- 1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Institute of Virology and Biotechnology, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Institute of Crop Science and Institute of Bioinformatics, Zhejiang University, Hangzhou, China
- 4Shanghai Key Laboratory of Plant Molecular Sciences, College of Life Sciences, Shanghai Normal University, Shanghai, China
- 5School of Biological Sciences, The University of Hong Kong, Hong Kong, Hong Kong SAR, China
Aminoacyl tRNA synthetases primarily function to attach specific amino acids to the corresponding tRNAs during protein translation. However, their roles in regulating plant growth and development still remain elusive. Here we reported a rice thermo-sensitive mutant yellow leaf chlorosis3 (ylc3) with reduced chlorophyll content, altered thylakoid structure, and substantially elevated levels of free aspartate, asparagine and glutamine in leaves under low temperature condition. Map-based cloning identified that YLC3 encodes an aspartyl-tRNA synthetase which is localized in cytosol and mitochondria. In addition, quantitative proteomics analysis revealed that both nuclear and chloroplast-encoded thylakoid proteins were significantly down-regulated in the mutant. On the other hand, proteins involved in amino acid metabolism and the process of protein synthesis were up-regulated in ylc3, particularly for key enzymes that convert aspartate to asparagine. Moreover, uncharged tRNA-Asp accumulation and phosphorylation of the translation initiation factor eIF2α was detected in the mutant, suggesting that YLC3 regulates the homeostasis of amino acid metabolism and chloroplast thylakoid development through modulation of processes during protein synthesis.
Introduction
Rice is one of the most important staple crops in the world, feeding more than half of Asia’s population. Photosynthesis efficiency is a major determinant of crop productivities. Chloroplasts are the location not only for photosynthesis, but also for biosynthesis of many important metabolites. Originated from endosymbiotic cyanobacteria approximately 1 billion years ago, chloroplasts are semi-autonomous organelles capable of independent transcription and translation (Dyall et al., 2004; Jarvis and López-Juez, 2013). Over a long period of symbiotic evolution, the majority of chloroplast genes had been transferred to the nuclear genome, and their encoded proteins are translated in cytosol and then translocated to chloroplasts (Woodson and Chory, 2008). Currently there are approximately 3,000 chloroplast proteins, among which only about 100 proteins are encoded by the chloroplast genome (Abdallah et al., 2000; Richardson et al., 2017). Signaling regulatory mechanisms exist between nuclear genes and chloroplast genes. There are numerous nuclear genes regulating the processes of transcription, post-transcriptional modification, and translation in chloroplasts (Fernández and Strand, 2008). On the other hand, chloroplasts are regulating nuclear gene expression through tetrapyrrole signals and its own redox status (Fernández and Strand, 2008). Meanwhile, mitochondria, as the cellular energy factories which generate a large amount of ATP, are important for chloroplast biogenesis and development. In the rice mutant wp3, mitochondrial functional deficiencies also resulted in inhibition of chloroplast development (Li et al., 2018). Hence, normal chloroplast development requires coordinated regulation by nucleus, chloroplasts, and mitochondria.
Homeostasis of amino acid metabolism is pivotal to cellular growth and development (Bröer and Bröer, 2017). During amino acid deficiencies, mammalian-specific protein kinases are activated and they phosphorylate the translation initiation factor eiF2α (Hinnebusch, 2005; Wek et al., 2006). Consequently, translation of most proteins is inhibited to reduce energy consumption. At the same time, the expression of genes encoding enzymes for amino acid biosynthesis is enhanced to ensure cellular survival by transcription factor GCN4 (General Control Non-derepressible-4) during nutritional deficiencies (Natarajan et al., 2001; Dever and Hinnebusch, 2005; Li and Lam, 2008). In yeast, the GCN2 (General Control Non-derepressible-2) kinase-mediated eIF2α phosphorylation is an important adaptation strategy for metabolic and physiological changes resulting from nutritional deficiencies (Dever et al., 1992). Interaction between GCN1 and GCN2 is a necessary condition for the activation of GCN2 (Marton et al., 1993; Sattlegger and Hinnebusch, 2005). The Arabidopsis AtGCN2 could complement the yeast gcn2 mutant phenotypes, implicating functional conservation (Zhang et al., 2003). In addition, chemically-induced amino acid starvation, UV light, wounding, chilling, and hormone treatments in Arabidopsis could all cause AtGCN2-dependent eIF2α phosphorylation which requires AtGCN1 and AtGCN2 interactions (Lageix et al., 2008; Zhang et al., 2008; Li et al., 2013; Wang et al., 2017). Moreover, Arabidopsis atgcn1-1 and atgcn1-2 mutants are cold-stress sensitive with retarded chloroplast development (Zhang et al., 2017). A recent study in Arabidopsis revealed that change in reactive oxygen species (ROS) levels in chloroplasts could also initiate AtGCN2-mediated eIF2α phosphorylation (Lokdarshi et al., 2020). These results indicated that eIF2a phosphorylation is a cold-stress tolerance mechanism in plants through inhibition of protein translation.
Aminoacyl-tRNA synthetases (AARSs) are one of the key enzymes involved in protein synthesis by linking amino acids with their specific tRNA (Ibba and Soll, 2000; O’Donoghue and Luthey-Schulten, 2003). Plant proteins are synthesized in cytosol, mitochondria and chloroplasts which all have a complete set of AARSs. In plants, AARSs are all nuclear-encoded, translated in cytosol, and then translocated to cytosol, mitochondria, or chloroplasts. There are 45 AARSs identified in Arabidopsis and some of them are translocated to different organelles to ensure normal protein synthesis (Duchêne et al., 2005). It can be perceived that sharing of AARSs between organelles is imperative for regulation of translation since the activities of AARSs and tRNA charged state are the major control points for prokaryotic and eukaryotic translational systems (Duchêne et al., 2005).
Gene cloning and functional characterizations of AARS mutants in tobacco, Arabidopsis, and rice revealed the involvement of some mitochondrial- and chloroplast-targeted AARSs chloroplast development and their mutations resulted in chlorotic or albino phenotypes (Kim et al., 2005; Liu et al., 2007; Wang et al., 2016; Zhang et al., 2017; Fang et al., 2020). The glutamyl-tRNA synthetase encoded by rice Os10g0369000 is localized in cytosol and mitochondria, showing anther-specific expression and participating in anther formation and development (Yang et al., 2018). In Arabidopsis, 21 out of the 45 known AARSs are necessary for endosperm formation and embryogenesis (Berg et al., 2005). For example, cysteinyl-tRNA synthetase is specifically expressed in central cells of female gametes, determining the fate of the adjacent accessory cells (Kägi et al., 2010). Meanwhile, the Arabidopsis aspartyl-tRNA synthetase (IBI1, Impaired in BABA-induced disease Immunity1) is a beta-aminobutyric acid-inducible receptor protein for broad-spectrum disease resistance. IBI1 is mainly localized in endoplasmic reticulum and cytoplasm. Its deficiency led to accumulation of uncharged tRNAs, which in turn promoted GCN2-dependent phosphorylation of eIF2α, thereby inhibiting plant growth and development (Luna et al., 2014). In addition, IBI1 can be localized in nucleus, interacting with the transcription factors VOZ1/2 (Vascular Plant One Zinc Finger1/2) and regulating the expression of cell-wall defense and abiotic stress-responsive genes through the abscisic acid signaling pathway (Schwarzenbacher et al., 2020). Plant AARSs are mainly affecting amino acid homeostasis, protein synthesis and abiotic stress (Luna et al., 2014; Yang et al., 2018; Schwarzenbacher et al., 2020). However, it remains largely elusive regarding the protein localization and expression pattern of most of the other AARSs and their roles in plant growth and development, particularly for those in monocot species like rice.
In the present study, we reported the function of a cytosol- and mitochondria-localized aspartyl-tRNA synthetase in rice. YLC3 functional deficiency disrupted amino acid homeostasis, initiated eIF2α phosphorylation, inhibited translation, and influenced chloroplast development under low temperature condition.
Results
Phenotypic Characterization of the ylc3 Mutant
From a population of ethyl methanesulfonate (EMS)-induced rice mutants, a yellow leaf mutant designated as ylc3 (yellow leaf chlorosis3) was identified. At 19°C, 2-week-old ylc3 mutant plants showed yellow leaves containing only 30% photosynthetic pigments compared to wild-type plants. The photosynthetic pigments increased to 50 and 80% of wild-type levels at 24 and 30°C, respectively. In addition, chlorophyll a content was inhibited more pronouncedly than chlorophyll b in the mutant. These data indicated that ylc3 is a thermo-sensitive mutant (Figure 1). In light growth chambers, plant growth was affected by different temperatures. Under field conditions, ylc3 plants were slightly shorter than wild-type plants while heading period and seeding rate were both normal (Figure 1). Transmission electronic microscopy analysis revealed the impaired development of chloroplasts in ylc3 growing at 19°C with substantially reduced number of thylakoid grana lamella (Figure 2).
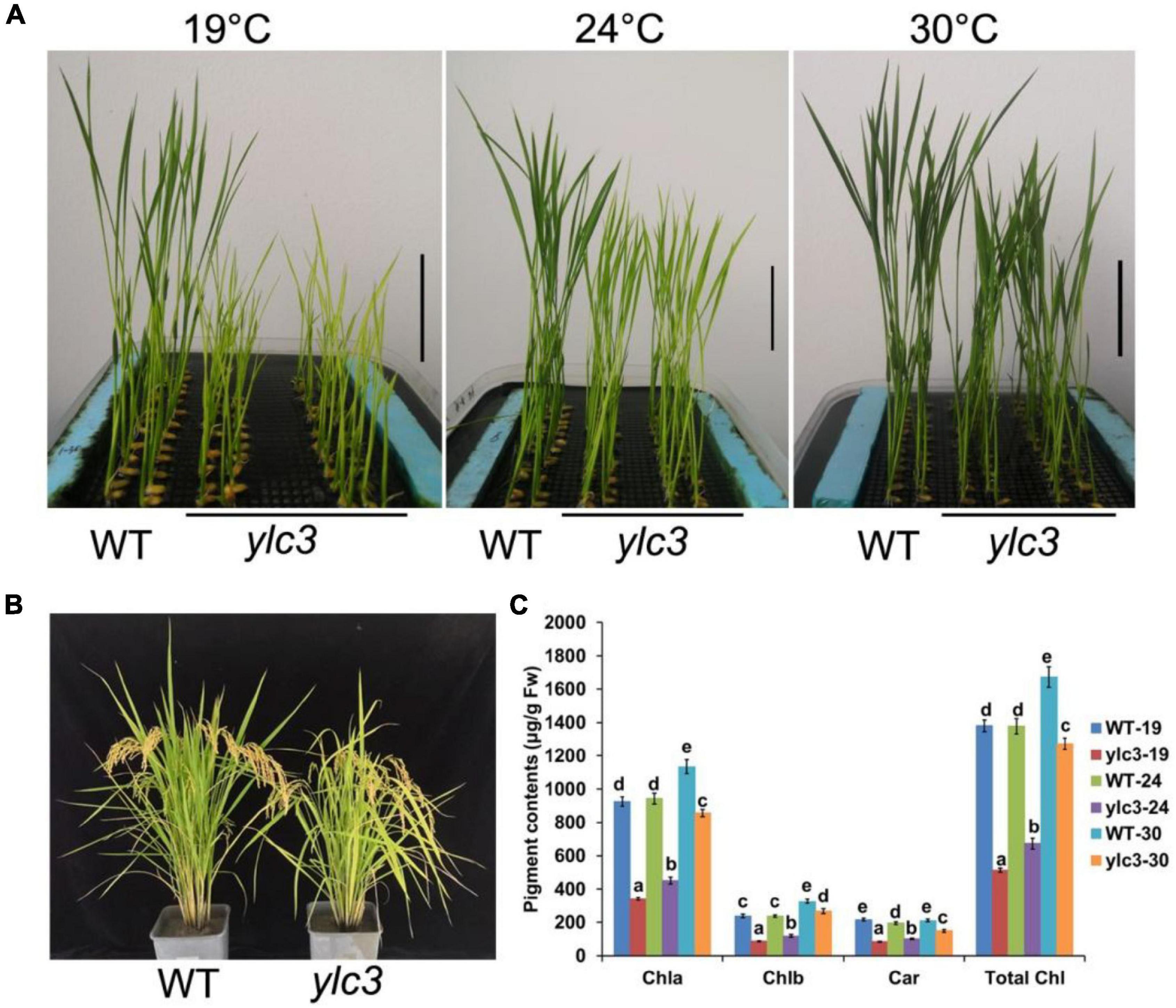
Figure 1. Phenotypic analyses of ylc3 mutant. (A) Phenotypes of the WT and ylc3 seedlings (10-day-old) grown at different temperatures. Bar = 5 cm. (B) Heading stage. (C) Photosynthetic pigments of WT and ylc3 (10-day-old) grown at different temperatures (°C). Error bars represent SD (n = 5). Bars with different letters indicate significant differences at P < 0.05, ANOVA.
