- 1MSU DOE-Plant Research Laboratory, Michigan State University, East Lansing, MI, United States
- 2Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, United States
- 3Department of Microbiology and Molecular Genetics, Michigan State University, East Lansing, MI, United States
Phytochromes (phy) are key regulators of photomorphogenesis in plants. Among the different phys characterized in higher plants (i.e., phyA to phyE), phyA and phyB primarily regulate phenotypic responses in plants under far-red (FR) and red (R) conditions, respectively. Recent findings suggest that some zinc finger proteins (ZFPs) are involved in plant light-modulated morphogenesis. However, the interaction(s) between phyA, phyB and ZFP homologs potentially involved in photomorphogenesis, as well as their phenotypic and molecular effects in Arabidopsis seedlings exposed to R and FR light remain to be elucidated fully. Prior analyses with phytochrome chromophore deficient lines indicated that ZFP6 expression is misregulated compared to levels in Col-0 wild type (WT). Here, we used plants with phytochrome chromophore or apoprotein (specifically phyA and phyB) deficiencies, lines with mutations in ZFP6 and ZFP6 HOMOLOG (ZFPH) genes, and plants overexpressing ZFP6 to examine regulatory interactions between phytochromes, ZFP6, and ZFPH. Our results indicate that phytochromes are required for downregulation of ZFP6 and ZFPH and suggest a role for light-regulated control of ZFP levels in phytochrome-dependent photomorphogenesis. Conversely, PHYB is downregulated in zfp6 mutants under R light. Analyses of a zfp6zfph double mutant confirmed disruption in photomorphogenic phenotypes, including the regulation of hypocotyl elongation in seedlings grown under FR light. In addition, PIF3 and PIF4 levels are transcriptionally regulated by ZFP6 and ZFPH in a gibberellic acid-dependent manner. ZFP6 overexpression resulted in opposite phenotypic responses to those observed in the zfp6 and zfph mutants grown in FR and R light, as well as a reduction in the rosette size of mature ZFP6 OX plants relative to WT under white light. Based on these observations, we provide insight into how phy and ZFPs interact to regulate specific aspects of light-dependent processes in Arabidopsis.
Introduction
Light controls multiple and critical processes throughout the plant life cycle. Aspects of plant growth and development regulated by light include seed germination, etiolation or de-etiolation behaviors in seedlings, responses to neighboring plants in competition for light, and the shift between vegetative and reproductive stages, among others (Fankhauser and Chory, 1997). These light-dependent growth and developmental processes are mediated by light perception by photoreceptors throughout the life cycle of plants, including phytochromes, cryptochromes, phototropins, and UVR8 (Legris et al., 2019). Phytochrome (phy) A (phyA) and phyB are the most extensively studied photoreceptors; they are the predominant phytochromes that control photomorphogenic responses in the presence of far-red (FR) and red (R) light, respectively (Li et al., 2011; Cheng et al., 2021; Kim et al., 2021). Encoded by genes in the nucleus, phy proteins are synthesized and the chromophore covalently attached in the cytoplasm; holophytochromes remain in the cytosol in their inactive form (Pr) if no activating light is present, or upon light-activated conversion to their active form (Pfr) are translocated into the nucleus (Kevei et al., 2007). In the nucleus, phytochromes control distinct classes of regulatory genes, including those encoding transcription factors.
Zinc finger proteins (ZFPs) are one class of transcription factor families that are widely distributed in plants. ZFPs participate in numerous biological processes, including flowering, light-mediated morphogenesis, disease suppression, and activation of defense mechanisms in response to abiotic stress (Feurtado et al., 2011; Noman et al., 2019; Xie et al., 2019). They are classified into nine families based on their conserved cysteine-histidine-amino acid motif, which coordinates with a zinc atom (Xie et al., 2019). The largest group comprises 176 C2H2-type ZFP proteins (Englbrecht et al., 2004). ZFPs have been shown to have DNA-binding activity in plants, indicating roles for these proteins in transcriptional regulation (Han et al., 2020). While one of the larger protein families, this group of regulatory proteins have been underexplored in planta (Fedotova et al., 2017). ZFP6 and closely related ZFP6 HOMOLOG (ZFPH) are of particular interest in this current research given their identification as differentially regulated genes in prior transcriptomic analyses of phytochrome-deficient plant lines (Oh et al., 2013).
Prior experimental analyses demonstrated that ZFP6 overexpression in 35S:ZFP6 transgenic lines led to an increased number of trichomes on the sepals of flowers, in addition to ectopic trichome formation on carpels in Arabidopsis (Zhou et al., 2013). Of note, exogenous gibberellic acid (GA) application induced significantly higher ZFP6 expression compared to untreated plants, indicating interaction between GA signaling and ZFP6 function (Zhou et al., 2013). The GA hormone is implicated in several plant development stages in Arabidopsis, including control of seed germination, promotion of stem elongation and leaf expansion, and the induction of flowering (Phillips, 1998). In addition to ZFP6, ZFP5 also aids in the GA pathway to induce trichome initiation on shoots in Arabidopsis (Zhou et al., 2011). Molecular-based approaches suggested that ZFP6 regulates ZFP5 expression (Zhou et al., 2013) and ZFP5 in turn induces GLABROUS INFLORESCENCE STEMS 1 (GIS1), GLABROUS INFLORESCENCE STEMS 2 (GIS2), and ZFP8 expression (Zhou et al., 2011). GIS genes also encode C2H2-type ZFPs. ZFPH was previously identified as GIS3 and also was demonstrated to increase trichome density when overexpressed (Sun et al., 2015). ZFPH/GIS3, however, exerts its impact on trichomes independent of ZFP6 and ZFP5; yet, impacts GIS1, GIS2, and ZFP8 similar to ZFP5 (Sun et al., 2015). Another ZFP family member, i.e., ZFP3, impacts seedling development, but through a distinct mechanism. Overexpression of ZFP3 interfered with the ABA signaling pathway in Arabidopsis, rendering the seeds unable to germinate. Additionally, seedlings overexpressing ZFP3 displayed shorter hypocotyls both in light and dark conditions (Joseph et al., 2014). Together, these results indicate multiple roles for ZFP homologs in plant growth and development, including some phenotypes that overlap with those controlled by light and phytochromes.
PIFs (Phytochrome-Interacting Factors) are phytochrome-dependent transcription factors that have been shown to physically interact with phytochromes and to activate organ-elongation genes and promote etiolation (Leivar and Monte, 2014), i.e., the dark-dependent development of seedlings with long stems and small, yellow-colored cotyledons. During de-etiolation, R light-dependent activation of phyB leads to degradation of PIFs and characteristic inhibition of stem elongation and promotion of leaf development and greening (Leivar and Monte, 2014). In addition to impacting PIFs, phyB inhibits the morphogenetic repressor COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1) in a light-dependent manner, restraining its ability to target the transcription factor ELONGATED HYPOCOTYL 5 (HY5) for proteasome-mediated degradation (Osterlund and Deng, 1998). Thus, HY5 accumulates in the light and promotes photomorphogenesis in plants (Osterlund et al., 2000; Shi et al., 2018). Conversely to its R-dependent movement into the nucleus, phyB remains in its inactive red-light absorbing form (Pr) in the cytoplasm under FR light conditions. When phyB remains in the cytosol in FR, PIF molecules are able to accumulate in the nucleus where they function to transcribe PIF target genes, including those that promote elongation (Ejaz et al., 2021). PIF proteins intersect with hormone-based regulation of growth as targets of the GA signaling pathway (Hernández-García et al., 2021). PIFs are targeted for inactivation by DELLA proteins, which are molecules that suppress growth (Kusnetsov et al., 2020). DELLAs restrain PIFs (PIF1, PIF3, PIF4, and PIF5) by targeting them for proteasome-mediated degradation in a light-independent manner (Li et al., 2016).
Given the prior associations of ZFP6 as a target of transcriptional regulation by phytochromes and GA regulation, as well as roles for both phytochromes and GA in light-dependent growth and development in Arabidopsis, we investigated light and phytochrome-dependent transcript accumulation for ZFP6 and ZFPH, light and GA-dependent phenotypic and molecular responses of zfp6 and zfph mutants, and the consequence of overexpressing the ZFP6 gene on light-dependent plant growth and development. To gain specific insights into the crosstalk between phytochromes, PIFs, and ZFP6 during the regulation of growth, we assessed expression of select genes within the phytochrome, GA, and organ elongation pathways. Considering these observations collectively, we describe specific aspects of phytochrome-dependent processes that are mediated via ZFP6 and closely related ZFPH in Arabidopsis.
Materials and Methods
Plant Materials
Col-0 wild type (WT) ecotype of Arabidopsis thaliana (hereafter Arabidopsis) was obtained from the Arabidopsis Biological Resource Center (ABRC).1 An Arabidopsis phyAphyB (PHYA: AT1G09570; PHYB: AT2G18790) double mutant line was previously constructed and described (Mayfield et al., 2007; Ruckle et al., 2007). zfp6 (SALK_200865; AT1G68360) and zfph (SALK_043793; AT1G68360) mutant lines were also obtained from ABRC, and the zfp6zfph double mutant was isolated from a genetic cross between the two single mutants. The production of transgenic BVR lines was previously described (Montgomery et al., 1999; Warnasooriya and Montgomery, 2009).
