- 1College of Life Science, Taizhou University, Taizhou, China
- 2National Key Laboratory for Rice Biology, Institute of Biotechnology, Zhejiang University, Hangzhou, China
E3 ubiquitin ligases are involved in many processes, regulating the response to biotic and abiotic stresses. In this study, 11 E3 ubiquitin ligase genes from Arabidopsis, which were hypothesized to function in response to biotic or abiotic stresses were selected, and the homologous genes in rice were found. Their functions were analyzed in rice. These 11 E3 ubiquitin ligase genes showed different patterns of expression under different treatments. The BMV:OsPUB39-infiltrated seedlings showed decreased resistance to Magnaporthe grisea (M. grisea) when compared with BMV:00-infiltrated seedlings, whereas the BMV:OsPUB34- and BMV:OsPUB33-infiltrated seedlings showed increased resistance. The involvement of these genes in the resistance against M. grisea may be attributed to the regulation of the accumulation of reactive oxygen species (ROS) and expression levels of defense-related genes. Seedlings infiltrated by BMV:OsATL69 showed decreased tolerance to drought stress, whereas BMV:OsPUB33-infiltraed seedlings showed increased tolerance, possibly through the regulation of proline content, sugar content, and expression of drought-responsive genes. BMV:OsATL32-infiltrated seedlings showed decreased tolerance to cold stress by regulating malondialdehyde (MDA) content and the expression of cold-responsive genes.
Introduction
Ubiquitination is a post-translational modification, involved in many processes. Ubiquitination is mediated by three sequential ubiquitin enzymes, E1 (the ubiquitin-activating enzyme), E2 (the ubiquitin-conjugating enzyme), and E3 (the ubiquitin ligase). The E3 ubiquitin ligase confers the specificity of the reaction and can either be single-subunit (including HECT, RING finger, and U-box domain family) or multi-subunit (such as SCF complex) (Morreale and Walden, 2016).
E3 ubiquitin ligases have been reported to function in several processes, including the responses to biotic and abiotic stresses (Ariani et al., 2017; Sun et al., 2019, 2022; Zhang et al., 2019; Du et al., 2021; Sharma et al., 2021). First, E3 ubiquitin ligases are involved in response to biotic stresses. The ATL subfamily that contains the conserved RING-H2 domain is activated by the elicitor and plays important roles in disease resistance, possibly through the regulation of the elicitor-signaling pathway, including chitin (Serrano and Guzman, 2004; Berrocal-Lobo et al., 2010; Ni et al., 2010; Chen et al., 2017; Deng et al., 2017). The U-box domain family of E3 ubiquitin ligases also plays important roles in defense response. The spl11 mutants show increased resistance to multiple fungal and bacterial pathogens (Yin et al., 2000; Zeng et al., 2004). AtPUB22, 23, and 24 act as negative regulators of biotic stresses (Cho et al., 2008; Trujillo et al., 2008). Moreover, E3 ubiquitin ligases are involved in the basic resistance of plants. Plants with overexpression of HUB1 have a thickened cell wall to increase the resistance to Botrytis cinerea (B. cinerea), whereas the knockout mutants show a thinned cell wall to decrease the resistance to B. cinerea and Alternaria brassicicola (Dhawan et al., 2009). The overexpression of OsBBI1 leads to increased accumulation of H2O2 and phenolic compounds, thicker cell wall, and increased resistance to M. oryzae (Li et al., 2011).
Secondly, E3 ubiquitin ligases are involved in response to abiotic stresses, including drought and cold stresses. E3 ubiquitin ligases function in response to drought stress, being either dependent on the ABA pathway (Zhang et al., 2005, 2015; Ko et al., 2006; Stone et al., 2006; Joo et al., 2016; Yang et al., 2016; Lim et al., 2017; Chapagain et al., 2018; Qin et al., 2020; Chen et al., 2021) or independent of it (Qin et al., 2008; Prasad et al., 2010; Suh and Kim, 2015; Wu et al., 2015). Moreover, E3 ubiquitin ligases also function in response to cold stress. In Arabidopsis, HOS1, AtATL78, and AtATL80 negatively regulated the tolerance to cold stress (Lee et al., 2001; Dong et al., 2006; Kim and Kim, 2013; Suh and Kim, 2015), whereas PUB25 and PUB26 positively regulated the tolerance to cold stress (Wang et al., 2019). OsDIRP1 positively regulated the tolerance to cold stress in rice (Cui et al., 2018) while OsATL38 negatively regulates the tolerance to cold in rice (Cui et al., 2022).
E3 ubiquitin ligases comprise a large protein family and are encoded by several genes (Bhaskar and Joemar, 2020); for example, more than 1,200 genes in Arabidopsis encode E3 ubiquitin ligases. This large number indicates the importance of E3 ubiquitin ligase. In this study, the functional analysis of 11 E3 ubiquitin ligase genes in rice was performed. The expression levels of some ubiquitin ligase genes were induced by one or several tested treatments, using different models. The silencing of the OsPUB34 and OsPUB33 led to increased resistance to M. oryzae, whereas the silencing OsPUB39 led to increased resistance. The silencing of OsPUB33 led to increased tolerance to drought stress, whereas that of OsATL69 led to decreased tolerance, possibly through regulation of the proline content, sugar content, and expression levels of drought-responsive genes. Finally, the silencing of OsATL32 led to decreased tolerance to cold stress, possibly through regulation of the MDA content and expression levels of cold-responsive genes.
Materials and Methods
Conditions for Plant Growth and Treatments Used
The rice cultivar Yuanfengzao, a pair of isogenic lines (H8R and H8S), and IR64 were used in this research for various purposes. The cultivar Yuanfengzao was used for the analysis of gene expression in response to hormone treatments and abiotic stress. H8R and H8S were used for the analysis of gene expression following inoculation with M. grisea. IR64 was used for VIGS infiltration. In the hormone treatment, 2-weeks-old Yuanfengzao seedlings were treated with 1.5 mM salicylic acid (SA, pH 6.5), 100 μM jasmonic acid (JA), 100 μM 1-amino cyclopropane-1-carboxylic acid (ACC), and 100 μM abscisic acid (ABA) (Sigma-Aldrich, St. Louis, United States). The leaves of the control group were sprayed with the same volume of water or 0.1% ethanol.
M. grisea (strain 85-14B1, race ZB1) was cultivated on oatmeal medium at 25°C for 10 days. The spores were collected and resuspended in water to a final concentration of 5 × 105 conidia/mL with 0.02% Tween-20. Then, the spore solution was sprayed on the leaves of H8R and H8S (Luo et al., 2005). Leaf samples were collected at the indicated time points and stored at −80°C until further use.
For extreme temperature stress, 3-week-old plants were subjected to temperatures of 42 and 4°C. For the drought stress, hydroponic 3-week-old plants were placed on the floor of the frame in the greenhouse for growth after water on their root surface was absorbed by filter paper. For salt stress, hydroponic 3-week-old plants were transferred to 200 mM NaCl solution. Then, the samples were collected at the indicated time points (Hong et al., 2016). All the above-mentioned seedlings were grown in a room at 28°C, with a photoperiod of 14 h light/10 h dark. IR64 was used for VIGS assays, and the infiltrated seedlings were placed in a room at 24°C, with a photoperiod of 14 h light/10 h dark.
Vector Construction and VIGS
The 200–400 bp fragments of target genes were constructed into BMV vector and confirmed by sequencing. The obtained recombinant plasmids were transformed into Agrobacterium tumefaciens strain C58C1 by electroporation using a GENE PULSER II Electroporation System. The agrobacteria confirmed by colony PCR were cultivated in a liquid YEP medium containing corresponding antibiotics at 28°C overnight. The bacteria were collected and resuspended in induction buffer (10 mM MgCl2, 10 mM MES, 200 μM acetosyringone, pH 5.7) and kept at 28°C for 5 h. The induction was stopped by centrifugation. The cells were then resuspended in an infiltration solution (10 mM MES, 10 mM MgCl2, 0.4 g/L L-cysteine, 0.15 g/L DTT, 0.75 mg/L silver nitrate, and 15 μL Silwet-77 in 10% YEP) and incubated at 28°C until the OD600 value reached 2.0. These treated cells were then mixed with the same volume of agrobacteria harboring pC13/F1 + 2 before vacuum infiltration. About 8–10-day-old IR64 seedlings were submerged completely in the mixed Agrobacterium suspension with vacuum infiltration for 7 min under a pressure of 20 kPa (model no. Rocker 410, Xiamen B&C Instrument Co., Ltd., China). Then, these plants were placed in a room at 24°C, with a photoperiod of 14 h light/10 h dark, and were recorded as BMV:target gene-infiltrated seedlings. The BMV:empty vector was transformed to seedlings as control, which were recorded as BMV:00-infiltrated seedlings (Zhang et al., 2021).
qRT-PCR
RNA was extracted using the Trizol reagent (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. cDNA was obtained using AMV reverse transcriptase (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The qRT-PCR was performed using SYBR Premix Ex Taq™ (TaKaRa, Dalian, China) in a CFX96 real-time PCR detection system (BioRad, Hercules, CA, United States) according to the manufacturer’s instructions. The sequence of primers used in this study can be found in Table 1.
Disease Assay of M. oryzae
About 5 μL of the M. oryzae spore solution was placed on the surface of leaves of 4-week-old silenced plants, which had been placed on a wet cheesecloth. Then, the inoculated leaves were kept in high humidity in a room for the growth of the silenced seedlings. After 7 days, photographs were taken, and the lesion sizes were recorded. For the analysis of the expression of defense-related genes and the quantification of M. oryzae in plants, we used the whole plant assay. It was carried out using the same method as done on the H8R and H8S seedlings.
