- 1Department of Biology, Swarthmore College, Swarthmore, PA, United States
- 2College of Life Sciences, Xinyang Normal University, Xinyang, China
Phospholipid biosynthesis is a core metabolic pathway that affects all aspects of plant growth and development. One of the earliest step in this pathway is mediated by choline/ethanolamine kinases (CEKs), enzymes in the Kennedy pathway that catalyze the synthesis of the polar head groups found on the most abundant plant phospholipids. The Arabidopsis genome encodes four CEKs. CEK1-3 have been well characterized using viable mutants while CEK4 encodes an essential gene, making it difficult to characterize its effects on plant development and responses to the environment. We have isolated an EMS-induced allele of CEK4 called bumpy stem (bst). bst plants are viable, allowing the effects of decreased CEK4 function to be characterized throughout the Arabidopsis life cycle. bst mutants have a range of developmental defects including ectopic stem growths at the base of their flowers, reduced fertility, and short roots and stems. They are also sensitive to cold temperatures. Supplementation with choline, phosphocholine, ethanolamine, and phosphoethanolamine rescues bst root phenotypes, highlighting the flow of metabolites between the choline and ethanolamine branches of the Kennedy pathway. The identification of bst and characterization of its phenotypes defines new roles for CEK4 that go beyond its established biochemical function as an ethanolamine kinase.
Introduction
Biological membranes compartmentalize cells and, in concert with embedded and associated protein complexes, regulate transport and signal transduction between cells and cellular compartments. Membranes are composed of a phospholipid bilayer consisting of a hydrophobic core sandwiched between hydrophilic head groups. Phosphatidylethanolamine (PtdEtn) and phosphatidylcholine (PtdCho) are the two most abundant phospholipids found in eukaryotic membranes. Their polar head groups, phosphoethanolamine and phosphocholine, are synthesized in the earliest steps of the Kennedy pathway when ethanolamine and choline, respectively, are phosphorylated by choline/ethanolamine kinases (CEKs). Later in the pathway these phosphorylated head groups are enzymatically coupled to the diacylglycerol to create PtdEtn and PtdCho which are subsequently incorporated into membranes (Gibellini and Smith, 2010). Mutations in CEKs and other Kennedy pathway enzymes result in a wide range of phenotypes in plants as well as heritable genetic diseases in humans, highlighting the importance of this core biosynthetic pathway in all eukaryotes (Kwon et al., 2012; Lin et al., 2015, 2020; Yunus et al., 2016; Tavasoli et al., 2020; Tannert et al., 2021). Extensive efforts to characterize the Kennedy pathway have revealed that most organisms have multiple CEK isoforms encoded by distinct CEK genes.
The Arabidopsis genome encodes four CEKs numbered CEK1-4. These genes are expressed at different levels compared to each other and exhibit tissue-specific and environmentally responsive expression patterns, suggesting that each enzyme plays a distinct role in choline and ethanolamine metabolism (Tasseva et al., 2004; Lin et al., 2020). Examples of sub-specialization in this gene family include diverse substrate specificities among the CEKs as well as their differential effects on plant growth and development. In vitro, CEK1 and CEK2 have both choline and ethanolamine kinase activity while CEK3 and CEK4 have substrate specificity for choline and ethanolamine alone, respectively. Neither CEK1 or CEK2 exhibited detectable ethanolamine kinase activity in vivo, suggesting that CEK4 is the principal ethanolamine kinase in Arabidopsis (Lin et al., 2019, 2020). Similar enzymatic specialization has also been described in Glycine max, suggesting that it is widespread in plants (Wharfe and Harwood, 1979). Complementing this biochemical approach, reverse genetics has also been used to reveal the developmental and biochemical roles of the Arabidopsis CEKs. cek2 and cek3 single mutants have short roots while cek1 mutant roots are normal. This shows that at least two of the Arabidopsis CEKs are required for normal growth and development. Additional insights into CEK function have come from a chemical rescue experiment. In this experiment cek3 seedlings were grown on media containing phosphocholine, which rescued the cek3 short-root phenotype. This experiment’s results are consistent with the biochemical characterization of CEK3 as a choline kinase. cek mutants have also been used to provide in vivo support for the in vitro biochemical characterization. cek1 single mutant and cek1 cek3 double mutant plants were shown to have decreased choline kinase activity in vivo (Lin et al., 2020).
Compared to CEK1-3, CEK4 remains relatively uncharacterized. This is because cek4 loss of-function mutants are embryo lethal (Meinke et al., 2008; Lin et al., 2015; Meinke, 2020). The viability of cek1, cek2, and cek3 T-DNA loss-of-function mutants coupled with the lack of detectable changes in lipid content in their single mutants suggests that cek1-3 have partially overlapping functions in lipid metabolism and plant development (Lin et al., 2015, 2020). CEK4 stands out from the other Arabidopsis CEKs as the functional alleles of CEK1, CEK2, and CEK3 in cek4 mutants cannot compensate for the loss of CEK4 activity. Fundamental differences between CEK4 and the other CEKs are also suggested by the observation that CEK4 is phylogenetically distant from the other Arabidopsis CEKs (Lin et al., 2015). Combined with CEK4’s ethanolamine substrate specificity in vitro, it appears that CEK4 plays unique and important roles in plant biochemistry and development. These roles are only partially understood. CEK4 is expressed throughout the plant, including throughout heart stage and later embryos. High expression levels are found in the reproductive organs and in pollen, roots, and trichomes (Honys and Twell, 2004; Marks et al., 2009; Gilding and Marks, 2010; Lin et al., 2015). A 35S:CEK4 line that complemented cek4 embryonic arrest was shown to result in increased levels of both PtdEtn and PtdCho in seedlings and increased levels of PtdCho in mature siliques, consistent with a predicted function as a CEK (Lin et al., 2015). No further phenotypes were reported for this overexpression line and, to the best of our knowledge, no roles in post-embryonic plant development or responses to the environment have been described for CEK4.
Here we describe the isolation of a viable EMS induced allele of CEK4 that appears to result in a partial loss of function. This novel allele reveals important post-embryonic developmental functions as well as a role in chilling responses for CEK4.
Materials and Methods
EMS Mutagenesis
EMS mutagenesis was performed as described in Silverblatt-Buser et al. (2018).
Plant Growth
Plants were grown on soil under standard long-day greenhouse conditions with supplemental lighting. Plants grown on plates were grown on 0.5× Murashige and Skoog (0.5× MS) media containing 1% sucrose at 22°C under constant light conditions in E-30B growth chambers (Percival Scientific).
bumpy stem Mapping and Cloning
DNA from 412 individuals with mutant phenotypes in a bst mapping population was prepared using a DNeasy Plant Maxi Kit (Qiagen). A NEBNext DNA library prep set for Illumina (NEB) was used to prepare the sequencing library that was sequenced using an Illumina HiSeq (Illumina). Reads were mapped to the TAIR10 reference genome using SHORE and bst was mapped using SHOREmap (Schneeberger et al., 2009).
Scanning Electron Microscopy
Fresh tissue was imaged using low vacuum mode on a Quanta 200 scanning electron microscope (FEI) equipped with a cooled stage.
Chemical Complementation
Ler and bst seeds were sterilized and plated on 0.5× MS media, stratified at 4°C for 2 days, and transferred to an incubator for vertical growth at 22°C. After 3 days, seedlings were transferred to treatment plates containing either 100 μM or 1 mM of choline chloride (Cho), phosphocholine chloride calcium salt tetrahydride (PCho), monoethanolamine (MEA), or O-phosphorylethanolamine (PEA) (Sigma, St. Louis, MO, United States). The position of root tips were marked at transplanting. Seven days after transplanting the plates were scanned using an Epson V600 scanner with a black piece of felt as a background and root growth was measured using ImageJ (Schneider et al., 2012). These experiments were repeated three times with 10–12 seedlings in each replicate.
