
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 10 March 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.841688
This article is part of the Research TopicOn the Frontier of a Plant's Dilemma: Exploring the Molecular Basis of Growth Versus Defense AntagonismView all 5 articles
Salicylic acid (SA) and N-hydroxypipecolic acid (NHP) are two central plant immune signals involved in both resistance at local sites of pathogen infection (basal resistance) and at distal uninfected sites after primary infection (systemic acquired resistance). Major discoveries and advances have led to deeper understanding of their biosynthesis and signaling during plant defense responses. In addition to their well-defined roles in immunity, recent research is emerging on their direct mechanistic impacts on plant growth and development. In this review, we will first provide an overview of how SA and NHP regulate local and systemic immune responses in plants. We will emphasize how these two signals are mutually potentiated and are convergent on multiple aspects—from biosynthesis to homeostasis, and from signaling to gene expression and phenotypic responses. We will then highlight how SA and NHP are emerging to be crucial regulators of the growth-defense balance, showcasing recent multi-faceted studies on their metabolism, receptor signaling and direct growth/development-related host targets. Overall, this article reflects current advances and provides future outlooks on SA/NHP biology and their functional significance as central signals for plant immunity and growth. Because global climate change will increasingly influence plant health and resilience, it is paramount to fundamentally understand how these two tightly linked plant signals are at the nexus of the growth-defense balance.
Plants rely on their two-tiered and interlinked innate immune system to initiate local responses against pathogenic attack (Jones and Dangl, 2006; Kim and Castroverde, 2020; Zhou and Zhang, 2020; Yuan et al., 2021). First, pattern-triggered immunity (PTI) is initiated after activation of cell surface pattern recognition receptors (PRRs) that typically recognize conserved pathogen-associated molecular patterns (PAMPs; Macho and Zipfel, 2014; Li et al., 2016; DeFalco and Zipfel, 2021). Second, a more robust effector-triggered immunity (ETI) is activated when pathogen effectors are recognized by intracellular nucleotide-binding leucine-rich repeat receptors (NLRs), often resulting in localized cell death (Zebell and Dong, 2015; Saur et al., 2021). Sustained immune activation at the local infection site primes unaffected systemic tissues against future biotic stress via systemic acquired resistance (SAR; Vlot et al., 2021; Zeier, 2021). Several key SAR inducers have been identified, including salicylic acid (SA), methyl SA, azelaic acid (AzA), glycerol-3-phosphate (G3P), dehydroabietinal (DA), nitric oxide (NO), reactive oxygen species (ROS), pipecolic acid (Pip), and N-hydroxypipecolic acid (NHP; Wendehenne et al., 2014; Singh et al., 2017; Hartmann et al., 2018).
A central regulator of local and systemic immunity is the plant hormone SA (Zhang and Li, 2019). Because it serves various roles, SA levels and metabolism are altered during immune responses to suit the plant’s needs (Dempsey et al., 2011). SA is produced via two independent pathways: isochorismate synthase (ICS) and phenylalanine ammonia lyase (PAL) pathways (Dempsey et al., 2011; Hartmann and Zeier, 2019; Zhang and Li, 2019; Huang et al., 2020a). In Arabidopsis, most of the pathogen-induced SA is produced through the ICS pathway involving pathogen-induced genes ISOCHORISMATE SYNTHASE 1 (ICS1), ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5), and AVRPPHB SUSCEPTIBLE 3 (PBS3; Chen et al., 2009; Huang et al., 2020a). Of the two Arabidopsis ICS paralogs, ICS1 plays a major role in SA synthesis following infection (Nawrath and Metraux, 1999; Wildermuth et al., 2001; Garcion et al., 2008). In plastids, ICS1 converts chorismate to isochorismate, which is transported by EDS5 to the cytosol (Garcion et al., 2008). PBS3 and EPS1 then catalyze the final conversions to SA (Rekhter et al., 2019; Torrens-Spence et al., 2019). Although low SA levels can be transported to systemic tissues during SAR, its long-distance mobility alone is not responsible for SAR establishment (Vernooij et al., 1994; Lim et al., 2020). It is proposed that SA contributes to systemic propagation of defenses alongside other signaling molecules (Lim et al., 2020; Vlot et al., 2021).
Another metabolite involved in plant immunity is NHP, a hydroxylated derivative of the non-protein amino acid Pip that can induce SA accumulation (Návarová et al., 2012; Hartmann et al., 2018; Wang et al., 2018). The NHP biosynthetic pathway is inducible by pathogens and leads to SAR (Hartmann et al., 2018). NHP can induce defense gene expression, amplify the resistance response, synergistically function with SA, and promote the hypersensitive response (Hartmann et al., 2018). Recent exciting studies have provided detailed insights into NHP biosynthesis and mobilization. Three pathogen-inducible genes are involved in NHP biosynthesis: AGD2-LIKE DEFENSE RESPONSE PROTEIN 1 (ALD1), SAR DEFICIENT 4 (SARD4), and FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1; Hartmann and Zeier, 2018). ALD1 is an L-Lys-α-aminotransferase that deaminates L-Lys, spontaneously leading to dehydropipecolic acid intermediates (Hartmann and Zeier, 2018). These are reduced by SARD4 to Pip, which is then converted by FMO1 to NHP (Hartmann and Zeier, 2018). The local and systemic accumulation of Pip and NHP after pathogen attack are necessary for SAR (Hartmann and Zeier, 2018).
Deployment of SA, NHP, and other defense responses must be balanced with the plants’ ability to grow and/or develop in order to optimize overall fitness (Huot et al., 2014). This “growth-defense equilibrium” paradigm has been postulated due to limited resources that must be balanced leading to reciprocal tradeoffs (Coley et al., 1985). Alternatively, this is due to interlinked and conditional coordination between growth and immune responses depending on the environment (Kliebenstein, 2016). In terms of SA and NHP, over-accumulating mutants exhibit decreased growth (Abreu and Munné-Bosch, 2009; Pastorczyk-Szlenkier and Bednarek, 2021), reflecting that SA/NHP mediate the delicate equilibrium between plant growth and immunity.
To understand the relationship between immunity and growth via the SA and NHP pathways, it is important to highlight the tight mechanistic linkage between these two central immune-activating metabolites (Figure 1; for detailed review, see Zeier, 2021). SA and NHP biosynthesis and downstream signaling are closely intertwined, relying on overlapping regulatory proteins and signaling components (Sun et al., 2015; Hartmann and Zeier, 2019; Ding and Ding, 2020). The SA pathway genes ICS1, EDS5, and PBS3 and the NHP biosynthetic genes ALD1, SARD4, and FMO1 are regulated via two partially redundant master transcription factors SAR DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60-LIKE G (CBP60g; Wang et al., 2011; Sun et al., 2015; Huang et al., 2020a). SARD1 and CBP60g activation by pathogen infection and/or immune elicitation leads to increased SA and NHP levels (Hartmann and Zeier, 2019; Huang et al., 2020a).
