- 1College of Forestry, Shandong Agricultural University, Tai’an, China
- 2Shandong Provincial Academy of Forestry, Jinan, China
- 3Shandong Taishan Forest Ecosystem Research Station, Shandong Agricultural University, Tai’an, China
- 4State Forestry and Grassland Administration Key Laboratory of Silviculture in Downstream Areas of the Yellow River, Shandong Agricultural University, Tai’an, China
- 5Culaishan Forest Farm, Tai’an, China
Soil salinization is a major environmental problem that seriously threatens the sustainable development of regional ecosystems and local economies. Fraxinus velutina Torr. is an excellent salt-tolerant tree species, which is widely planted in the saline-alkaline soils in China. A growing body of evidence shows that microRNAs (miRNAs) play important roles in the defense response of plants to salt stress; however, how miRNAs in F. velutina exert anti-salt stress remains unclear. We previously identified two contrasting F. velutina cuttings clones, salt-tolerant (R7) and salt-sensitive (S4) and found that R7 exhibits higher salt tolerance than S4. To identify salt-responsive miRNAs and their target genes, the leaves and roots of R7 and S4 exposed to salt stress were subjected to miRNA and degradome sequencing analysis. The results showed that compared with S4, R7 showed 89 and 138 differentially expressed miRNAs in leaves and roots, respectively. Specifically, in R7 leaves, miR164d, miR171b/c, miR396a, and miR160g targeting NAC1, SCL22, GRF1, and ARF18, respectively, were involved in salt tolerance. In R7 roots, miR396a, miR156a/b, miR8175, miR319a/d, and miR393a targeting TGA2.3, SBP14, GR-RBP, TCP2/4, and TIR1, respectively, participated in salt stress responses. Taken together, the findings presented here revealed the key regulatory network of miRNAs in R7 responding to salt stress, thereby providing new insights into improving salt tolerance of F. velutina through miRNA manipulation.
Introduction
Soil salinization is a major environmental problem. It is estimated that by the year 2050, more than half of global arable land will be saline contamination (Butcher et al., 2016). Salinized soils hinder the growth and development of plants, resulting in the loss of biomass production, and even the deterioration of regional ecosystem (Polle and Chen, 2015; Hossain and Dietz, 2016; Ondrasek and Rengel, 2021). What’s more serious is that increasing soil salinization is now threatening sustainable development of local economies (Chen and Mueller, 2018). How to protect and restore the fragile ecosystem in salinized areas has become an urgent global issue. The selection and cultivation of naturally salt-tolerant plants are currently considered as an economically feasible strategy for the problem (Litalien and Zeeb, 2020). Fraxinus velutina Torr. is an excellent salt-tolerant tree species, which is widely planted in the saline-alkaline soils in Yellow River Delta, China (Mao et al., 2017). However, the mechanisms underlying salt tolerance of F. velutina remain largely unclear.
MicroRNAs (miRNAs) are a class of endogenous small non-coding RNAs with 18–25 nucleotides (nt), playing key regulatory roles in the defense response of plants to salt stress by regulating their target genes at a post-transcriptional level (Kumar et al., 2018; Song S. et al., 2019; Xu et al., 2021). The overexpression of miR528 in rice (Oryza sativa) can increase the contents of ascorbic and abscisic acid, and decrease reactive oxygen species (ROS) accumulation, thereby enhancing rice salt tolerance (Wang et al., 2021). Constitutive expression of rice miR528 and miR396 in creeping bentgrass modulates the growth and development, and enhances the response to salinity stress (Yuan et al., 2015, 2019). In wheat (Triticum aestivum), miRNA408 has been found to act as a crucial mediator in the tolerance to Pi deprivation and salt stress through modulating multiple stresses related to physiological processes (Bai et al., 2018). In maize (Zea mays), it is evidenced that miR169/NF-YA module is a key regulatory mediator in the response to salt stress in leaves and roots (Luan et al., 2014, 2015). In Arabidopsis, it is confirmed that miRNA393-associated regulatory modules can enhance the salt stress resistance by mediating several biological processes, including auxin signaling, redox-related components, osmoregulation and increased Na+ exclusion (Iglesias et al., 2014; Chen Z. et al., 2015; Denver and Ullah, 2019). The overexpressed miR414c in cotton (Gossypium hirsutum) can negatively regulate iron superoxide dismutase gene, thereby enhancing plant tolerance to salinity stress (Wang et al., 2019). A miR156/SPL regulatory module increases tolerance to salinity stress via up-regulating MdWRKY100 in apple (Malus domestica) (Ma et al., 2021). However, little information is available about the regulatory role of miRNAs in F. velutina responding to salt stress.
As important regulatory molecules, plant miRNAs bind to their target mRNAs and regulate gene expression by direct cleavage and degradation of their targets (Djami-Tchatchou et al., 2017). To date, multiple approaches to study miRNAs, their target genes, and regulatory networks have been established (Sun et al., 2014). Of these approaches, the most commonly used is direct miRNA cloning and/or deep sequencing. Various experimental approaches like RLM-RACE and degradome sequencing have been developed to identify miRNA targets in plants (Shamimuzzaman and Vodkin, 2012; Wang C. et al., 2013). Degradome sequencing is a powerful approach that integrates a modified RLM-RACE and deep sequencing, which has been used to confirm miRNA targets globally in plants. Recently, the combination of miRNA and degradome sequencing has been widely used to identify the salt-responsive miRNAs and their target genes involved in regulating plant tolerance to salt stress (Yu et al., 2016; Kumar et al., 2017; Cervera-Seco et al., 2019; Zhang Y. et al., 2020; Xu et al., 2021).
In our previous study, we identified two contrasting cutting clones R7 (salt-tolerant) and S4 (salt-sensitive) of F. velutina in which R7 exhibits higher salt tolerance than S4. Meanwhile, we performed a comparative transcriptome analysis between R7 and S4, and identified some key genes and signaling pathways underlying high salt tolerance of R7 (Ma et al., 2022). In this study, to identify the salt-responsive miRNAs and their target genes involved in salt stress tolerance, an integration of miRNA and degradome sequencing analysis was performed on the leaves and roots of R7 and S4 with or without salt treatment. Our work revealed the key regulatory network of miRNAs in R7 responding to salt stress, thereby providing new insights into improving salt tolerance of F. velutina through miRNA manipulation.
Materials and Methods
Plant Materials and Culture Conditions
The salt-tolerant R7 and salt-sensitive S4 cutting clones of F. velutina were collected from the Experimental Base of Afforestation on Saline-Alkali Soil of Shandong Provincial Academy of Forestry, Shouguang city, China (118°42′9.18′′ E, 37°9′38.94′′ N). As previously described (Ma et al., 2022), 1-year-old cutting clones of both R7 and S4 were used in this study. After gently being removed rhizosphere soils, the clones were first pre-cultured in distilled water without any nutrient for 2 weeks, and then were transferred to a plastic container with 6 L of half-strength Hoagland’s solution for 4 weeks; the solution was refreshed every 7 days. All the experimental cutting clones were cultured in a growth incubator (LICHEN, Shanghai, China) with 25/20°C (day/night temperature), and 16 h light (1,200 μmol m–2 s–1)/8 h dark.
Salt Treatment and Sampling
After 6 weeks of acclimatization, the healthy clones with uniform size were selected and treated with half-strength Hoagland’s solution containing 250 mM NaCl for 12 h; the seedlings without NaCl treatment were considered as the control. After treatment, the leaves and roots of R7 and S4 were collected and stored at −80°C for further analysis. The symbols of leaves and roots samples were letters ‘L’ and ‘R,’ respectively. For instance, the samples R7SL and R7CL represented the leaves of R7 cutting clones with or without salt treatment, respectively.
Small RNA and Degradome Library Construction and Sequencing
Total of 24 samples (roots and leaves of R7 and S4, and two treatments with three biological replicates) were used small RNA and degradome library construction and sequencing. Total RNAs including small and large-size of tested samples were extracted using E.Z.N.A. Micro RNA Kit (Omega Bio-Tek, Norcross, GA, United States) following the manufacturer’s instructions. The quality and quantity of the isolated RNA were evaluated using a Bioanalyzer 2100 instrument (Agilent, Santa Clara, CA, United States) and an RNA 6000 Nano LabChip Kit (Agilent, Santa Clara, CA, United States), ensuring that the RNA integrity number value was higher than 8.0. For small RNA sequencing, the library of each sample was constructed using TruSeq Small RNA Library Preparation Kit (Illumina, San Diego, CA, United States), according to the manufacturer’s protocol. For degradome sequencing, two libraries from equally pooled samples of S7 and S4 were, respectively, constructed as previously described (German et al., 2008, 2009). Both small RNA and degradome libraries were sequenced on an Illumina Hiseq 4000 instrument (LC-Bio, Hangzhou, China) at single-end (50 bp).
Identification of miRNAs
The raw reads were first trimmed using Trimmomatic v.0.39 (Bolger et al., 2014) to remove the junk reads, adapters, and low-complexity sequences. The validated reads were subjected to remove 3′ adaptor (5′-TGGAATTCTCGGGTGCCAAGG-3′) and perform length filter using Cutadapt v.3.5 (Kechin et al., 2017). The sequences with 18–25 nt were then subjected to exclude mRNA and other non-coding RNAs (tRNAs, rRNAs, snoRNAs, and snRNAs) via aligning the sequences, respectively to F. velutina mRNA sequences1 (Kelly et al., 2020) and Rfam v.14.6 database2 using the BLAST tool (BLASTN) (Altschul et al., 1990). The remaining unique sequences were subsequently aligned to the available plant miRNAs sequences stored in miRBase v.22.13 to identify the conserved miRNAs in F. velutina using bowtie v1.3.0 (Langmead et al., 2009) with a maximum allowed mismatch ≤ 2.
