- MOE Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou, China
SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins assemble to drive the final membrane fusion step of membrane trafficking. Thus, SNAREs are essential for membrane fusion and vesicular trafficking, which are fundamental mechanisms for maintaining cellular homeostasis. In plants, SNAREs have been demonstrated to be located in different subcellular compartments and involved in a variety of fundamental processes, such as cytokinesis, cytoskeleton organization, symbiosis, and biotic and abiotic stress responses. In addition, SNAREs can also contribute to the normal growth and development of Arabidopsis. Here, we review recent progress in understanding the biological functions and signaling network of SNAREs in vesicle trafficking and the regulation of root growth and development in Arabidopsis.
Introduction
Plant cells contain multiple membrane-bound organelles, each of which contains a unique set of lipids and proteins that play different functions within and between cells. Membrane transport pathways connect these organelles, which are important for maintaining cell function and responding to various environmental stimuli. Vesicle trafficking involves vesicle formation, vesicle translocation, vesicle binding, and fusion of vesicles with target compartments. Vesicle formation is the process of vesicle bud formation from the donor compartment for cargo packaging and is mediated by the coat protein complex I (COPI), COPII and clathrin, and the small GTPases secretion-associated RAS super family 1 (Sar1) and ADP-ribosylation factor 1 (Arf1; Bremser et al., 1999; Bonifacino and Lippincott-Schwartz, 2003; Bonifacino and Glick, 2004). Vesicle translocation is mediated by motor proteins that propel vesicles along the cytoskeleton (Kamal and Goldstein, 2002). Tethering and Rab proteins regulate the docking of vesicles and receptor compartments (Grosshans et al., 2006; Verhage and Sørensen, 2008). Fusion is the final step of vesicle transport, mediated by the SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein family (Hong, 2005; Chen et al., 2021).
SNAREs are a highly conserved superfamily of proteins that mediate vesicle transport between endosomes and the trafficking to the plasma membrane of all eukaryotic cells (Jahn and Scheller, 2006). Some SNARE proteins in plants have been found in various intracellular trafficking pathways and are involved in other physiological processes (Uemura et al., 2004), such as cell cytokinesis (Lauber et al., 1997; Collins et al., 2003; El Kasmi et al., 2013;Park et al., 2018), defense responses (Kwon et al., 2008, 2020; Gu et al., 2017; Yun and Kwon, 2017; Kim et al., 2019; Wang et al., 2020), shoot and root gravitropism (Kato et al., 2002; Morita et al., 2002; Yano et al., 2003; Gu et al., 2021), osmotic stress tolerance (Zhu et al., 2002), salt stress responses (Salinas-Cornejo et al., 2021; Sun et al., 2021), and ion channel regulation (Honsbein et al., 2009; Zhang et al., 2015a, 2020; Waghmare et al., 2018).
Arabidopsis primary root growth and development are regulated by elongation and cell division (Beemster and Baskin, 1998; Beemster et al., 2002; Lavrekha et al., 2017; Vyplelová et al., 2017). SNAREs regulate vesicle trafficking and are necessary for root growth, for example, the vesicle trafficking of SNAREs is important for the transport of the auxin transporter PINFORMED (PIN) in roots (Shirakawa et al., 2010; Gu et al., 2021; Zhang et al., 2021). Cytokinesis, the final step of cell division, physically separates the two daughter cells (Frémont and Echard, 2018), and SNARE complexes of different components together mediate the membrane fusion of Arabidopsis cytokinesis (Zhang et al., 2011a; El Kasmi et al., 2013; Park et al., 2018). This review begins with a description of the SNARE protein and SNARE-related vesicle transport pathways. Then, we predominantly focus on recent insights into the regulation of Arabidopsis root growth and how SNAREs participate in cytokinesis.
Snare Proteins
The SNARE protein was first identified in the late 1980s and was quickly identified as a key element involved in membrane fusion (Oyler et al., 1989; Söllner et al., 1993). SNARE proteins form a superfamily of small proteins, there are 64 members in Arabidopsis, 25 members in Saccharomyces cerevisiae, and 36 members in humans (Jahn and Scheller, 2006; Zhang et al., 2020). The SNARE domain contains a SNARE motif with 60–70 amino acids and it consists of seven repeats that form a coiled-coil structure. Through hetero-oligomer interactions, SNAREs mediate the fusion of membranes and intracellular vesicle-related transport processes, which occur in vesicles, the inner membrane system of organelles, and the plasma membrane (PM; Lipka et al., 2007).
The relationship between SNAREs and the lipid bilayer occurs through the C-terminal transmembrane (TM) domain. Although most SNAREs are inserted into the cell membrane through transmembrane motifs, some SNAREs, such as Qbc-SNARE synaptosomal-associated 25 (SNAP25) and R-SNARE YKT6, are associated with posttranslational lipids of the peripheral junction membrane. In addition to the SNARE domain and C-terminal TM domain, many SNAREs also contain N-terminal regulatory motifs, which together with a series of accessory peptides control the activity of SNARE proteins in vivo (Fasshauer et al., 1998; Hong, 2005; Sutter et al., 2006; Lipka et al., 2007).
According to their subcellular localization, SNAREs are divided into two types: vesicle-associated (v-SNARE) and target membrane-associated (t-SNARE; Söllner et al., 1993; Jahn and Scheller, 2006). Depending on the glutamine (Q) or arginine (R) residue in the middle of the SNARE domain, it can be divided into Q- and R-SNAREs. In fact, t-SNAREs and v-SNAREs correspond to Q-SNAREs and R-SNAREs, respectively. R-SNAREs located on vesicles are usually called vesicle-associated membrane proteins (VAMPs). According to their sequence similarity, Q-SNAREs are divided into three subgroups: Qa-, Qb-, and Qc-SNAREs (Fasshauer et al., 1998; Jahn and Scheller, 2006; Lipka et al., 2007; Bock et al., 2011; Gu et al., 2020). Qa-SNAREs contain an autoregulation domain at the N-terminus, and this self-inhibitory domain is comprised of three helices, which are called Habc motifs in neuronal synapses. The Habc motif interacts with the SNARE domain in the same polypeptide. This intramolecular interaction is called the “close” conformation and prevents the assembly of Qa-SNARE with other SNAREs. Closed Qa-SNAREs unfold to form an active conformation (also called the “open”) and allow the formation of complexes. The Qa-Qb-Qc-cis SNARE complex and R-SNARE (v-SNARE) create a functional fusion of the trans-SNARE complex on the target membrane. After membrane fusion, SNARE complexes are transformed from a trans- to cis-configuration. Then, α-soluble NSF attachment protein (α-SNAP) mediates the breakdown of the SNARE complex by recruiting n-ethylmaleimide-sensitive factor (NSF) and activating its ATP enzyme activity, thereby releasing SNARE component units, and the cycle can then restart (Figure 1; Burgoyne and Morgan, 2007; Toonen and Verhage, 2007; Ryu et al., 2015; Zhao et al., 2015; Jun and Wickner, 2019; Chen et al., 2021; Song et al., 2021).
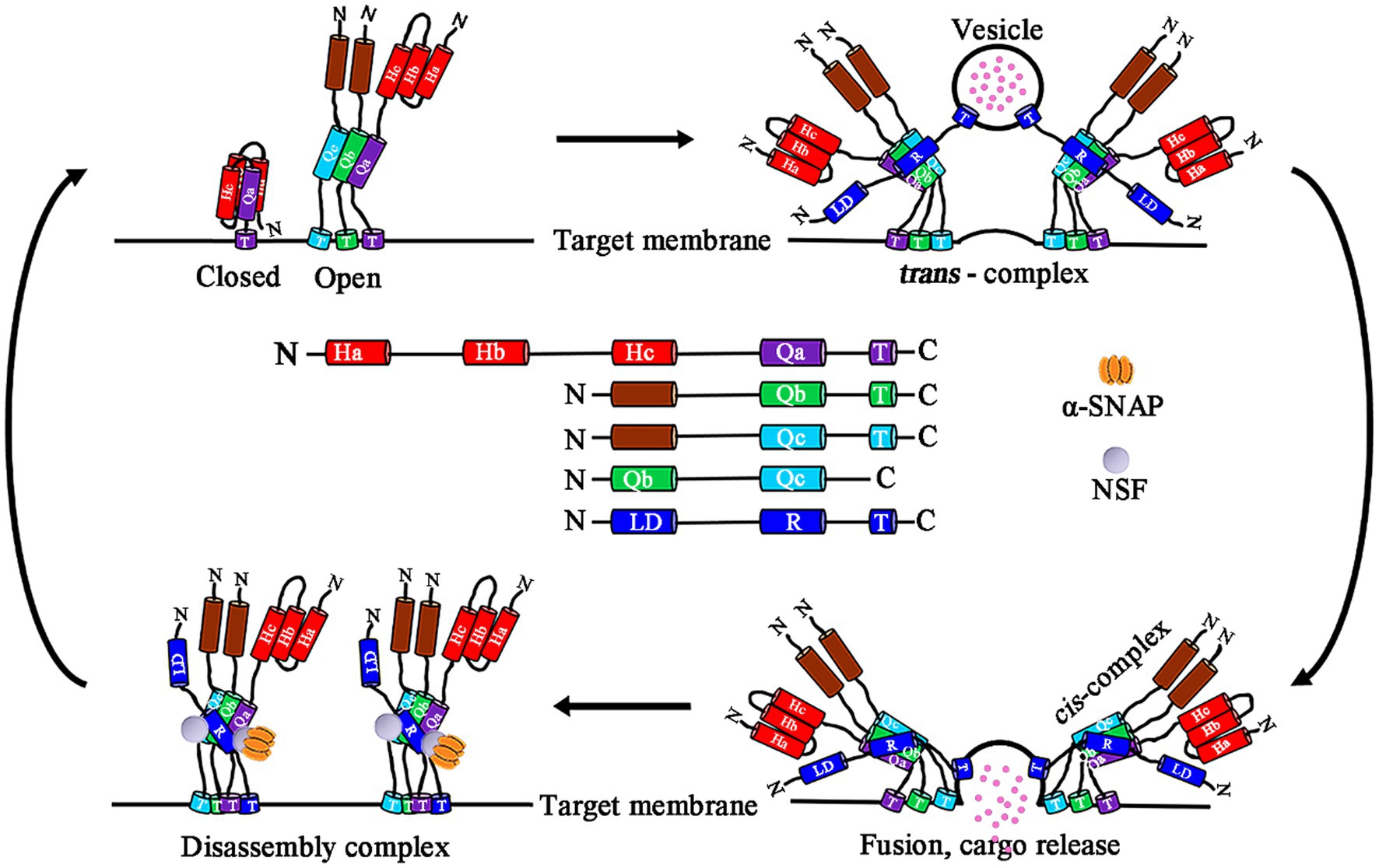
Figure 1. Schematic model domain structure of plant SNARE subfamilies and a model of SNARE complex vesicle fusion. Plant SNARE protein domains, including Qa, Qb, and Qc domains and regulatory N-terminal regions of Q-SNAREs, the R domain and the longin domain (LD) of R-SNAREs, and the C-terminal transmembrane helices (T), are present in most SNARE proteins. Qa-SNARE protein moving form a closed to open conformation that allows assembly with helices provided by membrane-associated Qb-, Qc-, and R-SNAREs. R-SNAREs reside in a vesicle. The assembly of a trans-SNARE complex is accompanied by an increase in the density of the core α-helical structure that transitions to the cis-complex and membrane fusion. Dissociation of the cis-complex requires the energy input of ATP hydrolysis and is achieved through the binding of α-SNAP and the NSF ATPase. Qa, Qb, Qc, and Q-SNARE three domains; R, R-SNARE domain; N, N-terminal; C, C-terminal; Ha, Hb, and Hc, the three helices of the Qa-SNARE N-terminal autoregulatory domain; LD, Longin domain; T, Transmembrane helices; α-SNAP, α-soluble NSF attachment protein; and NSF, n-ethylmaleimide-sensitive factor.
