- Key Laboratory of Forest Protection of National Forestry and Grassland Administration, Research Institute of Forest Ecology, Environment and Nature Conservation, Chinese Academy of Forestry, Beijing, China
The pine wood nematode (PWN), Bursaphelenchus xylophilus, is one of the most lethal nematode species, which causes pine wilt disease (PWD), a devastating forest disease. To date, no effective methods have been developed to control the disease; hence, rapid precise detection of B. xylophilus is of great significance. Traditional molecular diagnostic methods are time-consuming and require sophisticated instruments or skilled operators, which are unavailable in resource-limited settings. A specific, sensitive, and field-applicable diagnostic method is urgently needed. In this study, we developed a diagnostic method using recombinase polymerase amplification combined with lateral flow dipstick (RPA-LFD) for the rapid on-site detection of B. xylophilus. The false-positive signals from primer-dependent artifacts were eliminated using a probe, and base substitutions were included in the primer and probe. The entire detection process for the RPA-LFD assay can be completed under 38°C within approximately 30 min, including 15 min for crude nematode genomic DNA (gDNA) extraction and master mix preparation, 15 min for the RPA-LFD assay. This assay displayed high specificity toward B. xylophilus and showed no cross-reactions with closely related species, including Bursaphelenchus mucronatus and Bursaphelenchus doui. The sensitivity of this assay had a detection limit as low as 1 pg of B. xylophilus purified genomic DNA. Furthermore, the application of the RPA-LFD assay in simulated spiked pinewood samples showed accurate detection results. The RPA-LFD assay in this study successfully detected B. xylophilus in less than 30 min, providing a novel alternative for the simple, sensitive, and specific detection of B. xylophilus and showed potential for B. xylophilus point-of-care testing (POCT) in resource-limited areas or in field.
Introduction
Pine wilt disease (PWD) is a devastating forest disease caused by the pine wood nematode (PWN) (Mota and Vieira, 2008). The spread of PWNs is mainly caused by increased global trade. As PWD is regarded as an “incurable disease,” rapid and accurate detection, quarantine, and monitoring can manage disease spread (Futai, 2013; Yazaki et al., 2018; Kim et al., 2020). Therefore, the primary task in PWD control is to accurately detect PWN.
Various Bursaphelenchus xylophilus diagnostic methods have been developed, including morphological methods using microscopic observations and molecular diagnostic methods using diverse molecular markers (Inácio et al., 2015; Lee et al., 2021). However, due to the similar morphological characteristics of B. xylophilus and its closely related species, Bursaphelenchus mucronatus, accurately identifying B. xylophilus requires specialized knowledge and professional skills (Gu et al., 2011; Gu and Wang, 2011; Wang et al., 2012; Gu, 2014; Li et al., 2021). Furthermore, molecular methods require sophisticated equipment and are not suitable for implementation in the field, causing delayed detection and response to PWD pandemics (Sultana et al., 2013). Therefore, a specific, sensitive, and field-applicable diagnostic method is needed to improve the efficiency of B. xylophilus detection and quarantine.
Alternatively, isothermal DNA amplification techniques that do not require the use of thermal cycling apparatus have been applied to detect B. xylophilus. These include loop-mediated isothermal amplification (LAMP) (Kikuchi et al., 2009; Meng et al., 2018; Ahuja and Somvanshi, 2021), denaturation bubble-mediated strand exchange amplification (SEA) (Liu C. et al., 2019), and recombinase polymerase amplification (RPA) (Cha et al., 2019, 2020; Fang et al., 2021). RPA, an isothermal nucleic acid amplification method that has attracted much attention because it is sensitive, rapid, works at isothermal temperature (Piepenburg et al., 2006; Glökler et al., 2021). In addition, RPA assays have been developed to detect some plant-parasitic and animal-parasitic nematodes, including B. xylophilus (Cha et al., 2019, 2020), Meloidogyne enterolobii (Subbotin, 2019), Meloidogyne spp. (M. incognita, M. arenaria, M. javanica, and M. enterolobii) (Ju et al., 2019), Meloidogyne javanica (Chi et al., 2020), Meloidogyne hapla (Song et al., 2020; Subbotin and Burbridge, 2021), Angiostrongylus cantonensis (Jarvi et al., 2021), and Trichinella spp. (Li et al., 2019).
Recombinase polymerase amplification amplicons can be visualized using lateral flow dipstick (LFD), with results could be directly interpreted by the naked eye within a few minutes (Daher et al., 2016). LFDs are products based on lateral flow technology using gold nanoparticles and can promptly detect amplification products making results easy to interpret (Qi et al., 2018).
The LFD assay requires a special nfo probe with a fluorescein amidites (FAM) (carboxyfluorescein) antigen label at the 5′-end, a tetrahydrofuran (THF) spacer (abasic site) in the middle, a C3 spacer (amplification blocker) at the 3′-end, and reverse primer labeled with biotin at the 5′-end. RPA amplicons are amplified using two primers and a probe, which contains both biotin and FAM, and the mouse anti-FAM antibody is enveloped with AuNPs. After reaction mixture is added to the sample well, they are driven by capillary force to move across the conjugate pad and bind to the anti-FAM AuNPs. The test line, which is enveloped with streptavidin, captures molecules with a biotin label when the amplification products go through. The control line, which is used to validate LFD detection, only captures the anti-FAM antibody enveloped with AuNPs because the anti-FAM antibody is from a mouse (Figure 1; Dai et al., 2019; Miao et al., 2019).
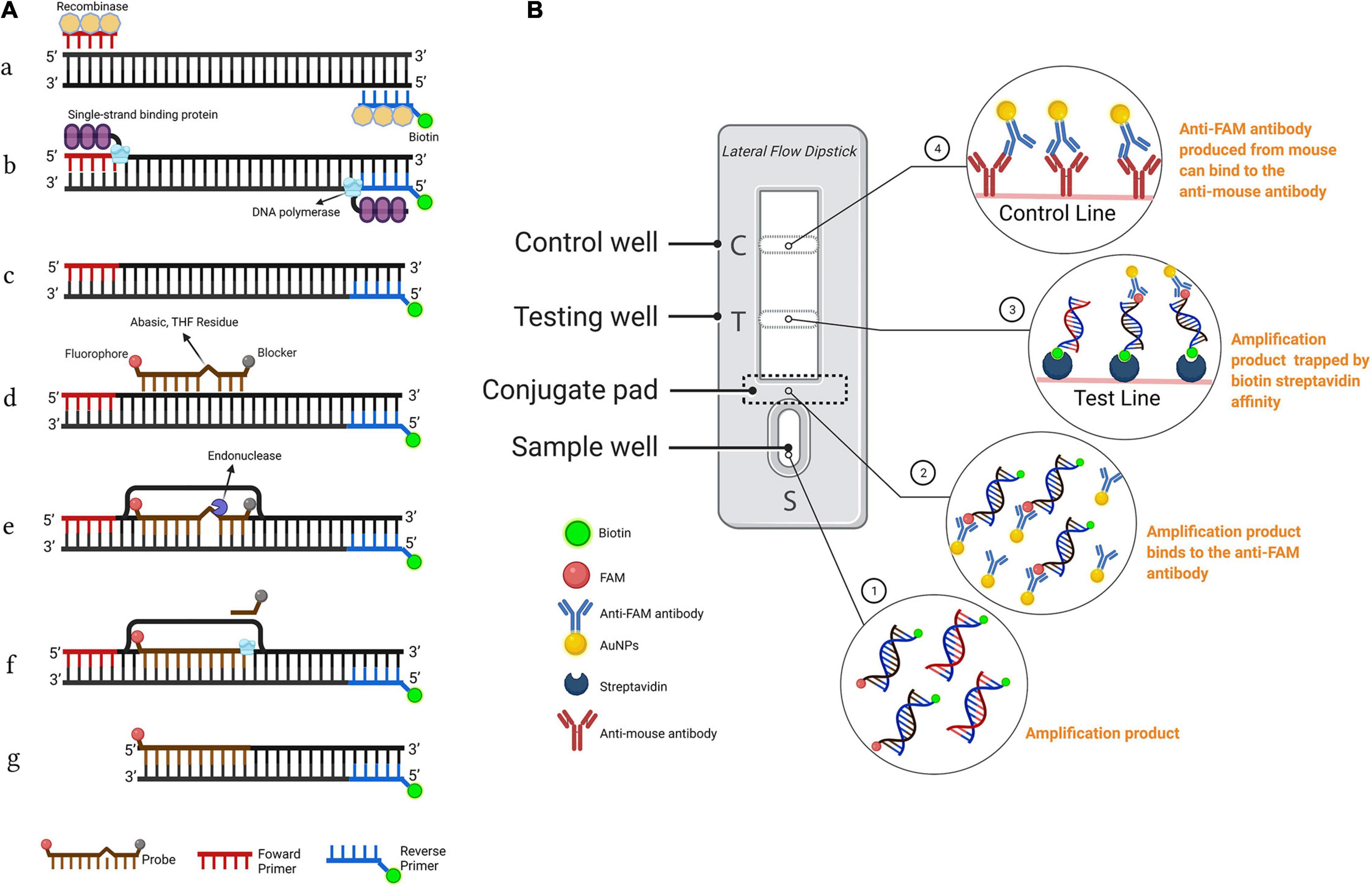
Figure 1. Schematic representation of the recombinase polymerase amplification combined with lateral flow dipstick (RPA-LFD) assay. (A) Principle of RPA. DNA strands are represented by horizontal lines, and base pairings are represented by short vertical lines between DNA strands. The forward primer (F), reverse primer (R), probe (P), nfo, single strand DNA-binding protein (SSB), polymerase, recombinase, fluorophore, blocker, and tetrahydrofuran (THF) residue are shown by various color and forms and text annotation. Created with BioRender.com. (B) Schematic representation of the lateral flow dipstick working principle. Lateral flow dipstick was painted after real ones. The segment names are displayed on the left of the dipstick drawing. The direction of liquid migration is from the sample well, through the conjugate pad, to the test and control wells. The coating material on each segment of the dipstick is shown in the image to the right. Molecules can be captured by the coating material on the test line and control line. Molecules and their symbolic shape are displayed in the lower-left corner. Dipstick created with BioRender.com.
It should be noted that false-positive signals from the self-linked primer–probe complex should be considered a vital inherent defect of the RPA-LFD assay (Wu et al., 2020). Without the temperature cycles in PCR-based methods, primer combination in RPA is performed at ambient temperature, which does not provide an opportunity for dissociation if mispairing occurs and may generate a false-positive signal. In addition, LFD could not differentiate the size of the amplification product that releases signal. Furthermore, the RPA-LFD assay is overly sensitive; thus, interference from false-positive signals due to primer-dependent artifacts must be avoided (Poritz and Ririe, 2014; Wang et al., 2020).
