- 1Research Institute of Climate Change and Agriculture, National Institute of Horticultural and Herbal Science, Rural Development Administration (RDA), Jeju, South Korea
- 2Interaction Laboratory, Future Convergence Engineering, Advanced Technology Research Center, Korea University of Technology and Education, Cheonan-si, South Korea
- 3Floriculture Research Division, RDA, Wanju-gun, South Korea
- 4CSSR and SRRM degree and PG College, Kadapa, India
- 5Department of Medical Biotechnology, Dongguk University, Seoul, South Korea
- 6Department of Life Science, College of Life Science and Biotechnology, Dongguk University, Seoul, South Korea
The CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein 9) method is a versatile technique that can be applied in crop refinement. Currently, the main reasons for declining agricultural yield are global warming, low rainfall, biotic and abiotic stresses, in addition to soil fertility issues caused by the use of harmful chemicals as fertilizers/additives. The declining yields can lead to inadequate supply of nutritional food as per global demand. Grains and horticultural crops including fruits, vegetables, and ornamental plants are crucial in sustaining human life. Genomic editing using CRISPR/Cas9 and nanotechnology has numerous advantages in crop development. Improving crop production using transgenic-free CRISPR/Cas9 technology and produced fertilizers, pesticides, and boosters for plants by adopting nanotechnology-based protocols can essentially overcome the universal food scarcity. This review briefly gives an overview on the potential applications of CRISPR/Cas9 and nanotechnology-based methods in developing the cultivation of major agricultural crops. In addition, the limitations and major challenges of genome editing in grains, vegetables, and fruits have been discussed in detail by emphasizing its applications in crop refinement strategy.
Introduction
In agroecology, the integrated approach in production of crops and conceptual marketing management draws holistic economic concern. As reported by “Food and Agriculture Organization” (FAO, 2017), the crop failure be falls mainly due to biotic and abiotic factors, significantly influencing the economic values of crops (Gautam and Kumar, 2020). Environmental and climate changes, causing frequent flood, droughts, temperature variations, higher soil salinity, use of harmful chemicals as additives/fertilizers, pathogen triggered diseases, deteriorate the plant health and affects directly to the crop quality and yield (Bing, 2020). Typically, breeders follow customary methods along with marker-assisted selection to introduce new traits in plants. Some chemical compounds and irradiation techniques are also employed to attain desirable traits; however, they often lead to random mutations in crop genomes.
The customary methods have certain drawbacks such as non-specificity and the generation of mutations with abundance of nucleotides (Mao et al., 2019). Natural chemical compounds from plant source that are essential to the pharmaceutical industry can only be acquired in limited quantity from normal plants compared to genomically edited plants. Moreover, the rational methods are time-consuming; consequently, breeders are unable to grow plants with desired trait in deadline. Supplying sufficient food and other plant-based chemical constituents to ever-growing population is challenging; and quite perplexing topic in near future. The CRISPR/Cas9 genome editing (GE) with nanotechnology-based protocols can potentially challenge these obstacles.
The targeted genomic engineering can be extremely beneficial to agriculture. If the function of a specific gene is known, it can be over expressed or suppressed to obtain the desired trait. Eventually, the crops that are capable of withstanding biotic and abiotic stresses can be easily developed using CRISPR/Ca9 and nanotechnology. Similarly, undesired genes can also be silenced by using these technologies, which would allow expression of only the desirable traits to eventually obtain higher yield. It will be an extremely difficult task to improve crop refinement without genomic engineering. Conventional breeding programs would require longer duration to introduce new quality-related traits or disease-tolerance traits. Therefore, genomic engineering is a striking technology for the future development of agricultural crops with nutrition. Nanotechnology-based protocols has new sets of advanced applications in agriculture and biomedicine; typically, the nano-sized particles are used to deliver the task obtain desirable results in crop development. Coalescing biotechnology and nanotechnology approaches, including GE, have more benefits than customary breeding to improve the development of food crops, which can naturally overcome biotic and abiotic stress along with enhancing the yield. This brief review mainly focuses the importance of genomic engineering in agriculture, and the progress in developing mutant plants using sequence-specific nucleases (SSNs). Furthermore, the procreation of genomically edited crops are already developed in agricultural biotechnology, the main objective, limitations and prospective challenges of GE are highlighted including the role of nanotechnology-based methods in crop refinement.
Generating Mutant Plants Using Sequence Specific Nucleases
Sequence specific nucleases are mainly used for precise gene editing in plants and animals. It can create mutations at desired loci in multiple genes via addition, deletion, and alternation of sequences (Songstad et al., 2017). To generate the mutant plants, firstly SSNs requires to be articulated in cells; subsequently, recognizing a specific DNA sequence to make the double stranded break. We can classify the SSNs into four major classes, namely (i) CRISPR-Cas9, (ii) Zinc finger nucleases (ZFN), (iii) Meganucleases, and (iv) Transcription activator-like effector nucleases (TALENs). These methods can be effectively utilized for the GE technique. CRISPR-Cas9 is derived from the adaptive immune systems of bacteria; in this mechanism, abounding components come into play to perform the GE. Zinc finger nucleases (ZFNs) are the enzymes that have been characterized 77 candidate two-finger modules (Urnov et al., 2005; Miller et al., 2007). Meganucleases are the microbial enzymes (Smith et al., 2006) that can 76 distinguish and more than 14 nucleotides for cleavage. TALENs have been developed by combining the FokI nuclease domain with TALE proteins of Xanthomonas (Christian et al., 2010). CRISPR/Cas9 is one among the four classes of SSNs that can be used for GE (Songstad et al., 2017). GE technologies by adopting CRISPR/Cas9 methodology was investigated in 1987 and its functional application in human cell was reconnoitered in 2013 (Mali et al., 2013). Hsu et al. (2014) discussed the detailed challenges and its future prospective. Consequently, the GE technology was successfully implemented in crop refinement of soybean plant (Cai et al., 2015). Once the SSNs construct is incorporated into the plant genome, they are expressed at a distal site, while the remaining construct is removed by crossing the plant to obtain a mutated plant with no transgene. For GE with the CRISPR/Cas9 system, it is essential to deliver sgRNA and Cas9 proteins into the target cells. Expression vectors or microinjected RNA/mRNA (for Cas9) are usually used to express the sgRNA and Cas9 protein in the plant cells. The CRISPR/CAS9 technology has been performed in plant cells by using electroporation, Agrobacterium-mediated transformation, shotgun methodologies, and polyethylene glycol-mediated routes (Jiang et al., 2013; Li et al., 2013; Mao et al., 2013; Nekrasov et al., 2013; Shan et al., 2013). The double stranded break can be fixed either by homology directed repair (HDR) or non-homologous end joining (NHEJ) (Hsu et al., 2014). Recently, RNA viruses have been used to deliver hairpin RNAs for gene silencing, which is another reported technique to incorporate SSNs into the plant cells (Lacomme, 2015). Before the integration into plant’s genomic DNA, SSNs are transiently expressed from viral vectors into mRNAs and its respective proteins. The ability to modify the genes to modulate specific traits and homologous recombination (HR) allows plant to metabolize in a manner that develops their resistance to biotic and abiotic stresses. Plants synthesized by fast-growing genome engineering, generally exhibit higher crop yields because of its higher ability for photosynthesis. Precise altering the DNA sequence is extremely important to comprehend the achievable challenges insynthetic biology (Abdallah et al., 2014; Carroll, 2014).
Components and CRISPR/Cas9 Mechanism
The guide RNA (gRNA) sequence comprising of twenty nucleotides that are essential to balance to the target DNA and the details of sgRNAs designing is well explained by Hussain et al. (2018). Similarly, the protein Cas9 has the catalytic activity and having the capability to cut the double standard DNA. When Cas9 and gRNA are combined to form a complex, cas9 immediately cuts the double-stranded DNA (Tang et al., 2019), and so forth the total gene sequence will be altered and specific protein synthesis will not befall by translation. There are two major pathways to repair the broken double-stranded DNA i.e., Non-homologous end joining (NHEJ) pathway and HDR (El-Mounadi et al., 2020).
Non-homologs End Joining Pathway
The lost DNA part cannot be recollected in this NHEJ pathway. In this repair pathway, the dimeric protein complex (Ku) binds at the end of the broken DNA and later to DNA protein kinase catalytic subunits (DNA-PKCs). Artemis proteins are also come in to play and bind at the DNA terminal to make a complex; allowing phosphorylation and eventually the synthesis of DNA begin. This double-stranded DNA converted to blunt-ended double-stranded DNA by catalytic DNA ligase reaction, forming covalent linkage of phosphodiesters (Gomez et al., 2017). This repair system allows insertions or deletions of nucleotide bases that occur during a process (Bernheim et al., 2017; Tang et al., 2019).
Homology Directed Repair Pathway
The HDR pathway use the autologous donor DNA sequences from sister chromatids or foreign DNA to create precise insertion and substitution between DNA double-strand break (DSB) sites for further alterations. Considerable research has been done previously on proteins involved in the HDR pathway, MRE11-Rad50-Nbs1 (MRN) complex binds at the 5′ end of the DSBs and forms the 3′ overhangs. Later the replication protein A (RPA) binds to the single-strand DNA to prevent the nuclease activity. RAD-51 protein involves in search of homologous DNA and eventually the invasion occurs to complete the homologs-directed repair (Tang et al., 2019). El-Mounadi et al. (2020) explained the CRISPR/Cas9 mechanism system was depicted in Figure 1.
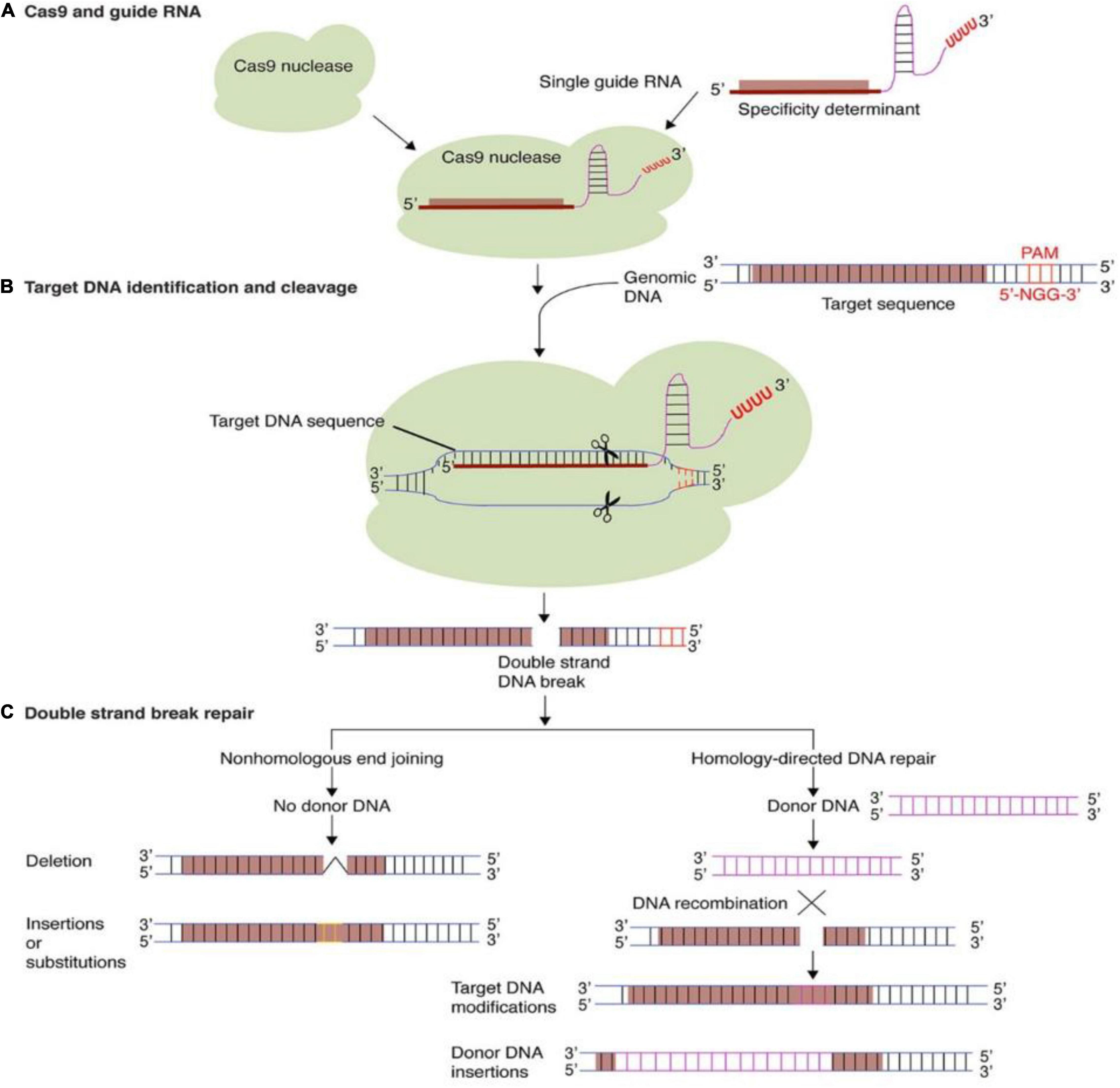
Figure 1. Targeted genome editing via CRISPR-Cas9. (A) The CRISPR-Cas9 system comprises of a Cas9 protein and guide RNA. Guide RNAs regulate the target DNA specificity by sequence complementarity. (B) gRNA and Cas9 protein form a binary complex that specifically cleaves target DNA creating a double-strand DNA break. (C) Cellular DNA repair mechanisms: non-homologous end joining (NHEJ) and homology-directed repair (HDR), repairs the double strand DNA break. In the process, short insertions, deletions, nucleotide substitutions, or gene insertion may occur. Reproduced with permission from El-Mounadi et al. (2020) Frontiers.
Importance of Genomic Engineering in Agriculture
Transgenesis profoundly involving genetic addition without changing the genetic pool to create specific traits for agricultural/agroeconomical benefits. In the present scenario, providing food security to a mounting populace is one of the major challenges in this modern world. Thenceforth, the food fructification needs to be increased over 80–90%. Furthermore, the food production is declining every year due to extreme weather conditions, climate change, global warming, farmland availability, and cumulative biotic and abiotic problems. To overcome these challenging obstacles, gene modification technique in food crops will be the superlative method to achieve the targeted tasks (FAO, 2017). The gene knockout step is a critical stage and it directly influence the phenotype. Wada et al. (2020) demonstrated genomic manipulation without the introduction of DSBs. In this modular approach, a dead Cas9 variant (dCas9) binds to the target sequence; however, it does not cleave the double-stranded DNA (Qi et al., 2013; Wang et al., 2016a; Adli, 2018). Generating gene knockouts using SSNs facilitates genetic analyses and the study of important gene functions, which will eventually help with crop improvement. The first mutation generated using SSNs were of the IPK1 gene in maize, which is catalyzed in the last step during phytate biosynthesis (Liang et al., 2014). Knocking out this gene helps in removing unwanted metabolites; eventually contributing to the accumulation of valuable biosynthetic intermediates. Prime editing is another significant approach for genomic manipulation that has been indicated in mammalian and yeast cells (Anzalone et al., 2019). Certain crops such as grains, vegetables, and fruits are vitally important to maintain global food securities and sustainable system.
Homologous recombination is a challenging process wherein chromosome can chasm by the nuclease that must be coordinated with the distribution of the DNA repair template. Initially, HR in plants was confirmed by the incorporation of marker genes at detailed chromosomal sites. Targeted transgene insertion into the euchromatin should provide promising results in plants with high transgene expression; therefore, it is a great improvement over the integration by the traditional Transgenesis (Neelendra et al., 2018). Moreover, the insertion of multiple genes at the same site will facilitate their transfer to a single mendelian locus, when the plant is crossed. Further efforts are required to familiarize plentiful genes into the germplasm by breeding. Targeted gene inclusion through HR using different SSNs has been verified in tobacco, maize, and rice (Abdallah et al., 2014; Wang et al., 2016b). In order to develop new plant varieties, some techniques have been used that are controlled by process-based regulatory frameworks. Most of the farmers, globally aim to cultivate crops that are tolerant to drought, high salinity, and diseases, with appreciable yield. Therefore, the researchers in agriculture domain are actively looking for transgenic technology and CRISPR/CAS9 to achieve targeted tasks. Guidelines and process-based regulations have been formulated and implemented by the United Nations Food and Agricultural Organization adopted by European and South American countries (FAO, 2017).
Applications of Clustered Regularly Interspaced Short Palindromic Repeats to Develop Cereal Crops
Globally, there is a renowned demand for Basmati rice because of its fragrance, long grains, and flavor texture. However, due to bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo), the yield and rice quality will be deprived from its authentic taste. By CRISPR/Cas9 GE technique, the genes namely OsSWEET11, OsSWEET13, and OsSWEET14 could be possibly edited to overcome the bacterial blight disease (Zafar et al., 2020). New Japonica rice is another prominent Asian rice variety, developed by editing Ehd1 (Early heading date 1) gene via agrobacterium mediated transformation. The mutated rice varieties exhibited prolonged basic vegetative growth (BVG) period at latitudes (Wu et al., 2020). It was documented that, OsBADH2 (betaine aldehyde dehydrogenase) gene was modified to develop the fragrance in non-aromatic rice grains for better marketing (Ashokkumar et al., 2020). Two endogenous genes, namely TaWaxy and TaMTL, were edited by using three different promotors (OsU6a, TaU3, and TaU6) for the development of haploid plants in wheat crops. Among these three promotors, TaU3 showed better results (Liu et al., 2020). TaPDS gene was edited in wheat by using Cas9 and Cpf1 (AsCpf1 and LbCpf1) nucleases in wheat (Kim et al., 2021). Lipoxygenases genes (GmLox1, GmLox2, and GmLox3) were edited by using CRISPR/Cas9 to afford lipoxygenase-free new mutant lines in soybean crop, so that it can be useful for human consumption with upsurge amount of protein for health benefits (Wang et al., 2020). Aside from maize, wheat, and rice, the fourth most highly consumable crop in the world is barley. The two mutated genes, HvHPT and HvHGGT in barley are mainly accountable for the dwindled grain size to weight ratios. Furthermore, these mutated barley lines showed reduced amount of tocotrienols assayed by HPLC (Zeng et al., 2020). Groundnut is the sixth most significant oil-seed crop in the world; this legume crop fixes the nitrogen via symbiotic relationship with rhizobia. In this process NFR (Nod Factor Receptor) play a major role in nitrogen fixation cycle. Thence, AhNFR1 and AhNFR5 genes were mutated by CRISPR/Cas9 to proliferate the root nodules via hairy root transformation system (Shu et al., 2020). CAD (cinnamyl alcohol dehydrogenase) and PDS (phytoene desaturase) genes were edited by CRISPR/Cas9 in sorghum via Biolistic bombardment resulted in enhanced biosynthesis of carotenoid and chlorophylls (Liu et al., 2019). SiMTL gene, which is orthologous to the maize MATRILINEAL/NOT-LIKE-DAD/PHOSPHOLIPASE A (MTL/NLD/ZmPLA) gene is edited by CRISPER/Cas9 with OsU3 promotor via Agrobacterium for the haploid induction in foxtail millet (Cheng et al., 2021). Please see Supplementary Table S1 (Fauser et al., 2014; Sugano et al., 2014; Baltes et al., 2015; Li et al., 2015, Li et al., 2016, 2018b,f; Li J. et al., 2017; Michno et al., 2015; Tsai and Xue, 2015; Xie et al., 2015; Zhou et al., 2015, 2016; Zhou X. et al., 2018; Baek et al., 2016; Duan et al., 2016; Feng et al., 2016; Gao et al., 2016, 2017; Iqbal et al., 2016; Osakabe et al., 2016; Pan L. et al., 2016; Pioneer, 2016; Pyott et al., 2016; Qi et al., 2016; Waltz, 2016; Watanabe et al., 2016; Zhang Y. et al., 2016; Zhao et al., 2016; Zhu et al., 2016; Chen et al., 2017; Kapusi et al., 2017; Mercx et al., 2017; Ordon et al., 2017; Wang et al., 2017b,c; Yang et al., 2017c; Che et al., 2018; Pankaj et al., 2018). For brief listing of prime cereal/food crop genes along with its specific functions modified via CRISPR/Cas9 system.
