- 1School of Life Sciences, Anhui Agricultural University, Hefei, China
- 2CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
BGLU β-glucosidases in glycoside hydrolase family 1 (GH1) are involved in many processes of plant secondary metabolism. In particular, its de-glycosylation function plays an important role in the transport of lignin monolignols. No comprehensive study of the BGLU family in Chinese pear (Pyrus bretschneideri Rehd.) has been reported yet. In this study, the 50 BGLU family members from Chinese white pear were identified. Three candidate genes, PbBGLU1, PbBGLU15, and PbBGLU16, that may be involved in lignin synthesis were screened by bioinformatics analysis and qRT-PCR. Subcellular localization showed that all three of these candidate genes were expressed in the extracellular region. Then, we analyzed the functions of PbBGLU1 and PbBGLU16. In situ hybridization analysis showed that PbBGLU1 transcripts were not only localized to some pulp cell walls, lignin deposition, and stone cell areas of a pear fruit, but that was also a small amount of enrichment in normal pear flesh cells. PbBGLU16 transcripts were only enriched in lignin deposition and stone cell areas of pear fruit. Enzyme activity analysis revealed that GST-PbBGLU1 and GST-PbBGLU16 had a stronger activity and higher catalytic efficiency for coniferin than syringin. In addition, GST-PbBGLU16 exhibited the higher activity and catalytic efficiency for the two substrates compared with GST-PbBGLU1. The transformation of PbBGLU1 and PbBGLU16 into Arabidopsis identified that the lignin contents of Arabidopsis BGLU-45 mutant, PbBGLU1-RE, and PbBGLU16-RE were not changed than that of wild-type. However, compared with wild-type Arabidopsis, the overexpression of the plant’s lignin increased in varying degrees. The effect of PbBGLU16 on the lignin increment was greater than that of PbBGLU1 in Arabidopsis. In pear fruits, with transient overexpression of PbBGLU1, the contents of lignin and stone cells were significantly higher (0.01 < P < 0.05) than those with empty vector injection pear fruits. After transient expression of PbBGLU16, lignin in pear fruit increased significantly (0.01 < P < 0.05) and stone cells showed a very significant difference (P < 0.01) compared with the control group. However, RNA interference silenced these two genes in pear fruit, which seemed to have no impression on lignin and stone cells. This study provides a molecular biological basis for improving pear fruit quality at the molecular level.
Introduction
Pear belongs to the Rosaceae family and is one of the most important deciduous fruit tree species worldwide (Wu J. et al., 2012). “Dangshan Su” pear (Pyrus bretschneideri cv. Dangshan Su) is a diploid pear variety that originated in Dangshan County, Anhui Province, China. It not only has good flavor and high nutritional value but also has medicinal value and is very popular among people. However, due to its high content of stone cells, its economic value is not high enough (Yan et al., 2014). Stone cells are a kind of specific cell in pear fruit, and they are among the important factors that affect pear fruit processing and fresh consumption. Current research shows that the diameter, number, and density of stone cells have significant effects on pear fruit quality. The greater the diameter and density of the cell mass is, the greater the stone cell content of pear fruit, and the worse the fruit quality is (Zhang et al., 2017). Stone cells are formed by secondary thickening of parenchyma cells, and lignin is one of the crucial components of stone cells (Cheng et al., 2019a; Su et al., 2019).
Lignin is a secondary metabolite. Lignin synthesis, transportation, and deposition are closely related to stone cells development during pear fruit development (Cai et al., 2010; Jin et al., 2013). Structurally, lignin is a phenylpropanoid polymer mainly derived from three kinds of monolignols (p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol) connected by different chemical bonds (Boerjan et al., 2003; Barros et al., 2015). Glycosylation of these three monolignols is catalyzed by UDP-glucosyltransferase (UGT) into monolignol glucosides (p-coumarin, coniferin, syringin) (Cheng et al., 2019b; Perkins et al., 2019), and glycosylation increases the solubility and stability of monolignols, which is beneficial to the transport and storage of monolignols (Gachon et al., 2005). Studies have found that a large number of monolignol glucosides can be stored in the vacuoles of different xylem cells (Steeves et al., 2001; Tsuji and Fukushima, 2004). In-depth research on the transport mechanism of monolignols is scarce, but the academic community currently believes that there are three transport mechanisms for the extracellular secretion of monolignols, namely, passive diffusion (PD), vesicle-related exocytosis, and the use of ABC transport factors or proton coupling to reverse the active transport of ATP (Miao and Liu, 2010; Alejandro et al., 2012; Barros et al., 2015). These monolignol glucosides are then transported to specific parts of the cell wall and hydrolyzed to lignin monolignols under the action of β-glucosidase, ultimately forming guaiacyl (G), syringyl (S), and p-hydroxyphenyl (H) units by laccase and peroxidase [dicotyledons have only guaiacyl (G) and syringyl (S) lignin] (Liu, 2012).
In recent years, an increasing number of species of the β-glucosidase family have been discovered, and the function of the β-glucosidase-encoding gene has become clearer. In 1994, coniferin-specific β-glucosidase was identified in the xylem of two Pinus species (Leinhos et al., 1994; Dharmawardhana et al., 1995). A study in 1999 found that coniferin β-glucosidase catalyzed the hydrolysis of monolignol glucosides to release cinnamyl alcohols for oxidative polymerization to lignin (Dharmawardhana et al., 1999). At1g61810 (AtBGLU45), At1g61820 (AtBGLU46), and At4g21760 (AtBGLU47) in Arabidopsis play an important role in the synthesis of lignin (Escamilla-Treviño et al., 2006; Chapelle et al., 2012). Os4BGlu14, Os4BGlu16, and Os4BGlu18, which are members of the rice β-glucosidase family, have been proven to be functional genes involved in rice lignin synthesis (Baiya et al., 2018). In addition, a β-glucosidase was identified from poplar and named “PtrBGLU6”; this gene plays an important role in poplar lignification and cell wall formation (Tsuyama and Takabe, 2015).
In our study, all β-glucosidase family members in pear had been identified and further three β-glucosidase (PbBGLU1, PbBGLU15, and PbBGLU16) candidate genes were considered as functional genes for pear lignin synthesis. We through a series of experiments including in situ hybridization, in vitro enzyme activity studies, stable expression in Arabidopsis thaliana and transient expression in pear fruit. We provided evidence that PbBGLU1 and PbBGLU16 can act in the formation of lignin and stone cell development in pear fruit.
Materials and Methods
Identification of BGLU Genes in Five Rosaceae Species
The Chinese white pear genome database was obtained from the Pear Genome Project.1 To identify putative BGLU genes from Chinese white pear, several approaches were employed. We downloaded the HMM profile (PF00232) from the Pfam website2 (Finn et al., 2010) and searched for candidate genes using the HMM with E-values < 1e-10 from the Chinese white pear genome (Zhou et al., 2016). After performing multiple sequence alignment, the retrieved domain sequences of the pear are aligned, and the HMM of the BGLU genes family of the pear is constructed again. Using the constructed new hidden Markov model with E-values < 1e-10 to blasted against the Chinese pear genome for carryout PbBGLU genes. The online site SMART3 (Letunic et al., 2012) was used to determine the domain PF00232 of BGLU. The online analysis tool EXPASY4 was used to predict and analyze the physical and chemical properties of the obtained BGLU amino acids sequences.
Phylogenetic Conserved Motif and Exon-Intron Structures Analyses of BGLU Genes in Pear
Sequence alignment of PbBGLUs proteins was done by using the MUSCLE method in MEGA 7.0 software. The phylogenetic tree was constructed with MEGA 7.0 software using the neighbor-joining (NJ) (bootstrap = 2,500) (Kumar et al., 2016). The conservative motifs were analyzed by MEME5 software the maximum value of the motif was set to 20, and the motif length was set between 6 and 50 (Bailey et al., 2015). The Gene Structure Server6 was used to generate exon-intron maps (Hu et al., 2015).
Phylogenetic analysis of BGLU gene family in Arabidopsis, P. bretschneideri, Oryza sative, and Populus. The software MEGA 7.0 was used to construct the phylogenetic tree. The amino-acid sequences of Arabidopsis, O. sative, and Populus were obtained from phytozome database.7
Plant Materials, Treatments, and Growth Conditions
The materials were from “Dangshan Su” pear (P. bretschneideri cv. Dangshan Su) with the same growing environment for 40 years in Dangshan County, Anhui Province, China. We collected the mature leaves, buds, stem segments, and flowers on March 30, 2021; and collected the fruit on April 15 (15 days after flowering), 2021; May 14 (39 DAF), 2021; May 22 (47 DAF), 2021; June 4 (55 DAF), 2021; June 12 (63 DAF), 2021; June 28 (79 DAF), 2021; and August 30 (145 DAF), 2021. Each sample was frozen in liquid nitrogen and brought back to the laboratory for use. All samples were stored at–80°C for subsequent RNA extraction.
Arabidopsis thaliana L. wild type was purchased from the American Arabidopsis Biological Resources Center. Arabidopsis At1g61810 bglu45-2 mutant seeds corresponding to the Salk_117269 flanking sequence tag were ordered from the Salk Institute Genomic Analysis Laboratory. Homozygous BGLU45-2 mutant plants were isolated by genotype analysis with Arabidopsis BGLU45 gene and T-DNA specific primers, as described by Chapelle et al. (2012). The Arabidopsis plants were cultivated in a growth chamber at 23°C under 16/8 h light/dark cycles (constant illumination 100 μEm-2s-1). Ten replicates of each line were planted, and three similarly growing plants were collected for further analysis.
RNA Extraction, Reverse Transcription PCR, and Real-Time PCR Analysis
The total RNA of all the used “Dangshan Su” pear fruits in this study was extracted using a Plant RNA extraction kit from Tiangen (Beijing, China). Total RNA (1 μg) from each sample was used in reverse transcription. First-strand cDNA was synthesized with a PrimeScript™ RT reagent kit with a gDNA Eraser kit (TaKaRa, Kyoto, Japan). All RT-qPCR primers were designed using Primer Premier 5.0 software (Supplementary Table 3). We chose the tubulin (No. AB239680.1) served as the reference gene for RT-qPCR analysis (Wu T. et al., 2012). The RT-qPCR was performed with a CFX96 Touch™ Real-Time PCR Detection System (BIO-RAD), each sample was subjected to three biological replicates. The relative changes in gene expression levels were calculated using the 2–ΔΔ CT method (Livak and Schmittgen, 2001).
Subcellular Localization of PbBGLU1, PbBGLU15, and PbBGLU16
The three genes RT-PCR primers were designed using Primer Premier 5.0 software (Supplementary Table 4). The full-length CDS of PbBGLU1, PbBGLU15, and PbBGLU16 without stop codon were cloned based on genomic information and constructed into pCAMBIA1305 vectors (Clontech, Beijing, country-region China) between the XbaI and BamHI (NEB) sites which have both CaMV35S promoter and GFP gene. After electroporation of these constructions into Agrobacterium tumefaciens EHA105, using pCAMBIA1305 vector as a negative control. The infection solution was injected into the epidermis of Nicotiana tabacum leaves, after culturing in the dark for 3 days, the glass slide was made and placed under the FV1200 laser confocal microscope (Olympus Corporation, Tokyo, Japan) to observe the distribution of fusion protein.