The mature miRNA sequences obtained were mapped to the F. velutina reference genome sequence (see Text Footnote 1) by bowtie software, and then the corresponding flanking sequences of these miRNAs were extracted and used to predict the hairpin RNA structures using UNAfold v3.8 (Markham and Zuker, 2008). The potential miRNA precursors must meet the criteria as following: (1) the miRNAs were located on the arms (3′ or 5′) of stem-loop hairpin structure; (2) the miRNAs cannot contain large loops or breaks; (3) a maximum mismatches ≤ 6 were allowed between the miRNAs and their opposite sequences; (4) the predicted miRNA precursor structures must have minimal free folding energy index (>0.8) and negative minimal folding free energy, to differentiate them from other RNAs (Zhang et al., 2006; Yin et al., 2008).
The remaining small RNA sequences were used to identify novel miRNAs. After mapped to the F. velutina reference genome sequence, the flanking sequences of small RNAs were retrieved to predict secondary structures as the above mentioned. Only the small RNAs exhibiting a perfect stem-loop structure and meeting the criteria for plant miRNAs were identified as candidate novel miRNAs (Meyers et al., 2008).
Degradome Sequencing Data Analysis and miRNA Targets Identification
After trimmed by Trimmomatic software, the validated degradome sequencing reads were subjected to predict the putative miRNA cleaved targets using the CleaveLand pipeline v.3.0 (Addo-Quaye et al., 2009). In brief, the sequencing reads were mapped to the F. velutina mRNA sequences downloaded from Ash Tree Genomes Database (see Text Footnote 1), and the perfect matching alignments were retained. The resulting tags with a 35–36 nt extended signature by adding 15 nt of upstream sequence were subsequently aligned to the identified mature miRNAs in this study, with a maximum allowed mismatch ≤ 5. Alignments where the degradome tag position coincided with the 10th or 11th nt of a given miRNA were kept and scored (Allen et al., 2005). According to the previously described, targets were classified into I, II, or III (Addo-Quaye et al., 2008). In addition, t-plots showing the distribution of signatures of miRNA cleaved targets were built using R package.
Identification of Salt-Responsive miRNAs and Their Targets
The expression levels of the miRNAs were determined and normalized by transcripts per kilobase million. The differentially expressed miRNAs (DEmiRs) between each group were identified using R package DEseq2 (Love et al., 2014), with | log2 fold change| ≥ 1 and P-value ≤ 0.05.
We previously performed a comparative transcriptome analysis between R7 and S4, and identified some key genes and signaling pathways underlying high salt tolerance of R7 (Ma et al., 2022). To narrow the targets of salt-responsive miRNAs, our previous transcriptome data were used. For each comparison, the miRNA-target pairs containing DEmiRs and differentially expressed genes (DEGs) were selected for further analysis, and for each selected pair, the expression patterns of DEmiR were opposite to that of DEGs.
Validation of the Identified miRNAs by Quantitative Real-Time PCR
To verify the results of small RNA sequencing, 15 DEmiRs including 12 conserved (ptc-miR160a, ptc-miR160g, mtr-miR164d, stu-MIR167d-p3_2ss6TC19CT, bna-MIR169c-p5_2ss12GC17TG, gma-miR169j-5p, mtr-MIR2592bj-p3_2ss12TC19AT, mtr-MIR2592bj-p3_2ss12TC19AT, mtr-miR393a_L+1, hbr-miR396a_R-1_2ss19TC20CT, hbr-miR396a_R-1_2ss19TC20CT, and gma-miR403a_R-1) and 3 novels (PC-3p-46517_176, PC-3p-56802_134, and PC-5p-55954_137) were randomly selected to perform quantitative real-time PCR (qRT-PCR) analysis (Supplementary Table 1). Total RNA isolated from leaves and roots using the E.Z.N.A. Micro RNA Kit (Omega Bio-Tek, Norcross, GA, United States) according to the manufacturer’s protocol. The primers were designed using Primer 5.0 software and synthesized by Sangon Biotech Co., Ltd. (Shanhai, China). The reverse transcription of miRNA was performed using Mir-X miRNA First-Strand Synthesis and TB Green qRT-PCR User Manual (Takara, Dalian, China) according to the manufacturer’s protocol.
The qRT-PCR was performed with the TB Green Premix Ex Taq II kit (Takara, Dalian, China) on a CFX Connect Real-Time instrument (Bio-Rad, Hercules, CA, United States). Each sample had three independent replicates. The 5.8s rRNA was used as an internal reference gene. The relative expression level of each miRNA was calculated based on the 2–Δ Δ Ct method (Livak and Schmittgen, 2001).
Statistical Analyses
Statistical data were presented as mean ± standard deviation (SD). Student’s t-test was used to compare the differences between two groups. A P-value ≤ 0.05 was considered to be significant difference. GraphPad Prism v.9.0 (GraphPad Software Inc., La Jolla, CA, United States) was used to perform statistical analysis.
Results
Identification of Conserved and Novel miRNAs in F. velutina
To identify miRNAs in F. velutina responding to salt stress, 24 small RNA sequencing libraries from leaf and root samples of R7 and S4 under salt treatment and control conditions were constructed and sequenced. In total, an average of 11.92 million raw reads for each sample were obtained. After removing low quality reads and adaptor sequences, approximately 2.13 million unique validated reads were obtained for each sample (Supplementary Table 2). After further excluding mRNA, tRNAs, rRNAs, snoRNAs, and snRNAs, an average of 2.12 million unique reads with length of 18–25 nt were generated for each sample (Supplementary Table 3).
To identify the conserved and novel miRNAs in F. velutina, the filtered unique reads were aligned to the miRBase v.22.1 database and F. velutina reference genome. A total of 987 miRNAs including 560 conserved and 427 novel miRNAs were identified. The majority of miRNAs were distributed between 20 and 24 nt, with 24 nt exhibiting the highest abundance (Figure 1A). On the basis of sequence similarity, these miRNAs were further classified into 50 miRNA families, with MIR156, MIR159, MIR166, and MIR396 presenting relatively high abundance (Supplementary Figure 1A and Supplementary Table 4).
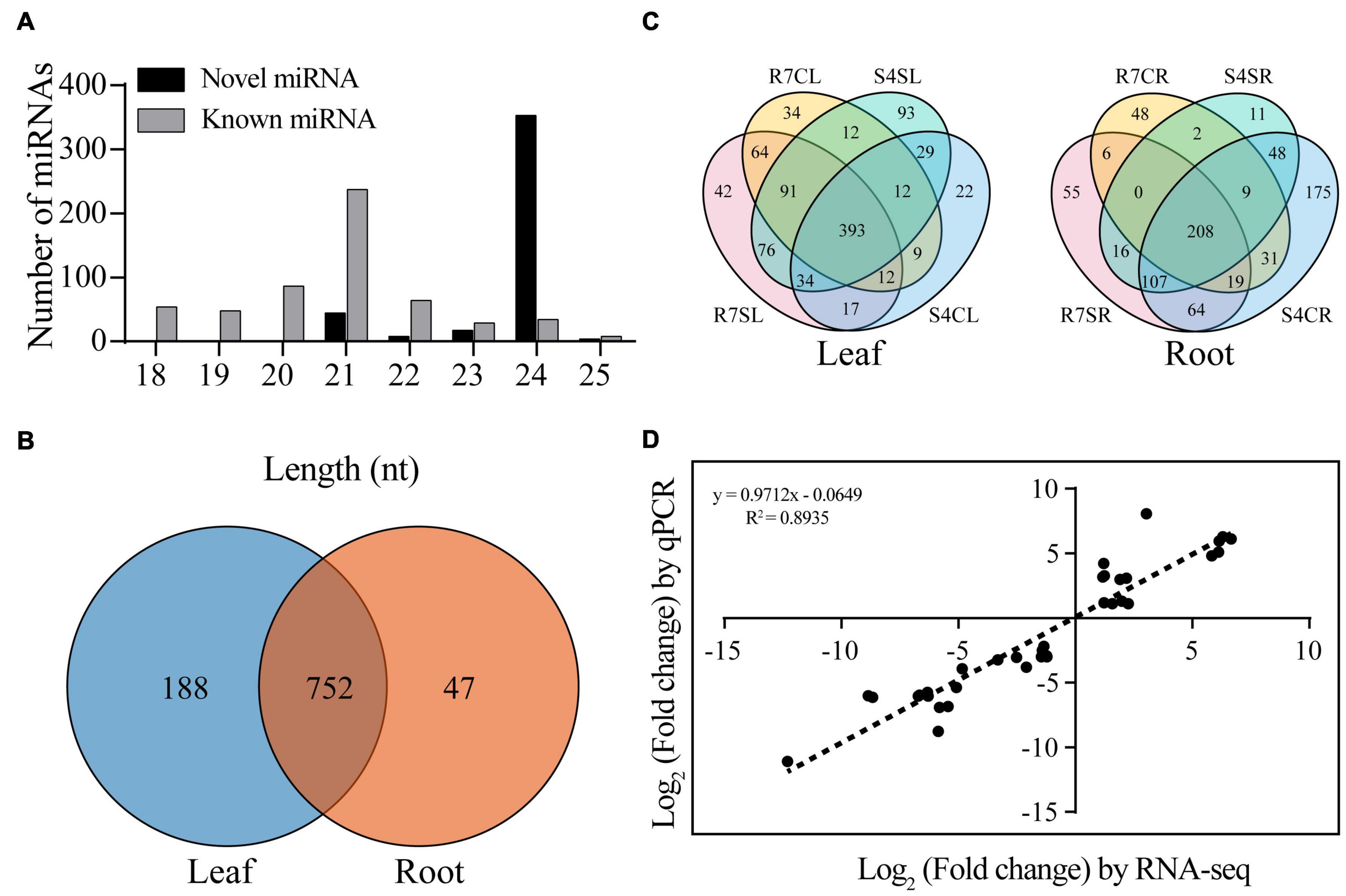
Figure 1. Identification of miRNAs from Fraxinus velutina tolerant (R7) and sensitive (S4) leaves and roots. (A) Length distribution of all miRNAs identified in this study. (B) The distribution of all miRNAs between leaf and root samples. (C) The distribution of expressed miRNAs of leaves and roots in R7 and S4 with or without salt treatment. (D) The correlation between small RNA sequencing (x-axis) and qRT-PCR (y-axis) results. R7SL, R7 leaves with salt treatment; R7CL, R7 leaves without salt treatment; R7SR, R7 roots with salt treatment; R7CR, R7 roots without salt treatment; S4SL, S4 leaves with salt treatment; S4CL, S4 leaves without salt treatment; S4SR, S4 roots with salt treatment; S4CR, S4 roots without salt treatment.
