
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 02 March 2022
Sec. Plant Membrane Traffic and Transport
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.841366
This article is part of the Research Topic Metal Transport in Plants, Volume II View all 8 articles
Iron (Fe) is an essential micronutrient element for all organisms including plants. Chlorosis of young leaves is a common symptom of Fe deficiency, reducing the efficiency of photosynthesis, and, ultimately, crop yield. Previous research revealed strong responsiveness of the putative key transcription factor ERF109 to the Fe regime. To elucidate the possible role of ERF109 in leaf Fe homeostasis and photosynthesis, we subjected Arabidopsis thaliana erf109 knockout lines and Col-0 wild-type plants to transcriptome profiling via RNA-seq. The transcriptome profile of Fe-sufficient erf109 leaves showed a 71% overlap with Fe-deficient Col-0 plants. On the other hand, genes that were differentially expressed between Fe-deficient and Fe-sufficient Col-0 plants remained unchanged in erf109 plants under conditions of Fe deficiency. Mutations in ERF109 increased the expression of the clade Ib bHLH proteins bHLH38, bHLH39, bHLH101, the nicotianamine synthase NAS4, and the Fe storage gene FER1. Moreover, mutations in ERF109 led to significant down-regulation of defense genes, including CML37, WRKY40, ERF13, and EXO70B2. Leaves of erf109 exhibited increased Fe levels under both Fe-sufficient and Fe-deficient conditions. Reduced Fv/Fm and Soil Plant Analysis Development (SPAD) values in erf109 lines under Fe deficiency indicate curtailed ability of photosynthesis relative to the wild-type. Our findings suggest that ERF109 is a negative regulator of the leaf response to Fe deficiency. It further appears that the function of ERF109 in the Fe response is critical for regulating pathogen defense and photosynthetic efficiency. Taken together, our study reveals a novel function of ERF109 and provides a systematic perspective on the intertwining of the immunity regulatory network and cellular Fe homeostasis.
By virtue of its ability to change valency, iron (Fe) is a critical component of photosynthesis and respiratory electron transport, a constituent of Fe-sulfur clusters, and a cofactor of a multitude of vital redox enzymes. Owing to the chemical characteristic of Fe, free Fe ions in plant cells are highly redox active and can react with H2O2 to produce the reactive hydroxyl (⋅OH) radical in the so-called Fenton reaction, which can cause oxidative stress and cell damage when produced in excess (Floyd and Lewis, 1983; Baker and Gebicki, 1986). The most significant symptom of Fe deficiency in plants is interveinal chlorosis of young leaves. In Fe-deficient plants, compromised chlorophyll production reduces the efficiency of photosynthesis and causes a decrease in fruit yield and quality (Àlvarez-Fernàndez et al., 2006; Rombolà and Tagliavini, 2006). Insufficient Fe supply decreases the level of the electron donor cytochrome c6 and was shown to partially block the electron transfer between PSII and PSI in the marine diatom Phaeodactylum tricornutum (Roncel et al., 2016). Similar to Fe deficiency, high levels of light irradiation cause oxidative stress in plant cells (Erickson et al., 2015). When the light intensity exceeds the photosynthetic capacity of the plant, excessive energy can induce light inhibition of photosynthesis and cause the formation of excessive ROS species, resulting in leaf cell death (Karpiński et al., 2013). Systemic Acquired Acclimation (SAA) is induced by the exposure of leaves to high light stress, which triggers systemic signaling and preacclimation of shaded leaves (Rossel et al., 2007).
ETHYLENE-RESPONSIVE TRANSCRIPTION FACTOR109 (ERF109), also described as REDOX-RESPONSIVE TRANSCRIPTION FACTOR1 (RRTF1), is a member of the ERF/AP2 transcription factor family that is involved in a multitude of abiotic and biotic stresses, such as salt (Bahieldin et al., 2016, 2018; Soliman and Meyer, 2019), high light (Vogel et al., 2014; Carmody et al., 2016; Huang et al., 2017), and oxidative stress (Khandelwal et al., 2008; Matsuo et al., 2015; Pospisil, 2016; Kong et al., 2018), infection with Alternaria brassicae (Vahabi et al., 2018), as well as in essential processes such as hormone signaling and root stem cell maintenance (Cai et al., 2014; Kong et al., 2018; Vahabi et al., 2018). It was confirmed that high light stress-induced chloroplast singlet oxygen stress is the beginning of a systemic domestication reaction, transmitting a signal to the cell nucleus that regulates ERF109 expression, and, subsequently, generates signal fluctuations between cells through the plasma membrane protein RBOHD/F (Carmody et al., 2016). Hence, the availability of Fe and the Fe nutritional status of the plant are important factors for light responsiveness. Furthermore, chloroplast retrograde and ethylene signaling were shown to be connected with Fe homeostasis (Balparda et al., 2020). Here, we attempt to validate the supposition that ERF109 plays a critical role in the coordination of Fe deficiency, light signaling, and pathogen defense. Transcriptome analysis revealed that lines harboring defects in ERF109 exhibited a similar pattern of a constitutively expressed subset of genes associated with the immune network, resembling the Fe-deficiency response of leaves of wild-type plants. In addition, erf109 mutant plants accumulated higher Fe levels than the wild-type and constitutively induced several Fe deficiency response genes in shoots, suggesting that ERF109 is a key node in the regulation of Fe-responsive genes in above-ground plant parts.
The Arabidopsis thaliana Col-0 ecotype and erf109 knock-out mutant lines (SALK_150614) were obtained from the Arabidopsis Biological Resource Center. Seeding holder was made by punching 96 holes on a foam board and filled by the hydroponic solution which solidified with 1% Agargel (A3301, Sigma, MO, United States) containing essential nutrients as described (Rodríguez-Celma et al., 2013), which included 5 mM KNO3, 2 mM MgSO4, 2 mM Ca(NO3)2, 2.5 mM KH2PO4, 70 μM H3BO3, 14 μM MnCl2, 1 μM ZnSO4, 0.5 μM CuSO4, 10 μM NaCl, 0.2 μM Na2MoO4, 40 μM FeEDTA, 4.7 mM MES, with pH 5.7. Then, the seeding holders were floated on the hydroponic solution inside boxes. Seeds were stratified on seeding holders at 4°C in the dark for 1 day and subsequently transferred to acclimatize under continuous light (90 μmol m–2 s–1) and were grown at 21°C in a growth chamber. The hydroponic solution was renewed every 3 days. Plants were transferred to Fe-sufficient or Fe-deficient (−Fe, 0 μM Fe2+ supplemented with 100 μM 3-(2-pyridyl)-5,6diphenyl-1,2,4-triazine sulfonate to chelate trace iron) hydroponic solution for treatment. Shoot tissue was harvested for all experiments.
For RNA-seq and quantitative RT-PCR (RT-qPCR) analysis, Col-0 and erf109 plants were grown in hydroponic solution for 10 days and transferred to Fe-sufficient or Fe-deficient hydroponic solution for an additional 3 days. Three replications for each treatment and genotype were applied for data collection. For Fe concentration analysis, plants were grown in hydroponic solution for 18 days under continuous light and transferred to Fe-sufficient or Fe-deficient solution under continuous light for 3 days. Two replications for each treatment were applied for data collection. For chlorophyll fluorescence and chlorophyll content measurements, plants were grown in Fe-sufficient hydroponic solution for 14 days and transferred to Fe-sufficient or Fe-deficient solution under continuous light for 3 days. Three replications for each treatment were applied for data collection.
For RNA-seq, RNA extraction, sequencing, and annotation were conducted as described (Rodríguez-Celma et al., 2013). Total RNA was extracted from shoots of Col-0 and erf109 plants using the RNeasy Plant Mini Kit (Qiagen), following the manufacturer’s instructions. The cDNA libraries were constructed with equal amounts of total RNA following the manufacturer’s protocol (Illumina, CA, United States), enriched by PCR amplification and subjected to paired-end sequencing on an Illumina Genome Analyzer II. Data collection was conducted as previously described (Mortazavi et al., 2008). Reads from RNA-seq were mapped to the Arabidopsis genome version TAIR10 by Bowtie2 (Langmead and Salzberg, 2012), other unmappable reads were mapped to Arabidopsis genome version TAIR10 using BLAT (Kent, 2002). Read counts were calculated and normalized by means of RackJ package1 and the TMM-quantile method (Robinson and Oshlack, 2010), respectively. Normalized read counts were further converted into RPKM (Reads Per Kilobase Million) values.
To obtain a comprehensive catalog of differentially expressed genes (DEGs), three subsets of DEGs were selected based on mean RPKM compared to control with a Z-test P-value < 0.05: “Col-Fe” indicates genes that were differentially expressed between Fe-deficient and Fe-sufficient Col-0 plants, “erf109-Fe” indicates genes that were differentially expressed between Fe-deficient and Fe-sufficient erf109 mutant plants, and “erf109-regulon” indicates genes that were differentially expressed between erf109 and Col-0 plants under Fe-sufficient conditions.
For DEG clustering, the DEGs of ‘‘Col-Fe,’’ ‘‘erf109-Fe,’’ and ‘‘erf109-regulon’’ were used as input for the MACCU software2 (Lin et al., 2011) to build co-expression networks. The network was based on co-expression relationships with a Pearson’s coefficient greater than or equal to 0.8. The network was visualized using Cytoscape software ver. 3.7.2.3 The color was set to show up-regulated (red) and down-regulated (blue) DEGs from log2 fold-change ranging from 2 to −2.
Differentially expressed genes with the same regulation pattern in “Col-Fe” and in the “erf109-regulon” were selected for functional analysis using the singular enrichment analysis tool in agriGO (Tian et al., 2017), which was applied with the default parameters for A. thaliana. The biological process result was visualized using the REVIGO (Supek et al., 2011) tool with the default parameters for A. thaliana GO terms. All the DEGs of “Col-Fe,” “erf109-Fe,” and “erf109-regulon” were put into the MapMan software (Thimm et al., 2004) with log2 fold-change values for visualizing functional categories. The “Ath_AGI_TAIR9_Jan2010” database in MapMan was applied for mapping. The heatmaps were plotted in R (version 4.1.0).
