- State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Nanjing University, Nanjing, China
Several angiosperm GRETCHEN HAGEN 3 (GH3) genes, including tomato SlGH3.4 and rice OsGH3.2 are induced during arbuscular mycorrhizal (AM) symbiosis, but their functions remain largely unclear. Recently, tomato SlGH3.4 was suggested to negatively regulate arbuscule incidence via decreasing auxin levels in colonized cells. In this study, by acquiring rice OsGH3.2pro:β-glucuronidase (GUS) transgenic plants and generating Osgh3.2 mutants via CRISPR/Cas9 technique, the roles of OsGH3.2 in modulating rice root morphology and affecting AM symbiosis were investigated through time course experiments. Unlike SlGH3.4, OsGH3.2 showed asymbiotic expression in rice young lateral roots, and its mutation resulted in a “shallow” root architecture. Such root morphological change was also observed under symbiotic condition and it likely promoted AM fungal colonization, as the mutants exhibited higher colonization levels and arbuscule incidence than wild-type at early stages. Similar to SlGH3.4, OsGH3.2 showed symbiotic expression in cortical cells that have formed mature arbuscules. At late stages of symbiosis, Osgh3.2 mutants showed elongated cortical cells and larger arbuscules than wild-type, indicating elevated auxin level in the colonized cells. Together, these results revealed both asymbiotic and symbiotic roles of OsGH3.2 in modulating rice root architecture and controlling auxin levels in arbusculated cells, which further affected colonization rate and arbuscule phenotype.
Introduction
Arbuscular mycorrhiza (AM) is a symbiosis formed between most land plants from bryophytes to angiosperms and soil fungi of the subphylum Glomeromycotina (Wang and Qiu, 2006; Smith and Read, 2008; Spatafora et al., 2016). Its establishment involves a series of complex and well-coordinated events. Before physical contact, plant roots secrete a variety of compounds to trigger nearby AM fungi (AMF) for hyphae elongation and branching (Akiyama et al., 2005; Nagahashi and Douds, 2011), while AMF also produce chitinaceous signals to pre-activate root cells (Maillet et al., 2011; Genre et al., 2013). After touching the root surface, AMF form adhesion structures called hyphopodia, then invade the roots via pre-penetration apparatuses (PPA), a tunnel structure assembled by plants (Genre et al., 2005, 2008, 2012). By reaching the inner cortex, fungal hyphae spread longitudinally and penetrate inside cortical cells, developing highly branched, bush-like structures called arbuscules (Genre et al., 2008). Arbuscules are surrounded by a plant-derived plasma membrane called the peri-arbuscular membrane (PAM). Through the interface of PAM and arbuscules, nutrient exchanges occur between symbiotic partners, where plants provide organic carbon to fungi and fungi supply mineral nutrients, especially phosphate and nitrate, to plants (Smith and Smith, 2011; Wang et al., 2017, 2020; Wipf et al., 2019).
As key regulators of plant physiological and developmental processes, many phytohormones have been found to play roles in AM symbiosis (Foo et al., 2013; Gutjahr, 2014; Pozo et al., 2015). Strigolactones (SLs), a class of carotenoid-derived hormone, are secreted by plant roots to promote hyphae branching and to accelerate AMF colonization (Akiyama et al., 2005; Gomez-Roldan et al., 2008); while salicylic acid (SA), a defense-related phytohormone, shows an antagonistic delay effect on AMF penetration and colonization (Herrera-Medina et al., 2003). Auxins, abscisic acid (ABA) and jasmonic acid (JA) have been all found to positively regulate arbuscule development, while gibberellins (GA) conversely inhibit arbuscule formation (Isayenkov et al., 2005; Herrera-Medina et al., 2007; Foo et al., 2013; Etemadi et al., 2014; Pozo et al., 2015). Large knowledge gaps still remain for the detailed regulation mechanisms by phytohormones in AM symbiosis.
Only recently, genetic and molecular studies started to uncover the regulatory roles of auxin in AM symbiosis. In tomato (Solanum lycopersicum), examining the auxin-signaling defective mutant diageotropia (dgt) and the auxin hyper-transporting mutant polycotyledon (pct) revealed striking differences in AM phenotype (Hanlon and Coenen, 2011). Growing no lateral roots, the root organ cultures (ROCs) of dgt mutant showed no colonization by AMF, and the fungal hyphae even curled away from the roots. By contrast, the ROCs of pct grew more lateral roots and had enhanced fungal colonization with more arbuscules. In pea, an auxin-deficient mutant bushy (bsh) showed reduced colonization, accompanied by a significant decrease of SL exudation and 10 times lower expression of a SL-biosynthesis gene, CCD8 (Foo, 2013). Moreover, a tomato gene SlIAA27 encoding an auxin response regulator was up-regulated at the pre-contact stage of AM symbiosis, and silencing this gene resulted in down-regulation of several SL-biosynthesis genes and low AMF colonization rate (Guillotin et al., 2017). Further evidence supporting a function of auxin signaling in arbuscular development came from a study of microRNA393 (miR393), which is a negative regulator of the plant auxin receptors TIR1 (transport inhibitor response1) and AFB (auxin-related F box) (Etemadi et al., 2014). Overexpression of miR393 resulted in strong defects in arbuscule formation in three different plant species, Medicago truncatula, rice (Oryza sativa) and tomato. A Direct-repeat5 (DR5)-β-glucuronidase (GUS) promoter serving as an indicator of auxin response was also activated in the arbuscule-containing cells (Etemadi et al., 2014). In Brachypodium distachyon, a mutant with higher endogenous auxin content also increased AMF colonization and arbuscule abundance (Buendia et al., 2019). Together, these studies revealed a coordinate role of auxin in regulating SL synthesis, thus promoting AMF colonization, and also supported a requirement of auxin signaling responses for arbuscular development during AM symbiosis.
In plant cells, free auxin levels are determined by a balance of transport, synthesis, storage, and degradation mechanisms, and indole-3-acetic acid (IAA) is the most abundant form (Casanova-Sáez and Voß, 2019). By conjugating IAA to amino acids for storage or degradation, members of the GRETCHEN HAGEN 3 (GH3) family encoding acyl acid-amido synthetases are critical for maintaining auxin homeostasis (Staswick et al., 2005). With its first member identified in soybean as an auxin rapid response gene (Hagen and Guilfoyle, 1985), GH3 family has a total of 19 and 13 genes in Arabidopsis thaliana and rice genomes, respectively, and these genes were phylogenetically divided into three groups (Staswick et al., 2002; Terol et al., 2006; Okrent and Wildermuth, 2011). In Arabidopsis, a GH3 group II member, AtGH3.6 (DFL1, dwarf in light 1) showed a function of negatively regulating shoot and hypocotyl cell elongation as well as lateral root formation under light conditions (Nakazawa et al., 2001), and in vitro experiments demonstrated that AtGH3.6 can conjugate IAA to several amino acids, especially Asp (Staswick et al., 2005). Another gene AtGH3.17 in group III also exhibited a function of inhibiting hypocotyl elongation, and mutation of AtGH3.17 increased free IAA levels in hypocotyl at the expense of IAA-Glu (Zheng et al., 2016). Both IAA-Asp and IAA-Glu result in IAA degradation in Arabidopsis (Ludwig-Müller, 2011; Casanova-Sáez and Voß, 2019). In rice, two GH3 group II genes, OsGH3.8 and OsGH3.2 were reported to suppress IAA-induced expressions of cell wall expansin genes, consequently enhancing plant basal immunity to pathogens (Ding et al., 2008; Fu et al., 2011). Overexpressing OsGH3.8 resulted in several-fold higher IAA-Asp in rice leaves, indicating a function of conjugating free IAA for degradation (Ding et al., 2008). Similarly, OsGH3.2 conjugates IAA to Asp in vitro and overexpressing OsGH3.2 in rice caused a dwarf phenotype associated with low IAA levels (Fu et al., 2011; Du et al., 2012).
Considering the critical roles of plant GH3 genes in maintaining auxin homeostasis and the positive roles of auxin in AM symbiosis, especially for arbuscule development, it was wondered whether some GH3 family members are involved in AM symbiosis. In tomato, one GH3 gene, SlGH3.4 was found strongly up-regulated by AMF inoculation and it was predominantly expressed in cells forming arbuscules (Liao et al., 2015). Transcriptome data for several other angiosperms including O. sativa (Fiorilli et al., 2015; Gutjahr et al., 2015; Roth et al., 2018), Sorghum bicolor (Watts-Williams et al., 2019), M. truncatula (Garcia et al., 2017; Luginbuehl et al., 2017), and Populus trichocarpa (Calabrese et al., 2017) also indicate that certain GH3 genes in these species are induced during AM symbiosis. Little is known about the exact roles of GH3 genes in AM symbiosis. Only recently, a study reported that tomato SlGH3.4 could negatively regulate AM symbiosis, especially arbuscule incidence in mycorrhizal roots, likely by maintaining auxin homeostasis in colonized cortical cells (Chen et al., 2021). More aspects remain to be investigated, though, such as asymbiotic expression patterns of GH3 genes and mutant phenotypes at different stages of symbiosis. In this study, by acquiring rice OsGH3.2pro:GUS transgenic plants and generating Osgh3.2 mutants via CRISPR/Cas9 technique, the roles of OsGH3.2 in modulating rice root morphology and also affecting AM symbiosis were revealed through time-course experiments.
Materials and Methods
Reconstructing a Phylogeny of Plant GH3 Gene Family
A consensus sequence of GH3 domain (Pfam03321) was downloaded from the NCBI Conserved Domain Database1 and used to run BLASTP searches against the proteomes of 26 representative green plants (for species list and detailed genome information, see Supplementary Table 1). Sequences showing an E-value < 10–4 and >50% length coverage were kept and further examined on Pfam2 to confirm the presence of GH3 domain. All the identified plant GH3 sequences were aligned by ClustalW and Muscle programs, which are implemented in MEGA 7.0 (Kumar et al., 2016). A phylogenetic tree of GH3 family was then reconstructed using the aligned protein sequences by adopting a maximum-likelihood method under the JTT + I + G4 model via IQ-TREE, and the robustness of internal branches was tested by calculating the Shimodaira–Hasegawa approximate likelihood ratio test (SH-aLRT) and by performing 1000 ultrafast bootstrap replicates (Trifinopoulos et al., 2016).
Plant Cultivation and the Inoculation With Rhizophagus irregularis
The rice (O. sativa ssp. japonica) wild-type and transgenic plants used in this study were in the cv. Nipponbare background. The rice seeds were surface-sterilized with 2.5% sodium hypochlorite solution for 20 min, washed extensively with sterile water and germinated on a modified 1/2 Murashige-Skoog (MS) medium. Two-week-old rice seedlings were planted in pots (12 cm in diameter, three seedlings/pot) containing a mixture of autoclaved sand and perlite (3:1, V/V). For AM colonization, about 150 spores of R. irregularis freshly extracted from carrot hairy root co-cultures were added to the rhizosphere of each seedling. For mock inoculation, only the washed distilled water without spores were added. Plants were placed in a growth room under a 16/8 h day/night cycle at 28°C/22°C and were fertilized twice a week with 1/2 Hoagland solution containing 20 μM phosphates. Plants were harvested at several time points, including 1-week post inoculation (wpi), 3-wpi, 5-wpi, 7-wpi, and 9-wpi. Normally, half collected root samples were used for staining/mycorrhizal phenotype examination and the other half were frozen in liquid nitrogen and kept at –80°C for RNA extraction.