Applications of Clustered Regularly Interspaced Short Palindromic Repeats to Develop Fruits/Vegetable Crops
Trans-Acting Small-interfering locus 4 (TAS4) and MYBA7 (Transcription factor) genes are edited by CRISPR/Cas9 via Agrobacterium to enhance the biotic and abiotic tolerance in grapes. These genes showed pronounced tolerance against the bacterium Xylella fastidiosa and Grapevine Red Blotch Virus (GRBV) causes Red Blotch Disease (Sunitha and Rock, 2020). Citrus canker is a dangerous disease caused by the bacterium Xanthomonas axonopodis. It is threatening to citrus family crops worldwide. The CsWRKY22 gene was edited by CRISPR/Cas9 with AtU6-1 promotor via Agrobacterium methodology to produce the Canker disease-free citrus in Wanjincheng orange plants. PDS (phytoene desaturase) gene was mutated for the development of Albino phenotype and carotenoid biosynthesis in banana crop (Otang Ntui et al., 2020). Fusarium oxysporum is a dangerous pathogen for watermelon. The editing of Clpsk1 gene that encode Phytosulfokine (PSK) precursor could be conferred to enhance resistance of Fusarium oxysporum to watermelon, the gene was efficiently edited by CRISPR/Cas9 system via Agrobacterium method to produce the Fusarium oxysporum resistant watermelons in appreciable yield (Zhang et al., 2020).
SlJAZ2 is a major co-receptor of coronatine (COR) in the stomatal guard cells of tomato fruit. This gene was edited for the development of bacterial speck disease resistance via Agrobacterium (Ortigosa et al., 2019). Broomrapes (Phelipanche aegyptiaca and Orobanche spp.), a kind of plant parasite can cause severe damage to the tomato plants. Thus, the CCD8 (Carotenoid Cleavage Dioxygenase 8) gene was edited to afford the Phelipanche aegyptiaca parasite resistant tomatoes (Bari et al., 2019). SlMlo1 and SlPelo genes were altered by CRISPR/Cas9 in tomato for resistant to yellow leaf curl virus and powdery mildew. These gene modifications successfully develop the pathogen-resistant tomatoes (Pramanik et al., 2021). Steroidal glycoalkaloids (SGAs) are plant secondary plant metabolites, better known for their toxic effects in humans and animals. High SGAs content can severely damage the potato quality. To diminish the SGAs content to optimum levels, StSSR2 (Sterol side chain reductase 2) gene, a key enzyme for the biosynthesis of SGAs was edited (Zheng et al., 2021). BoaCRTISO gene of Chinese kale was altered to increase the Carotenoid biosynthesis and the observed mutation rate was 81.25% (Sun B. et al., 2020). CsCRUC (Camelina sativa CRUCIFERIN C) gene encodes the seed proteins in Camelina sativa and this gene was edited by CRISPR/Cas9 to enhance the seed storage protein and enriched saturated fatty acids contents (Lyzenga et al., 2019). Powdery mildew is typically observed destructive disease that affect the leaves of wheat crop. This fungal leaf ailment can severely damage up to 40% of the crop under optimum ecological conditions (Gil-Humanes and Voytas, 2014). Thenceforth to overcome this problem MLO (MILDEW-RESISTANCE LOCUS) genes were modified in bread wheat (Wang et al., 2014). Assuredly, the bread wheat verities showed effective tolerance to powdery mildew. Bacterial blight disease is one of the most destructive afflictions, caused by Xanthomonas oryzae that can severely devastate for nutritional crop growth. In rice crops, the bacterial blight disease resistance was significantly improved by incorporating OsSWEET11, OsSWEET13, and OsSWEET14 genes (Zafar et al., 2020). To control the weed growth, the herbicide-resistant crops were instigated by OsPDS, OsPMS3, OsEPSPS genes (Zhang et al., 2014). The amylose content in rice endosperm was increased by the waxy (Wx) gene (Yunyan et al., 2019), thereby enhancing the grain number by OsSPL16 gene (Usman et al., 2021). Tiller-spreading phenotype of rice plants were improved by LAZY1 (LA1) gene (Miao et al., 2013). OsRR22 gene encodes 696-amino acid B-type response transcription factor that is intricate in cytokinin-signal transduction and metabolism, its loss of function suggestively upsurges the salt tolerance (Zhang A. et al., 2019) and the drought tolerance by OsNAC14 gene has been successfully developed CRISPR/Cas9 technique. For brief listing of prime fruits genes (see Table 1) and vegetable genes (see Supplementary Table S2; Ron et al., 2014; Butler et al., 2015; Ito et al., 2015; Lawrenson et al., 2015; Woo et al., 2015; Pan C. et al., 2016; Thomazella et al., 2016; Xu et al., 2016; Hayut et al., 2017; Hu et al., 2017; Klap et al., 2017; Koseoglou, 2017; Lang et al., 2017; Nonaka et al., 2017; Roldan et al., 2017; Soyk et al., 2017; Ueta et al., 2017; Yang et al., 2017b; Ye et al., 2017; Yu et al., 2017; Zhou et al., 2017) along with its specific functions modified via CRISPR/Cas9 system.
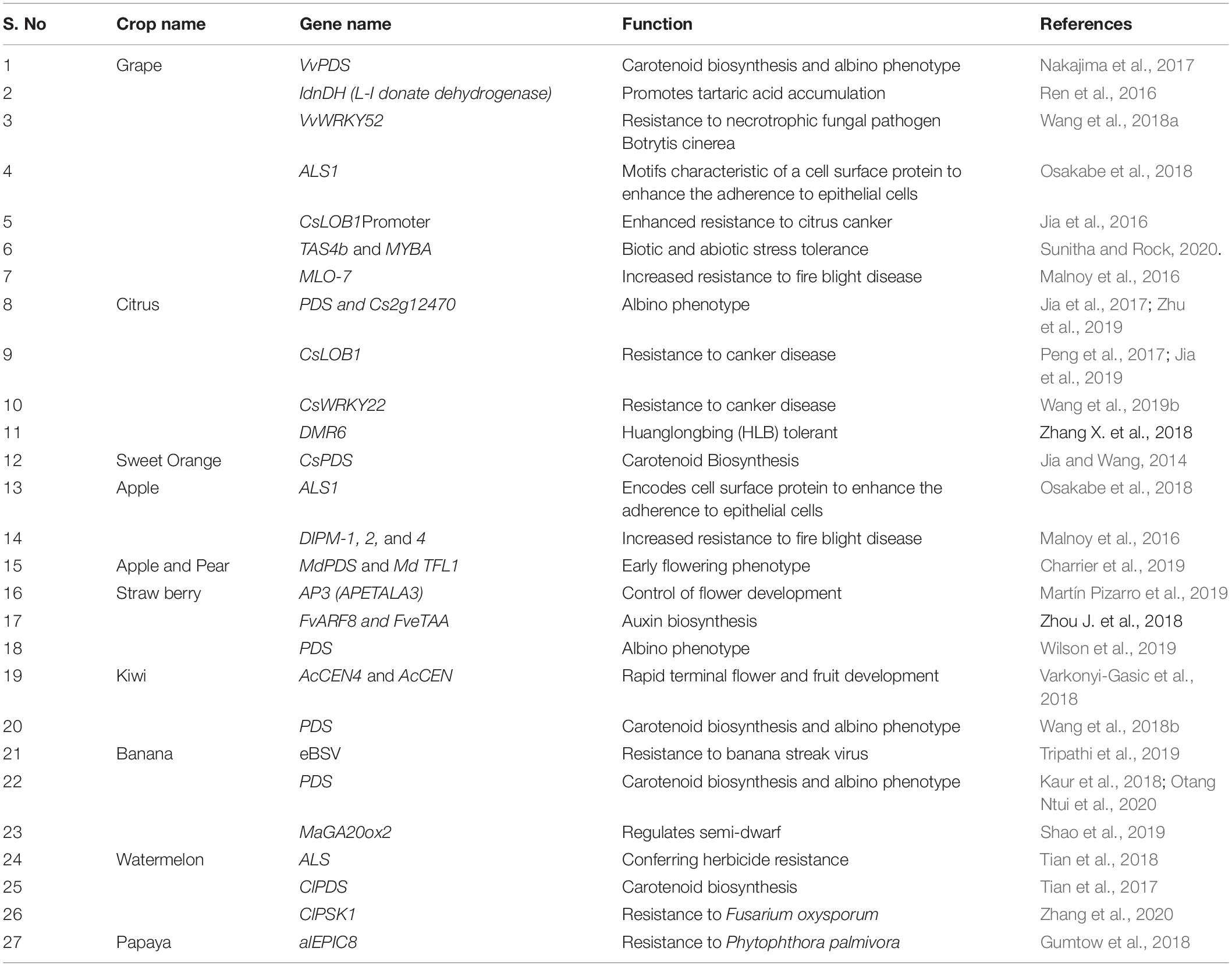
Table 1. Partial list of genes of fruit crops and its specific functions modified via CRISPR/Cas9 system.
Applications of Clustered Regularly Interspaced Short Palindromic Repeats to Develop Ornamental Crops
Ornamental plants usually grown for decoration aspirations and are often associated with commercial orientations in agroecological farming practices. These attractive flowering plants are customarily utilized in extracting perfumes and opens a profitable platform in fragrance market. Ornamental plants also a play a vital economic role in farming economy and sustainable agricultural business. Marigold flower and its extracts are used in poultry industry as feed additive, in order to enhance the quality of egg production and enticing yolk color. PhNR (Petunia Nitrate Reductase) gene was modified to check the nitrogen uptake and nitrate metabolism in petunia plants. Mutated plants showed better nitrogen uptake efficiency (Subburaj et al., 2016). CiPDS (chicory phytoene desaturase) gene was edited by under U6 promotor via Agrobacterium mediated and protoplast transfection methods. Among these two methods, Agrobacterium mediated route showed 31.25% transformation efficiency in chicory and the mutated chicory plants showed better Albino phenotype content (Bernard et al., 2019). MADS genes (MADS, MADS44, MADS36, and MADS8) from flowering plant of the orchid genus Phalaenopsis was using CRISPR/Cas9 for floral initiation and flower development (Tong et al., 2020). Lilium spp. is a genus of more than 100 species of flowering plants emergent from bulbs. To intensify the beauty and color shades, LpPDS gene was edited by CRISPR/Cas9 (Yan et al., 2019), to enforce to bloom attractive pale yellow to albino–green color shades. Table 2 gives the concise listing of prime genes of fruits and its specific functions modified via CRISPR/Cas9 system.
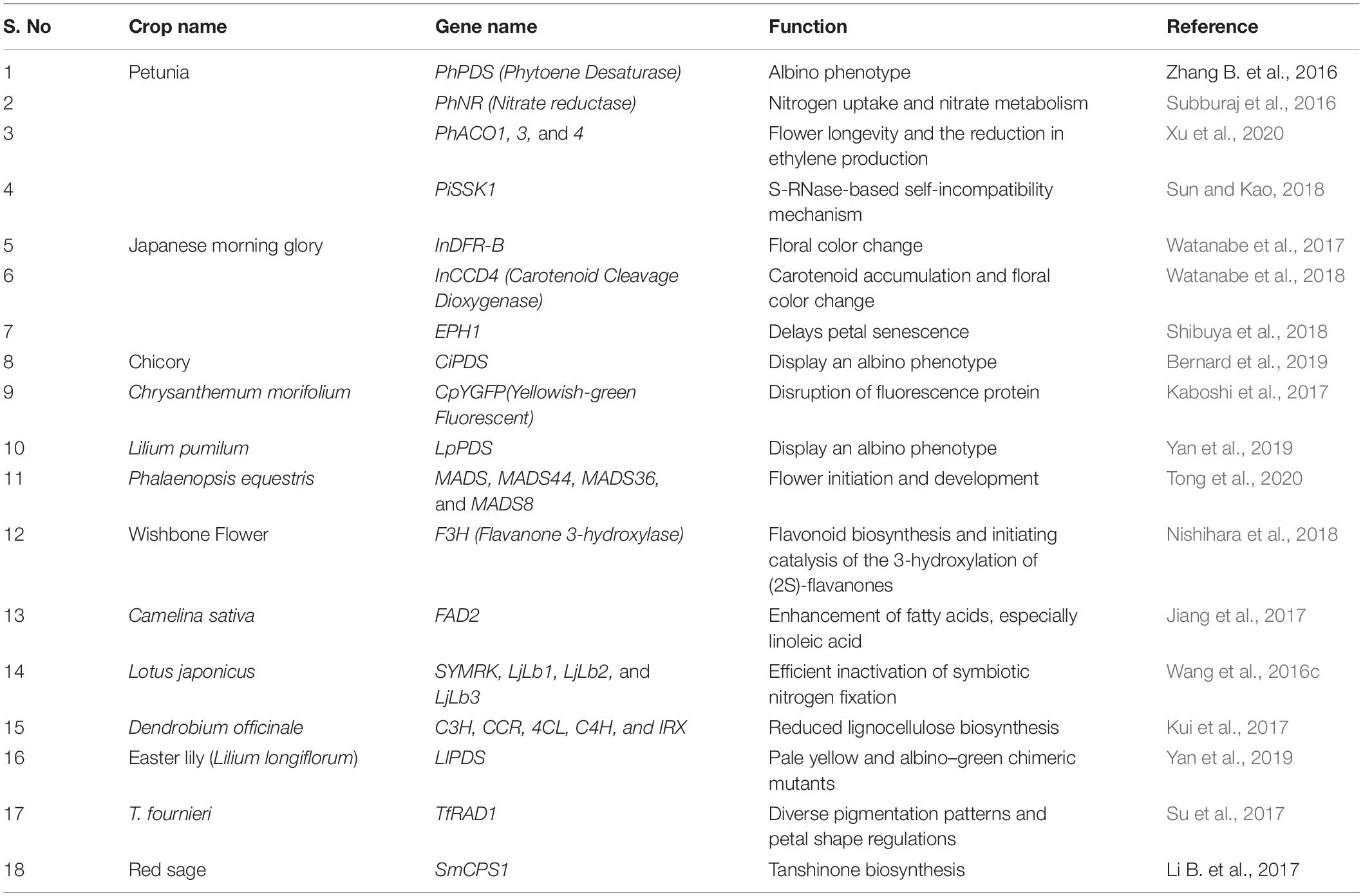
Table 2. Partial list of genes of ornamental plants and its specific functions modified via CRISPR/Cas9 system.
Important Considerations for CRISPR/Cas9 Genome Editing
Genomic engineering has been encountered broad range of applications to introduce targeted alterations of the plant’s genome to acquire desired function (Hsu et al., 2014). The NmeCas9 (Neisseria meningitidis) recognizes an 8-mer PAM (5′-NNNNGATT) sequence hence it can progress the target particularity, whereas SaCas9 recognizes a 6-mer PAM sequence (5′-NNGRRT) (Xing et al., 2014; Ma et al., 2015). This technique involves introducing mutations, and harnessing transgene supplementation for gene therapy and livestock improvement. CMV (Califlower mosaic Virus), AtUBO (Arabidopsis UBO) OsUBO (Oriza sativa UBO), LTR (Long terminal Repeat), OsnoRNA U3 (Oriza sativa snoRNA U3), AtU6 (Arabidopsis U6), OsUBI (Oriza sativa Ubiquitin), ZmUBI (Zea mays UBI), cauliflower mosaic virus 35S promoters have been used to promote Cas9 expression in plants and, more than 30 empty gRNA backbones in binary vectors was supplied by Addgene (Belhaj et al., 2013; Feng et al., 2013; Mao et al., 2013; Nekrasov et al., 2013; Upadhyay et al., 2013; Xie and Yang, 2013). Tissue specific promoters can also be used in CRISPR/Cas9 technology to edit genomes; for instance, Wang et al. (2015) used the promoter of the egg cell-specific EC1.2 gene, to initiative of Cas9. The germ line-specific SPOROCYTELESS and embryo-specific promotor DD45 (Mao et al., 2016). The promotor AtDMC1 involved in meiotic recombination (Xu et al., 2018). The pLAT52-GT for Pollen tissues (Mao et al., 2016), and pDD45-GT for egg cell-early embryo tissues (Mao et al., 2016), INCURVATA2 for dividing tissue-targeted site-directed mutagenesis (Hyun et al., 2015), and the YAO promoter for cell-division specific tissues (Yan et al., 2015). In order to knock out the expressions of multiple genes cassettes can be inserted into one plasmid, thereby guiding the Cas9 to different targets (Xing et al., 2014; Ma et al., 2015; Oz et al., 2021). Although this GE technology has key advantages, there are some negative shades involving ethical issues concerning to the disruption of ecological balance (Tavakoli et al., 2021). To overcome this, different methods including nanotechnology-based methods are being implemented to insert or silence the genes in plant cells (Nadakuduti and Enciso-Rodríguez, 2021).
Emphasis of CRISPR-Cas9 in Nutrition and Healthcare
Despite animal-based food consumption, the global ecosystem mainly be contingent to the agro-based crops including herbivorous animals. In concern to this, extensive research findings are mainly focusing on developing nutritional cereal/vegetables/fruits/nuts (FAO, 2017). Cultivating nutritional crops mainly depends on the nature and the fertility of the soil. Sources of soil nutrients are not same and is mainly depending on the presence of organic matter. In order to overcome this nutrient deficiency, CRISPR/Cas9 technology is extremely useful to grow cereal/vegetables/fruits/nuts with high nutritional values. Recently, genomic engineering utilizes TALENs, Cas9, dCas9, and Cre inserting into the cells. The use of these proteins in human cell lines have also been verified in vitro and in vivo (Zuris et al., 2015). HR (Sun et al., 2016) mediates the modifications made by the engineered nucleases. To date, plentiful of food-based crops have been modified to obtain good nutritional values in vegetables and fruits (Kathleen, 2015). Varieties of fruits comprised of different nutrients and biologically active compounds that are necessary in daily healthy diet. Recently, Dalla et al. (2019), Wan et al. (2021), and Xu et al. (2021) have discussed the importance of gene editing in fruits and vegetables to get nutritional rich cereal/vegetables/fruits/nuts, which are beneficial to maintain good health.