Total RNA was extracted with TRIzol™ Reagent (Invitrogen, MA, United States) following the manufacturer’s instructions. The HiScript II 1st Strand cDNA Synthesis Kit (Bionovas, Toronto, ON, Canada) was used for cDNA preparation. RT-qPCR was performed on the CFX Connect Real-Time PCR Detection System (Bio-Rad, CA, United States) using iQ™ SYBR ® Green Supermix (Bio-Rad, CA, United States). For each reaction, 0.3 μM of the forward and the reverse primers and 100 ng of cDNA template were added. All primers used in this study are listed in Supplementary Table 3. The relative gene expression level was determined according to the 2–ΔΔCt method (Livak and Schmittgen, 2001) and fold-changes were calculated relative to Col-0 values. For each sample, the mRNA abundances of the target genes were normalized to those of the UBIQTIN10 (At4g05320) gene. Three biological replicates were run for each gene. The final figures were plotted in R (version 4.1.0).
Shoots were rinsed with 1% HCl and at least three times with deionized water, dried at 100°C for 1 h, and kept at 70°C for 3 days. NIST SRM 1573a (tomato leaves) served as a standard. Dried shoots were weighed into a Teflon vessel and digested with 5 ml of 65% HNO3 (Merck, Darmstadt, Germany) and 2 ml H2O2 (Choneye Pure Chemicals, Taipei, Taiwan) in a MarsXpress microwave digestion system (MARS 5 Xpress; CEM). ICP-optical emission spectrometry (ICP-OES; PerkinElmer, MA, United States) was used for metal element analyses. Determination of metal concentration was conducted as described (Shanmugam et al., 2011). The final figures were plotted in R (version 4.1.0).
The plant material was acclimated in the dark for 20 min before the measurements. Chlorophyll fluorescence was determined at room temperature using a PAM-2100 (Heinz Walz GmbH, Effeltrich, Germany) fluorometer. Minimal (Fo) and maximal (Sies and Menck, 1992) fluorescence yields of dark-adapted samples were recorded after applying a saturating pulse of light. Maximal quantum yield of photosystem II (Fv/Fm) was calculated from the given Fo and Fm-values using the equation: Fv:Fm = (Fm−Fo):Fm. The effective quantum yield of photosystem II (yield) was calculated according to the equation: Y = (Fm′−Ft): Fm′ = ΔF: Fm′. A Soil Plant Analysis Development (SPAD) chlorophyll meter (Spectrum Technologies, Inc., IL, United States) was applied to measure the chlorophyll content in each line. The SPAD value represents the index of relative chlorophyll content (Richardson et al., 2002). The final figures were plotted in R (version 4.1.0).
Data were analyzed by applying a Student’s t-test with a P-value < 0.05.
To explore the effects of ERF109 on the Fe content and light responsiveness in leaves, we first collected phenotypic data of erf109 mutant plants. After 72 h of Fe deficiency, no visible differences between the mutant and its wild-type were observed (Figure 1A). However, the Fe concentration of both Col-0 and erf109 plants was decreased in response to Fe deficiency (Figure 1B). Notably, erf109 mutant plants exhibited significantly higher Fe levels than the wild-type under both growth conditions (Figure 1B). The maximum quantum yield of photosystem II (Fv/Fm) was reduced upon Fe-deficiency in both Col-0 and erf109 plants, with erf109 plants exhibiting significantly lower values than Col-0 under Fe-deficiency (Figure 1C). A similar pattern was observed for the quantum yield of PSI and yield, the difference between the erf109 and Fe-deficiency Col-0 was, however, not statistically significant (Figure 1D). SPAD values, providing a read-out for the chlorophyll content, were reduced in both genotypes under Fe-deficient conditions, with significantly lower values in erf109 plants under Fe-sufficient and Fe-deficient conditions (Figure 1E). Together, these results indicate that in erf109 plants the chlorophyll content and photosynthetic efficiency was decreased relative to the wild-type despite higher Fe concentrations in the leaves even under Fe-sufficient conditions, suggesting that the additional Fe may be not physiologically available.
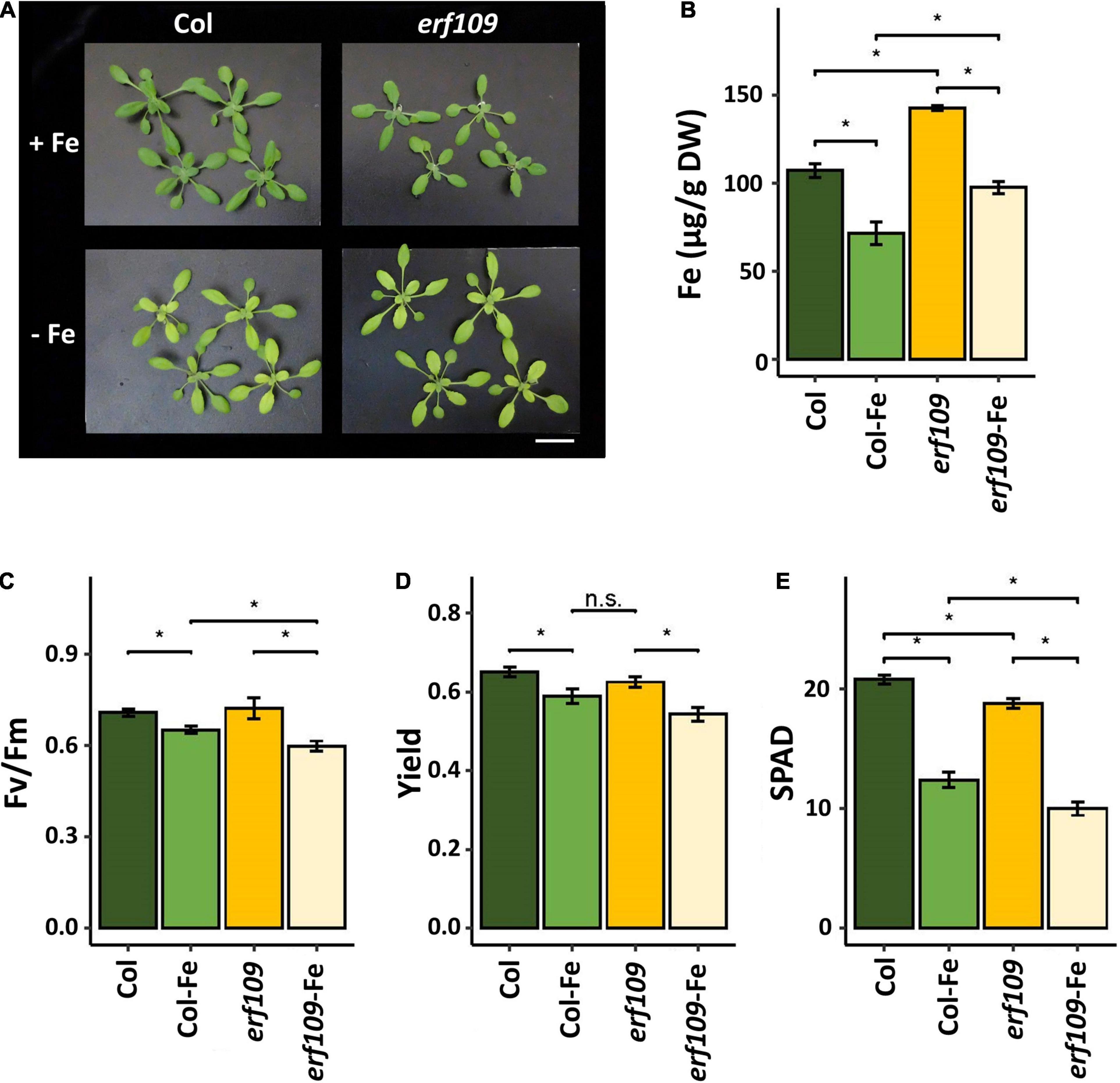
Figure 1. Phenotype of erf109 mutant plants and Col-0 wild-type. (A) Growth (21-day-old plants), (B) Fe concentration, (C) Fv/Fm ratio, (D) quantum yields of PSI, and (E) SPAD value of chlorophyll content of Col-0 and erf109 under Fe sufficiency (Col, erf109) or Fe deficiency (Col-Fe, erf109-Fe). Statistically significant difference was conducted by Student’s t-test at P < 0.05 between either two selected treatment or genotype, marked as asterisk. n.s., no significant; +Fe, iron sufficient condition; –Fe, iron-deficiency treatment. Bar, 5 cm.
To uncover a possible role of ERF109 in the Fe deficiency response of Arabidopsis shoots, we subjected wild-type plants and erf109 mutant lines to transcriptional profiling using RNA-seq technology. Genes that were differentially expressed between the mutant and its wild-type are here referred to as the erf109-regulon, while Fe-responsive genes of the wild-type and mutant plants were designated Col-Fe and erf109-Fe, respectively. To explore the global regulation pattern of the three DEG subsets, heatmaps of log fold-change values were generated (Figure 2A). Conspicuously, highly similar expression patterns were observed for Col-Fe and the erf109-regulon. A subset of 681 of the total 955 DEGs in the erf109-regulon overlapped with the Col-Fe DEGs (Figure 2B), with 149 up-regulated and 515 down-regulated genes in both Col-Fe and the erf109-regulon (Figure 2D). The significant overlap and the similar direction of gene regulation in Fe-deficient wild-type plants and erf109 mutants indicate a partial constitutive Fe deficiency response in shoots of erf109 mutant plants. Under Fe-deficient conditions, a subset of 416 out of 1,654 DEGs that responded to Fe starvation in the wild-type were also responsive to the Fe regime in erf109 mutants; the majority of Col-Fe genes (1,238 DEGs), however, were not affected in the mutant (Figure 2C). The subset of overlapping DEGs revealed a similar direction of expression in both Col-Fe and erf109-Fe, with 124 genes that were induced and 273 genes that were repressed under Fe starvation (Figure 2E). From the 1,238 Col-0-specific Fe-responsive DEGs that were not affected in erf109 mutant plants under Fe deficiency, 103 induced and 433 repressed genes were regulated similarly in the erf109-regulon but lost their differential expression in the erf109-Fe dataset (Figure 2F). The function of the 681 DEGs that were comprised in both the Col-Fe subset and the erf-109 regulon were further analyzed by their gene ontology. Among these genes, 149 up-regulated DEGs are involved in the response to abiotic stimulus and cellular nitrogen compound metabolism (Figure 2G), and 515 down-regulated DEGs are associated with the response to biotic stimulus, wounding, chitin, and the immune system (Figure 2H). These results indicate that knock-out of ERF109 induces gene expression patterns that were to a large part similar to those observed in Fe-deficient wild-type plants and predicted to be involved in establishing immunity.