Abiotic Stress Tolerance Assay
For the analysis of drought stress, 4-week-old seedlings of BMV:OsATL69-infiltrated plants and BMV:00-infiltrated plants in the same pot were withheld from water for 10 days before re-watering, whereas the BMV:OsPUB33-infiltrated plants were withheld for 15 days. After 12 days, the survival rate, water loss, proline content, and sugar content were measured (Bates et al., 1973; Hong et al., 2016). For the analysis of cold stress, 4-week-old seedlings of BMV:target gene-infiltrated plants and BMV:00-infiltrated plants in the sameed that their expression decreased ied that their expression decreased i pot were kept at 4°C for 2 days before recovering to normal growth condition (Huang et al., 2016). The survival rate, MDA content, electrolyte leakage, chlorophyll content, and the expression levels of cold-responsive genes were analyzed as reported before (Hong et al., 2016).
Results
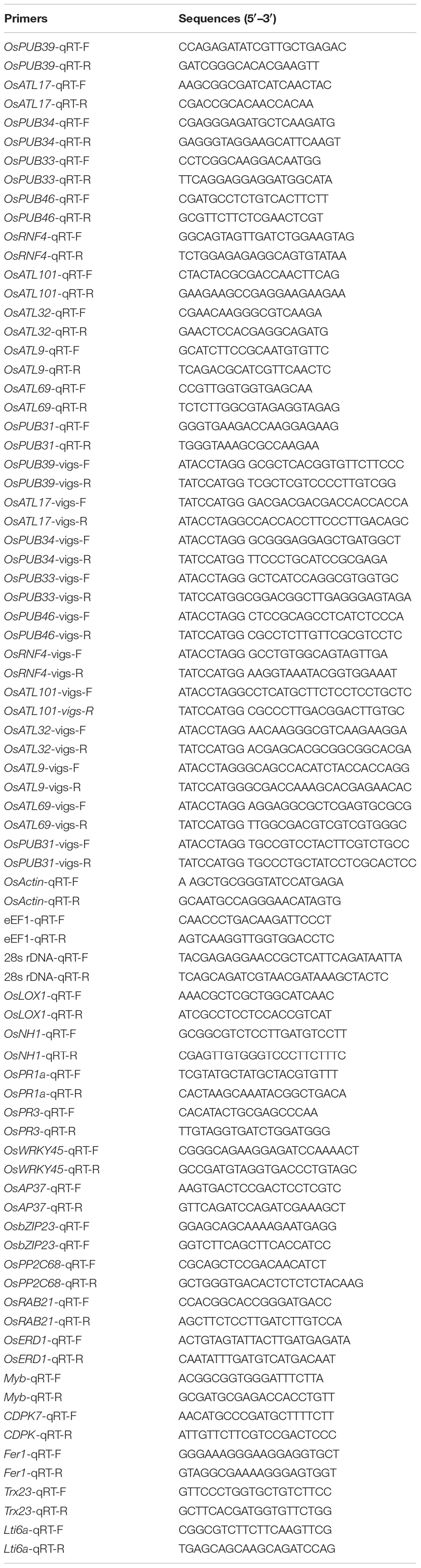
The Expression Levels of OsPUB39, OsPUB34, and OsPUB33 Were Strongly Induced by the Inoculation of M. girsea and Hormone Molecules
By conducting searches on BLAST-P of the rice genome database using the characterized 11 Arabidopsis genes as queries, the corresponding gene sequences were obtained (Supplementary Material). To test whether these 11 genes functioned in response to stress, the expression patterns of these genes were analyzed following inoculation with M. grisea and treatment with hormone molecules. The expression of OsPUB39, OsPUB34, and OsPUB33 was strongly induced by the inoculation of M. grisea in incompatible interaction, whereas the expression of other genes showed no significant difference between the treatment and control at the time point we test (Figure 1A). Furthermore, the expression levels of OsPUB39, OsPUB34, and OsPUB33 were strongly induced by the hormones SA, JA, and ACC, whereas the expression levels of other genes were unaffected (Figure 1B).
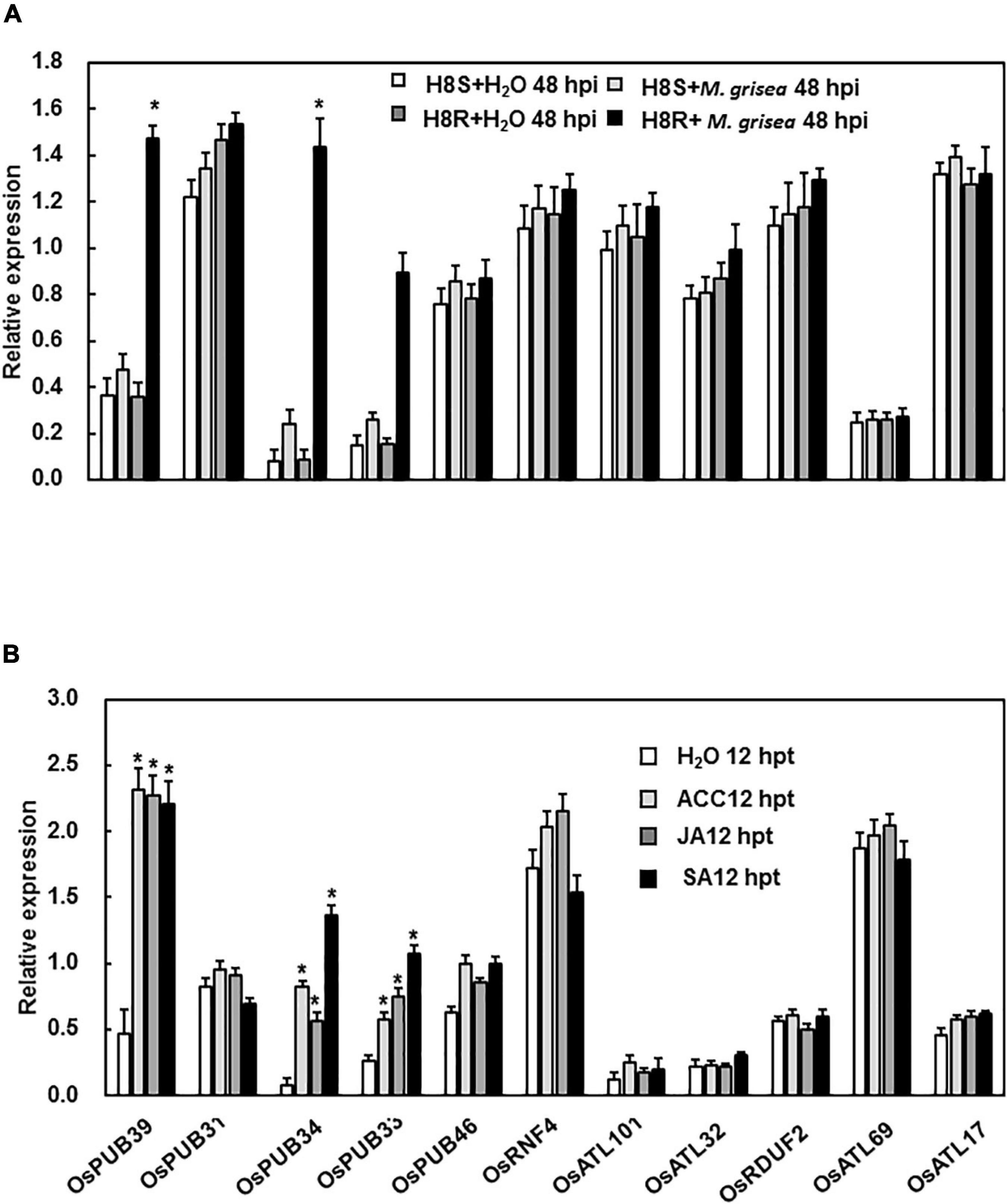
Figure 1. Expression patterns of E3 ubiquitin ligase genes in response to infection by M. grisea and hormone treatment. (A) Expression of E3 ubiquitin ligase genes in response to M. grisea. Leaves of H8R and H8S seedlings were sprayed with a solution of M. grisea spores. (B) Expression of E3 ubiquitin ligase genes in response to signaling hormones. Leaves of 4-week-old Yuanfengzao seedlings were treated with 1.5 mM SA, 100 μM JA, and 100 μM ACC solutions by spraying. JA and ACC solutions were prepared in 0.1% ethanol, and SA solution was prepared in water. The same volume of 0.1% ethanol or sterile distilled water was used as control. The samples were harvested at specific time points for the analysis of gene expression. Expression levels were presented as multiples of the OsActin expression level, which was used as standard. Data presented are the means ± SD from three independent experiments, and * above the columns indicate significant differences between the treated plants and control at p < 0.05 level.
