
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 25 March 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.852377
This article is part of the Research TopicDiscovery, Biosynthesis, Regulation, Transport, Release and Engineering of Plant Natural ProductsView all 13 articles
 Shuang Ye1†
Shuang Ye1† Lei Feng1†
Lei Feng1† Shiyu Zhang2,3
Shiyu Zhang2,3 Yingchun Lu1
Yingchun Lu1 Guisheng Xiang1
Guisheng Xiang1 Bo Nian1
Bo Nian1 Qian Wang1
Qian Wang1 Shuangyan Zhang1
Shuangyan Zhang1 Wanling Song1
Wanling Song1 Ling Yang1
Ling Yang1 Xiangyu Liu1
Xiangyu Liu1 Baowen Feng4
Baowen Feng4 Guanghui Zhang1
Guanghui Zhang1 Bing Hao1*
Bing Hao1* Shengchao Yang1*
Shengchao Yang1*
Gynostemma longipes contains an abundance of dammarane-type ginsenosides and gypenosides that exhibit extensive pharmacological activities. Increasing attention has been paid to the elucidation of cytochrome P450 monooxygenases (CYPs) and UDP-dependent glycosyltransferases (UGTs) that participate downstream of ginsenoside biosynthesis in the Panax genus. However, information on oxidosqualene cyclases (OSCs), the upstream genes responsible for the biosynthesis of different skeletons of ginsenoside and gypenosides, is rarely reported. Here, an integrative study of the metabolome and the transcriptome in the leaf, stolon, and rattan was conducted and the function of GlOSC1 was demonstrated. In total, 46 triterpenes were detected and found to be highly abundant in the stolon, whereas gene expression analysis indicated that the upstream OSC genes responsible for saponin skeleton biosynthesis were highly expressed in the leaf. These findings indicated that the saponin skeletons were mainly biosynthesized in the leaf by OSCs, and subsequently transferred to the stolon via CYPs and UGTs biosynthesis to form various ginsenoside and gypenosides. Additionally, a new dammarane-II synthase (DDS), GlOSC1, was identified by bioinformatics analysis, yeast expression assay, and enzyme assays. The results of the liquid chromatography–mass spectrometry (LC–MS) analysis proved that GlOSC1 could catalyze 2,3-oxidosqualene to form dammarenediol-II via cyclization. This work uncovered the biosynthetic mechanism of dammarenediol-II, an important starting substrate for ginsenoside and gypenosides biosynthesis, and may achieve the increased yield of valuable ginsenosides and gypenosides produced under excess substrate in a yeast cell factory through synthetic biology strategy.
Ginsenosides are valuable natural compounds found throughout species of the Panax genus that have the ability to treat cardiovascular and cerebrovascular diseases (Liu et al., 2017). Based on the different skeletons, three types of ginsenoside have been found in the Panax genus: dammarane-, ocotillo-, and oleanane-type ginsenosides (Christensen, 2008; Hou et al., 2021). Previous studies have shown that dammarane ginsenosides exist only in the Panax genus, such as Panax ginseng (ginseng), Panax quinquefolium (American ginseng), and Panax notoginseng (Sanqi), which are the three main species of the Panax genus cultivated worldwide. The global market demands for this high-value medical plant cannot be met due to the 4- to 7-year planting cycle and low yield (He et al., 2019).
Species in the Gynostemma genus, a member of the Cucurbitaceae family, have similar dammarane ginsenoside content to those in the Panax genus (Gantait et al., 2020). Gynostemma longipes has the highest content of dammarane ginsenoside, reaching 25%, which is even higher than Panax plants (Razmovski-Naumovski et al., 2005). Gypenosides in G. longipes have panaxadiol and 2α-hydroxypanaxadiol skeletons, which are similar to those of dammarane ginsenosides and have similar pharmacological effects, such as anticancer, cardioprotective, hepatoprotective, neuroprotective, and anti-inflammatory activities (Nagai et al., 1976; Li et al., 2019; Nguyen et al., 2021; Su et al., 2021). Unlike the medicinal root of Panax genus, the main medical parts of G. longipes are the stolon and the leaf, which can be harvested for many years from a single planting, indicating it was more convenient to obtain dammarane-type saponins from Gynostemma plants than from Panax species. G. longipes has become an alternative plant to support the deficiencies in the ginsenoside extract market (Ky et al., 2010). In China, 3.2 billion tons of G. longipes are processed for pharmaceutical extraction, with an annual production of 803 billion RMB in 2018 (Cheng and Cheng, 2019).
In the past decade, researchers have employed synthetic biology techniques to produce valuable natural compounds from microorganism that are difficult to product through chemical synthesis to satisfy the demands of the market. The ginsenosides R0, Rh2, Rg3, Rg1, and Rb1 have been biosynthesized in Escherichia coli and yeast (Wang et al., 2015; Lu et al., 2018; Tang et al., 2019, 2021). Previous studies have described the biosynthesis of dammarenediol-II from 2,3-oxidosqualene under the catalysis of dammarenediol-II synthase. PgCYP716A47 catalyzed the hydroxylation at the C-12 of dammarenediol to form protopanaxadiol (Han et al., 2011). After the formation of the protopanaxadiol skeleton, UDP-dependent glycosyltransferases add glucose to different positions to generate various ginsenosides: Pq-O-UGT1, PgUGT45, or PgUGT74AE2 catalyzed the glycosylation of protopanaxadiol to form ginsenoside Rh2; Pq-O-UGT2 and PgUGT94 catalyzed Rh2 to form Rg3 (Yang et al., 2020); Rd was formed under the catalysis of PgUGT71; PgUGT1 and PgUGT71A27 catalyzed protopanaxadiol to form compound K (CK), which was subsequently catalyzed by PgUGT74AE2, PgUGT71A27, Pq-O-UGT2, and PgUGT71A27 to produce F2 and Rd. PgUGTdGT and PgUGT71A29 could utilize Rd as substrate to generate Rb1 (Yang et al., 2018; Figure 1). These ginsenosides mentioned above were found in Gynostemma plants, indicating Gynostemma was the best resource for ginsenosides after Panax plants (Qin et al., 1992).
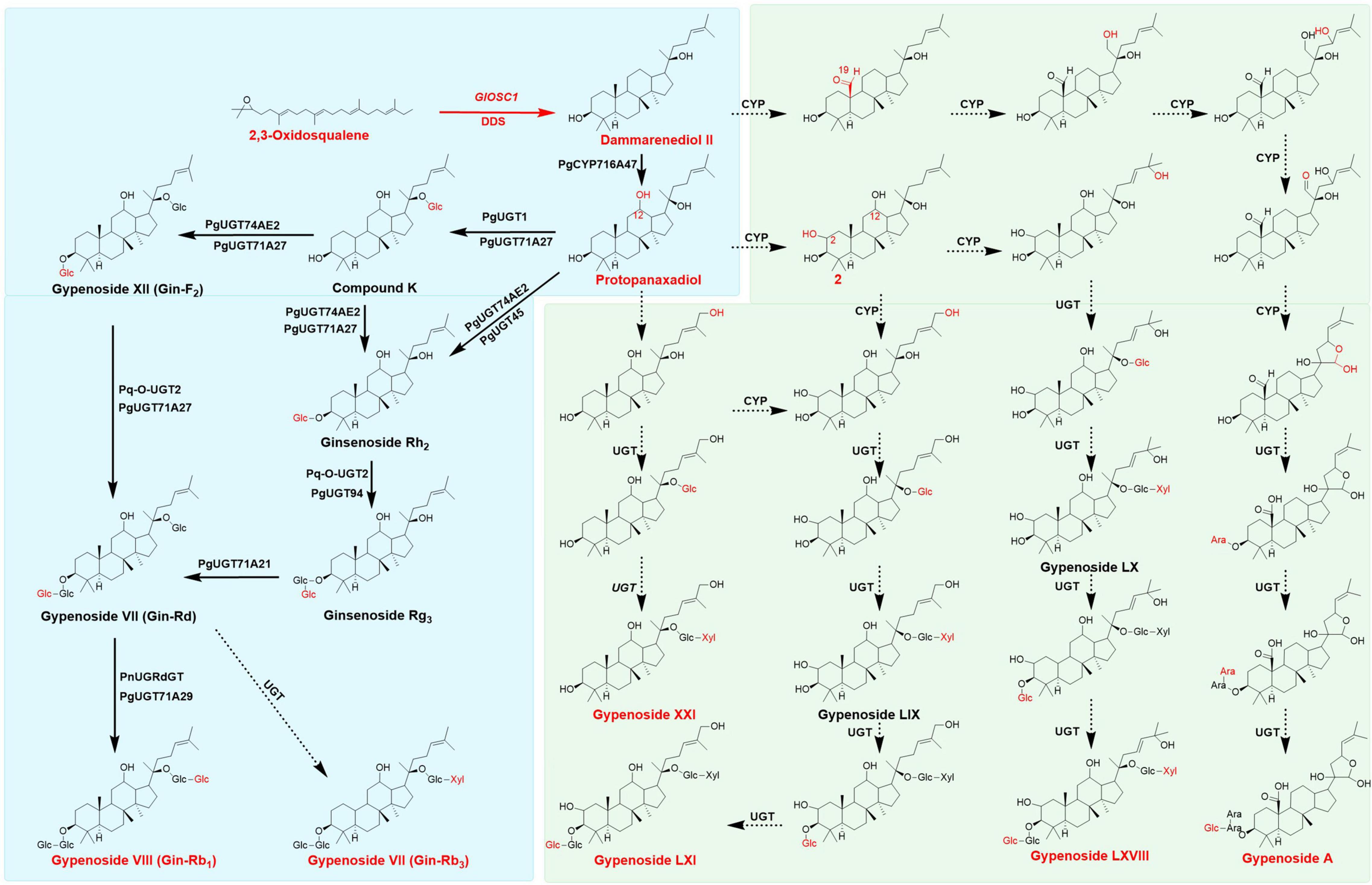
Figure 1. Proposed Biosynthesis pathway of gypenosides and ginsenosides, the main triterpenes were colored in red, the broad arrows presented the enzymes already identified, dotted arrows presented the assumed enzymes in gypenosides analysis. Pathways in green background only found in Gynostemma plants, pathways in blue back ground could appear both in Gynostemma plants and Panax plants.
