
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 25 April 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.844545
This article is part of the Research TopicAbiotic Stress: Molecular Genetics and Genomics, Volume IIView all 32 articles
 Hong-Ru Xu1,2†
Hong-Ru Xu1,2† Ying Liu2†
Ying Liu2† Tai-Fei Yu2†
Tai-Fei Yu2† Ze-Hao Hou2
Ze-Hao Hou2 Jia-Cheng Zheng3
Jia-Cheng Zheng3 Jun Chen2
Jun Chen2 Yong-Bin Zhou2
Yong-Bin Zhou2 Ming Chen2
Ming Chen2 Jin-Dong Fu2
Jin-Dong Fu2 You-Zhi Ma2
You-Zhi Ma2 Wen-Liang Wei1*
Wen-Liang Wei1* Zhao-Shi Xu2*
Zhao-Shi Xu2*Tubby-like proteins (TLPs) are transcription factors that are widely present in eukaryotes and generally participate in growth and developmental processes. Using genome databases, a total of 22 putative TLP genes were identified in the soybean genome, and unevenly distributed across 13 chromosomes. Phylogenetic analysis demonstrated that the predicted GmTLP proteins were divided into five groups (I-V). Gene structure, protein motifs, and conserved domains were analyzed to identify differences and common features among the GmTLPs. A three-dimensional protein model was built to show the typical structure of TLPs. Analysis of publicly available gene expression data showed that GmTLP genes were differentially expressed in response to abiotic stresses. Based on those data, GmTLP8 was selected to further explore the role of TLPs in soybean drought and salt stress responses. GmTLP8 overexpressors had improved tolerance to drought and salt stresses, whereas the opposite was true of GmTLP8-RNAi lines. 3,3-diaminobenzidine and nitro blue tetrazolium staining and physiological indexes also showed that overexpression of GmTLP8 enhanced the tolerance of soybean to drought and salt stresses; in addition, downstream stress-responsive genes were upregulated in response to drought and salt stresses. This study provides new insights into the function of GmTLPs in response to abiotic stresses.
The Tubby-like proteins (TLPs) are a class of eukaryotic transcription factors that were originally identified in obese mice (Kleyn et al., 1996; Liu, 2008). A typical TLP has a highly conserved tubular domain composed of 270 amino acids at the C-terminal, forming a β-barrel with a central hydrophobic α-helix and 12 antiparallel strands. It binds specific phosphatidylinositol 4,5-diphosphates to properly connect to the plasma membrane (Santagata et al., 2001; Mukhopadhyay and Jackson, 2011). TLPs have been widely studied in animals. For example, TULP3 is defined as a universal adapter for the transport of integral membrane proteins in the ciliary membrane to the cilia (Badgandi et al., 2017). Mutations of TLPs in humans lead to delayed obesity (Coleman and Eicher, 1990; Kleyn et al., 1996; Noben-Trauth et al., 1996; Kapeller et al., 1999; Borman et al., 2014), and mice with TLP mutations develop retinal degeneration, neurosensory hearing loss, and insulin resistance (Stretton et al., 2009).
In addition to the typical C-terminal tubular domain, plant TLPs have a conserved N-terminal F-box domain, which is not present in mammalian TLPs (Gagne et al., 2002; Lai et al., 2004). Previous studies have shown that TLPs have a variety of functions in plants, including growth, development, and disease resistance. TLPs may function in pollen grains, consistent with the fact that AtTLP6, AtTLP7, and AtTLP2 are mainly expressed in pollen grains of Arabidopsis (Bao et al., 2014). AtTLP2 is involved in the biosynthesis of homogalacturonic acid in Arabidopsis seed coat mucilage (Wang et al., 2019). Fourteen TLP genes have been identified in rice (OsTLPs), and differential expression analysis confirmed that the members of this group play important roles in processes related to physiological development (Liu, 2008). Expression of each OsTLP was induced by infection with Xanthomonas oryzae pv. oryzae, indicating that the OsTLP family is involved in host–pathogen interaction (Kou et al., 2009). OsTLP2 can bind to the OsWRKY13 promoter to regulate rice resistance to fungal plague and bacteria (Cai et al., 2008). Tomato SlTLP1 and SlTLP2 may have important roles in ethylene-dependent fruit ripening (Zhang et al., 2020); SlTLFP8 regulates cell size and stomatal density through endoreduplication, reduces water loss, and enhances water use efficiency (Li et al., 2020).
Previous reports have demonstrated the responses of TLPs to various abiotic stresses. During seed germination and seedling growth, AtTLP3 responds to abiotic stresses such as abscisic acid (ABA), NaCl, and mannitol (North et al., 1997; Bao et al., 2014). AtTLP9 regulates ABA sensitivity during seed germination and early seedling development (Lai et al., 2004; Chen et al., 2020). Overexpression of CaTLP1 in chickpeas can enhance tolerance to drought, salt stresses, and ABA (Bhushan et al., 2007). In apples, polyethylene glycol (PEG) treatment up-regulates expression of MdTLP1-MdTLP5 and MdTLP9 (Xu et al., 2016). Overexpression of apple MdTLP7 enhances the tolerance of Arabidopsis to osmotic, salt, and temperature stresses (Xu et al., 2019). ZmTLP2 and ZmTLP11 are significantly up-regulated in maize under drought stress (Chen et al., 2016). CsTLP8 plays a negative regulatory role in osmotic stress in cucumber, and its effects may be related to ABA (Li S. et al., 2021). Transcriptome analysis has shown that cotton GhTULPs are involved in abiotic stresses and tissue development. Overexpression of GhTULP34 was shown to decrease the germination rate of Arabidopsis seeds under salt stress, inhibit root development under osmotic stress, and lead to the closure of plant stomata (Li Z. et al., 2021). In summary, TLPs play key roles in plant growth and development and in responses to biotic and abiotic stresses.
Soybean (Glycine max) is one of the most economically important crops in the world, often used as a source of food for humans and livestock because of its rich oil and protein (Papiernik et al., 2005). As global climate change occurs, the adaptability of soybean to its living environment is gradually reduced, causing a demand for stress-tolerant soybean varieties. Further studies are needed to improve soybean tolerance to extreme environments, including various abiotic stresses such as drought and salt (Le et al., 2012). There is little published information about TLPs and their relationship with abiotic stress mechanisms in soybean. In this study, 22 TLP genes were identified in the soybean genome, and bioinformatic analyses were conducted to determine their chromosomal locations, gene structures, protein domains, conserved motifs, three-dimensional structures, and cis-acting elements. Based on RNA-Seq and quantitative Real-Time Polymerase Chain Reaction (qRT-PCR), we further investigated the role of GmTLP8 in drought and salt stress responses in soybean, and found that overexpression of GmTLP8 enhanced tolerance to drought and salt stresses in soybean. These findings provide insights into the function of GmTLP8, specifically in abiotic stress responses, and into the importance of GmTLPs more broadly in plant abiotic stress responses.
Soybean genome, protein, complementary DNA (cDNA) sequences, and gene annotation files were obtained from NCBI1 and the Phytozome database2 (Finn et al., 2011; Fernandez-Pozo et al., 2015). The Hidden Markov Model (HMM) profile corresponding to the TLP Tub domain (PF01167) from the Pfam protein family database3 was used to identify potential TLPs in the soybean genome (G. max Wm82. a2.v1) using HMMER v3 (Eddy, 1998; Mistry et al., 2021). Finally, the presence of the Tub domain in each TLP protein sequence was confirmed with the SMART tool4 (Letunic et al., 2012) and Pfam database. The molecular weight and isoelectric point data for GmTLPs were calculated by ExPASY5 (Artimo et al., 2012). Subcellular localization was predicted with WoLF PSORT6.
The full-length amino acid sequences of TLP members in rice (OsTLPs), Arabidopsis (AtTLPs), cotton (GhTLPs), maize (ZmTLPs), apple (MdTLPs), poplar (PtTLPs), wheat (TaTLPs), tomato (SlTLPs), and the newly identified GmTLPs were obtained from NCBI and Phytozome, respectively, and aligned with default parameters using ClustalW (Chenna et al., 2003). An unrooted phylogenetic tree was constructed using the neighbor-joining (NJ) method in MEGAX (version 10.1.8) (Tang et al., 2021) with the following parameters: pairwise deletion; Poisson model; 1000 bootstrap replications.
The amino acid sequence of 22 TLP proteins of soybean aligned using DNAMAN (version 6.0.3).
Chromosomal location data for GmTLPs were obtained from the Phytozome database. Intron insertion sites were identified by comparing the coding sequence of each TLP gene with the corresponding full-length sequence using the Gene Structure Display Server (GSDS) 2.07 (Hu et al., 2015). The conserved domain of the identified soybean GmTLP protein sequences were determined using MEME8 with the maximum number of motifs set to 10 (Bailey et al., 2009).
Protein sequences of the 22 GmTLPs were submitted to the SMART website9 to obtain data related to conserved protein domains, and GSDS 2.0 was used for visual analysis. Three-dimensional models of the Tub domain were built with SWISS-MODEL10 (Dong et al., 2019). Tub domain models were obtained for 20 GmTLPs with the protein sequence identity set to ≥30%.
Soybean gene expression files were downloaded from the Soybase website11 to analyze the expression patterns of 22 GmTLPs members in different tissues at different developmental stages under normal conditions, including young-leaf, flower, pod, pod shell, seed, root, and nodule. In the database file provided by Soybase website, only 18 members’ tissue differential expression information were found for further analysis. Transcriptome data for GmTLPs members under various abiotic stresses from our previous studies (Wang et al., 2020). 22 GmTLPs members were used for searching in transcriptome data, and their expression levels under normal condition, ABA treatment, drought and salt stresses were analyzed. Finally, the relevant information of 21 members was obtained. TBtools (version 1.075) (Chen et al., 2020) was used for visualization and cluster analysis of GmTLP expression patterns.
GmTLP sequences obtained from the Phytozome database were extracted in batches with TBtools, and the 2000 bp upstream promoter sequences of the 22 GmTLP genes were obtained and submitted to the online program PlantCARE12 to identify cis-acting elements. GSDS 2.0 was used for data visualization.
The soybean variety Zhonghuang39 was used for analysis of GmTLP gene expression in this study. Soybeans were grown in 1: 1 vermiculite: humus in a greenhouse with a 16/8 h light/dark cycle, day/night temperatures of 28/20°C, and a relative humidity of 70%. At 14 days, the seedlings at the four-leaf stage were stressed with drought or salt. Referring to previous research methods (Wang et al., 2021), soybean seedlings were removed from soil. For drought stress, the seedlings were placed on filter paper; for salt stress, the seedlings were immersed in 200 mM NaCl solution. The sampling time of drought or salt stress was 0, 0.5, 1, 2, 4, 8, 12, and 24 h. There were three biological replicates per treatment. After treatment, the leaves were frozen in liquid nitrogen and stored at –80°C before further analysis (Xu et al., 2008). These samples were used for qRT-PCR analysis of subsequent GmTLPs members.
Total RNA was extracted from soybean leaves using a plant RNA extraction kit following the manufacturer’s instructions (TIANGEN, Beijing, China). cDNA was synthesized using the PrimeScript™ RT Reagent Kit (TaKaRa, Shiga, Japan) following the manufacturer’s protocol. Primers (Supplementary Table 2) were designed using Primer Premier 5.0. The soybean Actin gene (U60506) was used as the internal control for quantitative real-time PCR (qRT-PCR). There were three technical replicates for each sample. Differential expression was determined from the relative gene expression data using the 2–Δ Δ CT method (Le et al., 2011).
We constructed an expression vector labeled with green fluorescent protein (hGFP) for subcellular localization analysis. The full-length cDNA sequence of GmTLP8 was fused to the N-terminal hGFP protein driven by the CaMV35S promoter (Xu et al., 2007). The 35S:GFP vector was used as a control. A PEG4000-mediated method was used to transform the GmTLP8-GFP recombinant plasmid into Arabidopsis protoplasts (He et al., 2016). After incubation for 18–20 h in the dark at 22°C, the nucleus of GmTLP8-GFP protoplasts were specifically stained with 4’, 6-diamidino-2-phenylindole (DAPI). The fluorescence signal was observed using a confocal laser scanning microscope (Zeiss LSM 700, Oberkochen, Germany). There were three technical replicates for each group.
