
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 21 March 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.848869
This article is part of the Research TopicFruit Ripening: From Present Knowledge to Future Development, Volume IIView all 9 articles
 Shengchao Liu1,2
Shengchao Liu1,2 Xincheng Liu1,2
Xincheng Liu1,2 Bangrui Gou1,2
Bangrui Gou1,2 Dengliang Wang3
Dengliang Wang3 Chunrong Liu3
Chunrong Liu3 Jun Sun4
Jun Sun4 Xueren Yin1,2,5
Xueren Yin1,2,5 Donald Grierson5,6
Donald Grierson5,6 Shaojia Li1,2,5*
Shaojia Li1,2,5* Kunsong Chen1,2,5
Kunsong Chen1,2,5Citric acid plays significant roles in numerous physiological processes in plants, including carbon metabolism, signal transduction, and tolerance to environmental stress. For fruits, it has a major effect on fruit organoleptic quality by directly influencing consumer taste. Citric acid in citrus is mainly regulated by the balance between synthesis, degradation, and vacuolar storage. The genetic and molecular regulations of citric acid synthesis and degradation have been comprehensively elucidated. However, the transporters for citric acid in fruits are less well understood. Here, an aluminum-activated malate transporter, CitALMT, was characterized. Transient overexpression and stable transformation of CitALMT significantly reduced citrate concentration in citrus fruits and transgenic callus. Correspondingly, transient RNA interference-induced silencing of CitALMT and increased citrate significantly, indicating that CitALMT plays an important role in regulating citrate concentration in citrus fruits. In addition, dual-luciferase assays indicated that CitMYB52 and CitbHLH2 could trans-activate the promoter of CitALMT. EMSA analysis showed that CitbHLH2 could physically interact with the E-box motif in the CitALMT promoter. Bimolecular fluorescence complementation assays, yeast two-hybrid, coimmunoprecipitation and transient overexpression, and RNAi assay indicated that the interaction between CitMYB52 and CitbHLH2 could synergistically trans-activate CitALMT to negatively regulate citrate accumulation.
In plants, organic acids are of fundamental importance and play significant roles in numerous physiological processes. They serve as respiratory substrates in the tricarboxylic acid (TCA) cycle, precursors for numerous primary metabolites, such as amino acids, fatty acids, and secondary metabolites, such as aromatics and terpenoids (Ladanyia, 2008). Changes in organic acids levels can be perceived and have a major impact on the abundance of nucleus-encoded transcripts involved in photosynthesis, protein synthesis, biotic and abiotic stress (Finkemeier et al., 2013; Dyson et al., 2016). In addition, organic acids are involved in chemical modification of proteins, such as lysine succinylation (Zhang et al., 2011; Weinert et al., 2013), and play a role in the hygroscopicity of inorganic salts, such as ammonium sulfate, sodium chloride, and nitrate (Wang et al., 2017, 2018 Wu et al., 2018).
Citrus is the world’s most cultivated fruit and citric acid is the most important organic acid in citrus fruit, which serves as a typical material for citric acid metabolism studies. Agronomic factors (temperature, water, light, mineral elements, etc.) greatly affect citrate concentrate during fruit development and postharvest stage. In “Ponkan” fruit, hot air treatment could effectively reduce citric acid concentration in postharvest citrus (Li et al., 2021). Iron deficiency induced a decrease in cytoplasmic aconitase and a concomitant increase in citrate levels (Shlizerman et al., 2007). However, intrinsic genetic characteristics determine the citric acid concentration in different citrus varieties and fruit development periods. In recent years, the genetic and molecular regulations of citric acid synthesis and degradation have been widely studied. Citrate concentration in fruit cells is codetermined by the balance between synthesis, degradation, and vacuolar storage (Zampini et al., 2008; Wang et al., 2014). In Malus xiaojinensis, when MxCS2 was introduced into Arabidopsis, citrate synthase was enhanced and citrate concentration was increased (Han et al., 2015). Genomic and transcriptomic analyses of wild mandarin populations and abundant mandarin varieties indicated that CitAco2 and CitAco3 are associated with lower citrate concentration (Wang et al., 2018), transient overexpression of CitAco3 in citrus leaves and fruits could significantly decrease the citrate concentration (Li et al., 2017).
In mature citrus fruits, most of the citric acid is stored in vacuoles and contributes to flavor, nutrition, and juice quality. Thus, vacuolar transport processes may play a major role in determining citric acid accumulation (Etienne et al., 2013). The transport of citrate is related to the electrochemical gradient and transport carriers across the tonoplast (Huang et al., 2021). Proton pumps cause a massive proton influx that generates an acidic vacuolar pH and electrochemical gradient, which provides a strong driving force for citrate transportation across the tonoplast. An H+-ATPase gene CsPH8 may regulate the accumulation of citric acid in citrus (Shi et al., 2015). Subsequent studies showed that the low acidity of citrus fruit is directly related to the downregulation of proton pump genes CitPH1 and CitPH5, which is associated with mutations that disrupt expression of MYB, HLH, and/or WRKY transcription factors homologous to those activating PH1 and PH5 in petunia (Strazzer et al., 2019). In addition, the transport of citrate is achieved by the manipulation of anion channels or transport carriers on the tonoplast (Huang et al., 2021). CsCit1 may act as a vacuolar citrate/H+ symporter in Citrus (Shimada et al., 2006), but the homolog in Arabidopsis, AttDT, was shown not to be the main tonoplast citrate carrier (Hurth et al., 2005). In citrus fruit, the major transport carriers of citric acid are poorly understood.
In the present study, an aluminum-activated malate transporter CitALMT was identified and functionally characterized by transient overexpression and RNA interference-induced silencing (RNAi) in citrus fruits and stable transgenic callus. Two transcription factors, CitMYB52 (Ciclev10001803m) and CitbHLH2 (Ciclev10005233m), trans-activated the CitALMT promoter. CitbHLH2 directly interacted with the E-box motif in the CitALMT promoter. Yeast two-hybrid assay, BiFC assay, and CoIP assay indicated that CitMYB52 and CitbHLH2 could interact with each other physically. Further transient dual overexpression and RNAi of CitMYB52 and CitbHLH2 in citrus fruits validated the role of this interaction in citrate accumulation by regulating transcript level of CitALMT. These findings provide new insights into the molecular basis of citrate modulation in citrus fruits.
“Ponkan” (Citrus reticulata Blanco cv. Ponkan) fruits were obtained from a commercial orchard in Quzhou (Zhejiang, China) at different developmental stages, 120, 135, 150, 165, and 180 days after full bloom (DAFB) in 2017. At each sampling point, the size and appearance of the collected fruits from three different trees for three biological replicates are uniform. The flesh samples were taken and frozen in liquid nitrogen and stored at −80°C for subsequent experiments. Citrus hot air treatment, “Ordinary Ponkan (OPK)” and its spontaneous early maturing mutant “Zaoshu Ponkan (ZPK)” were described in detail in our previous study (Li et al., 2016, 2021).
In the two previous reports, the genetic and molecular regulation of citric acid synthesis and degradation have been studied in high- and low-citrate containing fruits. The fruit flesh samples of “Gaocheng” (“GC”) and “Satsuma mandarin” (“SM”) were used for transcriptomic analysis to explore the inner mechanism of difference in acidity among different varieties (Lin et al., 2015a); the fruit flesh samples of different developmental stages of “Ponkan” (“PK”) fruits were used to study the relationship between genes and metabolites during fruit development (Lin et al., 2015b). Using RNA-seq results from our previous reports, we obtained the DEGs (differentially expressed genes) shared in the two data sets. After filtering out the DEGs with low expression value [max (RPKM) > 1], we took the genes common to the two datasets. By analyzing the correlation between gene expression and citric acid content, we obtained 260 DEGs (the threshold value > 0.7). Hierarchical cluster analysis was performed using these 260 DEGs data by the pheatmap package of R with the clustering method set to complete and average linkage with Euclidean distance. Considering the hierarchical clustering result of all samples and the physiological data, the data were divided into two subclasses on the dimension of samples. The mean value and the ratio of the two subclasses were calculated for each candidate and used to do the entropy weight method analysis (Liu et al., 2019), and a relative ranking of all the 260 DEGs was obtained. Transcription factors whose expression patterns were positively correlated to the CitALMT’s were identified by the calculation of the Pearson Correlation Coefficient (PCC > 0.6).
Citrate concentration in fruits and transgenic callus was measured according to Li et al. (2017). Samples were ground in liquid nitrogen, and 1.4 ml chromatographic methanol was added to 0.1 g sample; after vortex for three times, the sample was placed at 70°C for 15 min and then centrifuged at 11,000 g for 10 min at 4°C. 0.75 ml trichloromethane and 1.5 ml purified water were added to the upper phase. After vortex fully for 3 times and centrifuging, an internal standard was a 100 μl aliquot of each sample with 20 μl ribitol (0.2 mg/ml). After vacuum drying the sample, adding 60 μl of 20 mg/ml methoxyamine hydrochloride dissolved in pyridine to the residue and incubate for 1.5 h at 37°C with vortex. 40 μl Bis (trimethylsilyl) trifluoroacetamide (1% trimethylchlorosilane) was added to the sample and incubated for another 30 min at 37°C. Each sample was added to the injection bottle and injected into a GC–MS fitted with a fused-silica capillary column (30 m × 0.25 mm internal diameter, 0.25 μm DB-5MS stationary phase) with a split ratio of 10:1. The injector temperature was 250°C and the helium carrier gas flow rate was 1.0 ml/min. The column temperature was maintained at 100°C for 1 min, increased to 185°C at a speed of 3°C/min, then increased to 250°C at a speed of 15°C/min and maintained for 2 min. The MS operating parameters: ionization voltage 70 eV, ion source temperature 230°C, and interface temperature 280°C.