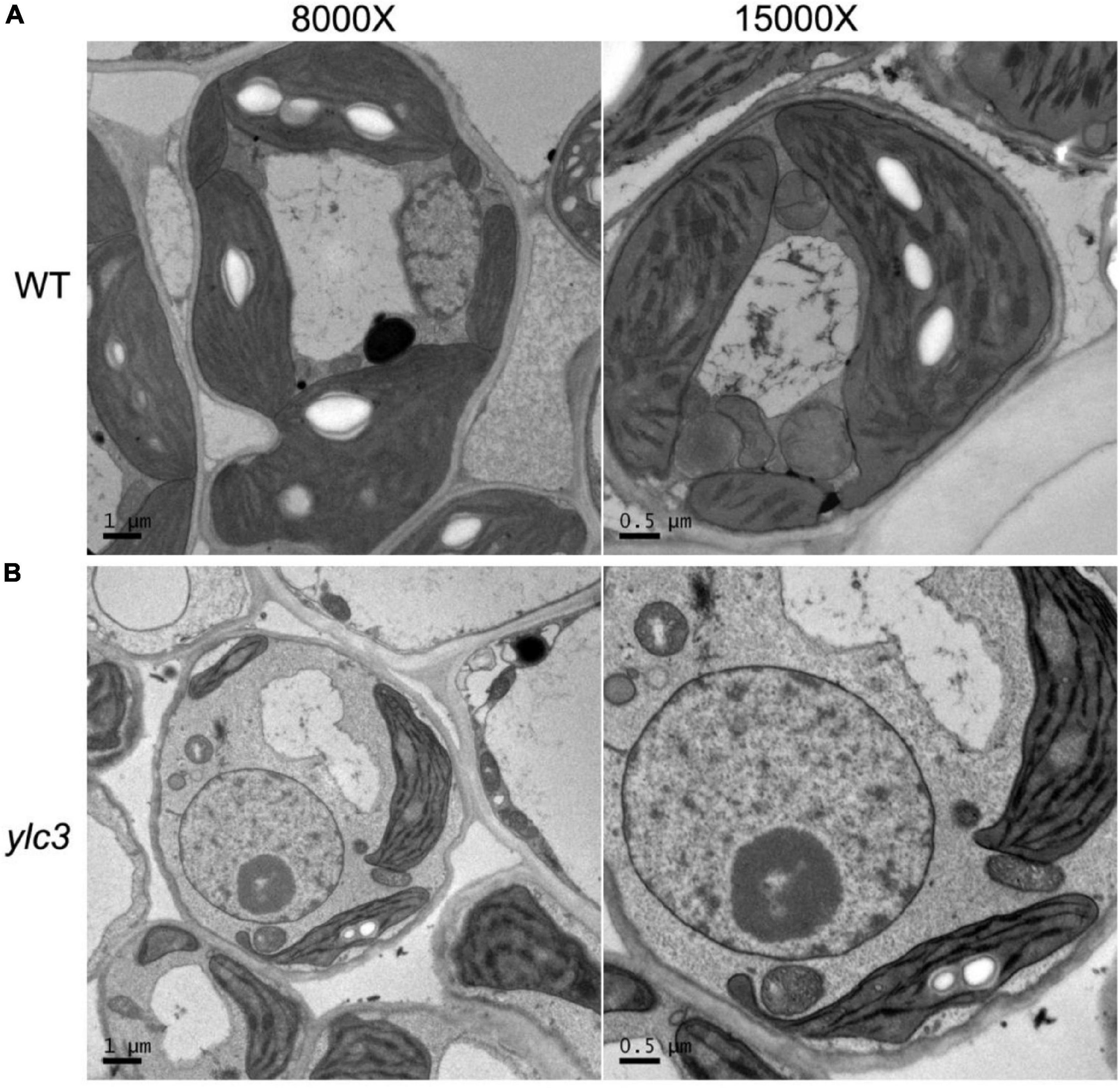
Figure 2. Impaired thylakoid development in ylc3 mutant. (A) WT chloroplasts showed well-developed thylakoids. (B) ylc3 chloroplasts showed reduced number of thylakoid grana lamella.
Map-Based Cloning of the YLC3 Gene
To study the genetics of the yellow leaf phenotype, ylc3 was crossed separately with Nipponbare or Kasalath wild-type plants. Under low temperature condition, the F2 population of the ylc3 × Nipponbare cross showed a segregation ratio of 95:330 for yellow leaf vs. green leaf(1:3 ratio, χ2 = 2.38 < χ20.05 = 3.84, P > 0.05); 129 of 409 plants showed yellow leaf phenotype in the F2 generation of ylc3 × Kasalath (1:3 ratio, χ2 = 0.30 < χ20.05 = 3.84, P > 0.05). The above results indicate that the yellow leaf phenotype is conferred by a single recessive nuclear gene. F2 population from the ylc3 × Kasalath cross was then used for genetic mapping. In the preliminary mapping, 94 F2 plants with yellow leaf phenotype were analyzed with 120 sequence-tagged site (STS) markers which are evenly distributed on 12 rice chromosomes. YLC3 was initially mapped within a 950-kb region between the molecular markers STS2 and STS3 on chromosome 2. At the same time, F2 population from the ylc3 and Nipponbare cross was subject to whole-genome sequencing-based MutMap method to map YLC3 (Abe et al., 2012; Lü et al., 2015). The mutation was identified to be a single-base substitution (G → A) in the aspartyl-tRNA synthetase-encoding gene located on chromosome 2 (LOC_Os02g46130), resulting in a single amino acid replacement (Arginine → Lysine) near the C-terminal region of YLC3 (Figure 3).
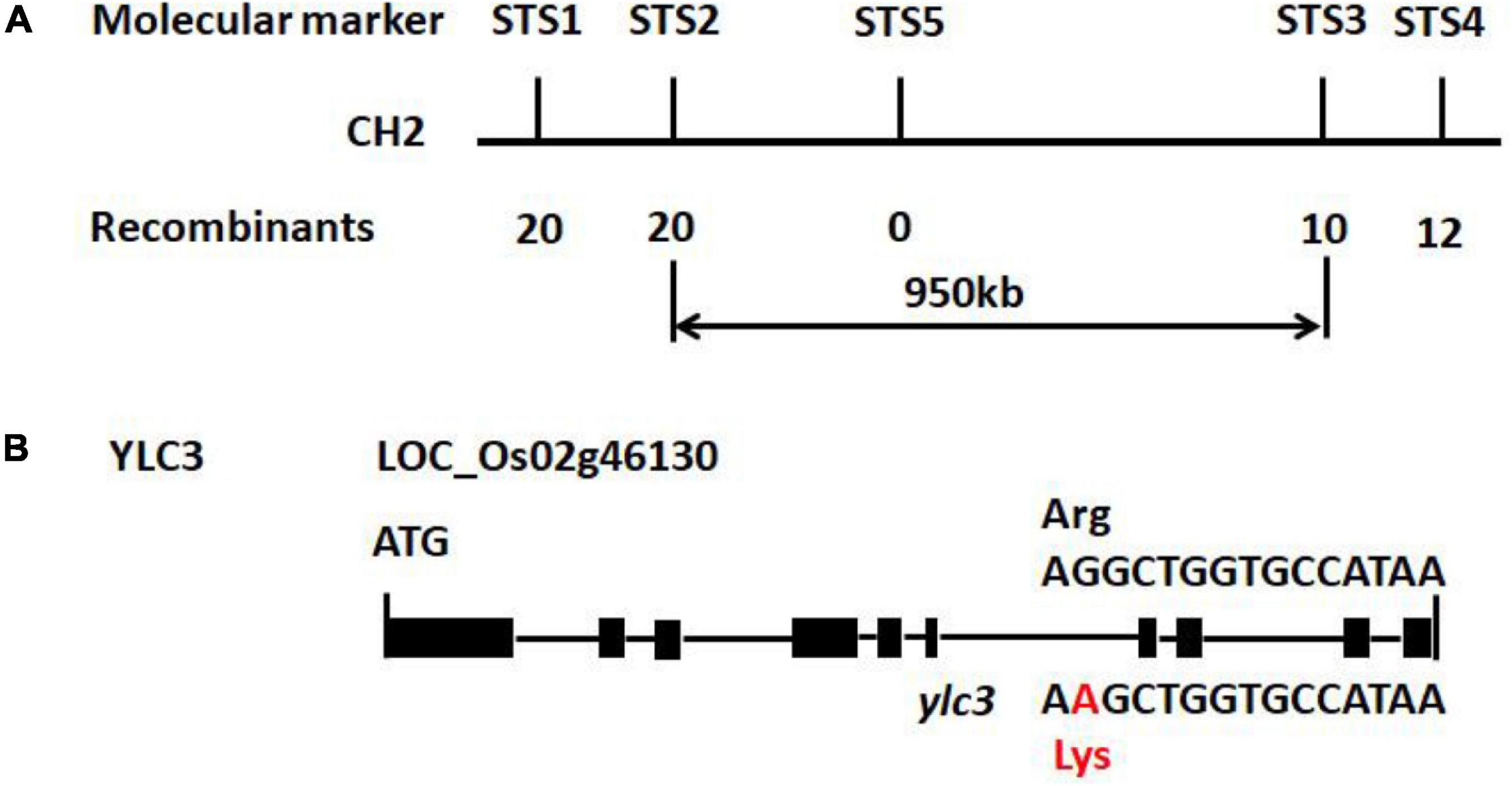
Figure 3. Preliminary gene mapping and ylc3 mutation. (A) InDel markers (STS1-5) used for preliminary mapping are indicated. Numbers of recombinants and F2 mutants (n) are shown. (B) Gene structure of YLC3 and the mutation site. The “A” in red indicates a one base-pair substitution in ylc3. Black rectangles represent exons.
To validate that LOC_Os02g46130 was the affected gene, we constructed the binary vector Pro:YLC3-NOS for ylc3 transformation. A total of 20 independent transformants were obtained with wild-type phenotypes. Meanwhile, a gene-editing vector was constructed for Nipponbare transformation. A total of 22 positive transformants were obtained, 9 of which were homozygous for the G → A substitution and they all showed yellow leaf phenotype at low temperature condition. Phenotyping and genotyping analyses revealed the yellow leaf phenotype in the T1 plants growing at lower temperatures. The above results established that the yellow leaf phenotype was conferred by the ylc3 mutation in LOC_Os02g46130 (Figure 4).
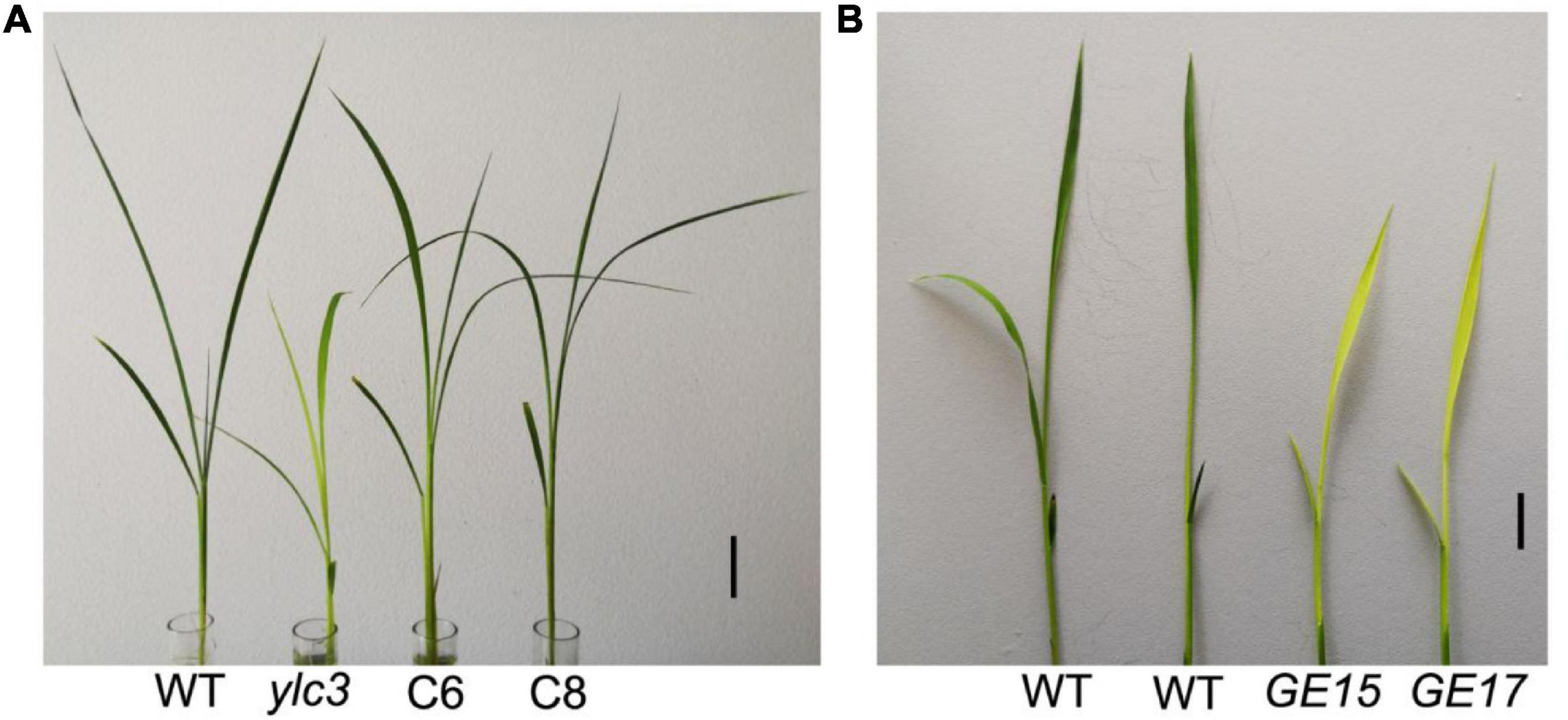
Figure 4. Complementation and YLC3 gene editing analysis. (A) The ylc3 mutation was complemented by YLC3 promoter expression of a full-length YLC3 coding sequence. Bar = 3 cm. (B) YLC3 gene-editing transgenic seedlings showed the yellow leaf phenotype at low temperature. Bar = 1 cm.