Rosette Size Measurements
Rosette sizes were measured from images using ImageJ (Schneider et al., 2012) by measuring the Feret diameter of the smallest oval that completely surrounded each rosette.
Results
bumpy stem Is a Recessive Mutant With Pleiotropic Developmental Phenotypes
bumpy stem (bst) was identified as a mutant with bumpy and twisted stem during a genetic screen unrelated to phospholipid head group biosynthesis. In this screen, bob1-3 mutants in a Ler background were mutagenized with EMS and screened for synthetic developmental phenotypes (Silverblatt-Buser et al., 2018). Although bst does not exhibit a genetic interaction with bob1-3, we decided to characterize and clone the mutant gene because it had such an unusual phenotype. We back crossed bst into both Col-0 and Ler five times in order to remove bob1-3 and other unlinked EMS induced mutations and their phenotypes from our analysis. bst phenotypes, while similar in both Col-0 and Ler backgrounds, are consistently stronger in Ler. F1 plants of bst/bst × +/+ crosses did not display bst phenotypes and bst phenotypes segregate in a mendelian fashion in F2 families. This suggests that they are caused by a single recessive mutation. The most striking phenotype in bst mutants is the abnormal patterning of the inflorescence stem.
bumpy stem inflorescence stems are irregularly swollen and bumpy compared to Col-0 inflorescence stems. In Col-0 plants the phyllotactic patterning of flowers on the flanks of the stem is regular (Figure 1A). The regular spacing between flowers observed in Col-0 plants is irregular in the mutant, resulting in clusters of flowers with disrupted phyllotactic patterning (Figure 1B). Closer inspection of the stem/pedicel boundary revealed a disruption of this boundary in bst plants. Col-0 plants have a distinct boundary between the stem and the floral pedicels. This boundary lines up with the lateral edge of the stem (Figure 1C). In bst plants this boundary is moved away from the edge of the stem. Unlike wild-type plants, bst plants have bumps of tissue that extend from the edge of the stem. The boundary between this ectopic tissue and the floral pedicels is a clearly visible notch separating the stem bumps and the pedicels. Instead of being in line with the edge of the stem, this boundary is displaced laterally (Figure 1D). In addition to flowers, Arabidopsis inflorescences also have secondary or axillary inflorescences that emerge from their flanks. The boundary between axillary inflorescences and the primary inflorescence stem is similar in bst and Col-0 plants with no ectopic growth subtending bst axillary inflorescences (Figures 1E,F).
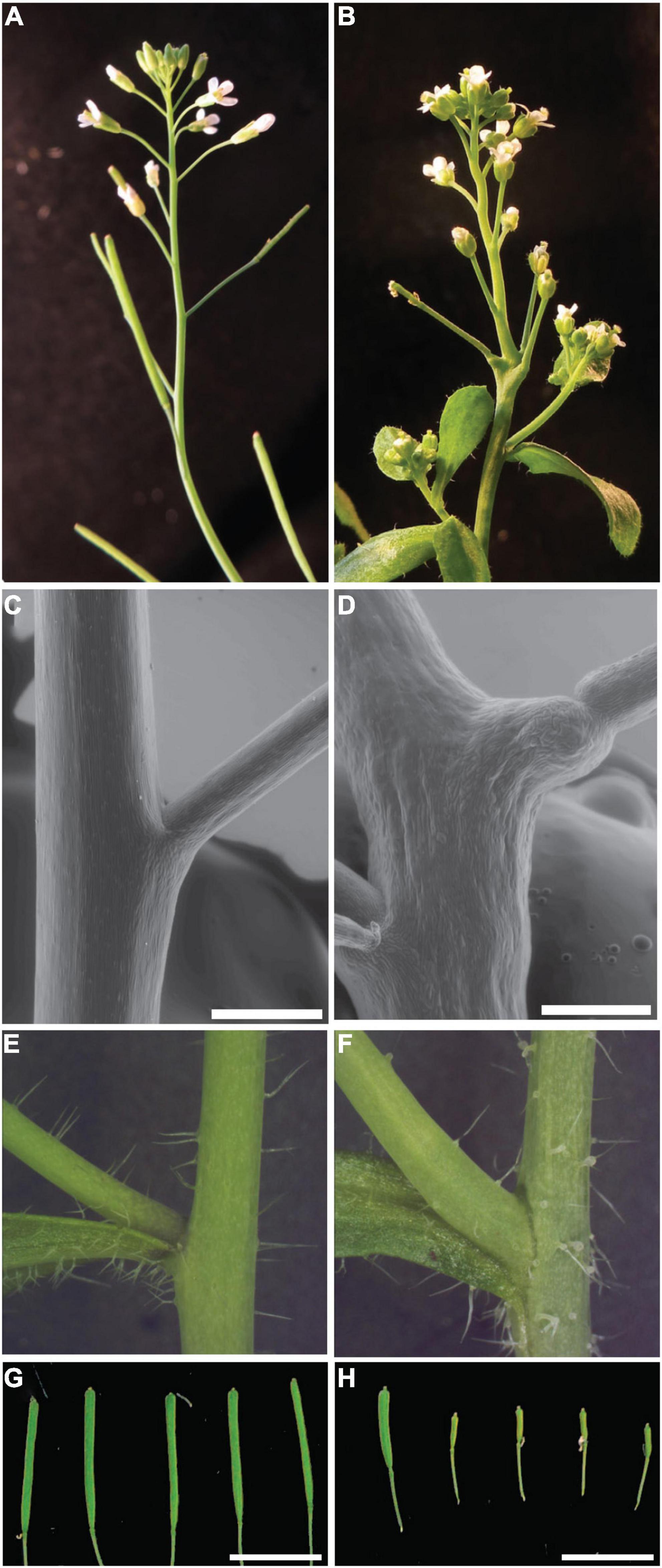
Figure 1. bumpy stem inflorescence phenotypes. Normal Col-0 inflorescences exhibit spiral phyllotaxy with flowers separated from each other along the inflorescence (A). bst flowers are found in clusters on the inflorescence and exhibit disrupted phyllotactic patterning (B). The boundary between the pedicel and stem is flush with the side of the stem in Col-0 plants (C). In bst mutants the notch defining the boundary is displaced, occurring at the tip of ectopic stem tissue that extends from the side of the stem (D). No displacement is observed at the boundary between the main stem and axillary stems (E,F). Compared to Col-0 trichomes (E), bst trichomes have swollen bases (F). Wild-type Ler plants are fertile and have filled and expanded siliques (G). bst mutants in a Ler background are semi sterile, resulting in short siliques containing few seeds (left silique) and many sterile unexpanded siliques (right siliques) (H). Scale bars are 0.5 mm in C,D and 1 mm in G,H.
bumpy stem plants have several other phenotypes associated with their inflorescences. Compared to the trichomes found on Col-0 stems bst trichomes have swollen bases (Figures 1E,F). bst plants are also semi-sterile. In the more severe Ler background only some siliques (16%, n = 93), most frequently from later flowers, produce seeds. The bst siliques that do make seeds and elongate are significantly shorter than Ler siliques (Ler 1.23 ± 0.15 cm, bst 0.82 ± 0.19 cm, two-tailed t-test p < 1 × 10–5, Figures 1G,H). In the less severe and more fertile Col-0 background all siliques contain seeds. However, bst siliques are shorter than wild-type siliques (Col-0 1.36 ± 0.18 cm, bst 0.57 ± 0.24 cm, two-tailed t-test p < 1 × 10–4). In addition to these phenotypes, bst mutants also have smaller leaves, shorter inflorescences, and shorter roots compared to normal plants (Figures 3, 4).
