- 1Hubei Key Laboratory of Food Crop Germplasm and Genetic Improvement, Institute of Food Crops, Hubei Academy of Agricultural Sciences, Wuhan, China
- 2Henan Institute of Crop Molecular Breeding, Henan Academy of Agricultural Sciences, Zhengzhou, China
- 3College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 4Department of Resources and Environment, Moutai Institute, Renhuai, China
Molybdenum (Mo) is an essential micronutrient for almost all organisms. Wheat, a major staple crop worldwide, is one of the main dietary sources of Mo. However, the genetic basis for the variation of Mo content in wheat grains remains largely unknown. Here, a genome-wide association study (GWAS) was performed on the Mo concentration in the grains of 207 wheat accessions to dissect the genetic basis of Mo accumulation in wheat grains. As a result, 77 SNPs were found to be significantly associated with Mo concentration in wheat grains, among which 52 were detected in at least two sets of data and distributed on chromosome 2A, 7B, and 7D. Moreover, 48 out of the 52 common SNPs were distributed in the 726,761,412–728,132,521 bp genomic region of chromosome 2A. Three putative candidate genes, including molybdate transporter 1;2 (TraesCS2A02G496200), molybdate transporter 1;1 (TraesCS2A02G496700), and molybdopterin biosynthesis protein CNX1 (TraesCS2A02G497200), were identified in this region. These findings provide new insights into the genetic basis for Mo accumulation in wheat grains and important information for further functional characterization and breeding to improve wheat grain quality.
Introduction
As a critical component of enzymes that catalyze key reactions in nitrogen, carbon, and sulfur metabolism, molybdenum (Mo) is an essential micronutrient required for the growth and development of plants and animals (Arnon and Stout, 1939; Turnlund, 2002; Mendel and Bittner, 2006). Higher plants and animals absorb or take in Mo as oxyanion molybdate, which becomes biologically active by binding to pterin to form Mo cofactor (Moco; Mendel and Bittner, 2006; Mendel, 2013). Thereafter, Moco participates in the synthesis of molybdate-dependent enzymes (Molybdoenzymes), including nitrate reductase, sulfite oxidase, xanthine oxidoreductase/dehydrogenase, aldehyde oxidase, and mitochondrial amidoxime reducing component (Hille et al., 2011; Bittner, 2014). To date, a number of molybdate transporters have been identified in plants, including AtMOT1;1 and AtMOT1;2 in Arabidopsis thaliana (Tomatsu et al., 2007; Baxter et al., 2008; Gasber et al., 2011), OsMOT1;1 and OsMOT1;2 in Oryza sativa (Yang et al., 2018; Huang et al., 2019; Ishikawa et al., 2021), FaMOT1 in strawberry (Liu et al., 2020b), LjMOT1 in Lotus japonicus (Gao et al., 2016; Duan et al., 2017), and MtMOT1.2 and MtMOT1.3 in Medicago truncatula (Tejada-Jimenez et al., 2017; Gil-Diez et al., 2019).
Mo deficiency frequently occurs in plants when grown in acidic soils with low Mo bioavailability. Mo-deficient plants generally show overall impaired plant growth and decrease in productivity (Kaiser et al., 2005). Moreover, Mo content in seeds may positively affect seedling vigor in acidic soils (Modi and Cairns, 1994; Modi, 2002; Norton et al., 2014). In humans, Moco deficiency (MoCD) will lead to metabolic defects in molybdoenzymes, giving rise to the accumulation of sulfite, taurine, S-sulfocysteine, and thiosulfate. MoCD is a rare genetic disease, which causes neurological disorders and ultimately early death (Johnson and Duran, 2001; Atwal and Scaglia, 2016). In addition, a molybdenum compound, tetrathiomolybdate, has been clinically used to treat the Wilson’s disease, a genetic disorder of copper metabolism (Brewer et al., 2009).
Wheat (Triticum aestivum L.) is a major staple food crop worldwide, providing ~25% of calories and nutrients in human diet (Zhou et al., 2021b). Food products of wheat grains are one of the primary sources of dietary Mo for adults (Novotny, 2011; Novotny and Peterson, 2018). However, Mo deficiency is a widespread problem of micronutrient deficiency in agriculture. In Australia, Mo deficiency generally occurs in large areas of cropland with acidic soils and has been identified as the second most common micronutrient deficiency after Zinc (Holloway et al., 2008). In China, more than 44.6 million ha of arable land is Mo-deficient (Zou et al., 2008; Nie et al., 2014). Wheat plants grown in these Mo-deficient soils exhibit severe Mo deficiency symptoms such as pale green leaves, chlorosis of seedlings, and lower yields (Yu et al., 2010). Therefore, dissecting the genetic basis and the molecular mechanism for Mo accumulation in wheat grains will greatly help to improve wheat yield and quality.
In previous studies, to understand the genetic mechanism for Mo accumulation in plants, a number of loci and genes related to Mo accumulation have been identified in several species using the genome-wide association study (GWAS) strategy. For example, molybdate transporter 1 (MOT1), which controls the natural variation of Mo concentration in A. thaliana leaves, was identified by GWA mapping (Shen et al., 2012; Forsberg et al., 2015). In rice, Norton et al. (2014) found a significant locus for Mo accumulation in grains on chromosome 8, which contains the MOT1 orthologue. Yang et al. (2018) analyzed the Mo concentration in rice grains by GWAS and demonstrated that variations of Mo concentration in rice grains can be attributed to variable expression of OsMOT1;1. Furthermore, Cobb et al. (2021) also found strong genetic signals for Mo concentration in the shoot in the genomic region of the OsMOT1;1 gene on rice chromosome 8 by GWAS analysis. In wheat, GWAS has been employed to analyze the concentrations of several minerals in the grains, such as zinc (Alomari et al., 2018; Velu et al., 2018; Cu et al., 2020; Zhou et al., 2020), iron (Alomari et al., 2019; Cu et al., 2020), and calcium (Alomari et al., 2017). However, no GWAS has been carried out on grain Mo concentration (GMoC) in wheat to our knowledge.
In this study, by using a panel of 207 wheat accessions, we aimed to: (i) explore the genetic variation of GMoC in wheat; (ii) identify the genomic regions associated with wheat GMoC using the GWAS approach; and (iii) identify the candidate genes for the variations of wheat GMoC. The results of the present study may facilitate the development of wheat varieties with improved nutritional quality.
Materials and Methods
Plant Materials and Growth Conditions
A total of 207 common wheat accessions were used in this study, including both elite cultivars and landraces mainly from China and seven other nations (Supplementary Table 1). These accessions were grown in Yuanyang (35°5′N, 113°97′E), Henan province, a main wheat growing area of China during the 2018–2019 and 2019–2020 cropping seasons in a randomized complete block design. Each genotype was planted in two rows of 2 m length with a 0.3 m row spacing. Field management was conducted based on standard agronomic practices.
Determination of Grain Mo Concentration
At maturity, wheat grains were harvested, dried at 55°C for 24 h, and milled to fine powders. Then, the milled samples were dried at 55°C for another 24 h. After that, 200 mg of dried powder for each sample was digested in 8.0 ml HNO3 in a microwave reactor with a gradient of temperature from 120°C to 180°C for 30 min. After dilution in deionized distilled water, the Mo concentration was measured by inductively coupled plasma mass spectrometry (ICP-MS, NexION 1,000, Perkin Elmer, United States).
Statistical Analysis
The best linear unbiased predictor (BLUP) of Mo concentration for each accession across the 2 years was calculated using the R package lme4 (Bates et al., 2015). The broad-sense heritability (H2) was calculated using the equation , where σ2g is the variance of genotype, σ2e is the variance of environment, and e is the number of years (e = 2 in this study). Pearson’s correlation coefficient (r) for the grain Mo trait across the 2 years was calculated in R version 4.1.1.