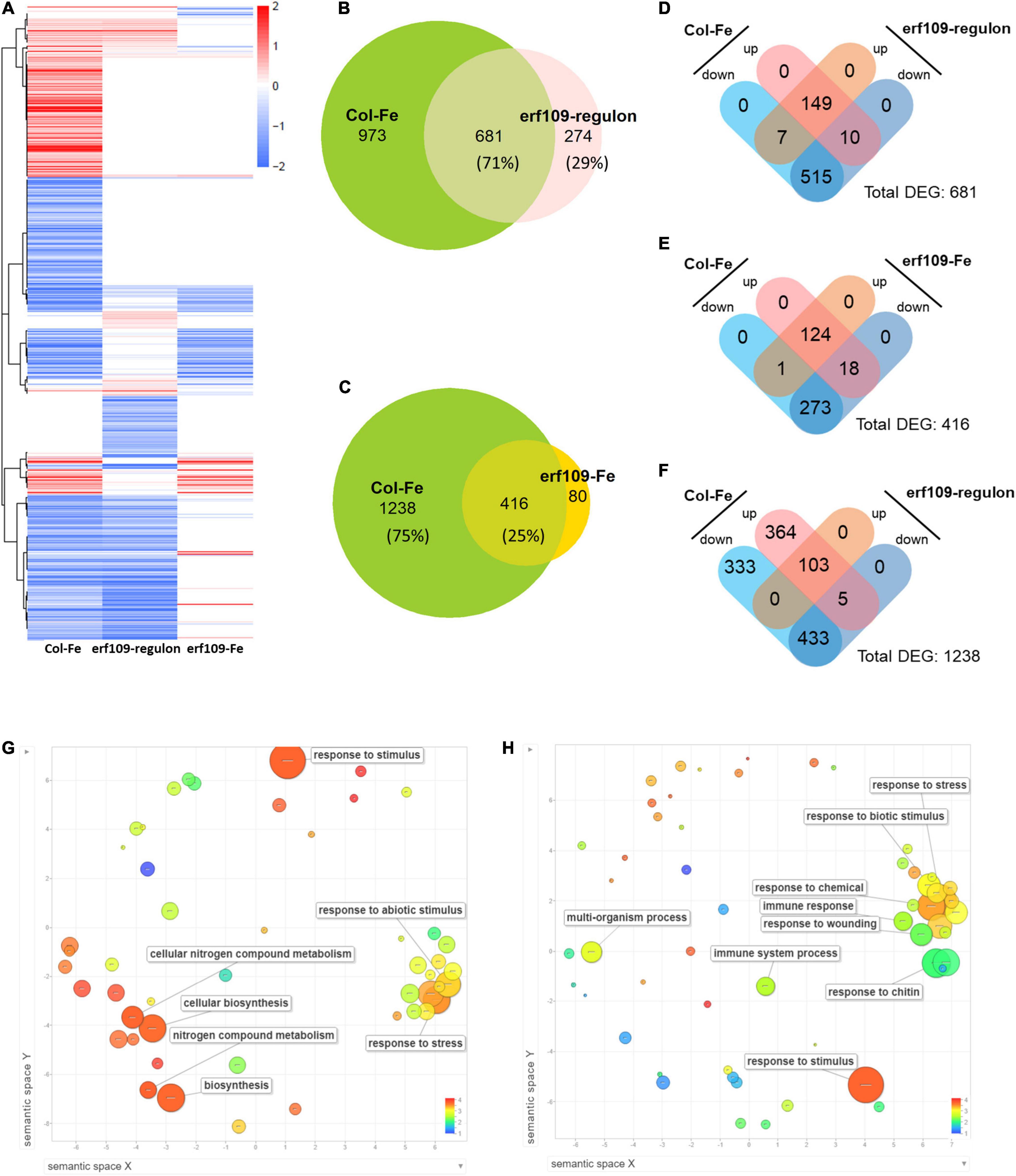
Figure 2. Differentially expressed transcripts, expression profiles and gene ontologies of Col-Fe, the erf109-regulon and erf109-Fe. (A) Heatmap of three transcriptomic profiles. Red and blue color indicates up- and down-regulated expression of DEGs with log2-fold change, respectively. (B) Venn diagram of DEGs from Col-Fe and the erf109-regulon. (C) Venn diagram of DEGs from Col-Fe and erf109-Fe. Distribution of up- and down-regulated DEGs overlapping between Col-Fe and the erf109-regulon (D), overlapping between Col-Fe and erf109-Fe (E), and erf109-regulon DEGs in specific of 1,238 Col-Fe DEGs (F). GO analysis (biological process) of 149 up- (G) and 515 down-regulated DEGs (H) overlapping in the Col-Fe and erf109-regulon data sets. Dot size indicates –log10 (P-value), the log value is labeled with the color bar from 1 to 4.
To further explore the changes in the transcriptome induced by either Fe starvation, dysfunctional ERF109, or both conditions, a co-expression network was constructed for the gene subsets Col-Fe, erf109-Fe, and erf109-regulon. Total DEGs were divided into two main clusters by their co-expression relationships: group 1 comprises mainly down-regulated genes, while group 2 comprises mostly up-regulated genes, which can be further subdivided into chloroplast-encoded and nucleus-encoded genes (Figure 3A). In the Col-Fe network, group 1 genes are connected to group 2 DEGs, which in turn is connected to a group of chloroplast-encoded DEGs which were significantly up-regulated (Figure 3A). The latter subset is not significantly regulated within the erf109-regulon (Figure 3B). Under Fe-deficient conditions, genes in erf109-Fe lost most of the response within both groups; some group 2 DEGs were even down-regulated (Figure 3C). Taken together, the network revealed that the overlap of the erf109-regulon and Col-Fe partly comprises the down-regulated genes in group 1, and the erf109-regulon completely abolishes the up-regulation of chloroplast-encoded genes. Moreover, defective ERF109 dramatically affected the gene expression pattern observed under Fe-deficient conditions in wild-type plants.
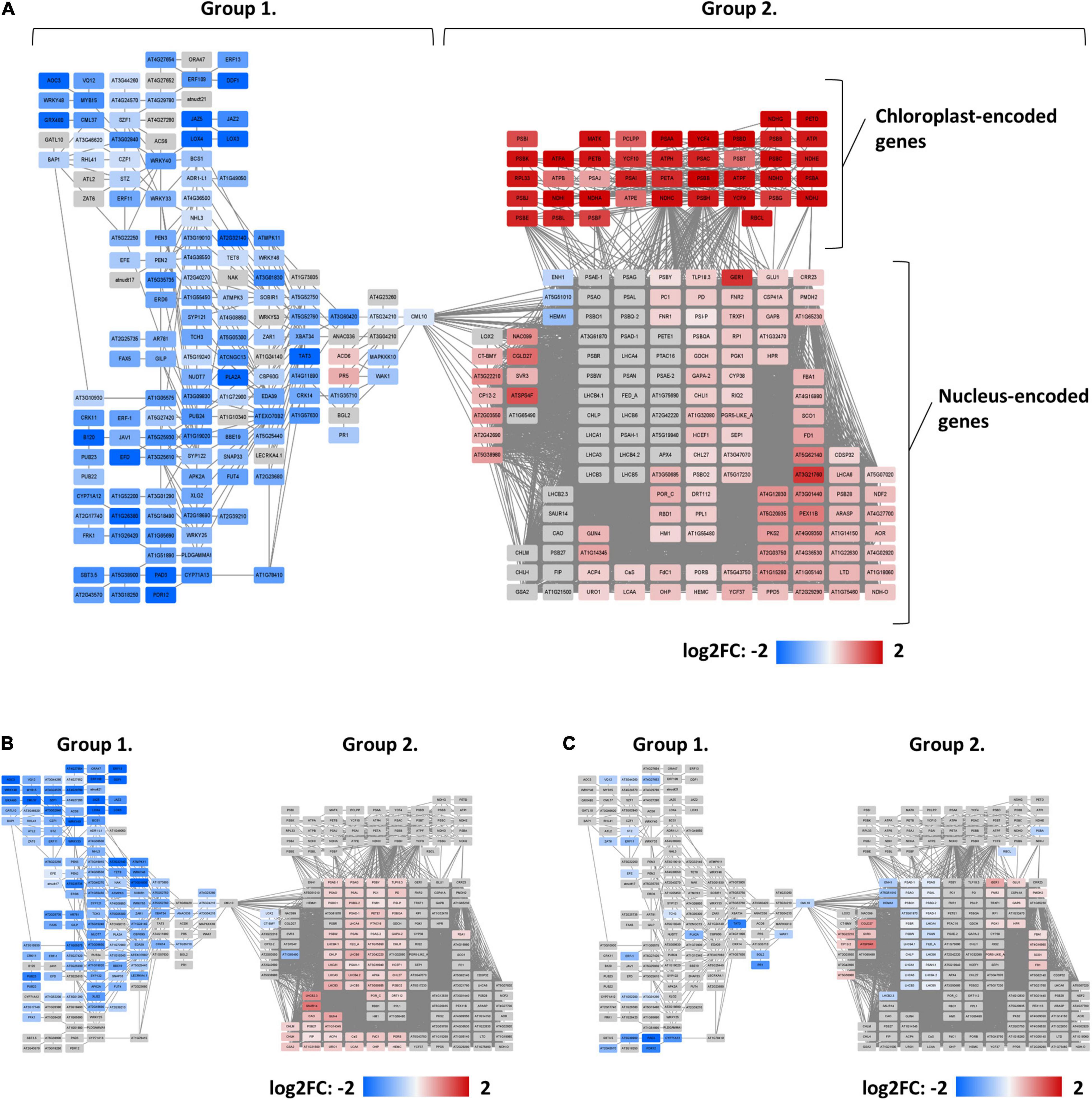
Figure 3. Co-expression networks of genes that were differentially expressed between genotypes or treatments. (A) Col-Fe, (B) erf109-regulon, and (C) erf109-Fe. Log2-fold change of DEGs are labeled by color: gray, no significant changes; red, up-regulated; blue, down-regulated.
