
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 07 September 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.847166
This article is part of the Research TopicAbiotic Stress: Molecular Genetics and Genomics, Volume IIView all 32 articles
Legume plants produce one-third of the total yield of primary crops and are important food sources for both humans and animals worldwide. Frequent exposure to abiotic stresses, such as drought, salt, and cold, greatly limits the production of legume crops. Several morphological, physiological, and molecular studies have been conducted to characterize the response and adaptation mechanism to abiotic stresses. The tolerant mechanisms of the model legume plant Medicago truncatula to abiotic stresses have been extensively studied. Although many potential genes and integrated networks underlying the M. truncatula in responding to abiotic stresses have been identified and described, a comprehensive summary of the tolerant mechanism is lacking. In this review, we provide a comprehensive summary of the adaptive mechanism by which M. truncatula responds to drought, salt, and cold stress. We also discuss future research that need to be explored to improve the abiotic tolerance of legume plants.
The climate change products a series of environmental factors which show negative effects to plants (Farooq et al., 2022). Among the environmental constraints, drought, salt, and cold are the main abiotic stresses that influence plants’ physiological and biochemical processes, ultimately reducing crop production (Rhaman et al., 2021; Farooq et al., 2022). Up to 45% of the world’s farmland faces frequent water scarcity (Rhaman et al., 2021), and 20–50% of irrigated lands are affected by salinity (Munns and Tester, 2008). Approximately 57 and 26% of the world’s land and rural areas are affected by cold stress, respectively (Cramer et al., 2011). Much work has been devoted to explore the mechanism by which plants respond and adapt to abiotic stresses, and these findings have meaningful implications for improving crop production.
Legume plants are particularly important sources of food for both humans and animals. Abiotic stresses affect their growth and development. There is an eager need to clarify the mechanism by which legumes respond to abiotic stresses, and such research will aid the breeding of climate-resilient varieties. The legume model plant M. truncatula has small genome, short life cycle, self-pollination ability, and high genetic transformation efficiency (Tang et al., 2014). So, M. truncatula has been widely used in genomic, genetic, and physiological studies. Many studies focus on elucidating the mechanism by which M. truncatula responds and adapts to abiotic stresses. In this review, we summarize the general morphological, physiological, and molecular features by which M. truncatula responds and adapts to drought, salt, and cold stress. We also incorporate the crosstalk between different abiotic stresses, and discuss the implications for breeding stress-tolerant legume crops.
Drought stress significantly reduces leaf water potential and stomatal closure of M. truncatula plants, resulting in reduced photosynthesis, which in turn restricts plants’ growth (Nunes et al., 2008; Luo et al., 2016). Drought also causes photooxidative damage to thylakoid membranes and reduces chlorophyll content and photosystem II activity (Luo et al., 2016). To cope with drought stress, M. truncatula plants have evolved various responses such as alterations in tissue architectures and expression patterns of functional genes (Figure 1). Indeed, 5-week-old M. truncatula R108 plants are still recoverable after withholding irrigation for 12 days (Luo et al., 2016).
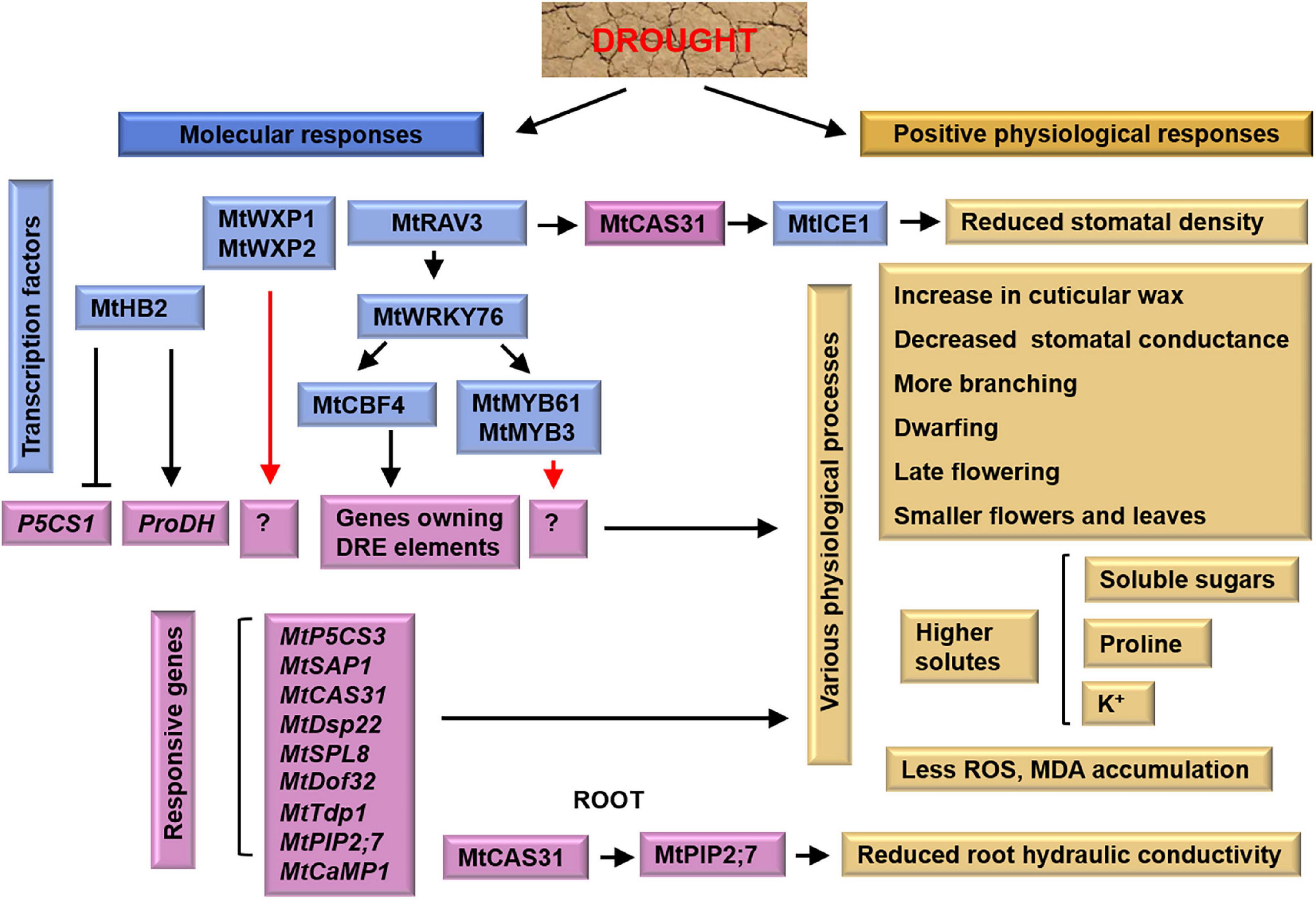
Figure 1. Tolerant mechanism of M. truncatula to drought stress. In shoots, MtRAV3 induces the expression of MtCAS31, whose encoded protein binds with MtICE1, resulting in reduced stomatal density. In roots, MtCAS31 facilitates the autophagic degradation of aquaporin MtPIP2; 7, and reduces root hydraulic conductivity. Blue boxes and purple boxes indicate transcription factors and responsive genes, respectively. Yellow boxes indicate physiological responses of M. truncatula. The black arrows represent the targets, the red arrows represent the unknown factors and the cross-lines mean suppression.
Plants’ shoot architecture is affected by drought stress and can be used as an indicator of drought adaptation (Farooq et al., 2009; Nguyen et al., 2013). Branching is a key determinant of shoot architecture. In Arabidopsis, AtSPL9 (squamosa promoter binding protein-like) controls the initiation of cauline leaf axillary meristems (Tian et al., 2014). In transgenic M. truncatula plants overexpressing MtSPL8 inhibits branching by directly suppressing axillary bud formation (Gou et al., 2018). Down-regulation MsSPL8 increases branch density and enhances drought and salt tolerance in transgenic alfalfa (Gou et al., 2018). Interestingly, M. truncatula plants with up-regulated MtRAV3 (related to ABI3/VP1 transcription factor) have the similar tolerance to drought and salt stress, but exhibiting dwarfing, late flowering, and smaller leaves (Wang S. et al., 2021). Thus, MtRAV3 and MtSPL8 might play different roles in morphological development. However, Guo et al. (2021) find that overexpression of MtDof32 (DNA-binding one zinc finger 32) in Arabidopsis results in reduced branches and enlarged leaves, but still have enhanced osmotic and salt tolerance. Although both MtDof32 and MtRAV3 enhance drought and salt tolerance in transgenic plants, they regulate different morphological development. Indeed, MtDof32 enhances plants’ stress tolerance by regulating the rosette numbers. Thus, how to improve the shoot structure under drought condition might be a useful way to improve crops’ drought tolerance.
In addition, regulation of stomatal conductance (Nunes et al., 2008) and epidermal wax accumulation (Zhang et al., 2005) is an effective way for M. truncatula plants to cope with drought stress. Furthermore, osmotic and oxidative regulation are also essential in the response of M. truncatula to drought stress (Yousfi et al., 2010; Luo et al., 2016). For instance, M. truncatula populations adapt to drought tolerance by accumulating solutes such as proline, soluble sugars, and K+ (Yousfi et al., 2010). The M. truncatula lines overexpressing the oat arginine decarboxylase gene produce more soluble polyamines (PAs), resulting in greater drought tolerance compared to wild-type plants (Duque et al., 2016). The PAs including spermidine (Spd), spermine (Spm), and putrescine (Put) are involved in plant responses to abiotic stresses (Alcázar et al., 2010). Pagano et al. (2022) found that desiccation induces the expression of Spm synthase MtSPMS and Spd synthase MtSPDS in over-primed seeds. As for the oxidative reactions, M. truncatula plants that accumulate less peroxide and malondialdehyde (MDA) are more tolerant to drought stress (Luo et al., 2016; Wang S. et al., 2021).