SNP Genotyping and GWAS
All wheat accessions were genotyped using the Wheat 660 K SNP array. To avoid spurious SNPs, the SNPs with minor allele frequency (MAF) < 0.05 and missing data >10% were removed. After filtering, a total of 224,706 SNPs were used for GWAS. The population structure and kinship matrix of the association panel were calculated as described in a previous study (Liu et al., 2020a).
GWAS for grain Mo concentration was performed by a mixed-model approach with the FaST-LMM (factored spectrally transformed linear mixed models) program (Lippert et al., 2011). The effective number of SNPs (n = 37,350 in this study) was calculated with the GEC software (Li et al., 2012). Accordingly, the Bonferroni value of p threshold of 2.68E-05 (p = 1/n; n is the effective number of SNPs) was used to determine significant SNPs. R package CMplot1 was used to visualize the Manhattan and quantile-quantile (QQ) plots.
Candidate Gene Identification
To explore the candidate genes responsible for GMoC, genes in the genomic region of 500 Kb upstream and downstream of each significant SNP consistently identified in two or more sets of data were screened based on published slow LD decay in common wheat (Sun et al., 2017; Zhou et al., 2021a). The potential candidate genes were selected based on their annotation information of IWGSC RefSeq v1.1 and functions of homologous genes in Arabidopsis and Oryza sativa. The phylogenetic tree was constructed by using the Neighbour-Joining (NJ) method in MEGA 11 with a bootstrap of 1,000 replicates (Tamura et al., 2021).
Gene Expression Level Analysis
As described in a previous study (Liu et al., 2020a), the grains at 20 days after pollination of each wheat accession were collected for RNA-sequencing. The expression levels (FPKM) of the potential candidate genes were extracted for gene expression level analysis. Moreover, the expression levels of these candidate genes in different tissues were downloaded from the Wheat Expression Brower,2 which was powered by expVIP platform (Borrill et al., 2016; Ramirez-Gonzalez et al., 2018). Heatmap of the expression levels of the candidate genes was visualized by R package pheatmap.
Results
Phenotype Variation of Grain Mo Concentration in Wheat
Two hundred and seven wheat accessions, including elite cultivars and landraces from eight nations representing abundant genetic diversity, were grown at Yuanyang, China in the year of 2019 and 2020 to evaluate the Mo concentration in mature grains (Supplementary Table 1). As a result, continuous and extensive variations in grain Mo concentration were observed among different accessions in the 2 years. As shown in Figure 1A, the GMoC in wheat ranged from 496.00 to 1941.58 ng/g, with an average of 1072.67 ng/g and a coefficient of variation (CV) of 24.05% in 2020 (Table 1 and Figure 1A). In the year of 2019, the GMoC ranged from 293.36 to 1324.41 ng/g, with an average of 729.66 ng/g and a CV of 24.67% (Table 1 and Figure 1A). In addition, the BLUP analysis revealed that the average GMoC was 901.16 ng/g and ranged from 528.13 to 1329.19 ng/g across the 2 years. GMoC in 2 years and the BLUP value all showed continuous variations and approximately normal distributions (Figure 1B; Supplementary Figure 1). In addition, the broad-sense heritability (H2) of GMoC across the 2 years was 0.497, and the Pearson’s correlation coefficient was 0.62 (p < 0.001; Figure 1C).
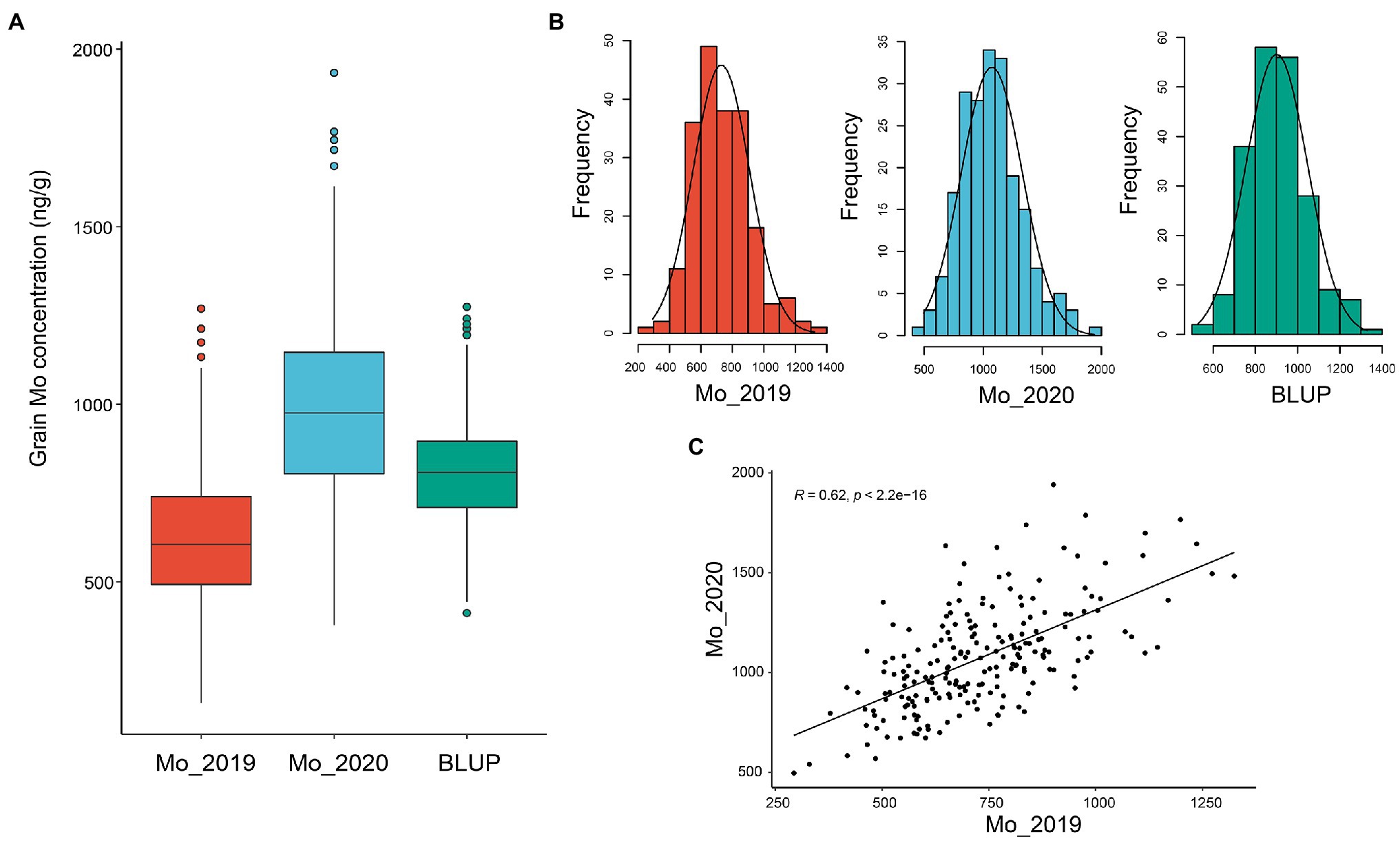
Figure 1. Grain Mo concentration of 207 wheat accessions in 2019 and 2020, and BLUP values. (A) Boxplots of grain Mo concentrations for two single years and BLUP values. (B) Phenotype distribution of grain Mo concentration (ng/g) in the single season (2019 and 2020) and BLUP value. (C) Pearson’s correlation between the two years.