In situ Hybridization
Segments of the 39 DAF fresh pear fruit were fixed in in situ hybridization fixed solution overnight. Pear pulp segments fixed: Take out the segment, wash, clean, put in the fixed fluid (DEPC) immediately to fix above 12 h. Dehydration: The segment is dehydrated by gradient alcohol, paraffin, embedding, and vacuum pumping in the dehydration process. Section: The paraffin is sliced through the slicer, the piece of the slicing machine, and the 62–degree oven roast for 2 h. Dewaxing and dehydration: Soak sections in 2 changes of xylene, 15 min each. Dehydrate in 2 changes of pure ethanol for 5 min each. Then, followed respectively by dehydrating in gradient ethanol of 85 and 75% ethanol 5 min each. Wash in DEPC dilution. Digestion: Mark the objective tissue with a liquid blocker pen, according to the characteristics of tissues, add proteinase K (20 ug/ml) working solution to cover objectives and incubate at 37°C for 22 min. Washing with pure water, then wash three times with PBS (pH 7.4) in a rocker device, 5 min each. Hybridization: Discard the pre-hybridization solution, add the probe hybridization solution, concentration, and incubate the section in a humidity chamber and hybridize overnight. Add the Alkaline Phosphatase-conjugated IgG Fraction Monoclonal Mouse Anti-Digoxin Antibody: (anti-DIG-AP): Remove the blocking solution and add anti-DIG-HRP. Incubate at 37°C for 40 min, then wash sections in TBS four times for 5 min each. NBT/BCIP developing: dry sections slightly, add fresh prepared NBT/BCIP (Thermo Scientific 1-Step NBT/BCIP) chromogenic reagent to marked tissue. Manage reaction time by observing in microscopy until positive expression appears shows blue-purple. Then stop developing reaction by a wash in running tap water. The probe sequence was unique to the PbBGLU1 and PbBGLU16 locus (Supplementary Table 4) and resulted in a single hit when used as a quarry to BLAST the Chinese white pear genome. In situ hybridization probes were synthesized by Sangon Bio Shang hai and labeled with Digoxigenin.
Construction of PbBGLUs Expression Vector and Induced Purification of Recombinant Protein
The RT-PCR primers were designed using Primer Premier 5.0 software (Supplementary Table 4). The full-length coding sequence of PbBGLU1 and PbBGLU16 were cloned and then expressed in pGEX4T-1 vector (GE Healthcare, Chicago, IL, United States) between the SamI and NotI (NEB) sites. Two recombinant pGEX4T-1-PbBGLUs were transformed into Escherichia coli BL21 (DE3) Competent Cells. Add Escherichia coli BL21 (DE3) cells carrying pGEX4T-1-PbBGLU1 and pGEX4T-1-PbBGLU16 plasmid to 100 ml of LB liquid containing ampicillin for shaking. Expand the culture to an OD value of about 0.6, add isopropyl-β-D-thioacetamide IPTG to a final concentration of 1 mM/L to induce the fusion expression of the recombinant protein, culture with shaking at 16°C for 24 h, and collect the cells by centrifugation at 4°C. The centrifuged cells were suspended in PBS (pH 7.2–7.4) buffer, using an ultrasonic cell disruptor for disruption, then centrifuge to take the supernatant and discard the pellet. Using affinity chromatography resin with GST tag GST⋅ Bind™ resin (Novagen) to Purify protein. The Elution Buffer (pH 8.0) for GST-Sefinose (TM) Resin (containing reducing glutathione reagent at a concentration of 5 mM) Eluent was used for elution.
Recombinant Protein Enzyme Assays
To analyze the activity of recombinant protein GST-PbBGLU1 and GST-PbBGLU16, 10 μg of purified protein was incubated at 35°C with 20 μL buffer (50 mM MgSO4, 200 mM KCL, 100 mM PBS pH 7.2–7.4) and 1 mM substrates (coniferin and syringin). The water was added to a final volume of 50 μL. After 1 h of reaction, add 50 μL methanol termination reaction. All reactions were supplemented with 0.1% (V/V) β- Mercaptoethanol. Then the reaction results were analyzed by HPLC.
To calculate the Km, Vmax, and kcat value of the recombinant protein, under the same original reaction conditions, the substrates (coniferin and syringin) were set to 10 concentrations (0.025, 0.05, 0.075, 0.1, 0.15, 0.2, 0.3, 0.4, 0.5, and 0.6 mM). Then the product concentration C is calculated by an external standard method, and then the reaction rate V is further calculated. Finally, use the double reciprocal method to plot to calculate Km and Vmax.
Genetic Transformation of Arabidopsis and Transient Transformation of Pear Fruit
The RT-PCR primers were designed using Primer Premier 5.0 software (Supplementary Table 4). The full-length coding sequence of PbBGLU1 and PbBGLU16 were cloned from “Dangshan Su” pear first-strand complementary DNA (cDNA) with PrimeSTAR® Max DNA Polymerase (Takara, Japan). The fragment was cloned into pCAMBIA1301-35S binary vectors between the BamHI and XbaI (NEB) sites and used in plant transformation. Wild type and Arabidopsis At1g61810 BGLU45-2 mutant were used for transformation with A. tumefaciens GV3101 carrying the above binary plasmid using the floral dip method.
The full-length coding fragment was cloned into pCAMBIA1301-35S binary vectors and used in the transient injection of pear fruit overexpression. Hairpin constructs were based on the pRNAiDE001 vector (Heneghan et al., 2007; Eastwood et al., 2008) where self-complementary sense (PbBGLU1: 905-1220bP; PbBGLU16: 832-1183bP) and antisense sequences are separated by a non-functional sequence (loop), then construct this fragment into the pCAMBIA1301 vector with the 35S promoter (Supplementary Figures 1, 2). Pear trees (40-year-old) and 39 days after flowering young fruits were selected for transient injection. The constructed pCAMBIA1301-35s binary vectors of PbBGLUs overexpression and PbBGLUs RNA interference gene were injected on the left of the fruit, and the empty pCAMBIA1301-35s binary vectors were injected on the right for control. The method followed a previously reported protocol (Zhang et al., 2021) nine fruits were injected with each construct in an experiment that was repeated three times independently. After 10 days, the transient injected fruits were picked up.
Lignin Determination and Histochemical Staining of Arabidopsis and Pear Fruits
We collected the Arabidopsis plants for approximately 6 weeks, removed the leaves, and dried them in an oven at 65°C for 48 h. The lignin content of the Arabidopsis plants was estimated following the method of Anderson et al. (Anderson et al., 2015).
We obtained the inflorescence stem segments (young stem regions) of the transgenic and wild-type Arabidopsis plants for approximately 6 weeks. The samples were placed in a solution containing 95% methanol, 70% (v/v) ethanol and glacial acetic acid for 12 h and embedded in paraffin for sectioning with the pathology slicer (RM 2018). Plant tissue sections were stained following standard phloroglucinol staining protocols (Pradhan and Loqué, 2014).
After the transiently injected pear fruits was brought back to the laboratory, samples 5 g pulp from 2.0 mm under the peel to 0.5 mm outside the core, was collected and frozen at –80°C for 24 h, homogenized with a high-speed homogenizer for 3 min, rotating speed was 20,000 RPM/min, added water and stood for a while, then poured out the upper suspension, repeated several times until the upper layer was clear, then filtered and dried stone cells were weighed, repeated for 3 times, stone cell content = measured stone cell dry weight/5 × 100%. The lignin content was measured using the Klason method (Raiskila et al., 2007). A small amount (0.2 g) of stone cells was extracted with 15 ml of 72% H2SO4 at 30°C for 1 h, combined with 115 ml of distilled water, and boiled for 1 h. The volume was kept constant during boiling. The liquid mixture was filtered and the residue was rinsed with 500 ml of hot water, air-dried and weighed. The lignin content was shown as a percentage (calculated lignin content/dry weight of stone cells × 100%) (Zhang et al., 2021).
Targeted Metabolite Determination
HPLC (Thermo Scientific UltiMate 3000 HPLC) analysis was carried out using a Columbus (Thermo Scientific 5-μm C18 column; 250×4.60 mm). Acetonitrile (solvent A) and H2O (solvent B) with gradient of 10–30% acetonitrile in water (all solutions contained 0.1% trifluoroacetic acid) with a flow rate at 1 ml min–1 over 35 min was used. Each peak on the chromatogram was scanned between 200 and 400 nm (photodiode array profile) and was integrated at 264, 280, and 324 nm. The data were acquired and analyzed using the software ChromQuest version. Extraction of tissue was carried out as described in Lanot et al. (2006).
Results
Identification of BGLU Genes in Pyrus bretschneideri
The BGLU family conserved domain (Pfam: PF00232) was used as a target query sequence, and the corresponding hidden Markov model was obtained from the Pfam website (see text footnote 2) which was used to identify the BGLU members. SMART (see text footnote 3) was used to check whether there are characteristic domains, with the redundant and repeated sequences ultimately removed. As a result, we identified 50 BGLU genes from Chinese white pear (P. bretschneideri). The detailed information on the gene ID, gene name, chromosomal location, protein structure, and characteristics of the corresponding BGLU proteins are listed in Supplementary Table 1.
Phylogenetic Analyses of the Conserved Motif and the Exon–Intron Structure of BGLU Genes in Chinese White Pear
Inclusive of all PbBGLU genes, a phylogenetic tree was constructed with MEGA 7.0 software using the neighbor-joining (NJ) method (bootstraps = 1,000), which divided them into 8 groups (I, II, III, IV, V-A, V-B, V-C, V-D) with supported bootstrap values (Figure 1). We used MEME online software to analyze the amino acid sequence of PbBGLU genes and identified 20 motifs among all the sequences (Supplementary Table 2 and Figure 1). All the PbBGLUs contain motif1 (CTGGBSATEPYJVAHHQLLAHAAAVKLYREKYQA), and only two genes do not contain motif9 (WFEPASES KEDKAAALRALDF), suggesting that motif1 and motif9 are conserved gene motifs of PbBGLUs family. Only group I, group II, and group III genes contain motif19, whereas the other groups do not, and no members of group V-D contain motif2, it is speculated that these motifs may have special significance. Previous studies implied that gene structural diversity can lead to the evolution of multi-gene families (Dong et al., 2019). To better characterize and understand the structural diversity of the PbBGLUs, gene exon-intron analysis was carried out (Figure 1). Notably, PbBGLU23 and PbBGLU24 have only one exon, whereas the others contain 7 or more exons; PbBGLU34 contains the most exons (21), and most genes had between 10 and 15 exons. These results indicate the possible occurrence of alternative splicing during gene evolution, which in turn leads to functional differences between these closely related PbBGLUs (Cao et al., 2017; Dong et al., 2019).