The Expression Patterns of E3 Ubiquitin Ligase Genes in Response to Abiotic Stress and Abscisic Acid
As reported previously, PUB genes were involved in response to abiotic stresses, including drought, cold, heat, and salinity, to different degrees (Lu et al., 2020). Hence, drought, salinity, cold, and heat stresses were selected for the analysis of the expression patterns under abiotic stress. In the drought stress, the expression patterns of all genes except two (OsATL69 and OsPUB33) showed no change. The expression of these two genes increased dramatically 2 h after treatment (Figure 2A). Under cold stress, the expression level of only OsATL32 was strongly induced 12 h after treatment (Figure 2B). Under heat and salinity stresses, the expression levels of none of the genes were significantly affected (Figures 2C,D). ABA is a well-known stress-related hormone in plants and is involved in the responses to abiotic stress. We tested the expression patterns of E3 ubiquitin ligase genes following ABA treatment and observed that expression levels of OsATL69 and OsATL32 were significantly affected by ABA treatment at the time point we test (Figure 2E).
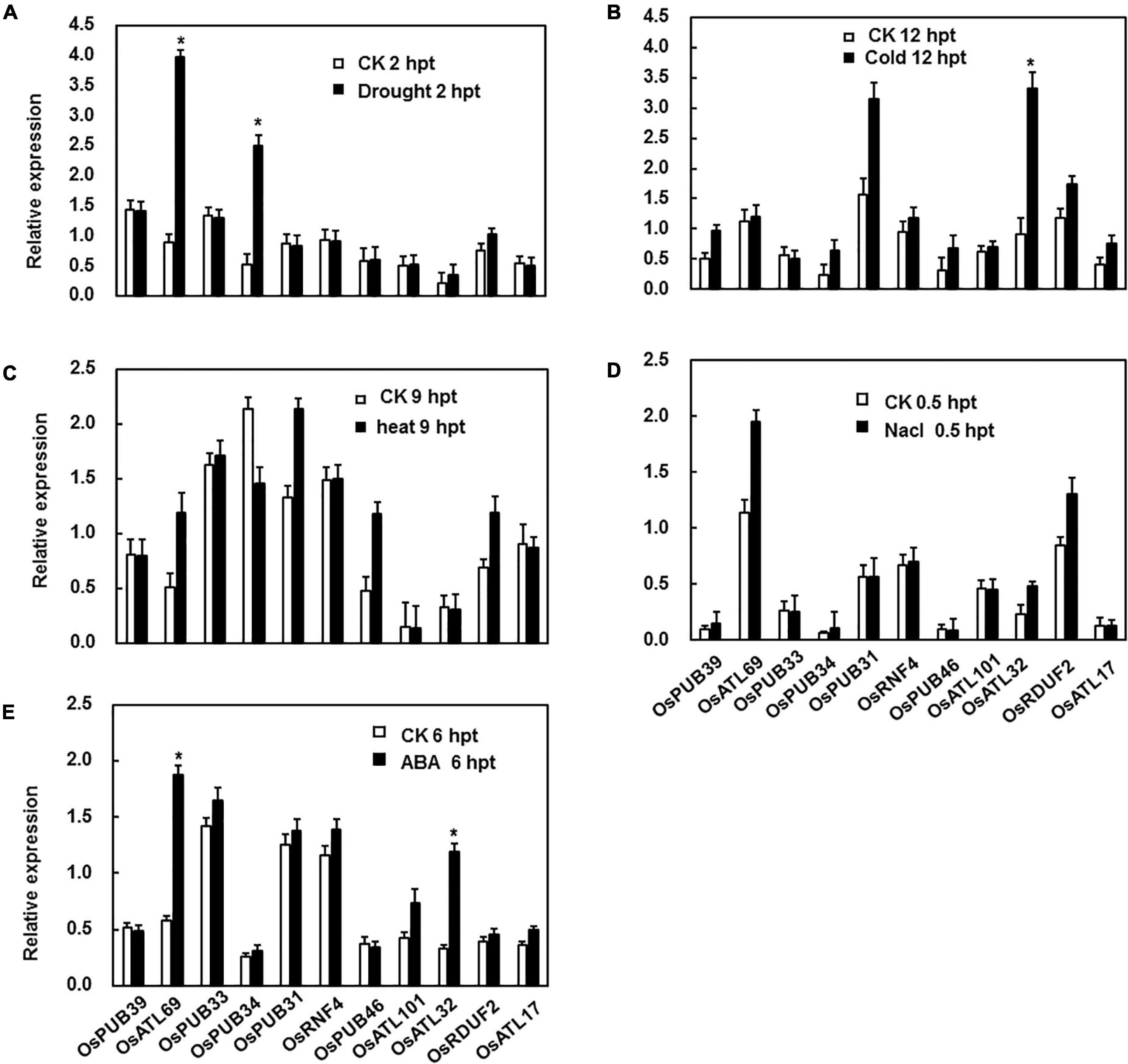
Figure 2. Expression patterns of E3 ubiquitin ligase genes in response to abiotic stress and ABA treatment. (A) For drought stress, the hydroponic 3-week-old plants were placed on the frame floor in the greenhouse after the water on the surface of the root was dried by filter paper. For cold (B) and heat stress (C), 3-week-old plants were placed in a climatic cabinet at 4°C and 42°C. (D) For salt stress, hydroponic 3-week-old plants were placed in a 200 mM NaCl solution. (E) Expression patterns of E3 ubiquitin ligase genes in response to ABA. Leaves of 4-week-old Yuanfengzao seedlings were sprayed with 100 μM ABA solution. Ethanol (0.1%) was used as the control. The samples were harvested at specific time points for the analysis of gene expression. Expression levels were presented as multiples of the OsActin expression level, which was used as a standard. Data presented are the means ± SD from three independent experiments, and * above the columns indicate significant differences between the treated and control plants at p < 0.05 level.
BMV:OsPUB39-Infiltrated Plants Showed Decreased Resistance to M. grisea When Compared With BMV:00-Infiltrated Plants, Whereas BMV:OsPUB34- and BMV:OsPUB33-Infiltrated Plants Showed Increased Resistance
We explored the possible role of the tested genes in the resistance to M. grisea by comparing the phenotype of BMV:target gene-infiltrated and BMV:00-infiltrated plants after the inoculation of M. grisea. The efficiency of silencing was tested before inoculation, and the most efficiently silencing plants were selected for the disease assay (Supplementary Material). After 7 days, the BMV:OsPUB39-infiltrated plants showed a more severe disease phenotype with larger lesion size and more fungi growth when compared with control, whereas the BMV:OsPUB34- and BMV:OsPUB33-infiltrated plants showed lighter disease phenotype with smaller lesion size and less fungal growth (Figure 3).
Figure 3. BMV:OsPUB34- and BMV:OsPUB33-infiltrated seedlings showed significantly increased resistance to M. grisea when compared with the control, whereas BMV:OsPUB39-infiltrated seedlings showed decreased resistance to M. oryzae. (A) The lesions on the rice leaves 7 days after inoculation. (B) The quantities of bacteria in leaves of BMV:target gene- and BMV:00-infiltrated seedlings. (C) The length and width of the lesions on BMV:target gene- and BMV:00-infiltrated seedlings. *Above the columns indicate significant differences between BMV:target gene- and BMV:00-infiltrated seedlings at p < 0.05 level.
To investigate the mechanism of function of OsPUB39, OsPUB34, and OsPUB33’s in the resistance to M. grisea, we analyzed the ROS accumulation and the expression levels of defense-related genes. There was no significant difference in ROS accumulation between BMV: OsPUB39-, BMV: OsPUB34-, BMV: OsPUB33-, and BMV:00-infiltrated seedlings before M. grisea inoculation. However, after inoculation with M. grisea, the BMV:OsPUB34- and BMV:OsPUB33-infiltrated seedlings accumulated less ROS than the BMV:00-infiltrated seedlings, whereas BMV:OsPUB39-infiltrated seedlings accumulated more ROS (Figure 4A). Similar results were observed in terms of H2O2 content. After M. grisea inoculation, H2O2 content in BMV:OsPUB34- and BMV:OsPUB33-infiltrated seedlings was lower than that in BMV:00-infiltrated seedlings and higher in BMV:OsPUB39-infiltrated seedlings (Figure 4B). The activities of SOD and CAT were analyzed to investigate the reason behind the changed H2O2 content in BMV: OsPUB39-, BMV: OsPUB34-, and BMV:OsPUB33-infiltrated seedlings. Before M. grisea inoculation, SOD activity and CAT activity in the BMV:target gene- and BMV:00-infiltrated seedlings were not significantly different (Figures 4C,D). After M. grisea inoculation, SOD activity in BMV:OsPUB34- and BMV:OsPUB33-infiltrated seedlings decreased, whereas CAT activity increased when compared with BMV:00-infiltrated seedlings. In contrast, SOD activity in BMV:OsPUB39-infiltrated seedlings increased, whereas CAT activity decreased when compared with BMV:00-infiltrated seedlings after M. grisea inoculation (Figures 4C,D).