For hydroponic culture, surface-sterilized rice seeds were germinated in sterile water for 2 days at 37°C, then transferred into hydroponic boxes (length 12.6 cm, width 8.5 cm, height 11 cm, 16 seedlings/box). The plants were grown in sterile water for 1 week, then in 1/4 Hoagland solution for another 2 weeks. To compare the root morphology between wild-type rice and the mutants, the root length of the longest crown root (CR), the total number of CRs, and the total number of large lateral roots (LLRs) were recorded each week. At the end of this experiment, the root samples of both wild-type and Osgh3.2-1 mutant were sent to measure the free IAA levels commercially via the AB Sciex QTRAP 6500 LC-MS/MS platform (MetWare, Wuhan, China).
RNA Extraction and Quantitative RT-PCR
In a time-course inoculation experiment, root samples of three wild-type rice individuals were collected at multiple time points and further used to examine the expression patterns of 13 GH3 genes. For either wild-type or the Osgh3.2 mutants, root samples of three individuals were also collected at different time points and used to evaluate the expression patterns of OsPT11 (phosphate transporter 11), OsAFB2 (auxin-related F box 2) and OsD17/CCD7 (dwarf 17/carotenoid cleavage dioxygenase 7) genes. Total RNAs of rice roots were extracted by the Trizol method. The RNA purity and integrity were examined by RNA electrophoresis in 2.5% (w/v) agarose gel. One μg RNA was treated with DNase I prior to cDNA synthesis using a Revert-Aid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, United States). Quantitative RT-PCR (qRT-PCR) was performed by measuring the intensity of SYBR Green Fluorescent dye conjugated to double stranded DNA molecules using a C1000 Thermal Cycler Real-Time PCR detection system (Bio-Rad, Hercules, CA, United States). Expression values of interested genes were normalized to rice ubiquitin 1 gene (OsUBI1, Chen et al., 2008), and analyzed with the 2–Δ CT method (Livak and Schmittgen, 2001). Statistical differences were evaluated via the Student’s t-test. All the qRT-PCR primers used are provided in Supplementary Table 2.
Constructing the OsGH3.2pro:GUS Vector and GUS Staining
A 2.0 kb fragment upstream of the OsGH3.2 start codon was amplified from rice genomic DNA (for primers, see Supplementary Table 2) using a high fidelity Fastpfu enzyme and cloned into pEASY-Blunt Zero vector by TA cloning (Transgene, Beijing, China). The fragment was then digested from the vector and ligated with a pre-digested linear binary vector pBI101.1. The constructed pBI101.1 vector containing the OsGH3.2pro:GUS reporter unit was introduced into rice via Agrobacterium tumefaciens-mediated transformation, as described in Liu et al. (2021). Positively transformed seedlings were identified by PCR using the primers OsGH3.2pro-F and PBI101.1-GUS-R (Supplementary Table 2) and selected for seed propagation. The transformed T1 plants were then used to examine the expression activities of the OsGH3.2 promoter in rice roots under both asymbiotic and symbiotic conditions. The rice roots were incubated at 37°C in GUS staining solution (100 mM sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.5mM K4[Fe(CN)6], 0.5 mM K3[Fe(CN)6], 0.1% Triton X-100, 20% (w/v) methanol and 0.5 mM X-Gluc) for 5 h, then fixed by FAA (5% formaldehyde, 5% ethanoic acid, and 50% ethanol) and dehydrated with ethanol. The treated roots were observed under a Nexcope-NE620 light microscope. To visualize the fungal structures, the roots were counterstained further by WGA-Alexa Fluor 488 as described below.
CRISPR/Cas9 Vector Construction and Obtaining Osgh3.2 Mutant Lines
To generate Osgh3.2 mutants, the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9 technique was employed. Single-guide RNA (sgRNA) targeting OsGH3.2 was designed using the CRISPRdirect3 online tool (Naito et al., 2015). Two best-scored sgRNAs (sgRNA1 and sgRNA2) showing the least off-target possibilities were selected, with their spacer sequences (upstream of the protospacer adjacent motif sites) matching nucleotides 2151–2170 and 2271–2290 in exon 3 of OsGH3.2, respectively (Supplementary Figure 3A). For each sgRNA, a pair of complementary primers with sticky ends were synthesized, annealed, and ligated with a linear CRISPR vector, pRGEB31, which was pre-digested by the restriction enzyme, BsaI. The pRGEB31-sgRNA1 and pRGEB31-sgRNA2 vectors were then introduced into rice calli. The edited alleles of OsGH3.2 were examined among transformed rice seedlings through PCR and sequencing with a pair of primers, OsGH3.2 edit-F/R, which cover both editing regions. In subsequent generations, plants carrying desired homozygous mutations without the pRGEB31 vector, which can be detected by pRGEB31-Cas9-F/R primers, were selected for reproduction. Finally, T2 or T3 generations of two mutant lines (Osgh3.2-1, -2) were used for further studies. All the primers used are provided in Supplementary Table 2.
Fungal Structure Staining, Quantification and Microscopic Imaging
For wheat germ agglutinin (WGA) staining (Schaffer and Peterson, 1993), fresh rice roots were fixed in 50% ethanol for >4 h and cleared in 20% KOH (w/v) for 2–3 days at room temperature. Next, the roots were acidified in 0.1 M HCl for 20 min and rinsed five times with phosphate-buffered saline (PBS) solution (pH 7.4). After incubating overnight in PBS solution containing 1 μg/mL WGA-Alexa Fluor 488 (Invitrogen, Carlsbad, CA, United States) or further with 10 μg/mL Propidium iodide (Sangon Biotech, Shanghai, China), the stained roots were examined under a confocal laser scanning microscope (ZEISS LSM980) and representative images were captured. For ink-vinegar staining (Vierheilig et al., 1998), rice roots were cleared with 20% KOH for 40–120 min at 65°C, acidified by 5% acetic acid for 5 min at room temperature, incubated in ink-vinegar solution for 30 min at 65°C, and decolored in tap water for 14 h. The stained roots were examined by a Nexcope-NE620 light microscope. To quantify the total colonization rate, a modified gridline intersect method (McGonigle et al., 1990) was used to score fungal structures.
Measuring Longitudinal Lengths of Cortical Cells and Arbuscules
After Ink-vinegar staining, the images of colonized root fragments (see an example in Supplementary Figure 7) were taken by JieDa microscopic imaging software and the longitudinal lengths of cortical cells and arbuscules were measured by using a ruler implemented in the software. Also, the total length of infection units from hyphopodia to the running edge of hyphae toward the root tip and the longitudinal length of neighboring non-colonized cortical cells were measured. In total, for either wild-type rice or the mutants, at least 15 root fragments from three individuals were measured at 3-wpi.
At 7- and 9-wpi, non-colonized cells were hard to be found since lateral roots had been heavily colonized. The longitudinal length of an array of colonized cortical cells and their contained arbuscules from individual infection site (hyphopodia) toward the root tip were measured in roots of wild-type rice and both Osgh3.2 mutants. The arbuscules population were further sorted into three size categories, small/degenerate (<30 μm), middle (30–50 μm), and large (>50 μm) according to their longitudinal length. For each genotype, three rice individuals were included and for each rice individual, 100 colonized cells from an average of 10 infection sites were measured.
Results
Multiple Arbuscular Mycorrhizal-Induced GH3 Genes Belong to the Subgroup II-A of GH3 Gene Family
To first explore the evolutionary history of GH3 gene family in plants, a total of 250 GH3 genes (Supplementary Table 1) were identified from 22 surveyed plant species, including 215 genes identified from 13 angiosperms. Based on their protein sequences, a phylogeny of GH3 family was then reconstructed (Figure 1A and Supplementary Figure 1). On the phylogeny, the algal GH3 sequences occupied the most basal position (Figure 1A and Supplementary Figure 1), suggesting that an ancestral GH3 gene had originated in the algal ancestor. This ancestral GH3 gene further divided into two separate lineages in bryophytes, as represented by two P. patens GH3 genes, PpGH3.1 (Pp3c24_16260) and PpGH3.2 (Pp3c10_20960), which share 43.4% identities at the protein level (Figure 1A and Supplementary Figure 1). More GH3 lineages further evolved in seed plants. To be largely in accordance with previous classification system covering Arabidopsis and rice GH3 genes (Staswick et al., 2002; Terol et al., 2006; Okrent and Wildermuth, 2011), three major angiosperm lineages (groups I, II, III) were distinguished, with each further separated into two sublineages (subgroups I-A, I-B, II-A, II-B, III-A and III-B) (Figure 1A and Supplementary Figure 1). It was reported that a tomato GH3 gene, SlGH3.4, is strongly induced in mycorrhizal roots (Liao et al., 2015). Based on the reconstructed phylogeny, SlGH3.4 belongs to the subgroup II-A (Figure 1B). Also, transcriptome data of AM symbioses are available for some other angiosperm species. Therefore, additional AM-induced GH3 genes were identified, including OsGH3.2 in rice, Sorbi003G306500 in S. bicolor, MtGH3.6 in M. truncatula, and Poptr007G050300 in P. trichocarpa. Interestingly, all these genes belong to the subgroup II-A (Figure 1B), suggesting that these genes may have similar function during AM symbiosis. To explore such function, in this study, we focused on the rice OsGH3.2 gene for further investigations.