Identification of Specifically Expressed miRNAs Between Salt-Tolerant R7 and Salt-Sensitive S4
Based on the normalized expression levels, the correlation analysis between the expression levels within each sample was analyzed. The results showed that the correlation coefficient, γ2 between three biological replicates in each group was 0.878, indicating the replicates were highly consistent (Supplementary Figure 1B). There were 188 and 47 specifically expressed miRNAs in leaf and root, respectively, and 752 shared miRNAs in both tissues (Figure 1B). There were 796 and 800 miRNAs were expressed in R7 and S4 leaves, respectively; 42 and 93 miRNAs were exclusively expressed in R7 and S4 leaves under salt stress, respectively (Figure 1C). In roots, there were 565 and 690 miRNAs expressed in R7 and S4, respectively; 55 and 11 miRNAs were exclusively expressed in R7 and S4 roots under salt treatment, respectively (Figure 1C).
To confirm the small RNA sequencing data, 15 DEmiRs were randomly selected to conduct qRT-PCR analysis. The results showed that these selected miRNAs exhibited the same expression patterns with that of small RNA sequencing data (Supplementary Figure 2). In addition, based on the log2 fold change of each comparison, the correlation analysis between qRT-PCR results and sequencing data was performed. The results revealed a high correlation coefficient (R2 = 0.8935) between sequencing data and qRT-PCR results, demonstrating that the sequencing data are accurate and reliable (Figure 1D).
Identification of Target Genes for miRNAs
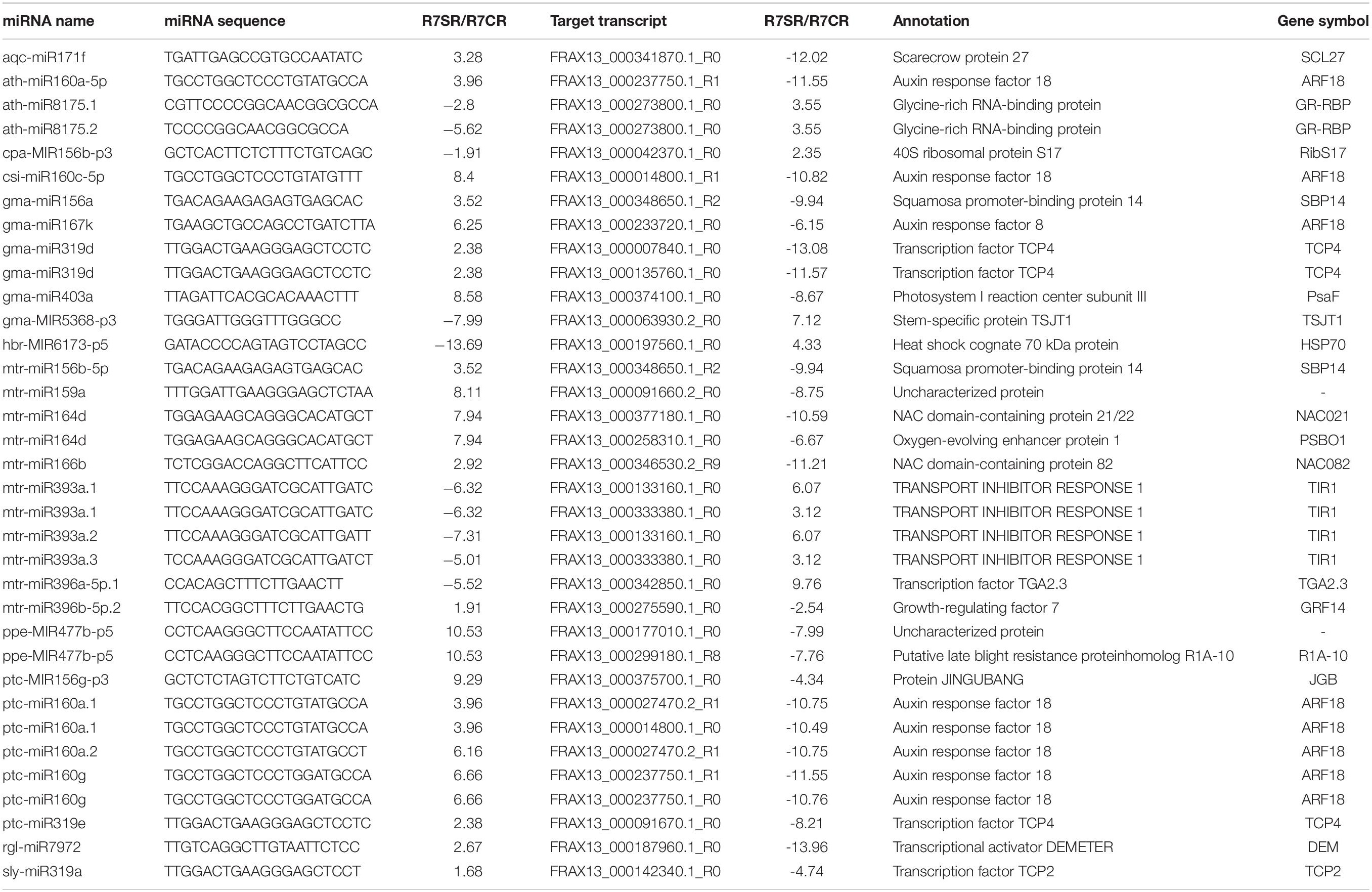
To investigate biological function of these miRNAs, degradome sequencing from R7 and S4 samples were performed to identify the putative target genes for the miRNAs. After trimmed and polished, 9719583 and 13021448 validated reads were generated from R7 and S4, respectively. The reads were further mapped to the F. velutina reference genome, and the results revealed that more than 99% of the reads were perfectly aligned back to the reference (Supplementary Table 5). After analyzing by CleaveL and, 247 miRNA-target pairs, including 194 miRNAs targeting 229 genes were identified in R7; 273 pairs, including 213 miRNAs targeting 239 genes were in S4 (Figure 2A and Supplementary Table 6). Among the miRNA-target pairs, 185 and 210 pairs were exclusively identified in R7 and S4, respectively; 62 pairs were overlapped in both R7 and S4 (Figure 2B). Among the targets, we found that there were 25 (10.92%) and 28 (11.72%) genes encoding transcription factors (TF) targeted by the miRNAs in R7 and S4, respectively (Figure 2C). For example, an ethylene-responsive transcription factor 4 (ERF4) was targeted by bna-MIR169c-p5_2ss12GC17TG in R7, and a transcription factor TGA2.3 (TGA2.3) was regulated by mtr-miR390_L-1 in S4. Additionally, we also found multiple shared miRNA-TF pairs in both plants, such as mtr-miR166c_1ss9GT-MYBS3 and mtr-miR164d_1ss13GA-NAC1 (Figure 2D).