The genes involved in biosynthesis of gypenosides were remain large unknown, few studies analyzed and predicted the possible candidate genes involved in gypenosides biosynthesis in G. pentaphyllum (Huang et al., 2021; Zhang et al., 2021). Recently, 5 UDP-glycosyltransferases were elucidated in ginsenoside biosynthesis which could form F1, Rh2, Rg3, CK, F2 and Rd with UDP-glucose as sugar donor in G. pentaphyllum (Le et al., 2021). The biosynthesis of gypenoside starts with the formation of 2,3-oxidosqualene through the mevalonate (MVA) pathway in the cytoplasm and the methylerythritol phosphate (MEP) pathway in plastids. 2,3-Oxidosqualene forms different triterpenoid skeletons under the catalysis of oxidosqualene cyclase (OSCs), which are subsequently modified by CYPs and UDP-dependent glycosyltransferases (UGTs), forming different ginsenosides (Kim et al., 2015; Yang et al., 2018). However, compared with ginsenosides, the structural diversity of gypenosides was more abundant owing to multiple levels of oxidation (CH2OH or CHO) at C-2 and C-19 sites, as well as the side chains in gypenosides (Figure 1; Yoshikawa, 1986; Razmovski-Naumovski et al., 2005; Nguyen et al., 2021). However, the specific genes involved gypenoside biosynthesis and their functions remain largely uncharacterized.
To elucidate the biosynthesis pathway of gypenosides in Gynostemma plants, an integrated analysis of transcriptomics and metabolomics was performed in this study to provide more information on various specific genes and gypenosides. The solon and leaf of Gynostemma longipes, the main resource for commercial gypenoside extraction in China, were collected and analyzed by high-throughput omics techniques. Three non-steroidal-type triterpene synthases that catalyze oxidosqualene through dammarenyl cations were found after comparison of amino acids and phylogenetic analysis. Twelve candidate CYPs were found to participate in gypenoside biosynthesis by performing gene co-expression analysis between CYPs and triterpenes. Moreover, a dammarenediol-II synthase that catalyzed 2,3-oxidosqualene to form dammarenediol-II was found and verified in yeast. These results provide insight into gypenoside biosynthesis in Gynostemma plants to allow better development of gypenosides using synthetic biology techniques.
cDNA libraries were constructed from the total RNA of the leaf, rattan, and stolon to characterize transcriptome of G. longipes. Three biological replicates of each tissue were processed and sequenced on an Illumina Hiseq™ 2000 platform. In total, 438,908,642 (65,385,856,136 bp) clean reads were generated with 44.5429% GC after filtering out adaptor sequences, ambiguous reads, and low-quality reads (Q20 < 20). All clean reads were assembled into 1,290,066 unigenes within a total length of 96,791,710 bp. The number order of unigenes contig N50 was 23114; the length was 1184 bp. The size of unigenes ranged from 201 bp to 16470 bp with an average length of 749 bp (Table 1). The high-quality reads in this research have been deposited in NCBI SRA database (accession number: PRJNA784129).
BLASTx was applied to annotate unigenes based on four databases: NCBI non-redundant protein (Nr), Kyoto Encyclopedia of Genes and Genomes (KEGG), the Clusters of Orthologous Groups/Clusters of orthologous groups for eukaryotic complete (COG/KOG), and the Swiss-Prot protein database. In total, 81,721 (63.32%) unigenes were annotated; among them, 78,398 (60.74%) unigenes were matched in the Nr database, 66,100 (51.21%) unigenes were matched in the KEGG database, 49,036 (37.99%) unigenes were matched in COG/KOG database, and 58,300 (45.17%) unigenes were matched in the Swiss-Prot database. Overall, 41,791 (32.38%) unigenes were matched in all databases (Table 2).
The comparison of unigenes from the three tissues identified 4883, 2374, and 4480 unigenes specifically annotated in the rattan, stolon, and leaf, respectively, with 15,015 unigenes annotated in all tissues (Supplementary Figure S1B). The results showed that 59,894 unigenes were classified in biological process ontology (GO: 008150), the major terms were metabolite process (15,454, 25.80%), cellular process (12,824, 21.41%), and single-organism process (9791, 16.35%); 26,224 unigenes belonged to cell component ontology (GO:0005575) (Supplementary Figure S1A). In this ontology analysis, catalytic activity (13,234, 50.47%) and binding (10380, 39.58%) were the major terms; 43,190 unigenes belonged to molecular function ontology (GO: 003647), cell (10701, 24.78%), cell part (10687, 24.74%), and organelle (6710, 15.53%) (Supplementary Figure S1C). Moreover, based on KEGG annotation, 8877 unigenes were divided into five categories, including organism system, cellular processing, environment information process, genetic information process, and metabolism. It is notable that 491 unigenes (3.5% of the metabolism category) participated in the metabolism of terpenoids and polyketides (Supplementary Figure S1A). These data provided basic information for mining candidate genes involved in gypenoside biosynthesis. The original data of RNA-seq annotation and expression could be downloaded in Medicinal Plants multi-Omics Database1 (He et al., 2022).
The metabolomic analyses of the leaf, stolon, and rattan detected 46 triterpenes in G. longipes. Seventeen triterpenes (ginsenoside Rd2 to 2-3-O-acetyl-3,12,23,24-tetrahydroxy-20,25-epoxydammarane-3-O-xylopranosyl-glucopyranoside) had the highest content in the leaf, followed by the rattan (*indicates isomers exist). For nine triterpenes (from gypenoside 16 to m-Gin-Rd), the content was higher in the rattan than in the stolon. The relative content of 20 triterpenes in the stolon was higher than in the rattan and the leaf (from gypenosides LXXVI to ginsenoside Rb2). The results showed that the content of 29 triterpenes was relatively high in the stolon, suggesting it might be a potential tissue in which triterpene accumulation occurs (Figure 2). Otherwise, the relative contents of gypenoside LXI, gypenoside A, and gypenoside LXVIII were relatively high in the whole plant, indicating these two triterpenes may be the main triterpenes in G. longipes.
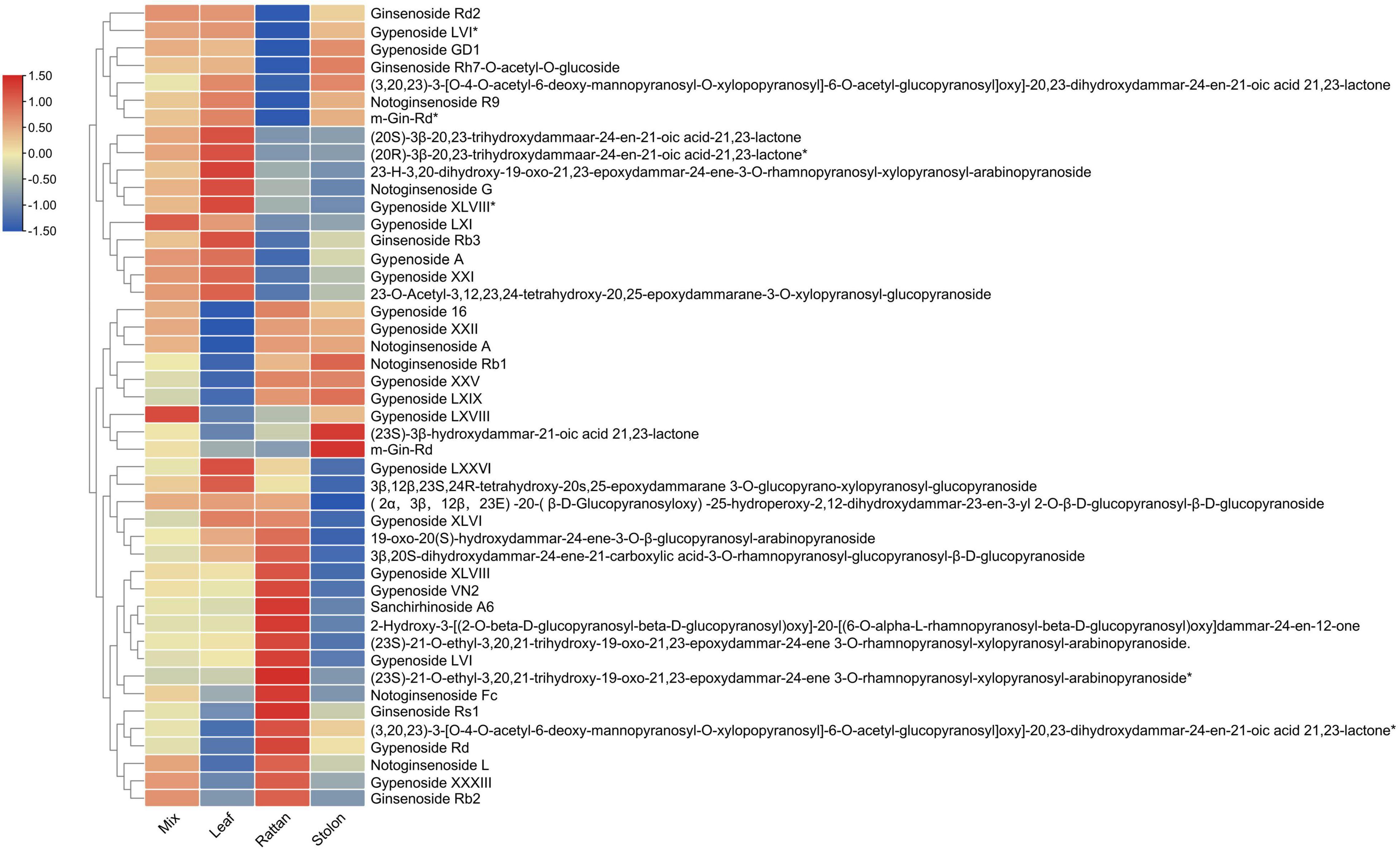
Figure 2. The heatmap of triterpene content level in leaf, rattan, stolon and whole plant of G. longipes.
From the transcriptome analysis, 16 genes were considered to encode key enzymes involved in triterpene biosynthesis through the MVA pathway: four HMGRs (3-hydroxy-3-methylglutaryl-coenzyme A reductases), one FPS (farnesyl diphosphate synthases), one SS (squalene synthase), four SQEs (squalene epoxidases), and six OSCs (2,3-oxidosqualene cyclases). The expression of SS, FPS, HMGR2, SQE1, SQE2, GlOSC1, and GlOSC2 was extremely high in the leaf, whereas GlOSC5, HMGR3, and GlOSC6 were highly expressed in the stolon. The expression of SQE3, SQE4, and GlOSC5 was highest in the rattan, followed by the stolon (Figure 3A). Genes in the triterpene biosynthesis pathway were co-expressed; the expressions of SS, FPS, HMGR2, SQE1, SQE2, GlOSC1, and GlOSC2 in the leaf were highly correlated and therefore the leaf is a possible tissue for the biosynthesis of triterpene skeletons. Real-time quantitative polymerase chain reaction (PCR) analysis was performed to uncover the expression of candidate genes in triterpenoid biosynthesis. The expression pattern of other 14 genes was similar to those in the transcriptome data, except HMGR5 and GlOSC4 (Figure 3B).