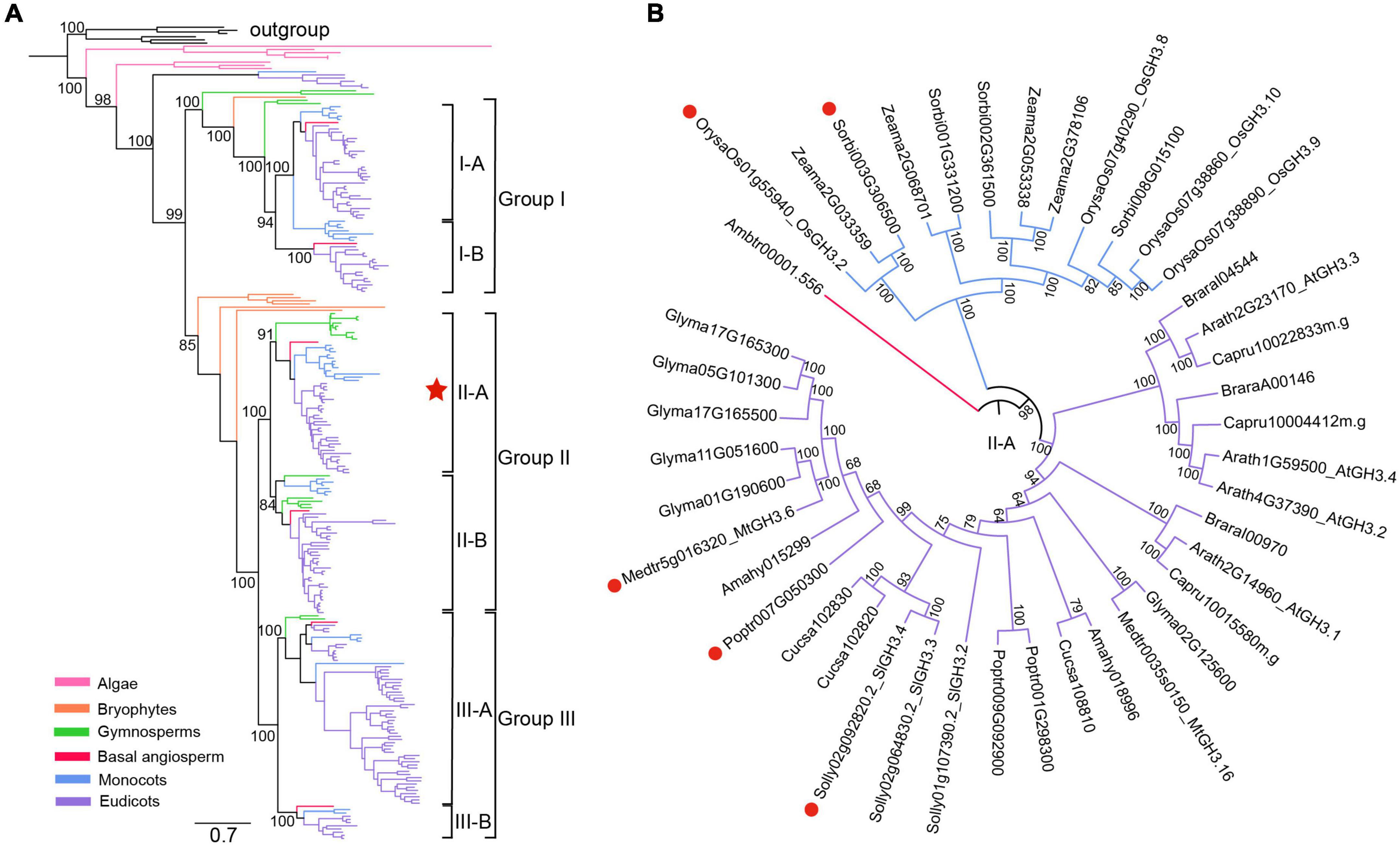
Figure 1. Phylogenetic analyses of plant GH3 genes. (A) A reconstructed phylogeny of plant GH3 gene family using a maximum-likelihood method under the JTT + I + G4 model via IQ-TREE (Trifinopoulos et al., 2016). The robustness of internal branches was evaluated by calculating the Shimodaira–Hasegawa approximate likelihood ratio test (SH-aLRT) and by performing 1000 ultrafast bootstrap replicates. Six bacterial GH3 genes were designated as the outgroup. A total of 250 GH3 genes (22 plant species) identified from algae, bryophytes, gymnosperms, basal angiosperm, monocots and eudicots were covered (for a complete tree showing all sequence names, please refer to Supplementary Figure 1). The angiosperm GH3 genes were classified into three major groups and six subgroups, with supporting values of ultrafast bootstrap test shown on branches of these clades. The bar represents number of nucleotide substitutions per site. (B) A circled phylogeny of subgroup II-A (labeled by a red star in (a) covering surveyed angiosperm species. The AM-induced GH3 genes from five angiosperms, as supported by transcriptome data, were indicated by red dots.
OsGH3.2 Showed Asymbiotic Expression in Rice Young Lateral Roots and Symbiotic Expression in Cortical Cells That Have Formed Mature Arbuscules
The RT-PCR results showed that seven out of 13 OsGH3 genes appear to be expressed in rice roots under both asymbiotic and symbiotic conditions (Supplementary Figure 2A). Quantitative RT-PCR (qRT-PCR) experiments were then performed for the seven GH3 genes. As Figure 2A showed, at 3-weeks post inoculation (wpi) by AM fungus Rhizophagus irregularis, OsGH3.2 expression level was only slightly higher in mycorrhizal roots than in control roots (1.3-fold, p = 0.13). At 5-wpi, however, OsGH3.2 showed a 2.7-fold induction in mycorrhizal roots (p < 0.01), thus confirming previous transcriptome data. As for the other six GH3 genes, their expression patterns were not altered at a statistically significant level at either 3- or 5-wpi by R. irregularis (Supplementary Figure 2B). The expression level of OsGH3.2 in mycorrhizal roots further increased at 7-wpi (6.6-fold to control, p < 0.01), then decreased at 9-wpi (2.4-fold to control, p < 0.05, Figure 2A). Such pattern seemed to be consistent with the arbuscule abundance level in mycorrhizal roots, as many arbuscules would become degenerated at late stage of symbiosis.
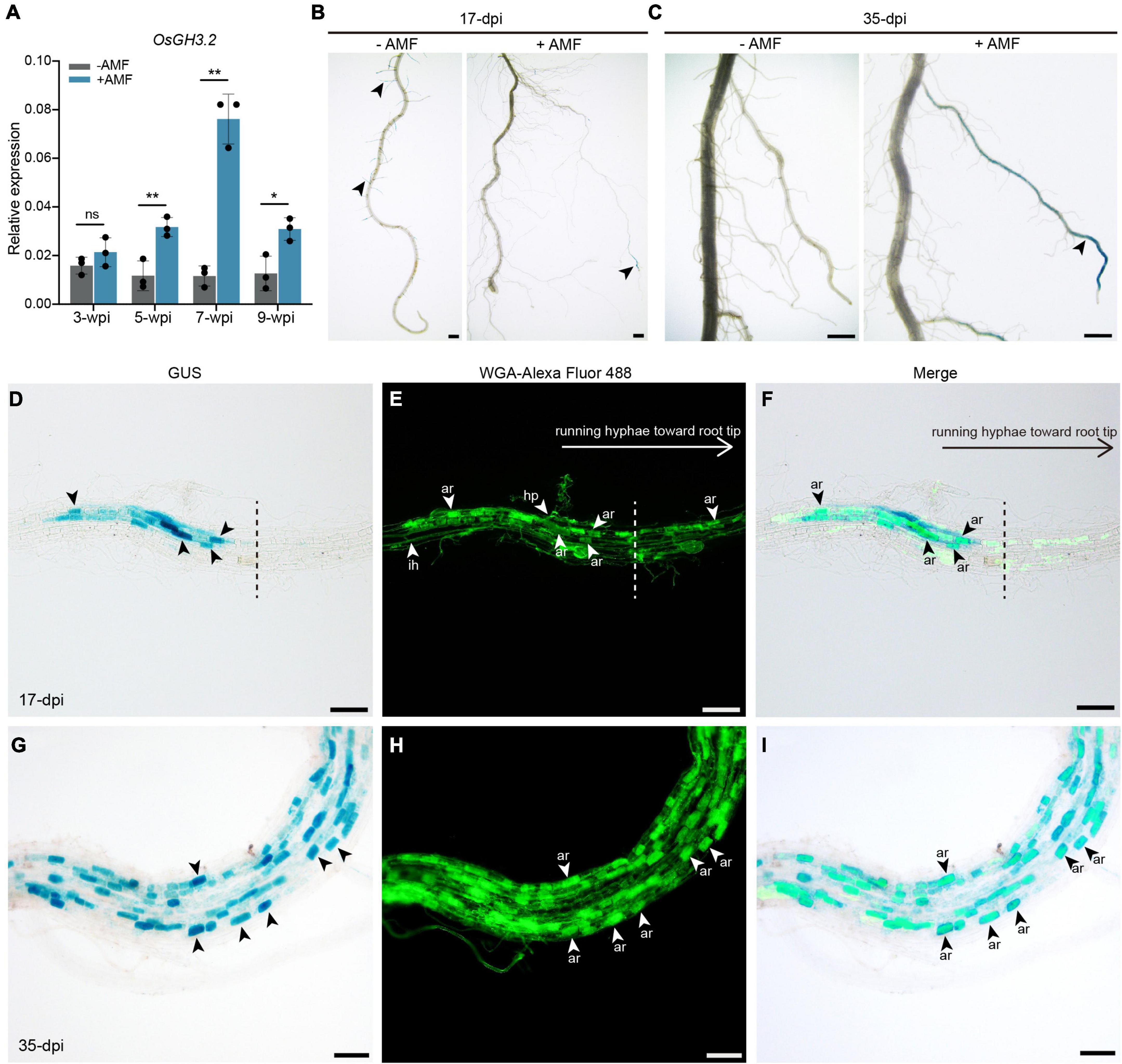
Figure 2. Promoter activity of OsGH3.2 in mock-inoculated and R. irregularis-inoculated rice roots. (A) The relative expression levels of OsGH3.2 in wild-type rice roots were measured by qRT-PCR at 3-, 5-, 7-, and 9-wpi by R. irregularis. A housekeeping gene, OsUBI1, was used for normalization. Data represent mean ± standard deviations (SD), n = 3. Statistical analysis was performed with Student’s t-test (ns, not significant; *p ≤ 0.05; **P ≤ 0.01); (B) A representative crown root with GUS activity detected in many young lateral roots of mock inoculated (–AMF) roots and in a large lateral root of R. irregularis inoculated (+AMF) roots at 17-dpi; (C) after 35-days inoculation, a representative mock inoculated crown root showed no more GUS activity in mature LLR, while the GUS activity were shown in more mature LLRs of R. irregularis inoculated roots. The roots showed GUS activity were then counterstained with WGA-Alexa Fluor 488 to visualize fungal structures at 17- and 35-days post inoculation (dpi) by R. irregularis. (D–F) GUS activities were detected in cortical cells containing mature arbuscules below or near the hyphopodia; at the side of running hyphae toward root tip (separate with a dashed line), no GUS activities were detected in colonized cells. (G–I) After 35-dpi, GUS activities were detected in more mature arbuscules. Arrowheads indicate various of fungal structures: hp, hyphopodia; ih, intraradical hyphae; ar, arbuscules. The scale bars represent 1 mm (B,C) and 100 μm (D–I).
To visualize the spatial expression pattern of OsGH3.2, an OsGH3.2pro:GUS vector was constructed and transformed into rice to obtain stable transgenic lines (see section “Materials and Methods”). The transgenic T1 rice plants were either inoculated with R. irregularis or with mock solution and the GUS activities were examined in roots at two time points. At 17-days post inoculation (dpi) by mock solution, blue stains were seen in rice young lateral roots (Figure 2B). Examination at a later time point (35-dpi) then showed no more asymbiotic expression of OsGH3.2 in rice mature large lateral roots (LLRs, Figure 2C). Differently, at 17- and 35-dpi by R. irregularis, GUS activity was mainly detected in rice mature LLRs (Figures 2B,C). To know whether fungal structures are associated with symbiotic expression of OsGH3.2, we then counterstained the mycorrhizal roots with WGA-Alexa Fluor 488. At 17-dpi, some fragments of LLR showed individual infection unit by AMF, and GUS activity was mainly detected in the cortical cells below or near the hyphopodia, which had formed mature arbuscules (Figures 2D–F). At the side of running hyphae toward root tip, however, no GUS activity was detected despite some cortical cells had formed mature arbuscules (Figures 2D–F). Such observation made at early symbiotic stage thus indicate that symbiotic induction of OsGH3.2 in cortical cells is not fully synchronical to arbuscule maturation. At 35-dpi, as a large fraction of LLRs had been colonized, GUS activity were detected in many more cortical cells that had formed mature arbuscules (Figures 2G–I).
CRISPR/Cas9-Mediated Mutagenesis of OsGH3.2 Gene
To explore the roles of OsGH3.2 in rice root development and in AM symbiosis, we tried to obtain null Osgh3.2 mutants via CRISPR/Cas9 technique. Regenerated rice seedlings were screened for edited Osgh3.2 alleles. A total of six mutated alleles were detected in the T0 generation and two of these, Osgh3.2-1 and Osgh3.2-2, were selected to reach homozygosity in subsequent generations, meanwhile eliminating the pRGEB31 vector (Supplementary Figure 3). By introducing a 2-nucleotide deletion, Osgh3.2-1 allele had altered reading frame, and consequently encoded a protein sharing no homology with the normal protein beyond amino acid site 299. Also, by introducing a 4-nucleotide deletion, Osgh3.2-2 allele had a premature stop codon and thus encoded a truncated protein only 298 aa in length. Therefore, for both mutated alleles, their encoded proteins only retain the N-terminal half of the GH3 domain (Supplementary Figure 3C). Since the proper function of GH3 enzymes requires both the N-terminal domain and the C-terminal domain (Westfall et al., 2012), it can be expected that the mutated Osgh3.2-1 and -2 alleles probably lost the function of OsGH3.2.