The CitALMT gene and relevant transcription factors were isolated based on the citrus genome database,1 with the primers listed in Supplementary Table S1. The amino acid sequence alignment analysis with several reported ALMT genes in fruit was carried out by DNAMAN (V6.0.3.99).
Total RNA was extracted according to our previous report (Li et al., 2017). The genomic DNA was degraded with RNase-free DNase I (Ambion), total of 1.0 μg RNA was used for cDNA synthesis. According to the manufacturer’s protocol, first-strand cDNA synthesis was initiated with GoScript™ Reverse Transcriptase (Promega) following the manufacturer’s protocol. The actin gene was used to adjust the concentration of the template. Three biological replicates were set for each sampling site.
Ssofast Eva Green Supermix Kit and CFX96 (Bio-RAD) were used for RT-qPCR. The PCR reaction mixture (total volume of 20 μl) consisted of 10 μl 2× real-time PCR mix (Bio-Rad), 1 μl for each primer (10 μM), 2 μl for diluted cDNA, and 6 μl for DEPC H2O. The initial step of the PCR program was to cycle 50 times at 95°C for 5 min, then at 95°C for 10 s, 60°C for 10 s, and 72°C for 15 s. At the end of each run, each gene was analyzed by melting curves. The actin gene was used as the internal control (Li et al., 2017). The 2(−△Ct) method was used to analyze the expression levels of genes. All the primers used for real-time PCR were in Supplementary Table S2.
Full-length CitALMT, CitMYB52, and CitbHLH2 were amplified with primers (listed in Supplementary Table S3) and inserted into the pGreen II 002962-SK vector (Hellens et al., 2005). Forward and reverse PCR-amplified cDNA fragments of CitALMT, CitMYB52, and CitbHLH2 were inserted into the pHB vector driven by 2× CaMV 35S (Zhang et al., 2020). All the constructs were electroporated into Agrobacterium tumefaciens GV3101. Fruits with the same size at 150 DAFB (with relative high citrate concentration) and 180 DAFB (with relative low-citrate concentration) were selected for overexpression and RNAi assays, separately. Agrobacterium-mediated transient transformation in fruit was based on previous reports with some modifications (Li et al., 2017). Uniform sections from the fruit were selected and infiltrated with either empty vector (SK or PHB, as control) or Agrobacterium culture containing the target gene. After infiltration for 5 days, the infiltrated sections were sampled for citrate and expression analysis.
For citrus callus transformation, the full-length of CitALMT was cloned into downstream of the CaMV 35S promoter of a modified pCAMBIA1301 vector with primers (listed in Supplementary Table S3). A GUS reporter gene following the CaMV 35S promoter in the pCAMBIA1301 vector was used for positive screening. The recombinant vector was then introduced into A. tumefaciens strain EHA105.
Valencia orange embryogenic calluses in tissue culture were used for transformation. The recombinant plasmid was introduced into the wild-type calluses using an Agrobacterium-mediated method, as described by Li et al., (2002) with minor modification. After 3 days of dark co-culture, the calluses were transferred into the MT selective medium containing 400 mg/L cefotaxime and 50 mg/L hygromycin at 25°C in the dark. The proliferated calluses were subcultured at 4-week intervals on fresh MT selective medium.
Transgenic callus lines overexpressing CitALMT were identified by RT-qPCR and GUS staining. RT-qPCR and citrate concentration analysis were performed as described above. Three biological replicates were performed for each line.
Histochemical staining confirmed that GUS reporter gene co-transformed with CitALMT. The GUS staining assay was carried out using the GUS staining kit (Real-Times Biotechnology, China) according to the manufacturer’s protocol.
Dual-luciferase assays were performed as described in our previous report (Xu et al., 2014). The promoter of the CitALMT was amplified with the primers described in Supplementary Table S1 and inserted into the pGreen II 0800-LUC vector. Full-length transcripts of the transcription factors were inserted into the pGreen II 002962-SK vector (SK) with the primers described in Supplementary Table S1.
All the recombinant vectors were electroporated into Agrobacterium tumefaciens GV3101. Osmotic buffer for Agrobacterium cultures (10 mM MES, 10 mM MgCl2, 150 μM acetosyringone, pH 5.6) was prepared to a concentration (OD600 = 0.75). A mixed Agrobacterium culture (v/v, 10:1) of transcription factors and promoters was infiltrated into tobacco leaves using a needleless syringe. Fluorescence intensity of Luc and Ren after infiltration for 3 days was measured using a dual-luciferase assay reagent (Promega) in the GlomaXTM 96 microplate luminescence apparatus (Promega). Tobacco plants were grown in a growth chamber, with 16:8 h light:dark cycles. Results were calculated from at least three separate experiments, each with at least five biological replicates.
The full-length of CitALMT, CitMYB52, and CitbHLH2 without termination codon were cloned into pCAMBIA1300-sGFP, the empty GFP vector was used as controls. By Agrobacterium-mediated infiltration (GV3101), the constructed GFP vectors were transiently expressed in transgenic N. benthamiana leaves (stably transformed with nuclear location signal, NLS-mCherry; Li et al., 2017). The 35S-CitALMT-GFP Agrobacterium culture was mixed with the RFP Agrobacterium culture (as a marker located on the tonoplast). A Zeiss LSM710NLO confocal laser scanning microscope was used to image the green fluorescent protein (GFP) fluorescence in tobacco leaves after infiltration for 2 days. Primers used for GFP construction are described in Supplementary Table S3.
The full-length of CitbHLH2 was inserted into pET-32a (Clontech) to express a CitbHLH2-His fusion protein. The recombinant vector was transformed into E.coli strain BL21 which was then incubated with LB medium at 37°C until OD600 = 0.5. 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the medium and then incubated at 37°C for 3 h. The E. coli cells were collected with 20 mM Tris–HCl buffer (0.1 M NaCl, pH = 8.0) and ultrasonicated on ice at 200 W with on/off at 3 s/2 s cycle for 15 min, then centrifuged at 9,000 g for 20 min at 4°C. Subsequently, the Ni-NTA resin (Transgene) was added to the supernatant to combine His-tagged proteins at 4°C for 1 h, which were eluted with elution buffers containing gradient imidazole (30, 300, and 500 mM).
EMSA detection was performed using the LightShift Chemiluminescent EMSA kit (Thermo) according to the manufacturer’s instructions. The probes were labeled from the 3′ biotin terminal by HuaGene (Shanghai, China), annealed with complementary oligonucleotide, heated at 95°C for 5 min, and then gradually reduced to 25°C at a rate of 0.1°C s−1, and transformed into a double-stranded DNA probe. The probes used for EMSA were listed in Supplementary Table S4. The binding specificity was tested by mutant probe and competition with unlabeled probes (50, 200, and 500-fold unlabeled oligonucleotides).
Yeast two-hybrid assays (Y2H) were performed using the MatchmakerTM gold yeast two-hybrid system (Clontech, United States) to analyze protein–protein interactions. The full-length of CitMYB52 and CitbHLH2 was separately subcloned into pGADT7AD vector and pGBKT7 BD vector (primers are listed in Supplementary Table S4). Meanwhile, pGBKT7-53 BD and pGBKT7-Lam BD separately combined with pGADT7-T AD was used as positive and negative control.
The full-length of CitMYB52 and CitbHLH2 were cloned into the C- and N-terminus of YFP vectors, respectively (Sainsbury et al., 2009), using the primers listed in Supplementary Table S4. All the recombinant constructs were transiently expressed in transgenic N. benthamiana leaves (stably transformed with nuclear location signal, NLS-mCherry) through Agrobacterium-mediated infiltration (GV3101) based on previous reports (Li et al., 2017). Two days after infiltration, the YFP fluorescence imaging of tobacco leaves was performed using a Zeiss LSM710NLO confocal laser scanning microscope.
The coding sequences of CitMYB52 and CitbHLH2 without stop codon were cloned with primers (listed in Supplementary Table S4) and fused with 4myc tags and 3HA tags separately, then ligated into pCAMBIA1300-221 vector. Recombinant vectors were electroporated into Agrobacterium tumefaciens GV3101 and then transiently expressed in N. benthamiana leaves (Li et al., 2018). Two d after infiltration, N. benthamiana leaves were ground in liquid nitrogen and proteins were extracted with extraction buffer (20 mM HEPES-KOH, pH 7.5, 40 mM KCl, 1 mM EDTA, 0.5% Triton X-100, and 1× protease inhibitors; Roche) in the proportion of 3 ml/g tissue powder. After centrifugation at 20,000 g for 10 min, 100 μl of supernatant was retained as input, and 1 ml supernatant was incubated with anti-HA agarose (Sigma-Aldrich) at 4°C for 2 h. The proteins retained on the beads were then washed for three times with wash buffer (20 mM HEPES-KOH, pH 7.5, 40 mM KCl, and 1% Triton X-100), boiled in 2% SDS sample buffer for 10 min for elution. Western blotting gel electrophoresis was used for analysis using anti-myc or anti-HA antibodies (Sigma-Aldrich).
Eluents and inputs in Co-IP assay were separated on 10% SDS-PAGE gels and transferred to a PVDF membrane. The membrane was blocked with 5% skim milk at room temperature for 2 h. e. Antibodies for protein tags (anti-myc and anti-HA) were added into the TBST buffer (Tris-buffered saline plus Tween20) at a ratio of 1:2,000 and incubated at room temperature for 2 h. The membranes were rinsed with TBST buffer for three times, 10 min each. The anti-rabbit/anti-mouse horseradish peroxidase secondary antibody was added at a ratio of 1:10,000 and incubated at room temperature for 1 h. After washing three times with TBST buffer, the membranes were observed with chemiluminescence system.
All experiments in this study included at least three biological replicates, and all data represented as the mean ± standard error. DPS 7.05 (Zhejiang University, Hangzhou, China) was used to calculate the least significant difference (LSD0.05). Student’s t-test was used to calculate the statistical significance of the difference with a confidence level of 95.0% (*p < 0.05), 99.0% (**p < 0.01), or 99.9% (***p < 0.001). The figures were drawn using Origin 8.0 (Microcal Software Inc).