For ZFP6 overexpression lines, ZFP6 cDNA was amplified from a cDNA clone for ZFP6 (U13157) from ABRC using forward primer 5′-ATGGCGACTGAAACATCTTCTT-3′ and reverse primer 5′-TCATGGCCCAAGGCTTAAAT-3′ and recombined into the pCR™8/GW/TOPO™ vector using a TA Cloning Kit according to manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, United States). Insertion of the full-length ZFP6 cDNA fragment into the vector was confirmed via EcoRI digestion and validated by DNA sequencing. The recombinant vector was cloned into One Shot™ TOP10 E. coli cells (Thermo Fisher Scientific, Waltham, MA, United States) following the manufacturer’s directions. Full-length ZFP6 cDNA was recombined into the 35S promoter-containing pEarlyGate 100 vector using LR Clonase II enzyme according to manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, United States) to generate the 35S:ZFP6 construct, which was introduced into GV3101 Agrobacterium to transform Col-0 WT plants via a standard floral-dip transformation protocol (Clough and Bent, 1998). ZFP6 overexpression was confirmed by RT-PCR as described below.
Plant Growth and Light Sources
Arabidopsis seeds were sterilized with 2.88% (v/v) sodium hypochlorite including 0.025% (v/v) SDS for 15 min. Chlorine was removed by rinsing seeds with sterilized ddH2O five times. Then, the seeds were planted on 0.5 × Murashige and Skoog (MS) medium (Caisson Laboratories, Smithfield, UT, United States) containing 0.9% (w/v) Phytoblend (Caisson Laboratories) and 1% (w/v) sucrose (Thermo Fisher Scientific, Waltham, MA, United States). Seeds were stratified on agar plates at 4°C for 4 days in darkness and then replicate plates were incubated in white (W), far-red (FR) and red (R) light for 7 days at 22°C. Additional treatments included germination and/or growth of seedlings on plates with 10 μM GA and GA biosynthesis inhibitor paclobutrazol (PAC) at a concentration of 100 nM. For W light, a Percival chamber model no. CU36LA irradiating light at 110 μmol m–2 s–1 was used; for the rest of the tested lights (see below), Percival LED chambers (model E30LED; Percival, Perry, IA, United States) were employed. For continuous FR (FR; λmax ∼735 nm) light, the light was emitted at 5 μmol m–2 s–1; for R conditions (λmax ∼670 nm), the fluence rate was ∼25 to 50 μmol m–2 s–1; and for blue (B) conditions (λmax ∼ 470 nm), the fluence rate was ∼50 μmol m–2 s–1.
Phenotypic Analyses
Hypocotyl and root lengths of 7-days-old zfp6, zfph, and zfp6zfph mutant seedlings were measured using the ruler tool in Photoshop 2021 or using Image J. Similar measurements were performed on single copy, homozygous lines overexpressing ZFP6 grown for 7 days in MS media containing 1% sucrose and 0.7% agar, pH 5.7, under FR (5 μmol m–2 s–1), R (50 μmol m–2 s–1), and blue (50 μmol m–2 s–1) lights at 22°C. To document additional phenotype characteristics of mature lines overexpressing ZFP6, Col-0 WT and 35S:ZFP6 overexpression lines were grown in soil for 21 days at 22°C in W at ∼125 μmol m–2 s–1 under a 16 h light/8 h dark cycle. Plants were photographed to evaluate rosette architecture and trichome formation.
RNA Extraction and RT-PCR
Seven-day-old seedlings incubated in R and FR light at 22°C were harvested in green light conditions, while those grown in W light were harvested under room light. Collected seedlings were immediately submerged in liquid nitrogen and stored at –70°C. Total RNA was extracted using the E.Z.N.A. Plant RNA Kit (Omega Bio-Tek, Norcross, GA, United States), following the manufacturer’s instructions and including the DNase I digestion protocol. An additional DNA digestion was performed using DNase I RNase-free (Thermo Fisher Scientific, Waltham, MA, United States) using 1 unit per microgram of RNA. Total RNA (500 ng) was reverse-transcribed using a qScript cDNA SuperMix kit (Quantabio, Beverly, MA, United States). Real-time quantitative PCR (qPCR) was performed on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). The primers (at a final concentration of 200–500 nm) and cycling conditions are specified in Supplementary Table 1. A melting curve protocol was performed at the end of the PCR starting at 60°C with increments of 0.5°C/20 s. Three biological replicates along with three technical replicates were used. The UBC21 gene was used for normalizing purposes and gene expression analyses were conducted using the 2–ΔCT method. To confirm overexpression of ZFP6, standard RT-PCR was performed with UBC21 as the internal control using primers (final primer concentration 400 nm) and cycling conditions indicated in Supplementary Table 1.
In silico Promoter Analyses
Analyses of the ZFP6 and ZFPH promoter regions were performed using the PlantCare database2 to identify conserved cis-elements potentially involved in gene regulation. Approximately 1,000 nucleotides upstream of the start codon of each gene were analyzed to search for transcription start (TS) sites using the neural network promoter prediction with a minimum promoter score of 0.93 and for predicted transcription factor binding sites using the PlantCare database. In parallel, the TF2Network database (Kulkarni et al., 2018)4 was used to investigate potential light- and/or phytochrome-dependent regulators for ZFP6 and ZFPH.
Statistical Analysis
ANOVA analysis was performed to examine significant differences in the means. A normal distribution of the data was evaluated by the Kolmogorov-Smirnov test. Data that did not follow a normal distribution were transformed using the Box-Cox algorithm. The Fisher test at p ≤ 0.05 was conducted to evaluate significant differences among population means. All statistical analyses and graphs were generated on OriginPro 2018.
Results
Phytochrome A and Phytochrome B Negatively Regulate ZFP6 and ZFPH Transcript Levels
Mining of previous transcriptomic data indicated that ZFP6 and ZFPH were differentially regulated in phytochrome chromophore-deficient transgenic BVR lines grown in FR light conditions (Oh et al., 2013). BVR (biliverdin IX reductase) inactivates the tetrapyrrole precursors required for synthesis of the phytochrome chromophore, phytochromobilin; thus, BVR induces a chromophore deficiency in transgenic plants (Montgomery et al., 1999). The mRNA levels for both ZFP6 and ZFPH were significantly higher in FR-grown CAB3:pBVR lines that lack the accumulation of photoactive phytochromes in mesophyll cells of leaves (Figures 1A,B; Oh et al., 2013). To confirm this finding for ZFP6, we assessed its expression by quantitative, real-time PCR (qRT-PCR) analysis. ZFP6 mRNA levels were ∼2.6-fold higher in a CAB3:pBVR line than in Col-0 WT grown in FR light (Figure 1C). As the lack of phytochrome chromophore results in a lack of all holophytochromes, we used a phyAphyB mutant lacking the two predominant phytochromes to confirm that it was the lack of phytochromes in the BVR line which directly contributed to a disruption in transcript accumulation for ZFP homologs. Consistent with the phenotype for chromophore-deficient BVR-expressing plants, ZFP6 transcripts levels also were increased ∼2.5 fold in a phyAphyB T-DNA mutant line (Figure 1D), compared to Col-0 WT. These findings suggest that phyA and phyB are the primary phytochromes required to downregulate ZFP6.