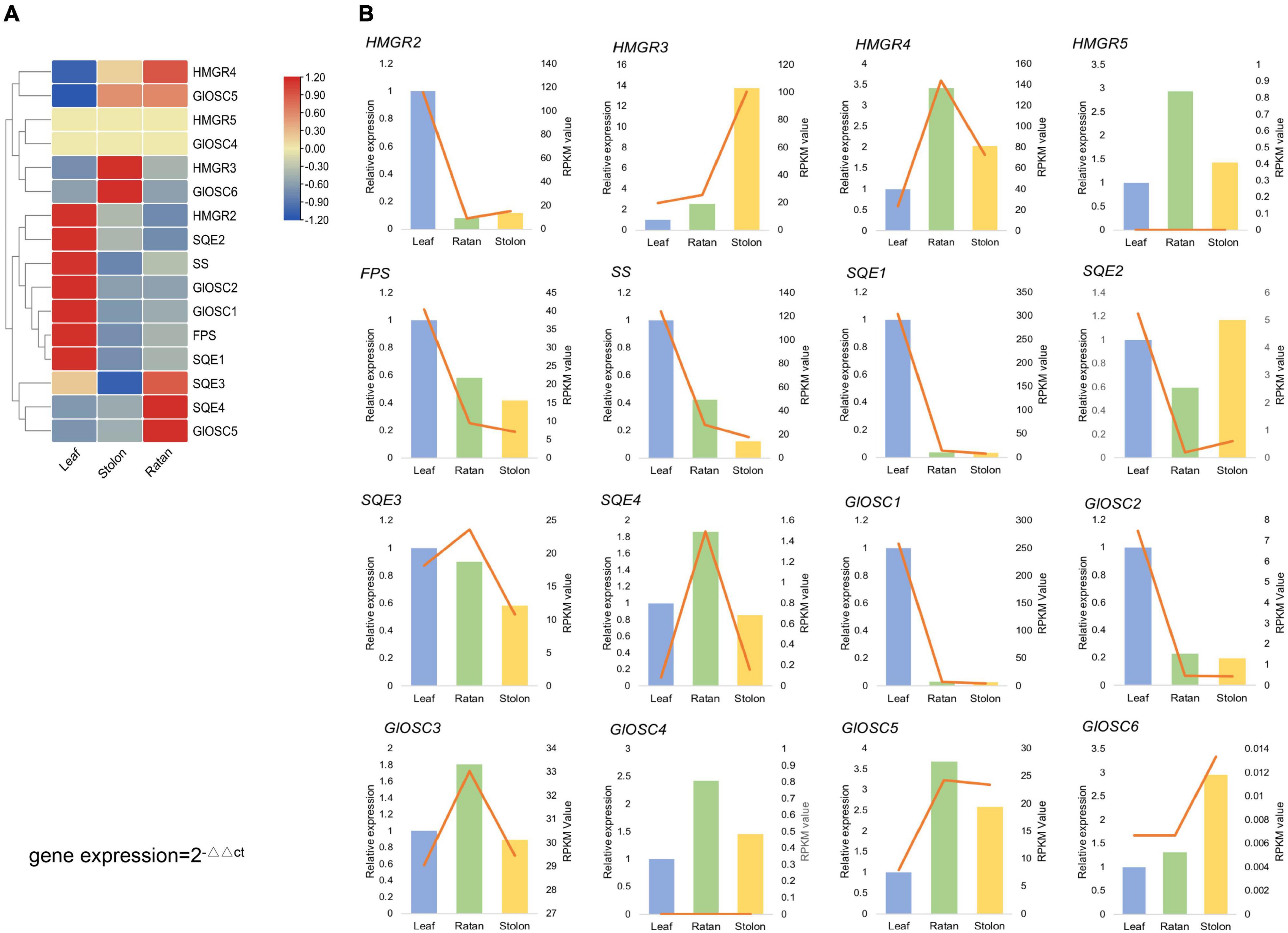
Figure 3. (A) Expression analysis of the candidate genes, expression level of HMGRs, GlOSCs, SSs, FPS, SE in leaf, rattan and stolon of G. longipes. (B) Real-time quantitative polymerase chain reaction (PCR) validation of candidate unigenes involved in gypenosides biosynthesis by RNA-seq. The histogram indicated the relative gene gypenosides biosynthesis by RNA-seq, the right y-axis was gene expression level calculated as RPKM value, the left y-axis was relative gene expression obtained via real-time PCR. The relative expression obtained from real-time PCR calculated by 2–ΔΔCt method.
While OSCs are the key enzymes that determine the skeleton diversity of triterpenes (Phillips et al., 2006). Seven candidate OSCs were obtained and named GlOSC1 to GlOSC7; the number of amino acids ranged from 777 to 583. The results of the alignment analysis showed that all candidates contained a DCTAE motif that is highly conserved in OSCs families and is responsible for initiating the cyclization reaction (Abe and Prestwich, 1995; Ito et al., 2013). GlOSC1, GlOSC5, and GlOSC7 had conserved Y257, T364, D474, and E556, whereas four others OSCs had conserved H257, C364, T474, and D556. These residues were found to differentiate the steroidal-type triterpene synthases from the non-steroidal-type (Corey et al., 1997; Forestier et al., 2019). In this case, GlOSC1, GlOSC5, and GlOSC7 were the most likely OSCs to cyclize oxidosqualene through reaction with the dammarenyl cation (Supplementary Figure S2).
The phylogenetic relationship between GlOSCs and known OSCs from plants was analyzed to support further prediction of the function of these OSCs. Two exogenous OSCs (RatLAS and BosLAS) were added for the construction of phylogenetic relationships (Figure 4A). Based on the different catalyzed cyclization reaction formation, the sequences were divided into two groups. The first group (GlOSC2, GlOSC3, GlOSC4, and GlOSC5) contained CAS (cycloartenol synthase), LAS (lanosterol synthase), CbQ (cucurbitadienol synthase), and PS (parkeol synthase), which were able to fold 2,3-oxidosqualene into the chair-boat-chair conformation through the protosteryl cation when initiating the cyclization reaction. The second group (GlOSC5, GlOSC1, and GlOSC7) contained βAS (β-amyrin synthase), DDS (dammarenediol II synthase), LUS (lupeol synthase), MS (mix-triterpene synthase), these were capable of folding substrates into chair–chair–chair (CCC) conformation through dammarenyl cation (Phillips et al., 2006; Thimmappa et al., 2014). This result was consistent with the results of the amino acid sequence comparison mentioned above. The first group was steroidal-type triterpene synthases; the second group was non-steroidal-type triterpene synthase.
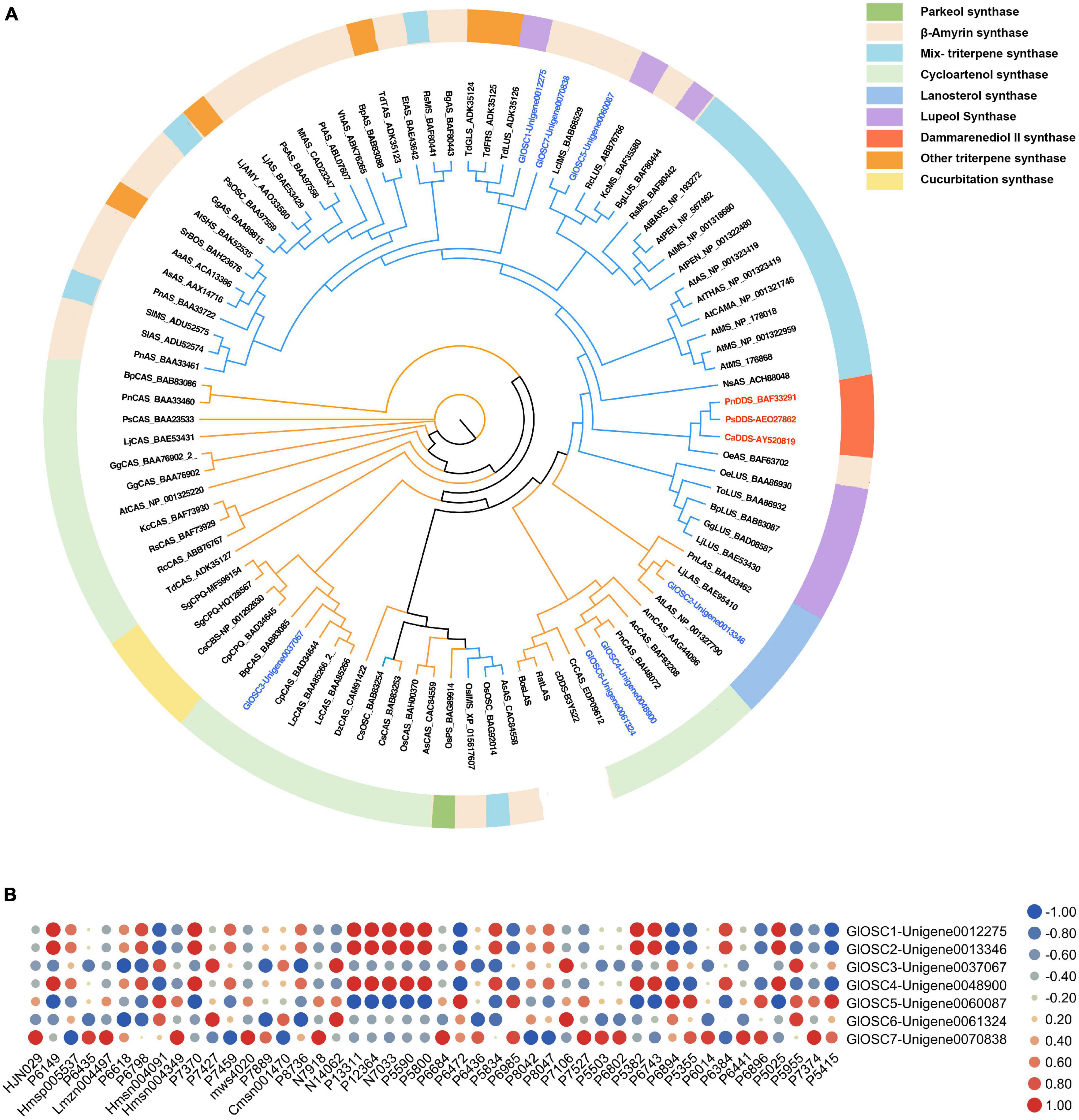
Figure 4. (A) Phylogenetic analyses of the GlOSCs from G. longipes and characterized OSCs from other plants. Their catalytic functions were classified in colors, green: parkeol synthase, light orange: β-amyrin synthase, light blue: Mix-triterpene synthase, light green: cycloartenol synthase, purple: lupeol synthase, red: dammarenediol synthase, orange: triterpene synthase with other functions, light yellow: cucurbitadienol synthase. (B) Pearson correlation analysis between expression level of GlOSCs and content level of triterpenes, the red cycle presented positive correlation, the blue cycle presented negative correlation, and size of cycle presented the degree of correlation.
