
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 28 February 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.843633
 Yong Zhang1†
Yong Zhang1† Jiling Song1†
Jiling Song1† Lei Wang1†
Lei Wang1† Mengping Yang1
Mengping Yang1 Kaifeng Hu1
Kaifeng Hu1 Weiwei Li1
Weiwei Li1 Xuhong Sun1
Xuhong Sun1 Hong Xue1
Hong Xue1 Quanzhong Dong1
Quanzhong Dong1 Mingming Zhang1
Mingming Zhang1 Shubao Lou1
Shubao Lou1 Xingyong Yang1
Xingyong Yang1 Hao Du2
Hao Du2 Yongli Li2
Yongli Li2 Lidong Dong2
Lidong Dong2 Zhijun Che3*
Zhijun Che3* Qun Cheng2*
Qun Cheng2*
Soybean mosaic virus (SMV) is detrimental to soybean (Glycine max) breeding, seed quality, and yield worldwide. Improving the basic resistance of host plants is the most effective and economical method to reduce damage from SMV. Therefore, it is necessary to identify and clone novel SMV resistance genes. Here, we report the characterization of two soybean cultivars, DN50 and XQD, with different levels of resistance to SMV. Compared with XQD, DN50 exhibits enhanced resistance to the SMV strain SC7. By combining bulked-segregant analysis (BSA)-seq and fine-mapping, we identified a novel resistance locus, RSMV-11, spanning an approximately 207-kb region on chromosome 11 and containing 25 annotated genes in the reference Williams 82 genome. Of these genes, we identified eleven with non-synonymous single-nucleotide polymorphisms (SNPs) or insertion-deletion mutations (InDels) in their coding regions between two parents. One gene, GmMATE68 (Glyma.11G028900), harbored a frameshift mutation. GmMATE68 encodes a multidrug and toxic compound extrusion (MATE) transporter that is expressed in all soybean tissues and is induced by SC7. Given that MATE transporter families have been reported to be linked with plant disease resistance, we suggest that GmMATE68 is responsible for SC7 resistance in DN50. Our results reveal a novel SMV-resistance locus, improving understanding of the genetics of soybean disease resistance and providing a potential new tool for marker-assisted selection breeding in soybean.
Soybean [Glycine max (L.) Merr.] is one of the most important sources of plant protein and vegetable oil, providing more than one-quarter of the world’s protein for food and animal feed (Hoeck et al., 2003; Boerma and Specht, 2004). Breeding soybean varieties with both high seed quality and high yield remains an important goal of breeders. Both traits can be strongly influenced by pathogen attack. For example, soybean mosaic virus (SMV), a single-stranded, positive-sense RNA virus of the genus Potyvirus, causes losses in soybean yields and seed quality worldwide (Ross, 1977; Hajimorad et al., 2018). Combined infection of soybean with SMV and bean pod mottle virus (BPMV) (genus comovirus, family comoviridae) further reduces yield, by up to 85% (Ross, 1968). Thus, SMV resistance is a critical trait in soybean. However, a series of Rsv loci have been reported in soybean accessions and introduced into commercial varieties: Resistance to SMV1 (Rsv1), Rsv3, Rsv4, and Rsv5 (Kiihl and Hartwig, 1979; Buss et al., 1997; Hayes et al., 2000; Klepadlo et al., 2017). Rsv1, Rsv3, and Rsv5 are SMV strain specific and are presumed to encode NB-LRR proteins (Cho and Goodman, 1979; Hajimorad and Hill, 2001; Suh et al., 2011; Klepadlo et al., 2017). Rsv1 resistance alleles have been found in the cultivars Ogden, York, Marshall, Kwanggyo, Raiden, Suweon97, PI 486355, PI 507389, and FT-10 (Moon et al., 2009; Tucker et al., 2009), and confer resistance to SMV G1–G3 strains (Chen et al., 1991). In contrast, Rsv3 confers resistance to SMV G5–G7strains (Gunduz et al., 2002). Interestingly, Rsv4 confers broad-spectrum SMV resistance through an atypical mechanism that delays viral proliferation (Gunduz et al., 2004). The Rsv4 locus was first reported in 1995, and its molecular characterization has recently been achieved: it encodes an RNase H family protein with double-stranded (ds) RNA-degrading activity that enters the viral replication compartment and degrades viral dsRNA (Ishibashi et al., 2019).
Over the past several decades, extensive efforts have been made to identify SMV resistance loci and genes in soybean. In China, 22 SMV strains (SC1–SC22) have been identified (Wang et al., 2003, 2011; Yan et al., 2015), along with several resistance loci. The resistance genes Rsa, Rn1, Rn3, Rsc7, Rsc8, Rsc9, Rsc13, and SC18-resistance gene derived from the cultivar Kefeng 1 were mapped to chromosome 2 (Wang et al., 2003; Cai et al., 2014; Yan et al., 2015; Li et al., 2015). The resistance gene Rsc15, derived from RN-9, was mapped to chromosome 6 (Yang and Gai, 2011). The genes Rsc3, Rsc11, Rsc12, and Rsc14 derived from Qihuang 1 were mapped to chromosome 13 (Li et al., 2010; Ma et al., 2011; Cai et al., 2014; Zheng et al., 2014). Finally, Rsc4 derived from Dabaima was mapped to chromosome 14 (Wang et al., 2011).
Despite the identification of several SMV resistance loci and genes over the past several decades, breeding for SMV resistance remains challenging, requiring many time-consuming and laborious artificial inoculation experiments in the greenhouse or field. Bulked-segregant analysis (BSA) is an effective method to identify DNA markers closely linked to the candidate gene for an extreme phenotype (Michelmore et al., 1991).
In this paper, we report a novel QTL for SC7 resistance, named RSMV-11 (Resistance locus to SMV on chromosome 11). This QTL was identified by whole-genome resequencing of two DNA bulks of progeny showing extreme phenotypic values. Through combined fine-mapping and qRT-PCR analysis, we isolated five possible genes associated with RSMV-11. Our data proves important information for use in marker-assisted selection in soybean resistance breeding.
The SC7-resistant soybean cultivar DN50 (Dongnong 50) used in this study was obtained from Northeast Agricultural University, Harbin. The SC7-susceptible soybean cultivar XQD (Xiaoqingdou) was obtained from the KeShan Branch of HeiLongJiang Academy of Agricultural Sciences, KeShan.
For SC7 inoculation, the seeds of DN50, XQD and near-isogenic inbred lines (NILs) of RSMV-11 were planted in a greenhouse with a 16 h light/8 h dark photoperiod and maintained at 25°C with 70% relative humidity. Soybean plants were inoculated with SMV strain SC7 following the methods described by Che et al. (2020). The control leaves were carried out with equivalent amounts of 0.01 M sodium phosphate buffer (PH 7.2–7.4). Foliar symptoms were monitored every three days after inoculation.
Primers were designed online using Primer 5 based on the Williams 82 reference genome. All primers used for fine-mapping and qRT-PCR assays for candidate genes are listed in Supplementary Table 1.
A single young leaf was collected from each plant at the V2 stage (one fully expanded trifoliate). Genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method (Saghaimaroof et al., 1984). Total RNA was isolated from 1 g soybean leaves using TRIzol (Invitrogen, Shanghai, China) according to the manufacturer’s protocol. The extracted DNA and RNA were quantified using a NanoDrop 2000C ultra-micro spectrophotometer (Sunnyvale, CA, United States) and 1.5% agarose gel electrophoresis.