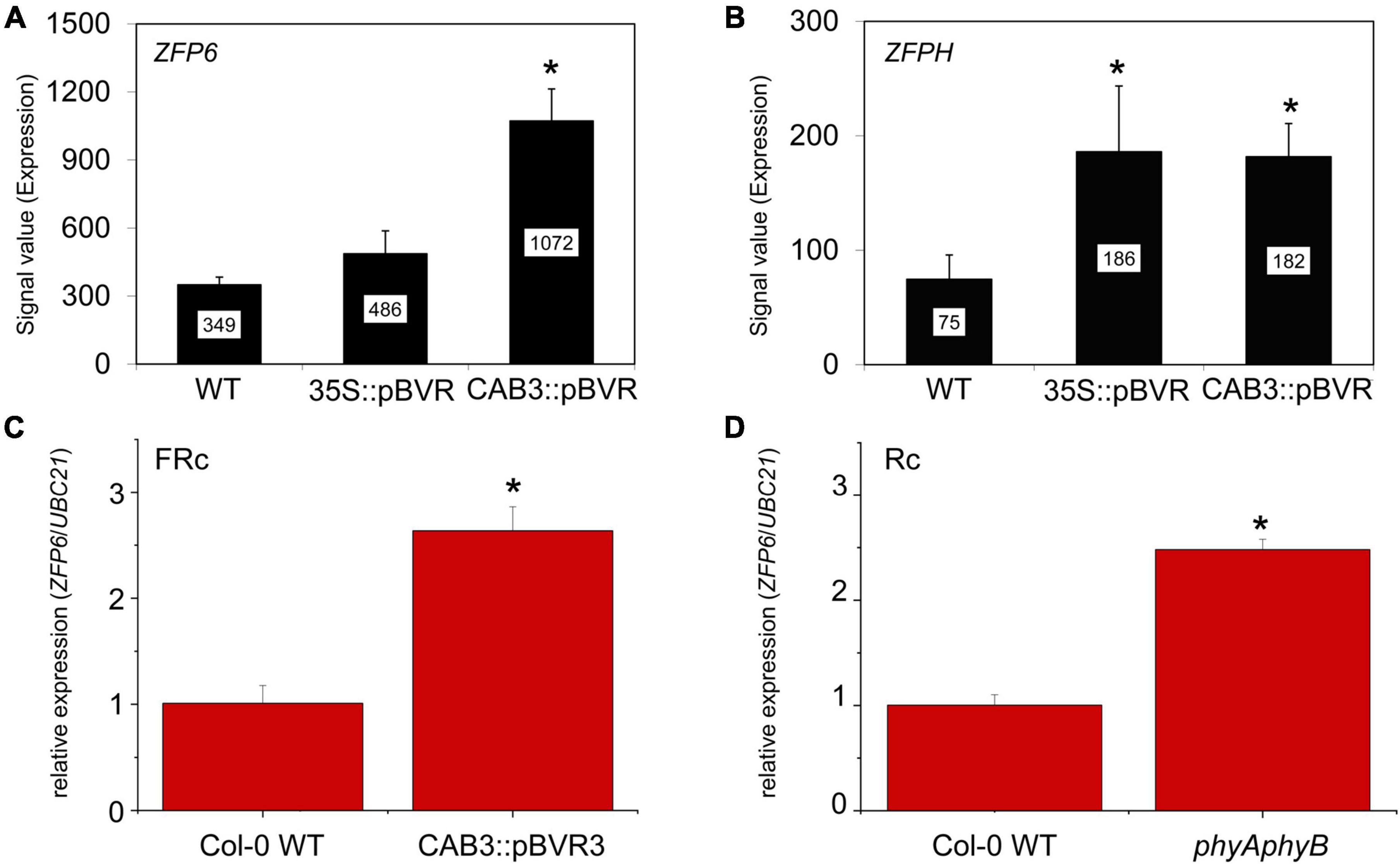
Figure 1. Relative expression of ZFP6 homologs in Arabidopsis seedlings. Expression levels (signal value) of (A) ZFP6 and (B) ZFPH in wild-type (WT), 35S:pBVR, and CAB3:pBVR seedlings grown under continuous far-red (FRc) from published data set (Oh et al., 2013) are shown (± SD, n = 3). Signal value indicates signal intensity on the ATH1 array as calculated by Affymetrix Microarray Suite (MAS). (C) ZFP6 relative expression in CAB3:pBVR3 line (Warnasooriya and Montgomery, 2009) in FRc light compared to WT. (D) ZFP6 relative expression in phyAphyB mutant in continuous red (Rc) light conditions compared to WT. (C,D) Seedlings were stratified at 4°C for 4 days in MS plates with 1% sucrose and then incubated in FRc or Rc light for 7 days. Gene expression data were obtained following the 2–ΔCT method using UBC21 as the reference gene. Means ± SD were calculated from at least three biological replicates. *p ≤ 0.05, relative to WT.
ZFP6 and ZFPH Are Expressed in Different Tissues and in Response to Distinct Light Conditions
Given the role of phys in regulating ZFP6 and ZFPH, we examined the expression of these genes and the closely related ZFP5 in different tissues and light conditions utilizing public microarray data for Col-0 WT from AtGenExpress5 (Figures 2A,B). We chose to examine expression of ZFP5 in parallel given that it is regulated by ZFP6 (Zhou et al., 2013), shares regulation of similar GA-dependent phenotypic responses as ZFP6 (Zhou et al., 2011), and controls expression of some of the same genes as ZFPH (Sun et al., 2015). ZFP5 shared some overlap with ZFP6 and ZFPH in terms of tissues in which it was expressed, including roots and hypocotyls; yet, ZFP5 was expressed to relatively higher levels in roots than either ZFP6 or ZFPH (Figure 2A). ZFP6 and ZFPH are highly expressed in roots, hypocotyls, and internodes, with ZFPH also exhibiting some expression in the shoot apex and inflorescence tissues (Figure 2A). We focused our subsequent analyses on the most closely related ZFP6 and ZFPH.
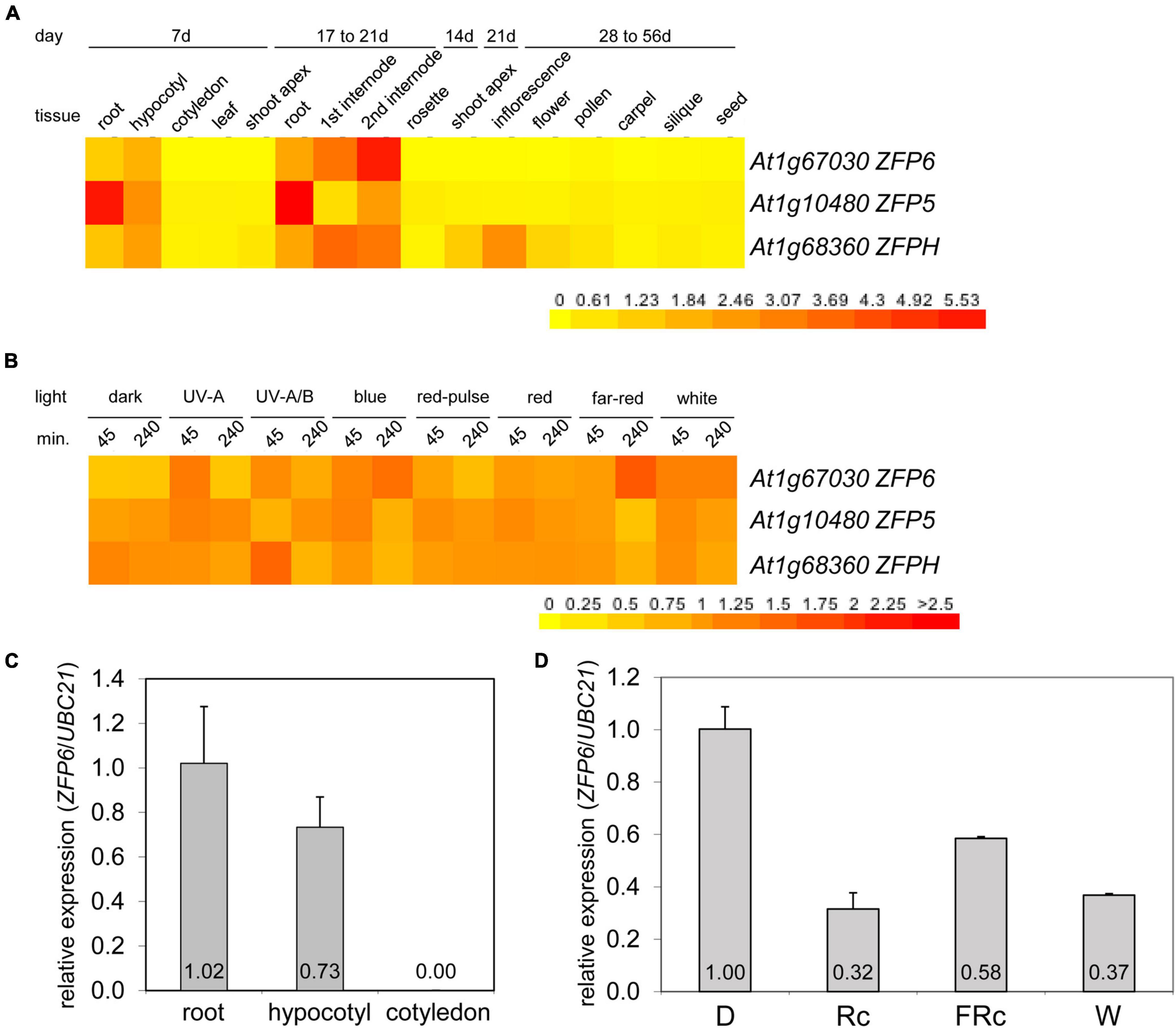
Figure 2. Expression of ZFP6, ZFP5, and ZFPH in different tissues and light conditions. Heat map showing the expression of ZFP6, ZFP5, and ZFPH in (A) different tissues or (B) different light conditions for Arabidopsis. For heat map, mean-normalized values of Col-0 WT from AtGenExpress expression library and BAR Heatmapper Plus (bar.utoronto.ca) were used. For light experiments in panel (B), aerial parts (hypocotyl and cotyledons) of 4-days-old Col-0 WT seedling grown on MS medium were treated with different light for either 45 or 240 min. (C) qRT-PCR analysis of ZFP6 expression in Col-0 WT root, hypocotyl, or cotyledon tissues from seedlings grown on MS medium containing 1% sucrose and 0.7% Phytoblend agar at 22°C for 7 days under white light (W) at 100 μmol m–2 s–1. (D) ZFP6 expression analyses in Col-0 WT grown as mentioned in (C) but under dark, continuous red (Rc; 50 μmol m–2 s–1), continuous far-red (FRc; 5 μmol m–2 s–1), or W (100 μmol m–2 s–1) light conditions. Relative ZFP6 expression level compared with UBC21 is shown (± SD, n = 3).