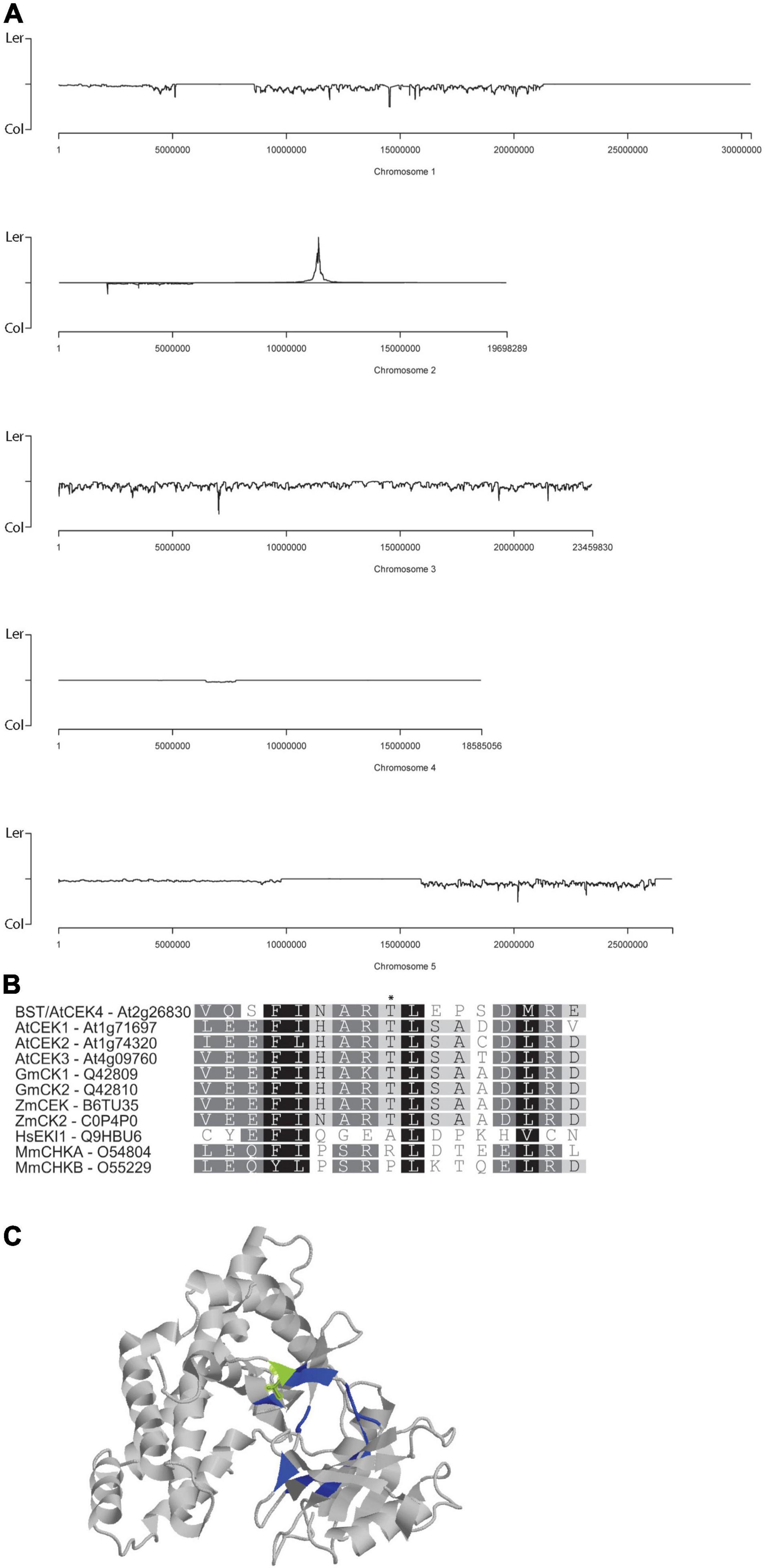
Figure 2. Cloning and characterization of bst. bst was simultaneously mapped and cloned using a NGS approach. bst was generated in a Ler background and its position on chromosome 2 can be seen as peak of enrichment in Ler SNPs (A). A protein alignment of CEKs from Arabidopsis (At), soybean (Gm), maize (Zm), humans (Hs), and mice (Mm) centered on T144 (indicated with a *) which is the site of the T144I mutation in bst (B). CEK4 structure with the ATP binding pocket visible. Residues that participate in ATP binding are shaded blue and T141 is shaded green. Image generated using the molecule viewer at bar.utoronto.ca/eplant (Waese et al., 2017) (C).
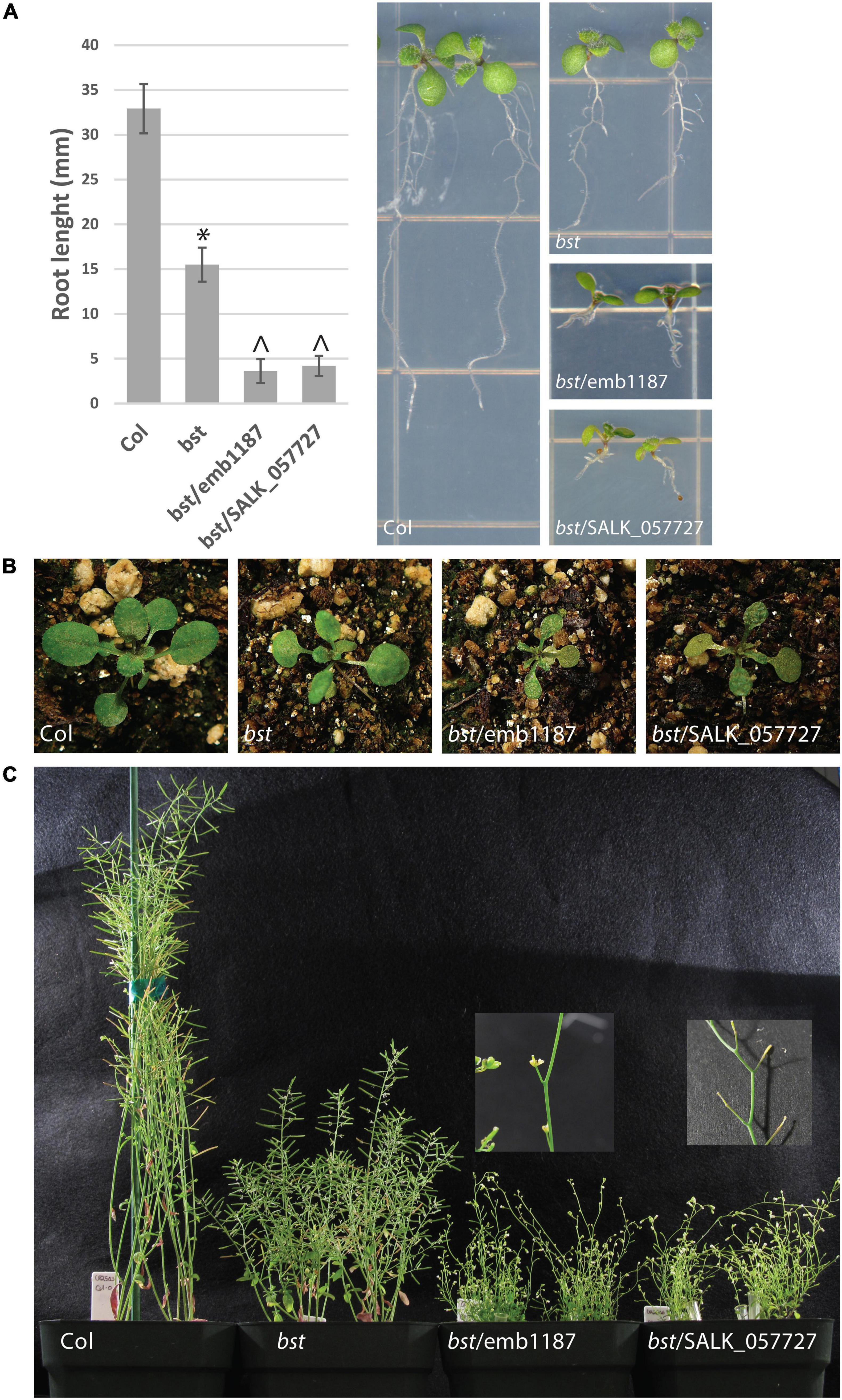
Figure 3. bumpy stem root and shoot phenotypes and genetic complementation. bst plants were crossed to heterozygous emb1187 and SALK_057727 plants, both of which contain T-DNA insertions in CEK4. Seven day old bst/bst plants were imaged and their root lengths were measured. Root lengths were compared using unpaired two-tailed t-tests. The symbol “*” indicates a difference from Col with a p-value < 10– 10. The symbol “^” indicates a difference from both Col and bst with a p-value < 10– 7. Error bars are SD (A). bst vegetative rosettes are smaller than wild-type. This phenotype is enhanced when the bst allele is combined with either emb1187 or SALK_057727 (B). The height of mature plants is reduced in bst plants and even more so in bst/emb1187 and bst/SALK_057727 plants. bst/emb1187 and bst/SALK_057727 plants are sterile (C).