Snares in Trafficking Pathways
SNAREs are involved in different vesicle trafficking pathways (Figure 2). The majority of Arabidopsis SNAREs is located in a specific intracellular compartment, but some SNAREs have multiple patterns of localization in two or more organelles (Uemura et al., 2004; Sanderfoot, 2007; Bassham et al., 2008; Kim and Brandizzi, 2012). Various subcellular parts of plant cells form specific SNARE complexes that mediate various transport events (Kim and Brandizzi, 2012). SNAREs are key molecules involved in vesicle transport and membrane fusion, and they are also involved in different processes of the vesicle transport pathway, e.g., ER-Golgi anterograde/retrograde trafficking, trans-Golgi network (TGN) and post-Golgi trafficking, and the plasma membrane (Martinière and Moreau, 2020). SNAREs regulate transport in a complex membrane system, including endocytosis, secretory, and vacuolar trafficking steps in Arabidopsis (Sanderfoot, 2007; Saito and Ueda, 2009).
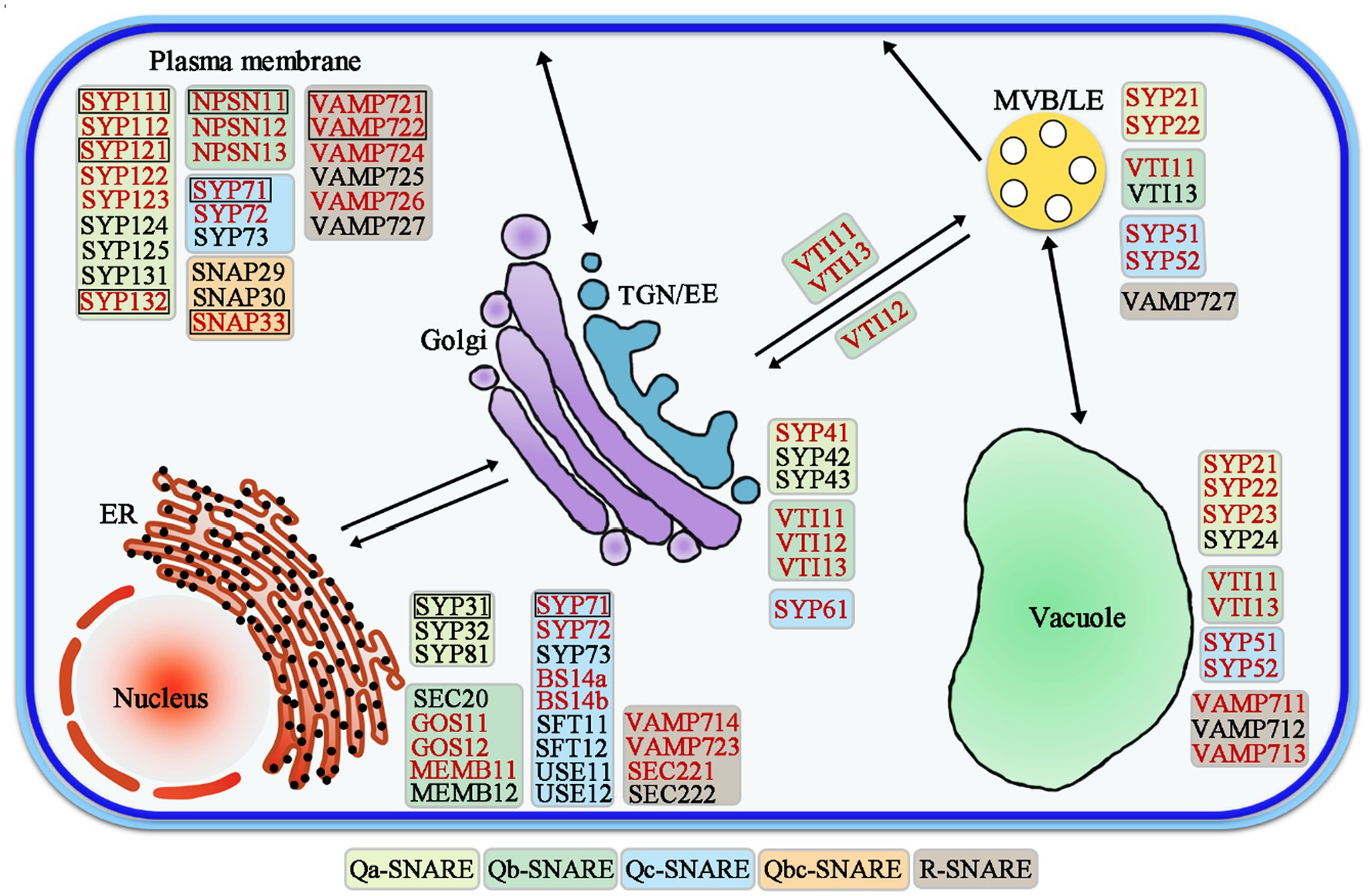
Figure 2. Schematic model of SNARE-mediated vesicle trafficking pathways in Arabidopsis. The genes highlighted in red are those that are highly expressed in roots. The black boxes mark the genes that are located on the cell plate. EE, Early endosome; LE, Late endosome; TGN, Trans-Golgi network; MVBs, Multivesicular bodies; and ER, Endoplasmic reticulum.
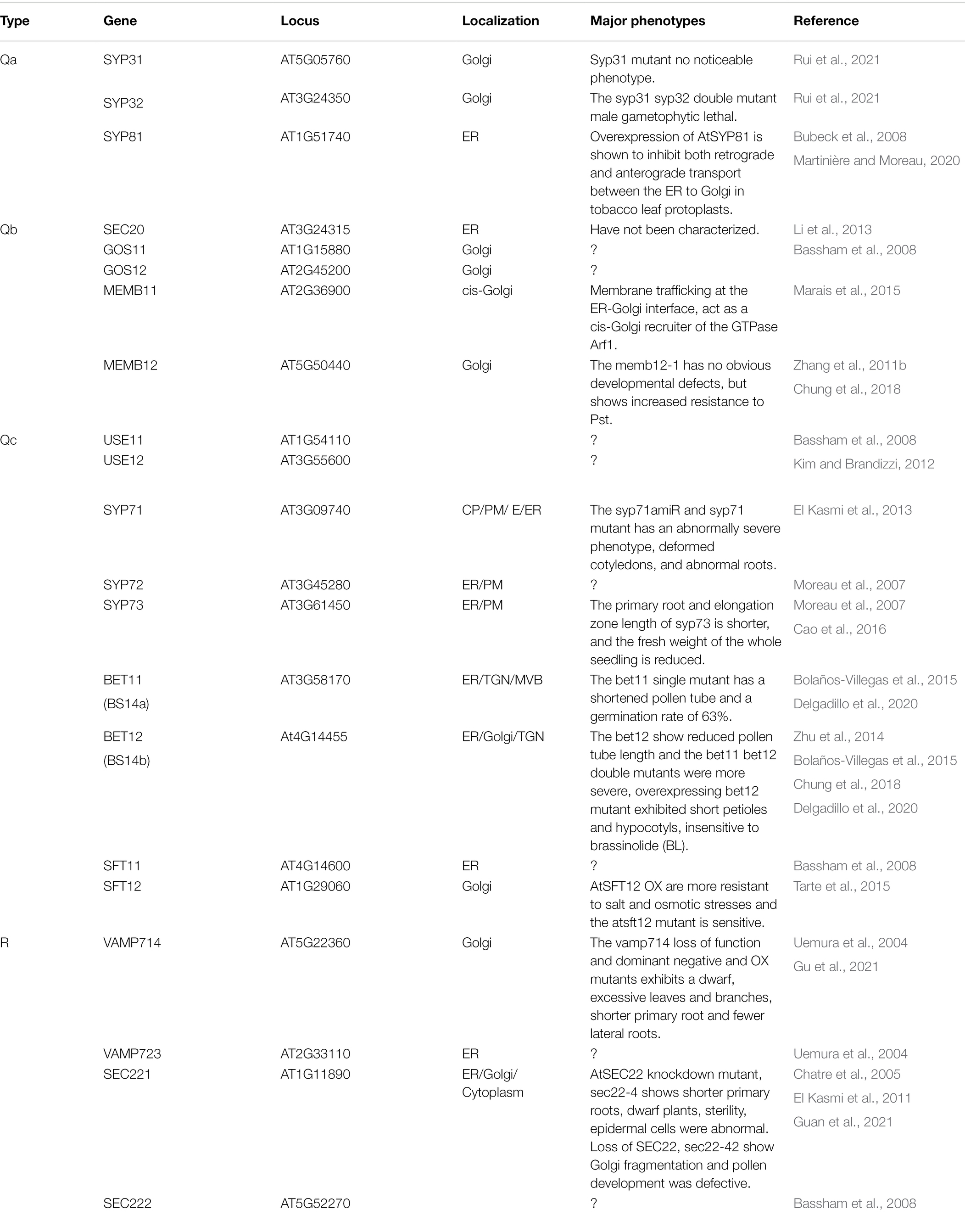
A total of 21 SNAREs are localized in the endoplasmic reticulum (ER) and Golgi (Figure 2; Table 1; Kim and Brandizzi, 2012). Qa-SNAREs syntaxin of plants 31 (SYP31) and SYP32 is located in the Golgi, and SYP31 is located at the formation cell plate in Arabidopsis (Sanderfoot et al., 2000; Rancour et al., 2002; Uemura et al., 2004). In pollen with syp31 syp32 double mutations, secretion of JIM7 (labeled highly methylesterified pectins)-positive vesicles from the Golgi/trans-Golgi network into the ectoplasm is blocked. Partial loss of the association of the endomembrane protein12 (EMP12) with the Golgi apparatus in syp31 syp32 double mutations pollen (Gao et al., 2012; Li et al., 2019; Rui et al., 2021). Furthermore, mCherry-HDEL trafficking between the ER and Golgi was disrupted, and it was mistargeted to vacuoles. SYP31 and SYP32 interact directly with conserved oligomeric Golgi 3 (COG3), which is a subunit of the COG complex and is responsible for its Golgi localization, indicating a role of SYP31/32 in intra-Golgi trafficking (Tan et al., 2016; Rui et al., 2021). Qc-SNARE BET12 localizes to both the Golgi and TGN, and it is involved in protein transport in the early secretory pathway. BET12 ectopic expression caused no inhibition in the ER-Golgi anterograde transport but caused intracellular accumulation of the antimicrobial protein PR1 (Pathogenesis-Related Gene 1). BET12 interacts with the Golgi-localized Qb-SNARE MEMB12, and MEMB12 overexpression accumulated PR1 in intracellular membranes. MEMB12 could be involved in retrograde protein trafficking from the Golgi back to the ER, and therefore, PR1 could be recycled to the ER instead of being secreted (Zhang et al., 2011b; Chung et al., 2018).