OsGH3.2 Has Developmental Roles in Modulating Rice Root Architecture, Panicle Traits, as Well as Grain Length
Under asymbiotic condition, the expression of OsGH3.2 in rice young lateral roots prompted us to explore its potential role in regulating rice root development. Seeds of wild-type rice and the Osgh3.2 mutants were germinated and cultured hydroponically (see section “Materials and Methods”). After 3 weeks, both Osgh3.2 mutants exhibited a “shallow” root architecture compared to wild-type (Figure 3A). The mutants had shorter maximum crown root (CR) length and grew more LLRs (∼3.4 times) and CRs (∼1.2 times) than wild-type (Figure 3B). To explore whether such phenotypic changes in mutant roots are related to altered auxin levels, free IAA levels were measured for root samples of wild-type rice and Osgh3.2-1. The mutant had an average of 4.1 ng IAA/g fresh root, which is indeed higher than that in wild-type rice (3.2 ng IAA/g fresh root, p < 0.05, Supplementary Figure 5A). Meanwhile, tryptophan, the IAA synthetic precursor, also accumulated more in Osgh3.2-1 roots (Supplementary Figure 5B).
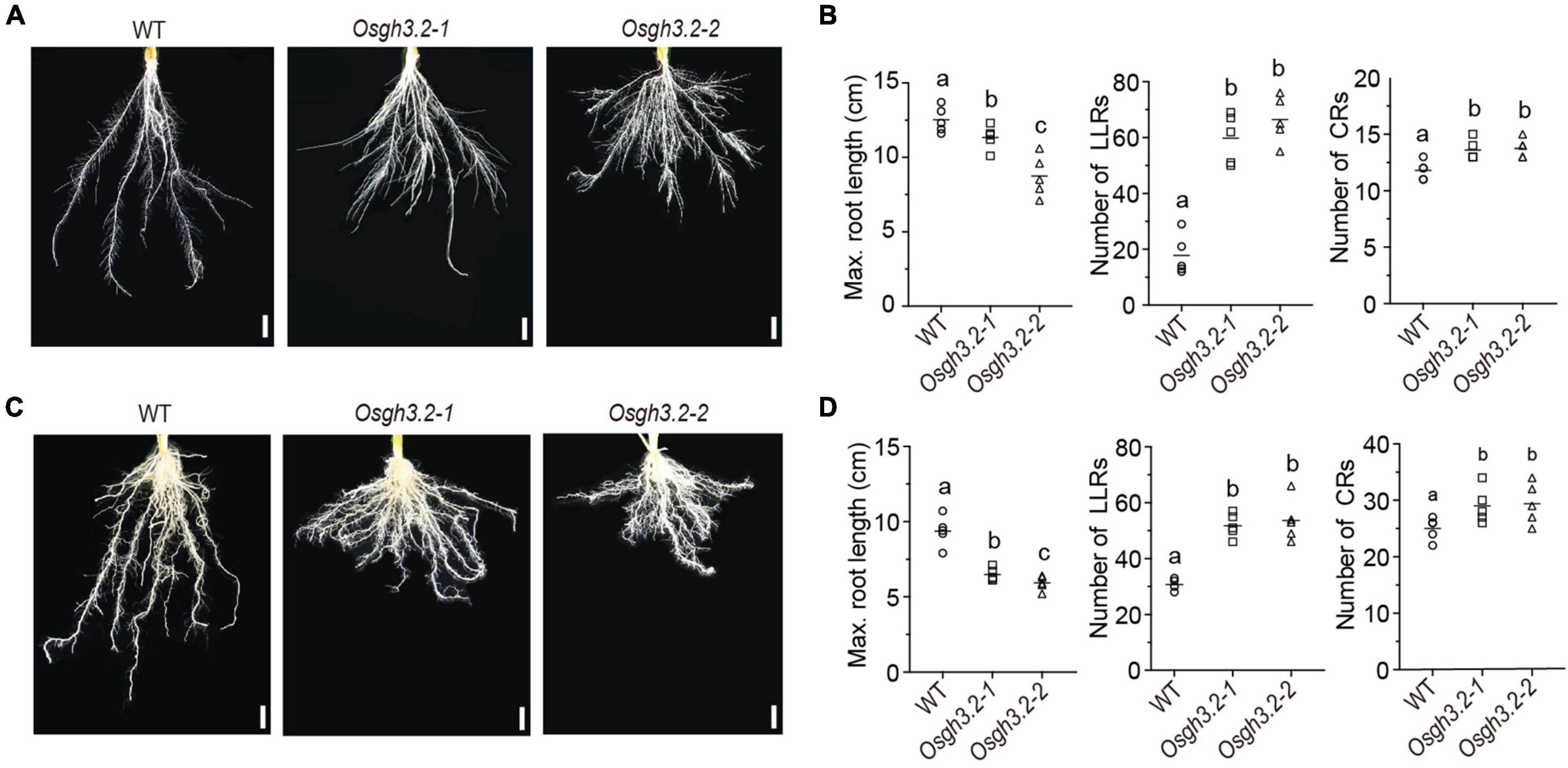
Figure 3. Osgh3.2 mutants showed an altered “shallow” root architecture. Rice roots of wild-type, Osgh3.2-1 and Osgh3.2-2 after hydroponic culture (A) or inoculation by R. irregularis (C) for 3 weeks. Scale bars, 1 cm. (B,D) Maximum length of crown roots (CRs), number of large lateral roots (LLRs) and number of CRs were compared between wild type rice and the Osgh3.2 mutants under two cultivation conditions, respectively. Data are individual values from five biological replicates and the horizontal line shows the mean value. Statistical analysis was performed with Student’s t-test. Different letters indicate statistically significant differences (P < 0.05).
We further investigated whether the alteration of root architecture can still be observed under symbiotic condition. As shown in Figures 3C,D, similar morphological changes also occurred in Osgh3.2 mutants after 3-weeks inoculation by R. irregularis, demonstrating that the developmental role of OsGH3.2 in modulating rice root architecture is independent of AMF inoculation. At intermediate time points (1- or 2-week), the root morphological changes could be also detected in Osgh3.2 mutants under either asymbiotic or symbiotic condition (Supplementary Figures 4A–D).
To see whether OsGH3.2 has a regulatory role in above-ground tissues, we also examined the panicle traits and grain sizes of wild-type rice and the Osgh3.2 mutants (Supplementary Figures 6A,B). Both mutants showed longer panicle length, more primary panicle branches, and Osgh3.2-2 also showed more secondary panicle branches than wild-type rice (Supplementary Figure 6C). As for grains, the mutants exhibited longer grain length, but showed no differences to wild-type rice in terms of grain width and thickness (Supplementary Figure 6D).
Under Symbiotic Condition, the Osgh3.2 Mutants Exhibited Higher Colonization Rates and Arbuscule Abundance Levels Than Wild-Type at Most Examined Time Points Except at 7-wpi
To explore how OsGH3.2 expressions would affect AM symbiosis, the mycorrhizal phenotypes of the two Osgh3.2 mutants were compared to wild-type in a time course experiment. After inoculation by R. irregularis for 1 week, both hyphopodia and intraradical fungal hyphae were observed in rice LLRs, with Osgh3.2-1 exhibiting a higher colonization rate than wild-type rice (6.4% vs. 3.5%, p < 0.05, Figure 4A). Nearly no arbuscules or vesicles were formed at this early stage (Figures 4B,C), so the colonization rate difference is unlikely to be attributed to symbiotic expression of OsGH3.2 in arbusculated cells. At 3-wpi, the colonization rate in wild-type rice LLRs reached ∼11%, with arbuscules seen in 3% root length and still no vesicle formed. Strikingly, both Osgh3.2 mutants showed > 25% colonization by AMF, with 20% and 2% root length forming arbuscules and vesicles, respectively (Figures 4A–C). The morphology of arbuscules formed in mutant roots was as normal as in wild-type, suggesting loss function of OsGH3.2 would not impair arbuscular development (Figure 4D). At 5-wpi, the total colonization rate in wild-type rice LLRs further increased to 66%, while in both mutants reached a higher rate of ∼77% (Figure 4A). The arbuscule abundance level was also higher in mutant roots, though comparable numbers of vesicles were observed between wild-type and the mutants (Figures 4B,C). Together, these results illustrated that the Osgh3.2 mutants were colonized by R. irregularis at higher rates than wild-type rice from 1- to 5-wpi.
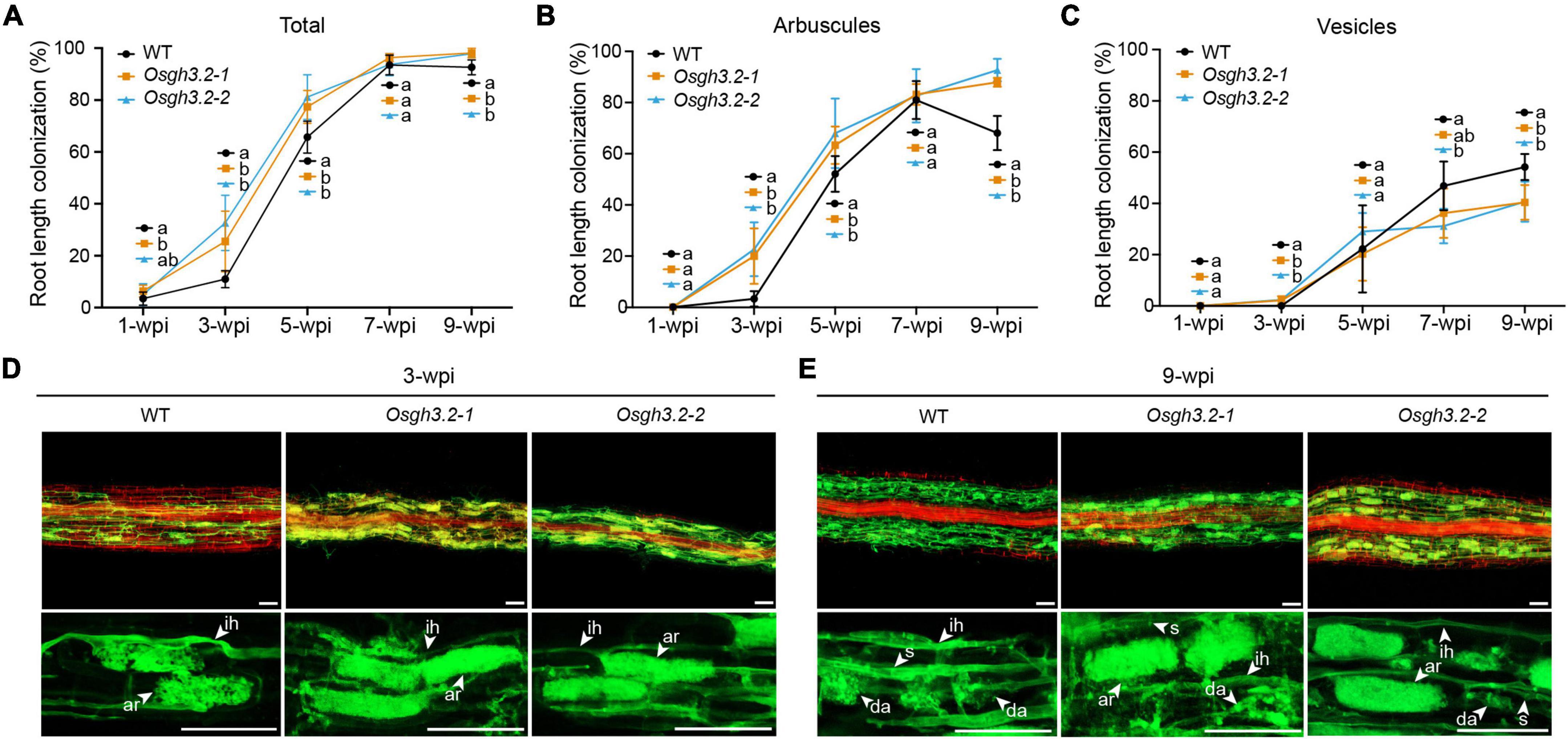
Figure 4. The mycorrhizal phenotypes of Osgh3.2 mutants. (A–C) For both wild-type rice and the Osgh3.2 mutants, the total root length colonization rates (%) as well as the rates of specific fungal structures including arbuscules, and vesicles were determined by the gridline intersect method at 1-, 3-, 5-, 7-, and 9-wpi by R. irregularis. Data represent mean ± standard deviations (SD), n = 4∼6. Different letters indicate statistically significant differences (Student’s t-test, P < 0.05). (D,E) Laser scanning confocal images showing fungal structures stained by WGA-Alexa Fluor 488 (green) and plant cell walls stained by propidium iodide (red) in wild-type rice and the Osgh3.2 mutants at 3- and 9-wpi, respectively; ih, intraradical hyphae; ar, arbuscules; da, degenerating arbuscules; s, septa. scale bar = 50 μm.