A cross was generated between the resistant cultivar DN50 and the susceptible XQD. After self-pollination of F1 plants, 355 F2 seeds were harvested. All individual F2 plants were grown in a field in KeShan, China, under natural conditions and a bi-parental F6 recombinant inbred lines (RIL) population was developed by single-seed descent. The F6 RILs were planted in the field in KeShan, China, in 2018 for mapping of the RSMV-11 locus.
Thirty highly resistant and 30 highly susceptible F6 individuals were screened to generate the R- and S-bulks, respectively. DNA samples of the parental lines and the two bulks were subjected to whole-genome resequencing using the Illumina HiSeq X Ten platform, followed by standard paired-end 150-bp sequencing library construction. Primer sequences of the markers for mapping are listed in Supplementary Table 1. For fine-mapping, 11 markers between positions 1,632,020 and 4,642,090 were identified. Six recombinants were identified in the fine-mapping population using 11 markers, and the SC7 resistance phenotype of their progeny was evaluated to delimit the genomic interval containing Rsmv-11. The genotypes of the Rsmv-11 allele were analyzed by tagging marker M24 or M28.
cDNA was synthesized from total RNA using an Oligo (dT) 18 primer and PrimeScript 1st strand cDNA Synthesis Kit (Takara, Dalian, China). qRT-PCR analysis was performed to determine the transcript abundance of candidate genes using LightCycler 480 SYBR Green I Master (Roche, Mannheim, Germany) in a LightCycler 480 system (Roche, Mannheim, Germany). The soybean housekeeping gene Tubulin was used as the internal control. The relative transcript level of the target gene was calculated using the 2–ΔΔCt method. Three biological replicates with three technical replicates each were performed. The expression data of candidate genes in different soybean tissues (leaf, stem, root, flower, seed, pod, and nodule) were obtained from the RNA-seq database (Machado et al., 2020). Primers used are listed in Supplementary Table 1.
To assess the variation in disease resistance between DN50 and XQD, we identified the phenotype of DN50 and XQD. The result domesticated soybean varieties DN50 and XQD, display clear differences in resistance to the SC7 viral strain in the field (Figures 1A–C). Under natural conditions, SC7 caused symptoms in up to 90% of leaves in XQD, but did not induce any symptoms in the leaves of DN50. Moreover, in the greenhouse, SC7-treated XQD exhibited enhanced rugosity, curling, and chlorosis symptoms compared with DN50 (Figures 1D,E). We also analyzed the relative accumulation of SC7 in top non-inoculated leaves at 21 days post inoculation (dpi) in the greenhouse. The accumulation of viral CP gene in DN50 was significantly lower than that in XQD (Figure 1F). Together, these results suggested that DN50 is more resistant than XQD to SC7.
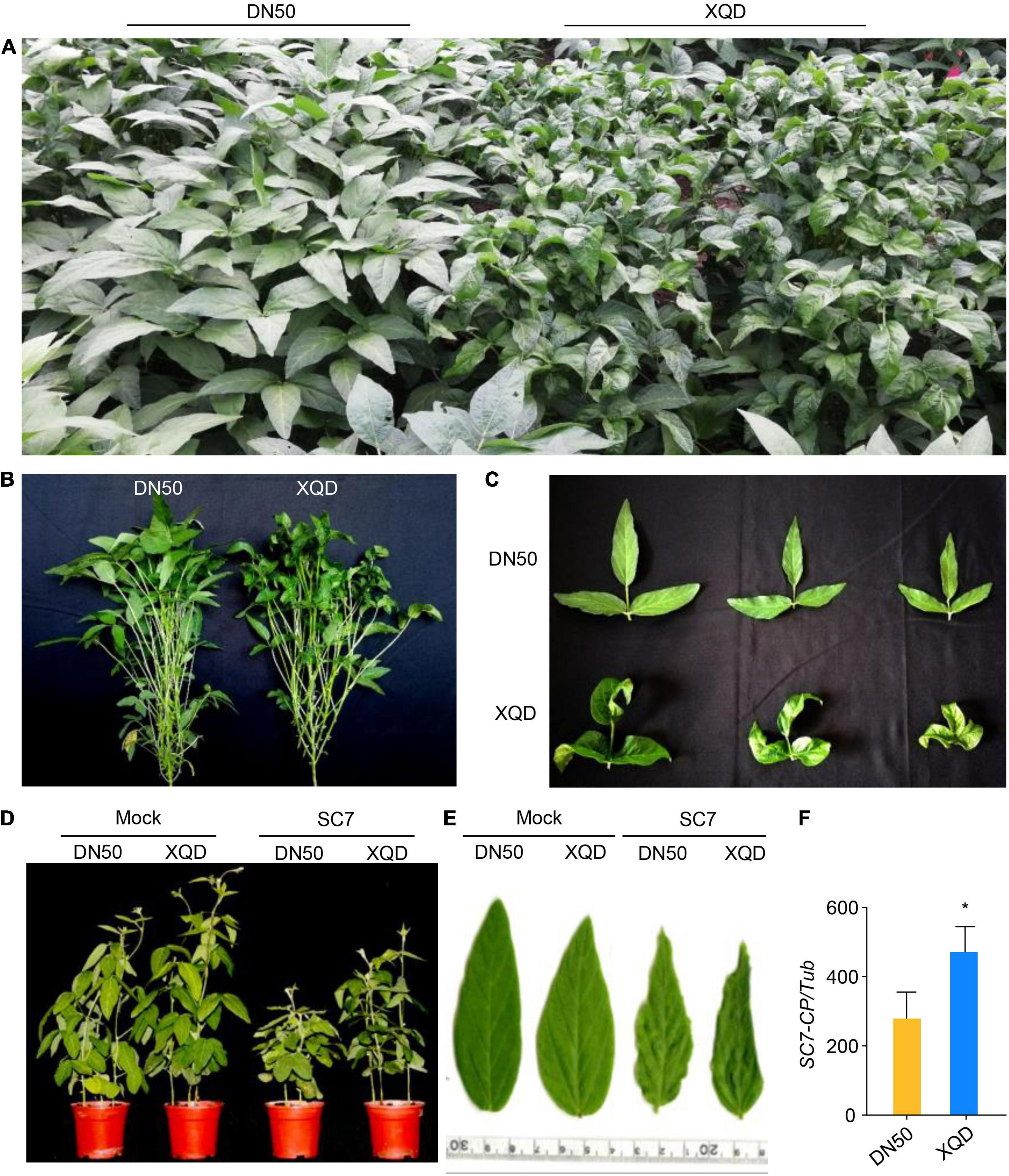
Figure 1. Phenotypes of two soybean cultivars, DN50 and XQD, inoculated with soybean mosaic virus strain SC7. (A,B) Phenotype of DN50 and XQD inoculated with SMV in the field. (C) Leaf phenotypes. DN50 shows partial resistance, whereas XQD is susceptible. (D) Phenotype of DN50 and XQD inoculated with SC7 in greenhouse. (E) Leaf phenotype. (F) Accumulation of SC7 in DN50 and XQD SC7-inoculated leaves. The amplification of soybean TUB (GmTubulin) was used as an internal control to normalize all data. Data from three biological replicates are shown, each with three technical replicates. Statistically significant differences were determined using Student’s t-test (*P < 0.05). Error bars indicate standard error of the mean.