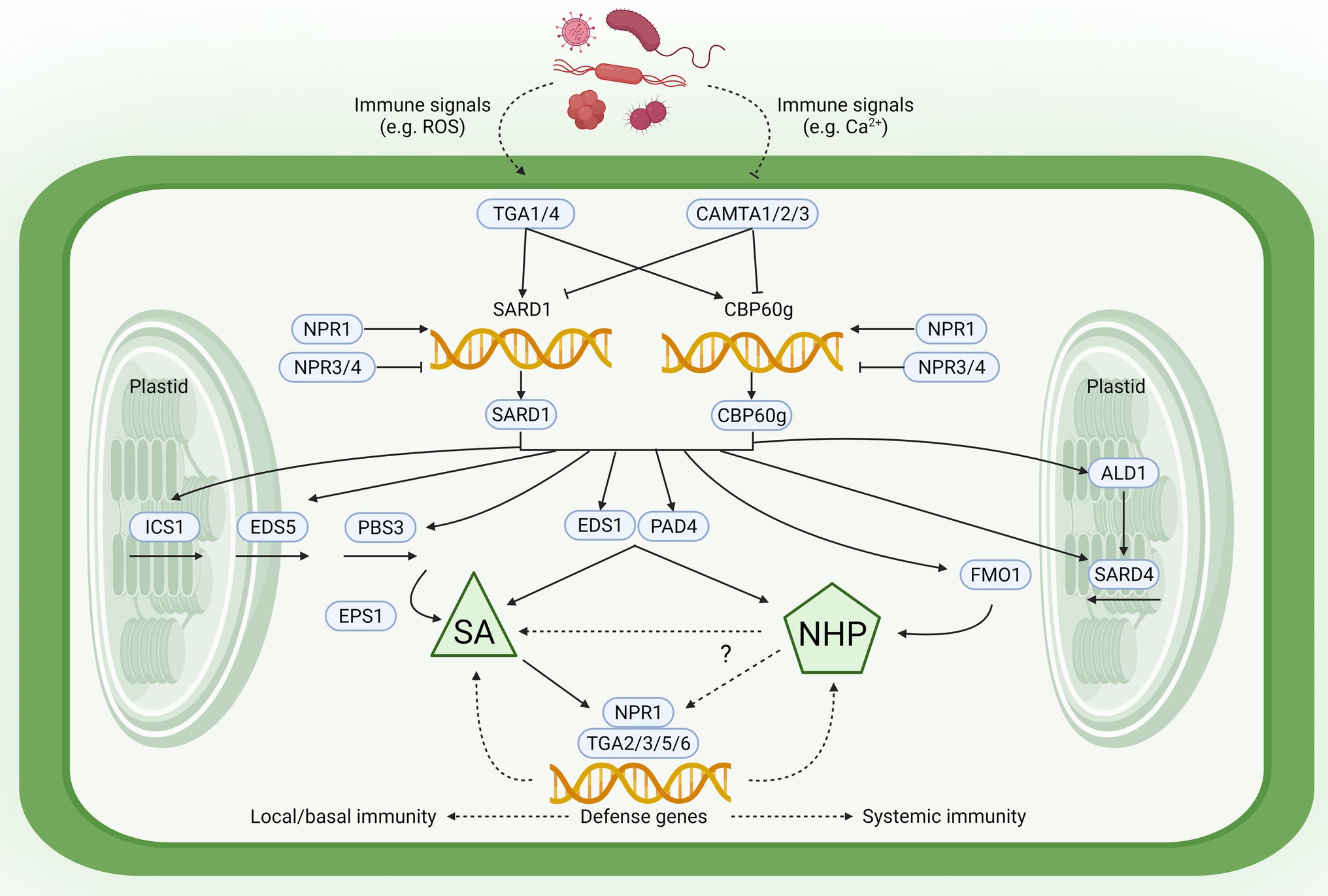
Figure 1. Regulatory convergence and mutual potentiation of salicylic acid (SA) and N-hydroxypipecolic acid (NHP) biosynthesis and signaling. Upstream immunity-associated signals [e.g., reactive oxygen species (ROS), Ca2+] lead to activation/repression of TGACG SEQUENCE-SPECIFIC BINDING PROTEIN 1 (TGA1)/4 transcriptional activators and CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR (CAMTA) transcriptional repressors. Along with the antagonistic SA receptors NONEXPRESSER OF PR GENES 1 (NPR1; co-activator) and NPR3/4 (co-repressors), TGA1/4 and CAMTA1/2/3 control expression of CALMODULIN-BINDING PROTEIN 60-LIKE G (CBP60g) and SAR DEFICIENT 1 (SARD1) that encode functionally redundant master transcription factors of plant immunity. SARD1 and CBP60g directly bind the promoters of SA biosynthetic (ICS1, EDS5, and PBS3) and NHP biosynthetic genes (ALD1, SARD4, and FMO1). Central immune regulators ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) and PHYTOALEXIN DEFICIENT 4 (PAD4; which mediate both pattern-triggered immunity and effector-triggered immunity) are also required for SA and NHP accumulation. Downstream of their biosynthesis, SA directly activates while NHP indirectly activates the SA receptor NPR1. NPR1 then promotes TGA-directed transcription of key defense genes for local/basal and systemic immune responses. Created with BioRender.com.
Full induction of SARD1 and CBP60g gene expression requires TGACG SEQUENCE-SPECIFIC BINDING PROTEIN 1 and 4 (TGA1 and TGA4) transcription factors, which modulate SA and NHP levels (Hartmann et al., 2018; Sun et al., 2018; Zhang and Li, 2019). TGA1 and TGA4 are paralogs of the TGA transcription factor family, which specifically bind variants of the palindromic sequence TGACGTCA in target gene promoters (Xiang et al., 1997). In addition to TGA1/4, other TGAs include TGA2/3/5/6, which are essential for responses to SA and NHP (Kesarwani et al., 2007; Nair et al., 2021). Higher-order tga mutants have significantly reduced sensitivity to SA and NHP (Zhang et al., 2003; Nair et al., 2021), which could potentially explain their SAR-deficient phenotypes (Zhang et al., 2003; Kesarwani et al., 2007). The requirement of these TGAs for SA- and NHP-mediated transcriptional reprogramming is expected since TGAs recruit the master coactivator and SA receptor NONEXPRESSER OF PR GENES 1 (NPR1), which is required for SA- and NHP-responsive expression (Ding et al., 2018; Nair et al., 2021). In addition to TGAs, SA, and NHP biosynthesis and signaling can be modulated by CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR (CAMTA) 1, 2, and 3—central transcriptional repressors in plant immunity that directly target CBP60g and SARD1 promoters (Sun et al., 2020).
In addition to transcription factors, other proteins also control SA/NHP accumulation. These include two lipase-like proteins ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) and PHYTOALEXIN DEFICIENT 4 (PAD4; Hartmann and Zeier, 2019; Zeier, 2021), which mediate both ETI and PTI. This potentially suggests the major importance of the SA and NHP pathways after immune activation. Interestingly, EDS1 and PAD4 are target genes of SARD1 and CBP60g (Sun et al., 2015), further reflecting the close mechanistic relationships of these immune regulators during SA/NHP production. Recent studies have identified another key component involved in local and systemic immunity—a Jumonji (JMJ) domain-containing H3K4 demethylase, JMJ14 (Li et al., 2020). In local leaves, JMJ14 positively regulates immunity by upregulating ALD1/FMO1 transcription and enhanced SA-responsiveness; in distal leaves, JMJ14 is vital for systemic NHP accumulation and SAR (Li et al., 2020). The jmj14 mutants exhibited reduced local and systemic defenses. Remarkably, JMJ14 positively regulates immunity-induced H3K4me3 histone enrichment in SA- and NHP-associated defense genes (Li et al., 2020). Altogether, these studies highlight the common and overlapping molecular players that impinge on the SA and NHP pathways.
