- 1School of Life Sciences, University of Bristol, Bristol, United Kingdom
- 2Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Liverpool, United Kingdom
The bread wheat (Triticum aestivum) pangenome is a patchwork of variable regions, including translocations and introgressions from progenitors and wild relatives. Although a large number of these have been documented, it is likely that many more remain unknown. To map these variable regions and make them more traceable in breeding programs, wheat accessions need to be genotyped or sequenced. The wheat genome is large and complex and consequently, sequencing efforts are often targeted through exome capture. In this study, we employed exome capture prior to sequencing 12 wheat varieties; 10 elite T. aestivum cultivars and two T. aestivum landrace accessions. Sequence coverage across chromosomes was greater toward distal regions of chromosome arms and lower in centromeric regions, reflecting the capture probe distribution which itself is determined by the known telomere to centromere gene gradient. Superimposed on this general pattern, numerous drops in sequence coverage were observed. Several of these corresponded with reported introgressions. Other drops in coverage could not be readily explained and may point to introgressions that have not, to date, been documented.
Introduction
The bread wheat (Triticum aestivum) pangenome is a patchwork containing translocations and introgressions from wheat’s wild relatives (Przewieslik-Allen et al., 2021) as well as numerous deletions. Some of these features may be present in only a handful of accessions coming from a limited geographic area whilst others may be prevalent and present in varying combinations across many accessions. Some variable regions may have occurred naturally by mutation or as a consequence of promiscuous pollination events between wheat and one of its primary relatives (He et al., 2019). Others are the result of breeding efforts (Schneider et al., 2008) using traditional methods to introduce segments from progenitors and close relatives or, more recently, using more advanced methods to perform wide crosses (Cseh et al., 2019; Devi et al., 2019; King et al., 2019; Xu et al., 2020). Regardless of their origin, the number of these variable regions that have been documented is probably not a genuine reflection of their true number; breeding companies may not have reported, and indeed may not know, all the introgressed regions in their elite lines, and chance events in landrace accessions are unlikely to have been documented at all. It would seem highly likely, therefore, that there are numerous unknown, introgressions present in modern wheat accessions (Przewieslik-Allen et al., 2021).
With this in mind, and with modern techniques allowing for wide crossing with increasing success, increasingly diverse wheat accessions are becoming available for pre-breeding (Hao et al., 2020). To be of use to research and breeding programs, such material needs to be tracked using either targeted molecular markers (Singh et al., 2018; Rasheed and Xia, 2019) or sequencing. The former has most frequently been used because it offers low cost and high throughput (Zhang J. et al., 2017; Zhang W. et al., 2017; Przewieslik-Allen et al., 2019). However, marker probes will only hybridize to, and so provide a signal for, the sequences for which they were designed. Thus, wheat genotyping markers intended for introgression detection need to be designed using sequences from a combination of wheat and the progenitors and relatives thought to have been the source of those introgressions (Wang et al., 2014; Zhang J. et al., 2017; Przewieslik-Allen et al., 2019). Where the source of introgressed material is unknown, and so not included in probe design, genotyping is unlikely to track such regions.
Sequencing, having no requirement for prior knowledge of the target, does not suffer from such a problem. However, the size and complexity of the wheat genome create problems in this regard. T. aestivum has a large (∼17 Gb) polyploid and highly repetitive genome of which the exome constitutes less than 5% (International Wheat Genome Sequencing Consortium [IWGSC], 2014). To sidestep these issues, targeted sequencing approaches, such as exome capture, are used (Kaur and Gaikwad, 2017). In wheat, several exome capture systems that incorporate capture probe sets derived from both hexaploid wheat and its relatives have been proposed (Winfield et al., 2012; Gardiner et al., 2019; He et al., 2019). The capture probes themselves can tolerate some degree of mismatch thus allowing the capture of sequences outside the immediate confines of the species from which they are derived. The Roche SeqCap EZ system can tolerate up to 10% (Roche pers com) and the Arbor Biosciences myBaits system can tolerate up to 20% divergence from the target sequence (Arbor Biosciences, 2021). This property is highly beneficial where the exact source of the material is unknown and has been exploited to capture sequences from diverse origins in the wild relative species of cotton (Salmon et al., 2012), cows (Cosart et al., 2011), and humans (Jin et al., 2012) as well as in wheat (Saintenac et al., 2011; Henry et al., 2014; He et al., 2019).
We recently described the variable sequence coverage of the wheat variety ‘Player’ when exome capture data were aligned to the ‘Chinese Spring’ reference sequence (Przewieslik-Allen et al., 2021). Distinct drops in sequence coverage were evident in chromosomes 2A and 2B which correlated with introgressions from Aegilops ventricosa and Triticum timopheevii, respectively. As the use of exome capture prior to sequencing followed by alignment to a standard reference is common practice, the potential for this to be disrupted by introgressions is a concern, especially as many interesting, rare, and novel alleles may be located in regions derived from wild relatives. This was investigated using 10 elite T. aestivum cultivars and 2 T. aestivum landrace accessions used in breeding.
Results
Sequence Coverage
Using gene and promoter sequence capture (Gardiner et al., 2019), 12 T. aestivum accessions (10 elite varieties and 2 landrace accessions) were sequenced and total coverage compared. Total reads were between 48,255,718 and 145,897,760 per accession; after quality trimming and alignment to the IWGSC RefSeq v1.0 ‘Chinese Spring’ reference (International Wheat Genome Sequencing Consortium [IWGSC], 2018), there were between 20,973,857 and 63,662,179 uniquely mapped, paired reads per accession (Table 1).
Table 1. Read statistics before and after trimming with alignment statistics for total mapped and uniquely mapped reads.
Sequence coverage across chromosomes displayed a characteristic pattern; that is, there was a greater depth of coverage toward the ends of chromosome arms and lower coverage across centromeres (Figure 1A). However, this overall pattern was, in places, interrupted by regions of pronounced reduction in sequence coverage. These regions were not seen on all chromosomes or simultaneously in all accessions (Supplementary File 1). The most pronounced of these reductions in coverage was observed in ‘Bacanora’, ‘Bobwhite’, and ‘KWS Kielder’ and extended across the whole of the short arm of chromosome 1B (c. 240 Mb; Figure 1B) in line with the well documented and prevalent 1RS/1BL Secale cereale translocation (Rabinovich, 1998). There was no reduction in capture probe density across 1BS (Figure 1C) and the nine accessions without the 1RS translocation do not show a reduction in read coverage across this chromosome arm (Supplementary File 1: 1B).
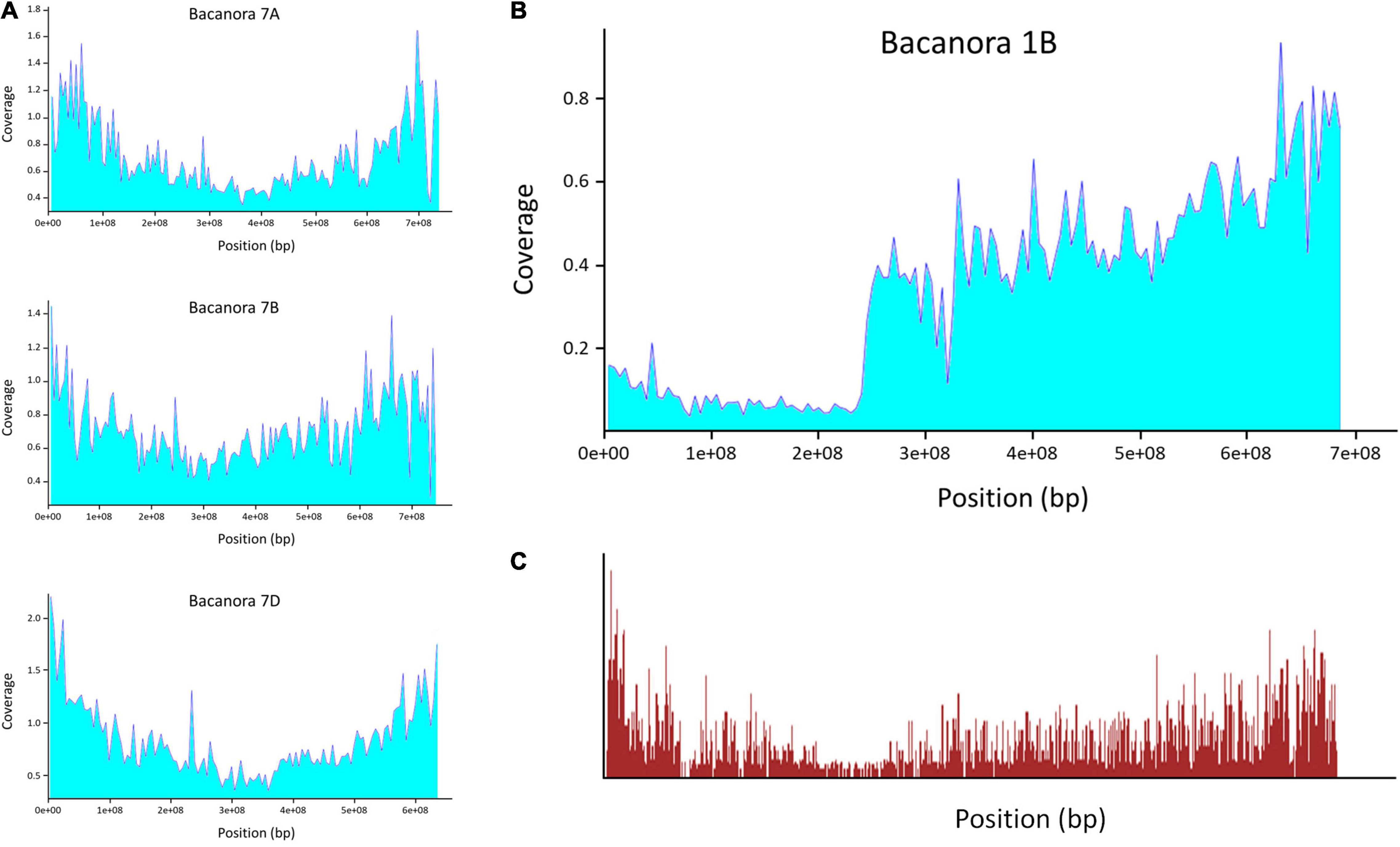
Figure 1. Read coverage for the accession ‘Bacanora’ after alignment to IWGSC ‘Chinese Spring’ assembly version 1.0. (A) Read coverage across chromosomes tended to be higher toward the telomeres and lower across the centromere. (B) Chromosome 1B shows a clear drop in coverage across the short arm (NB in all plots, chromosome short arms are on the left). (C) Location and density of capture probes across chromosome 1B (data from Gardiner et al., 2019).