Using RNA-seq results from our previous reports (Lin et al., 2015a,b), we identified 260 DEGs that are highly correlated with citrate and performed hierarchical clustering (the threshold value > 0.7; Supplementary Figure S1A). The data were divided into two subclasses on the dimension of samples which correspond to two sample sets with relatively high acid and low acid periods. We combined the maximum expression values and the fold change values of the subclasses using the entropy weight method and obtained a relative ranking of all the 260 DEGs (Supplementary Dataset S1). An aluminum-activated malate transporter named CitALMT (Ciclev10019573m) was ranked first (Supplementary Figure S1B). In addition, 22 transcription factors whose expression patterns were positively correlated to the CitALMT’s were identified (correlation coefficient > 0.6; Supplementary Figure S1C).
The amino acid sequence of CitALMT was compared to other ALMTs including MA1 (MDP0000252114), VvALMT9 (GSVIVT01008270001), Sl-ALMT9, Sl-ALMT5 (Solyc03g119640.2.1), Sl-ALMT4 (Solyc03g096820.2.1), AtALMT9 (AT3G18440.1). The results indicated that CitALMT contains a conserved transmembrane domain TMD5 that may perform a transport function (Zhang et al., 2013; Figure 1A). The subcellular localization assay showed that CitALMT co-localized with tonoplast markers (Figure 1B).
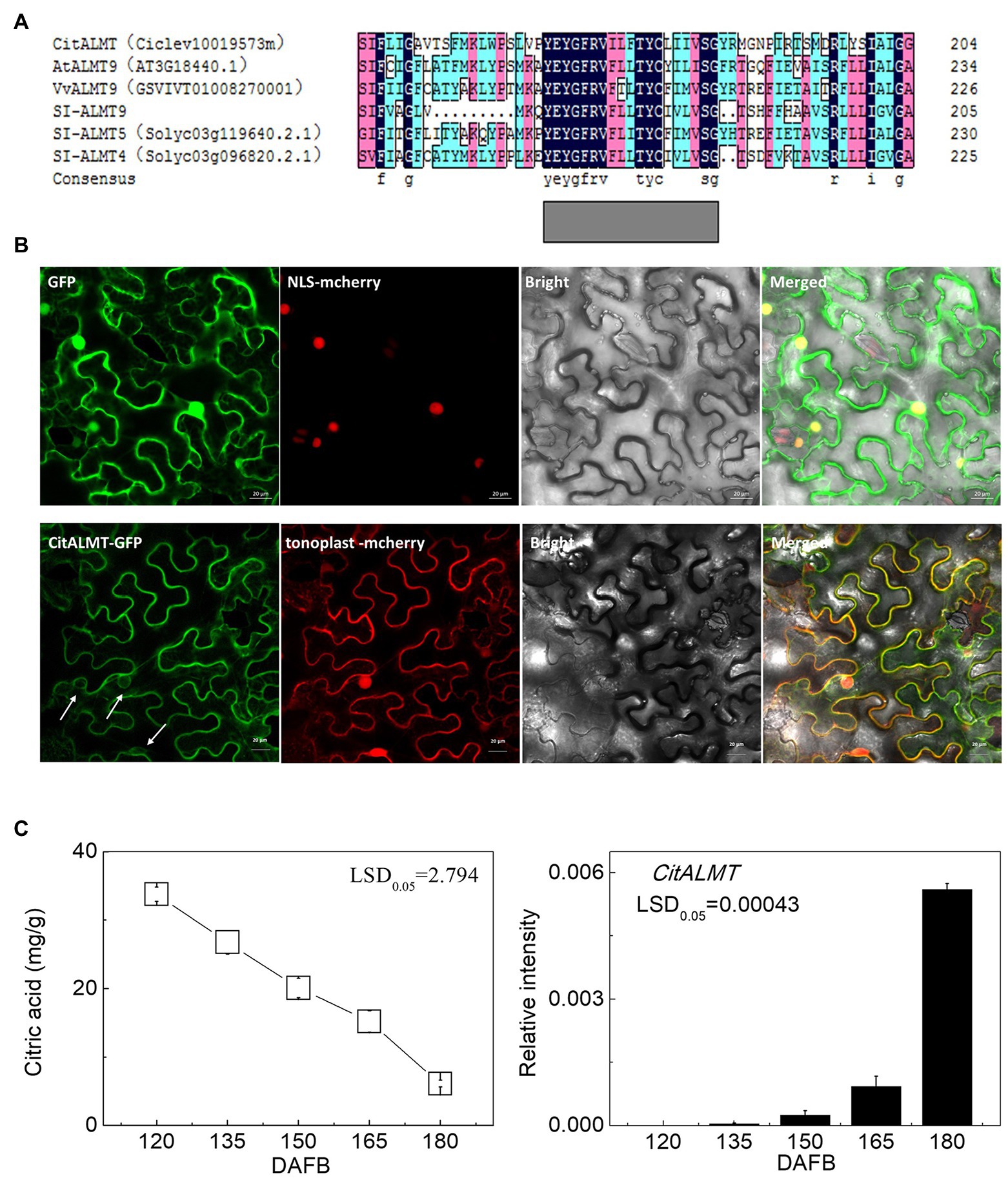
Figure 1. Identification of CitALMT. (A) The amino acid sequences analysis of CitALMT compared with some ALMTs reported previously. (B) Subcellular localization of CitALMT in tobacco leaves stably transformed with a red nuclear localization marker and agroinfiltrated with CitALMT-GFP and tonoplast-RFP marker. White arrows highlight co-localization of CitALMT with tonoplast marker. The empty vector control is on the top without tonoplast-RFP marker. The fluorescence was measured at 488 nm with a LSM780 microscope and photographed. Bars = 20 μm. (C) The citrate concentration and expression of the CitALMT genes in flesh of “Ponkan” fruits during the late stage of fruit development. DAFB, days after full blossom.
The relationship between CitALMT expression and citrate concentration was further analyzed in multiple sets of citrus fruit at different developmental stages or after different treatments. During “Ponkan” fruit development, citrate concentration decreased continuously from 33.77 to 6.11 mg/g from 120 to 180 DAFB, while CitALMT transcripts increased significantly (Figure 1C).
In our previous research, citrate concentration was shown to decrease under hot air treatment (Li et al., 2021). Using these materials, we found the expression of CitALMT in hot air-treated “Ordinary Ponkan” was significantly higher than non-treated control. Moreover, “Ordinary Ponkan (OPK)” and its spontaneous early maturing mutant “Zaoshu Ponkan (ZPK)” were also used (Li et al., 2016). Compared with OPK, ZPK fruit had lower citrate concentrations and significantly higher CitALMT transcript level (Supplementary Figure S2).
To investigate the function of CitALMT, a rapid and efficient transient overexpression assay was performed in citrus fruit. The citrate concentration declined from 14.44 mg/g (in control fruits) to 10.87 mg/g (in CitALMT-OE fruits), while the malate concentration increased from the control value of 0.4 mg/g to 0.51 mg/g. The expression level of CitALMT was significantly increased in overexpressing fruits compared to the control (Figure 2A). For a better understanding of CitALMT’s role, we performed transient RNAi inhibition in citrus fruits. The results showed that the transcript level of CitALMT in CitALMT-RNAi fruit was reduced to 55% compared to the control fruit, while the citrate showed a significant increase from 7.73 mg/g in control to 13.89 mg/g and malate showed a significant decrease (Figure 2B).
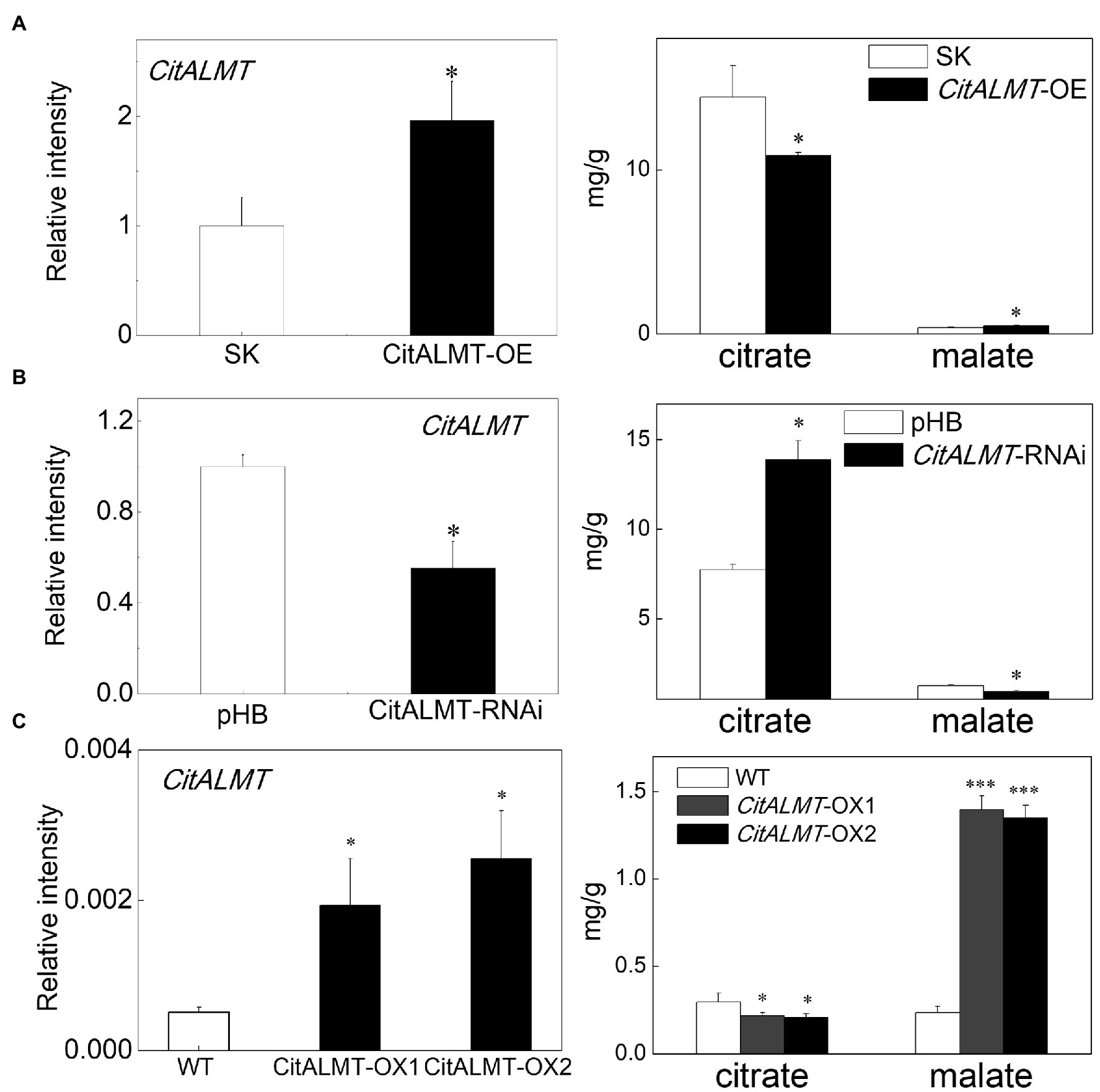
Figure 2. Gene function analysis of CitALMT. The transcript level of CitALMT (left) and organic acid concentration (right) were analyzed in citrus fruits and citrus callus. (A) Transient overexpression of CitALMT in citrus fruits. SK represents empty vector. (B) Transient RNAi of CitALMT in citrus fruits. pHB represents empty vector. (C) The analysis of transgenic CitALMT callus. Error bars indicate SE from three biological replicates. *Significant differences (p < 0.05), ***Significant differences (p < 0.001).
