
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 May 2022
Sec. Aquatic Photosynthetic Organisms
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.851151
Hybridization has fascinated biologists in recent centuries for its evolutionary importance, especially in plants. Hybrid zones are commonly located in regions across environmental gradients due to more opportunities to contact and ecological heterogeneity. For aquatic taxa, intrazonal character makes broad overlapping regions in intermediate environments between related species. However, we have limited information on the hybridization pattern of aquatic taxa in alpines, especially submerged macrophytes. In this study, we aimed to test the hypotheses that niche overlap and hybridization might be extensive in related aquatic plants across an altitudinal gradient. We evaluated the niche overlap in three related species pairs on the Qinghai-Tibetan Plateau and assessed the spatial pattern of hybrid populations. Obvious niche overlap and common hybridization were revealed in all three pairs of related aquatic plants. The plateau edge and river basins were broad areas for the sympatry of divergent taxa, where a large proportion of hybrid populations occurred. Hybrids are also discretely distributed in diverse habitats on the plateau. Differences in the extent of niche overlap, genetic incompatibility and phylogeographic history might lead to variation differences in hybridization patterns among the three species pairs. Our results suggested that plateau areas are a hotspot for ecologically divergent aquatic species to contact and mate and implied that hybridization may be important for the freshwater biodiversity of highlands.
Hybridization between related species or genetic lineages is a common phenomenon in nature. Generally, allopatric or ecologically isolated taxa with weak reproductive barrier contact and mate in limited regions of overlap, forming hybrid zones (Barton and Hewitt, 1985; Seehausen, 2004). Ecologists have regarded hybrid zones as natural laboratories in which to study evolutionary processes because hybridization provides a great contribution to the accumulation of genetic variation, the generation of novelty, and speciation, especially in plants (Barton and Hewitt, 1985; Arnold, 1997; Taylor and Larson, 2019). Swarms in hybrid zones could also be useful resources to explore adaptive traits from the perspective of genetic conservation and landscape management (Hamilton and Miller, 2016; Matthews et al., 2020).
For related species with ecological divergence, parents are typically located at opposite extremes, whereas their ranges can overlap in intermediate environments (Barton, 2001). As reviewed in Abbott (2017), a large proportion of hybrid zones occur across environmental gradients, which provide a favorable niche for both parental species. The structure and types of hybrid zones in these areas vary, depending on the fitness of hybrids to their parents in heterogeneous niches (Barton and Hewitt, 1985; Harrison, 1986; Abbott, 2017). Hence, various habitats may facilitate the contact of related species and the formation of hybrid populations (Yakimowski and Rieseberg, 2014). Exploring natural hybrid zones and studying their patterns in regions across environmental gradients are also hotspots in the research field of biodiversity.
Highlands are particularly good systems for examining interactions and interspecies gene flow between related species because steep changes in ecological variables facilitate the coexistence of taxa with different distribution characteristics (Abbott and Brennan, 2014). They may also serve as marginal or new areas for parental plants, and hybrids may have more opportunities to persist when benefiting from adaptation to unique environments and limited competition (Arnold, 1997; Barton, 2001; Arnold and Martin, 2010). According to relatively few case studies, hybrid zones across altitudinal gradients encompass all types of hybrid zone models and have the potential to contain high levels of phenotypic, genetic, and adaptative variation (Arnold, 1997; Abbott and Brennan, 2014; Wu et al., 2022).
In aquatic taxa, hybridization is also an important mechanism for the maintenance and amplification of variation (Barrett et al., 1993; Les and Philbrick, 1993). Many widespread aquatic macrophytes have closely related species with boreal distributions, and hybrids could be found in areas of their northern ranges (Santamaría, 2002). Diverse aquatic plants also occur in alpine regions (Wang, 2003). Considering the intrazonal character of aquatic flora, plateaus and their adjacent areas are expected to provide potential regions for the overlaps of ecologically divergent species as well, which may subsequently influence aquatic biodiversity regionally. However, we know little about the niche overlap of related aquatic plants in alpine ecosystems and its consequences on the spatial pattern of genetic variation. Here, we aimed to validate the hypotheses that (1) the niche overlap might be extensive in related aquatic plants in highland regions and (2) hybridization events were frequent, and the distribution of hybrid populations was related to the spatial pattern of ecologically divergent plants.
Our research was performed on the Qinghai-Tibetan Plateau (QTP). The QTP is the highest plateau in the world and an ideal region for research on hybridization because it provides a complex landscape and novel environments in which plants can persist, which is driven by elevational gradients (Shen et al., 2014; Sun H. et al., 2014). As the origin of many great rivers in Asia, the QTP is full of wetlands, which provide abundant habitats for aquatic life (Wang and Dou, 1998). To test the hypotheses, we chose three typical species pairs of submerged plants with ecological divergence. Hybridization between all species pairs has been reported previously (Hollingsworth et al., 1996; Moody and Les, 2002; Wiegleb et al., 2017). Based on previous studies and our initial work, they cooccurred on the QTP with different distribution patterns (Wang, 2003; Wu et al., 2015; Du and Wang, 2016). In the present study, we conducted an overall survey on the distribution of the three species pairs. We quantified the ecological niche and potential distribution of each species and measured the overlap between related plants. Molecular markers were used to complement the identification of their hybrid. We also conducted comparisons of the niches between hybrid populations and their parents and discussed the differences in hybridization patterns among the three species pairs.
Myriophyllum spicatum, also called Eurasian watermilfoil, is one of most important alien weeds in the world. The species is native to Eurasia and northern Africa and invaded North America in the 19th century (Aiken, 1981; Couch and Nelson, 1985). Now, it occurs on all continents except Antarctica. It is distributed in cold regions north to 60°N. Its sister species Myriophyllum sibiricum is native to Eurasia and North America and presents a circumboreal distribution, rarely extending south of the mean isotherm of 0°C in winter (Aiken, 1981; Ceska and Ceska, 1986; Moody and Les, 2002). Hybridization between the species has been reported in North America (Moody and Les, 2002, 2007) and East Asia (Wu et al., 2015). Morphologically, the two species can be distinguished by leaf segment numbers and lengths.
Stuckenia pectinata is a perennial submerged aquatic macrophyte with a cosmopolitan distribution that occurs on all continents except Antarctica and does not reside in polar regions (Kaplan, 2008). It occurs in a wide range of habitats, most often in lentic wetlands but also in running water. It tolerates brackish water (up to 20‰ salinity) and may grow in arid zones, on high mountains or at seasides (Nies and Reusch, 2005; Kaplan, 2008; Li et al., 2015). Stuckenia filiformis is widespread in the Northern Hemisphere and common in Siberia and the mountain regions of Central Asia (Kaplan, 2008). Stuckenia filiformis tends to grow in cold and shallow water and is also able to tolerate saline habitats (Kaplan, 2008). Both Stuckenia species are extremely variable in morphology and distinguishable by the structure of leaf sheaths, branching pattern, and fruit size (Hollingsworth et al., 1996). Hybrids between the two species have been identified in the northern half of Europe, Siberia, and China (Hollingsworth et al., 1996; Kaplan, 2008; Du and Wang, 2016) and named S. x suecicus.
Ranunculus trichophyllus is a perennial aquatic plant with a subcosmopolitan distribution. The species is morphologically and genetically polymorphic. The plant grows in various habitats, particularly in alkaline waters. The morphological overlap and hybridization of R. trichophyllus with many other R. section Batrachium species has been reported (Zalewska-Gałosz et al., 2015; Wiegleb et al., 2017). Ranunculus subrigidus is an Amphipacific species with an arid to subarctic distribution and tends to colonize inland habitats with hard or even brackish water. It is separated from R. trichophyllus by rigid leaves, length of adjacent internodes and flower size (Wiegleb et al., 2017). Hybridization between the two species occurred but has not been well-studied.
Field surveys on the QTP were performed from 2010 to 2015, and in total, 254 sites were examined (Supplementary Figure 1). Based on morphological identification, M. spicatum and M. sibiricum were found at 46 and 28 sites, S. pectinata and S. filiformis were found at 40 and 92 sites, and R. trichophyllus and R. subrigidus were found at 35 and 54 sites (Figure 1). In the summer of 2015, our field work was focused on mixed populations and plant material collection for molecular analysis. We did not detect new mixed Myriophyllum populations, and a total of 25 sites (Myriophyllum: 10 sites that have been included in our previous work, Wu et al., 2015; Stuckenia: 8 sites; Ranunculus: 7 sites) contained individuals of both parental species or individuals with morphological ambiguity (Figure 1 and Table 1). The plant materials were also sampled and preserved in silica gel for the molecular identification of hybrids.