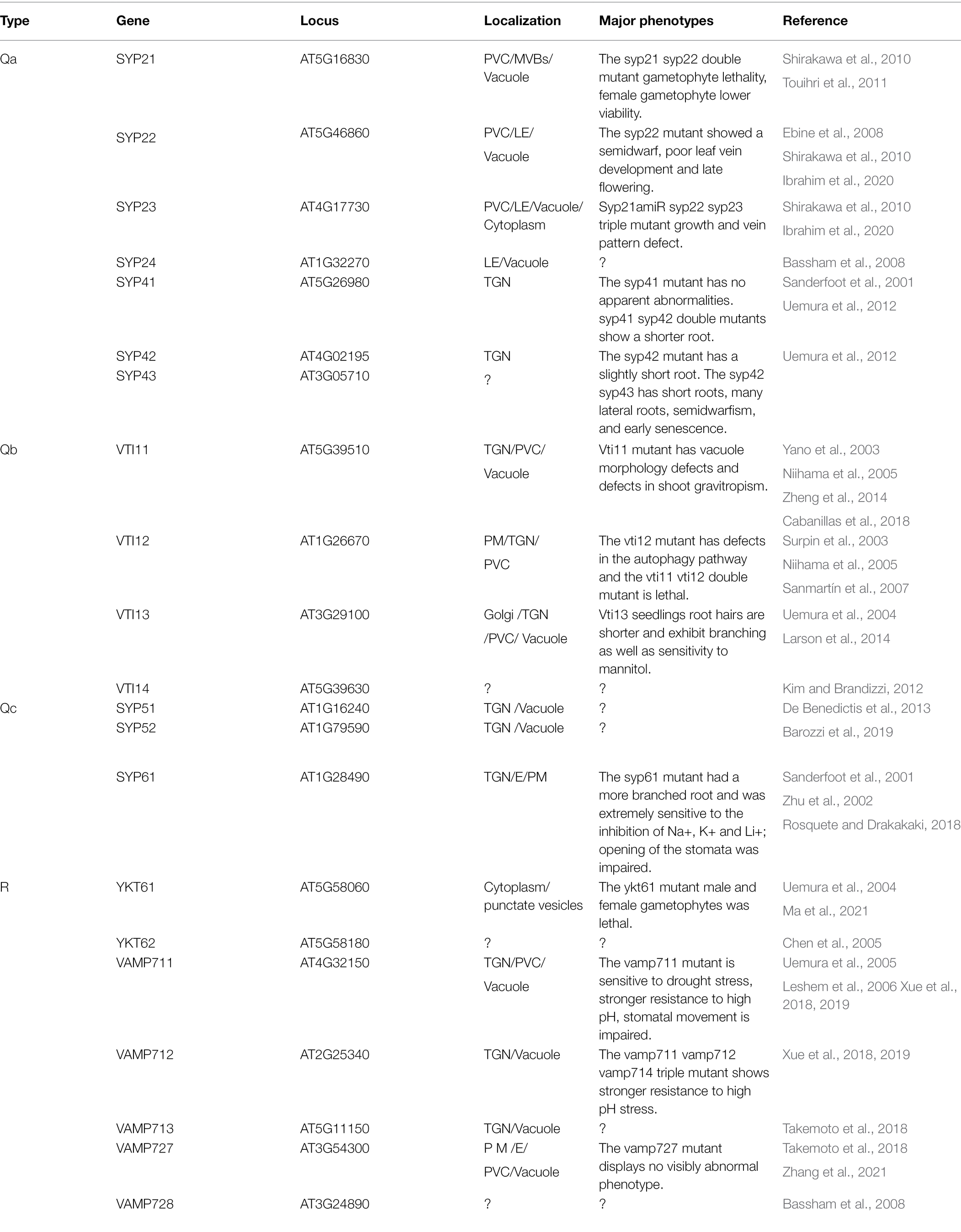
There are 21 SNAREs localized in the TGN, endosomes, and vacuoles (Figure 2; Table 2; Kim and Brandizzi, 2012; Ito and Boutté, 2020). R-SNAREs vesicle-associated membrane protein 721 (VAMP721) and VAMP722 have been confirmed to be located in the PM; TGN/EE, VAMP721, and VAMP722 are involved in endocytosis of FM4-64 and the secretion and recycling of the PINFORMED 2 (PIN2) transporter in PM (Lipka et al., 2007; Zhang et al., 2011a, 2021; Uemura et al., 2019). Arabidopsis tomosyn protein (AtTMS) localizes to the trans-Golgi network, PM, and cytosol, where it can interact with several Qa-SNAREs through its C-terminal R-SNARE-like motifs. In some cases, overexpressed AtTMS binds to syntaxins and blocks secretion during pollen development. Transmission electron microscopy showed irregular membrane structure aggregation under the PM, but the Golgi stack looks normal (Larson, 2019; Li et al., 2019). These results suggest that the R-SNAREs VAMP721, VAMP722, and AtTMS mediate post-Golgi trafficking. The negatively dominant form of SYP22 (SYP22ND) is in the cytoplasm, while SYP22 is in the cytoplasm and vesicle-like compartments in tobacco leaves. The syp22 vamp727−/+ double mutants and syp22nd mutants are not sensitive to BRASSINOSTEROID (BR) treatment and BRI1 recycling to the PM is defective in syp22nd plants. VAMP727 and SYP22 interact with BRASSINOSTEROID INSENSITIVE (BRI1; Jones et al., 2014; Zhang et al., 2019b). SYP22 and VAMP727 are involved in BR signaling via regulation of BRI1 trafficking and they regulate plant defense by controlling the abundance of BRI1 on the PM (Zhu et al., 2019; Zhang et al., 2019a). The syp22 vamp727 double mutant contains several small vacuoles instead of the large vacuoles that occur in the wild type and vamp727, syp22 mutants. SYP22 and VAMP727 can form a complex for membrane fusion between the prevacuolar compartment (PVC) and vacuoles (Sanderfoot et al., 1999; Ebine et al., 2008).
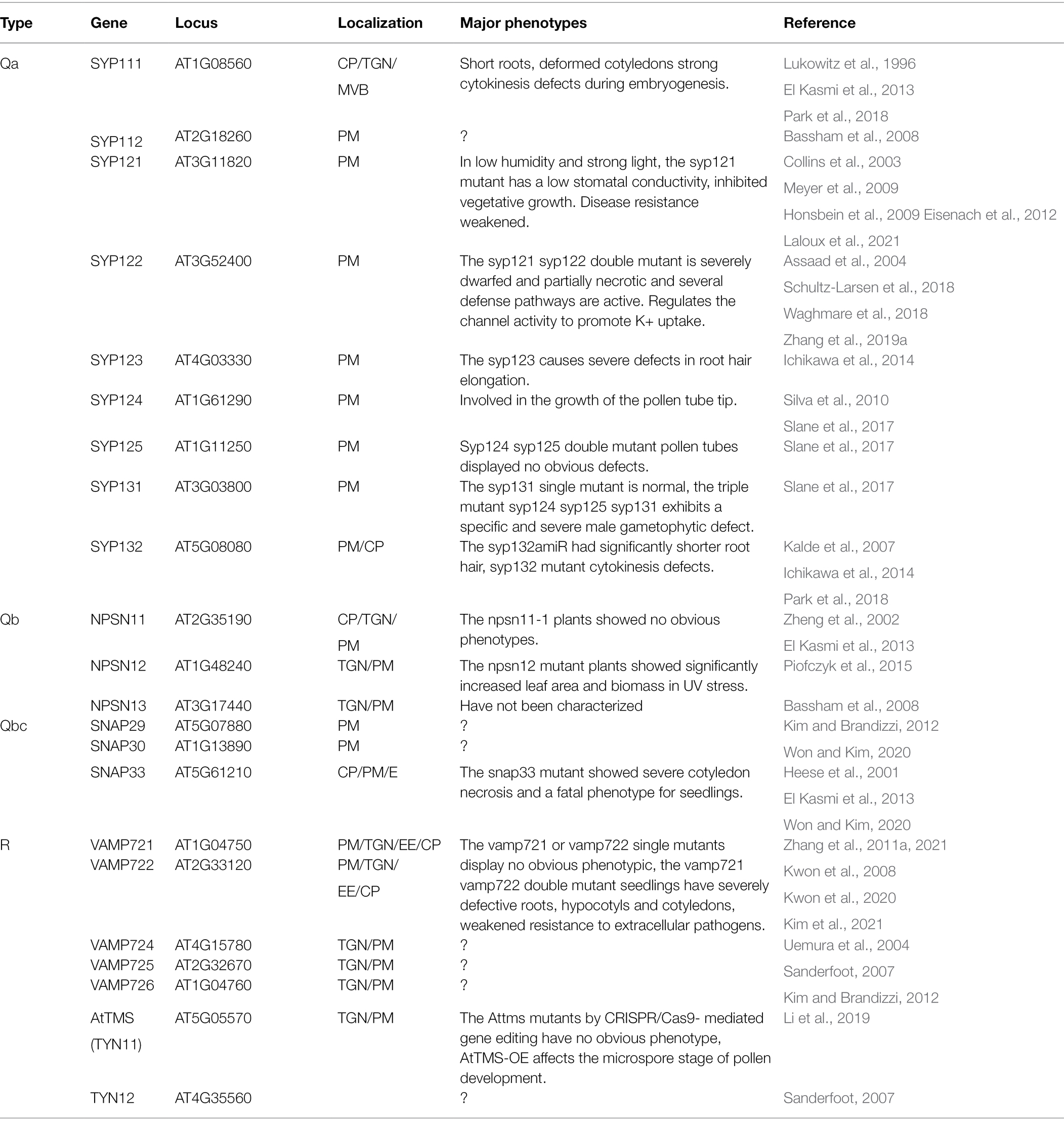
There are 22 SNAREs localized in the PM (Figure 2; Table 3; Kim and Brandizzi, 2012; Ruan et al., 2021). Syntaxin SYP121 is a plasma membrane Qa-SNARE and it consists of N, H, Q, and C four regions. The SYP121 sequence deletion shows that the C region contains the transmembrane domain and the H and Q regions contain the Habc and Qa-SNARE functional domains, interacting with plasma membrane intrinsic protein 2;7 (PIP2;7), which is involved in membrane fusion. SYP1s, except SYP112, and SYP121 orthologs interact with PIP2,7 (Hachez et al., 2014; Zhang et al., 2019a; Laloux et al., 2021). The proteins, PICALM1a and PICALM1b, which contain the ANTH domain are used as adapter proteins of the secretory vesicle-associated VAMP72 group clathrin-mediated endocytosis (CME). Retrieving VAMP721 from the PM requires PICALM1 and the loss of this function will result in the accumulation of VAMP721 in the PM (Fujimoto et al., 2020).
Snares Regulate Root Growth and Development
SNAREs Affecting Cell Elongation/Cell Growth
In plants, root systems are the underground organs that develop from the root apical meristem initiated during embryogenesis, and they respond to a variety of environmental obstacles and stimuli. Not only do the roots provide structural support for the plant’s aboveground parts, but they also absorb nutrients and water, both of which are required for plant growth (Motte et al., 2019). Thus, root growth and development are critical to overall plant survival. Cell elongation and cell division are used to promote root tip growth (De Smet et al., 2015). Intracellular vesicle transport from the ER to the Golgi apparatus, plasma membrane, vacuole or lysosome, and endocytosis plays important physiological functions in regulating plant development. Many proteins, such as ADP-ribosylation factor (ARF), ARF-guanine exchange factors (ARF-GEFs), ARF-GTPase-activating proteins (ARF-GAPs), Rab GTPase, and Rho-related GTPase of plants (ROPs), or their regulators, such as phospholipids, sterol kinases, and phosphatase, regulate root growth through vesicular trafficking (Yao and Xue, 2011). SNAREs play an important role in different pathways of vesicular trafficking and fusion with specific organelles to regulate root growth.