Sequence and Phylogenetic Analyses
Aminoacyl-tRNA synthetases are classified into two categories: eukaryotic-specific (clade I) and prokaryotic-originated (clade II). Sequence analysis of YLC3 revealed the presence of an N-terminal coiled coil functional domain and a C-terminal tRNA synthetase class II functional domain, hence YLC3 belongs to a clade II AARS. Clustering analysis further illustrated that YLC3 is very conserved among different species with highly homologous functional domain and motif. Among angiosperms, YLC3 shares higher homology with proteins from monocotyledons such as sorghum, barley and corn, with slightly lower homology with proteins from dicot species such as Arabidopsis, Brassica napus and soybean (Figure 5). Taken together, YLC3 is a highly conserved clade II aspartyl-tRNA synthetase.
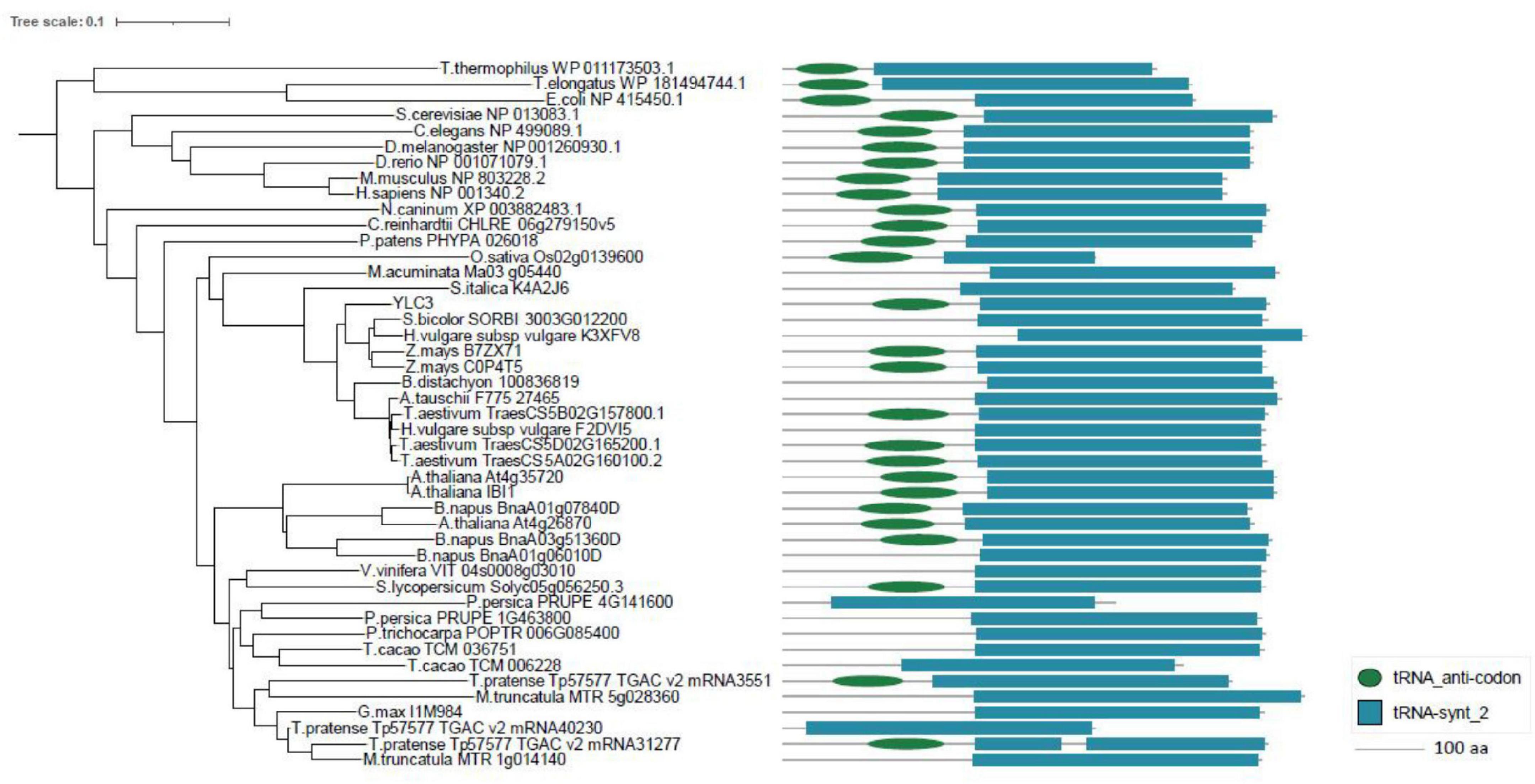
Figure 5. Phylogenetic analysis and motif alignment. A phylogenetic tree was constructed with aligned full-length sequences of homologs of YLC3. Amino acid sequences from regions 101 to 188 and 223 to 544 in YLC3 were used for motif alignment by MEGA.
Expression and Subcellular Localization Analyses
To examine the spatial expression of YLC3, a promoter: GUS binary vector was constructed and transformed into Nipponbare rice. GUS staining was performed in roots, stems, leaves, glumes, anthers, and pistils from the positive transformants. Results indicated that YLC3 was expressed in all tissues examined. GUS staining was stronger in roots, stems, and leaves but weaker in anthers and pistils (Figure 6). Hence, YLC3 is considered a constitutively expressed gene.
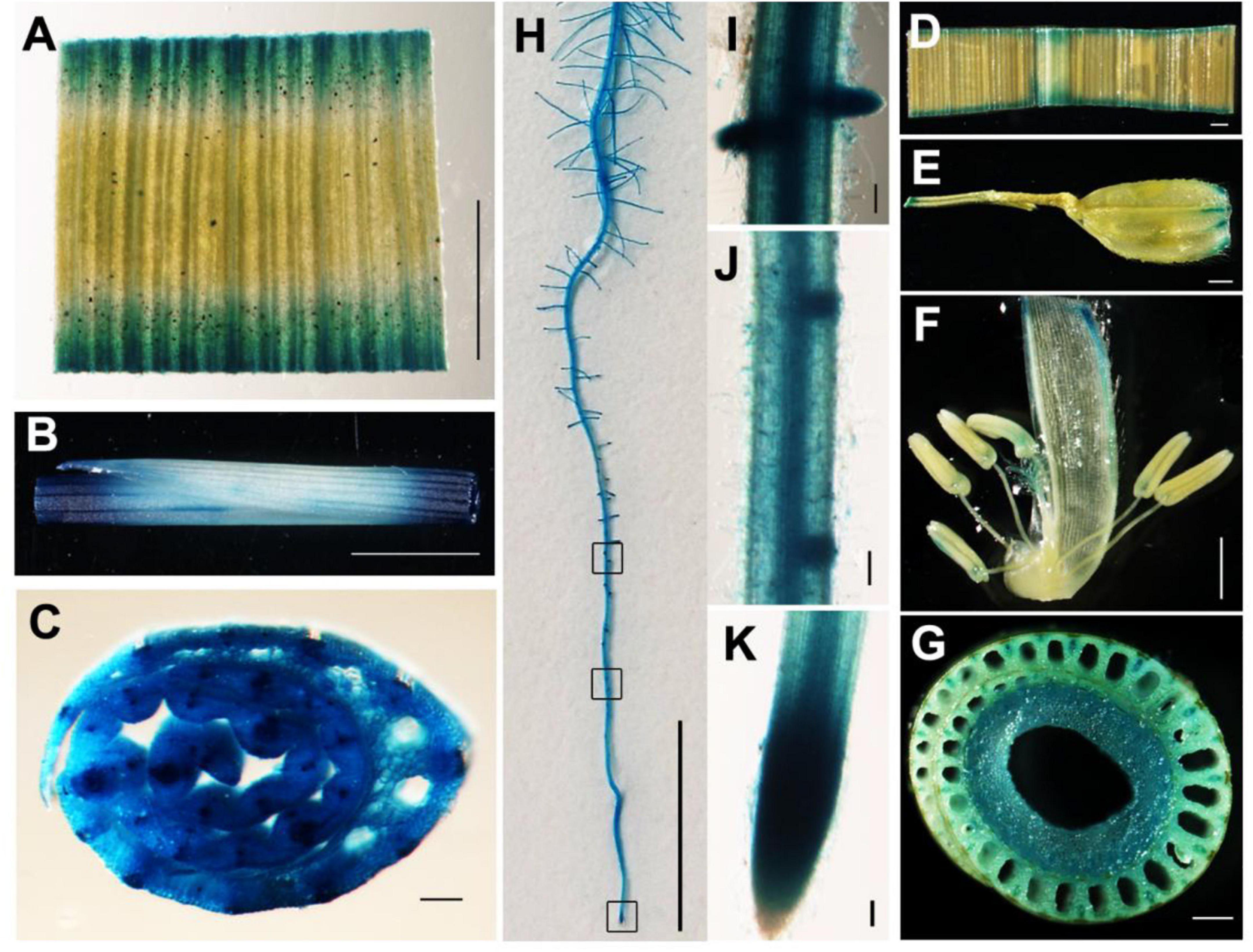
Figure 6. Tissue expression pattern of YLC3. (A–C,H–K) GUS staining of 7-day-old ProYLC3:GUS transgenic seedlings. (A) Leaf. (B) Stem. (C) Cross section of stem. (H) Primary root. (I) Lateral root. (J) Lateral root primordium. (K) Root tip. (D–G) GUS staining of the heading stage of ProYLC3:GUS transgenic plants. (D) Leaf. (E) Glume. (F) Anthers and pistils. (G) Cross section of stem. Scale bars are 100 μm in (C,D,E,I–K), 1 mm in (A,B,F,G), and 1 cm in (H).
Aminoacyl-tRNA synthetases are mainly participating in protein synthesis and there is a complete set of AARSs in cytosol, mitochondria, and chloroplasts. To examine the subcellular localization of YLC3, a 35S promoter-driven YLC3:sGFP fusion expression vector was constructed. Transient expression of YLC3:sGFP in rice protoplasts revealed green fluorescent signals mainly in cytosol (Figure 7). After staining the transfected protoplasts with a mitochondrial specific dye (MitoTracker, Invitrogen, Carlsbad, CA, United States), some YLC3:sGFP green fluorescent signals were found to overlap with the red mitochondrial signals (Figure 7). These observations indicated that YLC3 is mainly localized in cytosol and mitochondria, but not in chloroplasts.
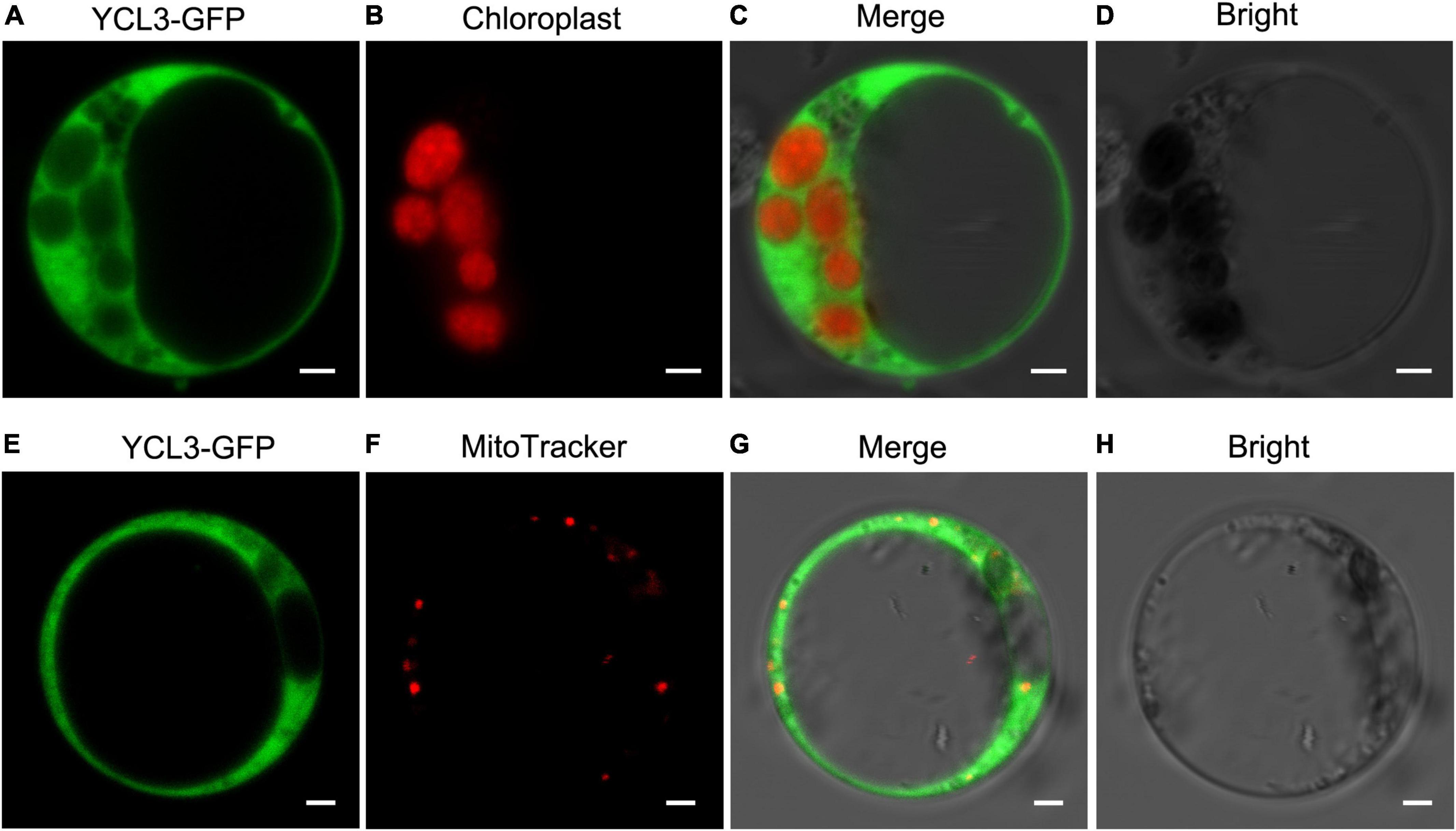
Figure 7. Subcellular localization of YLC3-sGFP. (A–D) Transient expression of YLC3:sGFP in rice protoplasts. (A) GFP. (B) Chlorophyll autofluorescence. (C) Merged images of (A,B). (D) Bright field. (E–H) Transient expression of YLC3:sGFP and MitoTracker staining of transfected protoplasts. (E) GFP. (F) Mitochondrial fluorescence staining by MitoTracker. (G) Merged images of (E,F). (H) Bright field.