Genome-wide association studies (GWAS) have been employed to identify specific locations in the genome that can anchorage polygenic diseases such as Alzheimer’s, diabetes, autism, and schizophrenia. This technique is crucial in biomedical field to treat various diseases, including the removal of HIV genome (Ebina et al., 2013; Liao et al., 2015; Kaminski et al., 2017). The ex vivo and in vivo GE of neurons, immune cells, and endothelial cells has been successfully reported in mice (Platt et al., 2014), which are challenging to modify the sensitive cells and its effective editing. Researchers have introduced resistance against malaria by editing the DNA in Anopheles mosquitoes (Gantz et al., 2015; Hammond et al., 2016; Macias et al., 2020). Cancer therapy strategies have also been conducted in biomedical field to treat cancerous cell lines (Yao et al., 2015; Khan et al., 2016; Yang H. et al., 2019). Furthermore, AIDS research is still ongoing to engraft Cas9-modified CCR5-human hematopoietic stem cells and progenitor cells (Mandal et al., 2014). GE techniques are gaining its impact in promising therapeutics in regenerative medicine. The primary route for disease treatment is direct GE in tissues; some reports have documents the correction of monogenic recessive genetic disorders, such as Duchenne muscular dystrophy (Ousterout et al., 2015), hemophilia (Park et al., 2016) cystic fibrosis (Schwank et al., 2013), and sickle cell anemia (Sun and Zhao, 2014). The Cas9 system has exhibited its efficacy in therapies through the insertion of SSNs into microbial populations using phages and conjugative plasmids (Citorik et al., 2014).
Developments and Possibilities for Genomic Engineering in Agriculture
The comprehensive study on rice (Shan et al., 2013), sorghum (Jiang et al., 2013), tobacco (Li et al., 2013), wheat (Wang et al., 2014), tomato (Brooks et al., 2014), maize (Liang et al., 2014), sweet orange (Jia and Wang, 2014), and Arabidopsis (Li et al., 2013), stretches the vast knowledge of specific gene handling and editing. Before SSNs, RNAi technology was used to study the gene function by knocking down the targeted genes, which was not as advantageous as SSNs. Certain characteristics discussed below encompasses the examples for certain horticultural and ornamental plants using the CRISPR system. The color and weight/size ratio of tomato fruit could be developed by the editing of PL and TBG4 genes (Wang et al., 2019a). The SlNPR1 and SlCBF1 genes corresponding to drought and cold tolerance can also be modified (Li et al., 2018b; Li R. et al., 2019) and the fruit ripening transcription factor RIN (Ripening Inhibitor) could be edited, so that the tomato with desired characteristics will be maintained. Albino phenotype and flowering characters could be modified in cabbage using FRI and PDS gene editing (Murovec et al., 2018). In addition, the biosynthesis of Carotenoid pigment can be enhanced in wild cabbage with BoaCRTISO (Carotenoid isomerase) gene editing (Sun B. et al., 2020). By editing DcF3H and DcPDS, DcMYB113 genes, the accumulation of acylated anthocyanins can be enriched in the roots of carrot to afford pigmented purple carrots (Chodacka et al., 2018; Xu et al., 2019). Cucumber mosaic virus (CMV-Z1) and Zucchini yellow mosaic virus (ZYMV) are two major rapidly affecting pathogens, which can severely damage the crop. To overcome this, the pathogenic resistance can be developed/enhanced by editing elF4EF gene (Chandrasekaran et al., 2016). Drought tolerance in an important commercial crop, hot pepper (Capsicum annuum L. syn. chilli) was developed by editing NAC72 gene (Joshi, 2019). Furthermore, to enhance the flower longevity of attractive petunia flowers can be edited using PhACO1, 3, and 4 gene (Xu et al., 2020). The color and the Carotenoid accumulation of Japanese morning glory flower can be edited by altering its related gene InCCD4 (Carotenoid Cleavage Dioxygenase) (Watanabe et al., 2018). Compatibly, the flavonoid biosynthesis could be enriched in Wishbone flower by editing F3H (Flavanone 3- hydroxylase) gene (Nishihara et al., 2018). Moreover, the AhFAD2A and AhFAD2B genes encoding fatty acid desaturases in groundnut have been reported to be edited (Yuan et al., 2019). The TYLCV-IR (Intergenic regions) gene has been modified to overcome the multiple viral diseases in Nicotiana benthamiana. After gene modification plant exhibited resistance to geminiviridaevirus, begomovirus, curtovirus, becurtovirus, eragrovirus, Turncurtovirus, and Topocuvirus (Ali et al., 2015a,b). The resistance against Phytophthora tropicalis in cacao has been overcome by altering the TcNPR3 gene (Fister et al., 2018). Giovannini et al., 2021 have identified the desirable phenotypic characters in ornamental flowers, including flowering induction, floral meristem initiation and organ development, as well as color, fragrance, and shelf life. Flower longevity has been induced in petunia by altering a group of Petunia hybrid 1-aminocyclopropane-1-carboxylate oxidase (PhACO, PhACO1, PhACO3, and PhACO4) genes (Xu et al., 2020). Canker and huanglongbing diseases are the major factors in reducing the productivity of citrus plants; this problem was overcome by modifying the CsLOB1, CsWRKY22, and DMR6 genes by CRISPR/Cas9 system (Peng et al., 2017; Zhang et al., 2018c; Wang et al., 2019b). Selectable marker gene (SMG) systems are critical and play a major role in the identification of transgenic crops. Nowadays, the scientists are considering the SMGs that can affect human and animal health. The gene transferred plants (GMP: Genetically Modified Plants) usually contain the antibiotic resistant gene, so that GM plants should survive and regenerate under antibiotic medium. Whereas, the non-gene transformed plants will not rejuvenate, eventually die under toxic proximity. Although antibiotics have positive health and life prospective in human/animal health, the negative impacts of antibiotic associated diarrhea and pseudomembranous colitis will proliferate the possibilities of subsequent diseases. By consuming those GMP (Fruits and vegetables) for prolonged usage can severely affect the human/animal health (Breyer et al., 2014). Thus, it is imperative to eliminate SMGs from transgenic crops by using CRISPR technology (Yau and Stewart, 2013). Arndell et al. (2019) edited the biosynthesis of 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) for the functional confirmation of EPSPS gene in wheat using CRISPR/Cas9. The soybean storage protein genes were also been edited successfully to observe the efficacy of CRISPR/Cas9 technique using Agrobacterium rhizogenes-mediated hairy root transformation method (Li C. et al., 2019). Acetolactate synthase (ALS) participates in amino acid biosynthesis; this amino acid is targeted by numerous herbicides. These two plant enzymes(EPSPS, ALS, ACCase), and BFP genes, confer herbicide tolerance in plants (Voytas and Gao, 2014; Sauer et al., 2016; Li et al., 2018e). Hybrid paddy are susceptible to bentazon and sulfonylureas and the BEL gene has been mutated using radiation. In the production of hybrid rice, these mutants can be used to prevent contamination in hybrid seed lots (Cantos et al., 2014). Here, the BEL gene was edited using CRISPR-Cas9 technology, and transformed into rice through A. tumefaciens (Xu et al., 2014). Nutrient values have also been increased in vegetables and fruits by knocking out genes using the CRISPR-Cas9 system. Visually attractive flowers possess pleasant aroma because of the presence of anthocyanin, whose expression is regulated by the MYB-bHLH-WD (MBW) complex (Albert et al., 2014; Lloyd et al., 2017). Gibberellin (GA) determines plant height and strigolactone (SL) affects branching of the shoot branching, both of which can be modulated by modifying the biosynthesis or signal transduction of GA and/or SL (D’Halluin et al., 2008).
Unwanted metabolites usually have negative impacts on the crop yield and its quality; the accumulation of these undesired metabolites can be avoided by using GE. Cyanide intoxication, ataxia or partial paralysis, and goiters are caused by cyanide, which is present in cassava (Padmaja, 1995). Glucosinolates, which produced by mustard and cabbage, also possess a high toxic content, were edited (Hannoufa et al., 2014). FAD2 and FAD3 genes produce high oleic acid and low linolenic acid in soybean; however, soybean oil allows the accumulation of monounsaturated fats and reduces the linolenic acid in the seeds (Pham et al., 2012). AtPDS3, AtFLS2, AtADH, AtFT, AtSPL4, and AtBRI1 genes are targeted in Arabidopsis with mutation rates (MRs) from 1.1 to as high as 84.8% in the first generation (Li et al., 2013). The OsPDS and OsBADH2 genes have been knocked out with MRs of 9.4 and 7.1% (Hinge et al., 2015). The DsRED2, DD20, and DD43 genes have been targeted in sorghum with MRs of 33, 59, and 76%, respectively. Similarly, the ZmIPK (13.1%), LIG1, MS26, MS45, and ALS1 genes have been edited in maize with MRs lower than 5% (Liang et al., 2014; Svitashev et al., 2015). TaMLO-A, TaMLO-B, and TaMLO-Dare three homeo alleles that confer powdery mildew resistance and have been edited with the same moderate MR of 5.6% (Wang et al., 2014). In BRI1, JAZ1, and GAI genes mutation frequencies of 26–84% have been observed (Feng et al., 2013). NtPDS and NtPDR6 have been mutated with MRs of 81.8 and 87.5%, respectively (Gao et al., 2015). The squamosa promoter binding protein-like 4 and Flowering Locus T (FT) have been mutated with an MR of 90%, which caused it to exhibit late flowering (Hyun et al., 2015).
Ma et al. (2015), have reported genome modifications at 46 target sites with an average of 85.4% mutations in monocot and dicot plants using either golden gate ligation or Gibson assembly. Using the sgRNA single, double and triple mutants have also been generated for CDKA2, CDKB1, and CDKB2 in rice (Endo et al., 2015). Dort et al. (2020) discussed forest pathosystems; some disease problems were solved using the CRISPR/Cas9 system. Zhang et al. (2014), have also reported mutations in young seedling albino (OsYSA) and OsROC5 genes with MRs of 65–66.7%. Similarly, Wang et al. (2016b), have edited the OsERF922 gene that encodes ERF transcription factors to develop resistance to rice blast disease. Transgenic poplar plants have been modified and phenotypically results revealed an MR of 51% (Fan et al., 2015). El-Mounadi et al. (2020) explained biosafety of genomically edited plants and the applications of CRISPR/CAS9 technology to enhance yield, quality, and nutritional values. The crops and seeds developed using CRISPR/Cas9 technology for cereals, vegetables, ornamental, and fruits plants are shown in Tables 1, 2 and Supplementary Tables S1, S2. Some multinational companies (DuPont, Monsanto, and BASF) had obtained licenses to develop new crops using CRISPR technology (Khurana and Rajarshi kumar, 2019). The sequential steps for CRISPR/Cas9 genetic transformation in plants was sketched in Figure 2 (Manghwar et al., 2019).
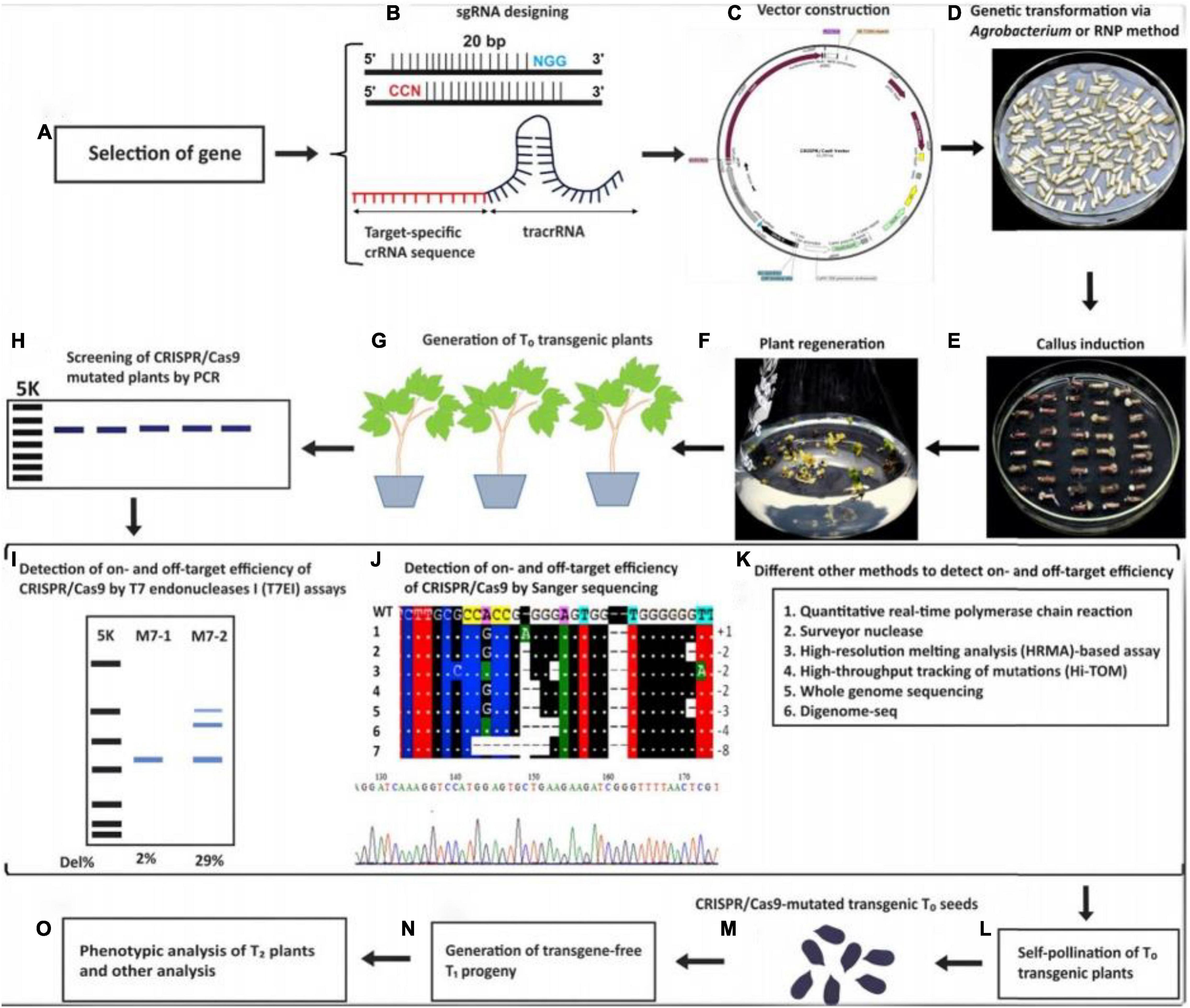
Figure 2. Schematic illustration of the steps involved in CRISPR/Cas9 Genetic Transformation; (A) Specific gene selection, (B) sgRNA designing for the specific gene, (C) vector construction, (D) transformation of the CRISPR/Cas9 system via Agrobacterium, (E) callus induction from agrobacterium infected explants, (F) plant regeneration from callus, (G) T0 CRISPR/Cas9 mutated transgenic plants, (H) screening of transgenic plants by PCR, (I) identification of mutated plants by T7E1, (J) detection of transgenics by sanger sequencing, (K) various techniques to detect transgenic plants, (L) self-pollination of T0 transgenic plants for generation of homozygous T1 plants, (M) mutated T0 seeds, (N) Generation of transgene- free T1 progeny, (O) Phenotypic analysis of T1 plants. Reproduced with permission from Manghwar et al. (2019) CellPress.
Role of Nanotechnology in Crop Refinement
Nanotechnology plays a prominent role in biological, medicinal, and pharmaceuticals including plant science. Increasing resilience to biotic and abiotic stress and improving the yield/quality of the crops via gene editing, nanotechnology shares its connotation with CRISPR/Cas9. Nano-fertilizers (Subramanian et al., 2015; Thul and Saragani, 2015) are used in horticultural plants, including vegetables and fruits, and implemented in food crops to enhance the growth, germination rate, and genetic manipulations (Lee et al., 2010; Sheykhbaglou et al., 2010; Dimkpa et al., 2012; Shang et al., 2019; Mittal et al., 2020; Aqsa et al., 2021; Rana et al., 2021). Similarly, plant growth, nutrient uptake from roots, flowering; have also been developed by metal and carbon based nanoparticles (Aqsa et al., 2021). Photosynthesis is an energy conversion process in plants, transforming light energy into chemical energy; however, it does not occur effectively under cloudy conditions and in sun-drenched plants during the rainy season. Consequently, cell mechanisms possibly down regulated. The gold nanoparticles could be beneficial to enhance the light-harvesting capacity, thereby promoting highly excited electron transfer in the chloroplast (So et al., 2015). Environmental factors (abiotic stress) cause biochemical and physiological changes in plants and these are more susceptible to stress. Even in stressful conditions, the use of metallic nanoparticles can increase the anti-oxidative enzyme levels in plants (Mittal et al., 2020; Zhao et al., 2020; Wu and Li, 2021) and reduce the reactive oxygen species (ROS) levels in the mitochondria and chloroplasts to protect the plant (Sun L. et al., 2020; Sun et al., 2021). However, the use of these nanoparticle fertilizers in crop fields not only increase the soil fertility, but also greatly influence the water resource contamination (Naderi and Abedi, 2012; Mittal et al., 2020). Fertilizers containing microorganisms are labeled as biofertilizers, which can activate the plant system and improve the nutrient uptake from soil (Manikandan and Subramanian, 2016). Nano-fertilizers have the similar benefits like biofertilizers (Elias et al., 2019). Moreover, metallic nanoparticles have anti-pathogenic, antifungal, and antibacterial properties (Kah and Hofmann, 2014; Servin et al., 2015), so that they can survive under pathogenic bout under the soil. Brief explanation of using metallic nanoparticles in farming and its benefit in sustainable agriculture is explained in Figure 3 (Mittal et al., 2020).