In this study, we developed a simple, rapid, specific, and sensitive RPA-LFD assay to detect B. xylophilus. This method eliminated false-positive signals from non-specific primer–probe complexes due to an elaborate design and strict screening of primers and probe, as well as by bringing in base substitutions on the reverse primer and probe. The specificity and sensitivity of the assay were also investigated. The practicability was also analyzed by detecting B. xylophilus in spiked pine wood samples.
Materials and Methods
Specimen Collection and DNA Extraction
Bursaphelenchus xylophilus, B. mucronatus, and Bursaphelenchus doui were provided by the Key Laboratory of Forest Protection of National Forestry and Grassland Administration, Chinese Academy of Forestry, China. A total of 17 nematode isolates representing three Bursaphelenchus species, collected from multiple sites in China, were used in this study (Table 1). Nematode isolates were reared on Botrytis cinerea mycelia cultured on potato dextrose agar (PDA) plates at 25°C for 5–7 days and were identified using morphological methods and molecular diagnostic methods. Nematodes were then separated using the Baermann funnel technique (Viglierchio and Schmitt, 1983), and 500 μl of nematodes were collected in a 1.5 ml Eppendorf tube after being washed three times in sterilized water and stored at 4°C for future use.
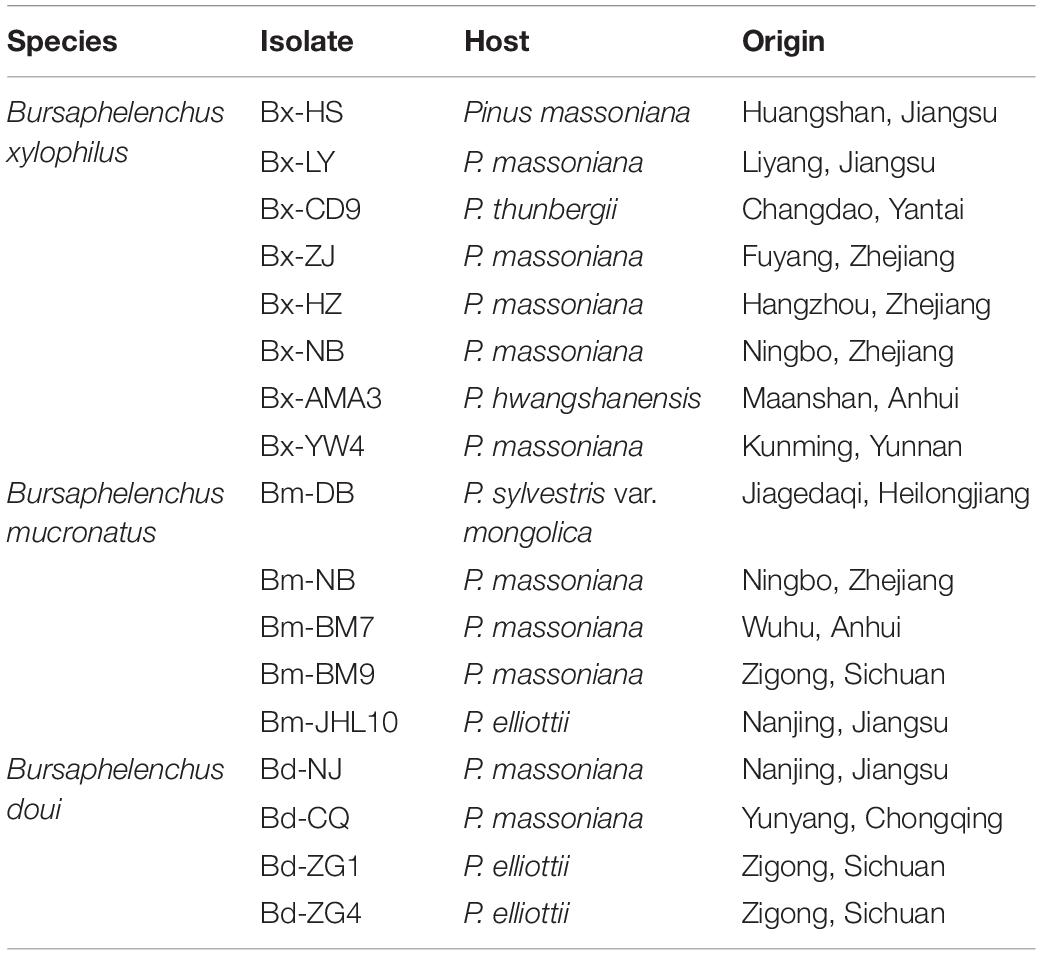
Table 1. Nematode species used in B. xylophilus recombinase polymerase amplification combined with lateral flow dipstick (RPA-LFD) detection assay.
Genomic DNA (gDNA) was extracted from approximately 500 nematodes of each nematode species (B. xylophilus, B. mucronatus, and B. doui) using the TIANamp Genomic DNA Kit [Tiangen Biotech (Beijing) Co. Ltd., Beijing, China] according to the manufacturer’s instructions. The DNA concentration and purity were quantified using a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). The DNA was diluted to 10 ng/μl with distilled water and stored at −20°C for future use.
Designing of Recombinase Polymerase Amplification Primers
To design specific primers for RPA, the previously identified conserved gene encoding synaptic guidepost protein (SYG-2), was selected as the target segment (Gou, 2014; Meng et al., 2018). The syg-2 gene part sequences were amplified using PCR primer pairs syg2-part-f/syg2-part-r, and the PCR cycle conditions were set as described by Gou (2014). The PCR products (25 μl) were electrophoresed on a 2% agarose gel, and the amplified products were recovered using a PCR Cleaning Kit [Tiangen Biotech (Beijing) Co., Ltd., Beijing, China] and sequenced [Sangon Biotech (Shanghai) Co. Ltd., Shanghai, China], and the sequences were aligned using the ClustalW program implemented in MEGA version 7.0.14. RPA-basic assays require forward and reverse primers; accordingly, the RPA primers were designed using Primer Premier 5.0 (Premier Biosoft, Palo Alto, CA, United States) according to the TwistDx instruction manual (Cambridge, United Kingdom). RPA primer length was 30–36 bp, the primer GC content was 30–70%, and the amplicon length was between 150 and 300 bp.
Recombinase Polymerase Amplification Procedure and Electrophoresis
The Basic RPA 50-μl reaction volume was prepared according to the instructions for the RPA-basic kit (#WLN8201KIT; AMP-Future Biotech Co. Ltd., Weifang, China). It consisted of 29.4 μl rehydration buffer, 2 μl forward primer, 2 μl reverse primer (primer concentration, 10 μM), one lyophilized enzyme pellet, 1 μl DNA template, and 13.1 μl nuclease-free water; the reaction was initiated by adding 2.5 μl magnesium acetate. After incubation at 38°C for 30 min, amplicons were purified using spin columns before electrophoresis on 1.5% agarose gels.
Probe Designing
The probes were designed using Primer Premier 5.0 software (Premier Biosoft). The size of the probe was set as 46–53 nt, GC content was 20–80%, and Tm set as 50–80°C. Maximum hairpin and primer dimer parameters were set at ≤4 bonds near 3′-end. Other settings were set as default. Following design, primers and probes were assessed using the OligoAnalyzer Tool (IDT) and screened depending on their binding score for continuous base pairings between the probe and reverse primer. The RPA primers and nfo probe were synthesized and provided by Sangon Biotech (Shanghai) Co. Ltd.
Recombinase Polymerase Amplification-Lateral Flow Dipstick Procedure
Recombinase polymerase amplification-lateral flow dipstick assays were carried out in a 50 μl reaction volume using the RPA nfo kit (#WLN8203KIT: AMP-Future Biotech Co. Ltd.) according to the manufacturer’s instructions. The reaction mixture comprised 29.4 μl rehydration buffer, 2.5 μl magnesium acetate, 2 μl each primer pair (10 μM), 0.6 μl nfo probe (10 μM), 12.5 μl nuclease-free water, and 1.0 μl DNA template. All reagents except the DNA template and magnesium acetate were prepared in a master mix, which was subsequently dispensed into reaction tubes containing a dried enzyme pellet provided with the kit. After adding 1 μl of the nucleic acid template to the tubes and pipetting 2.5 μl of magnesium acetate into the tube lids, the lids were closed carefully, and the tubes were inverted several times and briefly centrifuged at 6,000 rpm for 30 s.
Post-amplification results were visually interpreted using Hybridetect LFD (kit #WLFS8201; AMP-Future Biotech Co. Ltd.). Thereafter, 10 μl of reaction solution was added to a centrifuge tube containing 190 μl ddH2O, after which 50 μl diluted solution was pipetted onto the LFD sample well and incubated for 3–5 min at ambient temperature. If both test and control line are visible, it is positive; if only the control line is visible, it is negative; and if the control line is not visible, it is invalid. A repeat test would then be conducted using a new dipstick in an invalid situation.
Introduction of Base Substitutions to Probe and Reverse Primer
Recombinase polymerase amplification can tolerate several base mispairings in primers without noticeably influencing accuracy (Daher et al., 2015; Liu X. et al., 2019). Therefore, base substitutions were introduced into the probe and reverse primer according to previous research (Wu et al., 2020). The probe and primer with base substitution were evaluated for false-positive signals using no template control (NTC).
Optimization of Recombinase Polymerase Amplification-Lateral Flow Dipstick Assays
To investigate the optimal conditions of the RPA-LFD assay, different reaction temperatures (15, 20, 25, 30, 35, 40, and 45°C) and reaction times (5, 10, 15, 20, 25, 30, and 35 min) were assessed. For determination of optimal reaction temperature, all treatments were reacted for 30 min. After the reaction, tubes were immediately put on ice to stop the reaction.
Specificity of Recombinase Polymerase Amplification-Lateral Flow Dipstick Assay
To investigate the specificity of the assay, nematode genomic DNA was prepared from 17 nematode isolates, including eight B. xylophilus, five B. mucronatus, and four B. doui isolates, were used as DNA templates, as described above. Specificity tests were repeated three times.
Analysis of Recombinase Polymerase Amplification-Lateral Flow Dipstick Assay Sensitivity
The sensitivity of the RPA assay was investigated using the RPA nfo kit by testing a 10-fold serial dilution of gDNA extracted from B. xylophilus. Genomic DNA extracted from B. xylophilus was diluted into seven concentrations with sterile distilled water, including 10, 1, 10–1, 10–2, 10–3, 10–4, and 10–5 ng, respectively. Each dilution and sterile distilled water as the NTC, were used as templates in the RPA-LFD assay, respectively, as described previously. The sensitivity tests were repeated thrice.