Zhang et al. (2014) identified many drought-responsive genes in M. truncatula. The genes MtP5CS (encoding proline synthase) and MtProDH (encoding proline dehydrogenase) regulate proline accumulation coordinately in response to drought stress. Indeed, overexpression of P5CS in M. truncatula results in more proline accumulation and greater drought tolerance (Verdoy et al., 2006). Heterologous expression of calcium-binding protein gene MtCaMP1 in Arabidopsis induces P5CS1 and suppresses ProDH, making transgenic plants more tolerant to drought stress (Wang T. Z. et al., 2013). Whereas, plants with Tnt1 transposon insertion of MtP5CS3 accumulate less proline and are sensitivity to salt and drought stresses (Nguyen et al., 2013). In addition, cold-acclimation specific protein 31 (MtCAS31), a Y2K4-type dehydrin, interacts with AtICE1 (inducer of CBF expression 1) to regulate stomatal development. Overexpression of MtCAS31 in Arabidopsis reduces stomatal density and significantly enhances drought tolerance in transgenic plants (Xie et al., 2012). Li et al. (2018) generate the mtcas31 mutant by transcription activator-like effector nuclease (TALEN) technology, and identify that MtCAS31 interacts with leghemoglobin MtLb120-1 to regulate drought response. Moreover, in response to drought stress, MtCAS31 promotes the autophagic degradation of the aquaporin MtPIP2; 7, thereby reducing water loss and improving drought tolerance (Li et al., 2020). Recently, 39 autophagy−related (ATG) genes are identified in M. truncatula. Most of them are highly induced during seed development and drought stress, indicating that autophagy plays an important role in seed development and responses to drought stress in M. truncatula (Yang et al., 2021).
Plants overexpressing the stress-associated protein genes MtSAP1 accumulate more nitric oxide (NO), which is beneficial to plant growth under osmotic and salt stress (Charrier et al., 2012, 2013). In turn, NO interacts with reactive oxygen species (ROS) to affect the SAPs’ expression (Delledonne et al., 2001; Wendehenne et al., 2004; Qiao and Fan, 2008). Of the 17 MtSAPs, the MtSAP4, 6, 9, 11, 14, and 15 are induced by drought stress (Zhou et al., 2018). In addition, Macovei et al. (2010) find that MtTdp1, a tyrosyl-DNA phosphodiesterase gene, is up-regulated by PEG treatment suggesting a relationship between drought response and DNA repair pathway. While, MtTdp2α positively regulates M. truncatula in salt response due to strong antioxidant effects of transgenic plants (Confalonieri et al., 2019). Recently, Pagano et al. (2022) found that desiccation treatment on over-primed seeds alters rRNA accumulation, promotes signal molecule 3′−phosphoadenosine 5′−phosphate (PAP) production, and up−regulates genes involved in ribogenesis. In addition, early light-inducible proteins (ELIPs) and ELIP-like proteins are pigment-binding components that protect against photooxidative damage (Araújo et al., 2013). Transgenic plants overexpressing the ELIP-like gene CpDsp22 (desiccation stress protein 22 from Craterostigma plantagineum) recover faster from water deficit (Araújo et al., 2013). These results provide insights into NO and nucleic acid organization in response to oxidative stress caused by drought stress in M. truncatula.
Transcription factors (TFs) regulate the transcription of downstream genes by binding to their cis-elements in promoters playing important roles in response to various stresses (Oztur et al., 2002; Porto et al., 2014). For instance, the C-repeat binding factor 4 (MtCBF4), belonging to the APETALA2/EREBP (AP2-EREBP) family, binds to the dehydration responsive (DRE) element of downstream genes to regulate drought response (Li et al., 2011; Table 1). Overexpression of TF MtWRKY76 in M. truncatula promotes the expression of MtCAS31, MtCBF4, MtMYB61, and MtMYB3, and enhances drought tolerance in transgenic plants (Liu et al., 2016). Meanwhile, TF MtRAV3 up-regulates the expression of MtWRKY76, MtMYB61, MtCAS31, MtAOX1, and MtERF1 (Wang S. et al., 2021). In addition, the ethylene response factor (ERF) TFs MtWXP1 and MtWXP2 mediate cuticular wax production. Overexpression of MtWXP1 and MtWXP2 enhances transgenic plants’ drought tolerance (Zhang et al., 2005, 2007). These two wax genes are expected to have great potential for crop improvement through genetic modification. While, TF MtHB2 is a homeodomain leucine zipper (HD-Zip) protein that negatively regulates drought stress by affecting osmotic and oxidative responses (Song et al., 2012). Li et al. (2022) identifies 15 HD-ZIP ? genes in M. truncatula. In particular, MtHB7 and MtHB12 are positively associated with salt, osmotic stress, and abscisic acid (ABA), while MtHB13 and MtHB23 are negatively associated with these stresses. This genome-wide analysis of the HD-ZIP I TFs in M. truncatula provides valuable references for further research.
Plant growth regulators (PGRs) such as auxin, ABA, and ethylene regulate plants in response to abiotic stresses (Rhaman et al., 2021). Both PEG and ABA treatment induces the expression of 9-cis-epoxycarotenoid dioxygenase gene NCED5 leads to increased endogenous ABA content in M. truncatula (Planchet et al., 2011; Luo et al., 2016). Meanwhile, water deficit induces endogenous NO accumulation through an ABA-dependent pathway (Planchet et al., 2014). While, exogenous ABA addition induces asparagine and proline production contributing to osmotic adjustment under water deficit (Planchet et al., 2011). However, the modulation of proline metabolism is independent of NO production under water deficit (Planchet et al., 2014). So, exploring the central role of ABA in water-deficit tolerance could lead us to obtain more information on osmotic adjustment and nitrogen metabolism under adverse conditions.
Salt stress causes osmotic stress, ion toxicity, and oxidative damage to M. truncatula plants, resulting in reduced photosynthesis and biomass (Yousfi et al., 2010; Arraouadi et al., 2012; Luo et al., 2016; Gou et al., 2018; Zhang X. X. et al., 2019; Wang S. et al., 2021). M. truncatula minimizes these damages by regulating the production of osmolytes and antioxidants in cells, the extrusion of Na+ out of cells, and the reduction of Na+ in leaves (Figure 2). In fact, hydroponic M. truncatula R108 can tolerate 100 mM NaCl for nearly 1 week (Merchan et al., 2003; Zhang X. X. et al., 2019;). M. truncatula genotype TN1.11 is the most tolerant to salt stress among R108, Jemalong A17, TN6.18, and TN1.11.
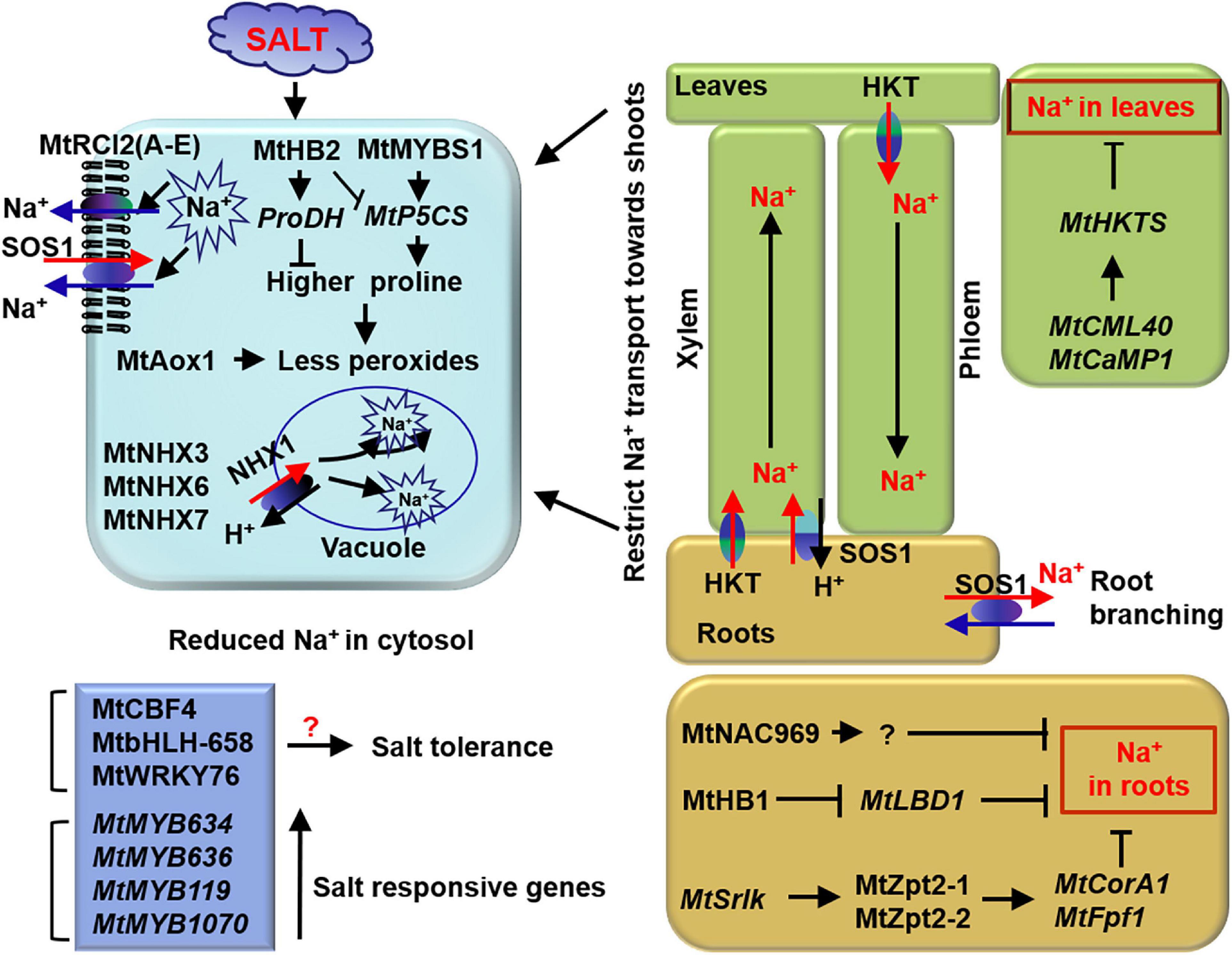
Figure 2. Tolerant mechanism of M. truncatula to salt stress. MtHB2 and MtMYBS1 and oxidase MtAox1 are involved in osmotic regulation. Membrane proteins MtRCI2 (A–E) and Na+/H+ exchangers MtNHX3, MtNHX6, and MtNHX7 play roles in reducing Na+ accumulation in cells. MtNAC969, MtHB1, MtZpt2-1, and MtZpt2-1 play roles in reducing Na+ accumulation in roots. While, Ca2+ sensor MtCML40 and MtCaMP1 participate in Na+ long-distance transportation regulation between roots and shoots. Light blue box represents cells. Blue boxes represent transcription factors related to salt stress. Green and yellow boxes mean shoots and roots, respectively. The black arrows represent the targets or directions, the red arrows represent transport of Na+ and the cross-lines mean suppression.