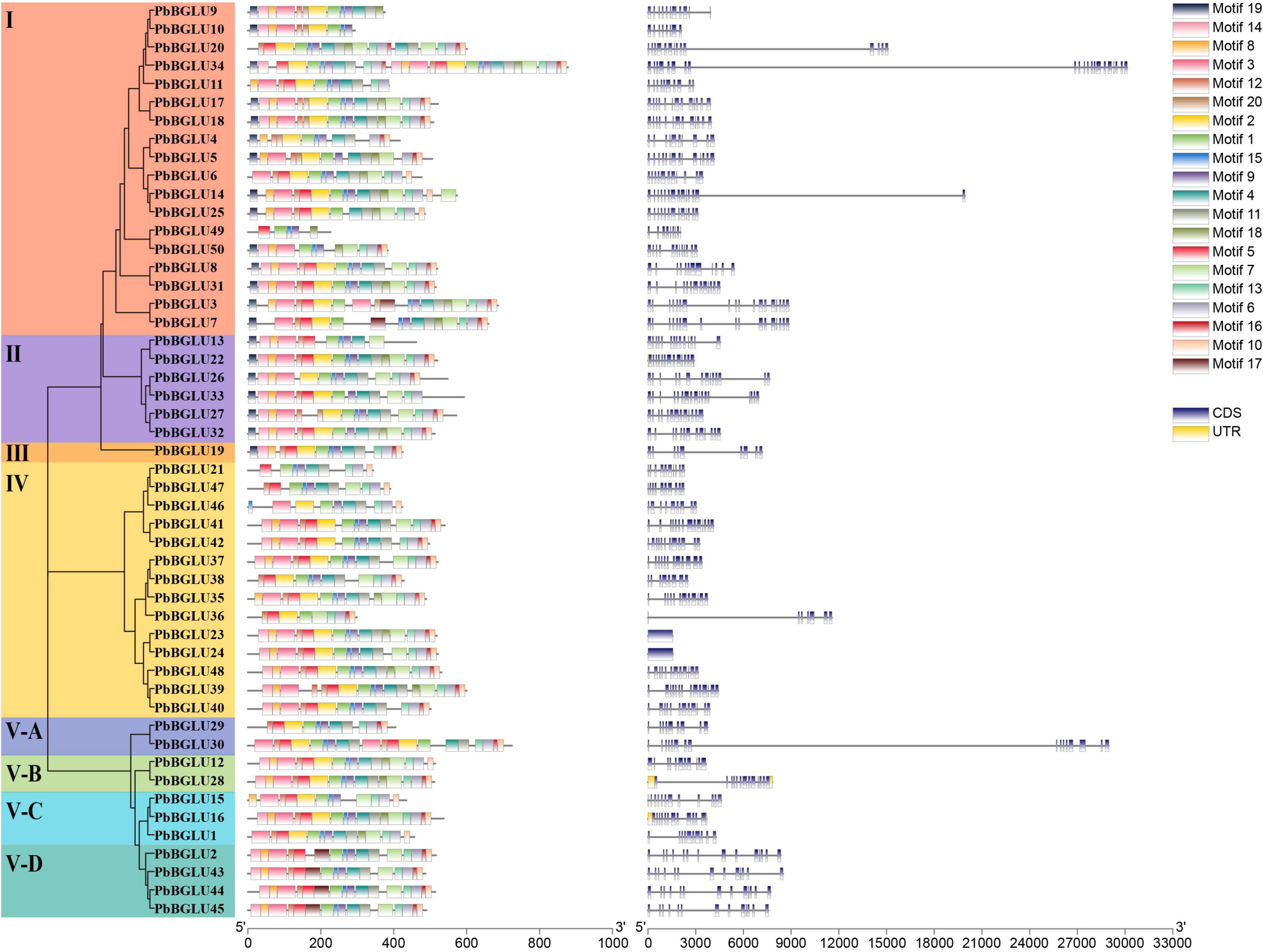
Figure 1. Phylogeny, conserved motifs and exon-intron structure of BGLU genes in pear. MEME tools were used to identify motifs. Different colors represent different motifs. GSDS online website was used to generate structure. The legend is located at the bottom right of the figure. Scale represents the length of the DNA sequence.
To further understand the evolutionary relationship and related functions of PbBGLUs, a phylogenetic tree comprising BGLUs from A. thaliana (Xu et al., 2004), O. sative (Opassiri et al., 2006), poplar (Tsuyama and Takabe, 2015), and P. bretschneideri was constructed (Figure 2). We marked the position of the eight groups in pear on this phylogenetic tree and found that group I, group II, and group IV contained only PbBGLUs but not AtBGLUs, Osbglus, and PtrBGLUs, indicating that some changes occurred among the BGLUs of different species during the evolutionary process (Cao et al., 2016a,b). Arabidopsis AtBGLU17 (Roepke et al., 2017) was related to the use of Arabidopsis flavonoids which was clustered together with PbBGLU19 (group III), and it is speculated that PbBGLU19 might be involved in the hydrolysis of flavonoid glycosides in pear. PbBGLU group V-D clustered together with AtBGLU42, and which encodes an MYB72-dependent key regulator of rhizobacterium-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots (Zamioudis et al., 2014). AtBGLU1-6, which plays a role in the accumulation of flavonoids in Arabidopsis (Ishihara et al., 2016), clustered together with PbBGLU group V-A genes, speculating that PbBGLU29 and PbBGLU30 may have similar functions in pear. The functions of AtBGLU45 and AtBGLU46 in Arabidopsis (Escamilla-Treviño et al., 2006; Chapelle et al., 2012); Os4BGlu14, Os4BGlu16, and Os4BGlu18 in O. sative (Baiya et al., 2014; Baiya et al., 2018); PtrBGLU6 in poplar (Tsuyama and Takabe, 2015) have been clarified, and their products catalyze the hydrolysis of monolignol glucosides during lignification. The three genes (PbBGLU1, PbBGLU15, and PbBGLU16) in PbBGLU group V-C clustered together with these functional genes, speculating that PbBGLU1, PbBGLU15, and PbBGLU16 may be able to affect lignification by catalyzing the hydrolysis of monolignol glucosides in pear fruit, which in turn affects the development of pear stone cells.
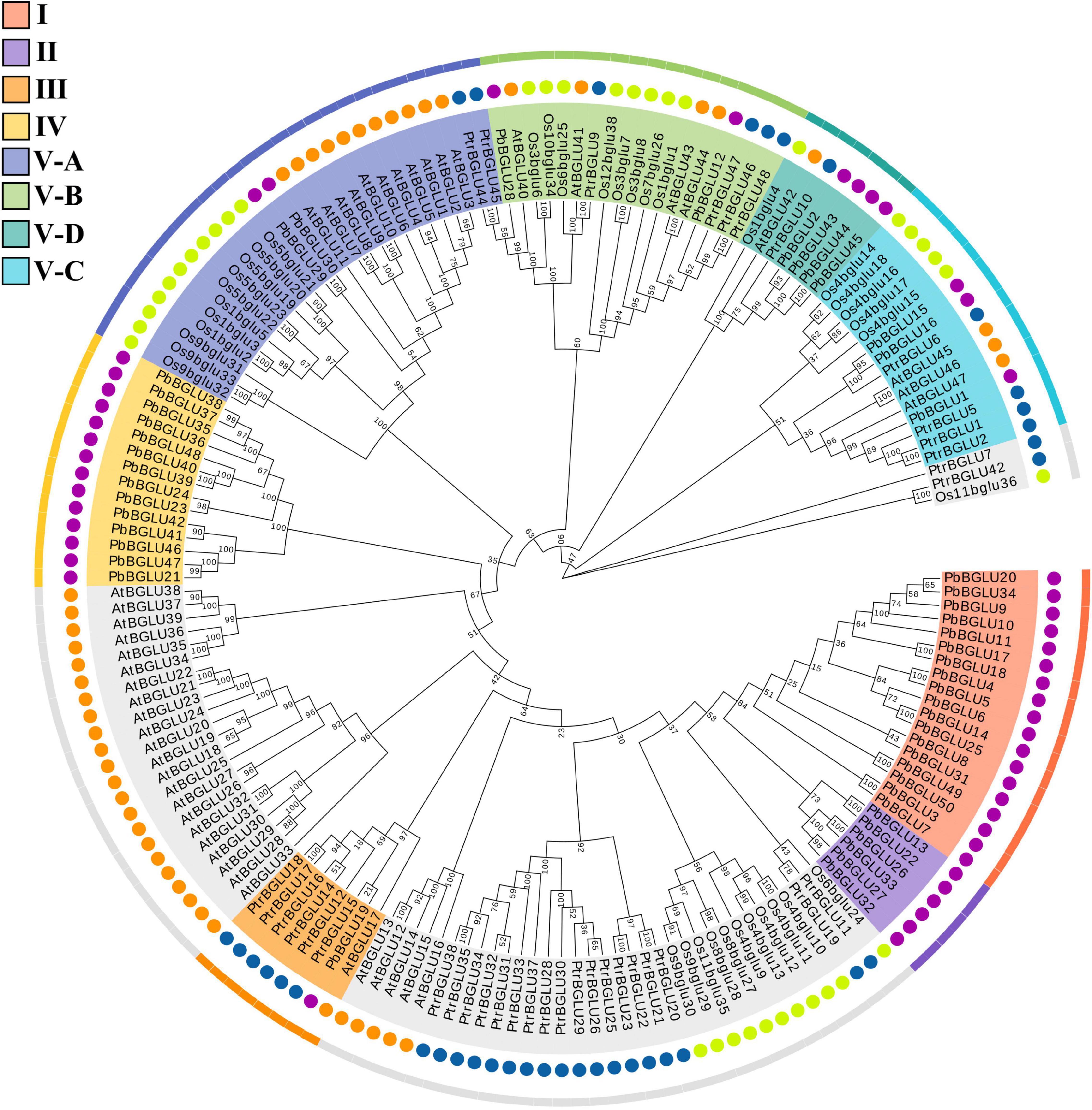
Figure 2. Phylogenetic analysis of BGLUs from Pyrus bretschneideri, Arabidopsis thaliana, Oryza sative, and poplar. The neighbor-joining (NJ) method was used to construct the phylogenetic tree. Pb: Pyrus bretschneideri; Os: Oryza sative; Ptr: Poplar L.; At: Arabidopsis thaliana.
Expression Characteristics of Chinese White Pear BGLU Genes
We constructed a phylogenetic tree comprising BGLUs genes from A. thaliana (Xu et al., 2004), O. sative (Opassiri et al., 2006), poplar (Tsuyama and Takabe, 2015), and P. bretschneideri, and found that PbBGLU1, PbBGLU15, and PbBGLU16 in the group V-C PbBGLUs of pear clustered together with AtBGLU45, AtBGLU46, Os4BGlu14, Os4BGlu16, Os4BGlu18, and PtrBGLU6 functional genes, which can hydrolyze monolignol glucosides during lignification (Escamilla-Treviño et al., 2006; Chapelle et al., 2012; Baiya et al., 2014; Tsuyama and Takabe, 2015; Baiya et al., 2018). To verify whether PbBGLU1, PbBGLU15, and PbBGLU16 have similar functions, we selected all PbBGLU genes in group V (a total of 11 PbBGLUs) for qRT-PCR analysis. The expression profiles of these genes in different tissues of pear (leaves, buds, stems, flowers) and pear fruit at different developmental stages (15, 39, 47, 55, 63, 79, and 145 days after flowering) were also investigated (Figure 3). It found that PbBGLU43 was not expressed in different tissue parts or at different developmental stages of the fruit. The expression levels of the other ten PbBGLUs were generally high in leaves, buds, flowers, and young fruits (15, 39, 47, and 55 days after flowering). This result was consistent with those of previous research showing that β-glucosidase can be found mainly in young vegetative parts (Kristoffersen et al., 2000; Biely et al., 2003). The expression levels of the PbBGLU1, PbBGLU15, and PbBGLU16 genes were relatively high in the leaves and flowers as compared to buds and stems. All three genes were expressed at different stages of fruit development, and their expression showed a trend of first increasing then decreasing, with a similar trend of the content for lignin and stone cells in “Dangshan Su” pear fruit (Cai et al., 2010; Jin et al., 2013). The expression patterns of these three genes were also similar to those of the key genes involved in the regulation of the lignin synthesis pathway (Xie et al., 2013). These results suggested that the three genes (PbBGLU1, PbBGLU15, and PbBGLU16) in the group V-C PbBGLUs might be functional genes involved in the hydrolysis of monolignol glucosides in the pear lignin synthesis pathway.
