- Department of Horticulture and Crop Science, The Ohio State University, Wooster, OH, United States
Developmental petal senescence is a type of programmed cell death (PCD), during which the production of ethylene is induced, the expression of PCD-related genes is upregulated, and nutrients are recycled. Autophagy is an intracellular mechanism involved in PCD modulation and nutrient cycling. As a central component of the autophagy pathway, Autophagy Gene 6 (ATG6) was previously shown as a negative regulator of petal senescence. To better understand the role of autophagy in ethylene biosynthesis and nutrient remobilization during petal senescence, we generated and characterized the knockout (KO) mutants of PhATG6 using CRISPR/Cas9 in Petunia × hybrida ‘Mitchell Diploid.’ PhATG6-KO lines exhibited decreased flower longevity when compared to the flowers of the wild-type or a non-mutated regenerative line (controls), confirming the negative regulatory role of ATG6 in petal senescence. Smaller capsules and fewer seeds per capsule were produced in the KO plants, indicating the crucial function of autophagy in seed production. Ethylene production and ethylene biosynthesis genes were upregulated earlier in the KO lines than the controls, indicating that autophagy affects flower longevity through ethylene. The transcript levels of petal PCD-related genes, including PhATG6, PhATG8d, PhPI3K (Phosphatidylinositol 3-Kinase), and a metacaspase gene PhMC1, were upregulated earlier in the corollas of PhATG6-KO lines, which supported the accelerated PCD in the KO plants. The remobilization of phosphorus was reduced in the KO lines, showing that nutrient recycling was compromised. Our study demonstrated the important role of autophagy in flower lifespan and seed production and supported the interactions between autophagy and various regulatory factors during developmental petal senescence.
Introduction
The senescence of flower petals is a gradual process of programmed cell death (PCD) (Rogers, 2006). Based on the morphological changes, petal senescence can be categorized as wilting, withering, or abscission of turgid petals (Van Doorn, 2001). In petunia (Petunia × hybrida), the flower petals are fused to form a corolla, which wilts during senescence (Jones et al., 2009). Petal senescence can be induced by different factors, including aging, pollination, abiotic stresses, and pathogens, where different forms of PCD are involved (Van Doorn and Woltering, 2008). A sequence of events that includes nuclear fragmentation, protein degradation, nutrient mobilization, and membrane leakage accompanies petal senescence (Jones et al., 2009). Various senescence-related genes are upregulated during petal senescence, including genes involved in macromolecule degradation, nutrient remobilization, hormone biosynthesis and signaling, and cell death regulation (Broderick et al., 2014; Tsanakas et al., 2014; Wang et al., 2018; Yang et al., 2019).
Autophagy is an intracellular process that is responsible for the transportation and degradation of toxic or damaged cellular components in eukaryotic cells (Su et al., 2020). Autophagy assists in maintaining cell homeostasis and modulates developmental PCD in plants (Üstün et al., 2017). The most prominent and well-studied autophagy pathway is macroautophagy (hitherto referred to as “autophagy”), where the cellular components are enclosed in double-membrane vesicles (i.e., autophagosomes) to be transported to the vacuole for degradation (Feng et al., 2014). Autophagosomes are constructed collaboratively by proteins encoded from a variety of autophagy genes (ATGs) (Su et al., 2020). ATGs are involved in the regulation of PCD during petal senescence (Shibuya, 2012). The expression of ATGs, including ATG1, ATG4, ATG5, ATG6, ATG7, ATG8a, b, d-f, ATG13, and Phosphatidylinositol 3-Kinase (PI3K), is upregulated during petal senescence in petunia, Japanese morning glory (Ipomoea nil), and hibiscus (Hibiscus rosa-sinensis) (Shibuya et al., 2011; Broderick et al., 2014; Trivellini et al., 2016; Quijia Pillajo et al., 2018). In our previous study, silencing ATG6 or PI3K in petunia results in accelerated petal senescence, as well as reduced flower number and biomass (Lin and Jones, 2021).
ATG6/VPS30/Beclin1 is a central component of the autophagy pathway and is involved in the regulation of PCD (Cao and Klionsky, 2007; Menon and Dhamija, 2018). ATG6 is a single copy gene in Arabidopsis (Arabidopsis thaliana), barley (Hordeum vulgare), and grape (Vitis vinifera), as well as tobacco (Nicotiana benthamiana) and tomato (Solanum lycopersicum), which are in the same family (Solanaceae) as petunia (Tang and Bassham, 2018). Although the copy number of ATG6 in petunia has not been confirmed, only one ATG6-like protein was found when blasting the petunia genomes with Arabidopsis ATG6 sequence.1 In addition to its function in petal senescence, ATG6 also plays an important role in the regulation of leaf developmental and stress-induced senescence. Barley leaves with silenced ATG6 are more susceptible to darkness, oxidative stress, nutrient deficiency, and salt stress (Zeng et al., 2017). Arabidopsis, tobacco, and wheat (Triticum aestivum) with suppressed ATG6 show early leaf senescence under normal conditions and enhanced pathogen-induced PCD in leaves when infected with viral, bacterial, or fungal pathogens (Liu et al., 2005; Patel and Dinesh-Kumar, 2008; Yue et al., 2015). Even though most of the studies of ATG6 in plants focus on leaf senescence, similarities have been identified between leaf and petal senescence, including the upregulation of autophagy genes (Price et al., 2008; Wagstaff et al., 2009). Understanding the regulatory mechanisms of leaf senescence can facilitate research on petal senescence, and vice versa.
Ethylene interacts with autophagy during petal senescence (Liao and Bassham, 2020). In ethylene-treated petunia flowers, the expression of ATG8a-d increases, while the application of an ethylene inhibitor suppresses ATG8a-d expression (Shibuya et al., 2013). Treating pollinated flowers with an ethylene inhibitor delays the induced expression of ATG8a-d (Shibuya et al., 2013). Ethylene-induced ATG expression in flowers is thought to be regulated by ethylene-responsive transcription factors (TF). Arabidopsis ethylene-responsive TFs EIL and AP2 interact with the promoters of ATG8a and ATG8h in yeast one-hybrid assays (Wang et al., 2020). In tomato, direct binding of ethylene-responsive TF ERF5 to the promoters of ATG8d and ATG18h has been shown in vitro (Zhu et al., 2018). Correlations are also found between autophagy and ethylene biosynthesis. The expression of a key ethylene biosynthesis gene, 1-aminocyclopropane-1-carboxylic acid synthase (ACC synthase, ACS), decreases in the senescing petals of ATG6- or PI3K-silenced petunias (Lin and Jones, 2021). In PI3K-overexpressing tobacco, the production of ethylene is enhanced as a result of increased expression of another ethylene biosynthesis gene ACC oxidase 1 (ACO1) despite decreases in the expression of ACO2 (Dek et al., 2017).
The remobilization of mineral nutrients is a critical process during petal senescence (Jones, 2013). Evolutionarily, the purpose of flower petals is to attract pollinators for sexual reproduction. Because it is energetically costly to maintain these elaborate structures, the flower petals senesce once the flower is pollinated, or when the stigma is no longer receptive to pollination (Jones et al., 2009). To preserve resources, nutrients are remobilized from the senescing petals to other tissues or organs for recycling (Borghi and Fernie, 2020). Nutrient remobilization during petal senescence is regulated by autophagy (Avila-Ospina et al., 2014). Nuclear fragmentation and DNA mass reduction are delayed in pollinated flowers treated with 3-methyladenine (3-MA), an inhibitor of autophagy protein PI3K (Yamada et al., 2009). The application of the autophagy inhibitor concanamycin A limits the growth of the ovary in pollinated flowers, suggesting that autophagy is a key regulator in pollination-mediated nutrient recycling (Shibuya et al., 2013). Phosphorus (P) and nitrogen (N) are two of the major nutrient elements remobilized during petal senescence (Jones, 2013), and an increase of N and P is found in the ovary as a sink organ after pollination-induced senescence (Hew et al., 1989). Ethylene plays an important role in autophagy-mediated nutrient remobilization during petal senescence (Jones, 2013). In both pollinated and unpollinated senescing flowers, the remobilization of N and P is reduced in ethylene-insensitive transgenic petunia compared to the ethylene-sensitive wild-type control (Jones, 2008).
The clustered regularly interspaced short palindromic repeats (CRISPR) and associated endonuclease 9 (Cas9) system is a powerful tool for reverse genetic analysis and horticulture crop improvement (Karkute et al., 2017). Mutagenesis of the ACO genes in petunia or a NAC transcription factor in Japanese morning glory using CRISPR/Cas9 successfully delays petal senescence in these plants (Shibuya et al., 2018; Xu et al., 2020, 2021). To better understand the function of PhATG6 in the regulation of corolla senescence and nutrient recycling in petunia and to characterize ethylene’s role in autophagy-mediated corolla senescence, we analyzed stable PhATG6-knockout (KO) mutants generated with CRISPR/Cas9-mediated gene editing.
Materials and Methods
Plant Materials
Seeds of petunia (Petunia × hybrida) inbred line ‘Mitchell Diploid’ (MD) were surface-sterilized with 95% ethanol for 1 min and 10% bleach for 10 min before rinsing with sterile water three times. The sterilized seeds were sown on ½ strength MS (Murashige and Skoog) media to generate sterile seedlings. Seedlings were grown under a 16-h light/8-h dark cycle at 25°C for 4–7 weeks until the leaves were ready to be used for transformation.
Plasmid Construction
The CRISPR plasmid for generating the PhATG6 knockout (KO) was constructed following a published protocol (Xing et al., 2014). The vector pKSE401 was from Qi-Jun Chen laboratory (plasmid #62202; Addgene, Watertown, MA, United States). This binary vector contains the sequence of Cas9 and an insert site for gRNA sequences, along with a kanamycin resistance selective marker. The genomic DNA sequence of PhATG6 (Subject ID: Peaxi162Scf00403) was obtained from the petunia genome sequence in the Sol Genome Network2 for gRNA design. Specifically, 23-bp target sites (including a 20-bp gRNA sequence followed by a 3-bp sequence NGG, a PAM for Cas9) were identified manually within the exons of the PhATG6 DNA sequence as potential gRNAs. These potential gRNA sequences were evaluated for the potential knockout accuracy3, and two gRNAs were selected from exon 3 of PhATG6 DNA (Figure 1). The gRNAs were inserted into the pKSE401 vector under an AtU6-26 promoter using a Golden Gate Assembly method (Xing et al., 2014).