Other large drops in coverage were seen on 2BL, 2DL, and 5BL (Figure 2) which extended over approximately 85, 45, and 40 Mb, respectively. Additional, smaller drops in coverage were also observed in telomeric regions, such as 2AS (Figure 3), 7DL, and an additional region in 2DL (Supplementary File 1).
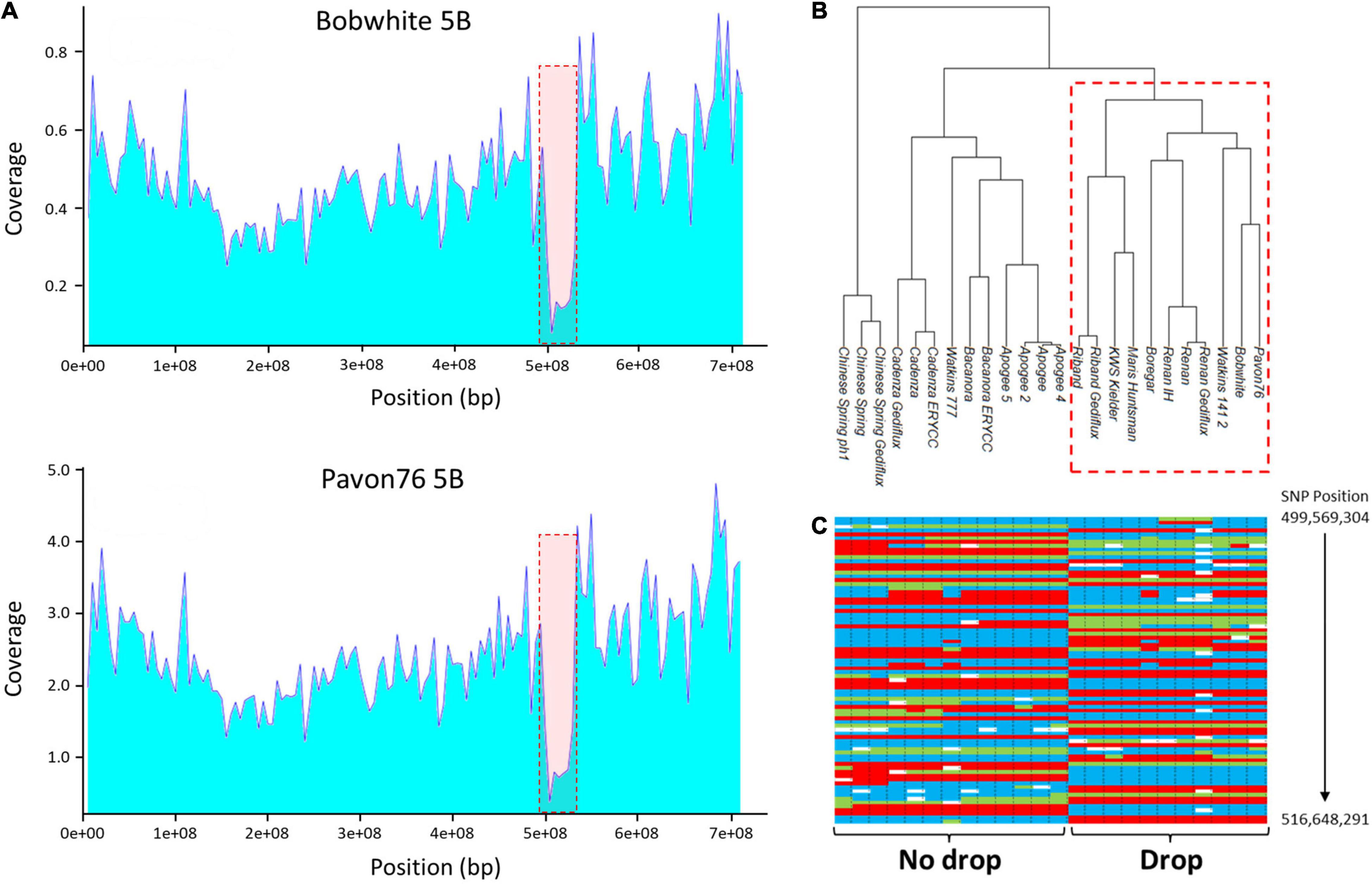
Figure 2. (A) Average depth of coverage for chromosome 5B in the accessions ‘Bobwhite’ and ‘Pavon 76’; both show a drop in read coverage on the long arm at approximately position 490,000,000–540,000,000. (B) Dendrogram based on the 1,749 Axiom markers mapped to chromosome 5B; the 8 varieties (‘Bobwhite’, ‘Boregar’, ‘KWS Kielder’, ‘Maris Huntsman’, ‘Pavon 76’, ‘Renan’, ‘Riband’, and ‘Watkins 141’) with the drop in read coverage cluster. (C) A sample of the SNP calls across the interval 499,569, 304–534,345,241 highlighting the difference between the two groups (blue and red are the alternative homozygote calls; green indicates heterozygote calls).
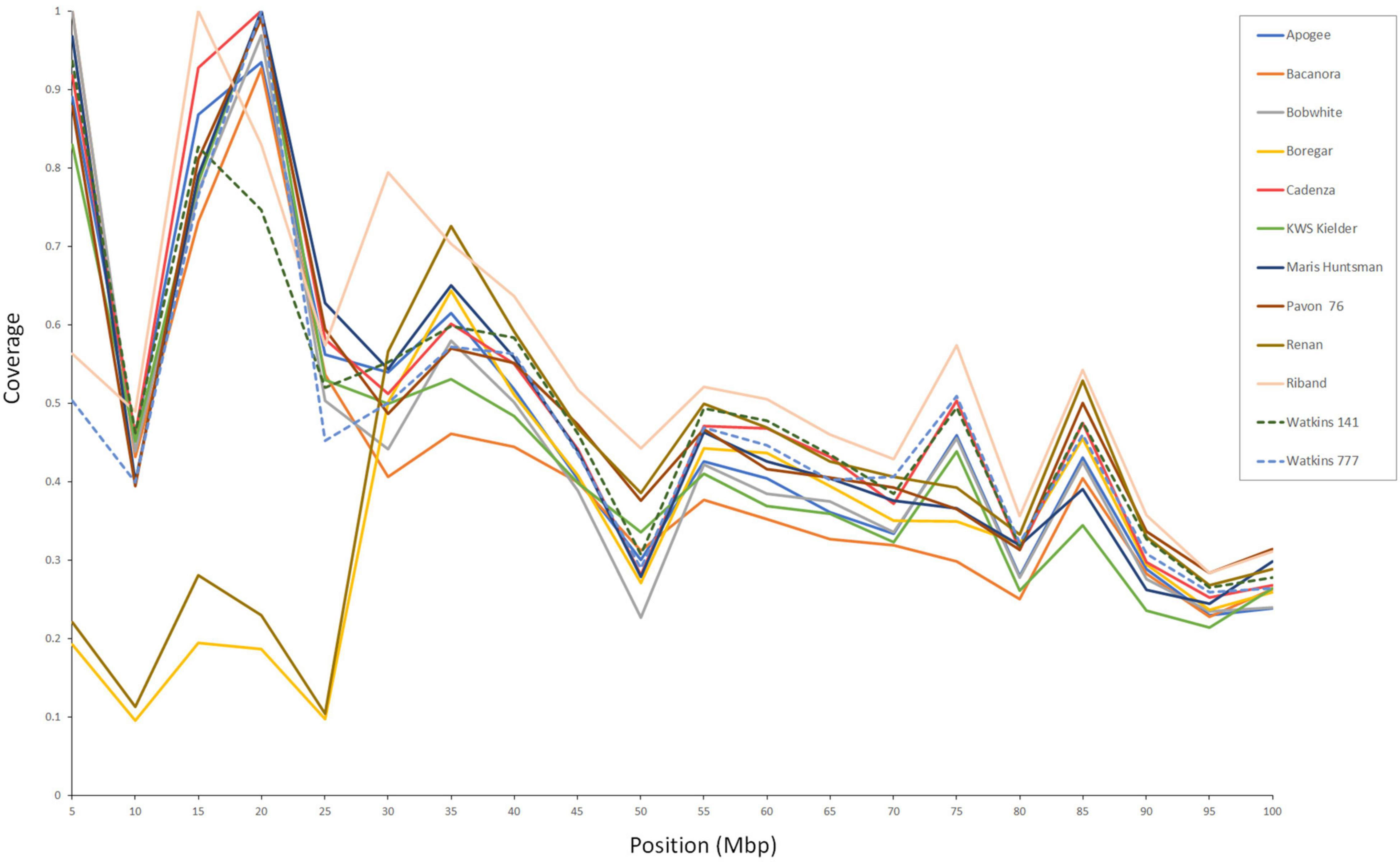
Figure 3. Sequence coverage across the first 100 Mb of chromosome 2AS. The two accessions, ‘Boregar’ and ‘Renan’, show reduced coverage across the first 25–30 Mb which corresponds with the size of the known introgression from Ae. ventricosa (Robert et al., 1999).