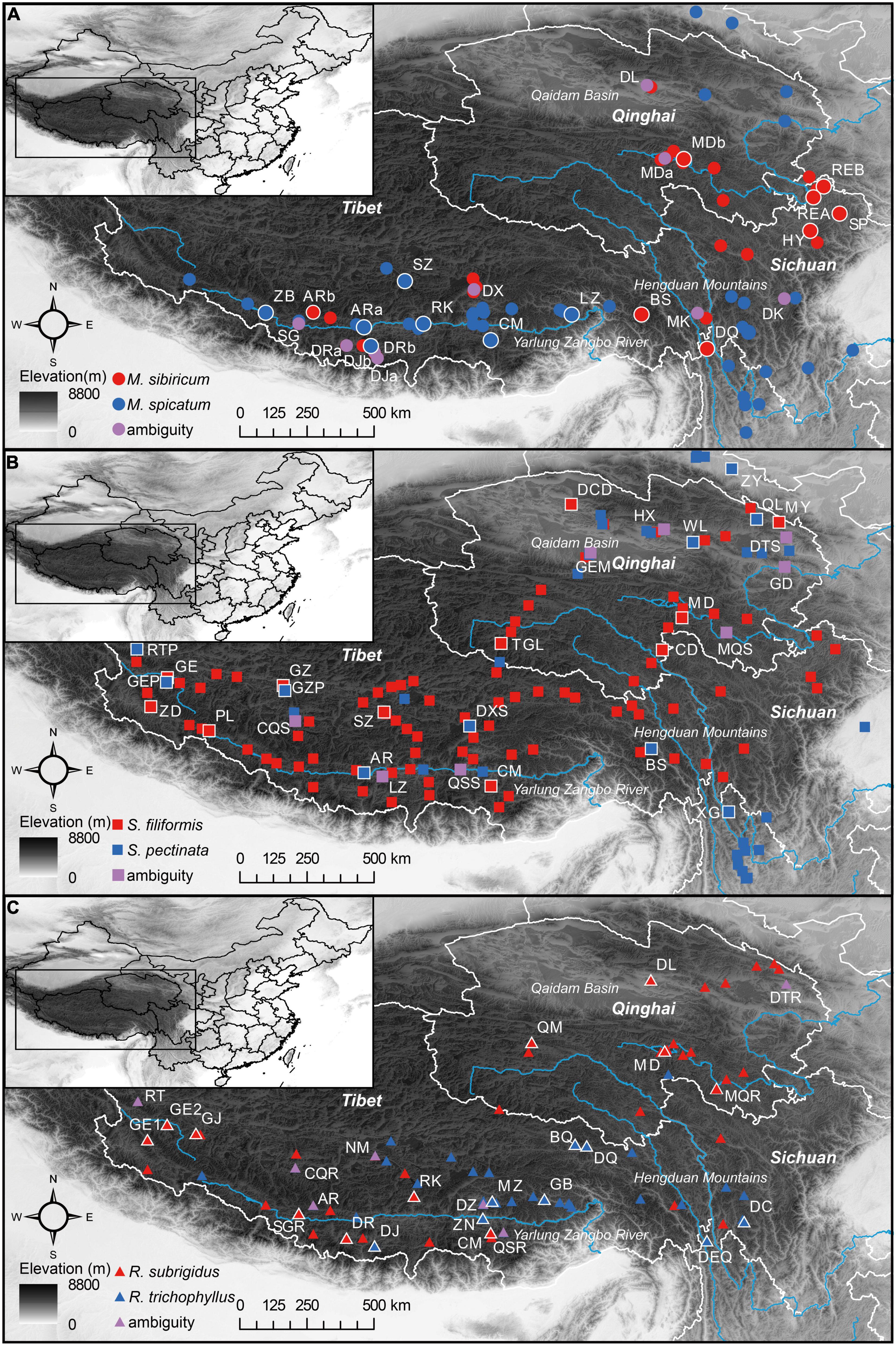
Figure 1. Geographic distribution of each parental species [(A) Myriophyllum, (B) Stuckenia, and (C) Ranunculus] and morphologically ambiguous populations on the Qinghai-Tibetan Plateau. The pure populations used in molecular identification are labeled and outlined in white.
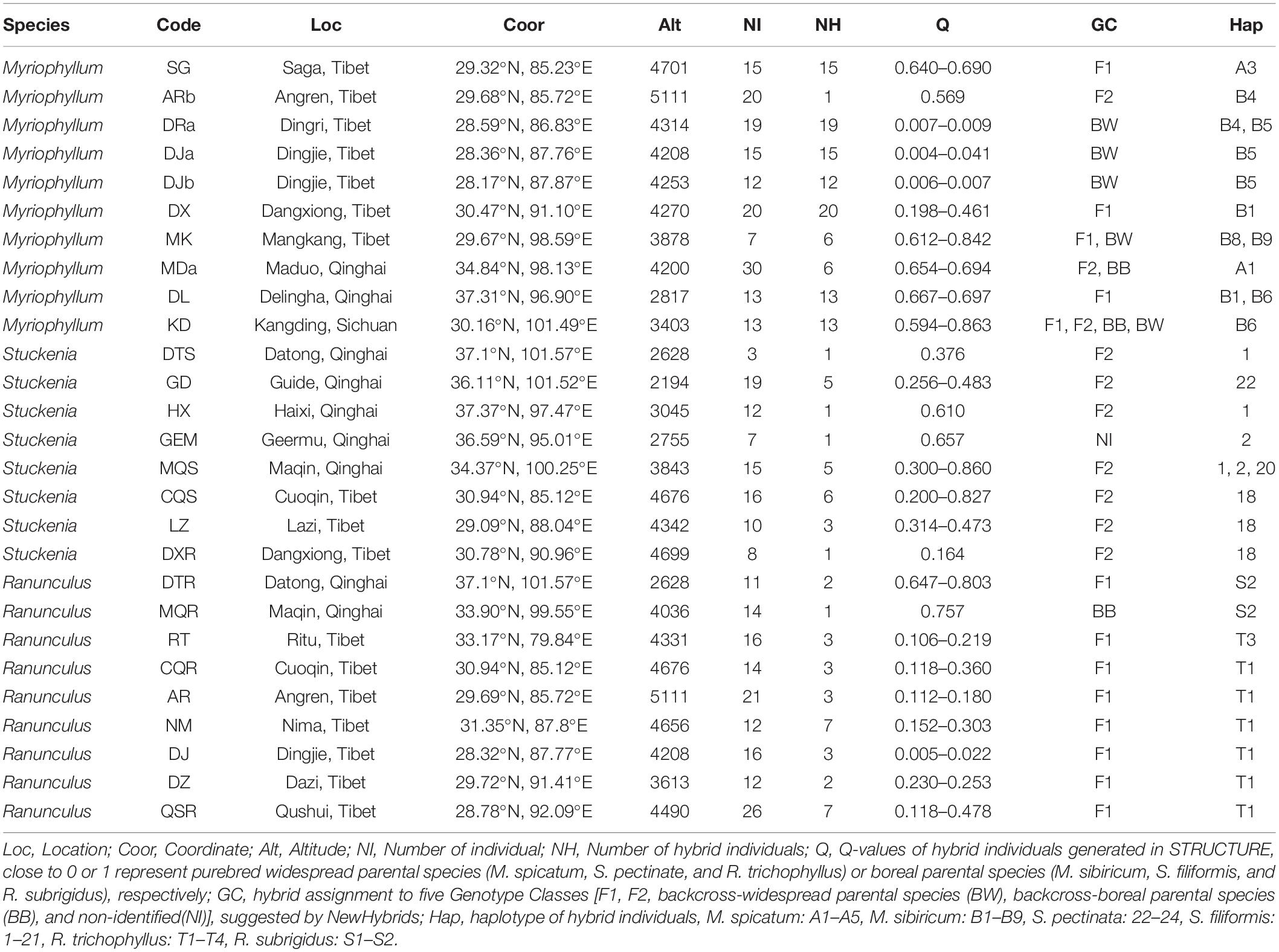
Table 1. Geographic locations, numbers of samples and hybrids, Q values of STRUCTURE and genotype class assignment of NewHybrids revealed by microsatellite markers, and cpDNA haplotypes for the populations involved in hybridization events.