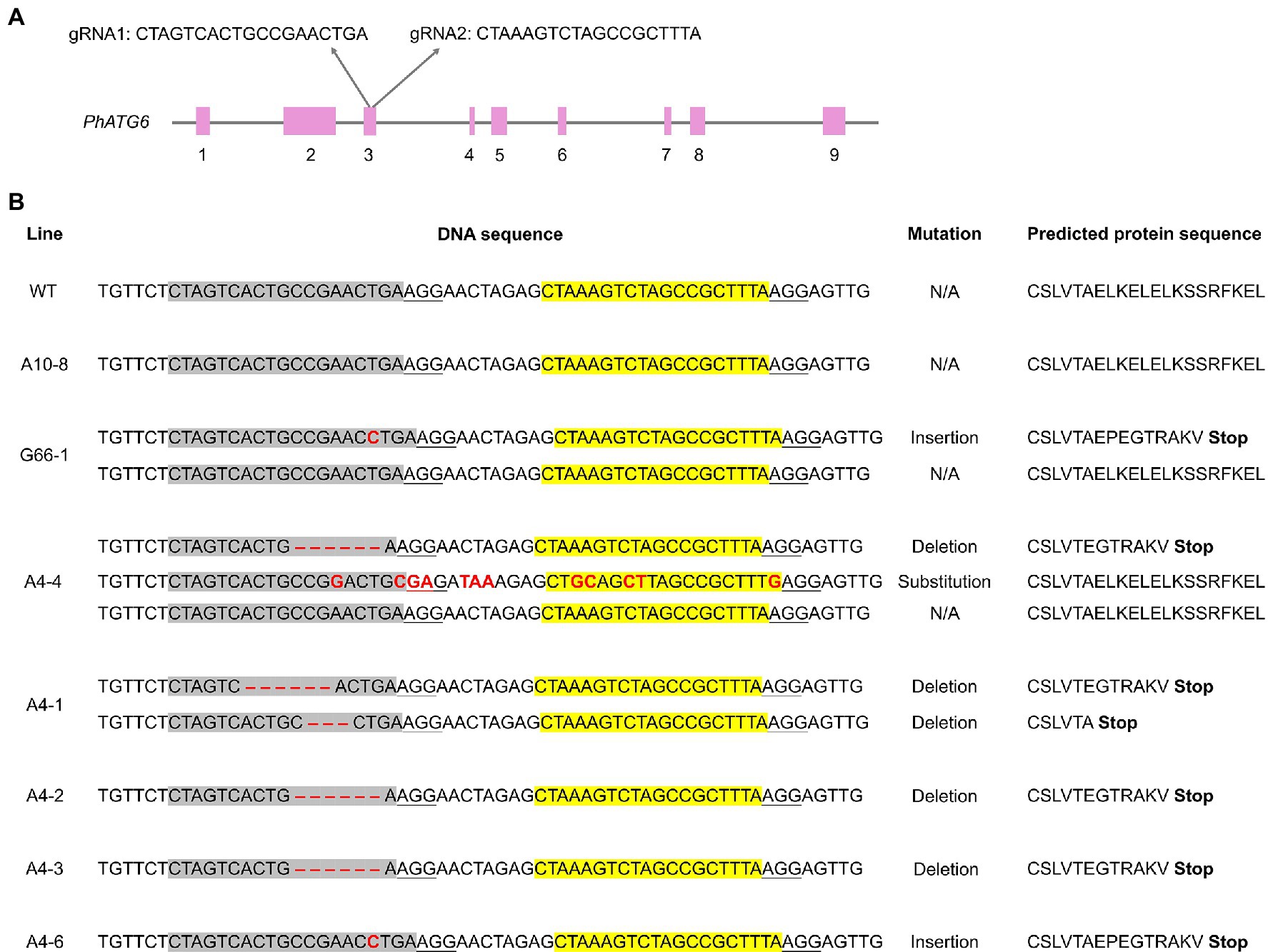
Figure 1. CRISPR/Cas9-edited sequences of Autophagy Gene 6 (PhATG6) in T0 Petunia × hybrida ‘Mitchell Diploid’ plants with mutated PhATG6 and wild-type control (WT). (A) Two guide RNAs (gRNAs) were used to target PhATG6 exon 3. (B) DNA sequences and predicted protein sequences of PhATG6 exon 3 in wild-type plant, non-mutated regenerated line A10-8, and different PhATG6-KO lines. Pink boxes represent exons 1–9 of PhATG6, grey sequences represent the targeted site of gRNA1, yellow sequences represent the targeted site of gRNA2, and red texts represent mutation sites. The protospacer adjacent motif (PAM) for Cas9 recognition (NGG) was underlined. N/A represents no mutation detected.
Generation of PhATG6-KO Lines
The constructed plasmids carrying both Cas9 and PhATG6 gRNA sequences were introduced into Agrobacterium tumefaciens GV3101 via electroporation for Agrobacterium-mediated plant transformation. The transformation was carried out using an established protocol modified from Jorgensen et al., 1996. Briefly, Agrobacterium was cultured on an LB plate with selective antibiotics kanamycin (50 mg L−1), gentamicin (25 mg L−1), and rifampicin (10 mg L−1). Single colonies were selected to culture in 6-ml liquid LB media with the same antibiotics overnight at 28°C. The overnight culture was used to inoculate 50 ml selective LB media at a 1:10 ratio and cultured at 28°C until OD600 = 0.8–1.0. The culture was diluted to OD600 = 0.15 with co-culture MS media including 6-benzylaminopurine (BAP, 2 mg L−1), indole-3-butyric acid (IBA, 5 μg L−1), and acetosyringone (0.2 mM). Young, fully expanded leaves were excised from petunia plants and cut into 5 × 5 mm2 pieces. Leaf explants were immersed in diluted Agrobacterium culture for 15 min, blotted dry using sterile filter paper, and placed on co-culture medium plates with the adaxial side facing down. The medium plates were sealed and incubated in the dark at room temperature for 3 days. After that, the explants were transferred and cultured on regeneration media (MS with 2 mg L−1 BAP, 5 μg L−1 IBA, 150 mg L−1 kanamycin, and 500 mg L−1 carbenicillin) to induce callus and shoots. The regenerated shoots were excised from the base without any callus and transferred to rooting media (MS with 150 mg L−1 kanamycin and 500 mg L−1 carbenicillin) for root induction.
Validation of PhATG6-KO Lines
Once the transformed plants were regenerated through tissue culture, DNA sequencing was used to verify whether the PhATG6 gene was mutated. Total DNA from each event was extracted from leaf tissue using a DNeasy kit (Qiagen, Hilden, Germany). Fragments of the PhATG6 sequences were amplified via PCR, and the PCR products were cloned into a sequencing vector pJET1.2 using CloneJET PCR Cloning Kit (Thermo Fisher Scientific, Waltham, MA, United States). The cloned products were transformed into chemically competent Escherichia coli using the Mix & Go E. coli Transformation Kit (Zymo Research, Irvine, CA, United States), and the transformation was verified with colony PCR. At least five colonies from each event were sent to Molecular Cloning Laboratories (MCLAB, South San Francisco, CA, United States) for sequencing to identify the specific PhATG6 gene edits. The DNA sequences of the mutated genes were used to determine the predicted protein sequences using Expasy Translate.4 Plants with mutated PhATG6 are named PhATG6-KO lines hereinafter, and a line identified without any mutation in PhATG6 is referred to as the non-mutated line.
Plant Phenotypic Evaluation
The regenerated transgenic plantlets were moved out of in vitro culture and transplanted into 6.4-cm pots with a peat-based potting mix (Promix BX; Premier Tech Horticulture, Quebec, Canada). Plants were placed in a clear bin with a lid that was opened over time to slowly acclimate the plants. The acclimated plants were moved into a greenhouse at The Ohio State University CFAES Wooster Campus (Wooster, OH, United States). Wild-type MD petunia seeds were sown in a peat-based germination mix (Promix PGX; Premier Tech Horticulture). Four-week-old seedlings were transplanted to 6.4-cm pots with a peat-based media (Promix BX; Premier Tech Horticulture) and moved into the greenhouse with the regenerated plants. One cutting was made from each of the mutants and the wild-type plant, and the new plants produced from the cuttings were transplanted into 3.8-L pots with the same peat-based media (Promix BX; Premier Tech Horticulture) and grown under 23°C day/18°C night temperature with 14-h light/10-h dark cycle. Plants were fertigated with 75 mg L−1 N from 15N-2.2P-12.5K-2.9Ca-1.2Mg water-soluble fertilizer (Jack’s Professional LX, JR Peters, Allentown, PA, United States).
The phenotypic evaluation of all plants included measurements of flower longevity, flower size, capsule weight, capsule maturation time, number of seeds per capsule, and 100-seed weight. To measure the longevity of flowers in planta, the anthers were removed from the flowers 1 day before opening (deanthered), and the dates of corolla opening and senescence were recorded. Flower longevity (n = 10) was calculated as the number of days from corolla opening to when it senesced (wilted but not dry). Flower size (n = 4) was determined by measuring the diameters of flowers using a ruler on the day of opening. To collect capsules, the anthers were removed from the flowers 1 day before opening, the flowers were pollinated with pollen from the same plant on the day of opening, and capsules were collected when they were mature (the capsules were brown and dried). The average number of days from flower pollination to capsule collection was recorded as capsule maturation time. Five-to-eight mature capsules were collected from each plant, placed in a 1.5-ml centrifuge tube, and dried in a chamber with desiccant for 1 month. The seeds in each capsule were isolated and counted. The weight of the dry capsules and 100-seed weight was measured using a balance (Adventurer Pro AV313; Ohaus, Pine Brook, NJ, United States).
Evans Blue Staining
The corolla limbs of the flowers were stained with Evans Blue (MP Biomedicals, Solon, OH, United States), a dye used to stain dead cells. The deanthered flowers were collected daily from the day before flower opening (day −1) to the day after the last day of senescence. Individual petal limbs were excised and submerged in Evans Blue solution (0.1% w/v) for 20 min, rinsed with distilled water three times, and blotted dry with filter paper. Photographs were taken immediately after the staining process.
Ethylene Production Measurements
The production of ethylene was measured using a Varian CP-3800 gas chromatograph (GC) equipped with a flame ionization detector (FID) and a HayeSep R packed column (Agilent Technologies, Santa Clara, CA, United States) following a previously published method (Broderick et al., 2014). The deanthered flowers were collected on day 0 and day 3 after flower opening, and daily starting on day 6 until the last day of senescence. The whole corollas were isolated and individually incubated in sealed 22-ml glass vials for 2 h. From each vial, a 1-ml gas sample was withdrawn and injected into the GC to determine ethylene concentration (nl). Ethylene production was calculated by dividing the ethylene concentration by the fresh weight of the corolla (g) and the incubation time (h). Four-to-six corollas were analyzed for each line at each time point.
Gene Expression Analysis
The expression of senescence-related genes in corollas was measured following a previously reported method (Lin and Jones, 2021). Briefly, deanthered flowers were collected daily from flower opening (day 0) through senescence from different plants, and the corollas were immediately frozen using liquid nitrogen and stored at −80°C. Total RNA was extracted from three finely ground corollas using TRIzol Reagent (Invitrogen, Carlsbad, CA, United States) and treated with RQ1 RNase-free DNase (Promega, Madison, WI, United States). A spectrophotometer (NanoDrop ND-1000; Thermo Fisher Scientific) was used to determine the concentration of RNA. Gene expression was measured via RT-qPCR (reverse transcription-quantitative PCR) using cDNA synthesized from 1 μg RNA with iScript Reverse Transcription Supermix (Bio-Rad, Hercules, CA, United States). The qPCR was conducted using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) in a C1000 Touch™ Thermo Cycler (Bio-Rad). The procedure for qPCR was 30 s at 95°C for denaturation, 40 cycles of 10 s denaturation at 95°C and 30 s annealing and extension at 50°C, followed by melt curve analysis. The relative expression was calculated using CFX Manager™ software (Bio-Rad). Both PhActin (Chapin and Jones, 2009) and PhSAND (Mallona et al., 2010) were used as reference genes. Primers used for qPCR are listed in Supplementary Table S1. Two biological replicates and two technical replicates were included for each gene expression analysis.