To understand the molecular basis of SC7 resistance, we aimed to detect genetic differences between SC7-resistant and -susceptible plants. To this end, we first generated an F2 population by crossing DN50 and XQD (Figure 2A). The F2 population by four rounds of self-fertilization generated an F6 population (Figure 2A). We selected two groups of F6 plants reflecting segregation of the SC7 resistance trait (phenotype of F6 populations revealed a 3:1 ratio of the resistance and susceptible to SC7): one group of 30 individuals showing resistance to SC7 and a second group containing 30 individuals susceptible to SC7 (Figure 2A).
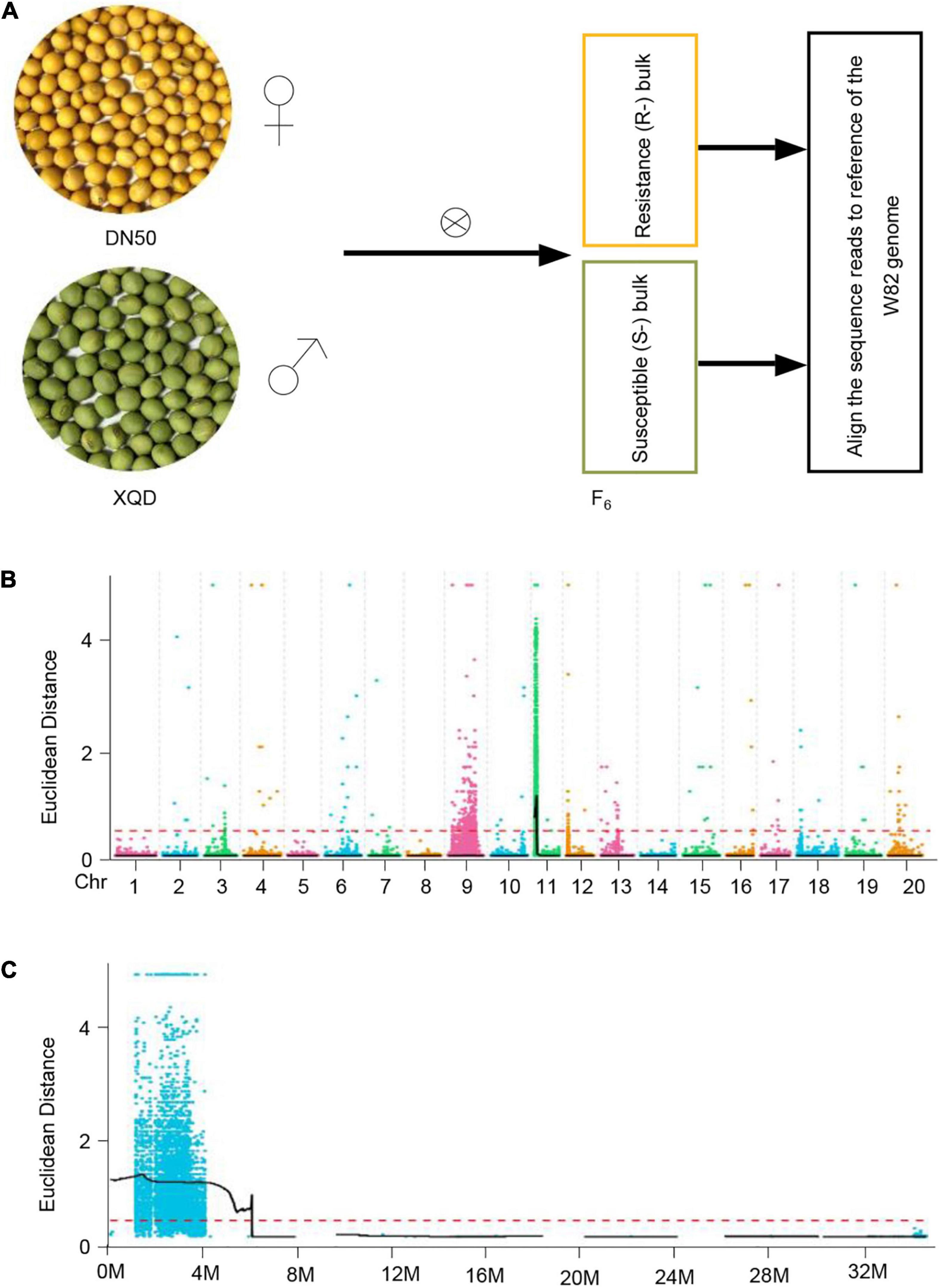
Figure 2. A simplified scheme of BSA-seq as applied to soybean. (A) Morphological difference in seed size between DN50 and XQD accessions. Two cultivars with different phenotypes are crossed to generate F6 progeny segregating for the trait value. Multiple progeny exhibiting resistance and susceptibility to SC7 are selected, and their DNA is bulked to produce “Resistant” and “Susceptible” bulks, respectively. (B) Examples of Euclidean distance (ED)-associated values on chromosomes. The color points represent the ED value of each single-nucleotide polymorphism (SNP) locus. The black line is the fitted ED value, and the red dotted line represents the significantly associated threshold. Higher ED values indicate stronger correlations. (C) The distribution of Euclidean distance (ED)-associated values on chromosome 11.
Next, we sequenced the two bulked DNAs (R- and S-bulks), along with single DNA samples of the two parents (DN50 and XQD), on the Illumina HiSeq X Ten platform and then resequenced the genomes of the two parents at 30× coverage each and the two bulked DNAs at 20× coverage each (∼100 Gb data). After filtering, a total of 2,005,612 bi-allelic single-nucleotide polymorphisms (SNPs) and a total of 432,197 bi-allelic short insertions and deletions (InDels) were identified in the two parents, and 113,258 bi-allelic SNPs and 46,172 bi-allelic InDels were identified in the two bulked DNAs. We then aligned the resequencing data of the two bulked sample groups (R- and S-bulks) to the Williams 82 reference genome (Glycine max Wm82.a2.v2; Schmutz et al., 2010).
Using the genotype data from the parents and the two sample groups (R and S), we used the Euclidean distance (ED) algorithm to locate the SC7 resistance QTL locus. Only one QTL at the beginning of chromosome 11 was significantly associated with resistance to SC7; this QTL was named RSMV-11 (Figures 2B,C). ED value analysis revealed that a physical region of 0.944 kb–3.418 Mb on chromosome 11 was possibly linked to SC7 resistance.
To more precisely map the RSMV-11 gene within the previously identified candidate regions, we used the F6 RIL population. We localized RSMV-11 to a 207-kb region between markers M24 and M28 (Figure 3A), adjacent to the RSMV-11 region mapped by BSA-seq.