Using qRT-PCR, we confirmed the differential accumulation of ZFP6 mRNA in roots and hypocotyls, but not in cotyledons (Figure 2C). Additionally, ZFP6 is upregulated by light, with significant upregulation after 4 h of FR (10 μmol m–2 s–1) exposure according to public microarray data (Figure 2B). By comparison, ZFPH exhibits more moderate light-dependent changes in expression, with UV-A/B having the most significant impact. Given the association of multiple wavelengths of light that are correlated with phytochrome activity having a greater impact on ZFP6 induction, we documented that ZFP6 expression was 3. 1-, 1. 72-, and 2.7-fold downregulated in Col-0 WT exposed to continuous R, FR, and W light conditions, respectively, compared to Col-0 WT grown in dark (Figure 2D).
ZFP6 and ZFPH-Deficient Lines Exhibit Defects in Light-Dependent Phenotypes
Aiming to evaluate the phenotypic impact conferred by ZFP6 and ZFPH, we identified homozygous T-DNA mutants for ZFP6 (i.e., zfp6) and ZFPH (i.e., zfph). We also created a homozygous zfp6zfph double mutant via a genetic cross. Given the regulation of ZFP6 and ZFPH mRNA accumulation by phytochromes and by light for ZFP6, we examined seedling photomorphogenic phenotypes in R and FR light grown seedlings. In FR light, the zfp6, zfph, and zfp6zfph mutant lines all exhibited significantly longer hypocotyls (∼1.2-fold longer) than Col-0 WT (Figure 3A). Although mutant seedlings trended longer that WT under R light conditions, hypocotyl elongation was not significantly different in R conditions (Figure 3B). Noted differences in hypocotyl elongation were light-specific as there was no difference among WT, zfp6, zfph, and zfp6zfph for seedlings grown in darkness (Supplementary Figure 1).

Figure 3. Hypocotyl measurements in FR and R light-exposed seedlings. Seedlings were stratified at 4°C for 4 days in dark on MS plates with 1% sucrose, and subsequently incubated in continuous far-red FR light or R light for 7 days at 22°C. (A,B) Untreated seedlings on MS plates exposed to (A) FR or (B) R light. (C,D) Seedlings on MS plates with (+) or without (–) the addition of 10 μM GA under (C) FR or (D) R light. Hypocotyl measurements were performed using Photoshop 2021 with 3 plates per treatment, each containing 10 seedlings (n = 30). Significant differences are highlighted with different letters (A,B) or asterisks (C,D) at p ≤ 0.05. Representative images of seedlings under each condition are shown to the right of bar graphs and the bar in the images equals 1 cm.
In addition to the impact of light, seedling growth is tightly regulated by plant hormones. For instance, auxins and GA promote plant growth while abscisic acid is generally known as a plant-growth inhibitor. Given the importance of GA in promoting elongation in seedlings and the prior report of GA regulation of ZFP6 (Zhou et al., 2013), we evaluated the effect of GA or inhibition of GA accumulation using the pharmacological agent paclobutrazol (PAC) on zfp6, zfph, and zfp6zfph mutant lines. Seedling hypocotyl length was significantly increased by ∼1.3-fold on average in zfp6, zfph, and zfp6zfph seedlings treated with GA compared to their corresponding untreated seedlings in FR, which was slightly less than the 1.4-fold longer seedlings observed for GA-treated WT seedlings (Figure 3C). Adding the GA biosynthesis inhibitor PAC disrupted the germination process in all seedlings exposed to FR light, including Col-0 WT. To overcome this, seeds were stratified on MS media without PAC and grown under W light for ∼2 d to allow germination, after which, they were exposed to PAC under FR conditions. All FR-grown, PAC-treated seedlings exhibited significantly shorter hypocotyls than their untreated counterparts, with no significant differences observed between WT and mutants (Supplementary Figure 2A). For R light-grown seedlings, we observed a moderate increase in hypocotyl lengths for GA-treated seedlings compared to control conditions for all lines including WT (Figure 3D), although these differences were not statistically significant as they were under FR. R-light, PAC-treated seedlings exhibited significantly shorter hypocotyls than untreated seedlings for all lines tested inclusive of WT (Supplementary Figure 2B). We also assessed hypocotyl lengths of seedlings under W light, where there were no significant changes in hypocotyl length observed among Col-0 WT and the zfp6 mutants (Supplementary Figure 3A). There were also no significant changes in root lengths for any of the seedlings lines grown in R, FR, or W light (Supplementary Figures 3B–D).
ZFP6 Overexpression Is Sufficient to Inhibit Hypocotyl Elongation in R and Far-Red Light-Grown Seedlings
An absence of ZFP6 and ZFPH expression resulted in longer hypocotyls in FR light compared to WT; hence, we hypothesized that overexpression of ZFP6 or ZFPH may inversely result in shorter hypocotyls in seedlings. As expected, elongated hypocotyls observed in zfp6 seedlings were inversely shortened in ZFP6-overexpression (OX) lines (Figure 4). In multiple transgenic lines exhibiting elevated levels of ZFP6 mRNA (Figure 4A), the inhibition of hypocotyl elongation was impacted compared to WT and vector control (VC) lines (Figures 4B,C), particularly under R and FR conditions. The strongest reduction occurred under R light conditions for the homozygous ZFP6 #7-2 OX line, compared to Col-0 WT and VC seedlings. Hypocotyl elongation phenotypes were not affected in ZFP6 OX seedlings treated with blue light, with the exception of a reduction observed for ZFP6 #7-2 OX in blue light, suggesting a direct interaction between phys and ZFP6 in the regulation of hypocotyl length. There were no differences for any lines grown in darkness.
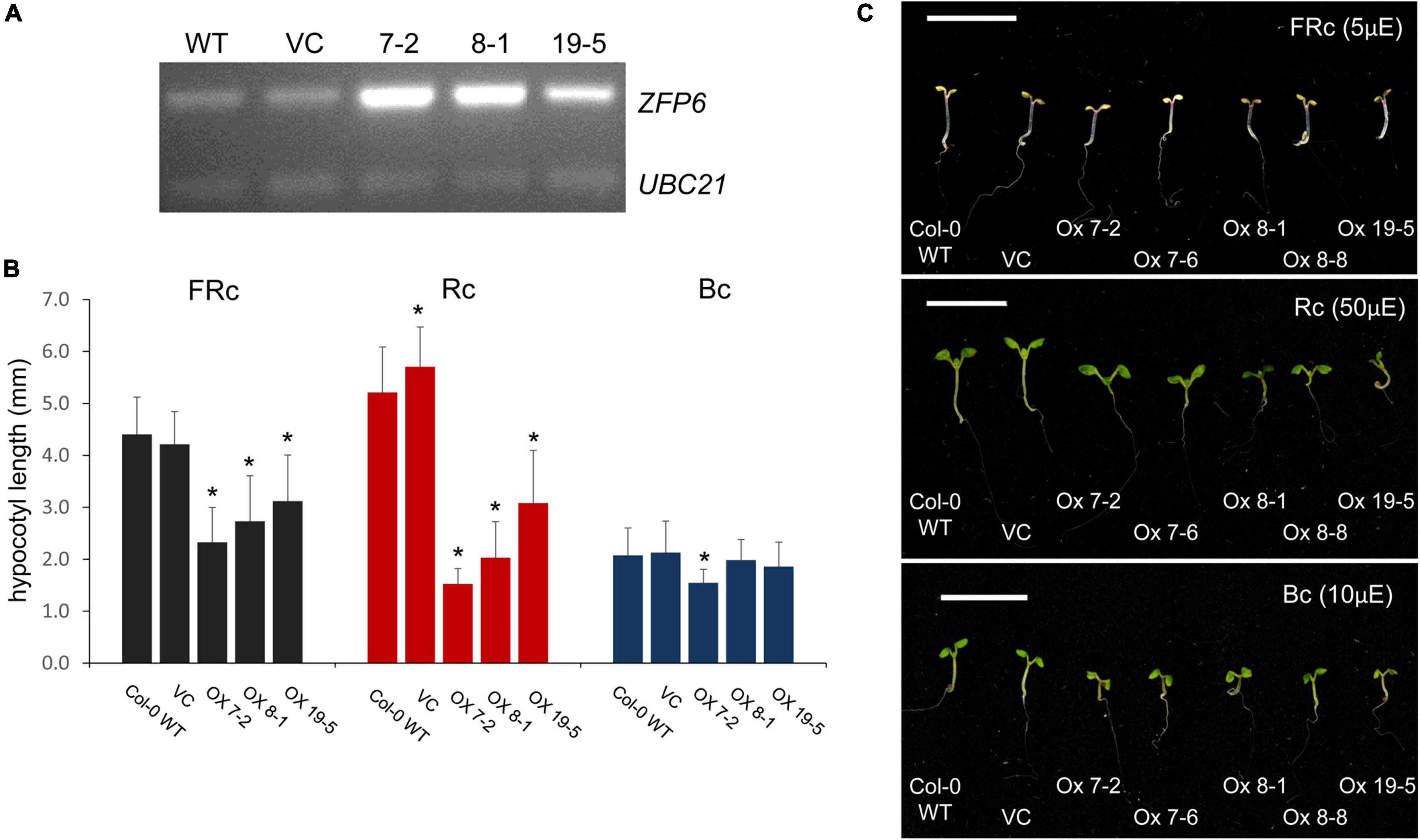
Figure 4. ZFP6 Overexpression and analyses of hypocotyl lengths of seedlings under different light conditions. (A) RT-PCR of ZFP6 mRNA levels relative to control gene UBC21 for wild-type, control lines transformed with an empty vector or vector control (VC), and three independent ZFP6 overexpression lines. (B,C) Seedlings were stratified at 4°C for 4 days in the dark, grown in MS plates with 1% sucrose, 0.7% agar, and subsequently incubated in different light conditions for 7 days at 22°C. Seedlings (Col-0 WT, VC lines, and different ZFP6 overexpression (OX) lines (i.e., OX 7-2, OX 7-6, OX 8-1, OX 8-8, and OX 19-5) were grown under continuous far-red (FRc; 5 μmol m–2 s–1 or μE), red (Rc; 50 μmol m–2 s–1 or μE), and blue (Bc; 10 μmol m–2 s–1 or μE) light. (B) Hypocotyl measurements were performed using Image J with at least 25 seedlings per line. Significant differences are shown with asterisks at p ≤ 0.05. (C) Representative images of seedlings are shown. Bar, 1 cm.