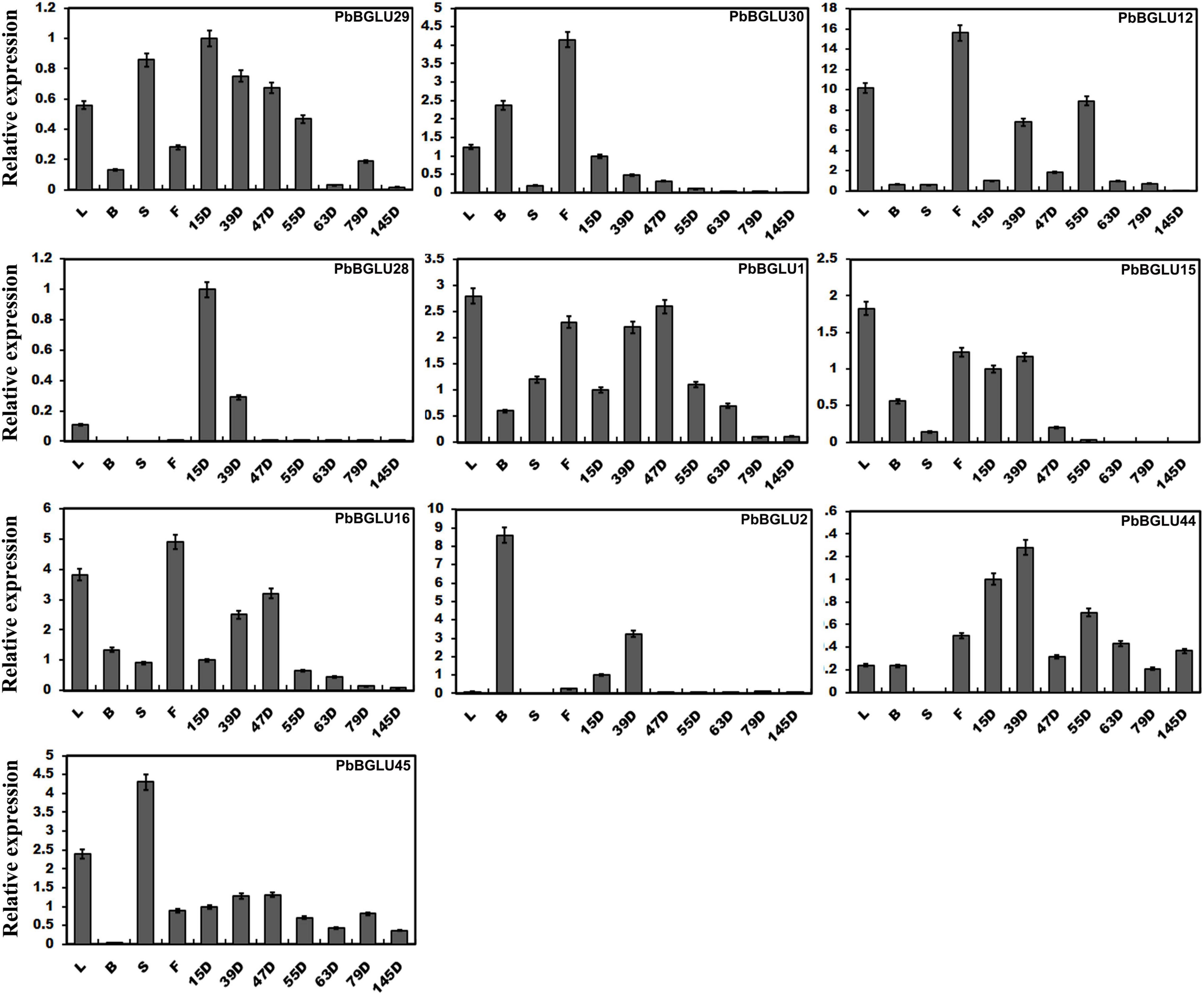
Figure 3. Expression levels of PbBGLU genes in different plant tissues. 15D, 39D, 47D, 55D, 63D, 79D, and 145D correspond to seven different developmental stages (days after flowering) of pear fruit. In addition, leaf, bud, stem, and flower were represented by L, B, S, and F, respectively. The value on the left Y-axis indicates the relative gene expression levels.
Subcellular Localization of PbBGLU1, PbBGLU15, and PbBGLU16
As early as 1995, it found that Pinus contorta β-glucosidase activity occurred in the cell wall of secondary xylem tissue (Dharmawardhana et al., 1995). AtBGLU45 and AtBGLU46 of A. thaliana were also expressed in the cell wall (Chapelle et al., 2012). The rice Os4BGlu16 tagged with a C-terminal GFP was transiently expressed in tobacco leaf epithelial cells, which revealed that the GFP signal was exclusively localized to the plasma membrane and extracellular space in both intact and plasmolyzed cells (Baiya et al., 2018). It is consistent with apoplastic or cell wall localization. In this study, to explore the subcellular localization of PbBGLU1, PbBGLU15, and PbBGLU16, PbBGLU-GFP expression vectors were constructed and transformed into N. tabacum. As shown in Figure 4, green fluorescence signals from the expressed PbBGLU1-GFP, PbBGLU15-GFP, and PbBGLU16-GFP fusion constructs were specifically distributed in the extracellular region.
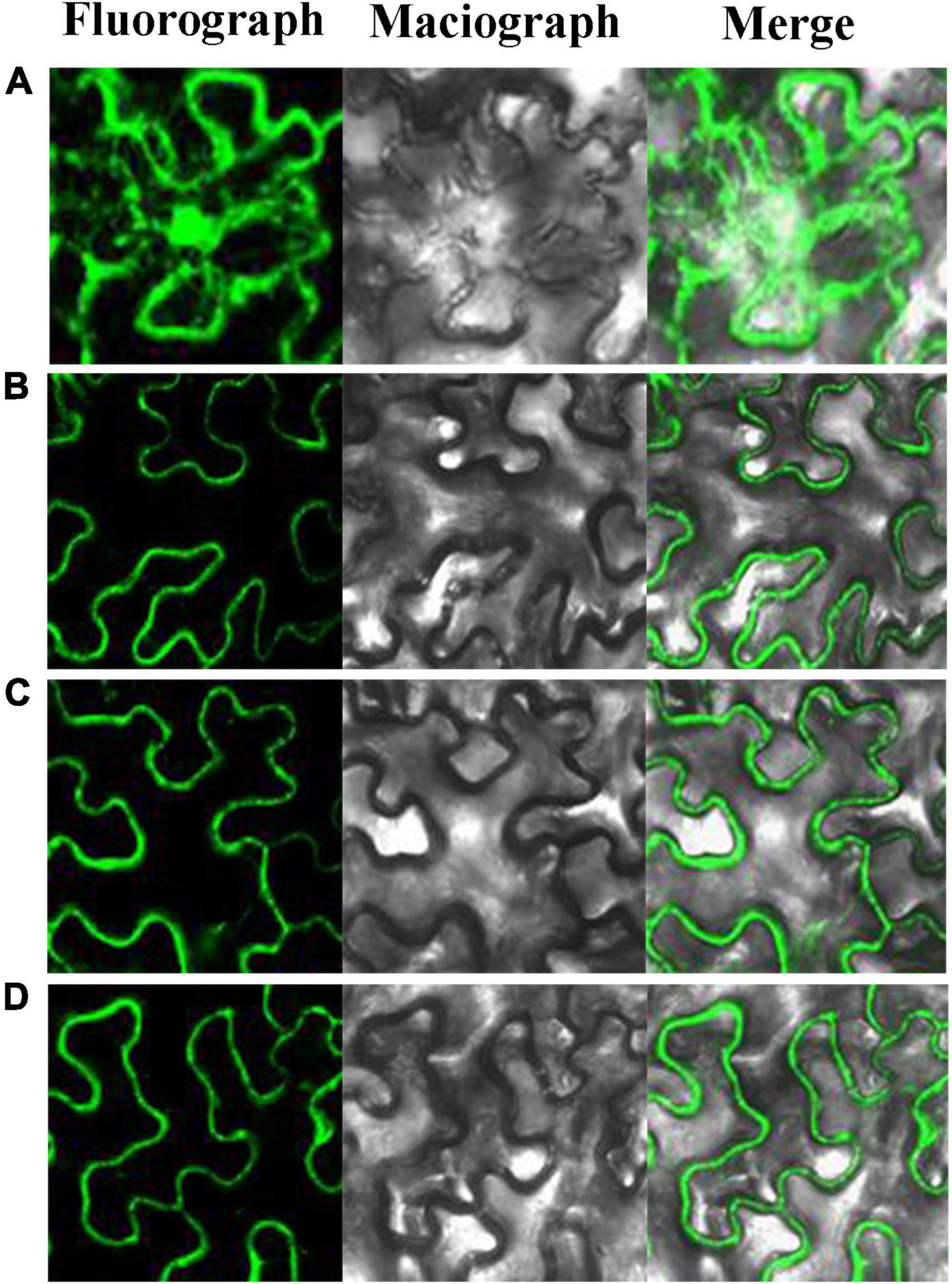
Figure 4. Subcellular localization of PbBGLU1-GFP, PbBGLU15-GFP, and PbBGLU16-GFP fusion protein. (A) GFP control; (B) Subcellular localization of PbBGLU1-GFP fusion protein; (C) Subcellular localization of PbBGLU15-GFP fusion protein; (D) Subcellular localization of PbBGLU16-GFP fusion protein.
In situ Hybridization Analysis of PbBGLU1 and PbBGLU16 in 39 DAF Pear Fruit
To confirm the PbBGLU1 and PbBGLU16 gene expression associated with those stone cells undergoing lignin deposition and secondary wall thickening, we carried out RNA in situ hybridization (Wu and Wagner, 2012) on 39 DAF (days after flowering) pear fruit. The results of PbBGLU1 transcripts in situ hybridization with PbBGLU1 anti-sense probe showed that PbBGLU1 transcripts were not only localized to some pulp cell walls, lignin deposition and stone cell areas of a pear fruit, there was also a small amount of enrichment in normal pear flesh cells (Figure 5A). This result shows that PbBGLU1 is not only involved in lignin biosynthesis but also may be involved in other metabolic activities during pear fruit development. However, PbBGLU16 transcripts were only enriched in lignin deposition and stone cell areas of pear fruit (Figure 5C), indicating that it may be a specific gene for lignin synthesis in pear fruit. Hybridization with control PbBGLU1 and PbBGLU16 sense probe showed no labeling as expected (Figures 5B,D).
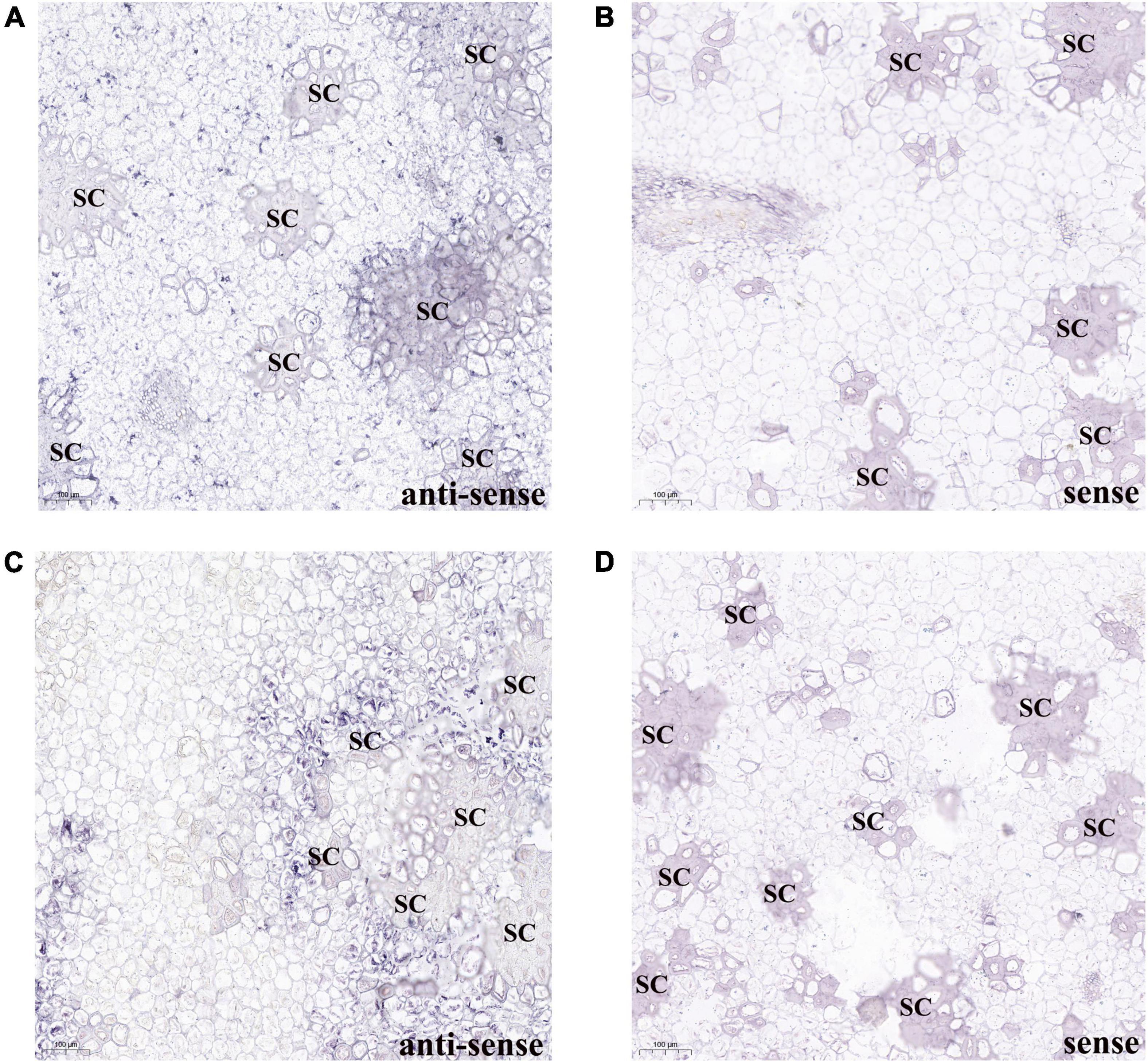
Figure 5. RNA in situ hybridization illustrating PbBGLU1 and PbBGLU16 transcript localization in 39 DAF (day after flower) pear fruit. (A,B) PbBGLU1 RNA in situ hybridization in 39 DAF (day after flower) pear fruit. (C,D) PbBGLU16 RNA in situ hybridization in 39 DAF (day after flower) pear fruit; Sections were taken through the flesh and probed with anti-sense (A,C) and sense probes (B,D) and imaged at 10 magnification. The positive expression of BCIP/NBT was blue-purple; the sense probes is for negative control; the bar = 100 μm; SC, stone cells.