Preparation of Pinewood Spiked Sample for Recombinase Polymerase Amplification-Lateral Flow Dipstick Evaluation
To investigate the interference from the DAP buffer or pine wood, which includes humic acid, ethanol, polysaccharides, and polyphenols (Cha et al., 2019). Pine wood chips (100 mg) from healthy pine trees were placed in a 2 ml tube filled with 1 ml DAP buffer (20 mM sodium hydroxide, 5% polyethylene glycol 200, and 5% dimethyl sulfoxide) (Cha et al., 2019), and 10 ng of pure gDNA of different B. xylophilus or Bursaphelenchus spp. isolates was spiked into the lysates. The tubes were then vibrantly mixed and incubated at ambient temperature for 10 min. Subsequently, 2.5 μl of lysate solution was used as the RPA template (the final concentration of gDNA was 25 pg in each RPA reaction).
Results
Primer Design and Screening
Using Primer Premier 5.0, six potential primer pairs were obtained (Table 2). They were screened using basic RPA (Figure 2). The image showed amplification of the syg-2 (synaptic guidepost protein) target fragments, which have a typical ladder pattern, comprised multimers of the amplicon-sized monomer, probably because the protocadherin gene family which syg-2 belongs to is arrayed in tandem on the chromosome (Figure 2; Yamagata et al., 2003). However, five of them showed primer dimer bands under specific amplification bands. Primer pair S1F/R amplified specific products and showed no obvious primer-dimer (Figure 2). In order to use LFD to interpret results, the reverse primer was labeled at the 5′-end with biotin.

Table 2. Sequences of the primers and probes used in B. xylophilus recombinase polymerase amplification (RPA) assay.
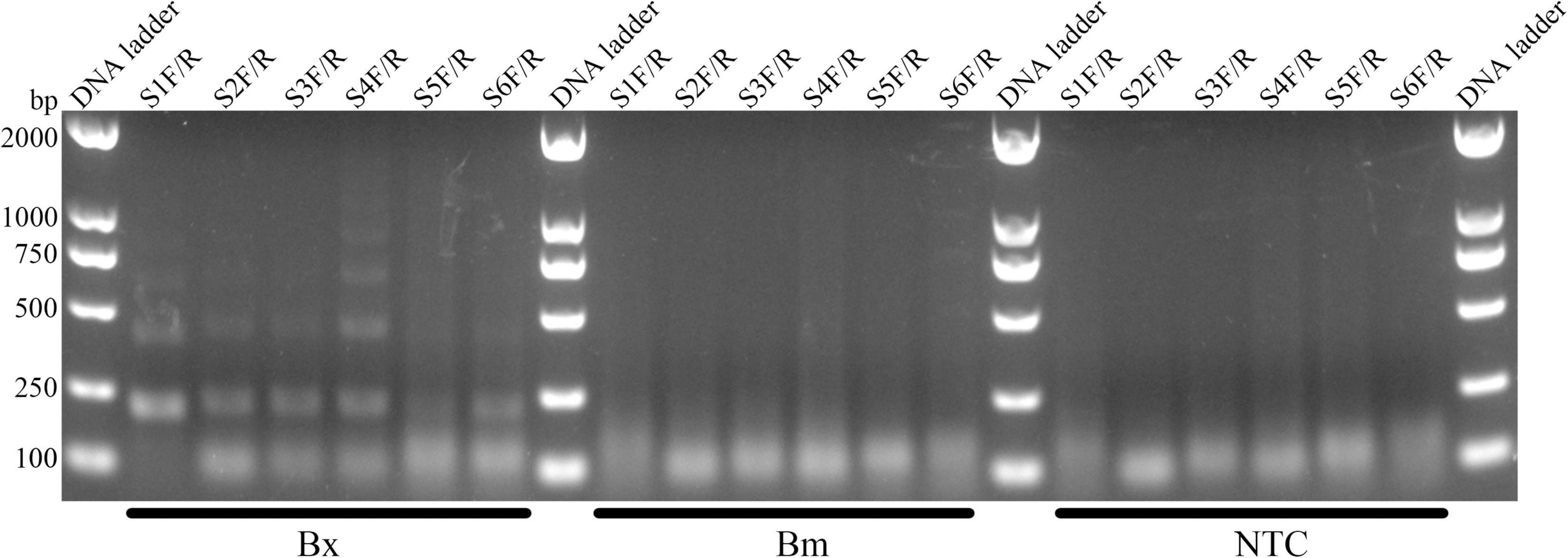
Figure 2. Screening of primer pairs by Basic-RPA. The image shows amplification bands targeting the syg-2 gene. The primer name is shown above each lane. The following Bx, Bm, and NTC lanes are B. xylophilus, B. mucronatus genomic DNA, and no template controls for corresponding primer pairs, respectively. Band sizes for DNA ladders are shown on the left.
Addition of a Probe Into the Recombinase Polymerase Amplification Reaction
The nfo probe (S1-nfo-P) was between the forward and reverse primers and comprised of an oligonucleotide with a 5′-FAM as antigenic label and a 3′-C3 spacer that could block extension. An THF spacer (tetrahydrofuran) was placed on the probe to replace a guanine. The nfo enzyme activated after bases flanking THF matching with complementary bases and would cut THF site, removing probe 3′-end blocker for extension (Figure 1A). The specific RPA primers, nfo probe, and their sequences are listed in Table 2.
For the newly added probe, the RPA-LFD assay was carried out to evaluate the amplification signal and false-positive signal. The amplification signal was evident for primer–probe set S1R-nfo/S1-nfo-P; however, the false-positive signal also existed for NTC (Figure 3A). Cross dimer analysis showed that there was still a continuous base match between the probe and primer. Furthermore, the matched bases were located on two sides of THF site, facilitating the nfo enzyme cutting activity (Figure 3B).
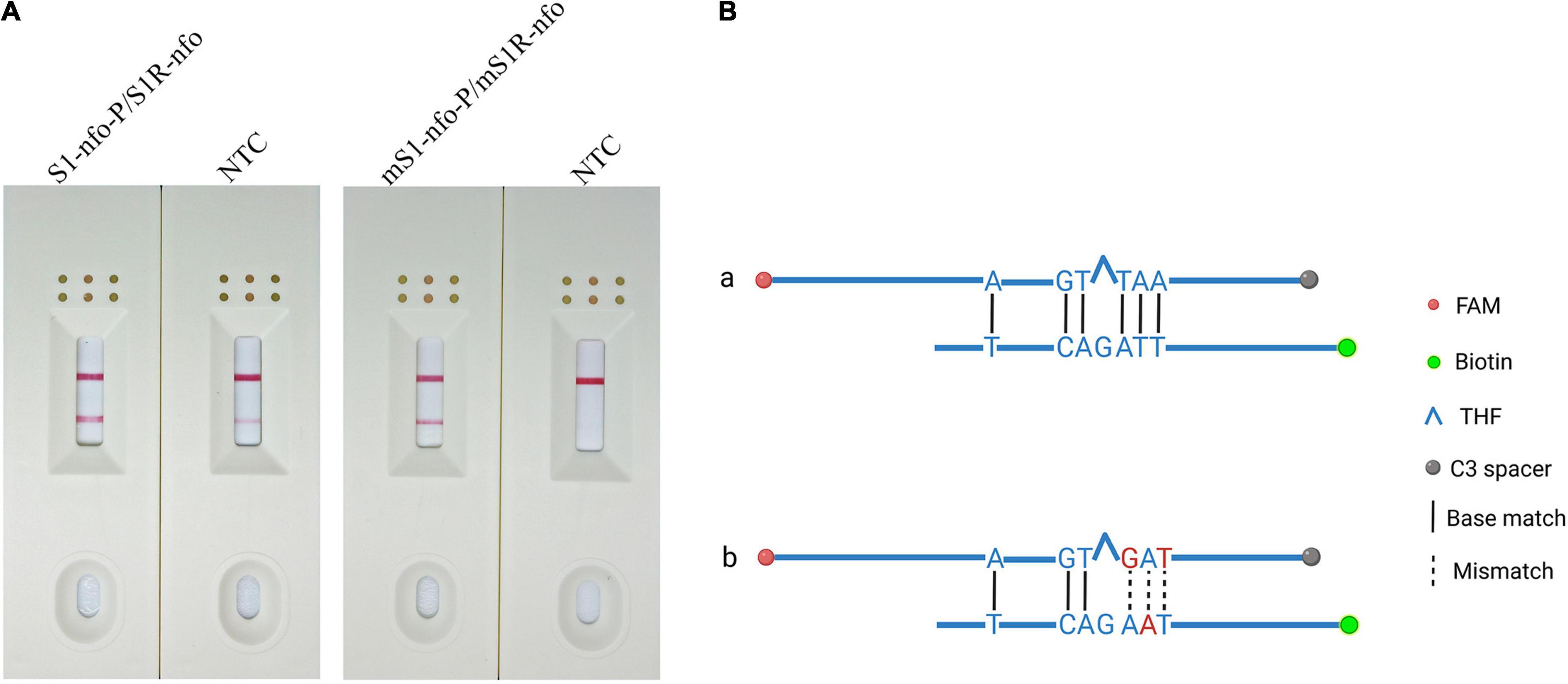
Figure 3. Preliminary test and analysis of the original primer–probe set and base-substituted primer–probe set. (A) The image shows the LFD results for RPA amplifications using original primer–probe set and base-substituted primer–probe set. The name of each set is displayed above dipstick. NTC is no template control tested for false-positive signals. The positions of test and control lines are marked on the right of the dipsticks image. The template was B. xylophilus gDNA. The reactions are carried out at 38°C for 10 min. (B) Analysis of the cross-dimer formation between the probe and reverse primer. The original probe has five-continuous base pairings with original reverse primer, which could lead to primer–probe complex, resulting in a false positive signal. After bringing in mispairings into the original self-ligating primer–probe set, one of which is on reverse primer and two on the probe, the formation of the probe-primer complex is artificially interrupted and signal is eliminated. Labels and modifications for DNA, nfo, base matches, and mismatches are represented by different shapes and colors, and a legend is given to the right of the figure. Created with BioRender.com.
Elimination of False-Positive Signals With Base Mismatches
The results showed that false-positive signal was eliminated by bringing mispairings on probe and reverse primer (mS1-nfo-P/mS1R-nfo) (Figures 3A,B, substituted bases in red). The sequences of the modified probe (mS1-nfo-P) and reverse primer (mS1R-nfo) are listed in Table 2. These base substitutions did not influence amplification accuracy and efficiency by observing band color density (Figure 3A). Therefore, this primer and probe were used for the following RPA-LFD reactions in this study.