Genome-Wide Association Study of GMoC
The GMoC data of 207 wheat accessions in 2019 (E1), 2020 (E2), and their BLUP values (B) were employed for GWAS with the 224,706 SNPs using the FaST-LMM program. The Bonferroni value of p threshold of 2.68E-05 (−log10 (p) = 4.57) was used to determine significant SNPs. A total of 45, 51 and 71 SNPs were determined to be significantly associated with GMoC in E1, E2, and BLUP, respectively (Figure 2; Supplementary Table 2). Interestingly, 39, 47, and 66 SNPs associated with GMoC in three analyses (E1, E2, and BLUP) were located in the region of 726,761,412–728,409,806 bp on chromosome 2A, which could explain 7.23–15.94% of the phenotypic variation (PVE). The remaining significantly associated SNPs were located on chromosome 7A, 7B, and 7D, explaining 4.46–13.88% of the phenotypic variation. In BLUP dataset, AX-108792390, AX-111609105, and AX-109440948 were the most significant SNPs on chromosome 2A, 7B, and 7D, respectively. The GMoC was significantly different among wheat accessions carrying different homozygous genotypes of these SNPs (Figure 3; Supplementary Table 3).
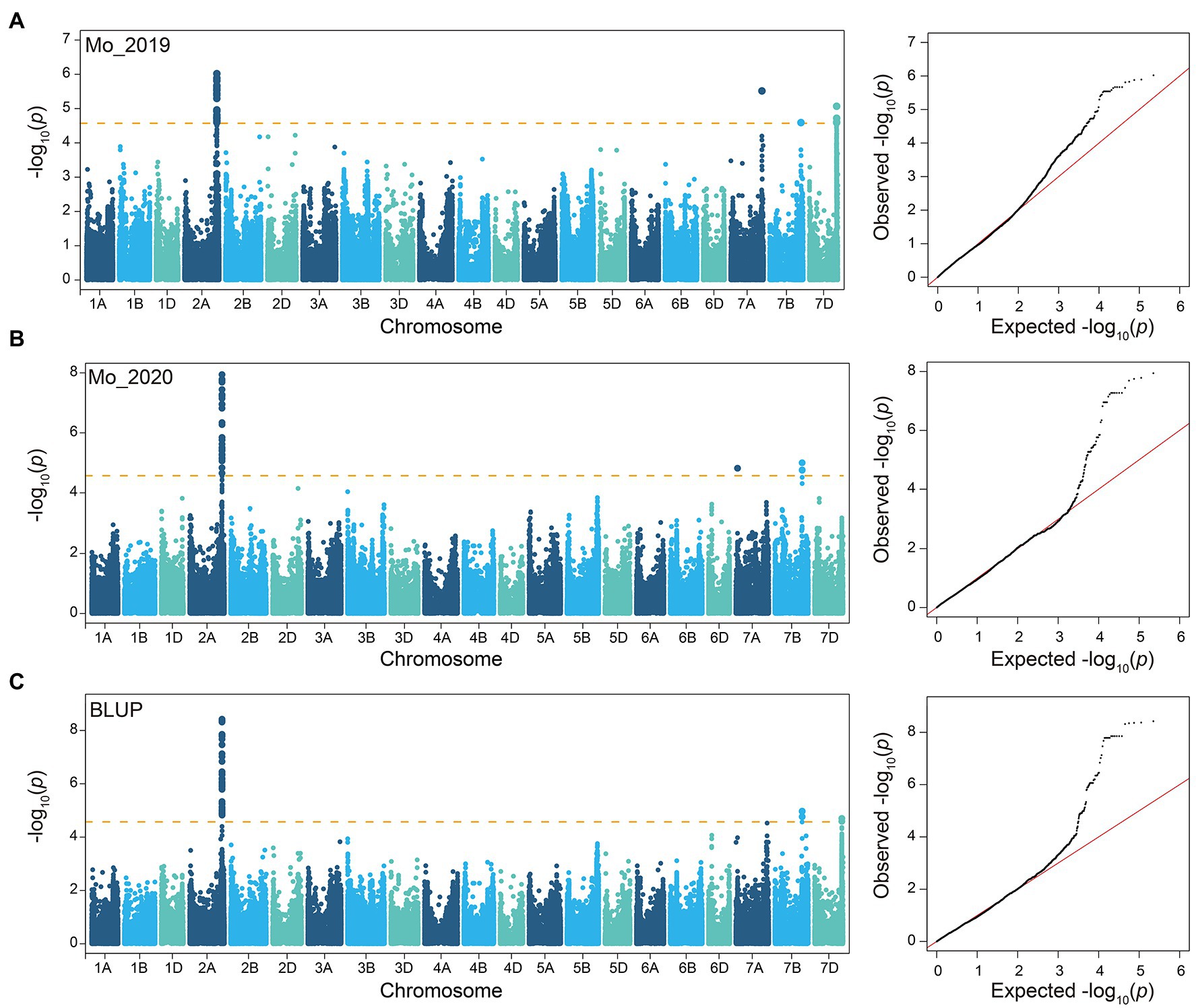
Figure 2. Manhattan plots and QQ plots based on GWAS of GMoC by FaST-LMM model. (A) 2019, (B) 2020, and (C) BLUP values.
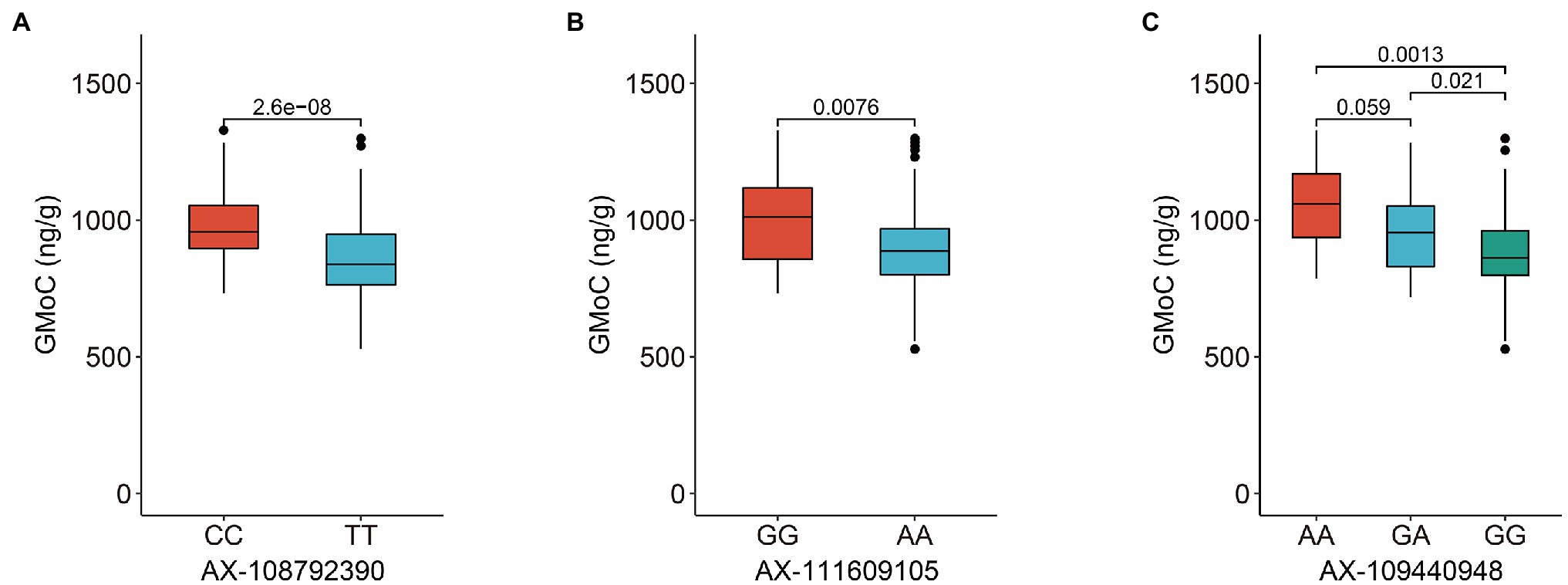
Figure 3. Phenotypic differences in grain Mo concentration of wheat accessions carrying different genotypes of three most significant SNPs on chromosome 2A, 7B, and 7D, respectively. (A) AX-108792390 at the position of Chr2A: 727,180,360 bp. (B) AX-111609105 at the position of Chr 7B: 611,944,380 bp. (C) AX-109440948 at the position of Chr 7D: 611,243,842 bp.