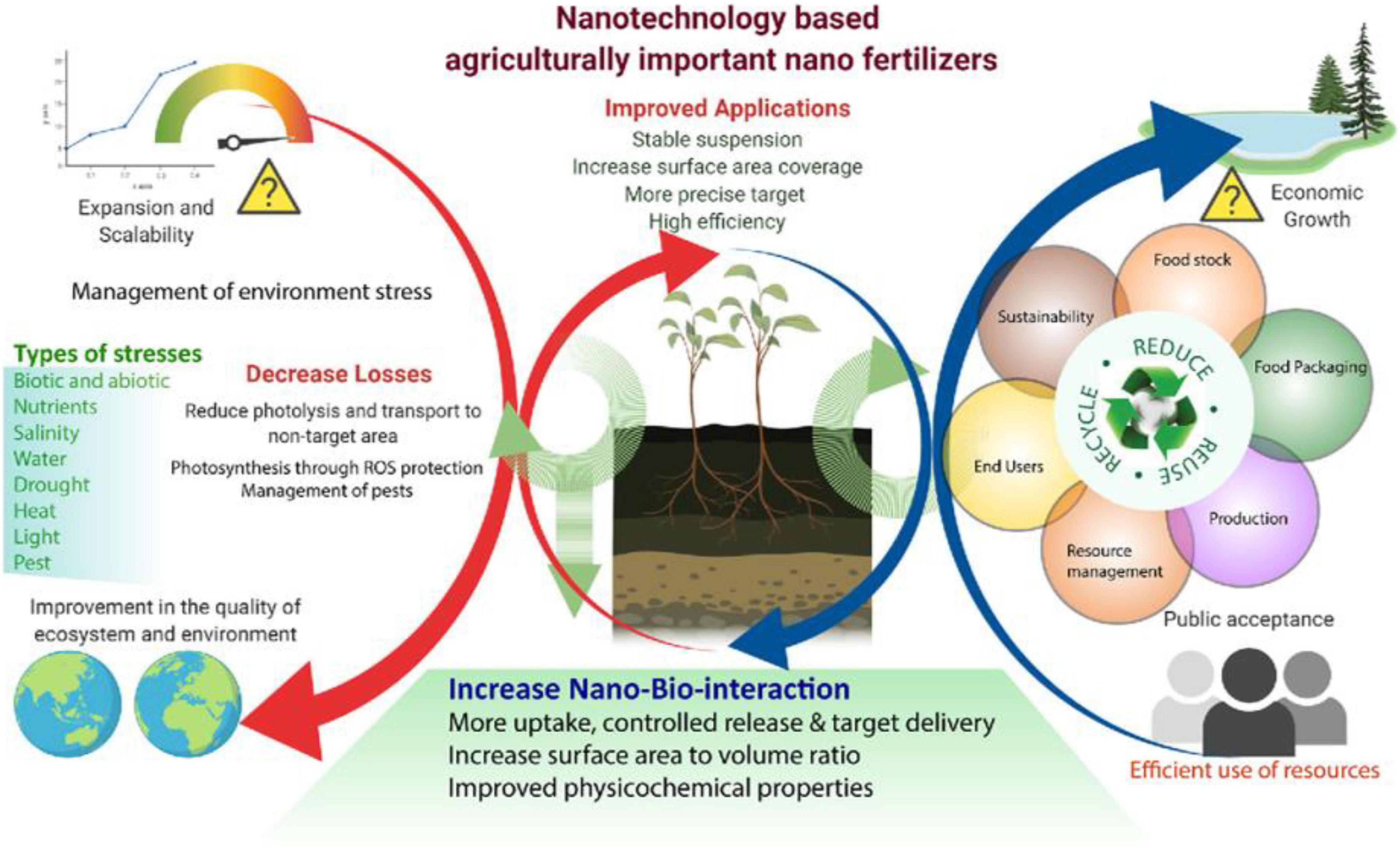
Figure 3. Nanotechnology-based agriculturally important nano-fertilizers, which are increasing the agronomic productivity, efficiency, and reduce environmental stress. Although showed the improved applications in agriculture by nanotechnology and Types of stresses overcome by nanotechnology. Reproduced with permission from Mittal et al. (2020) Frontiers.
Role of Nanoparticle-Based Genetic Modification of Crops
Nanoparticles acts as a carrier to deliver the necessary materials into plant cells, animal cells, and specific organs for cancer therapy, genetic disease treatment, and to obtain desirable traits in plants (Peng et al., 2019). Lv et al. (2020) and Ahmar et al. (2021) explained the method in which the gene transformation is performed using nanoparticles. Various types of nanoparticles have been used to deliver genetic material into the plant cells through different platforms. These reports have also detailed the merits and demerits of utilizing nanoparticles in gene transfer methods. Typically, the mesoporous silica nanoparticles, carbon nanotubes, gold, and magnetic nanoparticles have been used to deliver plasmid DNA, double-stranded RNA, and siRNA into plant protoplasts or other intact cell lines (Al-Babili and Bouwmeester, 2015; Yao et al., 2016; Peng et al., 2019; Huan et al., 2020; Tsveta et al., 2021). However, Lv et al. (2020) have demonstrated the gene silencing and gene editing in plants with the use of nanoparticles; magnetic nanoparticle-based pollen transformation was used to achieve the task. In this approach, the vector-magnetic nanoparticle complex has been associated with the pollen that is dropped onto the stigma of desired plant flowers. Finally, plants produce the desired seeds by transferring the vector-magnetic nanoparticle complex into the flower stigma. Later, these flowers are modified into fruits, and the seeds were screened on antibiotic plates. The speed breeding protocol was used to obtain T0, T1, and T2 generations of transgenic plants; this breeding program is inexpensive for editing plant genomes and is employed for various Brassica species (Ahmar et al., 2021). Similarly, dsRNA has been loaded into the layered double hydroxide (LDH) clay nanosheets that are non-degradable, non-toxic, and resistant to easy wash. Furthermore, when these nanoparticle-dsRNA complexes are sprayed onto the plant leaves, they immediately attach to the leaf surface and are absorbed by plant viruses to induce RNAi, eventually degrading the targeted plant pathogens or endogenous mRNA can be minimized/eliminated (Lv et al., 2020). Similarly, gene editing has also been demonstrated with small NPs-CRISPR/Cas9 vector complex that was microinjected into the leaves or any other plant parts, which can be proliferated further by tissue culture or other ease protocols (Lv et al., 2020; Duan et al., 2021). Carbon dots-siRNA complex has been used to silence the GFP in tobacco and tomato plants (Schwartz et al., 2020). Demirer et al. (2021) recently demonstrated genome editing in plants using the CRISPR/Cas9 system along with nanoparticles and explained the regeneration, and phenotypic/metabolic changes of genomically edited crops. The methods in which gene expression, silencing, editing, and other applications involving nanoparticles can account for the crop refinement and are briefly explained in the Figure 4 (Peng et al., 2019).
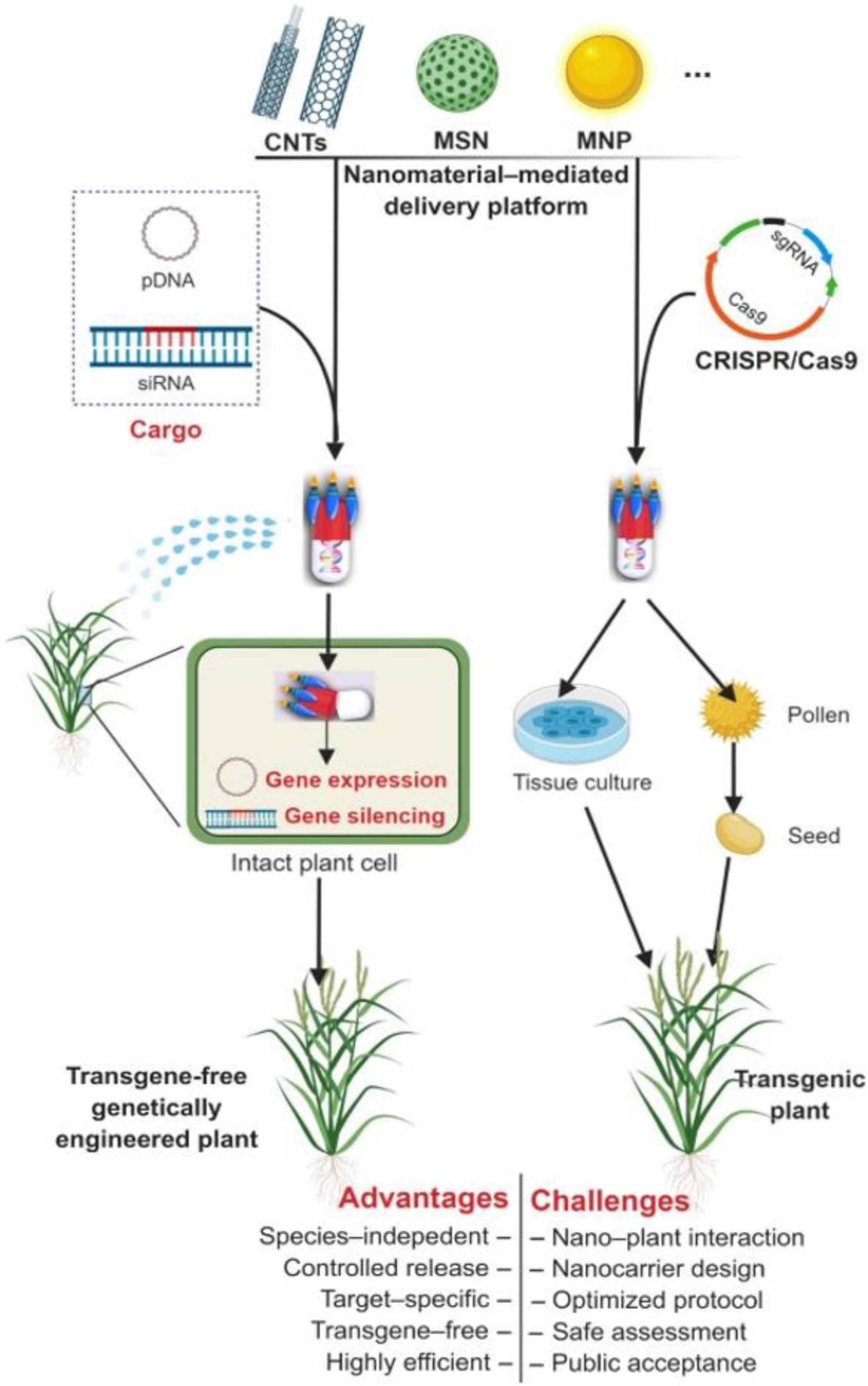
Figure 4. Nanomaterial-mediated plant genetic engineering. Functionalized nanomaterials can provide a delivery platform that is capable of traversing barriers (e.g., multilayered cell walls) to deliver exogenous plasmid DNA (pDNA) and siRNA into intact plant cells. CNTs, carbon nanotubes; MSN, mesoporous silica nanoparticles; MNP, magnet nanoparticles. Reproduced with permission from Peng et al. (2019) CellPress.
Applications and Limitations of CRISPR/Cas9 and Nanotechnology Systems in Modern Biology
The CRISPR/Cas9 and nanotechnology aspects were successfully implemented so that relevant solicitations in plant biotechnology, food industry, livestock improvement can be achieved (Islam et al., 2020). Biofuel production (Javed et al., 2019), and rectifications of genetic and metabolic diseases can be done by controlling bacterial and viral diseases (Robert et al., 2017; Zafar et al., 2020), controlling bacterial and viral diseases (Zafar et al., 2020).
(1) This GE technology has been used to treat hematologic diseases, infectious diseases, and malignant tumors via gene therapy.
(2) Gene pyramids and gene knockouts can be created by inserting foreign genes through this approach.
(3) Repression/activation of gene expression.
(4) Delivery of proteins to genomic loci.
(5) Gene expression can be regulated with this technology. It also plays a major role in the progress of new mutant diversities to help the breeders.
(6) Gene silencing is an important aspect in the crop improvement program based on CRISPR intrusion (CRISPRi) (Larson et al., 2013; Qi et al., 2013).
(7) It is a promising technique to study the new gene’s function.
(8) Plants that are lenient to biotic and abiotic stresses, as well as herbicides, can be easily developed.
(9) Researchers are already using this technology in gene therapy (Qian et al., 2019; Li et al., 2020).
(10) Currently, nanotechnology is being used to prepare nano-fertilizers, nano-pesticides, and to enhancing the abiotic stress tolerance, thereby protecting the crops.
(11) Different types of NPs have also been used for the transformation, editing, and silencing of genes to improve crop yield and quality.
CRISPR/Cas9 technology is among the best and most cost-effective methods for genomic editing in plants. However, some of its limitations are mentioned below.
(1) Genomic editing is inefficient and slow by the presence of larger protein sizes. Therefore, protein size should be small to ensure speedy and efficient genomic editing.
(2) It contains limited number of PAM sites at limited loci.
(3) CRISPR/CAS9 can introduce multiple random mutations, as well as mutations at unspecific loci.
(4) It has low HDR efficiency.
(5) It exhibits low efficiency against viruses.
(6) It has become difficult to commercialize transgenic crops developed using CRISPR/Cas9 technology in many countries because different countries have different rules and regulations.
(7) In backward and developing countries, there is a lack of proper understanding of pesticides, fertilizers, and other products developed by nanotechnology; therefore, it is impossible to grow better crops.
Challenges
At present, this technology is helping researchers in agriculture and breeders in developing crops that can overcome biotic and abiotic stress, with high nutritional values, and optimal yield parameters, thereby providing adequate food grains, vegetables, and fruits to current world population. Even with the availability of all types of biotechnological and bioinformatics tools, there are enduring challenges in developing genetically modified plants using the CRISPR system. Owing to unavailability of complete genome sequence information and the large genome size of some tropical, medicinally valuable crops, and fruits, some studies have been unable to edit genes to obtain desirable traits (Sanskriti et al., 2019; Zhou et al., 2019; Yang, 2020). Therefore, the biological pathways of genes and their interactions with environmental factors are still unknown (Haque et al., 2018). In order to discover new traits in tropical and other crops, one must be well-versed in the functions and regulatory elements of each gene. Moreover, in some crops, the transformation efficiency is extremely low; therefore, it takes a long time for the regeneration of explants, which is very difficult in some crops (Altpeter et al., 2016; Sanskriti et al., 2019). However, genetically modified crops require extensive field trials (Syed et al., 2020). Of all the challenges facing in the development of genetically modified plants, lack of proper public acceptance has become the biggest challenge (Yang, 2020), and believing to be accepted in forthcoming decades (Chao et al., 2021).
Conclusion and Future Perspective
Scientists and plant breeding researchers are working to develop high yielding and biotic/abiotic stress resistant variety. CRISPR/Cas9 GE in consolidation with nanotechnology have emerged as an important platform to improve the quality and desirable quality of agricultural crops with appreciable yield. These technologies are significantly susceptible and open new prospects in plant genetics. The nutritional values and quality of health prospects of plants and human could be enriched with broad spectrum of applications including in biomedical domain. The imminent fertility and diminishing useful prokaryotic microorganism issues in soil are delicate issues in addition to biotic problems. Relapsing the aforesaid issues could be feasible using CRISPR/Cas9 GE and nanotechnology aspects, it will be challenging and greatly influence to the crop refinement. Skillful editing of genetic sequence of plant genome could progressively improve the agronomic traits, photosynthetic capacity, and nutritional values. Furthermore, biotic and abiotic stress-induced issues in plants could be configured and minimized. The methodologies can also be diversified to characterize the individual gene functions, thereby improving the genomic sequences of agricultural crops to produce exceptional yield. These findings could point new strategies in agroecology facilitating the sustainable production of nutritional quality food to satisfy the increasing demand of ever-growing population.
CRISPR/Cas9 and nanoparticle complex system is an advanced innovative technique in agricultural crop development/refinement. Thus far, CRISPR system in connotation with nanotechnology has been used to improve the quality and yield of many valuable crops for future benefits. Using nanoparticle based fertilizers/additives; extinct nutrients could be regained, ensuring that the soil nutrients could be maintained. The green, non-toxic nanoparticles is a sagacious approach to increase micro and macronutrient levels in the soil for healthy growth of crops. Chemically derived nanoparticle usage should be eliminated; these can harm not only the crops but also to the environment. Zinc oxide-based nanocomposites and nano-fertilizers have shown appreciable results in crop growth; maintaining the soil salinity and fertility with robust yield. The green protocols to develop Zinc oxide-based green nanocomposites draws the attention of agro-scientific community and it should be given prime importance, so that toxic free effective nano-priming techniques can reduce the soil contamination and improves the seed quality. Metal organic frameworks (MOF) have been recently investigated as delivery systems for CRISPR/Cas9. Non-toxic and eco-friendly MOF 3-dimentional structures with biopolymer conjugates are potentially promising. Furthermore, CRISPR/Cas9-associated nanoparticle complex has been successfully utilized for transformation, silencing, and modification of genes to overcome the existing and expected critical biotic and abiotic issues, thereby producing nutritional-rich crops. This technology can be used to alter the metabolic pathways in plants to obtain desired high-quality secondary metabolites for future usage. Recently, revolutionary changes in this crop refinement program witnessed auspicious results. Nod factors have been shown to increase nitrogen efficiency in legume crops using CRISPR/Cas9 system with nano-technological contrivance. Modifying the Nod signaling pathway in cereal crops should eliminates the use of toxic inorganic fertilizers. The transformation efficiency by Agrobacterium tumefaciens is quite low in some specified plant tissues. Hence, an alternative bacterial system to gene transfer is necessary for ease genome editing in all crops by adopting CRISPR/Cas9-Nanotechnology system. Furthermore, the development of tissue culture free delivery protocols involving direct genomic editing in germplasms and meristematic cells of plants, can yield propitious results. To date, numerous crops have been developed with this combined CRISPR/Cas9-Nanoparticle complex system. It is advisable to framework on the products already developed by this method and ensure their recurrent use in agricultural locales. Extensive studies are recommended to elucidate the complete interactions (plant cell mechanisms) of nanoparticles/nanocomposites in all types of crops. We strongly believe that the products developed by conjoining these technologies will beneficially assist the agro-based researchers to bloom their ideas for innovative crop development/refinement.
Author Contributions
BN planned the manuscript outline, wrote the draft, and prepared the tables. GS contributed in writing, reshaping/editing the manuscript and modifying the tables. GS, S-CK, MM, CS, RP, and MK proofread the manuscript. S-YK acquired the funding. S-YK and S-HL supervised the study and revised the manuscript. All the authors reviewed and approved the final version of the manuscript.
Funding
This work was supported by the Institute of Information and Communications Technology Planning and Evaluation (IITP) grant, funded by the Korea Government (MSIT) (No. 2020-0-00594, Morphable Haptic Controller for Manipulating VR⋅AR Contents). Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1A6A1A03025526) also supported this work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge Cooperative Equipment Center at KoreaTech for formal discussions. The authors are gratified to KoreaTech for providing the research opportunity/facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.843575/full#supplementary-material
References
Abdallah, N. A., Prakash, C. S., and Hughen, A. G. (2014). Genome editing for crop improvement: challenges and opportunities. GM Crops Food 6, 183–205. doi: 10.1080/21645698.2015.1129937
Abe, K., Araki, E., Suzuki, Y., Toki, S., and Saika, H. (2018). Production of high oleic/low linoleic rice by genome editing. Plant Physiol. Biochem. 131, 58–62. doi: 10.1016/j.plaphy.2018.04.033
Adli, M. (2018). The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9:1911. doi: 10.1038/s41467-018-04252-2
Ahmar, S., Mahmood, T., Fiaz, S., Poblet, F., Shafique, M. S., Chattha, M. S., et al. (2021). Advantage of nanotechnology-based genome editing system and its application in crop improvement. Front. Plant Sci. 12:663849.
Al Amin, N., Ahmad, N., Wu, N., Pu, X., Ma, T., Du, Y., et al. (2019). CRISPR-Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max.L). BMC Biotechnol. 19:501.
