
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 11 April 2022
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.856187
This article is part of the Research Topic Insights in Plant Symbiotic Interactions: 2021 View all 15 articles
The nitrogen-fixing symbiosis between leguminous plants and soil bacteria collectively called rhizobia plays an important role in the global nitrogen cycle and is an essential component of sustainable agriculture. Genetic determinants directing the development and functioning of the interaction have been identified with the help of a very limited number of model plants and bacterial strains. Most of the information obtained from the study of model systems could be validated on crop plants and their partners. The investigation of soybean cultivars and different rhizobia, however, has revealed the existence of ineffective interactions between otherwise effective partners that resemble gene-for-gene interactions described for pathogenic systems. Since then, incompatible interactions between natural isolates of model plants, called ecotypes, and different bacterial partner strains have been reported. Moreover, diverse phenotypes of both bacterial mutants on different host plants and plant mutants with different bacterial strains have been described. Identification of the genetic factors behind the phenotypic differences did already and will reveal novel functions of known genes/proteins, the role of certain proteins in some interactions, and the fine regulation of the steps during nodule development.
Nitrogen is an essential macronutrient for plants and is required for the synthesis of nucleic acids, amino acids, and many other important metabolites. It is one of the most limiting elements for plant growth despite dinitrogen gas (N2) accounting for a large proportion (around 78%) of Earth’s atmosphere. Its strong chemical stability, however, makes it inaccessible for most organisms including plants, only certain prokaryotic microorganisms can fix nitrogen, i.e., to break the triple covalent bonds between the nitrogen atoms and produce ammonium (Vance, 2001).
Leguminous plants have the unique ability to grow in nitrogen-poor soils because they establish symbiosis (Suzaki et al., 2015) with a wide range of nitrogen-fixing Gram-negative α- and β-Proteobacteria collectively referred to as rhizobia (Masson-Boivin et al., 2009; Lindström and Mousavi, 2020). This interaction provides advantages for the participating partners. Legumes have access to reduced nitrogen, which they can metabolize, at the cost of energy and organic materials originating from photosynthesis. At the same time, bacteria are provided by a nutrient-rich environment in the symbiotic nodules formed on the roots or occasionally on the stems of the host plant (see later), where a much larger population of descendants than in soil can be established. As crop and pasture legumes can fix as much as 200–300 kg of nitrogen per hectare per year (Peoples et al., 1995), they have been an important element of crop rotation systems for a very long time and provide multiple benefits for agriculture sustainability (Stagnari et al., 2017). The ability of legumes to convert N2 into ammonia, as well as the first isolation and the morphological changes of rhizobia from and in nodules, were demonstrated in the 1880s, which was quickly followed by the market introduction of the first commercial Rhizobium inoculant (Nitragen) in 1895, almost 20 years earlier than performing industrial-scale ammonia synthesis by Carl Bosch (for a historical review, see Soumare et al., 2020). Since then, many inocula containing rhizobia for different legumes have been developed and commercialized to improve the yield of leguminous crops through symbiotic nitrogen fixation.
The development of the symbiosis between leguminous plants and rhizobia is a complex program including several interconnected developmental processes (bacterial infection and nodule organogenesis), multiple exchanges of signals (Gibson et al., 2008), and the activity and coordinated regulation of the expression of numerous genes (Mergaert et al., 2020; Roy et al., 2020). Legumes perceive the lack of nitrogen in soils and secrete flavonoids into the rhizosphere that are recognized as signals by rhizobia (Peters et al., 1986; Redmond et al., 1986) and—through their putative interaction with the NodD transcription factor (TF) proteins (Györgypál and Kondorosi, 1991; Györgypál et al., 1991; Peck et al., 2006), inducing the expression of nodulation (nod) genes. The proteins encoded by nod genes are essential for the synthesis and export of lipo-chitooligosaccharides called nodulation or Nod Factors (NFs) that have a core structure with 4-5 N-acetyl-glucosamine residues and an acyl chain conserved in all different species of rhizobia. Length and saturation level of the acyl chain, as well as the decoration of the backbone with several chemical modifications, such as methyl, acetyl, carbamoyl, or fucosyl groups are determined and mediated by enzymes encoded by the strain-specific nod genes and contribute to the specificity of the symbiosis between the partners (Dénarié et al., 1996; Long, 1996). Even in the absence of rhizobia (Truchet et al., 1991), the NFs induce quick ion fluxes through the membrane of root hairs (Felle et al., 1988), oscillations in calcium concentrations (calcium spiking) in the nuclei of epidermal cells (Ehrhardt et al., 1996), swelling and deformation of the root hairs as well as division of cortical cells (Catoira et al., 2000). The NFs are recognized by a membrane-anchored receptor complex and the perceived signal is transmitted through the so-called common symbiosis signaling (CSS) pathway, shared by another beneficial symbiosis that is established with arbuscular mycorrhizal fungi (Suzaki et al., 2015; Kronauer and Radutoiu, 2021) and, then, translated into gene expression changes by a network of transcription factors (Diedhiou and Diouf, 2018).
In the presence of NF-producing rhizobia and after the original electrophysiological changes, bacteria enter the root and then, invade the cells of the developing nodules (Figure 1). Depending on the interaction, there are two main ways for rhizobia to enter root tissues (Ibáñez et al., 2017): (i) For intercellular invasion, bacteria cross the epidermal layer between neighboring root hair cells or root hair and epidermal cells or through cracks/fissures, and spread through the cortex between cell walls or intercellular air spaces or by a progressive collapse of the invaded cells; (ii) In the model legume plants (Medicago truncatula and Lotus japonicus), as well as in most crop plants (such as soybean, bean, and pea), rhizobia invade roots through the transcellular infection threads (ITs), tubular structures that guide bacteria into the inner tissues of the nodule. In this latter mode, NF production by attached rhizobia induces continuous re-orientation of the root hair growth resulting in a shepherd’s crook-like curled root hair, which forms a so-called infection pocket and surrounds bacteria that establish a microcolony (Gage, 2004). In the infection pocket, the IT is initiated by cell wall degradation and invagination of the root hair membrane (Murray, 2011), and then, it extends by polar growth toward the base of the root hair cell, enters the cortical cell layers until it reaches the new cells produced by the nodule primordium. The IT polar growth requires the coordinated and dynamic action of several proteins that determine membrane domains, polarity, or involvement in the rearrangement of the cytoskeleton or the regulation of NF levels (Tsyganova et al., 2021). On the rhizobial side, the initiation and growth of ITs required from the rhizobia, which are topologically in the extracellular space and multiply in the growing ITs, the adaptation to the specific osmotic, pH, and ionic environment of ITs (Dylan et al., 1990; Putnoky et al., 1998), the regulation of NF levels (Malolepszy et al., 2018) and correct production of surface polysaccharides (López-Baena et al., 2016), such as extracellular polysaccharide (EPS), K-antigen capsular polysaccharide (KPS), or lipopolysaccharide (LPS). In L. japonicus, a receptor structurally similar to the NF receptors monitors, whether the EPS of the symbiotic bacteria has the correct structure, i.e., it prevents bacterial entry if Mesorhizobium loti produces truncated EPS, but it allows infection of bacteria that produce wild-type or no EPS (Kawaharada et al., 2015). In contrast, the Sinorhizobium meliloti strains that are defective in the production of the succinoglycan EPS are not able to infect Medicago roots (Leigh et al., 1985), unless they produce a second exopolysaccharide (galactoglycan EPS II) or KPS (Glazebrook and Walker, 1989; Putnoky et al., 1990; Pellock et al., 2000). In other legumes, such as Glycine or Phaseolus species, the correct structure of LPS is required for the successful infection program (Carlson et al., 1987; Stacey et al., 1991; Margaret et al., 2013).
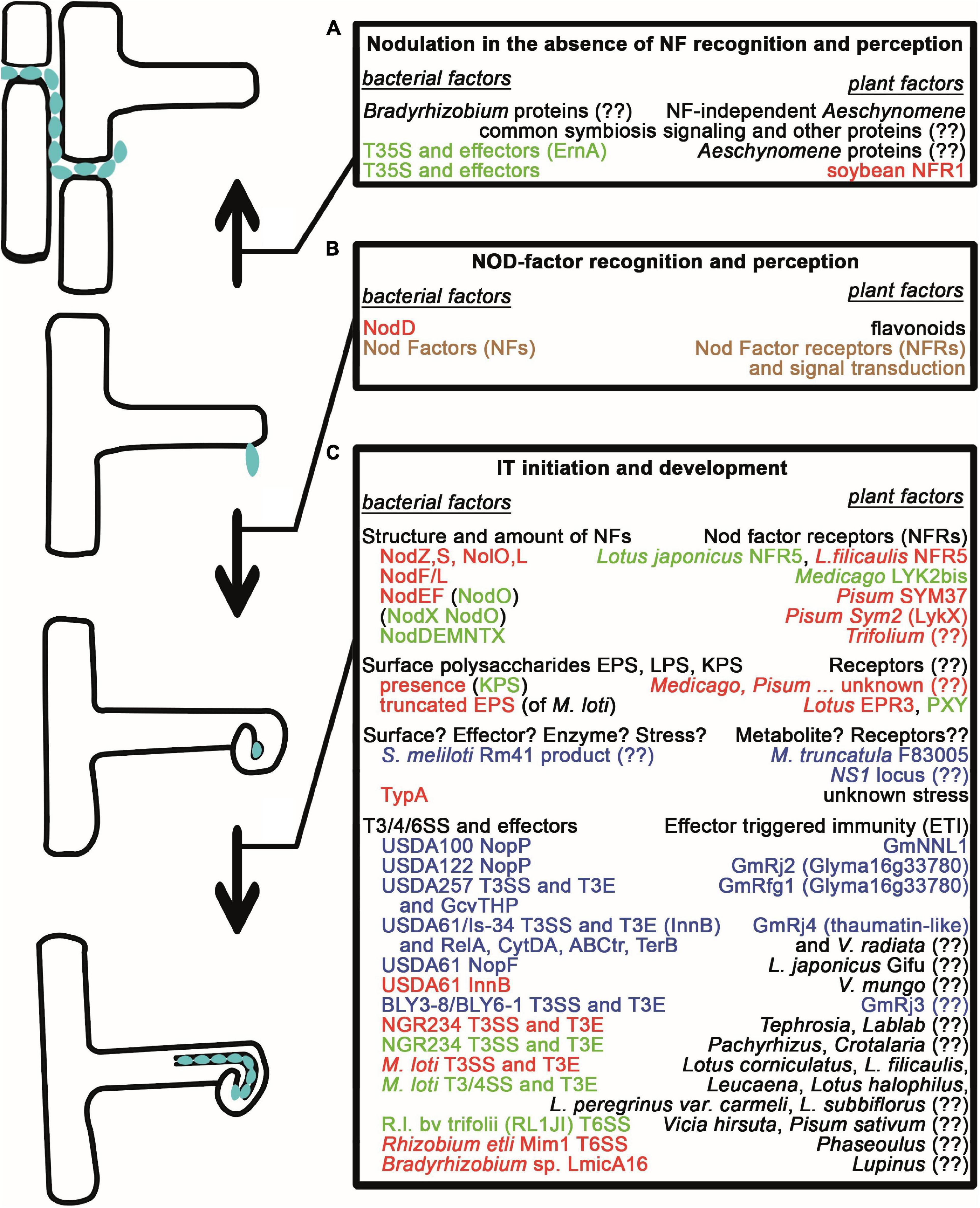
Figure 1. Natural variations affecting early nodule development and infection. (A) Nodule formation and intercellular infection in the absence of Nod factors and NF recognition/perception requires either an unknown mechanism or the activity of Type III secretion systems for the translocation of effector proteins in bradyrhizobia as well as the common symbiosis signaling pathway and other unknown proteins in Aeschynomene species and in soybean plants. (B) Nod Factor production of rhizobia and NF recognition/perception are required for the early symbiotic events such as root hair curling. (C) Initiation and progression of bacterial invasion through ITs are strictly controlled. The structure and amount of NFs determine whether NF receptors allow the initiation of IT development. Similarly, the presence of surface polysaccharides with correct structure is checked by the plants with help of receptors and unknown mechanisms. Unknown mechanisms and stress from the plants also might inhibit infection unless bacteria can deal with them, for example, by expressing the TypA stress protein. Bacterial effector molecules (Nops) resembling pathogen virulence factors transported into the plant cells and Effector Triggered Immunity often led to the restriction of infection, however, there are cases, when they have a positive effect on nodulation. Those bacterial macromolecules, whose lack or incorrect structure led to the arrest of the interaction with certain partners, and those plant proteins, which restrict the bacterial mutants or whose lack can be overturned, are shown in red. Plant molecules, as well as bacterial proteins/molecules, whose lack or presence (in parenthesis), that are able to overturn these defects, are shown in green. Those bacterial and corresponding plant proteins, that are responsible for incompatibility and a defect in either of them leads to compatibility, are shown in blue. If the compatibility needs both the plant and bacterial factors, the proteins are shown in brown. (??) denotes that the given factor has not been identified yet.
In the newly formed differentiating nodule cells, individual bacteria are released from the ITs through an endocytosis-like process (Figure 2) and become surrounded by a membrane of host origin called peribacteroid or symbiosome membrane (Roth and Stacey, 1989). These organelle-like structures, called symbiosomes, divide to fill the cytoplasm of the nodule cells and the rhizobia, thus, differentiating into their nitrogen-fixing form called bacteroids (Vasse et al., 1990). The parallel differentiation of bacteria and plant cells is accompanied by drastic physiological, metabolic, and gene expression changes. The new cells formed by the nodule meristem and being infected by rhizobia complete multiple cycles of endoreduplication resulting in enlargement of nuclear and cell volumes (Foucher and Kondorosi, 2000), change their expression profile in multiple waves (Maunoury et al., 2010) and adjust their metabolism and the cellular environment for nitrogen fixation (Udvardi and Poole, 2013). To protect the oxygen-sensitive nitrogenase enzyme and at the same time, to support bacterial respiration energizing nitrogen fixation, the free oxygen concentration is kept at a very low but constant level within the infected nodule cells. The low free oxygen concentration is achieved by a physical barrier in the outer cortical layer of the nodule and massive production of the oxygen-binding leghemoglobin protein (Rutten and Poole, 2019). This low oxygen concentration regulates the production of bacterial proteins that are involved in the reduction of nitrogen, in the metabolism and transport of the fixed nitrogen, as well as in the exchange of metabolites between the bacterial and plant cells (Terpolilli et al., 2012; Udvardi and Poole, 2013).