Enzymatic Assays of Recombinant PbBGLU1 and PbBGLU16 in vitro
PbBGLU1 and PbBGLU16 were cloned into pGEX4T-1 vector, and then the constructed vector was induced and expressed in BL21 (ED3) E. coli. A target protein GST-PbBGLU1 with a weight of 78.07 kDa and another target protein GST-PbBGLU16 with a weight of 87.23 kDa obtained (GST: 26kDa; PbBGLU1: 52.07 kDa; PbBGLU16: 61.23 kDa). The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to analysis and verification (Supplementary Figure 3). To verify whether the recombinant protein has the ability to glycosylate the monolignol glucosides coniferin, syringin. We added recombinant protein, coniferin, and syringin and buffer in the certain reaction system. After the reaction for a while, methanol was added to terminate the reaction and then put into the HPLC for detection. The standard of coniferyl alcohol, sinapyl alcohol, coniferin and syringin were put into the HPLC for reference (Figures 6A,B). The results showed that after the recombinant protein was added, two peaks of the substrate (coniferin and syringin) and product (coniferyl alcohol and sinapyl alcohol) were detected by HPLC. This phenomenon indicates that the recombinant protein of GST-PbBGLU1 and GST-PbBGLU16 were active against both substrates (coniferin and syringin). To calculate the kinetic parameters of the recombinant protein for coniferin and syringin enzyme, 10 substrates with different concentrations were set in this study. The Km values, Vmax and kcat of GST-PbBGLU1 and GST-PbBGLU16 were calculated through the double reciprocal method. The results are shown in Supplementary Figure 4 and Table 1. Compared with syringin, GST-PbBGLU1 and GST-PbBGLU16 had a stronger activity and faster catalytic efficiency for coniferin. In addition, compared with GST-PbBGLU1, GST-PbBGLU16 shows higher catalytic activity and catalytic efficiency for the two substrates.
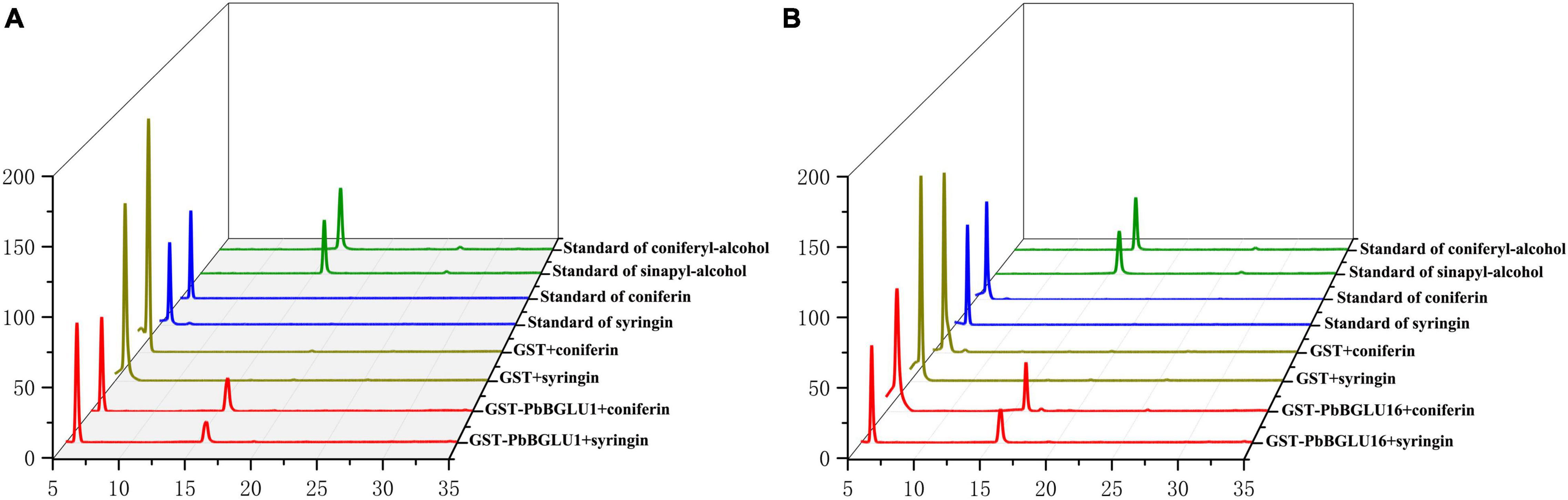
Figure 6. Enzyme activity of GST-PbBGLU1 and GST-PbBGLU16 recombinant protein toward coniferin and syringin. (A) HPLC analysis of standards and reaction products of GST-PbBGLU1 recombinant protein. (B) HPLC analysis of standards and reaction products of GST-PbBGLU16 recombinant protein. Reaction conditions: 10 μg of purified protein was incubated at 35°C with 20 μL buffer (50 mM MgSO4, 200 mM KCL, 100 mM PBS pH 7.2–7.4) and 1 mM substrates (coniferin and syringin), the water was added to a final volume of 50 μL. After 1 h of reaction, add 50 μL methanol termination reaction.
Analysis of Transgenic Arabidopsis With PbBGLU1 and PbBGLU16 Gene
The PbBGLU1 and PbBGLU16 were transferred into Arabidopsis BGLU-45 mutants (PbBGLU1-RE and PbBGLU16-RE) and wild-type Arabidopsis (PbBGLU1-OE and PbBGLU16-OE), reverse transcription PCR validation of Arabidopsis is shown in Figures 7A,B. To directly observe the in situ distribution of lignin in inflorescence stems of these Arabidopsis plants, we performed the Wiesner (phloroglucinol-HCl) histochemical staining (Pradhan and Loqué, 2014) of inflorescence stems of Arabidopsis plants. The Wiesner staining results indicated that the xylem and intravascular fibers in the stems of Arabidopsis BGLU-45 mutants showed no increase to any extent compared with wild-type Arabidopsis (Figures 7C-2,D-2), this result is consistent with the results of previous studies (Chapelle et al., 2012). Then, we observed the stained sections of PbBGLU1-OE and PbBGLU16-OE Arabidopsis and the Wiesner staining results indicated that the xylem and intravascular fibers in the stems of Arabidopsis PbBGLU1-OE and PbBGLU16-OE showed stronger phloroglucinol staining compared to the wild-type plants (Figures 7C-4,D-4). According to the staining results, we can also find that PbBGLU16-OE xylem and intravascular fibers Arabidopsis increase more strongly than PbBGLU1-OE Arabidopsis. Subsequently, we measured the lignin content of wild-type, Arabidopsis BGLU-45 mutant, PbBGLU1-RE, and PbBGLU16-RE, PbBGLU1-OE, and PbBGLU16-OE Arabidopsis by acetyl bromide method (Anderson et al., 2015; Figures 7E,F). It found that the lignin content of Arabidopsis BGLU-45 mutant, PbBGLU1-RE and PbBGLU16-RE was no change than that of wild-type. However, compared with wild-type Arabidopsis the overexpression plants lignin increased in varying degrees, the effect of PbBGLU16 on the increase of lignin in Arabidopsis is greater than that of PbBGLU1.
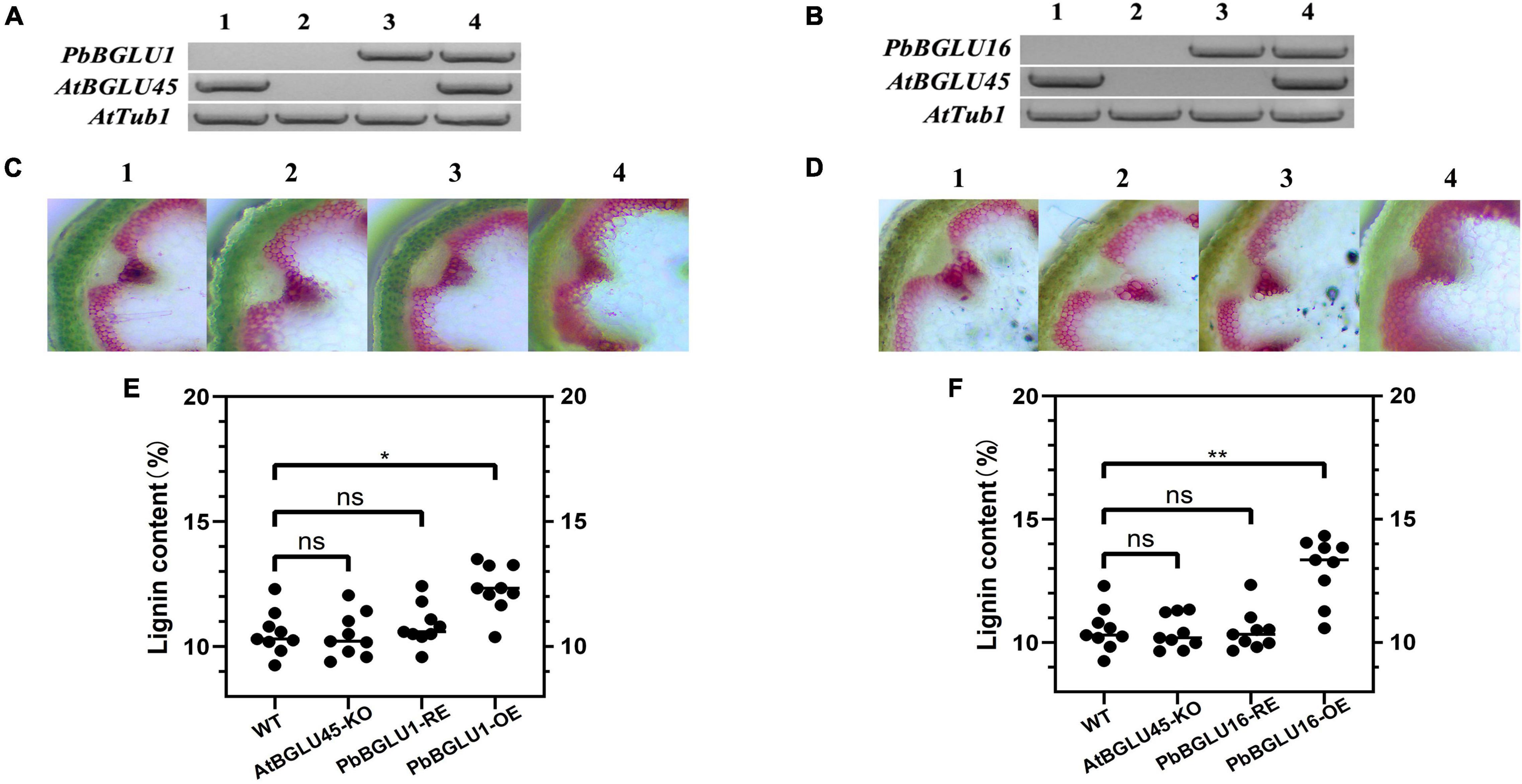
Figure 7. The lignin content in inflorescence stems of Arabidopsis. (A) PCR analysis of Arabidopsis thaliana wild type (A-1,B-1); AtBGLU-45 mutant (A-2,B-2); AtBGLU-45 mutant lines rescued by PbBGLU1 and PbBGLU16 (A-3,B-3); PbBGLU1 and PbBGLU16 overexpressing Arabidopsis (A-4,B-4). (C,D) Cross-sections of the inflorescence stem were stained with phloroglucinol–HCl; (C-1,D-1) Wild-type (WT) plants; (C-2,D-2) BGLU45 knock out mutant plants; (C-3,D-3) PbBGLU1 and PbBGLU16 rescue plants; (C-4,D-4) Over-expression PbBGLU1 and PbBGLU16 plants. (E,F) Measurement of the lignin content in inflorescence stems of Arabidopsis thaliana. The bar = 100 μm. (* P < 0.05, ** P < 0.01). KO, knock out; RE, rescue; OE, over expression.