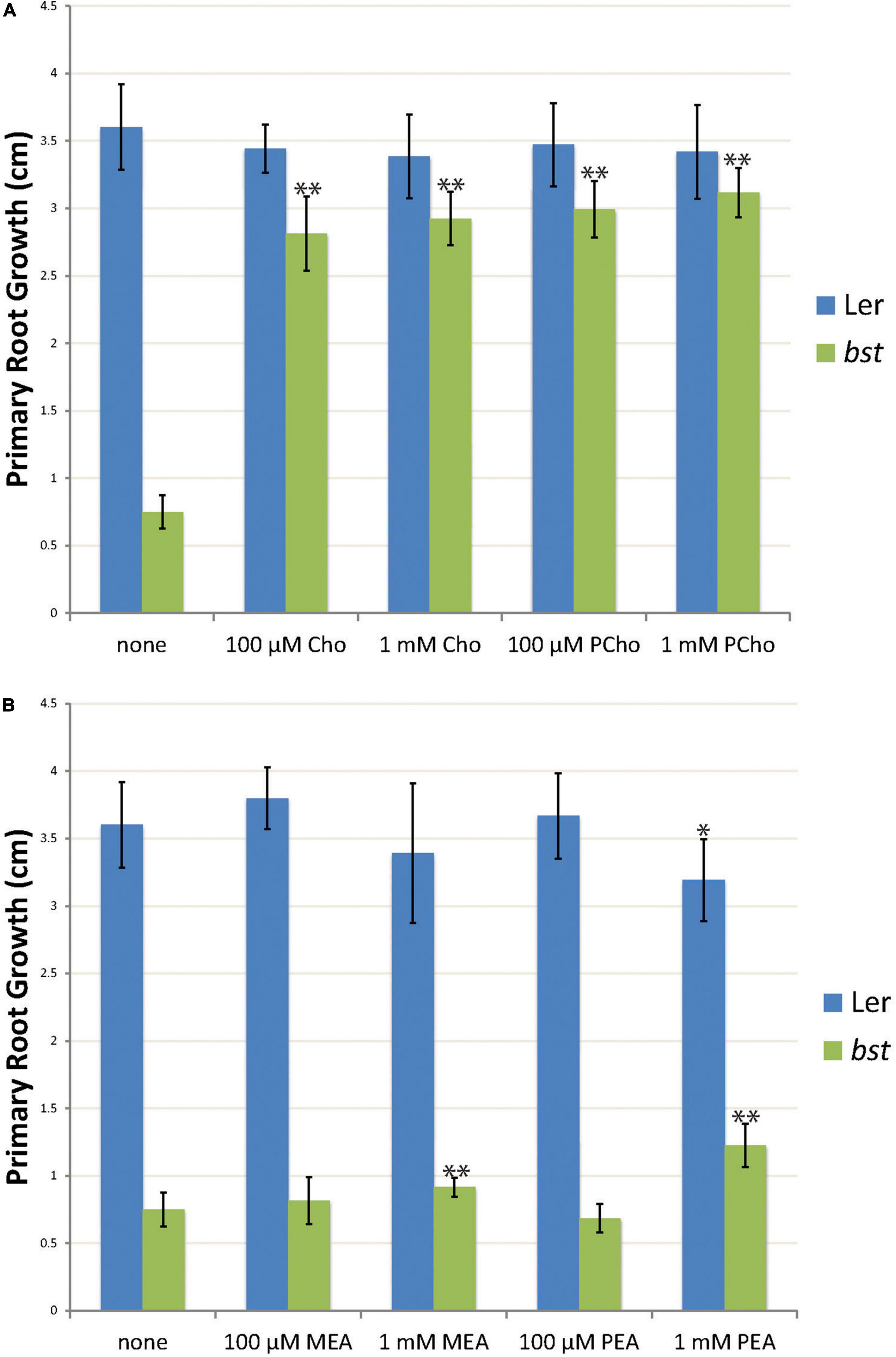
Figure 4. The effects of choline, phosphocholine, ethanolamine, and phosphoethanolamine supplementation on root growth. Three day old seedlings were transplanted to plates containing choline (Cho) or phosphocholine (PCho) (A) or ethanolamine (MEA) or phosphoethanolamine (PEA) (B). Primary root growth was measured after seven days of growth. The root length of treated plants was compared to untreated plants of the same genotype using unpaired t-tests. “*” indicates a p-value < 0.01 and “**” indicates a p-value < 0.001. Error bars are SD.
A Point Mutation in the Choline/Ethanolamine Kinase At2g26830/CEK4/emb1187 Causes bumpy stem Phenotypes
To map and clone bst we generated a mapping population by crossing bst twice into a Col-0 background. Pooled genomic DNA from 412 mutants isolated from this mapping population was sequenced to ∼100-fold coverage. SHOREmap (Schneeberger et al., 2009) was used to simultaneously map and identify putative causal mutations in the sequence data using Col-0/Ler SNPs as mapping markers. A single mapping peak was identified at 11.4 Mb on chromosome 2 (Figure 2A). Of the two non-synonymous mutations within 250 kb of the mapping peak, the 422C>T mutation in At2g26830 turned out to be the bst causal mutation (see below). The mutation results in a T141I substitution, converting the polar threonine residue at this position into isoleucine (which is hydrophobic). The threonine at position 141 in CEK4 is invariant in the four Arabidopsis CEKs as well as in corn (Zea mays) and soybean (G. max) CEKs (Figure 2B). It lies outside of the CEK catalytic domains d-6 and d-7 (Aoyama et al., 2004) at a position that directly interacts with ATP and forms part of the nucleotide binding pocket in the crystalized human choline kinase (Malito et al., 2006; Figure 2C).