The mycorrhizal phenotypes were observed continuously. At 7-wpi, the total colonization rate by R. irregularis in both wild-type rice and the Osgh3.2 mutants had reached > 90% and entered a plateau phase (Figure 4A). At this time point, the mutants showed similar arbuscular abundance levels to wild-type rice (Figure 4B), though less lipid-storage vesicles were observed in the Osgh3.2-2 mutant (Figure 4C). At 9-wpi, a majority of arbuscules observed in wild-type rice LLRs became degenerated (Figure 4E), so the arbuscule abundance level declined to 68% and the vesicle abundance level increased to 54% (Figures 4B,C). For both Osgh3.2 mutants, however, the arbuscular abundances were maintained at even higher levels than at 7-wpi, with many arbuscules still occupying the cortical cells rather fully (Figures 4B,E). Also, the vesicle abundance level in mutant roots was much lower than in wild-type (Figure 4C).
At Late Stage of Symbiosis, the Osgh3.2 Mutants Showed Elongated Colonized Cells as Well as Larger Arbuscules Than Wild-Type
To investigate whether OsGH3.2 can affect root cortical cell morphology, multiple root fragments containing individual infection units (n = 15–18) were firstly analyzed at 3-wpi (see section “Materials and Methods”). On average, the length of infection units in the mutant roots was twice longer than in wild-type roots (Figure 5A), reflecting a more efficient colonization by AMF in the mutants. Also, larger arbuscule mean size was observed in the mutants (Figure 5B). The longitudinal lengths of colonized cortical cells and their neighboring non-colonized cells were further measured. On average, for either wild-type or the mutants, the colonized cortical cells showed longer lengths than their neighboring non-colonized cells (Figure 5C), suggesting that fungal invasion and forming arbuscules would enlarge the infected cortical cells. However, between wild-type and the mutants, neither colonized nor non-colonized cortical cells showed mean length differences (Figure 5C), suggesting that loss function of OsGH3.2 had not caused obvious effect on the cortical cell length at this early symbiotic stage. This is consistent with the low induction level of OsGH3.2 observed at this time point (Figure 2A).
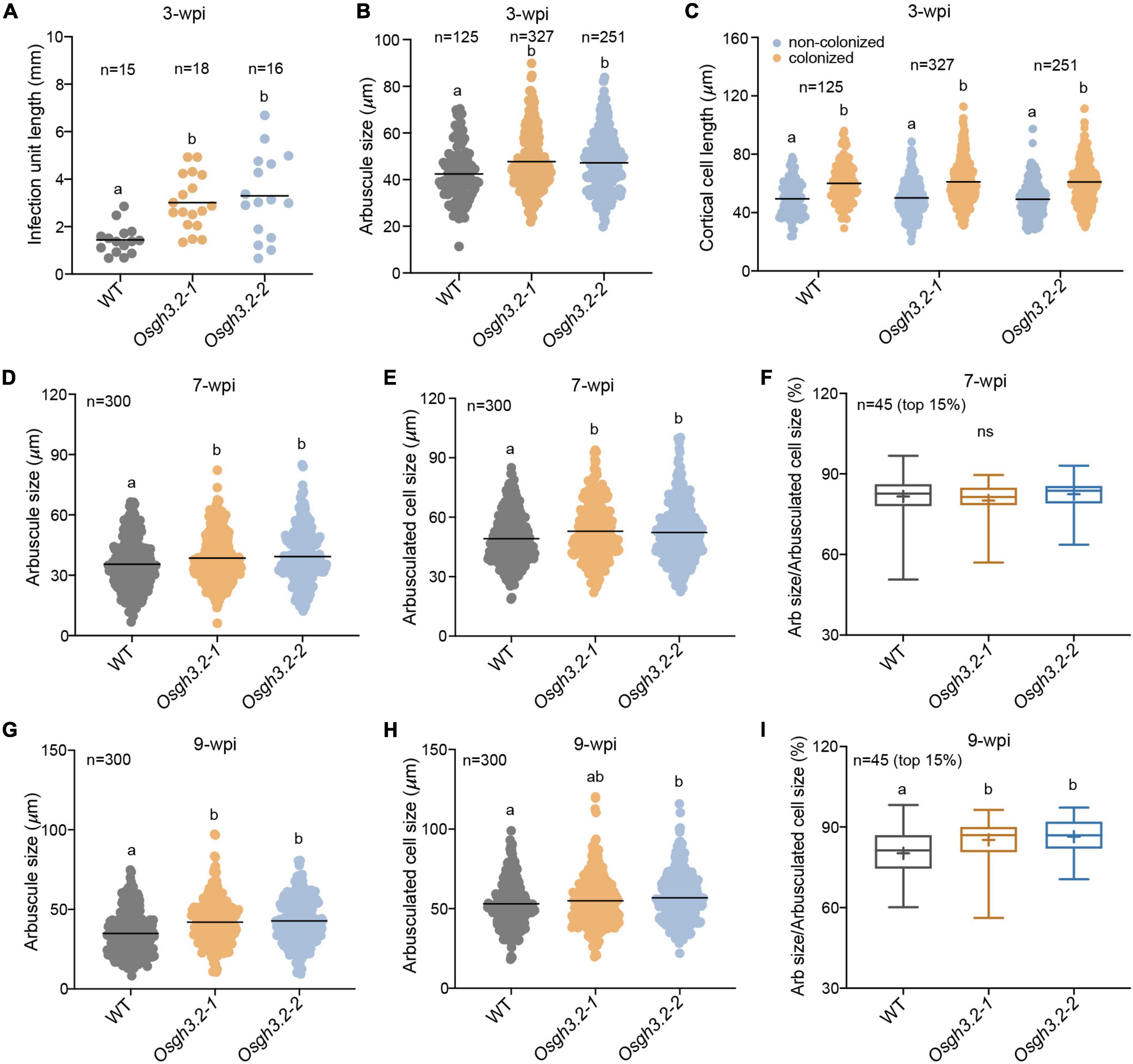
Figure 5. Osgh3.2 mutants showed larger arbuscules as well as enlarged arbuscule-containing cortical cells at 7- and 9-wpi. (A) Lengths of infection units in wild-type and the Osgh3.2 mutants at 3-wpi by R. irregularis. Data are individual values from at least 15 individual infection units from three biological replicate samples of wild-type and Osgh3.2 mutants. (B) Data represent arbuscule sizes (longitudinal length) of wild-type (125 arbuscules), Osgh3.2-1 (327 arbuscules), and Osgh3.2-2 (251 arbuscules) at 3-wpi from all the measured infection units in (A). (C) The measured longitudinal lengths of colonized and neighboring non-colonized cortical cells in wild-type (125 cells each), Osgh3.2-1 (327 cells each), and Osgh3.2-2 (251 cells each) at 3-wpi. (D,G) For either wild-type rice or the Osgh3.2 mutants, the longitudinal lengths of 300 arbuscules from three rice individuals were measured. (E,H) The longitudinal lengths of cortical cells containing arbuscules in wild-type and the Osgh3.2 mutants at 7- and 9-wpi, respectively. Data represent individual values of 300 colonized cells from three biological replicate samples of wild-type and Osgh3.2 mutants. (A–E,G,H) The horizontal line shows the mean value. Statistical analysis was performed with Student’s t-test. Different letters indicate statistically significant differences (P < 0.05). (F,I) When only the 15% largest arbuscules were considered, the longitudinal length ratios of arbuscules to cortical cells were shown in box plots and compared between wild-type and the mutants. The boxes represent the variation between the first and third quartiles; the “+” indicates mean value, and the horizontal line indicate the median. Statistical analysis was performed with Student’s t-test (ns, not significant; different letters indicate statistically significant differences, P < 0.05).
At 7-wpi, when OsGH3.2 had been extensively expressed in mycorrhizal roots (Figure 2A), we further measured the longitudinal lengths of colonized cortical cells from each genotype (see section “Materials and Methods”). It was found that on average, both Osgh3.2 mutants had elongated colonized cortical cells than wild-type (Figure 5E). Measuring the lengths of arbuscules within these cortical cells also revealed larger arbuscules in the mutants (Figure 5D). When only a subset of large arbuscules (top 15%) was considered, the relative ratio of arbuscule length to cell length then showed no difference between wild-type and the mutants (81.6% vs. 81.3% on average, Figure 5F), suggesting that the larger arbuscules observed in the mutants were most likely due to extra room provided in the enlarged cortical cells. At 9-wpi, the measurements were repeated again. Since many arbuscules had started degenerating in wild-type (Figure 4E), the mutants not only exhibited longer lengths of colonized cortical cells and arbuscules (Figures 5G,H), but also showed higher relative ratio of arbuscule to cell lengths (Figure 5I), indicating that the arbuscules in the mutants were less degenerated than in wild-type at this time point. We also sorted the arbuscule population into three size categories, small/degenerate (<30 μm), middle (30–50 μm), and large (>50 μm). At 9 wpi, both mutants had maintained significantly higher ratios of large arbuscules but lower rations of small/degenerate arbuscules than wild-type (Supplementary Figure 8B), indicating that the arbuscules are less degenerated in the mutants. While at 7 wpi, the ratio differences were not statistically significant for either large or middle size arbuscules (Supplementary Figure 8A). Even for small size arbuscules, only the Osgh3.2-2 but not Osgh3.2-1 mutant showed significant lower ratio than wild-type (Supplementary Figure 8A), likely because the arbuscules had not entered into degeneration stage largely at 7 wpi in wild-type.