Because of common overlapping SA and NHP regulators, it is not surprising that SA/NHP cooperatively and synergistically influence each other to induce SAR (Figure 1; for detailed review, see Zeier, 2021). This mutual amplification is best exemplified by their effect on each other’s biosynthetic genes. NHP biosynthetic enzymes ALD1 and FMO1 are required for systemic SA accumulation (Mishina and Zeier, 2006; Cecchini et al., 2015). Indeed, NHP treatment directly induces and also primes SA biosynthetic gene expression (ICS1, EDS5, and PBS3) and SA production, as elegantly demonstrated by Yildiz et al. (2021). Downstream of SA biosynthesis, NHP also primes SA-induced defense gene expression (Bernsdorff et al., 2016; Yildiz et al., 2021).
On the other hand, SA can enhance NHP-activated immunity and gene expression (Hartmann et al., 2018; Yildiz et al., 2021). In particular, both ALD1 and FMO1 gene expression can be directly upregulated by SA (Cecchini et al., 2015), although they also exhibit SA-independent expression (Bartsch et al., 2006; Bernsdorff et al., 2016). SA induction-deficient sid2 mutants are SAR-deficient, but not to the same extent as NHP-deficient ald1 and fmo1 mutants (Hartmann et al., 2018; Yildiz et al., 2021). Potentially, this could be due to basal SA levels present in sid2 mutants (Nair et al., 2021), but further genetic and molecular dissection is necessitated.
This mutual potentiation can be explained since SA- and NHP-mediated signaling both depend on the coactivator NPR1 (Návarová et al., 2012; Yildiz et al., 2021) and its paralogous corepressors NPR3 and NPR4, all of which can bind SA and regulate SAR (Fu et al., 2012; Wu et al., 2012; Fu and Dong, 2013; Ding et al., 2018; Liu et al., 2020). Both SA-induction of NHP biosynthetic genes and NHP-induction of SA-associated genes depend on the NPR1 regulatory module (Ding et al., 2018; Nair et al., 2021; Zeier, 2021). Overall, these demonstrate that SAR is dependent on mutual amplification of SA and NHP (Bernsdorff et al., 2016; Huang et al., 2020a; Nair et al., 2021; Yildiz et al., 2021), illustrating the cooperative interactions between these two central immune-activating metabolites.
Although SA is typically known as a defense hormone, it also affects plant growth and development (Figure 2) independently and/or via crosstalk with other hormones and signaling molecules (van Butselaar and Van den Ackerveken, 2020; Castroverde and Dina, 2021; Pokotylo et al., 2021; Saleem et al., 2021). SA-depleted Arabidopsis NahG transgenic plants are larger, while mutants with constitutively high SA levels such as acd6–1 are dwarfed (Rivas-San Vicente and Plasencia, 2011). SA can also delay or inhibit seed germination in Arabidopsis, possibly from the resulting oxidative stress (Rajjou et al., 2006). This interplay between SA and ROS positively affects cell division in the quiescent center (QC), directly linking SA to root phenotypes (Wang et al., 2021). In agreement, SA-accumulating mutants and/or exogenous SA treatment can increase cell division in the QC by promoting ROS generation (Wang et al., 2021). Reproductive development is also modulated by SA. In Arabidopsis, SA inhibits pollen tube tip growth, whereas methylated SA promotes tip growth (Rong et al., 2016). The enzymes that interconvert between SA and MeSA (MeSA methylesterase and SA methyltransferase) can be found at the pollen tube apical regions, implying localized pollen tip synthesis (Rong et al., 2016). There is also an antagonistic effect between SA and ethylene-mediated apical hook formation, which is essential for growth above soil after germination (Huang et al., 2020b). Apical hooks are promoted by ethylene and involve transcription factors ETHYLENE INSENSITIVE 3 (EIN3) and ETHYLENE INSENSITIVE 3-like 1 (EIL1; Huang et al., 2020b). SA activates NPR1 and inhibits EIN3 binding to target gene promoters, such as HLS1 (Huang et al., 2020b). Though varied, SA clearly has an impact on various growth and developmental processes, which are facilitated by the intricate crosstalk between SA and other signals (e.g., major growth hormones).
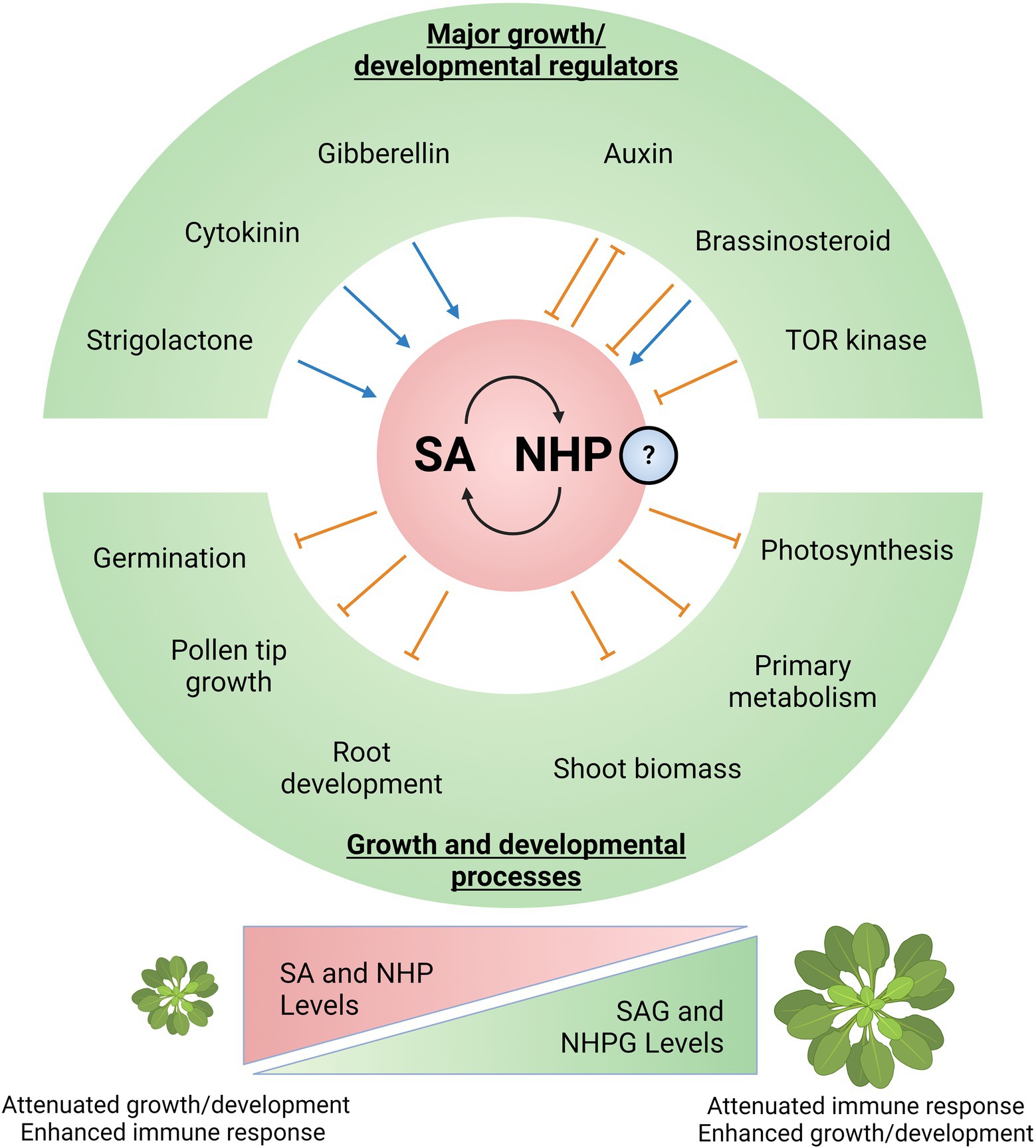
Figure 2. Salicylic acid and NHP at the nexus of the plant growth-defense balance. Major regulators of growth and development have synergistic and/or antagonistic relationships with SA and potentially with NHP. These include key plant hormones (auxin, brassinosteroid, gibberellin, cytokinin, and strigolactone) and the master regulatory kinase Target of Rapamycin (TOR). SA and potentially NHP could independently or synergistically impact various aspects of plant growth and development. In particular, SA has been shown to influence germination and apical hook development, pollen tip growth during floral development, root growth and patterning, shoot biomass accumulation, primary metabolism, and photosynthesis. Ultimately, levels and homeostasis between free bioactive SA/NHP and inactive storage forms (SAG/NHPG) allow plants to dynamically balance resources between growth and defense. Higher SA/NHP potentiates immune responses at the expense of growth, while lower SA/NHP promotes growth processes and modulates immunity. Created with BioRender.com.