Additionally, we performed Pearson correlation analysis between the candidate OSCs and all triterpenes using SPSS 26 (Figure 4B); the results showed that GlOSC1, GlOSC2, and GlOSC4 had the same correlation pattern between gene expression and triterpene content. The expression was highly positively correlated with the content of P7370, P1331, P12364, P5590, and P5800, and negatively correlated with the content of P6472. The expression of GlOSC5 was negatively correlated with the content of P1331, P12364, P5590, and P5800, however, positively correlated with the content of P6472 (Supplementary Table S3). The expression of GlOSC3 and GlOSC6 was negatively correlated with the content of P6618, but positively correlated with the content of N14062 and P7106. The correlation pattern between GlOSC7 expression and the content of triterpenes is different from others; the expression of GlOSC7 is positively correlated with the content of Lmzn004497, msw4020, and N7918, but negatively correlated with the content of P8042. All correlations mentioned above were under the conditions of a Pearson correlation coefficient (PCC) of ≥0.9 and a p-value of ≤0.05. These results suggest that GlOSC1, GlOSC2, and GlOSC4 are likely connected to the biosynthesis of triterpene skeletons in the leaf.
As the third largest family of plant genes, CYPs play an essential role in the formation and evolution of metabolism in plants (Nelson and Reichhart, 2011). CYPs are crucial enzymes in natural product biosynthesis and metabolism in plants; they catalyze a wide range of reactions, including hydroxylation, decarboxylation, epoxidation, C–C bond formation, N- and O-demethylation, and other oxidizing reactions (Jeffreys et al., 2018; Li et al., 2020). In terpenoid biosynthesis, CYPs determine the structural diversity and bioactivity of terpenoids upstream, with more than 97% of terpenoids modified under catalysis by CYPs (Guo et al., 2016; Xiao et al., 2018). In this study, 250 CYPs were identified by HMMER 3.0. The phylogenic tree was constructed with 250 candidate CYPs, 74 reference CYPs, and 245 AtCYPs by iqtree (Figure 5A). To date, 11 clans of plant CYPs have been found, and the CYPs of G. longipes are categorized into 10 clans. Seven unigenes between clan 97 and clan 711 and one unigene between clan 74 and clan 71 did not belong to any clans. Clan 71 contains the highest number of CYP unigenes involved in gypenoside biosynthesis. PgCYP716A47 and PgCYP716A53v2 are important enzymes that catalyze dammarenediol to protopanaxadiol and protopanaxadiol to protopanaxatriol, respectively (Figure 5B; Han et al., 2011, 2012). According to the results of the phylogenetic analysis, Unigene0005406 and Unigene0122930 had 51.8% identity to PgCYP716A53v2; thus, Unigene0005406 and Unigene0122930 are potential protopanaxadiol synthases in G. longipes.
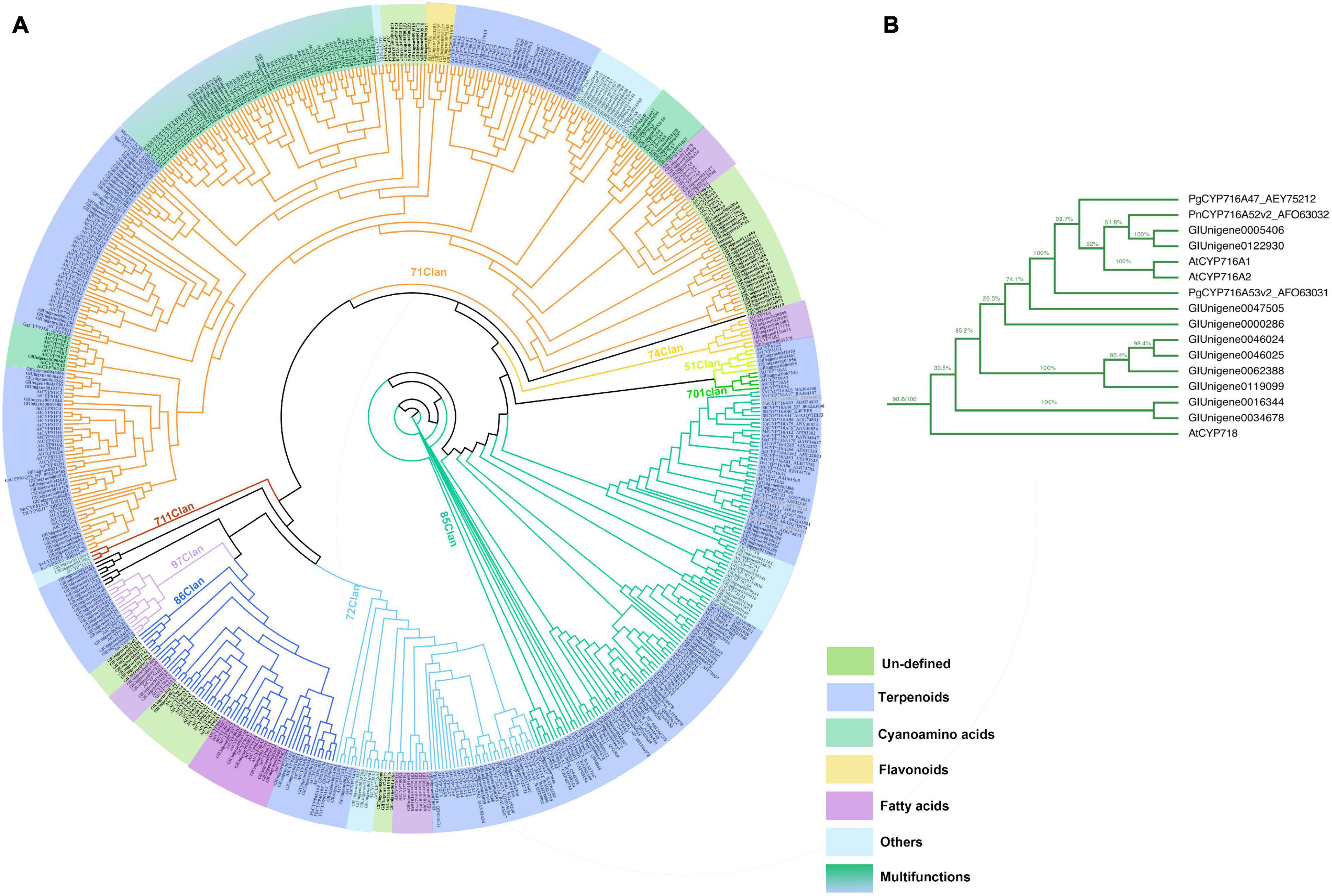
Figure 5. (A) Phylogenetic analyses of the GlCYPs from G. longipes and other reference CYPs, their catalytic functions were classified in colors, blue part related to terpenoids modification, green part related to cyanoamino acid modification, yellow part related to flavonoids modification, light blue part might have other functions, lawngreen part were unidentified. (B) Local zoom of Figure 5A. The phylogenetic analysis of several GlCYPs and PgCYP716As.
For a better understanding of the association between genes and gypenosides, the integrated transcriptome and metabolite data were analyzed by applying a correlation analysis, and the network was constructed. Pearson correlation analysis was conducted between the expression of 209 CYPs and the content of 46 triterpenes using SPSS 26; only correlation pairs satisfying the conditions of PCC ≥ 0.8 and p-value ≤ 0.05 were selected. In total, 214 pairs were identified and visualized by Cytoscape (Figure 6 and Supplementary Table S3). In the network, 91 nodes were connected by 214 edges. These 91 nodes included 38 triterpenes and 53 unigenes; here, 60 pairs were negatively correlated and 154 pairs were positively correlated. From the metabolism data, P6441 (gymnemaside III), P6014 (gypenoside LVI), and P5025 (gypenoside LXI) were all hydroxylated at the C-2 position. The results of the correlation analysis showed that the content of P6441 was positively correlated with the expression of Unigene0032271 (PCC = 0.818), Unigene0067788 (PCC = 0.908), and Unigene0081102 (PCC = 0.892); the content of P6014 was positively correlated with the expression of Unigene0114879; and the content of P5025 was positively correlated with the expression of Unigene0001688 (PCC = 0.803), Unigene0039203 (PCC = 0.801), and Unigene0086841 (PCC = 0.885). Thus, these unigenes are likely to catalyze the hydroxylation at the C-2 position. In addition, P6684 [(23S)-21-O-ethyl-3,20,21-trihydroxy-19-oxo-21,23-epoxydammar-24-ene-3-O-rhamnopyranosyl-xylopyranosyl-arabinopyranoside*] and P5382 (gypenoside XLVIII*) with an aldehyde group at the C-19 position, were also in a distinct position from the gypenosides. The content of P6684 was positively correlated with the expression of Unigene0043066 (PCC = 0.812), Unigene0067788 (PCC = 0.937), and Unigene0081102 (PCC = 0.926); the content of P5382 was positively correlated with the expression of Unigene0015374 (PCC = 0.818) (Supplementary Table S2). Therefore, these three unigenes were highly likely to be related to the oxidation at the C-19 position.