Flower Tissue Nutrient Analysis
Flower corollas were collected on the day of flower opening (non-senescing) and the last day of flower senescence (senescing). The samples were dried in a forced air-drying oven set at 60°C, ground into a powder that could pass a 2-mm sieve, and sent to the Service Testing and Research (STAR) Lab (The Ohio State University, Wooster, OH, United States) for nutrient analysis. Total nitrogen (N) was analyzed using a Vario Max combustion analyzer (Elementar Americas, Ronkonkoma, NY, United States) following the Dumas combustion method (Sweeney, 1989). Plant tissue was digested using an automated microwave digestion system (Discover SP-D, CEM, Matthews, NC, United States). The concentrations of other mineral nutrients including phosphorus (P) were determined using an inductively coupled plasma spectrometer (Agilent 5110 ICP-OES, Agilent Technologies, Santa Clara, CA, United States; Isaac and Johnson, 1985). The remobilized nutrients were determined by subtracting the nutrient concentrations that remained in the senescing corollas from the concentrations in the non-senescing corollas. Three replicates were included for each line at each time point.
Statistical Analysis
All statistical analyses were conducted following the model y = μ + Line + Replicate + e in R 3.3.1 (R Core Team, 2013). All the PhATG6-KO lines and the non-mutated regenerated line A10-8 were compared to the wild-type control using ANOVA-protected two-sided Dunnett’s test (multcomp package).
Results
PhATG6 Was Knocked Out Using CRISPR/Cas9
To understand the function of PhATG6, CRISPR/Cas9 was used to induce PhATG6 editing in petunia. Based on the DNA sequences of exon 3 of the PhATG6 gene in the wild-type and the regenerated plants, we identified 13 PhATG6-KO lines with mutated PhATG6 out of 23 regenerated plants, resulting in a mutation frequency of 57% (Table 1). Seven biallelic mutants, where both alleles of PhATG6 were mutated, and two monoallelic mutants, where only one allele of PhATG6 was mutated, were regenerated (Table 1). In addition, we also generated four chimeric lines (Table 1). Different mutations were detected in the KO lines, including deletions, insertions, and base substitutions in PhATG6 exon 3 (Figure 1). Based on the nucleotide sequences, the predicted amino acid sequences were determined. Lines with truncated protein sequences were considered to be loss-of-function PhATG6 mutants (Figure 1). Further characterizations were conducted using the PhATG6-KO lines including four biallelic mutants (A4-1, A4-2, A4-3, A4-6) with loss-of-function PhATG6, 1 monoallelic mutant (G66-1), and 1 chimeric mutant (A4-4). A non-mutated regenerated line (A10-8) and a wild-type plant (WT) were used as controls (Figure 1).
PhATG6-KO Lines Showed Decreased Flower Longevity
Flowers of all six PhATG6-KO lines had reduced flower longevity (i.e., accelerated corolla senescence) when compared to the controls (Figures 2, 3A; Supplementary Figure S1). Flowers of the control plants lasted an average of 10 days, while flowers of the KO lines lasted an average of 7–8 days (Figure 3A). Photographs of the corolla senescence process showed that the senescence of petunia corollas began with the collapsing of the neck connecting the limb and the tube and then proceeded to the wilting of the limb (Figure 2; Supplementary Figure S1). Evans blue staining of the flower petal limbs also showed that cell death (loss of membrane integrity) started in the neck and continued to move up to the rest of the limb (Figure 2; Supplementary Figure S1). The corolla senescence pattern was similar in flowers of the PhATG6-KO lines compared to the controls (Figure 2; Supplementary Figure S1). KO line G66-1 had smaller flowers than the controls (Figure 3B). Flowers produced by line G66-1 were 17% smaller than the flowers of line A10-8 and 13% smaller than wild-type flowers (Figure 3B).
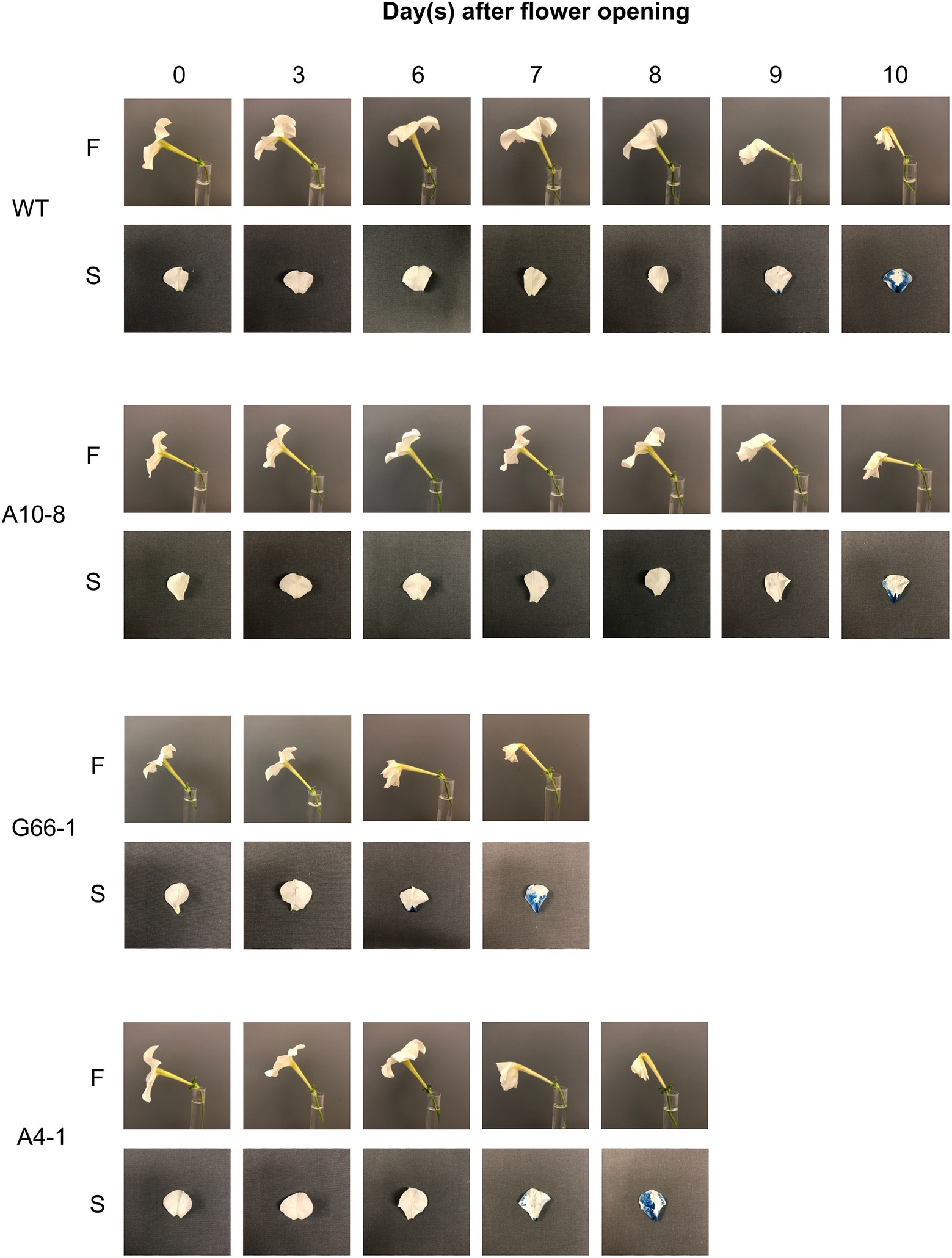
Figure 2. Corolla senescence in Petunia × hybrida ‘Mitchell Diploid’ wild-type plant (WT), non-mutated regenerated line A10-8, and PhATG6-KO lines (G66-1 and A4-1). Pictures of representative flowers (F) and Evans Blue-stained petal limbs (S) were taken from the day of flower opening (day 0) to the day of flower senescence. Additional timepoints and lines can be seen in Supplementary Figure S1.
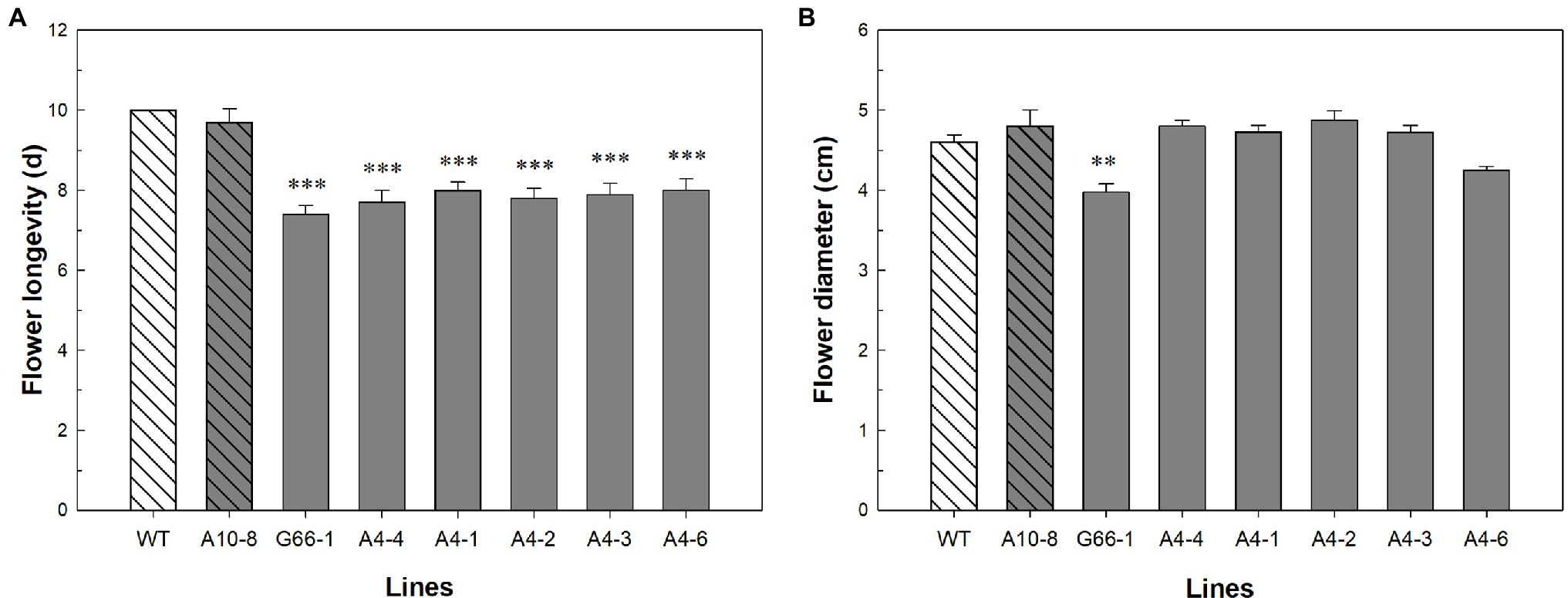
Figure 3. Flower phenotypic evaluation of Petunia × hybrida ‘Mitchell Diploid’ wild-type plant (WT), non-mutated regenerated line A10-8, and PhATG6-KO lines. (A) Flower longevity (n = 10). (B) Flower diameter (n = 4). Bars represent the means of the measurements ± standard error, and bars with stars indicate a significant difference compared to the WT: “***”p ≤ 0.001, “**”p ≤ 0.01.