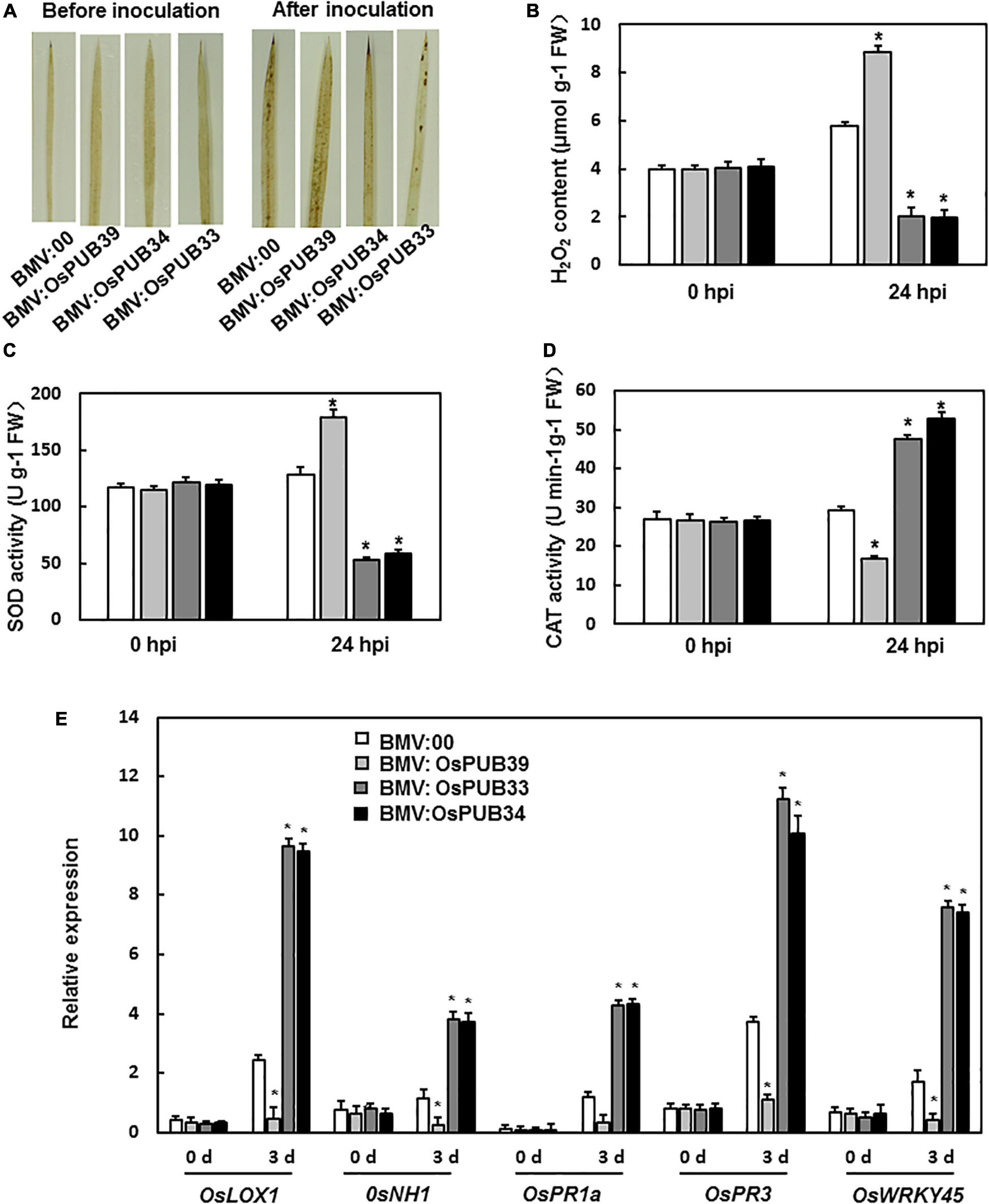
Figure 4. ROS accumulation and the expression levels of defense-related genes in BMV:target gene- and BMV:00-infiltrated seedlings before and after M. oryzae inoculation. (A) DAB staining of leaves from BMV:target gene- and BMV:00-infiltrated seedlings before and after M. oryzae inoculation. (B) H2O2 content in BMV:target gene- and BMV:00-infiltrated seedlings before and after M. oryzae inoculation. SOD activity (C) and CAT activity (D) in BMV:target gene- and BMV:00-infiltrated seedlings before and after M. oryzae inoculation. (E) Expression levels of defense-related genes were shown as multiples of the OsActin expression level, which was used as a standard. Data presented are the means ± SD from three independent experiments, and * above the columns indicate significant differences between BMV:target gene- and BMV:00-infiltrated seedlings at p < 0.05 level.
We also analyzed the expression levels of defense-related genes. After M. grisea inoculation, the expression levels of OsLOX1, OsPR3, OsNH1, OsPR1a, and OsWRKY45 decreased in BMV:OsPUB39-infiltrated plants and increased in BMV:OsPUB34- and BMV:OsPUB33-infiltrated seedlings when compared to the control (Figure 4E). These results indicated that OsPUB39, OsPUB34, and OsPUB33 were involved in the resistance to M. grisea, possibly through the regulation of the accumulation of ROS and expression of defense-related genes.
The BMV:OsPUB33-Infiltrated Plants Showed Increased Tolerance to Drought Stress, Whereas the BMV:OsATL69-Infiltrated Plants Showed Decreased Tolerance
To explore the possible function of the 11 genes in response to abiotic stress, we compared the phenotype of BMV:target gene- and BMV:00-infiltrated plants after exposure to abiotic stress. None of the plants showed a significant difference in tolerance to drought stress, except BMV:OsATL69- and BMV:OsPUB33-infiltrated plants. The BMV:OsPUB33-infiltrated plants showed increased tolerance to drought stress, whereas BMV:OsATL69-infiltrated plants showed decreased tolerance when compared with control (Figure 5A). The survival rate, water loss, proline content, and sugar content decreased significantly in BMV:OsATL69-infiltrated plants and increased significantly in BMV:OsPUB33-infiltrated plants (Figures 5B–E). We also tested the expression levels of drought-responsive genes and observed that their expression decreased in BMV:OsATL69-infiltrated plants and increased in BMV:OsPUB33- infiltrated plants (Figures 5F,G).
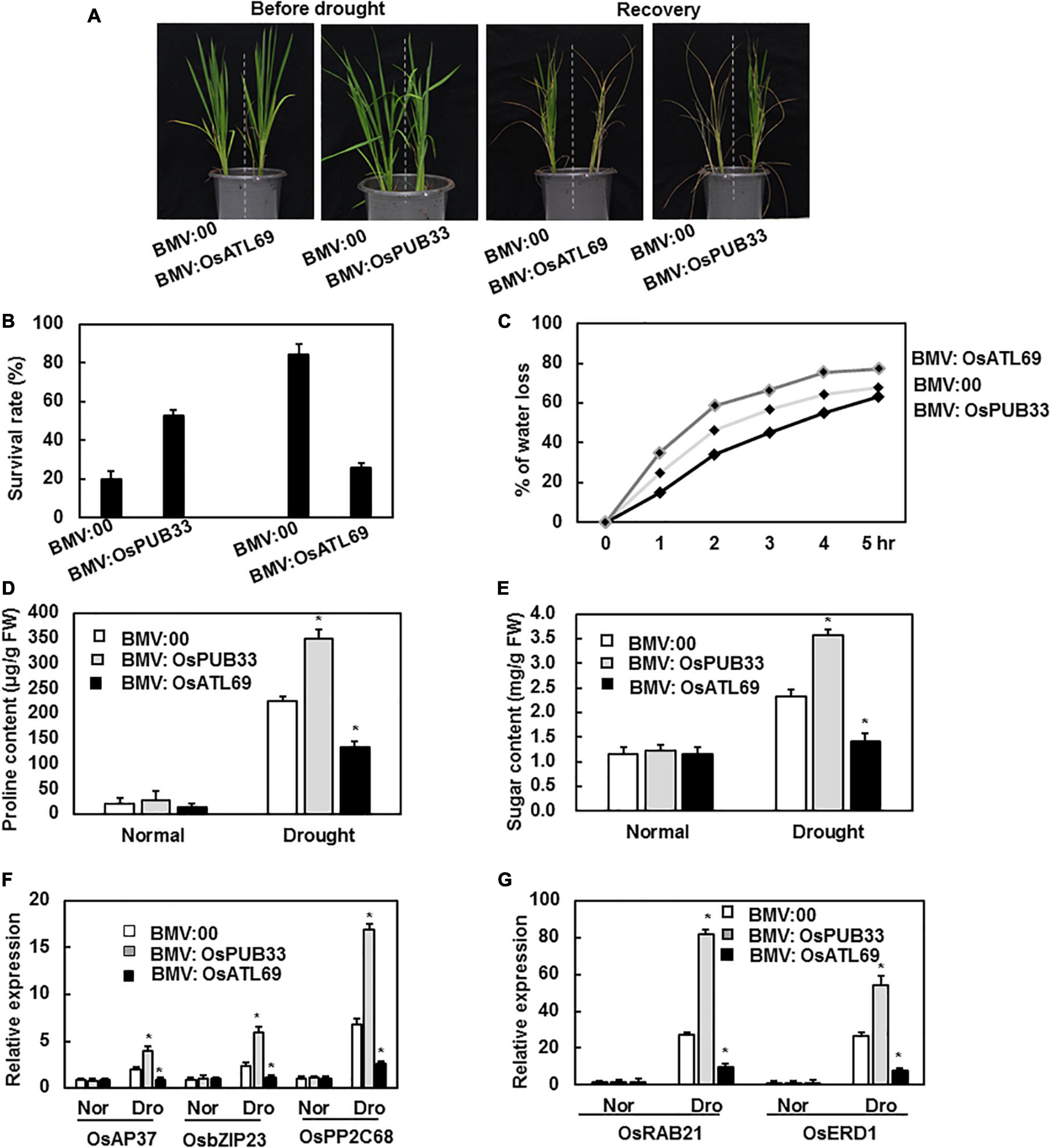
Figure 5. BMV:OsATL69-infiltrated seedlings showed decreased tolerance to drought stress while BMV:OsPUB33-infiltrated seedlings showed increased tolerance to drought stress. The BMV:OsATL69- and BMV:00-infiltrated seedlings in the same pot were withheld watering for 10 days and recovered with normal watering for another 12 days. The phenotype (A) and the survival rate (B) of BMV:target gene- and BMV:00-infiltrated seedlings after suffered drought stress. BMV:OsATL69- and BMV:00-infiltrated seedlings in the same pot were withheld watering for 10 days and recovered with normal watering for another 12 days. And BMV:OsPUB33- and BMV:00-infiltrated seedlings in the same pot which were withheld watering for 15 days and recovered with normal watering for another 12 days. (C) Water loss of BMV:OsATL69- and BMV:00-infiltrated seedlings after exposure to drought stress for different times. Proline content (D) and sugar content (E) of BMV: OsATL69-, BMV: OsPUB33-, and BMV:00-infiltrated seedlings after exposure to drought stress for 10 days. (F,G) The expression levels of drought-responsive genes were upregulated in BMV:OsPUB33-infiltrated plants and downregulated in BMV:OsATL69- infiltrated plants, when compared to the control after drought stress. Expression levels were shown as multiples of the OsActin expression level, which was used as a standard. Data presented are the means ± SD from three independent experiments, and * above the columns indicate significant differences between BMV:target genes- and BMV:00-infiltrated plants at p < 0.05 level.