Optimization of Recombinase Polymerase Amplification-Lateral Flow Dipstick Assay for Bursaphelenchus xylophilus Detection
The RPA-LFD assays were tested at temperature range from 15 to 45°C at 5°C intervals for 30 min. Results showed that test lines were visible from 20 to 45°C, color density of the test line did not increase noticeably from 35 to 45°C. Therefore, 38°C was selected as the optimal temperature (Figure 4A). Moreover, the RPA-LFD reaction time was tested from 5 to 35 min at 5 min intervals at 38°C. Results showed that test lines were visible when the reaction time was 10–35 min; therefore, 10 min was selected as the optimal time for the RPA-LFD assay of B. xylophilus detection considering rapid diagnostics (Figure 4B).
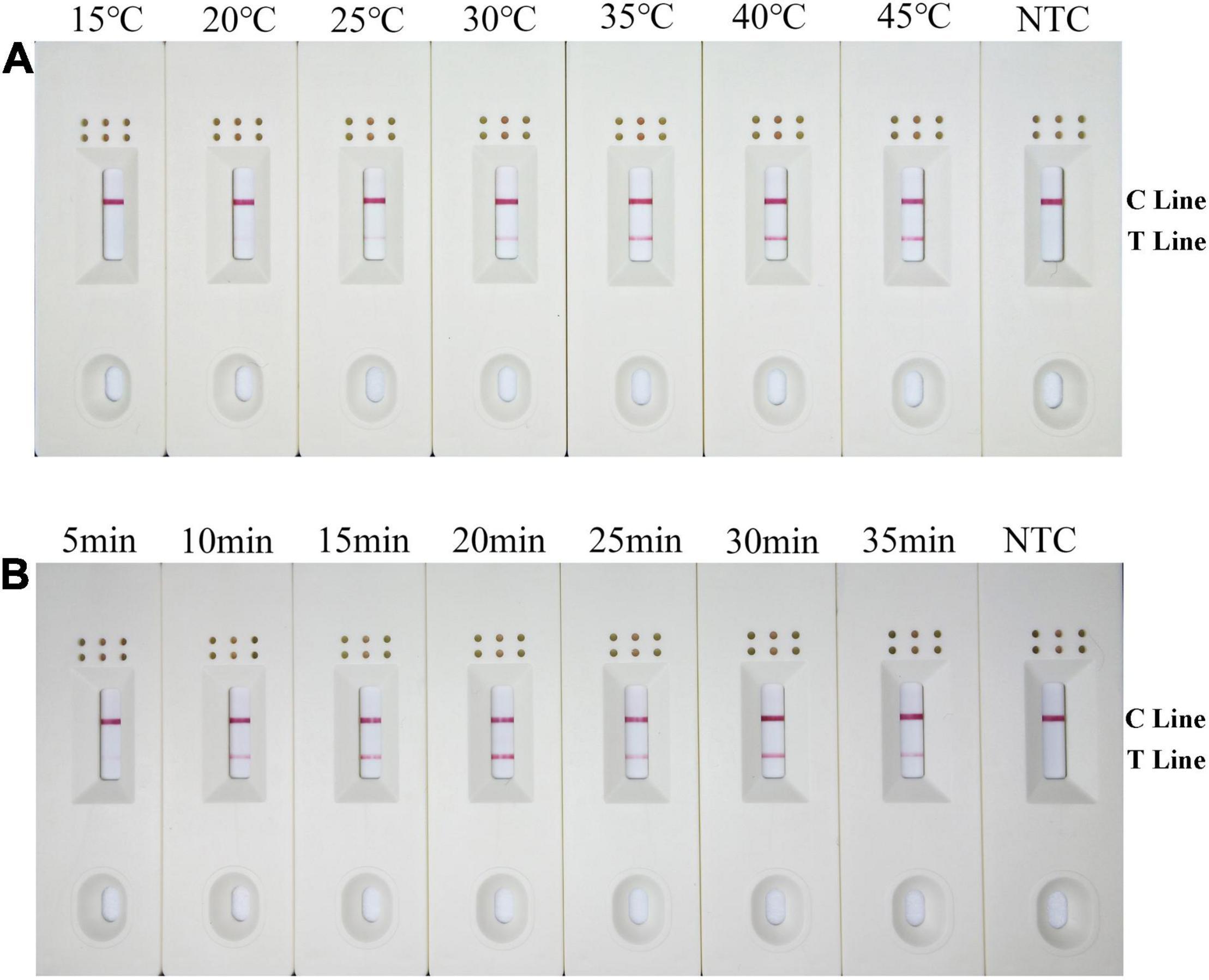
Figure 4. Optimization of the modified primer–probe set in RPA–LFD assay. (A) Optimal reaction temperature of the RPA–LFD assay. The figure shows the LFD results of RPA amplification at different temperatures. The temperature is displayed at the top of each dipstick. The amplification template is B. xylophilus gDNA. The NTC band is no template control carried out at 38°C. The positions of the control and test lines are shown on the right side of the dipstick. (B) Optimal reaction time of the RPA–LFD assay. The image shows the LFD results of RPA amplifications with different times. The time to perform the RPA reaction is displayed at the top of each dipstick. The amplification template is B. xylophilus gDNA. NTC dipsticks are performed for 10 min without template. The positions of the control and test lines are shown on the right side of the dipstick.
Analytical Specificity of Recombinase Polymerase Amplification-Lateral Flow Dipstick Assay
To assess the RPA-LFD analytical specificity, gDNA was prepared using several nematode species, including eight B. xylophilus, five B. mucronatus, and four B. doui isolates (Table 1). Only when B. xylophilus gDNA as DNA template can produce red test lines on dipsticks, indicating positive results. When other nematode species gDNA or NTC as DNA template, no red test lines were seen on dipsticks (Figure 5), indicating that this primer–probe combination had good specificity toward B. xylophilus and had no cross-reactions with other nematode species.
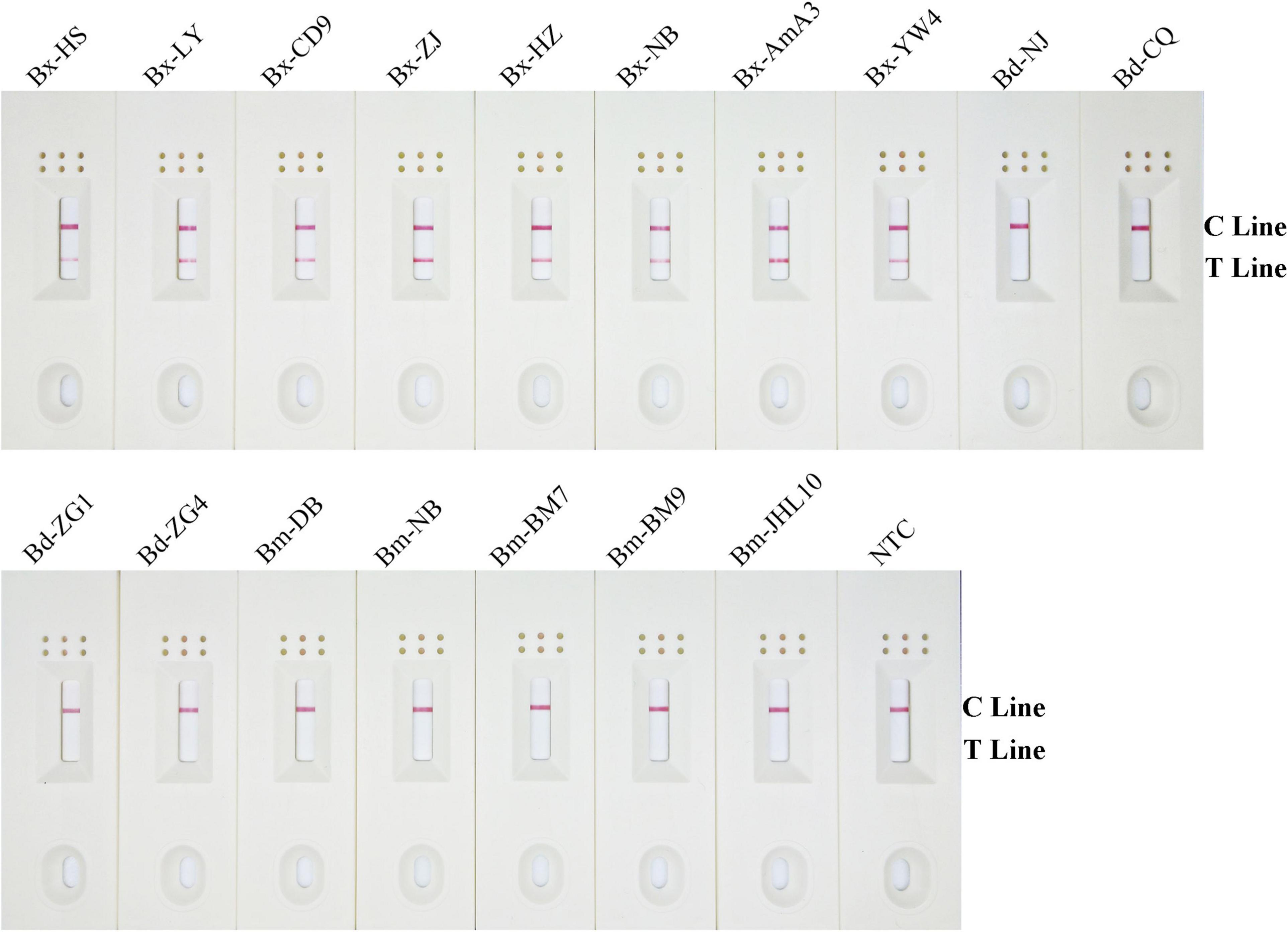
Figure 5. Detection specificity of the modified primer–probe set in RPA–LFD assay. Specificity determination of RPA-LFD using different nematode isolates gDNA templates. The isolate of nematode is marked above each dipstick. The NTC dipstick is no template control. The positions of test and control lines are shown on the right side of dipstick. Reactions are carried out at 38°C for 10 min.
Detection Limit of Recombinase Polymerase Amplification-Lateral Flow Dipstick Assay
Results showed that the LFD test and control lines were displayed from 10 to 10–3 ng gDNA. Furthermore, the red color of the test line lightened with decreasing B. xylophilus concentrations, and B. xylophilus gDNA as low as 1 pg (10–3 ng) could be detected (Figure 6). Therefore, the detection limit of the RPA-LFD assay for B. xylophilus was 1 pg of gDNA.