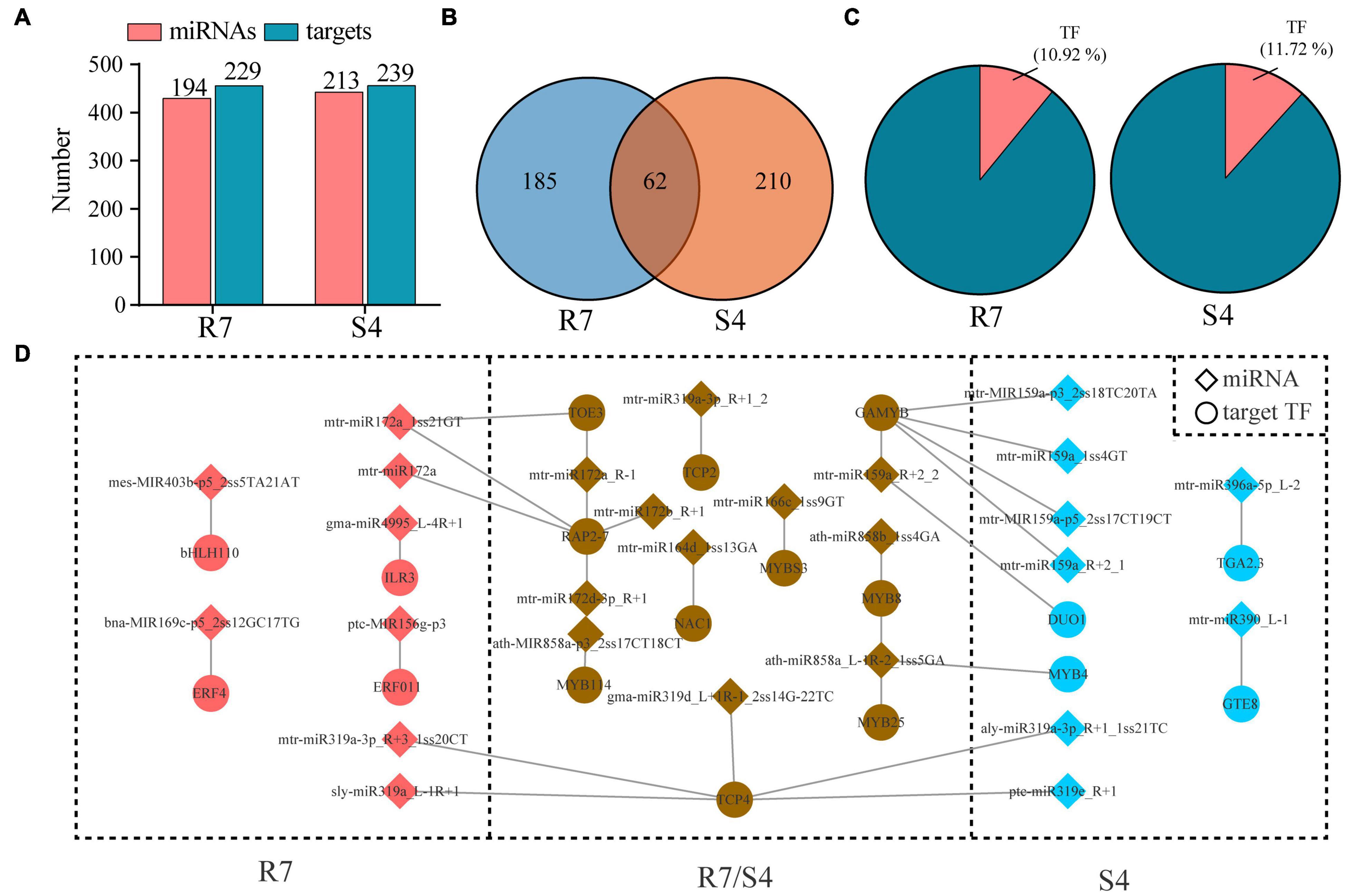
Figure 2. Prediction of miRNA targets based on degradome sequencing data. (A) Number of miRNAs and target genes in all miRNA-target pairs in Fraxinus velutina tolerant (R7) and sensitive (S4). (B) The distribution of miRNA-target pairs between R7 and S4. (C) The percentage of transcript factors (TFs) in all targets from R7 and S4. (D) Network of all miRNA-TF pairs in R7 and S4.
Identification of Salt Stress-Responsive miRNAs
To identify miRNAs involved in response to salt stress, the differentially expressed miRNAs (DEmiRs) was analyzed using DEseq2. The results revealed 141 (40 up- and 101 down-regulated miRNAs) and 192 (102 up- and 90 down-regulated miRNAs) DEmiRs in R7, and S4 leaves, respectively (Figure 3A). In roots, 262 DEmiRs including 124 up- and 138 down-regulated miRNAs were identified in R7, and 249 DEmiRs including 68 up- and 181 down-regulated miRNAs were in S4 (Figure 3A and Supplementary Table 7). Under salt stress, there were 66 and 172 specific DEmiRs in leaf and root samples, respectively, with 215 shared DEmiRs in both tissues (Figure 3B and Supplementary Table 7). Among these shared DEmiRs, 27 DEmiRs presented in all comparisons, in which only 8 DEmiRs showed the consistent or opposite expression patterns between these two tissues in two clones after salt treatment (Figure 3C). The distribution analysis of leaf DEmiRs revealed that 24 DEmiRs exhibited opposite expression patterns between R7 and S4 after salt treatment (Figure 3D). Among the root DEmiRs, 32 DEmiRs presented opposed regulations between R7 and S4 after salt stress (Figure 3E).
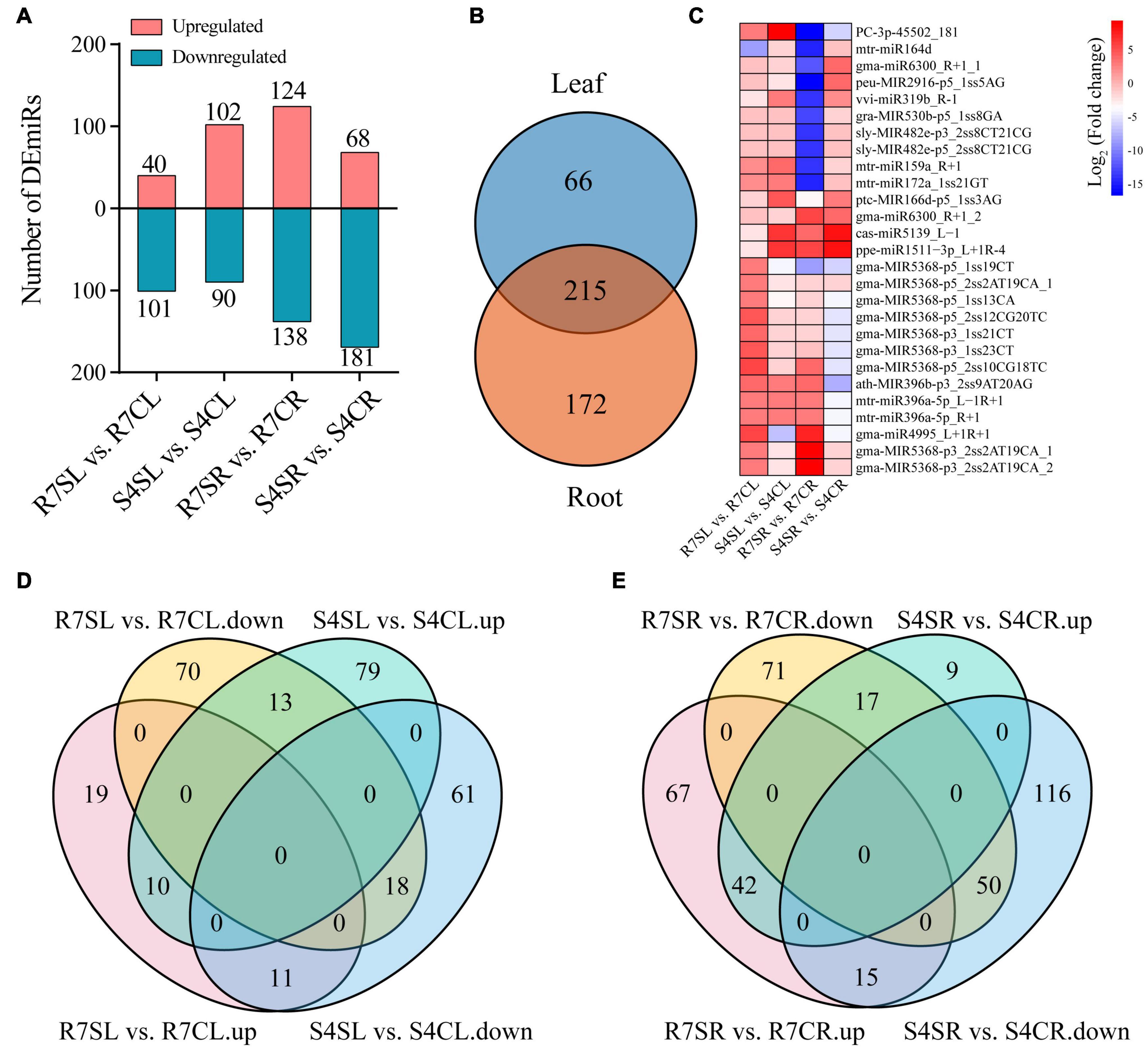
Figure 3. Identification of salt stress-responsive miRNAs. (A) Statistics of up- and downregulated differentially expressed miRNAs (DEmiRs) in leaves and roots. The red and green represented up- and down-regulated DEmiRs, respectively. (B) The distribution of DEmiRs between leaves and roots. (C) Expression patterns of the shared DEmiRs in all comparisons. From red to blue indicated the fold change from high to low. (D) The distribution of up- and downregulated DEmiR in R7 and S4 leaves. (E) The distribution of up- and down-regulated DEmiR in R7 and S4 roots. R7SL, R7 leaves with salt treatment; R7CL, R7 leaves without salt treatment; R7SR, R7 roots with salt treatment; R7CR, R7 roots without salt treatment; S4SL, S4 leaves with salt treatment; S4CL, S4 leaves without salt treatment; S4SR, S4 roots with salt treatment; S4CR, S4 roots without salt treatment.