ER and Golgi SNAREs in the Root
Eleven of the ER/Golgi pathway SNAREs are highly expressed in roots, including three Qb-SNAREs: GOS11, GOS12, and MEMB11; five Qc-SNAREs: SYP71, SYP72, SYP73, BS14a, and BS14b; and three R-SNAREs: VAMP714, VAMP723, and SEC221. These SNAREs are necessary for root growth and development of Arabidopsis (Table 1; Figure 2; Uemura et al., 2004; Lipka et al., 2007). After 3 days of germination, syp73 had a shorter primary root, a significantly reduced elongation zone, and a lower fresh weight of the whole seedling. Overexpression of SYP73 causes rearrangement of the ER over actin. In Arabidopsis, SYP73 anchors the ER to the actin cytoskeleton to maintain the integrity and fluidity of the ER (Moreau et al., 2007; Cao et al., 2016). R-SNARE SEC22 is located in the ER, Golgi apparatus, and cytoplasm (Guan et al., 2021). Sec22-4 shows shorter primary roots, dwarf plants, sterility, and abnormal epidermal cells. AtSEC22 plays an important role in membrane transport and cytoskeletal dynamics during plant development (Chatre et al., 2005; El Kasmi et al., 2011; de Marcos Lousa et al., 2016; Guan et al., 2021). The vamp714 transfer DNA (T-DNA) insertional mutants grown in soil develop a shorter primary root and fewer lateral roots, have abnormal gravity responses, and disordered root cell arrangements. In addition, VAMP714 overexpressing lines and dominant-negative mutants also show a reduced seedling root system. The Arabidopsis R-SNARE VAMP714 protein colocalized in the ER, Golgi vesicles, and PIN proteins on the plasma membrane. The auxin distribution pattern in the root tip of the vamp714 mutant is aberrant, and the content is low (Gu et al., 2021). VAMP714 is required for PIN1 and PIN2 recycling. The actin depolymerizing agent latrunculin B (Lat B) caused the accumulation of VAMP714 vesicles in the cell. VAMP714 is part of the extracellular vesicle transport pathway of the ER/Golgi and the actin-dependent endocytic circulation pathway, which together regulate the abundance of PIN proteins in the plasma membrane (Geldner et al., 2001; Gu et al., 2021). This suggests that VAMP714 regulates the endocytic circulation pathway of vesicle transport involved with the PIN proteins and that the circulation of endosomes is essential for root growth and development.
TGN/Endosome and Vacuolar SNAREs in the Root
Twelve of the TGN/endosome, vacuolar pathway SNAREs are highly expressed in roots: four Qa-SNAREs: SYP21, SYP22, SYP23, and SYP41; three Qb-SNAREs: VTI11, VTI12, and VTI13; three Qc-SNAREs: SYP51, SYP52, and SYP61; and two R-SNAREs: VAMP711 and VAMP713 (Table 2; Figure 2; Uemura et al., 2004; Lipka et al., 2007). The Qa-SNARE SYP4 group (SYP41, SYP42, and SYP43) is all located in the same TGN compartment and some function redundantly (Uemura et al., 2012). The root length of the syp42 mutant is slightly shorter than that of the wild type, but there is no obvious abnormality in the syp41 and syp43 mutants. The syp42 syp43 double mutant has short roots, a large number of lateral roots, semidwarfism, early senescence, the transport of defective PIN2-GFP to the vacuole, and visible accumulation of secGFP (a signal peptide added to a variant of GFP; Uemura et al., 2012, 2019). This suggests that SYP4s regulate multiple transport pathways in plants that are involved in root growth and extracellular disease resistance. The Qb-SNARE VTI13 localizes to the Golgi, TGN/EE, and vacuole membrane in root hair cells, the vti13 mutant has short and bifurcated root hairs, and osmotic stress can exacerbate this phenotype. VTI13 is involved not only in the transport of cargo to vacuoles but also in cell wall organization and has a role in root hair growth (Larson et al., 2014). Vacuolar SNARE VTI11 is required for auxin-dependent morphogenesis of vacuoles, and its loss of function makes cells significantly insensitive to the growth inhibitory effect of auxin (Löfke et al., 2015). Thus, SNARE-dependent vacuolar morphogenesis allows auxin to limit cell expansion and promote the growth rate of root organs. In Arabidopsis, R-SNARE VAMP7C: VAMP711-VAMP714 forms a complex that promotes the fusion of vesicles and tonoplasts (Uemura et al., 2004; Leshem et al., 2006). The vamp711 vamp712 vamp714 triple mutant showed greater resistance to high pH stress than the vamp711 mutant. However, the vamp711 mutant is also more resistant to high pH than the wild type. A luciferase complementation (LUC) assay showed that VAMP7C interacted with PM H+-ATPase 2 (AHA2) and was involved in the regulation of PM H+-ATPase activity (Xue et al., 2019). The tonoplast localized SNARE (VAMP711-YFP, SYP21-YFP, and SYP22-GFP) fluorescence intensity increases with increasing auxin biosynthesis or the administration of exogenous auxin.
PM-Associated SNAREs in the Root
Thirteen of the PM-associated SNAREs are highly expressed in roots: five Qa-SNAREs: SYP111, SYP112, SYP121, SYP122, and SYP132; three Qb-SNAREs: NPSN11, NPSN12, and NPSN13; one Qb + Qc-SNARE: SNAP33; and four R-SNAREs: VAMP721, VAMP722, VAMP724, and VAMP726 (Table 3; Figure 2; Uemura et al., 2004; Lipka et al., 2007). Double homozygous vamp721 vamp722 mutant seedlings stopped growing after 2 days of germination and produced extremely thick roots, hypocotyls, and cotyledons; the seedlings died after 10 days. In addition, compared with wild-type seedlings, the roots of the vamp721 vamp722 mutant showed disordered root tips, including abnormal meristematic cells and root caps (Zhang et al., 2011a). The vamp721 vamp722 mutant shows aberrant localization of PINs and AUX1 and an enlarged TGN structure at the root (Zhang et al., 2021). The Arabidopsis R-SNAREs VAMP721 and VAMP722 have been found in the PM, TGN/EE, and cell plate (Lipka et al., 2007; Zhang et al., 2011a; Uemura et al., 2019). VAMP721 and VAMP722 are involved in endocytosis of FM4-64 and the secretion and recycling of the PIN2 transporter in PM but not in vacuoles. The R-SNAREs VAMP721 and VAMP722 play important roles in mediating the post-Golgi trafficking of auxin transporters and seedling growth (Zhang et al., 2021). The small GTPase RABA2a recruits the ternary complex VAMP721/722-SYP121-SNAP33 and interacts with it to achieve membrane fusion. The plant-specific RABA2a-SNARE pathway is essential for maintaining potassium ion homeostasis (Pang et al., 2021). This suggests that the R-SNARE VAMP721/722 plays important roles in mediating vesicle trafficking to maintain Arabidopsis root growth. The Qa-SNARE SYP132 is primarily localized in the PM (Uemura et al., 2004; Xia et al., 2019). The root length of SYP132-overexpressing seedlings was shorter than that of the wild type. Auxin regulates SYP132 in root growth and the geotropic response. The expression of SYP132 is tightly regulated by auxin, and increased expression of SYP132 reduces the content of H+-ATPase protein on the plasma membrane and is involved in the endocytosis of plasma membrane H+-ATPase proteins, reducing their density and activity on the PM (Xia et al., 2019, 2020). The loss of function of SYP123 and SYP132 leads to serious defects in root hair elongation. SYP123, rather than SYP132, is located at the root hair tip region in an f-actin-dependent manner by circulating between brefeldin A (BFA) sensitive endosomes and the PM at the expanded root hair tip (Ichikawa et al., 2014). Cumulatively, SYP123 and SYP132 work together to mediate membrane transport at the root hair tip and to promote root hair tip growth.
SNARES Affecting Cytokinesis
Plant morphogenesis is regulated by cell division and expansion (Bednarek and Falbel, 2002). Cell division is a major biological process that has been extensively studied. Similar to other eukaryotic cells, plant cells form mitotic spindles to divide replicated chromosomes (sun et al., 2018). Mitosis of plant cells is a dynamic process controlled by the rearrangement of microtubules that gradually transition into distinct arrangements of microtubules during mitosis and cytokinesis. Before cell division, cortical microtubules gradually narrow to form the preprophase band (PPB), which determines the level of cell division (Rasmussen et al., 2011). After the nuclear envelope ruptures and the mitotic process begins, microtubules form the mitotic spindle, which is responsible for dividing chromosomes into daughter cells during mitosis (Vos et al., 2004; Marcus et al., 2005; Azimzadeh et al., 2008). The final stage of cell division is cytokinesis, in which the bipolar spindle turns into the phragmoplast and acts as a guide to cell plate assembly and the subsequent formation of a new cell wall (Lee and Liu, 2013), which divides the cytoplasm and organelles and completes the formation of membrane barriers between the daughter cells, separating them (Bednarek and Falbel, 2002; Segui-Simarro et al., 2004; Jürgens et al., 2015). In Arabidopsis, the SNARE protein can promote the formation of cell plates (Figure 2; Lukowitz et al., 1996). The Qa-SNAREs SYP132 and KNOLLE form two SNARE complexes to regulate cytokinesis in Arabidopsis (El Kasmi et al., 2013; Park et al., 2018).