In addition, stable transformation was performed in Valencia orange callus tissue. Two transgenic lines were obtained, as indicated by GUS staining (Supplementary Figure S4A) and RT-qPCR (Figure 2C). The expression of CitALMT in transgenic overexpressing lines OX-1 and OX-2 were 4-fold and 5-fold higher, respectively, than that of wild type. Citrate analysis showed that both transgenic lines had significantly lower citrate concentrations (0.21 mg/g and 0.20 mg/g, respectively) compared to wild-type callus (0.30 mg/g citrate). Meanwhile, the malate concentrations in transgenic callus were significantly enhanced approximately 5-fold (Figure 2C).
Citrate is degraded in the cytoplasm mainly through the GABA shunt. Thus, we analyzed the expression pattern of key genes (CitAco3-CitIDH3-CitGAD4) involved in the GABA shunt (Chen et al., 2012; Lin et al., 2016). The results indicated that the transcripts of these three genes in CitALMT-overexpressing fruits were significantly higher than in the control. In contrast, transcripts of these three genes in the GABA shunt were decreased in CitALMT-RNAi fruits (Supplementary Figure S3). RT-qPCR analysis indicated that both transgenic callus lines showed higher transcript levels of GABA shunt genes than wild type (Supplementary Figure S4B).
The regulatory effects of the 22 identified transcription factors on CitALMT were tested using dual-luciferase assays and the results showed that in the presence of CitMYB52 or CitbHLH2, the transcriptional activity of the CitALMT promoter was significantly increased, about 3.2-fold and 2.6-fold, respectively, while the other tested transcription factors showed no significant effects (Figure 3A). Furthermore, we found that the combination of CitMYB52 and CitbHLH2 resulted in 9.8-fold induction of CitALMT promoter activity compared to CitMYB52 or CitbHLH2 alone (2.5-fold, 2.4-fold, respectively; Figure 3B).
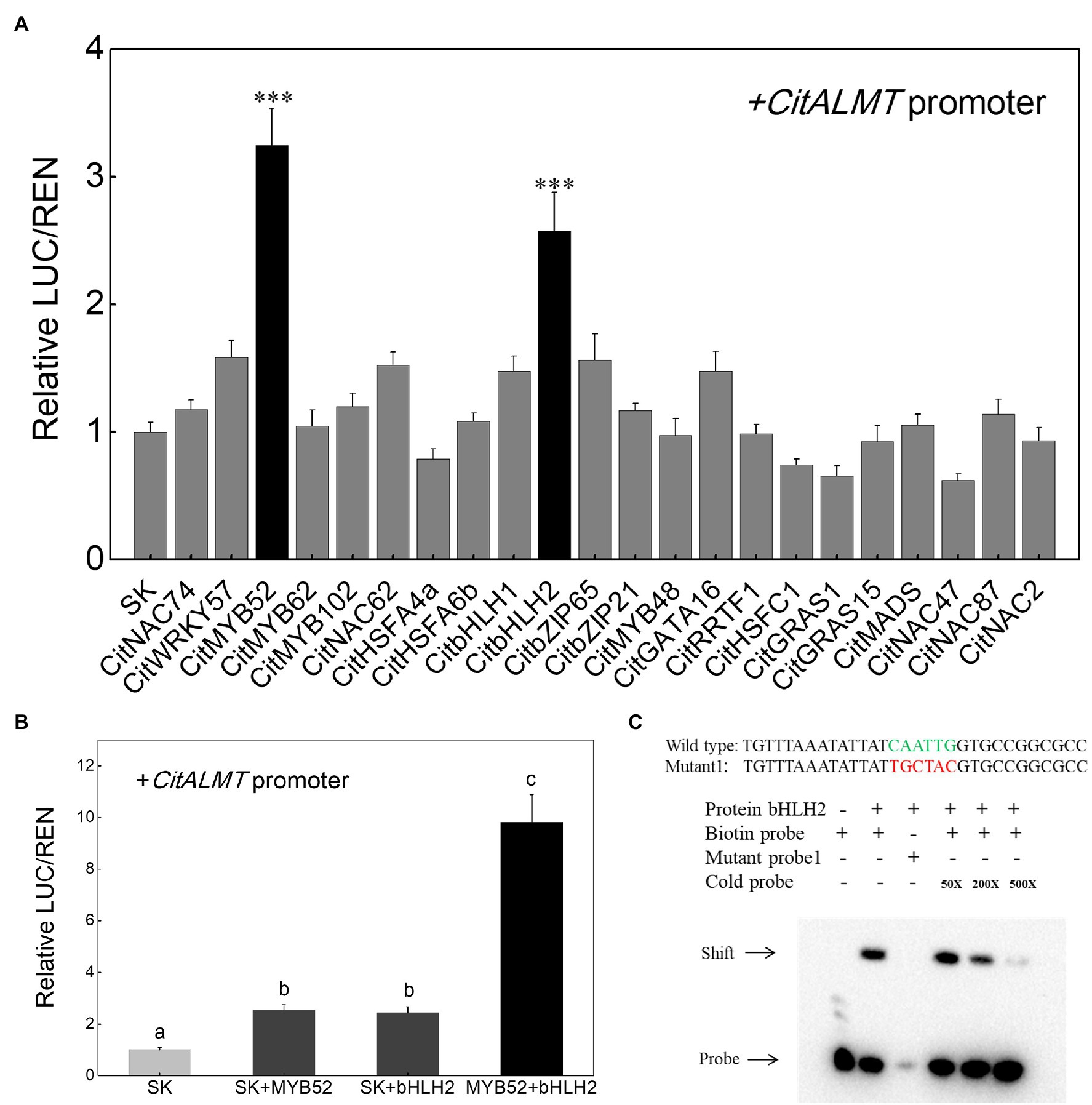
Figure 3. Interaction between transcription factors and CitALMT’s promoter. (A) In vivo interaction of transcription factors with the promoter of the CitALMT gene from “Ponkan” fruit. In vivo associations of the transcription factors and promoter were obtained by transient expression assays in tobacco leaves. The ratio of LUC/REN of the empty vector (SK) plus promoter was used as calibrator (set as 1). Error bars indicate SE from at least five replicates. ***Significant differences (p < 0.001). (B) Effect of the combination of CitMYB52 and CitbHLH2 on the CitALMT’ promoter. The ratio of LUC/REN of the empty vector (SK) plus promoter was used as calibrator (set as 1). Error bars indicate SEs from five biological replicates. Different letters above the columns represent significant differences (the combination effects were compared to two individual effects, p < 0.05). (C) Electrophoretic mobility shift assay (EMSA) of CitbHLH2 binding to the CitALMT promoter. Purified CitbHLH2 proteins and biotin-labeled DNA probe were mixed and analyzed on 6% native polyacrylamide gels. The presence (+) or absence (−) of specific probes is indicated. The concentration of the cold probe is shown; the biotinylated probe concentration was 1 nM.
Based on dual-luciferase assays (Figures 3A,B), the mechanism by which CitMYB52 and CitbHLH2 regulate the CitALMT promoter was further investigated. An EMSA assay showed that CitbHLH2 could physically bind to an E-box motif (CAATTG) in the CitALMT promoter. The specificity of the interaction was confirmed by mutated probes and cold probe competition experiments (Figure 3C). In contrast, CitMYB52 did not directly interact with the putative MYB-binding element on the promoter of CitALMT in this assay (Supplementary Figure S5). The promoter sequence and Cis-elements information of CitALMT are shown in Supplementary Table S5.
A subcellular localization assay was performed to visualize the locations of the two transcription factors and the results showed that CitMYB52 and CitbHLH2 were both located within the nucleus (Figure 4A).