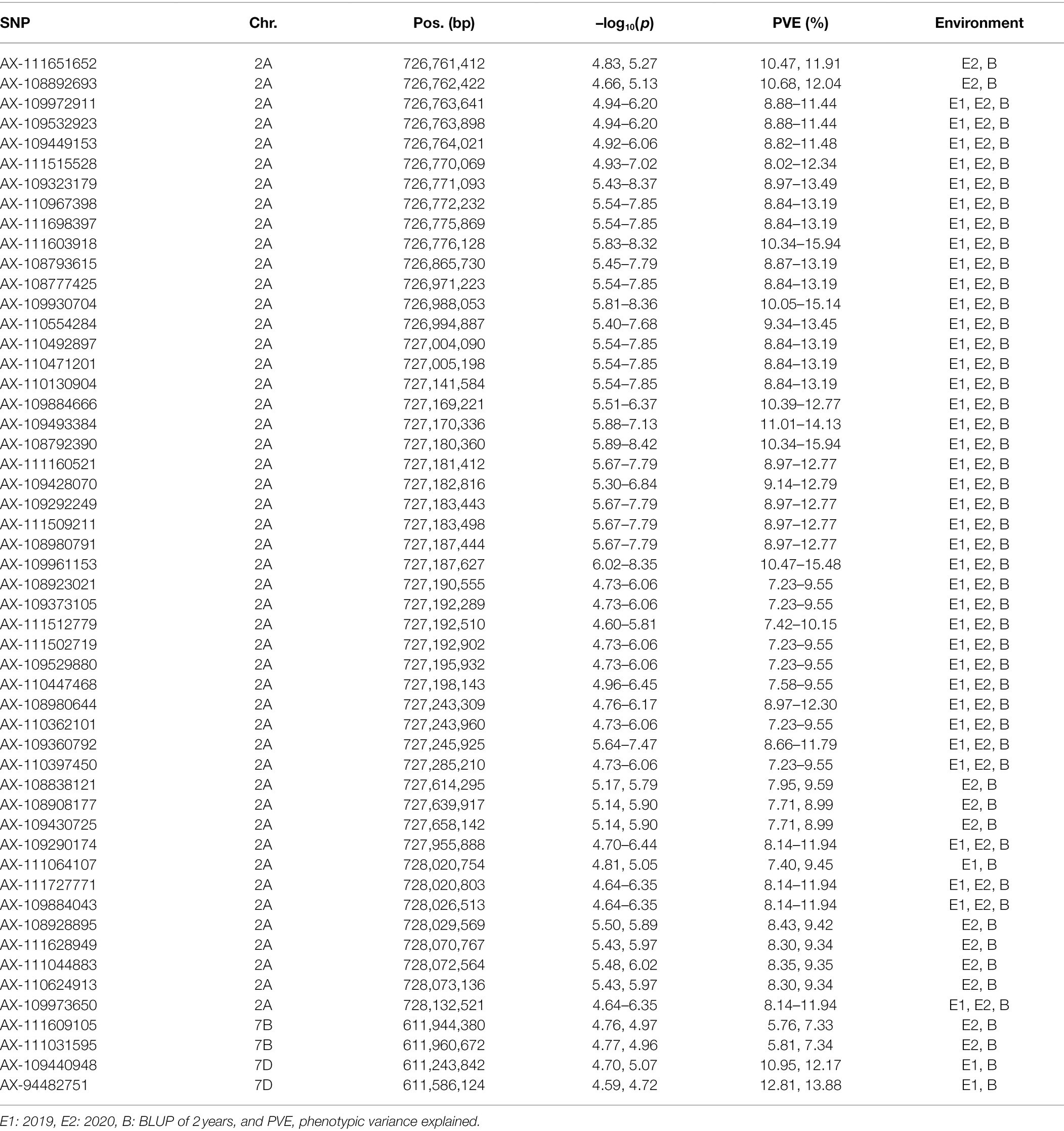
The significant SNPs detected in at least two analyses were defined as common SNPs and used for further exploration of candidate genes. Finally, 52 common SNPs were screened, which were distributed on the chromosome of 2A (48 SNPs), 7B (two SNPs), and 7D (two SNPs) with PVE of 5.76–15.94% (Table 2). The significant SNPs detected in the three sets of data were all located on chromosome 2A. The two common SNPs (AX-111609105 and AX-111031595) on chromosome 7B were detected in E2 and B, while the two common SNPs (AX-109440948 and AX-94482751) on chromosome 7D were identified in E1 and B. The GMoC was significantly different among wheat accessions carrying different genotypes at the 52 SNPs (Figure 3; Supplementary Figure 2).
Prediction of Candidate Genes for Grain Mo Concentration in Wheat
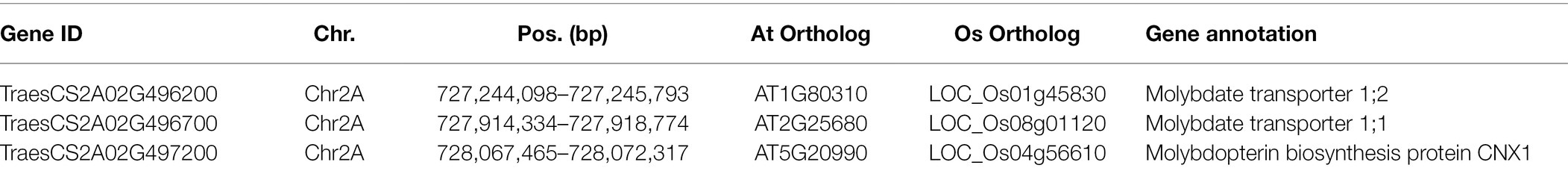
The genomic regions of 500 kb upstream and downstream of the 52 common significant SNPs were defined as candidate regions and used to explore the candidate genes for GMoC. Genes in three candidate regions on chromosomes 2A, 7B, and 7D were screened, which were located in the intervals of 726.26–728.63 Mb (2A), 611.44–612.46 Mb (7B), and 610.74–612.09 Mb (7D). The potential candidate genes were selected based on their annotation information and functions of the homologous genes in Arabidopsis and Oryza sativa. Finally, three potential candidate genes associated with GMoC were screened out, which were all distributed in the candidate region of chromosome 2A, including TraesCS2A02G496200, TraesCS2A02G496700, and TraesCS2A02G497200 at the position of 727,244,098–727,245,793 bp, 727,914,334–727,918,774 bp, and 728,067,465–728,072,317 bp, respectively (Table 3).
The gene TraesCS2A02G496200 encodes a molybdate transporter 1;2 protein associated with GMoC and was only 132 bp from the significant SNP marker AX-109360792 (Chr 2A: 727,245,925 bp). The gene TraesCS2A02G496700 is annotated as a molybdate transporter 1;1 associated with the GMoC and was close (37.1 kb) to the SNP marker AX-109290174 (Chr 2A: 727,955,888 bp). The SNP marker AX-111628949 (Chr 2A: 728,070,767 bp) on chromosome 2A was located in the intron of the TraesCS2A02G497200 gene encoding a molybdopterin biosynthesis protein CNX1, and the SNP marker AX-111044883 (Chr 2A: 728,072,564 bp) was close (247 bp) to the 5′-untranslated region.