Cluster Analysis of Accessions
To determine whether there was any relationship between the lines that shared read coverage profiles, cluster analysis was performed with Axiom 35K Wheat Breeders’ Array genotyping data (Allen et al., 2016). Analysis was performed on markers specific to the chromosomes 2B (2,083 markers), 2D (2,237 markers), and 5B (1,749 markers). Accessions showed a pattern of clustering that corresponded with the drops in coverage (Figure 2B and Supplementary File 2). For chromosome 5B, for example, the 12 accessions separated into two main clusters; the accessions thought to contain the deletion fell into one cluster while those with even sequence coverage fell into the other. The separation into two clusters was driven by the markers spanning the drop. Across the interval corresponding to the decline in read coverage on chromosome 5B (position 499,569,304–534,345,241), there were 141 single nucleotide polymorphism (SNP) markers; for these markers, the mean percentage similarity between the genotype calls for ‘Chinese Spring’ and those of the eight accessions displaying the drop in coverage was only 13.3%. This compares to a mean similarity of 59.1% for the SNP calls across the rest of the chromosome (Figure 2C).
Bibliographic Search for Introgressions
A number of wheat introgressions reported in the literature were assembled (Table 2) to determine whether there was any relationship between them and the patterns of reduced sequence coverage observed in this study. The large drop in coverage on 1BS, for example, is present in those varieties (Bacanora, Bobwhite, and KWS Kielder) known to possess a whole arm translocation from S. cereale; we have previously reported this ourselves based on genotyping results using the Axiom High-Density Array (Winfield et al., 2015). Other chromosomal regions with reduced read coverage were also related to regions of known introgressions. However, not all the reports of introgressions that we found in the literature had a corresponding drop in sequence coverage, and in some cases, there was a drop in sequence coverage for which no source was found. Notable deletions, such as that on 1DL of ‘Cadenza’, highlight the similarity between deletions and introgressions in sequence coverage.
Efficacy of Sequence Capture
The accessions containing the 1RS.1BL translocation (‘Bacanora’, ‘Bobwhite’, and ‘KWS Kielder’) displayed a clear drop in read coverage across the short arm of 1B; we hypothesized that this was due to capture efficacy in the different backgrounds. The potential efficacy of probes to capture sequences from either ‘Chinese Spring’ or S. cereale was assessed by BLASTing their sequences to their respective assemblies. Capture probe sequences for chromosome 1BS (26,985 sequences) were BLASTed against the 1B pseudomolecule of ‘Chinese Spring’ and 1R of S. cereale. This resulted in 29,652 hits to ‘Chinese Spring’ 1BS and 12,120 hits to S. cereale 1RS. To both assemblies, some probes had multiple hits. The number of probe sequences that had a hit was 26,222 and 8,419, respectively. Those with a single hit were 23,969 and 5,822, respectively (Figure 4A), and the percentage similarity between probe sequences and their target was 99.8 and 95.6%, respectively (Figure 4B). That is, a greater number of probes matched the ‘Chinese Spring’ sequence and with greater percentage similarity.
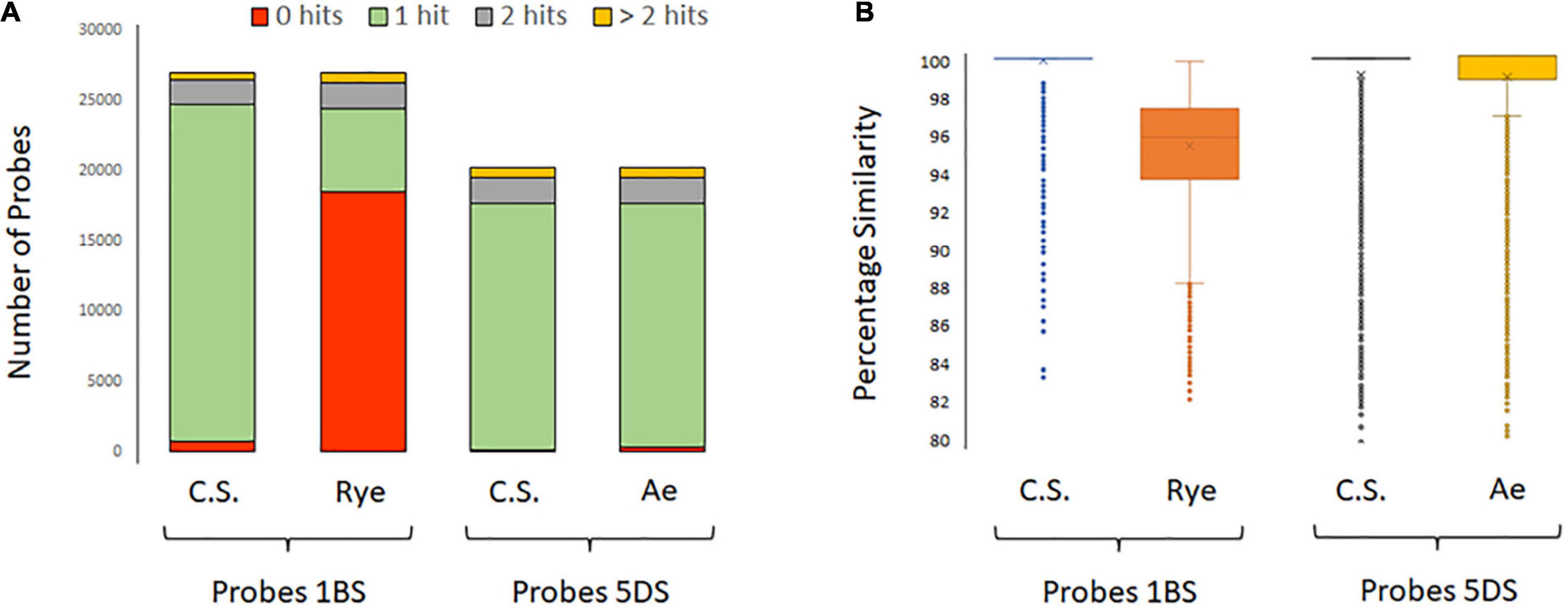
Figure 4. (A) Bar graphs showing the number of capture probes that had BLAST hits to ‘Chinese Spring’ chromosome 1BS (IWGSC v1), S. cereale chromosome 1RS (JADQCU000000000 v1 of the cultivar Weining), ‘Chinese Spring’ chromosome 5DS (IWGSC v1), and Ae. tauschii chromosome 5DS (PRJNA341983 assembly of Ae. tauschii subsp. strangulata). The number of probe sequences for chromosomes 1BS and 5DS was 26,985 and 20,253, respectively. The number of probes that produced a hit was 26,222 to ‘Chinese Spring’ 1BS, 8,419 to S. cereale 1RS, 20,082 to ‘Chinese Spring’ 5DS, and 19,872 to Ae. tauschii 5DS. There were more hits than probe sequences as some probes had multiple hits. (B) Box and whisker plots showing the percentage similarity between the probe sequences and their respective targets.
In contrast, the known Ae. tauschii introgression into 5DS of the variety ‘Maris Huntsman’ (Wang et al., 2005) was not evidenced by a drop in read coverage. The probe sequences for chromosome 5DS (20,253 sequences) were BLASTed against the assemblies of both ‘Chinese Spring’ and Ae. tauschii 5DS resulted in 24,300 hits to the former and 24,173 hits to the latter. The number of probe sequences that had a hit was 20,082 and 19,872, respectively. Those with a single hit were 17,550 and 17,358, respectively (Figure 4A). The percentage similarity between probe sequences and their target was 99.1 and 98.9%, respectively (Figure 4B). Thus, it would appear, the sequences of wheat and Ae. tauschii are sufficiently similar over this region that capture probes are equally efficient at capturing sequences from them. To confirm this hypothesis, the sequences surrounding Pm2, were compared. Based on the alignment, the ‘Chinese Spring’ and Ae. tauschii reference assemblies were highly similar across the 2 Mb of sequence centered on the Pm2 gene (99.1% similarity); in each, there were 21 annotated genes and synteny appears to be maintained apart from the presence of an inverted repeat of TraesCS5D02G044500 (position 43,382,967–43,386,355) to the upstream position 42,989015–42,992,511 – TraesCS5D02G043600 (Supplementary Table 1). The sequences from ‘Maris Huntsman’ also aligned well to both assemblies. However, within the coding sequence of the Pm2 gene itself, two indels, one particularly relevant, supported the hypothesis that ‘Maris Huntsman’ is more similar to Ae. tauschii than to ‘Chinese Spring’. That is, relative to ‘Chinese Spring’, both Ae. tauschii and ‘Maris Huntsman’ carry a 3 bp insertion at position 43,405,954 and a 7 bp insertion at position 43,407,045 (Figure 51).