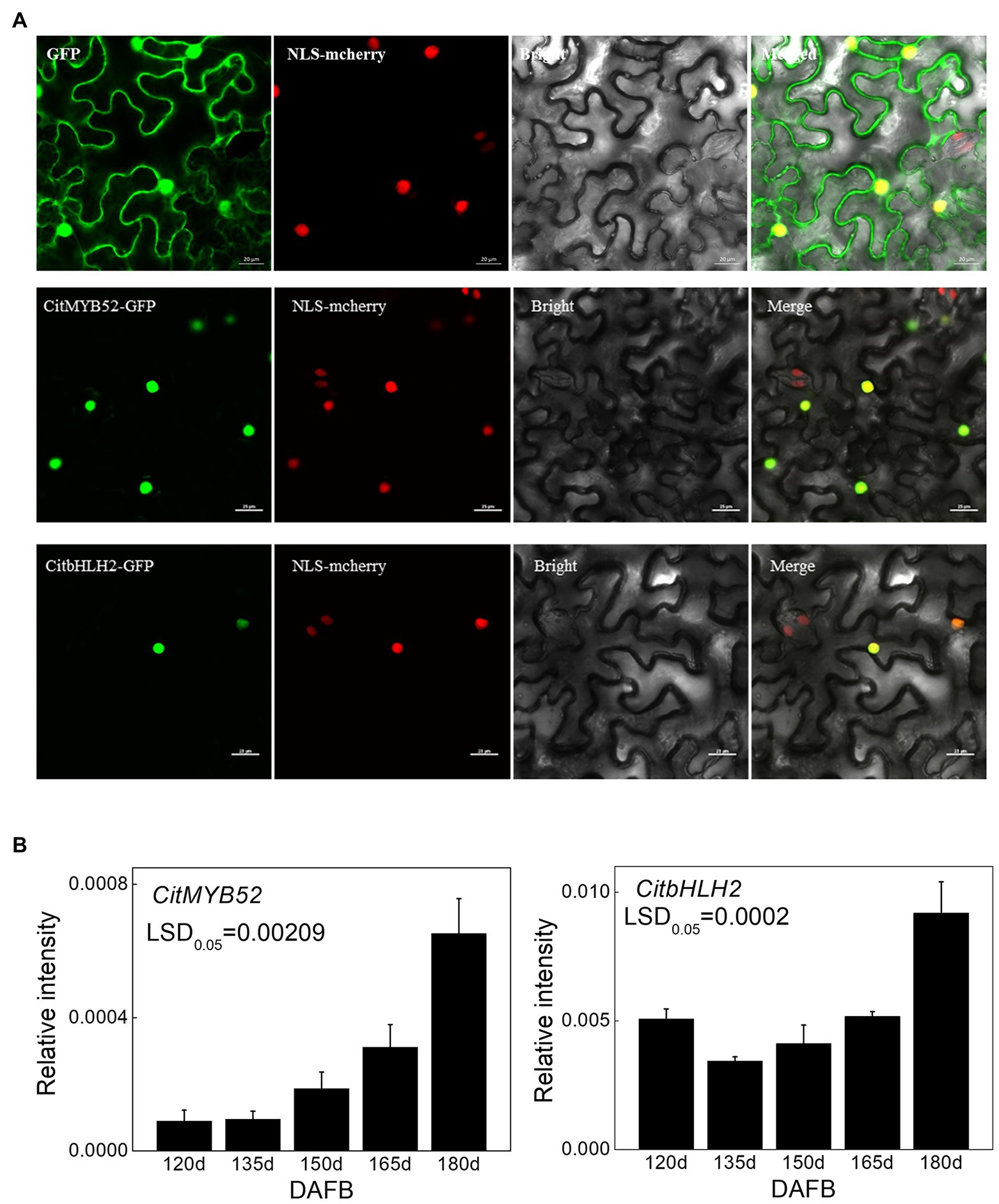
Figure 4. Characterization of CitMYB52 and CitbHLH2. (A) Subcellular localization analysis was performed in N. benthamiana leaves stably transformed with a red nuclear localization marker. GFP fluorescence of CitMYB52-GFP and CitbHLH2-GFP are indicated. The empty vector control is at the top. Bars = 25 μm. (B) Expression of the CitMYB52 and CitbHLH2 genes in the flesh of “Ponkan” fruits during the late stage of fruit development, DAFB, days after full blossom. Error bars represent SE (n = 3).
Both CitMYB52 and CitbHLH2 exhibited a rising trend in expression which was negatively correlating with citrate concentration, excluding the 120 DAFB in CitbHLH2 time point (Figure 4B).
Based on the EMSA results, the relationship between CitMYB52 and CitbHLH2 was further studied. BiFC assays showed that all the negative controls (empty vector and a combination of one gene with empty vector), including YFPN/CitMYB52-YFPC, CitbHLH2-YFPN/YFPC, and YFPN/YFPC, produced no fluorescence signal. Co-expression of CitMYB52-YFPC and CitbHLH2-YFPN, however, resulted in a positive signal in the nucleus (Figure 5A), indicating a protein–protein physical interaction between CitMYB52 and CitbHLH2. Furthermore, this interaction was verified by yeast two-hybrid assay and CoIP assay with HA and myc antibody (Figures 5B,C). Taken together, these results suggest that CitMYB52 and CitbHLH2 act cooperatively to increase CitALMT promoter activity.
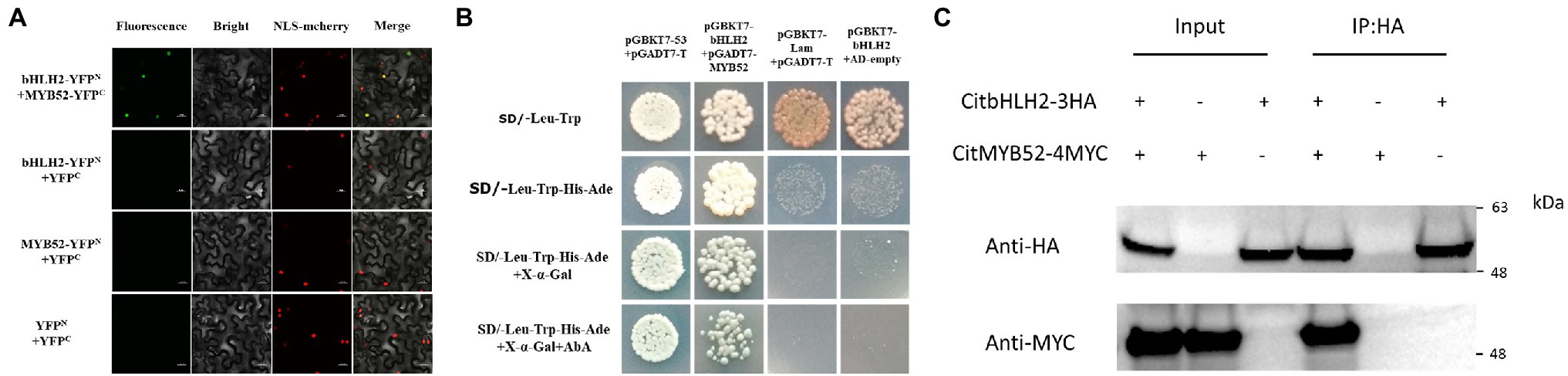
Figure 5. Protein–protein interaction between CitMYB52 and CitbHLH2. (A) In vivo interaction between CitMYB52 and CitbHLH2, determined using BiFC. N- and C-terminal fragments of YFP (indicated on the figure as YFPN and YFPC) were fused to the C terminus of CitbHLH2 and CitMYB52, respectively. The pairs of fusion proteins tested were CitbHLH2-YFPN+CitMYB52-YFPC. The other combinations were negative controls. Fluorescence of YFP represents protein–protein interaction. Bars = 50 μm. (B) Interaction between CitMYB52 and CitbHLH2 in yeast two-hybrid assays. Liquid cultures of double transformants were plated at OD600 = 0.01 dilutions on synthetic dropout selective media: (1) SD medium lacking Trp and Leu; (2) SD medium lacking Trp, Leu, His, and Ade; and (3) SD medium lacking Trp, Leu, His, and Ade, and supplemented with AbA. pGBKT7-p53 and pGADT7-T were used as positive controls, while pGBKT7-Lam and pGADT7-T were used as negative controls. (C) Interactions between CitMYB52 and CitbHLH2 measured by CoIP. Total protein extracts (Input) and protein complexes immunoprecipitated with anti-HA agarose (IP) were separated on gels and blotted. Anti-HA and anti-myc antibodies were used in Western blotting.
In order to investigate the effects of the two transcription factors on citrate, CitMYB52 and CitbHLH2 were introduced into citrus fruits by Agrobacterium-mediated transient transformation. The transcript levels of CitMYB52 and CitbHLH2 in overexpressing and RNAi fruits are indicated in Supplementary Figure S6 compared to the empty vector control. Transient overexpression of CitMYB52 and CitbHLH2 separately produced no difference in the citrate concentration in citrus fruits. However, the combined expression of CitMYB52 and CitbHLH2 significantly decreased the citrate concentration from 19.28 mg/g to 13.28 mg/g. In addition, the transcript abundance of CitALMT significantly increased 2.9-fold in CitMYB52 and CitbHLH2 dual overexpressing fruits (Figure 6A). Transient RNAi inhibition of both CitMYB52 and CitbHLH2 reduced the transcript level of CitALMT and increased the citrate concentration in citrus fruits (Figure 6B). These results indicated that these two transcription factors work together to influence citrate concentration by regulating the transcript level of CitALMT. In addition, the transcript levels of Aco3, IDH3, and GAD4 were in general higher in dual overexpressing fruits and lower in dual RNAi fruits (Supplementary Figure S7).