Analysis of Free Amino Acids in ylc3 Mutant
YLC3 encodes an aspartyl-tRNA synthetase which catalyzes the reaction between aspartate and its specific tRNA. If aspartyl-tRNA synthetase is functionally deficient, it may result in the accumulation of free aspartate and uncharged tRNAs. Wild type plants, ylc3 mutant plants and complementation lines were kept at low temperature (19°C) and high temperature (30°C) in light growth chambers and free amino acid contents in leaves were determined at 2-leaf stage. Under low temperature condition, ylc3 mutant leaves showed 92% increase in aspartate, 10-fold increase in glutamine, and 78-fold increase in asparagine, when compared to wild-type plants and complementation lines (Figure 8). Under high temperature condition, contents of aspartate, glutamine, and asparagine were restored to normal levels in the ylc3 mutant leaves (Figure 8). The above results demonstrated the changes in free amino acid contents in ylc3 leaves, especially asparagine, under low temperature condition.
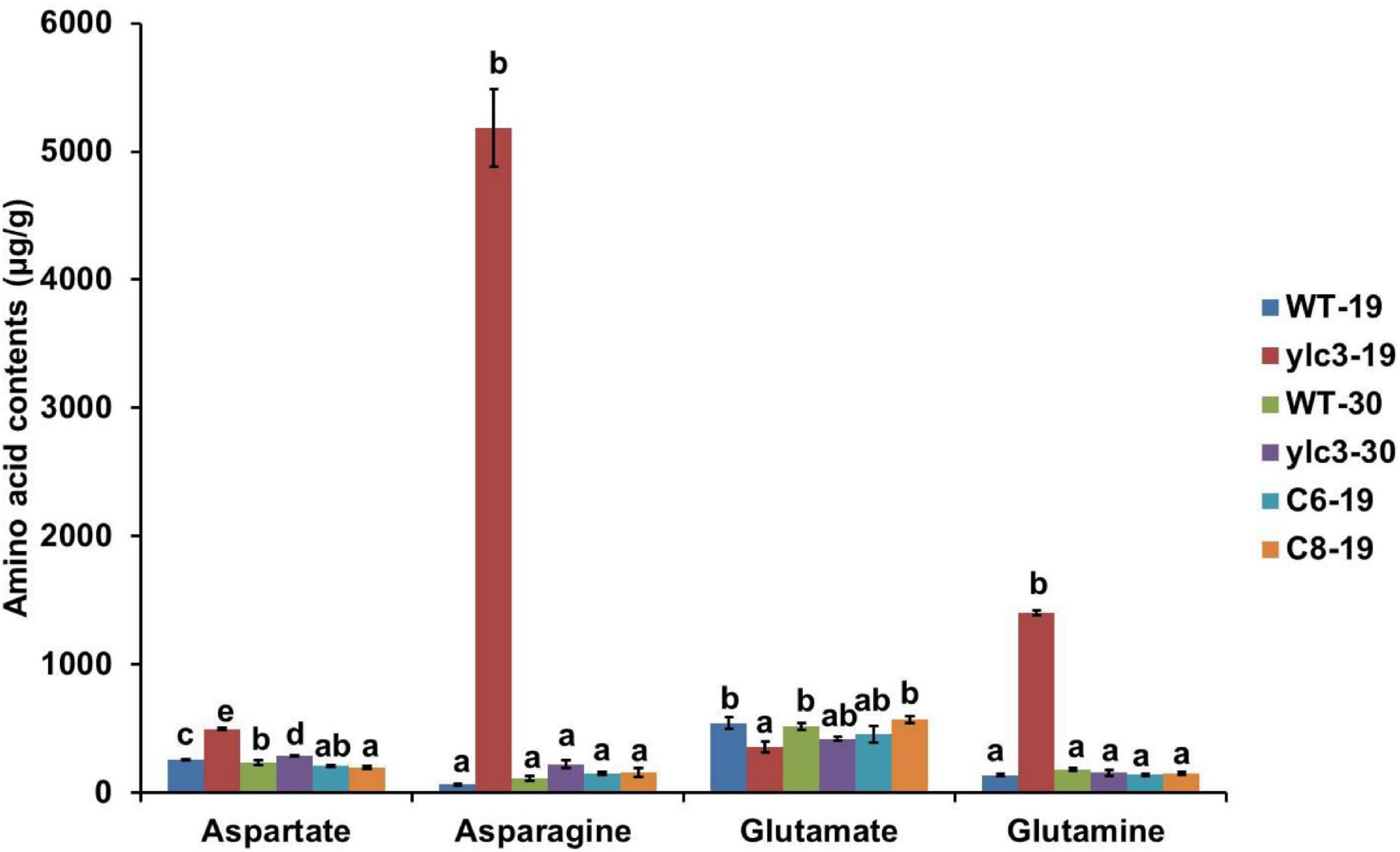
Figure 8. Free amino acid analysis of 10-day-old WT and ylc3 seedlings grown at different temperatures. Error bars represent SD (n = 3). Bars with different letters indicate significant differences at P < 0.05, ANOVA.
Quantitative Proteomics Analyses
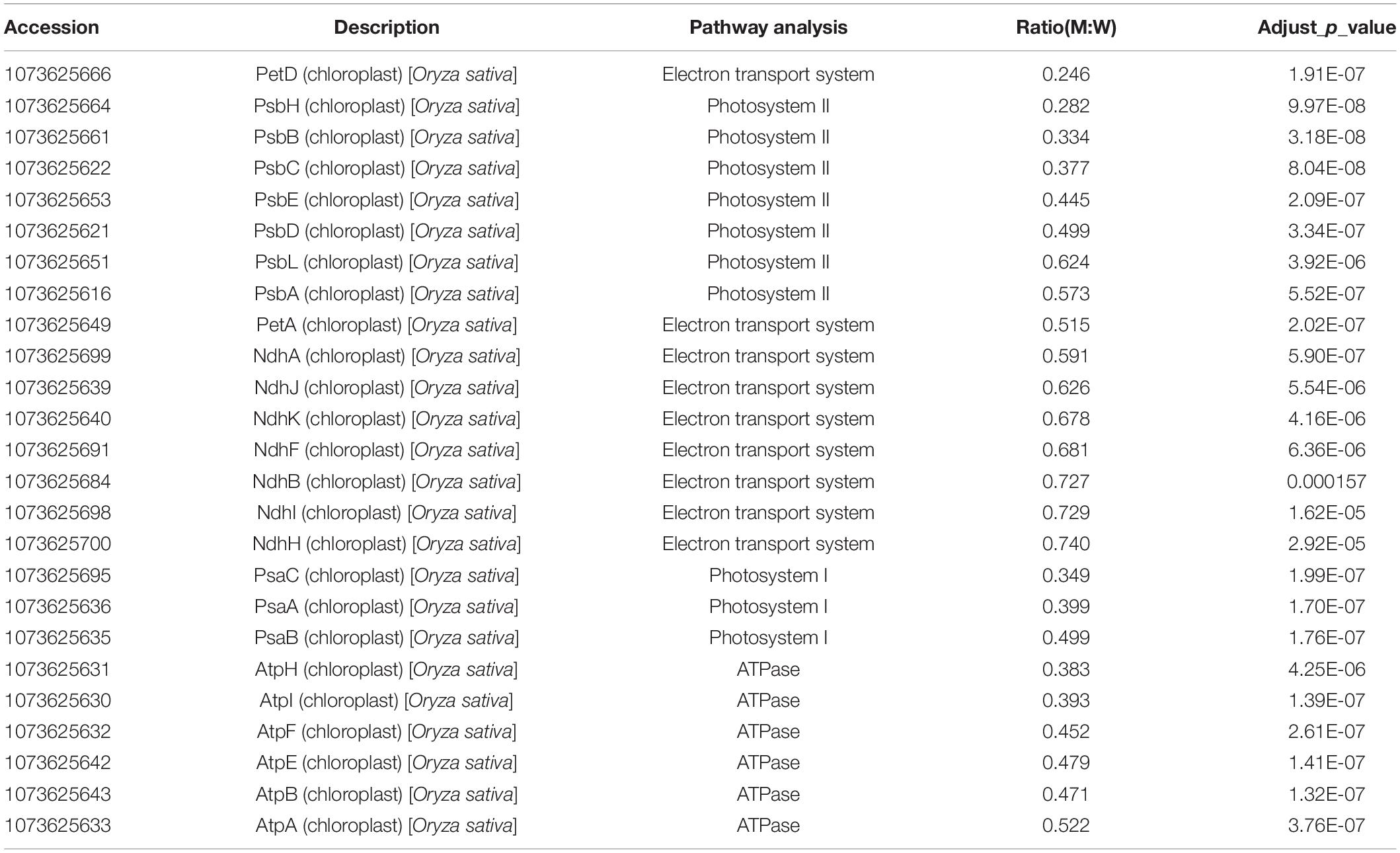
Since AARSs are playing pivotal roles in protein synthesis while ylc3 mutant showed severe inhibition of chlorophyll production and chloroplast development under low temperature condition, quantitative proteomics analyses were performed with ylc3 and wild-type seedlings growing at 19°C. Using the high-throughput tandem mass tag method, a total of 9,212 proteins comprising 9143 nuclear-encoded proteins, 51 chloroplast-encoded proteins, and 18 mitochondrial-encoded proteins were detected (Supplementary Table 1). Among the mitochondrial-encoded proteins, 11 proteins were up-regulated in ylc3 seedlings while the others were unaffected. All the 25 chloroplast-encoded proteins in different thylakoid protein complexes such as Photosystem I, Photosystem II, cytochrome b6f complex, and NADH dehydrogenase complex were down-regulated in the mutant (Table 1).