In order to confirm that the mutation in CEK4 was responsible for bst phenotypes we used two previously identified cek4 T-DNA alleles in a genetic complementation experiment. Both alleles have embryo-lethal phenotypes. The first, cek4-1 (SALK_057727), arrests at the heart stage (Lin et al., 2015). The second, emb1187 (CS16107), arrests at the globular stage (Meinke et al., 2008; Meinke, 2020). As both of these T-DNA alleles are in a Col-0 background, we crossed each to bst introgressed into Col-0. Both lines failed to complement bst mutant phenotypes including short roots, small rosettes, short inflorescences, and bumpy stems. Each of these phenotypes was more severe in bst/emb1187 and bst/SALK_057727 plants than in bst/bst plants and the transheterozygous plants were completely sterile (Figure 3). The enhancement of the bst phenotype by two loss of function T-DNA alleles demonstrates that genetically, bst is a partial loss of function or hypomorphic allele (Muller, 1932).
Supplementation With Choline, Phosphocholine, Ethanolamine, and Phosphoethanolamine Rescues bumpy stem Root Growth Phenotypes
CEK4 has ethanolamine kinase activity in vivo and does not appear to function as a choline kinase (Lin et al., 2020). We took advantage of the viability of bst plants to test the hypothesis that CEK4 functions as an ethanolamine kinase in plants using a chemical supplementation approach. The simplest model is that bst phenotypes are caused by a lack of ethanolamine kinase activity. This would make low phosphoethanolamine levels the proximal deficiency causing phenotypes. If this is the case then phosphoethanolamine supplementation should rescue bst phenotypes, just as phosphocholine can rescue cek3 root phenotypes and ethanolamine and choline can rescue serine decarboxylase 1 mutants (Kwon et al., 2012; Lin et al., 2020). We decided to test this by quantitating root growth rates in wild-type and bst seedlings after supplementing them with polar head group precursors. Seedlings in a Ler background were germinated and then transferred to plates containing either choline, phosphocholine, ethanolamine, or phosphoethanolamine. Treatment with 100 μM choline or phosphocholine resulted in significant increases in bst root growth rates, nearly completely rescuing the short root mutant phenotype. The same effect was observed at 1 mM concentrations of choline and phosphocholine, suggesting that 100 μM concentrations are sufficient for the observed phenotypic rescue (Figure 4A). In contrast, treatment with 100 μM phosphoethanolamine or ethanolamine did not significantly change bst root growth rates. Increasing the concentration of ethanolamine or phosphoethanolamine to 1 mM resulted in significant increases in bst root growth rates. However, the effect of ethanolamine or phosphoethanolamine supplementation at 1 mM was much smaller than that seen with either choline or phosphocholine treatment at either 100 μM or 1 mM (Figure 4B).
bumpy stem Mutants Have a Chilling Sensitive Phenotype
The composition of phospholipid bilayers is known to change as a compensatory response to a wide range of abiotic stresses. Temperature extremes, both high and low, result in alterations in membrane lipids (Welti et al., 2002). Gene expression profiling in response to a range of abiotic stresses showed that CEK4 is strongly induced by cold temperatures (4°C) but not by other abiotic stresses (Figure 5; Kilian et al., 2007). This suggests that CEK4 may have important functions in low temperature responses and that bst mutants might have chilling inducible phenotypes. To test this hypothesis we germinated and grew plants on soil for 15 days at 23°C before transferring them to 6°C for an additional 35 days. bst plants in both Col-0 and Ler backgrounds grew similarly to wild-type when grown at 23°C for 29 days. Differences between wild-type and bst plants were apparent after 35 days of growth at 6°C. After extended growth at low temperatures, bst plants were stunted compared to wild-type plants and their leaves were covered with cold induced lesions (Figure 5). While every Col-0 and Ler plant grown at 6°C continued to make new leaves that expanded and turned green, none of the bst plants in either genetic background did so. Instead, their apices became discolored and ceased to produce visible new leaves. The rosette sizes of cold grown plants were significantly smaller than those of untreated plants for all genotypes and growth reductions were larger in bst than in normal plants in both genetic backgrounds (Ler 55% reduction, bst^Ler 63% reduction, Col-0 46% reduction, bst^Col-0 55% reduction, two-tailed t-test p < 0.002 for all genotypes). The interaction between genotype and treatment on rosette size was significant in a Ler background [f(1) = 6.414, p = 0.015] but not in a Col background, consistent with the increased severity of other phenotypes in Ler.
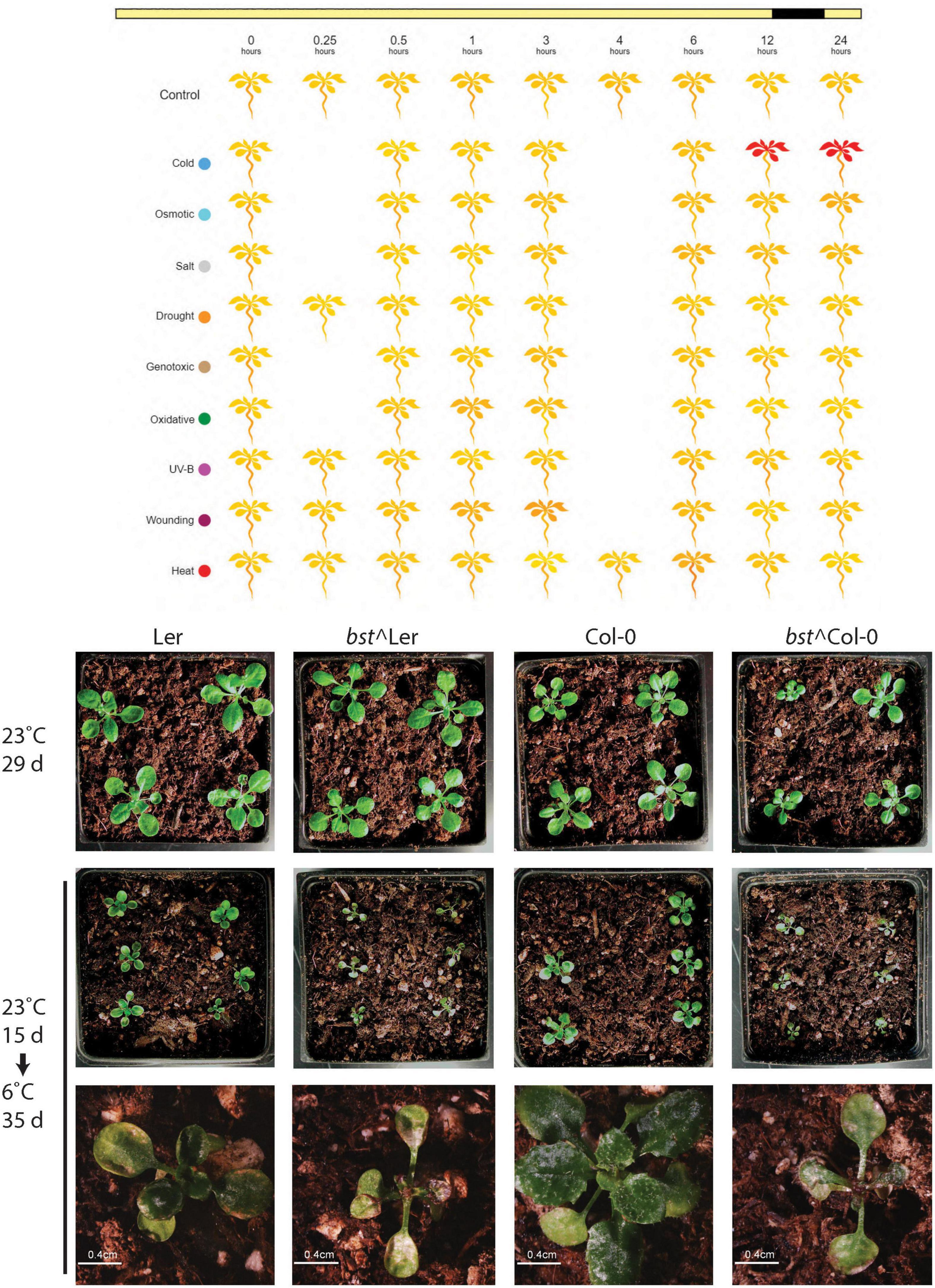
Figure 5. bumpy stem responses to low temperatures. bst expression is induced by cold temperatures but not by other abiotic stresses. Yellow indicates low expression and red indicates high expression [data from Kilian et al. (2007), image generated using eplant at bar.utoronto.ca/eplant (Waese et al., 2017)]. Normal plants and bst plants in both Ler or Col-0 backgrounds were grown for 15 days at 23°C. Untreated plants were grown for an additional 14 days at 23°C while cold treated plants were transferred to 6°C and grown for an additional 35 days.