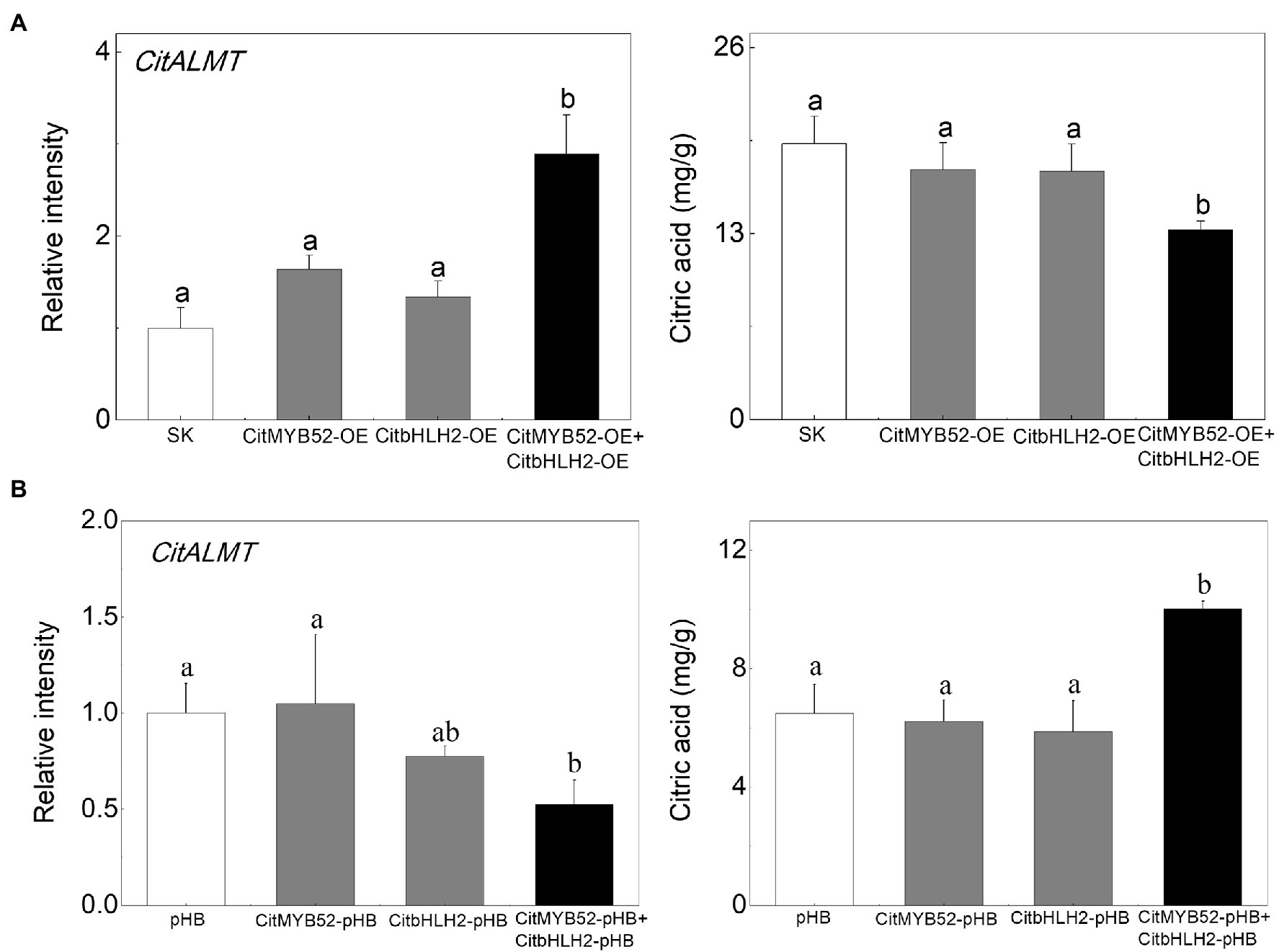
Figure 6. Effects on the citrate concentration of transient overexpression or RNAi inhibition of CitMYB52 and CitbHLH2. The transcript level of CitALMT (left) and citrate concentration (right) were analyzed in citrus fruits. (A) Transient overexpression of CitMYB52 and CitbHLH2 in citrus fruits. SK represents empty vector. (B) Transient RNAi of CitMYB52 and CitbHLH2 in citrus fruits. pHB represents empty vector. Error bars indicate SE from three biological replicates. Different letters above the columns represent significant differences (p < 0.05).
ALMT was originally characterized as a type of aluminum-activated malate transporter that is responsible for the exudation of malate in plant roots to chelate aluminum ions (Sasaki et al., 2004; Hoekenga et al., 2006). Subsequent research revealed that the physiological role of ALMT is not limited to the detoxification process of aluminum ions (Palmer et al., 2016; Sharma et al., 2016). In addition to malate, ALMTs may also mediate a bidirectional (i.e., efflux or influx) transport permeability and selectivity to both organic anions (i.e., citrate or tartrate) and inorganic anions (i.e., NO3− or Cl−; Ligaba et al., 2012; De Angeli et al., 2013; Sasaki et al., 2016). For example, ZmALMT2 exhibits a high permeability to malate and citrate in maize (Ligaba et al., 2012). HvALMT1 can utilize malate, fumarate, or citrate as the substrate for the transport process in barley (Gruber et al., 2010). VvALMT9 is thought to mediate the transport process of both tartrate and malate, leading to the accumulation of malate and malate in grapes (De Angeli et al., 2013). In fruits, it is widely known that organic acid transporters may regulate the accumulation of various organic acids simultaneously (Huang et al., 2021). Citrate transport through the tonoplast may occur via malate channels in most fleshy fruits (Etienne et al., 2013; Huang et al., 2021). In tomato fruit, Overexpression of SlTDT increased the malate concentration significantly, while the citrate concentration is reduced, indicating that SlTDT may play a crucial role in the distribution of malate and citrate in tomato vacuoles (Liu et al., 2017). However, unlike malate, due to the lack of reports on citrate transporters, the role of ALMT on the accumulation of citrate has not received enough attention in fruits, although a genome-wide association study (GWAS) using 775 tomato accessions has been reported that Sl-ALMT9 on tomato chromosome 6 was significantly associated with both malic acid and citric acid (Zhao et al., 2019).
In our study, the transcriptomic analysis and expression profiles showed that CitALMT was negatively correlated with citrate concentration (Figure 1C; Supplementary Figures S1, S2), indicating that CitALMT may be correlated with the reduction of citrate. Because of the challenges associated with obtaining transgenic fruiting citrus plants, transient overexpression and RNAi assay in citrus fruits and transgenic callus were conducted to analyze the role of CitALMT. Overexpression of CitALMT in citrus callus and fruits could effectively promote the accumulation of malate while CitALMT-RNAi fruits exhibited a decreased malate concentration, which was in keeping with previous suggestions that ALMT could promote malate accumulation in fruits. Moreover, overexpressing or decreasing CitALMT transcript level significantly reduced or increased citrate concentration, respectively (Figure 2). These results indicated that CitALMT could negatively regulate citrate accumulation but promoted malate accumulation. We proposed that CitALMT functions as a channel protein that may affect the reallocation of citrate and malate in vacuoles.
In mature citrus fruits, most of the citric acid is stored in vacuoles. During the late stage of fruit development, citric acid is moved from the vacuole to the cytoplasm, where excess citrate is degraded by activating the GABA shunt genes (Cercós et al., 2006). Our results indicate that the transcript levels of GABA shunt genes were upregulated in the CitALMT-overexpressing fruits and callus transgenic lines, while downregulated in the CitALMT-RNAi fruits (Supplementary Figures S3, S4). These results raised a possibility that CitALMT, as a channel protein, may function to regulate the reallocation of citrate, which in turn affects the transcript levels of GABA shunt genes to regulate the degradation process of citric acid.
Transcription factors regulate the structural genes and transporters in plants. Overexpressing SIAREB1 in tomato upregulated the expression of mCS and resulted in increased citrate and malate accumulation (Bastías et al., 2011); MdMYB1 was reported to be involved in malate accumulation by transcriptionally activating MdVHA-B1 and MdVHA-B2 in apple (Hu et al., 2016). In citrus, Noemi, a bHLH transcription factor, was essential for the production of flavonoid pigments and regulation of fruit acidity in citrus (Butelli et al., 2019). Our previous reports identified an ethylene response factor, CitERF13, which interacts with a vacuolar proton pump CitVHA-c4 to regulate citrate accumulation (Li et al., 2016); A heat shock transcription factor CitHsfA7 participates in citric acid degradation in citrus fruit via modulating CitAco3 (Li et al., 2021).
In recent years, some transcription factors that show regulatory effects on ALMT have been characterized. For instance, AtSTOP1, a C2H2 type transcription factor, is crucial to malate excretion by activating AtALMT1 expression in Arabidopsis (Iuchi et al., 2007). The apple MYB genes MYB1/73 are reported to regulate malate accumulation by directly transcriptionally activating the expression of several proton pump genes and MdALMT9 (Hu et al., 2016, 2017). A CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR2 was identified as an activator of AtALMT1 expression (Tokizawa et al., 2015), WRKY46 was shown to be a negative regulator of ALMT1, and the wrky46 mutant showed increased malic acid secretion and decreased accumulation of aluminum in the root apices (Ding et al., 2013). In tomato, Sl-WRKY42 acts as a transcriptional repressor of SlALMT9 (Ye et al., 2017). However, as with the research on the citrate transport process, the transcription factors involved in this process are also poorly studied. Here, the transcriptomic analysis identified 22 candidate transcription factors whose expression patterns were positively correlated to the CitALMT’s (Supplementary Figure S1C). Of these, CitMYB52 and CitbHLH2 showed significant regulatory activities on the CitALMT promoter (Figure 3A). This finding was further supported by associations between the expression of CitMYB52 and CitbHLH2 with CitALMT’s (Figure 4B). Moreover, EMSA analysis indicated that CitbHLH2 physically binds to E-box (CAATTG) within the CitALMT promoter (Figure 3C), which indicated the potential regulatory mechanism of the transcription factor by direct effects.
