
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 06 May 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.850949
This article is part of the Research Topic Plant Specialized Metabolism for Plant Protection: Genomics and Biotechnology View all 13 articles
Dendrobium is the second biggest genus in the Orchidaceae family, some of which have both ornamental and therapeutic values. Alkaloids are a group of active chemicals found in Dendrobium plants. Dendrobine has emerged specific pharmacological and therapeutic properties. Although Dendrobium alkaloids have been isolated and identified since the 1930s, the composition of alkaloids and their biosynthesis pathways, including metabolic intermediates, alkaloid transporters, concrete genes involved in downstream pathways, and associated gene clusters, have remained unresolved scientific issues. This paper comprehensively reviews currently identified and tentative alkaloids from the aspect of biogenic pathways or metabolic genes uncovered based on the genome annotations. The biosynthesis pathways of each class of alkaloids are highlighted. Moreover, advances of the high-throughput sequencing technologies in the discovery of Dendrobium alkaloid pathways have been addressed. Applications of synthetic biology in large-scale production of alkaloids are also described. This would serve as the basis for further investigation into Dendrobium alkaloids.
Dendrobium is the second biggest genus in the Orchidaceae family, and many of them have high medicinal and ornamental benefits (Lam et al., 2015). For almost 2,000 years, Dendrobium has been used as a prized traditional Chinese medicine. This can be traced back to the monograph “Shen Nong Ben Cao Jing.” The medicinal Dendrobium materials included in the Chinese Pharmacopoeia (11th Edition) are divided into two categories: one is the cultivated varieties from D. nobile, D. huoshanense, D. chrysotoxum, or D. fimbriatum Hook., and the fresh or dried stems of similar species in the same genus. The other is the dry stem of D. officinale. According to the records of the national general survey of key medicinal resources, the Ta-pieh Mountains in Anhui Province, as a natural wild progeny reserve, is home to numerous of wild Dendrobium species, including D. moniliforme, D. officinale, and D. huoshanense.
The chemical composition and categorization of different medicinal Dendrobium species vary massively due to the wide variety of medicinal Dendrobium species (Teixeira da Silva and Ng, 2017). At present, the chemical components isolated from Dendrobium spp. mainly included polysaccharides, alkaloids, amino acids, bibenzyls, phenanthrenes, sesquiterpenes, fluorenones, flavonoids, phenolic acids, phenylpropanoids, lignans, amides, alkaloids, steroids, etc. (Ng et al., 2012; Xu et al., 2013). Alkaloids were the first chemicals studied and structurally confirmed in Dendrobium. The total alkaloid content of D. nobile typically approached 0.5% (Wang et al., 2016). More than 40 sesquiterpene alkaloids and terpenes have been isolated and identified from D. nobile (Mou et al., 2021). D. officinale is one of the most frequently cultivated Dendrobium species in China. The chemical constituents have been thoroughly isolated and characterized (Wang et al., 2021). Polysaccharides, bibenzyl, phenanthrene, lignans, flavonoids, alkaloids, and other chemicals have been found in D. officinale. However, there have been few studies on the isolation and identification of alkaloids monomers from D. officinale. D. huoshanense is a second-class wild endangered plant in China, having extremely high medicinal and health benefits (Jin et al., 2016; Cakova et al., 2017; Yuan et al., 2018, 2019). The extraction, separation, and structural characterization of polysaccharides are the main focus of research on the active ingredients of D. huoshanense (Zha et al., 2017). There have also been some studies on the separation of other small molecule chemicals, such as flavonoids, terpenoids, and alkaloids (Chang et al., 2010; Song et al., 2020; Li et al., 2021). However, the biosynthesis pathway of alkaloids from D. huoshanense remain unclear.
Previous researches have indicated that D. officinale and D. huoshanense contain a substantial amount of nitrogen-containing compounds, including alkaloids, the composition of which varies greatly between the two Dendrobium species (Song et al., 2020). Aside from the complicated structure of alkaloids, the scarcity of high-quality genomes makes pinpointing key genes in the alkaloid biosynthesis pathway more challenging. Currently, the mining of alkaloid functional genes and the research of biosynthesis pathway in some model medicinal plants, such as Artemisia annua, Papaver somniferum, Catharanthus roseus, Camptotheca acuminata, and other species, has been carried out by using third-generation sequencing technologies like Pacbio and Oxford nanopore (Kellner et al., 2015a; Shen et al., 2018; Guo et al., 2018b; Kang et al., 2021). These findings have further spurred and enhanced research into the process of alkaloids production. Here, we highlighted recent progresses in the chemical identification and biosynthesis processes of Dendrobium alkaloids, and also discussed the alkaloid biosynthesis pathways across diverse biogenic pathways. Meanwhile, we deeply discussed the significance of high-throughput sequencing technology in uncovering the critical genes of Dendrobium alkaloid biosynthesis pathways. Biosynthetic gene clusters were exploited to construct the heterologous pathways for Dendrobium alkaloids biosynthesis based on the synthetic biology approaches, which could fulfill large-scale production and market demand.
Alkaloids are a class of nitrogen-containing alkaline organic compounds that exist in organisms (mostly plants; Facchini, 2001; Zhan et al., 2020). Most alkaloids have a complex ring structure, and nitrogen atom is mostly contained in ring. Alkaloids have a high level of biological activity and are one of the most important active substances in herbal medicines (Facchini and St-Pierre, 2005; Kiss et al., 2017). Chinese herbal medicines abundant in alkaloids have been discovered in more than 100 families with the most dicotyledonous plants, followed by monocots, gymnosperms, and ferns. Among them, Papaveraceae, Leguminosae, Fangchiaceae, Ranunculaceae, Apocynaceae, Solanaceae, and Amaryllidaceae are all rich in alkaloids (Kishimoto et al., 2016). Most alkaloids are derived from L-amino acids (such as tyrosine, phenylalanine, tryptophan, lysine, and arginine), or combined with steroids, secoiridoids, and other ligands (Kumar et al., 2018). Under the catalysis of a set of specific enzymes, natural amino acids can be converted into highly specialized alkaloid precursors via the TCA cycle (Lichman, 2021). Alkaloids are the first compounds to be isolated and validated in the structure, among which sesquiterpene alkaloids are even more studied (Kreis and Carreira, 2012). The isolation and identification of dendrobine first occurred in D. nobile as a significant therapeutic substance (Wang et al., 2016). Afterward, dozens of alkaloid monomers were identified from D. parishii Rchb.f, D. chrysanthum Wall, D. crepidatum Lind, D. findlayanum par.et Rchb.f, D. friedricksianum Lindl., D. hilderbrandii, and D. loddigesii Rolfe (Liu et al., 2007, 2020a; Zhang et al., 2017; Yang et al., 2018, 2020; Wang et al., 2019, 2020). Based on the chemical structure of the separated alkaloids, they were divided into sesquiterpene alkaloids, imidazole alkaloids, pyrrolidine alkaloids, phthalide alkaloids, indolizidine alkaloids, and other types (Mou et al., 2021; Wang et al., 2022). At least 60 alkaloids were characterized by structure, including 35 sesquiterpene alkaloids, 14 indolizidine alkaloids, five pyrrolidine alkaloids, four phthalide alkaloids, two organic amine alkaloids, one imidazole type, and one indole alkaloid (Figure 1). Among these alkaloids, the activities of total alkaloid extracts and several monomers have been verified in vivo or in vitro at various levels (Nie et al., 2018; Huang et al., 2019; Song et al., 2019; Hu et al., 2020). Some alkaloids have shown significant anti-inflammatory, anti-cancer, anti-viral, and neuroprotective properties (Table 1). Different types of alkaloids exhibit significantly different pharmacological and pharmacodynamic effects. The total alkaloids and monomers from D. nobile, such as dendrobine, have obvious therapeutic effects in repairing nerve and liver injuries, improving memory, suppressing LPS-induced apoptosis, and anti-virus activity (Li et al., 2011, 2017c; Nie et al., 2016; Zhou et al., 2020). In D. crepidatum, homocrepidines and dendrocrepidine F exhibit anti-inflammatory properties (Hu et al., 2016). Shihunine and related extracts from D. loddigesii have significant anti-inflammatory and diabetic symptom alleviation effects (Chen et al., 2018; Li et al., 2019). Dendrofindline A from D. findlayanum exhibits notable cytotoxic effects on human tumor cells while also promoting gastrointestinal motility (Liu et al., 2020a). Although several alkaloids have been identified in D. officinale, no relevant pharmacological effects have been reported for these substances.
Sesquiterpene alkaloids are the most abundant alkaloids found in Dendrobium plants. Dendrobine is one of the most rigorously studied alkaloids, with significant pharmacological effects in anti-tumor, cardiovascular and gastrointestinal inhibitory effects, and neuroprotection (Ohc and Claisen, 2012; Song et al., 2019; Zhou et al., 2020). Picrotoxane-type sesquiterpene lactone is the fundamental skeleton of this type of alkaloid. Most alkaloids have a nitrogen atom that is bonded to the C2 and C11 positions of a sesquiterpene to create a five-membered heterocyclic ring. The nitrogen atom is commonly linked to functional groups, such as methyl and isopentenyl (Morita et al., 2000; Guo et al., 2018a; Yang et al., 2019). This conformation is considered to be directly correlated with the biogenic origins of picrotoxin and tutin (Ma et al., 2019a). The isotope tracer test demonstrated that mevalonic acid is involved in the formation of the carbon skeleton of dendrobine by using substances containing radioactive isotopes 14C, 2H, and 3H to cultivate and feed D. nobile (Meng et al., 2017). The canonical precursor of all sesquiterpene alkaloids (2-trans-6-trans-farnesol) is initially determined by 2-trans-6-trans-farnesyl diphosphate under the catalysis of farnesyl pyrophosphate synthesase (FPS). After farnesol undergo a cycloaddition event to form germacradiene, the electrons of germacrene follow the HR transition from C11 to C1 to form a germacrene cation immediately. The reaction center is also shifted from C11 to C1. This stereoisomerization yields the diastereomers of the germacrene cation. Since the reaction center meets the cyclization conditions at the C1 position, the intermediate muurolane is achieved. The C2-C3 double bond is then broken, and the C3 and C7 bonds occur simultaneously, resulting in the tricyclic intermediate copaborneol. It is a common precursor in the biosynthesis of picrotoxin, tutin, and dendrobine (Wang et al., 2019). Following that, the C2-C3 bond of copaborneol is broken to form intermediate 1 (5-isopropyl-7a-methyl-1-methyleneoctahydro-1H-inden-4-yl) methanol, which is further oxidized to form intermediate 2 (5-isopropyl-7a-methyl-1-methyleneoctahydro-1H-indene-4-carboxylic acid), to which a H2O is incorporated to form intermediate 3. Dioxygenase catalyzes the hydroxylation of intermediate 3 at the C8 and C9 positions, resulting in the formation of picrotoxane (Li et al., 2017b). Picrotoxane esterifies to produce picrotoxane-lactone. Picrotoxin and tutin are synthesized by a series of oxidation reactions catalyzed by CYP450 enzymes (Figure 2). Furthermore, under the action of monooxygenase, aminotransferase, and methyltransferase, picrotoxane-lactone could ultimately be converted to dendrobine (Li et al., 2017b; Mou et al., 2021).