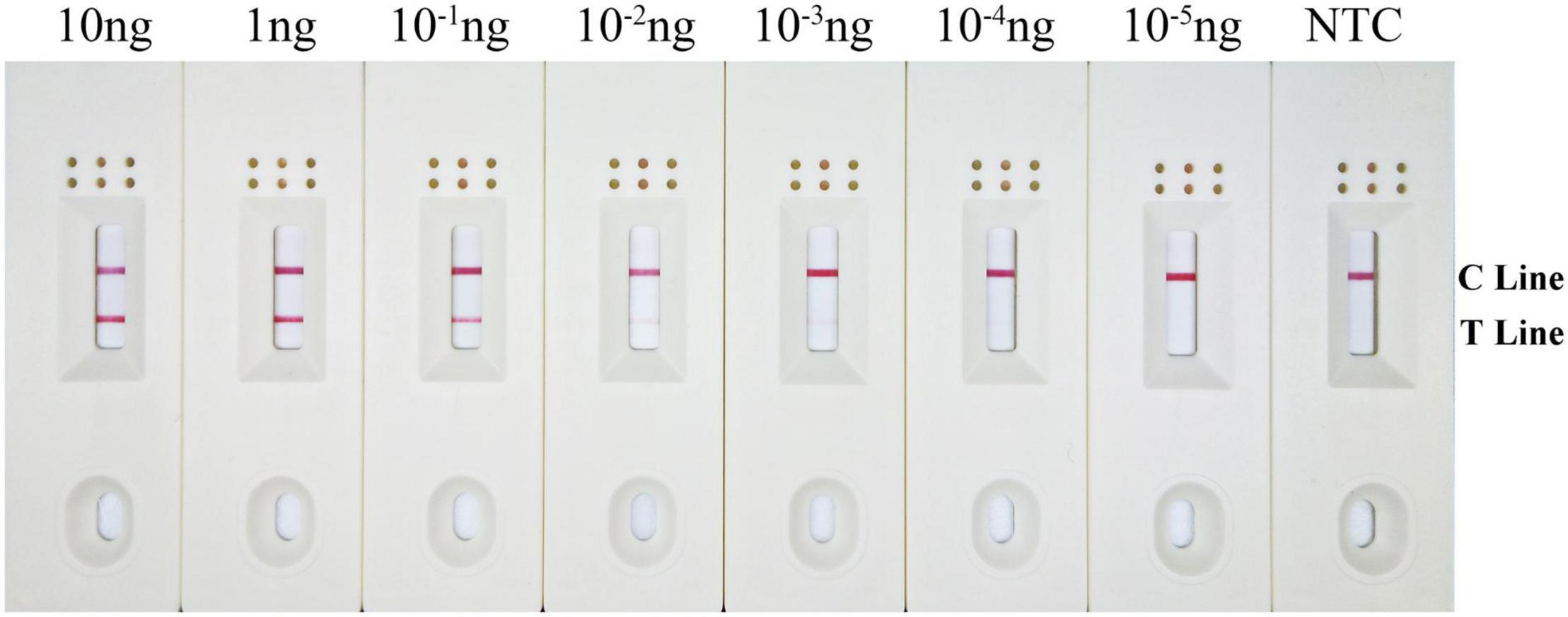
Figure 6. Detection limit of the RPA–LFD assay. The image shows the LFD results of RPA amplification with different concentrations of B. xylophilus gDNA. The DNA concentration used as template is shown above each dipstick. NTC is the no template control. The reactions are carried out at 38°C for 10 min. Test line and control lines are shown on the right side of dipstick.
Application Simulation of the Recombinase Polymerase Amplification-Lateral Flow Dipstick Test for Bursaphelenchus xylophilus Detection
Using an application simulation, the RPA-LFD test was applied for B. xylophilus detection using artificially spiked pine wood samples. Thirteen healthy pine wood samples were prepared, of which eight were artificially spiked with B. xylophilus gDNA, two with B. mucronatus gDNA, another two with B. doui gDNA, and one with sterilized distilled water as NTC. The results of the RPA-LFD assay were consistent with those of the spiked gDNA species and all eight B. xylophilus gDNA spiked pine wood samples were successfully detected (Figure 7).
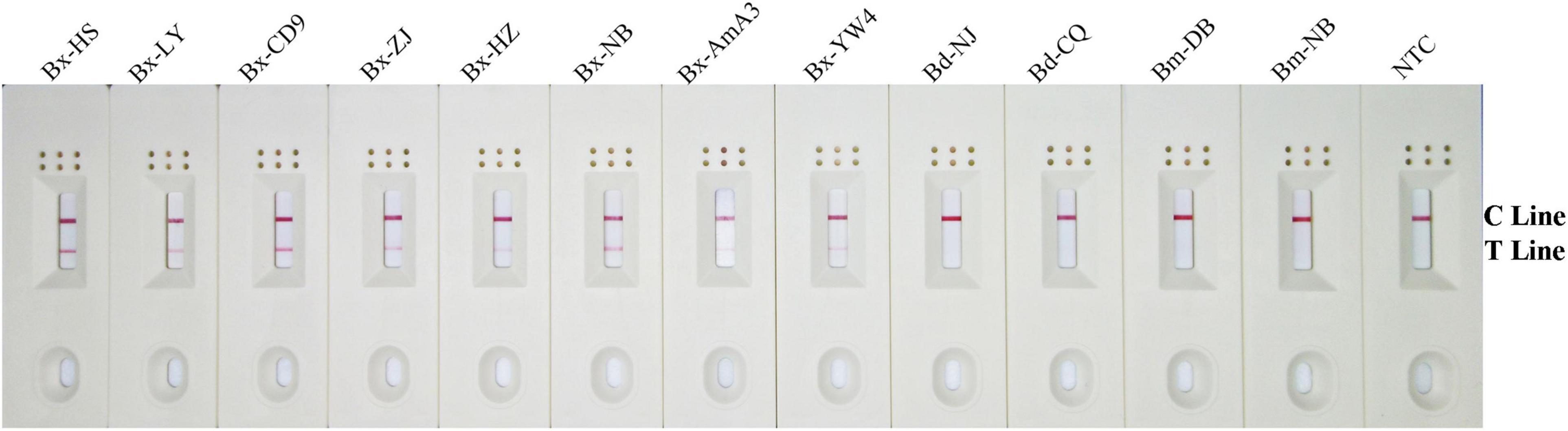
Figure 7. Detection of B. xylophilus purified gDNA in spiked pinewood samples. Images of the LFD results of RPA amplification of healthy pinewood samples spiked with gDNA of various nematode isolates. The spiked gDNA species are shown at the top of each dipstick. The double-distilled water was used as the no-template control, corresponding to the NTC dipstick. The positions of the control and test lines are shown on the right side of the image. The reactions are carried out at 38°C for 10 min.
Discussion
Bursaphelenchus xylophilus induced PWD has destroyed numerous pine trees and has spread to 18 provinces and 693 counties in China over the past 40 years, causing considerable economic losses of hundreds of billions of yuan [Announcement of the State Forestry and Grassland Administration (No. 6 of 2021), 2021]. Therefore, a primary procedure for disease prevention and quarantine comprising an accurate, rapid, sensitive, and convenient method for detecting B. xylophilus in pine wood is urgently needed.
Polymerase chain reaction-based amplification is considered the gold standard of molecular diagnostics; however, it requires trained personnel and specialized, expensive equipment generally unavailable outside laboratory settings (Patel et al., 2014). As an emerging isothermal amplification technique, RPA has displayed many strengths, such as short detection time, high sensitivity, easy operation, and minimum instrument needed. Besides, the RPA amplification results could be interpreted using LFD within a few mins. All these characteristics make RPA-LFD very suitable for on-site detection (James and Macdonald, 2015; Li et al., 2019).
The choice of detection target is crucial for specific diagnostics. Several genes have been selected as biomarkers for PWN detection, including 5S rRNA, hsp70, satellite DNA, ITS rRNA, topoisomerase I, syg-2, and pel-3 (Kang et al., 2004; Castagnone et al., 2005; Leal et al., 2007; Huang et al., 2010; Zhuo et al., 2011; Kang et al., 2015; Meng et al., 2018). Among these, the syg-2 gene has been reported to have superior performance in the LAMP method and was thus selected as the target in this study. To identify regions specific to B. xylophilus, the syg-2 gene sequence of B. xylophilus was amplified using the PCR primers designed by Gou (2014) and then compared with two closely related species, after which the diverged region specific to B. xylophilus was selected (Supplementary Figure 1). The diverged region was considered a possible target area for designing primer and probe in the assay. Since the protocadherin DNA family has been shown to consist of tandemly arranged repeats, the amplification of a ladder of multimers of the amplicon-sized monomer was observed in basic-RPA experiments using the designed primers (Figure 2; Hirano et al., 2003; Washbourne et al., 2004).
On the LFD, the control line is enveloped with anti-mouse antibody, and the test line was enveloped with streptavidin. The test line first captures products labeled with biotin when the reaction mixture passes through. If the products also had a FAM label, it will combine with the AuNPs, which are enveloped with anti-FAM antibody, led to the enrichment of AuNPs at the test line, showing a red line (Wang et al., 2020). The control line is used to validate LFD detection, only captures the anti-FAM antibody enveloped with AuNPs, because the anti-FAM antibody is from a mouse (Figure 1).
Nonetheless, the RPA-LFD method has a non-ignorable inherent defect of primer-dependent artifacts. In the RPA-basic-based LFD reaction, forward and reverse primers were labeled with FAM and biotin at 5′-end, respectively. Amplicons are labeled with both biotin and FAM, if primer dimers are formed, it also produces a positive signal (Figures 8A,B). No matter how cautious the screening of primer is, it is nearly impossible to prevent primer dimers in the DNA amplification process (Meagher et al., 2018). Thermal cycling strategies could prevent primer-dimer formation in PCR but are unsuitable for RPA because RPA works at an isothermal temperature of about 38°C, and mispaired primers are difficult to dissociate (Brownie et al., 1997). Furthermore, the LFD method cannot differentiate the size of the molecule emitting signal and treats each molecule as a positive signal (Safenkova et al., 2020). Considering the RPA-LFD method is highly sensitive (Miao et al., 2019), any interference from primer-dependent artifacts could result in false-positive detection.