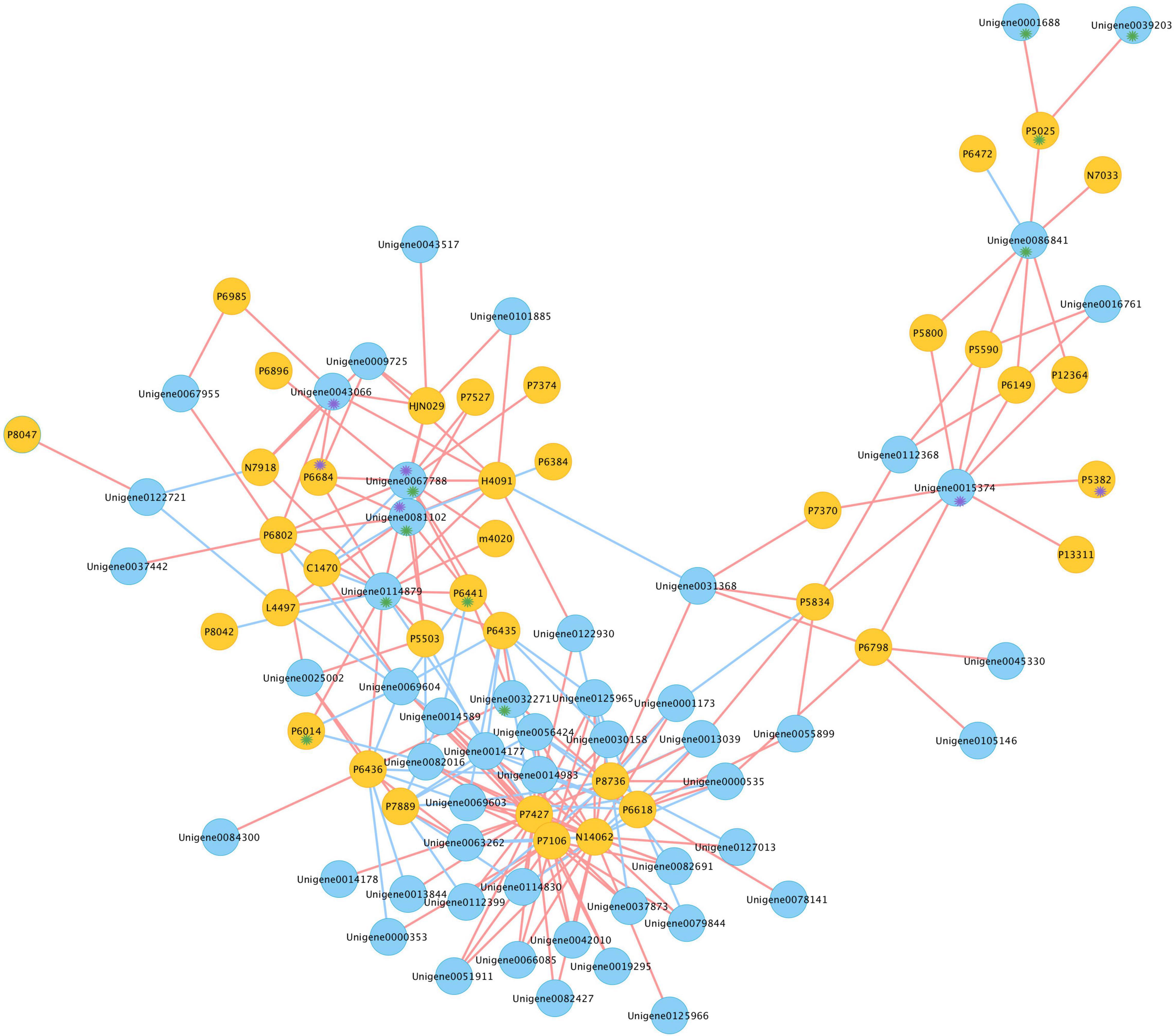
Figure 6. Co-expression network of candidates CYPs and triterpenes in G. longipes. Blue nodes represented genes, yellow nodes represented triterpenes, red edges presented positive correlation, blue edges presented negative correlation. Compounds marked with purple * had oxidization at C-19, unigene marked with purple * possibly catalyzed oxidization at C-19; compounds marked in green * had hydroxylation at C-2, unigene marked with green * possibly catalyzed hydroxylation at C-2.
The full-length open reading frame of GlOSC1 was successfully cloned from G. longipes cDNA and inserted in the pYES2 expression vector under the control of the GAL1 promoter. The transgene vectors were transformed into Saccharomyces cerevisiae BY4742 by addition of galactose. The final product of the yeast extracts was analyzed by LC chromatogram and liquid chromatography–mass spectrometry (LC–MS) after incubation for 24 h. The results showed that the extract from transgenic yeast expressing GlOSC1 appeared a peak at 23.56 min, which matched the retention time of the dammarenediol-II standard peak at 23.55 min. In contrast, the LC–MS of extracts from the empty vector showed no dammarenediol-II product (Figure 7). In addition, the LC–MS was performed to verify the molecular weight of the dammarenediol-II product. The spectrum of GlOSC1 transgenic yeast showed LC/APCIMS fragmentation ion values of the dammarenediol II product at m/z 427 [M+H–H2O]+, m/z 409 [M+H–2H2O]+, m/z 219, and m/z 191, (Supplementary Figures S3, S4) which were consistent with dammarenediol-II standard previously described (Han et al., 2006). The peak at 23.56 min from the extract of transgenic yeast GlOSC1 also had the same LC/APCIMS fragmentation ion values as those in authentic dammarenediol-II. These data indicated that the Unigene0012275 (GlOSC1) could catalyze 2,3-oxidosqualene to produce dammarenediol-II via cyclization.
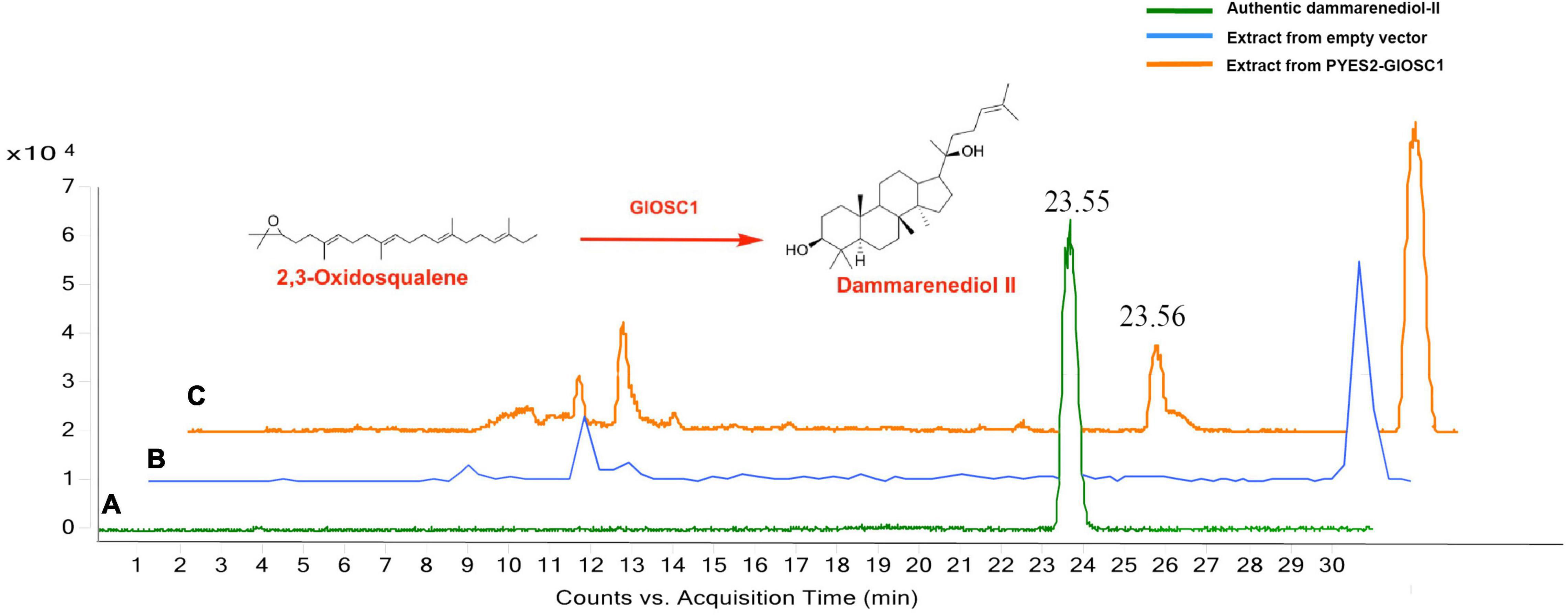
Figure 7. Three chromatograms of the analysis: (A) authentic dammarenediol II, (B) the extracts of the yeast with empty vector, and (C) dammarenediol II biosynthesized by GlOSC1 in yeast.
Triterpenoids are not only crucial compounds for plant development and growth, but are also important sources of drugs and antibiotic for humans and other creatures (Haralampidis et al., 2002). Owing to their various biological properties, researchers are interested in their molecular structures. Currently, more than 100 distinct skeletons of triterpenoids are found in plants (Xu et al., 2004). OSCs are the key enzymes that determine the diversity of triterpene skeletons (Phillips et al., 2006; Cardenas et al., 2019).
Previous transcriptome studies showed that FPS, SS, SE, and βAS were highly expressed in the leaf of G. pentaphyllum (Chen et al., 2016; Liang et al., 2019; Xu et al., 2020). In this study, based on gene expression profiling, the expression patterns of upstream genes (SS, FPS, HMGR2, SQE1, SQE2, OSC1, OSC2, and OSC4) in the biosynthesis of gypenosides were highly correlated in the leaf. This result indicated that the leaf was the main tissue for biosynthesis of terpene skeletons in G. pentaphyllum. Meanwhile, the metabolism profile indicated that the content of 17 triterpenes and 20 triterpenes was relatively high in the leaf and the stolon, respectively. In addition, the content of only nine triterpenes was high in the rattan; thus, the leaf and stolon may be the main tissues for biosynthesis and modification of triterpene skeletons and the accumulation of gypenosides.
Based on their functions, the OSCs can be classified as steroidal synthases or non-steroidal synthase (Thimmappa et al., 2014). In this study, seven OSCs were obtained from the G. longipes transcriptome; according to the phylogenetic analysis, all candidate OSCs had a conserved DCTAE motif that indicated triterpene synthase activity. Unigene0012275 (GlOSC1), Unigene0060087 (GlOSC5), and Unigene0070838 (GlOSC7) contained conserved residues Y257, T364, D474, and E556, which belong to non-steroidal-type triterpene synthases (Ito et al., 2013). Since 2006, PgDDS was first identified from P. ginseng (Tansakul et al., 2006), and PqDS, CaDDS, and PnDS were subsequently identified (Kim et al., 2009; Niu et al., 2014; Wang et al., 2014). Candidate OSCs in this research shared low homology with these four DDSs. A multi-functional triterpene synthase LsOSC1 was reported that could catalyze the formation of dammarenediol-II in Lactuca sativa. TcOSC1 and TcOSC2 were also identified in Taraxacum coreanum (Han et al., 2019; Choi et al., 2020). GlOSC1 showed high homology with LsOSC1, TcOSC1, and TcOSC2, and the results of the functional expression experiment proved that GlOSC1 had the ability to catalyze the formation of dammarenediol-II; this was the first OSC reported in the Gynostemma genus.