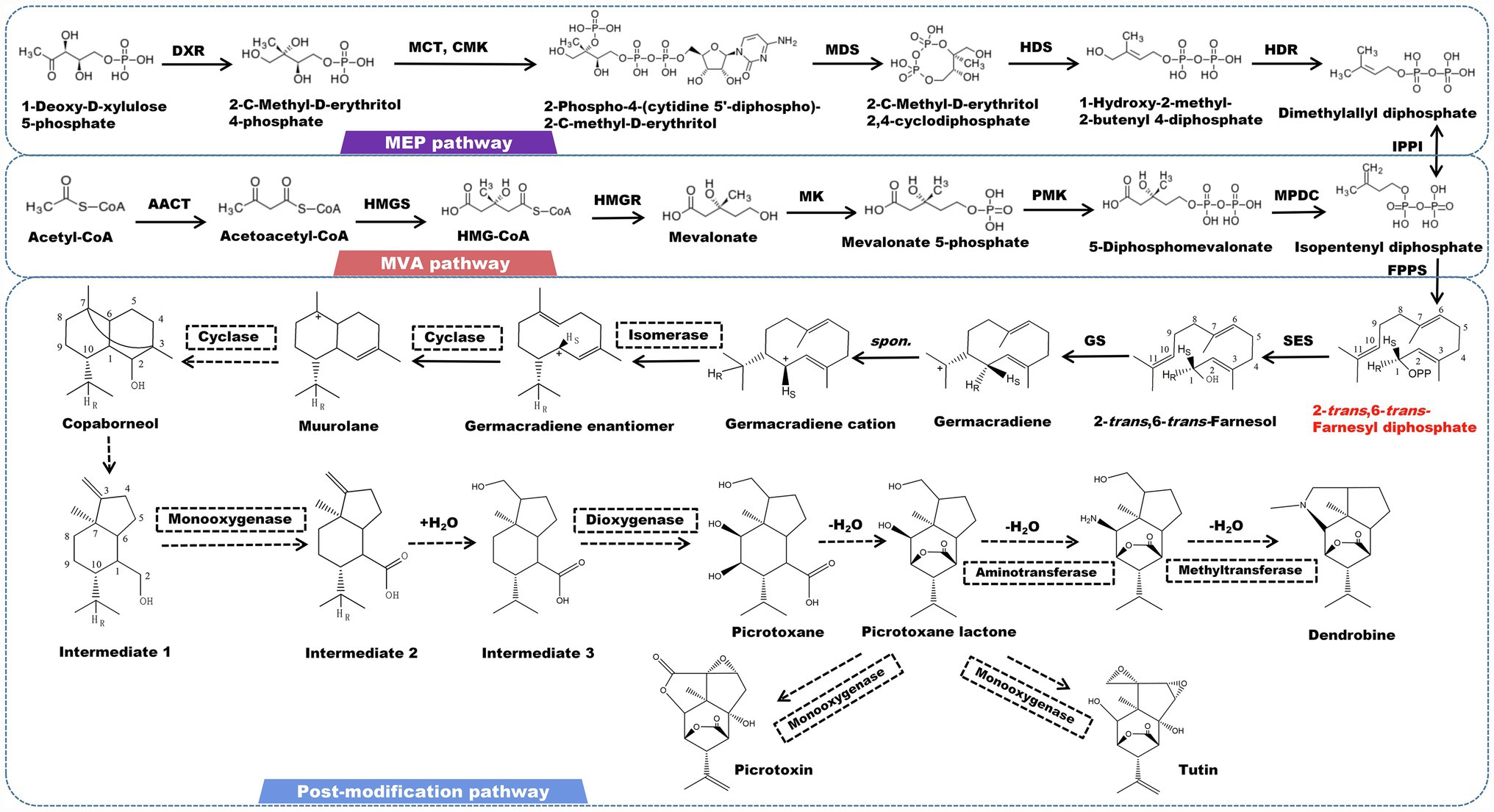
Figure 2. A plausible biosynthesis pathway of sesquiterpene alkaloids. The dashed box represents the putative enzymes. The dashed arrow represents the speculated pathway. Acetyl-CoA acetyltransferase (AACT), HMG-CoA synthase (HMGS), HMG-CoA reductase (HMGR), mevalonate kinase (MK), phosphomevalonate kinase (PMK), mevalonate diphosphate decarboxylase (MPDC), 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR), 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (MCT), 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK), 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MDS), 4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS), 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR), isopentenyl pyrophosphate: dimethylallyl pyrophosphate isomerase (IPPI), farnesyl diphosphate synthase (FPPS), sesquiterpene synthase (SES), germacradiene synthase (GS). SES catalyzes the formation of the precursor 2-trans-6-trans-farnesol by 2-trans-6-trans-farnesyl diphosphate.
Indolizidine alkaloids are a type of heterocyclic compounds with specific biological and pharmacological activities that are discovered mostly in the Apocynaceae, Convolvulaceae, Orchidaceae, Pandanaceae, and Leguminosae (Hu et al., 2018). Currently, a large number of indolizidine alkaloids have been found in marine and terrestrial animals and plants. The fundamental structure of indolizidine alkaloids is a 5-membered ring joined with a 6-membered ring sharing a nitrogen atom (Hu et al., 2020; Zhang et al., 2021a). The biogenic pathway of indolizidine alkaloids originates from L-lysine in terms of molecular structure. L-lysine undergoes a deamination in the lysine degradation pathway, which is catalyzed by saccharopine dehydrogenase (SD) to form saccharopine. Saccharopine is catalyzed by SD to form 2-aminoadipate 6-semialdehyde, which then cyclized to form 1-piperideine-6-L-carboxylate. Under the catalysis of L-pipecolate oxidase, 1-piperideine-6-L-carboxylate degrades into pipecolate (PO). Pipecolate and acetic acid are catalyzed by cyclase to yield 1-indolizidinone. 1-Indolizidinone undergoes a multi-step reaction involving CYP450 monooxygenase, reductase, and methyltransferase to form intermediate 3 (Figure 3). It is believed that the C2 and C3 in 1-indolizidinone came from acetate via malonate; however, the enzyme that catalyzes this step has not been found. Intermediate 3 is catalyzed by phenyltransferase (PT) and malonyl-CoA-ACP transacylase (MCAT) to generate crepidatumine A, crepidatumine D, and crepidine, based on the location of the malonyl group binding (Xu et al., 2019, 2020). Depending on CYP450 enzyme, crepidatumine A and crepidine polymerize to form two type of enantiomeric octahydroindolizine alkaloids—homocrepidine A and dendrocrepidamine A (Hu et al., 2016). Crepidatumine A was converted to dendroprimine via CYP450 and cyclase (Kobayashi et al., 2012). Crepidine underwent the cyclization to yield isocrepidamine, which then combined with intermediate 3 to form isodendrocrepine (Xu et al., 2019).
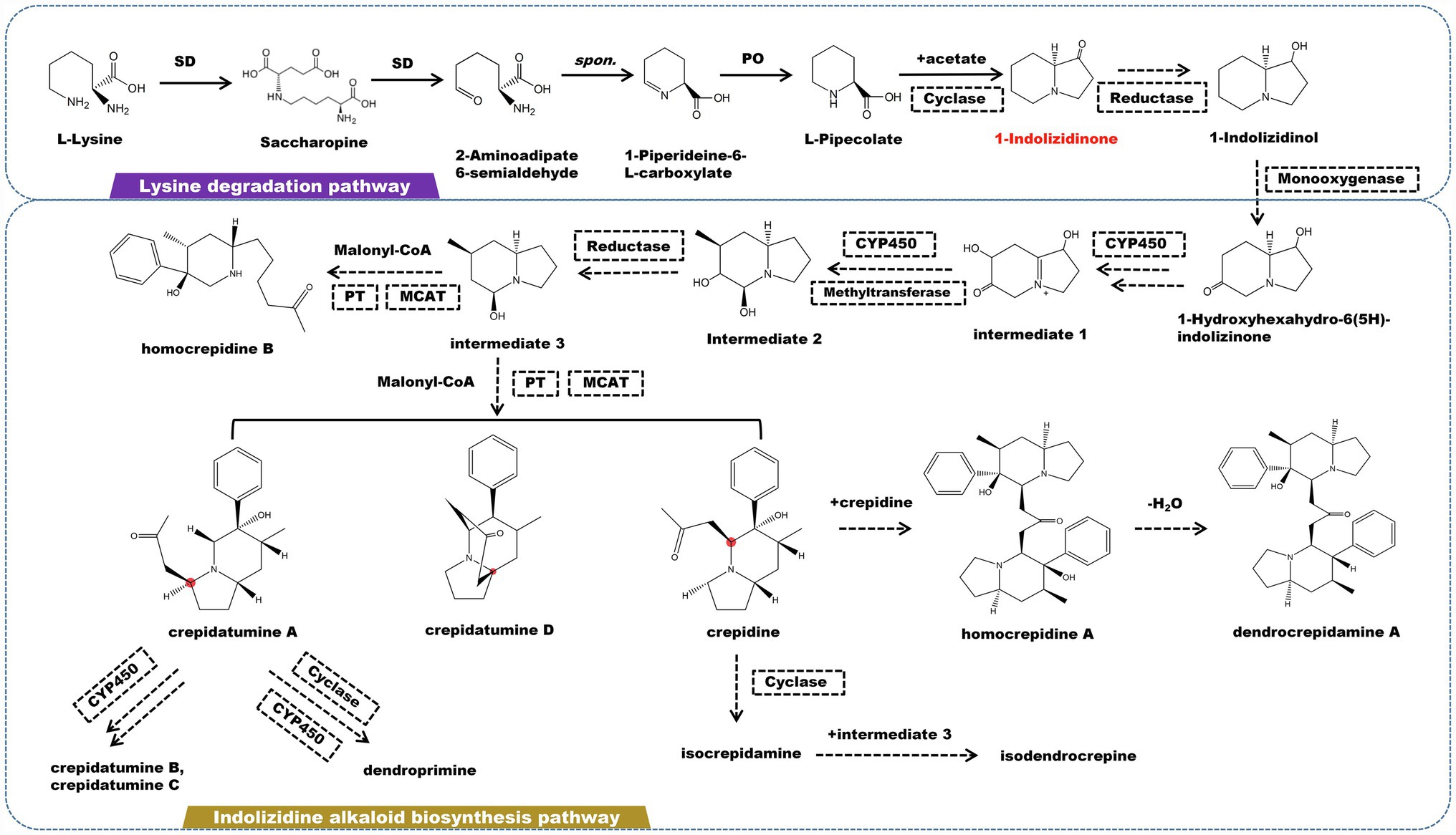
Figure 3. A plausible biosynthesis pathway of indolizidine alkaloids. The dashed box represents the putative enzymes. The dashed arrow represents the speculated pathway. The multiple arrows represent multi-step reactions. Saccharopine dehydrogenase (SD), L-pipecolate oxidase (PO), phenyltransferase (PT), malonyl-CoA-ACP transacylase (MCAT). Cyclase catalyzes the connection of pipecolate and acetic acid to produce the precursor 1-indolizidinone.
Pyrrolidine alkaloids are a family of structurally related substances found in Asteraceae, Leguminosae, and Boraginaceae. Some of them have pharmacological effects on liver toxicity to humans and livestock (Khosravi et al., 2019; Lichman, 2021). Pyrrolidine alkaloids are abundant in several edible plants, including Senecio, Crotalaria, and Heliotropium (Seigler, 1998). Some plants featuring pyrrolidine alkaloids are also utilized to make herbal remedies and medicinal teas. Japanese daisy tea, for example, is rich in pyrrolidine alkaloids. Currently, almost 700 pyrrolidine alkaloids have been identified from diverse plants. L-ornithine is the source of pyrrolidine alkaloids. L-ornithine is initially catalyzed by ornithine decarboxylase (ODC) to produce putrescine, which then catalyzed by putrescine N-methyltransferase (PMT) to obtain methylputrescine. Under the catalysis of primary amine: oxygen oxidoreductase (AOC), methylputrescine is deaminated and oxidized to form 1-methylpyrrolinium—the precursor of all pyrrolidine alkaloids (Figure 4). Depending on acetyl-CoA acetyltransferase (AACT), one molecule 1-methylpyrrolinium and two molecules of acetyl-CoA link to form two enantiomers (−)-hygrine and (+)-hygrine (Nguyen et al., 2015). In addition, (R)-2-acetoacetyl-CoA-1-methylpyrrolidine can also be combined with 1-methylpyrrolinium to form cuscohygrine. Cuscohygrine coupled with p-cinnamoyl-CoA form trans- and cis-dendrochrysine. In addition, (−)-hygrine and p-cinnamoyl-CoA condense to produce cis- and trans-dendrochrysanine (Yang et al., 2006).