Expression of Several Marker Genes at Different Symbiotic Stages
Finally, the relative expression levels of several rice genes were examined at different colonization stages between wild-type and the Osgh3.2 mutants, including OsPT11, an AM marker gene associated with arbuscular phosphate transferring, OsAFB2, an auxin response gene and OsD17, a gene involved in SL synthesis. The relative expression levels of OsPT11 were in good accordance with the arbuscule abundance levels (Figure 6A), thus supporting the observed arbuscular phenotype at different time points (Figure 4B). OsAFB2 in both mutants were expressed at significantly higher levels than in wild-type rice at 3-wpi and 7-wpi (less evident at 9-wpi), indicating more active auxin responses in the mutant roots (Figure 6B). The expression of OsD17 gene is relevant to the SL synthesis, and stronger expression of this gene in the mutants at 3-wpi (Figure 6C) suggested a possibility of synthesizing and secreting more SLs to attract AMF colonization. Also, OsD17 was expressed at comparable levels between wild-type and the mutants at 7-wpi, but at lower level in the wild-type at 9-wpi, reflecting a correlation with arbuscular abundance level like OsPT11 (Figure 6C).
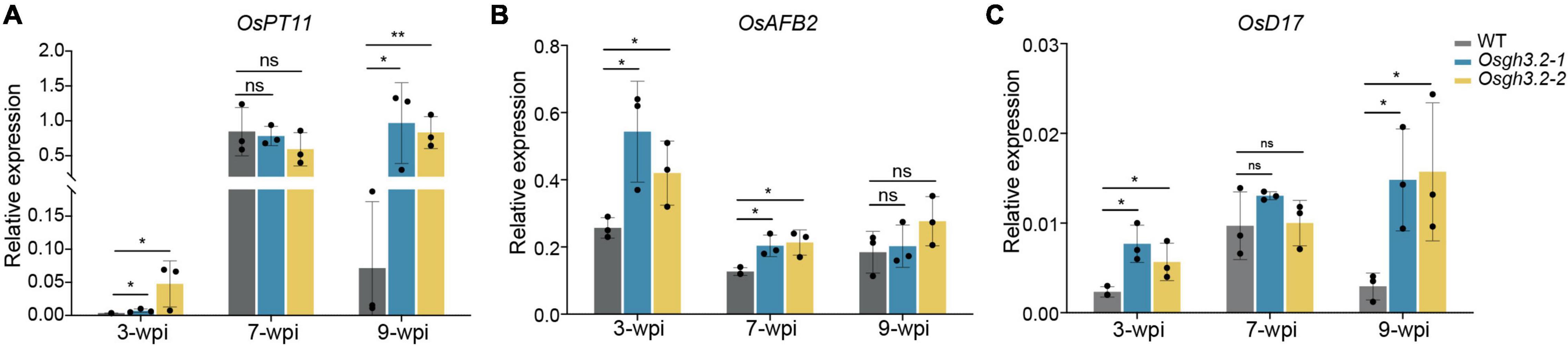
Figure 6. The expression patterns of OsPT11, OsAFB2, and OsD17 at 3-, 7- and 9-wpi by R. irregularis in roots of wild-type rice and the Osgh3.2 mutants. (A–C) Relative expression levels of OsPT11, OsAFB2, and OsD17 were measured by qRT-PCR experiments. A housekeeping gene, OsUBI1, was used for normalization. Data represent mean ± standard deviations (SD) of three biological replicates. Statistical analysis was performed with Student’s t-test (ns, not significant; *P ≤ 0.05; **P ≤ 0.01).
Discussion
The plant GH3 gene family contains members encoding acyl acid-amido synthetases that can modulate phytohormone levels by conjugating IAA, JA, or SA to amino acids (Staswick et al., 2005). To explore a full evolutionary history of this gene family, we identified a total of 250 GH3 genes from 22 green plant species and further reconstructed a phylogeny (Figure 1A and Supplementary Figure 1). It turned out that an ancestral GH3 gene had originated in the algal ancestors of land plants, and it further diverged into two lineages in bryophytes, as represented by two GH3 genes in the moss P. patens. Enzyme activity assays had revealed that both PpGH3.1 and PpGH3.2 can conjugate IAA and knock-out either PpGH3 gene increased plant sensitivity to auxin, with the mutants accumulating higher levels of free IAAs than wild type plants (Ludwig-Müller et al., 2009).
Among the 13 GH3 genes in rice genome, seven appeared to be expressed in roots (Supplementary Figure 2A), and qRT-PCR results not only confirmed previous transcriptome data that OsGH3.2 is induced during AM symbiosis (Fiorilli et al., 2015; Gutjahr et al., 2015; Roth et al., 2018), but further revealed that OsGH3.2 expression level is correlated with arbuscule abundance levels at different symbiotic stages (Figures 2A, 4B). In root fragments containing individual infection units at 17-dpi, OsGH3.2 promoter activity was mainly detected in colonized cortical cells that had formed mature arbuscules below or near to the hyphopodia (Figures 2D–F). It is interesting to observe no OsGH3.2 promoter activity at the foreside of running hyphae toward root tips, although some cortical cells had formed mature arbuscules (Figures 2D–F). Such observation indicated that the OsGH3.2 expression is not synchronical to arbuscule maturation, but is after mature arbuscules have been developed. It could therefore explain the low expression level of OsGH3.2 at 3-wpi (Figure 2A), at which time mature arbuscules were not extensively formed in wild-type rice roots (Figure 4B). Similar expression pattern was also revealed for MtGH3.6 in Medicago. According to the transcriptome data reported recently, MtGH3.6 showed no obvious induction at 8-dpi but was induced 64-fold at 23-dpi during AM symbiosis (Luginbuehl et al., 2017). It is possible that these GH3 genes are quickly induced after the free auxin level in the arbusculated cells reaches certain threshold. Previously, both SlGH3.4 and OsGH3.2 were revealed to be rapidly induced by exogenous IAA treatment (Fu et al., 2011; Liao et al., 2015).
In plants, auxin is a major regulator of root development, especially for lateral root formation (Du and Scheres, 2018). In Arabidopsis, AtGH3.6/DFL1 negatively regulates lateral root formation (Nakazawa et al., 2001), while AtGH3.2 mutant (ydk1-D) had a short primary root with a reduced number of lateral roots (Takase et al., 2004). In rice, overexpressing OsGH3.8 displayed shorter and fewer adventitious roots (Ding et al., 2008) and overexpression of OsGH3.2 resulted in fewer but longer crown roots (Du et al., 2012). In this study, two Osgh3.2 mutant lines were generated via CRISPR/Cas9-mediated mutagenesis (Supplementary Figure 3) and the developmental roles of OsGH3.2 were explored since this gene showed asymbiotic expression in rice young lateral roots (Figure 2B). After 3-weeks hydroponic experiment, both Osgh3.2 mutants showed altered root architecture, with more CRs and LLRs developed, while the maximum length of CR was inhibited (Figures 3A,B). Increased levels of free IAA and its precursor were further detected in roots of Osgh3.2-1 mutant (Supplementary Figure 5), supporting that the phenotypic changes are related to auxin activities. It is also in contrast with the phenotypes observed in OsGH3.2 over-expression lines, which had reduced IAA levels (Du et al., 2012). Interestingly, no remarkable effect on plant growth and root development were observed in tomato slgh3.4 mutants, and based on qRT-PCR data, the gene was neither expressed in roots nor in above-ground tissues under non-mycorrhizal condition (Chen et al., 2021). However, a duplicated gene SlGH3.2 showed expression in tomato roots (Liao et al., 2015). It is possible that gene duplications had resulted in subfunctionalization in tomato.
After inoculation by R. irregularis, the altered root morphology was still observed in the Osgh3.2 mutants (Figures 3C,D), indicating that the architectural change is independent of AMF colonization. Similar alterations were also observed for the panicles. On reaching maturity, the mutant panicles exhibited more primary branches and secondary branches, with the grain length also increased (Supplementary Figure 6).
We conducted a time-course inoculation experiment to investigate whether AM symbiosis would be affected in the Osgh3.2 mutants. It was revealed that both mutants exhibited higher colonization levels than wild-type from 1- to 5-wpi by R. irregularis (Figures 4A–C). The altered root morphology in Osgh3.2 mutants likely provided more potential colonization sites for fungal hyphae, thus promoted AMF colonization. Previously in tomato, the auxin hyper-transporting mutant pct also showed more lateral roots and enhanced fungal colonization (Hanlon and Coenen, 2011). Additionally, examining OsD17/CCD7 expressions at 3-wpi revealed that both Osgh3.2 mutants expressed this SL-biosynthesis gene several-fold higher than wild-type rice (Figure 6C). The mutants could therefore have more SLs to attract AMF and establish colonization more successfully. Interestingly, like OsPT11, OsD17 expression levels in both wild-type and Osgh3.2 mutants exhibited a good correlation with arbuscular abundance levels at later time points, indicating a potential role of OsD17 and SLs in arbuscule development. Previous study by Floss et al. (2008) showed that in Medicago, gene DXS2 [located at upstream of methylerythritol phosphate (MEP) pathway and SL biosynthesis] is mainly expressed in the arbusculated cells, suggesting that the arbuscule development would potentially need SLs. Further study would be required to explore whether OsD17 is also expressed in the arbusculated cells. Also, in pea, an auxin-deficient mutant bsh expressed another SL-biosynthesis gene, CCD8 at much lower level than wild-type and showed reduced colonization by AMF (Foo, 2013). It remains uncertain whether the altered root architecture, or the changed expression of SL-biosynthesis genes (or even both) leaded to the elevated colonization rate by AMF in the Osgh3.2 mutants, but it is unlikely caused by the symbiotic expression of OsGH3.2 in the colonized cortical cells, at least at early symbiotic stages (1- and 3-wpi) when OsGH3.2 had not been extensively expressed in mycorrhizal roots (Figure 2A). Such results would provide new thoughts on the tomato Slgh3.4 phenotype, which showed higher arbuscule incidence but comparable total colonization rates to wild-type at 5-wpi by AMF (Chen et al., 2021). Examining the symbiosis at earlier time points would help to elucidate when the arbuscule incidence started to increase in the Slgh3.4 mutants.
It has been demonstrated that arbuscular development in cortical cells requires auxin signaling responses (Etemadi et al., 2014). In tomato, over-expressing SlGH3.4 also resulted in underdeveloped arbuscules, likely due to insufficient cellular auxin levels (Chen et al., 2021). It is so far unclear on the fluctuations of free auxin levels in colonized cortical cells at different symbiotic stages. However, based on the demonstrated roles of OsGH3.2 in conjugating IAA and reducing free IAA levels (Fu et al., 2011) and the induction pattern of OsGH3.2 in cortical cells (Figure 2), we assume that the auxin levels in the colonized cortical cells would be down-regulated by OsGH3.2 to maintain a cellular auxin level homeostasis after mature arbuscule have developed. This assumed function explains why the expression level of OsGH3.2 is highly correlated with arbuscule abundance levels, which both peaked at 7-wpi (Figures 2A, 4B). At 9-wpi, when the arbuscules largely entered into degeneration phase in wild-type rice roots, the expression level of OsGH3.2 also decreased (Figures 2A, 4B). Loss function of OsGH3.2, although did not impair arbuscule development, however, seemed to affect (or delay) the degeneration phase, as arbuscule abundance level further increased in the Osgh3.2 mutants at 9-wpi, with fewer degenerating arbuscules and vesicles observed than wild-type (Figures 4B,C,E). Most likely, the Osgh3.2 mutants were not able to down-regulate auxin levels in the colonized cortical cells to the extend as in wild-type. However, it remains unclear why plants need to induce the GH3 genes to control the cellular auxin levels. One possibility is that elevated auxin levels may bring plants potential “hurt or danger,” e.g., pathogen invasion. Studies on OsGH3.8 and OsGH3.2 genes had revealed functions of suppressing IAA-induced expressions of cell wall expansin genes, consequently enhancing plant basal immunity to pathogens (Ding et al., 2008; Fu et al., 2011). Another possibility is that a lower auxin level would be advantageous to arbuscule degeneration, since previous studies have revealed that auxin signaling responses are required for arbuscule development in rice (Etemadi et al., 2014). Therefore, it is logical to deduce that the reduced IAA levels in the arbuscule-containing cells would hinder further arbuscule development and accelerate their degeneration.