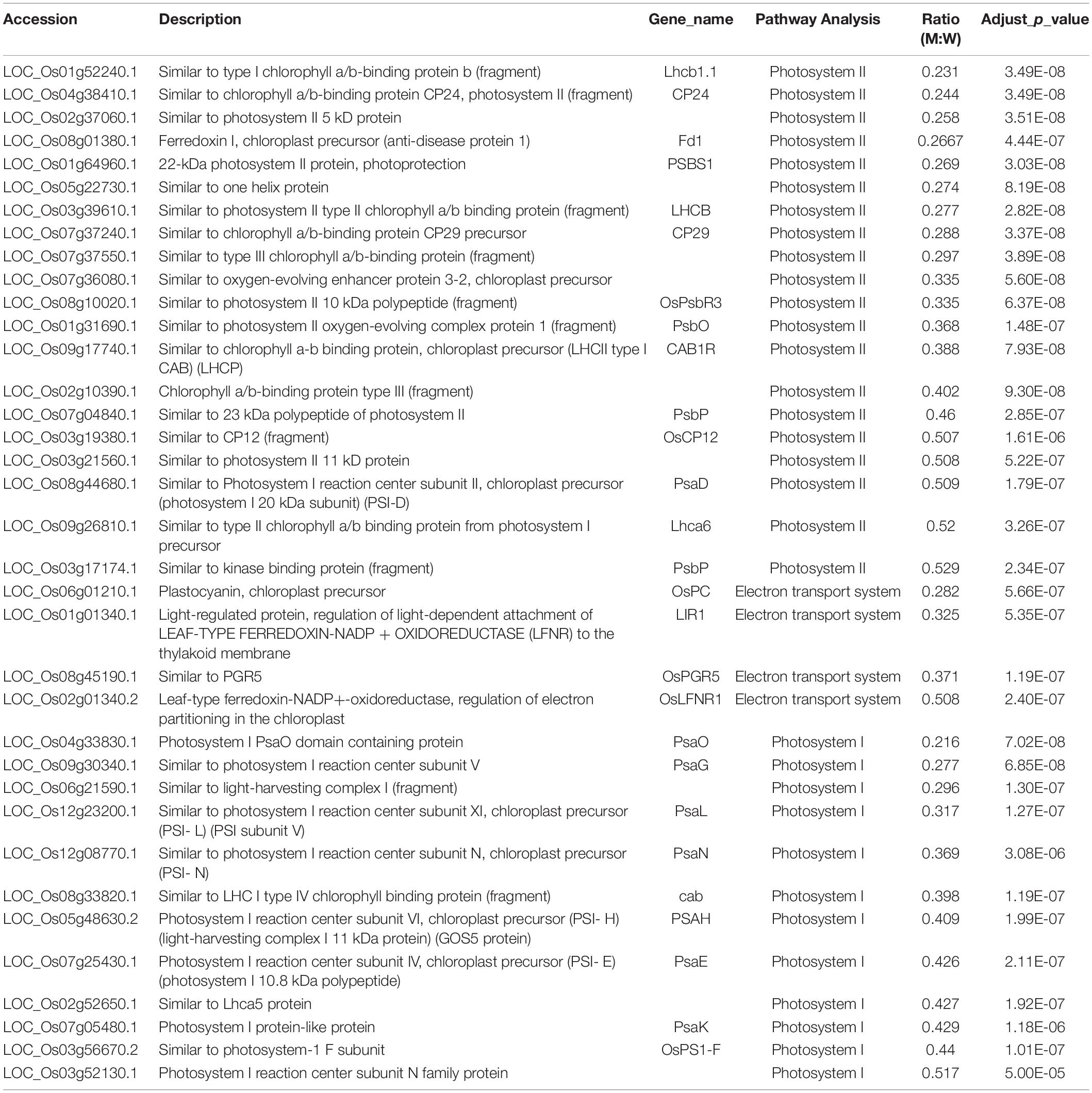
The KEGG analysis of 4,979 nuclear-encoded proteins revealed 1,384 differentially expressed proteins (>2-fold changes) with 154 proteins down-regulated and 1,230 proteins up-regulated (173 of them were up-regulated by more than threefold) in the ylc3 mutant (Supplementary Table 2). The down-regulated proteins are enriched in photosynthesis-related proteins including 13 photosynthetic antenna proteins, 20 photosystem-related proteins, 14 photosynthetic carbon fixation proteins, and 14 carbohydrate metabolism-related proteins (Table 2). Most of the nuclear-encoded and chloroplast-encoded thylakoid membranes were apparently down-regulated, which was probably the major cause for the reduced chlorophyll content, impaired thylakoid development, and yellow seedling phenotype in the ylc3 mutant.
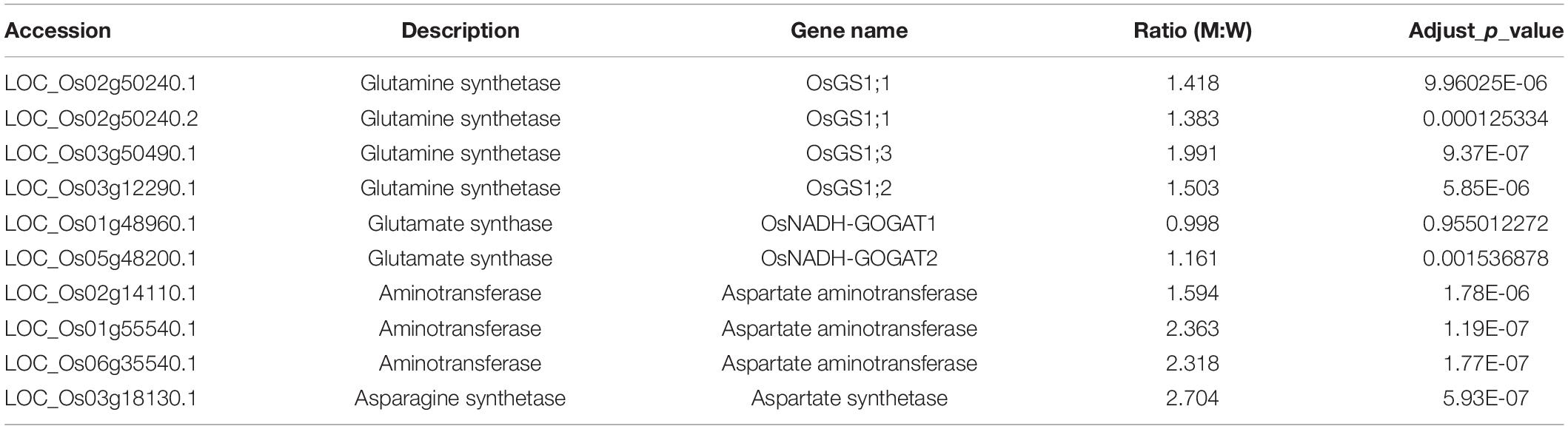
The KEEG analysis also identified 166 ribosomal proteins, 92 carbon metabolism-related proteins, 88 amino acid biosynthesis proteins, and 41 pyruvate metabolism-related proteins (Figure 9). Most of these proteins were significantly up-regulated in the ylc3 mutant. For the aspartate metabolic pathways, asparagine synthetase, glutamine synthetase, and aspartate aminotransferase were all up-regulated while glutamate synthetase did not show any significant changes (Table 3). The above results suggested that ribosomal protein translation efficiencies were increased and amino acid biosynthesis was enhanced, especially for asparagine and glutamine, in the ylc3 mutant under low temperature condition.
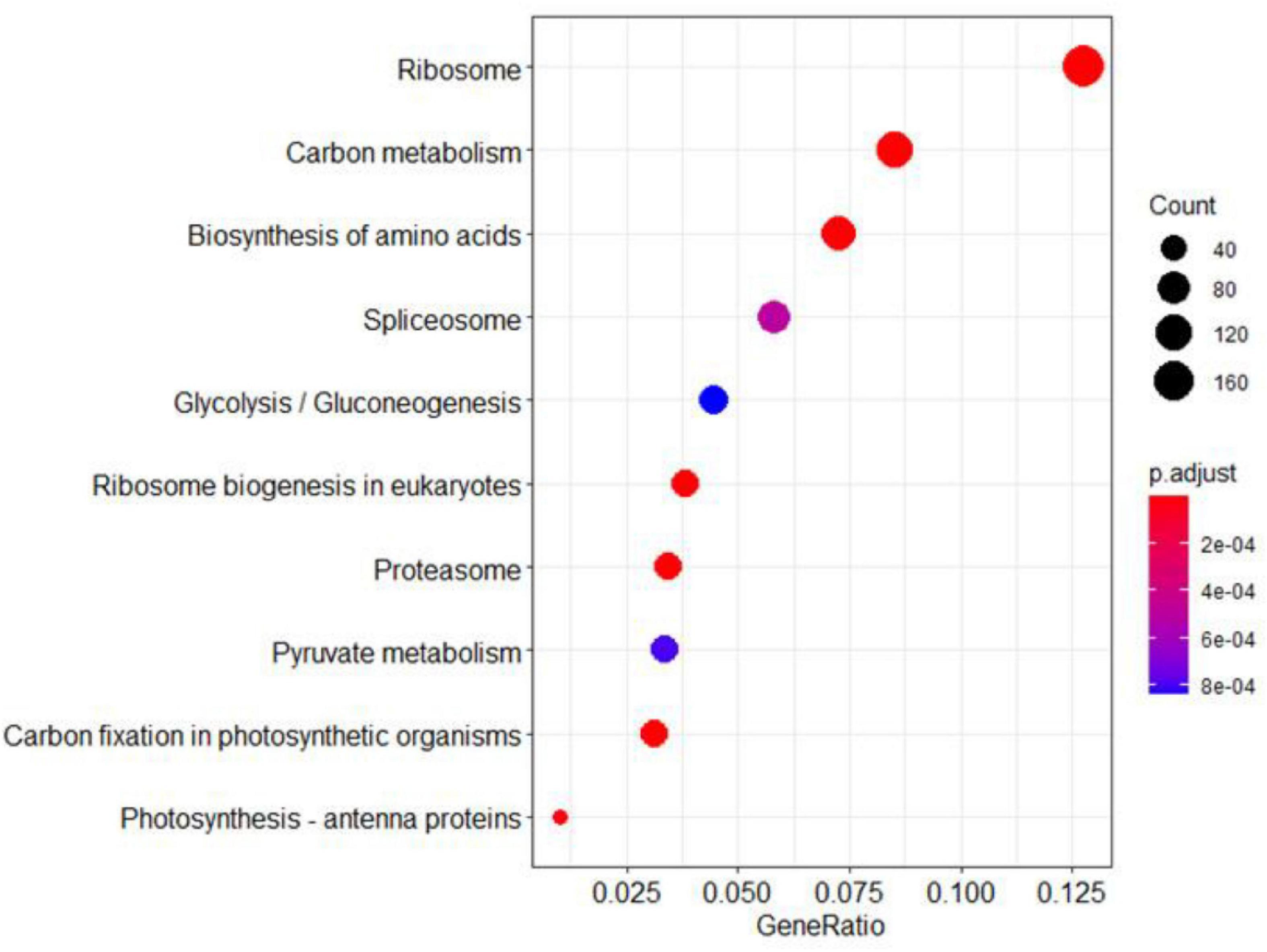
Figure 9. KEGG pathway analysis. Differentially regulated proteins were used to perform KEGG analysis on clusterProfiler. Significant differences between ylc3 and WT were detected for proteins related to photosynthesis. Pathways including carbon metabolism, pyruvate metabolism, carbon fixation in photosynthetic organisms and photosynthesis-antenna proteins were enriched (corrected P-value < 0.05).
Uncharged tRNA-Asp Accumulation and Immunoblot Analysis of eif2α Phosphorylation
In Arabidopsis, aspartyl-tRNA synthetase deficiency resulted in accumulation of aspartate and uncharged tRNA, which in turn interacted with AtGCN2 and activated its kinase activities. The activated AtGCN2 then phosphorylated eIF2a, reduced protein translation efficiencies, and inhibited plant growth and development (Luna et al., 2014). Two aspartyl-tRNA genes (trnD-GUC, Id: 29141347; trnD-GTC, Id: 3950710) in rice were retrieved from the NCBI database. To check uncharged tRNA-Asp levels, northern-blot analysis was performed. Our results demonstrated that tRNA-trnD-GUC was obviously increased in ylc3 seedlings under low temperature. However, we failed to detect the transcription of trnD-GTC in rice seedlings.
We speculated that the eif2α phosphorylation level might be increased in the ylc3 mutant under low temperature condition. Accordingly, AtEIF2α, AtGCN1, and AtGCN2 protein sequences were searched against the NCBI rice database to retrieve the homologous proteins in rice as OsEIF2α (LOC_Os03g18510), OsGCN1 (LOC_Os03g51140) and OsGCN2 (LOC_Os04g41530), respectively. These three proteins were found to be up-regulated by onefold from the above quantitative proteomics analysis. Since the phosphorylation sites are identical between OsEIF2α and AtEIF2α (as revealed by sequence alignment), eif2α-specific phosphorylation antibody (Luna et al., 2014; Wang et al., 2017) could be utilized for immunoblot detection in rice plants. We found that eif2α was apparently phosphorylated in the ylc3 mutant growing under low temperature condition when compared to the wild-type plants (Figure 10). Hence, under low temperature condition, ylc3 mutation resulted in eif2α phosphorylation which could inhibit the translation of thylakoid complex proteins, leading to impaired chloroplast development and yellow leaf phenotype.