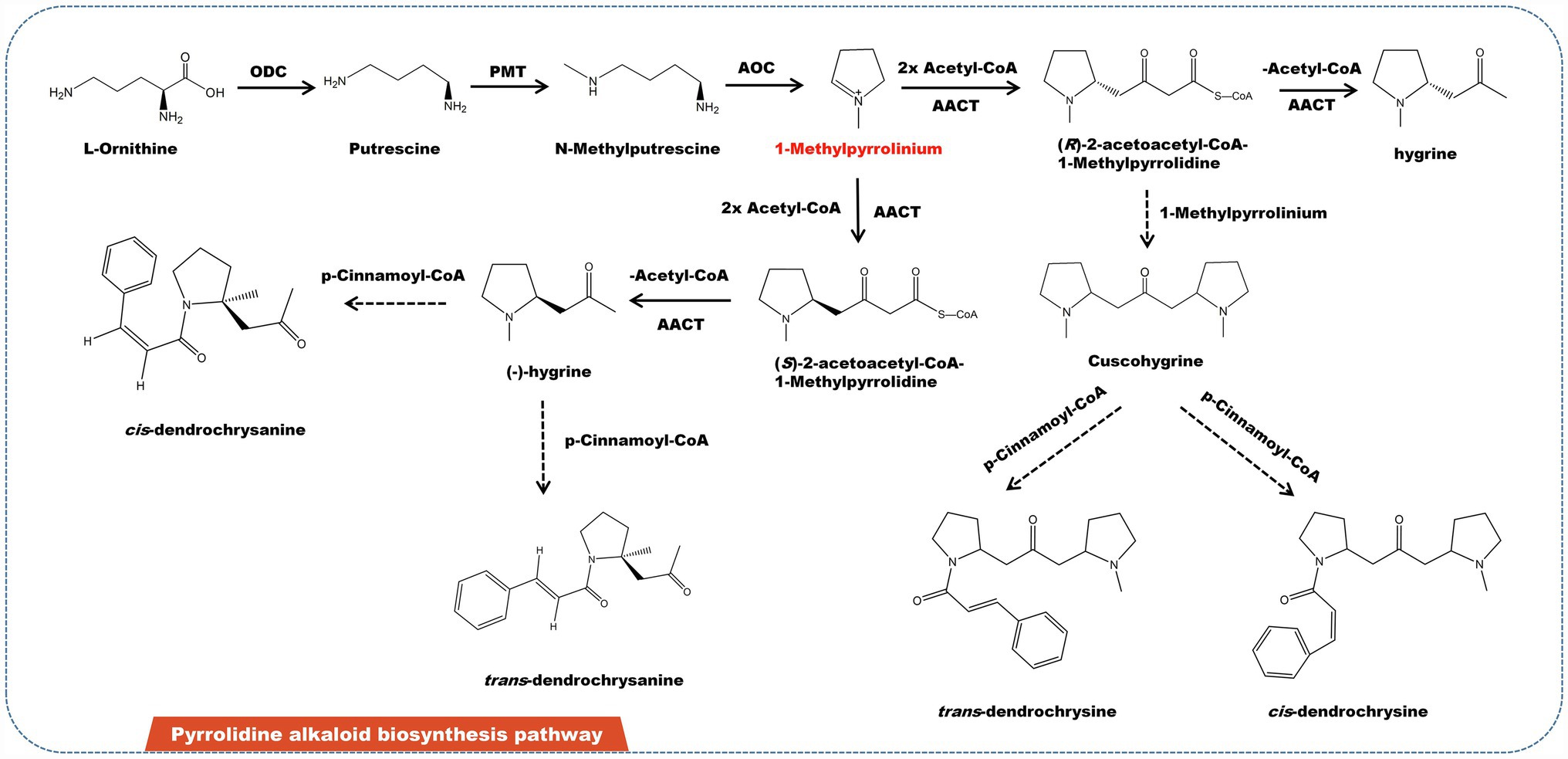
Figure 4. A plausible biosynthesis pathway of pyrrolidine alkaloids. The dashed box represents the putative enzymes. The dashed arrow represents the speculated pathway. Ornithine decarboxylase (ODC), putrescine N-methyltransferase (PMT), primary amine: oxygen oxidoreductase (AOC), acetyl-CoA acetyltransferase (AACT). Under the catalysis of AOC, methylputrescine is deaminated and oxidized to produce the precursor 1-methylpyrrolinium.
A rare imidazolium-type alkaloid called anosmine was isolated from D. nobile (Chen et al., 2018). L-lysine is the starting point for the production of this kind of alkaloids (Hemscheidt and Spenser, 1993). Lysine decarboxylase catalyzes the conversion of L-lysine to cadaverine. Simultaneously, saccharopine dehydrogenase (SD) and L-pipecolate oxidase (PO) catalyze the formation of L-pipecolate. Cadaverine is oxidized by amine oxidase to form 1-piperideinium. L-Pipecolate and 1-piperideinium combine to form [1,2′-bipiperidine]-2-carboxylic acid, which is dehydrated to form intermediates 1 and 2. The latter is subjected to a series of oxidation to produce anosmine (Figure 5A). Other alkaloids, such as terpenoid indole alkaloids (TIAs), are also found in some Dendrobium spp. However, there have been few studies on the isolation and identification of indole alkaloids, with only the β-carbolin-type indole alkaloid (S)-2-tryptoline-3-carboxylic acid ethyl ester being isolated and characterized. Based on the chemical structure, the biosynthetic pathway of (S)-2-tryptoline-3-carboxylic acid ethyl ester is assumed to have originated from tryptophan metabolism (Figure 5B). L-tryptophan is converted to tryptamine via the tryptophan degradation pathway, which is catalyzed by L-tryptophan decarboxylase (TDC). Based on Pictet–Spengler reaction, tryptamine is catalyzed by strictosidine synthase (STR) to form strictosidine, which is then cracked to become tetrahydroharman (Pressnitz et al., 2018; Sharma et al., 2018; Yang et al., 2021; You et al., 2021). Through a multi-step enzymatic reaction, tetrahydroharman finally creates (S)-2-tryptoline-3-carboxylic acid ethyl ester.
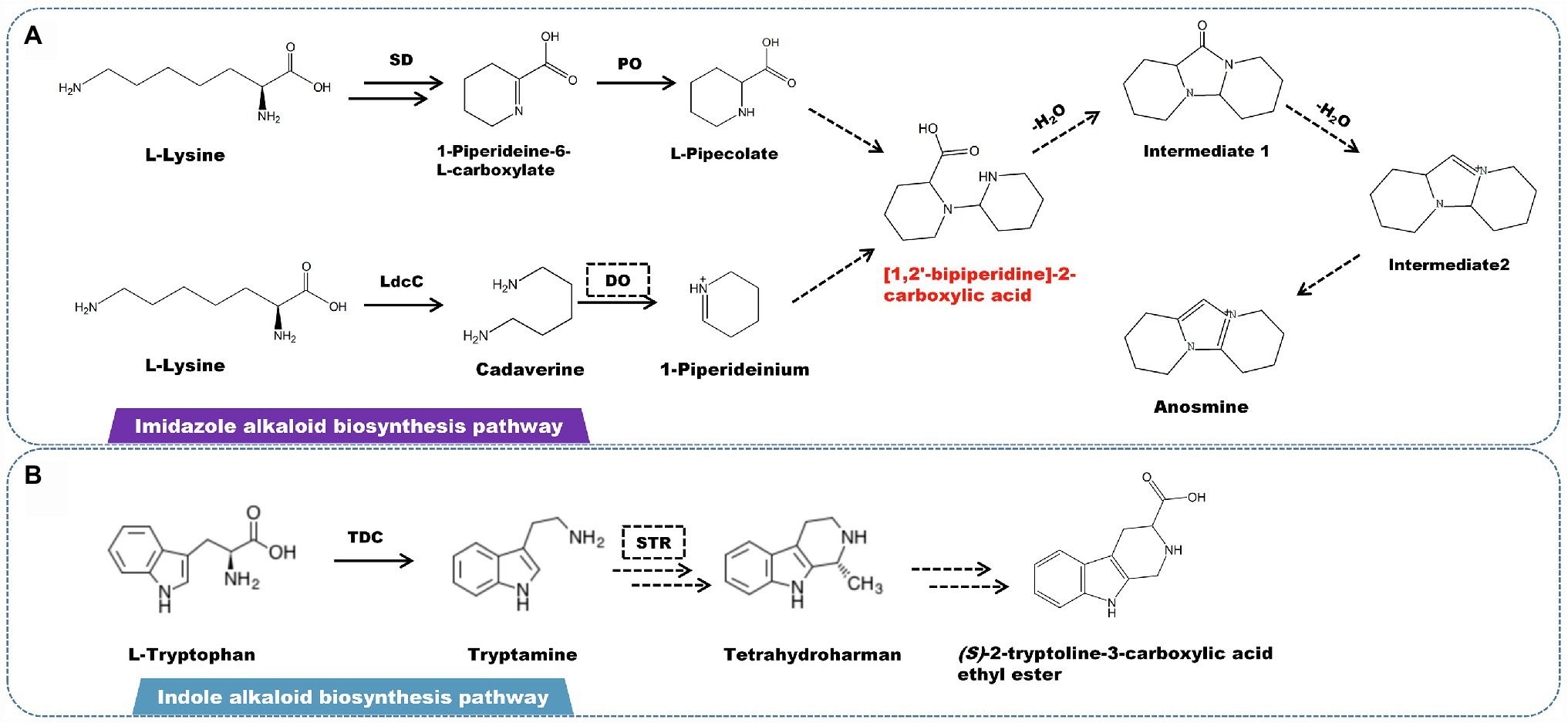
Figure 5. A plausible biosynthesis pathway of imidazole and indole alkaloids. (A) The imidazole alkaloid biosynthetic pathway. (B) The indole alkaloid biosynthetic pathway. The dashed box represents the putative enzymes. The dashed arrow represents the speculated pathway. The multiple arrows represent multi-step reactions. Saccharopine dehydrogenase (SD), L-pipecolate oxidase (PO), diamine oxidase (DO), lysine decarboxylase (LdcC), L-tryptophan decarboxylase (TDC), strictosidine synthase (STR). The precursor [1,2′-bipiperidine]-2-carboxylic acid is synthesized by linking L-pipecolate with 1-piperideinium derived from lysine.