The transformation was conducted to produce soybean hairy roots that were characterized by overexpression of GmTLP8 (GmTLP8-OE), RNA interference of GmTLP8 (GmTLP8-RNAi), or with the empty pCAMBIA3301 vector (EV-Control) (Chen et al., 2021). The CDS of GmTLP8 was amplified without stop codon using gene-specific primer pairs, under the control of the CaMV35S promoter, GmTLP8 cDNA was ligated into the plant transformation vector pCAMBIA3301 to generate GmTLP8-overexpressing (GmTLP8-OE) vector. In order to construct the RNAi vector, a 564 bp fragment including the first intron sequence and its reverse complement was synthesized (Biomed, Beijing, China) and inserted into pCAMBIA3301 to generate the pCAMBIA3301-GmTLP8-RNAi (GmTLP8-RNAi) vector. The recombinant construct and the empty pCAMBIA3301 (EV-Control) vector were transferred into A. rhizogenes strain K599, as previously described, then injected into soybean (G. max cv. Zhonghuang39) hypocotyl for A. rhizogenes-mediated transformation of soybean hairy roots (Wang et al., 2009; Du et al., 2018).
The injected plants were placed in a high-humidity greenhouse until hairy roots were generated at the infected site and had grown to ∼5 cm in length. After cutting off the original tap root 0.5 cm below the infected site, the seedlings were transplanted into fertilized soil and cultivated in a greenhouse at 25°C with a 16/8 h light/dark photoperiod for 7 days (Yu et al., 2021). The qRT-PCR analysis of GmTLP8 expression in GmTLP8-OE, EV-control and GmTLP8-RNAi transgenic hairy root plants before processing (Supplementary Figure 2G). Each of the hairy root-related experiments was replicated at least three times independently. The primers of GmTLP8-3301-F and GmTLP8-3301-R were listed in Supplementary Table 2.
Transgenic hairy root composite soybean plants were used in drought and salt stress assays after 7 days of normal growth. For drought treatment, soybean plants were grown for 7 days without watering; for NaCl treatment, soybean plants were treated with 150 mM NaCl for 3 days. Drought and salt treatment experiments were conducted a minimum of three times. Both the treated and untreated soybean hairy roots were washed with water prior to RNA isolation and physiological/biochemical experiments.
Several physiological parameters were measured in transgenic GmTLP8-OE, EV-Control, and GmTLP8-RNAi lines after the drought and NaCl treatments, namely levels of proline (Pro), malondialdehyde (MDA), hydrogen peroxide (H2O2), superoxide anion (O2–), and chlorophyll. Measurements were taken in soybean leaves using appropriate assay kits (Cominbio, Suzhou, China) following the manufacturer’s instructions. All measurements were performed in three biological replicates.
Leaves from the three transgenic lines were stained with 3,3-diaminobenzidine and nitro blue tetrazolium after drought or salt stress treatment. The leaves were immersed in DAB solution or NBT staining solution (Solarbio, Beijing, China) for 18 or 14 h, respectively. Samples were then destained in a boiling solution of 3: 1 anhydrous ethanol: glycerol until the leaves were white (Du et al., 2018). Images were taken using a Canon 50D camera (Canon, Tokyo, Japan). There were three biological replicates for each plant line–treatment group combination.
Twenty-two GmTLP family members were identified in this study. The SMART and Pfam databases were used to confirm the presence of the conserved Tub domain in all of the putative TLP proteins. Twenty-two GmTLP genes were unevenly distributed across 13 chromosomes of soybean. According to their positions on chromosomes, we named them GmTLP1 to GmTLP22. The details of TLPs in soybean, such as the coding sequence (CDS) length, amino acid length (aa), molecular weight (MW), isoelectric point (pI), and subcellular location are shown in Table 1.
Among the 22 GmTLPs, the protein length ranged from 183 (GmTLP5) to 454 amino acids (GmTLP21). The minimum protein MW was 20.8 kDa (GmTLP5), and the maximum was 50.6 kDa (GmTLP21). The pI ranged from 7.54 in GmTLP10 to 9.65 in GmTLP8. Twelve of the proteins were predicted to be located in the nucleus, five in the cytosol, four in the mitochondria, and two in the chloroplast (Table 1), with GmTLP1 predicted to be located in either the nucleus or cytosol.
A physical location map of the GmTLPs was drawn using physical location data from the soybean genome. The 22 GmTLP genes were distributed across 13 chromosomes, which were chromosome 1, 2, 7, 8, 10, 11, 12, 13, 14, 15, 16, 17, 20, respectively. There were three genes on chromosomes 2 and 13, two genes on chromosomes 7, 12, 15, 16, and 17, and only one gene each on chromosomes 1, 8, 10, 11, 14, and 20 (Figure 1).
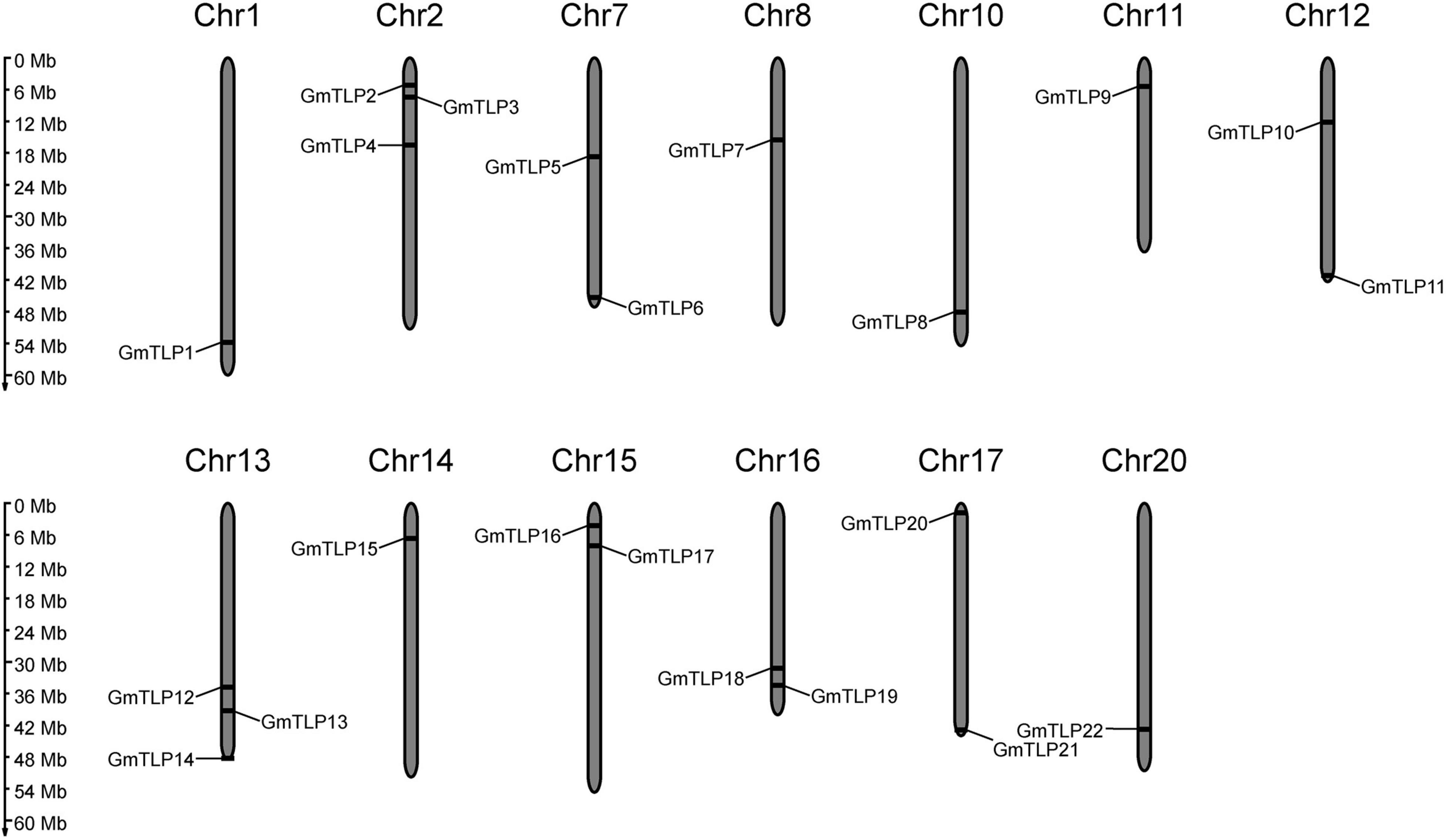
Figure 1. Chromosomal distribution of the 22 putative TLP genes identified in soybean. The scale bar at left indicates the size of the chromosomes.
To reveal the phylogenetic relationships between TLPs in different plant species, an unrooted phylogenetic tree was constructed by comparing the amino acid sequences for all of the known TLP members in several species, totaling 132 proteins. There were 11 from Arabidopsis (Lai et al., 2004), 15 from maize (Chen et al., 2016), 14 from rice (Liu, 2008), four from wheat (Hong et al., 2015), 11 from tomato (Zhang et al., 2020), nine from apple (Xu et al., 2016), 11 from poplar (Yang et al., 2008), and 35 from cotton (Li Z. et al., 2021) in addition to the 22 putative TLPs identified in soybean. Phylogenetic tree was divided into five groups based on protein homology, and there were one, two, five, six, and eight GmTLP members in groups I, II, III, IV, and V, respectively (Figure 2).
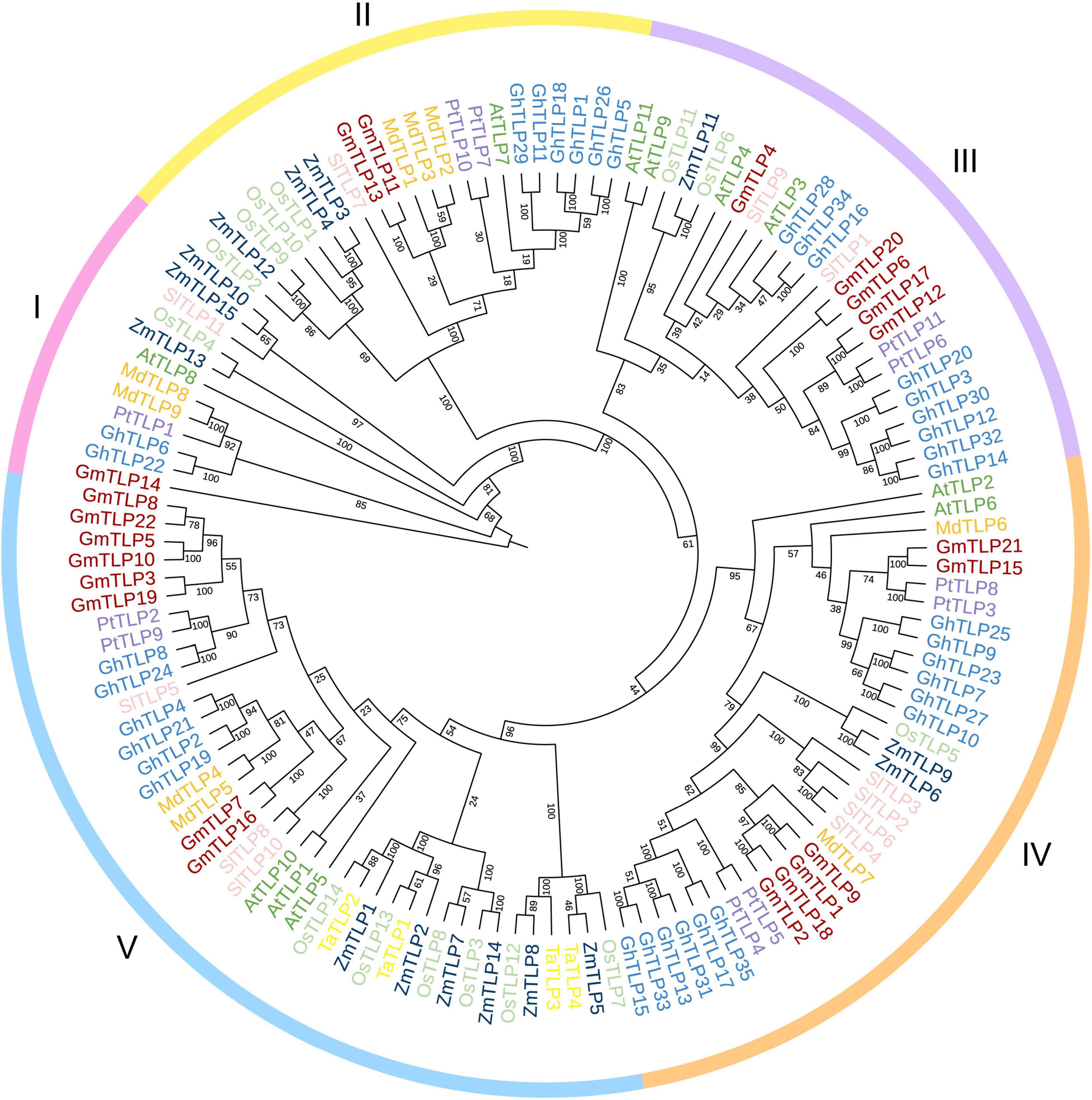
Figure 2. Phylogenetic analysis of TLP proteins. The full-length amino acid sequences of TLP proteins from Arabidopsis (AtTLPs), rice (OsTLPs), maize (ZmTLPs), tomato (SlTLPs), apple (MdTLPs), cotton (GhTLPs), poplar (PtTLPs), wheat (TaTLPs), and soybean (GmTLPs) were aligned using ClustalW. The phylogenetic tree was constructed using the NJ (Neighbor-joining) method with 1000 bootstrap replicates. Distinct subfamilies are marked with different colors.