Discussion
CEK4 is the least well characterized choline/ethanolamine kinase in Arabidopsis because it is required for embryonic viability and, up to now, only T-DNA loss of function alleles have been available. This has made it hard to establish what roles it plays in plant growth and development. In this study we isolated bst, a partial loss of function allele of CEK4, and used it to demonstrate that CEK4 plays roles in post-embryonic developmental patterning and cold responses.
CEK4 had previously been shown to be required for embryogenesis as cek4 T-DNA mutant embryos arrest early during development (Lin et al., 2015). While it is not surprising that a key enzyme in a core biosynthetic pathway would be an essential gene, there are three other CEK genes in the Arabidopsis genome which could compensate for a loss of CEK4 activity. The observation that they don’t suggests that CEK4 has specialized functions that overlap minimally, if at all, with those of the other CEKs. The bst allele of CEK4 behaves as a partial loss of function, demonstrated by the increased phenotypic severity when it is combined with a loss of function allele. bst is a point mutant, altering a key residue in the ATP binding pocket of the enzyme. The biochemical consequences of this change are uncharacterized but presumably the mutation results in decreased kinase activity due to altered ATP binding. This hypothesis could be tested in the future by directly assaying the effects of the bst T141I substitution on ATP binding and ethanolamine kinase activity, either in vitro or in vivo. We have not determined if bst mutant plants have alterations in their membranes. An analysis of any alterations may provide insights into the role that CEK4 plays in the biological pathways that are affected in bst mutants.
bumpy stem plants have pleiotropic phenotypes, consistent with CEK4 expression throughout plant development. These phenotypes include short roots and stems, swollen trichome bases, partial sterility, stem bumps at stem/pedicel junctions, and decreased chilling tolerance. These results suggest that the correct flux of metabolites through the Kennedy pathway is important not only for membrane biosynthesis but also for a variety of higher order processes in plants. Although we did not generate mechanistic insights into how bst results in decreased chilling tolerance, there are examples that illustrate the role of temperature dependent changes in phospholipid composition on temperature dependent responses. One example is temperature dependent flowering. The florigen FT has been shown to bind to membrane phospholipids at low temperatures, preventing it from moving and functioning as a signal. This serves as a mechanism that allows plants to delay flowering in response to chilling (Nakamura et al., 2014; Susila et al., 2021). Changes in the phospholipid composition of membranes could have direct functional effects on the protein machinery embedded in or associated with them which, in turn, may alter intra- and inter-cellular signaling (Champeyroux et al., 2020). In addition, there are many well established examples of lipids functioning directly as signaling molecules in plants (Wang and Chapman, 2013; Ruelland and Valentova, 2016). These examples include roles for lipids in responding to abiotic stresses such as chilling and freezing responses (Welti et al., 2002; Hou et al., 2016).
CEK4 has been shown to have ethanolamine kinase but no detectable choline kinase activity in vitro (Lin et al., 2020). This does not mean that mutations in CEK4 will only affect phosphoethanolamine production as phosphoethanolamine can be converted to phosphocholine by phosphoethanolamine N-methyltransferases (PMTs) (Figure 6). The observation that pmt1 mutants have short roots (Cruz-Ramírez et al., 2004) demonstrates that normal levels of phosphocholine, produced from phosphoethanolamine, are required for robust root growth. Arabidopsis pmt1 pmt3 double mutant plants have reduced phosphocholine levels and exhibit developmental phenotypes including small roots and shoots, but not bumpy stems, that can be rescued by choline or phosphocholine supplementation (Chen et al., 2018, 2019). pmt1 pmt2 pmt3 triple mutants are seedling lethal and exhibit up to 80% reductions in phosphocholine levels. This suggests that the bulk, if not all of, the phosphocholine found in Arabidopsis is produced from phosphoethanolamine and that decreased ethanolamine kinase activity will result in decreased phosphocholine levels (Liu et al., 2018, 2019; Chen et al., 2019). The conversion of phosphoethanolamine to phosphocholine by PMTs probably explains why CEK4 overexpression in Arabidopsis resulted in both increased PtdEth as well as increased PtdCho levels even though CEK4 only has ethanolamine kinase activity (Lin et al., 2015).