Seed Number Decreased While Individual Seed Weight Increased in PhATG6-KO Lines
The production of seeds was impacted in the PhATG6-KO lines. The weight of seed capsules (fruits) was 46–86% lower in the KO lines compared to the controls (Figure 4A). The capsules from the KO lines were visually smaller than the control capsules (Figure 4B). There was no difference in capsule maturation time (Figure 4C). The numbers of seeds were decreased by 46–93% in the KO lines compared to the controls (Figure 4D). In contrast, the 100-seed weight increased 23–26% in five of the PhATG6-KO lines compared to the controls, while line G66-1 did not show any statistical difference (Figure 4E). Compared to the other five PhATG6-KO lines, G66-1 had a smaller difference in capsule weight, seed number per capsule, and 100-seed weight compared to the controls (Figures 4A,D,E).
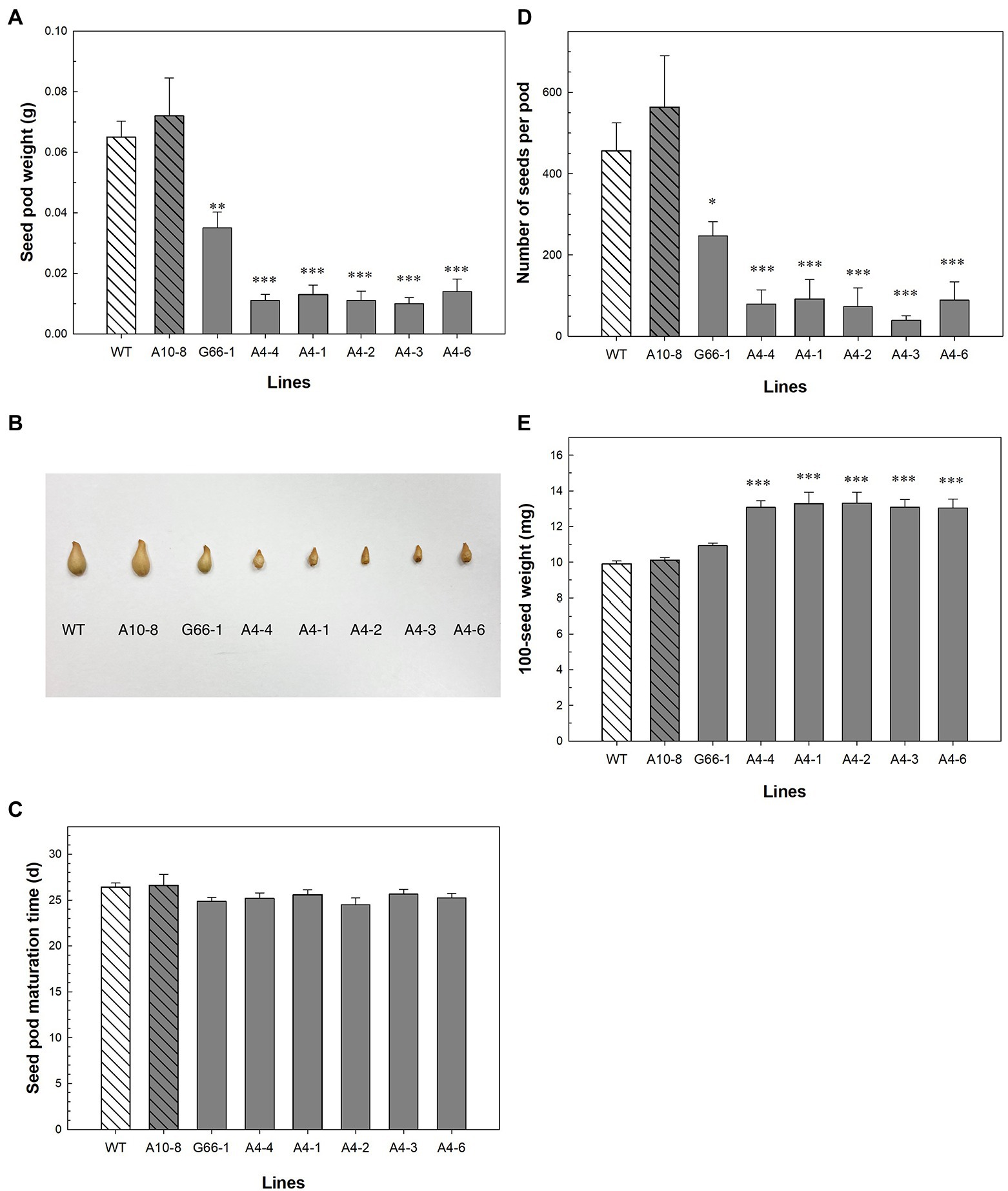
Figure 4. Seed phenotypic evaluation of Petunia × hybrida ‘Mitchell Diploid’ wild-type plant (WT), non-mutated regenerated line A10-8, and PhATG6-KO lines. (A) Capsule weight (n = 3). (B) Picture of representative capsules. (C) Capsule maturation time (n = 8). (D) Number of seeds per capsule (n = 3). (E) 100-seed weight (n = 4). Bars represent the means of the measurements ± standard error, and bars with stars indicate a significant difference compared to the WT: “***”p ≤ 0.001, “**”p ≤ 0.01, “*”p ≤ 0.05.
Ethylene Production Was Induced Earlier in PhATG6-KO Flowers
To determine whether ethylene is involved in the PhATG6-KO-accelerated corolla senescence, the production of ethylene, as well as the transcript levels of ethylene biosynthesis genes PhACS and PhACO1, was measured overtime after flower opening. The levels of ethylene production from the corollas increased over time and reached a peak 1–2 days before the last day of senescence in the controls and the KO lines (Figure 5A). Ethylene production was induced earlier in the PhATG6-KO corollas concomitant with their earlier senescence (Figure 5A). The amount of ethylene produced at the peaks varied among the KO lines, but was not different than that of the controls (Figure 5A). Two ethylene biosynthesis genes in petunia, PhACS and PhACO1, were similarly upregulated earlier in the KO corollas than the control corollas, corresponding with the earlier ethylene production (Figures 5B,C). The expression of PhACS in flower corollas was upregulated on day 8 in the KO lines and on day 10 in the controls (Figure 5B). Variation in PhACS transcript levels was found across the PhATG6-KO lines when they reached the maximum expression (Figure 5B). The expression of PhACO1 in corollas increased over time, reaching the maximum expression on day 7 or 8 in the KO lines, and on day 10 in the controls (Figure 5C). The maximum expression levels of PhACO1 were also variable among the PhATG6-KO lines (Figure 5C).
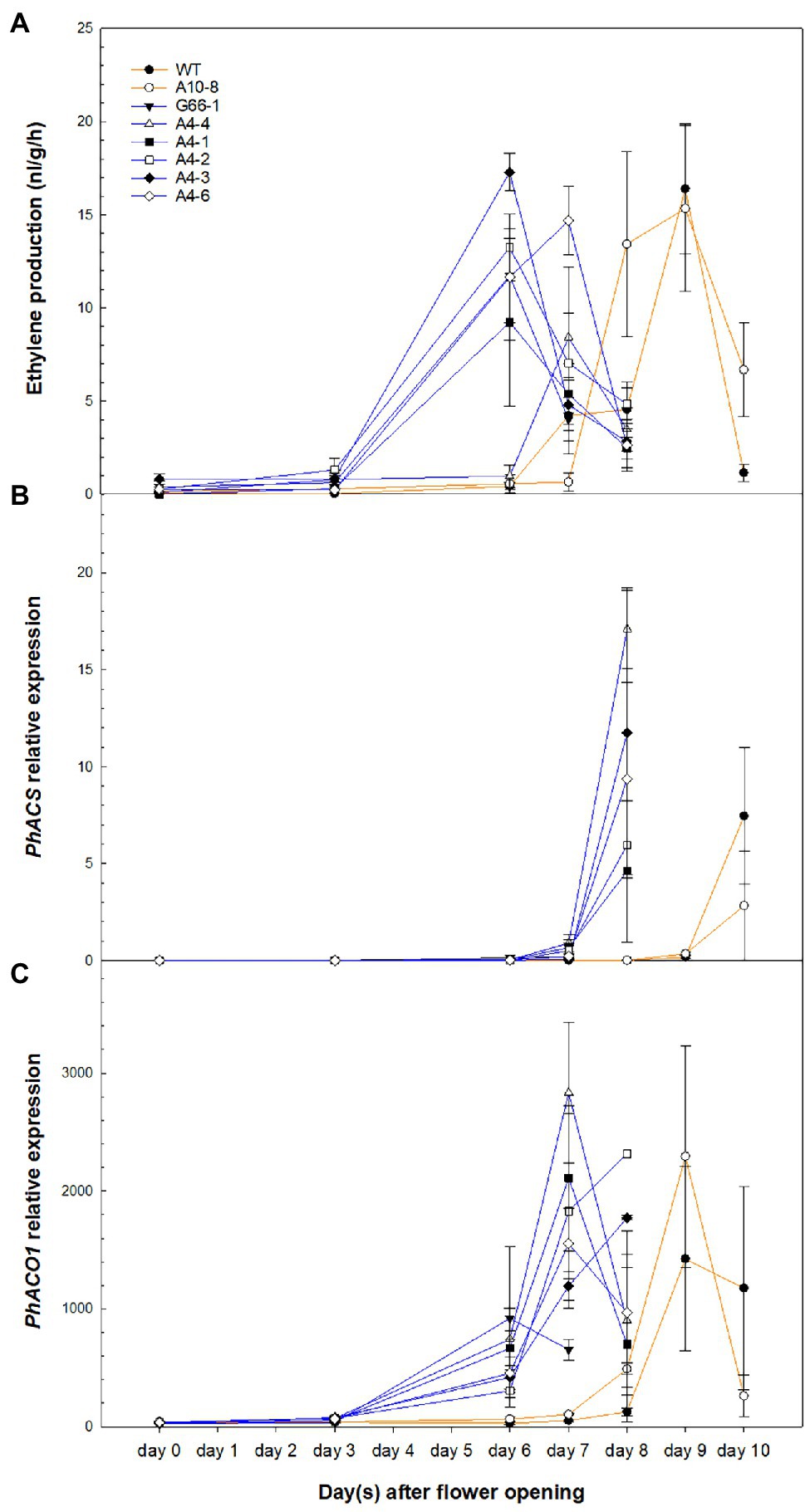
Figure 5. Ethylene production and ethylene biosynthesis gene expression in the corollas of Petunia × hybrida ‘Mitchell Diploid’ wild-type plant (WT), non-mutated regenerated line A10-8, and PhATG6-KO lines from flower opening to senescence. (A) Ethylene production (n = 6). (B) Relative expression of 1-Aminocyclopropane 1-Carboxylate Synthase (PhACS; n = 2). (C) Relative expression of 1-Aminocyclopropane 1-Carboxylate Oxidase 1 (PhACO1; n = 2). Orange lines represent the results of controls (WT and line A10-8), and blue lines represent the results of the PhATG6-KO lines. The relative expression was calculated based on the expression of reference genes PhActin and PhSAND. Data represent the means of the measurements ± standard error.