The results of multiple sequence alignment showed that the positions of F-box domain and Tub domain in GmTLP protein sequence were located in the front and rear segments of the sequence (Figure 3).
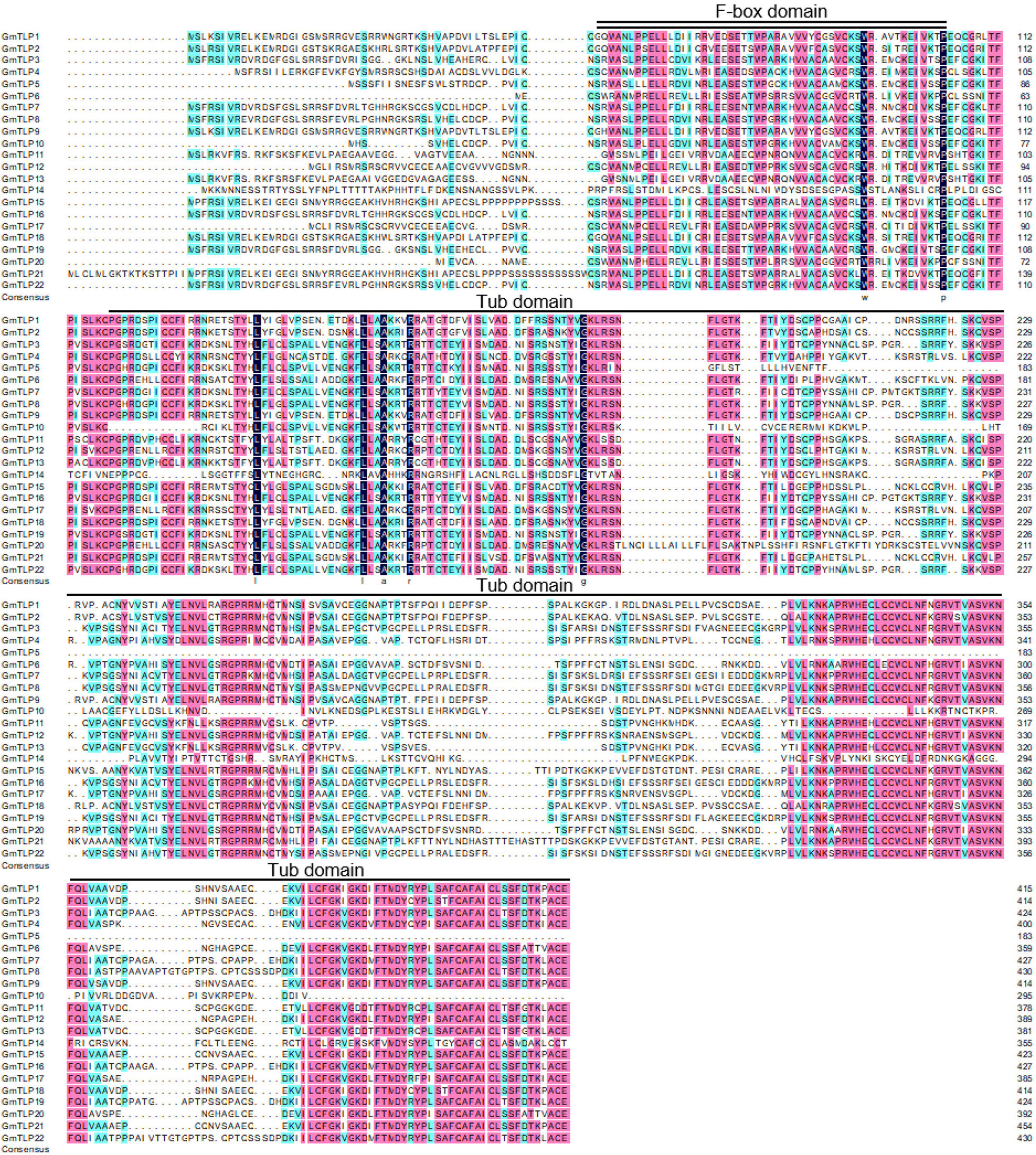
Figure 3. Multiple sequence alignment of GmTLP proteins from soybean using DNAMAN. Black, pink and light blue shading, respectively, represent amino acids with 100, ≥75, and 50% similarity of amino acids. The locations of the F-box domain and tubby domain are indicated with double and single solid lines above the sequences, respectively. The alignment is generated by the ClustalW program.
We analyzed the gene structure of the 22 GmTLPs using GSDS 2.0 online to determine the intron and exon distribution of each (Figure 4A). A total of 15 genes contained three introns and four exons each, and the other seven genes contained four introns and five exons.
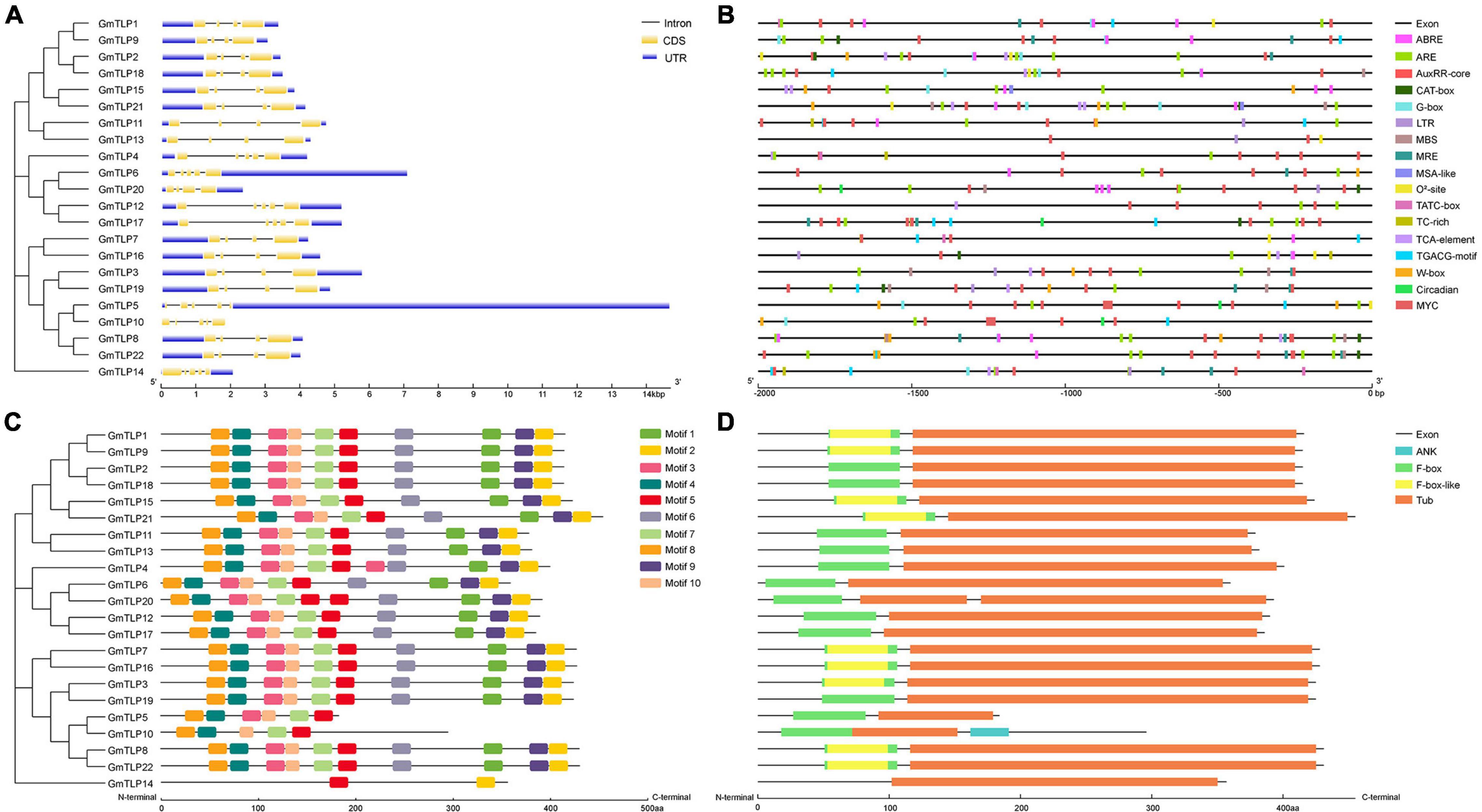
Figure 4. Gene structures, cis-acting elements, motifs and conserved domains analysis of GmTLPs. (A) Phylogenetic relationships (left) and gene structures (right) of GmTLPs. The phylogenetic tree was constructed using MEGAX; the different classes of TLP proteins make up separate clades. The schematic diagram shows gene structure. Introns and exons are indicated by black lines and yellow boxes, respectively. The lengths of introns and exons in each gene are displayed proportionally. (B) Predicted cis-acting elements in the GmTLP promoters. Distinct color blocks indicate different cis-elements, including ABRE, ARE, LTR, TC-rich, MBS, MRE, G-box, TCA-element, TGACG-motif, W-box, circadian, MSA-like, O2–-site, TATC-box, AuxRR-core, CAT-box, and MYC. The upstream distance from the translation start site can be estimated using the scale at the bottom. (C) Phylogenetic relationships (left) and putative motifs (right) of GmTLPs. Conserved motifs were identified using the MEME website and TBtools software. Ten putative motifs are indicated by colored boxes. (D) Conserved domain analyses of GmTLPs. The length of each protein can be estimated using the scale at the bottom.
A total of 10 conserved motifs (E ≤ 0.01) were analyzed using the MEME website to explore conservation and diversity of soybean TLPs. Among the 22 GmTLP family members, 19 contained all 10 motifs, with GmTLP4 containing two copies of motif 3 and GmTLP20 containing two copies of motif 5. GmTLP5, GmTLP10, and GmTLP14 contained six, five, and two motifs, respectively (Figure 4C). Consensus sequences for putative motifs are shown in Supplementary Figure 1.
From the Pfam database, we found that two conserved domains in GmTLPs were Tub (PF01167) and F-box (PF00646). We then analyzed the conserved protein domains using both the SMART website (Figure 4D) and homology modeling in SWISS-MODEL (Figure 5).