The root architecture of plants is affected by salt stress. de Zélicourt et al. (2012) find that shorter and less branched roots are beneficial for preventing Na+ uptake in M. truncatula (Ariel et al., 2010).
Multiple studies show that M. truncatula plants with high drought tolerance also display strong salt tolerance, suggesting some overlapping mechanism between them (Yousfi et al., 2010; Luo et al., 2016; Gou et al., 2018; Wang S. et al., 2021). Both salt and drought responses involve osmotic and oxidative regulation. López et al. (2008) find that accumulation of amino acids and sugars in shoot cells alleviates the adverse effects of Na+ in M. truncatula. Salt-tolerant M. truncatula genotypes accumulate more antioxidants and have strong peroxide scavenging ability (Mhadhbi et al., 2011, 2013; Amouri et al., 2018). In rice seedlings, PAs regulate cell membrane stability as ROS scavengers and antioxidants under salt stress (Ghosh et al., 2012). The sensitive M. truncatula cultivar TN6.18 has a lower (Spd + Spm)/Put ratio, indicating that this ratio may be related to oxidative status (Antoniou et al., 2021). Nevertheless, the PA levels are easily affected by plants’ condition and environment. So, the application of (Spd + Spm)/Put ratio and PA levels to asses salt tolerance in plants needs to be determined.
Salt stress causes ion toxicity in plant cells. Greater Na+ accumulation in M. truncatula leads to smaller root and shoot structures (Arraouadi et al., 2012; Zhang X. X. et al., 2019). When M. falcata and M. truncatula are subjected to salt shock, M. falcata shows stronger tolerance for its effective extrusion of Na+ out of cells (Liu et al., 2015). This result is in agreement with that M. truncatula lines with the highest salt stress tolerance have the lowest Na+ content in leaves (Aydi et al., 2008). In addition, legumes tend to restrict Na+ transport toward the shoots to keep a relatively low Na+ content in their photosynthetic organs (Winter and Läuchli, 1982). Transgenic M. truncatula lines overexpressing the calmodulin-like gene MtCML40 are more sensitive to salt stress because of the greater Na+ accumulation in their shoots (Zhang X. X. et al., 2019).
Several salt-responsive genes throughout leaf senescence are identified in M. truncatula (Dong et al., 2021). These genes are mainly related to protein and amino acid metabolism, photosynthesis, chlorophyll metabolism, and hormone signaling. Long et al. (2016, 2018) characterize the proteome-level changes associated with the salt stress response, which are consistent with previous studies (Kang et al., 2010; Dong et al., 2021). Hence, M. truncatula responds to salt stress by altering gene expression, biosynthesis of proteins and metabolites, and modifications in hormonal signaling, etc.
Several studies show that proline is related to the regulation of salt stress. Arabidopsis plants expressing MtHB2 are susceptible to salt stress due to lower proline and soluble sugar content in the cells. This is because MtHB2 may bind to the promoters of P5CS1 and P5CS2 to inhibit their expression (Song et al., 2012). Besides, De Lorenzo et al. (2009) identify a salt-induced gene Srlk in M. truncatula and RNA interference (RNAi) created Srlk mutants accumulate less Na+ in plants than in wild-type plants. Liu et al. (2015) find that the SOS (salt overly sensitive) system mediates cytosolic Na+ out of cells. The protein MtCaMP1 up-regulates the vacuolar Na+/H+ antiporter AtNHX1 and reduces Na+ content in transgenic Arabidopsis plants (Wang T. Z. et al., 2013). The NHX transporters sequester Na+ into vacuoles and decrease the Na+ damage to the organelles in the cytoplasm. Four distinct NHX isoforms (AtNHX1–AtNHX4) are confirmed in Arabidopsis, and their roles in vacuolar ion and pH homeostasis have been determined (Bassil et al., 2019). In M. truncatula, six MtNHXs are identified, and MtNHX3, MtNHX6, and MtNHX7 in roots are induced by salt stress (Sandhu et al., 2018). In addition, Du et al. (2021) identify several salt stress responsive CBL-CIPK genes in M. truncatula and M. sativa. Collectively, SOS pathway, CBL-CIPK family genes, and NHX genes play crucial roles in response to salt stress.
Gruber et al. (2009) identify many salt-responsive TFs in M. truncatula roots belonging to AP2/EREBP, HD-ZIP, and MYB families (Table 1). TF MtMYBS1 promotes the expression of P5CS and mitigates the restriction of root growth under salt stress (Dong et al., 2017). TF MtHB1 suppresses the expression of MtLBD1 (lateral organ boundaries gene), reducing lateral roots formation and Na+ uptake (Ariel et al., 2010). Furthermore, overexpression of MtNAC969 induces the formation of shorter and less branched roots, whereas RNAi-mediated MtNAC969 inactivation promotes lateral root formation. Interestingly, both root systems improved plant growth under salt stress (de Zélicourt et al., 2012). This discrepancy might because that MtNAC969 might participate in multiple pathways controlling root system adaptation to salt stress. In addition, overexpression of MtbHLH-658, MtRAV3, and MtWRKY76 improves root growth under salt stress in transgenic plants (Zahaf et al., 2012; Liu et al., 2016; Wang S. et al., 2021). Merchan et al. (2007) identify two salt responsive IIIA-like TFs MtZpt2-1 and MtZpt2-2. Overexpression each of them significantly improves root growth under salt stress (De Lorenzo et al., 2007). There exists many stress-related cis-elements in MtZpt2-1, allowing it to respond and adapt to abiotic stresses (Wang T. Z. et al., 2014). However, the target genes for most of these TFs have not been identified.
Epigenetic modifications play important “switch” roles in regulating gene expression, thereby affecting plant responses to abiotic stresses (Dong et al., 2018). The epigenetics refers to alterations in gene expressions caused by DNA methylation and histone modification (Saeed et al., 2022). Yaish et al. (2018) analyze the M. truncatula genome-wide DNA methylation in response to salt stress and find that the whole DNA methylation level is increased, and the 5-methylcytosine nucleotide (5-mC) landscape is remodeled under salt stress. More precisely, the DNA methylation and histone modification of MtMYBS1 are analyzed under salt stress. Indeed, the expression of MtMYBS1 is negatively correlated with its DNA methylation modification, and positively correlated with histone H3K9ac modification under salt stress (Dong et al., 2018). These studies provide critical theoretical guidance for further understanding of epigenetic regulation in response to salt stress in M. truncatula.
Bianco and Defez (2009) compared Mt-RD64 plants noduled by Sinorhizobium meliloti RD64, which have higher indole-3-acetic acid (IAA) content in nodules and roots, with the control plants. The results show that Mt-RD64 plants accumulate higher endogenous osmolyte in shoots and are more tolerant to salt stress (Bianco and Defez, 2009). Thus, exogenous IAA might be able to stimulate osmolyte production and positively affect plant development and differentiation under salt stress.
Cold stress includes chilling stress and freezing stress. When the temperature is low but above 0°C (i.e., chilling), membrane fluidity decreases. When it is below 0°C and is defined as freezing, ice formation might occur within tissues, resulting in membrane damage (Raza et al., 2021). The freezing tolerance of many plants is increased after exposure to low, non-freezing temperatures, which is referred to as cold acclimation (Xin and Browse, 2000; Figure 3). Cold acclimated M. truncatula A17 seedlings exposed to −10°C are still survivable (Zhang et al., 2011). In molecular terms, the cold responsive genes and CBF-dependent signaling pathways play roles to enhance the cold tolerance of M. truncatula (Figure 3).