Analysis of Salt Stress-Responsive miRNA Targets
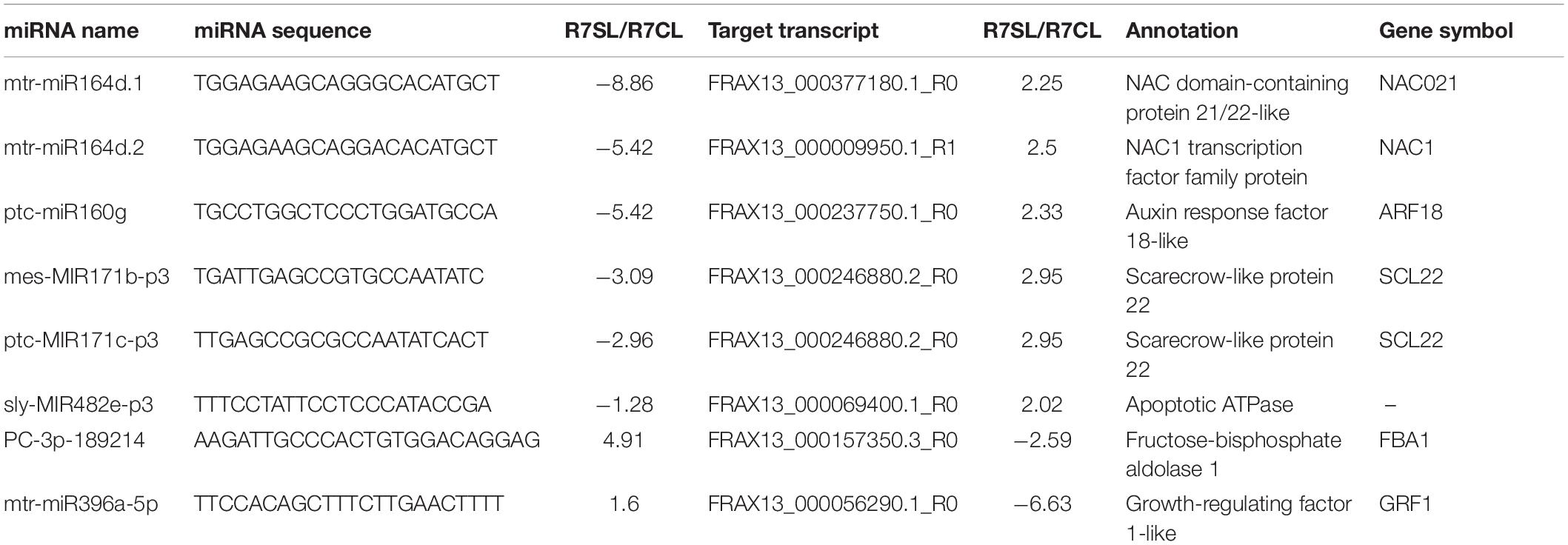
To explore regulatory roles of DEmiRs in F. velutina response to salt stress, our previous transcriptome data were used to narrow the miRNA-target pairs. The results showed that 65 DEmiR-DEG pairs were identified among the comparisons (Supplementary Table 8). To further investigate the mechanisms underlying enhanced salt tolerance in R7, the specific DEmiRs in R7 leaves (89 DEmiRs) and roots (138 DEmiRs) were further analyzed. In the leaves, 8 specific DEmiR-DEG pairs, including 8 DEmiRs targeting 7 DEGs were identified (Table 1). Among these pairs, multiple DEGs were related to the plant response to salt stress, such as mtr-miR164d_1ss13GA targeted NAC1 transcription factor (NAC1), ptc-miR160g targeted ARF18 (Figures 4A,B). In the roots, there were 35 specific DEmiR-DEG pairs including 29 DEmiRs targeting 29 DEGs (Table 2). Among these pairs, multiple DEGs were associated with the plant response to salt stress, such as mtr-miR396a-5p_L-2 targeted TGA2.3 and mtr-miR396b-5p_1ss7AG targeted growth-regulating factor 7 (GRF7) (Figures 4A,B).
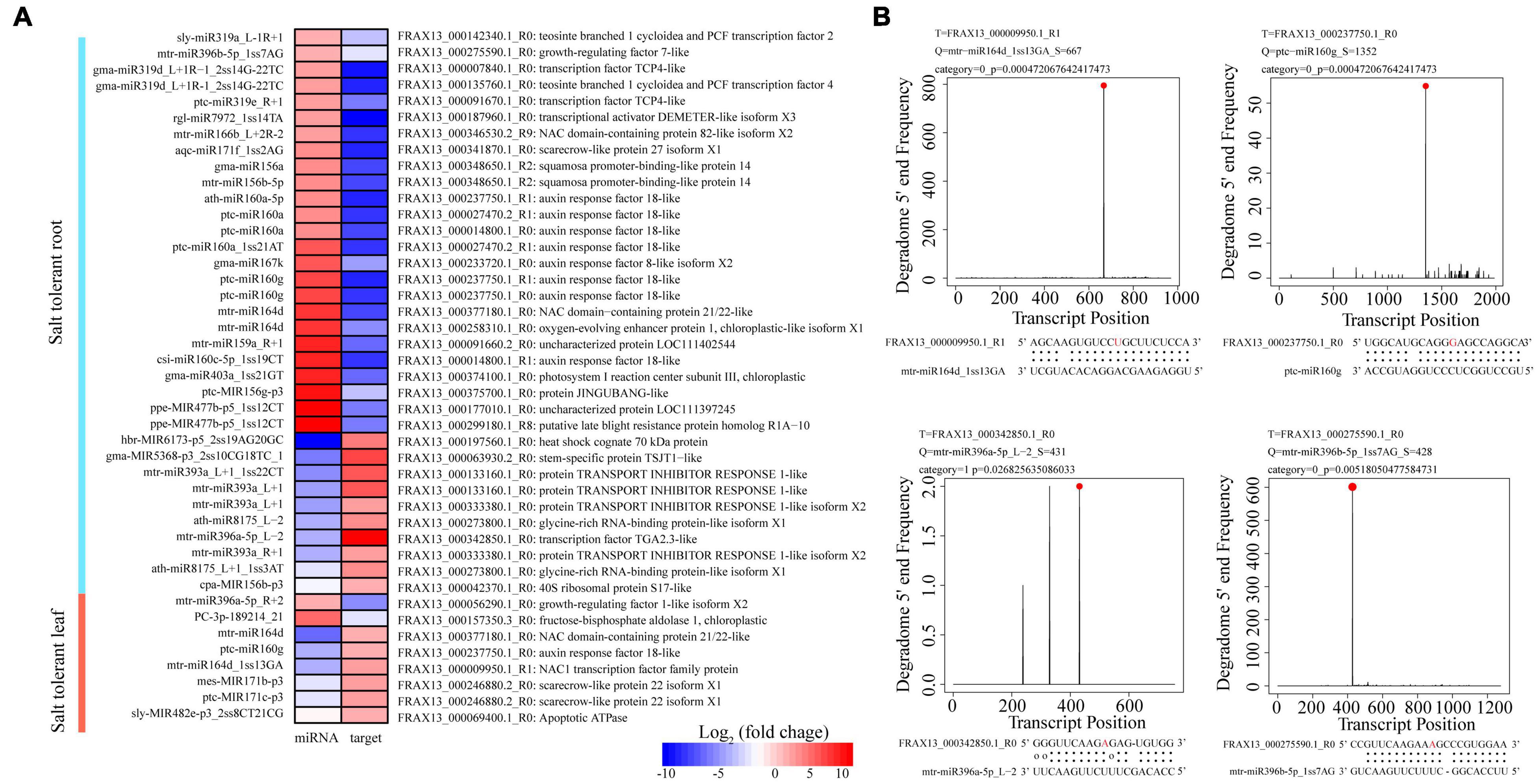
Figure 4. Analysis of salt stress-responsive miRNAs in F. velutina tolerant ‘R7’ leaves and roots. (A) Expression patterns of miRNAs and targets from specific DEmiR-DEG pairs in R7 leaves and roots. From red to blue represented the fold change from high to low. (B) Target plots (t-plots) of the selected DEmiR-DEG pairs in R7 leaves and roots confirmed by degradome sequencing. The red circle and letter represented slice site.
Discussion
In our previous study, we identified two F. velutina cuttings clones, salt-tolerant R7 and salt-sensitive S4 and found that R7 exhibits higher salt tolerance than S4. Meanwhile, we performed a comparative transcriptome analysis between R7 and S4, and identified several crucial genes and signaling pathways involved in high salt tolerance of R7 (Ma et al., 2022). In the present study, utilizing an integration of miRNA, mRNA, and degradome sequencing data analysis; we identified several key miRNA-target modules contributing to the high salt tolerance of R7. Specifically, the miRNA-target modules identified in R7 leaf were primarily related with antioxidant system and auxin signaling; while miRNA-target modules in R7 root mainly belonged to ion homeostasis and ROS scavenging.
