawi, M. A., and Alayafi, A. A. (2019). Overexpression of StDREB2 transcription factor enhances drought stress tolerance in cotton (Gossypium barbadense L.). Genes 10:142. doi: 10.3390/genes10020142
Fang, L., Su, L., Sun, X., Li, X., Sun, M., Karungo, S. K., et al. (2016). Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J. Exp. Bot. 67, 2829–2845. doi: 10.1093/jxb/erw122
Fichman, Y., and Mittler, R. (2020). Rapid systemic signaling during abiotic and biotic stresses: is the ROS wave master of all trades? Plant J. 102, 887–896. doi: 10.1111/tpj.14685
Giri, M. K., Swain, S., Gautam, J. K., Singh, S., Singh, N., Bhattacharjee, L., et al. (2014). The Arabidopsis thaliana At4g13040 gene, a unique member of the AP2/EREBP family, is a positive regulator for salicylic acid accumulation and basal defense against bacterial pathogens. J. Plant Physiol. 171, 860–867. doi: 10.1016/j.jplph.2013.12.015
Gururani, M. A., Venkatesh, J., and Tran, L. S. P. (2015). Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 8, 1304–1320. doi: 10.1016/j.molp.2015.05.005
Ji, H., Pardo, J. M., Batelli, G., Oosten, M. V., Bressan, R. A., and Li, X. (2013). The salt overly sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6, 275–286. doi: 10.1093/mp/sst017
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Li, H., Zhang, D., Li, X., Guan, K., and Yang, H. (2016). Novel DREB A-5 subgroup transcription factors from desert moss (Syntrichia caninervis) confers multiple abiotic stress tolerance to yeast. J. Plant Physiol. 194, 45–53. doi: 10.1016/j.jplph.2016.02.015
Li, X., Liang, Y., Gao, B., Mijiti, M., Bozorov, T. A., Yang, H., et al. (2019). ScDREB10, an A-5c type of DREB gene of the desert moss Syntrichia caninervis, confers osmotic and salt tolerances to Arabidopsis. Genes 10:146. doi: 10.3390/genes10020146
Li, X., Zhang, D., Gao, B., Liang, Y., Yang, H. L., Wang, Y., et al. (2017). Transcriptome-wide identification, classification, and characterization of AP2/ERF family genes in the desert moss Syntrichia caninervis. Front. Plant Sci. 8:262. doi: 10.3389/fpls.2017.00262
Li, X., Zhang, D., Li, H., Gao, B., Yang, H., Zhang, Y., et al. (2015). Characterization of reference genes for RT-qPCR in the desert moss Syntrichia caninervis in response to abiotic stress and desiccation/rehydration. Front. Plant Sci. 6:38. doi: 10.3389/fpls.2015.00038
Li, Y., Yang, X., and Li, X. (2019). Role of jasmonate signaling pathway in resistance to dehydration stress in Arabidopsis. Acta Physiol. Plant. 41:100. doi: 10.1007/s11738-019-2897-7
Liang, Y., Li, X., Zhang, D., Gao, B., Yang, H., and Wang, Y. (2017). ScDREB8, a novel A-5 type of DREB gene in the desert moss Syntrichia caninervis, confers salt tolerance to Arabidopsis. Plant Physiol. Biochem. 120, 242–251. doi: 10.1016/j.plaphy.2017.09.014
Lim, C. W., Han, S. W., Hwang, I. S., Kim, D. S., Hwang, B. K., Lee, S. C., et al. (2015). The pepper lipoxygenase CaLOX1 Plays a role in osmotic, drought and high salinity stress response. Plant Cell Physiol. 56, 930–942. doi: 10.1093/pcp/pcv020
Liu, N., Zhong, N., Wang, G., Li, L., Liu, X., He, Y., et al. (2007). Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from physcomitrella patens. Planta 226, 827–838. doi: 10.1007/s00425-007-0529-8
Ma, J., Yin, C., Guo, Q., Zhou, M., and Wu, Y. (2014). A novel DREB transcription factor from Halimodendron halodendron leads to enhance drought and salt tolerance in Arabidopsis. Biol. Plant. 59, 74–82. doi: 10.1007/s10535-014-0467-9
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283, 185–196. doi: 10.1007/s10973-006-7579-1
Mizoi, J., Ohori, T., Moriwaki, T., Kidokoro, S., Todaka, D., Maruyama, K., et al. (2013). GmDREB2A;2, a canonical dehydration-responsive element-binding protein2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 161, 346–361. doi: 10.1104/pp.112.204875