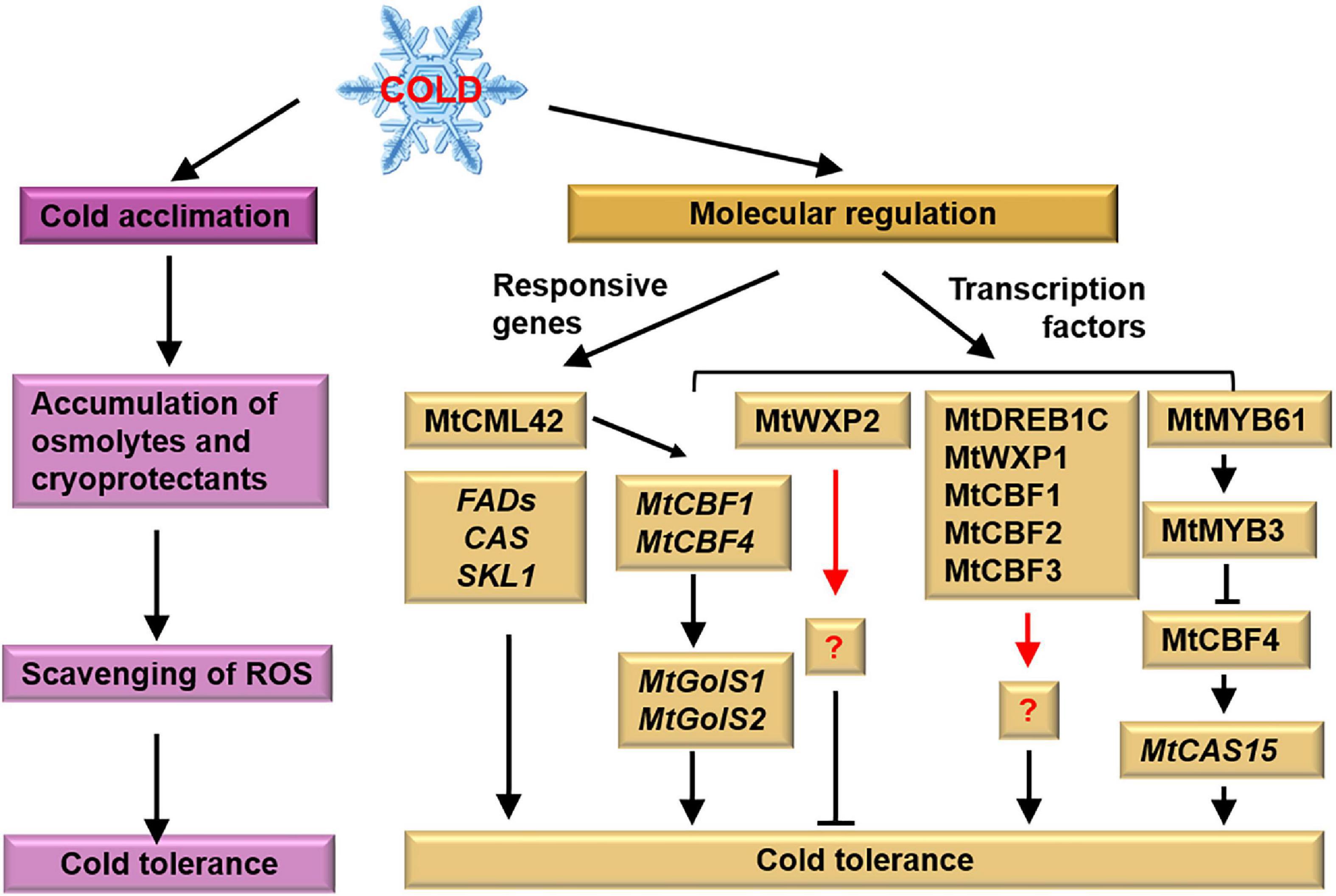
Figure 3. Tolerant mechanism of M. truncatula to cold stress. M. truncatula enhances its cold tolerance after cold acclimation. Meanwhile, cold responsive genes and transcription factors associated with cold tolerance have been identified. Pink and yellow boxes represent physiological and molecular responses, respectively. The black arrows represent the targets, the red arrows represent the unknown factors and the cross-lines mean suppression.
Cold-acclimated M. truncatula exhibits shorter stems, fewer leaves, smaller tissues, and higher root/shoot ratio compared to plants under normal condition (Thapa et al., 2008). While, the relationship between these phenotypes and cold tolerance remains vague. Pennycooke et al. (2008) find that cold acclimation does not significantly improve the survival rate of M. truncatula under freezing stress. However, Thapa et al. (2008) and Zhang et al. (2011) demonstrate that cold acclimation is able to improve the freezing tolerance of M. truncatula. The different cold acclimation regimes, 4°C in Zhang et al. (2011) and 2°C in Pennycooke et al. (2008), and different cultivars and ages of seedlings may explain this discrepancy. Thapa et al. (2008) propose that 3.5°C day/–1°C night for 1 week is the best regime for cold acclimation in M. truncatula. Cold acclimation induces the accumulation of sucrose and proline in M. falcata and M. truncatula (Zhang et al., 2011). Although MtP5CS3 is induced at 4°C, it is unlikely associated with cold tolerance (Nguyen et al., 2013). These data indicate that the cold tolerance might be independent with the proline concentration but positively correlated with the soluble sugar concentration.
Zhang et al. (2018) figure out 20 MtFAD genes (fatty acid desaturase genes) involved in chilling response. The FADs are involved in the desaturation of fatty acids affecting the function of the membrane system (Wallis and Browse, 2002). Another important agent of cold-related genes is the cold-responsive (COR) genes. Mohapatra et al. (1989) isolate three CORs specifically expressed during cold acclimation in Medicago referred to as CAS (cold acclimation specific). The CAS18 gene of M. falcata is positively correlated with freezing tolerance and its expression is much higher in cold-acclimated plants than in non-acclimated plants (Wolfraim et al., 1993). Pennycooke et al. (2008) find that the M. truncatula genome contains a single CAS31 gene, whereas the M. falcata genome contains multiple MfCAS30 and MfCAS31 genes. So, M. falcata is more tolerant to cold stress than M. truncatula. Zhang et al. (2011) clarify that MfCAS17 and MfCAS18 contribute to the stronger cold acclimation effects on M. alfalfa than on M. truncatula. Zhao et al. (2014) find that cold acclimation—induced the transcription of MtCAS15 is suppressed in the ethylene-insensitive mutant skl, indicating that MtSKL1 is required for cold acclimation.
Recently, the expression profiles of DREBs in M. truncatula and M. sativa are identified in the cold-stress response (Shu et al., 2016; Sheng et al., 2022). A cluster of DREB subfamily members on M. truncatula chromosome 6 is induced by both cold and freezing stress (Shu et al., 2016), and 33 MsDREBs are significantly upregulated by cold treatment (Sheng et al., 2022). The genome-wide identification of DREBs in Medicago species provides promising molecular targets for the improvement of cold tolerance in crops. Overexpression of MtDREB1C/MtCBF3 inhibits shoot growth and enhances the freezing tolerance of M. truncatula (Chen et al., 2010). Transgenic M. truncatula plants overexpressing MfERF1 show enhanced tolerance to both freezing and chilling stress through promoting PA turnover, antioxidant protection, and proline accumulation (Zhuo et al., 2018). Overexpression of WXP1 in M. truncatula enhances the plants’ freezing tolerance without altering growth and development. However, plants overexpressing WXP2 are more sensitive to freezing (Zhang et al., 2007). These results indicate that WXP1 is a useful candidate gene for improving plant freezing tolerance by genetic conduction.
The TFs MtCBF1, MtCBF2, and MtCBF3 have been shown to participate in cold acclimation in M. truncatula (Pennycooke et al., 2008; Zhang et al., 2011). TF MtCBF4 not only positively regulates cold acclimation and freezing tolerance but also enhances drought and salt tolerance (Li et al., 2011; Zhang et al., 2016). Although the differential response of MtCBFs to cold stress is unknown, the major components involved in CBF-dependent signaling pathways are illustrated under cold stress. TF MtMYB3 binds to the cis-elements of MtCBF4 promoter and represses its expression. TF MtCBF4 directly activates the transcription of MtCAS15. TF MtMYB61 relieve the inhibitory effect of MtMYB3 on MtCBF4 (Zhang et al., 2016). Besides, Qu et al. (2016) indicates that the MfNAC3 plays roles in response to cold stress by regulating the expression of MtCBF4. Recently, Sun et al. (2021) identifies that MtCML42 positively regulates the expression of MtCBF1 and MtCBF4, thereby upregulating the expression of the COR genes, MtGolS1 and MtGolS2, and leads to raffinose accumulation and improved cold tolerance.
Demethylases containing Jumonji (JMJ) C domain are involved in removal of methyl groups at lysine or arginine residues (Lu et al., 2008). In M. truncatula, MtJMJC5 undergoes cold-specifically induced alternative splicing, which is reversible depending on temperature (Shen et al., 2016). Previous studies show that AtJMJ30/JMJD5 is a component of the plant circadian clock (Lu et al., 2011). So, there may exist a MtJMJC5-dependent link between the circadian clock and ambient temperature fluctuation in M. truncatula.
Zhao et al. (2009) proposes that nitrate reductase (NR)-dependent NO production plays an important role in the cold acclimation-induced increase in freezing tolerance by modulating proline accumulation in Arabidopsis. In addition, the role of NO in cold acclimation through the regulation of glutathione (GSH) synthesis has been studied in M. falcata and M. truncatula (Zhang P. P. et al., 2019). Exogenous application of ethylene reduces cold acclimation-induced freezing tolerance (Zhao et al., 2014). These results indicate that there may have some relationships between NO and ethylene molecules and osmotic regulation in response to cold tolerance.
Legumes are a particularly important source of food for livestock worldwide (Wang T. Z. et al., 2021). The conventional breeding of crops is time-consuming, labor-intensive, and cost-inefficient (Mishra et al., 2021). An efficient solution is to generate stress-tolerant varieties with the help of information obtained in the lab. Thus, understanding the physiological and molecular processes of legumes in response to abiotic stresses is really important. M. truncatula is closely related to many legumes and forages (Figure 4). In this review, we summarize the mechanism by which M. truncatula responds and adapts to drought (Figure 1), salt (Figure 2), and cold stress (Figure 3) as well as crosstalk between them (Figure 5). These studies provide genetic resources and molecular markers that could be used in future studies.