We then analyzed the expression levels of these candidate genes in grains at 20 days after pollination in 207 wheat accessions. Surprisingly, TraesCS2A02G496700 showed extremely low expression in grains (Supplementary Figure 3) and exhibited no significant difference in expression among wheat accessions carrying different genotypes of the SNP AX-109290174 (Figure 4B; Supplementary Table 4). In contrast, both TraesCS2A02G496200 (AX-109360792) and TraesCS2A02G497200 (AX-111628949 and AX-111044883) showed significant differences in expression among different genotypes of the closely associated SNPs (Figure 4; Supplementary Table 4). GMoC tended to be consistent with TraesCS2A02G496200 expression at the AX-109360792 site and inverse to TraesCS2A02G497200 expression at the AX-111628949 and AX-111044883 site (Figure 4; Supplementary Figure 2).
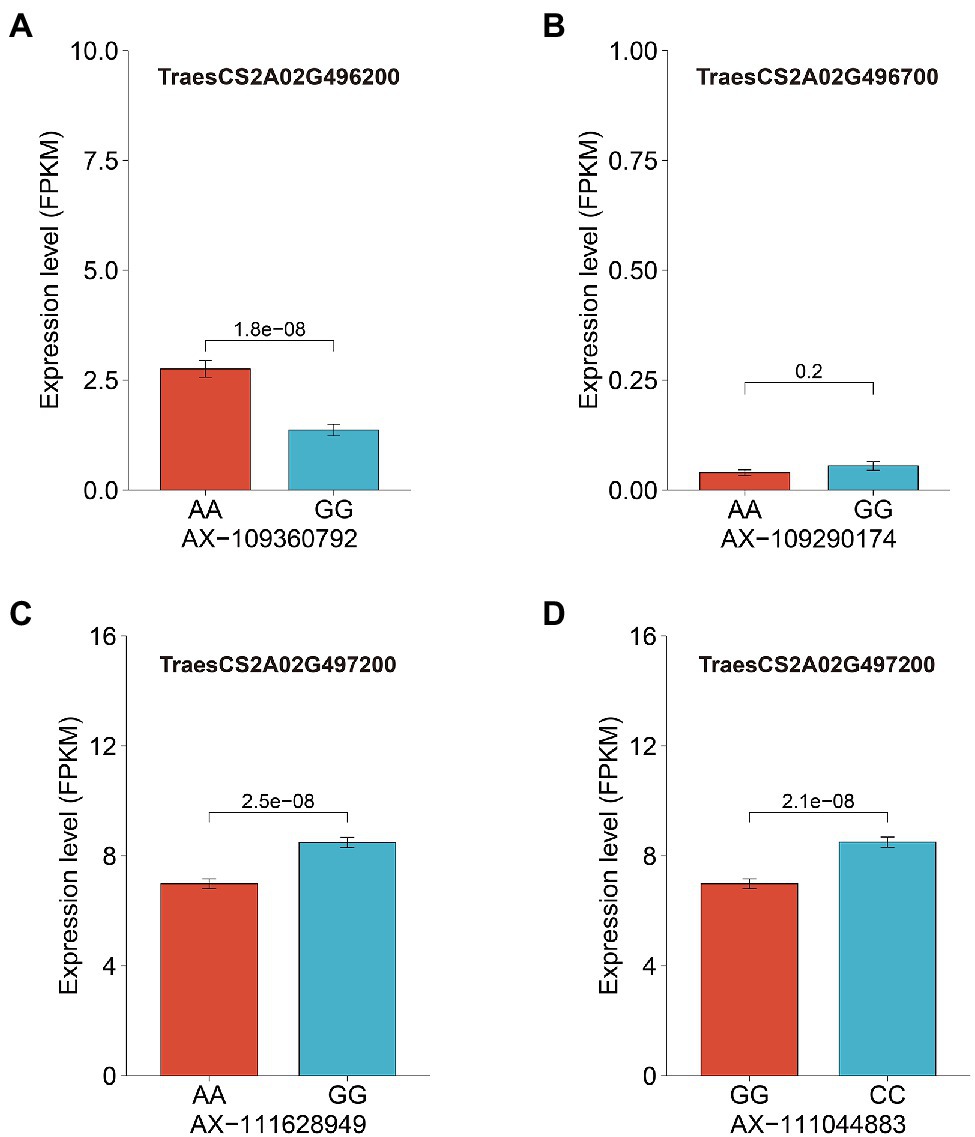
Figure 4. The average expression levels of three candidate genes of wheat accessions with different genotypes of its closely associated SNPs. (A): The expression level of TraesCS2A02G496200 between accessions with different genotypes at SNP AX-109360792; (B): The expression level of TraesCS2A02G496700 between accessions with different genotypes at SNP AX-109290174; and (C,D): The expression level of TraesCS2A02G497200 between accessions with different genotypes at SNPs AX-111628949 and AX-111044883.
Discussion
As an essential element, Mo is indispensable for nearly all living organisms (Mendel and Bittner, 2006). Several genes have been identified to be responsible for natural variations of Mo level in plants (Huang and Salt, 2016; Yang et al., 2018; Huang et al., 2019; Whitt et al., 2020). However, the genetic basis for natural variation of Mo level in wheat is still poorly understood. Thus, we carried out a GWAS analysis on the Mo concentration in grains among 207 wheat accessions grown in two consecutive years to dissect the genetic basis for Mo accumulation in wheat grains.
Wide Variations of Mo Concentration in Wheat Grains
When grown in acidic soils, wheat tends to have low grain yield, quality, and Mo content due to Mo efficiency (Yu et al., 1999; Chatterjee and Nautiyal, 2001; Modi, 2002; Kaiser et al., 2005). Compared with the application of Mo fertilizer, the application of wheat seeds with high Mo concentrations is more economical and environment-friendly to solve the problem of Mo deficiency in acidic soils (Brennan and Bolland, 2007). Therefore, breeding of wheat cultivars with high GMoC may be an effective approach to overcome the Mo deficiency for wheat in acidic soils. Our results showed that GMoC had great variations among the 207 wheat accessions, ranging from 293.36 to 1324.41 ng/g in 2019 and from 496.00 to 1941.58 ng/g in 2020 (Table 1). There was a moderate heritability (H2 = 0.497) between the 2 years, suggesting that GMoC is affected by both genetic and environmental factors. This seems to be consistent with previous findings in Arabidopsis (Baxter et al., 2008). Norton et al. (2014) and Yang et al. (2018) reported an even higher heritability of Mo accumulation in rice grains. Our results revealed that the GMoC between 2019 and 2020 was significantly positively correlated with each other (r = 0.621; p < 0.001), indicating that GMoC is relatively stable across different years and genetic factors are important determinants. Therefore, the wheat varieties with stably high GMoC like Taishan 5, Xinmai 18, and Bainong 160 have the potential to be utilized in future wheat breeding programs.
Identification of Potential Candidate Genes for Grain Mo Concentration in Wheat
The bi-parental QTL mapping in a previous study identified one QTL (Qgmo.tamu.3B.540) associated with GMoC on chromosome 3B (Yu et al., 2021). However, no GMoC-related genes have been identified by GWAS in common wheat so far. In the present study, we identified 52 common SNPs significantly associated with GMoC through GWAS, which are distributed on chromosome 2A, 7B, and 7D. Interestingly, 48 out of the 52 common SNPs were located on chromosome 2A and distributed in the region of 726,761,412–728,132,521 bp. Based on gene functional annotations, TraesCS2A02G496200, TraesCS2A02G496700, and TraesCS2A02G497200 on chromosome 2A were identified as potential candidate genes.