Auxin is important for growth and development (Lavy and Estelle, 2016); therefore, elucidating how SA impacts auxin biosynthesis/signaling is key to understanding the central role of SA in plant growth-defense balance. Since both SA and auxin biosynthetic pathways proceed from the precursor chorismate (product of the shikimate pathway), it is possible that one hormone shifts the shikimate pathway metabolic flux away from the other (Koo et al., 2020). SA can affect root meristem patterning, suggesting changes in auxin synthesis and transport (Pasternak et al., 2019). For example, exposure to low SA concentration (below 50 μM) promotes adventitious root formation in Arabidopsis, potentially by elevating root tip auxin levels to promote root meristem maturation (Pasternak et al., 2019). Because of this SA-auxin interplay, pathogens sometimes co-opt the auxin pathway to better infect plants (Pasternak et al., 2019). In response to pathogens, plants can use SA to repress the auxin pathway. SA can interact with and inhibit CATALASE2 (CAT2) to increase H2O2 levels, thereby repressing biosynthesis of the auxin precursor tryptophan by sulfenylating a key enzyme (Yuan et al., 2017). SA treatment also leads to increasing AUXIN RESISTANT/INDOLE-3-ACETIC ACID INDUCIBLE (Aux/IAA) repressor levels thereby repressing auxin-related gene transcription (Wang et al., 2007). In addition, SA can interfere with auxin transport by repressing clathrin-mediated endocytosis (Du et al., 2013). SA also antagonizes auxin by inhibiting protein phosphatase 2A resulting in auxin transporter PIN-FORMED 2 (PIN2) hyperphosphorylation, leading to attenuated root growth (Tan et al., 2020). Strikingly, SA can enhance adventitious root formation in cucumbers by competitively inhibiting the enzyme Cucumis sativus GRETCHEN HAGEN 3.5 (CsGH3.5), thereby increasing free auxin levels (Dong et al., 2020). Altogether, SA can influence aspects of plant growth and development by interfering with the auxin pathway.
Like auxins, gibberellins (GA) constitute another major class of hormones mediating growth and development (Emamverdian et al., 2020). During germination of the halophyte Limonium bicolor under salt stress, SA upregulated various genes involved in GA biosynthesis (Liu et al., 2019). Complementing this finding, exogenous GA increased expression of NPR1 and WRKY70, resulting in elevated SA (Alonso-Ramírez et al., 2009).
The impact of SA on growth and development is well-documented (Carviel et al., 2009; Rivas-San Vicente and Plasencia, 2011; Carella et al., 2014; van Butselaar and Van den Ackerveken, 2020; Pokotylo et al., 2021); however, the effect of NHP is only starting to be explored (Figure 2). For example, altering free NHP levels by inactivating UGT76B1-mediated glycosylation to NHPG can affect plant growth by decreasing rosette size and biomass (Bauer et al., 2021; Cai et al., 2021; Mohnike et al., 2021). Inhibited plant development and enhanced SAR was observed in the ugt76b1 mutant, while overexpression led to opposite phenotypes (Bauer et al., 2021; Cai et al., 2021; Mohnike et al., 2021). Since NHP activates SAR, UGT76B1 dictates NHP levels and thus the SAR response (Bauer et al., 2021; Cai et al., 2021; Holmes et al., 2021; Mohnike et al., 2021). Interestingly, UGT76B1 (along with glucosyltransferases UGT74F1/UGT74F2) also conjugates and inactivates SA to modulate disease resistance (Huang et al., 2020a; Bauer et al., 2021), further emphasizing the regulatory and metabolic convergence of NHP and SA. Complementing these studies, recent genetic analyses demonstrated that autoimmunity and growth suppression in the camta1/2/3 triple mutant can be reversed by mutations in the NHP biosynthetic genes ALD1 and FMO1 (Sun et al., 2020).
There are several major knowledge gaps regarding how NHP affects growth and development, particularly on its mechanistic impact on canonical growth hormones like auxin, GA, and brassinosteroid (BR). Although crosstalk with hormones is relatively uncharacterized, the NHP precursor Pip has been described as an osmoprotectant in both bacteria and plants (Gouesbet et al., 1994; Moulin et al., 2006; Pérez-García et al., 2019), and this could have profound consequences on overall plant physiology. Pip levels were found to increase under hyperosmotic conditions and decrease under hypo-osmotic conditions, although the authors did not measure growth phenotypes (Moulin et al., 2006). During osmotic stress, lysine-ketoglutarate reductase and saccharopine dehydrogenase can regulate L-lysine (Pip/NHP precursor) catabolism (Moulin et al., 2006). Under drought conditions, Pip accumulates in the roots/rhizosphere of sorghum, likely mediating root growth suppression (Caddell et al., 2020). Strawberry leaves with a stunted growth phenotype were also found to accumulate Pip after chilling or treatment with maleic hydrazide (Yatsu and Boynton, 1959).
Consistent with the negative impact of NHP on growth phenotypes, transcriptome analyses in Arabidopsis revealed that NHP-suppressed genes are associated with photosynthesis and primary metabolism, particularly those involved in fatty acid and amino acid biosynthesis (Yildiz et al., 2021). Close examination of their transcriptome data reveal that certain NHP-downregulated genes are associated with the auxin (IAAs and AUXIN RESPONSE FACTORS/ARFs), BR (BRASSINOSTEROID INSENSITIVE 1/BRI1 and BRI1-EMS-SUPPRESSOR 1/BES1), and GA pathways (DELLA, GA2OX). It is important to highlight that NHP-downregulation of these growth/development-related genes is less pronounced than in biologically induced SAR (Yildiz et al., 2021).