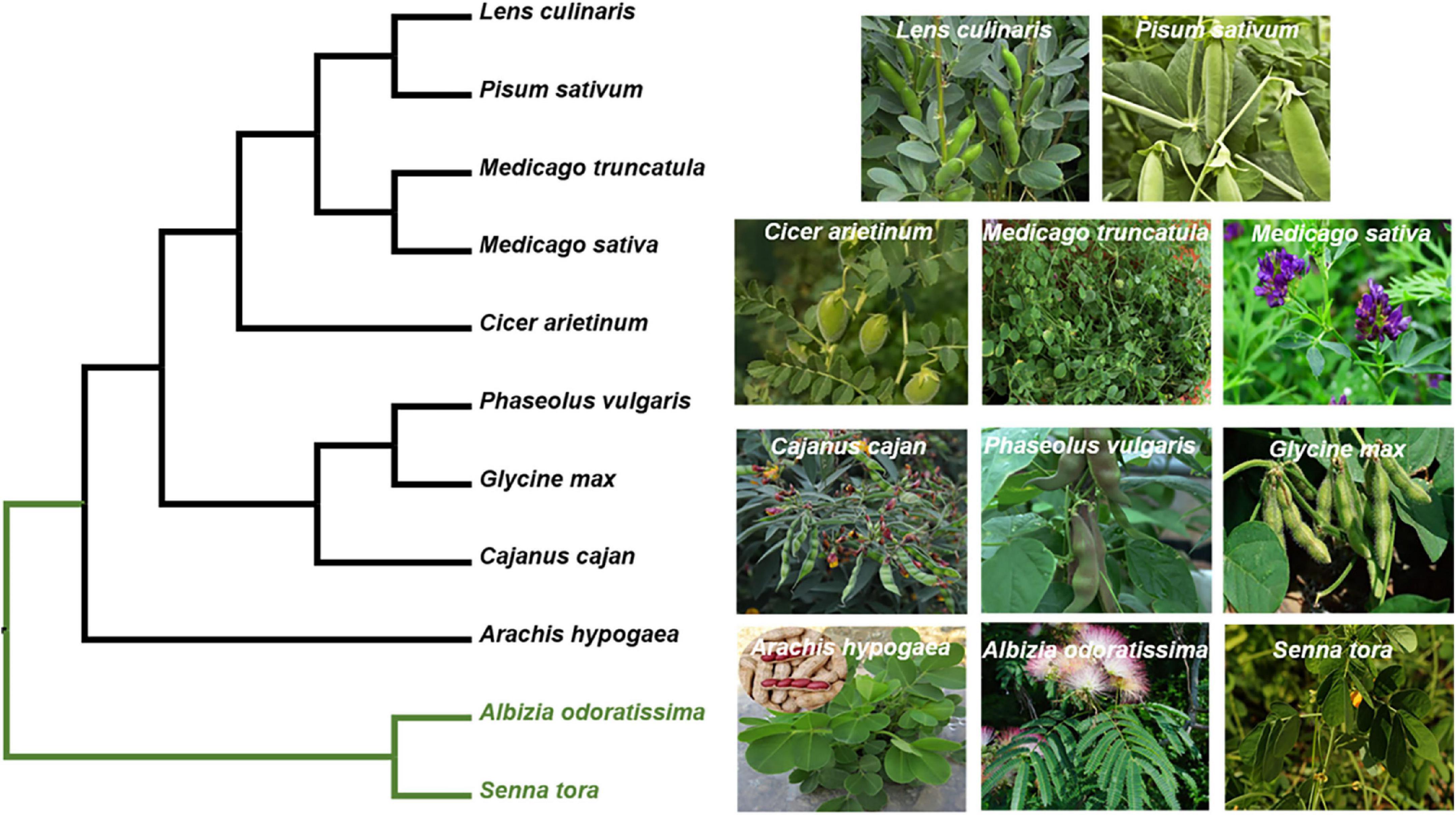
Figure 4. Phylogeny of M. truncatula with the other legumes. The phylogeny of the targeted species was reconstructed based on their plastomes. The data matrix of Azani et al. (2017) was used as a basic DNA matrix, from which we extract the sequences of the targeted species and outgroups (Albizia odoratissima and Senna tora). Then, these sequences were aligned with the complete plastomes of M. truncatula. The maximum likelihood (ML) phylogeny was reconstructed using RAxML version 8.2.12. Their accession number obtained from GenBank. Albizia odoratissima: NC_034987.1; Arachis hypogaea: NC_026676.1; Cajanus cajan: NC_031429.1; Cicer arietinum: NC_011163.1; Glycine max: NC_007942.1; Lens culinaris: NC_027152.1; Medicago sativa: KU321683.1; Medicago truncatula: JX512024.1; Phaseolus vulgaris: NC_009259.1; Pisum sativum: NC_014057.1; Senna tora: NC_030193.1.
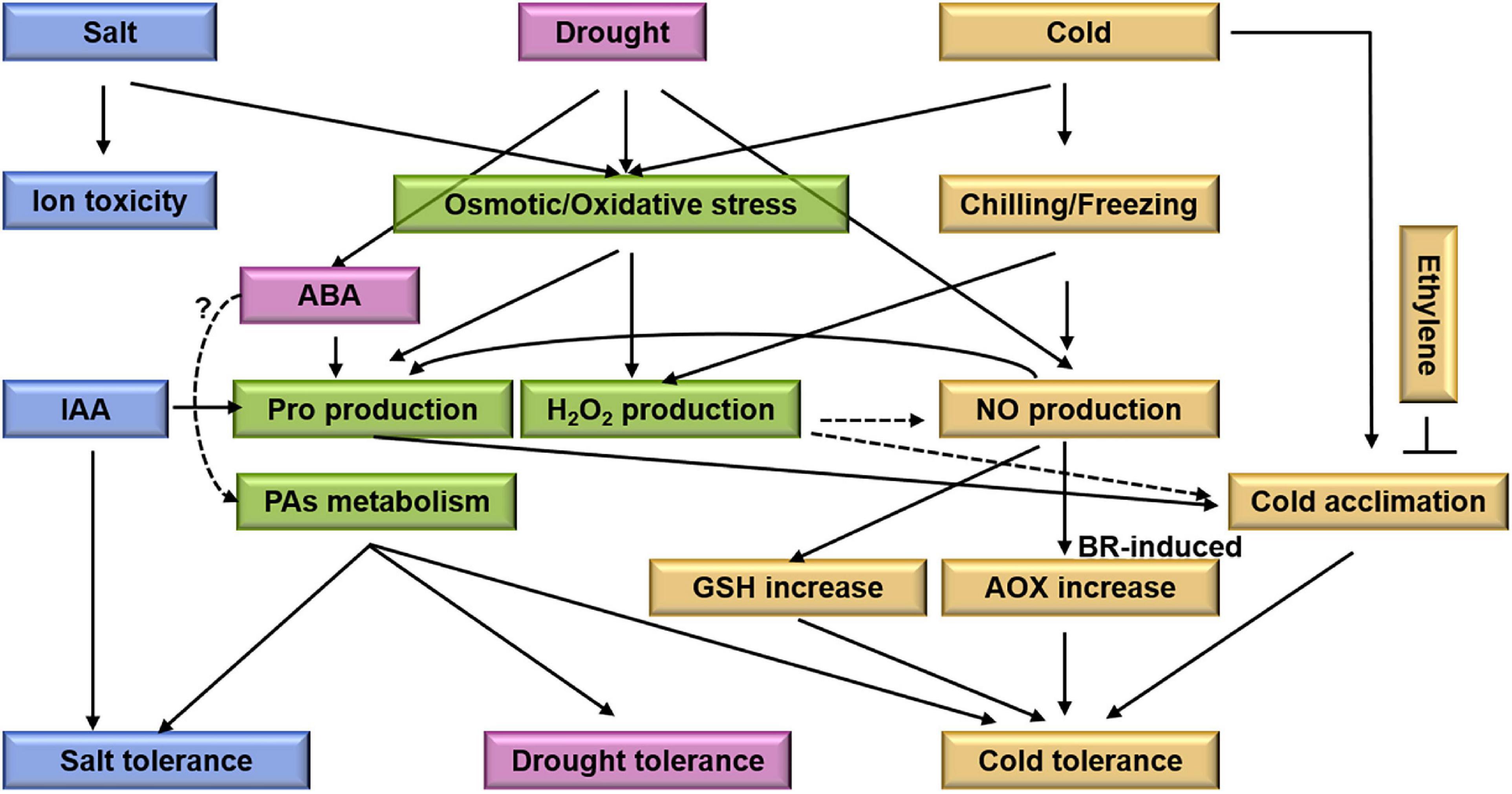
Figure 5. A schematic representation of cross-talks between drought, salt, and cold stress in M. truncatula. Drought, salt, and cold stress cause secondary stress including osmotic stress and oxidative stress. And then induce proline and H2O2 production. In M. truncatula, ABA could induce proline accumulation, contributing to osmotic adjustment under drought condition. And NO in cold acclimation through regulation on GSH synthesis and redox is associated with the differential cold tolerance. Meanwhile, ethylene reduces cold acclimation-induced freezing tolerance. IAA may have positive effects on their development and differentiation under salt stress. PAs as compound positively regulate M. truncatula in response to drought, salt, and cold stress. Ion toxicity and cold acclimation are specific process for salt stress and cold stress, respectively. Blue boxes indicate salt related elements. Pink and yellow boxes indicate drought and cold related elements, respectively. Green boxes indicate the central elements between drought, salt, and cold stress. The black arrows represent the targets, the dotted lines represent the possible regulatory targets and the cross-lines mean suppression.
All drought, salt and cold stress can induce osmotic and oxidative stress. Proline is involved in abiotic tolerance as a compatible osmolyte, molecular chaperone, and ROS scavenger (Szabados and Savouré, 2010). There are three MtP5CSs in M. truncatula. The MtP5CS1 is constitutively expressed and the MtP5CS3 participates in osmotic regulation (Armengaud et al., 2004; Kim and Nam, 2013). Both IAA and ABA induce proline accumulation but enhance the salt and drought tolerance, respectively, indicating that proline play different roles in salt and drought stress. Meanwhile, both drought and cold stress induce NO production, however, drought or cold induced NO production via different pathways (Planchet et al., 2014; Arfan et al., 2019). Complex crosstalk suggests that plants integrate hormones and signaling pathways to get better adaptation to abiotic stresses. With the help of modern molecular technologies, such as transgenic and CRISPR/Cas9 approaches, clarifying the functions of different factors is promising. Recently, Wang et al. (2022) using the CRISPR/Cas9 toolkit generates single and double knockout mutants in MtDMP8 or MtDMP9 and assesses their roles in haploid induction in M. truncatula. However, there is no report about tolerant mechanism to abiotic stresses using CRISPR/Cas9 technology.