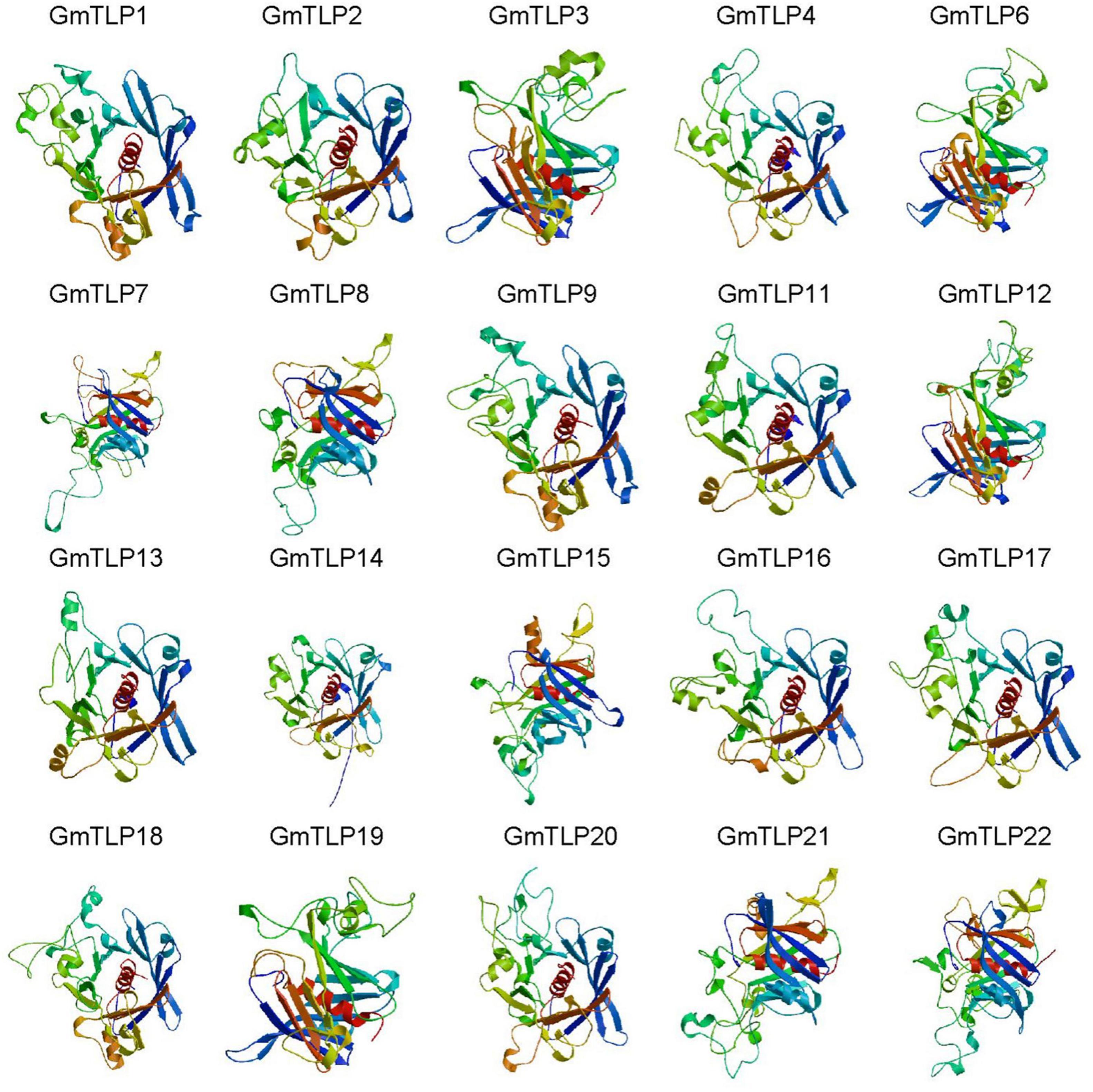
Figure 5. Homology modeling of the 3D structure of GmTLP Tub domains. The α-helices are shown in red, and β-barrels are shown in different colors surround the α-helices. GmTLP, soybean tubby-like protein.
The results of conserved domain analysis showed that 20 of the GmTLPs contained one F-box domain in the N-terminal region and one Tub domain in the C-terminal region. GmTLP4 had a Tub domain in the C-terminal region but no F-box domain in the N-terminal region, and GmTLP20 contained one F-box domain in the N-terminal region and two Tub domains in the C-terminal region. The analysis also revealed one ANK domain in the C-terminal region of GmTLP10, the function of which was not clear (Figure 4D).
The homology modeling was a useful tool for the prediction of protein structure, and protein structural information was often more valuable than sequence data alone in determining protein function. We generated three-dimensional (3D) models of the Tub domains for 20 of the GmTLPs. These models showed that the Tub domain of each GmTLP was closed by a β-barrel with 12 anti-parallel strands and a central hydrophobic α-helix, which is a typical structure for a Tub domain (Figure 5).
Cis-acting regulatory elements play an important role in modulating gene expression. To understand transcriptional regulation of GmTLPs, we identified cis-acting elements within the promoter region of each GmTLP gene, defined as the 2000 bp region upstream of the start codon (Figure 4B and Supplementary Table 1). Results showed that most cis-acting elements in GmTLP promoters were involved in hormone or stress responses.
The main hormone-related cis-acting elements identified were ABA response element (ABRE), TATC-box (gibberellin), AuxRR core (auxin), TCA element (salicylic acid), and the TGACG motif (methyl jasmonate). Among the GmTLP promoters, 14 genes contained ABRE, four contained a TATC-box, six contained the AuxRR core, 11 contained a TCA element, and 12 contained a TGACG motif. This indicated that the GmTLPs may be involved in hormone-related responses.
The abiotic stress cis-acting elements identified were as follows: anaerobic inducing element (ARE), low temperature response element (LTR), a MYB binding site involved in drought induction (MBS), a drought and salt response element (MYC), and a defense and stress response element (TC-rich element). MYC, which was previously reported to be involved in drought and salt stress-induced responses, was revealed to be distributed in all of the GmTLP promoter sequences. In addition, 20 GmTLP promoters contained ARE, seven contained LTR, seven contained MBS, and six contained TC-rich elements. The presence of these cis-acting elements related to abiotic stresses indicated that GmTLPs are abiotic stress-responsive.
Among the 22 GmTLPs, 14 contained ABREs, indicating that these genes can be regulated by ABA. Drought and salt response element MYCs were present in all 22 GmTLP promoter sequences, strongly suggesting that GmTLP members are involved in the responses to those stressors (Zuo et al., 2020). In total, cis-acting element analysis indicated that most members of the GmTLP family may be regulated by ABA in response to drought and salt stresses.
To understand expression patterns of TLPs during the growth and development of soybean and throughout different plant tissues, publicly available transcriptome sequencing data from the SoyBase database were analyzed. For 18 GmTLP members, we analyzed gene expression levels in different plant tissues including young-leaf, flower, pod, pod shell, seed, root, and nodule. The results showed that GmTLP7 was extremely high expressed in all seven tissues. GmTLP6, 10, 22, 11, and 21 were expressed at extremely low levels or not expressed in seven tissues. GmTLP2, 4, 9, 12, and 14 were expressed in some tissues, but not in others. GmTLP13, 17, 3, 5, 20, 15, and 16 were expressed in all seven tissues, with extremely high expression in some tissues and extremely low expression in others. Phylogenetic analysis divided the 18 GmTLP members into different groups, and members within each group shared similarities at the expression level in all seven tissues (Figure 6). The results showed a great deal of spatiotemporal difference in GmTLP expression levels.
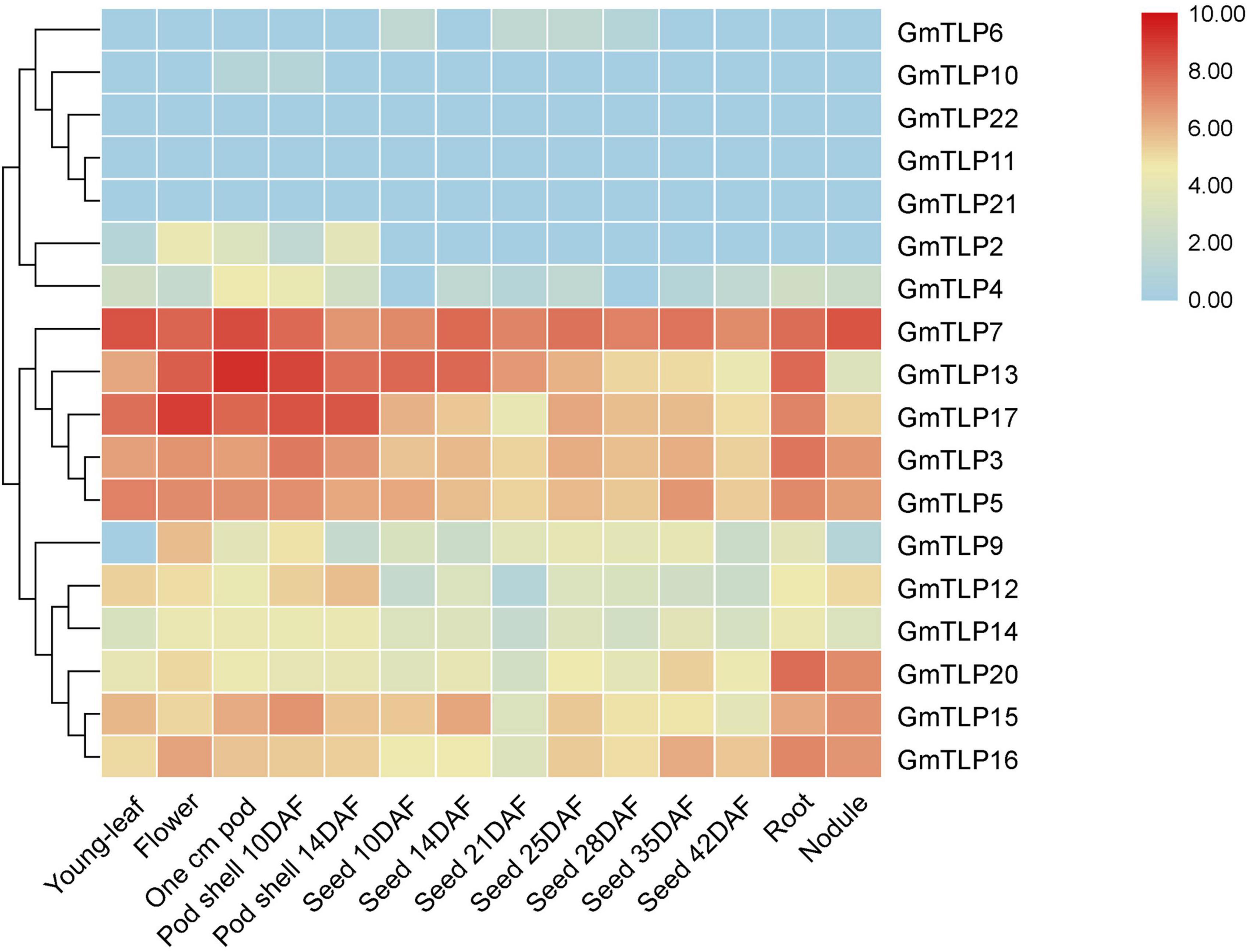
Figure 6. Expression profiles of GmTLPs from six soybean tissues. Gene expression was analyzed in soybean young-leaf, flower, pod, pod shell, seed, root, and nodule. The abundance of each transcript (in log10-based FPKM) is represented by the color bar. Red indicates higher and blue indicates lower expression levels.
We used a previously published transcriptome sequencing database to quantify the expression of GmTLPs under normal condition, ABA treatment, drought and salt stresses (Wang et al., 2020), and screened 21 GmTLP members (Figure 7). The results showed that GmTLP21 was up-regulated and GmTLP14 was down-regulated under ABA treatment. Under drought stress, six genes were up-regulated, and GmTLP3, 8, 11, 13, 19, and 22, four genes were down-regulated, and GmTLP14, 15, 18, and 20, respectively; Under salt stress, GmTLP8 was up-regulated and GmTLP14 was down-regulated. The P-value of data in abiotic stress expression profiles is shown in Supplementary Table 3.