Al-Babili, S., and Bouwmeester, H. J. (2015). Strigolactones, a novel carotenoid- derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186. doi: 10.1146/annurev-arplant-043014-114759
Albert, N. W., Davies, K. M., Lewis, D. H., Zhang, H., Montefiori, C., Boase, M. R., et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980. doi: 10.1105/tpc.113.122069
Ali, Z., Abul Faraj, A., Idris, A., Ali, S., Tashkandi, M., and Mahfouz, M. M. (2015a). CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16, 238–249. doi: 10.1186/s13059-015-0799-6
Ali, Z., Abul-faraj, A., Piatek, M., and Mahfouz, M. M. (2015b). Activity and specificity of TRV-mediated gene editing in plants. Plant Signal. Behav. 10:1044191. doi: 10.1080/15592324.2015.1044191
Altpeter, F., Springer, N. M., Bartler, L. E., Blechl, A. E., Brutnell, T. P., Citovsky, V., et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520. doi: 10.1105/tpc.16.00196
Andersson, M., Turesson, H., Olsson, N., Sofie, F. A., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR-Cas9 ribo nucleoprotein delivary. Physiol. Plant. 164, 378–384. doi: 10.1111/ppl.12731
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi: 10.1038/s41586-019-1711-4
Aqsa, S., Tahira, Shaheera, Q., and Waqas, M. B. (2021). Applications of nanobiotechnology in plant sciences. BJSTR 35, 27236–27240. doi: 10.26717/BJSTR.2021.35.005632
Arndell, T., Sharma, N., Langridge, P., Baumann, U., Watson-Haigh, N. S., and Whitford, R. (2019). gRNA validation for wheat genome editing with the CRISPR-Cas9 system. BMC Biotechnol. 30:71. doi: 10.1186/s12896-019-0565-z
Ashokkumar, S., Jaganathan, D., Ramanathan, V., Rahman, H., Palaniswamy, R., Kambale, R., et al. (2020). Creation of novel alleles of fragrance gene OsBADH2 in rice through CRISPR/Cas9 mediated gene editing. PLoS One 15:e0237018. doi: 10.1371/journal.pone.0237018
Baek, K., Kim, D. H., Jeong, J., Sim, S. J., Melis, A., Kim, J. S., et al. (2016). DNA-freetwo-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleo proteins. Sci. Rep. 6:30620. doi: 10.1038/srep30620
Baltes, N. J., Hummel, A. W., Konecna, E., Cegan, R., Bruns, A. N., Bisaro, D. M., et al. (2015). Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants 1:15145. doi: 10.1039/nplants.2015.145
Bao, A., Chen, H., Chen, L., Chen, S., Hao, Q., Guo, W., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 19:131. doi: 10.1186/s12870-019-1746-6
Bari, V. K., Nassar, J. A., Kheredin, S. M., Gal-On, A., Ron, M., Britt, A., et al. (2019). CRISPR/Cas9- mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci. Rep. 9:11438. doi: 10.1038/s41598-019-47893-z
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., and Nekrasov, V. (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9, 1–10. doi: 10.1186/1746-4811-9-39
Bernard, G., Gagneul, D., Alves Dos Santos, H., Etienne, A., Hilbert, J. L., and Rambaud, C. (2019). Efficient genome editing using CRISPR/Cas9 technology in chicory. Int. J. Mol. Sci. 20:1155. doi: 10.3390/ijms20051155
Bernheim, A., Calvo-Villamanan, A., Basier, C., Cui, L., Rocha, E. P. C., and Touchon, M. (2017). Inhibitions of NHEJ repair by type II-A CRISPR-Cas systems in bacteria. Nat. Commun. 8, 1–9. doi: 10.1038/s41467-017-02350-1
Bertier, L. D., Ron, M., Huo, H., Bradford, K. J., Britt, A. B., and Michelmore, R. W. (2018). High-resolution analysis of the efficiency, heritability, and editing outcomes of CRISPR/Cas9-induced modifications of NCED4 in lettuce (Lactuca sativa). G3 Genes Genomes Genet. 8, 1513–1521. doi: 10.1534/g3.117.300396
Bo, W., Zhaohui, Z., Huanhuan, Z., Xia, W., Binglin, L., Lijia, Y., et al. (2019). Targeted mutagenesis of NAC transcription factor gene, OsNAC041, leading to salt sensitivity in rice. Rice Sci. 26, 98–108. doi: 10.1016/j.rsci.2018.12.005
Breitler, J. C., Dechamp, E., Campa, C., Rodrigues, L. A. Z., Guyot, R., Marraccini, P., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis has the potential to accelerate the domestication of Coffea canephora. PCTOC 134, 383–394. doi: 10.1007/s11240-018-1429-2
Breyer, D., Kopertekh, L., and Reheul, D. (2014). Alternatives to antibiotic resistance marker genes for in vitro selection of genetically modified plants – Scientific developments, current use, operational access, and biosafety considerations. Crit. Rev. Plant Sci. 33, 286–330. doi: 10.1080/07352689.2013.870422
Brooks, C., Nekrasov, V., Lippman, Z. B., and Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Butler, N. M., Atkins, P. A., Voytas, D. F., and Douches, D. S. (2015). Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS One 10:e0144591. doi: 10.1371/journal.pone.0144591
Cai, Y., Chen, L., Liu, X., Guo, C., Sun, S., Wu, C., et al. (2018). CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 16, 176–185. doi: 10.1111/pbi.12758
Cai, Y., Chen, L., Liu, X., Sun, S., Wu, C., Jiang, B., et al. (2015). CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS One 10:e0136064. doi: 10.1371/journal.pone.0136064
Cantos, C., Francisco, P., Trijatmiko, K. R., Slamet-Loedin, I., and Chadha-Mohanty, P. K. (2014). Identification of “safe harbor” loci in indica rice genome by harnessing the property of zinc-finger nucleases to induce DNA damage and repair. Front. Plant Sci. 5:302. doi: 10.3389/fpls.2014.00302
Carroll, D. (2014). Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439. doi: 10.1146/annurev-biochem-060713-035418
Caterina, D., Adriana, L. S., and Giovanni, G. (2018). CRISPR/Cas9 editing of carotenoid genes in tomato. Transgenic Res. 27, 367–378. doi: 10.1007/s11248-018-0079-9
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Chao, L., and Baohong, Z. (2019). Genome editing in cotton using CRISPR/Cas9 system. Methods Mol. Biol. 1902, 95–104. doi: 10.1007/978-1-4939-8952-2_8
Chao, L., Eleanor, B., Hikmet, B., and Baohong, Z. (2021). CRISPR/Cas: a nobel prize award-winning precise genome editing technology for gene therapy and crop improvement. J. Zhejiang. Univ. Sci. B 22, 253–284. doi: 10.1631/jzus.B2100009
Chao, L., Mengyu, H., Wenxiang, W., Hui, W., Fan, C., Wen, C., et al. (2018). An efficient CRISPR/Cas9 platform for rapidly generating simultaneous mutagenesis of multiple gene homoeologs in allotetraploid oilseed rape. Front. Plant Sci. 9:442. doi: 10.3389/fpls.2018.00442
Chao, S., Cai, Y., Feng, B., Jiao, G., Sheng, Z., Luo, L., et al. (2019). Editing of rice Isoamylase gene ISA1 provides insights in to its function in starch formation. Rice Sci. 26, 77–87. doi: 10.1016/j.rsci.2018.07.001
Charrier, A., Vergne, E., Dousset, N. J. P., Richer, A., Petiteau, A., and Chevreau, E. (2019). Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 10:40. doi: 10.3389/fpls.2019.00040
Che, P., Anand, A., Wu, E., Sander, D., Simon, M., Zhu, W., et al. (2018). Developing a flexible, high efficiency Agrobacterium-mediated sorghum transformation system with broad application. Plant Biotechnol. J. 16, 1388–1395. doi: 10.1111/pbi.12879
Chen, R., Xu, Q., Liu, Y., Zhang, J., Ren, D., Wang, G., et al. (2018). Generation of transgene-free maize male sterile lines using the CRISPR/Cas9 system. Front. Plant Sci. 9:1180. doi: 10.3389/fpls.2018.01180
Chen, X., Lum, X., Shu, N., Wang, S., Wang, J., and Wang, D. (2017). Targeted mutagenesis in cotton (Gossypium hirsutum L.) using the CRISPR/Cas9 system. Sci. Rep. 7, 44304–44311. doi: 10.1038/srep44304
Chen, Y., Zhu, A., Xue, P., Wen, X., Cao, Y., Wang, B., et al. (2020). Effects of GS3 and GL3.1 for grain size editing by CRISPR/Cas9 in rice. Rice Sci. 27, 405–413. doi: 10.1016/j.rsci.2019.12.010
Cheng, Q., Dong, L., Su, T., Li, T., Gan, Z., Nan, H., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol. 19:562. doi: 10.1186/s12870-019-2145-8
Cheng, Z., Sun, Y., Yang, S., Zhi, H., Yin, T., Ma, X., et al. (2021). Establishing In planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italic). Plant Biotechnol. J. 19, 1089–1091. doi: 10.1111/pbi.13584
Chodacka, M. K., Oleszkiewicz, T., Lowder, L. G., Qi, Y., and Baranski, R. (2018). Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 37, 575–586. doi: 10.1007/s00299-018-2252-2
Christian, M., Cermak, T., Doyle, E. L., Schmidt, C., Zhang, F., Hummel, A., et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761. doi: 10.1534/genetics.110.120717
Citorik, R. J., Mimee, M., and Lu, T. K. (2014). Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32, 1141–1145. doi: 10.1038/nbt.3011
Curtin, S. J., Xiong, Y., Michno, J. M., Campbell, B. W., Stec, A. O., Cermák, T., et al. (2018). CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 16, 1125–1137. doi: 10.1111/pbi.12857
Dalla, C. L. E., Mahmoud, L., Moraes, T. S., Mou, Z., Grosser, J. W., and Dutt, M. (2019). Development of improved fruit, vegetable, and ornamental crops using the CRISPR/Cas9 genome editing technique. Plants 8:601. doi: 10.3390/plants8120601
Demirer, G. S., Silva, T. N., Jackson, C. T., Thomas, J. B., Ehrhardt, D., Rhee, S. Y., et al. (2021). Nanotechnology to advance CRISPR–Cas genetic engineering of plants. Nat. Nanotechnol. 16, 243–250. doi: 10.1038/s41565-021-00854-y
Deng, L. (2018). Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J. Genet. Genome 45, 51–54. doi: 10.1016/j.jgg.2017.10.002
D’Halluin, K., Vanderstraeten, C., Stals, E., Cornelissen, M., and Ruiter, R. (2008). Homologous recombination: a basis for targeted genome optimization in crop species such as maize. Plant Biotechnol. J. 6, 93–102. doi: 10.1111/J.1467-7652.2007.0035.X
Dimkpa, C. O., McLean, J. E., Latta, D. E., Manangón, E., and Britt, D. W. (2012). CuO and ZnO nanoparticles: phytotoxicity, metal speciation, and induction of oxidative stress in sand-grown wheat. J. Nanopart Res. 14, 1–15. doi: 10.1007/s11051-012-1125-9
Dort, E. N., Tanguay, P., and Hamelin, R. C. (2020). CRISPR/Cas9 gene editing: an unexplored frontier for forest pathology. Front. Plant Sci. 11:1126. doi: 10.3389/fpls.2020.01126
Duan, L., Ouyang, K., Xu, X., Xu, L., Wen, C., Zhou, X., et al. (2021). Nanoparticle delivery of CRISPR/Cas9 for genome editing. Front. Genet. 12:673286. doi: 10.3389/fgene.2021.673286
Duan, Y. B., Li, J., Qin, R. Y., Xu, R. F., Li, H., Yang, Y. C., et al. (2016). Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and insitu promoter analysis. Plant Mol. Biol. 90, 49–62. doi: 10.1007/s11103-015-0393-z
Ebina, H., Misawa, N., Kanemura, Y., and Koyanagi, Y. (2013). Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 3:2510. doi: 10.1038/srep02510
Elias, E. E., Ifeyinwa, M. U., Damian, C. O., and Olubukola, O. B. (2019). The role of nanotechnology in the fortification of plant nutrients and improvement of crop production. Appl. Sci. 9, 1–32. doi: 10.3390/app9030499
El-Mounadi, K., Morales-Floriano, M. L., and Garcia-Ruiz, H. (2020). Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front. Plant Sci. 11:56. doi: 10.3389/fpls.2020.00056
Endo, M., Mikami, M., and Toki, S. (2015). Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 56, 41–47.
Fan, D., Liu, T., Li, C., Jiao, B., Li, S., Hou, Y., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5, 12217–12223. doi: 10.1038/srep12217
Fauser, F., Schiml, S., and Puchta, H. (2014). Both CRISPR/Cas based nucleases and nickases can be used efficiently for genome engineering in Arabidopsisthaliana. Plant J. 79, 348–359. doi: 10.1111/tpj.12554
Feng, C., Su, H., Bai, H., Wang, R., Liu, Y., Guo, X., et al. (2018). High efficiency genome editing using a dmc1 promoter controlled CRISPR/Cas9 system in maize. Plant Biotechnol. J. 16, 1848–1857. doi: 10.1111/pbi.12920
Feng, C., Yuan, J., Wang, R., Liu, Y., Birchler, J. A., and Han, F. (2016). Efficient targeted genome modification in maize using CRISPR/Cas9 system. J. Genet. Genomics 43, 37–43. doi: 10.1016/j.jgg.2015.10.002
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D. L., Wei, P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. doi: 10.1038/cr.2013.114
Ferrara, M., Haidukowski, M., Logrieco, A., Leslie, J., and Mule, G. (2019). A CRISPR-Cas9 system for genome editing of Fusarium proliferatum. Sci. Rep. 9:19836. doi: 10.1038/s41598-019-56270-9
Fister, A. S., Landherr, L., Maximova, S. N., and Guiltinan, M. J. (2018). Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front. Plant Sci. 9:268. doi: 10.3389/fpls.2018.00268
Gantz, V. M., Jasinskiene, N., Tatarenkova, O., Fazekas, A., Macias, V. M., Bier, E., et al. (2015). Highly efficient Cas9-mediated gene drives for population modification of the malaria vector mosquito Anopheles stephensi. PNAS 112, 6736–6743. doi: 10.1073/pnas.1521077112
Gao, J., Wang, G., Ma, S., Xie, X., Wu, X., and Xia, Q. (2015). CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 87, 99–110. doi: 10.1007/s11103-014-0263-0
Gao, W., Long, L., Tian, X., Xu, F., Liu, J., Singh, P. K., et al. (2017). Genome editing in cotton with the CRISPR/Cas9 system. Front. Plant Sci. 8:1364. doi: 10.3389/fpls.2017.01364
Gao, X., Chen, J., Dai, X., Zhang, D., and Zhao, Y. (2016). An effective strategy for reliably isolating heritable and Cas9-Free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 171, 1794–1800. doi: 10.1104/pp.16.00663
Gasparis, S., Kała, M., Przyborowski, M., Łyżnik, L. A., Orczyk, W., and Nadolska-Orczyk, A. A. (2018). Simple and efficient CRISPR/Cas9 platform for induction of single and multiple, heritable mutations in barley (Hordeum vulgare L.). Plant Methods 14:111. doi: 10.1186/s13007-018-0382-8
Gautam, K., and Kumar, S. (2020). “Techniques for the detection, identification, and diagnosis of agricultural pathogens and diseases,” in Natural Remedies for Pest, Disease and Weed Control, eds C. Egbuna and B. Sawicka (Cambridge, MA: Academic Press), 135–142.
Gil-Humanes, J., and Voytas, D. F. (2014). Wheat rescued from fungal disease. Nat. Biotechnol. 32, 886–887. doi: 10.1038/nbt.3013
Giovannini, A., Laura, M., Nesi, B., Savona, M., and Cardi, T. (2021). Genes and genome editing tools for breeding desirable phenotypes in ornamentals. Plant Cell Rep. 40, 461–478. doi: 10.1007/s00299-020-02632-x
Gomez, M. A., Lin, Z. D., Moll, T., Luebbert, C., Chauhan, R. D., Vijayaraghavan, A., et al. (2017). Simultaneous CRISPR/Cas9-mediated editing of cassava EIF4E isoforms NCBP-1 and NCBP-2 confers elevated resistance to cassava brown streak disease. bioRxiv [Preprint]. doi: 10.1111/pbi.12987
Gumtow, R., Wu, D., Uchida, J., and Tian, M. (2018). A Phytophthora palmivora extracellular cystatin-like protease inhibitor targets papain to contribute to virulence on papaya. Mol. Plant Microbe Interact. 31, 363–373. doi: 10.1094/MPMI-06-17-0131-FI
Hammond, A., Galizi, R., Kyrou, K., Simoni, A., Siniscalchi, C., Katsanos, D., et al. (2016). A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83. doi: 10.1038/nbt.3439
Hannoufa, A., Pillai, B. V. S., and Chellamma, S. (2014). Genetic enhancement of Brassica napus seed quality. Transgenic Res. 23, 39–52. doi: 10.1007/s11248-013-9742-3
Haque, E., Taniguchi, H., Hassan, M. M., Bhowmik, P., Karim, M. R., Smiech, M., et al. (2018). Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: recent progress, prospects, and challenges. Front. Plant Sci. 9:617. doi: 10.3389/fpls.2018.00617
Hayut, F. S., Melamed Bessudo, C., and Levy, A. A. (2017). Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 8:15605. doi: 10.1038/ncomms15605
Hinge, V., Patil, H., and Nadaf, A. (2015). Comparative characterization of aroma volatiles and related gene expression analysis at vegetative and mature stages in basmati and non-basmati rice (Oryza sativa L.) cultivars. Appl. Biochem. Biotechnol. 178, 619–639. doi: 10.1007/s12010-015-1898-2
Howells, R. M., Craze, M., Bowden, S., and Wallington, E. (2018). Efficient generation of stable, heritable gene edits in wheat using CRISPR/Cas9. BMC Plant Biol. 18:6088. doi: 10.1186/s12870-018-1433-z
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278.
Hu, B., Li, D., Liu, X., Qi, J., Gao, D., Zhao, S., et al. (2017). Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Plant Mol. Biol. 10, 1575–1578. doi: 10.1016/j.molp.2017.09.005
Hu, J., Israeli, A., Ori, N., and Sun, T. (2018). The interaction between DELLA and ARF/IAA mediates crosstalk between gibberellin and auxin signaling to control fruit initiation in tomato. Plant Cell 30, 1710–1728. doi: 10.1105/tpc.18.00363
Hu, N., Xian, Z., Li, N., Liu, Y., Huang, W., Yan, F., et al. (2019). Rapid and user-friendly open-source CRISPR/Cas9 system for single- or multi-site editing of tomato genome. Hortic. Res. 6:7. doi: 10.1038/s41438-018-0082-6
Huan, Z., Honglu, Z., Gozde, S. D., Eduardo, G. G., Chunhai, F., and Markita, P. L. (2020). Engineering DNA nanostructures for SiRNA delivary in plants. Nat. Protoc. 15, 3064–3087. doi: 10.1038/s41596-020-0370-0
Huang, L., Zhang, R., Huang, G., Li, H., Melaku, G., Zhang, S., et al. (2018). Developing superior alleles of yield genes in rice by artificial mutagenesis using the CRISPR/Cas9 system. J. Crop Prod. 6, 475–481. doi: 10.1016/j.cj.2018.05.005
Huang, X. Z., Zeng, X. F., Li, J. R., and Zhao, D. (2017). Construction and analysis of tify1a and tify1b mutants in rice (Oryza sativa) based on CRISPR/Cas9 technology. J. Agric. Biotechnol. 25, 1003–1012.