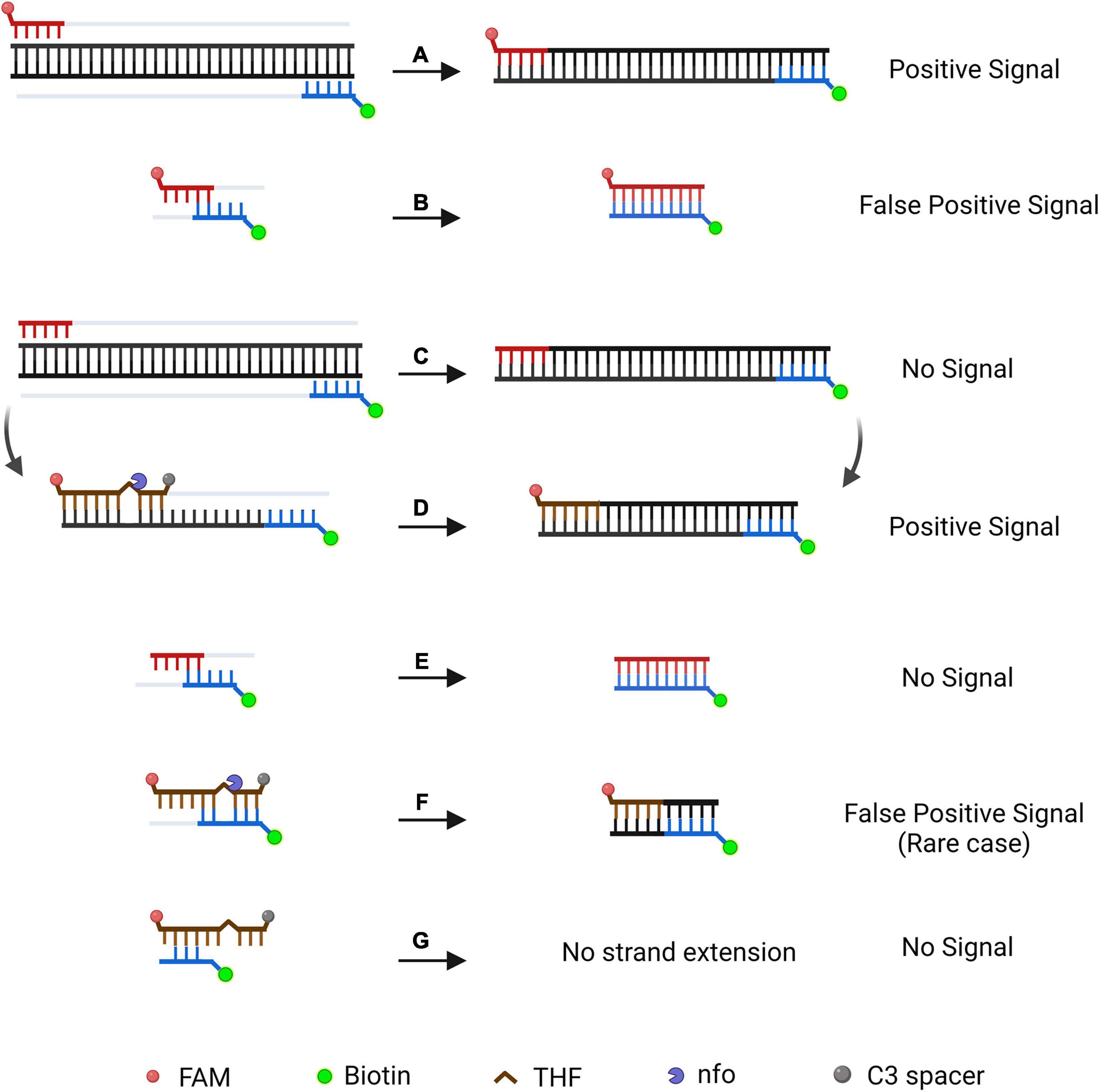
Figure 8. Schematic illustration of a specially designed probe that reduces false-positive signals from primer-dependent artifacts. In the RPA-Basic-based LFD reaction, both the amplification products (A) and the primer–dimers (B) can give positive signals. By using a probe in RPA reaction, amplification of the target DNA from the primers does not yield a positive signal (C). The amplification product is subjected to another round of amplification under the guidance of the probe and shows a positive signal (D). Primer dimers in the probe-based RPA reaction give no positive signal (E). In most cases, the primer–probe complex will not give a positive signal (G). The complex gives a positive signal when the primer and probe pairing multiple bases flanking the THF site (F). DNA strands are represented by horizontal lines, and matching bases are represented by short vertical lines between the DNA strands. The expected amplification of the DNA strand is indicated by the gray line. Modification group and nfo are represented by different colors and forms, and a legend is given at the bottom of the figure. Created with BioRender.com. Picture inspired by Wu et al. (2020).
Research has shown that introducing a probe in RPA would enhance specificity and decrease non-specific amplification products (Figures 8C,D; Piepenburg et al., 2006; Ivanov et al., 2021). FAM was labeled at the 5′-end of the probe, while the C3-spacer was labeled at the 3′-end of the probe that block extension. The THF site was placed on the probe to be cut by nfo enzyme. In this way, amplification led by primer pair would result in a product that only has biotin label, which would not generate a signal on the LFD (Figure 8C). The probe fully pairing amplified DNA strand can be cut by nfo at THF site (nfo was activated if bases flanking THF site paired with complementary bases), freeing the 3′-end for the extension to produce a positive signal on the LFD (Figure 8D). Using this probe-guided amplification, primer-dimers did not produce a signal (Figure 8E). The partly matched primer–probe complex could not be amplified (Figure 8G). Only in the rare case, the primer and probe paired several bases flanking the THF site (Figure 8F), and this would generate false-positive signal (Wu et al., 2020).
Therefore, elimination of the false-positive signal could not only count on using a probe, elaborate design and strict screening are still essential to prevent false-positive scenarios (Figure 8). Based on previous research, we inferred that the false-positive signal occurred in this study should be caused by the probe-primer complex since the labels were on the probe and reverse primer (Wu et al., 2020). By setting stringent parameters, probes were designed to have the lowest theoretical chance of primer pairing. However, cross-dimer analysis displayed many possible primer–probe dimer scenarios between the nfo probe and the reverse primer, and some cross-dimers fit the special case of false-positive signals. That is, the reverse primer formed mismatches with bases flanking the THF site on the probe, which could elucidate why the original primer–probe set (S1-nfo-P/S1R-nfo) displayed false-positive signals.
The probe needed to be in the middle of two primers. However, obtaining an ideal probe without any continuous bases pairing to the reverse primer was scarcely possible. Nonetheless, we could exploit the tolerance of RPA to certain base mispairings on primers against the template, and attempt to bring in base substitutions in RPA-LFD method (Daher et al., 2015; Liu X. et al., 2019). By imitating a previously described method of bringing in three mismatches (Wu et al., 2020) in which two are in the probe and one in the reverse primer, the probe-primer dimer ligation was artificially interrupted, and false-positive was avoided. In addition, the RPA-LFD efficacy was not noticeably influenced when compared the color difference between the S1-nfo-P/S1R-nfo line and mS1-nfo-P/mS1R-nfo line, as shown in Figure 3.
Primer-dependent artifacts were successfully eliminated, resulting in the establishment of the RPA-LFD method. The analytical specificity test showed that the assay displayed high specificity toward B. xylophilus and avoided cross-reactions with the other two Bursaphelenchus species tested. A previous study reported that the detection limit of the RPA assay is approximately 1.6 fg (1.6 × 10–6 ng) of gDNA from B. xylophilus (using pure gDNA) (Cha et al., 2020). However, in our RPA-LFD assay, a sensitivity test using B. xylophilus gDNA showed that the detection limit was 1 pg. This is probably because the RPA probe-based approach had a lower sensitivity than the approach with labeled primers (Ivanov et al., 2021).
The whole procedure for the RPA-LFD assay could be finished under 38°C within about 30 min, including 15 min for nematode gDNA extraction and master mix preparation, 15 min for the RPA-LFD assay (10 min for RPA reaction, 5 min for visual detection on the LFDs) (Figure 9). Compared with other diagnostic methods for B. xylophilus detection, the time needed for RPA-LFD detection was the shortest. Furthermore, RPA displayed tolerance to interference from pinewood (Figure 7), presenting an opportunity for further investigation into the prevalence of B. xylophilus, especially in new areas of occurrence.
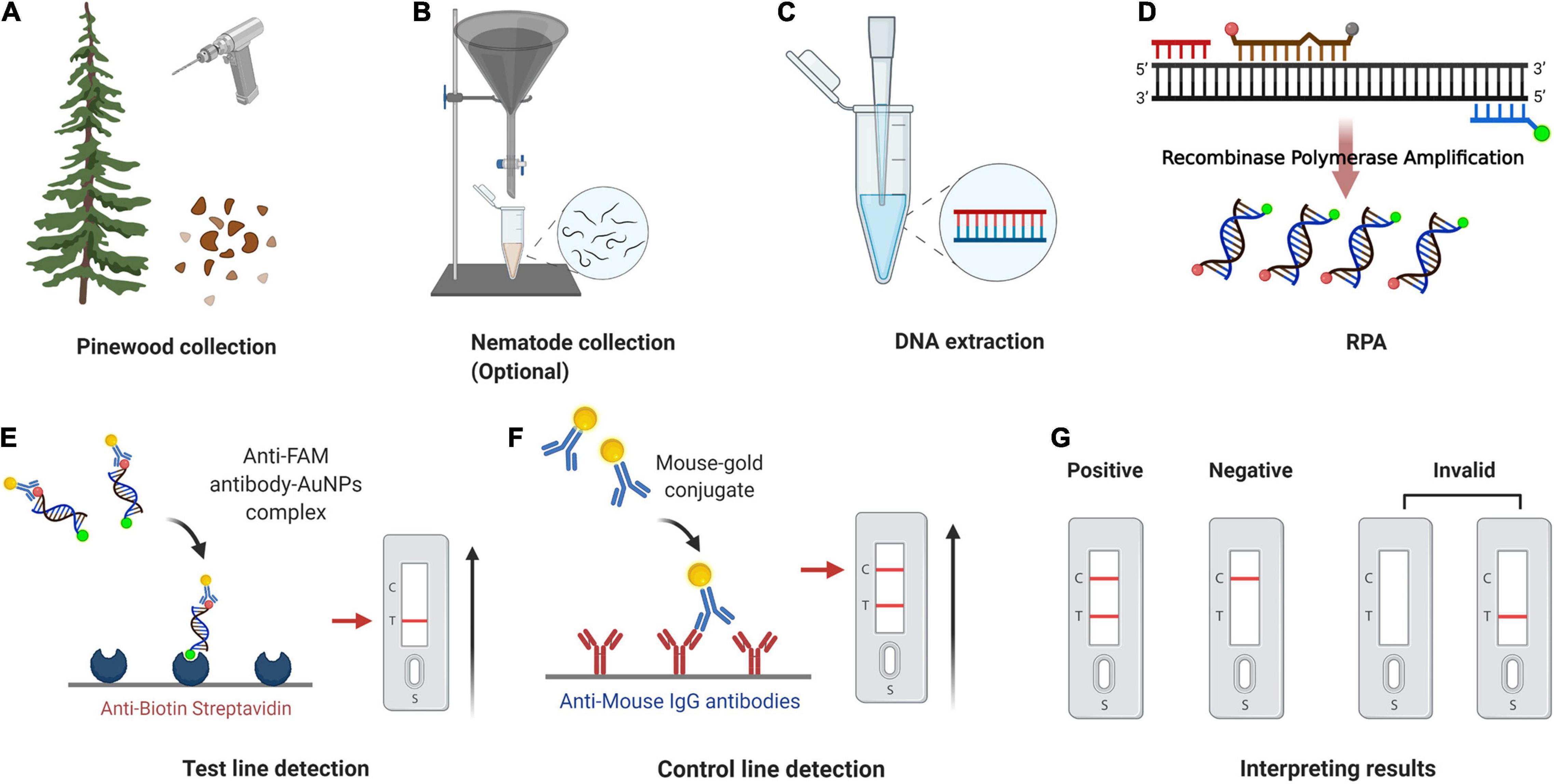
Figure 9. Point-of-Care testing workflow for pinewood nematode detection. (A) Pinewood chips are collected from trees using an electric drill or axe. (B) Nematodes are collected from pinewood chips using Baermann funnel method (optional). (C) gDNA is extracted from nematode/pinewood using DAP lysis buffer. (D) Recombinase polymerase amplification. (E) The sample enters testing well (T) and anti-FAM antibody-AuNPs complex binds to immobilized anti-biotin streptavidin. (F) Mouse antibody-gold conjugate binds to immobilized anti-mouse IgG antibodies. (G) Positive result: one dipstick each in C line and T line; Negative result: one dipstick in C line; Invalid result: no dipstick in C line despite the T line. The whole procedure for the RPA-LFD assay could be finished under 38°C within about 30 min, including 15 min for nematode gDNA extraction and master mix preparation, 15 min for the RPA-LFD assay (10 min for RPA reaction, 5 min for visual detection on the LFDs). Adapted from “COVID-19 Serologic Diagnostic Test through Antibody Detection,” by BioRender.com (2020). Retrieved from https://app.biorender.com/biorender-templates.