The variables climate, soil, and landcover were used as environmental factors for the niche analysis (Table 2). Environmental data for the entire studied region (25–40°N, 75–105°E) were obtained from open sources with a resolution of 1 km (climate: www.worldclim.org; soil: Harmonized World Soil Database, v 1.2; landcover: www.resdc.cn; Table 2). Because autocorrelated variables may bias the niche comparison analyses (Broennimann et al., 2012), we kept one of any set of highly correlated variables (Pearson’s r > 0.75). Thirteen variables were selected as evaluator variables, including five bioclimatic variables (Bio1, Bio2, Bio3, Bio4, and Bio12), altitude, solar radiation, average wind speed in the growing season, three soil parameters (texture type, pH, and salinity), and landcover (Table 2).
The ecological niche of each plant was quantified through both a species distribution model (SDM) and an ordination approach. 1. The most widely used SDM algorithm, maximum entropy (Maxent, Phillips et al., 2006), was chosen to model the potential distribution of each species based on the locations of field-observed occurrence. Through initial runs, Maxent performed better on our dataset than other generally used algorithms, such as generalized additive models, boosted regression trees and random forest, based on the AUC (area under the receiver operator curve) and TSS (maximizing true positive rate/sensitivity and true negative rate/specificity) values. One-fifth of the presence was used as testing data for cross-validation, and the threshold of occurrence was generated based on the sum of the sensitivity and specificity. The algorithm either runs 1,000 iterations or ends at convergence (threshold = 0.00001). 2. An ordination approach, such as principal components analysis (PCA), was used to identify the niches of species observations, with the available ecological data of the entire study area as the background (Broennimann et al., 2012). The observed occurrences were converted into smooth densities and represented using the first two PC axes, which was named the “pca-env” technique by Broennimann et al. (2012). It was proven to perform well in the subsequent niche overlap analysis among different ordination and niche model techniques (Broennimann et al., 2012). When executing the analysis, we used the proportions of sand and clay instead of the categorical variable soil texture.
The niche overlap of each species pair was measured with D metrics (Warren et al., 2008; Broennimann et al., 2012). Statistical tests of niche equivalency were performed for the null hypothesis that the niches of the parental species were identical in the study area. The overlaps of the pseudoreplicate model were estimated by randomly reassigning the presence sites to a pair of species and then compared to the observed niche overlap (Warren et al., 2008). We also performed background/niche similarity tests for the null hypothesis that the empirical overlap between two species is greater than the simulated overlap between the niche of one species and the backgrounds of the other (Warren et al., 2008). Contributor variables of PCA were used for canonical discriminants analysis (CDA), to compare the niche requirements among parent species and their hybrids. Wilks’ test was employed to evaluate the goodness of fit statistic and measure the model performance.
All the above analyses were conducted in R version 3.6.2. The SDM-based analyses were performed using the packages “dismo” (Hijmans et al., 2017) and “ENMTools” (Warren et al., 2010). The Java program Maxent v 3.3.3k was also implemented in “dismo” (Hijmans et al., 2017). The PCA-based analyses were performed in the package “ecospat” (Di Cola et al., 2017). The “candisc” package was used for CDA analyses (Friendly and Fox, 2021).
Our previous work identified hybrids in mixed Myriophyllum populations using microsatellite and cpDNA markers (Wu et al., 2015). Molecular studies have also been suggested for the identification of Stuckenia and Ranunculus hybrids because they are morphologically variable and difficult to distinguish from the parental species (Kaplan, 2008; Zalewska-Gałosz et al., 2015). We conducted a molecular analysis of a total of 303 individuals from all 15 indeterminate populations for the two species pairs (Stuckenia: 91 individuals of 8 populations; Ranunculus: 112 individuals of 7 populations) (Figure 1 and Table 1). We also included parental populations of each species pair in the molecular analysis as reference data (Figure 1). A total of 21 pure Stuckenia populations (75 individuals of S. pectinata and 131 individuals of S. filiformis) and 20 pure Ranunculus populations (110 individuals of R. trichophyllus and 215 individuals of R. subrigidus) were involved.
Total genomic DNA was extracted using the DNAsecure Plant Kit (Tiangen Biotech, Beijing, China). Microsatellite markers (7 loci for Stuckenia, Nies and Reusch, 2004; Wu et al., 2020; 17 loci for Ranunculus, Wu et al., 2017; Wu et al., 2019) and one cpDNA fragment (rpl32-trnL) were chosen to identify kinships. The primers “rpL32-F” and “trnL(UAG)” (Shaw et al., 2007) were used to amplify and sequence the region. The PCR conditions were performed according to the respective protocols.
Microsatellite genotyping was performed using GeneMarker v1.5 (SoftGenetics, State College, PA, United States). A Bayesian clustering method approach was used for individual genetic assignment, implemented in STRUCTURE v2.3.4 (Pritchard et al., 2000; Falush et al., 2003). Ten independent runs were performed at K = 2 with a burn-in period of 500,000 iterations and 2,000,000 MCMC iterations under the admixture model. The average Q value of all STRUCTURE runs was used to indicate individuals as parental species or hybrids, using Q < 0.10 or Q > 0.90 for pure individuals and 0.10 < Q < 0.90 for hybrids (Burgarella et al., 2009). We also assessed the optimal number of clusters for further genetic divergence (Evanno et al., 2005), when K = 2 was suggested for all species pairs.
To mutually verify hybrid identification and evaluate hybrid class assignment, a model-based Bayesian approach was applied using NewHybrids to further assign all individuals to one of six possible classes through evaluation of the Bayesian posterior probability of membership: two parental species, F1s, F2s, and backcrosses to both parents (Anderson and Thompson, 2002). In NewHybrids program, pure samples of the two species need not be specified for hybrids identification, because the allele frequencies of the two species are not required as a priori. The runs were performed using Jeffreys’ prior distribution, with a burn-in of 200,000 iterations and a run length of 2,000,000 iterations. The threshold of assignment probability was set to 75%. When Myriophyllum populations were analyzed, the same dataset was used as that in Wu et al. (2015). All cpDNA fragment sequences were aligned using the program Mafft v6.7 (Katoh and Toh, 2008). The cpDNA haplotypes were extracted using DNASP v5.10 (Librado and Rozas, 2009). The published rpl32-trnL sequences for each genus (for Ranunculus, R. section Batrachium species were included) were used to confirm that no other closely related species involved. We implemented a median-joining network for the genealogical relationships among the genotypes and haplotypes in NETWORK v10.1 (Bandelt et al., 1999).
The potential distribution of each species was quantified using Maxent. The AUC of the training values ranged from 0.935 to 0.957 for the SDM of the six parental species (Supplementary Table 1), indicating that the models of all species performed well and generated excellent evaluations. The thresholds of presence/absence for each species ranged from 0.211 to 0.276, as suggested by the TSS (Supplementary Table 1). We defined and represented the species potential habitats as “high potential” (>0.6), “moderate potential” (0.4–0.6), “low potential” (threshold-0.4) and “no potential” (<threshold), based on the probability of presence suggested by the models (Supplementary Figure 2).
Temperature, precipitation, and elevation were the most decisive environmental factors for the distribution of M. spicatum on the QTP (Table 2). Its highly suitable region on the QTP was found to include the edge of the plateau and the river valley areas (Supplementary Figure 2A). The distribution of M. sibiricum can be mainly attributed to the conditions of temperature and soil texture. The northern slope of the Himalayas and border areas among Qinghai, Sichuan, and Tibet were the main regions that were suitable for M. sibiricum (Supplementary Figure 2B). Elevation, temperature isothermality, and soil pH were the environmental variables that constrained the distribution of S. pectinata (Table 2). Its suitable areas were mainly at the northeast and southeast edges of the QTP (Supplementary Figure 2C). Elevation, annual temperature, and precipitation mainly influenced the distribution of S. filiformis (Table 2). The species had broad potential distribution areas over the plateau (Supplementary Figure 2D). Elevation, temperature isothermality, and soil texture were the main factors influencing the distribution of R. trichophyllus (Table 2), which was most likely to occur in central Tibet and western Sichuan (Supplementary Figure 2E). Annual temperature, landcover, and solar radiation greatly affected the distribution of S. subrigidus (Table 2). It was the second most common submerged plant on the QTP and had extensive suitable distribution areas (Supplementary Figure 2F).
By multiplying the presence distributions (>threshold) of related species, we obtained the potential areas for the coexistence of each species pair (Figure 2). For Myriophyllum, the two species could be separated by the conditions of temperature, seasonality, elevation, and solar radiation (Supplementary Figure 3). The potential sympatric areas for the two species were river valleys in southern Tibet and western Sichuan (Figure 2A). For Stuckenia, the temperature diurnal range and seasonality, elevation, wind speed, and soil texture were decisive for the coexistence of the two species (Supplementary Figure 4). Their potential sympatric regions were medium-elevation areas in north Qinghai and mosaic areas along the Yarlung Tsangpo River (Figure 2B). The annual temperature and elevation mainly separated the two Ranunculus species on the QTP (Supplementary Figure 5). They might cooccur in central and southern Tibet (Figure 2C).