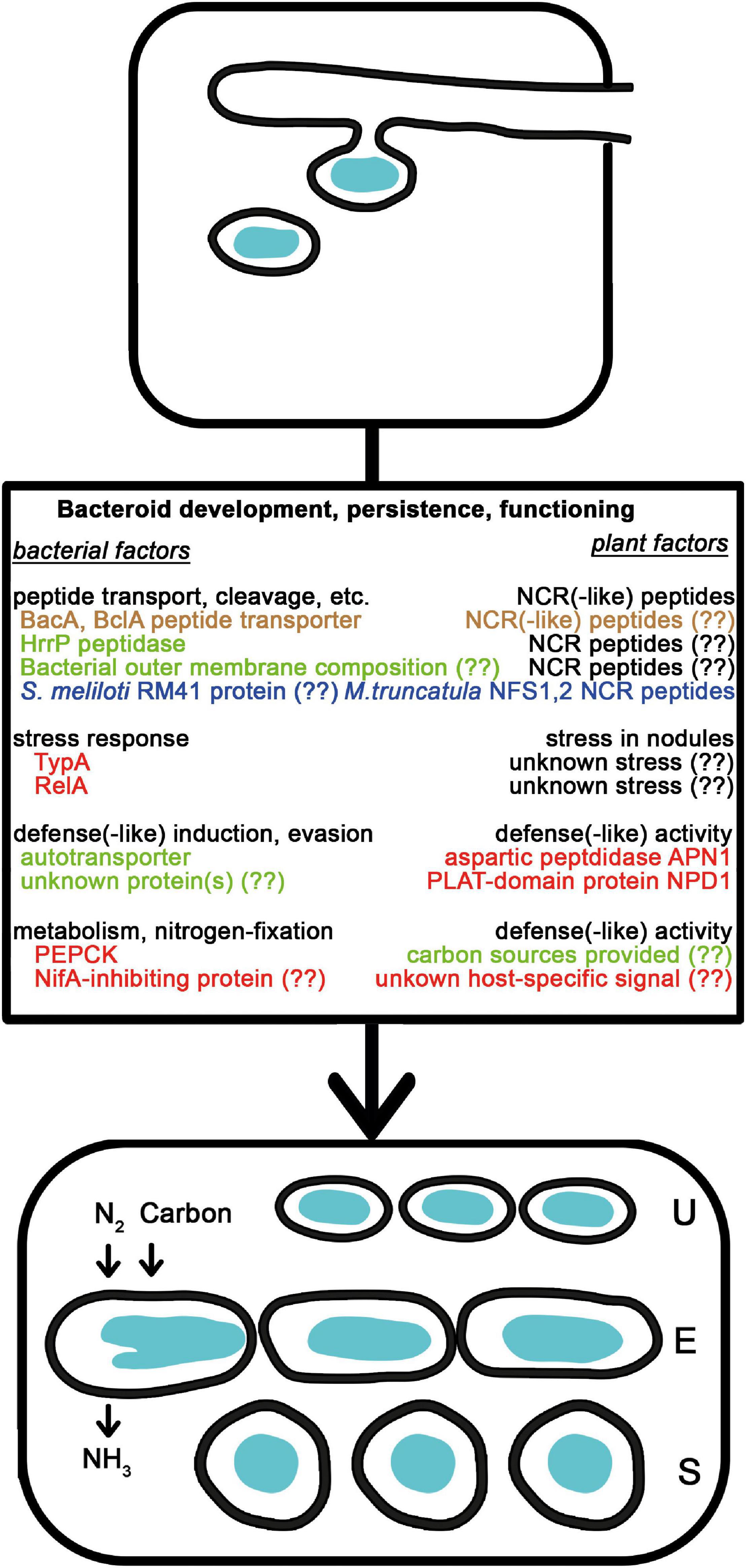
Figure 2. Natural variations affecting bacteroid development, persistence, and functioning. After the release of rhizobia from the ITs, bacteria differentiate into nitrogen-fixing bacteroids of unmodified (U), elongated (E), or spherical (S) morphotypes. The NCR peptides affecting bacterial membrane and intracellular functions are delivered into bacteria by peptide transporters and contribute to the terminal differentiation of E and U morphotype bacteroids. Bacterial outer membrane composition, peptidases and independently evolved transporters affect the success of bacteroid development. The NCR peptides are also involved in strain discrimination. The level of stress and the available carbon sources in the plants affect how bacteria with mutations in stress related genes or carbon metabolism perform. Bacteria also might induce and endure plant defense reactions and sense signals from certain plants that affect gene expression. Colors are the same as in Figure 1.
After reaching its peak, the nitrogen fixation in the nodule cells declines, and this is accompanied by the decrease of leghemoglobin concentration and nitrogenase activity by morphological changes of the cells, or lifestyle of the rhizobial population present. This complex and programmed process called nodule senescence is a normal stage of the symbiosis, however, it can also be induced by the transition from vegetative to the reproductive stage of plant development or by adverse environmental and physiological conditions (Zhou et al., 2021).
Based on their development and morphology that is determined by the plant, both the nodules and the bacteroids can be divided into two major types (Ferguson et al., 2010; Alunni and Gourion, 2016). Indeterminate nodules on the roots of, for example, the Medicago, Pisum, Coronilla, Leucaena, and Amorpha species have a cylindrical shape resulting from the activity of a persistent meristem, which continuously produces new cells. These new cells become infected with bacteria and develop into nitrogen fixation tissue. As a consequence, indeterminate nodules keep increasing in size and have a gradient of developmental stages recognized as meristematic (Zone I), infection (Zone II), differentiation (Interzone II-III), nitrogen fixation (Zone III), and in older nodules, the senescence (Zone IV) zones. In contrast, determinate nodules that form on legume species from, for example, the Lotus, Glycine, Phaseolus, Aeschynomene, or Arachys genera are spherical and the cell population in the inner tissues is relatively homogenous and does not form a developmental gradient. During the formation of determinate nodules, the meristem is not persistent, it ceases to divide at an early stage, and further nodule development takes place more-or-less synchronously.
Independently from the nodule morphology, the differentiation and the fate of bacteria also depend on the plant host (Mergaert et al., 2006): Bacteroids in the nodules of Lotus, Glycine, Phaseolus or Leucaena are similar to the free-living cells in shape, size, DNA content and retain their cell division capacity. In contrast, bacteroids in the nodule cells of Medicago, Pisum, Aeschynomene or Arachys species have increased size, membrane permeability and DNA content, different—spherical or elongated-(branched)—shape compared to their free-living siblings. These nodule bacteria also lost their cell division capacity, thus, they are considered terminally differentiated bacteroids (Alunni and Gourion, 2016). The ability to impose terminal bacteroid differentiation on the bacterial partner appeared at least five times during the evolution of the Papilionoideae legumes indicating a possible fitness benefit for the plant (Oono et al., 2010). Using bacteria, which can establish symbiosis with plant’s hosts governing different bacteroid fates, it was shown that terminally differentiated bacteroids have increased symbiotic efficiency as compared to unmodified bacteroids (Oono and Denison, 2010). Moreover, even the level of terminal differentiation correlates with nitrogen fixation efficiency, as in nodules of different Aeschynomene species, highly polyploid spherical bacteroids are more efficient than the elongated ones with lower ploidy level (Lamouche et al., 2018, 2019). Terminal bacteroid differentiation is induced by plant-derived molecules, termed as nodule-specific cysteine-rich (NCR and NCR-like) peptides (Mergaert et al., 2003; Van de Velde et al., 2010; Czernic et al., 2015), which are produced almost exclusively in the infected cells of nodules (Guefrachi et al., 2014) solely by those plants, which impose terminal bacteroid differentiation on their symbiont. The number and type of NCR peptides in the Inverted Repeat Lacking Clade (IRLC) of Papilionoideae legumes are highly variable and correlate with the morphotype of the bacteroids in the nodules of these species (Montiel et al., 2016, 2017). The NCR peptides were shown to interact with several bacterial proteins in M. truncatula, thus, affecting, for example, the transcription, translation, and cell cycle regulation (Tiricz et al., 2013; Farkas et al., 2014; Penterman et al., 2014). Despite their high number (over 700 genes in Medicago) and putatively redundant functions, individual NCR peptides were proven to be required for bacteroid development and persistence (Horváth et al., 2015; Kim et al., 2015).
In general, if the symbionts possess the above-reviewed tool kits, they can establish symbiosis with the partners in their cross-inoculation group to reduce atmospheric nitrogen and to support plant growth. However, it was recognized by early investigations performed mainly on crop plants that there exist symbiotic incompatibilities (i.e., no nodule formation or no nitrogen fixation) among symbionts that form effective interactions with other cultivars, or strains of the partner species (Bergersen and Nutman, 1957; Caldwell, 1966; Melino et al., 2012). It was also shown that when compatible interactions lead to nitrogen fixation, their efficiency, i.e., the plant benefit derived from the symbiosis is highly variable (Snyman and Srijdom, 1980; Terpolilli et al., 2008; Kazmierczak et al., 2017). To further complicate the picture, mutations may have different effects on the interaction depending on the partners investigated [for example (Wilson et al., 1987; Osteras et al., 1991; Rodpothong et al., 2009)].
It was recognized early after the discovery of NFs that the induction of root hair deformation and cortical cell division does not require the full and proper decoration of these signal molecules in contrast to root hair curling, as well as the initiation and growth of ITs (Ardourel et al., 1994). IT development in alfalfa root hairs was strongly inhibited by the nodL and nodEF mutants of Sinorhizobium meliloti lacking the O-acetylation and N-acylation with specific fatty acids, respectively, at the non-reducing end of NFs that was more pronounced with the double mutant. These observations led to the hypothesis that different receptor forms, i.e., less stringent signaling receptors for the initiation of root hair deformation and cortical cell division, and the entry receptors with more rigorous requirements for bacterial infection regulate the nodulation process.
Similar to M. sativa, most M. truncatula accessions form no nodule with the nodF/nodL double mutant S. meliloti strains, but M. truncatula ssp. tricycla R108 establishes an effective nitrogen-fixing symbiosis with the mutant as with the wild-type strain (Luu et al., Under review1). As the NF receptor proteins NFP and LYK3 (Arrighi et al., 2006; Smit et al., 2007) in ecotype Jemalong, and the NFP protein in R108 (Feng et al., 2019) were shown to be essential for NF perception and nodulation, the genomic sequence of R108 around the LYK3 gene was analyzed in more detail. Between LYK2 and LYK3, an additional gene designated as LYK2bis, which is a chimera of the two neighboring genes, was identified in R108 that could not be found in any other M. truncatula accessions with a sequenced genome. Using loss-of-function (lyk2bis mutant in R108) and gain-of-function (transforming LYK2bis into Jemalong) approaches, it was proven that the LYK2bis protein enables R108 to be nodulated and infected by the nodF/nodL mutant.
The nodE gene of Rhizobium leguminosarum was also shown to be a determinant of host specificity of the biovars and mutations in this gene’s generally decreased nodulation on pea (Surin and Downie, 1989). Interestingly, the nodE mutation in R. leguminosarum bv. viciae almost blocked the nodulation on certain pea lines but affected less severely the other accessions. Genetic analysis revealed that nodE-dependent nodulation is associated with a haplotype (Li et al., 2011) of the PsSym37 gene coding for a LysM-type receptor kinase, which is closely related to the LjNFR1/MtLYK3 Nod Factor receptors, and is essential for infection-thread initiation in pea (Zhukov et al., 2008). Wild isolates—commonly referred to as Afghan peas—from the Middle East (Afghanistan, Iran, Turkey, Israel, Uzbekistan, and Tajikistan) and known as the center of origin for peas, cannot be nodulated by rhizobia collected in Europe, but only by strains isolated from soils from the same region (Lie, 1984). In Afghan peas, cortical cell divisions are initiated upon induction by European R. leguminosarum bv. viciae strains but IT formation and bacterial invasion are blocked. This inhibition can be overcame either by growing the plants at elevated temperature (Kozik et al., 1995) or by the production of the NodX protein in rhizobia that adds an O-acetyl group to the C6 carbon of N-acetylglucosamine residue at the reducing end of the pentameric NF (Götz et al., 1985; Firmin et al., 1993). Genetic analysis of the locus called sym2, which is responsible for the incompatibility of Afghan peas, added a level of complexity. The sym2A allele of Afghan peas is dominant over the sym2C alleles of cultivated peas when R. leguminosarum bv. viciae strain PRE is used as inoculum, however, the dominance is changed when strains 248 or PF2, producing higher amount of Nfs, are applied (Kozik et al., 1995). Despite the long history of research on the specificity of Afghan peas, no direct evidence, such as changing the specificity of a cultivated line by transformation, about responsible for the strict partner selection has emerged. However, several genetic and bioinformatics data indicates that another LysM-type receptor kinase, termed PsLykX (for P. sativum LysM kinase eXclusive), probably interacting with the pea NFR1 (PsSym10) receptor may determine the trait (Sulima et al., 2017, 2019; Solovev et al., 2021). Interestingly, the nodO gene coding for a secreted protein, which was shown to form ion channels in membranes (Sutton et al., 1994), can compensate both for the nodE mutation (Walker and Downie, 2000) and partially or fully for the absence of the nodX gene in strain 248 on plants carrying the sym2A allele in homozygous or heterozygous forms, respectively (Geurts et al., 1997). The non-nodulating phenotype of the nodO mutant of strain 248 on the heterozygous plants, as compared to nodule formation by wild-type bacteria, might explain the strain-specific differences regarding the dominance of the sym2 alleles. It is possible that the genome of strain PRE does not code for a functional NodO protein for compensation. It is hypothesized that NodO might function either to bypass the NF receptor activity or to amplify a weak signal originating from the interaction of the not fully compatible NFs and receptors.
Similar variability in NF recognition and sensitivity might exist in Trifolium species. R. leguminosarum bv. trifolii strain TA is not able to nodulate T. subterraneum cv. Woogenellup at 22°C because of the arrest of IT development but forms an effective symbiosis with this cultivar at 28°C (Lewis-Henderson and Djordjevic, 1991a). This trait is determined by the recessive alleles of a gene named rwtl (resistance of Woogenellup to strain TA1). The nodulation deficiency of strain TA1 is overcome by mutations in genes that affect the structure (nodE and nodX), the amount (nodD, nodM, nodN, and probably, the negatively acting csn-1 (for cultivar-specific nodulation) of unknown identity and function), or transport (nodT) of Nod Factors (Lewis-Henderson and Djordjevic, 1991b).
The most numerous and most detailed investigations of NF recognition and perception have been performed in Lotus species, mostly in L. japonicus, serving as a model for determinate nodule development, in symbiosis with Mesorhizobium loti. This rhizobium produces mainly pentameric NFs with an acetylated fucosyl residue at the C6 carbon of the reducing sugar, a carbamoyl group, and a N-methylated 18:1 acyl chain at the C4 and C2 carbons, respectively, of the non-reducing sugar (López-Lara et al., 1995). The symbiotic capability of M. loti R7A mutants that fail to synthetize acetyl-fucosylated NFs depends on the Lotus species serving as partner (Rodpothong et al., 2009). Nodule formation by a nolL mutants failing to add the acetyl modification to the fucosyl moiety is delayed on all four Lotus species (L. japonicus, L. corniculatus, L. filicaulis, and L. burttii) tested. Moreover, IT formation in L. filicaulis is also arrested. Defects in the synthesis (in the nolK and noeL mutants) and transfer to the NF backbone (in the nodZ mutant) of the fucosyl residue prevent infection not only in L. filicaulis but also in L. corniculatus, revealing different stringency requirements in the Lotus-Mesorhizobium cross-inoculation group. An engineered symbiont of Lotus called strain DZL and was created by expressing an inducer-independent NodD protein, as well as the NodZ fucosyl-transferase and the NolL acetyltransferase in R. leguminosarum bv. viciae (Pacios Bras et al., 2000), produces NFs that only differ from those of M. loti in the decorations of the non-reducing sugar, i.e., the acyl chain on the C2 carbon is not methylated, the C6 carbon is not carbamoylated but the C5 carbon is acetylated. This strain effectively nodulated L. japonicus but failed to infect L. filicaulis. It was shown by domain and amino acid swaps between the Nfr receptors of the two species that a single amino acid difference in the M2 domain of Nfr5 is responsible for the differential recognition of NFs (Radutoiu et al., 2007).
Legumes rigorously choose their rhizobial partners by recognizing the chemical structure of the first signal molecules, the Nod Factors, via the activity of the NF receptors. Structural variations and extension of the NF receptor repertoire might both strengthen (NodX requirement by SYM2) or weaken (LYS2bis) the stringency of the identification process, which is also affected by the amount of NFs and temperature.
In the indeterminate nodule forming rhizobia, an efficient infection (thread development) requires the production of exopolysaccharides (EPSs), the lack of EPS resulting in no IT formation or in ITs aborted in the root hairs (Skorupska et al., 2006). In S. meliloti strain Rm41, however, EPS-deficient (exoB mutant) bacteria induce the formation of infected, nitrogen-fixing nodules on alfalfa and this phenotype is associated with its ability to produce a strain-specific K-antigen (KPS), a polymer of a disaccharide repeating units composed of glucuronic acid and N5-β-hydroxybutyryl-N7-acetyl-5,7-diamino-3,5,7,9-tetradeoxynonulosonic acid (pseudaminic acid) on the surface (Putnoky et al., 1990; Kereszt et al., 1998; Reuhs et al., 1998). Interestingly, if the lpsZ (rkpZ) gene, found in the Rm41 strain-specific rkp-3 gene cluster (Kiss et al., 2001), responsible for the synthesis of the pseudaminic acid precursor, as well as for the export of the KPS, is introduced into an EPS-deficient mutant of strain 1,021 producing a structurally different KPS, the transconjugant forms effective symbiosis with alfalfa (Williams et al., 1990). The LpsZ protein determines (decreases) the polymerization level of the KPS in both strains and enables the formation of the symbiotically efficient molecular weight form (Reuhs et al., 1995). The KPS of strain Rm41 can complement for the absence of EPS, not only on alfalfa (M. sativa), but also on other Medicago (M. media and M. varia) and Melilotus (M. albus and M. officinalis) species tested (Putnoky et al., 1990), however, not on M. truncatula (Hozbor et al., 2004; Liu et al., 2014). In S. meliloti strain 1,021, a third polysaccharide, the galactoglucan EPS II with a disaccharide repeating unit of a β-(1-3)-linked acetylated glucose and succinylated galactose can contribute to the infection process by replacing the succinoglycan EPS I during nodulation of M. sativa (Glazebrook and Walker, 1989). However, EPS II cannot function in place of EPS I on other investigated hosts such as M. coerulea, M. truncatula, Melilotus albus, and Trigonella foenum-graecum.