However, applying RPA-LFD detection to PWN diagnostics still faces some issues, with the expense being a major one. Reagent costs for RPA-LFD assays are approximately 50 RMB per reaction at present, even when using the RPA nfo kit manufactured locally, which are higher than those for polymerase chain reaction. However, when considering instrumentation costs, the RPA-LFD method becomes relatively more affordable. Even though PCR is currently considered the gold standard for nematode diagnostics, thermocycling instruments are expensive. Local forestry bureaus or quarantine departments are not commonly equipped with these, results usually take days to weeks to obtain due to the shortage of instruments. The minimal equipment needed for conducting RPA-LFD assays would allow reactions to be completed timely in the field or at quarantine departments for point-of-care diagnostics within minutes rather than days.
The relatively low cost of RPA-LFD assay can be used by more people to support extensive surveillance across the country, whether in areas known to have high levels of PWD infection, such as Zhejiang Province, or in areas with relatively low levels of infection regions, supporting decision-making. In addition, detection capability in remote areas with limited resources could drastically increase with the emergency deployment of RPA-LFD assay kits. Thus, the RPA-LFD assay developed here has potential applications in the field and areas with limited resources.
Conclusion
In conclusion, a rapid, on-site RPA-LFD assay for specific and sensitive detection of B. xylophilus was developed. The RPA-LFD assay prevents the risk of false-positive results from primer-dependent artifacts by using a probe and bringing base substitutions in the primer and probe. Furthermore, the assay successfully detected B. xylophilus with high specificity and sensitivity in less than 30 min at an isothermal temperature of 38°C. The developed RPA-LFD assay can provide a novel alternative for PWN point-of-care testing that has a simple read-out system and can be performed under field conditions without any special instrumentation or in areas with minimal laboratory infrastructure.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
QZ conceptualized and designed the research, analyzed the data, interpreted the results, and wrote the manuscript. QZ and YL performed the experiments. QL, YL, ZW, and HW participated in the discussion for experimental design. XZ helped funding acquisition. QL revised the manuscript and directed the project. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from Zhejiang Science and Technology Program (2020C02007) and Fundamental Research Funds of Chinese Academy of Forestry (CAFYBB2021ZG001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Fengmao Chen of Nanjing Forestry University, Zhenyu Liu of Shandong Agricultural University, Laifa Wang and Xizhuo Wang of Chinese Academy of Forestry for their kind support on nematode isolates of Bursaphelenchus species. We also thank Tingting Dai of Nanjing Forestry University for providing guidance on RPA-LFD methodology.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.856109/full#supplementary-material
Supplementary Figure 1 | Nucleotide sequence alignment of the syg-2 gene part sequence from Bursaphelenchus xylophilus (Bx), Bursaphelenchus mucronatus (Bm), and Bursaphelenchus doui (Bd). Location of S1F/R primers and S1-nfo-P are marked by black arrow lines, respectively. Gaps in sequence alignment were indicated by dots.
References
Ahuja, A., and Somvanshi, V. S. (2021). Diagnosis of plant-parasitic nematodes using loop-mediated isothermal amplification (LAMP): a review. Crop Protect. 147:105459. doi: 10.1016/j.cropro.2020.105459
Announcement of the State Forestry and Grassland Administration (No. 6 of 2021) (2021). Announcement of the State Forestry and Grassland Administration (No. 6 of 2021). Available online at: http://www.forestry.gov.cn/main/3457/20210329/152323508644481.html (accessed March 29, 2021).
Brownie, J., Shawcross, S., Theaker, J., Whitcombe, D., Ferrie, R., Newton, C., et al. (1997). The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 25, 3235–3241. doi: 10.1093/nar/25.16.3235
Castagnone, C., Abad, P., and Castagnone-Sereno, P. (2005). Satellite DNA-based species-specific identification of single individuals of the pinewood nematode Bursaphelenchus xylophilus (Nematoda: aphelenchoididae). Eur. J. Plant Pathol. 112, 191–193. doi: 10.1007/s10658-004-0580-2
Cha, D., Kim, D., Choi, W., Park, S., and Han, H. (2020). Point-of-care diagnostic (POCD) method for detecting Bursaphelenchus xylophilus in pinewood using recombinase polymerase amplification (RPA) with the portable optical isothermal device (POID). PLoS One 15:e0227476. doi: 10.1371/journal.pone.0227476
Cha, D., Kim, D., Lee, S., and Han, H. (2019). A new on-site detection method for Bursaphelenchus xylophilus in infected pine trees. For. Pathol. 49:e12503. doi: 10.1111/efp.12503
Chi, Y., Zhao, W., Ye, M., Ali, F., Wang, T., and Qi, R. (2020). Evaluation of recombinase polymerase amplification assay for detecting Meloidogyne Javanica. Plant Dis. 104, 801–807. doi: 10.1094/PDIS-07-19-1473-RE
Daher, R. K., Stewart, G., Boissinot, M., and Bergeron, M. G. (2016). Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 62, 947–958. doi: 10.1373/clinchem.2015.245829
Daher, R. K., Stewart, G., Boissinot, M., Boudreau, D. K., and Bergeron, M. G. (2015). Influence of sequence mismatches on the specificity of recombinase polymerase amplification technology. Mol. Cell. Probes 29, 116–121. doi: 10.1016/j.mcp.2014.11.005
Dai, T., Yang, X., Hu, T., Jiao, B., Xu, Y., Zheng, X., et al. (2019). Comparative evaluation of a novel recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay, LAMP, conventional PCR, and leaf-disc baiting methods for detection of Phytophthora sojae. Front. Microbiol. 10:1884. doi: 10.3389/fmicb.2019.01884
Fang, Y., Wu, X., Lin, Y., Wang, H., Wu, H., and Ju, Y. (2021). Duplex-RPA Detection for Bursaphelenchus xylophilus and Bursaphelenchus mucronatus. Biotechnol. Bull. 37:183. doi: 10.13560/j.cnki.biotech.bull.1985.2021-0579
Futai, K. (2013). Pine wood nematode, Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 51, 61–83. doi: 10.1146/annurev-phyto-081211-172910
Glökler, J., Lim, T. S., Ida, J., and Frohme, M. (2021). Isothermal amplifications–a comprehensive review on current methods. Crit. Rev. Biochem. Mol. Biol. 56, 543–586. doi: 10.1080/10409238.2021.1937927
Gou, D. (2014). A New Method to Extract Genomic DNA of Bursaphelenchus xylophilus from the Pine Wood and the Establishment of the LAMP Detection System. Master’s thesis. Sichuan: Sichuan Agricultural University.
Gu, J. (2014). Identification Techniques of Bursaphelenchus xylophilus and its Approximate Species. Xiamen: Xiamen University Press.
Gu, J., and Wang, J. (2011). Morphological and Molecular Identification of Bursaphelenchus Species. Xiamen: Xiamen University Press.
Gu, J., Wang, J., Braasch, H., Burgermeister, W., and Schröder, T. (2011). Morphological and molecular characterisation of mucronate isolates (‘M’form) of Bursaphelenchus xylophilus (Nematoda: aphelenchoididae). Russian J. Nematol. 19, 103–120.