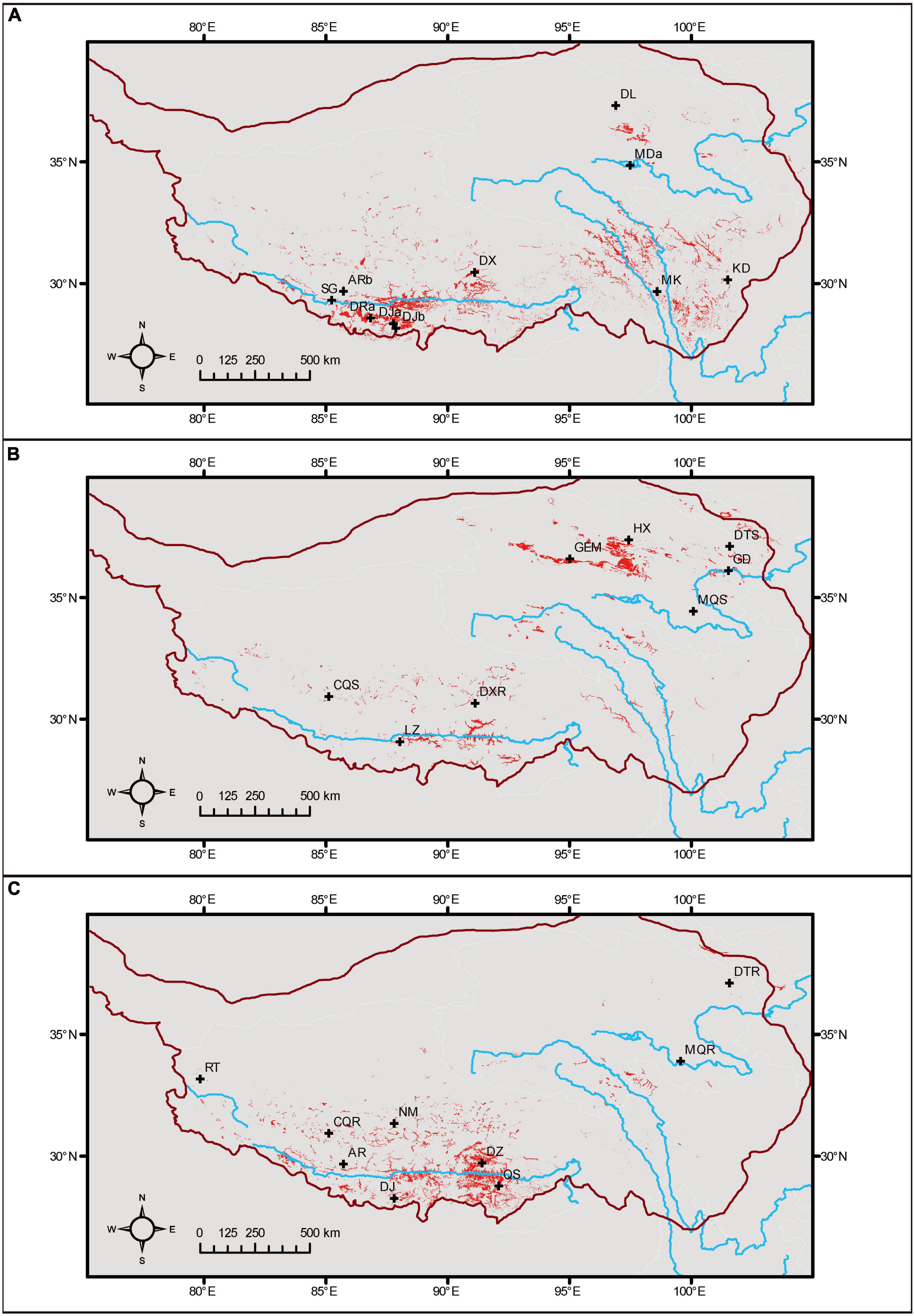
Figure 2. Potential coexistence regions for related species, based on the presence in species distribution models. (A) Myriophyllum, (B) Stuckenia, and (C) Ranunculus. Sites of hybrid populations were plotted, and the boundary of the QTP is shown.
Obvious niche overlap was suggested in all species pairs (Myriophyllum D = 0.594; Stuckenia D = 0.424; Ranunculus D = 0.545, Table 3). The null hypothesis of niche equivalency was rejected in all species pairs, indicating that the niches of ecologically isolated parental species did not shift to equivalence on the QTP (Table 3 and Supplementary Figure 6). The results of the background test in Myriophyllum and Ranunculus were significant, indicating that the niche overlap between the two parental species pairs was more obvious than that between either parent and random background points (Table 3 and Supplementary Figure 7).
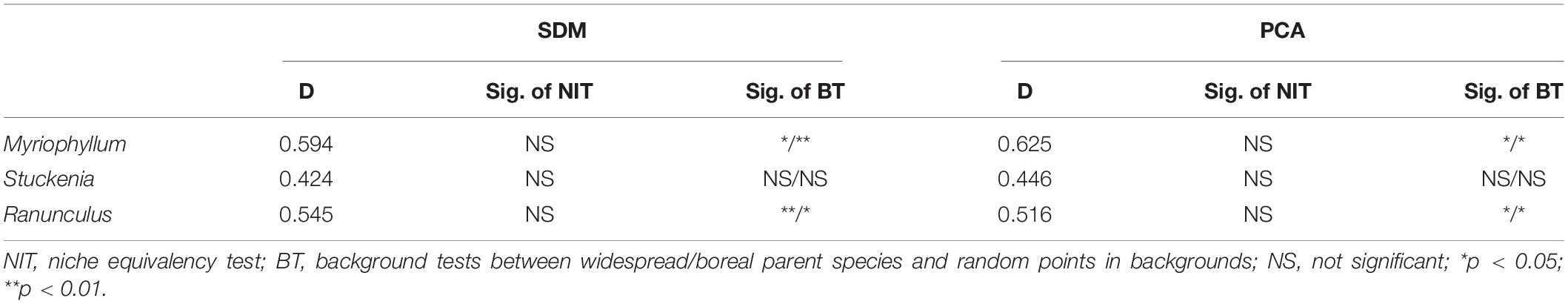
Table 3. Niche overlap index and significance in tests of niche equivalency and backgrounds for related species pairs.
The ecological niche was also measured by an ordination approach. The first two axes in PCA explained 76.61% of the variation in environmental factors (Supplementary Figure 8). Axis 1 was associated with the temperature variables, precipitation, soil texture, and wind speed, while axis 2 was mainly associated with the elevation and solar radiation (Supplementary Figure 8).
Obvious niche overlap was also identified when quantified using the ordination approach (Figure 3). Limited unique niches for each parental species and few changes in niche centers were found for Myriophyllum and Ranunculus (Figures 3A,C). The niche overlap between S. pectinata and S. filiformis was obvious as well, while the high-occurrence density areas of the two species were different (Figure 3B). The values of niche overlap between each species pair were 0.625 (Myriophyllum), 0.446 (Stuckenia), and 0.516 (Ranunculus) (Table 3 and Figure 3).