TraesCS2A02G496700 and TraesCS2A02G496200, which are annotated as molybdate transport 1;1 and molybdate transport 1;2, respectively, are two specific molybdate transporters and belong to the Molybdate Transporter 1 (MOT1) family. In eukaryotes, the MOT1 family mediates high-affinity and specific molybdate transport (Tejada-Jimenez et al., 2013). The orthologous genes of MOT1 have been identified in different species, such as Chlamydomonas reinhardtii (Tejada-Jimenez et al., 2007), A. thaliana (Tomatsu et al., 2007; Baxter et al., 2008; Gasber et al., 2011), rice (Norton et al., 2014; Yang et al., 2018; Huang et al., 2019; Wang et al., 2020; Ishikawa et al., 2021), maize (Asaro et al., 2016), strawberry (Liu et al., 2020b), Lotus japonicus (Gao et al., 2016; Duan et al., 2017), and Medicago truncatula (Tejada-Jimenez et al., 2017; Gil-Diez et al., 2019). The phylogenetic tree showed that TraesCS2A02G496700 is the homolog of LOC_Os08g01120 (OsMOT1;1) in rice, while TraesCS2A02G496200 is the homolog of LOC_Os01g45830 (OsMOT1;2) in rice and AT1G80310 (AtMOT1;2) in Arabidopsis (Supplementary Figure 4). MOT1 is responsible for molybdate uptake, translocation, and accumulation (Tomatsu et al., 2007; Baxter et al., 2008; Gasber et al., 2011; Tejada-Jimenez et al., 2013). Yang et al. (2018) attributed the variations of Mo accumulation in rice grains to changes in the expression level of OsMOT1;1. However, TraesCS2A02G496700 showed quite low expression in wheat grains (Supplementary Figure 3), which could be verified by the public transcript data from expVIP platform (Supplementary Figure 5). The public data demonstrate that TraesCS2A02G496700 is highly expressed in roots. Huang et al. (2019) revealed that the expression level of OsMOT1;1 in roots affects the Mo concentration in rice grains. Therefore, TraesCS2A02G496700 is considered as a candidate gene in this study.
In A. thaliana, AtMOT1;2 (formerly named AtMOT2) is a vacuolar molybdate transporter and involved in inter-organ Mo translocation as well as Mo accumulation in seeds (Gasber et al., 2011). Ishikawa et al. (2021) identified OsMOT1;2 as a vacuolar molybdate export protein that plays an important role in inter-organ Mo distribution in rice. The deletion of OsMOT1;2 decreased the grain Mo concentration in the rice mutant osmot1;2. In our study, TraesCS2A02G496200 is considered as the ortholog of MOT1;2 in wheat (Table 3; Supplementary Figure 4). At the AX-109360792 locus (Chr 2A: 727,245,925 bp; close to the upstream of TraesCS2A02G496200), the accessions with the AA genotype had both significantly higher GMoC and TraesCS2A02G496200 expression than those with the GG genotype (Figure 4A; Supplementary Figure 2), suggesting that it is a candidate gene of GMoC. However, the expression of TraesCS2A02G496200 was the highest in leaves and shoots, followed by roots, while the lowest in grains (Supplementary Figure 5). This is similar to the expression pattern of OsMOT1;2 in rice and AtMOT1;2 in A. thaliana, suggesting its role in inter-organ Mo translocation and distribution (Gasber et al., 2011; Ishikawa et al., 2021). Therefore, it is of great significance to explore the molecular mechanism for the regulatory effect of these two candidate genes (TraesCS2A02G496700 and TraesCS2A02G496200) on Mo accumulation in wheat grains in future studies.
The third potential candidate gene TraesCS2A02G497200 is annotated as molybdopterin biosynthesis protein CNX1 (cofactor for nitrate reductase and xanthine dehydrogenase 1), which is involved in Moco biosynthesis in plants by inserting Mo into molybdopterin (Schwarz et al., 2000; Kuper et al., 2003; Llamas et al., 2004, 2006; Tejada-Jimenez et al., 2018). Significant differences in phenotype and expression were observed for the genotypes at two SNPs (AX-111628949 and AX-111044883) closely associated with TraesCS2A02G497200. Interestingly, GMoC tended to have an inverse relationship with the gene expression at the two sites (Figure 4; Supplementary Figure 2). This phenomenon may be attributed to the conversion of more molybdate into Moco under the catalysis of CNX1, which leads to a decrease in GMoC. CNX1 has been found to be constitutively expressed in all organs of Arabidopsis plants (Schwarz et al., 2000). Similarly, the expression of TraesCS2A02G497200 was also found in almost all organs of wheat plants and was higher in roots than in grains (Supplementary Figure 5). This might be due to a large amount of Mo absorbed by roots from the soil, which is then used for MoCo synthesis. Thus, the above-mentioned three genes are considered as potential candidate genes for GMoC in wheat, which may be utilized in wheat breeding. However, further functional studies are required to verify their functions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZfZ, ZL, and ZW designed the experiment. CL conducted the field experiment. ZjZ and YP investigated the phenotype and revised and edited the manuscript. XJ and SY conducted the GWAS analysis. XJ and YP drafted the manuscript. All authors have read and approved the final manuscript.
Funding
This research was funded by National Natural Science Foundation of China (31701506), the key area projects of Guizhou education department (KY [2020]044), the Agriculture Research System of Henan Province (S2010-01), the Science-Technology Foundation for Excellent Youth Scholars of Henan Academy of Agricultural Sciences (2020YQ02), and Special fund projects of science and technology development of Henan Academy of Agricultural Sciences (YNK20177511).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.854966/full#supplementary-material
Footnotes
References
Alomari, D. Z., Eggert, K., von Wiren, N., Alqudah, A. M., Polley, A., Plieske, J., et al. (2018). Identifying candidate genes for enhancing grain Zn concentration in wheat. Front. Plant Sci. 9:1313. doi: 10.3389/fpls.2018.01313
Alomari, D. Z., Eggert, K., von Wiren, N., Pillen, K., and Roder, M. S. (2017). Genome-wide association study of calcium accumulation in grains of european wheat cultivars. Front. Plant Sci. 8:1797. doi: 10.3389/fpls.2017.01797
Alomari, D. Z., Eggert, K., von Wiren, N., Polley, A., Plieske, J., Ganal, M. W., et al. (2019). Whole-genome association mapping and genomic prediction for iron concentration in wheat grains. Int. J. Mol. Sci. 20:76. doi: 10.3390/ijms20010076
Arnon, D. I., and Stout, P. R. (1939). Molybdenum as an essential element for higher plants. Plant Physiol. 14, 599–602. doi: 10.1104/pp.14.3.599
Asaro, A., Ziegler, G., Ziyomo, C., Hoekenga, O. A., Dilkes, B. P., and Baxter, I. (2016). The interaction of genotype and environment determines variation in the maize kernel ionome. G3-genes Genom. Genet. 6, 4175–4183. doi: 10.1534/g3.116.034827