A cytokinesis-specific syntaxin KNOLLE (SYP111) is produced in the late G2/M phase, and it quickly reverses at the end of cytokinesis (Lauber et al., 1997; Völker et al., 2001; Reichardt et al., 2011). A newly synthesized KNOLLE protein is inserted into the ER membrane and travels along the secretory pathway to the plane of cell division via the Golgi stack and TGN, where it is endocytosed and directed to the vacuole via the multivesicular body (MVB) after cell plate formation (Reichardt et al., 2007; Karnahl et al., 2017). The cotyledons of all knolle seedlings are malformed, and the roots are short but clearly distinguishable. In weakly affected seedlings, the cotyledons are green and the roots have well-formed root hairs (Lukowitz et al., 1996). KNOLLE forms two SNARE complexes that are important in the cytokinesis process: a trimer complex comprised of KNOLLE and its companion Qbc-SNARE SNAP33 and R-SNARE VAMP721 or VAMP722 and a tetramer complex comprised of Qb-SNARE NOVEL PLANT SNARE 11 (NPSN11), Qc-SNARE SYP71, and R-SNARE VAMP721 or VAMP722 (El Kasmi et al., 2013; Karnahl et al., 2017; Park et al., 2018). KNOLLE, SNAP33, and NPSN11 are three Q-SNAREs found on the cell plate of dividing cells (Lauber et al., 1997; Heese et al., 2001; Zheng et al., 2002). Endogenous SYP71 accumulates on the cell plate, where it colocalizes with KNOLLE (El Kasmi et al., 2013). The snap33 mutant exhibits only minor cytokinesis defects during the seedling stage; however, severe cotyledon necrosis and seedling death occurred during seedling growth (Heese et al., 2001). There is no obvious cytokinesis defect in the npsn11 mutant, and homozygous plants are viable (Zheng et al., 2002). The roots of double mutant npsn11 syp71 seedlings are short, the root hair and hypocotyl grow well, and they may not have a cell division defect phenotype. The roots of snap33 syp71 double mutants are extremely short and exhibit a major cytokinetic defect, with dividing cells exhibiting abnormal morphology similar to that of knolle mutant (El Kasmi et al., 2013; Park et al., 2018). The vamp721 vamp722 double mutants had a defective cotyledon, vein patterning and root growth, and disordered root epidermis, cortex, and stele cell layer patterns, which displayed abnormal cell files (Zhang et al., 2011a, 2015b). Vamp721 vamp722 mutant seedlings exhibit cell wall stubs and incomplete cytokinesis and they inhibit the secretion of plasma membrane proteins. During cytokinesis, VAMP721 and VAMP722 are found on the cell plate (Zhang et al., 2011a; El Kasmi et al., 2013; Park et al., 2018). Concanamycin A (ConcA) treatment slows the expansion of the cell plate labeled with GFP-KNOLLE, resulting in cell wall stubs, as does intracellular accumulation of GFP-VAMP721 and GFP-VAMP722, so inhibiting trafficking at the TGN affects cell plate formation (Huss et al., 2002; Reichardt et al., 2007; Zhang et al., 2011a). VAMP721 and VAMP722 activities are required for secretory trafficking from the TGN to the cell plate in dividing cells and the plasma membrane (Zhang et al., 2011a). This suggests that the SNARE complex, which contains two types of KNOLLE, is functionally redundant and mediates cytokinesis in Arabidopsis.
SYP132 is a nonspecialized Qa-SNARE derived from an alga-like ancestor, and the SNARE partners for SYP132 and KNOLLE are the same. They have no overlapping functions in secretion, and the cellularization process of nourishing the embryonic endosperm is caused by the double fertilization unique to flowering plants (Park et al., 2018). In vitro and in vivo interactions occur between SYP132 and VAMP721/722, but not VAMP723 (Yun et al., 2013; Ichikawa et al., 2014). Qa-SNARE SYP132 is necessary for cytokinesis. SYP132 T-DNA insertion in the promoter region produces mutant syp132T seedlings that have abnormally formed adventitious roots instead of a single primary root. The SYP132 artificial microRNA mutant syp132amiR displays a disorganized shoot meristem and the hallmarks of defective cytokinesis as well as a very short root (Enami et al., 2009; Reichardt et al., 2011; Park et al., 2018). The combination of syp132amiR and syp132T alleles creates a SYP132 mutant with an enhanced mutant phenotype (syp132tam). Syp132tam mutant embryos and seedlings have a mutated phenotype that is difficult to distinguish from that of knolle mutant embryos and seedlings. Cytokinesis defects in syp132tam embryos include enlarged multinucleated cells and occasionally enlarged nuclei, and syp132tam embryos show bands of unfused vesicles (Weijers and Offringa, 2003; Park et al., 2018). Membrane vesicles delivered to the cell division plane fuse to form a partitioning membrane and require the SM protein KEULE to interact with KNOLLE in Arabidopsis cytokinesis. KEULE has the paralog SEC1B, which strongly preferentially interacts with the ancient Qa-SNARE SYP132, which is involved in secretion and cytokinesis, and KEULE also interacts with SYP132 (Karnahl et al., 2018). These results show that SYP132 is necessary for embryonic secretion and plays a role in embryonic cytokinesis. KNOLLE is necessary not only for somatic cytokinesis but also for cellularization of the endosperm.
Perspective
Recent studies have fully demonstrated that SNAREs are a multifunctional protein family with a wide range of biological functions in plants, and they not only participate in the normal growth and development of plants by mediating vesicle fusion, but they also regulate vesicle transport to response to the external environment by cooperating with other factors. Through signal transduction, the whole plant coordinates to adapt to environmental changes and continues to survive and reproduce. However, there is limited information in plant genetics research on the role of SNAREs. Some SNARE mutants do not show obvious phenotypes under normal conditions, which could be due to functional redundancy among related family members, or it could perform a specific function under a specific type of stress. This will provide an interesting perspective for future research into the functional diversity of different homologous genes in protein transport and plant stress tolerance. SNARE function requires sequential dynamic interactions between different SNAREs, and other proteins may be involved in SNARE function; for example, the Qa-SNARE KNOLLE interacts with Q-SNAREs SNAP33 and SYP71 to mediate membrane fusion in Arabidopsis cytokinesis. SM proteins and α-soluble NSF participate in SNARE complex assembly and disassembly (El Kasmi et al., 2013; Yoon and Munson, 2018). SNAREs and Rab GTPases have been shown to functionally promote vesicle fusion synergy and improve membrane fusion specificity and efficiency (Ohya et al., 2009; Ebine et al., 2011). SNAREs have been discovered for many years, and the mechanisms underlying SNARE-mediated membrane fusion are well understood. However, there are a few unanswered questions. Root development in Arabidopsis is a dynamic process involving complex interactions between transcriptional regulators and plant hormones. SNARE regulates auxin transport and contributes to root development, and what factors in addition to auxin are involved in Arabidopsis root growth? How is SNARE-mediated vesicular trafficking coordinated, thereby controlling root growth?
In the process of cytokinesis, KNOLLE and SYP132 have serious cytokinesis defects in the embryonic stage, but some of them can grow into seedlings, how is cell fate finely regulated in this process? What is the biological significance of two distinct SNARE complexes that mediate the same process of membrane fusion during cytokinesis? More SNARE complexes and more SNARE interaction factors remain to be discovered. With further research, it is expected that the molecular fine-regulation mechanism of SNARE-mediated vesicle transport will become clearer in the near future, thus providing a more comprehensive understanding of the important role of this protein family in plant growth and development.
Author Contributions
CL, YS, and YX wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31970195).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the editor and reviewers for their helpful comments and their input.
References
Assaad, F. F., Qiu, J. L., Youngs, H., Ehrhardt, D., Zimmerli, L., Kalde, M., et al. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15, 5118–5129. doi: 10.1091/mbc.e04-02-0140
Azimzadeh, J., Nacry, P., Christodoulidou, A., Drevensek, S., Camilleri, C., Amiour, N., et al. (2008). Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20, 2146–2159. doi: 10.1105/tpc.107.056812
Barozzi, F., Papadia, P., Stefano, G., Renna, L., Brandizzi, F., Migoni, D., et al. (2019). Variation in membrane trafficking linked to SNARE AtSYP51 interaction with aquaporin NIP1;1. Front. Plant Sci. 9:1949. doi: 10.3389/fpls.2018.01949
Bassham, D. C., Brandizzi, F., Otegui, M. S., and Sanderfoot, A. A. (2008). The secretory system of Arabidopsis. Arabidopsis Book 6:e0116. doi: 10.1199/tab.0116
Bednarek, S. Y., and Falbel, T. G. (2002). Membrane trafficking during plant cytokinesis. Traffic 3, 621–629. doi: 10.1034/j.1600-0854.2002.30904.x
Beemster, G. T., and Baskin, T. I. (1998). Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116, 1515–1526. doi: 10.1104/pp.116.4.1515
Beemster, G. T., De Vusser, K., De Tavernier, E., De Bock, K., and Inzé, D. (2002). Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol. 129, 854–864. doi: 10.1104/pp.002923
Bock, J. B., Matern, H. T., Peden, A. A., and Scheller, R. H. (2011). A genomic perspective on membrane compartment organization. Nature 409, 839–841. doi: 10.1038/35057024
Bolaños-Villegas, P., Guo, C. L., and Jauh, G. Y. (2015). Arabidopsis qc-SNARE genes BET11 and BET12 are required for fertility and pollen tube elongation. Bot. Stud. 56:21. doi: 10.1186/s40529-015-0102-x
Bonifacino, J. S., and Glick, B. S. (2004). The mechanisms of vesicle budding and fusion. Cell 116, 153–166. doi: 10.1016/s0092-8674(03)01079-1
Bonifacino, J. S., and Lippincott-Schwartz, J. (2003). Coat proteins: shaping membrane transport. Nat. Rev. Mol. Cell Biol. 4, 409–414. doi: 10.1038/nrm1099
Bubeck, J., Scheuring, D., Hummel, E., Langhans, M., Viotti, C., Foresti, O., et al. (2008). The syntaxins SYP31 and SYP81 control ER-Golgi trafficking in the plant secretory pathway. Traffic 9, 1629–1652. doi: 10.1111/j.1600-0854.2008.00803.x
Bremser, M., Nickel, W., Schweikert, M., Ravazzola, M., Amherdt, M., Hughes, C. A., et al. (1999). Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell 96, 495–506. doi: 10.1016/S0092-8674(00)80654-6
Burgoyne, R. D., and Morgan, A. (2007). Membrane trafficking: three steps to fusion. Curr. Biol. 17, R255–R258. doi: 10.1016/j.cub.2007.02.006
Cabanillas, D. G., Jiang, J., Movahed, N., Germain, H., Yamaji, Y., Zheng, H., et al. (2018). Turnip mosaic virus uses the SNARE protein VTI11 in an unconventional route for replication vesicle trafficking. Plant Cell 30, 2594–2615. doi: 10.1105/tpc.18.00281
Cao, P., Renna, L., Stefano, G., and Brandizzi, F. (2016). SYP73 anchors the ER to the actin cytoskeleton for maintenance of ER integrity and streaming in Arabidopsis. Curr. Biol. 26, 3245–3254. doi: 10.1016/j.cub.2016.10.024
Chatre, L., Brandizzi, F., Hocquellet, A., Hawes, C., and Moreau, P. (2005). Sec22 and Memb11 are v-SNAREs of the anterograde endoplasmic reticulum-Golgi pathway in tobacco leaf epidermal cells. Plant Physiol. 139, 1244–1254. doi: 10.1104/pp.105.067447