In the future, it would be interesting to conduct focused mechanistic studies on how NHP intercepts various growth hormone pathways and to determine whether common molecular components are targeted by both SA and NHP. Because of the known functional synergism between SA and NHP, it is intriguing to speculate that NHP influences these other hormones through similar mechanisms perturbed by SA. It is also unclear if the antagonistic effect of NHP on growth/development is dependent on or parallel with functional SA signaling. These potential directions will establish whether NHP is central to the growth-immunity balance just like SA.
Salicylic acid and possibly NHP can impact growth and developmental processes, sometimes directly regulating other hormone pathways. SA, in particular, has been well-demonstrated for its central role in the growth-immunity balance (Huot et al., 2014). It is not surprising that growth-related pathways (e.g., major growth hormones) can directly impinge on SA biosynthesis and signaling (Figure 2).
A well-demonstrated example is auxin signaling modulating the SA pathway (Wang et al., 2007). Lowering auxin levels via GH3.5 is associated with higher SA levels, contributing to this canonical plant tradeoff (Hagen and Guilfoyle, 2002). AUXIN SIGNALING F BOX PROTEIN 1 (AFB1) overexpression enhances auxin signaling, resulting in lower SA levels and increased host susceptibility (Robert-Seilaniantz et al., 2011). Auxin may also negatively impact the NHP pathway. NHP biosynthetic genes ALD1 and FMO1 are downregulated after treatment with the auxin indole-3-acetic acid as revealed by transcriptome datasets in the Gene Expression Atlas.1 However, further mechanistic investigations are still lacking.
Another class of hormones, BRs, have differential relationships with SA depending on the species (De Vleesschauwer et al., 2012). In rice, BR treatment represses SA signaling, while the opposite is observed in Arabidopsis (De Vleesschauwer et al., 2012). Like auxin, BR also antagonizes SA by blocking rice resistance. Specifically, the synthetic SA analog benzothiadiazole is less effective against the root oomycete pathogen Pythium graminicola after BR treatment (De Vleesschauwer et al., 2012). In contrast to BRs, exogenous GA promotes expression of ICS1 and NPR1, leading to increased SA levels in Arabidopsis (Alonso-Ramírez et al., 2009). The SA pathway is also influenced by another growth-related hormone, cytokinin (CK). The CK-associated type-B response regulator 2 (ARR2) directly interacts with TGA3 that regulates SA-responsive PR genes (O’Brien and Benková, 2013), thereby increasing Arabidopsis resistance against Hyaloperonospora arabidopsidis after CK treatment (Argueso et al., 2012). In rice, CK and SA synergistically activate PR gene expression against Magnaporthe oryzae infection (Jiang et al., 2013), although CK did not induce expression of SA signaling regulators NPR1 and WRKY45 (Jiang et al., 2010). Finally, it has been demonstrated that strigolactones can induce SA accumulation (Omoarelojie et al., 2019). How these hormones intercept NHP levels and signaling remain unclear.
Apart from major hormone pathways, the growth-defense balance can be regulated by the Target of Rapamycin (TOR) kinase (De Vleesschauwer et al., 2018). TOR is a broadly conserved eukaryotic master regulator of growth and development (Shi et al., 2018). In rice, TOR aids growth and development at the expense of immunity by antagonizing SA and suppressing PTI (De Vleesschauwer et al., 2018). Increased SA-dependent responses were observed after TOR disruption genetically or pharmacologically, while overexpressing TOR resulted in downregulated SA-associated genes (De Vleesschauwer et al., 2018). Currently, the impact of TOR on NHP biosynthesis/signaling is unknown.
These studies altogether suggest a model that growth and developmental processes mechanistically impact the SA pathway. It would be intriguing to investigate whether NHP biosynthesis and signaling are similarly impacted by major growth hormones and TOR, and whether this occurs dependently or independently of SA. It would not be surprising to discover direct functional linkage of growth/developmental processes on NHP biosynthesis and signaling, since growth suppression is associated with NHP over-accumulation (Pastorczyk-Szlenkier and Bednarek, 2021) and the NHP pathway exhibits close mechanistic connections to SA (Zeier, 2021).
Increased SA and NHP levels through mutual potentiation lead to effective plant immunity against biotrophic and hemibiotrophic pathogens (Vlot et al., 2021; Zeier, 2021). Optimal defenses can sometimes result in tradeoffs to growth and development (Huot et al., 2014). Indeed, higher SA and NHP levels lead to dwarfed plants (Rivas-San Vicente and Plasencia, 2011; Cai et al., 2021). However, further studies on the broad conservation and/or specificity of SA/NHP-growth antagonism should be performed in other plant taxa. Notably, the NHP pathway and its role in SAR has been demonstrated in various plant species (Schnake et al., 2020). Although there is intensive crosstalk between SA and NHP, the impact of elevated NHP levels on plant physiology is largely unexplored. The additional dimensions of plant-microbiome and plant-environment interactions (Lebeis et al., 2015; Nazar et al., 2015; Pluhařová et al., 2019; Conesa et al., 2020) via the SA and NHP pathways remain low-hanging fruits, which can be facilitated by recent global datasets on microbiota assembly and hormone interactomes (Altmann et al., 2020; Trivedi et al., 2020).
Ultimately, the dream goal would be to optimize the plant’s growth-defense balance to maximize both yield and immune resilience (Mathan et al., 2016; Kim et al., 2021). Apart from tunable calibration of SA levels and signaling (van Butselaar and Van den Ackerveken, 2020), a potential avenue to bypass the growth-defense tradeoff may be optimally manipulating the NHP levels (Cai et al., 2021). Nevertheless, targeted engineering of this pathway still needs to be fully demonstrated and whether unforeseen collateral damage result from bypassing growth-defense tradeoffs must be investigated. These open questions and future directions highlight the exciting promise of elucidating and dissecting the mechanisms underpinning the equilibrium between plant growth and immunity.
CDMC conceptualized the review, supervised the research, and acquired funding. AS and VS surveyed the literature and synthesized the sources. AS, VS, and CDMC wrote the final version of the paper. All authors contributed to the article and approved the submitted version.
We are grateful for research funding from the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant, Canada Foundation for Innovation, Ontario Research Fund, and the Faculty of Science at Wilfrid Laurier University (to CDMC). We also acknowledge funding from the Mitacs Research Training Award (to VS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank members of the Castroverde Lab for meaningful discussions. We apologize to all authors whose work we could not cite because of space limitations.