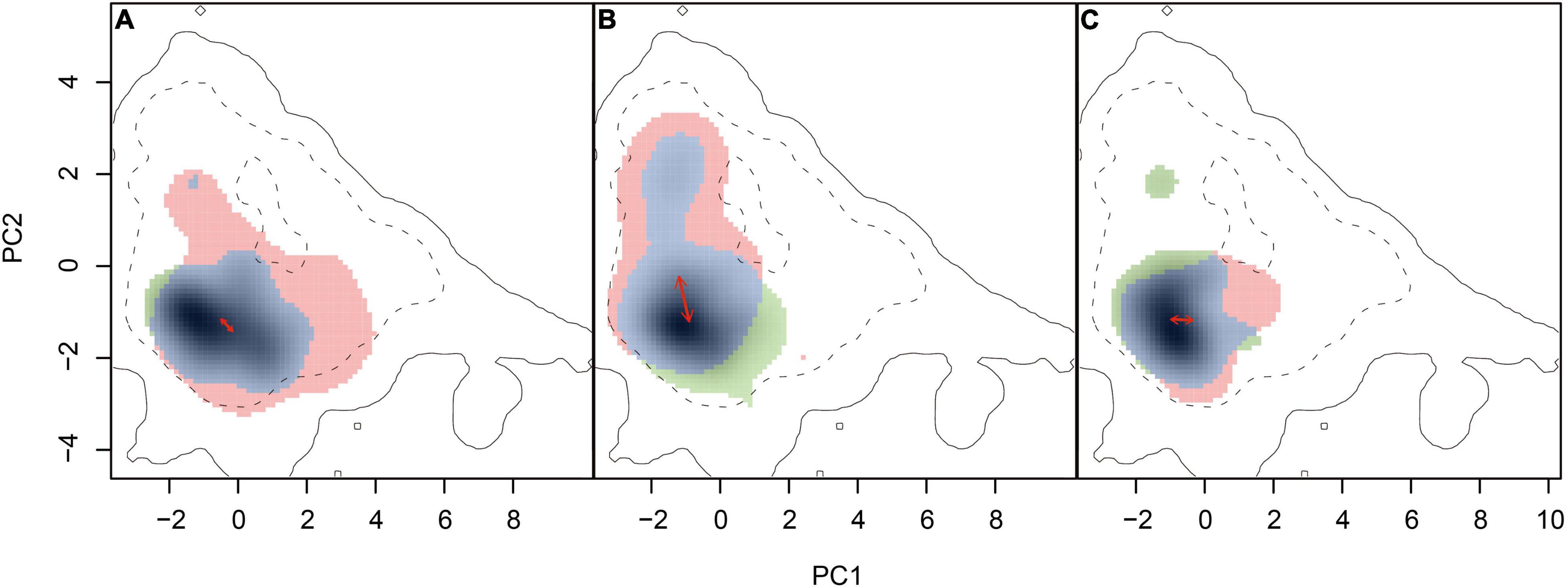
Figure 3. Niche overlap between related species. (A) Myriophyllum, (B) Stuckenia, and (C) Ranunculus. The solid and dashed contour lines indicate 100 and 50% of the available environmental data for the whole study region as backgrounds. The unique niches of widespread species (red: M. spicatum, S. pectinate, and R. trichophyllus), the unique niches of boreal species (green: M. sibiricum, S. filiformis, and R. subrigidus), and the overlap (blue) were represented, respectively. The solid arrows represent the differences in the niche density center between parental species.
Similar to the results based on SDM, no niches of species pairs were identical according to the niche equivalency tests (Table 3 and Supplementary Figure 9). The background tests showed that the niche similarity between Myriophyllum species and between Ranunculus species was higher than expected at random on the QTP (Table 3 and Supplementary Figure 10). Consistent with the results of background tests, CDA analyses yielded one discriminant function that explained 100% of the variance in PC contributors, showing ecological niche differences were significant between parent Stuckenia (Wilks’ λ = 0.669, p < 0.001), but not significant between parental Myriophyllum species (Wilks’ λ = 0.705, p = 0.054) or Ranunculus species (Wilks’ λ = 0.774, p = 0.053) (Supplementary Figure 11). While moderate Wilks’ λ values (close to 1) suggested all species pairs occupied some regions that were ecologically similar.
Totals of 82 and 126 multilocus genotypes were revealed in the ambiguous populations of Stuckenia and Ranunculus using microsatellite markers. Under the assumption that K = 2, the two genetic clusters suggested by STRUCTURE analysis corresponded to the parental species (Supplementary Figure 12). All the individuals from parental populations had high posterior probabilities close to 0 and 1, except for DXS of S. pectinata and MQR of R. subrigidus (Supplementary Figure 12). Of the eight ambiguous Stuckenia populations, most contained individuals who were genetically admixed, with a probability range from 0.200 to 0.860 (Table 1, Figure 1B, and Supplementary Figure 12A), while QS was composed of two pure parental species. Of the seven ambiguous Ranunculus populations, all contained hybrids, with a probability range from 0.106 to 0.803 (Table 1, Figure 1C, and Supplementary Figure 12B). Two populations (DXS and MQR) assigned to S. pectinata and R. subrigidus by morphology included hybrid individuals (Table 1, Figure 1C, and Supplementary Figure 12B). The PL of S. filiformis contained one individual of S. pectinata.
The hybrid individuals suggested by NewHybrids were consistent with the results of STRUCTURE with few exceptions (Table 1 and Figure 4). All genotype classes were found in the Myriophyllum hybrids. In MDa, more hybrids were suggested compared to the STRUCTURE results (Table 1 and Figure 4A). Stuckenia hybrids were rarely F1s. The hybrid individuals in GEM suggested by STRUCTURE were assigned to S. filiformis (Table 1 and Figure 4B). We assessed the STRUCTURE results under K = 3 in Stuckenia, which did not suggest an assignment of hybrids to independent cluster, but a split of S. filiformis consistent to genetic structure in Wu et al. (2020). For Ranunculus, most hybrids were F1s, while the hybrid individual in MQR was assigned to backcross-R. subrigidus. Additionally, three individuals with DJ were suggested to be F1s (Table 1 and Figure 4C).
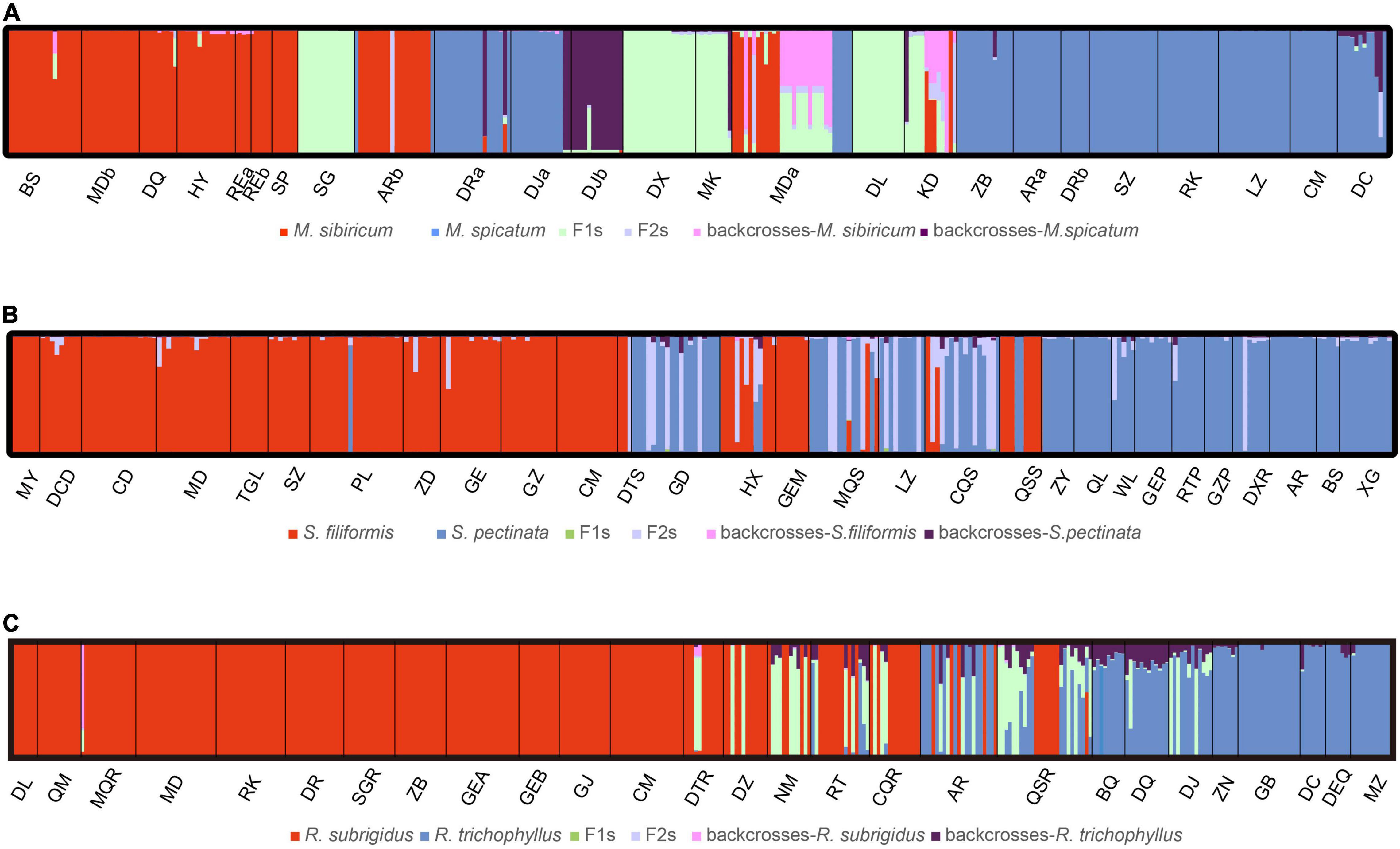
Figure 4. The results of the NewHybrids analysis for all species pairs based on microsatellite data. (A) Myriophyllum, (B) Stuckenia, and (C) Ranunculus. The vertical bars display the probability of assignment to six membership classes: two parents, F1s, F2s, and backcrosses to each parent.