Phthalide alkaloids mainly distributed in D. loddigesii (Li et al., 2019). The carbon skeleton structure is o-succinylbenzoate, and the shikimic acid/o-succinyl benzoate pathway is thought to be involved in the synthesis of this kind of alkaloid (Figure 6). O-succinylbenzoate (OSB) is also the precursor of menaquinones. A number of OSB-CoA synthetase family genes have been characterized in some bacteria (Lu et al., 2012). The isochorismic acid (derived from the shikimate pathway), α-ketoglutarate (originated from TCA cycle), and thiamine diphosphate (TPP) eventually form OSB under the successive reaction of 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1- carboxylic acid synthase (SEPHCHC synthase), 2-succinyl-6-hydroxy-2,4- cyclohexadiene-1-carboxylate synthase (SHCHC synthase) and o-succinylbenzoate synthetase (OSBS). This process lead to the release of one molecule of CO2 and one molecule of phosphoenolpyruvate (PEP), respectively (Thoden et al., 2004). Following that, OSB is reduced to aldehydes and ketones, which are subsequently catalyzed by aminotransferase to generate 2-(4-aminobutanoyl) benzoic acid. The carbonyl group undergoes a reduction and combines with the oxygen on the ortho carboxyl group to produce a lactone, which is accompanied by methylation on the nitrogen atom. Pierardine is the final outcome of the chemical reaction. Simultaneously, 2-(4-Aminobutanoyl)benzoic acid can be cyclized to form 2-(3,4-dihydro-2H-pyrrol-5-yl)benzoic acid. As a precursor, 2-(3,4-dihydro-2H-pyrrol-5-yl)benzoic acid is transformed into lactone by an addition reaction, followed by methylation on the nitrogen atom to yield shihunine (Chen et al., 2018). Shihunidine is structurally related to shihunine. It is hypothesized that the hydroxyl group on 2-(3,4-dihydro-2H-pyrrol-5-yl)benzoic acid is substituted by a phenyl group, which first becomes an aniline form and then cyclizes and methylates to produce shihunidine.
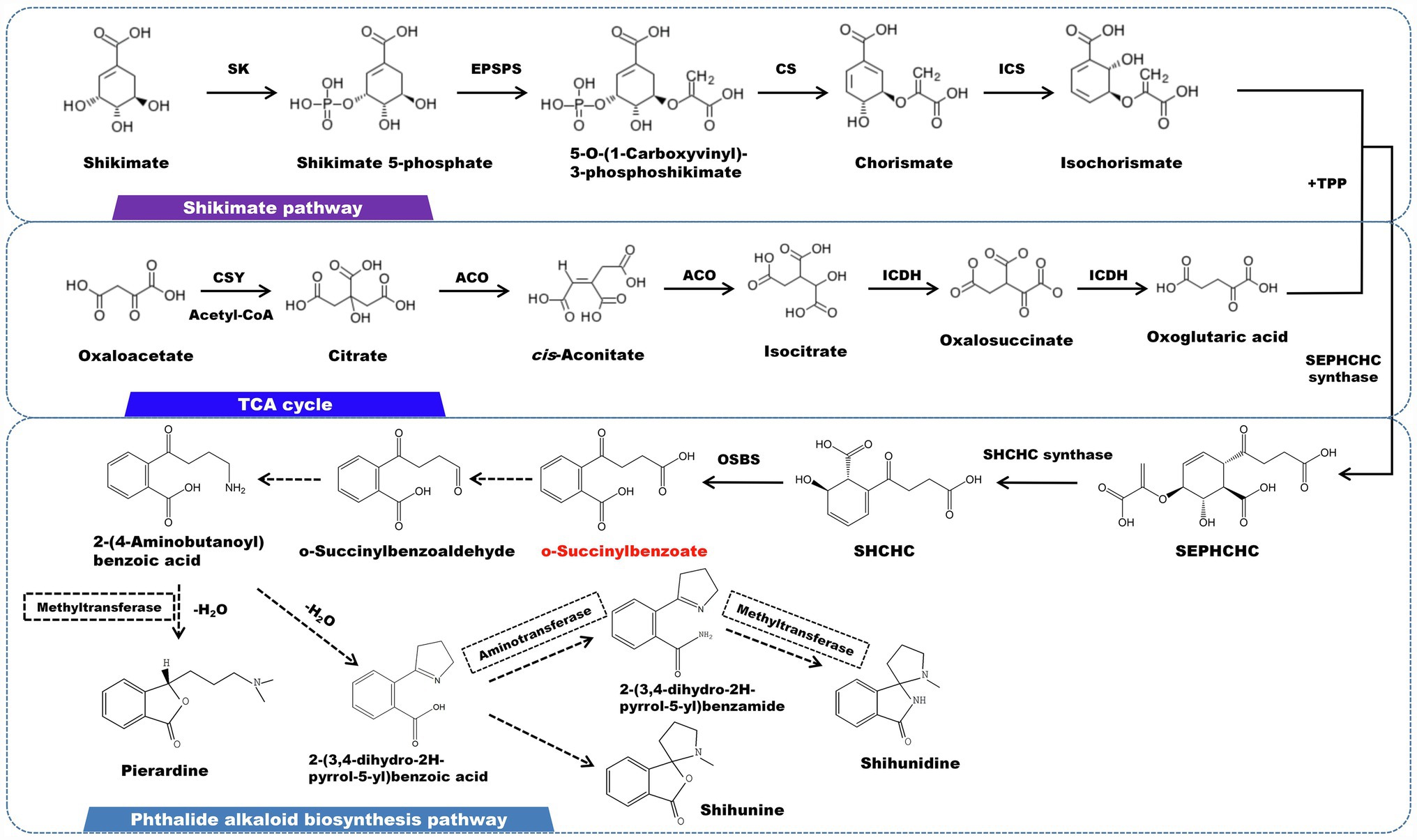
Figure 6. A plausible biosynthesis pathway of phthalide alkaloids. The dashed box represents the putative enzymes. The dashed arrow represents the speculated pathway. Shikimate kinase (SK), 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS), chorismate synthase (CS), isochorismate synthase (ICS), citrate synthase (CSY), aconitate hydratase (ACO), isocitrate dehydrogenase (ICDH),2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic acid synthase (SEPHCHC synthase), 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (SHCHC synthase), o-succinylbenzoate synthase (OSBS). O-succinylbenzoate served as the core structure for the biosynthesis of the phthalide alkaloid, which is produced via the catalysis of OSBS.
High-throughput sequencing has been widely employed in the discovery of novel genes and the investigation of gene structural variation in recent years (Balilashaki et al., 2019). By integrating two or more “omics,” the transcription regulators and hub genes that impact plant growth and secondary metabolism have been uncovered in many non-model plants (Kellner et al., 2015b; Saiman et al., 2018). D. officinale, D. huoshanense, D. chrysotoxum, and other varieties with significant medicinal benefit have undertaken de novo genome sequencing (Han et al., 2020; Niu et al., 2021; Zhang et al., 2021d). Combining with RNA-seq results, some researchers have systematically studied the regulatory mechanism of genes transcription and spatiotemporal characteristics under different conditions (Zheng et al., 2018; Yuan et al., 2020). The biosynthetic genes and metabolic pathways involved in Dendrobium alkaloids have been increasingly elucidated by the approaches described above (Zhang et al., 2021b). The genome of D. officinale has been upgraded in three versions, and the sequencing accuracy and assembly quality have massively improved (Yan et al., 2015; Zhang et al., 2016b; Niu et al., 2021). The polyneuridine-aldehyde esterase (PNAE) was tentatively identified based on the genomic annotation of D. officinale grown in Yunnan. Since PNAE is a crucial gene in the downstream pathway of monoterpene indole alkaloids, it is suggested that the alkaloid production of D. officinale may extend to the branch of 16-epivellosimine (Yan et al., 2015). Through KEGG functional enrichment on the transcriptome of D. officinale, 25 genes were identified as being involved in the formation of the alkaloid skeleton (Guo et al., 2013). The expression of five key enzyme genes, geraniol 10-hydroxylase (G10H), β-subunit of tryptophan synthase (TSB), tryptophan decarboxylase (TDC), secretoganin synthase (SCS), and strictosidine synthase (STR) suggested that monoterpene indole alkaloids are mainly produced in the leaves of D. officinale. Many studies have indicated that MeJA treatment could stimulate the expression of most genes in the MEP and MVA pathways, as well as the accumulation of total alkaloids (Jiao et al., 2018; Chen et al., 2019). A comparative transcriptome analysis of D. officinale tissues and protocorm-like bodies showed 42 genes involved in the alkaloid biosynthesis pathway, and dozens of alkaloid-coding genes were identified. The expressions of some aminotransferases and CYP450 genes were significantly higher in protocorm-like bodies than in leaves, and the total alkaloid content in protocorm-like bodies likewise exhibited a similar accumulation (Wang et al., 2021). The qRT-PCR method was used to examine the expression levels of monoterpene indole alkaloid downstream pathway genes STR, strictosidine-D-glucosidase (SG), PNAE, and vinorine synthase (VS), which also revealed that the expressions of these genes are higher in protocorm-like bodies than in leaves. Transcriptome sequencing on D. officinale yielded a total of 2,115 unigenes related to secondary metabolism, including 4 sequences related to the indole alkaloids biosynthetic pathway, 34 sequences related to isoquinoline alkaloids biosynthetic pathway, and 38 sequences related to tropine, pyridine, and pyridine alkaloids biosynthesis (Zhang et al., 2016b). Previous studies had indicated that DoSTR was localized in vacuoles and that its expression was regulated by hormones, such as MeJA and ABA (Zhu et al., 2020). Recent studies generated a high-quality D. officinale genome at the chromosomal level, demonstrating that the sequencing data contained a contig N50 of 1.44 Mb and a Hi-C anchored rate of 93.5% (Niu et al., 2021). A total of 98 alkaloid-related genes were identified, including 56 genes encoding 25 enzymes associated with the biosynthesis of sesquiterpene alkaloids, terpenoid indole alkaloids, and their upstream pathways, including the shikimate, MVA, and MEP pathways. These genes comprise primary amine oxidase (AOC), aspartate aminotransferase (AAT), tyrosine aminotransferase (TAT), tyrosine decarboxylase (TDC), 1-benzyl-1,2,3,4-tetrahydroisoquinoline N-methyltransferase (CNMT), norbelladine O-methyltransferase (N4OMT), tropinone reductase I (TR1), PNAE, etc. Transcriptome sequencing of D. nobile co-cultured with the endophytic fungus MF23 showed that a total of 16 genes were involved in the synthesis of terpenoid backbone or sesquiterpene alkaloids (Li et al., 2017b). The expression levels of PMK and MVD increased in the ninth week of co-cultivation, which was consistent with the accumulation pattern of dendrobine. The expression of TPS21 is negatively correlated with the biosynthesis of dendrobine, and the expression of AACT is positively correlated with the biosynthesis of dendrobine. Further analysis of 9 post-modification enzymes found that the expression levels of CYP1D10, METTL23, ATX4, and BCAT2 were higher than other culture stages after 9 weeks inoculation. The expression of ATX4 was at a low level at 6 weeks and was activated at 9 weeks. It demonstrates that ATX4 is essential in the modulation of the biosynthesis of Dendrobium alkaloids that is altered by MF23. Besides the crucial genes, some transcription factors have important regulatory roles in stress responses and secondary metabolite production in Dendrobium plants (Zhang et al., 2021d). DcTCP4 and DcTCP9 are involved in JA-dependent leaf developmental processes (Zhang et al., 2021b). In D. catenatum, the bHLH-MYB-WD40 module is deeply engaged in anthocyanin biosynthesis. DcTT8, a IIIf bHLH transcription factor, strongly regulates the expression of the anthocyanin-related genes F3’H and UFGT (Jia et al., 2021). By binding to the promoters of CHS, CHI, and F3H, a light-induced WD40-repeat TF DoTTG1 regulates their expression and promotes anthocyanin formation (Jia et al., 2021). MYB2 could interact with bHLH1 to enhance the expressions of DFR and ANS (Li et al., 2017a). WRKY TFs are well-studied transcriptional regulators that are mainly involved in the stress of D. officinale and the production of active components. Some WRKY genes commonly activated by cold stress and MeJA treatment, while others may be involved in polysaccharide biosynthesis and hydrolysis (He et al., 2017; Wang et al., 2018). DoMYB25 participated in the positive regulation of water-soluble polysaccharides (He et al., 2019). DcWRKY22, DcWRKY36, and DcWRKY45 are involved in plant drought, cold, and salt stress (Zhang et al., 2022). DobHLHs are induced by ABA and MeJA treatments. DobHLH4 positively regulates the expression of TPS10, therefore boosting linalool production (Wang and Liu, 2020; Yu et al., 2021). So far, there are limited reports on transcription factors that regulate dendrobium alkaloids biosynthesis. We previously isolated a MYC2 transcription factor from D. officinale. Overexpression of DoMYC2 in A. thaliana reduces the expression of HMGR2, FPS1, and FPS2, implying that DoMYC2 functions as a negative regulator to regulate the expression of JA-responsive genes. DoMYC2 typically binds to the E-box in the promoters to influence transcription (Zhu et al., 2017). We highlight the classification of alkaloids and main enzyme coding genes of different Dendrobium species found by high-throughput sequencing in recent years (Table 2).