BMV:OsATL32-Infiltrated Plants Decreased the Tolerance to Cold Stress
Under cold stress, BMV:OsATL32-infiltrated plants showed decreased resistance when compared to the control (Figure 6A). The survival rate of BMV:OsATL32- infiltrated plants was 19.62 and 83.65% in control (Figure 6B). The MDA content and electrolyte leakage in BMV:OsATL32-infiltrated plants increased when compared to control (Figures 6C,D), whereas the chlorophyll content in BMV:OsATL32- infiltrated plants decreased (Figure 6E). Next, the expression levels of cold-responsive genes were analyzed and were observed to decrease significantly in BMV:OsATL32-infiltrated plants when compared to the control (Figure 6F).
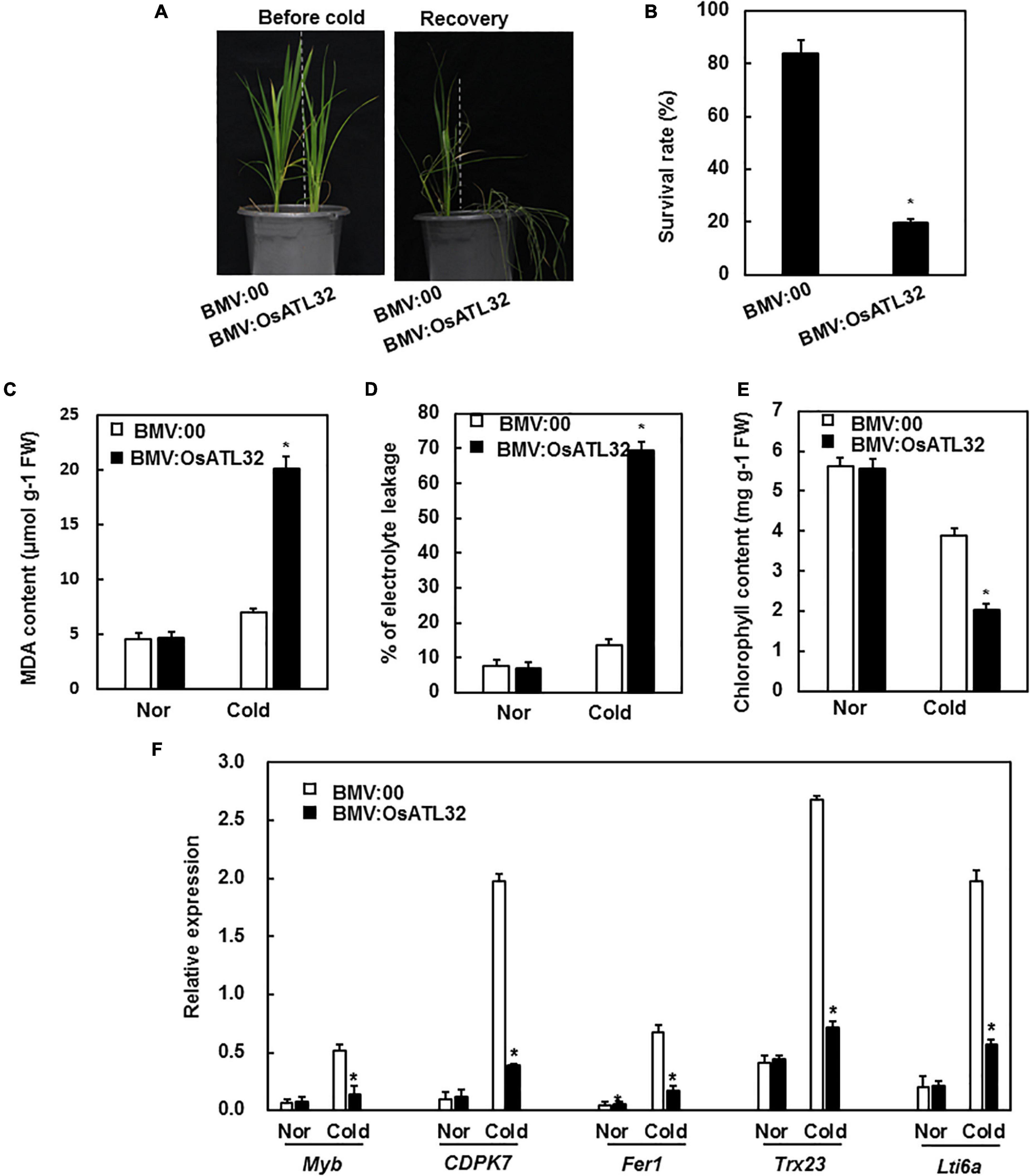
Figure 6. BMV:OsATL32-infiltrated seedlings showed decreased tolerance to cold stress when compared to the control. Four-week-old BMV:OsATL32- and BMV:00-infiltrated seedlings in the same pot were placed in a growth chamber at 4°C for 2 days, then transferred to normal growth conditions. Seven days later, the survival rate, MDA content, chlorophyll content, electrolyte leakage, and expression levels of cold-responsive genes were analyzed. (A) The phenotype of BMV: OsATL32-infiltrated plants after exposure to cold stress. (B) The survival rates were recorded. MDA content (C), electrolyte leakage (D), and chlorophyll content (E) were analyzed in BMV:OsATL32- and BMV:00-infiltrated seedlings with or without exposure to cold stress. (F) Expression of cold-responsive genes in BMV:OsATL32- and BMV:00-infiltrated seedlings with or without exposure to cold stress. Expression levels of cold-responsive genes were analyzed using qRT-PCR with gene-specific primers. Data presented are the means ± SD from three independent experiments, and * above the columns indicate significant differences between BMV:OsATL32- and BMV:00- infiltrated plants at p < 0.05 level.
Discussion
E3 ubiquitin ligases play an important role in the ubiquitin-proteasome pathway, which is among the most important protein degradation pathway in eukaryotic organisms (Pickart and Eddins, 2004; Wang and Deng, 2011). There are several E3 ubiquitin ligases in plants, for example, at least 60 in Arabidopsis. The study of E3 ubiquitin ligases is a topic of immense interest, and the functions of E3 ubiquitin ligases are explored regularly. However, the functions of E3 ubiquitin ligases in rice were rarely studied. In this research, 11 E3 ubiquitin ligase genes from Arabidopsis, which were hypothesized to function in response to biotic or abiotic stresses, were selected, and the homologous genes in rice were found.
Plants are unavoidably exposed to abiotic and biotic stresses and have formed sophisticated mechanisms to adapt to such adverse conditions. Phytohormones play an important role in helping plants to adapt to environmental situations (Verma et al., 2016). Several phytohormone signaling pathways are dependent on the ubiquitin-proteasome system, specifically E3 ubiquitin ligases that can perceive and initiate signal transduction (Kelley, 2018; Tal et al., 2020). Therefore, the expression patterns of these 11 genes following treatment with hormones such as JA, ACC, and SA were analyzed first. The expression levels of only three genes, OsPUB39, OsPUB34, and OsPUB33, were induced by treatment with JA, ACC, and SA (Figure 1B). This result indicates that these genes may function in response to biotic stress. Next, the functions of these genes in response to M. grisea were analyzed. The expression levels of OsPUB39, OsPUB34, and OsPUB33 were induced by M. grisea, and the silencing of these three genes altered the resistance to M. grisea (Figures 1A, 3). OsPUB34 and OsPUB33 negatively regulated the resistance to M. grisea. The functions of OsPUB34 and OsPUB33 in rice in response to biotic stress were similar to that of AtPUB22 and AtPUB23 in Arabidopsis, with both sets of genes being negative regulators of biotic stresses (Cho et al., 2008; Trujillo et al., 2008). However, OsPUB31 appeared not to affect the resistance to M. grisea. CMPG1, a highly related gene to Arabidopsis PUB20 and PUB21, positively regulated the response to disease resistance in tomato and tobacco (Gonzalez et al., 2006). The OsPUB39 probably positively regulated the resistance to M. grisea (Figure 3). These results further confirmed previous reports that E3 ubiquitin ligases regulated the resistance to biotic stress, either positively or negatively (Ni et al., 2010; He et al., 2015; Wang et al., 2016; You et al., 2016; Chen et al., 2017; Kim et al., 2021).