Determinate nodule formation and infection require the correct structure of LPS on the bacterial surface, while the production of exopolysaccharides by rhizobia infecting these nodules seems to be not important. In Lotus corniculatus and L. japonicus Gifu, however, bacteria, which do not produce EPS, can establish as effective symbiosis as the wild-type strain, whereas mutants affected in mid or late biosynthetic steps (e.g., exoU) and produce truncated form of the polysaccharide induced uninfected nodule primordia (Kelly et al., 2013). In contrast, L. japonicus MG20 is less stringent in its selection because it forms nitrogen-fixing nodules with mesorhizobia that are both producing no or truncated EPS. The incompatibility in L. japonicus Gifu is mediated by the LysM-type receptor EPR3 (Kawaharada et al., 2015), recognizing the diffusible octasaccharide monomer of EPS, not only from M. loti, but also from R. leguminosarum and S. meliloti (Wong et al., 2020). The PXY leucine-rich repeat receptor-like kinase in L. japonicus MG20, which also regulates stem vascular development, was identified by quantitative trait locus sequencing (QTL-seq) as a casual component of the differential and less stringent exoU response (Kawaharada et al., 2021).
The S. fredii strain HH103 establishes symbiosis with a wide variety of legumes forming indeterminate and determinate nodules (Margaret et al., 2011). Infection of the determinate nodules of soybean and pigeon pea (Cajanus cajan) by this bacterium necessitates the production of the 5-acetamido-3,5,7,9-tetradeoxy-7-(3-hydroxybutyramido)-L-glycero-L-manno-nonulosonic acid homopolymer K-antigen, however, KPS-deficient mutants of HH103 induce infected and nitrogen-fixing determinate nodules on cowpea (Parada et al., 2006; Hidalgo et al., 2010).
The presence of certain polysaccharides on the bacterial surface is essential for the infection process monitored by a currently unknown mechanism in the plants. Certain bacteria are able to produce alternative polysaccharides to compensate for the absence of the generally used ones, however, this compensation might not be effective on all hosts. Moreover, not only the presence, but the correct structure, for example, of the EPS is checked by another mechanism.
Large collections of legume hosts and bacteria isolated from nodules developed on plants in natural habitats have allowed the establishment of cross-inoculation groups and investigation of natural variations within cross-inoculation groups and species. Investigations, for example, with the model legumes L. japonicus and M. truncatula and/or species closely related to them, revealed large variations in symbiotic compatibility and efficiency (Snyman and Srijdom, 1980; Crook et al., 2012; Gossmann et al., 2012; Granada et al., 2014; Liu et al., 2014; Lorite et al., 2018). For example, experiments using a number of Medicago species and M. truncatula accessions in combination with different S. meliloti isolates revealed tremendous variation in nodulation capacity and nitrogen fixation specificity between different genotype-rhizobial combinations, 40–50% of all host-strain pairs resulted in an ineffective symbiosis (Crook et al., 2012; Liu et al., 2014). Detailed characterization accompanied with the identification of the genes and alleles determining these symbiotic variations beyond soybeans’ incompatibility (see later) has been very rare.
Liu et al. (2014) reported that S. meliloti strain Rm41 induced root hair curling and nodule primordium formation but failed to infect the roots of M. truncatula ecotype F83005.5, although both partners have the genetic capacity for nitrogen fixation. The infection process is arrested at the microcolony stage, either no or occasionally, aberrant ITs not entering the cortex can be observed on the roots. This phenotype is similar to the ones induced by EPS-deficient mutants of S. meliloti or by M. loti producing truncated EPS or by rhizobia incompatible with certain genotypes of soybean. This dominant trait in M. truncatula, however, must be independent of these bacterial effectors. EPS recognition can be ruled out because the strain produces wild-type EPS and its EPS-deficient derivatives cannot establish effective symbiosis with other ecotypes of the plant either. The possibility of effector-triggered immunity (ETI) responsible for the soybean incompatibilities (see later) can also be excluded because the genome of strain Rm41 does not code for any Type III, Type IV, and Type VI secretion systems implicated in ETI.
An R. leguminosarum strain (termed Norway) isolated from L. corniculatus nodules shows host-genotype specific differences when inoculated on wild-type plants of Lotus species and pea cultivars. It induced no nodules on L. japonicus Gifu and L. filicaulis, L. japonicus MG20 formed bumps and occasionally, very small and infected nodules, while L. japonicus Nepal showed broadened elongated infection zones. On L. glaber, the strains provoked the development of swellings and tumor-like structures, while L. burtii developed normal-sized and infected but inefficient nodules. The strain-induced ITs on Pisum sativum cv. Sparkle without the formation of nodule(-like) structures, whereas pea cultivar Little Marvel and Latyrus sativus plants had ineffective nodules (Gossmann et al., 2012; Liang et al., 2019). In Lotus species, the strain induced early and strong induction of a symbiosis-specific gene but no ITs, and rather intercellular accumulation of the bacteria through epidermal cracks could be observed. The strain could invade intact nodule cells where it formed symbiosomes, however, these infected cells exhibited the signs of early senescence (Liang et al., 2019). As the strain seems to possess all the genetic repertoire required for and still fails in the establishment of an effective sysmbiosis (Liang et al., 2018), it will be interesting to find one or more compatible hosts and determine which factors in the bacteria and plants cause the incompatibilities.
The incompatible interactions arrested at the infection stage indicate the presence of additional checkpoints beyond NF and polysaccharide recognition.
Bacteria secrete proteins and other (macro)molecules to modulate their interactions with the environments, especially when they are interacting with eukaryotic host organisms. In the case of Gram-negative bacteria, secretion requires translocation across both the inner and the outer membranes, and several different molecular machines have been elaborated for this purpose. Many proteins secreted by pathogens and symbionts are aimed to enter the host cells to modify the physiology of the partner, and, thus, several secretion systems include an apparatus to translocate proteins across the plasma membrane of the host also (Tseng et al., 2009). In this context, it is not surprising that rhizobia are also equipped with certain or all types of effector delivery machinery that may have special roles in their interactions with different legume partners.
Plants developed a multilayered defense system against microbes that acts both locally and systematically (Ngou et al., 2021) and its elements are also important during the interaction of legumes and rhizobia (Cao et al., 2017). The first line of defense detects the presence of microorganisms via the activity of receptors recognizing microbe/pathogen-associated molecular patterns (MAMPs/PAMPs), which are conserved motifs present on essential components of a microbe/pathogen. The binding of MAMPs by these pattern recognition receptors (PRRs) induces rapid changes in the cell—as can be observed upon the recognition of NFs—and leads to MAMP/PAMP-triggered immunity (MTI/PTI). Interestingly, rhizobial MAMPs investigated, so far, such as flagellin or LPS, differ from, for example, those of plant pathogenic or enteric bacteria and do not induce MTI (Tellström et al., 2007; Lopez-Gomez et al., 2012). The legume hosts also evolved recognition mechanisms to distinguish beneficial and harmful microorganisms. L. japonicus and M. truncatula have LysM pattern-recognition receptors that are related to the NF receptors to separate the perception of chitin oligomeric microbe-associated molecular patterns from the perception of NFs by the NFR1/NFR5 receptor complex (Bozsóki et al., 2017). Another LysM receptor in L. japonicus, EPR3 distinguishes M. loti cells producing no or normal EPS from those that produce a truncated one with a pentasaccharide repeat instead of an octasaccharide repeat (Kawaharada et al., 2015). Although MTI/PTI at the cell surface is very effective, microbes evolved virulence factors and apparatus for their delivery to the host cells to modify or attenuate the original immune responses. The type III secretion system (T3SS) of Gram-negative bacteria including pathogens and symbionts is a complex multiprotein secretion apparatus that actively exports effector proteins (T3 effectors) with diverse biochemical activities (Schreiber et al., 2021) through the lumen of these tubular structures and directly into the eukaryotic host cells. In rhizobia, proteins that are either extracellular components of or secreted by the T3SS apparatus are termed as Nops, nodulation outer proteins, and are produced upon NF induction (Staehelin and Krishnan, 2015). The T3 effector proteins can be detected by intracellular receptors of the plant immune system that are usually highly specific in the effectors they recognize, an observation that led to the gene-for-gene hypothesis (Flor, 1971). The recognition of the effector results in a very robust immune response called Effector Triggered Immunity (ETI), which is often culminated in programmed cell death called hypersensitive response to halt the spread of the pathogen.
The incompatibilities between soybean cultivars and specific rhizobium strains that have been studied and described as gene-for-gene interactions for a long time (Hayashi et al., 2012) seem true to be determined by ETI. Four dominant genes of soybean, Rj2 (Caldwell, 1966), Rj3 (Vest, 1970), Rj4 (Vest and Caldwell, 1972), and Rfg1 (Trese, 1985) were described to prevent nodulation with certain soybean-nodulating strains belonging to the Bradyrhizobium and Sinorhizobium genera. The block of these interactions takes place just after the first initial steps. Root hair deformation and/or curling and cortical cell division can be observed although in a lower number than in compatible interactions, but ITs and nodule meristems are not formed (Sadowsky et al., 1995; Yasuda et al., 2016). Analyses of rhizobial mutants have revealed the important role of T3 effectors in the determination of incompatibility with all the Rj2, Rj3, Rj4, and Rfg1 soybeans because mutants defective in the T3SS, or the production of certain effectors, could form functional nodules on the roots of their incompatible hosts (Meinhardt et al., 1993; Tsukui et al., 2013; Faruque et al., 2015; Tsurumaru et al., 2015; Shobudani et al., 2020; Ratu et al., 2021a).
The Rj2, Rj4, and Rfg1 genes of soybean were identified by map-based cloning with surprising results. The Rj4 gene codes for a thaumatin-like protein (TLP), which belongs to the PR-5 family of pathogenesis-related proteins (Tang et al., 2016), considered as effectors to biotic (for example, fungal attack) and abiotic (for example, osmotic shock) stresses. It was most probably evolved by recent local gene duplication and diversification because both Rj4 and rj4 plants harbor a gene coding for another thaumatin-like protein, which is highly similar to Rj4 (only 13 amino acid differences between the two proteins of 296 residues) but does not cause incompatibility with B. elkanii strain USDA61, while the second gene causing the incompatibility is present only in the Rj4 plants. The fact that the Rj4 protein is not a classical receptor protein is surprising and it will be intriguing to understand how this thaumatin-like protein is involved in ETI to regulate bacterial infection.
Another surprising and interesting result was that the allelic variants of the same gene, Glyma16g33780 coding for a toll-interleukin receptor/nucleotide-binding site/leucine-rich repeat (TIR-NBS-LRR) resistance (R) protein are responsible for the Rj2 and Rfg1 incompatibilities (Yang et al., 2010). It was shown that a single amino acid difference, isoleucine versus arginine at amino acid position 490 after the NBS domain in the Rj2 and rj2 alleles, respectively, determines symbiotic (in)compatibility (Sugawara et al., 2019). Polymorphism at five amino acids in and after the sixth LRR domain of this R protein differentiates the Rfg1 and rfg1 alleles (Yang et al., 2010), however, systematic investigation of these differences to reveal the role of the individual residues has not been conducted yet. All four possible alleles (rj2/rfg1, Rj2/rfg1, rj2/Rfg1, and Rj2/Rfg1) of the gene were constructed and shown to determine the expected compatibility profile when transformed into a non-restrictive (rj2/rfg1) soybean (Fan et al., 2017). A genome-wide association study (GWAS) to identify natural variants in key loci that regulate the compatibility between soybean and rhizobia, using Chinese landraces and improved cultivars and B. diazoefficiens strain USDA110 as inoculant, pinpointed and confirmed the Glyma.02G076900 gene termed G. max Nodule Number Locus 1 (GmNNL1) coding for another TIR–NBS–LRR receptor protein and carrying a SINE transposon in the compatible plants as the determinant of the incompatibility (Zhang et al., 2021).
As for the bacterial side, the T3 effector NopP protein of strains USDA122 and USDA110 were shown to be responsible for the incompatibility with Rj2 and GmNNL1-positive soybeans, respectively. Interestingly, if only three amino acids of USDA122 NopP are replaced by those of the compatible strain USDA110, the compatibility with Rj2 cultivars is restored (Sugawara et al., 2018). Such specificity has not been identified between the Rfg1 allele-encoded protein and any of the T3 effectors in strain USDA257. Lack of synthesis and/or secretion of the T3 effectors leads to compatibility with soybeans carrying the Rfg1 allele (Meinhardt et al., 1993) indicating that classical ETI affects compatibility. Moreover, mutations disrupting the gcvTHP operon coding for the elements of the glycine cleavage system enables strain USDA257 to form nitrogen-fixing nodules on Rfg1 soybeans (Lorio et al., 2010). The glycine cleavage system is involved in the generation of C1 units in the form of N5, N10-methylene tetrahydrofolate, that is used in a variety of biochemical reactions, including the synthesis of purines, histidine, thymine, and methionine as well as the formylation of tRNAfMet already loaded with methionine. The gcvTHP mutations do not affect the synthesis and secretion of the Nop proteins in general, thus, further studies are required to elucidate whether a putative decrease in the C1 pool has a direct or indirect role. This putative decrease might affect, for example, the T3SS/T3 effectors or decrease the amount of the innate immune system activating formyl-methionine containing oligopeptides released by damaged bacteria as described in mammalian systems (Zhang et al., 2010).
The incompatibilities of B. elkanii strain USDA61 and B. japonicum strain Is-34 with Rj4 soybeans are also dependent on the T3SS because these hosts form an effective symbiosis with the strains if their T3SS systems are non-functional (Faruque et al., 2015; Tsurumaru et al., 2015). Moreover, mutations in four genes coding for proteins (the RelA ppGpp synthetase; a cytosine deaminase; a TerB family tellurite resistance protein; a substrate-binding protein of an ABC transporter) that are not T3E effectors but might affect T3SS synthesis and/or function also restored compatibility (Nguyen et al., 2017). Indeed, it was recently shown that ppGpp synthetase deficient bradyrhizobia are not able to activate the T3SS (Pérez-Giménez et al., 2021).
The Nop proteins are indeed “double-edged swords” of rhizobia (Staehelin and Krishnan, 2015) because they can restrict nodulation on one host, while promoting interaction with another plant. At present, it is not known why bacteria are equipped with these effectors that can restrict their symbiotic interactions. One possible explanation, which would be worth investigating, is that these molecules facilitate the interactions of the given strain with legumes of the natural flora where the strain evolved, and their incompatibilities can be observed with plants with different spatial origin. The B. elkanii strain USDA61 is incompatible not only with Rj4 soybeans but also with Vigna radiata cultivar KPS1 and different Lotus accessions. As with Rj4 soybean plants, these hosts form effective symbiosis with USDA61 if its T3SS system and the above-mentioned four genes are non-functional. This similarity suggests that the proteins participate in a common mechanism contributing to nodulation restriction in these legumes. Intriguingly, the lack of another T3E Nop effector identified in the same screen as the four non-Nops, a protein of unknown function termed InnB restored the compatibility of the strain with Vigna radiata cultivar KPS1 but not with Rj4 soybean, meaning that different effector proteins might be recognized by the R proteins of the two plant species. To further complicate the picture, the InnB deficient strain proved to be less efficient than the wild-type bacterium when the host was V. mungo (Nguyen et al., 2018). Similarly, the T3SS had a positive impact on the symbiotic efficiency of the B. vignae strain ORS3257 in V. unguiculata and V. mungo, but it blocked symbiosis with V. radiata (Songwattana et al., 2021). In line with the observations on the host-dependent effect of InnB effector, T3SS-based incompatibility of strain USDA61, with the diverse Lotus accessions, arrests the interaction at different developmental stages. The L. japonicus Gifu inhibited infection while L. burttii inhibited nodule maturation at the post-infection stage, whereas both in L. burttii and L. japonicus MG-20, a nodule early senescence-like response can be observed (Kusakabe et al., 2020). It was shown that NopF and NopM were the effector proteins that triggered the inhibition of infection and the nodule early senescence–like response, respectively.