Next generation sequencing technology provided a strong foundation for the acquisition of high-quality genome datasets of the medicinal plants, which facilitated the investigation of crucial genes in the secondary metabolism (Goossens and Rischer, 2007; Liu et al., 2020b). However, the industry still faces constraints, such as lack of raw materials and a long cultivated period. Meanwhile, alkaloid contents in most Dendrobium spp. are generally limited, which makes it tricky to obtain a large quantity of alkaloids rapidly. The utilization of microbial fermentation plants and synthetic biology can provide technological support for the secondary development and sustainable utilization of Dendrobium resources (Ehrenworth and Peralta-Yahya, 2017; Li et al., 2018a). Based on existing studies, there are two main technological methods for using synthetic biology in natural products: one is the heterologous expression of synthetic gene clusters of natural products in suitable chassis cells (Ro et al., 2006; Dangel et al., 2010; Gwak et al., 2017); the other is to introduce positive regulatory regulatory elements or delete negative regulatory elements in the original host to achieve regulation or overexpression of related genes (Zhuang et al., 2017; Ma et al., 2018). Since the synthetic route of natural products is generally extended, the problem of heavy metabolic load and low yield of single bacteria are often stressful to overcome. The exploration of multicellular systems provides flexibility and personalization for the production of such complex substances (Li et al., 2018b). By rationally designing and constructing an artificial multi-cell culture system, the metabolic pathways could be dispersed and assembled into multiple independent cells, which can lower the metabolic burden of a single bacteria. The optimal module combination could be achieved by engineering and maximizing the metabolic capacity of a single chassis cell (Zhang and Wang, 2016). The goal of synthetic biology has gradually evolved from the synthesis of biological components and devices (the first stage of synthetic biology) to the construction of multicellular life systems (the second stage of synthetic biology; Paddon et al., 2013; Ma et al., 2019b). With the gradual development of gene editing and de novo synthesis technologies, the chassis of synthetic biology is becoming more abundant, mainly including microbial chassis (Saccharomyces cerevisiae and Escherichia coli, etc.) and plant chassis (tobacco, suspension cells, hairy roots, etc.). The heterologous synthesis of iridoids, isoflavin, and vinblastine in yeast and the heterologous synthesis of triterpenoids, such as artemisinin and its precursors, β-amyrin and oleanolic acid in tobacco, provide scientific reference for in vitro synthesis of dendrobium alkaloids (Farhi et al., 2011; Miettinen et al., 2014; Brown et al., 2015; Fuentes et al., 2016; Malhotra et al., 2016; Kumar et al., 2018; Saiman et al., 2018).
Recent research found that specialized metabolic enzymes usually clustered in a few chromosomes, forming metabolic gene clusters. These gene clusters are widely present in the biosynthesis pathways of secondary metabolites in dicotyledonous and monocotyledonous plants (Nützmann et al., 2016). Generally, genes in some gene clusters are arranged continuously and tightly on chromosomes (Polturak and Osbourn, 2021). The thaliano-synthetic gene cluster in over 80% of Arabidopsis ecotypes, the acylated tyramine gene cluster in rice, and the diterpene casbene gene cluster present a continuous arrangement (Zhan et al., 2020; Shen et al., 2021). Some gene clusters are scattered in distant areas, such as the genes that synthesize the sterol alkaloid α-solanine in tomato clusters on chromosome 7 (Itkin et al., 2013). This encompasses two upstream pathway genes, GAME11 and GAME6, as well as four glycosyltransferase genes, GAME1, GAME17, GAME18, and GAME2. The other two structural genes, GAME12 and GAME4, which encode aminotransferases, are adjacent to each other on chromosome 12. Based on genome and transcriptome co-expression analysis, it is feasible to carry out initial mining of gene clusters of specific metabolic pathways (Jeon et al., 2020; Liu et al., 2020c). Some software and algorithms are capable of rapidly identifying potential gene clusters in non-model plants. Plant Cluster Finder is a more developed tool that has studied 18 plants and identified approximately 12,000 possible biosynthetic gene clusters (Schläpfer et al., 2017). PlantiSMASH and PhytoClust employ a more accurate hidden Markov model to identify distinct biosynthetic enzymes and integrate the gene locations to predict candidate gene clusters (Kautsar et al., 2017; Töpfer et al., 2017). It should be noted that Dendrobium alkaloids are mostly composed of TIAs and sesquiterpene alkaloids (Table 1). By altering particular critical enzymes in the MEP and MVA pathways, heterologous expression strategies can supply richer terpene precursors for the production of these two kinds of alkaloids (Zhu et al., 2014).
With the rapid development of sequencing technology and various “omics” technologies, plant-specific metabolic biosynthesis has rapidly progressed from small-scale sequencing and functional identification of individual genes to the large-scale sequencing and comparative genomics studies. Transcriptomics and metabolomics-associated analysis, including co-expression and co-response analysis, have been successfully employed in the functional identification of unknown genes in many non-model plants. The completion of high-quality genome sequencing and re-sequencing of natural populations has also provided considerable prospects for the exploration of crop metabolic pathways and valuable Chinese herbal medicines. The metabolome genome-wide association study (mGWAS) is a new approach to decipher in-depth analyses of complicated metabolic pathways and their regulatory mechanisms, as well as other basic theoretical studies. However, this technology is rarely applied in the research of active specific metabolites of medicinal plants. The main reason for this is that medicinal plants, unlike commercial crops, do not typically have systematic classification and whole germplasm lines. Synthetic biology is the design or modification of living systems at the molecular level to meet certain purposes. Plants can transform light energy and CO2 from the environment into their own energy and carbon sources. The use of synthetic biology to rebuild metabolic networks in plant cells and reroute carbon flow to more useful plant-specific metabolites has enormous potential. The convergence of cutting-edge technologies, such as improved genome editing, sophisticated gene module assembly, tobacco transient expression technology, rapid and efficient genetic modification with magnetic nanoparticles as carriers, and computer-aided design, will propel synthetic biology into the next era in the future.
CS, QJ, and YC discussed the writing plan. CS and JM drafted the manuscript. CS, JM, GL, HP, and YZ edited the manuscript. BH and QJ acquired the funding. All authors have read, reviewed, and approved the submitted version.
This work was supported by China Agricultural Research System of MOF and MARA. The authors are also grateful for the financial support of National Industry Technology System of Traditional Chinese Medicine (CARS-21), Anhui University Collaborative Innovation Project (GXXT-2019-043 and GXXT-2019-049), High-level Talents Research Initiation Funding Project of West Anhui University (WGKQ2022025), and Natural Science Foundation of Anhui (2108085MC80).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Balilashaki, K., Zakizadeh, H., Olfati, J. A., Vahedi, M., Kumar, A., and Indracanti, M. (2019). Recent advances in phalaenopsis orchid improvement using omics approaches. Plant Tissue Cult. Biotechnol. 29, 133–149. doi: 10.3329/ptcb.v29i1.41986
Brown, S., Clastre, M., Courdavault, V., and O’Connor, S. E. (2015). De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. U. S. A. 112, 3205–3210. doi: 10.1073/pnas.1423555112
Cakova, V., Bonte, F., and Lobstein, A. (2017). Dendrobium: sources of active ingredients to treat age-related pathologies. Aging Dis. 8, 827–849. doi: 10.14336/AD.2017.0214
Cao, H., Ji, Y., Li, S., Lu, L., Tian, M., Yang, W., et al. (2019). Extensive metabolic profiles of leaves and stems from the medicinal plant Dendrobium officinale kimura et migo. Meta 9, 13–18. doi: 10.3390/metabo9100215
Chang, C. C., Ku, A. F., Tseng, Y. Y., Yang, W. B., Fang, J. M., and Wong, C. H. (2010). 6,8-Di-C-glycosyl flavonoids from dendrobium huoshanense. J. Nat. Prod. 73, 229–232. doi: 10.1021/np900252f
Chen, W., Cheng, X., Zhou, Z., Liu, J., and Wang, H. (2013). Molecular cloning and characterization of a tropinone reductase from Dendrobium nobile Lindl. Mol. Biol. Rep. 40, 1145–1154. doi: 10.1007/s11033-012-2156-0
Chen, H., Li, X., Xu, Y., Lo, K., Zheng, H., Hu, H., et al. (2018). Study on the polar extracts of Dendrobium nobile, D. officinale, D. loddigesii, and flickingeria fimbriata: metabolite identification, content evaluation, and bioactivity assay. Molecules 23:1185. doi: 10.3390/molecules23051185
Chen, Y., Wang, Y., Lyu, P., Chen, L., Shen, C., and Sun, C. (2019). Comparative transcriptomic analysis reveal the regulation mechanism underlying MeJA-induced accumulation of alkaloids in Dendrobium officinale. J. Plant Res. 132, 419–429. doi: 10.1007/s10265-019-01099-6
Cheng, X., Chen, W., Zhou, Z., Liu, J., and Wang, H. (2013). Functional characterization of a novel tropinone reductase-like gene in Dendrobium nobile Lindl. J. Plant Physiol. 170, 958–964. doi: 10.1016/j.jplph.2013.02.007