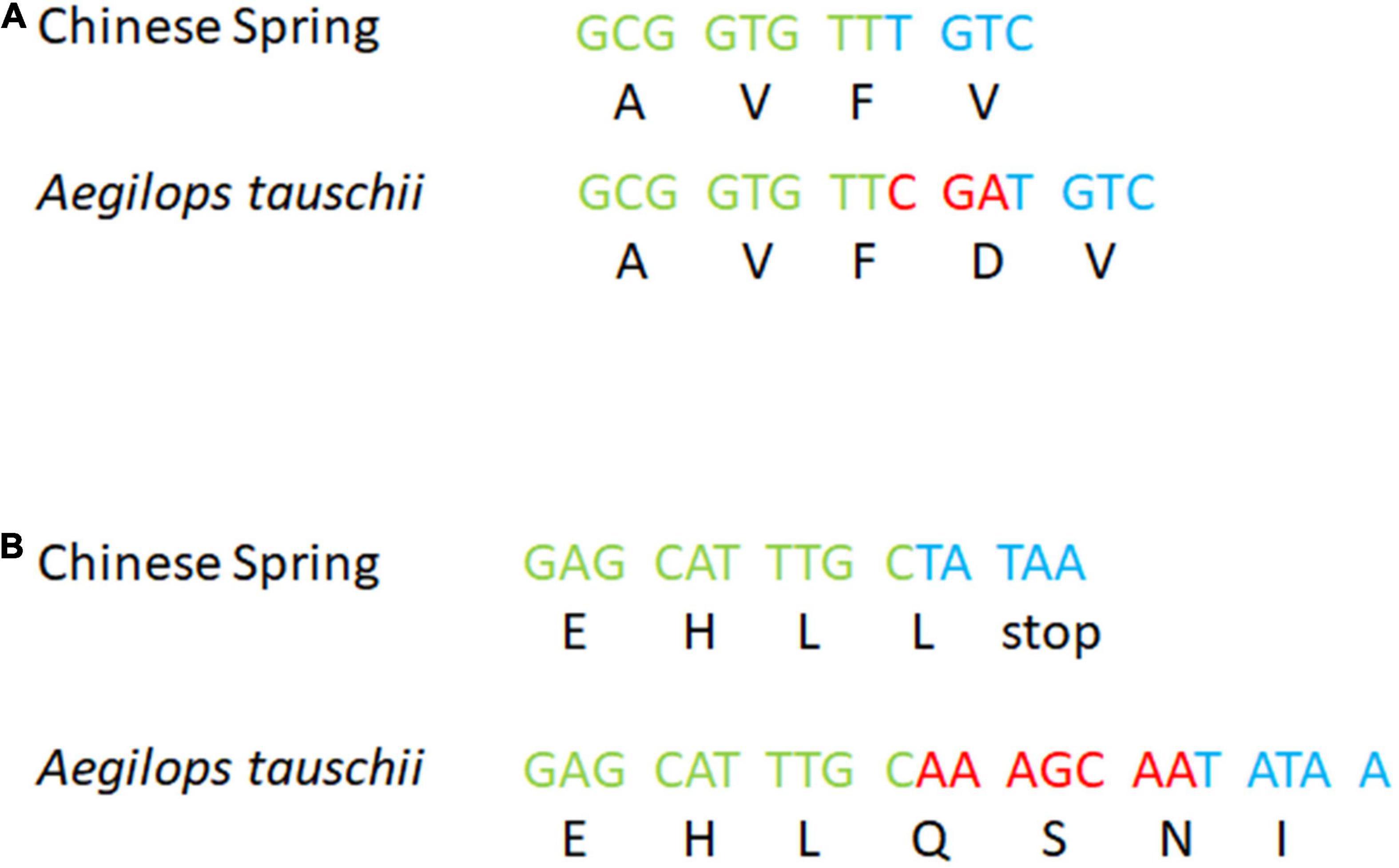
Figure 5. Details of the Pm2 gene in ‘Chinese Spring’ and Ae. tauschii: (A) a 3 bp insertion and (B) a 7 bp insertion. Respectively, green and blue bases are ‘Chinese Spring’ reference sequences before and after the indel. Red bases are the insertion (found in both Ae. tauschii and ‘Maris Huntsman’).
Efficacy of Alignment to the Reference Assembly
To further investigate the role of sequence alignment in the regions of reduced sequence coverage, a BLAST search was performed using the mapped and unmapped reads from ‘Bacanora’ against a database containing both T. aestivum and S. cereale sequences. Of the 1,959 unmapped reads, 709 (36.2%) hit sequences in the BLAST database: 654 (33.4%) to the S. cereale 1R sequence and 55 (2.8%) to the T. aestivum 1B sequence. Conversely, for the 1,421 reads that had successfully mapped to the T. aestivum ‘Chinese Spring’ reference sequence, there were only 167 (11.8%) hits to the S. cereale 1R sequence while 1,242 (87.4%) hits to the wheat 1B reference sequence.
For unknown introgressions, it is not possible to compare the unmapped reads to the source sequence. To better understand from where these reads came, an assembly of unmapped reads for all 12 accessions was created and then compared with a database of Poaceae/S. cereale protein sequences (Figure 6). The unmapped sequences were predominantly (62.1%) found in the progenitor accessions Triticum turgidum (AABB genome), Ae. tauschii (DD), and Triticum urartu (AA). There were also additional hits to the more distant relatives Hordeum vulgare (HH) and S. cereale (RR).
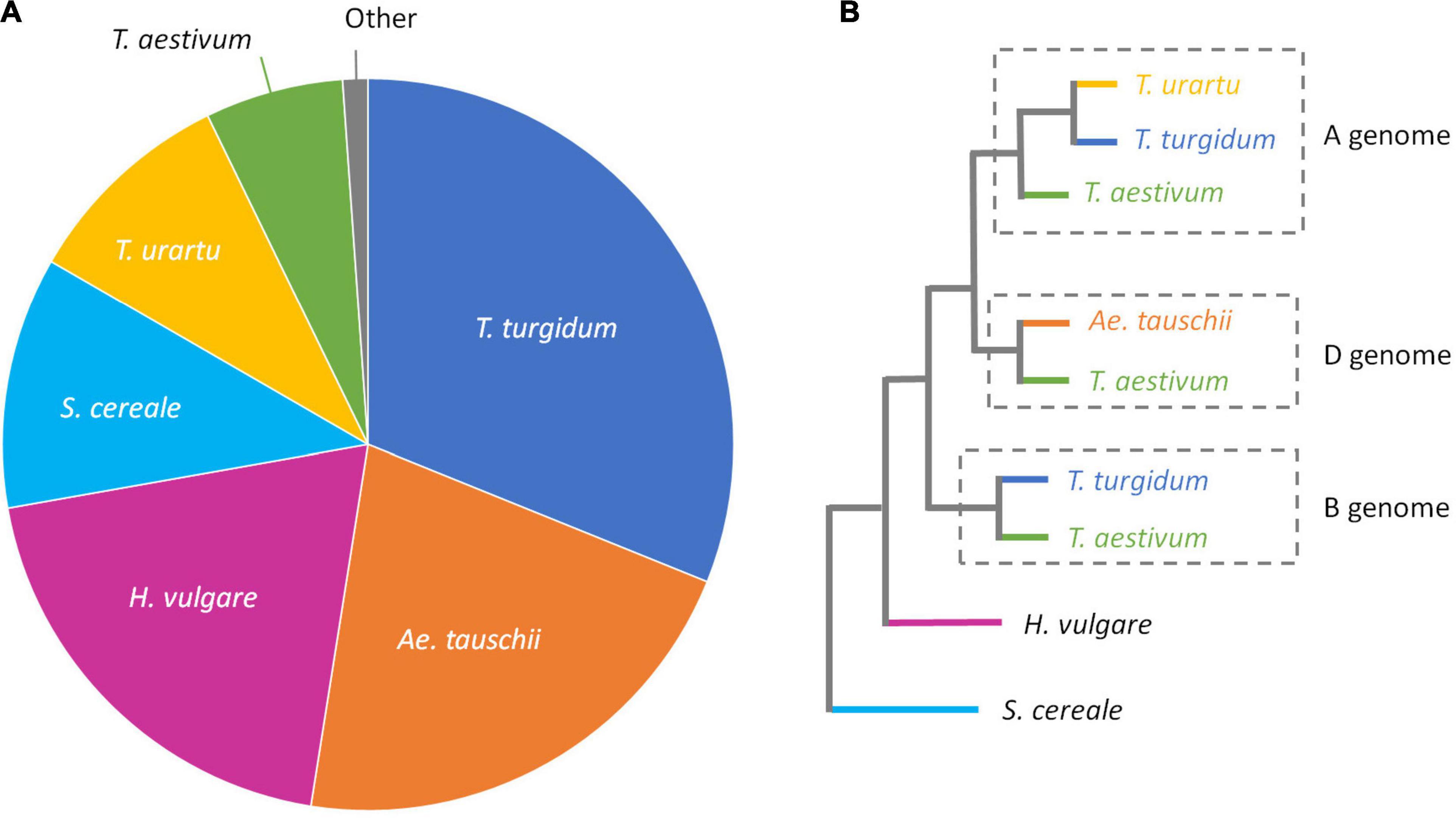
Figure 6. (A) Pie chart showing the best BLAST hits against a combined Poaceae/S. cereale database for captured reads that didn’t map to the IWGSC ‘Chinese Spring’ assembly v1. (B) Phylogenetic tree (redrawn from Zhou et al., 2017), showing the relationship of the species used in our Poaceae/S. cereale database.