Targeted Metabolite Determination
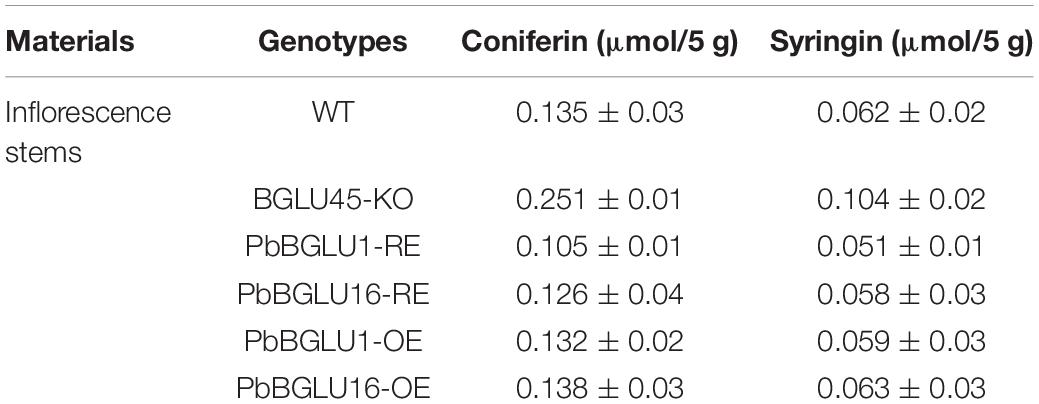
In order to further explore how BGLU-45 mutants and overexpression of PbBGLU1 and PbBGLU16 affect the lignin of A. thaliana, we measured the A. thaliana contents of coniferin and syringin by HPLC. The result showed that coniferin and syringin increased significantly in BGLU-45 mutant compared with the wild type (Table 2), but the lignin did not increase. This phenomenon occurred probably because coniferin and syringin are not precursors of lignin synthesis and usually exist in vacuoles as transport or storage forms. This is also probably the reason why there is no significant change in Arabidopsis lignin after knocking out BGLU45 in previous research (Chapelle et al., 2012; Baiya et al., 2018). Then, the contents of coniferin and syringin rising phenomenon have been restored through transferred PbBGLU1 and PbBGLU16 into Arabidopsis BGLU-45 mutant (Table 2). However, the lignin monolignol glycosides of transgenic Arabidopsis after overexpression of PbBGLU1 and PbBGLU16 decreased to some extent, but the decrease was not obvious, it may be because the decrease of lignin monolignol glycosides will increase the expression of UGT.
Transient Transformation of PbBGLU1 and PbBGLU16, PbBGLU1-RNAi and PbBGLU16-RNAi in Pear Fruit
Since the stable genetic transformation system of pears is in its infancy, it is difficult to perform a functional analysis of the PbBGLU1 and PbBGLU16 in pears. Therefore, we chose the method of transient expression by injection for analysis. We injected transiently the PbBGLU1, PbBGLU16, and PbBGLU1 and PbBGLU16 RNA interference in the 39 DAF (days after flowering) fruit of the pear tree. By Wiesner (phloroglucinol-HCl) histochemical staining (Pradhan and Loqué, 2014), the results show (Figures 8B,D) that when PbBGLU1 and PbBGLU16 were silenced by RNA interference, the change of pear lignin and stone cells was not obvious compared with the empty vector injection. But after PbBGLU1 and PbBGLU16 gene were overexpressed in the 39 DAF pear fruit, it found that the lignin and stone cells increased compared with the empty vector injection control in the pear fruit (Figures 8A,C). Moreover, compared with the transient expression of PbBGLU1, PbBGLU16 had a stronger effect on the lignin and expression cells of pear fruit. To explore the specific effects of overexpression and RNAi of PbBGLU1 and PbBGLU16 on stone cells and lignin of pear fruit, we measured the contents of stone cells and lignin in pear fruit after empty vector, overexpression, and RNAi injection. The results are shown in Figures 8B,D which indicate that when PbBGLU1 and PbBGLU16 were silenced, the contents of stone cells and lignin in pear fruit did not change compared with empty vector injection. However, in pear fruits with transient overexpression of PbBGLU1, the contents of lignin and stone cells in pear fruits were significantly higher (0.01 < P < 0.05) than those in pear fruits with empty vector injection. After transient expression of PbBGLU16, lignin in pear fruit increased significantly compared with the control group (0.01 < P < 0.05), while the content of stone cells in pear fruit injected with PbBGLU16 showed a very significant difference (P < 0.01) compared with the control group.
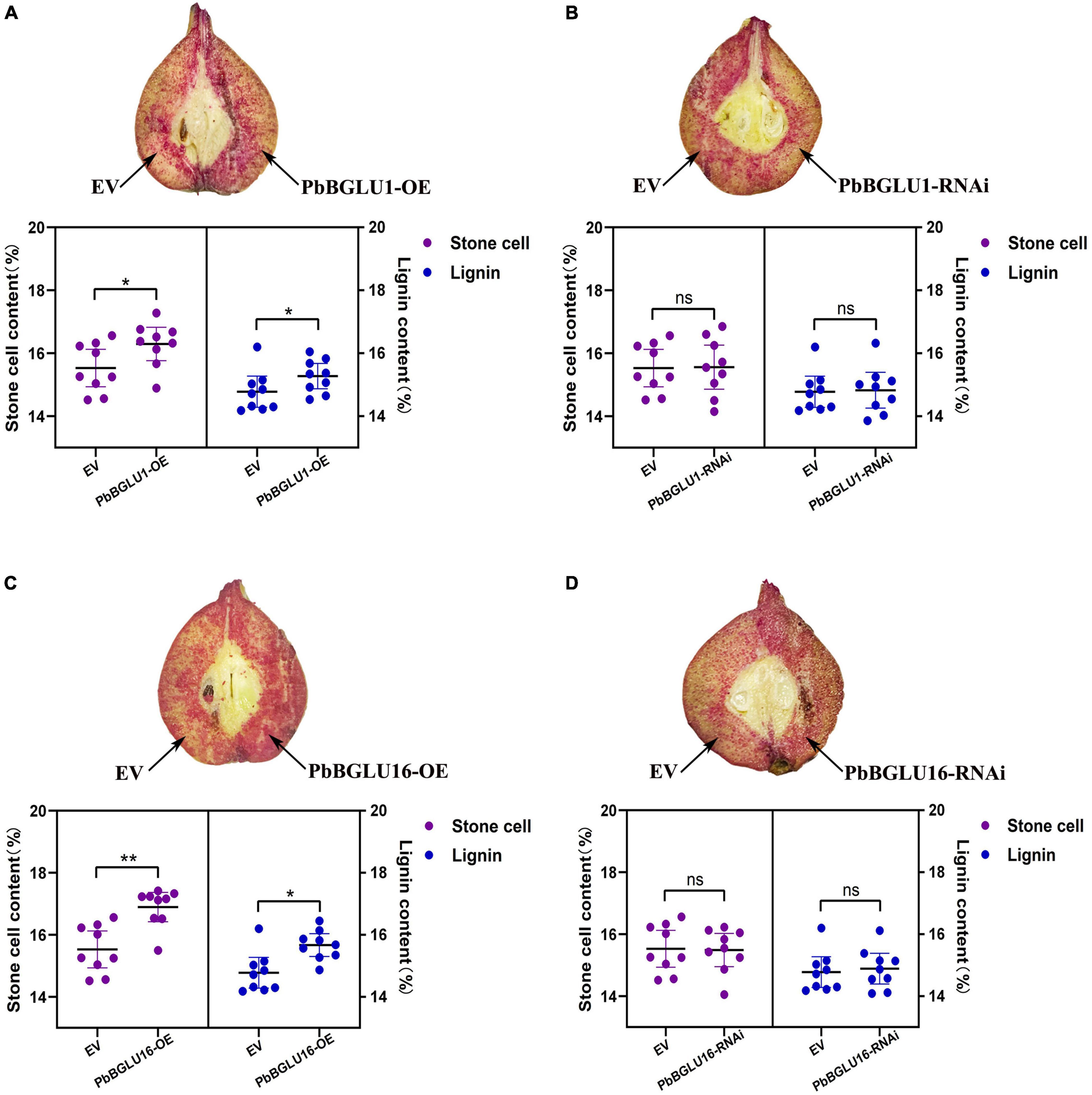
Figure 8. Transient assays using PbBGLU1 and PbBGLU16 silencing and overexpression constructs in “Dangshansuli” fruit at 39 days after flowering (DAF). Images were taken at 10 days after agro-infiltration. Cross-sections of PhBGLU1, PbBGL.U16 overexpression (A,C) and silencing (B,D) pear fruits were stained with phloroglucinol–HCI. Lignin and stone cell contents in PbBGLU1, PbBGLU16 overexpression (A,C) and silencing (B,D) flesh tissue around the infiltration sites, more than three fruits were injected with each construct in an experiment that was repeated three times. EV, empty vector; (* P < 0.05, ** P < 0.01).
Discussion
In plants, glycoside hydrolase (GH) family 1 β-glycosidases are believed to play an important role in many diverse processes, including chemical defense against herbivory, hydrolysis of cell wall-derived oligosaccharides during germination, control of active phytohormone levels, and lignification (Xu et al., 2004; Lee et al., 2006; Ahn et al., 2010; Xu et al., 2012; Zhou et al., 2012; Miyahara et al., 2013).
The complete sets of members of the GH1 BGLU genes family have been identified only in a few species. We identified a total 50 BGLUs in pear. The number was similar to that in Arabidopsis (48) (Xu et al., 2004) and poplar (48) (Tsuyama and Takabe, 2015), but more than that in rice (34) (Opassiri et al., 2006) and maize (26) (Gomez-Anduro et al., 2011). Previous studies have shown that the role of monolignol glucosides was necessary for transmembrane transport (Tsuyama and Takabe, 2015; Julien et al., 2016), monolignol glucosides form can increase the solubility and stability of monolignols, which is very beneficial for the transportation of lignin, ultimately to specific locations at the cell wall, after which point lignin begins to deglycosylate and polymerize (Liu, 2012). Many previous studies have shown that BGLU genes are localized to the cell wall and extracellular space (Dharmawardhana et al., 1995; Chapelle et al., 2012; Baiya et al., 2018). In this study, we found that green fluorescence signals from the expressed PbBGLU1-GFP, PbBGLU15-GFP, and PbBGLU16-GFP fusion constructs were specifically distributed in the extracellular region, which is consistent with previous research. Also, in our in situ hybridization analysis, it found that PbBGLU1 and PbBGLU16 transcripts were mainly located in lignin deposition and stone cell areas of pear fruit. This result further suggests that PbBGLU1 and PbBGLU16 may be functional genes in the process of lignin synthesis and stone cell development in pear.