To date, 328 dammarane-type saponins from Gynostemma plants have been reported (Nguyen et al., 2021). The downstream CYPs and UGTs genes were also determined to affect the diversity of gypenosides. During terpenoid biosynthesis processes in plants, more than 97% CYPs catalyzed the structural diversity of terpenoids via oxygenation reactions (Guo et al., 2016). A previous study showed that 25% of gypenosides shared similar structures to ginsenosides; in the case of panaxadiol and 2α-hydroxypanaxadiol, this was the basic structure of gypenosides (Nagai et al., 1976; Subramaniyam et al., 2011). In this study, GlCYPs were categorized according to their catalytic functions in metabolism (Liu et al., 2020). The results identified a total of 123 CYPs that could possibly catalyze the modification of terpenoids: 7 CYPs may be related to the catalysis of cyanoamino acid, 5 CYPs might catalyze the modification of flavonoids, 29 CYPs might be related to the catalysis of fatty acids, 28 CYPs may have other catalytic functions, and 11 CYPs might have multiple functions when catalyzing terpenoids in plants. The catalytic function of 47 CYPs are still unidentified.
The homologous gene of PgCYP716A47 that catalyzed dammarenediol-II to panaxadiol in G. longipes was the very first step in gypenoside biosynthesis. According to the phylogenetic analysis, Unigene0005406 and Unigene0122930 had higher homology with PgCYP716A47; thus, these two unigenes were responsible for catalyzation of the hydroxylation at C-12. Hydroxylation at C-2α and oxidization at C-19 are also important sites for gypenoside diversity. The integrated analysis of transcriptome and metabolism was performed to narrow down possible candidate genes that catalyzed the C-12 and C-19 positions. According to the co-expression network describing triterpenes content and gene expression, Unigene0032271, Unigene0067788, Unigene0081102, Unigene0114879, Unigene0001688, Unigene00392303, and Unigene0086841 were in the pool of candidate genes that could catalyze hydroxylation at C-2 position, whereas Unigene0043066, Unigene0067788, Unigene0081102, and Unigene0015374 were classified in the candidate gene pool for the catalysis of oxidization at C-19.
In the past decade, G. pentaphyllum has served as an alternate candidate for Panax genus to fulfill the market demands for these types of compounds. Recently, the noteworthy pharmacological effects of gypenosides have opened a brand-new market in China, resulting in increasing demand for gypenosides (Su et al., 2021). However, gypenoside biosynthesis and heterologous expression is rarely studied compared with ginsenosides. In this study, the key OSC and CYP enzymes involved in gypenoside biosynthesis were illustrated and an integrated transcriptome and metabolic analysis was conducted for gypenosides. A series of genes that participated in gypenoside biosynthesis was located, offering significant input to the further characterization of the molecular mechanisms of dammarenediol-type gypenoside biosynthesis in G. pentaphyllum. Moreover, the first GlOSC1 to catalyze the formation of dammarenediol-II in the Gynostemma genus was found. In summary, comprehensive information on the transcriptional regulation and metabolic content in G. pentaphyllum has been discovered, helping to advance the study of dammarenediol-type saponin biosynthesis and creating opportunities to engineer microorganisms for the de novo production of valuable gypenosides.
Wild G. longipes plants were collected from Ankang county, Shaanxi province (Supplementary Figure S5). The leaf, rattan, and stolon were collected separately, frozen in liquid nitrogen, and stored at −80°C. Three replicates of each tissue were collected and divided into two parts for metabolism and transcriptome analysis.
RNA from the leaf, rattan, and stolon was extracted and reversed transcribed into cDNA. A Poly(A) tail was added after purification of the cDNA fragment and Illumina sequencing adapter ligation was applied. The final sequences were sequenced using Illumina Hiseq™ 4000 by Gene Denovo Biotechnology Co. (Guangzhou, China). Three biological repetitions of each tissue sample were processed. The raw reads obtained from sequencing machines; after filtering out low-quality reads (those containing more than 40% of low-quality (Q-value ≤10) bases), the clean reads were assembled by Trinity using default parameters (Grabherr et al., 2011).
For the functional annotation of unigenes, the BLASTx program2 was used with an E-value threshold of 1e-5 in the NCBI non-redundant protein (Nr) database3, the Swiss-Prot protein database4 the Kyoto Encyclopedia of Gens and Genomes (KEGG) database5, and the COG/KOG database6. Based on the Nr annotation results, the GO annotation was analyzed by Blast2GO software. Then, the functional classification of unigenes was performed using WEGO software (Ashburner et al., 2000; Ye et al., 2006). The Nr, Swiss-Prot, KEGG, and COG/KOG databases, BLAST and ESTscan programs were used for protein coding sequence (CDS) prediction (Iseli et al., 1999).
The phylogenetic analysis of GlOSCs and GlCYPs was performed on the deduced amino acid sequences to avoid other referential sequences from plants and two exogeneous sequences from animals. All deduced acid sequences were aligned with Clustal X under default parameters (Jiang et al., 2014). The preferred IQ-TREE version 1.6.12 on alignments was used to calculate the best model according to the software instructions, as described by Kalyaanamoorthy et al. (2017). From the results of the modelfinder, LG+I+G4 was the most suitable model for the construction of phylogenetic trees for this study. Figtree version 1.4.4 was used for data visualization.
Multiple reaction monitoring (MRM) was performed by Metware Biotechnology Co. Lit (Wuhan, China). After the addition of zirconia beads, samples of the leaf, rattan, and stolon from G. longipes were processed, each tissues had three biological duplicates. The samples were freeze-dried and ground into a fine powder by a mixing mill (MM400, Restch, Haan, Germany). Then, 100 mg of sample powder was dissolved in 1 mL of 70% methanol. The sample was held at 4°C overnight and then centrifuged for 10 min at 12,000 × g to obtain the filtrate. The extracts were analyzed by UPLC-ESI-MS/MS (UPLC, SHIMADZU Nexera X27; MS, Applied Biosystems 4500 Q TRAP8). The following analysis conditions were used: UPLC column, Agilent SB-C18 (1.8 μm, 2.1 mm × 100 mm); mobile phase solvent A, water with 0.1% formic acid, solvent B acetonitrile with 0.1% formic acid; gradient program, starting at 95:5 (A:B), then at 9 min 5:95 (A:B) and held for 1 min, 95:5 (A:B) at 10 min, from 95:5 (A:B) from 11.1 to 14 min; flow rate 0.35 mL/min; temperature 40°C; and injection volume 4 μL. The effluent was connected to an ESI-triple quadrupole-linear ion trap (Q-TRAP)-MS. LIT and triple quadrupole (QQQ) scans were acquired on a triple quadrupole-linear ion trap mass spectrometer (Q TRAP), AB4500 Q TRAP UPLC–MS/MS System, equipped with an ESI Turbo Ion-Spray interface, operating in positive and negative ion mode and controlled by Analyst 1.6.3 software (AB Sciex). The ESI source operation parameters were as follows: ion source, turbo spray; source temperature 550°C; ion spray voltage (IS) 5500 V (positive ion mode)/−4500 V (negative ion mode); ion source gas I (GSI), gas II (GSII), curtain gas (CUR) pressure of 50, 60, and 25.0 psi, respectively; collision-activated dissociation (CAD), high. Instrument tuning and mass calibration were performed with 10 and 100 μmol/L polypropylene glycol solutions in QQQ and LIT modes, respectively. QQQ scans were acquired as MRM experiments with the collision gas (nitrogen) set to medium. The decluttering potential (DP) and collision energy (CE) for individual MRM transitions was performed with further DP and CE optimization. A specific set of MRM transitions was monitored for each period according to the metabolites eluted within this period. The metabolites were annotated by Metware datebase9).
In the transcriptome, the expression of unigenes was calculated and normalized to RPKM (reads per kb per million reads) values by BWA program (Mortazavi et al., 2008). TBtools version 1.0 was used for gene expression profiling (Chen et al., 2020).
Six GlOSCs and 10 other unigenes that participated in the MVA pathway were selected for RT-qPCR analysis relative to the reference gene ACT1. One microgram of RNA of each sample was reverse-transcribed to cDNA by using PrimeScript™ RT Reagent kit with gDNA Eraser (Takara, Dalian, China). The specific primers were designed through online Primer3Web version 4.0.010 (Supplementary Table S1). The quantitative reactions were performed in a volume of 20 μL, which comprised 1 μL of cDNA, 0.4 μL of primer1, 0.4 μL of primer2, 10 μL of 2 × Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) and 8.8 μL of ddH2O and analyzed by Quantstudio 5K Flex Real-Time polymerase chain reaction System (Applied Biosystems, Foster City, CA, United States) under the following conditions: 1 cycle of 95°C (30 s), 40 cycles of 95 (10 s) and 60°C (30 s), 1 cycle of 95°C (15 s), 60°C (60 s), and 95°C 15 s. The relative gene expression was calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
The Pearson model was used to determine the correlation analysis between gene expression and chemical content; the analysis was performed using SPSS version 26. Correlation pairs under the conditions of PCC ≥ 0.9 and p-value ≤ 0.05 were selected and visualized by Cytoscape 3.8.2.
The protein coding sequences (CDS) of GlOSC1 (Unigene0012275) were obtained from G. longipes transcriptome. The primers for fragment PCR were designed using SnapGene. The PCR reactions were performed with Phanta Super-Fidelity DNA Polymerase (Vazyme Biotech Co., Ltd. Nanjing, China), processed in accordance with the user manual11. The PCR fragments were ligated into the BamHI site of the PYES2 vector under the control of the GAL1 promoter by the ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, China) and then transferred into the mix buffer into Escherichia coli line DH5α (TransGen Biotech. Co., Ltd., Beijing, China) for cloning. After incubation at 37°C overnight in LB medium with ampicillin, the reconstructed plasmids were extracted by EastPure Hipure Plasmid Maxiprep Kit (TransGen Biotech. Co., Ltd., Beijing, China).