In general, the T3SS of a rhizobium strain may play both positive and negative role or can be indifferent during nodulation depending on the plant partner and, thus, may determine (in)compatibility and host-range (Marie et al., 2001; Nelson and Sadowsky, 2015; Songwattana et al., 2017). For example, in the case of S. fredii strain NGR234 with extreme broad host range (Pueppke and Broughton, 1999), abolition of the T3SS has no effect on nodulation of V. unguiculata, but increased nodule number on Pachyrhizus tuberosus and allowed to form effective symbiosis instead of empty pseudo-nodules on Crotalaria juncea, while the mutant strains formed fewer nodules on Tephrosia vogeli, Lablab purpureus, and Flemingia congesta roots (Viprey et al., 1998; Marie et al., 2003). Similarly, it was shown that the NopC effector protein of S. fredii strain HH103 is beneficial for the symbiosis with Glycine max and V. unguiculata (Jiménez-Guerrero et al., 2015), but blocks nodulation with L. japonicus (Jiménez-Guerrero et al., 2020). The T3SS of Mesorhizobium loti strain MAFF303099 inhibits the interaction with Leucaena leucocephala (Sánchez et al., 2009) and three Lotus species (L. peregrinus var. carmeli, L. subbiflorus, and L. halophilus), while having positive effect on the nodulation of L. corniculatus subsp. frondosus and L. filicaulis (Okazaki et al., 2010). In addition, the competitiveness of the T3SS mutants against the wild-type strain also depends on the host. L. tenuis cultivars INTA Pampa and Esmeralda have contrasting, while L. japonicus MG20 has no strain preference (Sánchez et al., 2012).
Besides T3SS, only Type IV (T4SS) and Type VI (T6SS) Secretion Systems have the ability to deliver effector proteins from bacteria into the cytosol of eukaryotic cells and, thus, to affect their interactions. Compared to T3SS, a relatively very low information is available about the potential influence of T4SS and T6SS of rhizobia on symbiotic performance and compatibility.
Most M. loti strains including R7A lack the genes coding for the elements and effectors of T3SS, but contain genes coding for a type 4 secretion system (T4SS) that can also deliver effectors into target cells. Interestingly, as reported on the T3SS mutants of M. loti strain MAFF303099, strain R7A T4SS mutants formed large and bacteroid-containing effective nodules on Leucaena leucocephala in contrast to the wild-type strain that could not infect the plant, were delayed in nodulation, and less competitive than the wild-type bacteria on L. corniculatus (Hubber et al., 2004). Based on this similarity in the phenotypes, the authors concluded that the type IV and type III system are inter-changeable and over the course of evolution, rhizobia can adopt either type. Among sinorhizobia nodulating, only Medicago, Trigonella and Melilotus species, the presence of T4SS is quite widespread, while T3SS have been detected in a small number of S. meliloti strains and no T6SS coding genes have been shown to be present in the S. meliloti and S. medicae genomes (Sugawara et al., 2013). Abolition of T4SS function in one strain of both species revealed no, as well as positive and negative, effects on the symbiosis depending on host inoculated.
The pRL1JI (pRle248a) symbiotic plasmid carries the nodulation and nitrogen fixation genes of R. leguminosarum bv. viciae strain 248, a symbiotic partner of Pisum, Vicia, and Lens species. Introduction of pRL1JI into R. leguminosarum bv. trifolii strain RCR5, from which its symbiotic plasmid had been cured, resulted in a strain, which could form effective symbiosis with Vicia sativa but failed to infect V. hirsuta and pea (Roest et al., 1997). A transposon insertion mutant that is able to establish nitrogen-fixing symbiosis with the two incompatible hosts led to the identification of a gene cluster that later turned out to code for the T6SS of R. leguminosarum bv. trifolii strain RCR5 (Bladergroen et al., 2003). In contrast to pea, the Type VI Secretion Systems of Rhizobium etli Mim1 (Salinero-Lanzarote et al., 2019) and Bradyrhizobium sp. LmicA16 (Tighilt et al., 2021) have a positive and essential role in developing effective symbiosis with Phaseolus and Lupinus species, respectively.
In the majority of cases, nodule development in legume plants is initiated by the bacterial NFs that are recognized by plasma membrane receptors of the plant. It was, however, shown that certain photosynthetic bradyrhizobia, such as Bradyrhizobium sp. BTAi1 and ORS278, do not produce NFs because they have no nodABC genes, but they are able to still form nodules on the stem of certain Aeschynomene species, such as A. indica, A. evenia, and A. sensitive, which are exclusively nodulated by photosynthetic bradyrhizobia (Giraud et al., 2007). This surprising discovery was conformed when a nodB mutant of Bradyrhizobium sp. ORS285 could form nodules on these plants, while it could not establish symbiosis with other host species (for example, A. afraspera) that are stem-nodulated by both non-photosynthetic and photosynthetic strains. Searching for the genetic determinants required to induce nodule formation in the absence of NFs by transposon insertion mutagenesis has not revealed any essential genes indicating the likely redundancy of currently unknown functions involved in nodule induction (Bonaldi et al., 2010). Despite the NF independent nodule formation, nodulation of A. evenia requires the activity of the common symbiosis signaling pathway because if the genes coding for the Symbiosis Receptor Kinase (SYMRK), the Ca2+/calmodulin-dependent kinase (DMI3) or the cytokinin receptor histidine kinase (LH1/CRE1) were silenced, nodule formation was inhibited (Fabre et al., 2015).
Even NF-dependent hosts can be nodulated in the absence of NFs and NF perception. NFR1-deficient mutants of soybean varieties Enrei and Clark formed nodules when inoculated with B. elkanii strain USDA61 even if its NF production was abolished by a mutation in the nodC gene, providing the T3SS was functional (Okazaki et al., 2013). Root-hair curling and ITs were not observed in the roots and nodules formed on the NF receptor mutants, indicating that T3SS is involved in crack entry or intercellular infection. Similarly, USDA61 could form (though ineffective) nodules on NF-independent Aeschynomene species in a T3SS-dependent manner, however, not all the species or all ecotypes of A. evenia were nodulated by USDA61. These results prompted an investigation to determine whether other non-photosynthetic bradyrhizobia are able to induce nodules on these Aeschynomene plants, and whether such nodulation depends on the T3SS (Okazaki et al., 2016). The authors observed a whole spectrum of responses from no nodule formation through uninfected and infected ineffective nodules to nitrogen-fixing interactions. while the invasion of nodules happened through both intercellular and root hair infection.
The efficiency of the more intimate interaction between strain ORS3257 and NF-independent Aeschynomene accessions showed plant-determined natural variations (such as disturbed bacteroid differentiation and nitrogen fixation) and the nodule formation also depended on T3SS activity. In contrast, the genome of most photosynthetic bradyrhizobia does not contain genes coding for the T3SS and T3Es, and if it contains, as the case with strain ORS285, this translocation system is not required for NF-independent symbiosis, meaning that two mechanisms to induce nodules exists; one depends on T3SS, while the other uses a so far unknown activity. The T3SS of ORS285, however, contributes to the NF-dependent nodulation although it affects the efficiency, but not equally in all plants. The T3SS mutation had no consequences on certain NF-dependent hosts, but had positive (more nodules) or negative (less nodules) effect on others. To determine which T3Es of strains ORS3257 and USDA61 are responsible for the induction of NF-independent nodule formation on Aeschynomene species and the nfr1 mutant soybean, respectively, mutants deficient in genes coding for the T3Es were inoculated on the corresponding partners. The ORS3257 mutants showed a wide variety of phenotypes (Teulet et al., 2019). One mutation increased both nodulation and nitrogen fixation, two mutations led to reduced number of nodules, some of which displayed large necrotic zones, and the nodules induced by two other mutants contained no bacteria. A strain with mutation in an effector gene termed ernA for “effector required for nodulation-A” and widely conserved among bradyrhizobia lost the capacity for nodule formation, not only in ORS3257 but also in strain USDA61. Introduction of the ernA gene into a Bradyrhizobium strain, which does not have an ernA gene and is unable to nodulate Aeschynomene species in an NF-independent manner, despite the presence of a functional T3SS, enabled the transconjugant to form nodules. Moreover, the nucleus targeted ErnA protein, if ectopically produced in A. indica roots, activated organogenesis of root- and nodule-like structures.
The Bel2-5 effector of strain USDA61 that resembles the XopD effector of the plant pathogen Xanthomonas campestris was shown by both loss-of-function and gain-of-function approaches to enable the nfr1 mutant of soybean cultivar Enrei to form nodules (Ratu et al., 2021b). The same effector causes restriction of nodulation on soybeans carrying the Rj4 allele, and it was shown by mutational analysis that most of its predicted domains were essential for both NF-independent nodulation and nodulation restriction (Ratu et al., 2021a).
As pathogenic bacteria, some rhizobia equipped themselves with effector molecules that can modify the functioning of plant cells and with the delivery machinery to translocate these effectors into the partners’ cells. These effector molecules sometimes facilitate more efficient interaction with certain plants and, thus, extend host-range or overcome the lack of NFs/NF recognition, but in other interactions, they trigger plant defenses, which arrest the interaction.
Legumes in the dalbergioid and inverted repeat-lacking clades of the Papilionoideae subfamily produce NCR and NCR-like peptides to impose terminal bacteroid differentiation on their bacterial partners (Figure 2). In the IRLC species, there was a positive correlation between the degree of bacteroid elongation and the number of the expressed NCRs, but the process can be affected by the inoculant. When the basal IRLC legume Glycyrrhiza uralensis producing seven NCR peptides is inoculated with Mesorhizobium tianshanense HAMBI3372, bacteroids show the signs of terminal bacteroid differentiation, such as increased cell size and decreased cell division capacity (Montiel et al., 2016), but the bacteroids of S. fredii strain HH103 are not terminally differentiated in the nodules of this plant (Crespo-Rivas et al., 2016). Interestingly, the electrophoretic profile of the LPSs, especially in the O-antigen containing S(mooth)-LPS region, was markedly different from that of cultured cells when the bacteroids were isolated from G. uralensis nodules, whereas it was indistinguishable when bacteroids were isolated from soybean or pigeon pea. Whether this LPS modification or/and the observed decreased sensitivity toward the antimicrobial activity of cationic NCR peptides are responsible for the lack of terminal bacteroid differentiation remains to be elucidated (Crespo-Rivas et al., 2016).
The bacA gene coding for a peptide transporter plays an essential role in bacteroid development and survival in those rhizobia that form nodules on NCR peptide producing IRLC legumes, such as pea and alfalfa. In contrast, the orthologous sequence is dispensable when plants such as Lotus not having NCRs are infected (Kereszt et al., 2011). Although the BacA proteins of different rhizobia play the same role during the terminal differentiation of bacteroids, it turned out that bacA genes from S. fredii or R. leguminosarum bv. Viciae 3841 failed to restore the Fix+ phenotype of an S. meliloti bacA mutant on M. sativa, however, they allowed for further developmental progression before a loss of viability (diCenzo et al., 2017). Interestingly, the same genes could complement the mutant when Melilotus albus was the host and the S. meliloti bacA gene could complement the symbiotic defect of the R. leguminosarum bv. Viciae mutants during nodule development on pea roots. The authors showed that the S. meliloti BacA has rapidly diverged from the other rhizobial proteins and suggested that it most probably has evolved toward a specific interaction with Medicago (NCR peptides). This suggestion is in agreement with the observation that, although NCR peptides have a single origin, their evolution has followed different routes in individual IRLC legume lineages (Montiel et al., 2017).
As described in a previous chapter, the Dalbergoid clade contains Aeschynomene species with either NF-dependent or NF-independent nodulation ability. A comparative study using the Bradyrhizobium strain ORS285, which can nodulate both types of plants, revealed that bacteria enter plant tissues via intercellular infection mechanism, and the division and development of infected founder cells give rise to the nodule tissues, where bacteria go through a terminal differentiation program (Bonaldi et al., 2011). Later it was shown that NCR-like peptides govern the terminal bacteroid differentiation in these species too, which is completed as spherical morphotype in the NF-independent species and as elongated bacteroids in NF-dependent species (Czernic et al., 2015). The differentiation of both S and E morphotype bacteroids requires the activity of the peptide transporter BclA, a homolog of S. meliloti BacA, both implicated in the transport of the NCR(-like) peptides, although it is required for the survival of bacteria only in the NF-independent plants (Guefrachi et al., 2015). Interestingly, B. diazoefficiens strain USDA110, the soybean model symbiont can nodulate the NF-dependent Aeschynomene species, however, the nodulation is associated with atypical bacteroid differentiation showing the signs of both the terminal and non-terminal developments and with suboptimal symbiotic efficiency (Barrière et al., 2017; Nicoud et al., 2021). The size and DNA content of the bacteroids in the Aeschynomene nodules and the requirement, more precisely, the indifference of the BclA protein during bacteroid differentiation were the same as in the soybean nodules, while the membrane permeability of the bacteroids is more similar to that of the ORS285 bacteroids.
The S. meliloti strains A145 and Rm41 form ineffective symbiosis with M. truncatula cv. Jemalong (Tirichine et al., 2000; Liu et al., 2014), while their interactions with other ecotypes, such as DZA315.16 or A20, is normal. Jemalong nodules are invaded by these strains, even bacteroid differentiation and nif gene induction takes place at 7 days post-inoculation (dpi), but the elongated bacteroids are eliminated (14 dpi), and only saprophytic bacteria remain in the older nodules at 21 dpi (Wang et al., 2017). Surprisingly, dominant allelic variants of two genes termed NFS1 and NFS2 (nitrogen fixation specificity) coding for NCR peptides function as a negative regulator of symbiotic persistence (Wang et al., 2017, 2018; Yang et al., 2017). The NCRs were shown not only to be positive regulators of bacteroid development and persistence (Van de Velde et al., 2010; Horváth et al., 2015; Kim et al., 2015), but the cationic peptides also have antimicrobial activity (Van de Velde et al., 2010; Tiricz et al., 2013). Although the NCR variants from the incompatible host possess stronger antimicrobial activity than those from the compatible plants, the differences in the bactericidal activity do not correlate with and are not responsible for the in planta function because the responses of a compatible strain are similar. The bacterial targets of the peptides and the genetic determinants of the incompatibility in the bacteria, however, remain to be identified.
Crook et al. (2012) described several accessory plasmids that restrict nodule development, cause impaired symbiotic nitrogen fixation, and enhance host invasion. One of these plasmids was reported to encode the HrrP metallopeptidase that is responsible for the symbiotic incompatibility with some host plants (Price et al., 2015). This enzyme was shown to cleave NCR peptides in vitro, although with different efficiency, suggesting some level of NCR peptide substrate selectivity. Bacteroids in the nodules—that are much smaller than normal, most probably because of the loss of meristem persistence—on the incompatible host plant develop normally and express the nif genes but in older cells, they appear to fragment and to degenerate. In addition, the saprophytic rhizobial population in both compatible and incompatible nodules is significantly larger. One possible explanation for these phenotypes is that the peptidase cleaves NCR peptides/peptide variants, both those that are required for bacteroid persistence and those that have antibacterial activity. This cleavage activity might be more efficient on the peptides of the incompatible ecotypes leading to bacteroid senescence. On the other hand, cleaving the cationic peptides that would restrict the cheaters might increase the saprophytic population.