Hirano, S., Suzuki, S. T., and Redies, C. (2003). The cadherin superfamily in neural development: diversity, function and interaction with other molecules. Front. Biosci. 8:d306–d355. doi: 10.2741/972
Huang, L., Ye, J., Wu, X., Xu, X., Sheng, J., and Zhou, Q. (2010). Detection of the pine wood nematode using a real-time PCR assay to target the DNA topoisomerase I gene. Eur. J. Plant Pathol. 127, 89–98. doi: 10.1007/s10658-009-9574-4
Inácio, M., Nobrega, F., Vieira, P., Bonifacio, L., Naves, P., Sousa, E., et al. (2015). First detection of Bursaphelenchus xylophilus associated with Pinus nigra in Portugal and in Europe. For. Pathol. 45, 235–238. doi: 10.1111/efp.12162
Ivanov, A. V., Safenkova, I. V., Zherdev, A. V., and Dzantiev, B. B. (2021). Recombinase polymerase amplification assay with and without nuclease-dependent-labeled oligonucleotide probe. Int. J. Mol. Sci. 22:11885. doi: 10.3390/ijms222111885
James, A., and Macdonald, J. (2015). Recombinase polymerase amplification: emergence as a critical molecular technology for rapid, low-resource diagnostics. Expert Rev. Mol. Diagnostics 15, 1475–1489. doi: 10.1586/14737159.2015.1090877
Jarvi, S. I., Atkinson, E. S., Kaluna, L. M., Snook, K. A., and Steel, A. (2021). Development of a recombinase polymerase amplification (RPA-EXO) and lateral flow assay (RPA-LFA) based on the ITS1 gene for the detection of Angiostrongylus cantonensis in gastropod intermediate hosts. Parasitology 148, 251–258. doi: 10.1017/S0031182020002139
Ju, Y., Lin, Y., Yang, G., Wu, H., and Pan, Y. (2019). Development of recombinase polymerase amplification assay for rapid detection of Meloidogyne incognita, M. javanica, M. arenaria, and M. enterolobii. Eur. J. Plant Pathol. 155, 1155–1163. doi: 10.1007/s10658-019-01844-6
Kang, J., Kim, A. Y., Han, H., Moon, Y., and Koh, Y. (2015). Development of two alternative Loop-mediated isothermal amplification tools for detecting pathogenic pine wood nematodes. For. Pathol. 45, 127–133. doi: 10.1111/efp.12147
Kang, J. S., Choi, K. S., Shin, S. C., Moon, I. S., Lee, S. G., and Lee, S. H. (2004). Development of an efficient PCR-based diagnosis protocol for the identification of the pinewood nematode, Bursaphelenchus xylophilus (Nematoda: aphelenchoididae). Nematology 6, 279–285. doi: 10.1163/1568541041217915
Kikuchi, T., Aikawa, T., Oeda, Y., Karim, N., and Kanzaki, N. (2009). A rapid and precise diagnostic method for detecting the pinewood nematode Bursaphelenchus xylophilus by loop-mediated isothermal amplification. Phytopathology 99, 1365–1369. doi: 10.1094/PHYTO-99-12-1365
Kim, B.-N., Kim, J. H., Ahn, J.-Y., Kim, S., Cho, B.-K., Kim, Y.-H., et al. (2020). A short review of the pinewood nematode, Bursaphelenchus xylophilus. Toxicol. Environ. Health Sci. 12, 1–8. doi: 10.1007/s13530-020-00068-0
Leal, I., Green, M., Allen, E., Humble, L., and Rott, M. (2007). Application of a real-time PCR method for the detection of pine wood nematode, Bursaphelenchus xylophilus, in wood samples from lodgepole pine. Nematology 9, 351–362. doi: 10.1163/156854107781352098
Lee, J. P., Sekhon, S. S., Kim, J. H., Kim, S. C., Cho, B. K., Ahn, J. Y., et al. (2021). The pine wood nematode Bursaphelenchus xylophilus and molecular diagnostic methods. Mol. Cell. Toxicol. 17, 1–13. doi: 10.1007/s13273-020-00110-9
Li, T. T., Wang, J. L., Zhang, N. Z., Li, W. H., Yan, H. B., Li, L., et al. (2019). Rapid and visual detection of Trichinella spp. using a lateral flow strip-based recombinase polymerase amplification (LF-RPA) assay. Front. Cell. Infect. Microbiol. 9:1. doi: 10.3389/fcimb.2019.00001
Li, Y. L., Fan, C. J., Jiang, X. H., Tian, X. Y., and Han, Z. M. (2021). Bursaphelenchus xylophilus: an important pathogenic factor of pine wilt disease and its relationship with bursaphelenchus mucronatus. Plant Dis. 105, 3055–3062. doi: 10.1094/PDIS-02-21-0396-RE
Liu, C., Hu, Z., Wang, X., Geng, Y., Ma, C., Wang, Z., et al. (2019). The rapid detection of the Bursaphelenchus xylophilus by denaturation bubble-mediated strand exchange amplification. Anal. Sci. 35, 449–453. doi: 10.2116/analsci.18P461
Liu, X., Yan, Q., Huang, J., Chen, J., Guo, Z., Liu, Z., et al. (2019). Influence of design probe and sequence mismatches on the efficiency of fluorescent RPA. World J. Microbiol. Biotechnol. 35, 1–11. doi: 10.1007/s11274-019-2620-2
Meagher, R. J., Priye, A., Light, Y. K., Huang, C., and Wang, E. (2018). Impact of primer dimers and self-amplifying hairpins on reverse transcription loop-mediated isothermal amplification detection of viral RNA. Analyst 143, 1924–1933. doi: 10.1039/C7AN01897E
Meng, F., Wang, X., Wang, L., Gou, D., Liu, H., Wang, Y., et al. (2018). A loop-mediated isothermal amplification-based method for detecting Bursaphelenchus xylophilus from Monochamus alternatus. For. Pathol. 48:e12404. doi: 10.1111/efp.12404
Miao, F., Zhang, J., Li, N., Chen, T., Wang, L., Zhang, F., et al. (2019). Rapid and sensitive recombinase polymerase amplification combined with lateral flow strip for detecting African swine fever virus. Front. Microbiol. 10:1004. doi: 10.3389/fmicb.2019.01004
Mota, M. M., and Vieira, P. (2008). Pine Wilt Disease: A Worldwide Threat to Forest Ecosystems. Berlin: Springer.
Patel, S. K., Pratap, C. B., Jain, A. K., Gulati, A. K., and Nath, G. (2014). Diagnosis of Helicobacter pylori: what should be the gold standard? World J. Gastroenterol.: WJG 20:12847. doi: 10.3748/wjg.v20.i36.12847
Piepenburg, O., Williams, C. H., Stemple, D. L., and Armes, N. A. (2006). DNA detection using recombination proteins. PLoS Biol. 4:e204. doi: 10.1371/journal.pbio.0040204
Poritz, M. A., and Ririe, K. M. (2014). Getting things backwards to prevent primer dimers. J. Mol. Diagnostics 16, 159–162. doi: 10.1016/j.jmoldx.2014.01.001
Qi, Y., Yin, Q., Shao, Y., Li, S., Chen, H., Shen, W., et al. (2018). Rapid and visual detection of Coxiella burnetii using recombinase polymerase amplification combined with lateral flow strips. BioMed Res. Int. 2018:6417354. doi: 10.1155/2018/6417354
Safenkova, I. V., Ivanov, A. V., Slutskaya, E. S., Samokhvalov, A. V., Zherdev, A. V., and Dzantiev, B. B. (2020). Key significance of DNA-target size in lateral flow assay coupled with recombinase polymerase amplification. Anal. Chim. Acta 1102, 109–118. doi: 10.1016/j.aca.2019.12.048
Song, Z., Yang, X., Zhang, X., Luan, M., Guo, B., Liu, C., et al. (2020). Rapid and visual detection of Meloidogyne hapla using recombinase polymerase amplification combined with a lateral flow dipstick (RPA-LFD) assay. Plant Dis. 105, 2697–2703. doi: 10.1094/PDIS-06-20-1345-RE
Subbotin, S. A. (2019). Recombinase polymerase amplification assay for rapid detection of the root-knot nematode Meloidogyne enterolobii. Nematology 21, 243–251. doi: 10.1163/15685411-00003210
Subbotin, S. A., and Burbridge, J. (2021). Sensitive, accurate and rapid detection of the northern root-knot nematode, meloidogyne hapla, using recombinase polymerase amplification assays. Plants 10:336. doi: 10.3390/plants10020336
Sultana, T., Han, H., and Park, J. K. (2013). Comparison of complete mitochondrial genomes of pine wilt nematode Bursaphelenchus xylophilus and Bursaphelenchus mucronatus (Nematoda: aphelenchoidea) and development of a molecular tool for species identification. Gene 520, 39–46. doi: 10.1016/j.gene.2013.02.006
Viglierchio, D., and Schmitt, R. V. (1983). On the methodology of nematode extraction from field samples: baermann funnel modifications. J. Nematol. 15:438.
Wang, J., Gu, J., Chen, X., and Duan, W. (2012). Identification of East Asian and European types of Bursaphelenchus mucronatus by morphological and ITS PCR-RFLP methods. Sci. Silvae Sin. 48, 113–117.
Wang, L., Zhao, P., Si, X., Li, J., Dai, X., Zhang, K., et al. (2020). Rapid and specific detection of Listeria monocytogenes with an isothermal amplification and lateral flow strip combined method that eliminates false-positive signals from primer–dimers. Front. Microbiol. 10:2959. doi: 10.3389/fmicb.2019.02959
Washbourne, P., Dityatev, A., Scheiffele, P., Biederer, T., Weiner, J. A., Christopherson, K. S., et al. (2004). Cell adhesion molecules in synapse formation. J. Neurosci. 24, 9244–9249. doi: 10.1523/JNEUROSCI.3339-04.2004
Wu, H., Zhao, P., Yang, X., Li, J., Zhang, J., Zhang, X., et al. (2020). A recombinase polymerase amplification and lateral flow strip combined method That Detects Salmonella enterica serotype typhimurium with no worry of primer-dependent artifacts. Front. Microbiol. 11:1015. doi: 10.3389/fmicb.2020.01015
Yamagata, M., Sanes, J. R., and Weiner, J. A. (2003). Synaptic adhesion molecules. Curr. Opinion Cell Biol. 15, 621–632. doi: 10.1016/S0955-0674(03)00107-8
Yazaki, K., Takanashi, T., Kanzaki, N., Komatsu, M., Levia, D. F., Kabeya, D., et al. (2018). Pine wilt disease causes cavitation around the resin canals and irrecoverable xylem conduit dysfunction. J. Exp. Bot. 69, 589–602. doi: 10.1093/jxb/erx417
Keywords: pine wilt disease, rapid diagnostic, recombinase polymerase amplification, lateral flow dipstick, Bursaphelenchus xylophilus, POCT, false positive
Citation: Zhou Q, Liu Y, Wang Z, Wang H, Zhang X and Lu Q (2022) Rapid On-Site Detection of the Bursaphelenchus xylophilus Using Recombinase Polymerase Amplification Combined With Lateral Flow Dipstick That Eliminates Interference From Primer-Dependent Artifacts. Front. Plant Sci. 13:856109. doi: 10.3389/fpls.2022.856109
Received: 16 January 2022; Accepted: 17 February 2022;
Published: 18 March 2022.
Edited by:
Anna Filipiak, Institute of Plant Protection – National Research Institute, PolandReviewed by:
Lee Robertson, National Institute of Agricultural and Food Research and Technology, SpainJu-Yeon Yoon, Jeonbuk National University, South Korea
Copyright © 2022 Zhou, Liu, Wang, Wang, Zhang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quan Lu, bHVxdWFuQGNhZi5hYy5jbg==
 Qinzheng Zhou
Qinzheng Zhou Ya Liu
Ya Liu Zheng Wang
Zheng Wang Quan Lu
Quan Lu