Chen, F., Chen, H., Chen, Y., Wei, W., Sun, Y., Zhang, L., et al. (2021). Dysfunction of the SNARE complex in neurological and psychiatric disorders. Pharmacol. Res. 165:105469. doi: 10.1016/j.phrs.2021.105469
Chen, Y., Shin, Y. K., and Bassham, D. C. (2005). YKT6 is a core constituent of membrane fusion machineries at the Arabidopsis trans-Golgi network. J. Mol. Biol. 350, 92–101. doi: 10.1016/j.jmb.2005.04.061
Chung, K. P., Zeng, Y., Li, Y., Ji, C., Xia, Y., and Jiang, L. (2018). Signal motif-dependent ER export of the qc-SNARE BET12 interacts with MEMB12 and affects PR1 trafficking in Arabidopsis. J. Cell Sci. 131:2838. doi: 10.1242/jcs.202838
Collins, N. C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J. L., et al. (2003). SNARE protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977. doi: 10.1038/nature02076
De Benedictis, M., Bleve, G., Faraco, M., Stigliano, E., Grieco, F., Piro, G., et al. (2013). AtSYP51/52 functions diverge in the post-Golgi traffic and differently affect vacuolar sorting. Mol. Plant 6, 916–930. doi: 10.1093/mp/sss117
de Marcos Lousa, C., Soubeyrand, E., Bolognese, P., Wattelet-Boyer, V., Bouyssou, G., Marais, C., et al. (2016). Subcellular localization and trafficking of phytolongins (non-SNARE longins) in the plant secretory pathway. J. Exp. Bot. 67, 2627–2639. doi: 10.1093/jxb/erw094
De Smet, S., Cuypers, A., Vangronsveld, J., and Remans, T. (2015). Gene networks involved in hormonal control of root development in Arabidopsis thaliana: a framework for studying its disturbance by metal stress. Int. J. Mol. Sci. 16, 19195–19224. doi: 10.3390/ijms160819195
Delgadillo, M. O., Ruano, G., Zouhar, J., Sauer, M., Shen, J., Lazarova, A., et al. (2020). MTV proteins unveil ER- and microtubule-associated compartments in the plant vacuolar trafficking pathway. Proc. Natl. Acad. Sci. U.S.A. 117, 9884–9895. doi: 10.1073/pnas.1919820117
Ebine, K., Fujimoto, M., Okatani, Y., Nishiyama, T., Goh, T., Ito, E., et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13, 853–859. doi: 10.1038/ncb2270
Ebine, K., Okatani, Y., Uemura, T., Goh, T., Shoda, K., Niihama, M., et al. (2008). A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20, 3006–3021. doi: 10.1105/tpc.107.057711
Eisenach, C., Chen, Z. H., Grefen, C., and Blatt, M. R. (2012). The trafficking protein SYP121 of Arabidopsis connects programmed stomatal closure and K+ channel activity with vegetative growth. Plant J. 69, 241–251. doi: 10.1111/j.1365-313X.2011.04786.x
El Kasmi, F., Krause, C., Hiller, U., Stierhof, Y. D., Mayer, U., Conner, L., et al. (2013). SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol. Biol. Cell 24, 1593–1601. doi: 10.1091/mbc.e13-02-0074
El Kasmi, F., Pacher, T., Strompen, G., Stierhof, Y. D., Muller, L. M., Koncz, C., et al. (2011). Arabidopsis SNARE protein SEC22 is essential for gametophyte development and maintenance of Golgi-stack integrity. Plant J. 66, 268–279. doi: 10.1111/j.1365-313x.2011.04487.x
Enami, K., Ichikawa, M., Uemura, T., Kutsuna, N., Hasezawa, S., Nakagawa, T., et al. (2009). Differential expression control and polarized distribution of plasma membrane-resident SYP1 SNAREs in Arabidopsis thaliana. Plant Cell Physiol. 50, 280–289. doi: 10.1093/pcp/pcn197
Fasshauer, D., Sutton, R. B., Brunger, A. T., and Jahn, R. (1998). Conserved structural features of the synaptic fusion complex SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. U.S.A. 95, 15781–15786. doi: 10.1073/pnas.95.26.15781
Frémont, S., and Echard, A. (2018). Membrane traffic in the late steps of cytokinesis. Curr. Biol. 28, R458–R470. doi: 10.1016/j.cub.2018.01.019
Fujimoto, M., Ebine, K., Nishimura, K., Tsutsumi, N., and Ueda, T. (2020). Longin R-SNARE is retrieved from the plasma membrane by ANTH domain-containing proteins in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 117, 25150–25158. doi: 10.1073/pnas.2011152117
Gao, C., Yu, C. K., Qu, S., San, M. W., Li, K. Y., Lo, S. W., et al. (2012). The Golgi-localized Arabidopsis endomembrane protein12 contains both endoplasmic reticulum export and Golgi retention signals at its C terminus. Plant Cell 24, 2086–2104. doi: 10.1105/tpc.112.096057
Geldner, N., Friml, J., Stierhof, Y. D., Jürgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428. doi: 10.1038/35096571
Grosshans, B. L., Ortiz, D., and Novick, P. (2006). Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U. S. A. 103, 11821–11827.
Gu, X., Brennan, A., Wei, W., Guo, G., and Lindsey, K. (2020). Vesicle transport in plants: a revised phylogeny of SNARE proteins. Evol. Bioinform. 16:56575. doi: 10.1177/1176934320956575
Gu, X., Fonseka, K., Agneessens, J., Casson, S. A., Smertenko, A., Guo, G., et al. (2021). The Arabidopsis R-SNARE VAMP714 is essential for polarisation of PIN proteins and auxin responses. New Phytol. 230, 550–566. doi: 10.1111/nph.17205
Gu, Y., Zavaliev, R., and Dong, X. (2017). Membrane trafficking in plant immunity. Mol. Plant 10, 1026–1034. doi: 10.1016/j.molp.2017.07.001
Guan, L., Yang, S., Li, S., Liu, Y., Liu, Y., Yang, Y., et al. (2021). AtSEC22 regulates cell morphogenesis via affecting cytoskeleton organization and stabilities. Front. Plant Sci. 12:635732. doi: 10.3389/fpls.2021.635732
Hachez, C., Laloux, T., Reinhardt, H., Cavez, D., Degand, H., Grefen, C., et al. (2014). Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26, 3132–3147. doi: 10.1105/tpc.114.127159
Heese, M., Gansel, X., Sticher, L., Wick, P., Grebe, M., Granier, F., et al. (2001). Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J. Cell Biol. 155, 239–249. doi: 10.1083/jcb.200107126
Hong, W. (2005). SNAREs and traffic. Biochim. Biophy. Acta. 1744, 120–144. doi: 10.1016/j.bbamcr.2005.03.014
Honsbein, A., Sokolovski, S., Grefen, C., Campanoni, P., Pratelli, R., Paneque, M., et al. (2009). A tripartite SNARE-K+ channel complex mediates in channel-dependent K+ nutrition in Arabidopsis. Plant Cell 21, 2859–2877. doi: 10.1105/tpc.109.066118
Huss, M., Ingenhorst, G., König, S., Gassel, M., Dröse, S., Zeeck, A., et al. (2002). Concanamycin A, the specific inhibitor of V-ATPases, binds to the VO subunit c. J. Biol. Chem. 277, 40544–40548. doi: 10.1074/jbc.M207345200
Ibrahim, A., Yang, X., Liu, C., Cooper, K. D., Bishop, B. A., Zhu, M., et al. (2020). Plant SNAREs SYP22 and SYP23 interact with tobacco mosaic virus 126 kDa protein and SYP2s are required for normal local virus accumulation and spread. Virology. 547, 57–71. doi: 10.1016/j.virol.2020.04.002
Ichikawa, M., Hirano, T., Enami, K., Fuselier, T., Kato, N., Kwon, C., et al. (2014). Syntaxin of plant proteins SYP123 and SYP132 mediate root hair tip growth in Arabidopsis thaliana. Plant Cell Physiol. 55, 790–800. doi: 10.1093/pcp/pcu048
Ito, Y., and Boutté, Y. (2020). Differentiation of trafficking pathways at golgi entry core compartments and post-golgi subdomains. Front. Plant Sci. 11:609516. doi: 10.3389/fpls.2020.609516
Jahn, R., and Scheller, R. H. (2006). SNAREs-engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643. doi: 10.1038/nrm2002
Jones, A. M., Xuan, Y., Xu, M., Wang, R. S., Ho, C. H., Lalonde, S., et al. (2014). Border control--a membrane-linked interactome of Arabidopsis. Science 344, 711–716. doi: 10.1126/science.1251358
Jun, Y., and Wickner, W. (2019). Sec17 (α-SNAP) and Sec18 (NSF) restrict membrane fusion to R-SNAREs, Q-SNAREs, and SM proteins from identical compartments. Proc. Natl. Acad. Sci. U.S.A. 116, 23573–23581. doi: 10.1073/pnas.1913985116
Jürgens, G., Park, M., Richter, S., Touihri, S., Krause, C., El Kasmi, F., et al. (2015). Plant cytokinesis: a tale of membrane traffic and fusion. Biochem. Soc. Trans. 43, 73–78. doi: 10.1042/BST20140246
Kalde, M., Nuhse, T., Findlay, K., and Peck, S. (2007). The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl. Acad. Sci. U.S.A. 104, 11850–11855. doi: 10.1073/pnas.0701083104
Karnahl, M., Park, M., Krause, C., Hiller, U., Mayer, U., Stierhof, Y. D., et al. (2018). Functional diversification of Arabidopsis SEC1-related SM proteins in cytokinetic and secretory membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 115, 6309–6314. doi: 10.1073/pnas.1722611115
Kamal, A., and Goldstein, L. S. (2002). Principles of cargo attachment to cytoplasmic motor proteins. Curr. Opin. Cell Biol. 14, 63–68. doi: 10.1016/s0955-0674(01)00295-2
Karnahl, M., Park, M., Mayer, U., Hiller, U., and Jürgens, G. (2017). ER assembly of SNARE complexes mediating formation of partitioning membrane in Arabidopsis cytokinesis. Elife 6:25327. doi: 10.7554/eLife.25327
Kato, T., Morita, M. T., Fukaki, H., Yamauchi, Y., Uehara, M., Niihama, M., et al. (2002). SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14, 33–46. doi: 10.1105/tpc.010215
Kim, S. J., and Brandizzi, F. (2012). News and views into the SNARE complexity in Arabidopsis. Front. Plant Sci. 3:28. doi: 10.3389/fpls.2012.00028
Kim, S., Choi, Y., Kwon, C., and Yun, H. S. (2019). Endoplasmic reticulum stress induced accumulation of VAMP721/722 requires CALRETICULIN 1 and CALRETICULIN 2 in Arabidopsis. J. Integr. Plant Biol. 61, 974–980. doi: 10.1111/jipb.12728
Kim, S., Kim, H., Park, K., Cho, D. J., Kim, M. K., Kwon, C., et al. (2021). Synaptotagmin 5 controls SYP132-VAMP721/722 interaction for Arabidopsis immunity to pseudomonas syringae pv tomato DC3000. Mol. Cells 44, 670–679. doi: 10.14348/molcells.2021.0100
Kwon, H., Cho, D. J., Lee, H., Nam, M. H., Kwon, C., and Yun, H. S. (2020). CCOAOMT1, a candidate cargo secreted via VAMP721/722 secretory vesicles in Arabidopsis. Biochem. Biophys. Res. Commun. 524, 977–982. doi: 10.1016/j.bbrc.2020.02.029
Kwon, C., Neu, C., Pajonk, S., Yun, H. S., Lipka, U., Humphry, M., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451, 835–840. doi: 10.1038/nature06545
Laloux, T., Matyjaszczyk, I., Beaudelot, S., Hachez, C., and Chaumont, F. (2021). Interaction between the SNARE SYP121 and the plasma membrane aquaporin PIP2;7 involves different protein domains. Front. Plant Sci. 11:631643. doi: 10.3389/fpls.2020.631643
Larson, E. R. (2019). Plant tomosyn is a negative regulator of SNARE-mediated secretion in pollen. Plant Physiol. 181, 843–844. doi: 10.1104/pp.19.01145
Larson, E. R., Domozych, D. S., and Tierney, M. L. (2014). SNARE VTI13 plays a unique role in endosomal trafficking pathways associated with the vacuole and is essential for cell wall organization and root hair growth in Arabidopsis. Ann. Bot. 114, 1147–1159. doi: 10.1093/aob/mcu041
Lauber, M. H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., et al. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485–1493. doi: 10.1083/jcb.139.6.1485
Lavrekha, V. V., Pasternak, T., Ivanov, V. B., Palme, K., and Mironova, V. V. (2017). 3D analysis of mitosis distribution highlights the longitudinal zonation and diarch symmetry in proliferation activity of the Arabidopsis thaliana root meristem. Plant J. doi: 10.1111/tpj.13720
Lee, Y.-R. J., and Liu, B. (2013). The rise and fall of the phragmoplast microtubule array. Curr. Opin. Plant Biol. 16, 757–763. doi: 10.1016/j.pbi.2013.10.008
Leshem, Y., Melamed-Book, N., Cagnac, O., Ronen, G., Nishri, Y., Solomon, M., et al. (2006). Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 103, 18008–18013. doi: 10.1073/pnas.0604421103
Li, B., Li, Y., Liu, F., Tan, X., Rui, Q., Tong, Y., et al. (2019). Overexpressed tomosyn binds syntaxins and blocks secretion during pollen development. Plant Physiol. 181, 1114–1126. doi: 10.1104/pp.19.00965