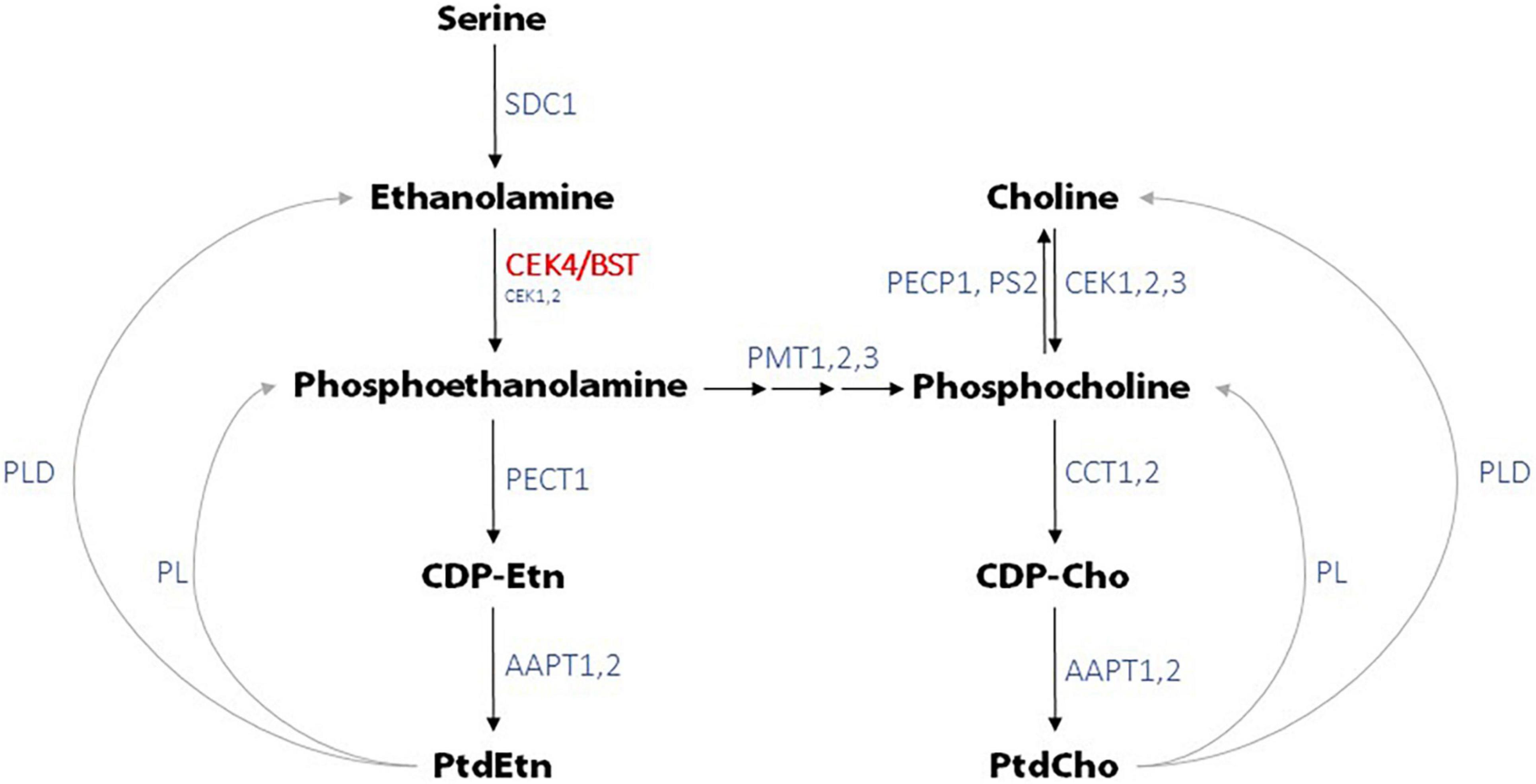
Figure 6. Phosphatidylethanolamine (PtdEtn) and phosphatidylcholine (PtdCho) biosynthetic pathways in Arabidopsis. CEK4/BST functions as the principal ethanolamine kinase, directly producing phosphoethanolamine which is used to generate PtdEth. Through the activity of PMTs, CEK4/BST is also responsible for the production of PtdCho and other choline containing compounds. Enzymes not defined in the text are SDC1, serine decarboxylase 1; PECT1, CTP:phosphorylethanolamine cytidylyltransferase 1; AAPT1,2, aminoalcohol aminophosphotransferase 1,2; PL, phospholipase; PLD, phospholipase D; PECP1, phosphoethanolamine/phosphocholine phosphatase 1; PS2, phosphate starvation-induced gene 2; CCT1,2, CTP:phosphorylcholine cytidylyltransferase 1,2. CDP-Cho is cytidine diphosphocholine and CDP-Etn is cytidine diphosphoethanolamine. Adapted from models presented in Lin et al. (2015), Chen et al. (2019), and Liu et al. (2019).
We tested the prediction that the bst short root phenotype would be rescued by supplementation with phosphoethanolamine, CEK4’s predicted product. Surprisingly, phosphoethanolamine only partially rescued bst root phenotypes and only did so at relatively high concentrations (1 mM). The same was true for ethanolamine, CEK4’s predicted substrate. We also supplemented bst seedlings with choline and phosphocholine. Rescue with both of these compounds was nearly complete at relatively low concentrations (100 μM). These unexpected results do not provide clear insights into bst’s root phenotypes; understanding them better will require a detailed metabolic analysis. One possible interpretation of these results is that, although CEK4 functions principally or solely as an ethanolamine kinase, much of the phosphoethanolamine it produces is subsequently used as a substrate for the choline branch of the Kennedy pathway (Figure 6). Since a reduction in PMT generated phosphocholine results in short roots (Cruz-Ramírez et al., 2004; Chen et al., 2019; Liu et al., 2019), the bst phenotype could be principally due to reduced levels of choline containing compounds as opposed to ethanolamine containing compounds. This could explain the rescue with lower concentrations of choline containing compounds than ethanolamine containing compounds. Presumably only some of the supplemental phosphoethanolamine would be converted to phosphocholine by PMTs, reducing the efficiency of rescue with ethanolamine containing compounds relative to choline containing compounds. This idea could be tested by supplementation with labeled compounds followed by a metabolic analysis. Combining such an analysis with chemical inhibition of PMTs (Liu et al., 2019) or by using bst pmt double mutants could further refine our understanding of the flux of metabolites in the Kennedy pathway on plant growth and development. When combined with mutations in other CEK genes, bst may also provide insights into the in planta functions of this important gene that are shared with the other CEKs.
The viability of the bst allele provides a useful tool for further investigation of CEK4 functions in development and responses to the environment. This study did not attempt to define the defects that underlie the developmental, reproductive, or chilling phenotypes we identified. Of special interest is the bumpy stem phenotype that this allele was named for. This phenotype is unusual and we are not aware of other Arabidopsis mutants that display this phenotype. Organ boundaries in plants are delineated by LOB gene expression (Shuai et al., 2002) and regulated by a complex genetic network that includes CUC and other genes (Hepworth and Pautot, 2015). Investigating the expression domains of LOB, CUC, and other boundary network genes in bst mutants could provide insights into the developmental signaling mechanisms that link phospholipid head group metabolism and the establishment of boundaries between organs.
Data Availability Statement
Strains described in this manuscript have been deposited at the ABRC with accession numbers CS73151 (bst^Col-0) and CS73152 (bst^Ler). The sequence data generated in the course of mapping and cloning bst have been deposited in the NCBI SRA as BioProject PRJNA796015.
Author Contributions
CR and MC conceived of and performed the experiments and edited the manuscript. NK conceived of and performed the experiments, supervised the work, wrote the original draft, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Research reported in this study was supported by the National Institute of General Medical Sciences of the National Institutes of Health award R15GM112173.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the staff of the Swarthmore Biology Department for their excellent support.