It is well reported that miR171/GRAS module is an important contributor to plant development and biotic and abiotic stress resistance in Medicago truncatula (Hirsch et al., 2009). In apple, miR171i/SCL26.1 module can enhance drought stress tolerance via regulating antioxidant system (Wang et al., 2020). In the present study, we found both miR171b (mes-MIR171b-p3) and miR171c (mes-MIR171b-p3) upregulated a GRAS transcription factor SCL22 (FRAX13_000246880.2_R0), suggesting that miR171b/c-SCL22 module is involved in regulating antioxidant system, resulting in the enhanced tolerance to salt stress. The miR396a-5p/GRF1 module enhances tobacco (Nicotiana tabacum) tolerance to salt stress (Chen L. et al., 2015). Under stress, GRF1 inhibits plant growth by regulating WRKY28 expression in Arabidopsis (Piya et al., 2020). The present study found that miR396a (mtr-miR396a-5p) over-expression down-regulated GRF1 (FRAX13_000056290.1_R0), further down-regulated WRKY28, implying that miR396a/GRF1/WRKY28 module enhances tolerance to salt stress by promoting plant growth. Thus, miR171b/c-SCL22 and miR396a/GRF1/WRKY28 modules are important regulators in enhancing salt stress resistance in F. velutina.
Auxin response factors (ARFs) have been confirmed to play crucial roles in plant tolerance to abiotic stress through regulating the auxin signaling pathway (Liu et al., 2018; Song X. et al., 2019; Cui et al., 2020). In poplar, miR390/TAS3/ARFs module has been confirmed to be a key regulator of lateral root growth of poplar (Populus spp.) plants under salt stress by modulating the auxin pathway (He et al., 2018). In the present study, we found miR160g (ptc-miR160g) up-regulated ARF18 (FRAX13_000237750.1_R0) expression, suggesting that the miR160g/ARF18 module contributes to salt tolerance via modulating auxin pathway. Previous study has shown that the overexpressed miR393 regulates rice salt and drought tolerance by inhibiting transport inhibitor response protein (TIR1) (Xia et al., 2012). In this study, we found that miR393a (mtr-miR393a.1, mtr-miR393a.2, and mtr-miR393a.3) down-regulated TIR1 (FRAX13_000333380.1_R0) under salt stress, indicating that miR393a/TIR1 module enhances plant tolerance to salt stress via mediating auxin signaling pathway. Together, miR160g/ARF18 and miR393a/TIR1 modules enhance tolerance to salt stress by regulating auxin signaling pathway in F. velutina.
Much evidence shows that NAC transcription factors play vital roles in plant development (Guo et al., 2005; Petricka et al., 2012), cell apoptosis (Lee et al., 2014), and abiotic stress tolerance (Tran et al., 2010). Overexpressed PeNAC1 in Arabidopsis enhances tolerance to salt stress by regulating Na+/K+ homeostasis (Wang J. Y. et al., 2013). In this study, we found that miR164d (mtr-miR164d) targeted NAC1 (FRAX13_000009950.1_R1) and up-regulated NAC1 expression in R7 subjected to salt stress, suggesting that miR164d/NAC1 module enhances salt tolerance through regulating Na+/K+ homeostasis. The increasing evidence has shown that the TGA (TGACG motif-binding factor) transcription factors, a basic leucine zipper (bZIP) gene subfamily, play crucial roles in response to salt stress (Du et al., 2014; Zhang et al., 2014). The overexpression of GmTGA13 enhances Arabidopsis and soybean (Glycine max) salt tolerance through regulating ion homeostasis (Ke et al., 2021). In the present study, we found TGA2.3 (FRAX13_000342850.1_R0) was targeted by miR396a (mtr-miR396a-5p.1), suggesting that miR396a/TGA2.3 module enhances salt tolerance via mediating ion homeostasis. Thus, miR164d/NAC1 and miR396a/TGA2.3 modules contribute to the enhanced salt tolerance in F. velutina.
Multiple studies have shown that squamosa-promoter binding protein box (SBP-box) can regulate the salt tolerance in many plants, such as Betula platyphylla Suk (Ning et al., 2017) and rice (Lan et al., 2019). Silencing CaSBP12 in pepper (Capsicum annuum) enhances tolerance to salt stress and reduces the ROS accumulation (Zhang H. X. et al., 2020). In this study, we found that miR156a/b (gma-miR156a and mtr-miR156b-5p) targeted SBP14 (FRAX13_000348650.1_R2) and up-regulated SBP14 expression under salt stress, suggesting that miR156a/b-SBP14 module increases ROS scavenging, thereafter leading to the enhanced salt tolerance. A previous study has shown that GR-RBPs is a positive regulatory molecule in regulating ROS accumulation to enhance salt tolerance in Arabidopsis (Tan et al., 2014). Our study showed that miR8175 (ath-miR8175.1 and ath-miR8175.2) directly bonded to GR-RBP (FRAX13_000273800.1_R0) and up-regulated GR-RBP expression in R7 subjected to salt stress, indicating that miR8175/GR-RBP module enhances tolerance to salt stress. Collectively, miR156a/b-SBP14 and miR8175/GR-RBP modules are key regulators, regulating ROS scavenging, of high salt tolerance in F. velutina.
Conclusion
In summary, this work revealed the key regulatory network of miRNAs in salt-tolerant clone R7 of F. velutina responding to salt stress. The small RNA and degradome sequencing data presented here allowed us to propose potential regulatory roles of miRNAs in the defense response of R7 to salt stress (Figure 5). Under salt stress, multiple miRNA/target modules were involved in R7 response to salt stress. In the leaf, miR164d/NAC1, miR171/SCL22, miR396a/GRF1/WRKY28, and miR160g/ARF18 modules were involved in plant response to salt stress by regulating multiple biological processes, such as antioxidant system and auxin signaling. In the root, miR396a/TGA2.3, miR156/SBP14, miR319/TCPs/ICKs, and miR393a/TIR1 modules enhanced plant tolerance to salt stress by regulating several processes including ROS scavenging, cell proliferation, and ion homeostasis. The miRNA-target modules identified here pave a novel avenue for improving the salt tolerance of F. velutina through miRNA manipulation.
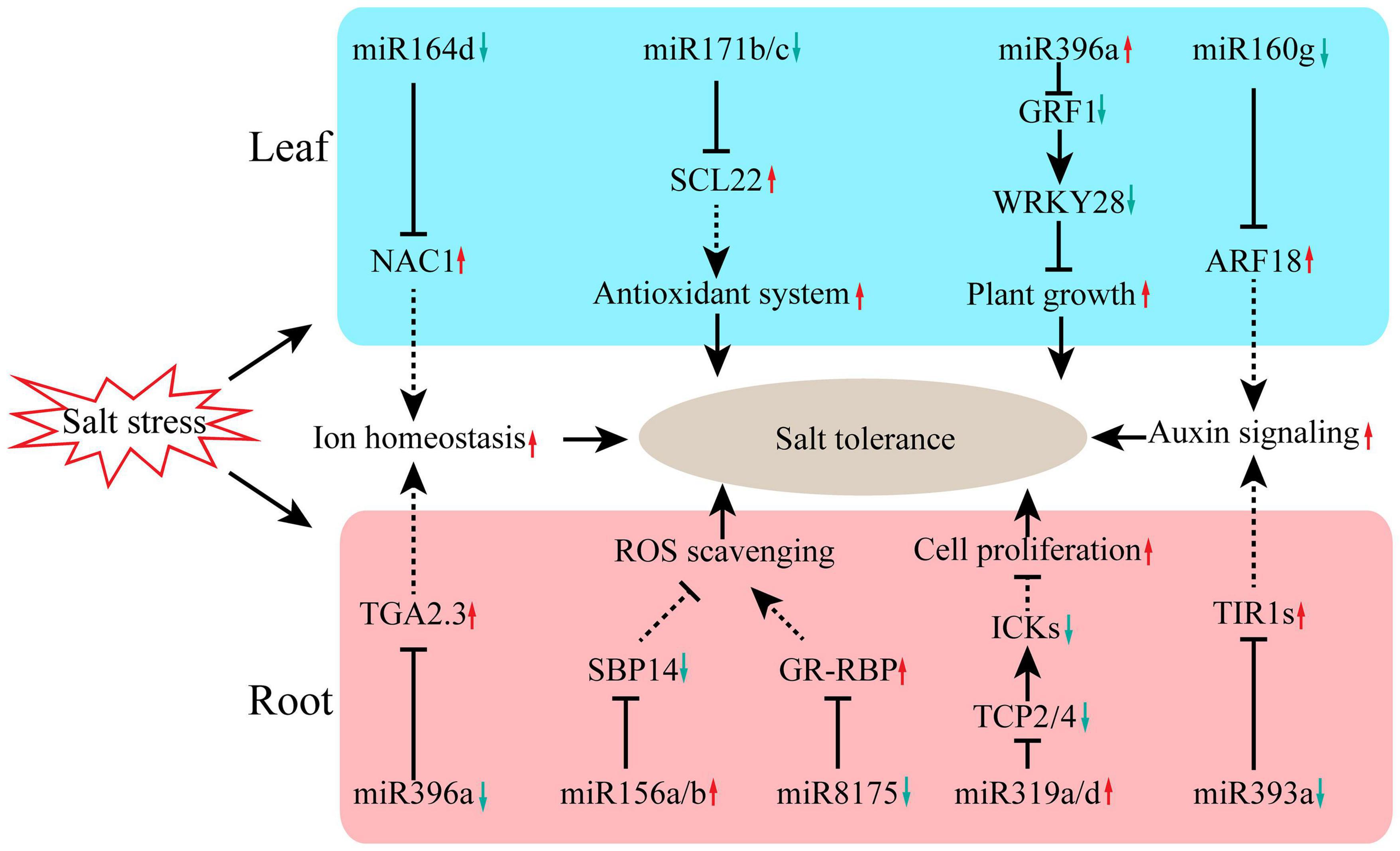
Figure 5. The miRNA-related molecular mechanisms for enhancing salt tolerance of F. velutina ‘R7’. Under salt stress, multiple miRNA/target modules were involved in R7 response to salt stress. In the leaf, miR164d/NAC1, miR171/SCL22, miR396a/GRF1/WRKY28, and miR160g/ARF18 modules were involved in plant response to salt stress by regulating multiple biological processes, such as antioxidant system and auxin signaling. In the root, miR396a/TGA2.3, miR156/SBP14, miR319/TCPs/ICKs, and miR393a/TIR1 modules enhanced plant tolerance to salt stress by regulating several processes including ROS scavenging, cell proliferation, and ion homeostasis.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: The small RNA and degradome sequencing data presented in this study can be available at the Sequence Read Archive under accession number PRJNA793056.