Abreu, M. E., and Munné-Bosch, S. (2009). Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J. Exp. Bot. 60, 1261–1271. doi: 10.1093/jxb/ern363
Alonso-Ramírez, A., Rodríguez, D., Reyes, D., Jiménez, J. A., Nicolás, G., López-Climent, M., et al. (2009). Evidence for a role of gibberellins in salicylic acid-modulated early plant responses to abiotic stress in Arabidopsis seeds. Plant Physiol. 150, 1335–1344. doi: 10.1104/pp.109.139352
Altmann, M., Altmann, S., Rodriguez, P. A., Weller, B., Elorduy Vergara, L., Palme, J., et al. (2020). Extensive signal integration by the phytohormone protein network. Nature 583, 271–276. doi: 10.1038/s41586-020-2460-0
Argueso, C. T., Ferreira, F. J., Epple, P., To, J. P. C., Hutchison, C. E., Schaller, G. E., et al. (2012). Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. 8:e1002448. doi: 10.1371/journal.pgen.1002448
Bartsch, M., Gobbato, E., Bednarek, P., Debey, S., Schultze, J. L., Bautor, J., et al. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18, 1038–1051. doi: 10.1105/tpc.105.039982
Bauer, S., Mekonnen, D. W., Hartmann, M., Yildiz, I., Janowski, R., Lange, B., et al. (2021). UGT76B1, a promiscuous hub of small molecule-based immune signaling, glucosylates N-hydroxypipecolic acid, and balances plant immunity. Plant Cell 33, 714–734. doi: 10.1093/plcell/koaa044
Bernsdorff, F., Döring, A.-C., Gruner, K., Schuck, S., Bräutigam, A., and Zeier, J. (2016). Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 28, 102–129. doi: 10.1105/tpc.15.00496
Caddell, D.F., Louie, K., Bowen, B., Sievert, J.A., Hollingsworth, J., Dahlberg, J., et al. (2020). Drought shifts sorghum root metabolite and microbiome profiles and enriches the stress response factor pipecolic acid. bioRxiv [Preprint]. doi: 10.1101/2020.11.08.373399
Cai, J., Jozwiak, A., Holoidovsky, L., Meijler, M. M., Meir, S., Rogachev, I., et al. (2021). Glycosylation of N-hydroxy-pipecolic acid equilibrates between systemic acquired resistance response and plant growth. Mol. Plant 14, 440–455. doi: 10.1016/j.molp.2020.12.018
Carella, P., Wilson, D. C., and Cameron, R. K. (2014). Some things get better with age: differences in salicylic acid accumulation and defense signaling in young and mature Arabidopsis. Front. Plant Sci. 5:775. doi: 10.3389/fpls.2014.00775
Carviel, J. L., Al-Daoud, F., Neumann, M., Mohammad, A., Provart, N. J., Moeder, W., et al. (2009). Forward and reverse genetics to identify genes involved in the age-related resistance response in Arabidopsis thaliana. Mol. Plant Pathol. 10, 621–634. doi: 10.1111/j.1364-3703.2009.00557.x
Castroverde, C. D. M., and Dina, D. (2021). Temperature regulation of plant hormone signaling during stress and development. J. Exp. Bot. 72, 7436–7458. doi: 10.1093/jxb/erab257
Cecchini, N. M., Jung, H. W., Engle, N. L., Tschaplinski, T. J., and Greenberg, J. T. (2015). Ald1 regulates basal immune components and early inducible defense responses in arabidopsis. Mol. Plant. Microbe. Interact. 28, 455–466. doi: 10.1094/MPMI-06-14-0187-R
Chen, Z., Zheng, Z., Huang, J., Lai, Z., and Fan, B. (2009). Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 4, 493–496. doi: 10.4161/psb.4.6.8392
Coley, P. D., Bryant, J. P., and Chapin, F. S. (1985). Resource availability and plant antiherbivore defense. Science 230, 895–900. doi: 10.1126/science.230.4728.895
Conesa, C. M., Saez, A., Navarro-Neila, S., de Lorenzo, L., Hunt, A. G., Sepúlveda, E. B., et al. (2020). Alternative polyadenylation and salicylic acid modulate root responses to low nitrogen availability. Plan. Theory 9:251. doi: 10.3390/plants9020251
De Vleesschauwer, D., Van Buyten, E., Satoh, K., Balidion, J., Mauleon, R., Choi, I.-R., et al. (2012). Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158, 1833–1846. doi: 10.1104/pp.112.193672
De Vleesschauwer, D., Filipe, O., Hoffman, G., Seifi, H. S., Haeck, A., Canlas, P., et al. (2018). Target of rapamycin signaling orchestrates growth–defense trade-offs in plants. New Phytol. 217, 305–319. doi: 10.1111/nph.14785
DeFalco, T. A., and Zipfel, C. (2021). Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell 81, 3449–3467. doi: 10.1016/j.molcel.2021.07.029
Dempsey, D. A., Vlot, A. C., Wildermuth, M. C., and Klessig, D. F. (2011). Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9:e0156. doi: 10.1199/tab.0156
Ding, P., and Ding, Y. (2020). Stories of salicylic acid: a plant defense hormone. Trends Plant Sci. 25, 549–565. doi: 10.1016/j.tplants.2020.01.004
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell. 173, 1454–1467.e15. doi: 10.1016/j.cell.2018.03.044
Dong, C.-J., Liu, X.-Y., Xie, L.-L., Wang, L.-L., and Shang, Q.-M. (2020). Salicylic acid regulates adventitious root formation via competitive inhibition of the auxin conjugation enzyme CsGH3.5 in cucumber hypocotyls. Planta 252:75. doi: 10.1007/s00425-020-03467-2
Du, Y., Tejos, R., Beck, M., Himschoot, E., Li, H., Robatzek, S., et al. (2013). Salicylic acid interferes with clathrin-mediated endocytic protein trafficking. Proc. Natl. Acad. Sci. U. S. A. 110, 7946–7951. doi: 10.1073/pnas.1220205110
Emamverdian, A., Ding, Y., and Mokhberdoran, F. (2020). The role of salicylic acid and gibberellin signaling in plant responses to abiotic stress with an emphasis on heavy metals. Plant Signal. Behav. 15:1777372. doi: 10.1080/15592324.2020.1777372
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Garcion, C., Lohmann, A., Lamodiere, E., Catinot, J., Buchala, A., Doermann, P., et al. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147, 1279–1287. doi: 10.1104/pp.108.119420
Gouesbet, G., Jebbar, M., Talibart, R., Bernard, T., and Blanco, C. (1994). Pipecolic acid is an osmoprotectant for Escherichia coli taken up by the general osmoporters ProU and ProP. Microbiology 140, 2415–2422. doi: 10.1099/13500872-140-9-2415
Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. doi: 10.1023/A:1015207114117
Hartmann, M., and Zeier, J. (2018). L-lysine metabolism to N-hydroxypipecolic acid: an integral immune-activating pathway in plants. Plant J. 96, 5–21. doi: 10.1111/tpj.14037
Hartmann, M., and Zeier, J. (2019). N-hydroxypipecolic acid and salicylic acid: a metabolic duo for systemic acquired resistance. Curr. Opin. Plant Biol. 50, 44–57. doi: 10.1016/j.pbi.2019.02.006
Hartmann, M., Zeier, T., Bernsdorff, F., Reichel-Deland, V., Kim, D., Hohmann, M., et al. (2018). Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173, 456.e16–469.e16. doi: 10.1016/j.cell.2018.02.049
Holmes, E. C., Chen, Y. C., Mudgett, M. B., and Sattely, E. S. (2021). Arabidopsis UGT76B1 glycosylates N-hydroxy-pipecolic acid and inactivates systemic acquired resistance in tomato. Plant Cell 33, 750–765. doi: 10.1093/plcell/koaa052