One cpDNA fragment (rpl32-trnL) was sequenced for kinship identification. The lengths of the rpl32-trnL alignment region were 731 bp for Stuckenia and 887 bp for Ranunculus. For Stuckenia, 26 polymorphic sites were found between the parental species (Supplementary Figure 13). Twenty-one haplotypes (H1–H21) were found in S. filiformis populations, and three haplotypes (H22–H24) corresponded to S. pectinata (Supplementary Figure 13A). Among the hybrids suggested by SSR data, only individuals from GD had the haplotype of S. pectinata, whereas cpDNA haplotypes of other hybrids were assigned to S. filiformis (Table 1). For Ranunculus, 21 polymorphic sites were found between the parental species (Supplementary Figure 13B). Two and four haplotypes were found in R. subrigidus (S1–S2) and R. trichophyllus (T1–T4), respectively (Supplementary Figure 13B). The hybrids identified by microsatellite loci from DTR and MQR had haplotypes of R. subrigidus, while other hybrids contained the cpDNA of R. trichophyllus (Table 1).
Overall, combining the microsatellite and cpDNA data revealed the distributions of hybrids. Twenty-three individuals from eight sites were identified as hybrids of Stuckenia (Table 1). The hybrids were suggested to be post-F1s, and S. filiformis tended to be the maternal species (Table 1). Half of the mixed populations contained moderate proportions of hybrid individuals (Table 1). Thirty-one hybrids from nine sites were found in Ranunculus, and they were likewise not dominant in mixed populations except NM (Table 1). Most hybrid individuals of Ranunculus were F1s, while R. trichophyllus was more frequent, and reciprocal hybridization occurred in Qinghai, where R. trichophyllus was rarely distributed (Table 1 and Figure 1).
We assessed the distribution suitability of parents at the locations of their hybrids based on SDM results. We found that most sites of hybrids were suitable for at least one parental species, except that DL of Myriophyllum, HX and MQS of Stuckenia, NM and RT of Ranunculus were rarely suitable for either parent (Supplementary Table 2). According to the PC scores of the hybrid populations in “pca-env” analysis, many were situated in the overlapping niches of parents, while the DL of Myriophyllum, HX, GEM and MQS of Stuckenia, DTR, MQR, RT, and NM of Ranunculus were distributed in the marginal niches of both parents (Supplementary Figure 14 and Supplementary Table 2).
In the present study, we found extensive niche overlap in related aquatic taxa on highlands, according to the tests of niche similarity based on different approaches. This result suggested that highlands and adjacent regions provided suitable niches for contact with related aquatic species. The results showed expansive potential coexistence regions between all three species pairs on the QTP. We subsequently performed molecular analysis for the ambiguous populations and found that hybridization events were frequent in all species pairs. Although infrequent hybridization might be possible (Chung et al., 2005), broad overlap in geographic distribution should be optimal for the formation of hybrid zones or hybrid swarms (Yakimowski and Rieseberg, 2014). Therefore, extensive overlap of ecologically divergent aquatic plants across altitude gradients might greatly contribute to their frequent hybridization. Hybridization plays an important role in the spectacular radiation and diversification of plants on the QTP and its adjacent regions (Wen et al., 2014; Yang et al., 2019), which contributes to a high diversity of plant species, including a large number of endemics, within the region, especially the southeast edge (Wu, 1988; Xing and Ree, 2017; Wu et al., 2022). High aquatic plant diversity was also observed on the QTP, especially the southern and eastern QTP, and the species of interest were the most common water plants there (Wang, 2003). Based on the results of this work, we strongly advocate for further investigation on whether hybridization events are prevalent and important in the diversification of other taxa, particularly species in floating or emerged life form.
The eastern edge of the plateau and the Yarlung Zangbo River basin were the main overlapping areas for related aquatic species, which were characterized by great environmental gradients along altitude. The areas also accommodated a large proportion of aquatic hybrid populations. Compared to the plateau edge, the relatively warm areas of river drainage basins that provided broad wetland habitats were more important for the sympatry of related aquatic taxa (Wang, 2003; Abbott and Brennan, 2014). We found that the overlapping regions and hybrid populations were not restricted at middle altitudes but occupied a large altitude range, probably because of the intrazonal distribution of parental species (Santamaría, 2002). This might be decisive for the high niche overlap of related aquatic plants in highland areas. In these areas, hybrids were mostly advanced offspring, and hybrid superiority was widely detected in Myriophyllum, suggesting the potential to generate genetic novelty through introgression (Abbott and Brennan, 2014).
Hybridization in land plants on the QTP has been widely reported (Sun Y. et al., 2014; Jiang et al., 2016; Li et al., 2020; Wu et al., 2022) and is more likely to follow the scenario of allopatric divergence in alpine and second contact via postglacial expansion (Wen et al., 2014; Li et al., 2020). In contrast to the situation in land species, the interaction between related aquatic plants was not constrained to a few narrow joint zones but was found in several discrete overlapping regions. Western and northern Tibet, the Qajdam Basin, and some other valleys at high altitudes (e.g., the Hengduan Mountains), which were away from the main contiguous transition areas, provided additional junctures that connected feasible habitats for parental plants. These areas were patchily distributed and ecologically diverse and characterized by more rigorous conditions of temperature, aridity, salinity, and isolation (Shen et al., 2014). In these areas, a certain number of genetically mixed individuals and various hybridization patterns were detected in all species pairs. This implied that local contact and independent interaction between pure species might facilitate genetic adaptation across heterogeneous habitats (Yakimowski and Rieseberg, 2014; Harrison and Larson, 2016), especially under bounded or novel environments (Burke and Arnold, 2001; Gompert et al., 2017; Pierce et al., 2017).
We suggested the following factors that might affect the hybridization pattern by comparing the three parental species pairs. (1) The extent of niche overlap. Relatively higher niche overlap and hybridization frequencies were found in Myriophyllum and Ranunculus species. More opportunities for sympatry would create diverse genotypes to maintain environmental selection and in competition with the parents (Ortego et al., 2014). (2) The genetic incompatibility. Genetic novelties in post-F1s and further backcross hybrids might facilitate adaptative introgression into highlands in Myriophyllum (Burke and Arnold, 2001; Wu et al., 2015), while the genetic incompatibility between parental Ranunculus and Stuckenia species was more severe; (3) Phylogeographic history. Alpine endemic genetic clades of S. filiformis and Ranunculus species were dated to occur in the Pliocene and early Pleistocene (Chen J.M. et al., 2014; Du and Wang, 2016). However, the split between M. sibiricum and M. spicatum did not occur earlier than the Quaternary (Chen L. et al., 2014), and no specific genetic clade was identified on the plateau (Wu et al., 2016). This implied that Myriophyllum colonized the plateau recently and probably benefited from adaptation and founder effects in new habitats.
Hybridization in all studied species pairs was not reported until recently (Kaplan, 2008; Wiegleb et al., 2017), but few examples of Stuckenia and Ranunculus have been deeply investigated (Hollingsworth et al., 1996; Zalewska-Gałosz et al., 2015). Hybrids between S. pectinata and S. filiformis were described as S. x suecicus in North Europe (Hollingsworth et al., 1996), but investigation was limited in the diversity center of the genus, Center Asia and adjacent regions (Kaplan, 2008, but see Du and Wang, 2016). We found a scattered distribution of S. pectinata in the hinterland of the QTP, and hybridization between the two species was not rare in this region. Independent and frequent hybridization events and high variation in hybrids were suggested by both previous studies and our results (Kaplan, 2008; Du and Wang, 2016). Hybrids between S. pectinata and S. filiformis were predicted to be sterile (Hollingsworth et al., 1996), whereas we considered that the possibility of sexual events in hybrids or ancient intercrosses could not be rejected (Taylor and Larson, 2019), because many hybrids were suggested to be post F1s, according to the results of molecular methods. Extensive hybridization between R. trichophyllus and R. subrigidus was first revealed in an alpine ecosystem (Wiegleb et al., 2017). The sterility of their hybrids was suggested, which might be attributed to the different ploidy pattern of two species (Wiegleb et al., 2017). The present study supported the importance of hybridization in the speciation of the genus Stuckenia and Ranunculus section Batrachium (Kaplan, 2008; Zalewska-Gałosz et al., 2015; Wiegleb et al., 2017).