Discussion
Exome Capture
The ‘Gene Capture v1’ and ‘Promoter Capture v1’ probes are based on sequences not only from T. aestivum but also Ae. tauschii and T. turgidum and, thus, should capture sequence from bread wheat and its progenitors (Gardiner et al., 2019). In this study, the exome capture protocol proved effective at capturing a representative genome sample from each of the 12 accessions examined with sequence coverage in distal regions of chromosomes being greater than that across centromeres (Figure 1A); this pattern reflects capture probe distribution which itself is determined by the known telomere to centromere gene gradient (Pingault et al., 2015; Gardiner et al., 2019). Probes appear to have successfully captured wheat sequence and that of introgressions from progenitors as demonstrated by the capture of sequence from the 5DS, Ae. tauschii introgression in ‘Maris Huntsman’ (Supplementary Table 1). A review of the literature reporting primary genepool introgression into bread wheat, further indicated that probes were effectively capturing sequence from these introgressions and, thus, resulting in even sequence coverage across such introgressions and the host sequences flanking them. For example, an introgression from T. turgidum subsp. carthlicum has been reported on 2AL of ‘Renan’ and ‘Riband’ (Chantret et al., 1999; United Kingdom Cereal Pathogen Virulence Survey [UKCVS], 2004); we saw no decrease in sequence coverage for either accession indicating successful capture and alignment. Importantly, this was not just the case for the primary relatives (T. turgidum and Ae. tauschii) that had been included in the design of the capture probes. An introgression from the primary relative, Triticum monococcum, has been reported to be present in 5AL of ‘Maris Huntsman’ (Chen et al., 2021; Supplementary File 1), a Triticum spelta introgression has been reported in 2BL of ‘Cadenza’ (Marchal et al., 2018; Supplementary File 1) and introgression from Triticum dicoccum has been reported in 3BS of ‘Pavon 76’ (Mago et al., 2014; Supplementary File 1) and none had a corresponding decrease in coverage suggesting adequate capture of these sequences.
The region associated with the Pm2 gene in ‘Maris Huntsman’ was used as a case study to confirm that sequence diversity present in regions with successful capture and alignment were, indeed, from a wild relative source. Alignment of the captured sequences from ‘Maris Huntsman’ to both the ‘Chinese Spring’ T. aestivum reference (IWGSC v1.0) and Ae. tauschii (Ae. tauschii v4.0 GCF_002575655.1) assemblies showed them to be highly similar. However, two small insertions, with respect to ‘Chinese Spring’, in ‘Maris Huntsman’ and Ae. tauschii give support to the hypothesis that ‘Maris Huntsman’ harbors an Ae. tauschii introgression (Figure 5). The successful capture of this region is hardly surprising considering that Ae. tauschii sequence was used to guide capture probe design (Gardiner et al., 2019) and given the high degree of similarity between the two species, T. aestivum and Ae. tauschii, across the Pm2 region. Indeed, capture probes designed exclusively from bread wheat sequence may well have proved equally efficacious at capturing sequence from this introgressed region.
The design of the probes, then, has allowed the capture of sequences beyond those belonging exclusively to T. aestivum. However, one must expect that beyond a certain level of sequence diversity, a reflection of the evolutionary distance of donors of introgressed segments, probes will no longer capture sequence. Such wide introgressions will not be captured, and coverage of the target will drop. This is a serious limitation if novel regions from more distant relatives are the aim of the capture sequencing and other sequencing methods will need to be employed.
Alignment to the Reference Assembly
In addition to successful capture and sequencing, one must be able to realign the sequence to the reference (in this case ‘Chinese Spring’ IWGSC v1) for it to be identified as present. There is the potential for the mapping parameters to under-utilize the available sequence as the stringency of the parameters used to align the captured sequences to the ‘Chinese Spring’ reference genome result in some successfully captured sequences being unable to align. Not all variation present in sequencing data is a true reflection of the sequence present and as the alignment stringency is relaxed, sequencing errors may enter the data. To preserve the high-quality sequences, it seems inevitable that diverse sequences will be lost by data processing.
Some mapping protocols, such as the mapping of non-unique hits, can allow for homoeologous sequences to mask gaps in coverage due to deletions or introgressions. In addition, as the mapping of zero in read coverage is not a standard protocol, the gaps seen as a result of diverse sequences are not made apparent (Supplementary Figure 1) and the inability to align diverse sequences to the reference is not reported.
Efficacy of Alignment to the Reference Assembly
For all 12 accessions, the captured sequences that could not be mapped to the reference were BLASTed against a Poaceae/S. cereale protein database (Figure 6). Of the sequences that had a hit to the protein database, 62.1% had a match to a sequence derived from a progenitor species (Figure 6). This indicates that some sequences were captured and sequenced but had no corresponding sequence in the ‘Chinese Spring’ reference. Given an alternative reference, some of these sequences may have aligned. The failure of almost 40% of the captured sequences that did not map to the reference probably reflects the limitations of the created Poaceae/S. cereale protein database since we recognize that there is limited sequence data available for many wheat relatives; the major crop species T. aestivum, T. turgidum, and H. vulgare are well represented in nucleotide databases, but this is not the case for wild relatives. Indeed, we chose to compare our un-mapped sequences to a protein database, rather than a nucleotide database, to maximize the amount of sequence data available. The Poaceae/S. cereale protein database contained 472,031 sequences. Through this approach, we were able to identify sequences potentially originating from secondary and tertiary genepool species. However, some sequences remained completely unidentified emphasizing that, probably, some diversity is regularly omitted from standard sequencing and alignment. As such, exome capture followed by alignment to a hexaploid reference is not a reliable tool for the identification of introgressions within hexaploid wheat. Where exome capture has been performed and an introgression is suspected, identification is limited by the current availability of wheat relative sequences.
Diverse sequences, such as the Ae. tauschii introgression, described in ‘Maris Huntsman’ were successfully captured, sequenced, and aligned in part due to the presence of Ae. tauschii sequences in the capture probe set and in part due to the similarity of the progenitor sequence to the D genome of the reference assembly. For the more distant wild relatives, both capture and alignment were less successful. The reduction in mapped sequences was most pronounced in the accessions containing the 1RS.1BL translocation (Figure 1). This is a known introgression that is from a tertiary source. When the 1BS capture probe sequences (26,985) were BLASTed against 1RS of the rye genome assembly (JADQCU000000000 v1), 31% had a hit (Figure 4A), suggesting that some capture would occur, but the percentage similarity between probe sequence and its target was lower in rye than in wheat, suggesting that it might not map back to the reference. This in silico assessment was reflected in the captured but un-mapped sequences. By performing a BLAST search against a T. aestivum and S. cereale database, a number of the unmapped reads in the 1RS containing accession ‘Bacanora’ were found to have matches to the S. cereale sequences (33.4%), considerably higher than the S. cereale sequences found within the mapped reads of the same accession (11.8%). This suggests that some of the unmapped reads were from regions of 1RS.1BL that were successfully captured but could not be successfully mapped back to the reference. As S. cereale sequences are poorly represented in the BLAST database (there were 25,214 out of 472,031 in total), the full extent of S. cereale sequences captured is not known and the ratio present may be higher. While it seems that this tertiary relative introgression was not captured to the same extent as a primary genepool relative, it is important to note that some sequences were captured despite the dissimilarity between S. cereale and the T. aestivum target but the presence of the sequenced further limited by alignment to the reference.
Each of the 12 accessions used in this study, showed reduced read coverage across some regions of at least one of its chromosomes. Most of these drops in coverage were common to several of the accessions studied and, in many cases, they co-located with documented introgressions or with regions where genotyping data had highlighted extensive variability. The accessions ‘Renan’ and ‘Boregar’ had reduced coverage at the end of the short arm of chromosome 2A corresponding to the known introgression from Ae. ventricosa associated with rust resistance (Lr37, Hanzalová et al., 2007; Yr17 Dedryver et al., 2009). The size of this introgression has been reported to be c. 33 Mb (Gao et al., 2021) which corresponds with the size of the decline in coverage observed in this study. The eyespot resistance gene, Pch1 located on the distal end of 7DL, also introduced from Ae. ventricosa (Leonard et al., 2007) corresponded to the terminal drop in coverage seen in ‘Boregar’ and ‘Renan’, both reported containing the Pch1 gene (Burt and Nicholson, 2011). The powdery mildew resistance gene Pm6 from T. timopheevii on 2BL was reported in both ‘Riband’ and ‘Maris Huntsman’ (United Kingdom Cereal Pathogen Virulence Survey [UKCVS], 1996; Wang et al., 2005) and reveals itself as a distinct decrease in coverage in both accessions. Interestingly, this dip is also found in ‘Boregar’ which hasn’t been reported to carry the 2BL introgression but, on the basis of evidence here, probably does. The presence of unreported introgressions is thought to be quite common. For example, several accessions (‘Bacanora’, ‘Boregar’, ‘Cadenza’, ‘KWS Kielder’, ‘Maris Huntsman’, ‘Renan’, and ‘Riband’) shared a large region (c. 45 Mb) with reduced read coverage, which we assume might indicate an introgression, but for which we could find no documentary evidence. This region spans over 640 genes with a range of functions, such as ion channel regulation, phosphorylation, and electron transfer (Supplementary File 4).
Here we demonstrate that there is a relationship between drops in sequence coverage and sequence similarity of the introgression sequence to the region it replaced. That is, introgressions from primary relatives, such as Ae. tauschii or T. dicoccum (Table 2), are unlikely to fail capture and thus be sequenced and aligned. On the other hand, introgressions from secondary and tertiary genepool species, such as S. cereale, Ae. ventricosa, and T. timopheevii, are likely to avoid capture (Figure 4) and, if captured, fail to align to the reference (Figure 6); such failures are characterized by reduced sequence coverage across the introgressions. The degree of sequence similarity between a wheat relative sequence and the T. aestivum equivalent reflects the evolutionary distance. The observations of this study agree with the study in which human exome capture probes were used to capture exome sequences in non-human primates; “specificity of the capture decreased as evolutionary divergence from humans increase” (Jin et al., 2012). Exome capture probes designed for T. aestivum efficiently captured genic sequences from the D genome progenitor species, Ae. tauschii, but performed much less well against S. cereale, an evolutionary more distant species belonging to the tertiary genome.