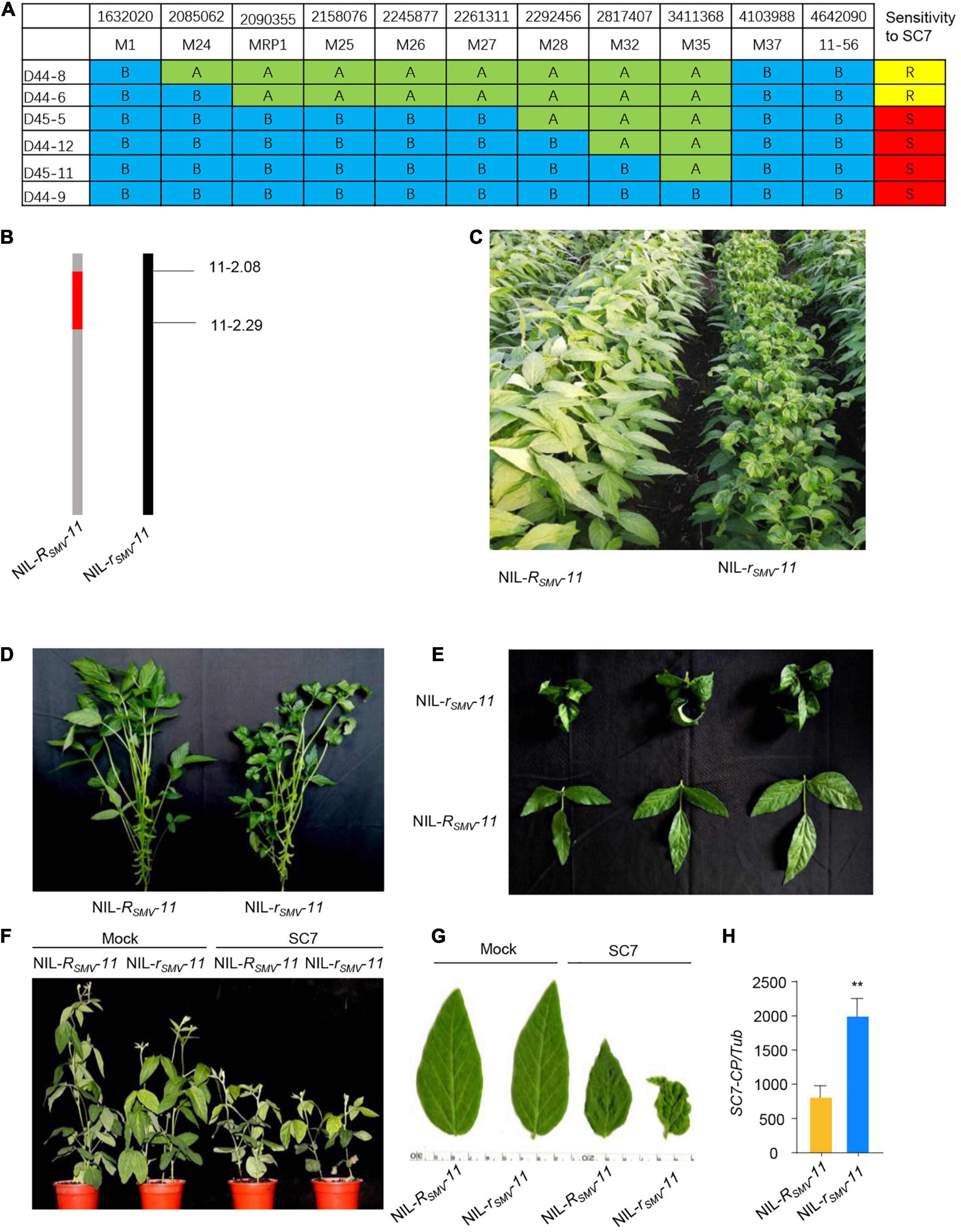
Figure 3. Fine-mapping of Rsmv-11. (A) Characterization of key recombinants from the F6 segregated population in the immediate vicinity of the Rsmv-11 locus (n = 36 plants). “A”, Rsmv-11 homozygous; “B”, rsmv-11 homozygous; “H”, heterozygous. “R”, Resistant to SC7; “S”, susceptible to SC7. (B) Chromosome maps of NIL-Rsmv-11 and NIL-rsmv-11. Red rectangles indicate the donor segment containing the Rsmv-11 locus. (C,D) Phenotype of NIL-Rsmv-11 and NIL-rsmv-11 inoculated with SC7 in the field. (E) Leaf phenotype. (F) Phenotype of NIL-Rsmv-11 and NIL-rsmv-11 inoculated with SC7 in the greenhouse for 2 weeks. (G) Leaf phenotype. (H) Accumulation of SC7 in leaves of NIL-Rsmv-11 and NIL-rsmv-11 treated with SC7. The amplification of soybean TUB (GmTubulin) gene was used as an internal control to normalize all data. Data from three biological replicates are shown, each with three technical replicates. Statistically significant differences were determined using Student’s t-test (*P < 0.05). Error bars indicate standard error of the mean.
To substantiate the relationship between the RSMV-11 QTL and SC7 resistance, we compared the phenotypes of two F6 near-isogenic lines (NILs) carrying either the functional RSMV-11 allele (NIL-RSMV-11) or the non-functional rSMV-11 allele (NIL-rSMV-11) (Figure 3B) in both the field and the greenhouse. The results showed that NIL-RSMV-11 had greater resistance to SC7 in the field than NIL-rSMV-11 (Figures 3C–E). Consistent with the field phenotypes, NIL-rSMV-11 exhibited enhanced curling symptoms and chlorosis compared with NIL- RSMV-11 in the greenhouse (Figures 3F,G). We also analyzed the relative biomass of SC7 in infected NIL-RSMV-11 and NIL-rSMV-11 soybean leaves at 21 days post infection (dpi) with SC7 in the greenhouse. The biomass of SC7 in NIL-RSMV-11 was significantly lower than in NIL-rSMV-11 (Figure 3H). These data confirm that the Rsmv-11 allele could greatly enhance resistance to SC7, and that the Rsmv-11 gene was in this candidate region.
In the 207-kb candidate mapping region, we found 25 genes (Figure 4A and Supplementary Table 2). We identified SNPs and InDels of all 25 genes in the two parents, and SNPs or InDels in 11 genes resulted in non-synonymous amino acid substitutions or frameshift mutation in the deduced protein sequences (Figure 4B). Of these 11 genes, only two (Glyma.11G028900 and Glyma.11G029900) harbored InDels predicted to cause frameshifts (Figure 4B and Supplementary Table 3).