Hummel, A. W., Chauhan, R. D., Cermak, T., Mutka, A. M., Vijayaraghavan, A., and Boyher, A. (2018). Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J. 16, 1275–1282. doi: 10.1111/pbi.12868
Hussain, B., Lucas, S. T., and Budak, H. (2018). CRISPR/Cas9 in plants: at play in the genome and at work for crop improvement. Brief Funct. Genomics 17, 319–328. doi: 10.1093/bfgp/ely016
Hyun, Y., Kim, J., Cho, S. W., Choi, Y., Kim, J. S., and Coupland, G. (2015). Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241, 271–284. doi: 10.1007/s00425-014-2180-5
Iqbal, Z., Sattar, M. N., and Shafiq, M. (2016). CRISPR/Cas9: a tool to circumscribe cotton leaf curl disease. Front. Plant Sci. 7:475. doi: 10.3389/fpls.2016.00475
Islam, M. D. A., Rony, S. A., Rahman, M. B., Cinar, M. U., Villena, J., Uddin, M. J., et al. (2020). Improvement of disease resistance in livestock: application of immuno genomics and CRISPR/Cas9 technology. Animals 10, 1–20. doi: 10.3390/ani10122236
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M., and Toki, S. (2015). CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467, 76–82. doi: 10.1016/j.bbrc.2015.09.117
Javed, R. M., Noman, M., Shahid, M., Ahmed, T., Khurshid, M., Rashid, M. H., et al. (2019). Current situation of biofuel production and its enhancement by CRISPR/Cas9 –mediated genome engineering of microbial cells. Microbiol. Res. 219, 1–11. doi: 10.1016/j.micres.2018.10.010
Jia, H., Orbovic, V., Jones, J. B., and Wang, N. (2016). Modification of the PthA4 effector binding elements in type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 14, 1291–1301. doi: 10.1111/pbi.12495
Jia, H., and Wang, N. (2014). Targeted genome editing of sweet orange using Cas9/sg RNA. PLoS One 9:e0093806. doi: 10.1371/journal.pone.0093806
Jia, H., Xu, J., Orbovic, V., Zhang, Y., and Wang, N. (2017). Editing citrus genome via SaCas9/sgRNA system. Front. Plant Sci. 8:2135. doi: 10.3389/fpls.2017.02135
Jia, H., Zhang, Y., Orbovic, V., Xu, J., White, F. F., Jones, J. B., et al. (2019). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. doi: 10.1111/pbi.12677
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, 188–188. doi: 10.1093/nar/gkt780
Jiang, W. Z., Henry, I. M., Lynagh, P. G., Comai, L., Cahoon, E. B., and Weeks, D. P. (2017). Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15, 648–657. doi: 10.1111/pbi.12663
Jie, J., Zhang, C., Sun, Z., Wang, L., Duanmu, D., and Fan, Q. (2019). Genome editing in cowpea Vigna unguiculata using CRISPR-Cas9. Int. J. Mol. Sci. 20:2471. doi: 10.3390/ijms20102471
Joshi, R. K. (2019). Genome editing in chili pepper using a CRISPR/Cas9 Cytidine base editing system. Asian J. Plant Sci. Res. 9:445.
Kaboshi, M., Aida, R., and Sasaki, K. (2017). Generation of gene-edited Chrysanthemum morifolium using multi-copy transgenes as targets and markers. Plant Cell Physiol. 58, 216–226. doi: 10.1093/pcp/pcw222
Kah, M., and Hofmann, T. J. E. I. (2014). Nanopesticide research: current trends and future priorities. Environ. Int. 63, 224–235. doi: 10.1016/j.envint.2013.11.015
Kaminski, R., Chen, Y., Fischer, T., Tedaldi, E., Napoli, A., Zhang, Y., et al. (2017). Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci. Rep. 6, 1–15. doi: 10.1038/srep22555
Kanazashi, Y., Hirose, A., Takahashi, I., Mikami, M., Endo, M., Hirose, S., et al. (2018). Simultaneous site directed mutagenesis of duplicated loci in soybean using a single guide RNA. Plant Cell Rep. 37, 553–563. doi: 10.1007/s00299-018-2251-3
Kapusi, E., Corcuera-Gómez, M., Melnik, S., and Stoger, E. (2017). Heritable genomic fragment deletions and small indels in the putative ENGase gene induced by CRISPR/Cas9 in barley. Front. Plant Sci. 8:540. doi: 10.3389/fpls.2017.00540
Kathleen, L. H. (2015). Nutritionally enhanced food crops; progress and perspectives. Int. J. Mol. Sci. 16, 3895–3914. doi: 10.3390/ijms16023895
Kaur, N., Alok, A., Shivani Kaur, N., Pandey, P., Awasthi, P., and Tiwari, S. (2018). CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genomics 18, 89–99. doi: 10.1007/s10142-017-0577-5
Khan, F. A., Pandupuspitasari, N. S., Chun-Jie, H., Ao, Z., Jamal, M., Zohaib, A., et al. (2016). CRISPR/Cas9 therapeutics: a cure for cancer and other genetic diseases. Oncotarget 7:52541. doi: 10.18632/oncotarget.9646
Khromov, A. V., Gushchin, V. A., Timerbaev, V. I, Kalinina, N. O., Taliansky, M. E., and Makarov, V. V. (2018). Guide RNA design for CRISPR/Cas9 mediated potato genome editing. Dokl. Biophys. Mol. Biol. 479, 90–94. doi: 10.1134/S1607672918020084
Khurana, S. M. P., and Rajarshi kumar, G. (2019). Plant Biotechnology: Progress in Genomic Era. Berlin: Springer Nature Singapore Pte Ltd.
Kim, D., Hager, M., Brant, E., and Budak, H. (2021). Efficient genome editing in wheat using Cas9 and Cpf1 (AsCpf1 and LbCpf1) nucleases. Funct. Integr. Genomics 21, 355–366. doi: 10.1007/s10142-021-00782-z
Kim, D., Kim, D., Alptekin, B., and Budak, H. (2018). CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genomics 18, 31–41. doi: 10.1007/s10142-017-0572-x
Kis, A., Hamar, É, Tholt, G., Bán, R., and Havelda, Z. (2019). Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 17, 1004–1006. doi: 10.1111/pbi.13077
Klap, C., Yeshayahou, E., Bolger, A. M., Arazi, T., Gupta, S. K., Shabtai, S., et al. (2017). Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 15, 634–647. doi: 10.1111/pbi.12662
Koseoglou, E. (2017). The Study of SlPMR4 CRISPR/Cas9-Mediated Tomato Allelic Series for Resistance Against Powdery Mildew. Wageningen: Wageningen University.
Kui, L., Chen, H., Zhang, W., He, S., Xiong, Z., Zhang, Y., et al. (2017). Building a genetic manipulation toolbox for orchid biology: identification of constitutive promoters and application of CRISPR/Cas9 in the orchid Dendrobiumofficinale. Front. Plant Sci. 7:2036. doi: 10.3389/fpls.2016.02036
Kumar, N., Galli, M., Ordon, J., Stuttmann, J., Kogel, K. H., and Imani, J. (2018). Further analysis of barley MORC1 using a highly efficient RNA-guided Cas9 gene-editing system. Plant Biotechnol. J. 16, 1892–1903.
Kusano, H., Ohnuma, M., Mutsuro-Aoki, H., Asahi, T., Ichinosawa, D., Onodera, H., et al. (2018). Establishment of a modified CRISPR/Cas9 system with increased mutagenesis frequency using the translational enhancer dMac3 and multiple guide RNAs in potato. Sci. Rep. 8:13753. doi: 10.1038/s41598-018-32049-2
Lacomme, C. (2015). Strategies for altering plant traits using virus-induced gene silencing technologies. Methods Mol. Biol. 1287, 25–41. doi: 10.1007/978-1-4939-2453-0_2
Lang, Z., Wang, Y., Tang, K., Tang, D., Datsenka, T., Cheng, J., et al. (2017). Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. U.S.A. 114, 4511–4519. doi: 10.1073/pnas.1705233114
Larson, M. H., Gilbert, L. A., Wang, X., Lim, W. A., Weissman, J. S., and Qi, L. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196. doi: 10.1038/nprot.2013.132
Lawrenson, T., and Harwood, W. A. (2019). Creating targeted gene knockouts in barley using CRISPR/Cas9. Methods Mol. Biol. 1900, 217–232. doi: 10.1007/978-1-4939-8944-7_14
Lawrenson, T., Shorinola, O., Stacey, N., Li, C., Østergaard, L., Patron, N., et al. (2015). Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16, 258–271. doi: 10.1186/s13059-015-0826-7
Lee, C. W., Mahendra, S., Zodrow, K., Li, D., and Tsai, Y. C. (2010). Developmental phytotoxicity of metal oxide nanoparticles to Arabidopsis thaliana. Environ. Toxicol. Chem. 29, 669–675. doi: 10.1002/etc.58
Lemmon, Z. H. (2018). Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770. doi: 10.1038/s41477-018-0259-x
Li, M., Li, X., Zhou, Z., Wu, P., Fang, M., and Pan, X. (2016). Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7:377. doi: 10.3389/fpls.2016.00377
Li, B., Cui, G., Shen, G., Zhan, Z., Huang, L., and Chen, J. (2017). Targeted mutagenesis in the medicinal plant Salvia miltiorrhiza. Sci. Rep. 7:43320. doi: 10.1038/srep43320
Li, J., Zhang, H., Si, X., Tian, Y., Chen, K., and Liu, J. (2017). Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. JGG 44, 465–468. doi: 10.1016/j.jgg.2017.02.002
Li, H., Yang, Y., Hong, W., Huang, M., Wu, M., and Zhao, X. (2020). Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct. Target. Ther. 5:1. doi: 10.1038/s41392-019-0089-y
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., and Bush, J. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Li, R., Li, R., Li, X., Fu, D., Zhu, B., Tian, H., et al. (2018a). Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ- aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 16, 415–427. doi: 10.1111/pbi.12781
Li, R., Zhang, L., Wang, L., Chen, L., Zhao, R., Sheng, J., et al. (2018b). Reduction of tomato-plant chilling tolerance by CRISPR-Cas9-mediated SlCBF1 mutagenesis. J. Agric. Food Chem. 66, 9042–9051. doi: 10.1021/acs.jafc.8b02177
Li, R., Zhu, D. F. B., Luo, Y., and Hu, H. (2018c). CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 4, 513–524. doi: 10.1111/tpj.13872
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., et al. (2018d). Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9:559. doi: 10.3389/fpls.2018.00559
Li, C., Zong, Y., Wang, Y., Jin, S., Zhang, D., Song, Q., et al. (2018e). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19:59. doi: 10.1186/s13059-018-1443-z
Li, A., Jia, S., Yobi, A., Ge, Z., Sato, S. J., Zhang, C., et al. (2018f). Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 177, 1425–1438. doi: 10.1104/pp.18.00200
Li, T., Yang, X., Yu, Y., Si, X., Zhai, X., Zhang, H., et al. (2018g). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163. doi: 10.1038/nbt.4273
Li, C., Nguyen, V., Liu, J., Fu, W., Chen, C., Yu, K., et al. (2019). Mutagenesis of seed storage protein genes in Soybean using CRISPR/Cas9. BMC Res. Notes 12:176. doi: 10.1186/s13104-019-4207-2
Li, R., Liu, C., Zhao, R., Wang, L., Chen, L., Yu, W., et al. (2019). CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 19:38. doi: 10.1186/s12870-018-1627-4
Li, Z. S., Liu, Z. B., Xing, A. Q., Moon, B. P., Koellhoffer, J. P., and Huang, L. X. (2015). Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 169, 960–970. doi: 10.1104/pp.15.00783
Liang, Z., Zhang, K., Chen, K., and Gao, C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41, 63–68. doi: 10.1016/j.jgg.2013.12.001
Liao, H. K., Gu, Y., Diaz, A., Marlett, J., Takahashi, Y., Li, M., et al. (2015). Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 6, 1–10. doi: 10.1038/ncomms7413
Liu, G., Li, J., and Godwin, I. D. (2019). Genome editing by CRISPR/Cas9 in sorghum through biolistic bombardment. Methods Mol. Biol. 1931, 169–183. doi: 10.1007/978-1-4939-9039-9_12
Liu, H., Wang, K., Jia, Z., Gong, Q., Lin, Z., Du, L., et al. (2020). Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR System. J. Exp. Bot. 71, 1337–1349. doi: 10.1093/jxb/erz529
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441. doi: 10.1093/pcp/pcx075
Lou, D., Wang, H., and Yu, D. (2018). The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice. BMC Plant Biol. 18:203. doi: 10.1186/s12870-018-1408-0
Lv, Z., Jiang, R., Chen, J., and Chen, W. (2020). Nanoparticle-mediated gene transformation strategies for plant genetic engineering. Plant J. 104, 880–891. doi: 10.1111/tpj.14973
Lyzenga, W. J., Harrington, M., Bekkaoui, D., Wigness, M., Dwayne, D., Hegedus, D. D., et al. (2019). CRISPR/Cas9 editing of three CRUCIFERIN C homoeologues alters the seed protein profile in Camelina sativa. BMC Plant Biol. 19:292. doi: 10.1186/s12870-019-1873-0
Ma, C., Liu, M., Li, Q., Si, J., Ren, X., and Song, H. (2019a). Efficient BoPDS gene editing in cabbage by the CRISPR/Cas9 System. Hortic. Plant J. 5, 164–169. doi: 10.1016/j.hpj.2019.04.001
Ma, C., Zhu, C., Zheng, M., Liu, M., Zhang, D., Liu, B., et al. (2019b). CRISPR/Cas9-mediated multiple gene editing in Brassica oleracea var. capitata using the endogenous tRNA-processing system. Hortic. Res. 6:20. doi: 10.1038/s41438-018-0107-1
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Macias, V. M., McKeand, S., Rodriguez, D. C., Hughes, G. L., Fazekas, A., Pujhari, S., et al. (2020). Cas9-Mediated gene-editing in the malaria mosquito Anopheles stephensi by remot control. G3 Genes Genomes Genetics 10, 1353–1360. doi: 10.1534/g3.120.401133
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Loedin, I. S., Čermák, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/pbi.12927
Maioli, A., Gianoglio, S., Moglia, A., Acquadro, A., Valentino, D., Milani, A. M., et al. (2020). Simultaneous CRISPR/Cas9 editing of three PPO genes reduces fruit flesh browning in Solanum melongena L. Front. Plant Sci. 11:607161. doi: 10.3389/fpls.2020.607161
Mali, P., Aach, J., Stranges, P. B., Esvelt, K. M., Moosburner, M., Kosuri, S., et al. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838. doi: 10.1038/nbt.2675
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleo proteins. Front. Plant Sci. 7:1904. doi: 10.3389/fpls.2016.01904
Mandal, P. K., Ferreira, L. M., Collins, R., Meissner, T. B., Boutwell, C. L., Friesen, M., et al. (2014). Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15, 643–652. doi: 10.1016/j.stem.2014.10.004
Manghwar, H., Lindsey, K., Zhang, X., and Jin, S. (2019). CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24, 1102–1125. doi: 10.1016/j.tplants.2019.09.006
Manikandan, A., and Subramanian, K. (2016). Evaluation of zeolite based nitrogen nano-fertilizers on maize growth, yield and quality on inceptisols and alfisols. I. J. Plant Soil Sci. 9, 1–9. doi: 10.9734/IJPSS/2016/22103
Mao, X., Zheng, Y., Xiao, K., Wei, Y., Zhu, Y., Cai, Q., et al. (2018). OsPRX2 contributes to stomatal closure and improves potassium deficiency tolerance in rice. Biochem. Biophys. Res. Commun. 495, 461–467. doi: 10.1016/j.bbrc.2017
Mao, Y., Botella, J. R., Liu, Y., and Zhu, J. K. (2019). Gene editing in plants: progress and challenges. Natl. Sci. Rev. 6, 421–437. doi: 10.1093/nsr/nwz005
Mao, Y., Zhang, H., Xu, N., Zhang, B., Gou, F., and Zhu, J. K. (2013). Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol. Plant. 6, 2008–2011. doi: 10.1093/mp/sst121
Mao, Y., Zhang, Z., Feng, Z., Wei, P., Zhang, H., Botella, J. R., et al. (2016). Development of germline specific CRISPR Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 14, 519–532. doi: 10.1111/pbi.12468
Martín Pizarro, C., Triviño, J. C., and Posé, D. (2019). Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 70, 885–895. doi: 10.1093/jxb/ery400
Mercx, S., Smargiasso, N., Chaumont, F., Pauw, E. D., Boutry, M., and Navarre, C. (2017). Inactivation of the b (1, 2)- xylosyltransferase and the a (1, 3)-fucosyltransferase genes in Nicotiana tabacum BY-2 cells by a multiplex CRISPR/Cas9 strategy results in glycoproteins without plant-specific glycans. Front. Plant Sci. 8:403. doi: 10.3389/fpls.2017.00403
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Michno, J. M., Wang, X., Liu, J., Curtin, S. J., Kono, T. J., and Stupar, R. M. (2015). CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6, 243–252. doi: 10.1080/21645698.2015.1106063
Miller, J. C., Holmes, M. C., Wang, J., Guschin, D. Y., Lee, Y. L., Rupniewski, I., et al. (2007). An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 25, 778–785. doi: 10.1038/nbt1319
Mittal, D., Kaur, G., Singh, P., Yadav, K., and Ali, S. A. (2020). Nanoparticle-based sustainable agriculture and food science: recent advances and future outlook. Front. Nanotechnol. 2:579954. doi: 10.3389/fnano.2020.579954
Murovec, J., Gucek, K., Bohanec, B., Avbelj, M., and Jerala, R. (2018). DNA-free genome editing of Brassica oleracea and B. Rapa protoplasts using CRISPR-Cas9 ribonucleoprotein complexes. Front. Plant Sci. 9:1594. doi: 10.3389/fpls.2018.01594
Nadakuduti, S. S., and Enciso-Rodríguez, F. (2021). Advances in genome editing with CRISPR systems and transformation technologies for plant DNA Manipulation. Front. Plant Sci. 11:637159. doi: 10.3389/fpls.2020.637159
Naderi, M. R., and Abedi, A. (2012). Application of nanotechnology in agriculture and refinement of environmental pollutants. J. Nanotechnol. 11, 18–26. doi: 10.4236/snl.2013.33008
Nakajima, I., Ban, Y., Azuma, A., Onoue, N., Moriguchi, T., Yamamoto, T., et al. (2017). CRISPR/Cas9- mediated targeted mutagenesis in grape. PLoS One 12:e0177966. doi: 10.1371/journal.pone
Nakayasu, M., Akiyama, R., Lee, H. J., Osakabe, K., Osakabe, Y., Watanabe, B., et al. (2018). Generation of solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. Biochem. 131, 70–77. doi: 10.1016/j.plaphy.2018
Neelendra, K., Galli, M., Ordon, J., Stuttmann, J., Kogel, K. H., and Imani, J. (2018). Further analysis of barley MORC1 using a highly efficient RNA-guided Cas9 gene-editing system. Plant Biotechnol. J. 16, 1892–1903. doi: 10.1111/pbi.12924
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482–486. doi: 10.1038/s41598-017-00578-x
Nishihara, M., Higuchi, A., Watanabe, A., and Tasaki, K. (2018). Application of the CRISPR/Cas9 system for modification of flower color in Torenia fournieri. BMC Plant Biol. 18:331. doi: 10.1186/s12870-018-1539-3
Nonaka, S., Arai, C., Takayama, M., Matsukura, C., and Ezura, H. (2017). Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 7:7057. doi: 10.1038/s41598-017-06400-y
Ordon, J., Gantner, J., Kemna, J., Schwalgun, L., Reschke, M., Streubel, J., et al. (2017). Generation of chromosomal deletions in dicotyledonous plants employing a user-friendly genome editing tool kit. Plant J. 89, 155–168. doi: 10.1111/tpj.13319
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., and Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 17, 665–673. doi: 10.1111/pbi.13006
Osakabe, Y., Liang, Z., Ren, C., Nishitani, C., Osakabe, K., Wada, M., et al. (2018). CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 13:2844. doi: 10.1038/s41596-018-0067-9
Osakabe, Y., Watanabe, T., Sugano, S. S., Ueta, R., Ishihara, R., Shinozaki, K., et al. (2016). Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 6:26685. doi: 10.1038/srep26685
Otang Ntui, V., Tripathi, J., and Tripathi, L. (2020). Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa spp.). Curr. Plant Biol. 21:100128. doi: 10.1016/j.cpb.2019.100128
Ousterout, D. G., Kabadi, A. M., Thakore, P. I., Majoros, W. H., Reddy, T. E., and Gersbach, C. A. (2015). Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 6:6244. doi: 10.1038/ncomms7244
Oz, M. T., Altpeter, A., Karan, R., Merotto, A., and Altpeter, F. (2021). CRISPR/Cas9-mediated multi-allelic gene targeting in sugarcane confers herbicide tolerance. Front. Genome Ed. 3:673566. doi: 10.3389/fgeed.2021.673566
Padmaja, G. (1995). Cyanide detoxification in cassava for food and feed uses. Crit. Rev. Food Sci. Nutr. 35, 299–339. doi: 10.1080/10408399509527703
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6:24765. doi: 10.1038/srep24765
Pan, L., Yan, J. L., Feng, J. Z., Gui, Z. Z., Xiao, Y. J., Hui, M. Y., et al. (2016). The Arabidopsis UDP-glycosyltransferasesUGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 89, 85–103. doi: 10.1111/tpj.13324
Pankaj, B., Ellison, E., Polley, B., Bollina, V., Kulkarni, M., Ghanbarnia, K., et al. (2018). Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Sci. Rep. 8:6502. doi: 10.1038/s41598-018-24690-8v
Park, C. Y., Lee, D. R., Sung, J. J., and Kim, D. W. (2016). Genome-editing technologies for gene correction of hemophilia. Hum. Genet. 15, 977–981. doi: 10.1007/s00439-016-1699-x
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker resistant plants through CRISPR/Cas9 targeted editing of the susceptibility gene CsLOB 1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Peng, W., Fang-Jie, Z., and Peter, M. K. (2019). Engineering crops without genome integration using nanotechnology. Trends Plant Sci. 24, 574–577.