In this study, we found obvious niche overlap between related aquatic species in the alpine environment, which was surmised to be an important cause of their hybridization. Hybrid populations are mainly located at the broad transition of parental species. The relatively warm drainage basins at high altitude and plateau edges were the high-potential regions for the sympatry and hybridization of closely related aquatic plants. These regions also accommodate high diversities of other aquatic taxa, and the influence of hybridization on diversification still needs additional empirical studies. The unique environment on the plateau also contributes to the hybridization of aquatic plants in discrete and novel habitats. We suggested that genetic compatibility, the extent of niche similarity and phylogeographic processes could influence hybridization patterns through comparisons among the three species pairs. Our findings highlighted that altitude gradients provided a suitable niche for extensive sympatry and the prevalence of hybridization in related aquatic species on the QTP. This result also implied the evolutionary importance of hybridization to the maintenance of aquatic plants in alpine environments. Genome-wide molecular markers are needed to finely evaluate the introgression patterns and evolutionary progress of aquatic taxa and to detect the corresponding adaptation mechanisms in future studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://datadryad.org/stash, doi: 10.5061/dryad.gxd2547md, https://www.ncbi.nlm.nih.gov/genbank/, MT262969–MT262989, and MZ456343–MZ456351.
ZWu, XX, TL, and JiZ designed the study. ZWu, ZWa, JuZ, PC, and XL performed the field work and experiments. ZWu, ZWa, and DX analyzed the data. ZWu, XX, and TL wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31000000) and the National Natural Science Foundation of China (31700190).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SF declared a shared affiliation with several of the authors ZWa, JuZ, PC, XL, and XX to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank to the members of Dan Yu’s lab for assistance on field work and the data resource from “National Earth System Science Data Center, National Science and Technology Infrastructure of China” (http://www.geodata.cn).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.851151/full#supplementary-material
Abbott, R. J. (2017). Plant speciation across environmental gradients and the occurrence and nature of hybrid zones. J. Syst. Evol. 55, 238–258. doi: 10.1111/jse.12267
Abbott, R. J., and Brennan, A. C. (2014). Altitudinal gradients, plant hybrid zones and evolutionary novelty. Philos. Trans. R. Soc. B Biol. Sci. 369:20130346. doi: 10.1098/rstb.2013.0346
Aiken, S. G. (1981). A conspectus of Myriophyllum (Haloragaceae) in North America. Brittonia 33, 57–69. doi: 10.2307/2806578
Anderson, E. C., and Thompson, E. A. (2002). A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160, 1217–1229. doi: 10.1093/genetics/160.3.1217
Arnold, M. L., and Martin, N. H. (2010). Hybrid fitness across time and habitats. Trends Ecol. Evol. 25, 530–536. doi: 10.1016/j.tree.2010.06.005
Bandelt, H. J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Barrett, S. C. H., Eckert, C. G., and Husband, B. C. (1993). Evolutionary processes in aquatic plant populations. Aquat. Bot. 44, 105–145. doi: 10.1016/0304-3770(93)90068-8
Barton, N. H. (2001). The role of hybridization in evolution. Mol. Ecol. 10, 551–568. doi: 10.1046/j.1365-294x.2001.01216.x
Barton, N. H., and Hewitt, G. M. (1985). Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16, 113–148. doi: 10.2307/2097045
Broennimann, O., Fitzpatrick, M. C., Pearman, P. B., Petitpierre, B., Pellissier, L., Yoccoz, N. G., et al. (2012). Measuring ecological niche overlap from occurrence and spatial environmental data. Glob. Ecol. Biogeogr. 21, 481–497. doi: 10.1111/j.1466-8238.2011.00698.x
Burgarella, C., Lorenzo, Z., Jabbour-Zahab, R., Lumaret, R., Guichoux, E., Petit, R. J., et al. (2009). Detection of hybrids in nature: application to oaks (Quercus suber and Q. ilex). Heredity 102, 442–452. doi: 10.1038/hdy.2009.8
Burke, J. M., and Arnold, M. L. (2001). Genetics and the fitness of hybrids. Annu. Rev. Genet. 35, 31–52. doi: 10.1146/annurev.genet.35.102401.085719
Ceska, A., and Ceska, O. (1986). Notes on Myriophyllum (Haloragaceae) in the Far East: the identity of Myriophyllum sibiricum Komarov. Taxon 35, 95–100. doi: 10.2307/1221038
Chen, J.-M., Du, Z.-Y., Yuan, Y.-Y., and Wang, Q.-F. (2014). Phylogeography of an alpine aquatic herb Ranunculus bungei (Ranunculaceae) on the Qinghai–Tibet Plateau. J. Syst. Evol. 52, 313–325. doi: 10.1111/jse.12093
Chen, L., Zhao, S., Mao, K., Les, D. H., Wang, Q., and Moody, M. L. (2014). Historical biogeography of Haloragaceae: an out-of-Australia hypothesis with multiple intercontinental dispersals. Mol. Phylogenet. Evol. 78, 87–95. doi: 10.1016/j.ympev.2014.04.030
Chung, M. Y., Nason, J. D., and Chung, M. G. (2005). Patterns of hybridization and population genetic structure in the terrestrial orchids Liparis kumokiri and Liparis makinoana (Orchidaceae) in sympatric populations. Mol. Ecol. 14, 4389–4402. doi: 10.1111/j.1365-294X.2005.02738.x
Couch, R., and Nelson, E. (1985). “Myriophyllum spicatum in North America,” in Proceedings of the 1st International Symposium on Watermilfoil (Myriophyllum spicatum) and Related Haloragaceae Species, ed. L. W. J. Anderson (Washington, DC: The Aquatic Plant Management Society, Inc), 8–18.
Di Cola, V., Broennimann, O., Petitpierre, B., Breiner, F. T., D’Amen, M., Randin, C., et al. (2017). ecospat: an R package to support spatial analyses and modeling of species niches and distributions. Ecography 40, 774–787. doi: 10.1111/ecog.02671
Du, Z.-Y., and Wang, Q.-F. (2016). Allopatric divergence of Stuckenia filiformis (Potamogetonaceae) on the Qinghai-Tibet Plateau and its comparative phylogeography with S. pectinata in China. Sci. Rep. 6:20883. doi: 10.1038/srep20883
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. doi: 10.1093/genetics/164.4.1567
Friendly, M., and Fox, J. (2021). Package ‘Candisc’-Visualizing Generalized Canonical Discriminant and Canonical Correlation Analysis. Version 0.8-6. R. Software.
Gompert, Z., Mandeville, E. G., and Buerkle, C. A. (2017). Analysis of population genomic data from hybrid zones. Annu. Rev. Ecol. Evol. Syst. 48, 207–229. doi: 10.1146/annurev-ecolsys-110316-022652
Hamilton, J. A., and Miller, J. M. (2016). Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 30, 33–41. doi: 10.1111/cobi.12574
Harrison, R. G. (1986). Pattern and process in a narrow hybrid zone. Heredity 56, 337–349. doi: 10.1038/hdy.1986.55
Harrison, R. G., and Larson, E. L. (2016). Heterogeneous genome divergence, differential introgression, and the origin and structure of hybrid zones. Mol. Ecol. 25, 2454–2466. doi: 10.1111/mec.13582
Hijmans, R. J., Phillips, S., Leathwick, J., and Elith, J. (2017). Package ‘dismo’-Species Distribution Modeling. V1.3-5. R. Software.