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Mohamed, H., Bettina, H., Elisabeth, E., Peter, N., and Michael, R. (2015). Increased tolerance to salt stress in OPDA-deficient rice ALLENE OXIDE CYCLASE mutants is linked to an increased ROS-scavenging activity. J. Exp. Bot. 66, 3339–3352. doi: 10.1093/jxb/erv142
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nakashima, K., Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2010). A nuclear gene, ERD1, encoding a chloroplast-targeted clp protease regulatory subunit homolog is not only induced by water stress but also developmentally up-regulated during senescence in Arabidopsis thaliana. Plant J. 12, 851–861. doi: 10.1046/j.1365-313X.1997.12040851.x
Nolan, T., Chen, J., and Yin, Y. (2017). Cross-talk of Brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 474, 2641–2661. doi: 10.1042/BCJ20160633
Oliver, M. J., Tuba, Z., and Mishler, B. D. (2000). The evolution of vegetative desiccation tolerance in land plants. Plant Ecol. 151, 85–100. doi: 10.1023/A:1026550808557
Pan, Z., Pitt, W. G., Zhang, Y. M., Wu, N., Tao, Y., and Truscott, T. T. (2016). The upside-down water collection system of Syntrichia caninervis. Nat. Plants 2:16076. doi: 10.1038/nplants.2016.76
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., and Tran, L. S. P. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 50, 54–69. doi: 10.1111/j.1365-313X.2007.03034.x
Raza, A., Charagh, S., Zahid, Z., Mubarik, M. S., Javed, R., Siddiqui, M. H., et al. (2021). Jasmonic acid: a key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 40, 1513–1541. doi: 10.1007/s00299-020-02614-z
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006a). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006b). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 18822–18827. doi: 10.1073/pnas.0605639103
Shah, W. H., Rasool, A., Saleem, S., Mushtaq, N. U., and Ul Rehman, R. (2021). Understanding the integrated pathways and mechanisms of transporters, protein kinases, and transcription factors in plants under salt stress. Int. J. Plant Genomics 2021:5578727. doi: 10.1155/2021/5578727
Sofo, A., Scopa, A., Nuzzaci, M., and Vitti, A. (2015). Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int. J. Mol. Sci. 16, 13561–13578. doi: 10.3390/ijms160613561
Song, R., Li, T., and Liu, W. (2021). Jasmonic acid impairs Arabidopsis seedling salt stress tolerance through MYC2-mediated repression of CAT2 expression. Front. Plant Sci. 12:730228. doi: 10.3389/fpls.2021.730228
Sun, W., Cao, Z., Li, Y., Zhao, Y., and Zhang, H. (2007). A simple and effective method for protein subcellular localization using Agrobacterium-mediated transformation of onion epidermal cells. Biologia 62, 529–532. doi: 10.2478/s11756-007-0104-6
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wang, J., Liu, Z., Liu, H., Peng, D., Zhang, J., and Chen, M. (2021). Linum usitatissimum FAD2A and FAD3A enhance seed polyunsaturated fatty acid accumulation and seedling cold tolerance in Arabidopsis thaliana. Plant Sci. 311:111014. doi: 10.1016/j.plantsci.2021.111014
Wang, J., Song, L., Gong, X., Xu, J. F., and Li, M. H. (2020a). Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 21:1446. doi: 10.3390/ijms21041446
Wang, J., Yang, H., Gao, B., Bozorov, T. A., Li, X., Zhang, D., et al. (2020b). Analysis and characterization of the aldehyde dehydrogenase (ALDH) gene superfamily in the desert moss Syntrichia caninervis in response to abiotic stress. Environ. Exp. Bot. 178:104176. doi: 10.1016/j.envexpbot.2020.104176
Wang, Z., Hong, Y., Li, Y., Shi, H., Yao, J., Liu, X., et al. (2020c). Natural variations in SlSOS1 contribute to the loss of salt tolerance during tomato domestication. Plant Biotechnol. J. 19, 20–22. doi: 10.1111/pbi.13443
Wasternack, C., and Song, S. (2017). Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68, 1303–1321. doi: 10.1093/jxb/erw443
Wood, A. J. (2007). The nature and distribution of vegetative desiccation-tolerance in hornworts, liverworts and mosses. Bryologist 110, 163–177.