Pham, A. T., Shannon, J. G., and Bilyeu, K. D. (2012). Combinations of mutant FAD2 and FAD3 genes to produce high oleic acid and low linolenic acid soybean oil. Theor. Appl. Genet. 125, 503–515. doi: 10.1007/s00122-012-1849-z
Pioneer (2016). DuPont Announces Intentions to Commercialize First CRISPR-Cas Product. Available online at: https://www.prweb.com/releases/dupont-pioneer-seed/crispr-cas-corn/prweb13349828.htm (accessed April 18, 2016).
Platt, R. J., Chen, S., Zhou, Y., Yim, M. J., Swiech, L., Kempton, H. R., et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455. doi: 10.1016/j.cell.2014.09.014
Pramanik, D., Shelake, R. M., Park, J., Kim, M. J., Hwang, I., Park, I., et al. (2021). CRISPR/Cas9-mediated generation of pathogen-resistant tomato against tomato yellow leaf curl virus and powdery mildew. Int. J. Mol. Sci. 22:1878. doi: 10.3390/ijms22041878
Prihatna, C., Barbetti, M. J., and Barker, S. J. (2018). A novel tomato Fusarium wilt tolerance gene. Front. Microbiol. 9:1226. doi: 10.3389/fmicb.2018.01226
Pyott, D. E., Sheehan, E., and Molnar, A. (2016). Engineering of CRISPR/Cas9- mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288. doi: 10.1111/mpp.12417
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183.
Qi, W., Zhu, T., Tian, Z., Li, C., Zhang, W., and Song, R. (2016). High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNAprocessing system-based strategy in maize. BMC Biotechnol. 16:58.
Qian, L., Zhou, Q., Qingnan, W., Ting, X., Yang, Y., and Zhiyao, H. (2019). Applications of genome editing technology in animal disease modeling and gene therapy. Comput. Struct. Biotechnol. J. 17, 689–698. doi: 10.1016/j.csbj.2019.05.006
Raitskin, O., Schudoma, C., West, A., and Patron, N. J. (2019). Comparison of efficiency and specificity of CRISPR-associated (Cas) nucleases in plants: An expanded toolkit for precision genome engineering. PLoS One 14:e0211598. doi: 10.1371/journal.pone.0211598
Rana, R. A., Siddiqui, M. N., Skalicky, M., Brestic, M., Hossain, A., Kayesh, E., et al. (2021). Prospects of nanotechnology in improving the productivity and quality of horticultural crops. Horticulturae 7:332. doi: 10.3390/horticulturae7100332
Ren, C., Liu, X., Zhang, Z., Wang, Y., Duan, W., Li, S., et al. (2016). CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitisvinifera L.). Sci. Rep. 6:32289. doi: 10.1038/srep32289
Robert, F., Huang, S., and Pelletier, J. (2017). CRISPR / Cas9 editing to facilitate and expand drug discovery. Curr. Gene Ther. 17, 275–285. doi: 10.2174/1566523217666171121164615
Rodriguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480. doi: 10.1016/j.cell.2017.08.030
Roldan, M. V. G., Perilleux, C., Morin, H., Huerga-Fernandez, S., Latrasse, D., Benhamed, M., et al. (2017). Natural and induced loss of function mutations in SlMBP21 MADS-box gene led to jointless-2 phenotype in tomato. Sci. Rep. 7:4402. doi: 10.1038/s41598-017-04556-1
Ron, M., Kajala, K., Pauluzzi, G., Wang, D., Reynoso, M. A., Zumstein, K., et al. (2014). Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 166, 455–469. doi: 10.1104/pp.114.239392
Sadanandom, A., Srivastava, A. K., and Zhang, C. (2019). Targeted mutagenesis of the SUMO protease, overly tolerant to salt1 in rice through CRISPR/Cas9-mediated genome editing reveals a major role of this SUMO protease in salt tolerance. BioRxiv [Preprint]. doi: 10.1101/555706
Sánchez-León, S., Gil-Humanes, J., Ozuna, C. V., Giménez, M. J., Sousa, C., Voytas, D. F., et al. (2018). Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910. doi: 10.1111/pbi.12837
Sanskriti, V., Surbhi, K., Virender, K., Gunvant, B. P., Trupti, J., Humira, S., et al. (2019). Genome editing in Plants: exploration of technological advancements and challenges. Cells 8:1386. doi: 10.3390/cells8111386
Sauer, N. J., Narváez-Vásquez, J., Mozoruk, J., Miller, R. B., Warburg, Z. J., and Woodward, M. J. (2016). Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 170, 1917–1928. doi: 10.1104/pp.15.01696
Schwank, G., Koo, B. K., Sasselli, V., Dekkers, J. F., Heo, I., and Demircan, T. (2013). Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658. doi: 10.1016/j.stem.2013.11.002
Schwartz, S. H., Hendrix, B., Hoffer, P., Sanders, R. A., and Zheng, W. (2020). Carbon dots for efficient sirna delivery and gene silencing in plants. Plant Physiol. 184, 647–657. doi: 10.1104/pp.20.00733
Servin, A., Elmer, W., Mukherjee, A., Torre Roche, R. D. L., and Hamdi, H. (2015). A review of the use of engineered nanomaterials to suppress plant disease and enhance crop yield. J. Nanopart Res. 17, 1–21. doi: 10.1007/s11051-015-2907-7
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Shang, Y., Hasan, M. K., Ahammed, G. J., Li, M., Yin, H., and Zhou, J. (2019). Applications of nanotechnology in plant growth and crop protection: a review. Molecules 24:2558. doi: 10.3390/molecules24142558
Shao, X., Wu, S., Dou, T., Zhu, H., Hu, C., Huo, H., et al. (2019). Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol. J. 18, 17–19. doi: 10.1111/pbi.13216
Shen, C., Que, Z., Xia, Y., Tang, N., Li, D., He, R., et al. (2017). Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J. Plant Biol. 60, 539–547. doi: 10.1007/s12374-016-0400-1
Sheykhbaglou, R., Sedghi, M., Shishevan, M. T., and Sharifi, R. S. (2010). Effects of nano-iron oxide particles on agronomic traits of soybean. Not. Sci. Biol. 2, 112–113. doi: 10.15835/nsb224667
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., and Yang, M. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Shibuya, K., Watanabe, K., and Ono, M. (2018). CRISP/Cas9-mediated mutagenesis of the EPHEMERAL1 locus that regulates petal senescence in Japanese morning glory. Plant Physiol. 131, 53–57. doi: 10.1016/j.plaphy.2018.04.036
Shim, J. S., Oh, N., Chung, P. J., Kim, Y. S., Choi, Y. D., and Kim, J. K. (2018). Over expression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 9:310. doi: 10.3389/fpls.2018.00310
Shu, H., Luo, Z., Peng, Z., and Wang, J. (2020). The application of CRISPR/Cas9 in hair roots to explore the functions of AhNFR1 and AhNFR5 genes during peanut nodulation. BMC Plant Biol. 20:417. doi: 10.1186/s12870-020-02614-x
Singh, M., Kumar, M., Albertsen, M. C., Young, J. K., and Cigan, A. M. (2018). Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aaestivum, L.). Plant Mol. Biol. 97, 371–383. doi: 10.1007/s11103-018-0749-2
Smith, J., Grizot, S., Arnould, S., Duclert, A., Epinat, J. C., Chames, P., et al. (2006). A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 34, 149–149. doi: 10.1093/nar/gkl720
So, M. C., Wiederrecht, G. P., Mondloch, J. E., Hupp, J. T., and Farha, O. K. (2015). Metal-organic framework materials for light-harvesting and energy transfer. Chem. Comm. 51, 3501–3510. doi: 10.1039/c4cc09596k
Songstad, D. D., Petolino, J. F., Voytas, D. F., and Reichert, N. A. (2017). Genome editing of plants. Crit. Rev. Plant Sci. 36, 1–23. doi: 10.1080/07352689.2017.1281663
Soyk, S., Muller, N. A., Park, S. J., Schmalenbach, I., Jiang, K., Hayama, R., et al. (2017). Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 49, 162–168. doi: 10.1038/ng.3733
Stajic, E., Kiełkowska, A., Murovec, J., and Bohanec, B. (2019). Deep sequencing analysis of CRISPR/Cas9 induced mutations by two delivery methods in target model genes and the CENH3 region of red cabbage (Brassica oleracea var. capitata f. rubra). PCTOC 139, 227–235. doi: 10.1007/s11240-019-01665-9
Su, S., Xiao, W., Guo, W., Yao, X., Xiao, J., Ye, Z., et al. (2017). The CYCLOIDEA-RADIALIS module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torenia fournieri (Linderniaceae). New Phytol. 215, 1582–1593. doi: 10.1111/nph.14673
Subburaj, S., Chung, S. J., Lee, C., Ryu, S. M., Kim, D. H., Kim, J. S., et al. (2016). Site-directed mutagenesis in Petunia hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleo proteins. Plant Cell Rep. 35, 1535–1544. doi: 10.1007/s00299-016-1937-7
Subramanian, K. S., Manikandan, A., Thirunavukkarasu, M., and Rahale, C. S. (2015). “Nano-fertilizers for balanced crop nutrition,” in Nanotechnologies in Food and Agriculture, eds M. Rai, C. Ribeiro, L. Mattoso, and N. Duran (Cham: Springer), 69–80. doi: 10.1007/s00210-021-02057-7
Sugano, S. S., Shirakawa, M., Takagi, J., Mastuda, Y., Shimada, T., Nishimura, I. H., et al. (2014). CRISPR/Cas9-Mediated targeted mutagenesis in Liverwort Marchantia polymorpha L. Plant Cell Physiol. 55, 475–481. doi: 10.1093/pcp/pcu014
Sun, B., Jiang, M., Zheng, H., Jian, Y., Huang, W. L., Yuan, Q., et al. (2020). Color-related chlorophyll and carotenoid concentrations of Chinese kale can be altered through CRISPR/Cas9 targeted editing of the carotenoid isomerase gene BoaCRTISO. Hortic. Res. 7:161. doi: 10.1038/s41438-020-00379-w
Sun, L., Song, F., Guo, J., Zhu, X., Liu, S., Liu, F., et al. (2020). Nano-ZnO-induced drought tolerance is associated with melatonin synthesis and metabolism in maize. Int. J. Mol. Sci. 21, 1–18. doi: 10.3390/ijms21030782
Sun, B., Zheng, A., Jiang, M., Xue, S., Yuan, Q., Jiang, L., et al. (2018). CRISPR/Cas9-mediated mutagenesis of homologous genes in Chinese kale. Sci. Rep. 8:16786. doi: 10.1038/s41598-018-34884-9
Sun, L., and Kao, T. H. (2018). CRISPR/Cas9-mediated knockout of PiSSK1 reveals essential role of S-locus F-box protein-containing SCF complexes in recognition of non-self S-RNases during cross-compatible pollination in self-incompatible Petunia inflata. Plant Reprod. 31, 129–143. doi: 10.1007/s00497-017-0314-1
Sun, L., Song, F., Zhu, X., Liu, S., Liu, F., Wang, Y., et al. (2021). Nano-ZnO alleviates drought stress via modulating the plant water use and carbohydrate metabolism in maize. Arch. Agro. Soil Sci. 67, 245–259. doi: 10.1080/03650340.2020.1723003
Sun, N., and Zhao, H. (2014). Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol. Bioeng. 111, 1048–1053. doi: 10.1002/bit.25018
Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., et al. (2017). Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 8:298. doi: 10.3389/fpls.2017.00298
Sun, Y., Li, J., and Xia, L. (2016). Precise genome modification via sequence-specific nucleases-mediated gene targeting for crop improvement. Front. Plant Sci. 7:1928. doi: 10.3389/fpls.2016.01928
Sunitha, S., and Rock, C. D. (2020). CRISPR/Cas9-mediated targeted mutagenesis of TAS4 and MYBA7 loci in grapevine rootstock 101-14. Transgenic Res. 29, 355–367. doi: 10.1007/s11248-020-00196-w
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., and Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Syed, S. A. Z., Ahmad, M., Herve, V., and Magdy, M. M. (2020). Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 21:289. doi: 10.1186/s13059-020-02204-y
Tang, L., Mao, B., Li, Y., Ly, Q., Zhang, L., Chen, C., et al. (2017). Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 7:14438. doi: 10.1038/s41598-017-14832-9
Tang, X. D., Gao, F., Liu, M. J., Fan, Q. L., Chen, D. K., and Ma, W. T. (2019). Methods for enhancing clustered regularly interspaced short palindromic repeats/Cas9-mediated homology-directed repair efficiency. Front. Genet. 10:551. doi: 10.3389/fgene.2019.00551
Tashkandi, M., Ali, Z., Aljedaani, F., Shami, A., and Mahfouz, M. M. (2018). Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 13:1525996. doi: 10.1080/15592324.2018
Tavakoli, K., Pour-Aboughadareh, A., Kianersi, F., Poczai, P., Etminan, A., and Shooshtari, L. (2021). Applications of CRISPR-Cas9 as an advanced genome editing system in life sciences. BioTech 10:14. doi: 10.3390/biotech10030014
Thomazella, D. P. T., Brail, Q., Dahlbeck, D., and Staskawicz, B. (2016). CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. BioRxiv [Preprint]. doi: 10.1101/064824
Thul, S. T., and Saragani, B. K. (2015). “Implications of nanotechnology on plant productivity and its rhizospheric environment,” in Nanotechnology and Plants: Nanoparticles and Their Impact On Plants, eds M. H. Siddiqui, M. H. Al-Whaibi, and M. Firoz (Cham: Springer), 37–53.