Modern elite wheat varieties carry numerous introgressions which provide genes of important agronomic traits (Table 2), but exome capture may limit the ability to sequence these novel and interesting regions. Introgressions from the primary genepool were successfully captured. Those from more distantly related species, members of the secondary and tertiary genepool, however, were poorly represented in the mapped sequences data (Table 2). While there was evidence that some sequences from secondary and tertiary genepool relatives were present amongst the captured sequences (Figure 6) their number was small and did not map to the reference. Localized reduction in sequence coverage was observed in all 12 accessions studied, including the landrace accessions. Many of these regions of low coverage were collocated with documented introgressions or deletions, while others remain unknown. The method of sequencing used here has essentially limited the diversity of sequence that could be reported. The careful design of capture probes is critically important as lack of capture probe diversity will lead to failure to capture sequence introgressed from distantly related species. The reference genome used will also strongly bias the sequences that can be aligned and so reported as present.
Experimental Procedures
Sample Preparation and Sequencing
Genomic DNA from 12 wheat accessions (14 days after germination) was extracted, RNase treated, and purified as described in Burridge et al. (2017).
Individual aliquots in a total volume of 55 μl were sheared to an average of 300 bp using an E220 Focused-ultrasonicator (Covaris, Woburn, MA, United States). SeqCap EZ HyperCap Workflow User’s Guide (Version 2.0) was used with the following modifications. The starting material was increased to 2 μg DNA. The A-tailing reaction was changed to 20°C for 30 min, followed by 65°C for 30 min. Size selection of the pre-capture libraries was replaced with a 0.9 bead: sample ratio. The precapture amplification was changed to nine cycles followed by immediate clean-up. COT human DNA was replaced with 1 μl of Developer Reagent Plant Capture Enhancer (NimbleGen) per 100 ng of DNA.
Exome capture was performed using ‘Gene Capture v1, 4000026820’ and ‘Promoter Capture v1, 4000030160’ wheat capture probes (Gardiner et al., 2019). Gene and Promoter capture probes were not lyophilized but capture reactions performed separately and products combined after post-capture amplification. For the capture wash, the first Wash Buffer I and both Stringent Wash Buffer steps used buffer preheated to 57°C. Fragment size distribution throughout was determined by TapeStation (Agilent) analysis.
Capture probe enriched sequencing libraries were sequenced at the Bristol Genomics Facility using NextSeq 500 and NextSeq500 2 × 150 bp High−Output v2 kit (Illumina). A final library concentration of 0.8 pM was used with a 5% PhiX control library. The full library preparation and capture method are described in detail in Supplementary File 3. All reads are available from the NCBI sequencing read archive using project ID: PRJNA789931.
Data Analysis
Fastq files for each wheat variety were subjected to quality control using FastQC1 (Babraham Bioinformatics, 2020) and were pre-processed using Fastp (Chen et al., 2018) to trim adaptor sequence and for quality filtering. Paired-end reads were aligned to the ‘Chinese Spring’ reference sequence (IWGSV v1.0) using Burrow-Wheeler Aligner (BWA) (Li and Durbin, 2009) (version 0.7.7-r441), and uniquely mapped reads were identified using sambamba (Tarasov et al., 2015) (version v0.4.4).
Coverage for each chromosome was calculated using samtools (Li et al., 2009) (version 0.1.19-44428cd) using the depth option. Custom perl scripts (available on request) were used to calculate the average depth of coverage for 5 million base pair bins across each chromosome and exome coverage graphs were generated using R (version 3.2.5) (R Core Team, 2013).
Capture probe coverage diagrams were generated with the R package chromPlot using unique location hits and including 0 reads (Verdugo and Oróstica, 2016).
All unmapped reads for the ‘Bacanora’ were extracted from the bam file using samtools (Version: 1.10-24-g383a31b), along with all reads that mapped to the chromosome 1B IWGSC v1.0 reference from physical mapping positions 1–230,000,000 bp (spanning the putative 1B/1RS introgression. These unmapped and mapped reads were then separately queried against a local BLAST database that contained the wheat 1B sequence and the S. cereale 1R sequence, using default BLASTN parameters. The top BLAST hit was then parsed from the BLAST output files using custom perl scripts.
Several gnome assemblies were required for this study: IWGSC v1 Chinese Spring assembly; Rye assembly of the Chinese rye cultivar Weining (Li et al., 2021); Ae. tauschii subsp. strangulata (Luo et al., 2017).
Exome Capture Probes to 1BS and 5DS
The browser extensible data (BED) file containing the genomic coordinates of the gene capture probes, Wheat_gene_capture_probes.bed, from Gardiner et al. (2019) was downloaded from the Grassroots Data Repository.2 From this file, the coordinates for the TGAC v1 probes to chromosomes 1BS and 5DS were extracted. Using the python package pysam, the sequences for these probes were extracted from the TGAC version 1 genome assembly of ‘Chinese Spring’ (Triticum_aestivum.TGACv1.30.dna.genome.fa). The gene capture probe sequences for chromosome 1BS were BLASTed against the chromosome 1BS sequence from the IWGSC v1 assembly and to the chromosome 1RS sequence of the genome assembly of the cultivar ‘Weining’ rye (JADQCU000000000 v1), an elite Chinese S. cereale variety (Li et al., 2021). Likewise, the capture probe sequences for chromosome 5DS were BLASTed against the chromosome 5DS sequence from the IWGSC v1 assembly and to the chromosome 5DS from Ae. tauschii subsp. strangulata assembly, Aet v4.0 (GCA_002575655.1).
Gene Sequences Surrounding Pm2 Gene
The putative 5D introgression in ‘Maris Huntsman’ containing the powdery mildew resistance gene Pm2, was used as the point of reference. The Pm2 gene (TraesCS5D02G044600.1) sequence downloaded from EnsemblPlants is 1,266 bp long and produces a protein of 421 aa. To obtain the Ae. tauschii homolog, the ‘Chinese Spring’ Pm2 sequence was BLASTed against the NCBI Triticeae database; the top hit, with 99.3% identity (1,255/1,264), was the Ae. tauschii subsp. strangulata sequence on 5D (sequence id MW538911.1). The full length of this sequence was 4,421 bp.
To compare sequence similarity of ‘Chinese Spring’ and Ae. tauschii coding sequences around the Pm2 gene, we identified, using the gff3 file for IWGSC v1 (Ensembl Plants genome browser), all the annotated genes within 1 Mb up- and down-stream; in ‘Chinese Spring’, 21 genes were present within this interval (Supplementary Table 1). The sequences of these 21 genes were BLASTed against the NCBI Triticeae database to obtain their homologs in Ae. tauschii. These were then BLASTED against the Ae. tauschii v 4.0 (GCF_002575655.1) assembly to find their positions.
Using BWA, (Li and Durbin, 2009) we aligned the ‘Maris Huntsman’ captured sequences against both ‘Chinese Spring’ (IWGSC v1.0) and Ae. tauschii (Aet V4.0) assemblies. Both assemblies and the ‘Maris Huntsman’ BAM files were indexed using Samtools. The gff3 file of the ‘Chinese Spring’ assembly was also downloaded. An equivalent gff3 file for Ae. tauschii was created based on the positions obtained by BLAST and the regions viewed in IGV (Robinson et al., 2011).
The ‘Maris Huntsman’ captured sequences were aligned, using BWA, to both CS and Aet the sequences around the Pm2 gene (TraesCS5D02G044600 in ‘Chinese Spring’ and AET5Gv20114600 in Ae. tauschii) in the accession ‘Maris Huntsman’ to that of the ‘Chinese Spring’ assembly (IWGSC v1.0) and Ae. tauschii v4.0. For both assemblies, using pysam, pulled out the sequence for the Pm2 gene (c. 4,420 bp) plus 1 Mb both up and downstream from it. In both assemblies, this region contains 21 genes.
We were interested to see whether the exome captured sequences from ‘Maris Huntsman’ 5DS had greater similarity to the gene sequences of ‘Chinese Spring’ or those of Ae. tauschii. Because we believed that the putative introgression contained the powdery mildew resistance gene Pm2, we used this gene as our point of reference. We began by pulling down the Pm2 gene (TraesCS5D02G044600.1) sequence from EnsemblPlants; this is 1,266 bp long and produces a protein of 421 aa. To obtain the homolog from Ae. tauschii, we BLASTed the ‘Chinese Spring’ Pm2 sequence (TraesCS5D02G044600) against the NCBI Triticeae database; the top hit was the homologous gene on 5D of Ae. tauschii subsp. strangulata (AET5Gv20114600). This sequence was then BLASTed against the NCBI Triticeae database. With 99.3% identity (1,255/1,264), it hit the Ae. tauschii Pm2 sequence (MW538911.1), which has a full-length functional gene of 4,421 bp.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA789931. The SNP markers and genotypes are available from the CerealsDB website (Wilkinson et al., 2016): https://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/array_info.php.
Author Contributions
AB prepared the samples and took the lead in writing the manuscript. MW carried out the computational analyses and contributed to the manuscript. PW and GB carried out the computational analyses. KE and GB secured the required funding and supervised the project. All authors helped to interpret the data and contributed to the final manuscript.
Funding
This study was funded by the Biotechnology and Biological Sciences Research Council (Grant BBS/E/C/000I0250) as part of the Designing Future Wheat (DFW) program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Sequencing was performed by the Bristol Genomics Facility. We would like to thank Karim Gharbi, Leah Catchpole, and Thomas Brabbs at the Earlham Institute for assistance and advice regarding the optimization of the exome capture protocol and Jane Coghill and Christy Waterfall at the Bristol Genomics Facility for assistance with library preparation optimization.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.841855/full#supplementary-material
Supplementary Figure 1 | Sequence coverage diagrams for chromosome 2B of the accession ‘Riband’ using different alignment parameters. (A) Average depth of coverage across 5 Mb bins using any good hit to the 2B reference. (B) Average depth of coverage across 5 Mb bins using only sequences that give a unique hit to the 2B reference and allowing the display of zero reads.