PCD-Related Gene Expression Was Upregulated Earlier in PhATG6-KO Flowers
To determine whether knocking out PhATG6 could affect the function of PCD-related genes in corolla senescence, the relative expression of PhATG6, PhPI3K, PhATG8d, and a metacaspase gene Metacaspase 1 (PhMC1) was measured and found to be upregulated earlier in the KO corollas than the controls, corresponding to the time of corolla senescence (Figure 6). PhATG6 transcript levels were upregulated on the last 2 days of the corolla senescence process in the controls and all the KO lines, with variations in the expression found across the different KO lines (Figure 6A). The relative expression of PhPI3K was also enhanced on the last 2 days of corolla senescence in every plant, and the transcript levels were variable among the different PhATG6-KO lines (Figure 6B). PhATG8d was upregulated on the last day of the senescence process in the corollas of the controls (day 10), and the PhATG6-KO plants (day 7 or 8), and the relative expression of PhATG8d varied among the KO lines (Figure 6C). Similarly, PhMC1 expression was upregulated on the last day of corolla senescence, and it was induced earlier in the PhATG6-KO flowers (day 7 or 8) than the controls (day 10) (Figure 6D). Variations in the transcript levels of PhMC1 were observed among the PhATG6-KO lines (Figure 6D).
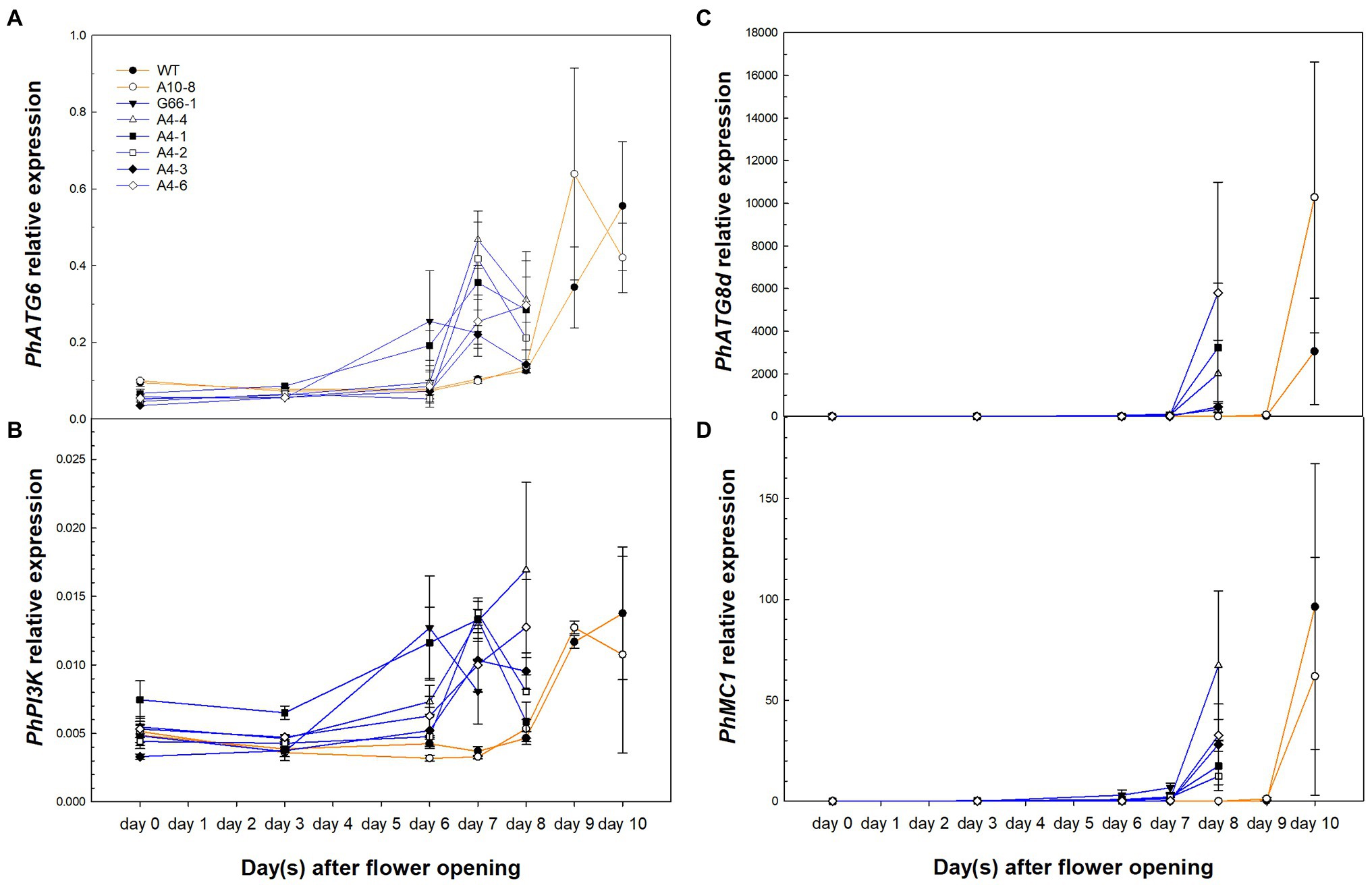
Figure 6. Relative expression of PCD-related genes Autophagy Gene 6 (PhATG6, A), Phosphoinositide 3-Kinase (PhPI3K, B), Autophagy Gene 8d (PhATG8d, C), and Metacaspase 1 (PhMC1, D) in the corollas of Petunia × hybrida ‘Mitchell Diploid’ wild-type plant (WT), non-mutated regenerated line A10-8, and PhATG6-KO lines from flower opening to senescence (n = 2). Orange lines represent the results of controls (WT and line A10-8), and blue lines represent the results of the PhATG6-KO lines. The relative expression was calculated based on the expression of reference genes PhActin and PhSAND. Data represent the means of the measurements ± standard error.
Nutrient Remobilization Was Impaired in PhATG6-KO Flowers
To characterize the effects of PhATG6 KO on nutrient remobilization during petal senescence, we measured the concentrations of different macro- and micro-nutrients in the corollas of KO lines and the controls. By comparing the total nutrient content of non-senescing and fully senescent corollas, the difference represents the nutrients that were remobilized from the corolla during the senescence process. The mobilization of phosphorus (P) was reduced in the PhATG6-KO lines compared to the controls (Figure 7A). From corolla opening to senescence, 64 and 65% of P was remobilized from the wild-type plant and line A10-8, respectively, while only 60, 58, 51, and 44% of P was remobilized from the KO lines A4-1, A4-2, A4-3, and A4-6 (Figure 7A). We did not observe reduced remobilization of nitrogen (N) or other macro-nutrients (potassium, magnesium, calcium, and sulfur), nor any micro-nutrient (aluminum, boron, copper, iron, manganese, molybdenum, and zinc) evaluated during corolla senescence in the PhATG6-KO lines compared to the controls (Figure 7B, data not shown). A senescence-associated cysteine protease, PhCP10, thought to be involved in protein degradation, was upregulated at the final stage of corolla senescence, earlier in the PhATG6-KO flowers than in the flowers of the controls (Figure 7C). Variations were found in the PhCP10 transcript levels among the PhATG6-KO lines; however, our results showed a trend of reduced PhCP10 expression in the KO flowers compared to the controls, even though the maximum expression levels were not statistically different (Figure 7C). The relative expression of a senescence-associated phosphate transporter PhPT1 was also upregulated on the last 2 days of corolla senescence (Figure 7D). Variations in PhPT1 expression were shown among the PhATG6-KO lines (Figure 7D).
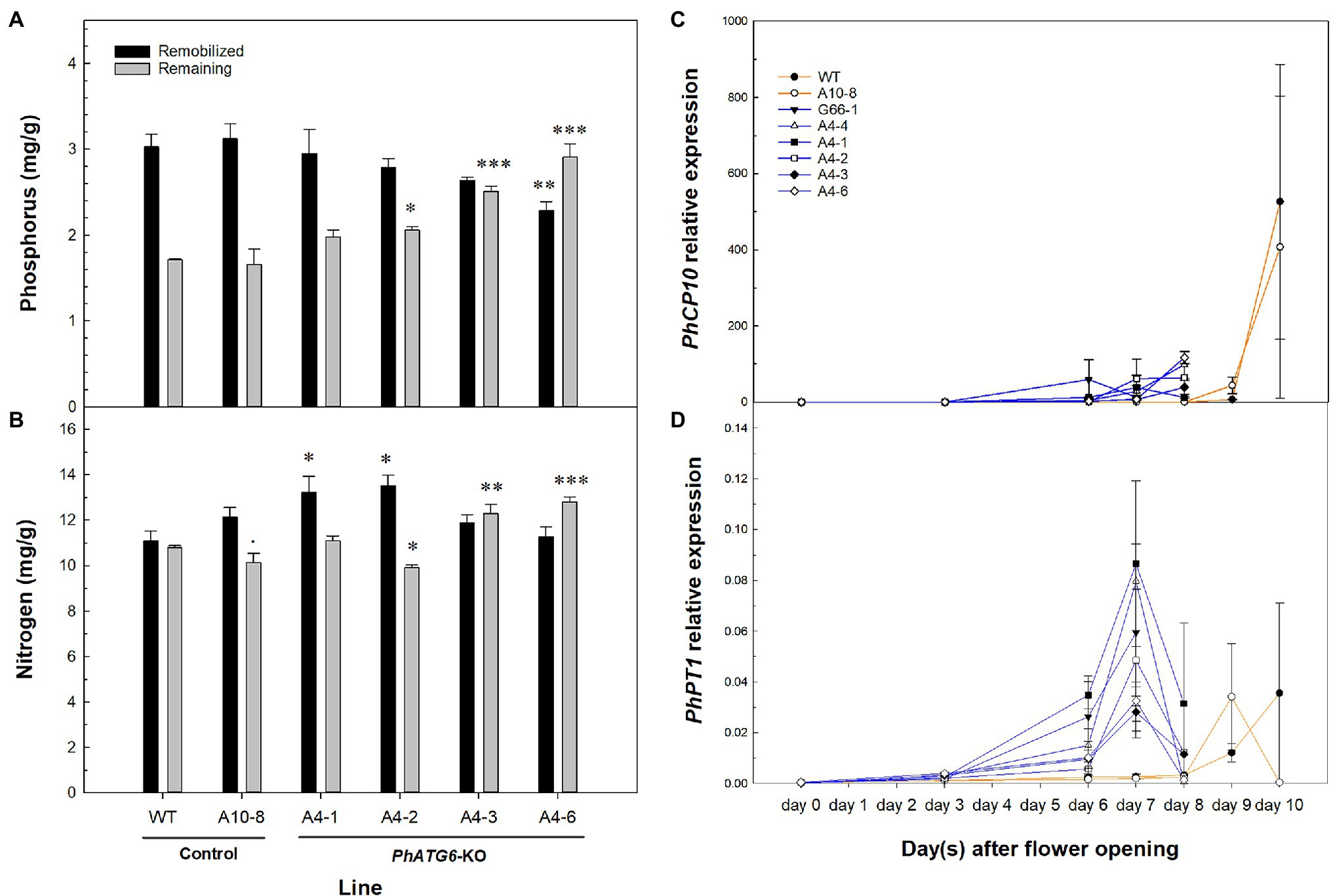
Figure 7. Nutrient mobilization from corollas of the Petunia × hybrida ‘Mitchell Diploid’ wild-type plant (WT), non-mutated regenerated line A10-8, and PhATG6-KO lines. Concentration of phosphorus (P) (A) and nitrogen (N) (B) that was remobilized (black) and remaining (grey) in the senescing corollas (n = 3). Relative expression of senescence-associated Cysteine Protease 10 (PhCP10) (C) and Phosphate Transporter 1 (PhPT1) (D) from corolla opening to senescence (n = 2). The remaining P and N is the concentration in senescing corollas. Remobilized N and P were calculated by subtracting the concentration of individual nutrients in the senescing corollas from the concentration in the non-senescing corollas. Orange lines represent the results of controls (WT and line A10-8), and blue lines represent the results of the PhATG6-KO lines. The relative expression was calculated based on the expression of reference genes PhActin and PhSAND. Data represent the means of the measurements ± standard error. Bars represent the means of the measurements ± standard error, and bars with stars indicate a significant difference compared to the WT: “***”p ≤ 0.001, “**”p ≤ 0.01, “*”p ≤ 0.05, “.”p ≤ 0.1.