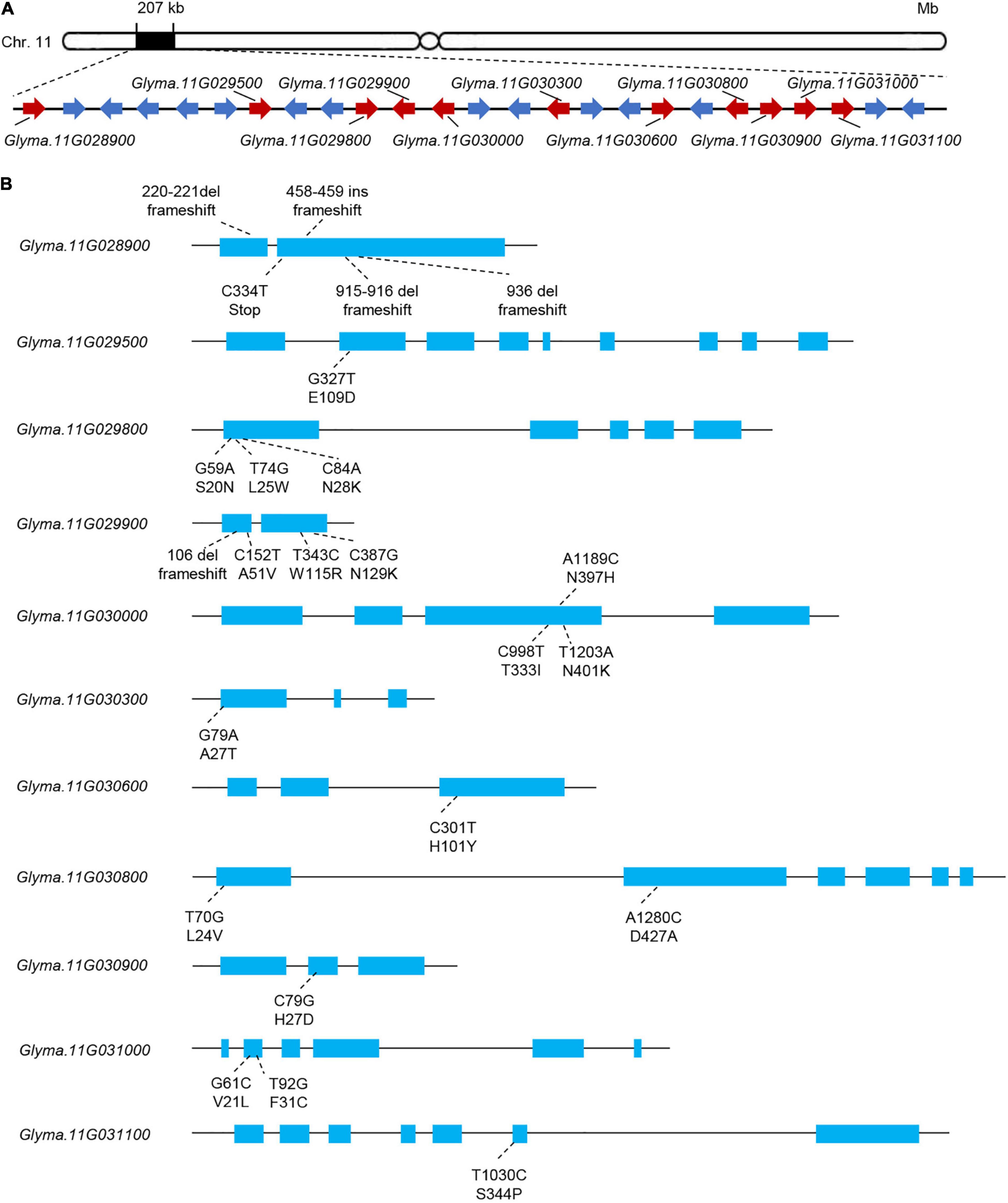
Figure 4. Polymorphism analysis of the candidate genes in the 207-kb region of interest. (A) Gene structure of Rsmv-11 showing the location of the candidate genes. Red represents genes with different sequences between parents. (B) Gene structure and variation information of candidate genes with different sequences between parents.
To further screen candidate genes, we searched an RNA-seq database and retrieved the expression data for 11 candidate genes with DNA sequences differing between DN50 and XQD. We analyzed data for seven tissues: flower, leaves, pod, stem, nodules, seed, and root (Machado et al., 2020). Seven genes were constitutively expressed in all tissues: Glyma.11G028900, Glyma.11G029500, Glyma.11G029800, Glyma.11G030000, Glyma.11G030600, Glyma.11G031100, and Glyma.11G031000 (Figures 5A–G). Glyma.11G030900 was highly expressed in all tissues except the seeds (Figure 5H). The other three genes displayed low transcript abundance in most tissues; however, Glyma.11G029900 showed high expression in the stem, Glyma.11G030300 was highly expressed in roots, and Glyma.11G030800 was abundant in nodules and seeds (Figures 5I–K). Given that Glyma.11G029900, Glyma.11G030300, and Glyma.11G030800 were barely expressed in leaves, we excluded these three genes from this candidate region and subsequent analyses.
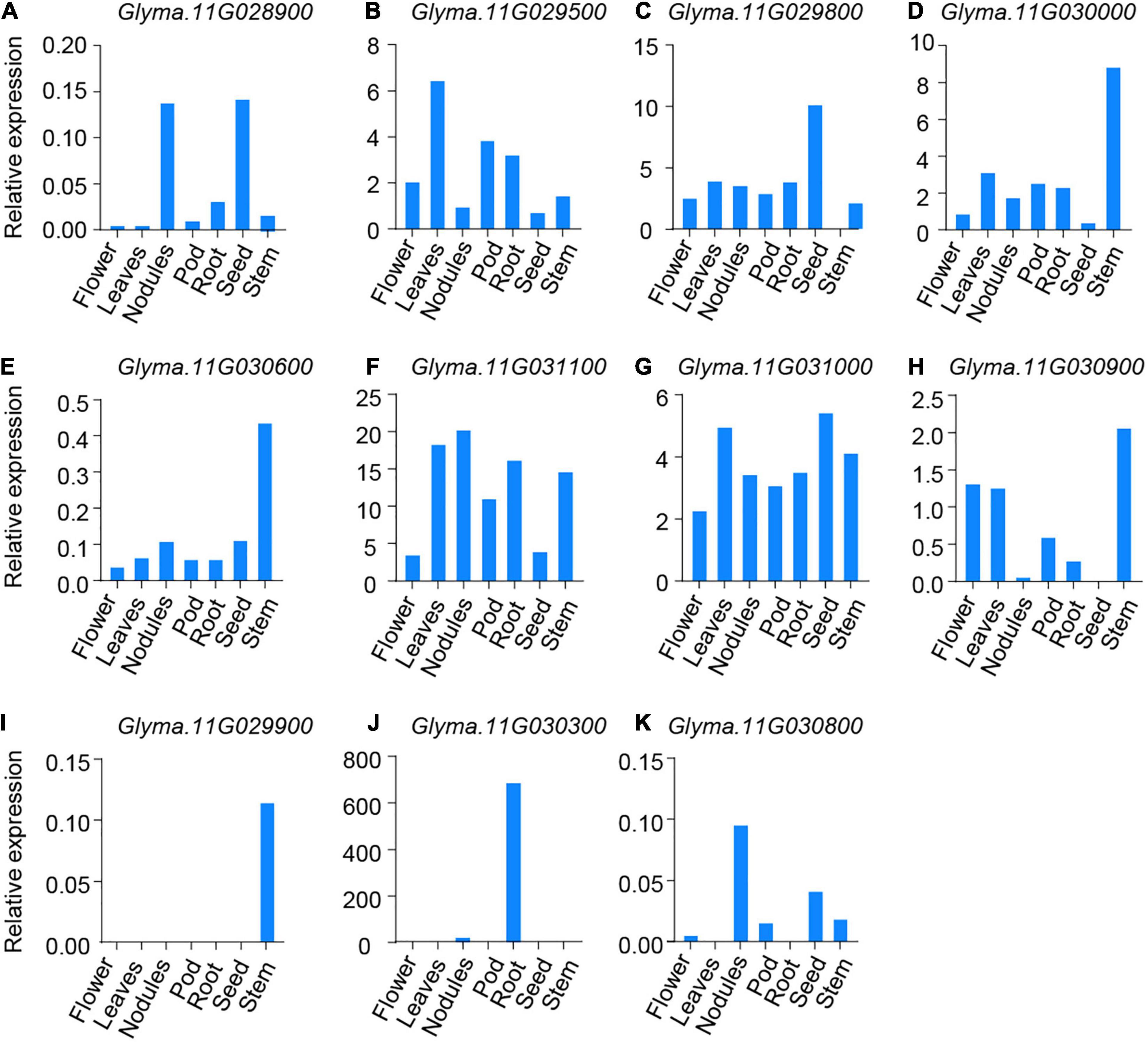
Figure 5. Expression of eleven candidate genes in different soybean tissues. (A–K) The expression of Glyma.11G028900, Glyma.11G029500, Glyma.11G029800, Glyma.11G030000, Glyma.11G030600, Glyma.11G031100, Glyma.11G031000, Glyma.11G030900, Glyma.11G029900, Glyma.11G030300, and Glyma.11G030800 in different soybean tissues (flower, leaves, pod, stem, nodule, seed, and root) obtained from an RNA-seq database. Values are mean ± s.e.m. (n = 3 biologically independent samples).