Besides the marked reductions in the lengths of hypocotyls observed for seedlings overexpressing ZFP6, we also noted other phenotypic changes for ZFP6 OX plants. Mature 21 day-old ZFP6 OX plants grown on soil at 22°C under long days (16 h light, 8 h dark) developed smaller rosettes than Col-0 WT or empty vector plants (Figure 5). Moreover, ZFP6 OX lines developed more trichomes on their rosette leaves than Col-0 WT plants, which is consistent with a previous report (Zhou et al., 2013).

Figure 5. Phenotypes of 35S:ZFP6 overexpression (OX) transgenic lines. (A) Rosette structure of Arabidopsis Col-0 WT, vector control (empty vector, i.e., V5-6), and the 35S:ZFP6 OX transgenic lines 7-2 and 8-1. Trichome formation in (B) Col-0 WT and (C) ZFP6 OX 7-2 line.
Light- and Growth-Responsive Genes Are Differentially Regulated in ZFP6- and ZFPH-Deficient Lines
Using qRT-PCR, we evaluated the expression of PIF3, PIF4, PHYB, and RGA1 (Repressor of GA1), key genes participating in the light- or hormone-dependent regulation of tissue growth (Figure 6; de Lucas et al., 2008; Leivar et al., 2008; Oh et al., 2014). RGA1 is one member of the DELLA family of proteins that binds to PIF3 and PIF4, inhibiting DNA binding activity of these PIF proteins and thus affecting the expression of PIF3- and PIF4-regulated genes (de Lucas et al., 2008; Feng et al., 2008). In R light, target gene PIF3 was ∼2.6-fold upregulated in Col-0 WT compared to W light-treated Col-0 WT (Figure 6A). Notably, PIF3 mRNA levels were significantly reduced in the zfph and zfp6zfph mutants (Figure 6A). This result indicated a positive role for ZFPH in R-dependent PIF3 mRNA accumulation.

Figure 6. PIF3, PIF4, PHYB, and RGA1 expression in seedlings grown in white, far-red, and red light conditions with or without GA. Seedlings were stratified at 4°C for 4 days in darkness and then incubated for 7 days at 22°C in white (W; 110 μmol m–2 s–1), continuous far-red (FRc; λmax ∼735 nm at 5 μmol m–2 s–1), or continuous red (Rc; λmax ∼670 nm at a fluence ∼25 μmol m–2 s–1) in the absence (A–D) or presence of GA (+ GA; E–H). UBC21 was used as the reference gene and the expression data was calculated using the 2–ΔCT method. Bars with different letters are significantly different.
In FR and R light conditions, PIF4 mRNA levels significantly increased in Col-0 WT by ∼3.7- and ∼5-fold, respectively, compared to W light (Figure 6B). These findings align with a previous report where PIF4 mRNA levels in Arabidopsis seedlings grown for 6 days in long days (16 h light, 8 h dark) increased ∼1.7–fold after the seedlings were incubated in continuous R light (Zhai et al., 2020), and prior reported upregulation of PIF4 in both R and FR (Huq and Quail, 2002). Under FR light, PIF4 expression was ∼1.5-fold reduced in zfp6 homolog mutant lines compared to Col-0 WT, although the difference was only significant for zfph. In zfp6 and zfph single mutant lines treated with R light, PIF4 transcripts were significantly upregulated by ∼1.3-fold compared to mRNA levels for R light-treated Col-0 WT seedlings.
PHYB expression levels were significantly higher in R light-treated seedlings than for W or FR light-exposed plants by an average of ∼3.1-fold (Figure 6C). It has been documented that PHY genes are generally constitutively expressed under different light conditions (Clack et al., 1994); however, another report suggests that PHYB is transcriptionally regulated (Somers and Quail, 1995). In R light, single and double zfp6 homolog mutants showed lower PHYB expression levels (∼1.7-fold reduction) than Col-0-WT, suggesting that ZFP6 is involved in upregulating PHYB.
RGA1 mRNA levels significantly increased in all R light-treated seedlings by ∼4.4-fold in comparison to W and FR light, with the exception of the zfp6zfph double mutant that had an increase but it was only marginally significant. However, no significant differences were detected among R light-treated Col-0 WT and R light-treated zfp6 mutant lines (Figure 6D).
Given that some ZFP genes exhibit cascade or reciprocal regulation and to facilitate interpretation of results for target genes, we tested whether ZFPH expression was impacted in a zfp6 mutant, as well as whether ZFP6 expression was impacted in the zfph mutant background. ZFP6 does not directly control ZFPH expression as ZFPH transcripts were present at near WT levels in zfp6 lines (Supplementary Figure 4). Likewise, ZFPH does not act upstream to impact ZFP6 as ZFP6 transcripts were present at near WT levels in zfph lines (Supplementary Figure 4).
Gibberellic Acid Modulates Light- Dependent mRNA Levels of Light- and Growth-Responsive Genes in ZFP6 and ZFPH Deficient Lines
Given the prior association of GA with an induction of ZFP6 expression and the noted impact of light and phytochromes on ZFP6 and ZFPH, we examined the impact of GA on the light- and growth-responsive genes assessed in WT, zfp6, zfph, and zfp6zfph lines. The addition of GA to the growth media resulted in a modulation of PIF3, PIF4, PHYB, and RGA1 expression levels in a light-dependent manner (Figure 6). The mRNA levels of PIF3 were not different among lines grown in the presence of GA; yet, this result in the presence of GA represents a loss of R light-associated induction of PIF3 mRNA levels in WT and zfp6 lines compared to growth in R light in the absence of GA (Figure 6A vs. Figure 6E). Thus, the R-induced accumulation of PIF3 appears to be dependent on both GA and ZFPH.
The mRNA levels of PIF4 were significantly upregulated in FR light-exposed seedlings compared to seedlings grown in R and W light in the presence of GA by ∼7.7- and 4-fold, respectively (Figure 6F). Notably, the pattern of PIF4 transcript levels in FR and R light-treated seedlings were inverted when the seedlings grew in the presence of GA (Figure 6B vs. Figure 6F). PIF4 mRNA levels were only significantly different in the zfp6zfph double mutant for GA-treated seedlings in FR light, suggesting a redundant role for the two factors under this condition.
By comparison, PHYB transcript levels were significantly lower in GA-treated zfp6, zfph, and zfp6zfph mutant seedlings under W light compared to WT (Figure 6G). While there were no significant differences in PHYB mRNA levels for any seedlings including WT when treated with GA under R light, PHYB levels were significantly reduced (∼4.8-fold) in GA-treated, R light-exposed seedlings compared to their untreated counterparts grown in R light (Figure 6C vs. Figure 6G). In FR light, PHYB was ∼2.6-fold upregulated in GA-treated zfph seedlings compared to GA-treated FR light-grown Col-0 WT, and also significantly upregulated in the zfp6zph double mutant (Figure 6G).