Hollingsworth, P. M., Preston, C. D., and Gornall, R. J. (1996). Isozyme evidence for the parentage and multiple origins of Potamogeton ×suecicus (P. pectinatus ×P. filiformis, Potamogetonaceae). Plant Syst. Evol. 202, 219–232. doi: 10.1007/bf00983384
Jiang, D., Feng, J., Dong, M., Wu, G., Mao, K., and Liu, J. (2016). Genetic origin and composition of a natural hybrid poplar Populus × jrtyschensis from two distantly related species. BMC Plant Biol. 16:89. doi: 10.1186/s12870-016-0776-6
Kaplan, Z. (2008). A taxonomic revision of Stuckenia (Potamogetonaceae) in Asia, with notes on the diversity and variation of the genus on a worldwide scale. Folia Geobot. 43, 159–234. doi: 10.1007/s12224-008-9010-0
Katoh, K., and Toh, H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298. doi: 10.1093/bib/bbn013
Les, D. H., and Philbrick, C. T. (1993). Studies of hybridization and chromosome number variation in aquatic angiosperms: evolutionary implications. Aquat. Bot. 44, 181–228. doi: 10.1016/0304-3770(93)90071-4
Li, J., Milne, R. I., Ru, D., Miao, J., Tao, W., Zhang, L., et al. (2020). Allopatric divergence and hybridization within Cupressus chengiana (Cupressaceae), a threatened conifer in the northern Hengduan Mountains of western China. Mol. Ecol. 29, 1250–1266. doi: 10.1111/mec.15407
Li, Z., Lu, W., Yang, L., Kong, X., and Deng, X. (2015). Seed weight and germination behavior of the submerged plant Potamogeton pectinatus in the arid zone of northwest China. Ecol. Evol. 5, 1504–1512. doi: 10.1002/ece3.1451
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Matthews, B., Jokela, J., Narwani, A., Räsänen, K., Pomati, F., Altermatt, F., et al. (2020). On biological evolution and environmental solutions. Sci. Total Environ. 724:138194. doi: 10.1016/j.scitotenv.2020.138194
Moody, M. L., and Les, D. H. (2002). Evidence of hybridity in invasive watermilfoil (Myriophyllum) populations. Proc. Natl. Acad. Sci. U.S.A. 99, 14867–14871. doi: 10.1073/pnas.172391499
Moody, M. L., and Les, D. H. (2007). Geographic distribution and genotypic composition of invasive hybrid watermilfoil (Myriophyllum spicatum × M. sibiricum) populations in North America. Biol. Invasions 9, 559–570. doi: 10.1007/s10530-006-9058-9
Nies, G., and Reusch, T. B. H. (2004). Nine polymorphic microsatellite loci for the fennel Pondweed Potamogeton pectinatus L. Mol. Ecol. Notes 4, 563–565. doi: 10.1111/j.1471-8286.2004.00734.x
Nies, G., and Reusch, T. B. H. (2005). Evolutionary divergence and possible incipient speciation in post-glacial populations of a cosmopolitan aquatic plant. J. Evol. Biol. 18, 19–26. doi: 10.1111/j.1420-9101.2004.00818.x
Ortego, J., Gugger, P. F., Riordan, E. C., and Sork, V. L. (2014). Influence of climatic niche suitability and geographical overlap on hybridization patterns among southern Californian oaks. J. Biogeogr. 41, 1895–1908. doi: 10.1111/jbi.12334
Phillips, S. J., Anderson, R. P., and Schapire, R. E. (2006). Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259. doi: 10.1016/j.ecolmodel.2005.03.026
Pierce, A. A., Gutierrez, R., Rice, A. M., and Pfennig, K. S. (2017). Genetic variation during range expansion: effects of habitat novelty and hybridization. Proc. R. Soc. B Biol. Sci. 284:20170007. doi: 10.1098/rspb.2017.0007
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Santamaría, L. (2002). Why are most aquatic plants widely distributed? Dispersal, clonal growth and small-scale heterogeneity in a stressful environment. Acta Oecol. 23, 137–154. doi: 10.1016/S1146-609X(02)01146-3
Seehausen, O. (2004). Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207. doi: 10.1016/j.tree.2004.01.003
Shaw, J., Lickey, E. B., Schilling, E. E., and Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94, 275–288. doi: 10.3732/ajb.94.3.275
Shen, M., Zhang, G., Cong, N., Wang, S., Kong, W., and Piao, S. (2014). Increasing altitudinal gradient of spring vegetation phenology during the last decade on the Qinghai–Tibetan Plateau. Agric. Forest Meteorol. 189-190, 71–80. doi: 10.1016/j.agrformet.2014.01.003
Sun, H., Niu, Y., Chen, Y.-S., Song, B., Liu, C.-Q., Peng, D.-L., et al. (2014). Survival and reproduction of plant species in the Qinghai–Tibet Plateau. J. Syst. Evol. 52, 378–396. doi: 10.1111/jse.12092
Sun, Y., Abbott, R. J., Li, L., Li, L., Zou, J., and Liu, J. (2014). Evolutionary history of Purple cone spruce (Picea purpurea) in the Qinghai–Tibet Plateau: homoploid hybrid origin and Pleistocene expansion. Mol. Ecol. 23, 343–359. doi: 10.1111/mec.12599
Taylor, S. A., and Larson, E. L. (2019). Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat. Ecol. Evol. 3, 170–177. doi: 10.1038/s41559-018-0777-y
Wang, D. (2003). The Geography of Aquatic Vascular Plants of Qinghai-Xizang (Tibet) Plateau. Ph.D. Dissertation. Wuhan: Wuhan University.
Warren, D. L., Glor, R. E., and Turelli, M. (2008). Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62, 2868–2883. doi: 10.1111/j.1558-5646.2008.00482.x
Warren, D. L., Glor, R. E., and Turelli, M. (2010). ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33, 607–611. doi: 10.1111/j.1600-0587.2009.06142.x
Wen, J., Zhang, J. Q., Nie, Z. L., Zhong, Y., and Sun, H. (2014). Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front. Genet. 5:4. doi: 10.3389/fgene.2014.00004
Wiegleb, G., Bobrov, A. A., and Zalewska-Gałosz, J. (2017). A taxonomic account of Ranunculus section Batrachium (Ranunculaceae). Phytotaxa 319:55. doi: 10.11646/phytotaxa.319.1.1
Wu, S., Wang, Y., Wang, Z., Shrestha, N., and Liu, J. (2022). Species divergence with gene flow and hybrid speciation on the Qinghai–Tibet Plateau. New Phytol. 234, 392–404. doi: 10.1111/nph.17956
Wu, Z., Ding, Z., Yu, D., and Xu, X. (2015). Influence of niche similarity on hybridization between Myriophyllum sibiricum and M. spicatum. J. Evol. Biol. 28, 1465–1475. doi: 10.1111/jeb.12667
Wu, Z., Li, X., Xie, D., Wang, H., Zhang, Z., Xu, X., et al. (2020). Spatial genetic structuring in a widespread wetland plant on a plateau: effects of elevation-driven geographic isolation and environmental heterogeneity. Freshw. Biol. 65, 1596–1607. doi: 10.1111/fwb.13525
Wu, Z., Wu, J., Wang, Y., and Hou, H. (2017). Development of EST-derived microsatellite markers in the aquatic macrophyte Ranunculus bungei (Ranunculaceae). Appl. Plant Sci. 5:1700022. doi: 10.3732/apps.1700022
Wu, Z., Xu, X., Zhang, J., Wiegleb, G., and Hou, H. (2019). Influence of environmental factors on the genetic variation of the aquatic macrophyte Ranunculus subrigidus on the Qinghai-Tibetan Plateau. BMC Evol. Biol. 19:228. doi: 10.1186/s12862-019-1559-0
Wu, Z., Yu, D., Li, X., and Xu, X. (2016). Influence of geography and environment on patterns of genetic differentiation in a widespread submerged macrophyte, Eurasian watermilfoil (Myriophyllum spicatum L., Haloragaceae). Ecol. Evol. 6, 460–468. doi: 10.1002/ece3.1882
Xing, Y., and Ree, R. H. (2017). Uplift-driven diversification in the Hengduan mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A. 114, E3444–E3451. doi: 10.1073/pnas.1616063114
Yakimowski, S. B., and Rieseberg, L. H. (2014). The role of homoploid hybridization in evolution: a century of studies synthesizing genetics and ecology. Am. J. Bot. 101, 1247–1258. doi: 10.3732/ajb.1400201
Yang, R., Folk, R., Zhang, N., and Gong, X. (2019). Homoploid hybridization of plants in the Hengduan mountains region. Ecol. Evol. 9, 8399–8410. doi: 10.1002/ece3.5393
Keywords: altitudinal gradient, hybrid zone, hydrophilous plant, introgression, population genetics, niche similarity, species distribution model
Citation: Wu Z, Wang Z, Xie D, Zhang J, Cai P, Li X, Xu X, Li T and Zhao J (2022) Extensive Sympatry and Frequent Hybridization of Ecologically Divergent Aquatic Plants on the Qinghai-Tibetan Plateau. Front. Plant Sci. 13:851151. doi: 10.3389/fpls.2022.851151
Received: 09 January 2022; Accepted: 13 April 2022;
Published: 12 May 2022.
Edited by:
Jianping Yu, National Renewable Energy Laboratory (DOE), United StatesReviewed by:
Jinming Chen, Wuhan Botanical Garden (CAS), ChinaCopyright © 2022 Wu, Wang, Xie, Zhang, Cai, Li, Xu, Li and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinwei Xu, eHV4d0B3aHUuZWR1LmNu; Tao Li, bGl0YW9AaWhiLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.