References
Aoyama, C., Liao, H., and Ishidate, K. (2004). Structure and function of choline kinase isoforms in mammalian cells. Prog. Lipid Res. 43, 266–281. doi: 10.1016/j.plipres.2003.12.001
Champeyroux, C., Stoof, C., and Rodriguez-Villalon, A. (2020). Signaling phospholipids in plant development: small couriers determining cell fate. Curr. Opin. Plant Biol. 57, 61–71. doi: 10.1016/J.PBI.2020.05.007
Chen, W., Salari, H., Taylor, M. C., Jost, R., Berkowitz, O., Barrow, R., et al. (2018). NMT1 and NMT3 N-Methyltransferase Activity Is Critical to Lipid Homeostasis, Morphogenesis, and Reproduction. Plant Physiol. 177, 1605–1628. doi: 10.1104/PP.18.00457
Chen, W., Taylor, M. C., Barrow, R. A., Croyal, M., and Masle, J. (2019). Loss of phosphoethanolamine n-methyltransferases abolishes phosphatidylcholine synthesis and is lethal. Plant Physiol. 179, 124–142. doi: 10.1104/pp.18.00694
Cruz-Ramírez, A., López-Bucio, J., Ramírez-Pimentel, G., Zurita-Silva, A., Sánchez-Calderon, L., Ramírez-Chávez, E., et al. (2004). The xipotl Mutant of Arabidopsis Reveals a Critical Role for Phospholipid Metabolism in Root System Development and Epidermal Cell Integrity. Plant Cell 16, 2020–2034. doi: 10.1105/TPC.103.018648
Gibellini, F., and Smith, T. K. (2010). The Kennedy pathway-De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62, 414–428. doi: 10.1002/iub.337
Gilding, E. K., and Marks, M. D. (2010). Analysis of purified glabra3-shapeshifter trichomes reveals a role for NOECK in regulating early trichome morphogenic events. Plant J. 64, 304–317. doi: 10.1111/J.1365-313X.2010.04329.X
Hepworth, S. R., and Pautot, V. A. (2015). Beyond the divide: boundaries for patterning and stem cell regulation in plants. Front. Plant Sci. 6:1052. doi: 10.3389/fpls.2015.01052
Honys, D., and Twell, D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5, 1–13. doi: 10.1186/GB-2004-5-11-R85
Hou, Q., Ufer, G., and Bartels, D. (2016). Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 39, 1029–1048. doi: 10.1111/PCE.12666
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313X.2007.03052.x
Kwon, Y., Yu, S., Lee, H., Yim, J. H., Zhu, J.-K. K., and Lee, B. H. (2012). Arabidopsis Serine Decarboxylase Mutants Implicate the Roles of Ethanolamine in Plant Growth and Development. Int. J. Mol. Sci. 13, 3176–3188. doi: 10.3390/ijms13033176
Lin, Y. C., Araguirang, G. E., Ngo, A. H., Lin, K. T., Angkawijaya, A. E., and Nakamura, Y. (2020). The four Arabidopsis choline/ethanolamine kinase isozymes play distinct roles in metabolism and development. Plant Physiol. 183, 152–166. doi: 10.1104/pp.19.01399
Lin, Y.-C., Kanehara, K., and Nakamura, Y. (2019). Arabidopsis CHOLINE/ETHANOLAMINE KINASE 1 (CEK1) is a primary choline kinase localized at the endoplasmic reticulum (ER) and involved in ER stress tolerance. New Phytol. 223, 1904–1917. doi: 10.1111/nph.15915
Lin, Y. C., Liu, Y. C., and Nakamura, Y. (2015). The choline/ethanolamine kinase family in arabidopsis: essential role of CEK4 in phospholipid biosynthesis and embryo development. Plant Cell 27, 1497–1511. doi: 10.1105/tpc.15.00207
Liu, Y. C., Lin, Y. C., Kanehara, K., and Nakamura, Y. (2018). A pair of phospho-base methyltransferases important for phosphatidylcholine biosynthesis in Arabidopsis. Plant J. 96, 1064–1075. doi: 10.1111/TPJ.14090
Liu, Y. C., Lin, Y. C., Kanehara, K., and Nakamura, Y. (2019). A methyltransferase trio essential for phosphatidylcholine biosynthesis and growth 1. Plant Physiol. 179, 433–445. doi: 10.1104/pp.18.01408
Malito, E., Sekulic, N., Too, W. C. S., Konrad, M., and Lavie, A. (2006). Elucidation of Human Choline Kinase Crystal Structures in Complex with the Products ADP or Phosphocholine. J. Mol. Biol. 364, 136–151. doi: 10.1016/J.JMB.2006.08.084
Marks, M. D., Wenger, J. P., Gilding, E., Jilk, R., and Dixon, R. A. (2009). Transcriptome Analysis of Arabidopsis Wild-Type and gl3–sst sim Trichomes Identifies Four Additional Genes Required for Trichome Development. Mol. Plant 2:803. doi: 10.1093/MP/SSP037
Meinke, D., Muralla, R., Sweeney, C., and Dickerman, A. (2008). Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci. 13, 483–491. doi: 10.1016/j.tplants.2008.06.003
Meinke, D. W. (2020). Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 226, 306–325. doi: 10.1111/nph.16071
Muller, H. J. (1932). Further studies on the nature and causes of gene mutations. Int. Congr. Genet. 61, 213–255.
Nakamura, Y., Andrés, F., Kanehara, K., Liu, Y. C., Dörmann, P., and Coupland, G. (2014). Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat. Commun. 5:3553. doi: 10.1038/ncomms4553
Ruelland, E., and Valentova, O. (2016). Editorial: lipid Signaling in Plant Development and Responses to Environmental Stresses. Front. Plant Sci. 7:324. doi: 10.3389/FPLS.2016.00324
Schneeberger, K., Ossowski, S., Lanz, C., Juul, T., Petersen, A. H., Nielsen, K. L., et al. (2009). SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6, 550–551. doi: 10.1038/nmeth0809-550
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Shuai, B., Reynaga-Pe— na, C. G., and Springer, P. S. (2002). The Lateral Organ Boundaries Gene Defines a Novel, Plant-Specific Gene Family. Plant Physiol. 129, 747–761. doi: 10.1104/PP.010926
Silverblatt-Buser, E. W., Frick, M. A., Rabeler, C., and Kaplinsky, N. J. (2018). Genetic interactions between BOB1 and multiple 26S proteasome subunits suggest a role for proteostasis in regulating Arabidopsis development. G3 Genes Genomes Genet. 8, 1379–1390. doi: 10.1534/g3.118.300496
Susila, H., Jurić, S., Liu, L., Gawarecka, K., Chung, K. S., Jin, S., et al. (2021). Florigen sequestration in cellular membranes modulates temperature-responsive flowering. Science 373, 1137–1142. doi: 10.1126/SCIENCE.ABH4054
Tannert, M., Balcke, G. U., Tissier, A., and Köck, M. (2021). At4g29530 is a phosphoethanolamine phosphatase homologous to PECP1 with a role in flowering time regulation. Plant J. 107, 1072–1083. doi: 10.1111/TPJ.15367
Tasseva, G., Richard, L., and Zachowski, A. (2004). Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 566, 115–120. doi: 10.1016/j.febslet.2004.04.015
Tavasoli, M., Lahire, S., Reid, T., Brodovsky, M., and McMaster, C. R. (2020). Genetic diseases of the Kennedy pathways for membrane synthesis. J. Biol. Chem. 295, 17877–17886. doi: 10.1074/JBC.REV120.013529
Waese, J., Fan, J., Pasha, A., Yu, H., Fucile, G., Shi, R., et al. (2017). ePlant: visualizing and Exploring Multiple Levels of Data for Hypothesis Generation in Plant Biology. Plant Cell 29, 1806–1821. doi: 10.1105/TPC.17.00073
Wang, X., and Chapman, K. D. (2013). Lipid signaling in plants. Front. Plant Sci. 4:216. doi: 10.3389/FPLS.2013.00216
Welti, R., Li, W., Li, M., Sang, Y., Biesiada, H., Zhou, H. E., et al. (2002). Profiling membrane lipids in plant stress responses: role of phospholipase Dα in freezing-induced lipid changes in arabidopsis. J. Biol. Chem. 277, 31994–32002. doi: 10.1074/jbc.M205375200
Wharfe, J., and Harwood, J. L. (1979). Lipid metabolism in germinating seeds: purification of ethanolamine kinase from soya bean. Biochim. Biophys. Acta Lipids Lipid Metab. 575, 102–111. doi: 10.1016/0005-2760(79)90135-8
Keywords: choline/ethanolamine kinase, CEK4, At2g26830, chilling, pedicel stem junction, phospholipid biosynthesis
Citation: Rabeler C, Chen M and Kaplinsky N (2022) BUMPY STEM Is an Arabidopsis Choline/Ethanolamine Kinase Required for Normal Development and Chilling Responses. Front. Plant Sci. 13:851960. doi: 10.3389/fpls.2022.851960
Received: 10 January 2022; Accepted: 17 March 2022;
Published: 29 April 2022.
Edited by:
Patricia Springer, University of California, Riverside, United StatesReviewed by:
Ying-Chen Lin, University of Oxford, United KingdomHai Anh Ngo, Institute of Plant and Microbial Biology, Academia Sinica, Taiwan
Copyright © 2022 Rabeler, Chen and Kaplinsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nick Kaplinsky, bmthcGxpbjFAc3dhcnRobW9yZS5lZHU=
 Christina Rabeler
Christina Rabeler Mingjie Chen2
Mingjie Chen2 Nick Kaplinsky
Nick Kaplinsky