Strains of R. leguminosarum bv. trifolii (such as ICC105), able to form effective nodules on Trifolium ambiguum (Caucasian clover), form bacteroid containing but ineffective nodules on T. repens (white clover), whereas strains (for example, NZP514) that form effective nodules on white clover usually do not nodulate Caucasian clover. It was shown (Miller et al., 2007) that the nifHDKEN, fixABCX, and, probably, the NifBfdxNfixU operons of strain ICC105 are not induced in the white clover nodules that were not caused by the lack of nifA expression or NifA function. Rather, it is supposed that the intergenic region between the nifH and fixA genes might be bound not only by the canonical transcription factors NifA and RpoN, but also by additional host-specific regulatory elements. This proposed protein might bind upstream of the NifA binding sites either in response to or in the absence of a host-specific signal and interacts with NifA to prevent it from activating the expression of the nif/fix operons.
Certain host plants impose terminal bacteroid differentiation on their rhizobial partners with the help of NCR(-like) peptides produced in the infected nodule cells. The NCRs are required not only for bacteroid differentiation, persistence, and functioning, but are also used for strain discrimination and elimination. Bacteria might counteract the effects of NCRs by producing a modified outer membrane or a peptidase cleaving the peptides. Some of these peptides possess antibacterial activity, which is attenuated by the BacA/BclA peptide transporters to a different extent in different hosts that show only limited interchangeability between bacteria infecting legumes with diverse peptide profiles. It is not known whether NCRs or other factors are responsible for the host-specific activation of the nitrogen-fixation genes in nodules of clovers.
The majority of the genes required for the development and function of nitrogen-fixing nodules were identified by forward genetics studies when the failure of the mutants in establishing effective symbiosis indicated their essentiality in the interaction. Intriguingly, there are cases, apart from bacterial T3SS effectors and ETI elements, when a mutation manifested in the arrest of the interaction at a certain developmental stage with one partner, whereas in no disturbance or halt in another stage with another partner(s).
The production of NFs and exopolysaccharides are controlled by complex regulatory networks including the stringent response, which induces a physiological change in response to adverse growth conditions/stress and can also control bacterial development, virulence using guanosine tetra- and penta-phosphate (ppGpp) as effector molecules (Wells and Long, 2002). The relA-deletion mutant of S. meliloti incapable of ppGpp synthesis fails to nodulate M. sativa because of the early arrest in IT development and meristem formation, but successfully infects M. truncatula although bacteroids have disorganized positioning within nodule cells and reduced nitrogen-fixation capacity in this host (Wippel and Long, 2019). The mutant seems to produce NFs and EPS in a higher amount than the wild-type cells and these elevated levels might be inhibitory for alfalfa and neutral or even stimulant in barrel medic. Transcriptomic changes in the mutant upon luteolin induction also pointed to other stress regulatory processes, such as osmoregulation, that might also affect the infection process. In the nodules of M. truncatula, the number of differentially expressed bacterial genes are rather low, however, the upregulation of numerous transcripts related to metabolism and transcriptional regulation depends on RelA, indicating that full metabolic adaptation to the nodule environment and, as a direct or indirect consequence, the normal bacteroid organization and nitrogen fixation might require RelA function.
Mutation in another stress-related gene of S. meliloti, typA, coding for a ribosome-binding GTPase acting at the level of protein synthesis, was shown to be involved not only in stress adaptation, but to be required for the establishment of nitrogen-fixing symbiosis on certain Medicago hosts (Kiss et al., 2004). The mutant formed efficient nodules on different M. sativa cultivars, with only ∼20% decrease in shoot dry weight of some lines and with M. truncatula ecotype DZA315.16. In contrast, it formed Fix– nodules on ecotypes Jemalong and F83005, however, the nodules on the former were small and devoid of bacteroids, while no obvious differences in nodule occupancy between the wild-type and mutant bacteria-induced F83005 nodules could be observed. The TypA and related proteins in other bacteria are required for the adaptation to several stresses under different conditions. Rhizobia must adapt to ever-changing physiological conditions such as pH, ion composition, osmotic concentration, oxidative stress, or plant defense reactions during nodule invasion and bacteroid differentiation. These conditions, which are most probably different in each host plant, may explain why TypA is not required equally on the investigated hosts.
Energy and carbon sources for bacteria in the nodule cells are provided in the forms of the TCA-cycle intermediary C4-dicarboxylates, succinate, fumarate, and malate that necessitate the activity of gluconeogenesis to generate intermediates for growth and metabolism. The first step of gluconeogenesis, the decarboxylation and phosphorylation of the TCA-cycle intermediate oxaloacetate is catalyzed by the phosphoenolpyruvate carboxykinase (PEPCK) enzyme. The effect of a mutation in the pck gene coding for PEPCK in S. fredii strain NGR234 on nodule development and nitrogen fixation was shown to depend on the host inoculated with the mutant (Osteras et al., 1991). The determinate nodule-forming plants, Macroptilium atropurpureum and V. unguiculata formed fewer nodules when inoculated with the mutant as compared to the wild-type that provided very low or no gain in plant biomass attributable to the symbiosis. On Leucaena leucocephala forming indeterminate nodules, the mutant induced an excess number of nodules that had 60% of wild-type efficiency of symbiotic nitrogen fixation. In all nodules harboring the mutant, the number of infected cells was lower compared to that observed in NGR234 induced nodules. One possible explanation for the variability of symbiotic efficiency is that different carbon source availability might be provided by a given plant during the invasion, bacteroid development, and nitrogen-fixation that would determine the relative importance of the bacterial gluconeogenesis pathway.
The forward genetic screen of M. truncatula Tnt1 insertion mutants using S. meliloti strain 1021, as inoculant identified the npd1-1 mutant (line NF4608), which formed Fix– nodules, arrested at an early developmental phase, just after bacteria are released from the ITs (Pislariu et al., 2019). The released rhizobia start to elongate but fail to differentiate into functional, nitrogen-fixing bacteroids, rather, they go through a degradative senescence process most probably caused by the activation of plant defense responses. Interestingly, strain Rm41 induces functional, nitrogen-fixing nodules that can support plant growth as in wild-type plants. The mutation abolishes the function of a symbiotically expressed gene termed NPD1 (Nodule-specific PLAT Domain) coding for a small protein localized in the endoplasmatic reticulum (ER) and symbiosomes, but not co-localized with Golgi. It is supposed that NPD1 with other ER-localized proteins suppresses rhizobium induced plant defense reactions. How and why the hosts discriminate against bacterial strains remains to be elucidated.
The L. japonicus apn1 (sym104) mutants, when inoculated with Mesorhiozobium loti strain TONO, display severe nitrogen deficiency symptoms and form small white nodules, in which the early senescence of nodule cells and the bacteroids therein can be observed (Yamaya-Ito et al., 2018). This senescence was manifested in a decrease in bacteroid density, abnormal enlargement, and irregular shapes of symbiosomes, the presence of lytic vacuoles, and disintegration of infected cells. Interestingly, not all the M. loti strains induce Fix– nodules on the mutant plants; out of the nine strains tested five bacteria, including strain MAFF303099, formed effective pink nodules. The mutations in Lotus disrupt a symbiosis-specific late nodulin gene coding for a secreted aspartic peptidase enzyme termed ASPARTIC PEPTIDASE NODULE-INDUCED 1 (APN1). Random mutagenesis of strain TONO identified the causal gene termed DCA1 (Determinant of nitrogen fixation Compatibility of APN1) causing the incompatibility that encodes an active autotransporter, one of the proteins also known as the Type V protein secretion system found in Gram-negative bacteria (Shimoda et al., 2020). The protein contains an N-terminal secretion signal peptide, a C-terminal autotransporter β-domain, and 43 divergent glycine-rich repeats of 44–53 amino acids. Autotransporters cleave a part of their protein known as the passenger domain and transport it through the outer membrane to the outside of the cell and the whole length of this passenger domain with all glycine-rich repeats is required for its activity. It was shown that the APN1 protein can degrade the DCA1 protein under the acidic conditions required for the activity of aspartic peptidases. The phenotypic dissimilarity caused by the different strains producing orthologous, basically identical autotransporter is determined by the expression differences of the gene in the bacteria, i.e., DCA1 promoter-GUS activity in wild-type and apn1 mutant nodules was negligible in strain MAFF303099 and high in strain TONO. It is proposed that strain TONO produces a higher amount of the effector protein than MAFF303099 that is degraded by the APN1 aspartic peptidase in the wild-type plant. In the mutant plants, however, the effector is not degraded and is able to induce the expression of cysteine protease genes leading to suppression of nodule maturation and initiation of senescence, while the amount of DCA1 from MAFF303099 is not enough for the induction.
Bacteria face diverse levels of stresses and defense responses, as well as metabolite availability and composition, in different wild-type and mutant plants. The ability of wild-type and mutant bacteria to respond and adapt to those conditions is not the same in different strains, and that is manifested in phenotypic variations. These phenotypic variations will allow us to reveal what causes the differences.
Exploring natural variations has already facilitated the recognition of important determinants of symbiotic nodule initiation, development, and function, such as the identification of the LysM2 domain of NFR5 as a potential binding site for the bacterial NFs or recognition of the role of T3SS, Type3 effectors, and ETI in partner selection or revealing that NCR peptides, not only direct the terminal differentiation and persistence of bacteroids, but also participate in partner discrimination and selection. These examples as well as the results with mutants showing multiple phenotypes depending on the hosts suggest that it is worth to reinvestigate the already existing mutants with other symbiotic partners and identify the genetic determinants behind the phenotypic differences in both wild-type and mutant interactions. The agricultural productivity of legume crops depends on their interaction, i.e., (in)compatibility with the rhizobia resident or introduced as inoculum in the soils that is also affected by the competition of the different strains. Similarly, engineering nitrogen-fixing non-legume crops in the future, as well as their cultivation on the field, requires the knowledge of the details and fine-regulation of the symbiotic process. The results of studies on natural variations will identify the determinants of symbiotic (in)compatibility and reveal the fine details that exist in the regulation of the interaction, thus, may contribute to the implementation and realization of the plans aiming at the development of novel crops for sustainable agriculture.
TW, BB, and SK collected, collated, organized the reviewed publications, and participated in the writing of the manuscript. AK conceived the topic of the review, supervised the co-authors’ work, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Work in the lab of AK was supported by the Hungarian National Office for Research, Development, and Innovation (NKFIH Grant Nos. K120122, K128486, and K134841), with contribution from the NKFIH Frontline Research project KKP129924 and the Balzan Research Grant to É. Kondorosi. SK has been a recipient of the Young Researcher Fellowship previously from the Hungarian Academy of Sciences, and currently from the Eötvös Loránd Research Network (ELKH).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alunni, B., and Gourion, B. (2016). Terminal bacteroid differentiation in the legume-rhizobium symbiosis: nodule-specific cysteine-rich peptides and beyond. New Phytol. 211, 411–417. doi: 10.1111/nph.14025
Ardourel, M., Demont, N., Debellé, F., Maillet, F., De Billy, F., Prome, J. C., et al. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6, 1357–1374. doi: 10.1105/tpc.6.10.1357
Arrighi, J. F., Barre, A., Ben Amor, B., Bersoult, A., Soriano, L. C., Mirabella, R., et al. (2006). The medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279. doi: 10.1104/pp.106.084657
Barrière, Q., Guefrachi, I., Gully, D., Lamouche, F., Pierre, O., Fardoux, J., et al. (2017). Integrated roles of BclA and DD-carboxypeptidase 1 in bradyrhizobium differentiation within NCR-producing and NCR-lacking root nodules. Sci. Rep. 7:9063. doi: 10.1038/s41598-017-08830-0
Bergersen, F. J., and Nutman, P. S. (1957). Symbiotic effectiveness in nodulated red clover. IV. the influence of the host factors i1 and ie upon nodule structure and cytology. Heredity 11, 175–184. doi: 10.1038/hdy.1957.15
Bladergroen, M. R., Badelt, K., and Spaink, H. P. (2003). Infection-blocking genes of a symbiotic rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe Int. 16, 53–64. doi: 10.1094/MPMI.2003.16.1.53
Bonaldi, K., Gargani, D., Prin, Y., Fardoux, J., Gully, D., Nouwen, N., et al. (2011). Nodulation of Aeschynomene afraspera and a. indica by photosynthetic bradyrhizobium sp. strain ORS285: the nod-dependent versus the nod-independent symbiotic interaction. Mol. Plant Microbe Int. 24, 1359–1371. doi: 10.1094/MPMI-04-11-0093
Bonaldi, K., Gourion, B., Fardoux, J., Hannibal, L., Cartieaux, F., Boursot, M., et al. (2010). Large-scale transposon mutagenesis of photosynthetic bradyrhizobium sp. strain ORS278 reveals new genetic loci putatively important for nod-independent symbiosis with aeschynomene indica. Mol. Plant Microbe Int. 23, 760–770. doi: 10.1094/MPMI-23-6-0760
Bozsóki, Z., Cheng, J., Feng, F., Gysel, K., Vinther, M., Andersen, K. R., et al. (2017). Receptor-mediated chitin perception in legume roots is functionally separable from nod factor perception. Proc. Natl. Acad. Sci. U.S.A. 114, E8118–E8127. doi: 10.1073/pnas.1706795114
Caldwell, B. E. (1966). Inheritance of a strain-specific ineffective nodulation in soybeans. Crop Sci. 6, 427–428. doi: 10.2135/cropsci1966.0011183x000600050010x