To further investigate the function of the DEGs in the erf109-regulon, a gene ontology analysis using the MapMan software was employed. The analysis revealed that the most significant down-regulated DEGs are involved in biotic and abiotic stress responses, including R genes that code for proteins recognizing specific pathogen effectors, induction of the respiratory burst, signaling, and defense genes such as PR-proteins. Genes putatively involved in processes related to biotic stress such as hormone signaling, cell wall, proteolysis, and transcription factors were significantly reduced in both Col-Fe and erf109-regulon (Figures 4A,B). These defense-related DEGs comprise transcription factors such as DWARF AND DELAYED FLOWERING (DDF), SG2-type R2R3-MYB (MYB), WRKY, a putative zinc-finger (RHL), and other ERF family proteins (Figure 4C) as well as signaling proteins such as calmodulin-like proteins (CML), MAP kinases (MPK), and other defense-related proteins including a cysteine-rich receptor-like protein kinase (CRK), a PLANT U-BOX (PUB) protein, PROTEIN KINASE 2A (APK2A), and SUPPRESSOR OF BIR1 (SOBIR1) (Figure 4D). The observation that these stress response-associated genes were commonly affected in Col-Fe and in the erf109-regulon suggests that ERF109 governs the regulation of Fe-responsive immunity-associated genes that are part of the Fe deficiency response.
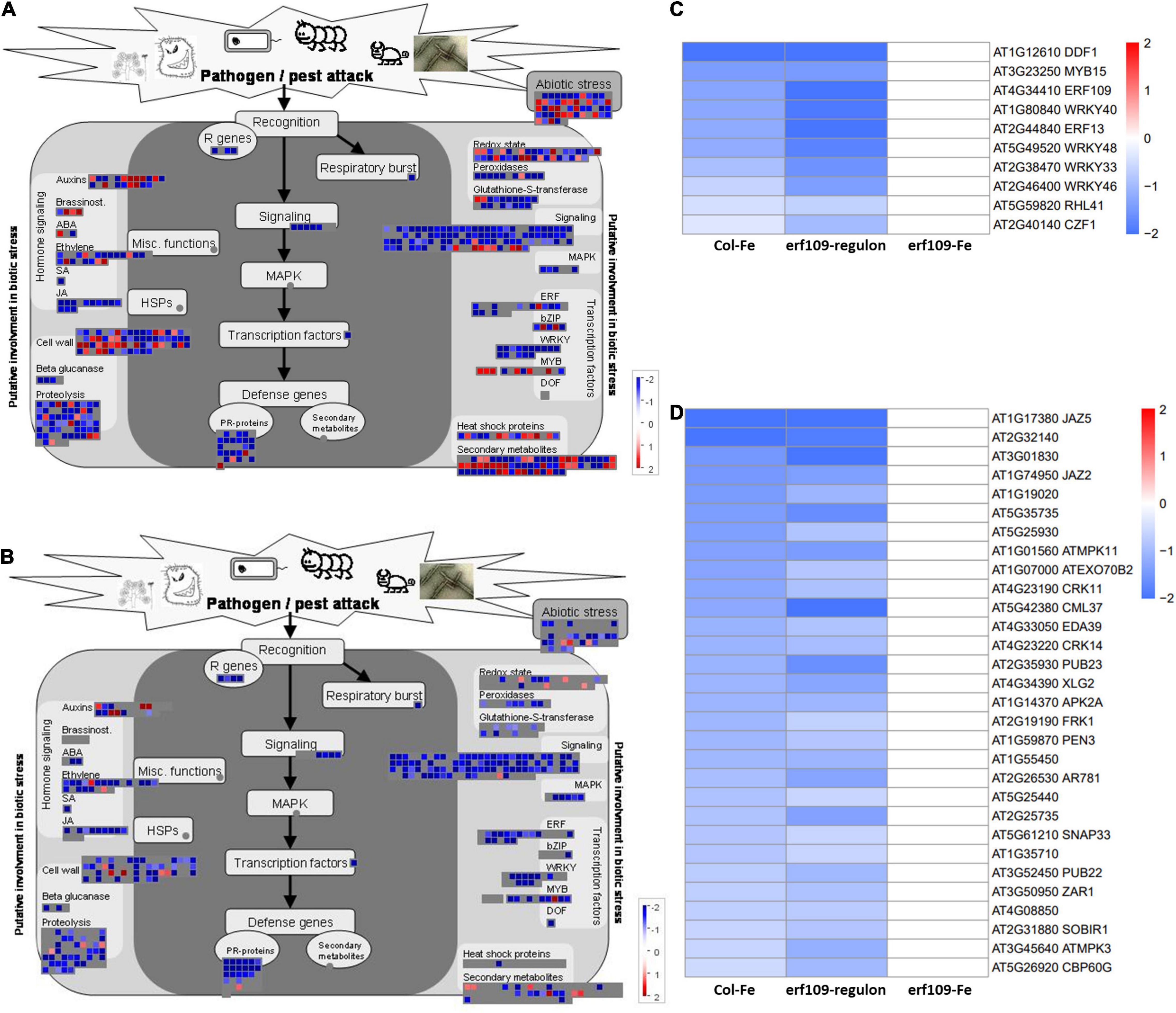
Figure 4. Functional visualization of DEGs involved in pathogen defense. (A) Col-Fe, (B) erf109-regulon. Graphs were generated with MapMan software (Thimm et al., 2004). Heatmap depicting log2 fold-changes of the transcription factors (C) and other functional DEGs (D) in group 1. Log2-fold changes of DEGs are labeled by color: white, no significant changes; red, up-regulated, blue: down-regulated.
In addition, induction of genes encoding photosystem subunits was observed in both Col-Fe and in the erf109-regulon, including a large fraction of genes encoding proteins involved in photosynthetic electron transport which showed increased transcription in Col-Fe and, to a lesser extent and partly different from those of Col-Fe, in erf109 plants (Figures 5A,B). In Col-Fe, the most highly expressed genes were encoded by plastidic DNA, including genes encoding proteins associated with the reaction centers of PSI and PSII, electron transfer proteins, NAD(P)H dehydrogenases, and ATP synthases (Figure 5C). By contrast, in the erf109-regulon, other genes associated with PSI and PSII, ferredoxin, and light-harvesting chlorophyll a/b-binding proteins were higher expressed than in the wild-type. When erf109 plants were subjected to Fe-deficient conditions, genes involved in these processes were rather repressed, showing a pattern that was clearly distinct from the other two transcriptomes (Figure 5C).
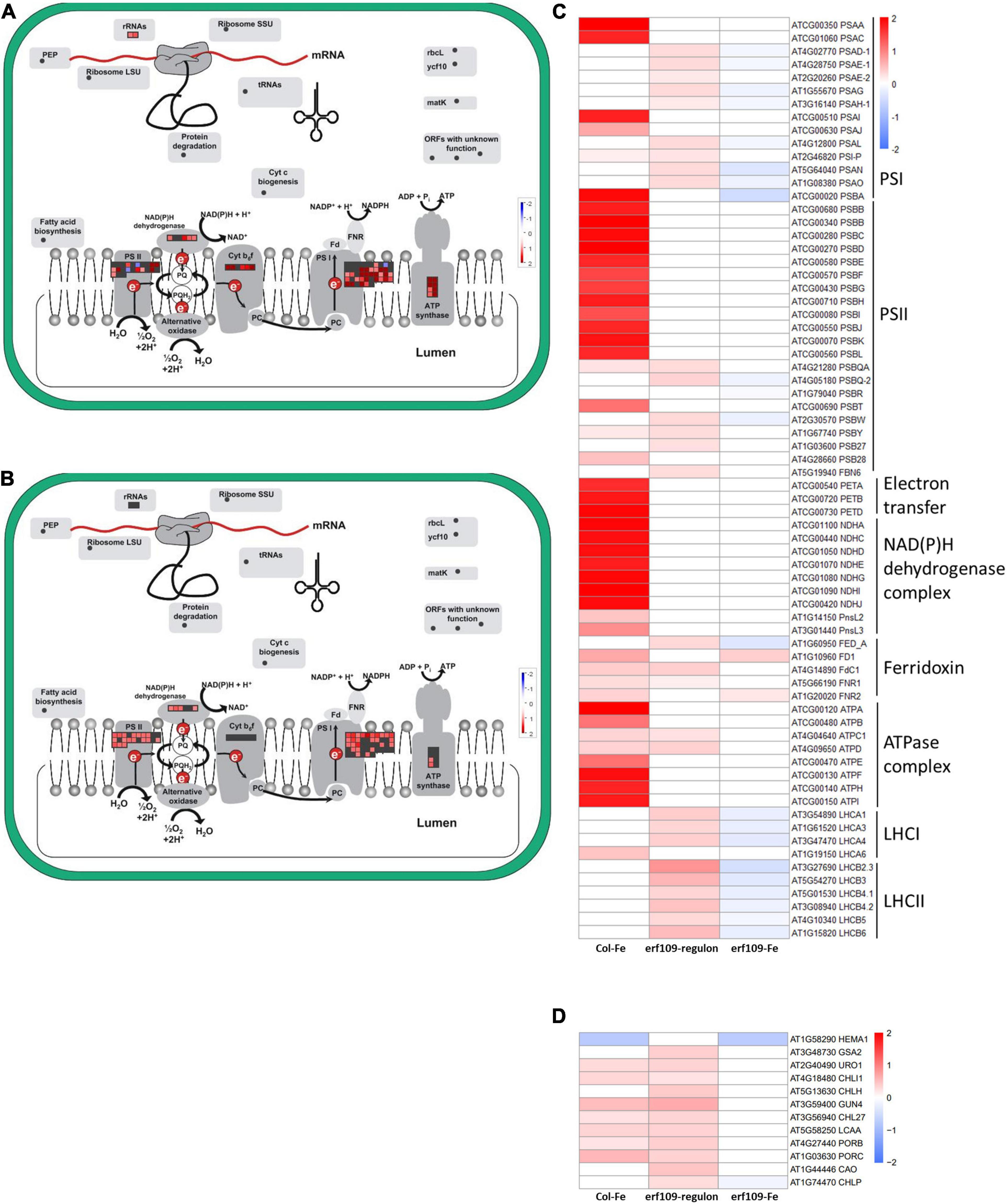
Figure 5. Differentially expressed genes encoding photosynthesis-related proteins. (A) Col-Fe, (B) erf109-regulon. Graphs were generated with MapMan software (Thimm et al., 2004). Heatmap depicting log2 fold-changes of genes encoding photosynthesis subunits (C) and genes involved in chlorophyll biosynthesis (D) in group 2. Log2-fold changes of DEGs are labeled by color: white, no significant changes; red, up-regulated; blue, down-regulated.