Li, L., Shimada, T., Takahashi, H., Koumoto, Y., Shirakawa, M., Takagi, J., et al. (2013). MAG2 and three MAG2-INTERACTING PROTEINs form an ER-localized complex to facilitate storage protein transport in Arabidopsis thaliana. Plant J. 76, 781–791. doi: 10.1111/tpj.12347
Lipka, V., Kwon, C., and Panstruga, R. (2007). SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu. Rev. Cell. Dev. Biol. 23, 147–174. doi: 10.1146/annurev.cellbio.23.090506.123529
Löfke, C., Dünser, K., Scheuring, D., and Kleine-Vehn, J. (2015). Auxin regulates SNARE-dependent vacuolar morphology restricting cell size. Elife 4:e05868. doi: 10.7554/eLife.05868
Lukowitz, W., Mayer, U., and Jürgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84, 61–71. doi: 10.1016/s0092-8674(00)80993-9
Ma, T., Li, E., Li, L. S., Li, S., and Zhang, Y. (2021). The Arabidopsis R-SNARE protein YKT61 is essential for gametophyte development. J. Integr. Plant Biol. 63, 676–694. doi: 10.1111/jipb.13017
Marais, C., Wattelet-Boyer, V., Bouyssou, G., Hocquellet, A., Dupuy, J. W., Batailler, B., et al. (2015). The Qb-SNARE Memb11 interacts specifically with arf1 in the Golgi apparatus of Arabidopsis thaliana. J. Exp. Bot. 66, 6665–6678. doi: 10.1093/jxb/erv373
Marcus, A. I., Dixit, R., and Cyr, R. J. (2005). Narrowing of the preprophase microtubule band is not required for cell division plane determination in cultured plant cells. Protoplasma 226, 169–174. doi: 10.1007/s00709-005-0119-1
Martinière, A., and Moreau, P. (2020). Complex roles of Rabs and SNAREs in the secretory pathway and plant development: a never-ending story. J. Microsc. 280, 140–157. doi: 10.1111/jmi.12952
Meyer, D., Pajonk, S., Micali, C., O'Connell, R., and Schulze-Lefert, P. (2009). Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 57, 986–999. doi: 10.1111/j.1365-313X.2008.03743.x
Moreau, P., Brandizzi, F., Hanton, S., Chatre, L., Melser, S., Hawes, C., et al. (2007). The plant ER-Golgi interface: a highly structured and dynamic membrane complex. J. Exp. Bot. 58, 49–64. doi: 10.1093/jxb/erl135
Morita, M. T., Kato, T., Nagafusa, K., Saito, C., Ueda, T., Nakano, A., et al. (2002). Involvement of the vacuoles of the endodermis in the early process of shoot gravitropism in Arabidopsis. Plant Cell 14, 47–56. doi: 10.1105/tpc.010216
Motte, H., Vanneste, S., and Beeckman, T. (2019). Molecular and environmental regulation of root development. Annu. Rev. Plant Biol. 70, 465–488. doi: 10.1146/annurev-arplant-050718-100423
Niihama, M., Uemura, T., Saito, C., Nakano, A., Sato, M. H., Tasaka, M., et al. (2005). Conversion of functional specificity in Qb-SNARE VTI1 homologues of Arabidopsis. Curr. Biol. 15, 555–560. doi: 10.1016/j.cub.2005.02.021
Ohya, T., Miaczynska, M., Coskun, U., Lommer, B., Runge, A., Drechsel, D., et al. (2009). Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature 459, 1091–1097. doi: 10.1038/nature08107
Oyler, G. A., Higgins, G. A., Hart, R. A., Battenberg, E., Billingsley, M., Bloom, F. E., et al. (1989). The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J. Cell Biol. 109, 3039–3052. doi: 10.1083/jcb.109.6.3039
Pang, L., Ma, Z., Zhang, X., Huang, Y., Li, R., Miao, Y., et al. (2021). The small GTPase RABA2a recruits SNARE proteins to regulate secretory pathway in parallel with the exocyst complex in Arabidopsis. Mol. Plant. [Epub ahead of print]. doi: 10.1016/j.molp.2021.11.008
Park, M., Krause, C., Karnahl, M., Reichardt, I., El Kasmi, F., Mayer, U., et al. (2018). Concerted action of evolutionarily ancient and novel SNARE complexes in flowering-plant cytokinesis. Dev. Cell 44, 500.e4–511.e4. doi: 10.1016/j.devcel.2017.12.027
Piofczyk, T., Jeena, G., and Pecinka, A. (2015). Arabidopsis thaliana natural variation reveals connections between UV radiation stress and plant pathogen-like defense responses. Plant Physiol. Biochem. 93, 34–43. doi: 10.1016/j.plaphy.2015.01.011
Rancour, D. M., Dickey, C. E., Park, S., and Bednarek, S. Y. (2002). Characterization of AtCDC48. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants. Plant Physiol. 130, 1241–1253. doi: 10.1104/pp.011742
Rasmussen, C. G., Humphries, J. A., and Smith, L. G. (2011). Determination of symmetric and asymmetric division planes in plant cells. Ann. Rev. Plant Biol. 62, 387–409. doi: 10.1146/annurev-arplant-042110-103802
Reichardt, I., Slane, D., el Kasmi, F., Knöll, C., Fuchs, R., Mayer, U., et al. (2011). Mechanisms of functional specificity among plasma membrane syntaxins in Arabidopsis. Traffic 12, 1269–1280. doi: 10.1111/j.1600-0854.2011.01222.x
Reichardt, I., Stierhof, Y. D., Mayer, U., Richter, S., Schwarz, H., Schumacher, K., et al. (2007). Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr. Biol. 17, 2047–2053. doi: 10.1016/j.cub.2007.10.040
Rosquete, M. R., and Drakakaki, G. (2018). Plant TGN in the stress response: a compartmentalized overview. Curr. Opin. Plant Biol. 46, 122–129. doi: 10.1016/j.pbi.2018.09.003
Ruan, H., Li, J., Wang, T., and Ren, H. (2021). Secretory vesicles targeted to plasma membrane During pollen germination and tube growth. Front. Cell Dev. Biol. 8:615447. doi: 10.3389/fcell.2020.615447
Rui, Q., Tan, X., Liu, F., Li, Y., Liu, X., Li, B., et al. (2021). Syntaxin of plants31 (SYP31) and SYP32 is essential for Golgi morphology maintenance and pollen development. Plant Physiol. 186, 330–343. doi: 10.1093/plphys/kiab049
Ryu, J. K., Min, D., Rah, S. H., Kim, S. J., Park, Y., Kim, H., et al. (2015). Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover. Science 347, 1485–1489. doi: 10.1126/science.aaa5267
Saito, C., and Ueda, T. (2009). Chapter 4: functions of RAB and SNARE proteins in plant life. Int. Rev. Cell Mol. Biol. 274, 183–233. doi: 10.1016/s1937-6448(08)02004-2
Salinas-Cornejo, J., Madrid-Espinoza, J., Verdugo, I., Pérez-Díaz, J., Martín-Davison, A. S., Norambuena, L., et al. (2021). The exocytosis associated SNAP25-type protein, SlSNAP33, increases salt stress tolerance by modulating endocytosis in tomato. Plants 10:1322. doi: 10.3390/plants10071322
Sanderfoot, A. (2007). Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 144, 6–17. doi: 10.1104/pp.106.092973
Sanderfoot, A. A., Assaad, F. F., and Raikhel, N. V. (2000). The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant physiol. 124, 1558–1569. doi: 10.1104/pp.124.4.1558
Sanderfoot, A. A., Kovaleva, V., Bassham, D. C., and Raikhel, N. V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell. 12, 3733–3743. doi: 10.1091/mbc.12.12.3733
Sanderfoot, A. A., Kovaleva, V., Zheng, H., and Raikhel, N. V. (1999). The t-SNARE AtVAM3p resides on the prevacuolar compartment in Arabidopsis root cells. Plant Physiol. 121, 929–938. doi: 10.1104/pp.121.3.929
Sanmartín, M., Ordóñez, A., Sohn, E. J., Robert, S., Sánchez-Serrano, J. J., Surpin, M. A., et al. (2007). Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 3645–3650. doi: 10.1073/pnas.0611147104
Schultz-Larsen, T., Lenk, A., Kalinowska, K., Vestergaard, L. K., Pedersen, C., Isono, E., et al. (2018). The AMSH3 ESCRT-III-associated deubiquitinase is essential for plant immunity. Cell Rep. 25, 2329–2338. doi: 10.1016/j.celrep.2018.11.011
Segui-Simarro, J. M., Austin, J. R. II, White, E. A., and Staehelin, L. A. (2004). Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell 16, 836–856. doi: 10.1105/tpc.017749
Shirakawa, M., Ueda, H., Shimada, T., Koumoto, Y., Shimada, T. L., Kondo, M., et al. (2010). Arabidopsis Qa-SNARE SYP2 proteins localized to different subcellular regions function redundantly in vacuolar protein sorting and plant development. Plant J. 64, 924–935. doi: 10.1111/j.1365-313X.2010.04394.x
Silva, P. A., Ul-Rehman, R., Rato, C., Di Sansebastiano, G. P., and Malho, R. (2010). Asymmetric localization of Arabidopsis SYP124 syntaxin at the pollen tube apical and sub-apical zones is involved in tip growth. BMC Plant Biol. 10:179. doi: 10.1186/1471-2229-10-179
Slane, D., Reichardt, I., El Kasmi, F., Bayer, M., and Jurgens, G. (2017). Evolutionarily diverse SYP1 Qa-SNAREs jointly sustain pollen tube growth in Arabidopsis. Plant J. 92, 375–385. doi: 10.1111/tpj.13659
Söllner, T., Whiteheart, S. W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P., et al. (1993). SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324. doi: 10.1038/362318a0
Song, H., Torng, T. L., Orr, A. S., Brunger, A. T., and Wickner, W. T. (2021). Sec17/Sec18 can support membrane fusion without help from completion of SNARE zippering. Elife 10:e67578. doi: 10.7554/eLife.67578
Sun, X., Cai, X., Yin, K., Gu, L., Shen, Y., Hu, B., et al. (2021). Wild soybean SNARE proteins BET1s mediate the subcellular localization of the cytoplasmic receptor-like kinases CRCK1s to modulate salt stress responses. Plant J. 105, 771–785. doi: 10.1111/tpj.15072
Sun, H., Furt, F., and Vidali, L. (2018). Myosin XI localizes at the mitotic spindle and along the cell plate during plant cell division in Physcomitrella patens. Biochem. Biophys. Res. Commun. 506, 409–421. doi: 10.1016/j.bbrc.2018.01.082
Surpin, M., Zheng, H., Morita, M. T., Saito, C., Avila, E., Blakeslee, J. J., et al. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15, 2885–2899. doi: 10.1105/tpc.016121
Sutter, J. U., Campanoni, P., Blatt, M. R., and Paneque, M. (2006). Setting SNAREs in a different wood. Traffic 7, 627–638. doi: 10.1111/j.1600-0854.2006.00414.x
Takemoto, K., Ebine, K., Askani, J. C., Kruger, F., Gonzalez, Z. A., Ito, E., et al. (2018). Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E2457–E2466. doi: 10.1073/pnas.1717839115
Tan, X., Cao, K., Liu, F., Li, Y., Li, P., Gao, C., et al. (2016). Arabidopsis COG complex subunits COG3 and COG8 modulate Golgi morphology, vesicle trafficking homeostasis and are essential for pollen tube growth. PLoS Genet. 12:e1006140. doi: 10.1371/journal.pgen.1006140
Tarte, V. N., Seok, H. Y., Woo, D. H., Le, D. H., Tran, H. T., Baik, J. W., et al. (2015). Arabidopsis Qc-SNARE gene AtSFT12 is involved in salt and osmotic stress responses and Na+ accumulation in vacuoles. Plant Cell Rep. 34, 1127–1138. doi: 10.1007/s00299-015-1771-3
Toonen, R. F., and Verhage, M. (2007). Munc18-1 in secretion: lonely Munc joins SNARE team and takes control. Trends Neurosci. 30, 564–572. doi: 10.1016/j.tins.2007.08.008
Touihri, S., Knoll, C., Stierhof, Y. D., Muller, I., Mayer, U., and Jurgens, G. (2011). Functional anatomy of the Arabidopsis cytokinesis-specific syntaxin KNOLLE. Plant J. 68, 755–764. doi: 10.1111/j.1365-313X.2011.04736.x
Uemura, T., Kim, H., Saito, C., Ebine, K., Ueda, T., Schulze-Lefert, P., et al. (2012). Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 1784–1789. doi: 10.1073/pnas.1115146109
Uemura, T., Nakano, R. T., Takagi, J., Wang, Y., Kramer, K., Finkemeier, I., et al. (2019). A golgi-released subpopulation of the trans-golgi network mediates protein secretion in Arabidopsis. Plant Physiol. 179, 519–532. doi: 10.1104/pp.18.01228
Uemura, T., Sato, M. H., and Takeyasu, K. (2005). The longin domain regulates subcellular targeting of VAMP7 in Arabidopsis thaliana. FEBS Lett. 579, 2842–2846. doi: 10.1016/j.febslet.2005.04.022
Uemura, T., Ueda, T., Ohniwa, R. L., Nakano, A., Takeyasu, K., and Sato, M. H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-golgi network in plant cells. Cell Struct. Funct. 29, 49–65. doi: 10.1247/csf.29.49
Verhage, M., and Sørensen, J. B. (2008). Vesicle docking in regulated exocytosis. Traffic 9, 1414–1424.