The heterologous expression of Arabidopsis BGLU45 and BGLU46 genes in Pichia pastoris revealed that BGLU45 was highly specific for the three monolignol glucosides (coniferin, syringin, and p-coumaryl alcohol 4-O-b-D-glucoside). By contrast, BGLU46 has p-coumaryl alcohol glucoside as the preferred substrate, but it displays a broad specificity toward various substrates (Xu et al., 2004; Escamilla-Treviño et al., 2006). The in vitro substrate specificity of BGLU45 and BGLU46 and the expression of their genes in stems (Escamilla-Treviño et al., 2006) support the hypothesis that these enzymes are involved in the lignification of Arabidopsis. To further, verify the functions of PbBGLU1 and PbBGLU16, we induced expression and purification recombinant protein, and selected coniferin and syringin as substrates (Dicotyledons mainly contain syringic units and guaiacyl unit lignin: G-S). Enzyme activity analysis showed that the two recombinant proteins could act on coniferin and syringin to deglycosylate them and produce coniferyl alcohol and sinapyl alcohol. Moreover, compared with GST-PbBGLU1 recombinant protein, GST-PbBGLU16 recombinant protein shows higher activity and catalytic efficiency for the two substrates.
In previous studies, the implication of monolignol glucosides in the lignification process has been repeatedly suggested (Boerjan et al., 2003). The hypothesis was that monolignol glucosides would be stored in the vacuole or transported from the cytosol to the cell. One of the most predominant questions about phenylpropanoid biosynthesis concerns the spatial distribution of both precursors and final products within the cells. Many phenylpropanoid molecules necessary for plant development and/or defense are also toxic and an important challenge for plants is how to successfully manage the production/storage of these potentially dangerous molecules (Väisänen et al., 2015). In previous studies, the knock-out of BGLU-45 and BGLU-46 in Arabidopsis can lead to a significant increase of monolignol glycosides, but the lignin content does not seem to change significantly (Chapelle et al., 2012; Baiya et al., 2018). We also obtained the same results in our experiment. After transferred PbBGLU1 and PbBGLU16 into BGLU-45 Arabidopsis mutant, we found that the rise of these two glycosides in BGLU-45 Arabidopsis mutant was some degree of recovery, which further proved the function of PbBGLU1 and PbBGLU16. When PbBGLU1 and PbBGLU16 were overexpressed into Arabidopsis, the lignin content of A. thaliana was significantly increased, and the increasing degree of PbBGLU16-OE was greater than that of PbBGLU1-OE. The same result also occurred in the experiment of instantaneous injection in pear fruit, when we silenced PbBGLU1 and PbBGLU16 by RNA interference, there was no significant difference in the contents of lignin and stone cells between pear and the control group, but the contents of stone cells and lignin were significantly increased when PbBGLU1 and PbBGLU16 were transiently overexpressed, and PbBGLU16 has a greater effect on lignin and stone cells than PbBGLU1. A past study also showed that the expression of BGLU45 and BGLU46 was deregulated under various biological stresses, suggesting that the monolignol storage pool might be used under pathogen attack in the fabrication of “defense lignin” and/or phytoalexins, cell, and vacuole disruption following pathogen attack, insect/herbivore feeding and potentially severe abiotic stress (cell freezing, desiccation) or during programmed cell death (xylem vessel) would lead to contact between themonolignol glucosides and β-glucosidases at the cell wall where PRXs and/or LACs could rapidly polymerize the aglycone forms (Morant et al., 2008; Chapelle et al., 2012). This also proves β-glucosidase can directly or indirectly increase lignin. However, monolignol glucosides had not been considered a dead-end product and its content is still regulated by many parties. Monolignol glucosides had often been detected in gymnosperms and its content in the differentiating xylems of gymnosperms peaks at around the cambium and decreases as lignification progresses (Fukushima et al., 1997; Tsuyama and Takabe, 2015). The regulation mechanism of lignin transport is very complex, which needs to be explored continuously. In conclusion, this study screened and proved the function of β-glucosidase during the process of lignin synthesis and stone cells development in pear fruit, and laid a foundation for improving pear fruit quality at the molecular level.
Conclusion
In order to analyze the BGLU β-glucosidases in the glycoside hydrolase family 1 (GH1) of pear, bioinformatics analysis, qRT-PCR, subcellular localization, in situ hybridization, enzyme activity analysis, the transformation of PbBGLU1 and PbBGLU16 into Arabidopsis, and transient injection PbBGLU1 and PbBGLU16 into pear fruit experiment were performed in this study. A total of 50 non-redundant BGLU family members were identified in Chinese white pears while three candidate genes PbBGLU1, PbBGLU15, and PbBGLU16 were identified that might be involved in lignin synthesis. Subcellular localization showed that all three genes were located on the extracellular region. In situ hybridization analysis revealed that PbBGLU1 and PbBGLU16 were involved in lignin synthesis and stone cell development in pear fruit. Enzyme activity analysis showed that GST-PbBGLU1 and GST-PbBGLU16 had activity on coniferin and syringin. The transformation of PbBGLU1 and PbBGLU16 into Arabidopsis demonstrated that as compare with wild-type Arabidopsis, the overexpression plants lignin increased in varying degrees and the effect of PbBGLU16 on the increase of lignin in Arabidopsis was greater than that of PbBGLU1. In pear fruits with transient overexpression of PbBGLU1 and PbBGLU16, the contents of PbBGLU1-OE and PbBGLU16-OE lignin and stone cells in pear fruits were significantly higher than those in pear fruits with empty vector injection, the effect of PbBGLU16 on the increase of lignin and stone cells was greater than that of PbBGLU1. In summary, the identification of 50 BGLUs and analysis of these two BGLUs gene functions in pear will help to improve the quality of pear fruit in the future. These results provide information that may facilitate further functional analyses of PbBGLU genes to elucidate their biological roles in pear.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
YoC conceived and designed the experiments. HW, YiZ, and XF performed the experiments. FP, MM, YaZ, WH, YuZ, JL, and YuC analyzed the data. HW wrote the manuscript. All authors reviewed and approved the final submission.
Funding
This study was supported by National Natural Science Foundation of China (No. 31640068), Anhui Agricultural University 2020 Graduate Innovation Foundation (2020ysj-50), and 2021 Postgraduate Research Project in Colleges and Universities in Anhui Province (YJS20210202).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank everyone who contributed to this article. This manuscript part of the data has been released as a pre-print at Research Square (Wang et al., 2020).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.852001/full#supplementary-material
Abbreviations
BGLU, β-glucosidase; GH1, glycoside hydrolase family 1; HMM, hidden Markov model; NJ, neighbor-joining; qRT-PCR, real-time PCR; ABC transporter, ATP-binding cassette transporter.
Footnotes
- ^ http://gigadb.org/dataset/10008
- ^ https://pfam.xfam.org/
- ^ http://smart.embl.de/
- ^ http://web.expasy.org/compute_pi/
- ^ https://meme-suite.org/meme/doc/meme.html
- ^ http://gsds.cbi.pku.edu.cn
- ^ https://phytozome.jgi.doe.gov/pz/portal.html#
References
Ahn, Y. O., Shimizu, B., Sakata, K., Gantulga, D., Zhou, C., Esen, A., et al. (2010). Scopolin-hydrolyzing beta-glucosidases in roots of Arabidopsis. Plant Cell Physiol. 51, 132–143. doi: 10.1093/pcp/pcp174
Alejandro, S., Lee, Y., Tohge, T., Sudre, D., Osorio, S., Martinoia, E., et al. (2012). AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 22, 1207–1212. doi: 10.1016/j.cub.2012.04.064
Anderson, N. A., Tobimatsu, Y., Ciesielski, P. N., Ximenes, E., Ralph, J., Donohoe, B. S., et al. (2015). Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell 27, 2195–2209. doi: 10.1105/tpc.15.00373
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Baiya, S., Hua, Y., Ekkhara, W., and Ketudat Cairns, J. R. (2014). Expression and enzymatic properties of rice (sativa L.) monolignol β-glucosidases. Plant Sci. 227, 101–109. doi: 10.1016/j.plantsci.2014.07.009
Baiya, S., Mahong, B., Lee, S. K., Jeon, J. S., and Ketudat Cairns, J. R. (2018). Demonstration of monolignol beta-glucosidase activity of rice Os4BGlu14, Os4BGlu16 and Os4BGlu18 in Arabidopsis thaliana bglu45 mutant. Plant Physiol. Biochem. 127, 223–230. doi: 10.1016/j.plaphy.2018.03.026
Barros, J., Serk, H., Granlund, I., and Pesquet, E. (2015). The cell biology of lignification in higher plants. Ann. Bot. 15, 1053–1074. doi: 10.1093/aob/mcv046
Biely, P., Ahlgren, J. A., Leathers, T. D., Greene, R. V., and Cotta, M. A. (2003). Aryl-glycosidase activities in germinating maize. Cereal Chem. 80, 144–147. doi: 10.1094/CCHEM.2003.80.2.144
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Cai, Y., Li, G., Nie, J., Lin, Y., Nie, F., Xu, Y., et al. (2010). Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 125, 374–379. doi: 10.1016/j.scienta.2010.04.029
Cao, Y., Han, Y., Li, D., Lin, Y., and Cai, Y. (2016a). MYB transcription factors in chinese pear (Pyrus bretschneideri Rehd.): genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 7:577. doi: 10.3389/fpls.2016.00577
Cao, Y., Li, D. H., Meng, D. D., Jin, Q., Cai, Y. P., and Lin, Y. (2016b). Structural, evolutionary, and functional analysis of the Class III peroxidase gene family in chinese pear (Pyrus bretschneideri). Front. Plant Sci. 7:1874. doi: 10.3389/fpls.2016.01874
Cao, Y., Han, Y., Meng, D., Li, D., Jiao, C., Cai, Y., et al. (2017). B-BOX genes: genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 17:156. doi: 10.1186/s12870-017-1105-4
Chapelle, A., Morreel, K., Vanholme, R., Le-Bris, P., Morin, H., Demont-Caulet, N., et al. (2012). Impact of the absence of stem-specific beta-glucosidases on lignin and monolignols. Plant Physiol. 160, 1204–1217. doi: 10.1104/pp.112.203364
Cheng, X., Li, G., Manzoor, M. A., Wang, H., Abdullah, M., Lin, Y., et al. (2019a). In silico genome-wide analysis of respiratory burst oxidase homolog (RBOH) family genes in five fruit-producing trees, and potential functional analysis on lignification of stone cells in Chinese White pear. Cells 8:520. doi: 10.3390/cells8060520
Cheng, X., Muhammad, A., Li, G., Zhang, J., Cheng, J., Lin, Y., et al. (2019b). Family-1 UDP glycosyltransferases in pear (Pyrus bretschneideri): molecular identification, phylogenomic characterization and expression profiling during stone cell formation. Mol. Biol. Rep. 46, 2153–2175. doi: 10.1007/s11033-019-04669-y
Dharmawardhana, D. P., Ellis, B. E., and Carlson, J. E. (1995). A beta-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin. Plant Physiol. 107, 331–339. doi: 10.2307/4276327
Dharmawardhana, D. P., Ellis, B. E., and Carlson, J. E. (1999). cDNA cloning and heterologous expression of coniferin β-glucosidase. Plant Mol. Biol. 40, 365–372. doi: 10.1023/A:1006226931512
Dong, X., Jiang, Y., and Hur, Y. (2019). Genome-wide analysis of glycoside hydrolase family 1 beta-glucosidase Genes in Brassica rapa and their potential role in pollen development. Int. J. Mol. Sci. 20:1663. doi: 10.3390/ijms20071663
Eastwood, D. C., Challen, M. P., Zhang, C., Jenkins, H., Henderson, J., and Burton, K. S. (2008). Hairpin-mediated down-regulation of the urea cycle enzyme argininosuccinatelyase in Agaricus bisporus. Mycol. Res. 112(Pt 6), 708–716. doi: 10.1016/j.mycres.2008.01.009
Escamilla-Treviño, L. L., Chen, W., Card, M. L., Shih, M. C., Cheng, C. L., Poulton, J. E., et al. (2006). Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides. Phytochemistry 67, 1651–1660. doi: 10.1016/j.phytochem.2006.05.022
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Bateman, A., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222. doi: 10.1093/nar/gkp985
Fukushima, K., Taguchi, S., Matsui, N., and Yasuda, S. (1997). Distribution and seasonal changes of monolignol glucosides in Pinus thunbergii. Mokuzai Gakkaishi 43, 254–259.