There was no significant impact of GA treatment on RGA mRNA levels in W or FR light conditions (Figure 6H). However, RGA1 mRNA levels were significantly reduced (∼7.5-fold) in GA-treated R-light exposed seedlings compared to R light-exposed seedlings grown without the addition of GA (Figure 6D vs. Figure 6H).
Together, these results indicate interactions between light, GA and, ZFP6/ZFPH in regulating the expression of some genes, including PIF3 and RGA1.
ZFP6 and ZFPH Promoters Contain Light-Responsive Elements and Are Potentially Regulated by Light-Induced Genes
Analyses of the ZFP6 and ZFPH promoter regions were conducted to identify cis-elements potentially involved in light responsiveness. Potential transcription start (TS) sites for ZFP6 and ZFPH were found at nucleotide –109 (score cutoff 0.98) and –80 (score cutoff 1.0), respectively, from the start codon. ZFP6 and ZFPH promoter region analyses resulted in the identification of different light-responsive motifs (Figure 7). For ZFP6, elements identified included G-Box, GA, GATA, GT1, and TCT motifs. The ZFPH promoter possessed two consensus sequences belonging to the TCT and Box 4 motifs. All these motifs have been previously documented as light-responsive elements (Shariatipour and Heidari, 2018).
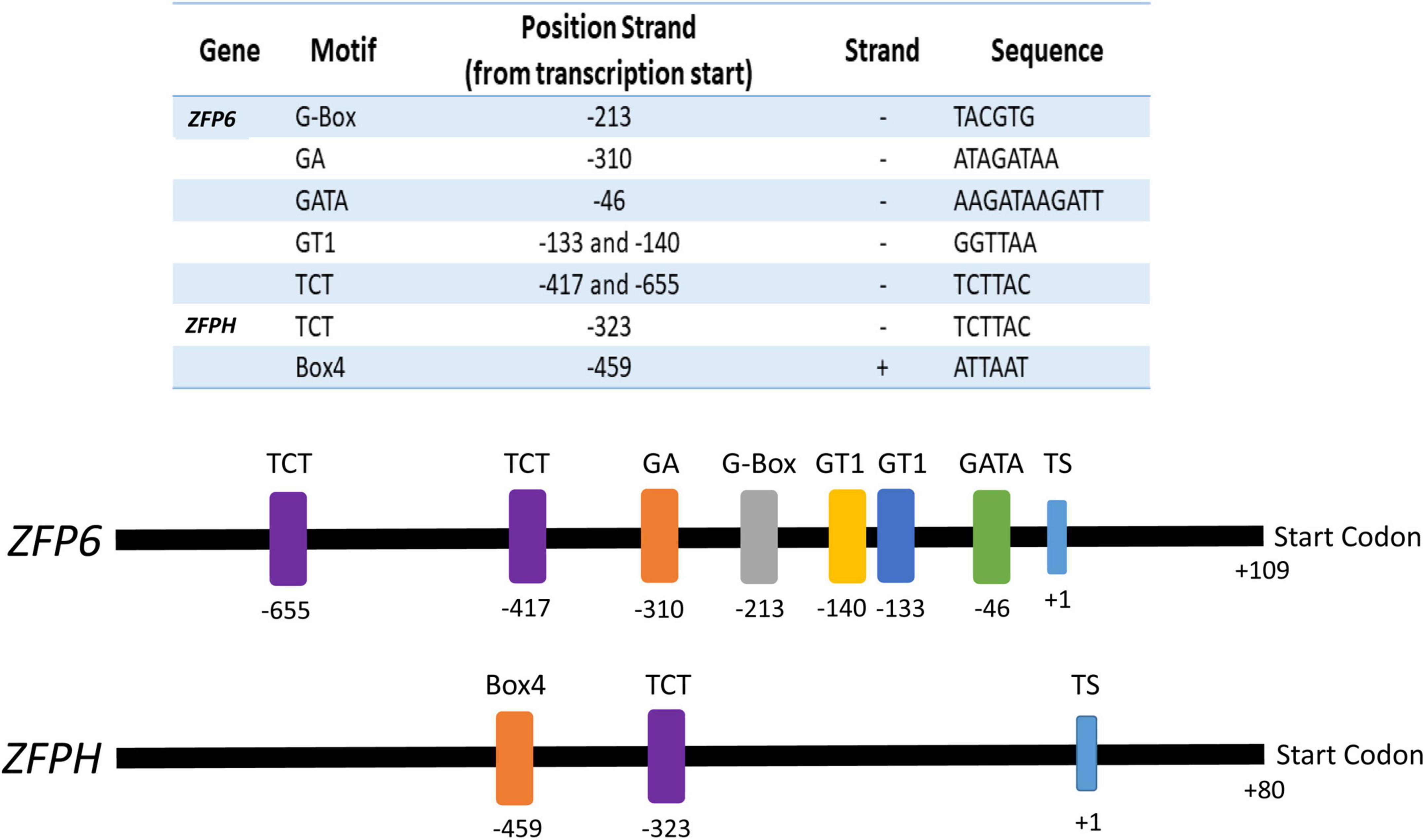
Figure 7. Promoter analyses for ZFP6 and ZFPH genes. The transcription start (TS) was identified using the neural network promoter prediction website and the putative light-responsive promoter regions were found using the PlantCare database (see text for details). Numbers indicate the motif and start codon positions. Sequence of the motif and the strand of DNA on which it is found (–, negative; +, positive) is indicated in the table.
To determine whether ZFP6 and ZFPH genes are potentially regulated by proteins encoded by light-responsive genes, an additional in silico analysis was performed using the TF2Network database (see text footnote 4). We compared the ZFP6 and ZFPH genes vs. 3,290 genes previously reported as light-responsive genes (Table 1; Bechtold et al., 2008; Shi et al., 2018). We found 9 and 17 genes that may encode proteins that bind and potentially regulate expression of ZFP6 and ZFPH, respectively. One notable factor predicted to regulate ZFPH is PIF4, which together with altered PIF4 levels in the zfph mutant in FR and R light conditions (Figure 6A) suggests an interesting potential feedback loop between ZFPH and PIF4. Many of the identified genes belong to the ZFP family, which indicates a cascade regulation among ZFP genes, as previously reported (Zhou et al., 2013). In addition, among genes predicted to encode factors that regulate ZFP6 and ZFPH are an overrepresentation of hormone-inducible genes, mainly those regulated by ABA (Supplementary Figure 2).
Discussion
Phytochromes negatively regulate ZFP6 and ZFPH expression. We, thus, investigated whether phytochrome-dependent regulation of ZFP6 and ZFPH is involved in controlling aspects of photomorphogenesis. To examine the interplay between phytochromes and ZFP6 and ZFPH during development, we analyzed the development of zfp6, zfph and zfp6zfph mutants under distinct light conditions. Given the prior association of ZFP6 induction by GA, we also examined the impact of modulating GA levels on development through treatment of seedlings with exogenous GA or a GA inhibitor. The zfp6, zfph, and zfp6zfph mutant lines exhibited significantly longer hypocotyls than Col-0 WT under FR light conditions. There was no specific effect of GA treatment or inhibition of GA accumulation on zfp6, zfph, or zfp6zfph seedling relative to WT in either R or FR light, indicating that the impacts of GA and phytochromes on these ZFP homologs may occur independently.
In FR light, PIFs escape phyB-mediated degradation as phyB remains in the cytosol and thus its transcriptional activity is blocked (Kevei et al., 2007); this FR-associated block of phyB translocation and a lack of associated phyB activity such as the downregulation of PIF4 in the nucleus promotes PIF4 accumulation and elongated hypocotyls (Fiorucci and Fankhauser, 2017). In line with this, we observed that PIF4 transcripts significantly increased in response to FR light in Col-0 WT (Figure 6B). As previously reported, PIF4 mRNA levels also increase in R light (Figure 6A; Zhai et al., 2020). Of note, the phytochrome-dependent regulation of a transcription factor that results in downregulation of PIF4 mRNA levels in FR light and upregulation in R light in deficient mutants was previously reported for sig2 mutants (Oh and Montgomery, 2013), which parallels the response noted here for zfp6 and zfph mutants. Of note, SIG2 is a regulatory factor also controlled by phyA and phyB and that impacts both PIF4 mRNA levels and hypocotyl elongation among other phenotypes (Oh and Montgomery, 2013). However, the regulation of PIF4 levels did not correspond with significantly longer hypocotyls in R or FR light for zfp6 and zfph mutants. Thus, although ZFP6 and ZFPH appear to exert positive transcriptional regulation on PIF4 under FR light and negative regulation under R light, this does not explain in full the significant disruption in hypocotyl elongation under FR. This finding may suggest that other members of the PIF family, or other factors altogether, may be involved in coordinating the observed etiolated responses in FR light where the hypocotyls of ZFP6 and ZFPH-deficient seedlings were significantly longer than WT. We also checked PIF3 mRNA levels in FR and its expression was not significantly changed under these conditions.