Atwal, P. S., and Scaglia, F. (2016). Molybdenum cofactor deficiency. Mol. Genet. Metab. 117, 1–4. doi: 10.1016/j.ymgme.2015.11.010
Bates, D., Machler, M., Bolker, B., and Walker, S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. doi: 10.18637/jss.v067.i01
Baxter, I., Muthukumar, B., Park, H. C., Buchner, P., Lahner, B., Danku, J., et al. (2008). Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1). PLoS Genet. 4:e1000004. doi: 10.1371/journal.pgen.1000004
Bittner, F. (2014). Molybdenum metabolism in plants and crosstalk to iron. Front. Plant Sci. 5:28. doi: 10.3389/fpls.2014.00028
Borrill, P., Ramirez-Gonzalez, R., and Uauy, C. (2016). expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol. 170, 2172–2186. doi: 10.1104/pp.15.01667
Brennan, R. F., and Bolland, M. D. A. (2007). Increased concentration of molybdenum in sown wheat seed decreases grain yield responses to applied molybdenum fertilizer in naturally acidic sandplain soils. J. Plant Nutr. 30, 2005–2019. doi: 10.1080/01904160701700442
Brewer, G. J., Askari, F., Dick, R. B., Sitterly, J., Fink, J. K., Carlson, M., et al. (2009). Treatment of Wilson's disease with tetrathiomolybdate: V. control of free copper by tetrathiomolybdate and a comparison with trientine. Transl. Res. 154, 70–77. doi: 10.1016/j.trsl.2009.05.002
Chatterjee, C., and Nautiyal, N. (2001). Molybdenum stress affects viability and vigor of wheat seeds. J. Plant Nutr. 24, 1377–1386. doi: 10.1081/PLN-100106988
Cobb, J. N., Chen, C., Shi, Y., Maron, L. G., Liu, D., Rutzke, M., et al. (2021). Genetic architecture of root and shoot ionomes in rice (Oryza sativa L.). Theor. Appl. Genet. 134, 2613–2637. doi: 10.1007/s00122-021-03848-5
Cu, S. T., Guild, G., Nicolson, A., Velu, G., Singh, R., and Stangoulis, J. (2020). Genetic dissection of zinc, iron, copper, manganese and phosphorus in wheat (Triticum aestivum L.) grain and rachis at two developmental stages. Plant Sci. 291:110338. doi: 10.1016/j.plantsci.2019.110338
Duan, G., Hakoyama, T., Kamiya, T., Miwa, H., Lombardo, F., Sato, S., et al. (2017). LjMOT1, a high-affinity molybdate transporter from Lotus japonicus, is essential for molybdate uptake, but not for the delivery to nodules. Plant J. 90, 1108–1119. doi: 10.1111/tpj.13532
Forsberg, S. K., Andreatta, M. E., Huang, X. Y., Danku, J., Salt, D. E., and Carlborg, O. (2015). The multi-allelic genetic architecture of a variance-heterogeneity locus for molybdenum concentration in leaves acts as a source of unexplained additive genetic variance. PLoS Genet. 11:e1005648. doi: 10.1371/journal.pgen.1005648
Gao, J. S., Wu, F. F., Shen, Z. L., Meng, Y., Cai, Y. P., and Lin, Y. (2016). A putative molybdate transporter LjMOT1 is required for molybdenum transport in Lotus japonicus. Physiol. Plant. 158, 331–340. doi: 10.1111/ppl.12489
Gasber, A., Klaumann, S., Trentmann, O., Trampczynska, A., Clemens, S., Schneider, S., et al. (2011). Identification of an Arabidopsis solute carrier critical for intracellular transport and inter-organ allocation of molybdate. Plant Biol. 13, 710–718. doi: 10.1111/j.1438-8677.2011.00448.x
Gil-Diez, P., Tejada-Jimenez, M., Leon-Mediavilla, J., Wen, J., Mysore, K. S., Imperial, J., et al. (2019). MtMOT1.2 is responsible for molybdate supply to Medicago truncatula nodules. Plant Cell Environ. 42, 310–320. doi: 10.1111/pce.13388
Hille, R., Nishino, T., and Bittner, F. (2011). Molybdenum enzymes in higher organisms. Coord. Chem. Rev. 255, 1179–1205. doi: 10.1016/j.ccr.2010.11.034
Holloway, R. E., Graham, R. D., and Stacey, S. P. (2008). “Micronutrient deficiencies in australian field crops” in Micronutrient Deficiencies in Global Crop Production. ed. B. J. Alloway (Dordrecht: Springer), 63–92.
Huang, X. Y., Liu, H., Zhu, Y. F., Pinson, S. R. M., Lin, H. X., Guerinot, M. L., et al. (2019). Natural variation in a molybdate transporter controls grain molybdenum concentration in rice. New Phytol. 221, 1983–1997. doi: 10.1111/nph.15546
Huang, X. Y., and Salt, D. E. (2016). Plant ionomics: from elemental profiling to environmental adaptation. Mol. Plant 9, 787–797. doi: 10.1016/j.molp.2016.05.003
Ishikawa, A. S., Hayashi, S., Tanikawa, H., Iino, M., Abe, T., Kuramata, M., et al. (2021). Tonoplast-localized OsMOT1;2 participates in interorgan molybdate distribution in rice. Plant Cell Physiol. 62, 913–921. doi: 10.1093/pcp/pcab050
Johnson, J. L., and Duran, M. (2001). “Molybdenum cofactor deficiency and isolated sulfite oxidase deficiency” in The Metabolic and Molecular Bases of Inherited Disease. eds. C. Scriver, A. Beaudet, W. Sly, and D. Valle (New York: McGraw-Hill), 3163–3177.
Kaiser, B. N., Gridley, K. L., Ngaire Brady, J., Phillips, T., and Tyerman, S. D. (2005). The role of molybdenum in agricultural plant production. Ann. Bot. 96, 745–754. doi: 10.1093/aob/mci226
Kuper, J., Winking, J., Hecht, H.-J., Mendel, R. R., and Schwarz, G. (2003). The active site of the molybdenum cofactor biosynthetic protein domain Cnx1G. Arch. Biochem. Biophys. 411, 36–46. doi: 10.1016/S0003-9861(02)00714-2
Li, M. X., Yeung, J. M., Cherny, S. S., and Sham, P. C. (2012). Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 131, 747–756. doi: 10.1007/s00439-011-1118-2
Lippert, C., Listgarten, J., Liu, Y., Kadie, C. M., Davidson, R. I., and Heckerman, D. (2011). FaST linear mixed models for genome-wide association studies. Nat. Methods 8, 833–835. doi: 10.1038/nmeth.1681
Liu, L., Shi, H., Li, S., Sun, M., Zhang, R., Wang, Y., et al. (2020b). Integrated analysis of molybdenum nutrition and nitrate metabolism in strawberry. Front. Plant Sci. 11:1117. doi: 10.3389/fpls.2020.01117
Liu, C., Zhou, Z., Wu, Z., Qin, M., Shi, X., Wang, Y., et al. (2020a). In-depth genetic analysis reveals conditioning of polyphenol oxidase activity in wheat grains by cis regulation of TaPPO2A-1 expression level. Genomics 112, 4690–4700. doi: 10.1016/j.ygeno.2020.08.019
Llamas, A., Mendel, R. R., and Schwarz, G. (2004). Synthesis of adenylated molybdopterin: an essential step for molybdenum insertion. J. Biol. Chem. 279, 55241–55246. doi: 10.1074/jbc.M409862200
Llamas, A., Otte, T., Multhaup, G., Mendel, R. R., and Schwarz, G. (2006). The mechanism of nucleotide-assisted molybdenum insertion into molybdopterin: a novel route toward metal cofactor assembly. J. Biol. Chem. 281, 18343–18350. doi: 10.1074/jbc.M601415200
Mendel, R. R. (2013). The molybdenum cofactor. J. Biol. Chem. 288, 13165–13172. doi: 10.1074/jbc.R113.455311
Mendel, R. R., and Bittner, F. (2006). Cell biology of molybdenum. Biochim. Biophys. Acta 1763, 621–635. doi: 10.1016/j.bbamcr.2006.03.013
Modi, A. T. (2002). Wheat seed quality in response to molybdenum and phosphorus. J. Plant Nutr. 25, 2409–2419. doi: 10.1081/PLN-120014703