Currently, many studies on M. truncatula are carried out in the culture room. So, more field experiments should be performed in future research. In field condition, plants often face several distinct environmental stresses simultaneously. For example, plants in arid regions often suffer from drought and heat stress (Iyer et al., 2013). In M. truncatula, interactive effects of ozone and drought have been well studied (Iyer et al., 2013). However, how combined occurrence of other kinds of abiotic stresses impact growth and development of M. truncatula is still not known yet and will be an important research topic in the future. In addition, M. truncatula is a cultivated species, some stress tolerance genes might have been lost during the domestication process (Wang T. Z. et al., 2021). M. ruthenica, a wild Medicago forage, retains these genes. Therefore, M. ruthenica provides a valuable model plant for studying the molecular mechanism of abiotic stresses tolerance in legumes.
XZ and TW conceived the concept of the work and wrote the manuscript. YS, XQ, HL, and IH revised the manuscript. All authors approved the final manuscript.
This research was co-funded by the National Natural Science Foundation of China (32101419 and 32070351) and the Science and Technology Program of Inner Mongolia, China (2021GG0372).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alcázar, R., Altabella, T., Marco, F., Bortolotti, C., Reymond, M., Koncz, C., et al. (2010). Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231, 1237–1249.
Amouri, A. A., González, E. M., and Aoul, S. H. (2018). Physiological and biochemical characterization of rootlets response to salt stress in two Medicago truncatula Gaertn. ecotypes. Plant Root 12, 1–10.
Antoniou, C., Zarza, X., Gohari, G., Panahirad, S., Filippou, P., Tiburcio, A. F., et al. (2021). Involvement of polyamine metabolism in the response of Medicago truncatula genotypes to salt stress. Plants 10:269. doi: 10.3390/plants10020269
Araújo, S. S., Duque, A. S., Silva, J. M., Santos, D., Silva, A. B., and Fevereiro, P. (2013). Water deficit and recovery response of Medicago truncatula plants expressing the ELIP-like DSP22. Biol. Plant. 57, 159–163.
Arfan, M., Zhang, D. W., Zou, L. J., Luo, S. S., Tan, W. R., Zhu, T., et al. (2019). Hydrogen peroxide and nitric oxide crosstalk mediates brassinosteroids induced cold stress tolerance in Medicago truncatula. Int. J. Mol. Sci. 20:144. doi: 10.3390/ijms20010144
Ariel, F., Diet, A., Verdenaud, M., Gruber, V., Frugier, F., Chan, R., et al. (2010). Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell 22, 2171–2183. doi: 10.1105/tpc.110.074823
Armengaud, P., Thiery, L., Buhot, N., Grenier de March, G., and Savouré, A. (2004). Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiol. Plant 120, 442–450. doi: 10.1111/j.0031-9317.2004.00251.x
Arraouadi, S., Badri, M., Abdelly, C., Huguet, T., and Aouani, M. E. (2012). QTL mapping of physiological traits associated with salt tolerance in Medicago truncatula recombinant inbred lines. Genomics 99, 118–125. doi: 10.1016/j.ygeno.2011.11.005
Aydi, S., Sassi, S., and Abdelly, C. (2008). Growth, nitrogen fixation and ion distribution in Medicago truncatula subjected to salt stress. Plant Soil 312, 59–67.
Azani, N., Babineau, M., Donovan Bailey, C., Banks, H., Barbosa, A. R., Barbosa Pinto, R., et al. (2017). A new subfamily classification of the leguminosae based on a taxonomically comprehensive phylogeny: the Legume Phylogeny Working Group (LPWG). Taxon 66, 44–77.
Bassil, E., Zhang, S., Gong, H. J., Tajima, H., and Blumwald, E. (2019). Cation specificity of vacuolar NHX-type cation/H+ antiporters. Plant Physiol. 179, 616–629. doi: 10.1104/pp.18.01103
Bianco, C., and Defez, R. (2009). Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J. Exp. Bot. 11, 3097–3107. doi: 10.1093/jxb/erp140
Charrier, A., Lelievre, E., Limami, A. M., and Planchet, E. (2013). Medicago truncatula stress associated protein 1 gene (MtSAP1) overexpression confers tolerance to abiotic stress and impacts proline accumulation in transgenic tobacco. J. Plant Physiol. 170, 874–877. doi: 10.1016/j.jplph.2013.01.008
Charrier, A., Planchet, E., Cerveau, D., Gimeno-Gilles, C., Verdu, I., Limami, A. M., et al. (2012). Overexpression of a Medicago truncatula stress-associated protein gene (MtSAP1) leads to nitric oxide accumulation and confers osmotic and salt stress tolerance in transgenic tobacco. Planta 236, 567–577. doi: 10.1007/s00425-012-1635-9
Chen, J. R., Lü, J. J., Liu, R., Xiong, X. Y., Wang, T. X., Chen, S. Y., et al. (2010). DREB1C from Medicago truncatula enhances freezing tolerance in transgenic M. truncatula and China Rose (Rosa chinensis Jacq.). Plant Growth Regul. 60, 199–211.
Confalonieri, M., Carelli, M., Tava, A., and Borrelli, L. (2019). Overexpression of MtTdp2α (tyrosyl-DNA phosphodiesterase 2) gene confers salt tolerance in transgenic Medicago truncatula. Plant Cell Tiss. Org. 137, 157–172.
Cramer, G. R., Urano, K., Delrot, S., Pezzotti, M., and Shinozaki, K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11:163. doi: 10.1186/1471-2229-11-163
De Lorenzo, L., Merchan, F., Blanchet, S., Megias, M., Frugier, F., Crespi, M., et al. (2007). Differential expression of the TFIIIA regulatory pathway in response to salt stress between Medicago truncatula genotypes. Plant Physiol. 145, 1521–1532. doi: 10.1104/pp.107.106146
De Lorenzo, L., Merchan, F., Laporte, P., Thompson, R., Clarke, J., Sousa, C., et al. (2009). A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. Plant Cell 21, 668–680. doi: 10.1105/tpc.108.059576
de Zélicourt, A., Diet, A., Marion, J., Laffont, C., Ariel, F., Moison, M., et al. (2012). Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J. 70, 220–230. doi: 10.1111/j.1365-313X.2011.04859.x
Delledonne, M., Zeier, J., Marocco, A., and Lamb, C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. U.S.A. 98, 13454–13459. doi: 10.1073/pnas.231178298
Dong, S., Sang, L., Xie, H., Chai, M., and Wang, Z. Y. (2021). Comparative transcriptome analysis of salt stress-induced leaf senescence in Medicago truncatula. Front. Plant Sci. 12:666660. doi: 10.3389/fpls.2021.666660
Dong, W., Liu, X. J., Gao, T. X., and Song, Y. G. (2018). Analyses of DNA methylation and histone modification of MtMYBS1 in Medicago truncatula under salinity stress. Plant Physiol. J. 54, 1596–1604.
Dong, W., Song, Y., Zhao, Z., Qiu, N. W., Liu, X., and Guo, W. (2017). The Medicago truncatula R2R3-MYB transcription factor gene MtMYBS1 enhances salinity tolerance when constitutively expressed in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 490, 225–230. doi: 10.1016/j.bbrc.2017.06.025
Du, W. X., Yang, J. F., Ma, L., Su, Q., and Pang, Y. Z. (2021). Identification and characterization of abiotic stress responsive CBL-CIPK family genes in Medicago. Int. J. Mol. Sci. 22:4634. doi: 10.3390/ijms22094634
Duque, A. S., López-Gómez, M., Kráčmarová, J., Gomes, C. N., Araújo, S. S., Lluch, C., et al. (2016). Genetic engineering of polyamine metabolism changes Medicago truncatula responses to water deficit. Plant Cell Tiss. Org. 127, 681–690.
Farooq, M., Wahid, A., Kobayashi, N., Fujita, D., and Basra, S. M. A. (2009). Plant drought stress: effects, mechanism and management. Agron. Sustain. Dev. 29, 185–212.
Farooq, M. S., Uzair, M., Raza, A., Habib, M., Xu, Y. L., Yousuf, M., et al. (2022). Uncovering the research gaps to alleviate the negative impacts of climate change on food security: a review. Front. Plant Sci. 13:927535. doi: 10.3389/fpls.2022.927535
Ghosh, N., Das, S. P., Mandal, C., Gupta, S., Das, K., Dey, N., et al. (2012). Variations of antioxidative responses in two rice cultivars with polyamine treatment under salinity stress. Physiol. Mol. Biol. Plants 18, 301–313. doi: 10.1007/s12298-012-0124-8
Gou, J., Debnath, S., Sun, L., Flanagan, A., Tang, Y., Jiang, Q., et al. (2018). From model to crop: functional characterization of SPL8 in M. truncatula led to genetic improvement of biomass yield and abiotic stress tolerance in alfalfa. Plant Biotechnol. J. 16, 951–962. doi: 10.1111/pbi.12841
Gruber, V., Blanchet, S., Diet, A., Zahaf, O., Boualem, A., Kakar, K., et al. (2009). Identification of transcription factors involved in root apex responses to salt stress in Medicago truncatula. Mol. Genet. Genomics 281, 55–66. doi: 10.1007/s00438-008-0392-8
Guo, T., Wang, S. M., Zhang, T. J., Xu, L. X., Li, Y. R. Z., Chao, Y. H., et al. (2021). Expression of the Medicago truncatula MtDof32 transcription factor regulates plant growth and enhances abiotic stress tolerances in transgenic Arabidopsis. Environ. Exp. Bot. 183:10433.
Iyer, N., Tang, Y. H., and Mahalingam, R. (2013). Physiological, biochemical and molecular responses to a combination of drought and ozone in Medicago truncatula. Plant Cell Environ. 36, 706–720. doi: 10.1111/pce.12008
Kang, J. M., Xie, W. W., Sun, Y., Yang, Q. C., and Wu, M. S. (2010). Identification of genes induced by salt stress from Medicago truncatula L. seedlings. Afr. J. Biotechnol. 9, 7589–7594.