Under R light, our results imply that ZFP6 may limit hypocotyl elongation in part in WT by blocking PIF4 mRNA accumulation in R light (Figure 8), taking into consideration previous research that has shown consistency at the level of transcript levels and protein accumulation for PIF (Lee et al., 2021) and DELLAs (Zentella et al., 2007; Achard et al., 2008). To demonstrate whether ZFP6 is sufficient to inhibit hypocotyl elongation in seedlings, we created transgenic plants overexpressing ZFP6. The ZFP6 OX plants exhibited shorter hypocotyls than Col-0 WT Arabidopsis seedlings, especially those exposed to FR and R light conditions (Figure 4). These results confirm a key regulatory role of ZFP6 in restraining tissue elongation.
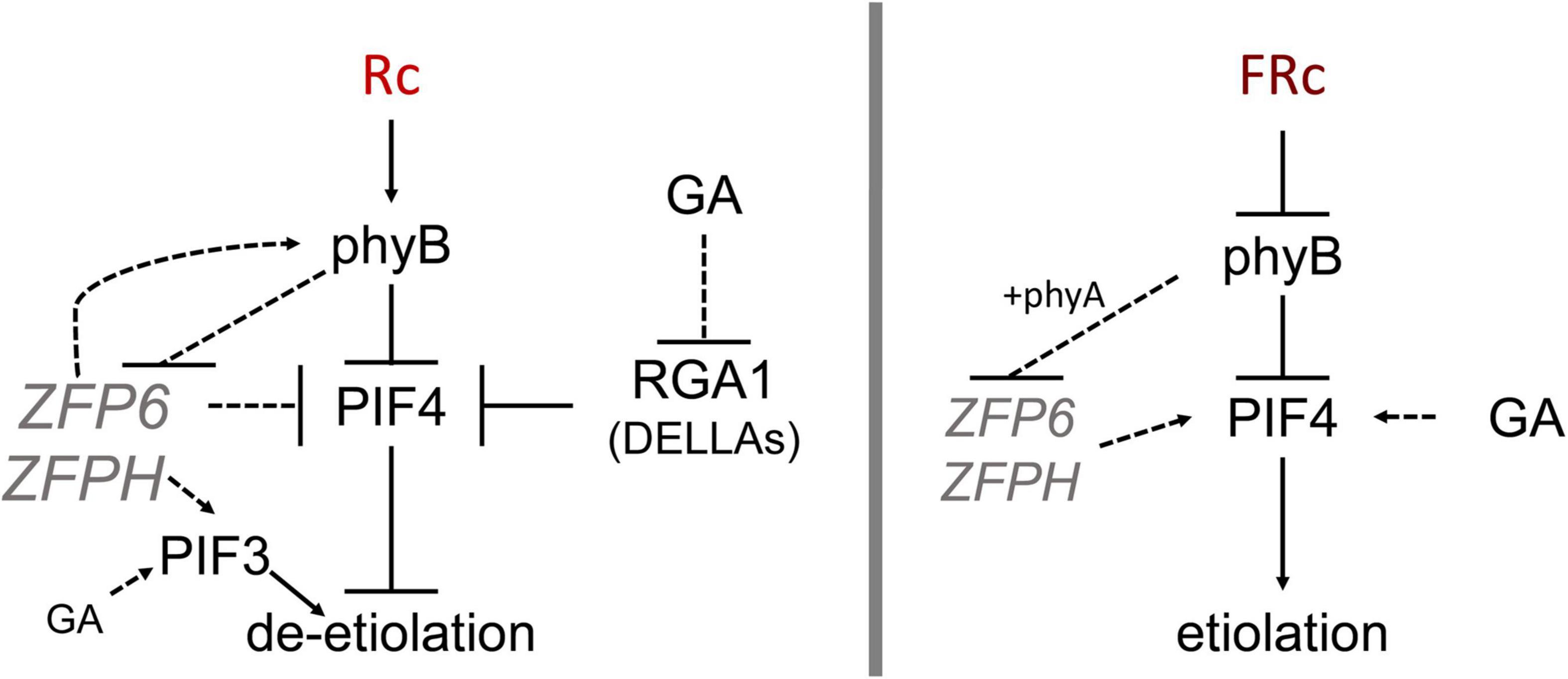
Figure 8. Model showing ZFP6 genetic interactions in red and far-red light. Published literature has demonstrated that phyB destabilizes PIFs while RGA1 (a DELLA family protein) blocks PIF4 (prior relationships represented with solid black lines). In red (R) light (left), phyB reduces ZFP6 expression. In turn, ZFP6 downregulates PIF4 and induces PHYB (relationships established in this work represented with dotted lines) in R light likely providing a feedback loop to aptly modulate hypocotyl lengths in response to light. ZFPH serves to promote PIF3 in R light, and PIF3 is also promoted by GA in R. In the presence of the growth-promoting GA hormone in R light, GA reduces RGA1 transcripts, which allows PIF4 accumulation and promotion of elongation in presence of GA. In far-red (FR) light (right), phyB together with phyA reduce ZFP6 (and ZFPH) expression. In the presence of the growth-promoting GA hormone in FR, GA promotes PIF4 accumulation leading to elongation typical of etiolation.
We observed elongated hypocotyls in all cases when GA was added. Additionally, Col-0 WT and all mutant seedlings treated with PAC displayed the same phenotypes independent of whether grown in R or FR light. These results indicate that DELLAs exert their impact on seedling elongation via an independent mechanism compared to ZFP6 and ZFPH, and that DELLAs likely serve as master regulators in response to GA.
As we observed increased ZFP6 mRNA levels for phyB mutants, we were also interested in evaluating the expression of PHYB in zfp6 mutant lines to test for reciprocal regulation. PHYB was downregulated in zfp6 and zfph mutant lines grown in R light, suggesting that ZFP6 is implicated in upregulating PHYB under these conditions (Figure 8). Indeed, by performing in silico analysis of the ZFP6 and ZFPH promoters, we identified several light-regulated motifs in the ZFP6 and ZFPH promoters. We also identified several proteins encoded by light-regulated genes that can potentially regulate ZFP6 and ZFPH. This finding aligns with prior analyses in which some members of the ZFP family have been previously associated with photomorphogenesis in plants (Ito et al., 2018).
Here, we report that hypocotyl elongation can be modulated at the seedling stage depending on ZFP6 and ZFPH phytochrome-dependent regulation. In addition to ZFP6 and ZFPH being regulated by light and phytochrome activity, ZFP6 and ZFPH regulate PHYB and PIF4 and PIF3, key components of the photomorphogenesis signaling cascade that can impact organ elongation genes. In mature plants, the rosette architecture is markedly reduced in lines overexpressing ZFP6, while the hairy trichomes become denser as previously reported (Zhou et al., 2013). Additional research is needed to fully elucidate the phytochrome and ZFP6/ZFPH-dependent regulatory network(s) that target organ-elongation genes and, ultimately, control light-dependent morphogenesis in planta.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
KC-R and SO designed and conducted the research, analyzed and interpreted data, and contributed to writing and editing the article. BM designed the research, analyzed and interpreted the data, and contributed to writing and editing article. All authors approved the submitted article.
Funding
This work was supported by the National Science Foundation (NSF; MCB-1243983 to BM) and the Office of Science of the U.S. Department of Energy (DE-FG02-91ER20021 to BM).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Hussien Alameldin for his assistance with genotyping experiments and growing plants.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.846262/full#supplementary-material
Footnotes
- ^ https://abrc.osu.edu/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ https://www.fruitfly.org/seq_tools/promoter.html
- ^ http://bioinformatics.psb.ugent.be/webtools/TF2Network/
- ^ https://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp
References
Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., and Genschik, P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. doi: 10.1105/tpc.108.058941
Bechtold, U., Richard, O., Zamboni, A., Gapper, C., Geisler, M., Pogson, B., et al. (2008). Impact of chloroplastic- and extracellular-sourced ROS on high light-responsive gene expression in Arabidopsis. J. Exp. Bot. 59, 121–133. doi: 10.1093/jxb/erm289
Cheng, M.-C., Kathare, P. K., Paik, I., and Huq, E. (2021). Phytochrome signaling networks. Annu. Rev. Plant Biol. 72, 217–244. doi: 10.1146/annurev-arplant-080620-024221
Clack, T., Mathews, S., and Sharrock, R. A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. doi: 10.1007/BF00043870
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313X.1998.00343.x
de Lucas, M., Daviere, J. M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. doi: 10.1038/nature06520
Ejaz, M., Bencivenga, S., Tavares, R., Bush, M., and Sablowski, R. (2021). Arabidopsis thaliana HOMEOBOX GENE 1 controls plant architecture by locally restricting environmental responses. Proc. Natl. Acad. Sci. U.S.A 118, 1–6. doi: 10.1073/pnas.2018615118
Englbrecht, C. C., Schoof, H., and Böhm, S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5:39. doi: 10.1186/1471-2164-5-39
Fankhauser, C., and Chory, J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13, 203–229. doi: 10.1146/annurev.cellbio.13.1.203
Fedotova, A. A., Bonchuk, A. N., Mogila, V. A., and Georgiev, P. G. (2017). C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Nat. 9, 47–58.