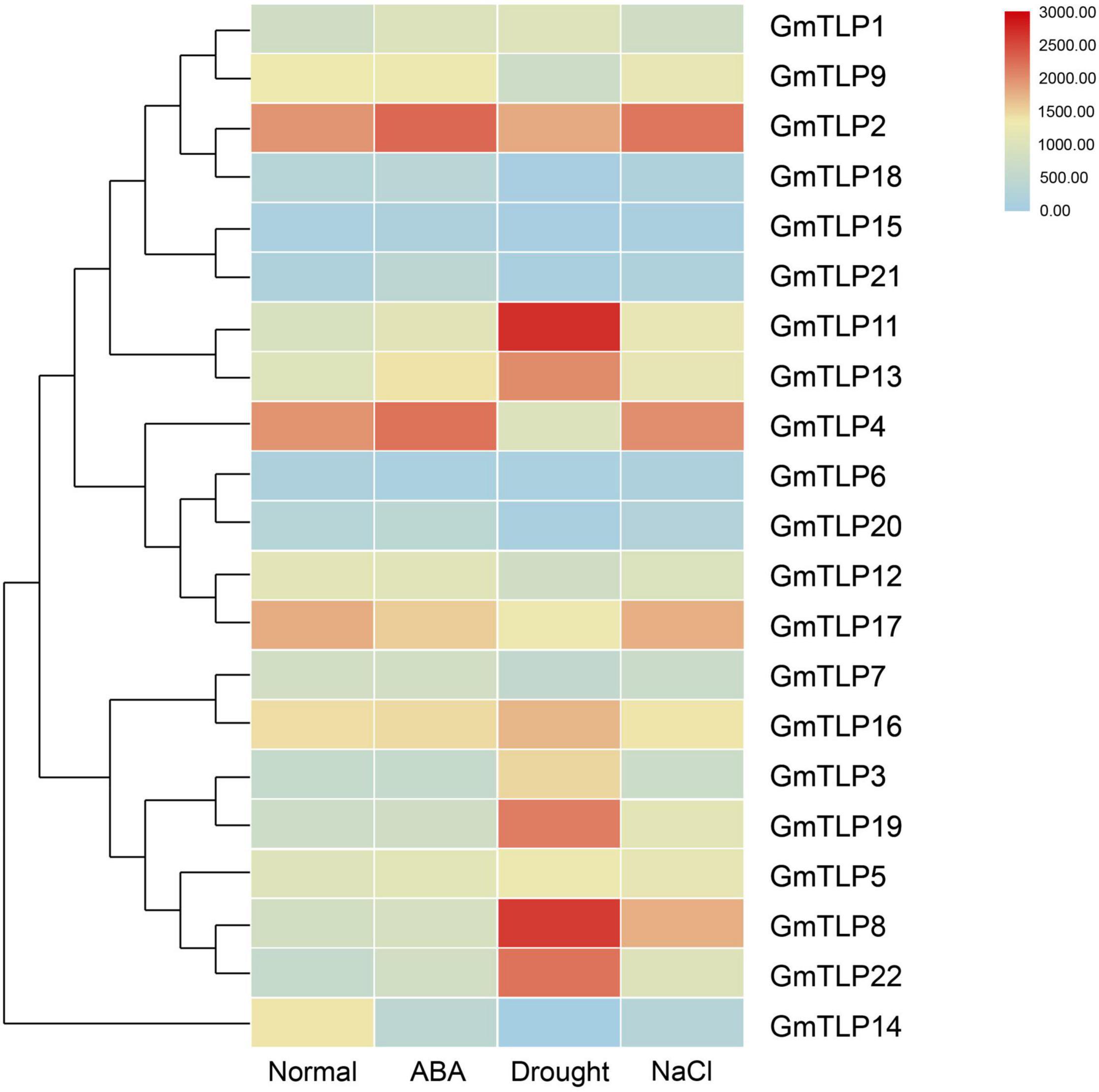
Figure 7. Expression profiles of GmTLPs under normal condition (Normal), ABA treatment (ABA), drought (Drought) and salt (NaCl) stresses. The expression abundance of each transcript (in log10-based FPKM) is represented by the color. Red indicates higher and blue indicates lower expression levels.
According to the expression profiles of GmTLPs under different abiotic stresses, five genes (GmTLP8, 11, 13, 19, and 22) up-regulated expression under drought stress and one gene (GmTLP8) under salt stress were selected for qRT-PCR analysis to further verify their relative expression levels under drought and NaCl treatments. The selected genes were GmTLP8, 11, 13, 19, and 22. Under drought treatment (Figure 8A), GmTLP8 expression peaked at 8 h (with an 8.5-fold increase compared to 0 h), GmTLP11 at 24 h (3.7-fold), GmTLP13 at 1 h (3.7-fold), GmTLP19 at 24 h (4.9-fold), and GmTLP22 at 8 h (4.8-fold). Under salt treatment (Figure 8B), GmTLP8 expression peaked at 8 h (7.3-fold), GmTLP11 at 2 h (4.5-fold), GmTLP13 at 24 h (1.8-fold), GmTLP19 at 12 h (4.4-fold), and GmTLP22 at 12 h (4.5-fold). These results showed that GmTLP8 was the most highly expressed in response to drought and salt treatments, and it was therefore selected for further study.
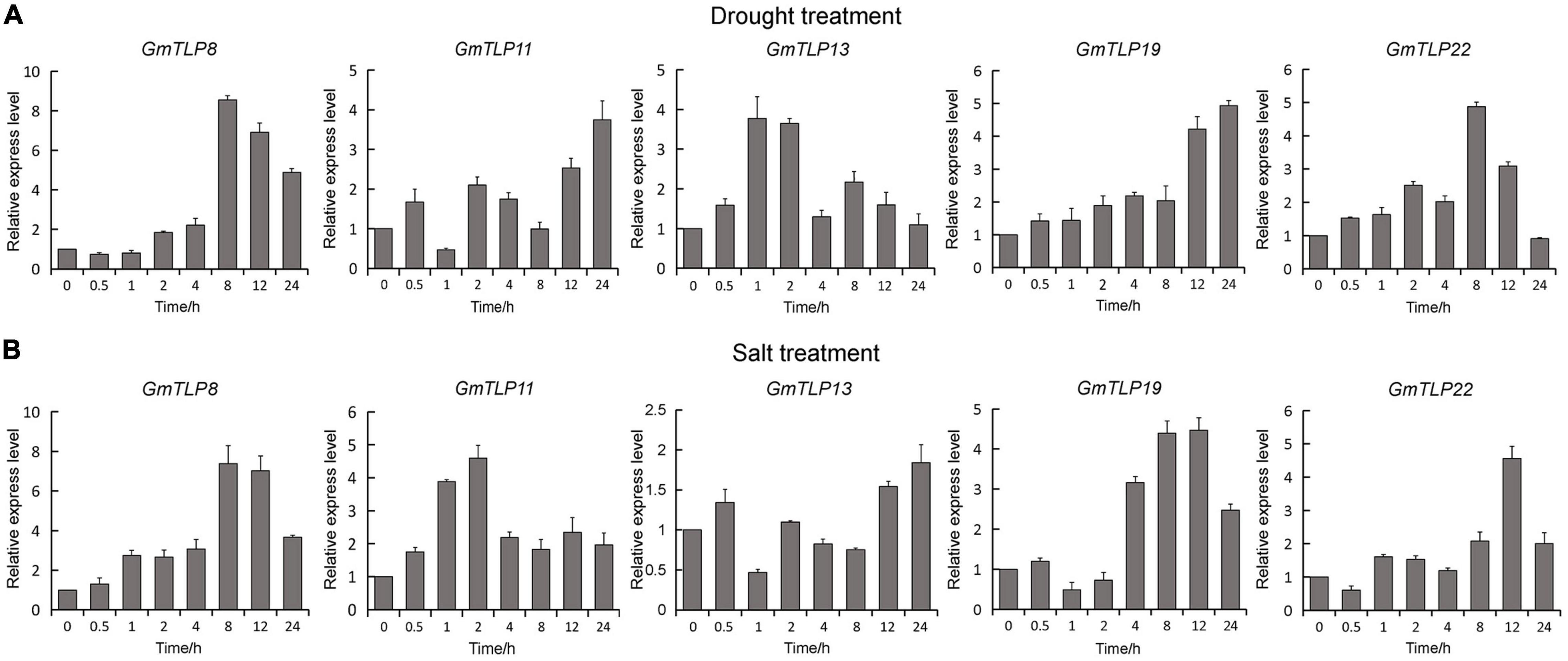
Figure 8. Expression patterns of GmTLPs under drought and salt treatments. GmTLP gene expression was measured in response to drought (A) and NaCl (B) treatments using qRT-PCR. The x-axes show the duration of treatment and y-axes depict relative expression level. The data are shown as mean ± standard deviation (SD) of three technical and three biological replicates.
To determine the subcellular localization of GmTLP8, the open reading frame (ORF) sequence (excluding the termination codon of GmTLP8) was fused with the N-terminal of the humanized green fluorescent protein (hGFP) reporter and co-transformed into Arabidopsis protoplasts. A 35S:hGFP as the control, the fluorescence signal in the cells was detected by confocal laser scanning microscopy. The fluorescence of GmTLP8 was detected in the nucleus and cytoplasm, while the fluorescence of the control 35S:hGFP was observed in the whole cell. DAPI staining also showed the localization of GmTLP8 in the nucleus (Figure 9). It suggests that GmTLP8 act as a transcription factor in the nucleus (Li S. et al., 2021).
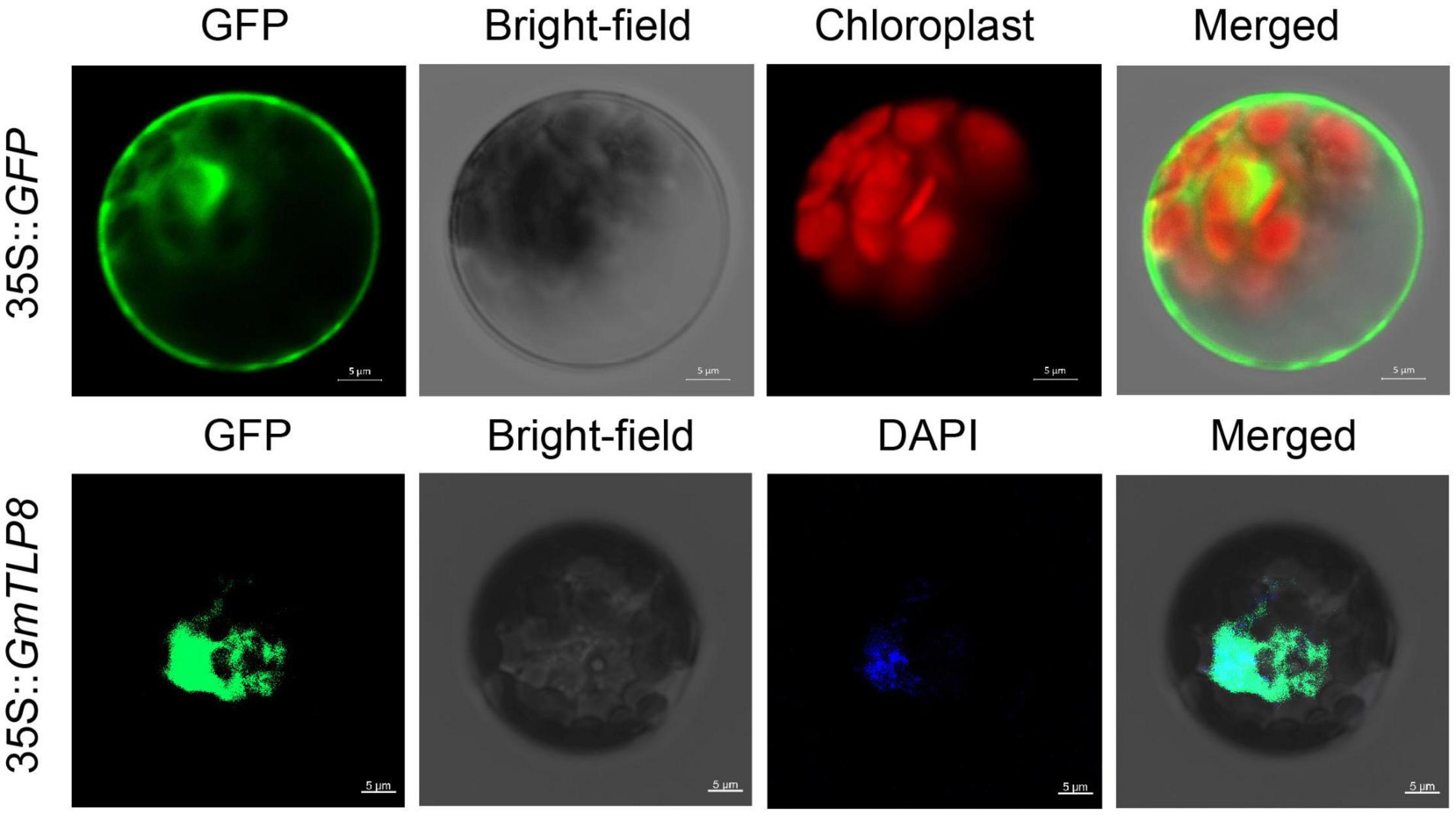
Figure 9. Subcellular localization of GmTLP8-hGFP fusion protein. 35S:GFP was used as the control. Scale bar = 5 μm.
The stress-tolerant effect of GmTLP8 in soybean was explored using transgenic soybean hairy root composite plants. The hairy roots of GmTLP8-OE, EV-Control and GmTLP8-RNAi transgenic lines were used to analyze the relative expression level of GmTLP8. qRT-PCR analysis showed that the expression level of GmTLP8-OE transgenic hairy roots was significantly higher than that in EV-Control, and the expression level of GmTLP8-RNAi transgenic hairy roots was lower than that in EV-Control (Supplementary Figure 2G). Under normal growth conditions, no significant differences were observed between GmTLP8-OE, the EV-Control, and GmTLP8-RNAi lines (Figure 10A). However, after exposure to drought (Figure 10B) and salt (Figure 10C) treatments, there were significant phenotypic differences between GmTLP8-OE, EV-Control, and GmTLP8-RNAi plants. Compared with EV-Control, GmTLP8-RNAi plants showed more severe leaf dehydration and wilting stress phenotype, whereas GmTLP8-OE showed fewer rolled leaves and a delayed leaf wilting phenotype. The survival rates of GmTLP8-OE, EV-Control, and GmTLP8-RNAi lines under drought stress were 93, 67, and 40%, respectively; these survival rates were comparable to those of salt-stressed plants (Figure 10H).