Wu, N., Zhang, Y., Downing, A., Zhang, J., and Yang, C. (2012). Membrane stability of the desert moss Syntrichia caninervis Mitt. during desiccation and rehydration. J. Bryol. 34, 1–8. doi: 10.1179/1743282011Y.0000000043
Xie, Z., Nolan, T. M., Jiang, H., and Yin, Y. (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 10:228. doi: 10.3389/fpls.2019.00228
Xu, P., Fang, S., Chen, H., and Cai, W. (2020). The brassinosteroid-responsive xyloglucan endotransglucosylase/hydrolase 19 (XTH19) and XTH23 genes are involved in lateral root development under salt stress in Arabidopsis. Plant J. 104, 59–75. doi: 10.1111/tpj.14905
Xu, Z., Chen, M., Li, L., and Ma, Y. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Yang, H., Zhang, D., Li, H., Dong, L., and Lan, H. (2015). Ectopic overexpression of the aldehyde dehydrogenase ALDH21 from Syntrichia caninervis in tobacco confers salt and drought stress tolerance. Plant Physiol. Biochem. 95, 83–91. doi: 10.1016/j.plaphy.2015.07.001
Yang, H., Zhang, D., Wang, J., Wood, A. J., and Zhang, Y. (2012). Molecular cloning of a stress-responsive aldehyde dehydrogenase gene ScALDH21 from the desiccation-tolerant moss Syntrichia caninervis and its responses to different stresses. Mol. Biol. Rep. 39, 2645–2652. doi: 10.1007/s11033-011-1017-6
Yang, Y., and Guo, Y. (2018). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
You, J., and Chan, Z. (2015). Ros regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6:1092. doi: 10.3389/fpls.2015.01092
Yu, P., Hochholdinger, F., and Li, C. (2019). Plasticity of lateral root branching in maize. Front. Plant Sci. 10:363. doi: 10.3389/fpls.2019.00363
Yu, Z., Duan, X., Luo, L., Dai, S., and Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Zhang, H., Zhu, J., Gong, Z., and Zhu, J. (2021). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Keywords: DREB transcription factor, Syntrichia caninervis, salt stress, jasmonic acid (JA) biosynthesis, ROS scavenging ability
Citation: Liu J, Yang R, Liang Y, Wang Y and Li X (2022) The DREB A-5 Transcription Factor ScDREB5 From Syntrichia caninervis Enhanced Salt Tolerance by Regulating Jasmonic Acid Biosynthesis in Transgenic Arabidopsis. Front. Plant Sci. 13:857396. doi: 10.3389/fpls.2022.857396
Received: 18 January 2022; Accepted: 28 February 2022;
Published: 06 April 2022.
Edited by:
Jian Li Yang, Zhejiang University, ChinaReviewed by:
Necla Pehlivan, Recep Tayyip Erdoðan University, TurkeyShuai Li, Qingdao Agricultural University, China
Copyright © 2022 Liu, Yang, Liang, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Wang, d2FuZ3lhbnhqdUAxMjYuY29t; Xiaoshuang Li, bGl4c0Btcy54amIuYWMuY24=
†These authors share first authorship
 Jinyuan Liu
Jinyuan Liu Ruirui Yang
Ruirui Yang Yuqing Liang
Yuqing Liang Yan Wang
Yan Wang Xiaoshuang Li
Xiaoshuang Li