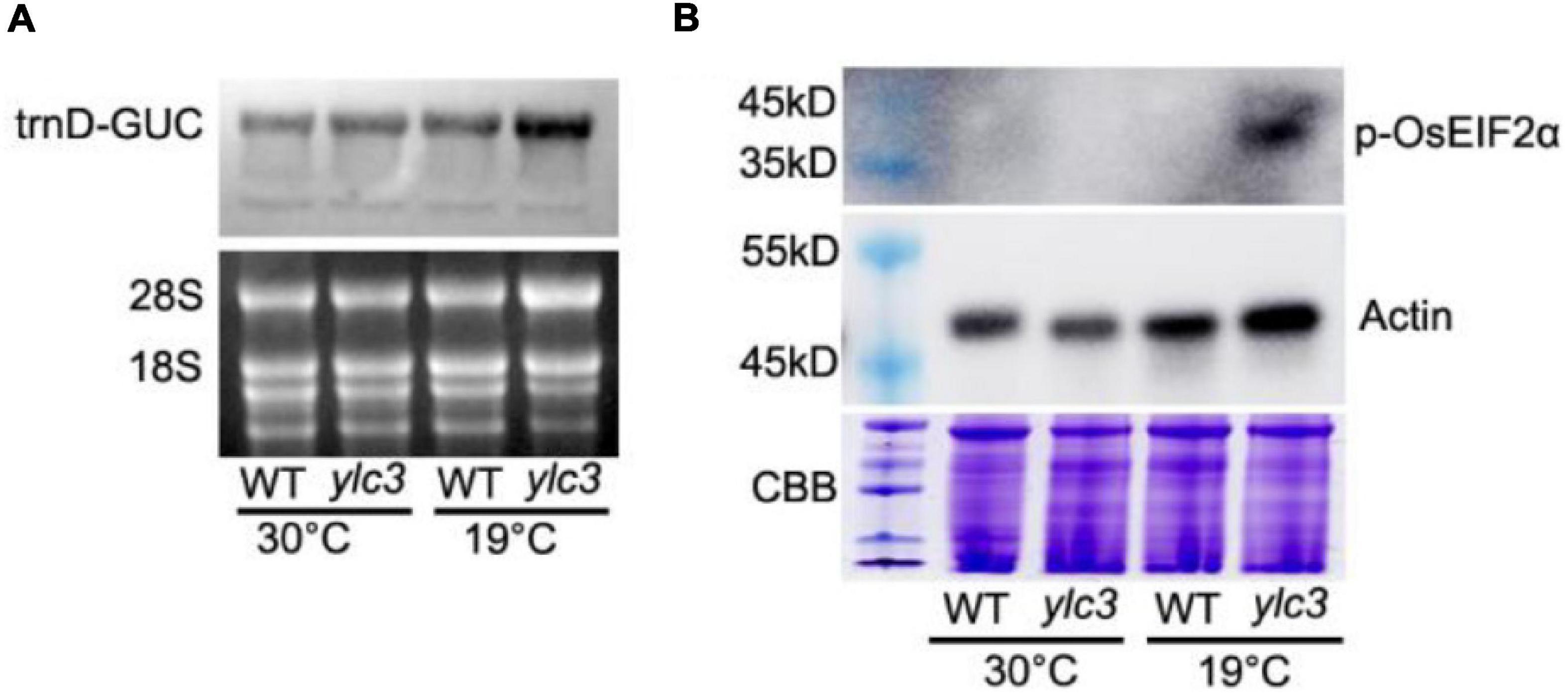
Figure 10. Uncharged tRNA-Asp accumulation and phosphorylation of OsEIF2α at different growth temperatures. (A) Uncharged tRNA-Asp levels in WT and ylc3 seedlings were analyzed by northern blot. The ethidium bromide-stained gel is shown as a loading reference. (B) Phosphorylation levels of OsEIF2α in WT and ylc3 seedlings were monitored by immunoblot analysis. Upper and lower panels represent immunoblot results using antibodies against phosphorylated eIF2α and actin, respectively. Coomassie Brilliant Blue (CBB) staining is shown as a loading reference. Protein size marker is indicated.
Discussion
Plant AARSs with different expression patterns and subcellular locations play key role in protein synthesis (Ibba and Soll, 2000; Duchêne et al., 2005). YLC3, a cytosol- and mitochondrial-localized aspartyl-tRNA synthetase, is required for free amino acid homeostasis in rice under low temperature condition. In addition, translation of thylakoid proteins is likely to be down-regulated by the GCN2-eif2α phosphorylation pathway in the ylc3 seedlings.
YLC3 Encodes an Aspartyl-tRNA Synthetase
YLC3 is a constitutively expressed gene with stronger expression in roots, stem and leaves but weaker expression in anthers and pistils (Figure 6). YLC3 contains the typical aspartyl-tRNA catalytic domain and a coiled coil functional domain. Phylogenetic analysis illustrated that YLC3 is highly conserved with reported aspartyl-tRNA synthetases including Arabidopsis IBI1 (Luna et al., 2014). Furthermore, free amino acid analyses revealed that aspartate level was increased in the ylc3 mutant. Collectively, the enhanced aspartate level together with the protein functional domain and phylogenetic analyses strongly suggested that YLC3 is an aspartyl-tRNA synthetase.
The rice ylc3 mutant is a thermo-sensitive chlorotic mutant and gene mapping identified a single amino acid substitution in the C-terminal region of YLC3. The substitution occurred outside the catalytic and coiled coil domains, indicating that the C-terminus is also critical for the activities of aspartyl-tRNA synthetase. The importance of this substitution was further supported by gene editing approach which generated transgenic rice lines with the same mutation and phenotype. On the other hand, chlorophyll and free amino acid contents are normal in ylc3 plants under high temperature condition, indicating that the mutant YLC3 protein can be functional. Alternatively, as there are at least 3 aspartyl-tRNA synthetases in rice, it remains to be investigated whether the other isozymes are involved in restoration of chlorophyll and free amino acid contents under high temperature conditions.
There is a lack of an N-terminal signal peptide in YLC3 which is localized in cytosol and mitochondria, but not chloroplasts (Figure 7). Currently, all the known aminoacyl-tRNA synthetases participating in chloroplast development are chloroplast-localized. The Arabidopsis aspartyl-tRNA synthetase IBI1 is localized in endoplasmic reticulum and cytosol. During pathogen attack or low temperature stress, IBI1 is translocated to nucleus, inducing immunity responses in Arabidopsis (Schwarzenbacher et al., 2020). It cannot be excluded that YLC3 is translocated to chloroplasts to participate in chloroplast development under low temperature condition.
Changes of Amino Acid Homeostasis in ylc3 Mutant
We have measured the 20 free amino acids in leaves of ylc3 mutant growing under low temperature condition. Surprisingly, asparagine was increased by 78-fold, glutamine was increased by 10-fold while aspartate was only increased by 92% compared to wild-type plants. Meanwhile, Arabidopsis ibi1 mutant showed 100% increase in aspartate (Luna et al., 2014). Rice osers1 mutant anthers contain 144% more glutamate, 76% more aspartate, and 168% more histidine (Yang et al., 2018). According to the aspartate metabolic pathway, aspartate aminotransferase converts aspartate to glutamate and oxaloacetate, then glutamine synthetase converts glutamate to glutamine, and finally asparagine synthetase converts glutamine and aspartate to asparagine (Figure 11). Consistently, quantitative proteomics analysis demonstrated the up-regulation of the above enzymes with asparagine synthetase (ASN1) being most elevated. A previous study indicated that asparagine synthesis is mainly dependent on ASN1 (Ohashi et al., 2015; Luo et al., 2018). As the free amino acids were restored to normal levels in the ylc3 complementation lines, the over-accumulation of glutamine and asparagine under low temperature condition represents a consequence of ylc3 mutation. Consistently, the mutant showed normal levels of free amino acids under high temperature condition.
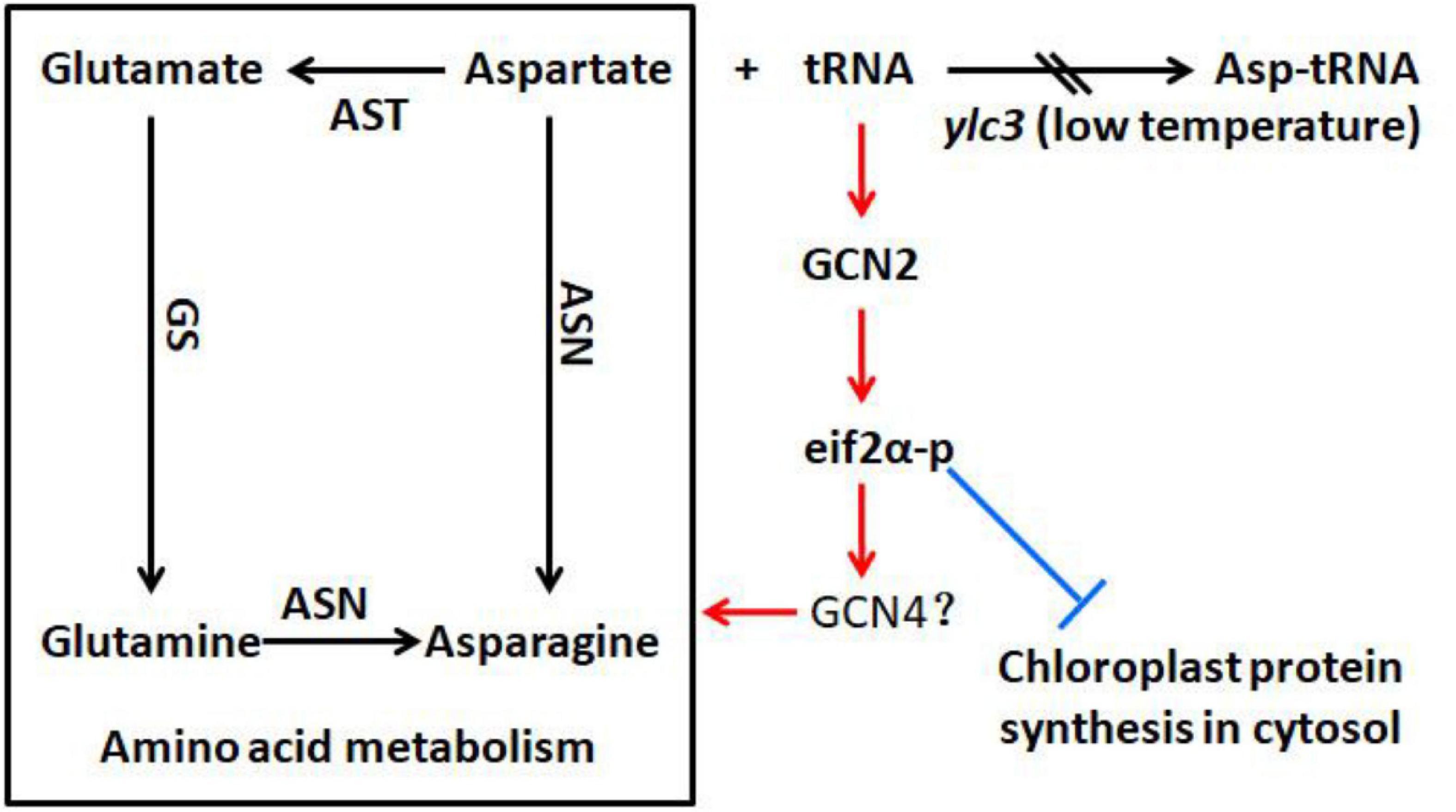
Figure 11. Possible involvement of YLC3 in amino acid homeostasis and chloroplast development. Loss of YLC3 function leads to accumulation of uncharged tRNAs, which promote GCN2-dependent phosphorylation of eIF2α (eIF2α-p), thereby up-regulating amino acid synthesis and inhibiting chloroplast development. Consequently, excess free aspartate is converted to glutamine and asparagine for storage. Positive and negative regulatory effects are indicated in red and blue colors, respectively. ASN, asparagine synthetase; AST, aspartate aminotransferase; GS, glutamine synthetase.
Apparently, the ylc3 mutation resulted in functional deficiency of the encoded aspartyl-tRNA synthetase under low temperature condition. Consequently, aspartate and uncharged tRNA-ASP levels would be elevated (Figure 10A). We speculate that most of the excess aspartate had been converted to glutamine and asparagine which are the major storage form of organic nitrogen, some of them could be stored in xylem and phloem sap (Urquhart and Joy, 1981; Lea et al., 2007; Gaufichon et al., 2010, 2016). On the other hand, the conversion may relieve the inhibitory effects of high concentrations of aspartate (Schultz et al., 1998). Meanwhile, it is unknown whether over-accumulation of asparagine and glutamine may affect chloroplast development. Functional investigation of aspartyl-tRNA synthetases in different plant species may reveal whether there are conserved mechanisms for regulating aspartate metabolism and whether there are relationships between changes in free amino acid levels and chloroplast development.
Differential Regulation of Protein Abundances in ylc3 Mutant
During amino acid starvation, the yeast GCN2 kinase inhibits protein translation and activates amino acid biosynthesis (Natarajan et al., 2001; Dever and Hinnebusch, 2005). This mechanism is at least partially conserved in the Arabidopsis homologous protein (Li et al., 2013; Luna et al., 2014). Amino acid starvation or AARS deficiencies could result in accumulation of uncharged tRNA. AtGCN2 interacts with uncharged tRNA and becomes activated, thereby phosphorylating the translation initiation factor AtEIF2α and leading to inhibition of protein translation (Luna et al., 2014). Under low temperature condition, aspartate accumulation in the ylc3 mutant could lead to uncharged tRNA accumulation (Figure 10A). Hence, OsGCN2 would become activated to phosphorylate eif2α, leading to inhibition of protein translation (Figure 10B). In fact, there was no a GCN4 homolog in Arabidopsis (Halford et al., 2004; Halford, 2006). It is unclear which OsGCN2-regulated proteins are involved in the adaptation to amino acid starvation in rice. Under low temperature condition, the up-regulation of enzymes involved in amino acid biosynthesis and related pathways, such as those for pyruvate metabolism in ylc3 mutant, is consistent with the transcriptional up-regulation of amino acid biosynthesis in yeast during amino acid starvation (Dever and Hinnebusch, 2005; Luna et al., 2014). Furthermore, up-regulation of aspartate metabolic enzymes could convert some of the excess aspartate to asparagine and glutamine for storage, implicating a precise regulating mechanism during over-accumulation of aspartate. There was no significant down-regulation of chlorophyll biosynthesis enzymes, suggesting that the reduced chlorophyll content in the ylc3 mutant was not due to their inhibition. On the other hand, thylakoid complex proteins are largely down-regulated in ylc3 mutant under low temperature condition.