ROS production is important for the activation of immune responses against pathogen infection, and the expression levels of defense-related genes are closely related to disease resistance (Lehmann et al., 2015; Qi et al., 2018; Segal and Wilson, 2018; Waszczak et al., 2018). E3 ubiquitin ligase also regulated the resistance to biotic stress in these two ways (Yaeno and Iba, 2008; Zhou and Zeng, 2018). Silencing of APIP6 resulted in reduced resistance to M. oryzae in rice by reducing the generation of flg22-induced ROS and suppressing the expression of defense-related genes (Park et al., 2012). In our study, ROS accumulation and the expression levels of defense-related genes were analyzed to explore the mechanism for the altered resistance by gene silencing. The silencing of OsPUB39 resulted in increased ROS accumulation and H2O2 content, while the silencing of OsPUB34 and OsPUB33 resulted in less ROS accumulation and H2O2 content (Figures 4A,B). Plants have a highly efficient system for maintaining ROS homeostasis (Mittler et al., 2004). They possess two mechanisms of scavenging ROS, one is by small molecules (including glutathione, ascorbic acid, flavonoids, alkaloids, and carotenoids), and the other is by detoxifying enzymes, including superoxide dismutase (SOD), catalase (CAT), peroxidase, and peroxiredoxins (Lehmann et al., 2009). The activities of SOD and CAT were analyzed to explain the altered ROS accumulation. The altered SOD and CAT activities in BMV:target gene-infiltrated seedlings may explain the change in ROS accumulation (Figures 4C,D). The expression levels of defense-related genes were also analyzed. LeATL6 regulated elicitor-activated defense responses via a JA-dependent signaling pathway in tomato (Hondo et al., 2007). ASK1/ASK2 and cullin-1 formed the SCF-ubiquitin ligase complex, as the signaling receptor of JA, through the degradation of the inhibitor JAZ of the JA pathway, to activate the expression of JA-response genes (Devoto et al., 2002; Xu et al., 2002; Ren et al., 2005; Chini et al., 2007; Thines et al., 2007). MdPUB29 increased the resistance to Botryosphaeria dothidea by the SA pathway (Han et al., 2019). Therefore, SA-responsive genes, JA-responsive genes, and OsWRKY45 (a positive regulator in response to fungal pathogens) were selected. The expression level of OsWRKY45 decreased in BMV:OsPUB39-infiltrated plants and increased in the BMV:OsPUB34- and BMV:OsPUB33-infiltrated plants. The expression levels of JA-responsive genes (OsLOX1 and OsPR3) and SA-responsive genes (OsNH1 and OsPR1a) were decreased in BMV:OsPUB39-infiltrated plants when compared to control and increased in BMV:OsPUB34- and BMV:OsPUB33-infiltrated plants (Figure 4E). These results indicate that OsPUB39, OsPUB34, and OsPUB33 were involved in response to M. grisea, possibly through SA and JA/ET pathways.
Drought and cold are major abiotic stresses that severely affect plant growth and productivity. E3 ubiquitin ligases have been reported to be positively or negatively involved in response to drought stress. OsiSAP7 negatively regulated ABA-stress signaling and imparted sensitivity to drought stress in Arabidopsis (Sharma et al., 2015). AIRP1 positively regulated the response to drought. Its overexpression in plants increased stomatal closure, ROS accumulation, expression of drought-responsive genes, and ABA-responsive bZIP transcript factor (Ryu et al., 2010). SDIR1 positively regulated the ABA pathway. The knockout mutants were less sensitive to ABA, whereas the overexpression plants showed increased stomatal closure and resistance to drought in Arabidopsis (Zhang et al., 2007). Similarly, the plants with overexpression of RHA2a/RHA2b were highly sensitive to ABA, with increased stomatal closure, decreased water loss, and increased resistance to drought (Bu et al., 2009; Li et al., 2011). Wheat TaPUB1 positively regulated the tolerance to drought stress by improving antioxidant capability (Zhang et al., 2017). Our study showed that the expression levels of OsATL69 and OsPUB33 were induced by drought stress (Figure 2A). OsATL69 positively regulated the tolerance to drought stress (Figure 5), which was in line with the previous reports that AtATL78 acted as positive regulators of drought stress (Cho et al., 2008; Trujillo et al., 2008). On the other hand, OsPUB33 negatively regulated the tolerance to drought stress (Figure 5), consistent with previous reports that AtPUB22, AtPUB23, AtPUB24, and OsPUB41 acted as negative regulators of drought stress (Cho et al., 2008; Trujillo et al., 2008; Seo et al., 2021). To explore the reason for the altered tolerance to drought stress due to the silencing of OsATL69 or OsPUB33, the proline content, sugar content, and expression levels of drought-responsive genes were analyzed. Proline content and sugar content in BMV:OsATL69-infiltrated plants decreased under drought stress when compared to the control but increased in BMV:OsPUB33-infiltrated plants (Figures 5D,E). The expression levels of drought-responsive genes increased in BMV:OsPUB33-infiltrated plants and decreased in BMV:OsATL69- infiltrated plants compared to the control (Figures 5F,G). These results indicated that OsATL69 and OsPUB33 conferred tolerance to drought stress, possibly through the regulation of proline content, sugar content, and the expression levels of drought-responsive genes.
E3 ubiquitin ligases are also involved in the tolerance to cold stress. AtATL78 and AtATL80 negatively regulated the tolerance to cold stress in Arabidopsis (Lee et al., 2001; Kim and Kim, 2013; Suh and Kim, 2015). OsDIRP1 positively regulated the tolerance to cold stress in rice (Cui et al., 2018). We observed that the expression of OsATL32 was induced by cold stress (Figure 2B). The BMV:OsATL32-infiltrated seedlings showed decreased resistance to cold resistance when compared to control (Figure 6A), with a lower survival rate and chlorophyll content (Figures 6B,E), but higher MDA content and electrolyte leakage (Figures 6C,D). Moreover, the expression of cold-responsive genes in BMV:OsATL32-infiltrated seedlings was downregulated when compared to the control (Figure 6E). These results indicate that OsATL32 regulated the tolerance to cold stress, possibly through MDA content and the expression levels of cold-responsive genes.
ABA is a critical signaling mediator that regulates diverse biological processes in various organisms (Kumar et al., 2019). Plants regulate response to abiotic stress via two pathways, one is ABA-dependent, and the other is ABA-independent. E3 ubiquitin ligases regulate the tolerance to abiotic stresses dependent or independent of ABA. AtARRE negatively regulates ABA signaling in Arabidopsis thaliana (Wang et al., 2018). PeCHYR1 elevates the tolerance to drought stress by ABA-induced stomatal closure via ROS production in Populus euphratica (He et al., 2018). Rma1H1 responds to drought by mediating the ubiquitination of water channel protein isoenzyme PIP2;1 to downregulate the expression of the water channel protein, independent of ABA (Lee et al., 2009; Son et al., 2009; Bae et al., 2011). Our results indicate that the expression levels of OsATL69 and OsATL32 were induced by ABA, whereas those of OsPUB33 were not (Figure 2E). This may indicate that the regulation of response by OsATL69 and OsATL32 to abiotic stress was ABA-dependent, whereas that of OsPUB33 was ABA-independent. This need further experiment by ABA content or ABA treatment on infiltrated seedlings.