Cao, Y., Halane, M. K., Gassmann, W., and Stacey, G. (2017). The role of plant innate imunity in the legume-rizobium smbiosis. Ann. Rev. Plant Biol. 68, 535–561. doi: 10.1146/annurev-arplant-042916-041030
Carlson, R. W., Kalembasa, S., Turowski, D., Pachori, P., and Noel, K. D. (1987). Characterization of the lipopolysaccharide from a rhizobium phaseoli mutant that is defective in infection thread development. J. Bacteriol. 169, 4923–4928. doi: 10.1128/jb.169.11.4923-4928.1987
Catoira, R., Galera, C., De Billy, F., Penmetsa, R. V., Journet, E. P., Maillet, F., et al. (2000). Four genes of medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12, 1647–1666. doi: 10.1105/tpc.12.9.1647
Crespo-Rivas, J. C., Guefrachi, I., Mok, K. C., Villaécija-Aguilar, J. A., Acosta-Jurado, S., Pierre, O., et al. (2016). Sinorhizobium fredii HH103 bacteroids are not terminally differentiated and show altered O-antigen in nodules of the inverted repeat-lacking clade legume glycyrrhiza uralensis. Environ. Microbiol. 18, 2392–2404. doi: 10.1111/1462-2920.13101
Crook, M. B., Lindsay, D. P., Biggs, M. B., Bentley, J. S., Price, J. C., Clement, S. C., et al. (2012). Rhizobial plasmids that cause impaired symbiotic nitrogen fixation and enhanced host invasion. Mol. Plant Microbe Int. 25, 1026–1033. doi: 10.1094/MPMI-02-12-0052-R
Czernic, P., Gully, D., Cartieaux, F., Moulin, L., Guefrachi, I., Patrel, D., et al. (2015). Convergent evolution of endosymbiont differentiation in dalbergioid and inverted repeat-lacking clade legumes mediated by nodule-specific cysteine-rich peptides. Plant Physiol. 169, 1254–1265. doi: 10.1104/pp.15.00584
Dénarié, J., Debellé, F., and Promé, J. C. (1996). Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Ann. Rev. Biochem. 65, 503–535. doi: 10.1146/annurev.bi.65.070196.002443
diCenzo, G. C., Zamani, M., Ludwig, H. N., and Finan, T. M. (2017). Heterologous complementation reveals a specialized activity for BacA in the medicago-sinorhizobium meliloti symbiosis. Mol. Plant Microbe Int. 30, 312–324. doi: 10.1094/MPMI-02-17-0030-R
Diedhiou, I., and Diouf, D. (2018). Transcription factors network in root endosymbiosis establishment and development. World J. Microbiol. Biotechnol. 34:37. doi: 10.1007/s11274-018-2418-7
Dylan, T., Nagpal, P., Helinski, D. R., and Ditta, G. S. (1990). Symbiotic pseudorevertants of Rhizobium meliloti ndv mutants. J. Bacteriol. 172, 1409–1417. doi: 10.1128/jb.172.3.1409-1417.1990
Ehrhardt, D. W., Wais, R., and Long, S. R. (1996). Calcium spiking in plant root hairs responding to rhizobium nodulation signals. Cell 85, 673–681. doi: 10.1016/s0092-8674(00)81234-9
Fabre, S., Gully, D., Poitout, A., Patrel, D., Arrighi, J. F., Giraud, E., et al. (2015). Nod factor-independent nodulation in aeschynomene evenia required the common plant-microbe symbiotic toolkit. Plant Physiol. 169, 2654–2664. doi: 10.1104/pp.15.01134
Fan, Y., Liu, J., Lyu, S., Wang, Q., Yang, S., and Zhu, H. (2017). The soybean Rfg1 gene restricts nodulation by Sinorhizobium fredii USDA193. Front. Plant Sci. 8:1548. doi: 10.3389/fpls.2017.01548
Farkas, A., Maróti, G., Dürgõ, H., Györgypál, Z., Lima, R. M., Medzihradszky, K. F., et al. (2014). Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc. Natl. Acad. Sci. U.S.A. 111, 5183–5188. doi: 10.1073/pnas.1404169111
Faruque, O. M., Miwa, H., Yasuda, M., Fujii, Y., Kaneko, T., Sato, S., et al. (2015). Identification of Bradyrhizobium elkanii genes involved in incompatibility with soybean plants carrying the Rj4 allele. Appl. Environ. Microbiol. 81, 6710–6717. doi: 10.1128/AEM.01942-15
Felle, H. H., and Kondorosi, É, Kondorosi, Á, and Schultze, M. (1988). The role of ion fluxes in nod factor signalling in Medicago sativa. Plant J. 13, 455–463. doi: 10.1046/j.1365-313X.1998.00041.x
Feng, F., Sun, J., Radhakrishnan, G. V., Lee, T., Bozsoki, Z., Fort, S., et al. (2019). A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in medicago truncatula. Nat. Commun. 10:5047. doi: 10.1038/s41467-019-12999-5
Ferguson, B. J., Indrasumunar, A., Hayashi, S., Lin, M. H., Lin, Y. H., Reid, D. E., et al. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76. doi: 10.1111/j.1744-7909.2010.00899.x
Firmin, J. L., Wilson, K. E., Carlson, R. W., Davies, A. E., and Downie, J. A. (1993). Resistance to nodulation of cv. afghanistan peas is overcome by nodX, which mediates an O-acetylation of the rhizobium leguminosarum lipo-oligosaccharide nodulation factor. Mol. Microbiol. 10, 351–360. doi: 10.1111/j.1365-2958.1993.tb01961.x
Flor, H. H. (1971). Current status of the gene-for-gene concept. Ann. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev-phyto-072910-095339
Foucher, F., and Kondorosi, É (2000). Cell cycle regulation in the course of nodule organogenesis in medicago. Plant Mol. Biol. 43, 773–786. doi: 10.1023/a:1006405029600
Gage, D. J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68, 280–300. doi: 10.1128/MMBR.68.2.280-300.2004
Geurts, R., Heidstra, R., Hadri, A. E., Downie, J. A., Franssen, H., Van Kammen, A., et al. (1997). Sym2 of pea is involved in a nodulation factor-perception mechanism that controls the infection process in the epidermis. Plant Physiol. 115, 351–359. doi: 10.1104/pp.115.2.351
Gibson, K. E., Kobayashi, H., and Walker, G. C. (2008). Molecular determinants of a symbiotic chronic infection. Ann. Rev. Gene.t 42, 413–441. doi: 10.1146/annurev.genet.42.110807.091427
Giraud, E., Moulin, L., Vallenet, D., Barbe, V., Cytryn, E., Avarre, J. C., et al. (2007). Legumes symbioses: absence of nod genes in photosynthetic bradyrhizobia. Science 316, 1307–1312. doi: 10.1126/science.1139548
Glazebrook, J., and Walker, G. C. (1989). A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56, 661–672. doi: 10.1016/0092-8674(89)90588-6
Gossmann, J. A., Markmann, K., Brachmann, A., Rose, L. E., and Parniske, M. (2012). Polymorphic infection and organogenesis patterns induced by a Rhizobium leguminosarum isolate from lotus root nodules are determined by the host genotype. New Phytol. 196, 561–573. doi: 10.1111/j.1469-8137.2012.04281.x
Götz, R., Evans, I. J., Downie, J. A., and Johnston, A. W. B. (1985). Identification of the host-range DNA which allows Rhizobium leguminosarum strain TOM to nodulate cv. afghanistan peas. Mol. General Genet. MGG 201, 296–300. doi: 10.1007/BF00425674
Granada, C. E., Strochein, M., Vargas, L. K., Bruxel, M., De Sa, E. L., and Passaglia, L. M. (2014). Genetic diversity and symbiotic compatibility among rhizobial strains and desmodium incanum and lotus spp. plants. Genet. Mol. Biol. 37, 396–405. doi: 10.1590/s1415-47572014000300012
Guefrachi, I., Nagymihály, M., Pislariu, C. I., Van De Velde, W., Ratet, P., Mars, M., et al. (2014). Extreme specificity of NCR gene expression in medicago truncatula. BMC Genom. 15:712. doi: 10.1186/1471-2164-15-712
Guefrachi, I., Pierre, O., Timchenko, T., Alunni, B., Barrière, Q., Czernic, P., et al. (2015). Bradyrhizobium BclA is a peptide transporter required for bacterial differentiation in symbiosis with aeschynomene legumes. Mol. Plant Microbe Int. 28, 1155–1166. doi: 10.1094/MPMI-04-15-0094-R
Györgypál, Z., Kiss, G. B., and Kondorosi, Á (1991). Transduction of plant signal molecules by the rhizobium NodD proteins. Bioessays 13, 575–581. doi: 10.1002/bies.950131106
Györgypál, Z., and Kondorosi, Á (1991). Homology of the ligand-binding regions of Rhizobium symbiotic regulatory protein NodD and vertebrate nuclear receptors. Mol. Gen. Genet. 226, 337–340. doi: 10.1007/BF00273624
Hayashi, M., Saeki, Y., Haga, M., Harada, K., Kouchi, H., and Umehara, Y. (2012). Rj (rj) genes involved in nitrogen-fixing root nodule formation in soybean. Breed Sci. 61, 544–553. doi: 10.1270/jsbbs.61.544
Hidalgo, A., Margaret, I., Crespo-Rivas, J. C., Parada, M., Murdoch, P. D. S., López, A., et al. (2010). The rkpU gene of Sinorhizobium fredii HH103 is required for bacterial K-antigen polysaccharide production and for efficient nodulation with soybean but not with cowpea. Microbiology 156, 3398–3411. doi: 10.1099/mic.0.042499-0
Horváth, B., Domonkos, Á, Kereszt, A., Szücs, A., Ábrahám, E., Ayaydin, F., et al. (2015). Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc. Natl. Acad. Sci. U.S.A. 112, 15232–15237. doi: 10.1073/pnas.1500777112
Hozbor, D. F., Pich Otero, A. J., Lodeiro, A. R., Del Papa, M. F., Pistorio, M., and Lagares, A. (2004). The symbiotic defect in a Sinorhizobium meliloti lipopolysaccharide mutant can be overcome by expression of other surface polysaccharides. Res. Microbiol. 155, 855–860. doi: 10.1016/j.resmic.2004.06.012
Hubber, A., Vergunst, A. C., Sullivan, J. T., Hooykaas, P. J., and Ronson, C. W. (2004). Symbiotic phenotypes and translocated effector proteins of the mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol. Microbiol. 54, 561–574. doi: 10.1111/j.1365-2958.2004.04292.x
Ibáñez, F., Wall, L., and Fabra, A. (2017). Starting points in plant-bacteria nitrogen-fixing symbioses: intercellular invasion of the roots. J. Exp. Bot. 68, 1905–1918. doi: 10.1093/jxb/erw387
Jiménez-Guerrero, I., Acosta-Jurado, S., Medina, C., Ollero, F. J., Alias-Villegas, C., Vinardell, J. M., et al. (2020). The Sinorhizobium fredii HH103 type III secretion system effector NopC blocks nodulation with Lotus japonicus gifu. J. Exp. Bot. 71, 6043–6056. doi: 10.1093/jxb/eraa297
Jiménez-Guerrero, I., Pérez-Montano, F., Medina, C., Ollero, F. J., and López-Baena, F. J. (2015). NopC is a rhizobium-specific type 3 secretion system effector secreted by sinorhizobium (ensifer) fredii HH103. PLoS One 10:e0142866. doi: 10.1371/journal.pone.0142866
Kawaharada, Y., Kelly, S., Nielsen, M. W., Hjuler, C. T., Gysel, K., Muszynski, A., et al. (2015). Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523, 308–312. doi: 10.1038/nature14611
Kawaharada, Y., Sandal, N., Gupta, V., Jin, H., Kawaharada, M., Taniuchi, M., et al. (2021). Natural variation identifies a pxy gene controlling vascular organisation and formation of nodules and lateral roots in lotus japonicus. New Phytol. 230, 2459–2473. doi: 10.1111/nph.17356
Kazmierczak, T., Nagymihály, M., Lamouche, F., Barrière, Q., Guefrachi, I., Alunni, B., et al. (2017). Specific host-responsive associations between medicago truncatula accessions and sinorhizobium strains. Mol. Plant Microbe Int. 30, 399–409. doi: 10.1094/MPMI-01-17-0009-R
Kelly, S. J., Muszynski, A., Kawaharada, Y., Hubber, A. M., Sullivan, J. T., Sandal, N., et al. (2013). Conditional requirement for exopolysaccharide in the mesorhizobium-lotus symbiosis. Mol. Plant Microbe Int. 26, 319–329. doi: 10.1094/MPMI-09-12-0227-R
Kereszt, A., Kiss, E., Reuhs, B. L., Carlson, R. W., and Kondorosi, Á, and Putnoky, P. (1998). Novel rkp gene clusters of Sinorhizobium meliloti involved in capsular polysaccharide production and invasion of the symbiotic nodule: the rkpK gene encodes a UDP-glucose dehydrogenase. J. Bacteriol. 180, 5426–5431. doi: 10.1128/JB.180.20.5426-5431.1998
Kereszt, A., Mergaert, P., Maróti, G., and Kondorosi, É (2011). Innate immunity effectors and virulence factors in symbiosis. Curr. Opin. Microbiol. 14, 76–81. doi: 10.1016/j.mib.2010.12.002
Kim, M., Chen, Y., Xi, J., Waters, C., Chen, R., and Wang, D. (2015). An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112, 15238–15243. doi: 10.1073/pnas.1500123112
Kiss, E., Huguet, T., Poinsot, V., and Batut, J. (2004). The typA gene is required for stress adaptation as well as for symbiosis of Sinorhizobium meliloti 1021 with certain Medicago truncatula lines. Mol. Plant Microbe Int. 17, 235–244. doi: 10.1094/MPMI.2004.17.3.235
Kiss, E., Kereszt, A., Barta, F., Stephens, S., Reuhs, B. L., Kondorosi, Á, et al. (2001). The rkp-3 gene region of Sinorhizobium meliloti Rm41 contains strain-specific genes that determine K antigen structure. Mol. Plant Microbe Int. 14, 1395–1403. doi: 10.1094/MPMI.2001.14.12.1395
Kozik, A., Heidstra, R., Horváth, B., Kulikova, O., Tikhonovich, I., Ellis, T. H. N., et al. (1995). Pea lines carrying Sym1 or Sym2 can be nodulated by rhizobium strains containing nodX - syml and sym2 are allelic. Plant Sci. 108, 41–49. doi: 10.1016/0168-9452(95)04123-C
Kronauer, C., and Radutoiu, S. (2021). Understanding nod factor signalling paves the way for targeted engineering in legumes and non-legumes. Curr. Opin. Plant Biol. 62:102026. doi: 10.1016/j.pbi.2021.102026
Kusakabe, S., Higasitani, N., Kaneko, T., Yasuda, M., Miwa, H., Okazaki, S., et al. (2020). Lotus accessions possess multiple checkpoints triggered by different type III secretion system effectors of the wide-host-range symbiont bradyrhizobium elkanii USDA61. Microbes Environ. 35:141. doi: 10.1264/jsme2.ME19141
Lamouche, F., Bonadé-Bottino, N., Mergaert, P., and Alunni, B. (2019). Symbiotic efficiency of spherical and elongated eacteroids in the aeschynomene-bradyrhizobium symbiosis. Front. Plant Sci. 10:377. doi: 10.3389/fpls.2019.00377
Lamouche, F., Gully, D., Chaumeret, A., Nouwen, N., Verly, C., Pierre, O., et al. (2018). Transcriptomic dissection of Bradyrhizobium sp. strain ORS285 in symbiosis with Aeschynomene spp. inducing different bacteroid morphotypes with contrasted symbiotic efficiency. Environ. Microbiol. 21, 3244–3258. doi: 10.1111/1462-2920.14292
Leigh, J. A., Signer, E. R., and Walker, G. C. (1985). Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. U.S.A. 82, 6231–6235. doi: 10.1073/pnas.82.18.6231
Lewis-Henderson, W. R., and Djordjevic, M. A. (1991a). A cultivar-specific interaction between Rhizobium leguminosarum bv. trifolii and subterranean clover is controlled by nodM, other bacterial cultivar specificity genes, and a single recessive host gene. J. Bacteriol. 173, 2791–2799. doi: 10.1128/jb.173.9.2791-2799.1991
Lewis-Henderson, W. R., and Djordjevic, M. A. (1991b). nodT, a positively-acting cultivar specificity determinant controlling nodulation of Trifolium subterraneum by Rhizobium leguminosarum biovar trifolii. Plant Mol. Biol. 16, 515–526. doi: 10.1007/BF00023418
Li, R., Knox, M. R., Edwards, A., Hogg, B., Ellis, T. H., Wei, G., et al. (2011). Natural variation in host-specific nodulation of pea is associated with a haplotype of the SYM37 LysM-type receptor-like kinase. Mol. Plant Microbe Int. 24, 1396–1403. doi: 10.1094/MPMI-01-11-0004
Liang, J., Hoffrichter, A., Brachmann, A., and Marin, M. (2018). Complete genome of Rhizobium leguminosarum norway, an ineffective lotus micro-symbiont. Stand Gen. Sci. 13:36. doi: 10.1186/s40793-018-0336-9
Liang, J., Klingl, A., Lin, Y. Y., Boul, E., Thomas-Oates, J., and Marin, M. (2019). A subcompatible rhizobium strain reveals infection duality in lotus. J. Exp. Bot. 70, 1903–1913. doi: 10.1093/jxb/erz057
Lie, T. A. (1984). Host genes in Pisum sativum L. conferring resistance to european rhizobium leguminosarum strains. Plant Soil 82, 415–425. doi: 10.1007/bf02184279
Lindström, K., and Mousavi, S. A. (2020). Effectiveness of nitrogen fixation in rhizobia. Microb Biotechnol. 13, 1314–1335. doi: 10.1111/1751-7915.13517
Liu, J., Yang, S., Zheng, Q., and Zhu, H. (2014). Identification of a dominant gene in Medicago truncatula that restricts nodulation by Sinorhizobium meliloti strain Rm41. BMC Plant Biol. 14:167. doi: 10.1186/1471-2229-14-167
Long, S. R. (1996). Rhizobium symbiosis: nod factors in perspective. Plant Cell 8, 1885–1898. doi: 10.1105/tpc.8.10.1885