GlOSC1-PYES2 and an empty vector yeast strain BY4742 (Miaoling Biotechnology Co., Ltd., Wuhan, China) were transformed by the lithium acetate method (Kushiro et al., 1998). A single clone was incubated in complete medium without uracil (CM-U) at 30°C with shaking at 220 g for 48 h; otherwise, 13 mg/L hemin and 5 g/L Tween 80 were included in CM-U. The glucose medium was replaced with galactose medium for induction for 48 h at 30°C, 220 g further incubated in 0.1 M potassium phosphate for 1 day. Finally, 50 mL of methyl alcohol was refluxed into the cells and then ultrasound treatment was applied for 30 min.
The yeast extracts were analyzed by UPLC-QTOF-MS performed on an Agilent UPLC time-of-flight mass spectrometer (UPLC/MS-TOF) under a fragment dissociation voltage of 135 V. A high performance liquid chromatography (HPLC) Agilent Zorbax SB-C18 column (250 mm × 4.6 mm, 5 μm) was used for chemical separation, with a column temperature of 25°C. The mobile phase solvent A was water with 0.1% formic acid, solvent B was acetonitrile, and the following linear gradient elution was applied: 0–20 min, 20%→10% A; 20–28 min; 10% A; 28–29 min, 20%→10% A; 29–31 min, 20% A; the flow rate was 0.5 mL/min, the wavelength was 210 nm, and the injection volume was 10 μL.
The saponins and related genes involved biosynthesis were analyzed by the integrated analysis of the metabolome and transcriptome in the leaf, stolon, and rattan of G. longipes. The saponin skeletons were mainly biosynthesized in the leaf by OSCs, and then modified via CYPs and UGTs to form various ginsenoside and gypenosides in the stolon. The catalytic function of GlOSC1 encoded by Unigene0012275 was heterologous expression in yeast to verify the ability of 2,3-oxidosqualene to catalyze the formation of dammarenediol-II via cyclization. The study has also provided essential information for the better utilization of G. longipes to produce valuable ginsenosides and gypenosides with excess substrate in a yeast cell factory through a synthetic biology strategy.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA784129.
BF, GZ, and SCY conceived the study. SY, LF, SYZ, QW, and WS performed the experiments. SY, LF, SYZ, and GZ the designed experiments. SY, GX, BN, LY, and XL analyzed the data. SY, SYZ, BH, and GZ drafted the manuscript. BH, GZ, and SCY reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Key R&D Plan (Grant No. 2017YFC1702500), Major Science and Technique Programs in Yunnan Province (Grant No. 2019ZF011-1), Science and Technology Innovation team of Yunnan (202105AE160011), Yunnan Provincial Key Programs of Eco-friendly Food International Cooperation Research Center Project (Grant No. 2019ZG00901), Natural Science Foundation of Yunnan Province (Grant No. 202001AT070125), and National Natural Science Foundation of China (Grant No. 31701854).
BF was employed by Honwin Pharma (Lianghe) Co., LTD.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.852377/full#supplementary-material
Supplementary Figure S1 | (A) Functional classification by KEGG. The number of genes annotated to pathways, including Cellular Processes, Environmental Information Processing, Genetic Information Processing, and Metabolism. (B) Venn diagram of the KEGG, SwissProt, Pfam, and GO results for the G. longipes transcriptome. (C) Functional classification by GO. The number of genes annotated into the three categories: Biological Process, Cellular Component, and Molecular Function.
Supplementary Figure S2 | Comparison of GlOSCs amino sequences; the sequences in the yellow box are the conserved motif (DCTAE) for all OSCs; the sequences in the green box are the amino acid sites to distinguish steroidal synthases and non-steroidal synthases.
Supplementary Figure S3 | LC–MS analysis of authentic dammarenediol-II.
Supplementary Figure S4 | LC–MS analysis of products from PYES2-GlOSC1.
Supplementary Figure S5 | Images of G. longipes and tissues used in this research are marked in white.
Supplementary Table S1 | Primer sequences used for gene expression by qPCR.
Supplementary Table S2 | Pearson correlation analysis between the levels of candidate triterpenes and the expression level of candidate genes.
Supplementary Table S3 | Comparison table between index and compounds names, and their relative content.
DDS, dammarenediol-II synthase; MVA, mevalonate; MEP, methylerythritol phosphate; OSC, oxidosqualene cyclase; CYP, cytochrome P450 monooxygenase; UGT, UDP-dependent glycosyltransferases; HMGR, 3-hydroxy-3-methylglutaryl-coenzyme A reductase; FPS, farnesyl diphosphate synthase; SS, squalene synthase; SQEs, squalene epoxidase; CAS, cycloartenol synthase; LAS, lanosterol synthase; CbQ, cucurbitadienol synthase; PS, parkeol synthase; β AS, β-amyrin synthase; LUS, lupeol synthase; MS, mix-triterpene synthase; CDS, protein coding sequences; PPD, protopanaxadiol.
Abe, I., and Prestwich, G. D. (1995). Identification of the active site of vertebrate oxidosqualene cyclase. Lipids 30, 231–234. doi: 10.1007/BF02537826
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Cardenas, P. D., Almeida, A., and Bak, S. (2019). Evolution of structural diversity of triterpenoids. Front. Plant Sci. 10:1523. doi: 10.3389/fpls.2019.01523
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Q., Ma, C., Qian, J., Lan, X., Chao, N., Sun, J., et al. (2016). Transcriptome sequencing of Gynostemma pentaphyllum to identify genes and enzymes involved in triterpenoid biosynthesis. Int. J. Genomics 2016, 7840914. doi: 10.1155/2016/7840914
Cheng, Y., and Cheng, X. (2019). Development Report of Gynostemma Industry in Ankang City. Available online at: https://www.ankang.gov.cn/Content-1537672.html (accessed December 24, 2019).
Choi, H. S., Han, J. Y., and Choi, Y. E. (2020). Identification of triterpenes and functional characterization of oxidosqualene cyclases involved in triterpene biosynthesis in lettuce (Lactuca sativa). Plant Sci. 301:110656. doi: 10.1016/j.plantsci.2020.110656
Christensen, L. P. (2008). Chapter 1 ginsenosides: chemistry, biosynthesis, analysis, and potential health effects. Adv. Food Nutr. Res. 55, 1–99. doi: 10.1016/S1043-4526(08)00401-4
Corey, E. J., Cheng, H., and Baker, C. H. (1997). Studies on the substrate binding segments and catalytic action of lanosterol synthase. Affinity labeling with carbocations derived from mechanism-based analogs of 2,3-oxidosqualene and site-directed mutagenesis probes. J.Am.Chem.Soc. 119, 1289–1296. doi: 10.1021/ja963228o
Forestier, E., Romero-Segura, C., Pateraki, I., Centeno, E., Compagnon, V., Preiss, M., et al. (2019). Distinct triterpene synthases in the laticifers of Euphorbia lathyris. Sci. Rep. 9:4840. doi: 10.1038/s41598-019-40905-y
Gantait, S., Mitra, M., and Chen, J. T. (2020). Biotechnological interventions for ginsenosides production. Biomolecules 10:538. doi: 10.3390/biom10040538
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, J., Ma, X., Cai, Y., Ma, Y., Zhan, Z., Zhou, Y. J., et al. (2016). Cytochrome P450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones. New Phytol. 210, 525–534. doi: 10.1111/nph.13790
Han, J. Y., Hwang, H. S., Choi, S. W., Kim, H. J., and Choi, Y. E. (2012). Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 53, 1535–1545. doi: 10.1093/pcp/pcs106
Han, J. Y., Jo, H. J., Kwon, E. K., and Choi, Y. E. (2019). Cloning and characterization of oxidosqualene cyclases involved in taraxasterol, taraxerol and bauerenol triterpene biosynthesis in Taraxacum coreanum. Plant Cell Physiol. 60, 1595–1603. doi: 10.1093/pcp/pcz062
Han, J. Y., Kim, H. J., Kwon, Y. S., and Choi, Y. E. (2011). The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 52, 2062–2073. doi: 10.1093/pcp/pcr150
Han, J. Y., Kwon, Y. S., Yang, D. C., Jung, Y. R., and Choi, Y. E. (2006). Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng. Plant Cell Physiol. 47, 1653–1662. doi: 10.1093/pcp/pcl032
Haralampidis, K., Trojanowska, M., and Osbourn, A. E. (2002). Biosynthesis of triterpenoid saponins in plants. Adv. Biochem. Eng. 75, 31–48. doi: 10.1007/3-540-44604-4_2
He, S., Yang, L., Ye, S., Lin, Y., Li, X., Wang, Y., et al. (2022). MPOD: applications of integrated multi-omics database for medicinal plants. Plant Biotechnol. J. doi: 10.1111/pbi.13769 [Epub ahead of print].
He, Y., Hu, Z., Li, A., Zhu, Z., Yang, N., Ying, Z., et al. (2019). Recent advances in biotransformation of saponins. Molecules 24:2365. doi: 10.3390/molecules24132365
Hou, M., Wang, R., Zhao, S., and Wang, Z. (2021). Ginsenosides in Panax genus and their biosynthesis. Acta Pharm. Sin. B 11, 1813–1834. doi: 10.1016/j.apsb.2020.12.017
Huang, D., Ming, R., Xu, S., Wang, J., Yao, S., Li, L., et al. (2021). Chromosome-level genome assembly of Gynostemma pentaphyllum provides insights into gypenoside biosynthesis. DNA Res. 28:dsab018. doi: 10.1093/dnares/dsab018
Iseli, C., Jongeneel, C. V., and Bucher, P. (1999). ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 99, 138–148.