Translation of cytosolic mRNA is regulated at both global and mRNA-specific levels. For example, uncharged tRNAs-ASP accumulated and activated AtGCN2 in the Arabidopsis ibi1 mutant (Li et al., 2013; Luna et al., 2014). Recently, inhibition of ribosome loading by activated GCN2 kinase was demonstrated for mRNAs functionally involved in mitochondrial ATP synthesis, chloroplast thylakoids, vesicle trafficking, and translation (Lokdarshi et al., 2020). In our study, the uncharged tRNAs-ASP level was increased and the translation initiation factor eIF2α was phosphorylated in ylc3 seedlings under low temperature condition (Figure 10). Quantitative proteomics data also indicated that cytosolic mRNA translation of thylakoid proteins was suppressed specifically in the mutant. Overall, our results suggested that YLC3 deficiency could promote the GCN2-eif2α phosphorylation and impaired chloroplast development by suppressing cytosolic mRNA translation in rice (Figure 11). Further investigations are necessary in order to fully understand the functions of YLC3 and other aspartyl-tRNA synthetases in rice under different stress conditions.
Materials and Methods
Plant Materials and Growth Conditions
The rice (Oryza sativa ssp. japonica cv. Nipponbare) ylc3 mutant was isolated from an EMS-induced mutant population. Wild-type and ylc3 mutant plants were grown in Kimura nutrient solution as described previously (Chen et al., 2013). They were kept in a light growth chamber (Panasonic MLR-352H-PC) with a 12 h-light/12 h-dark cycle at 70% relative humidity. Temperatures were set according to each specific treatment.
Genetic Analysis and Construction of F2 Mapping Populations
The ylc3 mutant was individually crossed with Nipponbare or Kasalath rice to generate the F1 progenies which were self-pollinated to obtain the F2 population. Using the F1 and F2 populations, genetic analysis and preliminary mapping were performed. From the F2 population of the ylc3 × Kasalath cross, 22 mutant plants were selected for preliminary mapping. The F2 mutant number was increased to 94 during fine mapping. At the same time, genome sequencing and gene cloning were performed using the F2 population from the ylc3 × Nipponbare cross (Abe et al., 2012).
TEM Analysis of Chloroplast Structures and GUS Staining of Rice Tissues
Fifteen-day-old wild-type and ylc3 seedlings kept in a light growth chamber at 19°C were used for chloroplast ultra-structural analysis. Leaves were cut into 2-mm sections and fixed using 2.5% glutaraldehyde in cacodylate buffer, following by secondary fixation in OsO4. The fixed tissues were dehydrated by ethanol, embedded in epoxy resin, and sectioned for examination under an Hitachi H7650 TEM electron microscope. GUS staining was performed according to Jefferson et al. (1987). Roots, stems, and leaves from 7-day-old seedlings grown at 26°C in a light growth chamber as well as stems, leaves, and panicles from heading stage of mature plants grown in paddy field were collected. Stems and leaves were sliced into 2-mm sections and placed in GUS staining solution for vacuum infiltration (5–10 times) until the samples were completely submerged. After staining at 37°C for 8 h, the tissues were decolored using 70% ethanol and then observed under a Nikon SMZ1000 stereomicroscope.
Complementation Analysis and Gene Editing Vector Construction
A complementation construct in the pCambia1300 binary vector harboring the YLC3 3.388-kb upstream sequence, the YLC3 1.846-kb cDNA containing the full coding region, and the NOS-3 terminator was generated and named as pCAMBIA1300-PR-YLC3-NOS. It was transformed into Agrobacterium tumefaciens EHA105 which was used to infect ylc3 calluses for 3 days. Afterwards, the calluses were selected on hygromycin plates, followed by differentiation and tissue regeneration (Lee et al., 1999). For construction of the base editing vector, a 19-bp target-specific oligonucleotide initialized by “G” and a 5-bp adaptor were synthesized, annealed and ligated to a BsaI-digested CBEmax-NGG vector (Wang et al., 2019). The constructed vector was confirmed by Sanger sequencing and used for Agrobacterium-mediated transformation of rice calluses. For the construction of YLC3 promoter-Gus vector, the 3.388-kb promoter region of YLC3 was fused with the GUS gene in the modified pCAMBIA1300-GUS vector. All primers used for constructing vectors are listed in Supplementary Table 3.
Subcellular Localization
The YLC3 coding sequence without a stop codon and fused in-frame with sGFP was cloned into the pCAMBIA1301-35S-NOS vector and transiently expressed in rice protoplasts. Rice seedlings cultured on MS media for 10 days were digested with cellulases for protoplast preparation as described previously (Zhang et al., 2011). Protoplasts (100 μl) were transfected with the vector (5–10 μg) and dark-incubated at room temperature, following by examination of green fluorescence signals under a Zeiss confocal laser scanning microscope. For mitochondria co-localization, transfected protoplasts were stained using a mitochondria fluorescent dye (Mitotracker, Invitrogen, Carlsbad, CA, United States).
Phylogenetic Analysis and KEGG Analysis
A phylogenetic tree was constructed using aligned full-length sequences of homologs of YLC3. MEGA (version 10.1.7) (Kumar et al., 2018) and the neighbor-joining methods were used with a p-distance model, pairwise deletion and bootstrap (1,000 replicates). The maximum parsimony method of MEGA also was used to support the neighbor-joining tree using the default parameters. Amino acid sequences from regions 101 to 188 and 223 to 544 in YLC3 were used for motif alignment by MEGA. For GO and KEGG analysis, the differentiated expressed proteins were enriched with rice pathways and GO terms using clusterProfiler (Wu et al., 2021) and org.Osativa.eg.db. (Xu, 2019). The filtering criteria of p value Cutoff 0.05 and qvalueCutoff 0.1 were used.
Measurements of Free Amino Acid Contents
Free amino acid contents in rice leaves were measured using an Hitachi LA8080 automatic amino acid analyzer. Leaves (0.05 g) were placed in 2-ml centrifuge tubes, followed by addition of 1 ml 4% sulfosalicylic acid and two zirconia beads. Bead-beating was performed for 1 h at 1 time per 2 min until the samples became slurry. The samples were allowed to settle for 1 h, followed by centrifugation at 4°C for 10 min. Supernatant (500 μl) was taken, mixed with 500 μl 0.2 M HCl, and filtered through a 0.22 μl millipore filter. Finally, 20 μl sample was used for free amino acid analysis.
Quantitative Proteomics Analysis of Rice Seedlings
Protein Isolation
Crude proteins from rice seedling tissues were extracted by the modified phenol-methanol method as described (Deng et al., 2007). Extracted proteins were dissolved in lysis buffer [8 M urea, 50 mM triethylammonium bicarbonate (TEAB), pH 8.0] and quantified using a 2-D Quant kit (GE Healthcare, Piscataway, NJ, United States) with bovine serum albumin as a standard.
Tryptic Digestion, Peptide Labeling and Fractionation, LC-MS/MS Analysis
Tryptic digestion, TMT labeling, peptide fractionation and LC-MS/MS were performed as described (Zhu et al., 2022) unless otherwise stated. After tris(2carboxyethyl)phosphin (TCEP) and dithiothreitol (DTT) treatments, proteins were precipitated and dissolved in 50 mM TEAB buffer. Each sample (25 μg) was mixed with its respective 6-plex TMT reagent and incubated for 1 h at room temperature. Three biological replicates were labeled for each sample group, in which the Nipponbare samples were labeled with TMT reagents 126, 127, and 128, while ylc3 samples were labeled with TMT reagents 129, 130, and 131, respectively. The labeling reactions were stopped by addition of hydroxylamine, then combined and dried by vacuum. The combined multiplexed TMT-labeled samples were separated on a Waters Acquity BEH C18 1.7 μm 2.1–100 mm column using H class UPLC system (Waters, Milford, MA, United States) at a flow rate of 300 μl/min. A total of 24 fractions were collected, then combined into 12 fractions and vacuum dried for LC-MS/MS analysis.
TMT-labeled samples were analyzed on an Ultimate 3000 nano UHPLC system (Thermo Scientific, Waltham, MA, United States) coupled online to a Q Exactive HF mass spectrometer (Thermo Scientific, Waltham, MA, United States) equipped with a Nanospray Flex Ion Source (Thermo Scientific, Waltham, MA, United States). Samples were separated by a binary buffer system of buffer A (0.1% formic acid) and buffer B (80% acetonitrile plus 0.1% formic acid). Peptides were eluted in a gradient of 5–8% solvent B in 2 min, 8–20% solvent B in 66 min, 20–40% solvent B in 33 min, and 40–90% solvent B in 4 min.
TMT Data Analysis
Raw data were processed using Proteome Discoverer 2.4.0.305 (Thermo Scientific, Waltham, MA, United States) with the SEQUEST HT search engine searching against a rice proteome database (Rice Genome Annotation Project1, version 7.0, total 66,338 entries). Searches were configured with static modifications for the TMT reagents on lysine and N-termini, carbamidomethyl on cysteine, dynamic modifications for oxidation of methionine and acetylation of protein N-termini, precursor mass tolerance of 10 ppm, fragment mass tolerance of 0.02 Da, and trypsin cleavage (max 2 missed cleavages). Searches used a reversed sequence decoy strategy to control peptide false discovery and identifications were validated by Percolator software. Protein groups, peptide groups and PSMs were accepted at a false discovery rate (FDR) < 1%. Normalization was applied for the grand total reporter ion intensity for each channel within the 6-plex experiment. Further downstream analysis of the results was performed in the R scripting and statistical environment, using the limma package from Bioconductor2. The basic statistics used for significance analysis is the moderated t-statistics. Significantly expressed proteins were filtered for an average fold-change > 1.3 or <0.77, with p-values adjusted for multiple testing correction by FDR (Benjamini–Hochberg).
RNA Isolation and Northern-Blot Analysis
Total RNAs were extracted from 15-day-old seedlings grown at different temperatures using TRIzol reagent (Invitrogen, Carlsbad, CA, United States). RNA was precipitated by ethanol overnight at −20°C. Northern blot was performed as previously described (Huang et al., 2018). The sequences of biotinylated oligomer probes were listed as follows: trnD-GUC (Id: 29141347), 5′-TTGTAGTTCAATTGGTCAGAGCACC-3′; trnD(GTC) (Id: 3950710) 5′-GAAATAGCTCAGTTGGTTAGAGTG-3′.
Total Protein Extraction and Immunoblot Analysis
The first leaves were collected from WT and ylc3 mutant seedlings grown at 19 or 30°C for immunoblot analysis. Total proteins were extracted using extraction buffer (containing 25 mM Tris-HCl [pH 7.5], 10 mM NaCl, 4 mM PMSF, 20 mM MG132, and protease inhibitor cocktail). Extracted proteins were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblotting. Immunoblotting was performed using a monoclonal antibody of phospho-eIF2α (Ser51) (Catalogue no. 9721, Cell Signalling, Danvers, MA, United States 1/1000 dilution) or a monoclonal antibody of Anti Plant Actin Mouse (Abbkine, A01050). Signals were detected using the Immobilon kit (Catalogue no WBKLS0500, Millipore) under standard conditions.
Data Availability Statement
The proteomics data have been deposited to the ProteomeXchange Consortium. Link: https://www.iprox.cn/page/project.html?id=IPX0004037000; ProteomeXchange ID:PXD031497. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (Ma et al., 2019) with the dataset identifier PXD031497.
Author Contributions
HL and CL designed the experiments. HL, XG, HD, JT, FW, WW, ZZ, RX, and HH performed the experiments. YS, JT, ZZ, HL, and CL analyzed the data. HL and CL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31571633).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Hui Zhang (Shanghai Normal University) for construction of the YLC3 gene editing vector, Yu Liu (Zhejiang University) for measurement of free amino acid contents.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.847364/full#supplementary-material
Supplementary Figure 1 | GUS staining of 7-day-old ProYLC3::GUS transgenic seedlings grown at 19°C. Two independent ProYLC3::GUS transgenic lines (pr-1 and pr-4) was used to gus staining test. Bar = 1 cm.
Supplementary Figure 2 | Transcriptional expression of YLC3 was performed by Real-time PCR. Gene-specific primers is list below: YLC3-qpF, 5′-CTCCCTCAGC AAGGAATCAA-3′; YLC3-qpR, 5′-CACCTGAATCTCCACCTGCT-3′; Actin-qpF, 5′-GTGTGACAATGGAACTGGCA-3′; Actin-qpR 5′-CCACGATACTAGGGAAAAC AGC-3′.
Supplementary Figure 3 | Subcellular localization analysis in YLC3::GFP transgenic rice. Two independent YLC3::GFP transgenic lines cultured on MS media for 10 days were digested with cellulases for protoplast preparation as described previously (Zhang et al., 2011). Green fluorescence signals was observed and captured by a Zeiss confocal laser scanning microscope. For mitochondria co-localization, protoplasts were stained in a mitochondria fluorescent dye (Mitotracker, Invitrogen, Carlsbad, CA, United States).
Supplementary Table 1 | 9,212 proteins from quantitative proteomics analysis.
Supplementary Table 2 | 4,979 proteins from KEGG analysis.
Supplementary Table 3 | Vector construction primers and indel primers.