Author Contributions
KY and DW conceptualized the research program. JL, XM, and LY finished the analysis of this study and wrote the manuscript. QL, HF, CW, and LW conducted the RNA sequencing data analysis. YD and ZC designed the qRT-PCR experiment and finished the operation. RZ, YB, XL, SG, and RC planted the material and finished the physiology analysis. KY, RC, and DW revised the manuscript. All authors discussed the results, commented on the manuscript, and approved the submitted version.
Funding
This work was supported by the Improved Variety Program of Shandong Province of China (2019LZGC009).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.847853/full#supplementary-material
Supplementary Figure 1 | Analysis of miRNA sequencing data. (A) Number of conserved miRNAs in each family. (B) Heatmap showed the correlation coefficient (r2) between each sample.
Supplementary Figure 2 | The qRT-PCR analysis was performed to verify the reliability of miRNA sequencing data. The heatmap showed the log2 fold change of 15 selected miRNAs in different comparisons of qRT-PCR results and sequencing data. From blue to red represented the value of log2 fold change from low to high.
Supplementary Table 1 | All primers used in this study.
Supplementary Table 2 | Summary of the small RNA sequences from each sample.
Supplementary Table 3 | Statistics of the small RNAs identified in each sample.
Supplementary Table 4 | The details of all miRNAs identified in this study.
Supplementary Table 5 | The overview of the degradome sequencing data.
Supplementary Table 6 | Identification of target genes of the miRNAs.
Supplementary Table 7 | Identification of the differentially expressed miRNAs.
Supplementary Table 8 | Identification of target genes of the differentially expressed miRNAs.
Footnotes
References
Addo-Quaye, C., Eshoo, T. W., Bartel, D. P., and Axtell, M. J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762. doi: 10.1016/j.cub.2008.04.042
Addo-Quaye, C., Miller, W., and Axtell, M. J. (2009). CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25, 130–131. doi: 10.1093/bioinformatics/btn604
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Bai, Q., Wang, X., Chen, X., Shi, G., Liu, Z., Guo, C., et al. (2018). Wheat miRNA taemiR408 acts as an essential mediator in plant tolerance to Pi deprivation and salt stress via modulating stress-associated physiological processes. Front. Plant Sci. 9:499. doi: 10.3389/fpls.2018.00499
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Butcher, K., Wick, A. F., DeSutter, T., Chatterjee, A., and Harmon, J. (2016). Soil salinity: a threat to global food security. Agron. J. 108, 2189–2200. doi: 10.2134/agronj2016.06.0368
Cervera-Seco, L., Marques, M. A. C., Sanz-Carbonell, A., Marquez-Molins, J., Carbonell, A., Darï, S. J., et al. (2019). Identification and characterization of stress-responsive TAS3-derived tasiRNAs in melon. Plant Cell Physiol. 60, 2382–2393. doi: 10.1093/pcp/pcz131
Chen, J., and Mueller, V. (2018). Coastal climate change, soil salinity and human migration in Bangladesh. Nat. Clim. Chang. 8, 981–985. doi: 10.1038/s41558-018-0313-8
Chen, L., Luan, Y., and Zhai, J. (2015). Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 34, 2013–2025. doi: 10.1007/s00299-015-1847-0
Chen, Z., Hu, L., Han, N., Hu, J., Yang, Y., Xiang, T., et al. (2015). Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in Arabidopsis thaliana. Plant Cell Physiol. 56, 73–83. doi: 10.1093/pcp/pcu149
Cui, J., Li, X., Li, J., Wang, C., Cheng, D., and Dai, C. (2020). Genome-wide sequence identification and expression analysis of ARF family in sugar beet (Beta vulgaris L.) under salinity stresses. PeerJ 8:e9131. doi: 10.7717/peerj.9131
Denver, J. B., and Ullah, H. (2019). miR393s regulate salt stress response pathway in Arabidopsis thaliana through scaffold protein RACK1A mediated ABA signaling pathways. Plant Signal. Behav. 14:1600394. doi: 10.1080/15592324.2019.1600394
Djami-Tchatchou, A. T., Sanan-Mishra, N., Ntushelo, K., and Dubery, I. A. (2017). Functional roles of microRNAs in agronomically important plants-potential as targets for crop improvement and protection. Front. Plant Sci. 8:378. doi: 10.3389/fpls.2017.00378
Du, X., Du, B., Chen, X., Zhang, S., Zhang, Z., and Qu, S. (2014). Overexpression of the MhTGA2 gene from crab apple (Malus hupehensis) confers increased tolerance to salt stress in transgenic apple (Malus domestica). J. Agric. Sci. 152, 634–641. doi: 10.1017/S0021859613000130
German, M. A., Luo, S., Schroth, G., Meyers, B. C., and Green, P. J. (2009). Construction of parallel analysis of RNA ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 4, 356–362. doi: 10.1038/nprot.2009.8
German, M. A., Pillay, M., Jeong, D. H., Hetawal, A., Luo, S., Janardhanan, P., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26, 941–946. doi: 10.1038/nbt1417
Guo, H. S., Xie, Q., Fei, J. F., and Chua, N. H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell 17, 1376–1386. doi: 10.1105/tpc.105.030841
He, F., Xu, C., Fu, X., Shen, Y., Guo, L., Leng, M., et al. (2018). The microRNA390/trans-acting short interfering RNA3 module mediates lateral root growth under salt stress via the auxin pathway. Plant Physiol. 177, 775–791. doi: 10.1104/pp.17.01559
Hirsch, S., Kim, J., Muñoz, A., Heckmann, A. B., Downie, J. A., and Oldroyd, G. E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21, 545–557. doi: 10.1105/tpc.108.064501
Hossain, M. S., and Dietz, K. J. (2016). Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Front. Plant Sci. 7:548. doi: 10.3389/fpls.2016.00548
Iglesias, M. J., Terrile, M. C., Windels, D., Lombardo, M. C., Bartoli, C. G., Vazquez, F., et al. (2014). MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS One 9:e107678. doi: 10.1371/journal.pone.0107678
Ke, D., He, Y., Fan, L., Niu, R., Cheng, L., Wang, L., et al. (2021). The soybean TGA transcription factor GmTGA13 plays important roles in the response to salinity stress. Plant Biol. (Stuttg) 24, 313–322. doi: 10.1111/plb.13360
Kechin, A., Boyarskikh, U., Kel, A., and Filipenko, M. (2017). cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 24, 1138–1143. doi: 10.1089/cmb.2017.0096
Kelly, L. J., Plumb, W. J., Carey, D. W., Mason, M. E., Cooper, E. D., Crowther, W., et al. (2020). Convergent molecular evolution among ash species resistant to the emerald ash borer. Nat. Ecol. Evol. 4, 1116–1128. doi: 10.1038/s41559-020-1209-3
Kumar, D., Dutta, S., Singh, D., Prabhu, K. V., Kumar, M., and Mukhopadhyay, K. (2017). Uncovering leaf rust responsive miRNAs in wheat (Triticum aestivum L.) using high-throughput sequencing and prediction of their targets through degradome analysis. Planta 245, 161–182. doi: 10.1007/s00425-016-2600-9
Kumar, V., Khare, T., Shriram, V., and Wani, S. H. (2018). Plant small RNAs: the essential epigenetic regulators of gene expression for salt-stress responses and tolerance. Plant Cell Rep. 37, 61–75. doi: 10.1007/s00299-017-2210-4
Lan, T., Zheng, Y., Su, Z., Yu, S., Song, H., Zheng, X., et al. (2019). OsSPL10, a SBP-box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L.). G3 (Bethesda) 9, 4107–4114. doi: 10.1534/g3.119.400700
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Lee, S., Lee, H. J., Huh, S. U., Paek, K. H., Ha, J. H., and Park, C. M. (2014). The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci. 227, 76–83. doi: 10.1016/j.plantsci.2014.07.003
Litalien, A., and Zeeb, B. (2020). Curing the earth: a review of anthropogenic soil salinization and plant-based strategies for sustainable mitigation. Sci. Total Environ. 698:134235. doi: 10.1016/j.scitotenv.2019.134235
Liu, N., Dong, L., Deng, X., Liu, D., Liu, Y., Li, M., et al. (2018). Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L. BMC Plant Biol. 18:336. doi: 10.1186/s12870-018-1559-z
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Luan, M., Xu, M., Lu, Y., Zhang, L., Fan, Y., and Wang, L. (2015). Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene 555, 178–185. doi: 10.1016/j.gene.2014.11.001