Dangel, V., Westrich, L., Smith, M. C. M., Heide, L., and Gust, B. (2010). Use of an inducible promoter for antibiotic production in a heterologous host. Appl. Microbiol. Biotechnol. 87, 261–269. doi: 10.1007/s00253-009-2435-4
Ehrenworth, A. M., and Peralta-Yahya, P. (2017). Accelerating the semisynthesis of alkaloid-based drugs through metabolic engineering. Nat. Chem. Biol. 13, 249–258. doi: 10.1038/nchembio.2308
Facchini, P. J. (2001). Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu. Rev. Plant Biol. 52, 29–66. doi: 10.1146/annurev.arplant.52.1.29
Facchini, P. J., and St-Pierre, B. (2005). Synthesis and trafficking of alkaloid biosynthetic enzymes. Curr. Opin. Plant Biol. 8, 657–666. doi: 10.1016/j.pbi.2005.09.008
Fan, H., Wu, Q., Wang, X., Wu, L., Cai, Y., and Lin, Y. (2016). Molecular cloning and expression of 1-deoxy-d-xylulose-5-phosphate synthase and 1-deoxy-d-xylulose-5-phosphate reductoisomerase in Dendrobium officinale. Plant Cell Tissue Organ Cult. 125, 381–385. doi: 10.1007/s11240-016-0945-1
Farhi, M., Marhevka, E., Ben-Ari, J., Algamas-Dimantov, A., Liang, Z., Zeevi, V., et al. (2011). Generation of the potent anti-malarial drug artemisinin in tobacco. Nat. Biotechnol. 29, 1072–1074. doi: 10.1038/nbt.2054
Fuentes, P., Zhou, F., Erban, A., Karcher, D., Kopka, J., and Bock, R. (2016). A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. elife 5, 1–26. doi: 10.7554/eLife.13664
Goossens, A., and Rischer, H. (2007). Implementation of functional genomics for gene discovery in alkaloid producing plants. Phytochem. Rev. 6, 35–49. doi: 10.1007/s11101-006-9018-0
Guo, L., Frey, W., and Plietker, B. (2018a). Catalytic enantioselective total synthesis of the Picrotoxane alkaloids (-)-Dendrobine, (-)-Mubironine B, and (-)-Dendroxine. Org. Lett. 20, 4328–4331. doi: 10.1021/acs.orglett.8b01782
Guo, X., Li, Y., Li, C., Luo, H., Wang, L., Qian, J., et al. (2013). Analysis of the Dendrobium officinale transcriptome reveals putative alkaloid biosynthetic genes and genetic markers. Gene 527, 131–138. doi: 10.1016/j.gene.2013.05.073
Guo, L., Winzer, T., Yang, X., Li, Y., Ning, Z., He, Z., et al. (2018b). The opium poppy genome and morphinan production. Science 347, 343–347. doi: 10.1126/science.aat4096
Gwak, Y. S., Han, J. Y., Adhikari, P. B., Ahn, C. H., and Choi, Y. E. (2017). Heterologous production of a ginsenoside saponin (compound K) and its precursors in transgenic tobacco impairs the vegetative and reproductive growth. Planta 245, 1105–1119. doi: 10.1007/s00425-017-2668-x
Han, B., Jing, Y., Dai, J., Zheng, T., Gu, F., Zhao, Q., et al. (2020). A chromosome-level genome assembly of Dendrobium Huoshanense using long reads and Hi-C data. Genome Biol. Evol. 12, 2486–2490. doi: 10.1093/gbe/evaa215
He, C., Teixeira da Silva, J. A., Tan, J., Zhang, J., Pan, X., Li, M., et al. (2017). A genome-wide identification of the WRKY family genes and a survey of potential WRKY target genes in Dendrobium officinale. Sci. Rep. 7, 1–14. doi: 10.1038/s41598-017-07872-8
He, C., Teixeira da Silva, J. A., Wang, H., Si, C., Zhang, M., Zhang, X., et al. (2019). Mining MYB transcription factors from the genomes of orchids (Phalaenopsis and Dendrobium) and characterization of an orchid R2R3-MYB gene involved in water-soluble polysaccharide biosynthesis. Sci. Rep. 9, 13818–13819. doi: 10.1038/s41598-019-49812-8
Hemscheidt, T., and Spenser, I. D. (1993). Biosynthesis of anosmine: incorporation of the intact six-carbon chain of lysine and of pipecolic acid. J. Nat. Prod. 56, 1281–1287. doi: 10.1021/np50098a012
Huang, S., Wu, Q., Liu, H., Ling, H., He, Y., Wang, C., et al. (2019). Alkaloids of dendrobium nobile lindl. altered hepatic lipid homeostasis via regulation of bile acids. J. Ethnopharmacol. 241:111976. doi: 10.1016/j.jep.2019.111976
Hu, Y., Ren, J., Wang, L., Zhao, X., Zhang, M., Shimizu, K., et al. (2018). Protective effects of total alkaloids from Dendrobium crepidatum against LPS-induced acute lung injury in mice and its chemical components. Phytochemistry 149, 12–23. doi: 10.1016/j.phytochem.2018.02.006
Hu, Y., Yang, H., Ding, X., Liu, J., Wang, X., Hu, L., et al. (2020). Anti-inflammatory octahydroindolizine alkaloid enantiomers from Dendrobium crepidatum. Bioorg. Chem. 100:103809. doi: 10.1016/j.bioorg.2020.103809
Hu, Y., Zhang, C., Zhao, X., Wang, Y., Feng, D., Zhang, M., et al. (2016). (±)-Homocrepidine A, a pair of anti-inflammatory enantiomeric octahydroindolizine alkaloid dimers from Dendrobium crepidatum. J. Nat. Prod. 79, 252–256. doi: 10.1021/acs.jnatprod.5b00801
Itkin, M., Heinig, U., Tzfadia, O., Bhide, A. J., Shinde, B., Cardenas, P. D., et al. (2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341, 175–179. doi: 10.1126/science.1240230
Jeon, J. E., Kim, J. G., Fischer, C. R., Mehta, N., Dufour-Schroif, C., Wemmer, K., et al. (2020). A pathogen-responsive gene cluster for highly modified fatty acids in tomato. Cell 180, 176–187.e19. doi: 10.1016/j.cell.2019.11.037
Jia, N., Wang, J. J., Liu, J., Jiang, J., Sun, J., Yan, P., et al. (2021). DcTT8, a bHLH transcription factor, regulates anthocyanin biosynthesis in Dendrobium candidum. Plant Physiol. Biochem. 162, 603–612. doi: 10.1016/j.plaphy.2021.03.006
Jia, N., Wang, J., Wang, Y., Ye, W., Liu, J., Jiang, J., et al. (2021). The light-induced WD40-repeat transcription factor DcTTG1 regulates anthocyanin biosynthesis in Dendrobium candidum. Front. Plant Sci. 12:633333. doi: 10.3389/fpls.2021.633333
Jiao, C., Song, C., Zheng, S., Zhu, Y., Jin, Q., Cai, Y., et al. (2018). Metabolic profiling of Dendrobium officinale in response to precursors and methyl jasmonate. Int. J. Mol. Sci. 19:728. doi: 10.3390/ijms19030728
Jin, Q., Jiao, C., Sun, S., Song, C., Cai, Y., Lin, Y., et al. (2016). Metabolic analysis of medicinal Dendrobium officinale and Dendrobium huoshanense during different growth years. PLoS One 11. doi: 10.1371/journal.pone.0146607
Kang, M., Fu, R., Zhang, P., Lou, S., Yang, X., Chen, Y., et al. (2021). A chromosome-level Camptotheca acuminata genome assembly provides insights into the evolutionary origin of camptothecin biosynthesis. Nat. Commun. 12, 3531–3512. doi: 10.1038/s41467-021-23872-9
Kautsar, S. A., Suarez Duran, H. G., Blin, K., Osbourn, A., and Medema, M. H. (2017). PlantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 45, W55–W63. doi: 10.1093/nar/gkx305
Kellner, F., Geu-Flores, F., Sherden, N. H., Brown, S., Foureau, E., Courdavault, V., et al. (2015a). Discovery of a P450-catalyzed step in vindoline biosynthesis: a link between the aspidosperma and eburnamine alkaloids. Chem. Commun. 51, 7626–7628. doi: 10.1039/c5cc01309g
Kellner, F., Kim, J., Clavijo, B. J., Hamilton, J. P., Childs, K. L., Vaillancourt, B., et al. (2015b). Genome-guided investigation of plant natural product biosynthesis. Plant J. 82, 680–692. doi: 10.1111/tpj.12827
Khosravi, E., Mousavi, A., Farhadpour, M., Ghashghaie, J., Ghanati, F., and Haghbeen, K. (2019). Pyrrolizidine alkaloids-free extract from the cell culture of Lithospermum officinale with high antioxidant capacity. Appl. Biochem. Biotechnol. 187, 744–752. doi: 10.1007/s12010-018-2830-3
Kishimoto, S., Sato, M., Tsunematsu, Y., and Watanabe, K. (2016). Evaluation of biosynthetic pathway and engineered biosynthesis of alkaloids. Molecules 21:1078. doi: 10.3390/molecules21081078
Kiss, T., Borcsa, B., Orvos, P., Tálosi, L., Hohmann, J., and Csupor, D. (2017). Diterpene lipo-alkaloids with selective activities on cardiac K + channels. Planta Med. 83, 1321–1328. doi: 10.1055/s-0043-109556
Kobayashi, T., Hasegawa, F., Hirose, Y., Tanaka, K., Mori, H., and Katsumura, S. (2012). Stereocontrolled synthesis of substituted chiral piperidines via one-pot asymmetric 6π-azaelectrocyclization: asymmetric syntheses of (−)-dendroprimine, (+)-7-epidendroprimine, (+)-5-epidendroprimine, and (+)-5,7-epidendroprimine. J. Org. Chem. 77, 1812–1832. doi: 10.1021/jo202350z
Kreis, L. M., and Carreira, E. M. (2012). Total synthesis of (−)-Dendrobine. Angew. Chemie 124, 3492–3495. doi: 10.1002/ange.201108564
Kumar, S. R., Shilpashree, H. B., and Nagegowda, D. A. (2018). Terpene moiety enhancement by overexpression of geranyl(Geranyl) diphosphate synthase and geraniol synthase elevates monomeric and dimeric monoterpene indole alkaloids in transgenic Catharanthus roseus. Front. Plant Sci. 9:942. doi: 10.3389/fpls.2018.00942
Lam, Y., Ng, T. B., Yao, R. M., Shi, J., Xu, K., Sze, S. C. W., et al. (2015). Evaluation of chemical constituents and important mechanism of pharmacological biology in Dendrobium plants. Evidence-based Complement. Altern. Med. 2015, 1–25. doi: 10.1155/2015/841752
Li, Q., Ding, G., Li, B., and Guo, S. X. (2017b). Transcriptome analysis of genes involved in dendrobine biosynthesis in Dendrobium nobile Lindl. infected with mycorrhizal fungus MF23 (Mycena sp.). Sci. Rep. 7, 1–16. doi: 10.1038/s41598-017-00445-9
Li, N., Dong, Y., Lv, M., Qian, L., Sun, X., Liu, L., et al. (2021). Combined analysis of volatile terpenoid metabolism and transcriptome reveals transcription factors related to terpene synthase in two cultivars of Dendrobium officinale flowers. Front. Genet. 12:661296. doi: 10.3389/fgene.2021.661296
Li, X. W., Huang, M., Lo, K., Chen, W. L., He, Y. Y., Xu, Y., et al. (2019). Anti-diabetic effect of a shihunine-rich extract of Dendrobium loddigesii on 3t3-l1 cells and db/db mice by up-regulating AMPK–GLUT4–PPARα. Molecules 24, 1–20. doi: 10.3390/molecules24142673
Li, S., Li, Y., and Smolke, C. D. (2018a). Strategies for microbial synthesis of high-value phytochemicals. Nat. Chem. 10, 395–404. doi: 10.1038/s41557-018-0013-z
Li, Y., Li, F., Gong, Q., Wu, Q., and Shi, J. (2011). Inhibitory effects of dendrobium alkaloids on memory impairment induced by lipopolysaccharide in rats. Planta Med. 77, 117–121. doi: 10.1055/s-0030-1250235