Huang, P., Dong, Z., Guo, P., Zhang, X., Qiu, Y., Li, B., et al. (2020b). Salicylic acid suppresses apical hook formation via NPR1-mediated repression of EIN3 and EIL1 in Arabidopsis. Plant Cell 32, 612–629. doi: 10.1105/tpc.19.00658
Huang, W., Wang, Y., Li, X., and Zhang, Y. (2020a). Biosynthesis and regulation of salicylic acid and N-hydroxypipecolic acid in plant immunity. Mol. Plant 13, 31–41. doi: 10.1016/j.molp.2019.12.008
Huot, B., Yao, J., Montgomery, B. L., and He, S. Y. (2014). Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287. doi: 10.1093/mp/ssu049
Jiang, C.-J., Shimono, M., Sugano, S., Kojima, M., Liu, X., Inoue, H., et al. (2013). Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Mol. Plant-Microbe Interact. 26, 287–296. doi: 10.1094/MPMI-06-12-0152-R
Jiang, C.-J., Shimono, M., Sugano, S., Kojima, M., Yazawa, K., Yoshida, R., et al. (2010). Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Mol. Plant-Microbe Interact. 23, 791–798. doi: 10.1094/MPMI-23-6-0791
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kesarwani, M., Yoo, J., and Dong, X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144, 336–346. doi: 10.1104/pp.106.095299
Kim, J. H., and Castroverde, C. D. M. (2020). Diversity, function and regulation of cell surface and intracellular immune receptors in Solanaceae. Plan. Theory 9:434. doi: 10.3390/plants9040434
Kim, J. H., Hilleary, R., Seroka, A., and He, S. Y. (2021). Crops of the future: building a climate-resilient plant immune system. Curr. Opin. Plant Biol. 60:101997. doi: 10.1016/j.pbi.2020.101997
Kliebenstein, D. J. (2016). False idolatry of the mythical growth versus immunity tradeoff in molecular systems plant pathology. Physiol. Mol. Plant Pathol. 95, 55–59. doi: 10.1016/j.pmpp.2016.02.004
Koo, Y. M., Heo, A. Y., and Choi, H. W. (2020). Salicylic acid as a safe plant protector and growth regulator. Plant Pathol. J. 36, 1–10. doi: 10.5423/PPJ.RW.12.2019.0295
Lavy, M., and Estelle, M. (2016). Mechanisms of auxin signaling. Development 143, 3226–3229. doi: 10.1242/dev.131870
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. doi: 10.1126/science.aaa8764
Li, D., Liu, R., Singh, D., Yuan, X., Kachroo, P., and Raina, R. (2020). JMJ14 encoded H3K4 demethylase modulates immune responses by regulating defence gene expression and pipecolic acid levels. New Phytol. 225, 2108–2121. doi: 10.1111/nph.16270
Li, B., Meng, X., Shan, L., and He, P. (2016). Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 19, 641–650. doi: 10.1016/j.chom.2016.04.011
Lim, G. H., Liu, H., Yu, K., Liu, R., Shine, M. B., Fernandez, J., et al. (2020). The plant cuticle regulates apoplastic transport of salicylic acid during systemic acquired resistance. Sci. Adv. 6:eaaz0478. doi: 10.1126/sciadv.aaz0478
Liu, J., Li, L., Yuan, F., and Chen, M. (2019). Exogenous salicylic acid improves the germination of Limonium bicolor seeds under salt stress. Plant Signal. Behav. 14:e1644595. doi: 10.1080/15592324.2019.1644595
Liu, Y., Sun, T., Sun, Y., Zhang, Y., Radojičić, A., Ding, Y., et al. (2020). Diverse roles of the salicylic acid receptors NPR1 and NPR3/NPR4 in plant immunity. Plant Cell 32, 4002–4016. doi: 10.1105/tpc.20.00499
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Mathan, J., Bhattacharya, J., and Ranjan, A. (2016). Enhancing crop yield by optimizing plant developmental features. Development 143, 3283–3294. doi: 10.1242/dev.134072
Mishina, T. E., and Zeier, J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141, 1666–1675. doi: 10.1104/pp.106.081257
Mohnike, L., Rekhter, D., Huang, W., Feussner, K., Tian, H., Herrfurth, C., et al. (2021). The glycosyltransferase UGT76B1 modulates N-hydroxy-pipecolic acid homeostasis and plant immunity. Plant Cell 33, 735–749. doi: 10.1093/plcell/koaa045
Moulin, M., Deleu, C., Larher, F., and Bouchereaum, A. (2006). The lysine-ketoglutarate reductase-saccharopine dehydrogenase is involved in the osmo-induced synthesis of pipecolic acid in rapeseed leaf tissues. Plant Physiol. Biochem. 44, 474–482. doi: 10.1016/j.plaphy.2006.08.005
Nair, A., Goyal, I., Voß, E., Mrozek, P., Prajapati, S., Thurow, C., et al. (2021). N-hydroxypipecolic acid-induced transcription requires the salicylic acid signaling pathway at basal SA levels. Plant Physiol. 187, 2803–2819. doi: 10.1093/plphys/kiab433
Návarová, H., Bernsdorff, F., Döring, A. C., and Zeier, J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24, 5123–5141. doi: 10.1105/tpc.112.103564
Nawrath, C., and Metraux, J. (1999). Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. doi: 10.1105/tpc.11.8.1393
Nazar, R., Umar, S., and Khan, N. A. (2015). Exogenous salicylic acid improves photosynthesis and growth through increase in ascorbate-glutathione metabolism and S assimilation in mustard under salt stress. Plant Signal. Behav. 10:e1003751. doi: 10.1080/15592324.2014.1003751
O’Brien, J. A., and Benková, E. (2013). Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 4:451. doi: 10.3389/fpls.2013.00451
Omoarelojie, L. O., Kulkarni, M. G., Finnie, J. F., and Van Staden, J. (2019). Strigolactones and their crosstalk with other phytohormones. Ann. Bot. 124, 749–767. doi: 10.1093/aob/mcz100
Pasternak, T., Groot, E. P., Kazantsev, F. V., Teale, W., Omelyanchuk, N., Kovrizhnykh, V., et al. (2019). Salicylic acid affects root meristem patterning via auxin distribution in a concentration-dependent manner. Plant Physiol. 180, 1725–1739. doi: 10.1104/pp.19.00130
Pastorczyk-Szlenkier, M., and Bednarek, P. (2021). UGT76B1 controls the growth-immunity trade-off during systemic acquired resistance. Mol. Plant 14, 544–546. doi: 10.1016/j.molp.2021.03.012
Pérez-García, F., Brito, L. F., and Wendisch, V. F. (2019). Function of L-pipecolic acid as compatible solute in Corynebacterium glutamicum as basis for its production under hyperosmolar conditions. Front. Microbiol. 10:340. doi: 10.3389/fmicb.2019.00340
Pluhařová, K., Leontovyčová, H., Stoudková, V., Pospíchalová, R., Maršík, P., Klouček, P., et al. (2019). “Salicylic acid mutant collection” as a tool to explore the role of salicylic acid in regulation of plant growth under a changing environment. Int. J. Mol. Sci. 20:6365. doi: 10.3390/ijms20246365
Pokotylo, I., Hodges, M., Kravets, V., and Ruelland, E. (2021). A ménage à trois: salicylic acid, growth inhibition, and immunity. Trends Plant Sci. doi: 10.1016/j.tplants.2021.11.008
Rajjou, L., Belghazi, M., Huguet, R., Robin, C., Moreau, A., Job, C., et al. (2006). Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol. 141, 910–923. doi: 10.1104/pp.106.082057
Rekhter, D., Lüdke, D., Ding, Y., Feussner, K., Zienkiewicz, K., Lipka, V., et al. (2019). Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 365, 498–502. doi: 10.1126/science.aaw1720