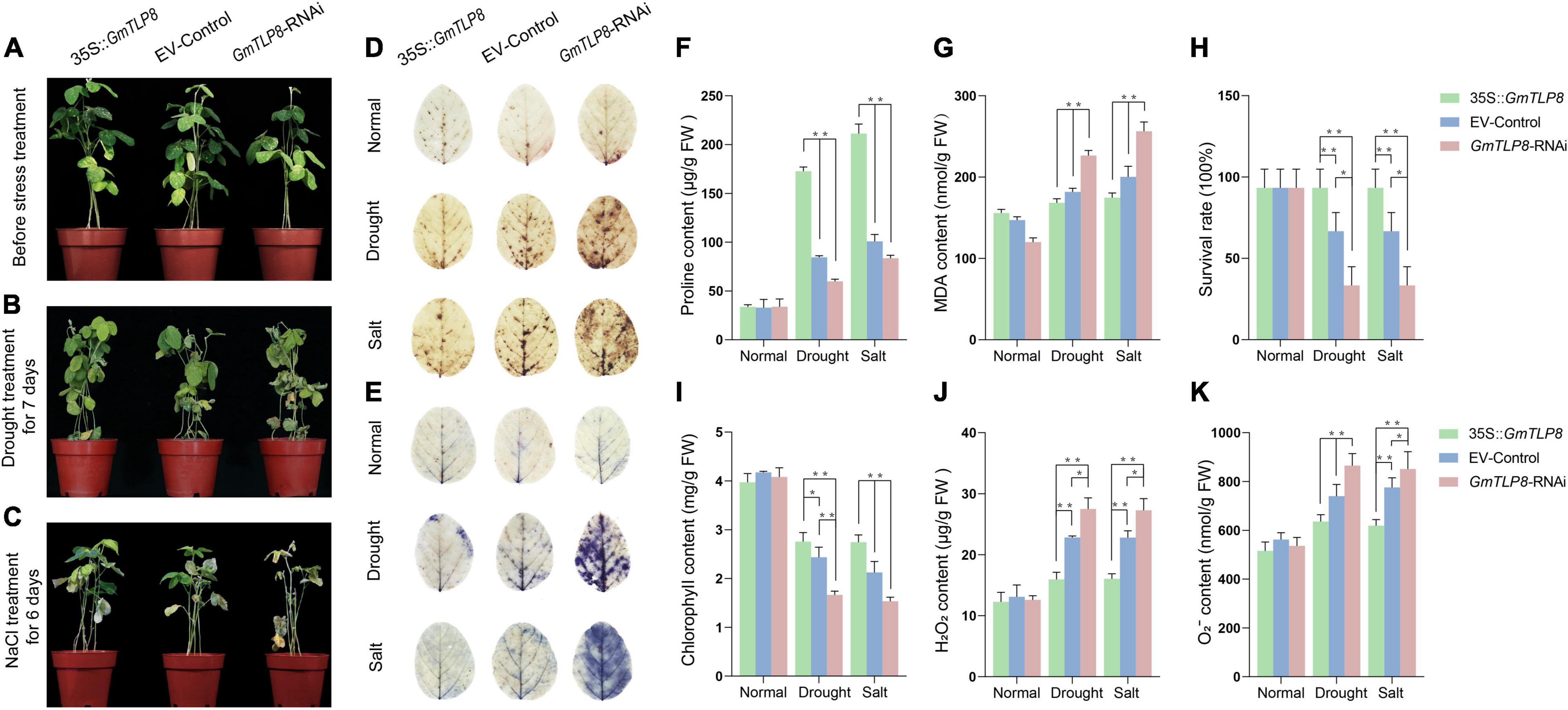
Figure 10. Analysis of the function of soybean GmTLP8. (A–C) Phenotypes of transgenic soybean hairy root composites GmTLP8-OE (35S:GmTLP8), EV-Control (empty plasmid), and GmTLP8-RNAi plants under (A) normal conditions, (B) drought stress, or (C) salt stress. (D) DAB and (E) NBT leaf staining of the GmTLP8-OE, EV-Control, and GmTLP8-RNAi lines under normal conditions and drought or salt stress. The depth of color corresponds to the concentrations of H2O2 and O2– in the leaves. (F) Proline (Pro) content, (G) malondialdehyde (MDA) content, (H) survival rate, (I) chlorophyll content, (J) H2O2 content, and (K) O2– content in transgenic soybean hairy root composite plants and EV-control plants under normal conditions and drought or salt stress. Vertical bars indicate ± SD of three technical and three biological replicates. *p < 0.05, **p < 0.01 (Student’ s t-test).
Proline (Pro), malondialdehyde (MDA), hydrogen peroxide (H2O2), and superoxide anion (O2–) levels are important indicators of the effects of abiotic stresses on plant growth (Leng et al., 2021). Proline is a protective agent against osmotic stress; MDA reflects the degree of lipid oxidative damage; H2O2 and O2– play immune and signal transduction roles, although excessive accumulation may lead to cell membrane damage (Du et al., 2018; Zhang et al., 2019). Chlorophyll levels are an important indicator of plant photosynthetic capacity (Tanaka and Tanaka, 2006). To further analyze the potential physiological mechanism of GmTLP8 in plant stress tolerance, we measured the levels of Pro, MDA, H2O2, O2–, and chlorophyll in the leaves of GmTLP8-OE, EV-Control, and GmTLP8-RNAi plants under normal growth conditions and under drought or salt stress (Figures 10F,G,I–K). Levels of Pro and chlorophyll were higher in GmTLP8-OE compared with EV-Control, whereas levels of MDA, H2O2, and O2– were lower. In contrast, the GmTLP8-RNAi lines had lower Pro and chlorophyll levels but higher MDA, H2O2, and O2– levels than EV-Control.
H2O2 and O2–, produced by the reactive oxygen species (ROS) pathway in leaf cells under abiotic stress, were measured to assess the degree of damage in leaf cells (Cui et al., 2019). This was done using DAB and NBT to stain the leaves of GmTLP8-OE, EV-Control, and GmTLP8-RNAi plants (Figures 10D,E). Under normal conditions, leaves from the GmTLP8-OE, EV-Control, and GmTLP8-RNAi lines showed minimal staining, with no significant difference between lines. Under drought and salt stresses, compared with EV-Control, GmTLP8-OE leaves showed shallow staining, whereas GmTLP8-RNAi showed deeper staining. These results demonstrated that the GmTLP8-OE line had lower levels of leaf damage and the GmTLP8-RNAi line had more severe leaf damage compared to EV-Control in response to exogenous abiotic stresses. The results of staining leaves with DAB and NBT were consistent with the physiological indexes of H2O2 and O2– contents.
To analyze the potential stress tolerance mechanism of GmTLP8, genes known to be involved in drought and salt stress responses were selected, namely GmDREB1 (Kasuga et al., 1999), GmDREB2 (Chen et al., 2007), GmNAC11 (An et al., 2018), GmNCED3 (Pandey and Gautam, 2020), GmSOS1 (Ma et al., 2020), and GmWRKY27 (Wang et al., 2015) (Figures 11A–F,H–M). Expression of these genes in the hairy roots of GmTLP8-OE, EV-Control, and GmTLP8-RNAi transgenic soybean lines were measured via qRT-PCR. Plants were drought-treated by withholding water for 7 days or salt-treated with 150 mM NaCl for 3 days. Under normal growth conditions, the selected stress responsive genes were expressed at lower levels in all three plant lines compared to plants that had been exposed to drought or salt stress (Supplementary Figures 2A–F and Figures 11A–F,H–M). In plants that had been stressed, compared with EV-Control, the six stress-related genes were significantly up-regulated in GmTLP8-OE plants and down-regulated in GmTLP8-RNAi plants. These results suggest that overexpression of GmTLP8 may activate expression of downstream drought- and salt-response genes.
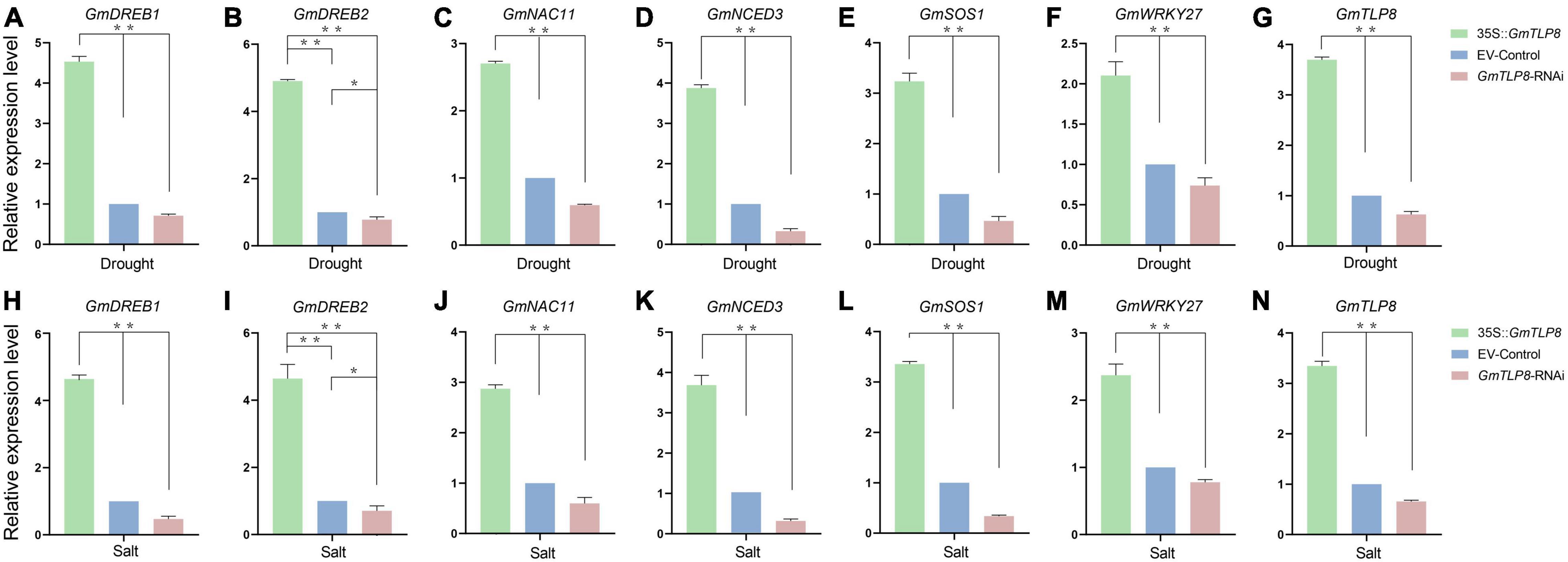
Figure 11. GmTLP8 regulates stress-responsive gene expression in transgenic soybean plants. (A–F) Expression levels of selected stress-related genes in transgenic soybean plants under drought stress. (H–M) Expression levels of selected stress-related genes in transgenic soybean plants under salt stress. (G) Expression levels of GmTLP8 in transgenic soybean plants under drought stress. (N) Expression levels of GmTLP8 in transgenic soybean plants under salt stress. Vertical bars indicate ± SD of three technical and three biological replicates. *p < 0.05, **p < 0.01 (Student’ s t-test).
Previous reports have proven that TLP family members participate in plant growth and development, response to abiotic stress, and can also be involved in the ABA signaling pathway (North et al., 1997; Lai et al., 2004; Bao et al., 2014; Chen et al., 2020). Also, reports have confirmed the resistance of TLPs members in Arabidopsis (Lai et al., 2004), maize (Chen et al., 2016), wheat (Hong et al., 2015), tomato (Zhang et al., 2020), apple (Xu et al., 2016), and cotton (Li Z. et al., 2021) to abiotic stress, but no report has been found in soybean. We used the NJ method to construct the phylogenetic tree of multiple species. According to the homology of protein sequences, they were divided into five groups, which were similar to the phylogenetic tree group in cotton previously reported (Li Z. et al., 2021). In group I, there is only one GmTLPs member, GmTLP14, which is the same as AtTLP8 in Arabidopsis previously reported (Lai et al., 2004). N-terminal of GmTLP14 and AtTLP8 do not contain F-Box domain, indicating that they may come from the same ancestor, so they are classified as the same group (Figure 2).