Völker, A., Stierhof, Y. D., and Jürgens, G. (2001). Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J. Cell Sci. 114, 3001–3012. doi: 10.1242/jcs.114.16.3001
Vos, J. W., Dogterom, M., and Emons, A. M. (2004). Microtubules become more dynamic but not shorter during preprophase band formation: a possible “search-and-capture” mechanism for microtubule translocation. Cell Motil. Cytoskeleton 57, 246–258. doi: 10.1002/cm.10169
Vyplelová, P, Ovecka, M, and Šamaj, J. (2017). Alfalfa root growth rate correlates with progression of microtubules during mitosis and cytokinesis as revealed by environmental light-sheet microscopy. Front. Plant Sci. 8:1870. doi: 10.3389/fpls.2017.01870
Waghmare, S., Lileikyte, E., Karnik, R., Goodman, J. K., Blatt, M. R., and Jones, A. M. E. (2018). SNAREs SYP121 and SYP122 mediate the secretion of distinct cargo subsets. Plant Physiol. 178, 1679–1688. doi: 10.1104/pp.18.00832
Wang, X., Xu, M., Gao, C., Zeng, Y., Cui, Y., Shen, W., et al. (2020). The roles of endomembrane trafficking in plant abiotic stress responses. J. Integr. Plant Biol. 62, 55–69. doi: 10.1111/jipb.12895
Weijers, D., and Offringa, R. (2003). Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol. 133, 1882–1892. doi: 10.1104/pp.103.030692
Won, K. H., and Kim, H. (2020). Functions of the plant Qbc SNARE SNAP25 in cytokinesis and biotic and abiotic stress responses. Mol. Cells 43, 313–322. doi: 10.14348/molcells.2020.2245
Xia, L., Mar Marques-Bueno, M., Bruce, C. G., and Karnik, R. (2019). Unusual roles of secretory SNARE SYP132 in plasma membrane H+-ATPase traffic and vegetative plant growth. Plant Physiol. 180, 837–858. doi: 10.1104/pp.19.00266
Xia, L., Mar Marques-Bueno, M., and Karnik, R. (2020). Trafficking SNARE SYP132 partakes in auxin-associated root growth. Plant Physiol. 182, 1836–1840. doi: 10.1104/pp.19.01301
Xue, Y., Yang, Y., Yang, Z., Wang, X., and Guo, Y. (2018). VAMP711 is aequired for abscisic acid-mediated inhibition of plasma membrane H+-ATPase activity. Plant Physiol. 178, 1332–1343. doi: 10.1104/pp.18.00499
Xue, Y., Zhao, S., Yang, Z., Guo, Y., and Yang, Y. (2019). Regulation of plasma membrane H+-ATPase activity by the members of the V-SNARE VAMP7C family in Arabidopsis thaliana. Plant Signal. Behav. 14:e1573097. doi: 10.1080/15592324.2019.1573097
Yun, H. S., and Kwon, C. (2017). Vesicle trafficking in plant immunity. Curr. Opin. Plant Biol. 40, 34–42. doi: 10.1016/j.pbi.2017.07.001
Yano, D., Sato, M., Saito, C., Sato, M. H., Morita, M. T., and Tasaka, M. (2003). A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc. Natl. Acad. Sci. U.S.A. 100, 8589–8594. doi: 10.1073/pnas.1430749100
Yao, H. Y., and Xue, H. W. (2011). Signals and mechanisms affecting vesicular trafficking during root growth. Curr. Opin. Plant Biol. 14, 571–579. doi: 10.1016/j.pbi.2011.06.009
Yoon, T. Y., and Munson, M. (2018). SNARE complex assembly and disassembly. Curr. Biol. 28, R397–R401. doi: 10.1016/j.cub.2018.01.005
Yun, H. S., Kwaaitaal, M., Kato, N., Yi, C., Park, S., Sato, M. H., et al. (2013). Requirement of vesicle-associated membrane protein 721 and 722 for sustained growth during immune responses in Arabidopsis. Mol. Cells 35, 481–488. doi: 10.1007/s10059-013-2130-2
Zhang, B., Karnik, R., Alvim, J., Donald, N., and Blatt, M. R. (2019a). Dual sites for SEC11 on the SNARE SYP121 implicate a binding exchange during secretory traffic. Plant Physiol. 180, 228–239. doi: 10.1104/pp.18.01315
Zhang, B., Karnik, R., Wang, Y., Wallmeroth, N., Blatt, M. R., and Grefen, C. (2015a). The Arabidopsis R-SNARE VAMP721 interacts with KAT1 and KC1 K+ channels to moderate K+ current at the plasma membrane. Plant Cell 27, 1697–1717. doi: 10.1105/tpc.15.00305
Zhang, L., Li, W., Wang, T., Zheng, F., and Li, J. (2015b). Requirement of R-SNAREs VAMP721 and VAMP722 for the gametophyte activity, embryogenesis and seedling root development in Arabidopsis. Plant Growth Regul. 77, 57–65. doi: 10.1007/s10725-015-0035-0
Zhang, L., Liu, Y., Zhu, X. F., Jung, J. H., Sun, Q., Li, T. Y., et al. (2019b). SYP22 and VAMP727 regulate BRI1 plasma membrane targeting to control plant growth in Arabidopsis. New Phytol. 223, 1059–1065. doi: 10.1111/nph.15759
Zhang, L., Ma, J., Liu, H., Yi, Q., Wang, Y., Xing, J., et al. (2021). SNARE proteins VAMP721 and VAMP722 mediate the post-Golgi trafficking required for auxin-mediated development in Arabidopsis. Plant J. 108, 426–440. doi: 10.1111/tpj.15450
Zhang, B., Wang, H., and Zhang, Y. (2020). SNARE proteins and their role in plant ion channel regulation. Plant Growth Regul. 92, 443–453. doi: 10.1007/s10725-020-00656-7
Zhang, L., Zhang, H., Liu, P., Hao, H., Jin, J. B., and Lin, J. (2011a). Arabidopsis R-SNARE proteins VAMP721 and VAMP722 are required for cell plate formation. PLoS One 6:e26129. doi: 10.1371/journal.pone.0026129
Zhang, X., Zhao, H., Gao, S., Wang, W. C., Katiyar-Agarwal, S., Huang, H. D., et al. (2011b). Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol. Cell 42, 356–366. doi: 10.1016/j.molcel.2011.04.010
Zhao, M., Wu, S., Zhou, Q., Vivona, S., Cipriano, D. J., Cheng, Y., et al. (2015). Mechanistic insights into the recycling machine of the SNARE complex. Nature 518, 61–67. doi: 10.1038/nature14148
Zheng, H., Bednarek, S. Y., Sanderfoot, A. A., Alonso, J., Ecker, J. R., and Raikhel, N. V. (2002). NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE. Plant Physiol. 129, 530–539. doi: 10.1104/pp.003970
Zheng, J., Han, S. W., Rodriguez-Welsh, M. F., and Rojas-Pierce, M. (2014). Homotypic vacuole fusion requires VTI11 and is regulated by phosphoinositides. Mol. Plant 7, 1026–1040. doi: 10.1093/mp/ssu019
Zhu, J., Gong, Z., Zhang, C., Song, C. P., Damsz, B., Inan, G., et al. (2002). OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 14, 3009–3028. doi: 10.1105/tpc.006981
Zhu, X. F., Liu, Y., Gai, X. T., Zhou, Y., Xia, Z. Y., Chen, L. J., et al. (2019). SNARE proteins SYP22 and VAMP727 negatively regulate plant defense. Plant Signal. Behav. 14:1610300. doi: 10.1080/15592324.2019.1610300
Keywords: Arabidopsis, vesicle trafficking, SNAREs, membrane fusion, root
Citation: Luo C, Shi Y and Xiang Y (2022) SNAREs Regulate Vesicle Trafficking During Root Growth and Development. Front. Plant Sci. 13:853251. doi: 10.3389/fpls.2022.853251
Edited by:
Hao Wang, South China Agricultural University, ChinaReviewed by:
Yonglun Zeng, The Chinese University of Hong Kong, ChinaXiangfeng Wang, China Agricultural University, China
Xiaohong Zhuang, The Chinese University of Hong Kong, China
Copyright © 2022 Luo, Shi and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Xiang, eGlhbmd5QGx6dS5lZHUuY24=
 Changxin Luo
Changxin Luo Yumei Shi
Yumei Shi Yun Xiang
Yun Xiang