Li, Y., Li, S., Thodey, K., Trenchard, I., Cravens, A., and Smolke, C. D. (2018b). Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U. S. A. 115, E3922–E3931. doi: 10.1073/pnas.1721469115
Li, R., Liu, T., Liu, M., Chen, F., Liu, S., and Yang, J. (2017c). Anti-influenza A virus activity of dendrobine and its mechanism of action. J. Agric. Food Chem. 65, 3665–3674. doi: 10.1021/acs.jafc.7b00276
Li, C., Qiu, J., Ding, L., Huang, M., Huang, S., Yang, G., et al. (2017a). Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals. Plant Physiol. Biochem. 112, 335–345. doi: 10.1016/j.plaphy.2017.01.019
Lichman, B. R. (2021). The scaffold-forming steps of plant alkaloid biosynthesis. Nat. Prod. Rep. 38, 103–129. doi: 10.1039/d0np00031k
Liu, Z., Cheema, J., Vigouroux, M., Hill, L., Reed, J., Paajanen, P., et al. (2020c). Formation and diversification of a paradigm biosynthetic gene cluster in plants. Nat. Commun. 11, 1–11. doi: 10.1038/s41467-020-19153-6
Liu, Y., Cui, Y., Lu, L., Gong, Y., Han, W., and Piao, G. (2020b). Natural indole-containing alkaloids and their antibacterial activities. Arch. Pharm. 353, e2000120–e2000110. doi: 10.1002/ardp.202000120
Liu, W. H., Hua, Y. F., and Zhan, Z. J. (2007). Moniline, a new alkaloid from Dendrobium moniliforme. J. Chem. Res. 38, 317–318. doi: 10.3184/030823407X218048
Liu, G. Y., Tan, L., Cheng, L., Ding, L. S., Zhou, Y., Deng, Y., et al. (2020a). Dendrobine-type alkaloids and bibenzyl derivatives from Dendrobium findlayanum. Fitoterapia 142:104497. doi: 10.1016/j.fitote.2020.104497
Lu, X., Zhou, R., Sharma, I., Li, X., Kumar, G., Swaminathan, S., et al. (2012). Stable analogues of OSB-AMP: potent inhibitors of MenE, the o-Succinylbenzoate-CoA Synthetase from bacterial Menaquinone biosynthesis. Chembiochem 13, 129–136. doi: 10.1002/cbic.201100585
Ma, C., Meng, C. W., Zhou, Q. M., Peng, C., Liu, F., Zhang, J. W., et al. (2019a). New sesquiterpenoids from the stems of Dendrobium nobile and their neuroprotective activities. Fitoterapia 138:104351. doi: 10.1016/j.fitote.2019.104351
Ma, D., Reichelt, M., Yoshida, K., Gershenzon, J., and Constabel, C. P. (2018). Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 96, 949–965. doi: 10.1111/tpj.14081
Ma, T., Shi, B., Ye, Z., Li, X., Liu, M., Chen, Y., et al. (2019b). Lipid engineering combined with systematic metabolic engineering of Saccharomyces cerevisiae for high-yield production of lycopene. Metab. Eng. 52, 134–142. doi: 10.1016/j.ymben.2018.11.009
Malhotra, K., Subramaniyan, M., Rawat, K., Kalamuddin, M., Qureshi, M. I., Malhotra, P., et al. (2016). Compartmentalized metabolic engineering for artemisinin biosynthesis and effective malaria treatment by oral delivery of plant cells. Mol. Plant 9, 1464–1477. doi: 10.1016/j.molp.2016.09.013
Meng, C. W., He, Y. L., Peng, C., Ding, X. J., Guo, L., and Xiong, L. (2017). Picrotoxane sesquiterpenoids from the stems of Dendrobium nobile and their absolute configurations and angiogenesis effect. Fitoterapia 121, 206–211. doi: 10.1016/j.fitote.2017.07.017
Miettinen, K., Dong, L., Navrot, N., Schneider, T., Burlat, V., Pollier, J., et al. (2014). The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 5:3606. doi: 10.1038/ncomms4606
Morita, H., Fujiwara, M., Yoshida, N., and Kobayashi, J. (2000). New Picrotoxinin-type and Dendrobine-type Sesquiterpenoids from Dendrobium Snowflake ‘Red Star’. Tetrahedron 56, 5801–5805. doi: 10.1016/S0040-4020(00)00530-5
Mou, Z., Zhao, Y., Ye, F., Shi, Y., Kennelly, E. J., Chen, S., et al. (2021). Identification, biological activities and biosynthetic pathway of Dendrobium alkaloids. Front. Pharmacol. 12:605994. doi: 10.3389/fphar.2021.605994
Ng, T. B., Liu, J., Wong, J. H., Ye, X., Wing Sze, S. C., Tong, Y., et al. (2012). Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 93, 1795–1803. doi: 10.1007/s00253-011-3829-7
Nguyen, T. K. O., Jamali, A., Lanoue, A., Gontier, E., and Dauwe, R. (2015). Unravelling the architecture and dynamics of tropane alkaloid biosynthesis pathways using metabolite correlation networks. Phytochemistry 116, 94–103. doi: 10.1016/j.phytochem.2015.03.005
Nie, J., Jiang, L. S., Zhang, Y., Tian, Y., Li, L. S., Lu, Y. L., et al. (2018). Dendrobium nobile Lindl. Alkaloids decreases the level of intracellular β-amyloid by improving impaired Autolysosomal proteolysis in APP/PS1 mice. Front. Pharmacol. 9:1479. doi: 10.3389/fphar.2018.01479
Nie, J., Tian, Y., Zhang, Y., Lu, Y. L., Li, L. S., and Shi, J. S. (2016). Dendrobium alkaloids prevent aß25-35-induced neuronal and synaptic loss via promoting neurotrophic factors expression in mice. PeerJ 4, 1–16. doi: 10.7717/peerj.2739
Niu, Z., Zhu, F., Fan, Y., Li, C., Zhang, B., Zhu, S., et al. (2021). The chromosome-level reference genome assembly for Dendrobium officinale and its utility of functional genomics research and molecular breeding study. Acta Pharm. Sin. B 11, 2080–2092. doi: 10.1016/j.apsb.2021.01.019
Nützmann, H. W., Huang, A., and Osbourn, A. (2016). Plant metabolic clusters – from genetics to genomics. New Phytol. 211, 771–789. doi: 10.1111/nph.13981
Ohc, C., and Claisen, I. (2012). Synthesis of (−)-Dendrobine. Synfacts 8:592. doi: 10.1055/s-0031-1291052
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., McPhee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi: 10.1038/nature12051
Polturak, G., and Osbourn, A. (2021). The emerging role of biosynthetic gene clusters in plant defense and plant interactions. PLoS Pathog. 17, e1009698–e1009611. doi: 10.1371/journal.ppat.1009698
Pressnitz, D., Fischereder, E. M., Pletz, J., Kofler, C., Hammerer, L., Hiebler, K., et al. (2018). Asymmetric synthesis of (R)-1-alkyl-substituted Tetrahydro-ß-carbolines catalyzed by Strictosidine synthases. Angew. Chem. Int. Ed. 57, 10683–10687. doi: 10.1002/anie.201803372
Ro, D. K., Paradise, E. M., Quellet, M., Fisher, K. J., Newman, K. L., Ndungu, J. M., et al. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943. doi: 10.1038/nature04640
Saiman, M. Z., Miettinen, K., Mustafa, N. R., Choi, Y. H., Verpoorte, R., and Schulte, A. E. (2018). Metabolic alteration of Catharanthus roseus cell suspension cultures overexpressing geraniol synthase in the plastids or cytosol. Plant Cell Tissue Organ Cult. 134, 41–53. doi: 10.1007/s11240-018-1398-5
Schläpfer, P., Zhang, P., Wang, C., Kim, T., Banf, M., Chae, L., et al. (2017). Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 173, 2041–2059. doi: 10.1104/pp.16.01942
Seigler, D. S. (1998). “Pyrrolizidine, quinolizidine, and indolizidine alkaloids,” Plant Secondary Metabolism. Boston, MA: Springer.
Sharma, A., Verma, P., Mathur, A., and Mathur, A. K. (2018). Overexpression of tryptophan decarboxylase and strictosidine synthase enhanced terpenoid indole alkaloid pathway activity and antineoplastic vinblastine biosynthesis in Catharanthus roseus. Protoplasma 255, 1281–1294. doi: 10.1007/s00709-018-1233-1
Shen, C., Guo, H., Chen, H., Shi, Y., Meng, Y., Lu, J., et al. (2017). Identification and analysis of genes associated with the synthesis of bioactive constituents in Dendrobium officinale using RNA-Seq. Sci. Rep. 7, 187–111. doi: 10.1038/s41598-017-00292-8
Shen, S., Peng, M., Fang, H., Wang, Z., Zhou, S., Jing, X., et al. (2021). An Oryza-specific hydroxycinnamoyl tyramine gene cluster contributes to enhanced disease resistance. Sci. Bull. 66, 2369–2380. doi: 10.1016/j.scib.2021.03.015
Shen, Q., Zhang, L., Liao, Z., Wang, S., Yan, T., Shi, P., et al. (2018). The genome of Artemisia annua provides insight into the evolution of Asteraceae Family and Artemisinin biosynthesis. Mol. Plant 11, 776–788. doi: 10.1016/j.molp.2018.03.015
Song, T. H., Chen, X. X., Lee, C. K. F., Sze, S. C. W., Feng, Y. B., Yang, Z. J., et al. (2019). Dendrobine targeting JNK stress signaling to sensitize chemotoxicity of cisplatin against non-small cell lung cancer cells in vitro and in vivo. Phytomedicine 53, 18–27. doi: 10.1016/j.phymed.2018.06.018
Song, C., Jiao, C., Jin, Q., Chen, C., Cai, Y., and Lin, Y. (2020). Metabolomics analysis of nitrogen-containing metabolites between two Dendrobium plants. Physiol. Mol. Biol. Plants. 26, 1425–1435. doi: 10.1007/s12298-020-00822-1
Teixeira da Silva, J. A., and Ng, T. B. (2017). The medicinal and pharmaceutical importance of Dendrobium species. Appl. Microbiol. Biotechnol. 101, 2227–2239. doi: 10.1007/s00253-017-8169-9
Thoden, J. B., Taylor Ringia, E. A., Garrett, J. B., Gerlt, J. A., Holden, H. M., and Rayment, I. (2004). Evolution of enzymatic activity in the Enolase superfamily: structural studies of the promiscuous o-Succinylbenzoate synthase from Amycolatopsis. Biochemistry 43, 5716–5727. doi: 10.1021/bi0497897
Töpfer, N., Fuchs, L. M., and Aharoni, A. (2017). The PhytoClust tool for metabolic gene clusters discovery in plant genomes. Nucleic Acids Res. 45, 7049–7063. doi: 10.1093/nar/gkx404
Wang, Y., Avula, B., Abe, N., Wei, F., Wang, M., Ma, S., et al. (2016). Tandem mass spectrometry for structural characterization of dendrobine-type sesquiterpene alkaloids from the stems of Dendrobium nobile using LC-MS/MS. Planta Med. 82, 662–670. doi: 10.1055/s-0042-103031
Wang, P., Chen, X., Cai, C. H., Kong, F. D., Huang, S. Z., Yuan, J. Z., et al. (2020). A new picrotoxane-type sesquiterpene from Dendrobium nobile Lindl. Nat. Prod. Res. 36, 2112–2117. doi: 10.1080/14786419.2020.1851224
Wang, P., Chen, X., Wang, H., Huang, S., Cai, C., Yuan, J., et al. (2019). Four new picrotoxane-type sesquiterpenes from Dendrobium nobile Lindl. Front. Chem. 7:812. doi: 10.3389/fchem.2019.00812
Wang, Q., Gong, Q., Wu, Q., and Shi, J. (2010). Neuroprotective effects of Dendrobium alkaloids on rat cortical neurons injured by oxygen-glucose deprivation and reperfusion. Phytomedicine 17, 108–115. doi: 10.1016/j.phymed.2009.05.010