Robert-Seilaniantz, A., MacLean, D., Jikumaru, Y., Hill, L., Yamaguchi, S., Kamiya, Y., et al. (2011). The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 67, 218–231. doi: 10.1111/j.1365-313X.2011.04591.x
Rong, D., Luo, N., Mollet, J. C., Liu, X., and Yang, Z. (2016). Salicylic acid regulates pollen tip growth through an NPR3/NPR4-independent pathway. Mol. Plant 9, 1478–1491. doi: 10.1016/j.molp.2016.07.010
Saleem, M., Fariduddin, Q., and Castroverde, C. D. M. (2021). Salicylic acid: a key regulator of redox signalling and plant immunity. Plant Physiol. Biochem. 168, 381–397. doi: 10.1016/j.plaphy.2021.10.011
Saur, I. M. L., Panstruga, R., and Schulze-Lefert, P. (2021). NOD-like receptor-mediated plant immunity: from structure to cell death. Nat. Rev. Immunol. 21, 305–318. doi: 10.1038/s41577-020-00473-z
Schnake, A., Hartmann, M., Schreiber, S., Malik, J., Brahmann, L., Yildiz, I., et al. (2020). Inducible biosynthesis and immune function of the systemic acquired resistance inducer N-hydroxypipecolic acid in monocotyledonous and dicotyledonous plants. J. Exp. Bot. 71, 6444–6459. doi: 10.1093/jxb/eraa317
Shi, L., Wu, Y., and Sheen, J. (2018). TOR signaling in plants: conservation and innovation. Development 145:dev160887. doi: 10.1242/dev.160887
Singh, A., Lim, G. H., and Kachroo, P. (2017). Transport of chemical signals in systemic acquired resistance. J. Integr. Plant Biol. 59, 336–344. doi: 10.1111/jipb.12537
Sun, T., Huang, J., Xu, Y., Verma, V., Jing, B., Sun, Y., et al. (2020). Redundant CAMTA transcription factors negatively regulate the biosynthesis of salicylic acid and N-hydroxypipecolic acid by modulating the expression of SARD1 and CBP60g. Mol. Plant 13, 144–156. doi: 10.1016/j.molp.2019.10.016
Sun, T., Li, Y., Zhang, Q., Ding, Y., Zhang, Y., and Zhang, Y. (2015). ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 6, 1–12. doi: 10.1038/ncomms10159
Sun, T., Liang, W., Zhang, Y., and Li, X. (2018). Negative regulation of resistance protein-mediated immunity by master transcription factors SARD1 and CBP60g. J. Integr. Plant Biol. 60, 1023–1027. doi: 10.1111/jipb.12698
Tan, S., Abas, M., Verstraeten, I., Glanc, M., Molnár, G., Hajný, J., et al. (2020). Salicylic acid targets protein phosphatase 2A to attenuate growth in plants. Curr. Biol. 30, 381.e8–395.e8. doi: 10.1016/j.cub.2019.11.058
Torrens-Spence, M. P., Bobokalonova, A., Carballo, V., Glinkerman, C. M., Pluskal, T., Shen, A., et al. (2019). PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Mol. Plant 12, 1577–1586. doi: 10.1016/j.molp.2019.11.005
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2020). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
van Butselaar, T., and Van den Ackerveken, G. (2020). Salicylic acid steers the growth-immunity tradeoff. Trends Plant Sci. 25, 566–576. doi: 10.1016/j.tplants.2020.02.002
Vernooij, B., Friedrich, L., Morse, A., Reist, R., Kolditz-Jawhar, R., Ward, E., et al. (1994). Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6, 959–965. doi: 10.1105/tpc.6.7.959
Vicente, M. R.-S., and Plasencia, J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. doi: 10.1093/jxb/err031
Vlot, A. C., Sales, J. H., Lenk, M., Bauer, K., Brambilla, A., Sommer, A., et al. (2021). Systemic propagation of immunity in plants. New Phytol. 229, 1234–1250. doi: 10.1111/nph.16953
Wang, C., Liu, R., Lim, G. H., de Lorenzo, L., Yu, K., Zhang, K., et al. (2018). Pipecolic acid confers systemic immunity by regulating free radicals. Sci. Adv. 4:eaar4509. doi: 10.1126/sciadv.aar4509
Wang, D., Pajerowska-Mukhtar, K., Culler, A. H., and Dong, X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790. doi: 10.1016/j.cub.2007.09.025
Wang, Z., Rong, D., Chen, D., Xiao, Y., Liu, R., Wu, S., et al. (2021). Salicylic acid promotes quiescent center cell division through ROS accumulation and down-regulation of PLT1, PLT2, and WOX5. J. Integr. Plant Biol. 63, 583–596. doi: 10.1111/jipb.13020
Wang, L., Tsuda, K., Truman, W., Sato, M., Nguyen, L., Katagiri, F., et al. (2011). CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 67, 1029–1041. doi: 10.1111/j.1365-313X.2011.04655.x
Wendehenne, D., Gao, Q. M., Kachroo, A., and Kachroo, P. (2014). Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 20, 127–134. doi: 10.1016/j.pbi.2014.05.012
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. doi: 10.1016/j.celrep.2012.05.008
Xiang, C., Miao, Z., and Lam, E. (1997). DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 34, 403–415. doi: 10.1023/A:1005873500238
Yatsu, L., and Boynton, D. (1959). Pipecolic acid in leaves of strawberry plant as influenced by treatments affecting growth. Science 130, 864–865. doi: 10.1126/science.130.3379.864
Yildiz, I., Mantz, M., Hartmann, M., Zeier, T., Kessel, J., Thurow, C., et al. (2021). The mobile SAR signal N-hydroxypipecolic acid induces NPR1-dependent transcriptional reprogramming and immune priming. Plant Physiol. 186, 1679–1705. doi: 10.1093/plphys/kiab166
Yuan, H. M., Liu, W. C., and Lu, Y. T. (2017). CATALASE2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe 21, 143–155. doi: 10.1016/j.chom.2017.01.007
Yuan, M., Ngou, B. P. M., Ding, P., and Xin, X. F. (2021). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62:102030. doi: 10.1016/j.pbi.2021.102030
Zebell, S. G., and Dong, X. (2015). Cell cycle regulators and cell death in immunity. Cell Host Microbe 18, 402–407. doi: 10.1016/j.chom.2015.10.001
Zeier, J. (2021). Metabolic regulation of systemic acquired resistance. Curr. Opin. Plant Biol. 62:102050. doi: 10.1016/j.pbi.2021.102050
Zhang, Y., and Li, X. (2019). Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 50, 29–36. doi: 10.1016/j.pbi.2019.02.004
Zhang, Y., Tessaro, M. J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15, 2647–2653. doi: 10.1105/tpc.014894
Keywords: salicylic acid, N-hydroxypipecolic acid, pipecolic acid, plant immunity, plant growth, plant development, plant hormone, growth-defense tradeoff
Citation: Shields A, Shivnauth V and Castroverde CDM (2022) Salicylic Acid and N-Hydroxypipecolic Acid at the Fulcrum of the Plant Immunity-Growth Equilibrium. Front. Plant Sci. 13:841688. doi: 10.3389/fpls.2022.841688
Received: 22 December 2021; Accepted: 14 February 2022;
Published: 10 March 2022.
Edited by:
Ian T. Major, Michigan State University, United StatesReviewed by:
Cristiana T. Argueso, Colorado State University, United StatesCopyright © 2022 Shields, Shivnauth and Castroverde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Danve M. Castroverde, ZGNhc3Ryb3ZlcmRlQHdsdS5jYQ==, orcid.org/0000-0002-9982-8451
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.