Gene structure analysis showed that each member of the GmTLPs had introns and exons, and their numbers were similar to those previously reported in Arabidopsis (Lai et al., 2004), indicating that the soybean TLPs was evolutionary conserved (Figure 4A). Motif analysis showed that except GmTLP14 containing two motifs, the number of motifs contained by other members was not less than five (Figure 4C). Analysis of protein conserved domains showed that except GmTLP14 had only one domain, other members contained two/three conserved domains (Figure 4D). Multiple sequence alignment marks the protein sites of two key conserved domains (Figure 3), consistent with the domain distribution shown in Figure 4D. Above results showed that the protein structures of other members of GmTLPs were similar except GmTLP14. Further analysis of transcriptome data showed that the up-regulated gene was GmTLP8 under drought and salt stresses, while the down-regulated gene was GmTLP14 (Figure 7). The down-regulated expression of GmTLP14 under drought and salt stresses might be due to the lack of N-terminal F-box domain (Figure 4D).
Two key conserved domains are in plant TLPs, the F-box at the N-terminal and the Tub domain at the C-terminal, and these differ from the conserved domains in mammalian TLPs. In mammals, TLPs are binary transcription factors; the N-terminal induces transcriptional activation, and the Tub domain binds to double-stranded DNA (Boggon et al., 1999; Li S. et al., 2021). In plants, the N-terminal F-box can participate in the formation of the Skp1-Cullin1-F-box (SCF) complex, which is an important part of E3 ubiquitin ligase and can participate in protein ubiquitination process (Gagne et al., 2002). It has been reported that GhTULP34 interacts with the subunit GhSKP1A of the SCF complex to form a functional SCF-type E3 ligase, which may be involved in the response of plants to abiotic stresses (Li Z. et al., 2021). Arabidopsis AtTLPs and wheat TaTULPs have been shown to interact with specific S-phase kinase-associated protein 1 (SKP1)-like proteins (Lai et al., 2004; Bao et al., 2014; Hong et al., 2015). These findings suggest that TLPs may play a role as subunits of the SCF complex in plants. Yeast two-hybrid assays showed that AtTLP7 and AtTLP11 interacted with NDR1/HIN1-like protein NHL6 (Song et al., 2019). Because both AtTLP11 and AtTLP7 are functional E3 ligases (Bao et al., 2014), it is possible that AtTLP11 and AtTLP7 redundantly manipulate the function of NHL6 by regulating its protein turnover (Bao et al., 2016). The above reports confirmed that the TLPs family, as F-box proteins, played a key role in protein ubiquitination, and may play a key role in plant response to various adverse environmental conditions. Based on these findings, it is speculated that GmTLP14 may be due to the lack of F-Box domain that affects the protein ubiquitination process and then down-regulates its expression under drought and salt stresses. However, the detailed functions of GmTLP14 gene need to be verified by related experiments. In this study, through RNA-Seq transcriptome data analysis and qRT-PCR verification, we determined the up-regulated expression of GmTLP8 under drought and salt stresses for subsequent studies (Figures 7, 8). Conserved domain analysis showed that GmTLP8 had two key conserved domains, namely, F-Box and Tub domains (Figure 4D), 3D modeling showed the integrity of GmTLP8 C-terminal tubby structure, which might play a role in the response of its to abiotic stress (Figure 5).
In this study, Agrobacterium rhizogenes-mediated transformation of soybean hairy roots was used to induce transgenic roots in soybean to study the function of GmTLP8 gene (Kereszt et al., 2007). Through phenotypic observation, leaf staining and physiological index analysis of soybean, it was confirmed that the overexpression of GmTLP8 enhanced the tolerance of soybean to drought and salt stresses (Figure 10). However, this genetic transformation mode is transient expression and cannot be stably inherited to the next generation through sexual reproduction. Therefore, further exploration of the application of GmTLP8 gene in transgenic drought-resistant and salt-resistant soybean needs further research on transformation.
Previous studies identified genes that play important roles in response to drought and salt stresses. To further analyze the molecular mechanism of GmTLP8 in regulating stress tolerance, we chose several confirmed stress-related genes (Figure 11 and Supplementary Figure 2). GmDREB1, GmDREB2, GmNAC11, and GmWRKY27 can specifically recognize and bind to cis-acting elements to up-regulate the expression of downstream stress-responsive genes, improving stress tolerance (Marè et al., 2004; Tran et al., 2004; Xu et al., 2011; Bouaziz et al., 2013, 2015; Sarkar et al., 2019). GmNCED3 is considered to be an important contributor to ABA synthesis and its overexpression enhances drought tolerance in seedlings (Li et al., 2019). GmSOS1 improves the salt tolerance of plants, potentially playing a role in Na+ extrusion out of the roots and regulation of Na+ transport from roots to shoots (Nie et al., 2015; Cao et al., 2018). These selected stress-related genes were up-regulated in GmTLP8-OE plants under drought and salt treatments. Taken together, these indicated that GmTLP8 responds to drought and salt stresses by activating stress-related transcription factors and the SOS pathway, which provides a scientific basis for further analysis of the function of GmTLP8 gene under drought and salt stresses. However, further studies were needed to fully elucidate its internal mechanism in abiotic stress response.
In the present study, we identified 22 TLP genes in the soybean genome. Based on expression patterns in response to abiotic stresses, we found that GmTLP14 showed different structural characteristics and expression patterns from most other members, but the function of GmTLP14 still needs further experimental verification. In this study, we selected GmTLP8 with complete structure and up-regulated expression under drought and salt stresses, and verified its expression level under abiotic stress by qRT-PCR. GmTLP8 was responsive to drought and salt stresses. Overexpression of GmTLP8 enhanced the tolerance of soybean to drought and salt stresses by activating downstream stress-responsive genes. These results improve understanding of the GmTLP family and provide a basis for further study of the molecular mechanism of GmTLP8 in soybean abiotic stress responses.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA694374.
Z-SX coordinated the project, conceived and designed the experiments, and edited the manuscript. H-RX performed the experiments and wrote the first draft. W-LW and Z-SX revised the manuscript. YL, Z-HH, JC, Y-BZ, and MC contributed to data analysis and managed reagents. T-FY, J-DF, J-CZ, and Y-ZM contributed with valuable discussions. All authors reviewed and approved the final manuscript.
This work was financially supported by the National Natural Science Foundation of China (32071967 and 31871624), the Agricultural Science and Technology Innovation Program (CAAS-ZDRW202109 and CAAS-ZDRW202002), and the Central Public-interest Scientific Institution Basal Research Fund (S2022ZD02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Lijuan Qiu and Shi Sun of the Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS) for kindly providing soybean seeds. Also, we are grateful to Wensheng Hou and Hui Zhang of the Institute of Crop Science, CAAS for kindly providing vectors and the protocol for high-efficiency A. rhizogenes-mediated transformation, respectively.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.844545/full#supplementary-material
TLP, tubby-like protein; ABA, abscisic acid; GFP, green fluorescent protein; MDA, malondialdehyde; PRO, proline; PEG, polyethylene glycol; DAB, 3, 3-diaminobenzidine; NBT, nitro blue tetrazolium; qRT-PCR, quantitative real-time PCR; ABRE, ABA-responsive element; MYC, drought and salt-responsive element; NCED, 9-cis-epoxycarotenoid dioxygenase; SCF-complex, Skp1-Cullin1-F-box complex; SKP1-like proteins, S-phase kinase-associated protein 1 like proteins; NHL, NDR1/HIN1-like gene.
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., and Hao, Y. J. (2018). An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J. Plant Physiol. 221, 74–80. doi: 10.1016/j.jplph.2017.12.009
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Badgandi, H. B., Hwang, S. H., Shimada, I. S., Loriot, E., and Mukhopadhyay, S. (2017). Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J. Cell Biol. 216, 743–760. doi: 10.1083/jcb.201607095
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bao, Y., Song, W. M., Jin, Y. L., Jiang, C. M., Yang, Y., Li, B., et al. (2014). Characterization of Arabidopsis tubby-like proteins and redundant function of AtTLP3 and AtTLP9 in plant response to ABA and osmotic stress. Plant Mol. Biol. 86, 471–483. doi: 10.1007/s11103-014-0241-6
Bao, Y., Song, W. M., Pan, J., Jiang, C. M., Srivastava, R., Li, B., et al. (2016). Overexpression of the NDR1/HIN1-like gene NHL6 modifies seed germination in response to abscisic acid and abiotic stresses in Arabidopsis. PLoS One 11:e0148572. doi: 10.1371/journal.pone.0148572
Bhushan, D., Pandey, A., Choudhary, M. K., Datta, A., Chakraborty, S., and Chakraborty, N. (2007). Comparative proteomics analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol. Cell Proteomics 6, 1868–1884. doi: 10.1074/mcp.M700015-MCP200
Boggon, T. J., Shan, W. S., Santagata, S., Myers, S. C., and Shapiro, L. (1999). Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 286, 2119–2125. doi: 10.1126/science.286.5447.2119
Borman, A. D., Pearce, L. R., Mackay, D. S., Nagel-Wolfrum, K., Davidson, A. E., Henderson, R., et al. (2014). A homozygous mutation in the TUB gene associated with retinal dystrophy and obesity. Hum. Mutat. 35, 289–293. doi: 10.1002/humu.22482
Bouaziz, D., Jbir, R., Charfeddine, S., Saidi, M. N., and Gargouri-Bouzid, R. (2015). The StDREB1 transcription factor is involved in oxidative stress response and enhances tolerance to salt stress. Plant Cell 121, 237–248. doi: 10.1007/s11240-014-0698-7
Bouaziz, D., Pirrello, J., Charfeddine, M., Hammami, A., Jbir, R., Dhieb, A., et al. (2013). Overexpression of StDREB1 transcription factor increases tolerance to salt in transgenic potato plants. Mol. Biotechnol. 54, 803–817. doi: 10.1007/s12033-012-9628-2
Cai, M., Qiu, D., Yuan, T., Ding, X., Li, H., Duan, L., et al. (2008). Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ. 31, 86–96. doi: 10.1111/j.1365-3040.2007.01739.x
Cao, D., Li, Y. Y., Liu, B. H., Kong, F. J., and Tran, L. P. (2018). Adaptive mechanisms of soybean grown on salt-affected soils. Land Degrad. Dev. 29, 1054–1064. doi: 10.1002/ldr.2754
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, M., Wang, Q. Y., Cheng, X. G., Xu, Z. S., Li, L. C., Ye, X. G., et al. (2007). GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 353, 299–305. doi: 10.1016/j.bbrc.2006.12.027
Chen, Y., Dai, W., Sun, B., Zhao, Y., and Ma, Q. (2016). Genome-wide identification and comparative analysis of the TUBBY-like protein gene family in maize. Genes Genomics 38, 25–36. doi: 10.1007/s13258-015-0338-6
Chen, Z. F., Ru, J. N., Sun, G. Z., Du, Y., Chen, J., Zhou, Y. B., et al. (2021). Genomic-wide analysis of the PLC family and detection of GmPI-PLC7 responses to drought and salt stresses in soybean. Front. Plant Sci. 12:631470. doi: 10.3389/fpls.2021.631470
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G., et al. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. doi: 10.1093/nar/gkg500
Coleman, D. L., and Eicher, E. M. (1990). Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J. Hered. 81, 424–427. doi: 10.1093/oxfordjournals.jhered.a111019
Cui, X. Y., Gao, Y., Guo, J., Yu, T. F., Zheng, W. J., Liu, Y. W., et al. (2019). BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 180, 605–620. doi: 10.1104/pp.19.00100
Dong, M. Y., Fan, X. W., Pang, X. Y., and Li, Y. Z. (2019). Decrypting tubby-like protein gene family of multiple functions in starch root crop cassava. AoB Plants 11:lz075. doi: 10.1093/aobpla/plz075
Du, Y. T., Zhao, M. J., Wang, C. T., Gao, Y., Wang, Y. X., Liu, Y. W., et al. (2018). Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 18:320. doi: 10.1186/s12870-018-1551-7
Eddy, S. R. (1998). Profile hidden Markov models. Bioinformatics 14, 755–763. doi: 10.1093/bioinformatics/14.9.755
Fernandez-Pozo, N., Menda, N., Edwards, J. D., Saha, S., Tecle, I. Y., Strickler, S. R., et al. (2015). The sol genomics network (SGN)–from genotype to phenotype to breeding. Nucleic Acids Res. 43, D1036–D1041. doi: 10.1093/nar/gku1195
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Gagne, J. M., Downes, B. P., Shiu, S. H., Durski, A. M., and Vierstra, R. D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 11519–11524. doi: 10.1073/pnas.162339999
He, G. H., Xu, J. Y., Wang, Y. X., Liu, J. M., Li, P. S., Chen, M., et al. (2016). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 16:116. doi: 10.1186/s12870-016-0806-4
Hong, M. J., Kim, D. Y., and Seo, Y. W. (2015). Interactions between wheat Tubby-like and SKP1 like proteins. Genes Genet. Syst. 90, 293–304. doi: 10.1266/ggs.14-00084
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Kapeller, R., Moriarty, A., Strauss, A., Stubdal, H., Theriault, K., Siebert, E., et al. (1999). Tyrosine phosphorylation of tub and its association with Src homology 2 domain-containing proteins implicate tub in intracellular signaling by insulin. J. Biol. Chem. 274, 24980–24986. doi: 10.1074/jbc.274.35.24980
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Kereszt, A., Li, D., Indrasumunar, A., Nguyen, C. D., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952. doi: 10.1038/nprot.2007.141
Kleyn, P. W., Fan, W., Kovats, S. G., Lee, J. J., Pulido, J. C., Wu, Y., et al. (1996). Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell 85, 281–290. doi: 10.1016/s0092-8674(00)81104-6
Kou, Y., Qiu, D., Wang, L., Li, X., and Wang, S. (2009). Molecular analyses of the rice tubby-like protein gene family and their response to bacterial infection. Plant Cell Rep. 28, 113–121. doi: 10.1104/pp.103.037820
Lai, C. P., Lee, C. L., Chen, P. H., Wu, S. H., Yang, C. C., and Shaw, J. F. (2004). Molecular analyses of the Arabidopsis TUBBY-like protein gene family. Plant Physiol. 134, 1586–1597.