Tian, S., Jiang, L., Cui, X., Zhang, J., Guo, S., Li, M., et al. (2018). Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 37, 1353–1356. doi: 10.1007/s00299-018-2299-0
Tian, S., Jiang, L., Gao, Q., Zhang, J., Zong, M., Zhang, H., et al. (2017). Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 36, 399–406. doi: 10.1007/s00299-016-2089-5
Tomlinson, L., Yang, Y., Emenecker, R., Smoker, M., Taylor, J., Perkins, S., et al. (2019). Using CRISPR/Cas9 genome editing in tomato to create a gibberellin-responsive dominant dwarf DELLA allele. Plant Biotechnol. J. 17, 132–140. doi: 10.1111/pbi.12952
Tong, C. G., Wu, F. H., Yuan, Y. H., Chen, Y. R., and Lin, C. S. (2020). High-efficiency CRISPR/Cas-based editing of Phalaenopsis orchid MADS genes. Plant Biotechnol. J. 18, 889–891. doi: 10.1111/pbi.13264
Tripathi, J. N., Ntui, V. O., Ron, M., Muiruri, S. K., Britt, A., and Tripathi, L. (2019). CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2, 1–11. doi: 10.1038/s42003-019-0288-7
Tsai, C. J., and Xue, L. J. (2015). CRISPRing into the woods. GM Crops Food 6, 206–215. doi: 10.1080/21645698.2015
Tsveta, T., Lidia, S., Lora, T., Atanas, A., and Ivelin, P. (2021). DNA-free gene editing in plants: a brief overview. Biotechnol. Biotechnol. Equip. 35, 131–138. doi: 10.1080/13102818.2020.1858159
Tuncel, A., Corbin, K. R., Ahn-Jarvis, J., Harris, S., Hawkins, E., Smedley, M. A., et al. (2019). Cas9-mediated mutagenesis of potato starch-branching enzymes generates a range of tuber starch phenotypes. Plant Biotechnol. J. 17, 2259–2271. doi: 10.1111/pbi.13137
Ueta, R., Abe, C., Watanabe, T., Sugano, S. S., Ishihara, R., Ezura, H., et al. (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7:507. doi: 10.1038/s41598-017-00501-4
Upadhyay, S. K., Kumar, J., Alok, A., and Tuli, R. (2013). RNA-guided genome editing for target gene mutations in wheat. G3 Genes Genomes Genet. 3, 2233–2238. doi: 10.1534/g3.113.008847
Urnov, F. D., Miller, J. C., Lee, Y. L., Beausejour, C. M., Rock, J. M., Augustus, S., et al. (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651. doi: 10.1038/nature03556
Usman, B., Nawaz, G., Zhao, N., Liao, S., Qin, B., Liu, F., et al. (2021). Programmed editing of rice (Oryza sativa L.) OsSPL16 gene using CRISPR/Cas9 improves grain yield by modulating the expression of pyruvate enzymes and cell cycle proteins. Int. J. Mol. Sci. 22:249. doi: 10.3390/ijms22010249
Varkonyi-Gasic, E., Wang, T. C., Jeon, S., Drummond, R. S. M., Gleave, A. P., and Allan, A. C. (2018). Mutagenesis of kiwifruit CENTRORADIALIS-like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnol. J. 17, 869–880. doi: 10.1111/pbi.13021
Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M. P., Guyon-Debast, A., Chauvin, J. E., et al. (2019). Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 20:402. doi: 10.3390/ijms20020402
Voytas, D. F., and Gao, C. (2014). Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 12:e1001877. doi: 10.1371/journal.pbio.1001877
Wada, N., Ueta, R., Osakabe, Y., and Osakabe, K. (2020). Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 20:234. doi: 10.1186/s12870-020-02385-5
Waltz, E. (2016). Gene-edited CRISPR mushroom escapes US regulation. Nature 532:293. doi: 10.1038/nature.2016.19754
Wan, L., Wang, Z., Tang, M., Hong, D., Sun, Y., Ren, J., et al. (2021). CRISPR-Cas9 gene editing for fruit and vegetable crops: strategies and prospects. Horticulturae 7:193. doi: 10.3390/horticulturae7070193
Wang, D., Samsulrizal, N. H., Yan, C., Allcock, N. S., Craigon, J., Ulate, B. B., et al. (2019a). Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 179, 544–557. doi: 10.1104/pp.18.01187
Wang, L., Chen, S., Peng, A., Xie, Z., He, Y., and Zou, X. (2019b). CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in wanjincheng orange (Citrus sinensis (L.) Osbeck). Plant Biotechnol. Rep. 13, 501–510. doi: 10.1007/s11816-019-00556-x
Wang, R., da Rocha Tavano, E. C., Lammers, M., Martinelli, A. P., Angenent, G. C., and de Maagd, R. A. (2019c). Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 9:1696. doi: 10.1038/s41598-018-38170-6
Wang, H., Wu, Y., Zhang, Y., Yang, J., Fan, W., Zhang, H., et al. (2019d). CRISPR/Cas9-based mutagenesis of starch biosynthetic genes in sweet potato (Ipomoea Batatas) for the improvement of starch quality. Int. J. Mol. Sci. 20:4702. doi: 10.3390/ijms20194702
Wang, B., Zhu, L., Zhao, B., Zhao, Y., Xie, Y., Zheng, Z., et al. (2019e). Development of a haploid- inducer mediated genome editing system for accelerating maize breeding. Mol. Plant 12, 597–602. doi: 10.1016/j.molp.2019.03.006
Wang, H., La Russa, M., and Qi, L. S. (2016a). CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264. doi: 10.1146/annurev-biochem-060815-014607
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., and Gao, Y. (2016b). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, L., Wang, L., Tan, Q., Fan, Q., Zhu, H., and Hong, Z. (2016c). Efficient inactivation of symbiotic nitrogen fixation related genes in Lotus japonicas using CRISPR-Cas9. Front. Plant Sci. 7:1333. doi: 10.3389/fpls.2016.01333
Wang, J., Kuang, H., Zhang, Z., Yang, Y., Yan, L., Zhang, M., et al. (2020). Generation of seed lipoxigenase free soyabean using CRISPR-Cas9. Crop J. 8, 432–439. doi: 10.1016/j.cj.2019.08.008
Wang, L., Chen, L., Li, R., Zhao, R., Yang, M., Sheng, J., et al. (2017a). Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 65, 8674–8682. doi: 10.1021/acs.jafc.7b02745
Wang, P., Zhang, J., Sun, L., Ma, Y., Xu, J., Liang, S., et al. (2017b). High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 16, 137–150. doi: 10.1111/pbi.12755
Wang, Y., Meng, Z., Liang, C., Meng, Z., Wang, Y., Sun, G., et al. (2017c). Increased lateral root formation by CRISPR/Cas9 mediated editing of arginase genes in cotton. Sci. China Life Sci. 60, 524–527. doi: 10.1007/s11427-017-9031-y
Wang, X., Tu, M., Wang, D., Liu, J., Li, Y., Li, Z., et al. (2018a). CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16, 844–855. doi: 10.1111/pbi.12832
Wang, Z., Wang, S., Li, D., Zhang, Q., Li, L., Zhong, C., et al. (2018b). Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. J. 16, 1424–1433. doi: 10.1111/pbi.12884
Wang, T., Deng, Z., Zhang, X., Wang, H., Wang, Y., Liu, X., et al. (2018c). Tomato DCL2b is required for the biosynthesis of 22-nt small RNAs, the resulting secondary siRNAs, and the host defense against ToMV. Hortic. Res. 5:62. doi: 10.1038/s41438-018-0073-7
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, Z. P., Xing, H. L., Dong, L., Zhang, H. Y., Han, C. Y., Wang, X. C., et al. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16:144. doi: 10.1186/s13059-015-0715-0
Watanabe, K., Breier, U., Hensel, G., Kumlehn, J., Schubert, I., and Reiss, B. (2016). Stable gene replacement in barley by targeted double-strand breaks induction. J. Exp. Bot. 67, 1433–1445. doi: 10.1093/jxb/erv537
Watanabe, K., Kobayashi, A., Endo, M., Sage-Ono, M., Toki, S., and Ono, M. (2017). CRISPR/Cas9- mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci. Rep. 7:10028. doi: 10.1038/s41598-017-10715-1
Watanabe, K., Oda-Yamamizo, C., Sage-Ono, K., Ohmiya, A., and Ono, M. (2018). Alteration of flower colour in Ipomoeanil through CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 4. Transgenic Res. 27, 25–38. doi: 10.1007/s11248-017-0051-0
Wilson, F. M., Harrison, K., Armitage, A. D., Simkin, A. J., and Harrison, R. J. (2019). CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 15:45. doi: 10.1186/s13007-019-0428-6
Woo, J. W., Kim, J., Kwon, S., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Wu, H., and Li, Z. (2021). Recent advances in nano-enabled agriculture for improving plant performance. Crop J. 10, 1–12. doi: 10.1016/j.cj.2021.06.002
Wu, M. J., Liu, H. Q., Lin, Y., Chen, J., Fu, Y., Luo, J., et al. (2020). In-frame and frame-shift editing of the Ehd1 gene to develop Japonica rice with prolonged basic vegetative growth periods. Front. Plant Sci. 11:307. doi: 10.3389/fpls.2020.00307
Xie, K., Minkenberg, B., and Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. PNAS 112, 3570–3575. doi: 10.1073/pnas.1420294112
Xie, K., and Yang, Y. (2013). RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 6, 1975–1983. doi: 10.1093/mp/sst119
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 tool kit for multiplex genome editing in plants. BMC Plant Biol. 14:327. doi: 10.1186/s12870-014-0327-y
Xiong, X., Liu, W., Jiang, J., Xu, L., Huang, L., and Cao, J. (2019). Efficient genome editing of Brassica campestris based on the CRISPR/Cas9 system. Mol. Genet. Genom. 294, 1251–1261. doi: 10.1007/s00438-019-01564-w
Xu, C., Park, S. J., Van Eck, J., and Lippman, Z. B. (2016). Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genet. Dev. 30, 2048–2061. doi: 10.1101/gad.288415.116
Xu, J., Kang, B. C., Naing, A. H., Bae, S. J., Kim, J. S., Kim, H., et al. (2020). CRISPR/Cas9 mediated editing of 1aminocyclopropane1carboxylate oxidase1 enhances Petunia flower longevity. Plant Biotechnol. J. 18, 287–297. doi: 10.1111/pbi.13197
Xu, P., Su, H., Chen, W., and Lu, P. (2018). The application of a meiocyte-specific CRISPR/Cas9 (MSC) system and a suicide-MSC system in generating inheritable and stable mutations in Arabidopsis. Front. Plant Sci. 13:1007. doi: 10.3389/fpls.2018.01007
Xu, R., Li, H., Qin, R., Wang, L., Li, L., Wei, P., et al. (2014). Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice 7:5. doi: 10.1186/s12284-014-0005-6
Xu, X., Yuan, Y., Feng, B., and Deng, W. (2021). CRISPR/Cas9-mediated gene-editing technology in fruit quality improvement. Food Qual. Saf. 4, 159–166. doi: 10.1093/fqsafe/fyaa028
Xu, Z. S., Feng, K., and Xiong, A. S. (2019). CRISPR/Cas9-mediated multiply targeted mutagenesis in orange and purple carrot plants. Mol. Biotechnol. 61, 191–199. doi: 10.1007/s12033-018-00150-6
Yan, L., Wei, S., Wu, Y., Li, H., Yang, W., and Xie, Q. (2015). High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 12, 1820–1823. doi: 10.1016/j.molp.2015.10.004
Yan, R., Wang, Z., Ren, Y., Li, H., Liu, N., and Sun, H. (2019). Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in Lilium pumilum DC. Fisch. and Lilium longiflorum white heaven. Int. J. Mol. Sci. 20:2920. doi: 10.3390/ijms20122920
Yang, H., Bailey, P., and Pilarsky, C. (2019). CRISPR Cas9 in pancreatic cancer research. Front. Cell Dev. Biol. 7:239. doi: 10.3389/fcell.2019.00239
Yang, Y., Tang, K., Datsenka, T. U., Liu, W., Lv, S., Lang, Z., et al. (2019). Critical function of DNA methyltransferase 1 in tomato development and regulation of the DNA methylome and transcriptome. J. Integr. Plant Biol. 61, 1224–1242. doi: 10.1111/jipb.12778
Yang, X., Chen, L., He, J., and Yu, W. (2017a). Knocking out of carotenoid catabolic genes in rice fails to boost carotenoid accumulation, but reveals a mutation in strigolactone biosynthesis. Plant Cell Rep. 36, 1533–1545. doi: 10.1007/s00299-017-2172-6
Yang, Y., Zhu, G., Li, R., Yan, S., Fu, D., Zhu, B., et al. (2017b). The RNA editing factor SlORRM4 is required for normal fruit ripening in tomato. Plant Physiol. 175, 1690–1702. doi: 10.1104/pp.17.01265
Yang, H., Wu, J. J., Tang, T., Liu, K. D., and Dai, C. (2017c). CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 7:7489. doi: 10.1038/s41598-017-07871-9
Yao, L., Zhang, Y., Liu, C., Liu, Y., Wang, Y., and Liang, D. (2018). OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 4, 530–533. doi: 10.1038/s41477-018-0193-y
Yao, N., Camacho, T. A., Chukmaitov, A. S., Fleming, S. T., and Anderson, R. T. (2015). Diabetes management before and after cancer diagnosis: missed opportunity. Ann. Transl. Med. 3, 1–10. doi: 10.3978/j.issn.2305-5839.2015.03.52
Yao, R., Ming, Z., Yan, L., Li, S., Wang, F., Ma, S., et al. (2016). DWARF 14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473. doi: 10.1038/nature19073
Yau, Y. Y., and Stewart, C. N. (2013). Less is more: strategies to remove marker genes from transgenic plants. BMC Biotechnol. 13:36. doi: 10.1186/1472-6750-13-36
Ye, J., Wang, X., Hu, T., Zhang, F., Wang, B., Li, C., et al. (2017). An in Del in the promoter of Al-activated malate transporter9 selected during tomato domestication determines fruit malate contents and Aluminum tolerance. Plant Cell 29, 2249–2268. doi: 10.1105/tpc.17.00211
Yin, Y., Qin, K., Song, X., Zhang, Q., Zhou, Y., Xia, X., et al. (2018). BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physiol. 59, 2239–2254. doi: 10.1093/pcp/pcy146
Yu, Q. H., Wang, B., Li, N., Tang, Y., Yang, S., Yang, T., et al. (2017). CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 7:11874. doi: 10.1038/s41598-017-12262-1
Yuan, M., Zhu, J., Gong, L., He, L., Lee, C., Han, S., et al. (2019). Mutagenesis of FAD2 genes in peanut with CRISPR/Cas9 based gene editing. BMC Biotechnol. 19:24. doi: 10.1186/s12896-019-0516-8
Yunyan, F., Jie, Y., Fangquan, W., Fangquan, F., Wenqi, L., Jun, W., et al. (2019). Production of two elite glutinous rice varieties by editing wx gene. Rice Sci. 26, 118–124. doi: 10.1016/j.rsci.2018.04.007
Yuste-Lisbona, F. J., Fernandez-Lozano, A., Pineda, B., Bretones, S., Ortiz-Atienza, A., Garcia-Sogo, B., et al. (2020). ENO regulates tomato fruit size through the floral meristem development network. PNAS 117, 8187–8195. doi: 10.1073/pnas.1913688117
Zafar, K., Khan, M. Z., Amin, I., Mukhtar, Z., Yasmin, S., Arif, M., et al. (2020). Precise CRISPR-Cas9 mediated genome editing in super sasmati rice for resistance against bacterial blight by targeting the major susceptibility gene. Front. Plant Sci. 11:575. doi: 10.3389/fpls.2020.00575
Zeng, Z., Han, N., Liu, C., Buerte, B., Zhou, C., Chen, J., et al. (2020). Functional dissection of HGGT and HPT barley vitamin E biosynthesis via CRISPR/Cas9-enabled genome editing. Ann. Bot. 126, 929–942. doi: 10.1093/aob/mcaa115
Zhang, A., Liu, Y., Wang, F., Li, T., Chen, Z., Kong, D., et al. (2019). Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 39:47. doi: 10.1007/s11032-019-0954-y
Zhang, Z., Hua, L., Gupta, A., Tricoli, D., Edwards, K. J., Yang, B., et al. (2019). Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 17, 1623–1635. doi: 10.1111/pbi.13088
Zhang, B., Yang, X., and Yang, C. (2016). Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in petunia. Sci. Rep. 6:20315. doi: 10.1038/srep20315
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7:12617. doi: 10.1038/ncomms12617
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014). The CRISPR/C as9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. doi: 10.1111/pbi.12200
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91:714. doi: 10.1111/tpj.13599
Zhang, M., Cao, Y., Wang, Z., Wang, Z. Q., Shi, J., Liang, X., et al. (2018a). A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+exclusion and salt tolerance in maize. New Phytol. 217, 1161–1176. doi: 10.1111/nph.14882
Zhang, S., Shi, Q., Duan, Y., Hall, D., Gupta, G., and Stover, E. (2018b). “Regulation of citrus DMR6 via RNA interference and CRISPR/Cas9-mediated gene editing to improve Huanglongbing tolerance,” in Proceedings of the Biotechnology and Genetic Enginneering-Odd, Fort Pierce, FL.
Zhang, Z., Ge, X., Luo, X., Wang, P., Fan, Q., Hu, G., et al. (2018c). Simultaneous editing of two copies of Gh14-3-3d confers enhanced transgene-clean plant defense against Verticillium dahliae in allotetraploid upland cotton. Front. Plant Sci. 9:842. doi: 10.3389/fpls.2018.00842
Zhang, M., Liu, Q., Yang, X., Xu, J., Liu, G., Yao, X., et al. (2020). CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f.sp. niveum. Plant Cell Rep. 39, 589–595. doi: 10.1007/s00299-020-02516-0
Zhang, S. (2018). Knockout of SlMAPK3 reduced disease resistance to Botrytis cinerea in tomato plants. J. Agric. Food Chem. 66, 8949–8956. doi: 10.1021/acs.jafc.8b02191
Zhao, D. S., Li, Q. F., Zhang, C. Q., Zhang, C., Yang, Q. Q., Pan, L. X., et al. (2018). GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9:1240. doi: 10.1038/s41467-018-03616-y
Zhao, L., Lu, L., Wang, A., Zhang, H., and Huang, M. (2020). Nano-biotechnology in agriculture: use of nanomaterials to promote plant growth and stress tolerance. J. Agric. Food Chem. 68, 1935–1947. doi: 10.1021/acs.jafc.9b06615
Zhao, Y., Zhang, C., Liu, W., Gao, W., Liu, C., Song, G., et al. (2016). An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep. 6:23890. doi: 10.1038/srep23890
Zheng, Z., Ye, G., Zhou, Y., Pu, X., Su, W., and Wang, J. (2021). Editing sterol side chain reductase 2 gene (StSSR2) via CRISPR/Cas9 reduces the total steroidal glycoalkaloids in potato. Agriculture 14, 401–413. doi: 10.1080/26895293.2021.1925358
Zhou, H., He, M., Li, J., Chen, L., Huang, Z., Zheng, S., et al. (2016). Development of commercial thermo sensitive genic male sterile rice accelerates hybrid rice breeding usingthe CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 6:37395. doi: 10.1038/srep37395
Zhou, J., Li, D., Wang, G., Wang, F., Kunjal, M., Joldersma, D., et al. (2019). Application and future perspective of CRISPR/Cas9 genome editing in fruit crops. J. Integr. Plant Biol. 62, 269–286. doi: 10.1111/jipb.1279
Zhou, J., Wang, G., and Liu, Z. (2018). Efficient genome editing of wild strawberry genes, vector development and validation. Plant Biotechnol. J. 16, 1868–1877. doi: 10.1111/pbi.12922
Zhou, X., Liao, H., Chern, M., Yin, J., Chen, Y., Wang, J., et al. (2018). Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. U.S.A. 115, 3174–3179. doi: 10.1073/pnas.1705927115
Zhou, X., Jacobs, T. B., Xue, L. J., Harding, S. A., and Tsai, C. J. (2015). Exploiting SNPs for biallelic CRISPR mutations in the out crossing woody perennial Populus reveals 4-coumarate CoA ligase specificity and redundancy. New Phytol. 208, 298–301. doi: 10.1111/nph.13470
Zhou, X., Zha, M., Li, L., Imran, M., and Zhang, C. (2017). StMYB44 negatively regulates phosphate transport by suppressing expression of PHOSPHATE1 in potato. J. Exp. Bot. 68, 1265–1281. doi: 10.1093/jxb/erx026
Zhu, C., Zheng, X., Huang, Y., Ye, J., Chen, P., Zhang, C., et al. (2019). Genome sequencing and CRISPR/Cas9 gene editing of an early flowering Mini-Citrus (Fortunella hindsii). Plant Biotechnol. J. 17, 2199–2210. doi: 10.1111/pbi.13132
Zhu, J., Song, N., Sun, S., Yang, W., Zhao, H., Song, W., et al. (2016). Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. JGG 43, 25–36. doi: 10.1016/j.jgg.2015.10.006
Zsogon, A., Čermák, T., and Naves, E. R. (2018). De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216. doi: 10.1038/nbt.4272
Zuo, Y., Feng, F., Qi, W., and Song, R. (2019). Dek42 encodes an RNA-binding protein that affects alternative Pre-MRNA splicing and maize kernel development. J. Integr. Plant Biol. 61, 728–748. doi: 10.1111/jipb.12798
Keywords: Cas9, biotic and abiotic stress, horticultural crops, nutritional value, nanoparticles, nano-fertilizers, Cas9 activators
Citation: Naik BJ, Shimoga G, Kim S-C, Manjulatha M, Subramanyam Reddy C, Palem RR, Kumar M, Kim S-Y and Lee S-H (2022) CRISPR/Cas9 and Nanotechnology Pertinence in Agricultural Crop Refinement. Front. Plant Sci. 13:843575. doi: 10.3389/fpls.2022.843575
Received: 26 December 2021; Accepted: 07 February 2022;
Published: 08 April 2022.
Edited by:
Ahmad M. Alqudah, Aarhus University, DenmarkReviewed by:
Asad Riaz, Zhejiang University, ChinaHikmet Budak, Montana Bioagriculture, Inc., United States
Copyright © 2022 Naik, Shimoga, Kim, Manjulatha, Subramanyam Reddy, Palem, Kumar, Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Youn Kim, c3lraW1Aa29yZWF0ZWNoLmFjLmty; Soo-Hong Lee, c29vaG9uZ0Bkb25nZ3VrLmVkdQ==
 Banavath Jayanna Naik
Banavath Jayanna Naik Ganesh Shimoga
Ganesh Shimoga Seong-Cheol Kim
Seong-Cheol Kim Mekapogu Manjulatha
Mekapogu Manjulatha Chinreddy Subramanyam Reddy
Chinreddy Subramanyam Reddy Ramasubba Reddy Palem5
Ramasubba Reddy Palem5 Manu Kumar
Manu Kumar Sang-Youn Kim
Sang-Youn Kim Soo-Hong Lee
Soo-Hong Lee