López-Baena, F. J., Ruiz-Sainz, J. E., Rodríguez-Carvajal, M. A., and Vinardell, J. M. (2016). Bacterial molecular signals in the sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 17:755. doi: 10.3390/ijms17050755
Lopez-Gomez, M., Sandal, N., Stougaard, J., and Boller, T. (2012). Interplay of flg22-induced defence responses and nodulation in lotus japonicus. J. Exp. Bot. 63, 393–401. doi: 10.1093/jxb/err291
López-Lara, I. M., Van Den Berg, J. D., Thomas-Oates, J. E., Glushka, J., Lugtenberg, B. J., and Spaink, H. P. (1995). Structural identification of the lipo-chitin oligosaccharide nodulation signals of rhizobium loti. Mol. Microbiol. 15, 627–638. doi: 10.1111/j.1365-2958.1995.tb02372.x
Lorio, J. C., Kim, W. S., Krishnan, A. H., and Krishnan, H. B. (2010). Disruption of the glycine cleavage system enables Sinorhizobium fredii USDA257 to form nitrogen-fixing nodules on agronomically improved north american soybean cultivars. Appl. Environ. Microbiol. 76, 4185–4193. doi: 10.1128/AEM.00437-10
Lorite, M. J., Estrella, M. J., Escaray, F. J., Sannazzaro, A., Videira, E. C. I. M., Monza, J., et al. (2018). The rhizobia-lotus symbioses: deeply specific and widely diverse. Front. Microbiol. 9:2055. doi: 10.3389/fmicb.2018.02055
Malolepszy, A., Kelly, S., Sørensen, K. K., James, E. K., Kalisch, C., Bozsoki, Z., et al. (2018). A plant chitinase controls cortical infection thread progression and nitrogen-fixing symbiosis. Elife 7, e38874. doi: 10.7554/eLife.38874
Margaret, I., Becker, A., Blom, J., Bonilla, I., Goesmann, A., Göttfert, M., et al. (2011). Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean. J. Biotechnol. 155, 11–19. doi: 10.1016/j.jbiotec.2011.03.016
Margaret, I., Lucas, M. M., Acosta-Jurado, S., Buendía-Clavería, A. M., Fedorova, E., Hidalgo, A., et al. (2013). The Sinorhizobium fredii HH103 lipopolysaccharide is not only relevant at early soybean nodulation stages but also for symbiosome stability in mature nodules. PLoS One 8:e74717. doi: 10.1371/journal.pone.0074717
Marie, C., Broughton, W. J., and Deakin, W. J. (2001). Rhizobium type III secretion systems: legume charmers or alarmers? Curr. Opin. Plant Biol. 4, 336–342. doi: 10.1016/s1369-5266(00)00182-5
Marie, C., Deakin, W. J., Viprey, V., Kopciñska, J., Golinowski, W., Krishnan, H. B., et al. (2003). Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant Microbe Int. 16, 743–751. doi: 10.1094/MPMI.2003.16.9.743
Masson-Boivin, C., Giraud, E., Perret, X., and Batut, J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 17, 458–466. doi: 10.1016/j.tim.2009.07.004
Maunoury, N., Redondo-Nieto, M., Bourcy, M., Van De Velde, W., Alunni, B., Laporte, P., et al. (2010). Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS One 5:e9519. doi: 10.1371/journal.pone.0009519
Meinhardt, L. W., Krishnan, H. B., Balatti, P. A., and Pueppke, S. G. (1993). Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 9, 17–29. doi: 10.1111/j.1365-2958.1993.tb01665.x
Melino, V. J., Drew, E. A., Ballard, R. A., Reeve, W. G., Thomson, G., White, R. G., et al. (2012). Identifying abnormalities in symbiotic development between Trifolium spp. and Rhizobium leguminosarum bv. trifolii leading to sub-optimal and ineffective nodule phenotypes. Ann. Bot. 110, 1559–1572. doi: 10.1093/aob/mcs206
Mergaert, P., Kereszt, A., and Kondorosi, É (2020). Gene expression in nitrogen-fixing symbiotic nodule cells in Medicago truncatula and other nodulating plants. Plant Cell 32, 42–68. doi: 10.1105/tpc.19.00494
Mergaert, P., Nikovics, K., Kelemen, Z., Maunoury, N., Vaubert, D., Kondorosi, Á, et al. (2003). A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 132, 161–173. doi: 10.1104/pp.102.018192
Mergaert, P., Uchiumi, T., Alunni, B., Evanno, G., Cheron, A., Catrice, O., et al. (2006). Eukaryotic control on bacterial cell cycle and differentiation in the rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 5230–5235. doi: 10.1073/pnas.0600912103
Miller, S. H., Elliot, R. M., Sullivan, J. T., and Ronson, C. W. (2007). Host-specific regulation of symbiotic nitrogen fixation in Rhizobium leguminosarum biovar trifolii. Microbiology 153, 3184–3195. doi: 10.1099/mic.0.2007/006924-0
Montiel, J., Downie, J. A., Farkas, A., Bihari, P., Herczeg, R., Bálint, B., et al. (2017). Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc. Natl. Acad. Sci. U.S.A. 114, 5041–5046. doi: 10.1073/pnas.1704217114
Montiel, J., Szücs, A., Boboescu, I. Z., Gherman, V. D., and Kondorosi, É, and Kereszt, A. (2016). Terminal bacteroid differentiation is associated with variable morphological changes in legume species belonging to the inverted repeat-lacking clade. Mol. Plant Microbe Int. 29, 210–219. doi: 10.1094/MPMI-09-15-0213-R
Murray, J. D. (2011). Invasion by invitation: rhizobial infection in legumes. Mol. Plant Microbe Int. 24, 631–639. doi: 10.1094/MPMI-08-10-0181
Nelson, M. S., and Sadowsky, M. J. (2015). Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 6:491. doi: 10.3389/fpls.2015.00491
Ngou, B. P. M., Jones, J. D. G., and Ding, P. (2021). Plant immune networks. Trends Plant Sci. 2021:12. doi: 10.1016/j.tplants.2021.08.012
Nguyen, H. P., Miwa, H., Kaneko, T., Sato, S., and Okazaki, S. (2017). Identification of Bradyrhizobium elkanii genes involved in incompatibility with vigna radiata. Genes (Basel) 8:374. doi: 10.3390/genes8120374
Nguyen, H. P., Ratu, S. T. N., Yasuda, M., Göttfert, M., and Okazaki, S. (2018). InnB, a novel type III effector of Bradyrhizobium elkanii USDA61, controls symbiosis with vigna species. Front. Microbiol. 9:3155. doi: 10.3389/fmicb.2018.03155
Nicoud, Q., Lamouche, F., Chaumeret, A., Balliau, T., Le Bars, R., Bourge, M., et al. (2021). Bradyrhizobium diazoefficiens USDA110 nodulation of Aeschynomene afraspera is associated with atypical terminal bacteroid differentiation and suboptimal symbiotic efficiency. mSystems 6, e01237-20. doi: 10.1128/mSystems.01237-20
Okazaki, S., Kaneko, T., Sato, S., and Saeki, K. (2013). Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 110, 17131–17136. doi: 10.1073/pnas.1302360110
Okazaki, S., Okabe, S., Higashi, M., Shimoda, Y., Sato, S., Tabata, S., et al. (2010). Identification and functional analysis of type III effector proteins in mesorhizobium loti. Mol. Plant Microbe Int. 23, 223–234. doi: 10.1094/MPMI-23-2-0223
Okazaki, S., Tittabutr, P., Teulet, A., Thouin, J., Fardoux, J., Chaintreuil, C., et al. (2016). Rhizobium-legume symbiosis in the absence of nod factors: two possible scenarios with or without the T3SS. ISME J. 10, 64–74. doi: 10.1038/ismej.2015.103
Oono, R., and Denison, R. F. (2010). Comparing symbiotic efficiency between swollen versus nonswollen rhizobial bacteroids. Plant Physiol. 154, 1541–1548. doi: 10.1104/pp.110.163436
Oono, R., Schmitt, I., Sprent, J. I., and Denison, R. F. (2010). Multiple evolutionary origins of legume traits leading to extreme rhizobial differentiation. New Phytol 187, 508–520. doi: 10.1111/j.1469-8137.2010.03261.x
Osteras, M., Finan, T. M., and Stanley, J. (1991). Site-directed mutagenesis and DNA sequence of pckA of rhizobium NGR234, encoding phosphoenolpyruvate carboxykinase: gluconeogenesis and host-dependent symbiotic phenotype. Mol. Gen. Genet. 230, 257–269. doi: 10.1007/BF00290676
Pacios Bras, C., Jorda, M. A., Wijfjes, A. H., Harteveld, M., Stuurman, N., Thomas-Oates, J. E., et al. (2000). A lotus japonicus nodulation system based on heterologous expression of the fucosyl transferase NodZ and the acetyl transferase NoIL in Rhizobium leguminosarum. Mol. Plant Microbe Int. 13, 475–479. doi: 10.1094/MPMI.2000.13.4.475
Parada, M., Vinardell, J. M., Ollero, F. J., Hidalgo, A., Gutiérrez, R., Buendía-Clavería, A. M., et al. (2006). Sinorhizobium fredii HH103 mutants affected in capsular polysaccharide (KPS) are impaired for nodulation with soybean and cajanus cajan. Mol. Plant Microbe Int. 19, 43–52. doi: 10.1094/MPMI-19-0043
Peck, M. C., Fisher, R. F., and Long, S. R. (2006). Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J. Bacteriol. 188, 5417–5427. doi: 10.1128/JB.00376-06
Pellock, B. J., Cheng, H. P., and Walker, G. C. (2000). Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182, 4310–4318. doi: 10.1128/JB.182.15.4310-4318.2000
Penterman, J., Abo, R. P., De Nisco, N. J., Arnold, M. F., Longhi, R., Zanda, M., et al. (2014). Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc. Natl. Acad. Sci. U.S.A. 111, 3561–3566. doi: 10.1073/pnas.1400450111
Peoples, M. B., Herridge, D. F., and Ladha, J. K. (1995). Biological nitrogen fixation: an efficient source of nitrogen for sustainable agricultural production? Plant Soil 174, 3–28. doi: 10.1007/BF00032239
Pérez-Giménez, J., Iturralde, E. T., Torres Tejerizo, G., Quelas, J. I., Krol, E., Borassi, C., et al. (2021). A stringent-response-defective Bradyrhizobium diazoefficiens strain does not activate the type 3 secretion system, elicits an early plant defense response, and circumvents NH4NO3-induced inhibition of nodulation. Appl. Environ. Microbiol. 87:e02989-20. doi: 10.1128/AEM.02989-20
Peters, N. K., Frost, J. W., and Long, S. R. (1986). A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233, 977–980. doi: 10.1126/science.3738520
Pislariu, C. I., Sinharoy, S., Torres-Jerez, I., Nakashima, J., Blancaflor, E. B., and Udvardi, M. K. (2019). The nodule-specific PLAT domain protein NPD1 is equired for nitrogen-fixing symbiosis. Plant Physiol. 180, 1480–1497. doi: 10.1104/pp.18.01613
Price, P. A., Tanner, H. R., Dillon, B. A., Shabab, M., Walker, G. C., and Griffitts, J. S. (2015). Rhizobial peptidase HrrP cleaves host-encoded signaling peptides and mediates symbiotic compatibility. Proc. Natl. Acad. Sci. U.S.A. 112, 15244–15249. doi: 10.1073/pnas.1417797112
Pueppke, S. G., and Broughton, W. J. (1999). Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant Microbe Int. 12, 293–318. doi: 10.1094/MPMI.1999.12.4.293
Putnoky, P., Kereszt, A., Nakamura, T., Endre, G., Grosskopf, E., Kiss, P., et al. (1998). The pha gene cluster of rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol. Microbiol. 28, 1091–1101. doi: 10.1046/j.1365-2958.1998.00868.x
Putnoky, P., Petrovics, G., Kereszt, A., Grosskopf, E., Ha, D. T., Bánfalvi, Z., et al. (1990). Rhizobium meliloti lipopolysaccharide and exopolysaccharide can have the same function in the plant-bacterium interaction. J. Bacteriol. 172, 5450–5458. doi: 10.1128/jb.172.9.5450-5458.1990
Radutoiu, S., Madsen, L. H., Madsen, E. B., Jurkiewicz, A., Fukai, E., Quistgaard, E. M., et al. (2007). LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 26, 3923–3935. doi: 10.1038/sj.emboj.7601826
Ratu, S. T. N., Hirata, A., Kalaw, C. O., Yasuda, M., Tabuchi, M., and Okazaki, S. (2021a). Multiple domains in the rhizobial type III effector Bel2-5 determine symbiotic efficiency with soybean. Front. Plant Sci. 12:689064. doi: 10.3389/fpls.2021.689064
Ratu, S. T. N., Teulet, A., Miwa, H., Masuda, S., Nguyen, H. P., Yasuda, M., et al. (2021b). Rhizobia use a pathogenic-like effector to hijack leguminous nodulation signalling. Sci. Rep. 11:2034. doi: 10.1038/s41598-021-81598-6
Redmond, J., Batley, M., Djordjevic, M. A., Innes, R. W., Kuempel, P. L., and Rolfe, B. G. (1986). Flavones induce expression of nodulation genes in rhizobium. Nature 323, 632–635. doi: 10.1038/323632a0
Reuhs, B. L., Geller, D. P., Kim, J. S., Fox, J. E., Kolli, V. S., and Pueppke, S. G. (1998). Sinorhizobium fredii and Sinorhizobium meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Appl. Environ. Microbiol. 64, 4930–4938. doi: 10.1128/AEM.64.12.4930-4938.1998
Reuhs, B. L., Williams, M. N., Kim, J. S., Carlson, R. W., and Côté, F. (1995). Suppression of the Fix- phenotype of rhizobium meliloti exoB mutants by lpsZ is correlated to a modified expression of the K polysaccharide. J. Bacteriol. 177, 4289–4296. doi: 10.1128/jb.177.15.4289-4296.1995
Rodpothong, P., Sullivan, J. T., Songsrirote, K., Sumpton, D., Cheung, K. W., Thomas-Oates, J., et al. (2009). Nodulation gene mutants of mesorhizobium loti R7A-nodZ and nolL mutants have host-specific phenotypes on lotus spp. Mol. Plant Microbe Int. 22, 1546–1554. doi: 10.1094/MPMI-22-12-1546
Roest, H. P., Mulders, I. H., Spaink, H. P., Wijffelman, C. A., and Lugtenberg, B. J. (1997). A rhizobium leguminosarum biovar trifolii locus not localized on the sym plasmid hinders effective nodulation on plants of the pea cross-inoculation group. Mol. Plant Microbe Int. 10, 938–941. doi: 10.1094/MPMI.1997.10.7.938
Roth, L. E., and Stacey, G. (1989). Bacterium release into host cells of nitrogen-fixing soybean nodules: the symbiosome membrane comes from three sources. Eur. J Cell Biol 49, 13–23.
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32, 15–41. doi: 10.1105/tpc.19.00279
Rutten, P. J., and Poole, P. S. (2019). Oxygen regulatory mechanisms of nitrogen fixation in rhizobia. Adv. Microb Physiol. 75, 325–389. doi: 10.1016/bs.ampbs.2019.08.001
Sadowsky, M. J., Kosslak, R. M., Madrzak, C. J., Golinska, B., and Cregan, P. B. (1995). Restriction of nodulation by Bradyrhizobium japonicum is mediated by factors present in the roots of glycine max. Appl. Environ. Microbiol. 61, 832–836. doi: 10.1128/aem.61.2.832-836.1995
Salinero-Lanzarote, A., Pacheco-Moreno, A., Domingo-Serrano, L., Durán, D., Ormeño-Orrillo, E., Martinez-Romero, E., et al. (2019). The type VI secretion system of Rhizobium etli mim1 has a positive effect in symbiosis. FEMS Microbiol. Ecol. 95:54. doi: 10.1093/femsec/fiz054
Sánchez, C., Iannino, F., Deakin, W. J., Ugalde, R. A., and Lepek, V. C. (2009). Characterization of the mesorhizobium loti MAFF303099 type-three protein secretion system. Mol. Plant Microbe Int. 22, 519–528. doi: 10.1094/MPMI-22-5-0519
Sánchez, C., Mercante, V., Babuin, M. F., and Lepek, V. C. (2012). Dual effect of Mesorhizobium loti T3SS functionality on the symbiotic process. FEMS Microbiol. Lett. 330, 148–156. doi: 10.1111/j.1574-6968.2012.02545.x
Schreiber, K. J., Chau-Ly, I. J., and Lewis, J. D. (2021). What the wild things do: mechanisms of plant host manipulation by bacterial type III-secreted effector proteins. Microorganisms 9:1029. doi: 10.3390/microorganisms9051029
Shimoda, Y., Nishigaya, Y., Yamaya-Ito, H., Inagaki, N., Umehara, Y., Hirakawa, H., et al. (2020). The rhizobial autotransporter determines the symbiotic nitrogen fixation activity of Lotus japonicus in a host-specific manner. Proc. Natl. Acad. Sci. U.S.A. 117, 1806–1815. doi: 10.1073/pnas.1913349117
Shobudani, M., Htwe, A. Z., Yamakawa, T., Ishibashi, M., and Tsurumaru, H. (2020). Mutants disrupted in the type III secretion system of Bradyrhizobium elkanii BLY3-8 overcame nodulation restriction by Rj3-genotype soybean. Microbes Environ. 35:151. doi: 10.1264/jsme2.ME19151
Skorupska, A., Janczarek, M., Marczak, M., Mazur, A., and Król, J. (2006). Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb Cell Fact. 5:7. doi: 10.1186/1475-2859-5-7
Smit, P., Limpens, E., Geurts, R., Fedorova, E., Dolgikh, E., Gough, C., et al. (2007). Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145, 183–191. doi: 10.1104/pp.107.100495
Snyman, C. P., and Srijdom, B. W. (1980). Symbiotic characteristics of lines and cultivars of Medicago truncatula inoculated with strains of Rhizobium meliloti. Phytophylatica 12, 173–176.