Luan, M., Xu, M., Lu, Y., Zhang, Q., Zhang, L., Zhang, C., et al. (2014). Family-wide survey of miR169s and NF-YAs and their expression profiles response to abiotic stress in maize roots. PLoS One 9:e91369. doi: 10.1371/journal.pone.0091369
Ma, X., Liu, J. N., Yan, L., Liang, Q., Fang, H., Wang, C., et al. (2022). Comparative transcriptome analysis unravels defense pathways of Fraxinus velutina Torr against salt stress. Front. Plant Sci. 2022:842726. doi: 10.3389/fpls.2022.842726
Ma, Y., Xue, H., Zhang, F., Jiang, Q., Yang, S., Yue, P., et al. (2021). The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 19, 311–323. doi: 10.1111/pbi.13464
Mao, P., Tang, Q., Cao, B., Liu, J., Shao, H., Cao, Z., et al. (2017). Eco-physiological adaptability in mixtures of Robinia pseudoacacia and Fraxinus velutina and coastal eco-engineering. Ecol. Eng. 106, 109–115. doi: 10.1016/j.ecoleng.2017.05.021
Markham, N. R., and Zuker, M. (2008). UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 453, 3–31. doi: 10.1007/978-1-60327-429-6_1
Meyers, B. C., Axtell, M. J., Bartel, B., Bartel, D. P., Baulcombe, D., Bowman, J. L., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20, 3186–3190. doi: 10.1105/tpc.108.064311
Ning, K., Chen, S., Huang, H., Jiang, J., Yuan, H., and Li, H. (2017). Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell Tissue Organ. Cult. (PCTOC) 130, 469–481. doi: 10.1007/s11240-017-1226-3
Ondrasek, G., and Rengel, Z. (2021). Environmental salinization processes: detection, implications & solutions. Sci. Total Environ. 754, 142432. doi: 10.1016/j.scitotenv.2020.142432
Petricka, J. J., Winter, C. M., and Benfey, P. N. (2012). Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63, 563–590. doi: 10.1146/annurev-arplant-042811-105501
Piya, S., Liu, J., Burch-Smith, T., Baum, T. J., and Hewezi, T. (2020). A role for Arabidopsis growth-regulating factors 1 and 3 in growth-stress antagonism. J. Exp. Bot. 71, 1402–1417. doi: 10.1093/jxb/erz502
Polle, A., and Chen, S. (2015). On the salty side of life: molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ. 38, 1794–1816. doi: 10.1111/pce.12440
Shamimuzzaman, M., and Vodkin, L. (2012). Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genomics 13:310. doi: 10.1186/1471-2164-13-310
Song, S., Hao, L., Zhao, P., Xu, Y., Zhong, N., Zhang, H., et al. (2019). Genome-wide identification, expression profiling and evolutionary analysis of auxin response factor gene family in potato (Solanum tuberosum Group Phureja). Sci. Rep. 9:1755. doi: 10.1038/s41598-018-37923-7
Song, X., Li, Y., Cao, X., and Qi, Y. (2019). MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Sun, X., Zhang, Y., Zhu, X., Korir, N. K., Tao, R., Wang, C., et al. (2014). Advances in identification and validation of plant microRNAs and their target genes. Physiol. Plant 152, 203–218. doi: 10.1111/ppl.12191
Tan, Y., Qin, Y., Li, Y., Li, M., and Ma, F. (2014). Overexpression of MpGR-RBP1, a glycine-rich RNA-binding protein gene from Malus prunifolia (Willd.) Borkh., confers salt stress tolerance and protects against oxidative stress in Arabidopsis. Plant Cell Tissue Organ. Cult. (PCTOC) 119, 635–646. doi: 10.1007/s11240-014-0563-8
Tran, L. S., Nishiyama, R., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2010). Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1, 32–39. doi: 10.4161/gmcr.1.1.10569
Wang, C., Han, J., Korir, N. K., Wang, X., Liu, H., Li, X., et al. (2013). Characterization of target mRNAs for grapevine microRNAs with an integrated strategy of modified RLM-RACE, newly developed PPM-RACE and qPCRs. J. Plant Physiol. 170, 943–957. doi: 10.1016/j.jplph.2013.02.005
Wang, J. Y., Wang, J. P., and He, Y. (2013). A Populus euphratica NAC protein regulating Na+/K+ homeostasis improves salt tolerance in Arabidopsis thaliana. Gene 521, 265–273. doi: 10.1016/j.gene.2013.03.068
Wang, M., Guo, W., Li, J., Pan, X., Pan, L., Zhao, J., et al. (2021). The miR528-AO module confers enhanced salt tolerance in rice by modulating the ascorbic acid and abscisic acid metabolism and ROS scavenging. J. Agric. Food Chem. 69, 8634–8648. doi: 10.1021/acs.jafc.1c01096
Wang, W., Liu, D., Chen, D., Cheng, Y., Zhang, X., Song, L., et al. (2019). MicroRNA414c affects salt tolerance of cotton by regulating reactive oxygen species metabolism under salinity stress. RNA Biol. 16, 362–375. doi: 10.1080/15476286.2019.1574163
Wang, Y., Feng, C., Zhai, Z., Peng, X., Wang, Y., Sun, Y., et al. (2020). The apple microR171i-SCARECROW-LIKE PROTEINS26.1 module enhances drought stress tolerance by integrating ascorbic acid metabolism. Plant Physiol. 184, 194–211. doi: 10.1104/pp.20.00476
Xia, K., Wang, R., Ou, X., Fang, Z., Tian, C., Duan, J., et al. (2012). OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One 7:e30039. doi: 10.1371/journal.pone.0030039
Xu, T., Zhang, L., Yang, Z., Wei, Y., and Dong, T. (2021). Identification and functional characterization of plant miRNA under salt stress shed light on salinity resistance improvement through miRNA manipulation in crops. Front. Plant Sci. 12:665439. doi: 10.3389/fpls.2021.665439
Yin, Z., Li, C., Han, X., and Shen, F. (2008). Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum). Gene 414, 60–66. doi: 10.1016/j.gene.2008.02.007
Yu, Y., Wu, G., Yuan, H., Cheng, L., Zhao, D., Huang, W., et al. (2016). Identification and characterization of miRNAs and targets in flax (Linum usitatissimum) under saline, alkaline, and saline-alkaline stresses. BMC Plant Biol. 16:124. doi: 10.1186/s12870-016-0808-2
Yuan, S., Li, Z., Li, D., Yuan, N., Hu, Q., and Luo, H. (2015). Constitutive expression of rice microRNA528 alters plant development and enhances tolerance to salinity stress and nitrogen starvation in creeping bentgrass. Plant Physiol. 169, 576–593. doi: 10.1104/pp.15.00899
Yuan, S., Zhao, J., Li, Z., Hu, Q., Yuan, N., Zhou, M., et al. (2019). MicroRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass. Hortic. Res. 6:48. doi: 10.1038/s41438-019-0130-x
Zhang, B. H., Pan, X. P., Cox, S. B., Cobb, G. P., and Anderson, T. A. (2006). Evidence that miRNAs are different from other RNAs. Cell Mol. Life Sci. 63, 246–254. doi: 10.1007/s00018-005-5467-7
Zhang, H. X., Zhu, W. C., Feng, X. H., Jin, J. H., Wei, A. M., and Gong, Z. H. (2020). Transcription factor CaSBP12 negatively regulates salt stress tolerance in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 21:444. doi: 10.3390/ijms21020444
Zhang, J. Y., Qu, S. C., Qiao, Y. S., Zhang, Z., and Guo, Z. R. (2014). Overexpression of the Malus hupehensis MhNPR1 gene increased tolerance to salt and osmotic stress in transgenic tobacco. Mol. Biol. Rep. 41, 1553–1561. doi: 10.1007/s11033-013-3001-9
Keywords: Fraxinus velutina Torr., salt stress, microRNA, degradome, defense response
Citation: Liu JN, Ma X, Yan L, Liang Q, Fang H, Wang C, Dong Y, Chai Z, Zhou R, Bao Y, Wang L, Gai S, Lang X, Yang KQ, Chen R and Wu D (2022) MicroRNA and Degradome Profiling Uncover Defense Response of Fraxinus velutina Torr. to Salt Stress. Front. Plant Sci. 13:847853. doi: 10.3389/fpls.2022.847853
Received: 03 January 2022; Accepted: 23 February 2022;
Published: 01 April 2022.
Edited by:
Rohini Garg, Shiv Nadar University, IndiaReviewed by:
Charu Lata, National Institute of Science Communication and Information Resources (CSIR), IndiaYouxiong Que, Fujian Agriculture and Forestry University, China
Copyright © 2022 Liu, Ma, Yan, Liang, Fang, Wang, Dong, Chai, Zhou, Bao, Wang, Gai, Lang, Yang, Chen and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ke Qiang Yang, eWFuZ3dlcmVAMTI2LmNvbQ==; Dejun Wu, c2Rsa3k0MTJ4QDE2My5jb20=; Rong Chen, Y3IxMzg1NDgwMDgwNUAxNjMuY29t
†These authors have contributed equally to this work
 Jian Ning Liu
Jian Ning Liu Xinmei Ma1†
Xinmei Ma1† Hongcheng Fang
Hongcheng Fang Yuhui Dong
Yuhui Dong Ke Qiang Yang
Ke Qiang Yang