A previous study in cucumber had revealed that the cortical cells containing the AM fungus were significantly larger than the corresponding cortical cells from non-mycorrhizal roots (Balestrini et al., 2005). Considering the role of auxin in cell expansion (Du et al., 2020), and OsGH3.2 likely has a symbiotic function of down-regulating auxin levels in colonized cortical cells, we wonder whether this gene, when mutated, would affect cell sizes as well as arbuscule sizes. At 3-wpi, we examined root fragments containing individual infection units and measured both colonized and neighboring non-colonized cortical cells (Figure 5C). Although a longer longitudinal length of colonized cells than non-colonized cells was detected in each genotype, there were no statistically significant differences for the mean longitudinal lengths of either colonized or non-colonized cortical cells between wild-type and the two mutants (Figure 5C). This is consistent with the rather low expression of OsGH3.2 among colonized cells at this early stage (Figure 2A). At 7- and 9-wpi, however, both Osgh3.2 mutants had increased cortical cell lengths (Figures 5E,H) and larger arbuscules (Figures 5D,G) than wild-type, likely because the mutants had no functional OsGH3.2 proteins to inhibit the auxin effects in the colonized cells. When only 15% of the largest arbuscules were considered, the relative ratio of arbuscule length to their cortical cell length showed no differences between wild-type and the mutants at 7-wpi (Figure 5F), therefore suggesting that arbuscular size change observed in the mutants is most likely an indirect consequence of enlarged cortical cell sizes. At 9-wpi, as more arbuscules were degenerating in wild-type, the size ratio of arbuscule to cortical cell also showed differences between wild-type and the mutants (Figure 5I). Taken together, these data suggested that AM symbiosis can induce the expression of OsGH3.2 in cortical cells that have formed mature arbuscules and further affect the longitudinal length of colonized cells by down-regulating cellular auxin levels. Previously, another study in M. truncatula had also revealed that AMF are able to modulate root cortical cell development by activating a GRAS-domain transcription factor, MIG1, which can determine both the longitudinal and radical sizes of cortical cells (Heck et al., 2016).
In summary, by paying attention to the plant GH3 gene family, this study revealed that OsGH3.2, a GH3 subgroup II-A member, can not only modulate rice root architecture and further affect colonization levels by AMF, but also can modulate colonized cortical cell longitudinal lengths and consequently affect arbuscule sizes.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
BW and C-CL planned and designed the research, wrote the manuscript. C-CL and Y-NL performed most of the experiments, analyzed the data, and contributed equally to this work. J-FC helped in quantifying colonization rates and measuring cell/arbuscule lengths. RG and LT helped in conducting fieldwork and analyzing data. All authors contributed to the article and approved the submitted version.
Funding
This research was financed by National Natural Science Foundation of China (31870203) to BW.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Many thanks are given to Dong Wang at University of Massachusetts for reading the manuscript and providing valuable comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.853435/full#supplementary-material
Supplementary Figure 1 | A phylogenetic tree of plant GH3 family containing 250 genes identified from 22 plant species. The phylogeny was reconstructed using a maximum-likelihood method under the JTT + I + G4 model via IQ-TREE (Trifinopoulos et al., 2016). The robustness of internal branches was evaluated by calculating the Shimodaira–Hasegawa approximate likelihood ratio test (SH-aLRT) and by performing 1000 ultrafast bootstrap replicates. Six bacterial GH3 genes were designated as the outgroup. The bar represents number of nucleotide substitutions per site.
Supplementary Figure 2 | The expression patterns of 13 OsGH3 genes in rice roots during AM symbiosis. The transcript abundances of 13 OsGH3 genes in rice roots at 3- and 5-wpi either by mock solution (–) or by R. irregularis (+) were measured by RT-PCR first. (A) Apart from OsGH3.2, six OsGH3 genes appear to be expressed in rice roots, including OsGH3.3, OsGH3.5, OsGH3.7, OsGH3.8, OsGH3.11, and OsGH3.13. (B) Relative expression levels of the six OsGH3 genes were further measured by quantitative RT-PCR. The gene expression values were normalized to the rice housekeeping gene ubiquitin 1 (OsUBI1). Data represent mean ± standard deviations (SD) of three biological replicates. Statistical analysis was performed with Student’s t-test (ns, not significant).
Supplementary Figure 3 | CRISPR/Cas9-mediated OsGH3.2 gene mutagenesis in rice. (A) The OsGH3.2 gene has three exons, with the GH3 domain (PF03321)-encoding region shown in orange. The target sites (red vertical rectangles) of two designed single-guide RNAs (sgRNAs) were located on the third exon. The red and green letters represent the target sequences and the protospacer adjacent motif (PAM) sites, respectively. (B) A total of six mutated alleles were identified, from which two alleles, Osgh3.2-1 and Osgh3.2-2, were chosen to further obtain their homozygotes. (C) The deduced proteins of wild-type OsGH3.2 and two mutated alleles via Pfam (http://pfam.xfam.org) were shown.
Supplementary Figure 4 | Osgh3.2 mutants showed altered root morphology. Rice roots of wild-type, Osgh3.2-1 and Osgh3.2-2 after hydroponic culture (A) or inoculation by R. irregularis (C) for 1 and 2 weeks. Scale bars, 1 cm. (B,D) Maximum length of crown roots (CRs), number of large lateral roots (LLRs) and number of CRs were compared between wild type rice and the Osgh3.2 mutants under two cultivation conditions, respectively. Data are individual values from 4 to 5 biological replicates. The horizontal lines indicate the mean values. Different letters indicate statistically significant differences (Student’s t-test, P < 0.05).
Supplementary Figure 5 | Osgh3.2 mutants showed higher free IAA and tryptophan levels. (A,B) Free IAA and tryptophan (the IAA synthetic precursor) levels were measured in roots of both wild-type rice and Osgh3.2-1 mutant after hydroponic culture, respectively. Data are individual values from three biological replicates and the horizontal line shows the mean value. Statistical analysis was performed with Student’s t-test. Different letters indicate statistically significant differences (P < 0.05).
Supplementary Figure 6 | Osgh3.2 mutants exhibited altered panicle architecture and longer grain length. (A,B) Images of mature panicles and grains of wild-type rice and the Osgh3.2 mutants. Scale bars, 1 cm. (C) Statistical analysis of panicle length, primary panicle branches and secondary panicle branches among wild -type and Osgh3.2 mutants, six biological replicates were included. (D) Statistical analysis of grain length, grain width and grain thickness among wild type and Osgh3.2 mutants. Data are individual values from six biological replicates; the horizontal lines indicate the mean values. Different letters indicate statistically significant differences (Student’s t-test, P < 0.05).
Supplementary Figure 7 | An exemplar image of rice root fragment colonized by R. irregularis after stained with ink-vinegar solution. The image showed how the longitudinal lengths of arbuscules and cells were measured. Scale bar = 50 μm.
Supplementary Figure 8 | Arbuscule populations at 7- and 9-wpi by R. irregularis in roots of wild-type rice and the Osgh3.2 mutants. (A,B) The longitudinal lengths of 300 arbuscules from three rice individuals were measured (100 arbuscules per individual) at 7- and 9-wpi, respectively. Arbuscules were classified into three categories according to the longitudinal length, small/degenerate (<30 μm), middle (30–50 μm), and large (>50 μm). Data represent means ± standard deviation (SD). Different letters indicate significant differences (Student’s t-test, P < 0.05).
Supplementary Table 1 | The 250 GH3 genes identified from 26 surveyed plant genomes.
Supplementary Table 2 | The primers used in this study.
Footnotes
References
Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. doi: 10.1038/nature03608
Balestrini, R., Cosgrove, D. J., and Bonfante, P. (2005). Differential location of α-expansin proteins during the accommodation of root cells to an arbuscular mycorrhizal fungus. Planta 220, 889–899. doi: 10.1007/s00425-004-1431-2
Buendia, L., Ribeyre, C., Bensmihen, S., and Lefebvre, B. (2019). Brachypodium distachyon tar2lhypo mutant shows reduced root developmental response to symbiotic signal but increased arbuscular mycorrhiza. Plant Signal. Behav. 14:e1651608. doi: 10.1080/15592324.2019.1651608
Calabrese, S., Kohler, A., Niehl, A., Veneault-Fourrey, C., Boller, T., and Courty, P. E. (2017). Transcriptome analysis of the Populus trichocarpa-Rhizophagus irregularis mycorrhizal symbiosis: regulation of plant and fungal transportomes under nitrogen starvation. Plant Cell Physiol. 58, 1003–1017. doi: 10.1093/pcp/pcx044
Casanova-Sáez, R., and Voß, U. (2019). Auxin metabolism controls developmental decisions in land plants. Trends Plant Sci. 24, 741–754. doi: 10.1016/j.tplants.2019.05.006
Chen, C. Y., Ané, J. M., and Zhu, H. Y. (2008). OsIPD3, an ortholog of the Medicago truncatula DMI3 interacting protein IPD3, is required for mycorrhizal symbiosis in rice. New Phytol. 180, 311–315. doi: 10.1111/j.1469-8137.2008.02612.x
Chen, X., Chen, J. D., Liao, D. H., Ye, H. H., Li, C., Luo, Z. Z., et al. (2021). Auxin-mediated regulation of arbuscular mycorrhizal symbiosis: a role of SlGH3. 4 in tomato. Plant Cell Environ. [Epub ahead of print], doi: 10.1111/pce.14210
Ding, X. H., Cao, Y. L., Huang, L. L., Zhao, J., Xu, C. G., Li, X. H., et al. (2008). Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20, 228–240. doi: 10.1105/tpc.107.055657
Du, H., Wu, N., Fu, J., Wang, S. P., Li, X. H., Xiao, J. H., et al. (2012). A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 63, 6467–6480. doi: 10.1093/jxb/ers300
Du, M., Spalding, E. P., and Gray, W. M. (2020). Rapid auxin-mediated cell expansion. Annu. Rev. Plant Biol. 71, 379–402. doi: 10.1146/annurev-arplant-073019-025907
Du, Y., and Scheres, B. (2018). Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 69, 155–167. doi: 10.1093/jxb/erx223
Etemadi, M., Gutjahr, C., Couzigou, J. M., Zouine, M., Lauressergues, D., Timmers, A., et al. (2014). Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 166, 281–292. doi: 10.1104/pp.114.246595
Fiorilli, V., Vallino, M., Biselli, C., Faccio, A., Bagnaresi, P., and Bonfante, P. (2015). Host and non-host roots in rice: cellular and molecular approaches reveal differential responses to arbuscular mycorrhizal fungi. Front. Plant Sci. 6:636. doi: 10.3389/fpls.2015.00636
Floss, D. S., Hause, B., Lange, P. R., Kuster, H., Strack, D., and Walter, M. H. (2008). Knock-down of the MEP pathway isogene 1-deoxy-d-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. Plant J. 56, 86–100. doi: 10.1111/j.1365-313X.2008.03575.x
Foo, E. (2013). Auxin influences strigolactones in pea mycorrhizal symbiosis. J. Plant Physiol. 170, 523–528. doi: 10.1016/j.jplph.2012.11.002
Foo, E., Ross, J. J., Jones, W. T., and Reid, J. B. (2013). Plant hormones in arbuscular mycorrhizal symbioses: an emerging role for gibberellins. Ann. Bot. 111, 769–779. doi: 10.1093/aob/mct041
Fu, J., Liu, H. B., Li, Y., Yu, H. H., Li, X. H., Xiao, J. H., et al. (2011). Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 155, 589–602. doi: 10.1104/pp.110.163774
Garcia, K., Chasman, D., Roy, S., and Aneì, J. M. (2017). Physiological responses and gene co-expression network of mycorrhizal roots under K+ deprivation. Plant Physiol. 173, 1811–1823. doi: 10.1104/pp.16.01959
Genre, A., Chabaud, M., Balzergue, C., Puech-Pagès, V., Novero, M., Rey, T., et al. (2013). Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 198, 179–189. doi: 10.1111/nph.12146
Genre, A., Chabaud, M., Faccio, A., Barker, D. G., and Bonfante, P. (2008). Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20, 1407–1420. doi: 10.1105/tpc.108.059014
Genre, A., Chabaud, M., Timmers, T., Bonfante, P., and Barker, D. G. (2005). Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago root epidermal cells before infection. Plant Cell 17, 3489–3499. doi: 10.1105/tpc.105.035410
Genre, A., Ivanov, S., Fendrych, M., Faccio, A., Zarsky, V., Bisseling, T., et al. (2012). Multiple exocytotic markers accumulate at the sites of perifungal membrane biogenesis in arbuscular mycorrhizas. Plant Cell Physiol. 53, 244–255. doi: 10.1093/pcp/pcr170
Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pagès, V., Dun, E. A., Pillot, J. P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455, 189–194.