We next tested the expression of the eight candidate genes in response to SC7 or mock inoculation of DN50 leaves. Five of eight genes showed altered expression patterns following SC7 treatment as compared to mock inoculation: the expression levels of Glyma.11G028900 and Glyma.11G030600 were induced at 4 h post-inoculation; Glyma.11G029500, Glyma.11G030000, and Glyma.11G030900 were upregulated at 24 h post-inoculation; however, transcript levels of Glyma.11G029800, Glyma.11G031000, and Glyma.11G031100 did not change after SC7 inoculation (Figures 6A–H). These results suggested that Glyma.11G028900, Glyma.11G030600, Glyma.11G029500, Glyma.11G030000, and Glyma.11G030900 were the main candidate genes for the RSMV-11 locus. Of these genes, only Glyma.11G028900 harbored a SNP and InDel variation in its coding sequence, resulting in a frameshift (Figure 2B). In addition, according to gene function annotation in the Phytozome database (Liu et al., 2016), Glyma.11G028900 corresponds to GmMATE68, a gene that encodes a multidrug and toxic compound extrusion (MATE) transporter. MATE transporters result in pleiotropic phenotypes, including enhanced plant disease resistance (Nawrath et al., 2002; Ishihara et al., 2008). Taken together, these results indicate that GmMATE68 is a likely candidate gene for the RSMV-11 locus.
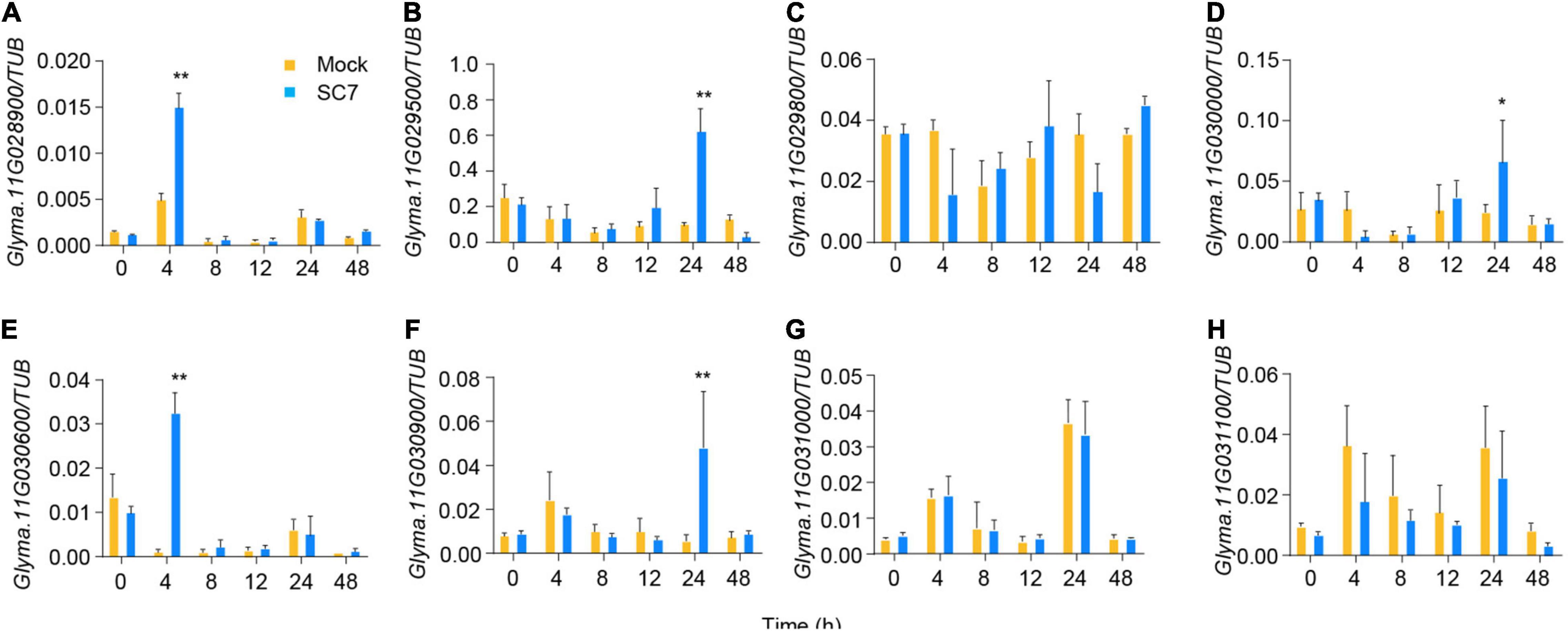
Figure 6. Expression analysis of eight candidate genes in leaves after SC7 inoculation. Values are mean ± s.e.m. (n = 3 biologically independent samples). A Student’s t-test was used to generate the P-values. *P < 0.05; **P < 0.01. Orange boxes represent mock treatment and blue boxes represent treat with SC7 stain in (A–H), respectively.
Race-specific resistance or basal resistance are major traits that affect soybean yield (Poland et al., 2009; Yasuda et al., 2015). Therefore, identifying resistance genes or resistance-associated QTLs is important for breeding crops with enhanced disease resistance. SMV is a serious threat to soybean production and seed quality worldwide (Adams et al., 2005); however, only a few QTLs linked with SMV resistance and only two resistance genes, Rsv4 and Rsc4, have been cloned (Ishibashi et al., 2019; Yin et al., 2021). Although forward genetics approaches such as positional cloning can successfully isolate candidate genes for diverse traits, they require considerable labor and time (Salvi and Tuberosa, 2005). As an alternative, the next-generation sequencing technology of BSA-seq can quickly detect QTLs for important agronomic traits over wide ranges of experimental variables. In this study, we employed BSA-seq to identify a new candidate QTL on soybean chromosome 11, RSMV-11. Our findings also confirmed that RSMV-11 confers basal resistance to SMV. Overall, our data provide valuable information regarding the genetic basis of SMV resistance in soybean.
Many putative genes have been identified in soybean by fine-mapping and genetic diversity (Lu et al., 2017; Li et al., 2018; Ishibashi et al., 2019; Dong et al., 2021). For example, J, a major classical locus conferring the long-juvenile trait, was cloned within a 239-kb region between markers M1 and M3. Only one gene, EARLY FLOWERING 3 (ELF3), differed in sequence between the parental lines in this region (Lu et al., 2017). Furthermore, the broad-spectrum SMV resistance gene Rsv4 was cloned by fine-mapping, and encodes an RNase H family protein with dsRNA-degrading activity (Ishibashi et al., 2019). In the present study, the soybean cultivar DN50 clearly showed improved SC7 resistance compared to XQD (Figure 1A). To clone the key gene associated with SC7 resistance, we produced an F2 population by crossing DN50 and XQD (Figure 2A). The F6 population was generated by self-fertilization, and analysis of this population localized RSMV-11 to a 207-kb region between markers M24 and M28 (Figure 3A), a region harboring 25 genes (Figure 4A). We thus identified a novel locus, RSMV-11, conferring resistance to SC7.