Le, D. T., Aldrich, D. L., Valliyodan, B., Watanabe, Y., Ha, C. V., Nishiyama, R., et al. (2012). Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS One 7:e46487. doi: 10.1371/journal.pone.0046487
Le, D. T., Nishiyama, R., Watanabe, Y., Mochida, K., Yamaguchi-Shinozaki, K., Shinozaki, K., et al. (2011). Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 18, 17–29. doi: 10.1093/dnares/dsq032
Leng, Z. X., Liu, Y., Chen, Z. Y., Guo, J., Chen, J., Zhou, Y. B., et al. (2021). Genome-wide analysis of the DUF4228 family in soybean and functional identification of GmDUF4228-70 in response to drought and salt stresses. Front. Plant Sci. 12:628299. doi: 10.3389/fpls.2021.628299
Letunic, I., Doerks, T., and Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Li, S., Wang, N., Ji, D., Zhang, W., Wang, Y., Yu, Y., et al. (2019). A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell 31, 2107–2130. doi: 10.1105/tpc.18.00662
Li, S., Wang, Z., Wang, F., Lv, H., Cao, M., Zhang, N., et al. (2021). A tubby-like protein CsTLP8 acts in the ABA signaling pathway and negatively regulates osmotic stresses tolerance during seed germination. BMC Plant Biol. 21:340. doi: 10.1186/s12870-021-03126-y
Li, S., Zhang, J., Liu, L., Wang, Z., Li, Y., Guo, L., et al. (2020). SlTLFP8 reduces water loss to improve water-use efficiency by modulating cell size and stomatal density via endoreduplication. Plant Cell Environ. 43, 2666–2679. doi: 10.1111/pce.13867
Li, Z., Wang, X., Cao, X., Chen, B., Ma, C., Lv, J., et al. (2021). GhTULP34, a member of tubby-like proteins, interacts with GhSKP1A to negatively regulate plant osmotic stress. Genomics 113(1 Pt 2) 462–474. doi: 10.1016/j.ygeno.2020.09.024
Liu, Q. (2008). Identifification of rice TUBBY-like genes and their evolution. FEBS J. 275, 163–171. doi: 10.1111/j.1742-4658.2007.06186.x
Ma, X. J., Fu, J. D., Tang, Y. M., Yu, T. F., Yin, Z. G., Chen, J., et al. (2020). GmNFYA13 improves salt and drought tolerance in transgenic soybean plants. Front. Plant Sci. 11:587244. doi: 10.3389/fpls.2020.587244
Marè, C., Mazzucotelli, E., Crosatti, C., Francia, E., Stanca, A. M., and Cattivelli, L. (2004). Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol. Biol. 55, 399–416. doi: 10.1007/s11103-004-0906-7
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Mukhopadhyay, S., and Jackson, P. K. (2011). The tubby family proteins. Genome Biol. 12:225. doi: 10.1186/gb-2011-12-6-225
Nie, W. X., Xu, L., and Yu, B. J. (2015). A putative soybean GmsSOS1 confers enhanced salt tolerance to transgenic Arabidopsis sos1-1 mutant. Protoplasma 252, 127–134. doi: 10.1007/s00709-014-0663-7
Noben-Trauth, K., Naggert, J. K., North, M. A., and Nishina, P. M. (1996). A candidate gene for the mouse mutation tubby. Nature 380, 534–538. doi: 10.1038/380534a0
North, M. A., Naggert, J. K., Yan, Y., Noben-Trauth, K., and Nishina, P. M. (1997). Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc. Natl. Acad. Sci. U.S.A. 94, 3128–3133. doi: 10.1073/pnas.94.7.3128
Pandey, A. K., and Gautam, A. (2020). Stress responsive gene regulation in relation to hydrogen sulfide in plants under abiotic stress. Physiol. Plant. 168, 511–525. doi: 10.1111/ppl.13064
Papiernik, S. K., Grieve, C. M., Lesch, S. M., and Yates, S. R. (2005). Effects of salinity, imazethapyr, and chlorimuron application on soybean growth and yield. Commun. Soil Sci. Plant Anal. 36, 951–967. doi: 10.1081/css-200050280
Santagata, S., Boggon, T. J., Baird, C. L., Gomez, C. A., Zhao, J., Shan, W. S., et al. (2001). G-protein signaling through tubby proteins. Science 292, 2041–2050. doi: 10.1126/science.1061233
Sarkar, T., Thankappan, R., Mishra, G. P., and Nawade, B. D. (2019). Advances in the development and use of DREB for improved abiotic stress tolerance in transgenic crop plants. Physiol. Mol. Biol. Plants. 25, 1323–1334. doi: 10.1007/s12298-019-00711-2
Song, W. M., Cheng, Z. H., Guo, X. T., Yu, C. Y., Wang, H. H., Wang, J., et al. (2019). Overexpression of NHL6 affects seed production in transgenic Arabidopsis plants. Plant Growth Regul. 88, 41–47. doi: 10.1007/s10725-019-00486-2
Stretton, C., Litherland, G. J., Moynihan, A., Hajduch, E., and Hundal, H. S. (2009). Expression and modulation of TUB by insulin and thyroid hormone in primary rat and murine 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 390, 1328–1333. doi: 10.1016/j.bbrc.2009.10.147
Tanaka, A., and Tanaka, R. (2006). Chlorophyll metabolism. Curr. Opin. Plant Biol. 9, 248–255. doi: 10.1016/j.pbi.2006.03.011
Tang, Y., Yan, J., Peng, Y., Weng, W., Yao, X., Gao, A., et al. (2021). First report of Botryosphaeria dothidea causing gray mold on tartary buckwheat in Southwest China. Plant Dis. 25, DIS–07–21–1403–DN. doi: 10.1094/PDIS-07-21-1403-PDN
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Wang, F., Chen, H. W., Li, Q. T., Wei, W., Li, W., Zhang, W. K., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. doi: 10.1111/tpj.12879
Wang, M., Xu, Z., Ahmed, R. I., Wang, Y., Hu, R., Zhou, G., et al. (2019). Tubby-like protein 2 regulates homogalacturonan biosynthesis in Arabidopsis seed coat mucilage. Plant Mol. Biol. 99, 421–436. doi: 10.1007/s11103-019-00827-9
Wang, T. T., Yu, T. F., Fu, J. D., Su, H. G., Chen, J., Zhou, Y. B., et al. (2020). Genome-wide analysis of the GRAS gene family and functional identification of GmGRAS37 in drought and salt tolerance. Front. Plant Sci. 11:604690. doi: 10.3389/fpls.2020.604690
Wang, X., Wang, Y., Tian, J., Lim, B. L., Yan, X., and Liao, H. (2009). Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol. 151, 233–240. doi: 10.1104/pp.109.138891
Wang, Z. Q., Yu, T. F., Sun, G. Z., Zheng, J. C., Chen, J., Zhou, Y. B., et al. (2021). Genome-wide analysis of the Catharanthus roseus RLK1-Like in soybean and GmCrRLK1L20 responds to drought and salt stresses. Front. Plant Sci. 12:614909. doi: 10.3389/fpls.2021.614909
Xu, J., Xing, S., Sun, Q., Zhan, C., Liu, X., Zhang, S., et al. (2019). The expression of a tubby-like protein from Malus domestica (MdTLP7) enhances abiotic stress tolerance in Arabidopsis. BMC Plant Biol. 19:60. doi: 10.1186/s12870-019-1662-9
Xu, J. N., Xing, S. S., Zhang, Z. R., Chen, X. S., and Wang, X. Y. (2016). Genome-wide identification and expression analysis of the tubby-like protein family in the Malus domestica genome. Front. Plant Sci. 7:1693. doi: 10.3389/fpls.2016.01693
Xu, Z. S., Chen, M., Li, L. C., and Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Xu, Z. S., Ni, Z. Y., Liu, L., Nie, L. N., Li, L. C., Chen, M., et al. (2008). Characterization of the TaAIDFa gene encoding a CRT/DRE-binding factor responsive to drought, high-salt, and cold stress in wheat. Mol. Genet. Genomics 280, 497–508. doi: 10.1007/s00438-008-0382-x
Xu, Z. S., Xia, L. Q., Chen, M., Cheng, X. G., Zhang, R. Y., Li, L. C., et al. (2007). Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 65, 719–732. doi: 10.1007/s11103-007-9237-9
Yang, Z., Zhou, Y., Wang, X., Gu, S., Yu, J., Liang, G., et al. (2008). Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis, rice, and poplar. Genomics 92, 246–253. doi: 10.1016/j.ygeno.2008.06.001
Yu, T. F., Liu, Y., Fu, J. D., Ma, J., Fang, Z. W., Chen, J., et al. (2021). The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol. J. 19, 2589–2605. doi: 10.1111/pbi.13684
Zhang, X. Z., Zheng, W. J., Cao, X. Y., Cui, X. Y., Zhao, S. P., Yu, T. F., et al. (2019). Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in Arabidopsis and soybean. Front. Plant Sci. 10:1453. doi: 10.3389/fpls.2019.01453
Zhang, Y., He, X., Su, D., Feng, Y., Zhao, H., Deng, H., et al. (2020). Comprehensive profiling of tubby-like protein expression uncovers ripening-related TLP genes in tomato (Solanum lycopersicum). Int. J. Mol. Sci. 21:1000. doi: 10.3390/ijms21031000
Zuo, Z. F., Kang, H. G., Hong, Q. C., Park, M. Y., Sun, H. J., Kim, J., et al. (2020). A novel basic helix-loop-helix transcription factor, ZjICE2 from Zoysia japonica confers abiotic stress tolerance to transgenic plants via activating the DREB/CBF regulon and enhancing ROS scavenging. Plant Mol. Biol. 102, 447–462. doi: 10.1007/s11103-019-00957-0
Keywords: tubby-like protein, genome-wide analysis, abiotic stress, responsive mechanism, soybean
Citation: Xu H-R, Liu Y, Yu T-F, Hou Z-H, Zheng J-C, Chen J, Zhou Y-B, Chen M, Fu J-D, Ma Y-Z, Wei W-L and Xu Z-S (2022) Comprehensive Profiling of Tubby-Like Proteins in Soybean and Roles of the GmTLP8 Gene in Abiotic Stress Responses. Front. Plant Sci. 13:844545. doi: 10.3389/fpls.2022.844545
Received: 28 December 2021; Accepted: 15 March 2022;
Published: 25 April 2022.
Edited by:
Prasanta Kumar Subudhi, Louisiana State University, United StatesReviewed by:
Ajit Ghosh, Shahjalal University of Science and Technology, BangladeshCopyright © 2022 Xu, Liu, Yu, Hou, Zheng, Chen, Zhou, Chen, Fu, Ma, Wei and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Liang Wei, d2h3ZW5saWFuZ0AxNjMuY29t; Zhao-Shi Xu, eHV6aGFvc2hpQGNhYXMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.