Chlorophyll plays an essential role in light harvesting and electron transfer in photosynthesis (Schreiber et al., 1995). The expression of genes encoding enzymes in tetrapyrrole biosynthesis such as HEME2, GUN4, CHLI1, and PORB was enhanced in both Col-Fe and in the erf109-regulon, but this induction was abolished in Fe-deficient erf109 plants (Figure 5D). A comparison between the genotypes suggests that ERF109 is mainly required for mounting defense responses under conditions of Fe deficiency. In addition, defects in ERF109 under Fe-deficiency is accompanied by partial repression of photosynthesis subunits and genes involved in chlorophyll biosynthesis.
To investigate how ERF109 influences cellular Fe homeostasis in leaves, RT-qPCR of a subset of Fe-responsive genes was selected, including four clade Ib bHLH proteins (bHLH38/39/100/101) that are critically required for the induction of all Fe deficiency responses (Wang et al., 2013), a gene encoding the Fe chelator nicotianamine (NICOTIANAMINE SYNTHASE4, NAS4) that is involved in metal translocation within the plant (Nozoye, 2018), and Fe storage protein FERRITIN1 (FER1) (Harrison and Arosio, 1996). The expression of bHLH38/39/101 was significantly increased in erf109 mutant plants and even more so after the Fe deficiency treatment (Figure 6). The expression of NAS4 and FER1 was higher in erf109 than in Col-0 plants in both Fe-sufficient and Fe-deficient conditions (Figure 6). Mutations in ERF109 increased bHLH38/39/101 and NAS4 expression, mimicking Fe deficiency. While FER1 was down-regulated in Fe-deficient Col-0 plants, FER1 transcript levels were increased in erf109 mutants under Fe-sufficient conditions, reflecting the higher Fe content in leaves of erf109 mutants (Figure 6).
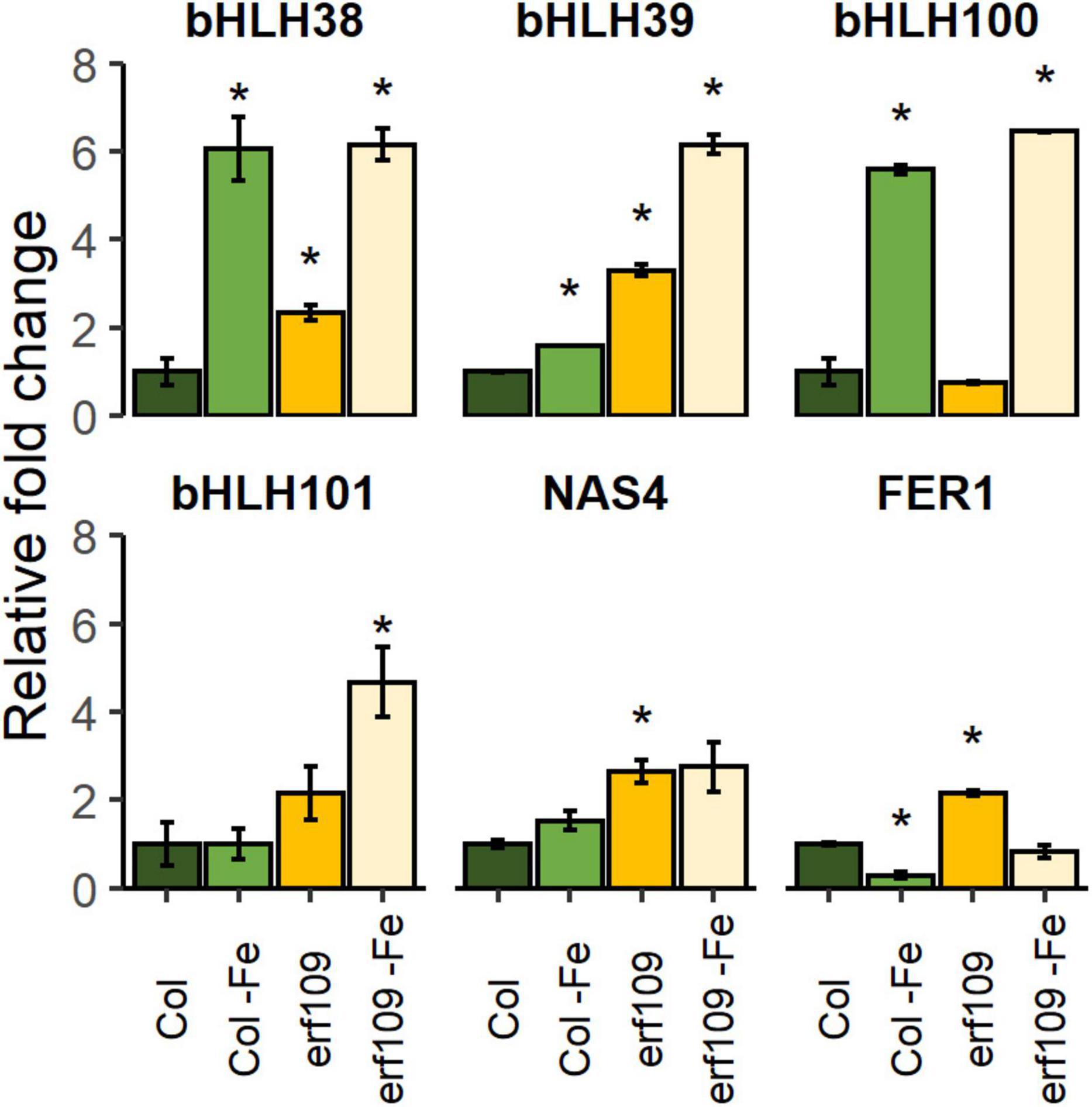
Figure 6. Quantitative RT-PCR analysis of relative fold change of Fe-responsive genes in Col-0 and erf109 plants under Fe-sufficient or Fe-deficient conditions. The level of transcript was normalized to Col-0 under Fe-sufficient conditions. Data are presented as mean ± SE from two biological replicates. Student’s t-test significantly different at P < 0.05 is marked with an asterisk.
To validate the differential expression pattern observed in the RNA-seq analysis, RT-qPCR of DEGs selected from the groups of defense-related, photosynthesis-related, and chloroplast-encoded genes was carried out. The criterion for the selection of these DEGs was based on the significance of the fold-changes and the molecular function of the genes. In wild-type plants, ERF109 was down-regulated in response to Fe-deficiency treatment; no ERF109 transcripts could be detected in the knock-out lines (Figure 7). Decreased expression of the calmodulin-like protein CML37, transcription factors involved in the defense response, in negotiating between abscisic acid (ABA) and ethylene (WRKY40, ERF13), and the exocyst subunit EXO70 family protein B2 (EXO70B2) was observed in Fe-deficient Col-0 plants and in erf109 mutant plants under both Fe-sufficiency and Fe-deficiency (Figure 7). By contrast, the expression of the ferredoxin-NADP reductase FNR2, photosystem II light-harvesting protein LHCB3, and the chlorophyll biosynthesis gene GUN4 was significantly increased upon Fe-deficiency in Col-0 but not in erf109 mutant plants (Figure 7). CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 (CGLD27) was up-regulated in both Fe-deficient Col-0 and erf109 plants (Figure 7). However, chloroplast-encoded genes such as the subunit of the chloroplast NAD(P)H dehydrogenase complex NDHI, photosystem II reaction center protein PSBH, and a protein required for photosynthesis I assembly and stability (YCF4) were only responsive to Fe deficiency in Col-0 plants (Figure 7). All expression patterns were in accordance with the RNA-seq data.
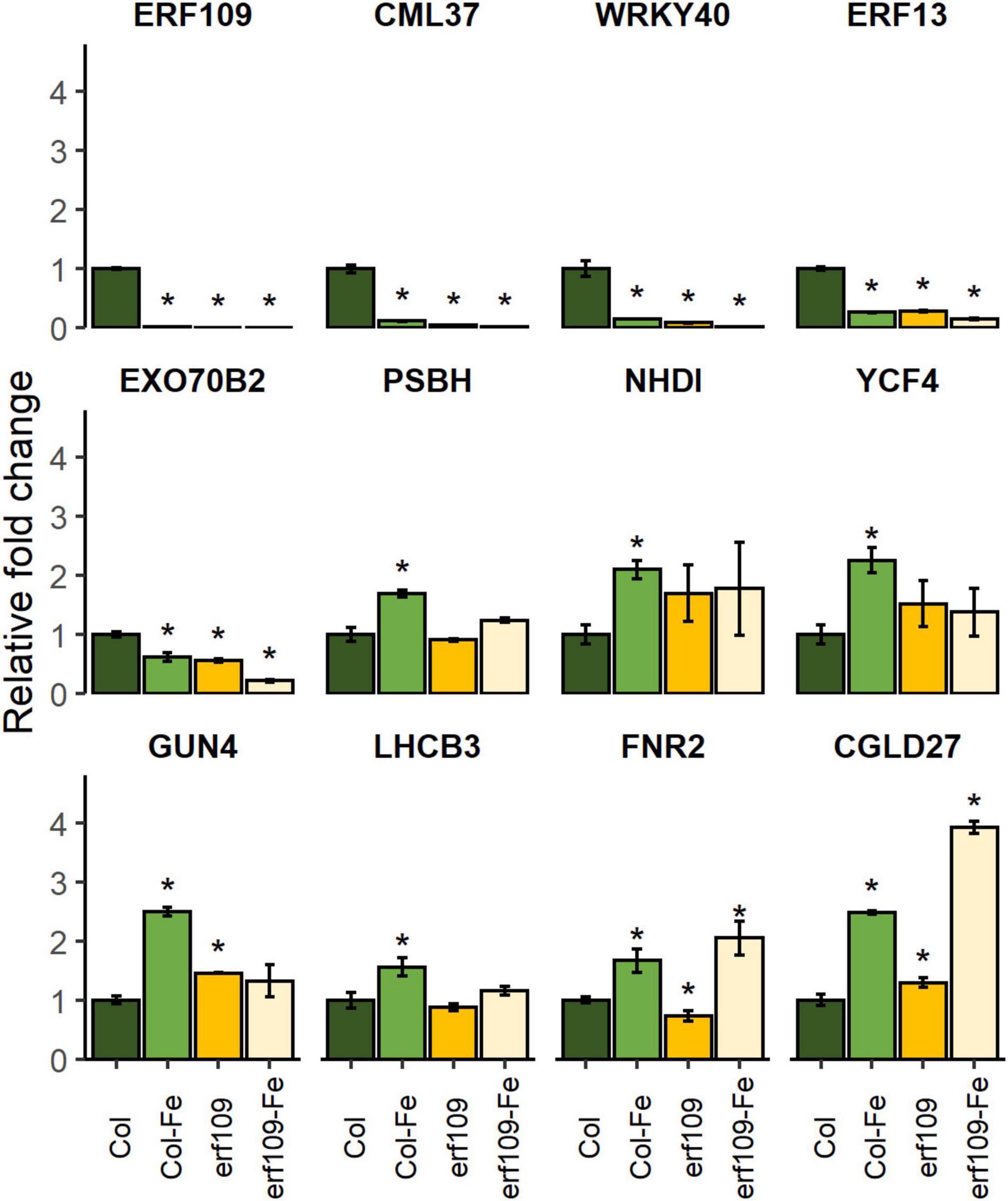
Figure 7. Quantitative RT-PCR analysis of relative fold change of DEGs in Col-0 and erf109 under Fe-sufficient (Col, erf109) or Fe-deficient conditions (Col-Fe, erf109-Fe). The level of transcript was normalized to Col-0 under Fe-sufficient conditions. Data are presented as mean ± SE from three biological replicates. Student’s t-test significantly different at P < 0.05 is marked with an asterisk.