Multidrug and toxic compound extrusion (MATE) family members are widely distributed in bacteria, fungi, mammals, and plants (Omote et al., 2006). MATE transporters are involved in a wide range of biological processes in plants, such as the transport of secondary metabolites (Shoji et al., 2009; Zhao and Dixon, 2009), detoxification of toxic compounds or metals (Li et al., 2002; Zhou et al., 2019), and regulation of disease resistance (Nawrath et al., 2002; Ishihara et al., 2008). For example, GmMATE75, GmMATE79, and GmMATE87 are plasma membrane–localized MATE family members whose overexpression resulted in aluminum-induced citrate efflux in soybean (Zhou et al., 2019). Moreover, EDS5, which is homologous to members of the MATE family, responds to salicylic acid–dependent signaling, and its overexpression leads to enhanced viral resistance in Arabidopsis (Nawrath et al., 2002; Ishihara et al., 2008). In this study, we analyzed the genetic variation, tissue-specific expression pattern, and SC7-induced expression of candidate genes located within the 207-kb region harboring RSMV-11. According to gene function annotation in the Phytozome database, Glyma.11G028900 (GmMATE68) encodes a MATE transporter (Liu et al., 2016). There were 22 SNPs and four InDel mutations detected in the GmMATE68 sequence of XQD, resulting in a protein frameshift (Figure 4B). Our data also demonstrated that GmMATE68 is expressed in all tissues and is induced following SC7 treatment (Figures 5A, 6A). Our results along with those from the literature suggest that GmMATE68 may be a strong candidate gene for RSMV-11.
Overall, our research provides an important strategy for the rapid identification of target genes controlling key agronomic traits, and our results will promote the development of functional genomics in crops.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
QC and ZC designed the experiments and wrote the article. YZ, JS, MY, HD, YL, LD, and KH performed the research. LW, XS, WL, XY, HX, QD, MZ, SL, and YL analyzed the data. All authors contributed to the article and approved the submitted version.
This work was supported by Evaluation, innovation, and excellent gene excavation for elite soybean cultivars between China and Europe (2019YFE0105900-02), Science and technology planning project of Guangzhou (202102010388 and 202102010389), and Natural Science Foundation of China (32001507).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Plant Editors (https://planteditors.com/) for its linguistic assistance during the preparation of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.843633/full#supplementary-material
Adams, M. J., Antoniw, J. F., and Fauquet, C. M . (2005). Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 150, 459–479. doi: 10.1007/s00705-004-0440-6
Boerma, H. R., and Specht, J. E . (eds) (2004). Soybeans: Improvement, Production and Uses, 3rd Edn. Madison, WI: American Society of Agronomy.
Buss, G. R., Ma, G., Chen, P., and Tolin, S. A . (1997). Registration of V94-5152 soybean germplasm resistant to Soybean mosaic potyvirus. Crop Sci. 37, 1987–1988. doi: 10.2135/cropsci1997.0011183X003700060068x
Cai, C., Jiang, X., Zhao, C., and Ma, J . (2014). Sequence analysis of the coat protein gene of Chinese Soybean mosaic virus strain SC7 and comparison with those of SMV strains from the USA. Chin. J. Virol. 30, 489–494.
Che, Z. J., Yan, H. L., Liu, H. L., Yang, H., Du, H. P., Yang, Y. M., et al. (2020). Genome-wide association study for Soybean mosaic virus SC3 resistance in soybean. Mol. Breed. 40:69. doi: 10.1007/s11032-020-01149-1
Chen, P., Buss, G. R., Roane, C. W., and Tolin, S. A . (1991). Allelism among genes for resistance to Soybean mosaic virus in strain-diff erential soybean cultivars. Crop Sci. 31, 305–309. doi: 10.2135/cropsci1991.0011183X003100020015x
Cho, E. K., and Goodman, R. M . (1979). Strains of soybean mosaic virus: classification based on virulence in resistant soybean cultivars. Phytopathology 69, 467–470. doi: 10.1094/Phyto-69-467
Dong, L. D., Fang, C., Cheng, Q., Su, T., Kou, K., Kong, L. P., et al. (2021). Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Common. 12:5445. doi: 10.1038/s41467-021-25800-3
Gunduz, I., Buss, G. R., Chen, P., and Tolin, S. A . (2002). Characterization of SMV resistance genes in tousan 140 and hourei soybean. Crop Sci. 42, 90–95. doi: 10.2135/cropsci2002.0090
Gunduz, I., Buss, G. R., Chen, P., and Tolin, S. A . (2004). Genetic and phenotypic analysis of Soybean mosaic virus resistance in PI 88788 soybean. Phytopathology 94, 687–692. doi: 10.1094/PHYTO.2004.94.7.687
Hajimorad, M. R., Domier, L. L., Tolin, S. A., Whitham, S. A., and Saghai Maroof, M. A . (2018). Soybean mosaic virus: a successful Potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 19, 1563–1579. doi: 10.1111/mpp.12644
Hajimorad, M. R., and Hill, J. H . (2001). Rsv1-mediated resistance against Soybean mosaic virus-N is hypersensitive response-independent at inoculation site, but has the potential to initiate a hypersensitive response-like mechanism. Mol. Plant Microbe Interact. 14, 587–598. doi: 10.1094/MPMI.2001.14.5.587
Hayes, A. J., Ma, G., Buss, G. R., and Maroof, M. A. S . (2000). Molecular marker mapping of RSV4, a gene conferring resistance to all known strains of Soybean mosaic virus. Crop Sci. 40, 1434–1437. doi: 10.2135/cropsci2000.4051434x
Hoeck, J. A., Fehr, W. R., Shoemaker, R. C., Welke, G. A., Johnson, S. L., and Cianzio, S. R . (2003). Molecular marker analysis of seed size in soybean. Crop Sci. 43, 68–74. doi: 10.2135/cropsci2003.6800
Ishibashi, K., Saruta, M., Shimizu, T., Shu, M., Anai, T., Komatsu, K., et al. (2019). Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat. Commun. 10:4033. doi: 10.1038/s41467-019-12052-5
Ishihara, T., Sekine, K. T., Hase, S., Kanayama, Y., Seo, S., Ohashi, Y., et al. (2008). Overexpression of the Arabidopsis thaliana EDS5 gene enhances resistance to viruses. Plant Biol. 10, 451–461. doi: 10.1111/j.1438-8677.2008.00050.x
Kiihl, R. A. S., and Hartwig, E. E . (1979). Inheritance of reaction to Soybean mosaic virus in soybeans1. Crop Sci. 19, 372–375. doi: 10.2135/cropsci1979.0011183x001900030024x
Klepadlo, M., Chen, P. Y., Shi, A., Mason, E. R., Korth, K. L., and Srivastava, V . (2017). Two tightly linked genes for Soybean mosaic virus resistance in soybean. Crop Sci. 57, 1844–1853. doi: 10.2135/cropsci2016.05.0290
Li, K., Ren, R., Adhimoolam, K., Gao, L., Yuan, Y., Liu, Z., et al. (2015). Genetic analysis and identification of two soybean mosaic virus resistance genes in soybean [Glycine max (L.) Merr]. Plant Breed. 134, 684–695. doi: 10.1111/pbr.12315