We proposed that a protein–protein interaction between CitMYB52 and CitbHLH2 exists, based on the co-regulatory effect in dual-luciferase assays (Figure 3B). MYB and bHLH proteins control multiple pathways in numerous biological processes. The well-characterized functions involve the formation of ternary complexes of MYB-bHLH-WD40 that participate in the biosynthesis of flavonoid and anthocyanin, such as poplar MYB165 and bHLH131, kiwifruit AcMYB123 and AcbHLH42 (Ma et al., 2018; Wang et al., 2019). Recent research suggested the potential role of MYB and bHLH proteins on vacuolar acidification. In petunia petals, PH4-AN1-AN11 complex (a MBW complex) is important for vacuolar acidification by recruiting WRKY factor PH3 to regulate proton pump gene CitPH5 (Verweij et al., 2016). In grape, VvWRKY26 enhances the activation effect of MBW complex on proton pump genes VvPH1 and VvPH5 (Amato et al., 2019). Mutations that disrupt the expression of homologous MYB, HLH, and/or WRKY transcription factors inhibited the expression of proton pump genes CitPH1 and CitPH5 to regulate citrus fruit acidity (Strazzer et al., 2019). This provides supporting evidence for the involvement of MYB-bHLH complex in regulating proton pump genes. Considering that the transport process of citric acid is also manipulated by channel proteins, however, the transcription complexes involved in this process are poorly identified.
Here, we identified two transcriptional activators CitMYB52 and CitbHLH2, which could both trans-activate CitALMT (Figure 3A). However, our transient overexpression assay suggested that overexpressing CitMYB52 or CitbHLH2 individually showed no significant effects on citrate concentration (Figure 6A). Y2H, BiFC, and CoIP assays confirmed a protein–protein interaction between CitMYB52 and CitbHLH2 exits (Figures 5A–C). Furthermore, this new CitMYB52-CitbHLH2 interaction synergistically trans-activated the promoter activity of CitALMT (Figures 3B, 6A) and significantly decreased citrate concentration (Figure 6A). Moreover, the RNAi assay verified that silencing of both CitMYB52 and CitbHLH2 suppressed the expression level of CitALMT, and thereby increased the citrate concentration in citrus fruits (Figure 6B). In addition, overexpression and silencing of both CitMYB52 and CitbHLH2 increased and reduced the expression levels of GABA shunt genes, respectively (Supplementary Figure S7), which corresponded to the transcript level of CitALMT. These results provided an example of MYB-bHLH mediated regulation of citrus fruit acidity by a carrier protein and important insights into the molecular mechanisms underlying acidity regulation in citrus fruits. Considering citrate is an important contributor to taste in citrus, the identification of this complex uncovers another instrumental step for organic acid accumulation.
Citric acid plays an important role in the flavor, nutrition, and beverage quality of citrus fruits. Here, the transcriptomic analysis identified an aluminum-activated malate transporter (CitALMT) and 22 transcription factors that showed a high correlation to citrate accumulation. Using transient overexpression, RNAi, and stable transformation, CitALMT was confirmed as a novel transporter that negatively regulates citrate accumulation. Moreover, two transcription factors, named CitMYB52 and CitbHLH2 could interact with each other to co-regulate citrate accumulation by regulating the expression of CitALMT. These findings provide an important clue and new insights into the underlying regulatory mechanism of CitALMT, which opens the way to develop molecular breeding for improved citrus fruit acidity.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
SjL and KC conceived the research plans and supervised the experiments. ScL and XL performed most of the experiments. CL and JS provided research material to ScL. BG and DW provided technical assistance to ScL. SjL, XY, and KC designed the experiments, analyzed the data, and wrote the article. DG revised the article with contributions of all the authors. All authors contributed to the article and approved the submitted version.
This research was supported by National Key Research and Development Program (2016YFD0400101), the National Natural Science Foundation of China (31801591), and the Fundamental Research Funds for the Central Universities (2018FZA6010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Harry Klee (University of Florida) for providing comments on the manuscript and Wenli Liu for data analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.848869/full#supplementary-material
Amato, A., Cavallini, E., Walker, A. R., Pezzotti, M., Bliek, M., Quattrocchio, F., et al. (2019). The MYB 5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J. 99, 1220–1241. doi: 10.1111/tpj.14419
Bastías, A., López-Climent, M., Valcárcel, M., Rosello, S., Gómez-Cadenas, A., and Casaretto, J. A. (2011). Modulation of organic acids and sugar content in tomato fruits by an abscisic acid-regulated transcription factor. Physiol. Plant. 141, 215–226. doi: 10.1111/j.1399-3054.2010.01435.x
Butelli, E., Licciardello, C., Ramadugu, C., Durand-Hulak, M., Celant, A., Reforgiato Recupero, G., et al. (2019). Noemi controls production of flavonoid pigments and fruit acidity and illustrates the domestication routes of modern citrus varieties. Curr. Biol. 29, 158.e2–164.e2. doi: 10.1016/j.cub.2018.11.040
Cercós, M., Soler, G., Iglesias, D. J., Gadea, J., Forment, J., and Talón, M. (2006). Global analysis of gene expression during development and ripening of citrus fruit flesh. A proposed mechanism for citric acid utilization. Plant Mol. Biol. 62, 513–527. doi: 10.1007/s11103-006-9037-7
Chen, M., Qian, J., Yin, X. R., Lin, Q., Chen, J. Y., Allan, A. C., et al. (2012). Effect of hot air treatment on organic acid-and sugar-metabolism in Ponkan (Citrus reticulata) fruit. Sci. Hortic. 147, 118–125. doi: 10.1016/j.scienta.2012.09.011
De Angeli, A., Baetz, U., Francisco, R., Zhang, J. B., Chaves, M. M., and Regalado, A. (2013). The vacuolar channel VvALMT9 mediates malate and tartrate accumulation in berries of Vitis vinifera. Planta 238, 283–291. doi: 10.1007/s00425-013-1888-y
Ding, Z. J., Yan, J. Y., Xu, X. Y., Li, G. X., and Zheng, S. J. (2013). WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 76, 825–835. doi: 10.1111/tpj.12337
Dyson, B. C., Miller, M. A., Feil, R., Rattray, N., Bowsher, C. G., Goodacre, R., et al. (2016). FUM2, a cytosolic fumarase, is essential for acclimation to low temperature in Arabidopsis thaliana. Plant Physiol. 172, 118–127. doi: 10.1104/pp.16.00852
Etienne, A., Génard, M., Lobit, P., Mbeguié-A-Mbéguié, D., and Bugaud, C. (2013). What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J. Exp. Bot. 64, 1451–1469. doi: 10.1093/jxb/ert035
Finkemeier, I., König, A. C., Heard, W., Nunes-Nesi, A., Pham, P. A., Leister, D., et al. (2013). Transcriptomic analysis of the role of carboxylic acids in metabolite signaling in Arabidopsis leaves. Plant Physiol. 162, 239–253. doi: 10.1104/pp.113.214114
Gruber, B. D., Ryan, P. R., Richardson, A. E., Tyerman, S. D., Ramesh, S., Hebb, D. M., et al. (2010). HvALMT1 from barley is involved in the transport of organic anions. J. Exp. Bot. 61, 1455–1467. doi: 10.1093/jxb/erq023
Han, D. G., Shi, Y., Wang, B., Liu, W., Yu, Z. Y., Lv, B. Y., et al. (2015). Isolation and preliminary functional analysis of MxCS2: a gene encoding a citrate synthase in Malus xiaojinensis. Plant Mol. Biol. Report. 33, 133–142. doi: 10.1007/s11105-014-0735-z
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 1–14. doi: 10.1186/1746-4811-1-13
Hoekenga, O. A., Maron, L. G., Piñeros, M. A., Cançado, G. M., Shaff, J., Kobayashi, Y., et al. (2006). AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 103, 9738–9743. doi: 10.1073/pnas.0602868103
Hu, D. G., Li, Y. Y., Zhang, Q. Y., Li, M., Sun, C. H., Yu, J. Q., et al. (2017). The R2R3-MYB transcription factor MdMYB73 is involved in malate accumulation and vacuolar acidification in apple. Plant J. 91, 443–454. doi: 10.1111/tpj.13579
Hu, D. G., Sun, C. H., Ma, Q. J., You, C. X., Cheng, L., and Hao, Y. J. (2016). MdMYB1 regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples. Plant Physiol. 170, 1315–1330. doi: 10.1104/pp.15.01333
Huang, X. Y., Wang, C. K., Zhao, Y. W., Sun, C. H., and Hu, D. G. (2021). Mechanisms and regulation of organic acid accumulation in plant vacuoles. Hortic Res. 8: 227. doi: 10.1038/s41438-021-00702-z
Hurth, M. A., Suh, S. J., Kretzschmar, T., Geis, T., Bregante, M., Gambale, F., et al. (2005). Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol. 137, 901–910. doi: 10.1104/pp.104.058453
Iuchi, S., Koyama, H., Iuchi, A., Kobayashi, Y., Kitabayashi, S., Kobayashi, Y., et al. (2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. U. S. A. 104, 9900–9905. doi: 10.1073/pnas.0700117104
Ladanyia, M. (2008). Citrus Fruit: Biology, Technology and Evaluation. USA: Academic Press, 135–137.