Gachon, C. M., Langlois-Meurinne, M., and Saindrenan, P. (2005). Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 10, 542–549. doi: 10.1016/j.tplants.2005.09.007
Gomez-Anduro, G., Ceniceros-Ojeda, E. A., Casados-Vazquez, L. E., Bencivenni, C., Sierra-Beltran, A., Tiessen, A., et al. (2011). Genome-wide analysis of the beta-glucosidase gene family in maize (Zea mays L. var B73). Plant Mol. Biol. 77, 159–183. doi: 10.1007/s11103-011-9800-2
Heneghan, M. N., Costa, A. M. S. B., Challen, M. P., Peter, R. M., Bailey, A., and Foste, G. D. (2007). A comparison of methods for successful triggering of gene silencing in Coprinopsis cinerea. Mol. Biotechnol. 35, 283–296. doi: 10.1007/BF02686014
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G., et al. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Ishihara, H., Tohge, T., Viehover, P., Fernie, A. R., Weisshaar, B., Stracke, R., et al. (2016). Natural variation in flavonol accumulation in Arabidopsis is determined by the flavonol glucosyltransferase BGLU6. J. Exp. Bot. 67, 1505–1517. doi: 10.1093/jxb/erv546
Jin, Q., Yan, C., Qiu, J., Zhang, N., Lin, Y., and Cai, Y. (2013). Structural characterization and deposition of stone cell lignin in Dangshan Su pear. Sci. Hortic. 155, 123–130. doi: 10.1016/j.scienta.2013.03.020
Julien, L. R., Brigitte, H., Anne, C., Simon, H., and Godfrey, N. (2016). Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 7:735. doi: 10.3389/fpls.2016.00735
Kristoffersen, P., Brzobohaty, B., Höhfeld, I., Bako, L., Melkonian, M., and Palme, K. (2000). Developmental regulation of the maize Zm-p60.1 gene encoding a beta-glucosidase located to plastids. Planta 210, 407–415. doi: 10.1007/PL00008149
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lanot, A., Hodge, D., Jackson, R. G., George, G. L., Elias, L., Lim, E. K., et al. (2006). The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 48, 286–295. doi: 10.1111/j.1365-313X.2006.02872.x
Lee, K. H., Piao, H. L., Kim, H. Y., Choi, S. M., Jiang, F., Hwang, I., et al. (2006). Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126, 1109–1120. doi: 10.1016/j.cell.2006.07.034
Leinhos, V., Udagama-Randeniya, P. V., and Savidge, R. A. (1994). Purification of an acidic coniferin-hydrolysing beta-glucosidase from developing xylem of Pinus banksiana. Phytochemistry 37, 11–15. doi: 10.1016/0031-9422(94)85053-4
Letunic, I., Doerks, T., and Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Liu, C. J. (2012). Deciphering the enigma of lignification: precursor transport, oxidation, and the topochemistry of lignin assembly. Mol. Plant 5, 304–317. doi: 10.1093/mp/ssr121
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001
Miao, Y. C., and Liu, C. J. (2010). ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. U.S. A. 107, 22728–22733. doi: 10.1073/pnas.1007747108
Miyahara, T., Sakiyama, R., Ozeki, Y., and Sasaki, N. (2013). Acyl-glucose-dependent glucosyltransferase catalyzes the final step of anthocyanin formation in Arabidopsis. J. Plant Physiol. 170, 619–624. doi: 10.1016/j.jplph.2012.12.001
Morant, A. V., Jorgensen, K., Jorgensen, C., Paquette, S. M., SanchezPerez, R., Moller, B. L., et al. (2008). beta-Glucosidases as detonators of plant chemical defense. Phytochemistry 69, 1795–1813. doi: 10.1016/j.phytochem.2008.03.006
Opassiri, R., Pomthong, B., Onkoksoong, T., Akiyama, T., Esen, A., and Ketudat Cairns, J. R. (2006). Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 beta-glucosidase. BMC Plant Biol. 6:33.
Perkins, M., Smith, R. A., and Samuels, L. (2019). The transport of monomers during lignification in plants: anything goes but how? Curr. Opin. Biotechnol. 56, 69–74. doi: 10.1016/j.copbio.2018.09.011
Pradhan, M. P., and Loqué, D. (2014). Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J. Vis. Exp. 87:51381. doi: 10.3791/51381
Raiskila, S., Pulkkinen, M., Laakso, T., Fagerstedt, K., Löija, M., Mahlberg, R., et al. (2007). FTIR spectroscopic prediction of Klason and acid soluble lignin variation in Norway spruce cutting clones. Silva Fenn. 41, 351–371. doi: 10.14214/sf.301
Roepke, J., Gordon, H. O. W., Neil, K. J. A., Gidda, S., Mullen, R. T., Freixas Coutin, J. A., et al. (2017). An apoplastic β-glucosidase is essential for the degradation of flavonol 3-O-β-glucoside-7-O-α-rhamnosides in Arabidopsis. Plant Cell Physiol. 58, 1030–1047. doi: 10.1093/pcp/pcx050
Steeves, V., Förster, H., Pommer, U., and Savidge, R. (2001). Coniferyl alcohol metabolism in conifers – I. Glucosidic turnover of cinnamyl aldehydes by UDPG: coniferyl alcohol glucosyltransferase from pine cambium. Phytochemistry 57, 1085–1093. doi: 10.1016/S0031-9422(01)00107-8
Su, X., Zhao, Y., Wang, H., Li, G., Cheng, X., Cai, Y., et al. (2019). Transcriptomic analysis of early fruit development in Chinese white pear (Pyrus bretschneideri Rehd.) and functional identification of PbCCR1 in lignin biosynthesis. BMC Plant Biol. 19:417. doi: 10.1186/s12870-019-2046-x
Tsuji, Y., and Fukushima, K. (2004). Behavior of monolignol glucosides in angiosperms. J. Agric. Food Chem. 52, 7651–7659. doi: 10.1021/jf0491198
Tsuyama, T., and Takabe, K. (2015). Coniferin β-glucosidase is ionically bound to cell wall in differentiating xylem of poplar. J. Wood Sci. 61, 438–444. doi: 10.1007/s10086-015-1486-7
Väisänen, E. E., Smeds, A. I., Fagerstedt, K. V., Teeri, T. H., Willför, S. M., and Kärkönen, A. (2015). Coniferyl alcohol hinders the growth of tobacco BY-2 cells and Nicotiana benthamiana seedlings. Planta 242, 747–760. doi: 10.1007/s00425-015-2348-7
Wang, H., Zhang, Y., Zhao, Y., Han, W., Lu, J., Cai, Y., et al. (2020). Genome-wide comparative analysis of the β-glucosidase family in five rosaceae species and their potential role on lignification of stone cells in Chinese white Pear. Res. Squ. [Preprint]. doi: 10.21203/rs.3.rs-46971/v1
Wu, J., Wang, Z., Shi, Z., Zhang, S., Ming, R., Zhang, S., et al. (2012). The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 23, 396–408. doi: 10.1101/gr.144311.112
Wu, M. F., and Wagner, D. (2012). “RNA in situ hybridization in Arabidopsis,” in RNA Abundance Analysis. Methods in Molecular Biology (Methods and Protocols), Vol. 883, eds H. Jin and W. Gassmann (Totowa, NJ: Humana Press).
Wu, T., Zhang, C., Wu, J., Wan, H., and Zhang, S. (2012). Evaluation of candidate reference genes for real time quantitative PCR normalization in pear fruit. Afr. J. Agric. Res. 7, 3701–3704. doi: 10.5897/AJAR11.1842
Xie, M., Huang, Y., Zhang, Y., Wang, X., Yang, H., Yu, O., et al. (2013). Transcriptome profiling of fruit development and maturation in chinese white pear (Pyrus bretschneideri rehd). BMC Genomics 14:823. doi: 10.1186/1471-2164-14-823
Xu, Z. Y., Lee, K. H., Dong, T., Jeong, J. C., Jin, J. B., Hwang, I., et al. (2012). A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24, 2184–2199. doi: 10.1105/tpc.112.095935
Xu, Z., Luis, L., Zeng, L. H., Mallikarjun, L., David, R., Cheng, C. L., et al. (2004). Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 55, 343–367. doi: 10.1007/s11103-004-0790-1
Yan, C., Yin, M., Zhang, N., Jin, Q., Fang, Z., Cai, Y., et al. (2014). Stone cell distribution and lignin structure in various pear varieties. Sci. Hortic. 174, 142–150. doi: 10.1016/j.scienta.2014.05.018
Zamioudis, C., Hanson, J., and Pieterse, C. M. J. (2014). β-glucosidase bglu42 is a myb72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 204, 368–379. doi: 10.1111/nph.12980
Zhang, J., Cheng, X., Jin, Q., Su, X., Li, M., Cai, Y., et al. (2017). Comparison of the transcriptomic analysis between two Chinese white pear (Pyrus bretschneideri Rehd.) genotypes of different stone cells contents. PLoS One 12:e0187114. doi: 10.1371/journal.pone.0187114
Zhang, M. Y., Xue, C., Hu, H., Li, J., Xue, Y., Wang, R., et al. (2021). Genome-wide association studies provide insights into the genetic determination of fruit traits of pear. Nat. Commun. 12:1144. doi: 10.1038/s41467-021-21378-y
Zhou, C., Tokuhisa, J. G., Bevan, D. R., and Esen, A. (2012). Properties of beta-thioglucoside hydrolases (TGG1 and TGG2) from leaves of Arabidopsis thaliana. Plant Sci. 191, 82–92. doi: 10.1016/j.plantsci.2012.02.004
Keywords: pear, β-glucosidase, gene function, lignin synthesis, stone cells
Citation: Wang H, Zhang Y, Feng X, Peng F, Mazoor MA, Zhang Y, Zhao Y, Han W, Lu J, Cao Y and Cai Y (2022) Analysis of the β-Glucosidase Family Reveals Genes Involved in the Lignification of Stone Cells in Chinese White Pear (Pyrus bretschneideri Rehd.). Front. Plant Sci. 13:852001. doi: 10.3389/fpls.2022.852001
Received: 10 January 2022; Accepted: 31 March 2022;
Published: 10 May 2022.
Edited by:
Yuexue Liu, Shenyang Agricultural University, ChinaReviewed by:
James Robert Ketudat Cairns, Suranaree University of Technology, ThailandZe Peng, South China Agricultural University, China
Copyright © 2022 Wang, Zhang, Feng, Peng, Mazoor, Zhang, Zhao, Han, Lu, Cao and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongping Cai, eXBjYWlhaEAxNjMuY29t
†These authors have contributed equally to this work
 Han Wang
Han Wang Yingjie Zhang1†
Yingjie Zhang1† Yunpeng Cao
Yunpeng Cao