Modi, A. T., and Cairns, A. L. P. (1994). Molybdenum deficiency in wheat results in lower dormancy levels via reduced ABA. Seed Sci. Res. 4, 329–333. doi: 10.1017/S0960258500002373
Nie, Z., Hu, C., Liu, H., Tan, Q., and Sun, X. (2014). Differential expression of molybdenum transport and assimilation genes between two winter wheat cultivars (Triticum aestivum). Plant Physiol. Biochem. 82, 27–33. doi: 10.1016/j.plaphy.2014.05.002
Norton, G. J., Douglas, A., Lahner, B., Yakubova, E., Guerinot, M. L., Pinson, S. R., et al. (2014). Genome wide association mapping of grain arsenic, copper, molybdenum and zinc in rice (Oryza sativa L.) grown at four international field sites. PLoS One 9:e89685. doi: 10.1371/journal.pone.0089685
Novotny, J. A. (2011). Molybdenum nutriture in humans. J. Evid. Based Comp. Altern. Med. 16, 164–168. doi: 10.1177/2156587211406732
Novotny, J. A., and Peterson, C. A. (2018). Molybdenum. Adv. Nutr. 9, 272–273. doi: 10.1093/advances/nmx0001
Ramirez-Gonzalez, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361:eaar6089. doi: 10.1126/science.aar6089
Schwarz, G., Schulze, J., Bittner, F., Eilers, T., Kuper, J., Bollmann, G., et al. (2000). The molybdenum cofactor biosynthetic protein Cnx1 complements molybdate-repairable mutants, transfers molybdenum to the metal binding pterin, and is associated with the cytoskeleton. Plant Cell 12:2455. doi: 10.1105/tpc.12.12.2455
Shen, X., Pettersson, M., Ronnegard, L., and Carlborg, O. (2012). Inheritance beyond plain heritability: variance-controlling genes in Arabidopsis thaliana. PLoS Genet. 8:e1002839. doi: 10.1371/journal.pgen.1002839
Sun, C., Zhang, F., Yan, X., Zhang, X., Dong, Z., Cui, D., et al. (2017). Genome-wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the yellow and huai valley of China. Plant Biotechnol. J. 15, 953–969. doi: 10.1111/pbi.12690
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tejada-Jimenez, M., Chamizo-Ampudia, A., Calatrava, V., Galvan, A., Fernandez, E., and Llamas, A. (2018). From the eukaryotic molybdenum cofactor biosynthesis to the moonlighting enzyme mARC. Molecules 23:3287. doi: 10.3390/molecules23123287
Tejada-Jimenez, M., Chamizo-Ampudia, A., Galvan, A., Fernandez, E., and Llamas, A. (2013). Molybdenum metabolism in plants. Metallomics 5, 1191–1203. doi: 10.1039/c3mt00078h
Tejada-Jimenez, M., Gil-Diez, P., Leon-Mediavilla, J., Wen, J., Mysore, K. S., Imperial, J., et al. (2017). Medicago truncatula Molybdate transporter type 1 (MtMOT1.3) is a plasma membrane molybdenum transporter required for nitrogenase activity in root nodules under molybdenum deficiency. New Phytol. 216, 1223–1235. doi: 10.1111/nph.14739
Tejada-Jimenez, M., Llamas, A., Sanz-Luque, E., Galvan, A., and Fernandez, E. (2007). A high-affinity molybdate transporter in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 104, 20126–20130. doi: 10.1073/pnas.0704646104
Tomatsu, H., Takano, J., Takahashi, H., Watanabe-Takahashi, A., Shibagaki, N., and Fujiwara, T. (2007). An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proc. Natl. Acad. Sci. U. S. A. 104, 18807–18812. doi: 10.1073/pnas.0706373104
Turnlund, J. R. (2002). Molybdenum metabolism and requirements in humans. Met. Ions Biol. Syst. 39, 727–739.
Velu, G., Singh, R. P., Crespo-Herrera, L., Juliana, P., Dreisigacker, S., Valluru, R., et al. (2018). Genetic dissection of grain zinc concentration in spring wheat for mainstreaming biofortification in CIMMYT wheat breeding. Sci. Rep. 8:13526. doi: 10.1038/s41598-018-31951-z
Wang, C., Tang, Z., Zhuang, J.-Y., Tang, Z., Huang, X.-Y., and Zhao, F.-J. (2020). Genetic mapping of ionomic quantitative trait loci in rice grain and straw reveals OsMOT1;1 as the putative causal gene for a molybdenum QTL qMo8. Mol. Gen. Genomics. 295, 391–407. doi: 10.1007/s00438-019-01632-1
Whitt, L., Ricachenevsky, F. K., Ziegler, G. Z., Clemens, S., Walker, E., Maathuis, F. J. M., et al. (2020). A curated list of genes that affect the plant ionome. Plant Direct. 4:e00272. doi: 10.1002/pld3.272
Yang, M., Lu, K., Zhao, F. J., Xie, W., Ramakrishna, P., Wang, G., et al. (2018). Genome-wide association studies reveal the genetic basis of ionomic variation in rice. Plant Cell 30, 2720–2740. doi: 10.1105/tpc.18.00375
Yu, S., Assanga, S. O., Awika, J. M., Ibrahim, A. M. H., Rudd, J. C., Xue, Q., et al. (2021). Genetic mapping of quantitative trait loci for end-use quality and grain minerals in hard red winter wheat. Agronomy 11:2519. doi: 10.3390/agronomy11122519
Yu, M., Hu, C., Sun, X., and Wang, Y. (2010). Influences of Mo on nitrate reductase, glutamine synthetase and nitrogen accumulation and utilization in mo-efficient and mo-inefficient winter wheat cultivars. Agr. Sci. China 9, 355–361. doi: 10.1016/S1671-2927(09)60104-8
Yu, M., Hu, C., and Wang, Y. (1999). Influences of seed molybdenum and molybdenum application on nitrate reductase activity, shoot dry matter, and grain yields of winter wheat cultivars. J. Plant Nutr. 22, 1433–1441. doi: 10.1080/01904169909365724
Zhou, Z., Guan, H., Liu, C., Zhang, Z., Geng, S., Qin, M., et al. (2021a). Identification of genomic regions affecting grain peroxidase activity in bread wheat using genome-wide association study. BMC Plant Biol. 21:523. doi: 10.1186/s12870-021-03299-6
Zhou, Z., Liu, C., Qin, M., Li, W., Hou, J., Shi, X., et al. (2021b). Promoter DNA hypermethylation of TaGli-γ-2.1 positively regulates gluten strength in bread wheat. J. Adv. Res. 36, 163–173. doi: 10.1016/j.jare.2021.06.021
Zhou, Z., Shi, X., Zhao, G., Qin, M., Ibba, M. I., Wang, Y., et al. (2020). Identification of novel genomic regions and superior alleles associated with Zn accumulation in wheat using a genome-wide association analysis method. Int. J. Mol. Sci. 21:1928. doi: 10.3390/ijms21061928
Keywords: common wheat, molybdenum, GWAS, SNP, candidate genes
Citation: Jin X, Zou Z, Wu Z, Liu C, Yan S, Peng Y, Lei Z and Zhou Z (2022) Genome-Wide Association Study Reveals Genomic Regions Associated With Molybdenum Accumulation in Wheat Grains. Front. Plant Sci. 13:854966. doi: 10.3389/fpls.2022.854966
Edited by:
Peter Michael Dracatos, The University of Sydney, AustraliaReviewed by:
Xin-Yuan Huang, Nanjing Agricultural University, ChinaShengguan Cai, Zhejiang University, China
Copyright © 2022 Jin, Zou, Wu, Liu, Yan, Peng, Lei and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanchun Peng, eWFuY2h1bnBlbmdAbWFpbC5oemF1LmVkdS5jbg==; Zhensheng Lei, emhlbnNoZW5nbGVpQDEyNi5jb20=; Zhengfu Zhou, emhvdXpmMjE1QDE2My5jb20=
 Xiaojie Jin
Xiaojie Jin Zhaojun Zou3
Zhaojun Zou3 Congcong Liu
Congcong Liu Songxian Yan
Songxian Yan Yanchun Peng
Yanchun Peng Zhengfu Zhou
Zhengfu Zhou