Discussion
Knocking out PhATG6 in petunia accelerated corolla senescence by 2–3 days (Figure 3A), indicating that autophagy is involved in delaying corolla senescence. This result supports our previous finding, where silencing PhATG6 using Virus-induced Gene Silencing (VIGS) results in decreased flower longevity by 3–4 days in petunia (Lin and Jones, 2021). Even though knocking out PhATG6 with CRISPR did not seem to reduce flower longevity as much as VIGS, this could be due to the difference in the plant growing environment and petunia cultivars. The PhATG6-KO lines (‘Mitchell Diploid’) were grown under greenhouse conditions, while the PhATG6-silenced petunias (‘Picobella Blue’) were grown in growth chambers. The proliferation of virus may also play a role by placing more stress on the VIGS plants, leading to a more severe reduction of flower longevity. In addition to ATG6, the genetic modifications of another autophagy gene, PI3K, also affect the longevity of flowers (Dek et al., 2017; Lin and Jones, 2021).
The morphological cell death processes in flower corollas vary in different species. In Japanese morning glory, corolla PCD starts from the edge of the petal limbs and gradually moves toward the base of the petals (Shibuya et al., 2014). Our results showed that petunia corolla PCD began in the corolla neck connecting the limb and tube and proceeded to the rest of the corolla, with the majority of petal cells maintaining intact plasma membranes until the last 1–2 days of the senescence process (as shown by Evans Blue staining; Figure 2). Similarly, in Alstroemeria (Alstroemeria peruviensis), the PCD in petals does not occur until the last stages of flower senescence, as demonstrated by the electrolyte leakage, DNA laddering, and protease activity in petals (Wagstaff et al., 2003). Cut daylily (Hemerocallis sp.) flowers also do not show reduced membrane stability until visible senescence symptoms appear (Chakrabarty et al., 2009). Our results provide further evidence that the loss of membrane integrity does not occur gradually but at the late stage of petal senescence.
PhATG6 is functionally involved in fruit (capsule) and seed production in petunia (Figure 4). Autophagy is the major regulator of nutrient recycling and therefore indirectly affects the nutrient supplies for seeds (Ren et al., 2014). In rice (Oryza sativa), the overexpression of ATG8a or ATG8b increases grain number and yield per plant, which are reduced in plants with RNAi-suppressed ATG8b compared to the wild-type plants (Yu et al., 2019; Fan et al., 2020). Rice atg7 mutants and ATG8b-RNAi plants also show reduced grain quality, demonstrated by the chalky appearance and the starch and protein composition of the grains (Sera et al., 2019; Fan et al., 2020). Arabidopsis mutants of ATG5 or ATG7 produce fewer seeds per silique, fewer siliques per plant, and lower seed yield than the wild-type plants (Guiboileau et al., 2012; Barros et al., 2017; Lornac et al., 2020). Arabidopsis plants with heterozygous PI3K (+/−) gene produce fewer numbers of seeds per silique (Lee et al., 2008). The number of siliques per plant is decreased in Arabidopsis heterozygous atg6 (+/−) mutants (Fujiki et al., 2007; Harrison-Lowe and Olsen, 2008). Similarly, we observed smaller capsules and fewer seeds per capsule in the PhATG6-KO petunias compared to the controls (Figures 4A,B,D). The average weight of individual seeds was higher in most of the PhATG6-KO lines than the control plants (Figure 4E), consistent with what is observed in Arabidopsis atg7 mutants (Barros et al., 2017). Although we did not find a difference in capsule maturation time between the PhATG6-KO plants and the controls (Figure 4C), a study in pepper (Capsicum annuum) shows an increase in autophagy gene expression, autophagy protein abundance, and autophagosome-like vesicles in mature fruit, suggesting that autophagy is involved in fruit ripening (López-Vidal et al., 2020).
Ethylene is involved in the regulation of accelerated corolla senescence in the PhATG6-KO lines. As a regulator for senescence in plants, ethylene plays a key role in autophagy-mediated petal senescence (Shibuya et al., 2013). The early elevation of ethylene levels was likely the reason for early corolla senescence in the PhATG6-KO lines. Our results suggested that autophagy could affect the timing of ethylene biosynthesis. However, in tobacco plants with overexpressed PI3K, even though the detached flower senescence is accelerated, and the ethylene production is higher in the entire detached flowers, no difference in the timing of ethylene production is observed (Dek et al., 2017). The difference in ethylene production timing may indicate different regulatory mechanisms of corolla senescence. The expression of ethylene biosynthesis genes PhACS and PhACO1 was also induced earlier in the PhATG6-KO corollas compared to the controls, but later than the elevated ethylene levels (Figures 5B,C). As shown in carnation flowers, ethylene treatment can induce the expression of ethylene biosynthesis genes (Jones, 2003). Therefore, the increase in ethylene biosynthesis gene expression in petunia corollas was likely induced by the earlier produced ethylene. Multiple ACS genes with different roles have been found in Arabidopsis, tobacco, carnation, orchid (Phalaenopsis sp.), tomato, and potato (Solanum tuberosum) (Yip et al., 1992; Destéfano-Beltrán et al., 1995; Liang et al., 1995; Bui and O’Neill, 1998; Jones and Woodson, 1999; Ge et al., 2000), while little is known about the function of additional ACS genes in petunia. It is possible that the ethylene produced earlier in the corollas of PhATG6-KO flowers was synthesized by another ACS member in petunia. Another possibility is that the ethylene could be synthesized using ACC translocated from other parts of the flowers, or the ethylene itself could be translocated, as shown in carnation and orchid (Reid et al., 1984; Woltering, 1990; O’Neill et al., 1993). Even though ACS expression increased at the end of corolla senescence, ethylene production decreased, indicating that ACS activity or stability could be affected by posttranscriptional regulation during corolla senescence.
The early upregulation of PCD-related gene expression supported early PCD in the senescing corollas of PhATG6-KO lines. ATG6 is an essential component of the PI3K protein complex in the autophagy pathway (McKnight and Yue, 2013). ATG6 serves as a binding hub for different interacting proteins to regulate the composition, subcellular localization, and/or function of the PI3K complex and consequently affect autophagy activity (He and Levine, 2010; Hill et al., 2019). Therefore, knocking out ATG6 could result in direct and/or indirect effects on autophagy and other PCD regulatory mechanisms. Our previous study shows that ATG6 expression is upregulated when PI3K is silenced, and ATG8d expression is downregulated when ATG6 is silenced (Lin and Jones, 2021). This study showed that PhATG6, PhPI3K, and PhATG8d expression was upregulated earlier in the PhATG6-KO corollas than the controls, correlated with the accelerated senescence (Figures 6A–C). The early induction of these genes was likely a result of the early increase of ethylene production, as ethylene treatment induces autophagy gene expression in petunia and Japanese morning glory (Shibuya et al., 2013; Quijia Pillajo et al., 2018). Apoptosis is another type of PCD modulated by caspases, and the interaction between autophagic PCD and apoptotic PCD is based on caspase-mediated cleavage of ATG6 (Djavaheri-Mergny et al., 2010; Wirawan et al., 2010). In plants, the apoptosis-like PCD is regulated by metacaspases, a group of cysteine proteases similar to mammalian caspases (Minina et al., 2013). RNAi-suppressed PhMC1 similarly causes accelerated corolla senescence in petunia (Chapin et al., 2017). The expression of PhMC1 was also upregulated earlier in PhATG6-KO lines (Figure 6D), suggesting accelerated PCD in the corollas and indicating a potential interaction between autophagy and metacaspases at the transcriptional level.
Knocking out PhATG6 in petunia negatively impacted nutrient remobilization during corolla senescence of unpollinated flowers. During leaf senescence, autophagy plays an important role in N remobilization as shown in the atg mutants of Arabidopsis, rice, and maize, where the remobilization efficiency of N is reduced (Guiboileau et al., 2012; Li et al., 2015; Wada et al., 2015; Yu et al., 2019; Fan et al., 2020). Even though the remobilization of N was not affected in the PhATG6-KO lines during corolla senescence, P remobilization was decreased in the KO lines (Figures 7A,B). The expression of two petal senescence-specific genes, a cysteine protease PhCP10 and a phosphate transporter PhPT1, was induced earlier in the PhATG6-KO lines, and PhCP10 showed a trend of downregulated expression in the KO plants (Figures 7C,D), indicating that autophagy affects the transcription of genes involved in nutrient degradation and transportation. Cysteine proteases play an important role in protein degradation during petal senescence (Jones et al., 1995; Wagstaff et al., 2002; Arora and Singh, 2004). PhCP10, a homolog to Arabidopsis Senescence-Associated Gene 12 (SAG12), is exclusively expressed in senescing petals and its expression is delayed in ethylene-insensitive petunias, corresponding to the delayed corolla senescence (Jones et al., 2005). PhPT1, a high-affinity phosphate transporter, belongs to the Phosphate Transporter1 family in petunia (Wegmüller et al., 2008; Chapin and Jones, 2009). Compared to the other petunia high-affinity phosphate transporters, PhPT1 is highly expressed in senescing petunia corollas (Chapin and Jones, 2009). Ethylene affects the expression of PhPT1, as the transcript levels of PhPT1 decrease in ethylene-insensitive plants during corolla senescence, and ethylene treatment induces PhPT1 expression in corollas (Chapin and Jones, 2009). The early induction of PhCP10 and PhPT1 expression in the PhATG6-KO lines was likely the result of the early increased ethylene production. Autophagy may affect nutrient remobilization via ethylene-mediated transcriptional regulation of genes involved in nutrient degradation and transportation.