Footnotes
References
Abdallah, F., Salamini, F., and Leister, D. (2000). A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 5, 141–142. doi: 10.1016/s1360-1385(00)01574-0
Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178. doi: 10.1038/nbt.2095
Berg, M., Rogers, R., Muralla, R., and Meinke, D. (2005). Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J. 44, 866–878. doi: 10.1111/j.1365-313X.2005.02580.x
Bröer, S., and Bröer, A. (2017). Amino acid homeostasis and signalling in mammalian cells and organisms. Biochem. J. 474, 1935–1963. doi: 10.1042/BCJ20160822
Chen, X., Shi, J., Hao, X., Liu, H., Shi, J., Wu, Y., et al. (2013). OsORC3 is required for lateral root development in rice. Plant J. 74, 339–350. doi: 10.1111/tpj.12126
Deng, Z., Zhang, X., Tang, W., Oses-Prieto, J. A., Suzuki, N., Gendron, J. M., et al. (2007). A proteomics study of brassinosteroid response in Arabidopsis. Mol. Cell Proteomics 6, 2058–2071. doi: 10.1074/mcp.M700123-MCP200
Dever, T. E., Feng, L., Wek, R. C., Cigan, A. M., Donahue, T. F., and Hinnebusch, A. G. (1992). Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68, 585–596. doi: 10.1016/0092-8674(92)90193-g
Dever, T. E., and Hinnebusch, A. G. (2005). GCN2 whets the appetite for amino acids. Mol Cell 18, 141–142. doi: 10.1016/j.molcel.2005.03.023
Duchêne, A. M., Giritch, A., Hoffmann, B., Cognat, V., Lancelin, D., Peeters, N. M., et al. (2005). Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 102, 16484–16489. doi: 10.1073/pnas.0504682102
Dyall, S. D., Brown, M. T., and Johnson, P. J. (2004). Ancient invasions: from endosymbionts to organelles. Science 304, 253–257. doi: 10.1126/science.1094884
Fang, G., Yang, S., Ruan, B., Liu, C., Zhang, A., Jiang, H., et al. (2020). Isolation of TSCD11 Gene for Early Chloroplast Development under High Temperature in Rice. Rice 13:49. doi: 10.1186/s12284-020-00411-6
Fernández, A. P., and Strand, A. (2008). Retrograde signaling and plant stress: plastid signals initiate cellular stress responses. Curr. Opin. Plant Biol. 11, 509–513. doi: 10.1016/j.pbi.2008.06.002
Gaufichon, L., Reisdorf-Cren, M., Rothstein, S. J., Chardon, F., and Suzukia, A. (2010). Biological functions of asparagine synthetase in plants. Plant Sci. 179, 141–153.
Gaufichon, L., Rothstein, S. J., and Suzuki, A. (2016). Asparagine Metabolic Pathways in Arabidopsis. Plant Cell Physiol. 57, 675–689. doi: 10.1093/pcp/pcv184
Halford, N. G. (2006). Regulation of carbon and amino acid metabolism, roles of sucrose nonfermenting-1-related protein kinase-1 and general control nonderepressible-2-related protein kinase. Adv. Bot. Res. 43, 93–142.
Halford, N. G., Hey, S., Jhurreea, J., Laurie, S., McKibbin, R. S., and Zhang, Y. (2004). Highly conserved protein kinases involved in the regulation of carbon and amino acid metabolism. J. Exp. Bot. 55, 35–42. doi: 10.1093/jxb/erh019
Hinnebusch, A. G. (2005). Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450. doi: 10.1146/annurev.micro.59.031805.133833
Huang, W., Zhu, Y., Wu, W., Li, X., Zhang, D., and Yin, P. (2018). The Pentatricopeptide Repeat Protein SOT5/EMB2279 Is Required for Plastid rpl2 and trnK Intron Splicing. Plant Physiol. 177, 684–697. doi: 10.1104/pp.18.00406
Ibba, M., and Soll, D. (2000). Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650. doi: 10.1146/annurev.biochem.69.1.617
Jarvis, P., and López-Juez, E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. doi: 10.1038/nrm3702
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Kägi, C., Baumann, N., Nielsen, N., Stierhof, Y. D., and Gross-Hardt, R. (2010). The gametic central cell of Arabidopsis determines the lifespan of adjacent accessory cells. Proc. Natl. Acad. Sci. U. S. A. 107, 22350–22355. doi: 10.1073/pnas.1012795108
Kim, Y. K., Lee, J. Y., Cho, H. S., Lee, S. S., Ha, H. J., Kim, S., et al. (2005). Inactivation of organellar glutamyl- and seryl-tRNA synthetases leads to developmental arrest of chloroplasts and mitochondria in higher plants. J. Biol. Chem. 280, 37098–37106. doi: 10.1074/jbc.M504805200
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lageix, S., Lanet, E., Pelissier, M. N., Espagnol, M. C., Robaglia, C., Deragon, J. M., et al. (2008). Arabidopsis eIF2alpha kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 8:134. doi: 10.1186/1471-2229-8-134
Lea, P. J., Sodek, L., Parry, M. A. J., Shewry, P. R., and Halford, N. G. (2007). Asparagine in plants. Ann. Appl. Biol. 150, 1–26.
Lee, S., Jeon, J. S., Jung, K. H., and An, G. (1999). Binary vectors for efficient transformation of rice. J. Plant Biol. 42, 310–316.
Li, H., Ji, G., Wang, Y., Qian, Q., Xu, J., and Sodmergen, et al. (2018). WHITE PANICLE3, a Novel Nucleus-Encoded Mitochondrial Protein, Is Essential for Proper Development and Maintenance of Chloroplasts and Mitochondria in Rice. Front. Plant Sci. 9:762. doi: 10.3389/fpls.2018.00762
Li, M. W., AuYeung, W. K., and Lam, H. M. (2013). The GCN2 homologue in Arabidopsis thaliana interacts with uncharged tRNA and uses Arabidopsis eIF2α molecules as direct substrates. Plant Biol. 15, 13–18. doi: 10.1111/j.1438-8677.2012.00606.x
Li, M. W., and Lam, H. M. (2008). Searching for nitrogen sensing systems in higher plants. Dyn. Soil Dyn. Plant 2, 13–22.
Liu, W., Fu, Y., Hu, G., Si, H., Zhu, L., Wu, C., et al. (2007). Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.). Planta 226, 785–795. doi: 10.1007/s00425-007-0525-z
Lokdarshi, A., Guan, J., Urquidi Camacho, R. A., Cho, S. K., Morgan, P. W., Leonard, M., et al. (2020). Light Activates the Translational Regulatory Kinase GCN2 via Reactive Oxygen Species Emanating from the Chloroplast. Plant Cell 32, 1161–1178. doi: 10.1105/tpc.19.00751
Lü, Y., Cui, X., Li, R., Huang, P., Zong, J., Yao, D., et al. (2015). Development of genome-wide insertion/deletion markers in rice based on graphic pipeline platform. J. Integr. Plant Biol. 57, 980–991. doi: 10.1111/jipb.12354
Luna, E., Van Hulten, M., Zhang, Y., Berkowitz, O., López, A., Pétriacq, P., et al. (2014). Plant perception of beta-aminobutyric acid is mediated by an aspartyl-tRNA synthetase. Nat. Chem. Biol. 10, 450–456. doi: 10.1038/nchembio.1520
Luo, L., Qin, R., Liu, T., Yu, M., Yang, T., and Xu, G. (2018). OsASN1 Plays a Critical Role in Asparagine-Dependent Rice Development. Int. J. Mol. Sci. 20:130. doi: 10.3390/ijms20010130
Ma, J., Chen, T., Wu, S., Yang, C., and Bai, M. Shu K., et al. (2019). iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217. doi: 10.1093/nar/gky869
Marton, M. J., Crouch, D., and Hinnebusch, A. G. (1993). GCN1, a translational activator of GCN4 in Saccharomyces cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell Biol. 13, 3541–3556. doi: 10.1128/mcb.13.6.3541-3556.1993
Natarajan, K., Meyer, M. R., Jackson, B. M., Slade, D., Roberts, C., Hinnebusch, A. G., et al. (2001). Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21, 4347–4368. doi: 10.1128/MCB.21.13.4347-4368
O’Donoghue, P., and Luthey-Schulten, Z. (2003). On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol. Mol. Biol. Rev. 67, 550–573. doi: 10.1128/MMBR.67.4.550-573
Ohashi, M., Ishiyama, K., Kojima, S., Konishi, N., Nakano, K., Kanno, K., et al. (2015). Asparagine synthetase1, but not asparagine synthetase2, is responsible for the biosynthesis of asparagine following the supply of ammonium to rice roots. Plant Cell Physiol. 56, 769–778. doi: 10.1093/pcp/pcv005
Richardson, L. G. L., Singhal, R., and Schnell, D. J. (2017). The integration of chloroplast protein targeting with plant developmental and stress responses. BMC Biol. 15:118. doi: 10.1186/s12915-017-0458-3
Sattlegger, E., and Hinnebusch, A. G. (2005). Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2{alpha} kinase GCN2 during amino acid starvation. J. Biol. Chem. 280, 16514–16521. doi: 10.1074/jbc.M414566200
Schultz, C. J., Hsu, M., Miesak, B., and Coruzzi, G. M. (1998). Arabidopsis mutants define an in vivo role for isoenzymes of aspartate aminotransferase in plant nitrogen assimilation. Genetics 149, 491–499. doi: 10.1093/genetics/149.2.491
Schwarzenbacher, R. E., Wardell, G., Stassen, J., Guest, E., Zhang, P., Luna, E., et al. (2020). The IBI1 Receptor of β-Aminobutyric Acid Interacts with VOZ Transcription Factors to Regulate Abscisic Acid Signaling and Callose-Associated Defense. Mol. Plant 13, 1455–1469. doi: 10.1016/j.molp.2020.07.010
Urquhart, A. A., and Joy, K. W. (1981). Use of Phloem exudate technique in the study of amino Acid transport in pea plants. Plant Physiol. 68, 750–754. doi: 10.1104/pp.68.3.750
Wang, L., Li, H., Zhao, C., Li, S., Kong, L., Wu, W., et al. (2017). The inhibition of protein translation mediated by AtGCN1 is essential for cold tolerance in Arabidopsis thaliana. Plant Cell Environ. 40, 56–68. doi: 10.1111/pce.12826
Wang, M., Wang, Z., Mao, Y., Lu, Y., Yang, R., and Tao, X. (2019). Optimizing base editors for improved efficiency and expanded editing scope in rice. Plant Biotechnol. J. 17, 1697–1699. doi: 10.1111/pbi.13124
Wang, Y., Wang, C., Zheng, M., Lyu, J., Xu, Y., Li, X., et al. (2016). WHITE PANICLE1, a Val-tRNA Synthetase Regulating Chloroplast Ribosome Biogenesis in Rice, Is Essential for Early Chloroplast Development. Plant Physiol. 170, 2110–2123. doi: 10.1104/pp.15.01949
Wek, R. C., Jiang, H. Y., and Anthony, T. G. (2006). Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11. doi: 10.1042/BST20060007
Woodson, J. D., and Chory, J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395. doi: 10.1038/nrg2348
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2:100141. doi: 10.1016/j.xinn.2021.100141
Xu, Z. (2019). xuzhougeng/org.Osativa.eg.db v0.01 (Version v0.01). Geneva: Zenodo. doi: 10.5281/zenodo.3374105
Yang, X., Li, G., Tian, Y., Song, Y., Liang, W., and Zhang, D. (2018). A Rice Glutamyl-tRNA Synthetase Modulates Early Anther Cell Division and Patterning. Plant Physiol. 177, 728–744. doi: 10.1104/pp.18.00110
Zhang, Y., Dickinson, J. R., Paul, M. J., and Halford, N. G. (2003). Molecular cloning of an Arabidopsis homologue of GCN2, a protein kinase involved in coordinated response to amino acid starvation. Planta 217, 668–675. doi: 10.1007/s00425-003-1025-4
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30. doi: 10.1186/1746-4811-7-30
Zhang, Y., Wang, Y., Kanyuka, K., Parry, M. A., Powers, S. J., and Halford, N. G. (2008). GCN2-dependent phosphorylation of eukaryotic translation initiation factor-2alpha in Arabidopsis. J. Exp. Bot. 59, 3131–3141. doi: 10.1093/jxb/ern169
Zhang, Y. Y., Hao, Y. Y., Wang, Y. H., Wang, C. M., Wang, Y. L., Long, W. H., et al. (2017). Lethal albinic seedling, encoding a threonyl-tRNA synthetase, is involved in development of plastid protein synthesis system in rice. Plant Cell Rep. 36, 1053–1064. doi: 10.1007/s00299-017-2136-x
Keywords: rice, aspartyl-tRNA synthetase, chloroplast, amino acid metabolism, eiF2α
Citation: Liu H, Gong X, Deng H, Tan J, Sun Y, Wang F, Wu W, Zhou Z, Xu R, He H and Lo C (2022) The Rice Aspartyl-tRNA Synthetase YLC3 Regulates Amino Acid Homeostasis and Chloroplast Development Under Low Temperature. Front. Plant Sci. 13:847364. doi: 10.3389/fpls.2022.847364
Received: 02 January 2022; Accepted: 31 January 2022;
Published: 04 March 2022.
Edited by:
Shaojun Dai, Shanghai Normal University, ChinaCopyright © 2022 Liu, Gong, Deng, Tan, Sun, Wang, Wu, Zhou, Xu, He and Lo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongjia Liu, bGhqemp1QGFsaXl1bi5jb20=
 Hongjia Liu
Hongjia Liu Xue Gong
Xue Gong Hui Deng
Hui Deng Jinjuan Tan
Jinjuan Tan Yanqing Sun
Yanqing Sun Fang Wang
Fang Wang Wenjuan Wu4
Wenjuan Wu4 Zhongjing Zhou
Zhongjing Zhou Clive Lo
Clive Lo