Ito, R., Masukawa, Y., and Hoshino, T. (2013). Purification, kinetics, inhibitors and CD for recombinant beta-amyrin synthase from Euphorbia tirucallane L and functional analysis of the DCTA motif, which is highly conserved among oxidosqualene cyclase. FEBS J. 280, 1267–1280. doi: 10.1111/febs.12119
Jeffreys, L. N., Girvan, H. M., McLean, K. J., and Munro, A. W. (2018). Characterization of cytochrome P450 enzymes and their applications in synthetic biology. Methods Enzymol. 608, 189–261. doi: 10.1016/bs.mie.2018.06.013
Jiang, N. H., Zhang, G. H., Zhang, J. J., Shu, L. P., Zhang, W., Long, G. Q., et al. (2014). Analysis of the transcriptome of Erigeron breviscapus uncovers putative scutellarin and chlorogenic acids biosynthetic genes and genetic markers. PLoS One 9:e100357. doi: 10.1371/journal.pone.0100357
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kim, O. T., Lee, J. W., Bang, K. H., Kim, Y. C., Hyun, D. Y., Cha, S. W., et al. (2009). Characterization of a dammarenediol synthase in Centella asiatica (L.) Urban. Plant Physiol. Biochem. 47, 998–1002. doi: 10.1016/j.plaphy.2009.08.001
Kim, Y. J., Zhang, D., and Yang, D. C. (2015). Biosynthesis and biotechnological production of ginsenosides. Biotechnol. Adv. 33(6 Pt 1), 717–735. doi: 10.1016/j.biotechadv.2015.03.001
Ky, P. T., Huong, P. T., My, T. K., Anh, P. T., Kiem, P. V., Minh, C. V., et al. (2010). Dammarane-type saponins from Gynostemma pentaphyllum. Phytochemistry 71, 994–1001. doi: 10.1016/j.phytochem.2010.03.009
Le, D. D., Kim, W., Lim, S., Kim, S. C., and Choi, G. (2021). Identification of three groups of ginsenoside biosynthetic UDP-glycosyltransferases from Gynostemma pentaphyllum. Plant Sci. 313:111069. doi: 10.1016/j.plantsci.2021.111069
Li, K., Ma, C., Li, H., Dev, S., He, J., and Qu, X. (2019). Medicinal value and potential therapeutic mechanisms of Gynostemma pentaphyllum (Thunb.) makino and its derivatives: an overview. Curr. Top. Med. Chem 19, 2855–2867. doi: 10.2174/1568026619666191114104718
Li, Z., Jiang, Y., Guengerich, F. P., Ma, L., Li, S., and Zhang, W. (2020). Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J. Biol. Chem. 295, 833–849. doi: 10.1074/jbc.REV119.008758
Liang, T., Zou, L., Sun, S., Kuang, X., Wei, J., Wang, L., et al. (2019). Hybrid sequencing of the Gynostemma pentaphyllum transcriptome provides new insights into gypenoside biosynthesis. BMC Genomics 20:632. doi: 10.1186/s12864-019-6000-y
Liu, J., Xu, Y., Yang, J., Wang, W., Zhang, J., Zhang, R., et al. (2017). Discovery, semisynthesis, biological activities, and metabolism of ocotillol-type saponins. J. Ginseng Res. 41, 373–378. doi: 10.1016/j.jgr.2017.01.001
Liu, X., Zhu, X., Wang, H., Liu, T., Cheng, J., and Jiang, H. (2020). Discovery and modification of cytochrome P450 for plant natural products biosynthesis. Synth. Syst. Biotechnol. 5, 187–199. doi: 10.1016/j.synbio.2020.06.008
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, J., Yao, L., Li, J. X., Liu, S. J., Hu, Y. Y., Wang, S. H., et al. (2018). Characterization of UDP-glycosyltransferase involved in biosynthesis of ginsenosides Rg1 and Rb1 and identification of critical conserved amino acid residues for its function. J. Agric. Food Chem. 66, 9446–9455. doi: 10.1021/acs.jafc.8b02544
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nagai, M., Nagumo, S., and Izawa, K. (1976). “Abstracts of papers,” in Prodeecings of the 23rd Meeting of the Japanese Society of Pharmacognosy, Hiroshima.
Nelson, D., and Reichhart, W. (2011). A P450-centric view of plant evolution. Plant J. 66, 194–211. doi: 10.1111/j.1365-313X.2011.04529.x
Nguyen, N. H., Ha, T. K. Q., Yang, J. L., Pham, H. T. T., and Oh, W. K. (2021). Triterpenoids from the genus Gynostemma: chemistry and pharmacological activities. J. Ethnopharmacol. 268:113574. doi: 10.1016/j.jep.2020.113574
Niu, Y., Luo, H., Sun, C., Yang, T. J., Dong, L., Huang, L., et al. (2014). Expression profiling of the triterpene saponin biosynthesis genes FPS, SS, SE, and DS in the medicinal plant Panax notoginseng. Gene 533, 295–303. doi: 10.1016/j.gene.2013.09.045
Phillips, D. R., Rasbery, J. M., Bartel, B., and Matsuda, S. P. (2006). Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9, 305–314. doi: 10.1016/j.pbi.2006.03.004
Qin, Z., Zhao, L., Bi, S., and You, L. (1992). Saponin constituents and resource of Gynostemma pentaphyllum. Tianran Chanwu Yanjiu Kaifa 4, 83–98.
Razmovski-Naumovski, V., Huang, T. H.-W., Tran, V. H., Li, G. Q., Duke, C. C., and Roufogalis, B. D. (2005). Chemistry and pharmacology of Gynostemma pentaphyllum. Phytochem. Rev. 4, 197–219. doi: 10.1007/s11101-005-3754-4
Su, C., Li, N., Ren, R., and Wang, Y. (2021). Progress in the medicinal value, bioactive compounds, and pharmacological activities of Gynostemma pentaphyllum. Molecules 202126:6249. doi: 10.3390/molecules26206249
Subramaniyam, S., Mathiyalagan, R., Jun Gyo, I., Bum-Soo, L., Sungyoung, L., and Deok Chun, Y. (2011). Transcriptome profiling and insilico analysis of Gynostemma pentaphyllum using a next generation sequencer. Plant Cell Rep. 30, 2075–2083. doi: 10.1007/s00299-011-1114-y
Tang, J. R., Chen, G., Lu, Y. C., Tang, Q. Y., Song, W. L., Lin, Y., et al. (2021). Identification of two UDP-glycosyltransferases involved in the main oleanane-type ginsenosides in Panax japonicus var. major. Planta 253:91. doi: 10.1007/s00425-021-03617-0
Tang, Q. Y., Chen, G., Song, W. L., Fan, W., Wei, K. H., He, S. M., et al. (2019). Transcriptome analysis of Panax zingiberensis identifies genes encoding oleanolic acid glucuronosyltransferase involved in the biosynthesis of oleanane-type ginsenosides. Planta 249, 393–406. doi: 10.1007/s00425-018-2995-6
Tansakul, P., Shibuya, M., Kushiro, T., and Ebizuka, Y. (2006). Dammarenediol-II synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng. FEBS Lett. 580, 5143–5149. doi: 10.1016/j.febslet.2006.08.044
Thimmappa, R., Geisler, K., Louveau, T., O’Maille, P., and Osbourn, A. (2014). Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 65, 225–257. doi: 10.1146/annurev-arplant-050312-120229
Wang, L., Zhao, S. J., Cao, H. J., and Sun, Y. (2014). The isolation and characterization of dammarenediol synthase gene from Panax quinquefolius and its heterologous co-expression with cytochrome P450 gene PqD12H in yeast. Funct. Integr. Genomics 14, 545–557. doi: 10.1007/s10142-014-0384-1
Wang, P., Wei, Y., Fan, Y., Liu, Q., Wei, W., Yang, C., et al. (2015). Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab. Eng. 29, 97–105. doi: 10.1016/j.ymben.2015.03.003
Xiao, H., Zhang, Y., and Wang, M. (2018). Discovery and engineering of cytochrome P450s for terpenoid biosynthesis. Trends Biotechnol. 37, 618–631. doi: 10.1016/j.tibtech.2018.11.008
Xu, R., Fazio, G. C., and Matsuda, S. P. (2004). On the origins of triterpenoid skeletal diversity. Phytochemistry 65, 261–291. doi: 10.1016/j.phytochem.2003.11.014
Xu, S., Yao, S., Huang, R., Tan, Y., and Huang, D. (2020). Transcriptome-wide analysis of the AP2/ERF transcription factor gene family involved in the regulation of gypenoside biosynthesis in Gynostemma pentaphyllum. Plant Physiol. Biochem. 154, 238–247. doi: 10.1016/j.plaphy.2020.05.040
Yang, C., Li, C., Wei, W., Wei, Y., Liu, Q., Zhao, G., et al. (2020). The unprecedented diversity of UGT94-family UDP-glycosyltransferases in Panax plants and their contribution to ginsenoside biosynthesis. Sci. Rep. 10:15394. doi: 10.1038/s41598-020-72278-y
Yang, J. L., Hu, Z. F., Zhang, T. T., Gu, A. D., Gong, T., and Zhu, P. (2018). Progress on the studies of the key enzymes of ginsenoside biosynthesis. Molecules 23:589. doi: 10.3390/molecules23030589
Ye, J., Fang, L., Zheng, H., Zhang, Y., Chen, J., Zhang, Z., et al. (2006). WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34, W293–W297. doi: 10.1093/nar/gkl031
Yoshikawa, K. (1986). Studies on the constituents of cucurbitaceae plants. XV on the saponin constituents of Gynostemma pentaphyllum MAKINO. Yakugaku Zasshi 106, 758–764. doi: 10.1248/yakushi1947.106.9_758
Zhang, Y., Chen, Q., Huang, Y., Zhao, R., Sun, J., Yuan, X., et al. (2021). Gene excavation and expression analysis of CYP and UGT related to the post modifying stage of gypenoside biosynthesis in Gynostemma pentaphyllum (Thunb.) Makino by comprehensive analysis of RNA and proteome sequencing. PLoS one 16:e0260027. doi: 10.1371/journal.pone.0260027
Keywords: gypenosides, oxidosqualene cyclase, cytochrome P450 monooxygenase, dammarenediol II synthase, Gynostemma longipes
Citation: Ye S, Feng L, Zhang S, Lu Y, Xiang G, Nian B, Wang Q, Zhang S, Song W, Yang L, Liu X, Feng B, Zhang G, Hao B and Yang S (2022) Integrated Metabolomic and Transcriptomic Analysis and Identification of Dammarenediol-II Synthase Involved in Saponin Biosynthesis in Gynostemma longipes. Front. Plant Sci. 13:852377. doi: 10.3389/fpls.2022.852377
Received: 11 January 2022; Accepted: 28 February 2022;
Published: 25 March 2022.
Edited by:
Pan Liao, Purdue University, United StatesReviewed by:
Qingzhu Li, Shanghai Academy of Agricultural Sciences, ChinaCopyright © 2022 Ye, Feng, Zhang, Lu, Xiang, Nian, Wang, Zhang, Song, Yang, Liu, Feng, Zhang, Hao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Hao, QmluZy5oYW9AaG90bWFpbC5jb20=; Shengchao Yang, c2hlbmdjaGFveWFuZ0AxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.