Knocking out a central autophagy gene PhATG6 in petunia led to accelerated petal senescence, decreased P remobilization, and reduced seed yield, likely due to early expression of senescence-related genes and early ethylene production. Our study contributes to the current understanding of autophagy in senescence and nutrient remobilization and provides information for the improvement of flower longevity in ornamental plants.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
YL and MLJ conceptualized the project and designed the experiments. YL conducted the experiments and led the writing of the manuscript. MLJ acquired resources and funding for the study, supervised the project, and edited the manuscript. All authors contributed to the manuscript revision and approved the submitted version.
Funding
The funding for this study was sponsored by the American Floral Endowment, the OSU D.C. Kiplinger Floriculture Endowment, and the OSU Alumni Grants for Graduate Research and Scholarship. Salaries and research support were provided in part by State and Federal funds appropriated to the College of Food, Agricultural and Environmental Sciences, The Ohio State University. Department of Horticulture and Crop Science Manuscript #21-15.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful for John Finer’s guidance and use of his laboratory for tissue culture. We thank Qi-Jun Chen for providing the pKSE401 vector and Laura Chapin for maintaining the petunia plants in the greenhouse.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.840218/full#supplementary-material
Footnotes
1. ^https://solgenomics.net, accessed on 31st March 2021.
2. ^https://solgenomics.net, accessed on November 24, 2018.
3. ^http://www.rgenome.net/cas-offinder/, accessed on December 17, 2018.
4. ^https://web.expasy.org/translate/, accessed on May 6, 2021.
References
Arora, A., and Singh, V. P. (2004). Cysteine protease gene expression and proteolytic activity during floral development and senescence in ethylene-insensitive Gladiolus grandiflora. J. Plant Biochem. Biotechnol. 13, 123–126. doi: 10.1007/BF03263206
Avila-Ospina, L., Moison, M., Yoshimoto, K., and Masclaux-Daubresse, C. (2014). Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 65, 3799–3811. doi: 10.1093/jxb/eru039
Barros, J. A., Cavalcanti, J. H. F., Medeiros, D. B., Nunes-Nesi, A., Avin-Wittenberg, T., Fernie, A. R., et al. (2017). Autophagy deficiency compromises alternative pathways of respiration following energy deprivation in Arabidopsis thaliana. Plant Physiol. 175, 62–76. doi: 10.1104/pp.16.01576
Borghi, M., and Fernie, A. R. (2020). Outstanding questions in flower metabolism. Plant J. 103, 1275–1288. doi: 10.1111/tpj.14814
Broderick, S. R., Wijeratne, S., Wijeratn, A. J., Chapin, L. J., Meulia, T., and Jones, M. L. (2014). RNA-sequencing reveals early, dynamic transcriptome changes in the corollas of pollinated petunias. BMC Plant Biol. 14:307. doi: 10.1186/s12870-014-0307-2
Bui, A. Q., and O’Neill, S. D. (1998). Three 1-aminocyclopropane-1-carboxylate synthase genes regulated by primary and secondary pollination signals in orchid flowers. Plant Physiol. 116, 419–428. doi: 10.1104/pp.116.1.419
Cao, Y., and Klionsky, D. J. (2007). Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 17, 839–849. doi: 10.1038/cr.2007.78
Chakrabarty, D., Verma, A. K., and Datta, S. K. (2009). Oxidative stress and antioxidant activity as the basis of senescence in Hemerocallis (daylily) flowers. J. Hortic. For. 1, 113–119. doi: 10.5897/JHF.9000081
Chapin, L. J., and Jones, M. L. (2009). Ethylene regulates phosphorus remobilization and expression of a phosphate transporter (PhPT1) during petunia corolla senescence. J. Exp. Bot. 60, 2179–2190. doi: 10.1093/jxb/erp092
Chapin, L. J., Moon, Y., and Jones, M. L. (2017). Downregulating a type I metacaspase in petunia accelerates flower senescence. J. Am. Soc. Hortic. Sci. 142, 405–414. doi: 10.21273/JASHS04204-17
Dek, M. S. P., Padmanabhan, P., Sherif, S., and Subramanian, J. (2017). Upregulation of Phosphatidylinositol 3-Kinase (PI3K) enhances ethylene biosynthesis and accelerates flower senescence in transgenic Nicotiana tabacum L. Int. J. Mol. Sci. 18:1533. doi: 10.3390/ijms18071533
Destéfano-Beltrán, L. J., Van Caeneghem, W., Gielen, J., Richard, L., Van Montagu, M., and Van Der Straeten, D. (1995). Characterization of three members of the ACC synthase gene family in Solanum tuberosum L. Mol. Gen. Genet. 246, 496–508. doi: 10.1007/BF00290453
Djavaheri-Mergny, M., Maiuri, M. C., and Kroemer, G. (2010). Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 29, 1717–1719. doi: 10.1038/onc.2009.519
Fan, T., Yang, W., Zeng, X., Xu, X., Xu, Y., Fan, X., et al. (2020). A rice autophagy gene OsATG8b is involved in nitrogen remobilization and control of grain quality. Front. Plant Sci. 11:588. doi: 10.3389/fpls.2020.00588
Feng, Y., He, D., Yao, Z., and Klionsky, D. J. (2014). The machinery of macroautophagy. Cell Res. 24, 24–41. doi: 10.1038/cr.2013.168
Fujiki, Y., Yoshimoto, K., and Ohsumi, Y. (2007). An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 143, 1132–1139. doi: 10.1104/pp.106.093864
Ge, L., Liu, J. Z., Wong, W. S., Hsiao, W. L. W., Chong, K., Xu, Z. K., et al. (2000). Identification of a novel multiple environmental factor-responsive 1-aminocyclopropane-1-carboxylate synthase gene, NT-ACS2, from tobacco. Plant Cell Environ. 23, 1169–1182. doi: 10.1046/j.1365-3040.2000.00618.x
Guiboileau, A., Yoshimoto, K., Soulay, F., Bataillé, M. P., Avice, J. C., and Masclaux-Daubresse, C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194, 732–740. doi: 10.1111/j.1469-8137.2012.04084.x
Harrison-Lowe, N. J., and Olsen, L. J. (2008). Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 4, 339–348. doi: 10.4161/auto.5629
He, C., and Levine, B. (2010). The beclin 1 interactome. Curr. Opin. Cell Biol. 22, 140–149. doi: 10.1016/j.ceb.2010.01.001
Hew, C. S., Tan, S. C., Chin, T. Y., and Ong, T. K. (1989). Influence of ethylene on enzyme activities and mobilization of materials in pollinated Arachnis orchid flowers. J. Plant Growth Regul. 8, 121–130. doi: 10.1007/BF02025279
Hill, S. M., Wrobel, L., and Rubinsztein, D. C. (2019). Post-translational modifications of Beclin 1 provide multiple strategies for autophagy regulation. Cell Death Differ. 26, 617–629. doi: 10.1038/s41418-018-0254-9
Isaac, R. A., and Johnson, W. C. (1985). Elemental analysis of plant tissue by plasma emission spectroscopy: collaborative study. J. Assoc. Off. Anal. Chem. 68, 499–505.
Jones, M. L. (2003). Ethylene biosynthetic genes are differentially regulated by ethylene and ACC in carnation styles. Plant Growth Regul. 40, 129–138. doi: 10.1023/A:1024241006254
Jones, M. L. (2008). Ethylene signaling is required for pollination-accelerated corolla senescence in petunias. Plant Sci. 175, 190–196. doi: 10.1016/j.plantsci.2008.03.011
Jones, M. L. (2013). Mineral nutrient remobilization during corolla senescence in ethylene-sensitive and-insensitive flowers. AoB Plants 5:plt023. doi: 10.1093/aobpla/plt023
Jones, M. L., Chaffin, G. S., Eason, J. R., and Clark, D. G. (2005). Ethylene-sensitivity regulates proteolytic activity and cysteine protease gene expression in petunia corollas. J. Exp. Bot. 56, 2733–2744. doi: 10.1093/jxb/eri266
Jones, M. L., Larsen, P. B., and Woodson, W. R. (1995). Ethylene-regulated expression of a carnation cysteine proteinase during flower petal senescence. Plant Mol. Biol. 28, 505–512. doi: 10.1007/BF00020397
Jones, M. L., Stead, A. D., and Clark, D. G. (2009). “Petunia flower senescence,” in Petunia. eds. Gerats, T., and Strommer, J. (New York, NY: Springer), 301–324.
Jones, M. L., and Woodson, W. R. (1999). Differential expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in carnation. Plant Physiol. 119, 755–764. doi: 10.1104/pp.119.2.755
Jorgensen, R. A., Cluster, P. D., English, J., Que, Q., and Napoli, C. A. (1996). Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol. Biol. 31, 957–973. doi: 10.1007/BF00040715
Karkute, S. G., Singh, A. K., Gupta, O. P., Singh, P. M., and Singh, B. (2017). CRISPR/Cas9 mediated genome engineering for improvement of horticultural crops. Front. Plant Sci. 8:1635. doi: 10.3389/fpls.2017.01635
Lee, Y., Kim, E. S., Choi, Y., Hwang, I., Staiger, C. J., Chung, Y. Y., et al. (2008). The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol. 147, 1886–1897. doi: 10.1104/pp.108.121590
Li, F., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389–1408. doi: 10.1105/tpc.15.00158
Liang, X., Oono, Y., Shen, N. F., Köhler, C., Li, K., Scolnik, P. A., et al. (1995). Characterization of two members (ACS1 and ACS3) of the 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Gene 167, 17–24. doi: 10.1016/0378-1119(95)00694-X
Liao, C. Y., and Bassham, D. C. (2020). Combating stress: the interplay between hormone signaling and autophagy in plants. J. Exp. Bot. 71, 1723–1733. doi: 10.1093/jxb/erz515
Lin, Y., and Jones, M. L. (2021). Silencing ATG6 and PI3K accelerates petal senescence and reduces flower number and shoot biomass in petunia. Plant Sci. 302:110713. doi: 10.1016/j.plantsci.2020.110713
Liu, Y., Schiff, M., Czymmek, K., Tallóczy, Z., Levine, B., and Dinesh-Kumar, S. P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577. doi: 10.1016/j.cell.2005.03.007
López-Vidal, O., Olmedilla, A., Sandalio, L. M., Sevilla, F., and Jiménez, A. (2020). Is autophagy involved in pepper fruit ripening? Cell 9:106. doi: 10.3390/cells9010106
Lornac, A., Havé, M., Chardon, F., Soulay, F., Clément, G., Avice, J. C., et al. (2020). Autophagy controls sulphur metabolism in the rosette leaves of Arabidopsis and facilitates S remobilization to the seeds. Cell 9:332. doi: 10.3390/cells9020332
Mallona, I., Lischewski, S., Weiss, J., Hause, B., and Egea-Cortines, M. (2010). Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 10:4. doi: 10.1186/1471-2229-10-4
McKnight, N. C., and Yue, Z. (2013). Beclin 1, an essential component and master regulator of PI3K-III in health and disease. Curr. Pathobiol. Rep. 1, 231–238. doi: 10.1007/s40139-013-0028-5
Menon, M. B., and Dhamija, S. (2018). Beclin 1 phosphorylation–at the center of autophagy regulation. Front. Cell Dev. Biol. 6:137. doi: 10.3389/fcell.2018.00137
Minina, E. A., Filonova, L. H., Fukada, K., Savenkov, E. I., Gogvadze, V., Clapham, D., et al. (2013). Autophagy and metacaspase determine the mode of cell death in plants. J. Cell Biol. 203, 917–927. doi: 10.1083/jcb.201307082
O’Neill, S. D., Nadeau, J. A., Zhang, X. S., Bui, A. Q., and Halevy, A. H. (1993). Interorgan regulation of ethylene biosynthetic genes by pollination. Plant Cell 5:419. doi: 10.1105/tpc.5.4.419
Patel, S., and Dinesh-Kumar, S. P. (2008). Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4, 20–27. doi: 10.4161/auto.5056
Price, A. M., Orellana, D. F. A., Salleh, F. M., Stevens, R., Acock, R., Buchanan-Wollaston, V., et al. (2008). A comparison of leaf and petal senescence in wallflower reveals common and distinct patterns of gene expression and physiology. Plant Physiol. 147, 1898–1912. doi: 10.1104/pp.108.120402
Quijia Pillajo, J. O., Chapin, L. J., and Jones, M. L. (2018). Senescence and abiotic stress induce expression of autophagy-related genes in Petunia. J. Am. Soc. Hortic. Sci. 143, 154–163. doi: 10.21273/JASHS04349-18
R Core Team (2013). R: A Language and Environment for Statistical Computing, Vienna, Austria: R Foundation for Statistical Computing.