Footnotes
- ^ https://plants.ensembl.org/Triticum_aestivum/Location/Compara_Alignments?align=9814–Aegilops_tauschii–5D:46999777-47002088;db=core;g=TraesCS5D02G044600;r=5D:43405783-43407148;t=TraesCS5D02G044600.1
- ^ https://opendata.earlham.ac.uk/wheat/under_license/toronto/Gardiner_2018-07-04_Wheat-gene-promoter-capture/
References
Allen, A. M., Winfield, M. O., Burridge, A. J., Downie, R. C., Benbow, H. R., and Barker, G. L. A. (2016). Characterization of a Wheat Breeders’ Array suitable for high-throughput SNP genotyping of global accessions of hexaploid wheat (Triticum aestivum). Plant Biotechnol. J. 15, 390–401. doi: 10.1111/pbi.12635
Arbor Biosciences (2021). myBaits® - Hybridization Capture for Targeted NGS – Manual v.5.01 - Long Insert Protocol. Ann Arbor, MI: Arbor Biosciences.
Arraiano, L. S., Chartrain, L., Bossolini, E., Slatter, H. N., Keller, B., and Brown, J. K. M. (2007). A gene in European wheat cultivars for resistance to an African isolate of Mycosphaerella graminicola. Plant Pathol. 56, 73–78.
Babraham Bioinformatics (2020). FastQC Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed June 9, 2020).
Bai, B., Du, J. Y., Lu, Q. L., He, C. Y., Zhang, L. J., Zhou, G., et al. (2014). Effective Resistance to Wheat Stripe Rust in a Region with High Disease Pressure. Plant Dis. 98, 891–897. doi: 10.1094/PDIS-09-13-0909-RE
Boyd, L. (2005). Can Robigus defeat an old enemy? – Yellow rust of wheat. J. Agricult. Sci. 143, 233–243. doi: 10.1017/S0021859605005095
Burridge, A. J., Winfield, M. O., Allen, A. M., Wilkinson, P. A., Barker, G. L., and Coghill, J. (2017). “High-Density SNP Genotyping Array for Hexaploid Wheat and Its Relatives,” in Wheat Biotechnology: Methods and Protocols, eds P. Bhalla and M. Singh (Totowa, NJ: Humana Press), 293–306. doi: 10.1007/978-1-4939-7337-8_19
Burt, C., and Nicholson, P. (2011). Exploiting co-linearity among grass species to map the Aegilops ventricosa-derived Pch1 eyespot resistance in wheat and establish its relationship to Pch2. Theoret. Appl. Genet. 123, 1387–1400. doi: 10.1007/s00122-011-1674-9
Chantret, N., Pavoine, M. T., and Doussinault, G. (1999). The race specific resistance gene to powdery mildew, MIRE, has a residual effect on adult plant resistance of winter wheat line RE714. Phytopathology 89, 533–539. doi: 10.1094/PHYTO.1999.89.7.533
Chen, S., Hegarty, J., Shen, T., Hua, L., Li, H., Luo, J., et al. (2021). Stripe rust resistance gene Yr34 (synonym Yr48) is located within a distal translocation of Triticum monococcum chromosome 5AmL into common wheat. Theoret. Appl. Genet. 134, 2197–2211. doi: 10.1007/s00122-021-03816-z
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cobo, N., Wanjugi, H., Lagudah, E., and Dubcovsky, J. (2019). A High-Resolution Map of Wheat QYr.ucw-1BL, an Adult Plant Stripe Rust Resistance Locus in the Same Chromosomal Region as Yr29. Plant Genome 12:180055. doi: 10.3835/plantgenome2018.08.0055
Cosart, T., Beja-Pereira, A., Chen, S., Ng, S. B., Shendure, J., and Luikart, G. (2011). Exome-wide DNA capture and next generation sequencing in domestic and wild species. BMC Genomics 12:347. doi: 10.1186/1471-2164-12-347
Cseh, A., Yang, C., Hubbart-Edwards, S., Scholefield, D., Ashling, S. S., and Burridge, A. J. (2019). Development and validation of an exome-based SNP marker set for identification of the St Jr and Jvs genomes of Thinopyrym intermedium in a wheat background. Theoret. Appl. Genet. 132, 1555–1570. doi: 10.1007/s00122-019-03300-9
Dedryver, F., Paillard, S., Mallard, S., Robert, O., Trottet, M., Nègre, S., et al. (2009). Characterization of genetic components involved in durable resistance to stripe rust in the bread wheat ‘Renan’. Phytopathology 99, 968–973. doi: 10.1094/PHYTO-99-8-0968
Devi, U., Grewal, S., Yang, Cy, Hubbart-Edwards, S., Scholefield, D., and Ashling, S. S. (2019). Development and characterisation of interspecific hybrid lines with genome-wide introgressions from Triticum timopheevii in a hexaploid wheat background. BMC Plant Biol. 19:183. doi: 10.1186/s12870-019-1785-z
Driever, S. M., Lawson, T., Andralojc, P. J., Raines, C. A., and Parry, M. A. J. (2014). Natural variation in photosynthetic capacity, growth, and yield in 64 field-grown wheat genotypes. J. Exp. Bot. 65, 4959–4973. doi: 10.1093/jxb/eru253
Durbin, H. J., Johnson, R., and Stubbs, R. W. (1989). Postulated genes to stripe rust in selected CIMMYT and related wheats. Plant Dis. 73, 472–475. doi: 10.1094/pd-73-0472
Gao, L., Koo, D. H., Juliana, P., Rife, T., Singh, D., Lemes, et al. (2021). The Aegilops ventricosa 2NvS segment in bread wheat: cytology, genomics and breeding. Theoret. Appl. Genet. 134, 529–542. doi: 10.1007/s00122-020-03712-y
Gardiner, L.-J., Brabbs, T., Akhunov, A., Jordan, K., Budak, H., Richmond, T., et al. (2019). Integrating genomic resources to present full gene and putative promoter capture probe sets for bread wheat. GigaScience 8:4. doi: 10.1093/gigascience/giz018
Hanzalová, A., Dumalasová, V., Sumíkova, T., and Bartoš, P. (2007). Rust resistance of the French wheat cultivar Renan. Czech J. Genet. Plant Breed. 43, 53–60. doi: 10.17221/1912-CJGPB
Hao, M., Zhang, L., Ning, S., Huang, L., Yuan, Z., Wu, B., et al. (2020). The Resurgence of Introgression Breeding, as Exemplified in Wheat Improvement. Front. Plant Sci. 11:252. doi: 10.3389/fpls.2020.00252
He, F., Pasam, R., Shi, F., Kant, S., Keeble-Gagnere, G., Kay, P., et al. (2019). Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet. 51, 896–904. doi: 10.1038/s41588-019-0382-2
Henry, I. M., Nagalakshmi, U., Lieberman, M. C., Ngo, K. J., Krasileva, K. V., and Vasquez-Gross, H. (2014). Efficient Genome-Wide Detection and Cataloging of EMS-Induced Mutations Using Exome Capture and Next-Generation Sequencing. Plant Cell 26, 1382–1397. doi: 10.1105/tpc.113.121590
International Wheat Genome Sequencing Consortium [IWGSC] (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 18:345. doi: 10.1126/science.1251788
International Wheat Genome Sequencing Consortium [IWGSC] (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 17:361. doi: 10.1126/science.aar7191
Jin, X., He, M., Ferguson, B., Meng, Y., Ouyang, L., Ren, J., et al. (2012). An effort to use human-based exome capture methods to analyze chimpanzee and macaque exomes. PLoS One 7:e40637. doi: 10.1371/journal.pone.0040637
Kanyuka, K., Lovell, D. J., Mitrofanova, O. P., Hammond-Kosack, K., and Adams, M. J. (2004). A controlled environment test for resistance to Soil-borne cereal mosaic virus (SBCMV) and its use to determine the mode of inheritance of resistance in wheat cv. Cadenza and for screening Triticum monococcum botypes for sources of SBCMV resistance. Plant Pathol. 53, 154–160. doi: 10.1111/j.0032-0862.2004.01000.x
Kaur, P., and Gaikwad, K. (2017). From genomes to GENE-omes: exome sequencing concept and applications in crop improvement. Front. Plant Sci. 8:2164. doi: 10.3389/fpls.2017.02164
King, J., Newell, C., Grewal, S., Hubbart-Edwards, S., Yang, C.-Y., Scholefield, D., et al. (2019). Development of Stable Homozygous Wheat/Amblyopyrum muticum (Aegilops mutica) Introgression Lines and Their Cytogenetic and Molecular Characterization. Front. Plant Sci. 10:34. doi: 10.3389/fpls.2019.00034
Kiseleva, A. A., Potokina, E. K., and Salina, E. A. (2007). Features of Ppd-B1 expression regulation and their impact on the flowering time of wheat near-isogenic lines. BMC Plant Biol. 17:172. doi: 10.1186/s12870-017-1126-z
Leonard, J. M., Watson, C. J. W., Carter, A. H., Hansen, J. L., Zemetra, R. S., and Santra, D. K. (2007). Identification of a candidate gene for the wheat endopeptidase Ep-D1 locus and two other STS markers linked to the eyespot resistance gene Pch1. Theoret. Appl. Genet. 116, 261–270. doi: 10.1007/s00122-007-0664-4
Li, G., Wang, L., Yang, J., He, H., Jin, H., Li, X., et al. (2021). A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 53, 574–584. doi: 10.1038/s41588-021-00808-z
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Luo, M. C., Gu, Y., Puiu, D., Wang, H., Twardziok, S. O., Deal, K. R., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502. doi: 10.1038/nature24486
Ma, J., Wingen, L. U., Orford, S., Fenwick, P., Wang, J. K., and Griffiths, S. (2015). Using the UK reference population Avalon x Cadenza as a platform to compare breeding strategies in elite Western European bread wheat. Mol. Breed. 35:70. doi: 10.1007/s11032-015-0268-7
Mago, R., Tabe, L., Vautrin, S., Šimková, H., Kubaláková, M., Upadhyaya, N., et al. (2014). Major haplotype divergence including multiple germin-like protein genes at the wheat Sr2 adult plant stem rust resistance locus. BMC Plant Biol. 14:379. doi: 10.1186/s12870-014-0379-z
Marchal, C., Zhang, J., Zhang, P., Fenwick, P., Steuernagel, B., and Adamski, N. M. (2018). BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat. Plants 4, 662–668. doi: 10.1038/s41477-018-0236-4
McIntosh, R. A., Wellings, C. R., and Park, R. F. (1995). Wheat Rusts: An Atlas of Resistance Genes. Dordrecht: Kluwer Academic Publishers.