Chlorosis on the young leaves is the most prominent symptom caused by Fe limitation. The chlorophyll content and the maximum quantum yield of photosystem II (Fv/Fm) serve as a phenotypic index of Fe deficiency in a variety of studies (Bertamini et al., 2002; Timperio et al., 2007; Kobayashi et al., 2013; Lei et al., 2014; Liu et al., 2017a; Rajniak et al., 2018; Becker et al., 2020; Tombuloglu et al., 2020). Upon Fe deficiency, wild-type plants showed a significant reduction in Fe levels, chlorophyll concentration, Fv/Fm, and quantum yield of PSI. Homozygous erf109 lines, on the other hand, exhibited elevated levels of Fe under both Fe-sufficient and Fe-deficient conditions when compared with Col-0. In addition, the erf109-regulon revealed a pattern of gene expression similar to that of Fe-deficient wild-type plants in leaves, suggesting that – independent of the external Fe supply – erf109 plants exhibit a constitutive Fe-deficiency response. One of the potential reasons for the pronounced chlorosis of erf109 plants lies in the increment of chlorophyll breakdown. ABA signaling can induce the decomposition of chlorophyll (Kuai et al., 2018; Asad et al., 2019). The expression of LHCB3 was significantly upregulated in wrky40 mutants, indicating that ABA positively regulated the expression of light-harvesting chlorophyll a/b-binding proteins via the WRKY40 transcriptional repressor (Liu et al., 2013). The results of the present study also showed that down-regulation of WRKY40 is accompanied by up-regulation of LHCB3 in erf109 mutant plants (Figure 7). The LHCB3 antenna subunit is a crucial participant in the modulation of PSII antenna size upon long-term acclimation to increased light levels in thylakoids (Albanese et al., 2016). Other research supported the assumption that LHCB family proteins are involved in ABA signaling by modulating ROS homeostasis (Xu et al., 2012). Another study also points out that WRKY40 is required for upregulation of ERF109 by stress stimuli and H2O2 (Matsuo et al., 2015). These data indicate that ERF109 potentially cooperates with hormones and the photosynthesis receptors to regulate immunity via WRKY40.
Several ethylene response factors such as ERF4 and ERF72 have been shown to exert negative effects on the Fe deficiency response in Arabidopsis (Liu et al., 2017a,b). DEGs in the erf109 transcriptome related to hormones are At5g35735 (auxin), ERF13, and ERF13/109 (ethylene), JAZ2/5, LOX2/3/4, and AOC3 (jasmonic acid; JA) (Supplementary Tables 1, 2). Both ERF13 and ERF109 are under the regulation of the JA-activated transcription factors MYC2/MYC3/MYC4 (Van Moerkercke et al., 2019). Jasmonate signaling has been observed in rice roots exposed to 3 h Fe deficiency (Kobayashi et al., 2016), exerting a repressing effect on Fe uptake (Cui et al., 2018). Although ERF109 was found to be up-regulated in response to a 6 h Fe-deficiency treatment (Hsieh et al., 2022), the present results revealed a repression of the gene after 72 h Fe deficiency in Col-0 leaves (Figure 7). It may thus be assumed that ERF109 and JA-signaling genes were inversely regulated in the early and later Fe deficiency response. The molecular mechanisms by which ERF109 and JA signaling interact on the Fe deficiency response remain, however, to be unraveled.
Previously, ERF109 was shown to serve as a major regulator of the light stress response and ROS homeostasis (Khandelwal et al., 2008; Matsuo et al., 2015; Carmody et al., 2016; Pospisil, 2016; Kong et al., 2018). In the present study, transcriptome and phenotypic data supported these findings and revealed that ERF109 is particularly important for balancing the Fe status in addition to immunity and the response to light stress. In addition, ERF109 was severed as a marker gene for high light-induced SAA (Carmody et al., 2016). In the present study, transcriptome analysis revealed that the majority of Fe-deficiency-induced DEGs in leaves were affected by a lack of functional ERF109. A co-expression network derived from genes that were differentially expressed between the growth- and genotypes revealed that the overlap of the erf109-regulon and Col-Fe consisted of defense-related and photosynthesis-related genes. Moreover, the mutation in ERF109 caused de-regulation of most Fe-related genes in erf109 plants under Fe-deficient conditions (Figure 3C). Moreover, Fe levels in erf109 plants were higher than in the Col-0 line (Figure 1B). From the lack of significant regulation of Fe-responsive genes in the erf109-Fe transcriptome and the increased Fe levels in erf109 mutant plants (Figures 1B, 3C), it may be speculated that the Fe deficiency response is constitutively induced in erf109 leaves. These results indicate that ERF109 potentially governs the defense response and, in part, photosynthesis and plays an important role in the Fe-deficiency response. Decreased expression of defense-related genes was the major change in the erf109-regulon and Col-Fe transcriptome, supporting the supposition that ERF109 controls the expression of immune networks in plants. Compromised Fe homeostasis was found to affect the immune system in plants. For instance, Fe aggregation was observed in corn, barley, oat, sorghum, and millet after attack by powdery mildew Blumeria graminis f. sp. tritici (Bgt) (Liu et al., 2007). Ferric Fe accumulation at the pathogen attack site together with intracellular Fe depletion was shown to promote the transcription of pathogenesis-related genes (Liu et al., 2007). Another report showed that the Fe nutritional status affected infection of maize by Colletotrichum graminicola (Ye et al., 2014). These studies revealed a close relationship between Fe homeostasis and the immune system of plants.
The immune system of plants is a complex network that integrates different regulative gene families such as stress-related genes, signaling, transcription factors, and response elements (Ngou et al., 2021). From our transcriptomic survey, it appears that ERF109 controls different defense-related signaling mechanisms and pathways associated with abiotic and biotic stresses. One of the best described signaling cascades is the Ca2+-based response. Ca2+ fluxes act as an intracellular secondary messenger, which is initiated by Ca2+ sensor proteins such as calmodulin and calmodulin-like proteins (Clapham, 2007; Bootman and Bultynck, 2020). CALMODULIN-LIKE PROTEIN (CML37) is highly expressed in younger leaves, and quickly (typically within 0.5–3 h) responds to wounding, osmotic stress, and drought (Vanderbeld and Snedden, 2007). It was shown that Arabidopsis lacking functional CML37 is highly susceptible to drought stress. Moreover, CML37 is a positive regulator of the plant hormone ABA (Scholz et al., 2015). In the present study, expression of CML37 was significantly decreased in Col-Fe, erf109-regulon, and erf109-Fe, suggesting that ERF109 participates in the regulation of intercellular Ca2+ signal transduction in response to Fe deficiency.
The WRKY transcription factor family plays essential roles in pathogen defense (Rushton et al., 2010; Chen et al., 2017). WRKY40 targets several downstream genes involved in the perception and transduction of microbial-associated and damage-associated molecular pattern-triggered immunity, the production of secondary indolic metabolites, and the modulating of distinct plant hormone pathways (Birkenbihl et al., 2017). Previous research revealed that ERF13 protein interacts with ERF109 and WRKY40, and is involved in the defense against herbivory (Chia, 2013; Miyamoto et al., 2019). All three genes are down-regulated in Col-Fe, erf109-regulon, and erf109-Fe, suggesting that ERF13, WRKY40, and ERF109 cooperate in the Fe deficiency response in immune signaling and hormone modulation.
Genes that robustly respond to the Fe regime across different transcriptomic datasets have been referred to as the “ferrome” (Schmidt and Buckhout, 2011; Hsieh et al., 2022). A subset of 12 genes that are within the erf109-regulon and the Col-Fe subset belong to the shoot ferrome (Hsieh et al., 2022), among them the ferritin FER1, a sensitive marker for the Fe status, the chlorophyll biosynthesis gene PORB, and the U-box type E3 ubiquitin ligase PUB23. Previous research demonstrated that PUB22, PUB23, and PUB24 negatively regulate PAMP-triggered responses (Trujillo et al., 2008). In PAMP-triggered responses, PUB22 targets a subunit of the exocyst complex encoded by EXO70B2, which mediates the process of vesicle tethering during exocytosis (Stegmann et al., 2012). EXO70B2 also regulates the receptor of bacterial flalso re or its immunogenic epitope flg22 at the plasma membrane, inhibiting the infection of bacterial pathogens by influencing the initiation of microbe-associated molecular pattern-triggered immunity (Wang et al., 2020). Decreased expression of EXO70B2 in Col-Fe, the erf109-regulon and erf109-Fe suggests compromised defense responses in erf109 mutant plants. From what has been mentioned above, the decline in expression of ERF109 and other defense DEGs in response to Fe deficiency suggests a link between Fe deficiency signaling and defense pathways.