Li, S. J., Liu, S. C., Lin, X. H., Grierson, D., Yin, X. R., and Chen, K. S. (2021). Citrus heat shock transcription factor CitHsfA7-mediated citric acid degradation in response to heat stress. Plant Cell Environ. 45, 95–104. doi: 10.1111/pce.14207
Li, D. D., Shi, W., and Deng, X. X. (2002). Agrobacterium-mediated transformation of embryogenic calluses of Ponkan mandarin and the regeneration of plants containing the chimeric ribonuclease gene. Plant Cell Rep. 21, 153–156. doi: 10.1007/s00299-002-0492-6
Li, S., Xu, H. J. L., Ju, Z., Cao, D. Y., Zhu, H. L., Fu, D. Q., et al. (2018). The RIN-MC fusion of MADS-box transcription factors has transcriptional activity and modulates expression of many ripening genes. Plant Physiol. 176, 891–909. doi: 10.1104/pp.17.01449
Li, S. J., Yin, X. R., Wang, W. L., Liu, X. F., Zhang, B., and Chen, K. S. (2017). Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. J. Exp. Bot. 68, 3419–3426. doi: 10.1093/jxb/erx187
Li, S. J., Yin, X. R., Xie, X. L., Allan, A. C., Ge, H., Shen, S. L., et al. (2016). The citrus transcription factor, CitERF13, regulates citric acid accumulation via a protein-protein interaction with the vacuolar proton pump, CitVHA-c4. Sci. Rep. 6: 20151. doi: 10.1038/srep20151
Ligaba, A., Maron, L., Shaff, J., Kochian, L., and Piñeros, M. (2012). Maize ZmALMT2 is a root anion transporter that mediates constitutive root malate efflux. Plant Cell Environ. 35, 1185–1200. doi: 10.1111/j.1365-3040.2011.02479.x
Lin, Q., Li, S. J., Dong, W. C., Feng, C., Yin, X. R., Xu, C. J., et al. (2015a). Involvement of CitCHX and CitDIC in developmental-related and postharvest-hot air driven citrate degradation in citrus fruits. PLoS One 10:e0119410. doi: 10.1371/journal.pone.0119410
Lin, Q., Qian, J., Zhao, C. N., Wang, D. L., Liu, C. R., Wang, Z. D., et al. (2016). Low temperature induced changes in citrate metabolism in ponkan (Citrus reticulata Blanco cv. Ponkan) fruit during maturation. PLoS One 11:e0156703. doi: 10.1371/journal.pone.0156703
Lin, Q., Wang, C. Y., Dong, W. C., Jiang, Q., Wang, D. L., Li, S. J., et al. (2015b). Transcriptome and metabolome analyses of sugar and organic acid metabolism in Ponkan (Citrus reticulata) fruit during fruit maturation. Gene 554, 64–74. doi: 10.1016/j.gene.2014.10.025
Liu, R. L., Li, B. Q., Qin, G. Z., Zhang, Z. Q., Tian, S. P., et al. (2017). Identification and functional characterization of a tonoplast dicarboxylate transporter in tomato (Solanum lycopersicum). Front. Plant Sci. 8:186. doi: 10.3389/fpls.2017.00186
Liu, W. L., Zhang, J., Jiao, C., Yin, X. R., Fei, Z. J., Wu, Q. B., et al. (2019). Transcriptome analysis provides insights into the regulation of metabolic processes during postharvest cold storage of loquat (Eriobotrya japonica) fruit. Hortic Res. 6: 49. doi: 10.1038/s41438-019-0131-9
Ma, D., Reichelt, M., Yoshida, K., Gershenzon, J., and Constabel, C. P. (2018). Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 96, 949–965. doi: 10.1111/tpj.14081
Palmer, A. J., Baker, A., and Muench, S. P. (2016). The varied functions of aluminium-activated malate transporters: much more than aluminium resistance. Biochem. Soc. Trans. 44, 856–862. doi: 10.1042/BST20160027
Sainsbury, F., Thuenemann, E. C., and Lomonossoff, G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693. doi: 10.1111/j.1467-7652.2009.00434.x
Sasaki, T., Tsuchiya, Y., Ariyoshi, M., Nakano, R., Ushijima, K., Kubo, Y., et al. (2016). Two members of the aluminum-activated malate transporter family, SlALMT4 and SlALMT5, are expressed during fruit development and the overexpression of SlALMT5 alters organic acid contents in seeds in tomato (Solanum lycopersicum). Plant Cell Physiol. 57, 2367–2379. doi: 10.1093/pcp/pcw157
Sasaki, T., Yamamoto, Y., Ezaki, B., Katsuhara, M., Ahn, S. J., Ryan, P. R., et al. (2004). A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37, 645–653. doi: 10.1111/j.1365-313x.2003.01991.x
Sharma, T., Dreyer, I., Kochian, L., and Piñeros, M. A. (2016). The ALMT family of organic acid transporters in plants and their involvement in detoxification and nutrient security. Front. Plant Sci. 7:1488. doi: 10.3389/fpls.2016.01488
Shi, C. Y., Song, R. Q., Hu, X. M., Liu, X., Jin, L. F., and Liu, Y. Z. (2015). Citrus PH5-like H(+)-ATPase genes: identification and transcript analysis to investigate their possible relationship with citrate accumulation in fruits. Front. Plant Sci. 6:135. doi: 10.3389/fpls.2015.00135
Shimada, T., Nakano, R., Shulaev, Y., Sadka, A., and Blumwald, E. (2006). Vacuolar citrate/H+ symporter of citrus juice cells. Planta 224, 472–480. doi: 10.1007/s00425-006-0223-2
Shlizerman, L., Marsh, K., Blumwald, E., and Sadka, A. (2007). Iron-shortage-induced increase in citric acid content and reduction of cytosolic aconitase activity in citrus fruit vesicles and calli. Physiol. Plant. 131, 72–79. doi: 10.1111/j.1399-3054.2007.00935.x
Strazzer, P., Spelt, C. E., Li, S., Bliek, M., Federici, C. T., Roose, M. L., et al. (2019). Hyperacidification of citrus fruits by a vacuolar proton-pumping P-ATPase complex. Nat. Commun. 10:744. doi: 10.1038/s41467-019-08516-3
Tokizawa, M., Kobayashi, Y., Saito, T., Kobayashi, M., Iuchi, S., Nomoto, M., et al. (2015). Sensitive to proton rhizotoxicity1, calmodulin binding transcription activator2, and other transcription factors are involved in aluminum-activated malate transporter1 expression. Plant Physiol. 167, 991–1003. doi: 10.1104/pp.114.256552
Verweij, W., Spelt, C. E., Bliek, M., de Vries, M., Wit, N., Faraco, M., et al. (2016). Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in Arabidopsis. Plant Cell 28, 786–803. doi: 10.1105/tpc.15.00608
Wang, L., He, F., Huang, Y., He, J. X., Yang, S. Z., Zeng, J. W., et al. (2018). Genome of wild mandarin and domestication history of mandarin. Mol. Plant 11, 1024–1037. doi: 10.1016/j.molp.2018.06.001
Wang, X. W., Jing, B., Tan, F., Ma, J. B., Zhang, Y. H., and Ge, M. F. (2017). Hygroscopic behavior and chemical composition evolution of internally mixed aerosols composed of oxalic acid and ammonium sulfate. Atmos. Chem. Phys. 17, 12797–12812. doi: 10.5194/acp-17-12797-2017
Wang, L. H., Tang, W., Hu, Y. W., Zhang, Y. B., Sun, J. Q., Guo, X. H., et al. (2019). A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 99, 359–378. doi: 10.1111/tpj.14330
Wang, X. Y., Wang, P., Qi, Y. P., Zhou, C. P., Yang, L. T., Liao, X. Y., et al. (2014). Effects of granulation on organic acid metabolism and its relation to mineral elements in Citrus grandis juice sacs. Food Chem. 145, 984–990. doi: 10.1016/j.foodchem.2013.09.021
Weinert, B. T., Schölz, C., Wagner, S. A., Iesmantavicius, V., Su, D., Daniel, J. A., et al. (2013). Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851. doi: 10.1016/j.celrep.2013.07.024
Wu, F. M., Wang, X. W., Pang, S. F., and Zhang, Y. H. (2018). Importance of water-soluble organic acid on the hygroscopicity of nitrate. Atmos. Environ. 219, 104–109. doi: 10.1016/j.saa.2019.04.034
Xu, Q., Yin, X. R., Zeng, J. K., Ge, H., Song, M., Xu, C. J., et al. (2014). Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 65, 4349–4359. doi: 10.1093/jxb/eru208
Ye, J., Wang, X., Hu, T. X., Zhang, F. X., Wang, B., Li, C. X., et al. (2017). An InDel in the promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell 29, 2249–2268. doi: 10.1105/tpc.17.00211
Zampini, M., Wantling, E., Phillips, N., and Spence, C. (2008). Multisensory flavor perception: assessing the influence of fruit acids and color cues on the perception of fruit-flavored beverages. Food Qual. Prefer. 19, 335–343. doi: 10.1016/j.foodqual.2007.11.001
Zhang, J. B., Baetz, U., Krügel, U., Martinoia, E., and De Angeli, A. (2013). Identification of a probable pore-forming domain in the multimeric vacuolar anion channel AtALMT9. Plant Physiol. 163, 830–843. doi: 10.1104/pp.113.219832
Zhang, Z. Y., Shi, Y. N., Ma, Y. C., Yang, X. F., Yin, X. R., Zhang, Y. Y., et al. (2020). The strawberry transcription factor farav1 positively regulates anthocyanin accumulation by activation of famyb10 and anthocyanin pathway genes. Plant Biotechnol. J. 18, 2267–2279. doi: 10.1111/pbi.13382
Zhang, Z. Z., Tan, M. J., Xie, Z. Y., Dai, L. Z., Chen, Y., and Zhao, Y. M. (2011). Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63. doi: 10.1038/nchembio.495
Keywords: citrus fruit, citric acid, aluminum-activated malate transporter, transcriptional regulation, MYB-bHLH protein complex
Citation: Liu S, Liu X, Gou B, Wang D, Liu C, Sun J, Yin X, Grierson D, Li S and Chen K (2022) The Interaction Between CitMYB52 and CitbHLH2 Negatively Regulates Citrate Accumulation by Activating CitALMT in Citrus Fruit. Front. Plant Sci. 13:848869. doi: 10.3389/fpls.2022.848869
Received: 05 January 2022; Accepted: 28 February 2022;
Published: 21 March 2022.
Edited by:
Luciano Freschi, University of São Paulo, BrazilReviewed by:
Manuel Talón, Instituto Valenciano de Investigaciones Agrarias, SpainCopyright © 2022 Liu, Liu, Gou, Wang, Liu, Sun, Yin, Grierson, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaojia Li, c2hhb2ppYWxpQHpqdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.