Kim, G. B., and Nam, Y. W. (2013). A novel Δ1-pyrroline-5-carboxylate synthetase gene of Medicago truncatula plays a predominant role in stress-induced proline accumulation during symbiotic nitrogen fixation. J. Plant Physiol. 170, 291–302. doi: 10.1016/j.jplph.2012.10.004
Li, D., Zhang, Y., Hu, X., Shen, X., Ma, L., Su, Z., et al. (2011). Transcriptional profiling of Medicago truncatula under salt stress identified a novel CBF transcription factor MtCBF4 that plays an important role in abiotic stress responses. BMC Plant Biol. 11:109. doi: 10.1186/1471-2229-11-109
Li, X., Feng, H., Wen, J. Q., Dong, J. L., and Wang, T. (2018). MtCAS31 aids symbiotic nitrogen fixation by protecting the leghemoglobin MtLb120-1 under drought stress in Medicago truncatula. Front. Plant Sci. 9:633. doi: 10.3389/fpls.2018.00633
Li, X., Hou, Y. Y., Zhang, F., Li, M. N., Yi, F. Y., Kang, J. M., et al. (2022). Identification and characterization of stress responsive homeodomain leucine zipper transcription factors in Medicago truncatula. Mol. Biol. Rep. 49, 3569–3581. doi: 10.1007/s11033-022-07197-4
Li, X., Liu, Q., Feng, H., Deng, J., Zhang, R., Wen, J., et al. (2020). Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 16, 862–877. doi: 10.1080/15548627.2019.1643656
Liu, L., Zhang, Z., Dong, J., and Wang, T. (2016). Overexpression of MtWRKY76 increases both salt and drought tolerance in Medicago truncatula. Environ. Exp. Bot. 123, 50–58.
Liu, M., Wang, T. Z., and Zhang, W. H. (2015). Sodium extrusion associated with enhanced expression of SOS1 underlies different salt tolerance between Medicago falcata and Medicago truncatula seedlings. Environ. Exp. Bot. 110, 46–55.
Long, R., Gao, Y., Sun, H., Zhang, T., Li, X., Li, M., et al. (2018). Quantitative proteomic analysis using iTRAQ to identify salt-responsive proteins during the germination stage of two Medicago species. Sci. Rep. 8:9553. doi: 10.1038/s41598-018-27935-8
Long, R., Li, M., Zhang, T., Kang, J., Sun, Y., Cong, L., et al. (2016). Comparative proteomic analysis reveals differential root proteins in Medicago sativa and Medicago truncatula in response to salt stress. Front. Plant Sci. 7:424. doi: 10.3389/fpls.2016.00424
López, M., Herrera-Cervera, J. A., Iribarne, C., Tejera, N. A., and Lluch, C. (2008). Growth and nitrogen fixation in Lotus japonicus and Medicago truncatula under NaCl stress: nodule carbon metabolism. J. Plant Physiol. 165, 641–650. doi: 10.1016/j.jplph.2007.05.009
Lu, F. L., Li, G. L., Cui, X., Liu, C. Y., Wang, X. J., and Cao, X. F. (2008). Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 50, 886–896. doi: 10.1111/j.1744-7909.2008.00692.x
Lu, S. X., Knowles, S. M., Webb, C. J., Celaya, R. B., Cha, C., Siu, J. P., et al. (2011). The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 155, 906–915. doi: 10.1104/pp.110.167015
Luo, S. S., Sun, Y. N., Zhou, X., Zhu, T., Zhu, L. S., Arfan, M., et al. (2016). Medicago truncatula genotypes Jemalong A17 and R108 show contrasting variations under drought stress. Plant Physiol. Biochem. 109, 190–198. doi: 10.1016/j.plaphy.2016.09.019
Macovei, A., Balestrazzi, A., Confalonieri, M., and Carbonera, D. (2010). The tyrosyl-DNA phosphodiesterase gene family in Medicago truncatula Gaertn.: bioinformatic investigation and expression profiles in response to copper- and PEG-mediated stress. Planta 232, 393–407. doi: 10.1007/s00425-010-1179-9
Merchan, F., Breda, C., Hormaeche, J. P., Sousa, C., Kondorosi, A., Aguilar, O., et al. (2003). A Krüppel-like transcription factor gene is involved in salt stress responses in Medicago spp. Plant Soil 257, 1–9.
Merchan, F., De Lorenzo, L., Rizzo, S. G., Niebel, A., Manyani, H., Frugier, F., et al. (2007). Identification of regulatory pathways involved in the reacquisition of root growth after salt stress in Medicago truncatula. Plant J. 1, 1–17. doi: 10.1111/j.1365-313X.2007.03117.x
Mhadhbi, H., Fotopoulos, V., Mylona, P. V., Jebara, M., Aouani, M. E., and Polidoros, A. N. (2011). Antioxidant gene-enzyme responses in Medicago truncatula genotypes with different degree of sensitivity to salinity. Physiol. Plant. 141, 201–214. doi: 10.1111/j.1399-3054.2010.01433.x
Mhadhbi, H., Fotopoulos, V., Mylona, P. V., Jebara, M., Aouani, M. E., and Polidoros, A. N. (2013). Alternative oxidase 1 (Aox1) gene expression in roots of Medicago truncatula is a genotype-specific component of salt stress tolerance. J. Plant Physiol. 170, 111–114. doi: 10.1016/j.jplph.2012.08.017
Mishra, D., Shekhar, S., Chakraborty, S., and Chakraborty, N. (2021). High temperature stress responses and wheat: impacts and alleviation strategies. Environ. Exp. Bot. 190, 1–13.
Mohapatra, S. S., Wolfraim, L., Poole, R. J., and Dhindsa, R. S. (1989). Molecular cloning and relationship to freezing tolerance of cold-acclimation-specific genes of Alfalfa. Plant Physiol. 89, 375–380. doi: 10.1104/pp.89.1.375
Munns, R., and Tester, M. (2008). Mechanism of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681.
Nguyen, M. L., Kim, G. B., Hyun, S. H., Lee, S. Y., Lee, C. Y., Choi, H. K., et al. (2013). Physiological and metabolo mic analysis of a knockout mutant suggests a critical role of MtP5CS3 gene in osmotic stress tolerance of Medicago truncatula. Euphytica 193, 101–120.
Nunes, C., de Sousa Araújo, S., da Silva, J. M., Fevereiro, M. P. S., and da Silva, A. B. (2008). Physiological responses of the legume model Medicago truncatula cv. Jemalong to water deficit. Environ. Exp. Bot. 63, 289–296.
Oztur, Z. N., Talame, V., Deyholos, M., Michalowski, C. B., Galbraith, D. W., Gozukirmizi, N., et al. (2002). Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol. Biol. 48, 551–573. doi: 10.1023/a:1014875215580
Pagano, A., Zannino, L., Pagano, P., Doria, E., Dondi, D., Macovei, A., et al. (2022). Changes in genotoxic stress response, ribogenesis and PAP (3′-phosphoadenosine 5′-phosphate) levels are associated with loss of desiccation tolerance in overprimed Medicago truncatula seeds. Plant Cell Environ. 45, 1457–1473. doi: 10.1111/pce.14295
Pennycooke, J. C., Cheng, H., and Stockinger, E. J. (2008). Comparative genomic sequence and expression analyses of Medicago truncatula and alfalfa subspecies falcata COLD-ACCLIMATION-SPECIFIC genes. Plant Physiol. 146, 1242–1254. doi: 10.1104/pp.107.108779
Planchet, E., Rannou, O., Ricoult, C., Boutet-Mercey, S., Maia-Grondard, A., and Limami, A. M. (2011). Nitrogen metabolism responses to water deficit act through both abscisic acid (ABA)-dependent and independent pathways in Medicago truncatula during post-germination. J. Exp. Bot. 2, 605–615. doi: 10.1093/jxb/erq294
Planchet, E., Verdu, I., Delahaie, J., Cukier, C., Girard, C., Morère-Le Paven, M., et al. (2014). Abscisic acid-induced nitric oxide and proline accumulation in independent pathways under water-deficit stress during seedling establishment in Medicago truncatula. J. Exp. Bot. 8, 2161–2170. doi: 10.1093/jxb/eru088
Porto, M. S., Pinheiro, M. P., Batista, V. G., dos Santos, R. C., Filho Pde, A., and de Lima, L. M. (2014). Plant promoters: an approach of structure and function. Mol. Biotechnol. 56, 38–49.
Qiao, W., and Fan, L. M. (2008). Nitric oxide signaling in plant responses to abiotic stresses. J. Int. Plant Biol. 50, 1238–1246.
Qu, Y., Duan, M., Zhang, Z., Dong, J., and Wang, T. (2016). Overexpression of the Medicago falcata NAC transcription factor MfNAC3 enhances cold tolerance in Medicago truncatula. Environ. Exp. Bot. 129, 67–76.
Raza, A., Tabassum, J., Kudapa, H., and Varshney, R. K. (2021). Can omics deliver temperature resilient ready-to-grow crops? Crit. Rev. Biotechnol. 41, 1209–1232. doi: 10.1080/07388551.2021.1898332
Rhaman, M. S., Imran, S., Karim, M. M., Chakrobortty, J., Mahamud, M. A., Sarker, P., et al. (2021). 5-aminolevulinic acid-mediated plant adaptive responses to abiotic stress. Plant Cell Rep. 40, 1451–1469. doi: 10.1007/s00299-021-02690-9
Saeed, F., Chaudhry, U. K., Bakhsh, A., Raza, A., Saeed, Y., Bohra, A., et al. (2022). Moving beyond DNA sequence to improve plant stress responses. Front. Genet. 13:874648. doi: 10.3389/fgene.2022.874648
Sandhu, D., Pudussery, M. V., Kaundal, R., Suarez, D. L., Kaundal, A., and Sekhon, R. S. (2018). Molecular characterization and expression analysis of the Na+/H+ exchanger gene family in Medicago truncatula. Funct. Integr. Genomics 18, 141–153.