E3 ubiquitin ligases have been reported to function in response to heat stress. AtSAP5 conferred tolerance to heat stress (Kim et al., 2015). AtPPRT1 increased the tolerance to heat stress in Arabidopsis (Liu et al., 2020). SlSIZ1 positively regulated the tolerance to heat stress in tomato (Zhang et al., 2018), whereas HTD1 negatively regulated thermotolerance in Arabidopsis (Kim et al., 2014). However, in our study, the expression levels of none of the E3 ubiquitin ligase genes were observed to be induced by heat stress (Figure 2C). Also, because of the elimination of VIGS by high temperature, we could not research its function in response to heat stress. The possible roles of these 11 E3 ubiquitin ligase genes in the resistance to heat stress can be explored using transgenic lines.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Author Contributions
FS conceived the study. HZ and MJ designed the experiments. HZ and DZ performed the experiments. HZ and FS analyzed the data. MJ drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Taizhou Municipal Science and Technology Project (20ny18), the Science Foundation for Distinguished Young Scholars of Taizhou University (2019JQ001), and the Zhejiang Provincial Natural Science Foundation of China (LQ20C130005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.840360/full#supplementary-material
References
Ariani, P., Vandelle, E., Wong, D., Giorgetti, A., Porceddu, A., Camiolo, S., et al. (2017). Comprehensive workflow for the genome-wide identification and expression meta-analysis of the ATL E3 ubiquitin ligase gene family in grapevine. J. Vis. Exp. 130:56626. doi: 10.3791/56626
Bae, H., Kim, S., Cho, S., Kang, B., and Kim, W. (2011). Overexpression of OsRDCP1, a rice RING domain-containing E3 ubiquitin ligase, increased tolerance to drought stress in rice (Oryza sativa L.). Plant Sci. 180, 775–782. doi: 10.1016/j.plantsci.2011.02.008
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Berrocal-Lobo, M., Stone, S., Yang, X., Antico, J., Callis, J., Ramonell, K. M., et al. (2010). ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase-mediated defense responses. PLoS One 5:e14426. doi: 10.1371/journal.pone.0014426
Bhaskar, S., and Joemar, T. (2020). Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 10:9581. doi: 10.1038/s41598-020-66553-1
Bu, Q., Li, H., Zhao, Q., Jiang, H., Zhai, Q., Zhang, J., et al. (2009). The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol. 150, 463–481. doi: 10.1104/pp.109.135269
Chapagain, S., Park, Y. C., Kim, J. H., and Jang, C. S. (2018). Oryza sativa salt-induced RING E3 ligase 2 (OsSIRP2) acts as a positive regulator of transketolase in plant response to salinity and osmotic stress. Planta 247, 925–939. doi: 10.1007/s00425-017-2838-x
Chen, S. C., Zhao, H. J., Wang, M. M., Li, J. D., Wang, Z. H., Wang, F. H., et al. (2017). Overexpression of E3 ubiquitin ligase gene AdBiL contributes to resistance against chilling stress and leaf mold disease in tomato. Front. Plant Sci. 8:1109. doi: 10.3389/fpls.2017.01109
Chen, X., Wang, T., Rehman, A., Wang, Y., Qi, J., Li, Z., et al. (2021). Arabidopsis U-box E3 ubiquitin ligase PUB11 negatively regulates drought tolerance by degrading the receptor-like protein kinases LRR1 and KIN7. J. Integr. Plant Biol. 63, 494–509. doi: 10.1111/jipb.13058
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448, 666–671. doi: 10.1038/nature06006
Cho, S. K., Ryu, M. Y., Song, C., Kwak, J. M., and Kim, W. T. (2008). Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20, 1899–1914. doi: 10.1105/tpc.108.060699
Cui, L., Min, H., Yu, S., Byun, M., Oh, Tae, Lee, A., et al. (2022). OsATL38 mediates mono-ubiquitination of the 14-3-3 protein OsGF14d and negatively regulates the cold stress response in rice. J. Exp. Bot. 73, 307–323. doi: 10.1093/jxb/erab392
Cui, L. H., Min, H. J., Byun, M. Y., Oh, H. G., and Kim, W. T. (2018). OsDIRP1, a putative RING E3 ligase, plays an opposite role in drought and cold stress responses as a negative and positive factor, respectively, in rice (Oryza sativa L.). Front. Plant Sci. 9:1797. doi: 10.3389/fpls.2018.01797
Deng, F., Guo, T., Lefebvre, M., Scaglione, S., Antico, C. J., Jing, T., et al. (2017). Expression and regulation of ATL9, an E3 ubiquitin ligase involved in plant defense. PLoS One 12:e0188458. doi: 10.1371/journal.pone.0188458
Devoto, A., Nieto-Rostro, M., Xie, D., Ellis, C., Harmston, R., Patrick, E., et al. (2002). COI1 links jasmonate signaling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32, 457–466. doi: 10.1046/j.1365-313x.2002.01432.x
Dhawan, R., Luo, H., Foerster, A. M., Abuqamar, S., Du, H. N., Briggs, S. D., et al. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21, 1000–1019. doi: 10.1105/tpc.108.062364
Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 103, 8281–8286. doi: 10.1073/pnas
Du, B., Nie, N., Sun, S., Hu, Y., Bai, Y., He, S., et al. (2021). A novel sweetpotato RING-H2 type E3 ubiquitin ligase gene IbATL38 enhances salt tolerance in transgenic Arabidopsis. Plant Sci. 304:110802. doi: 10.1016/j.plantsci.2020.110802
Gonzalez, L., Tsitsigiannis, D., Ludwig, A., Panicot, M., Shirasu, K., and Jones, J. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18, 1067–1083. doi: 10.1105/tpc.106.040998
Han, P., Dong, Y., Gu, K., Yu, J., Hu, D., and Hao, Y. (2019). The apple U-box E3 ubiquitin ligase MdPUB29 contributes to activate plant immune response to the fungal pathogen Botryosphaeria dothidea. Planta 249, 1177–1188. doi: 10.1007/s00425-018-03069-z
He, F., Wang, H., Li, H., Su, Y., Li, S., Yang, Y., et al. (2018). PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA-induced stomatal closure by ROS production in Populus. Plant Biotechnol. J. 16, 1514–1528. doi: 10.1111/pbi.12893
He, Q., McLellan, H., Boevink, P., Sadanandom, A., Xie, C., Birch, P., et al. (2015). U-box E3 ubiquitin ligase PUB17 acts in the nucleus to promote specific immune pathways triggered by Phytophthora infestans. J. Exp. Bot. 66, 3189–3199. doi: 10.1093/jxb/erv128
Hondo, D., Hase, S., Kanayama, Y., Yoshikawa, N., Takenaka, S., and Takahashi, H. (2007). The LeATL6-associated ubiquitin/proteasome system may contribute to fungal elicitor-activated defense response via the jasmonic acid-dependent signaling pathway in tomato. Mol. Plant Microbe Interact. 20, 72–81. doi: 10.1094/MPMI-20-0072
Hong, Y., Zhang, H., Huang, L., Li, D., and Song, F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7:4. doi: 10.3389/fpls.2016.00004
Huang, L., Hong, Y., Zhang, H., Li, D., and Song, F. (2016). Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 16:203. doi: 10.1186/s12870-016-0897-y
Joo, H., Lim, C., and Lee, S. (2016). Identification and functional expression of the pepper RING type E3 ligase, CaDTR1, involved in drought stress tolerance via ABA-mediated signaling. Sci. Rep. 6:30097. doi: 10.1038/srep30097
Kelley, D. (2018). E3 ubiquitin ligases: key regulators of hormone signaling in plants. Mol. Cell Proteomics 17, 1047–1054. doi: 10.1074/mcp.MR117.000476
Kim, G., Cho, Y., and Yoo, S. (2015). Regulatory functions of evolutionarily conserved AN1/A20-like Zinc finger family proteins in Arabidopsis stress responses under high temperature. Biochem. Biophys. Res. Commun. 457, 213–220. doi: 10.1016/j.bbrc.2014.12.090
Kim, M., Kang, K., and Cho, Y. (2021). Molecular and functional analysis of U-box E3 ubiquitin ligase gene family in rice (Oryzasativa). Int. J. Mol. Sci. 22:12088. doi: 10.3390/ijms222112088
Kim, S., Lee, J., Seo, K., Ryu, B., Sung, Y., Chung, T., et al. (2014). Characterization of a novel DWD protein that participates in heat stress response in Arabidopsis. Mol. Cells 37, 833–840. doi: 10.14348/molcells.2014.0224
Kim, S. J., and Kim, W. T. (2013). Suppression of Arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 587, 2584–2590. doi: 10.1016/j.febslet.2013.06.038
Ko, J. H., Yang, S. H., and Han, K. H. (2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47, 343–355. doi: 10.1111/j.1365-313X.2006.02782.x
Kumar, M., Kesawat, M., Ali, A., Lee, S., Gill, S., and Kim, A. (2019). Integration of abscisic acid signaling with other signaling pathways in plant stress responses and development. Plants 8:592. doi: 10.3390/plants8120592
Lee, H., Xiong, L., Gong, Z., Ishitani, M., Stevenson, B., and Zhu, J. K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15, 912–924. doi: 10.1101/gad.866801
Lee, H. K., Cho, S. K., Son, O., Xu, Z., Hwang, I., and Kim, W. T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21, 622–641. doi: 10.1105/tpc.108.061994
Lehmann, M., Schwarzlander, M., Obata, T., Sirikantaramas, S., Burow, M., Olsen, C. E., et al. (2009). The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol. Plant 2, 390–406. doi: 10.1093/mp/ssn080
Lehmann, S., Serrano, M., Haridon, F., Tjamos, S., and Metraux, J. (2015). Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 112, 54–62. doi: 10.1016/j.phytochem.2014.08.027
Li, H., Jiang, H., Bu, Q., Zhao, Q., Sun, J., Xie, Q., et al. (2011). The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol. 156, 550–563. doi: 10.1104/pp.111.176214
Lim, C., Park, C., Kim, J., Joo, H., Hong, E., and Lee, S. (2017). Pepper CaREL1, a ubiquitin E3 ligase, regulates drought tolerance via the ABA-signaling pathway. Sci. Rep. 7:477. doi: 10.1038/s41598-017-00490-4