The reasons for the increase of the Fe content in erf109 mutant leaves were investigated through the determination of the expression of six typical Fe deficiency-response genes. Although the four Ib subgroup bHLH proteins show functional redundancy in the Fe deficiency responses, multiple mutant lines of these bHLH genes in Arabidopsis exhibited differential degrees of chlorosis and IRT1 and FRO2 gene expression levels (Wang et al., 2013), suggesting that these bHLH proteins play distinct roles in regulating Fe uptake (Wang et al., 2013). All four clade Ib bHLH proteins form heterodimers with FIT to enhance the transcription of Fe uptake transporters in root, but the genes are also induced by Fe deficiency in leaves (Shen et al., 2016). RT-qPCR analysis revealed robust induction of bHLH38/39 expression in erf109 leaves under Fe-sufficient conditions (Figure 6), suggesting that ERF109 is repressing the Fe deficiency response. High Fe levels in the tomato mutant chloronerva was found to be caused by compromised biosynthesis of nicotianamine, which resulted in excessive Fe uptake and precipitation of Fe in the form of insoluble ferric phosphate compounds, protecting the cells from Fe overload (Becker et al., 1995). The nicotianamine synthase NAS4 showed increased expression in erf109 mutants induction under Fe-sufficient conditions compared to Col-0 wild-type plants (Figure 6), supporting the supposition that ERF109 is negatively regulating cellular Fe homeostasis. The regulation of FER1 corresponded to the Fe content in erf109 lines and Col-0 plants under the various Fe regimes (Figure 6). Together, these data support the hypothesis that compromised expression of ERF109 perturbs Fe homeostasis at an early stage of the signaling cascade by repressing the Fe acquisition machinery.
To sum up, the highly similar transcriptome expression patterns of Fe-sufficient erf109 mutants and Fe-deficient wild-type plants suggest that ERF109 is an upstream regulator of the Fe deficiency-induced immunity response. Moreover, higher Fe levels in erf109 leaves and reduced photosynthetic efficiency indicate that ERF109 attunes the physiologically available Fe for photosynthesis. We uncovered that the immunity network induced by Fe deficiency is the major target of ERF109 in shoots, which included down-regulation of CML37, WRKY40, ERF13, and EXO70B2. Moreover, dysfunctional ERF109 reduced the photosynthetic efficiency under Fe-deficient conditions. We provided evidence for a regulatory role of ERF109 in plant Fe homeostasis and set the stage for future studies and crop breeding to generate germplasms with improved systemic immunity and Fe uptake efficiency.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA793283.
C-LY wrote the manuscript and analyzed the data. Y-TH carried out the experiments. WS participated in experimental design and manuscript writing. PK participated in data analysis. M-TC conceived the original idea. I-CP contributed to experimental design, project management, and coordination and drafted the manuscript. All authors read and approved the final manuscript.
This work was supported by grants from the Ministry of Science and Technology (Nos. MOST 107-2313-B-005-015-MY3 and MOST 105-2313-B-005-007-MY2), and the Council of Agriculture, Agriculture and Food Agency (111AS-1.4.1-ST-a5), Taiwan. This work was supported in part by the Ministry of Education, Taiwan, R.O.C. under the Higher Education Sprout Project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the High Throughput Sequencing Core hosted in the Biodiversity Research Center at Academia Sinica for performing the NGS experiments. The core facility is funded by Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-108-114). We thank Kuo-Chen Yeh (Agricultural Biotechnology Research Center, Academia Sinica) for supporting iron content detection by ICP-optical emission spectrometry.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.841366/full#supplementary-material
Albanese, P., Manfredi, M., Meneghesso, A., Marengo, E., Saracco, G., Barber, J., et al. (2016). Dynamic reorganization of photosystem II supercomplexes in response to variations in light intensities. Biochim. Biophys. Acta 1857, 1651–1660. doi: 10.1016/j.bbabio.2016.06.011
Àlvarez-Fernàndez, A., Abadía, J., and Abadía, A. (2006). “Iron deficiency, fruit yield and fruit quality,” in Iron Nutrition in Plants and Rhizospheric Microorganisms, eds L. L. Barton and J. Abadia (Dordrecht: Springer Press), 85–101.
Asad, M. A. U., Zakari, S. A., Zhao, Q., Zhou, L., Ye, Y., and Cheng, F. (2019). Abiotic stresses intervene with ABA signaling to induce destructive metabolic pathways leading to death: premature leaf senescence in plants. Int. J. Mol. Sci. 20:256. doi: 10.3390/ijms20020256
Bahieldin, A., Atef, A., Edris, S., Gadalla, N. O., Ali, H. M., Hassan, S. M., et al. (2016). Ethylene responsive transcription factor ERF109 retards PCD and improves salt tolerance in plant. BMC Plant Biol. 16:216. doi: 10.1186/s12870-016-0908-z
Bahieldin, A., Atef, A., Edris, S., Gadalla, N. O., Ramadan, A. M., Hassan, S. M., et al. (2018). Multifunctional activities of ERF109 as affected by salt stress in Arabidopsis. Sci. Rep. 8:6403. doi: 10.1038/s41598-018-24452-6
Baker, M. S., and Gebicki, J. M. (1986). The effect of pH on yields of hydroxyl radicals produced from superoxide by potential biological iron chelators. Arch. Biochem. Biophys. 246, 581–588. doi: 10.1016/0003-9861(86)90313-9
Balparda, M., Armas, A. M., Estavillo, G. M., Roschzttardtz, H., Pagani, M. A., and Gomez-Casati, D. F. (2020). The PAP/SAL1 retrograde signaling pathway is involved in iron homeostasis. Plant Mol. Biol. 102, 323–337. doi: 10.1007/s11103-019-00950-7
Becker, M., Ngo, N. S., and Schenk, M. K. A. (2020). Silicon reduces the iron uptake in rice and induces iron homeostasis related genes. Sci. Rep. 10:5079. doi: 10.1038/s41598-020-61718-4
Becker, R., Fritz, E., and Manteuffel, R. (1995). Subcellular localization and characterization of excessive iron in the nicotianamine-less tomato mutant chloronerva. Plant Physiol. 108, 269–275. doi: 10.1104/pp.108.1.269
Bertamini, M., Muthuchelian, K., and Nedunchezhian, N. (2002). Iron deficiency induced changes on the donor side of PS II in field grown grapevine (Vitis vinifera L. cv. Pinot noir) leaves. Plant Sci. 162, 599–605. doi: 10.1016/S0168-9452(01)00604-5
Birkenbihl, R. P., Kracher, B., Roccaro, M., and Somssich, I. E. (2017). Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 29, 20–38. doi: 10.1105/tpc.16.00681
Bootman, M. D., and Bultynck, G. (2020). Fundamentals of cellular calcium signaling: a primer. Cold Spring Harbor Perspect. Biol. 12:a038802. doi: 10.1101/cshperspect.a038802
Cai, X. T., Xu, P., Zhao, P. X., Liu, R., Yu, L. H., and Xiang, C. B. (2014). Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 5:5833. doi: 10.1038/ncomms6833
Carmody, M., Crisp, P. A., d’Alessandro, S., Ganguly, D., Gordon, M., Havaux, M., et al. (2016). Uncoupling high light responses from singlet oxygen retrograde signaling and spatial-temporal systemic acquired acclimation. Plant Physiol. 171, 1734–1749. doi: 10.1104/pp.16.00404
Chen, F., Hu, Y., Vannozzi, A., Wu, K., Cai, H., Qin, Y., et al. (2017). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36, 311–335. doi: 10.1080/07352689.2018.1441103
Chia, K. F. (2013). The Arabidopsis Transcription Factor ERF13 Negatively Regulates Defense Against Pseudomonas syringae. San Diego: University of California Press.