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Feurtado, J. A., Huang, D., Wicki-Stordeur, L., Hemstock, L. E., Potentier, M. S., Tsang, E. W. T., et al. (2011). The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23, 1772–1794. doi: 10.1105/tpc.111.085134
Fiorucci, A. S., and Fankhauser, C. (2017). Plant strategies for enhancing access to sunlight. Curr. Biol. 27, R931–R940. doi: 10.1016/j.cub.2017.05.085
Han, G., Lu, C., Guo, J., Qiao, Z., Sui, N., Qiu, N., et al. (2020). C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci. 11:115. doi: 10.3389/fpls.2020.00115
Hernández-García, J., Briones-Moreno, A., and Blázquez, M. A. (2021). Origin and evolution of gibberellin signaling and metabolism in plants. Semin. Cell Dev. Biol. 109, 46–54. doi: 10.1016/j.semcdb.2020.04.009
Huq, E., and Quail, P. H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21, 2441–2450. doi: 10.1093/emboj/21.10.2441
Ito, T., Okada, K., Fukazawa, J., and Takahashi, Y. (2018). DELLA-dependent and -independent gibberellin signaling. Plant Signal. Behav. 13:e1445933. doi: 10.1080/15592324.2018.1445933
Joseph, M. P., Papdi, C., Kozma-Bognár, L., Nagy, I., López-Carbonell, M., Rigó, G., et al. (2014). The Arabidopsis ZINC FINGER PROTEIN3 interferes with abscisic acid and light signaling in seed germination and plant development. Plant Physiol. 165, 1203–1220. doi: 10.1104/pp.113.234294
Kevei, E., Schafer, E., and Nagy, F. (2007). Light-regulated nucleo-cytoplasmic partitioning of phytochromes. J. Exp. Bot. 58, 3113–3124. doi: 10.1093/jxb/erm145
Kim, J. Y., Lee, J. H., and Park, C. M. (2021). A multifaceted action of phytochrome B in plant environmental adaptation. Front. Plant Sci. 12:659712. doi: 10.3389/fpls.2021.659712
Kulkarni, S. R., Vaneechoutte, D., Van De Velde, J., and Vandepoele, K. (2018). TF2Network: predicting transcription factor regulators and gene regulatory networks in Arabidopsis using publicly available binding site information. Nucleic Acids Res. 46:e31. doi: 10.1093/nar/gkx1279
Kusnetsov, V. V., Doroshenko, A. S., Kudryakova, N. V., and Danilova, M. N. (2020). Role of phytohormones and light in de-etiolation. Russ. J. Plant Physiol. 67, 971–984. doi: 10.1134/S1021443720060102
Lee, S., Wang, W., and Huq, E. (2021). Spatial regulation of thermomorphogenesis by HY5 and PIF4 in Arabidopsis. Nat. Commun. 12:3656. doi: 10.1038/s41467-021-24018-7
Legris, M., Ince, Y. Ç, and Fankhauser, C. (2019). Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 10:5219. doi: 10.1038/s41467-019-13045-0
Leivar, P., and Monte, E. (2014). PIFs: systems integrators in plant development. Plant Cell 26, 56–78. doi: 10.1105/tpc.113.120857
Leivar, P., Monte, E., Oka, Y., Liu, T., Carle, C., Castillon, A., et al. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823. doi: 10.1016/j.cub.2008.10.058
Li, J., Li, G., Wang, H., and Deng, X. W. (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9:e0148. doi: 10.1199/tab.0148
Li, K., Yu, R., Fan, L.-M., Wei, N., Chen, H., and Deng, X. W. (2016). DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 7:11868. doi: 10.1038/ncomms11868
Mayfield, J. D., Folta, K. M., Paul, A.-L., and Ferl, R. J. (2007). The 14-3-3 proteins μ and υ influence transition to flowering and early phytochrome response. Plant Physiol. 145, 1692–1702. doi: 10.1104/pp.107.108654
Montgomery, B. L., Yeh, K.-C., Crepeau, M. W., and Lagarias, J. C. (1999). Modification of distinct aspects of photomorphogenesis via targeted expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Physiol. 121, 629–640. doi: 10.1104/pp.121.2.629
Noman, A., Aqeel, M., Khalid, N., Islam, W., Sanaullah, T., Anwar, M., et al. (2019). Zinc finger protein transcription factors: integrated line of action for plant antimicrobial activity. Microb. Pathog. 32, 141–149. doi: 10.1016/j.micpath.2019.04.042
Oh, E., Zhu, J.-Y., Bai, M.-Y., Arenhart, R. A., Sun, Y., and Wang, Z.-Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 3:e03031. doi: 10.7554/eLife.03031
Oh, S., and Montgomery, B. L. (2013). Phytochrome-induced SIG2 expression contributes to photoregulation of phytochrome signaling and photomorphogenesis in Arabidopsis thaliana. J. Exp. Bot. 64, 5457–5472. doi: 10.1093/jxb/ert308
Oh, S., Warnasooriya, S. N., and Montgomery, B. L. (2013). Downstream effectors of light- and phytochrome-dependent regulation of hypocotyl elongation in Arabidopsis thaliana. Plant Mol. Biol. 81, 627–640. doi: 10.1007/s11103-013-0029-0
Osterlund, M. T., and Deng, X.-W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16, 201–208. doi: 10.1046/j.1365-313x.1998.00290.x
Osterlund, M. T., Hardtke, C. S., Wei, N., and Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462. doi: 10.1038/35013076
Phillips, A. L. (1998). Gibberellins in Arabidopsis. Plant Physiol. Biochem. 36, 115–124. doi: 10.1016/S0981-9428(98)80096-X
Ruckle, M. E., DeMarco, S. M., and Larkin, R. M. (2007). Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19, 3944–3960. doi: 10.1105/tpc.107.054312
Shariatipour, N., and Heidari, B. (2018). Investigation of drought and salinity tolerance related genes and their regulatory mechanisms in Arabidopsis (Arabidopsis thaliana). Open Bioinform. J. 11, 12–28. doi: 10.2174/1875036201811010012
Shi, H., Lyu, M., Luo, Y., Liu, S., Li, Y., He, H., et al. (2018). Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc. Natl. Acad. Sci. U.S.A 115, 6482–6487. doi: 10.1073/pnas.1803861115
Somers, D. E., and Quail, P. H. (1995). Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J. 7, 413–427. doi: 10.1046/j.1365-313X.1995.7030413.x
Sun, L., Zhang, A., Zhou, Z., Zhao, Y., Yan, A., Bao, S., et al. (2015). GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol. 206, 220–230. doi: 10.1111/nph.13218
Warnasooriya, S. N., and Montgomery, B. L. (2009). Detection of spatial-specific phytochrome responses using targeted expression of biliverdin reductase in Arabidopsis. Plant Physiol. 149, 424–433. doi: 10.1104/pp.108.127050
Xie, M., Sun, J., Gong, D., and Kong, Y. (2019). The roles of Arabidopsis C1-2i subclass of C2H2-type zinc-finger transcription factors. Genes 10:653. doi: 10.3390/genes10090653
Zentella, R., Zhang, Z.-L., Park, M., Thomas, S. G., Endo, A., Murase, K., et al. (2007). Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037–3057. doi: 10.1105/tpc.107.054999
Zhai, H., Xiong, L., Li, H., Lyu, X., Yang, G., Zhao, T., et al. (2020). Cryptochrome 1 inhibits shoot branching by repressing the self-activated transciption loop of PIF4 in Arabidopsis. Plant Commun. 1:100042. doi: 10.1016/j.xplc.2020.100042
Zhou, Z., An, L., Sun, L., Zhu, S., Xi, W., Broun, P., et al. (2011). Zinc finger protein5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis. Plant Physiol. 157, 673–682. doi: 10.1104/pp.111.180281
Keywords: phytochrome, ZFP6, ZFPH, gibberellic acid, PIF, DELLA, far-red light
Citation: Cota-Ruiz K, Oh S and Montgomery BL (2022) Phytochrome-Dependent Regulation of ZFP6 and ZFPH Impacts Photomorphogenesis in Arabidopsis thaliana. Front. Plant Sci. 13:846262. doi: 10.3389/fpls.2022.846262
Received: 31 December 2021; Accepted: 10 May 2022;
Published: 01 June 2022.
Edited by:
Gabriela Toledo-Ortiz, Lancaster University, United KingdomReviewed by:
Miguel De Lucas, Durham University, United KingdomEugenio Gómez Minguet, Polytechnic University of Valencia, Spain
Copyright © 2022 Cota-Ruiz, Oh and Montgomery. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beronda L. Montgomery, bW9udGcxMzNAbXN1LmVkdQ==
 Keni Cota-Ruiz
Keni Cota-Ruiz Sookyung Oh1
Sookyung Oh1 Beronda L. Montgomery
Beronda L. Montgomery