Liu, Y., Xiao, S., Sun, H., Pei, L., Liu, Y., Peng, L., et al. (2020). AtPPRT1, an E3 ubiquitin ligase, enhances the thermotolerance in Arabidopsis. Plants 9:1074. doi: 10.3390/plants9091074
Lu, X., Shu, N., Wang, D., Wang, J., Chen, X., Zhang, B., et al. (2020). Genome-wide identification and expression analysis of PUB genes in cotton. BMC Genomics 21:213. doi: 10.1186/s12864-020-6638-5
Luo, H., Song, F., and Zheng, Z. (2005). Overexpression in transgenic tobacco reveals different roles for the rice homeodomain gene OsBIHD1 in biotic and abiotic stress responses. J. Exp. Bot. 56, 2673–2682. doi: 10.1093/jxb/eri260
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Morreale, F., and Walden, H. (2016). Types of ubiquitin ligases. Cell 165, 248–248. doi: 10.1016/j.cell.2016.03.003
Ni, X., Tian, Z., Liu, J., Song, B., and Xie, C. (2010). Cloning and molecular characterization of the potato RING finger protein gene StRFP1and its function in potato broad-spectrum resistance against Phytophthorainfestans. J. Plant Physiol. 167, 488–496. doi: 10.1016/j.jplph.2009.10.019
Park, C., Chen, S., Shirsekar, G., Zhou, B., Khang, C., Songkumarn, P., et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Pickart, C., and Eddins, M. (2004). Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72. doi: 10.1016/j.bbamcr.2004.09.019
Prasad, M., Schofield, A., Lyzenga, W., Liu, H., and Stone, S. (2010). Arabidopsis RING E3 ligase XBAT32 regulates lateral root production through its role in ethylene biosynthesis. Plant Physiol. 153, 1587–1596. doi: 10.1104/pp.110.156976
Qi, J., Song, C., Wang, B., Zhou, J., Kangasjärvi, J., Zhu, J., et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. doi: 10.1111/jipb.12654
Qin, F., Sakuma, Y., Tran, L., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20, 1693–1707. doi: 10.1105/tpc.107.057380
Qin, Q., Wang, Y., Huang, L., Du, F., Zhao, X., Li, Z., et al. (2020). A U-box E3 ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol. Biol. 102, 89–107. doi: 10.1007/s11103-019-00933-8
Ren, C., Pan, J., Peng, W., Genschik, P., Hobbie, L., Hellmann, H., et al. (2005). Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 42, 514–524. doi: 10.1111/j.1365-313X.2005.02394.x
Ryu, M., Cho, S., and Kim, W. (2010). The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol. 154, 1983–1997. doi: 10.1104/pp.110.164749
Segal, L., and Wilson, R. (2018). Reactive oxygen species metabolism and plant-fungal interactions. Fungal Genet. Biol. 110, 1–9. doi: 10.1016/j.fgb.2017.12.003
Seo, D., Lee, S., Yu, S., Cui, L., Min, H., Lee, S., et al. (2021). OsPUB41, a U-box E3 ubiquitin ligase, acts as a negative regulator of drought stress response in rice (Oryza sativa L.). Plant Mol. Biol. 106, 463–477. doi: 10.1007/s11103-021-01158-4
Serrano, M., and Guzman, P. (2004). Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics 167, 919–929. doi: 10.1534/genetics.104.028043
Sharma, B., Saxena, H., and Negi, H. (2021). Genome-wide analysis of HECT E3 ubiquitin ligase gene family in Solanum lycopersicum. Sci. Rep. 11:15891. doi: 10.1038/s41598-021-95436-2
Sharma, G., Giri, J., and Tyagi, A. (2015). Rice OsiSAP7 negatively regulates ABA stress signaling and imparts sensitivity to water-deficit stress in Arabidopsis. Plant Sci. 237, 80–92. doi: 10.1016/j.plantsci.2015.05.011
Son, O., Cho, S., Kim, E., and Kim, W. (2009). Characterization of three Arabidopsis homologs of human RING membrane anchor E3 ubiquitin ligase. Plant Cell Rep. 28, 561–569. doi: 10.1007/s00299-009-0680-8
Stone, S., Williams, L., Farmer, L., Vierstra, R., and Callis, J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428. doi: 10.1105/tpc.106.046532
Suh, J., and Kim, W. (2015). Arabidopsis RING E3 ubiquitin ligase AtATL80 is negatively involved in phosphate mobilization and cold stress response in sufficient phosphate growth conditions. Biochem. Biophys. Res. Commun. 463, 793–799. doi: 10.1016/j.bbrc.2015.06.015
Sun, H., Li, J., Li, X., Lv, Q., Chen, L., Wang, B., et al. (2022). RING E3 ubiquitin ligase TaSADR1 negatively regulates drought resistance in transgenic Arabidopsis. Plant Physiol. Biochem. 170, 255–265. doi: 10.1016/j.plaphy.2021.12.004
Sun, J., Sun, Y., Ahmed, R., Ren, A., and Xie, M. (2019). Research progress on plant RING-finger proteins. Genes 10:973. doi: 10.3390/genes10120973
Tal, L., Gil, M., Guercio, A., and Shabek, N. (2020). Structural aspects of plant hormone signal perception and regulation by ubiquitin ligases. Plant Physiol. 182, 1537–1544. doi: 10.1104/pp.19.01282
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF (COI1) complex during jasmonate signaling. Nature 448, 661–665. doi: 10.1038/nature05960
Trujillo, M., Ichimura, K., Casais, C., and Shirasu, K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18, 1396–1401. doi: 10.1016/j.cub.2008.07.085
Verma, V., Ravindran, P., and Kumar, P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wang, B., Li, C., Kong, X., Li, Y., Liu, Z., Wang, J., et al. (2018). AtARRE, an E3 ubiquitin ligase, negatively regulates ABA signaling in Arabidopsis thaliana. Plant Cell Rep. 37, 1269–1278. doi: 10.1007/s00299-018-2311-8
Wang, F., and Deng, X. (2011). Plant ubiquitin-proteasome pathway and its role in gibberellin signaling. Cell Res. 21, 1286–1294. doi: 10.1038/cr.2011.118
Wang, T., Chang, C., Gu, C., Tang, S., Xie, Q., and Shen, Q. (2016). An E3 ligase affects the NLR receptor stability and immunity to Powdery Mildew. Plant Physiol. 172, 2504–2515. doi: 10.1104/pp.16.01520
Wang, X., Ding, Y., Li, Z., Shi, Y., Wang, J., Hua, J., et al. (2019). PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15. Dev. Cell 51, 222–235. doi: 10.1016/j.devcel.2019.08.008
Waszczak, C., Carmody, M., and Kangasjärvi, J. (2018). Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 69, 209–236. doi: 10.1146/annurev-arplant-042817-040322
Wu, H., Ye, H., Yao, R., Zhang, T., and Xiong, L. (2015). OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 232, 1–12. doi: 10.1016/j.plantsci.2014.12.010
Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W. L., Ma, H., et al. (2002). The SCF (COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. doi: 10.1105/tpc.003368
Yaeno, T., and Iba, K. (2008). BAH1/NLA, a RING-type ubiquitin E3 ligase, regulates the accumulation of salicylic acid and immune responses to Pseudomonas syringae DC3000. Plant Physiol. 148, 1032–1041. doi: 10.1104/pp.108.124529
Yang, L., Liu, Q., Liu, Z., Yang, H., Wang, J., Li, X., et al. (2016). Arabidopsis C3HC4-RING finger E3 ubiquitin ligase AtAIRP4 positively regulates stress-responsive abscisic acid signaling. J. Integr. Plant Biol. 58, 67–80. doi: 10.1111/jipb.12364
Yin, Z., Chen, J., Zeng, L., Goh, M., Leung, H., Khush, G., et al. (2000). Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 13, 869–876. doi: 10.1094/MPMI.2000.13.8.869
You, Q., Zhai, K., Yang, D., Yang, W., Wu, J., Liu, J., et al. (2016). An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 20, 758–769. doi: 10.1016/j.chom.2016.10.023
Zeng, L., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., et al. (2004). Spotted leaf 11, a negative regulator of plant cell death and defence, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16, 2795–2808. doi: 10.1105/tpc.104.025171
Zhang, C., Hao, Z., Ning, Y., and Wang, G. (2019). SINA E3 ubiquitin ligases: versatile moderators of plant growth and stress response. Mol. Plant 12, 610–612. doi: 10.1016/j.molp.2019.03.013
Zhang, G., Zhang, M., Zhao, Z., Ren, Y., Li, Q., and Wang, W. (2017). Wheat TaPUB1 modulates plant drought stress resistance by improving antioxidant capability. Sci. Rep. 7:7549. doi: 10.1038/s41598-017-08181-w
Zhang, H., Liu, J., He, F., Wang, Z., Ning, Y., and Wang, G. (2015). OsHUB1 and OsHUB2 interact with SPIN6 and form homo- and hetero-dimers in rice. Plant Signal. Behav. 10:e1039212. doi: 10.1080/15592324.2015.1039212
Zhang, H., Zheng, D., Yin, L., Song, F., and Jiang, M. (2021). Functional analysis of OsMED16 and OsMED25 in response to biotic and abiotic stresses in rice. Front. Plant Sci. 12:652453. doi: 10.3389/fpls.2021.652453
Zhang, S., Wang, S., Lv, J., Liu, Z., Wang, Y., Ma, N., et al. (2018). SUMO E3 ligase SlSIZ1 facilitates heat tolerance in tomato. Plant Cell Physiol. 59, 58–71. doi: 10.1093/pcp/pcx160
Zhang, X., Garreton, V., and Chua, N. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532–1543.
Zhang, Y., Yang, C., Li, Y., Zheng, N., Chen, H., Zhao, Q., et al. (2007). SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19, 1912–1929. doi: 10.1105/tpc.106.048488
Keywords: rice, E3 ubiquitin ligases, biotic stress, abiotic stress, ROS, expression patterns
Citation: Zhang H, Zheng D, Song F and Jiang M (2022) Expression Patterns and Functional Analysis of 11 E3 Ubiquitin Ligase Genes in Rice. Front. Plant Sci. 13:840360. doi: 10.3389/fpls.2022.840360
Received: 21 December 2021; Accepted: 10 February 2022;
Published: 02 March 2022.
Edited by:
Sang-Wook Han, Chung-Ang University, South KoreaReviewed by:
Chunliu Zhuo, University of North Texas, United StatesJi Huang, Nanjing Agricultural University, China
Copyright © 2022 Zhang, Zheng, Song and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Jiang, amlhbmdtaW5nMTk3M0AxMzkuY29t
 Huijuan Zhang
Huijuan Zhang Dewei Zheng
Dewei Zheng Fengming Song
Fengming Song Ming Jiang
Ming Jiang