Guillotin, B., Etemadi, M., Audran, C., Bouzayen, M., Bécard, G., and Combier, J. P. (2017). Sl-IAA27 regulates strigolactone biosynthesis and mycorrhization in tomato (var. MicroTom). New Phytol. 213, 1124–1132. doi: 10.1111/nph.14246
Gutjahr, C. (2014). Phytohormone signaling in arbuscular mycorhiza development. Curr. Opin. Plant Biol. 20, 26–34. doi: 10.1016/j.pbi.2014.04.003
Gutjahr, C., Sawers, R. J. H., Marti, G., Andrés-Hernández, L., Yang, S. Y., Casieri, L., et al. (2015). Transcriptome diversity among rice root types during asymbiosis and interaction with arbuscular mycorrhizal fungi. Proc. Natl. Acad. Sci. 112, 6754–6759. doi: 10.1073/pnas.1504142112
Hagen, G., and Guilfoyle, T. J. (1985). Rapid induction of selective transcription by auxins. Mol. Cell. Biol. 5, 1197–1203. doi: 10.1128/mcb.5.6.1197-1203.1985
Hanlon, M. T., and Coenen, C. (2011). Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytol. 189, 701–709. doi: 10.1111/j.1469-8137.2010.03567.x
Heck, C., Kuhn, H., Heidt, S., Walter, S., Rieger, N., and Requena, N. (2016). Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr. Biol. 26, 2770–2778. doi: 10.1016/j.cub.2016.07.059
Herrera-Medina, M. J., Gagnon, H., Piché, Y., Ocampo, J. A., Garrido, J. M. G., and Vierheilig, H. (2003). Root colonization by arbuscular mycorrhizal fungi is affected by the salicylic acid content of the plant. Plant Sci. 164, 993–998. doi: 10.1016/s0168-9452(03)00083-9
Herrera-Medina, M. J., Steinkellner, S., Vierheilig, H., Bote, J. A. O., and Garrido, J. M. G. (2007). Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol. 175, 554–564. doi: 10.1111/j.1469-8137.2007.02107.x
Isayenkov, S., Mrosk, C., Stenzel, I., Strack, D., and Hause, B. (2005). Suppression of allene oxide cyclase in hairy roots of Medicago truncatula reduces jasmonate levels and the degree of mycorrhization with Glomus intraradices. Plant Physiol. 139, 1401–1410. doi: 10.1104/pp.105.069054
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liao, D. H., Chen, X., Chen, A. Q., Wang, H. M., Liu, J. J., Liu, J. L., et al. (2015). The characterization of six auxin-induced tomato GH3 genes uncovers a member, SlGH3.4, strongly responsive to arbuscular mycorrhizal symbiosis. Plant Cell Physiol. 56, 674–687. doi: 10.1093/pcp/pcu212
Liu, Y. N., Liu, C. C., Zhu, A. Q., Niu, K. X., Guo, R., Tian, L., et al. (2021). OsRAM2 function in lipid biosynthesis is required for arbuscular mycorrhizal symbiosis in rice. Mol. Plant Microbe Interact. doi: 10.1094/MPMI-04-21-0097-R
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative gene expression data using real-time quantitative PCR and the 2–Δ Δ CT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ludwig-Müller, J. (2011). Auxin conjugates: their role for plant development and in the evolution of land plants. J. Exp. Bot. 62, 1757–1773. doi: 10.1093/jxb/erq412
Ludwig-Müller, J., Jülke, S., Bierfreund, N. M., Decker, E. L., and Reski, R. (2009). Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New phytol. 181, 323–338. doi: 10.1111/j.1469-8137.2008.02677.x
Luginbuehl, L. H., Menard, G. N., Kurup, S., Van Erp, H., Radhakrishnan, G. V., Breakspear, A., et al. (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178. doi: 10.1126/science.aan0081
Maillet, F., Poinsot, V., Andre, O., Puech-Pages, V., Haouy, A., Gueunier, M., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–63. doi: 10.1038/nature09622
McGonigle, T. P., Miller, M. H., Evans, D. G., Fairchild, G. L., and Swan, J. A. (1990). A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115, 495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x
Nagahashi, G., and Douds, D. D. (2011). The effects of hydroxy fatty acids on the hyphal branching of germinated spores of AM fungi. Fungal Biol. 115, 351–358. doi: 10.1016/j.funbio.2011.01.006
Naito, Y., Hino, K., Bono, H., and Ui-Tei, K. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123. doi: 10.1093/bioinformatics/btu743
Nakazawa, M., Yabe, N., Ichikawa, T., Yamamoto, Y. Y., Yoshizumi, T., Hasunuma, K., et al. (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25, 213–221. doi: 10.1046/j.1365-313x.2001.00957.x
Okrent, R. A., and Wildermuth, M. C. (2011). Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Mol. Biol. 76, 489–505. doi: 10.1007/s11103-011-9776-y
Pozo, M. J., López-Ráez, J. A., Azcón-Aguilar, C., and García-Garrido, J. M. (2015). Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol. 205, 1431–1436. doi: 10.1111/nph.13252
Roth, R., Chiapello, M., Montero, H., Gehrig, P., Grossmann, J., O’Holleran, K., et al. (2018). A rice Serine/Threonine receptor-like kinase regulates arbuscular mycorrhizal symbiosis at the peri-arbuscular membrane. Nat. Commun. 9:4677. doi: 10.1038/s41467-018-06865-z
Schaffer, G. F., and Peterson, R. L. (1993). Modifications to clearing methods used in combination with vital staining of roots colonized with vesicular-arbuscular mycorrhizal fungi. Mycorrhiza 4, 29–35. doi: 10.1007/bf00203248
Smith, S. E., and Smith, F. A. (2011). Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 62, 227–250. doi: 10.1146/annurev-arplant-042110-103846
Spatafora, J. W., Chang, Y., Benny, G. L., Lazarus, K., Smith, M. E., Berbee, M. L., et al. (2016). A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108, 1028–1046. doi: 10.3852/16-042
Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M. T., Maldonado, M. C., et al. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. The Plant Cell 17, 616–627. doi: 10.1105/tpc.104.026690
Staswick, P. E., Tiryaki, I., and Rowe, M. L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. doi: 10.1105/tpc.000885
Takase, T., Nakazawa, M., Ishikawa, A., Kawashima, M., Ichikawa, T., Takahashi, N., et al. (2004). ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 37, 471–483. doi: 10.1046/j.1365-313x.2003.01973.x
Terol, J., Domingo, C., and Talón, M. (2006). The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371, 279–290. doi: 10.1016/j.gene.2005.12.014
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Vierheilig, H., Coughlan, A. P., Wyss, U., and Piche, Y. (1998). Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 64, 5004–5007. doi: 10.1128/AEM.64.12.5004-5007.1998
Wang, B., and Qiu, Y. L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16, 299–363. doi: 10.1007/s00572-005-0033-6
Wang, S. S., Chen, A. Q., Xie, K., Yang, X. F., Luo, Z. Z., Chen, J. D., et al. (2020). Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc. Natl. Acad. Sci. U.S.A. 117, 16649–16659. doi: 10.1073/pnas.2000926117
Wang, W. X., Shi, J. C., Xie, Q. J., Jiang, Y. N., Yu, N., and Wang, E. T. (2017). Nutrient exchange and regulation in arbuscular mycorrhizal symbiosis. Mol. Plant 10, 1147–1158. doi: 10.1016/j.molp.2017.07.012
Watts-Williams, S. J., Emmett, B. D., Levesque-Tremblay, V., MacLean, A. M., Sun, X., Satterlee, J. W., et al. (2019). Diverse Sorghum bicolor accessions show marked variation in growth and transcriptional responses to arbuscular mycorrhizal fungi. Plant Cell Environ. 42, 1758–1774. doi: 10.1111/pce.13509
Westfall, C. S., Zubieta, C., Herrmann, J., Kapp, U., Nanao, M. H., and Jez, J. M. (2012). Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 336, 1708–1711. doi: 10.1126/science.1221863
Wipf, D., Krajinski, F., van Tuinen, D., Recorbet, G., and Courty, P. E. (2019). Trading on the arbuscular mycorrhiza market: from arbuscules to common mycorrhizal networks. New Phytol. 223, 1127–1142. doi: 10.1111/nph.15775
Keywords: rice (Oryza sativa), arbuscular mycorrhiza (AM), OsGH3.2, auxin, root architecture, cortical cell length, arbuscular size
Citation: Liu C-C, Liu Y-N, Cheng J-F, Guo R, Tian L and Wang B (2022) Dual Roles of OsGH3.2 in Modulating Rice Root Morphology and Affecting Arbuscular Mycorrhizal Symbiosis. Front. Plant Sci. 13:853435. doi: 10.3389/fpls.2022.853435
Received: 12 January 2022; Accepted: 14 March 2022;
Published: 11 April 2022.
Edited by:
Andrea Genre, University of Turin, ItalyReviewed by:
Benoit Lefebvre, Institut National de la Recherche Agronomique de Toulouse, FranceJose Manuel García-Garrido, Department of Soil Microbiology and Symbiotic Systems, Experimental Station of Zaidín (CSIC), Spain
Copyright © 2022 Liu, Liu, Cheng, Guo, Tian and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Wang, Ymlud2FuZ0BuanUuZWR1LmNu
†These authors have contributed equally to this work
 Cheng-Chen Liu†
Cheng-Chen Liu† Bin Wang
Bin Wang