Reid, M. S., Fujino, D. W., Hoffman, N. E., and Whitehead, C. S. (1984). 1-aminocyclopropane-l-carboxylic acid (ACC)—The transmitted stimulus in pollinated flowers? J. Plant Growth Regul. 3, 189–196.
Ren, C., Liu, J., and Gong, Q. (2014). Functions of autophagy in plant carbon and nitrogen metabolism. Front. Plant Sci. 5:301. doi: 10.3389/fpls.2014.00301
Rogers, H. J. (2006). Programmed cell death in floral organs: how and why do flowers die? Ann. Bot. 97, 309–315. doi: 10.1093/aob/mcj051
Sera, Y., Hanamata, S., Sakamoto, S., Ono, S., Kaneko, K., Mitsui, Y., et al. (2019). Essential roles of autophagy in metabolic regulation in endosperm development during rice seed maturation. Sci. Rep. 9:18544. doi: 10.1038/s41598-019-54361-1
Shibuya, K. (2012). Molecular mechanisms of petal senescence in ornamental plants. J. Jap. Soc. Hortic. Sci. 81, 140–149. doi: 10.2503/jjshs1.81.140
Shibuya, K., Niki, T., and Ichimura, K. (2013). Pollination induces autophagy in petunia petals via ethylene. J. Exp. Bot. 64, 1111–1120. doi: 10.1093/jxb/ers395
Shibuya, K., Shimizu, K., Niki, T., and Ichimura, K. (2014). Identification of a NAC transcription factor, EPHEMERAL 1, that controls petal senescence in Japanese morning glory. Plant J. 79, 1044–1051. doi: 10.1111/tpj.12605
Shibuya, K., Shimizu, K., Yamada, T., and Ichimura, K. (2011). Expression of autophagy-associated ATG8 genes during petal senescence in Japanese morning glory. J. Jap. Soc. Hortic. Sci. 80, 89–95. doi: 10.2503/jjshs1.80.89
Shibuya, K., Watanabe, K., and Ono, M. (2018). CRISPR/Cas9-mediated mutagenesis of the EPHEMERAL1 locus that regulates petal senescence in Japanese morning glory. Plant Physiol. Biochem. 131, 53–57. doi: 10.1016/j.plaphy.2018.04.036
Su, T., Li, X., Yang, M., Shao, Q., Zhao, Y., Ma, C., et al. (2020). Autophagy: an intracellular degradation pathway regulating plant survival and stress response. Front. Plant Sci. 11:164. doi: 10.3389/fpls.2020.00164
Sweeney, R. A. (1989). Generic combustion method for determination of crude protein in feeds: collaborative study. J. Assoc. Off. Anal. Chem. 72, 770–774.
Tang, J., and Bassham, D. C. (2018). Autophagy in crop plants: what’s new beyond Arabidopsis? R. Soc. Open Biol. 8:180162. doi: 10.1098/rsob.180162
Trivellini, A., Cocetta, G., Hunter, D. A., Vernieri, P., and Ferrante, A. (2016). Spatial and temporal transcriptome changes occurring during flower opening and senescence of the ephemeral hibiscus flower, Hibiscus rosa-sinensis. J. Exp. Bot. 67, 5919–5931. doi: 10.1093/jxb/erw295
Tsanakas, G. F., Manioudaki, M. E., Economou, A. S., and Kalaitzis, P. (2014). De novo transcriptome analysis of petal senescence in Gardenia jasminoides Ellis. BMC Genomics 15:554. doi: 10.1186/1471-2164-15-554
Üstün, S., Hafrén, A., and Hofius, D. (2017). Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 40, 122–130. doi: 10.1016/j.pbi.2017.08.011
Van Doorn, W. G. (2001). Categories of petal senescence and abscission: a re-evaluation. Ann. Bot. 87, 447–456. doi: 10.1006/anbo.2000.1357
Van Doorn, W. G., and Woltering, E. J. (2008). Physiology and molecular biology of petal senescence. J. Exp. Bot. 59, 453–480. doi: 10.1093/jxb/erm356
Wada, S., Hayashida, Y., Izumi, M., Kurusu, T., Hanamata, S., Kanno, K., et al. (2015). Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 168, 60–73. doi: 10.1104/pp.15.00242
Wagstaff, C., Leverentz, M. K., Griffiths, G., Thomas, B., Chanasut, U., Stead, A. D., et al. (2002). Cysteine protease gene expression and proteolytic activity during senescence of Alstroemeria petals. J. Exp. Bot. 53, 233–240. doi: 10.1093/jexbot/53.367.233
Wagstaff, C., Malcolm, P., Rafiq, A., Leverentz, M., Griffiths, G., Thomas, B., et al. (2003). Programmed cell death (PCD) processes begin extremely early in Alstroemeria petal senescence. New Phytol. 160, 49–59. doi: 10.1046/j.1469-8137.2003.00853.x
Wagstaff, C., Yang, T. J., Stead, A. D., Buchanan-Wollaston, V., and Roberts, J. A. (2009). A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J. 57, 690–705. doi: 10.1111/j.1365-313X.2008.03722.x
Wang, H., Chang, X., Lin, J., Chang, Y., Chen, J. C., Reid, M. S., et al. (2018). Transcriptome profiling reveals regulatory mechanisms underlying corolla senescence in petunia. Hortic. Res. 5:16. doi: 10.1038/s41438-018-0018-1
Wang, P., Nolan, T. M., Yin, Y., and Bassham, D. C. (2020). Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 16, 123–139. doi: 10.1080/15548627.2019.1598753
Wegmüller, S., Svistoonoff, S., Reinhardt, D., Stuurman, J., Amrhein, N., and Bucher, M. (2008). A transgenic dTph1 insertional mutagenesis system for forward genetics in mycorrhizal phosphate transport of Petunia. Plant J. 54, 1115–1127. doi: 10.1111/j.1365-313X.2008.03474.x
Wirawan, E., Walle, L. V., Kersse, K., Cornelis, S., Claerhout, S., Vanoverberghe, I., et al. (2010). Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 1:e18. doi: 10.1038/cddis.2009.16
Woltering, E. J. (1990). Interorgan translocation of 1-aminocyclopropane-1-carboxylic acid and ethylene coordinates senescence in emasculated cymbidium flowers. Plant Physiol. 92, 837–845. doi: 10.1104/pp.92.3.837
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327. doi: 10.1186/s12870-014-0327-y
Xu, J., Kang, B. C., Naing, A. H., Bae, S. J., Kim, J. S., Kim, H., et al. (2020). CRISPR/Cas9-mediated editing of 1-aminocyclopropane-1-carboxylate oxidase1 enhances Petunia flower longevity. Plant Biotechnol. J. 18, 287–297. doi: 10.1111/pbi.13197
Xu, J., Naing, A. H., Bunch, H., Jeong, J., Kim, H., and Kim, C. K. (2021). Enhancement of the flower longevity of petunia by CRISPR/Cas9-mediated targeted editing of ethylene biosynthesis genes. Postharvest Biol. Technol. 174:111460. doi: 10.1016/j.postharvbio.2020.111460
Yamada, T., Ichimura, K., Kanekatsu, M., and Van Doorn, W. G. (2009). Homologs of genes associated with programmed cell death in animal cells are differentially expressed during senescence of Ipomoea nil petals. Plant Cell Physiol. 50, 610–625. doi: 10.1093/pcp/pcp019
Yang, C. P., Xia, Z. Q., Hu, J., Zhuang, Y. F., Pan, Y. W., and Liu, J. P. (2019). Transcriptome analysis of Oncidium petals provides new insights into the initiation of petal senescence. J. Hortic. Sci. Biotechnol. 94, 12–23. doi: 10.1080/14620316.2018.1432297
Yip, W. K., Moore, T., and Yang, S. F. (1992). Differential accumulation of transcripts for four tomato 1-aminocyclopropane-1-carboxylate synthase homologs under various conditions. Proc. Natl. Acad. Sci. U. S. A. 89, 2475–2479. doi: 10.1073/pnas.89.6.2475
Yu, J., Zhen, X., Li, X., Li, N., and Xu, F. (2019). Increased autophagy of rice can increase yield and nitrogen use efficiency (NUE). Front. Plant Sci. 10:584. doi: 10.3389/fpls.2019.00584
Yue, J., Sun, H., Zhang, W., Pei, D., He, Y., and Wang, H. (2015). Wheat homologs of yeast ATG6 function in autophagy and are implicated in powdery mildew immunity. BMC Plant Biol. 15:95. doi: 10.1186/s12870-015-0472-y
Zeng, X., Zeng, Z., Liu, C., Yuan, W., Hou, N., Bian, H., et al. (2017). A barley homolog of yeast ATG6 is involved in multiple abiotic stress responses and stress resistance regulation. Plant Physiol. Biochem. 115, 97–106. doi: 10.1016/j.plaphy.2017.03.013
Keywords: Beclin1, CRISPR, nutrient recycling, programmed cell death, petal senescence
Citation: Lin Y and Jones ML (2022) CRISPR/Cas9-Mediated Editing of Autophagy Gene 6 in Petunia Decreases Flower Longevity, Seed Yield, and Phosphorus Remobilization by Accelerating Ethylene Production and Senescence-Related Gene Expression. Front. Plant Sci. 13:840218. doi: 10.3389/fpls.2022.840218
Edited by:
Lara Reale, University of Perugia, ItalyReviewed by:
Rongfang Guo, Fujian Agriculture and Forestry University, ChinaDaniele Rosellini, University of Perugia, Italy
Copyright © 2022 Lin and Jones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michelle L. Jones, am9uZXMuMTk2OEBvc3UuZWR1
 Yiyun Lin
Yiyun Lin Michelle L. Jones
Michelle L. Jones