Cui, Y., Chen, C. L., Cui, M., Zhou, W. J., Wu, H. L., and Ling, H. Q. (2018). Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Mol. Plant 11, 1166–1183. doi: 10.1016/j.molp.2018.06.005
Erickson, E., Wakao, S., and Niyogi, K. K. (2015). Light stress and photoprotection in Chlamydomonas reinhardtii. Plant J. 82, 449–465. doi: 10.1111/tpj.12825
Floyd, R. A., and Lewis, C. A. (1983). Hydroxyl free radical formation from hydrogen peroxide by ferrous iron-nucleotide complexes. Biochemistry 22, 2645–2649. doi: 10.1021/bi00280a008
Harrison, P. M., and Arosio, P. (1996). The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta. 1275, 161–203. doi: 10.1016/0005-2728(96)00022-9
Hsieh, E. J., Lin, W., and Schmidt, W. (2022). Genomically hardwired regulation of gene activity orchestrates cellular iron homeostasis in Arabidopsis. RNA Biol. 19, 143–161. doi: 10.1080/15476286.2021.2024024
Huang, D., Lin, W., Deng, B., Ren, Y., and Miao, Y. (2017). Dual-located WHIRLY1 interacting with LHCA1 alters photochemical activities of photosystem I and is involved in light adaptation in Arabidopsis. Int. J. Mol. Sci. 18:2352. doi: 10.3390/ijms18112352
Karpiński, S., Szechyńska-Hebda, M., Wituszyńska, W., and Burdiak, P. (2013). Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 36, 736–744. doi: 10.1111/pce.12018
Kent, W. J. (2002). BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664. doi: 10.1101/gr.229202
Khandelwal, A., Elvitigala, T., Ghosh, B., and Quatrano, R. S. (2008). Arabidopsis transcriptome reveals control circuits regulating redox homeostasis and the role of an AP2 transcription factor. Plant Physiol. 148, 2050–2058. doi: 10.1104/pp.108.128488
Kobayashi, T., Itai, R. N., Senoura, T., Oikawa, T., Ishimaru, Y., Ueda, M., et al. (2016). Jasmonate signaling is activated in the very early stages of iron deficiency responses in rice roots. Plant Mol. Biol. 91, 533–547. doi: 10.1007/s11103-016-0486-3
Kobayashi, T., Nagasaka, S., Senoura, T., Itai, R. N., Nakanishi, H., and Nishizawa, N. K. (2013). Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 4:2792. doi: 10.1038/ncomms3792
Kong, X., Tian, H., Yu, Q., Zhang, F., Wang, R., Gao, S., et al. (2018). PHB3 maintains root stem cell niche identity through ROS-responsive AP2/ERF transcription factors in Arabidopsis. Cell Rep. 22, 1350–1363. doi: 10.1016/j.celrep.2017.12.105
Kuai, B., Chen, J., and Hörtensteiner, S. (2018). The biochemistry and molecular biology of chlorophyll breakdown. J. Exp. Bot. 69, 751–767. doi: 10.1093/jxb/erx322
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/NMETH.1923
Lei, G. J., Zhu, X. F., Wang, Z. W., Dong, F., Dong, N. Y., and Zheng, S. J. (2014). Abscisic acid alleviates iron deficiency by promoting root iron reutilization and transport from root to shoot in Arabidopsis. Plant Cell Environ. 37, 852–863. doi: 10.1111/pce.12203
Lin, W. D., Liao, Y. Y., Yang, T. J., Pan, C. Y., Buckhout, T. J., and Schmidt, W. (2011). Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiol. 155, 1383–1402. doi: 10.1104/pp.110.166520
Liu, G., Greenshields, D. L., Sammynaiken, R., Hirji, R. N., Selvaraj, G., and Wei, Y. (2007). Targeted alterations in iron homeostasis underlie plant defense responses. J. Cell Sci. 120, 596–605. doi: 10.1242/jcs.001362
Liu, R., Xu, Y.-H., Jiang, S.-C., Lu, K., Lu, Y.-F., Feng, X.-J., et al. (2013). Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. J. Exp. Bot. 64, 5443–5456. doi: 10.1093/jxb/ert307
Liu, W., Karemera, N. U., Wu, T., Yang, Y., Zhang, X., Xu, X., et al. (2017a). The ethylene response factor AtERF4 negatively regulates the iron deficiency response in Arabidopsis thaliana. PLoS One 12:e0186580. doi: 10.1371/journal.pone.0186580
Liu, W., Li, Q., Wang, Y., Wu, T., Yang, Y., Zhang, X., et al. (2017b). Ethylene response factor AtERF72 negatively regulates Arabidopsis thaliana response to iron deficiency. Biochem. Biophys. Res. Commun. 491, 862–868. doi: 10.1016/j.bbrc.2017.04.014
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Matsuo, M., Johnson, J. M., Hieno, A., Tokizawa, M., Nomoto, M., Tada, Y., et al. (2015). High redox responsive transcription factor1 levels result in accumulation of reactive oxygen species in Arabidopsis thaliana shoots and roots. Mol. Plant 8, 1253–1273. doi: 10.1016/j.molp.2015.03.011
Miyamoto, T., Uemura, T., Nemoto, K., Daito, M., Nozawa, A., Sawasaki, T., et al. (2019). Tyrosine kinase-dependent defense responses against herbivory in Arabidopsis. Front. Plant Sci. 10:776. doi: 10.3389/fpls.2019.00776
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Ngou, B. P. M., Ahn, H.-K., Ding, P., and Jones, J. D. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Nozoye, T. (2018). The nicotianamine synthase gene is a useful candidate for improving the nutritional qualities and Fe-deficiency tolerance of various crops. Front. Plant Sci. 9:340. doi: 10.3389/fpls.2018.00340
Pospisil, P. (2016). Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front. Plant Sci. 7:1950. doi: 10.3389/fpls.2016.01950
Rajniak, J., Giehl, R. F. H., Chang, E., Murgia, I., von Wiren, N., and Sattely, E. S. (2018). Biosynthesis of redox-active metabolites in response to iron deficiency in plants. Nat. Chem. Biol. 14, 442–450. doi: 10.1038/s41589-018-0019-2
Richardson, A. D., Duigan, S. P., and Berlyn, G. P. (2002). An evaluation of noninvasive methods to estimate foliar chlorophyll content. New Phytol. 153, 185–194. doi: 10.1046/j.0028-646X.2001.00289.x
Robinson, M. D., and Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:r25. doi: 10.1186/gb-2010-11-3-r25
Rodríguez-Celma, J., Pan, I., Li, W. D., Lan, P. D., Buckhout, T. J., and Schmidt, W. (2013). The transcriptional response of Arabidopsis leaves to Fe deficiency. Front. Plant Sci. 4:276. doi: 10.3389/fpls.2013.00276
Rombolà, A. D., and Tagliavini, M. (2006). “Iron nutrition of fruit tree crops,” in Iron Nutrition in Plants and Rhizospheric Microorganisms, eds L. L. Barton and J. Abadia (Dordrecht: Springer Press), 61–83.
Roncel, M., Gonzalez-Rodriguez, A. A., Naranjo, B., Bernal-Bayard, P., Lindahl, A. M., Hervas, M., et al. (2016). Iron Deficiency induces a partial inhibition of the photosynthetic electron transport and a high sensitivity to light in the diatom Phaeodactylum tricornutum. Front. Plant Sci. 7:1050. doi: 10.3389/fpls.2016.01050
Rossel, J. B., Wilson, P. B., Hussain, D., Woo, N. S., Gordon, M. J., Mewett, O. P., et al. (2007). Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19, 4091–4110. doi: 10.1105/tpc.106.045898
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends. Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Schmidt, W., and Buckhout, T. J. (2011). A hitchhiker’s guide to the Arabidopsis ferrome. Plant Physiol. Biochem. 49, 462–470. doi: 10.1016/j.plaphy.2010.12.001
Scholz, S. S., Reichelt, M., Vadassery, J., and Mithöfer, A. (2015). Calmodulin-like protein CML37 is a positive regulator of ABA during drought stress in Arabidopsis. Plant Signal. Behav. 10:e1011951. doi: 10.1080/15592324.2015.1011951
Schreiber, U., Bilger, W., and Neubauer, C. (1995). “Chlorophyll fluorescence as a nonintrusive indicator for rapid assessment of in vivo photosynthesis,” in Ecophysiology of Photosynthesis, eds E. D. Schulze and M. M. Caldwell (Berlin: Springer Press), 49–70.
Shanmugam, V., Lo, J. C., Wu, C. L., Wang, S. L., Lai, C. C., Connolly, E. L., et al. (2011). Differential expression and regulation of iron-regulated metal transporters in Arabidopsis halleri and Arabidopsis thaliana–the role in zinc tolerance. New Phytol. 190, 125–137. doi: 10.1111/j.1469-8137.2010.03606.x
Shen, C., Yang, Y., Liu, K., Zhang, L., Guo, H., Sun, T., et al. (2016). Involvement of endogenous salicylic acid in iron-deficiency responses in Arabidopsis. J. Exp. Bot. 67, 4179–4193. doi: 10.1093/jxb/erw196
Sies, H., and Menck, C. F. (1992). Singlet oxygen induced DNA damage. Mutat. Res. 275, 367–375. doi: 10.1016/0921-8734(92)90039-R
Soliman, E. R. S., and Meyer, P. (2019). Responsiveness and adaptation to salt stress of the redox-responsive transcription factor 1 (RRTF1) gene are controlled by its promoter. Mol. Biotechnol. 61, 254–260. doi: 10.1007/s12033-019-00155-9
Stegmann, M., Anderson, R. G., Ichimura, K., Pecenkova, T., Reuter, P., Žárský, V., et al. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24, 4703–4716. doi: 10.1105/tpc.112.104463
Supek, F., Bošnjak, M., Škunca, N., and Šmuc, T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. doi: 10.1371/journal.pone.0021800
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Timperio, A. M., D’Amici, G. M., Barta, C., Loreto, F., and Zolla, L. (2007). Proteomics, pigment composition, and organization of thylakoid membranes in iron-deficient spinach leaves. J. Exp. Bot. 58, 3695–3710. doi: 10.1093/jxb/erm219
Tombuloglu, H., Slimani, Y., AlShammari, T. M., Bargouti, M., Ozdemir, M., Tombuloglu, G., et al. (2020). Uptake, translocation, and physiological effects of hematite (α-Fe2O3) nanoparticles in barley (Hordeum vulgare L.). Environ. Pollut. 266:115391. doi: 10.1016/j.envpol.2020.115391
Trujillo, M., Ichimura, K., Casais, C., and Shirasu, K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18, 1396–1401. doi: 10.1016/j.cub.2008.07.085
Vahabi, K., Reichelt, M., Scholz, S. S., Furch, A. C. U., Matsuo, M., Johnson, J. M., et al. (2018). Alternaria brassicae induces systemic jasmonate responses in Arabidopsis which travel to neighboring plants via a Piriformsopora indica hyphal network and activate abscisic acid responses. Front. Plant Sci. 9:626. doi: 10.3389/fpls.2018.00626
Van Moerkercke, A., Duncan, O., Zander, M., Šimura, J., Broda, M., Bossche, R. V., et al. (2019). A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. U.S.A. 116, 23345–23356. doi: 10.1073/pnas.1911758116
Vanderbeld, B., and Snedden, W. A. (2007). Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39. Plant Mol. Biol. 64, 683–697. doi: 10.1007/s11103-007-9189-0
Vogel, M. O., Moore, M., Konig, K., Pecher, P., Alsharafa, K., Lee, J., et al. (2014). Fast retrograde signaling in response to high light involves metabolite export, mitogen-activated protein kinase6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell 26, 1151–1165. doi: 10.1105/tpc.113.121061
Wang, N., Cui, Y., Liu, Y., Fan, H., Du, J., Huang, Z., et al. (2013). Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 6, 503–513. doi: 10.1093/mp/sss089
Wang, W., Liu, N., Gao, C., Cai, H., Romeis, T., and Tang, D. (2020). The Arabidopsis exocyst subunits EXO70B1 and EXO70B2 regulate FLS2 homeostasis at the plasma membrane. New Phytol. 227, 529–544. doi: 10.1111/nph.16515
Xu, Y.-H., Liu, R., Yan, L., Liu, Z.-Q., Jiang, S.-C., Shen, Y.-Y., et al. (2012). Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 63, 1095–1106. doi: 10.1093/jxb/err315
Keywords: ERF109, RRTF1, iron deficiency, immunity, photosynthesis, transcriptome
Citation: Yang C-L, Huang Y-T, Schmidt W, Klein P, Chan M-T and Pan I-C (2022) Ethylene Response Factor109 Attunes Immunity, Photosynthesis, and Iron Homeostasis in Arabidopsis Leaves. Front. Plant Sci. 13:841366. doi: 10.3389/fpls.2022.841366
Received: 22 December 2021; Accepted: 09 February 2022;
Published: 02 March 2022.
Edited by:
Seçkin Eroğlu, Middle East Technical University, TurkeyReviewed by:
Hannetz Roschzttardtz, Pontificia Universidad Católica de Chile, ChileCopyright © 2022 Yang, Huang, Schmidt, Klein, Chan and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: I-Chun Pan, aWNwQGRyYWdvbi5uY2h1LmVkdS50dw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.