Wang, Z., Jiang, W., Liu, Y., Meng, X., Su, X., Cao, M., et al. (2021). Putative genes in alkaloid biosynthesis identified in Dendrobium officinale by correlating the contents of major bioactive metabolites with genes expression between Protocorm-like bodies and leaves. BMC Genomics 22, 579–518. doi: 10.1186/s12864-021-07887-6
Wang, Y., and Liu, A. (2020). Genomic characterization and expression analysis of basic helix-loop-helix (BHLH) family genes in traditional chinese herb Dendrobium officinale. Plan. Theory 9, 1–23. doi: 10.3390/plants9081044
Wang, T., Song, Z., Wei, L., and Li, L. (2018). Molecular characterization and expression analysis of WRKY family genes in Dendrobium officinale. Genes Genomics 40, 265–279. doi: 10.1007/s13258-017-0602-z
Wang, Y., Tong, Y., Adejobi, O. I., Wang, Y., and Liu, A. (2022). Research advances in multi-Omics on the traditional Chinese herb Dendrobium officinale. Front. Plant Sci. 12:808228. doi: 10.3389/fpls.2021.808228
Xu, X., Chen, X., Yang, R., Li, Z., Zhou, H., Bai, Y., et al. (2020). Crepidtumines A and B, two novel Indolizidine alkaloids from Dendrobium crepidatum. Sci. Rep. 10, 9564–9568. doi: 10.1038/s41598-020-66552-2
Xu, J., Han, Q. B., Li, S. L., Chen, X. J., Wang, X. N., Zhao, Z. Z., et al. (2013). Chemistry, bioactivity and quality control of Dendrobium, a commonly used tonic herb in traditional Chinese medicine. Phytochem. Rev. 12, 341–367. doi: 10.1007/s11101-013-9310-8
Xu, X., Li, Z., Yang, R., Zhou, H., Bai, Y., Yu, M., et al. (2019). Crepidatumines C and D, two new indolizidine alkaloids from Dendrobium crepidatum lindl. ex Paxt. Molecules 24, 2–10. doi: 10.3390/molecules24173071
Xu, Y. Y., Xu, Y. S., Wang, Y., Wu, Q., Lu, Y. F., Liu, J., et al. (2017). Dendrobium nobile Lindl. alkaloids regulate metabolism gene expression in livers of mice. J. Pharm. Pharmacol. 69, 1409–1417. doi: 10.1111/jphp.12778
Yan, L., Wang, X., Liu, H., Tian, Y., Lian, J., Yang, R., et al. (2015). The genome of Dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol. Plant 8, 922–934. doi: 10.1016/j.molp.2014.12.011
Yang, D., Cheng, Z. Q., Hou, B., Yang, L., Zi, C. T., Dong, F. W., et al. (2020). Two unusual dendrobine-type alkaloids from Dendrobium findlayanum. Fitoterapia 144:104607. doi: 10.1016/j.fitote.2020.104607
Yang, D., Cheng, Z. Q., Yang, L., Hou, B., Yang, J., Li, X. N., et al. (2018). Seco-Dendrobine-type alkaloids and bioactive Phenolics from Dendrobium findlayanum. J. Nat. Prod. 81, 227–235. doi: 10.1021/acs.jnatprod.7b00150
Yang, D., Cheng, Z. Q., Zi, C. T., Yang, L., Dong, F. W., Hu, J. M., et al. (2019). Four new sesquiterpene derivatives from Dendrobium findlayanum. Chin. J. Nat. Med. 17, 900–905. doi: 10.1016/S1875-5364(19)30110-4
Yang, S., Gong, Q., Wu, Q., Li, F., Lu, Y., and Shi, J. (2014). Alkaloids enriched extract from Dendrobium nobile Lindl. attenuates tau protein hyperphosphorylation and apoptosis induced by lipopolysaccharide in rat brain. Phytomedicine 21, 712–716. doi: 10.1016/j.phymed.2013.10.026
Yang, L., Qin, L. H., Bligh, S. W. A., Bashall, A., Zhang, C. F., Zhang, M., et al. (2006). A new phenanthrene with a spirolactone from Dendrobium chrysanthum and its anti-inflammatory activities. Bioorgan. Med. Chem. 14, 3496–3501. doi: 10.1016/j.bmc.2006.01.004
Yang, M., Wang, Q., Liu, Y., Hao, X., Wang, C., Liang, Y., et al. (2021). Divergent camptothecin biosynthetic pathway in Ophiorrhiza pumila. BMC Biol. 19, 122–116. doi: 10.1186/s12915-021-01051-y
You, D., Feng, Y., Wang, C., Sun, C., Wang, Y., Zhao, D., et al. (2021). Cloning, characterization, and enzymatic identification of a new tryptophan decarboxylase from Ophiorrhiza pumila. Biotechnol. Appl. Biochem. 68, 381–389. doi: 10.1002/bab.1935
Yu, Z., Zhang, G., Teixeira da Silva, J. A., Zhao, C., and Duan, J. (2021). The methyl jasmonate-responsive transcription factor DobHLH4 promotes DoTPS10, which is involved in linalool biosynthesis in Dendrobium officinale during floral development. Plant Sci. 309:110952. doi: 10.1016/j.plantsci.2021.110952
Yu, Z., Zhao, C., Zhang, G., Teixeira da Silva, J. A., and Duan, J. (2020). Genome-wide identification and expression profile of tps gene family in Dendrobium officinale and the role of dotps10 in linalool biosynthesis. Int. J. Mol. Sci. 21, 1–22. doi: 10.3390/ijms21155419
Yuan, Y., Liu, X., Wang, J., and Zhang, J. (2019). Morphological and microscopic identification of three major medicinal Dendrobium species in Ta-Pieh Mountains area. Microsc. Res. Tech. 82, 483–493. doi: 10.1002/jemt.23191
Yuan, Y., Yu, M., Jia, Z., Song, X., Liang, Y., and Zhang, J. (2018). Analysis of Dendrobium huoshanense transcriptome unveils putative genes associated with active ingredients synthesis. BMC Genomics 19, 978–916. doi: 10.1186/s12864-018-5305-6
Yuan, Y., Zhang, B., Tang, X., Zhang, J., and Lin, J. (2020). Comparative transcriptome analysis of different dendrobium species reveals active ingredients-related genes and pathways. Int. J. Mol. Sci. 21:861. doi: 10.3390/ijms21030861
Zhan, C., Lei, L., Liu, Z., Zhou, S., Yang, C., Zhu, X., et al. (2020). Selection of a subspecies-specific diterpene gene cluster implicated in rice disease resistance. Nat. Plants 6, 1447–1454. doi: 10.1038/s41477-020-00816-7
Zhang, J., He, C., Wu, K., Teixeira da Silva, J. A., Zeng, S., Zhang, X., et al. (2016b). Transcriptome analysis of Dendrobium officinale and its application to the identification of genes associated with polysaccharide synthesis. Front. Plant Sci. 7:5. doi: 10.3389/fpls.2016.00005
Zhang, L., Jiao, C., Cao, Y., Cheng, X., Wang, J., Jin, Q., et al. (2020). Comparative analysis and expression patterns of the PLP_dec genes in Dendrobium officinale. Int. J. Mol. Sci. 21:54. doi: 10.3390/ijms21010054
Zhang, L., Li, C., Yang, D., Wang, Y., Yang, Y., and Sun, X. (2021b). Genome-wide analysis of the tcp transcription factor genes in Dendrobium catenatum lindl. Int. J. Mol. Sci. 22:10269. doi: 10.3390/ijms221910269
Zhang, C., Liu, S. J., Yang, L., Yuan, M. Y., Li, J. Y., Hou, B., et al. (2017). Sesquiterpene amino ether and cytotoxic phenols from Dendrobium wardianum Warner. Fitoterapia 122, 76–79. doi: 10.1016/j.fitote.2017.08.015
Zhang, J., Morris-Natschke, S. L., Ma, D., Shang, X. F., Yang, C. J., Liu, Y. Q., et al. (2021a). Biologically active indolizidine alkaloids. Med. Res. Rev. 41, 928–960. doi: 10.1002/med.21747
Zhang, G. Q., Xu, Q., Bian, C., Tsai, W. C., Yeh, C. M., Liu, K. W., et al. (2016a). The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 6, 1–10. doi: 10.1038/srep19029
Zhang, H., and Wang, X. (2016). Modular co-culture engineering, a new approach for metabolic engineering. Metab. Eng. 37, 114–121. doi: 10.1016/j.ymben.2016.05.007
Zhang, T., Xu, Y., Ding, Y., Yu, W., Wang, J., Lai, H., et al. (2022). Identification and expression analysis of WRKY gene family in response to abiotic stress in Dendrobium catenatum. Front. Genet. 13:800019. doi: 10.3389/fgene.2022.800019
Zhang, M., Yu, Z., Zeng, D., Si, C., Zhao, C., Wang, H., et al. (2021c). Transcriptome and metabolome reveal salt-stress responses of leaf tissues from Dendrobium officinale. Biomol. Ther. 11, 1–16. doi: 10.3390/biom11050736
Zhang, Y., Zhang, G. Q., Zhang, D., Liu, X. D., Xu, X. Y., Sun, W. H., et al. (2021d). Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic. Res. 8:183. doi: 10.1038/s41438-021-00621-z
Zha, X. Q., Deng, Y. Y., Li, X. L., Wang, J. F., Pan, L. H., and Luo, J. P. (2017). The core structure of a Dendrobium huoshanense polysaccharide required for the inhibition of human lens epithelial cell apoptosis. Carbohydr. Polym. 155, 252–260. doi: 10.1016/j.carbpol.2016.08.087
Zheng, S.-g., Hu, Y.-d., Zhao, R.-x., Yan, S., Zhang, X.-q., Zhao, T.-m., et al. (2018). Genome-wide researches and applications on Dendrobium. Planta 248, 769–784. doi: 10.1007/s00425-018-2960-4
Zhou, J., Zhang, Y., Li, S., Zhou, Q., Lu, Y., Shi, J., et al. (2020). Dendrobium nobile Lindl. alkaloids-mediated protection against CCl4-induced liver mitochondrial oxidative damage is dependent on the activation of Nrf2 signaling pathway. Biomed. Pharmacother. 129:110351. doi: 10.1016/j.biopha.2020.110351
Zhu, Y. F., Fan, H. H., Li, D. H., Jin, Q., Zhang, C. M., Zhu, L. Q., et al. (2020). Molecular cloning, bioinformation analysis and expression of the strictosidine synthase in Dendrobium officinale. Acta Sci. Pol. Hortorum Cultus 19, 111–124. doi: 10.24326/asphc.2020.3.10
Zhu, Y., Meng, C., Zhu, L., Li, D., Jin, Q., Song, C., et al. (2017). Cloning and characterization of DoMYC2 from Dendrobium officinale. Plant Cell. Tissue Organ Cult. 129, 533–541. doi: 10.1007/s11240-017-1198-3
Zhu, X., Zeng, X., Sun, C., and Chen, S. (2014). Biosynthetic pathway of terpenoid indole alkaloids in Catharanthus roseus. Front. Med. China 8, 285–293. doi: 10.1007/s11684-014-0350-2
Zhuang, Y., Yang, G. Y., Chen, X., Liu, Q., Zhang, X., Deng, Z., et al. (2017). Biosynthesis of plant-derived ginsenoside Rh2 in yeast via repurposing a key promiscuous microbial enzyme. Metab. Eng. 42, 25–32. doi: 10.1016/j.ymben.2017.04.009
Keywords: alkaloid, Dendrobium, secondary metabolism, chemicals, pathway
Citation: Song C, Ma J, Li G, Pan H, Zhu Y, Jin Q, Cai Y and Han B (2022) Natural Composition and Biosynthetic Pathways of Alkaloids in Medicinal Dendrobium Species. Front. Plant Sci. 13:850949. doi: 10.3389/fpls.2022.850949
Received: 08 January 2022; Accepted: 30 March 2022;
Published: 06 May 2022.
Edited by:
Zhihua Liao, Southwest University, ChinaReviewed by:
Jungui Dai, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaCopyright © 2022 Song, Ma, Li, Pan, Zhu, Jin, Cai and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Jin, cWluZ2ppbkBhaGF1LmVkdS5jbg==; Yongping Cai, eXBjYWlhaEAxNjMuY29t; Bangxing Han, aGFuYngxOTc4QHNpbmEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.