Pathan, A. K., and Park, R. F. (2006). Evaluation of seedling and adult plant resistance to leaf rust in European wheat cultivars. Euphytica 149, 327–342. doi: 10.1007/s10681-005-9081-4
Pingault, L., Choulet, F., Alberti, A., Glover, N., Wincker, P., Feuillet, C., et al. (2015). Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biol. 16:29. doi: 10.1186/s13059-015-0601-9
Przewieslik-Allen, A. M., Burridge, A. J., Wilkinson, P. A., Winfield, M. O., and Shaw, D. S. (2019). Developing a High-Throughput SNP-Based Marker System to Facilitate the Introgression of Traits from Aegilops Species into Bread Wheat (Triticum aestivum). Front. Plant Sci. 9:1993. doi: 10.3389/fpls.2018.01993
Przewieslik-Allen, A. M., Wilkinson, P. A., Burridge, A., Winfield, M., Dai, X., and Beaumont, M. (2021). The role of gene flow and chromosomal instability in shaping the bread wheat genome. Nat. Plants 7, 172–183. doi: 10.1038/s41477-020-00845-2
R Core Team (2013). R: A language and environment for statistical computing R Foundation for Statistical Computing Vienna Austria. Austria: R Core Team.
Rabinovich, S. V. (1998). Importance of wheat-rye translocations for breeding modern cultivars of Triticum aestivum L. Euphytica 100, 323–340.
Rasheed, A., and Xia, X. (2019). From markers to genome-based breeding in wheat. Theoret. Appl. Genet. 132, 767–784. doi: 10.1007/s00122-019-03286-4
Robert, O., Abelard, C., and Dedryver, F. (1999). Identification of molecular markers for the detection of the yellow rust resistance gene Yr17 in wheat. Mol. Breed. 5, 167–175. doi: 10.1023/A:1009672021411
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., and Getz, G. (2011). Intergrative Genomics Viewer. Nat. Biotechnol. 29, 24–26.
Saintenac, C., Jiang, D., and Akhunov, E. D. (2011). Targeted analysis of nucleotide and copy number variation by exon capture in allotetraploid wheat genome. Genome Biol. 12:R88. doi: 10.1186/gb-2011-12-9-r88
Salmon, A., Udall, J. A., Jeddeloh, J. A., and Wendel, J. (2012). Targeted capture of homoeologous coding and noncoding sequence in polyploid cotton. G3 2, 921–930. doi: 10.1534/g3.112.003392
Schneider, A., Molnár, I., and Molnár-Láng, M. (2008). Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 163, 1–19. doi: 10.1007/s10681-007-9624-y
Singh, R. P., and Rajaram, S. (1991). Resistance to Puccinia recondita f.sp. tritici in 50 Mexican bread wheat cultivars. Crop Sci. 31, 1472–1479. doi: 10.2135/cropsci1991.0011183x003100060016x
Singh, S., Vikram, P., Sehgal, D., Burgueño, J., Sharma, A., Singh, S. K., et al. (2018). Harnessing genetic potential of wheat germplasm banks through impact-oriented-prebreeding for future food and nutritional security. Scient. Rep. 21:12527. doi: 10.1038/s41598-018-30667-4
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J., and Prins, P. (2015). Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034. doi: 10.1093/bioinformatics/btv098
United Kingdom Cereal Pathogen Virulence Survey [UKCVS] (1996). Annual Report. Accessed from AHDB archives: ahdb.org.uk/ukcpvs. Telford, UK: UKCVS.
United Kingdom Cereal Pathogen Virulence Survey [UKCVS] (2004). Annual Report. Accessed from AHDB archives. Telford, UK: UKCVS.
Verdugo, R. A., and Oróstica, K. Y. (2016). chromPlot: Global visualization tool of genomic data R package version 1160. Bioinformatics 32, 2366–2368. doi: 10.1093/bioinformatics/btw137
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genome diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wang, Z. L., Li, L. H., He, Z. H., Duan, X. Y., Zhou, Y. L., Chen, X. M., et al. (2005). Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 89, 457–463. doi: 10.1094/PD-89-0457
Warburton, M., Skovmand, B., and Mujeeb-Kazi, A. (2002). The molecular genetic characterization of the ‘Bobwhite’ bread wheat family using AFLPs and the effect of the T1BL.1RS translocation. Theoret. Appl. Genet. 104, 868–873. doi: 10.1007/s00122-001-0816-x
Wellings, C. R. (1986). Host: Pathogen Studies of Wheat Stripe Rust in Australia. Ph.D thesis, Australia: University of Sydney.
Wilkinson, P. A., Winfield, M. O., Barker, G. L. A., Tyrell, S., Bian, X., Allen, A. M., et al. (2016). CerealsDB 3.0: expansion of resources and data integration. BMC Bioinform. 17:256. doi: 10.1186/s12859-0161139-x
William, M., Singh, R. P., Huerta-Espino, J., Islas, S. O., and Hoisington, D. (2003). Molecular marker mapping of leaf rust resistance gene Lr46 and its association with stripe rust resistance gene Yr29 in wheat. Phytopathology 93, 153–159. doi: 10.1094/PHYTO.2003.93.2.153
Winfield, M. O., Allen, A. M., Burridge, A. J., Barker, G. L. A., Benbow, H. R., and Wilkinson, P. A. (2015). High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 14, 1195–1206. doi: 10.1111/pbi.12485
Winfield, M. O., Wilkinson, P. A., Allen, A. M., Barker, G. L. A., Coghill, J. A., and Burridge, A. (2012). Targeted re-sequencing of the allohexaploid wheat exome. Plant Biotechnol. J. 10, 733–742. doi: 10.1111/j.1467-7652.2012.00713.x
Worland, A. J., Korzun, V., Roder, M. S., Ganal, M. W., and Law, C. N. (1998). Genetic analysis of the dwarfing gene Rht8 in wheat. Part II. The distribution and adaptive significance of allelic variants at the Rht8 locus of wheat as revealed by microsatellite screening. Theoret. Appl. Genet. 96, 1110–1120. doi: 10.1007/s001220050846
Xu, J., Wang, L., Deal, K. R., Zhu, T., Ramasamy, R. K., Luo, Mc, et al. (2020). Genome-wide introgression from a bread wheat×Lophopyrum elongatum amphiploid into wheat. Theoret. Appl. Genet. 133, 1227–1241. doi: 10.1007/s00122-020-03544-w
Zhang, J., Liu, W., Lu, Y., Liu, Q., Yang, X., Li, X., et al. (2017). A resource of large-scale molecular markers for monitoring Agropyron cristatum chromatin introgression in wheat background based on transcriptome sequences. Scient. Rep. 7:11942. doi: 10.1038/s41598-017-12219-4
Zhang, W., Cao, Y., Zhang, M., Zhu, X., Ren, S., Long, Y., et al. (2017). Meiotic Homoeologous Recombination-Based Alien Gene Introgression in the Genomics Era of Wheat. Crop Sci. 57, 1189–1198. doi: 10.2135/cropsci2016.09.0819
Zhou, S., Yan, B., Li, F., Zhang, J., Zhang, J., Ma, H., et al. (2017). RNA-Seq Analysis Provides the First Insights into the Phylogenetic Relationship and Interspecific Variation between Agropyron cristatum and Wheat. Front. Plant Sci. 8:1644. doi: 10.3389/fpls.2017.01644
Keywords: wheat, Triticum aestivum, introgression, exome capture, exome capture sequencing, sequence variation
Citation: Burridge AJ, Winfield MO, Wilkinson PA, Przewieslik-Allen AM, Edwards KJ and Barker GLA (2022) The Use and Limitations of Exome Capture to Detect Novel Variation in the Hexaploid Wheat Genome. Front. Plant Sci. 13:841855. doi: 10.3389/fpls.2022.841855
Received: 22 December 2021; Accepted: 28 February 2022;
Published: 12 April 2022.
Edited by:
Surya Saha, Boyce Thompson Institute (BTI), United StatesReviewed by:
Weilong Kong, Wuhan University, ChinaZhenyang Liao, Agricultural Genomics Institute at Shenzhen (CAAS), China
Dong Xu, Laboratory of Genome Analysis, Agricultural Genomics Institute at Shenzhen (CAAS), China
Copyright © 2022 Burridge, Winfield, Wilkinson, Przewieslik-Allen, Edwards and Barker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amanda J. Burridge, YW1hbmRhLmJ1cnJpZGdlQGJyaXN0b2wuYWMudWs=
 Amanda J. Burridge
Amanda J. Burridge Mark O. Winfield
Mark O. Winfield Paul A. Wilkinson
Paul A. Wilkinson Alexandra M. Przewieslik-Allen
Alexandra M. Przewieslik-Allen Keith J. Edwards1
Keith J. Edwards1 Gary L. A. Barker
Gary L. A. Barker