Li, K., Yang, Q. H., Zhi, H. J., and Gai, J. Y . (2010). Identification and distribution of Soybean mosaic virus strains in southern China. Plant Dis. 94, 351–357. doi: 10.1094/PDIS-94-3-0351
Li, L. G., He, Z. Y., Pandey, G. K., Tsuchiya, T., and Luan, S . (2002). Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 277, 5360–5368. doi: 10.1074/jbc.M108777200
Li, Z. F., Guo, Y., Ou, L., Hong, H. L., Wang, J., Liu, Z. X., et al. (2018). Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by sequencing and linkage analysis. Theor. Appl. Genet. 131, 1001–1016. doi: 10.1007/s00122-017-3044-8
Liu, J. G., Li, Y., Wang, W., Gai, J. Y., and Li, Y . (2016). Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics 17:223. doi: 10.1186/s12864-016-2559-8
Lu, S. J., Zhao, X. H., Hu, Y. L., Liu, S. L., Nan, H. Y., Li, X. M., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779. doi: 10.1038/ng.3819
Ma, Y., Wang, D. G., Li, H. C., Zheng, G. J., Yang, Y. Q., Li, H. W., et al. (2011). Fine mapping of the RSC14Q locus for resistance to Soybean mosaic virus in soybean. Euphytica 181, 127–135. doi: 10.1007/s10681-011-0457-3
Machado, F. B., Moharana, K. C., Fabricio, A. S., Rajesh, K. G., and Thiago, M. V . (2020). Systematic analysis of 1,298 RNA-Seq samples and construction of a comprehensive soybean (Glycine max) expression atlas. Plant J. 103, 1894–1909. doi: 10.1101/2019.12.23.886853
Michelmore, R. W., Paran, I., and Kesseli, R . (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Moon, J. K., Jeong, S. C., Van, K., Saghai Maroof, M. A., and Lee, S. H . (2009). Marker-assisted identify cation of resistance genes to Soybean mosaic virus in soybean lines. Euphytica 169, 375–385. doi: 10.1007/s10681-009-9970-z
Nawrath, C., Heck, S., Parinthawong, N., and Métraux, J. P . (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286. doi: 10.1105/tpc.010376
Omote, H., Hiasa, M., Matsumoto, T., Otsuka, M., and Moriyama, Y . (2006). The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 27, 587–593. doi: 10.1016/j.tips.2006.09.001
Poland, J. A., Balint-Kurti, P. J., Wisser, R. J., Pratt, R. C., and Nelson, R. J . (2009). Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14, 21–29. doi: 10.1016/j.tplants.2008.10.006
Ross, J. P . (1968). Effect of single and double infections of soybean mosaic and bean pod mottle viruses on soybean yield and seed characters. Plant Dis. Rep. 52, 344–348.
Ross, J. P . (1977). Effect of aphid-transmitted Soybean mosaic virus on yields of closely related resistant and susceptible soybean lines. Crop Sci. 17, 869–872. doi: 10.2135/cropsci1977.0011183x001700060014x
Saghaimaroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W . (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8018. doi: 10.1073/pnas.81.24.8014
Salvi, S., and Tuberosa, R . (2005). To clone or not to clone plant QTLs: present and future challenges. Trends Plant Sci. 10, 297–304. doi: 10.1016/j.tplants.2005.04.008
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J. X., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Shoji, T., Inai, K., Yazaki, Y., Sato, Y., Takase, H., Shitan, N., et al. (2009). Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol. 149, 708–718. doi: 10.1104/pp.108.132811
Suh, S. J., Bowman, B. C., Jeong, N., Yang, K., Kastl, C., Tolin, S. A., et al. (2011). The Rsv3 locus conferring resistance to Soybean mosaic virus is associated with a cluster of coiled-coil nucleotide-binding leucine-rich repeat genes. Plant Genome 4, 55–64. doi: 10.3835/plantgenome2010.11.0024
Tucker, D. M., Saghai Maroof, M. A., Jeong, S. C., Buss, G. R., and Tolin, S. A . (2009). Validation and interaction of the Soybean mosaic virus lethal necrosis allele, Rsv1-n, in PI 507389. Crop Sci. 49, 1277–1283. doi: 10.2135/cropsci2008.08.0477
Wang, D., Ma, Y., Liu, N., Yang, Z., Zheng, G., and Zhi, H . (2011). Fine mapping and identification of the soybean RSC4 resistance candidate gene to Soybean mosaic virus. Plant Breed. 130, 653–659. doi: 10.1111/j.1439-0523.2011.01888.x
Wang, X., Gai, J., and Pu, Z . (2003). Classification and distribution of strains of Soybean mosaic virus in middle and lower Huanghuai and Changjiang river valleys. Soybean Sci. 22, 102–107.
Yan, H., Wang, H., Cheng, H., Hu, Z., Chu, S., Zhang, G., et al. (2015). Detection and fine-mapping of SC7 resistance genes via linkage and association analysis in soybean. J. Integr. Plant Biol. 57, 722–729. doi: 10.1111/jipb.12323
Yang, Q. H., and Gai, J. Y . (2011). Identification, inheritance and gene mapping of resistance to a virulent Soybean mosaic virus strain SC15 in soybean. Plant Breed. 130, 128–132. doi: 10.1111/j.1439-0523.2010.01797.x
Yasuda, N., Mitsunaga, T., Hayashi, K., Koizumi, S., and Fujita, Y . (2015). Effects of pyramiding quantitative resistance genes Pi21, Pi34, and Pi35 on rice leaf blast disease. Plant Dis. 99, 904–909. doi: 10.1094/pdis-02-14-0214-re
Yin, J., Wang, L., Jin, T., Nie, Y., Liu, H., Qiu, Y., et al. (2021). A cell wall-localized NLR confers resistance to soybean mosaic virus by recognizing viral-encoded cylindrical inclusion protein. Mol. Plant 14, 1–20. doi: 10.1016/j.molp.2021.07.013
Zhao, J., and Dixon, R. A . (2009). MATE transporters facilitate vacuolar uptake of epicatechin 3’-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21, 2323–2340. doi: 10.1105/tpc.109.067819
Zheng, G. J., Yang, Y. Q., Ma, Y., Yang, X. F., Chen, S. Y., Ren, R., et al. (2014). Fine mapping and candidate gene analysis of resistance gene RSC3Q to Soybean mosaic virus in Qihuang 1. J. Integr. Agric. 13, 2608–2615. doi: 10.1016/S2095-3119(13)60738-8
Keywords: soybean, QTL, Soybean mosaic virus, SC7 strain, QTL-seq, MATE transporter
Citation: Zhang Y, Song J, Wang L, Yang M, Hu K, Li W, Sun X, Xue H, Dong Q, Zhang M, Lou S, Yang X, Du H, Li Y, Dong L, Che Z and Cheng Q (2022) Identifying Quantitative Trait Loci and Candidate Genes Conferring Resistance to Soybean Mosaic Virus SC7 by Quantitative Trait Loci-Sequencing in Soybean. Front. Plant Sci. 13:843633. doi: 10.3389/fpls.2022.843633
Received: 26 December 2021; Accepted: 09 February 2022;
Published: 28 February 2022.
Edited by:
Abraham J. Escobar-Gutiérrez, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Qingshan Chen, Northeast Agricultural University, ChinaCopyright © 2022 Zhang, Song, Wang, Yang, Hu, Li, Sun, Xue, Dong, Zhang, Lou, Yang, Du, Li, Dong, Che and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijun Che, Y2hlempAZ3podS5lZHUuY24=; Qun Cheng, Y2hlbmdxdW4wMTE4QGd6aHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.