Solovev, Y. V., Igolkina, A. A., Kuliaev, P. O., Sulima, A. S., Zhukov, V. A., Porozov, Y. B., et al. (2021). Towards understanding afghanistan pea symbiotic phenotype through the molecular modeling of the interaction between LykX-Sym10 receptor heterodimer and nod factors. Front. Plant Sci. 12:642591. doi: 10.3389/fpls.2021.642591
Songwattana, P., Chaintreuil, C., Wongdee, J., Teulet, A., Mbaye, M., Piromyou, P., et al. (2021). Identification of type III effectors modulating the symbiotic properties of Bradyrhizobium vignae strain ORS3257 with various vigna species. Sci. Rep. 11:4874. doi: 10.1038/s41598-021-84205-w
Songwattana, P., Noisangiam, R., Teamtisong, K., Prakamhang, J., Teulet, A., Tittabutr, P., et al. (2017). Type 3 secretion system (T3SS) of Bradyrhizobium sp. DOA9 and its roles in legume symbiosis and rice endophytic association. Front. Microbiol. 8:1810. doi: 10.3389/fmicb.2017.01810
Soumare, A., Diedhiou, A. G., Thuita, M., Hafidi, M., Ouhdouch, Y., Gopalakrishnan, S., et al. (2020). Exploiting biological nitrogen fixation: a route towards a sustainable agriculture. Plants 9:1011. doi: 10.3390/plants9081011
Stacey, G., So, J. S., Roth, L. E., Lakshmi, S. B., and Carlson, R. W. (1991). A lipopolysaccharide mutant of Bradyrhizobium japonicum that uncouples plant from bacterial differentiation. Mol. Plant Microbe Int. 4, 332–340. doi: 10.1094/mpmi-4-332
Staehelin, C., and Krishnan, H. B. (2015). Nodulation outer proteins: double-edged swords of symbiotic rhizobia. Biochem J. 470, 263–274. doi: 10.1042/BJ20150518
Stagnari, F., Maggio, A., Galieni, A., and Pisante, M. (2017). Multiple benefits of legumes for agriculture sustainability: an overview. Chem. Biol. Technol. Agric. 4. doi: 10.1186/s40538-016-0085-1
Sugawara, M., Epstein, B., Badgley, B. D., Unno, T., Xu, L., Reese, J., et al. (2013). Comparative genomics of the core and accessory genomes of 48 sinorhizobium strains comprising five genospecies. Genome Biol. 14:R17. doi: 10.1186/gb-2013-14-2-r17
Sugawara, M., Takahashi, S., Umehara, Y., Iwano, H., Tsurumaru, H., Odake, H., et al. (2018). Variation in bradyrhizobial NopP effector determines symbiotic incompatibility with Rj2-soybeans via effector-triggered immunity. Nat. Commun. 9:3139. doi: 10.1038/s41467-018-05663-x
Sugawara, M., Umehara, Y., Kaga, A., Hayashi, M., Ishimoto, M., Sato, S., et al. (2019). Symbiotic incompatibility between soybean and bradyrhizobium arises from one amino acid determinant in soybean Rj2 protein. PLoS One 14:e0222469. doi: 10.1371/journal.pone.0222469
Sulima, A. S., Zhukov, V. A., Afonin, A. A., Zhernakov, A. I., Tikhonovich, I. A., and Lutova, L. A. (2017). Selection signatures in the first exon of paralogous receptor kinase genes from the Sym2 region of the Pisum sativum L. genome. Front. Plant Sci. 8:1957. doi: 10.3389/fpls.2017.01957
Sulima, A. S., Zhukov, V. A., Kulaeva, O. A., Vasileva, E. N., Borisov, A. Y., and Tikhonovich, I. A. (2019). New sources of Sym2(A) allele in the pea (Pisum sativum L.) carry the unique variant of candidate LysM-RLK gene LykX. PeerJ 7:e8070. doi: 10.7717/peerj.8070
Surin, B. P., and Downie, J. A. (1989). Rhizobium leguminosarum genes required for expression and transfer of host specific nodulation. Plant Mol. Biol. 12, 19–29. doi: 10.1007/BF00017444
Sutton, J. M., Lea, E. J., and Downie, J. A. (1994). The nodulation-signaling protein NodO from Rhizobium leguminosarum biovar viciae forms ion channels in membranes. Proc. Natl. Acad. Sci. U.S.A. 91, 9990–9994. doi: 10.1073/pnas.91.21.9990
Suzaki, T., Yoro, E., and Kawaguchi, M. (2015). Leguminous plants: inventors of root nodules to accommodate symbiotic bacteria. Int. Rev. Cell. Mol. Biol. 316, 111–158. doi: 10.1016/bs.ircmb.2015.01.004
Tang, F., Yang, S., Liu, J., and Zhu, H. (2016). Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol. 170, 26–32. doi: 10.1104/pp.15.01661
Tellström, V., Usadel, B., Thimm, O., Stitt, M., Küster, H., and Niehaus, K. (2007). The lipopolysaccharide of Sinorhizobium meliloti suppresses defense-associated gene expression in cell cultures of the host plant Medicago truncatula. Plant Physiol. 143, 825–837. doi: 10.1104/pp.106.090985
Terpolilli, J. J., Hood, G. A., and Poole, P. S. (2012). What determines the efficiency of N(2)-fixing rhizobium-legume symbioses? Adv. Microb Physiol. 60, 325–389. doi: 10.1016/B978-0-12-398264-3.00005-X
Terpolilli, J. J., O’hara, G. W., Tiwari, R. P., Dilworth, M. J., and Howieson, J. G. (2008). The model legume medicago truncatula A17 is poorly matched for N2 fixation with the sequenced microsymbiont sinorhizobium meliloti 1021. New Phytol. 179, 62–66. doi: 10.1111/j.1469-8137.2008.02464.x
Teulet, A., Busset, N., Fardoux, J., Gully, D., Chaintreuil, C., Cartieaux, F., et al. (2019). The rhizobial type III effector ErnA confers the ability to form nodules in legumes. Proc. Natl. Acad. Sci. U.S.A. 116, 21758–21768. doi: 10.1073/pnas.1904456116
Tighilt, L., Boulila, F., De Sousa, B. F. S., Giraud, E., Ruiz-Argüeso, T., Palacios, J. M., et al. (2021). The bradyrhizobium sp. LmicA16 type VI secretion system is required for efficient nodulation of Lupinus spp. Microb Ecol. doi: 10.1007/s00248-021-01892-8 [Epub ahead of print].
Tirichine, L., De Billy, F., and Huguet, T. (2000). Mtsym6, a gene conditioning sinorhizobium strain-specific nitrogen fixation in medicago truncatula. Plant Physiol. 123, 845–851. doi: 10.1104/pp.123.3.845
Tiricz, H., Szücs, A., Farkas, A., Pap, B., Lima, R. M., Maróti, G., et al. (2013). Antimicrobial nodule-specific cysteine-rich peptides induce membrane depolarization-associated changes in the transcriptome of sinorhizobium meliloti. Appl. Environ. Microbiol. 79, 6737–6746. doi: 10.1128/AEM.01791-13
Trese, A. T. (1985). A single dominant gene in McCall soybean prevents effective nodulation with rhizobium fredii USDA257. Euphytica 81, 279–282. doi: 10.1007/bf00025618
Truchet, G., Roche, P., Lerouge, P., Vasse, J., Camut, S., De Billy, F., et al. (1991). Sulphated lipo-oligosaccharide signals of rhizobium meliloti elicit root nodule organogenesis in alfalfa. Nature 351, 670–673. doi: 10.1038/351670a0
Tseng, T. T., Tyler, B. M., and Setubal, J. C. (2009). Protein secretion systems in bacterial-host associations, and their description in the gene ontology. BMC Microbiol 9:S2. doi: 10.1186/1471-2180-9-S1-S2
Tsukui, T., Eda, S., Kaneko, T., Sato, S., Okazaki, S., Kakizaki-Chiba, K., et al. (2013). The type III secretion system of Bradyrhizobium japonicum USDA122 mediates symbiotic incompatibility with Rj2 soybean plants. Appl. Environ. Microbiol. 79, 1048–1051. doi: 10.1128/AEM.03297-12
Tsurumaru, H., Hashimoto, S., Okizaki, K., Kanesaki, Y., Yoshikawa, H., and Yamakawa, T. (2015). A putative type III secretion system effector encoded by the MA20_12780 gene in Bradyrhizobium japonicum is-34 causes incompatibility with Rj4 genotype soybeans. Appl. Environ. Microbiol. 81, 5812–5819. doi: 10.1128/AEM.00823-15
Tsyganova, A. V., Brewin, N. J., and Tsyganov, V. E. (2021). Structure and development of the legume-rhizobial symbiotic interface in infection threads. Cells 10:1050. doi: 10.3390/cells10051050
Udvardi, M., and Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Ann. Rev. Plant Biol. 64, 781–805. doi: 10.1146/annurev-arplant-050312-120235
Van de Velde, W., Zehirov, G., Szatmári, A., Debreczeny, M., Ishihara, H., Kevei, Z., et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126. doi: 10.1126/science.1184057
Vance, C. P. (2001). Symbiotic nitrogen fixation and phosphorus acquisition. plant nutrition in a world of declining renewable resources. Plant Physiol. 127, 390–397. doi: 10.1104/pp.010331
Vasse, J., De Billy, F., Camut, S., and Truchet, G. (1990). Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172, 4295–4306. doi: 10.1128/jb.172.8.4295-4306.1990
Vest, G. (1970). Rj3 - a gene conditioning ineffective nodulation in soybean. Crop Sci. 10, 34–35. doi: 10.2135/cropsci1970.0011183X001000010013x
Vest, G., and Caldwell, B. E. (1972). Rj4 - a gene conditioning ineffective nodulation in soybean. Crop Sci. 12, 692–693. doi: 10.1104/pp.94.3.899
Viprey, V., Del Greco, A., Golinowski, W., Broughton, W. J., and Perret, X. (1998). Symbiotic implications of type III protein secretion machinery in rhizobium. Mol. Microbiol. 28, 1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x
Walker, S. A., and Downie, J. A. (2000). Entry of Rhizobium leguminosarum bv.viciae into root hairs requires minimal nod factor specificity, but subsequent infection thread growth requires nodO or nodE. Mol. Plant Microbe Int. 13, 754–762. doi: 10.1094/MPMI.2000.13.7.754
Wang, Q., Liu, J., Li, H., Yang, S., Körmöczi, P., Kereszt, A., et al. (2018). Nodule-specific cysteine-rich peptides negatively regulate nitrogen-fixing symbiosis in a strain-specific manner in Medicago truncatula. Mol. Plant Microbe Int. 31, 240–248. doi: 10.1094/MPMI-08-17-0207-R
Wang, Q., Yang, S., Liu, J., Terecskei, K., Ábrahám, E., Gombár, A., et al. (2017). Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 114, 6854–6859. doi: 10.1073/pnas.1700715114
Wells, D. H., and Long, S. R. (2002). The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 43, 1115–1127. doi: 10.1046/j.1365-2958.2002.02826.x
Williams, M. N., Hollingsworth, R. I., Klein, S., and Signer, E. R. (1990). The symbiotic defect of rhizobium meliloti exopolysaccharide mutants is suppressed by lpsZ+, a gene involved in lipopolysaccharide biosynthesis. J. Bacteriol. 172, 2622–2632. doi: 10.1128/jb.172.5.2622-2632.1990
Wilson, K. J., Anjaiah, V., Nambiar, P. T., and Ausubel, F. M. (1987). Isolation and characterization of symbiotic mutants of Bradyrhizobium sp. (Arachis) strain NC92: mutants with host-specific defects in nodulation and nitrogen fixation. J. Bacteriol. 169, 2177–2186. doi: 10.1128/jb.169.5.2177-2186.1987
Wippel, K., and Long, S. R. (2019). Symbiotic performance of Sinorhizobium meliloti lacking ppGpp depends on the medicago host species. Mol. Plant Microbe Int. 32, 717–728. doi: 10.1094/MPMI-11-18-0306-R
Wong, J., Gysel, K., Birkefeldt, T. G., Vinther, M., Muszynski, A., Azadi, P., et al. (2020). Structural signatures in EPR3 define a unique class of plant carbohydrate receptors. Nat. Commun. 11:3797. doi: 10.1038/s41467-020-17568-9
Yamaya-Ito, H., Shimoda, Y., Hakoyama, T., Sato, S., Kaneko, T., Hossain, M. S., et al. (2018). Loss-of-function of ASPARTIC PEPTIDASE NODULE-INDUCED 1 (APN1) in lotus japonicus restricts efficient nitrogen-fixing symbiosis with specific mesorhizobium loti strains. Plant J. 93, 5–16. doi: 10.1111/tpj.13759
Yang, S., Tang, F., Gao, M., Krishnan, H. B., and Zhu, H. (2010). R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. U.S.A. 107, 18735–18740. doi: 10.1073/pnas.1011957107
Yang, S., Wang, Q., Fedorova, E., Liu, J., Qin, Q., Zheng, Q., et al. (2017). Microsymbiont discrimination mediated by a host-secreted peptide in medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 114, 6848–6853. doi: 10.1073/pnas.1700460114
Yasuda, M., Miwa, H., Masuda, S., Takebayashi, Y., Sakakibara, H., and Okazaki, S. (2016). Effector-triggered immunity determines host genotype-specific incompatibility in legume-rhizobium symbiosis. Plant Cell Physiol. 57, 1791–1800. doi: 10.1093/pcp/pcw104
Zhang, B., Wang, M., Sun, Y., Zhao, P., Liu, C., Qing, K., et al. (2021). Glycine max NNL1 restricts symbiotic compatibility with widely distributed bradyrhizobia via root hair infection. Nat. Plants 7, 73–86. doi: 10.1038/s41477-020-00832-7
Zhang, Q., Raoof, M., Chen, Y., Sumi, Y., Sursal, T., Junger, W., et al. (2010). Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464, 104–107. doi: 10.1038/nature08780
Zhou, S., Zhang, C., Huang, Y., Chen, H., Yuan, S., and Zhou, X. (2021). Characteristics and research progress of legume nodule senescence. Plants 10:1103. doi: 10.3390/plants10061103
Keywords: symbiotic nitrogen fixation, rhizobium strains, legume ecotypes/cultivars, symbiotic incompatibility, partner dependent mutation manifestation
Citation: Wang T, Balla B, Kovács S and Kereszt A (2022) Varietas Delectat: Exploring Natural Variations in Nitrogen-Fixing Symbiosis Research. Front. Plant Sci. 13:856187. doi: 10.3389/fpls.2022.856187
Received: 16 January 2022; Accepted: 08 March 2022;
Published: 11 April 2022.
Edited by:
Katharina Pawlowski, Stockholm University, SwedenReviewed by:
Ulrike Mathesius, Australian National University, AustraliaCopyright © 2022 Wang, Balla, Kovács and Kereszt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Attila Kereszt, a2VyZXN6dGFAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.