Shen, Y. F., Wu, X. P., Liu, D. M., Song, S. J., Liu, D. C., and Wang, H. Q. (2016). Cold-dependent alternative splicing of a Jumonji C domain-containing gene MtJMJC5 in Medicago truncatula. Biochem. Biophys. Res. Commun. 474, 271–276. doi: 10.1016/j.bbrc.2016.04.062
Sheng, S., Guo, X. Y., Wu, C. Z., Xiang, Y. C., Duan, S. H., Yang, W. Q., et al. (2022). Genome-wide identification and expression analysis of DREB genes in alfalfa (Medicago sativa) in response to cold stress. Plant Signal. Behav. 17:e2081420. doi: 10.1080/15592324.2022.2081420
Shu, Y., Liu, Y., Zhang, J., Song, L., and Guo, C. (2016). Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant Sci. 6:1247. doi: 10.3389/fpls.2015.01247
Song, S. Y., Chen, Y., Zhao, M. G., and Zhang, W. H. (2012). A novel Medicago truncatula HD-Zip gene, MtHB2, is involved in abiotic stress responses. Environ. Exp. Bot. 80, 1–9.
Sun, Q., Huang, R., Zhu, H. F., Sun, Y. M., and Guo, Z. F. (2021). A novel Medicago truncatula calmodulin-like protein (MtCML42) regulates cold tolerance and flowering time. Plant Physiol. 108, 1069–1082. doi: 10.1111/tpj.15494
Szabados, L., and Savouré, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97.
Tang, H., Krishnakumar, V., Bidwell, S., Rosen, B., Chan, A., Zhou, S., et al. (2014). An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15:312. doi: 10.1186/1471-2164-15-312
Thapa, B., Arora, R., Knapp, A. D., and Brummer, E. C. (2008). Applying freezing test to quantify cold acclimation in Medicago truncatula. J. Amer. Soc. Hort. Sci. 133, 684–691.
Tian, C., Zhang, X., He, J., Yu, H., Wang, Y., Shi, B., et al. (2014). An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 10:755. doi: 10.15252/msb.20145470
Verdoy, D., Coba De La Pena, T., Redondo, F. J., Lucas, M. M., and Pueyo, J. J. (2006). Transgenic Medicago truncatula plants that accumulate proline display nitrogen-fixing activity with enhanced tolerance to osmotic stress. Plant Cell Environ. 29, 1913–1923. doi: 10.1111/j.1365-3040.2006.01567.x
Wallis, J. G., and Browse, J. (2002). Mutants of Arabidopsis reveal many roles for membrane lipids. Prog. Lipid Res. 41, 254–278. doi: 10.1016/s0163-7827(01)00027-3
Wang, N., Xia, X. Z., Jiang, T., Li, L. L., Zhang, P. C., Niu, L. F., et al. (2022). In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula. Plant Biotechnol. J. 20, 22–24. doi: 10.1111/pbi.13740
Wang, S., Guo, T., Shen, Y., Wang, Z., Kang, J., Zhang, J., et al. (2021). Overexpression of MtRAV3 enhances osmotic and salt tolerance and inhibits growth of Medicago truncatula. Plant Physiol. Biochem. 163, 154–165. doi: 10.1016/j.plaphy.2021.04.003
Wang, T. Z., Ren, L. F., Li, C. H., Zhang, D., Zhang, X. X., Zhou, G., et al. (2021). The genome of a wild Medicago species provides insights into the tolerant mechanism of legume forage to environmental stress. BMC Biol. 19:96. doi: 10.1186/s12915-021-01033-0
Wang, T. Z., Tian, Q. Y., Wang, B. L., Zhao, M. G., and Zhang, W. H. (2014). Genome variations account for different response to three mineral elements between Medicago truncatula ecotypes Jemalong A17 and R108. BMC Plant Biol. 14:122. doi: 10.1186/1471-2229-14-122
Wang, T. Z., Zhang, J. L., Tian, Q. Y., Zhao, M. G., Zhang, W. H., and Diane, B. (2013). A Medicago truncatula EF-Hand family gene, MtCaMP1, is involved in drought and salt stress tolerance. PLoS One 8:e58952. doi: 10.1371/journal.pone.0058952
Wendehenne, D., Durner, J., and Klessig, D. F. (2004). Nitric oxide: a new player in plant signaling and defense responses. Curr. Opin. Plant Biol. 7, 449–455.
Winter, E., and Läuchli, A. (1982). Salt tolerance of Trifolium alexandrinum L. I. comparison of the salt response of T. alexandrinum and T. pratense. Funct. Plant Biol. 9, 221–226.
Wolfraim, L. A., Langis, R., Tyson, H., and Dhindsa, R. S. (1993). cDNA sequence, expression, and transcript stability of a cold acclimation-specific gene, cas18, of alfalfa (Medicago falcata) cells. Plant Physiol. 101, 1275–1282. doi: 10.1104/pp.101.4.1275
Xie, C., Zhang, R., Qu, Y., Miao, Z., Zhang, Y., Shen, X., et al. (2012). Overexpression of MtCAS31 enhances drought tolerance in transgenic Arabidopsis by reducing stomatal density. New Phytol. 195, 124–135. doi: 10.1111/j.1469-8137.2012.04136.x
Xin, Z. G., and Browse, J. (2000). Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ. 23, 893–902.
Yaish, M. W., Al-Lawati, A., Al-Harrasi, I., and Patankar, H. V. (2018). Genome wide DNA methylation analysis in response to salinity in the model plant caliph medic (Medicago truncatula). BMC Genom. 19:78. doi: 10.1186/s12864-018-4484-5
Yang, M. K., Wang, L. P., Chen, C. M., Guo, X., Lin, C. L., Huang, W., et al. (2021). Genome-wide analysis of autophagy-related genes in Medicago truncatula highlights their roles in seed development and response to drought stress. Sci. Rep. 11:22933. doi: 10.1038/s41598-021-02239-6
Yousfi, N., Slama, I., Ghnaya, T., Savoure, A., and Abdelly, C. (2010). Effects of water deficit stress on growth, water relations and osmolyte accumulation in Medicago truncatula and M. laciniata populations. C. R. Biol. 333, 205–213. doi: 10.1016/j.crvi.2009.12.010
Zahaf, O., Blanchet, S., de Zélicourt, A., Alunni, B., Plet, J., Laffont, C., et al. (2012). Comparative transcriptomic analysis of salt adaptation in roots of contrasting Medicago truncatula genotypes. Mol. Plant 5, 1068–1081. doi: 10.1093/mp/sss009
Zhang, J. Y., Broeckling, C. D., Blancaflor, E. B., Sledge, M. K., Sumner, L. W., and Wang, Z. Y. (2005). Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 42, 689–707. doi: 10.1111/j.1365-313X.2005.02405.x
Zhang, J. Y., Broeckling, C. D., Sumner, L. W., and Wang, Z. Y. (2007). Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 64, 265–278. doi: 10.1007/s11103-007-9150-2
Zhang, J. Y., Cruz, D. E. C. M. H., Torres-Jerez, I., Kang, Y., Allen, S. N., Huhman, D. V., et al. (2014). Global reprogramming of transcription and metabolism in Medicago truncatula during progressive drought and after rewatering. Plant Cell Environ. 37, 2553–2576. doi: 10.1111/pce.12328
Zhang, L. L., Zhao, M. G., Tian, Q. Y., and Zhang, W. H. (2011). Comparative studies on tolerance of Medicago truncatula and Medicago falcata to freezing. Planta 234, 445–457.
Zhang, X. X., Wang, T. Z., Liu, M., Sun, W., and Zhang, W. H. (2019). Calmodulin-like gene MtCML40 is involved in salt tolerance by regulating MtHKTs transporters in Medicago truncatula. Environ. Exp. Bot. 157, 79–90.
Zhang, P. P., Li, S. S., Guo, Z. F., and Lu, S. Y. (2019). Nitric oxide regulates glutathione synthesis and cold tolerance in forage legumes. Environ. Exp. Bot. 167, 103851–103858.
Zhang, Z., Hu, X., Zhang, Y., Miao, Z., Xie, C., Meng, X., et al. (2016). Opposing control by transcription factors MYB61 and MYB3 increases freezing tolerance by relieving C-repeat binding factor suppression. Plant Physiol. 172, 1306–1323. doi: 10.1104/pp.16.00051
Zhang, Z., Wei, X., Liu, W., Min, X., Jin, X., Ndayambaza, B., et al. (2018). Genome-wide identification and expression analysis of the fatty acid desaturase genes in Medicago truncatula. Biochem. Biophys. Res. Commun. 499, 361–367. doi: 10.1016/j.bbrc.2018.03.165
Zhao, M. G., Chen, L., Zhang, L. L., and Zhang, W. H. (2009). Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiol. 151, 755–767. doi: 10.1104/pp.109.140996
Zhao, M. G., Liu, W. J., Xia, X. Z., Wang, T. Z., and Zhang, W. H. (2014). Cold acclimation-induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene. Physiol. Plant. 152, 115–129. doi: 10.1111/ppl.12161
Zhou, Y., Zeng, L., Chen, R., Wang, Y., and Song, J. (2018). Genome-wide identification and characterization of stress-associated protein (SAP) gene family encoding A20/AN1 zinc-finger proteins in Medicago truncatula. Arch. Biol. Sci. 70, 87–98.
Keywords: Medicago truncatula, abiotic stresses, morphological regulation, physiological regulation, functional genes, transcription factors
Citation: Zhang X, Sun Y, Qiu X, Lu H, Hwang I and Wang T (2022) Tolerant mechanism of model legume plant Medicago truncatula to drought, salt, and cold stresses. Front. Plant Sci. 13:847166. doi: 10.3389/fpls.2022.847166
Received: 01 January 2022; Accepted: 15 August 2022;
Published: 07 September 2022.
Edited by:
Mukesh Jain, Jawaharlal Nehru University, IndiaReviewed by:
Fuai Sun, University of California, Davis, United StatesCopyright © 2022 Zhang, Sun, Qiu, Lu, Hwang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianzuo Wang, dHp3YW5nQGliY2FzLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.